Abstract
Breast cancer is the most prevalent cancer diagnosed in women and the major malignancy that threatens women health, thus we explored the role of long noncoding RNA LINC01605 in triple‐negative breast cancer (TNBC). We collected tissue samples from TNBC patients and cultured breast cancer cells to detect LINC01605 levels by RT‐PCR. We then constructed LINC01605 knockdown and LINC01605 overexpressed TNBC cell lines, cell proliferation was measured by CCK‐8 and colony formation assays, cell migration and invasion were measured by Transwell assay, and aerobic glycolysis of cells was detected. Furthermore, a downstream target gene was found, and its role was confirmed by mouse allogeneic tumor formation. It discovered that LINC01605 expression was significantly increased in TNBC patients, and its high expression predicted a low survival prognosis for TNBC patients. Stable knockdown of LINC01605 remarkably inhibited cell proliferation, migration, and invasion, as well as aerobic glycolysis by inhibiting lactate dehydrogenase A in TNBC cell lines. Notably, knockdown of LINC01605 suppressed in vivo tumor formation and migration in TNBC transplanted mice. In conclusion, targeting long noncoding RNA LINC01605 might serve as a therapeutic candidate strategy to treat patients with TNBC.
Keywords: aerobic glycolysis, breast cancer, LDHA, LINC01605, TNBC
Knockdown of LINC01605 suppressed in vivo tumor formation and migration in triple‐negative breast cancer (TNBC) transplanted mice. Targeting long noncoding RNA LINC01605 might serve as a therapeutic candidate strategy to treat patients with TNBC.

Abbreviations
- ceRNA
competing endogenous RNA
- ECAR
extracellular acidification rate
- HER2
human epidermal growth factor receptor 2
- lncRNA
long noncoding RNA
- LDHA
lactate dehydrogenase A
- miR/miRNA
microRNA
- TNBC
triple‐negative breast cancer
1. INTRODUCTION
Breast cancer occurs in the breast tissue due to the uncontrolled growth of the breast cells. Breast cancer is the most prevalent cancer diagnosed in women and also the major malignancy that threatens women’s health, accounting for 25% of all cancer incidences. 1 Although breast cancer can occur in both men and women, it is far more common and harmful to women. Globally, millions of women develop breast cancer and more than 400,000 women die of breast cancer every year. 2 Due to the substantial support for breast cancer awareness and research on the mechanism of breast cancer, helping create advances in the diagnosis and treatment of breast cancer, breast cancer survival rates have increased, and the number of deaths associated with this disease is steadily declining.
Epidemiological data have indicated that the cumulative exposure of estrogen for a long time, such as early premature menstruation, delayed menopause, and hormone replacement therapy, caused genetic and epigenetic changes that are important factors in inducing breast cancer. Approximately 70% of patients with breast cancer are clinically positive for estrogen receptor. 3 , 4 , 5 But approximately 10%–20% of breast cancer patients are diagnosed with TNBC, which is characterized by no expression of the three markers in breast cancer, namely estrogen receptor, progesterone receptor, and HER2. 6 , 7 , 8 , 9 Unlike hormone receptor‐positive or HER2‐positive breast cancer, TNBC is often accompanied by lymph node metastasis and higher breast cancer grades. 10 Additionally, until now, effective targeted drugs for TNBC have been relatively scarce, because hormone therapies target one of these three receptors. Therefore, compared to other breast cancer subtypes, TNBCs often require combination therapies with more toxic chemotherapy. However, drug resistance is prone to appear after surgical intervention and approximately 40%–80% of patients will relapse within 3 years 11 , 12 , 13 , 14 ; therefore, it has important theoretical and clinical significance to explore the detailed mechanism of TNBC. In recent years, increasing evidence showed that noncoding RNAs play an important role in the development of tumor, 15 and also participate in the regulation of tumor metabolism. 16 , 17 Therefore, in our study, we explored the role and mechanism of noncoding RNA LINC01605 in TNBC.
2. MATERIALS AND METHODS
2.1. Human specimens
Breast cancer tissues were collected from the patients with their written consent. The study was approved by the Department of General Surgery, Hainan General Hospital.
2.2. Establishment of stable cell clones
The knockdown of LINC01605 was achieved using shRNA, which was wrapped with lentivirus purchased from GenScript. The cells were infected with the lentivirus according to the manufacturer’s instructions. The cells were selected by puromycin (1 μg/ml) for 2 weeks.
2.3. Reverse transcription–PCR
Total RNA was extracted from patients’ tissue or breast cancer cells using TRIzol reagent (Invitrogen), then reverse transcripted into cDNA by PrimeScript RT reagent Kit (Takara). Polymerase chain reaction was carried out using a SYBR Green Real‐time OCR Master Mix in an ABI PCR system (Applied Biosystems). The quantitative value was expressed using the 2−ΔΔCt method and normalized to GAPDH.
The primer sequences were: LINC01605‐F, GACAGGAGACAGAGGCAGTTG; LINC01605‐R, TCCAGACAGGTGTCAGACAAG; LDHA‐F, GTGAAGGTGACTCTGACTTC; LDHA‐R, GTGAAATGATATGACATCAG; GAPDH‐F, TGGCACCGTCAAGGCTGAG; and GAPDH‐R, CAGCCTTCTCCATGGTGGTG.
2.4. Cell proliferation assay
Breast cancer cells in the logarithmic phase of growth were digested and seeded into a 96‐well plate at the density of 1000 cells/well. After 96 h, cell proliferation was determined using CCK‐8 (Dojindo Laboratories) in accordance with the manufacturer’s instructions.
2.5. Colony formation assay
Cells were seeded into 6‐well plates at a density of 1000 cells/well, and 2 weeks later, visible colony was formed. Medium was removed following three‐time wash with PBS, then cells were fixed in 4% paraformaldehyde for 1 h and stained with 0.1 mg/ml crystal violet solution. After washing, stained colonies were photographed using microscope (Leica). More than 50 cells were determined as a colony, and total number of colonies was counted.
