Abstract
Background
Cell invasion is a crucial step of tumor metastasis, finding new regulators of which offers potential drug targets for cancer therapy. Aberrant GLYAT expression is associated with human cancers, yet its role in cancer remains unknown. This study aims to understand the function and mechanism of Drosophila GLYAT in cell invasion.
Results
We found that dGLYAT regulates Gadd45-mediated JNK pathway activation and cell invasion. Firstly, loss of dGLYAT suppressed scrib depletion- or Egr overexpression-induced JNK pathway activation and invasive cell migration. Secondary, mRNA-seq analysis identified Gadd45 as a potential transcriptional target of dGLYAT, as depletion of dGLYAT decreased Gadd45 mRNA level. Finally, Gadd45 knockdown suppressed scrib depletion-induced JNK pathway activation and cell invasion.
Conclusions
These evidences reveal the role of dGLYAT and Gadd45 in JNK-dependent cell invasion, and provide insight for the roles of their human homologs in cancers.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13008-022-00080-5.
Keywords: Drosophila, Cell invasion, JNK, dGLYAT, Gadd45
Background
Tumor metastasis, rather than primary tumor formation, is the main cause of fatality in cancer patients [1]. Therefore, identifying additional factors involved in tumor cell invasion and metastasis is of great significance to develop novel strategies for cancer prevention and therapies [2]. Various approaches and model systems have been employed to comprehend the mechanisms underlying cancer metastasis, among which the fruit fly, Drosophila melanogaster, has emerged as an excellent model organism to dissect different cancer processes [3]. For example, depletion of cell polarity genes or C-terminal SRC kinase (Csk) in Drosophila larval wing disc epithelia induces epithelia-mesenchymal transition (EMT)-like cell migration [4].
The Jun N-terminal Kinase (JNK) signaling is evolutionarily conserved from Drosophila to human, and plays critical roles in cancer initiation and progression [5]. In Drosophila, the JNK pathway consists of a kinase cascade including the JNK kinase kinase such as dTAK1 [6], the JNK kinase hemipterous (Hep) [7], and the sole fly JNK Basket (Bsk) [8]. Upon activation of the kinase cascade by sequential phosphorylation, Bsk phosphorylates and activates downstream transcription factors including Jun and Fos, which form the AP-1 heterodimers [9]. puckered (puc), a transcriptional target of JNK signaling, encodes a JNK phosphatase that inhibits Bsk activity in a negative feed-back manner [10]. Previous studies in Drosophila have found that JNK signaling plays pivotal roles in cell proliferation, migration and apoptosis in normal development, and promotes tumorigenic cell death and invasion in a context dependent manner [11].
Human glycine N-acyltransferases (GLYAT, GLYATL1, GLYATL2, and GLYATL3) promote conjugation of carboxylic acids to glycine and glutamine, which play crucial roles in the detoxification of endogenous and exogenous acyl-CoA. Previous studies suggest potential roles of GLYAT family members in various cancers, for instance, aberrant GLYAT expression has been associated with hepatocellular carcinomas and breast cancer [12], and reduced GLYATL1 expression is correlated with short overall survival in hepatocellular carcinoma patients [13]. On the other hand, data from the Oncomine Platform (https://www.oncomine.org) show that GLYATL1 mRNA expression is up-regulated in colorectal and prostate cancers, but is down-regulated in kidney and liver cancers. Despite these association, the exact functions of GLYAT family proteins in cancers remain elusive. Our previous work found a Drosophila homolog of GLYAT (dGLYAT) is required for JNK-mediated cell death [14], yet two critical questions remain unanswered. Whether dGLYAT is required for other in vivo functions of JNK pathway, and by which mechanism does dGLYAT regulate JNK signaling?
To address the above questions, we took advantage of the well-established Drosophila cell invasion model. In this model, knockdown of a cell polarity gene, e.g. scrib, lgl or dlg, along the anterior/posterior (A/P) compartment boundary in 3rd instar larval wing imaginal discs by ptc-Gal4, induces JNK-dependent invasive cell migration [15]. To assess whether dGLYAT contributes to JNK–mediated cell invasion, we depleted dGLYAT by mutation or RNAi-mediated knockdown, and found dGLYAT is required for JNK-dependent cell invasion. In particular, loss of dGLYAT suppresses scrib depletion- or Egr overexpression-induced JNK-dependent cell invasion, and impedes scrib knockdown-triggered JNK pathway activation. To investigate the mechanism by which dGLYAT regulates JNK signaling, we performed mRNA-seq analysis, and found a significant reduction of Gadd45 (Growth Arrest and DNA Damage-inducible 45) mRNA level upon dGLYAT depletion. Finally, knockdown of Gadd45 suppresses loss-of-scrib-induced JNK activation and cell invasion. Thus, these data provide the first in vivo evidence that dGLYAT modulates JNK-dependent cell invasion through Gadd45.
