Abstract
Many microbial communities, including those involved in chronic human infections, are patterned at the micron scale. In this Review, we summarize recent work that has defined the spatial arrangement of microorganisms in infection and begun to demonstrate how changes in spatial patterning correlate with disease. Advances in microscopy have refined our understanding of microbial micron-scale biogeography in samples from humans. These findings then serve as a benchmark for studying the role of spatial patterning in preclinical models, which provide experimental versatility to investigate the interplay between biogeography and pathogenesis. Experimentation using preclinical models has begun to show how spatial patterning influences the interactions between cells, their ability to coexist, their virulence and their recalcitrance to treatment. Future work to study the role of biogeography in infection and the functional biogeography of microorganisms will further refine our understanding of the interplay of spatial patterning, pathogen virulence and disease outcomes.
It has been proposed that up to 80% of bacteria live in spatially structured communities1–4. This biogeography has been studied on numerous scales, revealing clear evidence for microbial patterning, often reminiscent of the patterning observed in macro-organisms5,6. At the micron scale within a single environment, spatial patterning of microorganisms (microbiogeography) influences their physiology and behaviour by shaping the interactions they can form with surrounding cells and with the abiotic microenvironment (FIG. 1).
Fig. 1 |. Polymicrobial biogeography can determine the severity of disease and treatment outcomes.
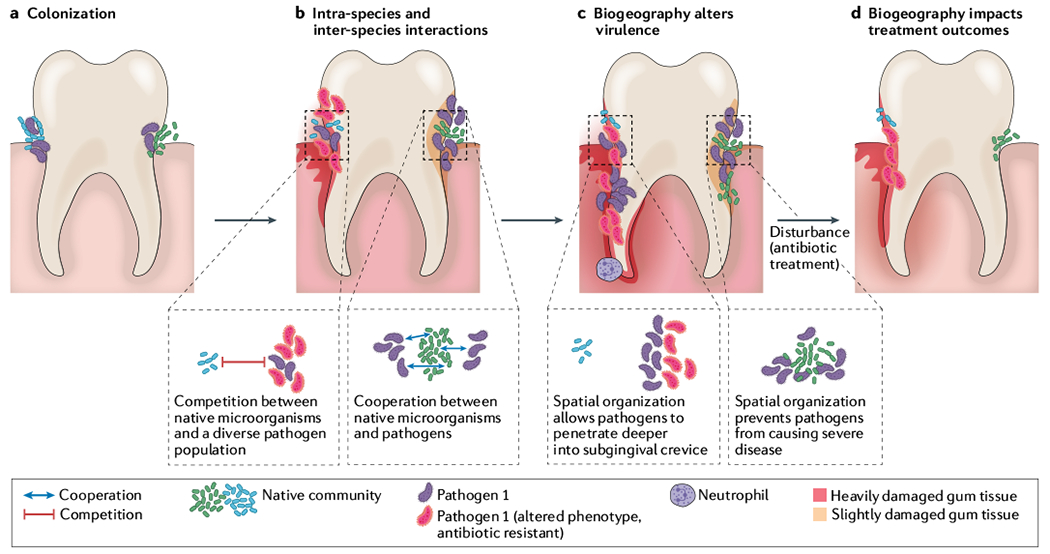
Spatial patterning of bacterial pathogens in relation to other microorganisms can alter the environment, microbial fitness and host immune responses. a | Colonization and aggregate formation by a pathogen (purple), along with the native microbial community (green and blue cells) in the subgingival pocket of the tooth. b | Expansion of the pathogen population can alter the environment by either limiting available nutrients or releasing exoproducts. Inter-species competitive (blue and purple cells) or cooperative (green and purple cells) dynamics determine infection biogeography. In addition, phenotypically and genetically diverse subpopulations can emerge (red cells); this heterogeneity can further alter the nature of intra-species interactions. c | Differential spatial patterning results in changes in virulence. Left side of tooth: localized expansion of pathogen leads to increased virulence, damaging the enamel (red enamel, root and gums) and resulting in periodontitis. This activity amplifies inflammatory signals and recruits immune cells such as neutrophils. In addition, increased pathogen abundance reduces available nutrients, decreasing fitness of the original microbial community. Right side of tooth: dispersed aggregates of the pathogen allow for increased fitness of the original microbial community, preventing both pathogen expansion and heavy damage to the subgingival pocket. d | Antimicrobial treatment is one of the most common disturbances for polymicrobial infections. Outcome of antimicrobial treatment is impacted by biogeography. On left side of tooth, spatial patterning protects the community from antimicrobial treatment. Further, the antibiotic increases fitness of an antibiotic-resistant pathogen (red cells), leading to niche expansion and increased damage. On right side of tooth, spatial patterning allows the pathogen to be susceptible to antibiotic treatment, eliminating the pathogen and reducing abundance of the original microbial community (green cells). This reduces virulence of the community and results in less damage to the tooth.
Microbiogeography is influenced by numerous factors, including the nutritional and physiochemical landscape and the ability of microorganisms to attach to surfaces. In addition, microbial interactions influence, and are influenced by, the community’s biogeography. Bacteria are equipped with various regulatory mechanisms to sense and respond to chemical and physical environmental cues7–10. Many chemical cues are diffusible molecules produced by neighbouring microorganisms that are within ‘earshot’, and include metabolites such as lactic acid11,12 as well as dedicated signals such as acyl-homoserine lactone quorum sensing signals13–15. Other cues require physical contact between microorganisms or result from the interaction of microorganisms with a host16–19. Whether mediated by diffusible molecules or direct physical contact, these interactions alter the patterning of microorganisms as well as their growth and fitness in an environment. We previously reviewed several environmental cues and biological properties that shape the microbiogeography of infection20.
Despite the presumed importance of biofilms and their spatial patterning, we have a limited understanding of these structures at the micron scale. Studies of microbiogeography are relatively rare, particularly those using intact samples. In this Review, we highlight recent findings from studies using human- derived samples of chronic polymicrobial infection environments and microbial communities in the oral cavity, as these areas have seen recent advancement in quantifying biogeography and its physiological consequences (FIG. 2). Chronic polymicrobial infections, such as those in the lungs of people with cystic fibrosis or within chronic wounds, are persistent infections colonized by several species of bacteria, whose interactions with each other limit treatment efficacy. The diverse communities of microorganisms in the human oral cavity are present during health, but also cause caries and gum disease. Studies of chronic polymicrobial infections, oral infections and microorganisms colonizing the oral cavity have revealed that microorganisms are spatially structured at the micron scale, with defined arrangements of multiple strains or species21–24. In addition, microorganisms often form aggregates or biofilms in these infections with defined spatial patterning relative to host cells. These findings form a framework for studying and conceptualizing the biogeography of diverse polymicrobial sites, including all sites that are colonized by the microbiota.
Fig. 2 |. Microbiogeography of human-associated microbial communities is the benchmark for in vitro preclinical models.
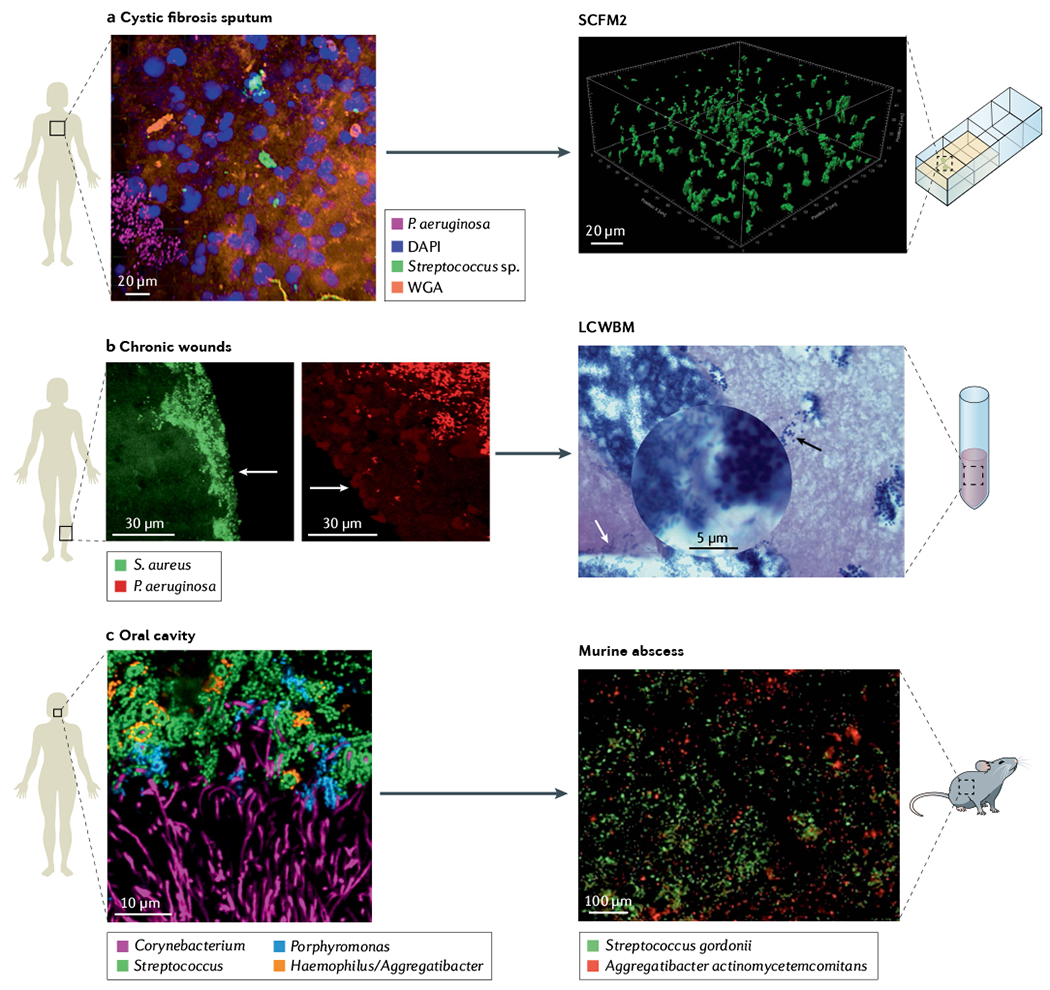
a | Spatial arrangement of Pseudomonas aeruginosa (magenta) and streptococci (green) within a cystic fibrosis sputum sample visualized using MiPACT (microbial identification after passive clarity technique) and hybridization chain reaction46. Structural context of the environment is visible using DAPI labelling of host DNA (blue) and wheat germ agglutinin (WGA) labelling of mucin (orange). Evaluation of cystic fibrosis sputum chemical composition and structure enable development of synthetic cystic fibrosis sputum medium (SCFM2), a medium that recapitulates the physical and chemical properties of cystic fibrosis sputum, where surface-detached aggregates of P. aeruginosa strain PAO1 in SCFM2 have similar spatial patterning as in cystic fibrosis sputum samples. b | P. aeruginosa (red) and Staphylococcus aureus (green) in two chronic wound biopsies visualized by peptide nucleic acid fluorescence in situ hybridization (PNA-FISH)54. Images show the spatial patterning of these two pathogens in wounds is not random. P. aeruginosa aggregates are localized deeper in wounds (50–60 μm from the surface) in comparison with S. aureus aggregates localized close to the wound surface (within 20–30 μm of the wound surface). Arrows show edge of wounds. Lubbock chronic wound biofilm model (LCWBM) provides a chemical environment similar to chronic wounds. However, spatial structure is dependent on microbial activities. Spatial arrangement of P. aeruginosa (white arrow) and S. aureus (black arrow) in this model is dependent on coagulase activity of S. aureus to turn soluble fibrinogen in plasma into insoluble fibrin, forming a fibrin network that gives gel-like properties to the media. In this model, S. aureus and P. aeruginosa can coexist in adjacent aggregates93. c | Combinatorial labelling and spectral imaging FISH (CLASI-FISH) image of microbial communities from human supragingival plaque sampled from the human oral cavity showing a spatially patterned consortia of microorganisms in complex corncob structures60. Investigations using a murine abscess model of infection demonstrated that precise spatial arrangement between Aggregatibacter actinomycetemcomitans and Streptococcus gordonii promotes virulence11. Panel a cystic fibrosis sputum adapted from REF.46, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Panel b chronic wound adapted with permission from REF.54, ASM. Panel b LCWBM adapted with permission from REF.93, ASM. Panel c oral cavity adapted with permission from REF.60, PNAS. Panel c murine abscess adapted with permission from REF.11, PNAS.
