Abstract
Lysogenic bacteriophages are major vehicles for the transfer of genetic information between bacteria, including pathogenicity and/or virulence determinants. In the enteric pathogen Escherichia coli O157:H7, which causes hemorrhagic colitis and hemolytic-uremic syndrome, Shiga toxins 1 and 2 (Stx1 and Stx2) are phage encoded. The sequence and analysis of the Stx2 phage 933W is presented here. We find evidence that the toxin genes are part of a late-phage transcript, suggesting that toxin production may be coupled with, if not dependent upon, phage release during lytic growth. Another phage gene, stk, encodes a product resembling eukaryotic serine/threonine protein kinases. Based on its position in the sequence, Stk may be produced by the prophage in the lysogenic state, and, like the YpkA protein of Yersinia species, it may interfere with the signal transduction pathway of the mammalian host. Three novel tRNA genes present in the phage genome may serve to increase the availability of rare tRNA species associated with efficient expression of pathogenicity determinants: both the Shiga toxin and serine/threonine kinase genes contain rare isoleucine and arginine codons. 933W also has homology to lom, encoding a member of a family of outer membrane proteins associated with virulence by conferring the ability to survive in macrophages, and bor, implicated in serum resistance.
The production of one or more forms of Shiga toxin (Stx) is a defining characteristic of enterohemorrhagic Escherichia coli (EHEC), along with the capacity to evoke attaching-and-effacing intestinal lesions and the presence of a characteristic large plasmid (50). These strains, particularly E. coli serotype O157:H7, have emerged as an important public health concern worldwide as the causative agents of a severe bloody diarrheal syndrome, hemorrhagic colitis, and an acute renal disease, hemolytic-uremic syndrome. E. coli O157:H7 is the subject of a recent text (44), as well as a novel (21) and a nonfiction first-hand account of HUS (40). The potent cytotoxins produced by these bacteria are similar or nearly identical to those produced by Shigella dysenteriae (59, 69). Although the terms “Shiga-like toxins (SLT)” and “verotoxins (VT)” are still encountered, the term “Shiga toxin (Stx)” refers to the entire family of related toxins (16, 47); the Stx family contains two subgroups, Stx1 and Stx2, which are distinguishable serologically.
In the EHEC strains, as well as many other Stx-producing E. coli (STEC) strains, the toxins are encoded by lysogenic bacteriophages (70, 71, 90, 94). The EHEC O157:H7 strain EDL933 produces both Stx variants Stx1 and Stx2: Stx2 is encoded by the temperate bacteriophage 933W, while Stx1 is thought to be encoded by a cryptic prophage (70, 71). The isolation of Stx1-encoding phages from this strain has been reported, but the phage called 933J was apparently a contaminant (70); other isolates, less well characterized, seem to be 933W variants that have exchanged the Stx1 structural genes for the Stx2 genes, perhaps via a rare recombination event (79).
We have sequenced the entire Stx2 toxin-converting phage 933W, and we describe our initial analysis below. This project is part of an ongoing effort to sequence the entire genome of the EHEC O157:H7 strain EDL933; the sequences of the large virulence plasmid pO157 and the chromosomal pathogenicity island LEE have also been completed and are described elsewhere (15, 73).
MATERIALS AND METHODS
Strains and media.
EHEC EDL933 was obtained from C. W. Kaspar (Food Microbiology & Toxicology, University of Wisconsin—Madison), who obtained it from the American Type Culture Collection (ATCC 43895). Phage 933W was routinely prepared from overnight cultures of EDL933 as spontaneously released phage, separated from cells by centrifugation and filtration. The initial phage titers were ∼105 PFU ml−1, but the titers fell more than 20-fold after overnight storage at 4°C despite supplementation with 10 mM CaCl2, 10 mM MgCl2, and/or 0.1% (wt/vol) gelatin. Attempts to propagate the phage on E. coli K-12 strains in liquid culture were unsuccessful, but plate lysates were prepared on lawns of E. coli K-12. Phage were purified by precipitation with polyethylene glycol and/or by equilibrium centrifugation in CsCl density gradients, using standard techniques (3). Modified Luria-Bertani agar and broth were supplemented with 10 mM CaCl2 (71); phage titers were determined by using E. coli K-12 strain LE392 or K802.
Electron microscopy.
CsCl-banded phage in 10 mM MgCl2 were adsorbed to Pioloform-coated 400-mesh copper grids and negatively stained with 1% (wt/vol) ammonium molybdate. Negatively stained samples were viewed on a Philips CM120 STEM instrument at 60 kV.
Preparation and sequencing of nucleic acids.
Viral DNA was isolated from CsCl-banded phage, sheared by nebulization (53), and shotgun cloned into M13 Janus (14). The “933 lysate” shotgun was prepared from polyethylene glycol-precipitated cleared culture fluid of an EDL933 overnight growth. The majority of the sequencing was carried out with Sequenase and 35S label; additional data was collected with Prism fluorescent dye terminators on ABI373 and ABI377 automated fluorescence sequencers. Further sequence data was collected from a whole-genome shotgun of the original-source lysogen EDL933 as part of an ongoing effort to sequence-scan the entire genome of this pathogen. Sequence covering two final ambiguous areas was collected by PCR amplification from EDL933 genomic DNA. Sequence data was assembled and edited as described previously (22) to yield a circular duplex sequence of 61,663 bp.
Sequence analysis.
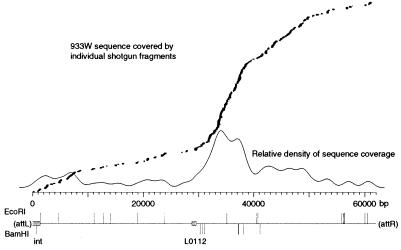
Open reading frame (ORF) identification, homology searches, and other analyses were carried out as described for the E. coli K-12 genome (11). While a number of database search and sequence alignment tools were used in the analysis, the percent identity values reported here are from the implementation of the Clustal method in MegAlign (DNASTAR) (24); unless explicitly stated otherwise, sequence comparisons are for amino acid sequences. For some comparisons, E values from BLAST 2.0 (2) are also noted. tRNAs were initially found by visual inspection of the sequence and verified with tRNAscan-SE (52). Control sequences for the tRNA search included E. coli K-12 (86 tRNAs and 2 “pseudo-tRNAs”) (11), bacteriophage T4 (8 tRNAs) (49), and the non-tRNA-containing bacteriophage sequences of lambda (85) and φ80 (74). The sequence coverage density in Fig. 3 was calculated and plotted by using S-PLUS (MathSoft, Inc., Seattle, Wash.).
FIG. 3.
Location of the endpoints of virion DNA. Each of the 222 sequence reads from the 933-lysate shotgun library was aligned with the completed 933W prophage sequence, and their location is shown above the base pair scale; the vertical offset is only to allow the display of overlapping sequences. Superimposed over that graph is plotted the relative density of sequence coverage (calculated as the midpoint value for each of 200 windows, using a kernel-density smoother with a Gaussian kernel). Below the scale are indicated the positions of EcoRI and BamHI sites in the sequence, as well as the positions of the int (integrase) and L0112 (terminase?) genes.
Nucleotide sequence accession number.
The 933W sequence has been deposited in GenBank (accession no. AF125520) as a 61,670-bp linear prophage sequence delimited by copies of the 7-bp att core sequence.
RESULTS
Morphology of the 933W virion.
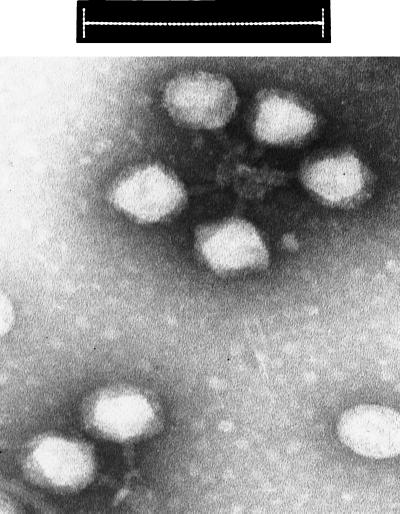
We examined the phages spontaneously released from EDL933, and confirmed the shape and dimensions reported for 933W and other Stx-converting phages from serotype O157:H7 or O157:H− strains (70, 79, 98). As shown in Fig. 1, the phage have regular hexagonal heads, about 70 nm wide. They have been variously reported as having no tails, very short tails, or short contractile tails, apparently depending on how the virions land on the grid. In many of our images, the phage particles exhibit clumping by some sort of tail-tail interaction. In such cases, the tails were more readily discerned as short contractile tails, about 27 nm long and 13 nm wide. There are indications of a baseplate-like structure as well, but no details could be made out.
FIG. 1.
Transmission electron micrograph of bacteriophage 933W. Bar, 200 nm.
Sequence of the 933W genome.
The toxin genes of 933W were previously sequenced and are the basis of several diagnostic sequence probes (13, 36, 45, 46, 75). A few other sequences from 933W have been previously reported as well (23, 88).
