Abstract
Several virulence-related genes have been described for prototype enteroaggregative Escherichia coli (EAEC) strain 042, which has been shown to cause diarrhea in human volunteers. Among these factors are the enterotoxins Pet and EAST and the fimbrial antigen aggregative adherence fimbria II (AAF/II), all of which are encoded on the 65-MDa virulence plasmid pAA2. Using nucleotide sequence analysis and insertional mutagenesis, we have found that the genes required for the expression of each of these factors, as well as the transcriptional activator of fimbrial expression AggR, map to a distinct cluster on the pAA2 plasmid map. The cluster is 23 kb in length and includes two regions required for expression of the AAF/II fimbria. These fimbrial biogenesis genes feature a unique organization in which the chaperone, subunit, and transcriptional activator lie in one cluster, whereas the second, unlinked cluster comprises a silent chaperone gene, usher, and invasin reminiscent of Dr family fimbrial clusters. This plasmid-borne virulence locus may represent an important set of virulence determinants in EAEC strains.
Enteroaggregative Escherichia coli (EAEC) is an emerging enteropathogen associated with infantile diarrhea in developing countries (2, 8, 27, 47) and has more recently been associated with food-borne diarrhea outbreaks in developed countries (21, 43). This E. coli pathotype is defined by aggregative adherence (AA) to HEp-2 cells, where bacteria display adherence to the cell surface and also to the intervening substratum in a stacked-brick configuration (27). Potential virulence factors, such as adhesins and toxins, have been identified in EAEC strains (32). However, the complete picture of EAEC pathogenesis remains unclear.
The AA phenotype is associated with the presence of a plasmid 60 to 65 MDa in size and with the expression of one of two distinct aggregative adherence fimbria (AAF/I and AAF/II). AAF/I is expressed in EAEC strain 17-2, is 2 to 3 nm in diameter, and is arranged in flexible bundles (28). The AAF/I biogenesis genes are organized as two unlinked plasmid-borne regions separated by 9 kb (29). Region 1 comprises a contiguous cluster of fimbrial structural genes, in the order chaperone-usher-putative invasin-pilin genes (41). These roles have been assigned on the basis of nucleotide and organizational homology with members of the Dr family of adhesins, a group of fimbrial and nonfimbrial adherence factors which recognize the Dra blood group antigen and which have been associated with uropathogenic and possibly diarrheagenic E. coli (18, 22, 36, 50). AAF/I region 2 encodes a regulator termed AggR, which is a member of the AraC family of DNA binding proteins and which is necessary for AAF/I fimbrial expression in strain 17-2 (30). Other Dr adhesins have not been shown to require AraC-homologous regulators.
We have recently described a second EAEC fimbrial antigen, designated AAF/II, in prototype EAEC strain 042 (9), a strain which caused diarrhea in a volunteer study (31). The AAF/II fimbriae are 5 nm in diameter and are arranged in semirigid bundles of filaments. The plasmid-borne fimbrial subunit of AAF/II (aafA) has been cloned, sequenced, and mutated; an aafA mutant lost the ability to adhere to human intestinal tissue in culture, suggesting a role for AAF/II as a human intestinal colonization factor (9). The AAF/I and AAF/II fimbrial subunits are 25% identical and are related to those of the Dr family.
In this work we describe the complete AAF/II fimbrial gene organization in strain 042 and show that the genes define a plasmid-borne virulence gene cluster. We characterize the genes essential for AAF/II biogenesis and demonstrate the role of the AggR transcriptional activator in AAF/II expression. Our data reveal a novel fimbrial genetic organization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
EAEC strain 042 (O44:H18) used in this study was originally isolated from a child with diarrhea in Lima, Peru. 042 adheres to HEp-2 cells in an aggregative pattern and harbors a 65-MDa adherence-encoding plasmid designated pAA2 (9). 042 expresses AAF/II but not AAF/I. Also used in this study was HB101 transformed with plasmid pAA2 [designated HB101(pAA2)], which is able to adhere to HEp-2 cells in an AA pattern (9).
Characteristics of strains and vectors used in this work are listed in Table 1. All strains were grown at 37°C on Luria (L) agar or in L broth. Ampicillin (100 μg ml−1), streptomycin (30 μg ml−1), tetracycline (15 μg ml−1), kanamycin (30 μg ml−1), chloramphenicol (20 μg ml−1), nalidixic acid (50 μg ml−1), 5-bromo-4-chloro-3-indolylphosphate (X-P; 30 μg ml−1), 5-bromo-4-chloro-3-indolylgalactoside (X-Gal; 20 μg ml−1), and isopropyl-β-d-thiogalactopyranoside (1 mM) were added when indicated.
TABLE 1.
Characteristics and source of host strains and plasmid vectors used in this study
| Strain or plasmid | Relevant characteristicsa | Reference or source |
|---|---|---|
| E. coli strains | ||
| 042 | Prototype EAEC strain (O44:H18) | 31 |
| HB101 | E. coli B/K-12 hybrid | 6 |
| HB101(pAA2) | HB101 carrying the native pAA2 plasmid from 042 | 9 |
| DH5α | supE44ΔlacU169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 39 |
| DH5α(λpir) | DH5α transduced with λpir | 13 |
| SM10(λpir) | thi thr leu tonA lacY supE recA::RP4-2-Tc::Mu (Km) | 42 |
| S17-1(λpir) | pro res mod+ RP4-2, Tc::Mu-Km::Tn7 | 42 |
| Plasmids | ||
| pBluescript II SK | 2.9-kb high-copy-number phagemid (Ap) | Stratagene |
| pSPORT1 | 4.1-kb high-copy-number phagemid (Ap) | Gibco BRL |
| pRK415 | 10-kb RK290-derived vector (Tc) | 23 |
| pJRD215 | 10.2-kb RSF1010-derived vector (Km, Sm) | 11 |
| pJP5603 | 3.1-kb R6K-based suicide vector (Km) | 37 |
| pHP45 | Ω interposon-bearing plasmid (Sm) | 38 |
| pSLM852 | 390-bp PstI fragment of pSSS1 cloned into pBluescript (Ap) | 4 |
| pRT733 | TnphoA-bearing plasmid (Km) | 44 |
| pCVD301 | 21.5-kb cosmid vector with RK290 replicon (Tc) | 10 |
Ap, ampicillin resistance; Km, kanamycin resistance; Sm, streptomycin resistance; Tc, tetracycline resistance.
