Abstract
The naphthalene dioxygenase enzyme system carries out the first step in the aerobic degradation of naphthalene by Pseudomonas sp. strain NCIB 9816-4. The crystal structure of naphthalene dioxygenase (B. Kauppi, K. Lee, E. Carredano, R. E. Parales, D. T. Gibson, H. Eklund, and S. Ramaswamy, Structure 6:571–586, 1998) indicates that aspartate 205 may provide the most direct route of electron transfer between the Rieske [2Fe-2S] center of one α subunit and mononuclear iron in the adjacent α subunit. In this study, we constructed four site-directed mutations that changed aspartate 205 to alanine, glutamate, asparagine, or glutamine to test whether this residue is essential for naphthalene dioxygenase activity. The mutant proteins were very inefficient in oxidizing naphthalene to cis-naphthalene dihydrodiol, and oxygen uptake in the presence of naphthalene was below detectable levels. The purified mutant protein with glutamine in place of aspartate 205 had identical spectral properties to wild-type naphthalene dioxygenase and was reduced by NADH in the presence of catalytic amounts of ferredoxinNAP and reductaseNAP. Benzene, an effective uncoupler of oxygen consumption in purified naphthalene dioxygenase, did not elicit oxygen uptake by the mutant protein. These results indicate that electron transfer from NADH to the Rieske center in the mutant oxygenase is intact, a finding consistent with the proposal that aspartate 205 is a necessary residue in the major pathway of electron transfer to mononuclear iron at the active site.
Bacterial multicomponent dioxygenase enzyme systems carry out the first reaction in the aerobic degradation of a variety of aromatic compounds. These enzymes have been of interest to researchers for several reasons. Since they initiate the removal from the environment of toxic and carcinogenic compounds such as benzene, toluene, naphthalene, polychlorinated biphenyls, polycyclic aromatic hydrocarbons, and nitroaromatics, they have been the subject of intense study by environmental microbiologists. The ability to produce enantiomerically pure products from a wide range of substrates has brought dioxygenases to the attention of organic chemists for the production of chiral synthons used in the preparation of biologically active chemicals and pharmaceuticals (for reviews, see references 7, 10, 20, 29, and 42). During the past three decades, the purification and characterization of the components of more than 15 of these related enzyme systems have been reported. In addition, more than 40 sets of dioxygenase genes have been cloned and their nucleotide sequences have been determined. However, we do not yet have a clear understanding of the catalytic mechanism used by these enzymes to add molecular oxygen to an aromatic ring.
Aromatic ring-hydroxylating dioxygenases consist of two or three protein components that form a short electron transport chain to transfer electrons from NAD(P)H to the oxygenase (8). The reduced oxygenase catalyzes the stereospecific addition of both atoms of molecular oxygen to the aromatic nucleus of the substrate. Some oxygenases are homomultimers consisting of a single subunit type (α), while others are heteromultimers consisting of large and small subunits (α and β). In each case, the α subunit contains a Rieske [2Fe-2S] center and mononuclear iron. The mononuclear iron is believed to be the site of oxygen activation (8). Based on amino acid sequence alignments, four conserved amino acids in the α subunit of toluene dioxygenase were proposed to be iron ligands (23). When these amino acids (Glu-214, Asp-219, His-222, and His-228) were changed to alanines by site-directed mutagenesis, the resulting proteins were completely inactive (23). The corresponding amino acids in naphthalene dioxygenase (NDO) are Glu-200, Asp-205, His-208, and His-213.
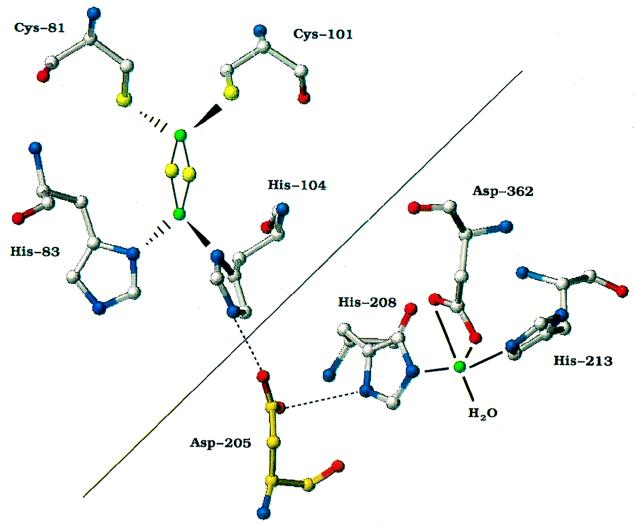
With the completion of the three-dimensional crystal structure of NDO at 2.25-Å resolution (24), the role of each of these amino acids in catalysis can now be clarified. The crystal structure revealed that the enzyme is an α3β3 hexamer. Each α subunit contains a Rieske [2Fe-2S] center and mononuclear nonheme iron. Histidine residues 208 and 213 in NDO were confirmed by the NDO structure to be ligands to the active-site iron as previously proposed (Fig. 1). Asp-362 was also found to be a mononuclear iron ligand in NDO (Fig. 1). This amino acid is conserved in alignments of the α subunits of 30 heteromultimeric oxygenases but only 2 of 9 homomultimeric oxygenases (37). This type of 2-His–1-carboxylate facial triad appears to be a common structural motif for the coordination of iron(II) that is also found in isopenicillin synthetase, the ring cleavage enzyme 2,3-dihydroxybiphenyl dioxygenase, and tyrosine hydroxylase (25).
FIG. 1.
