BCL10 mutations in diffuse large B-cell lymphoma (DLBCL) fall into two classes, with both conferring BTK inhibitor resistance but only truncation mutations conferring MALT1 dependency, suggesting use of these mutation classes as precision therapy biomarkers.
Abstract
Activated B cell–like diffuse large B-cell lymphomas (ABC-DLBCL) have unfavorable outcomes and chronic activation of CARD11–BCL10–MALT1 (CBM) signal amplification complexes that form due to polymerization of BCL10 subunits, which is affected by recurrent somatic mutations in ABC-DLBCLs. Herein, we show that BCL10 mutants fall into at least two functionally distinct classes: missense mutations of the BCL10 CARD domain and truncation of its C-terminal tail. Truncating mutations abrogated a motif through which MALT1 inhibits BCL10 polymerization, trapping MALT1 in its activated filament-bound state. CARD missense mutations enhanced BCL10 filament formation, forming glutamine network structures that stabilize BCL10 filaments. Mutant forms of BCL10 were less dependent on upstream CARD11 activation and thus manifested resistance to BTK inhibitors, whereas BCL10 truncating but not CARD mutants were hypersensitive to MALT1 inhibitors. Therefore, BCL10 mutations are potential biomarkers for BTK inhibitor resistance in ABC-DLBCL, and further precision can be achieved by selecting therapy based on specific biochemical effects of distinct mutation classes.
Significance:
ABC-DLBCLs feature frequent mutations of signaling mediators that converge on the CBM complex. We use structure–function approaches to reveal that BCL10 mutations fall into two distinct biochemical classes. Both classes confer resistance to BTK inhibitors, whereas BCL10 truncations confer hyperresponsiveness to MALT1 inhibitors, providing a road map for precision therapies in ABC-DLBCLs.
See related commentary by Phelan and Oellerich, p. 1844.
This article is highlighted in the In This Issue feature, p. 1825
INTRODUCTION
The CARD11–BCL10–MALT1 (CBM) complex plays a critical role in integrating signaling pathways involved in immunity and inflammation in a broad repertoire of cell types. In B cells and T cells, the CBM complex is activated downstream of B-cell receptor (BCR) or T-cell receptor (TCR) signaling and serves to amplify such signals, leading to powerful phenotype responses conferred by critical downstream mediators (1, 2). Accordingly, aberrant CBM function has been shown to play critical roles in diseases such as B-cell lymphoma and autoimmunity (3, 4). Upon antigen receptor engagement, the CARD11 subunit is phosphorylated by protein kinase C (PKC), which activates its function by reducing the interaction of its autoinhibitory coiled-coil domain to its CARD domain (5–8). The activated form of CARD11 then can interact with BCL10 and facilitate its forming of large macromolecular filaments, providing a large scaffold for binding and activation of MALT1, which is the enzymatic paracaspase subunit of the CBM complex that results in the further downstream activation of a variety of effector molecules (9, 10).
Like other supramolecular organizing center (SMOC)–mediated signaling transduction, such as Toll-like receptor (TLR) triggering Myddosome (11, 12), RIG-I–like receptor sensing intracellular viral RNA, and activating mitochondrial antiviral signaling protein filament formation (13), the BCL10 filament formation is also critical for BCR/TCR signaling amplification and robust downstream NF-κB activation (10). BCL10 is composed of an N-terminal CARD domain and a long C-terminal unstructured region containing a distal serine (Ser)- and threonine (Thr)-rich region. Structure-guided studies showed that BCL10 filament polymerizes in a unidirectional manner through CARD–CARD interactions, providing a surface for the cooperative binding of MALT1 through its N-terminal death domain (DD; refs. 10, 14). Upon BCL10 filament binding, MALT1 is immediately dimerized and incorporates TRAF6 to form a higher ordered assembly leading to all-or-none activation of downstream pathways including NF-κB and JNK (10, 14). Binding to BCL10 also activates MALT1 paracaspase activity and cleavage of substrate proteins. BCL10 filament formation is dynamic in activated T lymphocytes and precisely regulated by disassembly and degradation through BCL10 K63 polyubiquitination and p62-dependent selective autophagy–lysosomal proteolysis system (15, 16). Hence, dynamic BCL10 filament turnover might be critical to precisely tune its effect on downstream signaling pathways such as NF-κB.
Chronic active NF-κB signaling is a hallmark of the highly aggressive activated B cell–like diffuse large B-cell lymphomas (ABC-DLBCL), due to somatic mutations of BCR and TLR subunit genes such as CD79b and MYD88 (17–19), as well as activating mutations of CARD11 and amplifications of MALT1 (20–22). Collectively, these mutations induce chronic activation of the CBM complex to maintain robust and sustained NF-κB and other downstream pathway activation. The involvement of these signaling pathways in highly aggressive tumors has inspired the development of targeted therapies disrupting oncogenic BCR/TLR activity. However, the position where mutations happen in the BCR pathway may be critical for assigning potential precision therapy to patients. For example, mutations in the most upstream BCR proteins like CD79B confer sensitivity to BTK inhibitors, whereas downstream mutations like PLCγ2 and CARD11 confer resistance (23–26). Hence, a mechanistic study of oncogenic mutations is beneficial to guide targeted therapy in B-cell lymphomas.
Recent genomic sequencing studies in DLBCLs and other lymphomas have revealed recurrent and widely spread somatic mutations of BCL10 (27–30). However, the functionality and mechanism of BCL10 mutations in DLBCL have not been studied. Whereas it is evident how mutations causing the constitutive activation of CARD11 or increased abundance of MALT1 might result in enhanced CBM function (20–22), it is not immediately clear how these BCL10 mutations might function. Therefore, we set out to explore the structure and function of BCL10 mutations in DLBCL, identifying distinct classes of mutant proteins with different biochemical effects and distinct impact on response to targeted therapies. These studies have critical implications for selecting targeted therapy agents for lymphoma precision therapy.
RESULTS
BCL10 Mutations Are Genetic Drivers and Occur in Two Broad Classes
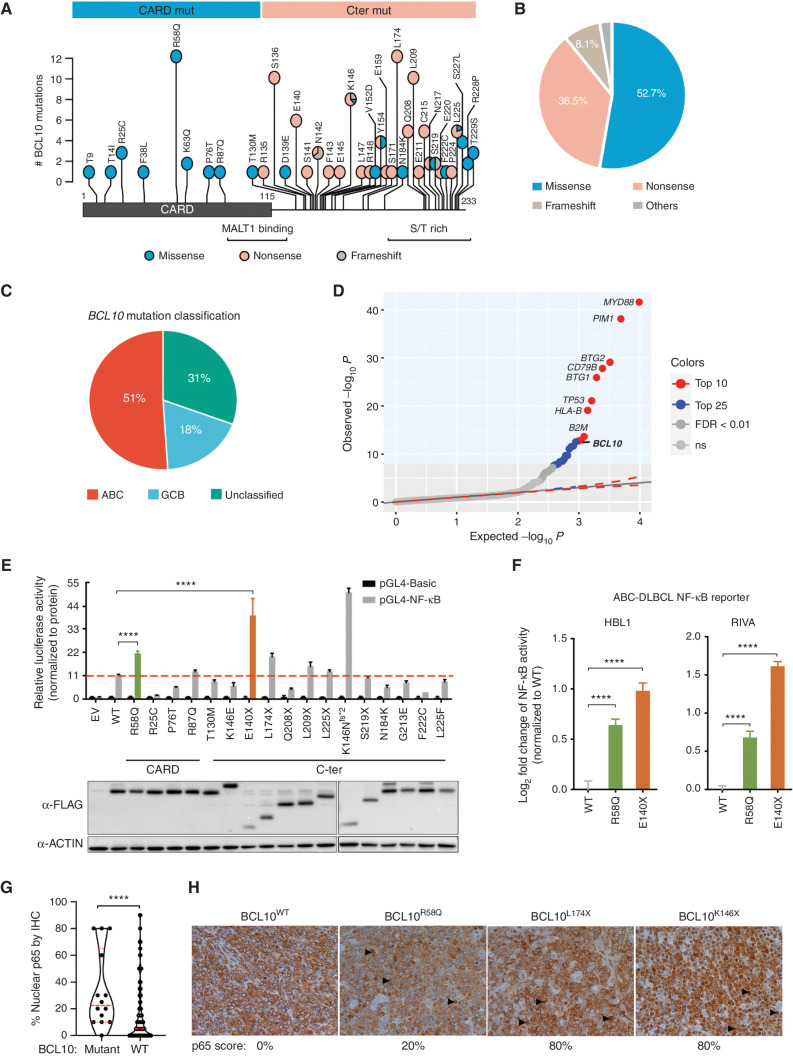
As a first approach to explore the structure–function of BCL10 mutations, we merged DLBCL sequencing databases (refs. 27, 29–32; n = 2,255) and identified 75 patients with BCL10 mutations. BCL10 contains a structured CARD domain at its N-terminal half that mediates interaction with CARD11 and polymerization of BCL10 into fibrils. The C-terminal region is unstructured and contains Ser and Thr residues targeted by posttranslational modifications (9, 33). BCL10 mutations affected both regions. Although mutations in the CARD were all missense mutations with a prominent hotspot at arginine 58, a majority of those in the C-terminal region were truncating (nonsense or frameshift) mutations with a number of hotspot residues observed (Fig. 1A and B).
Figure 1.
Characterization of human BCL10 mutations in DLBCL. A, BCL10 protein domain and the locations of all BCL10 mutations in DLBCL identified and reported from the literature and open databases. S/T, serine/threonine. B, Proportion of mutation types among patients with BCL10 mutations. C, Cell-of-origin classification of DLBCL patients with BCL10 mutations. D, Quantile–quantile plot showing the P values for BCL10 single-nucleotide variants across 243 patients with ABC-DLBCL. E, NF-κB activity measured by NF-κB-RE luciferase reporter 24 hours after transfection of different BCL10 mutations into 293T cells. Luciferase activity was normalized to the expression level of each mutation. EV, empty vector. β-Actin was used as an internal control. ****, P < 0.0001. F, NF-κB reporter activity in lymphoma cells expressing mutations. ****, P < 0.0001. G, Statistical comparison of nuclear p65 staining scores between BCL10 WT and BCL10-mutant tumors in tissue microarray for patients with DLBCL (n = 298). ****, Mann–Whitney P < 0.0001. H, Representative images of p65 IHC staining in BCL10 WT and BCL10-mutant DLBCLs in G. Images were taken under magnification of 400×. Black arrowheads point to examples of p65 nuclear staining.
Examining BCL10 mutations in a cohort of patients with rigorous cell-of-origin and genetic cluster information (29, 34), we observed that 51% occurred in ABC-DLBCLs, 31% in unclassifiable cases, and 18% in germinal center B cell–like DLBCLs (GCB-DLBCL; Fig. 1C). However, the incidence of BCL10 mutation was highest in the unclassifiable patients, followed by those with ABC-DLBCL (Supplementary Fig. S1A). As reported previously (35) using the LymphGen classification system, most BCL10 mutations were observed in the BN2 class of DLBCLs (Supplementary Fig. S1B), which had the highest incidence of these lesions (35% of patients; Supplementary Fig. S1C). BCL10 expression was also highest among the BN2 cases (Supplementary Fig. S1D).
To determine whether BCL10 somatic mutations were likely to be robust genetic drivers of ABC-DLBCLs, we performed a rigorous genomic covariate “fishhook” analysis (36) controlling for gene size, as well as GCB-cell gene expression profiles, activating promoter histone marks, chromatin accessibility profiles, and others. This analysis captured BCL10 as one of the top 10 driver mutations in ABC-DLBCL, along with genes such as MYD88, CD79B, PIM1, and TP53 (FDR < 0.01; Fig. 1D). BCL10 mutations were still among the top 15 drivers when considering all DLBCLs (Supplementary Fig. S1E).
