Abstract
Proteins of the Wiskott-Aldrich syndrome protein (WASP) family play a central role in regulating actin cytoskeletal dynamics in a wide range of cellular processes. Genetic mutations or misregulation of these proteins are tightly associated with many diseases. The WASP-family proteins act by transmitting various upstream signals to their conserved WH2-Central-Acidic (WCA) peptide sequence at the C-terminus, which in turn binds to the Arp2/3 complex to stimulate the formation of branched actin networks at membranes. Despite this common feature, the regulatory mechanisms and cellular functions of distinct WASP-family proteins are very different. Here, we summarize and clarify our current understanding of WASP-family proteins and how disruption of their functions is related to human disease.
Introduction
Dynamic rearrangements of the actin cytoskeleton are essential to all eukaryotic organisms. Polymerization of actin provides locomotive forces to drive fundamental processes that involve membrane deformation, such as cell migration, neuron growth, and vesicle trafficking (Blanchoin et al., 2014; Luo, 2002; Pollard and Cooper, 2009). The rate limiting step of actin polymerization is the formation of a nucleus formed by several actin monomers, which creates a barbed end that can spontaneously grow a new actin filament. Efficient actin polymerization in the cell requires a regulatory factor to facilitate the nucleation process (Pollard, 2016). One central factor that promotes actin nucleation is the Arp2/3 complex (actin-related-protein 2/3 complex), which acts by binding to the side of an existing actin filament to generate a new, branched filament (Blanchoin et al., 2000; Machesky et al., 1994). Basally inactive, the Arp2/3 complex needs to be activated by proteins collectively known as nucleation promoting factors, or NPFs, through direct protein-protein interaction (Goley and Welch, 2006; Higgs and Pollard, 2001; Rotty et al., 2013). Proteins of the Wiskott-Aldrich syndrome protein (WASP) family are major NPFs, playing an essential role in directing a large diversity of upstream signals to Arp2/3-mediated actin polymerization in many important processes throughout the cell (Figure 1).
Figure 1. WASP-family protein domain structure and cellular function.
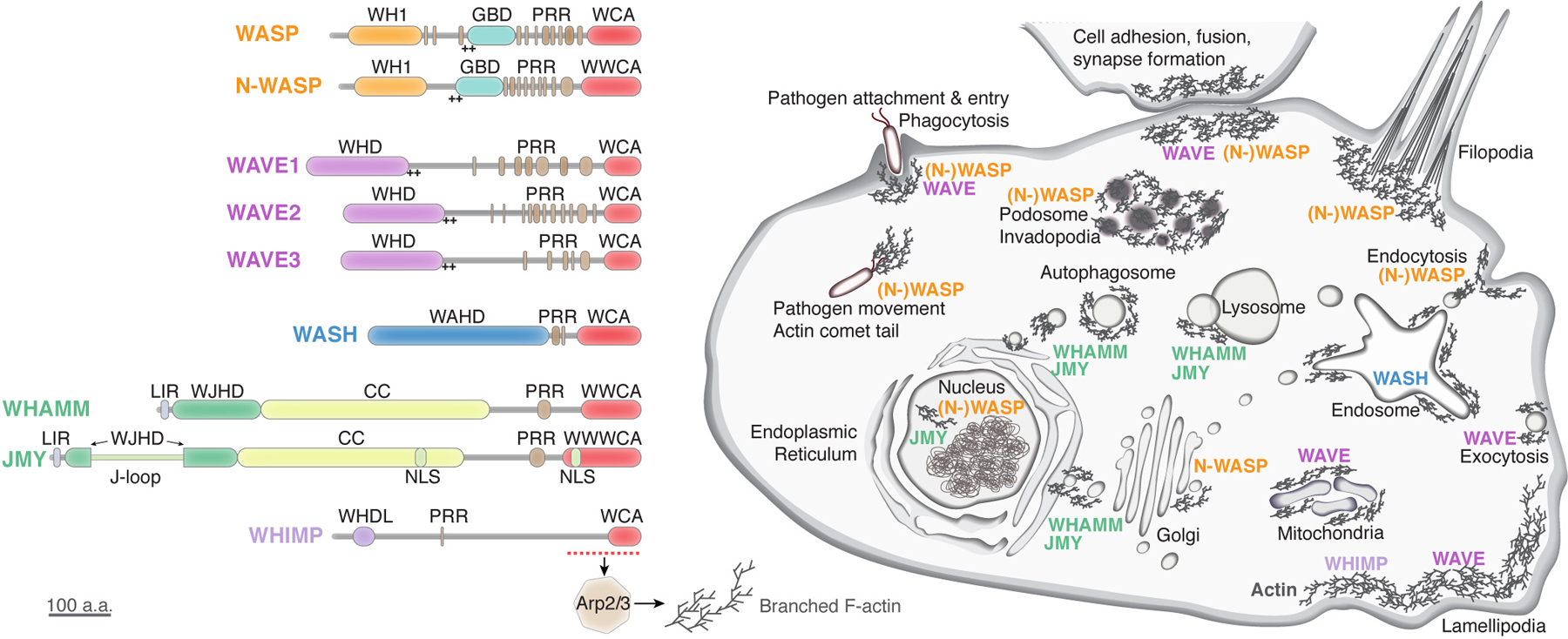
Schematic showing domain organization of different WASP-family proteins found in mammals (left) and their main functions and localizations in the cell (right). Domain structures are drawn to scale. Based on the structural homology from AlphaFold 2 prediction shown in Figure 5, we re-define WHAMM and JMY N-terminal domain as WJHD (WHAMM and JMY homology domain). WH1: WASP homology 1; GBD: GTPase binding domain; PRR: proline-rich region; WCA: WH2-central-acidic domain; WHD: WAVE homology domain; WAHD: WASH homology domain; LIR: LC3-interacting region; WJHD: WHAMM and JMY homology domain; J-loop: JMY-specific loop; CC: coiled coil domain; NLS: nuclear localization signal; WHDL: WAVE homology domain-like. “++” indicates positively charged sequence.
WASP-family proteins in mammals include nine different members, falling into five different groups based on their sequence similarity: WASP and N-WASP (neuronal-WASP); WAVE1, WAVE2, and WAVE3 (Wiskott Aldrich syndrome protein and verprolin homologue, also known as SCAR for suppressor of cAMP receptor); WASH (Wiskott-Aldrich syndrome protein and SCAR homologue); WHAMM (WASP homolog-associated protein with actin, membranes and microtubules) and JMY (junction-mediating and -regulatory protein); and the recently discovered WHIMP (WAVE homology in membrane protrusions) (Alekhina et al., 2017; Campellone et al., 2008b; Derry et al., 1994; Kabrawala et al., 2020; Kollmar et al., 2012; Linardopoulou et al., 2007; Miki et al., 1998a, 1996; Shikama et al., 1999; Suetsugu et al., 1999; Zuchero et al., 2012). The feature that defines all WASP family proteins is their conserved C-terminal WCA (WASP homology 2 (WH2), central, acidic) sequence, which is able to bind to and activate the Arp2/3 complex (Figure 1, left) (Higgs et al., 1999; Machesky and Insall, 1998). The WH2 motif—sometimes multiple WH2 motifs in a row—binds to actin monomers and is necessary for recruiting actin to the Arp2/3 complex. The WH2 motif is also found in a variety of other actin regulators, such as Spire, Cobl, WIP, and VopL/VopF (effector proteins from bacterial pathogen Vibrio) (Husson et al., 2011; Paunola et al., 2002; Pernier et al., 2013; Quinlan et al., 2005; Vaduva et al., 1999). The CA sequence in the WCA is responsible for binding Arp2/3. Two individual CA sequences can bind to two distinct locations on Arp2/3, which induces conformational changes necessary for initiating actin polymerization (Marchand et al., 2001; Padrick et al., 2011, 2008; Padrick and Rosen, 2010; Panchal et al., 2003; Shaaban et al., 2020; Zimmet et al., 2020). Except this common WCA sequence, WASP-family proteins contain distinct N-terminal domains and linker sequences, which dictate their differences in activity regulation, membrane localization, ligand interaction, and cellular functions (Figure 1). In this review, we summarize what is known of the structure and regulation of each WASP-family protein and the different ways in which they are associated with disease. We also provide analysis showing WHAMM and JMY are homologous proteins and may share a similar mechanism for regulation.
WASP and N-WASP
WASP and N-WASP are the founding members of the WASP protein family and are the best understood compared to other WASP-family proteins (Imai et al., 2003; Snapper and Rosen, 1999; Thrasher and Burns, 2010). Here, we discuss them together as the two proteins share similar domain architectures, regulatory mechanisms, and cellular functions. One major distinction between the two proteins is that WASP is specifically expressed in hematopoietic cells and N-WASP is ubiquitously expressed in human tissues (Miki et al., 1996; Uhlén et al., 2015). Therefore, mutations in WASP usually have a profound impact on the immune system, while N-WASP is involved in broader physiological processes.
WASP was first discovered in 1994 as the gene product responsible for Wiskott-Aldrich syndrome (WAS), an X-linked immune disorder affecting roughly 1 in 300000 males and characterized by the “trifecta” of eczema, thrombocytopenia, immune deficiencies, and chronic, recurring infections (Derry et al., 1994; Massaad et al., 2013; Notarangelo et al., 2008; Orange et al., 2002). N-WASP was soon discovered in 1996 as a Grb2-binding protein from brain lysate, which shared ~50% sequence homology with WASP (Miki et al., 1996). Even before the discovery of WASP, the connection between WAS and the actin cytoskeleton was noticed from anomalies associated with the actin cytoskeleton in the white blood cells of WAS patients, including reduced microvilli structure on neutrophils, defects in the actin cytoskeleton of T-cells and platelets, and reduced actin polymerization when treated with antigen (Facchetti et al., 1998; Gallego et al., 1997; Kenney et al., 1986; Molina et al., 1992). WASP was soon found to bind the Rho-family GTPase Cdc42 and use its C-terminal 59 amino acids (now known as the WCA sequence) to promote actin polymerization through the Arp2/3 complex in cells (Marchand et al., 2001; Miki and Takenawa, 1998; Panchal et al., 2003; Symons et al., 1996). Since then, a plethora of biochemical, structural, and cellular work over the past ~25 years has revealed how the activity of WASP and N-WASP is finely regulated by several layers of control.
WASP and N-WASP contain three domains, an N-terminal WASP homology 1 (WH1) domain, a central GTPase binding domain (GBD), and the C-terminal WCA domain (with N-WASP containing an additional W element, herein denoted as WWCA). Connecting these domains are long, unstructured sequences with important regulatory roles, including the proline-rich region (PRR) between GBD and WCA, and the polybasic sequence next to the GBD. WASP/N-WASP are autoinhibited in the basal state, and Cdc42 plays a central role in relieving the inhibition and activating WASP/N-WASP towards the Arp2/3 complex (Abdul-Manan et al., 1999; Higgs and Pollard, 2000; Kim et al., 2000; Rohatgi R. et al., 1999). In the autoinhibited state, the C-helix in WCA is associated with the GBD, which harbors the Cdc42/Rac Interacting Binding (CRIB) motif. This conformation keeps WCA sequestered from accessing Arp2/3 (Figure 2A, left). Cdc42 binding to the CRIB motif leads to a dramatic structural rearrangement in GBD, which both eliminates the inhibited conformation and occludes the surface that is required for sequestering WCA. Thus, Cdc42 uses a direct competition mechanism to release WCA from GBD, which in turn can bind Arp2/3 to trigger actin polymerization (Figure 2A, middle) (Prehoda et al., 2000; Rohatgi et al., 2000).
Figure 2. WASP and N-WASP.
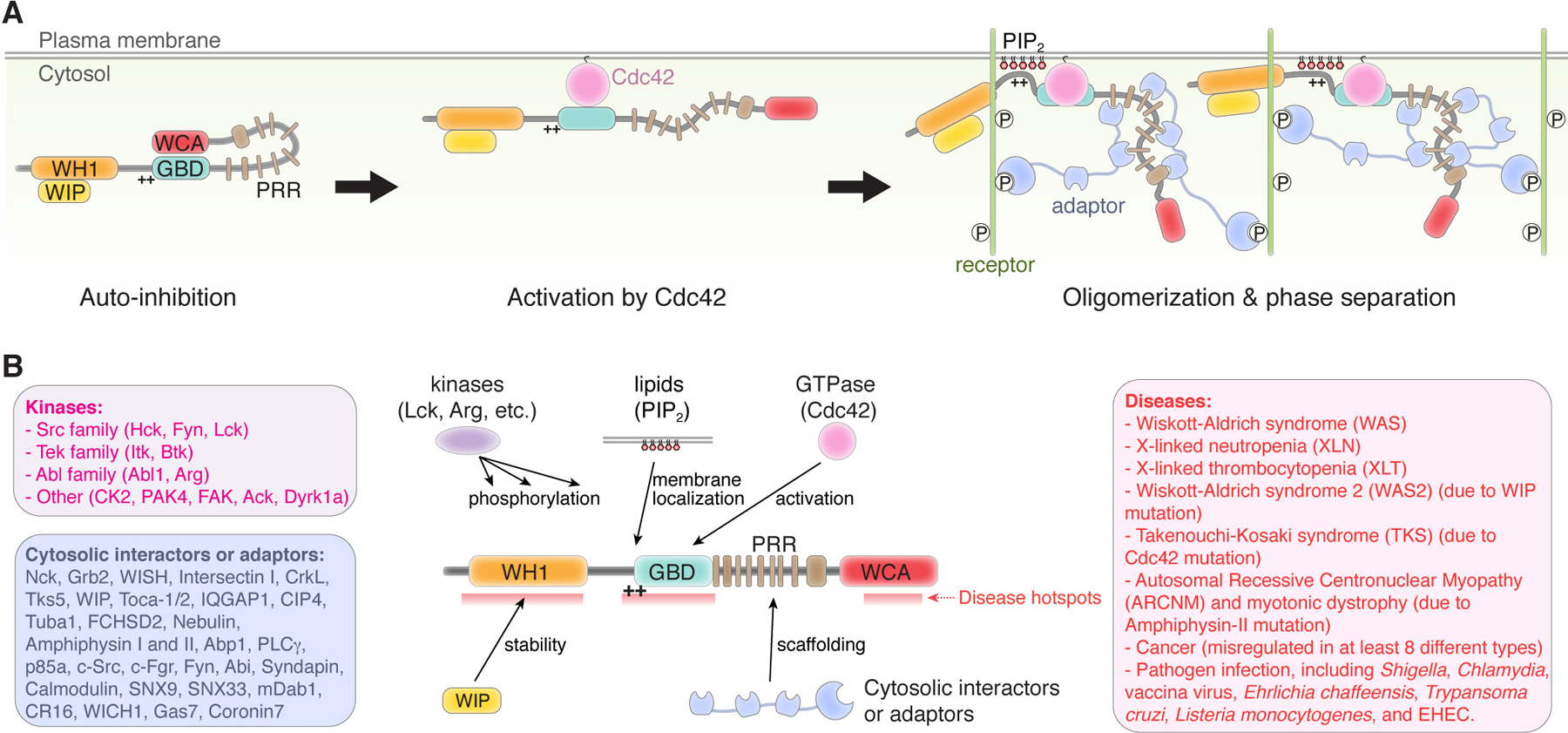
(A) Major mechanisms underlying WASP and N-WASP auto-inhibition, activation, membrane localization, and oligomerization. (B) Schematic showing how different regulatory ligands interact with WASP and N-WASP. Text boxes show representative ligands in indicated category and diseases caused by or associated with WASP and N-WASP. Hotspots in WASP where most missense mutations in patients are clustered are indicated.
Many other mechanisms often act cooperatively with Cdc42 to promote WASP/N-WASP activation. These include phosphorylation, acidic phospholipid binding, and adaptor protein binding (Figure 2). Phosphorylation of WASP by various families of kinases, including Src (Hck, Fyn, and Lck), Tek (Itk and Btk, CK2, PAK4, FAK, Ack), Abl (Abl1 and Arg), and DYRK (Dyrk1a), is clustered in the WH1, GBD, PPR, and WCA regions (Banin et al., 1996; Bunnell et al., 1996; Cory et al., 2003, 2002; Finan et al., 1996; Guinamard et al., 1998; Hornbeck et al., 2015; Labno et al., 2003; Miller et al., 2010; Park et al., 2012; Suetsugu et al., 2002; Wu et al., 2004; Yokoyama et al., 2005; Zhao et al., 2017). Phosphorylation in the WH1 domain (e.g., Y102 in WASP) can disrupt WIP-WASP interaction to destabilize WASP and reduce WASP protein level and activity. By comparison, phosphorylation in the GBD (e.g., Y291 in WASP) and the WCA (e.g., S483, S501 in WASP) typically enhances WASP activity, either by relieving the GBD-WCA auto-inhibition or by increasing the affinity of WCA to Arp2/3 (Badour et al., 2004; Cory et al., 2002; Guinamard et al., 1998; Hornbeck et al., 2015; Thrasher & Burns, 2010). It is worth noting that phosphorylated Y291 in WASP can in turn provide a docking site for SH2 domain-containing proteins such as Src-family kinases, which can further stabilize the activated conformation by sterically blocking WCA binding (Torres and Rosen, 2003). Inositol phospholipids (e.g., PIP2) interact with the polybasic sequences near the GBD domain, which can facilitate WASP/N-WASP activation likely through two distinct mechanisms simultaneously: 1) increasing membrane localization and local protein concentration of WASP/N-WASP and 2) influencing GBD-WCA stability and shifting it from the autoinhibited to activated conformation (Higgs and Pollard, 2000; Papayannopoulos et al., 2005).
Various multiple-domain scaffolding proteins, including WASP Interaction Protein (WIP), Toca-1/2, NCK, IQGAP1, Grb2, CIP4, Intersectin, Tuba, FCHSD2, Nebulin, and Amphiphysins I and II provide another important layer of control over the localization and organization of WASP/N-WASP at cell membranes (Figure 2) (Almeida-Souza et al., 2018; Bu et al., 2009; Carlier et al., 2000; Falcone et al., 2014; Ho et al., 2004; Hussain et al., 2001; Le Clainche et al., 2007; Ramesh et al., 1997; Rohatgi et al., 2001; Salazar et al., 2003; Takano et al., 2010; Yamada et al., 2009). Among them, WIP, and possibly its homologues CR16 and WICH, plays a unique role. It forms a stable complex with the WH1 domain of WASP/N-WASP in the cell (Antón et al., 2007; Ho et al., 2001; Kato et al., 2002). This interaction does not directly contribute to WASP/N-WASP activation but plays an essential role in protecting WASP/N-WASP from degradation. WIP can also link WASP/N-WASP to other adaptor proteins, such as CrkL and Zap70, which are key adaptors for recruiting WASP to the immunological synapse (Figure 2) (Sasahara et al., 2002; Volkman et al., 2004; Zettl and Way, 2002).
Many adaptor proteins, such as Nck, Grb2, WISH, Intersectin I, CrkL, and Tks5, contain multiple SH3 domains, and they can regulate WASP/N-WASP activity by simultaneously binding many SH3-binding motifs located in the PRR of WASP/N-WASP (Figure 2A, right) (Fukuoka et al., 2001; Hussain et al., 2001; Oda et al., 2001; Oikawa et al., 2008; Ramesh et al., 1997; Rivero-Lezcano et al., 1995; She et al., 1997). The multivalent interactions between SH3-containing proteins and the PRR of WASP/N-WASP promote protein oligomerization, which can concomitantly trigger liquid-liquid phase separation (LLPS). Through LLPS, relevant signaling molecules are quickly confined and organized in condensed protein droplets, which can produce a sharp increase in stimulating Arp2/3-mediated actin polymerization (Banjade and Rosen, 2014; Li et al., 2012). In addition to their ability to promote LLPS, these SH3-containing adaptors often harbor other protein-protein interaction domains, which can connect WASP/N-WASP to upstream signaling molecules through additional multivalent interactions. One of the most commonly seen domains is the SH2 domain, which binds to phosphorylated tyrosine residues in the intracellular domains (ICD) of membrane proteins, such as nephrin (an adhesin molecule in kidney podocytes) and LAT (a membrane protein important to T-cell receptor, or TCR, signaling). These ICDs usually contain multiple tyrosine-phosphorylation sites, which can be quickly phosphorylated/dephosphorylated in response to kinase signaling, allowing for docking multiple SH2-containing adaptors in a highly tunable manner. These additional multivalent interactions, plus the clustering of membrane proteins, are important not only for further enhancing the nonlinear LLPS behavior of WASP/N-WASP on the surface of lipid bilayers, but also for linking specific signaling events on the membrane to WASP/N-WASP-mediated actin polymerization. Together, the above mechanisms provide highly precise and tunable spatiotemporal control of actin polymerization at the plasma membrane to support a particular process, such as TCR signaling and kidney podocyte foot process formation. Such adaptor protein-mediated signaling has now emerged as a common theme for the regulation of WASP and N-WASP activity in the cell (Figure 2A, right) (Banani et al., 2017; Banjade et al., 2015; Case et al., 2019b, 2019a; Ditlev et al., 2019; Kim et al., 2019).
WASP expression is limited to hematopoietic cells, and WASP-mediated actin polymerization plays a key role in T-cell receptor signaling in the formation of the immunological synapse between T cells or natural killer (NK) cells and antigen presenting cells (APC) (Blundell et al., 2010; Burns et al., 2004; Calvez et al., 2011; Dupré et al., 2002). This explains the profound impact of WASP mutations on the immune system (Massaad et al., 2013). Disruption of WASP activity impairs immunological synapse formation, which will reduce infection clearance and antibody production (De Meester et al., 2010; Gismondi et al., 2004; Orange et al., 2002). WASP is also important for cell motility, intercellular trafficking, phagocytosis, chemotaxis of macrophages, and the ability of NK cells to screen and clear infected or malignant cells, which can contribute to lymphoreticular tumors and leukemia malignancies (Burns et al., 2004; Derry et al., 1994; Menotti et al., 2019; Murga-Zamalloa et al., 2017; Snapper et al., 2005). In addition, WASP (with N-WASP and WAVE as well) plays a role in autoimmunity, where it dampens B-cell signaling by triggering actin polymerization to prevent B-cell receptor (BCR) clustering and by stimulating the removal of activated BCR from the membrane through endocytosis (Liu et al., 2013; Massaad et al., 2013; Recher et al., 2012; Volpi et al., 2016; Westerberg et al., 2012). Besides its function in the cytosol, WASP also plays an important role in promoting Arp2/3-mediated actin polymerization in the nucleus, which is required for driving double-strand breaks into discrete sub-nuclear clusters to facilitate homology-directed DNA repair (Schrank et al., 2018).
Mutations in WASP are the cause of Wiskott-Aldrich syndrome and related syndromes, including X-linked thrombocytopenia (XLT) and X-linked neutropenia (XLN). Since the discovery of WAS, sequences from over 400 different patients have been studied, leading to a wealth of data on mutations in WASP that can cause disease (reviewed in Jin et al., 2004 and compiled online at http://pidj.rcai.riken.jp/waspbase/). The mutations include nonsense, frameshift, and splicing site mutations, as well as over 50 individual missense mutations across the entire sequence. While nonsense and frameshift mutations diminish the production of functional full-length protein, missense mutations can have effects ranging from protein level to regulation (Kolluri et al., 1995; MacCarthy-Morrogh et al., 1998). In general, mutations that result in the loss of WASP protein cause more severe symptoms than those that leave WASP protein levels reduced or unaffected (Imai et al., 2003).
The missense mutations found in human patients are primarily localized to the WH1 domain (with some scattered in the GBD and WCA region, as indicated by “hotspots” in Figure 2B), which is believed to impair the interaction between WASP and WIP. Consequently, this leads to the degradation of WASP and disrupts its localization at the immunological synapse (Antón et al., 2007; Jin et al., 2004; Volkman et al., 2004). By comparison, missense mutations in the GBD or the WCA domain may affect the inhibition or activation of WASP and its ability to promote Arp2/3 activation. While most mutations impair WASP activity, several mutations have an opposite, stimulatory effect. For example, a mutation in the GBD, L270P, which was found in patients suffering from X-linked Severe Congenital Neutropenia (XLN), was shown to disrupt the autoinhibition of the GBD-WCA interaction to produce a constitutively active WASP (Devriendt et al., 2001). The different, albeit similarly pathogenic, effects of disruptive vs. stimulatory mutations speak for the importance for the cell to have a precise control of WASP activity level. In addition to diseases caused by genetic mutations in WASP, mutations in WASP regulators like WIP and Cdc42 are also often associated with diseases closely related to WAS. For example, nonsense mutations in WIP cause Wiskott-Aldrich syndrome 2 (WAS2), likely due to the loss of WIP expression and the subsequent destabilization of WASP (Schwinger et al., 2018). Many missense mutations in Cdc42 lead to Takenouchi-Kosaki syndrome (TKS), which is a complex congenital developmental disorder affecting multiple organ systems, including the psychomotor, cardiac, and hematologic/lymphatic system, as well as recurrent infections (Martinelli et al., 2018).
The only effective therapy for WAS to date is hematopoietic stem cell transplantation from non-affected donors (Burroughs et al., 2020; Massaad et al., 2013). Wiskostatin, a small-molecule inhibitor of WASP that binds to the GBD to stabilize the autoinhibited conformation, has a potential for treating conditions caused by hyperactive WASP, but there is still a long way to go before its clinical application (Guerriero and Weisz, 2007; Peterson et al., 2004).
By comparison, N-WASP is ubiquitously expressed and plays a wide variety of roles in physiology, even though no genetic mutations have been reported to directly link N-WASP to a particular disease. On the cellular level, N-WASP has an essential role in endocytosis, where its activity is required for assembling actin filaments at clathrin-coated pits, which is important for pinching off the pits and propelling the internalized vesicle from the plasma membrane into the cytosol (Benesch et al., 2005; Hussain et al., 2001; Innocenti et al., 2005; Qualmann et al., 1999; Salazar et al., 2003; Shin et al., 2008; Taunton et al., 2000; Yamada et al., 2009; Zhang et al., 2009). Moreover, N-WASP is important to a variety of cellular structures, including the formation of filopodia (finger-like protrusions at the leading edge of migrating cells or growing neurons), podosomes, invadopodia, tight junctions, and the maintenance of Golgi morphology – including anterograde trafficking from the trans-Golgi network (TGN) to the endoplasmic reticulum (ER) and dispersal of the Golgi network after DNA damage (Figure 1) (Bhattacharya et al., 2016; Ivanov et al., 2005; Kovacs et al., 2011; Linder et al., 1999; Luna et al., 2002; Mizutani et al., 2002; Taunton et al., 2000; Tsuboi et al., 2006; Wen et al., 2020; Yamaguchi et al., 2005). Similar to WASP, N-WASP also plays an important role in the nucleus, where it can interact with the PSF-NonO complex and promote nuclear actin polymerization to regulate RNA polymerase II transcription activity (Wu et al., 2006).
On the tissue level, N-WASP is required for the proper differentiation and/or development of many different cell types, including hematopoietic cells, fibroblasts, muscle cells, and neurons. In hematopoietic cells, N-WASP function is partially redundant with WASP in the control of immunological synapse development, B-cell development and signaling, antigen uptake, and chemotaxis (Jain and Thanabalu, 2015; Liu et al., 2013; Westerberg et al., 2012). Fibroblasts lacking N-WASP expression fail to form the hallmark actin fibers and show reduced contractility, adhesion, and spreading (Cai et al., 2012; Misra et al., 2007). N-WASP is also required for the function and development of muscle cells, in which N-WASP is recruited to Z disks of myofibrils to induce actin polymerization through the PRR-SH3 interaction with the 900-kDa actin-binding protein Nebulin (Takano et al., 2010). N-WASP is also important to the proper positioning of nuclei in muscle cells through the PRR-SH3 interaction with Amphiphysin-II (Falcone et al., 2014). Mutations in Amphiphysin-II that have been linked to the muscle disorders Autosomal Recessive Centronuclear Myopathy (ARCNM) and myotonic dystrophy are found in the SH3 domain and likely act by disrupting its interaction with N-WASP, which affected both the localization and protein levels of N-WASP in muscle cells (Falcone et al., 2014). In neurons, N-WASP participates in various aspects of neuron development, including neurite outgrowth, formation of dendritic spines and synapses, and myelination (Irie and Yamaguchi, 2002; Katanov et al., 2020; Pinyol et al., 2007; Shekarabi et al., 2005; Suetsugu et al., 2004, 2002; Wegner et al., 2008; You and Lin-Chao, 2010). Elevated levels of N-WASP have been found in the brains of patients with Alzheimer’s and intractable epilepsy, both diseases characterized by aberrant sprouting of neurites (Kitamura et al., 2003; Xiao et al., 2008).
As a major driver of actin polymerization, N-WASP levels are often mis-regulated in cancer cells (reviewed in Biber et al., 2020). Changed expression levels of N-WASP are correlated with reduced survival in several cancer types, including pancreatic ductal adenocarcinoma (PDAC), hepatocellular carcinoma (HCC), renal cell carcinoma, nasopharyngeal carcinoma, and lung, breast, cervical, and gastric cancers (Frugtniet et al., 2017; Guo et al., 2014; Hou et al., 2017; Jin et al., 2013; Martin et al., 2012, 2008; Sanchez et al., 2010; Wang et al., 2010; Yanagawa et al., 2001; Yang et al., 2020; Yu et al., 2012). The roles of N-WASP in cancer progression vary depending on the cancer type, but there are several common factors. One of the most commonly accepted factors is the role of N-WASP in the formation and maintenance of invadopodia, filopodia-like structures that extend into the extracellular matrix (ECM) to promote ECM degradation by matrix metalloproteinases (MMPs), which in turn promotes cancer cell migration, invasion, and metastasis (DesMarais et al., 2009; Gligorijevic et al., 2012; Lorenz et al., 2004; Martin et al., 2008; Mizutani et al., 2002; Oser et al., 2009; Sanchez et al., 2010; Sarmiento et al., 2008; Shortrede et al., 2016; Yamaguchi et al., 2005). In parallel, N-WASP also promotes the delivery of MMPs to and maintenance of MMP levels at the tip of invadopodia (Yu et al., 2012). While reduced N-WASP levels could be ameliorated by gene therapy or retrovirus-mediated expression of N-WASP in cancer cells, overactive protein could be targeted using the inhibitor wiskostatin and its derivatives, as well as nanobodies raised against the N-WASP WCA (Hebbrecht et al., 2017; Peterson et al., 2004).
It is remarkable that many pathogens have co-opted the N-WASP-mediated signaling to facilitate their infection, either by driving their attachment to or entry into target cells or by facilitating their intercellular movement (Snapper et al., 2001). The intracellular bacterial pathogen Shigella flexneri, which is the main cause of dysentery, utilizes its effector protein IcsA (or VirG) to recruit N-WASP to the bacterial surface, where IcsA binds to the N-WASP GBD to competitively release WCA and drive actin-based motility throughout the host cytoplasm (Egile et al., 1999; Mauricio et al., 2017; Suzuki et al., 1998). Similarly, Chlamydia trachomatis, a common sexually transmitted infection, uses its effector protein TmeA to bind the N-WASP GBD, which is important for recruiting N-WASP to the site of invasion (Faris et al., 2020; Keb et al., 2021). The vaccinia virus uses its membrane protein A36R to recruit Nck, WIP, and N-WASP to drive actin-based motility (Frischknecht et al., 1999; Moreau et al., 2000). Other intracellular pathogens, such as the bacteria Ehrlichia chaffeensis (causative agent of the life-threatening tick-borne disease monocytic ehrlichiosis) and the protozoan parasite Trypansoma cruzi (causative agent of Chagas disease, which affects 6–7 million people worldwide) recruit N-WASP (sometimes WAVE as well) to the sites of invasion, but the molecular mechanisms remain unknown (Bonfim-Melo et al., 2018; Kumar et al., 2015). The parasite Listeria monocytogenes, a leading food borne pathogen causing meningitis and death in pregnant women, infants, and the elderly, is known to use ActA to coopt the host cell actin machinery to propel itself within host cytosol. In order to spread between cells, the protein IniC is secreted and binds to the SH3 domain of the scaffolding protein Tuba. This prevents Tuba from interacting with N-WASP, leading to weakened tight junctions and allowing for easier passage between cells for the bacteria (Cossart and Bierne, 2001; Rajabian et al., 2009). Furthermore, the extracellular pathogen, enterohaemorrhagic Escherichia coli (EHEC), a major cause of hemorrhagic colitis and pediatric kidney failure in developed countries, uses its effector protein EspFU injected into the host epithelial cells to activate N-WASP, which locally stimulates the formation of actin pedestals, an actin-rich membrane protrusion underneath the EHEC attachment site (Campellone et al., 2004; Hartland and Leong, 2013; Kalman et al., 1999; Lommel et al., 2004, 2001). EspFU promotes N-WASP activation through at least two mechanisms in parallel: 1) it binds to GBD of WASP with high affinity, which directly competes off WCA to drive WASP activation, and 2) it uses multiple repeats of this GBD binding sequence to achieve high efficiency, likely through clustering WASP at membranes (Campellone et al., 2008a; Cheng et al., 2008; Sallee et al., 2008).
In summary, WASP and N-WASP play important roles in many fundamental processes. Even though studies in the past two decades have revealed many important mechanisms underlying the regulation and function of WASP and N-WASP on the molecular, cellular, and organismal levels, new mechanisms and functions are still emerging, and many questions remain to be answered.
WAVE and WAVE regulatory complex (WRC)
The second group of WASP-family proteins, WAVE, contains three orthologs in vertebrates: WAVE1, WAVE2, and WAVE3. The three proteins differ mainly in their middle, unstructured regions encompassing the PRR sequence, while their N-terminal and C-terminal structured regions share ~80% sequence homology (Figure 1, left) (Suetsugu et al., 1999). One other major difference between the three proteins is their expression pattern, with WAVE1 and WAVE3 enriched in the brain and WAVE2 widely expressed in all tissues and particularly in peripheral blood leukocytes (Suetsugu et al., 1999; Uhlén et al., 2015).
WAVE1 was discovered as a WASP-related protein by multiple laboratories at nearly the same time in 1998. One approach identified it as a suppressor of cAMP receptor signaling in Dictyostelium, which led to its first name SCAR (suppressor of cAMP receptor) and the initial indication of its human homologs through a sequence database search (Bear et al., 1998). In the meantime, two other approaches, one searching for Arp2/3-interaction partners through a yeast-two-hybrid assay and the other searching for novel WH2-containing proteins in the human sequence database, identified the human homolog SCAR1 and introduced the name WAVE (WASP-family Verprolin-homologous protein) (Machesky and Insall, 1998; Miki et al., 1998b). Shortly after that, WAVE2 and WAVE3 were cloned and shown to have similar properties in driving Arp2/3-mediated actin polymerization (Suetsugu et al., 1999). Since then, the name WAVE has been widely used in vertebrates, while SCAR is used more often in invertebrates and plants. For clarity, we herein use WAVE to refer to all WAVE orthologs, unless an isoform is specifically mentioned.
