Abstract
Diffuse large B‐cell lymphoma (DLBCL) is the most common lymphoid malignancy with a high relapse rate of up to 40%. The prognosis of the disease needs improvement and requires a understanding of its molecular mechanism. We investigated the mechanisms of DLBCL development and its sensitivity to chemotherapy by focusing on circPCBP2/miR‐33a/b/PD‐L1 axis. Human DLBCL specimens and cultured cancer cell lines were used. Features of circPCBP2 were systematically characterized through Sanger sequencing, Actinomycin D, RNase R treatment, and FISH. The expression levels of circPCBP2, miR‐33a/b, PD‐L1, stemness‐related markers, ERK/AKT and JAK2/STAT3 signaling were measured using qRT‐PCR, western blotting, and immunohistochemistry. Stemness of DLBCL cells was assessed through spheroid formation assay and flow cytometry. Cell viability and apoptosis upon cyclophosphamide, doxorubicin, vincristine, and prednisone (CHOP) treatment were determined using MTT assay and flow cytometry, respectively. Interactions of circPCBP2‐miR‐33a/b and miR‐33a/b‐PD‐L1 were validated using dual luciferase activity assay and RNA‐RIP. Nude mouse xenograft model was used to assess the function of circPCBP2 in DLBCL growth in vivo. circPCBP2 was upregulated in human DLBCL specimens and cultured DLBCL cells while miR‐33a/b was reduced. Knockdown of circPCBP2 or miR‐33a/b overexpression inhibited the stemness of DLBCL cells and promoted cancer cell apoptosis upon CHOP treatment. circPCBP2 directly bound with miR‐33a/b while miR‐33a/b targeted PD‐L1 3’‐UTR. circPCBP2 disinhibited PD‐L1 signaling via sponging miR‐33a/b. miR‐33a/b inhibitor and activating PD‐L1 reversed the effects of circPCBP2 knockdown and miR‐33a/b mimics, respectively. circPBCP2 knockdown restrained DLBCL growth in vivo and potentiated the anti‐tumor effects of CHOP. In conclusion, circPCBP2 enhances DLBCL cell stemness but suppresses its sensitivity to CHOP via sponging miR‐33a/b to disinhibit PD‐L1 expression. circPCBP2/miR‐33a/b/PD‐L1 axis could serve as a diagnosis marker or therapeutic target for DLBCL.
Keywords: CHOP, CircPCBP2, DLBCL, miR‐33a/b, PD‐L1
1)CircPCBP2 was elevated while miR‐33a/b was reduced in DLBCL specimens and cells. 2)Knockdown of circPCBP2 or miR‐33a/b overexpression suppressed DLBCL cell stemness and enhanced their sensitivity to CHOP.
Abbreviations
- circRNA
Circular RNA
- DLBCL
diffuse large B‐cell lymphoma
- IHC
immunohistochemistry
- NHL
non‐Hodgkin lymphoma
- PD‐1
programmed cell death 1
- PD‐L1
programmed cell death 1 ligand 1
- R‐CHOP
rituximab plus cyclophosphamide, hydroxydaunorubicin, vincristine sulfate, and prednisolone
- RIP
RNA immunoprecipitation
1. INTRODUCTION
DLBCL is the most prevalent B‐cell type of NHL, making up 30%–40% of NHL cases. 1 It is an extremely aggressive malignancy with high heterogeneity and progression rate. DLBCL is caused by mature B cells at various stages of differentiation. 2 The first‐line treatment for the disease is a combination of chemotherapy consisting of rituximab plus cyclophosphamide, hydroxydaunorubicin, vincristine sulfate, and prednisolone (R‐CHOP). 3 Nevertheless, nearly 40% DLBCL patients develop drug resistance to the treatment afterward and suffer from the relapsed disease. 2 , 3 There is a necessity to uncover the molecular mechanisms of DLBCL development, particularly the mechanisms underlying CHOP resistance, to find new avenues to treat the disease.
Programmed cell death 1 (PD‐1)/programmed cell death 1 ligand 1 (PD‐L1) or PD‐L2 signaling is one of the most critical central checkpoints in immune evasion during tumor progression. 4 , 5 PD‐1 is highly expressed in tumor‐specific T cells and functions to suppresses both adaptive and innate immune responses. 4 , 5 However, tumor cells can upregulate the level of PD‐L1 as a mechanism to induce T‐cell exhaustion and escape T‐cell‐induced anti‐tumor responses. Therefore, the PD‐1/PD‐L1 axis has been shown to play an important role in regulating the stemness of cancer cells, as well as their sensitivity and resistance to chemotherapy/radiotherapy. 5 In addition, PD‐1/PD‐L1 axis can be regulated using various signaling cascades, including PI3K/AKT and MAPK signaling pathways, and these signaling pathways in turn can significantly contribute to tumorigenesis. 4 , 5 Previous studies have reported an increase of PD‐L1 in DLBCL cells. Also, much research has indicated a crucial role of PD‐1/PD‐L1 in cancer progression. 6 , 7 , 8 Nevertheless, the detailed mechanisms underlying the upregulation remain elusive.
Circular RNAs (circRNAs) are a newly discovered and underappreciated class of endogenous noncoding RNAs. 9 They are very stable due to the closed structure. Recently, numerous evidence indicates that circRNAs play important roles in many physiological processes, as well as diseases, including cancers. 10 , 11 In DLBCL, many circRNAs have been implicated in cancer development and progression, such as circ_APC and has_circ_0000877. 12 A recent study reported an elevation of has_circ_0026652 (circPCBP2) in human DLBCL tissues. 12 Nevertheless, the function of this increase in DLBCL is largely unknown. The majority of circRNAs exerts functions by sponging microRNAs (miRNAs), which primarily function to suppress gene expression. 9 Dysregulated miRNAs have been shown as a key mechanism underlying DLBCL progression, as well as the development of drug resistance. 13 For instance, miR‐33a/b, believed to act as tumor suppressors, was significantly reduced in DLBCL patients with primary refractory disease following R‐CHOP treatment compared with those with complete remission receiving the same therapy, 14 suggesting that miR‐33a/b is involved in the R‐CHOP sensitivity of DLBCL patients. Notably, previous studies have indicated that miR‐33a/b suppressed tumor progression through suppressing the stemness of cancer cells. 15 In addition, our bioinformatic analysis uncovered complementary binding sites between circPCBP2 and miR‐33a/b, as well as between miR‐33a/b and PD‐L1 mRNA. We, therefore, speculated that circPCBP2 might participate in DLBCL development through miR‐33a/b and PD‐L1.
In the present study, we sought to characterize the role of circPCBP2/miR‐33a/b axis in DLBCL, with a focus on the sensitivity of cancer cells to CHOP. We first observed the aberrant expression of circPCBP2 and miR‐33a/b in human DLBCL tissues and cultured DLBCL cells. Subsequent functional studies revealed that knockdown of circPCBP2 suppressed the stemness of DLBCL cells, increased their sensitivity to CHOP treatment, and inhibited tumor growth in vivo via targeting miR‐33a/b/PD‐L1 signaling. Altogether, our findings uncover an essential role of circPCBP2/miR‐33a/b/PD‐L1 axis in DLBCL development and progression, which sheds light onto the mechanisms of cancer progression, as well as provide potential targets to develop therapy for DLBCL.
2. MATERIALS AND METHODS
2.1. Human DLBCL specimens
Human DLBCL tissues were collected from 60 diagnosed DLBCL patients without preoperative treatment at the Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital (Zhengzhou, China). Here, 60 samples of reactive lymph node hyperplasia (RLH) tissues from patients were collected as controls. All specimens were snap frozen and stored at −80°C. The study was approved by the ethics committee of the Henan Cancer Hospital (Zhengzhou, China). All patients were informed of the study and consented to the study.
2.2. Cell culture and treatment
Human DLBCL cell lines (U2932, OCI‐LY3, and SU‐DHL‐3) and one normal human lymphoblastoid cell line (GM12878) were purchased from BeNa Culture Collection (Suzhou, China). NU‐DUL‐1 was obtained from the American Type Culture Collection (ATCC, USA). Cells were cultured in RPMI 1640 medium (Gibco) replenished with 15% FBS (Gibco) and 1% penicillin/streptomycin in a standard cell incubator at 37°C with 5% CO2.