Recombinant DNA techniques.
DNA analysis and cloning were performed as described by Sambrook et al. (39). A plasmid Midi kit (Qiagen, Inc., Chatsworth, Calif.) was used for large-scale DNA preparation. Restriction fragments were isolated by using Prep-A-Gene (Bio-Rad Laboratories, Hercules, Calif.). Plasmid DNA was introduced into host cells by the calcium chloride method (19) or by electroporation (2.5 kV, 25 μF, and 200 Ω). DNA fragments used in hybridization were labeled with [α-32P]dATP by random priming using a Prime-It kit and protocols recommended by the manufacturer (Boehringer Mannheim, Indianapolis, Ind.).
Transposon mutagenesis.
Strain 042 was mutagenized by using transposon TnphoA delivered on the suicide vector pRT733 (44), following the methodology described by Nataro et al. (28). Alkaline phosphatase-producing mutants were selected on L-agar plates containing kanamycin and X-P. Mutants lacking HEp-2 adherence were selected for further cloning and sequencing analysis. The sites of TnphoA insertion were determined by cloning of genomic fragments conferring kanamycin resistance and subsequent nucleotide sequencing of the flanking DNA.
Cosmid library construction.
Plasmid DNA from strain 042 was purified and was partially digested with Sau3a to yield 15- to 30-kb fragments. After separation on a 0.7% agarose gel, fragments were extracted and were ligated with BamHI-digested cosmid vector pCVD301. Ligated DNA was packaged into phage by using the Gigapack II packaging extract (Stratagene, La Jolla, Calif.) and then transfected into strain HB101, and cosmid clones were selected on L-agar plates containing tetracycline. The cosmid clones were screened for the AA pattern in the HEp-2 cell adherence assay. Clones exhibiting HEp-2 adherence were selected for further analysis.
pBluescript pAA2 library construction.
Purified plasmid DNA from strain 042 was sonicated to generate fragments 1 to 5 kb in length. After separation in a 0.7% agarose gel, 1.5- to 2.5-kb fragments were eluted from the gel and were blunt ended by using Pfu DNA polymerase as instructed by the manufacturer (Stratagene). Fragments were ligated into SmaI-digested pBluescript. Ligated DNA was transformed into strain DH5α, selecting for ampicillin resistance.
DNA sequencing and analysis.
DNA sequencing was performed in the Biopolymer Laboratory, Department of Microbiology and Immunology, University of Maryland School of Medicine. Double-stranded plasmid DNA was sequenced by the chain terminator method (40) with an Applied Biosystem model 373A sequencer. Sequence analysis was performed through Genepro sequence analysis software (version 5.00; Riverside Scientific, Bainbridge Island, Wash.), DNASIS version 2.10, and the Wisconsin sequence analysis package (Genetics Computer Group), available through the Center of Marine Biotechnology, University of Maryland, and programs available through the National Center for Biotechnology Information.
Directed insertional mutagenesis.
To identify open reading frames (ORFs) related to AAF/II fimbrial biogenesis, the following strategy was used. Internal segments of the target genes were prepared by PCR, and primers were designed such that KpnI (5′)-SacI (3′) restriction sites were engineered at the ends of each fragment. PCR products were digested with KpnI and SacI and were cloned into the corresponding sites in suicide vector pJP5603. PCRs were performed with an Opti-Prime PCR optimization kit (Stratagene), plasmid pAA2 as the DNA template, and the PCR primers listed in Table 2. For cloning, PCR products were resolved by agarose gel electrophoresis and were eluted by using a Prep-A-Gene kit (Bio-Rad). After ligation and transformation into DH5α(λpir), transformants were selected on L-agar plates containing kanamycin and X-Gal; plasmid DNA of transformants harboring the correct insert was transformed into strain S17-1(λpir) for mobilization into strain 042 (nalidixic acid resistant) via filter mating on cellulose nitrate membranes. Transconjugants were selected on L-agar plates containing kanamycin and nalidixic acid, and their identity was confirmed by agglutination with anti-O44 antisera (Unipath, Inc., Ogdensburg, N.Y.). The site of integration was confirmed by restriction mapping. To terminate transcription, the 2.0-kb Ω interposon (38) with BamHI ends was cloned into the corresponding site of pJP5603 downstream of the PCR product internal to aafD′. The construction was mobilized into strain 042, and transconjugants were selected by using the technique described above.
TABLE 2.
Oligonucleotides used in PCRs
| Gene | Primer | Primer sequence |
|---|---|---|
| aafA | 5′ | ACATGCATGCAAAAAATCAGAATGTTTGTT |
| 3′ | CGGGATCCATTTGTCACAAGCTCAGC | |
| aafB | 5′ | ATGGTACCATGCTGTCGGTATCCTGCG |
| 3′ | ATGAGCTCACTCTCCAGGCTTTGCTC | |
| aafC | 5′ | ATGGTACCCGTTATAGCTCGCACATATTC |
| 3′ | ATGAGCTCTGCAGACTGATAATGCTC | |
| aafD′ | 5′ | ATGGTACCCGTATTTTCCCTTCATCTG |
| 3′ | ATGAGCTCTACCGTTCCCACCATTAG | |
| aafD | 5′ | ATGGTACCAAATCAACAGTCCTCTGGG |
| 3′ | ATGAGCTCGGGTATCAAATACTCATAAGAG | |
| aggR | 5′ | TAGGTACCTAATTGTACAATCGATGTA |
| 3′ | CAGAGCTCAAGTAAGTAATTCTTGAAT |
RNA analysis.
Total RNA was extracted from log-phase L-broth cultures of strains 042, 042 aggR, and HB101, using the Ultraspec RNA isolation system (Biotecx Laboratories, Houston, Tex.) according to the manufacturer’s protocol. 042 aggR, also called 042::pWPE20, was constructed as described above by insertion of an aggR internal fragment into pJP5603 and integration into the aggR gene of pAA2. After extraction, RNA samples were treated with DNase I-amplification grade (Gibco BRL, Gaithersburg, Md.), and 20-μg aliquots of total RNA were denatured and blotted onto nylon transfer membranes (Schleicher & Schuell, Keene, N.H.). As a control, each sample was blotted in duplicate, with one blot treated with 80 U of RNase (Promega, Inc., Madison, Wis.) for 30 min at 37°C. Membranes were hybridized with aafA, aafB, aafC, aafD, or aafD′ α-32P-labeled DNA probes (obtained by PCR amplification using the primers described in Table 2). Membranes were hybridized, washed, and visualized by autoradiography as described by Sambrook et al. (39). Only autoradiographic signals that were abolished by RNase treatment were considered to represent mRNA transcripts.
