ABSTRACT
Most motile cilia have a stereotyped structure of nine microtubule outer doublets and a single central pair of microtubules. The central pair of microtubules are surrounded by a set of proteins, termed the central pair apparatus. A specific kinesin, Klp1 projects from the central pair and contributes to ciliary motility in Chlamydomonas. The vertebrate ortholog, Kif9, is required for beating in mouse sperm flagella, but the mechanism of Kif9/Klp1 function remains poorly defined. Here, using Xenopus epidermal multiciliated cells, we show that Kif9 is necessary for ciliary motility and the proper distal localization of not only central pair proteins, but also radial spokes and dynein arms. In addition, single-molecule assays in vitro reveal that Xenopus Kif9 is a long-range processive motor, although it does not mediate long-range movement in ciliary axonemes in vivo. Together, our data suggest that Kif9 is integral for ciliary beating and is necessary for proper axonemal distal end integrity.
KEY WORDS: Cilia, Kif9, Kinesin, Central pair
Summary: An understudied kinesin, Kif9, is necessary for proximal to distal patterning of the motile cilium.
INTRODUCTION
Motile cilia are essential for fluid flow across epithelia and for cell motility in a variety of contexts in the animal kingdom (Brooks and Wallingford, 2014; Spassky and Meunier, 2017; Ishikawa, 2017). Most motile cilia have a conserved radial structure of nine microtubule outer doublets and a single central pair of microtubules. Ciliary motility relies on several different protein complexes that are organized throughout the axoneme in a specific fashion. These include the inner and outer dynein arms on the outer microtubule doublets and radial spokes that connect to inner dynein arms and project towards to central pair (Ishikawa, 2017). In addition, studies indicate that ∼50 different proteins surround the central pair to form the central pair apparatus (CA) (Loreng and Smith, 2017; Dai et al., 2020; Zhao et al., 2019). More-recent studies on the structure of the active central pair highlight the role of a specific kinesin, Klp1 in Chlamydomonas or Kif9 in vertebrates, in beating and ciliary waveform through the central pair (Han et al., 2022 preprint). All of these elements work in concert to orchestrate proper ciliary beat frequency and waveform.
In addition to the radial patterning of motile axonemes, a complex proximal to distal patterning has been extensively studied in Chlamydomonas and Tetrahymena (Pedersen et al., 2003; Bui et al., 2012; Louka et al., 2018). Although less well defined, such proximodistal patterning is also crucial in vertebrate multiciliated cells (Fliegauf et al., 2005; Loges et al., 2008; Yamamoto et al., 2010). The function and composition of the distal tip of motile cilia is of particular interest, as it is thought to be critical to protect the axoneme from disintegration due to the massive forces associated with beating. The distal tip differs in structure wildly across ciliary types (Soares et al., 2019), but enrichment of the microtubule end-binding proteins Eb1 and Eb3 (also known MAPRE1 and MAPRE3, respectively) as in distal regions is conserved from algae to vertebrates (Brooks and Wallingford, 2012; Schrøder et al., 2011; Pedersen et al., 2003). Additionally, we and others have identified that Spef1, a microtubule-bundling protein, is also highly enriched in the distal tip of motile axonemes in Xenopus (Gray et al., 2009; Schrøder et al., 2011, 1). Interestingly, Spef1 also contributes to central pair formation in mouse multiciliated cells. Whether Spef1 and the CA contribute to tip integrity in multiciliated cells remains unclear.
Here, we examined the function of Kif9, the vertebrate homolog of the Chlamydomonas kinesin Klp1, which projects from the C2 microtubule of the central pair and is required for ciliary beating (Bernstein et al., 1994, 1; Miyata et al., 2020; Yokoyama et al., 2004; Lechtreck and Witman, 2007). A recent report describes improper waveforms and disrupted ciliary beat in sperm from Kif9 mutant mice (Miyata et al., 2020), and, as in Chlamydomonas, Kif9 mutant sperm do not display overt defects in the assembly of the central pair microtubules (Yokoyama et al., 2004; Miyata et al., 2020). Thus, the mechanisms by which loss of Klp1/Kif9 disrupts cilia beating remain unclear.
Recently, a cryo-EM structure of the active CA in Chlamydomonas has been reported, and this shows that Klp1, does not participate in long-range transport along microtubules, but instead is stably bound to the central pair C2 microtubule and produces 16 nm swings between ATP-bound states (Han et al., 2022 preprint). This result further suggests Kif9 is necessary for ciliary beating in a manner similar to that of axonemal dyneins, and that it does not use its motor for trafficking of cargo but instead to mediate beating. Here, we find that loss of the vertebrate ortholog of Klp1, Kif9 in Xenopus results in defects in ciliary beating and cilia length related to specific disruption of the distal tip of the axoneme. Strikingly, we observed loss not only of central pair proteins, but also of radial spokes and dynein arms from distal axonemes after Kif9 knockdown. Finally, we show that Kif9 is a slowly processive microtubule motor in vitro, although it displays only negligible long-range movement in the axoneme. These data provide new insights into the central pair contribution to ciliary beating and so might contribute to our understanding of human motile ciliopathies.
RESULTS
Kif9 localizes to the axonemes of multiciliated cells in Xenopus and human airways
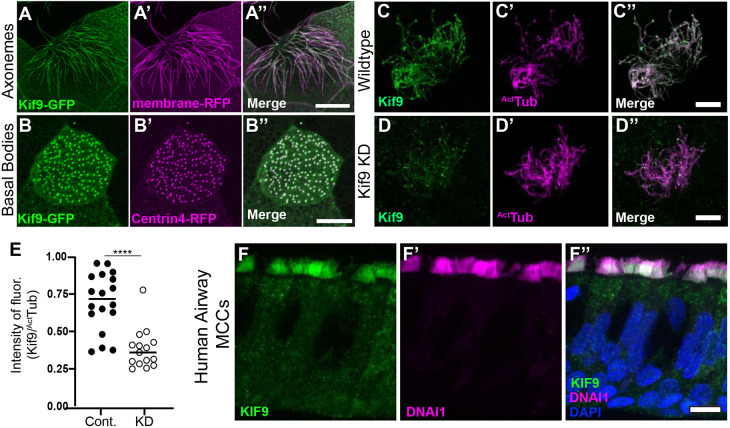
Xenopus tadpoles are a powerful model for studying the formation and function of multiciliated cells (Walentek and Quigley, 2017), so we used this system to examine the localization of Kif9 in multiciliated cells. En face confocal imaging of Kif9–GFP mRNA-injected whole-mounted embryos revealed localization to cilia and basal bodies of multiciliated cells (Fig. 1A–B″). We confirmed endogenous Kif9 localization in ciliary axonemes using immunofluorescence and co-staining with acetylated tubulin (Fig. 1C–C″). This localization is also evolutionarily conserved, as Kif9 immunostaining of human airway multiciliated cells also revealed localization to cilia (Fig. 1F–F″). Thus, Kif9 is expressed in Xenopus and human airway multiciliated cells, where it localizes to ciliary axonemes.
Fig. 1.
Kif9 localization to motile cilia. (A–A″) Kif9–GFP construct localizes to Xenopus multiciliated cells in the axonemes. Kif9–GFP is in green (A), membrane–RFP is in magenta (A′). (B–B″) The Kif9–GFP construct localizes to basal bodies of multiciliated cells in Xenopus. Kif9–GFP in green (B), Centrin4–RFP in magenta (B′). (C–C″) Kif9 antibody (green) confirms localization to Xenopus axonemes. Kif9 in green (C) and acetylated tubulin (ActTub) in magenta (C′). (D–D″) Knockdown of Kif9 with 15 ng of MO reduces the level of Kif9 (green) in the cilium. (E) Quantification of Kif9 staining intensity over the intensity of acetylated tubulin in control and MO-injected frogs. (F–F″) Kif9 immunostaining in human airway multiciliated cells (MCCs). (F) Kif9 immunostaining (green). (F′) DNAI immunostaining (magenta). (F″) Merge of F–F′ with DAPI. Images are representative of 30 different cells imaged from three independent experiments with at least 10 different embryos imaged for each replicate. Scale bars: 10 µm.
Kif9 is necessary for ciliary beating in multiciliated cells
Multiciliated cells generate fluid flow across several different epithelia (Brooks and Wallingford, 2014; Spassky and Meunier, 2017) and the Xenopus epidermis offers an accessible model for studying those flows (Walentek and Quigley, 2017). In order to knockdown (KD) expression of Kif9 and study effects of its loss on cilia-mediated fluid flow, we designed splice-blocking morpholino (MO) oligonucleotides against the first exon of Kif9 in Xenopus. Kif9 mRNA knockdown was confirmed by RT-PCR (Fig. S1A). Crucially, imaging of Kif9 immunostaining in knockdown cilia also revealed reduced levels of Kif9 protein in cilia (Fig. 1D–D″,E).