Next, to survey whether the different classes of BCL10 mutations had a functional impact on NF-κB signaling, we expressed a panel of CARD and C-terminal mutants, as well as wild-type (WT) BCL10 together with an NF-κB luciferase reporter in 293T cells. As expected, WT BCL10 was able to induce NF-κB activity (refs. 37, 38; Fig. 1E). Most of the CARD and C-terminal mutations also induced NF-κB activity. However, the hotspot missense mutant BCL10R58Q showed significantly higher NF-κB activity compared with WT BCL10, as did the truncating mutants BCL10E140X and BCL10K146Nfs*2 (Fig. 1E). Focusing our studies on representative CARD and C-terminal mutations, we showed that this markedly increased NF-κB reporter induction even occurred in ABC-DLBCL cells that already have chronic BCR activation including both MCD (HBL1) and BN2-DLBCL cells (RIVA; Fig. 1F), also validating that this reporter reacts as expected to disruption of NF-κB signaling (Supplementary Fig. S1F). Hence, both missense and truncating BCL10 mutations yield a significant gain-of-function effect on NF-κB activation. Using an additional MCD cell line, TMD8, we also noted reduction in IκBα abundance in cells expressing BCL10E140X truncating mutant (Supplementary Fig. S1G). Finally, to determine if such findings could be validated in primary human DLBCLs, we performed IHC staining of p65 in a set of tissue microarrays containing biopsies from 298 genetically annotated DLBCL patients. We found that BCL10-mutant DLBCLs manifested significantly increased p65 nuclear staining scores compared with BCL10 WT cases (Mann–Whitney P < 0.0001; Fig. 1G). BCL10 truncating mutants were associated with the highest nuclear p65 scores, whereas BCL10R58Q CARD mutants had less abundance of nuclear p65 that was still higher than in BCL10 WT cases (Fig. 1H).
BCL10 Polymerization Is Greatly and Moderately Enhanced, Respectively, by E140X and R58Q Mutants
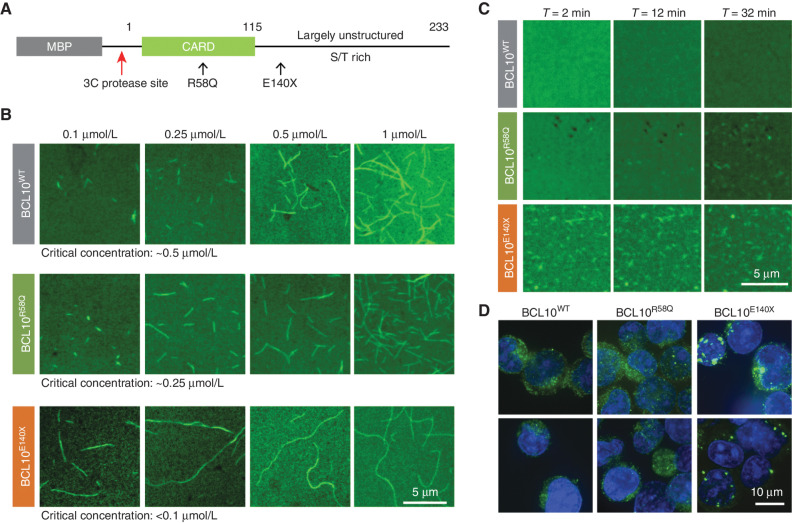
To determine the mechanism through which BCL10 mutants might confer a biochemical gain of function, we expressed and purified full-length BCL10WT, BCL10E140X, and BCL10R58Q fused to a 3C protease–cleavable maltose binding protein (MBP) at the N-terminus to keep the proteins in a monomeric state (Fig. 2A). The purified proteins were labeled with Alexa488 through cysteine residues in the unstructured C-terminal region. Spontaneous filament formation was then assessed by confocal fluorescence microscopy as a function of BCL10 concentration 4 hours after mixing with the 3C protease to remove the MBP tag. Judging from these images, BCL10WT had a critical concentration of polymerization at ∼0.5 μmol/L, while BCL10R58Q initiated filament formation at a somewhat lower concentration of ∼0.25 μmol/L (Fig. 2B). Strikingly, BCL10E140X started to form filaments even at the lowest concentration tested of 0.1 μmol/L, indicating a greatly enhanced ability to polymerize spontaneously (Fig. 2B). Of note, even MBP-fused BCL10E140X, in which MBP should suppress filament formation sterically, had a significant polymerized fraction during protein purification; in contrast, we did not observe this phenomenon for BCL10WT or BCL10R58Q (Supplementary Fig. S2). We further imaged the kinetics of filament formation by these proteins at 1 μmol/L concentration, which is above the critical concentration of polymerization for WT and these mutant BCL10 proteins using time-lapse confocal fluorescence microscopy. We found that BCL10E140X was already extensively polymerized at 2 minutes after mixing with 3C protease—the earliest time point we could image (Fig. 2C; Supplementary Movies S1–S3). Although filament formation kinetics of BCL10WT and BCL10R58Q were both slower than that of BCL10E140X, BCL10R58Q showed more apparent filament formation at 32 minutes after the addition of the 3C protease than BCL10WT (Fig. 2C; Supplementary Movies S1–S3), suggesting that BCL10E140X and to a lesser extent BCL10R58Q accelerated BCL10 polymerization threshold and kinetics.
Figure 2.
Representative missense and truncating mutant BCL10 enabled a faster polarization rate. A, Domain organization of the MBP-BCL10 construct. S/T, serine/threonine. B, Critical concentration determination of WT, E140X, and R58Q based on confocal images of BCL10 filaments formed at a concentration range between 0.1 and 1 μmol/L, imaged 4 hours after MBP cleavage. C, Confocal time lapse of WT, E140X, and R58Q imaged at 1 μmol/L for 30 minutes. E140X exhibits the fastest polymerization rate in comparison with WT and R58Q. D, Confocal images of HBL1 cells stably expressing FLAG-tagged WT, E140X, and R58Q, visualized by IF for BCL10 and DNA (Hoechst dye, blue).
Activated CBM complexes manifest as puncta when visualized through confocal microscopy of living cells (20, 39, 40). We therefore imaged ABC-DLBCL cells (HBL1) engineered for the constitutive expression of FLAG-tagged BCL10WT, BCL10E140X, and BCL10R58Q, respectively. These experiments revealed large, striking aggregates of BCL10E140X in comparison with the much smaller puncta of BCL10WT or BCL10R58Q (Fig. 2D), supporting the in vitro findings using recombinant proteins. These aggregates are reminiscent of the large puncta of an oncogenic, gain-of-function CARD11 mutant (20, 41), suggesting that these aggregates represent active CBM complexes.
The R58Q Mutant Forms Filaments with a Glutamine Ladder, Enhanced Stability, and Tendency to Bundle
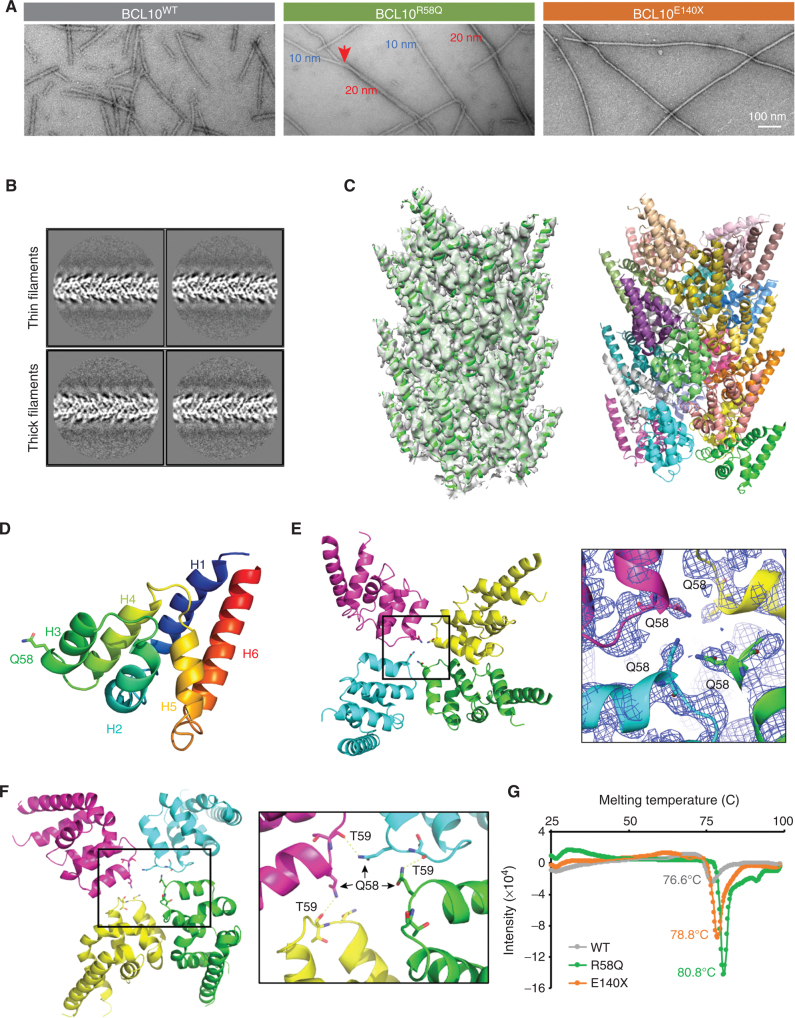
To better visualize these filaments, we purified 3C protease treatment—induced filaments of BCL10WT, BCL10R58Q, and BCL10E140X by collecting the void fraction (i.e., polymerized BCL10) from a Superose 6 gel filtration column, and imaged them using scanning electron microscopy (EM; Fig. 3A). Surprisingly, whereas BCL10WT and BCL10E140X formed the classic 10 nm filaments of CARDs (10), BCL10R58Q formed both 10 nm filaments and thicker 20 nm filaments (Fig. 3A). Closer inspection of the 20 nm filaments suggested that they are bundled 10 nm filaments because these thinner filaments were observed to merge into thicker filaments (Fig. 3A).
Figure 3.
Cryo-EM structure of the BCL10R58Q filament. A, Negative stained EM micrographs of WT, R58Q, and E140X filaments. R58Q formed a mixture of thin (10 nmol/L) and thick (20 nmol/L) filaments, whereas E140X formed only thin filaments similar to WT. Scale bar, 100 nmol/L. B, Representative 2D classes of BCL10R58Q thin and thick filaments. C, Cryo-EM structure of BCL10R58Q at 4.6 Å. Left, BCL10R58Q filament fitted into the cryo-EM map. Right, BCL10R58Q 16-mer filament in which each subunit is colored differently. D, BCL10R58Q monomer. 58Q residue is labeled as stick. E, BCL10R58Q layer, showing 58Q residues facing one another for stabilizing type III intrastrand interface (left). Zoom-in view of BCL10 Q58 fitted into cryo-EM density (right). F, Potential hydrogen bonding network formed by the Q58 side chain of one protomer and the carbonyl oxygen of T59 from the next protomer in the helical spiral, shown on four consecutive R58Q subunits in the filament (left), and as a zoom-in view (right). These interactions stabilize the type III intrastrand interface. G, Filament thermal stability assay performed by thermal shift assay for WT, E140X, and R58Q purified filaments. E140X and R58Q filaments showed a significant shift in 2.1°C and 4.2°C, respectively, in comparison with WT filaments.
To determine if the BCL10R58Q filaments are structurally different from BCL10WT or BCL10E140X filaments and how the bundling occurs, we collected cryo-EM data using an Arctica microscope operating at 200 keV and a K2 electron counting direct detection camera (Supplementary Fig. S3A). We manually selected thin and thick filaments from the cryo-EM micrographs and noted that 2D classification revealed average filaments of similar thickness (Fig. 3B). These data suggested that the association between filaments within the bundle is not specific, so only one thin filament within each thick filament could be aligned and the other filament was averaged out. Thus, the thick filaments consist of randomly bundled thin filaments (Fig. 3B).
