Abstract
It is well known that de-identified research brain images from MRI and CT can potentially be re-identified using face recognition; however, this has not been examined for PET images. We generated face reconstruction images of 182 volunteers using amyloid, tau, and FDG PET scans, and we measured how accurately commercial face recognition software (Microsoft Azure’s Face API) automatically matched them with the individual participants’ face photographs. We then compared this accuracy with the same experiments using participants’ CT and MRI. Face reconstructions from PET images from PET/CT scanners were correctly matched at rates of 42% (FDG), 35% (tau), and 32% (amyloid), while CT were matched at 78% and MRI at 97–98%. We propose that these recognition rates are high enough that research studies should consider using face de-identification (“de-facing”) software on PET images, in addition to CT and structural MRI, before data sharing. We also updated our mri_reface de-identification software with extended functionality to replace face imagery in PET and CT images. Rates of face recognition on de-faced images were reduced to 0–4% for PET, 5% for CT, and 8% for MRI. We measured the effects of de-facing on regional amyloid PET measurements from two different measurement pipelines (PETSurfer/FreeSurfer 6.0, and one in-house method based on SPM12 and ANTs), and these effects were small: ICC values between de-faced and original images were > 0.98, biases were <2%, and median relative errors were <2%. Effects on global amyloid PET SUVR measurements were even smaller: ICC values were 1.00, biases were <0.5%, and median relative errors were also <0.5%.
Keywords: Face recognition, De-facing, De-identification, Anonymization, PET/CT
1. Introduction
We have recently shown that modern commercial face recognition software now has very high accuracy for matching faces reconstructed from brain magnetic resonance imaging (MRI) with face photographs, potentially allowing re-identification of anonymous research participants from de-identified, publicly shared research datasets (Schwarz et al., 2019a, 2021a). Typical data use agreements for public datasets require a downloader to agrvvee that they will not attempt to re-identify participants, but these offer no direct protection for participants. The U.S. Health Insurance Portability and Accountability Act (HIPAA) specifies that de-identified data must remove, among other items, “Full face photographic images and any comparable images” (U.S. Department of Health and Human Services Office for Civil Rights, 2013). Our previous findings suggest that advancing technology may increasingly give brain images “comparable” identifiability. Software for automatic face-deidentification or “de-facing” MRI (automatically removing facial features or replacing them with another face, which is sometimes called “re-facing”) can greatly reduce this possibility of re-identification, but further work is needed to develop better defacing tools that more effectively prevent face recognition and further minimize effects on measurements from de-faced data (Buimer et al., 2021; de Sitter et al., 2020; Gao et al., 2022; Schwarz et al., 2021a).
Earlier works have also shown that computed tomography (CT) images, like MRI, are identifiable via both visual (human) and automated face recognition (Mazura et al., 2012; Chen et al., 2014; Parks and Monson, 2017). Positron Emission Tomography (PET) images of the brain are commonly shared by imaging research studies (Jagust et al., 2015; LaMontagne et al., 2019; Sperling et al., 2014), especially those of aging and dementia because amyloid and tau PET are typically used for the research diagnosis and staging of Alzheimer’s disease (Jack et al., 2018). However, the potential of these images for re-identification via captured facial imagery has not been studied, perhaps due to the belief that brain PET images typically lack sufficient resolution and field of view (FOV) for face reconstruction. Our aims for this study were: 1) measure performance of automated face recognition to match face photographs with face reconstructions derived from brain PET scans; and 2) measure performance of our automated mri_reface software for modifying the PET images, both in terms of how well it prevents face recognition, and the effects of its image modifications on automated brain biomarker measurements.
2. Materials and methods
2.1. Dataset for Aim 1 (face recognition)
Participants:
Our face recognition dataset included 157 individuals from our previous face recognition study (Schwarz et al., 2021a), as well as 25 additional volunteers from continuing recruitment. In total, we recruited 182 volunteers (ages 34–93, mean=63.2, median=65, SD=15.5), stratified by sex and age-decade, who had previous brain imaging within six months as part of their existing enrollment in the Mayo Clinic Study of Aging (MCSA) (Petersen et al., 2010; Roberts et al., 2008). All participants provided informed consent for this specific study, which was approved by the Mayo Clinic Institutional Review Board.
Photographs:
We photographed each participant’s face under indoor lighting conditions using standard iPads (Apple Inc., Cupertino, CA; models Air 2 and 6th generation). Participants were instructed to look directly at the camera, and then approximately 10 degrees up, down, left, and right, for a total of five photos. This design intended to provide five suitable, somewhat-unique photos of each individual with minimal participant burden, because we assume that an individual motivated to re-identify a study participant would be able to find more than one suitable photograph of their face. Photos were manually cropped loosely around the head and converted to grayscale to better match MRI (which does not capture color). This cropping retained the head, hair, and ears, removing only distant background and torso to reduce image size and speed up repeated image uploading during testing.
Brain imaging:
All 182 participants underwent a head MRI protocol on Siemens Prisma scanners including a magnetization prepared rapid gradient echo (MPRAGE) sequence (TR/TE/TI = 2300/3.14/945 ms, flip angle 9°, 0.8 mm isotropic resolution), and a sagittal 3D FLAIR sequence (TR/TE/TI = 4800/441/1650, resolution 1.0 × 1.0 × 1.2mm) matching the ADNI3 (Alzheimer’s disease Neuroimaging Initiative) protocol. Among these participants, 167 had previously undergone imaging with Pittsburgh Compound B (PiB) Amyloid PET (Klunk et al., 2004) and Flortaucipir (FTP) tau PET (Xia et al., 2013), and 129 had previously undergone imaging with Fludeoxyglucose (FDG) PET, all using GE PET/CT scanners (models Discovery 690XT and Discovery MI). We refer to these PET scanners as “older”, representing the generation of clinical PET/CT that are currently most prevalent in imaging research datasets. A small fraction of these participants (14 with FDG, 20 with PiB, and 19 with FTP) were also scanned at later visits with a Siemens Biograph64 Vision 600. This scanner creates images with higher signal to noise ratio than older generation models due to faster coincidence timing resolution, digital scintillation detectors, and ability to leverage this improved time-of-flight information during reconstructions. We show data from both scanners for that subset of individuals. For all PET/CT scans, a low-dose CT scan was acquired for use in attenuation correction, and PET images were reconstructed on-scanner using OSEM iterative algorithms with a 5mm Gaussian post-reconstruction filter and standard corrections for attenuation, scatter, random coincidences, and decay. All PET protocols acquired four late-uptake dynamic frames, which were co-registered and summed to produce static images from post-injection minutes 30–45 for FDG, 40–60 for PiB, and 80–100 for FTP. The low-dose CT scans were separately also used for testing face reconstruction and recognition from CT, but aside from their use in on-scanner attenuation correction, they were not used when testing face recognition from PET images.
2.2. Methods for Aim 1 (face recognition)
Face reconstruction:
Details of our methods for creating face reconstructions from structural MRI have been previously published (Schwarz et al., 2021a, 2019a). We used our previously described “standard” face reconstruction method. Briefly, we apply automated thresholding using Otsu’s method (Otsu, 1979) to binarize the image, remove any regions of suprathreshold voxels that are not spatially connected with the largest region, and attempt to re-attach any aliased nose-parts behind the head (which not occur in PET or CT and thus this step does nothing). After this preprocessing, we construct an isosurface using the isosurface function in Matlab, and create a render from this using Surf_Ice (Rorden, n.d.). For PET/CT images from older scanners, we also tested our “advanced” face reconstructions that match each input image with an average template of the same modality and use this template to “fill in” missing regions. The templates used for this process are the same average face templates we use in mri_reface (see below). For older PET/CT images, the advanced reconstructions replaced parts of nose and mouth that were outside the acquisition field of view, enabling face detection and recognition using the imaged regions (e.g. eye and forehead) despite their frequently missing nose and mouth. Both methods, originally designed for MRI, were applied for PET and CT without any modifications except altering some multipliers to automated hresholds to account for different contrast levels across image types.
De-identification (de-facing):
We have previously published a description and validation of mri_reface, our software for replacing face information in MRI to prevent potential re-identification via face recognition (Schwarz et al., 2021a). The software registers each input image to an average template image of the same modality and transforms the template to the native input image space (linearly in the face/ears and nonlinearly in the rest of the image)performs global and local intensity matching, then replaces face imagery with the linearly transformed average face from the template. Since that publication, we have publicly released this software, free for use by the research community (https://www.nitrc.org/projects/mri_reface). We have also improved its performance for preventing face recognition by replacing a larger area of the eyebrow ridge and forehead, because the eyebrow ridge (periocular region) is the most important area for modern automatic face recognition methods (Juefei-Xu et al., 2015, 2014, 2011; Juefei-Xu and Savvides, 2016; Le et al., 2014; Woodard et al., 2010). These changes will be described and validated in more detail as part of an upcoming separate publication exploring the potential for face recognition in additional MRI sequences.
