Summary
The extracellular RNA communication consortium (ERCC) is an NIH-funded program aiming to promote the development of new technologies, resources, and knowledge about exRNAs and their carriers. After Phase 1 (2013–2018), Phase 2 of the program (ERCC2, 2019–2023) aims to fill critical gaps in knowledge and technology to enable rigorous and reproducible methods for separation and characterization of both bulk populations of exRNA carriers and single EVs. ERCC2 investigators are also developing new bioinformatic pipelines to promote data integration through the exRNA atlas database. ERCC2 has established several Working Groups (Resource Sharing, Reagent Development, Data Analysis and Coordination, Technology Development, nomenclature, and Scientific Outreach) to promote collaboration between ERCC2 members and the broader scientific community. We expect that ERCC2’s current and future achievements will significantly improve our understanding of exRNA biology and the development of accurate and efficient exRNA-based diagnostic, prognostic, and theranostic biomarker assays.
Subject areas: Biological sciences, Biochemistry, Molecular biology, Cell biology
Graphical abstract
Biological sciences; Biochemistry; Molecular biology; Cell biology
Introduction
The NIH Common Fund identifies and supports emerging scientific disciplines by developing cross-cutting programs to address critical barriers and/or to stimulate new and potentially high-impact areas of research. All Common Fund programs are goal-driven, and as such are expected to be catalytic – producing foundational tools, technology, and/or data to be shared with the broader research community to accelerate progress in the field. The primary objective of the NIH Common Fund Extracellular RNA Communication Consortium (ERCC) Program is to support the development of a comprehensive set of resources to advance research in exRNA biology and enable the development of exRNA-based biomarkers and therapeutics. The presence of exRNAs and their carriers in biofluids (blood, saliva, urine, breast milk, cerebrospinal fluid, amniotic fluid, ascites, pleural effusions, aqueous humor, ascites, bile, bronchial lavage fluid, gastric fluid, seminal fluid, sputum, synovial fluid, sweat, tear fluid) suggests the exciting possibility that they can provide a real-time window into fundamental biological systems involved in human health and diseases (Max et al., 2018; Yan et al., 2020; Zhou et al., 2019), and perhaps serve as a means to modulate them. However, progress in this field is still hindered by significant gaps in the available methods, technologies, and resources for the analysis of exRNA molecules and their carriers.
The heterogeneity in the relative abundance of carrier subclasses, not only among different biofluids and source cell types, but also from donor-to-donor and experiment-to-experiment, has been a major impediment to progress toward understanding exRNA biogenesis and function (Gruner and McManus, 2021), and applying exRNAs as clinical biomarkers. Of the three major classes of exRNA carriers — extracellular vesicles (EVs), lipoprotein particles (LPPs), and free ribonucleoproteins (RNPs) — EVs have so far received the most attention, although a substantial fraction of the exRNAs present in biofluids is associated with non-EV carriers (Arroyo et al., 2011; Chevillet et al., 2014; Jeppesen et al., 2019; Turchinovich et al., 2011; Zhang et al., 2021). Moreover, within each class of carriers, multiple subclasses display tremendous diversity by physical characteristics (e.g., size, density, and surface charge), molecular composition, and cellular origin. Figure 1 illustrates several attributes of the diverse range of exRNA carriers that are the focus of the ERCC2 efforts.
Figure 1.
Diversity of exRNA carriers in biofluids
exRNA are associated with a plethora of carriers ranging from a few nanometers to several microns. Although EVs are the most studied exRNA carriers, RNPs, exomeres, supermeres, lipoproteins, and extracellular organelles are associated with a high proportion of exRNA in biofluids. Their relative abundance may explain the specific exRNA biotype composition observed in distinct biofluids. During viral infection, RNA viruses also constitute a significant exRNA carrier in specific biofluids, a property exploited for diagnostics. Here, all known exRNA carriers are represented at scale thereby illustrating their high size heterogeneity, and the challenge to fractionate them in parallel. Acronyms: RNP: ribonucleoprotein; HDL. High-density lipoprotein; LDL: Low-density lipoprotein; VLDL: Very-Low Density Lipoprotein; EVs: extracellular vesicles; SARS-Cov-2: severe acute respiratory syndrome coronavirus two; HIV-1: Human immunodeficiency virus 1.
Among the smallest exRNA carriers, RNPs are relatively homogeneous in size (average 4–30 nm), and there are several established techniques for their purification, quantification, and characterization based on their chemical, physical, or functional properties. However, beyond a few RNPs that have been extensively characterized as exRNA carriers (Fabbiano et al., 2020), there is still no study that has systematically validated the global impact of RNPs on the exRNA profile of different biofluids. Importantly, it is known that RNPs can assemble with RNA into granular structures initiated by phase-transition or aggregation mechanisms (Lin et al., 2015). It is possible that these structures could represent a form of carrier protecting RNA from degradation in the extracellular space. On the other end of the size spectrum, large EVs (such as oncosomes (Morello et al., 2013)), chylomicrons, and organelles (such as mitochondria (Al Amir Dache et al., 2020)) are large enough (>500 nm) to visualize by light microscopy or flow cytometry, allowing facile verification of purity/enrichment after fractionation, and sufficient material for characterizing their molecular composition. In between, a plethora of diverse exRNA carriers ranging in size from 10 nm (e.g., HDL) to 500 nm (e.g., EVs/LDL/VLDL), with most being smaller than 200 nm, are difficult to separate and characterize using standard methods. Layered on this carrier subclass heterogeneity, biofluids contain contributions from multiple cell types from a variety of tissues. The contributions of these cells and tissues to the exRNA pool can vary from donor-to-donor and be influenced by both normal physiological variables (e.g., diet, age, time of day) and disease state. Importantly, viral infections not only impact EV profiles from host cells (Martin-Jaular et al., 2021), but also lead to the accumulation of virion particles in biofluids. As virions encapsulate not only viral nucleic acids but also cellular RNA (Berkowitz et al., 1996), they can also serve as exRNA carriers. Interestingly, the recent emphasis on development of diagnostic tests for viral infections has led to technical innovations in enzyme-based viral nucleic acid and immunoassay-based viral protein detection techniques (Cassedy et al., 2021; Suea-Ngam et al., 2020) that can be adapted to exRNA carrier separation and characterization.
There is a critical need to develop comprehensive inventories of the cargo of exRNA carrier subclasses (for which subclass assignments are verified by their physical and molecular characteristics) to enable the development and validation of efficient and effective experimental and computational approaches to account for carrier subclass heterogeneity. This is because even reference-free deconvolution methods that do not require reference profiles to be defined before the deconvolution step (Decamps et al., 2020; Murillo et al., 2019) must be experimentally validated (Avila Cobos et al., 2020; Murillo et al., 2019). Although the availability of RNA-seq datasets capturing small and total RNA transcriptomes from diverse human tissues and cell types (deRie et al., 2017; Lorenzi et al., 2021; Melé et al., 2015), biofluids (Hulstaert et al., 2020; Murillo et al., 2019), and human organs at the single cell level (Regev et al., 2017; Uhlén et al., 2015), can be useful, dedicated datasets comprised comprehensive cargo profiling from multiple carrier classes rigorously separated to high purity from the same biofluid source are needed for rigorous validation.
It is important to ensure that studies aimed at studying specific carriers are not confounded by unintended co-isolation of different classes of carriers, or contamination by endogenous or exogenous RNAs that are introduced during the processes of carrier separation, exRNA isolation, or analysis. Approaches to overcome this hurdle include: bulk isolation and characterization of each carrier subclass; single-carrier characterization; and experimental or computational estimation of the abundance of each carrier subclass. However, for most of these approaches, this requires knowledge and/or technologies that are not readily available, or that are not applicable for clinical samples because of their high variability, complexity, and/or their requirement of high input volume of biofluids. These approaches can be divided into bulk and single-carrier analyses, each of which involves unique technical challenges.
Bulk separation and analysis of carrier subclasses require reproducible and scalable methods for efficient separation of carriers according to their physical and/or molecular properties and accurate low-input molecular profiling methods. This is challenging because of large differences in the biophysical characteristics of exRNA carriers in biofluids. A major limitation of bulk separation methods (e.g., size exclusion chromatography, differential centrifugation, density gradient ultracentrifugation) is their inability to cleanly separate even the major exRNA carrier classes. However, ongoing efforts to combine several of these methods within integrated exRNA carrier isolation pipelines have resulted in improved separation (Benedikter et al., 2017; Li et al., 2018a; Wei et al., 2020), but at the cost of lengthy procedures requiring large amounts of starting material, making them difficult to implement in a high-throughput manner. Alternatively, affinity capture approaches can enrich specific exRNA carriers based on molecular markers, are amenable to automation, and can operate on low-volume samples. However, the current lack of validated affinity reagents for isolating even the major exRNA carriers, and difficulties in multiplexing these assays from a single biofluid source limit their widespread application. Such challenges may be addressed by highly promising microfluidic microfluidics technologies that incorporate carrier separation and/or exRNA detection in a single device (Meng et al., 2021).
