Summary
Background
The mechanism of missense alteration at EGFR L792F in patients with non-small cell lung cancer resistant to osimertinib has not been sufficiently clarified. We aimed to explore the critical molecular events and coping strategies in osimertinib resistance due to acquired L792F mutation.
Methods
Circulating tumor DNA-based sequencing data of 1153 patients with osimertinib resistance were collected to illustrate the prevalence of EGFR L792F mutation. Sensitivity to osimertinib was tested with constructed EGFR 19Del/T790M-cis-L792F cell lines in vitro and in vivo. The correlation and linked pathways between M2 macrophage polarization and EGFR L792Fcis-induced osimertinib resistance were investigated. Possible interventions to suppress osimertinib resistance by targeting IL-4 or STAT3 were explored.
Findings
The concomitant EGFR L792F was identified as an independent mutation following the acquisition of T790M after osimertinib resistance, in that 5 of the 946 patients with osimertinib resistance harbored EGFR T790M-cis-L792F mutation. Transfected EGFR 19Del/T790M-cis-L792F in cell lines had decreased sensitivity to osimertinib and enhanced infiltrating macrophage with M2 polarization. Silico analyses confirmed the role of M2 polarization in osimertinib resistance induced by EGFR T790M-cis-L792F mutation. EGFR T790M-cis-L792F mutation upregulated phosphorylation of STAT3 Tyr705 and promoted its specific binding to IL4 promoter, enhancing IL-4 expression and secretion and inducing macrophage M2 polarization. Furthermore, blockade of STAT3/IL-4 (SH-4-54 or dupilumab) suppressed macrophage M2 polarization and regressed tumor sensitivity to osimertinib.
Interpretation
Our results proved that targeting EGFR T790M-cis-L792F/STAT3 Tyr705/IL-4 pathway could be a potential strategy to suppress osimertinib resistance in NSCLC.
Funding
This work was supported by the National Natural Science Foundation of China (81871889, 82072586, 81902910), Beijing Natural Science Foundation (7212084, 7214249), the China National Natural Science Foundation Key Program (81630071), the National Key Research and Development Project (2019YFC1315704), CAMS Innovation Fund for Medical Sciences (CIFMS 2021-1-I2M-012), Aiyou Foundation (KY201701) and CAMS Key Laboratory of translational research on lung cancer (2018PT31035).
Keywords: Non-small cell lung cancer, EGFR, L792F mutation, Osimertinib resistance, M2 macrophage polarization
Research in context.
Evidence before this study
With the success of osimertinib, the first third-generation epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor (TKI) in lung cancers, acquired resistance is a growing clinical challenge. Previous studies have reported acquired mutations, including EGFR C797S and M766Q induced osimertinib resistance. Also, mutations at EGFR L792 have been found in tumor tissues and circulating tumor DNA (ctDNA) of patients resistant to osimertinib. However, the specific molecular mechanism by which EGFR L792F mutation induced resistance to osimertinib has not been demonstrated, and a corresponding strategy for overcoming osimertinib resistance has not been explored.
Added value of this study
We identified EGFR T790M-cis-L792F mutation in ctDNA from Chinese lung cancer patients resistant to osimertinib. EGFR T790M-cis-L792F-mut cell lines were constructed and observed resistance to osimertinib in vitro and in vivo. We discovered that EGFR T790M-cis-L792F mutation activated phosphorylation of STAT3 Tyr705 and thus promoted its specific binding to IL4 promoter, which induced resistance to osimertinib. Enhancing IL-4 synthesis and secretion facilitated CD206+ M2 macrophage polarization. Knocking down STAT3 or IL-4 or blockade with SH-4-54 or dupilumab could inhibit the proliferation of T790M-cis-L792F-mut cells and tumor growth of corresponding xenografts under osimertinib treatment.