HEp-2 cell adherence assay.
The HEp-2 adherence assay was performed as described by Vial et al. (45). Briefly, HEp-2 cells were grown to 50% confluent monolayers on glass coverslips in a 24-well tissue culture plate. After a gentle wash with phosphate-buffered saline (PBS), 1 ml of fresh Eagle’s minimal essential medium with 1% d-mannose was added to each well with 40 μl of an overnight L-broth bacterial culture. The assay mixture was incubated for 3 h (for 042 and its derivatives) or 6 h (for HB101 and derivatives) at 37°C in 5% CO2. After the incubation period, cells were washed several times with PBS, fixed with methanol, stained with 10% Giemsa stain for 15 min, and examined by light microscopy. The tests were considered positive for the AA phenotype when bacteria displayed the stacked-brick aggregates attached to HEp-2 cells and to the glass substratum between cells.
Polystyrene adherence.
Bacterial strains were grown overnight at 37°C in L broth in 24-well polystyrene cell culture dishes (Falcon Industries, Becton-Dickinson, Lincoln Park, N.J.). Plates were then gently washed twice with PBS, dried, and stained with 10% Giemsa stain. The plates were analyzed visually for adherent bacteria present as a film on the plastic surface (20).
Electron microscopy.
Strains were grown statically in L broth in 24-well cell culture dishes. Cultures were passaged twice for 24 h each time at 37°C. Transmission electron microscopy of negatively stained specimens was performed by standard methodology (25). Grids were examined with a JEOL JEM 1200 EX II transmission electron microscope. AAF/II was detected by immunogold electron microscopy as previously described (9).
Nucleotide sequence accession numbers.
The sequences derived from the analyses performed in this study are deposited in GenBank under accession no. AF114827 and AF114828.
RESULTS
TnphoA mutagenesis of strain 042.
To localize genes required for the biogenesis of AAF/II, we performed transposon TnphoA mutagenesis on strain 042 and screened 480 insertion mutants for adherence to polystyrene culture dishes and to HEp-2 cells; these two phenotypes have been shown to correlate with AAF/II expression (9). Twelve nonsibling mutants that were deficient in adherence to both cells and plastic were identified. By nucleotide sequence and Southern blot analysis, 11 of these insertions were localized to the previously described AAF/II fimbrial subunit, aafA, which is located on the pAA2 plasmid (9) (GenBank accession no. AF012835). The remaining site identified by sequence analysis was found to lie within the chromosomal dsbA gene, which encodes a periplasmic enzyme catalyzing disulfide bond formation (1) (Genbank accession no. M77746). Like many fimbrial subunits, the aafA gene features two cysteine residues with the potential to form a disulfide-stabilized loop.
Localization of AAF/II biogenesis genes on plasmid pAA2.
In a further effort to define the complete AAF/II biogenesis cluster, we constructed a cosmid pCVD301 library of Sau3a partially digested fragments derived from plasmid pAA2. This approach was used because the biogenesis cluster of AAF/I fimbriae had been shown to comprise a primary biogenesis cluster regulated by an unlinked transcription activator (AggR) encoded approximately 9 kb from the other required genes (29, 30). The cosmid library was screened for clones that expressed polystyrene adhesion and aggregative adherence to HEp-2 cells. Five cosmid clones (designated C9, D6, F9, E6, and I10) were found to confer adhesion in these assays. The insert DNA from each of these cosmids was analyzed by restriction analysis, and the cosmids were found to have in common a region of approximately 23 kb. This region featured a 13-kb MluI fragment, which we have shown previously to encode the enterotoxins Pet and EAST, and the AafA fimbrial subunit (14).
After serial in vitro passage in the course of characterizing these cosmids, only one of them, D6, manifested stable expression of the AAF/II fimbrial phenotypes; all others became negative for adherence despite continued presence of the cosmid clones under tetracycline pressure. Only one of these latter cosmids, C9, displayed evidence of genomic deletion. Using a restriction fragment derived from the aggR gene, we found that 042 yielded a strong hybridization signal on a colony blot. Notably, Southern analysis using this probe localized the gene to plasmid pAA2, and of the adherent cosmids only D6 hybridized with this aggR fragment probe.
We also observed by colony hybridization that 042 and other AAF/II producers hybridized with the diffusely adherent E. coli (DAEC) probe, which is derived from the usher gene of the F1845 fimbriae (4). By Southern analysis, the DAEC probe fragment hybridized with insert DNA from each of the adherence-conferring cosmids.
Sequence analysis of the fimbrial biogenesis loci.
The nucleotide sequence of the DNA corresponding to the inserts of each of the adherence-positive cosmid clones was determined by use of a pBluescript II SK library. Selected restriction fragments derived from the pCVD301 cosmid clones were used to hybridize a pAA2 gene bank constructed in pBluescript II SK; the inserts of these pBluescript clones were then sequenced from universal primers. These experiments yielded 25 kb of contiguous sequence that comprised the complete adherence-encoding region.