Results
dGLYATis required for cell polarity disruption-induced cell invasion
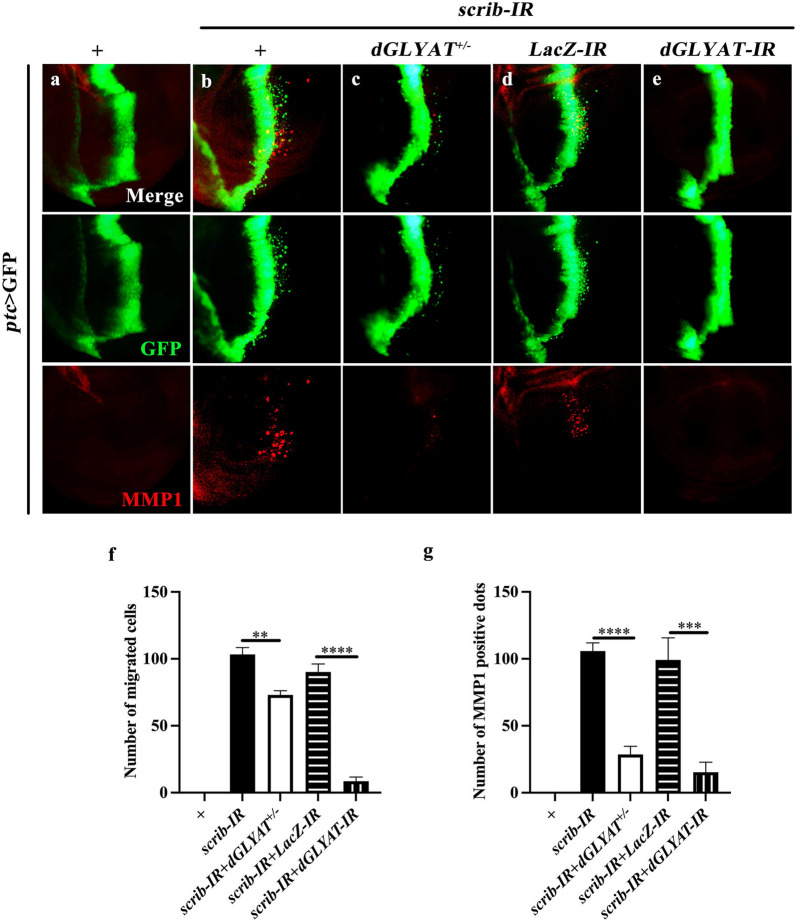
It is well known that disrupting cell polarity by depleting scrib along the anterior/posterior (A/P) compartment boundary of the wing discs leads to JNK-dependent invasive cell migration [16], in which cells migrate away from the A/P boundary with up-regulated expression of matrix metalloprotease 1 (MMP1) that is able to degrade the basement membrane [17]. Using this well-established in vivo cell invasion model, we examined the role of dGLYAT in cell polarity disruption-induced cell invasion. Compared with ptc > GFP control (Fig. 1a), knockdown of scrib induced extensive cell migration and MMP1 up-regulation (Fig. 1b, quantified in Fig. 1f, g). Both phenotypes were significantly suppressed in heterozygous dGLYATc02982 mutants (Fig. 1c), which has a piggyBac insertion into the second exon that deletes the critical Gcn5-related N-acetyltransferases (GNAT) domain [14]. To corroborate this result, we depleted dGLYAT by RNAi-mediated knockdown (Additional file 2: Figure S1), and confirmed that expression of a dGLYAT-IR dramatically inhibited scrib depletion-induced cell migration and MMP1 elevation, compared with a LacZ RNAi as a negative control (Fig. 1d-g).
Fig. 1.
dGLYAT is required for cell polarity disruption-induced cell invasion. Fluorescent micrographs of Drosophila third instar larval wing discs are shown (a-e). Compared with the ptc > GFP control (a), ptc > scrib-IR induced cell migration from A/P boundary toward posterior and elevated MMP1 expression (b). Both phenotypes were significantly suppressed by heterozygous mutation (c) or RNAi expression of dGLYAT (e). LacZ RNAi served as a negative control (d). f Statistic of number of migrated cells is shown (left to right: n = 10, n = 8, n = 5. n = 8, n = 10). g Statistic of number of MMP1 positive dots is shown (left to right: n = 10, n = 12, n = 5. n = 5, n = 10). t-test was used to compute P-values. **P < 0.01, ***P < 0.001, ****P < 0.0001. Detailed genotypes: a ptc-Gal4 UAS-GFP/+, b ptc-Gal4 UAS-GFP UAS-scrib-IR/+, c ptc-Gal4 UAS-GFP UAS-scrib-IR/dGLYATc02982, d ptc-Gal4 UAS-GFP UAS-scrib-IR/+; UAS-LacZ-IR/+, e ptc-Gal4 UAS-GFP UAS-scrib-IR/+; UAS-dGLYAT-IR/+
To further confirm the effect of dGLYAT on cell invasion, we checked other markers of epithelial-mesenchymal transition (EMT), a critical process in malignant transformation [18]. During the process of EMT, expressions of cell-cell junction proteins, such as E-cadherin, are switched off in epithelial cells [19]. Consistently, loss-of-scrib resulted in reduced expression of E-cadherin, as the fluorescent signal of E-cadherin antibody is significantly weakened along the A/P compartment boundary in the wing pouch area (Fig. 2a, b). This phenotype was suppressed by expression of dGLYAT-IR, but not LacZ-IR (Fig. 2c, d). Taken together, these data suggest that dGLYAT is required for cell polarity disruption-induced EMT-like cell migration.
Fig. 2.
dGLYAT is necessary for cell polarity disruption-induced EMT-like phenotype. Fluorescent micrographs of third instar larval wing discs are shown (a-d). Compared with the ptc > GFP control (a), ptc > scrib-IR induced cell migration and reduced E-cadherin expression in the A/P boundary (b, arrow), which were suppressed by expressing dGLYAT RNAi (d), but not LacZ RNAi (c). Detailed genotypes: a ptc-Gal4 UAS-GFP/+, b ptc-Gal4 UAS-GFP UAS-scrib-IR/+, c ptc-Gal4 UAS-GFP UAS-scrib-IR/+; UAS-LacZ-IR/+, d ptc-Gal4 UAS-GFP UAS-scrib-IR/+; UAS-dGLYAT-IR/+
dGLYATis necessary for Egr-JNK pathway-triggered invasive cell migration
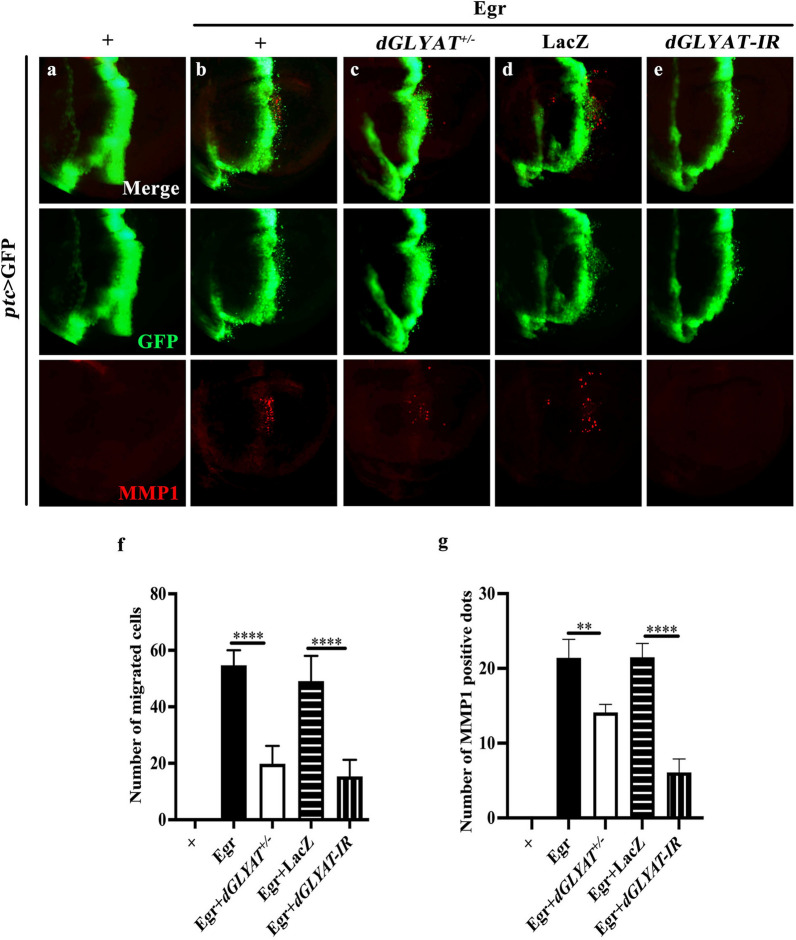
