To the Editor —
The epigenomics community, including the NIH 4D Nucleome (4DN) Consortium [1], is generating numerous genomic and imaging datasets for studying nuclear structure and function. Integrating primary data with secondary analysis results will be required to create a comprehensive reference map of nuclear organization. Currently no existing tool can effectively integrate such multimodal data for scalable and interactive visualization (Supplementary Table S1 and Supplementary Note A). Here, we introduce the Nucleome Browser (http://www.nucleome.org), an integrative and interactive multimodal data visualization and exploration platform that accelerates access, utilization, and sharing of 4DN data (see Supplementary Table S2 for a collection of resources supporting the Nucleome Browser). Our platform has two unique capabilities: (1) simultaneous and synchronized visualization of multiomic datasets (including Hi-C contact maps, 1D genomic signal tracks, and even single-cell data), imaging datasets, and 3D genome structure models; (2) scalable integration of custom datasets with existing data portals (e.g., UCSC Genome Browser [2], WashU Epigenome Browser [3], HiGlass [4], OMERO imaging data server [5], and Image Data Resource (IDR) [6]) in a unified, interactive platform. Nucleome Browser currently hosts 2,292 genomic tracks and 732 image datasets (Supplementary Table S3 and Supplementary Note B.1).
Nucleome Browser consists of a series of web components that can be flexibly arranged and customized by the user (i.e., composable and configurable) and that offer synchronized operations for integrative visualization of heterogeneous datasets or different views from the same data modality (Fig. 1a–b, Supplementary Note C, and Supplementary Note B.2). These web components of Nucleome Browser include: (1) a genome browser for visualizing both 1D (e.g., Repli-seq and TSA-seq) and 2D (e.g., Hi-C contact maps) genomic data; (2) a display component for 3D genome structure models; (3) extensible, customized web applications tailored towards specific data types (Fig. 1a–c and Supplementary Note C for details). Each web component is self-encapsulated into a panel coupled with an adaptive communication mechanism. Users can combine multiple linked panels for multiomic comparisons (Supplement Fig. S1) or multimodal data exploration (Fig. 1b). Importantly, the layout of panels can be saved into sessions, enabling management and sharing of the browser’s views.
Figure 1:
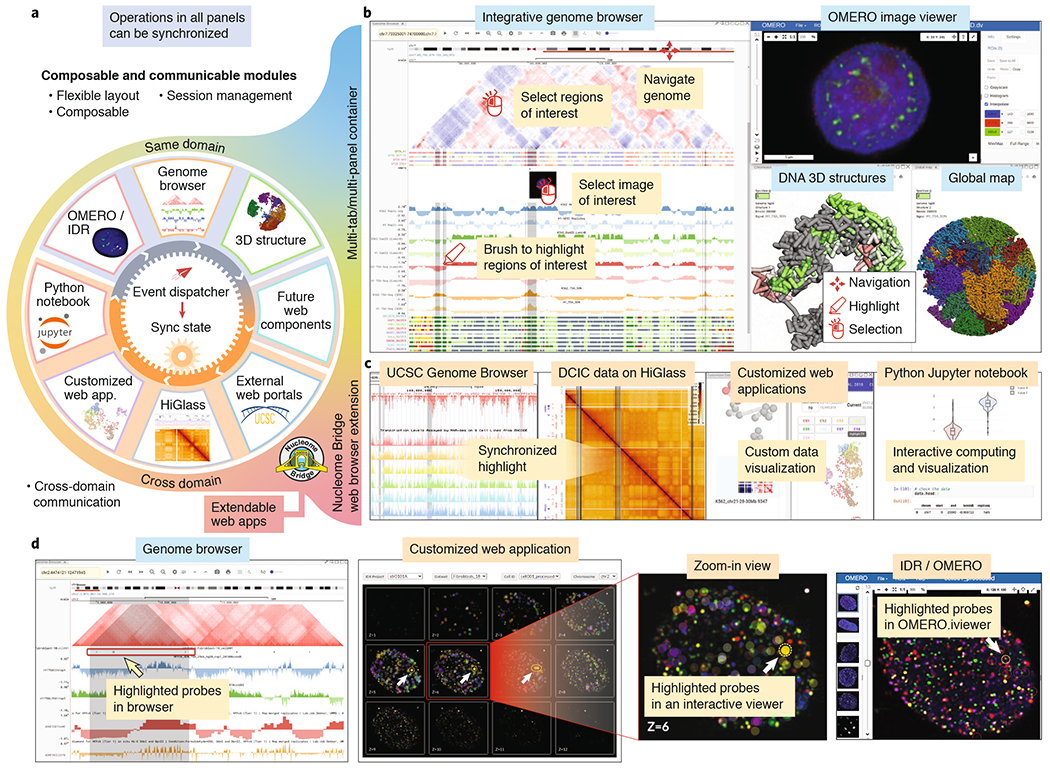
Nucleome Browser facilitates integrative and multimodal data navigation. (a) Overall design of Nucleome Browser. Nucleome Browser consists of a series of web components with synchronized operations, each of which is focused on a specific data type and can be combined in a flexible manner. A unified hierarchical event-dispatch system allows modules (outer circle) from either the same domain or different domains to synchronize their status based on genomic coordinates and highlighted regions. Operations (e.g., navigation to or highlight on the region(s)) initiated in any component will automatically dispatch