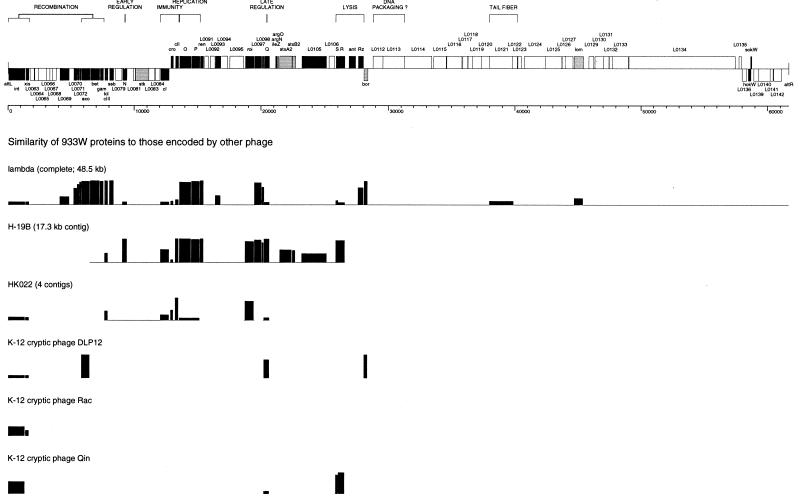
Although we expected a strong overall similarity between phage 933W and lambda (70), this was not the case for the majority of the genome. Nonetheless, despite a virion morphology quite distinct from both the “classic” lambda phages and the P22 family, 933W has similarities to lambdoid phages at both the sequence and gene organization levels. Examination of the sequence reveals a divergent arrangement of ORFs and other features reminiscent of bacteriophage lambda and its relatives. Similarities to several different lambdoid phages can be noted at both the DNA and protein levels, so that 933W can be described as a mosaic of different phages. Within this backbone of common phage elements, several known or potential pathogenicity determinants are inserted into the so-called dispensable, nonessential, or accessory regions. A map based on the sequence of 933W is presented in Fig. 2, and Table 1 lists the annotated genes. The various features of the sequence are described below.
FIG. 2.
Bacteriophage 933W map and comparison to selected other lambdoid phages. (Top) A scale drawing of the molecular map is presented above a base pair scale, arranged in the prophage orientation (attL to attR) as integrated into the E. coli O157:H7 genome. Genes and ORFs with homology to other lambdoid phages are indicated in black. Known or potential virulence factors are indicated in gray (including homologs of the lambda genes lom and bor, members of gene families that have been implicated in virulence in other contexts). All other ORFs, including those with no assigned function, are indicated in white. Gene clusters with related functions, where proposed, are indicated by brackets. (Bottom) Comparison of 933W to other lambdoid phages. Amino acid sequences were aligned with MegAlign (DNASTAR), using the Clustal method with the PAM 250 residue weight table. The percent identity is plotted as a histogram.
TABLE 1.
Genes (ORFs and tRNAs) of bacteriophage 933W
| Label | Gene | Start posi-tion | Stop posi-tion | Orien-tation | Length of product (amino acids) | Comments |
|---|---|---|---|---|---|---|
| L0061 | int | 1363 | 29 | < | 444 | Integrase |
| L0062 | xis | 1691 | 1392 | < | 99 | Excisionase? |
| L0063 | 2073 | 1762 | < | 103 | ||
| L0064 | 2498 | 2133 | < | 121 | ||
| L0065 | 3033 | 2410 | < | 207 | ||
| L0066 | 3324 | 3037 | < | 95 | ||
| L0067 | 3544 | 3326 | < | 72 | ||
| L0068 | 3833 | 3546 | < | 95 | ||
| L0069 | 4876 | 4103 | < | 257 | Ehly homolog (λ Ea22) | |
| L0070 | 5474 | 5193 | < | 93 | λ orf-61 homolog | |
| L0071 | 5676 | 5485 | < | 63 | λ orf-63 homolog | |
| L0072 | 5837 | 5649 | < | 62 | λ orf-60a homolog | |
| L0073 | exo | 6508 | 5831 | < | 225 | |
| L0074 | bet | 7290 | 6505 | < | 261 | |
| L0075 | gam | 7592 | 7296 | < | 98 | |
| L0076 | kil | 7937 | 7668 | < | 89 | |
| L0077 | cIII | 7944 | 7780 | < | 54 | |
| L0078 | ssb | 8385 | 8017 | < | 122 | |
| L0079 | 9006 | 8536 | < | 156 | ||
| L0080 | N | 9448 | 9065 | < | 127 | |
| L0081 | 10101 | 9937 | < | 54 | ||
| L0082 | stk | 11150 | 10104 | < | 348 | |
| L0083 | 11605 | 11144 | < | 153 | ||
| L0084 | 12014 | 11673 | < | 113 | ||
| L0085 | cI | 12782 | 12075 | < | 235 | |
| L0086 | cro | 12861 | 13088 | > | 75 | |
| L0087 | cII | 13227 | 13523 | > | 98 | |
| L0088 | O | 13556 | 14494 | > | 312 | |
| L0089 | P | 14491 | 15192 | > | 233 | |
| L0090 | ren | 15189 | 15479 | > | 96 | |
| L0091 | 15550 | 15828 | > | 92 | ||
| L0092 | 15997 | 16356 | > | 119 | ||
| L0093 | 16380 | 16826 | > | 148 | λ nin orf-146 homolog | |
| L0094 | 16823 | 17350 | > | 175 | DNA methylase? | |
| L0095 | 17532 | 18581 | > | 349 | ||
| L0096 | roi | 18727 | 19455 | > | 242 | |
| L0097 | 19455 | 20060 | > | 201 | λ nin orf-204 homolog | |
| L0098 | 20057 | 20251 | > | 64 | λ nin orf-68 homolog | |
| L0099 | Q | 20205 | 20678 | > | 157 | |
| L0100 | ileZ | 21120 | 21195 | > | Proposed tRNA-Ile | |
| L0101 | argN | 21205 | 21281 | > | Proposed tRNA-Arg | |
| L0102 | argO | 21295 | 21371 | > | Proposed tRNA-Arg | |
| L0103 | stxA2 | 21462 | 22421 | > | 319 | Shiga toxin 2 subunit A; also sltIIA |
| L0104 | stxB2 | 22433 | 22702 | > | 89 | Shiga toxin 2 subunit B; also sltIIB |
| L0105 | 23189 | 25126 | > | 645 | E. coli yjhS homolog | |
| L0106 | 25307 | 25753 | > | 148 | ||
| L0107 | S | 25830 | 26045 | > | 71 | |
| L0108 | R | 26050 | 26583 | > | 177 | |
| L0109 | ant | 26854 | 27423 | > | 189 | |
| L0110 | Rz | 27577 | 28041 | > | 154 | |
| Rz1 | 27797 | 27982 | > | 61 | Homolog of λ ORF overlapping Rz | |
| L0111 | bor | 28366 | 28073 | < | 97 | |
| L0112 | 28775 | 29581 | > | 268 | Terminase, small subunit? | |
| L0113 | 29562 | 31268 | > | 568 | Terminase, large subunit? | |
| L0114 | 31268 | 33412 | > | 714 | Portal protein? | |
| L0115 | 33570 | 34577 | > | 335 | ||
| L0116 | 34601 | 35815 | > | 404 | ||
| L0117 | 35871 | 36260 | > | 129 | ||
| L0118 | 36283 | 36771 | > | 162 | ||
| L0119 | 36647 | 37318 | > | 223 | ||
| L0120 | 37318 | 37968 | > | 216 | ||
| L0121 | 37965 | 39902 | > | 645 | Tail fiber | |
| L0122 | 39784 | 40173 | > | 129 | ||
| L0123 | 40220 | 40501 | > | 93 | ||
| L0124 | 40718 | 42421 | > | 567 | ||
| L0125 | 42418 | 43686 | > | 422 | ||
| L0126 | 43752 | 43979 | > | 75 | ||
| L0127 | 43985 | 44602 | > | 205 | ||
| L0128 | lom | 44693 | 45427 | > | 244 | |
| L0129 | 45857 | 46258 | > | 133 | ||
| L0130 | 46352 | 47008 | > | 218 | ||
| L0131 | 47011 | 47457 | > | 148 | ||
| L0132 | 47467 | 47718 | > | 83 | ||
| L0133 | 47729 | 48994 | > | 421 | ||
| L0134 | 49025 | 57445 | > | 2,806 | ||
| L0135 | 57728 | 57916 | > | 62 | ||
| L0136 | 58340 | 57996 | < | 114 | ||
| L0137 | hokW | 58615 | 58460 | < | 51 | gef-hokC-relF homolog |
| mokW | 58672 | 58460 | < | 70 | Longer ORF, same frame as hokW | |
| L0138 | sokW | 58664 | 58734 | > | Antisense RNA | |
| L0139 | 59376 | 58906 | < | 156 | ||
| L0140 | 60200 | 59301 | < | 299 | ||
| L0141 | 60418 | 60197 | < | 73 | ||
| L0142 | 61095 | 60466 | < | 209 |
Identification of the prophage attachment site.
Like coliphage lambda, many temperate bacteriophages integrate into their host genomes via a site-specific recombination event between short common-core sequences within the phage (attP) and bacterial (attB) attachment sites, generating two composite core sequences (attL and attR) flanking the integrated linear prophage genome. Excision of the prophage involves a similar site-specific recombination event between attL and attR, to generate a circular phage genome.
