Abstract
Protein export in Escherichia coli is mediated by translocase, a multisubunit membrane protein complex with SecA as the peripheral subunit and the SecY, SecE, and SecG proteins as the integral membrane domain. In the gram-positive bacterium Bacillus subtilis, SecA, SecY, and SecE have been identified through genetic analysis. Sequence comparison of the Bacillus chromosome identified a potential homologue of SecG, termed YvaL. A chromosomal disruption of the yvaL gene results in mild cold sensitivity and causes a β-lactamase secretion defect. The cold sensitivity is exacerbated by overexpression of the secretory protein α-amylase, whereas growth and β-lactamase secretion are restored by coexpression of yvaL or the E. coli secG gene. These results indicate that the yvaL gene codes for a protein that is functionally homologous to SecG.
Bacillus subtilis, a gram-positive bacterium, has arisen next to Escherichia coli as a paradigm for studies on protein secretion primarily because bacilli have a high capacity for the production of exoenzymes. Protein secretion across the cytoplasmic membrane of B. subtilis is thought to be catalyzed by a system that is homologous to the precursor protein translocase of E. coli (22, 34). In E. coli, precursor protein translocation is mediated by a cytosolic chaperone, SecB; the translocation ATPase, SecA; and a large integral membrane protein complex with SecY, SecE, and SecG (9). SecD and SecF are accessory subunits that are not essential for translocation but that add to the fidelity and, possibly, the specificity of the reaction. Only SecA, SecE, and SecY are essential for viability, and homologues have been identified genetically in B. subtilis. SecA is encoded by the divA gene (1, 24) and was originally found in a set of mutants conditionally defective in division or unable to sporulate. The integral membrane proteins SecY (5, 26, 30) and SecE (11) were identified after nucleotide sequence analysis of the chromosomal regions that contain the ribosomal spc operon and nusG, respectively. The analogous regions in E. coli contain secY and secE, respectively. Complete sequence analysis of the B. subtilis chromosome has also revealed the presence of a single protein that is homologous to both SecD and SecF (3), but a homologue of the SecB protein has not been found (17).
In addition to the high similarity of the precursor protein translocases of B. subtilis and E. coli, some marked differences have been noted. B. subtilis contains not one but multiple signal peptidases with different specificity towards various secretory proteins (2). PrsA, a protein that is membrane bound through the presence of an amino-terminal fatty acyl anchor and is itself a secretory protein, has a profound effect on the secretion of some proteins in B. subtilis but is absent in E. coli (15, 16). PrsA is thought to function as a peptidyl-prolyl isomerase, but this activity has not yet been demonstrated in vitro. Another point of interest is the degree of host specificity of the components of the secretory apparatus. E. coli SecA cannot complement a B. subtilis divA mutant (28), whereas B. subtilis SecA can complement conditionally lethal secA mutations but only under specific sets of conditions (2, 13, 18, 28). Also, the E. coli and B. subtilis SecY proteins do not appear to be exchangeable (26).
The purification of the E. coli precursor protein translocase (6) has, in addition to SecY and SecE, given rise to the copurification of a protein termed band 1 (8). This protein is identical to P12, which was identified as a proteinaceous factor that stimulates SecYE-mediated protein translocation in vitro (19). The gene for P12 has been cloned via reverse genetics, and its chromosomal inactivation renders some E. coli strains cold sensitive for growth (20). Suppressor mutations have been found that are linked to this gene and that rescue cells from the toxic effects of the expression of heterologous mammalian secretory proteins (4). Based on these observations, the gene coding for P12 or band 1 was termed secG. SecG is the third, but nonessential, component of the heterotrimeric integral membrane domain of the precursor protein translocase. SecD and SecF can functionally replace SecG (10). SecG harbors two transmembrane segments that are thought to reverse their topology when SecA initiates translocation at the expense of ATP (21). SecG has been proposed to facilitate the membrane insertion of SecA, more or less acting as grease, which might explain why secG is not an essential gene under all conditions.
Homologues of secG have been found in other gram-negative bacteria, but none have been demonstrated in gram-positive bacteria. Since secG codes for a nonessential component of the precursor protein translocase, its genetic identification is complicated. Recently, the sequencing of the B. subtilis chromosome has been completed (17). We now report on the identification of an open reading frame, yvaL, that bears significant sequence similarity to the E. coli secG gene. Our data demonstrate that yvaL codes for a protein that is functionally homologous to SecG.
MATERIALS AND METHODS
Bacterial strains and growth media.
Strains were grown in Luria-Bertani broth or on Luria-Bertani agar. When necessary, the medium was supplemented with relevant antibiotics as indicated. Construction of vectors was done with E. coli DH5α [supE44 ΔlacU169 (φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1]). Chromosomal deletions and growth experiments were done with B. subtilis DB104 (nprE18 aprEΔ3) (35) or E. coli KN370 (20).
Construction of plasmids.