We also extended mri_reface to support PET and CT by creating new average template images for PiB and Florbetapir (FBP) amyloid PET, Flortaucipir tau PET, and FDG PET, and by disabling replacement of air in regions in front of and behind the head, because PET and CT do not have MRI’s “aliasing” artifacts that may contain identifiable features. We show two examples of mri_reface on PET images in Fig. 1. We created templates for CT, FDG PET, PiB PET, and Flortaucipir PET using our previously published techniques (Schwarz et al., 2021a) with ANTs software (Avants et al., 2010) from in-house scans of participants in the MCSA, Mayo Clinic Alzheimer’s Disease Research Center, and Mayo Clinic DLB Consortium studies (n=192 FDG, n=200 PiB, n=200 FTP, n=200 CT) scanned on a Siemens Biograph64 Vision600 PET/CT scanner (the highest quality PET data available to us). Group-wise registration for PET from PET/CT was performed using corresponding CT images, and these parameters were used to transform the corresponding PET before averaging. To create the Florbetapir PET template, we used similar techniques with scans of 120 participants from PET/MRI scanners in the public OASIS-3 dataset (LaMontagne et al., 2019), except these PET images were group-co-registered using PET images directly, since no CT was available. After group-co-registration, templates were warped (nonlinearly) to match existing MRI-based templates, using ANTs (Avants et al., 2008). Prior to template creation, we intensity-normalized all PET scans to the standardized uptake value ratio (SUVR), using corresponding MRI. SUVR for amyloid and tau tracers used the bilateral cerebellar crus as a reference region; cerebellar reference regions are the typical choice for amyloid and tau PET because the cerebellum is not typically affected by Alzheimer’s pathology until very late stages (Klunk et al., 2004), and we used the crus sub-region to avoid bleed-in from cortical signal across the tentorium (Baker et al., 2017; Lowe et al., 2018). For FDG, we used the pons as a reference region, which is a typical choice because cerebellar cortex metabolism is affected by age and disease (Nugent et al., 2020). We present images of these templates later, in the Results section.
Fig. 1.
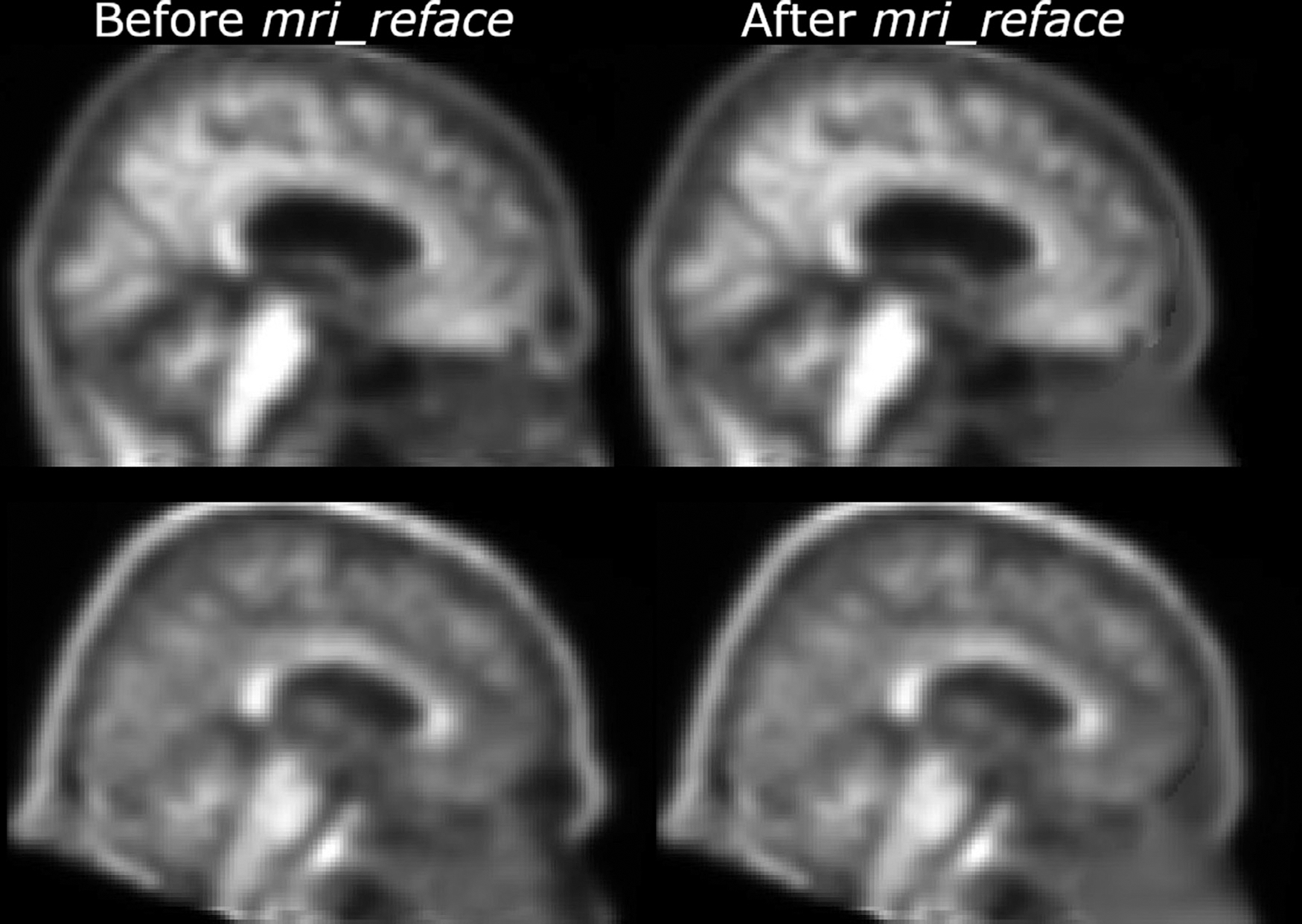
Two example PET images Florbetapir PET scans from ADNI (with all standard ADNI pre-processing), both before (left) and after (right) replacing face imagery with mri_reface.
Face recognition (matching):
Details of our methods for using the Microsoft Azure Face API to automatically match MRI-based face reconstructions with participant photographs have been previously published (Schwarz et al., 2021a, 2019a). Briefly, our testing paradigm measures whether a motivated individual, who has reason to believe that participant’s brain images exist in a particular dataset, could correctly identify that participant using their photographs with face recognition software. We generated face reconstructions (renders) from each brain image (see Face Reconstruction, above), and we used these as the training set for a face recognition classifier. When given an input face image (a participant’s face photograph, in our usage), this classifier returns a ranking of the faces in the training set (reconstructions from brain images, in our usage) by their similarity to the input face photograph. For each participant in the dataset, we input standard face photographs and recorded which face reconstruction in the dataset was chosen by the classifier as the best match. When the correct face (reconstructed from de-identified imaging) was the software’s top-ranked match for the face photograph (e.g. identified photos that would be available to someone trying to re-identify a specific participant), we counted this as a successful re-identification. When brain images from a particular modality or sequence were not available for a participant, we did not input their photographs, because there is no correct match for them in the dataset. Our approach could have been applied to MRI, PET, and CT images in the current study without any modification; however, we took the opportunity to update our testing software to use Microsoft’s latest pre-trained models for face detection (detection_03) and face recognition (recognition_04), updated from detection_01 and recognition_02 in our most recent earlier work (Schwarz et al., 2021a). These algorithms are proprietary Microsoft products, and their underlying methodologies have not been published.
Outcome measure:
We statistically compared matching proportions using a Pearson’s chi-squared test implemented by prop.test in R version 3.6.2 (R Development Core Team, 2008).
2.3. Dataset for Aim 2 (effects of de-facing on measurements)
Note that this aim does not require that participants have facial photographs available. We re-used a cross-sectional dataset of 300 individuals (100 scanned using each MRI vendor, each including 50 cognitively unimpaired (CU) + 50 with clinically diagnosed Alzheimer’s disease (AD)) from the Alzheimer’s Disease Neuroimaging Initiative (ADNI), previously constructed for our earlier work (Schwarz et al., 2021a). 244 of these individuals had available PET scans with Florbetapir, and we used this subset of 244 individuals for the current work. Among those 244 (119 CU, 125 with clinical AD), 85 had GE MRI, 78 had Phillips MRI, and 81 had Siemens MRI. PET scanner manufacturers were: 89 GE, 26 Philips, 126 Siemens, and 4 CPS Innovations. Details of ADNI PET acquisitions have been previously published (Jagust et al., 2010). We used the maximally preprocessed (co-registered, averaged, standardized image and voxel size, uniform resolution) images provided by ADNI (Joshi et al., 2009), which are considered homogenized against technical sources of variability across PET scanners. We have also previously shown that differing MRI vendors have only negligible effects on computing regional PET SUVR measurements (Schwarz et al., 2019b).