Similar to WASP/N-WASP acting downstream of the Rho-GTPase Cdc42, WAVE acts downstream of another Rho-GTPase, Rac1 (Miki et al., 1998b). Distinct from WASP/N-WASP, however, WAVE did not contain a recognizable GTPase binding domain, nor could it directly bind Rac1 (Miki et al., 1998b). Furthermore, the recombinantly purified WAVE had high basal activity towards the Arp2/3 complex, suggesting it is not autoinhibited as a standalone protein as WASP/N-WASP are (Innocenti et al., 2004). Instead, the biochemical purification of WAVE from various animal tissues or cultured cells and subsequent recombinant reconstitution studies established that WAVE was constitutively incorporated in a large complex of ~400 kDa that consisted of five different protein subunits: Sra1 (Cyfip1, or its ortholog Pir121/Cyfip2), Nap1 (Hem2, or its ortholog Hem1), Abi2 (or its orthologs Abi1 and Abi3/Nesh), HSPC300 (BRICK1), and WAVE1 (SCAR1, or its orthologs WAVE2 and WAVE3) (Derivery et al., 2009a; Eden et al., 2002; Gautreau et al., 2004; Innocenti et al., 2004; Ismail et al., 2009; Lebensohn and Kirschner, 2009). This complex was thereafter named the WAVE Regulatory Complex (WRC or WAVE complex) (Figure 3). The identification of the WRC explained the previously observed links between Rac1 and WRC components and their importance in the formation of lamellipodia, thin sheet-like membrane ruffling often found at the leading edge of migrating cells (Kobayashi et al., 1998; Nobes and Hall, 1995; Ridley et al., 1992; Rogers et al., 2003; Scita et al., 1999). These studies also reconciled the original debates about WRC activity regulation, establishing that the WRC is basally inhibited in the cytosol and is recruited to and activated at the plasma membrane by various ligands to promote Arp2/3-mediated actin polymerization (Figure 3A) (Kurisu and Takenawa, 2009; Rottner et al., 2021).
Figure 3. WAVE regulatory complex (WRC).
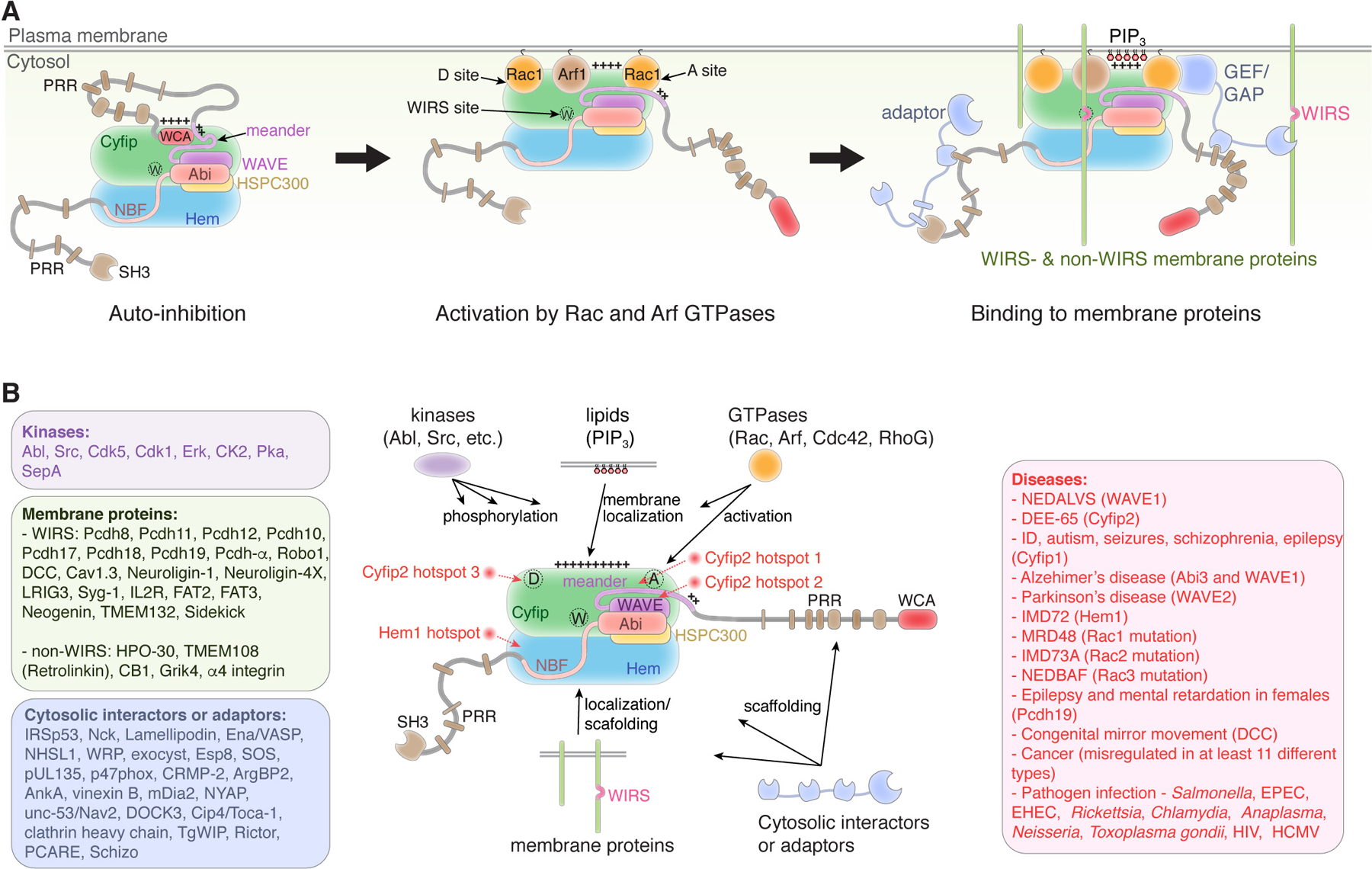
(A) Schematic showing mechanisms by which the WRC keeps WAVE auto-inhibited in the basal state, becomes activated by GTPase binding, and translocated to the membrane by directly interacting with membrane proteins and acidic phospholipids. “++++” indicates the positively charged side of the WRC. WAVE for WAVE1/2/3, Abi for Abi1/2/3, Hem for Hem1/Nap2, Cyfip for Sra1/Cyfip2. NBF: Nap1 binding fragment. (B) Schematic showing how different regulatory ligands interact with the WRC. Text boxes show representative ligands in indicated category and diseases caused by or associated with WRC subunits. Hotspots in the WRC where most missense mutations in patients are clustered are indicated. Ligands that bind to individual subunits of the WRC, but do not bind to the fully assembled WRC are not listed, such as N-WASP, FMRP, and eIF4E.
Biochemical and structural studies have now elucidated the mechanism underlying the assembly and autoinhibition of the WRC and have started to explain how the WRC interacts with various ligands to promote activation and membrane recruitment (Rottner et al., 2021). The WRC can be viewed as an assembly of a large, elongated dimer formed by Sra1 and Nap1, and a smaller trimer formed by WAVE1, Abi2, and HSPC300. Sra1 and Nap1 are predominately alpha helical, whereas the N-termini of WAVE1 (i.e., part of its WHD domain), Abi2, and HSPC300 form a four-helix bundle aligning along the long axis of the Sra1-Nap1 dimer (Figure 3). Following the helical bundle, WAVE1 and Abi2 extend long tails that have important regulatory roles. The long tail of WAVE1 consists of three parts: a “meander” region of ~90 amino acids (a.a.), which “meanders” across the surface of Sra1 and is critical for the inhibition and activation of the WRC; a long, unstructured sequence of ~300 a.a. that contains multiple PRRs; and the C-terminal WCA region of ~75 a.a. The long tail of Abi2 also consists of three parts: a sequence of ~ 40 a.a. named the Nap1 binding fragment (NBF), which “crawls” on the surface of Nap1 and is critical for WRC assembly; a long, unstructured sequence of ~250 a.a. that contains multiple PRRs; and a C-terminal SH3 domain. The WRC keeps WAVE1 inhibited in trans by sequestering the W and C elements of WCA to a conserved surface formed by both Sra1 and the meander region of WAVE1 (B. Chen et al., 2014a; Chen et al., 2010a). This is distinct from WASP/N-WASP, in which the WCA is sequestered in cis by binding to the GBD in the same polypeptide chain. It is believed other WRC assemblies, such as WRCs containing Cyfip2, Hem1, or WAVE2, are similarly assembled and autoinhibited, although their regulatory mechanisms may have some difference (X. J. Chen et al., 2014; Cook et al., 2020; Derivery et al., 2009a; Gautreau et al., 2004; Polesskaya et al., 2021).
A large number of interacting ligands of the WRC have been identified, and the list is still rapidly growing (Rottner et al., 2021). Like WASP/N-WASP, WRC directly interacts with small G proteins (Rho-family and Arf-family GTPases), inositol phospholipids (e.g., PIP3), various kinases (Abl, Src, Cdk5, Cdk1, Erk, CK2, Pka, SepA) and many cytosolic or adaptor proteins, such as IRSp53, Nck, Lamellipodin, Ena/VASP, NHSL1, and WRP (Ardern et al., 2006; X. J. Chen et al., 2014; Dai and Pendergast, 1995; Danson et al., 2007; Kim et al., 2006; Kitamura et al., 1996; Kobayashi et al., 1998; V. Koronakis et al., 2011; Law et al., 2021, 2013; Leng et al., 2005; Mendoza, 2013; Miki et al., 2000; Miyamoto et al., 2008; Nakanishi et al., 2007; Oikawa et al., 2004; Pocha and Cory, 2009; Shi et al., 2021; S. P. Singh et al., 2020; Soderling et al., 2002; Stuart et al., 2006; Ura et al., 2012; Westphal et al., 2000; Xu and Quinn, 2012; Yamashita et al., 2011). Unlike WASP/N-WASP, which are indirectly linked to membrane proteins through various cytosolic adaptors, the WRC directly interacts with many membrane proteins. Together, these interactions suggest the WRC acts as an important signaling hub, through which various membrane signals can be directly funneled down to the regulation of the actin cytoskeleton (Rottner et al., 2021).
Despite the long list of WRC ligands, most of their interaction mechanisms remain unsolved largely due to 1) technical difficulties caused by the large size and complexity of the WRC and 2) the cooperative nature and weak affinity of many ligands. Another complication is that only the fully assembled WRC exists as a stable, functional unit in the cell and as well-behaving material in biochemical studies. Recombinantly produced isolated subunits or subcomplexes of the WRC are prone to protein aggregation and could even cause “artificial” interactions through surfaces that would not be available in the assembled complex. Moreover, although the five subunits have co-existed through evolution and show a strong interdependence in cellular expression, it is likely some subunits may have evolved extra functions outside the WRC (Blagg et al., 2003; Echarri et al., 2004; Kunda et al., 2003; Rogers et al., 2003; Steffen et al., 2004; Stephan et al., 2011; Veltman and Insall, 2010). Examples include Sra1 binding to the Fragile X mental retardation protein FMRP and the translation initiation factor eIF4E, and Abi binding to N-WASP (Innocenti et al., 2005; Napoli et al., 2008; Schenck et al., 2001). Therefore, it is important to rigorously validate WRC-related interactions that were identified using isolated subunits. These interactions should be carefully evaluated based on fully assembled WRC both in vitro and in cells.
Among the WRC ligands, the Rho-GTPase Rac1 ubiquitously plays a central role in activating the WRC. Recent biochemical reconstitution and structural work has led to the identification of two separate Rac1 binding sites on the WRC, which are located on opposite ends of the elongated Sra1 subunit (Figure 3). The site adjacent to the WCA binding site was named the A site, and the site distant to WCA named the D site (Chen et al., 2017, 2010a). The two sites have ~40-fold difference in the affinity for Rac1, but both sites played a key role in activating the WRC in vitro (Chen et al., 2017). The weak binding site (i.e., A site) seemed to be more important for WRC-mediated lamellipodia formation in cells (Schaks et al., 2018). Without a high-resolution structure of the WRC bound to both sites, the exact mechanism underlying WRC activation is still unknown. Nevertheless, since both Rac1-binding sites are distinct from the WCA binding site, WRC activation must use an allosteric mechanism to destabilize the sequestered WCA, in contrast to the direct competition mechanism in WASP/N-WASP activation by Cdc42 (Figure 3A). Why the WRC contains two Rac1 binding sites is unknown, but there are two possible explanations: 1) the two sites are cooperative—Rac1 binding to D site could enhance A site binding, and 2) when Rac1 concentration is low, D site binding can “prime” the WRC on the membrane without causing activation, and only when Rac1 concentration is high can it bind to the A site and trigger activation (Chen et al., 2017). It is remarkable that a recently identified protein, Fam49/CYRI (Cyfip-related Rac1 interacting), uses a structure homologous to the A site to sequester Rac1 from activating the WRC and locally suppress actin polymerization and membrane protrusions (Fort et al., 2018).
While Rac1 is the canonical activator of the WRC, other molecules can contribute to WRC activation and membrane localization, and they often act cooperatively with Rac1 (Figure 3B). These include the Arf-family GTPases, Arf1 and Arf6, other Rho-family GTPases (Cdc42 and RhoG), different kinases, acidic phospholipids (PIP3), and various cytosolic and membrane proteins (Vassilis Koronakis et al., 2011; Rottner et al., 2021; Schaks et al., 2021; V. Singh et al., 2020). Arf1 and Arf6 were shown to act synergistically with Rac1 to promote WRC activation both in vitro and in various cellular processes (Anitei et al., 2010; Vassilis Koronakis et al., 2011). These interactions may provide additional control of the WRC and link the WRC to particular processes, such as intracellular trafficking and pathogen invasion. Exactly how Arf GTPases interact with the WRC or how Arf cooperates with Rac1 is unknown. Phosphorylation on the WRC by various kinases (e.g., Cdk5, Erk, Abl, Ck2, and Src) mainly occurs in the meander and WCA regions of WAVE, and the unstructured PRR sequences of both WAVE and Abi (Ardern et al., 2006; Danson et al., 2007; Kim et al., 2006; Mendoza, 2013; Nakanishi et al., 2007; Pocha and Cory, 2009; Sossey-Alaoui et al., 2007a). Particularly, phosphorylation in the meander sequence, including Y125 (by Src), T138 (by Cdk5), and Y151 (by Abl), has been shown to destabilize meander binding to Sra1, which in turn causes WCA release and WRC activation (Chen et al., 2010a). Phosphorylation in the WCA of WAVE and PRR regions of WAVE and Abi may influence WRC activity in the cell by tuning the interactions with PRR-binding proteins (e.g., SH3 domain-containing proteins) and Arp2/3. Similar to the importance of inositol phospholipids PIP2 to WASP/N-WASP activation, PIP3 are important to WRC activation likely through two distinct mechanisms: 1) increasing membrane localization and local protein concentration of the WRC by binding to the positively charged face of the complex, and 2) directly contributing to WCA release by interacting with the positively charged helix 6 in the meander sequence.
The long PRR sequences in WAVE and Abi are the most variable regions among different WAVE and Abi orthologs (Figure 1, left). They provide a rich environment for recruiting different adaptor proteins, such as SH3-domain or EVH1-domain containing proteins (e.g., Ena/VASP). Similarly, the SH3 domain at the C-terminus of Abi can recruit certain PRR-containing proteins, such as the Abl kinase. While these interactions may not directly contribute to WRC activation, they can play an important role in connecting various upstream signals to the WRC (Miki et al., 2000; Soderling et al., 2002; Stuart et al., 2006; Takenawa and Suetsugu, 2007). In principle, binding to multivalent adaptor proteins could induce LLPS, as is seen for WASP/N-WASP, but LLPS has not yet been reported for the WRC. It has been shown that the WRC forms high-order, wave-like assemblies on the plasma membrane, but the underlying mechanisms driving this organization are unknown (Pipathsouk et al., 2021; Weiner et al., 2007). It has been proposed that the N-terminal helix 1 of Sra1 could bind to a neighboring complex, which could potentially polymerize the WRC, but no experimental evidence has been found to confirm or deny this (Chen et al., 2010b).
In addition to interactions mediated by PRR sequences or the SH3 domain of WAVE and Abi, many other ligands either use an unidentified interaction mechanism or were found to interact with structured, non-PRR regions of the WRC. Here, we describe several ligands that link the WRC to various unique processes. The exocyst complex was shown to directly interact with the Cyfip and Abi subunits, which provides a mechanism to coordinate polarized exocytosis mediated by exocyst with cell migration mediated by the WRC (Biondini et al., 2016; Zago et al., 2018). PCARE (photoreceptor cilium actin regulator) was found to recruit the WAVE3-containing WRC to the cilia of photoreceptor cells, where the WRC played a key role in promoting the actin polymerization required for the outer segment disk formation (Corral-Serrano et al., 2020). Moreover, the neuronal protein Pancortin/Noelin was found to recruit WAVE1 to the surface of mitochondria after ischemic stroke, where the WAVE1-Pancortin complex sequestered Bcl-xL to facilitate cytochrome C release and apoptosis (Cheng et al., 2007). In leukemia cells, where Bcl2 is overexpressed to protect cells from apoptosis, WAVE1 was found to interact with Bcl2 and promote its localization to the mitochondria to enhance its anti-apoptotic activity (Kang et al., 2010). The general importance of the WRC to mitochondria is also seen in another study, in which WAVE1 was important to mitochondria positioning in neural dendrites (Sung et al., 2008).
Besides the above ligands, the WRC directly interacts with a large variety of transmembrane or membrane associated proteins. The majority of these membrane proteins (>100 in the human genome by prediction) can bind the WRC using a short peptide motif named the WRC interacting receptor sequence (WIRS) in their intracellular domain (ICD), which is defined as Φ-x-T/S-F-x-x (Φ for bulky hydrophobic residues and x for any residues) (B. Chen et al., 2014a). The WIRS proteins include various cell adhesion molecules (e.g., protocadherins, Robo1, neuroligins, and Syg-1), ion channels, and GPCRs (B. Chen et al., 2014a). This WIRS interaction is strictly conserved throughout animals, from human to sponge, and is not found in other species such as plants, suggesting the importance of WIRS-WRC interactions to processes unique to animals, like in the nervous system. Indeed, many studies have shown different neuronal receptors use the WIRS-WRC interaction to recruit the WRC to their sites of action and provide actin polymerization to support various neuronal activities, such as axon pathfinding, branching, and synapse formation (Chaudhari et al., 2021; Chia et al., 2014; Fan et al., 2018; Lee et al., 2016; Xing et al., 2018), as well as tissue morphogenesis (Lee et al., 2016; Malin et al., 2022; Squarr et al., 2016). It is remarkable that the WIRS binding pocket (indicated by the “W” site in Figure 3) is formed by both Sra and Abi subunits, highlighting the importance of fully assembled WRC to its function. Together, the diverse array of WIRS receptors provides the cell a versatile means to recruit the WRC to the membrane in response to many different upstream signals and in different cell types. In addition to serving as a membrane localization signal, WIRS proteins can also use the sequence flanking the WIRS motif to modulate—either inhibit or further promote—WRC activity (B. Chen et al., 2014a). Besides WIRS-containing proteins, membrane proteins that interact with the WRC without using a WIRS motif are emerging, such as HPO-30, Retrolinkin, and CB1, but the exactly interaction mechanisms remain to be solved, and whether there are more non-WIRS receptors for the WRC remain to be explored (Monday et al., 2020; Xu et al., 2016; Zou et al., 2018). It is intriguing that many ICDs, either directly interacting with the WRC or not, may use other protein-protein interaction motifs to recruit additional adaptor proteins, including various GEFs and GAPs for Rac1 and Arf1 (Kong et al., 2015; Lucas and Hardin, 2017; Lundström et al., 2004; Paskus et al., 2019; Stavoe and Colón-Ramos, 2012; Villanueva et al., 2021; Woolfrey et al., 2009; Yang and Bashaw, 2006). These interactions could bridge related signaling molecules to the WRC to provide more spatiotemporal control of WRC activation (Zou et al., 2018). Finally, the importance of membrane localization to WRC activity is further supported by a recent observation that the force generated by actin filament elongation could dissociate the WRC from the lamellipodia tip to decrease WRC dwell time and activity, which may provide a negative feedback loop to fine tune membrane protrusions (Mehidi et al., 2021).
The exact composition of the WRC subunits can provide an additional layer of control of WRC activity. Although in principle the WRC can be similarly assembled by various orthologs of each subunit through a “mix-and-match” process, different WRCs may have distinct interaction partners, respond to the same ligand differently, or have different activity output. For example, WAVE2-containing WRC, but not WAVE1-containing WRC, requires the adaptor protein IRSp53 for optimal activation (Miki et al., 2000; Suetsugu et al., 2006). WAVE1- and WAVE2-containing WRCs were also shown to play overlapping, but distinct roles in promoting membrane protrusions, cell migration, and actin network architecture (Suetsugu et al., 2003; Sweeney et al., 2015; Tang et al., 2020). Moreover, Cyfip2-containing WRC was shown to be less responsive to Rac1 activation than Cyfip1-containing WRC (Polesskaya et al., 2021). Similarly, Hem1-containing WRC could not be activated by Rac1 in vitro in identical conditions that could activate Nap1-containing WRC (Cook et al., 2020; Polesskaya et al., 2021). The biochemical and structural mechanisms underlying the difference between different WRC isoforms remain to be explored.
The WRC plays an essential role in many fundamental processes, including lamellipodia formation, cell migration, adhesion, and fusion (Gromnitza et al., 2018; Kim et al., 2015; Kunda et al., 2003; Nowak et al., 2009; O’Leary et al., 2017; Suetsugu et al., 2003; Yamazaki et al., 2007). The broad diversity of its ligands links the WRC to various physiological systems, particularly the nervous system and the immune system (Rottner et al., 2021; Yamazaki et al., 2003). Because of the interdependence of all five subunits in maintaining WRC integrity and function, genetic mutations disrupting any subunit can potentially disrupt WRC function and lead to disease.
The WRC, especially the WAVE1- and WAVE3-containing WRCs, are enriched in the brain and play an essential role in neural morphogenesis, axon growth, dendrite branching, synapse formation, and synaptic transmission and plasticity (Nozumi et al., 2003; Soderling et al., 2007, 2003; Sung et al., 2008). This explains the broad implication of the WRC in many types of neurological conditions (Rottner et al., 2021). Over a dozen de novo mutations in WAVE1 have been identified as the cause of a newly defined neurological syndrome named Neurodevelopmental Disorder with Absent Language and Variable Seizures (NEDALVS), which exhibits features of intellectual disability (ID), autism, and epilepsy. The mutations include different nonsense or frameshift mutations clustered in the WCA region, which would lead to a truncated WCA and loss of WAVE1 activity towards the Arp2/3 complex (Ito et al., 2018; Shimojima Yamamoto et al., 2021; Srivastava et al., 2021). Copy number variant (CNV) mutations caused by microdeletion of WAVE1-containing regions from chromosome 6q21 were also observed, which would cause haploinsufficiency of WAVE1 and reduce overall WRC activity in the brain (Srivastava et al., 2021). In addition, four cases of missense mutations (W161C/R or K172E) were identified in NEDALVS patients (Srivastava et al., 2021; Zhao et al., 2021). Located in helix 6 in the meander region, both W161 and K172 make contacts with the C helix to keep WCA sequestered. Therefore, W161C/R or K171E would lead to a constitutively active WRC, instead of loss of function as seen in other NEDALVS patients. That the two opposite effects lead to the same syndrome emphasizes the importance of precisely controlling WRC activity in vivo.
In addition to WAVE1, Cyfip1 and Cyfip2 are frequently mutated in human patients. Many mutations in Cyfip2, including CNVs caused by chromosome 5q33 microdeletion, nonsense or frameshift mutations, and over 25 different missense mutations have been identified as the cause of a neurodevelopmental disorder named Developmental and Epileptic Encephalopathy-65 (DEE-65), which involves ID, Early-Onset Epileptic Encephalopathy (EOEE), seizures, muscular hypotonia, West syndrome, eating disorders (in mouse), and altered drug addiction (in mouse) (Begemann et al., 2021; Kirkpatrick et al., 2017; Kumar et al., 2013; Zweier et al., 2019). The missense mutations are spatially clustered around three different “hotspots” in the WRC structure (Figure 3B) (Rottner et al., 2021). Hotspot 1 contains the meander and WCA binding surface and the A site. These mutations would disrupt WCA inhibition to increase WRC activity (Nakashima et al., 2018). For example, one of the frequently mutated residues, R87, plays a key structural role in stabilizing WAVE1 Y151 binding. Phosphorylation of Y151 by Abl kinase or mutating R87 or Y151 caused constitutive activation of the WRC (Chen et al., 2010a; Schaks et al., 2020; Stuart et al., 2006; Zweier et al., 2019). Hotspot 2 is buried in the WRC, where mutations would disrupt protein folding and stability to reduce WRC activity. Hotspot 3 is located immediately underneath the D site, where mutations have been shown to both destabilize WRC in neurons and enhance Rac1 binding and WRC activation in vitro. Again, similar to WASP/N-WASP and WAVE1, both loss-of-function mutations (which cause gene loss or disrupt protein folding) or gain-of-function mutations (which constitutively activate WRC) in Cyfip2 lead to similar syndromes, reiterating the importance of finely controlling WRC activity in the cell. In contrast to Cyfip2, no missense mutations in Cyfip1/Sra1 have been associated with disease, but the 15q11.2 region of the chromosome, which contains Cyfip1, is a hotspot of chromosomal microdeletions and duplications. CNVs of 15q11.2 are heavily associated with various neurological disorders, including ID, autism, seizures, schizophrenia, and epilepsy (Yoon et al., 2014). These mutations would act by altering the protein and activity level of the Cyfip1-containing WRC in the brain.
Accumulating evidence suggests the WRC also plays a role in neurodegenerative diseases, such as Alzheimer’s disease (AD) and Parkinson’s disease (PD), perhaps through its role in regulating migration and phagocytosis of microglia, the immune cells in the brain. WAVE was found to co-aggregate with the pathogenic hyper-phosphorylated Tau and CRMP2 proteins in neurofibrillary tangles and abnormal neurites of the AD brain (Takata et al., 2009). A rare variant of Abi3, S209F, was identified to be a risk factor for late-onset AD (Conway et al., 2018; Olive et al., 2020; Sims et al., 2017). The importance of Abi3 in AD pathogenesis was further confirmed by a recent mouse model, in which knocking out Abi3 disrupted microglia migration and phagocytosis, increased amyloid β (Aβ) accumulation, decreased microglia clustering around the Aβ plaques, and impaired long-term potentiation (Karahan et al., 2021). In addition, WAVE1 expression forms a negative feedback loop with Aβ production: the amyloid precursor protein (APP) binds to the promoter of WAVE1 to suppress WAVE1 expression, whereas reducing WAVE1 expression significantly reduced Aβ levels and restored memory deficits in a mouse AD model (Ceglia et al., 2015). Both elevated and decreased expression level of WAVE1 in AD patient brains was reported, suggesting the mechanism is complicated and may also depend on disease stage (Ceglia et al., 2015; Kitamura et al., 2003). In addition, WAVE2 was found to have a genetic interaction with LRRK2 (Leucine-rich repeat kinase-2), one of the commonly mutated proteins in Parkinson’s disease (PD). This interaction increased the lifetime of the WAVE2 protein and increased the phagocytic activity of microglial cells to increase neuron death, mimicking the cell death commonly seen in Parkinson’s (Kim et al., 2018).
Hem1-containing WRC, which usually contains WAVE2, is specifically expressed in hematopoietic cells and plays an essential role in immune system processes, including immune cell chemotaxis, phagocytosis, T-cell activation, immunological synapse formation, integrin-mediated adhesion, and B cell development and homeostasis (Castro et al., 2020; Cook et al., 2020; Nolz et al., 2006; Park et al., 2010; Salzer et al., 2020; Stahnke et al., 2021; Weiner et al., 2006; Zipfel et al., 2006). Recently, several missense mutations and an exon-deleting mutation in Hem1 have been identified to be the cause of a new immunological syndrome named Immunodeficiency-72 with an Autoinflammation (IMD72), which involves immunodeficiency and recurring infections mixed with atopy, lymphoproliferation, and cytokine overproduction (Castro et al., 2020; Cook et al., 2020; Salzer et al., 2020). All mutations are spatially clustered at a hotspot in the WRC proximal to the Rac1-binding D site, which was named Hem1 hotspot (Figure 3B) (Rottner et al., 2021). Most of the mutations are buried in the WRC structure and were shown to disrupt protein folding and stability, while the only mutation on a surface residue, M371V, seemed to affect Arf1-mediated but not Rac1-mediated WRC activation. Exactly how M371V disrupt Arf1-WRC interaction remains to be addressed. Loss of Hem1 also seemed to disrupt mTORC2 activation, leading to impaired Akt signaling, cytokine secretion, and T cell proliferation. The linkage of the Hem1-/WAVE2-containing WRC to mTOR signaling was further supported by a recent conditional WAVE2 knockout mouse model (Liu et al., 2021).
As a major driver of cell motility, the WRC is heavily involved in many types of cancers, including HCC, leukemia, and breast, gastric, prostate, ovarian, bladder, pancreatic, lung, colorectal, and cervical cancers (Biber et al., 2020; Kurisu and Takenawa, 2010; Miki et al., 1998b; Nozumi et al., 2003; Sossey-Alaoui et al., 2005, 2007b; Suetsugu et al., 1999, 2003; Yan et al., 2003). Overall, the mechanism of how WRC misregulation is linked to cancer is largely unclear, but in general overexpression (and sometimes deletion) of WRC components, including WAVE, Abi, HSPC300, Nap, and Cyfip is frequently observed in cancer cells and often associated with an increase in the motility and invasiveness of cancer cells, which is often accompanied with poor prognosis (Carmona et al., 2016; Huang et al., 2018; Jia et al., 2014; Taniuchi et al., 2018). This is likely due to transcriptional rewiring in cancer cells that elevates WRC expression and activity to help cancer cells alter the actin cytoskeleton and promote cell migration. WAVE was also found to affect the epithelial to mesenchymal transmission (EMT), which is critical for cancer metastasis and prognosis (Park and Kim, 2017; Taniuchi et al., 2018; Taylor et al., 2013). In addition to regulating cell migration, WAVE also regulates the expression level of matrix metalloproteinases MMP1 and MMP9, which localize to invadopodia and degrade ECM components (Sossey-Alaoui et al., 2005). The generally elevated expression of the WRC suggests inhibitors targeting WRC activity or inducing WRC degradation could be a potential therapeutic for difficult to treat cancers, such as prostate and triple-negative breast cancers (Cowell et al., 2017; Limaye et al., 2022; Loveless and Teng, 2021; Teng et al., 2016).
Various pathogens hijack WRC signaling to facilitate their infection. For example, Salmonella relies on WAVE2-containing WRC for effective internalization into epithelial cells (Shi et al., 2005). Salmonella does so through two signaling pathways, both merging on the WRC. First, the bacteria secrete its own Rac1-GEF, SopE/SopE2, into the host cell, which promotes Rac1 activation to recruit the WRC to the invading site. Second, the bacteria simultaneously recruit the host Arf-GEF, ARNO, to the invading site, which promotes Arf1 activation. Both Rac1 and Arf1 act cooperatively on the WRC to promote actin polymerization at the invading site and trigger bacterial entry through micropinocytosis (Humphreys et al., 2013, 2012). Interestingly, the cooperativity between Rac1 and Arf1 in promoting WRC activation is also hijacked, albeit in the opposite way, by two extracellular bacterial pathogens, enteropathogenic (EPEC) and enterohaemorrhagic (EHEC) Escherichia coli. Both EPEC and EHEC can secrete an effector protein EspG into host cells, which sequesters both Arf6 and Arf1 to reduce WRC activation, allowing the pathogen to evade WRC-dependent phagocytosis (Humphreys et al., 2016). Other intracellular bacterial pathogens also rely on Rac1-WRC signaling to promote their internalization, including Chlamydia trachomatis, Anaplasma phagocytophilum, the tick-born bacterial pathogen Rickettsia, and various Gram-negative bacteria (such as Neisseria gonorrhoeae) that rely on EACAM3-mediated phagocytosis (Carabeo et al., 2007; Lin et al., 2007; Pils et al., 2012; Reed et al., 2012). Viruses can also act through the WRC to promote their infection. During HIV internalization, WAVE2-containing WRC and its associated ligands, including Rac1, Arp2/3, and Abl kinase, are required for the viral envelope-mediated membrane fusion, entry, and infection (Harmon et al., 2010; Spear et al., 2014). During human cytomegalovirus (HCMV) infection, the viral protein pUL135 directly binds to Abi and recruits the WRC to the plasma membrane, which, instead of promoting actin polymerization, reduces the efficiency of immune synapse (IS) formation to help infected cells escape immune surveillance (Rak et al., 2018; Stanton et al., 2014). In addition, the protozoan parasite, Toxoplasma gondii, one of the most prevalent parasites on earth and the cause of toxoplasmosis, was found to rely on a novel parasite protein, TgWIP (T. gondii WRC interaction protein), to help the parasites disseminate from the primary infection site to distant organs (Sangaré et al., 2019). TgWIP contains a WIRS motif, which is believed to be important for binding the WRC and redirecting WRC-mediated actin polymerization in infected cells.
In addition to the WRC itself, mutations in various WRC ligands are also frequently associated with diseases that resemble the symptoms of WRC-associated disorders. For example, missense mutations in Rac1/2/3 cause autosomal dominant mental retardation-48 (MRD48), immunodeficiency-73A/B/C (IMD73A), and neurodevelopmental disorder with structural brain anomalies and dysmorphic facies (NEDBAF), respectively (Alkhairy et al., 2015; Ambruso et al., 2000; Costain et al., 2019; Hsu et al., 2019; Reijnders et al., 2017; White et al., 2018; Williams et al., 2000). Misregulation of Rac1 activity, including that caused by several hotspot missense mutations (such as P29S), was identified in human melanoma and lung, liver, and breast cancers (Bauer et al., 2007; Dokmanovic et al., 2009; Hodis et al., 2012; Kawazu et al., 2013; Krauthammer et al., 2012; Liu et al., 2008; Schnelzer et al., 2000; Stallings-Mann et al., 2012). Among them, P29S was shown to increase the Rac1 binding affinity to various downstream effectors, including the WRC (Chen et al., 2017; Hodis et al., 2012; Krauthammer et al., 2012). In addition, mutations in various WIRS-containing receptors, such as protocadherin19 and DCC, cause neurological/developmental disorders, such as epilepsy and mental retardation in females (EFMR) and congenital mirror movement, respectively (B. Chen et al., 2014b; Depienne et al., 2011; Depienne and Leguern, 2012).