Importantly, current exRNA profiling methods allow for profiling of small and long exRNA fragments from total biofluids, the major carrier classes (EVs, RNPs, and lipoproteins), and the more recently described exomere and supermeres (Hulstaert et al., 2021; Murillo et al., 2019; Rodosthenous et al., 2020; Srinivasan et al., 2019; Zhang et al., 2021). However, profiling of exRNAs associated with less abundant subclasses, or isolated from low volume biofluid samples, will require further improvements in the sensitivity of exRNA profiling methods. The development of next-generation RNA sequencing kits designed for ultra-low or single-cell RNA input material (Picelli et al., 2014), and their adaptation for sequencing EV-associated long RNAs (Rodosthenous et al., 2020) are key advances toward this goal. The use of Unique Molecular identifiers (UMIs) and spike-in synthetic RNA sequences, are proven approaches for enabling more rigorous quality control and quantitative analysis of the resulting RNA sequencing data (Hulstaert et al., 2020; Munchel et al., 2020; Rasmussen et al., 2022). Strategies for enrichment or depletion of specific RNA biotypes (e.g., mRNA, microRNA, tRNA, snRNA, snoRNA, piRNA), using either sequence-based (Hulstaert et al., 2021; Munchel et al., 2020; Rasmussen et al., 2022) or carrier-based (Arroyo et al., 2011) approaches have been shown to improve the quality of profiling data for various biotypes. In addition to the more commonly studied miRNA and mRNA biotypes, techniques enabling measurement of circular RNA (circRNA) in biofluids have been recently developed (Chen et al., 2020; Hulstaert et al., 2020; Hutchins et al., 2021; Kölling et al., 2019). circRNAs are particularly interesting targets for biomarker studies because the absence of 5′ and 3′ ends protects them from degradation by circulating exonucleases, possibly explaining why they are markedly enriched in biofluids in comparison to tissues (Hulstaert et al., 2020).
Recent developments in single-particle/single-molecule imaging and flow cytometry approaches are relevant to single-carrier analysis, with most efforts focused on the study of EVs, rather than other exRNA carriers. Even with this relatively large and experimentally tractable carrier subclass, single-EV analysis is impeded by low numbers of target molecules on a per-EV basis. Experimental measurement of carrier subclass distribution in a complex biofluid is also limited by the lack of reagents (e.g., antibodies or dyes) and methods for accurate quantification of specific biomolecules, particularly biomolecules specific to a given carrier subclass.
The ERCC2 initiative: Decoding exRNA complexity in biofluids
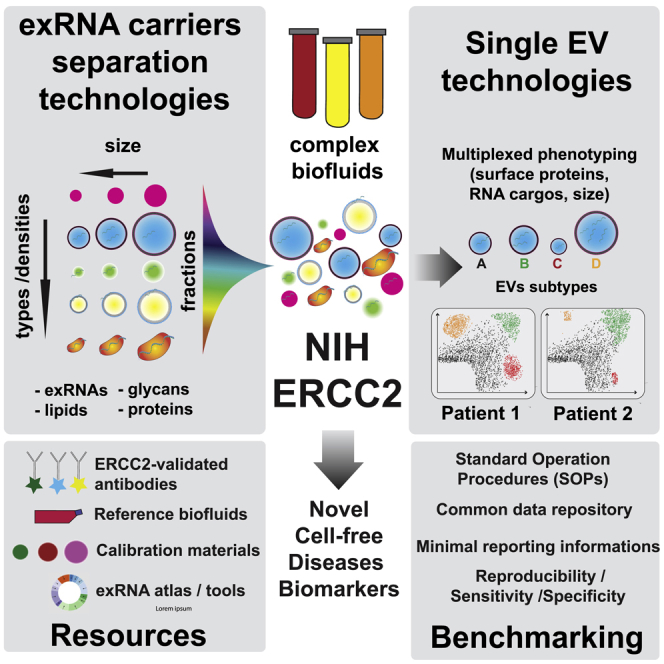
Phase 1 of the ERCC Program (ERCC1, 2013–2018) was designed to: (1) better understand the basic mechanisms responsible for exRNA biogenesis, export from source cells, and uptake and function in recipient cells; (2) establish reproducible methods for exRNA isolation and profiling from various human biofluids; (3) build a computational framework to enable efficient analysis and distribution of exRNA profiling data; and (4) demonstrate the utility of exRNAs as biomarkers and therapeutics (Das et al., 2019). and its extension Phase 2 of the ERCC Program (ERCC2) was launched in September 2019, and is focused on three goals to address the outstanding bottlenecks identified in the course of Phase 1: First, is the further development of a Data Management and Resource Repository, including the extension of the exRNA Atlas Resource to accommodate nano flow cytometry data. Second, is the development of improved separation techniques for sorting exRNA carrier vehicles and their associated molecular cargo. Third, is the development and validation of improved single EV isolation and analysis tools. How these technologies apply to single EV analysis and/or carrier separation is illustrated in Figure 2.
Figure 2.
ERCC2 Integrated technological development for exRNA separation and detection
The aim of ERCC2 is not only to combine and refine existing methods, but also to develop innovative technologies, in order to improve the multiplexed characterization of cargo-specific exRNA signatures from clinical samples, toward their implementation in disease biomarkers strategies. AF4, Asymmetric flow field flow fractionation; HF-TFF, hollow fiber tangential flow filtration; GF, gel filtration; F, filtration; RPS, resistive pulse sensing; NTA, nanoparticle tracking analysis; SERS, Surface-enhanced Raman spectroscopy; FLUO, fluorescence; DC, differential centrifugation; DGUC, density-gradient ultracentrifugation
The ERCC2-based effort, involving more than 30 research groups across more than 25 institutions, continues to follow the ERCC1 model, including the establishment of cross-project working groups and coordination of large-scale studies to address shared challenges. Moreover, ERCC2 operates in close coordination with scientific societies, including the American Society of Exosomes and Microvesicles (ASEMV, www.asemv.org) and the International Society of Extracellular Vesicles (ISEV, www.isev.org), in order to update existing exRNA-related protocols, metadata, and data recommendations and guidelines (Mateescu et al., 2017; Théry et al., 2018; Welsh et al., 2020), promote scientific exchange, and emphasize the need for rigor and reproducibility in the field of EV and exRNA research. Although this article is focused on ERCC2 priorities and efforts, it is important to note that ERCC2 operates within the rapidly growing field of exRNA/EV research, where groups across the world are developing innovative technologies for EV bead-based capture (Mitchell et al., 2021), exRNA sequencing (Hulstaert et al., 2021; Munchel et al., 2020), and automated EV enrichment (Y. Chen et al., 2021).
Data Management and Resource Repository (DMRR) for extracellular RNA
The overall programmatic goal of the DMRR is to integrate the efforts of all funded components of the ERCC and serve as a community-wide resource for extracellular RNA (exRNA) standards, protocols, and data.
The generation of high-volume sequencing data has become a routine part of genome research, genetics, and the nascent field of precision medicine. With increasing interest in exRNA biology, the ability of researchers to make discoveries and extract clinically relevant knowledge from these data requires access to new types of tools and analytical approaches. However, the specialized tools and the expertise required to develop and implement effective informatics solutions are beyond the reach of most researchers.
To address this problem, a team led by Aleksandar Milosavljevic, Ph.D. (Baylor College of Medicine, contact PI); David J. Galas, Ph.D. (Pacific Northwest Research Institute); Mark Bender Gerstein, Ph.D. (Yale University); and Matthew E. Roth, Ph.D. (Administrative Core of the DMRR, Baylor College of Medicine) continue to develop and host exRNA portal resources and tools for exRNA biology. The DMRR efforts of ERCC2 build on the infrastructure and computational tools developed in ERCC1 and aim to explore novel mechanisms to collect, identify, and characterize exRNA biotypes. The DMRR team, in collaboration with ERCC2 investigators, explores the use of computational deconvolution to complement physical separation and isolation methodologies, and to better characterize exRNA carriers and their cargo. By further extending the metadata standards established in ERCC1, the DMRR’s exRNA Atlas will continue to expand to become a unique data repository utilizing the emerging federated Data Commons based upon Findable, Accessible, Interoperable, and Reusable (FAIR) principles (Wilkinson et al., 2016). ERCC data are linked with geographically distributed resources, thereby contributing to a continuous cycle of expanding knowledge and free flow of information. An agile DMRR governance structure ensures responsiveness to the input from all ERCC2 members and community stakeholders. Outreach and training is targeted to various levels of expertise to ensure an ongoing engagement with a diverse community of users and contributors. By accomplishing these goals, the DMRR empowers researchers beyond the ERCC to make discoveries using exRNA datasets hosted within the exRNA Atlas and to extract from these datasets new knowledge to improve human health.
Advancing extracellular RNA (exRNA) communication research: Improved isolation and analysis of exRNA carrier subclasses
Circulating extracellular RNAs (exRNAs) can act as intracellular signaling molecules, both locally and systemically, but very little is known about specific components of these communication pathways. One reason for the paucity of understanding about exRNAs is that they are transported in body fluids in association with a variety of carrier vehicles of varying complexity, including EVs, RNPs, and lipoproteins (LPPs). These distinct carriers protect exRNAs from degradation and are thought to contribute to the biodistribution, uptake, and functional impact of exRNAs in target cells. The following projects in ERCC2 aim to develop and evaluate innovative separation tools, technologies, and approaches that will enable the scientific community to rapidly and reproducibly sort complex biofluids into homogeneous carrier populations of EVs (including EV subsets), RNPs, and LPPs, and that will also support high-throughput isolation and analysis of their exRNA content and associated exRNA cargo.
A team led by Hsueh-Chia Chang (University of Notre Dame, contact PI) aims to develop and integrate a suite of high-throughput microfluidic technologies that will encompass the entire analysis process – separation and isolation of exRNA carriers, and sensitive/selective quantification of carriers with specific surface markers and their RNAs, proteins, and lipids. The goal is to be able to complete the analysis for a minimal volume of human plasma in a few hours. The first carrier isolation module is an asymmetric nanopore membrane (ANM) ultrafiltration technology with tangential cross-flow for selective isolation of 50–200 nm EVs from other nanocarriers with >85% purity. ANM is an ion-track membrane with a conic pore geometry that increases throughput, reduces protein fouling, and minimizes EV lysing/fusion (Wang et al., 2021a). Downstream units include a continuous isoelectric fractionation (CIF) module with a pH actuating bipolar membrane (Cheng and Chang, 2014) and a magnetic nanopore membrane (MNM) module for superparamagnetic bead immunocapture. They separate other nanocarriers (LLP and RNP) and fractionate additional subsets of each carrier. Downstream sensor technologies for the various fractionated streams include a membrane sensor for high-abundance exRNA/protein (>106 copies per 100 µL) and LLP/EV (>103 particles per 100 µL) (Ramshani et al., 2019) and digital droplet/microwell platforms (L. Chen et al., 2021; Pan et al., 2018) for lower concentrations.