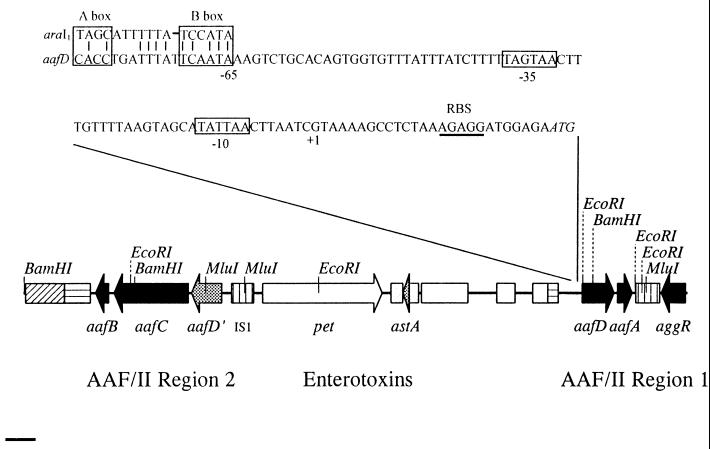
The position of the aafA gene is as shown in Fig. 1. The determined sequence was identical to that previously published (9) (accession no. AF012835). The closest homolog of the predicted protein AafA is the AggA protein, the structural subunit of the AAF/I fimbriae (at 25% identity [9]). Our analysis showed that the aafA gene was part of a cluster of potential fimbrial biogenesis genes. Upstream from the aafA gene, and in the same orientation, was a gene that we have designated aafD, which is 747 bp in length, yielding a predicted product of 28.7 kDa. This protein features a typical Sec-dependent signal sequence and demonstrates significant homology to the chaperone genes of other fimbrial operons. Not surprisingly, the closest homolog was the aggD gene of the AAF/I cluster, at 62.4% identity and 88.7% similarity. Immediately downstream of the aafA gene is an apparently complete IS1 element (98% identity to IS1 from the afa-3 cluster [accession no. X76688]). Flanking the IS1 element, 957 nucleotides downstream from the aafA gene and in opposite orientation, lies aggR, the sequence of which is 99% identical at the nucleotide level to the aggR allele from the AAF/I-producing strain 17-2 (30). This gene of 795 bp encodes a 30.6-kDa protein belonging to the AraC family of transcriptional activators (15). Like other members of the AraC family, the predicted protein encoded by the aggR gene features two helix-turn-helix motifs.
FIG. 1.
Map of the virulence region of pAA2. Black arrows represent AAF-related reading frames discussed in this paper. The pet and astA genes are enterotoxins previously published. IS-like elements are indicated by boxes; the boxes with horizontal lines represent directly repeated IS-like elements that are homologous to IS1353. Boxes with vertical stripes represent complete IS1 elements. Line, 1 kb. The inset at the top shows the upstream region of the aafD gene. Indicated are the predicted ATG start (italics), ribosomal binding site, and −10 and −35 promoter regions. Upstream from the predicted aafD promoter lies a region with high homology to the araI1 half-site, shown to be the binding site of the AraC protein (35). This region is thus a candidate binding site of the AraC homolog AggR, under which the aafD gene is positively regulated (see text).
No other fimbria-related genes were found immediately upstream or downstream of this fimbrial gene cluster. This locus, herein termed AAF/II region 1, is therefore distinct from other fimbrial biogenesis clusters in that it encodes a fimbrial subunit, chaperone, and regulator but lacks a fimbrial usher protein. As expected, we found that clones expressing only the aafD-aafA-aggR cluster did not confer fimbrial expression on HB101, suggesting that cosmids such as D6 indeed harbored other fimbrial genes.
Notably, all AAF/II-encoding cosmid clones carried genes for the enterotoxins Pet and EAST1 (astA); the positions of these genes are shown in Fig. 1. The 25-kb sequenced plasmid region was found to include the previously sequenced pet locus (accession no. AF056581). The genetic segment between pet and region 1 was found to harbor multiple insertion sequence (IS)-like elements (see reference 14). The first gene downstream of pet, convergently transcribed, encodes a transposase homolog (similar to accession no. U44824), carrying the EAST enterotoxin embedded within the coding strand (in the same sense but out of frame with the transposase). Recently, the sequence of pMT1 of Yersinia pestis has been deposited in the database (26). An IS-like element from pMT1 (accession no. AF053947) is now the closest homolog of the EAST putative transposase, at 77% identity over the full predicted length. Interestingly, however, the Y. pestis transposase includes the full EAST gene but includes a stop codon in place of one of the EAST cysteines. Three other IS elements are predicted to lie downstream of the pet gene. These have been described previously (14).
Examination of the nucleotide sequence revealed another unlinked cluster of potential fimbrial biogenesis genes (Fig. 1). This cluster, designated AAF/II region 2, began 2,477 bp upstream from the start of the pet enterotoxin gene (14) but in the divergent orientation. Just upstream of the region 2 cluster lies a locus exhibiting significant homology (57% nucleotide identity) to the aafD gene and other members of the Pap chaperone family of proteins. Interestingly, however, this locus (designated aafD′) featured several frameshifts, such that conventional translation from a predicted mRNA product would not generate a functional chaperone protein. Also of note, the closest homolog to aafD′ (61.2% identity) was the afaB gene, required for expression of the AFA-III afimbrial adhesin of uropathogenic E. coli (accession no. X76688).
Fifty-seven base pairs downstream from the aafD′ locus and in the same orientation lies a gene that we have designated aafC. aafC is 2,583 nucleotides in length and yields a predicted 94-kDa product which is 80% identical at the amino acid level and similar in size to the AfaC protein, the usher protein of AFA-III (accession no. X76688). The predicted aafC product was only 67% identical to the AggC usher protein of the AAF/I biogenesis cluster (accession no. U12894). The predicted amino acid sequence encoded by aafC features a fimbrial usher protein signature between amino acids 321 and 331.
Downstream from the aafC protein was an ORF of 438 bp transcribed in the same orientation, designated aafB. The predicted aafB product is a protein of 16.1 kDa, including a cleavable signal sequence, and bears closest homology (59% identity) to the putative invasin protein (AfaD) common to members of the Dr family of adhesins (22) (accession no. X76688). AafB is 56% identical to the AggB protein of the AAF/I biogenesis cluster (accession no. U12894).
An ORF immediately downstream of aafB yielded a predicted protein of as much as 300 amino acid residues (assuming a CTG start site) and did not include a cleavable signal sequence. This ORF was 60% identical over 282 amino acids with putative transposase IS1353 (U42226) from the Tn21 element of Shigella flexneri. This latter ORF is also postulated to feature an alternative start codon. Notably, a truncated copy (91% identical) of this IS-like element was found immediately upstream of aafD in region 1. Also noteworthy is the fact that the IS1353-like element (97% identical to that found here) is also present upstream of CS1 and CS5 fimbrial operons of enterotoxigenic E. coli. Downstream of the IS1353-like element flanking region 2 is an apparently complete IS629 element, originally identified in Shigella sonnei. At the other end of region 2, immediately upstream of the aafD′ locus, is a complete IS1 element 99% identical to that found in the afa3 cluster (accession no. X76688).
Insertional mutagenesis of fimbrial biogenesis genes.
