Abstract
BRCA1-associated protein 1 (BAP1) is a tumor suppressor gene that is mutated in cancer, including uveal melanoma. Loss-of-function BAP1 mutations are associated with uveal melanoma metastasis and poor prognosis, but the mechanisms underlying these effects remain unclear. Upregulation of cell–cell adhesion proteins is involved with collective migration and metastatic seeding of cancer cells. Here, we show that BAP1 loss in uveal melanoma patient samples is associated with upregulated gene expression of multiple cell adhesion molecules (CAM), including E-cadherin (CDH1), cell adhesion molecule 1 (CADM1), and syndecan-2 (SDC2). Similar findings were observed in uveal melanoma cell lines and single-cell RNA-sequencing data from uveal melanoma patient samples. BAP1 reexpression in uveal melanoma cells reduced E-cadherin and CADM1 levels. Functionally, knockdown of E-cadherin decreased spheroid cluster formation and knockdown of CADM1 decreased growth of BAP1-mutant uveal melanoma cells. Together, our findings demonstrate that BAP1 regulates the expression of CAMs which may regulate metastatic traits.
Introduction
The BRCA1-associated protein 1 (BAP1) gene is located on chromosome 3 and encodes a ubiquitin carboxy-terminal hydrolase enzyme (1). BAP1 associates with DNA damage repair proteins as part of the BRCA1-BARD E3 ligase complex (2). In addition, BAP1 functions as an epigenetic modulator and alters gene transcription through the formation of the Polycomb repressive deubiquitinase complex in concert with transcription factors FOXK1/K2 (3) and the cell-cycle regulator host-cell factor 1 (4). BAP1 also influences apoptotic signaling (5), ferroptosis regulation (6), and cellular metabolism (7), suggesting its diverse and context-dependent role as a tumor suppressor (8).
BAP1-inactivating mutations are associated with several cancers, including uveal melanoma, malignant mesothelioma, renal cell carcinoma, and cholangiocarcinoma (8). There is contrasting evidence regarding the role of BAP1 in cancer progression and prognosis. In malignant mesothelioma, BAP1 mutations are associated with a better prognosis (9). On the contrary, BAP1 mutations in uveal melanoma and renal cell carcinoma are associated with metastasis and lower overall survival (9). Despite contrasting prognoses between malignant mesothelioma and uveal melanoma, BAP1 loss is associated with an epithelioid cellular phenotype in non–epithelium-derived tissues of origin (10). Overall, it is key to investigate BAP1 functions in the appropriate tumor types and physiologic contexts.
Uveal melanoma is the most common primary intraocular malignancy in adults (11). Despite successful treatment of primary uveal melanoma with radioactive therapy or enucleation, approximately 50% of patients will ultimately develop metastases, predominantly in the liver (12). Loss-of-function BAP1 mutations are detected in up to 90% of metastatic uveal melanoma cases (13, 14), and there are no effective treatment strategies that target the effects of BAP1 loss (15). Loss of BAP1 is known to result in a more epithelial, stem cell–like phenotype (10); however, it remains unclear why BAP1 mutations are associated with metastatic uveal melanoma. It is important to understand the biological functions of BAP1 to improve treatment options and disease outcomes for patients with uveal melanoma.
Metastasis is a multistep process involving the primary tumor acquiring traits that promote invasion, migration, extravasation, and seeding into the metastatic site (16). In epithelial-based tumors, loss of cell adhesion molecules (CAM), including E-cadherin and cell adhesion molecule 1 (CADM1), is associated with epithelial–mesenchymal transition (EMT), which initiates the metastatic process (17). Although melanoma is not derived from epithelial tissue and does not undergo EMT, melanoma cells have a mesenchymal-like phenotype (18). E-cadherin and CADM1 are negatively regulated by EMT-inducing transcription factors such as Twist1, ZEB1, and Snail (19, 20). In renal cell carcinoma, loss of BAP1 downregulated Snail expression and facilitated a mesenchymal–epithelial transition, a reverse process to EMT (21). However, BAP1’s role in the metastatic progression of uveal melanoma, including induction of an EMT-like state, remains unclear.
In this study, we undertook a multi-omics approach to identify molecular changes associated with BAP1 mutational status in the context of uveal melanoma. Transcriptomic data from multiple, independent patient and cell line datasets indicate that loss of BAP1 correlates with upregulation of cell–cell adhesion genes, CDH1, CADM1, and SDC2. Proteomic analysis of BAP1 wild-type and mutant cells was consistent with these results. Knockdown of E-cadherin and CADM1 decreased cell growth, migration, and cell–cell adhesion of BAP1-mutant uveal melanoma cells. Knockdown of SDC2 did not have any overt functional effect. These findings suggest a role for E-cadherin and CADM1 in facilitating cancer cell survival and clustering for patients with BAP1-mutant uveal melanoma.
Materials and Methods
Bulk RNA-sequencing analysis
RNA-sequencing (RNA-seq) data were aligned to the human reference genome (GRCh 38) using Star aligner (22) and GEN-CODE (23) annotations. RSEM (24) was used to quantify gene and transcript-level expression. Gene differential expression analysis was performed using DESeq2 (25). Data analyses were performed in R (v3.5.1 http://www.R-project.org/). Gene set enrichment analysis (GSEA; ref. 26) was used to calculate normalized expression scores (NES) and to determine enriched pathways in the C2CP Kyoto Encyclopedia of Genes and Genomes (KEGG; ref. 27) and Hallmark gene set collections from the Molecular Signatures Database (MSigDB; ref. 28). The GSEA preranked method was performed using DESeq2 Wald test statistic values after filtering out zero and “NA” values.
Analysis of The Cancer Genome Atlas data
The Cancer Genome Atlas (TCGA) uveal melanoma RNA-seq data were retrieved from the Broad GDAC Firehose data run (stddata 2016_01_28). The RTCGA package (version 1.10.0) was used to gather mutation and copy-number data, and samples with a BAP1 copy-number segment mean less than 0.5 were classified as BAP1 mutants. Gene differential expression analysis and GSEA were performed between BAP1 mutant (n 40) and wild-type (n 40) groups as described previously (7). Evaluation of patient survival for TCGA uveal melanoma cohort was performed using cBioPortal (29). Survival outcome and gene expression data originated from TCGA Pan-Cancer Clinical Data Resource (30) and TCGA Pan-Cancer Atlas (31), respectively. Patients were stratified into high and low groups by median RNA expression. The log-rank test was used to determine statistical significance for progression-free survival.
Chromosome 8q copy-number call data were obtained from ref. 32. Multivariable survival analysis methods were used to determine the association between progression-free survival, SDC2 expression, and number of chromosome 8q copies. The Cox proportional hazards regression model was used with assumptions validated by a test of Schoenfeld residuals. Analyses were performed via the survival package (v 3.1.12 https://CRAN.R-project.org/packagesurvival). The ggplot2 package (v3.3.5 https://ggplot2.tidyverse.org) was used to generate box plots. Data analyses were conducted in R (v4.1.0 http://www.R-project.org/).
Cell lines and modifications
Details on acquirement and maintenance of cell lines MM66, MP46, MM28 MP65, MP38, and PDX4 cell lines have been reported previously (7, 33). UM001, UM004, OMM1.3, WM3618F, and 92.1 were acquired and maintained as described previously (34). Maintenance of the UM002B cell line is described previously (35). All cell lines were routinely tested for Mycoplasma and authenticated by short tandem repeat analysis every 6 months. For BAP1 expression, wild-type BAP1 plasmid, pLVX-M-Flag-BAP1 was used; it was a gift from BoyiGan (Addgene plasmid # 125840; http://n2t.net/addgene:125840; RRID: Addgene_125840; ref. 6). MP46 cells were incubated in the presence of BAP1 or GFP lentiviral supernatant for 72 hours. Transduced cell populations were then selected with puromycin.
RNA-seq sample processing
Bulk sequencing: total RNA was quantified using the Quant-iT RiboGreen RNA Assay Kit. Samples were prepared using the Illumina TruSeq Stranded mRNA Sample Preparation Kit, and the resultant 400 bp cDNA was then processed for dual-indexed library preparation. Sequencing was performed to generate 101 bp paired-end reads with eight-base index barcodes from a library with pooled samples using the NovaSeq 6000 according to the manufacturer’s protocols. Demultiplexing and data aggregation were performed using the Broad Picard Pipeline as described previously (7). Gene differential analysis and GSEA were performed between BAP1 mutant and wild-type samples as described above. RNA-seq data have been deposited to the Gene Expression Omnibus (GEO) database with accession codes GSE149920 and GSE181194.