Structure of the NDO active site and proposed electron transfer pathway from the Rieske center of one α subunit through Asp-205 (gold) to the mononuclear iron in the adjacent α subunit. Amino acids above the diagonal line are located in the Rieske domain of one α subunit. Amino acids below the diagonal line are in the catalytic domain of the adjacent α subunit. Atom colors: red, oxygen; blue, nitrogen; yellow, sulfur; green, iron.
In NDO, the Rieske [2Fe-2S] center is located 43.5 Å from the mononuclear iron center within a single α subunit but only 12 Å from the mononuclear iron in an adjacent α subunit within the hexamer (24). It has been proposed that the active site is located at the junction of two α subunits and that electrons are transferred from the Rieske center of one subunit to the mononuclear iron of the adjacent α subunit. At each α-α junction, Asp-205 is hydrogen bonded to His-208 in the same α subunit and to His-104 (a ligand to the Rieske center) in the adjacent α subunit. Asp-205 (which corresponds to Asp-219 in toluene dioxygenase) is not an iron ligand (Fig. 1) but could provide the most direct route for electron transfer between the Rieske center and the mononuclear iron in adjacent α subunits (24). Asp-205 is thus likely to be a very important amino acid for efficient catalysis. In this study, we constructed and characterized four variants of NDO with amino acid changes at position 205 to determine the role played by this residue in NDO catalysis.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are listed in Table 1. Escherichia coli DH5α and JM109(DE3) were used for subcloning and gene expression experiments, respectively. Competent E. coli ES1301 and JM109 were purchased from Promega Corp., Madison, Wis., and used in the site-directed mutagenesis procedure described below.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | F− Δ(lacZYA-argF)U169 hsdR17 relA1 supE44 endA1 recA1 thi-1 gyrA96 φ80dlacZΔM15 | Life Technologies, Gaithersburg, Md. |
| JM109 | endA1 recA1 gyrA96 thi hsdR17 relA1 supE44 Δ(lac-proAB) mcrA [F′, traD36 proAB+ lacIqZΔM15] | 47 |
| JM109(DE3) | endA1 recA1 gyrA96 thi hsdR17 relA1 supE44 Δ(lac-proAB) mcrA [F′, traD36 proAB+ lacIqZΔM15], λ(DE3) | Promega Corp., Madison, Wis. |
| ES1301 mutS | Kmr, lacZ53 mutS201::Tn5 thyA36 rha-5 metB1 deoC IN(rrnD-rrnE) | Promega Corp., Madison, Wis. |
| Plasmids | ||
| pALTER-1 | Tcr, mutagenesis vector | Promega Corp., Madison, Wis. |
| pMASTER-1 | Tcr, pALTER-1 carrying the KpnI-XbaI fragment of pDTG141 (nahAc′Ad) | This study |
| pDTG141 | Apr, nahAaAbAcAd under the control of the T7 promoter of pT7-5 | 44 |
| pDTG155A | Apr, pT7-5 carrying nahAaAb (constructed by deleting the SmaI fragment carrying nahAcAd from pDTG141) | This study |
| pDTG172 | Apr, Tcs, pMASTER-1 with D205E mutation | This study |
| pDTG173 | Apr, D205E mutation in pDTG141 | This study |
| pDTG174 | Apr, Tcs, pMASTER-1 with D205A mutation | This study |
| pDTG175 | Apr, D205A mutation in pDTG141 | This study |
| pDTG176 | Apr, Tcs, pMASTER-1 with D205N mutation | This study |
| pDTG177 | Apr, D205N mutation in pDTG141 | This study |
| pDTG178 | Apr, Tcs, pMASTER-1 with D205Q mutation | This study |
| pDTG179 | Apr, D205Q mutation in pDTG141 | This study |
Apr, ampicillin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; nahAa encodes reductaseNAP; nahAb encodes ferredoxinNAP; nahAc encodes the oxygenase α subunit; nahAd encodes the oxygenase β subunit.
Media and growth conditions.
E. coli strains were grown at 30 or 37°C in Luria-Bertani (LB) medium (12), or Terrific Broth (TB) medium (28). Antibiotics were added to the following final concentrations as appropriate: ampicillin, 150 μg/ml; tetracycline, 20 μg/ml. JM109(DE3) strains carrying plasmids of interest were maintained on minimal medium plates (MSB) (43) containing 10 mM glucose, 0.1 mM thiamine, and ampicillin. For plates, MSB was solidified with 1.8% Agar Noble (Difco Laboratories, Detroit, Mich.) and LB medium was solidified with 1.5% Bacto Agar (Difco Laboratories). For small-scale preparation of cell extracts, JM109(DE3) containing pDTG141 or the mutant derivatives were cultured at 30°C in TB medium containing ampicillin. Dioxygenase genes were induced by addition of 100 μM isopropyl-β-d-thiogalactopyranoside (IPTG) when the culture turbidity at 660 nm reached approximately 0.7. The incubation temperature was reduced to 27°C at the time of induction. Cells were harvested by centrifugation 2 h after addition of IPTG and stored at −70°C. The control strains JM109(DE3)(pT7-5) and JM109(DE3)(pDTG155A) were grown, induced, harvested, and stored in the same way. To obtain cells for protein purification, JM109(DE3)(pDTG179) and JM109(DE3)(pDTG141) (Table 1) were grown at 27°C in MSB in a 10-liter Biostat B fermentor (B. Braun Biotech International, Melsungen, Germany). Automated addition of NH4OH was used to maintain the pH at 7.3, and a slow glucose feed was used to maintain the dissolved-O2 concentration at approximately 25% saturation. The culture was induced for 3 h with 150 μM IPTG when the optical density of the culture at 660 nm reached 0.8. Cells were harvested by centrifugation, resuspended in BTGD buffer (50 mM bis-Tris [pH 6.8], 5% glycerol, 1 mM sodium dithiothreitol), and stored at −70°C.