2.6. Transwell migration assay
Cells were digested and seeded at a density of 50,000 cells/chamber into Transwell chamber which is a 6.5‐µm chamber with an 8‐µm pore, and placed in the upper of 24‐well plates. The lower chamber was filled with medium and 10% FBS (HyClone Laboratories). After incubation for 1 day, the migratory cells under the chamber surface were fixed in 4% paraformaldehyde, stained with 0.1 mg/ml crystal violet solution, counted under microscope with five random fields for each chamber.
2.7. Transwell invasion assay
Cells were plated at a density of 1 × 106 cells/chamber into 10% Matrigel (BD Biosciences) coated chamber, which were plated in the upper of 24‐well plates. The lower chamber was also filled with medium containing FBS. After incubation for 24 h, invasive cells on the lower membrane surface were fixed, stained, and counted using Transwell migration assay.
2.8. Analysis of glucose uptake and ECAR
Cells were treated with 10 mM fluorescent 2‐DG analog 2‐NBDG for 1 h, and glucose uptake was determined by flow cytometry (BD Biosciences) after washing twice with PBS. For the analysis of lactate production, cells were digested and seeded into XF 96‐well plate in pyruvate‐free DMEM overnight. One hour before measurement, medium was changed to XF medium, and 10 mM glucose, 1 mM ATP synthase inhibitor oligomycin, and 50 mM glycolysis inhibitor 2‐DG were added at indicated times, according to the manufacturer’s instructions. Lactate production in the medium was measured using Seahorse XF glycolytic rate assay kit by a Seahorse Bioscience XF96 analyzer. The ECAR was analyzed.
2.9. Western blot analysis
Protein was extracted from breast cancer cells and western blot analysis was carried out as previously described. The LDAH and β‐actin Abs were purchased from Abcam.
2.10. Measurement of mRNA decay
When seeded cells were adhesive to 6‐well plates, actinomycin D (10 µg/ml) was added to the medium, then RNA was extracted at 0, 2, 4, 6, and 8 h after adding actinomycin D.
2.11. In vivo mouse xenograft experiment
Animal experiments were carried out under the guidelines of the Department of General Surgery at Hainan General Hospital, and all animal procedures were approved by the same department. Female nude mice were purchased from GemPharmatech.
MDA‐MB‐231 cells with or without LINC01605 knockdown were suspended in DMEM. Single cell suspension (5 × 105) was mixed with 0.15 ml Matrigel (50% v/v, Corning Matrigel), and subcutaneously injected into both sides of the breast of 4‐week‐old nude mice. Tumor volume was measured every 4 days using the formula: π/6 × length × width × thickness (mm3). Tumors were collected on day 30, weighed, and cut into halves. One half was used to extract RNA, and the other half was fixed in formalin following embedding in paraffin.
2.12. Immunohistochemistry
Five micrometer thick sections from paraffin‐embedded tissue underwent antigen retrieval by Diva Declonaker RTU (Biocare Medical) for 10 min at room temperature, then incubated with primary anti‐LDHA or Ki‐67 (Abcam) for 1 h, followed by the incubation of secondary Ab (Invitrogen). Sections were washed and slides were photographed using a microscope (Leica) as previously described. 18
2.13. Hematoxylin–eosin staining
One million MDA‐MB‐231 cells with or without LINC01605 knockdown in 100 µl PBS were injected into 4‐week‐old nude mice through the tail vein. After 1 month, mice were killed and lungs were collected, fixed in 4% formalin, embedded in paraffin, and cut into 5‐µm thick sections. Lung sections were then stained with H&E as previously described. 19
2.14. Statistical analysis
Data were analyzed using Student’s t‐test or the one‐ or two‐way ANOVA method followed by a post‐hoc test by Prism 7.0, and are represented as mean ± SD. Kaplan–Meier analysis was applied using SPSS.
3. RESULT
3.1. LINC01605 level was associated with TNBC
To assess the role of LINC01605 in TNBC, we first measured LINC01605 expression in patients with TNBC. LINC01605 level was increased in TNBC cancer tissue in comparison with normal tissue in patients (Figure 1A). Next, we revealed that LINC01605 expression was much higher in some TNBC‐derived epithelial cells, namely MDA‐MB‐231, SUM159, BT549, and MDA‐MB‐468, than in normal epithelial cells from breast, including MCF‐7, T47D, BT474, and SKBR3 (Figure 1B). For the following studies, we chose three breast cancer cells that had higher expression of LINC01605, which were MDA‐MB‐231, SUM159, and BT549. In addition, the Kaplan–Meier analysis showed that LINC01605 expression is correlated with both overall survival and relapse‐free survival of breast cancer patients (Figure 1C,D). Overall, LINC01605 expression was associated with TNBC patients.
FIGURE 1.