All of the relevant plasmids are listed in Table 1. The E. coli secG and B. subtilis yvaL genes, including suitable ribosome binding sites, were amplified as BamHI-XbaI cassettes by PCR from chromosomal DNA of strains DH5α and DB104, respectively, and cloned into pBluescript SK+ by using the primers listed in Table 2. The sequences of both open reading frames were determined and compared against the relevant databases. For expression in E. coli, the genes were cloned into pET324 (31), yielding pET304 (E. coli secG) and pET820 (B. subtilis yvaL).
TABLE 1.
Plasmids used in this study
| Plasmid | Replicon(s) | Resistance(s) | Relevant expression |
|---|---|---|---|
| pDELG2 | ColE1 | Ampr Camr | —a |
| pPR111 | ColE1, repR | Ampr Phler | — |
| pET470 | ColE1, repR | Ampr Phler | E. coli SecG |
| pET471 | ColE1, repR | Ampr Phler | B. subtilis YvaL |
| pET468 | repA | Eryr | AmyQ |
| pET472 | repA | Eryr | AmyQ, E. coli SecG |
| pET473 | repA | Eryr | AmyQ, B. subtilis YvaL |
| pET304 | ColE1 | Ampr | E. coli SecG |
| pET820 | ColE1 | Ampr | B. subtilis YvaL |
—, deletion vector.
TABLE 2.
PCR amplification primers used in this study
| Primer | Sequence (restriction enzyme)a |
|---|---|
| B. amyloliquefaciens amyQ forward | CGCCATGGTTCAAAAACGAAAGC (NcoI) |
| B. amyloliquefaciens amyQ reverse | GCGGATCCTTATTTCTGAACATA (BamHI) |
| B. subtilis yvaL forward | AAAGGATCCTAGTCTGGAGGTGTATGGGATGC (BamHI) |
| B. subtilis yvaL reverse | AAATCTAGATTCTCGAGCCCTATAGGATATAAGCAAGC (XbaI) |
| E. coli secG forward | CCCGGATCCGGAGGTTTTAATTCATGTATGAAGCTCTTT (BamHI) |
| E. coli secG reverse | CCCTCTAGACTCGAGTTAGTTCGGGATATCGC (XbaI) |
Recognition sites of the restriction enzymes used are underlined. Ribosome binding sites and start and stop codons are in boldface.
Vectors pPR111, a pUB110 derivative (7), and pBEY13 a gift from R. Breitling (4a), are shuttle vectors using a ColE1 origin for replication in E. coli and repR for replication in gram-positive organisms. These plasmids encode ampicillin resistance (Ampr) markers for E. coli and phleomycin resistance (Phler) markers for B. subtilis. Vector pBEY13 expresses the B. subtilis secY and secE genes from the constitutive staphylococcal sak promoter. Plasmids pET470 and pET471 were formed by replacing the secYE cassette with E. coli secG and B. subtilis yvaL, respectively.
Vector pAMP21 is a pGK13 (14)-based broad-host-range vector containing the p32 promoter derived from Lactococcus lactis (32) with a synthetic ribosome binding site and an NcoI site overlapping the start codon. The B. amyloliquefaciens α-amylase gene was isolated by PCR from plasmid pKTH10 (23) as an NcoI-BamHI cassette and ligated into NcoI-BamHI-digested pAMP21. The resulting vector, named pET468, harbors the amyQ gene under the control of the constitutive p32 promoter. Vectors pET472 and pET473 were generated by ligating the secG and yvaL gene-containing BamHI-BssHII fragments, respectively, from the pBluescript derivates into BamHI-MluI-digested pET468. The resulting vectors express B. amyloliquefaciens α-amylase and secG or yvaL as a tandem operon from the single p32 promoter.
Disruption of the yvaL gene.
The yvaL gene was disrupted in B. subtilis DB104 as follows. Regions immediately upstream and downstream of yvaL were amplified from chromosomal DNA from strain DB104 as BamHI-XbaI and KpnI-HincII cassettes, respectively, and cloned into pBluescript SK+. Subsequently, a BglII-PvuII digested chloramphenicol resistance (Camr) marker was placed between the BamHI and HincII sites, yielding pDELG2. This vector contains the DB104 chromosomal region with the yvaL gene replaced with the Camr marker. Vector pDELG2 was linearized with PvuII to yield a 2.8-kb fragment containing the yvaL::cam region and subsequently transformed into B. subtilis DB104 by natural competence (36). Camr colonies resulting from a double crossover were selected. The correct position of the chromosomal replacement was confirmed by PCR. In the resulting strain, DB104ΔyvaL, the Camr-encoding gene replaced the yvaL gene while leaving the flanking regions intact. Since the mutations cause a complete deletion, no selective pressure is needed after the initial selection.
Growth experiments.