Scrib depletion triggers Eiger-JNK pathway-mediated invasive cell migration [15]. To investigate whether dGLYAT is involved in Egr-JNK pathway-activated cell migration, we used ptc-Gal4 to drive ectopic Egr expression. Compared with the control (Fig. 3a), ectopic expression of Egr driven by ptc-Gal4 resulted in invasive cell migration, accompanied by up-regulated MMP1 expression (Fig. 3b). Both phenotypes were considerably suppressed in heterozygous dGLYAT mutants, or by expressing dGLYAT-IR, but not LacZ (Fig. 3c-e, quantified in Fig. 3f, g). As MMP1 also serves as a reporter of JNK signaling, these results suggest that dGLYAT is necessary for Egr-JNK pathway-triggered invasive cell migration.
Fig. 3.
dGLYAT is necessary for Eiger-JNK pathway-induced invasive cell migration. Fluorescent micrographs of third instar larval wing discs are shown (a-e). Compared with the ptc > GFP control (a), ptc > Egr-induced cell migration and up-regulated MMP1 expression (b) were suppressed by dGLYAT mutation (c) or RNAi (e), but not by LacZ expression (d). f Statistic of number of migrated cells is shown (left to right: n = 10, n = 12, n = 10. n = 12, n = 16). g Statistic of number of MMP1 positive dots is shown (left to right: n = 10, n = 22, n = 26. n = 27, n = 25). t-test was used to compute P-values. **P < 0.01, ****P < 0.0001. Detailed genotypes: a ptc-Gal4 UAS-GFP/+, b ptc-Gal4 UAS-GFP UAS-Egr Regg1/+, c ptc-Gal4 UAS-GFP UAS-EgrRegg1/dGLYATc02982, d ptc-Gal4 UAS-GFP UAS-EgrRegg1/+; UAS-LacZ/+, e ptc-Gal4 UAS-GFP UAS-EgrRegg1/+; UAS-dGLYAT-IR/+
dGLYAT is required for cell polarity disruption-induced JNK pathway activation
As loss-of-cell polarity-induced cell invasion depends on JNK signaling, and depletion of dGLYAT impedes ptc > scrib-IR-induced cell invasion and MMP1 expression, a transcriptional target of JNK signaling, we proposed that dGLYAT might be required for cell polarity disruption-triggered JNK pathway activation. To test this possibility, we examined the expression of two well-recognized JNK pathway reporters, puc-LacZ and TRE-RFP [20]. In agreement with previous reports, knockdown of scrib along the A/P compartment boundary triggered JNK pathway activation, as indicated by elevated expression of puc-LacZ (Fig. 4a, b) and TRE-RFP (Fig. 4d, e), both of which were significantly suppressed by knockdown of dGLYAT (Fig. 4c, f). Collectively, these results suggest that dGLYAT is required for loss-of-cell polarity-induced JNK pathway activation. dGLYAT regulates Gadd45 transcription.
Fig. 4.
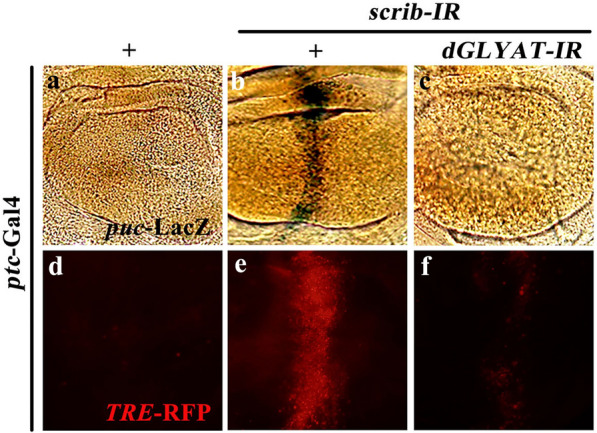
dGLYAT is required for cell polarity disruption-induced JNK pathway activation. Light micrographs of third instar wing discs with X-gal staining (a-c) and fluorescent micrographs of third instar wing discs (d-f) are shown. Compared with the ptc-Gal4 control (a, d), ptc > scrib-IR activated puc-LacZ (b) and TRE-RFP (e) expression, which were suppressed by dGLYAT RNAi (c, f). Detailed genotypes: a ptc-Gal4 UAS-GFP/+; puc-LacZ/+, b ptc-Gal4 UAS-GFP UAS-scrib-IR/+; puc-LacZ/+, c ptc-Gal4 UAS-GFP UAS-scrib-IR/+; puc-LacZ/UAS-dGLYAT-IR, d ptc-Gal4 UAS-GFP/TRE-RFP, e ptc-Gal4 UAS-GFP UAS-scrib-IR/TRE-RFP, f ptc-Gal4 UAS-GFP UAS-scrib-IR/TRE-RFP; UAS-dGLYAT-IR/+
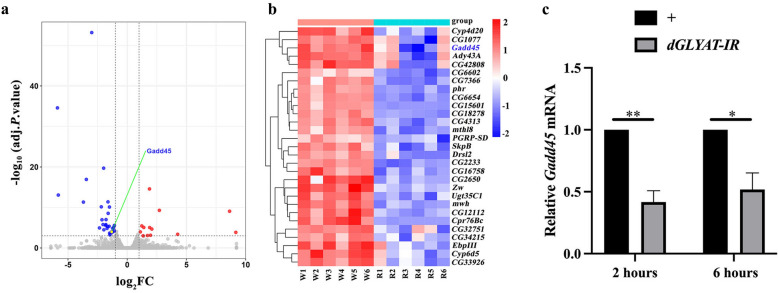