For CHOP stimulation, CHOP (Sigma‐Aldrich) at indicated concentrations (10, 20, 80, 160, 320 ng/ml) was directly added into the cell culture medium for 2 days. For rPD‐1 treatment, cultured cells were treated with human recombinant PD‐1 (R&D Systems, Minneapolis, USA, 8 μg/ml) for 24 h followed by CHOP treatment for an additional 2 days.
2.3. Cell transfection
miR‐33a/b mimics, miR‐33a/b inhibitor, and mimics negative control (NC), inhibitor NC were obtained from Genepharma (Shanghai, China). shRNA specifically targeting circPCBP2 and the control shRNA‐NC were synthesized and purchased from Geenseed Biotech (Guangzhou, China). Lipofectamine 3000 (Invitrogen) was used to deliver plasmids into cells as the manufacturer's instructions described. To make lentivirus, sh‐circPCBP3 and the negative control were cloned into the lentivirus vector pLV‐CMV. The constructs were transfected into HEK‐293 T cells together with pSPAX2 (#12260, Addgene, Watertown, USA) and pMD2G (#12259, Addgene, Watertown, USA). The virus‐containing medium was collected twice (24 h and 48 h after transfection). Lentivirus was concentrated through centrifugation at 25,000 g for 2 h at 4°C. The virus was resuspended in PBS and then used to infect cancer cells for 48 h.
2.4. Subcellular fractionation
Nuclear and cytoplasmic fractions were isolated first using a previously described method. 16 , 17 Cultured cells were homogenized in homogenization buffer and centrifuged at 800 g for 10 min. The pellet was saved as the nuclear fraction and subsequently lysed with TRIzol to extract total RNA. The supernatant was collected and centrifuged at 17,000 g for another 20 min. The supernatant was collected as the cytoplasm. TRIzol was added directly for RNA extraction.
2.5. Quantitative real‐time PCR (qRT‐PCR)
TRIzol (Invitrogen) containing DNase I was used to extract total RNA from both tissues and cultured cells using the manufacturer's protocol. Reverse transcription was performed with the cDNA synthesis kit (Thermo Fisher) to generate cDNAs. SYBR Green Master Mix (Invitrogen) was then used for quantitative PCR. For miRNAs, TaqMan microRNA assays were utilized for RT‐PCR (Thermo Fisher). Relative expression levels of circ_PCBP2, miR‐33a/b and PD‐L1 mRNA were calculated using the 2−ΔΔCt method through the normalization to U6 or GAPDH mRNA level, respectively. The primers used were from Guangzhou RiboBio Co., Ltd. (Guangdong, China):
circPCBP2 forward primer (FP): 5′‐AAAGGCGGGTGTAAGATCAAAG‐3′;
circPCBP2 reverse primer (RP): 5′‐GGCAAATCTGCTTGACACACTC‐3′;
miR‐33a FP: 5′‐GGCAAATCTGCTTGACACACTC‐3′;
miR‐33a RP: 5′‐GAACATGTCTGCGTATCTC‐3′;
miR‐33b FP: 5′‐ GCGTGCATTGCTGTTGCAT‐3′;
miR‐33b RP: 5′‐GTGCAGGGTCCGAGGT‐3′;
PD‐L1 FP: 5′‐CCAAGGCGCAGATCAAAGAGA‐3′;
PD‐L1 RP: 5′‐AGGACCCAGACTAGCAGCA‐3′;
U6 FP: 5′‐CTCGCTTCGGCAGCACA‐3′;
U6 RP: 5′‐AACGCTTCACGAATTTGCGT‐3′;
GAPDH FP: 5′‐ CCAGGTGGTCTCCTCTGA‐3′;
GAPDH RP: 5′‐ GCTGTAGCCAAATCGTTGT‐3′.
2.6. RNA FISH
Here, 4% paraformaldehyde (PFA) was directly added to cultured DLBCL cells for fixation at room temperature (RT) for 10–12 min followed by permeabilization with 1% Triton X‐100 for 3–5 min at RT. Fluorescence‐labeled specific probes for circ_PCBP2 (sequence: 5′‐TGAACTCACCATTCCAAACGATTCCCCCCCGAAGGGCGTGACCATCCCGT‐3′) were incubated with cells at 37°C overnight. The cells were washed off by PBS and then mounted on slices using DAPI‐containing mounting media followed by imaging with confocal microscope.
2.7. Actinomycin D and RNase R treatment
The circPCBP2 sequence was acquired by specific divergent primers and subsequently analyzed through Sanger sequencing (Sangon). Total RNA (3 μg) was treated with or without 2 U/μg of RNase R (Lucigen) for 1 h at 37 °C. Actinomycin D (1.5 mg/ml) or vehicle dimethyl sulphoxide (DMSO, Sigma‐Aldrich) was directly added to the culture medium for indicated periods. After treatment with corresponding chemicals, qRT‐PCR was performed to measure the levels of linear PCBP2 and circPCBP2.
2.8. MTT assay
Transfected DLBCL cells were first treated with CHOP (Sigma‐Aldrich) at concentrations of 20, 40, 80, 160, 320 ng/ml for 48 h and then 15 μl of MTT (5 mg/ml) was added to incubate with cells for 3 h at 37°C. Afterward, 150 μl dimethyl sulfoxide (Sigma‐Aldrich) was added to end the reaction and dissolve the formazan product. The absorbance at 590 nm in each condition was analyzed through plate reader.
2.9. Spheroid formation assay
A single‐cell population of transfected DLBCL cells was resuspended in culture medium supplemented with 20 ng/ml recombinant human epidermal growth factor (EGF; Abcam) and 10 ng/ml recombinant human basic fibroblast growth factor (FGF; R&D Systems) and then grown in 24‐well plates until spheroids formed. The number of spheres was counted through the microscope.
2.10. Flow cytometry
Transfected DLBCL cells were treated with or without rPD‐1 first and then CHOP stimulation was performed as described above. Afterward, treated cells were harvested and suspended in PBS followed by cell number counting. Anti‐CD133 (Thermo Fisher Scientific) was added to incubate with cells for 15 min on ice followed by centrifugation. Cell pellets were then washed with PBS twice and resuspended in PBS. The cell suspension was finally analyzed with flow cytometer (FACSAria II, BD Biosciences).
2.11. Cell apoptosis assay
Transfected DLBCL cells were harvested in binding buffer (Sigma‐Aldrich) followed by PBS wash. Annexin V‐FITC and propidium (PI) (Sigma‐Aldrich) were added to incubate with the suspension for 15 min on ice followed by a PBS wash. A flow cytometer (FACSAria II, BD Biosciences, Frankin Lakes, USA) was used to analyze the percentage of different populations of cells: annexin V‐FITC(−) and PI(−) were defined as living cells; annexin V‐FITC(+) and PI(−) cells were defined as early apoptotic cells; annexin V‐FITC(+) and PI(+) were defined as late apoptotic cells.
2.12. Dual luciferase assay
The wild type (WT) sequences of PD‐L1 3′UTR/circPCBP2 (WT‐PD‐L1/WT‐circPCBP2), or the sequences with the binding sites for miR‐33a/b mutated (MUT‐PD‐L1/MUT‐circPCBP2) were subcloned into the psiCHECK2 luciferase reporter vector (Promega). These constructs were then transfected into cultured DLBCL cells together with miR‐33a/b mimics or mimics‐NC using lipofectamine 3000 (Invitrogen). At 2 days later, the cells were harvested to measure relative luciferase activities using the Dual Luciferase Reporter Assay System (Promega).