In summary, the WRC acts as a central signaling hub that links a large array of ligands at membranes to the actin cytoskeleton in many different normal and disease related processes. Exactly how the WRC interacts with different ligands and how the interactions modulate WRC localization and activity is still largely unknown and requires rigorous biochemical, structural, and cell biological studies to elucidate the underlying mechanisms.
WASH and WASH Regulatory Complex (SHRC)
WASH was identified as the third WASP-family protein about 10 years after the discovery of WASP/N-WASP and WAVE. In 2007, when examining the subtelomeric region of human chromosomes, once considered a “genetic junkyard” filled with duplications and variations, Linardopoulou et al. found the most telomerically duplicated human genes, MGC52000, coded a protein homologous to WASP/N-WASP and WAVE, and named it WASH (Linardopoulou et al., 2007). These genes had been reported in other non-primate animals as a single-copy gene without a known function (Gianfrancesco et al., 2001; Hansen et al., 2005). Interestingly, the WASH gene was found to be extensively duplicated in primates. Even the six human individuals examined in their study were different from each other, having 15–20 copy numbers and 16 different chromosomal locations (Linardopoulou et al., 2007). The duplicated genes contained pseudogenes, different truncation variants, and full-length, intact ORFs. The full-length ORFs contained significant numbers of amino acid substitutions, with ≥ 95.8% identity among them. It was speculated that the subtelomeric variations in the human population might give slightly different functions, which might contribute to phenotypic differences between human individuals (Linardopoulou et al., 2007).
Soon after the discovery of WASH, a series of cellular and biochemical studies found that, similar to WAVE, WASH also existed in a large protein complex of ~500 kDa, thereafter named the WASH Regulatory Complex (SHRC, pronounced “shark”) (Derivery et al., 2009b; Gomez and Billadeau, 2009; Jia et al., 2010). Consisting of five core subunits, SWIP (KIAA1033, Strumpellin- and WASH-interacting protein), Strumpellin (KIAA0196), Fam21 (Fam21A/B and Fam21C), CCDC53, and WASH, the SHRC was found to resemble the WRC in many ways. First, both isolated WAVE and WASH proteins are constitutively active towards the Arp2/3 complex, while their corresponding complex keeps WAVE or WASH basally inhibited (Jia et al., 2010). Second, although the two complexes share less than 15% sequence identity (aside from the WCA domain of WASH and WAVE), a more advanced profile search by HHPred revealed that four subunits in the SHRC shared distant, but significant homology with a corresponding subunit in the WRC. SWIP is homologous to Sra1, Strumpellin to Nap1, the N-terminal helix of CCDC53 to HSPC300, and the N-terminal helix of WASH to the N-terminal helix of WAVE (Hildebrand et al., 2009; Jia et al., 2010). Although HHPred could not identify homology between the N-terminal helices of Fam21 and Abi2, they might share structural homology that was below the detection threshold. Third, the distant homology between SHRC and WRC subunits suggests the two complexes may have a similar structural organization (Figure 4). It is possible that the two large subunits, SWIP and Strumpellin, form a large, elongated dimer platform similar to the Sra1-Nap1 dimer, along which aligns a helix bundle formed by the N-terminal helices of Fam21, CCDC53, and WASH (similar to the Abi2-HSPC300-WAVE helix bundle). The overall structural resemblance was supported by both structure-function analysis in cells and negative stain EM analysis of purified SHRC, but exactly how the SHRC is assembled and how it keeps WASH basally inhibited remain unknown (Jia et al., 2010).
Figure 4. WASH regulatory complex (SHRC).
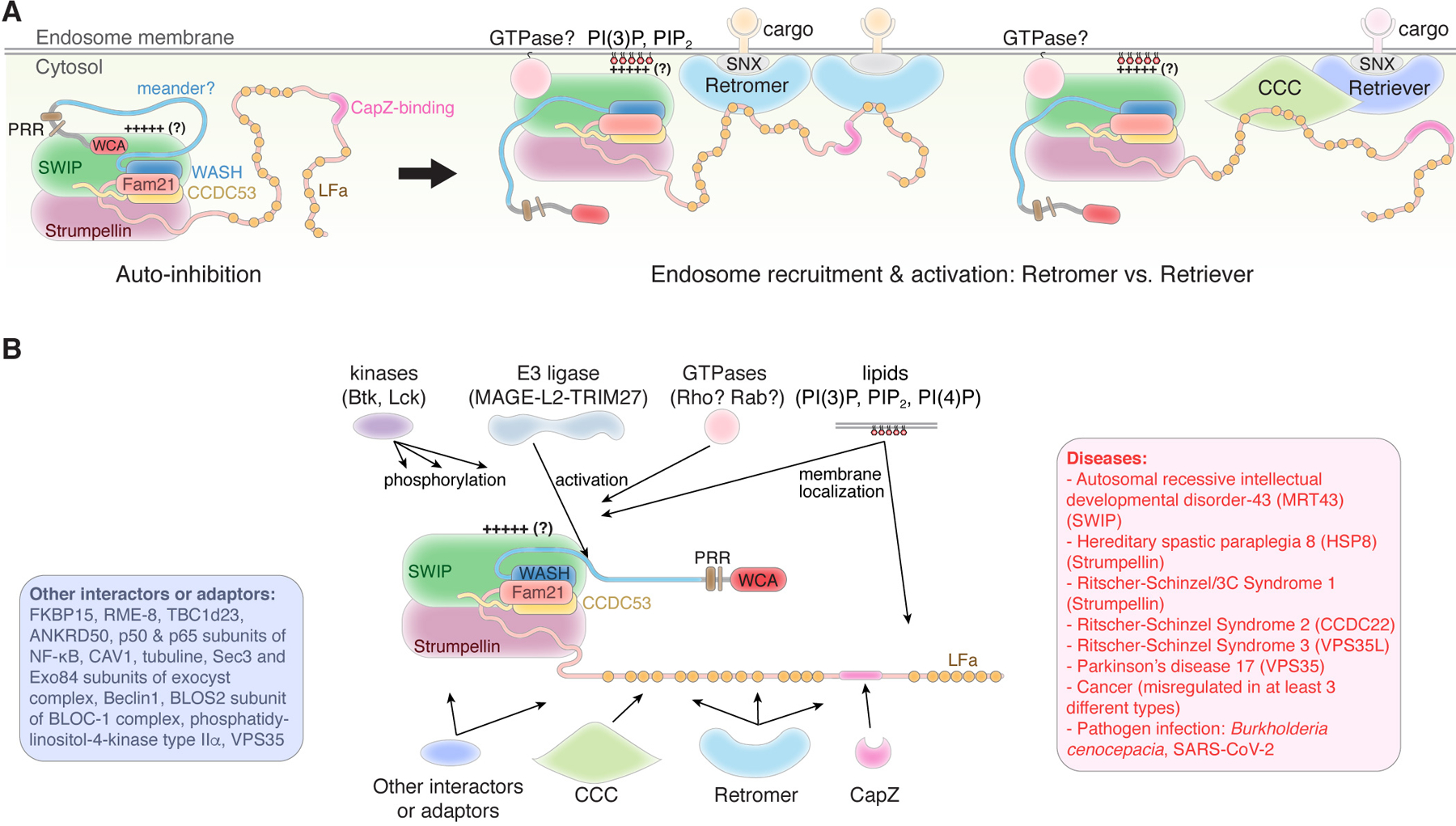
(A) Schematic showing mechanisms by which the SHRC may keep WASH auto-inhibited in the basal state and be activated and recruited to the endosomal membrane to regulate retromer- and CCC-retriever-mediated cargo sorting. The relative position of each subunit in the SHRC is based on its homology to the WRC. “++++?” indicates a positively charged surface possibly existing in the SHRC based on its resemblance to the WRC. “GTPase?” indicates the uncertainty of whether and what GTPase directly binds to the SHRC to induce activation. LFa: LF-(D/E)3–10-LF sequence, which directly binds to VPS35 in retromer. (B) Schematic showing how different regulatory ligands interact with the SHRC. Text boxes show representative ligands in indicated category and diseases caused by or associated with SHRC subunits.
Similar to Abi and WAVE, Fam21 and WASH extend long, unstructured sequences immediately following their N-terminal helices (Figure 4). These extended sequences play an important role in SHRC assembly and regulation. CCDC53 also contains an extended sequence at the C-terminus, but unlike its N-terminal helix, this extended sequence did not seem to be critical for the assembly or function of the SHRC (Gomez and Billadeau, 2009; Jia et al., 2010). The N-terminus of WASH contains two loosely defined conserved regions named WHD1 (WASH homology domain 1, a.a. 1–167) and WHD2 (a.a. 168–304), which was collectively named WAHD (WASH homology domain) (Gomez and Billadeau, 2009; Linardopoulou et al., 2007). The N-terminal part of WHD1 (a.a. 29–79) contains the helix homologous to the N-terminal helix of WAVE, which is presumably important for forming a helix bundle with the N-terminal helices of Fam21 and CCDC53. Deleting this helical sequence, or the N-terminal helix of Fam21 or CCDC53, abolished SHRC formation (Gomez and Billadeau, 2009; Jia et al., 2010). The remaining part of WAHD, including the C-terminal part of WHD1 and the entire WHD2, does not contain well defined secondary structures and possibly resembles the meander region of WAVE, which could interact with the SWIP-Strumpellin dimer and control SHRC inhibition (Figure 4). Indeed, WHD2 was not essential for assembling the SHRC, but deleting WHD2 abolished SHRC function in retromer-mediated cargo sorting, suggesting WHD2 plays an important regulatory role (Gomez and Billadeau, 2009). Following WAHD is a small stretch of PRR and then the C-terminal WCA sequence. It is unknown what proteins directly interact with the PRR sequence.
The C-terminal tail of Fam21 is over 1,000 a.a. long and harbors many protein-protein interaction sequences. First, structure-function analysis suggested the sequence of ~120 a.a. immediately following the N-terminal helix of Fam21 plays an essential role in maintaining SHRC assembly, likely through a mechanism similar to the NBF region of Abi2 binding to Nap1 in the WRC (Jia et al., 2010). Following this sequence, the long C-terminal tail Fam21 contains 21 copies of a unique L-F-[D/E]3–10-L-F motif (LFa, Figure 4), each motif being able to directly interact, with different affinities, with VPS35, a subunit in the cargo-selective complex named retromer (Harbour et al., 2012, 2010; Helfer et al., 2013; Jia et al., 2012). This multivalent interaction tightly links SHRC to retromer-mediated endosomal sorting and likely provides a mechanism for the cell to finely tune SHRC membrane recruitment based on the retromer density on endosome membranes. The C-terminal tail of Fam21 also contains a conserved capping protein binding motif, which binds to CapZ in the cell and inhibits its capping activity (Derivery et al., 2009b; Gomez and Billadeau, 2009; Jia et al., 2010). This binding activity was recently shown to competitively remove CapZ from the dynactin complex. As a result, the “de-capped” dynactin could provide an actin mini-filament that could elongate and prime SHRC-Arp2/3-mediated actin polymerization (Fokin et al., 2021; Fokin and Gautreau, 2021). The Fam21 tail also interacts with many other molecules, but the exact interaction mechanism is not known. For example, the N-terminal part of the Fam21 tail (a.a. 356–600) directly interacts with the C-termini of CCDC22 and CCDC93 of the COMMD/CCDC22/CCDC93 (CCC) complex, a key regulator of endosomal recycling. This interaction links SHRC to the CCC-retriever-mediated, retromer-independent cargo sorting (Figure 4A, right) (Harbour et al., 2012; McNally et al., 2017; Phillips-Krawczak et al., 2015). TBC1d23, a protein essential for endosome-to-Golgi trafficking, was shown to bind to the Fam21 tail, linking endosomal trafficking to the Trans-Golgi network (Wenjie et al., 2019). In addition, the C-terminus of Fam21 tail (a.a. 937–1341) interacted with various acidic phospholipids in vitro, in particular PI(3,5)P2 (which is enriched in early endosomes) and PI4P (which is enriched in the Golgi), which might serve as a retromer-independent mechanism for recruiting the SHRC to endosome membranes (Buckley et al., 2016; Gomez and Billadeau, 2009; McNally et al., 2017). The Fam21 tail also binds to FKBP15 and recruits it to endosome membranes, and binds to RME-8 to coordinate with the membrane-tubulating function of the sorting nexins (Freeman et al., 2014; Harbour et al., 2012). The N-terminus of Fam21 (a.a. 1–356) was shown to bind ANKRD50, but we speculate the binding region is located in the unstructured sequence (a.a. 120–356) (Kvainickas et al., 2017). Moreover, the Fam21 tail contains both a nuclear localization signal sequence (NLS) and a nuclear export signal (NES), and binds multiple components of the nuclear factor κB (NF-κB) pathway (such as the p50 and p65 (RelA) NF-κB subunits), suggesting the isolated Fam21 has a distinct role in the nucleus (Deng et al., 2015). The Fam21 C-terminal also tail contains many phosphorylation sites, which may fine tune protein-protein or protein-lipids interactions (Hornbeck et al., 2015).
While the aforementioned ligands that bind to the C-terminal tail of Fam21 play important roles in linking SHRC to endosomal membranes and other regulators of endosomal trafficking, they don’t directly bind to the core, structured part of the SHRC and, therefore, may not directly contribute to SHRC activation. Analogous to WASP/N-WASP and the WRC, several other mechanisms could directly contribute to SHRC activation through the core structured region and likely act in a cooperatively manner. These include post-translational modifications (phosphorylation and ubiquitination), binding to GTPases, and perhaps binding to novel ligands. WASH contains several phosphorylation sites in the WAHD region, which may promote SHRC activation analogous to the phosphorylation of WAVE meander region (Hornbeck et al., 2015). For example, Y141 in the WHD1 was shown to be phosphorylated by Lck in NK cells, and Y262 (Y261 in mouse WASH) in the WHD2 was phosphorylated by Btk in both Drosophila and mouse models (Huang et al., 2016; Tsarouhas et al., 2019). Y141F inhibited trafficking of lytic granules to the immune synapse, leading to impaired cytotoxicity of NK cells, while Y262 phosphorylation caused SHRC activation. It remains to be tested whether phosphorylation alone is sufficient to activate the SHRC. In addition to phosphorylation, ubiquitination at K220 in the WHD2 region by the MAGE-L2-TRIM27 E3 ubiquitin ligase was shown to be important for SHRC-mediated actin polymerization on endosomes and sufficient to activate the SHRC in vitro (Hao et al., 2013). Ubiquitination-mediated activation might not be unique to SHRC, as ubiquitination in the WAVE meander region (e.g., K161 in WAVE2 and K162 WAVE3) was observed in multiple high-throughput proteomic studies (Hornbeck et al., 2015).
Small GTPases have a central role in activating WASP/N-WASP (Cdc42) and the WRC (Rac1 and Arf). Does SHRC activation similarly require a GTPase? The answer is currently unknown, but a few clues suggest it is likely “yes”. First, small GTPases, especially the large number of Arf (~30 in human) and Rab GTPases (~60 in human), are found all over the endomembrane system (Donaldson and Jackson, 2011; Stenmark, 2009). They play important roles in virtually every step of intracellular trafficking (including endosomal trafficking) where they regulate biogenesis, sorting, tethering, fusion, tubulation, and fission of various organelles (Molendijk et al., 2004). It is possible one or a group of them activates the SHRC, but the interaction may be weak or transient and cooperative with other membrane ligands, similar to the interaction between Rac1 or Arf1 and the WRC, which can elude conventional identification methods. Second, a study in Drosophila showed the GTPase Rho directly interacted with WASH in a nucleotide-dependent manner (Liu et al., 2009; Jeffrey M Verboon et al., 2015). Other studies using purified human proteins, however, could not detect the interaction between SHRC and RhoA, but detected nucleotide-independent interaction between SHRC and Rac1 (Jia et al., 2010). Neither Rac1, RhoA, nor Cdc42 was able to activate SHRC in vitro, suggesting some other ligands or conditions may be involved (Jia et al., 2010). Third, by using an in situ APEX2-mediated proximal labeling method, a recent study found Rab21 and Rab7 interacted with the SHRC and revealed the importance of Rab21 in endosomal cargo sorting (Del Olmo et al., 2019). In addition, Rab9 was also linked to SHRC-mediated retrograde trafficking (Dong et al., 2013). It is, however, unknown if these interactions are direct and how they contribute to SHRC activation. In vitro reconstitution methods will be important for validating and characterizing these potential interactions.
Other ligands may also interact with the core structure of SHRC. It is possible that, similar to the WRC, SHRC may contain a positively charged surface to directly bind to inositol phospholipids, independent of the lipid-binding activity of Fam21 C-terminal tail (Figure 4) (Derivery et al., 2009b; Gomez and Billadeau, 2009). Strumpellin-SWIP were shown to interact with the caveolar protein CAV1, which was important for maintaining CAV1 homeostasis required for integrin-mediated cell adhesion (Lee et al., 2020). The WAHD region of WASH is implicated in many interactions, which may not only link SHRC to other ligands, but also potentially contribute to SHRC activation. The WHD2 region of WASH directly interacted with tubulin, which may link microtubules to the actin cytoskeleton in endosome trafficking (Gomez and Billadeau, 2009; Liu et al., 2009). The overall WAHD region of WASH was reported to interact with two subunits of the exocyst complex, Sec3 and Exo84, to regulate exocytosis of transmembrane type 1 matrix metalloproteinase (MT1-MMP) at invadopodia (Monteiro et al., 2013). The middle region of WAHD was found to directly interact with Beclin1, which provided a mechanism to suppress autophagy (Xia et al., 2013). In addition, SHRC interacted with Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type IIα, likely through a direction interaction between the WAHD region of WASH and the BLOS2 subunit of BLOC-1 complex (Monfregola et al., 2010; Ryder et al., 2013). WASH may also directly interact with VPS35, which could further facilitate retromer binding (Harbour et al., 2012). Again, most of the above interaction mechanisms and their effects on SHRC activity remain largely unknown.
Unlike WASP/N-WASP and the WRC, which mainly regulate actin polymerization at the cell membrane, the SHRC mainly regulates actin assembly at early and recycling endosome membranes (Figure 1, right) (Duleh and Welch, 2010; Fokin and Gautreau, 2021; Seaman et al., 2013; Wang et al., 2018). The activity of the SHRC in promoting actin assembly plays a key role in maintaining tubular structures of the recycling endosomes and facilitating protein sorting (Derivery et al., 2009b; Fokin et al., 2021; Gomez et al., 2012; Gomez and Billadeau, 2009). SHRC-dependent actin networks are often found as discrete patches at endosomal membranes (Derivery et al., 2012, 2009b; Gomez and Billadeau, 2009). These actin-rich domains promote tubulation of membrane structures where specific cargo proteins are clustered, and they further facilitate dynamin-mediated membrane scission to release the vesicles (Puthenveedu et al., 2010). Loss of SHRC activity collapses endo-lysosomal membranes and impedes receptor recycling, leading to excessive degradation by the lysosome, which affect homeostasis of numerous important membrane proteins, such as CI-MPR, TfnR, EGFR, β2AR, TCR, CD28, LFA-1, GLUT1, LDLR, SR-BI, ATP7A, CAV1, α5β1-integrin, and ACE2 (the entry receptor for SARS-CoV-2 infection) (Bartuzi et al., 2016; Courtland et al., 2021; Gomez et al., 2012; Gomez and Billadeau, 2009; Lee et al., 2020; Phillips-Krawczak et al., 2015; Piotrowski et al., 2013; Puthenveedu et al., 2010; Temkin et al., 2011; Wijers et al., 2019; Zech et al., 2011; Zhu et al., 2021).
In addition, new roles of the SHRC in regulating autophagy are emerging, where SHRC is required for trafficking of autophagy proteins, suppressing autophagy, and driving V-ATPase removal from lysosomes and lysosome neutralization, which is required for efficient phagocytic and autophagic clearance (Carnell et al., 2011; King et al., 2013; Nagel et al., 2017; Park et al., 2013; Xia et al., 2014, 2013; Zavodszky et al., 2014).
Given the fundamental role of the SHRC in endolysosomal biology, it is not surprising that disruption of SHRC function has a profound impact on homeostasis and signal transduction in various physiological processes, including cellular signaling, cholesterol clearance, and immune function (Simonetti and Cullen, 2019). Complete loss of the SHRC in animals severely interrupts development and leads to early embryonic death (Gomez et al., 2012; Linardopoulou et al., 2007). Mutations or aberrant activities of the SHRC and its closely related partners, including retromer, retriever, and the CCC complex, are the cause of various complicated developmental/neurological disorders, including hereditary spastic paraplegia 8 (HSP8), Ritscher-Schinzel syndrome, autosomal recessive intellectual developmental disorder-43 (MRT43), and Parkinson’s disease 17 (PARK17) (Gangfuß et al., 2022; Ginanneschi et al., 2020; Gjerulfsen et al., 2021; Rahman and Morrison, 2019). Without a high-resolution structure of the SHRC, it is currently unknown exactly how various missense mutations cause disease.
At least 4 types of homozygous and heterozygous missense mutations in SWIP, including P1019R, Q442*/D1048G, K1079R/H503R, and Y1014C, have been identified as the cause of autosomal recessive intellectual developmental disorder-43 (MRT43), which shows severely impaired developmental and intellectual development, poor learning and motor skills, short stature, dysmorphic features, and recurrent infections (Assoum et al., 2020; Gangfuß et al., 2022; Ropers et al., 2011). Among them, the P1019R mutation may interfere with protein folding, as the patient cells showed significantly reduced expression of SWIP, Strumpellin, and WASH, which resulted in loss of SHRC function (Ropers et al., 2011). Similar phenotypes were shown in a recent mouse model, in which SWIPP1019R was found to reduce SHRC level and significantly disrupt both endosomal and lysosomal pathways (Courtland et al., 2021). By contrast, Y1014C did not seem to affect SHRC expression and therefore may be located in a region important for SHRC activity regulation or ligand binding (Gangfuß et al., 2022).
Many mutations in Strumpellin have been associated with hereditary spastic paraplegia 8 (HSP8) and Ritscher-Schinzel Syndrome (Ginanneschi et al., 2020; Gjerulfsen et al., 2021). HSP8 is an autosomal dominant neurologic disorder characterized by late onset lower limb spasticity and hyperreflexia. At least 12 different mutations in Strumpellin have been identified in HSP8 patients, including various missense mutations, an internal exon, and a frameshift mutation (de Bot et al., 2013; Ginanneschi et al., 2020; Valdmanis et al., 2007). Cellular and biochemical studies of N471D, L619F, and V626F did not detect clear defects in protein expression, ligand binding, or endosomal trafficking (Freeman et al., 2013; Jia et al., 2010). A recent mouse model of StrumpellinN471D showed this mutation recapitulated HSP8 phenotypes in human patients and mildly altered the brain proteome, albeit without affecting protein levels of SHRC or related ligands, suggesting this mutation (or other mutations) might disrupt ligand binding (Clemen et al., 2021).
In addition to mutations that lead to HSP8, a splice site mutation (c.3335+2T-A) in Strumpellin is a cause of Ritscher-Schinzel Syndrome, also known as cranio-cerebello-cardiac syndrome or 3C syndrome, which is characterized by craniofacial abnormalities, congenital heart defects, and cerebellar malformations. This mutation causes frameshift and premature termination of Strumpellin and significantly reduces protein expression level (Elliott et al., 2013).
It is worth noting that mutations in closely related SHRC ligands, including CCDC22 (a subunit of the CCC complex) and VPS35L (a subunit of retriever), were the cause of two subtypes of Ritscher-Schinzel Syndrome (Gjerulfsen et al., 2021; Kato et al., 2020; Kolanczyk et al., 2015). Additionally, the D620N mutation in VPS35 (a subunit of retromer) is the cause of Parkinson’s disease 17 (PARK17) (Rahman and Morrison, 2019). The mutation was found to impair VPS35 binding to Fam21 and subsequently disrupt endosome-to-TGN transport of CI-MPR and the turnover of mitochondrial DLP1 complex causing mitochondrial fragmentation and dysfunction in neurons (McGough et al., 2014; Wang et al., 2016). The above suggests disrupting SHRC-CCC-retriever vs. SHRC-retromer recycling pathways results in separate disease conditions, supporting the notion that the two pathways play distinct roles in the cell (Figure 4A) (McNally et al., 2017; Phillips-Krawczak et al., 2015).
SHRC is also involved in cancer, mainly through its role in regulating the recycling of cell adhesion molecules (e.g., α5β1-integrin) and transmembrane matrix metalloproteinase (e.g., MT1-MMP), which are required for migration and invasion of cancer cells (Biber et al., 2020; MacDonald et al., 2018; Monteiro et al., 2013; Porkka et al., 2004; Zech et al., 2011). Finally, the intracellular bacterial pathogen, Burkholderia cenocepacia, the cause of severe pulmonary infections in cystic fibrosis and chronic granulomatous disease patients, was found to hijack SHRC-mediated endolysosomal recycling to escape phagosome maturation and facilitate survival and infection (Walpole et al., 2020). It is possible this mechanism is also used by other intracellular pathogens. Recently, a genome-wide CRISPR screen identified SHRC, the CCC complex, and retromer as important host factors that regulate the entry of SARS-CoV-2 virus (Zhu et al., 2021). Loss of any of these components led to a significant reduction of surface ACE2, the entry receptor critical for SARS-CoV-2 infection (Zhu et al., 2021).
In summary, the SHRC is a central regulator of the actin cytoskeleton on endosome membranes to support endolysosomal membrane trafficking. Disturbance of the function of SHRC and its closely related ligands has profound impact on many different signaling pathways associated with diseases. Compared to WASP/N-WASP and the WRC, most mechanistic questions of how the SHRC interacts with different ligands and how its membrane recruitment and activity are regulated still need to be answered.
WHAMM and JMY
We discuss WHAMM and JMY together here as they share similar domain structures and partially overlapping cellular functions (Figure 1 & 5, and see below) (Campellone et al., 2008b; Rottner et al., 2010). One major difference between WHAMM/JMY and aforementioned WASP/N-WASP, WAVEs, and WASH is that WHAMM and JMY are basally active in stimulating Arp2/3-mediated actin polymerization. Their WCA sequences are not autoinhibited, neither in cis in a single polypeptide chain as for WASP/N-WASP, nor in trans in a large protein complex as for WAVE and WASH (Figure 5) (Campellone et al., 2008b; Kabrawala et al., 2020; Zuchero et al., 2009). Nevertheless, their actin-nucleation activity is regulated in the cell by ligand binding and by control of their cellular localization, although very little is currently known about the underlying biochemical or structural mechanisms.
Figure 5. WHAMM and JMY.
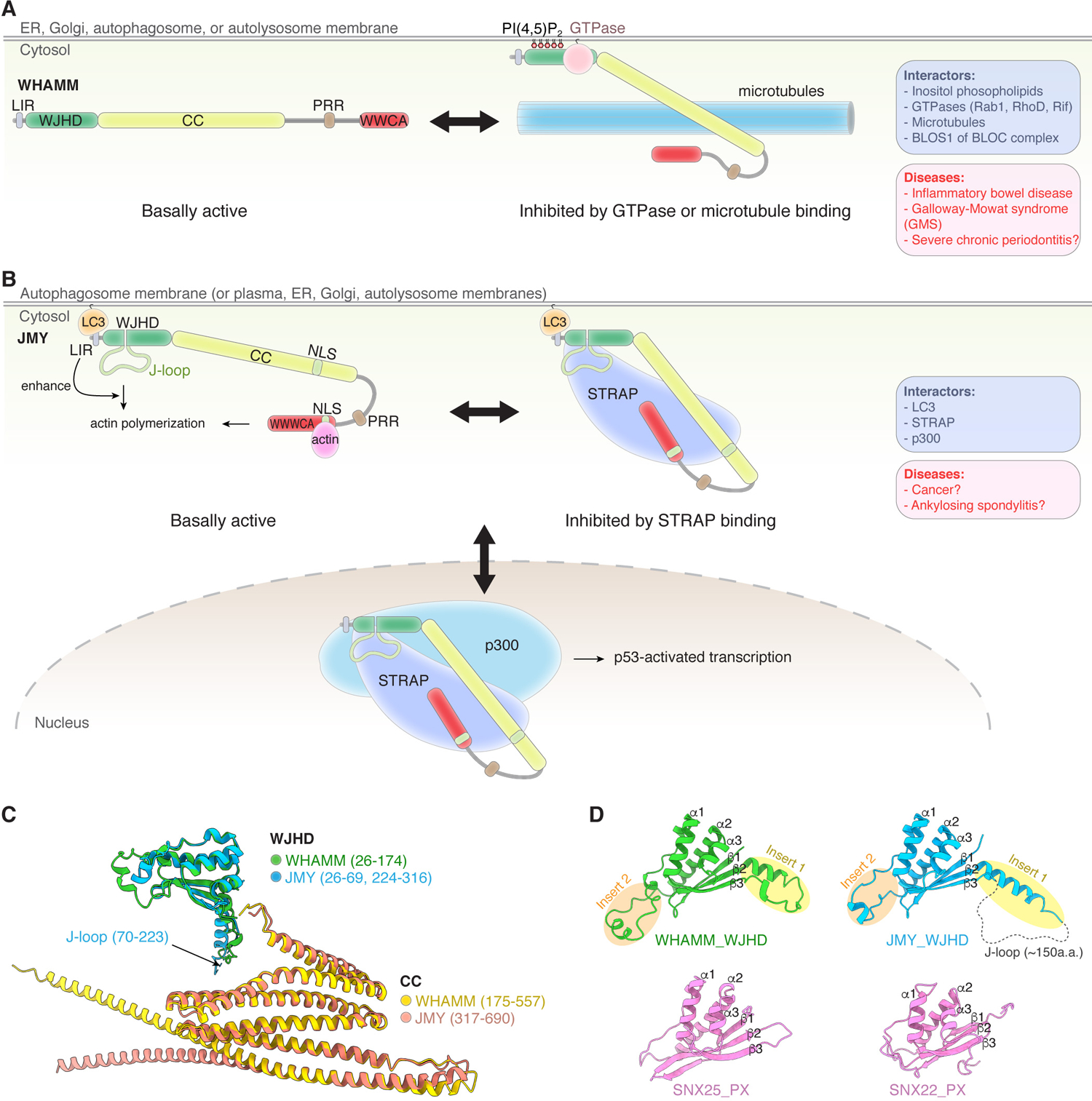
(A-B) Mechanisms underlying WHAMM and JMY membrane localization and inhibition, respectively. Text boxes show known interacting ligands and associated diseases. (C) Overlay of the N-terminal structures of WHAMM and JMY predicted by AlphaFold 2. As the relative orientation between the two domains is different for WHAMM and JMY, shown in the cartoon is the two domains of JMY separately aligned to WHAMM. For clarity, unstructured sequences are removed from the presentation, including the J-loop insertion in the WJHD of JMY. (D) Comparison of WJHD with two representative PX domains, one from SNX25 (sorting nexin 25, PDB: 5WOE) and the other from SNX22 (PDB: 2ETT). The core PX folding is indicated by similar secondary structural elements. Also indicated are the structural inserts protruding from the core, which are unique to the WJHD and not found in PX domains. The J-loop unique to JMY is indicated by a dotted line.
WHAMM was discovered in 2008 from a search in the human proteome for WCA-containing proteins (Campellone et al., 2008b). By contrast, JMY was initially discovered in 1999 as a cofactor for p53, where in response to DNA damage it accumulated in the nucleus and activated p53-mediated transcription in a complex with p300 and STRAP (Demonacos et al., 2001; Shikama et al., 1999). JMY was not recognized as a WASP-family protein until a genome database search in 2009 for WH2-containing proteins (Zuchero et al., 2009). WHAMM and JMY share ~27–35% amino acid identity throughout the entire sequence (Campellone et al., 2008b). WHAMM contains two consecutive WH2 motifs (making a WWCA domain at the C-terminus), while JMY contains three WH2 motifs (making a WWWCA domain at its C-terminus) (Figure 1, left). The domain assignment of their N-terminal sequences, however, has been ambiguous (Campellone et al., 2008b; Dai et al., 2019; Zuchero et al., 2009). In one commonly used assignment, WHAMM contains a WHAMM membrane-binding domain (WMD, a.a. 1–260) and a central coiled coil domain (CC, a.a. 260–570). Similarly, JMY also contains an NT domain (a.a. 1–392) and a CC domain (a.a. 393–793) (Campellone et al., 2008b; Dai et al., 2019).
At the writing of this review, protein structure prediction by AlphaFold 2 had just achieved tremendous success (Jumper et al., 2021). We thereby analyzed the structure models generated by AlphaFold 2 in an attempt to have a better definition of the structural organization of WHAMM and JMY. To our surprise, we found striking structural similarity between the N-terminal regions of JMY and WHAMM. The structural models of both proteins revealed two clearly defined domains, which were different from the domain organization previously derived from sequence analysis (Figure 5C). The relative orientation between the two domains differed between JMY and WHAMM models, but the individual domains aligned very well between the two models, giving a root-mean-square deviation (r.m.s.d.) of less than 1 Å (Figure 5C). This suggests 1) the two regions are likely independent domains, and 2) the N-terminal regions of JMY and WHAMM have very similar tertiary structures despite their relatively low sequence identity.
The first domain, formed by a.a. 26–174 in WHAMM and a.a. 26–69 and a.a. 224–316 in JMY, contains a core globular structure consisting of a three-helix bundle packed against a three anti-parallel β-strands (Figure 5D). Protruding from the core are two insertions, one between β2 and β3 and the other between α1 and α2, giving a L-shaped molecule (Figure 5D). We herein name this first domain as the WHAMM and JMY homology domain (WJHD) (Figure 1). Note in the insertion between β2 and β3 (insert 1 in Figure 5D), JMY contains a long, unstructured loop of ~150 a.a., which we name J-loop (for JMY-specific loop). This loop is not present in WHAMM and may carry functions unique to JMY. Remarkably, a.a. 1–314 in JMY was shown to have a cryptic actin-nucleation activity without a known mechanism (Hu and Mullins, 2019), while such an activity has not been reported for the corresponding region in WHAMM. It will be interesting to see if this activity comes from the structured WJHD domain or the unstructured J-loop.
Searching for structures similar to the WJHD in the PDB database using PDBeFold identified many different PH (phox-homology) domains, which aligned to the core structure, but not including the two inserts protruding from the core (see two examples in Figure 5D) (Krissinel and Henrick, 2004). The structural similarity between the WJHD and the PX domain is consistent with the functions of WHAMM and JMY, as PX domains usually bind inositol phospholipids and are found in various proteins involved in membrane binding, vesicle trafficking, cell signaling, and lipid metabolism (Chandra and Collins, 2019). As discussed below, WHAMM and JMY are key actin regulators in various membrane-associated processes in the endomembrane system. In addition, the previously defined WMD region of WHAMM, which covers the entire WJHD, was shown to bind different inositol phospholipids, especially PI(4,5)P2. It is possible the WJHD of both WHAMM and JMY acts as a PX domain to bind inositol phospholipids and facilitate their membrane localization. In addition to binding lipids, the WJHD may also mediate protein-protein interactions, which is sometimes seen for PX domains. The two inserts protruding from the core structure were not found in canonical PX domains, which may alter the function or add new functions to the WJHD, including novel protein-protein interactions or activity regulation.