The ex vivo analyses of samples from humans also serve as a benchmark for studying bacterial biogeography in experimental systems in the laboratory. Advancements in understanding the role of biogeography in infection are driven by the development of preclinical models that promote spatial patterning and enable hypothesis testing using genetic mutants and the manipulation of biogeography (BOX 1). Preclinical models are important to uncover the mechanisms controlling spatial patterning and the impact of microbiogeography on bacterial function. Although much is still unknown, these studies have started to reveal the mechanisms of the intricate, micron-scale interactions between microorganisms and neighbouring microbial or host cells. Importantly, these studies contribute to the idea that micron-scale biogeography affects the fitness of the microbial constituents as well as the severity of disease25–29.
Box 1 |. Framework of preclinical infection model development.
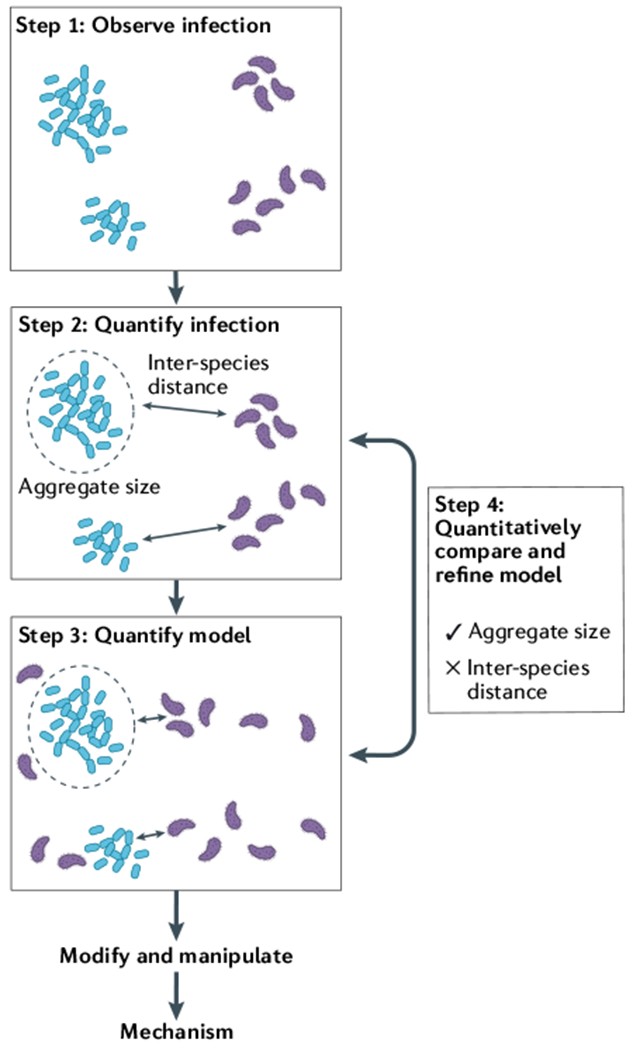
Preclinical models are essential for studying the processes that shape micron-scale biogeography in infection and for defining the role of biogeography in pathogenesis. Hundreds of laboratory models exist for studying infection, and more are constantly developed, but there is no gold standard for the qualities of a ‘good’ model for studying biogeography. The ideal model will vary depending on the focal microorganism or microbial community, the infection type and the stage of infection. The four stages proposed here (see the figure) offer a framework for using the human infection environment as a guide in the development and improvement of preclinical models of infection, ultimately leading to the elucidation of the mechanisms that shape microbiogeography and pathogenesis. Further, although this framework is focused on research that explicitly is studying biogeography, any study that investigates bacterial physiology in chronic infections will benefit from considering the spatial components of their experimental model system.
Step 1: observe infection
The key to developing a robust model to is to observe the infection environment one is trying to replicate. For polymicrobial infections, microscopic assessments of biogeography in human- derived samples show how microorganisms are spatially structured relative to each other and the host environment.
Step 2: quantify infection
The next step is to quantify the spatial patterning. BOX 2 summarizes methods to quantify properties such as the ratio of planktonic to aggregated cells, the size of aggregates, the density of biomass and the relationships between cell types. In addition, the function of microorganisms can be measured across the landscape of the infection using specialized microscopic and sequencing approaches, and abiotic factors such as the nutritional environment, pH, temperature and oxygen can be quantified with chemical analytical techniques.
Step 3: quantify model
Using the observation (step 1) and quantification (step 2) of the infection biogeography as a benchmark, one can then choose or design a model that seeks to recapitulate the spatial patterning in the host. The biogeography of microorganism(s) of interest grown in the model can then be quantified, using the same metrics used to study the human infection environment.
Step 4: quantitatively compare and refine model
Finally, the biogeography in the model is compared with that in the human infection. This process identifies aspects of the model that are accurately captured and, therefore, appropriate for future study. In addition, any differences in the spatial patterning between the model and the human infection can inspire further refinement of the model system in an iterative process in which models are repeatedly altered and compared back with the human infection environment. For instance, in our example, aggregate size is well captured by the model, whereas inter-species distance is not. Thus, this model could be used to study the role of aggregate size in pathogenesis, and further refinement could work to more accurately capture inter-species distance, for example, by altering the physical or chemical environment.
Modify and manipulate
Once the model system is established, it can be modified or manipulated to provide insights into the role of biogeography in infection. For example, defined bacterial mutants can be used to study how specific functions contribute to the establishment or maintenance of biogeographic patterns, or the addition of antibiotics can test how biogeography influences antibiotic resistance and how biogeography changes in response to antibiotics. Again, understanding these manipulations will rely on the ability to observe and quantify the spatial arrangement of microorganisms.
In this Review, we detail the recent findings that advance our understanding of microbial micron-scale biogeography, both in human specimens and in preclinical models. Additionally, much of the recent microbiogeography research has been driven by technological advances that have provided an unprecedented view of the spatial structure of microbial communities in infections. Thus, we also highlight the importance of recently developed approaches including computational quantification of micron-scale spatial structure (BOX 2) and cutting-edge imaging technologies. Finally, we discuss future approaches for understanding microbial biogeography including spatial assessment of microbial function.
Box 2 |. Quantitative image analyses.
To describe microbial biogeographic patterns and their changes over space and time, robust methods to quantify micrometre-scale spatial patterning are crucial. This work has been inspired by methods in landscape ecology that describe the spatial patterning of macroscopic organisms. However, quantification of microbial biogeography requires high-resolution microscopic images of the native environment and the models used to study it. Only a few image analysis pipelines are currently used to quantify microbial density and spatial patterning.
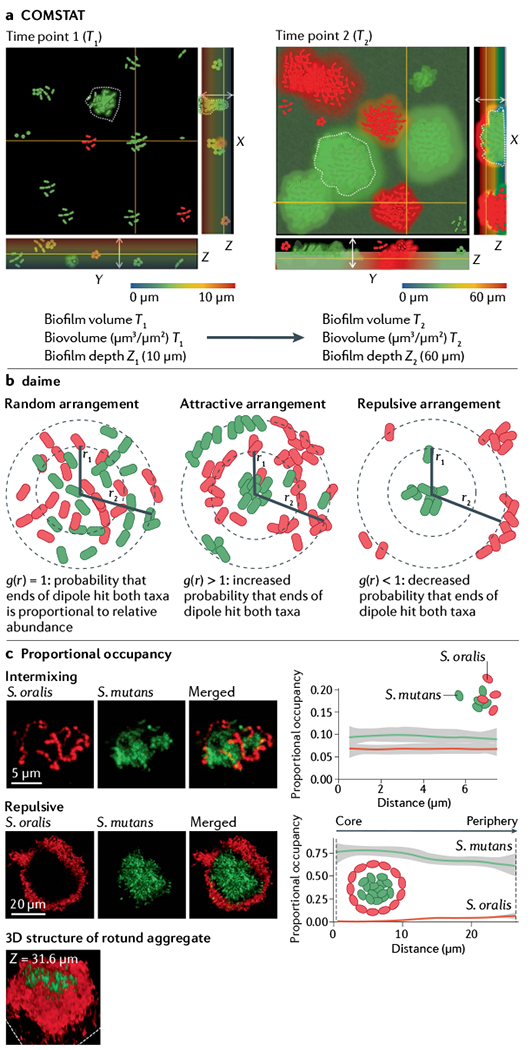
COMSTAT is one of the first image analysis tools for microorganisms, enabling the study of bacterial community development over space and time132. COMSTAT is an automated program developed for morphological analysis of communities formed in flow chambers. Changes in biofilms can be assessed by measuring the number of occupied pixels in a series of 3D images acquired by confocal laser scanning microscopy, using a MATLAB developed script. COMSTAT provides information on the total biomass (biovolume), area occupied by bacterial cells, thickness of biofilms (Z1 and Z2), distribution of microcolonies, roughness coefficient, diffusion distance and surface to volume ratio over time (see the figure, panel a). COMSTAT analyses have been instrumental in studying several aspects of biogeography, including spatial and temporal dynamics, with in vitro microbial communities across various growth environments, stressors and community compositions133–135.