RNA-seq mutation calling
RNA-seq data were aligned to the human reference genome (GRCh 38) using GENCODE (23) annotations and the Star aligner (22) two-pass method. The Genome Analysis Toolkit (v4.1.2) RNA-seq short variant discovery workflow (36) and EnsemblVariant Effect Predictor (37) were used to identify and annotate BAP1 mutations, respectively.
Single-cell RNA-seq analysis
Uveal melanoma single-cell RNA-seq (scRNA-seq) data were obtained from the GEO database (accession GSE139829). The Seurat package (v3.1.4; ref. 38) was used to analyze the data. Malignant cells were identified using the methods described previously (39). Raw count data were normalized using the SCTransform method (40) with regression based on the percent mitochondrial content. The ggplot2 package (v3.3.2 https://ggplot2.tidyverse.org) was used to generate dot plots. Data analyses were performed in R (v3.6.0 http://www.R-project.org/).
Reverse phase protein array analysis
Cells were plated in 6-well dishes at 4 × 105 cells per well, washed twice in ice-cold PBS, and then lysed in 50 mL of RPPA lysis buffer [1% Triton X-100, 50 mmol/L HEPES (pH 7.4), 150 mmol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 100 mmol/L NaF, 10 mmol/L NaPPI, 1 mmol/L Na3VO4, 10% glycerol, protease and phosphatase inhibitors (Boehringer/Roche)] for 20 minutes with occasional shaking on ice. Lysates were centrifuged for 10 minutes at 14,000 rpm, and the supernatant collected. Protein concentration was determined by Bradford assay. Lysates were analyzed at the MD Anderson Functional Proteomic core facility (Houston, TX), where antibodies are extensively validated before being included in the panel. Serial dilutions of samples were arrayed on nitrocellulose-coated slides and run against 456 validated antibodies. A 3,30-diaminobenzidine colorimetric reaction for a tyramide-based signal amplification approach was used to produce stained slides. The slides were scanned on a Huron Tissue-Scope scanner, and spot densities were determined using Array-Pro Analyzer. Relative protein levels were quantified using SuperCurve fitting and normalized for protein loading. Differential expression analysis between BAP1 mutant and wild-type cell lines was performed using the limma package (v 3.44.3; ref. 41). The Benjamin–Hochberg FDR (BHFDR) method was used to determine statistical significance. Labels were chosen for the top and bottom proteins by sorting the products of log2 fold change and negative log10(BHFDR) values.
Scatter plots were generated using the ggplot2 package (v 3.3.2). Data analyses were performed in R (v3.5.1 http://www.R-project.org/). The reverse phase protein array (RPPA) results were validated by Western blotting for key targets.
Patient microarrays survival analysis
Microarray data for 63 primary uveal melanoma patient tumors were obtained from the GEO database (accession GSE22138). Raw .CEL files were processed using a BrainArray (42) Entrez gene ID custom CDF file. Data were read into R using the affy package (v 1.66.0; ref. 43). Patient metadata was accessed using the GEOquery package (v 2.56.0; ref. 44). The log-rank test was used to determine statistical significance for time-to-metastasis using the survival package (v 3.1.12 https://CRAN.R-project.org/packagesurvival). Data analyses were performed in R (v4.0.2 http://www.R-project.org/).
Cell line microarrays analysis
Microarray data for 20 uveal melanoma cell line samples were obtained from the GEO database (accession GSE78033). Raw .CEL files were processed using a BrainArray (42) Entrez gene ID custom CDF file. Data were read into R using the affy package (v 1.66.0; ref. 43). Sample metadata was accessed using the GEOquery package (v 2.56.0; ref. 44) and Supplementary Files from Laurent and colleagues 2013 (45). Cell lines with a BAP1 mutation and chromosome 3 loss were defined as BAP1 mutants, while samples with no mutation and no chromosome 3 loss were defined as BAP1 wild-type. Differential expression analysis was performed using the limma package (v3.48.1; ref. 41). Data analyses were performed in R (v4.1.0 http://www.R-project.org/).
BAP1 reexpression and shBAP1 RNA-seq datasets
RNA-seq reads for BAP1 mutant and reexpressing UPMM3 cell line samples were obtained from the Sequence Read Archive (SRA) under accession number SRP102735. RNA-seq reads for shRNA targeting BAP1 (shBAP1) and BAP1 wild-type expressing 92.1 and MEL202 cell line samples were accessed under accession number SRP193943. Both datasets were obtained using the SRA toolkit (v 2.10.4; ref. 46). Data were processed as described above. Gene differential expression results between BAP1 mutant and reexpressing UM22 cell line samples were obtained from Karlsson and colleagues 2020 (14).
siRNA transfections
Cells were transfected for 72 hours with the indicated siRNAs at a final concentration of 25 nmol/L using Lipofectamine RNAiMAX (Invitrogen). Nontargeting control (50-UGGUUUACAUGUCGA-CUAA-30), CDH1 #2, #5 (D-003877-02, D-003877-05), and CADM1 #1, #2 (D-016565-01, D-016565-02) from Dharmacon were used.
Western blot analysis
Protein lysates were lysed in Laemmli sample buffer, separated by SDS-PAGE, and proteins were transferred to polyvinylidene difluoride membranes. The following primary antibodies were used: BAP1 (#13271, RRID:AB_2798168), E-cadherin (#5296, RRID: AB_10706939), phosphop44/42 MAPK (ERK1/2) (Thr202/Tyr204) (#9101, RRID:AB_331646), S6 ribosomal protein (#2217, RRID: AB_331355), phospho-S6 Ribosomal Protein (Ser235/236) (#4857, RRID:AB_2181035) from Cell Signaling Technology; CADM1 (ab138697) from Abcam; b-actin (#A2066, RRID:AB_476693) from Sigma-Aldrich, phospho-Cdk1 (pTpY14/15) (#44-686G, RRID: AB_1491072) from Invitrogen; cyclin B1 (GNS-1) (#sc-245, RRID:AB_627338) and ERK1 (#sc-93, RRID:AB_631453) from Santa Cruz Biotechnology. Immunoreactivity was detected using horseradish peroxidase–conjugated secondary antibodies from (CalBioTech) and chemiluminescence substrate from ThermoFisher Scientific on a Versadoc Imaging System (Bio-Rad).
IncuCyte live-cell growth assay
Cells were trypsinized and seeded at 400,000 cells per well onto a 6-well plate. Photomicrographs were taken every 2 hours using an IncuCyte Live Cell Analysis Imaging System using a 10 objective (Essen Biosciences). Plate confluence was measured using IncuCyte software and presented as a fold change of percent confluency compared with day 0 confluency.
Migration assay
Cells treated for 72 hours with appropriate siRNA were seeded (1.2 × 105) into 8.0 mmol/L cell culture inserts from BD Biosciences. The migration assay protocol was completed as described previously (19). Images were taken with an inverted fluorescence microscope (Nikon Ti-E), excluding areas of the membrane with a clear edge effect. Quantification was performed utilizing ImageJ. The experiment was carried out with at least five replicates.
Cell adhesion assay
To determine whether knockdown of E-cadherin and CADM1 affected cell–cell adhesion, the Vybrant cell adhesion assay kit protocol was adapted (Invitrogen Inc). A monolayer of MP38 cells (1.2 × 105) were cultured on a 96-well plate (Corning) for 24 hours. Cells treated with siRNA for 72 hours (1.0 × 106) were loaded with calcein AM (5 mmol/L) for 30 minutes, washed with PBS, and then resuspended in a serum-free medium. Calcein AM–labeled cells (5.0 × 104) were added to the 96-well plate coated with a layer of confluent cells (media removed) and incubated for 2 hours. Nonadherent cells were either washed off with PBS. PBS was added to both unwashed (total fluorescence) and washed plates (adherent cell fluorescence), and fluorescence was measured using a GloMax Discover Microplate Reader (Promega). The fluorescent filter was set at 494 nm and maximum emission of 517 nm. Adhesion percent is calculated as (adherent cell fluorescence)/(total cell fluorescence).