Preparation of cell extracts.
Frozen cells were thawed and suspended in BTGD buffer. DNase I was added to a final concentration of approximately 1 μg/ml. The cells were broken by two cycles through a chilled French pressure cell, and extracts were centrifuged at 150,000 × g at 4°C for 1 h to remove cell debris and membranes. The supernatant was drop-frozen in liquid nitrogen and stored at −70°C.
Molecular techniques.
Plasmid DNA was isolated as described previously (28) or by using the Midi kit (Qiagen, Inc., Chatsworth, Calif.). For sequencing, DNA was further purified with a Centricon-100 filter unit (Amicon, Inc., Beverly, Mass.). Restriction digests were performed as suggested by the enzyme suppliers (New England Biolabs, Inc., Beverly, Mass.; Promega Corp.). DNA fragments were purified from gel slices with the GeneClean spin kit as specified by the manufacturer (BIO 101, Vista, Calif.). Ligation reactions, transformation of E. coli strains, and agarose gel electrophoresis were performed by standard procedures (41).
Site-directed mutagenesis.
Mutagenesis of nahAc was carried out with the Altered Sites II in vitro mutagenesis system as specified by the manufacturer (Promega Corp.). A 1.5-kb KpnI-XbaI fragment carrying the 3′ half of the nahAc gene and the complete nahAd gene from pDTG141 was cloned into KpnI-XbaI-digested pALTER-1. The resulting plasmid, designated pMASTER-1, was used as the template for mutagenesis. Each mutagenic oligonucleotide was designed with a silent mutation that altered the restriction pattern of the plasmid (Table 2) to facilitate screening for clones carrying the desired mutation. Phosphorylated oligonucleotides used for mutagenesis were synthesized by Genosys Biotechnologies Inc., Midland, Tex. The nucleotide sequences of both strands of the entire insertion in pMASTER-1 were determined for each mutant. Fluorescent automated DNA sequencing was carried out by the University of Iowa DNA Facility with an Applied Biosystems 373A automated DNA sequencer. After verification of each mutation by restriction digestion and sequence analysis, the 1.5-kb KpnI-XbaI fragments carrying each mutation were individually cloned into KpnI-XbaI-digested pDTG141 and the resulting plasmids were introduced into JM109(DE3) for expression studies. After this subcloning step, the presence of each mutation was verified by restriction and sequence analysis.
TABLE 2.
Oligonucleotides used for site-directed mutagenesis
| Mutation | Mutagenic oligonucleotidea | Restriction site change |
|---|---|---|
| D205A | 5′-ACTTTGTGGGAGCTGCATACCACGT-3′ | Eliminate NsiI |
| D205E | 5′-ACTTTGTGGGAGAGGCATACCACGT-3′ | Eliminate NsiI |
| D205N | 5′-AAACTTTGTGGGAAACGCATACCACGTGG-3′ | Eliminate NsiI |
| D205Q | 5′-AAACTTTGTGGGACAGGCATACCACGTGG-3′ | Eliminate NsiI |
Underlined bases indicate the position of the eliminated restriction site. Base changes are in bold.
Indigo formation.
JM109(DE3) strains carrying plasmids of interest were grown overnight at 37°C on nitrocellulose filters placed on MSB agar plates containing glucose, thiamine, and ampicillin. Dried Whatman no. 1 filter papers that had been soaked in a 10% solution of indole dissolved in acetone were placed in the petri dish covers after colony formation. Production of indigo from indole vapor was observed as colonies turned blue. No induction was carried out for these studies.
Enzyme assays.
NDO activity was determined by measuring the accumulation of nonvolatile metabolites from [14C]naphthalene (Sigma Chemical Co., St. Louis, Mo.) by using a modification of the procedure described by Ensley et al. (14). Reaction mixtures (0.5 ml) contained 50 mM 2-N-morpholinoethanesulfonic acid (MES) buffer (pH 6.8), 12 μg of purified reductaseNAP, 12 μg of purified ferredoxinNAP, 0.4 mM NADH, and 1 mM ferrous ammonium sulfate. Reactions were initiated by the addition of 20 μl of [14C]naphthalene in methanol (5.55 × 104 dpm/μl; final concentration, 0.2 mM). After 30, 60, 90, and 300 s, aliquots (10 μl) were mixed with 10 μl of quench solution (10 mM unlabelled naphthalene in methanol). Samples were applied to thin-layer chromatography (TLC) squares (1.0 cm2) and air dried to remove all remaining [14C]naphthalene. The amount of nonvolatile cis-naphthalene dihydrodiol formed was determined in a scintillation counter. Reactions were carried out in triplicate. Specific activity is defined as micromoles of product formed per minute per milligram of protein. Protein concentrations were determined by the method of Bradford (6) with bovine serum albumin as the standard.
Detection of reaction products.
TLC followed by autoradiography was used to detect 14C-labelled reaction products. Modified [14C]naphthalene assay mixtures containing no added ferredoxinNAP or reductaseNAP (these are present in extracts since genes encoding both proteins are carried on pDTG141 and its mutant derivatives) were incubated for 45 min. Aliquots (8 μl) were applied to the origin of a TLC plate (silica gel 60 F254, 0.2 mm thick; EM Science, Gibbstown, N.J.) and developed with chloroform-acetone (80:20, vol/vol). Purified cis-naphthalene dihydrodiol was used as a standard (21, 22) and was visualized on TLC plates by observing quenching of fluorescence under UV light (254 nm). The X-ray film was exposed for 5 days prior to development.