Implications of all the available evidence
Our study demonstrated that EGFR T790M-cis-L792F mutation could induce osimertinib resistance via activating STAT Tyr705/IL-4 and subsequently promoting M2 macrophages polarization. Blockade of STAT3 or IL-4 could partly restore the sensitivity to osimertinib, providing insights about molecular mechanisms of osimertinib resistance and potentially clinical strategies coping it in lung cancers. In addition, we bridged the correlation of EGFR L792Fcis in manipulating macrophages’ switch, which also provided a chance to reverse the acquired osimertinib resistance by interfering with the tumor microenvironment of NSCLC. Although the mutation frequency of EGFR L792X, including L792F/H/P/V, was 2.46% in this study, considering the substantial NSCLC patient base in China and worldwide, the number of patients with L792X mutations and resistance to osimertinib might be pretty huge. Overcoming L792X mutations-induced resistance is vital to this large patient population. Our discoveries may also provide insights into EGFR TKI resistance induced by other endogenous mutations.
Alt-text: Unlabelled box
Introduction
Epidermal growth factor receptor (EGFR) mutations are vital driver mutations in non-small-cell lung cancers (NSCLC). Tyrosine kinase inhibitors (TKIs) targeting mutant EGFR have brought great survival benefits to patients. With the progresses of drug development, third-generation TKIs, including osimertinib, aumolertinib and furmonertinib, have been recommended as the first-line setting for EGFR mutant NSCLC patients,1, 2, 3 as salvage treatment for T790M mutation-positive patients after resistance to first- and second-generation EGFR TKIs4, 5, 6, 7 and as adjuvant therapy for post-surgery EGFR-mutant NSCLCs.8 Other developing third-generation TKIs, including lazertinib,9 abivertinib,10 and rezivertinib11 have shown powerful anti-tumor potentials in T790M-positive NSCLC patients. However, with widespread clinical applications of osimertinib and other third-generation TKIs, acquired resistance is a growing clinical challenge.12
Several studies have described a range of acquired resistance on target mutations hypothesized to be involved in osimertinib resistance, including EGFR C797S, L792X, G724S, and M766Q mutations.13, 14, 15, 16, 17, 18 Classified as off-target mutations, functional alterations in MET and other driver genes and transformation to small-cell lung cancer also play vital roles in osimertinib resistance.19 Acquired EGFR C797S or G724S mutation has mediated osimertinib resistance.16,17 However, its specific underlying mechanisms remain poorly understood.
EGFR L792X, mainly L792F/H, is found in post-osimertinib plasma cell-free DNA,13, 14, 15 suggesting the potential roles of L792X in osimertinib resistance. The mutation allele frequency (AF) of a single L792 mutation ranges from 0.5% to 8.8%.13,15,19 Interestingly, the occurrences of L792F/Y/H mutations were all on the same sequencing reads with T790M but were completely exclusive from C797S,13 which prompted L792X driving osimertinib resistance independent of C797S. Moreover, according to the University of Texas MD Anderson Lung Cancer Moonshot GEMINI and Moffitt Cancer Center lung cancer databases, up to 26% of L792X mutants were presented in T790M-preserved cases treated with osimertinib.19 Furthermore, in vitro experiments have validated that L792F/Y/H alone or concurrent with T790M could confer osimertinib resistance in 19Del- or L858R-mutated cells lines.14,15 In addition, predicting the structural collision of EGFR L792-mutated residue and osimertinib complex, L792H interrupted osimertinib binding to EGFR.15 These findings suggest that L792 mutations contribute to osimertinib resistance. Nevertheless, whether L792F/Y/H induces osimertinib resistance in vivo remains unclear. Moreover, specific mechanisms need to be elucidated. To the best of our knowledge, studies determining the functional cell behaviors of EGFR L792 mutations in osimertinib-tolerant cells have not been conducted yet. Therefore, our study emphasized the internal mechanism in TKI resistance of this phenotype.