Spheroid formation and viability assay
Cells treated for 72 hours with appropriate siRNA were seeded in a 96-well culture dish (Corning) coated in 1.5% agarose at 5,000 cells per well. Seventy-two hours after seeding, spheroid sizes were assessed by bright-field microscopy and quantified using ImageJ. A time course was also collected with the IncuCyte Live Cell Analysis Imaging System. Cell viability was assessed in three-dimensional (3D) cultures using the CellTiter-Glo 3D Cell Viability Assay (Promega) according to the manufacturer’s instructions. Briefly, after 72 hours of incubation 100 mL medium was removed from each well, and 100 mL CellTiter-Glo 3D was added. All procedures were performed at room temperature. The plate was incubated on a shaker in the dark for 5 minutes. After a further 25-minute incubation in the dark, luminescence was recorded using a GloMax Discover Plate Reader (Promega).
Statistical analyses
Data were analyzed using the two-sided Student t test with Microsoft Excel software (P < 0.05; P < 0.01; and P < 0.001).
Data availability
The data generated in this study are publicly available in GEO at GSE149920 and GSE181194, within the article, and its Supplementary Data files. The publicly available data analyzed in this study were obtained from GEO at GSE139829, GSE22138, and GSE78033, and the SRA at SRP102735 and SRP193943.
Results
Gene expression profiling inversely correlates BAP1 status with the expression of CAMs
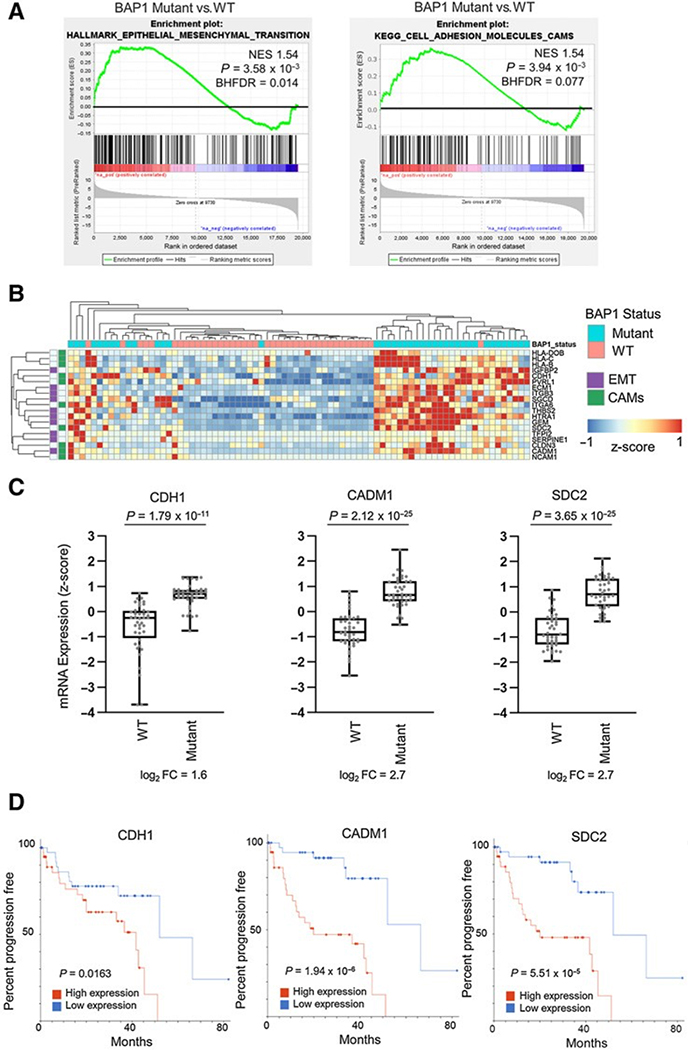
Given that the loss of BAP1 is detected frequently in high-risk metastatic uveal melanoma cases (13), we sought to study molecular differences that may explain disease progression. First, we analyzed uveal melanoma cases in TCGA separated into BAP1 mutant (n = 40) and wild-type (n = 40) groups (Supplementary Table S1) and utilized the Hallmark gene sets collection (28) to conduct GSEA (26). Our analysis identified that the Hallmark EMT gene set was significantly enriched (NES > 1.5, BHFDR < 0.05) in the BAP1 mutant compared with BAP1 wild-type uveal melanoma samples (Fig. 1A, left; Supplementary Table S2). There was no prior subsetting of the Hallmark gene sets; however, EMT is a known metastasis-related process (47, 48). Therefore, we were particularly interested when we saw statistically significant enrichment of the EMT gene set. We further conducted GSEA on the KEGG database (27) gene set collection and observed the CAMs pathway gene set to be among the top 20 most positively enriched (Fig. 1A, right; Supplementary Table S3). While investigating the top 10 enriched genes from the CAMs and EMT gene sets, CADM1 was the only one present in both (Fig. 1B; Supplementary Tables S4–S6).
Figure 1.
BAP1 mutations are correlated with increased expression of CAMs CDH1, CADM1, and SDC2. A, On the basis of BAP1 mutation and copy loss, TCGA uveal melanoma samples were stratified into BAP1 mutant and wild-type groups. Differential expression analysis was performed between BAP1 mutant (n ¼ 40) and wild-type (n ¼ 40) samples and used for performing GSEA. GSEA enrichment plots of the Hallmark epithelial to mesenchymal and KEGG CAMs) gene sets in BAP1 mutant versus wild-type groups are shown. B, A heatmap showing z-score values for the top 10 genes from the Hallmark EMT and KEGG CAMs gene sets in TCGA uveal melanoma patient tumor samples. C, CDH1, CADM1, and SDC2 gene expression from TCGA RNA-seq data in BAP1 wild-type (n ¼ 40) and mutant samples (n¼40). D, Analysis of TCGAdata for uveal melanoma patient progression-free survival according to CDH1, CADM1, and SDC2 expression, stratified by high or low median RNA expression. Log-rank test was used to determine the significance of progression-free survival (https://wiki.nci.nih.gov/plugins/servlet/mobile#content/view/24279961).
Next, we prioritized the top three enriched genes in the KEGG CAMs pathway between BAP1 mutant and wild-type groups for further evaluation. These genes were CDH1 (log2 fold change: 1.617), CADM1 (log2 fold change: 2.708), and SDC2 (log2 fold change: 2.702; Fig. 1C). High expression of CDH1, CADM1, and SDC2 was significantly associated with worse progression-free survival of patients with uveal melanoma irrespective of BAP1 mutational status (Fig. 1D; Supplementary Table S7). Additional analyses of microarray data from 63 patient tumor samples determined that high expression of CADM1 and SDC2 significantly correlated with earlier onset of metastasis (Supplementary Fig. S1A; Supplementary Table S8). Altogether, these findings indicate that BAP1-mutant uveal melanoma displays increased expression of CAMs, including CDH1, CADM1, and SDC2, which are associated with worse progression-free survival of patients with uveal melanoma.
Amplification of the q-arm of chromosome 8 (Chr8q) has been previously associated with earlier onset of metastatic disease (32), and high copy gains have been predominantly observed in metastatic tumors (49). Because SDC2 is located on Chr8q, we investigated whether the number of Chr8q copies or the expression level of SDC2 correlated better with progression-free survival. The expression level of SDC2 was more correlated with progression-free survival (Supplementary Fig. S1B). We also observed a positive correlation in the number of Chr8q copies and increase in SDC2 expression in TCGA data and increased expression of SDC2 in BAP1-mutant cases independent of Chr8q status (Supplementary Fig. S1C).