to all connected web components through the event-dispatch system. Nucleome Bridge, a web browser extension, empowers various external data portals (e.g., the UCSC Genome Browser) to communicate with other modules in the platform. (b) A screenshot of Nucleome Browser illustrates interactive exploration of multimodal 4DN data. Users can use the mouse to navigate to or highlight region-of-interest on 1D and 2D tracks in a genome browser web component. OMERO image viewer shows the details of the image on the selected locus on the image track. 3D genome structure model web component shows a 3D view of chromosome structure with colors indicating the signals of genomic data. (c) The web browser extension named Nucleome Bridge further transmits both genomic coordinates and highlighted regions to other web applications including UCSC Genome Browser, HiGlass viewer, custom data viewers, or Jupyter Notebook to generate plots in real-time. (d) A customized web application can connect Nucleome Browser to the in situ genome sequencing (IGS) data hosted on the IDR platform. Users can navigate the IGS dataset to select an individual cell and this web application will automatically fetch images from IDR and mark the location of probes as circles. As users select regions on the genome browser panel on the left, probes overlapping with the highlighted regions will also be highlighted. Clicking a probe will lead users to the IDR portal and open an OMERO.iviewer for further examination.
The core component of our adaptive communication design is a multi-channel hierarchical event-dispatch hub (Supplementary Fig. S2 and Supplementary Note C), which automatically recognizes a user’s operations triggered in any web component, and then synchronously updates the status of web components or data portals from different domains. For example, when a user navigates to or highlights a region-of-interest (ROI) in a genome browser panel, all other panels/portals (including the UCSC Genome Browser, the customized HiGlass viewer, and the Python Jupyter Notebook) synchronously change to display and highlight the corresponding region, initiate the OMERO image viewer, or update plots in the Jupyter Notebook accordingly (Fig. 1a–c, also see Supplementary Note B.3 and B.4). Multi-way communications are achieved through the “Nucleome Bridge” web browser extension (Supplementary Fig. S3 and Supplementary Note C). Users alternatively can divide panels into specific channels such that synchronization is restricted to those panels within the same channel (Supplementary Fig. S4). Users can efficiently perform integrative comparisons of nuclear organization in different cellular contexts (Supplementary Fig. S5 and Supplementary Note B.5). A scatterplot analysis tool allows global comparison between two bigWig tracks (Supplementary Fig. S5). We can take the same approach to comparing single-cell 3D genome data [7] (Supplementary Fig. S6 and Supplementary Note B.6). Nucleome Browser is the first platform that supports such multi-domain and multi-channel interactive visualization, which greatly enhances heterogeneous and multimodal data exploration.
