Abstract
Sarcomas produce an abnormal extracellular matrix (ECM), which in turn provides instructive cues for cell growth and invasion. Neural EGF like-like molecule 1 (NELL1) is a secreted glycoprotein characterized by its non-neoplastic osteoinductive effects, yet it is highly expressed in skeletal sarcomas. Here, we show that genetic deletion of NELL1 markedly reduces invasive behavior across human osteosarcoma (OS) cell lines. NELL1 deletion resulted in reduced OS disease progression, inhibiting metastasis and improving survival in a xenograft mouse model. These observations were recapitulated with Nell1 conditional knockout in mouse models of p53/Rb-driven sarcomagenesis, which reduced tumor frequency and extended tumor-free survival. Transcriptomic and phosphoproteomic analyses demonstrated that NELL1 loss skews the expression of matricellular proteins associated with reduced FAK signaling. Culturing NELL1 knockout sarcoma cells on wild-type OS-enriched matricellular proteins reversed the phenotypic and signaling changes induced by NELL1 deficiency. In sarcoma patients, high expression of NELL1 correlated with decreased overall survival. These findings in mouse and human models suggest that NELL1 expression alters the sarcoma ECM, thereby modulating cellular invasive potential and prognosis. Disruption of NELL1 signaling may represent a novel therapeutic approach to short-circuit sarcoma disease progression.
Keywords: NELL-1, Nel-like protein 1, Neural epidermal growth factor-like 1, osteosarcoma, osteosarcomagenesis, bone tumor, tumor invasion, FAK signaling
Introduction
Sarcomas are a broad group of malignant neoplasms that arise from mesenchymal cells origin in soft tissue and bone (1,2). Emerging molecular findings suggest that mechanical and chemical properties of the tumor microenvironment act together to accelerate sarcoma disease progression (3). Within the tumor microenvironment, structural extracellular matrix (ECM) proteins play crucial role in cell surface receptor signaling (4,5), which in turn may regulate sarcoma cell invasive and metastatic potential (6–8). Osteosarcoma (OS), the most common primary malignancy of bone (9,10), is defined by its pathogenic osseous ECM (11). Several studies have demonstrated OS matrix is extensively altered, including changes in collagens and proteoglycans (11,12). In particular, high expression of several sarcomatous matrix proteins has been associated with poor response to chemotherapy and poor prognosis in clinical studies of OS (13,14).
NELL1 (Neural EGFL like 1) is a secreted osteoinductive protein which has bone anabolic (15,16) and anti-osteoclastic effects (17,18). NELL1 can significantly stimulate mesenchymal stromal cell proliferation and osteogenic differentiation, until recently NELL1 was simply known to regulate skeletal patterning and induce bone defect healing in small and large animals (19–21). Earlier studies revealed that loss of Nell1 expression or function in mice reduces expression of numerous ECM proteins, as well as proteins involved in cell adhesion and cellular communication (22). Notably, NELL1 expression has been detected in bone tumors and cartilage-forming tumors (23). With these recent insights, it is likely that the cellular effects of NELL1 are much more pleiotropic than once understood. Nevertheless the biologic implications of NELL1 signaling in osteosarcomagenesis and/or disease progression have been until this point unknown.
Here, we sought to define the regulatory role of NELL1 in sarcoma pathogenesis. CRISPR/Cas9 mediated NELL1 gene deletion markedly reduced the malignant phenotype across human OS cell lines. NELL1 knockout significantly slowed OS disease progression, blunted metastatic potential, and improved survival in a xenograft model. Similar observations were identified with Nell1 gene deletion in a p53/Rb driven osteosarcomagenesis mouse model. Transcriptomic and proteomic analysis demonstrated NELL1 knockout skews the expression of key matricellular proteins associated with reduced FAK signaling. These findings in mouse and human osteosarcoma suggest that NELL1 signaling positively regulates multiple aspects of OS disease progression in part via alterations in the sarcomatous ECM, and that disruption of NELL1 signaling may represent a novel therapeutic approach.
Materials and Methods
Mice
All animal experiments were performed according to approved protocols (MO21M112) of the Animal Care and Use Committee (ACUC) at Johns Hopkins University (JHU). NOD scid mice (Stock No: 001303) were purchased from The Jackson Laboratory (Bar Harbor, ME, USA). OC-Cre animals were a kind gift from the Clemens laboratory at JHU (JAX Stock No. 019509). p53fl/fl;Rbfl/fl mice were a kind gift from the Jones laboratory at University of Utah (JAX Stock No. 008462 and 026563). Nell1fl/fl mice were supplied by the Ting/Zhang laboratories at UCLA (24). In order to achieve spontaneous osteosarcoma formation, p53fl/fl;Rbfl/fl (control) mice or p53fl/fl;Rbfl/fl;Nell1fl/fl (Nell1OC cKO) mice were crossbred with OC-Cre animals. Rates of OS tumor formation within p53fl/fl;Rbfl/fl;OC-Cre animals were in line with prior reports (25). Analyses were performed by investigators blinded to the mouse genotype.
Cell isolation, culture and characterization
Human osteosarcoma cell lines were purchased from American Type Culture Collection (Manassas, VA), including 143B (ATCC®-CRL-8303™), Saos-2 (ATCC® HTB-85™), HOS (ATCC® CRL-1543™), KHOS/NP (ATCC® CRL-1544™), KHOS-312H (ATCC® CRL-1546™), and G-292 (ATCC® CRL-1423™). Primary human OS cells were derived from human OS resection samples (n=4) under IRB approval at JHU with a waiver of informed consent. The pathologic diagnosis of high grade conventional osteosarcoma was confirmed by two independent bone pathologists (E.F.M. and A.W.J.). Immediately following resection, specimens of human OS were collected under sterile conditions, washed in PBS and dissected into small fragments (<1 mm3). Primary OS tumor cells were derived as per previous descriptions (26), and culture expanded for 3 to 5 passages before use. Characterization of primary tumor cells was performed by flow cytometry, in vitro differentiation and cytogenetic analyses. Karyotyping was performed by the Division of Molecular Pathology at JHU clinical laboratories. Human primary OS cells at passage 3–5 were used for characterization by flow cytometry, including mesenchymal (CD44, CD73, CD90, CD105), endothelial (CD31) and hematopoietic markers (CD45) (see Supplementary Table S1 for all antibody information). Multilineage differentiation of tumor cells was performed for osteogenic and chondrogenic differentiation, with assessments of mineralization at 10 d and glycosaminoglycan at 14 d (27,28). Mouse bone marrow mesenchymal stromal cells (BMSCs) were isolated from tibial and femoral bone marrow of either p53fl/fl;Rbfl/fl (DKO) or p53fl/fl;Rbfl/fl;Nell1fl/fl (TKO) mice (n=4) under sterile conditions (29). Briefly, bones were dissected from 8–12 week old female mice. Bone marrow was flushed, resuspended in PBS, and filtered through a 70-mm filter. Bone marrow cells were cultured in 10-cm culture dishes and incubated at 37 °C with 5% CO2 in a humidified chamber. After 3 h, non-adherent cells were removed. All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, Gibco, Grand Island, NY) supplemented with 15% fetal bovine serum (FBS, Gibco), 100 U/ml penicillin and 100 μg/ml streptomycin (Gibco) in a humidified incubator with 5% CO2 at 37°C.
NELL1 gene deletion and isolation of cell clones
CRISPR/Cas9 gene deletion of NELL1 in human OS cell lines and primary cells was achieved using a plasmid with a guide RNA (gRNA) sequence combined with U6gRNA-Cas9–2A-RFP vector (Sigma-Aldrich). The specific gRNA sequence of knockout and negative control plasmids were as in Supplementary Table S2. The transient plasmid DNA transfection was performed using TransIT®-LT1 Transfection Reagent (Mirus, Madisom, WI). Briefly, cells were plated at an initial density of 3×105 cells/ml in 6-well cell culture plates and incubated for 24 h. Cells were then transfected with TransIT-LT1 Reagent-plasmid DNA complex containing 250 μl of Opti-MEM Reduced-Serum Medium (Gibco), 2.5 μg plasmid DNA and 7.5 μl TransIT-LT1 Reagent and incubated for 72 h. Next, NELL1 KO single cell colonies were established by FACS. Viable RFP-positive cells from NELL1 KO-transfected cells and control-transfected cells were sorted into wells of a 96-well microtiter plate by using a Dako Cytomation MoFlo cell-sorter (Beckman, Indianapolis, IN). For single cell deposition, cells were sorted using the 70-mm nozzle with the sheath pressure set at 70 PSI using the sort precision mode set at single cell. After confirmation of gene deletion by qRT-PCR, n=6 colonies were expanded for use. Finally, to achieve Nell1 gene deletion of in murine cells, BMSCs derived from either p53fl/fl;Rbfl/fl (DKO) or p53fl/fl;Rbfl/fl;Nell1fl/fl (TKO) mice and were treated with either Ad-GFP-2A-iCre (Vector Biolabs, Cat No. 1772) or Ad-CMV-GFP (Vector Biolabs, Cat No. 1060) at 200 MOI.
T7 Endonuclease I Assay
NELL1 KO and control-transfected cells were collected for the detection of the mutation using the T7 Endonuclease I Assay Kit (Genocopoeis, Rockville, MD). DNA was extracted from the samples using Quick-DNA™ Miniprep Kit (Zymo Research, Irvine, CA). See Supplementary Table S2 for primer information. The PCR reactions were performed according to the manufacturer’s protocol. The PCR product was then digested using T7E1 enzyme at 37°C for 60 min, followed by separation using 1.5% agarose gel electrophoresis.
In vitro functional assays
Firstly, cell proliferation was assessed using the CellTiter 96® AQueous Non-Radioactive MTS Cell Proliferation Assay (Promega, Madison, WI). 2 × 103 cells were plated in 96-well plates and incubated for 24, 48, and 72 h. The optical density at 490 nm was measured on a microplate spectrophotometer (BioTek, Winooski, VT). Second, cell attachment was measured by crystal violet staining. 2 × 105 cells were plated in 24-well plates and incubated for 1, 3 and 5 h. Cells were washed with cold PBS and fixed with 100% methanol, then stained with 0.5% crystal violet solution for 10 min at RT. Stained cells were lysed in 10% acetic acid and absorbance was measured at 570 nm. In select experiments, Nell1 signaling was rescued by adenoviral NELL1 (Ad-NELL1) or control (Ad-LacZ). In select experiments, tissue culture plates were pre-coated with 5 μg/cm2 of either Fibronectin (Sigma-Millipore) or Laminin (Sigma-Millipore). Thirdly, migration assays were carried out using the Culture-Insert 2 Well (Ibidi, Martinsried, Germany). 4 × 104 cells were added into each well and incubated overnight. Next, the insert wells were gently removed and the plate replenished with growth medium. The cells were incubated for 12 and 24 h and photographs were taken under an inverted microscope (Olympus, Tokyo, Japan). The width of the cell-free gap was measured by using ImageJ software (Version 1.49 v, NIH, Bethesda, MD).
