Abstract
CspA, CspB, and CspG, the major cold shock proteins of Escherichia coli, are dramatically induced upon temperature downshift. In this report, we examined the effects of kanamycin and chloramphenicol, inhibitors of protein synthesis, on cold shock inducibility of these proteins. Cell growth was completely blocked at 37°C in the presence of kanamycin (100 μg/ml) or chloramphenicol (200 μg/ml). After 10 min of incubation with the antibiotics at 37°C, cells were cold shocked at 15°C and labeled with [35S]methionine at 30 min after the cold shock. Surprisingly, the synthesis of all these cold shock proteins was induced at a significantly high level virtually in the absence of synthesis of any other protein, indicating that the cold shock proteins are able to bypass the inhibitory effect of the antibiotics. Possible bypass mechanisms are discussed. The levels of cspA and cspB mRNAs for the first hour at 15°C were hardly affected in the absence of new protein synthesis caused either by antibiotics or by amino acid starvation.
Cold shock response in Escherichia coli is triggered by a sudden temperature downshift, which causes a transient inhibition of synthesis of most cellular proteins, resulting in a growth lag period called the acclimation phase. During this acclimation phase, at least 15 different cold shock proteins are significantly induced, and some of them are essential for cell growth at low temperature (8–11). Among these proteins, CspA, CspB, and CspG are termed the major cold shock proteins on the basis of their levels of induction (18). It has been shown that the induction of CspA is caused mainly by dramatic stabilization of its mRNA at low temperature (2, 4, 5). We have recently shown that the downstream box, which is a translational enhancer, also plays a crucial role in the expression of CspA and CspB at low temperature (12). Previously, we have reported that the replacement of the cspA promoter with the constitutive promoter of the lpp gene does not change the cold shock inducibility of cspA (4). Therefore, unlike with the heat shock response, a specific sigma factor is not required for the induction of CspA. However, it has not yet been established whether any new protein factor(s) is required for the stabilization of the major cold shock mRNAs at low temperature. Herein, we examine the effects of the protein synthesis inhibitors chloramphenicol and kanamycin on the cold shock induction of CspA, CspB, and CspG.
MATERIALS AND METHODS
Strains and media.
E. coli SB221 (lpp hsdR trpE5 lacY recA/F′ lacIq lac+ pro+) was used in this study (13). For protein labeling with [35S]methionine, cultures were grown in M9 medium supplemented with 19 amino acids (no methionine) or supplemented with 17 amino acids (no methionine but tryptophan and leucine) as described by Wanner et al. (17). Cell growth was monitored spectrophotometrically at an optical density at 600 nm (OD600).
Protein pulse-labeling.
Steady-state cultures of E. coli SB221 were grown at 37°C to an OD600 of approximately 0.4. At this time, chloramphenicol or kanamycin was added to a final concentration of 0.1 or 0.2 mg/ml, respectively. After 10 min, the cultures were shifted to 15°C and 1-ml samples before (time zero) and 30 min after the shift were taken for pulse-labeling. Each sample was pulse-labeled for 15 min with 100 μCi of [trans-35S]methionine (1,175 Ci/mmol; NEN Life Science Products Inc.). Cell extracts were prepared and analyzed by two-dimensional gel electrophoresis according to the procedure described by VanBogelen et al. (16).
At mid-log phase (OD600 = 0.4), E. coli SB221 cells grown under the same conditions described above were collected by centrifugation and washed twice with M9 medium containing 17 amino acids (no Met but Trp and Leu). Samples were pulse-labeled as described above at 37°C 30, 60, and 120 min after the temperature shift to 15°C.
Primer extensions.
Total RNA from E. coli SB221 was isolated at different time points before and after a temperature shift from 37 to 15°C by the hot-phenol method described by Sarmientos et al. (14). Primer extension assays were carried out with avian myeloblastoma virus-reverse transcriptase as previously described (12).
RESULTS
Cold shock induction of CspA, CspB, and CspG in the presence of protein synthesis inhibitors.
