Abstract
Background:
To clarify the mechanisms underlying physical activity (PA)-related cardioprotection, we examined the association of PA with plasma bioactive lipids (BALs) and cardiovascular disease (CVD) events. We additionally performed genome-wide associations.
Methods:
PA-BAL associations were examined in VITAL-CTSC (NCT01169259; N=1,032) and validated in JUPITER-NC (NCT00239681; N=589), using linear models adjusted for age, sex, race, LDL-C, total-C, and smoking. Significant BALs were carried over to examine associations with incident CVD in two nested CVD case-control studies: VITAL-CVD (741 case-control pairs) and JUPITER-CVD (415 case-control pairs; validation).
Results:
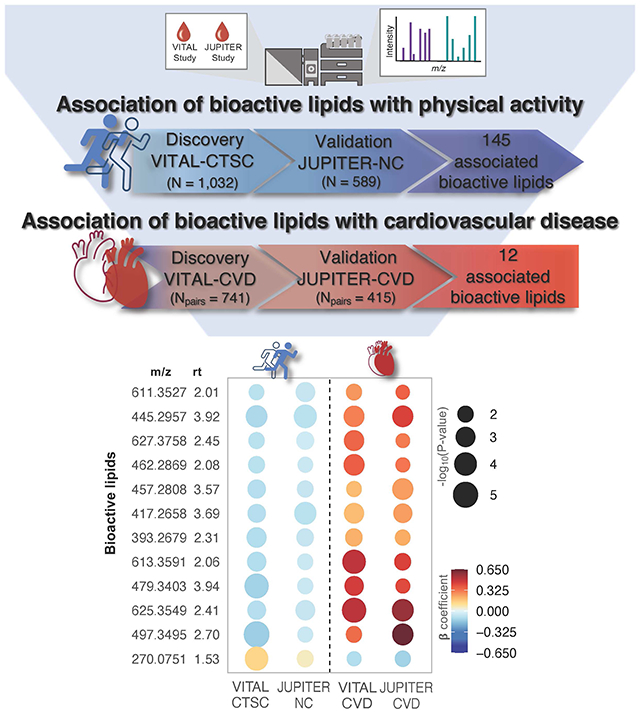
We detected 145 PA-BAL validated associations (FDR < 0.1). Annotations were found for 6 of these BALs: 12,13-diHOME, 9,10-diHOME, lysoPC(15:0), oxymorphone-3b-D-glucuronide, cortisone, and oleoyl-glycerol. Genetic analysis within JUPITER-NC showed associations of 32 PA-related BALs with 22 SNPs. From PA-related BALs, 12 associated with CVD.
Conclusion:
We identified a PA-related bioactive lipidome profile out of which 12 BALs also had opposite associations with incident CVD events.
Keywords: Metabolites, Metabolism, Exercise, Bioactive Lipids, Cardioprotection, Genetic analysis
Graphical Abstract
INTRODUCTION
The long-term effects of regular physical activity (PA) are of such importance in the context of non-communicable diseases burden that achievement of minimum PA levels is a global recommendation 1. There is undisputable evidence regarding PA-related benefits and many markers have been studied. Yet, the specific biomolecular mechanisms that underlie the protective effects of PA are still uncertain. Bioactive lipids (BAL) compose a diverse group of vital endogenous intra- and intercellular signaling molecules involved in a wide array of biological processes. These molecules play a pivotal role in immune regulation, modulation of inflammation, and maintenance of tissue homeostasis at a regulatory level 2–5. They can either influence or be susceptible to physiological stressors and adaptations, such as exercise.
Exercise workloads induce perturbations in inflammation response and immune function 2,3, processes that involve BALs and play an essential role in the pathogenesis of cardiovascular disease (CVD), described as a state of defective resolution of inflammation 6. In addition, several exercise-related physiological phenomena may reflect BAL activity, such as the quantity of red blood cells 7, capacity to secrete catecholamines 8, nitric oxide availability 9, and others. The identification of PA-related benefits from a bioactive lipidome standpoint is the main interest of this study. It has the potential to provide a scaffold to effectively evaluate factors related to PA lifestyle on biological and molecular mechanisms underlying cardiometabolic risk 10. Although evidence suggests a higher overall volume of PA/exercise is associated with better cardiometabolic health 11–13, habitual PA aspects on metabolic changes and its implications on specific outcomes, such as CVD, are still not fully explored.
Gaps in knowledge mentioned above have motivated us to evaluate the association of PA on the BAL profile and subsequent cardiovascular disease (CVD) events. The objective of this study was threefold: 1) to examine the association of habitual PA levels with circulating plasma levels of known and novel BALs, 2) to analyze the association of PA-related BALs with incident CVD in two independent cohorts, and 3) to provide insights into potential biological mechanisms related to PA- and CVD-associated BALs by performing genome-wide genetic association analysis.
METHODS
Data Availability
Study Populations
We assayed baseline BALs within baseline blood samples of two independent studies: The VITamin D and OmegA-3 TriaL (VITAL; NCT 01169259) 14 and the Justification for the Use of statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER; NCT 00239681) 15. Both VITAL and JUPITER were randomized, double-blinded, and placebo-controlled trials. Briefly, VITAL examined 25,871 participants, testing the effects of marine n-3 fatty acids and/or vitamin D (2000 IU/d) supplements vs. placebo in the prevention of CVD and cancer. JUPITER investigated the effects of 20 mg/day rosuvastatin vs. placebo on the rate of cardiovascular outcomes in 17,802 participants with average to low levels of low-density lipoprotein cholesterol (LDL-C < 130 mg/dl) and elevated high-sensitivity C-reactive protein (hs-CRP > 2 mg/L). Participants from both studies provided written informed consent at the time of enrollment. Institutional review board approvals for both studies were obtained from Partners HealthCare (Boston, MA). First and senior authors had full access to all data in the study and take responsibility for their integrity and data analysis.
From VITAL, we examined assays collected as part of two sub-studies: 1) VITAL-CTSC, a subsample (N=1,032) that had in-person clinical evaluations at the Brigham and Women’s Hospital Clinical Translational Science Center – CTSC 16, and 2) VITAL-CVD, a nested case-control sub-study for incident CVD events matched on age, sex, and fasting status (N case-control pairs=741). Only 46 participants from VITAL-CTSC were also included in VITAL-CVD sub-study (24 cases and 22 controls).
Two sub-studies from JUPITER were analyzed for validation: 1) JUPITER-NC, a non-cases subsample randomly selected amongst participants with European ancestry with genetic analysis (N=589), and 2) JUPITER-CVD, a nested case-control sub-study for incident CVD matched on age and sex (N case-control pairs=415). Among JUPITER sub-studies, there were no overlapping participants.
Study design
We had a two-stage study design (Fig. 1). First, we used discovery in VITAL-CTSC and validation in JUPITER-NC, to evaluate PA-BAL baseline cross-sectional associations. Second, we used a similar approach to investigate whether PA-related BALs were associated with future risk of CVD. This relationship was first examined in the VITAL-CVD case-control study, then validated in the JUPITER-CVD case-control study.
Fig. 1. Diagram of the two-stage analysis.
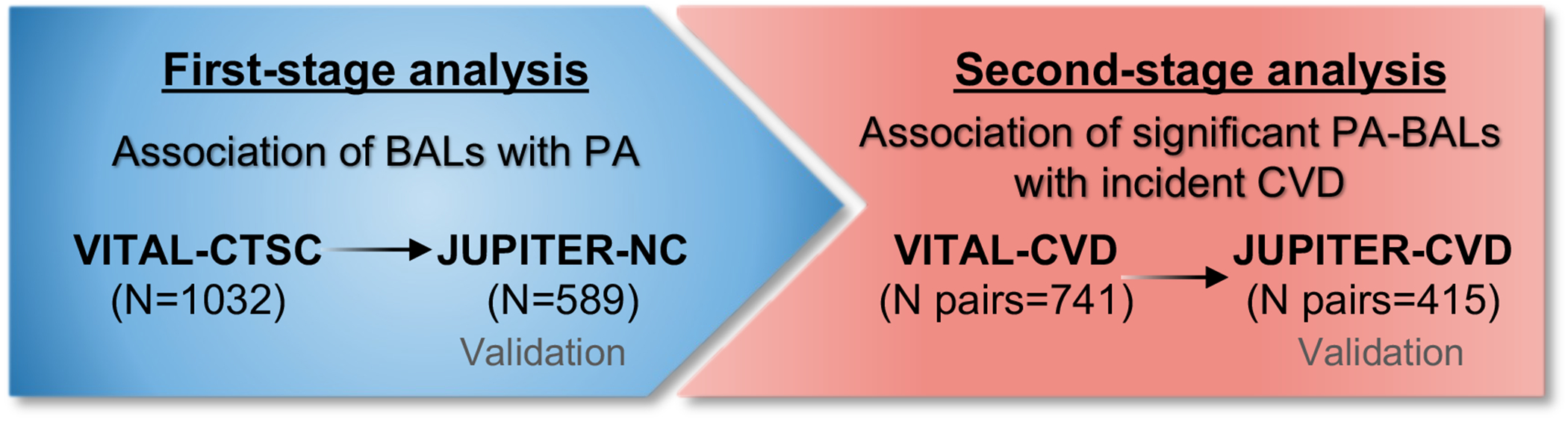
The left-side box shows the first-stage analysis performed to examine associations between PA and BAL in the discovery cohort, VITAL-CTSC, and validated in JUPITER-NC. BALs significantly associated with PA were carried over to the second-stage analysis (right-side box), in which their association with incident CVD was investigated in VITAL-CVD and validated in JUPITER-CVD.
Biomarker profiling
Blood samples from both main studies, all collected at baseline and before randomization, were assayed at the University of California, San Diego. Approximately 11,000 features were extracted using a directed non-targeted high-throughput liquid chromatography-mass spectrometry (LC-MS) approach to measure the relative intensities of small molecules 17. VITAL-CTSC and JUPITER-NC samples were run with respective study’s case and control samples. Corresponding cases and controls were placed in tandem wells on a plate (in random order) to avoid batch effects and were blindly assayed. To avoid experimental biases, blinded quality control (QC) samples were additionally embedded throughout the experiment within each of the batches. Moreover, the performing laboratory had a multi-tiered QC approach that allows for close monitoring of sample-to-sample variation and preparation, as well as system performance and any potential system drift over the sample run. Details of the blood sample collection, LC-MS method, and annotation process are provided in the Supplemental material. Features detected in the discovery cohorts were mapped in the validation cohorts by peaks alignment based on retention time, accurate mass, and tandem MS fragmentation pattern17.
PA assessment
In VITAL, PA was assessed through a self-administrated questionnaire with questions regarding the average amount of time during the last 12 months spent in activities such as: jogging, aerobic exercises, tennis, swimming (activities that require ≥ 6 metabolic equivalent [METs]); walking, bicycling, yoga, weight lifting (activities requiring 3 to 5.9 METs); and slow walking (light activity – < 3METs). Details of activities and intensities are provided in the Supplemental material. The total reported PA was calculated as weekly energy expenditure in METs (METs-hr/wk) by multiplying the intensity attributed to each type of PA (MET) and time spent in the activity according to the questionnaire. Participants from VITAL-CTSC with self-reported MET-hrs/wk ≥ 3SD from the average (24.5 MET-hr/wk) were considered outliers and excluded from the analysis (N=17). In VITAL-CTSC, PA was treated as a continuous variable which values were shifted and rescaled to mean 0 and SD = 1 (21.8 MET-hr/wk) for better comparison across studies and more interpretable results.
In JUPITER, PA was treated as an ordinal variable ranging from 1 to 6, according to the category of self-reported frequency per week: 1) Rarely/Never, 2) less than once a week, 3) once a week, 4) 2-3 times a week, 5) 4-6 times a week, or 6) daily.
Cardiovascular disease outcomes
VITAL-CVD cases were defined as an adjudicated composite endpoint of confirmed myocardial infarction, stroke, coronary revascularization, or cardiovascular death. In JUPITER-CVD, cases were defined as an adjudicated composite endpoint of confirmed myocardial infarction, stroke, coronary revascularization, unstable angina requiring hospitalization, or death.
Genetic data
To further inform the mass spectrometry data, we performed a genome-wide association study (GWAS) for replicated BALs using JUPITER-NC dataset. Detailed genotyping and analysis procedure was described by Chasman et al. 18. Briefly, all participants involved in the present study provided consent for genetic analyses, as part of the JUPITER main trial protocol, and had DNA available for genotyping. Genome-wide genotyping was performed on a fee-for-service basis by Illumina (San Diego, CA) using the Illumina Omni 1M Quad platform, and raw genotype intensity data were reduced to genotype calls using the Illumina Genome Studio (v. 1.6.2) software. Single nucleotide polymorphism (SNP) clusters were initially defined automatically using data from the JUPITER sample. Approximately 1% of the SNPs were identified for poor clustering on the basis of quality measures including Hardy-Weinberg equilibrium (HWE; P<10−6) or call rate (< 95%), and were visually inspected and annotated, removed, or clustered again with manual intervention. Re-clustered SNPs were retained if genotypes could be called in >95% of the samples. After these procedures, 99.71% of the loci met the preceding quality criteria. Included participants had self-reported European ancestry verified by multidimensional scaling procedures in PLINK 19 applied to 1067 unlinked ancestry informative SNPs selected from HapMap3. The relatedness (pi_hat) was estimated from identity-by-state in PLINK 19.