Fourth, invasion assays were carried out using Corning® BioCoat™ Matrigel® Invasion Chambers (Corning, Bedford, MA). Briefly, 2.5 × 105 cells in serum-free medium were added to the upper chamber, while 500 μl growth medium was added to the lower chamber. After 24 h incubation, cells were fixed with cold 100% methanol and stained with 0.5% crystal violet solution. Photographs of five random regions were taken under an inverted microscope (Olympus) and the number of invaded cells was enumerated. Fifth, tumorsphere assays were performed using primary OS cells with or without NELL1 KO which were trypsinized to generate a single cell suspension and seeded into 6-well ultralow attachment plates (Corning) at a density of 5 × 105 cells, followed by culturing in defined sphere medium (30). Spheres were imaged on day 7 using an inverted microscope (Olympus) and the number of spheres were manually counted.
qRT-PCR
Total RNA was extracted from cultured cells of equal passage number and density using TRIzol Reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. 1 μg of total RNA was used for reverse transcription with iScript cDNA synthesis kit (Bio-Rad). Real-time PCR was performed using SYBR Green PCR Master Mix (Thermo Scientific, Waltham, MA). Relative gene expression was calculated using a 2−ΔΔCt method by normalization with GAPDH. The primer sequences were listed in Supplemental Table S2.
Western blot
Cells were lysed in RIPA buffer (Thermo Scientific) with protease inhibitor cocktail (Cell Signaling Technology, Danvers, MA, USA). Proteins concentration were determined by BCA assay (Thermo Scientific). Cell lysates were separated by SDS-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. Protein extraction was blocked with 5% bovine serum albumin and incubated with primary antibodies at 4°C overnight. Finally, membranes were incubated with a horseradish-peroxidase (HRP)-conjugated secondary antibody and detected with ChemiDoc XRS+ System (Bio-rad). The antibodies were listed in Supplemental Table S1.
Total RNA Sequencing
Gene expression was detected by total RNA sequencing using the Illumina NextSeq 500 platform (Illumina, San Diego, CA). Briefly, total RNA was extract from clonal 143B cells with or without NELL1 KO by Trizol (Life Technologies Corporation, Gaithersburg, MD). The total RNA samples were sent to the JHMI Deep Sequencing and Microarray core (JHU) for sequencing. Data analysis were performed using software packages including CLC Genomics Server and Workbench (RRID: SCR_017396 and RRID: SCR_011853), Partek Genomics Suite (RRID: SCR_011860), Spotfire DecisopnSite with Functional Genomics (RRID: SCR_008858), and QIAGEN Ingenuity Pathway Analysis (RRID: SCR_008653).
Reverse phase protein analysis
Protein lysates were isolated from control and NELL1 KO clonal 143B cells (4 biological replicates). Modified RIPA buffer containing proteinase and phosphatase inhibitor was used for the lysis. Cellular proteins were denatured by 1% SDS with β-mercaptoethanol and analyzed by the Reverse Phase Protein Array (RPPA) Core at MD Anderson Cancer Center (https://www.mdanderson.org/research/research-resources/core-facilities/functional-proteomics-rppa-core.html). Briefly, protein lysates were diluted in five two-fold serial dilutions (undiluted, 1:2, 1:4, 1:8; 1:16) and arrayed on nitrocellulose-coated slides (Grace Bio Lab). Each slide was probed with a validated primary antibody plus a biotin-conjugated secondary antibody. The RPPA slides for 453 unique antibodies were scanned, analyzed, and quantified using a customized-software to generate spot intensity. Relative protein levels for each sample were determined by interpolating each dilution curve produced from the densities of the 5-dilution sample spots using a standard curve for each antibody. Relative protein levels are designated as log2 values. The protein concentrations of each set of slides were then normalized for protein loading (31).
Osteosarcoma implantation and assessments
All animal studies were performed with institutional ACUC approval within Johns Hopkins University, complying with all relevant ethical regulations. Experimental details and animal numbers are also summarized in Supplementary Table S3. For all 143B OS cell implantation, 8–10 week old, male and female, NOD scid mice were used. 1 × 106 clonal 143B cells with or without NELL1 KO in 50 μl PBS were injected subperiosteally within the proximal tibia metaphysis. Tumor size was measured by caliper twice weekly for 4 wks, and tumor volume was calculated. Experiments were replicated with a second cell clone. Magnetic resonance imaging (MRI) was performed as previously described (32) at 14 and 28 d post injection, and tumor size was calculated using ImageJ. Primary tumors and lungs were harvested at 28 d post-injection for histologic analysis. In a separate study, overall survival was evaluated from the first day of tumor cell injection until death using the Kaplan-Meier estimate. In a separate study, 143B cells with or without NELL1 knockout were injected systemically by tail vein (1.5 × 106 clonal 143B cells in 100 μl PBS). Mice were monitored three time weekly and sacrificed 28 d after injection for assessment of pulmonary burden of disease. In a separate study, 1 × 106 murine BMSCs in 50 μl PBS (p53fl/fl;Rbfl/fl or p53fl/fl;Rbfl/fl;Nell1fl/fl genotypes 2 d after Ad-Cre mediated transformation) were injected subperiosteally within the proximal tibia metaphysis. Tumor formation was monitored three times weekly and tumor free survival was recorded. Samples were harvested 28 d after tumor formation was observed. Analyses were performed by investigators blinded to the tumor type.
Spontaneous osteosarcomagenesis and assessments
The spontaneous osteosarcoma mice model is based on osteoblast-restricted deletion of p53 and pRb (33). Either p53fl/fl;Rbfl/fl mice or p53fl/fl;Rbfl/fl;Nell1fl/fl mice were crossed with p53fl/+;Rbfl/+;OC-Cre mice or p53fl/+;Rbfl/+;Nell1fl/+;OC-Cre mice, respectively. The offspring with genotype of p53fl/fl;Rbfl/fl; OC-Cre (control) and p53fl/fl;Rbfl/fl;Nell1fl/fl;OC-Cre (Nell1OC cKO) were used in this study. Tumor formation was monitored three times weekly and tumor free survival was recorded. Tumors were harvested 28 d after tumor formation was observed. Analyses were performed by investigators blinded to the mouse genotype.
Histology and Immunohistochemistry
Primary tumor samples and lungs were fixed in 4% paraformaldehyde (PFA) at 4°C for 24 h and decalcified in 14% EDTA (Sigma-Aldrich) for up to 56 d at 4°C. Samples were then cryoprotected in 30% sucrose overnight at 4°C and embedded in optimal cutting temperature compound (OCT, Tissue-Tek 4583, Torrance, CA) for cryosectioning at 10 μm thickness. Routine H&E staining was performed on the entire lung tissues sectioned in a coronal plane to assess pulmonary burden, and number of metastasis foci were manually counted. For immunohistochemistry, sagittal sections of primary or metastatic tumors were washed in PBS × 3 for 10 min and permeabilized with 0.5% Triton-X for 30 min. Next, 5% normal goat serum (S-1000, Vector Laboratories, Burlingame, CA) was applied for 30 min, then incubated in primary antibodies overnight at 4°C. The following day, slides were washed in PBS, incubated in the secondary antibody for 1 h at 25°C, and then mounted with DAPI mounting solution (Vectashield H-1500, Vector Laboratories). Digital images of these sections were captured with 10–100 × objectives using upright fluorescent microscopy (Leica DM6, Leica Microsystems Inc., Buffalo Grove, IL). Analyses were performed by investigators blinded to the sample identification.
Statistical analysis
Results are expressed as the mean ± 1 SD. A Shapiro-Wilk test for normality was performed on all datasets. Homogeneity was confirmed by a comparison of variances test. Parametric data was analyzed using either a Student’s t test for a two group comparison, or one-way analysis of variance when more than two groups were compared, followed by a post hoc Tukey test (Graphpad Software 7.0). *P<0.05 and **P<0.01 were considered significant. For in vivo 143B implantation studies, the sample size was calculated based on an anticipated effect size of 2.0 based on our in vitro studies comparing NELL1 KO and vector control. For this scenario, with eight replicates per group will provide 80% power to detect effect sizes of at least 1.5, assuming a two-sided 0.05 level of significance.
Study approval
All animals were housed and procedures performed under a protocol approved by the IACUC of Johns Hopkins University (protocol MO21M112). Human samples were used under a written informed consent for tissue banking and Institutional Review Board (IRB) approval (protocol number IRB00119905).
Data availability
RNA sequencing data is freely available within the NCBI GEO database (GSE182703). Proteomic data is submitted as a separate Excel file.