2.13. Western blot analysis
Protease inhibitor containing RIPA lysis buffer (Abcam) was used to extract total proteins from cultured DLBCL cells. The protein concentration was measured using the DC Protein Assay Kit (Bio‐Rad). The same amount of protein was loaded for electrophoresis and then transferred to PVDF membranes (Bio‐Rad). The membranes were blocked in blocking buffer for 0.5 h at RT and subsequently incubated with primary antibodies overnight at 4°C. Antibodies were washed off with TBST buffer at RT and subsequently stained with the corresponding secondary antibodies for 1 h at RT. Protein signals were detected using the commercial ECL kit (Bio‐Rad). Primary antibodies used in our study were the following: anti‐p‐ERK (1:3000; #3192, Cell Signaling Technology); anti‐ERK (1:5000, #9102, Cell Signaling Technology); anti‐p‐AKT (1:2500, #9271, Cell Signaling Technology), anti‐AKT (1:5000, #9272, Cell Signaling Technology); anti‐PD‐L1(1:3000, ab205921, Abcam, UK); anti‐pJAK2 (1:1000, #3771, Cell Signaling Technology); anti‐JAK2 (1:2000, #3230, Cell Signaling Technology); anti‐pSTAT3 (1:800, #9145, Cell Signaling Technology); anti‐STAT3 (1:1500, #9139, Cell Signaling Technology); and anti‐GAPDH (1:5000, ab8245, Abcam).
2.14. RNA Immunoprecipitation (RIP) assay
Lysis buffer supplemented with 1% Triton X‐100 and RNase and protease inhibitor (Thermo Fisher Scientific) was utilized to lyse transfected cells. The protein concentration was quantified and equal amounts of protein samples were used to incubate with the corresponding primary antibodies (anti‐Ago2 and IgG as control) (Abcam) at 4°C overnight. The antibody‐conjugated samples were then incubated with protein A Sepharose beads (Sigma‐Aldrich) for 4 h at 4°C. The beads were washed with lysis buffer and then eluted with proteinase K (Sangon). The elution was subjected to RNA extraction with TRIzol reagent (Invitrogen) followed by qRT‐PCR.
2.15. Nude mice xenograft experiments
All animal experiments were reviewed and approved by the Animal Care and Use Committee of the Henan Cancer Hospital (Zhengzhou, China). The experiments were carried out under the guidance of the committee. Here, 8‐week‐old male BALB/C nude mice (SJA Laboratory Animal Co., Ltd. [Hunan, China]) were randomly divided into four groups. Mice were anesthetized first and then injected subcutaneously with 1 × 107 transfected DLBCL cells (sh‐NC group, sh‐circPCBP2 group) on the right side. For CHOP treatment, mice were intravenously injected with cyclophosphamide (40 mg/kg, i.v., day 1), hydroxydoxorubicin (3.3 mg/kg, i.v., day 1), oncovin (0.5 mg/kg, i.v., day 1), prednisone (0.2 mg/kg, orally for 5 days) for four 1‐week cycles following the injection. Saline was used for sham injection. Mice were monitored on a daily base to observe the tumor growth for 30 days. Tumor volume (V) was calculated as V (mm3) = 0.5 × (W)2 × (L) (L: tumor length; W: width). At the end of the experiments, tumors were dissected out and weighed.
2.16. Tissue histopathology analysis
Tumor tissues were fixed in 4% PFA overnight at 4°C and washed with PBS. The fixed tissues were embedded in paraffin followed by sectioning to 4‐μm thick slices. The slices were stained with hematoxylin (0.5%, 5 min) and eosin (1%, 3 min) (H&E) according to the manufacturer's protocol.
For immunohistochemical (IHC) staining, paraffin sections were rinsed in 80% methanol containing 0.3% H2O2 for 20 min at RT and then blocked in 3% normal goal serum at RT for 1 h. Primary antibodies (anti‐Ki67 [1:500, ab15580, Abcam]; anti‐PD‐L1 [1:500, ab205921, Abcam]; anti‐CD133 [1:500, #14–1331‐82, Thermo Fisher Scientific]) were incubated with slices overnight at 4°C. The sections were washed again with PBS and stained with HRP‐labeled secondary antibodies (Abcam) at RT for 1 h. Secondary antibodies were washed off with PBS and the signal was detected using 3,3′‐diaminobenzidine substrate. The stained tissues were mounted on the slices using the DAPI‐containing mounting medium (Invitrogen) followed by imaging with confocal microscope.
2.17. Statistical analysis
All plots and statistical analyses were performed in GraphPad Prism 8. Statistical p‐values were calculated using an unpaired Student t‐test (two groups) or one‐way ANOVA (more than two groups). Kaplan–Meier estimator was used to examine the correlation between survival rate and expression level of specific molecules. Pearson correlation analysis was performed to analyze the correlation between two parameters. The data were presented as mean ± SD (standard deviation). *p < 0.05, **p < 0.01, ***p < 0.001.
3. RESULTS
3.1. circPCBP2 was elevated in human DLBCL tissue and cultured cells
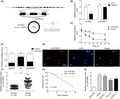
We first characterized the circular RNA features of circPCBP2. circPCBP2 is derived from exons 8–13 of the PCBP2 gene that is located on chromosome 12 (Figure 1A). Using the combination of divergent primers and Sanger sequencing, we validated the conjunction site of circPCBP2 and confirmed the length of circPCBP2 (378 bp) (Figure 1A). As expected, RNase R treatment markedly decreased the level of linear PCBP2, but did not affect circPCBP2 level (Figure 1B). Furthermore, circPCBP2 level in cells was consistently and significantly higher than the linear PCBP2 level after actinomycin D was added (Figure 1C). We also examined the subcellular localization of circPCBP2. First, we isolated nuclear and cytosol fractions using the subcellular fractionation method, and found that circPCBP2, similar to GAPDH mRNA, was primarily detected in the cytoplasm while U6, as expected, was mainly found in the nucleus (Figure 1D). Consistently, the FISH results showed that the majority of circPCBP2 was located in the cytoplasm (Figure 1E). These findings supported the notion that circPCBP2 is a stable circRNA that primarily resides in the cytoplasm.
FIGURE 1.
circPCBP2 was grown in human DLBCL tissue and cultured cells. (A) Structure of circPCBP2. (B) qRT‐PCR to measure expression levels of circPCBP2 and linear PCBP2 mRNA in cells treated with RNase R or control. (C) Expression levels of circPCBP2 and linear PCBP2 mRNA upon actinomycin D treatment. (D) Relative levels of GAPDH mRNA, U6 and circPCBP2 in cytoplasm and nuclear fractions. (E) Representative FISH images of circPCBP2 and DAPI staining. (F) Relative levels of circPCBP2 in human DLBCL tissues. (G) Overall survival rates of DLBCL patients of low and high circPCBP2 level analyzed by Kaplan–Meier analysis. (H) Relative levels of circPCBP2 in cultured DLBCL cell lines
To study the role of circPCBP2 in DLBCL, we measured its level in human DLBCL tissues and found a significant increase (Figure 1F). Furthermore, we observed that the survival rate of patients with high circPCBP2 was much lower than patients with low circPCBP2 (Figure 1G). As shown in Table 1, the number of patients with high circPCBP2 expression was higher in clinical stages III–IV, and patients with higher circPCBP2 level had an abnormal LDH level. Nevertheless, the expression of circPCBP2 did not correlate with other features (Table 1). Consistent with in vivo results, we observed a significant elevation of circPCBP2 in several DLBCL cell lines compared with normal lymphoblastoid cells (Figure 1H). Taken together, these data showed that circPCBP2 is upregulated in both DLBCL tissues and cells. We chose U2932 and NU‐DUL‐1 cell lines for subsequent studies because the level of circPCBP2 was higher in these two cell lines than the rest.
TABLE 1.
Association between circPCBP2 expression levels and clinicopathological features of DLBCL patients
| Clinical parameters | Cases (n) | circPCBP2 expression | p‐value (*p < 0.05) | |
|---|---|---|---|---|
| High (n) | Low (n) | |||
| Gender | ||||
| Male | 37 | 20 | 17 | 0.426 |
| Female | 23 | 10 | 13 | |
| Age | ||||
| <60 | 31 | 17 | 14 | 0.438 |
| ≥60 | 29 | 13 | 16 | |
| Stages | ||||
| I–II | 27 | 9 | 18 | 0.019* |
| III–IV | 33 | 21 | 12 | |
| LDH | ||||
| Normal | 24 | 8 | 16 | 0.035* |
| Elevated | 36 | 22 | 14 | |
| IPI score | ||||
| Low (0–2) | 24 | 14 | 10 | 0.292 |
| High (3–5) | 36 | 16 | 20 | |
| Hans classification | ||||
| GCB | 20 | 9 | 11 | 0.584 |
| Non‐GCB | 40 | 21 | 19 | |
| EBER | ||||
| Negative | 43 | 23 | 20 | 0.738 |
| Positive | 17 | 7 | 10 | |
Abbreviations: DLBCL, diffuse large B‐cell lymphoma; EBER, EBV‐encoded RNA; GCB, germinal center B‐like; IPI, International Prognostic Index; LDH, lactate dehydrogenase.