Elevated expression of CAMs is selective to BAP1-mutant uveal melanoma cells
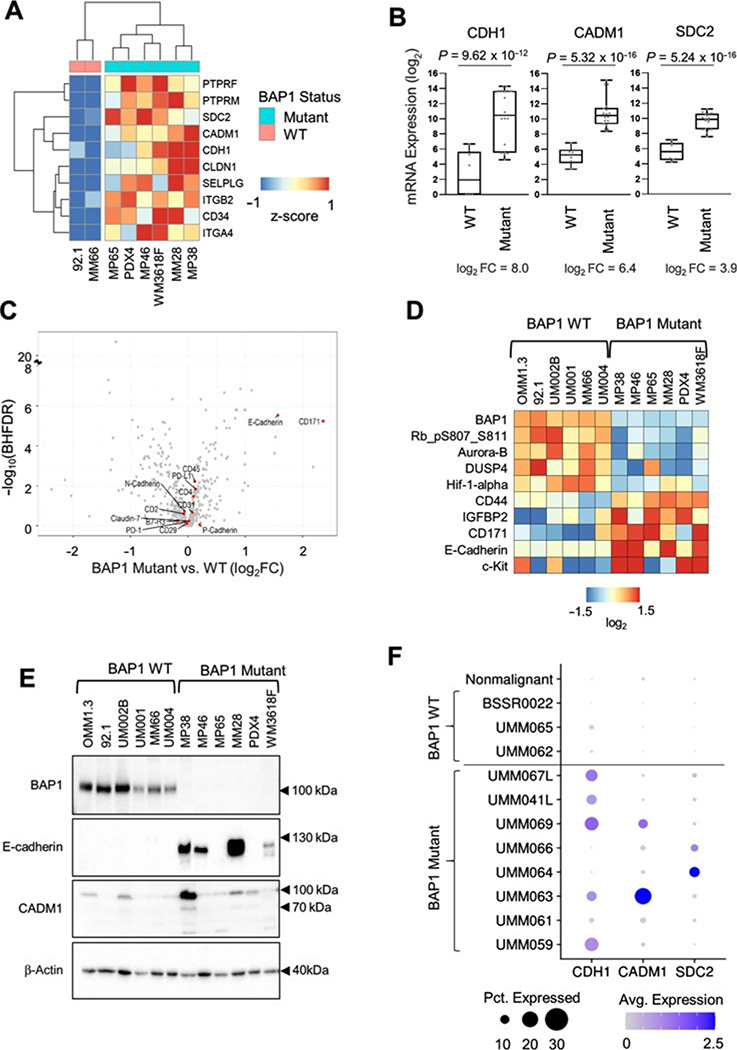
To further evaluate the association between CAM expression and BAP1 mutational status, we performed RNA-seq on eight uveal melanoma cell lines (MM28, MP46, MP65, PDX4, WM3618F, 92.1, and MM66). The uveal melanoma cell line, MP46, did not express BAP1 at the protein level, but no BAP1 mutation has previously been identified (50). Mutation calling on the RNA-seq data from MP46 cells showed that no BAP1 transcript levels were detected, and there was no detectable mRNA expression of the flanking gene, PHF7, which is located on the opposite strand and has a transcription start site within 600 bases of BAP1 (Supplementary Fig. S2A). This finding suggests that there may be a deletion spanning the transcription start sites of BAP1 and PHF7. Thus, we classified MP46 as a BAP1 mutant cell line. We compared the transcriptomic profiles between six BAP1 mutant cell lines (MM28, MP46, MP65, PDX4, and WM3618F) and two BAP1 wild-type cell lines (92.1 and MM66). GSEA revealed a positive enrichment of the EMT and KEGG CAMs pathways in BAP1 mutant cells versus wild-type cells (Supplementary Fig. S2B; Supplementary Tables S9–S11). CDH1, SDC2, and CADM1 were among the top 10 enriched genes from the KEGG CAMs pathway (Fig. 2A). Consistent with TCGA patient data (Fig. 1C), increased expression of CDH1, SDC2, and CADM1 was observed in the majority of BAP1 mutant compared with BAP1 wild-type cell lines (Fig. 2B). We followed this up by analyzing an independent dataset of uveal melanoma cell line samples where we discovered that CDH1, SDC2, and CADM1 were significantly enriched in the BAP1 mutant group (Supplementary Fig. S2D).
Figure 2.
BAP1 mutant and wild-type uveal melanoma cell line panel shows differential expression of CDH1, CADM1, and SDC2. Differential expression analysis of RNA-seq data from triplicates of six BAP1 mutant (MM28, MP38, MP46, MP65, WM3618F, and PDX4) and two BAP1 wild-type cell lines (92.1 and MM66) was performed using DESeq2. A, A heatmap showing average expression of six BAP1 mutant and two wild-type cell lines for the top 10 positively enriched KEGG CAMs pathway genes. B, Box plots of CDH1, CADM1, and SDC2 mRNA expression in BAP1 mutant (n ¼ 18) versus wild-type (n ¼ 6) cell line samples. C, A volcano plot showing antibodies targeting genes from the KEGG CAM pathway from the RPPA data. D, A heatmap of median-centered, log2-transformed RPPA data for the top and bottom five proteins ranked by the product of the fold change and log10(Benjamini–Hochberg FDR), when comparing BAP1 mutant versus wild-type cell lines. Each cell line was done in triplicate. Proteins are ordered on the basis of hierarchical clustering. E, Uveal melanoma cell line panel showing expression of E-cadherin and CADM1 in BAP1 wild-type versus mutant cell lines. F, Dot plot showing the average expression and percent of cells expressing CDH1, CADM1, and SDC2 from patient tumor scRNA-seq data. Cells were separated into on malignant and tumor-specific malignant cell groups.
Next, we sought to determine the extent to which CAMs were also elevated at the protein level. We performed RPPA analysis across a panel of six BAP1 mutant and six BAP1 wild-type cell lines. We observed significant upregulation of E-cadherin in BAP1 mutant cell lines among all KEGG CAMs pathway targets (Fig. 2C). When ranked by fold change and statistical significance, E-cadherin was one of the most differentially expressed targets, along with BAP1, HIF-1a, and c-Kit (Fig. 2D). CADM1 and syndecan-2 were not part of the RPPA analysis. Western blotting validated the RPPA results (Fig. 2E), showing elevated E-cadherin levels in BAP1 mutant cells such as MP38, MP46, MM28, and WM3618F. We probed for CADM1 and observed that expression was upregulated in MP38 but not other BAP1 mutant cell lines (Fig. 2E). These data indicate heterogeneity in the expression of CAMs between BAP1 mutant cells. We could not analyze syndecan-2 in uveal melanoma cell lines by Western blotting due to technical reasons. Syndecan-2 is heavily glycosylated (51, 52), but the use of heparinase I-III to digest glycosylated moieties did not resolve our issues.
To confirm whether CAM transcripts were detectable in uveal melanoma patient tumors, we investigated scRNA-seq data from BAP1 mutant and wild-type patient tumors (32). This analysis showed that BAP1-mutant uveal melanoma cells displayed high expression of CDH1, CADM1, and SDC2 compared with BAP1 wild-type cells and that expression of these CAMs occurs predominantly in cancerous cells (Fig. 2F; Supplementary Fig. S2C). These data support that upregulation of CAMs occurs in BAP1-mutant uveal melanoma tumors.