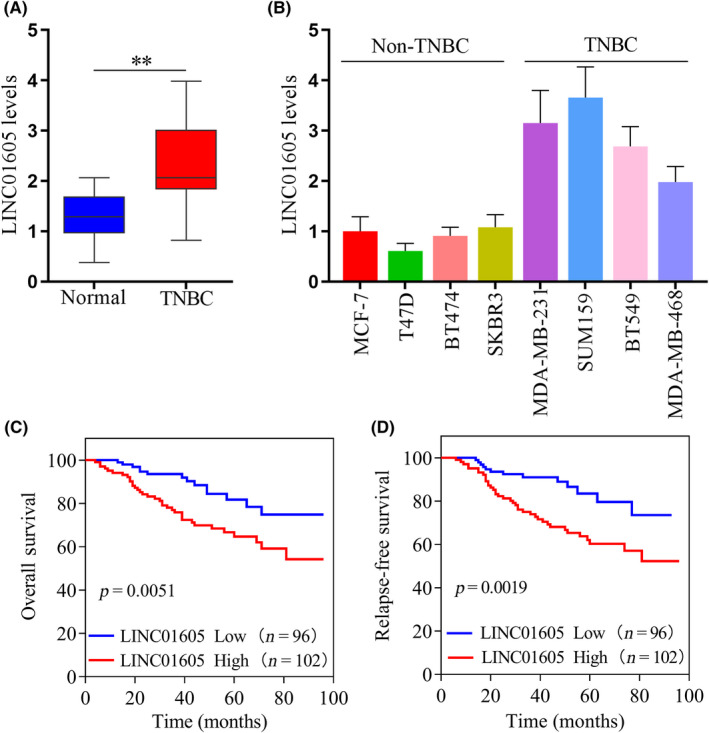
Increased LINC01605 expression predicts poor prognosis in triple‐negative breast cancer (TNBC) patients. (A) LINC01605 levels were measured by quantitative RT‐PCR (qRT‐PCR) and normalized to the level of GAPDH in TNBC cancer tissues and normal tissues. (B) LINC01605 levels were measured by qRT‐PCR in breast cancer cells. GAPDH was used as a control. (C) Kaplan–Meier analysis of the correlation between LINC01605 expression and overall survival of breast cancer patients. (D) Kaplan–Meier analysis of the correlation between LINC01605 expression and relapse‐free survival of breast cancer patients. **p < 0.01
3.2. LINC01605 promoted proliferation, migration, and invasion of TNBC cells in vitro
To further explore the role of LINC01605 in TNBC, we established cell lines that stably knocked down LINC01605 in three TNBC cell lines (Figure 2A). The proliferation of breast cancer cells detected by CCK‐8 assay (Figure 2B) and cell colony formation (Figure 2C) was significantly reduced after LINC01605 knockdown in MDA‐MB‐231, SUM159, and BT549 cells. Moreover, LINC01605 KD cells displayed remarkably decreased abilities of migration (Figure 2D) and invasion (Figure 2E) compared to normal TNBC cells. Therefore, LINC01605 knockdown significantly reduced proliferation, migration, and invasion of breast cancer cells, which suggested that LINC01605 promoted the abilities of migration, proliferation, and invasion in breast cancer cells.
FIGURE 2.
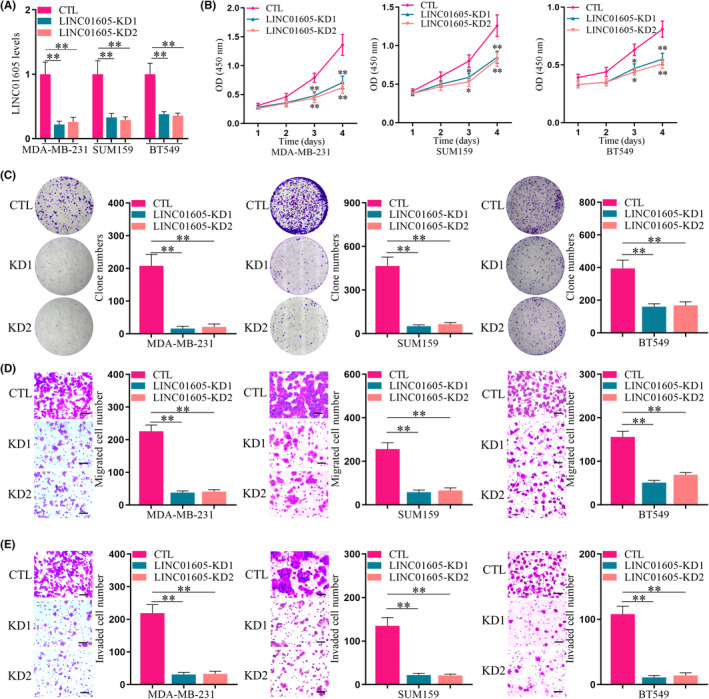
LINC01605 knockdown (KD) inhibits the proliferation, migration, and invasion of triple‐negative breast cancer cells in vitro. (A) Expression of LINC01605 in breast cancer cell lines was detected by quantitative PCR. GAPDH was used as control (CTL). (B) Proliferation of breast cancer cells was determined by the CCK‐8 assay after plating for 96 h with or without LINC01605‐KD. (C) Breast cancer cells were subjected to cell colony formation assay with or without LINC01605‐KD for 2 weeks. (D) Breast cancer cells were subjected to Transwell migration assay. (E) Breast cancer cells were subjected to Transwell invasion assay. Scale bar, 100 μm. **p < 0.01
Furthermore, we established cell lines that stably overexpressed LINC01605 in three TNBC cell lines (Figure 3A). The proliferation of breast cancer cells detected by CCK‐8 assay (Figure 3B) and cell colony formation (Figure 3C) were significantly increased after LINC01605 overexpression in MDA‐MB‐231, SUM159, and BT549 cell lines. Moreover, cells with LINC01605 overexpression displayed remarkably increased abilities of migration (Figure 3D) and invasion (Figure 3E) compared to normal TNBC cells. Therefore, LINC01605 knockdown significantly reduced proliferation, migration, and invasion of breast cancer cells and LINC01605 overexpression significantly increased proliferation, migration, and invasion of breast cancer cells, which suggested that LINC01605 promoted the abilities of migration, proliferation, and invasion in breast cancer cells.
FIGURE 3.