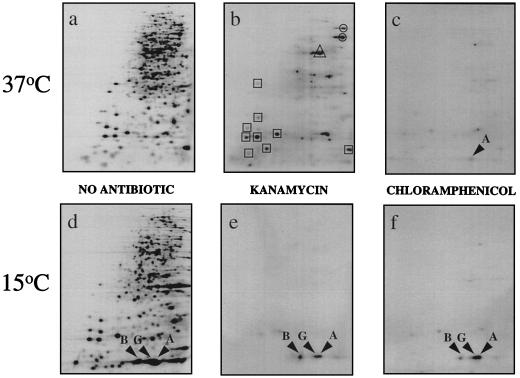
E. coli SB221 cells were grown at 37°C in a labeling medium as described previously (3). Chloramphenicol (0.2 mg/ml) or kanamycin (0.1 mg/ml) was added at the mid-log phase, and after 10 min, the cultures were shifted to 15°C. Cells were pulse-labeled for 15 min with 100 μCi of [trans-35S]methionine (NEN Life Science Products Inc.) before (time zero) and 30 min after the temperature shift. Cell extracts were prepared and analyzed by two-dimensional gel electrophoresis as described previously (16). Figure 1a shows the protein expression pattern at 37°C in the absence of antibiotics. Figure 1b and c shows the protein expression patterns at 37°C in the presence of kanamycin and chloramphenicol, respectively. Surprisingly, at 37°C in the presence of kanamycin (Fig. 1b), unlike in the presence of chloramphenicol (Fig. 1c), some proteins were still synthesized: EF-Tu and some ribosomal and heat shock proteins. Differential inhibitory effects by various antibiotics on cellular proteins have been previously described (7). Figure 1d shows the proteins synthesized at 30 min after the temperature downshift. It is evident that in the presence of kanamycin (Fig. 1e) or chloramphenicol (Fig. 1f) at 15°C, with the exception of the synthesis of CspA, CspB, and CspG, most if not all cellular protein synthesis was blocked. This result is consistent with the earlier finding that, after a temperature shift from 37 to 6°C, the major cold shock proteins, CspA, CspB, and CspG, were still produced in spite of the fact that the synthesis of all the other cellular proteins was completely inhibited (3). By densitometric analysis of Fig. 1d to f, the levels of production of CspA, CspB, and CspG decreased to 20, 40, and 18% in the presence of kanamycin, respectively, and to 30, 25, and 36% in the presence of chloramphenicol, respectively. Notably, CspB synthesis was more resistant to kanamycin than CspA and CspG synthesis (Fig. 1e) while CspB synthesis was more sensitive to chloramphenicol than CspA and CspG synthesis (Fig. 1f). These different effects may be related to the fact that these genes are differentially regulated, as judged from their induction patterns at low temperatures (3).
FIG. 1.
Protein expression patterns before and after cold shock at 15°C, in the absence and presence of kanamycin (0.1 mg/ml) or chloramphenicol (0.2 mg/ml). Two-dimensional gel electrophoresis shows protein synthesis from pIs 3 to 10 (left to right) in the absence of antibiotics at 37°C (a), in the absence of antibiotics at 15°C (d), in the presence of 0.1 mg of kanamycin per ml (b), and in the presence of 0.2 mg of chloramphenicol per ml (c). Cells were labeled with 100 μCi of [35S]methionine per ml for 15 min after incubation with kanamycin or chloramphenicol. Panels e and f are the same as panels b and c, respectively, except that cells were first incubated in the presence of antibiotics for 10 min at 37°C and further incubated for 30 min at 15°C. Cells were then labeled for 15 min with the same amount of [35S]methionine as was used in the experiment at 37°C. The arrows indicate the positions of CspA, CspB, and CspG (labeled A, B, and G, respectively). Circles, triangles, and squares enclose the heat shock proteins (DnaK and GroEL), elongation factor Tu, and ribosomal proteins, respectively.
Amounts of cspA and cspB mRNAs in the presence of antibiotics at low temperature.