B. subtilis DB104 and DB104ΔyvaL were transformed with each of six plasmids constructed for testing, i.e., pPR111, pET470, pET471, pET468, pET472, and pET473. After transformation, plates were incubated at 30°C overnight. Selective pressure using the appropriate antibiotics was applied from this point onward. No chloramphenicol was used at this stage. A single colony was picked for each transformant and cultured overnight at 30°C in liquid medium. Subsequently, cells were streaked on plates and incubated at temperatures ranging from 15 to 30°C. Plates were inspected daily, and the occurrence and size of the colonies were noted and scored when the wild-type strain reached a diameter of several millimeters.
For expression in E. coli, plasmids pET304 and pET820 were transformed to E. coli KN370 (secG::kan) as described before (20) and assayed for the formation of single colonies on agar plates at either 20 or 37°C, with or without induction by 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
Vesicle preparation and Western blotting.
Overnight cultures of E. coli KN370 transformed with pET304 or pET820 were diluted 1:50 into fresh medium and grown to an optical density at 600 nm (OD600) of 0.6, at which point 1 mM IPTG was added and growth was allowed to resume for another 3 h. Cells were harvested by centrifugation, resuspended in TN buffer (25 mM Tris-Cl [pH 7.5], 100 mM NaCl), and subjected to French pressure treatment (three times at 8,000 lb/in2). Cells debris was removed by centrifugation at 10,000 × g for 10 min, and vesicles were collected by centrifugation at 150,000 × g for 45 min. Vesicles were resuspended in TN buffer and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using antibodies directed against SecG and YvaL.
Analysis of cellular and secreted proteins.
B. subtilis DB104 and DB104ΔyvaL were grown overnight at 30°C in liquid medium. The overnight cultures were diluted 1:50 into fresh medium and grown to mid-logarithmic phase at different temperatures. Cultures were cooled on ice and fractionated into cellular and medium fractions by centrifugation. The medium fraction was precipitated with 10% (wt/vol) (final concentration) trichloroacetic acid, washed twice with cold acetone, and analyzed by SDS-PAGE. Cellular pellets were resuspended in sample buffer, sonicated, and analyzed by SDS-PAGE.
β-Lactamase activity was determined in strain DB104 transformed with plasmid pPR111 and in strain DB104ΔyvaL transformed with pPR111, pET470, or pET471. Overnight cultures of transformants were diluted 1:50 into fresh medium, and cells were grown to mid-logarithmic phase at 30°C. Cells were removed by centrifugation, and the culture supernatants were used for determination of β-lactamase activity with nitrocefin (25). Aliquots of 100 μl of culture supernatant were added to a reaction mixture (100 mM potassium phosphate [pH 7.0], 0.5-mg/ml nitrocefin), which was then incubated at 20°C. The OD486 was measured after 5 and 250 min.
Miscellaneous methods.
A peptide polyclonal antibody directed against an internal YvaL sequence (39Ala-Glu-Gln-Leu-Phe-Gly-Lys-Gln-Lys-Ala-Arg-Gly-Leu-Asp52) with an amino-terminal Tyr for coupling to keyhole limpet hemocyanin was produced in rabbits in accordance with standard procedures by NEOSYSTEM, Strasbourg, France. A peptide polyclonal antibody directed against the internal SecG sequence 89Ala-Pro-Ala-Lys-Thr-Glu-Gln-Thr-Gln-Pro98 was produced in rabbits in accordance with standard procedures by Research Genetics, Huntsville, Ala.
The yvaL gene was found in the Subtilist database by using the Blast search program included in reference 10a; other searches were done by using the Blast server at the National Center for Biotechnology Information (18a). Sequence alignments were done with ClustalX (29).
RESULTS
Identification of a secG homologue in B. subtilis.
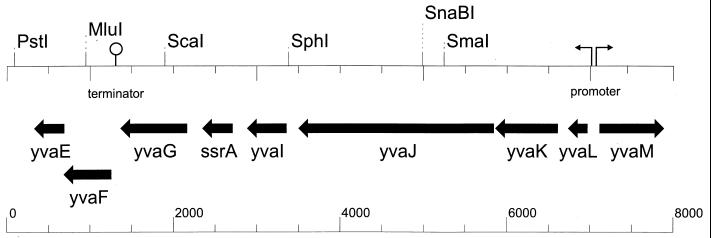
The Subtilist database of the B. subtilis chromosome was scanned with the E. coli secG gene by using the Blast search program included in reference 10a. This search yielded the yvaL gene (accession no. BG14067) as the only likely candidate (Fig. 1), with an E value of 2.4 × 10−8. The E value of the next best score was 0.65. YvaL is a 228-bp gene located at 295° of the genetic map of the B. subtilis chromosome (Fig. 2), in a region that bears many genes whose functions are unknown. yvaL seems to be the first gene in an operon, since the upstream gene yvaM is transcribed from the opposite strand. Downstream of yvaL, five genes can be identified without obvious promoter or terminator sequences in between but followed by a clear terminator structure. YvaL codes for an integral membrane protein of 76 amino acids that, in analogy to SecG, is predicted to span the membrane twice. It is 33% identical and 57% similar to E. coli SecG. Further searches with the B. subtilis yvaL and E. coli secG genes using the Blast server at reference 18a revealed the presence of homologues in other gram-positive bacteria. A multiple sequence alignment of these putative SecG proteins is shown in Fig. 1. Overall, the putative SecG homologues of gram-positive bacteria appear to be shorter than their counterparts in gram-negative bacteria. Although the Mycobacterium leprae secG gene is indicated in the data banks as such, no functional evidence is available that this open reading frame is, indeed, functionally homologous to SecG.
