SUMMARY
Spatially and temporally varying patterns of morphogen signals during development drive cell fate specification at the proper location and time. However, current in vitro methods typically do not allow for precise, dynamic spatiotemporal control of morphogen signaling and are thus insufficient to readily study how morphogen dynamics affect cell behavior. Here, we show that optogenetic Wnt/β-catenin pathway activation can be controlled at user-defined intensities, temporal sequences, and spatial patterns using engineered illumination devices for optogenetic photostimulation and light activation at variable amplitudes (LAVA). By patterning human embryonic stem cell (hESC) cultures with varying light intensities, LAVA devices enabled dose-responsive control of optoWnt activation and Brachyury expression. Furthermore, time-varying and spatially localized patterns of light revealed tissue patterning that models the embryonic presentation of Wnt signals in vitro. LAVA devices thus provide a low-cost, user-friendly method for high-throughput and spatiotemporal optogenetic control of cell signaling for applications in developmental and cell biology.
Graphical Abstract
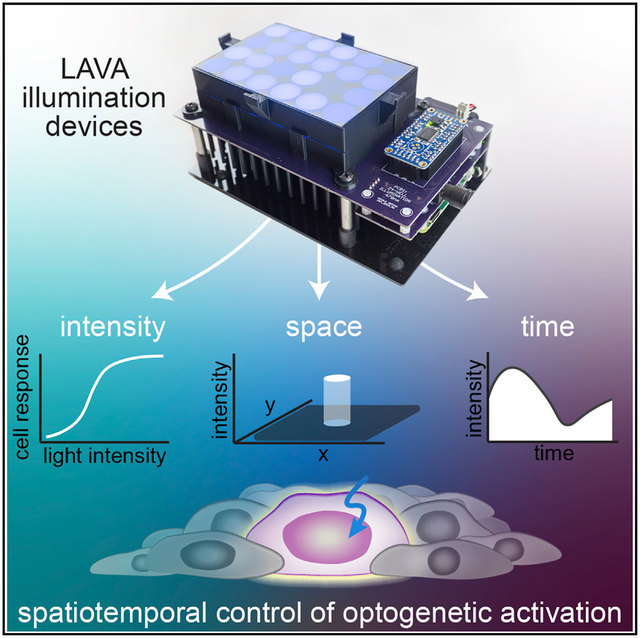
In Brief
With optogenetics, protein-protein interactions and cell signaling can be controlled with light. To allow user-defined illumination patterns to be projected onto cell cultures, Repina et al. designed programmable photostimulation devices called LAVA devices and use them to optogenetically induce patterns of Wnt signaling in human embryonic stem cells.
INTRODUCTION
Cell fate decisions are governed by dynamic spatially and temporally varying signals from the cellular environment. In particular, during development, morphogen gradients orchestrate the dynamic coordinated movement and differentiation of embryonic cell populations (Arnold and Robertson, 2009). Genetic perturbation and biomolecular treatment with pathway agonists or inhibitors have provided insight into the critical regulators of embryogenesis, yet spatially varying interactions between cell subpopulations and time-varying signal dynamics and duration thresholds remain largely unstudied due to a lack of tools to perturb signaling dynamics in biological systems (Arnold and Robertson, 2009; Oates et al., 2009).
Optogenetic methods aim to address this need for techniques that manipulate or perturb cell signaling dynamics. With optogenetics, light-responsive proteins from plants, algae, and bacteria have been adapted to control protein-protein interactions and signaling in mammalian cells (Boyden et al., 2005; Nagel et al., 2003; Hegemann and Nagel, 2013). In particular, a variety of photosensory domains have been discovered, optimized, and repur-posed to place intracellular signaling pathways under light control, capabilities that offer the unique advantage of using light patterns to stimulate signaling in a specific location and at a specific time (Repina et al., 2017). Within mammalian cells, optogenetic tools that control protein-protein interactions have, for example, been developed for diverse signaling pathways such as Wnt/β-catenin (β-CAT) (Bugaj et al., 2013; Repina et al., 2019), Ras/Raf/Mek/Erk (Toettcher et al., 2011; Johnson and Toettcher, 2019; Toettcher et al., 2013; Johnson et al., 2017), fibroblast growth factor (FGF) (Kainrath et al., 2017), receptor tyrosine kinases (Bugaj et al., 2015), and Rho-family guanosine triphosphatase (GTPase) signaling (Levskaya et al., 2009; Yazawa et al., 2009; Strickland et al., 2012; Guntas et al., 2015; Bugaj et al., 2013; Wang et al., 2010). Optogenetic methods have also been applied recently to studies in developmental biology in key model organisms (Johnson and Toettcher, 2018). For example, in Drosophila and zebrafish embryos, the dynamic control of signaling elucidated the spatiotemporal limits of ERK (Johnson et al., 2017; Johnson and Toettcher, 2019), Nodal (Sako et al., 2016), and Bicoid (Huang et al., 2017) patterning and how the local modulation of cell migration and contractility drives tissue morphogenesis (Izquierdo et al., 2018; Guglielmi et al., 2015; Čapek et al., 2019). Furthermore, in embryonic stem cell models for mammalian development, we have recently achieved optogenetic control of canonical Wnt signaling and elucidated mechanisms of tissue self-organization during mesendoderm differentiation (Bugaj et al., 2013; Repina et al., 2019).
To activate the variety of photosensory domains developed for cellular signaling pathway control, there must be complementary development of optical tools for cell culture illumination. However, the current optical methods lack practical applicability to routine cell culture stimulation and/or lack critical characterization and functionality. In particular, microscope-based systems widely used for optogenetic photostimulation implement one- or two-photon excitation to scan a single diffraction-limited spot (Packer et al., 2013, 2015; Prakash et al., 2012; Carrillo-Reid et al., 2016; Nikolenko et al., 2007) or project multi-spot light patterns (Papagiakoumou et al., 2008, 2010; Pégard et al., 2017; Hernandez et al., 2016) onto the biological sample. While such systems are essential for high-resolution in vivo experiments and precise manipulation of neural circuits, their application to intracellular signal pathway activation in cell cultures is limited by the complexity, low throughput, high cost, and need for continuous environmental control of the system (Yizhar et al., 2011). For manipulating signaling pathways in which speed and single-cell spatial resolution are often less critical, the paramount photostimulation needs are high-throughput control of multiple parallel biological conditions, defined illumination parameters, and compatibility with established cell culture formats and assays. Several methods have been developed for such photostimulation of signal pathways in cell cultures, although they can lack critical characterization and functionality. For example, a simple panel of light-emitting diodes (LEDs) enables optogenetic activation of a tissue culture (TC) plate, but lacks multiwell, high-throughput control (Müller et al., 2014; Shao et al., 2018) and spatiotemporal patterning capability (Tucker et al., 2014). Multiwell control can be achieved by incorporating a microcontroller and LED drivers that set user-defined intensities for separate wells (Olson et al., 2014; Gerhardt et al., 2016; Bugaj and Lim, 2019). Although such multiwell devices have advanced the throughput of optogenetic studies, current implementations have not characterized key performance parameters such as uniformity of illumination, spatial or temporal light-patterning resolution, and quantification of device overheating and cell toxicity (Gerhardt et al., 2016; Davidson et al., 2013; Lee et al., 2013; Hannanta-Anan and Chow, 2016; Bugaj et al., 2018; Hennemann et al., 2018; Richter et al., 2015; Olson et al., 2014). Several such designs also rely on the modification of expensive instrumentation (Richter et al., 2015) or focus on bacterial culture tubes (Olson et al., 2014).
We have designed a programmable illumination system for the photostimulation of multiwell plates that can be readily incorporated into the workflow of routine cell culture and allow controlled and quantitative spatiotemporal light patterning. Specifically, we engineer cell culture illumination devices for light activation at variable amplitudes (LAVA). We optimize the LAVA device optical configuration for illumination uniformity and achieve programmable photostimulation of independent wells of 24-well or 96-well culture plates kept in standard 37°C TC incubators. Each well can be wirelessly programmed through a graphical user interface (GUI) at user-defined intensities (0–20 μWmm–2, 0.005 μWmm–2 resolution), temporal sequences (10 ms resolution), and spatial patterns (100 μm resolution). Wedemonstrate LAVA device performance by modulating the intensity, timing, and spatial location of canonical Wnt/β-catenin signaling in human embryonic stem cell (hESC) cultures using the optoWnt optogenetic system (Bugaj et al., 2013; Repina et al., 2019). In particular, we show that Wnt pathway activation and hESC differentiation is dose responsive to light intensity and duration of illumination and that spatial patterning can be used to simulate the embryonic, spatially polarized presentation of the Wnt ligand. Lastly, we provide a detailed protocol for LAVA board assembly, which takes ~8 h and <$500 to fabricate and assemble and requires no previous experience with electronics.
RESULTS
Design Requirements for Spatiotemporal Photostimulation of Cell Cultures
A number of design considerations should be considered to ensure controlled, long-term illumination of mammalian cell cultures. The primary requirement is the illumination of cells with defined intensities of light and with illumination patterns that can vary in space and time. Also, the intensity range should be sufficient to activate the photosensory protein of interest, which can vary widely from continuous illumination at 1 μWmm–2 (Repina et al., 2019) to short pulses at 1,000–10,000 μWmm–2 (Yizhar et al., 2011). Moreover, the required temporal resolution also depends on the photosensory domain and experimental application: 1- to 100-ms pulses are typically used to control neural circuits (Yizhar et al., 2011), whereas signal pathway dynamics are controlled on the hour to multiday timescales (Repina et al., 2019; Johnson and Toettcher, 2018). Furthermore, for spatial resolution requirements, the optimal resolution for subcellular stimulation would be 0.2–2 μm, but such resolutions are infeasible without complex optical systems. Since the typical size of single cells is 10–20 μm, multicellular resolution can be sufficient for many applications at 50–200 μm.
Beyond the requirement for light delivery, the illumination device must also be compatible with routine stem cell culture experiments. As a result, the electronics must be compatible with the warm (37°C) and humid environment used for mammalian TC. In addition, the device should also not induce cell damage from overheating or phototoxicity, which is a significant concern during optogenetic stimulation since such processes alter cell physiology and/or cause non-specific activation of signaling pathways (Tyssowski and Gray, 2019; Acker et al., 2016; Allen et al., 2015; Yizhar et al., 2011; Owen et al., 2019). Illumination uniformity across a region of interest is critical as well. Since the strength of induced signaling is dependent on light dosage, nonuniformity can give rise to signal variation and heterogeneity across the region of optogenetic stimulation. Lastly, cost, functionality, and ease of use can be significant barriers for optogenetic studies, so simple user-programmable control of many simultaneous illumination conditions is a significant advantage for complex and high-throughput biological experiments.
Design Overview of Engineered LAVA Illumination Devices
To enable precise control over the intensity, timing, and location of optogenetic stimulation, we engineered illumination devices—LAVA devices—that incorporate into the workflow of stem cell culture (Figure 1A). LAVA devices project user-defined, programmable light patterns onto 24-well or 96-well TC plates maintained inside a standard TC incubator (Figures 1B, 1C, and S1A–S1C). In brief, light emitted by LEDs located underneath the multiwell culture plate passes through diffusive optical elements that ensure the uniform illumination of each well. For the stimulation of cells expressing optoWnt, a system we previously engineered for the optogenetic control of Wnt signaling (Bugaj et al., 2013; Repina et al., 2019), we chose blue LEDs emitting at a central wavelength of 470 nm to match the Arabidopsis thaliana Cryptochrome 2 (Cry2) photosensory domain absorption spectrum (Figure S1D), although we note that LEDs of diverse colors can readily be substituted for use with other optogenetic systems. The LAVA board electronics are designed for the programmable control of illumination intensity and temporal sequences, with independent control of each well. In addition, spatial precision is conveyed through an intensity mask attached to the culture plate. The hardware design also includes a cooling system and vibration isolation to ensure cell viability. Lastly, for ease of use, we developed a GUI to wirelessly program the illumination intensity and temporal sequences for each well (Figures S1E–S1G).