*represents the p‐value < 0.05.
3.2. Knockdown of circPCBP2 decreased the stemness of DLBCL cells and enhanced their sensitivity to CHOP
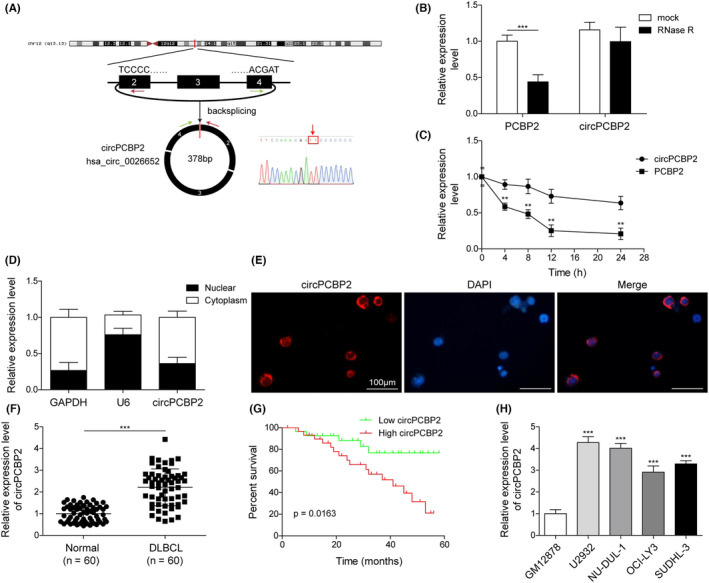
To further investigate the role of circPCBP2 in DLBCL, we reduced the expression of circPCBP2 in DLBCL cells through the use of sh‐circPCBP2 and examined the ensuing effects on cancer progression. As expected, transfection of DLBCL cells with sh‐circPCBP2 greatly diminished circPCBP2 levels (Figure 2A), confirming the high efficiency of the construct. Concomitantly, we found that sh‐circPCBP2 significantly decreased the number of cell spheroids formed (Figure 2B,C). We next examined how circPCBP2 regulated the stemness of the cancer cells by examining the levels of several pluripotent transcription factors including OCT4, Nanog, SOX2, and ALDH1A1 as they are well acknowledged stem cell markers. 18 We found that the levels of all those markers were greatly reduced in sh‐circPCBP2 transfected cells compared with control sh‐NC transfected cells (Figure 2D–G). We also measured how circPCBP2 regulated the sensitivity of DLBCL cells to CHOP, a widely used anthracycline‐based combinatorial chemotherapy regimen. As shown in Figure 2H, knockdown of circPCBP2 consistently and significantly decreased the viability of DLBCL cells upon CHOP treatment at various concentrations. In contrast, we observed a higher percentage of apoptotic cells in sh‐circPCBP2 transfected cells compared with cells transfected with control sh‐NC (Figure 2I,J). Altogether, these results demonstrated that knockdown of circPCBP2 suppressed the stemness of DLBCL cells but enhanced their sensitivity to CHOP treatment.
FIGURE 2.
Knockdown of circPCBP2 decreased the stemness of DLBCL cells and enhanced their sensitivity to CHOP. (A) qRT‐PCR to measure expression levels of circPCBP2 in sh‐NC or sh‐circPCBP2 transfected DLBCL cells. (B) Representative images of spheroids formed by DLBCL cells transfected with indicated plasmids. (C) Quantification of number of spheroids formed by transfected DLBCL cells. (D–G) Relative levels of OCT4 mRNA, Nanog mRNA, SOX2 mRNA, and ALDH1A1 mRNA in sh‐NC or sh‐circPCBP2 transfected DLBCL cells. (H) Cell viability of transfected DLBCL cells upon CHOP treatment assessed by MTT. (I, J) Percentage of apoptotic DLBCL cells following CHOP treatment analyzed by flow cytometry
3.3. circPCBP2 directly bound with miR‐33a/b and regulated PD‐L1 signaling
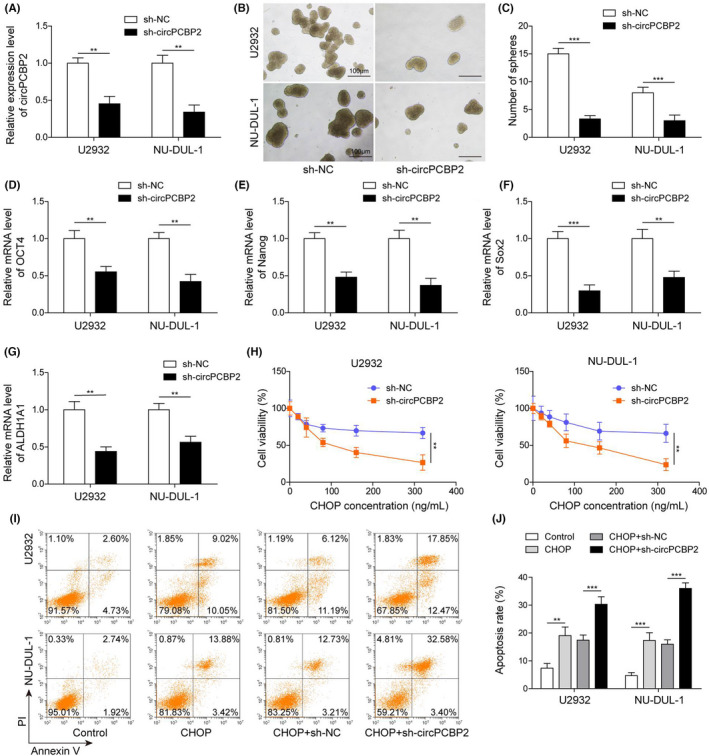
miR‐33a/b has been implicated in DLBCL. 14 In human DLBCL tissues we observed a significant reduction in miR‐33a/b compared with control specimens (Figure 3A,B). In addition, we detected a negative correlation between circPCBP2 level and miR‐33a/b level (Figure 3C,D). In cultured DLBCL cells, circPCBP2 knockdown increased the miR‐33a/b level (Figure 3E), suggesting that circPCBP2 might negatively regulate miR‐33a/b expression. We then used a bioinformatic tool (Starbase) to analyze whether they could bind. The analysis revealed complementary binding sites between circPCBP2 and miR‐33a/b. To further validate the interaction, we performed a dual luciferase activity assay. miR‐33a/b mimics greatly diminished the activity of circPCBP2‐WT but not circPCBP2‐MUT with the predicted binding sites mutated (Figure 3G). RNA‐RIP results indicated that Ago2 antibody enriched significantly more circPCBP2 and miR‐33a/b compared with IgG (Figure 3H). These results confirmed that circPCBP2 directly binds to miR‐33a/b. Previous studies have shown that PD‐L1 levels were largely associated with stemness of cancer cells. 5 We therefore examined whether circPCBP2 would regulate PD‐L1 expression, given that circPCBP2 modulated the stemness of DLBCL cells. We found that knockdown of circPCBP2 markedly reduced PD‐L1 mRNA levels in DLBCL cells (Figure 3I). Western blotting results showed that cells transfected with sh‐circPCBP2 had lower levels of PD‐L1, as well as p‐ERK, p‐AKT, p‐JAK2, and p‐STAT3 (Figure 3J,K) compared with sh‐NC transfected control cells. Therefore, circPCBP2 knockdown inhibits PD‐L1 expression and suppresses the JAK2/STAT3 and ERK/AKT pathways.