The location of the 933W prophage in EDL933 was determined by examination of the data from a whole-genome shotgun library of that strain and confirmed by PCR across the attL and attR junctions. The prophage sequence is flanked by two copies of a 7-bp repeat (GTTTCAA) present only once in E. coli K-12 and only once in the circular phage sequence, which we conclude is the core of the 933W att sites. Integration of 933W disrupts the wrbA gene, which encodes the Trp repressor-binding protein, WrbA. During stationary phase, E. coli K-12 cells deficient in WrbA are less efficient than wild-type cells in their ability to repress the trp promoter (99). It was proposed that the WrbA protein functions as an accessory element in blocking TrpR-specific transcriptional processes that might be physiologically disadvantageous in the stationary phase of the bacterial life cycle. Of course, it is not known what the physiological situation might be in the intestinal tract. In a 933W lysogen, translation starting from the wild-type wrbA initiator codon could yield only a 20-residue peptide, containing the first 18 amino acids of WrbA. At the other end of the prophage, a phage-encoded start codon overlapping the stop codon of the integrase (ATGA) might allow the synthesis of a 192-amino-acid product retaining most of the WrbA sequence, although the first 13 residues of the wild-type protein would be replaced by a different 7-amino-acid sequence. This sequence alteration would truncate a conserved domain noted in a “WrbA family” of proteins (domain 4535 of ProDom; 38 residues), but the impact, if any, upon function is unknown.
Identification of the virion DNA endpoints.
Since the majority of our sequence data for 933W was determined by using a shotgun library of DNA derived from phage particles, if the packaged phage DNA had specific endpoints (i.e., like bacteriophage lambda), we should have generated a unique linear sequence upon its assembly. Moreover, based on our experience with nebulized shotgun libraries derived from other linear DNAs, the actual endpoints of the source DNA would be expected to be proportionally overrepresented, presumably because the end repair of enzymatically generated ends is much more efficient than the repair of ends generated by physical shearing. Instead, assembly of the sequence data yielded a partial concatamer with no unique ends or pileups. The identification of the integrated prophage endpoints indicated that the assembled sequence was a circular permutation of the prophage sequence. The 61,670-bp sequence is presented here in the prophage state, starting and ending with the 7-bp att core.
The lack of definite endpoints in the assembly of virion DNA sequences might indicate the absence of a defined cos-type end (although cohesive ends might have annealed to generate circles or linear concatamers prior to nebulization). The full data set used to complete the sequence is anything but random, since specific data was collected in a directed manner to deal with gaps and ambiguities. To more specifically address the question of the virion DNA endpoints, all of the sequence data collected from the “933 lysate” shotgun library—which does represent a “random” data set—was aligned with the consensus prophage sequence. As shown in Fig. 3, our data supports a conclusion that the virion endpoints of this phage are not fixed in the genome but instead are distributed over a region of several kilobase pairs. We suggest that this is the result of a headful packaging of sequences longer than one full genome, as demonstrated for bacteriophages P22, P1, and T1 (8).
Earlier estimates of the 933W genome size, actually estimates of the virion DNA length, were longer than the sequence we have determined (70, 98). These estimates were based on the lengths of restriction fragments, which might also shed light on the unique-versus-headful-packaging question. In our experience, 933W virion DNA was recalcitrant to restriction digestion, but complete digestion with EcoRI or BamHI could be achieved with prolonged incubation; the positions of the EcoRI and BamHI sites in the sequence are shown in Fig. 3. The restriction fragments (data not shown) are entirely consistent with a circular form of the completed sequence and are indistinguishable from those reported previously. The discrepancies in calculated lengths can be accounted for by the inherent margin of error in determining fragment lengths based on electrophoretic mobility; for example, the reported BamHI fragments of >23, 6.3, 3.05, 0.88, and 0.39 kbp (98) can be correlated with sequence-derived lengths of 29,558 and 21,247, 6,161, 3,031, 890, and 389 and 387 bp. These results suggest either a circular virion DNA or a collection of circularly permuted linear DNAs where submolar or minority fragments from individual molecules (with one end generated by packaging instead of a restriction cut) do not appear as bands.
Descriptions of selected ORFs.
Genes with functional assignments are named, usually after the equivalent lambda genes when possible. ORFs with no functional assignment or only tentative assignments based on gene arrangement and location are designated only by ORF numbers. Genes (ORFs and RNAs) are labelled L0061 to L0142, from left to right as shown in Fig. 2; these labels are from a series of unique identifiers for the genes of EDL933. References to genes from E. coli K-12 include the corresponding identifier labels (b numbers) assigned to those genes in the complete genome sequence of that strain (11).
In all cases for which data is available, integrative recombination is mediated by a phage-encoded recombinase (integrase, or Int protein) which catalyzes the strand cleavage and rejoining, and excision usually requires the cooperative action of Int and a second phage-encoded excision protein (Xis). We have designated L0061, the ORF following attL in the prophage, as int; sequence comparisons suggest that it is a member of the integrase-recombinase family, although very distant from lambda, P22, and other lambdoid phages. The closest homologs are the putative integrases of the E. coli K-12 cryptic prophages Rac (40.1% identical to the product of ORF b1345, IntR) and Qin (52.0% identical to the product of ORF b1579, IntQ). Despite the sequence diversity exhibited by known Int proteins, all of these recombinases can be aligned in their C-terminal halves to reveal a conserved region implicated as the active site of this family (1, 4). The proposed 933W Int shows some similarity (10.5 to 15.3% identity; BLAST E values, 2 × 10−3 to 3 × 10−40) to a great number of other Int sequences, and examination of the alignments reveals that with a single exception, it contains the conserved residues, including the tyrosine residue identified as the active amino acid involved in a transient phosphodiester linkage to the DNA during strand cleavage and rejoining. The exception, a tyrosine residue instead of a histidine in the highly conserved His-X-X-Arg motif, is also present in the Rac and Qin integrases. Given the observation that in the P1 Cre integrase this precise substitution reduced the recombination activity about sevenfold in vivo (1), the question arises of how efficient these putative integrases are. However, this substitution is also present in the integrase of the Streptomyces lividans SLP1 element (12), while the putative integrase of the Pseudomonas aeruginosa phage φCTX contains an N (asparagine) residue instead of the H (39).
By analogy to lambda and other temperate phages, the 933W ORF L0062, immediately upstream of int, may encode the phage excisionase. A similar sequence is found in the Rac prophage (25.3% identical to YdaQ, b1346), which is known to be excisable, but not in Qin, where an IS2 insertion near intQ is accompanied or followed by a deletion of flanking sequences. With a few exceptions, Xis sequences show little homology to one another, and even such generalizations as the lambdoid Xis proteins being basic while those from gram-positive bacteria are often acidic (58) are rife with exceptions. The suggestion has been made that temperate phage excisionases have a helix-turn-helix motif, as scored by the metric of Dodd and Egan (26) for recognizing such motifs (83). This metric generates SD scores, which are standard deviation units relative to the appropriate mean; scores ≥ 2.5 SD are indicative of a helix-turn-helix motif. While most of the lambdoid Xis proteins do not score well with the program (25), the best-scoring regions of 933W Xis (score of 0.37 SD at position 33) and YdaQ (score of −0.34 SD at position 19) do align with the motif pointed out by Salmi et al. (83), suggesting that some helix-turn-helix character may be involved in these proteins as well. However, in the absence of any experimental data, the possibility must be noted that excision of 933W (and perhaps Rac) does not require an excisionase, as such. Excision of the Staphylococcus aureus phages φ13 and φ42 requires no phage-encoded product other than the integrase (17), and for coliphage 186 the transcriptional repressor protein Apl also serves as the phage-encoded excision factor (78).
ORF L0069 is homologous (89.7% identity) to “Ehly2,” the product of an ORF associated with an enterohemolysin 2 activity encoded by phage C3208 in E. coli O26:H11 (6). Both of these hypothetical proteins are also similar to lambda Ea22 (L0069, 35.2% identity; Ehly2, 30.2% identity) and P22 EaD (L0069, 24.9% identity; Ehly2, 25.3% identity), whose genes occur in analogous positions within their respective genomes. In the absence of any demonstrated hemolysin activity by the Ehly2 protein itself, this similarity is best viewed as an indication that phage C3208 is also a member of the lambdoid family. If this protein does have a cytotoxic effect, it may contribute to the virulence of O157:H7.
The 933W sequence spanning ORFs L0073 to L0078 is similar to the recombination region of lambda (97% identity over 3,698 bp), with both the gene order and predicted amino acid sequences of individual genes highly conserved; these ORFs are therefore designated exo (97.3% identity), bet (99.6% identity), gam (97.0% identity), kil (98.9% identity), cIII (98.1% identity), and ssb (99.2% identity).
ORF L0080 was initially identified as a candidate for an analog of the lambda regulatory gene N largely on the basis of its position within the sequence (its product shows 14.0% identity to lambda N; 29.0% identity to P22 gene 24 protein). It was subsequently found to be very similar (96.9% identity) to the N gene of H19-B.
The predicted product of ORF L0082 resembles the family of eukaryotic serine/threonine protein kinases (12.6 to 20.1% identity to more than 100 distinct serine/threonine kinases from a variety of organisms; BLAST E values, 2 × 10−4 to 3 × 10−20), and we have designated the gene stk (for “serine/threonine kinase”). The sequence similarities span the conserved regions in the catalytic domain of the eukaryotic protein kinases, including both the ATP binding and active sites. The Stk sequence is more similar to eukaryotic serine/threonine protein kinases (e.g., 17.2% identity to STE20 of Saccharomyces cerevisiae) than to other prokaryotic protein kinases, including those of Mycobacterium tuberculosis (10.7% identity), Streptomyces coelicolor (11.3% identity), Myxococcus xanthus (12.6% identity), Bacillus subtilis (12.7% identity), and Yersinia pseudotuberculosis and Y. enterocolitica (11.0% identity).