Covariates
Baseline questionnaires were used to collect sex, age, race/ethnicity, body weight and height, use of non-randomized supplements or medications, smoking, and other relevant aspects of health history. VITAL-CTSC participants also had their body weight and height measured by trained staff. VITAL-CTSC and VITAL-CVD blood samples were sent to Atherotech Diagnostics (Birmingham, AL) and Quest Diagnostics Nichols Institute (San Juan Capistrano, CA) for standard lipid panels (plasma). High-sensitivity C-reactive protein (hs-CRP) was assayed by latex-enhanced immunonephelometric assay 20. In JUPITER-NC and JUPITER-CVD, standard lipid panels were measured in a central laboratory as part of the clinical trial. LDL-C concentrations were calculated by the Friedewald equation when triglycerides were <400 mg/dL and measured by ultracentrifugation when ≥400 mg/dL. A high-sensitivity assay (Behring Nephelometer) was used for hs-CRP measurement 21.
Statistical analysis
Variables distributions were expressed as median and interquartile ranges. Mann–Whitney U test was used to examine contrasts between cases and controls in VITAL-CVD and JUPITER-CVD, and differences in categorical variables were compared with crosstabulation and subsequent X2 testing.
Prior to statistical analysis, we performed data preparation. We detected and removed carbon-13 (13C) isotopes – features with correlation coefficient > 0.95, up to ± 0.01 minutes retention time (rt) from each other, and difference in mass to charge ratio (m/z) equal to one or more 13C atom mass – and typically found adducts to reduce redundancy of assayed BALs.
Only BAL features with less than 20% of missing values were considered for this analysis. To remove other redundant fragments (due to contaminants, fragments, or other MS artifacts) 22, we examined remaining highly correlated BALs (r>0.95). From each pairwise comparison, we removed those with lower median relative intensity. Missing values were then imputed to 0.25 of the lowest observed value. All BALs were log-transformed prior to outlier detection (mean ± 3 standard deviations [SD]), and those exceeding these limits were trimmed to ± 3 SD of mean values. Next, to remove experimental biases, VITAL-CTSC and JUPITER-NC all measurements were adjusted for batch and run-day effects using ComBat (R-package sva) 23. No plate effects correction was performed in VITAL-CVD and JUPITER-CVD, as cases and controls were placed in wells next to each other. All values were rescaled to median of 0 and SD = 1.
PA-BAL cross-sectional associations were performed on baseline data collected before the randomization process for each trial. We used linear regressions adjusted for age and sex (model 1), and model 1 plus race/ethnicity, LDL-cholesterol, total cholesterol, and current smoking (model 2). A sensitivity analysis was conducted to investigated associations between validated BALs and continuous PA in subgroup analysis for participants being above and below the median for both PA and BMI. An additional model (model 3) was fitted, which included adjustments for categorized BMI (< 25, 25–29.9, or ≥ 30 kg/m2) and HDL-cholesterol.
Supplementary analyses were performed to meet model assumptions and ensure statistical rigor. First, the normality of linear regression residuals for BALs was checked using the Kolmogorov-Smirnov’s test. Then, BALs for which the residuals normality assumption was rejected were analyzed by the Wilcoxon test (BAL measures in PA levels above the median vs. PA below the median). Finally, BALs for which the Wilcoxon test null hypothesis was not rejected, we performed a bootstrap resampling procedure with 2000 bootstrap replicates to provide consistent estimates of confidence intervals.
For associations with incident CVD, we used conditional logistic regressions with similar models 1 and 2 (excluding sex, which was used as case-control matching criteria), plus the randomized treatment assignment to account for the interventions involved in the clinical trials. CVD outcome was included in the model as the dependent variable and PA-related BALs as independent variables.
To account for multiple comparisons in all regression analyses, we used the Benjamini-Hochberg approach 24,25 controlling for False Discovery Rate (FDR) < 0.1. The criteria for validation in JUPITER-NC and JUPITER-CVD were FDR < 0.1 and β-coefficient with the same directionality.
Common genetic variation that was not measured directly in the parent JUPITER data from which JUPITER-NC was derived (above) were imputed using the Markov Chain Haplotyping algorithm (MaCH 1.0, developed by Li et al.26) and the 1000 Genomes Project reference panel (phase 1, v. 3). Imputation was performed among JUPITER participants with verified European ancestry including 979,089 genotyped SNPs in HWE (P≥10−6) and without restriction by minor allele frequency. Analysis was limited to 6,528,605 SNPs with imputation quality r2 < 0.8 and with a minor allele frequency <5%, implying at least 55 copies of the heterozygote genotype given the sample size under Hardy-Weinberg equilibrium. For genetic associations analysis, each validated BAL related to PA (145 PA-associated BALs obtained using model 2) was further investigated for genetic associations at a genome-wide level of statistical significance (P < 5x10−8) by performing multivariable linear regression assuming an additive genetic model, adjusted for age, sex, and 10 principal components of sub-European ancestry. Index SNPs within independent loci reaching genome-wide significance were identified from the genome-wide scans across all PA-associated BALs using a clumping procedure that grouped SNPs within 250 kb and linkage disequilibrium (LD) r2 > 0.10 from the index SNP, with a significance threshold for index SNPs of 7.3x10−10 (genome-wide P-value adjusted for the effective number of independent tests [N=69], derived from Gao et al. 27 using a cutoff of 90%). We used the web-based application SNPnexus 28 to annotate index SNPs for gene and variant type based on data from the University of California Santa Cruz (UCSC) and Ensembl genome databases (human genome version hg19).
Additionally, two-sample inverse-variance weighted (IVW) Mendelian randomization (MR) 29 was implemented to examine potential causal associations between BALs significantly associated with PA and coronary artery disease (CAD). We used estimates for index genetic variants herein found to be associated with PA-related BALs with summary statistics for CAD from CARDIoGRAMplusC4D 1000 Genomes-based GWAS meta-analysis 30 (available at http://www.cardiogramplusc4d.org/data-downloads/). Mendelian randomization was performed using the R package “MendelianRandomization” (available at https://CRAN.R-project.org/package=MendelianRandomization) and the significance threshold was 0.002 (0.05/32 BALs).
In order to put our findings in a more clinical context, we also performed Spearman correlation analyses among PA-associated BALs (with annotations) with clinical factors such as body mass index (BMI), hs-CRP, and standard lipids. Additionally, the correlation of all significant BALs in VITAL-CVD and JUPITER-CVD with the same clinical variables was also examined. We used the Jennrich test 31 for the equality of two correlation matrices.
Finally, in addition to quantifying the associations of PA with BALs, we examined the extent to which baseline BMI may be a mediator in the associations of BALs with PA or incident CVD. Therefore, we performed mediation analysis using the method proposed by Valeri and VanderWeele 32, controlling for the same covariates used in model 2. Using the same method, we further explored the role of BALs as mediators in the PA- CVD relationship in VITAL-CVD.
Statistical analyses were performed using R software version 3.6.0 R (R Foundation for Statistical Computing, Vienna, Austria. URL: https://www.R-project.org/) and PLINK.
RESULTS
Baseline characteristics
Table 1 displays baseline demographic characteristics and clinical biomarkers in the sub-cohorts. In VITAL-CTSC (N=1,032; discovery cohort), the median energy expenditure during PA was 20 MET-hrs/wk, median age of 64.5 years old, and 48.8% were women. In JUPITER-NC (N=589; validation cohort), 43% of participants exercised rarely/never, followed by 2-3 times per week (23.8%), and the median age was 67 years old, and 49.9% were women.
Table 1.
Baseline characteristics of participants in VITAL-CTSC and JUPITER-NC sub-studies.
| VITAL-CTSC (Discovery Cohort) | JUPITER-NC (Validation Cohort) | |
|---|---|---|
| N | 1,032 | 589 |
| Age (years) * | 64.4 (60.4, 68.8) | 67.0 (62.0, 72.0) |
| Women (%) | 48.9 | 49.9 |
| White (%) | 82.8 | 100.0§ |
| Current smokers (%) | 5.4 | 11.2 |
| BMI (kg/m2) * | 26.4 (23.9, 29.7) | 28.6 (25.3, 32.5) |
| BMI categories | ||
| < 25 kg/m2 (%) | 35.0 | 23.3 |
| 25 to 29.9 kg/m2 (%) | 42.4 | 36.2 |
| ≥ 30 kg/m2 (%) | 22.6 | 40.6 |
| HDL-C (mg/dL) * | 51.0 (42.0, 63.0) | 52.0 (43.0, 62.0) |
| LDL-C (mg/dL) * | 111.0 (93.0, 131.0) | 110.0 (96.0, 120.0) |
| Total-C (mg/dL) * | 187.0 (166.0, 207.0) | 189.0 (176.0, 202.0) |
| Triglycerides (mg/dL) * | 94.0 (72.0, 131.0) | 120.0 (89.0, 172.0) |
| hs-CRP (mg/L) * | 1.2 (0.5, 2.5) ‡ | 4.0 (2.7, 6.5) |
| Main study intervention assignment (%) | 49.8 | 47.4 |
| Physical activity † | 20.0 (6.3, 35.5) | Rarely/Never (%): 43.0 Less than once a week (%): 5.6 Once a week (%): 6.3 2-3 times a week (%): 23.8 4-6 times a week (%): 12.7 Daily (%): 8.7 |
Median (IQR).
Physical activity is given in MET-hrs/wk (median, IQR) for VITAL-CTSC and in percentage for each category in JUPITER-NC.
Data available for 1015 participants.
JUPITER-NC sub-cohort was chosen to be 100% white for the genetic analyses. The parent JUPITER study was multiethnic.
Abbreviations: BMI – body mass index; LDL-C – low density lipoprotein cholesterol; HDL-C – high density lipoprotein cholesterol; Total-C – total cholesterol; hs-CRP – high-sensitivity C-reactive protein.
Table 2 shows baseline demographic characteristics and clinical biomarkers of the two CVD nested case-control sub-studies. VITAL-CVD (N = 741 case-control pairs) cases had a higher frequency of smokers, slightly higher BMI, and lower HDL-C cholesterol than controls. In JUPITER-CVD (N=415 case-control pairs), cases had slightly lower body mass index (BMI), lower frequency of whites, and more smokers.(Table 3)
Table 2.
Baseline characteristics of participants in VITAL-CVD case-control substudy.
| VITAL-CVD | Controls | Cases | P-value† |
|---|---|---|---|
| N | 741 | 741 | |
| Age (years) * | 69.5 (65.6, 74.9) | 70.2 (65.7, 76.2) | 1.7e-1 |
| Women (%) | 40.9 | 40.9 | 1.0 |
| White (%) | 87.2 | 88.5 | 1.0 |
| Current smokers (%) | 4.4 | 8.3 | 5.3e-3 |
| BMI (kg/m2) * | 26.6 (24.1, 30.1) | 27.4 (24.4, 31.0) | 8.0e-3 |
| BMI categories | 3.4e-1 | ||
| < 25 kg/m2 (%) | 33.5 | 27.8 | |
| 25 to 29.9 kg/m2 (%) | 41.1 | 42.2 | |
| ≥ 30 kg/m2 (%) | 25.4 | 30.0 | |
| HDL-C (mg/dL) * | 50.0 (39.0, 63.0) | 46.0 (38.0, 59.0) | 3.7e-4 |
| LDL-C (mg/dL) * | 123.0 (101.2, 145.0) | 119.0 (97.0, 145.0) | 2.7e-1 |
| Total-C (mg/dL) * | 202.0 (176.0, 228.0) | 196.0 (171.0, 227.0) | 1.3e-1 |
| Triglycerides (mg/dL) * | 113.0 (86.5, 158.0) | 121.0 (92.0, 177.0) | 7.0e-3 |
| hs-CRP (mg/L) *‡ | 1.7 (0.9, 3.9) | 2.1 (1.1, 5.3) | 1.5e-1 |
| Main study intervention assignment (%) | 48.9 | 51.0 | 2.0e-3 |
| Physical activity * | 14.6 (4.3, 30.0) | 12.9 (3.0, 28.0) | 4.1e-1 |
Median (IQR).
P-value for statistical test comparing controls vs. cases.
Data available only for 97 cases and 89 controls.