According to the Flybase (http://flybase.org/reports/FBgn0054010), dGLYAT encodes a Gcn5-related N-acetyltransferases (GNAT) family member that catalyzes the transfer of an acetyl moiety from Coenzyme A (Ac-CoA) to diverse substrates [21]. GNATs are evolutionarily conserved regulators that acetylate key amine of small molecules and proteins involved in numerous cellular processes. Some members of GNAT superfamily are involved in histone acetylation [22], which affects gene transcription [23]. Based on these information, we proposed that dGLYAT might be required for the transcriptional activation of JNK pathway positive regulator(s). In this scenario, loss of dGLYAT would compromise JNK pathway activation via reduced expression of the positive regulator(s). To find the potential regulator(s), we conducted the whole genome mRNA-seq assays, and analyzed differentially expressed genes (DEGs) between control and dGLYAT-depleted groups. We crossed hs-Gal4 with w1118 or dGLYAT RNAi lines, which serves as the control group or the experimental group, respectively. Third instar larvae were subjected to heat shock at 37 °C for 30 min, recovered at 29 °C for 6 h, and dissected for transcriptome sequencing. We found that 13 genes were up-regulated and 29 genes down-regulated upon dGLYAT knockdown (Fig. 5a, log2FC ≥ 1.0 or log2FC ≤ − 1.0, adjusted P value < 0.001 and Additional file 1: Table S1). We paid specific attention to the 29 down-regulated genes, whose expression profiles are presented (Fig. 5b). Intriguingly, among the 29 down-regulated genes, Gadd45 has been previously reported as a positive regulator of JNK pathway in apoptosis and egg asymmetric development [24, 25]. The mammalian GADD45 family consists of three members, GADD45a, GADD45b, and GADD45G. which interacts with cellular proteins in response to physiological or environmental stressors, and participates in cell cycle arrest, DNA repair, apoptosis, cell survival and senescence [26]. We performed reverse transcription-quantitative polymerase chain reaction (RT-qPCR) assay to validate the mRNA-seq results, and confirmed that Gadd45 mRNA level was significantly reduced 2 and 6 h after heat shock-induced dGLYAT knockdown (Fig. 5c). The relative short time interval (2 h) between dGLYAT knockdown and Gadd45 mRNA reduction implies dGLYAT may directly affect Gadd45 transcription.
Fig. 5.
Gadd45 mRNA level is reduced upon dGLYAT depletion. a Volcano plot between dGLYAT RNAi and control groups. Compared with the hs-Gal4 control groups, there are 29 down-regulated genes (indicated by blue points) and 13 up-regulated genes (showed by red points) in the hs > dGLYAT-IR groups, when log2FC > 1.0 or <-1.0 and adjusted P value < 0.001. Grey dotted lines show thresholds of log2FC (1.0 and − 1.0) and adjusted P value (0.001). b Heat map of 29 down-regulated genes between dGLYAT RNAi (R1-R6) and control (W1-W6) groups when log2FC<-1.0 and adjusted P value < 0.001. Genes showing significant changes are listed in Additional file 1: Table S1. c RT-qPCR data showed that Gadd45 mRNA level was decreased upon dGLYAT depletion. *P < 0.05, **P < 0.01. Third instar larvae were heat shocked at 37℃ for 30 min and recovered at 29℃ for 2 h (c) or 6 h (a-c) prior to tissues dissection and RNA extraction
Loss of Gadd45 suppresses scrib depletion-induced JNK activation and cell invasion
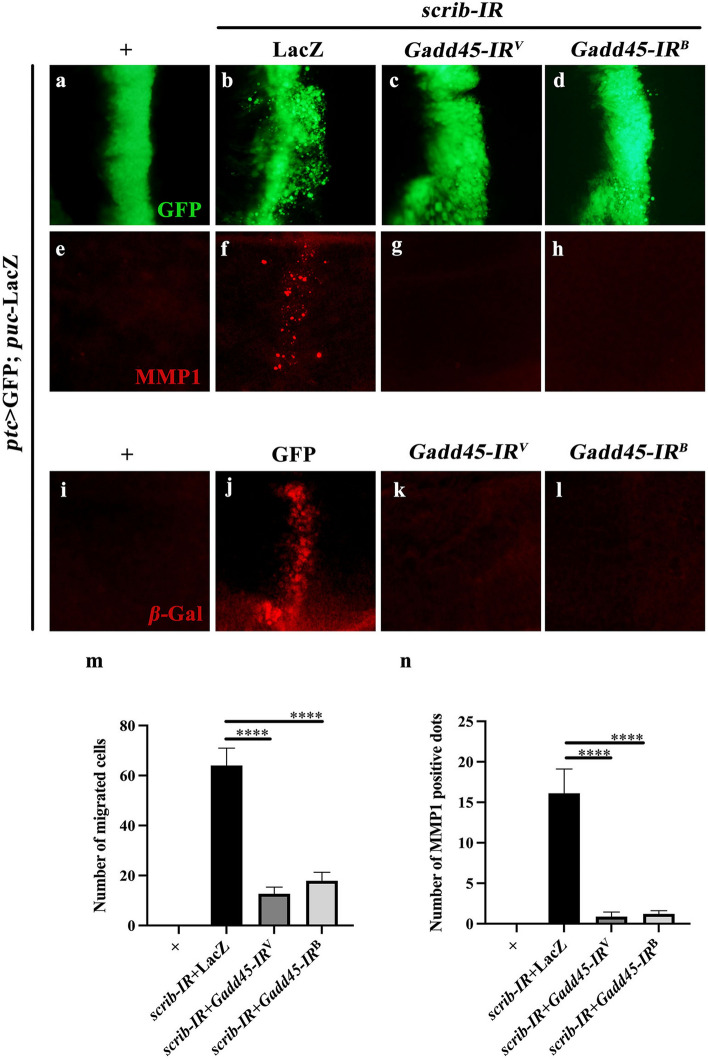
Given that loss of dGLYAT decreased Gadd45 mRNA expression and suppressed JNK-dependent cell invasion, we hypothesized that dGLYAT regulates JNK-dependent cell invasion through Gadd45. In agreement with this assumption, ptc > scrib-IR-induced invasive cell migration was significantly suppressed by knockdown of Gadd45 with two independent RNAi lines (Fig. 6a-d, quantified in Fig. 6 m). The knockdown efficiencies of Gadd45 RNAi lines were verified by RT-qPCR analysis (Additional file 2: Figure S1). Furthermore, scrib depletion-induced MMP1 upregulation was impeded by knockdown of Gadd45 (Fig. 6e-h, quantified in Fig. 6n). We also checked the expression of another JNK pathway reporter puc-LacZ, represented by anti--Gal immunostaining. We found that scrib depletion-elevated puc-LacZ expression was blocked by loss of Gadd45 (Fig. 6i-l).