In bacteriophages lambda and φ80, the NinR regions contain orf-221, which encodes a phosphoprotein phosphatase resembling those of mammalian origin (20). The function of this phage-encoded activity is unknown, although it presumably acts to modulate the signal transduction pathways of the E. coli host in a manner similar to the E. coli PrpA and PrpB phosphatases, described by Missiakas and Raina (60). While 933W apparently encodes a protein kinase, the phage does not encode a homolog of this phosphatase. There is some suggestion that the Yersinia protein kinase (YpkA) is involved in virulence by interfering with the signal transduction pathway of the mammalian host (38), and bacteriophage 933W may interfere with the host systems in the same manner. The location of stk is analogous to the position of the rexAB genes in lambda, suggesting that it could be expressed in the lysogen.
ORF L0085 encodes the CI repressor of 933W, which appears to be a hybrid of two species of repressors. The amino-terminal 89 amino acids most closely resemble the repressor of phage HK022 (37.4% similarity), while the rest of the sequence is almost identical to that of H19-B at both the nucleotide (95.2% identity) and amino acid (96.6% identity) levels. In the well-characterized lambda repressor, the amino-terminal residues contain the DNA binding helix-turn-helix motif that interacts with the operator sequence, while the carboxy-terminal domain of the protein is involved in dimerization. A similar “hybrid” repressor was recently noted in a comparison of the lysogeny modules from two temperate Streptococcus thermophilus bacteriophages (66).
ORF L0086 is the 933W cro homolog and encodes a protein most similar to that of HK022 (43.4% identity). This is consistent with the cI structure described above, given that both CI and Cro must recognize the same operator sequences. ORF L0087 is the cII homolog, encoding a protein similar to those of HK022 (91.9% identity) and H19-B (98.0% identity).
Confirming earlier hybridization and partial-sequence data (23), the replication origin and replication genes of 933W are nearly identical to those of lambda (94.0% nucleotide sequence identity), as well as to H19-B (96.7% nucleotide sequence identity). Sequence similarities allow assignment of L0088 and L0089 as the replication genes O (98.0% identity to lambda; 98.7% identity to H19-B) and P (96.6% identity to lambda; 95.7% identity to H19-B), respectively, and L0090 as ren (99.0% identity to lambda; 97.9% identity to H19-B). The replication origins of 933W and H19-B have a 39-bp insert relative to that of lambda, containing two additional iterons. This insert results in an in-frame insertion of 13 amino acids in the replication protein O of both Stx phages. An in vitro readthrough of the UAG termination codon of the O gene has been found in bacteriophage lambda (100); the conservation of the O sequences includes this extended carboxy-terminal region.
The 933W sequence to the right of the replication origin contains homologs of several lambda and P22 Nin region ORFs: L0093 (39.0% identity to the product of lambda orf-146; 39.3% identity to P22 NinB), L0097 (91.6% identity to the product of lambda orf-204; 88.1% identity to P22 NinG), and L0098 (73.8% identity to the product of lambda orf-68; 66.2% identity to P22 NinH), as well as HK022 Roi (L0096, 78.9% identity). These ORFs occupy analogous positions in the different phage genomes, and, except for L0093, there are also homologs in H19-B.
ORF L0094 encodes a protein similar to a hypothetical methylase of bacteriophage HP1 (30.1% identity) and the DNA (N6-adenine) methyltransferase of bacteriophage T1 (22.3% identity). Although no functional characterization of this protein is available, such a DNA modification might explain the difficulties we experienced when attempting to digest 933W DNA with a number of restriction enzymes.
ORF L0099 was identified as the homolog of the late regulatory gene Q, most closely resembling the functional analog of lambda Q from phage DLP12 (b0551, YbcQ; 77.2% identity); it was subsequently found to be almost identical (96.5% identity) to the Q gene product of H19-B.
The Stx2 subunits are encoded by stx2A and stx2B (aka sltIIA and sltIIB) and were previously characterized (43, 87, 89).
Downstream of the stx genes and seemingly part of the same transcript, ORF L0105 encodes a protein similar (50.8% identity) to E. coli K-12 YjhS (b4309). Examination of the H19-B sequence reveals an unannotated 849-bp ORF downstream from the stx1 genes (accession no. AF034975, bases 14651 to 15499), whose product is also similar to YjhS (20.5% identity). The function of these genes is unknown, but yjhS is part of a fimbrial synthesis and iron transport region which K-12 may have acquired by horizontal transfer.
ORF L0107 is the 933W analog of the lysis (holin) gene S, resembling those from the Qin prophage (79.2% identity) and H19-B (91.3% identity). Like a number of other lambdoid holin genes (9), this gene has two Met start codons separated by one or two codons. However, in 933W neither of the codons between the alternate starts is an arginine or lysine codon, and it is not clear whether a dual-start motif is involved in the regulation of 933W lysis.
L0108 is the R gene (endolysin) analog, also resembling that of Qin (88.8% identity); although no R gene is annotated in the H19-B sequence, the insertion of 4 bases near the end of that sequence (C between bases 16927 and 16928; AA between 17254 and 17255, and G between 17303 and 17304) would create an R homolog running off the end of the entry, 91.4% identical to L0108.
In other lambdoid phages, S and R are followed by Rz, and 933W ORF L0110 is an Rz homolog most like that from lambda (71.2% identity); a homolog of the overlapping Rz1 reading frame (48) is also present. 933W has an additional ORF, L0109, inserted between R and Rz, whose product resembles the P22 Ant antirepressor (34.2% identity). The function of this protein in 933W is unknown, although its location suggests a possible regulatory role in lysis of the host cell.
The end of the lysis region of 933W is similar to that of lambda (91.0% identity over 940 bp). In addition to Rz (and Rz1), this region contains a homolog (L0111; 96.9% identity) of the lambda bor gene. In lambda lysogens, bor expression has been implicated in serum resistance (5), which may confer a selective advantage to cells carrying the prophage.
Analogy to other phages was useful in the analysis of the first “half” of the 933W genome. However, if the ORFs in the remainder of this genome encode the virion structural and morphogenic proteins, as continued analogies would argue, very few can be even tentatively identified on the basis of sequence comparisons. A number of complete or partial phage sequences have been determined, and comparisons reveal homologies between different phages for genes encoding enzymes, regulatory proteins, replication proteins, and various “accessory” products. However, the genes encoding the actual structural proteins that comprise the virion seem to be drawn from a much larger pool of potential sequences—as if the possible ways to build morphologically similar phage particles are myriad and our sampling has merely skimmed the surface of the gene pool.
Based on the 933W virion DNA endpoint analysis, one might expect the region starting with ORF L0112 to contain the genes involved in DNA packaging. Although the analogous genes from a number of phages show little sequence homology, there is a conservation of gene position and relative size (8, 29, 92). We have tentatively assigned L0112 to L0114 as follows, with “informative” database matches indicated: L0112, the terminase small subunit (bacteriophage P1 PacA, BLAST E value 0.78); L0113, the terminase large subunit (bacteriophage T4 gp17; BLAST E value 0.005); and L0114, the prohead portal protein (bacteriophage P22 gp1; BLAST E value 0.002). The sequence matches have very poor scores, and these assignments must be thought of as provisional.
One of the few structural-gene candidates that can be identified is L0121, a putative tail fiber gene; its product shows 17.0% identity to the Stf tail fiber of lambda and contains motifs described in various phage tail fiber proteins (84). This ORF is also one of several phage genes encoding structural components of bacteriophage virions in which the characteristic collagen-like repeats (Gly-X-Y)n have been noted (91). The predicted 933W tail fiber protein displays extensive homologies to collagen (e.g., 26.8% identity to human alpha type I collagen), with stretches of 40 and 38 repeats of the collagen motif. The repeats have the bias toward proline at the second and third positions of the motif that has long been known to occur in vertebrate collagens. Furthermore, the collagen sequences in 933W tail fibers may well have the triple-helix structure found in animal collagen: all well-characterized phage tail fiber proteins are trimeric (18), including the phage λ fibers that have sequence similarity to the 933W protein. We note that the position of L0121 in the genome of 933W is not analogous to that of the lambda stf gene, but, given the difference in tail morphology between these phages, analysis by analogy is probably at its weakest for tail structural and morphogenic genes.
In addition to the bor homolog in the lysis region, two other 933W ORFs may be involved in virulence. L0128 encodes a homolog (28.6% identity) of the lambda Lom protein (77, 82), which encodes a member of a family of outer membrane proteins associated with virulence in two species. Expressed in lysogens, these proteins confer the ability to survive in macrophages.