FIG. 1.
Multiple-sequence alignment of secG genes and potential homologues. Organisms are indicated as follows: Bs, B. subtilis (EMBL accession no. E1186051); Ml, Mycobacterium leprae (SwissProt accession no. P38388); Mt, M. tuberculosis (EMBL accession no. Z95844); Cg, Corynebacterium glutamicum. (GenBank accession no. M25819); Ec, E. coli (PIR accession no. S40402); Hi, Haemophilus influenzae (PIR accession no. H64068); Ps, Pseudomonas syringae; (EMBL accession no. U85643); Af, Aquiflex aeolicus (TREMBLNEW accession no. G2982840). Conserved residues are shaded according to the number of sequences in which the residue is conserved, and potential transmembrane segments (tms) are underlined.
FIG. 2.
Analyses of the chromosomal region containing yvaL. The sequence runs from bp 3447500 to bp 3455500 of the B. subtilis chromosome. Open reading frames are represented by straight arrows and named as in the Subtilist database. The promoter of the yvaL operon is depicted as a broken arrow, and the terminator is shown as a loop.
Deletion of yvaL causes mild cold sensitivity of growth.
Disruption of the secG gene has been shown to result in a cold-sensitive phenotype of E. coli MC4100-derived strains (20) at temperatures of 25°C and below. Assuming that SecG and YvaL function in the same manner, deletion of yvaL is expected to render B. subtilis cold sensitive as well. Therefore, the yvaL gene was deleted completely from the chromosome of B. subtilis DB104 by homologous recombination and replaced with a Camr marker. The correct position of the chromosomal replacement was confirmed by PCR. The resulting strain, DB104ΔyvaL, was normally viable at 37°C when grown on either rich or minimal medium. Incubation below 20°C revealed mild cold sensitivity, and the strain showed progressively slower growth than DB104 (data not shown). The cold-sensitive phenotype is not absolute. Compared to that of the wild type, growth was retarded more severely when the temperature was further lowered, but after the cells were shifted again to higher temperatures, growth resumed at a rate comparable to that of the wild type.
To analyze in more detail the phenotype of the deletion strain compared to that of the wild type, cells were transformed with plasmids expressing E. coli SecG or B. subtilis YvaL, as well as a control plasmid (Table 3). After preincubation at temperatures that do not affect the growth of the deletion strain, cells were plated and incubated at various temperatures. Growth of the colonies was monitored over a period of sev-eral days. Wild-type cells were not affected, and mutant cells transformed with the control plasmid behaved like their nontransformed counterparts, showing retarded growth but not a complete stop at lower temperatures. Transformation of the deletion strain with pET471 expressing the yvaL gene product relieved the retardation of growth, showing that the phenotype of the mutant was caused not by any polar effects but by the deletion of yvaL alone. Surprisingly, when the mutant was transformed with pET470 expressing E. coli SecG, growth stopped completely at temperatures of 20°C or lower. Also in wild-type cells, expression of E. coli SecG caused some interference with growth at low temperatures, possibly due to competition for SecYE with YvaL. These data indicate that disruption of yvaL from the B. subtilis chromosome causes mild cold sensitivity of growth. However, the effect is much weaker than that reported for E. coli KN370 (20).
TABLE 3.
Results of growth experiments
| Strain | Expression | Growtha
at:
|
||
|---|---|---|---|---|
| 15°C | 20°C | 25°C | ||
| DB104::111 | ++ | ++ | ++ | |
| DB104::470 | E. coli SecG | ± | ± | ++ |
| DB104::471 | B. subtilis YvaL | ++ | ++ | ++ |
| ΔyvaL::111 | ± | ± | ++ | |
| ΔyvaL::470 | E. coli SecG | − | − | ++ |
| ΔyvaL::471 | B. subtilis YvaL | ++ | ++ | ++ |
| DB104::468 | α-Amylase | ++ | ++ | ++ |
| DB104::472 | α-Amylase, E. coli SecG | ± | ± | ++ |
| DB104::473 | α-Amylase, B. subtilis YvaL | ++ | ++ | ++ |
| ΔyvaL::468 | α-Amylase | − | − | ++ |
| ΔyvaL::472 | α-Amylase, E. coli SecG | ± | ± | ± |
| ΔyvaL::473 | α-Amylase, B. subtilis YvaL | ± | ± | ± |
Growth was scored as follows: ++, growth like that of the reference; ±, growth slower than that of the reference; −, no growth.