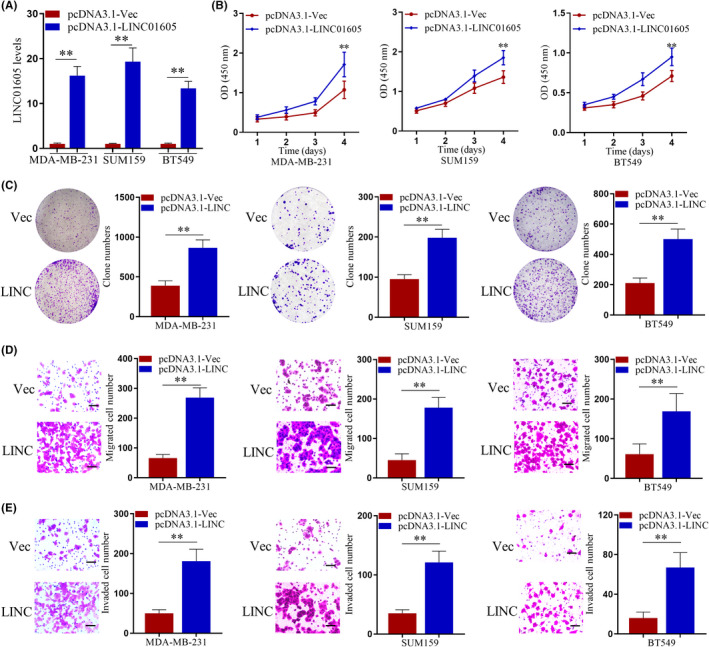
LINC01605 overexpression promotes the proliferation, migration, and invasion of triple negative breast cancer cells in vitro. (A) Expression of LINC01605 in breast cancer cell line were detected by quantitative PCR. GAPDH was used as control. (B) Proliferation of breast cancer cells was determined by CCK‐8 assay after plating for 96 h with or without LINC01605 overexpression. OD, optical density. (C) Breast cancer cells were subjected to cell colony formation assay with or without LINC01605 (LINC) overexpression for 2 weeks. (D) Breast cancer cells were subjected to Transwell migration assay. (E) Breast cancer cells were subjected to Transwell invasion assay. Scale bar, 100 μm. **p < 0.01. Vec, vector
3.3. LINC01605 enhanced aerobic glycolysis in breast cancer cells
In recent years, studies have shown that LINC01605 can function as ceRNA, and this study found that LINC01605 can regulate cell metabolism. Next, we evaluated the role of LINC01605 in the aerobic glycolysis and metabolism of breast cancer cells. Glucose uptake was significantly decreased in LINC01605‐KD breast cancer cells compared to breast cancer cells (Figure 4A). Moreover, lactate production in breast cancer cells were also remarkably reduced after knockdown of LINC01605 (Figure 4B). Glucose uptake and lactate production were increased in breast cancer cells with LINC01605 overexpression compared to breast cancer cells (Figure 4C,D). Not surprisingly, ECAR, which indicates the glycolytic rate of cells, was also significantly downregulated in LINC01605‐KD breast cancer cells and upregulated in LINC01605 overexpressed breast cancer cells in comparison with breast cancer cells (Figure 4E,F). Taken together, these data suggested that LINC01605 enhanced aerobic glycolysis in breast cancer cells.
FIGURE 4.
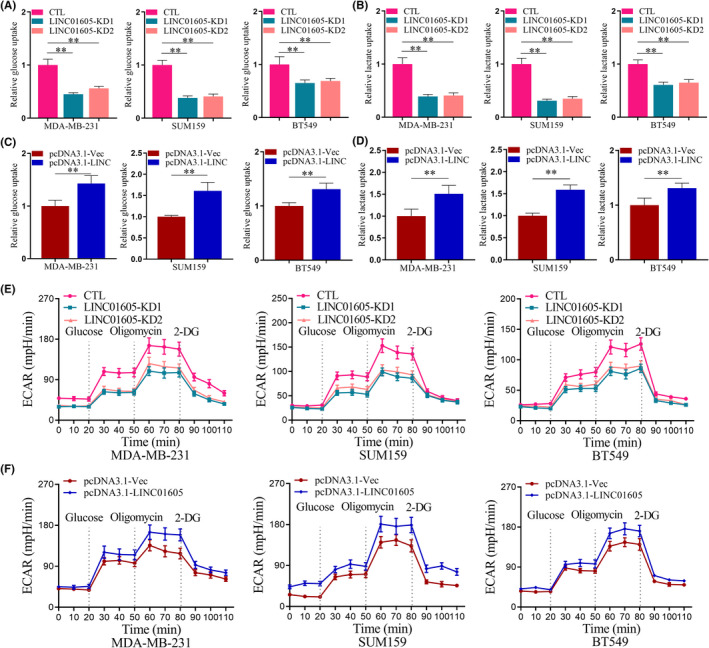
LINC01605 promotes aerobic glycolysis in breast cancer cells. (A) Relative glucose uptake measured in breast cancer cells with or without LINC01605 knockdown (KD). (B) Relative glucose uptake measured in breast cancer cells with or without LINC01605 overexpression. (C) Lactate production in breast cancer cells with or without LINC01605‐KD. (D) Lactate production in breast cancer cells with or without LINC01605 over‐expression. (E) Extracellular acidification rate (ECAR) of breast cancer cells with or without LINC01605‐KD were measured by the Seahorse Bioscience XF96 analyzer. (F) ECAR of breast cancer cells with or without LINC01605 overexpression were measured by the Seahorse Bioscience XF96 analyzer. Glucose (10 mM), ATP synthase inhibitor oligomycin (1 μM), and glycolysis inhibitor 2‐DG (50 mM) were added to the cells at the indicated time points. **p < 0.001. CTL, control
3.4. Lactate dehydrogenase A was the target of LINC01605
Lactate dehydrogenase A is a key metabolic enzyme that catalyzes the interconversion of pyruvate and L‐lactate with concomitant interconversion of NADH and NAD, which plays an important role in anaerobic metabolism. Bioinformatics analysis showed that LINC01605 and metabolism‐related gene LDHA had the same miR‐34a‐5p target site (Figure 5A). Dual luciferase reporter assay experiment had shown that miRNA can inhibit LDHA and LINC01605 (Figure 5B,C). Therefore, we guessed that LINC01605 regulated aerobic glycolysis by LDHA. First, we discovered that the mRNA level of LDHA was reduced after LINC01605 knockdown and increased after LINC01605 overexpression in breast cancer cells (Figure 5D,E). Moreover, the half‐life of LDHA was shortened by LINC01605 knockdown and extended by LINC01605 overexpression in three breast cancer cell lines (Figure 5F,G). Protein level of LDHA was also reduced after LINC01605 knockdown and increased after LINC01605 overexpression in breast cancer cells (Figure 5H,I). This suggested that LINC01605 increased aerobic glycolysis by inhibiting the degradation of LDHA and increasing the expression of LDHA in breast cancer cells.
FIGURE 5.