L0137 encodes a member of the hok-gef-relF family of killer proteins from “toxin-antitoxin” systems (90.4% identity to RelF; 78.4% identity to Gef). In these systems, the best characterized of which are involved in plasmid maintenance, an unstable antisense RNA prevents expression of the lethal protein by binding to the more stable mRNA (33, 76). Examination of the sequence surrounding L0137 reveals that all of the sequence elements and potential secondary-structure features described for these systems (30, 35) are conserved in 933W. By analogy to the Hok system, L0137 is designated hokW (for “host killing, 933W”) and the antisense RNA is designated sokW (for “suppression of killing, 933W”). If these genes are expressed in the lysogenic state, loss of the prophage would be selected against in a manner similar to the selection against loss of plasmids. Interestingly, in the O157:H7 strain EDL933, at least four such systems are present: in addition to phage 933W, the large virulence plasmid pO157 carries a Hok system homolog (15), and data from the genomic sequence indicates that Gef and RelF homologs are also present (10). The target specificity of these systems resides in the interactions between the antisense RNA and the mRNA, and for the plasmid and phage sequences these seem to be different. Whether the chromosomal loci interact with either of those systems to reinforce the maintenance of the various pathogenicity determinants is unknown.
tRNAs.
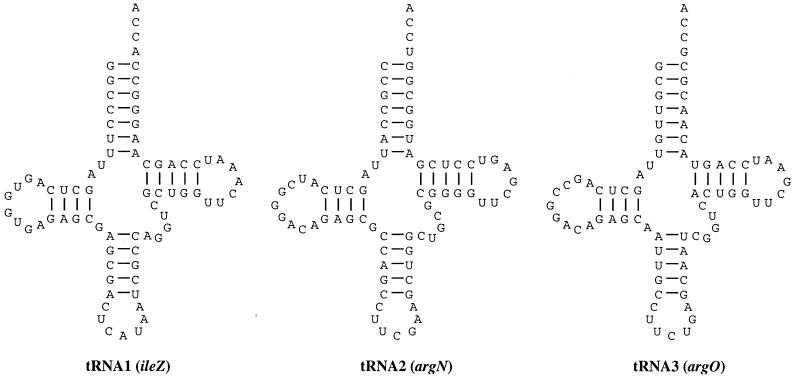
In the region between the late regulatory gene Q and the Shiga toxin genes, a tRNAIle-like sequence was previously found (88). Our reexamination of the sequence reveals genes for two additional tRNA sequences, and the proposed cloverleaf secondary structures of all three tRNAs are shown in Fig. 4. Most invariant or semi-invariant eubacterial tRNA residues are present in these sequences, and they may encode functional tRNAs although this has not been demonstrated.
FIG. 4.
Bacteriophage 933W apparently encodes three tRNAs. Each of the tRNA-like sequences is shown folded into the cloverleaf tRNA secondary structure.
tRNA1 (designated ileZ) has the anticodon CAU. The sequence of the tRNA closely resembles that of the tRNAIle species encoded by the E. coli K-12 genes ileY (92.1% identical) and ileX (89.5% identical), and none of the differences affect residues known to be involved in the function or identity of the tRNAs. The identification of these as Ile tRNAs rests on the experimental characterization of the chromosomal ileX locus. If the wobble position C34 remained unmodified, the CAU anticodon would correspond to the Met codon AUG. However, in the tRNAIle encoded by bacteriophage T4 and E. coli ileX (as well as a number of other organisms), the C34 is modified to lysidine (2-lysylcytidine, k2C). This modification bestows AUA (Ile) decoding capacity and is required for recognition by the isoleucyl-tRNA synthetase (54, 62, 63). The other known determinants for E. coli tRNAIle are present in ileZ: the anticodon loop bases A37 and A38, the discriminator base A73, and the C4 · G69, U12 · A23, and C29 · G41 base pairs (67, 72).
tRNA2 has the anticodon UCG, which is not found in any known E. coli tRNAs, and in fact the sequence is not similar to any other specific tRNA species. On the basis of the anticodon, this sequence is designated argN. Uridine at the first position of the anticodon is modified in all cases so far sequenced at the RNA level in E. coli (97, 101); therefore, modification of U34 may restrict recognition by this species to a subset of the CGN family of Arg codons. In E. coli K-12, the CGU, CGC, and CGA codons are read by the ICG anticodon while the CGG codon is read by CCG. This sequence does contain the A20 and C35 determinants for E. coli tRNAArg, but the residue at position 73 is U instead of A or G (57, 72). In addition, the almost invariant base pair R15 · Y48 is replaced by T15 · G48, which might prevent formation of the “Levitt pair” involved in tRNA tertiary structure (51)—although the E. coli tRNACys has G15 · G48 (55), which is an identity element for this tRNA (56).
tRNA3 (argO) has the anticodon UCU; with an unmodified U as the first nucleotide of the anticodon, this could correspond to the codons AGA (Arg), AGG (Arg), AGU (Ser), and possibly AGC (Ser). As with tRNA2, this sequence does not closely resemble any tRNA sequences in the sequence databases. However, the tRNAArg species encoded by bacteriophage T4, E. coli argU (dnaY), and Salmonella argU (fimU) all have UCU anticodons, and these species favor the arginine codon AGA due to modification of U34 to 5-methoxycarbonylmethyluridine (mcm5U) (93). The argO sequence contains the A20 and C35 determinants for E. coli tRNAArg; A73 is replaced G73, but this has been observed in some tRNAArg species (57).
It was suggested (88) that the tRNAIle sequence was related to the integration site of the 933W prophage into the E. coli chromosome, since a number of other bacteriophages and pathogenicity-associated islands are inserted at or near tRNA genes (19, 37). This is not the case for 933W, although the similar E. coli K-12 chromosomal ileY locus is the insertion target of coliphage 186 (78). However, the proximity of the tRNA genes to the stx genes might reflect the outcome of another recombination event during which these sequences were initially acquired by the phage. If this is the case, are the tRNAs functional or are they just along for the ride? Examination of codon usage (Table 2) suggests that the phage-encoded tRNAs could serve to supplement the host tRNA pool, allowing the rare codons to be more efficiently decoded (88). This may provide sufficient selective advantage to retain the tRNAs, regardless of their origin. There are no double “killer arginines” (102, 103) in the 933W genome, and so it seems unlikely that the phage makes any explicit regulatory use of differential tRNA availability. However, the alteration of tRNA base modifications has been reported to affect virulence factor expression in Shigella flexneri (27, 28) and Agrobacterium tumefaciens (34), and this may actually be the result of an alteration in the efficient translation of key proteins.
TABLE 2.
Isoleucine and arginine codon usagea
| Gene | Codon usageb
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ile (AUU) | Ile (AUC) | Ile (AUA) | Arg (CGC) | Arg (CGU) | Arg (CGG) | Arg (CGA) | Arg (AGA) | Arg (AGG) | |
| E. coli K-12 genome | 30.3 | 25.1 | 4.4 | 22.0 | 20.9 | 5.4 | 3.6 | 2.1 | 1.2 |
| E. coli K-12 all ORFs | 30.8 | 24.5 | 5.2 | 21.4 | 20.3 | 5.7 | 4.0 | 2.7 | 1.7 |
| 933W genome | 22.9 | 19.3 | 10.1 | 15.2 | 19.2 | 8.9 | 6.5 | 9.9 | 6.0 |
| 933W all ORFs | 24.2 | 23.2 | 12.5 | 14.5 | 14.0 | 9.0 | 7.9 | 11.6 | 6.6 |
| 933W stxA | 9.4 | 6.3 | 46.9 | 6.3 | 28.1 | 9.4 | 6.3 | 18.8 | 6.3 |
| 933W stxB | 11.1 | 11.1 | 0.0 | 11.1 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 933W stk | 22.9 | 2.9 | 48.7 | 0.0 | 8.6 | 5.7 | 0.0 | 25.8 | 11.5 |
For E. coli K-12 (4,290 ORFs, 1,363,501 codons) and bacteriophage 933W (78 ORFs, 18,246 codons), codon usage was calculated for each genome (i.e., all codons) and for each ORF individually.
Values are expressed as codon frequency per 1,000 codons; for “all ORFs,” the mean of the individual ORF values is shown.
Promoters, operators, and terminators.
A number of other noncoding sequence elements were examined in the 933W sequence, especially where homologs in better-characterized phages allowed “analysis by analogy.” These features are annotated in the GenBank entry, and a few are briefly noted here.
The 933W N-L0081 intergenic region should contain tM, nutL, OL, and pL. A candidate for tM is present immediately downstream (to the left) of L0081, in a short sequence of dyad symmetry conserved between 933W and H19-B; no assignment could be made for OL or pL. Since the H19-B and 933W N genes are almost identical and nut sites are N specific, the nut sites should be within the sequences conserved between the two phages; a nutL candidate was identified, although it is not clear which of two “boxB”-like sequences would be involved.
The cI-cro intergenic region should include OR, pRM, and pR. Given the similarity of the amino-terminal domains of the 933W and HK022 repressors, sequence comparisons with the equivalent region from HK022 (68) allowed us to tentatively identify all three features in 933W. The ready identification of OR makes the failure to detect OL all the more puzzling, and it may be that regulation of transcription of the left and right arms of 933W are achieved by entirely unrelated means.
The cro-cII intergenic region should include nutR, pRE (pE), and tR1. This region is 100% conserved between H19-B and 933W. pRE, by analogy to other lambdoid phages, would be activated by 933W (and H19-B) CII; however, the recognition site for this protein is not known, and no candidates are proposed. A nutR candidate was found, and, as was the case for nutL, there are two candidate “boxB” sequences; perhaps in 933W and H19-B the N recognition determinants involve an extended sequence relative to those in other lambdoid systems (31, 32, 86).