2.4. Methods for Aim 2 (effects of de-facing on measurements)
Regional Standardized Uptake Value Ratios (SUVRs) were measured for each PET image, before and after de-facing, using two different pipelines that each use corresponding T1-weighted MRIs for spatial normalization. For de-faced PET images, we also de-faced the corresponding MRIs, since we expect that shared datasets with de-faced PET would also de-face MRI. The pipelines were: 1) a previously published in-house cross-sectional method (Schwarz et al., 2021b) based on SPM12 (Ashburner, 2009), the Mayo Clinic Adult Lifespan Template (MCALT; https://www.nitrc.org/projects/mcalt/) (Schwarz et al., 2017), and ANTs (Avants et al., 2008), and 2) PETSurfer (Greve et al., 2016) from FreeSurfer version 6.0 (Fischl, 2012). The in-house pipeline performs a rigid registration of each PET to the corresponding T1-weighted MRI, then re-samples and performs regional calculations in this native MRI space with SPM12-based MRI segmentation and an ANTS transform of the MCALT_ADIR122 atlas to native MRI space. PETSurfer performs a rigid registration of each PET to the corresponding MRI using mri_coreg, then transforms these PET images to its standard MNI-based template space and performs regional calculations in this space using the Desikan-Killiany atlas (Desikan et al., 2006). We did not use partial volume correction (PVC) for either approach, to minimize effects of MRI on PET measurements. SUVR intensity normalization used the cerebellar crus for the in-house pipeline in order to avoid potential bleed-in from cortical uptake across the tentorium into the superior cerebellum. This option is not available for PETSurfer, so we used the standard cerebellar gray matter reference.
Outcome measures:
We compared regional SUVR values from each pipeline before and after de-facing using three metrics: 1) Intraclass Correlation Coefficient (ICC, a measure of non-systematic or random error), 2) bias (a measure of systematic error), and 3) median relative error (a measure with more intuitive units, including both systematic and non-systematic error). For ICC, we used the ICC function from the pysch package in R (Revelle, 2019) to calculate the fixed-raters ICC3 variant that is not sensitive to differences in means between raters (i.e. is not sensitive to systematic error). We then separately measured the systematic error (bias) of the de-faced image measurements as the percent difference between the identity line and a linear least-squares fit (lm function) of the original vs. de-faced measurements, taken at the “centercept” point (mean value across the x axis, i.e. all measurements from the unmodified images) (Wainer, 2000). We then separately calculated median relative error as a percentage by subtracting each pair of original and de-faced SUVR measurements for each region for each scan, taking the absolute value, normalizing by the SUVR from the original scan, and taking the median across scans (Eq. 1).
| (1) |
Finally, we then summarized these ICC, bias, and median relative error values (across all regions, within each combination of pipeline and de-facing method) using median values and boxplots. For readers who may prefer root mean square error instead of median relative error, we also provide this measurement in the supplementary table.
3. Results
3.1. Validation criterion 1: protection from face recognition
Human, visual face recognition assessment:
First, we present examples of photographs and face reconstructions from four volunteers, in Fig, 2. These images, from participants who generously consented to their publication, allow visual comparison of face identifiability across modalities and across PET scanner generations. While skin pigmentation, hair, and eyewear are not imaged by these modalities, many structural features of the faces were preserved. The older-generation, currently prevalent PET scanners have worse resolution, a lower signal to noise ratio (i.e. reduced sensitivity and contrast), and smaller field of view (i.e. the mouth is often omitted) than newer scanners, but the shape of the eyebrow ridge (the most important feature for modern algorithmic face recognition) was still present. Recognizability was visually similar across all three PET tracers. Artifacts around the mouth due to dental fillings or implants were present on MRI and CT in the two right-most participants, but not on PET. On the CT, the two left-most participants also had parts of face masks, worn due to COVID-19, visible across the nose bridge. Next in Fig. 3, we also show the average head template images for each image type that we constructed for use during face replacement with mri_reface. This figure is provided for additional visual comparison across modalities, including average brain slices for comparison of contrast properties (which we cannot show for individual participants), but because images were averaged across multiple scans from multiple individuals, they have higher image quality than most individual participant scans.
Fig. 2.
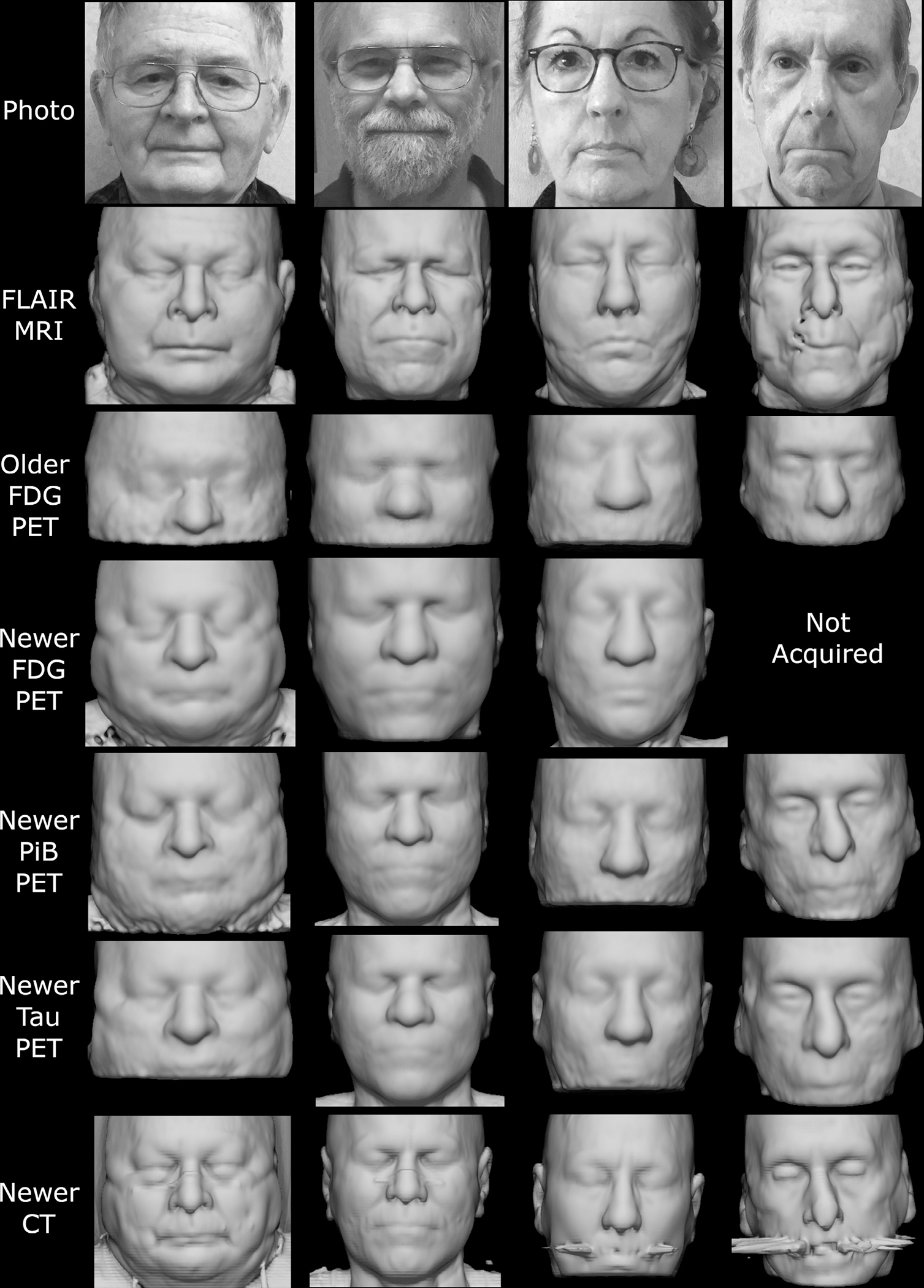
Example face reconstructions from PET and CT, for visual comparison with photographs and MRI. PET scans from the newer model of clinical PET/CT scanner showed many identifiable features, across all tracers. CT scans (from PET/CT) were also highly identifiable despite some dental artifacts and face mask nose bridges. PET from older-generation scanners had lower quality and a smaller field of view but retained some identifiable features. These participants specifically consented to allow publication of their photographs and face reconstructions. Note that although positioning and head restraints in the scanner distort the lower face in the facial reconstructions, the brow ridge, which is a dominant feature in facial recognition, is minimally affected.
Fig. 3.