Abbreviations: BMI – body mass index; LDL-C – low density lipoprotein cholesterol; HDL-C – high density lipoprotein cholesterol; Total-C – total cholesterol; hs-CRP – high-sensitivity C-reactive protein.
Table 3.
Baseline characteristics of participants in JUPITER-CVD case-control substudy.
| JUPITER-CVD | Controls | Cases | P-value† |
|---|---|---|---|
| N | 415 | 415 | |
| Age (years) * | 70.0 (64.0, 75.0) | 70.0 (64.0, 76.0) | 7.2e-1 |
| Women (%) | 28.4 | 28.4 | 1.0 |
| White (%) | 91.3 | 83.9* | 1.0e-3 |
| Current smokers (%) | 11.1 | 23.4* | 4.0e-6 |
| BMI (kg/m2) * | 28.4 (25.6, 31.3) | 27.2 (24.0, 30.9) † | 2.0e-3 |
| BMI categories | 1.0e-3 | ||
| < 25 kg/m2 (%) | 20.8 | 32.0 | |
| 25 to 29.9 kg/m2 (%) | 44.9 | 38.6 | |
| ≥ 30 kg/m2 (%) | 34.3 | 29.4 | |
| HDL-C (mg/dL) * | 49.0 (40.0, 61.0) | 47.0 (40.0, 59.0)† | 3.4e-2 |
| LDL-C (mg/dL) * | 109.0 (94.0, 120.0) | 107.0 (93.0, 119.0) | 4.0e-1 |
| Total-C (mg/dL) * | 187.0 (167.5, 201.0) | 184.0 (166.0, 198.0) | 1.5e-1 |
| Triglycerides (mg/dL) * | 116.0 (82.0, 162.5) | 118.0 (86.0, 166.5) | 4.0e-1 |
| hs-CRP (mg/L) * | 4.3 (2.9, 6.9) | 4.7 (3.0, 8.2) | 8.3e-2 |
| Main study intervention assignment (%) | 46.7 | 39.8† | 4.2e-2 |
| Physical activity | 1.5e-1 | ||
| Rarely/Never (%) | 47.0 | 56.9 | |
| Less than once a week (%) | 4.8 | 3.9 | |
| Once a week (%) | 7.0 | 5.5 | |
| 2-3 times a week (%) | 18.8 | 14.9 | |
| 4-6 times a week (%) | 10.4 | 8.9 | |
| Daily (%) | 12.0 | 9.9 |
Median (IQR).
P-value for statistical test comparing controls vs. cases.
Data available only for 97 cases and 89 controls.
Abbreviations: BMI – body mass index; LDL-C – low density lipoprotein cholesterol; HDL-C – high density lipoprotein cholesterol; Total-C – total cholesterol; hs-CRP – high-sensitivity C-reactive protein.
Association between PA and BALs
After 13C isotopes and adducts removal, VITAL-CTSC and JUPITER-NC BAL final datasets comprised 7,743 and 8,060 BAL features, respectively (1,809 and 2,309 features removed, respectively). In model 1 (adjusted for age and sex), PA was significantly associated with 1,644 BALs, of which 179 were validated in JUPITER-NC. Model 2 (model 1 plus race/ethnicity, LDL-C, total-C, and smoking) identified 1,257 significant BALs in VITAL-CTSC, of which 145 were validated in JUPITER-NC: 97 positive and 48 negative associations. Of those, annotations were found for six: 12,13 and 9,10 dihydroxyoctadecenoic (12,13-diHOME and 9,10-diHOME), 1-pentadecanoyl-glycerol-3-phosphocholine (LysoPC(15:0)), oxymorphone 3b-D-glucoronide (putative), cortisone, and oleoyl-glycerol (Figure 2). All multiple comparisons were adjusted at the level of False Discovery Rate (FDR) 24 < 0.1. Figure 2 additionally shows β-coefficients for the associations of PA and BALs in both VITAL-CTSC and JUPITER-NC, displaying all BALs with annotations and the top 20 BALs (ranked based on unadjusted P-values in the validation cohort) without annotations. The full set of 145 PA-BAL validated associations in both discovery and validation cohorts are in Table S1.
Fig. 2. First-stage analysis: Baseline cross-sectional association of BALs with PA, linear model adjusted for age, sex, race/ethnicity, LDL-C, total-C, and smoking.
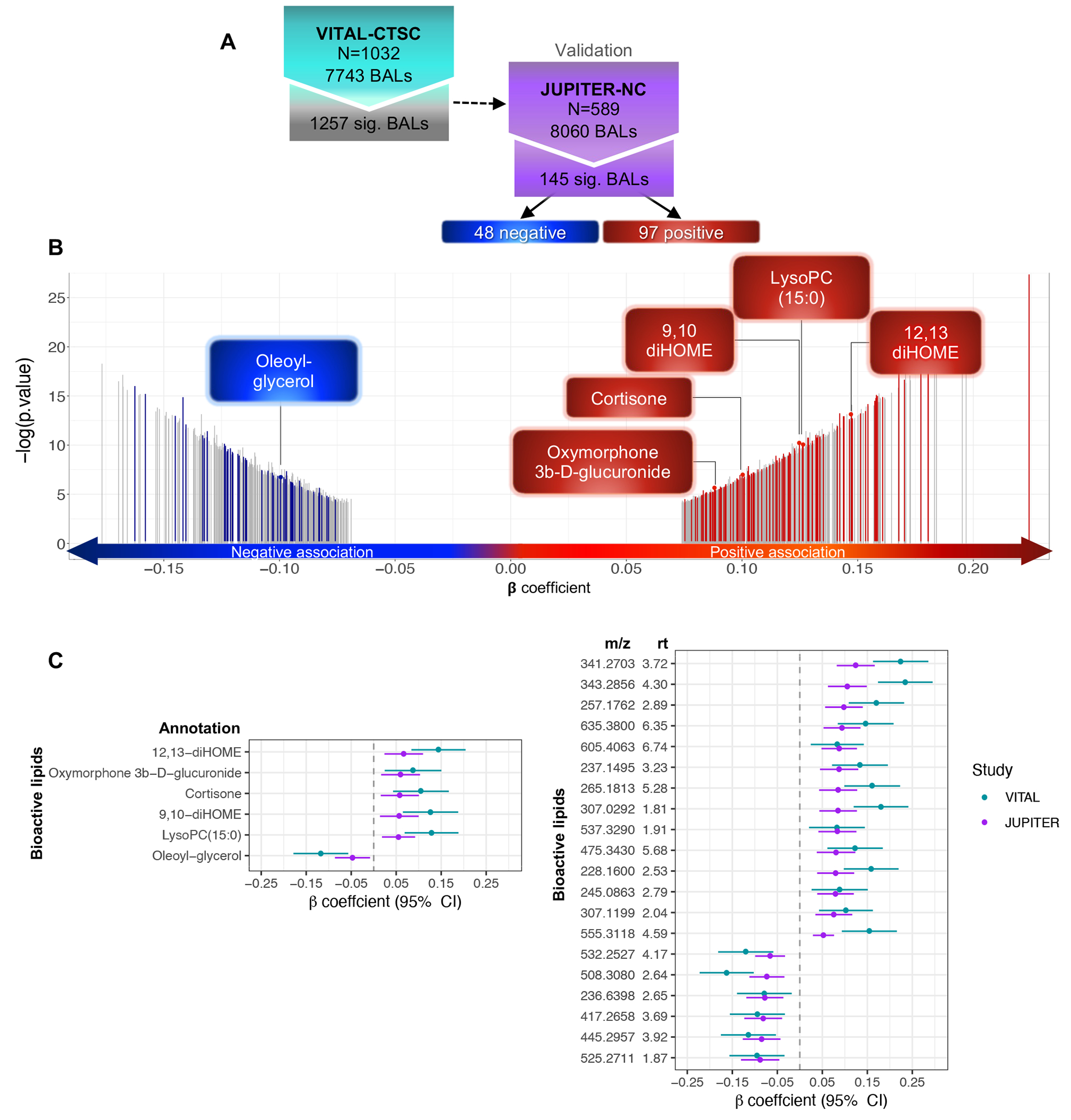
A) Flowchart of PA-BAL associations in VITAL-CTSC with validation in JUPITER-NC; B) Distribution of 1,257 BALs associated with baseline PA in VITAL-CTSC (FDR < 0.1), according to β-coefficients for SD change in BAL per exercise SD=21.8 MET-hrs/wk. Of those, 145 BALs validated in JUPITER-NC (FDR < 0.1 and the same directionality). Negative (blue) and positive associations (red) are highlighted, and gray bars represent significant BALs in VITAL-CTSC not validated in JUPITER-NC. Annotations are indicated. C) PA-BAL significant associations (FDR < 0.1) in both VITAL-CTSC and JUPITER-NC. Symbols with error bars are β -coefficients per SD of PA in VITAL-CTSC (blue) or per category in JUPITER-NC (purple), with 95% confidence intervals (CIs). Annotated BALs are presented, as well as the top 20 novel BAL (ranked based on unadjusted P-values in the validation cohort). Abbreviations: m/z – mass to charge ratio; rt – retention time.
In both discovery and validation cohorts, the residuals for the majority of the 145 validated BALs were normally distributed (124 and 126 BALs, respectively). For the remaining, the Wilcoxon test for BALs contrasts between PA levels above and below the median did not reject the null hypothesis for 6 BALS (3 in each cohort). For these 6 BALs, we calculated bootstrapped confidence intervals, which were only slightly different from the original model, and no significant change was observed (Table S2).
In a sensitivity analysis, we investigated associations between validated BALs and continuous PA in subgroup analysis for participants being above and below median for PA (20 MET-hr/wk) and BMI (26.4 kg/m2). Among participants below the median for both PA and BMI (N = 201), 26 BALs showed nominally significant association with PA (adjusted for age, sex, race, LDL-C, total-C, and smoking – model 2). In the group above the median for both variables (N = 213), 6 BALs were nominally significant. All had FDR ≥ 0.1.
Model 3 (model 2 plus categorized BMI and HDL) identified 220 BALs PA-associated BALs in VITAL-CTSC (FDR < 0.1) of which 45 validated in JUPITER-NC (nominal P < 0.05 and the same directionality) (Table S3). Models 2 and 3 overlapped in 33 BALs (Fig. S1), including 12,13-diHOME.
Associations of baseline BALs with incident CVD
After 13C isotopes removal from VITAL-CVD and JUPITER-CVD (respectively, 2,758 and 2,508 BALs), 8,564 BALs were examined in the discovery and 8,060 in the validation case-control substudies. In the minimally adjusted model (age, and matching strata defined by caliper age and sex), out of the 179 PA-significant BALs, 54 associated with CVD in VITAL-CVD. Of these, 19 were validated in JUPITER-CVD (Fig. S2). BALs that were associated with higher levels of PA tended to be associated with lower risk of CVD, and vice versa.
In model 2 (model 1 plus race/ethnicity, smoking, total-C, LDL-C, and randomized treatment assignment), out of 145 PA-related BALs, 38 were significant for CVD in VITAL-CVD (FDR < 0.1; odds ratio [OR] ranging from 0.67 to 1.64) and 12 validated in JUPITER-CVD (FDR < 0.1) with ORs ranging from 0.88 to 1.91. Consistent with model 1 results, all positive CVD associations (OR = 1.25 to 1.91) were negative with PA, whereas one negative relationship with CVD (OR = 0.88) was positively associated with PA (Fig. 3).
Fig. 3. Second-stage analysis: Association of baseline PA-related BALs with incident CVD, conditional logistic regression model adjusted for age, matching strata defined by caliper age and sex, race/ethnicity, LDL-C, total-C, smoking, and randomized treatment assignment.