Cold sensitivity of the growth of a B. subtilis ΔyvaL strain is exacerbated by overexpression of preAmyQ.
Since no complete cold sensitivity could be demonstrated for the DB104ΔyvaL strain, cells were transformed with high-copy plasmid PET468 and derivatives. These plasmids express the precursor form of α-amylase (preAmyQ) to high levels, thereby invoking secretory stress. Derivatives pET472 and pET473 express preAmyQ in combination with SecG or YvaL, respectively. Expression of preAmyQ did not retard the growth of the deletion mutant at 30°C, the temperature used to preculture the cells. The level of secreted α-amylase was the same for the wild type and the deletion mutant, as judged by halo formation on starch-containing plates and analysis of culture supernatants (data not shown). When pET468 transformants of B. subtilis DB104ΔyvaL were shifted to lower temperatures, clear and complete cold sensitivity was evident. Already at 20°C, cells stopped growing completely (Table 3), and when the bacteria were transferred back to the permissive temperature of 30°C after prolonged incubation at 20°C, growth was not resumed. Apparently, the deletion mutant is capable of sustaining a basic level of secretion even at lower temperatures but cannot handle the overexpression of a secretory protein within a broad temperature range. When coexpressed, both secG and yvaL complemented the deletion mutant, albeit the growth level of the transformants did not reach that of the wild type. Also, in this case, expression of SecG, but not that of YvaL, interferes with the growth of the wild type in a temperature-dependent way.
Effect of the yvaL deletion on the secretion of proteins.
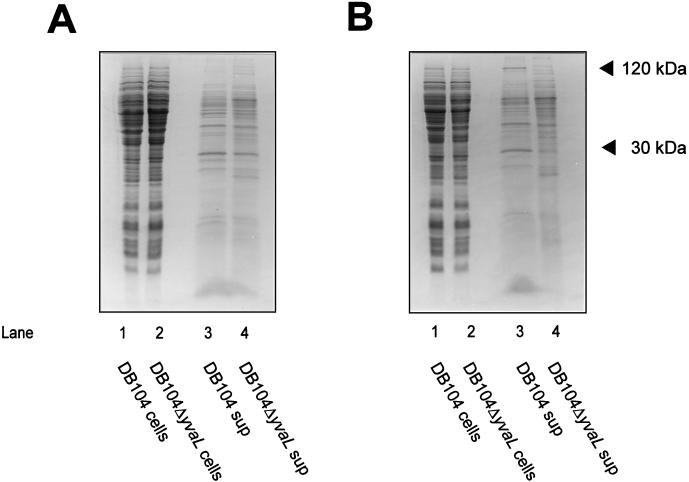
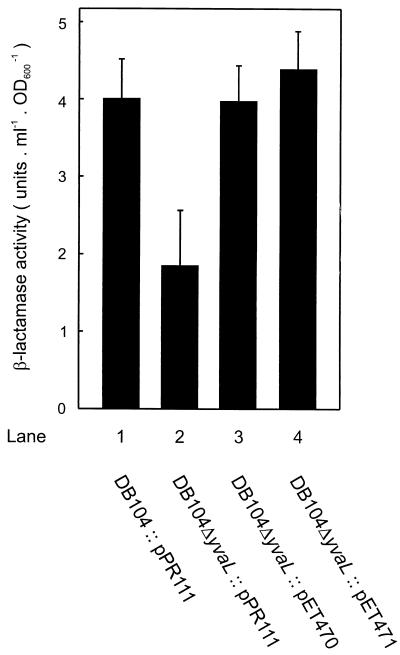
To investigate more directly the involvement of the precursor protein translocase in the cold-sensitive phenotype of the ΔyvaL deletion strain, the polypeptide patterns of wild-type and mutant cells were analyzed. In the culture supernatants of cells grown at 37°C, the yields of secreted proteins appeared generally to be similar for the wild type and mutants (Fig. 3A), although some cell lysis seems to occur in the deletion strain. However, the supernatant of cultures grown at 20°C showed some differences in the polypeptide pattern (Fig. 3B), e.g., at 30 and 120 kDa. Also, some secreted but cell-associated proteins appeared to be absent even at 37°C in the deletion mutant, as judged by proteinase K accessibility (data not shown). Since some, but not all, extracellular proteins are absent in the yvaL deletion strain, it seems that YvaL is needed for the secretion of a specific subset of proteins. The size of the 30-kDa protein may correspond to that of mature endogenous β-lactamase. The β-lactamase activity was about two- to threefold lower in the culture supernatant of strain DB104ΔyvaL than in that of strain DB104 (Fig. 4). Interestingly, the β-lactamase activity could be restored to normal levels by the expression of either B. subtilis YvaL or E. coli SecG. It is important to note that the E. coli β-lactamase present as an Ampr marker on the plasmids used is not expressed in B. subtilis (33). The β-lactamase activity of strain DB104 was the same with or without plasmid pPR111. These data strongly suggest that the B. subtilis yvaL deletion strain is impaired in the secretion of some proteins.