Using 3D reconstruction, we determined the cryo-EM structure of the BCL10R58Q filament at 4.6 Å resolution assessed by gold-standard Fourier shell correlation (FSC; Supplementary Fig. S3B). The overall structure was similar to that of the BCL10WT filament (10), with only the CARD domain ordered (Fig. 3C). Q58 resides in helix 3 (H3) of the six-helical bundle fold of the CARD domain (Fig. 3D). Within the BCL10R58Q filament structure, Q58 localizes near the center and its side chain density is well-defined (Fig. 3E), which contrasts with the poor density of the equivalent WT R58 in the BCL10WT filament structure (Supplementary Fig. S3C). Although the resolution of the structure is limited, it is tempting to speculate that Q58 residues at the center of the filament formed stacks of glutamine residues, with direct and possibly water-mediated interactions. Indeed in the center of the filament, we found that the Q58 side chain (NE2 atom) of one protomer in the filament forms a potential hydrogen bond with the carbonyl oxygen of T59 of the next protomer in the helical filament (Fig. 3F). To further demonstrate the potential role of the hydrogen bonding network in the BCL10 filament assembly in the R58Q mutant, we generated the R58E mutant, which does not have the NE2 atom (e.g., Q58 side chain) for hydrogen bond formation, and characterized its biochemical and biophysical properties. We found that there was no enhancement of filament formation for R58E (Supplementary Fig. S3D), and the critical concentration for R58E polymerization, ∼1 μmol/L, was even higher than WT BCL10 (Supplementary Fig. S3E), confirming that the hydrogen bond formed by Q58 is critical for its filament formation.
These Q58-mediated interactions prompted us to ask whether there might be a difference in the stability of the BCL10R58Q filament in comparison with BCL10WT and BCL10E140X filaments. To assess this possibility, we performed thermal melt assays on purified BCL10WT, BCL10R58Q, and BCL10E140X filaments, which revealed that BCL10R58Q, and to a lesser degree BCL10E140X, yielded more stable filaments, with thermal melting temperatures of 80.8°C and 78.8°C, respectively, in comparison with 76.6°C for the BCL10WT (Fig. 3G). Thus, although BCL10E140X shows enhanced polymerization, BCL10R58Q forms more stable filaments, which may explain its tendency to bundle.
Loss of Basal MALT1 Binding Promotes Spontaneous Polymerization of BCL10E140X
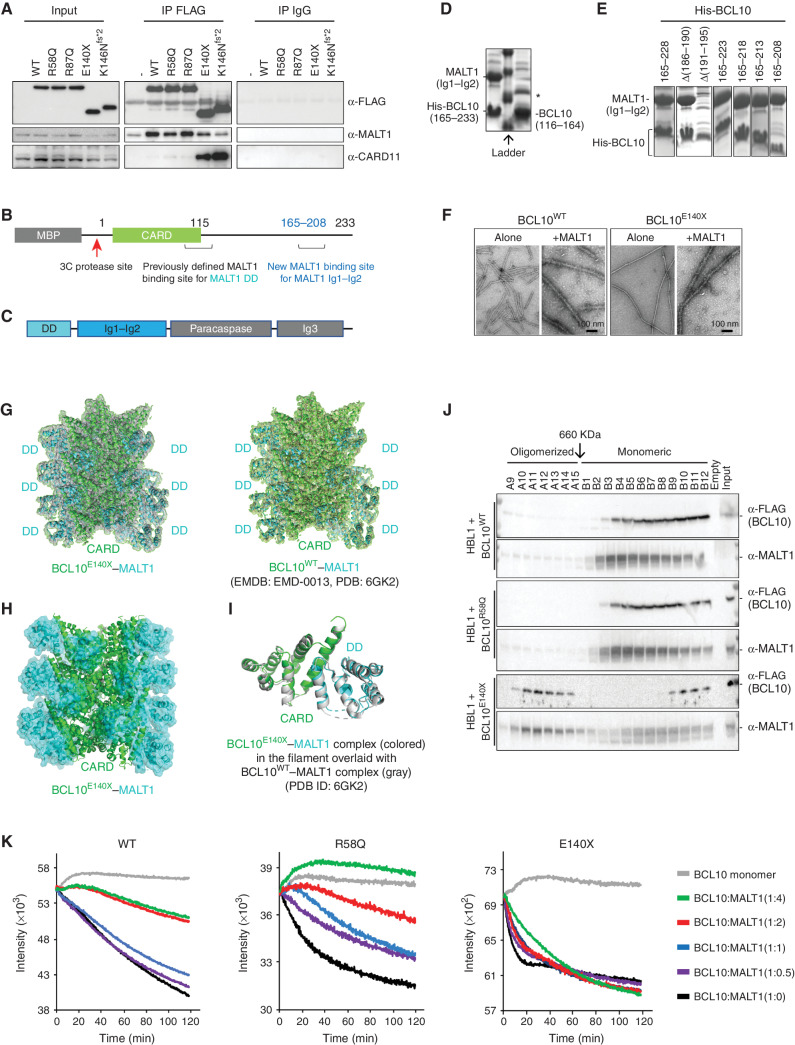
To investigate how the truncation mutants might affect CBM complex formation, we expressed FLAG-tagged WT and mutant forms of BCL10 in Raji cells, which lack constitutive BCR signaling. Performing anti-FLAG coimmunoprecipitations, we observed equivalent enrichment for MALT1 in WT and BCL10R58Q as well as in another CARD missense mutant BCL10R87Q. In contrast, there was less binding of MALT1 to BCL10E140X, as well as the similar BCL10K146Nfs*2 truncation mutant (Fig. 4A). Although BCL10 CARD mutants interacted with CARD11 marginally better than BCL10WT, the C-terminal mutants manifested much greater CARD11 interaction, which is likely due to the increased spontaneous polymerization of truncated BCL10, as BCL10 polymers were shown to enhance interaction with CARD11 (42). The observed weaker recruitment of MALT1 by BCL10 truncation mutants was surprising, as previous studies have mapped BCL10 CARD as a MALT1 interacting domain (Fig. 4B) shown by mutagenesis and cryo-EM structure of the BCL10−MALT1 filamentous complex (14, 38).
Figure 4.
Cryo-EM structure of the BCL10E140X filament. A, Immunoblot analysis of BCL10 interactors performed by coimmunoprecipitating with anti-FLAG antibody in Raji cells overexpressing either FLAG-BCL10WT or FLAG-BCL10mutant protein. Samples were blotted for anti-FLAG, anti-MALT1, and anti-CARD11. Input was loaded with 1% of total cell lysate used for immunoprecipitation (IP). Anti-IgG antibody was used as a negative control for coimmunoprecipitation. B, Domain organization of the MBP-human BCL10 construct mapped with MALT1 previously defined and new binding sites. C, Domain organization of the human MALT1 construct. D, SDS-PAGE of MALT1 (Ig1–Ig2) pulldown by His-tagged BCL10 (165–233; left) and His-tagged BCL10 (116–164; right). *, A contaminant. E, SDS-PAGE of MALT1 (Ig1–Ig2) pulldown by different truncations of His-tagged BCL10. F, Negative stained EM micrographs of purified BCL10 WT filaments alone and with MALT1 (left) in comparison with BCL10E140X filaments alone and with MALT1 filaments (right) resulted in similar filaments. G, Cryo-EM structure of BCL10E140X–MALT1 DD filament at 4.3 Å fitted into the cryo-EM density map (left). The 4.3 Å structure is similar to the previously published BCL10WT CARD-MALT1 DD structure at 4.9 Å (right). However, BCL10E140X–MALT1 DD shows improved density for the MALT1 DD domain. EMDB, Electron Microscopy Data Bank; PDB, Protein Data Bank. H, Cryo-EM structure of BCL10E140X CARD and MALT1 DD (cyan) filament, emphasizing EM density for MALT1 DD. I, Monomeric BCL10E140X CARD–MALT1 DD (cyan) align to published monomeric BCL10WT CARD–MALT1 DD. J, Western blot for gel filtration fractions from HBL1 cells stably expressing FLAG-tagged BCL10 WT, R58Q and E140X. Different fractions were blotted with anti-FLAG and anti-MALT1 antibodies. BCL10E140X formed highly ordered oligomers migrating together with MALT1. K, MALT1 inhibits BCL10 filament formation through the BCL10 C-terminal binding site. Quenching polymerization was measured for purified Alexa488-labeled BCL10 WT, E140X, and R58Q at 3 μmol/L in the presence of increasing amounts of MALT1 (0, 1.5, 3, 6, and 12 μmol/L). The assay was initiated upon the addition of the 3C protease in order to remove the MBP tag from BCL10 WT, E140X, and R58Q for allowing filament polymerization. Quenching was monitored for 2 hours with 30-second intervals using a Neo BioTek plate reader and performed with three biological replicates. Titration of increasing doses of MALT1 suppressed filament polymerization of fluorescently labeled BCL10 WT and R58Q. However, increasing doses of MALT1 had very little effect on E140X filament polymerization.
MALT1 has multiple domains (Fig. 4C), and in the reported cryo-EM structure, only the MALT1 DD was ordered and interacted with the BCL10 CARD (residues 1–115), whereas the MALT1 immunoglobulin-like domains (Ig1–Ig2) and the paracaspase domain were not visible (14). Given that a previous mapping study suggested that the Ig1–Ig2 domains of MALT1 also interact with BCL10 (43), we wondered if there were additional MALT1 binding sites on BCL10 at its largely unstructured C-terminus. We thus divided the C-terminal region into two halves and found that the second half (residues 165–233), but not the first half (residues 116–164), pulled down the Ig1–Ig2 construct of MALT1 when coexpressed in Escherichia coli shown by Coomassie blue–stained SDS-PAGE gel (Fig. 4D). Further truncations of the BCL10 165–233 fragment showed that a construct containing residues 165–208 was sufficient to pull down MALT1 Ig1–Ig2 (Fig. 4E), confirming this second MALT1 binding site (Fig. 4B). Of note, unlike the interaction between BCL10 CARD and MALT1 DD in the filamentous form, the interaction between the BCL10 C-terminal region and MALT1 Ig1–Ig2 is monomeric as assessed by gel filtration chromatography of the complex (Supplementary Fig. S4A).
Given that this MALT1 binding domain is deleted from BCL10E140X, we next explored whether there was any impairment in MALT1 recruitment to BCL10 filaments by generating BCL10WT and BCL10E140X filaments in vitro and incubating them with purified, full-length MALT1, followed by negative staining EM. These experiments showed equivalent patterns of MALT1 decorating the surface of WT and BCL10E140X filaments, suggesting that MALT1 recruitment was intact in each case (Fig. 4F). However, given that this analysis does not have sufficient resolution to show whether MALT1 distribution on BCL10 polymers was altered, we next collected cryo-EM data on a Titan Krios microscope operating at 300 keV and equipped with a Falcon II direct electron detector and determined the cryo-EM structure of BCL10E140X filaments in complex with MALT1 at 4.3 Å resolution. The structure of the BCL10E140X filament with MALT1 was highly similar to that of the BCL10WT filament with MALT1 at 4.9 Å resolution (14), in which the DDs of MALT1 bind the CARD of BCL10 and decorate the outside of the core CARD filament (Fig. 4G–I). The conserved structure confirmed that the truncation did not affect the association of BCL10 filaments with MALT1 but that BCL10E140X is nonetheless defective in interacting with monomeric MALT1.
To further investigate these associations, we performed gel filtration analysis from lysates of ABC-DLBCL cells expressing FLAG-tagged BCL10WT, BCL10R58Q, and BCL10E140X, respectively (Supplementary Fig. S4B). Cell fractionation of these lysates revealed a relatively small proportion of BCL10WT or BCL10R58Q in high-molecular-weight fractions corresponding to filaments, along with a small fraction of MALT1, whereas most BCL10 and MALT1 proteins were in low-molecular-weight fractions (Fig. 4J). In marked contrast, BCL10E140X was present at higher abundance in high-molecular-weight fractions, with corresponding enrichment of MALT1 (Fig. 4J). Reciprocally, there was a reduced abundance of BCL10E140X and a notable reduction of MALT1 in the lower molecular weight complexes. Similar findings were observed by performing sucrose gradient experiments (Supplementary Fig. S4C).