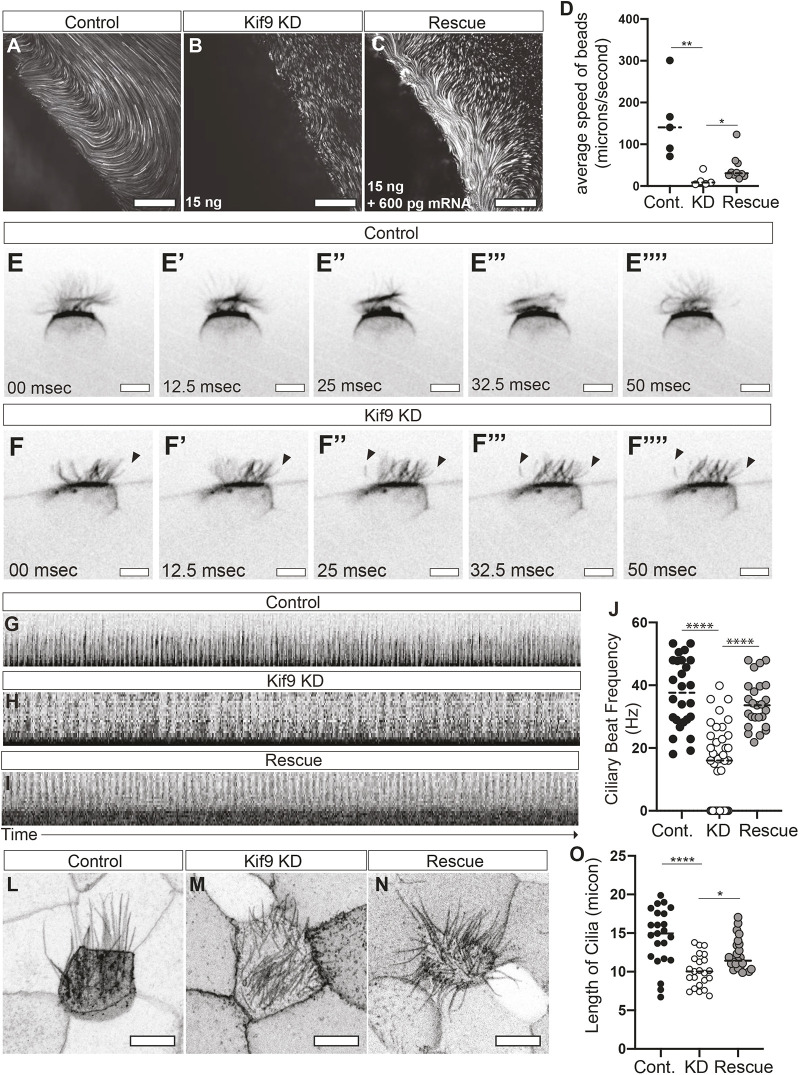
To visualize flows, we whole-mounted embryos with beads mixed into the culture medium and imaged at high speeds with confocal microscopy. Using Flowtrace analysis (Gilpin et al., 2017), we highlighted tracks created by beads across frames (Movies 1, 2 and 3). We then used particle image velocimetry (PIV) analysis to quantify the ciliary beating-induced fluid flow speeds. In control embryos, we observed average fluid flow speeds of ∼154 µm/s, whereas MO-injected embryos showed a 10-fold reduction in flow (P-value=0.0094; Fig. 2A,B,D). Reintroduction of Kif9–GFP mRNA into knockdown embryos rescued the flow defects, restoring flow rates to ∼42 µm/s, thereby supporting the specificity of this phenotype (Fig. 2C,D, P-value=0.0280).
Fig. 2.
Kif9 is necessary for ciliary beating and ciliary length. (A–C) Still images from bead flow movies subjected to Flowtrace analysis. (A) Control injected, (B) 15 ng Kif9 MO injected (KD), and (C) 15 ng MO with 600 pg Kif9 mRNA (Rescue). A is a still frame of Movie 1, B is a selected still frame from Movie 2, and C is a still frame from Movie 3. Note Movie 3 shows a different flow plane than Movies 1 and 2 hence the different direction of flow in Movie 3, but this was most representative of average flow rescues. Scale bars: 50 µm. (D) Graph of bead velocity averages from collected movies. **P=0.0094 and *P=0.0280 (one-way ANOVA with multiple comparisons and test for normality). (E–F″″) Still frames of ciliary beating in control (E–E″″) and Kif9 KD (F–F″″) multiciliated cells. Arrowheads point to stalled or paralyzed cilia frequently observed in Kif9 KD embryos. Scale bars: 10 µm. (G–I) Representative kymographs of ciliary beating in Control (G), Kif9 KD (H), and Rescue (I) injected embryos. The images represent 15 µm (height) and 10 s (width). (J) Graph of ciliary beat frequency calculated from kymographs of 27 different movies of cilia beating in Hz. (L–N) Representative images of ciliary length marked with membrane–GFP in control (L), Kif9 morpholino (M), and rescue (N) injected embryos. Note: the variability in cilia numbers in these images reflect the normally high variability seen in Xenopus multiciliated cells. Scale bars: 10 µm. (O) Graph of ciliary length from control, morpholino or rescue in µm. ****P<0.0001 and *P=0.0174 (one-way ANOVA with multiple comparisons and test for normality). Images are representative of 30 different cells imaged from three independent replicates with 10 embryos imaged for each replicate.
To ask how Kif9 knockdown affects ciliary beating, we used high-speed imaging of cilia labeled with a CAAX–GFP probe (membrane–GFP; Movies 4, 5 and 6). Compared to controls, Kif9 KD embryos displayed beat defects that were restored in the rescued embryos (Fig. 2E–F″″). These beat defects ranged from complete paralysis of cilia, to reduced beat frequency and disrupted waveforms. We also frequently observed cilia that stall during their beat cycle (Fig. 2F–F″″, black arrowheads).
Kymographs further highlighted the deficiencies in Kif9 KD embryos, including reveling the irregular beat (Fig. 2G,H). Both the speed and regularity of beating was restored in rescued embryos (Fig. 2I). Quantification revealed a mean beat frequency of ∼37 Hz for controls, and this was reduced by half in Kif9 knockdowns, with cells displaying a bimodal distribution of speeds (some paralyzed and others with reduced beat frequencies). Notably, even those cilia that did beat did so more slowly (Fig. 2J). In rescue embryos, the ciliary beat was restored to 34.02 Hz (P<0.0001; Fig. 2J).
Finally, in addition to ciliary beat defects, Kif9 KD embryos also had shorter cilia than controls (P<0.0001, Fig. 2L,M,O). This phenotype was also rescued by expression of Kif9–GFP (P=0.0174; Fig. 2N,O). These findings are consistent with the finding that axoneme length is often affected in central pair mutants in Chlamydomonas (Lechtreck et al., 2013). Together, these data demonstrate that Kif9 is necessary for normal ciliary length and beating in vertebrate multiciliated cells.
Kif9 is necessary for central pair protein localization along the proximal-distal axis of the axoneme
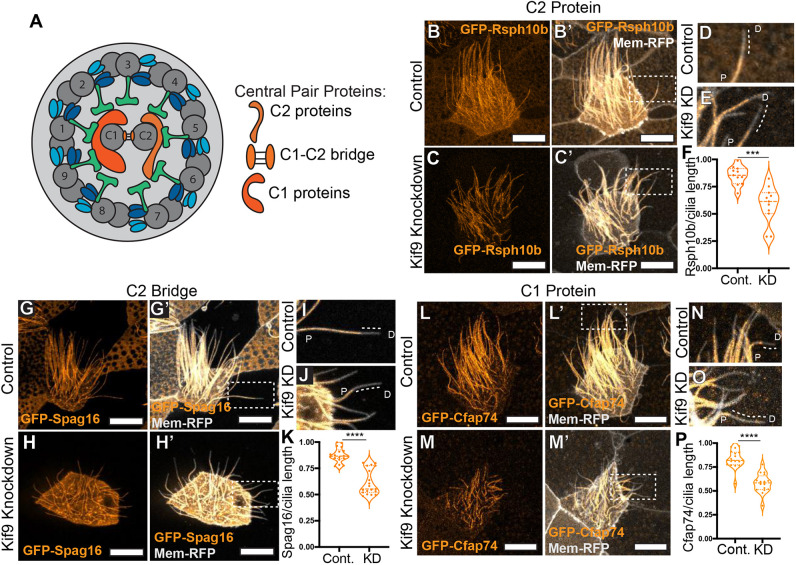
We wished to assess how loss of Kif9 affects other components of the central pair (Fig. 3A). We first considered Fap266, as proteomic data in Chlamydomonas suggest this central pair protein may interact with Klp1/Kif9 (Dai et al., 2020; Zhao et al., 2019). The vertebrate ortholog of Fap266 has not been established, but Rsph10b is the most likely candidate and a recent paper has demonstrated that Rsph10b is not present in mouse radial spokes, further suggesting it may be a central pair protein (Zheng et al., 2021). We therefore generated a GFP fusions to Rsph10b and found that its normal axonemal localization was disrupted after Kif9 KD (Fig. 3B–C′).
Fig. 3.