Results
Cell autonomous effects of NELL1 gene deletion in osteosarcoma
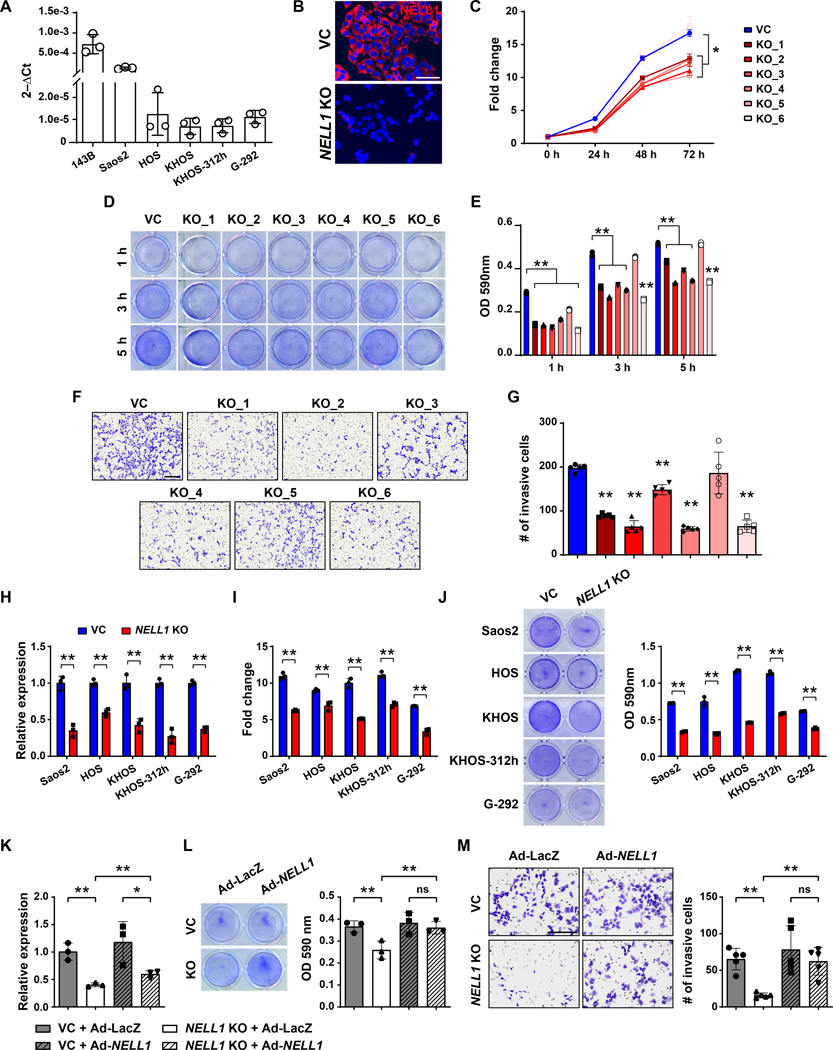
First, expression of NELL1 transcripts was observed to be a conserved feature across all human OS cell lines examined (Fig. 1A). Gene deletion studies were first performed using the 143B cell line, generating six NELL1 KO clones using CRISPR/Cas9. Knockout was confirmed using qRT-PCR, T7 endonuclease I assay (Supplementary Fig. S1) and immunocytochemistry (Fig. 1B). In comparison to vector control, multiple cellular effects were observed among NELL1 KO cell clones, including reduced proliferation (Fig. 1C, mean 34.9% reduction at 72 h), attachment (Fig. 1D, E, mean 31.9% reduction at 5 h), and invasion (Fig. 1F, G, 25.1–69.1% reduction across clones). Similar findings were observed with analysis of a polyclonal NELL1 KO 143B cell population (Supplementary Fig. S2), and indeed similar assessments across five other available OS cell lines recapitulated these findings (Fig. 1H–J). Overexpression studies were performed, in which adenoviral delivery (Ad-NELL1) led to partial restoration of gene expression and near complete restoration of attachment and invasion potential among NELL1 KO 143B sarcoma cells (Fig. 1K–M). These data confirmed that the NELL1 gene plays a crucial role in maintaining cellular proliferation, attachment, and invasion potential in OS cells.
Figure 1. Cell autonomous effects of CRISPR/Cas9-mediated NELL1 gene deletion in human osteosarcoma cell lines.
(A) NELL1 expression by qRT-PCR in osteosarcoma cell lines. (B-G) Effects of NELL1 gene deletion in 143B cells. (B) Confirmation of NELL1 knockout (KO) in single cell clones by representative immunocytochemical staining, in comparison to vector control (VC). Additional data shown in Supplementary Figure S1. (C) MTS proliferation assay among NELL1 KO single cell clones at 24, 48, and 72 h. (D, E) Attachment assay at 3 and 5h assessed by (D) crystal violet staining and (E) quantification. (F, G) Transwell invasion assay at 24 h, including (F) representative images and (G) quantification. (H-J) Effects of CRISPR-mediated NELL1 gene deletion across other human OS cell lines, including Saos2, HOS, KHOS, KHOS312h, and G292 cells. (H) Confirmation of NELL1 KO by qRT-PCR. (I) MTS proliferation assay at 72 h. (J) Attachment by crystal violet staining and quantification at 5 h. (K-M) Effects of adenoviral NELL1 (Ad-NELL1) among 143B cells with or without NELL1 gene deletion. Experiments performed in comparison to Ad-LacZ control. (K) NELL1 gene expression by qRT-PCR. (L) Attachment assay by crystal violet staining and quantification at 5 h. (M) Transwell invasion assay and quantification at 24 h. Data shown as mean ± 1 SD, with dots representing individual datapoints. All experiments performed in triplicate replicates, with results from a single replicate shown. *P<0.05; **P<0.01; ns: non-significant. Scale bars: 500 μm. A two-tail Student’s t test was used for two group comparisons (Fig. H, I, J), and one-way ANOVA with post hoc Tukey test when more than two groups were compared (Fig. C, E, G, K, L, M).
NELL1 knockout mitigates OS disease progression
NELL1 KO 143B clones were next implanted in an orthotopic xenograft model in NOD scid mice (Fig. 2A). In relation to control, tumor growth was impeded as assessed by serial caliper measurements (Fig. 2B, 66.9% reduction in tumor size at study endpoint). Findings were confirmed in an independent clone of knockout tumor cells (Supplementary Fig. S3, 89.2% reduction in tumor size). MRI confirmed a 57.1% and 66.1% reduction in tumor size at 14 and 28 d post-implantation (Fig. 2C). Histologic examination of primary tumors demonstrated complete loss of NELL1 immunoreactivity among NELL1 KO xenograft tumor cells (Fig. 2D), accompanied by a reduction in the proliferative index of NELL1 KO implants (Fig. 2E, 67.5% reduction in Ki67 labeling). Tumor associated angiogenesis also showed a notable reduction among NELL1 KO implants (Fig. 2F, 74.9% reduction in CD31-positive vessel area), in line with prior reports of the angiogenic effects of NELL1 signaling (34). In order to confirm the impact of NELL1 on invasion and metastasis, pulmonary metastatic burden was assessed by serial sections of lung fields. Results indicated complete absence of lung metastasis among NELL1 KO tumors at 28 d post-inoculation (Fig. 2G, H). In a separate cohort of animals, mice with NELL1 KO tumors demonstrated prolonged overall survival (Fig. 2I, 89.6% increase in median overall survival). Reduction in metastatic potential among NELL1 KO sarcoma cells was further verified using tail vein injection, in which an 81.1% reduction in pulmonary burden was observed (Fig. 2J, K). Together, our data demonstrates that NELL1 gene deletion slows OS tumor growth and mitigates metastasis in a xenograft model.
Figure 2. NELL1 knockout mitigates OS disease progression in 143B xenograft model.
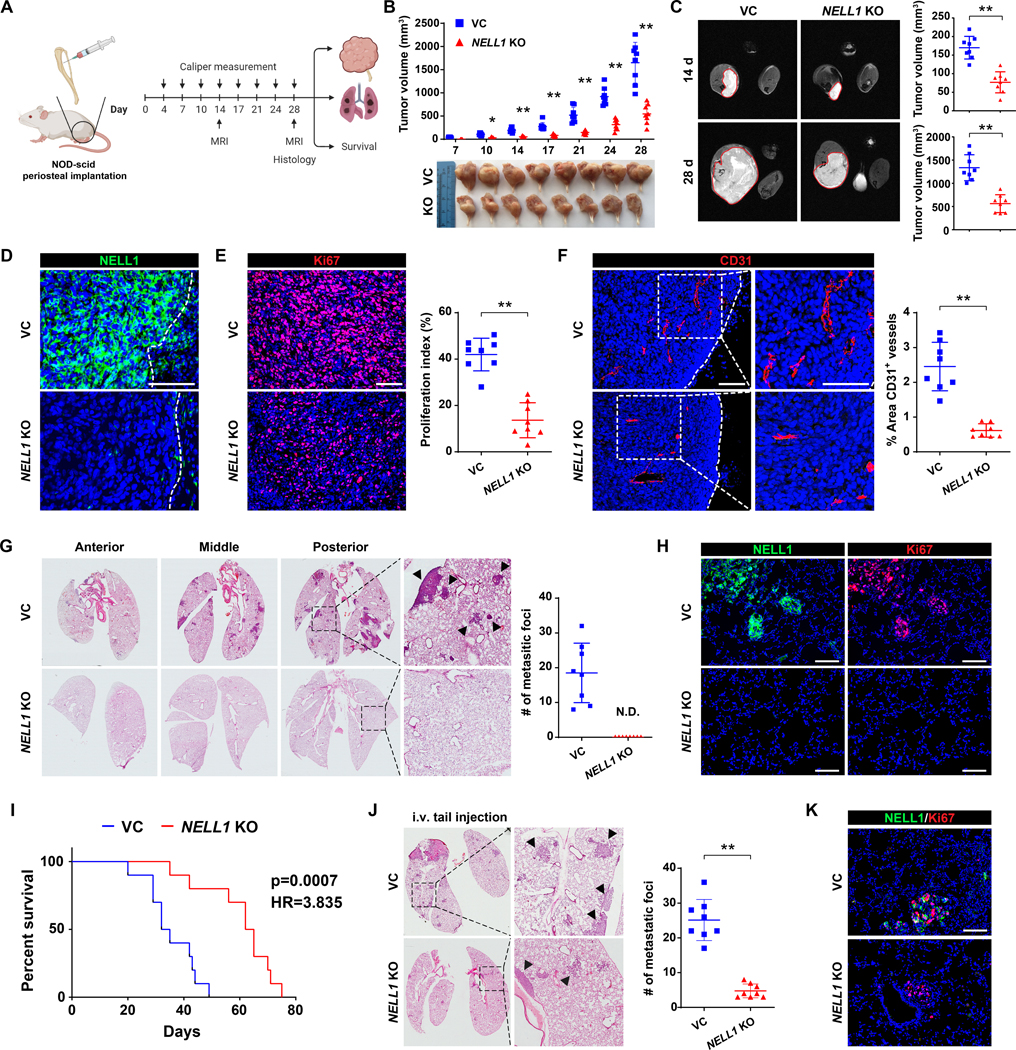
Orthotopic implantation of NELL1 KO or vector control clonal 143B cells within the proximal tibia of female NOD-Scid mice (n=8 mice per group, 1 × 106 per implant). Biological replicate shown in Supplementary Fig. S4. (A) Schematic diagram of study design (Created in Biorender.com). (B) Tumor volume, calculated by caliper measurements twice weekly until 28 d post-injection (above) and gross pathology of all tumors (below). (C) Representative MRI imaging at 14 and 28 d post-injection (left) and tumor volume (right). (D) Confirmation of NELL1 KO in xenograft tumors by immunostaining. Dashed line indicates edge of tumor. (E) Ki67 immunostaining (left) and quantification (right). (F) CD31 immunostaining (left) and quantification (right). Dashed line indicates edge of tumor. (G) Cross-sections of pulmonary fields (left) and quantification of metastatic foci (right). (H) Representative NELL1 and Ki67 immunostaining in lung metastatic foci from the same sample slide. (I) Overall survival, evaluated using Kaplan–Meier curves (n=10 mice per group, 1 × 106 per implant). (J) NELL1 gene deletion mitigates OS lung metastasis after systemic injection. Tail vein injection of NELL1 KO or vector control 143B cells in female NOD scid mice, followed by analysis after 28 d (1.5 × 106 cells / mouse, n=8 mice per group). Cross-sections of pulmonary fields (left) and quantification of metastatic foci (right). (K) Representative NELL1 and Ki67 immunostaining in lung metastatic foci. Scale bar: 100 μm. Individual dots in scatterplots represent values from single animal measurements, while mean and one SD are indicated by crosshairs and whiskers. *P<0.05; **P<0.01. ND: Not detected. A two-tailed student’s t test was used for all comparisons. Survival was assessed using Kaplan–Meier curves and log-rank analysis (Fig. I).