Fig. 6.
Gadd45 knockdown suppresses scrib depletion-induced JNK activation and cell invasion. Compared with the ptc-Gal4 control (a, e, i), scrib RNAi-induced cell migration (b), elevated expression of MMP1 (f) and puc-LacZ (j) were significantly suppressed by expressing two independent Gadd45 RNAi (c, d, g, h and k, l). m Statistic of number of migrated cells is shown (left to right: n = 10, n = 12, n = 9. n = 10). n Statistic of number of MMP1 positive dots is shown (left to right: n = 9, n = 9, n = 9, n = 9). One-way ANOVA was used to compute P-values, ****P < 0.0001. Larvae were reared at 25 °C. Detailed genotypes: a, e, i ptc-Gal4 UAS-GFP/+; puc-LacZ/+, b, f ptc-Gal4 UAS-GFP UAS-scrib-IR; puc-LacZ /UAS-LacZ, c, g, k ptc-Gal4 UAS-GFP UAS-scrib-IR/UAS-Gadd45-IRV100413; puc-LacZ/+, d, h, l ptc-Gal4 UAS-GFP UAS-scrib-IR/UAS-Gadd45-IRBL35023; puc-LacZ/+, j ptc > GFP UAS-scrib-IR; puc-LacZ/UAS-GFP
JNK signaling also regulates other in vivo functions, in particular, cell death. For instance, overexpression of Egr driven by GMR-Gal4 (GMR > Egr) in the developing eyes triggers cell death in the larval eye discs, indicated by AO staining (Additional file 2: Figure S2g, h), and generates a small eye phenotype in the adults (Additional file 2: Figure S2a, b). Both phenotypes were significantly suppressed by knockdown of dGLYAT or Gadd45, while expression of a dominant-negative form of Basket (BskDN) served as a positive control (Additional file 2: Figure S2c–f, i–l, quantified in Additional file 2: Figure S2m, n). Collectively, these results suggest that dGLYAT and Gadd45 are broadly required for JNK signaling in vivo.
Discussion
In this study we found that loss of Drosophila GLYAT suppresses cell polarity disruption-induced JNK-dependent cell invasion. Furthermore, loss of dGLYAT impedes Egr-JNK pathway-triggered invasive cell migration. Moreover, dGLYAT regulates the transcription of Gadd45, a positive regulator of JNK signaling in apoptosis and egg development. Finally, depletion of Gadd45 blocks loss-of-cell polarity-triggered JNK activation and cell invasion. Thus, dGLYAT regulates Gadd45-mediated JNK activation and EMT-like cell migration in Drosophila.
Consistent with our fly data, in the UALCAN cancer database containing TCGA data, the expression of GLYATL1 and GADD45G are elevated in breast invasive carcinoma, compared with normal tissues (Additional file 2: Figure S3a, b). In addition, there is a positive correlation between GLYATL1 and GADD45G mRNA expression in GEPIA database (Additional file 2: Figure S3d). More importantly, GEPIA confirmed that higher GLYATL1 mRNA is associated with poor overall survival in breast cancer patients (Additional file 2: Figure S3c). These data imply that the role of dGLYAT and Gadd45 in cell invasion might be conserved by their human orthologs in breast cancer.
The role of GADD45 in P38 and JNK signaling have been extensively studied, yet divergent mechanisms were reported. GADD45 proteins activate MTK1 by promoting its dimerization and autophosphorylation [27], or bind and activate MTK1 MAPKKK to regulate P38 and JNK signaling upon environmental stress [28]. In addition, GADD45 could interact directly with P38 and regulates oncogene-induced growth [29], while GADD45 interacts with ASK1 and MKK7 to regulate both JNK and P38 [30–32]. GADD45 proteins could execute either tumor suppressor or tumor promoter function in tumor initiation [33], yet their roles in tumor invasion have not been explored previously. The Drosophila genome encodes only one GADD45 member, Gadd45, which is involved in inflammatory response, egg development, wing disc regeneration and lifespan [24, 25, 34]. Gadd45 has been shown as a positive regulator of JNK signaling in germline development and cell death [24, 25], yet its role in JNK-mediated cell invasion has not been known previously. In this study, we found that Gadd45 is required for cell polarity disruption-induced JNK-mediated cell invasion in Drosophila, suggesting a potential role of GADD45 in tumor invasion and cancer progression, which deserves further investigation.
Although we found that dGLYAT regulates Gadd45 mRNA expression, the underlying mechanism remains unknown. We hypothesize that dGLYAT, which contains a GNAT domain, may promote histone acetylation at the promoter of Gadd45. In support of this assumption, the yeast GNAT domain-containing protein Gcn5 (general control nonderepressible-5) is reported to regulate PHO5 transcription by promoting histone acetylation at its promoter [35], and this histone acetylation activity has been conserved by Gcn5 homologs in Tetrahymena and human [36]. Future studies are needed to address whether GLYAT family proteins promote histone acetylation, and whether this activity is required for their roles in cancer initiation and progression.