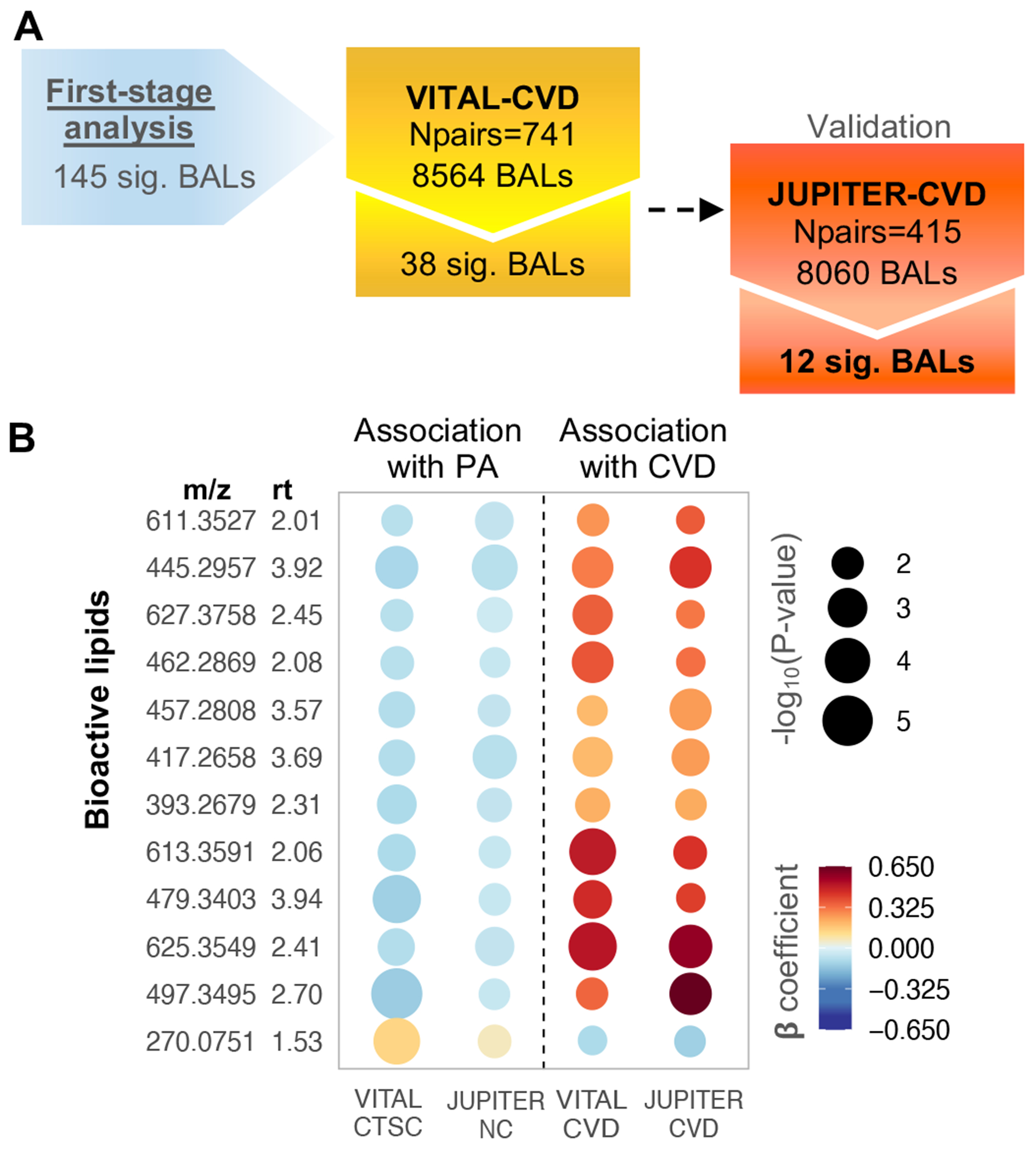
A) Flowchart showing BAL-CVD associations in VITAL-CVD with validation in JUPITER-CVD (FDR < 0.1), from 145 BALs cross-sectionally associated with baseline PA. B) Rainplot representing β-coefficients for 12 BALs validated with incident CVD (FDR < 0.1) and respective associations with PA (FDR < 0.1). Dashed line separates PA-BAL (left side) from BAL-CVD (right side) associations. Abbreviations: m/z – mass to charge ratio; rt – retention time.
Genetic association analysis
The 145 plasma BALs significantly associated with PA were further examined for genetic associations in JUPITER-NC (N = 589). This analysis identified 22 independent genome-wide-significant single nucleotide polymorphism (SNP) associated with 32 BALs (P-value < 7.3x10−10 [= 5x10−8/69, the effective number of BALs]; Manhattan plots in Fig. S3). Figure 4 displays a 2D clustered loci-by-BAL plot using the top SNP hit at each locus from the imputed genome-wide association study (GWAS) data alongside their respective results for associations with PA and CVD. We observed an effect size estimation ranging from −0.93 to 0.83 standard deviation (SD) change in BALs per minor allele (z-score from −9.66 to 11.50). We found 14 SNP positions overlapping gene locations: coding genes for UDP-glucuronosyltransferases, UGT1A8, UGT3A1, and UGT2B7; coding genes for cytochrome P450, CYP2C18, CYP4V2, and CYP39A1; coding gene for solute carrier organic anion transporter 1B1, SLCO1B1; coding gene for HEAT repeat-containing protein 4, HEATR4; coding gene for Acyl-CoA 6-desaturase, FADS2; coding gene for HHIP Antisense RNA 1, HHIP-AS1; coding gene for receptor expression-enhancing protein 3, REEP3; coding gene for DBH-like monooxygenase protein 1, MOXD1; coding gene for D-dopachrome decarboxylase-like protein, DDTL; and coding gene for the von Willebrand factor A domain-containing protein 3B, VWA3B (source: https://www.uniprot.org).
Fig. 4. 2-D clustered loci-by-BALs grid using the top SNP hit at each locus.
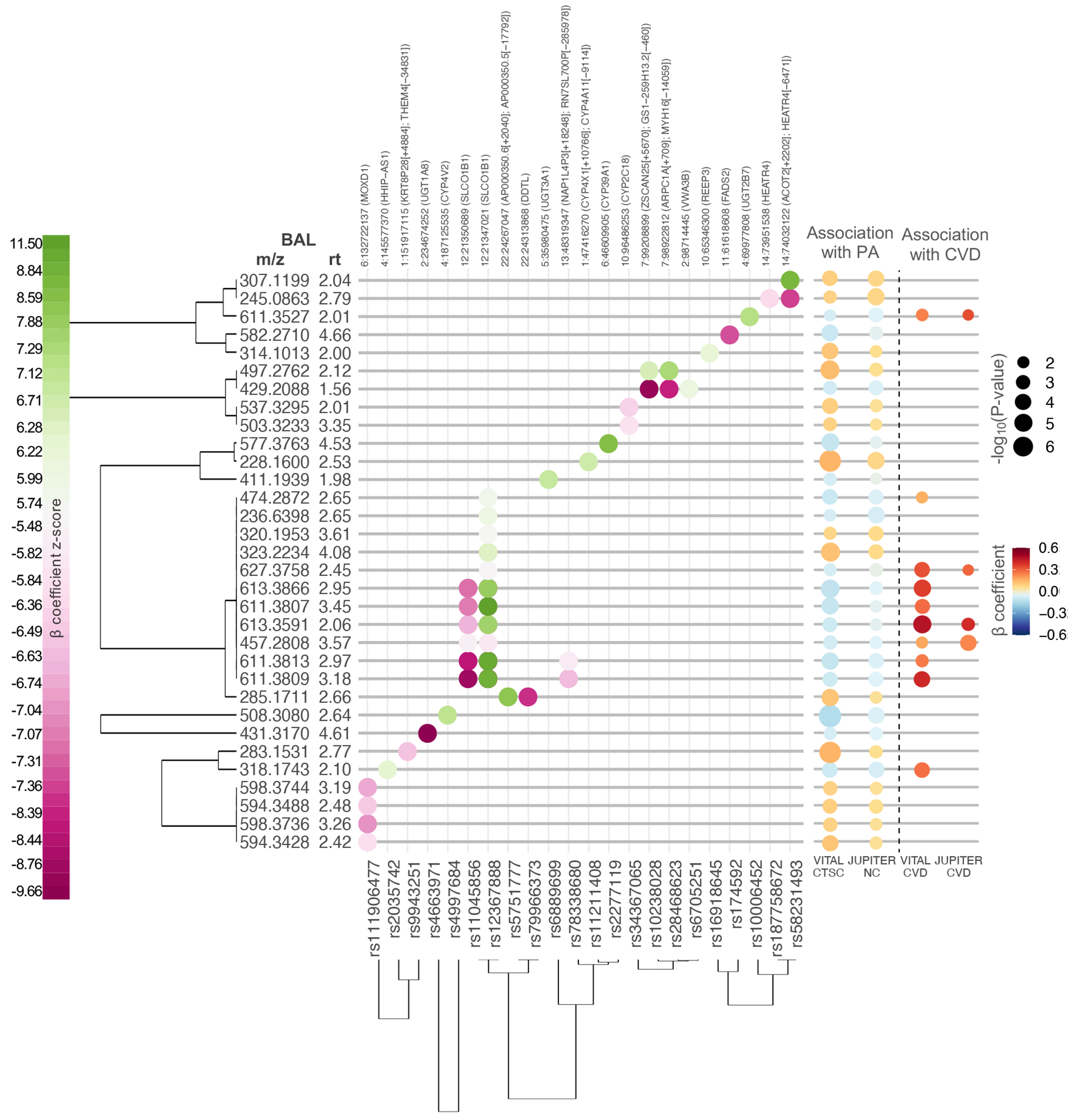
Significant SNPs associations, through minor allele frequency, for 32 BALs at a genome-wide level of statistical significance (P<5x10−8) in JUPITER-NC dataset. BAL-SNP associations are shown on the left-side panel, color coded according to β-coefficient z-score (β-coefficient/standard error). SNP position and closest gene are indicated at the top – chromosome number: position (overlapped gene or nearest downstream gene [+base pair distance]; nearest upstream gene [−base pair distance]). The right-side panel shows BALs’ respective association with baseline PA and incident CVD (linear regression model adjusted for age, matching strata defined by caliper age and sex, race/ethnicity, LDL-C, total-C, smoking, and randomized treatment assignment). β-coefficient is given per SD change in BAL per exercise SD (21.8 MET-h/wk) in VITAL-CTSC / exercise category in JUPITER-NC. BALs and SNPs were clustered through hierarchical clustering performed using a distance function based on the absolute value of the beta-coefficients. Abbreviations: m/z – mass to ratio; rt – retention time.
Among the 22 independent loci, 10 have not been previously reported regarding an association with metabolites, while 12 loci contained SNPs associated with metabolites/lipid species in large cohorts 33–52 (Supplementary data 1). As expected, based on the pathway analysis, variants mapping to the FADS1-3 gene cluster (rs174592) showed several hits, especially with phosphatidylcholines, 1-alkyl,2-acylglycero-3-phosphocholines, lysophosphatidylcholines, and 1-acyl-sn-glycero-3-phosphocholines. As well, SNPs close to the SLCO1B1 gene (rs12367888) showed association with multiple metabolites, mainly within the class of steroids and steroid derivatives, glycerophospholipids, and fatty acyls. Metabolite associations with the UGT1A8 (rs4663971) gene show results consistent with the hyperbilirubinemia pathway (Table S6).Of the 32 BALs associated with variants, four (m/z = 611.3527, 627.3758, 613.3591, 457.2808) were validated for association with CVD in JUPITER-CVD, the second-stage analysis of our study design. No variant was genome-wide significantly mapped in CARDIoGRAMplusC4D 1000 for coronary artery disease (CAD) 30 (available at http://www.cardiogramplusc4d.org/data-downloads/).
Mendelian randomization (MR) 29 assessing potential causality on CAD (from CARDIoGRAMC4D) of 32 BALs that had genome-wide significant SNP associations is presented in Table S4. Of the instrumental associations reaching significance after accounting for multiple testing (P-value < 0.05/32), all were driven solely by variants proximal to highly pleiotropic gene functions (Tables S4 and S5), and therefore unlikely to support causal roles of these BALs in CAD. Table S5 displays information on previously known SNP-trait associations reported in the GWAS catalog 53 retrieved for all 22 index SNPs and nearby SNPs in high linkage disequilibrium (LD r2>0.8). We observed that almost all index SNPs were associated with multiple phenotypes with a preponderance of phenotypes related to metabolism. Pathway analysis was also performed via SNPnexus 28 for significant SNPs and, similarly, highlighted clear concentration in known metabolic pathways, such as metabolism of alpha-linolenic and linoleic acid, steroids, bile acid and bile salt metabolism, and glucuronidation (Table S6).
Correlation analyses
Baseline PA-associated BALs with annotations were correlated with clinical factors such as BMI, high-sensitivity C-reactive protein (hs-CRP), and standard lipids. VITAL-CTSC and JUPITER-NC substudies both showed significant and consistent correlations for: oleoyl-glycerol with HDL-C (r = −0.25 and −0.20) and triglycerides (r = 0.50 and 0.43); 12,13-diHOME and HDL-C (r = 0.23 and 0.16); 9,10-diHOME with HDL-C (r = 0.22 and 0.21); LysoPC(15:0) with hs-CRP (r = −0.29 and −0.21), BMI (r = −0.31 and −0.18), and HDL-C (r = 0.26 and 0.1); and cortisone and HDL-C (r = 029 and 0.38), total-C (r = 0.08 and 0.14), triglycerides (r = −0.27 and −0.25), and BMI (r = −0.37 and −0.44) (Fig. S4).
As for CVD positive associations, seven out of 11 BALs negatively correlated with HDL-C, LDL-C, and total-C in both case-control subsamples. We also observed six positive correlations with triglycerides and three with hs-CRP (Fig. S5).