Oxygen uptake.
Oxygen uptake by NDO in the presence of naphthalene was measured at 30°C with a Clark-type oxygen electrode (Rank Brothers, Cambridge, England). Reactions (total volume, 1 ml) were carried out in air-saturated 50 mM MES buffer (pH 6.8) containing 1 mM ferrous ammonium sulfate, 12 μg of ferredoxinNAP, 12 μg of reductaseNAP, 0.5 mM NADH, and an appropriate amount of cell extract to give a linear O2 uptake rate. Reactions were initiated by the addition of naphthalene or benzene (final concentration, 0.1 mM). All rates were corrected for endogenous respiration.
Gel electrophoresis and Western blot analyses.
Proteins in cell extracts prepared from E. coli transformants expressing either the mutant or wild-type nahAc genes were separated on duplicate sodium dodecyl sulfate (SDS)–12% polyacrylamide gels (1). One gel was stained with Coomassie blue R-250 to verify that approximately equal amounts of protein were loaded in each lane. The second gel was subjected to Western blotting as described previously (19, 30). The monoclonal antibody raised against the NDO α subunit (36) was used to detect the presence of this protein in cell extracts. Antigens were visualized with alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Pierce, Rockford, Ill.). Native gels (8% polyacrylamide) were run in a similar manner without SDS (1).
Purification of the mutant NDO protein.
NDO and NDO-D205Q (NDO with Asp-205 replaced by glutamine) were purified from JM109(DE3)(pDTG141) and JM109(DE3)(pDTG179) cell extracts, respectively, by the procedure described by Lee et al. (27). The mutant protein was identified by its brown color and absorption spectrum, which are typical of wild-type NDO (13).
Iron determinations.
The iron content was analyzed as previously described (48).
Spectroscopy.
Absorption spectra were recorded on an Aminco DW-2000 UV-visible spectrophotometer. Reductions of NDO and NDO-D205Q by NADH in the presence of catalytic amounts of purified ferredoxinNAP and reductaseNAP were carried out under anaerobic conditions. Electron paramagnetic resonance (EPR) spectra of the oxidized and reduced forms of NDO-D205Q were recorded at 77 K in a Bruker model ESP 300 spectrometer (ESR Facility, University of Iowa). The settings used were 5.05-mW microwave power, 3,600-G centerfield, 9.29-GHz microwave modulation frequency, 42-s sweep time, and 1.0 × 105 receiver gain. The protein was completely reduced by the addition of excess sodium dithionite.
RESULTS
Site-directed mutagenesis of Asp-205 and initial analysis of the mutant NDO enzymes.
Asp-205 in the NDO α subunit was changed to alanine, asparagine, glutamate, or glutamine by using the pMASTER-1 plasmid (Table 1) and the oligonucleotides shown in Table 2. JM109(DE3) strains carrying each pDTG141 derivative (pDTG173, pDTG175, pDTG177, and pDTG179) were grown on plates and exposed to indole vapor as described in Materials and Methods. Indigo was not formed from indole by any of these strains, in contrast to colonies of JM109(DE3)(pDTG141), which turned dark blue. This result indicated that the mutant enzymes either were not produced or were severely defective in their ability to convert indole to indigo.
Production of wild-type and mutant NDO enzymes.
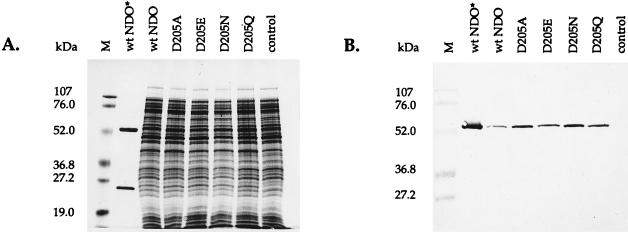
The wild-type and mutant NDO enzymes were produced by JM109(DE3) carrying pDTG141 or its mutant derivatives (Table 1). SDS-polyacrylamide gel electrophoresis and Western blot analyses of crude cell extracts were used to verify the production of approximately equivalent levels of full-length wild-type and mutant α subunits (Fig. 2). A protein band in the crude cell extracts corresponding to the calculated 49.6-kDa molecular mass of the NDO α subunit was identified in the Western blot by using the anti-NDOα monoclonal antibody. No band was observed in the extract of the negative-control strain JM109(DE3)(pT7-5).
FIG. 2.
SDS-polyacrylamide gel electrophoresis and Western blot analysis of E. coli cell extracts expressing wild-type and mutant NDO enzymes. (A) SDS–12% polyacrylamide gel stained with Coomassie blue R250. (B) Western blot analysis of a gel similar to that in panel A. A monoclonal antibody specific for the α subunit of NDO was used as described in Materials and Methods. Lanes: M, prestained molecular mass markers; wt NDO*, purified wild-type NDO (2.2 μg of protein); wt NDO, JM109(DE3)(pDTG141) extract; D205A, JM109(DE3)(pDTG175) extract; D205E, JM109(DE3)(pDTG173) extract; D205N, JM109(DE3)(pDTG177) extract; D205Q, JM109(DE3)(pDTG179) extract; control, JM109(DE3)(pT7-5) extract. For extracts, approximately 50 μg of protein was loaded per lane.
Activity of wild-type and mutant NDO enzymes.