Sequence analysis of cosmid D6 suggested that the AAF/II biogenesis genes may be arranged as two unlinked clusters with a unique organization. Of particular interest was the presence of the two chaperone-homologous regions and the apparently incomplete nature of each cluster. We undertook an insertional mutagenesis strategy to determine conclusively which of the sequenced ORFs was required for expression of the AAF/II fimbriae. An internal fragment of each of the putative biogenesis ORFs (aafB, aafC, aafD, aafD′, and aggR [Table 2]) was cloned into the multiple cloning site of the suicide vector pJP5603. These constructions were then independently mobilized into parent strain 042, and integration into the homologous genes was confirmed. An aafA mutant had been obtained previously by TnphoA mutagenesis (9). All mutants were screened for adherence to plastic and to HEp-2 cells and for the presence of AAF/II fimbriae by electron microscopy. The latter was determined by transforming the mutated pAA2 plasmids into HB101 to avoid confusion with other surface fimbriae of 042. Phenotypic analyses showed perfect correlation between adherence and fimbrial expression. Insertional inactivation of aafA, aafC, aafD, and aggR resulted in loss of fimbrial expression, whereas inactivation of the aafD′ and aafB reading frames did not result in loss of these phenotypes.
To rule out the possibility of polar effects from insertional inactivation, we complemented in trans (in an HB101 background) each of the insertional mutants that resulted in loss of fimbrial expression and adherence. We found that introduction of clones expressing only the inactivated genes restored expression of fimbriae for aafA (reported previously), aafC, aafD, and aggR. These data confirm that each of these plasmid genes, but apparently only these, are necessary and sufficient for expression of the AAF/II fimbriae. Thus, the data implicate two discontinuous plasmid regions in the expression of AAF/II fimbriae; the presence of the enterotoxins Pet and EAST between the two fimbrial gene clusters suggests that the AAF/II-encoding regions delimit a virulence gene cluster of EAEC.
Transcription of AAF/II-related genes.
Our data suggest that the aafD′ gene is a pseudogene derived from a fimbrial chaperone homolog. Data from Bilge et al. (3) have suggested that Dr fimbrial clusters, to which the AAF/II region 2 bears organizational and nucleotide homology, are transcribed from a single major promoter upstream from the first gene in the polycistronic message. We therefore asked whether the promoter of the region 2 cluster was located upstream of the silent aafD′ gene or whether instead it was located immediately upstream of the aafC gene. We constructed a mutation in the aafD′ locus by integration of pJP5603 (in strain 042) exactly as described above except that in addition to the internal aafD′ fragment, we also inserted an Ω interposon to terminate transcription at the site of integration. As for the previous mutation in the aafD′ gene, this construction was able to express functional fimbriae indistinguishable from those of the wild-type parent. These data suggest that the promoter for the region 2 cluster lies upstream from aafC and not from aafD′ as would be expected from published observations of other Dr fimbriae (5).
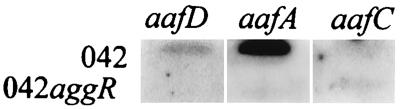
As noted above, we found that of the AA-expressing cosmid clones, only the stably AA+ clone D6 harbored aggR. Moreover, mutagenesis of aggR in 042 (see above) abolished fimbrial expression. Although we had previously shown that aggR is required for expression of the AAF/I fimbrial cluster by acting as a transcriptional activator of the fimbrial subunit gene, aggA (30), the location of the AggR-dependent promoter was not determined. We therefore asked whether aggR was required for the expression of the aafA fimbrial subunit gene and whether other genes of the fimbrial biogenesis apparatus were also under aggR control. Dot blots of mRNA from wild-type 042 and from 042 aggR were hybridized with DNA fragments representing each of the putative AAF/II genes (aafA, -B, -C, -D, and -D′). No transcripts were observed in dot blots using aafB or aafD′ probes. As seen in Fig. 2, the aafA transcript was abundant in wild-type 042 grown in L broth, yet this transcript was absent from a similarly grown 042 aggR construct. In vitro growth curves of the wild-type and mutant bacteria were similar (not shown). In wild-type 042, the aafD transcript produced a much lighter signal compared to the aafA transcript, but like aafA, aafD was undetectable in the aggR mutant. The transcript of the aafC gene produced a light but detectable hybridization signal in 042 but was present in similar amounts in both wild-type and aggR mutant blots. Our RNA dot blot data thus suggest that region 1 is under AggR control but that region 2 is not. Indeed, we noted a potential AraC-like recognition half-site (35) upstream of a potential promoter for aafD (Fig. 1); no such motif is found immediately upstream of aafA or aafC.
FIG. 2.
RNA dot blots of aafD, aafA, and aafC transcripts in the presence and absence of a functional aggR gene product. Bacterial RNA prepared from L-broth cultures of 042 or 042 aggR was blotted to nitrocellulose and hybridized with probes generated by PCR. Details are presented in Materials and Methods. aafA and aafD transcripts were clearly increased in the presence of AggR; the aafC transcript was barely detectable but did not exhibit AggR dependence.
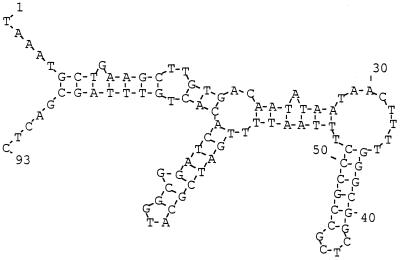
We had found that the cloned aafA gene was capable of complementation of an aafA null mutation in 042 (9); however, this complementation was weak even when the aafA gene was supplied in high copy number. Analysis of the region between the aafD and aafA genes did not reveal a strongly predicted promoter signature (as determined by inspection and by use of the NNPP promoter prediction algorithm). This observation would suggest that the abundance of the aafA transcript may be mediated by other parameters, such as an enhanced stability of the aafA transcript. Such a situation has been reported for the fimbrial subunit gene of the related Dr adhesin F1845 (3). For F1845, stability of the subunit transcript has been shown to be related to a stable downstream hairpin motif. For AAF/II, an extended, highly favorable hairpin region (ΔG = −32.2 kcal/mol) was predicted to lie immediately downstream of the aafA gene (Fig. 3).
FIG. 3.
Predicted secondary structure of the aggA downstream region. Nucleotide 1 is the first base of the aafA stop codon. The sequence was analyzed by using the Zuker-Stiegler algorithm processed with DNASIS version 2.10 software.