Conclusions
In summary, we found dGLYAT is required for JNK-mediated cell invasion. Through analyzing mRNA-seq results, we found Gadd45 mRNA expression is reduced upon dGLYAT depletion. Downregulation of Gadd45 also suppressed JNK-mediated cell invasion and cell death.
Methods
Drosophila genetics and stocks
Fly stocks were raised on standard cornmeal and agar medium at 25 °C. For cell migration assay, larvae were reared at 29 °C unless indicated. Fly strains used in this study are as follow: w1118, ptc-Gal4, UAS-GFP, UAS-scrib-RNAi, pucE69, TRE-RFP, UAS-EgrRegg1, dGLYATc02982 and UAS-dGLYAT-IR have been described previously [14]. UAS-Gadd45-RNAi (V100413 and BL35023) were gifts from Dr. Erjun Ling (Chinese Academy of Sciences, China).
Immunohistochemistry
Antibody staining was performed according to standard procedures. The following primary antibodies were used: mouse anti-MMP1 (3A6B4, 1:200, Developmental Studies Hybridoma Bank, DSHB), Rat anti-E-cad (DCAD2-c, 1:100, Developmental Studies Hybridoma Bank, DSHB) and mouse anti -gal (40-1a, 1:500, Developmental Studies Hybridoma Bank, DSHB). The following secondary antibodies were used: anti-mouse CY3 (A11032, 1:1000, Cell Signaling Technology) and anti-Rat CY3 (104,086, 1:500, Jackson Immuno research).
X-gal staining
Wing discs were dissected from third instar larvae in PBST (1× PBS pH 7.0, 0.1% Triton X-100) and stained for β-galactosidase activity as described [37].
RNA library preparation and data analysis
Third instar larvae were subjected to heat shock at 37 °C for 30 min and recovered at 29 °C for 6 h prior to tissues dissection. RNA extraction, library construction and sequencing were performed by Hua Gen Biotechnology (Shanghai, China). The sequencing platform is Illumina. The quality of sequenced raw reads was controlled by the FastQ Screen program. The reference genome sequence is Drosophila melanogaster (BDGP6.28) downloaded from Ensembl. (http://www.ensembl.org/info/data/ftp/index.html). Differentially expressed genes (DEGs) were identified by comparing the expression levels of genes between hs > dGLYAT RNAi and hs-Gal4 control groups. The threshold for DEGs was set at P- adjusted < 0.001 and log2 fold change (log2FC) ≥ 1.0 or ≤ − 1.0.
RT-qPCR
Total RNAs were extracted from fifteen third instar larval tissues of indicated genotypes with Trizol (Ambion, Life Technologies, Carlsbad, CA, USA) following the protocol of RNA preparation kit, and quantitative polymerase chain reaction (qPCR) was performed using SYBR Green PCR Premix Kit (TaKaRa). The primers used are as follow:
| Gene | Forward primer | Reverse primer |
|---|---|---|
| rp49 | TCTCCTTGCGCTTCTTGGA | TACAGGCCCAAGATCGTGAA |
| dGLYAT | ATACCATTAAGGGAACCCCAGA | TGACCCAAATTCAGCCAATATGC |
| Gadd45 | ATCGGACGCACCATCAAGTC | TGTCGTTCTCGTAGCAAAAGG |
UALCAN and GEPIA cancer databases
The UALCAN cancer database (http://ualcan.path.uab.edu/analysis.html) is a comprehensive web source that provide the data from The Cancer Genome Atlas (TCGA) [38]. The GEPIA database (http://gepia2.cancer-pku.cn/#index) contains data from TCGA and Genotype-Tissue Expression (GTEx) project [39].
Supplementary Information
Additional file 1: Table S1. mRNA-seq data between control and dGLYAT-depleted groups.
Additional file 2: Figure S1. The knock-down efficiencies of RNAi lines. The efficiencies of dGLYAT and GADD45 RNAi lines were measured by RT-qPCR. dGLYAT mRNA and Gadd45 mRNA levels were significantly downregulated by the expression of their corresponding RNAi. Third instar larvae were subjected to heat shock at 37°C for 30 minutes in the water bath and recovered for 2 hours at 29°C. Larval discs were dissected for RT-qPCR. Error bars represents standard deviation from three independent experiments. One-way ANOVA test was used to compute P-values, ****P<0.0001. Figure S2. Depletion of dGLYAT or Gadd45 suppresses GMR>Egr-induced cell death. Light micrographs of Drosophila adult eyes (a–f) and fluorescent micrographs of third instar larval eye discs (g–l) are shown. Compared with the GMR-Gal4 controls (a, g), GMR>Egr induces a small eye phenotype in adults (b) and massive cell death in third instar larval eye discs with AO staining (h). Both phenotypes were suppressed by knockdown of dGLYAT or Gadd45 (c-e and i-k). BskDN serves as a positive control (f, l). (m) Statistic of eyes size is shown (from left to right: n =7, n=10, n=10, n=10, n=9, n=5). (n) Statistic of AO-positive cell number is shown (from left to right: n =10, n=11, n=13, n=12, n=7, n=10), One-way ANOVA test was used to compute P-values, ****P<0.0001. Figure S3. Characterization of GLYATL1 and GADD45G in breast cancer. (a, b) Transcriptome sequencing of breast cancer. Expression of GLYATL1 and GADD45G in normal and tumor tissues were measured in transcript per million utilizing the TCGA data set. The tumor tissues show higher expression of GLYATL1 and GADD45G than normal tissues. (c) Survival analysis of GLYATL1 in breast cancer patients. The survival of breast cancer patients with higher GLYATL1 expression was significantly worse (P<0.05). (d) The expression relationship between GLYATL1 and GADD45G in breast cancer using GEPIA database: a positive correlation between expression of GLYATL1 and GADD45G (P=7.5e-08, R=0.15).
Acknowledgements
We thank the Bloomington Drosophila Stock Center, Tsinghua Fly Center and Dr. Erjun Ling for fly stocks, and Dr. Xin Xie for advice.