E. coli crude cell extracts containing wild-type and mutant NDO enzymes were each analyzed for NDO activity in the presence of additional ferredoxinNAP and reductaseNAP by the procedure described in Materials and Methods. Crude cell extracts containing wild-type NDO formed 314 ± 76 nmol of cis-naphthalene dihydrodiol min−1 mg of protein−1. When Asp-205 was replaced by alanine, asparagine, glutamate, or glutamine, no NDO activity was detected.
Oxygen uptake by wild-type and mutant NDO enzymes.
E. coli crude cell extracts containing wild-type and mutant NDO enzymes were each analyzed for the ability to take up oxygen in response to the addition of naphthalene as described in Materials and Methods. Extract containing wild-type NDO catalyzed the consumption of 274 ± 35 nmol of O2 min−1 mg of protein−1 after correction for endogenous O2 uptake. Naphthalene and O2 consumption were stoichiometric. Specific activities calculated based on cis-naphthalene dihydrodiol formation and O2 consumption were within the limits of experimental error. No O2 uptake by extracts containing any of the Asp-205 mutant enzymes was detected. This result confirms that NDO activity is undetectable in the Asp-205 mutants and suggests that O2 uptake is not uncoupled from product formation in these mutant enzymes.
Detection of reaction products.
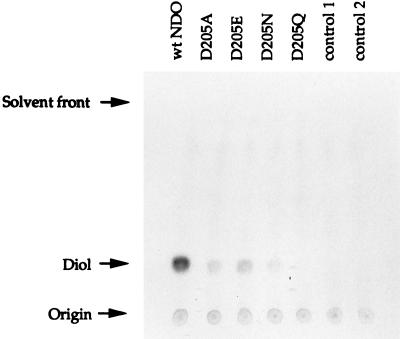
To detect the possibility that low levels of cis-naphthalene dihydrodiol were formed by the mutant enzymes, reaction mixtures were incubated for 45 min prior to TLC and autoradiography as described in Materials and Methods. The results show that replacement of Asp-205 with alanine, asparagine, or glutamate severely reduced the formation of cis-naphthalene dihydrodiol from naphthalene and replacement with glutamine completely eliminated all activity (Fig. 3).
FIG. 3.
Autoradiogram of TLC products showing the formation of cis-naphthalene dihydrodiol from [14C]naphthalene by extracts of cells expressing wild-type and mutant NDO enzymes as described in Materials and Methods. The lanes are as in Fig. 2, except that lane control 1 contains JM109(DE3)(pT7-5) extract and lane control 2 contains JM109(DE3)(pDTG155A) extract.
Quaternary structure of the mutant NDO enzymes.
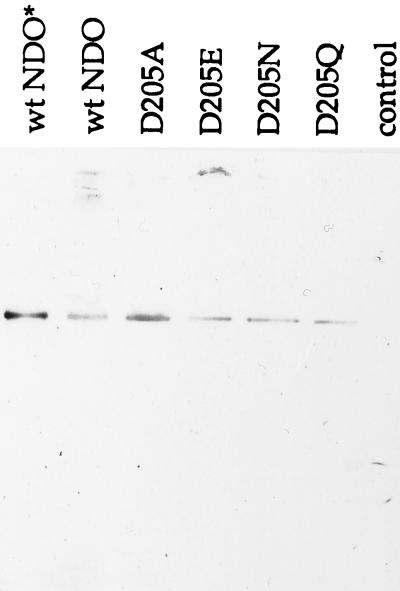
The conformational structures of the wild-type and mutant NDO enzymes were compared by native gel electrophoresis and Western blot analysis of crude cell extracts containing the various NDO derivatives. Only one band was detected in all extracts when using either a polyclonal antibody generated against purified NDO (reference 36 and data not shown) or a monoclonal antibody specific for the NDO α subunit (Fig. 4). Each band corresponded in mobility to the single band observed for purified NDO (Fig. 4, lane 1). This result indicates that each mutant NDO enzyme formed the native α3β3 conformation and suggests that reduced enzyme activities are not due to the inability to form proper quaternary protein structures.
FIG. 4.
Western blot of a native 8% polyacrylamide gel. Monoclonal antibody specific for the α subunit of NDO was used for detection of the native form of wild-type and mutant NDO enzymes. Lanes are as in Fig. 2. For extracts, approximately 100 μg of protein was loaded per lane.
Purification and characteristics of NDO-D205Q.
The least active mutant protein, NDO-D205Q, was purified by the same procedure used to purify wild-type NDO (27). Approximately 30 mg of purified NDO-D205Q was obtained from 75 g (wet weight) of cells. Purified NDO-D205Q had no detectable activity in assays with [14C]naphthalene (Table 3) or in O2 uptake assays. Benzene is an uncoupler of O2 uptake by NDO (26). The addition of 100 nmol of benzene to reaction mixtures containing purified wild-type NDO resulted in the uptake of almost 200 nmol of O2. In contrast, no O2 uptake was observed when benzene was added to reaction mixtures containing purified NDO-D205Q. This result provides further evidence that electrons are not being transferred to the mononuclear iron at the active site. Iron assays indicated that approximately equal amounts of iron were present in NDO and NDO-D205Q protein preparations (Table 3). The absorption spectrum of NDO-D205Q showed maxima at 330 and 462 nm and a shoulder at 558 nm, similar to the results obtained with wild-type NDO (13). The extinction coefficients for NDO and NDO-D205Q at 330 and 462 nm were not significantly different (Table 3). Reduced NDO-D205Q gave an EPR spectrum characteristic of a Rieske protein (11, 15), with g values very similar to those obtained with wild-type NDO (Table 3). The Rieske center of NDO-D205Q was reduced in the presence of NADH and catalytic amounts of ferredoxinNAP and reductaseNAP under anaerobic conditions. This treatment resulted in the loss of absorption at 462 and 558 nm and in new maxima at 380 and 524 nm, as was observed previously with wild-type NDO (13).