Figure 1. Overview of Illumination Device LAVA for Optogenetic Stimulation of hESC Cultures.
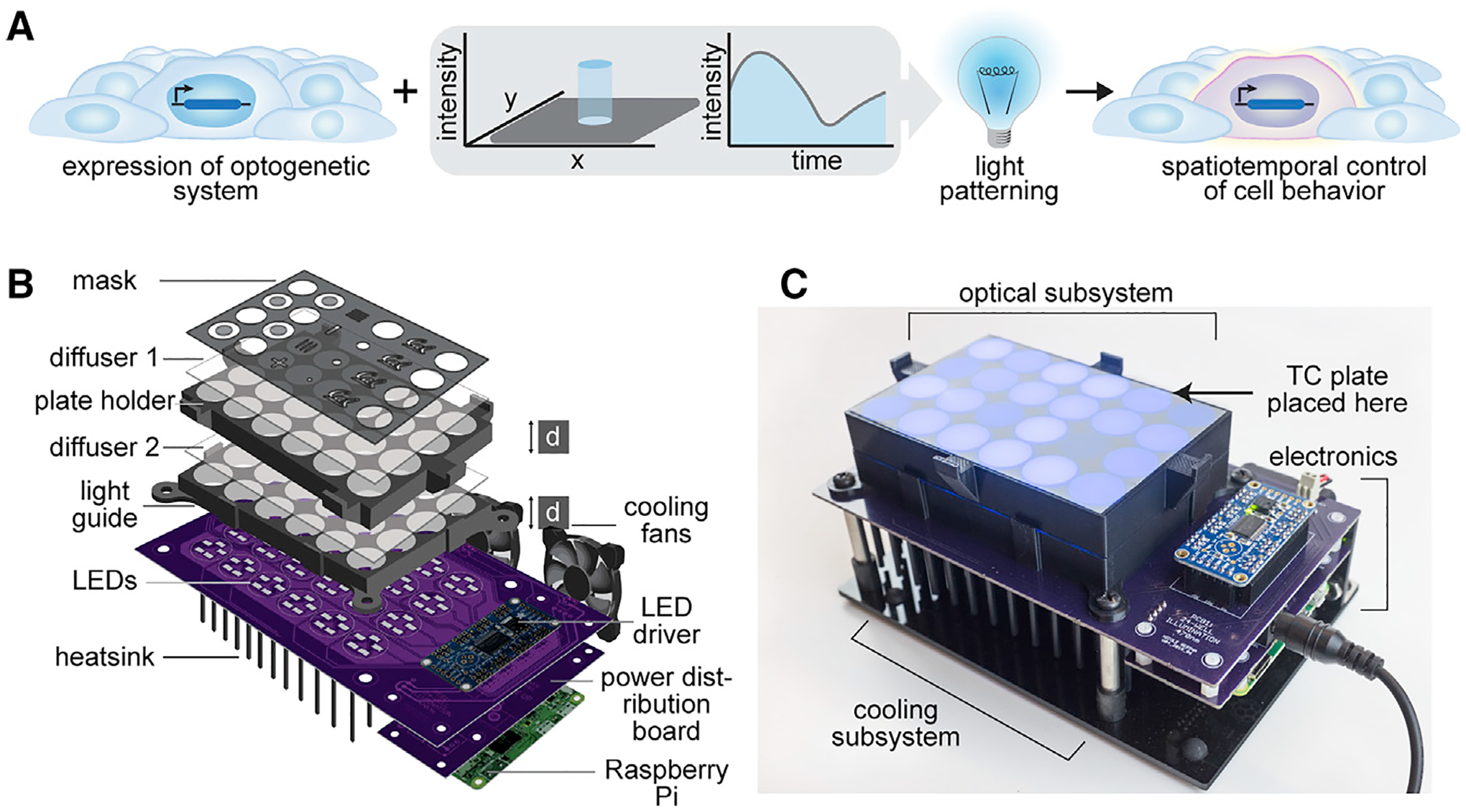
(A) Schematic of typical optogenetic experiment, in which spatiotemporal precision is conferred through light patterning.
(B) Diagram of LAVA illumination device design. LEDs illuminate a TC plate placed on top of the device, with light passing through a series of 2 light guides, 2 optical diffusers, and a die-cut mask. LEDs are programmed through a Raspberry Pi and LED driver and are cooled with a heat sink and cooling fans.
(C) Image of assembled 24-well LAVA board, with optical, cooling, and electronics subsystems highlighted. For the 96-well LAVA board, refer to Figure S1.
Optimization and Characterization of Illumination Uniformity
We established the LAVA optical system design by modeling the optical configuration of a single well in the optical ray tracing software Zemax OpticStudio (Figures S2A–S2H). We then optimized the optical configuration for uniform well illumination (Figure 2A). Due to the spatial constraints of a TC incubator, we used optical diffusers rather than lenses, in addition to optical scattering from the 3-dimensional (3D)-printed light guides to ensure illumination uniformity. In the Zemax model, parameters such as the LED position on the circuit board, diffuser strength, and light guide dimensions were optimized to reduce intensity drop-off at the well edge (Figures S2A–S2H and S3A–S3E). Modeling results showed that the parameter with the strongest effect on uniformity was the axial thickness, d, of the two 3D-printed light guides (labeled in Figure 1B). Based on these modeling results, we fabricated LAVA devices and experimentally validated the resulting well uniformity by imaging LAVA wells under a low-magnification microscope (Figures 2B and S4A–S4C). The measurement of light intensity as a function of radial distance confirmed the improved illumination uniformity, which came at the expense of maximum illumination intensity (Figure 2B). Increasing d from 1 to 1.5 cm attenuated the intensity decrease at the well edge from 20.4% to 16.9%, a roughly 20% improvement in uniformity. A larger d also resulted in a 2-fold improvement in well-to-well variability between the 24 independent wells (2.6% versus 1.2% coefficient of variation) (Figure S4D). For experimental applications in which the intensity of illumination is paramount to well uniformity, a lower d could be used to achieve higher illumination intensities. Thus, the LAVA devices can be used in 2 hardware configurations, which are summarized as follows: (1) a low-intensity, high-precision configuration at d = 1.5 cm, for which intensity can be programmed from 0 to 10 μW/mm2 in 0.0024 μW/mm2 increments, with high illumination uniformity and low well-to-well variability, and (2) a high-intensity, low-precision configuration at d = 1 cm, for which well intensity resolution, variability, and uniformity are sacrificed to achieve a doubling in light intensity (0–20 μW/mm2 in 0.005 μW/mm2 increments) (Figure 2C).
Figure 2. Optical Design for Illumination Uniformity of TC Plate Wells.
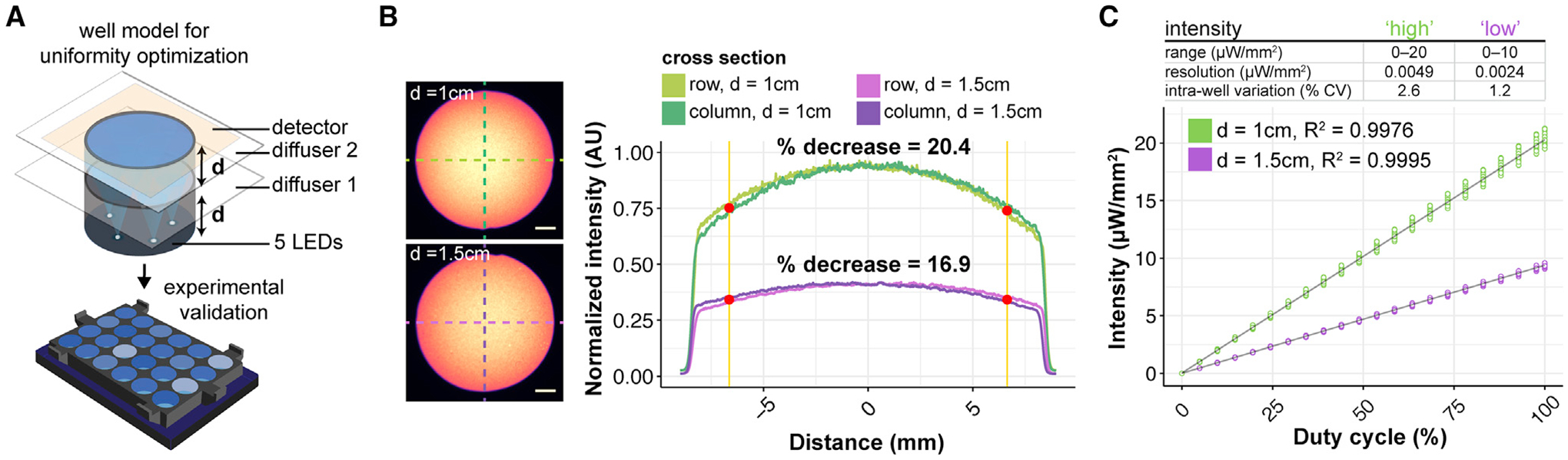
(A) Schematic of Zemax model used for LAVA optical system optimization.
(B) Bright-field images of LAVA wells (left) and graph of intensity line scans along indicated cross-sections (right) characterize the intensity uniformity of the 24-well LAVA device under 2 configurations, in which light guide thickness, d, is either (1) 1 cm (top, green) or (2) 1.5 cm (bottom, purple). The percentage of decrease is calculated between the intensity at the center of the well and the intensity at the highlighted red point, which indicates the location of the well edge of a 24-well culture plate. The graph shows mean normalized intensity over 4 independent wells. Scale bar, 2.5 mm.
(C) Measured light intensity in response to the programmed duty cycle of the LED pulse-width modulation signal. The graph shows the measured intensity from each well of a 24-well LAVA device and curve fit to a linear regression model.
Intensity Control of Optogenetic Stimulation Reveals Brachyury (BRA) Expression Level Is Correlated with LRP6 Oligomer Number and Size
During mammalian embryonic development, gradients in Wnt signal intensity control the progression of cell lineage commitment and axis patterning (Zeng et al., 1997; Liu et al., 1999; Kimura-Yoshida et al., 2005; Arnold and Robertson, 2009). In hESCs, the strength of Wnt signaling similarly modulates cell lineage commitment and differentiation potential (Davidson et al., 2012; Blauwkamp et al., 2012; Sumi et al., 2008; Bernardo et al., 2011). Equipped with a method for the optogenetic stimulation of cell cultures, we used the LAVA devices to activate canonical Wnt/β-CAT signaling in a clonal hESC line expressing the optoWnt system (Repina et al., 2019). In addition to on and off control of Wnt signaling, we sought to determine whether optoWnt could be activated in a dose-responsive manner to better mimic the signal gradients present during development.