The near identity between the 933W and H19-B Q genes extends 293 bp beyond the coding sequences (i.e., ending upstream of the tRNA genes), defining the region where the pR′ promoter and qut site should be located. The A(N)3T(S)2–3 motif in the nontranscribed DNA strand, noted by Ring and Roberts (80), can be found in several locations downstream of the presumptive pR′ promoter, but only one of these occurs in close proximity to the promoter and before the first potential terminator. We propose that this is part of the qut site for both 933W and H19-B. Once Q protein modifies the RNA polymerase complex at qut, sequence divergence should not alter its antitermination activity for the late transcripts; therefore, the late regions of both phages are likely to be under Q regulation.
In E. coli, tRNA genes are often found in clusters with typical prokaryotic −35 and −10 promoter elements as well as a GC-rich discriminator domain common to all E. coli genes subject to stringent control, and downstream of almost all tRNA genes is found a rho-independent terminator-like structure (42). Both of these features are found flanking the 933W ilvZ-argN-argO cluster, and the predicted transcript would be a trimeric precursor RNA resembling those readily processed by the E. coli RNA processing machinery.
DISCUSSION
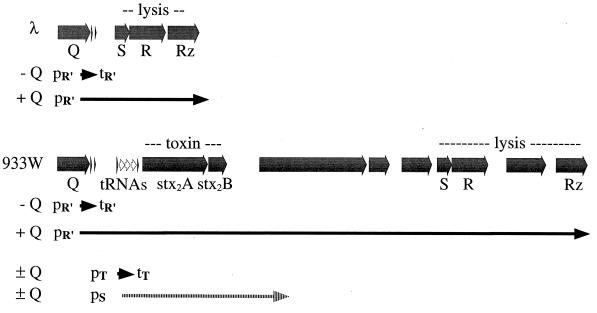
The Q gene product of lambdoid phages functions as a transcription antiterminator that regulates the expression of late phage genes by modifying the transcription complex initiated at the late promoter pR′. The protein acts at the qut site overlapping the promoter, and the Q analogs of different phages are specific for their own qut sites (81). Based on the arrangement of the 933W genes, the stx2 genes are part of an apparent Q-dependent late transcript, as diagrammed in Fig. 5. If this is the case, the toxin would be expressed only (or at least maximally) during lytic growth of the phage.
FIG. 5.
The stx genes of O157:H7 are integrated into the late operon of a lambdoid bacteriophage. The late operons of lambda and 933W are diagrammed for comparison, indicating ORFs, tRNAs, and real or predicted promoters (p) and terminators (t). Transcripts (known for lambda, proposed for 933W) in the presence or absence of the cognate gpQ antitermination activity are indicated by arrows below the gene maps. The proposed tRNA promoter (pT) is not associated with a qut site, and transcripts presumably terminate at tT. The reported Stx promoter (pS) also lacks a qut site, and the length of that transcript is unknown.
Mühldorfer et al. (61) examined the regulation of the stx2 operon in experiments involving a low-copy-number plasmid carrying a translational fusion of stx2A to a phoA reporter gene. They concluded that a phage factor played a positive regulatory role in the expression of stx2 and that this factor could be provided in trans by either 933W or H19-B but not lambda. The increased expression was mitomycin C and recA+ dependent, as expected for a mechanism requiring prophage induction. These results are entirely consistent with the phage factor being the Q gene: when provided in trans by the phage after induction and transition to lytic growth, Q could act to antiterminate pR′ transcripts on the reporter plasmid (the constructs included the entire Q-stx2A intergenic region). The similarity of the 933W and H19-B genes is such that we would expect the H19-B Q gene to function as well as that from 933W in this system.
The results of our analyses seem to be at odds with the report by Sung et al. (95) that a promoter for stx2 was located only 118 bp upstream of the stx2A coding sequence, which would put the promoter (pSlt-II or pStx2) within argO. It may be that some constitutive level of Stx2 expression is provided by that promoter but that Stx2 production is significantly increased after phage induction as a more efficient promoter becomes available.
A number of pathogenicity factors, including several toxins, are encoded by lysogenic phages (7, 19). A linking of toxin production to prophage induction in such cases might open another means of increasing the toxin yield. Infections are unlikely to occur in monoculture, and while other bacteria already carrying the prophage (uninduced) would be immune to superinfection, nonlysogens in the vicinity could be infected and produce additional phage (and toxin) in what can be envisioned as an amplification by recruitment. It has been shown that the cholera toxin-encoding phage CTXφ infects Vibrio cholerae more efficiently within the gastrointestinal tracts of mice than under laboratory conditions (96). A coupling of toxin release with phage release might also favor DNA transfer events, including the acquisition of new pathogenicity determinants, by other bacteria under conditions where the bacteria would be more likely to be subsequently released into the environment.
Bacteriophage H-19B, isolated from E. coli O26:H11 strain H19 (90), is morphologically quite distinct from 933W and more closely resembles lambda (98). This phage also displays greater sequence homologies to lambda (as detected by hybridization), and the virion DNA has cohesive termini (41). Nonetheless, both 933W and H-19B have stx genes in analogous positions and have similar regulatory elements. The cryptic Stx1 phage in EDL933 is still essentially uncharacterized, and there remains a possibility that it resembles H-19B. If the Stx prophages in the EDL933 genome have overlapping regulatory specificities, the coexistence of both elements could present an interesting additional layer of complexity.
ACKNOWLEDGMENTS
This work was supported by NIH grant AI41329-01 and by a research grant from the Ronald McDonald House Charities.
We thank the technical staff of the University of Wisconsin Genomes Project for help with sequencing, Randall Massey and Grayson Scott of the University of Wisconsin Medical School Electron Microscope Facility for electron microscopy, and Bill McClain for useful discussions about the tRNAs. For bearing with his return to the bench after too many years at the computer, one of us (G.P.) also thanks Heather Kirkpatrick of this laboratory.
ADDENDUM
Sequence data from the Stx1-encoding lambdoid bacteriophage H19-B (64) was compared to our sequence in the course of our analyses, and similarities are noted above. While this paper was being revised, Neely and Friedman (65) published their analysis of this H-19B sequence. They reached conclusions similar to our own regarding the regulation of Stx production and phage release and showed that the H-19B Q gene product can activate the expression of 933W stx2 genes as well as its own stx1 genes.
Footnotes
Paper 3519 from the Laboratory of Genetics.
REFERENCES
- 1.Abremski K E, Hoess R H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992;5:87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arber W, Enquist L, Hohn B, Murray N E, Murray K. Experimental methods for use with lambda. In: Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1983. pp. 433–466. [Google Scholar]
- 4.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Pierson III L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barondess J J, Beckwith J. A bacterial virulence determinant encoded by lysogenic coliphage lambda. Nature. 1990;346:871–874. doi: 10.1038/346871a0. [DOI] [PubMed] [Google Scholar]
- 6.Beutin L, Stroeher U H, Manning P A. Isolation of enterohemolysin-(Ehly2) associated sequence encoded on temperate phages of Escherichia coli. Gene. 1993;132:95–99. doi: 10.1016/0378-1119(93)90519-9. [DOI] [PubMed] [Google Scholar]
- 7.Bishai W R, Murphy J R. Bacteriophage gene products that cause human disease. In: Calender R, editor. The bacteriophages. Vol. 2. New York, N.Y: Plenum Press; 1988. pp. 683–724. [Google Scholar]
- 8.Black L W. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–292. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 9.Blasi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 10.Blattner, F. R. Unpublished data.
- 11.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 12.Brasch M A, Pettis G S, Lee S C, Cohen S N. Localization and nucleotide sequences of genes mediating site-specific recombination of the SLP1 element in Streptomyces lividans. J Bacteriol. 1993;175:3067–3074. doi: 10.1128/jb.175.10.3067-3074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brian M J, Frosolono M, Murray B E, Miranda A, Lopez E L, Gomez H F, Cleary T G. Polymerase chain reaction for diagnosis of enterohemorrhagic Escherichia coli infection and hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1801–1806. doi: 10.1128/jcm.30.7.1801-1806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burland V, Daniels D L, Plunkett III G, Blattner F R. Genome sequencing on both strands: the Janus strategy. Nucleic Acids Res. 1993;21:3385–3390. doi: 10.1093/nar/21.15.3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burland V, Shao Y, Perna N T, Plunkett III G, Sofia H J, Blattner F R. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 1998;26:4196–4204. doi: 10.1093/nar/26.18.4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calderwood S B, Acheson D W K, Keusch G T, Barrett T J, Griffin P M, Strockbine N A, Swaminathan B, Kaper J B, Levine M M, Kaplan B S, Karch H, O’Brien A D, Obrig T G, Takeda Y, Tarr P I, Wachsmuth I K. Proposed nomenclature for SLT (VT) family. ASM News. 1996;62:118–119. [Google Scholar]
- 17.Carroll D, Kehoe M A, Cavanagh D, Coleman D C. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages phi 13 and phi 42. Mol Microbiol. 1995;16:877–893. doi: 10.1111/j.1365-2958.1995.tb02315.x. [DOI] [PubMed] [Google Scholar]
- 18.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calender R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 15–91. [Google Scholar]
- 19.Cheetham B F, Katz M E. A role for bacteriophages in the evolution and transfer of bacterial virulence determinants. Mol Microbiol. 1995;18:201–208. doi: 10.1111/j.1365-2958.1995.mmi_18020201.x. [DOI] [PubMed] [Google Scholar]
- 20.Cohen P T, Cohen P. Discovery of a protein phosphatase activity encoded in the genome of bacteriophage lambda. Probable identity with open reading frame 221. Biochem J. 1989;260:931–934. doi: 10.1042/bj2600931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook R. Toxin. New York, N.Y: G. P. Putnam’s Sons; 1998. [Google Scholar]
- 22.Daniels D L, Plunkett III G, Burland V, Blattner F R. Analysis of the Escherichia coli genome: DNA sequence of the region from 84.5 to 86.5 minutes. Science. 1992;257:771–778. doi: 10.1126/science.1379743. [DOI] [PubMed] [Google Scholar]
- 23.Datz M, Janetzki-Mittmann C, Franke S, Gunzer F, Schmidt H, Karch H. Analysis of the enterohemorrhagic Escherichia coli O157 DNA region containing lambdoid phage gene P and Shiga-like toxin structural genes. Appl Environ Microbiol. 1996;62:791–797. doi: 10.1128/aem.62.3.791-797.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.DNASTAR Inc. LASERGENE for Macintosh User’s Guide. Madison, Wis: DNASTAR, Inc.; 1997. [Google Scholar]
- 25.Dodd, I. Personal communication.