The digital image analysis in microbial ecology (daime) program is an image analysis platform used to quantify spatial patterning136. daime first performs segmentation to differentiate bacterial cells from the background, including using a watershed segmentation function to differentiate cells in close contact in aggregates. daime can also measure fluorescence intensity, biovolume and spatial arrangement. The spatial arrangement measurements are especially important for studying microbial biogeography, and the method in daime determines whether two microbial populations are arranged randomly, or show attraction or repulsion. At a distance (r), daime determines the cross-correlation functions g(r) for the two populations (see the figure, panel b). If the distribution of the community is random, the probability of a dipole of length r hitting the two populations normalized to the densities of the two populations, g(r), is one. However, if the communities are attractive, then g(r) will be greater than one, and if the communities are repulsive, then g(r) will be less than one. By repeating this approach across a range of distances (for example, r1 and r2), daime determines how spatial arrangement varies over space.
An additional open-source pipeline for calculating spatial arrangement relies on proportional occupancy in 3D structures, which determines the proportion of the volume occupied by one bacterial population relative to a focal cell of a second bacterial population across discreet distance intervals65. Thus, proportional occupancy demonstrates how the microbial community around a focal object changes as one moves away from the focal object. This pipeline has been used to determine the dynamics of spatial arrangements in human dental caries and a preclinical model that uses two streptococci (Streptococcus oralis and Streptococcus mutans) that form rotund structures on the tooth surface over time65 (see the figure, panel c). The proportional occupancy calculations showed that the bacteria were initially intermixed, as the proportional occupancy of each Streptococcus sp. relative to the other did not vary across space. By contrast, at later time points, the community is segregated; using the centre of the corona-like structure as the focal point, the proportional occupancy calculation demonstrated that S. mutans occupies ~75% of the volume up to 30 μm away from the centre, whereas S. oralis is not detected at the centre of the structure but its proportional occupancy increases slightly at 30 μm.
Inspired by existing approaches, BiofilmQ is newly developed automated image analysis software that provides a platform to quantify internal architecture of microbial biofilms, alongside phenotypic heterogeneity137.
Although these approaches have been instrumental in quantifying bacterial biogeography during infection, there are exciting opportunities to quantify other aspects of micrometre-scale spatial patterning in the future. For example, open questions include how aggregate shape affects virulence; how patchy microorganisms are in an infection and how connected are the patches; and how we can better capture changes in these metrics over time and space, or across conditions. Part c proportional occupancy adapted from REF.65, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Biogeography in human infection
To understand the importance of biogeography in human infections, it is essential to assess the spatial arrangements of microorganisms in human- derived samples. Without this information, it is virtually impossible to formally assess the relevance of most laboratory studies to the human infection they are meant to mimic. For example, virtually any two microorganisms will interact in the laboratory when mixed, but understanding whether these interactions occur in human infections and affect community function and biogeography requires benchmark data from the actual infection. Historically, electron microscopy was used to visualize human-associated microorganisms with a focus on chronic bacterial infections and microorganisms in the oral cavity. However, electron microscopy does not provide information on the identity of microbial community members and live bacteria cannot be visualized. Despite these challenges, researchers used electron microscopy almost 50 years ago to study the spatial patterning of multispecies human oral communities, demonstrating that bacteria in the oral cavity are surface-associated and surrounded by an extracellular matrix30–33.
The advent of DNA sequencing technologies has fostered a renaissance in our understanding of the identity of microorganisms associated with the human body. These technologies enable the design of species-specific probes to directly visualize microorganisms by fluorescence in situ hybridization (FISH)34,35. In FISH, fluorescently conjugated DNA or peptide nucleic acid (PNA) probes are designed to hybridize with a specific sequence in the microorganisms of interest. Technical modifications in fixing procedures, probe hybridization steps and use of several probes led to methods such as signal-amplified catalysed reporter deposition FISH (CARD-FISH)36, double-labelling of oligonucleotide probes FISH (DOPE-FISH)37 and combinatorial labelling and spectral imaging FISH (CLASI-FISH)38. Despite advances in FISH imaging to study microbiogeography in infection, the technique usually only provides a ‘snapshot’ and thus has limitations for studying the spatiotemporal dynamics of microorganisms during chronic infections. In addition, FISH requires a priori knowledge of the microorganisms in the infection site, and although CLASI-FISH can label up to 120 microbial taxa, most FISH studies only describe the spatial patterning of a few taxa39.
Below we discuss the bacterial micron-scale biogeography in three human polymicrobial infection environments that have been visualized using FISH techniques.
Cystic fibrosis lung infections
The lungs of people with cystic fibrosis are often a site of persistent and damaging microbial infection. Several factors characterize the airway environment in cystic fibrosis, including a high number of neutrophils, high levels of proinflammatory cytokines and entrapment of several microbial species in a thick mucus layer40,41. Delineating the factors involved in the spatiotemporal dynamics of microorganisms within the cystic fibrosis airway is challenging due to limitations in sample collection and microbial infections that persist for decades. An early study used PNA-FISH imaging of explanted cystic fibrosis lungs to describe the spatial patterning of the pathogen Pseudomonas aeruginosa in the cystic fibrosis airways42. This study identified both aggregates and planktonic cells of P. aeruginosa in airways and respiratory zones, surrounded by alginate, extracellular DNA (eDNA) and polymorphonuclear leukocytes. Another study expanded on this work to determine the source of eDNA surrounding P. aeruginosa aggregates in explanted cystic fibrosis lung tissues, using PNA-FISH to label the bacteria and histone-specific antibody staining to label human- derived eDNA43. Although bacterial aggregates were surrounded by human-derived eDNA, this was not the result of NETosis of polymorphonuclear leukocytes, and, interestingly, no eDNA of host origin was detected within P. aeruginosa aggregates. Further, the authors hypothesized that the eDNA provides a barrier protecting the bacterial aggregates against antibiotics and antimicrobial components of the immune system. These findings highlight the role of eDNA and other extracellular polymers in microbiogeography.
Although FISH techniques are valuable for identifying microbial spatial patterning, autofluorescence and sample opacity can hinder analysis. Recent tissue clearing methods such as the passive clarity technique (PACT) have tackled this problem in human-derived samples44,45. A PACT method termed MiPACT (microbial identification after PACT) reduces the opacity of human expectorated cystic fibrosis sputum samples, thus making the sputum sample transparent and facilitating imaging46 (FIG. 2a). Following MiPACT, microorganisms were labelled with FISH, and the eDNA and mucin were detected by immunofluorescence staining. Additionally, to determine the physiological activity of the pathogen Staphylococcus aureus, the authors used the hybridization chain reaction, which enables the specific detection and quantification of transcript levels. The authors found differential growth rates across subpopulations of S. aureus, uncovering intra-population phenotypic heterogeneity in situ. Although these results were tremendously informative, it should be noted that this technique leads to an increase in bacterial volume (approximately fivefold); thus, the microbial communities visualized in this study have limited use for studying micron-scale spatial structure.
These studies of cystic fibrosis were instrumental in assessing how bacterial biogeography could potentially impact important pathogen functions, such as resistance to antibiotics. However, most human- derived samples were obtained from end-stage cystic fibrosis lung disease and do not provide information on the microbiogeography of pathogens during disease progression. Thus, the development of preclinical models and the acquisition of temporal sputum samples are required to assess the emergent properties of microbial spatial patterning in the cystic fibrosis lung. These studies will likely be hampered by the reduced ability of individuals with cystic fibrosis to spontaneously produce sputum, a result of the incredible success of new therapeutics47.
Chronic wound infections
Chronic wound infections are a substantial healthcare burden worldwide, and polymicrobial chronic wound infections display high levels of antibiotic resistance and result in delayed wound healing9,48–52. Although PNA-FISH methods have been used to examine the biogeography of bacteria infecting chronic human wounds, there has been considerably less work in this area than with cystic fibrosis lung infections. As was observed in human cystic fibrosis infection samples, microorganisms inhabiting chronic wounds are present primarily in aggregates, although planktonic cells were also observed53. Studies aimed at examining the spatial structure of wounds co-infected with P. aeruginosa and S. aureus revealed precise biogeography, with S. aureus present closer to the surface of the wound (within 30 μm) and P. aeruginosa deeper in the wound bed (>40 μm from the surface)53,54 (FIG. 2b). Studies of acute burn wounds in healthy volunteers also found medium to large aggregates of coagulase-negative Staphylococcus at the wound edges but no bacteria in the wound bed 4 days after wounding55,56. Thus, it is clear that wounds display precise biogeographical patterns, defined by segregation of taxa between the surface and the bed of the wounds. The next steps involve understanding the microbial functions required for establishing these patterns and the importance of these patterns for disease outcomes. For example, these processes my be influenced by functions such as respiration rate, the ability to consume certain metabolites and the modulation of host immune responses.
Oral cavity
As described above, spatial structure analysis has been a key component of human oral microbiology research for more than 50 years, and the oral cavity remains the leader in our understanding of the biogeography of both colonizing and infecting communities57. The tooth surface is covered with a diverse microbial community, and early microbiological studies led to a model of ecological succession24,58. However, although the acquisition of oral biofilms is often easier than many other human infections, the sheer number of microorganisms in the oral cavity presents challenges for most approaches aimed at identifying the spatial patterning of diverse genera or species. The development of CLASI-FISH has largely overcome this roadblock, and recent studies with CLASI-FISH using human dental plaque have shown precise species-patterning within the multispecies ‘corncob’ structures that were first identified with electron microscopy more than 40 years ago59,60 (FIG. 2c). In addition, a recent study showed highly structured microbial communities on the tongue dorsum. Quantification of spatial relationships revealed habitat specialists and correlated taxa, and the analysis of the shape of single-taxon patches indicated clonal range expansion and contraction relative to the distance from host epithelial cells61,62. These studies substantially increase our understanding of the ecology of oral in vivo biofilms from healthy sites, suggest additional functional roles and interactions, and serve as an example for investigations of diseased sites.
Whereas it is clear that oral communities form precise spatial structure via specific microorganism–microorganism and microorganism–host interactions57,63,64, the mechanisms that regulate the formation and stability of these spatial patterns over time are largely unknown. A recent study tackled this problem by imaging the microbial communities on intact teeth extracted from children with high levels of caries65. Dental caries is caused by a polymicrobial infection of the tooth surface, and the dynamics between commensal microorganisms and pathogens plays a crucial role in the progression of tooth decay. By localizing the microbial communities above carious lesions, the researchers identified multiple species arranged in a 3D corona-like structure containing an inner core of Streptococcus mutans, the causative agent of caries, surrounded by outer layers of other bacteria. Using an in vitro polymicrobial model that recapitulated the corona structure, they discovered that this architecture creates localized regions of acidic pH that lead to caries formation on the tooth surface, providing a causal link between micron-scale biogeography and disease. Further study revealed that corona formation is an active process initiated by production of an extracellular scaffold by S. mutans that directs precise assembly of other bacteria (BOX 2). This study provides a basic framework for establishing causal links between biogeography and microbial function that can be applied to other human infection studies and indicates the power of combining human infection data with relevant preclinical models.