Mediation analysis
To examine the extent to which the associations of BALs with PA or CVD may be mediated through BMI, we performed mediation analysis on 145 cross-sectionally PA-related BALs, revealing 89 mediated (indirect effect P < 0.05; proportion mediated ranging from 10.3% to 64.4%) and 56 non-mediated associations (indirect effect P ≥ 0.05). Only mediated PA-BAL associations revealed significant relationships with CVD in the validation cohort (11 BMI-mediated and 4 non-mediated). Although BMI plays a predominant role in the PA-BAL-CVD relationship, non-mediated effects were also detected (Fig. S6).
We further explored the role of the 145 validated PA-BALs as mediators in the relationship between PA and incident CVD (Fig. S7). In VITAL-CVD, the total effect β estimate was −0.034 (95%CI = −0.156, 0.089), and 14 BALs, all unannotated, showed nominally significant indirect effect. Three out of these 14 BALs presented nominally significant indirect mediation effect in the relationship between PA and CVD in JUPITER-CVD (total effect: β = −0.059; 95%CI = −0.135, 0.017) (Table S7).
DISCUSSION
The present findings support our hypothesis that a bioactive lipidome signature related to the overall volume of habitual PA is also associated with risk of CVD events. While evidence of the myriad physiological benefits conferred by regular PA is undisputable, its mechanisms of action are still not completely understood 10,54. Our results suggest a pattern for the associations of habitual PA with 145 BALs (33 independent of BMI and HDL), of which 12 had opposite associations with CVD, providing further insights into biological and molecular mechanisms related to PA physiological aspects that can be involved in the human overall health.
Of the 145 BALs associated and validated with PA, five annotated BALs were positively associated with PA, two of which are dihydroxyoctadecenoic (diHOME) – 12,13-diHOME and 9,10-diHOME – stable and abundant oxylipin products in human plasma, also called leukotoxins. They are produced by inflammatory leukocytes such as neutrophils and macrophages and have wide-ranging effects, playing a role in suppressing neutrophil respiratory burst activity, vasodilation, and cellular apoptosis 55,56. Nieman et al. 57 reported an increase in both 12,13-diHOME and 9,10-diHOME intensities immediately post-cycling time trial, and 1.5-h after exercise in competitive male cyclists. Specifically, 12,13-diHOME has been demonstrated as a novel exercised-induced lipokine that regulates brown adipocyte biology and may contribute to the metabolic changes that occur with physical exercise 58. Lipokines are a relatively recently identified class of lipids that act as signaling molecules linking adipose tissue to systemic metabolism 58,59 and their increase has been associated with improved metabolic health 59. Interestingly, our results show that the association of PA with these BALs was both BMI-independent and BMI-non-mediated. Stanford et al. 58 found that physically active people not only had significantly higher concentrations of circulating 12,13-diHOME at baseline than those with a sedentary lifestyle, but also showed a greater increment immediately after an acute bout of exercise. The authors additionally found that the brown adipose tissue is the source of exercise-induced increase in circulating 12,13-diHOME in animal models.
LysoPC(15:0), or 1-pentadecanoyl-glycero-3-phosphocholine, another BAL positively associated with PA in the current study, belongs to lysophosphatidylcholine (LysoPC or LPC) species. This family is described as an important component of oxidized LDL-C 60 and paradoxically seem to possess both pro- and antiatherogenic properties: on the one hand, LysoPC species have been reported to increase the release of arachidonic acid, inflammatory cytokines and growth factor release 61, whereas on the other hand, their lower levels were related either with increased arterial stiffness or worse endothelial function 62. Plasma levels of other several LysoPCs (14:0, 16:0, 16:1, 18:0, 18:1) from the same class of organic compounds were found to be increased after aerobic exercise training 63. Specifically regarding LysoPC(15:0), a negative correlation with hs-CRP (r= −0.27) and BMI (r= −0.36) was found in obese participants 64, results very similar to ours (Fig. S3). Furthermore, lower LysoPC (15:0) intensities were observed among patients with chronic heart failure as compared to those without the disease 65, adding more evidence to the potential benefits of this metabolite on clinical biomarkers and health outcomes.
Oxymorphone-3b-D-glucuronide is the main phase II glucuronide conjugated metabolite from the parent oxymorphone, a semisynthetic μ-opioid agonist approved to treat acute and chronic pain 66. This conjugated metabolite is a product of specific degradation pathways and is not a commercially available drug 67.
Cortisone, the inactive metabolite from the physiologically active steroid hormone cortisol, is described to have an effect through changes in the hypothalamic-pituitary hormonal system caused by exercise 68. An acute increment in its levels has been described after a prolonged exercise effort (marathon race) 69, and after maximal graded exercise test in healthy, endurance-trained men 70. Moreover, Gouarné et al. 68 reported a training effect on overnight urinary cortisone excretion, with higher levels in trained triathletes compared with untrained pairs, even in conditions with similar cortisol production. This finding suggests a higher inactivation of cortisol into cortisone in well trained men, and the authors highlight it as a potential mechanism developed through training to protect tissues against the effects of exercise-induced increased cortisol secretion.
The negative association of PA and oleyl-glycerol seems physiological plausible as it is a non-ionizable fusogenic lipid, that can stimulate the fusion of human erythrocytes and is associated with the degradation of some membrane proteins 71. This result is in line with a negative correlation with HDL-C and positive with triglycerides obtained by our additional analysis (Fig. S3).
In the current study, most of PA-associated and all CVD-associated BALs were unknown metabolites without putative annotations. Since the lipidome composition reflects the networks of enzymatic pathways encoded within the genome, as well as the interplay of developmental processes and a changing environment over the lifetime of the organism, we performed genetic analysis to further insight into biology underlying the mass spectrometry data. Four BALs negatively related to PA but positively to CVD showed significant genetic associations, three of which associated variants are near the gene SLCO1B1 (solute carrier organic anion transporter family member 1A2). This gene encodes a sodium independent, organic anion transporter of compounds including bilirubin, 17-beta-glucuronosyl estradiol, and leukotriene C4. This transporter is also involved in the removal of drug compounds such as statins from the blood into the hepatocytes 72. Reports on metabolites previously related to variants near this gene, such as LPE(20:4), hexadecanedioate, and glycocholenate sulfate (Supplementary data 1) showed positive association with inflammation 73, blood pressure and mortality 74, and atrial fibrillation 75, respectively.
Variants near coding genes for cytochrome P450 enzymes, CYP2C18, CYP4V2, and CYP39A1, that catalyze the metabolism of cholesterol, steroids, and other lipids as well as some drugs were detected. Specifically, CYP39A1 protein is reported to participate in the conversion of cholesterol to bile acids 76. BAL-SNP associations were also found near coding genes of enzymes in the class of UDP-glycosyltransferases, UGT1A8, UGT3A1, and UGT2B7, that catalyze reactions making lipophilic substrates more water-soluble, enhancing excretion into either the urine or bile77.
One variant mapping to the FADS1-3 gene cluster, but nearest to FADS2, associated with one BAL. This gene encodes delta-6 desaturase that catalyzes one of two rate-limiting enzymes that convert α-linolenic (18:3n-3) and linoleic acid (18:2n-6) to their respective metabolites. A previous study 78 observed that FADS2 was the closest gene to a variant associated with 9- and 13-hydroxyoctadecadienoic acids, products from linoleic acid pathway. Several other reports found metabolites associated with variants near the FADS1-3 cluster (Supplementary data 1), such as LysoPC(20:4), which has been described as a pathophysiological biomarker of gestational diabetes 79. Phosphatidylcholine(38:3), also associated with SNPs in this cluster, was identified in atherosclerotic lesions and showed a strong association with CVD 80. Furthermore, this gene was associated with risk of cardiovascular disease 81.
Information on previously known SNP-trait associations (Table S5) shows evidence of highly pleiotropic SNPs, as they were associated with a variety of phenotypes including those other than blood and serum metabolites levels, e.g., glycated hemoglobin levels and glycemic traits, red cell distribution width, lymphocyte counts, and resting heart rate. Since these SNPs are not specific to phenotypes, hence highly pleiotropic, causal inference for CAD based on MR analysis was not reliable, even though the results were significant for some of the BALs that were associated both with habitual PA and with CVD (Table S4). However, it must be noted that although MR could not support causality, it is not evidence of causal effect absence. Future studies may inform whether other instruments or larger sample sizes can provide different findings.
The congruence of underlying physiological phenomena of annotated BALs, the correlation between BALs and clinical biomarkers, the genetic findings, and further relationship with risk of CVD provides biological insights into how PA may create a favorable environment for a protective cardiometabolic profile. Although a cross-sectional analysis between PA and BALs does not allow to establish a causal relationship between these factors, we suppose that shifts observed on some of these BALs may be a consequence of PA. Therefore, we postulate that a 21.8 MET-hrs-wk increase in habitual PA (PA standard deviation in VITAL-CTSC) may promote modifications in PA-dependent BALs. From a practical standpoint, this is roughly equivalent and proportional to an additional effort of walking at a moderate pace for 1h every day (~3METs) 82.
Through mediation analysis, we observed that significant results with CVD occurred only among BMI-mediated PA-BAL associations. It is known that the relationship between PA and BMI is complex and bi-directional, and our results are in line with previous results showing significant BMI mediation on the relationship between cardiorespiratory fitness and cardiometabolic risk variables in children 83. Further, we found that although BMI-mediated effects were predominant in BAL-CVD associations, non-mediated effects were also detected, suggesting that more than one pathway may play a role in the PA-CVD relationship (Fig. S5).
Our study had several strengths, including large sample sizes, validation in independent datasets, a study design that minimized laboratory drifts, and correction for multiple testing. Moreover, this study is the first to investigate further PA-associated BAL with incident CVD, especially in two independent and clinically different cohorts. Finally, addressing biological aspects related to PA is of contemporary importance, given guidelines on PA for the primary prevention of CVD.
This study also had limitations. First, self-reported PA measurements might make the data prone to measurement error, however, our results are in line with previous findings using an objective measure of PA ( accelerometer) 12. Second, PA variable was a continuous variable in VITAL, whereas it was ordinal in JUPITER; nevertheless, significant findings in discovery were replicated. Third, many of the LC-MS BAL detected in the current study are unknown, but the results for PA and incident CVD suggest consistency and provide informative insights about their effects on cardiovascular health-related aspects. Finally, the validation cohort for PA-BAL associations (i.e., JUPITER-NC) was composed of white participants with European ancestry. This approach was used as this subset had genetic data to provide additional information to our findings. The genetic analysis results may not be generalizable to populations with other ancestries. However, the main goal of this study was to present biomolecular aspects associated with PA that can potentially explain its effects on the cardiometabolic health as the first step toward a better understanding of underlying physiological-related mechanisms, and these analyses were conducted among individuals with multiethnic ancestry. Moreover, the sample selection for each study was determined by the eligibility criteria of the two studies which may also limit the generalizability beyond the study populations, but the consistency and independent replication of the findings adds rigor to the results and is a strength of the study. We emphasize that further investigation in different ethnicity or ancestry populations with genetic data are needed to compare with present results. Additionally, future work can establish causal relationships, and answer questions such as whether PA/exercise has a differential impact on the metabolome of those susceptible to CVD or whether the disease-associated metabolomic changes outweigh those caused by PA.
In summary, higher PA was associated with 145 BAL markers, and we postulate that these BALs may be related to a better cardiometabolic profile and might partly explain the inverse relationship between PA and CVD. In addition, the genetic analysis revealed associations with SNPs, identifying specific metabolic pathways that regulates the BAL markers. Our findings inform mechanistic understanding of the underlying biological and physiological factors relating PA to health, that are potentially the means through which cardiovascular diseases can be prevented.
Supplementary Material
Novelty and Significance.
What is known?
Physical activity (PA) has several health-related benefits that can prevent from cardiovascular disease (CVD).
Bioactive lipids can either influence or be susceptible to physiological stressors and adaptations, such as exercise.
Inflammation and immune function play an essential role in the development of CVD.
What new information does this article contribute?
We examined physiological-related aspects associated with PA from a biomolecular standpoint, analyzing bioactive lipids.
Higher PA was associated with increased concentration of signaling molecules with cardioprotective properties related to metabolic health, such as brown adipocyte regulation, inactivation of cortisol, and endothelial function.
Bioactive lipids related to both PA and CVD showed opposite directions of association.
The exact mechanisms linking PA to cardiometabolic health and CVD prevention are still being explored but known to be related to bioactive lipids functions. Bioactive lipids are signaling molecules produced by lipids metabolism which are involved in several functions, such as immunity, inflammation, and tissue homeostasis. In this study, we provide a comprehensive and mechanistic overview of biological phenomena, showing that part of these molecules associated with PA also presented a relationship with CVD, but in the opposite way: bioactive lipids that are increased with more exercise are likely to reduce the risk of CVDs (myocardial infarction, stroke, coronary revascularization, or cardiovascular death) and vice-versa. Understanding the behavior of these bioactive lipids can potentially enhance the knowledge about the benefits of exercising and how the body uses these factors towards a favorable environment for good cardiometabolic health.