FIG. 3.
Coomassie-stained SDS-PAGE of cellular and medium fractions of B. subtilis DB104 and DB104ΔyvaL cultures grown to mid-logarithmic phase at 37°C (A) or 20°C (B). Positions of 30- and 120-kDa proteins absent in the supernatant of the yvaL mutant are indicated.
FIG. 4.
Restoration of β-lactamase secretion by YvaL and SecG. β-lactamase activities were determined in culture supernatants as described in Materials and Methods. One unit of activity is defined as the amount of enzyme needed to give an increase in OD600 of 10−3/min. Means and standard errors from three experiments are shown.
YvaL does not complement the cold sensitivity of the E. coli secG null strain.
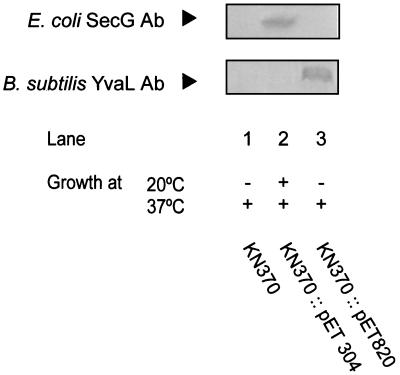
The secG disruption mutant E. coli KN370 shows a cold-sensitive phenotype (20). At 37°C, no growth defect is observed, while at 20°C, the strain is no longer able to form single colonies on agar plates. Upon induction with IPTG, plasmid pET304 expressing E. coli SecG was able to restore growth at the nonpermissive temperature of 20°C (Fig. 5). However, when E. coli KN370 was transformed with pET820 containing yvaL, no growth restoration was observed at the nonpermissive temperature, not even when the expression was induced by IPTG. On the other hand, growth was normal at the permissive temperature of 37°C. These data demonstrate that YvaL cannot functionally replace SecG in E. coli.
FIG. 5.
Complementation of the growth of E. coli KN370 by SecG or YvaL. The top panels show immunodetection of SecG or YvaL in E. coli KN370 bearing the plasmids indicated and after induction with IPTG. Results of the growth experiments at the indicated temperatures and in the presence of IPTG are listed below. Ab, antibody.
DISCUSSION
To facilitate functional studies on the precursor protein translocase of the gram-positive bacterium B. subtilis, we have cloned a homologue of SecG termed YvaL. The gene was identified on the basis of sequence similarity with its gram-negative counterpart. Although the overall identity is low, there is clear similarity between the YvaL and SecG proteins. Both proteins harbor two putative transmembrane segments that are connected via a glycine-rich loop. The YvaL protein is shorter than the SecG protein and lacks the carboxyl-terminal extension. This property is shared with other SecG homologues of gram-positive bacteria present in the databases. Like that of secG in E. coli (20), disruption of the yvaL gene in the chromosome of B. subtilis DB104 results in a cold-sensitive growth defect. However, this effect is mild compared to that in the E. coli secG null strain but is elevated when secretory stress is imposed by overexpression of the precursor form of α-amylase. The cold sensitivity can be overcome by expression of YvaL or SecG in trans, although growth is not restored to the level observed with the wild type only. Despite the care that was taken to disrupt only the yvaL open reading frame, the integration of the resistance marker could modulate the expression of the downstream genes and thereby affect physiology. It has been noted that the cold sensitivity of E. coli growth is strain dependent (10, 20). The reason for this is not entirely clear, but it may well relate to differences in growth physiology, secretion demand, and/or the level of translocase components in the various strains. Analysis of the profile of secreted proteins in wild-type B. subtilis and the ΔyvaL strain most notably reveals that two major proteins are absent in the latter strain. However, in the culture supernatant containing the secreted proteins, only certain polypeptides are affected. Therefore, it appears that deletion of yvaL does not result in a strong pleiotropic secretion defect but rather affects the secretion of a subset of proteins. No direct analyses of total secreted proteins has been performed with the E. coli secG null strain, but the in vitro translocation of the precursors proOmpA and proOmpF-Lpp demonstrates a clear difference in SecG dependence (19). Direct evidence that SecG and YvaL have the same function is provided by the observation that the secretion of β-lactamase in the B. subtilis ΔyvaL strain is restored not only by expression of YvaL but also by that of SecG. On the other hand, YvaL cannot complement the E. coli secG null strain. In conclusion, our results demonstrate that B. subtilis YvaL is a functional homologue of E. coli SecG. It is concluded that the heterotrimeric organization of the integral membrane domain of the translocase is conserved well in bacteria.
ACKNOWLEDGMENTS
We thank Reinhard Breitling for plasmids pPR111 and pBEY13 and Antonia Picón for plasmid pAMP21. We are grateful to Hadjime Tokuda for providing strain KN370. We thank Paulien Neefe for technical assistance.
These investigations were supported by CEC Biotech grants BIO2 CT 930254 and BIO4 CT 960097.