Mechanistic insights from preclinical models
Studies of human- derived specimens have been instrumental in describing the biogeography of infection but are limited by access to human samples and the technological challenges of analysing these samples. In addition, manipulation of biogeography or temporal assessment of microbial spatial structure is often not possible in ex vivo human samples, limiting the ability to study the clinical implications and emergent properties of spatial patterning. To fill this gap in knowledge, preclinical models of infection have been developed to uncover the mechanisms driving biogeography and provide a link between the spatial patterning of microbial communities and pathogenesis. Although preclinical models generally provide considerable versatility compared with human- derived samples, the relevance of these models to the human infection environment is often unknown. A key aspect of preclinical models designed to study biogeography is that they must allow microorganisms to spatially organize, which is not the case in traditional well-mixed laboratory cultures. Thus, human biogeography studies play a critical role as benchmarks to assess the strengths and weaknesses of preclinical models (BOX 1).
Preclinical infection models can either be in vitro or in vivo, and these models can differ substantially in both their utility and their relevance to the human infection environment. There has been progress across numerous infection types to derive mechanistic insights into the role of micron-scale biogeography in infection using these models. Here, we again focus on models meant to mimic the cystic fibrosis lung, chronic wounds and the oral cavity, as spatial patterning has been most studied in these human sites. Together, this work has demonstrated the relationship of pathogenesis and spatial patterning, providing a framework for broadly understanding the biogeography of infection.
In vitro preclinical models
Although it is generally thought that animal models of infection, particularly mouse models, are more relevant to human infection than in vitro models, this is not always the case (BOX 1). Mice are, of course, not humans, and often have limitations on the duration of the infection, require high numbers of infecting microorganisms to establish an infection and differ substantially from humans in their immune system25–29. In addition, mouse models are often constrained experimentally and are not ideal for studying questions that require, for example, extensive temporal data. These considerations have led to the development of in vitro preclinical models specifically designed to mimic the human infection environment.
In vitro preclinical models for studying biogeography retain the flexibility, low cost and ease of in vitro studies, with the addition of specific disease-relevant environmental factors that enable spatial structure to develop. Many preclinical models arise by adapting standard in vitro models to incorporate a clinically relevant substrate (for example, tooth enamel or catheter material), media supplements that reflect in vivo conditions (for example, saliva, plasma or mucin) and/or host cells or tissue, as detailed below. Typically, these models study biogeography using bacterial strains expressing fluorescent proteins of interest that can be visualized using various imaging techniques, often in real time. These approaches can also provide mechanistic insights into the emergent aspects of microbial community spatial structure by using defined mutants that have either been genetically altered or strains that have evolved particular traits of interest. However, alternative quantitative approaches such as Raman microscopy, atomic force microscopy66, surface plasmon resonance imaging, scanning electrochemical microscopy67,68 and imaging mass spectrometry69 have also been used in in vitro preclinical models70–74.
In vitro models that include surfaces.
Biofilms are an important feature of some human infections, and a large number of preclinical models have been developed to study the mechanisms involved in biofilm formation. Preclinical models have been instrumental in defining the mechanisms that are required for bacteria to form spatially structured biofilms and resist killing by antimicrobials. For example, the study of the biogeography of polymicrobial biofilms involved in dental caries has leveraged in vitro flow cell and static biofilm systems. Use of hydroxyapatite-coated surfaces with sterile human saliva as a component of the growth medium to mimic the oral environment has provided insights into how oral microorganisms interact at the tooth surface to produce precise spatial structures12. One of the primary findings from these studies is that spatial structure is impacted by metabolic exchange between bacteria. For instance, S. mutans and the oral bacterium Veillonella parvula form precise spatial structures when grown as an in vitro biofilm. This patterning likely forms due to metabolic interactions mediated by S. mutans-produced lactic acid that is used by V. parvula as a preferred carbon source and results in high resistance to the antimicrobial chlorhexidine75,76.
However, as discussed above, many human infections are dominated by small aggregates instead of the large surface-attached biofilms, such as those that form in once-flow-through flow cells and in static biofilm culture systems. Thus, although these approaches are still used and their ease of use has garnered widespread acceptance, we propose that they have limited utility for studying microbial functions in many chronic human infections such as those in the cystic fibrosis lung and in chronic wounds. Subsequently, there is an urgent need to increase the biological relevance of these methods for many infection types77. We recently proposed a quantitative methodology for improving model systems10,78,79 and propose that these sorts of approaches are crucial for improving the relevance of most biofilm models (BOX 1).
In vitro models containing host polymers.
Host polymers (such as mucin, eDNA, collagen, fibrin and so on) are a key component of virtually every infection environment, and microorganisms have evolved complex responses to these biopolymers and, in many cases, have high-affinity receptors for them80–83. Thus, the addition of biopolymers to in vitro growth media has been a common approach for developing more relevant model systems. Specifically, the addition of biopolymers often promotes and supports the formation of aggregates of tens to thousands of bacterial cells, similar to what is observed in many human infections, enabling researchers to study the development and function of this biogeographic patterning42,84,85. In combination, the work detailed below shows how host polymers can alter interactions and highlights the importance of measuring microbiogeography over time.
Researchers have used the preclinical model synthetic cystic fibrosis sputum medium (SCFM) and SCFM2 to study chronic bacterial infections of the cystic fibrosis lung (FIGS 2a,3). This growth medium contains ions, carbohydrates, amino acids, small acids and lipids at concentrations found in expectorated cystic fibrosis sputum from stable patients86,87. In addition, this model contains the host polymers mucin and eDNA. SCFM2 has been validated as a relevant model for studying human cystic fibrosis lung infection due to its viscosity, which is imparted by mucin, its promotion of antibiotic-resistant bacterial aggregates and its ability to replicate gene expression patterns and gene essentiality profiles for multiple bacterial pathogens78,79,85–90. SCFM2 was recently shown to be the most accurate preclinical model system for recapitulating the gene expression of P. aeruginosa observed in the cystic fibrosis lung78, outperforming several models including a mouse acute infection lung model. Although encouraging, SCFM2 is outperformed by other models for particular functions; thus, it is crucial that researchers choose preclinical models based on the functions of interest and not ease of use or experience with a model.
Fig. 3 |. Preclinical in vitro model SCFM2 provides a platform to study biological implications of microbiogeography.
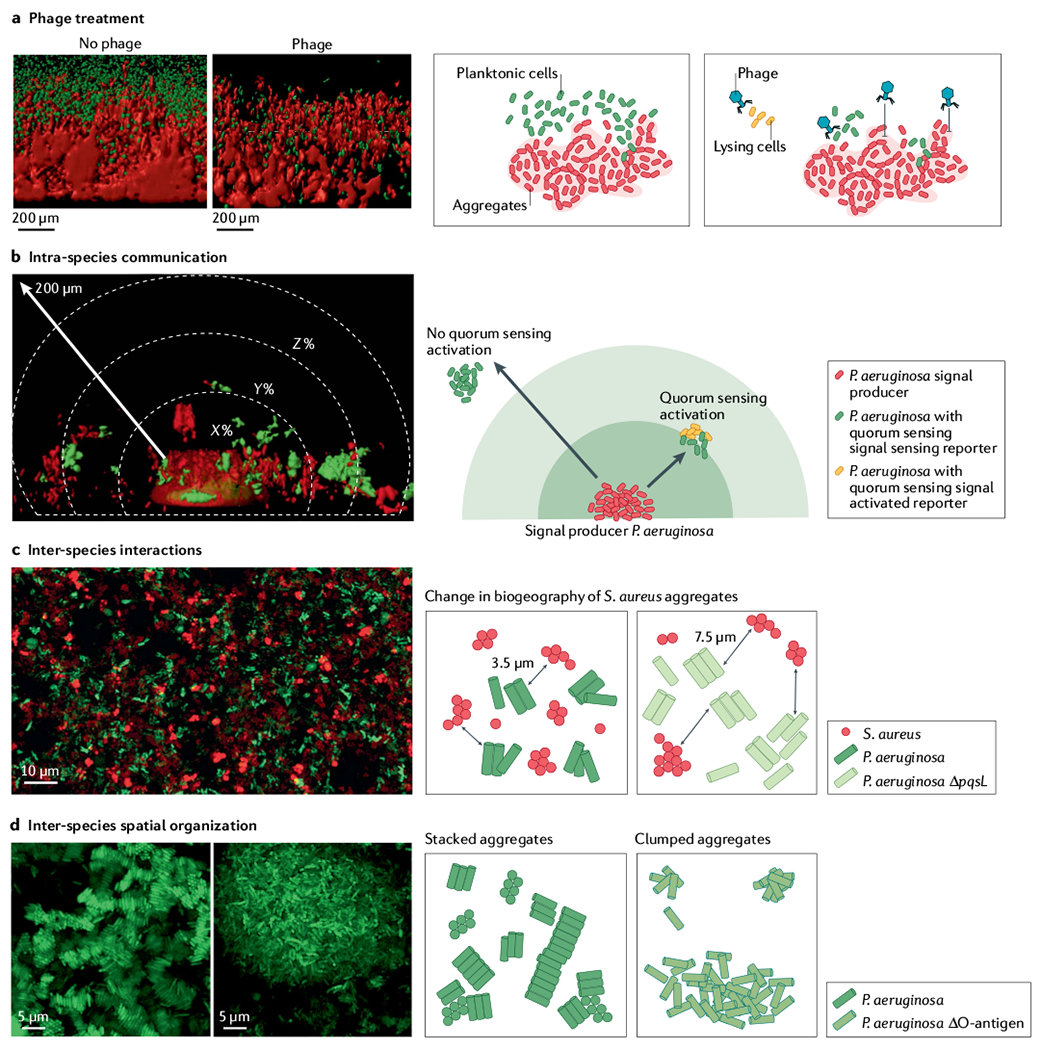
Synthetic cystic fibrosis sputum medium (SCFM2) preclinical model of cystic fibrosis sputum has been a leader in linking biogeography with microbial interactions and fitness. a | Assessment of phage treatment efficacy showed that in SCFM2, planktonic migrant cells are sensitive to phage killing, whereas cells in aggregates and encased in exopolysaccharide matrix limit phage attachment and lysis88. b | SCFM2 provides a structured environment to determine that a Pseudomonas aeruginosa aggregate of ~5,000 cells can signal to neighbouring aggregates up to 176 μm away, although the response to quorum sensing signalling can be heterogeneous85,131. In this experiment, only the aggregate in the bottom centre, confined in a micro-3D-printed trap, can produce the quorum sensing signal. When surrounding aggregates sense the quorum sensing signal, they express a fluorescent reporter gene (red in microscopy image, orange in schematic view). The quorum sensing signal is not detected in surrounding aggregates shown in green. c | Spatially structured environment of SCFM2 enables P. aeruginosa and Staphylococcus aureus aggregates to coexist, unlike in well-mixed environments. In this model, S. aureus aggregates are enriched at a distance of 3.5 μm from P. aeruginosa aggregates. S. aureus fitness did not change when co-cultured with the P. aeruginosa ΔpqsL mutant, which does not produce an anti-staphylococcal molecule. However, the average distance between S. aureus and P. aeruginosa ΔpqsL aggregates increased to 7.6 μm and antibiotic susceptibility of S. aureus increased, showing how changes in inter-species interactions and biogeography can impact S. aureus survival during infection89. d | Loss of O-antigen is a common adaptation of P. aeruginosa to cystic fibrosis airways. This adaptation leads to changes in cell surface hydrophobicity, altering spatial patterning of cells in SCFM2. Cells with O-antigen are stacked, whereas those lacking O-antigen assemble into clumped aggregates91, indicating that genetic variants of P. aeruginosa in cystic fibrosis airways can alter the biogeography of infection. Panel a adapted from REF.88, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Panel b reprinted from REF.85, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Panel c adapted from REF.89, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Panel d reprinted from REF.91, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
Biogeography studies using SCFM2 have revealed how the spatial patterns of P. aeruginosa alter interactions with phage and govern aggregate to aggregate communication85,88,89,91. In the early stages of growth in SCFM2, P. aeruginosa planktonic cells form aggregates that increase in size over time through bacterial growth. At later time points, migrant cells disperse from aggregates and colonize new areas, seeding new aggregates. In the presence of phage, aggregates are protected from killing because of exopolysaccharide production. However, seeding of new aggregates by migrants is inhibited88 (FIG. 3a). Studies in SCFM2 have also provided insights into how spatial structure affects P. aeruginosa signalling via quorum sensing, showing that aggregates of ~2,000 cells do not communicate with neighbouring aggregates, whereas P. aeruginosa aggregates greater than 5,000 cells can communicate with neighbouring aggregates as far away as 176 μm (REF.85) (FIG. 3b). These studies, enabled by the use of SCFM2, provided the first insights into the role of spatial structure in inter-aggregate signalling using aggregates of biologically relevant sizes. Finally, recent studies of P. aeruginosa aggregate assembly provided insight into how changes in bacterial genotype can alter both inter-species and intra-species microbial biogeography in the structured environment of SCFM2 (REFS89,91) (FIG. 3c,d).