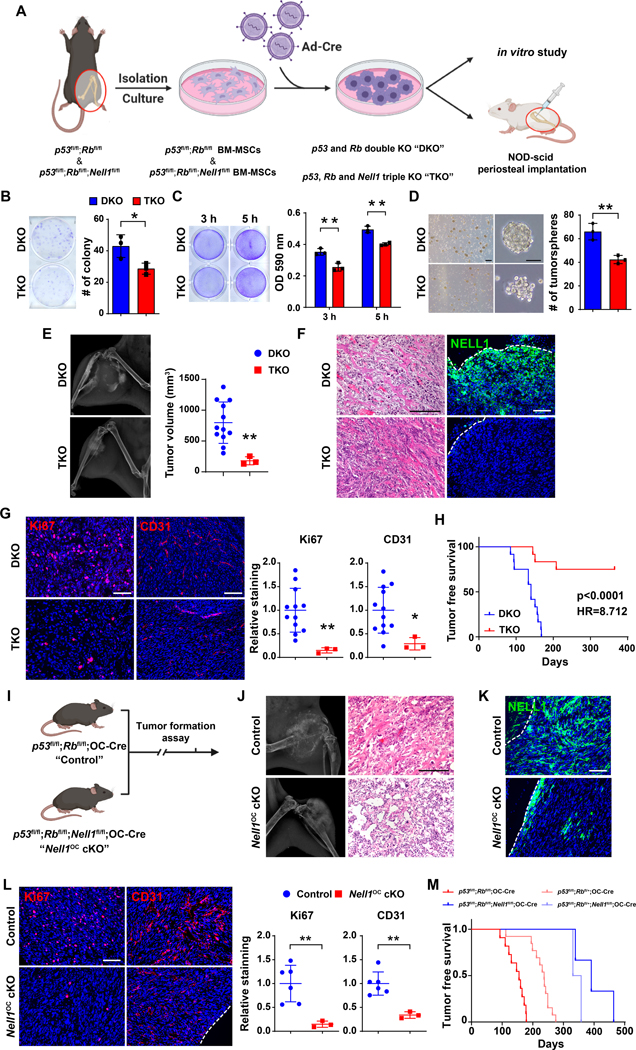
NELL1 gene deletion impedes osteosarcomagenesis
The consequences of NELL1 gene deletion were next examined in two murine OS models (33,35). Firstly, mouse BMSCs were isolated from p53fl/fl;Rbfl/fl (DKO) or p53fl/fl;Rbfl/fl;Nell1fl/fl (TKO) mice, recombination performed via adenovirus-mediated Cre delivery and the consequences examined in vitro and after periosteal transplantation (Fig. 3A). As expected, gene knockout of p53 and Rb resulted in an increased proliferative and invasive phenotype in comparison to vector control (Supplementary Fig. S4). In comparison to DKO tumor cells, TKO cells demonstrated reduced colony formation (Fig. 3B, 33.3% reduction), reduced cellular attachment (Fig. 3C, 27.5% and 19.8% reduction at 3 and 5 h, respectively), and reduced tumorsphere formation efficiency (Fig. 3D, 35.9% reduction at 5 d). DKO and TKO cells were next orthotopically implanted in NOD scid mice, and sarcomagenesis and growth were monitored (Fig. 3E–H). Tumor formation rates were significantly reduced among TKO implants (25.0% incidence in comparison to 100% incidence among DKO implants, Supplementary Table S4). Among those implants with tumor formation, tumor size was significantly lower in TKO cell implants (Fig. 3E, 76.7% reduction in tumor size). Histological analysis confirmed an osteosarcoma-like appearance of all tumors, and confirmed complete loss of NELL1 immunoreactivity among TKO tumor cells (Fig. 3F). In similarity to prior findings, Nell1 gene deletion demonstrated a reduction in proliferative index and vascularity among tumors, assessed by Ki67 and CD31 immunostaining (Fig. 3G, 86.9% reduction in Ki67, and 71.1% reduction in CD31 immunostaining). Moreover, tumor-free survival was significantly prolonged in mice with TKO implants, with a hazard ratio (HR) of 0.115 (Fig. 3H).
Figure 3. Nell1 gene deletion slows osteosarcomagenesis in mouse lines.
(A) Schematic diagram of study design (Created in Biorender.com). (B-D) Effects of Adeno-Cre treatment in p53fl/fl;Rbfl/fl (DKO) or p53fl/fl;Rbfl/fl;Nell1fl/fl (TKO) mouse bone marrow mesenchymal stromal cells. (B) Colony-forming unit (CFU) assay, assessed by crystal violet staining (left) and quantification (right) at 10 d. (C) Attachment assay assessed by crystal violet staining (left) and quantification (right), at 3 and 5 h. (D) Tumorsphere formation assay images (left) and quantification (right) at 5 d. (E) Osteosarcomagenesis visualized by X-ray of representative tumors (left) and tumor volume calculated by caliper measurements (right). (F) Representative histologic appearance by routine H&E staining and NELL1 immunostaining among DKO and TKO tumor tissue. (G) Representative Ki67 and CD31 immunostaining (left) and quantification (right) among DKO and TKO tumor tissues. (H) Tumor-free survival among DKO and TKO implanted mice, evaluated using Kaplan–Meier curves (n=12 mice per group, 1 × 106 per implant). (I-M) Effects of Nell1 gene deletion in spontaneous osteosarcomagenesis. (I) Schematic diagram of spontaneous osteosarcomagenesis among control (p53fl/fl;Rbfl/fl;OC-Cre) and Nell1OC cKO animals (p53fl/fl;Rbfl/fl;Nell1fl/fl;OC-Cre). (J) Representative X-ray images (left) and histologic appearance by routine H&E staining (right). (K) Representative NELL1 immunostaining among control and Nell1OC cKO tumor tissue. (L) Representative Ki67 and CD31 immunostaining (left) and quantification (right) among control and Nell1OC cKO tumors. (M) Tumor-free survival, evaluated using Kaplan–Meier curves. Scale bars: 100 μm. For (Fig. B-D), data shown as mean ± 1 SD with dots representing individual measurement, performed in triplicate experimental replicates. For (Fig. E, G, L), dots in scatterplots represent values from individual animal measurements, while mean and one SD are indicated by crosshairs and whiskers. *P<0.05; **P<0.01. A two-tailed student’s t test was used for all comparisons. For (Fig. H, M), survival was assessed using Kaplan–Meier curves and log-rank analysis.
Secondly, the same transgenes were used to monitor spontaneous osteosarcomagenesis, now utilizing the osteoblast specific Osteocalcin-Cre line (25). Animals with conditional knockout of p53 and Rb (p53fl/fl;Rbfl/fl;OC-Cre, referred to as control) were compared to animals with conditional knockout of p53, Rb and Nell1 (p53fl/fl;Rbfl/fl;Nell1fl/fl;OC-Cre, referred to as Nell1OC cKO, Fig. 3I). Mice heterozygote for Rb were also examined for tumor incidence. Tumor formation rates were significantly reduced among Nell1OC cKO animals (15.8% incidence in Nell1OC cKO animals in comparison to 52.4% incidence among control animals, Supplementary Table S5). Radiographic and histologic appearance across tumors was consistent with osteosarcoma (Fig. 3J, tumor locations summarized in Supplementary Table S6). NELL1 immunohistochemical staining confirmed loss of NELL1 within sarcoma cells (Fig. 3K). Immunohistochemical analyses again confirmed reduction in the proliferative index and tumor associated vasculature among tumors in Nell1OC cKO animals (Fig. 3L, 85.2% reduction in Ki67, and 66.1% reduction in CD31 immunostaining). Nell1OC cKO animals showed prolonged tumor-free survival (Fig. 3M, Supplementary Table S5, median tumor-free survival of 398 days in Nell1OC cKO versus 146.5 days in controls). Collectively, Nell1 loss impeded p53/Rb induced OS sarcomagenesis, but also reduced tumor growth and improved disease free survival in mouse models.
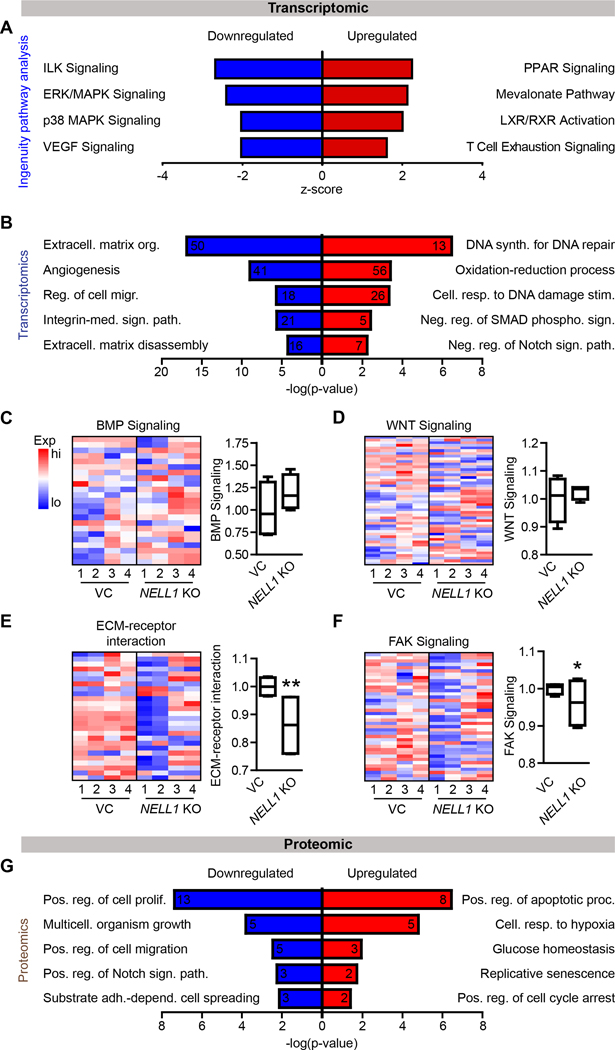
NELL1 gene deletion shifts the sarcoma matrisome
Differences in gene expression profile were next investigated using bulk RNA sequencing across 4 clones of NELL1 KO and VC 143B cells. Clear separation between gene expression profiles was observed between NELL1 KO and control cells, as assessed by principal component analysis and volcano plot (Supplementary Fig. S5A, B). QIAGEN Ingenuity Pathway Analysis (IPA) showed that multiple signaling pathways were downregulated in NELL1 KO cells, including for instance integrin-linked kinase (ILK), ERK/MAPK and VEGF signaling (Fig. 4A, see Supplementary Tables S7, 8 for a complete list). Next, pathway analysis of differentially expressed genes (DEGs) revealed Gene ontology (GO) term enrichment in extracellular matrix (ECM) organization, angiogenesis, integrin-mediated signaling pathway, and ECM disassembly were downregulated in NELL1 KO cells (Fig. 4B, see Supplementary Table S9, 10 for most downregulated and upregulated GO terms). Heat maps revealed differences in expression profile of OS related genes among NELL1 KO and VC cells, including for example MDM2, CDK4, and CD44 among others (Supplementary Fig. S5C, Supplementary Table S11) (36–38). Next, deletion of NELL1 in OS cells was further validated by downstream signaling-related gene expression, shown by heatmap and cumulative module scoring (Fig. 4C–F). Although in other contexts canonical BMP and Wnt signaling pathway activation are known to be downstream of NELL1 signaling (16,39), no significant change in either pathway was observed with NELL1 gene deletion (Fig. 4C, D). Instead, significant reductions in gene modules related to ECM-receptor interaction and FAK signaling were observed among NELL1 KO sarcoma cells (Fig. 4E, F). Consistent with our transcriptomic analysis, multiple NELL1 KO osteosarcoma cell lines showed decreased activation of FAK by Western blot (Supplementary Fig. S6, S7).