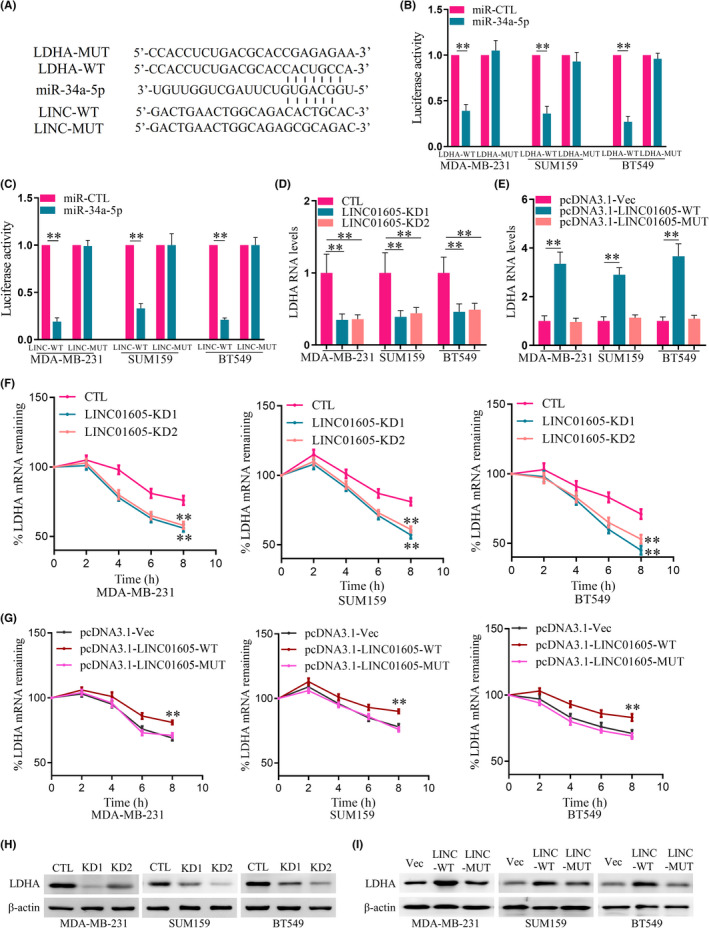
LINC01605 knockdown (KD) decreases lactate dehydrogenase A (LDHA) level. (A) Putative microRNA (miR)‐34a‐5p target sites in LDHA and LINC01605 are shown in the schematic. (B, C) A dual‐luciferase reporter assay was carried out to investigate the interactions between miR‐34a‐5p LDHA and LINC01605. Renilla luciferase activity was normalized to firefly activity and shown as a relative activity. (D) Quantitative RT‐PCR (qRT‐PCR) analyses of LDHA RNA expression in LINC01605‐KD and control (CTL) cells. GAPDH was used as an endogenous control. (E) qRT‐PCR analyses of LDHA RNA expression in LINC01605 overexpression and control cells. GAPDH was used as an endogenous control. (F) Breast cancer cells were treated with actinomycin D (10 μg/ml) for the indicated times. LDHA mRNA levels were measured by qRT‐PCR in LINC01605‐KD and control cells. GAPDH was used as an endogenous control. (G) Breast cancer cells were treated with actinomycin D (10 μg/ml) for the indicated times. LDHA mRNA levels were measured by qRT‐PCR in LINC01605 overexpression and control cells. GAPDH was used as an endogenous control. (H) Western blot analyses of LDHA protein expression in LINC01605‐KD and control cells. (I) Western blot analyses of LDHA protein expression in LINC01605 overexpression and control cells. **p < 0.01. MUT, mutant; Vec, vector
3.5. LINC01605 promoted proliferation and metastasis of TNBC in vivo
To further confirm the role of LINC01605 in TNBC, we implanted normal breast cancer cells or LINC01605‐KD cells into female nude mice. Implanted cells formed tumor in mice, however, the tumor volume (Figure 6A) and tumor weight (Figure 6B) were both decreased in the mice implanted with LINC01605‐KD cells with a low expression of LINC01605 (Figure 6C). LINC01605‐KD cell‐formed tumors also had decreased expression of LDHA and Ki‐67 (Figure 6D). MDA‐MB‐231 cells were injected into mice through the tail vein, the mice were killed 1 month later, and lung tissue was collected and examined. Therefore, we also determined lung metastases in these mice and H&E staining showed that LINC01605‐KD cell‐implanted mice displayed reduced lung metastases (Figure 6E). We then implanted normal breast cells or LINC01605‐OE cells into female nude mice. Implanted cells formed tumors in mice. Tumor volume (Figure 6F) and tumor weight (Figure 6G) were both increased in the mice implanted with LINC01605‐OE cells with a high expression of LINC01605 (Figure 6H). LINC01605‐OE cell‐formed tumors also had increased expression of LDHA and Ki‐67 (Figure 6I). MDA‐MB‐231 cells were injected into mice through the tail vein, and the mice were killed 3 weeks later; lung tissue was collected and examined. Lung metastases were found in five of the six mice in the control group, while lung metastases were detected in all six mice in the LINC01605‐OE group. Compared with the control group, each mouse in the overexpression group formed more and larger metastases (Figure 6J). Altogether, LINC01605 promoted TNBC in mice.
FIGURE 6.