Bob Coffey (Vanderbilt University Medical Center, contact PI), Al Charest (Beth Israel Deaconess Medical Center, Harvard Medical School, co-PI), et al. plan to uncover exRNA and protein determinants of secreted vesicles and nanoparticles (exomeres and supermeres) by serial ultracentrifugation and optimized density gradient centrifugation (Jeppesen et al., 2019; Zhang et al., 2021; Zhang et al., 2019) and flow cytometric methodologies (Jeppesen et al., 2019; Zhang et al., 2019). They will utilize fluorescence-activated vesicle sorting (FAVS) (Higginbotham et al., 2016) to analyze and sort specific EV subpopulations on a per vesicle basis. RNA carriers will be divided into classic exosomal EVs, non-exosomal EVs, as well as other nanoparticle constituents, such as exomeres and supermeres. The project will compare colorectal cancer (CRC)- and Glioblastoma (GBM)-secreted EV and nanoparticle heterogeneity. The Coffey group has demonstrated the capability of FAVS to purify small EVs derived from CRC and GBM models, patient-derived xenografts (PDXs) and plasma. The team intends to optimize the FAVS pipeline by: (1) validating pre-processing steps separating EVs based on their physical heterogeneity; (2) testing new candidate reagents for use with FAVS that more clearly delineate EV and nanoparticle subsets; and (3) uncovering markers of EVs and nanoparticles. Antibodies to total and conformationally active EGFR, as well as other targets, will be used to purify secreted subsets from these cancers.
A multi-institutional team led by Louise Laurent (University of California San Diego, contact PI) will focus on using immunoaffinity separation (IMS) for isolation of exRNA carrier subclasses (CSs). This transdisciplinary team, with expertise in exRNA and lipoprotein biology (Li et al., 2018b), exRNA biomarker discovery (Chen-Plotkin et al., 2018; Srinivasan et al., 2020; Xiao et al., 2017), exRNA sequencing (Rodosthenous et al., 2020; Srinivasan et al., 2019), low-input proteomics (Tsai et al., 2021), lipidomics (Wang et al., 2021b), and flow cytometry (Görgens and Nolan, 2020; Nolan and Duggan, 2018; Welsh et al., 2020), will work together to develop and apply a rigorous, reproducible, efficient, scalable, and cost-effective workflow for preparative isolation of subtypes of EVs, RNPs, and LPPs for downstream-omics analysis. This team’s initial approach will be to use antibodies conjugated to magnetic nanoparticles, but they will also explore the use of multiplex bead-based flow sorting for even more efficient separation of CSs. The cargo of these CSs will then be characterized using small and long RNAseq, proteomics, and lipidomics. Thus, this program will both develop a rigorous workflow for the separation of exRNA CSs that reproducibly and rapidly produces fractions that are highly enriched for the desired CSs with minimal contamination by other CSs in a cost-effective manner on clinically feasible volumes of input material and generate comprehensive reference profiling data on the RNA, protein, and lipid cargo carried by previously known and novel CSs.
Bogdan Mateescu (University of Zürich) is leading the Purification of exRNA by Immuno-capture and Sorting using Microfluidics (PRISM) project together with Robert Raffai (University of California), Kendall Van Keuren-Jensen (TGEN), and Andrew deMello (ETH Zürich). This transdisciplinary team with expertise in lipoproteins, RNA and EVs biology (Li et al., 2018a, 2018b; Mateescu et al., 2017), exRNA sequencing (Yeri et al., 2017), and microfluidics (Hess et al., 2021; Holzner et al., 2021) aims to improve the sensitivity and specificity of exRNA-based diagnostic assays. First, the team will develop viscoelastic microfluidics platforms allowing the size-based separation of sub-micron particles to promote facile enrichment of exRNA nanocarriers from unprocessed biofluids. Second, the team will establish a discovery pipeline combining bioinformatics, biochemical, and genetic approaches to identify and purify new extracellular RNPs (exRBPs) stabilizing specific exRNA sequences or biotypes (e.g., mRNA and miRNA) in biofluids. Finally, the team will combine microfluidics and multiplexed bead pull-down assays for the parallel isolation of multiple exRNA carriers (exRBPs, EVs, LPPs) from the same biofluid sample, with the objective to increase the discovery rate of informative RNA biomarkers. The efficiency and yield of PRISM technologies will be monitored by molecular analysis, imaging and nano-flow cytometry after spiking biofluids with fluorescent exRNA carriers specifically engineered as part of the project, and compared to gold standard exRNA carrier separation approaches.
An international group led by Kenneth Witwer (Johns Hopkins University) is developing new strategies and controls to ensure that comparisons of exRNA carriers are not confounded by co-isolation or contamination. The team includes experts on EVs, LPPs, and RNPs, along with experts in cutting-edge separation and characterization methods (Arab et al., 2021; Gámbaro et al., 2020; Martin-Jaular et al., 2021; Zhu et al., 2019). They will use combinations of physical and biochemical separation methods to separate up to eight subtypes of exRNA carriers from the same biological samples and with the best achievable purity and then produce “gold standard” proteomic, lipidomic, glycomic, and RNomic datasets for each. Carefully designed “process” controls will identify contaminants and other artifacts. The team will also test and refine techniques using asymmetric field-flow fractionation (AF4) and affinity capture platforms to improve the speed, resolution, and purity. In a follow-on phase, validation studies will be followed by assessments of biological factors such as diet and neurodegeneration. Throughout, attention will be paid to stability of RNA species (Tosar et al., 2021).
Advancing extracellular RNA (exRNA) communication research: Toward single extracellular vesicle (EV) sorting, isolation, and analysis of cargo
The ability to isolate and analyze single EVs from human biofluids would provide a unique opportunity to understand the cell or tissue from which their respective exRNA cargos originate (i.e., cell type-associated EV heterogeneity) and, importantly, add significant depth to our understanding of exRNA communication. However, several critical limitations exist with all available methods for individual EV isolation and analysis. Current approaches to the isolation and study of EVs lack the necessary sensitivity and precision to fully characterize and understand the make-up and the distribution of various EV subpopulations that may be present. Most current EV isolation methodologies, including differential centrifugation, affinity-based isolation, polymer-based precipitation, and size exclusion chromatography, are prone to contamination; for example, affinity methods can be adversely affected by non-specific interactions that can cause co-precipitation of contaminants with EVs. In addition, these bulk techniques can only report information that is averaged over many millions of EVs, and thus cannot be used to understand the high degree of EV heterogeneity. Yet, understanding the diversity of EVs and the cargos they carry is an essential step toward gaining a better understanding of the precise roles EVs play in both physiological and pathophysiological processes. This understanding, in turn, is important toward realizing the potential of EVs in diagnostics (e.g., as a new class of biomarkers) and therapeutics (e.g., as drug carriers).
Thus, the common goal of the following ERCC2 projects is to develop and demonstrate innovative technologies and reagents toward isolating single EVs and to characterize the exRNA cargos associated with specific EV subpopulations based on the cell of origin and their intended target cell. Shedding light on the diversity of exRNAs carried by EVs will allow for a better understanding of the precise role of exRNAs as signaling molecules for both physiological and pathophysiological processes, ultimately accelerating the development of exRNAs as diagnostics and therapeutics.
Daniel T. Chiu (University of Washington, contact PI) et al. are applying new single-molecule technologies to single EV sorting and analysis. These new techniques include a nanoscale flow analyzer and flow sorter for the analysis and sorting of individual EVs with high sensitivity and high throughput as well as a digital PCR approach for quantifying the nucleic-acid contents of EVs. Specifically, the single-molecule nanoscale flow analyzer has been demonstrated to offer unprecedented sensitivity and resolution in sizing and immuno-phenotyping of single EVs and in determining the copy number of surface markers on individual EVs. As a first demonstration, the new instrument has sized and quantified the tetraspanin distribution on individual semen exosomes. The assembled team is well positioned to develop and apply next-generation techniques for deep EV analysis, with expertise in single-molecule technologies (Andronico et al., 2021; Jiang et al., 2021), EV purification and biology (Jauregui et al., 2018; Vojtech et al., 2019), and RNA sequencing and nucleic-acid analysis (Giraldez et al., 2018, 2019).
Saumya Das (Massachusetts General Hospital, contact PI) et al. seek to leverage a novel quantitative single-molecule localization microscopy (qSMLM) approach combined with novel murine models and organ-on-chips for molecular phenotyping of EVs to define their origin and targets. This collaborative multi-PI program includes Tijana Jovanovic-Talisman and Kendall Van Keuren-Jensen, and has expertise in EV RNAseq (Rodosthenous et al., 2020), models to study EV-mediated signaling (Li et al., 2021), and molecular imaging (Lennon et al., 2019). Specifically, this team will identify cell/tissue-specific markers using computational analysis, transcriptomics, and EV-tracking in genetic mouse models. Individual EVs and EV-RNAs will be characterized with qSMLM and nano-flow cytometry to quantify the number of affinity isolated EVs, their size, and key RNA content using molecular beacons. The identified tissue-specific EV-markers and EV-RNAs will be used for (1) determining cellular/tissue contribution to EV-RNAs from Tissue-Chip effluents and (2) assessing dynamic changes in EVs from human plasma from subjects with acute disease or physiological processes. These proposed experiments will help determine the contribution of different tissues to the plasma biofluid EV-RNA landscape at baseline and in response to physiological or disease stressors and enable the use of tissue-specific EV-RNAs to probe disease states with higher sensitivity and fidelity compared to currently available technologies.