REFERENCES
- 1.Asai K, Kawamura F, Sadaie Y, Takahashi H. Isolation and characterization of a sporulation initiation mutation in the Bacillus subtilis secAgene. J Bacteriol. 1997;179:544–547. doi: 10.1128/jb.179.2.544-547.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolhuis A, Sorokin A, Azevedo V, Ehrlich S D, Braun P G, de Jong A, Venema G, Bron S, van Dijl J M. Bacillus subtilis can modulate its capacity and specificity for protein secretion through temporally controlled expression of the sipSgene for signal peptidase I. Mol Microbiol. 1996;22:605–618. doi: 10.1046/j.1365-2958.1996.d01-4676.x. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis A, Broekhuizen C P, Sorokin A, van Roosmalen M, Venema G, Bron S, Quax W J, van Dijl J M. SecDF of Bacillus subtilis, a molecular siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;274:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 4.Bost S, Belin D. A new genetic selection identifies essential residues in SecG a component of the Escherichia coliprotein export machinery. EMBO J. 1995;14:4412–4421. doi: 10.1002/j.1460-2075.1995.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Breitling, R. Unpublished data.
- 5.Breitling R, Schlott B, Behnke D. Modulation of the spc operon affects growth and protein secretion in Bacillus subtilis. J Basic Microbiol. 1994;34:145–155. doi: 10.1002/jobm.3620340303. [DOI] [PubMed] [Google Scholar]
- 6.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. The purified Escherichia coliintegral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 7.Diderichsen B, Poulsen G B, Jorgensen S T. A useful cloning vector for Bacillus subtilis. Plasmid. 1993;30:312–315. doi: 10.1006/plas.1993.1066. [DOI] [PubMed] [Google Scholar]
- 8.Douville K, Leonard M, Brundage L, Nishiyama K, Tokuda H, Mizushima S, Wickner W. Band 1 subunit of Escherichia colipreprotein translocase and integral membrane export factor P12 are the same protein. J Biol Chem. 1994;269:18705–18707. [PubMed] [Google Scholar]
- 9.Driessen A J M. Translocation of proteins across the bacterial cytoplasmic membrane. In: Konings W N, Kaback H R, Lolkema J S, editors. Handbook of biophysics. 2. Transport processes in membranes. Amsterdam, The Netherlands: Elsevier; 1996. pp. 759–790. [Google Scholar]
- 10.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Institut Pasteur Website. 2 January 1999, copyright date. [Online.] Institut Pasteur. http://www.pasteur.fr/Bio/SubtiList.html. [2 January 1999, last date accessed.]
- 11.Jeong S M, Yoshikawa H, Takahashi H. Isolation and characterization of the secE homologue gene of Bacillus subtilis. Mol Microbiol. 1993;10:133–142. doi: 10.1111/j.1365-2958.1993.tb00910.x. [DOI] [PubMed] [Google Scholar]
- 12.Klein M, Meens J, Freudl R. Functional characterization of the Staphylococcus carnosus SecA protein in Escherichia coli and Bacillus subtilis secAmutant strains. FEMS Microbiol Lett. 1995;131:271–277. doi: 10.1016/0378-1097(95)00267-9. [DOI] [PubMed] [Google Scholar]
- 13.Klose M, Schimp K-L, van der Wolk J, Driessen A J M, Freudl R. Lysine 106 of the putative catalytic ATP-binding site of the Bacillus subtilis SecA protein is required for functional complementation of Escherichia coli secA mutants in vivo. J Biol Chem. 1993;268:4504–4510. [PubMed] [Google Scholar]
- 14.Kok J, van der Vossen J M B M, Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kontinen V P, Saris P, Sarvas M. A gene (prsA) of Bacillus subtilisinvolved in a novel, late stage of protein export. Mol Microbiol. 1991;5:1273–1283. doi: 10.1111/j.1365-2958.1991.tb01901.x. [DOI] [PubMed] [Google Scholar]
- 16.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilisand sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 17.Kunst F, Ogasawara A M, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero V, Bessières P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Brignell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S-K, Codani J-J, Connerton I F, Cummings N J, Daniel R A, Denizot F, Devine K M, Düsterhöft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S-Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Hénaut A, Hilbert H, Holsappel S, Hosono S, Hullo M-F, Itaya M, Jones L, Joris B, Karamata D, Kasahara Y, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauber J, Lazarevic V, Lee S-M, Levine A, Liu H, Masuda S, Mauël C, Médigue C, Medina N, Mellado R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S-H, Parro V, Pohl T M, Portelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B-S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognoni A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H-F, Zumstein E, Yoshikawa H, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 18.McNicholas P, Rajapandi T, Oliver D. SecA proteins of Bacillus subtilis and Escherichia colipossess homologous amino-terminal ATP-binding domains regulating integration into the plasma membrane. J Bacteriol. 1995;177:7231–7237. doi: 10.1128/jb.177.24.7231-7237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.National Center for Biotechnology Information Website. 2 January 1999, copyright date. [Online.] National Center for Biotechnology Information. http://www.ncbi.nlm.nih.gov/Blast. [2 January 1999, last date accessed.]