- 26.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Durand J M, Bjork G R, Kuwae A, Yoshikawa M, Sasakawa C. The modified nucleoside 2-methylthio-N6-isopentenyladenosine in tRNA of Shigella flexneri is required for expression of virulence genes. J Bacteriol. 1997;179:5777–5782. doi: 10.1128/jb.179.18.5777-5782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Durand J M, Okada N, Tobe T, Watarai M, Fukuda I, Suzuki T, Nakata N, Komatsu K, Yoshikawa M, Sasakawa C. vacC, a virulence-associated chromosomal locus of Shigella flexneri, is homologous to tgt, a gene encoding tRNA-guanine transglycosylase (Tgt) of Escherichia coli K-12. J Bacteriol. 1994;176:4627–4634. doi: 10.1128/jb.176.15.4627-4634.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eppler K, Wyckoff E, Goates J, Parr R, Casjens S. Nucleotide sequence of the bacteriophage P22 genes required for DNA packaging. Virology. 1991;183:519–538. doi: 10.1016/0042-6822(91)90981-g. [DOI] [PubMed] [Google Scholar]
- 30.Franch T, Gultyaev A P, Gerdes K. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J Mol Biol. 1997;273:38–51. doi: 10.1006/jmbi.1997.1294. [DOI] [PubMed] [Google Scholar]
- 31.Franklin N C. Conservation of genome form but not sequence in the transcription antitermination determinants of bacteriophages lambda, phi 21 and P22. J Mol Biol. 1985;181:75–84. doi: 10.1016/0022-2836(85)90325-0. [DOI] [PubMed] [Google Scholar]
- 32.Franklin N C. N transcription antitermination proteins of bacteriophages lambda, phi 21 and P22. J Mol Biol. 1985;181:85–91. doi: 10.1016/0022-2836(85)90326-2. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes K, Poulsen L K, Thisted T, Nielsen A K, Martinussen J, Andreasen P H. The hok killer gene family in gram-negative bacteria. New Biol. 1990;2:946–956. [PubMed] [Google Scholar]
- 34.Gray J, Wang J, Gelvin S B. Mutation of the miaA gene of Agrobacterium tumefaciens results in reduced vir gene expression. J Bacteriol. 1992;174:1086–1098. doi: 10.1128/jb.174.4.1086-1098.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gultyaev A P, Franch T, Gerdes K. Programmed cell death by hok/sok of plasmid R1: coupled nucleotide covariations reveal a phylogenetically conserved folding pathway in the hok family of mRNAs. J Mol Biol. 1997;273:26–37. doi: 10.1006/jmbi.1997.1295. [DOI] [PubMed] [Google Scholar]
- 36.Gunzer F, Bohm H, Russmann H, Bitzan M, Aleksic S, Karch H. Molecular detection of sorbitol-fermenting Escherichia coli O157 in patients with hemolytic-uremic syndrome. J Clin Microbiol. 1992;30:1807–1810. doi: 10.1128/jcm.30.7.1807-1810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 38.Hakansson S, Galyov E E, Rosqvist R, Wolf-Watz H. The Yersinia YpkA Ser/Thr kinase is translocated and subsequently targeted to the inner surface of the HeLa cell plasma membrane. Mol Microbiol. 1996;20:593–603. doi: 10.1046/j.1365-2958.1996.5251051.x. [DOI] [PubMed] [Google Scholar]
- 39.Hayashi T, Matsumoto H, Ohnishi M, Terawaki Y. Molecular analysis of a cytotoxin-converting phage, φCTX, of Pseudomonas aeruginosa: structure of the attP-cos-ctx region and integration into the serine tRNA gene. Mol Microbiol. 1993;7:657–667. doi: 10.1111/j.1365-2958.1993.tb01157.x. [DOI] [PubMed] [Google Scholar]
- 40.Heersink M. E. coli O157: the true story of a mother’s battle with a killer microbe. Far Hills, N.J: New Horizon Press; 1996. [Google Scholar]
- 41.Huang A, Friesen J, Brunton J. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J Bacteriol. 1987;169:4308–4312. doi: 10.1128/jb.169.9.4308-4312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Inokuchi H, Yamao F. Structure and expression of prokaryotic tRNA genes. In: Söll D, RajBhandary U, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 17–30. [Google Scholar]
- 43.Jackson M P, Neill R J, O’Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 44.Kaper J B, O’Brien A D. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM Press; 1998. [Google Scholar]
- 45.Karch H, Meyer T. Evaluation of oligonucleotide probes for identification of shiga-like-toxin-producing Escherichia coli. J Clin Microbiol. 1989;27:1180–1186. doi: 10.1128/jcm.27.6.1180-1186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karch H, Meyer T. Single primer pair for amplifying segments of distinct Shiga-like-toxin genes by polymerase chain reaction. J Clin Microbiol. 1989;27:2751–2757. doi: 10.1128/jcm.27.12.2751-2757.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karmali M A, Lingwood C A, Petric M, Brunton J, Gyles C. Maintaining the existing phenotype nomenclatures for E. coli cytotoxins. ASM News. 1996;62:167–169. [Google Scholar]
- 48.Kedzierska S, Wawrzynow A, Taylor A. The Rz1 gene product of bacteriophage lambda is a lipoprotein localized in the outer membrane of Escherichia coli. Gene. 1996;168:1–8. doi: 10.1016/0378-1119(95)00712-1. [DOI] [PubMed] [Google Scholar]
- 49.Kutter E, Stidham T, Guttman B, Kutter E, Batts D, Peterson S, Djavakhishvili T, Arisaka F, Mesyanzhinov V, Rüger W, Mosig G. Genomic map of bacteriophage T4. In: Karam J D, editor. Bacteriophage T4. Washington, D.C: ASM Press; 1994. pp. 491–519. [Google Scholar]
- 50.Levine M M. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J Infect Dis. 1987;155:377–389. doi: 10.1093/infdis/155.3.377. [DOI] [PubMed] [Google Scholar]
- 51.Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969;224:759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 52.Lowe T M, Eddy S R. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mahillon J, Kirkpatrick H A, Kijenski H L, Bloch C A, Rode C K, Mayhew G F, Rose D J, Plunkett III G, Burland V, Blattner F R. Subdivision of the Escherichia coli K-12 genome for sequencing: manipulation and DNA sequence of transposable elements introducing unique restriction sites. Gene. 1998;223:47–54. doi: 10.1016/s0378-1119(98)00365-5. [DOI] [PubMed] [Google Scholar]
- 54.Matsugi J, Murao K, Ishikura H. Characterization of a B. subtilis minor isoleucine tRNA deduced from tDNA having a methionine anticodon CAT. J Biochem. 1996;119:811–816. doi: 10.1093/oxfordjournals.jbchem.a021312. [DOI] [PubMed] [Google Scholar]
- 55.Mazzara G P, McClain W H. Cysteine transfer RNA of Escherichia coli: nucleotide sequence and unusual metabolic properties of the 3′ C-C-A terminus. J Mol Biol. 1977;117:1061–1079. doi: 10.1016/s0022-2836(77)80013-2. [DOI] [PubMed] [Google Scholar]
- 56.McClain W H. Identity of Escherichia coli tRNA(Cys) determined by nucleotides in three regions of tRNA tertiary structure. J Biol Chem. 1993;268:19398–19402. [PubMed] [Google Scholar]
- 57.McClain W H, Foss K, Jenkins R A, Schneider J. Nucleotides that determine Escherichia coli tRNAArg and tRNALys identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci USA. 1990;87:9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McShan W M, Tang Y-F, Ferretti J J. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol Microbiol. 1997;23:719–728. doi: 10.1046/j.1365-2958.1997.2591616.x. [DOI] [PubMed] [Google Scholar]
- 59.Melton-Celsa A R, O’Brien A D. Structure, biology, and relative toxicity of Shiga toxin family members for cells and animals. In: Kaper J B, O’Brien A D, editors. Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. Washington, D.C: ASM Press; 1998. pp. 121–128. [Google Scholar]
- 60.Missiakas D, Raina S. Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E. coli: role of two new phosphoprotein phosphatases PrpA and PrpB. EMBO J. 1997;16:1670–1685. doi: 10.1093/emboj/16.7.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mühldorfer I, Hacker J, Keusch G T, Acheson D W, Tschäpe H, Kane A V, Ritter A, Ölschläger T, Donohue-Rolfe A. Regulation of the Shiga-like toxin II operon in Escherichia coli. Infect Immun. 1996;64:495–502. doi: 10.1128/iai.64.2.495-502.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muramatsu T, Nishikawa K, Nemoto F, Kuchino Y, Nishimura S, Miyazawa T, Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988;336:179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- 63.Muramatsu T, Yokoyama S, Horie N, Matsuda A, Ueda T, Yamaizumi Z, Kuchino Y, Nishimura S, Miyazawa T. A novel lysine-substituted nucleoside in the first position of the anticodon of minor isoleucine tRNA from Escherichia coli. J Biol Chem. 1988;263:9261–9267. doi: 10.1351/pac198961030573. [DOI] [PubMed] [Google Scholar]
- 64.Neely, M. N., and D. I. Friedman. 1997. Direct submission to DDBJ/EMBL/GenBank databases. Accession no. AF034975.