Abbreviations
- Csk
C-terminal SRC kinase
- JNK
c-Jun N-terminal kinase
- GLYAT
Glycine N-acyltransferases
- Gadd45
Growth Arrest and DNA Damage-inducible 45
- MMP1
Matrix metalloprotease 1
- EMT
Epithelial-mesenchymal transition
- GNAT
Gcn5-related N-acetyltransferases
- Ac-CoA
Acetyl moiety from Coenzyme A
- DEGs
Differentially expressed genes
- RT-qPCR
Reverse transcription-quantitative polymerase chain reaction
- TCGA
The Cancer Genome Atlas
Author contributions
M.X., P.R. W.L. and L.X. conceived and designed the experiments. M.X., P.R, J.T., L.X., P.H., P. C. and W.L. conducted experiments. P. R., W.L. and L.X. supervised/advised on the study, M.X., P.R, W.L. and L.X. analyzed the data, M.X. and L.X. wrote the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (31970536), Shanghai Committee of Science and Technology (09DZ2260100), and the Fundamental Research Funds for the Central Universities (22120180549).
Availability of data and materials
The data that support the finding of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors have read and agreed to the final version of the manuscript.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Pu Ren, Email: ren_pu123@qq.com.
Wenzhe Li, Email: lwz@tongji.edu.cn.
Lei Xue, Email: lei.xue@tongji.edu.cn.
References
- 1.Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res. 2019;79(12):3011–27. doi: 10.1158/0008-5472.CAN-19-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stoletov K, Beatty PH, Lewis JD. Novel therapeutic targets for cancer metastasis. Expert Rev Anticancer Ther. 2020;20(2):97–109. doi: 10.1080/14737140.2020.1718496. [DOI] [PubMed] [Google Scholar]
- 3.Miles WO, Dyson NJ, Walker JA. Modeling tumor invasion and metastasis in Drosophila. Dis Model Mech. 2011;4(6):753–61. doi: 10.1242/dmm.006908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vidal M, Larson DE, Cagan RL. Csk-deficient boundary cells are eliminated from normal Drosophila epithelia by exclusion, migration, and apoptosis. Dev Cell. 2006;10(1):33–44. doi: 10.1016/j.devcel.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Shizu R, Min J, Sobhany M, Pedersen LC, Mutoh S, Negishi M. Interaction of the phosphorylated DNA-binding domain in nuclear receptor CAR with its ligand-binding domain regulates CAR activation. J Biol Chem. 2018;293(1):333–44. doi: 10.1074/jbc.M117.806604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takatsu Y, Nakamura M, Stapleton M, Danos MC, Matsumoto K, O’Connor MB, et al. TAK1 participates in c-Jun N-terminal kinase signaling during Drosophila development. Mol Cell Biol. 2000;20(9):3015–26. doi: 10.1128/MCB.20.9.3015-3026.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glise B, Bourbon H, Noselli S. hemipterous encodes a novel Drosophila MAP kinase kinase, required for epithelial cell sheet movement. Cell. 1995;83(3):451–61. doi: 10.1016/0092-8674(95)90123-X. [DOI] [PubMed] [Google Scholar]
- 8.Chen HW, Marinissen MJ, Oh SW, Chen X, Melnick M, Perrimon N, et al. CKA, a novel multidomain protein, regulates the JUN N-terminal kinase signal transduction pathway in Drosophila. Mol Cell Biol. 2002;22(6):1792–803. doi: 10.1128/MCB.22.6.1792-1803.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkins KK, Dailey GM, Tjian R. Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. The EMBO journal. 1988;7(13):4265–73. doi: 10.1002/j.1460-2075.1988.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 1998;12(4):557–70. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.La Marca JE, Richardson HE, Two-Faced Roles of JNK signalling during tumourigenesis in the Drosophila model. Front Cell Dev Biol. 2020;8:42. doi: 10.3389/fcell.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuo M, Terai K, Kameda N, Matsumoto A, Kurokawa Y, Funase Y, et al. Designation of enzyme activity of glycine-N-acyltransferase family genes and depression of glycine-N-acyltransferase in human hepatocellular carcinoma. Biochem Biophys Res Commun. 2012;420(4):901–6. doi: 10.1016/j.bbrc.2012.03.099. [DOI] [PubMed] [Google Scholar]
- 13.Guan R, Hong W, Huang J, Peng T, Zhao Z, Lin Y, et al. The expression and prognostic value of GLYATL1 and its potential role in hepatocellular carcinoma. J Gastrointest Oncol. 2020;11(6):1305–21. doi: 10.21037/jgo-20-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren P, Li W, Xue L. GLYAT regulates JNK-mediated cell death in Drosophila. Sci Rep. 2017;7(1):5183. doi: 10.1038/s41598-017-05482-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C, Ding X, Li Z, Huang Y, Xu Q, Zou R, et al. CtBP modulates Snail-mediated tumor invasion in Drosophila. Cell Death Discov. 2021;7(1):202. doi: 10.1038/s41420-021-00516-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang S, Guo X, Wu H, Sun Y, Ma X, Li J, et al. Wingless modulates activator protein-1-mediated tumor invasion. Oncogene. 2019;38(20):3871–85. doi: 10.1038/s41388-018-0629-x. [DOI] [PubMed] [Google Scholar]
- 17.Sun Y, Zhang D, Guo X, Li W, Li C, Luo J, et al. MKK3 modulates JNK-dependent cell migration and invasion. Cell Death Dis. 2019;10(3):1–11. doi: 10.1038/s41419-019-1350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 19.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulton JS, Cuningham JC, Peifer M. Acentrosomal Drosophila epithelial cells exhibit abnormal cell division, leading to cell death and compensatory proliferation. Dev Cell. 2014;30(6):731–45. doi: 10.1016/j.devcel.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salah Ud-Din AI, Tikhomirova A, Roujeinikova A. Structure and functional diversity of GCN5-related N-acetyltransferases (GNAT) Int J Mol Sci. 2016;17(7):1018. doi: 10.3390/ijms17071018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vetting MW, LP SdC, Yu M, Hegde SS, Magnet S, Roderick SL, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433(1):212–26. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 23.Mizzen CA, Allis CD. Linking histone acetylation to transcriptional regulation. Cell Mol Life Sci CMLS. 1998;54(1):6–20. doi: 10.1007/s000180050121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camilleri-Robles C, Serras F, Corominas M. Role of D-GADD45 in JNK-dependent apoptosis and regeneration in Drosophila. Genes. 2019;10(5):378. doi: 10.3390/genes10050378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peretz G, Bakhrat A, Abdu U. Expression of the Drosophila melanogaster GADD45 homolog (CG11086) affects egg asymmetric development that is mediated by the c-Jun N-terminal kinase pathway. Genetics. 2007;177(3):1691–702. doi: 10.1534/genetics.107.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salvador JM, Brown-Clay JD, Fornace AJ. Jr. Gadd45 in stress signaling, cell cycle control, and apoptosis. Adv Exp Med Biol. 2013;793:1–19. doi: 10.1007/978-1-4614-8289-5_1. [DOI] [PubMed] [Google Scholar]
- 27.Miyake Z, Takekawa M, Ge Q, Saito H. Activation of MTK1/MEKK4 by GADD45 through induced N-C dissociation and dimerization-mediated trans autophosphorylation of the MTK1 kinase domain. Mol Cell Biol. 2007;27(7):2765–76. doi: 10.1128/MCB.01435-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takekawa M, Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95(4):521–30. doi: 10.1016/S0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 29.Bulavin DV, Kovalsky O, Hollander MC, Fornace AJ. Jr. Loss of oncogenic H-ras-induced cell cycle arrest and p38 mitogen-activated protein kinase activation by disruption of Gadd45a. Mol Cell Biol. 2003;23(11):3859–71. doi: 10.1128/MCB.23.11.3859-3871.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papa S, Zazzeroni F, Bubici C, Jayawardena S, Alvarez K, Matsuda S, et al. Gadd45 beta mediates the NF-kappa B suppression of JNK signalling by targeting MKK7/JNKK2. Nat Cell Biol. 2004;6(2):146–53. doi: 10.1038/ncb1093. [DOI] [PubMed] [Google Scholar]
- 31.Papa S, Monti SM, Vitale RM, Bubici C, Jayawardena S, Alvarez K, et al. Insights into the structural basis of the GADD45beta-mediated inactivation of the JNK kinase, MKK7/JNKK2. J Biol Chem. 2007;282(26):19029–41. doi: 10.1074/jbc.M703112200. [DOI] [PubMed] [Google Scholar]
- 32.Ueda T, Kohama Y, Kuge A, Kido E, Sakurai H. GADD45 family proteins suppress JNK signaling by targeting MKK7. Arch Biochem Biophys. 2017;635:1–7. doi: 10.1016/j.abb.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Yang Z, Liu Y. GADD45 proteins: roles in cellular senescence and tumor development. Exp Biol Med (Maywood) 2014;239(7):773–8. doi: 10.1177/1535370214531879. [DOI] [PubMed] [Google Scholar]
- 34.Plyusnina EN, Shaposhnikov MV, Moskalev AA. Increase of Drosophila melanogaster lifespan due to D-GADD45 overexpression in the nervous system. Biogerontology. 2011;12(3):211–26. doi: 10.1007/s10522-010-9311-6. [DOI] [PubMed] [Google Scholar]
- 35.Gregory PD, Schmid A, Zavari M, Lui L, Berger SL, Horz W. Absence of Gcn5 HAT activity defines a novel state in the opening of chromatin at the PHO5 promoter in yeast. Mol Cell. 1998;1(4):495–505. doi: 10.1016/S1097-2765(00)80050-7. [DOI] [PubMed] [Google Scholar]
- 36.Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, et al. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16(2):593–602. doi: 10.1128/MCB.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu C, Li Z, Ding X, Guo X, Sun Y, Wang X, et al. Snail modulates JNK-mediated cell death in Drosophila. Cell Death Dis. 2019;10(12):893. doi: 10.1038/s41419-019-2135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–58. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98-W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. mRNA-seq data between control and dGLYAT-depleted groups.
Additional file 2: Figure S1. The knock-down efficiencies of RNAi lines. The efficiencies of dGLYAT and GADD45 RNAi lines were measured by RT-qPCR. dGLYAT mRNA and Gadd45 mRNA levels were significantly downregulated by the expression of their corresponding RNAi. Third instar larvae were subjected to heat shock at 37°C for 30 minutes in the water bath and recovered for 2 hours at 29°C. Larval discs were dissected for RT-qPCR. Error bars represents standard deviation from three independent experiments. One-way ANOVA test was used to compute P-values, ****P<0.0001. Figure S2. Depletion of dGLYAT or Gadd45 suppresses GMR>Egr-induced cell death. Light micrographs of Drosophila adult eyes (a–f) and fluorescent micrographs of third instar larval eye discs (g–l) are shown. Compared with the GMR-Gal4 controls (a, g), GMR>Egr induces a small eye phenotype in adults (b) and massive cell death in third instar larval eye discs with AO staining (h). Both phenotypes were suppressed by knockdown of dGLYAT or Gadd45 (c-e and i-k). BskDN serves as a positive control (f, l). (m) Statistic of eyes size is shown (from left to right: n =7, n=10, n=10, n=10, n=9, n=5). (n) Statistic of AO-positive cell number is shown (from left to right: n =10, n=11, n=13, n=12, n=7, n=10), One-way ANOVA test was used to compute P-values, ****P<0.0001. Figure S3. Characterization of GLYATL1 and GADD45G in breast cancer. (a, b) Transcriptome sequencing of breast cancer. Expression of GLYATL1 and GADD45G in normal and tumor tissues were measured in transcript per million utilizing the TCGA data set. The tumor tissues show higher expression of GLYATL1 and GADD45G than normal tissues. (c) Survival analysis of GLYATL1 in breast cancer patients. The survival of breast cancer patients with higher GLYATL1 expression was significantly worse (P<0.05). (d) The expression relationship between GLYATL1 and GADD45G in breast cancer using GEPIA database: a positive correlation between expression of GLYATL1 and GADD45G (P=7.5e-08, R=0.15).
Data Availability Statement
The data that support the finding of this study are available from the corresponding author upon reasonable request.