Further, spatial patterning has been shown to impact inter-species microorganism–microorganism interactions using the Lubbock chronic wound biofilm model (LCWBM), developed to study chronic human wound infections. Unlike SCFM2, which adds polymers to the growth medium to enhance the biological relevance, the LCWBM relies on the biological activity of infecting bacteria to promote polymer formation. The LCWBM is constructed by the addition of plasma and red blood cells to a complex microbiological medium (Bolton broth) that supports growth of several microorganisms often found in human chronic wounds92 (FIG. 2b). Although the LCWBM begins as a liquid medium, inoculation with S. aureus or any microorganism that produces the enzyme coagulase activates the coagulation cascade, turning soluble fibrinogen in the plasma into insoluble fibrin. The production of fibrin imparts gel-like properties to the medium and provides a scaffold for the spatial organization of bacteria. Using this model, researchers showed that co-cultures of the common wound bacteria P. aeruginosa and S. aureus coexist as discreet aggregate biofilms, and the morphology of these clusters, as well as the host matrix surrounding them, was similar to that seen in sections from human and mouse wounds92,93. Importantly, co-culture increases the antibiotic tolerance of the bacteria, unlike in many other in vitro growth conditions but similar to human infection93–96.
Host cell models.
Models that incorporate immortalized mammalian cell lines and immune cells offer the opportunity to study biogeography in the context of host–microorganism interactions, although spatial information is generally ignored in these models as they have primarily been used to study the molecular interactions between host and microbial cells. One example in which a cell line model has been used to assess biogeography involves the use of differentiated cystic fibrosis human bronchial epithelial cell lines that are grown at the air–liquid interface (ALI). This model enables real-time imaging of microbial interactions with host cells as well as the introduction of environmental disturbances to assess their effect on microbial biogeography. Recent studies aimed at quantifying the accuracy of the ALI model indicate that it promotes gene expression of P. aeruginosa that closely resembles that in the cystic fibrosis lung78, indicating that this model has advantages for studying host–pathogen interactions. Use of ALI-differentiated cystic fibrosis bronchial epithelial cells showed that rhinovirus infection affects the pathogenesis and biogeography of P. aeruginosa97. The viral infection increased hydrogen peroxide (H2O2) production by cystic fibrosis cells, leading to the dispersal of mucoid P. aeruginosa aggregates to the basal surface of the epithelial cells. Infection of ALI cystic fibrosis airway cells with respiratory syncytial virus (RSV) also altered the aggregate assembly of both P. aeruginosa and S. aureus98,99. Interestingly, RSV infection of bronchial epithelial cells activates interferon signalling, increasing secretion of transferrin to the epithelial apical surface. This disruption in iron homeostasis then enhances the growth and biofilm formation of P. aeruginosa at the apical surface. By contrast, increased S. aureus biofilm formation on RSV-infected cystic fibrosis epithelial cells appears to be mediated by increased levels of nutrients other than iron.
Keratinocyte cell lines such as the HaCat cell line have been used to study microbial biogeography in wound infections. Infection of keratinocytes with of P. aeruginosa showed that the fucose-binding lectin LecB binds to the plasma membrane of epithelial cells and inhibits the wound healing process by changing cell attachment and growth signalling pathways50,100. Infection of keratinocytes with P. aeruginosa and S. aureus in the presence of other commensal bacteria decreased the level of biofilm formation and resulting damage to keratinocytes by these pathogens101. This decrease in biofilm formation was proposed to be mediated by the microbiogeography of the community in which the commensals form a layer between the pathogens and keratinocytes. Further, the bacterial inoculum ratios and the timing of the inoculations altered the thickness of the pathogen biofilm layer.
Together, this work builds on our understanding of the relationships between spatial patterning, microbial interactions, host cell physiology and pathogen fitness. Future work additionally has the opportunity to exploit 3D organoid models and organ-on-a-chip methods, which have been used extensively in recent years to study host–pathogen interaction dynamics (reviewed elsewhere102,103). For example, a recent 3D organoid airway model to mimic the cystic fibrosis airway environment is derived from cells isolated from people with cystic fibrosis104. Although models such as this have not been used to study the biogeography of infection, they provide a promising platform for studying microbial biogeography within complex human cell arrangements and at a large scale in the future.
In vivo preclinical models
Although in vitro models offer flexibility and ease of use, these models are often limited when studying complex interactions between the host and microorganisms and the role of biogeography in mediating these interactions. Animal models have been a cornerstone for studies aimed at examining host–microorganism interactions, and recent studies have begun to assess biogeography in these models.
Non-mammalian infection models.
Non-mammalian models offer the benefits of high-throughput analysis, genetic tractability and access to publicly available strains and reagents, all generally with low expense and low infrastructure requirements. Many types of invertebrates are used to study polymicrobial infection biogeography, including Drosophila melanogaster (fruit fly), Caenorhabditis elegans (nematode), larval and adult Danio rerio (zebrafish) and various species of plants105. Although there are clear advantages in using these models, interpretation of the findings in the context of human disease can be challenging (BOX 1). For example, immune system differences with regard to humans should be considered as well as any environmental differences. However, these models can be useful to test hypotheses such as the impact of gut movement on shaping the spatial organization of microorganisms106 or how synergistic interactions between pathogens alter niche colonization107.
D. melanogaster has been commonly used to study biofilm formation, virulence and host interactions of several pathogens such as P. aeruginosa, S. aureus and Candida albicans. In P. aeruginosa infection of D. melanogaster, the bacteria were found as antibiotic-resistant aggregates, and aggregate formation required pel exopolysaccharide expression108. pel mutants that did not form biofilms were hypervirulent and inhibited the expression of antimicrobial peptides by the fly. Further, the D. melanogaster infection model was used to show that the P. aeruginosa quorum sensing receptor RhlR is required for neutralizing the cellular immune response and for aggregate formation109.
C. elegans infections are a useful system for microbial biogeography studies due to the ease in imaging the entire nematode110. For example, Staphylococcus epidermidis infection required synthesis of the biofilm-associated polysaccharide intercellular adhesin in wild-type C. elegans, but not in immunocompromised nematodes111. This study shows the power of non-mammalian infection models to combine microscopy with both bacterial and host mutants. Imaging of C. elegans infections has also been used to determine virulence factors of P. aeruginosa, including quorum sensing genes and common goods such as iron chelators112. When co-infected, quorum sensing mutants co-localize with wild type cells, but there is high variability in spatial patterning and abundance of cells across individual nematodes. In addition, a study using C. elegans as an infection model showed that biogeographical history impacts virulence113. The authors found that P. aeruginosa cells recently dispersed from biofilms were more virulent than cells grown planktonically. As these studies and other similar studies are highly informative, it is surprising that C. elegans has not been used more broadly to assess the role of biogeography in polymicrobial infections. We propose that this model provides a platform for assessing polymicrobial biogeography in high throughput in the future.
Mammalian models.
Although mammalian models are often considered the gold standard in vivo models for studying infection, biogeography studies in these models are rare. The studies that do exist generally use microorganisms that express fluorescent proteins or have been labelled after tissue fixation using FISH. Imaging infection samples from mammalian models can be challenging, as the levels of autofluorescence are often high in tissue, thus making it difficult to identify microorganisms with high accuracy. Additionally, imaging mass spectrometry has also been used in mouse models to assess the chemical environment of infections114,115. Below we discuss some notable examples of how biogeographical studies using mammalian models can provide new insights into infection dynamics.
One of the first studies to assess the role of micron-scale spatial patterning in pathogenesis involved imaging of the oral bacteria Streptococcus gordonii and Aggregatibacter actinomycetemcomitans in a murine abscess model of infection11 (FIG. 2c). Quantification of the spatial structure of this polymicrobial abscess infection revealed precise spatial patterning, with A. actinomycetemcomitans enriched within 4–13 μm of S. gordonii aggregates. This enrichment was, in part, mediated by perception of the streptococcal metabolite H2O2 by A. actinomycetemcomitans, which, at high concentrations, triggers dispersal of this pathogenic bacterium. Elimination of the ability of A. actinomycetemcomitans to sense and respond to H2O2 altered the spatial structure of the community with A. actinomycetemcomitans enriched in areas immediately adjacent (≤4 μm) to S. gordonii. This altered spacing resulted in decreased virulence, thus providing a causal link between biogeography and pathogenesis. Although this is, to our knowledge, the only study that has linked micron-scale spatial patterning of microbial communities and pathogenesis, we expect that this is a common occurrence in polymicrobial infections.
The mouse has been a valuable model for our understanding of the gut microbiota, and both sequencing and microscopy approaches have established that microbial taxa vary along the length of the intestine, between the lumen and the crypts, and across different diets116,117. For example, to study the role of succession on niche colonization of the intestinal gland, a study in a mouse model used differentially labelled Helicobacter pylori118. The authors found intra-species competition over niche occupation, where colonization of H. pylori in the intestine is clonal and there are no mixed populations inhabiting the same micro-niche. This colonization required bacterial chemotaxis as H. pylori chemotaxis mutants could not prevent secondary infection of their resident gland and had a severe defect in the ability to spread to new glands. Thus, H. pylori establishes spatial patterning within glands, likely mediated by unknown host factors, and primary colonization prevents secondary colonization of the gland by the same species.