Kif9 contributes to central pair proteins and distal tip compartmentalization. (A) Schematic of 9+2 motile ciliary structure with central pair components schematized by tubule association. (B–C′) Confocal images of the localization of the radial spoke head component GFP–Rsph10B (orange) in control (B,B′) and Kif9 MO-injected (C,C′) embryos. (D,E) Magnified views of cilia from area indicated in B′,C′ for control (D) and (E) Kif9 KD cilia showing loss of GFP–Rsph10B (orange) from the distal ends of cilia. Dotted line outlines lack of protein at distal ends. (F) Quantification of GFP–Rsph10B length over total ciliary length in control and Kif9 KD embryos. (G–H′) Confocal images of GFP–Spag16 (orange) and membrane–RFP (gray) localization in Xenopus multiciliated cells in control (G,G′) and Kif9 MO (H,H′) injected embryos. (I,J) Magnified views of distal tips of cilia from area indicated in G′,H′ for control (I) and Kif9 KD (J) cilia showing loss of GFP–Spag16 (orange) from the distal tip of cilia. Dotted line outlining lack of protein at distal ends. (K) Quantification of GFP–Spag16 length over total ciliary length in control and Kif9 KD injected embryos. (L–M′) Confocal images of GFP–Cfap74 localization in control (L,L′) and Kif9 MO (M,M′) injected embryos. (N–O) Magnified views of distal tips of cilia from area indicated in L′,M′ for control (N) and Kif9 KD (O) cilia showing loss of GFP–Cfap74 (orange) from the distal tip of cilia. Dotted line outlining lack of protein at distal ends. (P) Quantification of GFP-Cfap74 length over total ciliary length in Control and Kif9 knockdown embryos. Results in F,K,P are shown as violin plots with the median and quartiles marked by dashed lines. ***P<0.001, ****P<0.0001 (unpaired two-tailed t-test). P, proximal; D, distal. Scale bars: 10 µm.
Intriguingly, this defect was restricted to the distal ends of the cilia (Fig. 3B–E). Live confocal imaging of embryos injected with membrane (Mem)–RFP (gray) and GFP–Rsph10b (orange) revealed a striking reduction in localization of Rsph10b in the distal axoneme. This loss was not simply a consequence of reduced cilia length, as we measured the length of cilia containing Rsph10b protein and normalized it to total cilia length as indicated by membrane–RFP; this quantification revealed that the proportion of the cilium occupied by Rsph10b was reduced (Fig. 3F). Moreover, when we quantified fluorescence intensity along individual axonemes, we observed that fluorescence intensity of Rsph10b was increased in the proximal regions of Kif9 KD axonemes (Fig. S2E).
Because the exact orthology of Rsph10b and Fap266 remains poorly defined, we next examined bona fide vertebrate CA proteins. Kif9 is present in the C2 region of the CA, and we found that the C2 ‘bridge’ protein Spag16 (Pf20 in Chlamydomonas) was also lost specifically from the distal axonemes (Fig. 3G–J). So too was the C1-associated ciliopathy protein Cfap74 (Dai et al., 2020; Zhao et al., 2019; Sha et al., 2020) (Fig. 3L–P; Fig. S2C,D). However, unlike Rsph10b, neither Spag16 nor Cfap74 was increased in the proximal axonemes (Fig. S2B,D,F). Together, these data suggest that loss of Kif9 leads to complex alterations in CA protein localization in motile cilia.
Loss of Kif9 affects the proximal to distal patterning of radial spokes, inner dynein arms and outer dynein arms in the cilium
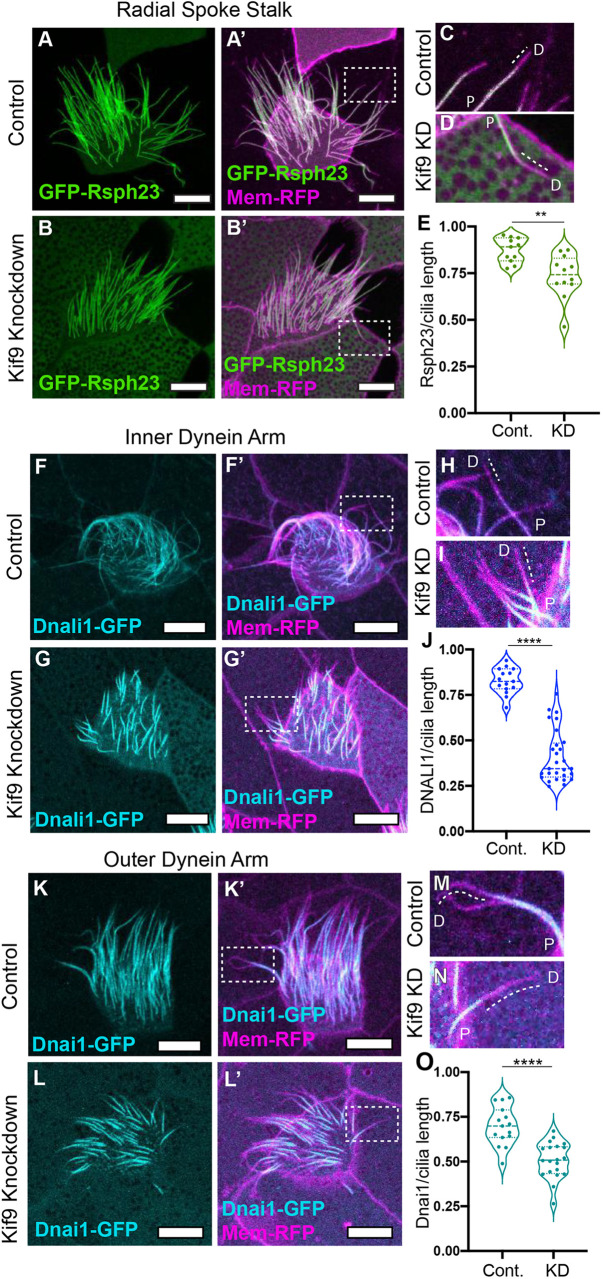
Previous studies suggest, but have not demonstrated, links between the Kif9/Klp1 and radial spokes in Chlamydomonas (Yokoyama et al., 2004; Lechtrek, 2007). We therefore examined the localization of Rsph23 (also known as Nme5), a radial spoke stalk or ‘neck’ protein that is out of direct contact from the CA (Cho et al., 2020). Live confocal imaging of control and Kif9 KD embryos revealed a striking reduction in Rsph23 along the axonemes (Fig. 4A–B′), stemming specifically from a loss of Rsph23 from the distal ends of the axonemes (Fig. 4C,D, P=0.0023; Fig. S2G,H).
Fig. 4.
Kif9 is necessary for radial spoke, inner dynein arm and outer dynein arm placement in the distal ends of cilia. (A–B′) Confocal images of radial spoke stalk component GFP–Rsph23 (green) localization in control (A,A′) and Kif9 MO (B,B′) injected embryos. (C,D) Magnified views of cilia from area indicated in A′,B′ for control (C) and Kif9 KD (D) cilia showing loss of GFP–Rsph23 from the distal ends of cilia. Dotted line outlining lack of protein at distal ends. (E) Quantification of GFP–Rsph23 length over total ciliary length in control and Kif9 KD embryos. (F–G′) Confocal images of inner dynein arm Dnali1–GFP localization in control (F,F′) and Kif9 MO (G,G′) injected embryos. (H,I) Magnified views of cilia from area indicated in F′,G′ for control (H) and Kif9 KD (I) cilia showing loss of Dnali1–GFP from the distal ends of cilia. Dotted line outlining lack of protein at distal ends. (J) Quantification of GFP–Dnai1 length over total ciliary length in control and Kif9 KD embryos. (K–L′) Confocal images of outer dynein arm GFP–Dnai1 (cyan) localization in control (K,K′) and Kif9 MO (L,L′) injected embryos. (M,N) Magnified views of cilia from area indicated in K′,L′ for control (M) and Kif9 KD (N) cilia showing loss of GFP–Dnai1 (cyan) from the distal ends of cilia. Dotted line outlining lack of protein at distal ends. (O) Quantification of GFP–Dnai1 length over total ciliary length in control and Kif9 KD embryos. Results in E,J,O are shown as violin plots with the median and quartiles marked by dashed lines. **P<0.01, ****P<0.0001 (unpaired two-tailed t-test). P, proximal; D, distal. Scale bars: 10 µm.
Radial spoke proteins control the activity of axonemal dyneins, and radial spoke mutants in many species display paralyzed flagella (Viswanadha et al., 2017). We therefore asked whether loss of Kif9 also affected inner and outer dynein arm localization in the axoneme by examining GFP–Dnali1 or GFP–Dnai1, respectively. Strikingly, we observed that both inner dynein arm Dnali1 and outer dynein arm Dnai1 were reduced in distal axonemes (Fig. 4F–O).