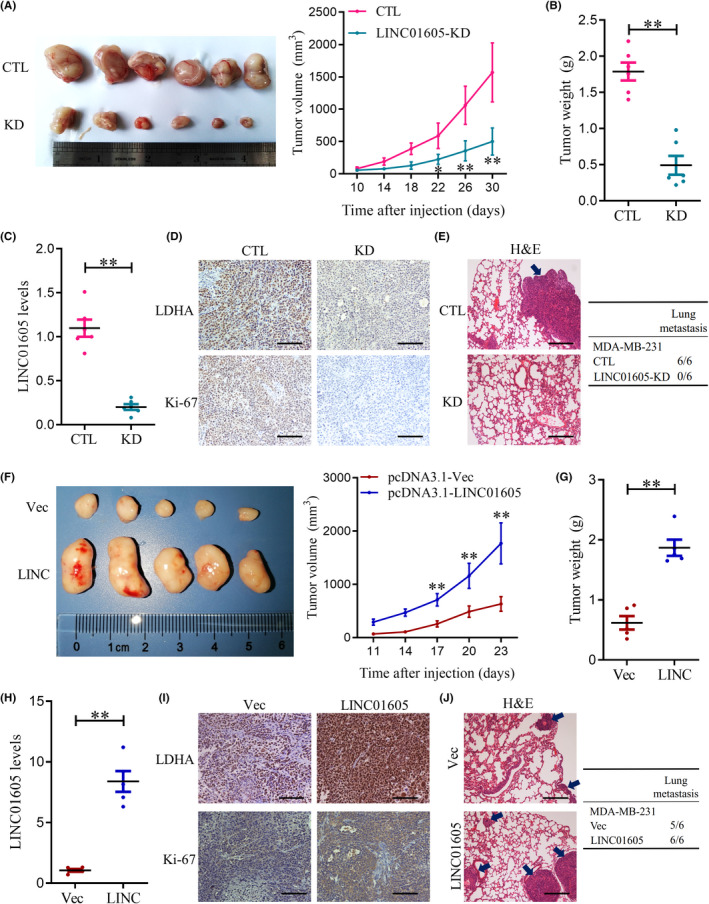
LINC01605 promotes triple‐negative breast cancer proliferation and metastasis in vivo. (A) Nude mice were implanted with 5 × 105 control (CTL) or LINC01605 ‐KD MDA‐MB‐231 cells. Tumor volumes were measured every 4 days. (B) At day 30, before tumors were excised, tumor weight was measured. (C) LINC01605 levels were measured by quantitative RT‐PCR in breast cancer cells. GAPDH was used as control. (D) Quantifications of lactate dehydrogenase A (LDHA) and Ki‐67 count of tumor by immunohistochemistry. (E) Examples (H&E) of lung metastases from mice. (F) Tumor volume, (G) tumor weight in the experimental groups. (H) Expression of LINC01605, (I) the expressions of LDHA and Ki‐67, and (J) the metastase in the LINC01605‐OE cells‐formed tumors. Scale bar, 100 μm. **p < 0.01. Vec, vector
3.6. Knockdown of LDHA rescues the phenotype induced by LINC01605 knockdown
To study the function of LDHA in mediating the oncogenic role of LINC01605 in breast cancer cells, here we further knocked down LDHA in LINC01605‐OE breast cancer cell lines. As shown in Figure 7A,B, LINC01605 overexpression stimulated cell viability, whereas knockdown of LDHA in LINC01605‐OE cells rescued the promoted effects in breast cancer cells. Likewise, migration and invasion assays showed that cell migration and invasion in LDHA knockdown cells were significantly restored in comparison with LINC01605 overexpression (Figure 7C,D). As shown in Figure 7E–G, LINC01605 overexpression increased cell glucose uptake, lactate production, and ECAR, whereas knockdown of LDHA in LINC01605‐OE cells rescued the promoted effects in breast cancer cells. Our results highlighted the predominant role of LDHA downstream of LINC01605 in breast cancer.
FIGURE 7.
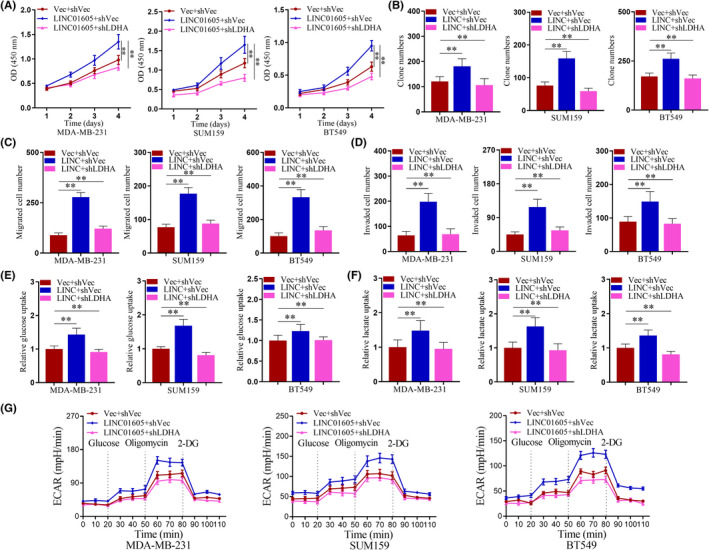
Knockdown of lactate dehydrogenase A (LDHA) rescues the phenotype induced by LINC01605 (LINC) overexpression. (A) Cell viability of breast cancer cells was evaluated by CCK‐8 assay. (B) Breast cancer cells were subjected to cell colony formation assay for 2 weeks. (C) Breast cancer cells were subjected to Transwell migration assay. (D) Breast cancer cells were subjected to Transwell invasion assay. (E) Relative glucose uptake measured in breast cancer cells. (F) Lactate production in breast cancer cells. (G) Extracellular acidification rate (ECAR) of breast cancer cells was measured by the Seahorse Bioscience XF96 analyzer. **p < 0.001. LDHA, lactate dehydrogenase A; Vec, vector
4. DISCUSSION
Our study showed for the first time that lncRNA LINC01605 was correlated with TNBC as well as the survival rate and relapse‐free survival of breast cancer patients. Knockdown of LINC01605 inhibited migration, proliferation, and invasion of breast cancer cells to delay the development of TNBC. Furthermore, we proved that LINC01605 regulated breast cancer by mediating aerobic glycolysis through targeting LDHA.
Long noncoding RNAs are a type of noncoding RNAs with transcriptional length of over 200 nucleotides. Accumulating evidence has indicated that lncRNAs are involved in multiple physiological and pathophysiological processes, 20 especially in tumor progression. 21 , 22 , 23 Many lncRNAs were proved to be biomarkers of cancers, and lncRNA‐based risk scoring systems were used to predict prognosis of alcohol‐related hepatocellular carcinoma. 24 Among the lncRNAs involved in the risk scoring systems, LINC01605 is poorly studied, but all the publications about LINC01605 have indicated that it exerted a certain impact on tumor progression. For example, LINC01605 accelerated laryngeal squamous cell carcinoma by targeting miR493‐3p. 25 Moreover, LINC01605 aggravated bladder cancer by enhancing MMP9. 26 Therefore, due to its high correlation with tumors, we explored the role of LINC01605 in breast cancer, which had not been studied. After skin cancer, breast cancer is the most common cancer diagnosed in women in the United States, which greatly threatens women’s health. Therefore, we explored the role of LINC01605 in breast cancer, especially TNBC, and our study for the first time discovered the relationship between the high expression of LINC01605 and TNBC.