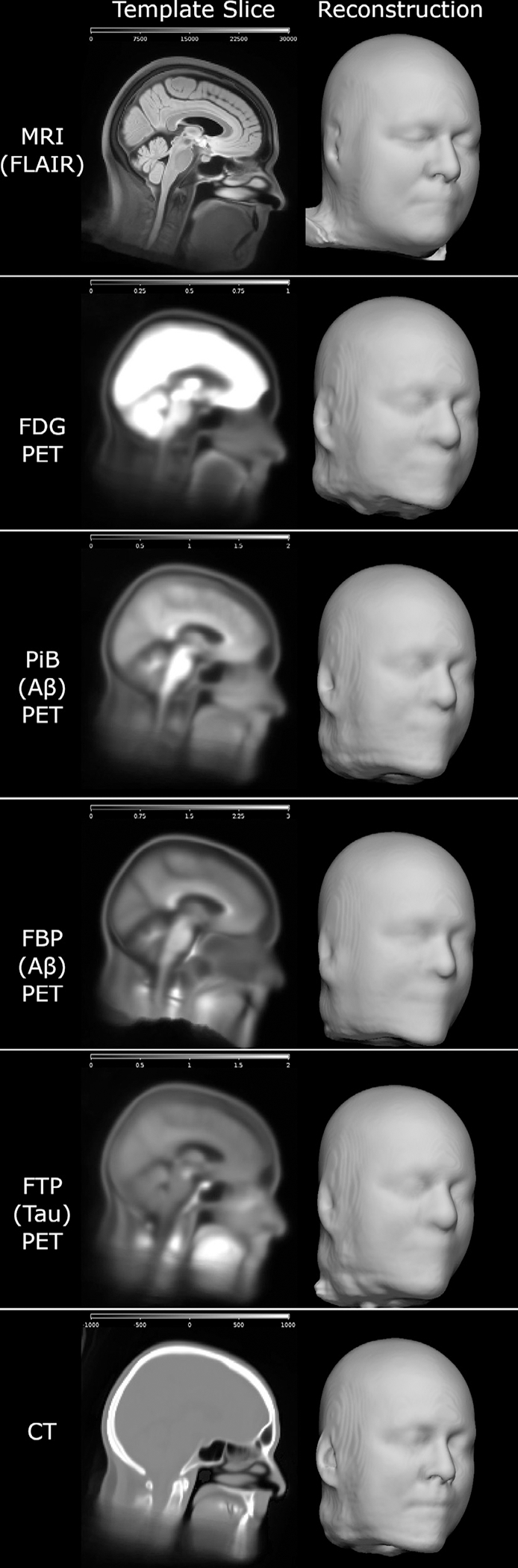
Average brain image templates from each image type, constructed for replacing the face with our mri_reface software. These illustrate the contrast properties of each modality and their relative potential for face reconstruction, but they are average images and thus have higher quality than individual participant scans. Scales were adjusted for best visibility for each image type. For FDG, the brain was intentionally oversaturated to allow visibility of the relatively dark face contour.
Automatic, algorithmic face recognition:
In Table 1, we present the results of automated face recognition testing from using the Microsoft Azure Face API to attempt to match participants’ face photos with their face reconstructions from brain imaging. T1-weighted and FLAIR MRI had the highest recognition rates at 97% and 98% respectively. This was followed by CT (from low-dose attenuation correction CT scans for PET/CT) at 78%. PET scans from older-generation, more prevalent PET/CT scanners had higher match rates when using our “advanced” face reconstruction methods (which replaced missing nose and mouth typically outside their field of view with average nose and mouth to enable face detection) than the “standard” reconstructions that used only their imaged FOV. Using these advanced reconstructions, match rates were highest for FDG (42%), while those of PiB and Tau PET were both smaller and comparable to each other (32% and 35% respectively). However, sample sizes were smaller with FDG (129) than PiB and Tau (both 167), and these differences across modalities were not significant (p=0.12 for FDG vs. PiB). Although our sample sizes for newer-generation PET scanners were very small (<=20), differences within-tracer across scanners were significant (all <0.001), but because matching problems are inherently easier with fewer candidates, their match percentages (85%–100%) are likely overestimated vs. what would be expected with a larger sample and they should not be directly compared to the percentages from other table rows with larger samples. De-facing each scan with mri_reface (replacing each face with a different, average face) greatly reduced recognition rates for all image types. Among modalities with data from at least 100 participants, the highest de-faced match rates were 8%, with MRI (both T1-w and FLAIR), which was reduced from 97–98%. Match rates with older PET scanners were reduced from 32–42% to 0–4%, and rates with CT were reduced from 78% to 5%. Match rates with newer PET scanners were also greatly reduced (85–100% to 15–21%), but due to their small sample sizes we suggest caution in interpreting their likely-inflated percentages.
Table 1.
Rates of automatically matching 5 photos of each participant to their correct corresponding imaging-based face reconstruction, using the Microsoft Azure Face API, before and after each de-facing technique.
| Standard Face Reconstruction (using the input image only, with minimal preprocessing) | Advanced Face Reconstruction (missing nose and mouth automatically replaced with those from an average template) | After Re-facing with mri_reface | |
|---|---|---|---|
| FLAIR MRI | 178/182 (98%) | N/A | 15/182 (8%) |
| T1-w MRI | 176/182 (97%) | N/A | 14/182 (8%) |
| Older FDG PET | 44/129 (34%) | 54/129 (42%) | 0/129 (0%) |
| Older PiB PET | 41/167 (25%) | 54/167 (32%) | 6/167 (4%) |
| Older Tau PET | 48/167 (29%) | 59/167 (35%) | 3/167 (2%) |
| Newer FDG PET | 14/14 (100%*) | N/A | 3/14 (21%) |
| Newer PiB PET | 17/20 (85%*) | N/A | 3/20 (15%) |
| Newer Tau PET | 18/19 (95%*) | N/A | 4/19 (21%) |
| CT (from older PET/CT) | 131/167 (78%) | N/A | 8/167 (5%) |
A * marks percentages with very low sample sizes that are likely overestimated and should not be directly compared with other rows.
3.2. Validation criterion 2: minimizing effects on brain measurements
We compared regional SUVR values computed from 244 Florbetapir amyloid PET scans from ADNI, each measured before and after de-facing with mri_reface, using both: 1) an in-house pipeline based on SPM12, MCALT, and ANTs, and 2) PETSurfer from FreeSurfer 6.0. For de-faced images, both the PET and the T1-weighted MRI (used for normalization) were each de-faced individually. FreeSurfer failed to produce segmentation results for one participant’s MRI after de-facing, and this participant was omitted from analyses of results from both pipelines. Effects on the standard global amyloid region (combined bilateral prefrontal, orbitofrontal, parietal, temporal, anterior cingulate, and posterior cingulate/precuneus regions) (Klunk et al., 2004) are presented in Fig. 4. Overall, effects of de-facing on this large, global region were very small: ICC values were 1.00, biases were <0.5%, and median relative errors were also <0.5%, and difference magnitudes were consistent across the range of amyloid levels. Across these global SUVR measurements (486 total), only 3/486 (0.6%) deviated >3% from their values with unmodified images. The largest differences overall were 5.5% with FreeSurfer and 3.1% with Mayo pipelines (on different images). Effects on individual regions are presented in Fig. 5. For all regions, ICC values were >0.98, biases were <2%, and median relative errors were also <2%.
Fig. 4.