Wnt co-receptors naturally oligomerize upon the addition of Wnt ligand, and we previously reported that this process can be emulated using light-induced oligomerization of Cry2 fused to the C terminus Wnt co-receptor LRP6 to activate canonical Wnt signaling upon blue light illumination (Bugaj et al., 2013). Since Cry2 oligomerization is a dynamic, reversible process wherein clustering is triggered upon photon absorption (Duan et al., 2017; Bugaj et al., 2013), we reasoned that the fraction of photostimulated Cry2 proteins per cell could be controlled with light intensity (Figure 3A). Using the LAVA boards, we were able to set independent wells to different light intensities using pulse width modulation (PWM) (Figure 2C). Upon continuous photostimulation at variable intensities, we observed that the number and size of visible LRP6 oligomers per cell increased monotonically with illumination intensity (Figures 3B, S5A, and S5B). To determine whether increased Cry2 oligomerization translated to a stronger Wnt signal intensity, we probed for the expression of BRA (also known by its gene name, T), a direct transcriptional target of Wnt signaling and regulator of mesendoderm and primitive streak differentiation (Rivera-Pérez and Magnuson, 2005; Yamaguchi et al., 1999; Arnold et al., 2000; Lindsley et al., 2006). Following a similar trend in Cry2 oligomerization, the mean intensity of BRA immunostaining increased with light intensity, suggesting that an increased number and size of LRP6 clusters correlate with a stronger differentiation signal (Figures 3C and S5C).
Figure 3. Optogenetic Induction of BRA Expression Is Light Dose Dependent.
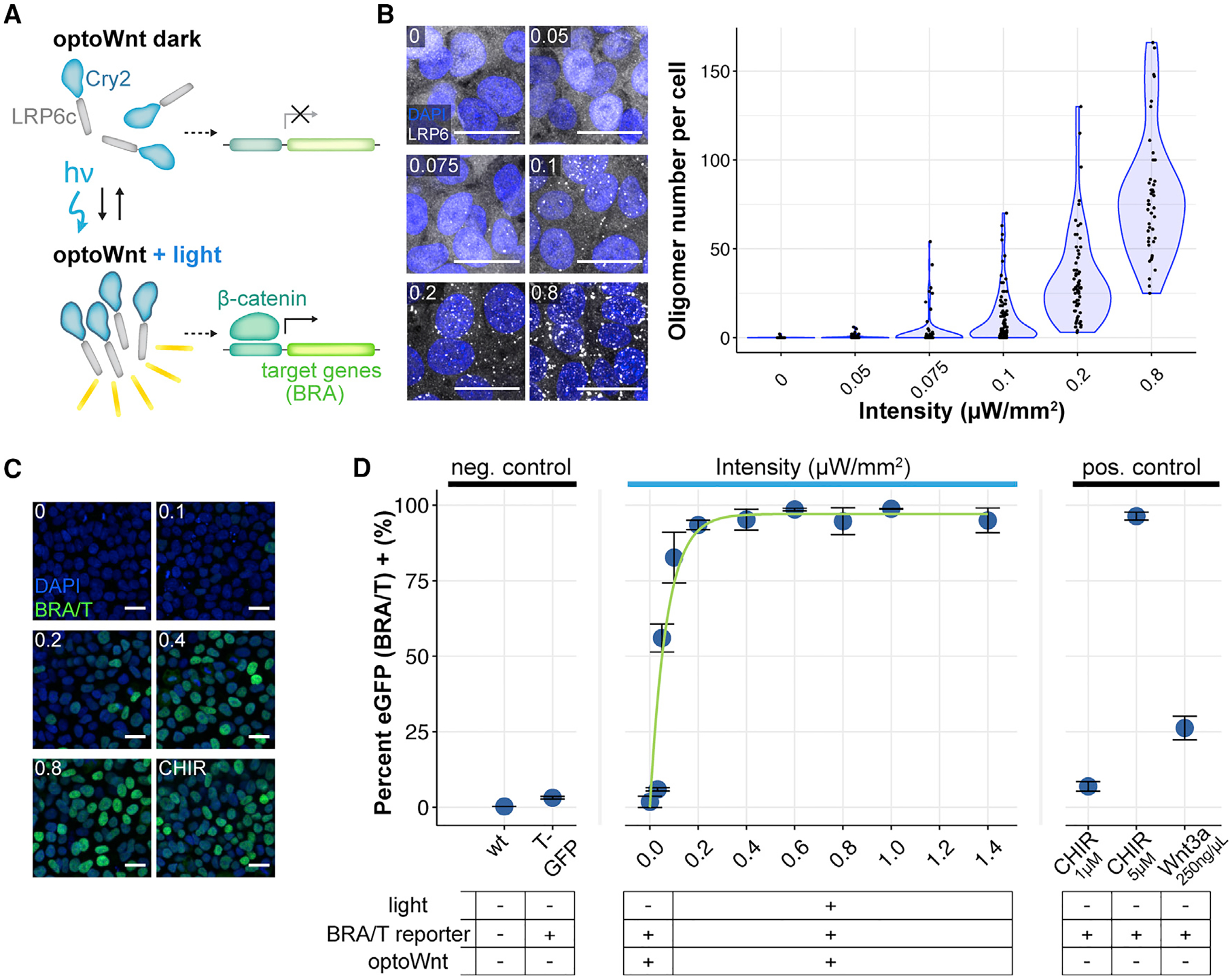
(A) Schematic of optoWnt system. In the dark, the Cry2 photosensory domain is diffuse. Illumination induces LRP6c oligomerization and the transcription of β-catenin target genes.
(B) Immunostaining for LRP6 (left) and quantification of cluster number per hESC in response to increasing light intensity after 1-h illumination. The graph shows individual cell quantification, with each point representing a single cell. Scale bar, 25 μm.
(C) Immunostaining for BRA in response to increasing light intensity after 24 h illumination or 3 μM CHIR treatment. Scale bar, 25 μm.
(D) Flow cytometry of optoWnt hESCs expressing EGFP reporter for BRA/T treated with varying light intensities or with Wnt pathway agonists (Wnt3a recombinant protein or CHIR). The graph shows the percentage of EGFP+ cells and nonlinear least-squares fit to increasing exponential decay curve. A subset of the data is reproduced from Repina et al. (2019). Graph shows means ± 1 SDs, n = 3 biological replicates.
To better quantify BRA expression at a single-cell level, we used a clonal hESC cell line co-expressing the optoWnt system and an EGFP reporter knocked into the endogenous T locus (referred to as T/EGFP) (Repina et al., 2019). Live-cell analysis with flow cytometry showed a heterogeneous response to Wnt stimulation at both low (0.1 μWmm–2) and high (1.4 μWmm–2) light intensities, confirming BRA immunostaining results (Figures S5C and S5D). Maximal light-induced activation increased BRA expression by ~33-fold over unilluminated optoWnt hESCs, which notably showed no detectable activation in the dark. Quantification of the percentage EGFP+ cells exhibited an exponential increase with light intensity that greatly exceeded activation achieved with recombinant Wnt3a protein (Figure 3D). Fitting to an increasing exponential decay function showed a very rapid increase in BRA expression at low light intensities (time constant τ = 0.07 μWmm–2 divided by the percentage of EGFP+, see Method Details), with saturation reached at ~0.4 μWmm–2. The high sensitivity at lower light intensities suggests a binary switch for the onset of BRA expression above a signaling threshold (>~0.05 μWmm–2), followed by a monotonic increase in BRA expression levels in a light dose-dependent manner.
Analysis of Phototoxicity Reveals No Detectable Effects on hESC Viability below a 1-μWmm–2 Illumination Intensity Threshold
Since high-intensity light can induce phototoxicity in cells (Tyssowski and Gray, 2019; Acker et al., 2016; Allen et al., 2015; Yizhar et al., 2011; Owen et al., 2019), we used the LAVA boards to analyze the phototoxicity threshold for hESC cultures. Inefficiencies in LED semiconductors cause heating, and although we incorporated a heat sink, cooling fans, and electrical vias in the circuit board design to conduct heat away from the cell culture plate, we observed heating of the LED dyes at continuous operation at high intensities (Figure 4A). Within the intensity range in which we did not observe significant heating (0–2 μWmm–2), we assessed potential phototoxic effects on hESC cultures after 48 h of continuous illumination. Above 1 μWmm–2, we observed a significant increase in apoptosis and membrane integrity markers Annexin IV and propidium iodide (PI), as well as a corresponding increase in cell debris and edge roughness of hESC colonies (Figures 4B and 4C). Thus, for all optogenetic stimulation, we used light intensity under 1 μWmm–2 (specifically, 0.8 μWmm–2). At this intensity, we saw no changes in cell proliferation after 48 h of illumination of wild-type hESCs (Figure 4D); no decrease in pluripotency markers SOX2, NANOG, and OCT4 (Figures S6A–S6C); and no spontaneous BRA+ mesendoderm differentiation (Figures S6A–S6D). Notably, since saturation in the percentage of BRA+ optoWnt cells occurred at ~0.4 μWmm–2 (Figure 3D), the optoWnt operating range falls below the 1 μWmm–2 phototoxicity threshold.
Figure 4. Low Phototoxicity during Continuous Optogenetic Stimulation of hESC Cultures.
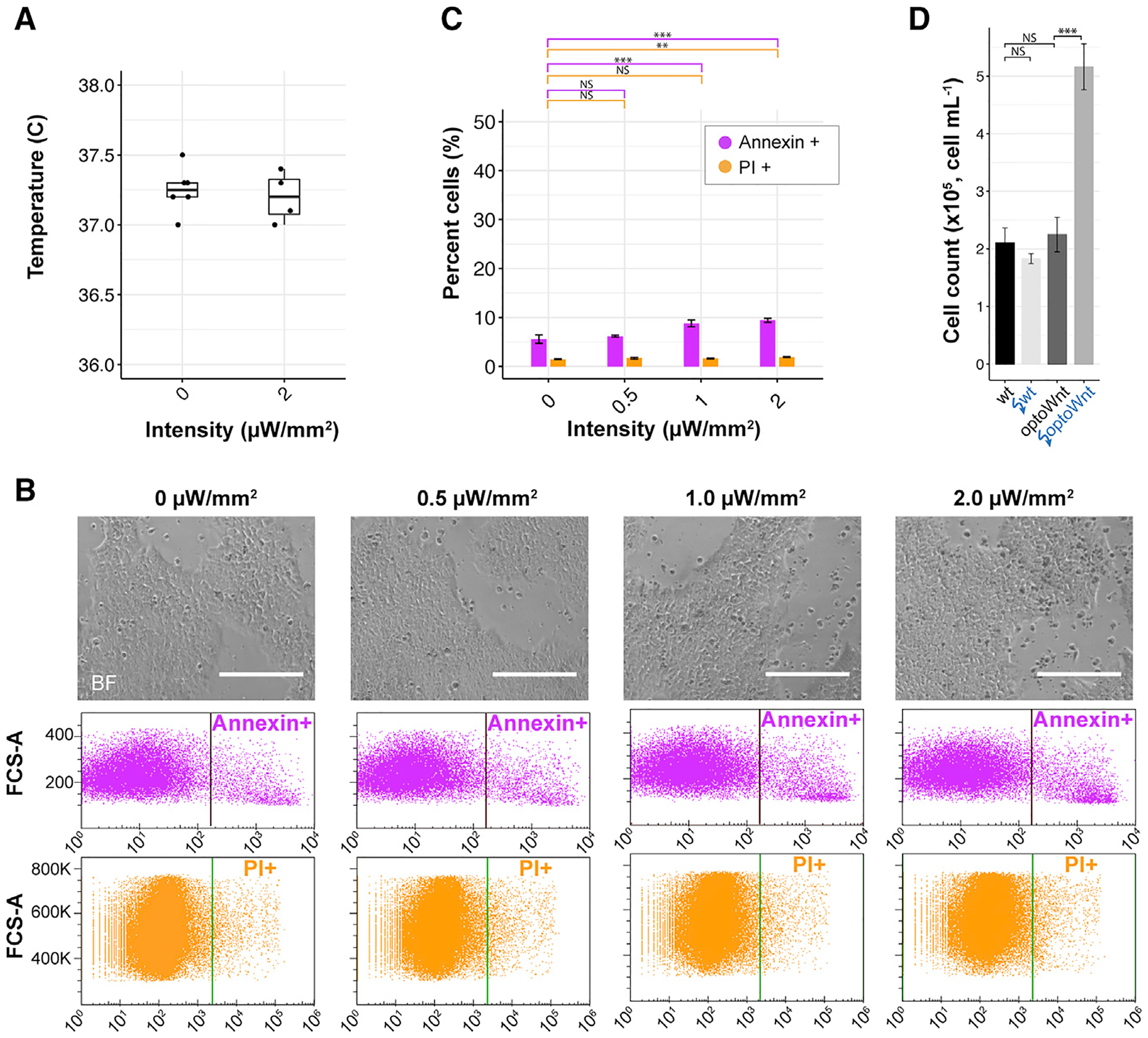
(A) Temperature of media after 24 h of continuous illumination at indicated light intensities. Each point represents the measurement from a single well.