Aerobic glycolysis is a 10‐step reaction wherein oxygen is required to reoxidize NADH and glucose into NAD+ and pyruvate. 27 The energy metabolism of cancer cells is characterized as increased aerobic glycolysis, referred TO as the Warburg effect. Otto Warburg first discovered this phenomenon in 1920. 28 Even in an environment with sufficient oxygen, cancer cells still quickly convert oxygen into energy through high glycolytic metabolism. Because high aerobic glycolysis is a hallmark of cancer cell metabolism, targeting it could be a therapeutic strategy for cancer therapy. Therefore, in recent decades, emerging evidence confirmed the role of aerobic glycolysis in breast cancer. 29 Molecular targeted therapies show many advantages for the treatment of various cancers in comparison with traditional chemotherapies. 30 In particular, targeting key enzymes of glycolysis, including hexokinase, pyruvate kinase, and phosphofructokinase, and glucose transporters were all reported as antibreast cancer therapeutic strategies. Furthermore, a wide range of factors that might serve as therapeutic targets suppressed breast cancer by regulating aerobic glycolysis, including betulinic acid, 31 miRNA‐186‐3p, 32 tumor‐derived macrophages, 33 and zoptarelin doxorubicin. 34 Our current study showed that LINC01605 promoted breast cancer by targeting LDHA, which is also a crucial enzyme in aerobic glycolysis. Although this is the first report of the role of LINC01605 in breast cancer by regulating aerobic glycolysis, noncoding RNAs have been extensively explored and proven as key mediators of aerobic glycolysis in breast cancer for a long time. 35
Here, we showed that LINC01605 targeted mRNA of LDHA to regulate aerobic glycolysis, leading to the accelerated breast cancer. Our data discovered that knockdown of LINC01605 could shorten the half‐life of LDHA mRNA, but how LINC01605 affected the mRNA of LDHA still remains unclear. Long noncoding RNAs always act as ceRNA to regulate other RNA transcription by competing for shared miRNAs. A previous publication revealed that LINC01605 promoted laryngeal squamous cell carcinoma by targeting miR‐493‐3p, and LINC01605 acted as ceRNA with miR‐293‐3p. 25 Additionally, LDHA and programmed death ligand 1 were completely binding to miRNA‐34a to act as ceRNA in TNBC. 36 These publications gave us a hint that LINC01605 might regulate LDHA in a ceRNA way, but the microRNA that was involved in this process was unknown. Therefore, we might further explore the detailed mechanism behind how LINC01605 regulates LDHA, and whether some specific miRNAs are involved.
In summary, our study discovered that LINC01605 was highly expressed in patients with TNBC, and its high expression was also correlated with overall survival rate and relapse‐free survival of breast cancer patients, which might suggest that, like other lncRNAs, LINC01605 might also serve as a diagnostic biomarker of TNBC in the clinic. Notably, due to the complexity of TNBC, in that the high expression of the three receptors that were always used as biomarkers of breast cancer cannot be utilized for TNBC, a diagnostic biomarker for TNBC is required. Our study could provide a novel insight into a diagnostic biomarker for TNBC. Furthermore, our data also suggested that LINC01605 might serve as a therapeutic target for patients with TNBC.
Our study indicated that lncNA LINC01605 was correlated with the survival rate and relapse‐free survival of TNBC patients. Knockdown of LINC01605 inhibited proliferation, migration, and invasion of breast cancer cells to delay the development of TNBC. Furthermore, we proved that LINC01605 regulated breast cancer by mediating aerobic glycolysis through targeting LDHA. Therefore, LINC01605 might serve as a potential diagnostic biomarker and therapeutic target for patients with TNBC.
DISCLOSURE
The authors have no conflict of interest.
ACKNOWLEDGMENT
This work was supported by Hainan Provincial Natural Science Foundation of China (820MS139).
Wang W, He X, Wang Y, et al. LINC01605 promotes aerobic glycolysis through lactate dehydrogenase A in triple‐negative breast cancer. Cancer Sci. 2022;113:2484–2495. doi: 10.1111/cas.15370
Wei Wang and Xionghui He contributed equally to this work.
Contributor Information
Shaowei Mo, Email: moshaowei3458@163.com.
Dong Chen, Email: daxrmyycd@163.com.