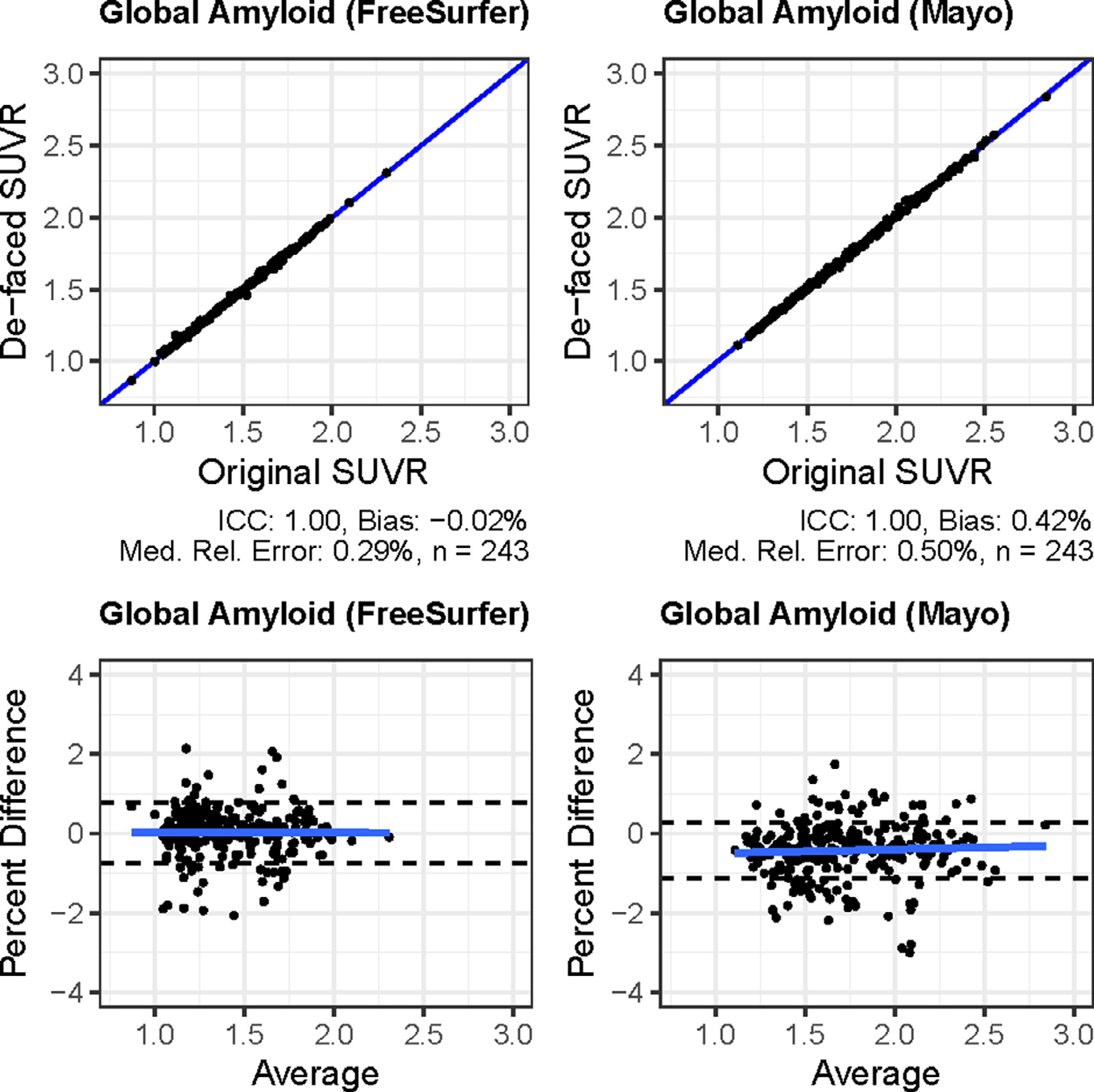
Effects of de-facing PET and MRI with mri_reface on global amyloid PET measurements from the FreeSurfer/PETSurfer pipeline (left) and the in-house pipeline (right). The top row are scatterplots, and the bottom row are Bland-Altman plots of percent differences from the same data. On the Bland-Altman plots, dashed lines show the 95% limits of agreement (mean ± (1.96 * SD)).
Fig. 5.
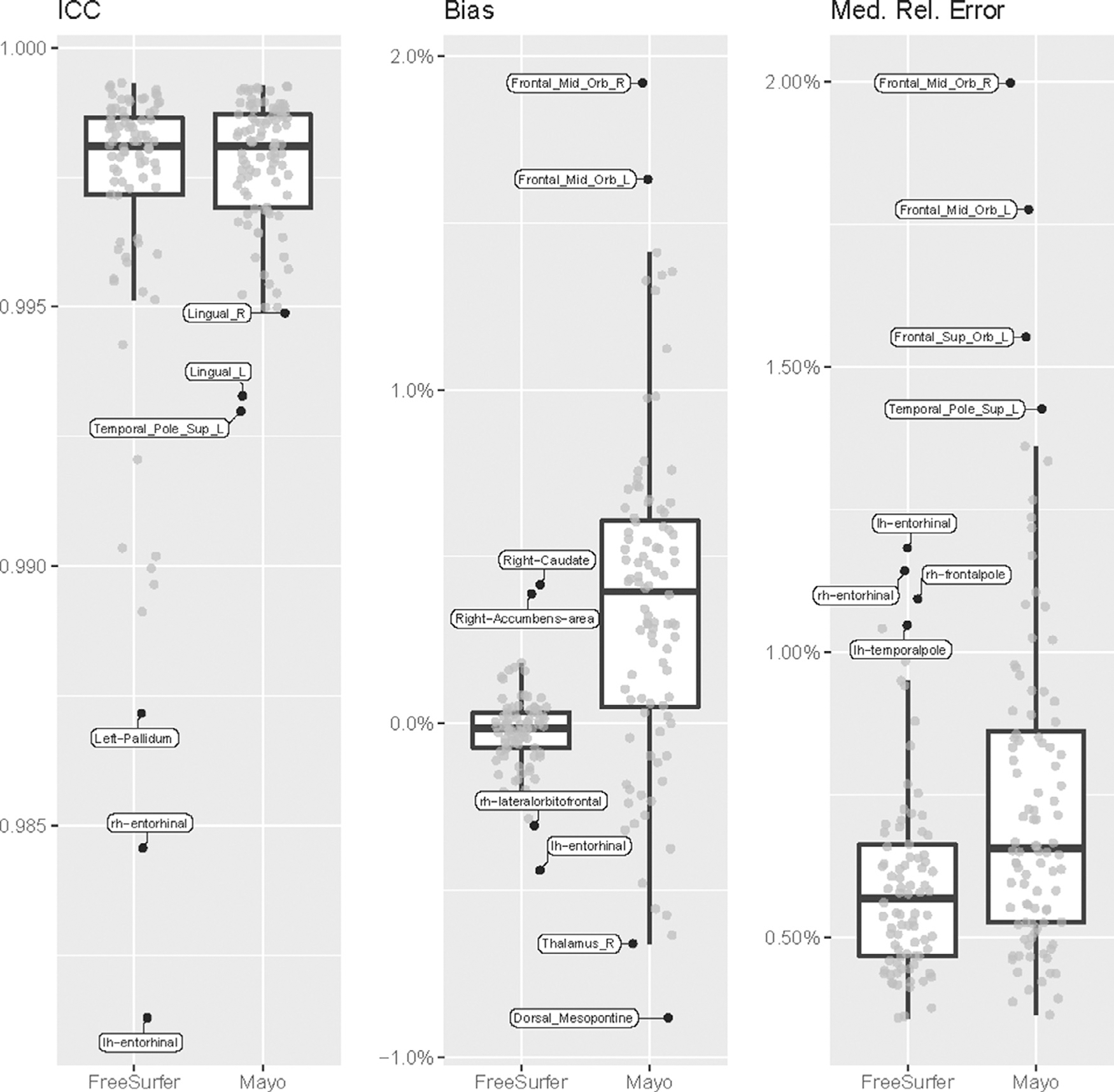
Effects of de-facing PET and MRI with mri_reface on regional amyloid PET measurements. The most extreme values on each plot are labelled. Complete data tables are available in supplementary material.
4. Discussion
4.1. Face recognition rates
Contrary to our initial hypothesis, we found that brain PET images have a substantial risk for re-identification. We hypothesized that identifiable face reconstructions would not be feasible because the spatial resolution of clinical PET scanners (roughly 5mm or more) would be insufficient. Instead, we found that images from newer PET scanners can produce very high-quality face reconstructions, and that even scans from older, more-prevalent PET scanners could be recognized at rates up to 42%, which we consider sufficient to warrant de-identification. Among the three tested PET tracers (FDG, PiB, and FTP), we hypothesized that face recognition rates would be smallest with FDG because the amount of signal relative to the brain, in typical attenuation-corrected images, is the smallest. Instead, we found that FDG had the highest recognition rates at 42%, compared to 32% and 35% for PiB and FTP respectively. These differences were not statistically significant, but still we were incorrect that recognition with FDG would be significantly smaller. This may be because there is some on-target uptake of glucose in facial muscle, whereas PiB and FTP binding in these regions is off-target with uncertain mechanisms that may vary across scans or be spatially nearby but different from the ideal recognizable face contour.
Our finding of high face recognition match rates for CT images (78%) was expected based on previous works (Mazura et al., 2012; Chen et al., 2014; Parks and Monson, 2017). We expected these rates to be higher and more comparable with MRI (97%–98%), but their reduced recognizability may be because our CT images are low-dose scans from PET/CT scanners that were designed only to provide data for attenuation correction during PET image reconstruction, and these have a lower signal to noise ratio (and thus lower contrast), and likely lower spatial resolution, than standard-dose head CT scans from dedicated CT scanners, which were used in these earlier studies. Dental fillings and implants also cause more prominent effects on CT than structural MRI, and these are likely more prevalent in our sample of older individuals than the younger populations used in earlier studies.