Figure 4. Bulk RNA sequencing and Reverse Phase Protein Arrays (RPPA) among NELL1 KO 143B sarcoma cells.
(A) Ingenuity Pathway Analysis (IPA) analysis of downregulated pathways (Z-score < 0; blue color) or upregulated pathways (Z-score > 0; red color) among NELL1 KO cells in comparison to vector control (VC). (B) Gene Ontology (GO) enrichment analysis identified representative pathways that were downregulated (blue color) or upregulated (red color) among NELL1 KO cells. (C-F) Heat map demonstrating expression levels of genes involved in NELL1 downstream signaling and corresponding module score among NELL1 KO as compared to VC cells, including (C) BMP signaling, (D) Wnt signaling, (E) ECM-receptor interaction, and (F) FAK signaling. (G) GO enrichment analysis identified representative pathways that were downregulated (blue color) or upregulated (red color) among NELL1 KO cells as derived from RPPA. n=4 biological replicates of VC and NELL1 KO cells. Gene module scores are shown as boxplot. *P<0.05; **P<0.01. A two-tailed student’s t test was used for all comparisons.
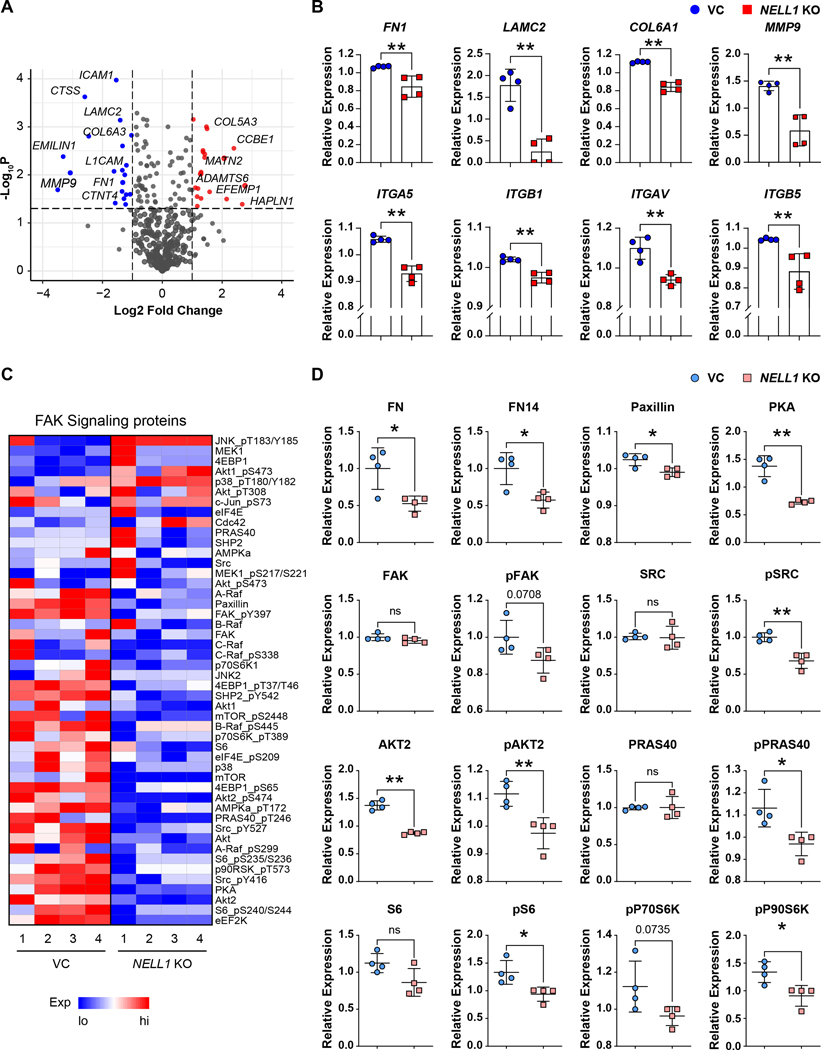
In order to further evaluate differences with NELL1 gene deletion, a reverse phase protein array was performed. A clear separation between protein profiles was observed when comparing NELL1 KO and VC cells, as assessed by principal component analysis and volcano plot (Supplementary Fig. S8A, B). Again, pathway analysis of downregulated DEGs revealed GO term enrichment in NELL1 KO cells associated with the regulation of cellular proliferation, migration, adhesion and cell spreading (Fig. 4G, Supplementary Fig. S8C. See Supplementary Table S12, 13 for most downregulated and upregulated GO terms). Having observed prominent shifts in terms related to cell-ECM interaction within the transcriptome and proteome of NELL1 KO sarcoma cells, we next examined specific misexpressed ECM transcripts within NELL1 KO cells (Fig. 5A, B). Analysis showed significant decrease in expression across several ECM components including for example FN1 (Fibronectin 1), LAMC2 (Laminin subunit gamma 2), and COL6A3 (Collagen type VI alpha 3 chain) with NELL1 gene deletion (Fig. 5A), as well as ITGA5 (Integrin alpha 5), ITGB1 (Integrin beta 1), ITGAV (Integrin alpha V), and ITGB5 (Integrin beta 5) (Fig. 5B). Heatmap revealed decreased phosphorylation of proteins downstream of FAK (Fig 5C). Proteomic analysis confirmed a downregulation of FN1 and FN14, as well as Paxillin and PKA in NELL1 KO cells (Fig 5D). Moreover, phosphorylation of SRC, AKT2, PRAS40, S6 and P90S6K were significantly reduced (Fig. 5D). Interestingly, increased JNK phosphorylation was observed in NELL1 KO cells (Supplementary Fig. S9). Thus, a transcriptomic-proteomic analysis suggested that deletion of NELL1 in sarcoma cells alters the ECM profile, including prominent reductions in FN1 and Laminin, alterations in pathways associated with cell-ECM interaction, and overall a reduction FAK and downstream signaling activity.
Figure 5. Transcriptomic and proteomic revealed downregulation of ECM genes and decreased phosphorylation of FAK signaling related proteins.
(A) Volcano plot demonstrating transcripts of ECM components that were downregulated (blue color) or upregulated (red color) in NELL1 KO compared with VC genes derived from transcriptomics. (B) ECM gene expression among NELL1 KO compared with VC, including FN1, LAMC2, COL6A1, MMP9, ITGA5, ITGB1, ITGAV, and ITGB5. Log2 (FKPM+1) values were median centered. (C) Heat map demonstrating level of proteins involved in FAK signaling. (D) Protein expression of FAK signaling components derived from RPPA among NELL1 KO cells. RowLog2 expression values were median centered and then transformed to linear value. n=4 biological replicates of VC and NELL1 KO cells. Dots in scatterplots represent values from individual measurement, while mean and one SD are indicated by crosshairs and whiskers. *P<0.05; **P<0.01; ns: not significant. A two-tailed student’s t test was used for all comparisons.
Matrix components can reverse phenotypic changes with NELL1 gene deletion
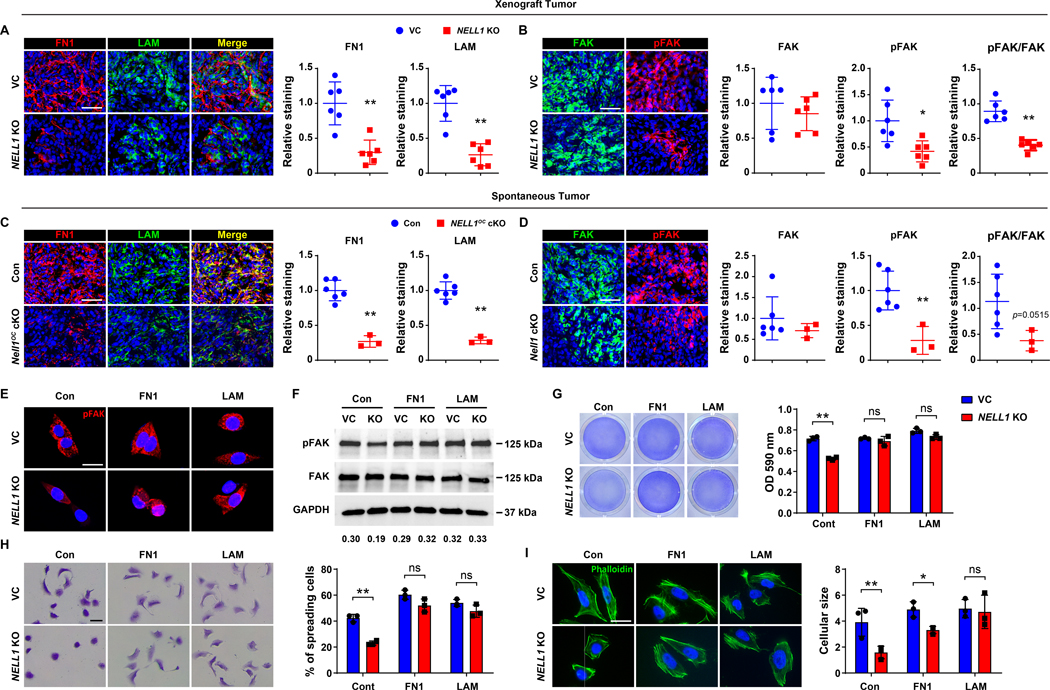
Having observed that NELL1 gene deletion among 143B sarcoma cells resulted in deficits in key matricellular proteins and reduced FAK signaling activation, we next sought to confirm these findings across in vivo models. In a candidate fashion, fibronectin (FN1) and laminin (LAM) expression among 143B xenografts was assessed by immunofluorescent staining (Fig. 6A). Immunostaining showed diffuse FN1 and LAM immunoreactivity in control tumors, staining for both proteins was significantly reduced in NELL1 KO tumors (Fig. 6A, 69.4% and 73.7% reduction in FN1 and LAM, respectively). Corresponding activation of FAK signaling was next accessed by immunofluorescent staining for FAK and pFAK across 143B sarcoma samples. Among NELL1 KO sarcoma sections, decreased pFAK and retained FAK immunoreactivity was observed in comparison to control tumors (Fig. 6B, no significant differences in FAK, 58.3% reduction in pFAK, and 54.9% reduction in pFAK/FAK staining). These findings regarding altered sarcoma matrix were likewise observed in the murine spontaneous OS model induced by p53/Rb deletion (Fig. 6C, D). Here, sarcomas among NELL1OC cKO animals demonstrated a 74.9% and 71.6% reduction in FN1 and LAM immunofluorescent staining across serial sections (Fig. 6C), associated with a 71.6% reduction in pFAK staining and 66.7% reduction in pFAK/FAK immunostaining in relation to control tumor tissues (Fig. 6D). This data further correlated NELL1 gene deletion with shifts in sarcoma associated ECM composition and a corresponding reduction in FAK signaling.