Another study examined the colonization of the gnotobiotic mouse gut with a consortium of 15 human gut commensal bacteria reconstructed from the healthy human gut119. Colonization of the bacterial consortia was imaged in the proximal colon after 14 days. To preserve the structure, the tissues were initially encased in agarose, and then fixed and embedded in a glycol methacrylate resin. Results from this study revealed that the colon may be better conceptualized as an incompletely mixed bioreactor, rather than an environment that has sharply stratified compartments.
Ex vivo preclinical models
Ex vivo models offer some of the versatility of in vitro preclinical models, while providing high fidelity to many of the nutritional and micron-scale structural aspects of the infection environment. These models have been especially powerful for demonstrating how spatial patterning develops over time and affects pathogen fitness. For example, in the ex vivo porcine lung model, the dissected bronchioles alone with surrounding alveolar tissue or in combination with SCFM86,90 provide a structured environment with physical and chemical properties similar to cystic fibrosis airways. In this model, infections of P. aeruginosa and S. aureus are formed in the small airway and can be studied for up to 7 days120–122. This model provides a platform to investigate social interactions, antimicrobial resistance and aggregate assembly over time. Cystic fibrosis P. aeruginosa isolates have increased antimicrobial resistance in the ex vivo porcine lung model compared with the standard antimicrobial susceptibility testing, suggesting that this model can be used to predict in vivo antimicrobial susceptibility levels123. In addition, a recently developed ex vivo human chronic wound model uses human skin to create 6-mm excisional wounds within a 12-mm biopsy of full-thickness tissue124. By infecting these wounds with microbial communities and imaging with electron microscopy, a study found that C. albicans serves as a scaffold, shaping the spatial patterning of S. aureus and Citrobacter freundii. Further, microorganism–microorganism interactions and priority effects in this system increased inflammation and neutrophil killing.
The future: functional biogeography
Microscopy-driven approaches using fluorescently labelled cells have enabled substantial advances in determining the identity and biogeography of microorganisms during human infection. Although the success of these studies required technological advancements in sample preparation and image analysis, there is a need to combine these approaches with those aimed at defining the functions of individual cells in situ. A small set of pioneering studies have adapted techniques derived from imaging eukaryotic cells to probe the functional biogeography of microorganisms at the micron scale. As discussed above, hybridization chain reaction probes designed for genes of interest were used to distinguish between different physiologies of S. aureus in cystic fibrosis sputum samples46. This approach was also used to spatially localize P. aeruginosa subpopulations expressing the exopolysaccharide alginate in response to varying oxygen levels125. A second technique called parallel sequential FISH (par-seqFISH), which uses sequential, combinatorial labelling of mRNA molecules, was recently adapted to study the expression of 105 genes across individual P. aeruginosa cells126. In this study, monoculture biofilms grown in vitro in SCFM showed spatial heterogeneity in their expression of metabolic and virulence-related genes. These and similar approaches will provide an immediate way forward in linking biogeography with bacterial single-cell functions.
Two sequencing-based approaches developed to map mRNA transcripts in eukaryotic systems also provide inspiration for how to study the spatial dynamics of global microbial gene expression during infection. One study developed the 3D intact tissue RNA sequencing method STARmap (spatially resolved transcript amplicon readout mapping)127,128. In STARmap, targeted, barcoded cDNA SNAIL probes (specific amplification of nucleic acid via intramolecular ligation probes) are hybridized to intracellular mRNA in solid tissues. cDNA is amplified in situ and the amplicons are embedded within a hydrogel. Then, the barcodes on each amplicon are sequenced in situ using fluorescent probes, creating a map of gene expression. A second approach for combining biogeography and sequencing analyses is spatial transcriptomics, a novel method that is high throughput, untargeted and simple to use and allows for the identification of functional differences across a tissue specimen129. These advancements in transcriptome spatial maps in eukaryotic organisms are introducing novel approaches that can be adapted to microorganisms to visualize the physiology of bacterial communities during infection.
Summary and conclusion
Chronic polymicrobial infections impose a high burden on healthcare systems and are often resistant to antimicrobial therapy130. Thus, understanding the mechanisms and dynamics of host–pathogen interactions and antimicrobial resistance during chronic infections is crucial. There is increasing evidence that spatial patterning is a key component of infection, especially polymicrobial infections. Preclinical models have demonstrated that biogeography can impact important functions including virulence, antibiotic resistance, epithelial cell damage and bacterial communication. However, although this Review has highlighted data from select human-associated polymicrobial communities, there remains much to learn about the similarities and differences in microbial biogeography at these and other sites, including the respiratory tract, the skin and the female genital tract. We believe that the framework proposed in BOX 1 can aid in these studies. Together, this work will result in the ability to answer fundamental questions in microbiogeography.
Thus, across the human microbiota and chronic infections, much work remains to elucidate the mechanisms that link microbial ecology, biogeography and disease. The ultimate goal is to understand how perturbing the spatial patterning of microbial communities can advance treatment strategies. For example, novel interventions could interfere with microbial interactions, altering the spatial patterning and, thus, pathogenesis. The additional development of efficient and high-throughput methods to study the spatial dynamics of microorganisms over the course of polymicrobial infection will further elucidate how to prevent and treat chronic infections.
Acknowledgements
The authors thank the Cystic Fibrosis foundation for a post-doctoral fellowship to S.A. (AZIMI18F0); the National Institutes of Health (NIH) for funding to G.R.L. (F32DE027281 and K99DE031018); and NIH Grants R01DE023193, R01DE020100 and 1R01GM116547 and a grant from the Shurl and Kay Curci Foundation to M.W.
Glossary
- Biogeography
The spatial assembly and distribution of various Organisms in an environment through time.
- Microbiogeography
The spatial patterning of microorganisms at the micron scale within a single environment.
- Quorum sensing
The detection of an increased concentration of small signal molecules at high cell density, which can control gene expression in bacterial populations.
- Cystic fibrosis
A disorder that is caused by mutation(s) in the cystic fibrosis transmembrane conductance regulator gene, which affects cells that produce mucus, sweat and digestive enzymes. in people with cystic fibrosis, the build-up of mucus in the airways results in chronic polymicrobial lung infections.
- Function
A physiological property of a bacterium, such as glucose catabolism or motility.
- Catalysed reporter deposition FISH
(CARD-FISH). In CARD-FISH, the nucleotide probe is conjugated to horseradish peroxidase that amplifies the fluorescence in situ hybridization (FISH) signal. This method is used for bacterial taxa or bacterial functions with inherently low signal, for example bacteria with low ribosome content.
- Double labelling of oligonucleotide probes FISH
(DOPE-FISH). DOPE-FISH uses 5′ and 3′ double-labelled oligonucleotide probes to increase the fluorescence in situ hybridization (FISH) signal.
- Combinatorial labelling and spectral imaging FISH
(CLASI-FISH). Simultaneous fluorescence in situ hybridization (FISH) labelling of individual taxa using multiple fluorophores. By mixing the fluorophore combinations used for each taxon, this method enables the detection of tens to hundreds of taxa concurrently. Spectral imaging is usually followed by linear unmixing analysis to differentiate between taxa.
- Alginate
An exopolysaccharide produced by Pseudomonas aeruginosa that helps cells to attach to surfaces and form biofilms.
- Extracellular DNA
(eDNA). eDNA can be produced by host cells or bacteria. it is one of the main components of the biofilm matrix that can provide structural scaffold for bacterial cells and may play a role in protecting bacterial cells in response to host cells and antibiotics.
- NETosis
Neutrophil extracellular traps (NETs) are nucleic acid and intracellular components that are forced out of polymorphonuclear leukocytes in response to microorganisms and proinflammatory cytokines. NETosis is a type of programmed cell death in polymorphonuclear leukocytes such as neutrophils that functions as cellular defence. During NETosis, cells force out their chromatin, forming sticky traps that can trap bacterial cells.
- Mucin
A type of high molecular weight glycosylated protein produced by epithelial cells in animals that forms gels and is found in high levels in lung infections. Bovine and porcine mucins can be purchased commercially which has led to their use in numerous preclinical models.
- Hybridization chain reaction
The detection and quantification of RNA transcripts using exogenously added fluorophore-conjugated DNA hairpins. These hairpins self-assemble, amplifying their signal.
- Coagulase
An enzyme produced by several types of bacteria that converts the soluble protein fibrinogen in blood to insoluble fibrin, leading to clot formation.
- Preclinical infection models
In vitro and in vivo models used to study microbial infection.
- Raman microscopy
A combination of laser-based imaging and Raman spectroscopy to detect the differential excitation levels of photons. This method is used to identify certain molecules without disturbing the spatial arrangement of the samples.
- Atomic force microscopy
A powerful imaging technique that uses a fine and specific tip attached to a cantilever that scans along the surface of a sample. The changes in contact forces between the tip and the surface of the sample are recorded by a laser beam that generates an accurate topographic image of the surface at nanometre resolution.
- Surface plasmon resonance imaging
A label-free imaging method that is used to detect and measure the attachment level and surface properties of bacterial biofilms. This method analyses changes in the angle of reflected light of a surface Covered with the sample of interest, compared with a control surface.
- Scanning electrochemical microscopy
A label-free microscopy method using probes that detect redox reactions and can provide an electrochemical map of an environment.
- Imaging mass spectrometry
Mass spectrometry performed across a spatial plane to build a map of detected chemicals.
- Hydroxyapatite
A mineral form of calcium apatite that is the main component of tooth enamel and bones.
- Fibrin
A glycosylated protein in blood formed by the enzymatic action of a serine protease on soluble fibrinogen. Polymerized fibrin leads to clotting of the blood.
- Transferrin
A glycoprotein that binds to iron and transports iron through the bloodstream.
- Keratinocyte
A cell that forms the outer layer of skin and produces keratin to form a protective barrier.
- Organoid
A small, differentiated collection of cells containing similar cell types and functions as an organ. Organoids are produced in vitro using stem cells derived from the organ of interest that are cultured in a medium containing growth factors and extracellular matrix.
- Organ-on-a-chip
A cell culture device that contains a multichannel 3D microfluidics chip, designed to mimic the physical and chemical properties of an organ. Organ-on-a-chip models can provide a structured microenvironment for high-throughput assessment of bacterial–host interactions, for instance in response to various stimuli.