Rates of face recognition after mri_reface (Table 1) were still higher than the rate of recognition (correct identification) by chance alone of 1/n (1 over sample size). However, they were far lower than the original rates before de-facing, lower than the 30% we previously found for MRI with our older version of mri_reface (Schwarz et al., 2021a), and lower than the 28%–38% we previously found for MRI with other competing software (Schwarz et al., 2021a). It is important to note that recognition rates remaining above chance after mri_reface were not caused by “failures” for any subset of the images: for all images, the face was completely replaced, as designed. We believe the face recognition software is exploiting remaining information from the proportions of the head, through the replacement face that is transformed only linearly to match the rest of the head. For all modalities, we will continue to improve mri_reface and release updated software at https://www.nitrc.org/projects/mri_reface, but we propose that the current performance is acceptable and worth using until newer versions are developed.
4.2. Effects of de-facing on brain measurements
Effects of de-facing both PET and MRI with mri_reface on global amyloid PET measurements (Figure 4) were very small: ICC values were 1.00, biases were <0.5%, and median relative errors were also <0.5%. Only 0.5% of measurements deviated >3% from their values with unmodified images. Estimates of scan-rescan error on global measurements from amyloid PET images range from 3% (Schwarz et al., 2018) to as high as 8% (Tolboom et al., 2009). Thus, only 0.5% of de-faced global amyloid SUVR measurements had differences due to de-facing that exceeded even the smallest estimates of test-retest error (3%), and the maximum difference (5.5%) was still well below the largest estimates (8%). Effects on individual regions were larger than the global meta-region, consistent with previous findings that measurements from larger PET regions tend to be more stable (Schwarz et al., 2021b). These effects were still very small, with ICC values > 0.98, biases <2%, and median relative errors also <2%. We believe these effects are small enough to be acceptable, but they are not perfect (where de-facing has no measurable effects on brain measurements). It is important to note that these effects were not caused by any alteration of brain regions or voxels by mri_reface; rather, quantification of brain regions by popular software is affected by alteration of non-brain voxels in the image. For example, most methods include linear registration of the entire image to a template, and alterations anywhere in the image can cause small effects on this registration that affects quantification globally. Many methods also use relative intensity during segmentation, e.g. comparing brain regions to other parts of the image, and thus altering non-brain regions can affect segmentation of brain regions (Buimer et al., 2021; de Sitter et al., 2020; Gao et al., 2022; Schwarz et al., 2021a). Again, we will continue to improve mri_reface and release updated software, but we propose that the current performance is acceptable and worth using until newer versions are developed. The regions with the largest effects (Figure 5) frequently included frontal areas most proximal to the face, and areas in the basal ganglia, consistent with previous findings showing that these were most affected by de-facing MRI (Buimer et al., 2021; Schwarz et al., 2021a). For PET images, instability in the basal ganglia may also be explained by the relatively low counts (i.e. noisier signal after boosting by attenuation correction) in these regions. These also included relatively small regions like the entorhinal cortex, accumbens, and dorsal mesopontine areas, which were not relatively poor performers in de-faced MRI, but their worse performance in de-faced PET is consistent with the principle that smaller regions typically have less-stable PET measurements.
4.3. Strengths and limitations of current study
Strengths:
While previous works have examined face identifiability of brain images from MRI and for CT, this is, to the best of our knowledge, the first to examine this question for PET images. We tested face recognition performance using leading, publicly available face recognition software, which we believe best represents the types of scenarios in which a motivated individual might attempt to re-identify research participants from de-identified data. We have previously published extensive discussion of the strengths and limitations of our face recognition testing approach (Schwarz et al., 2019a, 2021a). We believe this work is also the first to compare identifiability of MRI, PET, and CT directly, and our results were largely consistent across three different classes of PET tracers. We validated our de-facing software both for preventing face identification and minimizing its impacts on brain measurements from de-faced images, and we provide this software free to the research community.
Limitations:
Our sample sizes for newer PET scanners were very small (<=20), but our major conclusions were well supported without this data, and the limited data from these newer scanners suggests that face recognizability from PET images will only become a larger threat as data from these scanners becomes more widely available. We will re-examine these conclusions with a larger sample in future work as we continue to acquire data from newer scanners. We did not test face recognition with additional amyloid or tau tracers, but since our results were very consistent across FDG, PiB, and FTP, we hypothesize that other amyloid and tau tracers would behave similarly. We also did not test additional MRI modalities, but this is a focus of our future work. We also did not test for any relationships between brain PET signal and potential for face recognition (e.g. frontal lobe cortical signal spilling into the eyebrow region), but as our cohort grows we will have more participants with enough amyloid and tau tracer uptake allow this in future work. We also did not examine the effects of different levels of post-reconstruction smoothing filters on identifiability of PET images; it is possible that less smoothing could allow reconstruction of smaller identifiable features but also retain more noise that could impede reconstruction. We only tested our own de-facing software (mri_reface) because we believe it is the only one that has been adapted for PET imaging data. It would be theoretically possible to run programs designed for MRI or CT using PET images instead, but since they would be registered to MRI or CT templates with very different contrast and resolution properties, it is uncertain how well this would work. We tested effects of de-facing on SUVR measurements using amyloid PET (Florbetapir) images from ADNI because it is a free, public dataset that could allow replication of our findings. We did not perform similar testing using other PET tracers because these were not available in sufficient numbers from our re-used dataset, but the quantification techniques used for these modalities are identical to those used for amyloid, Still, we will examine this in future work. We hypothesize that results would be very similar, because we have shown that estimates of test-retest error and software factors that affect measurement precision are remarkably similar across amyloid and tau PET tracers (Schwarz et al., 2021b, 2018). Our datasets were both sampled from studies of aging and contain only images from older adults. Others have found that that de-facing software, primarily designed for images of older adults, may perform worse in images from younger individuals (Buimer et al., 2021), and we will also explore this in future work.
Our face recognition testing paradigm measures the success rates at which a motivated individual might be able to re-identify a participant whom they have reason to believe exists within a dataset. We think this is a plausible and scenario, but it is a much easier recognition problem than an opposite paradigm where someone attempts to identify brain-imaging-based face reconstructions by matching them to large databases of identified photos from theoretically all people on Earth. We are unable to quantify this paradigm due to limited legally obtained public datasets with face photographs of older participants, and limitations of the consent signed by our participants. However, our 98% recognition rates with the current paradigm (with MRI) suggests that the performance of matching MRIs with photos is rapidly approaching that of photos to photos, for which modern face recognition algorithms can successfully identify photos from databases of >12 million with <1% failure rates (Grother et al., 2018), so we hypothesize that the opposite paradigm may already be plausible with current or near-future technology.
5. Conclusions
To the best of our knowledge, this study is the first to demonstrate that de-identified research PET images may be re-identifiable via face recognition. Images from the currently prevalent generation of PET/CT scanners were automatically matched with identified face photos at rates of 32–42%. These rates were lower than MRI (97–98%) and CT (78%), but still high enough that research studies should consider using face de-identification software on PET images in addition to CT and structural MRI. We also showed preliminary evidence that the newest generations of PET/CT scanners can produce even better face reconstructions than older scanners, and the need for de-facing PET images may increase as these scanners become more prevalent. We tested FDG, amyloid (PiB) and tau (FTP) PET, and all showed similar rates of identifiability (32%–42%). We also extended our mri_reface software to support de-facing of PET and CT images, and we validated its effects. Rates of face recognition on de-faced images were reduced to 0–4% for PET, 5% for CT, and 8% for MRI. Effects of de-facing on regional amyloid PET SUVR measurements were small: ICC values were >0.98, biases were <2%, and median relative errors were <2%. Effects on global amyloid PET SUVR measurements were even smaller: ICC values were 1.00, biases were <0.5%, and median relative errors were also <0.5%.
Supplementary Material
Acknowledgments
The authors give their thanks to all the volunteers, participants, and coordinators who contributed to this research, with special thanks to Steven M. Smith, Josie M. Williams, and Paul D. Lewis, and to Bradley Kemp for assistance with PET scanner model descriptions. We gratefully thank our funding sources: NIH grants R01 AG068206, U01 AG006786, P50 AG016574, R01 AG034676, R37 AG011378, R01 AG041851, R01 NS097495, R01 AG056366, U01 NS100620; The GHR Foundation; The Elsie and Marvin Dekelboum Family Foundation; The Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Clinic; The Liston Award; The Schuler Foundation; and The Mayo Foundation for Medical Education and Research. We also gratefully acknowledge the support of NVIDIA Corporation with the donation of a Quadro P6000 GPU used in generating 3D facial reconstructions for this research. We also thank AVID Radiopharmaceuticals, Inc., for their support in supplying Flortaucipir precursor, chemistry production advice, and FDA regulatory cross-filing permission and documentation needed for this work. Images used for production of the Florbetapir PET template were provided by OASIS-3: Principal Investigators: T. Benzinger, D. Marcus, J. Morris; NIH P50 AG00561, P30 NS09857781, P01 AG026276, P01 AG003991, R01 AG043434, UL1 TR000448, R01 EB009352. Florbetapir doses in OASIS-3 were provided by Avid Radiopharmaceuticals, a wholly owned subsidiary of Eli Lilly.