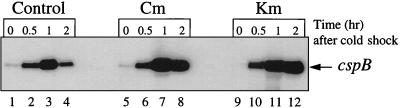
Next we examined the effects of antibiotics on cspA and cspB mRNAs. E. coli SB221 cells were grown in Luria-Bertani medium at 37°C. At mid-log phase, chloramphenicol or kanamycin was added to a final concentration of 200 μg/ml, and after 10 min, the cultures were shifted to 15°C. Total RNA was isolated by the hot-phenol method (14) 10 min after the addition of the antibiotics at 37°C (time zero) and 0.5, 1, and 2 h after the temperature shift. Primer extension was carried out as described previously (12) to detect the cspB mRNA. As shown in Fig. 2, the amounts of cspB mRNA were three- and fourfold higher in the presence of chloramphenicol (lane 8) and kanamycin (lane 12), respectively, than those in the absence of antibiotic. In the control experiment without the antibiotics, the levels of cspB mRNA decreased more than twofold at 2 h after the temperature downshift, while in the experiment with the antibiotics, the levels of the cspB mRNA at 2 h remained as high as that at 1 h after the cold shock. In the presence of kanamycin the amount of the cspB mRNA at 2 h was even higher (1.2-fold) than at 1 h (compare lanes 12 and 11). A similar pattern was observed for the cspA mRNA (data not shown). These results indicate that mRNAs for CspA and CspB are transcribed at 15°C in the presence of the antibiotics at levels similar to those reached in the absence of antibiotics and that the mRNAs in the presence of antibiotics are probably more stable than in the absence of antibiotics. It has been shown that CspA negatively regulates cspA and cspB at the level of transcription elongation (1). Therefore, it is possible that the cspA and cspB mRNAs were maintained at high levels at 15°C even after 2 h of incubation at 15°C because the CspA concentration could not increase to a level high enough to block cspA and cspB transcription under conditions that blocked the synthesis of protein. In addition, the mRNA stability of cspA and cspB may be increased when protein synthesis is blocked at low temperatures.
FIG. 2.
cspB mRNA production before and after cold shock at 15°C and in the absence and presence of 0.2 mg of kanamycin or chloramphenicol per ml. Primer extension was carried out as described previously. The results show the cspB mRNA amounts at 37°C (time zero) and 0.5, 1, and 2 h after the shift to 15°C in the absence (lanes 1 to 4) and the presence of 0.2 mg of chloramphenicol (Cm) (lanes 5 to 8) or kanamycin (Km) (lanes 9 to 12) per ml.
cspA and cspB mRNAs under amino acid starvation at low temperature.
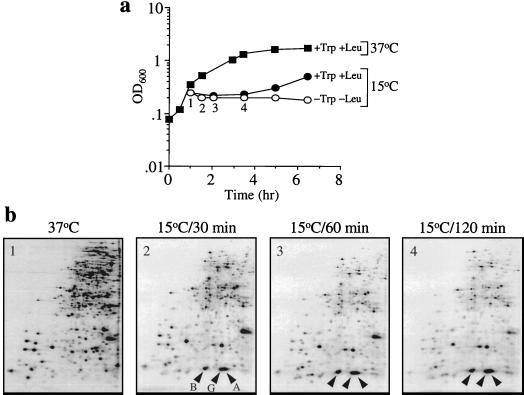
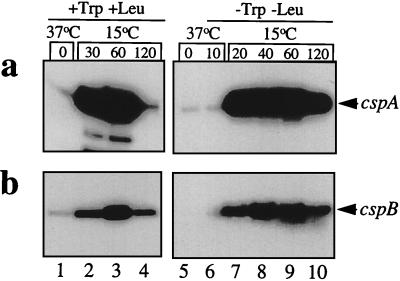
In order to confirm the above notion, we examined the levels of cspA and cspB mRNAs at 15°C in cells when protein synthesis was blocked by amino acid starvation rather than by antibiotics. E. coli SB221, which requires tryptophan and leucine for growth, was used (13). Cultures were grown at 37°C in M9-CAA medium. At mid-log phase, cells were collected by centrifugation and washed with M9 medium containing all amino acids except tryptophan and leucine. No cell growth was observed after amino acid starvation as judged by the ODs of the culture at both 37 and 15°C (Fig. 3a). Total RNA was obtained as described previously (14). Figure 4 shows the primer extensions of the cspA (Fig. 4a) and cspB (Fig. 4b) mRNAs before and after 10 min of amino acid starvation at 37°C (lanes 5 and 6, respectively) and 20, 40, 60, and 120 min after the temperature downshift (lanes 7, 8, 9, and 10, respectively). The amino acid starvation did not affect the levels of cspA and cspB mRNAs for up to 60 min at 15°C, as was apparent from a comparison with the levels in the control experiments in the presence of tryptophan and leucine (Fig. 4, lanes 2 and 3). Interestingly, at 120 min after the cold shock, cspA transcription was repressed in the presence of Trp and Leu (Fig. 4a, lane 4), as was shown previously (6), while the level of cspA mRNA remained very high in the absence of Trp and Leu (Fig. 4a, lane 10). The amount of cspA mRNA in lane 10 is eight times higher than that in lane 4. Similarly, the amount of the cspB transcript was 1.8 times higher in the absence of tryptophan and leucine (Fig. 4b, lane 10) than in the presence of these amino acids (Fig. 4b, lane 4). Poor repression of cspA and cspB transcription after 120 min of cold shock in the absence of protein synthesis (lane 10) is likely due to the absence of the production of a repressor(s) and/or stabilization of the mRNAs.