Recently developed multiplexed immuno-FISH imaging methods provide a direct, whole-genome visualization of 3D chromatin localization that genomic mapping cannot provide. We built customized web applications to demonstrate that users can interactively compare genomic data hosted on the Nucleome Browser with single-cell 3D genome structure models derived from multiplexed OligoSTORM [8] and OligoDNA-PAINT [9] data (Supplementary Fig. S3a–c) as well as in situ genome sequencing (IGS) [10] imaging data hosted on the IDR [6] (Fig. 1d). For the IGS data, users can explore a single-cell image dataset using the OMERO.iviewer with the markers of FISH probes highlighted on the images. Highlighting genomic regions in the genome browser also highlights FISH probes in the image overlapping with these genomic regions, allowing interactive comparison of sequencing-based genomic features with their 3D spatial nuclear localization (Fig. 1d). Our design supports integrative visualization of multimodal datasets across multiple domains while facilitating future extensions to support new data types as they become available.
Nucleome Browser represents the next-generation design of integrative and multimodal data visualization for multiscale nuclear organization. We anticipate that Nucleome Browser will serve as an important portal for users to fully utilize different types of 4DN data and diverse 3D epigenomics datasets to navigate multiscale nuclear structure and function in a wide range of biological contexts, enabling hypothesis generation and data sharing with the broad community.
Supplementary Material
Acknowledgement
Nucleome Browser was first introduced to the community during the 2018 Annual Meeting of the NIH 4D Nucleome Consortium (San Diego, CA). The authors would like to thank members of the 4D Nucleome Consortium and the broad 3D genome community for feedback and suggestions that have improved the tool. The authors are also grateful to members of the Ma lab for helpful discussions and comments on the manuscript. This work was supported by the National Institutes of Health Common Fund 4D Nucleome Program grants U54DK107965 (A.S.B. and J.M.), U54DK107965-05S1 (J.R.S. and J.M.), and UM1HG011593 (A.S.B., J.R.S., and J.M.). J.M. is additionally supported by a Guggenheim Fellowship from the John Simon Guggenheim Memorial Foundation.
Competing Interests
J.R.S. is affiliated with Glencoe Software, a commercial company that builds, delivers, supports and integrates image data management systems across academic, biotech and pharmaceutical industries. The remaining authors declare no competing interests.
Footnotes
Code Availability
The code of Nucleome Browser is available at: https://github.com/nucleome
Nucleome Browser can be accessed at: http://www.nucleome.org
Documentation and tutorials are available at: https://nb-docs.readthedocs.io/en/latest
Video tutorials are available at: https://tinyurl.com/nb-video-tutorial
Data Availability
All datasets available on Nucleome Browser were collected from published work or data portal from public consortium projects. For the public 4DN data, we developed an automatic workflow to download from the 4DN Data Portal (https://data.4dnucleome.org) periodically. We then further curated the meta-information of the data tracks for visualization. Supplementary Table S4 lists the detailed sources of all datasets used in Nucleome Browser (latest version updated in November 2021).
References
- [1].Dekker J et al. Nature 549, 219–226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kent WJ et al. Genome Research 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Zhou X et al. Nature Methods 10, 375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kerpedjiev P et al. Genome Biology 19, 125 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Allan C et al. Nature Methods 9, 245–253 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Williams E et al. Nature Methods 14, 775–781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Stevens TJ et al. Nature 544, 59–64 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Bintu B et al. Science 362, eaau1783 (2018).30361340 [Google Scholar]
- [9].Nir G et al. PLoS Genetics 14, e1007872 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Payne AC, et al. Science 371, eaay3446 (2021).33384301 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets available on Nucleome Browser were collected from published work or data portal from public consortium projects. For the public 4DN data, we developed an automatic workflow to download from the 4DN Data Portal (https://data.4dnucleome.org) periodically. We then further curated the meta-information of the data tracks for visualization. Supplementary Table S4 lists the detailed sources of all datasets used in Nucleome Browser (latest version updated in November 2021).