Footnotes
- Dr. Schwarz receives funding from the National Institutes of Health, related and unrelated to this study.
- Dr. Kremers received grant funding from NIH for this study, and from NIH, DOD, AstraZeneca, Biogen and Roche unrelated to this study.
- Dr. Lowe consults for Bayer Schering Pharma, Piramal Life Sciences, Eisai, Inc., and Merck Research and receives research support from GE Healthcare, Siemens Molecular Imaging, AVID Radiopharmaceuticals and the NIH (NIA, NCI).
- Dr. Savvides reports no disclosures.
- Dr. Gunter receives funding from the NIH.
- Mr. Senjem owns or has owned stock in medical related companies, unrelated to the current work, within the past 36 months: Align Technology, Inc., Inovio Pharmaceuticals, Inc., Mesa Laboratories, Inc., Nvidia, Inc., Johnson and Johnson, LHC Group, Inc., Natus Medical Inc., Varex Imaging Corporation.
- Dr. Vemuri receives funding from the NIH.
- Dr. Kantarci consults for Biogen Inc., receives research support from Avid Radiopharmaceuticals and Eli Lilly, and receives funding from NIH and Alzheimer’s Drug Discovery Foundation.
- Dr. Knopman served on a Data Safety Monitoring Board for the DIAN study. He served on a Data Safety monitoring Board for a tau therapeutic for Biogen but received no personal compensation. He is an investigator in clinical trials sponsored by Biogen, Lilly Pharmaceuticals and the University of Southern California. He serves as a consultant for Samus Therapeutics, Roche, Magellan Health and Alzeca Biosciences but receives no personal compensation. He receives research support from the NIH.
- Dr. Petersen is a consultant for Roche, Inc., Merck, Inc., Biogen, Inc., Nestle, Inc., and Eisai, Inc., served on a DSMB for Genentech, Inc.; receives royalties from publishing Mild Cognitive Impairment (Oxford University Press, 2003) and UpToDate; and receives research support from the NIH (P30 AG062677 (PI) and U01-AG006786 (PI), R01-AG011378 (Co-I), U24 AG057437 (Co-PI), UF1 NS125417 (C0-PI) and U01–024904 (Co-I)).
- Dr. Jack serves on an independent data monitoring board for Roche, has served as a speaker for Eisai, and consulted for Biogen, but he receives no personal compensation from any commercial entity. He receives research support from NIH and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Clinic.
Credit authorship contribution statement
Christopher G. Schwarz: Conceptualization, Methodology, Software, Validation, Formal analysis, Investigation, Data curation, Writing – original draft, Writing – review & editing, Visualization, Project administration, Funding acquisition. Walter K. Kremers: Methodology, Writing – review & editing. Val J. Lowe: Resources, Data curation, Writing – review & editing. Marios Savvides: Methodology, Writing – review & editing. Jeffrey L. Gunter: Software, Writing – review & editing. Matthew L. Senjem: Software, Writing – review & editing. Prashanthi Vemuri: Resources, Writing – review & editing, Funding acquisition. Kejal Kantarci: Resources, Writing – review & editing, Funding acquisition. David S. Knopman: Resources, Writing – review & editing, Funding acquisition. Ronald C. Petersen: Resources, Writing – review & editing, Funding acquisition. Clifford R. Jack Jr.: Conceptualization, Resources, Writing – review & editing, Funding acquisition.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2022.119357.
Data and code availability statement
ICC, bias, and median relative error calculations for every regional measurement from each pipeline are provided in supplementary material. ADNI images are available directly through ADNI (http://adni.loni.usc.edu/). The face recognition dataset contains participant photos, which are considered primary identifiers; to protect participant privacy and comply with their signed consent forms, these data cannot be shared. MRI, PET, and other data from the Mayo Clinic Study of Aging and the Alzheimer’s Disease Research Center are available to qualified academic and industry researchers by request to the MCSA and ADRC Executive Committee. Our mri_reface software is available at https://www.nitrc.org/projects/mri_reface. The Microsoft Azure Face API is a proprietary cloud-based service by Microsoft.
References
- Ashburner J, 2009. Computational anatomy with the SPM software. Magn. Reson. Imaging 27, 1163–1174. doi: 10.1016/j.mri.2009.01.006. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12, 26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC, 2010. The optimal template effect in hippocampus studies of diseased populations. Neuroimage 49, 2457–2466. doi: 10.1016/j.neuroimage.2009.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker SL, Maass A, Jagust WJ, 2017. Considerations and code for partial volume correcting [18F]-AV-1451 tau PET data. Data Brief 15, 648–657. doi: 10.1016/j.dib.2017.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buimer EEL, Schnack HG, Caspi Y, Haren NEMV, Milchenko M, Pas P, Brouwer RM, 2021. De-identification procedures for magnetic resonance images and the impact on structural brain measures at different ages. Hum. Brain Mapp 1–13. doi: 10.1002/hbm.25459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJS, Juluru K, Morgan T, Moffitt R, Siddiqui KM, Siegel EL, 2014. Implications of surface-rendered facial CT images in patient privacy. Am. J. Roentgenol 202, 1267–1271. doi: 10.2214/AJR.13.10608. [DOI] [PubMed] [Google Scholar]
- de Sitter A, Visser M, Brouwer I, Cover KS, van Schijndel RA, Eijgelaar RS, Müller DMJ, Ropele S, Kappos L, Rovira, Filippi M, Enzinger C, Frederiksen J, Ciccarelli O, Guttmann CRG, Wattjes MP, Witte MG, de Witt Hamer PC, Barkhof F, Vrenken H, 2020. Facing privacy in neuroimaging: removing facial features degrades performance of image analysis methods. Eur. Radiol 30, 1062–1074. doi: 10.1007/s00330-019-06459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ, 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 31, 968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. NeuroImage 62, 774–781. doi: 10.1016/j.neuroimage.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao C, Jin L, Prince JL, Carass A, 2022. Effects of defacing whole head MRI on neuroanalysis. In: Išgum I, Colliot O (Eds.), MEDICAL IMAGING 2022: IMAGE PROCESSING. PRESENTED AT THE IMAGE PROCESSING SPIE, San Diego, United States, p. 139. doi: 10.1117/12.2613175. [DOI] [Google Scholar]
- Greve DN, Salat DH, Bowen SL, Izquierdo-Garcia D, Schultz AP, Catana C, Becker JA, Svarer C, Knudsen G, Sperling RA, Johnson KA, 2016. Different partial volume correction methods lead to different conclusions: An 18F-FDG PET Study of aging. Neuroimage 132, 334–343. doi: 10.1016/j.neuroimage.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grother P, Ngan M, Hanaoka K, 2018. Ongoing face recognition vendor test (frvt) part 2 National Institute of Standards and Technology, Gaithersburg, MD: doi: 10.6028/NIST.IR.8238. [DOI] [Google Scholar]
- Jack CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, Holtzman DM, Jagust W, Jessen F, Karlawish J, Liu E, Molinuevo JL, Montine T, Phelps C, Rankin KP, Rowe CC, Scheltens P, Siemers E, Snyder HM, Sperling R, Elliott C, Masliah E, Ryan L, Silverberg N, 2018. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimer’s Dementia 14, 535–562. doi: 10.1016/j.jalz.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Bandy D, Chen K, Foster NL, Landau SM, Mathis CA, Price JC, Reiman EM, Skovronsky D, Koeppe RA, 2010. The Alzheimer’s Disease Neuroimaging Initiative positron emission tomography core. Alzheimer’s Dementia 6, 221–229. doi: 10.1016/j.jalz.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagust WJ, Landau SM, Koeppe RA, Reiman EM, Chen K, Mathis CA, Price JC, Foster NL, Wang AY, 2015. The Alzheimer’s Disease neuroimaging initiative 2 PET Core: 2015. Alzheimer’s and Dementia 11, 757–771. 10.1016/j.jalz.2015.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A, Koeppe RA, Fessler JA, 2009. Reducing between scanner differences in multi-center PET studies. Neuroimage 46, 154–159. doi: 10.1016/j.neuroimage.2009.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juefei-Xu F, Luu K, Savvides M, 2015. Spartans: single-sample periocular-based alignment-robust recognition technique applied to non-frontal scenarios. IEEE Trans. Image Process 24, 4780–4795. doi: 10.1109/TIP.2015.2468173. [DOI] [PubMed] [Google Scholar]