FIGURE 3.
circPCBP2 directly bound with miR‐33a/b and regulated PD‐L1 signaling. (A, B) qRT‐PCR to assess miR‐33a/b levels in human DLBCL specimens. (C, D) The relationship between miR‐33a/b levels and circPCBP2 level in DLBCL tissues. (E) Relative circPCBP2 levels in sh‐NC or sh‐circPCBP2 transfected DLBCL cells. (F) Predicted binding sites between circPCBP2 and miR‐33a/b. (G) Relative luciferase activities of circPCBP2‐WT and circPCBP2‐MUT in transfected DLBCL cells. (H) Relative enrichment of circPCBP2 and miR‐33a/b following immunoprecipitation with Ago2 or IgG antibody. (I) Relative PD‐L1 mRNA levels in DLBCL cells transfected with sh‐NC or sh‐circPCBP2. (J, K) Protein levels of p‐JAK2, JAK2, p‐STAT3, STAT3, p‐ERK, ERK, p‐AKT, AKT, and PD‐L1 in transfected DLBCL cells measured by western blotting
3.4. Inhibition of miR‐33a/b or activation of PD‐L1 reversed the effects of circPCBP2 knockdown on DLBCL cell properties
We next examined the function of the circPCBP2/miR‐33a/b/PD‐L1 axis in DLBCL. miR‐33a/b inhibitor suppressed the reduction in spheroid formation of DLBCL cells caused by circPCBP2 knockdown (Figure 4A,B). Activation of PD1/PD‐L1 with recombinant human PD‐1 (rPD‐1) also inhibited the reduction (Figure 4A,B). Furthermore, the decreased levels of pluripotent transcription factors induced by sh‐circPCBP2 were reversed by both miR‐33a/b inhibitors and rPD‐1 (Figure 4C–F). Flow cytometry results indicated that circPCBP2 knockdown reduced the number of CD133‐positive cells, whereas miR‐33a/b inhibitors and rPD‐1 restored the number (Figure 4G). Similarly, the hypersensitivity to CHOP induced by sh‐circPCBP2 including decreased cell viability and increased apoptotic cells, was significantly suppressed by miR‐33a/b inhibitors and rPD‐1 (Figure 4I–K). Taken together, these findings demonstrated that both inhibition of miR‐33a/b and activation of PD‐1/PD‐L1 signaling greatly attenuated the effects of circPCBP2 knockdown.
FIGURE 4.
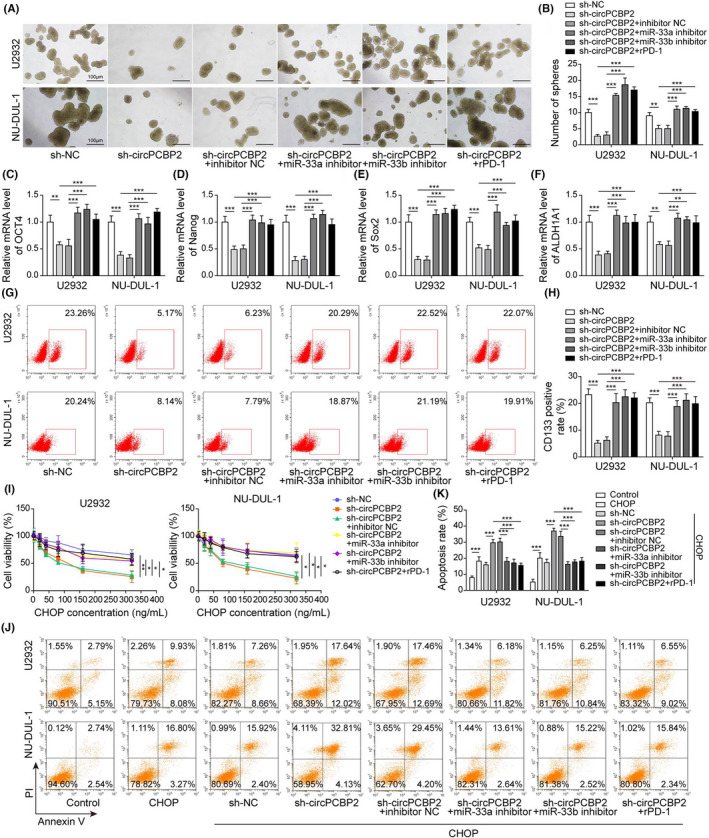
Inhibition of miR‐33a/b or activation of PD‐L1 reversed the effects of circPCBP2 knockdown on DLBCL properties. (A) Representative images of spheroids formed by DLBCL cells transfected with indicated plasmids. (B) Quantification of number of spheroids formed by transfected DLBCL cells. (C–F) Relative levels of OCT4 mRNA, Nanog mRNA, SOX2 mRNA, and ALDH1A1 mRNA in sh‐NC or sh‐circPCBP2 transfected DLBCL cells. (G, H) Flow cytometry analysis of the number of CD133‐positive cells following transfection. (I) Cell viability of transfected DLBCL cells upon CHOP treatment. (J, K) Percentage of apoptotic DLBCL cells following CHOP treatment
3.5. miR‐33a/b directly targeted PD‐L1 mRNA to suppress its expression
To investigate the relationship between miR‐33a/b and PD‐L1, we measured PD‐L1 expression levels in DLBCL tissues and cells. We found a significant elevation of PD‐L1 mRNA and a negative correlation between PD‐L1 mRNA levels and miR‐33a/b levels in human DLBCL tissues (Figure 5A,B). Notably, our bioinformatic analysis (Starbase) revealed a potential binding between miR‐33a/b and PD‐L1 mRNA (Figure 5C), suggesting that miR‐33a/b might target PD‐L1 mRNA. We then used a dual luciferase assay to directly verify this finding. The results indicated that miR‐33a/b mimics greatly decreased WT‐PD‐L1 activity, but had no effect on MUT‐PD‐L1 activity (Figure 5D), indicating that miR‐33a/b directly targeted PD‐L1 mRNA. Consistently, we found that miR‐33a/b mimics decreased PD‐L1 mRNA and protein levels in DLBCL cells, whereas miR‐33a/b inhibitor increased them (Figure 5E–G). Furthermore, the levels of p‐ERK, p‐AKT, p‐JAK2, and p‐STAT3 were reduced in miR‐33a/b mimics‐transfected DLBCL cells but were raised in miR‐33a/b inhibitor‐transfected DLBCL cells (Figure 5F,G). Taken together, we concluded that miR‐33a/b directly targeted PD‐L1 mRNA to suppress PD‐L1 signaling.
FIGURE 5.