The association between enhanced polymerization and lack of MALT1 monomeric interaction of BCL10E140X prompted us to hypothesize that MALT1 binding to the C-terminal region of BCL10 might inhibit BCL10 polymerization. To investigate whether this was the case, we incubated Alexa488-labeled, MBP-fused BCL10WT, BCL10R58Q, or BCL10E140X with increasing concentrations of purified MALT1, treated the reactions with C3 protease to remove the MBP moiety, and monitored polymerization kinetics using fluorescence quenching (Fig. 4K). For BCL10WT and BCL10R58Q, increasing doses of MALT1 suppressed BCL10 filament formation in a dose-dependent manner. By contrast, there was little suppression of BCL10E140X polymerization by MALT1 at any dose. Taken together, these data suggest that BCL10E140X, and likely other similar truncation mutations, favor BCL10 polymerization in cells not only by reducing its intrinsic critical concentration threshold (Fig. 2B and C) but also by abrogating a novel MALT1 inhibitory effect mediated through interaction with the novel BCL10 C-terminal region binding site. By the same token, loss of C-terminal tail binding would increase the pool of MALT1 that is available to bind to BCL10 polymers, suggesting that the end result would be a potent enhancement of BCL10 filament formation and MALT1 activity.
Differential Activation of MALT1 by BCL10E140X versus BCL10R58Q
Because MALT1 dimerization on BCL10 filaments activates its proteolytic function, we wondered whether skewing of MALT1 cellular pools toward the BCL10 polymer-bound state in the BCL10E140X setting might lead to higher cellular levels of MALT1 activity. We therefore examined the effect of BCL10 mutants on MALT1 activity within cells in the absence of basal BCR signaling. For this, we performed MALT1 enzymatic reporter assays, using a GloSensor protein construct engineered with a specific MALT1 cleavage site (44) in Raji cells. Raji GloSensor cells were then engineered to express BCL10WT, BCL10E140X, or BCL10R58Q, and we observed significantly greater MALT1 enzymatic activation in BCL10E140X cells compared with either BCL10WT or BCL10R58Q (Supplementary Fig. S4D and S4E). There was also greater MALT1 activity when comparing BCL10R58Q to BCL10WT, albeit to lesser extent, consistent with the slightly enhanced polymerization kinetics of BCL10R58Q. Finally, we performed similar MALT1 protease reporter assays in ABC-DLBCL cell lines expressing BCL10WT, BCL10E140X, or BCL10R58Q, in which there is constitutive activation of signaling to the CBM complex. Again, BCL10E140X generally yielded the strongest MALT1 activation (Supplementary Fig. S4F and S4G), and BCL10R58Q generally yielded greater MALT1 activity than WT BCL10. Collectively, these data indicate that both BCL10R58Q and especially BCL10E140X, through distinct mechanisms, lead to aberrantly increased MALT1 activity.
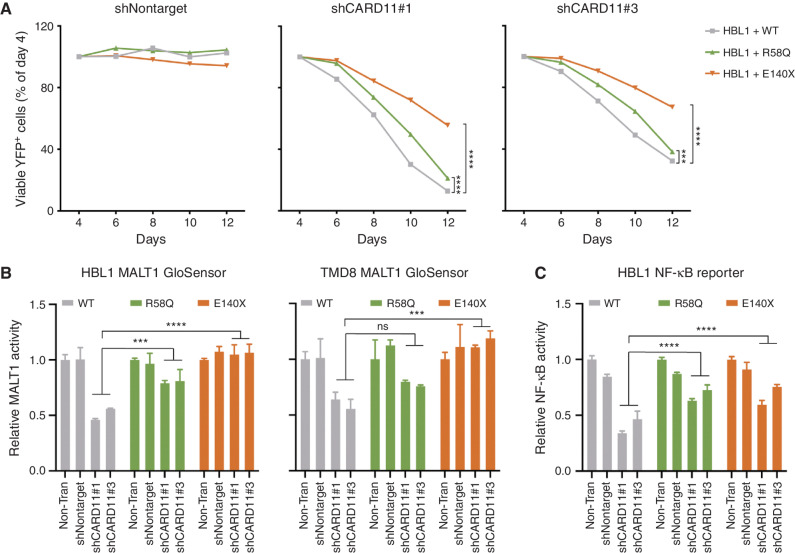
BCL10E140X Confers Reduced Dependency on CARD11 for CBM Activation
Normally, active CARD11 is required to nucleate the formation of BCL10 filaments (9, 10). However, we wondered whether the requirement for CARD11 might be diminished in ABC-DLBCL cells expressing BCL10E140X given its greater tendency to polymerize and loss of MALT1 inhibitory interactions. We therefore performed CARD11 short hairpin RNA (shRNA) knockdown experiments in isogenic ABC-DLBCL cells expressing BCL10WT, BCL10R58Q, and BCL10E140X. CARD11 depletion is known to cause proliferation arrest of ABC-DLBCL cells (20), and, accordingly, we observed significant growth suppression induced by CARD11 knockdown in the presence of WT BCL10 (Fig. 5A; Supplementary Fig. S5A–S5C). However, this effect was significantly blunted in the presence of BCL10E140X and to a lesser extent by BCL10R58Q. To determine how this effect might relate to CBM complex function, we tested the impact of CARD11 knockdown on MALT1 activity using GloSensor reporter assays. MALT1 activity was highly impaired after CARD11 knockdown in the presence of WT BCL10, whereas this effect was completely rescued in BCL10E140X cells and partially rescued by BCL10R58Q (Fig. 5B). CARD11 knockdown also reduced NF-κB reporter activation in BCL10WT ABC-DLBCL cells, an effect that was blunted in CARD11-depleted ABC-DLBCL cells expressing BCL10R58Q and BCL10E140X (Fig. 5C). Hence, an additional perturbation explaining BCL10E140X activation of MALT1 may link to its reduced requirement for CARD11 to induce filament formation, consistent with our data showing markedly greater activity in unstimulated B cells. BCL10R58Q, which does not polymerize as readily as BCL10E140X and is still inhibited by MALT1 monomers, retains a greater degree of CARD11 dependency.
Figure 5.
BCL10 mutations are less dependent on upstream CARD11. A, Viability of HBL1 lymphoma cell lines transduced to express shRNA targeting CARD11 with two independent hairpins or nontargeting control. The indicated lines stably expressing WT and mutant BCL10 were transduced with lentiviruses expressing CARD11 shRNA along with YFP. The relative number of YFP+ live cells was plotted by normalizing them to day 4 (the YFP+ peak). ***, P < 0.001; ****, P < 0.0001. B, MALT1 activity using the MALT1 GloSensor reporter cells with CARD11 knockdown. The indicated MALT1 GloSensor cell lines were stably expressing WT and mutant BCL10, and then transduced with lentiviruses expressing nontargeting or two independent CARD11 hairpins coexpressing a YFP reporter. At day 4, cells were harvested for a MALT1 activity assay. Error bars, SEM with four biological replicates. ***, P < 0.001; ****, P < 0.0001; ns, not significant. C, NF-κB activity in lymphoma reporter cells with shCARD11. The HBL1 NF-κB reporter cells were stably expressing WT and mutant BCL10, and then transduced with lentiviruses expressing nontargeting or two independent CARD11 hairpins coexpressing a YFP reporter. NF-κB activity was measured 72 hours posttransduction. Error bars, SEM with four biological replicates. ****, P < 0.0001.
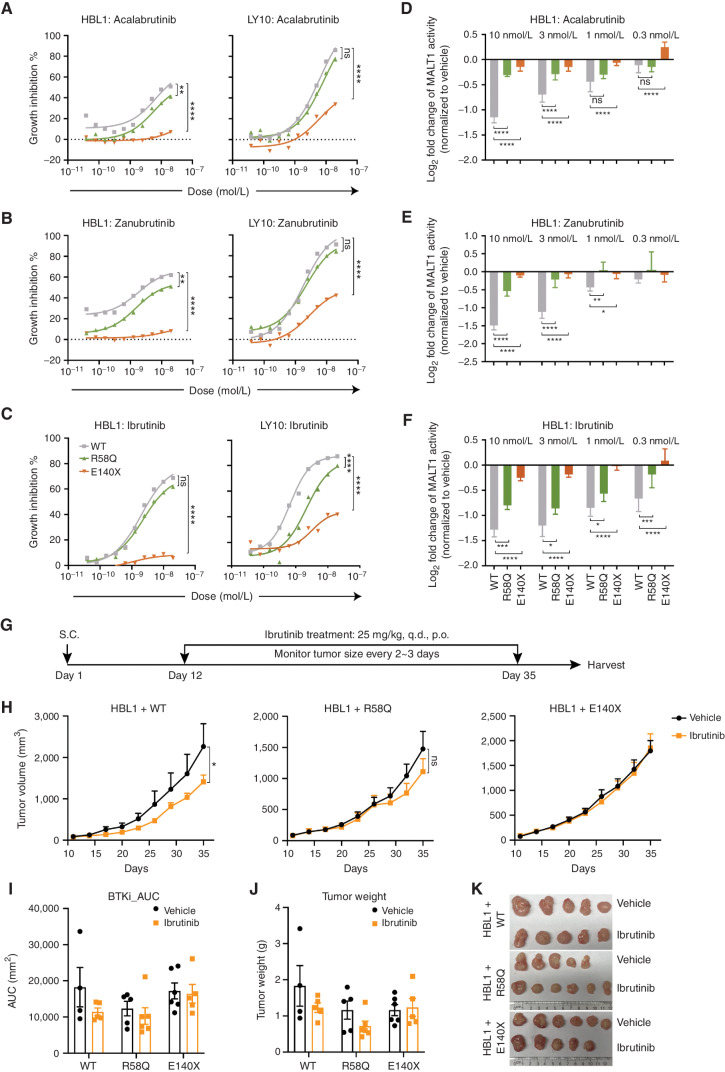
BCL10R58Q and BCL10E140X Confer Distinct Levels of Resistance to Ibrutinib
BTK inhibitors have emerged as a precision therapy modality for ABC-DLBCLs (23, 45). However, lymphoma cells with inherent or acquired mutations in activating proteins downstream of BTK (e.g., CARD11 mutations that induce potent MALT1 activation) are often resistant to such treatments (23, 26). Given the distinct functional profiles, CARD11 dependencies, and MALT1 activation effects of BCL10 CARD and truncation mutants, we wondered whether and to what extent they might confer BTK inhibitor resistance. We treated our isogenic ABC-DLBCL cells expressing BCL10WT, BCL10R58Q, and BCL10E140X proteins with escalating doses of three chemically distinct covalent BTK inhibitors—ibrutinib, acalabrutinib, or zanubrutinib—and tested their proliferation rates using an ATP fluorescence assay after 96 hours of drug exposure. BCL10R58Q conferred at least a modest and often significant reduction in response to these drugs (Fig. 6A–C; Supplementary Fig. S6A). In contrast, BCL10E140X conferred far more dramatic resistance in almost all cases.
Figure 6.
BCL10 gain-of-function mutant lymphomas are resistant to upstream BTK inhibitors. A–C, A growth inhibition assay of lymphoma cells expressing WT or mutant BCL10 in response to BTK inhibitors. X-axis, concentration of compound (mol/L); y-axis, inhibition of cell growth normalized to vehicle-treated cells.**, P < 0.01; ****, P < 0.0001; ns, not significant. D–F, Luciferase activity measured in the MALT1 GloSensor reporter cell lines with BTK inhibitor treatment. Indicated MALT1 GloSensor reporter lines were stably expressing WT and mutant BCL10, and then treated with BTK inhibitors of different ranges (10–0.3 nmol/L). NF-κB activity was performed 24 hours after treatment. Error bars, SEM with four biological replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. G, Experimental design of xenografts with ibrutinib treatment. p.o., orally; q.d., every day; S.C., subcutaneous. H, Tumor growth curve for xenografts of HBL1 cells expressing WT and mutant BCL10 treated with vehicle and ibrutinib (n = 5–6/group). Mice were treated orally with 25 mg/kg ibrutinib once per day for 24 consecutive days. *, P < 0.05; ns, not significant. I, Analysis of area under the curve (AUC) in H. AUC was calculated with Prism. BTKi, BTK inhibitor. J, Tumor weight measured at the endpoint. K, Representative photos of tumors harvested at the endpoint.