TABLE 3.
Characteristics of purified NDO-D205Q
| Enzyme | Sp acta | Iron content (g-atoms/αβ heterodimer) | Extinction coefficient (ɛ) (mM−1 cm−1)b at:
|
EPR spectrumc
|
||
|---|---|---|---|---|---|---|
| 330 nm | 462 nm | Oxidized | Reduced | |||
| NDO | 5.25 | 2.69 ± 0.04 | 13.4 ± 1.1 | 7.6 ± 0.6 | EPR silent | gx, 2.01 |
| gy, 1.91 | ||||||
| gz, 1.80 | ||||||
| NDO-D205Q | —d | 2.66 ± 0.03 | 13.6 ± 1.1 | 8.2 ± 0.6 | EPR silent | gx, 2.02 |
| gy, 1.91 | ||||||
| gz, 1.79 | ||||||
Specific activity was measured by the formation of [14C]naphthalene dihydrodiol as described in Materials and Methods and is defined as micromoles of product formed per minute per milligram of protein.
Per αβ heterodimer.
EPR data for wild-type NDO are from reference 45.
—, none detectable.
DISCUSSION
Biophysical studies have demonstrated the presence of a Rieske [2Fe-2S] center and mononuclear iron(II) in phthalate dioxygenase, benzene dioxygenase, and 4-methoxybenzoate O-demethylase (5, 16, 17, 46; reviewed in references 8, 31, and 40), enzymes that are evolutionarily related to NDO (18, 34). The mononuclear iron has been proposed to accept electrons from the Rieske center and activate oxygen by successive electron transfers (46).
It has been proposed that in NDO electrons travel from the Rieske center of one α subunit to the mononuclear iron of another α subunit, a distance of approximately 12 Å (24). The likelihood of electron transfer between two redox centers rapidly decreases with distance, and 12 Å is well within the typical range for electron transfer between redox centers (9, 32, 33). The Rieske [2Fe-2S] center and mononuclear iron within a single α subunit are 43.5 Å apart, a distance that has been reported to be too far for electron transfer to occur (33). It is generally accepted that electrons travel most efficiently through covalent bonds, less efficiently through hydrogen bonds, and least efficiently through space (2–4, 35). In electron transfer proteins, there is typically a preferred pathway for electron transfer between redox centers that is highly efficient. However, electrons can also travel via many minor pathways that may be much less efficient (2). The crystal structure of NDO revealed that Asp-205 is positioned to provide the most direct route for electron transfer from the Rieske center to the mononuclear iron through covalent and hydrogen bonds (24). Amino acid replacements at position 205 severely diminished the activity of the modified NDO enzymes, confirming that Asp-205 is indeed important for the catalytic reaction to occur. The detection of extremely low enzyme activity in three of the mutant proteins in which Asp-205 has been replaced is consistent with the possibility that the major route of electron transfer has been interrupted and at least one alternate, although highly inefficient, route of electron transfer is available.
The physical properties of the least active enzyme, NDO-D205Q (Table 3), suggest that the mutation does not significantly affect the structure of the protein. Measured extinction coefficients and EPR spectra of wild-type NDO and NDO-D205Q do not differ significantly. Furthermore, similar amounts of iron remain bound to purified wild-type and mutant proteins. Reduction of the Rieske center in NDO-D205Q by NADH in the presence of ferredoxinNAP and reductaseNAP is demonstrated by the change in the UV-visible spectrum. Thus, ferredoxinNAP can interact with NDO-D205Q in a manner similar to its interaction with wild-type NDO. This result, together with those from native gel electrophoresis studies with extracts (Fig. 4), suggests that contacts formed between adjacent α subunits in the mutant enzymes are similar to contacts in the wild-type enzyme and that the quaternary structure is not affected in the mutant proteins. The reduction of the Rieske center of NDO-D205Q demonstrates that the pathway of electron transfer is intact from NADH to the Rieske center of the mutant oxygenase. We have demonstrated that the substrate analog benzene, which has been shown to uncouple the reaction with the wild-type enzyme (26), does not stimulate any O2 uptake by NDO-D205Q, suggesting that electrons are not being transferred from the Rieske center to the mononuclear iron in this protein. It is difficult to directly monitor electron transfer from the Rieske center to the mononuclear iron, because of the spectroscopic inaccessibility of Fe(II) (39) and the fact that signals from the Rieske [2Fe-2S] center dominate in many spectroscopic analyses (38). Therefore, we must rely on assays that measure product formation or substrate removal; these do not differentiate individual electron transfer steps. All the results presented here are consistent with the participation of Asp-205 in electron transfer from the Rieske center to the active-site iron. However, we cannot rule out the possibility that mutations at Asp-205 affect some other aspect of catalysis.
With the crystal structure of NDO now available, site-directed mutagenesis studies such as these combined with biophysical analyses should allow the elucidation of the catalytic mechanism of this interesting class of iron-containing enzymes, the aromatic ring-hydroxylating dioxygenases.
ACKNOWLEDGMENTS
This work was supported by U.S. Public Health Service grant GM29909 from the National Institute of General Medical Sciences.