Kif9 contributes to distal tip integrity in motile axonemes
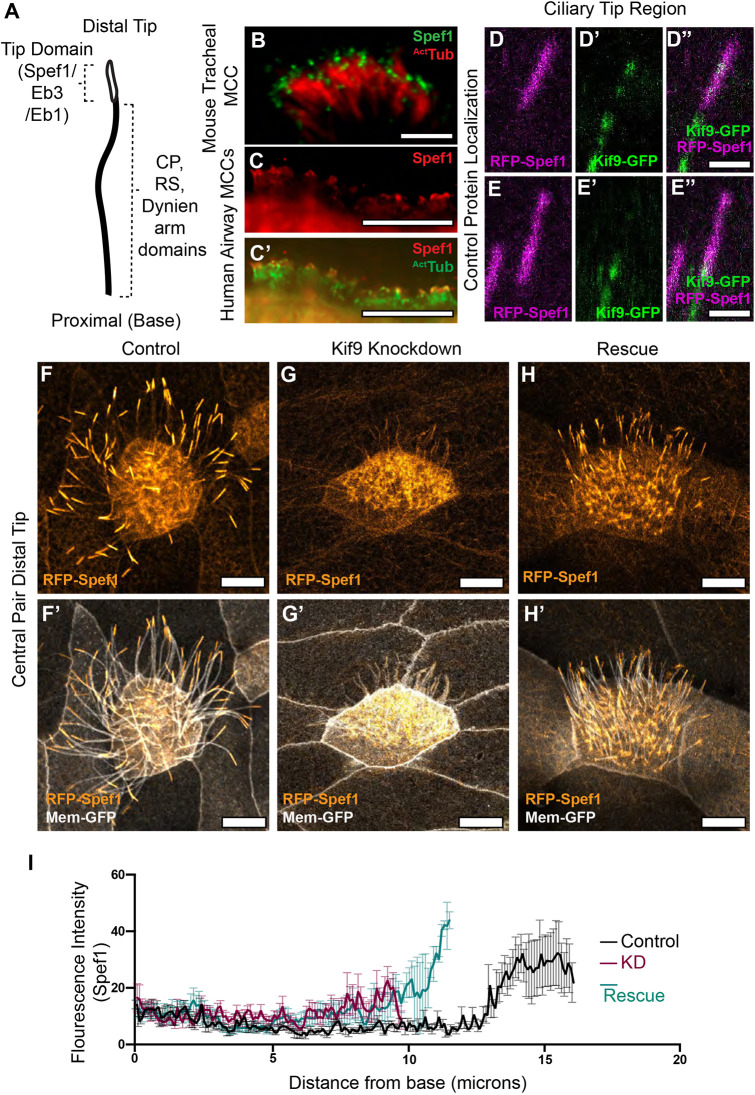
The specific, distal loss of CA, radial spoke, and axonemal dynein components led us to ask whether Kif9 might contribute to the specialized structures of the axoneme distal tip (Soares et al., 2019). We therefore examined the localization of Spef1, a microtubule-bundling protein that is both enriched in the distal tips of Xenopus multiciliated cilia and is required for central pair assembly in mammals (Gray et al., 2009; Chan et al., 2005; Zheng et al., 2019). First, we showed that Spef1 does indeed localize to multiciliated cell ciliary tips in mouse tracheal and human airway multiciliated cells (Fig. 5B–C′). This confirms our previous observation on the localization of Spef1 in frog multiciliated cells using RFP–Spef1 (Gray et al., 2009). Interestingly, when we looked at Kif9 and Spef1 localization in the tip, we found that, unlike other beating machinery, Kif9 protein extended distally in the region marked by enriched Spef1. Unlike Spef1, however, Kif9 was not enriched in this region, but rather displayed a speckled mosaic pattern that was not consistent from cilium to cilium (Fig. 5D–E″).
Fig. 5.
Kif9 contributes to distal tip integrity. (A) Schematic of proximal to distal patterning of the motile cilium. Tip domain schematized with proteins known to localize to that domain, as well as proteins localizing to the rest of the axonemes. (B) Images of immunostaining of Spef1 in mouse tracheal multiciliated cells (MCCs). Spef1 in green, acetylated tubulin (ActTub) in red. (C,C′) Images of immunostaining of human airway multiciliated cells. Spef1 in red, acetylated tubulin in green. (D–E″) Confocal images of RFP–Spef1 (magenta) and Kif9–GFP (green) colocalization in the distal tips of the cilium. (F–H′) Confocal images of RFP–Spef1 (orange) and Membrane–GFP (gray) in control (F,F′), Kif9 MO-injected (G,G′), and rescue (H,H′) embryos. (I) Plot showing RFP–Spef1 fluorescence intensity as a function of axoneme length. Note the higher intensity for RFP–Spef1 at the distal end of control but not Kif9-KD axonemes. Note too that both axoneme length and distal Spef1 enrichment are partially rescued by re-expression of Kif9 (N=45 control axonemes; 45 Kif9-KD axonemes; 42 rescue axonemes; data compiled from 15 embryos across three independent experiments). Scale bars: 1 µm in B; 10 µm in C,C′,F–H′; and 2 µm in D–E″.
More importantly, upon co-expressing RFP–Spef1 with membrane–GFP we observed a striking loss of the Spef1-enriched tip domain in Kif9 KD cilia (Fig. 5F–G′). This phenotype was specific, as it was rescued by re-introduction of Kif9 mRNA (Fig. 5H,H′). We quantified this loss using fluorescence intensity traces along the cilium (Fig. 5I).
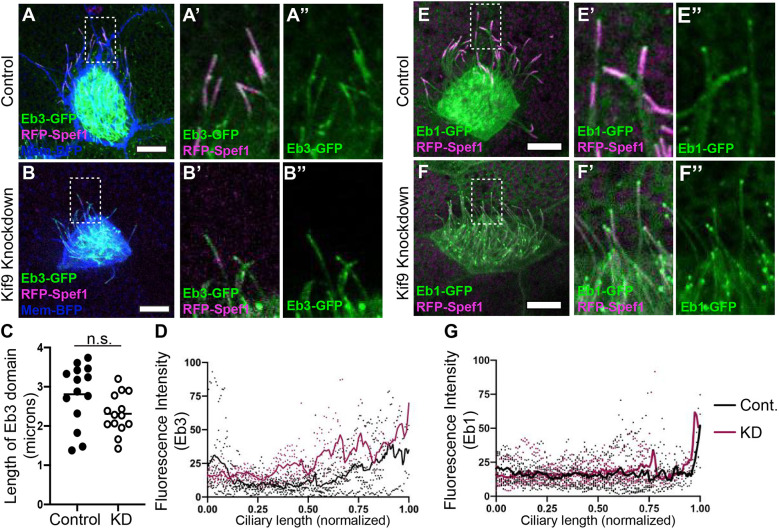
Finally, this phenotype did not reflect a general defect in the distal cilium tip, as other proteins that decorate this region were unaffected after Kif9 loss. For example, the microtubule plus-end binding protein Eb3 displayed a broad distal enrichment similar to Spef1 (Fig. 6), whereas Eb1 displayed a more concentrated enrichment at the extreme distal end of the cilium (Fig. 6). Both Eb1 and Eb3 remained distally enriched after Kif9 loss. These data argue that Kif9 contributes to the localization of specific elements of the distal tip of motile axonemes.
Fig. 6.
The microtubule plus-end-binding proteins Eb1 and Eb3 remain unaffected in the distal tip upon knockdown of Kif9. (A–B″) Confocal images of control (A–A″) and Kif9 MO-injected embryos (B–B″) with Eb3–GFP (green), RFP–Spef1 (magenta), and Membrane–BFP (blue). (A′,B′) Magnified views of area highlighted in A,B of ciliary tips with Eb3–GFP (green) and RFP–Spef1 (magenta) in control and KD cilia. (A″,B″) Magnified views of area highlighted in A,B of Eb3 alone in control and KD cilia. (C) Length measurements of Eb3 domain in control and Kif9 KD embryos. n.s., not significant (P=0.056) (unpaired two-tailed t-test). (D) Graph of line traces of fluorescence intensities of Eb3 in control and knockdown cilia. (E–F″) Confocal images of control (E–E″) and Kif9 MO-injected embryos (F–F″) with Eb1–GFP (green), RFP–Spef1 (magenta) and Membrane–BFP (blue). (E′,F′) Magnified views of area highlighted in E,F of ciliary tips with Eb1-GFP (green) an RFP–Spef1 (magenta) in Control and Knockdown cilia. (E″–F″) Magnified views of area highlighted in E,F of Eb1 alone in control and KD cilia. (G) Graph of line traces of fluorescence intensities of Eb1 in control and KD cilia. Images are representative of 30 different axonemes quantified across three independent replicates with 10 different embryos imaged for each replicate. Scale bars: 10 µm.
Kif9 can bind microtubules and has slow processivity in vitro but displays negligible movement in axonemes
Kinesins play diverse cellular roles. Whereas most kinesins acts as motors for intracellular trafficking, others function as microtubule-associated proteins or microtubule-depolymerizing proteins (Konjikusic et al., 2021). Klp1 binds microtubules in vitro (Yokoyama et al., 2004), but its motor activity and the microtubule interactions of its vertebrate ortholog Kif9 remain unknown. We therefore turned to single-molecule motility assays to assess Kif9 and its behavior on microtubules. Full-length Xenopus Kif9 showed no ability to bind microtubules in vitro (Fig. 7A,B–B″), consistent with the fact that full-length kinesin motors are often autoinhibited (Verhey and Hammond, 2009; Brunnbauer et al., 2010). By contrast, a truncated version of Kif9 containing only the motor domain and a portion of the coiled-coiled domain bound strongly to microtubules (Fig. 7A′,C–C″). Moreover, live time-lapse imaging revealed processive movement of Kif9 along microtubules in a unidirectional manner (Movie 7; Fig. 7D–D″, arrow following a Kif9 punctum). Kymographs confirmed processivity in vitro, albeit at relatively slow rates – 7.2 nm/s with an average of 790 nm run lengths and 122 s dwell times, respectively (Fig. 7E; Fig. S3A–C).
Fig. 7.