It is worth noting that the 26-a.a. unstructured sequence N-terminal of the WJHD contains a conserved LC3-interacting region (LIR) (Figure 1 & 5). LC3 is a protein specifically found on autophagosome membranes. The LIR in JMY was previously identified based on the consensus sequence found in various LC3-binding proteins, which was W/F/Y-x-x-L/I/V flanked by acidic residues and an S or T (Coutts and La Thangue, 2015). Mutating the conserved W and V in the LIR (ETLESDWVAVRP) abolished the autophagosome localization of JMY. It is not known if WHAMM also binds to LC3, but WHAMM contains a similar LIR (DSLEGWVPVRE) in the same region and also plays a role in autophagosome development and function (Campellone et al., 2008b; Dai et al., 2019; Kast et al., 2015). It is possible the juxtaposed LIR sequence and the WJHD act synergistically to facilitate the membrane recruitment of WHAMM and JMY to autophagosome.
The second domain, formed by a.a. 175–557 in WHAMM and a.a. 317–690 in JMY, contains coiled-coils stacked by 6 helices, for which we follow the previous name as the coiled coil domain (CC). Searching for similar structures did not give meaningful results due to the low-complexity nature of helical bundles. It is possible the predicted CC structure is either novel folding or inaccurate. In spite of this, these predicted structural models are exciting in providing new insights into the structure-function relationship in WHAMM and JMY. Experimental validation of these structural predictions will be important in future studies. Below we discuss features specific to each protein.
Purified WHAMM is a monomer and is constitutively active towards the Arp2/3 complex (Campellone et al., 2008b; Kast et al., 2015). The previously defined WMD directly binds to different inositol phospholipids, especially PI(4,5)P2, which is responsible for recruiting WHAMM to various intracellular compartments, including the cis-Golgi, the ER-Golgi intermediate compartment, autophagosomes, and autolysosomes (Campellone et al., 2008b; Dai et al., 2019; Kast et al., 2015). The CC domain directly binds to microtubules (Campellone et al., 2008b; Liu et al., 2017; Shen et al., 2012). The WWCA domain of WHAMM is competent in promoting Arp2/3-mediated actin polymerization. This activity is not absolutely required for ER to Golgi transport, but is important for the elongation and stabilization of WHAMM-associated tubular membranes, ER to autophagosome biogenesis and trafficking, and autophagic lysosome reformation (Campellone et al., 2008b; Dai et al., 2019; Kast et al., 2015).
Only a few WHAMM-binding ligands have been identified, including inositol phospholipids, microtubules, and GTPases (Figure 5A). It is interesting that although purified WHAMM is constitutively active in promoting Arp2/3-mediated actin polymerization, several ligands inhibit this activity in vitro, suggesting WHAMM activity in the cell is dynamically regulated depending on the cellular context. For example, microtubule binding inhibited WHAMM activity in a dose-dependent manner, suggesting that binding microtubules and promoting actin polymerization are mutually exclusive for WHAMM function (Shen et al., 2012). Both activities, however, were required for proper function of WHAMM in facilitating membrane tubulation and anterograde transport (Campellone et al., 2008b). It is therefore possible that two spatiotemporally regulated populations of WHAMM exist to coordinate the activities at the interfaces between various endomembranes and the two cytoskeletal systems (Shen et al., 2012). Different GTPases, including Rab1, RhoD, and Rif, were found to bind the WMD region of WHAMM in a nucleotide dependent manner (Gad et al., 2012; Russo et al., 2016). Among them, the prenylated Rab1 (but not the unprenylated form) directly interacted with the WMD. This interaction inhibited WHAMM-mediated actin polymerization in vitro in a dose- and nucleotide-dependent manner (Russo et al., 2016). In addition, the BLOS1 subunit of the BLOC complex might interact with WHAMM in the initiation of autolysosomal tubulation (Wu et al., 2021). Without detailed biochemistry or high-resolution structures, it remains unknown how ligand binding to N-terminal domains inhibits the activity of WWCA, since the two parts are separated by a long unstructured sequence (Liu et al., 2017). It is also unknown if any posttranslational modifications are important for WHAMM regulation (Hornbeck et al., 2015).
WHAMM is only found in vertebrates and is broadly expressed, particularly in the brain (Campellone et al., 2008b). WHAMM plays an important role in regulating various endomembrane systems, including Golgi positioning and morphology, ER to Golgi transport, ER to autophagosome biogenesis and trafficking, and autophagic lysosome formation (Campellone et al., 2008b; Dai et al., 2019; Kast et al., 2015). In addition, WHAMM and its activity in promoting Arp2/3-mediated actin polymerization is required for apoptosis, cell adhesion, and migration (Gad et al., 2012; King et al., 2021). Furthermore, WHAMM plays an essential role in spindle actin polymerization, spindle formation and migration, and asymmetric cytokinesis in mouse oocytes (Huang et al., 2013; Jo et al., 2021).
WHAMM mutations have been associated with several types of diseases. A missense mutation, R725W, in WHAMM was identified as a pathogenic factor in patients with inflammatory bowel disease (Ben-Yosef et al., 2021). In another case, homozygous frameshift mutations in WDR73 together with closely linked WHAMM were identified as the cause of Galloway-Mowat syndrome (GMS), a nephrocerebellar syndrome characterized by microcephaly and nephrosis (Jinks et al., 2015; Mathiowetz et al., 2017). WHAMM was also suggested as a candidate gene associated with severe chronic periodontitis in genome-wide association studies, but the results remained inconclusive (Rhodin et al., 2014; Shang et al., 2015).
JMY shares many similarities with WHAMM, but is also unique among the WASP-family proteins in that it can polymerize actin in an Arp2/3-dependent and -independent manner (Firat-Karalar et al., 2011; Zuchero et al., 2009). In the presence of Arp2/3, JMY uses its WCA sequence to produce branched actin filaments similar to other WASP-family proteins, while in the absence of Arp2/3, JMY uses its tandem WH2 sequences to produce unbranched actin filaments using a mechanism similar to actin nucleation factors that also contain multiple WH2 sequences, such as Spire (Dominguez, 2016; Firat-Karalar et al., 2011; Zuchero et al., 2009). Also unique to JMY is that its N-terminal WJHD does not contain any identifiable actin-binding motifs, but is sufficient to promote actin polymerization in vitro independent of WWWCA or Arp2/3 (Figure 5B) (Hu and Mullins, 2019). This region also bound the autophagy regulator LC3, with the interaction enhancing its actin nucleation activity (Hu and Mullins, 2019).
Similar to WHAMM, purified JMY was constitutively active in promoting actin polymerization and did not seem to exist in a large regulatory complex (Zuchero et al., 2009). Inside the cell, JMY activity seemed to be suppressed (Figure 5B) (Firat-Karalar et al., 2011). The mechanisms suppressing its actin polymerization activity remain unclear, but cellular localization likely plays an important role. Unlike its homolog WHAMM, which is primarily located at endomembranes, JMY was found primarily in the nucleus, cytosol, and the leading edge of motile cells, depending on the cell type and conditions (Firat-Karalar et al., 2011; Zuchero et al., 2009). JMY is dynamically shuttled between the cytosol and the nucleus to balance its activity in promoting actin polymerization and in augmenting gene transcription of proteins involved in cell motility, such as RhoD and cadherins (Coutts et al., 2009; King et al., 2021). It is remarkable that the localization of JMY to the leading edge was correlated with cell motility, and its role in promoting cell migration required its Arp2/3-activating activity (Firat-Karalar et al., 2011; Zuchero et al., 2009). Shuttling JMY between the cytosol and the nucleus depended on its second nuclear localization sequence (NLS), which spanned the C-terminal part of the first WH2 and the linker that leads to the second WH2 (Figure 5B). Mutating these two WH2 domains to abolish their actin binding capacity or depleting monomeric actin from the cytosol dramatically shifted JMY to the nucleus, suggesting actin binding to WH2 competed against its nuclear import (Zuchero et al., 2012). This presents a novel mechanism of using monomeric actin to balance the activity of JMY in promoting actin polymerization in the cytosol and transcription in the nucleus.
JMY is widely expressed in various tissues, particularly in the brain, heart, and testes (Firat-Karalar et al., 2011; Shikama et al., 1999). Its cellular functions often overlap with its homolog WHAMM, including apoptosis, anterograde vesicle trafficking, autophagosome formation, and spindle actin polymerization and asymmetric cytokinesis in oocytes (Figure 1, right) (Coutts and La Thangue, 2015; Hu and Mullins, 2019; King et al., 2021; Liu et al., 2012; Schlüter et al., 2014; Sun et al., 2011). JMY also plays an important role in cell migration and neurite outgrowth, oligodendrocyte differentiation and maturation, and spermatogenesis (Azevedo et al., 2018; Coutts et al., 2009; Firat-Karalar et al., 2011; Liu et al., 2020; Zuchero et al., 2009).
Recent studies have started to reveal how JMY is localized to different membranes and how its activity is regulated (Coutts and La Thangue, 2015; Hu and Mullins, 2019). The N-terminal WJHD of JMY has two functions: binding to LC3 using the LIR sequence and promoting actin polymerization using an unknown mechanism independent of the C-terminal WWWCA region (Figure 5B). Binding to LC3 provided a mechanism to recruit JMY to LC3-containing autophagosomes. In the meantime, LC3 binding enhanced the actin-nucleating activity of the N-terminal sequence to promote autophagosome formation and maturation (Figure 5B) (Coutts and La Thangue, 2015; Hu and Mullins, 2019). Interestingly, STRAP, the nuclear partner of JMY, also exists in the cytosol to bind JMY and regulate autophagy. The interaction between STRAP and JMY potently inhibited the overall actin nucleation activity of JMY and counteracted the effect of LC3 (Hu and Mullins, 2019). In the nucleus, STRAP, JMY, and p300 form a complex to prevent JMY degradation by MDM2 and facilitate p53 activation (Demonacos et al., 2001). The interaction between STRAP and JMY was shown to involve N-terminal, central, and C-terminal sequences of JMY (Demonacos et al., 2001). It is plausible that the cytoplasmic STRAP inhibits the actin-nucleation activity of JMY by sequestering both its the N-terminal WJHD and C-terminal WWWCA (Figure 5B). It remains an open question exactly how LC3 and STRAP interact with JMY to regulate its activity and how the WJHD or the J-loop of JMY promotes actin polymerization.
It is worth noting that, although the dual function of JMY in the cytosol and the nucleus is a remarkable feature of JMY, other WASP-family proteins, actin, and Arp2/3 have also been shown to have important roles in the nucleus, either dependent on or independent of their activity in Arp2/3-mediated actin polymerization. Their nuclear functions are a separate topic of great interest and are not the focus of this review (Caridi et al., 2018; Kluge et al., 2018; Miyamoto et al., 2013; Schrank et al., 2018; Shikama et al., 1999; Taylor et al., 2010; Jeffrey M. Verboon et al., 2015; Weston et al., 2012; Wu et al., 2006).
The connection of JMY with human diseases is not yet established, although its role in augmenting the activity of the tumor suppressor p53 in response to DNA damage and its role in regulating cell migration suggests JMY could play a role in tumor invasiveness. In addition, JMY was found to be a candidate gene for susceptibility of ankylosing spondylitis in genome-wide analysis (Chai et al., 2013).
In conclusion, WHAMM and JMY are homologous proteins and form a distinct group of the WASP-family protein. They are not autoinhibited in the basal state as is seen for WASP/N-WASP, WAVEs, and WASH. They link the Arp2/3 complex to a diverse array of important functions in the cell that cover various endomembrane compartments, intracellular trafficking pathways, and the plasma membrane. Their activity is dynamically regulated in the cell, although currently very little is known on the cellular, biochemical, and structural levels about how the regulation is achieved.
WHIMP
The newest member of the WASP-family proteins, WHIMP, was identified in 2020 from a genomic sequence search for WH2-domain containing proteins (Kabrawala et al., 2020). WHIMP seems to be the least conserved of the WASP-family proteins, because even though it exists in many examined mammals it was not identified in the human genome. Furthermore, sequence conservation between different animals was also relatively low (Kabrawala et al., 2020). This suggests WHIMP may have appeared recently in evolution and may have special functions in animals. The N-terminal domain of WHIMP is distantly related to part of the WHD region of WAVE, which we herein name as WHD-like domain (WHDL) (Figure 1, left). WHIMP does not contain a PRR, which is found in all other WASP-family proteins. The activity of its C-terminal WCA sequence is relatively weak towards Arp2/3-mediated actin polymerization both in vitro and in cells (Kabrawala et al., 2020). WHIMP is ubiquitously expressed in mouse tissues with low expression in the brain. It is enriched in membrane ruffles and the leading edge of migrating cells, sharing the same localization with Arp2/3, N-WASP, and WAVE. WHIMP expression induced formation of large, highly dynamic membrane ruffles and non-selective micropinocytosis, promoted cell migration, and increased Src-mediated phosphorylation at membranes (Kabrawala et al., 2020). The increased membrane ruffling was dependent on the WHIMP WCA, Arp2/3, and Rac1-WRC signaling. Increased Src phosphorylation is believed to provide positive feedback to facilitate Rac1-WRC and/or Cdc42-N-WASP signaling to enhance actin polymerization and membrane protrusions (Kabrawala et al., 2020). It remains uncertain whether WHIMP also exists in humans and how its activity is regulated in the cell.
Summary and Prospects
Since the initial discovery of WASP, two decades of work has now expanded this protein family to 9 members in mammals and has greatly advanced our understanding of their cellular functions and regulation mechanisms (Alekhina et al., 2017; Padrick and Rosen, 2010; Takenawa and Suetsugu, 2007; Veltman and Insall, 2010). Even though all WASP-family proteins are able to use their C-terminal WCA domain to stimulate actin polymerization through the Arp2/3 complex, it is fascinating how they use various N-terminal sequences to regulate their cellular localization and biochemical activity and how these regulatory sequences allow them to channel a vast diversity of upstream signals to the remodeling of the actin cytoskeleton to serve different membrane-associated processes throughout the cell. The fundamental roles of WASP family proteins in various processes explain their broad involvement in many diseases. The fact that genetic mutations disrupting or enhancing their activity often lead to similar disease conditions emphasizes the need for precise control of their activity in cells.
Despite many major achievements in this field, important questions remain open, and new, exciting functions and regulatory mechanisms are rapidly emerging. One major knowledge gap is the understanding, or in many cases clarification, of the regulatory mechanisms of these proteins. Filling this gap will require a combination of advanced tools in gene editing, cell imaging, proteomics, in vitro reconstitution, quantitative biochemistry, and structural biology. The other major knowledge gap is the discovery of new functions and regulatory mechanisms, both in vitro and in different physiological processes in vivo. Advanced gene editing, such as conditional knock-out and knock-in in both cultured cells and live animals, is becoming a powerful tool to approach these physiological questions. Meanwhile, advances in large-scale clinical genomics and high-throughput, quantitative proteomics and interactomics will provide an unprecedented wealth of information for uncovering the structure-function mechanisms of disease-causing mutations and identifying new regulatory molecules. At the same time, structural bioinformatics and targeted drug design will in parallel promote the identification of novel regulatory molecules useful both in research and in medical interventions.
Due to space limit, this review is only focused on WASP-family proteins in mammals, particularly in humans, and only focused on their canonical functions in regulating the Arp2/3-mediated actin polymerization. Many new functions and mechanisms in other organisms, such as plants, or those independent of Arp2/3 or actin are also rapidly emerging (Ali et al., 2020; Chin et al., 2021; Facette et al., 2015; Gavrin et al., 2020; Miyamoto et al., 2013; Taylor et al., 2010; Weston et al., 2012). These exciting fields await rigorous cellular, biochemical, and structural studies to address many mechanistic questions.
Acknowledgements
B.C. was supported by funding from the National Institutes of Health (R35 GM128786 and R33 DA050837), the American Heart Association (19IPLOI34660134), and the National Science Foundation (CAREER MCB-2047640 and MCB-2148534).
References
- Abdul-Manan N, Aghazadeh B, Liu GA, Majumdar A, Ouerfelli O, Simlnovitch KA, Rosen MK, 1999. Structure of Cdc42 in complex with the GTPase-binding domain of the “Wiskott-Aldrich syndrome” protein. Nature 399, 379–383. 10.1038/20726 [DOI] [PubMed] [Google Scholar]
- Alekhina O, Burstein E, Billadeau DD, 2017. Cellular functions of WASP family proteins at a glance. J. Cell Sci 130, 2235–2241. 10.1242/jcs.199570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali MF, Fatema U, Peng X, Hacker SW, Maruyama D, Sun MX, Kawashima T, 2020. ARP2/3-independent WAVE/SCAR pathway and class XI myosin control sperm nuclear migration in flowering plants. Proc. Natl. Acad. Sci. U. S. A 117, 32757–32763. 10.1073/pnas.2015550117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkhairy OK, Rezaei N, Graham RR, Abolhassani H, Borte S, Hultenby K, Wu C, Aghamohammadi A, Williams DA, Behrens TW, Hammarström L, Pan-Hammarström Q, 2015. RAC2 loss-of-function mutation in 2 siblings with characteristics of common variable immunodeficiency. J. Allergy Clin. Immunol 135, 1380–1384.e5. 10.1016/j.jaci.2014.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida-Souza L, Frank RAW, García-Nafría J, Colussi A, Gunawardana N, Johnson CM, Yu M, Howard G, Andrews B, Vallis Y, McMahon HT, 2018. A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits. Cell 174, 325–337.e14. 10.1016/j.cell.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambruso DR, Knall C, Abell AN, Panepinto J, Kurkchubasche A, Thurman G, Gonzalez-Aller C, Hiester A, DeBoer M, Harbeck RJ, Oyer R, Johnson GL, Roos D, 2000. Human neutrophil immunodeficiency syndrome is associated with an inhibitory Rac2 mutation. Proc. Natl. Acad. Sci. U. S. A 97, 4654–4659. 10.1073/pnas.080074897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitei M, Stange C, Parshina I, Baust T, Schenck A, Raposo G, Kirchhausen T, Hoflack B, 2010. Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat. Cell Biol 12, 330–340. 10.1038/ncb2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antón IM, Jones GE, Wandosell F, Geha R, Ramesh N, 2007. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 17, 555–562. 10.1016/j.tcb.2007.08.005 [DOI] [PubMed] [Google Scholar]
- Ardern H, Sandilands E, Machesky LM, Timpson P, Frame MC, Brunton VG, 2006. Src-dependent phosphorylation of Scar1 promotes its association with the Arp2/3 complex. Cell Motil. Cytoskeleton 63, 6–13. 10.1002/cm.20101 [DOI] [PubMed] [Google Scholar]
- Assoum M, Bruel A-L, Crenshaw ML, Delanne J, Wentzensen IM, McWalter K, Dent KM, Vitobello A, Kuentz P, Thevenon J, Duffourd Y, Thauvin-Robinet C, Faivre L, 2020. Novel KIAA1033/WASHC4 mutations in three patients with syndromic intellectual disability and a review of the literature. Am. J. Med. Genet. A 182, 792–797. 10.1002/ajmg.a.61487 [DOI] [PubMed] [Google Scholar]
- Azevedo MM, Domingues HS, Cordelières FP, Sampaio P, Seixas AI, Relvas JB, 2018. Jmy regulates oligodendrocyte differentiation via modulation of actin cytoskeleton dynamics. Glia 66, 1826–1844. 10.1002/glia.23342 [DOI] [PubMed] [Google Scholar]
- Badour K, Zhang J, Shi F, Leng Y, Collins M, Siminovitch KA, 2004. Fyn and PTP-PEST-mediated Regulation of Wiskott-Aldrich Syndrome Protein (WASp) Tyrosine Phosphorylation Is Required for Coupling T Cell Antigen Receptor Engagement to WASp Effector Function and T Cell Activation. J. Exp. Med 199, 99–111. 10.1084/jem.20030976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banani SF, Lee HO, Hyman AA, Rosen MK, 2017. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol 18, 285–298. 10.1038/nrm.2017.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banin S, Truong O, Katz DR, Waterfield MD, Brickell PM, Gout I, 1996. Wiskott-Aldrich syndrome protein (WASp) is a binding partner for c-Src family protein-tyrosine kinases. Curr. Biol 6, 981–988. 10.1016/S0960-9822(02)00642-5 [DOI] [PubMed] [Google Scholar]
- Banjade S, Rosen MK, 2014. Phase transitions of multivalent proteins can promote clustering of membrane receptors. Elife 3, 1–24. 10.7554/eLife.04123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banjade S, Wu Q, Mittal A, Peeples WB, Pappu RV, Rosen MK, 2015. Conserved interdomain linker promotes phase separation of the multivalent adaptor protein Nck. Proc. Natl. Acad. Sci. U. S. A 112, E6426–E6435. 10.1073/pnas.1508778112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartuzi P, Billadeau DD, Favier R, Rong S, Dekker D, Fedoseienko A, Fieten H, Wijers M, Levels JH, Huijkman N, Kloosterhuis N, van der Molen H, Brufau G, Groen AK, Elliott AM, Kuivenhoven JA, Plecko B, Grangl G, McGaughran J, Horton JD, Burstein E, Hofker MH, van de Sluis B, 2016. CCC- and WASH-mediated endosomal sorting of LDLR is required for normal clearance of circulating LDL. Nat. Commun 7, 10961. 10.1038/ncomms10961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer NN, Chen YW, Samant RS, Shevde LA, Fodstad O, 2007. Rac1 activity regulates proliferation of aggressive metastatic melanoma. Exp. Cell Res 313, 3832–3839. 10.1016/j.yexcr.2007.08.017 [DOI] [PubMed] [Google Scholar]
- Bear JE, Rawls JF Iii, C. LS, 1998. SCAR, a WASP-related Protein, Isolated as a Suppressor of Receptor Defects in Late. Cell 142, 1325–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann A, Sticht H, Begtrup A, Vitobello A, Faivre L, Banka S, Alhaddad B, Asadollahi R, Becker J, Bierhals T, Brown KE, Bruel AL, Brunet T, Carneiro M, Cremer K, Day R, Denommé-Pichon AS, Dyment DA, Engels H, Fisher R, Goh ES, Hajianpour MJ, Haertel LRM, Hauer N, Hempel M, Herget T, Johannsen J, Kraus C, Le Guyader G, Lesca G, Mau-Them FT, McDermott JH, McWalter K, Meyer P, Õunap K, Popp B, Reimand T, Riedhammer KM, Russo M, Sadleir LG, Saenz M, Schiff M, Schuler E, Syrbe S, Van der Ven AT, Verloes A, Willems M, Zweier C, Steindl K, Zweier M, Rauch A, 2021. New insights into the clinical and molecular spectrum of the novel CYFIP2-related neurodevelopmental disorder and impairment of the WRC-mediated actin dynamics. Genet. Med 23, 543–554. 10.1038/s41436-020-01011-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yosef N, Frampton M, Schiff ER, Daher S, Abu Baker F, Safadi R, Israeli E, Segal AW, Levine AP, 2021. Genetic analysis of four consanguineous multiplex families with inflammatory bowel disease. Gastroenterol. Rep 9, 521–532. 10.1093/gastro/goab007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch S, Polo S, Lai FP, Anderson KI, Stradal TEB, Wehland J, Rottner K, 2005. N-WASP deficiency impairs EGF internalization and actin assembly at clathrin-coated pits. J. Cell Sci 118, 3103–3115. 10.1242/jcs.02444 [DOI] [PubMed] [Google Scholar]
- Bhattacharya K, Swaminathan K, Peche VS, Clemen CS, Knyphausen P, Lammers M, Noegel AA, Rastetter RH, 2016. Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology. Sci. Rep 6, 25411. 10.1038/srep25411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biber G, Ben-Shmuel A, Sabag B, Barda-Saad M, 2020. Actin regulators in cancer progression and metastases: From structure and function to cytoskeletal dynamics, 1st ed, International Review of Cell and Molecular Biology. Elsevier Inc. 10.1016/bs.ircmb.2020.05.006 [DOI] [PubMed] [Google Scholar]
- Biber G, Ben-Shmuel A, Sabag B, Barda-Saad M, 2020. Actin regulators in cancer progression and metastases: From structure and function to cytoskeletal dynamics. Int. Rev. Cell Mol. Biol 356. [DOI] [PubMed] [Google Scholar]
- Biondini M, Sadou-Dubourgnoux A, Paul-Gilloteaux P, Zago G, Arslanhan MD, Waharte F, Formstecher E, Hertzog M, Yu J, Guerois R, Gautreau A, Scita G, Camonis J, Parrini MC, 2016. Direct interaction between exocyst and Wave complexes promotes cell protrusions and motility. J. Cell Sci 129, 3756–3769. 10.1242/jcs.187336 [DOI] [PubMed] [Google Scholar]
- Blagg SL, Stewart M, Sambles C, Insall RH, 2003. PIR121 Regulates Pseudopod Dynamics and SCAR Activity in Dictyostelium. Curr. Biol 13, 1480–1487. 10.1016/S [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Amann KJ, Higgs HN, Marchand J, Kaiser DA, Pollard TD, 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins 171, 1007–1011. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J, 2014. Actin dynamics, architecture, and mechanics in cell motility. Physiol. Rev 94, 235–263. 10.1152/physrev.00018.2013 [DOI] [PubMed] [Google Scholar]
- Blundell MP, Worth A, Bouma G, Thrasher AJ, 2010. The Wiskott-Aldrich syndrome: The actin cytoskeleton and immune cell function. Dis. Markers 29, 157–175. 10.3233/DMA-2010-0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonfim-Melo A, Ferreira ÉR, Mortara RA, 2018. Rac1/WAVE2 and Cdc42/N-WASP participation in actin-dependent host cell invasion by extracellular amastigotes of Trypanosoma cruzi. Front. Microbiol 9, 1–14. 10.3389/fmicb.2018.00360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu W, Chou AM, Lim KB, Sudhaharan T, Ahmed S, 2009. The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem 284, 11622–11636. 10.1074/jbc.M805940200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckley CM, Gopaldass N, Bosmani C, Johnston SA, Soldati T, Insall RH, King JS, 2016. WASH drives early recycling from macropinosomes and phagosomes to maintain surface phagocytic receptors. Proc. Natl. Acad. Sci 113, E5906–E5915. 10.1073/pnas.1524532113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnell SC, Henry PA, Kolluri R, Kirchhausen T, Rickles RJ, Berg LJ, 1996. Identification of Itk/Tsk Src homology 3 domain ligands. J. Biol. Chem 271, 25646–25656. 10.1074/jbc.271.41.25646 [DOI] [PubMed] [Google Scholar]
- Burns S, Cory GO, Vainchenker W, Thrasher AJ, 2004. Mechanisms of WASp-mediated hematologic and immunologic disease. Blood 104, 3454–3462. 10.1182/blood-2004-04-1678 [DOI] [PubMed] [Google Scholar]
- Burroughs LM, Petrovic A, Brazauskas R, Liu X, Griffith LM, Ochs HD, Bleesing JJ, Edwards S, Dvorak CC, Chaudhury S, Prockop SE, Quinones R, Goldman FD, Quigg TC, Chandrakasan S, Smith AR, Parikh S, Dávila Saldaña BJ, Thakar MS, Phelan R, Shenoy S, Forbes LR, Martinez C, Chellapandian D, Shereck E, Miller HK, Kapoor N, Barnum JL, Chong H, Shyr DC, Chen K, Abu-Arja R, Shah AJ, Weinacht KG, Moore TB, Joshi A, DeSantes KB, Gillio AP, Cuvelier GDE, Keller MD, Rozmus J, Torgerson T, Pulsipher MA, Haddad E, Sullivan KE, Logan BR, Kohn DB, Puck JM, Notarangelo LD, Pai SY, Rawlings DJ, Cowan MJ, 2020. Excellent outcomes following hematopoietic cell transplantation for Wiskott-Aldrich syndrome: A PIDTC report. Blood 135, 2094–2105. 10.1182/blood.2019002939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai GQ, Chou CF, Hu M, Zheng A, Reichardt LF, Guan JL, Fang H, Luckhardt TR, Zhou Y, Thannickal VJ, Ding Q, 2012. Neuronal Wiskott-Aldrich syndrome protein (N-WASP) is critical for formation of α-smooth muscle action filaments during myofibroblast differentiation. Am. J. Physiol. - Lung Cell. Mol. Physiol 303, 692–702. 10.1152/ajplung.00390.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvez R, Lafouresse F, de Meester J, Galy A, Valitutti S, Dupré L, 2011. The wiskott-aldrich syndrome protein permits assembly of a focused immunological synapse enabling sustained T-cell receptor signaling. Haematologica 96, 1415–1423. 10.3324/haematol.2011.040204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Cheng HC, Robbins D, Siripala AD, McGhie EJ, Hayward RD, Welch MD, Rosen MK, Koronakis V, Leong JM, 2008a. Repetitive N-WASP-binding elements of the enterohemorrhagic Escherichia coli effector EspFU synergistically activate actin assembly. PLoS Pathog. 4. 10.1371/journal.ppat.1000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Robbins D, Leong JM, 2004. EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7, 217–228. 10.1016/j.devcel.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Campellone KG, Webb NJ, Znameroski EA, Welch MD, 2008b. WHAMM Is an Arp2/3 Complex Activator That Binds Microtubules and Functions in ER to Golgi Transport. Cell 134, 148–161. 10.1016/j.cell.2008.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo RA, Dooley CA, Grieshaber SS, Hackstadt T, 2007. Rac interacts with Abi-1 and WAVE2 to promote an Arp2/3-dependent actin recruitment during chlamydial invasion. Cell. Microbiol 9, 2278–2288. 10.1111/j.1462-5822.2007.00958.x [DOI] [PubMed] [Google Scholar]
- Caridi CP, D’agostino C, Ryu T, Zapotoczny G, Delabaere L, Li X, Khodaverdian VY, Amaral N, Lin E, Rau AR, Chiolo I, 2018. Nuclear F-actin and myosins drive relocalization of heterochromatic breaks. Nature 559, 54–60. 10.1038/s41586-018-0242-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Nioche P, Broutin-L’hermite I, Boujemaa R, Le Clainche C, Egile C, Garbay C, Ducruix A, Sansonetti P, Pantaloni D, 2000. GRB2 links signaling to actin assembly by enhancing interaction of neural wiskott-aldrich syndrome protein (N-WASp) with actin-related protein (ARP2/3) complex. J. Biol. Chem 275, 21946–21952. 10.1074/jbc.M000687200 [DOI] [PubMed] [Google Scholar]