Alteration of CAMs depends on BAP1 functional status
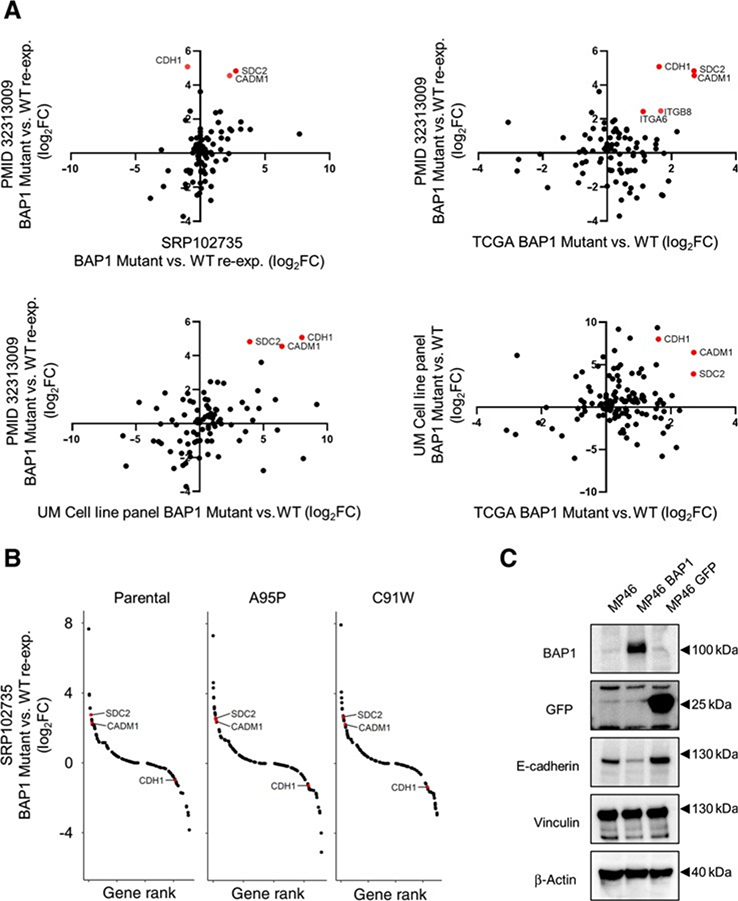
To further support our evidence of BAP1-associated differences in CAM expression, we cross-referenced RNA-seq data from four independent datasets: the uveal melanoma TCGA dataset, two BAP1 reexpression cell lines (Karlsson and colleagues, 2020 and Moore and colleagues, 2018; refs. 14, 53), and a uveal melanoma cell line panel. Differential gene expression analysis revealed CDH1, CADM1, and SDC2 were most upregulated within the KEGG CAMs pathway in BAP1 mutant versus BAP1 reexpressing cells with Karlsson and colleagues 2020 data, and that CADM1 and SDC2 were consistently increased in the Moore and colleagues 2018 dataset (Fig. 3A). When cross-referencing differential gene expression analysis of BAP1 mutant versus wild-type patient tumors from TCGA (32), a BAP1 mutant versus wild-type uveal melanoma cell line panel, and BAP1 mutant versus BAP1 reexpressing cells from Karlsson and colleagues, 2020, we observed consistent upregulation of CDH1, CADM1, and SDC2 in BAP1 null groups (Fig. 3A). Comparison of catalytically inactive (A95P or C91W) BAP1 reexpression to wild-type BAP1 reexpression samples in UPMM3 cells also showed high expression of CADM1 and SDC2 in BAP1 inactive samples (Fig. 3B). To determine whether CAM regulation via BAP1 is evident at the protein level, we expressed WT BAP1 in a BAP1 mutant cell line, MP46, by lentiviral transduction. Reduced E-cadherin expression was observed in MP46-BAP1 compared with parental and unaltered with the introduction of GFP as a negative control (Fig. 3C). We next utilized publicly available BAP1 knockdown datasets (54). Knockdown of BAP1 in the Mel202 cell line increased CDH1 and CADM1 but not SDC2 expression. Conversely, BAP1 short hairpin RNA knockdown in 92.1 cells increased SDC2 expression but not CDH1 and CADM1 levels (Supplementary Fig. S3). These data suggest that BAP1 alters CAM gene expression in uveal melanoma although selective changes may occur in different uveal melanoma–derived cell lines.
Figure 3.
Changes in CAMs gene expression due to BAP1 reexpression. A, Scatter plots showing fold change mRNA expression values for BAP1 mutant versus BAP1 reexpression or BAP1 mutant versus BAP1 wild-type comparisons from four independent datasets (14, 53) for the KEGG CAMs gene set. Red dots indicate genes with log2 fold changes greater than 1 and BHFDR value of less than 0.05. B, A BAP1-mutant uveal melanoma cell line (53) was transfected with wild-type or mutant BAP1 to produce proficient and deficient reexpressing samples, respectively. Each of the mutant reexpressing and parental samples were compared against the wild-type reexpressing samples. Rank list plots showing the fold change values for genes in the KEGG CAMs pathway for parental (left), mutant A95P reexpressing (middle), and mutant C91W reexpressing (right) samples compared against BAP1 wild-type reexpressing samples. C, Expression of BAP1 in BAP1 mutant cell cline, MP46, was confirmed by Western blot analysis, and protein expression of E-cadherin decreased in MP46-BAP1 cells. MP46-GFP was used as a control. Representative Western blots from triplicate experiments are shown.
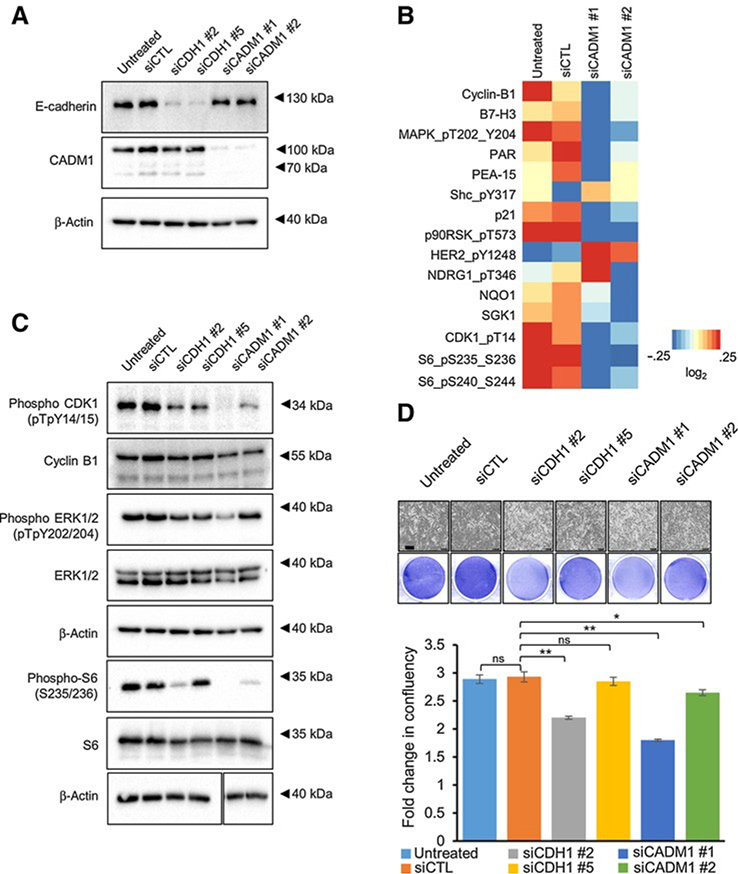
Depletion of CADM1 reduces cell-cycle proteins and proliferation
Because E-cadherin, CADM1, and syndecan-2 levels were upregulated with loss of BAP1 and BAP1 mutations are associated with a worse prognosis in uveal melanoma, we hypothesized that these CAMs promote malignant traits. We performed depletion experiments in BAP1-mutant MP38 cells in which both E-cadherin and CADM1 are highly expressed. E-cadherin and CADM1 were effectively knocked down with multiple independent siRNAs to reduce concerns about off-target effects (Fig. 4A; Supplementary Fig. S4A). Because of the aforementioned challenges faced for protein detection, the functional effects of syndecan-2 were not studied further. RPPA analysis revealed that proteins associated with cell-cycle progression were downregulated following depletion of CADM1 (Fig. 4B). In CADM1 knockdown cells, decreased expression of phospho-CDK1, cyclin B1, and phospho-S6 (Ser235/Ser 236), which are involved in cell cycle and protein translation, was validated by Western blot analysis (Fig. 4C). These data show that CADM1 depletion in BAP1 mutant cells decreases cell-cycle and cell growth protein expression levels. In vitro IncuCyte assays showed that in both MP38 and MM28 cells, E-cadherin knockdown elicited small to modest effects on cell growth (3%–25% inhibition), whereas knockdown of CADM1 significantly inhibited growth in MP38 cells (10%–40% inhibition) and to a lesser extent in MM28 cells (7%–21% inhibition; Fig. 4D; Supplementary Fig. S4B). These results suggest that in BAP1 mutant cells, proliferation is facilitated by high levels of CADM1.
Figure 4.
Knockdown of CDH1 and CADM1 decreases cell cycle– and cell growth–associated protein expression. A, Western blot analysis was used to validate E-cadherin and CADM1 expression levels with either control, CDH1, or CADM1 siRNAs in MP38 cells. B, A heatmap of median-centered, log2-transformed RPPA data for the top proteins (P < 0.05, log2fc > 0.32193) when comparing siCADM1 cells with untreated and siCTL. Each lysate was collected in triplicate (n 3). C, Representative Western blot analysis showing the effect of E-cadherin and CADM1 knockdown on cell-cycle and cell growth proteins. Identical lysates from A were used for the first four blots (phosphoCDK1, cyclin B1, phosphoERK1/2, and ERK1/2), and so the top b-actin loading control was duplicated from A. D, Effect of siCDH1 and siCADM1 on MP38 cell growth was analyzed using the IncuCyte Live Cell Analysis Imaging System. Scale bars, 300 mmol/L. Fold change was calculated as percent confluency compared with day 0, % inhibition was calculated as fold change difference as compared with control, P < 0.05, and P < 0.01 as determined by t test, and error bars are ±SEM.