We thank H. Jiang for carrying out iron determinations, K. Lee for providing purified ReductaseNAP and constructing pMASTER-1, G. Buettner for determining the EPR spectra at the University of Iowa ESR Facility, and Maja Ivkovic-Jensen for critical reading of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1993. [Google Scholar]
- 2.Beratan D N, Onuchic J N. The protein bridge between redox centers. In: Bendall D S, editor. Protein electron transfer. Oxford, United Kingdom: Bios Scientific; 1996. pp. 23–42. [Google Scholar]
- 3.Beratan D N, Onuchic J N, Winkler J R, Gray H B. Electron-tunneling pathways in proteins. Science. 1992;258:1740–1741. doi: 10.1126/science.1334572. [DOI] [PubMed] [Google Scholar]
- 4.Beratan D N, Skourtis S S. Electron transfer mechanisms. Curr Opin Chem Biol. 1998;2:235–243. doi: 10.1016/s1367-5931(98)80065-3. [DOI] [PubMed] [Google Scholar]
- 5.Bill E, Bernhardt F-H, Trautwein A X, Winkler H. Mössbauer investigation of the cofactor iron of putidamonooxin. Eur J Biochem. 1985;147:177–182. doi: 10.1111/j.1432-1033.1985.tb08734.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Brown S M, Hudlicky T. The use of arene-cis-diols in synthesis. In: Hudlicky T, editor. Organic synthesis: theory and applications. Greenwich, Conn: JAI Press, Inc.; 1993. pp. 113–176. [Google Scholar]
- 8.Butler C S, Mason J R. Structure-function analysis of the bacterial aromatic ring-hydroxylating dioxygenases. Adv Microb Physiol. 1997;38:47–84. doi: 10.1016/s0065-2911(08)60155-1. [DOI] [PubMed] [Google Scholar]
- 9.Canters G W, Dennison C. Biological electron transfer: structural and mechanistic studies. Biochimie. 1995;77:506–515. doi: 10.1016/0300-9084(96)88167-3. [DOI] [PubMed] [Google Scholar]
- 10.Carless H A J. The use of cyclohexa-3,5-diene-1,2-diols in enantiospecific synthesis. Tetrahedron Asymmetry. 1992;3:795–826. [Google Scholar]
- 11.Cline J F, Hoffman B M, Mims W B, LaHaie E, Ballou D P, Fee J A. Evidence for N coordination to Fe in the [2Fe-2S] clusters of Thermus Rieske protein and phthalate dioxygenase from Pseudomonas. J Biol Chem. 1985;260:3251–3254. [PubMed] [Google Scholar]
- 12.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 13.Ensley B D, Gibson D T. Naphthalene dioxygenase: purification and properties of a terminal oxygenase component. J Bacteriol. 1983;155:505–511. doi: 10.1128/jb.155.2.505-511.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ensley B D, Gibson D T, Laborde A L. Oxidation of naphthalene by a multicomponent enzyme system from Pseudomonas sp. strain NCIB 9816. J Bacteriol. 1982;149:948–954. doi: 10.1128/jb.149.3.948-954.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fee J A, Findling K L, Yoshida T, Hille R, Tarr G E, Hearshen D O, Dunham W R, Day E P, Kent T A, Münck E. Purification and characterization of the Rieske iron-sulfur protein from Thermus thermophilus. J Biol Chem. 1984;259:124–133. [PubMed] [Google Scholar]
- 16.Gassner G T, Ballou D P, Landrum G A, Whittaker J W. Magnetic circular dichroism studies on the mononuclear ferrous active site of phthalate dioxygenase from Pseudomonas cepacia show a change of ligation state on substrate binding. Biochemistry. 1993;32:4820–4825. doi: 10.1021/bi00069a017. [DOI] [PubMed] [Google Scholar]
- 17.Geary P J, Saboowalla F, Patil D, Cammack R. An investigation of the iron-sulphur proteins of benzene dioxygenase from Pseudomonas putida by electron-spin-resonance spectroscopy. Biochem J. 1984;217:667–673. doi: 10.1042/bj2170667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harayama S, Kok M, Neidle E L. Functional and evolutionary relationships among diverse oxygenases. Annu Rev Microbiol. 1992;46:565–601. doi: 10.1146/annurev.mi.46.100192.003025. [DOI] [PubMed] [Google Scholar]
- 19.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 20.Hudlicky T, Reed J W. An evolutionary perspective of microbial oxidations of aromatic compounds in enantioselective synthesis: history, current status, and perspectives. In: Hassner A, editor. Advances in asymmetric synthesis. Greenwich, Conn: JAI Press Inc.; 1995. pp. 271–312. [Google Scholar]
- 21.Jeffrey A M, Yeh H J C, Jerina D M, Patel T R, Davey J F, Gibson D T. Initial reactions in the oxidation of naphthalene by Pseudomonas putida. Biochemistry. 1975;14:575–583. doi: 10.1021/bi00674a018. [DOI] [PubMed] [Google Scholar]
- 22.Jerina D M, Daly J W, Jeffrey A M, Gibson D T. cis-1,2-Dihydroxy-1,2-dihydronaphthalene: a bacterial metabolite from naphthalene. Arch Biochem Biophys. 1971;142:394–396. doi: 10.1016/0003-9861(71)90298-0. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H, Parales R E, Lynch N A, Gibson D T. Site-directed mutagenesis of conserved amino acids in the alpha subunit of toluene dioxygenase: potential mononuclear non-heme iron coordination sites. J Bacteriol. 1996;178:3133–3139. doi: 10.1128/jb.178.11.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kauppi B, Lee K, Carredano E, Parales R E, Gibson D T, Eklund H, Ramaswamy S. Structure of an aromatic ring-hydroxylating dioxygenase—naphthalene 1,2-dioxygenase. Structure. 1998;6:571–586. doi: 10.1016/s0969-2126(98)00059-8. [DOI] [PubMed] [Google Scholar]