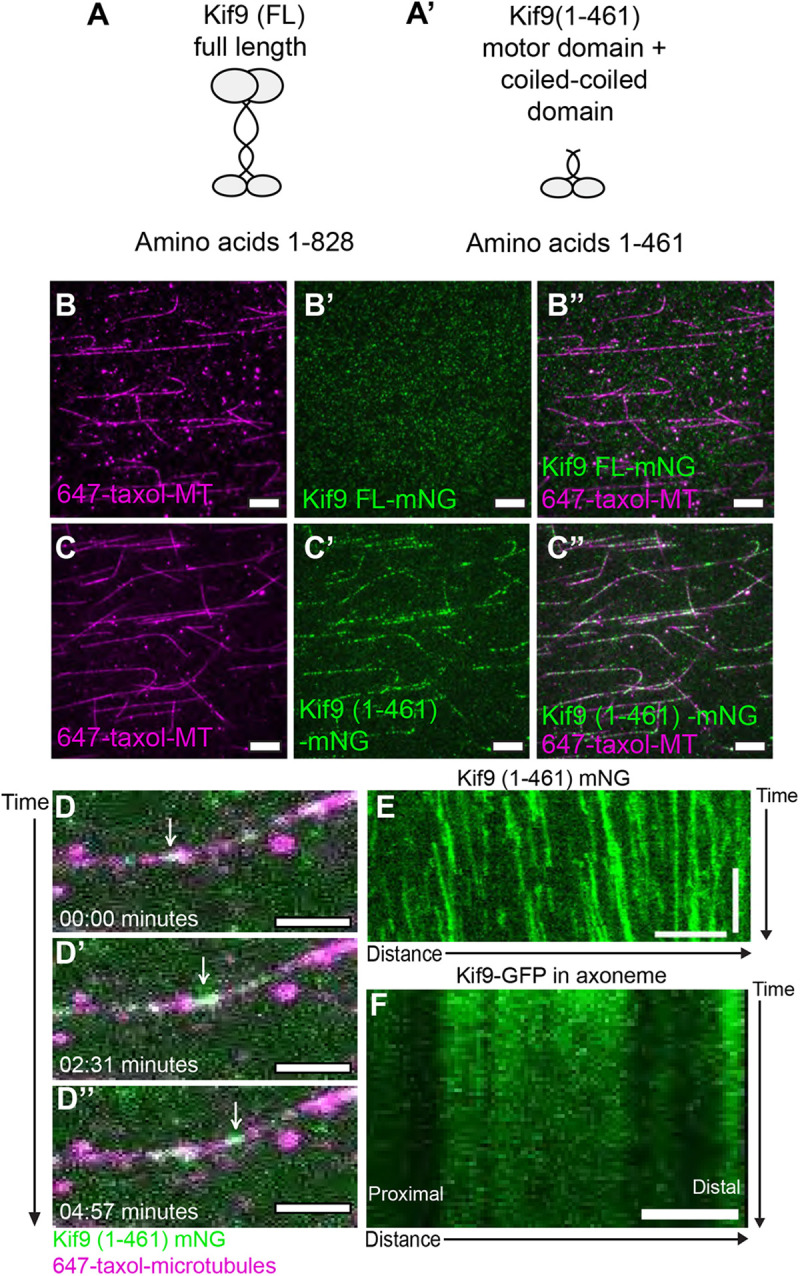
Motor activity of Xenopus Kif9. (A,A′) Schematic of Xenopus laevis Kif9 constructs made for single-molecule assays. (B–B″) Images from single-molecule microtubule binding assays for full-length Kif9 [Kif9 (FL), green) on Taxol-stabilized microtubules (magenta). (C–C″) Images from single-molecule microtubule-binding assays for a truncated version of Kif9 [Kif9 (1-461), green] on Taxol-stabilized microtubules (magenta). Scale bars: 10 µm. (D–D″) Still frames from live imaging of Kif9 processivity on microtubules in minutes. White arrow follows a Kif9 particle (green) moving along a microtubule (magenta). Scale bars: 2 μm. (E) Kymograph of live imaging of Kif9 (1-461)-mNG in vitro on microtubules. Scale bars: 2 min (vertical) and 5 µm (horizontal). (F) Kymograph of live imaging of Kif9–GFP construct in vivo in Xenopus laevis multiciliated cell axonemes. Images are representative of 30 different cells imaged across three independent replicates with at least 10 experiments imaged for each replicate. Scale bar: 2 μm
As an active motor, Kif9 could function in the axoneme either as a traditional transport motor [i.e. analogous to an intraflagellar transport (IFT) kinesin] or in an active array (i.e. analogous to an axonemal dynein). To explore these possibilities, we expressed Kif9–GFP in motile axonemes, performed time-lapse imaging, and examined kymographs. In the vast majority of nearly 100 axonemes imaged, we observed negligible long-range movement of Kif9–GFP (Fig. 7F; Movie 8, Fig. S3B). In rare cases, however, we did observe faint traces indicating movement of Kif9–GFP (Fig. S3C, arrows). Curiously, these traces indicated both anterograde and retrograde movement, with calculated transport rates of ∼600 nm/s, a rate similar to that of IFT transport in the axoneme (Wingfield et al., 2018; Craft et al., 2015; Brooks and Wallingford, 2012). Thus, we conclude that the vast majority of Kif9 in the axoneme is stationary and suggest that Kif9 may be transported in the axoneme by IFT.
DISCUSSION
The normal and effective beating of the 9+2 motile cilia on multiciliated cells, requires the action of the inner and outer dynein arms, the radial spokes and the central pair, but how these systems work in concert remains poorly understood. Here, we extend previous studies of the central pair kinesin Klp1 in Chlamydomonas by showing that the vertebrate ortholog Kif9 is not only required for ciliary beating, but also has processive motor activity and is required for the organization of the distal axoneme in multiciliated cells. This work provides several new insights into the cell biology of ciliary beating.
First, the function of the central pair has been postulated to control beat and waveform through mechanical links to radial spoke and inner and outer dynein arms (Oda et al., 2014). Several studies have shown a physical link between CA and radial spoke head proteins that occurs when cilia are undergoing ‘bending’ during beat (Goodenough and Heuser, 1985; Oda et al., 2014; Warner, 1970). However, which proteins create this link is unknown. One hypothesis is that a microtubule motor protein, such as a kinesin motor, could serve this function, as it could bind the central pair of microtubules and adjacent proteins and use its motor activity to finetune beat and waveform through motor action, much like the action of axonemal dyneins (Viswanadha et al., 2017). Our data are consistent with this model.
We found that Kif9 is processive in vitro, but only very rarely showed any processive movement in multiciliated cell axonemes in vivo; the vast majority was restricted from long-range movement within the axonemes (97/100 kymographs examined). This result is exciting for its similarity to that of axonemal dyneins, which also display processivity in vitro, but are not processive in vivo and participate strictly in force generation for effecting ciliary beating (Sakakibara et al., 1999). Indeed, a recent of the cryo-EM structure of the active Chlamydomonas CA further confirms that the Kif9 ortholog Klp1 forms active motor arrays on the C2 microtubule, suggesting a role in ciliary beating (Han et al., 2022 preprint). Our data therefore suggest that a similar mechanism of action is conserved in vertebrate motile cilia. Moreover, of the infrequent transport events observed for Kif9 in vivo (3/100 kymographs), it is necessary to point out two important notes: (1) it is bidirectional in the axoneme, suggesting that Kif9 motor activity does not drive such movement, as an N-type kinesin Kif9 should only move towards plus ends; and (2) it is near the rate of IFT transport reported in axonemes previously (Wingfield et al., 2018; Craft et al., 2015; Brooks and Wallingford, 2012). This suggest that these few captured instances of Kif9 movement may actually be IFT transport of Kif9 into and out of the axoneme and not Kif9 movement along axonemes itself. On this note, it will be interesting then to directly test the possibility that Kif9 may be a cargo of the IFT system in vertebrate multiciliated cells.
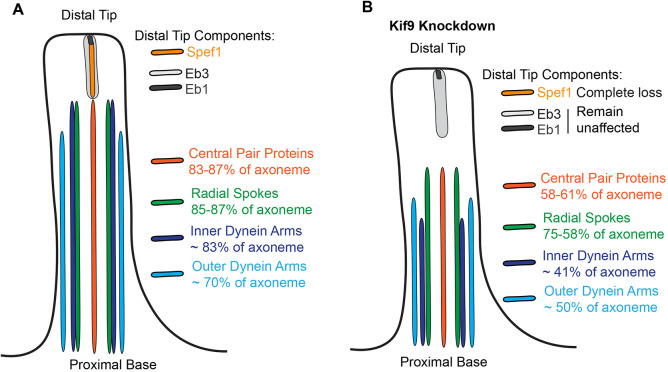
Motile axonemes also display a specific proximodistal patterning, and our data suggest a role, though likely indirect, for Kif9 in this patterning. Dynein arms, radial spoke and central pair proteins decorate the majority of multiciliated cell motile cilia, but are excluded from a specific tip domain characterized by several cell-type specific ‘capping’ structures (Soares et al., 2019; Dentler and Lecluyse, 1982; Portman et al., 1987; LeCluyse and Dentler, 1984). What proteins contribute to these structures remains unclear, but in Xenopus, Spef1 is enriched (Gray et al., 2009; Fig. 5A). Spef1 is necessary for central pair formation in mouse testes and ependymal cell culture, and functions in vitro as a microtubule-bundling protein (Chan et al., 2005; Zheng et al., 2019), and we show here that it is also enriched in the distal tip of mouse and human airway multiciliated cells. Moreover, we now show that loss of Kif9 leads to loss of the Spef1 distal tip domain, although two other distal tip proteins, Eb1 and Eb3 remain unaffected (Fig. 8B). Interestingly, several different central pair mutants in Chlamydomonas have shorter cilia (Lechtreck et al., 2013), and we speculate this decrease in length might arise from loss of distal tip integrity in these mutants. Additionally, although the beating defects seen upon Kif9 knockdown in our system appear generally similar to those seen on the Chlamydomonas knockdown of Klp1 (Yokoyama et al., 2004), these phenotypes differ slightly from the more subtle phenotypes in mouse sperm flagella (Miyata et al., 2020). We conjecture that these differences might arise due to distinct tip structures and/or axoneal components among the different ciliary subtypes (Soares et al., 2019).