REFERENCES
- 1. Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277‐300. [DOI] [PubMed] [Google Scholar]
- 2. Coughlin SS, Ekwueme DU. Breast cancer as a global health concern. Cancer Epidemiol. 2009;33:315‐318. [DOI] [PubMed] [Google Scholar]
- 3. Simpson ER, Misso M, Hewitt KN, et al. Estrogen–the good, the bad, and the unexpected. Endocr Rev. 2005;26:322‐330. [DOI] [PubMed] [Google Scholar]
- 4. Tyson JJ, Baumann WT, Chen C, et al. Dynamic modelling of oestrogen signalling and cell fate in breast cancer cells. Nat Rev Cancer. 2011;11:523‐532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jeruss JS, Mittendorf EA, Tucker SL, et al. Staging of breast cancer in the neoadjuvant setting. Cancer Res. 2008;68:6477‐6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yam C, Mani SA, Moulder SL. Targeting the molecular subtypes of triple negative breast cancer: understanding the diversity to progress the field. Oncologist. 2017;22:1086‐1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Morris GJ, Naidu S, Topham AK, et al. Differences in breast carcinoma characteristics in newly diagnosed African‐American and Caucasian patients: a single‐institution compilation compared with the National Cancer Institute's Surveillance, Epidemiology, and End Results database. Cancer. 2007;110:876‐884. [DOI] [PubMed] [Google Scholar]
- 8. Onitilo AA, Engel JM, Greenlee RT, Mukesh BN. Breast cancer subtypes based on ER/PR and Her2 expression: comparison of clinicopathologic features and survival. Clin Med Res. 2009;7:4‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)‐negative, progesterone receptor (PR)‐negative, and HER2‐negative invasive breast cancer, the so‐called triple‐negative phenotype: a population‐based study from the California cancer Registry. Cancer. 2007;109:1721‐1728. [DOI] [PubMed] [Google Scholar]
- 10. Ray CE Jr, Gupta AK, Shenoy SS. Left gastric artery arising from the superior mesenteric artery–case reports. Angiology. 1998;49:1017‐1021. [DOI] [PubMed] [Google Scholar]
- 11. Dent R, Trudeau M, Pritchard KI, et al. Triple‐negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13:4429‐4434. [DOI] [PubMed] [Google Scholar]
- 12. Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13:2329‐2334. [DOI] [PubMed] [Google Scholar]
- 13. Haffty BG, Yang Q, Reiss M, et al. Locoregional relapse and distant metastasis in conservatively managed triple negative early‐stage breast cancer. J Clin Oncol. 2006;24:5652‐5657. [DOI] [PubMed] [Google Scholar]
- 14. Symmans WF, Wei C, Gould R, et al. Long‐term prognostic risk after neoadjuvant chemotherapy associated with residual cancer burden and breast cancer subtype. J Clin Oncol. 2017;35:1049‐1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balas MM, Johnson AM. Exploring the mechanisms behind long noncoding RNAs and cancer. Noncoding RNA Res. 2018;3:108‐117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu J, Liu Z‐X, Wu Q‐N, et al. Long noncoding RNA AGPG regulates PFKFB3‐mediated tumor glycolytic reprogramming. Nat Commun. 2020;11:1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Malakar P, Stein I, Saragovi A, et al. Long noncoding RNA MALAT1 regulates cancer glucose metabolism by enhancing mTOR‐mediated translation of TCF7L2. Cancer Res. 2019;79:2480‐2493. [DOI] [PubMed] [Google Scholar]
- 18. Xue Y, Meehan B, Fu Z, et al. SMARCA4 loss is synthetic lethal with CDK4/6 inhibition in non‐small cell lung cancer. Nat Commun. 2019;10:557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pein M, Insua‐Rodríguez J, Hongu T, et al. Metastasis‐initiating cells induce and exploit a fibroblast niche to fuel malignant colonization of the lungs. Nat Commun. 2020;11:1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mercer TR, Dinger ME, Mattick JS. Long non‐coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155‐159. [DOI] [PubMed] [Google Scholar]
- 21. Sanchez Calle A, Kawamura Y, Yamamoto Y, Takeshita F, Ochiya T. Emerging roles of long non‐coding RNA in cancer. Cancer Sci. 2018;109:2093‐2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chi Y, Wang D, Wang J, Yu W, Yang J. Long non‐coding RNA in the pathogenesis of cancers. Cells. 2019;8(9):1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gutschner T, Diederichs S. The hallmarks of cancer: a long non‐coding RNA point of view. RNA Biol. 2012;9:703‐719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Luo Y, Ye J, Wei J, Zhang J, Li Y. Long non‐coding RNA‐based risk scoring system predicts prognosis of alcohol‐related hepatocellular carcinoma. Mol Med Rep. 2020;22:997‐1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang XY, Wang L, Xu PC, et al. LINC01605 promotes the proliferation of laryngeal squamous cell carcinoma through targeting miR‐493‐3p. Eur Rev Med Pharmacol Sci. 2019;23:10379‐10386. [DOI] [PubMed] [Google Scholar]
- 26. Qin Z, Wang YI, Tang J, et al. High LINC01605 expression predicts poor prognosis and promotes tumor progression via up‐regulation of MMP9 in bladder cancer. Biosci Rep. 2018;38:BSR20180562 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27. Dashty M. A quick look at biochemistry: carbohydrate metabolism. Clin Biochem. 2013;46:1339‐1352. [DOI] [PubMed] [Google Scholar]
- 28. Ganapathy‐Kanniappan S. Taming tumor glycolysis and potential implications for immunotherapy. Front Oncol. 2017;7:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Z, Wu J, Zhao Q, Fu S, Jin J. Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol. 2020;22:631‐646. [DOI] [PubMed] [Google Scholar]
- 30. Wang Y, Chen Z. Mutation detection and molecular targeted tumor therapies. STEMedicine. 2020;1:e11. [Google Scholar]
- 31. Zheng Y, Liu P, Wang N, et al. Betulinic acid suppresses breast cancer metastasis by targeting GRP78‐mediated glycolysis and ER stress apoptotic pathway. Oxid Med Cell Longev. 2019;2019:8781690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. He M, Jin Q, Chen C, et al. The miR‐186‐3p/EREG axis orchestrates tamoxifen resistance and aerobic glycolysis in breast cancer cells. Oncogene. 2019;38:5551‐5565. [DOI] [PubMed] [Google Scholar]
- 33. Chen F, Chen J, Yang L, et al. Extracellular vesicle‐packaged HIF‐1α‐stabilizing lncRNA from tumour‐associated macrophages regulates aerobic glycolysis of breast cancer cells. Nat Cell Biol. 2019;21:498‐510. [DOI] [PubMed] [Google Scholar]
- 34. Gründker C, Wokoun U, Hellriegel M, Emons G. Inhibition of aerobic glycolysis enhances the anti‐tumor efficacy of Zoptarelin Doxorubicin in triple‐negative breast cancer cells. J Obstet Gynaecol Res. 2019;45:1334‐1342. [DOI] [PubMed] [Google Scholar]
- 35. Xia M, Feng S, Chen Z, Wen G, Zu X, Zhong J. Non‐coding RNAs: Key regulators of aerobic glycolysis in breast cancer. Life Sci. 2020;250:117579. [DOI] [PubMed] [Google Scholar]
- 36. Huang X, Xie X, Wang H, et al. PDL1 And LDHA act as ceRNAs in triple negative breast cancer by regulating miR‐34a. J Exp Clin Cancer Res. 2017;36:129. [DOI] [PMC free article] [PubMed] [Google Scholar]