DISCUSSION
The mechanism of EAEC pathogenicity is still not completely elucidated, but several potential virulence factors have been described (32). These factors include several putative fimbrial and nonfimbrial adhesins (12, 24, 28, 46). The fimbrial adhesin AAF/II, expressed in human-virulent strain 042, has been shown to confer aggregative adherence and adherence to the human colonic mucosa (9). Although the fimbrial subunit gene (aafA) has been cloned and sequenced, the complete genetic organization of AAF/II has not been described.
In this work we have characterized the genes necessary for AAF/II expression. Our mutagenesis data demonstrate that dsbA and two plasmid-borne loci are required for expression of this adherence factor. The periplasmic thiol:disulfide oxidoreductase enzyme encoded by dsbA has been demonstrated to be necessary for the expression of other fimbrial antigens (49), presumably by catalyzing the formation of a disulfide loop in the fimbrial subunit protein.
We report here that the plasmid-borne AAF/II biogenesis genes feature a unique organization. The fimbrial genes are organized in two clusters separated by a 12-kb segment; the latter contains the genes encoding two enterotoxins, Pet and EAST, and a series of IS elements (9, 14). Analysis of AAF/II genes indicate that each is closely related to genes encoding the Dr family of adhesins (36). This family includes the nonfimbrial uropathogenic E. coli adhesins AFA-I, AFA-III, and Dr hemagglutinin (49); the F1845 fimbriae of DAEC (4); and more distantly AAF/I, whose fimbrial subunit is closest to that of AAF/II (9). However, all members of this family thus far described feature a conserved genetic arrangement, chaperone-usher-invasin-pilin, thought to be transcribed as a polycistronic message (3). Despite being a relative of this family, the AAF/II fimbria displays a markedly different organization.
In a genetic segment that we have designated AAF/II region 1, we localized the aafD, aafA, and aggR genes, encoding the chaperone, the pilin, and the transcriptional activator, respectively. Homology analysis demonstrates high similarity between each of the genes of this region and the AAF/I biogenesis genes. We have previously shown that AafA is the fimbrial subunit and that its closest homolog is AggA, the pilin subunit of AAF/I (9). The closest homolog of AafD is AggD of AAF/I (41); lesser homology is seen with other periplasmic chaperone proteins of Dr adhesins implicated in adhesin assembly. The aggR gene downstream of aafA displays almost complete homology at the nucleotide level to aggR of the AAF/I operon (30). aggR encodes a protein which belongs to the AraC binding-protein family and is required for AAF/I expression in strain 17-2 (30). We have found that for AAF/II, this gene is located only 956 bp downstream from aafA, while in AAF/I it is located 9 kb away from the fimbrial gene cluster.
The remaining AAF/II biogenesis genes are situated in region 2, comprising aafD′ (a silent chaperone-like locus), aafC, and aafB, which apparently encode an usher and the putative Dr invasin (16), respectively. Notably, the cryohemagglutinin described by Yamamoto et al. (48) appears to be AAF/II, as the inactivating insertion site reported by those authors maps to the aafC sequence (33). Our data do not allow us to draw inferences regarding a possible role for the aafB gene, since it does not appear to be required for the expression of functional AAF/II fimbriae or for wild-type adherence to HEp-2 cells.
The most notable feature of the AAF/II organization is the separation of the usher and chaperone genes in unlinked clusters. Nucleotide homology studies have allowed us to postulate one possible mechanism for the origin of this unique arrangement. Both the region 1 and region 2 clusters have some overall similarities in organization to the highly conserved Dr family biogenesis operons: region 1 begins with a chaperone homolog and ends with a subunit gene; region 2 begins with a chaperone locus (though silent) and then progresses, like other Dr adhesins, to the usher and then the putative invasin gene before truncating short of the expected pilin gene. Interestingly, whereas the closest homologs of the region 1 genes are the AAF/I genes, the closest homologs of the region 2 genes are genes reported for the Dr adhesin AFA-III. These data suggest that the two regions originated independently and not as the result of duplication. Moreover, it is notable that both region 1 and region 2 are flanked on one side by apparently complete IS1 transposase genes (region 1 downstream only and region 2 upstream only). It has been reported that the AFA-III cluster is flanked by intact IS1 elements which are capable of mediating transposition of the entire afa3 operon (17). Our data are thus most compatible with the following model: region 1 is likely to have been the original adhesin gene cluster, since it is encoded upon a plasmid that, overall, is highly homologous to that encoding AAF/I (33). We hypothesize that a Dr adhesin transposed onto the plasmid, which then would have harbored more than one copy of the chaperone, usher, invasin, and, most likely, a second pilin. Since this redundancy was apparently not adaptive, the organism evolved to lose one copy of each gene: the pilin from region 2 by deletion and the usher and invasin from region 1, also by deletion. The redundant chaperone in region 2 was apparently lost by point mutations. Analysis of G+C content of region 1 and region 2 genes supports this model of AAF/II evolution: G+C content of the region 1 genes aafA (41.2%) and aafD (37.5%) averages 37.6%, while that of the region 2 genes aafC (53.7%) and aafB (44.7%) and the pseudogene aafD′ (45.4%) averages a substantially higher 51.5%. The G+C contents of AAF/I genes aggA (37.6%), aggB (39.7%), aggC (41.7%), aggD (36.4%), and aggR (30.8%) are markedly lower than that of afa genes, which are in the range of 56 to 61% (accession no. X76688). Despite these differences, our data suggest that the accessory biogenesis genes of the two regions act in concert, and the ability of Dr accessory proteins to substitute for each other in the expression of Dr adhesin subunits has been demonstrated previously (7).
The presence of IS elements may play an important role in dissemination of Dr fimbrial genes. The presence of these elements apparently not only is conserved among Dr adhesins but also extends to their relatives, the AAFs, as well. On their sides lacking the IS1 elements, both AAF/II region 1 and region 2 have evidence of a gene that is also found upstream of the CS1 and CS5 fimbrial clusters and which also displays features of an insertion sequence. Whether this gene plays a role in the dissemination of fimbrial gene clusters remains to be determined.