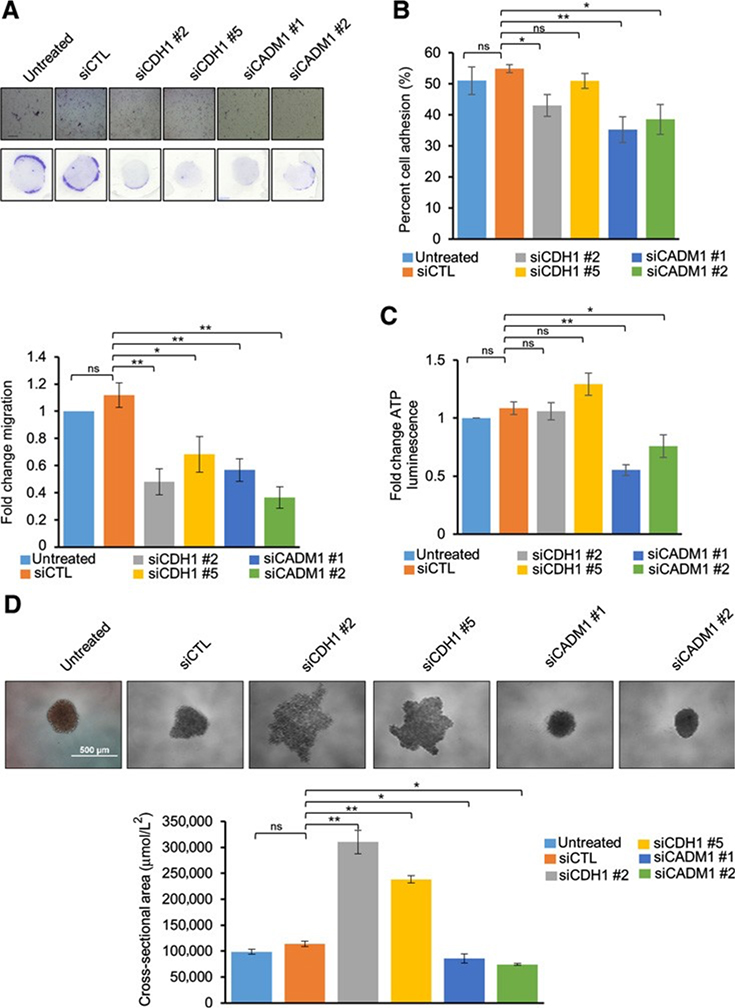
Knockdown of E-cadherin and CADM1 results in phenotypic changes in migration, adhesion, cluster formation, and cell viability
With either E-cadherin or CADM1 knockdown, we observed a significant decrease in MP38 cell migration, as measured by the Boyden chamber assay (Fig. 5A). In testing functionality, knockdown of either E-cadherin or CADM1 reduced cell–cell adhesion, as measured by the percentage of cells adhering to a monolayer of unaltered MP38 cells (Fig. 5B). To further understand the function of each CAM, we used a 3D spheroid model that recapitulates cell–cell interactions and maintains tumor-like growth characteristics (55). Knockdown of E-cadherin in both BAP1 mutant cell lines, MP38 and MM28, did not alter cell viability, as measured by ATP luminescence, but did disrupt the ability of cells to cluster and form tight spheroids (Fig. 5C and 5D; Supplementary Fig. S4C and S4D). Conversely, CADM1 knockdown decreased cell viability and resulted in the formation of smaller spheroids (Fig. 5C and 5D; Supplementary Fig. S4C and S4D). The inhibition of cell clustering to form spheroids following E-cadherin knockdown was observed over time after being plated on a low attachment plate (Supplementary Fig. S5). These results indicate that E-cadherin and CADM1 play differential roles in 3D; E-cadherin maintained cluster formation while CADM1 regulated cell growth.
Figure 5.
Functional effects of CDH1 and CADM1 knockdown on MP38 cells: Migration of MP38 cells after treatment with CDH1 and CADM1 siRNAs for 72 hours (A). Cells were trypsinized and subjected to Boyden chamber-based, serum-directed migration assays. Quantification of migrated cells and representative images are shown. Scale bars, 250 mmol/L. Data are graphed as fold change in cell migration compared with untreated cells from at least five independent experiments, P < 0.05, and P < 0.01 as determined by t test, and error bars are SEM. B, Effect of siCDH1 and siCADM1 knockdown on cell adhesion. Percent cell adhesion is calculated as (adherent cell fluorescence)/(total cell fluorescence) from four independent experiments, P < 0.05, and P < 0.01 as determined by t test, and error bars are SEM. C, Cell viability as measured by ATP luminescence (Cell Titer Glo) was studied after siCDH1 and siCADM1. Cells were treated with CDH1 and CADM1 siRNAs for 72 hours, trypsinized, and cultured for 72 hours in low attachment conditions. Fold change compared with control was calculated for each replicate, P < 0.05, and P < 0.01 as determined by t test, and error bars are SEM. Data were collected from three independent experiments. D, Effect of siCDH1 and siCADM1 on spheroid size and cluster formation after being cultured on low attachment conditions for 72 hours. Representative microscopy images of MP38 spheroids from six independent biological replicates were taken after 72 hours. Quantitation of spheroid size was determined by Image J, P < 0.05, and P < 0.01 as determined by t test, and error bars are SEM. Scale bars, 500mmol/L.
Discussion
BAP1 mutations co-occurring with monosomy 3 are associated with worse prognosis in uveal melanoma and are found in 90% of metastatic cases (13, 14). Although BAP1 is a known tumor suppressor, there is a gap in understanding how BAP1 loss promotes metastasis. By integrating transcriptomic data from independent patient sets and transcriptomic and proteomic data from BAP1 mutant versus wild-type cell lines, we identified BAP1-dependent molecular alterations. We found BAP1-mutant uveal melanoma exhibited upregulated expression of CAMs compared with BAP1 wild-type uveal melanoma. Consistent with patient evidence, individual datasets in uveal melanoma cell lines showed increased expression of CAMs such as CDH1, CADM1, and SDC2 resulting from BAP1 loss. Although CAMs expression is elevated generally in BAP1-mutant uveal melanoma patient tumors and cell lines, there is intertumoral and intratumoral heterogeneity in the expression of these genes at the single-cell level and between cell lines. For example, there are two tumors in the scRNA-seq data that have cells more frequently expressing CADM1 than the other tumors (Fig. 2F). This suggests that BAP1 regulation is context dependent, and not all BAP1 mutant cells within the tumor express elevated levels of E-cadherin or CADM1. Recent articles published within our lab have also determined heterogeneous BAP1 regulation of metabolic pathways (7, 56, 57). These differences could carry over to the expression of cell–cell adhesion molecules.
Functionally, knockdown of CADM1 decreased cell growth, and knockdown of either E-cadherin or CADM1 decreased migration and cell–cell adhesion in a two-dimensional (2D) model. Effects on growth were not as dramatic in the BAP1 mutant MM28 cell line, possibly due to decreased basal expression of CADM1 (Fig. 2E). When using a 3D model, differential effects were observed. Knockdown of E-cadherin led to poor spheroid formation, likely due to reduced cell–cell adhesion and cell clustering. In contrast, knockdown of CADM1 caused the formation of smaller spheroids consistent with decreased viability. The reduction in spheroid size differed from effects in 2D, which showed a decrease in cell–cell adhesion after the knockdown of CADM1. These findings can be reconciled with an understanding that CADM1 largely plays a role in cell growth while E-cadherin is responsible for cluster formation. Future experiments will include generating BAP1 reexpressing cells that express high levels of CADM1 and investigating tumor growth in vitro and in vivo.