(B) Bright-field images (top) of live wild-type hESC cultures illuminated at indicated light intensities after 48 h. Flow cytometry results (bottom) for Annexin V and propidium iodide (PI) stain of hESCs following 48 h of illumination. Positive gating indicated by vertical line. Scale bar, 250 μm.
(C) Quantification of Annexin V and PI shows a significant increase in apoptosis and cell death >1 μW/mm2 illumination intensity (pA = 0.0008, pPI = 0.39 at 0 versus 1 μW/mm2; pA = 0.0002, pPI = 0.009 at 0 versus 2 μW/mm2). No difference was observed between 0 and 0.5 μW/mm2 (pA = 0.61, pPI = 0.18). ANOVA followed by Tukey test. The graph shows means ± 1 SDs, n = 3 biological replicates.
(D) Endpoint cell count of wild-type and optoWnt hESCs after 48 h of continuous illumination at 0 or 0.8 μW/mm2. The graph shows means ± 1 SDs, n = 3 replicates. ANOVA followed by Tukey test.
Temporal Control of OptoWnt Using LAVA Devices
In addition to intensity control, LAVA boards enable the temporal control of illumination patterns (Figure 5A). Given the reversible oligomerization of Cry2 (Bugaj et al., 2013), we reasoned that the optoWnt system could be readily applied to the regulation of Wnt temporal dynamics.
Figure 5. Characterization of Temporal Control Using LAVA Devices.
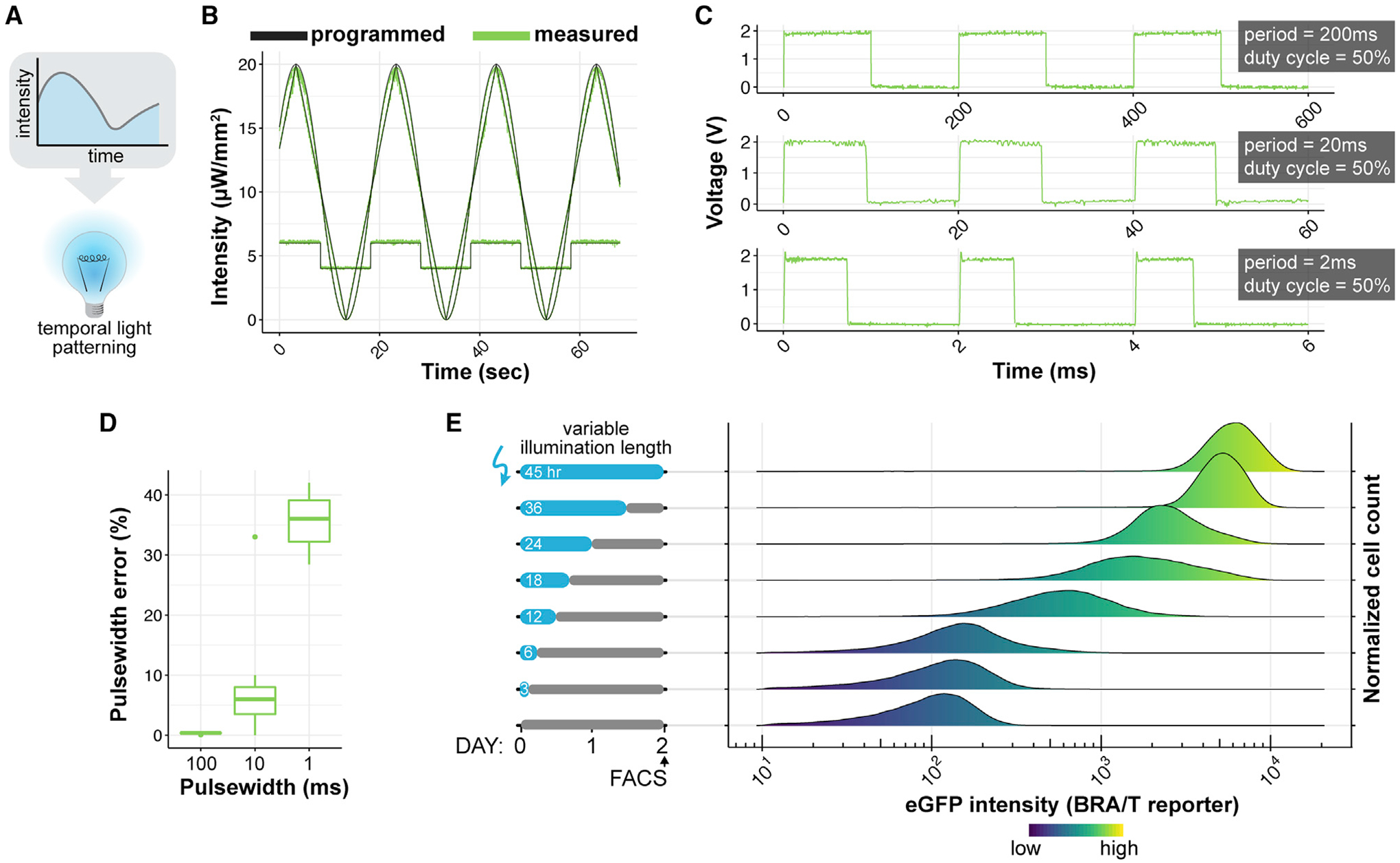
(A) Schematic of temporal light patterning in optogenetics.
(B) LAVA board well intensity as a function of time of various waveforms programmed through LAVA GUI. Programmed values shown in black and measured intensities shown in green.
(C) Measured illumination intensity during programmed blink sequences shows signal integrity at various frequencies. The voltage signal is directly measured from the optical power meter output with an oscilloscope and is proportional to illumination intensity. Blinking sequences were programmed through the LAVA GUI at a 50% duty cycle and at indicated pulse width on times.
(D) Percentage error in measured pulse width relative to programmed pulse width for blinking sequences shown in (C).
(E) OptoWnt hESCs containing an EGFP reporter for endogenous BRA/T activity were illuminated for varying lengths of time and analyzed by flow cytometry at a fixed endpoint. The graph shows histograms of EGFP expression at each illumination condition. Cell count histograms are normalized to total cells per condition (~30,000 cells), n = 3 biological replicates.
We designed a LAVA board GUI to allow users to input the desired temporal light patterns for each independent well (Figures S1E–S1G). Upon user input of the illumination parameters, the GUI wirelessly transmits the parameters to the LAVA device in the form of a JSON file. The file is parsed by the on-board Raspberry Pi microcontroller, which then sets LED intensities and temporal patterns for the duration of the experiment. The user can set each of the 24 wells to 1 of 3 modes: (1) constant illumination at a specified intensity; (2) blinking at a specified intensity, duty cycle, and period; or (3) a series of linear or sinusoidal functions at specified function parameters. Multiple piecewise functions can be programmed in sequence, enabling an immense variety of complex, time-varying light patterns (Figure 5B; Video S1). The shortest possible blink (i.e., the temporal resolution of the device) is set to 1 ms in firmware, although we observed a significant decrease in the accuracy and precision of stimulation at pulse widths <10 ms (Figures 5C and 5D).
We next asked whether sustained optoWnt activation is required to maintain BRA expression during hESC differentiation. We thus illuminated optoWnt cells expressing the T/EGFP reporter with varying durations of light and quantified EGFP fluorescence with flow cytometry at a fixed endpoint (Figure 5E). The duration of optoWnt stimulation determined endpoint EGFP intensity, suggesting that sustained illumination and optoWnt activation are necessary for a sustained β-CAT transcriptional response in this system.
Spatial Localization of Wnt Signaling and hESC Differentiation as a Model for Early Embryonic Wnt Patterning
Another advantage of optogenetic control is the ability to manipulate the spatial location of signal activation (Figure 6A). Although precise patterned illumination can be achieved with confocal scanning or the use of spatial light modulators (Repina et al., 2017), the cost and complexity of such microscope systems are restrictive, particularly for longer biological processes and/or large sample sizes. Instead, the majority of optogenetic studies leverage genetic specificity to target cell types of interest (Yizhar et al., 2011) or subcellular tagging to specific cell compartments (Benedetti et al., 2018) to achieve higher spatial resolutions. To readily incorporate spatial light patterning during cell culture, we designed die-cut photomasks that can be adhered to TC plates during illumination (Figure S7A). The mask feature size was limited by the cutting resolution of the die cutter to ~150 μm (Figure S7B). Using such photomasks, we were able to illuminate wells with arbitrary light patterns and induce optoWnt clustering only in illuminated regions (Figure 6B). Since the mask was placed underneath the TC plate, we anticipated that there would be light scattering through the plate bottom (170 μm-thick coverglass) that would compromise the mask resolution (Figure S7C). To quantify the extent of light scattering, we measured the number of LRP6 clusters as a function of distance beyond the mask edge and found that clusters were induced ~50 μm from the mask edge (Figure 6C). Thus, the photomask feature size was limited to ~150 μm, while patterning resolution (full width at half-maximum, see Method Details) was ~100 μm (Figure 6C).
Figure 6. Spatial Light Patterning with LAVA Devices for Localized OptoWnt Activation.