FIG. 3.
Protein expression pattern after a cold shock at 15°C in the absence of tryptophan and leucine. E. coli SB221 cells were grown as described in the legend to Fig. 4. In the growth curve (a) the filled squares and circles represent cell densities (OD600 values) in the presence of Trp and Leu and the open circles show cell growth at 15°C in the absence of Trp and Leu. Cell extracts for the two-dimensional gel electrophoresis were prepared as described previously (16) at 37°C 30, 60, and 120 min after the shift to 15°C (b). The number at the top left in each two-dimensional gel indicates the time point shown in the growth curve (open circles). B, G, and A, CspB, CspG, and CspA, respectively.
FIG. 4.
Amounts of the cspA and cspB mRNAs before and after the cold shock at 15°C, in the presence and in the absence of tryptophan and leucine. E. coli SB221 cells were washed twice at 37°C with M9 medium lacking tryptophan and leucine, and after 10 min the culture was shifted to 15°C. Total RNA was extracted at 37°C before (time zero) and after 10 min of Trp and Leu starvation; at 30, 60, and 120 min in the presence of Trp and Leu; and at 20, 40, 60, and 120 min in the absence of Trp and Leu. Primer extension was carried out for cspA mRNA (a) and cspB mRNA (b) as described previously (3).
Synthesis of CspA, CspB, and CspG under amino acid starvation.
In order to see if the cspA and cspB mRNAs are translated under conditions of cold shock in the absence of new protein synthesis due to amino acid starvation, cells were labeled at different time points (30, 60, and 120 min) after a temperature downshift from 37 to 15°C. E. coli SB221 cells were labeled for 10 min at 37°C or 15 min at 15°C with 100 μCi of [trans-35S]methionine. Cell extracts were prepared and analyzed by two-dimensional gel electrophoresis as described previously (16). Figure 3b shows the protein expression patterns at 37°C and at various time points after the shift to 15°C in the absence of Trp and Leu. The production of CspA, CspB, and CspG can still be observed even at 2 h after the cold shock, indicating that their mRNAs can be efficiently translated at a low temperature in the absence of cell growth. However, it should be noted that under these conditions some other cellular proteins were also synthesized at significantly high levels, probably because of reutilization of Trp and Leu generated by protein degradation.
DISCUSSION
Our results demonstrate that transcription and/or mRNA stability of cspA and cspB at a low temperature is hardly affected by inhibition of protein synthesis caused either by protein synthesis inhibitors or by amino acid starvation. Furthermore, their mRNAs can be efficiently translated under these conditions. In addition, these results provide compelling evidence that the induction of the major cold shock proteins at a low temperature does not require the synthesis of any new protein(s) such as a specific cold shock sigma factor(s). It is particularly interesting how the cold shock mRNAs can be efficiently translated at a low temperature even if the cellular protein synthesis is almost completely blocked by kanamycin or chloramphenicol. Since all the mRNAs for the cold shock proteins contain the downstream box (12), which is considered to enhance translation initiation (15), it is possible that the cold shock mRNAs may be able to bypass the inhibitory actions of the antibiotics. Alternatively, the synthesis of CspA, CspB, and CspG, which consist of only 70, 71, and 70 residues, respectively, may be less vulnerable to the antibiotic inhibitory effect. In this regard, it is interesting that the biosynthesis of the major outer membrane lipoprotein of 58 residues is resistant to puromycin (6). The fact that the biosynthesis of the cold shock proteins is further resistant to stresses caused by antibiotics and amino acid starvation is rather remarkable and may have an evolutionary significance for stress proteins.
ACKNOWLEDGMENTS
We thank S. Phadtare for a critical reading of the manuscript.
This work was supported by a grant from the National Institutes of Health (GM19043).