Fig. 8.
Summary of effect of Kif9 knockdown on proximal to distal patterning of the motile cilium. (A) Schematic of wild-type motile cilia in Xenopus with domains schematized based off findings presented here. (B) Schematic of Kif9 KD cilia and changes to domains observed.
Finally, the disruption of patterning of all major motility complexes in Kif9 KD cilia (Fig. 8), and the shorter overall length of these cilia lead us to speculate that IFT transport may be defective in Kif9 KD cilia. In Chlamydomonas, impairment of IFT has been observed in several central pair mutants (Lechtreck et al., 2013), and we can envision three different mechanisms by which IFT could be disrupted by loss of Kif9. In one scenario, a general derangement of the axoneme could impair normal IFT by simple structural means. Alternatively, defects in the distal tip could disrupt the normal re-organization of IFT particles as they shift from antero- to retrograde transport, resulting in disrupted IFT. Finally, loss of the distal tip could lead to shorter cilia in general through axonemal disassembly mechanisms and might also lead to derangement of IFT. Future experiments exploring these possibilities will be of interest.
In conclusion, we show here that vertebrate Kif9 is required for ciliary beating in multiciliated cells, and for the normal localization of three major motility complexes in motile axonemes. Moreover, Kif9 is a processive motor in vitro, and, consistent with data from Chlamydomonas, most likely acts to drive microtubule sliding in the central pair, thereby facilitating ciliary beating.
METHODS AND MATERIALS
Xenopus embryo manipulations
Xenopus embryo manipulations were carried out using the standard protocols. Briefly, female adult Xenopus were induced to ovulate by injection of hCG (human chorionic gonadotropin). In vitro fertilization was carried out by homogenizing a small fraction of a testis in 1/3× Marc's modified Ringer's (MMR). Embryos were de-jellied in 1/3× MMR with 2.5–3% L-cysteine (pH 7.8–8). Embryos were microinjected with mRNA and MOs in 2% Ficoll (w/v) in 1/3× MMR. Embryos were washed with 1/3× MMR 1 h post injection. Each injection was repeated 3–4 times across several days and clutches of embryos. All animal experiments were performed according to approved guidelines.
Microinjections
Gene sequences were obtained from Xenbase (http://www.xenbase.org). Open reading frames (ORFs) of genes were amplified from the Xenopus cDNA library by PCR, and then inserted into a pCS vectors containing a fluorescence tag. The following constructs were cloned into the pCS10R vector (made by C. Lee in the Wallingford Lab; modified from cs107 vector from the Harland lab at UC Berkeley, USA): GFP–Dnai1, GFP–Nme5 (Rsph23), RFP-clamp (Spef1), GFP–Spag16, GFP–Cfap74, GFP–Rsph10b, GFP–dnali1, Eb3–GFP (as described in Shindo et al., 2008), Eb1–GFP, membrane–RFP, membrane–GFP, membrane–BFP, Centrin4–RFP and Kif9–GFP. These constructs were linearized and capped mRNAs were synthesized using mMESSAGE/mMACHINE SP6 transcription kit. Between 50–100 pg of any given mRNA was injected into the two ventral blastomeres at the four-cell stage. Kif9–GFP DNA was injected at 35 pg. For rescue experiments, 600 pg of kif9 mRNA was injected. The kif9 MO was designed as a splice blocking MO to exon 1 of the kif9 mRNA sequence. The working concentration was 15 ng and sequence as follows: 5′-ATTCTCATATTCAATAGTCTTACCT-3′.
Imaging and analysis
Xenopus embryos were grown up to stage 22–25 and mounted onto an imaging chamber in 1/3× MMR and live imaged immediately. The bead flow movies and live cilia beat videos were taken on a Nikon W1 Spinning Disc Confocal at an image rate of 12.5 ms per frame. Live still images of axonemes were captured with a Zeiss LSM700 laser scanning confocal microscope with a 63× oil objective. Each experiment was repeated three or four times across several days and clutches of embryos. Approximately 15 different images (3–5 embryos) were taken for quantification; five different cilia per cell and at least three cells per image were quantified.
Beads (or microspheres) of size 1 µm were added to the fluid medium for the purpose of tracing/visualizing and quantifying the fluid flow induced by the beating cilia. Bead flow movies were subjected to Flowtrace (Gilpin et al., 2017) for visualization of bead flow patterns across the epidermis. For this Flowtrace analysis, we selected an interrogation window size of 50 frames.
To quantify the fluid flow speeds, we carried out a particle image velocimetry (PIV) analysis using the MATLAB-based PIVlab package (Thielicke and Stamhuis, 2014). First, we extracted individual frames from the time-lapse datasets (movies) and we carried out the image preprocessing step by selecting the contrast limited adaptive histogram equalization (CLAHE) option (window size 20 px). Next, we proceeded with the PIV analysis using these settings: FFT window deformation, Interrogation area 128×128 pixels with 50% overlap (Pass 1), and with interrogation area of 64×64 pixels with 50% overlap (Pass 2). After this step, the vector validation was performed by choosing suitable velocity limits, standard deviation filters and interpolation of missing data. We carefully selected a rectangular region just adjacent and parallel to the ciliary beating surface on the embryo. We quantified the average speed in this selected region over time, and the error bars represent the standard deviation. This procedure was utilized for each individual dataset for the control, Kif9 and rescue cases. To obtain a characteristic speed for each case (control, Kif9 and rescue), further averaging was carried out: for control and Kif9 (N=5 datasets), and for rescue (N=10 datasets).
Quantification of cilia length, length of each construct along axonemes, and fluorescence intensity profiles were all taken in FIJI software. Kymographs of cilia beat were also generated using FIJI software. Briefly, lines were drawn, taking length measurements and fluorescence intensity profiles. Outputs were placed into GraphPad Prism 8 and graphs were generated. All statistical analyses were done in GraphPad Prism 8 with either unpaired two-tailed t-tests or one-way ANOVA with an multiple comparisons and normality post-hoc test where appropriately comparing three different groups of data.
Xenopus immunostaining
For immunostaining, protocols previously described in Brooks and Wallingford (2015) were followed. Briefly, wild-type and MO-injected embryos were fixed with 4% paraformaldehyde (PFA) at stages 22–25 for 1 h at room temperature. They were then washed with PBS plus 0.1% Tween 20 (PBST), and subsequently washed in methanol. They were dehydrated and permeabilized in 100% methanol at −20°C for 30 min to 1 h. Embryos were then rehydrated and washed with PBST. Embryos were then blocked for 1 h at room temperature in 10% normal goat serum (NGS) and 5% DMSO in 1× PBST. Primary antibodies were incubated overnight and embryos were washed 3× in PBST, then incubated with secondary antibody for 1 h at room temperature. A rabbit anti-Kif9 antibody was used at 1:100 dilution from Atlas Antibodies (HPA022033). Mouse anti-acetylated tubulin antibody was used at 1:1000 from Sigma (6-11B-1). Secondaries anti-rabbit-IgG conjugated to Alexa Fluor 488 and anti-mouse-IgG conjugated to Alexa Fluor 647 were used at 1:1000 dilutions.
RT-PCR
To verify the efficiency of our Kif9 MO, it was injected into all for cells at the four-cell stage, and total RNA was isolated using the TRIreagent (Thermo Fisher Scientific) at stage 23. 1 µg of mRNA was used to synthesize cDNA using iScript (Bio-Rad). Kif9 and β-actin cDNAs were amplified by GoTag Green Master Mix (Promega) with the following primers: β-actin F1, 5′-GCCCGCATAGAAAGGAGACAG-3′; β-actin F2, 5′-CCAAACCTCGCTCAGTGACC-3′; β-actin R1, 5′-TCATCCCAGTTGGTGACAATGC-3′; β-actin R2, 5′-TCCCATTCCAACCATGACACC-3′; Kif9 F1, 5′-GAGACGGGATAGTTTACACACAGC-3′; Kif9 R1, 5′-TGGAGGCAAGGTTTAGGGATAAGC-3′; Kif9 F2, 5′-CGCTGAAGCCAAGAGCTGAAC-3′; Kif9 R2, 5′-GCATCTGGAACAGTGGAAAGGAG-3′.
Airway epithelial cell culture and immunostaining
Human airway tracheas were retrieved from surgical excess of tracheobronchial segments of lungs donated for transplantation. Human tracheobronchial epithelial cells (hTEC) were isolated from sections of normal human trachea obtained from non-smoking donors lacking respiratory pathology. These unidentified tissues are exempt from regulation by HHS regulation 45 CFR Part 46. hTEC cells were expanded in vitro and allowed to differentiate into ciliated cells using air-liquid interface (ALI) conditions on supported membranes (Transwell, Corning Inc., Corning, NY), as previously described (You et al., 2002). Paraffin-embedded tracheal sections or cultured primary airway cells were fixed and immunostained as previously described (Pan et al., 2007). Nuclei were stained using DAPI (Vector Laboratories, Burlingame, CA, USA). KIF9 was detected using primary antibodies obtained from MilliporeSigma (St. Louis, MO, USA; HPA022033, 1:100). Basal bodies were detected using antibodies against Centrin (clone 20H5, MilliporeSigma; 1:4000). DNAI1, a marker of outer dynein arm, was detected using NeuroMab (UC Davis, CA, USA; clone UNC 65.56.18.11; 1:1000). Images were acquired using an epifluorescence microscope interfaced with imaging software (LAS X; Leica) and adjusted globally for brightness and contrast using Affinity Photo (Serif Ltd, Nottinghamshire, UK).