By combining publicly available datasets, we found that BAP1 mutations are associated with an increase in CAMs, specifically CDH1, CADM1, and SDC2. These findings are similar to clear-cell renal cell carcinoma and prostate cancer in which loss of BAP1 resulted in a mesenchymal–epithelial transition and increased expression of E-cadherin (21, 58). The promoter regions of negative transcriptional regulators of E-cadherin, including SNAI1, were associated with histone H2A ubiquitylation (indicating gene suppression) following BAP1 loss and EMT (21). Results from genome-wide binding analysis of BAP1 in pancreatic cancer showed significant peaks in the promoter region of SNAI1 in both normal and cancerous pancreatic cell line samples, suggesting BAP1 might be involved in transcriptional regulation of SNAI1 (59). In contrast, loss of BAP1 in liver organoids resulted in the reduction of epithelial identity and decreased E-cadherin (60). These findings highlight the context-dependent role that BAP1 plays in different cancer and tissue types. Our data suggest that BAP1 may regulate gene expression of E-cadherin either directly or via additional transcriptional regulators. For example, increased H2A ubiquitylation due to loss of BAP1 could repress negative transcriptional regulators of E-cadherin or, in some cases, increase the expression of select genes (6). Alternatively, BAP1 could act in conjunction with YY1 or other transcriptional regulators at the promoter region of genes encoding for CAMs and repress transcriptional activity directly (61). A ubiquitylation screen could be conducted to identify novel targets of BAP1 in uveal melanoma. Future work is necessary to determine the mechanism by which BAP1 regulates CAMs in uveal melanoma and whether this could be dependent on BAP1’s deubiquitylating activity or intracellular localization.
Even though melanomas are not derived from epithelial tissue, phenotypic switching to an EMT-like state is necessary for metastasis. Decreased expression of E-cadherin and CADM1 is associated an EMT-like state in cutaneous melanoma and gain of invasive properties (19, 62). In our data, knockdown of E-cadherin and CADM1 led to decreased growth and/or migration in BAP1-mutant uveal melanoma cells, suggesting that these adhesion proteins play a role in uveal melanoma progression. Increased expression of E-cadherin and CADM1 with BAP1 loss is consistent with previous reports that link the increased expression of CADM1 to monosomy 3 tumors (63) and increased expression of CDH1 to high metastatic risk, class 2 uveal melanoma (64). There is emerging evidence for the requirement of these CAMs during the metastatic cascade (65). During initial extravasation and subsequent migration through the bloodstream, E-cadherin mediates adaptation to detached surface growth through anoikis resistance (66) and collective cell migration in breast and colorectal cancer (67, 68). Our studies show that E-cadherin and CADM1 loss resulted in decreased cell–cell adhesion and growth. One possibility is that increased CAM expression allows disseminated cells to adapt and survive as they reach distant sites. In addition, E-cadherin is necessary for metastatic seeding in distant organs (69). A limitation of our work is the lack of metastasis models. Future studies necessitate the use of optimized in vivo uveal melanoma metastasis models with inducible knockdown of E-cadherin and CADM1 to understand the metastatic process and the relevance of adhesion proteins. One such experiment is to inject fluorescently tagged BAP1-mutant uveal melanoma cells in the chick chorioallantoic membrane or zebrafish model and detect hematogenous migration to distant sites (70, 71). By targeting CAMs in BAP1-mutant uveal melanoma, there is potential to inhibit cluster formation and cell survival which are associated with worse patient survival in other cancer types (68).
In conclusion, we show BAP1-dependent alterations of gene expression in uveal melanoma. The use of multiple independent datasets provides rigorous evidence from patient samples and cell line–based studies. We provide novel insight into the role BAP1 plays in inversely regulating E-cadherin and CADM1. Although canonically, E-cadherin and CADM1 are thought to be tumor suppressors, our results suggest they are upregulated with BAP1 loss, which is associated with aggressive tumor formation and metastasis. They also may play alternative roles in increasing collective migration and growth in uveal melanoma.
Supplementary Material
Implications:
BAP1 mutations and increased metastasis may be due to upregulation of CAMs.
Acknowledgments
We thank Dr. Claudia Capparelli, Dr. Nicole Wilski, and Signe Caksa for valuable feedback during the preparation of this article. This work was supported by NIH/NCI grants R01s, CA253977 and CA257505, and P01 CA114046 project 4 to A.E. Aplin. In addition, the work was supported by NIH/NCI R00 CA207855 and the W.W. Smith Charitable Trusts grants to E.J. Hartsough and by the Melanoma Research Foundation Medical Student Award 2020 to U. Baqai. The Sidney Kimmel Cancer Center Flow Cytometry and Translational Pathology Core Facilities are supported by NIH/NCI (P30 CA056036). The RPPA studies were performed at the Functional Proteomics Core Facility at The University of Texas, MD Anderson Cancer Center, which is supported by an NCI Cancer Center Support Grant (P30 CA16672).
Authors’ Disclosures
U. Baqai reports grants from Melanoma Research Foundation Medical Student Award during the conduct of the study. E.J. Hartsough reports grants from NIH/NCI, DoD/CDMRP, W.W. Smith Charitable Trusts, American Cancer Society-IRG, and Sidney Kimmel Cancer Center during the conduct of the study. J.W. Harbour reports grants from Melanoma Research Alliance during the conduct of the study; grants from NCI, other support from Castle Biosciences and Immunoncore outside the submitted work. A.E. Aplin reports grants from NIH outside the submitted work; and A.E. Aplin reports receiving a commercial research grant from Pfizer, Inc. (2013–2017) and has ownership interest in patent number 9880150. No disclosures were reported by the other authors.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Research Online (http://mcr.aacrjournals.org/).
References
- 1.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, et al. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998;16: 1097–112. [DOI] [PubMed] [Google Scholar]
- 2.Ventii KH, Devi NS, Friedrich KL, Chernova TA, Tighiouart M, Van Meir EG, et al. BRCA1-associated protein-1 is a tumor suppressor that requires deubiquitinating activity and nuclear localization. Cancer Res 2008;68:6953–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campagne A, Lee M, Zielinski D, Michaud A, Le Corre S, Dingli F, et al. BAP1 complex promotes transcription by opposing PRC1-mediated H2A ubiquitylation. Nat Commun 2019;10:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machida YJ, Machida Y, Vashisht AA, Wohlschlegel JA, Dutta A. The deubiquitinating enzyme BAP1 regulates cell growth via interaction with HCF-1. J Biol Chem 2009;284:34179–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bononi A, Giorgi C, Patergnani S, Larson D, Verbruggen K, Tanji M, et al. BAP1 regulates IP3R3-mediated Ca2þ flux to mitochondria suppressing cell transformation. Nature 2017;546:549–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y, Shi J, Liu X, Feng L, Gong Z, Koppula P, et al. BAP1 links metabolic regulation of ferroptosis to tumour suppression. Nat Cell Biol 2018;20:1181–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Han A, Purwin TJ, Bechtel N, Liao C, Chua V, Seifert E, et al. BAP1 mutant uveal melanoma is stratified by metabolic phenotypes with distinct vulnerability to metabolic inhibitors. Oncogene 2021;40:618–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbone M, Yang H, Pass HI, Krausz T, Testa JR, Gaudino G. BAP1 and cancer. Nat Rev Cancer 2013;13:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carbone M, Harbour JW, Brugarolas J, Bononi A, Pagano I, Dey A, et al. Biological mechanisms and clinical significance of BAP1 mutations in human cancer. Cancer Discov 2020;10:1103–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matatall KA, Agapova OA, Onken MD, Worley LA, Bowcock AM, Harbour JW. BAP1 deficiency causes loss of melanocytic cell identity in uveal melanoma. BMC Cancer 2013;13:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang AE, Karnell LH, Menck HR. The national cancer data base report on cutaneous and noncutaneous melanoma: a summary of 84,836 cases from the past decade. The American College of Surgeons Commission on Cancer and the American Cancer Society. Cancer 1998;83:1664–78. [DOI] [PubMed] [Google Scholar]
- 12.Bakalian S, Marshall J-C, Logan P, Faingold D, Maloney S, Di Cesare S, et al. Molecular pathways mediating liver metastasis in patients with uveal melanoma. Clin Cancer Res 2008;14:951–6. [DOI] [PubMed] [Google Scholar]