Ionita Ghiran (Harvard Medical School, contact PI) et al. from NIH (Jennifer Jones) and BCM (Aleksandar Milosavljevic) seek to apply an integrative, multi-parametric approach to the characterization of the EV surface protein and nucleic acid landscape. These studies will utilize a collaborative approach aimed at streamlining EV analyses and improving antigen detection by: (1) detection of specific RNA/ssDNA molecules in EV populations by combining nano-flow cytometry and molecular beacons (Oliveira et al., 2020), (2) the use of molecular nano-tags for EV antigen detection, using nano-flow cytometry (Morales-Kastresana et al., 2020), and (3) the integration of RNA and protein multidimensional analyses by a dedicated cloud-based, free, bioinformatics pipeline. The results of this effort seek to provide the scientific community with: (1) new methods for EV sorting, detection of specific proteins, RNA/ssDNA molecules on EV subpopulations with a sensitivity currently unattained by any large-scale technique; (2) and protocols necessary for standardization across labs and to translation to clinical practice; and (3) bioinformatics infrastructure necessary for the extraction of subtle but relevant data present in multi-parametric analyses.
Eduardo Reátegui (The Ohio State University, contact PI) et al. are leveraging their expertise in microfluidics, EV characterization, molecular imaging, glioma diagnosis, and RNA profiling (Hu et al., 2017; Reátegui et al., 2018) for developing microfluidic arrays for EV sorting, isolation, and RNA analysis. The system will utilize size exclusion chromatography to first sort biofluid EVs into defined size-based subpopulations, and then distribute each subpopulation into a set of parallel microfluidic channels where each channel is patterned with several microdomains tethered to antibodies for enriched isolating/capturing of specific membrane protein single EVs in the subpopulation. Molecular beacons will be added to detect specific RNA targets via RNA hybridization. Finally, fluorescence-labeled antibodies will be incorporated into each microchannel to quantify the target membrane protein content of the captured single EVs. Development and proof-of-concept demonstrations of this novel technology will be conducted with a small-scale array for selected RNA and protein targets using EVs released from glioblastoma (GBM) cell lines. Once validated, the project will scale up the biochip system design for high-throughput analysis of EVs from serum and cerebrospinal fluid (CSF) from GBM patients, seeking to identify GBM EV subpopulations that may be involved in immunosuppression and/or associated with worse clinical outcomes.
David Routenberg (Meso Scale Diagnostics, contact PI) and his team are developing multi-marker EV screening and isolation techniques to identify and purify specific populations of EVs. These will be combined with advanced flow analyzer instrumentation to measure the distribution of proteins and RNAs within EV subpopulations. The team is developing a new highly multiplexed proximity-based immunosequencing assay for rapid combinatorial analysis of surface markers co-localized on EV populations of interest. This approach will be used to identify unique EV surface signatures representative of EVs from specific cell and tissue types. The screening methods will work in tandem with novel immunoaffinity isolation methods being developed to selectively capture EVs having specific multi-marker signatures. The goal is to produce highly enriched populations of EVs from specific cell and tissue types to facilitate more targeted studies of their molecular contents using single EV analytical methods. The team is also developing advanced flow analyzer instrumentation to improve detection sensitivity for small EVs and their molecular cargo.
David Wong (University of California Los Angeles, contact PI) et al. will develop an innovative technology, AcoustoFluidic Separation (AFS) coupling with Surface Enhanced Raman Spectroscopy (SERS), toward single EV isolation to characterize exRNA cargos associated with EV subpopulations based on the cells of origin, intended target cells, and biofluids. This team will develop: (1) AFS technology as a standard operating procedure (SOP) (Hao et al., 2020a, 2020b; Wang et al., 2020; Xie et al., 2020); (2) SERS for EV fingerprinting and co-localization of exRNA targets to single EVs (Yan et al., 2019; Yu et al., 2020); (3) two independent “Rigor and Reproducibility Labs (R&R Labs)” to evaluate the AFS SOP; and (4) approaches to share strategies, protocols, tools with the broader scientific community. These steps will enable the team to optimize and scale up the AFS SOP; perform sorting, tracking of validated salivary exRNA biomarkers for gastric cancer from cells of origin, to blood, to salivary glands, and to saliva; identify “R&R Labs” to optimize AFS technology; and share strategies, protocols, tools with the broader scientific community. These steps will constitute the foundation of “exRNA Saliva Liquid Biopsy (exRNA-SLB)” where the diagnostic and therapeutic functionality of exRNAs can be fully realized by analyzing with machine learning algorithms for the range of EV subpopulations harboring molecular cargos associated with pathology.
The ERCC2 benchmarking initiative
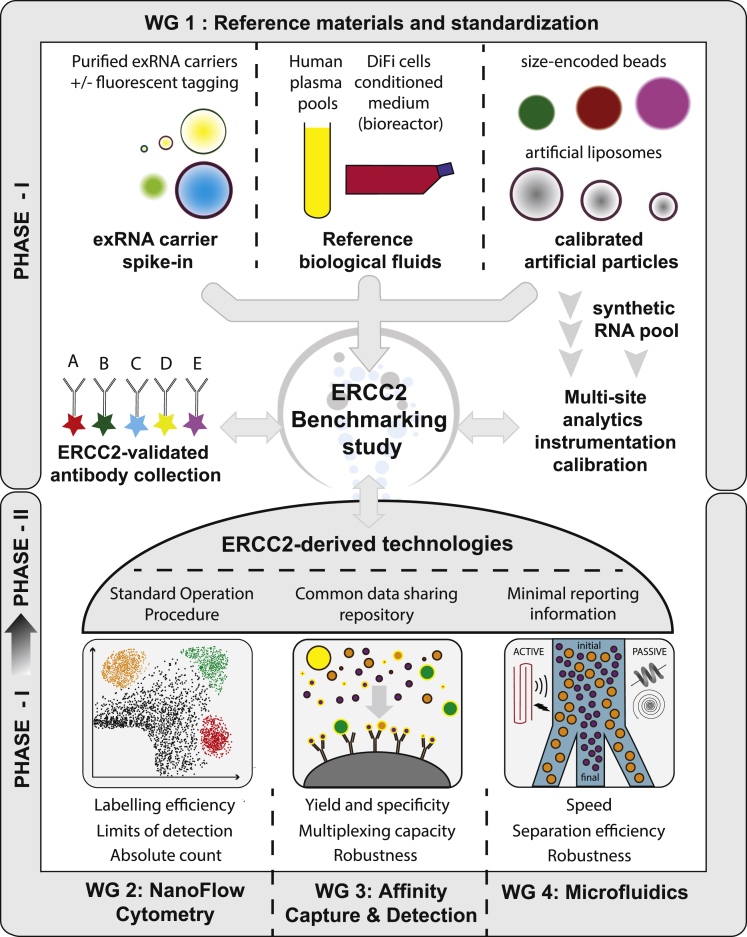
As a means of demonstrating the impact of consortium efforts on improving methods for single EV analysis and exRNA carrier separation, a benchmarking effort is being undertaken within the ERCC2 groups of teams (Figure 3). To improve the resolution, robustness, and reproducibility of single EV studies, Working Group 1 (WG1, Reference Material and Standardization) is responsible for coordinating the assembly and validation of reference materials and reagents, as well as for preparing standardization protocols for use across the Consortium, as well as public dissemination. The activities of WG1 include generation and distribution of large volumes of pooled reference plasma samples prepared by the Laurent group, and conditioned cell culture media and EVs produced from the DiFi colorectal cancer cell line in a hollow-fiber bioreactor by the Coffey group. The Mateescu and Raffai groups have jointly produced a series of highly purified fluorescently labeled exRNA carriers (lipoproteins, EVs, RNA granules, etc.) that can be used as spike-in reagents for monitoring exRNA carrier separation efficiency. WG2 (NanoFlow Cytometry) is focused on benchmarking the performance of nano-flow cytometry methods for phenotyping of single EVs. Working together with WG1, WG2 is validating exRNA carriers and synthetic particles for calibrating instrumentation among participating sites, with the primary objective of establishing standard operating procedures (SOPs) and analysis methods for determining the limit of detection and absolute count of single EVs, as well as for benchmarking all nano-flow cytometry approaches developed within ERCC2. WG3 (Affinity Capture & Detection) is testing and validating a series of reagents for capturing and detecting exRNA carriers. They will establish benchmarks for measuring yield, reproducibility, sensitivity, and specificity of exRNA carrier separation and analysis approaches used across the ERCC2. Finally, WG4 (Microfluidics) is focusing on microfluidics-based exRNA carrier separation and detection. In coordination with WG1 and WG2, WG4 is leveraging selected reference materials to benchmark the performance (e.g., speed, sorting efficiency, and limit of detection limit) of the microfluidics technologies developed by ERCC2 members. Rigor and reproducibility efforts in the ERCC2 include the transfer of instruments and protocols to laboratories within and outside the Consortium for validation studies.
Figure 3.
The ERCC2 benchmarking initiative
The ERCC2 groups are working together on a benchmarking study aiming at evaluating the performance of the ERCC2-driven technologies. This initiative not only aims at homogenizing practice and calibration tools, but also to promote rigor and reproducibility within ERCC2 and members of the scientific community at large.
ERCC2: Early impact
To date, the ERCC2 effort has already contributed to the body of knowledge about EVs and other exRNA carriers and produced several resources for the field. These include not only advances in instrumentation and the development of new methods, but also the identification of functional biological signatures for a variety of diseases and physiological states.