Single-molecule microtubule assays
Plasmid
To generate the pMT-xlKIF9(1-461)-mNeonGreen plasmid, the coiled-coil prediction of full-length Xenopus laevis (xl)KIF9 was carried out in Marcoil (Delorenzi and Speed, 2002) and COILS (Lupas, 1996). The xlKIF9(1-461) truncation, which includes the first two coiled coil domains and is expected to form a dimer, was amplified by PCR from full-length xlKIF9 and subcloned into pMT-mNeonGreen vector [pMT-mNeonGreen was generated from the pMT-DmKHC-BFP plasmid made by Dr Vladimir Gelfand's lab. The Gelfand lab used pMT/V5-His A vector (Invitrogen, V412020) to generate the plasmid] by NEBuilder HiFi DNA assembly cloning kit. The plasmid was verified by DNA sequencing.
Cell culture, transfection and lysate preparation
Drosophila S2 cells were cultured in Schneider's Drosophila medium (Gibco) supplemented with 10% (v/v) fetal bovine serum (FBS; HyClone) at 26°C. The plasmid for expression of xlKIF9(1-461)–mNeonGreen in the pMT vector was transfected into S2 cells using Lipofectamine LTX Reagent with PLUS Reagent (Invitrogen) according to the manufacturer's instructions. Expression of xlKIF9(1-461)–mNeonGreen was induced by adding 1 mM CuSO4 to the medium after 4–5 h transfection.
To prepare cell lysate for single-molecule assays, S2 cells were harvested after 48 h transfection. The cells were centrifuged at low speed (100 g for 3 min) at 4°C. The cell pellet was washed with PBS buffer and resuspended in ice-cold lysis buffer (25 mM HEPES/KOH, 115 mM potassium acetate, 5 mM sodium acetate, 5 mM MgCl2, 0.5 mM EGTA, and 1% Triton X-100, pH 7.4) freshly supplemented with 1 mM ATP, 1 mM PMSF, 1 mM DTT and protease inhibitors (Sigma-Aldrich). After the cell lysate was clarified by centrifugation at full-speed (21,000 g for 10 min) at 4°C, aliquots of the supernatant were snap frozen in liquid nitrogen and stored at −80°C until further use.
Single-molecule motility assays
HiLyte647-labeled microtubules were polymerized from purified tubulin including 10% Hily647-labeled tubulin (Cytoskeleton) in BRB80 buffer (80 mM Pipes/KOH pH 6.8, 1 mM MgCl2, and 1 mM EGTA) supplemented with 1 mM GTP and 2.5 mM MgCl2 at 37°C for 30 min. 20 μM Taxol in prewarmed BRB80 buffer was added and incubated at 37°C for additional 30 min to stabilize microtubules. Microtubules were stored in the dark at room temperature for further use. A flow cell (∼10 μl volume) was assembled by attaching a clean #1.5 coverslip (Thermo Fisher Scientific) to a glass slide (Thermo Fisher Scientific) with two strips of double-sided tape. Polymerized microtubules were diluted in BRB80 buffer supplemented with 10 μM Taxol and then were infused into flow cells and incubated for 5 min at room temperature for nonspecific adsorption to the coverslips. Subsequently, blocking buffer (15 mg/ml BSA and 10 μM Taxol in P12 buffer) was infused and incubated for 5 min. Finally, kinesin motors in the motility mixture [2 mM ATP, 0.4 mg/ml casein, 6 mg/ml BSA, 10 μM taxol, and oxygen scavenging (1 mM DTT, 1 mM MgCl2, 10 mM glucose, 0.2 mg/ml glucose oxidase, and 0.08 mg/ml catalase) in P12 buffer] was added to the flow cells. The flow-cell was sealed with molten paraffin wax.
Images for the single-molecule assay were acquired by TIRF microscopy using an inverted microscope Ti-E/B (Nikon) equipped with the perfect focus system (Nikon), a 100×1.49 NA oil immersion TIRF objective (Nikon), three 20-mW diode lasers (488 nm, 561 nm and 640 nm) and an electron-multiplying charge-coupled device detector (iXon X3DU897; Andor Technology). Image acquisition was controlled using Nikon Elements software and all assays were performed at room temperature. Images were acquired continuously every 5 s for 5 min. Maximum intensity projections were generated and the kymographs were produced by drawing along tracks of motors (width=3 pixels) using Fiji/ImageJ2 software.
Supplementary Material
Acknowledgements
We thank members of the Wallingford and Gray Labs for critical comments and thoughtful feedback.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: M.J.K., R.S.G., J.B.W.; Methodology: M.J.K., C.L., Y.Y., B.D.S., A.M.N., A.H., S.B., V.N.P., R.S.G., K.J.V., J.B.W.; Software: B.D.S., V.N.P.; Validation: M.J.K., Y.Y., A.M.N., A.H., S.B.; Formal analysis: M.J.K., Y.Y.; Investigation: M.J.K.; Resources: C.L.; Data curation: M.J.K., C.L., Y.Y., A.M.N.; Writing - original draft: M.J.K., J.B.W.; Writing - review & editing: M.J.K., C.L., Y.Y., A.M.N., A.H., S.B., V.N.P., R.S.G., K.J.V., J.B.W.; Visualization: M.J.K.; Supervision: A.H., S.B., V.N.P., R.S.G., K.J.V., J.B.W.; Project administration: A.H., S.B., V.N.P., R.S.G., K.J.V., J.B.W.; Funding acquisition: A.H., S.B., V.N.P., R.S.G., K.J.V., J.B.W.
Funding
This work was supported by grant funding. M.J.K. was supported by Provosts Graduate Excellence Fellowshipfrom University of Texas at Austin. R.S.G. was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS) R01AR072009. A.H. was supported by K08HL150223. S.B. was supported by HL128370. K.J.V. was supported by National Institutes of Health (NIH) R35GM131744. J.B.W. was supported by R01HL117164. Deposited in PMC for release after 12 months.
References
- Bernstein, M., Beech, P. L., Katz, S. G. and Rosenbaum, J. L. (1994). A new kinesin-like protein (Klp1) localized to a single microtubule of the Chlamydomonas flagellum. J. Cell Biol. 125, 1313-1326. 10.1083/jcb.125.6.1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, E. R. and Wallingford, J. B. (2012). Control of vertebrate intraflagellar transport by the planar cell polarity effector Fuz. J. Cell Biol. 198, 37-45. 10.1083/jcb.201204072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, E. R. and Wallingford, J. B. (2014). Multiciliated cells. Curr. Biol. 24, R973-R982. 10.1016/j.cub.2014.08.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks, E. R. and Wallingford, J. B. (2015). Chapter 7 - In vivo investigation of cilia structure and function using Xenopus. In Methods in Cell Biology (ed. Basto R. and Marshall W. F.), pp. 131-159. Academic Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunnbauer, M., Mueller-Planitz, F., Kösem, S., Ho, T. H., Dombi, R., Gebhardt, J. C. M., Rief, M. and Ökten, Z. (2010). Regulation of a heterodimeric kinesin-2 through an unprocessive motor domain that is turned processive by its partner. Proc. Natl. Acad. Sci. USA 107, 10460-10465. 10.1073/pnas.1005177107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui, K. H., Yagi, T., Yamamoto, R., Kamiya, R. and Ishikawa, T. (2012). Polarity and asymmetry in the arrangement of dynein and related structures in the Chlamydomonas axoneme. J. Cell Biol. 198, 913-925. 10.1083/jcb.201201120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. W., Fowler, K. J., Choo, K. H. A. and Kalitsis, P. (2005). Spef1, a conserved novel testis protein found in mouse sperm flagella. Gene 353, 189-199. 10.1016/j.gene.2005.04.025 [DOI] [PubMed] [Google Scholar]
- Cho, E. H., Huh, H. J., Jeong, I., Lee, N. Y., Koh, W.-J., Park, H.-C. and Ki, C.-S. (2020). A nonsense variant in NME5 causes human primary ciliary dyskinesia with radial spoke defects. Clin. Genet. 98, 64-68. 10.1111/cge.13742 [DOI] [PubMed] [Google Scholar]
- Craft, J. M., Harris, J. A., Hyman, S., Kner, P. and Lechtreck, K. F. (2015). Tubulin transport by IFT is upregulated during ciliary growth by a cilium-autonomous mechanism. J. Cell Biol. 208, 223-237. 10.1083/jcb.201409036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai, D., Ichikawa, M., Peri, K., Rebinsky, R. and Bui, K. H. (2020). Identification and mapping of central pair proteins by proteomic analysis. Biophys. Physicobiol. 17, 71-85. 10.2142/biophysico.BSJ-2019048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentler, W. L. and Lecluyse, E. L. (1982). Microtubule capping structures at the tips of tracheal cilia: evidence for their firm attachment during ciliary bend formation and the restriction of microtubule sliding. Cell Motil. 2, 549-572. 10.1002/cm.970020605 [DOI] [PubMed] [Google Scholar]
- Fliegauf, M., Olbrich, H., Horvath, J., Wildhaber, J. H., Zariwala, M. A., Kennedy, M., Knowles, M. R. and Omran, H. (2005). Mislocalization of DNAH5 and DNAH9 in respiratory cells from patients with primary ciliary dyskinesia. Am. J. Respir. Crit. Care. Med. 171, 1343-1349. 10.1164/rccm.200411-1583OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin, W., Prakash, V. N. and Prakash, M. (2017). Flowtrace: simple visualization of coherent structures in biological fluid flows. J. Exp. Biol. 220, 3411-3418. 10.1242/jeb.162511 [DOI] [PubMed] [Google Scholar]