- 13.Harbour JW, Onken MD, Roberson EDO, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 2010; 330:1410–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karlsson J, Nilsson LM, Mitra S, Als’en S, Shelke GV, Sah VR, et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat Commun 2020;11:1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chua V, Mattei J, Han A, Johnston L, LiPira K, Selig SM, et al. The latest on uveal melanoma research and clinical trials: updates from the cure ocular melanoma (CURE OM) science meeting (2019). Clin Cancer Res 2021;27:28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Q, Zhang H, Jiang X, Qian C, Liu Z, Luo D. Factors involved in cancer metastasis: a better understanding to “seed and soil” hypothesis. Mol Cancer 2017;16:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brabletz T, Kalluri R, Nieto MA, Weinberg RA. EMT in cancer. Nat Rev Cancer 2018;18:128–34. [DOI] [PubMed] [Google Scholar]
- 18.Li FZ, Dhillon AS, Anderson RL, McArthur G, Ferrao PT. Phenotype switching in melanoma: implications for progression and therapy. Front Oncol 2015;5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartsough EJ, Weiss MB, Heilman SA, Purwin TJ, Kugel CH, Rosenbaum SR, et al. CADM1 is a TWIST1-regulated suppressor of invasion and survival. Cell Death Dis 2019;10:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nieto MA, Huang RYJ, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166: 21–45. [DOI] [PubMed] [Google Scholar]
- 21.Chen P, Wang H, Zhang W, Chen Y, Lv Y, Wu D, et al. Loss of BAP1 results in growth inhibition and enhances mesenchymal–epithelial transition in kidney tumor cells. Mol Cell Proteomics 2019;18:1320–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 2019;47:D766–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34:267–73. [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liberzon A, Birger C, Thorvaldsd’ottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, et al. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 2018;173:400–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017;32:204–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paradis JS, Acosta M, Saddawi-Konefka R, Kishore A, Gomes F, Arang N, et al. Synthetic lethal screens reveal cotargeting FAK and MEK as a multimodal precision therapy for GNAQ-driven uveal melanoma. Clin Cancer Res 2021;27: 3190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teh JLF, Purwin TJ, Han A, Chua V, Patel P, Baqai U, et al. Metabolic adaptations to MEK and CDK4/6 cotargeting in uveal melanoma. Mol Cancer Ther 2020;19: 1719–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lapadula D, Farias E, Randolph CE, Purwin TJ, McGrath D, Charpentier TH, et al. Effects of oncogenic Gaq and Ga11 inhibition by FR900359 in uveal melanoma. Mol Cancer Res 2019;17:963–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinforma 2013; 43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol 2016;17:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Durante MA, Rodriguez DA, Kurtenbach S, Kuznetsov JN, Sanchez MI, Decatur CL, et al. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat Commun 2020;11:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 2005;33:e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gautier L, Cope L, Bolstad BM, Irizarry RA. Affyanalysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004;20:307–15. [DOI] [PubMed] [Google Scholar]
- 44.Sean D, Meltzer PS. GEOquery: a bridge between the gene expression omnibus (GEO) and BioConductor. Bioinformatics 2007;23:1846–7. [DOI] [PubMed] [Google Scholar]
- 45.Laurent C, Gentien D, Piperno-Neumann S, N’emati F, Nicolas A, Tesson B, et al. Patient-derived xenografts recapitulate molecular features of human uveal melanomas. Mol Oncol 2013;7:625–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res 2011;39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alonso SR, Tracey L, Ortiz P, P’erez-G’omez B, Palacios J, Poll’an M, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res 2007;67:3450–60. [DOI] [PubMed] [Google Scholar]
- 48.Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell 2019;49:361–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shain AH, Bagger MM, Yu R, Chang D, Liu S, Vemula S, et al. The genetic evolution of metastatic uveal melanoma. Nat Genet 2019;51:1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amirouchene-Angelozzi N, Nemati F, Gentien D, Nicolas A, Dumont A, Carita G, et al. Establishment of novel cell lines recapitulating the genetic landscape of uveal melanoma and preclinical validation of mTOR as a therapeutic target. Mol Oncol 2014;8:1508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vicente CM, Ricci R, Nader HB, Toma L. Syndecan-2 is upregulated in colorectal cancer cells through interactions with extracellular matrix produced by stromal fibroblasts. BMC Cell Biol 2013;14:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corti F, Wang Y, Rhodes JM, Atri D, Archer-Hartmann S, Zhang J, et al. N-terminal syndecan-2 domain selectively enhances 6-O heparan sulfate chains sulfation and promotes VEGFA165-dependent neovascularization. Nat Commun 2019;10:1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moore AR, Ran L, Guan Y, Sher JJ, Hitchman TD, Zhang JQ, et al. GNA11 Q209L mouse model reveals RasGRP3 as an essential signaling node in uveal melanoma. Cell Rep 2018;22:2455–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuznetsoff JN, Owens DA, Lopez A, Rodriguez DA, Chee NT, Kurtenbach S, et al. Dual screen for efficacy and toxicity identifies HDAC inhibitor with distinctive activity spectrum for BAP1-mutant uveal melanoma. Mol Cancer Res 2021;19:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Aughton K, Shahidipour H, Djirackor L, Coupland SE, Kalirai H. Characterization of uveal melanoma cell lines and primary tumor samples in 3D culture. Transl Vis Sci Technol 2020;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chua V, Han A, Bechtel N, Purwin TJ, Hunter E, Liao C, et al. The AMP-dependent kinase pathway is upregulated in BAP1 mutant uveal melanoma. Pigment Cell Melanoma Res 2022;35:78–87. [DOI] [PubMed] [Google Scholar]
- 57.Chattopadhyay C, Oba J, Roszik J, Marszalek JR, Chen K, Qi Y, et al. Elevated endogenous SDHA drives pathological metabolism in highly metastatic uveal melanoma. Invest Ophthalmol Vis Sci 2019;60:4187–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Park CM, Lee JE, Kim JH. BAP1 functions as a tumor promoter in prostate cancer cells through EMT regulation. Genet Mol Biol 2020;43:e20190328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Perkail S, Andricovich J, Kai Y, Tzatsos A. BAP1 is a haploinsufficient tumor suppressor linking chronic pancreatitis to pancreatic cancer in mice. Nat Commun 2020;11:3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Artegiani B, van Voorthuijsen L, Lindeboom RGH, Seinstra D, Heo I, Tapia P, et al. Probing the tumor suppressor function of BAP1 in CRISPR-engineered human liver organoids. Cell Stem Cell 2019;24:927–43. [DOI] [PubMed] [Google Scholar]
- 61.Ishii Y, Kolluri KK, Pennycuick A, Zhang X, Nigro E, Alrifai D, et al. BAP1 and YY1 regulate expression of death receptors in malignant pleural mesothelioma. J Biol Chem 2021;297:101223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smart JA, Oleksak JE, Hartsough EJ. Cell adhesion molecules in plasticity and metastasis. Mol Cancer Res 2021;19:25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCannel TA, Burgess BL, Rao NP, Nelson SF, Straatsma BR. Identification of candidate tumor oncogenes by integrative molecular analysis of choroidal melanoma fine-needle aspiration biopsy specimens. Arch Ophthalmol 2010; 128:1170–7. [DOI] [PubMed] [Google Scholar]
- 64.Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional gene expression analysis uncovers phenotypic switch in aggressive uveal melanomas. Cancer Res 2006;66:4602–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Janiszewska M, Primi MC, Izard T. Cell adhesion in cancer: beyond the migration of single cells. J Biol Chem 2020;295:2495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rayavarapu RR, Heiden B, Pagani N, Shaw MM, Shuff S, Zhang S, et al. The role of multicellular aggregation in the survival of ErbB2-positive breast cancer cells during extracellular matrix detachment. J Biol Chem 2015;290: 8722–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aiello NM, Maddipati R, Norgard RJ, Balli D, Li J, Yuan S, et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell 2018;45:681–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, et al. Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 2014;158:1110–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Padmanaban V, Krol I, Suhail Y, Szczerba BM, Aceto N, Bader JS, et al. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019;573:439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pandiani C, Strub T, Nottet N, Cheli Y, Gambi G, Bille K, et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ 2021;28: 1990–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Perez DE, Henle AM, Amsterdam A, Hagen HR, Lees JA. Uveal melanoma driver mutations in GNAQ/11 yield numerous changes in melanocyte biology. Pigment Cell Melanoma Res 2018;31:604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are publicly available in GEO at GSE149920 and GSE181194, within the article, and its Supplementary Data files. The publicly available data analyzed in this study were obtained from GEO at GSE139829, GSE22138, and GSE78033, and the SRA at SRP102735 and SRP193943.