Acknowledgments:
We are grateful to all VITAL and JUPITER studies participants and staff.
Sources of funding:
Dr. Hoshi is supported by the Lemann Foundation Cardiovascular Research Postdoctoral Fellowship. The VITamin D and OmegA-3 TriaL (VITAL) study (currently R01 AT011729) was supported by grants U01 CA138962 and R01 CA138962 that included support from the National Cancer Institute, National Heart, Lung and Blood Institute, Office of Dietary Supplements, National Institute of Neurological Disorders and Stroke and the National Center for Complementary and Integrative Health. The parent JUPITER trial was funded by AstraZeneca (Wilmington, DE), which had no role in this current study. Dr. Okereke was supported by the research grant R01MH091448. Dr. Demler was supported by a research grant from the National Heart, Lung and Blood Institute (1K01HL135342-01) and BWH Lerner Research Award. Dr. Mora was supported by research grants from the National Institute of Diabetes and Digestive and Kidney Diseases (DK112940), National Heart, Lung, and Blood Institute (R01HL134811 and K24 HL136852, R01HL134168, and 1R01HL143227). The funding sources had no role in the design and conduct of this study or the interpretation of the data. The opinions expressed in the manuscript are those of the study authors.
Disclosures:
Dr. Ridker is listed as a co-inventor on patents held by the Brigham and Women’s Hospital that relate to the use of inflammatory biomarkers in cardiovascular disease that have been licensed to AstraZeneca and Seimens. Dr. Demler received support from Kowa, not related to the current work. Dr. Mora received research grant support from Atherotech Diagnostics for work outside the current study, the Molino Family Trust, and NIH, and has served as consultant to Pfizer and Quest Diagnostics for work outside the current study. Dr. Tiwari holds a position at Sapient Bioanalytics. The remaining authors have no disclosures to report. Quest Diagnostics conducted ion mobility assays at no additional charge and Atherotech Diagnostics conducted Vertical Auto Profile tests (VAP) at no additional charge to the VITAL study.
Non-standard Abbreviations and Acronyms
- BAL
Bioactive Lipid
- CAD
Coronary artery disease
- CTSC
Clinical Translational Science Center
- CVD
Cardiovascular disease
- FDR
False Discovery Rate
- hs-CRP
High-sensitivity C-reactive protein
- IVW
Inverse-variance weighted
- JUPITER
Justification for the Use of statins in Prevention: an Intervention Trial Evaluating Rosuvastatin
- LC-MS
Liquid chromatography-mass spectrometry
- MET
Metabolic Equivalent of Task
- MR
Mendelian randomization
- NC
Non-cases
- OR
Odds ratio
- PA
Physical activity
- SNP
Single-nucleotide polymorphism
- UCSC
University of California Santa Cruz
- VITAL
VITamin D and OmegA-3 TriaL
REFERENCES
- 1.Bull FC, Al-Ansari SS, Biddle S, Borodulin K, Buman MP, Cardon G, Carty C, Chaput JP, Chastin S, Chou R, et al. World Health Organization 2020 guidelines on physical activity and sedentary behaviour. British journal of sports medicine. 2020;54:1451–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markworth JF, Maddipati KR, Cameron-Smith D. Emerging roles of pro-resolving lipid mediators in immunological and adaptive responses to exercise-induced muscle injury. Exercise Immunology Review. 2016;22:110–134. [PubMed] [Google Scholar]
- 3.Nieman DC, Wentz LM. The compelling link between physical activity and the body’s defense system. Journal of sport and health science. 2019;8:201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chiurchiù V, Leuti A, Maccarrone M. Bioactive lipids and chronic inflammation: Managing the fire within. Frontiers in Immunology. 2018;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Demler O, Liu Y, Luttmann-Gibson H, Watrous JD, Lagerborg KA, Dashti H, Giulianini F, Heath M, Camargo CA, Harris WS, et al. One-year effects of omega-3 treatment on fatty acids, oxylipins, and related bioactive lipids and their associations with clinical lipid and inflammatory biomarkers: Findings from a substudy of the vitamin d and omega-3 trial (vital). Metabolites. 2020;10:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sansbury BE, Spite M. Resolution of Acute Inflammation and the Role of Resolvins in Immunity, Thrombosis, and Vascular Biology. Circulation Research. 2016;119:113–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jiang H, Anderson GD, McGiff JC. The red blood cell participates in regulation of the circulation by producing and releasing epoxyeicosatrienoic acids. Prostaglandins & other lipid mediators. 2012;98:91–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alanko J, Riutta A, Vapaatalo H. Effects of catecholamines on eicosanoid synthesis with special reference to prostanoid/leukotriene ratio. Free radical biology & medicine. 1992;13:677–88. [DOI] [PubMed] [Google Scholar]
- 9.Nosarev A, Smagliy L, Anfinogenova Y, Popov S, Kapilevich L. Exercise and NO production: relevance and implications in the cardiopulmonary system. Frontiers in cell and developmental biology. 2014;2:73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelly RS, Kelly MP, Kelly P. Metabolomics, physical activity, exercise and health: A review of the current evidence. Biochimica et Biophysica Acta - Molecular Basis of Disease. 2020;1866:165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding M, Zeleznik OA, Guasch-Ferre M, Hu J, Lasky-Su J, Lee IM, Jackson RD, Shadyab AH, Lamonte MJ, Clish C, et al. Metabolome-Wide Association Study of the Relationship Between Habitual Physical Activity and Plasma Metabolite Levels. American Journal of Epidemiology. 2019;188:1932–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao Q, Moore SC, Keadle SK, Xiang YB, Zheng W, Peters TM, Leitzmann MF, Ji BT, Sampson JN, Shu XO, Matthews CE. Objectively measured physical activity and plasma metabolomics in the Shanghai Physical Activity Study. International Journal ofEpidemiology. 2016;45:1433–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mora S, Cook N, Buring JE, Ridker PM, Lee I-M. Physical Activity and Reduced Risk of Cardiovascular Events. Circulation. 2007;116:2110–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manson JE, Cook NR, Lee I-M, Christen W, Bassuk SS, Mora S, Gibson H, Gordon D, Copeland T, D’Agostino D, et al. Vitamin D Supplements and Prevention of Cancer and Cardiovascular Disease. New England Journal of Medicine. 2019;380:33–44.30415629 [Google Scholar]
- 15.Ridker PM, Danielson E, Fonseca FAH, Genest J, Gotto AM, Kastelein JJP, Koenig W, Libby P, Lorenzatti AJ, MacFadyen JG, et al. Rosuvastatin to Prevent Vascular Events in Men and Women with Elevated C-Reactive Protein. New England Journal of Medicine. 2008;359:2195–2207. [DOI] [PubMed] [Google Scholar]
- 16.Bassuk SS, Manson JAE, Lee IM, Cook NR, Christen WG, Bubes VY, Gordon DS, Copeland T, Friedenberg G, D’Agostino DM, et al. Baseline characteristics of participants in the VITamin D and OmegA-3 TriaL (VITAL). Contemporary Clinical Trials. 2016;47:235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watrous JD, Niiranen TJ, Lagerborg KA, Henglin M, Xu Y-J, Rong J, Sharma S, Vasan RS, Larson MG, Armando A, et al. Directed Non-targeted Mass Spectrometry and Chemical Networking for Discovery of Eicosanoids and Related Oxylipins. Cell Chemical Biology. 2019;26:433–442.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chasman DI, Giulianini F, MacFadyen J, Barratt BJ, Nyberg F, Ridker PM. Genetic Determinants of Statin-Induced Low-Density Lipoprotein Cholesterol Reduction. Circulation: Cardiovascular Genetics. 2012;5:257–264. [DOI] [PubMed] [Google Scholar]
- 19.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Mailer J, Sklar P, de Bakker PIW, Daly MJ, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. American Journal of Human Genetics. 2007;81:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costenbader KH, MacFarlane LA, Lee I-M, Buring JE, Mora S, Bubes V, Kotler G, Camargo CA Jr., Manson JE, Cook NR. Effects of One Year of Vitamin D and Marine Omega-3 Fatty Acid Supplementation on Biomarkers of Systemic Inflammation in Older US Adults. Clinical Chemistry. 2019;65:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mora S, Glynn RJ, Matthij s Boekholdt S, Nordestgaard BG, Kastelein JJP, Ridker PM. On-Treatment Non-HDL Cholesterol, Apolipoprotein B, Triglycerides, and Lipid Ratios in Relation to Residual Vascular Risk after Treatment with Potent Statin Therapy: The JUPITER Trial. Journal of the American College of Cardiology. 2012;59:1521–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouřil Š, Sousa J, Václavík J, Friedecký D, Adam T. CROP: correlation-based reduction of feature multiplicities in untargeted metabolomic data. Wren J, ed. Bioinformatics. 2020;36:2941–2942. [DOI] [PubMed] [Google Scholar]
- 23.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The SVA package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society: Series B (Methodological). 1995;57:289–300. [Google Scholar]
- 25.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. [Google Scholar]
- 26.Li Y, Wilier CJ, Ding J, Scheet P, Abecasis GR. MaCH: Using Sequence and Genotype Data to Estimate Haplotypes and Unobserved Genotypes. Genetic epidemiology. 2010;34:816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genetic Epidemiology. 2008;32:361–369. [DOI] [PubMed] [Google Scholar]
- 28.Oscanoa J, Sivapalan L, Gadaleta E, Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of human genome sequence variation (2020 update). Nucleic Acids Research. 2020;48:W185–W192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess S, Butterworth A, Thompson SG. Mendelian Randomization Analysis With Multiple Genetic Variants Using Summarized Data. Genetic Epidemiology. 2013;37:658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nikpey M, Goel A, Won H, Hall L, Willenborg C, Kanoni S, Saleheen D, Kyriakou T. A comprehensive 1000 Genomes–based genome-wide association meta-analysis of coronary artery disease. Nature Genetics. 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jennrich RI. An asymptotic χ2 test for the equality of two correlation matrices. Journal of the American Statistical Association. 1970;65:904–912. [Google Scholar]
- 32.Valeri L, VanderWeele TJ. Mediation analysis allowing for exposure-mediator interactions and causal interpretation: Theoretical assumptions and implementation with SAS and SPSS macros. Psychological Methods. 2013;18:137–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lotta LA, Pietzner M, Stewart ID, Wittemans LBL, Li C, Bonelli R, Raffler J, Biggs EK, Oliver-Williams C, Auyeung VPW, Luan J, et al. A cross-platform approach identifies genetic regulators of human metabolism and health. Nature Genetics. 2021;53:54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlosser P, Li Y, Sekula P, Raffler J, Grundner-Culemann F, Pietzner M, Cheng Y, Wuttke M, Steinbrenner I, Schultheiss UT, Kotsis F, et al. Genetic studies of urinary metabolites illuminate mechanisms of detoxification and excretion in humans. Nature Genetics. 2020;52:167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long T, Hicks M, Yu HC, Biggs WH, Kirkness EF, Menni C, Zierer J, Small KS, Mangino M, Messier H, et al. Whole-genome sequencing identifies common-to-rare variants associated with human blood metabolites. Nature Genetics. 2017;49:568–578. [DOI] [PubMed] [Google Scholar]