The Witwer team has found that inhibition of extracellular RNases dramatically alters extracellular RNA profiles, stabilizing (non-vesicular) extracellular ribosomes and tRNAs (Tosar et al., 2020). They demonstrated the extracellular biogenesis of non-vesicular rRNA and tRNA-derived fragments, with strong implications in our understanding of RNA release mechanisms (Tosar et al., 2021). This also highlights the importance of considering extracellular RNA stability when using non-vesicular RNAs as biomarkers (Tosar and Cayota, 2020). Additional ERCC2 work from the Witwer group has been published relating to extracellular ribosomes and tRNA-derived fragments (Gámbaro et al., 2020); transcriptomic analysis of EVs released from retinal epithelial cells (Ahn et al., 2021) and during HIV infection (Zhao et al., 2020); in-depth proteomic analysis of EVs during HIV-1 infection as part of a collaboration with the Thery lab (Martin-Jaular et al., 2021), and the development and comparison of methods for EV and lipoprotein separation and characterization (Arab et al., 2021; Huang et al., 2020; Royo et al., 2020; Zheng et al., 2021)
The Coffey group recently published the characterization of a new class of exRNA carriers, named supermeres (supernatant of exomeres) (Zhang et al., 2021), which are distinct from the previously characterized exomeres, lipoproteins and EVs. Importantly, they discovered that more exRNAs are associated with supermeres than small EVs and exomeres, and that the cargo of supermeres is characterized by highly enriched RNAs (e.g., miR-1246) and proteins (e.g., TGFBI), therefore opening new possibilities for exRNA-based biomarker analysis.
The Reátegui team has published manuscripts relating to three key technology components that would enable the molecular analysis of single EVs, including an immunomagnetic sequential ultrafiltration-based EV purification method (Zhang et al., 2021), antibody-based microdomains for microfluidic channels (Rima et al., 2020), and a large-scale generation of RNA encapsulated extracellular vesicles through cellular nano-electroporation (Yang et al., 2020).
In the field of cardiovascular disease, Das and Van Keuren-Jensen have reported new methods and pipelines for performing long RNAseq (transcriptome) from plasma and EVs with a goal of identifying tissue-specific transcripts (Rodosthenous et al., 2020), and further demonstrated a role for RBC-derived EVs in a mouse model of ischemic heart disease (Das et al., 2019).
The Mateescu team has reported new technological developments and the potential impact of specific exRNA carriers. Robert Raffai’s group gained insight into the function of macrophage-exosomes as potent modulators of inflammation via their microRNA cargo (Bouchareychas et al., 2020, 2021). Exosomes produced by macrophages cultured with the anti-inflammatory cytokine IL-4 or in the presence of elevated glucose levels were enriched with miRNAs that were responsible for controlling excessive hematopoiesis, monocytosis, and atherosclerotic lesion inflammation in mice with hyperlipidemia. Ongoing studies are exploring the value of macrophage exosomes as biomarkers of cardiovascular disease severity and as therapeutics for other inflammatory diseases. Andrew de-Mello’s group has developed a novel microfluidics technology based on oscillatory viscoelastic focusing to separate nanoscale species, and in particular EVs, from a complex size mixture (Asghari et al., 2020). Building on similar principles, they are now developing a next-generation microfluidics device to enable fractionation of unprocessed biofluids for exRNA analysis.
Also, in the field of microfluidics, the Wong and Huang teams have published a study using an acoustofluidic platform for EV isolation, including saliva. Specifically, the Huang group specializes in the use of surface acoustic waves to manipulate and isolate biological nanoparticles based on differences in their size, density, and compressibility. They demonstrated that salivary exosomes, packaged with HPV-associated biomarkers, are efficiently enriched by the acoustofluidic separation of salivary exosomes for enhanced diagnostic and prognostic performance for HPV-associated oropharyngeal cancer (OPC) (Wang et al., 2020). The acoustofluidic platform has achieved high-purity and high-yield salivary exosome isolation for downstream salivary exosome-based liquid biopsy applications. Another study demonstrated the use of such acoustofluidic methods for separation based on the biophysical properties of density and compressibility, irrespective of sizes (Xie et al., 2020). Finally, integration of zinc oxide nanoarrays into the acoustofluidic devices has enabled the use of ZnO-based surface-enhanced Raman spectroscopy (SERS) detection, for further enrichment of other attributes and types of molecules, including DNA oligonucleotides, exosomes, and E. coli bacteria (Hao et al., 2020a; 2020b).
Translational impact of ERCC2 program
The primary motivation behind the ERCC2 projects is to translate advances in basic exRNA research into clinical diagnostic and therapeutic applications. At a fundamental level, ERCC2 seeks to develop, test, and share tools that will improve characterization of those extracellular materials, by improving accuracy, calibration, and reporting. As described above, this focus is exemplified in one form by the ERCC2-wide EV flow cytometry efforts to produce widely available tools and results, and in another form by the development of an ERCC2 database of antibodies toward the harmonization of assay results across laboratories. Both of these examples will lead to open web-based resources for the wider research community. Similarly, large scale production of DiFi cell culture conditioned media and EVs from hollow fiber bioreactor cultures provides a robust source of EVs with canonical tumor-associated EV markers, such as EGFR, to evaluate and validate cross-Consortium harmonization efforts. The development of new technologies is leading in some cases toward improvement of our basic laboratory EV assay capabilities (such as single molecule/single EV detection and enumeration technologies), and in other cases toward more immediate production of clinical laboratory workflow-compatible devices (such as microfluidic devices), EV isolation and characterization tools, and other tools for performing EV and EV cargo measurements with more rapid timeframes, lower volumes, point-of-care ease of use, and lower cost than conventional or legacy methods. The dynamic interplay between these basic and applied goals and sources of expertise in the ERCC2 group are required to achieve rigor and impact of the ERCC2 efforts within the Consortium and promote the use of ERCC2 products as the foundation for the next stages of translational clinical diagnostic and therapeutic uses of exRNAs and exRNA carriers in the years to come.
Conclusion
As translational research teams worldwide seek to identify exRNA biomarkers of health and disease in biofluids, with publications relating to this topic expanding nearly exponentially in the past decade, technical challenges still remain in accurate characterization, sorting, and analysis of extracellular vesicles and other macromolecular RNA carriers in those biofluids. In Phase 2 of the Extracellular RNA Communication Consortium (ERCC2), participating investigators continue to work to generate foundational knowledge and develop and disseminate technologies and resources to advance the exRNA field. In contrast to ERCC1, which was comprised of studies focused on discovering the mechanisms of exRNA biogenesis, identification of exRNA biomarkers, and development of exRNA-based therapeutics, ERCC2 aims to develop technologies for separation of exRNA carrier subclasses and single-vesicle analysis. ERCC2 also aims to expand the computational tools and resources available through the exRNA Portal developed during ERCC1, by providing public access to additional datasets and incorporating new tools for meta-analyses of data from multiple datasets utilizing individual sample-level raw data. ERCC2 investigators have initiated collaborative efforts to develop standard biofluid samples to compare outputs from diverse Carrier Subclass separation and characterization technologies, as well as to perform rigorous benchmarking studies, and invite members of the broader exRNA community to participate in these initiatives.
Acknowledgments
We acknowledge program leadership by members of the NIH Extracellular RNA Communication Workgroup, especially Kevin Howcroft, Danilo Tagle, John Satterlee, Patricia Labosky, Christine Happel, Nic Johnston, Lillian L. Kuo, Tania B. Lombo, George A. McKie, Margaret Ochocinska, Daniel Shaughnessy, Dobrila Rudnicki, Anna Sadusky, Pothur R. Srinivas, Shurjo K. Sen, Alexandra Ainsztein, Angela Ambrosi, Preethi Chander, Aniruddha Ganguly, Daniel Gossett, Max Guo, Dinah S. Singer, Susmita Sahoo, Smita Krishnaswamy and Joni L. Rutter. We also acknowledge valuable feedback from the program’s External Scientific Panel members: Xandra Breakefield, Michael Davis, Scott Fraser, Hakho Lee, Tushar Patel, Marina Sirota, and Siyang Zheng. We would like to thank Elke Norwig-Eastaugh for her valuable contribution in organizing the ERCC investigator meetings. We would like to thank all of the teams engaged in the launch of ERCC2, and the investigators and contributors to the initial ERCC programs that formed the foundation of these next stage studies. The projects described in this article are supported by the NIH Common Fund through the ERCC Program awards 5UG3CA241703, 5UG3CA241687, 5UG3CA241684, 5UG3TR002874, 5UG3CA241685, 5UG3TR002878, 5UG3TR002881, 5U54DA049098, 5UG3TR002884, 5UG3TR002886, 5UG3CA241694, and 5UG3TR002978.
Declaration of interests
Crislyn D’Souza-Schorey, Satyajyoti Senapati and Hsueh-Chia Chang hold equity in AgenDx Biosciences. Hsueh-Chia Chang is a cofounder of Aopia Biosciences and holds equity. Daniel T. Chiu has financial interest and is a scientific founder and/or board member of the following companies and their respective affiliates: Micareo, Inc, Lamprogen, Inc, Cellectricon AB, and Fluicell AB. Saumya Das is a founding member of Dyrnamix Inc. that did not play any role in this study. Kendal Van Keuren-Jensen is a board member of Dyrnamix Inc. and HTG that did not play any role in this study. Aleksandar Milosavljevic is a founder of IP Genesis, Inc. that did not play any role in this study. Louise C. Laurent has stock in Illumina, Inc. Anil K. Sood has the following disclosures: Consultant (Merck, Kiyatec), shareholder (BioPath), research funding (MTrap). John Nolan holds equity in Cellarcus Biosciences Inc. L. James Lee holds equity at Spot Biosystems. David A. Routenberg, Sigal Shachar and Alexander K. Tucker-Schwartz are employees of Meso Scale Diagnostics, LLC. Andrey Turchinovich is founder of SciBerg e.Kfm, Mannheim, Germany. Angela M. Zivkovic is cofounder and board member at Innate Biology, Inc.
Contributor Information
Bogdan Mateescu, Email: bogdan.mateescu@uzh.ch.
Jennifer C. Jones, Email: jennifer.jones2@nih.gov.
Louise C. Laurent, Email: llaurent@health.ucsd.edu.