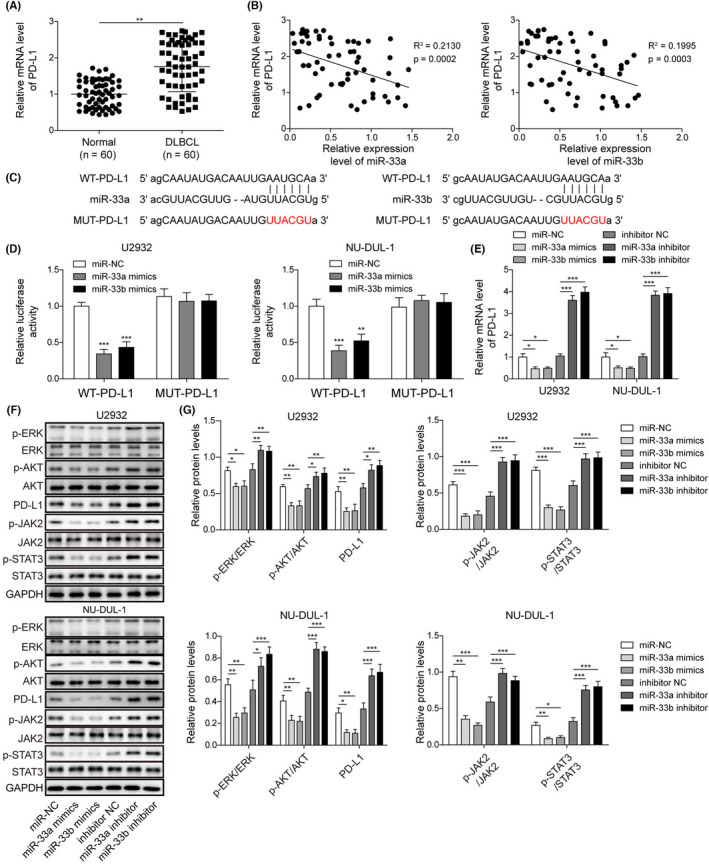
miR‐33a/b directly targeted PD‐L1 to suppress its expression. (A) qRT‐PCT to measure PD‐L1 mRNA levels in human DLBCL specimens. (B) The relationship between miR‐33a/b levels and PD‐L1 mRNA level in DLBCL tissues. (C) Predicted binding sites between PD‐L1 mRNA and miR‐33a/b. (D) Relative luciferase activities of PD‐L1‐WT and PD‐L1‐MUT in transfected DLBCL cells. (E) Relative PD‐L1 mRNA levels in transfected DLBCL cells assessed. (F, G) Protein levels of p‐JAK2, JAK2, p‐STAT3, STAT3, p‐ERK, ERK, p‐AKT, AKT, and PD‐L1 in transfected DLBCL cells
3.6. Overexpression of miR‐33a/b inhibited the stemness of DLBCL cells and enhanced their sensitivity to CHOP via PD‐L1
We next investigated whether miR‐33a/b regulated the DLBCL cell properties via PD‐L1. Consistently, overexpression of miR‐33a/b markedly suppressed the cell spheroid formation of DLBCL cells (Figure 6A,B). Moreover, the levels of OCT4, Nanog, SOX2 and ALDH1A1 were significantly decreased in DLBCL cells transfected with miR‐33a/b mimics compared with control transfected cells (Figure 6C–F). However, rPD‐1 reversed those effects caused by miR‐33a/b mimics (Figure 6A–F). Flow cytometry results revealed that miR‐33a/b overexpression greatly decreased the number of CD133‐positive cells, but rPD‐1 treatment restored this number (Figure 6G,H). Moreover, overexpression of miR‐33a/b greatly reduced cancer cell viability and increased the percentage of apoptotic cancer cells in response to CHOP treatment, whereas again these numbers were recovered by rPD‐1 (Figure 6I–K). These results demonstrated that miR‐33a/b overexpression suppressed the stemness of DLBCL cells and enhances their sensitivity to CHOP treatment through targeting PD‐L1.
FIGURE 6.
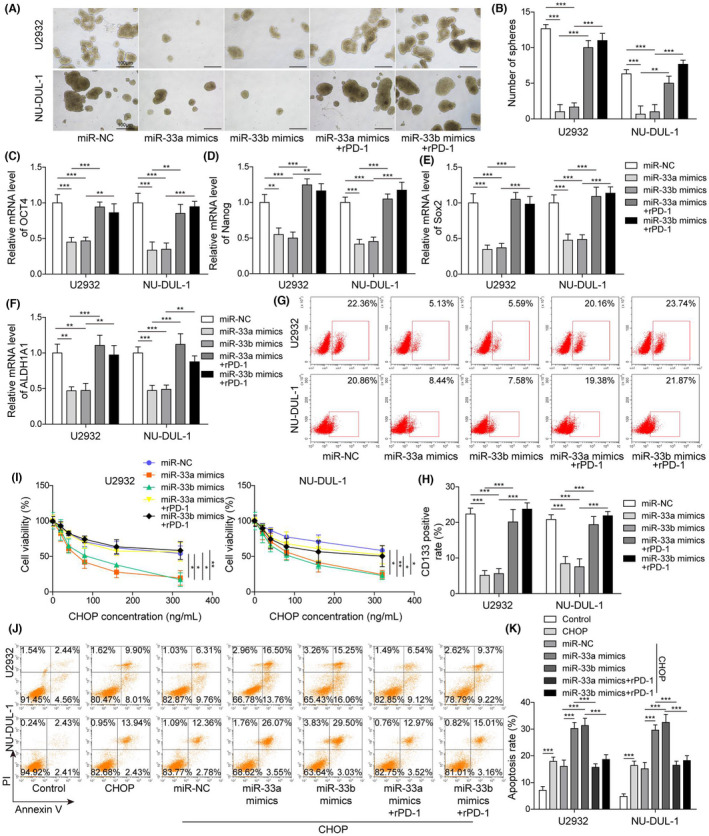
Overexpression of miR‐33a/b inhibited the stemness of DLBCL cells and enhanced their sensitivity to CHOP via PD‐L1. (A) Representative images of spheroids formed by transfected DLBCL cells. (B) The number of spheroids formed by transfected DLBCL cells. (C–F) Relative levels of OCT4 mRNA, Nanog mRNA, SOX2 mRNA, and ALDH1A1 mRNA in transfected DLBCL cells. (G, H) Flow cytometry analysis of the number of CD133‐positive cells following transfection. (I) Cell viability of transfected DLBCL cells upon CHOP treatment. (J, K) Percentage of apoptotic DLBCL cells following CHOP treatment
3.7. circPCBP2 knockdown suppressed DLBCL growth and increased its sensitivity of CHOP in vivo
Finally, we evaluated the role of circPCBP2 in DLBCL in vivo using the nude mouse xenograft model. Control DLBCL cells or cells transfected with sh‐circPCBP2 were implanted onto nude mice. The tumor grew rapidly in the mice implanted with control DLBCL cells (Figure 7A–C). However, knockdown of circPCBP2 or CHOP treatment significantly decreased the tumor volume and weight in vivo (Figure 7A–C). Notably, with a combination of circPCBP2 knockdown and CHOP treatment, the tumor volume and weight were further reduced (Figure 7A–C). At the molecular level, we found that circPCBP2 knockdown or CHOP treatment suppressed circPCBP2 and PD‐L1 expression, but promoted miR‐33a/b expression (Figure 7D–G). A combination of circPCBP2 knockdown and CHOP treatment further downregulated circPCBP2 and PD‐L1 levels and upregulated miR‐33a/b levels (Figure 7D–G). IHC results showed that circPCBP2 knockdown or CHOP treatment greatly decreased Ki‐67, PD‐L1, and CD133 signals in the tumor, whereas the combination of both further decreased those levels (Figure 7H). Consistently, western blotting data revealed that sh‐circPCBP2 or CHOP treatment significantly reduced the protein levels of PD‐L1, p‐AKT, p‐ERK, p‐JAK2, and p‐STAT3, whereas a combination of both further diminished the levels (Figure 7I,J). Therefore, we concluded that circPCBP2 knockdown inhibited DLBCL tumor growth in mice and potentiated the anti‐tumor effect of CHOP.
FIGURE 7.
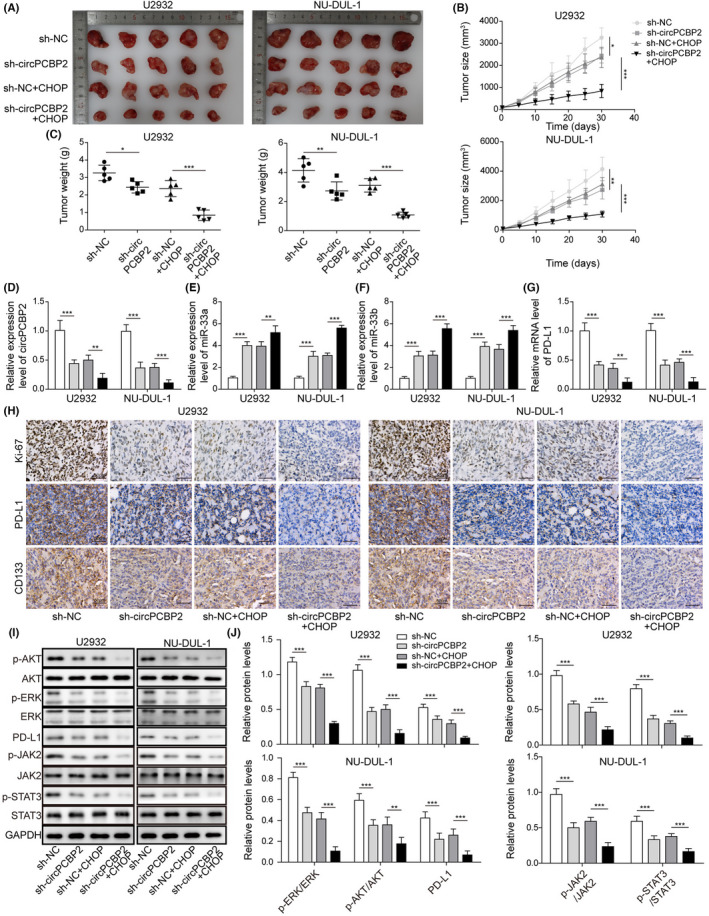
circPCBP2 knockdown suppressed DLBCL growth and increased its sensitivity of CHOP in vivo. (A) Representative tumor images in each group. (B) Tumor volume with time. (C) Tumor weight in each group at different time. (D) qRT‐PCR analysis of circPCBP2 levels in tumors of each group. (E, F) Relative miR‐33a/b levels in tumors of each group. (G) Relative PD‐L1 mRNA levels in tumors of each group. (H) IHC staining to measure Ki‐67, PD‐L1, and CD133 levels in the tumors of each group. Scale bar, 50 μm. (I, J) Protein levels of p‐JAK2, JAK2, p‐STAT3, STAT3, p‐ERK, ERK, p‐AKT, AKT, and PD‐L1 in tumor tissues
4. DISCUSSION
As the most common subtype of aggressive NHLs, DLBCL affects numerous people. The prognosis of the disease is poor and needs improvement. 19 , 20 One major challenge is the development of resistance to CHOP or the R‐CHOP regime in many patients and the underlying molecular mechanisms are poorly understood. 21 In the study, we observed aberrant expression of circPCBP2 and miR‐33a/b in DLBCL. We further demonstrated that circPCBP2 knockdown or miR‐33a/b overexpression suppressed the stemness of DLBCL cells and improved the pro‐apoptotic effects of CHOP, leading to less tumor growth. Mechanistically, we showed that circPCBP2 sponges miR‐33a/b to uninhibit PD‐L1 expression and thereby activate the ERK and AKT signaling pathways (Figure 8). Altogether, our study revealed that circPCBP2 promoted DLBCL progression by targeting miR‐33a/b to upregulate PD‐L1 expression.