After osimertinib resistance, only 7.5–14% of the patients presented with MET alterations, suggesting that the other independent mechanisms of EGFR secondary resistant mutations should be further identified.14,19 Therefore, this study aimed to identify other functional mechanisms of osimertinib resistance and develop strategies to overcome osimertinib resistance. We determined the prevalence of acquired EGFR L792F mutation in a Chinese NSCLC cohort with disease progression after osimertinib treatment. We further tested the sensitivity to osimertinib with constructed 19Del/T790M-cis-L792F mutation and control cell lines in vitro and in vivo. The correlation and linked pathways between M2 macrophage polarization and EGFR 19Del/T790M-cis-L792F-induced osimertinib resistance were investigated. In addition, possible interventions to reverse osimertinib resistance by targeting interleukin (IL)-4 or signal transducer and activator of transcription 3 (STAT3) were explored.
Methods
Patient case and cohort
A 60-year-old patient provided informed consent for peripheral blood and tumor tissue collection for next-generation sequencing according to the protocol approved by the Ethics Committee of Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College.
Peripheral blood circulating tumor DNA (ctDNA) sequencing data of 1383 samples from 1153 patients receiving first-line (236 samples from 207 patients) or second-line (1147 samples from 946 patients) osimertinib treatment from 2016 to 2021 were collected from Geneplus Technology database. The baseline characteristics of these patients are presented in Supplemental Table S1. All patients provided informed consent for tissue and peripheral blood collection for next-generation sequencing according to the protocol approved Cancer Hospital, Chinese Academy of Medical Sciences (reference number: NCC2019C-156).
Gene expression and lentivirus preparation
Oligonucleotide sequences encoding short hairpin RNA (shRNA) targeting EGFR (Supplementary Table S2) inserted into pLenti6-vector-Blasticidin were identified. pLenti6-U6 vector carrying NTC sequence was used as the negative control. The mutation sequences of EGFR (19Del, L792F, 19Del-cis-L792F, or 19Del/T790M-cis-L792F) were constructed into pLENs vector with Zocin selection, whose targeting shRNA oligonucleotide sequences were replaced by synonymous codon. Small guide RNAs (sgRNAs) of STAT3 and IL4 (Supplementary Table S2) designed using the Benchling tool (https://benchling.com) were inserted into SaCas9-2A-GFP into the BsaI sites after annealing. According to the manufacturer's instructions, various lentiviruses were packaged in 293T cells by Lipofectamine 2000 (Invitrogen, Grand Island, NY, USA), as described in our previous study.20,21
Cell lines
The Madison lung cell line of syngeneic BALB/c mice (M109) cells was grown in Roswell Park Memorial Institute (RPMI) 1640 medium with 10% fetal bovine serum (FBS). To establish osimertinib-resistant (OR) subclones of M109, cells were cultured with various concentrations of osimertinib for 12 weeks and constantly incubated with 10 µmol/L osimertinib. Surviving resistant subcloned cells were passaged four times, which was conducted prior to performing the experiments. Tohoku Hospital Pediatrics-1 (THP-1, RRID: CVCL_0006) and the human lung adenocarcinoma cell lines H1975 (RRID ID: CVCL_1511) and PC-9 (RRID ID: CVCL_B260) were purchased from the American Type Culture Collection. All cells were cultured in RPMI 1640 medium supplemented with 10% (v/v) FBS from Invitrogen. The PC-9 and H1975 cell lines were characterized by Genetic Testing Biotechnology Corporation (Suzhou, China) using short tandem repeat (STR) markers. The THP-1 and M109 cell lines were identified with STR markers by Guangzhou Cellcook Biotech and Beijing Miroread Genetics, respectively. Mycoplasma contamination testing was performed with negative results, and plasmon was added into the medium for two weeks before experiments.
Circulating tumor DNA-based next-generation sequencing
Peripheral blood samples were collected in Streck tubes (Streck, Omaha, NE, USA) and centrifuged within 72h to separate the plasma and cells. Circulating tumor DNA (ctDNA) was extracted from plasma samples using QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany). Genomic DNA (gDNA) was extracted from peripheral blood cells using QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany).