- Glycol methacrylate resin
An ester form of epoxy resin that can be used instead of paraffin for embedding biological samples for better quality imaging.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Hall-Stoodley L, Costerton JW & Stoodley P Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol 2, 95–108 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC & Wuertz S Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol 17, 247–260 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Azimi S, Klementiev AD, Whiteley M & Diggle SP Bacterial quorum sensing during infection. Annu. Rev. Microbiol 74, 201–219 (2020). [DOI] [PubMed] [Google Scholar]
- 4.Ibberson CB & Whiteley M The social life of microbes in chronic infection. Curr. Opin. Microbiol 53, 44–50 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martiny JB et al. Microbial biogeography: putting microorganisms on the map. Nat. Rev. Microbiol 4, 102–112 (2006). [DOI] [PubMed] [Google Scholar]
- 6.Nemergut DR et al. Global patterns in the biogeography of bacterial taxa. Env. Microbiol 13, 135–144 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whiteley M, Lee KM & Greenberg EP Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 96, 13904–13909 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giraudo AT, Mansilla C, Chan A, Raspanti C & Nagel R Studies on the expression of regulatory locus sae in Staphylococcus aureus. Curr. Microbiol 46, 246–250 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Ibberson CB et al. Co-infecting microorganisms dramatically alter pathogen gene essentiality during polymicrobial infection. Nat. Microbiol 2, 17079 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornforth DM et al. Pseudomonas aeruginosa transcriptome during human infection. Proc. Natl Acad. Sci. USA 115, E5125–E5134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stacy A et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA 111, 7819–7824 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this pivotal study, the authors show that precise spatial patterning impacts virulence and characterize the metabolic and genetic factors involved in this relationship.
- 12.Kolenbrander PE, Egland PG, Diaz PI & Palmer RJ Jr. Genome–genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13, 11–15 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Whiteley M, Diggle SP & Greenberg EP Progress in and promise of bacterial quorum sensing research. Nature 551, 313–320 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bassler BL How bacteria talk to each other: regulation of gene expression by quorum sensing. Curr. Opin. Microbiol 2, 582–587 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Williams P et al. Quorum sensing and the population-dependent control of virulence. Phil. Trans. R. Soc. Lond. B 355, 667–680 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegtmeyer N, Wessler S & Backert S Role of the cag-pathogenicity island encoded type IV secretion system in Helicobacter pylori pathogenesis. FEBS J. 278, 1190–1202 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou Y et al. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun 80, 1243–1251 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aubert DF et al. A Burkholderia type VI effector deamidates Rho GTPases to activate the pyrin inflammasome and trigger inflammation. Cell Host Microbe 19, 664–674 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Shalom G, Shaw JG & Thomas MS In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153, 2689–2699 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Stacy A, McNally L, Darch SE, Brown SP & Whiteley M The biogeography of polymicrobial infection. Nat. Rev. Microbiol 14, 93–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This review provides a primer on the factors that drive microbiogeography during infection, including attachment, the physiochemical environment, host factors and polymicrobial interactions.
- 21.Alhede M et al. Combination of microscopic techniques reveals a comprehensive visual impression of biofilm structure and composition. FEMS Immunol. Med. Microbiol 65, 335–342 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Hughes CV, Kolenbrander PE, Andersen RN & Moore LV Coaggregation properties of human oral Veillonella spp.: relationship to colonization site and oral ecology. Appl. Env. Microbiol 54, 1957–1963 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Werner E et al. Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Env. Microbiol 70, 6188–6196 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers JD, Palmer RJ Jr., Kolenbrander PE & Scannapieco FA Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun 69, 7046–7056 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelichio MJ, Spector J, Waldor MK & Camilli A Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun 67, 3733–3739 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Earle KA et al. Quantitative imaging of gut microbiota spatial organization. Cell Host Microbe 18, 478–488 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAlester G, O’Gara F & Morrissey JP Signal-mediated interactions between Pseudomonas aeruginosa and Candida albicans. J. Med. Microbiol 57, 563–569 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Reddinger RM, Luke-Marshall NR, Sauberan SL, Hakansson AP & Campagnari AA Streptococcus pneumoniae modulates Staphylococcus aureus biofilm dispersion and the transition from colonization to invasive disease. mBio 9, e02089–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schulte M & Hensel M Models of intestinal infection by Salmonella enterica: introduction of a new neonate mouse model. F1000Res. 5, 1498 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frank RM & Houver G in Dental Plaque (ed. McHugh WD) 85–108 (Livingstone S, 1970).
- 31.Kharazmi A, Giwercman B & Hoiby N Robbins device in biofilm research. Methods Enzymol. 310, 207–215 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Marrie TJ & Costerton JW Mode of growth of bacterial pathogens in chronic polymicrobial human osteomyelitis. J. Clin. Microbiol 22, 924–933 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gristina AG & Costerton JW Bacterial adherence to biomaterials and tissue. The significance of its role in clinical sepsis. J. Bone Jt. Surg. Am 67, 264–273 (1985). [PubMed] [Google Scholar]
- 34.DeLong EF, Taylor LT, Marsh TL & Preston CM Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl. Env. Microbiol 65, 5554–5563 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeLong EF, Wickham GS & Pace NR Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243, 1360–1363 (1989). [DOI] [PubMed] [Google Scholar]
- 36.Pernthaler A, Pernthaler J & Amann R Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Env. Microbiol 68, 3094–3101 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoecker K, Dorninger C, Daims H & Wagner M Double labeling of oligonucleotide probes for fluorescence in situ hybridization (DOPE-FISH) improves signal intensity and increases rRNA accessibility. Appl. Env. Microbiol 76, 922–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valm AM et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valm AM, Oldenbourg R & Borisy GG Multiplexed spectral imaging of 120 different fluorescent labels. PLoS ONE 11, e0158495 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cohen-Cymberknoh M, Kerem E, Ferkol T & Elizur A Airway inflammation in cystic fibrosis: molecular mechanisms and clinical implications. Thorax 68, 1157–1162 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Boucher RC Airway surface dehydration in cystic fibrosis: pathogenesis and therapy. Annu. Rev. Med 58, 157–170 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Bjarnsholt T et al. Pseudomonas aeruginosa biofilms in the respiratory tract of cystic fibrosis patients. Pediatr. Pulmonol 44, 547–558 (2009). [DOI] [PubMed] [Google Scholar]; This study combines numerous techniques to visualize P. aeruginosa in the cystic fibrosis lung and in sputum, relative to immune cells.
- 43.Alhede M et al. The origin of extracellular DNA in bacterial biofilm infections in vivo. Pathog. Dis 78, ftaa018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Treweek JB et al. Whole-body tissue stabilization and selective extractions via tissue–hydrogel hybrids for high-resolution intact circuit mapping and phenotyping. Nat. Protoc 10, 1860–1896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang B et al. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DePas WH et al. Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio 7, e00796–1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that the use of tissue clearing and hybridization chain reaction probes allows for detecting physiological heterogeneity and spatial organization of multiple species in cystic fibrosis sputum.
- 47.Rogers GB, Taylor SL, Hoffman LR & Burr LD The impact of cFTR modulator therapies on CF airway microbiology. J. Cyst. Fibros 19, 359–364 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao G et al. Delayed wound healing in diabetic (db/db) mice with Pseudomonas aeruginosa biofilm challenge: a model for the study of chronic wounds. Wound Repair Regen. 18, 467–477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjarnsholt T et al. Why chronic wounds will not heal: a novel hypothesis. Wound Repair Regen. 16, 2–10 (2008). [DOI] [PubMed] [Google Scholar]
- 50.Thuenauer R et al. The Pseudomonas aeruginosa lectin LecB causes integrin internalization and inhibits epithelial wound healing. mBio 11, e03260–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brothers KM et al. Putting on the brakes: bacterial impediment of wound healing. Sci. Rep 5, 14003 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen CK Human wounds and its burden: an updated compendium of estimates. Adv. Wound Care 8, 39–48 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirketerp-Moller K et al. Distribution, organization, and ecology of bacteria in chronic wounds. J. Clin. Microbiol 46, 2717–2722 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fazli M et al. Nonrandom distribution of Pseudomonas aeruginosa and Staphylococcus aureus in chronic wounds. J. Clin. Microbiol 47, 4084–4089 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bay L et al. Bacterial aggregates establish at the edges of acute epidermal wounds. Adv. Wound Care 7, 105–113 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.MacLeod AS & Mansbridge JN The innate immune system in acute and chronic wounds. Adv. Wound Care 5, 65–78 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mark Welch JL, Ramirez-Puebla ST & Borisy GG Oral microbiome geography: micron-scale habitat and niche. Cell Host Microbe 28, 160–168 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Palmer RJ Jr. et al. Retrieval of biofilms from the oral cavity. Methods Enzymol. 337, 393–403 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Jones SJ A special relationship between spherical and filamentous microorganisms in mature human dental plaque. Arch. Oral. Biol 17, 613–616 (1972). [DOI] [PubMed] [Google Scholar]
- 60.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE & Borisy GG Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this article, the authors use CLASI-FISH imaging to visualize the complex microbial consortium in human dental plaque and propose a model of the plaque microbial community microbiogeography.
- 61.Wilbert SA, Mark Welch JL & Borisy GG Spatial ecology of the human tongue dorsum microbiome. Cell Rep. 30, 4003–4015 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mark Welch JL, Dewhirst FE & Borisy GG Biogeography of the oral microbiome: the site-specialist hypothesis. Annu. Rev. Microbiol 73, 335–358 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Carpenter GH Salivary factors that maintain the normal oral commensal microflora. J. Dent. Res 99, 644–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kolenbrander PE et al. Bacterial interactions and successions during plaque development. Periodontol 2000 42, 47–79 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Kim D et al. Spatial mapping of polymicrobial communities reveals a precise biogeography associated with human dental caries. Proc. Natl Acad. Sci. USA 117, 12375–12386 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study combines imaging of specimens from the human oral cavity, preclinical models and quantification of microbiogeography to link spatial patterning with disease.
- 66.Dufrene YF et al. Imaging modes of atomic force microscopy for application in molecular and cell biology. Nat. Nanotechnol 12, 295–307 (2017). [DOI] [PubMed] [Google Scholar]
- 67.Connell JL, Kim J, Shear JB, Bard AJ & Whiteley M Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc. Natl Acad. Sci. USA 111, 18255–18260 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klementiev AD, Jin Z & Whiteley M Micron scale spatial measurement of the O2 gradient surrounding a bacterial biofilm in real time. mBio 11, e02536–20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garg N et al. Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host Microbe 22, 705–716.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wagner M & Horn H Optical coherence tomography in biofilm research: a comprehensive review. Biotechnol. Bioeng 114, 1386–1402 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Sussulini A, Becker JS & Becker JS Laser ablation ICP-MS: application in biomedical research. Mass. Spectrom. Rev 36, 47–57 (2017). [DOI] [PubMed] [Google Scholar]
- 72.Zhang P, Chen YP, Qiu JH, Dai YZ & Feng B Imaging the microprocesses in biofilm matrices. Trends Biotechnol. 37, 214–226 (2019). [DOI] [PubMed] [Google Scholar]
- 73.Caniglia G & Kranz C Scanning electrochemical microscopy and its potential for studying biofilms and antimicrobial coatings. Anal. Bioanal. Chem 412, 6133–6148 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skaar EP Imaging infection across scales of size: from whole animals to single molecules. Annu. Rev. Microbiol 75, 407–426 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This review provides an overview of imaging methods for various infection models.