All three BTK inhibitors yielded potent and dose-dependent suppression of MALT1 activity in BCL10WT ABC-DLBCL cells (Fig. 6D–F; Supplementary Fig. S6B–S6D). However, isogenic BCL10R58Q and BCL10E140X ABC-DLBCL cells manifested significantly less impact on MALT1, especially in the case of BCL10E140X. Analyzing the further downstream impact of the BTK inhibitors on NF-κB reporter activity revealed significant impairment in BCL10WT, which was significantly blunted in the presence of BCL10R58Q and BCL10E140X (Supplementary Fig. S6E–S6G). Finally, we administered ibrutinib (37.5 mg/kg, oral gavage, every day) to mice bearing BCL10WT-, BCL10R58Q-, and BCL10E140X-expressing ABC-DLBCL xenografts (Fig. 6G). Ibrutinib yielded the expected growth suppression of BCL10WT ABC-DLBCL tumors but had no significant antitumor effect against the two mutant forms, which was most clearly evident in the case of BCL10E140X (Fig. 6H–K). Collectively, BCL10R58Q and BCL10E140X confer distinct levels of resistance to BTK inhibition, consistent with their different mechanisms of action and impact on MALT1 activation.
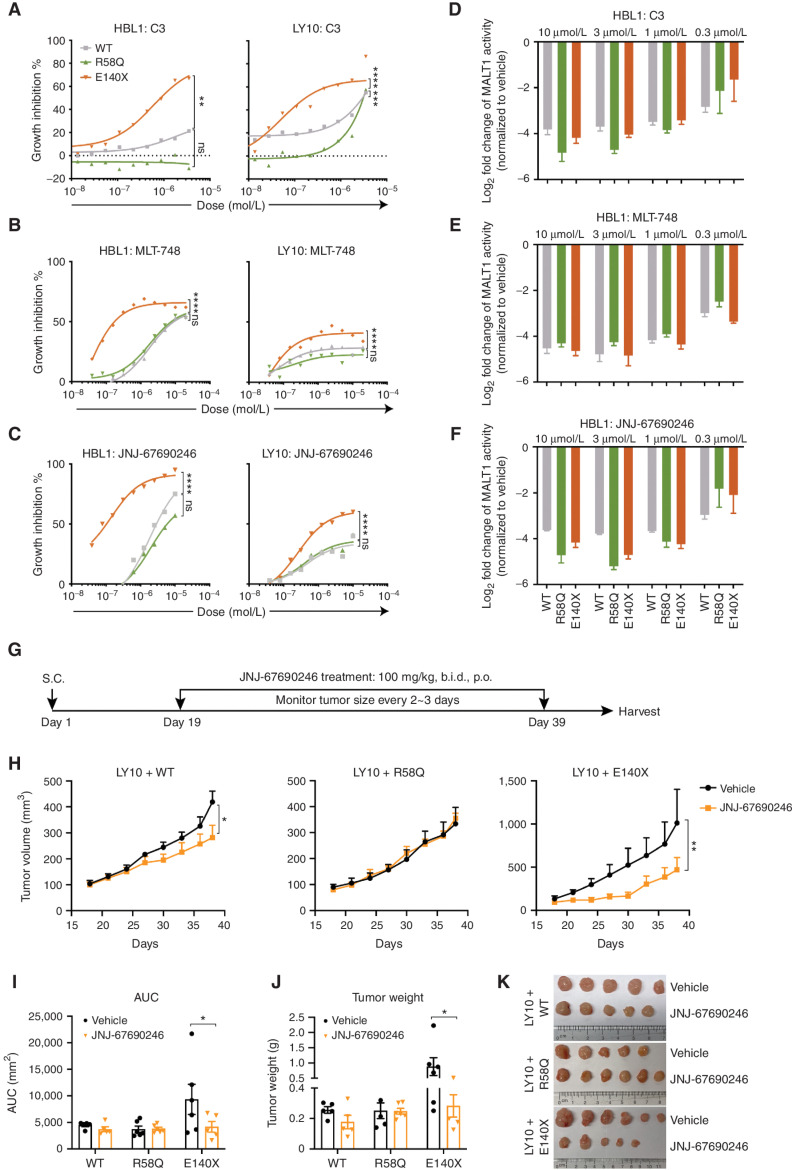
BCL10 Truncating Mutant Lymphomas Are Hypersensitive to the MALT1 Protease Inhibitor
The fact that BCL10 truncating mutants drive potent MALT1 activation even in the absence of CARD11 and confer reduced response to ibrutinib led us to hypothesize that these cells might be especially dependent on MALT1 and hence highly responsive to MALT1 inhibitors. To explore this question, we tested the impact of three chemically and mechanistically distinct MALT1 inhibitors against our set of isogenic ABC-DLBCL cells. These included C3, a potent and specific compound that covalently inactivates the MALT1 catalytic pocket (44); MLT-748, a reversible allosteric compound that binds MALT1 in the Trp580 side chain, thus to lock the protease inactive (46); and JNJ-67690246, an allosteric MALT1 inhibitor (Supplementary Fig. S7A; ref. 47). JNJ-67690246 potently inhibits MALT1 enzymatic activity (IC50 = 15 nmol/L) in biochemical assays and cytokine secretion of IL6/10 (IC50 = 60 nmol/L) in OCI-Ly3 cellular assays (Supplementary Fig. S7B).
Isogenic BCL10WT, BCL10R58Q, and BCL10E140X ABC-DLBCLs were exposed to increasing concentrations of each of these compounds for 96 hours, revealing striking differences in the response profiles of BCL10E140X versus BCL10WT and BCL10R58Q (Fig. 7A–C; Supplementary Fig. S7C). Both BCL10WT and BCL10R58Q were generally sensitive to the allosteric inhibitors, whereas BCL10R58Q cells were less sensitive than BCL10WT to C3. In marked contrast, BCL10E140X manifested a significantly greater response to all three MALT1 inhibitors. This differential effect was not due to variation in the degree of MALT1 inhibition, as MALT1 activity was equivalently suppressed by all three drugs in BCL10WT, BCL10R58Q, and BCL10E140X ABC-DLBCL cells (Fig. 7D–F; Supplementary Fig. S7D–S7F). Analysis of NF-κB reporter activity showed significantly greater impairment in MALT1 inhibitor–treated BCL10E140X cells as compared with either BCL10WT or BCL10R58Q (Supplementary Fig. S7G–S7I), with the latter even showing less impairment of NF-κB activity than in WT cells, suggesting that some other pathway may be maintaining NF-κB and hence conferring less dependency on MALT1 than in BCL10E140X cells. Finally, we treated BCL10WT, BCL10R58Q, and BCL10E140X ABC-DLBCL xenografts with JNJ-67690246 in vivo, using the OCI-Ly10 cell line, which is generally less sensitive to MALT1 inhibition, for greater stringency (Fig. 7A–C and G). We observed a significant reduction in the growth of BCL10E140X but not BCL10WT or BCL10R58Q lymphomas (Fig. 7H–K), confirming their increased dependency on MALT1 activity and the potential for MALT1 inhibitors to be most useful for patients bearing such mutations.
Figure 7.
BCL10 truncating mutant lymphomas are hypersensitive to MALT1 protease inhibitors. A–C, A growth inhibition assay of lymphoma cells expressing WT or mutant BCL10 in response to MALT1 inhibitors. X-axis, concentration of compound (mol/L); y-axis, inhibition of cell growth normalized to vehicle-treated cells. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant. D–F, Luciferase activity measured in the MALT1 GloSensor reporter cell lines with MALT1 inhibitor treatment. Indicated MALT1 GloSensor reporter lines were stably expressing wild-type and mutant BCL10, and then treated with MALT1 inhibitors of different ranges (10–0.3 μmol/L). NF-κB activity was performed 24 hours after treatment. Error bars, SEM with four biological replicates. G, Experimental design of xenografts with JNJ-67690246 treatment. b.i.d., twice per day; p.o., orally; S.C., subcutaneous. H, Tumor growth curve for xenografts of HBL1 cells expressing WT and mutant BCL10 treated with vehicle and JNJ-67690246 (n = 3–6/group). Mice were treated orally with 100 mg/kg twice per day for 19 consecutive days. *, P < 0.05; **, P < 0.01. I, Analysis of area under the curve (AUC) in H. AUC is calculated with Prism. *, P < 0.05. J, Tumor weight measured at the endpoint. *, P < 0.05. K, Representative photos of tumors harvested at the endpoint.
DISCUSSION
Herein, we show that BCL10, one of the most frequently mutated genes in DLBCL, is a bona fide genetic driver of lymphomagenesis. Importantly, our structure–function studies reveal that these mutations occur in at least two biochemically distinct classes: missense mutations of the CARD domain and truncation mutations of the C-terminal tail. These classes of mutations seem to affect distinct aspects of BCL10 functionality and lead to biologically distinct outcomes as indicated by their differential downstream effects on MALT1 and NF-κB signaling as well as vulnerability to targeted therapies. Many of the BCL10 truncating mutations cluster between AA 135 and 174, and representatives of these mutations manifested the most powerful activation of NF-κB activity. BCL10 truncation mutants such as BCL10E140X manifested a striking increase in its ability to polymerize into its filamentous form, accompanied by potent activation of MALT1 protease activity. This tendency to polymerize, indicated for example by its lower critical concentration threshold, may help to explain the reduced CARD11 dependency of lymphoma cells expressing BCL10E140X and is consistent with previous studies showing that the BCL10 CARD domain alone can undergo spontaneous polymerization in vitro (9, 48). Although it is generally understood that CARD11 serves to nucleate BCL10 polymerization, it has been suggested that CARD11 association with BCL10 filaments is further stabilized by nascent helical BCL10 polymers (42). This may explain why we observed an increased association of BCL10 and CARD11 in lymphoma cells expressing BCL10E140X, given its greater tendency to polymerize, whereas at the same time being consistent with its reduced requirement for CARD11 to induce filament formation and reduced biological dependency on CARD11 in BCL10E140X-expressing DLBCL cells.
The binding of the MALT1 DD to BCL10 was unperturbed in filaments composed of BCL10E140X, which is not surprising because this molecular association is mediated through the BCL10 CARD domain and proximal regions that are not affected by truncation mutation (14, 43). Yet coimmunoprecipitation experiments paradoxically indicated reduced interaction between BCL10 and MALT1. This prompted us to examine other modes of BCL10–MALT1 association, leading to the identification of a novel direct interaction site between the MALT1 Ig1–Ig2 region and BCL10 AAs 165–208, a region that is lost in a majority of truncation mutants except for a cluster deleting the extreme C-terminal Ser/Thr-rich tail. Importantly, we show that MALT1 impairs BCL10 polymerization through this interaction surface, thus constituting a novel CBM-negative regulatory mechanism preventing spurious polymerization of BCL10. This in turn likely explains the dramatically increased filament formation by BCL10E140Xin vitro, and its greatly enhanced ability to recruit MALT1 into the polymerized CBM complex, given that loss of monomeric BCL10 and MALT1 binding would increase the pool of MALT1 to associate with filaments. These events occur due to the presence of BCL10E140X and, thus, appear to constitute a positive feedback loop that ultimately causes potent MALT1 protease activation and biological dependency on MALT1 catalytic function. The more distal set of C-terminal truncating mutations such as Q208X, L209X, and L225X retain the MALT1 Ig1–Ig2 interacting region and hence would not be expected to escape from this MALT1 inhibitory binding. Accordingly, when expressed in cells, they did not induce greater NF-κB activity than WT BCL10, suggesting that there may be additional ways in which BCL10 function could be perturbed, perhaps due to specific loss of certain as of yet undiscovered posttranslational modifications. Interestingly, L225X produces a truncated form of BCL10 similar to the MALT1 protease–dependent cleavage form of BCL10 R228X observed in activated T cells as well as in ABC-DLBCL cells with chronic active BCR signaling (49, 50) and showed similar functional effects to WT BCL10 overexpression in NF-κB reporter assays, perhaps due to retaining both intact MALT1 binding sites. Notably, the cleaved BCL10 R228X form was shown to mediate migratory function in T cells (49), pointing to the need for further studies of these BCL10 mutations. BCL10 truncating mutations, translocation, and amplification were also shown to occur in marginal zone lymphomas including mucosa-associated lymphoid tissue (MALT) lymphomas (51–54). However, the impact of BCL10 mutations in marginal zone lymphomas has not been explored and warrants further study.