We demonstrate that AggR regulates transcription of aafA and aafD in region 1. AggR apparently acts directly to regulate the aafD promoter; however, the role of AggR in aafA transcription is not clear. Bilge et al. have suggested that the daaE (major adhesin) transcript of the Dr fimbria F1845 is present in greater abundance than those of upstream accessory genes, despite polycistronic transcription from a common upstream promoter (3). The mechanism of this effect was reported to be enhanced stability of the daaE transcript mediated by a downstream hairpin structure. We have identified a candidate hairpin downstream of aafA, but in addition, our prior work complementing an aafA mutant suggests that there may be an independent, perhaps AggR-regulated, promoter upstream of aafA (9). Given that AggR was shown to be required for aggA expression in AAF/I-producing strain 17-2 (30) and that AraC homologs are not known to be involved in AFA regulation, these data support our hypothetical model of AAF/II evolution.
Virulence in EAEC strains has been associated with the presence of a high-molecular-weight plasmid which seems to be conserved among different strains (33). The 23-kb DNA sequence of pAA2 described here defines a plasmid-borne virulence gene cluster. In this large plasmid region are localized the genes encoding the Pet and EAST enterotoxins and the proteins related to AAF/II biogenesis. This gene cluster seems to be an important virulence-related element in pAA2, since both AAF/II and Pet have been associated with EAEC effects on intestinal tissue (9, 34). Further studies will test whether these genes are both necessary and sufficient for EAEC pathogenesis.
ACKNOWLEDGMENTS
This work was supported by grant AI33096 from the National Institutes of Health to J.P.N. W.P.E. was supported in part by a scholarship from the Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (Brazil).
REFERENCES
- 1.Bardwell J C A, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 2.Bhan M K, Raj P, Levine M M, Kaper J B, Bhandari N, Srivastava R, Kumar R, Sazawal S. Enteroaggregative Escherichia coli associated with persistent diarrhea in a cohort of rural children in India. J Infect Dis. 1989;159:1061–1064. doi: 10.1093/infdis/159.6.1061. [DOI] [PubMed] [Google Scholar]
- 3.Bilge S M, Apostol J M, Jr, Aldape M A, Moseley S L. mRNA processing independent of Rnase III and Rnase E in the expression of the F1845 fimbrial adhesin of Escherichia coli. Proc Natl Acad Sci USA. 1993;90:1455–1459. doi: 10.1073/pnas.90.4.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilge S M, Clausen C R, Lau W, Moseley S L. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol. 1989;171:4281–4289. doi: 10.1128/jb.171.8.4281-4289.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilge S M, Apostol J M, Jr, Fullner K J, Moseley S L. Transcriptional organization of the F1845 fimbrial adhesin determinant of Escherichia coli. Mol Microbiol. 1993;7:993–1006. doi: 10.1111/j.1365-2958.1993.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Carnoy C, Moseley S L. Mutational analysis of receptor binding mediated by the Dr family of Escherichia coli adhesins. Mol Microbiol. 1997;23:365–379. doi: 10.1046/j.1365-2958.1997.2231590.x. [DOI] [PubMed] [Google Scholar]
- 8.Cravioto A, Tello A, Navarro A, Ruiz J, Villafan H, Uribe F, Eslava C. Association of Escherichia coli HEp-2 adherence patterns with type and duration of diarrhoea. Lancet. 1991;337:262–264. doi: 10.1016/0140-6736(91)90868-p. [DOI] [PubMed] [Google Scholar]
- 9.Czeczulin J, Balepur S, Hicks S, Phillips A D, Hall R, Kothary M H, Navarro-Garcia F, Nataro J P. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect Immun. 1997;65:4135–4145. doi: 10.1128/iai.65.10.4135-4145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta A R, Kaper J B, MacQuillan A M. Shuttle cloning vectors for the marine bacterium Vibrio parahaemolyticus. J Bacteriol. 1984;160:801–811. doi: 10.1128/jb.160.2.808-811.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davison J, Heusterspreute M, Chevalier N, Ha-Thi V, Brunel F. Vectors with restriction site banks. V. pJRD215, a wide-host-range cosmid vector with multiple cloning sites. Gene. 1987;51:275–280. doi: 10.1016/0378-1119(87)90316-7. [DOI] [PubMed] [Google Scholar]
- 12.Debroy C, Yealy J, Wilson R A, Bhan M K, Kumar R. Antibodies raised against the outer membrane protein interrupt adherence of enteroaggregative Escherichia coli. Infect Immun. 1995;63:2873–2879. doi: 10.1128/iai.63.8.2873-2879.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elliott S J, Kaper J B. Role of type 1 fimbriae in EPEC infections. Microb Pathog. 1997;23:113–118. doi: 10.1006/mpat.1997.0135. [DOI] [PubMed] [Google Scholar]
- 14.Eslava C, Navarro-Garcia F, Czeczulin J R, Henderson I R, Cravioto A, Nataro J P. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3155–3163. doi: 10.1128/iai.66.7.3155-3163.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallegos M-T, Schleif R, Bairoch A, Hofmann K, Ramos J L. AraC/XylS family of transcriptional regulators. Microbiol Mol Biol Rev. 1997;61:393–410. doi: 10.1128/mmbr.61.4.393-410.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia M-I, Gounon P, Courcoux P, Labigne A, Le Bouguénec C. The fimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol Microbiol. 1996;19:683–693. doi: 10.1046/j.1365-2958.1996.394935.x. [DOI] [PubMed] [Google Scholar]
- 17.Garcia M-I, Labigne A, Le Bouguénec C. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 sequences. J Bacteriol. 1994;176:7601–7613. doi: 10.1128/jb.176.24.7601-7613.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germani Y, Begaud E, Duval P, Le Bouguenec C. Prevalence of enteropathogenic, enteroaggregative, and diffusely adherent Escherichia coli among isolates from children with diarrhea in New Caledonia. J Infect Dis. 1996;174:1124–1126. doi: 10.1093/infdis/174.5.1124. [DOI] [PubMed] [Google Scholar]
- 19.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 20.Heilmann C, Schweitzer O, Gerke C, Vanittanakom N, Mack D, Götz F. Molecular basis of intercellular adhesion in the biofilm-forming Staphylococcus epidermidis. Mol Microbiol. 1996;20:1083–1091. doi: 10.1111/j.1365-2958.1996.tb02548.x. [DOI] [PubMed] [Google Scholar]