REFERENCES
- 1.Bae W, Jones P G, Inouye M. CspA, the major cold shock protein of Escherichia coli, negatively regulates its own gene expression. J Bacteriol. 1997;179:7081–7088. doi: 10.1128/jb.179.22.7081-7088.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandi A, Pietroni P, Gualerzi C O, Pon C L. Posttranscriptional regulation of CspA expression in Escherichia coli. Mol Microbiol. 1996;19:231–240. doi: 10.1046/j.1365-2958.1996.362897.x. [DOI] [PubMed] [Google Scholar]
- 3.Etchegaray J-P, Jones P, Inouye M. Differential thermoregulation of two highly homologous cold-shock genes, cspA and cspB, of Escherichia coli. Genes Cells. 1996;1:171–178. doi: 10.1046/j.1365-2443.1996.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 4.Fang L, Jiang W, Bae W, Inouye M. Promoter-independent cold-shock induction of cspA and its derepression at 37°C by mRNA stabilization. Mol Microbiol. 1997;23:355–364. doi: 10.1046/j.1365-2958.1997.2351592.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldenberg D, Azar I, Oppenheim A B. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol Microbiol. 1996;19:241–248. doi: 10.1046/j.1365-2958.1996.363898.x. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein J, Pollitt N S, Inouye M. Major cold-shock protein of Escherichia coli. Proc Natl Acad Sci USA. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirashima A, Childs G, Inouye M. Differential inhibitory effects of antibiotics on the biosynthesis of envelope proteins of Escherichia coli. J Mol Biol. 1973;79:373–389. doi: 10.1016/0022-2836(73)90012-0. [DOI] [PubMed] [Google Scholar]
- 8.Jones P G, VanBogelen R A, Neidhardt F C. Induction of proteins in response to low temperature in Escherichia coli. J Bacteriol. 1987;169:2092–2095. doi: 10.1128/jb.169.5.2092-2095.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones P G, Krah R, Tafuri S R, Wolffe A P. DNA gyrase, CS7.4, and the cold-shock response in Escherichia coli. J Bacteriol. 1992;174:5798–5802. doi: 10.1128/jb.174.18.5798-5802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones P G, Inouye M. RbfA, a 30S ribosomal binding factor, is a cold-shock protein whose absence triggers the cold-shock response. Mol Microbiol. 1996;21:1207–1218. doi: 10.1111/j.1365-2958.1996.tb02582.x. [DOI] [PubMed] [Google Scholar]
- 11.La Teana A, Brandi A, Falcony M, Spurio R, Pon C L, Gualerzi C O. Identification of a cold-shock transcriptional enhancer of the Escherichia coli gene encoding nucleoid protein H-NS. Proc Natl Acad Sci USA. 1991;88:10907–10911. doi: 10.1073/pnas.88.23.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mitta M, Fang L, Inouye M. Deletion analysis of cspA of Escherichia coli: requirement of the AT-rich UP element for cspA transcription and the downstream box in the coding region for its cold-shock induction. Mol Microbiol. 1997;26:321–335. doi: 10.1046/j.1365-2958.1997.5771943.x. [DOI] [PubMed] [Google Scholar]
- 13.Nakamura K, Masui Y, Inouye M. Use of a lac promoter-operator fragment as a transcriptional control switch for expression of the constitutive lpp gene in Escherichia coli. J Mol Appl Genet. 1982;1:289–299. [PubMed] [Google Scholar]
- 14.Sarmientos P, Sylvester J E, Contente S, Cashel M. Differential stringent control of the tandem E. col: ribosomal RNA promoters from RNA operon expressed in vivo in multicopy plasmids. Cell. 1983;32:1337–1346. doi: 10.1016/0092-8674(83)90314-8. [DOI] [PubMed] [Google Scholar]
- 15.Sprengart M L, Fuchs E, Porter A G. The downstream box: an efficient and independent translation signal in Escherichia coli. EMBO J. 1996;15:665–674. [PMC free article] [PubMed] [Google Scholar]
- 16.VanBogelen R A, Hutton M E, Neidhardt F C. Gene-protein database of Escherichia coli K-12: edition 3. Electrophoresis. 1990;11:1131–1166. doi: 10.1002/elps.1150111205. [DOI] [PubMed] [Google Scholar]
- 17.Wanner B L, Kodaira R, Neidhardt F C. Physiological regulation of a decontrolled lac operon. J Bacteriol. 1977;130:212–222. doi: 10.1128/jb.130.1.212-222.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka K, Fang L, Inouye M. The CspA family in Escherichia coli: multiple gene duplication for stress adaptation. Mol Microbiol. 1998;27:247–255. doi: 10.1046/j.1365-2958.1998.00683.x. [DOI] [PubMed] [Google Scholar]