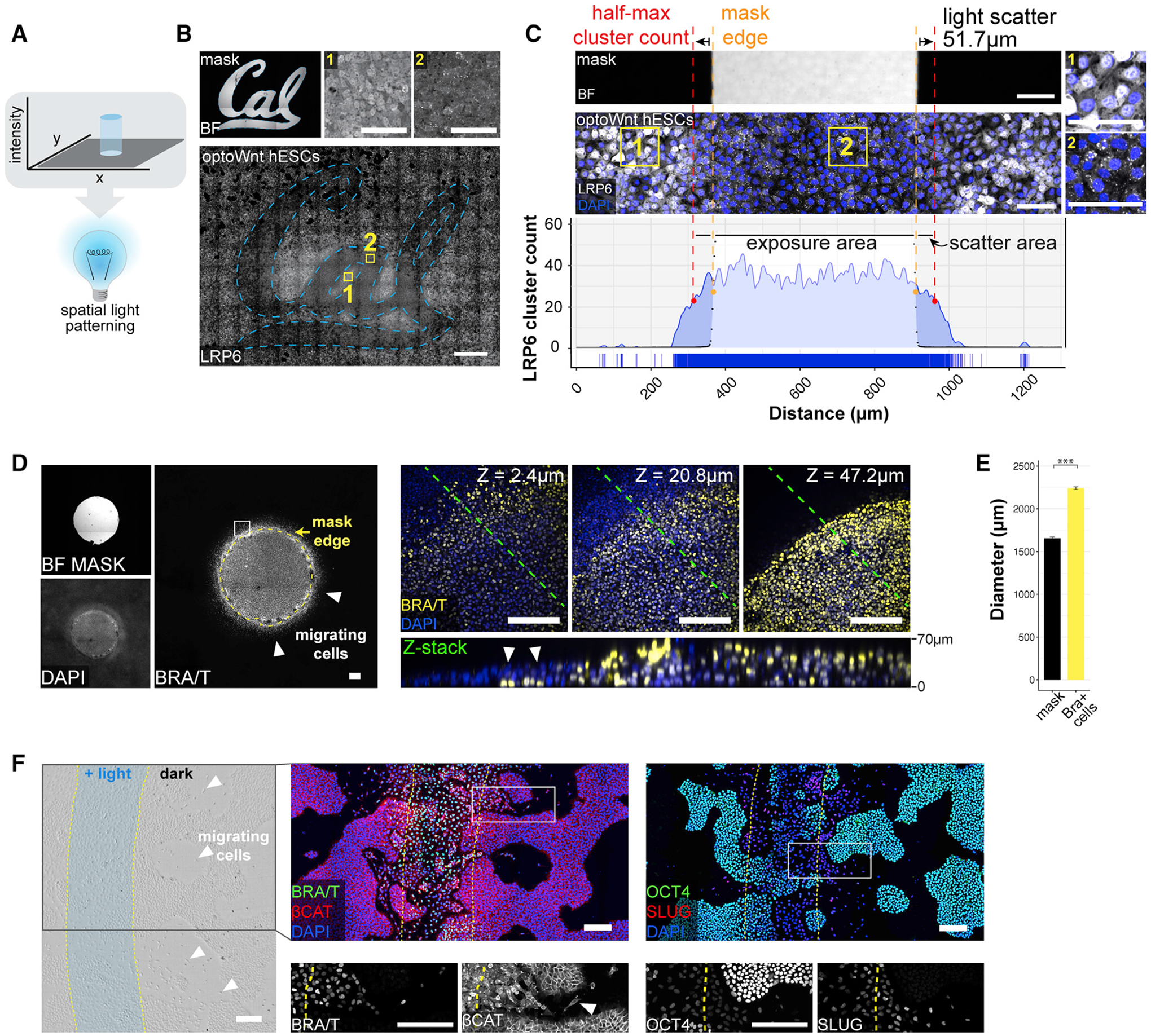
(A) Schematic of spatial light patterning in optogenetic experiments.
(B) Stitched bright-field and fluorescence confocal images of optoWnt hESCs illuminated with University of California, Berkeley (Cal) logo photomask. Immunostaining for LRP6 oligomers, with representative image of masked region (1) and illuminated region (2), as shown. Scale bar, 100 μm (top) and 1 mm (bottom).
(C) Quantification of light scattering through the bottom of a TC plate shows a ~50-μm spread (full width at half-maximal, red line) of optoWnt oligomers beyond photomask edge (orange line). Bright-field image of a photomask (top), fluorescence image of immunostaining for LRP6 (center), and quantification of LRP6 cluster count (bottom). Insets (1) and (2) show masked and illuminated regions, respectively. Scale bars, 100 μm.
(D) Patterned illumination with a 1.5-mm diameter circle of light. BRA immunostaining with photomask overlay shown in the left panel. Confocal z stacks of bottom (closest to coverslip, z = 2.4 μm), center (z = 20.8 μm), and top (z = 47.2 μm) cell layers show BRA+ cells localized beyond the photomask boundary and under the epithelial cell layer (white arrows). The bottom panel shows z slice through the cross-section highlighted with a green line. Scale bars, 100 μm.
(E) Quantification of BRA+ cell localization beyond the photomask edge. The mean diameter of the circular photomask pattern was quantified using the bright-field image channel, and the mean diameter of the BRA+ cell pattern was quantified through immunostaining, as shown in (D). The graph shows mean measured diameters ± 1 SDs, n = 3 biological replicates. Student’s t test (2-tailed). Scale bars, 200 μm.
(F) Patterned illumination with stripe of light, 500 μm in width. Bright-field image (left panel) with overlay of light pattern shows cells with mesenchymal morphology localized beyond the region of illumination (white arrows). Immunostaining for BRA and β-CAT (center panel), and OCT4 and SLUG (right panel). Overlay of light pattern is highlighted in yellow, and magnification (white box) shown below. Scale bars, 200 μm.
During mammalian development, a gradient of Wnt signaling emerges across the epiblast (Arnold and Robertson, 2009; Liu et al., 1999; Rivera-Pérez and Magnuson, 2005). Subsequently, cells in the posterior epiblast break away from the epithelial cell layer, migrating as single cells, which results in spatial rearrangement and morphological symmetry breaking of the epiblast in the region of the primitive streak (Thiery and Sleeman, 2006; Thiery et al., 2009; Williams et al., 2012). We modeled this spatial pattern of Wnt signaling by activating optoWnt hESCs in defined regions within the culture. To determine whether optoWnt activation could trigger cell migration outside the area of illumination, we used the LAVA devices to illuminate cells with a 1.5 mm diameter circular light pattern (Figure 6D). Immunostaining showed that BRA+ cells localized both within the illuminated region and up to ~300 μm beyond the illumination boundary (Figure 6E). Given the ~50 μm photomask scattering (Figure 6C), we excluded the possibility that cells were activated by scattered light. In addition, single-cell migration and tracking studies showed that illuminated optoWnt cells and wild-type cells treated with the Wnt agonist CHIR99021 are similarly capable of migrating >500 μm in the span of a 48 h time course experiment (Figures S7D–S7F; Videos S2 and S3), and we observed no spontaneous cell differentiation that may account for the observed patterning (Figures 3C, 3D, and S6D). Within the illuminated circle, BRA+ cells grew vertically upward and piled 4–6 cell diameters in height (~70 μm). In contrast, BRA+ cells beyond the illumination boundary localized underneath the epithelial cell layer (Figure 6D). These cells likely underwent an epithelial-to-mesenchymal transition (EMT) and migrated below the hESC colony, reflecting a migratory phenotype that has been analogously observed in hESCs undergoing EMT in conditions of high cell density and confinement (Warmflash et al., 2014).
Next, we illuminated optoWnt cultures with a stripe of light to mimic the spatial geometry of Wnt gradients established across the epiblast (Rivera-Pérez and Magnuson, 2005). Cells in the illuminated stripe broke away from the hESC colonies, adopted a mesenchymal morphology, and localized up to 500 μm beyond the boundary of the light pattern (Figure 6F). Immunostaining confirmed that these migratory cells expressed BRA, while the surrounding unilluminated cells retained epithelial morphology with no detectable BRA expression. In addition, migratory cells showed a decrease in OCT4 expression, a shift in β-CAT localization away from the plasma membrane, and an increase in snail family zinc finger 2 (SLUG) expression, all consistent with cells undergoing an EMT (Figure 6F). These data show that optogenetic Wnt activation is sufficient for inducing a migratory cell phenotype and that patterned illumination can be used as a tool to further study Wnt patterning and gastrulation-like events in ESC culture.
DISCUSSION
Spatial and temporal morphogen gradients have long been known to exist during development, although there has been a paucity of tools to create and perturb such signaling patterns. In this study, we combined the optogenetic control of Wnt signaling with engineered LAVA illumination devices to dynamically control hESC morphogen signaling in intensity, space, and time. Using this precise manipulation of morphogen signaling, we show that optogenetic Wnt patterning can be used to mimic embryonic morphogen patterning in vitro. This platform will enable future studies that elucidate spatial and temporal Wnt signaling thresholds and mechanisms of cell patterning in a variety of biological systems.
LAVA Devices for Quantitative, Spatiotemporal Control of Wnt Signaling Dynamics
With the continued development of new optogenetic proteins that perturb cell signaling and protein-protein interactions, there is a need for illumination devices that allow high-throughput intensity, spatial, and temporal control of stimulation. Previous illumination technologies for multiwell plate photostimulation have helped advance optogenetic studies, but have addressed only some of these individual design criteria, and do not fully characterize intensity parameters, uniformity, spatial and temporal resolution, or undesired heating and toxicity (Gerhardt et al., 2016; Davidson et al., 2013; Lee et al., 2013; Hannanta-Anan and Chow, 2016; Bugaj et al., 2018; Hennemann et al., 2018). Thus, in designing LAVA boards, we addressed such concerns and engineered devices that programmatically control photostimulation intensity, timing, and location at 0.005 μWmm–2, 10 ms, and 100 μm resolution, respectively. As optical system cost and ease of use can be significant barriers for the adoption of optogenetic studies, we also developed a GUI for simple configuration and wireless upload of desired intensity patterns to LAVA devices from a personal computer. Furthermore, to make optogenetic studies more accessible, we provide a detailed protocol, design files, and software source code for both 24-well and 96-well plate LAVA board assembly. Each device takes ~8 h and <$500 to fabricate and assemble.
One of the major optical design parameters we optimized during LAVA board design was illumination uniformity. Since the strength of induced Wnt signaling is dependent on light dosage (Figure 3D), nonuniformity can give rise to signal variation across the region of optogenetic stimulation. Specifically, the optoWnt dose response showed that Wnt activation can be sensitive to up to 0.025 μWmm–2 differences in intensity, especially at photostimulation near 0.1 μWmm–2 (Figure 3D). Despite this high sensitivity, the optimized LAVA board design is equipped to stimulate wells at sufficiently uniform levels via the optimization of LED placement and diffuser design. Illumination at 0.1 μWmm–2, for example, would have a 0.017 μWmm–2 intensity drop-off at the well edge (16.9% decrease) (Figure 2B), which is below the 0.025 μWmm–2 design criterion and thus results in uniform Wnt signal activation within the well. Although characterization of uniformity is critical for controlled signal activation, this design parameter has not been addressed in previous illumination systems (Gerhardt et al., 2016; Olson et al., 2014; Bugaj and Lim, 2019).
Equally important to illumination uniformity is the programmable control of intensity for dose-responsive and time-varying signal activation. Because we observed dose-dependent Wnt signal activation in response to increasing light intensities (Figure 3D), the LAVA boards combined with the optoWnt system can be readily applied to various biological systems in which Wnt signal strength directly regulates cell fate outcome. For example, different Wnt signal intensities can drive dose-dependent or divergent lineage specification during neural fate commitment (Kirkeby et al., 2012; Kiecker and Niehrs, 2001), hematopoiesis (Luis et al., 2011), and mesoderm differentiation (Davidson et al., 2012; Kempf et al., 2016). In addition, the optoWnt dose response suggests a role for LRP6 oligomer size in regulating β-CAT degradation (Kim et al., 2013; Li et al., 2012), which can be used to further shed light on the molecular regulation of Wnt signaling. In parallel, temporal modulation with LAVA devices opens a range of possible studies of Wnt signal dynamics (Figure 5). Combined with optoWnt reversibility (Bugaj et al., 2013) (Figure 5E), intricate studies of Wnt signaling thresholds and timing of signaling oscillations observed during development (Bao et al., 2017; Aulehla et al., 2003; Lian et al., 2012; Yu et al, 2008) can now be performed with much greater ease compared with microfluidic approaches (Sonnen et al., 2018) or manual pipetting (Massey et al., 2019).