- Carmona G, Perera U, Gillett C, Naba A, Law AL, Sharma VP, Wang J, Wyckoff J, Balsamo M, Mosis F, De Piano M, Monypenny J, Woodman N, McConnell RE, Mouneimne G, Van Hemelrijck M, Cao Y, Condeelis J, Hynes RO, Gertler FB, Krause M, 2016. Lamellipodin promotes invasive 3D cancer cell migration via regulated interactions with Ena/VASP and SCAR/WAVE. Oncogene 35, 5155–5169. 10.1038/onc.2016.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnell M, Zech T, Calaminus SD, Ura S, Hagedorn M, Johnston SA, May RC, Soldati T, Machesky LM, Insall RH, 2011. Actin polymerization driven by WASH causes V-ATPase retrieval and vesicle neutralization before exocytosis. J. Cell Biol 193, 831–839. 10.1083/jcb.201009119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Ditlev JA, Rosen MK, 2019a. Regulation of Transmembrane Signaling by Phase Separation. Annu. Rev. Biophys 48, 465–494. 10.1146/annurev-biophys-052118-115534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case LB, Zhang X, Ditlev JA, Rosen MK, 2019b. Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science (80-.) 363, 1093–1097. 10.1126/science.aau6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro CN, Rosenzwajg M, Carapito R, Shahrooei M, Konantz M, Khan A, Miao Z, Groß M, Tranchant T, Radosavljevic M, Paul N, Stemmelen T, Pitoiset F, Hirschler A, Nespola B, Molitor A, Rolli V, Pichot A, Faletti LE, Rinaldi B, Friant S, Mednikov M, Karauzum H, Aman MJ, Carapito C, Lengerke C, Ziaee V, Eyaid W, Ehl S, Alroqi F, Parvaneh N, Bahram S, 2020. NCKAP1L defects lead to a novel syndrome combining immunodeficiency, lymphoproliferation, and hyperinflammation. J. Exp. Med 217. 10.1084/JEM.20192275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceglia I, Reitz C, Gresack J, Ahn JH, Bustos V, Bleck M, Zhang X, Martin G, Simon SM, Nairn AC, Greengard P, Kim Y, 2015. APP intracellular domain-WAVE1 pathway reduces amyloid-β production. Nat. Med 21, 1054–1059. 10.1038/nm.3924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai W, Lian Z, Chen C, Liu J, Shi LL, Wang Y, 2013. JARID1A, JMY, and PTGER4 Polymorphisms Are Related to Ankylosing Spondylitis in Chinese Han Patients: A Case-Control Study. PLoS One 8, 1–8. 10.1371/journal.pone.0074794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra M, Collins BM, 2019. The Phox Homology (PX) Domain BT - Protein Reviews – Purinergic Receptors: Volume 20, in: Atassi MZ (Ed.), . Springer International Publishing, Cham, pp. 1–17. 10.1007/5584_2018_185 [DOI] [Google Scholar]
- Chaudhari K, Gorla M, Chang C, Kania A, Bashaw GJ, 2021. Robo recruitment of the wave regulatory complex plays an essential and conserved role in midline repulsion. Elife 10, 1–35. 10.7554/ELIFE.64474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK, 2014a. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195–207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Brinkmann K, Chen Z, Pak CW, Liao Y, Shi S, Henry L, Grishin NV, Bogdan S, Rosen MK, 2014b. The WAVE regulatory complex links diverse receptors to the actin cytoskeleton. Cell 156, 195–207. 10.1016/j.cell.2013.11.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Chou H-T, Brautigam CA, Xing W, Yang S, Henry L, Doolittle LK, Walz T, Rosen MK, 2017. Rac1 GTPase activates the WAVE regulatory complex through two distinct binding sites. Elife 6, 1–22. 10.7554/elife.29795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Squarr AJ, Stephan R, Chen B, Higgins TE, Barry DJ, Martin MC, Rosen MK, Bogdan S, Way M, 2014. Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton. Dev. Cell 30, 569–584. 10.1016/j.devcel.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK, 2010a. Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Borek D, Padrick SB, Gomez TS, Metlagel Z, Ismail AM, Umetani J, Billadeau DD, Otwinowski Z, Rosen MK, 2010b. Structure and control of the actin regulatory WAVE complex. Nature 468, 533–538. 10.1038/nature09623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng A, Arumugam TV, Liu D, Khatri RG, Mustafa K, Kwak S, Ling HP, Gonzales C, Xin O, Jo DG, Guo Z, Mark RJ, Mattson MP, 2007. Pancortin-2 interacts with WAVE1 and Bcl-xL in a mitochondria-associated protein complex that mediates ischemic neuronal death. J. Neurosci 27, 1519–1528. 10.1523/JNEUROSCI.5154-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HC, Skehan BM, Campellone KG, Leong JM, Rosen MK, 2008. Structural mechanism of WASP activation by the enterohaemorrhagic E. coli effector EspFU. Nature 454, 1009–1013. 10.1038/nature07160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia PH, Chen B, Li P, Rosen MK, Shen K, 2014. Local F-actin network links synapse formation and axon branching. Cell 156, 208–220. 10.1016/j.cell.2013.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin S, Kwon T, Khan BR, Alan Sparks J, Mallery EL, Szymanski DB, Blancaflor EB, 2021. Spatial and temporal localization of SPIRRIG and WAVE/SCAR reveal roles for these proteins in actin-mediated root hair development. Plant Cell 33, 2131–2148. 10.1093/plcell/koab115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemen CS, Schmidt A, Winter L, Canneva F, Wittig I, Becker L, Coras R, Berwanger C, Hofmann A, Eggers B, Marcus K, Gailus-Durner V, Fuchs H, de Angelis MH, Krüger M, von Hörsten S, Eichinger L, Schröder R, 2021. N471D WASH complex subunit strumpellin knock-in mice display mild motor and cardiac abnormalities and BPTF and KLHL11 dysregulation in brain tissue. Neuropathol. Appl. Neurobiol 10.1111/nan.12750 [DOI] [PubMed] [Google Scholar]
- Conway OJ, Carrasquillo MM, Wang X, Bredenberg JM, Reddy JS, Strickland SL, Younkin CS, Burgess JD, Allen M, Lincoln SJ, Nguyen T, Malphrus KG, Soto AI, Walton RL, Boeve BF, Petersen RC, Lucas JA, Ferman TJ, Cheshire WP, van Gerpen JA, Uitti RJ, Wszolek ZK, Ross OA, Dickson DW, Graff-Radford NR, Ertekin-Taner N, 2018. ABI3 and PLCG2 missense variants as risk factors for neurodegenerative diseases in Caucasians and African Americans. Mol. Neurodegener 13, 1–12. 10.1186/s13024-018-0289-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook SA, Comrie WA, Poli MC, Similuk M, Oler AJ, Faruqi AJ, Kuhns DB, Yang S, Vargas-Hernández A, Carisey AF, Fournier B, Anderson DE, Price S, Smelkinson M, Chahla WA, Forbes LR, Mace EM, Cao TN, Coban-Akdemir ZH, Jhangiani SN, Muzny DM, Gibbs RA, Lupski JR, Orange JS, Cuvelier GDE, Al Hassani M, Al Kaabi N, Al Yafei Z, Jyonouchi S, Raje N, Caldwell JW, Huang Y, Burkhardt JK, Latour S, Chen B, El Ghazali G, Rao VK, Chinn IK, Lenardo MJ, 2020. HEM1 deficiency disrupts mTORC2 and F-actin control in inherited immunodysregulatory disease. Science (80-.) 369, 202–207. 10.1126/science.aay5663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Serrano JC, Lamers IJC, Van Reeuwijk J, Duijkers L, Hoogendoorn ADM, Yildirim A, Argyrou N, Ruigrok RAA, Letteboer SJF, Butcher R, Van Essen MD, Sakami S, Van Beersum SEC, Palczewski K, Cheetham ME, Liu Q, Boldt K, Wolfrum U, Ueffing M, Garanto A, Roepman R, Collin RWJ, 2020. PCARE and WASF3 regulate ciliary F-actin assembly that is required for the initiation of photoreceptor outer segment disk formation. Proc. Natl. Acad. Sci. U. S. A 117, 9922–9931. 10.1073/pnas.1903125117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory GOC, Cramer R, Blanchoin L, Ridley AJ, 2003. Phosphorylation of the WASP-VCA domain increases its affinity for the Arp2/3 complex and enhances actin polymerization by WASP. Mol. Cell 11, 1229–1239. 10.1016/S1097-2765(03)00172-2 [DOI] [PubMed] [Google Scholar]
- Cory GOC, Garg R, Cramer R, Ridley AJ, 2002. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. J. Biol. Chem 277, 45115–45121. 10.1074/jbc.M203346200 [DOI] [PubMed] [Google Scholar]
- Cossart P, Bierne H, 2001. Host cell machinery and Listeria monocytogenes pathogenesis. Curr. Opin. Immunol 13, 96–103. [DOI] [PubMed] [Google Scholar]
- Costain G, Callewaert B, Gabriel H, Tan TY, Walker S, Christodoulou J, Lazar T, Menten B, Orkin J, Sadedin S, Snell M, Vanlander A, Vergult S, White SM, Scherer SW, Hayeems RZ, Blaser S, Wodak SJ, Chitayat D, Marshall CR, Meyn MS, 2019. De novo missense variants in RAC3 cause a novel neurodevelopmental syndrome. Genet. Med 21, 1021–1026. 10.1038/s41436-018-0323-y [DOI] [PubMed] [Google Scholar]
- Courtland JL, Bradshaw TW, Waitt G, Soderblom EJ, Ho T, Rajab A, Vancini R, Kim IH, Soderling SH, 2021. Genetic disruption of WASHC4 drives endo-lysosomal dysfunction and cognitive-movement impairments in mice and humans. Elife 10. 10.7554/eLife.61590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, La Thangue NB, 2015. Actin nucleation by WH2 domains at the autophagosome. Nat. Commun 6. 10.1038/ncomms8888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutts AS, Weston L, La Thangue NB, 2009. A transcription co-factor integrates cell adhesion and motility with the p53 response. Proc. Natl. Acad. Sci. U. S. A 106, 19872–19877. 10.1073/pnas.0906785106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell JK, Teng Y, Bendzunas NG, Ara R, Arbab AS, Kennedy EJ, 2017. Suppression of Breast Cancer Metastasis Using Stapled Peptides Targeting the WASF Regulatory Complex. Cancer Growth Metastasis 10, 117906441771319. 10.1177/1179064417713197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai A, Yu L, Wang HW, 2019. WHAMM initiates autolysosome tubulation by promoting actin polymerization on autolysosomes. Nat. Commun 10. 10.1038/s41467-019-11694-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z, Pendergast AM, 1995. Abi-2, a novel SH3-containing protein interacts with the c-Abl tyrosine kinase and modulates c-Abl transforming activity. Genes Dev. 9, 2569–2582. 10.1101/gad.9.21.2569 [DOI] [PubMed] [Google Scholar]
- Danson CM, Pocha SM, Bloomberg GB, Cory GO, 2007. Phosphorylation of WAVE2 by MAP kinases regulates persistent cell migration and polarity. J. Cell Sci 120, 4144–4154. 10.1242/jcs.013714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bot ST, Vermeer S, Buijsman W, Heister A, Voorendt M, Verrips A, Scheffer H, Kremer HPH, van de Warrenburg BPC, Kamsteeg E-J, 2013. Pure adult-onset spastic paraplegia caused by a novel mutation in the KIAA0196 (SPG8) gene. J. Neurol 260, 1765–1769. 10.1007/s00415-013-6870-x [DOI] [PubMed] [Google Scholar]
- De Meester J, Calvez R, Valitutti S, Dupré L, 2010. The Wiskott-Aldrich syndrome protein regulates CTL cytotoxicity and is required for efficient killing of B cell lymphoma targets. J. Leukoc. Biol 88, 1031–1040. 10.1189/jlb.0410197 [DOI] [PubMed] [Google Scholar]
- Del Olmo T, Lauzier A, Normandin C, Larcher R, Lecours M, Jean D, Lessard L, Steinberg F, Boisvert F-M, Jean S, 2019. APEX2-mediated RAB proximity labeling identifies a role for RAB21 in clathrin-independent cargo sorting. EMBO Rep. 20. 10.15252/embr.201847192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demonacos C, Krstic-Demonacos M, La Thangue NB, 2001. A TPR motif cofactor contributes to p300 activity in the p53 response. Mol. Cell 8, 71–84. 10.1016/S1097-2765(01)00277-5 [DOI] [PubMed] [Google Scholar]
- Deng Z-H, Gomez TS, Osborne DG, Phillips-Krawczak CA, Zhang J-S, Billadeau DD, 2015. Nuclear FAM21 participates in NF-κB-dependent gene regulation in pancreatic cancer cells. J. Cell Sci 128, 373–384. 10.1242/jcs.161513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depienne C, Cincotta M, Billot S, Bouteiller D, Groppa S, Brochard V, Flamand C, Hubsch C, Meunier S, Giovannelli F, Klebe S, Corvol JC, Vidailhet M, Brice A, Roze E, 2011. A novel DCC mutation and genetic heterogeneity in congenital mirror movements. Neurology 76, 260 LP–264. 10.1212/WNL.0b013e318207b1e0 [DOI] [PubMed] [Google Scholar]
- Depienne C, Leguern E, 2012. PCDH19-related infantile epileptic encephalopathy: An unusual X-linked inheritance disorder. Hum. Mutat 33, 627–634. 10.1002/humu.22029 [DOI] [PubMed] [Google Scholar]
- Derivery E, Helfer E, Henriot V, Gautreau A, 2012. Actin polymerization controls the organization of WASH domains at the surface of endosomes. PLoS One 7, e39774. 10.1371/journal.pone.0039774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derivery E, Lombard B, Loew D, Gautreau A, 2009a. The wave complex is intrinsically inactive. Cell Motil. Cytoskeleton 66, 777–790. 10.1002/cm.20342 [DOI] [PubMed] [Google Scholar]
- Derivery E, Sousa C, Gautier JJ, Lombard B, Loew D, Gautreau A, 2009b. The Arp2/3 Activator WASH Controls the Fission of Endosomes through a Large Multiprotein Complex. Dev. Cell 17, 712–723. 10.1016/j.devcel.2009.09.010 [DOI] [PubMed] [Google Scholar]
- Derry JM, Ochs HD, Francke U, 1994. Isolation of a novel gene mutated in Wiskott-Aldrich syndrome. Cell 78, 635–644. [DOI] [PubMed] [Google Scholar]
- DesMarais V, Yamaguchi H, Oser M, Soon L, Mouneimne G, Sarmiento C, Eddy R, Condeelis J, 2009. N-WASP and cortactin are involved in invadopodium-dependent chemotaxis to EGF in breast tumor cells. Cell Motil. Cytoskeleton 66, 303–316. 10.1002/cm.20361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Kim AS, Mathijs G, Frints SGM, Schwartz M, Van Den Oord JJ, Verhoef GEG, Boogaerts MA, Fryns JP, You D, Rosen MK, Vandenberghe P, 2001. Constitutively activating mutation in WASP causes X-linked severe congenital neutropenia. Nat. Genet 27, 313–317. 10.1038/85886 [DOI] [PubMed] [Google Scholar]
- Ditlev JA, Vega AR, Köster DV, Su X, Tani T, Lakoduk AM, Vale RD, Mayor S, Jaqaman K, Rosen MK, 2019. A composition-dependent molecular clutch between T cell signaling condensates and actin. Elife 8, 1–44. 10.7554/eLife.42695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokmanovic M, Hirsch DS, Shen Y, Wen JW, 2009. Rac1 contributes to trastuzumab resistance of breast cancer cells: Rac1 as a potential therapeutic target for the treatment of trastuzumab-resistant breast cancer. Mol. Cancer Ther 8, 1557–1569. 10.1158/1535-7163.MCT-09-0140 [DOI] [PubMed] [Google Scholar]
- Dominguez R, 2016. The WH2 Domain and Actin Nucleation-Necessary but Insufficient. Trends Biochem Sci 41, 478–490. 10.1016/j.tibs.2016.03.004.The [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL, 2011. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Biol 12, 362–375. 10.1038/nrm3117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong B, Kakihara K, Otani T, Wada H, Hayashi S, 2013. Rab9 and retromer regulate retrograde trafficking of luminal protein required for epithelial tube length control. Nat. Commun 4, 1358. 10.1038/ncomms2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duleh SN, Welch MD, 2010. WASH and the Arp2/3 complex regulate endosome shape and trafficking. Cytoskeleton (Hoboken). 67, 193–206. 10.1002/cm.20437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupré L, Aiuti A, Trifari S, Martino S, Saracco P, Bordignon C, Roncarolo MG, 2002. Wiskott-Aldrich syndrome protein regulates lipid raft dynamics during immunological synapse formation. Immunity 17, 157–166. 10.1016/S1074-7613(02)00360-6 [DOI] [PubMed] [Google Scholar]
- Echarri A, Lai MJ, Robinson MR, Pendergast AM, 2004. Abl Interactor 1 (Abi-1) Wave-Binding and SNARE Domains Regulate Its Nucleocytoplasmic Shuttling, Lamellipodium Localization, and Wave-1 Levels. Mol. Cell. Biol 24, 4979–4993. 10.1128/mcb.24.11.4979-4993.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eden S, Rohatgi R, Podtelejnikov AV, Mann M, Kirschner MW, 2002. Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793. 10.1038/nature00859 [DOI] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, Li R, Pantaloni D, Sansonetti PJ, Carlier M, 1999. Protein Promotes Actin Nucleation by Arp2 / 3 Complex and Bacterial. J. Cell Biol 146, 1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott AM, Simard LR, Coghlan G, Chudley AE, Chodirker BN, Greenberg CR, Burch T, Ly V, Hatch GM, Zelinski T, 2013. A novel mutation in KIAA0196: Identification of a gene involved in Ritscher-Schinzel/3C syndrome in a First Nations cohort. J. Med. Genet 50, 819–822. 10.1136/jmedgenet-2013-101715 [DOI] [PubMed] [Google Scholar]
- Facchetti F, Blanzuoli L, Vermi W, Notarangelo LD, Giliani S, Fiorini M, Fasth A, Stewart DM, Nelson DL, 1998. Defective actin polymerization in EBV-transformed B-cell lines from patients with the Wiscott-Aldrich syndrome. J. Pathol 185, 99–107. [DOI] [PubMed] [Google Scholar]
- Facette MR, Park Y, Sutimantanapi D, Luo A, Cartwright HN, Yang B, Bennett EJ, Sylvester AW, Smith LG, 2015. The SCAR/WAVE complex polarizes PAN receptors and promotes division asymmetry in maize. Nat. Plants 1. 10.1038/nplants.2014.24 [DOI] [PubMed] [Google Scholar]
- Falcone S, Roman W, Hnia K, Gache V, Didier N, Lainé J, Auradé F, Marty I, Nishino I, Charlet‐Berguerand N, Romero NB, Marazzi G, Sassoon D, Laporte J, Gomes ER, 2014. N‐ WASP is required for Amphiphysin - 2/ BIN 1 - dependent nuclear positioning and triad organization in skeletal muscle and is involved in the pathophysiology of centronuclear myopathy. EMBO Mol. Med 6, 1455–1475. 10.15252/emmm.201404436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Lu Y, Shen X, Shao H, Suo L, Wu Q, 2018. Alpha protocadherins and Pyk2 kinase regulate cortical neuron migration and cytoskeletal dynamics via rac1 GTPase and WAVE complex in mice. Elife 7, 1–26. 10.7554/eLife.35242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris R, McCullough A, Andersen SE, Moninger TO, Weber MM, 2020. The chlamydia trachomatis secreted effector TmeA hijacks the N-WASP-ARP2/3 actin remodeling axis to facilitate cellular invasion. PLoS Pathog. 16, 1–20. 10.1371/journal.ppat.1008878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finan PM, Soames CJ, Wilson L, Nelson DL, Stewart DM, Truong O, Hsuan JJ, Kellie S, 1996. Identification of regions of the Wiskott-Aldrich syndrome protein responsible for association with selected Src homology 3 domains. J. Biol. Chem 271, 26291–26295. 10.1074/jbc.271.42.26291 [DOI] [PubMed] [Google Scholar]
- Firat-Karalar EN, Hsiue PP, Welch MD, 2011. The actin nucleation factor JMY is a negative regulator of neuritogenesis. Mol. Biol. Cell 22, 4563–4574. 10.1091/mbc.E11-06-0585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin AI, David V, Oguievetskaia K, Derivery E, Stone CE, Cao L, Rocques N, Molinie N, Henriot V, Aumont-Nicaise M, Hinckelmann M-V, Saudou F, Le Clainche C, Carter AP, Romet-Lemonne G, Gautreau AM, 2021. The Arp1/11 minifilament of dynactin primes the endosomal Arp2/3 complex. Sci. Adv 7. 10.1126/sciadv.abd5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fokin AI, Gautreau AM, 2021. Assembly and Activity of the WASH Molecular Machine: Distinctive Features at the Crossroads of the Actin and Microtubule Cytoskeletons. Front. cell Dev. Biol 9, 658865. 10.3389/fcell.2021.658865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort L, Batista JM, Thomason PA, Spence HJ, Whitelaw JA, Tweedy L, Greaves J, Martin KJ, Anderson KI, Brown P, Lilla S, Neilson MP, Tafelmeyer P, Zanivan S, Ismail S, Bryant DM, Tomkinson NCO, Chamberlain LH, Mastick GS, Insall RH, Machesky LM, 2018. Fam49/CYRI interacts with Rac1 and locally suppresses protrusions. Nat. Cell Biol 20, 1159–1171. 10.1038/s41556-018-0198-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman C, Seaman MNJ, Reid E, 2013. The hereditary spastic paraplegia protein strumpellin: characterisation in neurons and of the effect of disease mutations on WASH complex assembly and function. Biochim. Biophys. Acta 1832, 160–173. 10.1016/j.bbadis.2012.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman CL, Hesketh G, Seaman MNJ, 2014. RME-8 coordinates the activity of the WASH complex with the function of the retromer SNX dimer to control endosomal tubulation. J. Cell Sci 127, 2053–2070. 10.1242/jcs.144659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht F, Moreau V, Röttger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M, 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401, 926–929. 10.1038/44860 [DOI] [PubMed] [Google Scholar]
- Frugtniet BA, Martin TA, Zhang L, Jiang WG, 2017. Neural Wiskott-Aldrich syndrome protein (nWASP) is implicated in human lung cancer invasion. BMC Cancer 17, 1–11. 10.1186/s12885-017-3219-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuoka M, Suetsugu S, Miki H, Fukami K, Endo T, Takenawa T, 2001. A novel neural Wiskott-Aldrich syndrome protein (N-WASP) binding protein, WISH, induces Arp2/3 complex activation independent of CDc42. J. Cell Biol 153, 471–482. 10.1083/jcb.152.3.471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gad AKB, Nehru V, Ruusala A, Aspenström P, 2012. RhoD regulates cytoskeletal dynamics via the actin nucleation-promoting factor WASp homologue associated with actin Golgi membranes and microtubules. Mol. Biol. Cell 23, 4807–4819. 10.1091/mbc.E12-07-0555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego MD, Santamaría M, Peña J, Molina IJ, 1997. Defective actin reorganization and polymerization of Wiskott-Aldrich T cells in response to CD3-mediated stimulation. Blood 90, 3089–3097. 10.1182/blood.v90.8.3089 [DOI] [PubMed] [Google Scholar]
- Gangfuß A, Czech A, Hentschel A, Münchberg U, Horvath R, Töpf A, O’Heir E, Lochmüller H, Stehling F, Kiewert C, Sickmann A, Kuechler A, Kaiser FJ, Kölbel H, Christiansen J, Schara-Schmidt U, Roos A, 2022. Homozygous WASHC4 variant in two sisters causes a syndromic phenotype defined by dysmorphisms, intellectual disability, profound developmental disorder, and skeletal muscle involvement. J. Pathol 256, 93–107. 10.1002/path.5812 [DOI] [PubMed] [Google Scholar]
- Gautreau A, Ho H. -y. H., Li J, Steen H, Gygi SP, Kirschner MW, 2004. Purification and architecture of the ubiquitous Wave complex. Proc. Natl. Acad. Sci 101, 4379–4383. 10.1073/pnas.0400628101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrin A, Rey T, Torode TA, Toulotte J, Chatterjee A, Kaplan JL, Evangelisti E, Takagi H, Charoensawan V, Rengel D, Journet EP, Debellé F, de Carvalho-Niebel F, Terauchi R, Braybrook S, Schornack S, 2020. Developmental Modulation of Root Cell Wall Architecture Confers Resistance to an Oomycete Pathogen. Curr. Biol 30, 4165–4176.e5. 10.1016/j.cub.2020.08.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianfrancesco F, Falco G, Esposito T, Rocchi M, D’Urso M, 2001. Characterization of the murine orthologue of a novel human subtelomeric multigene family. Cytogenet. Cell Genet 94, 98–100. 10.1159/000048796 [DOI] [PubMed] [Google Scholar]
- Ginanneschi F, D’Amore A, Barghigiani M, Tessa A, Rossi A, Santorelli FM, 2020. SPG8 mutations in Italian families: clinical data and literature review. Neurol. Sci 41, 699–703. 10.1007/s10072-019-04180-z [DOI] [PubMed] [Google Scholar]
- Gismondi A, Cifaldi L, Mazza C, Giliani S, Parolini S, Morrone S, Jacobelli J, Bandiera E, Notarangelo L, Santoni A, 2004. Impaired natural and CD16-mediated NK cell cytotoxicity in patients with WAS and XLT: Ability of IL-2 to correct NK cell functional defect. Blood 104, 436–443. 10.1182/blood-2003-07-2621 [DOI] [PubMed] [Google Scholar]
- Gjerulfsen CE, Møller RS, Fenger CD, Hammer TB, Bayat A, 2021. Expansion of the CCDC22 associated Ritscher-Schinzel/3C syndrome and review of the literature: Should the minimal diagnostic criteria be revised? Eur. J. Med. Genet 64, 104246. 10.1016/j.ejmg.2021.104246 [DOI] [PubMed] [Google Scholar]
- Gligorijevic B, Wyckoff J, Yamaguchi H, Wang Y, Roussos ET, Condeelis J, 2012. N-WASP-mediated invadopodium formation is involved in intravasation and lung metastasis of mammary tumors. J. Cell Sci 125, 724–734. 10.1242/jcs.092726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goley ED, Welch MD, 2006. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol 7, 713–726. 10.1038/nrm2026 [DOI] [PubMed] [Google Scholar]
- Gomez TS, Billadeau DD, 2009. A FAM21-containing WASH complex regulates retromer-dependent sorting. Dev. Cell 17, 699–711. 10.1016/j.devcel.2009.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez TS, Gorman JA, de Narvajas AA-M, Koenig AO, Billadeau DD, 2012. Trafficking defects in WASH-knockout fibroblasts originate from collapsed endosomal and lysosomal networks. Mol. Biol. Cell 23, 3215–3228. 10.1091/mbc.E12-02-0101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromnitza S, Lepa C, Weide T, Schwab A, Pavenstädt H, George B, 2018. Tropomyosin-related kinase C (TrkC) enhances podocyte migration by ERK-mediated WAVE2 activation. FASEB J. 32, 1665–1676. 10.1096/fj.201700703R [DOI] [PubMed] [Google Scholar]
- Guerriero CJ, Weisz OA, 2007. N-WASP inhibitor wiskostatin nonselectively perturbs membrane transport by decreasing cellular ATP levels. Am. J. Physiol. - Cell Physiol 292, 1562–1566. 10.1152/ajpcell.00426.2006 [DOI] [PubMed] [Google Scholar]
- Guinamard R, Aspenström P, Fougereau M, Chavrier P, Guillemot JC, 1998. Tyrosine phosphorylation of the Wiskott-Aldrich Syndrome protein by Lyn and Btk is regulated by CDC42. FEBS Lett. 434, 431–436. 10.1016/S0014-5793(98)01016-3 [DOI] [PubMed] [Google Scholar]
- Guo JC, Li J, Zhao YP, Zhou L, Cui QC, Zhou WX, Zhang TP, You L, Shu H, 2014. N-WASP in pancreatic ductal adenocarcinoma: Associations with perineural invasion and poor prognosis. World J. Surg 38, 2126–2131. 10.1007/s00268-014-2500-8 [DOI] [PubMed] [Google Scholar]
- Hansen M, Hsu A-L, Dillin A, Kenyon C, 2005. New genes tied to endocrine, metabolic, and dietary regulation of lifespan from a Caenorhabditis elegans genomic RNAi screen. PLoS Genet. 1, 119–128. 10.1371/journal.pgen.0010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y-H, Doyle JM, Ramanathan S, Gomez TS, Jia D, Xu M, Chen ZJ, Billadeau DD, Rosen MK, Potts PR, 2013. Regulation of WASH-dependent actin polymerization and protein trafficking by ubiquitination. Cell 152, 1051–1064. 10.1016/j.cell.2013.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SY, Seaman MNJ, 2012. Recruitment of the endosomal WASH complex is mediated by the extended “tail” of Fam21 binding to the retromer protein Vps35. Biochem. J 442, 209–220. 10.1042/BJ20111761 [DOI] [PubMed] [Google Scholar]
- Harbour ME, Breusegem SYA, Antrobus R, Freeman C, Reid E, Seaman MNJ, 2010. The cargo-selective retromer complex is a recruiting hub for protein complexes that regulate endosomal tubule dynamics. J. Cell Sci 123, 3703–3717. 10.1242/jcs.071472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmon B, Campbell N, Ratner L, 2010. Role of Abl kinase and the wave2 signaling complex in HIV-1 entry at a post-hemifusion step. PLoS Pathog. 6. 10.1371/journal.ppat.1000956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartland EL, Leong JM, 2013. Enteropathogenic and enterohemorrhagic E. Coli: Ecology, pathogenesis, and evolution. Front. Cell. Infect. Microbiol 4, 2011–2013. 10.3389/fcimb.2013.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbrecht T, Van Audenhove I, Zwaenepoel O, Verhelle A, Gettemans J, 2017. VCA nanobodies target N-WASp to reduce invadopodium formation and functioning. PLoS One 12, 1–19. 10.1371/journal.pone.0185076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfer E, Harbour ME, Henriot V, Lakisic G, Sousa-Blin C, Volceanov L, Seaman MNJ, Gautreau A, 2013. Endosomal recruitment of the WASH complex: active sequences and mutations impairing interaction with the retromer. Biol. cell 105, 191–207. 10.1111/boc.201200038 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Blanchoin L, Pollard TD, 1999. Influence of the C terminus of Wiskott-Aldrich syndrome protein (WASp) and the Arp2/3 complex on actin polymerization. Biochemistry 38, 15212–15222. 10.1021/bi991843+ [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD, 2001. Regulation of actin filament network formation through Arp2/3 complex: Activation by a diverse array of proteins. Annu. Rev. Biochem 70, 649–676. 10.1146/annurev.biochem.70.1.649 [DOI] [PubMed] [Google Scholar]
- Higgs HN, Pollard TD, 2000. Activation by Cdc42 and PIP2 of Wiskott-Aldrich Syndrome protein (WASp) stimulates actin nucleation by Arp2/3 complex. J. Cell Biol 150, 1311–1320. 10.1083/jcb.150.6.1311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand A, Remmert M, Biegert A, Söding J, 2009. Fast and accurate automatic structure prediction with HHpred. Proteins 77 Suppl 9, 128–132. 10.1002/prot.22499 [DOI] [PubMed] [Google Scholar]
- Ho H-YH, Rohatgi R, Lebensohn A, Ma L, Li J, Gygi S, Kirschner MW, 2004. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell 118, 203–216. [DOI] [PubMed] [Google Scholar]
- Ho HYH, Rohatgi R, Ma L, Kirschner MW, 2001. CR16 forms a complex with N-WASP in brain and is a novel member of a conserved proline-rich actin-binding protein family. Proc. Natl. Acad. Sci. U. S. A 98, 11306–11311. 10.1073/pnas.211420498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DSB, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L, 2012. A landscape of driver mutations in melanoma. Cell 150, 251–263. 10.1016/j.cell.2012.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E, 2015. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 43, D512–D520. 10.1093/nar/gku1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou JX, Yang H, Huang X, Leng XH, Zhou F, Xie C, Zhou YF, Xu Y, 2017. N-WASP promotes invasion and migration of cervical cancer cells through regulating p38 MAPKs signaling pathway. Am. J. Transl. Res 9, 403–415. [PMC free article] [PubMed] [Google Scholar]
- Hsu AP, Donkó A, Arrington ME, Swamydas M, Fink D, Das A, Escobedo O, Bonagura V, Szabolcs P, Steinberg HN, Bergerson J, Skoskiewicz A, Makhija M, Davis J, Foruraghi L, Palmer C, Fuleihan RL, Church JA, Bhandoola A, Lionakis MS, Campbell S, Leto TL, Kuhns DB, Holland SM, 2019. Dominant activating RAC2 mutation with lymphopenia, immunodeficiency, and cytoskeletal defects. Blood 133, 1977–1988. 10.1182/blood-2018-11-886028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Mullins RD, 2019. LC3 and STRAP regulate actin filament assembly by JMY during autophagosome formation. J. Cell Biol 218, 251–266. 10.1083/jcb.201802157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Zhu P, Xia P, Fan Z, 2016. WASH has a critical role in NK cell cytotoxicity through Lck-mediated phosphorylation. Cell Death Dis. 7, e2301. 10.1038/cddis.2016.212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Huang C, Chen W, Liu Y, Yin X, Lai J, Liang L, Wang Q, Wang A, Zheng C, 2018. WAVE3 promotes proliferation, migration and invasion via the AKT pathway in pancreatic cancer. Int. J. Oncol 53, 672–684. 10.3892/ijo.2018.4421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Ding L, Pan R, Ma PF, Cheng PP, Zhang CH, Shen YT, Xu L, Liu Y, He XQ, Qi ZQ, Wang HL, 2013. WHAMM is required for meiotic spindle migration and asymmetric cytokinesis in mouse oocytes. Histochem. Cell Biol 139, 525–534. 10.1007/s00418-012-1051-z [DOI] [PubMed] [Google Scholar]
- Humphreys D, Davidson A, Hume PJ, Koronakis V, 2012. Salmonella virulence effector SopE and host GEF ARNO cooperate to recruit and activate WAVE to trigger bacterial invasion. Cell Host Microbe 11, 129–139. 10.1016/j.chom.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D, Davidson AC, Hume PJ, Makin LE, Koronakis V, 2013. Arf6 coordinates actin assembly through the WAVE complex, a mechanism usurped by Salmonella to invade host cells. Proc. Natl. Acad. Sci 110, 16880–16885. 10.1073/pnas.1311680110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D, Singh V, Koronakis V, 2016. Inhibition of WAVE Regulatory Complex Activation by a Bacterial Virulence Effector Counteracts Pathogen Phagocytosis. Cell Rep. 17, 697–707. 10.1016/j.celrep.2016.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, Antonarakis SE, Kay BK, Stossel TP, Lamarche-Vane N, McPherson PS, 2001. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat. Cell Biol 3, 927–932. 10.1038/ncb1001-927 [DOI] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier MF, 2011. Cordon-Bleu Uses WH2 Domains as Multifunctional Dynamizers of Actin Filament Assembly. Mol. Cell 43, 464–477. 10.1016/j.molcel.2011.07.010 [DOI] [PubMed] [Google Scholar]
- Imai K, Nonoyama S, Ochs HD, 2003. WASP (Wiskott-Aldrich syndrome protein) gene mutations and phenotype. Curr. Opin. Allergy Clin. Immunol 3, 427–436. 10.1097/00130832-200312000-00003 [DOI] [PubMed] [Google Scholar]
- Innocenti M, Gerboth S, Rottner K, Lai FPL, Hertzog M, Stradal TEB, Frittoli E, Didry D, Polo S, Disanza A, Benesch S, Di Fiore PP, Carlier MF, Scita G, 2005. Abi1 regulates the activity of N-WASP and WAVE in distinct actin-based processes. Nat. Cell Biol 7, 969–976. 10.1038/ncb1304 [DOI] [PubMed] [Google Scholar]