In contrast, missense mutations of the BCL10 CARD domain seem to have distinct functional effects. Many of the BCL10 residues affected by mutations (e.g., R58 and K63) are localized within the core of BCL10 helical structures where they make important intrastrand (type III) and interstrand (type I) interactions that are critical for filament formation (10, 42). Along these lines, an R53Q mutation affecting type III interactions might be predicted to disrupt intrastrand interactions and caused a severe defect in BCL10 CARD domain polymerization. This was, however, proven to be incorrect, and the sole CARD domain hotspot mutant residue Q58 engages in a novel form of type III interaction resulting in the apparent formation of a hydrogen-bonded glutamine network. The consequence is a shift in BCL10 polymerization kinetics, favoring the polymerized state but without dramatically altering binding to CARD11 or MALT1 and yielding a more modest gain-of-function phenotype. Missense mutations such as K63Q might functionally resemble R58Q because they also locate at the central core of the BCL10 CARD filament and have positively charged residues switched to glutamine to potentially enhance rather than disrupt interactions.
These biochemical features of the BCL10R58Q mutant result in a hypomorphic phenotype compared with the truncation mutant in which they induced less potent MALT1 and NF-κB activation than BCL10E140X. However, our results also suggest that BCL10R58Q may engage in additional gain-of-function effects. This is suggested by the fact that cells expressing BCL10R58Q seemed relatively resistant to loss of NF-κB activity upon exposure to MALT1 inhibitors as compared with that on BCL10WT or BCL10E140X, whereas in contrast, reduction in NF-κB activity in response to the upstream BTK inhibitors was similar between BCL10R58Q and BCL10E140X. These data prompt us to hypothesize that other functions may derive from the stability or bundling of the R58Q filament. For example, more stable BCL10 filaments might enhance MALT1 recruitment and activation of TRAF6, which could partially support NF-κB independently from the MALT1 proteolytic function (10), or activate NF-κB through other alternative means such as linear ubiquitylation (LUBAC)–associated mechanisms (55, 56). Engagement of these or other MALT1 paracaspase–independent biochemical effects would be consistent with BCL10R58Q DLBCL cells retaining greater dependency on CARD11 and hence upstream signaling and responsiveness to BTK inhibitors.
Overall, our data spotlight the complexities involved in developing precision therapies for DLBCLs and other tumors. The identification of chronic active BCR signaling as a characteristic of ABC-DLBCLs has led to intense efforts to integrate BTK inhibitors into multimodality regimens. However, to date, it is clear that many patients still do not benefit from the addition of such compounds. Our data point to biochemical mechanisms that might help to explain this, as exemplified most clearly by the BTK inhibitor resistance conferred by BCL10 truncation mutants and this suggests that such patients should not be treated with BTK inhibitor–containing regimens. Instead, patients with BCL10 truncation mutations would likely best be served by incorporating MALT1 inhibitors. This concept is feasible because several MALT1 inhibitors are already in clinical trials. Although these findings could also be relevant to acquired BTK inhibitor resistance (25, 26), as of yet BCL10 mutations have not been identified in this setting. Similar considerations may apply to other (but not all) mutations downstream of BTK, as exemplified by the case of CARD11 coiled-coil domain mutants, which induce resistance to BTK inhibitors but not MALT1 inhibitors. However, the fact that BCL10 CARD domain mutations may still retain BTK inhibitor responsiveness further underlines the need for rigorous study of signaling pathway mutations such as these, and perhaps eventually the need for targeted sequencing studies to provide a precision therapy “map” of these tumors allowing selection (or even combination) of the targeted therapies (e.g., BTK inhibitors vs. MALT1 inhibitors) appropriate to their specific signaling scenarios.
METHODS
Cell Culture
Raji, HBL1, TMD8, and RIVA were cultured in RPMI supplemented with 10% FBS, 2 mmol/L L-glutamine, and 10 mmol/L HEPES. OCI-Ly10 was cultured in Iscove's medium supplemented with 20% FBS and 2 mmol/L L-glutamine. 293T was cultured in Dulbecco's modified Eagle medium with 10% FBS. All cell lines were authenticated by the University of Arizona Genetic Core, and grown in the presence of 1% penicillin G and streptomycin at 37°C in a humidified atmosphere of 5% CO2. HBL1 and RIVA were obtained from Jose A. Martinez-Climent (Universidad de Navarra, Pamplona, Spain); TMD8 was obtained from Louis M. Staudt (NCI, Bethesda, MD); and the OCI-Ly10 cell lines were obtained from the Ontario Cancer Institute (OCI).
Virus Production and Transduction
Lentiviruses were produced in 293T cells by cotransfecting shRNA (short hairpin sequences were: shNontargeting: CAACAAGATGAAGAGCACCAA; shCARD11#1: GGACGACAACTACAACTTAGC; shCARD11#3: TGGTCAAGAAGCTGACGATTC) or overexpression vectors with packaging vectors psPax2 (Addgene; #12260, RRID: Addgene_12260) and psMD2.g (Addgene; #12259, RRID: Addgene_12259) at a 4:3:1 ratio in serum-free media. The supernatant containing virus particles was harvested 48 and 72 hours after transfection, filtered through a 0.45-μm filter, and then concentrated with PEG-it according to the manufacturer's instructions (LV825A-1, System Biosciences). Virus was resuspended with PBS containing 25 μmol/L HEPES and added to the cells for overnight infection. Cells were selected 24 hours after transfection by adding puromycin (Sigma), blasticidin (InvivoGen), or G418 (Life Technologies) for at least 48 hours.
Xenograft
All mouse experiments were approved by Institutional Animal Care and Use Committee (IACUC) at Weill Cornell Medicine and were performed following the IACUC guidelines. Eight- to 10-week-old, female NOD.Cg-prkdcscid Il2rgtm1Wjl/SzJ (NSG, RRID: IMSR_JAX: 005557) mice were obtained from The Research Animal Resource Center at Weill Cornell Medicine. HBL1 (5 × 106) or OCI-Ly10 (107) and their derived engineered cells were resuspended with PBS/Matrigel (1:1) and subcutaneously injected to the right flank of mice. Treatments were started when tumor volume reached an average of 100 mm3. Ibrutinib was prepared in corn oil with 10% (v/v) DMSO or 0.5% methylcellulose in water and administrated orally at 25 or 37.5 mg/kg once per day. JNJ-67690246 was prepared in PEG400 with 10% (w/v) PVPVA64 and administrated orally at 100 mg/kg twice per day. Tumor volume was monitored 2 to 3 times/week with a digital caliper and calculated using the following formula: smallest diameter2 × largest diameter × 0.5.
Growth Inhibition Assay
DLBCL cell lines were cultured in exponential conditions, and the cell growth was determined by CellTiter-Glo (Promega). Three thousand to 5,000 cells were seeded and cultured in each well of a 384-well plate for 96 hours and treated with compounds every 48 hours. Luminescence was read at the endpoint with the Synergy NEO microplate reader (BioTek). The value of compound-treated cells was normalized to their vehicle-treated controls and then used to calculate IC50 in GraphPad Prism (RRID:SCR_002798).
NF-kB Reporter Assay
For the NF-κB reporter assay in 293T, the plasmids expressing different BCL10 mutations were transiently transfected into 293T cells using Lipofectamine (Invitrogen). Renilla luciferase plasmid was cotransfected as an internal control. Twenty-four hours after transfection, cells were collected, and luciferase activities were measured in the Synergy NEO microplate reader (BioTek) with the Dual Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions and normalized to Renilla luciferase activity.
To generate stable NF-κB reporter cells, the lentivirus expressing 3xNF-κB response element followed by firefly luciferase was made and infected the parental cells. Puromycin was then added for antibiotic selection 24 hours after infection. Reporter cells were further validated by BTK inhibitor and MALT1 protease inhibitor treatment and phorbol 12-myristate 13-acetate/ionomycin (PMA/IO) stimulation. NF-κB reporter cells expressing BCL10 were generated by infecting reporter cells with different lentiviral BCL10 isoforms (coexpressing GFP), followed by sorting out GFP+ cells. Stable NF-κB reporter cells were harvested at the indicated conditions and lysed with 1× passive lysis buffer at room temperature (RT) for 20 minutes. The lysate was briefly centrifuged, and the supernatant was collected for luciferase activity. All the assays were presented as mean ± SEM of three independent experiments.
MALT1 GloSensor Assay
The generation of the Raji MALT1 GloSensor reporter cell has been previously described (44). All other GloSensor reporter cells were generated by infecting parental cells with lentiviral MALT1 GloSensor (pLex306 backbone), followed by antibiotic (blasticidin) selection. All derived GloSensor cells were further validated by MALT1 protease inhibitor treatment and PMA/IO stimulation.
Immunoprecipitation
Lymphoma cells (108) were collected, washed with cold PBS, and resuspended with lysis buffer (1% NP40, 10% glycerol, 150 mmol/L NaCl, 20 mmol/L Tris-HCL pH 7.5, and freshly added protease inhibitors). The lysates were centrifuged at 15,000 × g at 4°C for 15 minutes, and the supernatant was then collected and incubated with 50 μL equilibrated anti-FLAG magnetic beads (Sigma-Aldrich; cat. #M8823, RRID:AB_2637089) at 4°C for 3 hours. The beads were washed 3 times with lysis buffer and followed by 3 times washing with the lysis buffer without NP40. SDS loading buffer without nonreducing reagent was added and boiled at 95°C for 5 minutes. The elution was added for β-mercaptoethanol (final 10%) and ready to run Western blot after boiling at 95°C for 5 minutes.
Western Blotting
Whole-cell lysates extracted with RIPA buffer or immunoprecipitation elution were separated by SDS-PAGE gels and followed by transferring to PVDF membranes. Membranes were incubated with the indicated primary antibodies: anti-FLAG (Sigma-Aldrich; cat. #F3165, RRID:AB_259529), anti-MALT1 (Santa Cruz Biotechnology; cat. #sc-46677, RRID:AB_627909), anti-CARD11 (Abcam; cat. #ab113409, RRID:AB_10861854), anti–β-Actin (AC-15, Sigma-Aldrich), and then mouse/rabbit peroxidase-conjugated secondary antibodies (Cell Signaling Technology). Protein intensity was detected with enhanced chemiluminescence using the ChemiDoc imaging system (Bio Rad).
Driver Mutation Analysis
The driver mutation analysis is performed using fishhook (https://github.com/mskilab/fishHook) on a total of 243 ABC-DLBCL cases from the NCI cohort. Fishhook is a model built with mutational calls, a set of hypothesis intervals, eligible genomic ranges, and a set of genomic covariates that identifies the depletion and enrichment of the genomic interval statistically. The model used a gamma-Poisson regression to implement the maximum likelihood approximation with consideration of user-assigned covariates and expected mutation density to the hypothesis. With this approach, the model helped us to identify enriched mutations with considerations like chromatin features, sequence context composition, and gene expression.
The eligible region is defined using genecode v19 and fractional coverage of hg19 positions provided by Agilent exome coverage. We also fed the model covariates that define B cell–specific transcriptional states and chromatin state information for the model. The covariates of ABC-specific transcriptional states were generated by the number of the overlap between the transcription start site of genes with TPM >2 in half the ABC-DLBCL cases from the same NCI cohort within the 10-kb eligible regions. The covariates of B cell–specific chromatin states are generated by the number of the overlap between H3K27Ac peaks that were previously reported in the B cells within 100 kb of the eligible regions and the Assay for Transposase-Accessible Chromatin (ATAC) peaks of the B cells within 10 kb of the eligible regions. A total of three covariates were fed to the fishhook model. Here we noted genes of FDR < 0.05 and BCL10 have an FDR of 1.4e−10. A quantile–quantile (QQ) plot is plotted by pairing observed −log10-transformed quantiles of observed P values (y-axis) with their corresponding −log10-transformed quantiles from the uniform distribution (x-axis).