- 75.Kara D, Luppens SB, van Marle J, Özok R & ten Cate JM Microstructural differences between single-species and dual-species biofilms of Streptococcus mutans and Veillonella parvula, before and after exposure to chlorhexidine. FEMS Microbiol. Lett 271, 90–97 (2007). [DOI] [PubMed] [Google Scholar]
- 76.Palmer RJ Jr., Diaz PI & Kolenbrander PE Rapid succession within the Veillonella population of a developing human oral biofilm in situ. J. Bacteriol 188, 4117–4124 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Roberts AE, Kragh KN, Bjarnsholt T & Diggle SP The limitations of in vitro experimentation in understanding biofilms and chronic infection. J. Mol. Biol 427, 3646–3661 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Cornforth DM, Diggle FL, Melvin JA, Bomberger JM & Whiteley M Quantitative framework for model evaluation in microbiology research using Pseudomonas aeruginosa and cystic fibrosis infection as a test case. mBio 11, e03042–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study, the authors provide a framework for quantifying the accuracy of preclinical models, relative to human infections.
- 79.Ibberson CB & Whiteley M The Staphylococcus aureus transcriptome during cystic fibrosis lung infection. mBio 10, e02774–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwiecinski JM & Horswill AR Staphylococcus aureus bloodstream infections: pathogenesis and regulatory mechanisms. Curr. Opin. Microbiol 53, 51–60 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paulsson M, Su YC, Ringwood T, Udden F & Riesbeck K Pseudomonas aeruginosa uses multiple receptors for adherence to laminin during infection of the respiratory tract and skin wounds. Sci. Rep 9, 18168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kavanaugh JS et al. Identification of extracellular DNA-binding proteins in the biofilm matrix. mBio 10, e01137–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moscoso M, Garcia E & Lopez R Biofilm formation by Streptococcus pneumoniae: role of choline, extracellular DNA, and capsular polysaccharide in microbial accretion. J. Bacteriol 188, 7785–7795 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moser C et al. Novel experimental Pseudomonas aeruginosa lung infection model mimicking long-term host–pathogen interactions in cystic fibrosis. APMIS 117, 95–107 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Darch SE et al. Spatial determinants of quorum signaling in a Pseudomonas aeruginosa infection model. Proc. Natl Acad. Sci. USA 115, 4779–4784 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Palmer KL, Aye LM & Whiteley M Nutritional cues control Pseudomonas aeruginosa multicellular behavior in cystic fibrosis sputum. J. Bacteriol 189, 8079–8087 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Turner KH, Wessel AK, Palmer GC, Murray JL & Whiteley M Essential genome of Pseudomonas aeruginosa in cystic fibrosis sputum. Proc. Natl Acad. Sci. USA 112, 4110–4115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Darch SE et al. Phage inhibit pathogen dissemination by targeting bacterial migrants in a chronic infection model. mBio 8, e00240–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barraza JP & Whiteley M A Pseudomonas aeruginosa antimicrobial affects the biogeography but not fitness of Staphylococcus aureus during coculture. mBio 12, e00047–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Palmer KL, Mashburn LM, Singh PK & Whiteley M Cystic fibrosis sputum supports growth and cues key aspects of Pseudomonas aeruginosa physiology. J. Bacteriol 187, 5267–5277 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Azimi S et al. O-Specific antigen-dependent surface hydrophobicity mediates aggregate assembly type in Pseudomonas aeruginosa. mBio 12, e00860–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Y, Dowd SE, Smith E, Rhoads DD & Wolcott RD In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 16, 805–813 (2008). [DOI] [PubMed] [Google Scholar]
- 93.DeLeon S et al. Synergistic interactions of Pseudomonas aeruginosa and Staphylococcus aureus in an in vitro wound model. Infect. Immun 82, 4718–4728 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Machan ZA, Taylor GW, Pitt TL, Cole PJ & Wilson R 2-Heptyl-4-hydroxyquinoline N-oxide, an antistaphylococcal agent produced by Pseudomonas aeruginosa. J. Antimicrob. Chemother 30, 615–623 (1992). [DOI] [PubMed] [Google Scholar]
- 95.Hoffman LR et al. Selection for Staphylococcus aureus small-colony variants due to growth in the presence of Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA 103, 19890–19895 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hotterbeekx A, Kumar-Singh S, Goossens H & Malhotra-Kumar S In vivo and in vitro interactions between Pseudomonas aeruginosa and Staphylococcus spp. Front. Cell Infect. Microbiol 7, 106 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chattoraj SS et al. Rhinovirus infection liberates planktonic bacteria from biofilm and increases chemokine responses in cystic fibrosis airway epithelial cells. Thorax 66, 333–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hendricks MR et al. Respiratory syncytial virus infection enhances Pseudomonas aeruginosa biofilm growth through dysregulation of nutritional immunity. Proc. Natl Acad. Sci. USA 113, 1642–1647 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kiedrowski MR et al. Staphylococcus aureus biofilm growth on cystic fibrosis airway epithelial cells is enhanced during respiratory syncytial virus coinfection. mSphere 3, e00341–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Landi A et al. Pseudomonas aeruginosa lectin LecB impairs keratinocyte fitness by abrogating growth factor signalling. Life Sci. Alliance 2, e201900422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jordana-Lluch E et al. A simple polymicrobial biofilm keratinocyte colonization model for exploring interactions between commensals, pathogens and antimicrobials. Front. Microbiol 11, 291 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Barrila J et al. Modeling host–pathogen interactions in the context of the microenvironment: three-dimensional cell culture comes of age. Infect. Immun 86, e00282–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Bartfeld S Modeling infectious diseases and host-microbe interactions in gastrointestinal organoids. Dev. Biol 420, 262–270 (2016). [DOI] [PubMed] [Google Scholar]
- 104.Sachs N et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Edwards S & Kjellerup BV Exploring the applications of invertebrate host–pathogen models for in vivo biofilm infections. FEMS Immunol. Med. Microbiol 65, 205–214 (2012). [DOI] [PubMed] [Google Scholar]
- 106.Wiles TJ et al. Host gut motility promotes competitive exclusion within a model intestinal microbiota. PLoS Biol. 14, e1002517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bergeron AC et al. Candida albicans and Pseudomonas aeruginosa interact to enhance virulence of mucosal infection in transparent zebrafish. Infect. Immun 85, e00475–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Mulcahy H, Sibley CD, Surette MG & Lewenza S Drosophila melanogaster as an animal model for the study of Pseudomonas aeruginosa biofilm infections in vivo. PLoS Pathog. 7, e1002299 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Limmer S et al. Pseudomonas aeruginosa RhlR is required to neutralize the cellular immune response in a Drosophila melanogaster oral infection model. Proc. Natl Acad. Sci. USA 108, 17378–17383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Garsin DA et al. A simple model host for identifying Gram-positive virulence factors. Proc. Natl Acad. Sci. USA 98, 10892–10897 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Begun J et al. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3, e57 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rezzoagli C, Granato ET & Kummerli R In-vivo microscopy reveals the impact of Pseudomonas aeruginosa social interactions on host colonization. ISME J. 13, 2403–2414 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chua SL et al. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun 5, 4462 (2014). [DOI] [PubMed] [Google Scholar]
- 114.Cassat JE et al. Integrated molecular imaging reveals tissue heterogeneity driving host-pathogen interactions. Sci. Transl Med 10, eaan6361 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Perry WJ et al. Staphylococcus aureus exhibits heterogeneous siderophore production within the vertebrate host. Proc. Natl Acad. Sci. USA 116, 21980–21982 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tropini C, Earle KA, Huang KC & Sonnenburg JL The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21, 433–442 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nava GM, Friedrichsen HJ & Stappenbeck TS Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J. 5, 627–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Keilberg D, Zavros Y, Shepherd B, Salama NR & Ottemann KM Spatial and temporal shifts in bacterial biogeography and gland occupation during the development of a chronic infection. mBio 7, e01705–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI & Borisy GG Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc. Natl Acad. Sci. USA 114, E9105–E9114 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Harrison F & Diggle SP An ex vivo lung model to study bronchioles infected with Pseudomonas aeruginosa biofilms. Microbiology 162, 1755–1760 (2016). [DOI] [PubMed] [Google Scholar]
- 121.Harrison F, Muruli A, Higgins S & Diggle SP Development of an ex vivo porcine lung model for studying growth, virulence, and signaling of Pseudomonas aeruginosa. Infect. Immun 82, 3312–3323 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sweeney E et al. An ex vivo cystic fibrosis model recapitulates key clinical aspects of chronic Staphylococcus aureus infection. Microbiology 167, 000987 (2021). [DOI] [PubMed] [Google Scholar]
- 123.Hassan MM, Harrington NE, Sweeney E & Harrison F. Predicting antibiotic-associated virulence of Pseudomonas aeruginosa using an ex vivo lung biofilm model. Front. Microbiol 11, 568510 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cheong JZA et al. Priority effects dictate community structure and alter virulence of fungal–bacterial biofilms. ISME J. 17, 2012–2027 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Jorth P, Spero MA, Livingston J & Newman DK Quantitative visualization of gene expression in mucoid and nonmucoid Pseudomonas aeruginosa aggregates reveals localized peak expression of alginate in the hypoxic zone. mBio 10, e02622–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dar D, Dar N, Cai L & Newman DK Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study assesses the gene expression heterogeneity in spatially organized aggregates of P. aeruginosa at the single-cell level by developing the par-seqFISH technique.
- 127.Wang X et al. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science 361, eaat5691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study develops and uses the single-cell approach STARmap to determine changes in the transcriptional profile across space.
- 128.Shah S et al. Single-molecule RNA detection at depth by hybridization chain reaction and tissue hydrogel embedding and clearing. Development 143, 2862–2867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Stahl PL et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 130.Järbrink K et al. The humanistic and economic burden of chronic wounds: a protocol for a systematic review. Syst. Rev 6, 15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Connell JL et al. Probing prokaryotic social behaviors with bacterial “lobster traps”. mBio 1, e00202–10 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Heydorn A et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146, 2395–2407 (2000). [DOI] [PubMed] [Google Scholar]
- 133.Nilsson M, Givskov M, Twetman S & Tolker-Nielsen T Inactivation of the pgmA gene in Streptococcus mutans significantly decreases biofilm-associated antimicrobial tolerance. Microorganisms 7, 310 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Beaudoin T et al. Staphylococcus aureus interaction with Pseudomonas aeruginosa biofilm enhances tobramycin resistance. npj Biofilms Microbiomes 3, 25 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ciofu O, Mandsberg LF, Wang H & Hoiby N Phenotypes selected during chronic lung infection in cystic fibrosis patients: implications for the treatment of Pseudomonas aeruginosa biofilm infections. FEMS Immunol. Med. Microbiol 65, 215–225 (2012). [DOI] [PubMed] [Google Scholar]
- 136.Daims H, Lucker S & Wagner M daime, a novel image analysis program for microbial ecology and biofilm research. Env. Microbiol 8, 200–213 (2006). [DOI] [PubMed] [Google Scholar]
- 137.Hartmann R et al. Quantitative image analysis of microbial communities with BiofilmQ. Nat. Microbiol 6, 151–156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]