- Innocenti M, Zucconi A, Disanza A, Frittoli E, Areces LB, Steffen A, Stradal TEB, Di Fiore PP, Carlier MF, Scita G, 2004. Abi1 is essential for the formation and activation of a WAVE2 signalling complex. Nat. Cell Biol 6, 319–327. 10.1038/ncb1105 [DOI] [PubMed] [Google Scholar]
- Irie F, Yamaguchi Y, 2002. EphB receptors regulate dendritic spine development via intersectin, Cdc42 and N-WASP. Nat. Neurosci 5, 1117–1118. 10.1038/nn964 [DOI] [PubMed] [Google Scholar]
- Ismail AM, Padrick SB, Chen B, Umetani J, Rosen MK, 2009. The WAVE regulatory complex is inhibited. Nat. Struct. Mol. Biol 16, 561–563. 10.1038/nsmb.1587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Carss KJ, Duarte ST, Hartley T, Keren B, Kurian MA, Marey I, Charles P, Mendonça C, Nava C, Pfundt R, Sanchis-Juan A, van Bokhoven H, van Essen A, van Ravenswaaij-Arts C, Aitman T, Bennett D, Caulfield M, Chinnery P, Gale D, Koziell A, Kuijpers TW, Laffan MA, Maher E, Markus HS, Morrell NW, Ouwehand WH, Perry DJ, Raymond FL, Roberts I, Smith KGC, Thrasher A, Watkins H, Williamson C, Woods G, Ashford S, Bradley JR, Fletcher D, Hammerton T, James R, Kingston N, Penkett CJ, Stirrups K, Veltman M, Young T, Brown M, Clements-Brod N, Davis J, Dewhurst E, Dolling H, Erwood M, Frary A, Linger R, Martin JM, Papadia S, Rehnstrom K, Stark H, Allsup D, Austin S, Bakchoul T, Bariana TK, Bolton-Maggs P, Chalmers E, Collins J, Collins P, Erber WN, Everington T, Favier R, Freson K, Furie B, Gattens M, Gebhart J, Gomez K, Greene D, Greinacher A, Gresele P, Hart D, Heemskerk JWM, Henskens Y, Kazmi R, Keeling D, Kelly AM, Lambert MP, Lentaigne C, Liesner R, Makris M, Mangles S, Mathias M, Millar CM, Mumford A, Nurden P, Payne J, Pasi J, Peerlinck K, Revel-Vilk S, Richards M, Rondina M, Roughley C, Schulman S, Schulze H, Scully M, Sivapalaratnam S, Stubbs M, Tait RC, Talks K, Thachil J, Toh CH, Turro E, Van Geet C, De Vries M, Warner TQ, Watson H, Westbury S, Furnell A, Mapeta R, Rayner-Matthews P, Simeoni I, Staines S, Stephens J, Watt C, Whitehorn D, Attwood A, Daugherty L, Deevi SVV, Halmagyi C, Hu F, Matser V, Meacham S, Megy K, Shamardina O, Titterton C, Tuna S, Yu P, von Ziegenweldt J, Astle W, Bleda M, Carss KJ, Gräf S, Haimel M, Lango-Allen H, Richardson S, Calleja P, Rankin S, Turek W, Anderson J, Bryson C, Carmichael J, McJannet C, Stock S, Allen L, Ambegaonkar G, Armstrong R, Arno G, Bitner-Glindzicz M, Brady A, Canham N, Chitre M, Clement E, Clowes V, Deegan P, Deshpande C, Doffinger R, Firth H, Flinter F, French C, Gardham A, Ghali N, Gissen P, Grozeva D, Henderson R, Hensiek A, Holden S, Holder M, Holder S, Hurst J, Josifova D, Krishnakumar D, Kurian MA, Lees M, MacLaren R, Maw A, Mehta S, Michaelides M, Moore A, Murphy E, Park SM, Parker A, Patch C, Paterson J, Rankin J, Reid E, Rosser E, Sanchis-Juan A, Sandford R, Santra S, Scott R, Sohal A, Stein P, Thomas E, Thompson D, Tischkowitz M, Vogt J, Wakeling E, Wassmer E, Webster A, Ali Sonia, Ali Souad, Boggard HJ, Church C, Coghlan G, Cookson V, Corris PA, Creaser-Myers A, DaCosta R, Dormand N, Eyries M, Gall H, Ghataorhe PK, Ghio S, Ghofrani A, Gibbs JSR, Girerd B, Greenhalgh A, Hadinnapola C, Houweling AC, Humbert M, in’t Veld AH, Kennedy F, Kiely DG, Kovacs G, Lawrie A, Ross RVM, Machado R, Masati L, Meehan S, Moledina S, Montani D, Othman S, Peacock AJ, Pepke-Zaba J, Pollock V, Polwarth G, Ranganathan L, Rhodes CJ, Rue-Albrecht K, Schotte G, Shipley D, Soubrier F, Southgate L, Scelsi L, Suntharalingam J, Tan Y, Toshner M, Treacy CM, Trembath R, Noordegraaf AV, Walker Sara, Wanjiku I, Wharton J, Wilkins M, Wort SJ, Yates K, Alachkar H, Antrobus R, Arumugakani G, Bacchelli C, Baxendale H, Bethune C, Bibi S, Booth C, Browning M, Burns S, Chandra A, Cooper N, Davies S, Devlin L, Drewe E, Edgar D, Egner W, Ghurye R, Gilmour K, Goddard S, Gordins P, Grigoriadou S, Hackett S, Hague R, Harper L, Hayman G, Herwadkar A, Huissoon A, Jolles S, Kelleher P, Kumararatne D, Lear S, Longhurst H, Lorenzo L, Maimaris J, Manson A, McDermott E, Murng S, Nejentsev S, Noorani S, Oksenhendler E, Ponsford M, Qasim W, Quinti I, Richter A, Samarghitean C, Sargur R, Savic S, Seneviratne S, Sewell C, Staples E, Stauss H, Thaventhiran J, Thomas M, Welch S, Willcocks L, Yeatman N, Yong P, Ancliff P, Babbs C, Layton M, Louka E, McGowan S, Mead A, Roy N, Chambers J, Dixon P, Estiu C, Hague B, Marschall HU, Simpson M, Chong S, Emmerson I, Ginsberg L, Gosal D, Hadden R, Horvath R, Mahdi-Rogers M, Manzur A, Marshall A, Matthews E, McCarthy M, Reilly M, Renton T, Rice A, Themistocleous A, Vale T, Van Zuydam N, Walker Suellen, Ormondroyd L, Hudson G, Wei W, Yu Wai Man P, Whitworth J, Afzal M, Colby E, Saleem M, Alavijeh OS, Cook HT, Johnson S, Levine AP, Wong EKS, Tan R, Boycott KM, MacKenzie A, Majewski J, Brudno M, Bulman D, Dyment D, Boycott KM, Kernohan KD, Dyack S, Raymond FL, 2018. De Novo Truncating Mutations in WASF1 Cause Intellectual Disability with Seizures. Am. J. Hum. Genet 103, 144–153. 10.1016/j.ajhg.2018.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov AI, Hunt D, Utech M, Nusrat A, Parkos CA, 2005. Differential Roles for Actin Polymerization and a Myosin II Motor in Assembly of the Epithelial Apical Junction Complex. Mol. Biol. Cell 16, 5356–5372. 10.1091/mbc.E05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N, Thanabalu T, 2015. Molecular difference between WASP and N-WASP critical for chemotaxis of T-cells towards SDF-1α. Sci. Rep 5, 1–12. 10.1038/srep15031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Billadeau DD, Rosen MK, 2012. Multiple repeat elements within the FAM21 tail link the WASH actin regulatory complex to the retromer. Mol. Biol. Cell 23, 2352–2361. 10.1091/mbc.E11-12-1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia D, Gomez TS, Metlagel Z, Umetani J, Otwinowski Z, Rosen MK, Billadeau DD, 2010. WASH and WAVE actin regulators of the Wiskott-Aldrich syndrome protein (WASP) family are controlled by analogous structurally related complexes. Proc. Natl. Acad. Sci 107, 10442–10447. 10.1073/pnas.0913293107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Jia Y, Weeks HP, Ruge F, Feng X, Ma R, Ji J, Ren J, Jiang WG, 2014. Down-regulation of WAVE2, WASP family verprolin-homologous protein 2, in gastric cancer indicates lymph node metastasis and cell migration. Anticancer Res. 34, 2185–2194. [PubMed] [Google Scholar]
- Jin KM, Lu M, Liu FF, Gu J, Du XJ, Xing BC, 2013. N-WASP is highly expressed in hepatocellular carcinoma and associated with poor prognosis. Surg. (United States) 153, 518–525. 10.1016/j.surg.2012.08.067 [DOI] [PubMed] [Google Scholar]
- Jin Y, Mazza C, Christie JR, Giliani S, Fiorini M, Mella P, Gandellini F, Stewart DM, Zhu Q, Nelson DL, Notarangelo LD, Ochs HD, 2004. Mutations of the Wiskott-Aldrich Syndrome Protein (WASP): Hotspots, effect on transcription, and translation and phenotype/genotype correlation. Blood 104, 4010–4019. 10.1182/blood-2003-05-1592 [DOI] [PubMed] [Google Scholar]
- Jinks RN, Puffenberger EG, Baple E, Harding B, Crino P, Fogo AB, Wenger O, Xin B, Koehler AE, McGlincy MH, Provencher MM, Smith JD, Tran L, Al Turki S, Chioza BA, Cross H, Harlalka GV, Hurles ME, Maroofian R, Heaps AD, Morton MC, Stempak L, Hildebrandt F, Sadowski CE, Zaritsky J, Campellone K, Morton DH, Wang H, Crosby A, Strauss KA, 2015. Recessive nephrocerebellar syndrome on the Galloway-Mowat syndrome spectrum is caused by homozygous protein-truncating mutations of WDR73. Brain 138, 2173–2190. 10.1093/brain/awv153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo YJ, Kwon J, Jin ZL, Namgoong S, Kwon T, Yoon S. Bin, Lee DH, Kim JS, Kim NH, 2021. WHAMM is essential for spindle formation and spindle actin polymerization in maturing mouse oocytes. Cell Cycle 20, 225–235. 10.1080/15384101.2020.1867791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, Hassabis D, 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabrawala S, Zimmer MD, Campellone KG, 2020. WHIMP links the actin nucleation machinery to Src-family kinase signaling during protrusion and motility, PLoS Genetics. 10.1371/journal.pgen.1008694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman D, Weiner OD, Goosney DL, Sedat JW, Finaly BB, Abo A, Bishop JM, 1999. Enteropathogenic E. coli acts through WASP and Arp2/3 complex to form actin pedestals. Nat. Cell Biol 1, 389–391. 10.1038/14087.Enteropathogenic [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Tang D, Yu Y, Wang Z, Hu T, Wang H, Cao L, 2010. WAVE1 regulates Bcl-2 localization and phosphorylation in leukemia cells. Leukemia 24, 177–186. 10.1038/leu.2009.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karahan H, Smith DC, Kim B, Dabin LC, Al-Amin MM, Sagara Wijeratne HR, Pennington T, Prisco G.V. di, McCord B, Lin PBC, Li Y, Peng J, Oblak AL, Chu S, Atwood BK, Kim J, 2021. Deletion of Abi3 gene locus exacerbates neuropathological features of Alzheimer’s disease in a mouse model of Aβ amyloidosis. Sci. Adv 7, 1–20. 10.1126/sciadv.abe3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kast DJ, Zajac AL, Holzbaur ELF, Ostap EM, Dominguez R, 2015. WHAMM Directs the Arp2/3 Complex to the ER for Autophagosome Biogenesis through an Actin Comet Tail Mechanism. Curr. Biol 25, 1791–1797. 10.1016/j.cub.2015.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katanov C, Novak N, Vainshtein A, Golani O, Dupree JL, Peles E, 2020. N-wasp regulates oligodendrocyte myelination. J. Neurosci 40, 6103–6111. 10.1523/JNEUROSCI.0912-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Oka Y, Muramatsu H, Vasilev FF, Otomo T, Oishi H, Kawano Y, Kidokoro H, Nakazawa Y, Ogi T, Takahashi Y, Saitoh S, 2020. Biallelic VPS35L pathogenic variants cause 3C/Ritscher-Schinzel-like syndrome through dysfunction of retriever complex. J. Med. Genet 57, 245–253. 10.1136/jmedgenet-2019-106213 [DOI] [PubMed] [Google Scholar]
- Kato M, Miki H, Kurita S, Endo T, Nakagawa H, Miyamoto S, Takenawa T, 2002. WICH, a novel verprolin homology domain-containing protein that functions cooperatively with N-WASP in actin-microspike formation. Biochem. Biophys. Res. Commun 291, 41–47. 10.1006/bbrc.2002.6406 [DOI] [PubMed] [Google Scholar]
- Kawazu M, Ueno T, Kontani K, Ogita Y, Ando M, Fukumura K, Yamato A, Soda M, Takeuchi K, Miki Y, Yamaguchi H, Yasuda T, Naoe T, Yamashita Y, Katada T, Choi YL, Mano H, 2013. Transforming mutations of RAC guanosine triphosphatases in human cancers. Proc. Natl. Acad. Sci. U. S. A 110, 3029–3034. 10.1073/pnas.1216141110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keb G, Ferrell J, Scanlon KR, Jewett TJ, Fields KA, 2021. Chlamydia trachomatis tmea directly activates N-WASP to promote actin polymerization and functions synergistically with TarP during invasion. MBio 12, 1–18. 10.1128/mBio.02861-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney D, Cairns L, Remold-O’Donnell E, Peterson J, Rosen FS, Parkman R, 1986. Morphological abnormalities in the lymphocytes of patients with the Wiskott-Aldrich Syndrome. Blood 68, 1329–1332. 10.1182/blood.v68.6.1329.1329 [DOI] [PubMed] [Google Scholar]
- Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK, 2000. Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein. Nature 404, 151–158. 10.1038/35004513 [DOI] [PubMed] [Google Scholar]
- Kim JH, Jin P, Duan R, Chen EH, 2015. Mechanisms of myoblast fusion during muscle development. Curr. Opin. Genet. Dev 32, 162–170. 10.1016/j.gde.2015.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KS, Marcogliese PC, Yang J, Callaghan SM, Resende V, Abdel-Messih E, Marras C, Visanji NP, Huang J, Schlossmacher MG, Trinkle-Mulcahy L, Slack RS, Lang AE, Park DS, 2018. Regulation of myeloid cell phagocytosis by LRRK2 via WAVE2 complex stabilization is altered in Parkinson’s disease. Proc. Natl. Acad. Sci. U. S. A 115, E5164–E5173. 10.1073/pnas.1718946115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Kalappurakkal JM, Mayor S, Rosen MK, 2019. Phosphorylation of nephrin induces phase separated domains that move through actomyosin contraction. Mol. Biol. Cell 30, 2996–3012. 10.1091/mbc.E18-12-0823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sung JY, Ceglia I, Lee KW, Ahn JH, Halford JM, Kim AM, Kwak SP, Park JB, Ho Ryu S, Schenck A, Bardoni B, Scott JD, Nairn AC, Greengard P, 2006. Phosphorylation of WAVE1 regulates actin polymerization and dendritic spine morphology. Nature 442, 814–817. 10.1038/nature04976 [DOI] [PubMed] [Google Scholar]
- King JS, Gueho A, Hagedorn M, Gopaldass N, Leuba F, Soldati T, Insall RH, 2013. WASH is required for lysosomal recycling and efficient autophagic and phagocytic digestion. Mol. Biol. Cell 24, 2714–2726. 10.1091/mbc.E13-02-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King VL, Leclair NK, Coulter AM, Campellone KG, 2021. The actin nucleation factors JMY and WHAMM enable a rapid Arp2/3 complex-mediated intrinsic pathway of apoptosis, PLoS Genetics. 10.1371/journal.pgen.1009512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick SL, Goldberg LR, Yazdani N, Babbs RK, Wu J, Reed ER, Jenkins DF, Bolgioni AF, Landaverde KI, Luttik KP, Mitchell KS, Kumar V, Johnson WE, Mulligan MK, Cottone P, Bryant CD, 2017. Cytoplasmic FMR1-Interacting Protein 2 Is a Major Genetic Factor Underlying Binge Eating. Biol. Psychiatry 81, 757–769. 10.1016/j.biopsych.2016.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kitamura Y, Yonezawa K, Totty NF, Gout I, Hara K, Waterfield MD, Sakaue M, Ogawa W, Kasuga M, 1996. Molecular cloning of p125Nap1, a protein that associates with an SH3 domain of Nck. Biochem. Biophys. Res. Commun 219, 509–514. 10.1006/bbrc.1996.0264 [DOI] [PubMed] [Google Scholar]
- Kitamura Y, Tsuchiya D, Takata K, Shibagaki K, Taniguchi T, Smith MA, Perry G, Miki H, Takenawa T, Shimohama S, 2003. Possible involvement of Wiskott-Aldrich syndrome protein family in aberrant neuronal sprouting in Alzheimer’s disease. Neurosci. Lett 346, 149–152. 10.1016/S0304-3940(03)00506-8 [DOI] [PubMed] [Google Scholar]
- Kluge F, Weissbach J, Weber A, Stradal T, Posern G, 2018. Regulation of MRTF-A by JMY via a nucleation-independent mechanism. Cell Commun. Signal 16, 1–14. 10.1186/s12964-018-0299-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Kuroda S, Fukata M, Nakamura T, Nagase T, Nomura N, Matsuura Y, Yoshida-Kubomura N, Iwamatsu A, Kaibuchi K, 1998. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J. Biol. Chem 273, 291–295. 10.1074/jbc.273.1.291 [DOI] [PubMed] [Google Scholar]
- Kolanczyk M, Krawitz P, Hecht J, Hupalowska A, Miaczynska M, Marschner K, Schlack C, Emmerich D, Kobus K, Kornak U, Robinson PN, Plecko B, Grangl G, Uhrig S, Mundlos S, Horn D, 2015. Missense variant in CCDC22 causes X-linked recessive intellectual disability with features of Ritscher-Schinzel/3C syndrome. Eur. J. Hum. Genet 23, 633–638. 10.1038/ejhg.2014.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollmar M, Lbik D, Enge S, 2012. Evolution of the eukaryotic ARP2/3 activators of the WASP family: WASP, WAVE, WASH, and WHAMM, and the proposed new family members WAWH and WAML. BMC Res. Notes 5. 10.1186/1756-0500-5-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolluri R, Shehabeldin A, Peacocke M, 1995. Identification of WASP mutations in patients with Wiskott-Aldrich syndrome and isolated thrombocytopenia reveals allelic heterogeneity at the WAS locus. Hum. Mol. Genet 4, 1119–1126. [DOI] [PubMed] [Google Scholar]
- Kong R, Yi F, Wen P, Liu J, Chen X, Ren J, Li X, Shang Y, Nie Y, Wu K, Fan D, Zhu L, Feng W, Wu JY, 2015. Myo9b is a key player in SLIT/ROBO-mediated lung tumor suppression. J. Clin. Invest 125, 4407–4420. 10.1172/JCI81673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis V, Hume PJ, Humphreys D, Liu T, Horning O, Jensen ON, McGhie EJ, 2011. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci 108, 14449–14454. 10.1073/pnas.1107666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koronakis Vassilis, Hume PJ, Humphreys D, Liu T, Hørning O, Jensen ON, McGhie EJ, 2011. WAVE regulatory complex activation by cooperating GTPases Arf and Rac1. Proc. Natl. Acad. Sci. U. S. A 108, 14449–14454. 10.1073/pnas.1107666108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs EM, Verma S, Ali RG, Ratheesh A, Hamilton NA, Akhmanova A, Yap AS, 2011. N-WASP regulates the epithelial junctional actin cytoskeleton through a non-canonical post-nucleation pathway. Nat. Cell Biol 13, 934–943. 10.1038/ncb2290 [DOI] [PubMed] [Google Scholar]
- Krauthammer M, Kong Y, Ha BH, Evans P, Bacchiocchi A, McCusker JP, Cheng E, Davis MJ, Goh G, Choi M, Ariyan S, Narayan D, Dutton-Regester K, Capatana A, Holman EC, Bosenberg M, Sznol M, Kluger HM, Brash DE, Stern DF, Materin MA, Lo RS, Mane S, Ma S, Kidd KK, Hayward NK, Lifton RP, Schlessinger J, Boggon TJ, Halaban R, 2012. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet 44, 1006–1014. 10.1038/ng.2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K, 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D Biol. Crystallogr 60, 2256–2268. 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- Kumar DM, Lin M, Xiong Q, Webber MJ, Kural C, Rikihisa Y, 2015. EtpE binding to dnase X induces ehrlichial entry via CD147 and hnRNP-K recruitment, followed by mobilization of N-WASP and actin. MBio 6, 1–12. 10.1128/mBio.01541-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Kim K, Joseph C, Kourrich S, Yoo S-H, Huang HC, Vitaterna MH, de Villena FP-M, Churchill G, Bonci A, Takahashi JS, 2013. C57BL/6N Mutation in Cytoplasmic FMRP interacting protein 2 Regulates Cocaine Response. Science (80-.) 342, 1508–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P, Craig G, Dominguez V, Baum B, 2003. Abi, Sra1, and Kette Control the Stability and Localization of SCAR/WAVE to Regulate the Formation of Actin-Based Protrusions. Curr. Biol 13, 1867–1875. 10.1016/j.cub.2003.10.005 [DOI] [PubMed] [Google Scholar]
- Kurisu S, Takenawa T, 2010. WASP and WAVE family proteins: Friends or foes in cancer invasion? Cancer Sci. 101, 2093–2104. 10.1111/j.1349-7006.2010.01654.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurisu S, Takenawa T, 2009. The WASP and WAVE family proteins. Genome Biol. 10, 226. 10.1186/gb-2009-10-6-226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvainickas A, Orgaz AJ, Nägele H, Diedrich B, Heesom KJ, Dengjel J, Cullen PJ, Steinberg F, 2017. Retromer- and WASH-dependent sorting of nutrient transporters requires a multivalent interaction network with ANKRD50. J. Cell Sci 130, 382–395. 10.1242/jcs.196758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno CM, Lewis CM, You D, Leung DW, Takesono A, Kamberos N, Seth A, Finkelstein LD, Rosen MK, Schwartzberg PL, Burkhardt JK, 2003. Itk Functions to Control Actin Polymerization at the Immune Synapse through Localized Activation of Cdc42 and WASP. Curr. Biol 13, 1619–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AL, Jalal S, Pallett T, Mosis F, Guni A, Brayford S, Yolland L, Marcotti S, Levitt JA, Poland SP, Rowe-Sampson M, Jandke A, Köchl R, Pula G, Ameer-Beg SM, Stramer BM, Krause M, 2021. Nance-Horan Syndrome-like 1 protein negatively regulates Scar/WAVE-Arp2/3 activity and inhibits lamellipodia stability and cell migration. Nat. Commun 12. 10.1038/s41467-021-25916-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AL, Vehlow A, Kotini M, Dodgson L, Soong D, Theveneau E, Bodo C, Taylor E, Navarro C, Perera U, Michael M, Dunn GA, Bennett D, Mayor R, Krause M, 2013. Lamellipodin and the Scar/WAVE complex cooperate to promote cell migration in vivo. J. Cell Biol 203, 673–689. 10.1083/jcb.201304051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clainche C, Schlaepfer D, Ferrari A, Klingauf M, Grohmanova K, Veligodskiy A, Didry D, Le D, Egile C, Carlier MF, Kroschewski R, 2007. IQGAP1 stimulates actin assembly through the N-wasp-Arp2/3 pathway. J. Biol. Chem 282, 426–435. 10.1074/jbc.M607711200 [DOI] [PubMed] [Google Scholar]
- Lebensohn AM, Kirschner MW, 2009. Activation of the WAVE Complex by Coincident Signals Controls Actin Assembly. Mol. Cell 36, 512–524. 10.1016/j.molcel.2009.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee NK, Fok KW, White A, Wilson NH, O’Leary CJ, Cox HL, Michael M, Yap AS, Cooper HM, 2016. Neogenin recruitment of the WAVE regulatory complex maintains adherens junction stability and tension. Nat. Commun 7. 10.1038/ncomms11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Park H, Zhu P-P, Jung S-Y, Blackstone C, Chang J, 2020. Hereditary spastic paraplegia SPG8 mutations impair CAV1-dependent, integrin-mediated cell adhesion. Sci. Signal 13, eaau7500. 10.1126/scisignal.aau7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Zhang J, Badour K, Arpaia E, Freeman S, Cheung P, Siu M, Siminovitch K, 2005. Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphory lation required for WAVE2 activation. Proc. Natl. Acad. Sci. U. S. A 102, 1098–1103. 10.1073/pnas.0409120102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Banjade S, Cheng HC, Kim S, Chen B, Guo L, Llaguno M, Hollingsworth JV, King DS, Banani SF, Russo PS, Jiang QX, Nixon BT, Rosen MK, 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature 483, 336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye AJ, Bendzunas GN, Whittaker MK, LeClair TJ, Helton LG, Kennedy EJ, 2022. In Silico Optimized Stapled Peptides Targeting WASF3 in Breast Cancer. ACS Med. Chem. Lett 1–7. 10.1021/acsmedchemlett.1c00627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, den Dulk-Ras A, Hooykaas PJJ, Rikihisa Y, 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol 9, 2644–2657. 10.1111/j.1462-5822.2007.00985.x [DOI] [PubMed] [Google Scholar]
- Linardopoulou EV, Parghi SS, Friedman C, Osborn GE, Parkhurst SM, Trask BJ, 2007. Human subtelomeric WASH genes encode a new subclass of the WASP family. PLoS Genet. 3, 2477–2485. 10.1371/journal.pgen.0030237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder S, Nelson D, Weiss M, Aepfelbacher M, 1999. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc. Natl. Acad. Sci. U. S. A 96, 9648–9653. 10.1073/pnas.96.17.9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Bai X, Wu J, Sharma S, Upadhyaya A, Dahlberg CIM, Westerberg LS, Snapper SB, Zhao X, Song W, 2013. N-WASP Is Essential for the Negative Regulation of B Cell Receptor Signaling. PLoS Biol. 11. 10.1371/journal.pbio.1001704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang QC, Wang F, Duan X, Dai XX, Wang T, Liu HL, Cui XS, Kim NH, Sun SC, 2012. Nucleation Promoting Factors Regulate the Expression and Localization of Arp2/3 Complex during Meiosis of Mouse Oocytes. PLoS One 7, 3–9. 10.1371/journal.pone.0052277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Zhang J, Pinder BD, Liu Q, Wang D, Yao H, Gao Y, Toker A, Gao J, Peterson A, Qu J, Siminovitch KA, 2021. WAVE2 suppresses mTOR activation to maintain T cell homeostasis and prevent autoimmunity. Science (80-.) 371. 10.1126/science.aaz4544 [DOI] [PubMed] [Google Scholar]
- Liu R, Abreu-Blanco MT, Barry KC, Linardopoulou EV, Osborn GE, Parkhurst SM, 2009. Wash functions downstream of Rho and links linear and branched actin nucleation factors. Development 136, 2849–2860. 10.1242/dev.035246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Yu M, He Y, Xiao L, Wang F, Song C, Sun S, Ling C, Xu Z, 2008. Melittin prevents liver cancer cell metastasis through inhibition of the Rac1-dependent pathway. Hepatology 47, 1964–1973. 10.1002/hep.22240 [DOI] [PubMed] [Google Scholar]
- Liu T, Dai A, Cao Y, Zhang R, Dong MQ, Wang HW, 2017. Structural Insights of WHAMM’s Interaction with Microtubules by Cryo-EM. J. Mol. Biol 429, 1352–1363. 10.1016/j.jmb.2017.03.022 [DOI] [PubMed] [Google Scholar]
- Liu Y, Fan J, Yan Y, Dang X, Zhao R, Xu Y, Ding Z, 2020. JMY expression by Sertoli cells contributes to mediating spermatogenesis in mice. FEBS J. 287, 5478–5497. 10.1111/febs.15328 [DOI] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rohde M, Wehland J, Rottner K, 2004. Enterohaemorrhagic and enteropathogenic Escherichia coli use different mechanisms for actin pedestal formation that converge on N-WASP. Cell. Microbiol 6, 243–254. 10.1111/j.1462-5822.2004.00364.x [DOI] [PubMed] [Google Scholar]
- Lommel S, Benesch S, Rottner K, Franz T, Wehland J, Kühn R, 2001. Actin pedestal formation by enteropathogenic Escherichia coli and intracellular motility of Shigella flexneri are abolished in N-WASP-defective cells. EMBO Rep. 2, 850–857. 10.1093/embo-reports/kve197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Yamaguchi H, Wang Y, Singer RH, Condeelis J, 2004. Imaging Sites of N-WASP Activity in Lamellipodia and Invadopodia of Carcinoma Cells. Curr. Biol 14, 697–703. 10.1016/j [DOI] [PubMed] [Google Scholar]
- Loveless R, Teng Y, 2021. Targeting WASF3 signaling in metastatic cancer. Int. J. Mol. Sci 22, 1–14. 10.3390/ijms22020836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas B, Hardin J, 2017. Mind the (sr)GAP - roles of Slit-Robo GAPs in neurons, brains and beyond. J. Cell Sci 130, 3695–3974. 10.1242/jcs.215392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna A, Matas OB, Martínez-Menárguez JA, Mato E, Durán JM, Ballesta J, Way M, Egea G, 2002. Regulation of protein transport from the golgi complex to the endoplasmic reticulum by CDC42 and N-WASP. Mol. Biol. Cell 13, 866–879. 10.1091/mbc.01-12-0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström A, Gallio M, Englund C, Steneberg P, Hemphälä J, Aspenström P, Keleman K, Falileeva L, Dickson BJ, Samakovlis C, 2004. Vilse, a conserved Rac/Cdc42 GAP mediating Robo repulsion in tracheal cells and axons. Genes Dev. 18, 2161–2171. 10.1101/gad.310204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, 2002. Actin cytoskeleton regulation in neuronal morphogenesis and structural plasticity. Annu. Rev. Cell Dev. Biol 18, 601–635. 10.1146/annurev.cellbio.18.031802.150501 [DOI] [PubMed] [Google Scholar]
- MacCarthy-Morrogh L, Gaspar HB, Wang YC, Katz F, Thompson L, Layton M, Jones AM, Kinnon C, 1998. Absence of expression of the Wiskott-Aldrich syndrome protein in peripheral blood cells of Wiskott-Aldrich syndrome patients. Clin. Immunol. Immunopathol 88, 22–27. 10.1006/clin.1998.4557 [DOI] [PubMed] [Google Scholar]
- MacDonald E, Brown L, Selvais A, Liu H, Waring T, Newman D, Bithell J, Grimes D, Urbé S, Clague MJ, Zech T, 2018. HRS-WASH axis governs actin-mediated endosomal recycling and cell invasion. J. Cell Biol 217, 2549–2564. 10.1083/jcb.201710051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Atkinson SJ, Ampe C, Vandekercldaove J, 1994. Purification of a Cortical Complex Containing Two Unconventional Actins from 127, 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Insall RH, 1998. Scar1 and the related Wiskott-Aldrich syndrome protein, WASP, regulate the actin cytoskeleton through the Arp2/3 complex. Curr. Biol 8, 1347–1356. 10.1016/S0960-9822(98)00015-3 [DOI] [PubMed] [Google Scholar]
- Malin J, Rosa Birriel C, Astigarraga S, Treisman JE, Hatini V, 2022. Sidekick dynamically rebalances contractile and protrusive forces to control tissue morphogenesis. J. Cell Biol 221, e202107035. 10.1083/jcb.202107035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand JB, Kaiser DA, Pollard TD, Higgs HN, 2001. Interaction of WASP/Scar proteins with actin and vertebrate Arp2/3 complex. Nat. Cell Biol 3, 76–82. 10.1038/35050590 [DOI] [PubMed] [Google Scholar]
- Martin TA, Pereira G, Watkins G, Mansel RE, Jiang WG, 2008. N-WASP is a putative tumour suppressor in breast cancer cells, in vitro and in vivo, and is associated with clinical outcome in patients with breast cancer. Clin. Exp. Metastasis 25, 97–108. 10.1007/s10585-007-9120-8 [DOI] [PubMed] [Google Scholar]
- Martin TA, Toms A, Davies LM, Cheng S, Jiang WG, 2012. The clinical and biological implications of N-WASP expression in human colorectal cancer. Transl. Gastrointest. Cancer 1, 9–20. 10.3978/j.issn.2224-4778.2011.10.01 [DOI] [Google Scholar]
- Martinelli S, Krumbach OHF, Pantaleoni F, Coppola S, Amin E, Pannone L, Nouri K, Farina L, Dvorsky R, Lepri F, Buchholzer M, Konopatzki R, Walsh L, Payne K, Pierpont ME, Vergano SS, Langley KG, Larsen D, Farwell KD, Tang S, Mroske C, Gallotta I, Di Schiavi E, della Monica M, Lugli L, Rossi C, Seri M, Cocchi G, Henderson L, Baskin B, Alders M, Mendoza-Londono R, Dupuis L, Nickerson DA, Chong JX, Meeks N, Brown K, Causey T, Cho MT, Demuth S, Digilio MC, Gelb BD, Bamshad MJ, Zenker M, Ahmadian MR, Hennekam RC, Tartaglia M, Mirzaa GM, 2018. Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am. J. Hum. Genet 102, 309–320. 10.1016/j.ajhg.2017.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massaad MJ, Ramesh N, Geha RS, 2013. Wiskott-Aldrich syndrome: A comprehensive review. Ann. N. Y. Acad. Sci 1285, 26–43. 10.1111/nyas.12049 [DOI] [PubMed] [Google Scholar]
- Mathiowetz AJ, Baple E, Russo AJ, Coulter AM, Carrano E, Brown JD, Jinks RN, Crosby AH, Campellone KG, 2017. An Amish founder mutation disrupts a PI(3)P-WHAMM-Arp2/3 complex-driven autophagosomal remodeling pathway. Mol. Biol. Cell 28, 2492–2507. 10.1091/mbc.E17-01-0022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio RPM, Jeffries CM, Svergun DI, Deane JE, 2017. The Shigella virulence factor IcsA relieves N-WASP autoinhibition by displacing the verprolin homology/cofilin/acidic (VCA) domain. J. Biol. Chem 292, 134–145. 10.1074/jbc.M116.758003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGough IJ, Steinberg F, Jia D, Barbuti PA, McMillan KJ, Heesom KJ, Whone AL, Caldwell MA, Billadeau DD, Rosen MK, Cullen PJ, 2014. Retromer binding to FAM21 and the WASH complex is perturbed by the Parkinson disease-linked VPS35(D620N) mutation. Curr. Biol 24, 1670–1676. 10.1016/j.cub.2014.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KE, Faulkner R, Steinberg F, Gallon M, Ghai R, Pim D, Langton P, Pearson N, Danson CM, Nägele H, Morris LL, Singla A, Overlee BL, Heesom KJ, Sessions R, Banks L, Collins BM, Berger I, Billadeau DD, Burstein E, Cullen PJ, 2017. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol 19, 1214–1225. 10.1038/ncb3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehidi A, Kage F, Karatas Z, Cercy M, Schaks M, Polesskaya A, Sainlos M, Gautreau AM, Rossier O, Rottner K, Giannone G, 2021. Forces generated by lamellipodial actin filament elongation regulate the WAVE complex during cell migration. Nat. Cell Biol 23, 1148–1162. 10.1038/s41556-021-00786-8 [DOI] [PubMed] [Google Scholar]
- Mendoza MC, 2013. Phosphoregulation of the WAVE regulatory complex and signal integration. Semin. Cell Dev. Biol 24, 272–279. 10.1016/j.semcdb.2013.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menotti M, Ambrogio C, Cheong TC, Pighi C, Mota I, Cassel SH, Compagno M, Wang Q, Dall’Olio R, Minero VG, Poggio T, Sharma GG, Patrucco E, Mastini C, Choudhari R, Pich A, Zamo A, Piva R, Giliani S, Mologni L, Collings CK, Kadoch C, Gambacorti-Passerini C, Notarangelo LD, Anton IM, Voena C, Chiarle R, 2019. Wiskott–Aldrich syndrome protein (WASP) is a tumor suppressor in T cell lymphoma. Nat. Med 25, 130–140. 10.1038/s41591-018-0262-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Miura K, Takenawa T, 1996. N-WASP, a novel actin-depolymerizing protein, regulates the cortical cytoskeletal rearrangement in a PIP2-dependent manner downstream of tyrosine kinases. EMBO J. 15, 5326–5335. 10.1002/j.1460-2075.1996.tb00917.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Sasaki T, Takai Y, Takenawa T, 1998a. Induction of filopodium formation by a WASP-related actin- depolymerizing protein N-WASP. Nature 391, 93–96. 10.1038/34208 [DOI] [PubMed] [Google Scholar]