- 65.Neely M N, Friedman D I. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 66.Neve H, Zenz K I, Desiere F, Koch A, Heller K J, Brüssow H. Comparison of the lysogeny modules from the temperate Streptococcus thermophilus bacteriophages TP-J34 and Sfi21: implications for the modular theory of phage evolution. Virology. 1998;264:61–72. doi: 10.1006/viro.1997.8960. [DOI] [PubMed] [Google Scholar]
- 67.Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giege R, Yokoyama S. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J Mol Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 68.Oberto J, Weisberg R A, Gottesman M E. Structure and function of the nun gene and the immunity region of the lambdoid phage HK022. J Mol Biol. 1989;207:675–693. doi: 10.1016/0022-2836(89)90237-4. [DOI] [PubMed] [Google Scholar]
- 69.O’Brien A D, LaVeck G D, Thompson M R, Formal S B. Production of Shigella dysenteriae type 1-like cytotoxin by Escherichia coli. J Infect Dis. 1982;146:763–769. doi: 10.1093/infdis/146.6.763. [DOI] [PubMed] [Google Scholar]
- 70.O’Brien A D, Marques L R, Kerry C F, Newland J W, Holmes R K. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb Pathog. 1989;6:381–390. doi: 10.1016/0882-4010(89)90080-6. [DOI] [PubMed] [Google Scholar]
- 71.O’Brien A D, Newland J W, Miller S F, Holmes R K, Smith H W, Formal S B. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 72.Pallanck L, Pak M, Schulman L H. tRNA discrimination in aminoacylation. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 371–394. [Google Scholar]
- 73.Perna N T, Mayhew G F, Posfai G, Elliot S, Donnenberg M S, Kaper J B, Blattner F R. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Plunkett, G., III, and F. R. Blattner. Unpublished data.
- 75.Pollard D R, Johnson W M, Lior H, Tyler S D, Rozee K R. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol. 1990;28:540–545. doi: 10.1128/jcm.28.3.540-545.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Poulsen L K, Larsen N W, Molin S, Andersson P. A family of genes encoding a cell-killing function may be conserved in all gram-negative bacteria. Mol Microbiol. 1989;3:1463–1472. doi: 10.1111/j.1365-2958.1989.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 77.Pulkkinen W S, Miller S I. A Salmonella typhimurium virulence protein is similar to a Yersinia enterocolitica invasion protein and a bacteriophage lambda outer membrane protein. J Bacteriol. 1991;173:86–93. doi: 10.1128/jb.173.1.86-93.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reed M R, Shearwin K E, Pell L M, Egan J B. The dual role of Apl in prophage induction of coliphage 186. Mol Microbiol. 1997;23:669–681. doi: 10.1046/j.1365-2958.1997.2521620.x. [DOI] [PubMed] [Google Scholar]
- 79.Rietra P J G M, Willshaw G A, Smith H R, Field A M, Scotland S M, Rowe B. Comparison of Vero-cytotoxin-encoding phages from Escherichia coli of human and bovine origin. J Gen Microbiol. 1989;135:2307–2318. doi: 10.1099/00221287-135-8-2307. [DOI] [PubMed] [Google Scholar]
- 80.Ring B Z, Roberts J W. Function of a nontranscribed DNA strand site in transcription elongation. Cell. 1994;78:317–324. doi: 10.1016/0092-8674(94)90300-x. [DOI] [PubMed] [Google Scholar]
- 81.Roberts J. Antitermination and the control of transcription elongation. In: McKnight S L, Yamamoto K R, editors. Transcriptional regulation. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 389–406. [Google Scholar]
- 82.Rybchin V N. Genetics of bacteriophage phi 80—a review. Gene. 1984;27:3–11. doi: 10.1016/0378-1119(84)90233-6. [DOI] [PubMed] [Google Scholar]
- 83.Salmi D, Magrini V, Hartzell P L, Youderian P. Genetic determinants of immunity and integration of temperate Myxococcus xanthus phage Mx8. J Bacteriol. 1998;189:614–621. doi: 10.1128/jb.180.3.614-621.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sandmeier H. Acquisition and rearrangement of sequence motifs in the evolution of bacteriophage tail fibers. Mol Microbiol. 1994;12:343–350. doi: 10.1111/j.1365-2958.1994.tb01023.x. [DOI] [PubMed] [Google Scholar]
- 85.Sanger F, Coulson A R, Hong G F, Hill D F, Petersen G B. Nucleotide sequence of bacteriophage λ DNA. J Mol Biol. 1982;162:729–773. doi: 10.1016/0022-2836(82)90546-0. [DOI] [PubMed] [Google Scholar]
- 86.Schicklmaier P, Schmieger H. Sequence comparison of the genes for immunity, DNA replication, and cell lysis of the P22-related Salmonella phages ES18 and L. Gene. 1997;195:93–100. doi: 10.1016/s0378-1119(97)00182-0. [DOI] [PubMed] [Google Scholar]
- 87.Schmidt, H. 1995. Direct submission to DDBJ/EMBL/GenBank databases. Accession no. X83722.
- 88.Schmidt H, Scheef J, Janetzki-Mittmann C, Datz M, Karch H. An ileX tRNA gene is located close to the Shiga toxin II operon in enterohemorrhagic Escherichia coli O157 and non-O157 strains. FEMS Microbiol Lett. 1997;149:39–44. doi: 10.1111/j.1574-6968.1997.tb10305.x. [DOI] [PubMed] [Google Scholar]
- 89.Schmitt C K, McKee M L, O’Brien A D. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect Immun. 1991;59:1065–1073. doi: 10.1128/iai.59.3.1065-1073.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith H R, Green P, Parsell Z. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J Gen Microbiol. 1983;129:3121–3137. doi: 10.1099/00221287-129-10-3121. [DOI] [PubMed] [Google Scholar]
- 91.Smith M C, Burns N, Sayers J R, Sorrell J A, Casjens S R, Hendrix R W. Bacteriophage collagen. Science. 1998;279:1834. doi: 10.1126/science.279.5358.1831g. . (Letter.) [DOI] [PubMed] [Google Scholar]
- 92.Smith M P, Feiss M. Sites and gene products involved in lambdoid phage DNA packaging. J Bacteriol. 1993;175:2393–2399. doi: 10.1128/jb.175.8.2393-2399.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spanjaard R A, Chen K, Walker J R, van Duin J. Frameshift suppression at tandem AGA and AGG codons by cloned tRNA genes: assigning a codon to argU tRNA and T4 tRNAArg. Nucleic Acids Res. 1990;18:5031–5036. doi: 10.1093/nar/18.17.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Strockbine N A, Marques L R, Newland J W, Smith H W, Holmes R K, O’Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sung L M, Jackson M P, O’Brien A D, Holmes R K. Transcription of the Shiga-like toxin type II and Shiga-like toxin type II variant operons of Escherichia coli. J Bacteriol. 1990;172:6386–6395. doi: 10.1128/jb.172.11.6386-6395.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 97.Watanabe K, Osawa S. tRNA sequences and variations in the genetic code. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 225–250. [Google Scholar]
- 98.Willshaw G A, Smith H R, Scotland S M, Field A M, Rowe B. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J Gen Microbiol. 1987;133:1309–1317. doi: 10.1099/00221287-133-5-1309. [DOI] [PubMed] [Google Scholar]
- 99.Yang W, Ni L, Somerville R L. A stationary-phase protein of Escherichia coli that affects the mode of association between the trp repressor protein and operator-bearing DNA. Proc Natl Acad Sci USA. 1993;90:5796–5800. doi: 10.1073/pnas.90.12.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yates J L, Gette W R, Furth M E, Nomura M. Effects of ribosomal mutations on the read-through of a chain termination signal: studies on the synthesis of bacteriophage lambda O gene protein in vitro. Proc Natl Acad Sci USA. 1977;74:689–693. doi: 10.1073/pnas.74.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yokoyama S, Nishimura S. Modified nucleosides and codon recognition. In: Söll D, RajBhandary U L, editors. tRNA: structure, biosynthesis, and function. Washington, D.C: ASM Press; 1995. pp. 207–223. [Google Scholar]
- 102.Zahn K. Overexpression of an mRNA dependent on rare codons inhibits protein synthesis and cell growth. J Bacteriol. 1996;178:2926–2933. doi: 10.1128/jb.178.10.2926-2933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zahn K, Landy A. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol Microbiol. 1996;21:69–76. doi: 10.1046/j.1365-2958.1996.6201335.x. [DOI] [PubMed] [Google Scholar]