References
- Ahn J.Y., Datta S., Bandeira E., Cano M., Mallick E., Rai U., Powell B., Tian J., Witwer K.W., Handa J.T., Paulaitis M.E. Release of extracellular vesicle miR-494-3p by ARPE-19 cells with impaired mitochondria. Biochim. Biophys. Acta Gen. Subj. 2021;1865:129598. doi: 10.1016/j.bbagen.2020.129598. [DOI] [PubMed] [Google Scholar]
- Al Amir Dache Z., Otandault A., Tanos R., Pastor B., Meddeb R., Sanchez C., Arena G., Lasorsa L., Bennett A., Grange T., et al. Blood contains circulating cell-free respiratory competent mitochondria. FASEB. J. 2020;34:3616–3630. doi: 10.1096/fj.201901917RR. [DOI] [PubMed] [Google Scholar]
- Andronico L.A., Jiang Y., Jung S.-R., Fujimoto B.S., Vojtech L., Chiu D.T. Sizing extracellular vesicles using membrane dyes and a single molecule-sensitive flow analyzer. Anal. Chem. 2021;93:5897–5905. doi: 10.1021/acs.analchem.1c00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab T., Mallick E.R., Huang Y., Dong L., Liao Z., Zhao Z., Gololobova O., Smith B., Haughey N.J., Pienta K.J., et al. Characterization of extracellular vesicles and synthetic nanoparticles with four orthogonal single-particle analysis platforms. J. Extracell. Vesicles. 2021;10:e12079. doi: 10.1101/2020.08.04.237156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asghari M., Cao X., Mateescu B., van Leeuwen D., Aslan M.K., Stavrakis S., deMello A.J. Oscillatory viscoelastic microfluidics for efficient focusing and separation of nanoscale species. ACS Nano. 2020;14:422–433. doi: 10.1021/acsnano.9b06123. [DOI] [PubMed] [Google Scholar]
- Avila Cobos F., Alquicira-Hernandez J., Powell J.E., Mestdagh P., De Preter K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 2020;11:5650. doi: 10.1038/s41467-020-19015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedikter B.J., Bouwman F.G., Vajen T., Heinzmann A.C.A., Grauls G., Mariman E.C., Wouters E.F.M., Savelkoul P.H., Lopez-Iglesias C., Koenen R.R., et al. Ultrafiltration combined with size exclusion chromatography efficiently isolates extracellular vesicles from cell culture media for compositional and functional studies. Sci. Rep. 2017;7:15297. doi: 10.1038/s41598-017-15717-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkowitz R., Fisher J., Goff S.P. RNA packaging. Curr. Top. Microbiol. Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- Bouchareychas L., Duong P., Covarrubias S., Alsop E., Phu T.A., Chung A., Gomes M., Wong D., Meechoovet B., Capili A., et al. Macrophage exosomes resolve atherosclerosis by regulating hematopoiesis and inflammation via MicroRNA cargo. Cell Rep. 2020;32:107881. doi: 10.1016/j.celrep.2020.107881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchareychas L., Duong P., Phu T.A., Alsop E., Meechoovet B., Reiman R., Ng M., Yamamoto R., Nakauchi H., Gasper W.J., et al. High glucose macrophage exosomes enhance atherosclerosis by driving cellular proliferation & hematopoiesis. iScience. 2021;24:102847. doi: 10.1016/j.isci.2021.102847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassedy A., Parle-McDermott A., O’Kennedy R. Virus detection: a review of the current and emerging molecular and immunological methods. Front. Mol. Biosci. 2021;8:637559. doi: 10.3389/fmolb.2021.637559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Huang C., Wu Q., Jiang L., Chen S., Chen L. Circular RNAs expression profiles in plasma exosomes from early-stage lung adenocarcinoma and the potential biomarkers. J. Cell. Biochem. 2020;121:2525–2533. doi: 10.1002/jcb.29475. [DOI] [PubMed] [Google Scholar]
- Cheng L.-J., Chang H.-C. Switchable pH actuators and 3D integrated salt bridges as new strategies for reconfigurable microfluidic free-flow electrophoretic separation. Lab Chip. 2014;14:979–987. doi: 10.1039/c3lc51023a. [DOI] [PubMed] [Google Scholar]
- Chen L., Yadav V., Zhang C., Huo X., Wang C., Senapati S., Chang H.-C. Elliptical pipette generated large microdroplets for POC visual ddPCR quantification of low viral load. Anal. Chem. 2021;93:6456–6462. doi: 10.1021/acs.analchem.1c00192. [DOI] [PubMed] [Google Scholar]
- Chen-Plotkin A.S., Albin R., Alcalay R., Babcock D., Bajaj V., Bowman D., Buko A., Cedarbaum J., Chelsky D., Cookson M.R., et al. Finding useful biomarkers for Parkinson’s disease. Sci. Transl. Med. 2018;10 doi: 10.1126/scitranslmed.aam6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Zhu Q., Cheng L., Wang Y., Li M., Yang Q., Hu L., Lou D., Li J., Dong X., et al. Exosome detection via the ultrafast-isolation system: EXODUS. Nat. Methods. 2021;18:212–218. doi: 10.1038/s41592-020-01034-x. [DOI] [PubMed] [Google Scholar]
- Chevillet J.R., Kang Q., Ruf I.K., Briggs H.A., Vojtech L.N., Hughes S.M., Cheng H.H., Arroyo J.D., Meredith E.K., Gallichotte E.N., et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. USA. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A., Valkov N., Salvador A.M., Kur I., Ziegler O., Yeri A., Garcia F.C., Lu S., Khamesra A., Xiao C., et al. Red blood cell-derived extracellular vesicles mediate intercellular communication in ischemic heart failure. Cold Spring Harb. 2019 doi: 10.1101/624841. [DOI] [Google Scholar]
- Das S., Extracellular RNA Communication Consortium. Ansel K.M., Bitzer M., Breakefield X.O., Charest A., Galas D.J., Gerstein M.B., Gupta M., et al. The extracellular RNA communication consortium: establishing foundational knowledge and technologies for extracellular RNA research. Cell. 2019;177:231–242. doi: 10.1016/j.cell.2019.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decamps C., Privé F., Bacher R., Jost D., Waguet A., HADACA consortium. Houseman E.A., Lurie E., Lutsik P., Milosavljevic A., et al. Guidelines for cell-type heterogeneity quantification based on a comparative analysis of reference-free DNA methylation deconvolution software. BMC Bioinformatics. 2020;21:16. doi: 10.1186/s12859-019-3307-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rie D., Abugessaisa I., Alam T., Arner E., Arner P., Ashoor H., Åström G., Babina M., Bertin N., Burroughs A.M., et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017;35:872–878. doi: 10.1038/nbt.3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbiano F., Corsi J., Gurrieri E., Trevisan C., Notarangelo M., D’Agostino V.G. RNA packaging into extracellular vesicles: an orchestra of RNA-binding proteins? J. Extracell. Vesicles. 2020;10:e12043. doi: 10.1002/jev2.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gámbaro F., Li Calzi M., Fagúndez P., Costa B., Greif G., Mallick E., Lyons S., Ivanov P., Witwer K., Cayota A., Tosar J.P. Stable tRNA halves can be sorted into extracellular vesicles and delivered to recipient cells in a concentration-dependent manner. RNA Biol. 2020;17:1168–1182. doi: 10.1080/15476286.2019.1708548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez M.D., Spengler R.M., Etheridge A., Godoy P.M., Barczak A.J., Srinivasan S., De Hoff P.L., Tanriverdi K., Courtright A., Lu S., et al. Comprehensive multi-center assessment of small RNA-seq methods for quantitative miRNA profiling. Nat. Biotechnol. 2018;36:746–757. doi: 10.1038/nbt.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez M.D., Spengler R.M., Etheridge A., Goicochea A.J., Tuck M., Choi S.W., Galas D.J., Tewari M. Phospho-RNA-seq: a modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 2019;38 doi: 10.15252/embj.2019101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görgens A., Nolan J.P. Aiming to compare Apples to Apples: analysis of extracellular vesicles and other nanosized particles by flow cytometry. Cytometry. 2020;97:566–568. doi: 10.1002/cyto.a.24173. [DOI] [PubMed] [Google Scholar]
- Gruner H.N., McManus M.T. Examining the evidence for extracellular RNA function in mammals. Nat. Rev. Genet. 2021;22:448–458. doi: 10.1038/s41576-021-00346-8. [DOI] [PubMed] [Google Scholar]
- Hao N., Liu P., Bachman H., Pei Z., Zhang P., Rufo J., Wang Z., Zhao S., Huang T.J. Acoustofluidics-Assisted engineering of multifunctional three-dimensional zinc oxide nanoarrays. ACS Nano. 2020;14:6150–6163. doi: 10.1021/acsnano.0c02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N., Pei Z., Liu P., Bachman H., Naquin T.D., Zhang P., Zhang J., Shen L., Yang S., Yang K., et al. Acoustofluidics-Assisted fluorescence-SERS bimodal biosensors. Small. 2020;16:e2005179. doi: 10.1002/smll.202005179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess D., Dockalova V., Kokkonen P., Bednar D., Damborsky J., deMello A., Prokop Z., Stavrakis S. Exploring mechanism of enzyme catalysis by on-chip transient kinetics coupled with global data analysis and molecular modeling. Chem. 2021;7:1066–1079. doi: 10.1016/j.chempr.2021.02.011. [DOI] [Google Scholar]
- Higginbotham J.N., Zhang Q., Jeppesen D.K., Scott A.M., Manning H.C., Ochieng J., Franklin J.L., Coffey R.J. Identification and characterization of EGF receptor in individual exosomes by fluorescence-activated vesicle sorting. J. Extracell. Vesicles. 2016;5:29254. doi: 10.3402/jev.v5.29254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzner G., Mateescu B., van Leeuwen D., Cereghetti G., Dechant R., Stavrakis S., deMello A. High-throughput multiparametric imaging flow cytometry: toward diffraction-limited sub-cellular detection and monitoring of sub-cellular processes. Cell Rep. 2021;34:108824. doi: 10.1016/j.celrep.2021.108824. [DOI] [PubMed] [Google Scholar]