Figure 6. Sarcoma-associated ECM proteins positively regulate FAK signaling and adhesive features in NELL1 KO sarcoma cells.
(A, B) Immunohistochemical analysis of 143B OS cell implants among VC and NELL1 KO tumor tissue, 28 d post-implantation. (A) Representative Fibronectin (FN1) and laminin (LAM) immunostaining (left) and quantification (right). (B) Representative FAK and pFAK immunostaining (left) and quantification (right). (C, D) Immunohistochemical analysis of mouse spontaneous OS tumor tissues from control (p53fl/fl;Rbfl/fl;OC-Cre) or Nell1OC cKO (p53fl/fl;Rbfl/fl;Nell1fl/fl;OC-Cre) animals. (C) Representative FN1 and LAM immunostaining (left) and quantification (right). (D) Representative FAK and pFAK immunostaining (left) and quantification (right) among control and Nell1OC cKO tumors. (E-I) Effects of sarcoma ECM components fibronectin (FN1) or laminin (LAM) on 143B OS cells with or without NELL1 KO. Comparisons made to vector control (VC) treated cells. Pre-coated wells include FN1 (10 μg) and LAM (10 μg). (E) Representative pFAK immunostaining at 3 h post-seeding. (F) Western blot of pFAK and FAK at 5 h post-seeding. Ratio of pFAK/FAK listed below. (G) Attachment assay assessed by crystal violet staining (left) and quantification (right) at 5 h post-seeding. (H) Cell spreading assay assessed by crystal violet staining (left) and quantification (right) at 3 h. (I) Representative phalloidin staining (left) and quantification (right) at 3 h. Scale bar: 50 μm (A-D), and 20 μm (E, H, I). In (A-D), dots in scatterplots represent values from individual animal measurements, while mean and one SD are indicated by crosshairs and whiskers. In (H-I), data shown as mean ± 1 SD, with dots representing individual wells, performed in triplicate experimental replicates. *P<0.05; **P<0.01. ns: non-significant. A two-tailed student’s t test was used for all comparisons.
Next, the extent to which the matricellular proteins FN1 and LAM could restore FAK signaling and alter cellular behavior among NELL1 KO 143B cells was assessed (Fig. 6E–I). Here, cell culture flasks were pre-coated with FN1 or LAM (5 μg/cm2). First, FAK activation was assessed in NELL1 KO cells by pFAK immunocytochemistry and western blot (Fig. 6E, F). Results confirmed a decrease in FAK phosphorylation among NELL1 KO cells in relation to vector control, which was observed to be restored by supplementation with FN1 or LAM coating (Fig. 6E). This finding was further validated by western blot and semi-quantitation (Fig. 6F, Supplementary Fig. S10). Further, restoration of FAK signaling activity with either FN1 or LAM coating led to alterations in cell attachment profiles among NELL1 KO sarcoma cells. Here, complete restoration of attachment rate was observed among NELL1 KO sarcoma cells on FN1 or LAM substrates (Fig. 6G). Correspondingly, a similar increase in adhesion-dependent cell spreading (Fig. 6H) and cytoskeletal size (Fig. 6I) was observed among NELL1 KO sarcoma cells when placed on either matrix component. Moreover, FN1 substrate restored the attachment rate among additional other NELL1 KO sarcoma cell lines, including HOS, Saos2, G292 and 143.98.2 (Supplementary Fig. S11). In sum, these data indicate that alterations in the sarcoma matrix composition mediate FAK signaling and tumor cell attachment potential, and implicate NELL1 as an important regulator of this process.
NELL1 expression typifies an aggressive phenotype in human osteosarcoma
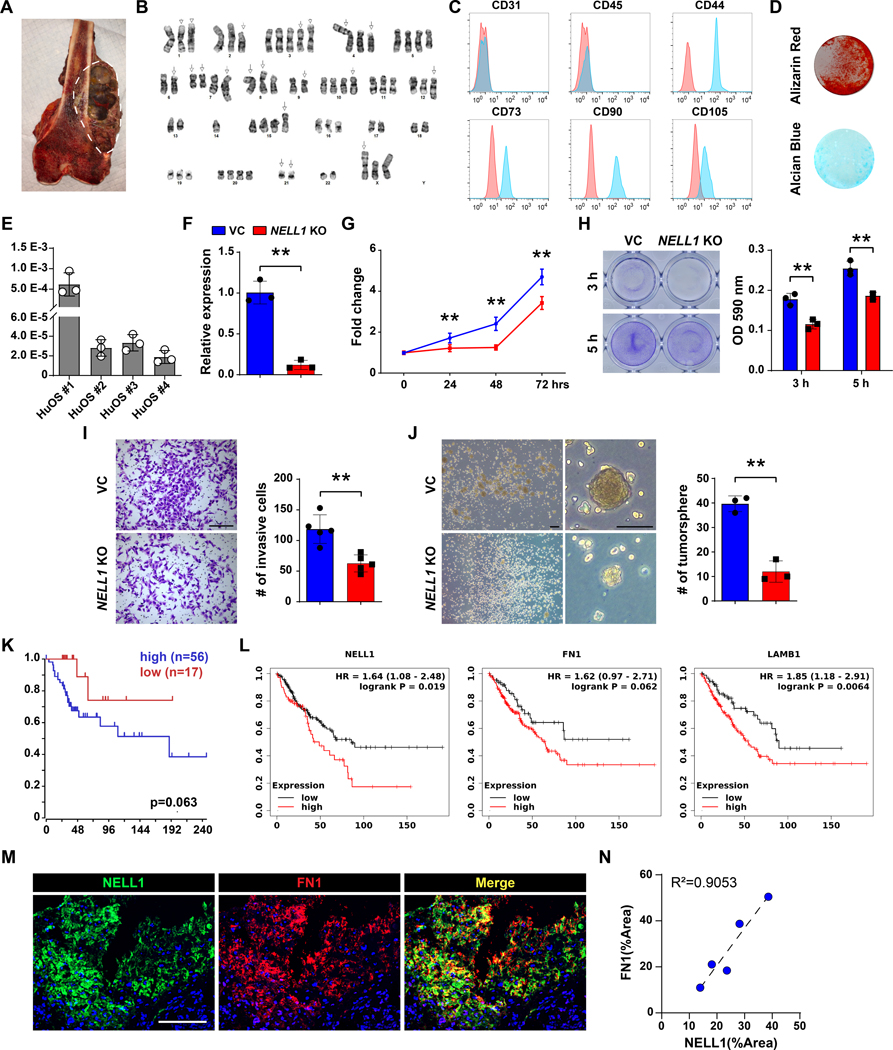
Finally, the critical role of NELL1 signaling in primary human osteosarcoma cells and tissues was examined. First, patient-derived human osteosarcoma cells (HuOS) were isolated and extensive karyotypic abnormalities were observed in the resulting cell isolates (Fig. 7A, B). Further, flow cytometry analysis confirmed absence of endothelial and hematopoietic markers (CD31, CD45) and presence of mesenchymal markers (CD44, CD73, CD90 and CD105) and among cell isolates (Fig. 7C, Supplementary Fig. S12). In addition, all isolated osteosarcoma cells underwent both osteogenic and chondrogenic differentiation (Fig. 7D). Basal expression of NELL1 was observed across all HuOS cells (Fig. 7E). Gene deletion studies were then performed among patient-derived OS cells using CRISPR/Cas9. Knockout was confirmed using qRT-PCR (Fig. 7F, 88.1% reduction). In comparison to vector control, multiple cellular effects were observed among HuOS, including reduced proliferation (Fig. 7G. 46.3% reduction at 72 h), attachment (Fig. 7H. 34.9% and 24.7% reduction at 3 and 5 h, respectively), invasion (Fig. 7I, 47.2% reduction at 24 h), and tumorsphere formation (Fig. 7J, 69.7% reduction at 5 d).
Figure 7. NELL1 regulates invasive potential in primary human OS cells and is associated with disease progression in human patients.
(A) Representative image of bisected osteosarcoma sample. White dashed lines indicate tumor. (B) Representative karyotype of primary osteosarcoma cells. (C) Representative flow cytometry analysis of patient-derived primary osteosarcoma cells. Frequency of expression is shown (blue) in relation to isotype control (red). (D) Representative Alizarin red (AR) at d 10 of osteogenic differentiation, and Alcian blue staining at d 14 of chondrogenic differentiation among human OS cells. (E) NELL1 expression in primary osteosarcoma cells by qRT-PCR. (F-J) Effects of NELL1 gene deletion in primary osteosarcoma cells. (F) Confirmation of NELL1 gene deletion efficiency by qRT-PCR. (G) MTS proliferation assay among NELL1 KO primary osteosarcoma cells at 24, 48, and 72 h. (H) Attachment assay assessed by crystal violet staining and quantification, at 3 and 5 h. (I) Transwell invasion assay and quantification, at 24 h. Representative images with number of invasive cells shown. Scale bars: 500 μm. (J) Tumorsphere formation assay and quantification at 5 d. (K) Prognostic value of NELL1 in patients with high grade osteosarcoma. Kaplan–Meier curve generated by R2 genomics analysis and visualization platform. (L) Prognostic value of NELL1, FN1, and LAMB1 in high grade sarcoma patients. Kaplan–Meier curve generated by Kaplan-Meier (KM) plotter pan-caner RNA seq. (M) Representative NELL1 and Fibronectin immunofluorescent staining in biopsy samples of high grade conventional osteosarcoma. (N) Correlation of NELL1 and Fibronectin staining across N=5 biopsy samples of high grade conventional osteosarcoma. Scale bars: 100 μm. Data shown as mean ± 1 SD, and represent triplicate experimental replicates. *P<0.05; **P<0.01. A two-tail Student’s t test was used for two group comparisons.