- 36.Nag A, Kurushima Y, Bowyer RC, Wells PM, Weiss S, Pietzner M, Kocher T, Raffler J, Völker U, Mangino M, et al. Genome-wide scan identifies novel genetic loci regulating salivary metabolite levels. Human Molecular Genetics. 2020;29:864–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsepilov YA, Sharapov SZ, Zaytseva OO, Krumsek J, Prehn C, Adamski J, Kastenmuller G, Wang-Sattler R, Strauch K, Gieger C, et al. A network-based conditional genetic association analysis of the human metabolome. GigaScience. 2018;7:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Draisma HHM, Pool R, Kobl M, Jansen R, Petersen AK, Vaarhorst AAM, Yet I, Haller T, Demirkan A, Esko T, et al. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nature Communications 2015. 6:1. 2015;6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson Å, Rudan I, Aulchenko YS, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genetics. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Illig T, Gieger C, Zhai G, Römisch-Margl W, Wang-Sattler R, Prehn C, Altmaier E, Kastenmüller G, Kato BS, Mewes HW, et al. A genome-wide perspective of genetic variation in human metabolism. Nat. Genet. 2010;42:137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gieger C, Geistlinger L, Altmaier E, de Angelis MH, Kronenberg F, Meitinger T, Mewes HW, Wichmann HE, Weinberger KM, et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rhee EP, Ho JE, Chen M, Shen D, Cheng S, Larson MG, Ghorbani A, Shi X, Helenius IT, O’Donnell CJ, et al. A Genome-wide Association Study of the Human Metabolome in a Community-Based Cohort. Cell Metabolism. 2013;18:130–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. An atlas of genetic influences on human blood metabolites. Nat. Genet. 2014;46:543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yet I, Menni C, Shin SY, Mangino M, Soranzo N, Adamski J, Suhre K, Spector TD, Kastenmüller G, Bell JT. Genetic influences on metabolite levels: A comparison across metabolomic platforms. PLoS ONE. 2016;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wägele B, Altmaier E, Deloukas P, Erdmann J, Grundberg E, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477:54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hysi PG, Mangino M, Christofidou P, Falchi M, Karoly ED, Mohney RP, Valdes AM, Spector TD, Menni C. Metabolome Genome-Wide Association Study Identifies 74 Novel Genomic Regions Influencing Plasma Metabolites Levels. Metabolites. 2022;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krumsiek J, Suhre K, Evans AM, Mitchell MW, Mohney RP, Milburn M v., Wägele B, Römisch-Margl W, Illig T, Adamski J, et al. Mining the Unknown: A Systems Approach to Metabolite Identification Combining Genetic and Metabolic Information. PLoS Genetics. 2012;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yousri NA, Fakhro KA, Robay A, Rodriguez-Flores JL, Mohney RP, Zeriri H, Odeh T, Kader SA, Aldous EK, Thareja G, et al. Whole-exome sequencing identifies common and rare variant metabolic QTLs in a Middle Eastern population. Nature Communications. 2018;9:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hong MG, Karlsson R, Magnusson PKE, Lewis MR, Isaacs W, Zheng LS, Xu J, Grönberg H, Ingelsson E, Pawitan Y, et al. A Genome-Wide Assessment of Variability in Human Serum Metabolism. Human Mutation. 2013;34:515–524. [DOI] [PubMed] [Google Scholar]
- 50.Harshfield EL, Fauman EB, Stacey D, Paul DS, Ziemek D, Ong RMY, Danesh J, Butterworth AS, Rasheed A, Sattar T, et al. Genome-wide analysis of blood lipid metabolites in over 5000 South Asians reveals biological insights at cardiometabolic disease loci. BMC Medicine. 2021;19:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tabassum R, Rämö JT, Ripatti P, Koskela JT, Kurki M, Karjalainen J, Palta P, Hassan S, Nunez-Fontarnau J, Kiiskinen TTJ, et al. Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nature Communications. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Coltell O, Sorlí J v., Asensio EM, Barragán R, González JI, Giménez-Alba IM, Zanón-Moreno V, Estruch R, Ramírez-Sabio JB, et al. Genome-Wide Association Study for Serum Omega-3 and Omega-6 Polyunsaturated Fatty Acids: Exploratory Analysis of the Sex-Specific Effects and Dietary Modulation in Mediterranean Subjects with Metabolic Syndrome. Nutrients. 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, Klemm A, Flicek P, Manolio T, Hindorff L, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Research. 2014;42:D1001–D1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neufer PD, Bamman MM, Muoio DM, Bouchard C, Cooper DM, Goodpaster BH, Booth FW, Kohrt WM, Gerszten RE, Mattson MP, et al. Understanding the Cellular and Molecular Mechanisms of Physical Activity-Induced Health Benefits. Cell Metabolism. 2015;22:4–11. [DOI] [PubMed] [Google Scholar]
- 55.Vella L, Markworth JF, Farnfield MM, Maddipati KR, Russell AP, Cameron-Smith D. Intramuscular inflammatory and resolving lipid profile responses to an acute bout of resistance exercise in men. Physiological Reports. 2019;7:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson DA, Hammock BD. Dihydroxyoctadecamonoenoate esters inhibit the neutrophil respiratory burst. Journal of Biosciences. 2007;32:279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nieman DC, Shanely RA, Luo B, Meaney MP, Dew DA, Pappan KL. Metabolomics approach to assessing plasma 13- and 9-hydroxy-octadecadienoic acid and linoleic acid metabolite responses to 75-km cycling. American Journal of Physiology - Regulatory Integrative and Comparative Physiology. 2014;307:68–74. [DOI] [PubMed] [Google Scholar]
- 58.Stanford KI, Lynes MD, Takahashi H, Baer LA, Peter J, May FJ, Lehnig AC, Middelbeek RJW, Richard JJ, So K, et al. 12,13-diHOME: An Exercise-Induced Lipokine that Increases Skeletal Muscle Fatty Acid Uptake. Cell metabolism. 2018;27:1111–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lynes MD, Leiria LO, Lundh M, Bartelt A, Shamsi F, Huang TL, Takahashi H, Hirshman MF, Schlein C, Lee A, et al. The cold-induced lipokine 12,13-diHOME promotes fatty acid transport into brown adipose tissue. Nature Medicine. 2017;23:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schmitz G, Ruebsaamen K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 2010;208:10–18. [DOI] [PubMed] [Google Scholar]
- 61.Aiyar N, Disa J, Ao Z, Ju H, Nerurkar S, Willette RN, Macphee CH, Johns DG, Douglas SA. Lysophosphatidylcholine induces inflammatory activation of human coronary artery smooth muscle cells. Molecular and Cellular Biochemistry. 2007;295:113–120. [DOI] [PubMed] [Google Scholar]
- 62.Paapstel K, Kals J, Eha J, Tootsi K, Ottas A, Piir A, Jakobson M, Lieberg J, Zilmer M. Inverse relations of serum phosphatidylcholines and lysophosphatidylcholines with vascular damage and heart rate in patients with atherosclerosis. Nutrition, Metabolism and Cardiovascular Diseases. 2018;28:44–52. [DOI] [PubMed] [Google Scholar]
- 63.Felder TK, Ring-Dimitriou S, Auer S, Soyal SM, Kedenko L, Rinnerthaler M, Cadamuro J, Haschke-Becher E, Aigner E, Paulweber B, et al. Specific circulating phospholipids, acylcarnitines, amino acids and biogenic amines are aerobic exercise markers. Journal of Science and Medicine in Sport. 2017;20:700–705. [DOI] [PubMed] [Google Scholar]
- 64.Heimerl S, Fischer M, Baessler A, Liebisch G, Sigruener A, Wallner S, Schmitz G. Alterations of plasma lysophosphatidylcholine species in obesity and weight loss. PLoS ONE. 2014;9:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marcinkiewicz-Siemion M, Ciborowski M, Ptaszynska-Kopczynska K, Szpakowicz A, Lisowska A, Jasiewicz M, Waszkiewicz E, Kretowski A, Musial WJ, Kaminski KA. LC–MS-based serum fingerprinting reveals significant dysregulation of phospholipids in chronic heart failure. Journal of Pharmaceutical and Biomedical Analysis. 2018;154:354–363. [DOI] [PubMed] [Google Scholar]
- 66.Prommer E Oxymorphone: A review. Supportive Care in Cancer. 2006;14:109–115. [DOI] [PubMed] [Google Scholar]
- 67.Grabenauer M, Bynum ND, Moore KN, White RM, Mitchell JM, Hayes ED, Flegel R. Detection and quantification of codeine- 6-glucuronide, hydromorphone-3-glucuronide, oxymorphone-3-glucuronide, morphine 3-glucuronide and morphine-6-glucuronide in human hair from opioid users by LC-MS-MS. Journal of Analytical Toxicology. 2018;42:115–125. [DOI] [PubMed] [Google Scholar]
- 68.Gouarné C, Groussard C, Gratas-Delamarche A, Delamarche P, Duclos M. Overnight urinary cortisol and cortisone add new insights into adaptation to training. Medicine and Science in Sports and Exercise. 2005;37:1157–1167. [DOI] [PubMed] [Google Scholar]
- 69.Bae YJ, Kratzsch J, Zeidler R, Fikenzer S, Werner C, Herm J, Jungehülsing GJ, Endres M, Haeusler KG, Thiery J, et al. Unraveling the steroid hormone response in male marathon runners: Correlation of running time with aldosterone and progesterone. Journal of Steroid Biochemistry and Molecular Biology. 2019;195:105473. [DOI] [PubMed] [Google Scholar]
- 70.del Corral P, Schurman RC, Kinza SS, Fitzgerald MJ, Kordick CA, Rusch JL, Nadolski JB. Salivary but not plasma cortisone tracks the plasma cortisol response to exercise: Effect of time of day. Journal of Endocrinological Investigation. 2016;39:315–322. [DOI] [PubMed] [Google Scholar]
- 71.Quirk SJ, Ahkong QF, Botham GM, Vos J, Lucy JA. Membrane proteins in human erythrocytes during cell fusion induced by oleoylglycerol. Biochemical Journal. 1978;176:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Voora D, Shah SH, Spasojevic I, Ali S, Reed CR, Salisbury BA, Ginsburg GS. The SLCO1B1*5 Genetic Variant is Associated with Statin-Induced Side Effects. Journal of the American College of Cardiology. 2009;54:1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meikle PJ, Wong G, Tsorotes D, Barlow CK, Weir JM, Christopher MJ, MacIntosh GL, Goudey B, Stern L, Kowalczyk A, et al. Plasma lipidomic analysis of stable and unstable coronary artery disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2011;31:2723–2732. [DOI] [PubMed] [Google Scholar]
- 74.Menni C, Graham D, Kastenmüller G, Alharbi NHJ, Alsanosi SM, Mcbride M, Mangino M, Titcombe P, Shin SY, Psatha M, et al. Metabolomic Identification of a Novel Pathway of Blood Pressure Regulation Involving Hexadecanedioate. Hypertension. 2015;66:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alonso A, Yu B, Qureshi WT, Grams ME, Selvin E, Soliman EZ, Loehr LR, Chen LY, Agarwal SK, Alexander D, et al. Metabolomics and Incidence of Atrial Fibrillation in African Americans: The Atherosclerosis Risk in Communities (ARIC) Study. PloS one. 2015;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grabovec I, Smolskaya S, Baranovsky A, Zhabinskii V, Dichenko Y, Shabunya P, Usanov S, Strushkevich N. Ligand-binding properties and catalytic activity of the purified human 24-hydroxycholesterol 7α-hydroxylase, CYP39A1. Journal of Steroid Biochemistry and Molecular Biology. 2019;193:105416. [DOI] [PubMed] [Google Scholar]
- 77.Mackenzie PI, Rogers A, Treloar J, Jorgensen BR, Miners JO, Meech R. Identification of UDP Glycosyltransferase 3A1 as a UDP N-Acetylglucosaminyltransferase. The Journal of Biological Chemistry. 2008;283:36205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Feofanova EV, Yu B, Metcalf GA, Liu X, Muzny D, Below JE, Wagenknecht LE, Gibbs RA, Morrison AC, Boerwinkle E. Sequence-based analysis of lipid-related metabolites in a multiethnic study. Genetics. 2018;209:607–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lehmann R, Friedrich T, Krebiehl G, Sonntag D, Häring HU, Fritsche A, Hennige AM. Metabolic Profiles during an Oral Glucose Tolerance Test in Pregnant Women with and without Gestational Diabetes. Experimental and Clinical Endocrinology and Diabetes. 2015;123:433–438. [DOI] [PubMed] [Google Scholar]
- 80.Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, Menni C, Moayyeri A, Santer P, Rungger G, et al. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based bruneck study. Circulation. 2014;129:1821–1831. [DOI] [PubMed] [Google Scholar]
- 81.Yang Q, Yin R-X, Cao X-L, Wu D-F, Chen W-X, Zhou Y-J. Association of two polymorphisms in the FADS1/FADS2 gene cluster and the risk of coronary artery disease and ischemic stroke. International Journal of Clinical and Experimental Pathology. 2015;8:7318. [PMC free article] [PubMed] [Google Scholar]
- 82.Ainsworth BE, Haskell WL, Whitt MC, Irwin ML, Swartz AM, Strath SJ, O’Brien WL, Bassett J, Schmitz KH, Emplaincourt PO, et al. Compendium of physical activities: An update of activity codes and MET intensities. Medicine and Science in Sports and Exercise. 2000;32. [DOI] [PubMed] [Google Scholar]
- 83.Díez-Fernández A, Sánchez-López M, Mora-Rodríguez R, Notario-Pacheco B, Torrijos-Niño C, Martínez-Vizcaíno V. Obesity as a mediator of the inf luence of cardiorespiratory fitness on cardiometabolic risk: A mediation analysis. Diabetes Care. 2014;37:855–862. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Study Populations
We assayed baseline BALs within baseline blood samples of two independent studies: The VITamin D and OmegA-3 TriaL (VITAL; NCT 01169259) 14 and the Justification for the Use of statins in Prevention: an Intervention Trial Evaluating Rosuvastatin (JUPITER; NCT 00239681) 15. Both VITAL and JUPITER were randomized, double-blinded, and placebo-controlled trials. Briefly, VITAL examined 25,871 participants, testing the effects of marine n-3 fatty acids and/or vitamin D (2000 IU/d) supplements vs. placebo in the prevention of CVD and cancer. JUPITER investigated the effects of 20 mg/day rosuvastatin vs. placebo on the rate of cardiovascular outcomes in 17,802 participants with average to low levels of low-density lipoprotein cholesterol (LDL-C < 130 mg/dl) and elevated high-sensitivity C-reactive protein (hs-CRP > 2 mg/L). Participants from both studies provided written informed consent at the time of enrollment. Institutional review board approvals for both studies were obtained from Partners HealthCare (Boston, MA). First and senior authors had full access to all data in the study and take responsibility for their integrity and data analysis.