- 19.Nishiyama K-I, Mizushima S, Tokuda H. A novel gene involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishiyama K-I, Hanada M, Tokuda H. Disruption of the gene encoding p12 (secG) reveals the direct involvement and important function of SecG in protein translocation of Escherichia coliat low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyma K-I, Suzuki T, Tokuda H. Inversion of the membrane topology of SecG coupled with SecA-dependent preprotein translocation. Cell. 1996;85:71–81. doi: 10.1016/s0092-8674(00)81083-1. [DOI] [PubMed] [Google Scholar]
- 22.Overhoff B, Klein M, Spies M, Freudl R. Identification of a gene fragment which codes for the 364 amino-terminal amino acid residues of a SecA homologue from Bacillus subtilis: further evidence for the conservation of the protein export apparatus in gram-positive and gram-negative bacteria. Mol Gen Genet. 1991;228:417–423. doi: 10.1007/BF00260635. [DOI] [PubMed] [Google Scholar]
- 23.Palva I. Molecular cloning of alpha-amylase gene from Bacillus amyloliquefaciens and its expression in B. subtilis. Gene. 1982;1:81–87. doi: 10.1016/0378-1119(82)90191-3. [DOI] [PubMed] [Google Scholar]
- 24.Sadaie Y, Takamatsu H, Nakamura K, Yamane K. Sequencing reveals similarity of the wild-type div+ gene of Bacillus subtilis to the Escherichia coli secAgene. Gene. 1991;98:101–105. doi: 10.1016/0378-1119(91)90110-w. [DOI] [PubMed] [Google Scholar]
- 25.Shannon K, Phillips I. Beta-lactamase detection by three simple methods: intralactam, nitrocefin and acidimetric. J Antimicrob Chemother. 1980;6:617–621. doi: 10.1093/jac/6.5.617. [DOI] [PubMed] [Google Scholar]
- 26.Suh J W, Boylan S A, Thomas S M, Dolan K M, Oliver D B, Price C W. Isolation of a secY homologue from Bacillus subtilis: evidence for a common protein export pathway in eubacteria. Mol Microbiol. 1990;4:305–314. doi: 10.1111/j.1365-2958.1990.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 27.Takamatsu H, Fuma S, Nakamura K, Sadaie Y, Shinkai A, Matsuyama S, Mizushima S, Yamane K. In vivo and in vitro characterization of the secA gene product of Bacillus subtilis. J Bacteriol. 1992;174:4308–4316. doi: 10.1128/jb.174.13.4308-4316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takamatsu H, Nakane A, Oguro A, Sadaie Y, Nakamura K, Yamane K A. Truncated Bacillus subtilis SecA protein consisting of the N-terminal 234 amino acid residues forms a complex with Escherichia coliSecA51(ts) protein and complements the protein translocation defect of the secA51 mutant. J Biochem Tokyo. 1994;116:1287–1294. doi: 10.1093/oxfordjournals.jbchem.a124677. [DOI] [PubMed] [Google Scholar]
- 29.Thompson J D, Gibson T J, Plewniak F, Jeanmougin F, Higgins D G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tschauder S, Driessen A J M, Freudl R. Cloning and molecular characterization of the secY genes from Bacillus licheniformis and Staphylococcus carnosus: comparative analysis of nine members of the SecY family. Mol Gen Genet. 1992;235:147–152. doi: 10.1007/BF00286192. [DOI] [PubMed] [Google Scholar]
- 31.Van der Does C, den Blaauwen T, de Wit J G, Manting E H, Groot N A, Fekkes P, Driessen A J M. SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol Microbiol. 1996;22:619–629. doi: 10.1046/j.1365-2958.1996.d01-1712.x. [DOI] [PubMed] [Google Scholar]
- 32.Van der Vossen J M, van der Lelie D, Venema G. Isolation and characterization of Streptococcus cremorisWg2-specific promoters. Appl Environ Microbiol. 1987;10:2452–2457. doi: 10.1128/aem.53.10.2452-2457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dijl, J. M. 1998. Unpublished results.
- 34.Van Wely K H M, Swaving J, Driessen A J M. Translocation of the precursor of α-amylase into Bacillus subtilis membrane vesicles. Eur J Biochem. 1998;255:690–697. doi: 10.1046/j.1432-1327.1998.2550690.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang M Y, Ferrari E, Henner D J. Cloning of the neutral protease gene of Bacillus subtilisand the use of the cloned gene to create an in vitro-derived deletion mutation. J Bacteriol. 1984;160:15–21. doi: 10.1128/jb.160.1.15-21.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young F E. Competence in Bacillus subtilistransformation system. Nature. 1967;213:773–775. doi: 10.1038/213773a0. [DOI] [PubMed] [Google Scholar]