Addendum to: EMBO Mol Med (2019) 11: e11115. DOI 10.15252/emmm.201911115 ¦ Published online 3 November 2019
The authors recently contacted the journal to inform the editor of changes they discovered in the isogenic control (HCMrep) clone derived from the hypertrophic cardiomyopathy (HCM)‐specific hiPSC clone.
After the initial publication, the authors performed a new genomic sequencing of the HCM and HCMrep hiPSC clones, which revealed in the HCMrep an additional point mutation (T>C, blue in Fig 1) next to the CRISPR/Cas9‐mediated repair of the HCM gene variant (T>C, violet). This heterozygous mutation (c.697+2T>C) is an on‐target artifact of CRISPR/Cas9 gene editing and is located in the donor splice site of ACTN2 intron 8.
Figure 1. Validation of the HCM and HCMrep hiPSC lines.
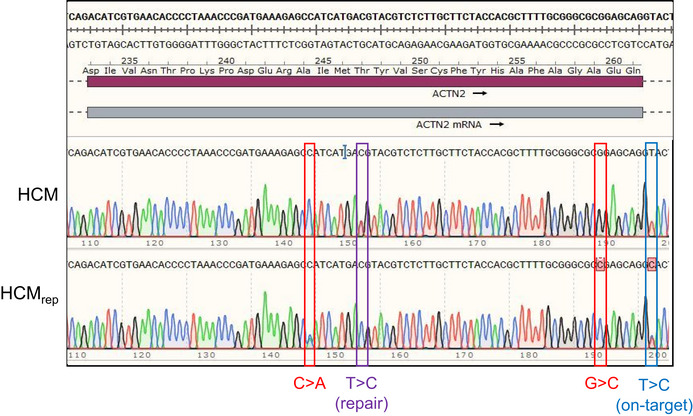
Sequencing of the HCM and HCMrep hiPSC clones. The HCMrep clone revealed on the +strand the repaired mutation (T>C (repair), violet), the 2 silent mutations from the repair template (C>A and G>C, red), and the on‐target defect (T>C (on‐target), blue). Note that the SnapGene software sometimes underlined the presence of a mutation with a red square at the nucleotide site, but sometimes did not.
The authors evaluated whether this additional c.697+2T>C transition may affect the mRNA and protein pattern in cultured HCMrep hiPSC‐derived cardiomyocytes (Fig 2). RT–PCR revealed additional bands after treatment with emetine, which blocks translation and prevents the degradation of nonsense mRNAs (Fig 2A). Sequencing of emetine‐treated RT–PCR subclones revealed wild‐type mRNA (Fig 2B) in 72% of cases and two nonsense mRNAs (Fig 2C and D) in 28% of cases. Both mutant mRNAs give rise to a frameshift and a premature termination codon, leading to C‐terminal‐truncated proteins. Western blot for ACTN2 revealed only one band without any truncated protein at the estimated molecular weight of about 30 kDa in HCMrep hiPSC‐derived cardiomyocytes (Fig 2E).
Figure 2. Evaluation of mRNAs and proteins from HCMrep hiPSC‐derived cardiomyocytes.
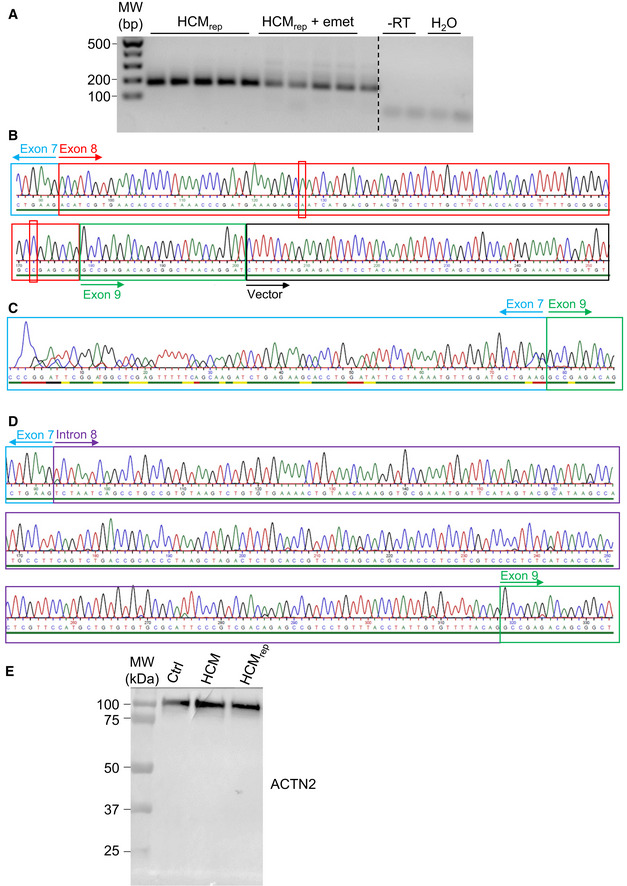
HCMrep hiPSC‐derived cardiomyocytes were cultured for 5 days and treated or not with emetine (600 μg/ml for 4 h).
-
AAgarose gel electrophoresis of RT–PCR performed between ACTN2 exons 7 and 9.
-
BSanger sequencing of wild‐type RT–PCR subclone containing the two silent mutations (C>A and G>C, red) in exon 8.
-
CSanger sequencing of RT–PCR mutant‐1 subclone deleted of exon 8.
-
DSanger sequencing of RT–PCR mutant‐2 subclone deleted of exon 8 and retaining 226 bp of intron 8.
-
ECrude protein fractions from control (Ctrl), HCM, and HCMrep hiPSC cardiomyocytes were processed by Western blot for ACTN2 on an 8% SDS–PAGE. Abbreviations: emet, emetine; −RT, without reverse transcriptase.
These data showed that HCMrep hiPSC cardiomyocytes exhibit only wild‐type ACTN2 mRNA in the absence of emetine and only the wild‐type full‐length ACTN2 protein. This addendum indicates that the original data are not affected by this newly discovered Cas9‐mediated on‐target mutation.