- Goodenough, U. W. and Heuser, J. E. (1985). Substructure of inner dynein arms, radial spokes, and the central pair/projection complex of cilia and flagella. J. Cell Biol. 100, 2008-2018. 10.1083/jcb.100.6.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, R. S., Abitua, P. B., Wlodarczyk, B. J., Szabo-Rogers, H. L., Blanchard, O., Lee, I., Weiss, G. S., Liu, K. J., Marcotte, E. M., Wallingford, J. B.et al. (2009). The planar cell polarity effector Fuz is essential for targeted membrane trafficking, ciliogenesis and mouse embryonic development. Nat. Cell Biol. 11, 1225-1232. 10.1038/ncb1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han, L., Rao, Q., Yang, R., Wang, Y., Chai, P., Xiong, Y. and Zhang, K. (2022). Cryo-EM structure of an active central apparatus. bioRxiv. 10.1101/2022.01.23.477438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa, T. (2017). Axoneme structure from Motile Cilia. Cold Spring Harb. Perspect. Biol. 9, a028076. 10.1101/cshperspect.a028076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konjikusic, M. J., Gray, R. S. and Wallingford, J. B. (2021). The developmental biology of kinesins. Dev. Biol. 469, 26-36. 10.1016/j.ydbio.2020.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F. and Witman, G. B. (2007). Chlamydomonas reinhardtii hydin is a central pair protein required for flagellar motility. J. Cell Biol. 176, 473-482. 10.1083/jcb.200611115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K.-F., Gould, T. J. and Witman, G. B. (2013). Flagellar central pair assembly in Chlamydomonas reinhardtii. Cilia 2, 15. 10.1186/2046-2530-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCluyse, E. L. and Dentler, W. L. (1984). Asymmetrical microtubule capping structures in frog palate cilia. J. Ultrastruct. Res. 86, 75-85. 10.1016/S0022-5320(84)90097-2 [DOI] [PubMed] [Google Scholar]
- Loges, N. T., Olbrich, H., Fenske, L., Mussaffi, H., Horvath, J., Fliegauf, M., Kuhl, H., Baktai, G., Peterffy, E., Chodhari, R.et al. (2008). DNAI2 mutations cause primary ciliary dyskinesia with defects in the outer dynein arm. Am. J. Hum. Genet. 83, 547-558. 10.1016/j.ajhg.2008.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loreng, T. D. and Smith, E. F. (2017). The central apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb. Perspect. Biol. 9, a028118. 10.1101/cshperspect.a028118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louka, P., Vasudevan, K. K., Guha, M., Joachimiak, E., Wloga, D., Tomasi, R. F.-X., Baroud, C. N., Dupuis-Williams, P., Galati, D. F., Pearson, C. G.et al. (2018). Proteins that control the geometry of microtubules at the ends of cilia. J. Cell Biol. 217, 4298-4313. 10.1083/jcb.201804141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata, H., Shimada, K., Morohoshi, A., Oura, S., Matsumura, T., Xu, Z., Oyama, Y. and Ikawa, M. (2020). Testis-enriched kinesin KIF9 is important for progressive motility in mouse spermatozoa. FASEB J. 34, 5389-5400. 10.1096/fj.201902755R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda, T., Yanagisawa, H., Yagi, T. and Kikkawa, M. (2014). Mechanosignaling between central apparatus and radial spokes controls axonemal dynein activity. J. Cell Biol. 204, 807-819. 10.1083/jcb.201312014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, J., You, Y., Huang, T. and Brody, S. L. (2007). RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J. Cell Sci. 120, 1868-1876. 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- Pedersen, L. B., Geimer, S., Sloboda, R. D. and Rosenbaum, J. L. (2003). The microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr. Biol. 13, 1969-1974. 10.1016/j.cub.2003.10.058 [DOI] [PubMed] [Google Scholar]
- Portman, R. W., LeCluyse, E. L. and Dentler, W. L. (1987). Development of microtubule capping structures in ciliated epithelial cells. J. Cell Sci. 87, 85-94. 10.1242/jcs.87.1.85 [DOI] [PubMed] [Google Scholar]
- Sakakibara, H., Kojima, H., Sakai, Y., Katayama, E. and Oiwa, K. (1999). Inner-arm dynein c of Chlamydomonas flagella is a single-headed processive motor. Nature 400, 586-590. 10.1038/23066 [DOI] [PubMed] [Google Scholar]
- Schrøder, J. M., Larsen, J., Komarova, Y., Akhmanova, A., Thorsteinsson, R. I., Grigoriev, I., Manguso, R., Christensen, S. T., Pedersen, S. F., Geimer, S.et al. (2011). EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J. Cell Sci. 124, 2539-2551. 10.1242/jcs.085852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha, Y., Wei, X., Ding, L., Ji, Z., Mei, L., Huang, X., Su, Z., Wang, W., Zhang, X. and Lin, S. (2020). Biallelic mutations of CFAP74 may cause human primary ciliary dyskinesia and MMAF phenotype. J. Hum. Genet. 65, 961-969. 10.1038/s10038-020-0790-2 [DOI] [PubMed] [Google Scholar]
- Shindo, A., Yamamoto, T. S. and Ueno, N. (2008). Coordination of cell polarity during xenopus gastrulation. PLoS ONE 3, e1600. 10.1371/journal.pone.0001600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares, H., Carmona, B., Nolasco, S., Viseu Melo, L. and Gonçalves, J. (2019). Cilia distal domain: diversity in evolutionarily conserved structures. Cells 8, 160. 10.3390/cells8020160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky, N. and Meunier, A. (2017). The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 18, 423-436. 10.1038/nrm.2017.21 [DOI] [PubMed] [Google Scholar]
- Thielicke, W. and Stamhuis, E. (2014). PIVlab – towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. 2, e30. 10.5334/jors.bl [DOI] [Google Scholar]
- Verhey, K. J. and Hammond, J. W. (2009). Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10, 765-777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Viswanadha, R., Sale, W. S. and Porter, M. E. (2017). Ciliary motility: regulation of axonemal dynein motors. Cold Spring Harb. Perspect. Biol. 9, a018325. 10.1101/cshperspect.a018325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentek, P. and Quigley, I. K. (2017). What we can learn from a tadpole about ciliopathies and airway diseases: using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis 55, e23001. 10.1002/dvg.23001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner, F. D. (1970). New observations on flagellar fine structure: the relationship between matrix structure and the microtubule component of the axoneme. J. Cell Biol. 47, 159-182. 10.1083/jcb.47.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingfield, J. L., Lechtreck, K.-F. and Lorentzen, E. (2018). Trafficking of ciliary membrane proteins by the intraflagellar transport/BBSome machinery. Essays Biochem. 62, 753-763. 10.1042/EBC20180030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, R., Hirono, M. and Kamiya, R. (2010). Discrete PIH proteins function in the cytoplasmic preassembly of different subsets of axonemal dyneins. J. Cell Biol. 190, 65-71. 10.1083/jcb.201002081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama, R., O'Toole, E., Ghosh, S. and Mitchell, D. R. (2004). Regulation of flagellar dynein activity by a central pair kinesin. Proc. Natl. Acad. Sci. USA 101, 17398-17403. 10.1073/pnas.0406817101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- You, Y., Richer, E. J., Huang, T. and Brody, S. L. (2002). Growth and differentiation of mouse tracheal epithelial cells: selection of a proliferative population. Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L1315-L1321. 10.1152/ajplung.00169.2002 [DOI] [PubMed] [Google Scholar]
- Zhao, L., Hou, Y., Picariello, T., Craige, B. and Witman, G. B. (2019). Proteome of the central apparatus of a ciliary axoneme. J. Cell Biol. 218, 2051-2070. 10.1083/jcb.201902017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, J., Liu, H., Zhu, L., Chen, Y., Zhao, H., Zhang, W., Li, F., Xie, L., Yan, X. and Zhu, X. (2019). Microtubule-bundling protein Spef1 enables mammalian ciliary central apparatus formation. J. Mol. Cell Biol. 11, 67-77. 10.1093/jmcb/mjy014 [DOI] [PubMed] [Google Scholar]
- Zheng, W., Li, F., Ding, Z., Liu, H., Zhu, L., Xu, C., Li, J., Gao, Q., Wang, Y., Fu, Z.et al. (2021). Distinct architecture and composition of mouse axonemal radial spoke head revealed by cryo-EM. Proc. Natl. Acad. Sci. USA 118. e2021180118. 10.1073/pnas.2021180118 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.