FIGURE 8.
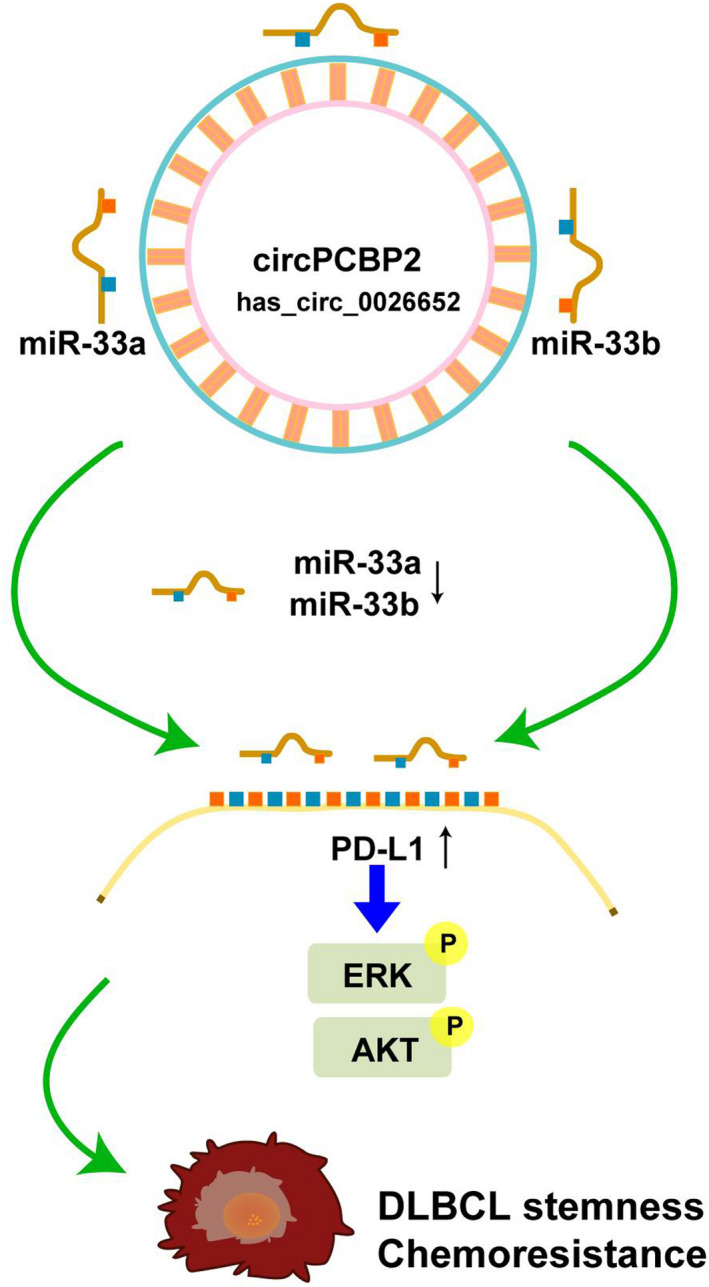
Schematic diagram of the circPCBP2/miR‐33a/b/PD‐L1 axis in DLBCL
Accumulating evidence indicated that circRNAs have a critical role in both normal physiological processes and pathological diseases, particularly in cancers. 10 , 22 circRNAs can regulate cancer cell proliferation and migration, the stemness of cancer cells, as well as their sensitivity to drugs, including radiotherapy and chemotherapy treatments. 22 , 23 , 24 Moreover, dysregulated circRNAs have been implicated in the development and progression of many kinds of cancers such as hepatocellular carcinoma, breast cancer, and ovarian cancers. 25 , 26 , 27 However, the number of studies of circRNAs in DLBCL is few and the function of circRNAs in DLBCL remains elusive. Our elucidation of the function of circRNAs in DLBCL helps fill the gap, not only shedding light on the mechanism of the disease, but also providing avenues to develop diagnosis or therapy strategies in the sense that circRNAs have high stability. 11 Recently, a study revealed substantial changes in many circRNAs in DLBCL using circRNA microarray expression profiling and one of those circRNAs was circPCBP2. 12 However, its function is largely unknown. Here, we demonstrated that circPCBP2 functions as a pro‐tumor molecule in DLBCL and that knockdown of circPCBP2 markedly suppressed DLBCL growth and progression. These results indicated that circPCBP2 can serve as a diagnosis marker or therapeutic target for DLBCL. The function of circPCBP2 is understudied. Whether it has similar pro‐tumor roles in other cancers or functions differently requires additional investigation. Moreover, it will be interesting to examine the role of circPCBP2 in normal physiological processes such as cell development, proliferation, and migration. Also, as mentioned before, many circRNAs have been shown to be altered in DLBCL cells, but their functions are not well understood. Future studies are necessary to fill this gap.
miRNAs have important roles in diseases such as cancers. 28 , 29 Many circRNAs exert functions by sponging miRNAs. Here, we uncovered miR‐33a/b as novel partners of circPCBP2 in DLBCL. miR‐33a/b functions as a tumor suppressor in various types of cancers such as breast cancer, prostate cancer, and lung cancer. 30 , 31 , 32 , 33 However, the detailed function of miR‐33a/b in DLBCL is poorly understood, although miR‐33a/b has been associated with the responses of DLBCL patients to R‐CHOP. 14 We showed that miR‐33a/b inhibited the stemness of DLBCL cells and potentiated their responses to CHOP treatment through targeting PD‐L1. Our study provides evidence that miR‐33a/b has a conserved role in cancers. However, it remains to be further exploration whether miR‐33a/b regulates other properties of DLBCL cells, such as cell migration or invasion abilities. In addition, whether other targets of miR‐33a/b are involved in DLBCL cells requires additional investigation. For instance, proteins including HMGA2, SALL4, and Twist1 have been shown to be regulated by miR‐33a/b during cancer metastasis. 15 It might be interesting to examine their roles in DLBCL.