Two existing datasets were used to first examine whether high expression of NELL1 serves as a prognostic marker in OS (Fig. 7K, L). Among a mixed population of 73 cases of high grade osteosarcoma, high NELL1 transcripts were associated with a reduced overall survival (Fig. 7K, median overall survival of 34 months in NELL1high versus 60 months in NELL1low cohort). Analyses performed in 259 cases of high grade sarcomas of soft tissue and bone showed similarly that transcripts of NELL1, FN1, or LAMB1 were all associated with decreased overall survival (Fig. 7L, median overall survival of 49, 23, and 54 months in NELL1high versus 86, 38, and 90 months in NELL1low cohort, respectively). Finally, 5 biopsies of conventional osteosarcoma prior to adjuvant chemotherapy were assessed for the expression of NELL1 and fibronectin (Fig. 7M, N, Supplementary Table S14). Results demonstrated immunoreactivity for both NELL1 and FN1 across all samples, a high degree of co-localization between antigens within biopsy tissue, and a relative correlation between the degree of NELL1 and FN1 immunostaining across samples ((Fig. 7M, N). Collectively, NELL1 is an OS related protein essential for sarcoma cell function via regulation of cell-matrix interactions, and represents a poor prognostic indicator across human sarcomas.
Discussion
In this study, we demonstrated that the sarcoma associated protein NELL1 plays an integral role in positively regulating both sarcomagenesis as well as features of aggressive sarcoma behavior on cellular and tumoral levels. In part, this is explained by a modulation of the sarcomatous ECM, which in turn modulates cell-ECM interaction and FAK/Src signaling activation.
The sarcomatous ECM displays distinct histologic, proteomic, and biomechanical properties (40–42). In some instances, sarcomatous ECM proteins have been linked to metastasis (41), subsequent growth of metastatic foci (43), or chemo-resistance (11,40,44). Bone and soft tissue sarcomas are exceptionally diverse (45,46), and even within osteosarcoma the matrix components may vary widely in composition (11,12). Nevertheless, NELL1 expression appears to be present across the histologic spectrum of human osteosarcoma (23). Prior observations have found NELL1 to be a key protein in the regulation of matrix components during morphogenesis, regeneration and tissue engineering. For example, mice globally deficient in Nell1 have altered ECM protein expression associated with skeletal developmental anomalies (22,24). In the context of tissue engineering and regeneration, Qi et al showed recombinant NELL1 treatment induces expression of the structural matrix protein Type II Collagen and promotes the differentiation of chondrocytes (24). In other contexts, NELL1 enhances the production of the molecule dentin matrix protein 1 (DMP1) accompanied by induction of odontoblastic differentiation (47). Our study observed similarly striking regulation of sarcoma associated matrix proteins by NELL1, which among other findings altered FAK signaling activity and modulated sarcoma cell functional characteristics. Of note, future studies are needed to demonstrate the precise regulatory roles that NELL1 exerts on ECM formation and remodeling.
An important consideration toward the positive regulatory roles of NELL1 signaling in sarcoma disease progression is the promotion of angiogenesis. In tissue engineering contexts, NELL1 is known to promote angiogenesis during non-neoplastic bone formation through induction of VEGFA expression (34,48). Here, NELL1 gene deletion led to a down-regulation of VEGF signaling in sarcoma cells, and potent reduction in sarcoma-associated blood vessels. Indeed, VEGF expression correlates with prognosis in patients with sarcoma (49). Beyond changes in angiogenic factors, it is likely that altered ECM associated with NELL1 KO may also have predisposed to lack of vascular ingrowth. NELL1 deficient sarcoma cells displayed changes in genes and proteins associated with ECM remodeling, including a significant downregulation in MMP9. Matrix metalloproteases (MMPs) are defined by their role in matrix degradation but also in their secondary stimulation of tumor vasculature (50). For instance, MMP9 renders normal islets into angiogenic islets by mobilizing VEGF during pancreatic carcinogenesis (51). Thus, changes in sarcoma-associated angiogenesis could be a result of combinatorial factors including NELL1 signaling induced transcription of angiogenic factors as well as altered ECM remodeling.
NELL1 is by no means solely expressed by osteosarcoma. Here, we observed NELL1 expression to serve as a prognostic biomarker among high grade sarcomas of bone (osteosarcoma and Ewings sarcoma), or even mixed diagnostic population of high grade sarcomas of bone and soft tissue. This is consistent with a prior study which implicated NELL1 in migration and invasion of rhabdomyosarcoma (RMS) cells, and negatively correlated NELL1 expression with RMS prognosis (52). Interestingly, a clear divide seems to be present in the literature between the potential roles of NELL1 in epithelial versus mesenchymal malignant tumors. For example, a GWAS study of non-small cell lung cancer (NSCLC) identified NELL1 as a tumor suppressor gene (53). The protective role of NELL1 was confirmed by functional studies conducted in colon cancer, renal cell carcinoma, lung cancer and esophageal adenocarcinoma cell lines and models (54–56). Among the numerous differences between carcinomas and sarcomas, our study suggests that the matrix itself may be an important distinguishing factor. Sarcoma cells are often loosely and often haphazardly arranged their sarcomatous ECM (57), while epithelial cancer cells are closely interacting cells often adherent to one another, but by and large without mesenchymal matrix production. These key cytologic and histologic features serve as future catalysts toward the understanding of the matrix regulating protein NELL1 as a differential disease modifier in carcinoma and sarcoma.
Finally, a number of key biological processes were altered in sarcoma cells lacking NELL1. Notably, reductions in cell proliferation were observed across cell lines and in vivo models. We acknowledge that defects in sarcoma cell proliferation would affect other in vitro assays and in vivo endpoints. Nevertheless, a larger scale reduction in migration, invasion and attachment was observed across NELL1 knockout cell lines than changes in proliferation. For this reason, we believe it less likely that the overall phenotypes observed are derivative impacts on changes to cell proliferation alone.
In combination, our aggregate data suggest that NELL1 is a novel protein highly expressed among human osteosarcoma which positively regulates multiple aspects of OS disease progression. Use of gene editing technology, or alternatively small molecule-mediated disruption of NELL1 signaling, may represent an adjunctive tool to improve survival in osteosarcoma and other sarcoma subtypes.
Supplementary Material
Significance.
NELL1 modulates the sarcoma matrisome to promote tumor growth, invasion, and metastasis, identifying the matrix-associated protein as an orchestrator of cell-ECM interactions in sarcomagenesis and disease progression.
Acknowledgments
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health or Department of Defense. We thank the JHU microscopy core facility, JHMI deep sequencing and microarray core facility, division of molecular pathology and Hao Zhang within the JHU Bloomberg Flow Cytometry and Immunology Core.
Financial support: American Cancer Society (Research Scholar Grant, RSG-18-027-01-CSM), NIH/NIAMS (R01 AR070773, R01 AR079171), Department of Defense (USAMRAA W81XWH-18-1-0336, W81XWH-18-1-0121, W81XWH-20-1-0795, W81XWH-20-1-0302), and the Maryland Stem Cell Research Foundation.
Footnotes
Conflict of interest: A.W.J. is a paid consultant for Novadip and Lifesprout LLC. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies. All other authors declare no potential conflicts of interest.