Biochemical Evaluation of MALT1 Protease Activity
MALT1 protease activity was assessed in vitro using full-length MALT1 protein [Strep-MALT1(1-824)-His] purified from baculovirus-infected insect cells. The tetrapeptide LRSR was coupled to 7-amino-4-methylcoumarin (AMC) and provided a quenched, fluorescent substrate for the MALT1 protease (SM Biochemicals). Cleavage of AMC from the arginine residue resulted in an increase in coumarin fluorescence measured at 460 nmol/L (excitation 355 nmol/L). Diluted compounds were preincubated with the MALT1 enzyme for 50 minutes at RT. Substrate was added subsequently, and the reaction was then incubated for 4 hours at RT, after which fluorescence was measured.
IL6/10 Secretion Assay
Secretion of the IL6 and IL10 cytokines by OCI-Ly3 ABC-DLBCL cells was measured using a Mesoscale assay (MSD). MALT1 inhibition results in a decrease of IL6/10 secretion. OCI-Ly3 cells were treated with diluted compounds for 24 hours at 37°C and 5% CO2. After 24 hours of incubation, 50 μL of the supernatant was transferred to an MSD plate [V-Plex Proinflammation Panel 1 (human) kit] and incubated for 2 hours at RT, followed by a 2-hour incubation with IL6/10 antibody solution. Plates were read on a SECTOR imager.
Protein Expression and Purification
All constructs of BCL10 and MALT1 were from human sequences. Full-length WT and mutant BCL10 constructs with N-terminal MBP tag were generated in vector pDB-His-MBP with a 3C protease site between MBP and BCL10. Full-length His-tagged MALT1 cloned into pET29b was purchased from Addgene (RRID:Addgene_48968) and was expressed in E. coli.
All proteins were purified by either Ni-NTA resin (Qiagen) or Amylose resin, followed by gel filtration chromatography (Superdex 200 10/300 GL, GE Healthcare). BCL10 full-length and mutant filaments were purified by an MBP affinity column in binding buffer containing 25 mmol/L Tris at pH 7.5, 300 mmol/L NaCl, and 1 mmol/L TCEP, followed by Superdex 200 gel filtration chromatography in buffer containing 20 mmol/L Tris at pH 7.5, 150 mmol/L NaCl, and 1 mmol/L TCEP, resulted in isolation of a monomeric fraction of BCL10. Then, monomeric MBP-BCL10 was cleaved by 3C protease and incubated at RT for 2 hours in order to allow filament formation. This step was followed by another Superdex 200 gel filtration chromatography, and BCL10 filaments were isolated at the void peak for structure determination and thermostability assay. Isolation of the BCL10 (1–140)–MALT1 complex was performed in a similar way in which BCL10 and MALT1 were purified separately. BCL10 preformed filaments after 3C cleavage were added and mixed together with full-length MALT1 to form a complex.
Negative Stained EM
Copper grids coated with layers of plastic and thin carbon film were glow-charged before 5 μL of purified complexes were applied. Samples were left on the grids for 1 minute followed by negative staining with 1% uranyl formate for 30 seconds and air-dried. In vitro BCL10 WT and mutants, BCL10/MALT1, and CBM were imaged with JEOL 1200EX or Tecnai G2 Spirit BioTWIN at Harvard Medical School EM facility operating at 80 keV.
Cryo-EM Data Collection
Cryo grids for BCL10 R58Q and BCL10 E140X/MALT1 filaments were prepared by applying 3 μL of protein sample on a c-flat (1.2/1.3) 300 mesh grids. Grids were plunged using vitrobot (FEI) at 4°C with 3-second blotting and force 4. For BCL10 R58Q data collection, 3,439 movies were collected at super-resolution mode using an Arctica microscope at the UMASS facility, operated at 200 kv facility with a k2 camera. The movies were collected automatically using SerialEM data collection at a nominal magnification of 36,000 and a pixel size of 0.435 Å, with a total dose of 38 e/Å2, which was fractionated into 40 movie frames, with a defocus range of −1 to −2.5 μm. For BCL10 E140X–MALT1 collection, 700 movies were collected at super-resolution mode using 300 kV FEI Titan Krios microscope equipped with an FEI Falcon II detector at the Pacific Northwest Center for Cryo-EM (PNCC), operated at 300 kv with a Falcon3 camera. The movies were collected automatically using SerialEM data collection at a nominal magnification of 47,000 and a pixel size of 0.4 Å, with a total dose of 55 e/Å2, which was fractionated into 40 movie frames.
Cryo-Electron Data Processing
For the helical reconstruction of BCL10 R58Q and BCL10 E140X/MALT1, Motioncor2 was used for drift correction, and micrographs were CTF-corrected using CTFFIND4. Data were processed using Relion (3.1). The resolutions of the reconstruction were determined by FSC to 4.6 Å and 4.3 Å, respectively. Model building was performed in program Coot36. Refinement was performed using Phenix refine50. Structural presentations were generated using Pymol (DeLano Scientific) and Chimera (57).
Confocal Imaging
Time-lapsed movies of full-length labeled Alexa488-BCL10 full length, BCL10 R58Q, and BCL10 E140X were recorded using a Nikon spinning disk confocal microscope at the Harvard MicRoN imaging core facility for 30 minutes to 1 hour with 1-minute intervals, with ×100 objective. 3C was added at a submolar ratio to allow MBP cleavage to occur within 2 to 3 minutes in order to provide ample time for setting up the microscope and starting the recording.
For critical concentration determination, labeled Alexa488-BCL10 full-length, R58Q, and E140X filaments were formed at increasing concentrations ranging between 0.1 and 1 μmol/L. 4 hours after cleavage with 3C and incubation at RT, samples were placed on a 35-mm bottom glass dish and imaged using a spinning disk confocal microscope with ×100 objective.
Fluorescence Quenching Assay
Purified full-length MBP-BCL10, BCL10 R58Q and BCL10 E140X were mixed with a 5-fold molar excess of Alexa 488-C5-maleimide (Invitrogen) and incubated at 4°C overnight. Gel filtration chromatography (Superdex 200, GE Healthcare) was used to remove free dyes. A fluorescence polarization assay was performed at 18°C in buffer containing 20 mmol/L Tris at pH 7.5, 150 mmol/L NaCl, and 0.5 mmol/L TCEP and in 20 μL volume. Labeled MBP-BCL10 (3 μmol/L) was cleaved with 3C in the presence of an increasing amount of MALT1. The fluorescence quenching was measured right after the 3C addition for 2 hours using a NEO plate reader (BioTek) using excitation/emission wavelengths of 495 nm/519 nm.
Protein Stability
BCL10 full-length, R58Q, and E140X filaments purified from the void peak of Superdex200 were mixed with 1-fold protein thermal shift dye (Thermo Fisher Scientific). Thermal scanning (25°C to 95°C at 1°C/minute) was performed, and melting curves were recorded on a StepOne RT-PCR machine. Data analysis was done by Protein Thermal Shift Software (Thermo Fisher Scientific).
Immunofluorescence
HBL1 cells cultured in fresh media were mixed in a 1:1 ratio with cytospin. Cells were spun at 800 × g for 5 minutes. Cell pellets were resuspended with cytospin and plated on CELLview 4-compartment dishes (Greiner Bio-One). Cells were left at RT overnight and were fixed with 100% cold methanol for 5 minutes at −20°C, followed by cell permeabilization with 0.1% Triton X-100 in PBS-Tween (PBST) for 10 minutes. Cells were incubated with blocking buffer containing 3% BSA for 3 hours to minimize nonspecific binding. After blocking, cells were incubated overnight at 4°C with FLAG primary antibody (Sigma-Aldrich; cat. #F1804, RRID:AB_262044). After incubation, cells were washed with PBST 3 times and incubated with AlexaFluor488-conjugated anti-mouse IgG (Abcam; cat. #ab150113, RRID:AB_2576208) for 1 hour at RT. After incubation, cells were washed with PBS and then stained with Hoechst for 10 minutes (1:500, Immunochemistry Technologies; cat. #639). Cells were imaged using a spinning disk confocal microscope with ×100 objective.
IHC Staining of p65
Formalin-fixed, paraffin-embedded tissue sections of 4 μm thickness were cut from a tissue microarray composed of duplicate 0.6-mm cores from 298 cases of de novo DLBCL. Slides were processed using standard IHC protocols and stained with an antibody against NF-κB p65 (Cell Signaling Technology; cat. #8242, RRID:AB_10859369, 1:500 dilution). Appropriate staining was verified in sections of benign tonsil, heart, and liver. Stained slides were assessed by an expert hematopathologist for the nuclear expression of p65 in DLBCL tumor cells and scored as a percentage of tumor cell nuclei.
Data Availability
The cryo-EM structures have been deposited in the Electron Microscopy Data Bank with the accession numbers EMD-27095 (BCL10 CARD R58Q) and EMD-27100 (BCL10 E140X-MALT1 DD). The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the accession numbers 8CZD (BCL10 CARD R58Q) and 8CZO (BCL10 E140X-MALT1 DD). All other data are available from the corresponding authors upon reasonable request.
Supplementary Material
Acknowledgments
We thank the Melnick lab and Wu lab for extremely helpful discussions and suggestions, as well as Janssen Pharmaceuticals for providing allosteric MALT1 inhibitors and recommendations on the in vivo drug treatment regimen. We thank Drs. Chen Xu and KangKang Song at the UMASS Cryo-EM core facility and Dr. Janette Myers and the Pacific Northwest Center for Cryo-EM at Oregon Health & Science University (OHSU) for CryoEM screening and data collection. We thank Dr. Paula Montero Llopis and Microscopy Resources on the North Quad (MicRoN) core at Harvard Medical School for microscope use and training as well as Maria Ericsson from the Harvard Medical School EM Core for microscope use of transmission electron microscopy imaging. We acknowledge the technical support from the Research Animal Resource Center from Weill Cornell Medicine. A.M. Melnick is supported by NCI/NIH R35 CA220499, LLS-SCOR 7012-16, the Samuel Waxman Cancer Research Foundation, the Follicular Lymphoma Consortium, and The Chemotherapy Foundation. M. Xia is funded by the Leukemia & Lymphoma Society Career Development Program (5483-19). L. Cerchietti and H. Wu are funded by NCI/NIH R01 CA249843. A portion of this research is supported by NIH grant U24GM129547 and was performed at the PNCC at OHSU and accessed through the Environmental Molecular Sciences Laboratory (EMSL; grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. L. Fontan is funded by the MSK Lymphoma SPORE Developmental Research Program (P50 CA192937-03).
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
M. Xia reports grants from Janssen outside the submitted work, as well as a patent for use of BCL10 mutations as biomarkers for targeted therapy in B-cell lymphomas pending. J. Gutierrez reports nonfinancial support from Janssen Oncology during the conduct of the study. C. Meydan reports personal fees from Thorne HealthTech outside the submitted work. G.W. Slack reports personal fees from Seagen outside the submitted work. D.W. Scott reports personal fees from AbbVie, AstraZeneca, Celgene, and Incyte, grants and personal fees from Janssen, and grants from NanoString and Roche/Genentech outside the submitted work, as well as a patent for method for subtyping lymphoma types by means of expression profiling issued and licensed to NanoString. L. Cerchietti reports grants from the NCI during the conduct of the study, as well as grants from Celgene outside the submitted work. L. Fontan reports other support from Janssen R&D and grants from the Department of Defense during the conduct of the study; other support from Janssen R&D outside the submitted work; and patent 11248007 issued to Cornell University and Dana-Farber Cancer Institute, patent 10711036 issued to Cornell University, Dana-Farber Cancer Institute, and Children's Medical Center Corporation, and patent 10689366 issued to Cornell University and Dana-Farber Cancer Institute. H. Wu reports a patent for a MALT1 inhibitor issued. A.M. Melnick reports grants from Janssen, Epizyme, and AstraZeneca, and personal fees from Treeline and Exo-Therapeutics outside the submitted work, as well as a patent for 7602-03-PC issued. No disclosures were reported by the other authors.