- Huang Y., Cheng L., Turchinovich A., Mahairaki V., Troncoso J.C., Pletniková O., Haughey N.J., Vella L.J., Hill A.F., Zheng L., Witwer K.W. Influence of species and processing parameters on recovery and content of brain tissue-derived extracellular vesicles. J. Extracell. Vesicles. 2020;9:1785746. doi: 10.1080/20013078.2020.1785746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Sheng Y., Kwak K.J., Shi J., Yu B., Lee L.J. A signal-amplifiable biochip quantifies extracellular vesicle-associated RNAs for early cancer detection. Nat. Commun. 2017;8:1683. doi: 10.1038/s41467-017-01942-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert E., Decock A., Morlion A., Everaert C., Verniers K., Nuytens J., Nijs N., Schroth G.P., Kuersten S., Gross S.M., et al. Messenger RNA capture sequencing of extracellular RNA from human biofluids using a comprehensive set of spike-in controls. STAR Protoc. 2021;2:100475. doi: 10.1016/j.xpro.2021.100475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulstaert E., Morlion A., Avila Cobos F., Verniers K., Nuytens J., Vanden Eynde E., Yigit N., Anckaert J., Geerts A., Hindryckx P., et al. Charting extracellular transcriptomes in the human biofluid RNA atlas. Cell Rep. 2020;33:108552. doi: 10.1016/j.celrep.2020.108552. [DOI] [PubMed] [Google Scholar]
- Hutchins E., Reiman R., Winarta J., Beecroft T., Richholt R., De Both M., Shahbander K., Carlson E., Janss A., Siniard A., et al. Extracellular circular RNA profiles in plasma and urine of healthy, male college athletes. Sci. Data. 2021;8:276. doi: 10.1038/s41597-021-01056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jauregui R., Srinivasan S., Vojtech L.N., Gammill H.S., Chiu D.T., Hladik F., Stayton P.S., Lai J.J. Temperature-responsive magnetic nanoparticles for enabling affinity separation of extracellular vesicles. ACS Appl. Mater. Interfaces. 2018;10:33847–33856. doi: 10.1021/acsami.8b09751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen D.K., Fenix A.M., Franklin J.L., Higginbotham J.N., Zhang Q., Zimmerman L.J., Liebler D.C., Ping J., Liu Q., Evans R., et al. Reassessment of exosome composition. Cell. 2019;177:428–445.e18. doi: 10.1016/j.cell.2019.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Andronico L.A., Jung S.-R., Chen H., Fujimoto B., Vojtech L., Chiu D.T. High-throughput counting and superresolution mapping of tetraspanins on exosomes using a single-molecule sensitive flow technique and transistor-like semiconducting polymer dots. Angew. Chem., Int. Ed. Engl. 2021;60:13470–13475. doi: 10.1002/anie.202103282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölling M., Haddad G., Wegmann U., Kistler A., Bosakova A., Seeger H., Hübel K., Haller H., Mueller T., Wüthrich R.P., Lorenzen J.M. Circular RNAs in urine of kidney transplant patients with acute T cell-mediated Allograft rejection. Clin. Chem. 2019;65:1287–1294. doi: 10.1373/clinchem.2019.305854. [DOI] [PubMed] [Google Scholar]
- Lennon K.M., Wakefield D.L., Maddox A.L., Brehove M.S., Willner A.N., Garcia-Mansfield K., Meechoovet B., Reiman R., Hutchins E., Miller M.M., et al. Single molecule characterization of individual extracellular vesicles from pancreatic cancer. J. Extracell. Vesicles. 2019;8:1685634. doi: 10.1080/20013078.2019.1685634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Salvador A.M., Li G., Valkov N., Ziegler O., Yeri A., Yang Xiao C., Meechoovet B., Alsop E., Rodosthenous R.S., et al. Mir-30d regulates cardiac remodeling by intracellular and paracrine signaling. Circ. Res. 2021;128:e1–e23. doi: 10.1161/CIRCRESAHA.120.317244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wong D.K., Hong K.Y., Raffai R.L. Cushioned-density gradient ultracentrifugation (C-DGUC): a refined and high performance method for the isolation, characterization, and use of exosomes. Methods Mol. Biol. 2018;1740:69–83. doi: 10.1007/978-1-4939-7652-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K., Wong D.K., Luk F.S., Kim R.Y., Raffai R.L. Isolation of plasma lipoproteins as a source of extracellular RNA. Methods Mol. Biol. 2018;1740:139–153. doi: 10.1007/978-1-4939-7652-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S.W., Rosen M.K., Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol. Cell. 2015;60:208–219. doi: 10.1016/j.molcel.2015.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzi L., Chiu H.-S., Avila Cobos F., Gross S., Volders P.-J., Cannoodt R., Nuytens J., Vanderheyden K., Anckaert J., Lefever S., et al. The RNA Atlas expands the catalog of human non-coding RNAs. Nat. Biotechnol. 2021;39:1467. doi: 10.1038/s41587-021-00936-1. [DOI] [PubMed] [Google Scholar]
- Martin-Jaular L., Nevo N., Schessner J.P., Tkach M., Jouve M., Dingli F., Loew D., Witwer K.W., Ostrowski M., Borner G.H.H., Théry C. Unbiased proteomic profiling of host cell extracellular vesicle composition and dynamics upon HIV-1 infection. EMBO J. 2021;40:e105492. doi: 10.15252/embj.2020105492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateescu B., Kowal E.J.K., van Balkom B.W.M., Bartel S., Bhattacharyya S.N., Buzás E.I., Buck A.H., de Candia P., Chow F.W.N., Das S., et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA - an ISEV position paper. J. Extracell. Vesicles. 2017;6:1286095. doi: 10.1080/20013078.2017.1286095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Max K.E.A., Bertram K., Akat K.M., Bogardus K.A., Li J., Morozov P., Ben-Dov I.Z., Li X., Weiss Z.R., Azizian A., et al. Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc. Natl. Acad. Sci. USA. 2018;115:E5334–E5343. doi: 10.1073/pnas.1714397115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., Johnson R., Segrè A.V., Djebali S., Niarchou A., GTEx Consortium. Wright F.A., Lappalainen T., Calvo M., Getz G., Dermitzakis E.T., Ardlie K.G., Guigó R. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y., Asghari M., Aslan M.K., Yilmaz A., Mateescu B., Stavrakis S., deMello A.J. Microfluidics for extracellular vesicle separation and mimetic synthesis: recent advances and future perspectives. Chem. Eng. J. 2021;404:126110. doi: 10.1016/j.cej.2020.126110. [DOI] [Google Scholar]
- Mitchell M.I., Ben-Dov I.Z., Liu C., Ye K., Chow K., Kramer Y., Gangadharan A., Park S., Fitzgerald S., Ramnauth A., et al. Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): a customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs. J. Extracell. Vesicles. 2021;10:e12110. doi: 10.1002/jev2.12110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Kastresana A., Welsh J.A., Jones J.C. Detection and sorting of extracellular vesicles and viruses using nanoFACS. Curr. Protoc. Cytom. 2020;95:e81. doi: 10.1002/cpcy.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morello M., Minciacchi V.R., de Candia P., Yang J., Posadas E., Kim H., Griffiths D., Bhowmick N., Chung L.W.K., Gandellini P., et al. Large oncosomes mediate intercellular transfer of functional microRNA. Cell Cycle. 2013;12:3526–3536. doi: 10.4161/cc.26539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munchel S., Rohrback S., Randise-Hinchliff C., Kinnings S., Deshmukh S., Alla N., Tan C., Kia A., Greene G., Leety L., et al. Circulating transcripts in maternal blood reflect a molecular signature of early-onset preeclampsia. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aaz0131. [DOI] [PubMed] [Google Scholar]
- Murillo O.D., Thistlethwaite W., Rozowsky J., Subramanian S.L., Lucero R., Shah N., Jackson A.R., Srinivasan S., Chung A., Laurent C.D., et al. exRNA atlas analysis reveals distinct extracellular RNA cargo types and their carriers present across human biofluids. Cell. 2019;177:463–477.e15. doi: 10.1016/j.cell.2019.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan J.P., Duggan E. Analysis of individual extracellular vesicles by flow cytometry. Methods Mol. Biol. 2018;1678:79–92. doi: 10.1007/978-1-4939-7346-0_5. [DOI] [PubMed] [Google Scholar]
- Oliveira G.P., Jr., Zigon E., Rogers G., Davodian D., Lu S., Jovanovic-Talisman T., Jones J., Tigges J., Tyagi S., Ghiran I.C. Detection of extracellular vesicle RNA using molecular beacons. iScience. 2020;23:100782. doi: 10.1016/j.isci.2019.100782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z., Men Y., Senapati S., Chang H.-C. Immersed AC electrospray (iACE) for monodispersed aqueous droplet generation. Biomicrofluidics. 2018;12:044113. doi: 10.1063/1.5048307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S., Faridani O.R., Björklund A.K., Winberg G., Sagasser S., Sandberg R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- Ramshani Z., Zhang C., Richards K., Chen L., Xu G., Stiles B.L., Hill R., Senapati S., Go D.B., Chang H.-C. Extracellular vesicle microRNA quantification from plasma using an integrated microfluidic device. Commun. Biol. 2019;2:189. doi: 10.1038/s42003-019-0435-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen M., Reddy M., Nolan R., Camunas-Soler J., Khodursky A., Scheller N.M., Cantonwine D.E., Engelbrechtsen L., Mi J.D., Dutta A., et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature. 2022;601:422–427. doi: 10.1038/s41586-021-04249-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reátegui E., van der Vos K.E., Lai C.P., Zeinali M., Atai N.A., Aldikacti B., Floyd F.P., H Khankhel A., Thapar V., Hochberg F.H., et al. Engineered nanointerfaces for microfluidic isolation and molecular profiling of tumor-specific extracellular vesicles. Nat. Commun. 2018;9:175. doi: 10.1038/s41467-017-02261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M., et al. The human cell atlas. Elife. 2017:6. doi: 10.7554/eLife.27041. Human Cell Atlas Meeting Participants. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rima X.Y., Walters N., Nguyen L.T.H., Reátegui E. Surface engineering within a microchannel for hydrodynamic and self-assembled cell patterning. Biomicrofluidics. 2020;14:014104. doi: 10.1063/1.5126608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodosthenous R.S., Hutchins E., Reiman R., Yeri A.S., Srinivasan S., Whitsett T.G., Ghiran I., Silverman M.G., Laurent L.C., Van Keuren-Jensen K., Das S. Profiling extracellular long RNA transcriptome in human plasma and extracellular vesicles for biomarker discovery. iScience. 2020;23:101182. doi: 10.1016/j.isci.2020.101182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo F., Théry C., Falcón-Pérez J.M., Nieuwland R., Witwer K.W. Methods for separation and characterization of extracellular vesicles: results of a worldwide survey performed by the ISEV rigor and standardization subcommittee. Cells. 2020:9. doi: 10.3390/cells9091955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Treacy R., Herrero T., Olsen R., Leonardo T.R., Zhang X., DeHoff P., To C., Poling L.G., Fernando A., et al. Discovery and verification of extracellular miRNA biomarkers for non-invasive prediction of pre-eclampsia in asymptomatic women. Cell Rep. Med. 2020;1 doi: 10.1016/j.xcrm.2020.100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Yeri A., Cheah P.S., Chung A., Danielson K., De Hoff P., Filant J., Laurent C.D., Laurent L.D., Magee R., et al. Small RNA sequencing across diverse biofluids identifies optimal methods for exRNA isolation. Cell. 2019;177:446–462.e16. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suea-Ngam A., Bezinge L., Mateescu B., Howes P.D., deMello A.J., Richards D.A. Enzyme-Assisted nucleic acid detection for infectious disease diagnostics: moving toward the point-of-care. ACS Sens. 2020;5:2701–2723. doi: 10.1021/acssensors.0c01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Cayota A. Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 2020;17:1149–1167. doi: 10.1080/15476286.2020.1729584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Segovia M., Castellano M., Gámbaro F., Akiyama Y., Fagúndez P., Olivera Á., Costa B., Possi T., Hill M., et al. Fragmentation of extracellular ribosomes and tRNAs shapes the extracellular RNAome. Nucleic Acids Res. 2020;48:12874–12888. doi: 10.1093/nar/gkaa674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosar J.P., Witwer K., Cayota A. Revisiting extracellular RNA release, processing, and function. Trends Biochem. Sci. 2021;46:438–445. doi: 10.1016/j.tibs.2020.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai C.-F., Zhang P., Scholten D., Martin K., Wang Y.-T., Zhao R., Chrisler W.B., Patel D.B., Dou M., Jia Y., et al. Surfactant-assisted one-pot sample preparation for label-free single-cell proteomics. Commun. Biol. 2021;4:265. doi: 10.1038/s42003-021-01797-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Weiz L., Langheinz A., Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–7233. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlén M., Fagerberg L., Hallström B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson Å., Kampf C., Sjöstedt E., Asplund A., et al. Proteomics.Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- Vojtech L., Zhang M., Davé V., Levy C., Hughes S.M., Wang R., Calienes F., Prlic M., Nance E., Hladik F. Extracellular vesicles in human semen modulate antigen-presenting cell function and decrease downstream antiviral T cell responses. PLoS One. 2019;14:e0223901. doi: 10.1371/journal.pone.0223901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Sensale S., Pan Z., Senapati S., Chang H.-C. Slowing down DNA translocation through solid-state nanopores by edge-field leakage. Nat. Commun. 2021;12:140. doi: 10.1038/s41467-020-20409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L.-B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., Rykunov D., Colaprico A., Rothstein J.H., Hong R., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528.e20. doi: 10.1016/j.ccell.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Li F., Rufo J., Chen C., Yang S., Li L., Zhang J., Cheng J., Kim Y., Wu M., et al. Acoustofluidic salivary exosome isolation: a liquid biopsy compatible approach for human papillomavirus-associated oropharyngeal cancer detection. J. Mol. Diagn. 2020;22:50–59. doi: 10.1016/j.jmoldx.2019.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei R., Zhao L., Kong G., Liu X., Zhu S., Zhang S., Min L. Combination of size-exclusion chromatography and ultracentrifugation improves the proteomic profiling of plasma-derived small extracellular vesicles. Biol. Proced. Online. 2020;22:12. doi: 10.1186/s12575-020-00125-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J.A., Van Der Pol E., Arkesteijn G.J.A., Bremer M., Brisson A., Coumans F., Dignat-George F., Duggan E., Ghiran I., Giebel B., et al. MIFlowCyt-EV: a framework for standardized reporting of extracellular vesicle flow cytometry experiments. J. Extracell. Vesicles. 2020;9:1713526. doi: 10.1080/20013078.2020.1713526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson M.D., Dumontier M., Aalbersberg I.J.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.-W., da Silva Santos L.B., Bourne P.E., et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Gao R., Bei Y., Zhou Q., Zhou Y., Zhang H., Jin M., Wei S., Wang K., Xu X., et al. Circulating miR-30d predicts survival in patients with acute heart failure. Cell. Physiol. Biochem. 2017;41:865–874. doi: 10.1159/000459899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Mao Z., Bachman H., Li P., Zhang P., Ren L., Wu M., Huang T.J. Acoustic cell separation based on biophysical properties. J. Biomech. Eng. 2020;142:0310051–0310059. doi: 10.1115/1.4046180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Shi J., Xie J., Wang Y., Sun J., Liu T., Zhao Y., Zhao X., Wang X., Ma Y., et al. Large-scale generation of functional mRNA-encapsulating exosomes via cellular nanoporation. Nat. Biomed. Eng. 2020;4:69–83. doi: 10.1038/s41551-019-0485-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Z., Dutta S., Liu Z., Yu X., Mesgarzadeh N., Ji F., Bitan G., Xie Y.-H. A label-free platform for identification of exosomes from different sources. ACS Sens. 2019;4:488–497. doi: 10.1021/acssensors.8b01564. [DOI] [PubMed] [Google Scholar]
- Yan Z., Zhou Z., Wu Q., Chen Z.B., Koo E.H., Zhong S. Presymptomatic increase of an extracellular RNA in blood plasma associates with the development of alzheimer’s disease. Curr.Biol. 2020;30:1771–1782.e3. doi: 10.1016/j.cub.2020.02.084. [DOI] [PubMed] [Google Scholar]
- Yeri A., Courtright A., Reiman R., Carlson E., Beecroft T., Janss A., Siniard A., Richholt R., Balak C., Rozowsky J., et al. Total extracellular small RNA profiles from plasma, saliva, and urine of healthy subjects. Sci. Rep. 2017;7:44061. doi: 10.1038/srep44061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Hayden E.Y., Wang P., Xia M., Liang O., Bai Y., Teplow D.B., Xie Y.-H. Ultrasensitive amyloid β-protein quantification with high dynamic range using a hybrid graphene-gold surface-enhanced Raman spectroscopy platform. J. Raman Spectrosc. 2020;51:432–441. doi: 10.1002/jrs.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nguyen L.T.H., Hickey R., Walters N., Wang X., Kwak K.J., Lee L.J., Palmer A.F., Reátegui E. Immunomagnetic sequential ultrafiltration (iSUF) platform for enrichment and purification of extracellular vesicles from biofluids. Sci. Rep. 2021;11:8034. doi: 10.1038/s41598-021-86910-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Higginbotham J.N., Jeppesen D.K., Yang Y.-P., Li W., McKinley E.T., Graves-Deal R., Ping J., Britain C.M., Dorsett K.A., et al. Transfer of functional cargo in exomeres. Cell Rep. 2019;27:940–954.e6. doi: 10.1016/j.celrep.2019.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Jeppesen D.K., Higginbotham J.N., Graves-Deal R., Trinh V.Q., Ramirez M.A., Sohn Y., Neininger A.C., Taneja N., McKinley E.T., et al. Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets. Nat. Cell Biol. 2021;23:1240–1254. doi: 10.1038/s41556-021-00805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z., Muth D.C., Mulka K., Liao Z., Powell B.H., Hancock G.V., Metcalf Pate K.A., Witwer K.W. miRNA profiling of primate cervicovaginal lavage and extracellular vesicles reveals miR-186-5p as a potential antiretroviral factor in macrophages. FEBS Open Bio. 2020;10:2021–2039. doi: 10.1002/2211-5463.12952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.J., Agus J.K., Hong B.V., Tang X., Rhodes C.H., Houts H.E., Zhu C., Kang J.W., Wong M., Xie Y., et al. Isolation of HDL by sequential flotation ultracentrifugation followed by size exclusion chromatography reveals size-based enrichment of HDL-associated proteins. Sci. Rep. 2021;11:16086. doi: 10.1038/s41598-021-95451-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z., Wu Q., Yan Z., Zheng H., Chen C.-J., Liu Y., Qi Z., Calandrelli R., Chen Z., Chien S., et al. Extracellular RNA in a single droplet of human serum reflects physiologic and disease states. Proc. Natl. Acad. Sci. USA. 2019;116:19200–19208. doi: 10.1073/pnas.1908252116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Wong M., Li Q., Sawrey-Kubicek L., Beals E., Rhodes C.H., Sacchi R., Lebrilla C.B., Zivkovic A.M. Site-specific glycoprofiles of HDL-associated ApoE are correlated with HDL functional capacity and unaffected by short-term diet. J. Proteome Res. 2019;18:3977–3984. doi: 10.1021/acs.jproteome.9b00450. [DOI] [PMC free article] [PubMed] [Google Scholar]