References
- 1.Dancsok AR, Asleh-Aburaya K, Nielsen TO. Advances in sarcoma diagnostics and treatment. Oncotarget 2017;8:7068–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van der Laan P, van Houdt WJ, van den Broek D, Steeghs N, van der Graaf WTA. Liquid Biopsies in Sarcoma Clinical Practice: Where Do We Stand? Biomedicines 2021;9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis DM, Pruitt H, Jain N, Ciccaglione M, McCaffery JM, Xia Z, et al. A Feedback Loop between Hypoxia and Matrix Stress Relaxation Increases Oxygen-Axis Migration and Metastasis in Sarcoma. Cancer Res 2019;79:1981–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frantz C, Stewart KM, Weaver VM. The extracellular matrix at a glance. J Cell Sci 2010;123:4195–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Multhaupt HA, Leitinger B, Gullberg D, Couchman JR. Extracellular matrix component signaling in cancer. Adv Drug Deliv Rev 2016;97:28–40 [DOI] [PubMed] [Google Scholar]
- 6.Levental KR, Yu H, Kass L, Lakins JN, Egeblad M, Erler JT, et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 2009;139:891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ivanov V, Ivanova S, Roomi MW, Kalinovsky T, Niedzwiecki A, Rath M. Naturally produced extracellular matrix inhibits growth rate and invasiveness of human osteosarcoma cancer cells. Med Oncol 2007;24:209–17 [DOI] [PubMed] [Google Scholar]
- 8.Brosicke N, Sallouh M, Prior LM, Job A, Weberskirch R, Faissner A. Extracellular Matrix Glycoprotein-Derived Synthetic Peptides Differentially Modulate Glioma and Sarcoma Cell Migration. Cell Mol Neurobiol 2015;35:741–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qiu S, Tao L, Zhu Y. Marital Status and Survival in Osteosarcoma Patients: An Analysis of the Surveillance, Epidemiology, and End Results (SEER) Database. Med Sci Monit 2019;25:8190–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Song K, Song J, Lin K, Chen F, Ma X, Jiang J, et al. Survival analysis of patients with metastatic osteosarcoma: a Surveillance, Epidemiology, and End Results population-based study. Int Orthop 2019;43:1983–91 [DOI] [PubMed] [Google Scholar]
- 11.Cui J, Dean D, Hornicek FJ, Chen Z, Duan Z. The role of extracelluar matrix in osteosarcoma progression and metastasis. J Exp Clin Cancer Res 2020;39:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikitovic D, Kavasi RM, Berdiaki A, Papachristou DJ, Tsiaoussis J, Spandidos DA, et al. Parathyroid hormone/parathyroid hormone-related peptide regulate osteosarcoma cell functions: Focus on the extracellular matrix (Review). Oncol Rep 2016;36:1787–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Na KY, Bacchini P, Bertoni F, Kim YW, Park YK. Syndecan-4 and fibronectin in osteosarcoma. Pathology 2012;44:325–30 [DOI] [PubMed] [Google Scholar]
- 14.De Vita A, Recine F, Miserocchi G, Pieri F, Spadazzi C, Cocchi C, et al. The potential role of the extracellular matrix in the activity of trabectedin in UPS and L-sarcoma: evidences from a patient-derived primary culture case series in tridimensional and zebrafish models. J Exp Clin Cancer Res 2021;40:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tanjaya J, Lord EL, Wang C, Zhang Y, Kim JK, Nguyen A, et al. The Effects of Systemic Therapy of PEGylated NEL-Like Protein 1 (NELL-1) on Fracture Healing in Mice. Am J Pathol 2018;188:715–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen J, James AW, Zhang X, Pang S, Zara JN, Asatrian G, et al. Novel Wnt Regulator NEL-Like Molecule-1 Antagonizes Adipogenesis and Augments Osteogenesis Induced by Bone Morphogenetic Protein 2. Am J Pathol 2016;186:419–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.James AW, Shen J, Zhang X, Asatrian G, Goyal R, Kwak JH, et al. NELL-1 in the treatment of osteoporotic bone loss. Nat Commun 2015;6:7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwak J, Zara JN, Chiang M, Ngo R, Shen J, James AW, et al. NELL-1 injection maintains long-bone quantity and quality in an ovariectomy-induced osteoporotic senile rat model. Tissue Eng Part A 2013;19:426–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ting K, Vastardis H, Mulliken JB, Soo C, Tieu A, Do H, et al. Human NELL-1 expressed in unilateral coronal synostosis. J Bone Miner Res 1999;14:80–9 [DOI] [PubMed] [Google Scholar]
- 20.Li W, Zara JN, Siu RK, Lee M, Aghaloo T, Zhang X, et al. Nell-1 enhances bone regeneration in a rat critical-sized femoral segmental defect model. Plast Reconstr Surg 2011;127:580–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James AW, Chiang M, Asatrian G, Shen J, Goyal R, Chung CG, et al. Vertebral Implantation of NELL-1 Enhances Bone Formation in an Osteoporotic Sheep Model. Tissue Eng Part A 2016;22:840–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai J, Shannon ME, Johnson MD, Ruff DW, Hughes LA, Kerley MK, et al. Nell1-deficient mice have reduced expression of extracellular matrix proteins causing cranial and vertebral defects. Hum Mol Genet 2006;15:1329–41 [DOI] [PubMed] [Google Scholar]
- 23.Shen J, LaChaud G, Khadarian K, Shrestha S, Zhang X, Soo C, et al. NELL-1 expression in benign and malignant bone tumors. Biochem Biophys Res Commun 2015;460:368–74 [DOI] [PubMed] [Google Scholar]
- 24.Qi HC, Kim JK, Ha P, Chen XY, Chen E, Chen Y, et al. Inactivation of Nell-1 in Chondrocytes Significantly Impedes Appendicular Skeletogenesis. Journal of Bone and Mineral Research 2019;34:533–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quist T, Jin H, Zhu JF, Smith-Fry K, Capecchi MR, Jones KB. The impact of osteoblastic differentiation on osteosarcomagenesis in the mouse. Oncogene 2015;34:4278–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maniscalco L, Iussich S, Morello E, Martano M, Biolatti B, Riondato F, et al. PDGFs and PDGFRs in canine osteosarcoma: new targets for innovative therapeutic strategies in comparative oncology. Vet J 2013;195:41–7 [DOI] [PubMed] [Google Scholar]
- 27.Wang Y, Xu J, Meyers CA, Gao Y, Tian Y, Broderick K, et al. PDGFRalpha marks distinct perivascular populations with different osteogenic potential within adipose tissue. Stem Cells 2020;38:276–90 [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Wang Y, Hsu CY, Negri S, Tower RJ, Gao Y, et al. Lysosomal protein surface expression discriminates fat- from bone-forming human mesenchymal precursor cells. Elife 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soleimani M, Nadri S. A protocol for isolation and culture of mesenchymal stem cells from mouse bone marrow. Nature Protocols 2009;4:102–6 [DOI] [PubMed] [Google Scholar]
- 30.Chen YY, Wang T, Huang MX, Liu Q, Hu C, Wang B, et al. MAFB Promotes Cancer Stemness and Tumorigenesis in Osteosarcoma through a Sox9-Mediated Positive Feedback Loop. Cancer Research 2020;80:2472–83 [DOI] [PubMed] [Google Scholar]
- 31.Sun C, Fang Y, Yin J, Chen J, Ju Z, Zhang D, et al. Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foroutan P, Kreahling JM, Morse DL, Grove O, Lloyd MC, Reed D, et al. Diffusion MRI and novel texture analysis in osteosarcoma xenotransplants predicts response to anti-checkpoint therapy. PLoS One 2013;8:e82875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman SD, Calo E, Landman AS, Danielian PS, Miller ES, West JC, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A 2008;105:11851–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fahmy-Garcia S, van Driel M, Witte-Buoma J, Walles H, van Leeuwen JPTM, van Osch GJVM, et al. NELL-1, HMGB1, and CCN2 Enhance Migration and Vasculogenesis, But Not Osteogenic Differentiation Compared to BMP2. Tissue Eng Pt A 2018;24:207–18 [DOI] [PubMed] [Google Scholar]
- 35.Walkley CR, Qudsi R, Sankaran VG, Perry JA, Gostissa M, Roth SI, et al. Conditional mouse osteosarcoma, dependent on p53 loss and potentiated by loss of Rb, mimics the human disease. Genes Dev 2008;22:1662–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilbao-Aldaiturriaga N, Askaiturrieta Z, Granado-Tajada I, Goricar K, Dolzan V, For The Slovenian Osteosarcoma Study G, et al. A systematic review and meta-analysis of MDM2 polymorphisms in osteosarcoma susceptibility. Pediatr Res 2016;80:472–9 [DOI] [PubMed] [Google Scholar]
- 37.Iwata S, Tatsumi Y, Yonemoto T, Araki A, Itami M, Kamoda H, et al. CDK4 overexpression is a predictive biomarker for resistance to conventional chemotherapy in patients with osteosarcoma. Oncology Reports 2021;46. [DOI] [PubMed] [Google Scholar]
- 38.Gao Y, Feng Y, Shen JK, Lin M, Choy E, Cote GM, et al. CD44 is a direct target of miR-199a-3p and contributes to aggressive progression in osteosarcoma. Sci Rep-Uk 2015;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen J, James AW, Zara JN, Asatrian G, Khadarian K, Zhang JB, et al. BMP2-Induced Inflammation Can Be Suppressed by the Osteoinductive Growth Factor NELL-1. Tissue Eng Pt A 2013;19:2390–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai C, Yang M, Fan Z, Li S, Gao T, Fang Z. Associations of chemo- and radio-resistant phenotypes with the gap junction, adhesion and extracellular matrix in a three-dimensional culture model of soft sarcoma. J Exp Clin Cancer Res 2015;34:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hawkins AG, Basrur V, da Veiga Leprevost F, Pedersen E, Sperring C, Nesvizhskii AI, et al. The Ewing Sarcoma Secretome and Its Response to Activation of Wnt/beta-catenin Signaling. Mol Cell Proteomics 2018;17:901–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saggioro M, D’Angelo E, Bisogno G, Agostini M, Pozzobon M. Carcinoma and Sarcoma Microenvironment at a Glance: Where We Are. Frontiers in Oncology 2020;10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen PC, Cheng HC, Yang SF, Lin CW, Tang CH. The CCN family proteins: modulators of bone development and novel targets in bone-associated tumors. Biomed Res Int 2014;2014:437096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao Z, Wan J, Nur AA, Dou P, Mankin H, Liu T, et al. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells. Cell Physiol Biochem 2018;51:1879–93 [DOI] [PubMed] [Google Scholar]
- 45.Kondo T. Current status of proteomics of soft tissue sarcomas. Expert Rev Proteomics 2017;14:1131–40 [DOI] [PubMed] [Google Scholar]
- 46.Ingley KM, Maleddu A, Grange FL, Gerrand C, Bleyer A, Yasmin E, et al. Current approaches to management of bone sarcoma in adolescent and young adult patients. Pediatr Blood Cancer 2021:e29442 [DOI] [PubMed] [Google Scholar]
- 47.Liu M, Wang Q, Tang R, Cao R, Wang X. Nel-like Molecule 1 Contributes to the Odontoblastic Differentiation of Human Dental Pulp Cells. J Endod 2016;42:95–100 [DOI] [PubMed] [Google Scholar]
- 48.Zhang X, Peault B, Chen W, Li W, Corselli M, James AW, et al. The Nell-1 growth factor stimulates bone formation by purified human perivascular cells. Tissue Eng Part A 2011;17:2497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DuBois S, Demetri G. Markers of angiogenesis and clinical features in patients with sarcoma. Cancer 2007;109:813–9 [DOI] [PubMed] [Google Scholar]
- 50.Jacob A, Prekeris R. The regulation of MMP targeting to invadopodia during cancer metastasis. Front Cell Dev Biol 2015;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, et al. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol 2000;2:737–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tombolan L, Poli E, Martini P, Zin A, Romualdi C, Bisogno G, et al. NELL1, whose high expression correlates with negative outcomes, has different methylation patterns in alveolar and embryonal rhabdomyosarcoma. Oncotarget 2017;8:33086–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu XF, Wang L, Ye YQ, Aakre JA, Pu X, Chang GC, et al. Genome-Wide Association Study of Genetic Predictors of Overall Survival for Non-Small Cell Lung Cancer in Never Smokers. Cancer Research 2013;73:4028–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin Z, Mori Y, Yang J, Sato F, Ito T, Cheng Y, et al. Hypermethylation of the nel-like 1 gene is a common and early event and is associated with poor prognosis in early-stage esophageal adenocarcinoma. Oncogene 2007;26:6332–40 [DOI] [PubMed] [Google Scholar]
- 55.Nakamura R, Oyama T, Tajiri R, Mizokami A, Namiki M, Nakamoto M, et al. Expression and regulatory effects on cancer cell behavior of NELL1 and NELL2 in human renal cell carcinoma. Cancer Sci 2015;106:656–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhai Y, Wei R, Sha S, Lin C, Wang H, Jiang X, et al. Effect of NELL1 on lung cancer stemlike cell differentiation. Oncol Rep 2019;41:1817–26 [DOI] [PubMed] [Google Scholar]
- 57.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data is freely available within the NCBI GEO database (GSE182703). Proteomic data is submitted as a separate Excel file.