Authors’ Contributions
M. Xia: Conceptualization, resources, data curation, formal analysis, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. David: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Teater: Resources, data curation, formal analysis, visualization, methodology. J. Gutierrez: Formal analysis, validation, investigation, visualization, methodology. X. Wang: Data curation, formal analysis, validation, visualization, methodology. C. Meydan: Data curation, formal analysis, visualization, methodology. A. Lytle: Resources, data curation, formal analysis, investigation, methodology. G.W. Slack: Resources, data curation, formal analysis, investigation, methodology. D.W. Scott: Resources, data curation, formal analysis, investigation, methodology. R.D. Morin: Resources, data curation, formal analysis, investigation, methodology. O. Onder: Data curation, formal analysis, investigation, visualization, methodology. K.S.J. Elenitoba-Johnson: Data curation, formal analysis, investigation, visualization, methodology. N. Zamponi: Software, formal analysis, validation, investigation, visualization, methodology. L. Cerchietti: Formal analysis, funding acquisition, methodology, writing–review and editing. T. Lu: Resources, data curation, investigation, methodology. U. Philippar: Resources, investigation, methodology, writing–review and editing. L. Fontan: Conceptualization, formal analysis, funding acquisition, investigation, methodology, writing–review and editing. H. Wu: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, writing–review and editing. A.M. Melnick: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Thome M, Charton JE, Pelzer C, Hailfinger S. Antigen receptor signaling to NF-kappaB via CARMA1, BCL10, and MALT1. Cold Spring Harb Perspect Biol 2010;2:a003004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Meininger I, Krappmann D. Lymphocyte signaling and activation by the CARMA1-BCL10-MALT1 signalosome. Biol Chem 2016;397:1315–33. [DOI] [PubMed] [Google Scholar]
- 3. Juilland M, Thome M. Role of the CARMA1/BCL10/MALT1 complex in lymphoid malignancies. Curr Opin Hematol 2016;23:402–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Turvey SE, Durandy A, Fischer A, Fung SY, Geha RS, Gewies A, et al. The CARD11-BCL10-MALT1 (CBM) signalosome complex: stepping into the limelight of human primary immunodeficiency. J Allergy Clin Immunol 2014;134:276–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lamason RL, McCully RR, Lew SM, Pomerantz JL. Oncogenic CARD11 mutations induce hyperactive signaling by disrupting autoinhibition by the PKC-responsive inhibitory domain. Biochemistry 2010;49:8240–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chan W, Schaffer TB, Pomerantz JL. A quantitative signaling screen identifies CARD11 mutations in the CARD and LATCH domains that induce Bcl10 ubiquitination and human lymphoma cell survival. Mol Cell Biol 2013;33:429–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCully RR, Pomerantz JL. The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol Cell Biol 2008;28:5668–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shinohara H, Maeda S, Watarai H, Kurosaki T. IkappaB kinase beta-induced phosphorylation of CARMA1 contributes to CARMA1 Bcl10 MALT1 complex formation in B cells. J Exp Med 2007;204:3285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Qiao Q, Yang C, Zheng C, Fontan L, David L, Yu X, et al. Structural architecture of the CARMA1/Bcl10/MALT1 signalosome: nucleation-induced filamentous assembly. Mol Cell 2013;51:766–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. David L, Li Y, Ma J, Garner E, Zhang X, Wu H. Assembly mechanism of the CARMA1-BCL10-MALT1-TRAF6 signalosome. Proc Natl Acad Sci U S A 2018;115:1499–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Netea MG, Wijmenga C, O'Neill LA. Genetic variation in Toll-like receptors and disease susceptibility. Nat Immunol 2012;13:535–42. [DOI] [PubMed] [Google Scholar]
- 12. Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol Rev 2012;250:216–29. [DOI] [PubMed] [Google Scholar]
- 13. Rawling DC, Pyle AM. Parts, assembly and operation of the RIG-I family of motors. Curr Opin Struct Biol 2014;25:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlauderer F, Seeholzer T, Desfosses A, Gehring T, Strauss M, Hopfner KP, et al. Molecular architecture and regulation of BCL10-MALT1 filaments. Nat Commun 2018;9:4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paul S, Kashyap AK, Jia W, He YW, Schaefer BC. Selective autophagy of the adaptor protein Bcl10 modulates T cell receptor activation of NF-kappaB. Immunity 2012;36:947–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Campanello L, Traver MK, Shroff H, Schaefer BC, Losert W. Signaling through polymerization and degradation: analysis and simulations of T cell activation mediated by Bcl10. PLoS Comput Biol 2021;17:e1007986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Young RM, Shaffer AL 3rd, Phelan JD, Staudt LM. B-cell receptor signaling in diffuse large B-cell lymphoma. Semin Hematol 2015;52:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2010;463:88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ngo VN, Young RM, Schmitz R, Jhavar S, Xiao W, Lim KH, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature 2011;470:115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, et al. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 2008;319:1676–9. [DOI] [PubMed] [Google Scholar]
- 21. Sanchez-Izquierdo D, Buchonnet G, Siebert R, Gascoyne RD, Climent J, Karran L, et al. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood 2003;101:4539–46. [DOI] [PubMed] [Google Scholar]
- 22. Vicente-Duenas C, Fontan L, Gonzalez-Herrero I, Romero-Camarero I, Segura V, Aznar MA, et al. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc Natl Acad Sci U S A 2012;109:10534–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wilson WH, Young RM, Schmitz R, Yang Y, Pittaluga S, Wright G, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat Med 2015;21:922–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendriks RW, Yuvaraj S, Kil LP. Targeting Bruton's tyrosine kinase in B cell malignancies. Nat Rev Cancer 2014;14:219–32. [DOI] [PubMed] [Google Scholar]
- 25. Woyach JA, Furman RR, Liu TM, Ozer HG, Zapatka M, Ruppert AS, et al. Resistance mechanisms for the Bruton's tyrosine kinase inhibitor ibrutinib. N Engl J Med 2014;370:2286–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caeser R, Walker I, Gao J, Shah N, Rasso-Barnett L, Anand S, et al. Acquired CARD11 mutation promotes BCR independence in diffuse large B cell lymphoma. JCO Precis Oncol 2021;5:145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 2017;171:481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmitz R, Wright GW, Huang DW, Johnson CA, Phelan JD, Wang JQ, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med 2018;378:1396–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood 2020;135:1759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Karube K, Enjuanes A, Dlouhy I, Jares P, Martin-Garcia D, Nadeu F, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia 2018;32:675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morin RD, Mungall K, Pleasance E, Mungall AJ, Goya R, Huff RD, et al. Mutational and structural analysis of diffuse large B-cell lymphoma using whole-genome sequencing. Blood 2013;122:1256–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Thys A, Douanne T, Bidere N. Post-translational modifications of the CARMA1-BCL10-MALT1 complex in lymphocytes and activated B-cell like subtype of diffuse large B-cell lymphoma. Front Oncol 2018;8:498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000;403:503–11. [DOI] [PubMed] [Google Scholar]
- 35. Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020;37:551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Imielinski M, Guo G, Meyerson M. Insertions and deletions target lineage-defining genes in human cancers. Cell 2017;168:460–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, et al. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 2004;427:167–71. [DOI] [PubMed] [Google Scholar]
- 38. Koseki T, Inohara N, Chen S, Carrio R, Merino J, Hottiger MO, et al. CIPER, a novel NF kappaB-activating protein containing a caspase recruitment domain with homology to herpesvirus-2 protein E10. J Biol Chem 1999;274:9955–61. [DOI] [PubMed] [Google Scholar]
- 39. Rossman JS, Stoicheva NG, Langel FD, Patterson GH, Lippincott-Schwartz J, Schaefer BC. POLKADOTS are foci of functional interactions in T-Cell receptor-mediated signaling to NF-kappaB. Mol Biol Cell 2006;17:2166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Traver MK, Paul S, Schaefer BC. T cell receptor activation of NF-kappaB in effector T cells: visualizing signaling events within and beyond the cytoplasmic domain of the immunological synapse. Methods Mol Biol 2017;1584:101–27. [DOI] [PubMed] [Google Scholar]
- 41. Stinson JR, Dorjbal B, McDaniel DP, David L, Wu H, Snow AL. Gain-of-function mutations in CARD11 promote enhanced aggregation and idiosyncratic signalosome assembly. Cell Immunol 2020;353:104129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cheng J, Maurer LM, Kang H, Lucas PC, McAllister-Lucas LM. Critical protein-protein interactions within the CARMA1-BCL10-MALT1 complex: take-home points for the cell biologist. Cell Immunol 2020;355:104158. [DOI] [PubMed] [Google Scholar]
- 43. Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, et al. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem 2001;276:19012–9. [DOI] [PubMed] [Google Scholar]
- 44. Fontan L, Qiao Q, Hatcher JM, Casalena G, Us I, Teater M, et al. Specific covalent inhibition of MALT1 paracaspase suppresses B cell lymphoma growth. J Clin Invest 2018;128:4397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aalipour A, Advani RH. Bruton's tyrosine kinase inhibitors and their clinical potential in the treatment of B-cell malignancies: focus on ibrutinib. Ther Adv Hematol 2014;5:121–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Quancard J, Klein T, Fung SY, Renatus M, Hughes N, Israel L, et al. An allosteric MALT1 inhibitor is a molecular corrector rescuing function in an immunodeficient patient. Nat Chem Biol 2019;15:304–13. [DOI] [PubMed] [Google Scholar]
- 47. Philippar U, Lu TB, Vloemans N, Bekkers M, Van Nuffel L, Gaudiano M, et al. Discovery of JNJ-67856633: a novel, first-in-class MALT1 protease inhibitor for the treatment of B cell lymphomas. Cancer Res 2020;80:5690. [Google Scholar]
- 48. Yan M, Lee J, Schilbach S, Goddard A, Dixit V. mE10, a novel caspase recruitment domain-containing proapoptotic molecule. J Biol Chem 1999;274:10287–92. [DOI] [PubMed] [Google Scholar]
- 49. Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol 2008;9:272–81. [DOI] [PubMed] [Google Scholar]
- 50. Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 2009;106:19946–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, et al. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 1999;96:35–45. [DOI] [PubMed] [Google Scholar]
- 52. Johansson P, Klein-Hitpass L, Grabellus F, Arnold G, Klapper W, Pfortner R, et al. Recurrent mutations in NF-kappaB pathway components, KMT2D, and NOTCH1/2 in ocular adnexal MALT-type marginal zone lymphomas. Oncotarget 2016;7:62627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vela V, Juskevicius D, Gerlach MM, Meyer P, Graber A, Cathomas G, et al. High throughput sequencing reveals high specificity of TNFAIP3 mutations in ocular adnexal marginal zone B-cell lymphomas. Hematol Oncol 2020;38:284–92. [DOI] [PubMed] [Google Scholar]
- 54. Du MQ, Peng H, Liu H, Hamoudi RA, Diss TC, Willis TG, et al. BCL10 gene mutation in lymphoma. Blood 2000;95:3885–90. [PubMed] [Google Scholar]
- 55. Yang Y, Schmitz R, Mitala J, Whiting A, Xiao W, Ceribelli M, et al. Essential role of the linear ubiquitin chain assembly complex in lymphoma revealed by rare germline polymorphisms. Cancer Discov 2014;4:480–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dubois SM, Alexia C, Wu Y, Leclair HM, Leveau C, Schol E, et al. A catalytic-independent role for the LUBAC in NF-kappaB activation upon antigen receptor engagement and in lymphoma cells. Blood 2014;123:2199–203. [DOI] [PubMed] [Google Scholar]
- 57. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 2004;25:1605–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM structures have been deposited in the Electron Microscopy Data Bank with the accession numbers EMD-27095 (BCL10 CARD R58Q) and EMD-27100 (BCL10 E140X-MALT1 DD). The atomic coordinates have been deposited in the Protein Data Bank (PDB) with the accession numbers 8CZD (BCL10 CARD R58Q) and 8CZO (BCL10 E140X-MALT1 DD). All other data are available from the corresponding authors upon reasonable request.