- 21.Itoh Y, Nagano I, Kunishima M, Ezaki T. Laboratory investigation of enteroaggregative Escherichia coli associated with a massive outbreak of gastrointestinal illness. J Clin Microbiol. 1997;35:2546–2550. doi: 10.1128/jcm.35.10.2546-2550.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jouve M, Garcia M-I, Courcoux P, Labigne A, Gounon P, Le Bouguénec C. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect Immun. 1997;65:4082–4089. doi: 10.1128/iai.65.10.4082-4089.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.Knutton S, Shaw R K, Bhan M K, Smith H R, McConnell M M, Cheasty T, Williams P H, Baldwin T J. Ability of enteroaggregative Escherichia coli strains to adhere in vitro to human intestinal mucosa. Infect Immun. 1992;60:2083–2091. doi: 10.1128/iai.60.5.2083-2091.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levine M M, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements M L, Cheney C, Patnaik R. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect Immun. 1984;44:409–420. doi: 10.1128/iai.44.2.409-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindler L E, Plano G V, Burland V, Mayhew G F, Blattner F R. Complete DNA sequence and detailed analysis of the Yersinia pestis KIM5 plasmid encoding murine toxin and capsular antigen. Infect Immun. 1998;66:5731–5742. doi: 10.1128/iai.66.12.5731-5742.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nataro J P, Kaper J B, Robins-Browne R, Prado V, Vial P A, Levine M M. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. J Pediatr Infect Dis. 1987;16:829–831. doi: 10.1097/00006454-198709000-00008. [DOI] [PubMed] [Google Scholar]
- 28.Nataro J P, Deng Y, Maneval D R, German A L, Martin W C, Levine M M. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect Immun. 1992;60:2297–2304. doi: 10.1128/iai.60.6.2297-2304.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nataro J P, Yikang D, Giron J A, Savarino S J, Kothary M H, Hall R. Aggregative adherence fimbria I expression in enteroaggregative Escherichia coli requires two unlinked plasmid regions. Infect Immun. 1993;61:1126–1131. doi: 10.1128/iai.61.3.1126-1131.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nataro J P, Yikang D, Yingkang D, Walker K. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J Bacteriol. 1994;176:4691–4699. doi: 10.1128/jb.176.15.4691-4699.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nataro J P, Deng Y, Cookson S, Cravioto A, Savarino S J, Guers L D, Levine M M, Tacket C O. Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J Infect Dis. 1995;171:465–468. doi: 10.1093/infdis/171.2.465. [DOI] [PubMed] [Google Scholar]
- 32.Nataro J P, Steiner T, Guerrant R L. Enteroaggregative Escherichia coli. Emerg Infect Dis. 1998;4:251–261. doi: 10.3201/eid0402.980212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nataro, J. P., T. Whittam, and J. Czeczulin. Unpublished data.
- 34.Navarro-Garcia F, Eslava C, Villaseca J M, López-Revilla R, Czeczulin J R, Srinivas S, Nataro J P, Cravioto A. In vitro effects of a high-molecular-weight heat-labile enterotoxin from enteroaggregative Escherichia coli. Infect Immun. 1998;66:3149–3154. doi: 10.1128/iai.66.7.3149-3154.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niland P, Hühne R, Müller-Hill B. How AraC interacts specifically with its target DNAs. J Mol Biol. 1996;264:667–674. doi: 10.1006/jmbi.1996.0668. [DOI] [PubMed] [Google Scholar]
- 36.Nowicki B, Labigne A, Moseley S, Hull R, Hull S, Moulds J. The Dr hemagglutinin, afimbrial adhesins AFA-I and AFA-III, and F1845 fimbriae of uropathogenic and diarrhea-associated Escherichia coli belong to a family of hemagglutinins with Dr receptor recognition. Infect Immun. 1990;58:279–281. doi: 10.1128/iai.58.1.279-281.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Penfold R J, Pemberton J M. An improved suicide vector for construction of chromosomal insertion mutations in bacteria. Gene. 1992;118:145–146. doi: 10.1016/0378-1119(92)90263-o. [DOI] [PubMed] [Google Scholar]
- 38.Prentki P, Krisch H M. In vitro mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Savarino S J, Fox P, Yikang D, Nataro J P. Identification and characterization of a gene cluster mediating enteroaggregative Escherichia coli aggregative adherence fimbria I biogenesis. J Bacteriol. 1994;176:4949–4957. doi: 10.1128/jb.176.16.4949-4957.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 43.Smith H R, Cheasty T, Rowe B. Enteroaggregative Escherichia coli and outbreaks of gastroenteritis in UK. Lancet. 1997;350:814–815. doi: 10.1016/s0140-6736(05)62611-6. [DOI] [PubMed] [Google Scholar]
- 44.Taylor R K, Manoil C, Mekalanos J J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989;171:1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vial P A, Mathewson J J, DuPont H L, Guers L, Levine M M. Comparison of two assay methods for patterns of adherence to HEp-2 cells of Escherichia coli from patients with diarrhea. J Clin Microbiol. 1990;28:882–885. doi: 10.1128/jcm.28.5.882-885.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wai S N, Takade A, Amako K. The hydrophobic surface protein layer of enteroaggregative Escherichia coli strains. FEMS Microbiol Lett. 1996;135:17–22. doi: 10.1111/j.1574-6968.1996.tb07960.x. [DOI] [PubMed] [Google Scholar]
- 47.Wanke C A, Schorling J B, Barret L J, de Souza M A, Guerrant R L. Potential role of adherence traits of Escherichia coli in persistent diarrhea in an urban Brazilian slum. Pediatr Infect Dis J. 1991;10:746–751. doi: 10.1097/00006454-199110000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Yamamoto T, Wakisaka N, Kakae T. A novel cryohemagglutinin associated with adherence of enteroaggregative Escherichia coli. Infect Immun. 1997;65:3478–3484. doi: 10.1128/iai.65.8.3478-3484.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang H-Z, Donnenberg M S. DsbA is required for stability of the type IV pilin of enteropathogenic Escherichia coli. Mol Microbiol. 1996;21:787–797. doi: 10.1046/j.1365-2958.1996.431403.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Foxman B, Tallman P, Cladera E, Le Bouguénec C, Marrs C F. Distribution of drb genes coding for Dr binding adhesins among uropathogenic and fecal Escherichia coli isolates and identification of new subtypes. Infect Immun. 1997;65:2011–2018. doi: 10.1128/iai.65.6.2011-2018.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]