Lastly, for spatial control, we implement a die-cut photomask to achieve spatial patterning at 100 μm resolution (Figure 6C), an improvement relative to previous innovative photomask approaches (Müller et al., 2014; Shao et al., 2018). Patterned illumination thus enables us to “paint” Wnt activation onto the target cells of interest and study how the shape, size, and intensity of spatial patterns influences differentiation and morphogenesis. Elegant optogenetic approaches for studying spatial signaling thresholds, such as patterned ERK signaling during early Drosophila embryogenesis (Johnson and Toettcher, 2019; Johnson et al., 2017), can now be extended to morphogen patterning in stem cell-based and organoid models for development.
Limitations
Having established proof-of-principle applications for LAVA devices and the optoWnt system, we want to comment on their limitations and on considerations for their generalizability to other optogenetic systems. Many hardware components of LAVA boards are exchangeable, so that LEDs of other colors can readily be incorporated to activate red or UV-based light sensors. Presently, the hardware design allows independent control of a single color, but it can be extended to multicolor stimulation by modifying the electronics design to incorporate more LED drivers.
To stimulate photosensory proteins, the light delivered by LAVA devices must be of sufficiently high intensity. We have found that the achieved illumination intensities (0–20 μWmm–2) are sufficient for optoWnt photostimulation, which requires merely 0.5–1 μWmm–2 for maximal activation (Figure 3D). However, LAVA board intensities may be insufficient for less-sensitive photosensory domains, which can require pulses of up to 1,000–10,000 μWmm–2 (Yizhar et al., 2011). LAVA board intensities can be increased by minimizing the light guide thickness, d (see Figure 2), but the maximal illumination intensity is still limited by the maximum forward current of LED dyes and by the phototoxicity of light at higher intensities (Tyssowski and Gray, 2019; Acker et al., 2016; Allen et al., 2015; Yizhar et al., 2011; Owen et al., 2019). Thus, for high-intensity applications, microscope-based systems and laser illumination would be more suitable, although phototoxicity should always be assessed.
Lastly, spatial patterning with a photomask is limited to a static, 2D light pattern, with the resolution limited by light scattering through the TC plate bottom surface. In the future, the pattern feature size can be improved by using chrome masks or laser-plotted mylar masks, which are limited to submicron and ~5–10 μm features, respectively (Folch, 2016). Such methods can also be used to achieve grayscale modulation (Folch, 2016).
In summary, LAVA devices are versatile tools for the photostimulation of cell cultures and can be applied to many biological systems for the study of cell signaling dynamics, high-throughput screens, and control of protein-protein interactions.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David V. Schaffer (schaffer@berkeley.edu).
Materials availability
Design files generated in this study have been deposited to the LAVAboard GitHub repository at https://github.com/LAVAboard/LAVA
Data and code availability
The code and graphical user interface files generated in this study have been deposited to the LAVAboard GitHub repository at https://github.com/LAVAboard/LAVA
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Embryonic stem cell culture
For routine culture and maintenance, all optogenetic and wild-type hESCs (H9, WiCell) (Thomson et al., 1998) lines were grown on Matrigel (Corning, lot # 7268012, 7275006) coated plates in mTeSR1 medium (STEMCELL Technologies) and 1% penicillin/streptomycin (Life Technologies) at 37°C and 5% CO2 with daily media changes. Optogenetic cells were cultured with hood lights off. For illumination experiments, cells were singularized with Accutase (STEMCELL Technologies) at 37°C for 5min and seeded onto Matrigel-coated 24-well plates (0030741021, Eppendorf, black-walled with 170 μm coverglass bottom) in E8 media (STEMCELL Technologies) with 5 μM ROCK inhibitor Y-27632 (Selleckchem) at a density of 35k – 70k cell cm−2. Wnt agonist CHIR99021 (Stemgent) or Wnt3a protein (StemRD) was diluted in E8 media and added to cells. Clonal optoWnt knock-in lines and clonal BRA/T reporter lines were generated through CRISPR/Cas9-mediated recombination as previously described (Repina et al., 2019).
METHOD DETAILS
Zemax modeling and uniformity optimization
The ray-tracing software Zemax OpticStudio was used in Non-Sequential mode to model illumination of a 24-well plate. Based on modeling results, the optimized configuration parameters are as follows: 5 surface-mount LEDs are symmetrically radially distributed around a 5mm-radius circle; the radius of each light guide is 8.25 mm; one 80°circular optical diffuser is placed between the two light guides and another onto the top light guide (i.e., between light guide and TC plate); the thickness of each light guide is 1.5 cm; and light guides are manufactured from black 3D-printed acrylic.
LAVA device construction
LAVA devices are constructed using two custom printed circuit boards (PCB) designed in EAGLE (Autodesk). PCB1 contains electronics for LED control while PCB2 is the power distribution board. For 24-well illumination, PCB1 contains solder pads for a circular array of 5 LEDs per well, which are connected in series and illuminate each well through two 3D-printed light guides and a series of diffusers (optical configuration optimized in Zemax, see below). For 96-well illumination, PCB1 contains solder pads for 1 LED per well of a 96-well plate; given the 24-channel LED driver, independent illumination control is possible for each group of 4 wells. For each channel, the ground wire connects to TLC5947 driver and is modulated with pulse-width modulation, while the positive terminal connects to the power plane of PCB1. PCB1 also contains headers for electrical connection to cooling fans. A heatsink mounts onto the bottom of PCB1, using thermally conductive adhesive (Arctic Silver, ASTA-7G), in a region without silk screen and thermally conductive electrical vias that draw heat away from surface-mount LEDs.
In PCB2, a power supply connects through a barrel power jack to power the LEDs through an LED driver (TLC5947, Adafruit). Power is also supplied to three fans and the Raspberry Pi microcontroller through switching voltage regulators.
On top of PCB1, optical assembly and TC plate is mounted in such a way that TC plate is illuminated from the bottom. It is critical that TC plate is made of black, opaque plastic with a thin, 170 μm coverslip bottom (Eppendorf Cell Imaging Plate, 24-well) to avoid light bleed-through between wells and thus enhance high spatial patterning resolution. The LED driver, PCB2, and the Raspberry Pi microcontroller are all mounted and electrically connected to PCB1, and the entire assembly is mounted onto an acrylic laser-cut base through vibration-dampening mounts. The base contains rubber footpegs to reduce static or electrical shorting with the TC incubator racks.
For details of LAVA board design and fabrication, see the LAVA GitHib repository which includes a detailed protocol, design files, and software source code for 24-well and 96-well plate LAVA board assembly. In brief, LAVA boards were constructed in the CITRIS Invention Lab, a UC Berkeley rapid prototyping facility, using the following equipment: 3D printer (Ultimaker 3), laser cutter (Speedy 400, Trotec), and standard soldering tools. Photomasks were dye-cut on a vinyl cutter (CAMM-1 GS-24, Roland).
LAVA software control and graphical user interface
The LEDs are controlled by an Adafruit 24-Channel 12-bit PWM LED driver with an SLI interface to a Raspberry Pi Zero W. The 12-bit PWM resolution allows 4086 unique illumination intensity levels over the LED operating range. For ease of use, a GUI has been written in Java and is conveniently packed into an executable file. This interface allows for independent control of each of the 24 channels. To accommodate the variety of experimental conditions, each LED can be programmed to a constant illumination, a blinking pattern, or a series of linear and sinusoidal patterns. Since each board has slightly different intensity characteristics, the intensity to PWM calibration parameters are input at runtime. Sinusoidal and linear functions are interpolated at a frequency of 1 Hz whereas blinking patterns have been tested up to 100 Hz. Since the LED board’s USB port may be inaccessible during certain experiments, it is possible to wirelessly upload new illumination settings from any Wi-Fi capable computer. The Java program parses the illumination settings and PWM calibration parameters, packages them into a JSON file, and transmits these settings to the Pi over an SFTP channel.
Upon booting the Raspberry Pi, a C++ script executes, checks the device for previous illumination settings and resumes the patterned illumination if found. The Pi polls for changes in the JSON file every few seconds, so the changes of a newly uploaded pattern will be reflected without an additional reboot. It should be noted that the decision to use C++ was motivated by a desire to break through certain speed limitations posed by an interpreted language’s rate of execution. A Python implementation was completed and included on the Pi operating system to easily generate custom pattern scripts, but only the compiled C++ version is able to drive the 24 channels at the desired refresh rate.
LAVA device intensity, uniformity, heating, and spectral characterization
For LAVA board characterization, light intensity was measured with a power meter (PM100D, Thorlabs) with a photodiode power sensor (S121C, Thorlabs) at 470 nm. For high-resolution measurement of temporal light patterns, the Thorlabs power meter was connected to an oscilloscope. Well uniformity measurements were performed by imaging wells on a Nikon Z100 microscope with a wide-field lens (Nicole AZ-Plan Apo 1x) and sCMOS monochrome camera (pco.edge 5.5). Image intensity was quantified as a function of distance in Fiji (Schindelin et al., 2012). Well temperature was measured with a digital multimeter (87-V, Fluke) with a Type-K thermocouple probe (80BK-A, Fluke). LED emission spectra were measured with a spectrometer (Red Tide USB650, Ocean Optics).
Immunostaining and imaging
Cells were fixed with 3% PBS – paraformaldehyde (ThermoFisher) for 20 min at room temperature and subsequently washed three times with PBS. Blocking and permeabilization was done with 5% donkey serum (D9663, Sigma-Aldrich) and 0.3% Triton X-100 (Fisher Scientific) (PBS-DT) for 1 hour. Cells were incubated with primary antibodies (Table S1) at 4°C overnight, then washed three times with PBS, and incubated with fluorescently conjugated secondary antibodies (Invitrogen) at 1:250 dilution for 1 hour at room temperature. Both primary and secondary antibodies were diluted in PBS-DT. Cells were washed with PBS and stained with 0.1 μg mL −1 DAPI nuclear stain (ThermoFisher) prior to imaging. Confocal imaging was performed on a Perkin Elmer Opera Phenix system (QB3 High-Throughput Screening Facility). Brightfield and widefield fluorescence imaging was performed on a Zeiss AxioObserver epi-fluorescent microscope (CIRM/QB3 Shared Stem Cell Facility).
Image analysis
Microscopy image processing, including stitching and z-slice projection, was performed in Fiji (Schindelin et al., 2012) and image quantification was performed in CellProfiler (Carpenter et al., 2006) with custom analysis pipelines detailed below.
For quantification of BRA, SOX2, OCT4, and NANOG nuclear fluorescence intensity, nuclei stained with DAPI were identified and used to generate a binary mask applied to the appropriate fluorescence channel. The mean fluorescence intensity per cell nucleus was calculated for each cell in a given field of view. A threshold defining ‘positive’ cells was determined from signal intensity of negative control wells. Quantification of LRP6 oligomer size was similarly performed in CellProfiler using the IdentifyPrimaryObjects module.