- Miki H, Suetsugu S, Takenawa T, 1998b. WAVE, a novel WASP-family protein involved in actin reorganization induced by Rac. EMBO J. 17, 6932–6941. 10.1093/emboj/17.23.6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Takenawa T, 1998. Direct binding of the verprolin-homology domain in N-WASP to actin is essential for cytoskeletal reorganization. Biochem. Biophys. Res. Commun 243, 73–78. 10.1006/bbrc.1997.8064 [DOI] [PubMed] [Google Scholar]
- Miki H, Yamaguchi H, Suetsugu S, Takenawa T, 2000. IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732–735. 10.1038/35047107 [DOI] [PubMed] [Google Scholar]
- Miller MM, Lapetina S, MacGrath SM, Sfakianos MK, Pollard TD, Koleske AJ, 2010. Regulation of actin polymerization and adhesion-dependent cell edge protrusion by the abl-related gene (Arg) tyrosine kinase and N-WASp. Biochemistry 49, 2227–2234. 10.1021/bi901721u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra A, Lim RPZ, Wu Z, Thanabalu T, 2007. N-WASP plays a critical role in fibroblast adhesion and spreading. Biochem. Biophys. Res. Commun 364, 908–912. 10.1016/j.bbrc.2007.10.086 [DOI] [PubMed] [Google Scholar]
- Miyamoto K, Teperek M, Yusa K, Allen GE, Bradshaw CR, Gurdon JB, 2013. Nuclear wave1 is required for reprogramming transcription in oocytes and for normal development. Science (80-.) 341, 1002–1005. 10.1126/science.1240376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Tanoue A, 2008. Cdk5 phosphorylation of WAVE2 regulates oligodendrocyte precursor cell migration through nonreceptor tyrosine kinase fyn. J. Neurosci 28, 8326–8337. 10.1523/JNEUROSCI.1482-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizutani K, Miki H, Takenawa T, Maruta H, Takenawa T, He H, Maruta H, 2002. Essential role of neural Wiskott-Aldrich syndrome protein in podosome formation and degradation of extracellular matrix in src-transformed fibroblasts. Cancer Res. 62, 669–674. [PubMed] [Google Scholar]
- Molendijk AJ, Ruperti B, Palme K, 2004. Small GTPases in vesicle trafficking. Curr. Opin. Plant Biol 7, 694–700. https://doi.org/ 10.1016/j.pbi.2004.09.014 [DOI] [PubMed] [Google Scholar]
- Molina IJ, Kenney DM, Rosen FS, Remold-O’donnell E, 1992. T cell lines characterize events in the pathogenesis of the wiskott-aldrich syndrome. J. Exp. Med 176, 867–874. 10.1084/jem.176.3.867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monday HR, Bourdenx M, Jordan BA, Castillo PE, 2020. Cb1-receptor-mediated inhibitory ltd triggers presynaptic remodeling via protein synthesis and ubiquitination. Elife 9, 1–25. 10.7554/ELIFE.54812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monfregola J, Napolitano G, D’Urso M, Lappalainen P, Ursini MV, 2010. Functional characterization of Wiskott-Aldrich syndrome protein and scar homolog (WASH), a bi-modular nucleation-promoting factor able to interact with biogenesis of lysosome-related organelle subunit 2 (BLOS2) and gamma-tubulin. J. Biol. Chem 285, 16951–16957. 10.1074/jbc.M109.078501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P, Rossé C, Castro-Castro A, Irondelle M, Lagoutte E, Paul-Gilloteaux P, Desnos C, Formstecher E, Darchen F, Perrais D, Gautreau A, Hertzog M, Chavrier P, 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol 203, 1063–1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M, 2000. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat. Cell Biol 2, 441–448. 10.1038/35017080 [DOI] [PubMed] [Google Scholar]
- Murga-Zamalloa CA, Mendoza-Reinoso V, Sahasrabuddhe AA, Rolland D, Hwang SR, McDonnell SRP, Sciallis AP, Wilcox RA, Bashur V, Elenitoba-Johnson K, Lim MS, 2017. NPM-ALK phosphorylates WASp Y102 and contributes to oncogenesis of anaplastic large cell lymphoma. Oncogene 36, 2085–2094. 10.1038/onc.2016.366 [DOI] [PubMed] [Google Scholar]
- Nagel BM, Bechtold M, Rodriguez LG, Bogdan S, 2017. Drosophila WASH is required for integrin-mediated cell adhesion, cell motility and lysosomal neutralization. J. Cell Sci 130, 344–359. 10.1242/jcs.193086 [DOI] [PubMed] [Google Scholar]
- Nakanishi O, Suetsugu S, Yamazaki D, Takenawa T, 2007. Effect of WAVE2 phosphorylation on activation of the Arp2/3 complex. J. Biochem 141, 319–325. 10.1093/jb/mvm034 [DOI] [PubMed] [Google Scholar]
- Nakashima M, Kato M, Aoto K, Shiina M, Belal H, Mukaida S, Kumada S, Sato A, Zerem A, Lerman-Sagie T, Lev D, Leong HY, Tsurusaki Y, Mizuguchi T, Miyatake S, Miyake N, Ogata K, Saitsu H, Matsumoto N, 2018. De novo hotspot variants in CYFIP2 cause early-onset epileptic encephalopathy. Ann. Neurol 83, 794–806. 10.1002/ana.25208 [DOI] [PubMed] [Google Scholar]
- Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, Witke W, Costa-Mattioli M, Sonenberg N, Achsel T, Bagni C, 2008. The Fragile X Syndrome Protein Represses Activity-Dependent Translation through CYFIP1, a New 4E-BP. Cell 134, 1042–1054. 10.1016/j.cell.2008.07.031 [DOI] [PubMed] [Google Scholar]
- Nobes CD, Hall A, 1995. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 81, 53–62. 10.1016/0092-8674(95)90370-4 [DOI] [PubMed] [Google Scholar]
- Nolz JC, Gomez TS, Zhu P, Li S, Medeiros RB, Shimizu Y, Burkhardt JK, Freedman BD, Billadeau DD, 2006. The WAVE2 complex regulates actin cytoskeletal reorganization and CRAC-mediated calcium entry during T cell activation. Curr. Biol 16, 24–34. 10.1016/j.cub.2005.11.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notarangelo LD, Miao CG, Ochs HD, 2008. Wiskott-Aldrich Syndrome. Curr. Opin. Hematol 15, 30–36. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Nahirney PC, Hadjantonakis AK, Baylies MK, 2009. Nap1-mediated actin remodeling is essential for mammalian myoblast fusion. J. Cell Sci 122, 3282–3293. 10.1242/jcs.047597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozumi M, Nakagawa H, Miki H, Takenawa T, Miyamoto S, 2003. Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci 116, 239–246. 10.1242/jcs.00233 [DOI] [PubMed] [Google Scholar]
- O’Leary CJ, Nourse CC, Lee NK, White A, Langford M, Sempert K, Cole SJ, Cooper HM, 2017. Neogenin Recruitment of the WAVE Regulatory Complex to Ependymal and Radial Progenitor Adherens Junctions Prevents Hydrocephalus. Cell Rep. 20, 370–383. 10.1016/j.celrep.2017.06.051 [DOI] [PubMed] [Google Scholar]
- Oda A, Ochs HD, Lasky LA, Spencer S, Ozaki K, Fujihara M, Handa M, Ikebuchi K, Ikeda H, 2001. CrkL is an adapter for Wiskott-Aldrich syndrome protein and Syk. Blood 97, 2633–2639. 10.1182/blood.V97.9.2633 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Itoh T, Takenawa T, 2008. Sequential signals toward podosome formation in NIH-src cells. J. Cell Biol 182, 157–169. 10.1083/jcb.200801042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikawa T, Yamaguchi H, Itoh T, Kato M, Ijuin T, Yamazaki D, Suetsugu S, Takenawa T, 2004. Ptdlns(3,4,5)P 3 binding is necessary for WAVE2-induced formation of lamellipodia. Nat. Cell Biol 6, 420–426. 10.1038/ncb1125 [DOI] [PubMed] [Google Scholar]
- Olive C, Ibanez L, Farias FHG, Wang F, John P, Norton JB, Gentsch J, Morris JC, Li Z, Del-aguila J, Bergmann K, Bradley J, Benitez BA, Harari O, Fagan A, Ances B, Cruchaga C, Victoria M, 2020. Examination of the Effect of Rare Variants in TREM2, ABI3, and PLCG2 in LOAD Through Multiple Phenotypes. J. Alzheimers Dis 77, 1469–1482. 10.3233/JAD-200019.Examination [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orange JS, Ramesh N, Remold-O’Donnell E, Sasahara Y, Koopman L, Byrne M, Bonilla FA, Rosen FS, Geha RS, Strominger JL, 2002. Wiskott-Aldrich syndrome protein is required for NK cell cytotoxicity and colocalizes with actin to NK cell-activating immunologic synapses. Proc. Natl. Acad. Sci. U. S. A 99, 11351–11356. 10.1073/pnas.162376099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oser M, Yamaguchi H, Mader CC, Bravo-Cordero JJ, Arias M, Chen X, DesMarais V, Van Rheenen J, Koleske AJ, Condeelis J, 2009. Cortactin regulates cofilin and N-WASp activities to control the stages of invadopodium assembly and maturation. J. Cell Biol 186, 571–587. 10.1083/jcb.200812176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Cheng HC, Ismail AM, Panchal SC, Doolittle LK, Kim S, Skehan BM, Umetani J, Brautigam CA, Leong JM, Rosen MK, 2008. Hierarchical Regulation of WASP/WAVE Proteins. Mol. Cell 32, 426–438. 10.1016/j.molcel.2008.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Doolittle LK, Brautigam CA, King DS, Rosen MK, 2011. Arp2/3 complex is bound and activated by two WASP proteins. Proc. Natl. Acad. Sci 108, E472–E479. 10.1073/pnas.1100236108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padrick SB, Rosen MK, 2010. Physical mechanisms of signal integration by WASP family proteins. Annu. Rev. Biochem 79, 707–735. 10.1146/annurev.biochem.77.060407.135452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchal SC, Kaiser DA, Torres E, Pollard TD, Rosen MK, 2003. A conserved amphipathic helix in WASP/scar proteins is essential for activation of Arp2/3 complex. Nat. Struct. Biol 10, 591–598. 10.1038/nsb952 [DOI] [PubMed] [Google Scholar]
- Papayannopoulos V, Co C, Prehoda KE, Snapper S, Taunton J, Lim WA, 2005. A polybasic motif allows N-WASP to act as a sensor of PIP2 density. Mol. Cell 17, 181–191. 10.1016/j.molcel.2004.11.054 [DOI] [PubMed] [Google Scholar]
- Park G. Bin, Kim D, 2017. TLR5/7-mediated PI3K activation triggers epithelial-mesenchymal transition of ovarian cancer cells through WAVE3-dependent mesothelin or OCT4/SOX2 expression. Oncol. Rep 38, 3167–3176. 10.3892/or.2017.5941 [DOI] [PubMed] [Google Scholar]
- Park H, Chan MM, Iritani BM, 2010. Hem-1: Putting the “WAVE” into actin polymerization during an immune response. FEBS Lett. 584, 4923–4932. 10.1016/j.febslet.2010.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Joongkyu, Sung JY, Park Joohyun, Song WJ, Chang S, Chung KC, 2012. Dyrk1a negatively regulates the actin cytoskeleton through threonine phosphorylation of N-WASP. J. Cell Sci 125, 67–80. 10.1242/jcs.086124 [DOI] [PubMed] [Google Scholar]
- Park L, Thomason PA, Zech T, King JS, Veltman DM, Carnell M, Ura S, Machesky LM, Insall RH, 2013. Cyclical action of the WASH complex: FAM21 and capping protein drive WASH recycling, not initial recruitment. Dev. Cell 24, 169–181. 10.1016/j.devcel.2012.12.014 [DOI] [PubMed] [Google Scholar]
- Paskus JD, Tian C, Fingleton E, Shen C, Chen X, Li Y, Myers SA, Badger JD, Bemben MA, Herring BE, Roche KW, 2019. Synaptic Kalirin-7 and Trio Interactomes Reveal a GEF Protein-Dependent Neuroligin-1 Mechanism of Action. Cell Rep. 29, 2944–2952.e5. 10.1016/j.celrep.2019.10.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paunola E, Mattila PK, Lappalainen P, 2002. WH2 domain: A small, versatile adapter for actin monomers. FEBS Lett. 513, 92–97. 10.1016/S0014-5793(01)03242-2 [DOI] [PubMed] [Google Scholar]
- Pernier J, Orban J, Avvaru BS, Jégou A, Romet-Lemonne G, Guichard B, Carlier MF, 2013. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat. Struct. Mol. Biol 20, 1069–1076. 10.1038/nsmb.2639 [DOI] [PubMed] [Google Scholar]
- Peterson JR, Bickford LC, Morgan D, Kim AS, Ouerfelli O, Kirschner MW, Rosen MK, 2004. Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat. Struct. Mol. Biol 11, 747–755. 10.1038/nsmb796 [DOI] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang J-S, Dai H, Sifuentes-Dominguez LF, Geng LN, Kaufmann SH, Hein MY, Wallis M, McGaughran J, Gecz J, Sluis B. van de, Billadeau DD, Burstein E, 2015. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol. Biol. Cell 26, 91–103. 10.1091/mbc.E14-06-1073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pils S, Kopp K, Peterson L, Tascón JD, Nyffenegger-Jann NJ, Hauck CR, 2012. The adaptor molecule Nck localizes the WAVE complex to promote actin polymerization during CEACAM3-mediated phagocytosis of bacteria. PLoS One 7. 10.1371/journal.pone.0032808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinyol R, Haeckel A, Ritter A, Qualmann B, Kessels MM, 2007. Regulation of N-WASP and the Arp2/3 complex by Abp1 controls neuronal morphology. PLoS One 2. 10.1371/journal.pone.0000400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowski JT, Gomez TS, Schoon RA, Mangalam AK, Billadeau DD, 2013. WASH knockout T cells demonstrate defective receptor trafficking, proliferation, and effector function. Mol. Cell. Biol 33, 958–973. 10.1128/MCB.01288-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipathsouk A, Brunetti RM, Town JP, Graziano BR, Breuer A, Pellett PA, Marchuk K, Tran NHT, Krummel MF, Stamou D, Weiner OD, 2021. The wave complex associates with sites of saddle membrane curvature. J. Cell Biol 220. 10.1083/jcb.202003086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocha SM, Cory GO, 2009. WAVE2 is regulated by multiple phosphorylation events within its VCA domain. Cell Motil. Cytoskeleton 66, 36–47. 10.1002/cm.20323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesskaya A, Boutillon A, Yang S, Wang Y, Romero S, Liu Y, Lavielle M, Vacher S, Schnitzler A, Molinie N, Rocques N, Fokin A, Guérois R, Bièche I, Chen B, David NB, Gautreau AM, 2021. CYFIP2-containing WAVE complexes inhibit cell migration by a competition mechanism. bioRxiv 2020.07.02.184655. [Google Scholar]
- Pollard TD, 2016. Actin and Actin-Binding Proteins. Cold Spring Harb. Perspect. Biol 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Cooper JA, 2009. Actin, a central player in cell shape and movement. Science (80-.) 326, 1208–1212. 10.1126/science.1175862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Tammela TLJ, Vessella RL, Visakorpi T, 2004. RAD21 and KIAA0196 at 8q24 Are Amplified and Overexpressed in Prostate Cancer. Genes Chromosom. Cancer 39, 1–10. 10.1002/gcc.10289 [DOI] [PubMed] [Google Scholar]
- Prehoda KE, Scott JA, Mullins RD, Lim WA, 2000. Integration of multiple signals through cooperative regulation of the N-WASP-Arp2/3 complex. Science (80-.) 290, 801–806. 10.1126/science.290.5492.801 [DOI] [PubMed] [Google Scholar]
- Puthenveedu MA, Lauffer B, Temkin P, Vistein R, Carlton P, Thorn K, Taunton J, Weiner OD, Parton RG, von Zastrow M, 2010. Sequence-dependent sorting of recycling proteins by actin-stabilized endosomal microdomains. Cell 143, 761–773. 10.1016/j.cell.2010.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualmann B, Roos J, DiGregorio PJ, Kelly RB, 1999. Syndapin I, a synaptic dynamin-binding protein that associates with the neural Wiskott-Aldrich syndrome protein. Mol. Biol. Cell 10, 501–513. 10.1091/mbc.10.2.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD, 2005. Drosophila Spire is an actin nucleation factor. Nature 433, 382–388. 10.1038/nature03241 [DOI] [PubMed] [Google Scholar]
- Rahman AA, Morrison BE, 2019. Contributions of VPS35 Mutations to Parkinson’s Disease. Neuroscience 401, 1–10. 10.1016/j.neuroscience.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajabian T, Gavicherla B, Heisig M, Müller-Altrock S, Goebel W, Gray-Owen SD, Ireton K, 2009. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol 11, 1212–1218. 10.1038/ncb1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak MA, Buehler J, Zeltzer S, Reitsma J, Molina B, Terhune S, 2018. Human Cytomegalovirus UL135 Interacts with Host Adaptor Proteins To Regulate Epidermal Growth Factor Receptor and Reactivation from Latency Michael. J. Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramesh N, Antón IM, Hartwig JH, Geha RS, 1997. WIP, a protein associated with Wiskott-Aldrich syndrome protein, induces actin polymerization and redistribution in lymphoid cells. Proc. Natl. Acad. Sci. U. S. A 94, 14671–14676. 10.1073/pnas.94.26.14671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recher M, Burns SO, Fuente M.A. De, Volpi S, Dahlberg C, Walter JE, Moffitt K, Mathew D, Honke N, Lang PA, Patrizi L, Falet H, Meszei M, Mizui M, Csizmadia E, Candotti F, Nadeau K, Bouma G, Delmonte OM, Frugoni F, Fomin ABF, Buchbinder D, Lundequist EM, Massaad MJ, Tsokos GC, Hartwig J, Manis J, Terhorst C, Geha RS, Snapper SB, Lang KS, Malley R, Westerberg LS, Thrasher AJ, Notarangelo LD, 2012. B cell – intrinsic deficiency of the Wiskott - Aldrich syndrome protein (WASp) causes severe abnormalities of the peripheral B - cell compartment in mice. Blood 119, 2819–2828. 10.1182/blood [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SCO, Serio AW, Welch MD, 2012. Rickettsia parkeri invasion of diverse host cells involves an Arp2/3 complex, WAVE complex and Rho-family GTPase-dependent pathway. Cell. Microbiol 14, 529–545. 10.1111/j.1462-5822.2011.01739.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnders MRF, Ansor NM, Kousi M, Yue WW, Tan PL, Clarkson K, Clayton-Smith J, Corning K, Jones JR, Lam WWK, Mancini GMS, Marcelis C, Mohammed S, Pfundt R, Roifman M, Cohn R, Chitayat D, Millard TH, Katsanis N, Brunner HG, Banka S, 2017. RAC1 Missense Mutations in Developmental Disorders with Diverse Phenotypes. Am. J. Hum. Genet 101, 466–477. 10.1016/j.ajhg.2017.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodin K, Divaris K, North KE, Barros SP, Moss K, Beck JD, Offenbacher S, 2014. Chronic periodontitis genome-wide association studies: Gene-centric and gene set enrichment analyses. J. Dent. Res 93, 882–890. 10.1177/0022034514544506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A, 1992. The small GTP-binding protein rac regulates growth factor-induced membrane ruffling. Cell 70, 401–410. 10.1016/0092-8674(92)90164-8 [DOI] [PubMed] [Google Scholar]
- Rivero-Lezcano OM, Marcilla A, Sameshima JH, Robbins KC, 1995. Wiskott-Aldrich syndrome protein physically associates with Nck through Src homology 3 domains. Mol. Cell. Biol 15, 5725–5731. 10.1128/mcb.15.10.5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers SL, Wiedemann U, Stuurman N, Vale RD, 2003. Molecular requirements for actin-based lamella formation in Drosophila S2 cells. J. Cell Biol 162, 1079–1088. 10.1083/jcb.200303023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ho HYH, Kirschner MW, 2000. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4,5-bisphosphate. J. Cell Biol 150, 1299–1309. 10.1083/jcb.150.6.1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R, Ma L, Miki H, Lopez M, Kirchhausen T, Takenawa T, Kirschner MW, 1999. N-WASP activates the Arp2/3 complex and links Cdc42 and phosphoinositide signals to actin assembly. Mol. Biol. Cell 10, 122A. [DOI] [PubMed] [Google Scholar]
- Rohatgi R, Nollau P, Henry Ho HY, Kirschner MW, Mayer BJ, 2001. Nck and Phosphatidylinositol 4,5-Bisphosphate Synergistically Activate Actin Polymerization through the N-WASP-Arp2/3 Pathway. J. Biol. Chem 276, 26448–26452. 10.1074/jbc.M103856200 [DOI] [PubMed] [Google Scholar]
- Ropers F, Derivery E, Hu H, Garshasbi M, Karbasiyan M, Herold M, Nürnberg G, Ullmann R, Gautreau A, Sperling K, Varon R, Rajab A, 2011. Identification of a novel candidate gene for non-syndromic autosomal recessive intellectual disability: The WASH complex member swip. Hum. Mol. Genet 20, 2585–2590. 10.1093/hmg/ddr158 [DOI] [PubMed] [Google Scholar]
- Rottner K, Hänisch J, Campellone KG, 2010. WASH, WHAMM and JMY: Regulation of Arp2/3 complex and beyond. Trends Cell Biol. 20, 650–661. 10.1016/j.tcb.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Rottner K, Stradal TEB, Chen B, 2021. WAVE regulatory complex. Curr. Biol 31, R512–R517. 10.1016/j.cub.2021.01.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE, 2013. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat. Rev. Mol. Cell Biol 14, 7–12. 10.1038/nrm3492 [DOI] [PubMed] [Google Scholar]
- Russo AJ, Mathiowetz AJ, Hong S, Welch MD, Campellone KG, 2016. Rab1 recruits WHAMM during membrane remodeling but limits actin nucleation. Mol. Biol. Cell 27, 967–978. 10.1091/mbc.E15-07-0508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryder PV, Vistein R, Gokhale A, Seaman MN, Puthenveedu MA, Faundez V, 2013. The WASH complex, an endosomal Arp2/3 activator, interacts with the Hermansky-Pudlak syndrome complex BLOC-1 and its cargo phosphatidylinositol-4-kinase type II. Mol. Biol. Cell 24, 2269–2284. 10.1091/mbc.E13-02-0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar MA, Kwiatkowski AV, Pellegrini L, Cestra G, Butler MH, Rossman KL, Serna DM, Sondek J, Gertler FB, De Camilli P, 2003. Tuba, a novel protein containing bin/amphiphysin/Rvs and Dbl homology domains, links dynamin to regulation of the actin cytoskeleton. J. Biol. Chem 278, 49031–49043. 10.1074/jbc.M308104200 [DOI] [PubMed] [Google Scholar]
- Sallee NA, Rivera GM, Dueber JE, Vasilescu D, Mullins RD, Mayer BJ, Lim WA, 2008. The pathogen protein EspFU hijacks actin polymerization using mimicry and multivalency. Nature 454, 1005–1008. 10.1038/nature07170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzer E, Zoghi S, Kiss MG, Kage F, Rashkova C, Stahnke S, Haimel M, Platzer R, Caldera M, Ardy RC, Hoeger B, Block J, Medgyesi D, Sin C, Shahkarami S, Kain R, Ziaee V, Hammerl P, Bock C, Menche J, Dupré L, Huppa JB, Sixt M, Lomakin A, Rottner K, Binder CJ, Stradal TEB, Rezaei N, Boztug K, 2020. The cytoskeletal regulator HEM1 governs B cell development and prevents autoimmunity. Sci. Immunol 5. 10.1126/sciimmunol.abc3979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AM, Flamini MI, Baldacci C, Goglia L, Genazzani AR, Simoncini T, 2010. Estrogen receptor-α promotes breast cancer cell motility and invasion via focal adhesion kinase and N-WASP. Mol. Endocrinol 24, 2114–2125. 10.1210/me.2010-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangaré LO, Ólafsson EB, Wang Y, Yang N, Julien L, Camejo A, Pesavento P, Sidik SM, Lourido S, Barragan A, Saeij JPJ, 2019. In Vivo CRISPR Screen Identifies TgWIP as a Toxoplasma Modulator of Dendritic Cell Migration. Cell Host Microbe 26, 478–492.e8. 10.1016/j.chom.2019.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmiento C, Wang W, Dovas A, Yamaguchi H, Sidani M, El-Sibai M, DesMarais V, Holman HA, Kitchen S, Backer JM, Alberts A, Condeelis J, 2008. WASP family members and formin proteins coordinate regulation of cell protrusions in carcinoma cells. J. Cell Biol 180, 1245–1260. 10.1083/jcb.200708123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasahara Y, Rachid R, Byrne MJ, De la Fuente MA, Abraham RT, Ramesh N, Geha RS, 2002. Mechanism of recruitment of WASP to the immunological synapse and of its activation following TCR ligation. Mol. Cell 10, 1269–1281. 10.1016/S1097-2765(02)00728-1 [DOI] [PubMed] [Google Scholar]
- Schaks M, Döring H, Kage F, Steffen A, Klünemann T, Blankenfeldt W, Stradal T, Rottner K, 2021. RhoG and Cdc42 can contribute to Rac-dependent lamellipodia formation through WAVE regulatory complex-binding. Small GTPases 12, 122–132. 10.1080/21541248.2019.1657755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks M, Reinke M, Witke W, Rottner K, 2020. Molecular Dissection of Neurodevelopmental Disorder-Causing Mutations in CYFIP2. Cells 9, 1–14. 10.3390/cells9061355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaks M, Singh SP, Kage F, Thomason P, Klünemann T, Steffen A, Blankenfeldt W, Stradal TE, Insall RH, Rottner K, 2018. Distinct Interaction Sites of Rac GTPase with WAVE Regulatory Complex Have Non-redundant Functions in Vivo. Curr. Biol 28, 3674–3684.e6. 10.1016/j.cub.2018.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenck A, Bardoni B, Moro A, Bagni C, Mandel JL, 2001. A highly conserved protein family interacting with the fragile X mental retardation protein (FMRP) and displaying selective interactions with FMRP-related proteins FXR1P and FXR2P. Proc. Natl. Acad. Sci. U. S. A 98, 8844–8849. 10.1073/pnas.151231598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlüter K, Waschbüsch D, Anft M, Hügging D, Kind S, Hänisch J, Lakisic G, Gautreau A, Barnekow A, Stradal TEB, 2014. JMY is involved in anterograde vesicle trafficking from the trans-Golgi network. Eur. J. Cell Biol 93, 194–204. 10.1016/j.ejcb.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Schnelzer A, Prechtel D, Knaus U, Dehne K, Gerhard M, Graeff H, Harbeck N, Schmitt M, Lengyel E, 2000. Rac1 in human breast cancer: Overexpression, mutation analysis, and characterization of a new isoform, Rac1b. Oncogene 19, 3013–3020. 10.1038/sj.onc.1203621 [DOI] [PubMed] [Google Scholar]
- Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, Gottesman ME, Gautier J, 2018. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature 559, 61–66. 10.1038/s41586-018-0237-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwinger W, Urban C, Ulreich R, Sperl D, Karastaneva A, Strenger V, Lackner H, Boztug K, Albert MH, Benesch M, Seidel MG, 2018. The phenotype and treatment of WIP deficiency: Literature Synopsis and Review of a Patient With Pre-transplant Serial Donor Lymphocyte Infusions to Eliminate CMV. Front. Immunol 9, 1–7. 10.3389/fimmu.2018.02554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scita G, Nordstrom J, Carbone R, Tenca P, Giardina G, Gutkind S, Bjarnegård M, Betsholtz C, Di Fiore PP, 1999. EPS8 and E3B1 transduce signals from Ras to Rac. Nature 401, 290–293. 10.1038/45822 [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Gautreau A, Billadeau DD, 2013. Retromer-mediated endosomal protein sorting: All WASHed up! Trends Cell Biol. 23, 522–528. 10.1016/j.tcb.2013.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaaban M, Chowdhury S, Nolen BJ, 2020. Cryo-EM reveals the transition of Arp2/3 complex from inactive to nucleation-competent state. Nat. Struct. Mol. Biol 27, 1009–1016. 10.1038/s41594-020-0481-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang D, Dong L, Zeng L, Yang R, Xu J, Wu Y, Xu R, Tao H, Zhang N, 2015. Two-stage comprehensive evaluation of genetic susceptibility of common variants in FBXO38, AP3B2 and WHAMM to severe chronic periodontitis. Sci. Rep 5, 1–7. 10.1038/srep17882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- She HY, Rockow S, Tang J, Nishimura R, Skolnik EY, Chen M, Margolis B, Li W, 1997. Wiskott-Aldrich syndrome protein is associated with the adapter protein Grb2 and the epidermal growth factor receptor in living cells. Mol. Biol. Cell 8, 1709–1721. 10.1091/mbc.8.9.1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shekarabi M, Moore SW, Tritsch NX, Morris SJ, Bouchard JF, Kennedy TE, 2005. Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. J. Neurosci 25, 3132–3141. 10.1523/JNEUROSCI.1920-04.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen QT, Hsiue PP, Sindelar CV, Welch MD, Campellone KG, Wang HW, 2012. Structural insights into WHAMM-mediated cytoskeletal coordination during membrane remodeling. J. Cell Biol 199, 111–124. 10.1083/jcb.201204010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Scita G, Casanova JE, 2005. WAVE2 signaling mediates invasion of polarized epithelial cells by Salmonella typhimurium. J. Biol. Chem 280, 29849–29855. 10.1074/jbc.M500617200 [DOI] [PubMed] [Google Scholar]
- Shi R, Kramer DA, Chen B, Shen K, 2021. A two-step actin polymerization mechanism drives dendrite branching. Neural Dev. 16, 1–16. 10.1186/s13064-021-00154-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikama N, Lee CW, France S, Delavaine L, Lyon J, Krstic-Demonacos M, La Thangue NB, 1999. A novel cofactor for p300 that regulates the p53 response. Mol. Cell 4, 365–376. 10.1016/S1097-2765(00)80338-X [DOI] [PubMed] [Google Scholar]
- Shimojima Yamamoto K, Yanagishita T, Yamamoto H, Miyamoto Y, Nagata M, Ishihara Y, Miyashita Y, Asano Y, Sakata Y, Yamamoto T, 2021. Recurrent de novo pathogenic variant of WASF1 in a Japanese patient with neurodevelopmental disorder with absent language and variable seizures. Hum. Genome Var 8, 2–4. 10.1038/s41439-021-00176-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin N, Ahn N, Chang-Ileto B, Park J, Takei K, Ahn SG, Kim SA, Di Paolo G, Chang S, 2008. SNX9 regulates tubular invagination of the plasma membrane through interaction with actin cytoskeleton and dynamin 2. J. Cell Sci 121, 1252–1263. 10.1242/jcs.016709 [DOI] [PubMed] [Google Scholar]
- Shortrede JE, Uzair ID, Neira FJ, Flamini MI, Sanchez AM, 2016. Paxillin, a novel controller in the signaling of estrogen to FAK/N-WASP/Arp2/3 complex in breast cancer cells. Mol. Cell. Endocrinol 430, 56–67. 10.1016/j.mce.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Simonetti B, Cullen PJ, 2019. Actin-dependent endosomal receptor recycling. Curr. Opin. Cell Biol 56, 22–33. 10.1016/j.ceb.2018.08.006 [DOI] [PubMed] [Google Scholar]
- Sims R, Van Der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, Martin ER, Grenier-Boley B, Heilmann-Heimbach S, Chouraki V, Kuzma AB, Sleegers K, Vronskaya M, Ruiz A, Graham RR, Olaso R, Hoffmann P, Grove ML, Vardarajan BN, Hiltunen M, Nöthen MM, White CC, Hamilton-Nelson KL, Epelbaum J, Maier W, Choi SH, Beecham GW, Dulary C, Herms S, Smith AV, Funk CC, Derbois C, Forstner AJ, Ahmad S, Li H, Bacq D, Harold D, Satizabal CL, Valladares O, Squassina A, Thomas R, Brody JA, Qu L, Sánchez-Juan P, Morgan T, Wolters FJ, Zhao Y, Garcia FS, Denning N, Fornage M, Malamon J, Naranjo MCD, Majounie E, Mosley TH, Dombroski B, Wallon D, Lupton MK, Dupuis J, Whitehead P, Fratiglioni L, Medway C, Jian X, Mukherjee S, Keller L, Brown K, Lin H, Cantwell LB, Panza F, McGuinness B, Moreno-Grau S, Burgess JD, Solfrizzi V, Proitsi P, Adams HH, Allen M, Seripa D, Pastor P, Cupples LA, Price ND, Hannequin D, Frank-García A, Levy D, Chakrabarty P, Caffarra P, Giegling I, Beiser AS, Giedraitis V, Hampel H, Garcia ME, Wang X, Lannfelt L, Mecocci P, Eiriksdottir G, Crane PK, Pasquier F, Boccardi V, Henández I, Barber RC, Scherer M, Tarraga L, Adams PM, Leber M, Chen Y, Albert MS, Riedel-Heller S, Emilsson V, Beekly D, Braae A, Schmidt R, Blacker D, Masullo C, Schmidt H, Doody RS, Spalletta G, Jr WTL, Fairchild TJ, Bossù P, Lopez OL, Frosch MP, Sacchinelli E, Ghetti B, Yang Q, Huebinger RM, Jessen F, Li S, Kamboh MI, Morris JC, Sotolongo-Grau O, Katz MJ, Corcoran C, Dunstan M, Braddel A, Thomas C, Meggy A, Marshall R, Gerrish A, Chapman J, Aguilar M, Taylor S, Hill M, Fairén MD, Hodges A, Vellas B, Soininen H, Kloszewska I, Daniilidou M, Uphill J, Patel Y, Hughes JT, Lord J, Turton J, Hartmann AM, Cecchetti R, Fenoglio C, Serpente M, Arcaro M, Caltagirone C, Orfei MD, Ciaramella A, Pichler S, Mayhaus M, Gu W, Lleó A, Fortea J, Blesa R, Barber IS, Brookes K, Cupidi C, Maletta RG, Carrell D, Sorbi S, Moebus S, Urbano M, Pilotto A, Kornhuber J, Bosco P, Todd S, Craig D, Johnston J, Gill M, Lawlor B, Lynch A, Fox NC, Hardy J, Albin RL, Apostolova LG, Arnold SE, Asthana S, Atwood CS, Baldwin CT, Barnes LL, Barral S, Beach TG, Becker JT, Bigio EH, Bird TD, Boeve BF, Bowen JD, Boxer A, Burke JR, Burns JM, Buxbaum JD, Cairns NJ, Cao C, Carlson CS, Carlsson CM, Carney RM, Carrasquillo MM, Carroll SL, Diaz CC, Chui HC, Clark DG, Cribbs DH, Crocco EA, Decarli C, Dick M, Duara R, Evans DA, Faber KM, Fallon KB, Fardo DW, Farlow MR, Ferris S, Foroud TM, Galasko DR, Gearing M, Geschwind DH, Gilbert JR, Graff-Radford NR, Green RC, Growdon JH, Hamilton RL, Harrell LE, Honig LS, Huentelman MJ, Hulette CM, Hyman BT, Jarvik GP, Abner E, Jin LW, Jun G, Karydas A, Kaye JA, Kim R, Kowall NW, Kramer JH, Laferla FM, Lah JJ, Leverenz JB, Levey AI, Li G, Lieberman AP, Lunetta KL, Lyketsos CG, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Murrell JR, Myers AJ, O’Bryant S, Olichney JM, Pankratz VS, Parisi JE, Paulson HL, Perry W, Peskind E, Pierce A, Poon WW, Potter H, Quinn JF, Raj A, Raskind M, Reisberg B, Reitz C, Ringman JM, Roberson ED, Rogaeva E, Rosen HJ, Rosenberg RN, Sager MA, Saykin AJ, Schneider JA, Schneider LS, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tanzi RE, Thornton-Wells TA, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Garzia F, Golamaully F, Septier G, Engelborghs S, Vandenberghe R, De Deyn PP, Fernadez CM, Benito YA, Thonberg H, Forsell C, Lilius L, Kinhult-Stählbom A, Kilander L, Brundin R, Concari L, Helisalmi S, Koivisto AM, Haapasalo A, Dermecourt V, Fievet N, Hanon O, Dufouil C, Brice A, Ritchie K, Dubois B, Himali JJ, Keene CD, Tschanz J, Fitzpatrick AL, Kukull WA, Norton M, Aspelund T, Larson EB, Munger R, Rotter JI, Lipton RB, Bullido MJ, Hofman A, Montine TJ, Coto E, Boerwinkle E, Petersen RC, Alvarez V, Rivadeneira F, Reiman EM, Gallo M, O’Donnell CJ, Reisch JS, Bruni AC, Royall DR, Dichgans M, Sano M, Galimberti D, St George-Hyslop P, Scarpini E, Tsuang DW, Mancuso M, Bonuccelli U, Winslow AR, Daniele A, Wu CK, Peters O, Nacmias B, Riemenschneider M, Heun R, Brayne C, Rubinsztein DC, Bras J, Guerreiro R, Al-Chalabi A, Shaw CE, Collinge J, Tsolaki M, Clarimón J, Sussams R, Lovestone S, O’Donovan MC, Owen MJ, Behrens TW, Mead S, Uitterlinden AG, Holmes C, Cruchaga C, Ingelsson M, Bennett DA, Powell J, Golde TE, Graff C, De Jager PL, Morgan K, Ertekin-Taner N, Combarros O, Psaty BM, Passmore P, Younkin SG, Berr C, Gudnason V, Rujescu D, Dickson DW, Dartigues JF, Destefano AL, Ortega-Cubero S, Hakonarson H, Campion D, Boada M, Kauwe JK, Farrer LA, Van Broeckhoven C, Ikram MA, Jones L, Haines JL, Tzourio C, Tzourio C, Escott-Price V, Mayeux R, Deleuze JF, Amin N, Goate AM, Pericak-Vance MA, Amouyel P, Van Duijn CM, Ramirez A, Wang LS, Lambert JC, Seshadri S, Williams J, Schellenberg GD, 2017. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet 49, 1373–1384. 10.1038/ng.3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SP, Thomason PA, Lilla S, Schaks M, Tang Q, Goodei BL, MacHesky LM, Rottner K, Insall RH, 2020. Cell-substrate adhesion drives scar/wave activation and phosphorylation by a ste20-family kinase, which controls pseudopod lifetime, PLoS Biology. 10.1371/JOURNAL.PBIO.3000774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Davidson AC, Hume PJ, Koronakis V, 2020. Arf6 can trigger wave regulatory complex-dependent actin assembly independent of arno. Int. J. Mol. Sci 21. 10.3390/ijms21072457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper SB, Meelu P, Nguyen D, Stockton BM, Bozza P, Alt FW, Rosen FS, von Andrian UH, Klein C, 2005. WASP deficiency leads to global defects of directed leukocyte migration in vitro and in vivo. J. Leukoc. Biol 77, 993–998. 10.1189/jlb.0804444 [DOI] [PubMed] [Google Scholar]