- Juefei-Xu F, Luu K, Savvides M, Bui TD, Suen CY, 2011. Investigating age invariant face recognition based on periocular biometrics 2011 International Joint Conference on Biometrics, IJCB 2011. IEEE doi: 10.1109/IJCB.2011.6117600. [DOI] [Google Scholar]
- Juefei-Xu F, Pal DK, Savvides M, 2014. Hallucinating the full face from the periocular region via dimensionally weighted K-SVD In: 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops. Presented at the 2014 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW). IEEE, Columbus, OH, USA, pp. 1–8. doi: 10.1109/CVPRW.2014.6. [DOI] [Google Scholar]
- Juefei-Xu F, Savvides M, 2016. Fastfood dictionary learning for periocular-based full face hallucination In: 2016 IEEE 8th International Conference on Biometrics Theory, Applications and Systems, BTAS 2016. IEEE, pp. 1–6. doi: 10.1109/BTAS.2016.7791174. [DOI] [Google Scholar]
- Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergström M, Savitcheva I, Huang GF, Estrada S, Ausén B, Debnath ML, Barletta J, Price JC, Sandell J, Lopresti BJ, Wall A, Koivisto P, Antoni G, Mathis CA, L\aangström B, 2004. Imaging Brain Amyloid in Alzheimer’s Disease with Pittsburgh Compound-B. Ann. Neurol 55, 306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- LaMontagne PJ, Benzinger TLS, Morris JC, Keefe S, Hornbeck R, Xiong C, Grant E, Hassenstab J, Moulder K, Vlassenko AG, Raichle ME, Cruchaga C, Marcus D, 2019. OASIS-3: longitudinal neuroimaging, clinical, and cognitive dataset for normal aging and Alzheimer Disease (preprint). Radiol. Imaging doi: 10.1101/2019.12.13.19014902. [DOI] [Google Scholar]
- Le THN, Prabhu U, Savvides M, 2014. A novel eyebrow segmentation and eyebrow shape-based identification In: IEEE International Joint Conference on Biometrics. Presented at the 2014 IEEE International Joint Conference on Biometrics (IJCB). IEEE, Clearwater, FL, USA, pp. 1–8. doi: 10.1109/BTAS.2014.6996262. [DOI] [Google Scholar]
- Lowe VJ, Wiste HJ, Senjem ML, Weigand SD, Therneau TM, Boeve BF, Josephs KA, Fang P, Pandey MK, Murray ME, Kantarci K, Jones DT, Vemuri P, Graff-Radford J, Schwarz CG, Machulda MM, Mielke MM, Roberts RO, Knopman DS, Petersen RC, Jack CR, 2018. Widespread brain tau and its association with ageing, Braak stage and Alzheimer’s dementia. Brain 141, 271–287. doi: 10.1093/brain/awx320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazura JC, Juluru K, Chen JJ, Morgan TA, John M, Siegel EL, 2012. Facial recognition software success rates for the identification of 3D surface reconstructed facial images: Implications for patient privacy and security. J. Digit. Imaging 25, 347–351. doi: 10.1007/s10278-011-9429-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent S, Croteau E, Potvin O, Castellano C-A, Dieumegarde L, Cunnane SC, Duchesne S, 2020. Selection of the optimal intensity normalization region for FDG-PET studies of normal aging and Alzheimer’s disease. Sci. Rep 10, 9261. doi: 10.1038/s41598-020-65957-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsu N, 1979. A threshold selection method from Gray-level histograms. IEEE Trans. Syst. Man Cybern 9, 62–66. doi: 10.1109/TSMC.1979.4310076. [DOI] [Google Scholar]
- Parks CL, Monson KL, 2017. Automated facial recognition of computed tomography-derived facial images: patient privacy implications. J. Digit. Imaging 30, 204–214. doi: 10.1007/s10278-016-9932-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Roberts RO, Knopman DS, Geda YE, Cha RH, Pankratz VS, Boeve BF, Tangalos EG, Ivnik RJ, Rocca WA, 2010. Prevalence of mild cognitive impairment is higher in men. The Mayo Clinic Study of Aging. Neurology 75, 889–897. doi: 10.1212/WNL.0b013e3181f11d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team, 2008. R: A Language and Environment for Statistical Computing Vienna, Austria. [Google Scholar]
- Revelle W, 2019. psych: Procedures for Psychological. Psychometri. Personal. Res. [Google Scholar]
- Roberts RO, Geda YE, Knopman DS, Cha RH, Pankratz VS, Boeve BF, Ivnik RJ, Tangalos EG, Petersen RC, Rocca WA, 2008. The Mayo Clinic Study of Aging: design and sampling, participation, baseline measures and sample characteristics. Neuroepidemiology 30, 58–69. doi: 10.1159/000115751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorden C (Ed.), 2022. n.d. Surf Ice. [WWW Document] URL https://www.nitrc.org/projects/surfice/. (accessed 12.11.18).
- Schwarz CG, Gunter JL, Lowe VJ, Weigand S, Vemuri P, Senjem ML, Petersen RC, Knopman DS, Jack CR, 2018. A comparison of partial volume correction techniques for measuring change in serial amyloid PET SUVR. J. Alzheimer’s Disease 67, 181–195. doi: 10.3233/JAD-180749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Gunter JL, Ward CP, Vemuri P, Senjem ML, Wiste HJ, Petersen RC, Knopman DS, Jack CR, 2017. The Mayo clinic adult lifespan template: better quantification across the lifespan. Alzheimer’s Dementia 13, P792. doi: 10.1016/j.jalz.2017.06.1071. [DOI] [Google Scholar]
- Schwarz CG, Kremers WK, Therneau TM, Sharp RR, Gunter JL, Vemuri P, Arani A, Spychalla AJ, Kantarci K, Knopman DS, Jack CR, 2019a. Identification of anonymous mri research participants with face recognition software. N. Engl. J. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Kremers WK, Wiste HJ, Gunter JL, Vemuri P, Spychalla AJ, Kantarci K, Schultz AP, Sperling RA, Knopman DS, Petersen RC, Jack CR, 2021a. Changing the face of neuroimaging research: comparing a new MRI de-facing technique with popular alternatives. Neuroimage 231. doi: 10.1016/j.neuroimage.2021.117845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Therneau TM, Weigand SD, Gunter JL, Lowe VJ, Przybelski SA, Senjem ML, Botha H, Vemuri P, Kantarci K, Boeve BF, Whitwell JL, Josephs KA, Petersen RC, Knopman DS, Jack CR, 2021b. Selecting software pipelines for change in flortaucipir SUVR: Balancing repeatability and group separation. Neuroimage 238, 118259. doi: 10.1016/j.neuroimage.2021.118259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz CG, Wiste HJ, Gunter JL, Senjem ML, Lowe VJ, Petersen RC, Knopman DS, Jack CR, 2019b. Variability in MRI and PET Measurements Introduced By Change in MRI Vendor. Alzheimer’s Dementia Wiley P104–P105. doi: 10.1016/j.jalz.2019.06.4241. [DOI] [Google Scholar]
- Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, Aisen P, 2014. The A4 study: Stopping AD before symptoms begin? Sci. Transl. Med 6, 4–7. doi: 10.1126/scitranslmed.3007941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolboom N, Yaqub M, Boellaard R, Luurtsema G, Windhorst AD, Scheltens P, Lammertsma AA, Van Berckel BNM, 2009. Test-retest variability of quantitative [11C]PIB studies in Alzheimer’s disease. Eur. J. Nucl. Med. Mol. Imaging 36, 1629–1638. doi: 10.1007/s00259-009-1129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services Office for Civil Rights, 2013. HIPAA Administrative Simplification: Regulation Text
- Wainer H, 2000. The centercept: an estimable and meaningful regression parameter. Psychol. Sci 11, 434–436. doi: 10.1111/1467-9280.00284. [DOI] [PubMed] [Google Scholar]
- Woodard DL, Pundlik SJ, Lyle JR, Miller PE, 2010. Periocular region appearance cues for biometric identification 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition - Workshops, CVPRW 2010 162–169 doi: 10.1109/CVPRW.2010.5544621. [DOI] [Google Scholar]
- Xia C-F, Arteaga J, Chen G, Gangadharmath U, Gomez LF, Kasi D, Lam C, Liang Q, Liu C, Mocharla VP, Mu F, Sinha A, Su H, Szardenings AK, Walsh JC, Wang E, Yu C, Zhang W, Zhao T, Kolb HC, 2013. [(18)F]T807, a novel tau positron emission tomography imaging agent for Alzheimer’s disease. Alzheimer’s Dementia 9, 666–676. doi: 10.1016/j.jalz.2012.11.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ICC, bias, and median relative error calculations for every regional measurement from each pipeline are provided in supplementary material. ADNI images are available directly through ADNI (http://adni.loni.usc.edu/). The face recognition dataset contains participant photos, which are considered primary identifiers; to protect participant privacy and comply with their signed consent forms, these data cannot be shared. MRI, PET, and other data from the Mayo Clinic Study of Aging and the Alzheimer’s Disease Research Center are available to qualified academic and industry researchers by request to the MCSA and ADRC Executive Committee. Our mri_reface software is available at https://www.nitrc.org/projects/mri_reface. The Microsoft Azure Face API is a proprietary cloud-based service by Microsoft.