The PD‐1/PD‐L1 axis is believed to be a pivotal signaling in mediating cancer immune escape and therefore has significant impacts on cancer therapy. 4 , 5 Increased PD‐L1 expression in cancer cells has been shown to promote stemness of cancer cells, as well as their resistance to radiotherapy and chemotherapy. 34 , 35 , 36 In addition, the intrinsic PD‐L1 signal in tumor cells greatly contributes to epithelial‐to‐mesenchymal transition (EMT), tumor invasion, cancer cell stemness, and cancer cell chemoresistance. However, conversely, EMT inducers could activate PD‐L1 expression and signaling. 37 In this study, consistently, we demonstrated that inhibiting PD‐L1 markedly decreased the stemness of cancer cells. Notably we showed that the circPCBP2/miR‐33a/b axis was a key upstream signaler that mediated the change in PD‐L1 in DLBCL cells. Therefore, modulating circPCBP2/miR‐33a/b can be used to change the PD‐1/PD‐L1 axis in DLBCL. It might be interesting to examine whether this regulatory network is conserved in other types of cancers or normal physiological processes.
In conclusion, our work proved that circPCBP2 increases DLBCL cell stemness and decreases its sensitivity to CHOP via sponging miR‐33a/b to uninhibit PD‐L1 expression. The characterization of this molecular network uncovers a key mechanism underlying DLBCL development and progression, which could provide promising targets for the development of future diagnosis or therapy strategies for DLBCL.
DISCLOSURE
The authors have no conflict of interest.
ETHICS APPROVAL
The study was approved by the ethic committee of the Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital (Zhengzhou, China).
The animal experiment was approved by the Animal Care and Use Committee of the Affiliated Cancer Hospital of Zhengzhou University, Henan Cancer Hospital (Zhengzhou, China).
ACKNOWLEDGMENTS
This work was supported by Joint Construction Project of Henan Medical Science and Technology Research Plan (LHGJ20190643).
Dong L, Huang J, Gao X, Du J, Wang Y, Zhao L. CircPCBP2 promotes the stemness and chemoresistance of DLBCL via targeting miR‐33a/b to disinhibit PD‐L1 . Cancer Sci. 2022;113:2888‐2903. doi: 10.1111/cas.15402
REFERENCES
- 1. Yin X, Xu A, Fan F, et al. Incidence and mortality trends and risk prediction nomogram for Extranodal diffuse large B‐cell lymphoma: an analysis of the surveillance, epidemiology, and end results database. Front Oncol. 2019;9:1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horvat M, Zadnik V, Juznic Setina T, et al. Diffuse large B‐cell lymphoma: 10 years' real‐world clinical experience with rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisolone. Oncol Lett. 2018;15:3602‐3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang L, Li LR. R‐CHOP resistance in diffuse large B‐cell lymphoma: biological and molecular mechanisms. Chin Med J (Engl). 2020;134:253‐260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han Y, Liu D, Li L. PD‐1/PD‐L1 pathway: current researches in cancer. Am J Cancer Res. 2020;10:727‐742. [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang Y, Chen M, Nie H, Yuan Y. PD‐1 and PD‐L1 in cancer immunotherapy: clinical implications and future considerations. Hum Vaccin Immunother. 2019;15:1111‐1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu‐Monette ZY, Zhou J, Young KH. PD‐1 expression and clinical PD‐1 blockade in B‐cell lymphomas. Blood. 2018;131:68‐83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Song MK, Park BB, Uhm J. Understanding immune evasion and therapeutic targeting associated with PD‐1/PD‐L1 pathway in diffuse large B‐cell lymphoma. Int J Mol Sci. 2019;20:1326‐1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Godfrey J, Tumuluru S, Bao R, et al. PD‐L1 gene alterations identify a subset of diffuse large B‐cell lymphoma harboring a T‐cell‐inflamed phenotype. Blood. 2019;133:2279‐2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Haddad G, Lorenzen JM. Biogenesis and function of circular RNAs in health and in disease. Front Pharmacol. 2019;10:428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829‐1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hu Y, Zhao Y, Shi C, et al. A circular RNA from APC inhibits the proliferation of diffuse large B‐cell lymphoma by inactivating Wnt/beta‐catenin signaling via interacting with TET1 and miR‐888. Aging (Albany NY). 2019;11:8068‐8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Getaneh Z, Asrie F, Melku M. MicroRNA profiles in B‐cell non‐Hodgkin lymphoma. EJIFCC. 2019;30:195‐214. [PMC free article] [PubMed] [Google Scholar]
- 14. Song G, Gu L, Li J, et al. Serum microRNA expression profiling predict response to R‐CHOP treatment in diffuse large B cell lymphoma patients. Ann Hematol. 2014;93:1735‐1743. [DOI] [PubMed] [Google Scholar]
- 15. Lin Y, Liu AY, Fan C, et al. MicroRNA‐33b inhibits breast cancer metastasis by targeting HMGA2, SALL4 and Twist1. Sci Rep. 2015;5:9995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tan HL, Chiu SL, Zhu Q, Huganir RL. GRIP1 regulates synaptic plasticity and learning and memory. Proc Natl Acad Sci U S A. 2020;117:25085‐25091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tan HL, Queenan BN, Huganir RL. GRIP1 is required for homeostatic regulation of AMPAR trafficking. Proc Natl Acad Sci U S A. 2015;112:10026‐10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhao W, Li Y, Zhang X. Stemness‐related markers in cancer. Cancer Transl Med. 2017;3:87‐95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martelli M, Ferreri AJ, Agostinelli C, Di Rocco A, Pfreundschuh M, Pileri SA. Diffuse large B‐cell lymphoma. Crit Rev Oncol Hematol. 2013;87:146‐171. [DOI] [PubMed] [Google Scholar]
- 20. Rovira J, Valera A, Colomo L, et al. Prognosis of patients with diffuse large B cell lymphoma not reaching complete response or relapsing after frontline chemotherapy or immunochemotherapy. Ann Hematol. 2015;94:803‐812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coiffier B, Sarkozy C. Diffuse large B‐cell lymphoma: R‐CHOP failure‐what to do? Hematology Am Soc Hematol Educ Program. 2016;2016:366‐378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui C, Yang J, Li X, Liu D, Fu L, Wang X. Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer. 2020;19:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ma S, Kong S, Wang F, Ju S. CircRNAs: biogenesis, functions, and role in drug‐resistant Tumours. Mol Cancer. 2020;19:119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Qiu L, Wang T, Ge Q, et al. Circular RNA Signature in Hepatocellular Carcinoma. J Cancer. 2019;10:3361‐3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li Z, Chen Z, Hu G, Jiang Y. Roles of circular RNA in breast cancer: present and future. Am J Transl Res. 2019;11:3945‐3954. [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang C, Ma L, Niu Y, et al. Circular RNA in lung cancer research: biogenesis, functions, and roles. Int J Biol Sci. 2020;16:803‐814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui M, Wang H, Yao X, et al. Circulating MicroRNAs in cancer: potential and challenge. Front Genet. 2019;10:626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ono K. Functions of microRNA‐33a/b and microRNA therapeutics. J Cardiol. 2016;67:28‐33. [DOI] [PubMed] [Google Scholar]
- 31. Weihua Z, Guorong Z, Xiaolong C, Weizhan L. MiR‐33a functions as a tumor suppressor in triple‐negative breast cancer by targeting EZH2. Cancer Cell Int. 2020;20:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Karatas OF, Wang J, Shao L, et al. miR‐33a is a tumor suppressor microRNA that is decreased in prostate cancer. Oncotarget. 2017;8:60243‐60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pan J, Zhou C, Zhao X, et al. A two‐miRNA signature (miR‐33a‐5p and miR‐128‐3p) in whole blood as potential biomarker for early diagnosis of lung cancer. Sci Rep. 2018;8:16699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yi M, Niu M, Xu L, Luo S, Wu K. Regulation of PD‐L1 expression in the tumor microenvironment. J Hematol Oncol. 2021;14:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhang JY, Yan YY, Li JJ, Adhikari R, Fu LW. PD‐1/PD‐L1 based combinational cancer therapy: icing on the cake. Front Pharmacol. 2020;11:722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Akinleye A, Rasool Z. Immune checkpoint inhibitors of PD‐L1 as cancer therapeutics. J Hematol Oncol. 2019;12:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dong P, Xiong Y, Yue J, Hanley SJB, Watari H. Tumor‐intrinsic PD‐L1 signaling in cancer initiation, development and treatment: beyond immune evasion. Front Oncol. 2018;8:386. [DOI] [PMC free article] [PubMed] [Google Scholar]