From VITAL, we examined assays collected as part of two sub-studies: 1) VITAL-CTSC, a subsample (N=1,032) that had in-person clinical evaluations at the Brigham and Women’s Hospital Clinical Translational Science Center – CTSC 16, and 2) VITAL-CVD, a nested case-control sub-study for incident CVD events matched on age, sex, and fasting status (N case-control pairs=741). Only 46 participants from VITAL-CTSC were also included in VITAL-CVD sub-study (24 cases and 22 controls).
Two sub-studies from JUPITER were analyzed for validation: 1) JUPITER-NC, a non-cases subsample randomly selected amongst participants with European ancestry with genetic analysis (N=589), and 2) JUPITER-CVD, a nested case-control sub-study for incident CVD matched on age and sex (N case-control pairs=415). Among JUPITER sub-studies, there were no overlapping participants.
Study design
We had a two-stage study design (Fig. 1). First, we used discovery in VITAL-CTSC and validation in JUPITER-NC, to evaluate PA-BAL baseline cross-sectional associations. Second, we used a similar approach to investigate whether PA-related BALs were associated with future risk of CVD. This relationship was first examined in the VITAL-CVD case-control study, then validated in the JUPITER-CVD case-control study.
Fig. 1. Diagram of the two-stage analysis.

The left-side box shows the first-stage analysis performed to examine associations between PA and BAL in the discovery cohort, VITAL-CTSC, and validated in JUPITER-NC. BALs significantly associated with PA were carried over to the second-stage analysis (right-side box), in which their association with incident CVD was investigated in VITAL-CVD and validated in JUPITER-CVD.
Biomarker profiling
Blood samples from both main studies, all collected at baseline and before randomization, were assayed at the University of California, San Diego. Approximately 11,000 features were extracted using a directed non-targeted high-throughput liquid chromatography-mass spectrometry (LC-MS) approach to measure the relative intensities of small molecules 17. VITAL-CTSC and JUPITER-NC samples were run with respective study’s case and control samples. Corresponding cases and controls were placed in tandem wells on a plate (in random order) to avoid batch effects and were blindly assayed. To avoid experimental biases, blinded quality control (QC) samples were additionally embedded throughout the experiment within each of the batches. Moreover, the performing laboratory had a multi-tiered QC approach that allows for close monitoring of sample-to-sample variation and preparation, as well as system performance and any potential system drift over the sample run. Details of the blood sample collection, LC-MS method, and annotation process are provided in the Supplemental material. Features detected in the discovery cohorts were mapped in the validation cohorts by peaks alignment based on retention time, accurate mass, and tandem MS fragmentation pattern17.
PA assessment
In VITAL, PA was assessed through a self-administrated questionnaire with questions regarding the average amount of time during the last 12 months spent in activities such as: jogging, aerobic exercises, tennis, swimming (activities that require ≥ 6 metabolic equivalent [METs]); walking, bicycling, yoga, weight lifting (activities requiring 3 to 5.9 METs); and slow walking (light activity – < 3METs). Details of activities and intensities are provided in the Supplemental material. The total reported PA was calculated as weekly energy expenditure in METs (METs-hr/wk) by multiplying the intensity attributed to each type of PA (MET) and time spent in the activity according to the questionnaire. Participants from VITAL-CTSC with self-reported MET-hrs/wk ≥ 3SD from the average (24.5 MET-hr/wk) were considered outliers and excluded from the analysis (N=17). In VITAL-CTSC, PA was treated as a continuous variable which values were shifted and rescaled to mean 0 and SD = 1 (21.8 MET-hr/wk) for better comparison across studies and more interpretable results.
In JUPITER, PA was treated as an ordinal variable ranging from 1 to 6, according to the category of self-reported frequency per week: 1) Rarely/Never, 2) less than once a week, 3) once a week, 4) 2-3 times a week, 5) 4-6 times a week, or 6) daily.
Cardiovascular disease outcomes
VITAL-CVD cases were defined as an adjudicated composite endpoint of confirmed myocardial infarction, stroke, coronary revascularization, or cardiovascular death. In JUPITER-CVD, cases were defined as an adjudicated composite endpoint of confirmed myocardial infarction, stroke, coronary revascularization, unstable angina requiring hospitalization, or death.
Genetic data
To further inform the mass spectrometry data, we performed a genome-wide association study (GWAS) for replicated BALs using JUPITER-NC dataset. Detailed genotyping and analysis procedure was described by Chasman et al. 18. Briefly, all participants involved in the present study provided consent for genetic analyses, as part of the JUPITER main trial protocol, and had DNA available for genotyping. Genome-wide genotyping was performed on a fee-for-service basis by Illumina (San Diego, CA) using the Illumina Omni 1M Quad platform, and raw genotype intensity data were reduced to genotype calls using the Illumina Genome Studio (v. 1.6.2) software. Single nucleotide polymorphism (SNP) clusters were initially defined automatically using data from the JUPITER sample. Approximately 1% of the SNPs were identified for poor clustering on the basis of quality measures including Hardy-Weinberg equilibrium (HWE; P<10−6) or call rate (< 95%), and were visually inspected and annotated, removed, or clustered again with manual intervention. Re-clustered SNPs were retained if genotypes could be called in >95% of the samples. After these procedures, 99.71% of the loci met the preceding quality criteria. Included participants had self-reported European ancestry verified by multidimensional scaling procedures in PLINK 19 applied to 1067 unlinked ancestry informative SNPs selected from HapMap3. The relatedness (pi_hat) was estimated from identity-by-state in PLINK 19.
Covariates
Baseline questionnaires were used to collect sex, age, race/ethnicity, body weight and height, use of non-randomized supplements or medications, smoking, and other relevant aspects of health history. VITAL-CTSC participants also had their body weight and height measured by trained staff. VITAL-CTSC and VITAL-CVD blood samples were sent to Atherotech Diagnostics (Birmingham, AL) and Quest Diagnostics Nichols Institute (San Juan Capistrano, CA) for standard lipid panels (plasma). High-sensitivity C-reactive protein (hs-CRP) was assayed by latex-enhanced immunonephelometric assay 20. In JUPITER-NC and JUPITER-CVD, standard lipid panels were measured in a central laboratory as part of the clinical trial. LDL-C concentrations were calculated by the Friedewald equation when triglycerides were <400 mg/dL and measured by ultracentrifugation when ≥400 mg/dL. A high-sensitivity assay (Behring Nephelometer) was used for hs-CRP measurement 21.
Statistical analysis
Variables distributions were expressed as median and interquartile ranges. Mann–Whitney U test was used to examine contrasts between cases and controls in VITAL-CVD and JUPITER-CVD, and differences in categorical variables were compared with crosstabulation and subsequent X2 testing.
Prior to statistical analysis, we performed data preparation. We detected and removed carbon-13 (13C) isotopes – features with correlation coefficient > 0.95, up to ± 0.01 minutes retention time (rt) from each other, and difference in mass to charge ratio (m/z) equal to one or more 13C atom mass – and typically found adducts to reduce redundancy of assayed BALs.
Only BAL features with less than 20% of missing values were considered for this analysis. To remove other redundant fragments (due to contaminants, fragments, or other MS artifacts) 22, we examined remaining highly correlated BALs (r>0.95). From each pairwise comparison, we removed those with lower median relative intensity. Missing values were then imputed to 0.25 of the lowest observed value. All BALs were log-transformed prior to outlier detection (mean ± 3 standard deviations [SD]), and those exceeding these limits were trimmed to ± 3 SD of mean values. Next, to remove experimental biases, VITAL-CTSC and JUPITER-NC all measurements were adjusted for batch and run-day effects using ComBat (R-package sva) 23. No plate effects correction was performed in VITAL-CVD and JUPITER-CVD, as cases and controls were placed in wells next to each other. All values were rescaled to median of 0 and SD = 1.
PA-BAL cross-sectional associations were performed on baseline data collected before the randomization process for each trial. We used linear regressions adjusted for age and sex (model 1), and model 1 plus race/ethnicity, LDL-cholesterol, total cholesterol, and current smoking (model 2). A sensitivity analysis was conducted to investigated associations between validated BALs and continuous PA in subgroup analysis for participants being above and below the median for both PA and BMI. An additional model (model 3) was fitted, which included adjustments for categorized BMI (< 25, 25–29.9, or ≥ 30 kg/m2) and HDL-cholesterol.
Supplementary analyses were performed to meet model assumptions and ensure statistical rigor. First, the normality of linear regression residuals for BALs was checked using the Kolmogorov-Smirnov’s test. Then, BALs for which the residuals normality assumption was rejected were analyzed by the Wilcoxon test (BAL measures in PA levels above the median vs. PA below the median). Finally, BALs for which the Wilcoxon test null hypothesis was not rejected, we performed a bootstrap resampling procedure with 2000 bootstrap replicates to provide consistent estimates of confidence intervals.
For associations with incident CVD, we used conditional logistic regressions with similar models 1 and 2 (excluding sex, which was used as case-control matching criteria), plus the randomized treatment assignment to account for the interventions involved in the clinical trials. CVD outcome was included in the model as the dependent variable and PA-related BALs as independent variables.
To account for multiple comparisons in all regression analyses, we used the Benjamini-Hochberg approach 24,25 controlling for False Discovery Rate (FDR) < 0.1. The criteria for validation in JUPITER-NC and JUPITER-CVD were FDR < 0.1 and β-coefficient with the same directionality.
Common genetic variation that was not measured directly in the parent JUPITER data from which JUPITER-NC was derived (above) were imputed using the Markov Chain Haplotyping algorithm (MaCH 1.0, developed by Li et al.26) and the 1000 Genomes Project reference panel (phase 1, v. 3). Imputation was performed among JUPITER participants with verified European ancestry including 979,089 genotyped SNPs in HWE (P≥10−6) and without restriction by minor allele frequency. Analysis was limited to 6,528,605 SNPs with imputation quality r2 < 0.8 and with a minor allele frequency <5%, implying at least 55 copies of the heterozygote genotype given the sample size under Hardy-Weinberg equilibrium. For genetic associations analysis, each validated BAL related to PA (145 PA-associated BALs obtained using model 2) was further investigated for genetic associations at a genome-wide level of statistical significance (P < 5x10−8) by performing multivariable linear regression assuming an additive genetic model, adjusted for age, sex, and 10 principal components of sub-European ancestry. Index SNPs within independent loci reaching genome-wide significance were identified from the genome-wide scans across all PA-associated BALs using a clumping procedure that grouped SNPs within 250 kb and linkage disequilibrium (LD) r2 > 0.10 from the index SNP, with a significance threshold for index SNPs of 7.3x10−10 (genome-wide P-value adjusted for the effective number of independent tests [N=69], derived from Gao et al. 27 using a cutoff of 90%). We used the web-based application SNPnexus 28 to annotate index SNPs for gene and variant type based on data from the University of California Santa Cruz (UCSC) and Ensembl genome databases (human genome version hg19).
Additionally, two-sample inverse-variance weighted (IVW) Mendelian randomization (MR) 29 was implemented to examine potential causal associations between BALs significantly associated with PA and coronary artery disease (CAD). We used estimates for index genetic variants herein found to be associated with PA-related BALs with summary statistics for CAD from CARDIoGRAMplusC4D 1000 Genomes-based GWAS meta-analysis 30 (available at http://www.cardiogramplusc4d.org/data-downloads/). Mendelian randomization was performed using the R package “MendelianRandomization” (available at https://CRAN.R-project.org/package=MendelianRandomization) and the significance threshold was 0.002 (0.05/32 BALs).
In order to put our findings in a more clinical context, we also performed Spearman correlation analyses among PA-associated BALs (with annotations) with clinical factors such as body mass index (BMI), hs-CRP, and standard lipids. Additionally, the correlation of all significant BALs in VITAL-CVD and JUPITER-CVD with the same clinical variables was also examined. We used the Jennrich test 31 for the equality of two correlation matrices.
Finally, in addition to quantifying the associations of PA with BALs, we examined the extent to which baseline BMI may be a mediator in the associations of BALs with PA or incident CVD. Therefore, we performed mediation analysis using the method proposed by Valeri and VanderWeele 32, controlling for the same covariates used in model 2. Using the same method, we further explored the role of BALs as mediators in the PA- CVD relationship in VITAL-CVD.
Statistical analyses were performed using R software version 3.6.0 R (R Foundation for Statistical Computing, Vienna, Austria. URL: https://www.R-project.org/) and PLINK.