Spatial light patterning resolution was quantified by illuminating optoWnt hESC wells with a dye-cut photomask for 1 hr at 1 μWmm−2and immunostaining for LRP6. With mask attached to plate, wells were imaged in brightfield at varying focus positions, to capture images of both the photomask and cells. The mask was subsequently removed, and cells were imaged in brightfield and fluorescence channels (with 5x and 60x water-immersion objectives on a Perkin Elmer Opera Phenix confocal system). The mask image was aligned to the LRP6 fluorescence channel using the brightfield channel in Fiji (Schindelin et al., 2012). An identical alignment procedure was used for all spatial patterning data shown. For quantification of light scattering, multiple fields of view were stitched together and oligomers were manually counted in Fiji based on the LRP6 fluorescence channel. The pixel location of each identified LRP6 oligomer was recorded. Since the photomask pattern was a vertical stripe, the oligomer counts were summed vertically over multiple fields of view to quantify the spatial distribution of oligomers relative to the photomask. The spatial extent of photostimulation due to light scatter was calculated by determining the full width half maximum (FWHM) of the LRP6 oligomer distribution outside of the photomask.
Live single-cell imaging and tracking
Wild-type hESCs were seeded at a density of 35,000 cells cm−2, with 10% of cells labeled with GFP expression driven by CAG promoter knocked in to the AAVS1 locus. OptoWnt hESCs were also seeded at a density of 35,000 cells cm−2, with 10% of cells labeled with nuclearly-localized mVenus. Such sparse labeling of cells enabled tracking accuracy. A sealing membrane (Breathe-Easy, Sigma-Aldrich) was applied to plates prior to imaging. Plates were imaged on a Molecular Devices ImageXpress Micro (IXM) imaging system with environmental control (37°C, 5% CO2, and humidity control) using a 10x objective. For each condition, 4 sites were imaged at 12 min intervals. Optogenetic stimulation was delivered from the fluorescence light source (SOLA Light Engine, Lumencor) set to 5% intensity, passing through the 10x objective and GFP filter set (472/30nm). Measured power at the sample was 2.82 mW. Optogenetic stimulation was delivered for 3 min at each site prior to imaging of each time point (i.e., for 3 min every 12 min) in a sequence of short light pulses (500 ms on-pulse, 10 s off-pulse). Single-cell tracking analysis was performed with Fiji using the Manual Tracking plugin.
Flow cytometry and analysis
Cells were lifted with Accutase at 37°C for 5 min, centrifuged, and resuspended in flow buffer (0.5% bovine serum albumin in PBS (w/v)) for analysis. For phototoxicity experiments, centrifuged cells were resuspended in PBS and stained with AlexaFluor 488-conjugated annexin V and 1 μg mL−1 propidium iodide (PI) as per kit manufacturer recommendations (V13241, ThermoFisher). Cells were analyzed on a BD LSR Fortessa X20 (UC Berkeley Cancer Research Laboratory Flow Cytometry Facility). Cell counting for proliferation assays was performed using a ThermoFisher Attune (UC Berkeley CIRM/QB3 Shared Stem Cell Facility) by measuring the number of cells per unit volume.
Data analysis was performed with FlowJo 10 software. To determine the fraction of BRA-eGFP+ cells after light treatment, gating was set such that less than 0.5% of undifferentiated wild-type hESCs were positive for eGFP and mCherry. Data were fit to an increasing exponential decay curve using a nonlinear least-squares model in R:
where y represents the percent BRA-eGFP+ cells and x is light intensity (μWmm−2). The curve asymptotically approaches the maximum percentage of BRA-eGFP+ cells (Amax = 97.1%) with time constant τ = 0.07 μWmm−2 / percent eGFP+.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are presented as mean ± 1 standard deviation (s.d.) unless otherwise specified. Statistical significance was determined by Student’s t test (two-tail) between two groups, and three or more groups were analyzed by one-way analysis of variance (ANOVA) followed by Tukey test. p < 0.05 was considered statistically significant (NS p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001).
Additional resources
Detailed assembly protocols, bill of materials, design files, and code for 24-well and 96-well LAVA board assembly can be found at the GitHub repository at https://github.com/LAVAboard/LAVA.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| LRP6 | Abcam | Cat#134146 |
| BRACHYURY | Santa Cruz | Cat#20109; RRID:AB_2255702 |
| Beta-CATENIN | Cell Signaling | Cat#8480; RRID:AB_11127855 |
| SLUG | Cell Signaling | Cat#9585; RRID:AB_2239535 |
| OCT4 | Santa Cruz | Cat#5279; RRID:AB_628051 |
| SOX2 | Abcam | Cat#97959; RRID:AB_2341193 |
| NANOG | Abcam | Cat#62734; RRID:AB_956161 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Matrigel | Corning | Cat#354277 |
| Y-27632 | Selleckchem | Cat#S1049 |
| CHIR99021 | Stemgent | Cat#04-0004 |
| Wnt3a recombinant protein | StemRD | Cat#w3a-h-005 |
| Deposited Data | ||
| Design files, BOM, and code | GitHub | https://github.com/LAVAboard/LAVA |
| Experimental Models: Cell Lines | ||
| H9 Human Embryonic Stem Cells | WiCell | N/A |
| Software and Algorithms | ||
| LAVA graphical user interface | GitHub | https://github.com/LAVAboard/LAVA |
Highlights.
Design of illumination devices for optogenetic photostimulation of multiwell plates
Spatiotemporal control of canonical Wnt signaling in human embryonic stem cells
Dose-dependent optogenetic Wnt activation
Spatial light patterns model embryonic presentation of Wnt signals in vitro
ACKNOWLEDGMENTS
We thank Laura Waller for discussions on optical design, as well as Nick Antipa and Zachary Phillips for helpful discussions on Zemax modeling and uniformity characterization. We are grateful to Christopher Myers, Mitchell Karchemsky, and Rundong Tian at the University of California, Berkeley Center for Information Technology Research in the Interest of Society (CITRIS) Invention Lab for assistance and discussions of rapid prototyping design and techniques. We also thank members of the Schaffer lab for helpful discussions. We are grateful to Mary West from the QB3 High-Throughput Screening Facility and the California Institute for Regenerative Medicine (CIRM)/QB3 Shared Stem Cell Facility and Hector Nolla from the Cancer Research Laboratory (CRL) Flow Cytometry Facility for technical assistance. Funding supporting this work was provided by the US National Institutes of Health (R01NS087253, to D.V.S. and R.S.K), the US National Science Foundation (to N.A.R.), and the Siebel Scholars Foundation (to N.A.R.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2020.107737.
DECLARATION OF INTERESTS
N.A.R., T.M., and D.V.S. are co-inventors on related intellectual property.
SUPPORTING CITATIONS
The following references appear in the Supplemental Information: Banerjee et al., 2007.
REFERENCES
- Acker L, Pino EN, Boyden ES, and Desimone R (2016). FEF inactivation with improved optogenetic methods. Proc. Natl. Acad. Sci. USA 113, E7297–E7306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen BD, Singer AC, and Boyden ES (2015). Principles of designing interpretable optogenetic behavior experiments. Learn. Mem 22, 232–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold SJ, and Robertson EJ (2009). Making a commitment: cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol 10, 91–103. [DOI] [PubMed] [Google Scholar]
- Arnold SJ, Stappert J, Bauer A, Kispert A, Herrmann BG, and Kemler R (2000). Brachyury is a target gene of the Wnt/β-catenin signaling pathway. Mech. Dev 91, 249–258. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Wehrle C, Brand-Saberi B, Kemler R, Gossler A, Kanzler B, and Herrmann BG (2003). Wnt3a plays a major role in the segmentation clock controlling somitogenesis. Dev. Cell 4, 395–406. [DOI] [PubMed] [Google Scholar]
- Banerjee R, Schleicher E, Meier S, Viana RM, Pokorny R, Ahmad M, Bittl R, and Batschauer A (2007). The signaling state of Arabidopsis cryptochrome 2 contains flavin semiquinone. J. Biol. Chem 282, 14916–14922. [DOI] [PubMed] [Google Scholar]
- Bao X, Lian X, Hacker TA, Schmuck EG, Qian T, Bhute VJ, Han T, Shi M, Drowley L, Plowright AT, et al. (2017). Long-term self-renewing human epicardial cells generated from pluripotent stem cells under defined xeno-free conditions. Nat. Biomed. Eng 1, 0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedetti L, Barentine AES, Messa M, Wheeler H, Bewersdorf J, and De Camilli P (2018). Light-activated protein interaction with high spatial subcellular confinement. Proc. Natl. Acad. Sci. USA 115, E2238–E2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, and Nusse R (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat. Commun 3, 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, and Deisseroth K (2005). Millisecond-timescale, genetically targeted optical control of neural activity. Nat. Neurosci 8, 1263–1268. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, and Lim WA (2019). High-throughput multicolor optogenetics in microwell plates. Nat. Protoc 14, 2205–2228. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, Choksi AT, Mesuda CK, Kane RS, and Schaffer DV (2013). Optogenetic protein clustering and signaling activation in mammalian cells. Nat. Methods 10, 249–252. [DOI] [PubMed] [Google Scholar]
- Bugaj LJ, Spelke DP, Mesuda CK, Varedi M, Kane RS, and Schaffer DV (2015). Regulation of endogenous transmembrane receptors through optogenetic Cry2 clustering. Nat. Commun 6, 6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugaj LJ, Sabnis AJ, Mitchell AJ, Garbarino E, Toettcher JE, Bivona TG, and Lim WA (2018). Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, eaao3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čapek D, Smutny M, Tichy AM, Morri M, Janovjak H, and Heisenberg CP (2019). Light-activated Frizzled7 reveals a permissive role of non-canonical wnt signaling in mesendoderm cell migration. eLife 8, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo-Reid L, Yang W, Bando Y, Peterka DS, and Yuste R (2016). Imprinting and recalling cortical ensembles. Science 353, 691–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson KC, Adams AM, Goodson JM, McDonald CE, Potter JC, Berndt JD, Biechele TL, Taylor RJ, and Moon RT (2012). Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc. Natl. Acad. Sci. USA 109, 4485–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EA, Basu AS, and Bayer TS (2013). Programming microbes using pulse width modulation of optical signals. J. Mol. Biol 425, 4161–4166. [DOI] [PubMed] [Google Scholar]
- Duan L, Hope J, Ong Q, Lou HY, Kim N, McCarthy C, Acero V, Lin MZ, and Cui B (2017). Understanding CRY2 interactions for optical control of intracellular signaling. Nat. Commun 8, 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch A (2016). Introduction to BioMEMS, First Edition (CRC Press; ). [Google Scholar]
- Gerhardt KP, Olson EJ, Castillo-Hair SM, Hartsough LA, Landry BP, Ekness F, Yokoo R, Gomez EJ, Ramakrishnan P, Suh J, et al. (2016). An open-hardware platform for optogenetics and photobiology. Sci. Rep 6, 35363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guglielmi G, Barry JD, Huber W, and De Renzis S (2015). An Optogenetic Method to Modulate Cell Contractility during Tissue Morphogenesis. Dev. Cell 35, 646–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guntas G, Hallett RA, Zimmerman SP, Williams T, Yumerefendi H, Bear JE, and Kuhlman B (2015). Engineering an improved light-induced dimer (iLID) for controlling the localization and activity of signaling proteins. Proc. Natl. Acad. Sci. USA 112, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannanta-Anan P, and Chow BY (2016). Optogenetic Control of Calcium Oscillation Waveform Defines NFAT as an Integrator of Calcium Load. Cell Syst 2, 283–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann P, and Nagel G (2013). From channelrhodopsins to optogenetics. EMBO Mol. Med 5, 173–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennemann J, Iwasaki RS, Grund TN, Diensthuber RP, Richter F, and Möglich A (2018). Optogenetic Control by Pulsed Illumination. ChemBio-Chem 19, 1296–1304. [DOI] [PubMed] [Google Scholar]