- Snapper SB, Rosen FS, 1999. The Wiskott-Aldrich Syndrome Protein (WASP): Roles in signaling and cytoskeletal organization. Annu. Rev. Immunol 17, 905–929. 10.1146/annurev.immunol.17.1.905 [DOI] [PubMed] [Google Scholar]
- Snapper SB, Takeshima F, Antón I, Liu CH, Thomas SM, Nguyen D, Dudley D, Fraser H, Purich D, Lopez-Ilasaca M, Klein C, Davidson L, Bronson R, Mulligan RC, Southwick F, Geha R, Goldberg MB, Rosen FS, Hartwig JH, Alt FW, 2001. N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol 3, 897–904. 10.1038/ncb1001-897 [DOI] [PubMed] [Google Scholar]
- Soderling SH, Binns KL, Wayman GA, Davee SM, Ong SH, Pawson T, Scott JD, 2002. The WRP component of the WAVE-1 complex attenuates Rac-mediated signalling. Nat. Cell Biol 4, 970–975. 10.1038/ncb886 [DOI] [PubMed] [Google Scholar]
- Soderling SH, Guire ES, Kaech S, White J, Zhang F, Schutz K, Langeberg LK, Banker G, Raber J, Scott JD, 2007. A WAVE-1 and WRP Signaling Complex Regulates Spine Density, Synaptic Plasticity, and Memory. J. Neurosci 27, 355–365. 10.1523/jneurosci.3209-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderling SH, Langeberg LK, Soderling JA, Davee SM, Simerly R, Raber J, Scott JD, 2003. Loss of WAVE-1 causes sensorimotor retardation and reduced learning and memory in mice. Proc. Natl. Acad. Sci 100, 1723–1728. 10.1073/pnas.0438033100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Li X, Cowell JK, 2007a. c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem 282, 26257–26265. 10.1074/jbc.M701484200 [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Ranalli TA, Li X, Bakin AV, Cowell JK, 2005. WAVE3 promotes cell motility and invasion through the regulation of MMP-1, MMP-3, and MMP-9 expression. Exp. Cell Res 308, 135–145. 10.1016/j.yexcr.2005.04.011 [DOI] [PubMed] [Google Scholar]
- Sossey-Alaoui K, Safina A, Li X, Vaughan MM, Hicks DG, Bakin AV, Cowell JK, 2007b. Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am. J. Pathol 170, 2112–2121. 10.2353/ajpath.2007.060975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear M, Guo J, Turner A, Yu D, Wang W, Meltzer B, He S, Hu X, Shang H, Kuhn J, Wu Y, 2014. HIV-1 triggers WAVE2 phosphorylation in primary CD4 T cells and macrophages, mediating Arp2/3-dependent nuclear migration. J. Biol. Chem 289, 6949–6959. 10.1074/jbc.M113.492132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squarr AJ, Brinkmann K, Chen B, Steinbacher T, Ebnet K, Rosen MK, Bogdan S, 2016. Fat2 acts through the WAVE regulatory complex to drive collective cell migration during tissue rotation. J. Cell Biol 212, 591–603. 10.1083/jcb.201508081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S, Macke EL, Swanson LC, Coulter D, Klee EW, Mullegama SV, Xie Y, Lanpher BC, Bedoukian EC, Skraban CM, Villard L, Milh M, Leppert MLO, Cohen JS, 2021. Expansion of the genotypic and phenotypic spectrum of wasf1-related neurodevelopmental disorder. Brain Sci. 11, 1–10. 10.3390/brainsci11070931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahnke S, Döring H, Kusch C, de Gorter DJJ, Dütting S, Guledani A, Pleines I, Schnoor M, Sixt M, Geffers R, Rohde M, Müsken M, Kage F, Steffen A, Faix J, Nieswandt B, Rottner K, Stradal TEB, 2021. Loss of Hem1 disrupts macrophage function and impacts migration, phagocytosis, and integrin-mediated adhesion. Curr. Biol 31, 2051–2064.e8. 10.1016/j.cub.2021.02.043 [DOI] [PubMed] [Google Scholar]
- Stallings-Mann ML, Waldmann J, Zhang Y, Miller E, Gauthier ML, Visscher DW, Downey GP, Radisky ES, Fields AP, Radisky DC, 2012. Matrix metalloproteinase induction of Rac1b, a key effector of lung cancer progression. Sci. Transl. Med 4. 10.1126/scitranslmed.3004062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton RJ, Prod’Homme V, Purbhoo MA, Moore M, Aicheler RJ, Heinzmann M, Bailer SM, Haas J, Antrobus R, Weekes MP, Lehner PJ, Vojtesek B, Miners KL, Man S, Wilkie GS, Davison AJ, Wang ECY, Tomasec P, Wilkinson GWG, 2014. HCMV pUL135 remodels the actin cytoskeleton to impair immune recognition of infected cells. Cell Host Microbe 16, 201–214. 10.1016/j.chom.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavoe AKH, Colón-Ramos DA, 2012. Netrin instructs synaptic vesicle clustering through Rac GTPase, MIG-10, and the actin cytoskeleton. J. Cell Biol 197, 75–88. 10.1083/jcb.201110127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen A, Rottner K, Ehinger J, Innocenti M, Scita G, Wehland J, Stradal TEB, 2004. Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759. 10.1038/sj.emboj.7600084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H, 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol 10, 513. [DOI] [PubMed] [Google Scholar]
- Stephan R, Gohl C, Fleige A, Klämbt C, Bogdan S, 2011. Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila. Mol. Biol. Cell 22, 4079–4092. 10.1091/mbc.E11-02-0121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JR, Gonzalez FH, Kawai H, Yuan Z-M, 2006. c-Abl Interacts with the WAVE2 Signaling Complex to Induce Membrane Ruffling and Cell Spreading. J. Biol. Chem 281, 31290–31297. 10.1016/s0021-9258(19)84041-3 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Hattori M, Miki H, Tezuka T, Yamamoto T, Mikoshiba K, Takenawa T, 2002. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev. Cell 3, 645–658. 10.1016/S1534-5807(02)00324-6 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Kurisu S, Oikawa T, Yamazaki D, Oda A, Takenawa T, 2006. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP3, and Rac. J. Cell Biol 173, 571–585. 10.1083/jcb.200509067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Miki H, Takenawa T, 1999. Identification of two human WAVE/SCAR homologues as general actin regulatory molecules which associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun 260, 296–302. 10.1006/bbrc.1999.0894 [DOI] [PubMed] [Google Scholar]
- Suetsugu S, Tezuka T, Morimura T, Hattori M, Mikoshiba K, Yamamoto T, Takenawa T, 2004. Regulation of actin cytoskeleton by mDab1 through N-WASP and ubiquitination of mDab1. Biochem. J 384, 1–8. 10.1042/BJ20041103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Yamazaki D, Kurisu S, Takenawa T, 2003. Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 5, 595–609. 10.1016/S1534-5807(03)00297-1 [DOI] [PubMed] [Google Scholar]
- Sun SC, Sun QY, Kim NH, 2011. JMY is required for asymmetric division and cytokinesis in mouse oocytes. Mol. Hum. Reprod 17, 296–304. 10.1093/molehr/gar006 [DOI] [PubMed] [Google Scholar]
- Sung JY, Engmann O, Teylan MA, Nairn AC, Greengard P, Kim Y, 2008. WAVE1 controls neuronal activity-induced mitochondrial distribution in dendritic spines. Proc. Natl. Acad. Sci. U. S. A 105, 3112–3116. 10.1073/pnas.0712180105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miki H, Takenawa T, Sasakawa C, 1998. Neural Wiskott-Aldrich syndrome protein is implicated in the actin-based motility of Shigella flexneri. EMBO J. 17, 2767–2776. 10.1093/emboj/17.10.2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney MO, Collins A, Padrick SB, Goode BL, 2015. A novel role for WAVE1 in controlling actin network growth rate and architecture. Mol. Biol. Cell 26, 495–505. 10.1091/mbc.E14-10-1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A, 1996. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell 84, 723–734. 10.1016/S0092-8674(00)81050-8 [DOI] [PubMed] [Google Scholar]
- Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, Karatsu T, Takenawa T, Endo T, 2010. Nebulin and N-WASP Cooperate to Cause IGF-1–Induced Sarcomeric Actin Filament Formation. Science (80-.) 330. [DOI] [PubMed] [Google Scholar]
- Takata K, Kitamura Y, Nakata Y, Matsuoka Y, Tomimoto H, Taniguchi T, Shimohama S, 2009. Involvement of WAVE accumulation in Aβ/APP pathology-dependent tangle modification in Alzheimer’s disease. Am. J. Pathol 175, 17–24. 10.2353/ajpath.2009.080908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takenawa T, Suetsugu S, 2007. The WASP-WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol 8, 37–48. 10.1038/nrm2069 [DOI] [PubMed] [Google Scholar]
- Tang Q, Schaks M, Koundinya N, Yang C, Pollard LW, Svitkina TM, Rottner K, Goode BL, 2020. WAVE1 and WAVE2 have distinct and overlapping roles in controlling actin assembly at the leading edge. Mol. Biol. Cell 31, 2168–2178. 10.1091/MBC.E19-12-0705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniuchi K, Furihata M, Naganuma S, Saibara T, 2018. WAVE2 is associated with poor prognosis in pancreatic cancers and promotes cell motility and invasiveness via binding to ACTN4. Cancer Med. 7, 5733–5751. 10.1002/cam4.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA, 2000. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol 148, 519–530. 10.1083/jcb.148.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MA, Davuluri G, Parvani JG, Schiemann BJ, Wendt MK, Plow EF, Schiemann WP, Sossey-Alaoui K, 2013. Upregulated WAVE3 expression is essential for TGF-B-mediated EMT and metastasis of triple negative breast cancer cells. Breast Cancer Res. Treat 142, 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D’Silva S, Selvakumar A, Candotti F, Vyas YM, 2010. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci. Transl. Med 2. 10.1126/scitranslmed.3000813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temkin P, Lauffer B, Jäger S, Cimermancic P, Krogan NJ, von Zastrow M, 2011. SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol 13, 715–721. 10.1038/ncb2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng Y, Bahassan A, Dong D, Hanold LE, Ren X, Kennedy EJ, Cowell JK, 2016. Targeting the WASF3-CYFIP1 complex using stapled peptides suppresses cancer cell invasion. Cancer Res. 76, 965–973. 10.1158/0008-5472.CAN-15-1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrasher AJ, Burns SO, 2010. WASP: A key immunological multitasker. Nat. Rev. Immunol 10, 182–192. 10.1038/nri2724 [DOI] [PubMed] [Google Scholar]
- Torres E, Rosen MK, 2003. Contingent phosphorylation/dephosphorylation provides a mechanism of molecular memory in WASP. Mol. Cell 11, 1215–1227. 10.1016/S1097-2765(03)00139-4 [DOI] [PubMed] [Google Scholar]
- Tsarouhas V, Liu D, Tsikala G, Fedoseienko A, Zinn K, Matsuda R, Billadeau DD, Samakovlis C, 2019. WASH phosphorylation balances endosomal versus cortical actin network integrities during epithelial morphogenesis. Nat. Commun 10, 2193. 10.1038/s41467-019-10229-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboi S, Nonoyama S, Ochs HD, 2006. Wiskott-Aldrich syndrome protein is involved in αIIbβ3-mediated cell adhesion. EMBO Rep. 7, 506–511. 10.1038/sj.embor.7400665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Olsson IM, Edlund K, Lundberg E, Navani S, Szigyarto CAK, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, Von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, Von Heijne G, Nielsen J, Pontén F, 2015. Tissue-based map of the human proteome. Science (80-.) 347. 10.1126/science.1260419 [DOI] [PubMed] [Google Scholar]
- Ura S, Pollitt AY, Veltman DM, Morrice NA, MacHesky LM, Insall RH, 2012. Pseudopod growth and evolution during cell movement is controlled through SCAR/WAVE dephosphorylation. Curr. Biol 22, 553–561. 10.1016/j.cub.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaduva G, Martinez-Quiles N, Anton IM, Martin NC, Geha RS, Hopper AK, Ramesh N, 1999. The human WASP-interacting protein, WIP, activates the cell polarity pathway in yeast. J. Biol. Chem 274, 17103–17108. 10.1074/jbc.274.24.17103 [DOI] [PubMed] [Google Scholar]
- Valdmanis PN, Meijer IA, Reynolds A, Lei A, MacLeod P, Schlesinger D, Zatz M, Reid E, Dion PA, Drapeau P, Rouleau GA, 2007. Mutations in the KIAA0196 gene at the SPG8 locus cause hereditary spastic paraplegia. Am. J. Hum. Genet 80, 152–161. 10.1086/510782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman DM, Insall RH, 2010. WASP family proteins: Their evolution and its physiological implications. Mol. Biol. Cell 21, 2880–2893. 10.1091/mbc.E10-04-0372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon Jeffrey M, Rahe TK, Rodriguez-Mesa E, Parkhurst SM, 2015. Wash functions downstream of Rho1 GTPase in a subset of Drosophila immune cell developmental migrations. Mol. Biol. Cell 26, 1665–1674. 10.1091/mbc.E14-08-1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verboon Jeffrey M., Rincon-Arano H, Werwie TR, Delrow JJ, Scalzo D, Nandakumar V, Groudine M, Parkhurst SM, 2015. Wash interacts with lamin and affects global nuclear organization. Curr. Biol 25, 804–810. 10.1016/j.cub.2015.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva AA, Sanchez-Gomez P, Muñoz-Palma E, Puvogel S, Casas BS, Arriagada C, Peña-Villalobos I, Lois P, Ramírez Orellana M, Lubieniecki F, Casco Claro F, Gallegos I, García-Castro J, Torres VA, Palma V, 2021. The Netrin-1-Neogenin-1 signaling axis controls neuroblastoma cell migration via integrin-β1 and focal adhesion kinase activation. Cell Adhes. Migr 15, 58–73. 10.1080/19336918.2021.1892397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA, 2004. Structure of the N-WASP EVH1 Domain-WIP Complex. Cell 111, 565–576. 10.1016/s0092-8674(02)01076-0 [DOI] [PubMed] [Google Scholar]
- Volpi S, Santori E, Abernathy K, Mizui M, Dahlberg CIM, Recher M, Capuder K, Csizmadia E, Ryan D, Mathew D, Tsokos GC, Snapper SB, Westerberg LS, Thrasher AJ, Candotti F, Notarangelo LD, 2016. N-WASP is required for B-cell – mediated autoimmunity in Wiskott-Aldrich syndrome. Blood 127, 216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walpole GFW, Plumb JD, Chung D, Tang B, Boulay B, Osborne DG, Piotrowski JT, Catz SD, Billadeau DD, Grinstein S, Jaumouillé V, 2020. Inactivation of Rho GTPases by Burkholderia cenocepacia Induces a WASH-Mediated Actin Polymerization that Delays Phagosome Maturation. Cell Rep. 31, 107721. 10.1016/j.celrep.2020.107721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Fedoseienko A, Chen B, Burstein E, Jia D, Billadeau DD, 2018. Endosomal receptor trafficking: Retromer and beyond. Traffic 19, 578–590. 10.1111/tra.12574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. Sen, Zhong HJ, Xiao DW, Huang X, Liao L. Di, Xie ZF, Xu XE, Shen ZY, Xu LY, Li EM, 2010. The expression of CFL1 and N-WASP in esophageal squamous cell carcinoma and its correlation with clinicopathological features. Dis. Esophagus 23, 512–521. 10.1111/j.1442-2050.2009.01035.x [DOI] [PubMed] [Google Scholar]
- Wang W, Wang X, Fujioka H, Hoppel C, Whone AL, Caldwell MA, Cullen PJ, Liu J, Zhu X, 2016. Parkinson’s disease-associated mutant VPS35 causes mitochondrial dysfunction by recycling DLP1 complexes. Nat. Med 22, 54–63. 10.1038/nm.3983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegner AM, Nebhan CA, Hu L, Majumdar D, Meier KM, Weaver AM, Webb DJ, 2008. N-WASP and the Arp2/3 complex are critical regulators of actin in the development of dendritic spines and synapses. J. Biol. Chem 283, 15912–15920. 10.1074/jbc.M801555200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW, 2007. An actin-based wave generator organizes cell motility. PLoS Biol. 5, 2053–2063. 10.1371/journal.pbio.0050221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner OD, Rentel MC, Ott A, Brown GE, Jedrychowski M, Yaffe MB, Gygi SP, Cantley LC, Bourne HR, Kirschner MW, 2006. Hem-1 complexes are essential for Rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 4, 186–199. 10.1371/journal.pbio.0040038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen KK, Han SS, Vyas YM, 2020. Wiskott-Aldrich syndrome protein senses irradiation-induced DNA damage to coordinate the cell-protective Golgi dispersal response in human T and B lymphocytes. J. Allergy Clin. Immunol 145, 324–334. 10.1016/j.jaci.2019.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenjie H, Zhe L, Fan Y, Huan Z, Xin Y, Xiaoyu Y, Yifei Z, Lijia X, Yihong Z, Dingdong L, Wentong M, Wenming Z, Xiaohu Z, Xiaofei S, Qingxiang S, Li L, Cong M, Yuquan W, D. BD, Xianming M, Da J, 2019. Structural and functional studies of TBC1D23 C-terminal domain provide a link between endosomal trafficking and PCH. Proc. Natl. Acad. Sci 116, 22598–22608. 10.1073/pnas.1909316116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerberg LS, Dahlberg C, Baptista M, Moran CJ, Detre C, Keszei M, Eston MA, Alt FW, Terhorst C, Notarangelo LD, Snapper SB, 2012. Wiskott-Aldrich syndrome protein (WASP) and N-WASP are critical for peripheral B-cell development and function. Blood 119, 3966–3974. 10.1182/blood-2010-09-308197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston L, Coutts AS, La Thangué NB, 2012. Actin nucleators in the nucleus: An emerging theme. J. Cell Sci 125, 3519–3527. 10.1242/jcs.099523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westphal RS, Soderling SH, Alto NM, Langeberg LK, Scott JD, 2000. Scar / WAVE-1, a Wiskott Aldrich syndrome protein, assembles an actin-associated multi-kinase scaffold. EMBO J. 19, 4589–4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JJ, Mazzeu JF, Coban-Akdemir Z, Bayram Y, Bahrambeigi V, Hoischen A, van Bon BWM, Gezdirici A, Gulec EY, Ramond F, Touraine R, Thevenon J, Shinawi M, Beaver E, Heeley J, Hoover-Fong J, Durmaz CD, Karabulut HG, Marzioglu-Ozdemir E, Cayir A, Duz MB, Seven M, Price S, Ferreira BM, Vianna-Morgante AM, Ellard S, Parrish A, Stals K, Flores-Daboub J, Jhangiani SN, Gibbs RA, Brunner HG, Sutton VR, Lupski JR, Carvalho CMB, 2018. WNT Signaling Perturbations Underlie the Genetic Heterogeneity of Robinow Syndrome. Am. J. Hum. Genet 102, 27–43. 10.1016/j.ajhg.2017.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijers M, Zanoni P, Liv N, Vos DY, Jäckstein MY, Smit M, Wilbrink S, Wolters JC, van der Veen YT, Huijkman N, Dekker D, Kloosterhuis N, van Dijk TH, Billadeau DD, Kuipers F, Klumperman J, von Eckardstein A, Kuivenhoven JA, van de Sluis B, 2019. The hepatic WASH complex is required for efficient plasma LDL and HDL cholesterol clearance. JCI insight 4. 10.1172/jci.insight.126462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams DA, Tao W, Yang F, Kim C, Gu Y, Mansfield P, Levine JE, Petryniak B, Derrow CW, Harris C, Jia B, Zheng Y, Ambruso DR, Lowe JB, Atkinson SJ, Dinauer MC, Boxer L, 2000. Dominant negative mutation of the hematopoietic-specific Rho GTPase, Rac2, is associated with a human phagocyte immunodeficiency. Blood 96, 1646–1654. 10.1182/blood.V96.5.1646 [DOI] [PubMed] [Google Scholar]
- Woolfrey KM, Srivastava DP, Photowala H, Yamashita M, Barbolina MV, Cahill ME, Xie Z, Jones KA, Quilliam LA, Prakriya M, Penzes P, 2009. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat. Neurosci 12, 1275–1284. 10.1038/nn.2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K, Seylani A, Wu J, Wu X, Bleck CKE, Sack MN, 2021. BLOC1S1/GCN5L1/BORCS1 is a critical mediator for the initiation of autolysosomal tubulation. Autophagy 17, 3707–3724. 10.1080/15548627.2021.1894759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Suetsugu S, Cooper LA, Takenawa T, Guan JL, 2004. Focal Adhesion Kinase Regulation of N-WASP Subcellular Localization and Function. J. Biol. Chem 279, 9565–9576. 10.1074/jbc.M310739200 [DOI] [PubMed] [Google Scholar]
- Wu X, Yoo Y, Okuhama NN, Tucker PW, Liu G, Guan JL, 2006. Regulation of RNA-polymerase-II-dependent transcription by N-WASP and its nuclear-binding partners. Nat. Cell Biol 8, 756–763. 10.1038/ncb1433 [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Du Y, Zhao Z, Shi L, Sun L, Huang G, Ye B, Li C, Dai Z, Hou N, Cheng X, Sun Q, Li L, Yang X, Fan Z, 2013. WASH inhibits autophagy through suppression of Beclin 1 ubiquitination. EMBO J. 32, 2685–2696. 10.1038/emboj.2013.189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia P, Wang S, Huang G, Du Y, Zhu P, Li M, Fan Z, 2014. RNF2 is recruited by WASH to ubiquitinate AMBRA1 leading to downregulation of autophagy. Cell Res. 24, 943–958. 10.1038/cr.2014.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Wang X. feng, Li J. mei, Xi Z. qin, Lu Y, Wang L, Zeng Y, Guan L. feng, Yuan J, 2008. Overexpression of N-WASP in the brain of human epilepsy. Brain Res. 1233, 168–175. 10.1016/j.brainres.2008.07.101 [DOI] [PubMed] [Google Scholar]
- Xing G, Li M, Sun Y, Rui M, Zhuang Y, Lv H, Han J, Jia Z, Xie W, 2018. Neurexin–neuroligin 1 regulates synaptic morphology and functions via the WAVE regulatory complex in Drosophila neuromuscular junction. Elife 7, 1–23. 10.7554/eLife.30457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Fu X, Zhu S, Liu J-J, 2016. Retrolinkin recruits the WAVE1 protein complex to facilitate BDNF-induced TrkB endocytosis and dendrite outgrowth. Mol. Biol. Cell 27, 3342–3356. 10.1091/mbc.e16-05-0326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Quinn CC, 2012. MIG-10 Functions with ABI-1 to Mediate the UNC-6 and SLT-1 Axon Guidance Signaling Pathways. PLoS Genet. 8. 10.1371/journal.pgen.1003054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Padilla-Parra S, Park SJ, Itoh T, Chaineau M, Monaldi I, Cremona O, Benfenati F, De Camilli P, Coppey-Moisan M, Tramier M, Galli T, Takei K, 2009. Dynamic interaction of amphiphysin with N-WASP regulates actin assembly. J. Biol. Chem 284, 34244–34256. 10.1074/jbc.M109.064204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J, 2005. Molecular mechanisms of invadopodium formation: The role of the N-WASP-Arp2/3 complex pathway and cofilin. J. Cell Biol 168, 441–452. 10.1083/jcb.200407076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Ueda K, Kioka N, 2011. WAVE2 forms a complex with PKA and is involved in PKA enhancement of membrane protrusions. J. Biol. Chem 286, 3907–3914. 10.1074/jbc.M110.145409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki D, Oikawa T, Takenawa T, 2007. Rac-WAVE-mediated actin reorganization is required for organization and maintenance of cell-cell adhesion. J. Cell Sci 120, 86–100. 10.1242/jcs.03311 [DOI] [PubMed] [Google Scholar]
- Yamazaki D, Suetsugu S, Miki H, Kataoka Y, Nishikawa S-I, Fujiwara T, Yoshida N, Takenawa T, 2003. WAVE2 is required for directed cell migration and cardiovascular development. Nature 424, 452–456. 10.1038/nature01822 [DOI] [PubMed] [Google Scholar]
- Yan C, Martinez-Quiles N, Eden S, Shibata T, Takeshima F, Shinkura R, Fujiwara Y, Bronson R, Snapper SB, Kirschner MW, Geha R, Rosen FS, Alt FW, 2003. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22, 3602–3612. 10.1093/emboj/cdg350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagawa R, Furukawa Y, Tsunoda T, Kitahara O, Kameyama M, Murata K, Ishikawa O, Nakamura Y, 2001. Genome-wide screening of genes showing altered expression in liver metastases of human colorectal cancers by cDNA microarray. Neoplasia 3, 395–401. 10.1038/sj.neo.7900185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Bashaw GJ, 2006. Son of Sevenless Directly Links the Robo Receptor to Rac Activation to Control Axon Repulsion at the Midline. Neuron 52, 595–607. 10.1016/j.neuron.2006.09.039 [DOI] [PubMed] [Google Scholar]
- Yang W, Wu P. fei, Ma J. xing, Liao M. jun, Xu L. shan, Yi L, 2020. TRPV4 activates the Cdc42/N-wasp pathway to promote glioblastoma invasion by altering cellular protrusions. Sci. Rep 10, 1–15. 10.1038/s41598-020-70822-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama N, Lougheed J, Miller WT, 2005. Phosphorylation of WASP by the Cdc42-associated kinase ACK1: Dual hydroxyamino acid specificity in a tyrosine kinase. J. Biol. Chem 280, 42219–42226. 10.1074/jbc.M506996200 [DOI] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, Chung R, Pekle E, Zhang C, Towe M, Hussaini SMQ, Lee Y, Rujescu D, St. Clair D, Kleinman JE, Hyde TM, Krauss G, Christian KM, Rapoport JL, Weinberger DR, Song H, Ming GL, 2014. Modeling a genetic risk for schizophrenia in iPSCs and Mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell 15, 79–91. 10.1016/j.stem.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JJ, Lin-Chao S, 2010. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J. Biol. Chem 285, 11652–11666. 10.1074/jbc.M109.051094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Zech T, McDonald L, Gonzalez EG, Li A, Macpherson I, Schwarz JP, Spence H, Futó K, Timpson P, Nixon C, Ma Y, Anton IM, Visegrády B, Insall RH, Oien K, Blyth K, Norman JC, Machesky LM, 2012. N-WASP coordinates the delivery and F-actin-mediated capture of MT1-MMP at invasive pseudopods. J. Cell Biol 199, 527–544. 10.1083/jcb.201203025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zago G, Veith I, Singh MK, Fuhrmann L, De Beco S, Remorino A, Takaoka S, Palmeri M, Berger F, Brandon N, El Marjou A, Vincent-Salomon A, Camonis J, Coppey M, Parrini MC, 2018. Ralb directly triggers invasion downstream ras by mobilizing the wave complex. Elife 7, 1–23. 10.7554/eLife.40474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zavodszky E, Seaman MNJ, Moreau K, Jimenez-Sanchez M, Breusegem SY, Harbour ME, Rubinsztein DC, 2014. Mutation in VPS35 associated with Parkinson’s disease impairs WASH complex association and inhibits autophagy. Nat. Commun 5, 1–16. 10.1038/ncomms4828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zech T, Calaminus SDJ, Caswell P, Spence HJ, Carnell M, Insall RH, Norman J, Machesky LM, 2011. The Arp2/3 activator WASH regulates 5 1-integrin-mediated invasive migration. J. Cell Sci 124, 3753–3759. 10.1242/jcs.080986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zettl M, Way M, 2002. The WH1 and EVH1 domains of WASP and Ena/VASP family members bind distinct sequence motifs. Curr. Biol 12, 1617–1622. 10.1016/S0960-9822(02)01112-0 [DOI] [PubMed] [Google Scholar]
- Zhang J, Zhang X, Guo Y, Xu L, Pei D, 2009. Sorting nexin 33 induces mammalian cell micronucleated phenotype and actin polymerization by interacting with Wiskott-Aldrich syndrome protein. J. Biol. Chem 284, 21659–21669. 10.1074/jbc.M109.007278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao A, Zhou R, Gu Q, Liu M, Zhang B, Huang J, Yang B, Yao R, Wang J, Lv H, Wang J, Shen Y, Wang H, Chen X, 2021. Trio exome sequencing identified a novel de novo WASF1 missense variant leading to recurrent site substitution in a Chinese patient with developmental delay, microcephaly, and early-onset seizures: a mutational hotspot p.Trp161 and literature review. Clin. Chim. Acta 10.1016/j.cca.2021.08.030 [DOI] [PubMed] [Google Scholar]
- Zhao M, Spiess M, Johansson HJ, Olofsson H, Hu J, Lehtiö J, Strömblad S, 2017. Identification of the PAK4 interactome reveals PAK4 phosphorylation of N-WASP and promotion of Arp2/3-dependent actin polymerization. Oncotarget 8, 77061–77074. 10.18632/oncotarget.20352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Yunkai, Feng F, Hu G, Wang Y, Yu Y, Zhu Yuanfei, Xu W, Cai X, Sun Z, Han W, Ye R, Qu D, Ding Q, Huang X, Chen H, Xu W, Xie Y, Cai Q, Yuan Z, Zhang R, 2021. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat. Commun 12, 961. 10.1038/s41467-021-21213-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmet A, Van Eeuwen T, Boczkowska M, Rebowski G, Murakami K, Dominguez R, 2020. Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism. Sci. Adv 6, 1–14. 10.1126/sciadv.aaz7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel PA, Bunnell SC, Witherow DS, Gu JJ, Chislock EM, Ring C, Pendergast AM, 2006. Role for the Abi/Wave protein complex in T cell receptor-mediated proliferation and cytoskeletal remodeling. Curr. Biol 16, 35–46. 10.1016/j.cub.2005.12.024 [DOI] [PubMed] [Google Scholar]
- Zou W, Dong X, Broederdorf TR, Shen A, Kramer DA, Shi R, Liang X, Miller DM, Xiang YK, Yasuda R, Chen B, Shen K, 2018. A Dendritic Guidance Receptor Complex Brings Together Distinct Actin Regulators to Drive Efficient F-Actin Assembly and Branching. Dev. Cell 45, 362–375.e3. 10.1016/j.devcel.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Belin B, Mullins RD, 2012. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol. Biol. Cell 23, 853–863. 10.1091/mbc.E11-12-0992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Coutts AS, Quinlan ME, La Thangue NB, Mullins RD, 2009. p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol 11, 451–459. 10.1038/ncb1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweier M, Begemann A, McWalter K, Cho MT, Abela L, Banka S, Behring B, Berger A, Brown CW, Carneiro M, Chen J, Cooper GM, Finnila CR, Guillen Sacoto MJ, Henderson A, Hüffmeier U, Joset P, Kerr B, Lesca G, Leszinski GS, McDermott JH, Meltzer MR, Monaghan KG, Mostafavi R, Õunap K, Plecko B, Powis Z, Purcarin G, Reimand T, Riedhammer KM, Schreiber JM, Sirsi D, Wierenga KJ, Wojcik MH, Papuc SM, Steindl K, Sticht H, Rauch A, 2019. Spatially clustering de novo variants in CYFIP2, encoding the cytoplasmic FMRP interacting protein 2, cause intellectual disability and seizures. Eur. J. Hum. Genet 27, 747–759. 10.1038/s41431-018-0331-z [DOI] [PMC free article] [PubMed] [Google Scholar]