- 25.Lange S J, Que L., Jr Oxygen activating nonheme iron oxygenases. Curr Opin Chem Biol. 1998;2:159–172. doi: 10.1016/s1367-5931(98)80057-4. [DOI] [PubMed] [Google Scholar]
- 26.Lee K. Biochemical studies on toluene and naphthalene dioxygenases. Ph.D. thesis. Iowa City: The University of Iowa; 1995. [Google Scholar]
- 27.Lee K, Kauppi B, Parales R E, Gibson D T, Ramaswamy S. Purification and crystallization of the oxygenase component of naphthalene dioxygenase in native and selenomethionine derivatized forms. Biochem Biophys Res Commun. 1997;241:553–557. doi: 10.1006/bbrc.1997.7863. [DOI] [PubMed] [Google Scholar]
- 28.Lee S-Y, Rasheed S. A simple procedure for maximum yield of high-quality plasmid DNA. BioTechniques. 1990;9:676–679. [PubMed] [Google Scholar]
- 29.Ley S V. Stereoselective synthesis of inositol phosphates. Pure Appl Chem. 1990;62:2031–2034. [Google Scholar]
- 30.Lynch N A, Jiang H, Gibson D T. Rapid purification of the oxygenase component of toluene dioxygenase from a polyol-responsive monoclonal antibody. Appl Environ Microbiol. 1996;62:2133–2137. doi: 10.1128/aem.62.6.2133-2137.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason J R, Cammack R. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu Rev Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 32.Moser C C, Dutton P L. Outline of theory of protein electron transfer. In: Bendall D S, editor. Protein electron transfer. Oxford, United Kingdom: Bios Scientific; 1996. pp. 1–21. [Google Scholar]
- 33.Moser C C, Keske J M, Warncke K, Farid R S, Dutton P L. Nature of biological electron transfer. Nature. 1992;355:796–802. doi: 10.1038/355796a0. [DOI] [PubMed] [Google Scholar]
- 34.Nakatsu C H, Straus N A, Wyndham R C. The nucleotide sequence of the Tn5271 3-chlorobenzoate 3,4-dioxygenase genes (cbaAB) unites the class IA oxygenases in a single lineage. Microbiology. 1995;141:485–495. doi: 10.1099/13500872-141-2-485. [DOI] [PubMed] [Google Scholar]
- 35.Onuchic J N, Beratan D N, Winkler J R, Gray H B. Pathway analysis of protein electron-transfer reactions. Annu Rev Biophys Biomol Struct. 1992;21:349–377. doi: 10.1146/annurev.bb.21.060192.002025. [DOI] [PubMed] [Google Scholar]
- 36.Parales R E, Emig M D, Lynch N A, Gibson D T. Substrate specificities of hybrid naphthalene and 2,4-dinitrotoluene dioxygenase enzyme systems. J Bacteriol. 1998;180:2337–2344. doi: 10.1128/jb.180.9.2337-2344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parales, R. E., and D. T. Gibson. Unpublished data.
- 38.Pavel E G, Martins L J, Ellis W R J, Solomon E I. Magnetic circular dichroism studies of exogenous ligand and substrate binding to the non-heme ferrous active site in phthalate dioxygenase. Chem Biol. 1994;1:173–183. doi: 10.1016/1074-5521(94)90007-8. [DOI] [PubMed] [Google Scholar]
- 39.Que L J. Oxygen activation at nonheme iron centers. In: Reedijk J, editor. Bioinorganic catalysis. New York, N.Y: Marcel Dekker, Inc.; 1993. pp. 347–393. [Google Scholar]
- 40.Que L J, Ho R Y N. Dioxygen activation by enzymes with mononuclear non-heme iron active sites. Chem Rev. 1996;96:2607–2624. doi: 10.1021/cr960039f. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 42.Sheldrake G N. Biologically derived arene cis-dihydrodiols as synthetic building blocks. In: Collins A N, Sheldrake G N, Crosby J, editors. Chirality in industry: the commercial manufacture and application of optically active compounds. Chichester, United Kingdom: John Wiley & Sons Ltd.; 1992. pp. 127–166. [Google Scholar]
- 43.Stanier R Y, Palleroni N J, Doudoroff M. The aerobic pseudomonads: a taxonomic study. J Gen Microbiol. 1966;43:159–271. doi: 10.1099/00221287-43-2-159. [DOI] [PubMed] [Google Scholar]
- 44.Suen W-C. Gene expression of naphthalene dioxygenase from Pseudomonas sp. NCIB 9816-4 in Escherichia coli. Ph.D. thesis. Iowa City: The University of Iowa; 1991. [Google Scholar]
- 45.Suen W-C, Gibson D T. Recombinant Escherichia coli strains synthesize active forms of naphthalene dioxygenase and its individual α and β subunits. Gene. 1994;143:67–71. doi: 10.1016/0378-1119(94)90606-8. [DOI] [PubMed] [Google Scholar]
- 46.Twilfer H, Bernhardt F-H, Gersonde K. Dioxygen-activating iron center in putidamonooxin: electron spin resonance investigation of the nitrosylated putidamonooxin. Eur J Biochem. 1985;147:171–176. doi: 10.1111/j.1432-1033.1985.tb08733.x. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 48.Zabinski R, Münck E, Champion P M, Wood J M. Kinetic and Mössbauer studies on the mechanism of protocatechuic acid 4,5-oxygenase. Biochemistry. 1972;11:3212–3219. doi: 10.1021/bi00767a012. [DOI] [PubMed] [Google Scholar]