- Hernandez O, Papagiakoumou E, Tanese D, Fidelin K, Wyart C, and Emiliani V (2016). Three-dimensional spatiotemporal focusing of holographic patterns. Nat. Commun 7, 11928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang A, Amourda C, Zhang S, Tolwinski NS, and Saunders TE (2017). Decoding temporal interpretation of the morphogen Bicoid in the early Drosophila embryo. eLife 6, e26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo E, Quinkler T, and De Renzis S (2018). Guided morphogenesis through optogenetic activation of Rho signalling during early Drosophila embryogenesis. Nat. Commun 9, 2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, and Toettcher JE (2018). Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol 52, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, and Toettcher JE (2019). Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev. Cell 48, 361–370.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson HE, Goyal Y, Pannucci NL, Schüpbach T, Shvartsman SY, and Toettcher JE (2017). The Spatiotemporal Limits of Developmental Erk Signaling. Dev. Cell 40, 185–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kainrath S, Stadler M, Reichhart E, Distel M, and Janovjak H (2017). Green-Light-Induced Inactivation of Receptor Signaling Using Cobalamin-Binding Domains. Angew. Chem. Int. Ed. Engl 56, 4608–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H, Olmer R, Haase A, Franke A, Bolesani E, Schwanke K, Robles-Diaz D, Coffee M, Göhring G, Dräger G, et al. (2016). Bulk cell density and Wnt/TGFbeta signalling regulate mesendodermal patterning of human pluripotent stem cells. Nat. Commun 7, 13602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiecker C, and Niehrs C (2001). A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development 128, 4189–4201. [DOI] [PubMed] [Google Scholar]
- Kim S-E, Huang H, Zhao M, Zhang X, Zhang A, Semonov MV, Mac-Donald BT, Zhang X, Garcia Abreu J, Peng L, and He X (2013). Wnt stabilization of β-catenin reveals principles for morphogen receptor-scaffold assemblies. Science 340, 867–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura-Yoshida C, Nakano H, Okamura D, Nakao K, Yonemura S, Belo JA, Aizawa S, Matsui Y, and Matsuo I (2005). Canonical Wnt signaling and its antagonist regulate anterior-posterior axis polarization by guiding cell migration in mouse visceral endoderm. Dev. Cell 9, 639–650. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, and Parmar M (2012). Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Rep 1, 703–714. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee J, Kim T, and Lee SK (2013). Switchable gene expression in Escherichia coli using a miniaturized photobioreactor. PLoS One 8, e52382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levskaya A, Weiner OD, Lim WA, and Voigt CA (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li VSW, Ng SS, Boersema PJ, Low TY, Karthaus WR, Gerlach JP, Mohammed S, Heck AJ, Maurice MM, Mahmoudi T, and Clevers H (2012). Wnt signaling through inhibition of β-catenin degradation in an intact Axin1 complex. Cell 149, 1245–1256. [DOI] [PubMed] [Google Scholar]
- Lian X, Hsiao C, Wilson G, Zhu K, Hazeltine LB, Azarin SM, Raval KK, Zhang J, Kamp TJ, and Palecek SP (2012). Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl. Acad. Sci. USA 109, E1848–E1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley RC, Gill JG, Kyba M, Murphy TL, and Murphy KM (2006). Canonical Wnt signaling is required for development of embryonic stem cell-derived mesoderm. Development 133, 3787–3796. [DOI] [PubMed] [Google Scholar]
- Liu P, Wakamiya M, Shea MJ, Albrecht U, Behringer RR, and Bradley A (1999). Requirement for Wnt3 in vertebrate axis formation. Nat. Genet 22, 361–365. [DOI] [PubMed] [Google Scholar]
- Luis TC, Naber BA, Roozen PP, Brugman MH, de Haas EF, Ghazvini M, Fibbe WE, van Dongen JJ, Fodde R, and Staal FJ (2011). Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 9, 345–356. [DOI] [PubMed] [Google Scholar]
- Massey J, Liu Y, Alvarenga O, Saez T, Schmerer M, and Warmflash A (2019). Synergy with TGFβ ligands switches WNT pathway dynamics from transient to sustained during human pluripotent cell differentiation. Proc. Natl. Acad. Sci. USA 116, 4989–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller K, Zurbriggen MD, and Weber W (2014). Control of gene expression using a red- and far-red light-responsive bi-stable toggle switch. Nat. Protoc 9, 622–632. [DOI] [PubMed] [Google Scholar]
- Nagel G, Szellas T, Huhn W, Kateriya S, Adeishvili N, Berthold P, Ollig D, Hegemann P, and Bamberg E (2003). Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl. Acad. Sci. USA 100, 13940–13945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolenko V, Poskanzer KE, and Yuste R (2007). Two-photon photostimulation and imaging of neural circuits. Nat. Methods 4, 943–950. [DOI] [PubMed] [Google Scholar]
- Oates AC, Gorfinkiel N, González-Gaitán M, and Heisenberg CP (2009). Quantitative approaches in developmental biology. Nat. Rev. Genet 10, 517–530. [DOI] [PubMed] [Google Scholar]
- Olson EJ, Hartsough LA, Landry BP, Shroff R, and Tabor JJ (2014). Characterizing bacterial gene circuit dynamics with optically programmed gene expression signals. Nat. Methods 11, 449–455. [DOI] [PubMed] [Google Scholar]
- Owen SF, Liu MH, and Kreitzer AC (2019). Thermal constraints on in vivo optogenetic manipulations. Nat. Neurosci 22, 1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Roska B, and Häusser M (2013). Targeting neurons and photons for optogenetics. Nat. Neurosci 16, 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packer AM, Russell LE, Dalgleish HW, and Häusser M (2015). Simultaneous all-optical manipulation and recording of neural circuit activity with cellular resolution in vivo. Nat. Methods 12, 140–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiakoumou E, de Sars V, Oron D, and Emiliani V (2008). Patterned two-photon illumination by spatiotemporal shaping of ultrashort pulses. Opt. Express 16, 22039–22047. [DOI] [PubMed] [Google Scholar]
- Papagiakoumou E, Anselmi F, Bègue A, de Sars V, Glückstad J, Isacoff EY, and Emiliani V (2010). Scanless two-photon excitation of channelrhodopsin-2. Nat. Methods 7, 848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pégard NC, Mardinly AR, Oldenburg IA, Sridharan S, Waller L, and Adesnik H (2017). Three-dimensional scanless holographic optogenetics with temporal focusing (3D-SHOT). Nat. Commun 8, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Yizhar O, Grewe B, Ramakrishnan C, Wang N, Goshen I, Packer AM, Peterka DS, Yuste R, Schnitzer MJ, and Deisseroth K (2012). Two-photon optogenetic toolbox for fast inhibition, excitation and bi-stable modulation. Nat. Methods 9, 1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repina NA, Rosenbloom A, Mukherjee A, Schaffer DV, and Kane RS (2017). At Light Speed: Advances in Optogenetic Systems for Regulating Cell Signaling and Behavior. Annu. Rev. Chem. Biomol. Eng 8, 13–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repina NA, Bao X, Zimmermann JA, Joy DA, Kane RS, and Schaffer DV (2019). Optogenetic control of Wnt signaling for modeling early embryo-genic patterning with human pluripotent stem cells. bioRxiv 8, 665695. [Google Scholar]
- Richter F, Scheib US, Mehlhorn J, Schubert R, Wietek J, Gernetzki O, Hegemann P, Mathes T, and Möglich A (2015). Upgrading a microplate reader for photobiology and all-optical experiments. Photochem. Photobiol. Sci 14, 270–279. [DOI] [PubMed] [Google Scholar]
- Rivera-Pérez JA, and Magnuson T (2005). Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Dev. Biol 288, 363–371. [DOI] [PubMed] [Google Scholar]
- Sako K, Pradhan SJ, Barone V, Inglés-Prieto Á, Müller P, Ruprecht V, Čapek D, Galande S, Janovjak H, and Heisenberg CP (2016). Optogenetic Control of Nodal Signaling Reveals a Temporal Pattern of Nodal Signaling Regulating Cell Fate Specification during Gastrulation. Cell Rep 16, 866–877. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Wang M, Yu G, Zhu S, Yu Y, Heng BC, Wu J, and Ye H (2018). Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc. Natl. Acad. Sci. USA 115, E6722–E6730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Lauschke VM, Uraji J, Falk HJ, Petersen Y, Funk MC, Beaupeux M, François P, Merten CA, and Aulehla A (2018). Modulation of Phase Shift between Wnt and Notch Signaling Oscillations Controls Mesoderm Segmentation. Cell 172, 1079–1090.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland D, Lin Y, Wagner E, Hope CM, Zayner J, Antoniou C, Sos-nick TR, Weiss EL, and Glotzer M (2012). TULIPs: tunable, light-controlled interacting protein tags for cell biology. Nat. Methods 9, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, and Suemori H (2008). Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development 135, 2969–2979. [DOI] [PubMed] [Google Scholar]
- Thiery JP, and Sleeman JP (2006). Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol 7, 131–142. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, and Nieto MA (2009). Epithelialmesenchymal transitions in development and disease. Cell 139, 871–890. [DOI] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Toettcher JE, Gong D, Lim WA, and Weiner OD (2011). Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods 8, 837–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toettcher JE, Weiner OD, and Lim WA (2013). Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker CL, Vrana JD, and Kennedy MJ (2014). Tools for controlling protein interactions using light. Curr. Protoc. Cell Biol 64, 17.16.1–17.16.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyssowski KM, and Gray JM (2019). Blue light induces neuronal-activity-regulated gene expression in the absence of optogenetic proteins. bioRxiv 12, 572370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, He L, Wu YI, Hahn KM, and Montell DJ (2010). Light-mediated activation reveals a key role for Rac in collective guidance of cell movement in vivo. Nat. Cell Biol 12, 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warmflash A, Sorre B, Etoc F, Siggia ED, and Brivanlou AH (2014). A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods 11, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams M, Burdsal C, Periasamy A, Lewandoski M, and Sutherland A (2012). Mouse primitive streak forms in situ by initiation of epithelial to mesenchymal transition without migration of a cell population. Dev. Dyn 241, 270–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi TP, Takada S, Yoshikawa Y, Wu N, and McMahon AP (1999). T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev 13, 3185–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M, Sadaghiani AM, Hsueh B, and Dolmetsch RE (2009). Induction of protein-protein interactions in live cells using light. Nat. Biotechnol 27, 941–945. [DOI] [PubMed] [Google Scholar]
- Yizhar O, Fenno LE, Davidson TJ, Mogri M, and Deisseroth K (2011). Optogenetics in neural systems. Neuron 71, 9–34. [DOI] [PubMed] [Google Scholar]
- Yu W, McDonnell K, Taketo MM, and Bai CB (2008). Wnt signaling determines ventral spinal cord cell fates in a time-dependent manner. Development 135, 3687–3696. [DOI] [PubMed] [Google Scholar]
- Zeng L, Fagotto F, Zhang T, Hsu W, Vasicek TJ, Perry WL 3rd, Lee JJ, Tilghman SM, Gumbiner BM, and Costantini F (1997). The mouse Fused locus encodes Axin, an inhibitor of the Wnt signaling pathway that regulates embryonic axis formation. Cell 90, 181–192. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code and graphical user interface files generated in this study have been deposited to the LAVAboard GitHub repository at https://github.com/LAVAboard/LAVA