Among all 1383 samples, 167 of them were sequenced with a 59-genes panel, 265 of them were sequenced with a 73-genes panel, and 951 of them were sequenced with a 1021-genes panel. The corresponding genes contained in different panels were displayed in Supplemental Table S3. Sequencing libraries of both ctDNA and gDNA were constructed using the KAPA DNA Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA), according to the manufacturer's protocol. The capture probe was designed to cover coding sequencing or hot exons of corresponding genes frequently mutated in solid tumors. Libraries were hybridized to custom-designed biotinylated oligonucleotide probes (Integrated DNA Technologies, Iowa, IA, USA). DNA sequencing was performed using the HiSeq 3000 Sequencing System (Illumina, San Diego, CA) with 2 × 101-bp paired-end reads.
From raw data, terminal adaptor sequences and low-quality reads were removed. BWA (version 0.7.12-r1039) was employed to align the clean reads to the reference human genome (hg19). Picard (version 1.98) was used to mark polymerase chain reaction (PCR) duplicates. Realignment and recalibration were performed using GATK (version 3.4-46-gbc02625). Single nucleotide variants were called using MuTect (version 1.1.4) and NChot. Software was developed in-house to review hotspot variants (24). GATK was called small insertions and deletions (indels). Somatic copy-number alterations were identified with CONTRA (version 2.0.8). Significant copy number of variations was expressed as the ratio of adjusted depth between ctDNA and control gDNA. The final candidate variants were all manually verified in the Integrative Genomics Viewer.
Cell proliferation assay
Aliquots of 1.0–3.0 × 103 cells/mL cells were plated in a 96-well microplate (Costar, USA). Cells were treated with different doses of osimertinib, gefitinib, cisplatin, paclitaxel, and pemetrexed, which were kept cultured at 37℃ under 5% CO2 for a further 48 or 72 h. Subsequently, cells were exposed to the MTS for 1 h and measured spectrophotometrically at a wavelength of 450 ± 2 nm.
Multiplex fluorescence flow cytometry analysis
Fluorochrome-labeled antibodies used for staining tumor-associated T cells, macrophages, monocytes, dendritic cells, and neutrophils were anti-CD3e-BV605, anti-CD45.2-APC-Cy7, anti-CD11b-BV421, anti-Ly6C-PE-Cy7, anti-F4-80-PE, anti-CD206-Alexa 674, anti-CD86-BV650, anti-CD27-BV510, anti-CD49B-PE-CF5947, and 7AAD-Percp-Cy5.5. The epithelial cell of human cells was separated from xenografted tissues using anti-CD326-BB515. Unless otherwise indicated, all antibodies and reagents were purchased from BD Biosciences (San Jose, CA) and are described in Supplemental Table S4. All antibodies have been validated by their manufactory. All antibodies RRID tags were displayed in Supplemental Table S4.
Multiplex immunohistochemistry (mIF) and hematoxylin and eosin (H&E) staining
Five-μm formation-fixed paraffin-embedded slides were prepared for multiplex immunohistochemistry staining. Briefly, sections were deparaffinized and subjected to heat-induced epitope retrieval in Tris buffer (pH 8.0). Next, a 5-plex panel (CD206, F4-80, iNOS, SMA, and DAPI) was designed, which is described in Supplemental Table S4. Anti-rabbit TSA HRP (Panovue) was used as the secondary antibody. Each biomarker was visualized and performed using tyramide signal amplification-conjugated fluorophores (PerkinElmer). The number of non-overlapping regions of interest per slide ranged from 25 to 36, as previously reported.22
For immunohistochemistry and hematoxylin and eosin staining, tissues were fixed, and staining was performed based on the protocol described previously. In addition, the primary antibody CD206 (Abcam) was used as a macrophage M2 marker. All antibodies RRID tags were displayed in Supplemental Table S4.
Animal xenograft tumor formation
All animals were randomized assigned to experimental or control group with randomized numbers. For the tumorigenicity assay, each 100 μL mixture containing 106 cells was injected into the back of 4–6-week-old BALB/c mice with innate immune responses or immunodeficient, athymic BALB/c-nude mice (Vital River, Beijing, China). Tumor formation was monitored every 3 days. Mice were treated with osimertinib (10 mg/kg, p.o., 5 days a week) or a vehicle (osimertinib diluted solution) or STAT3 inhibitor SH-4-54 (10 mg/kg p.o. twice a week, Selleck) or IL-4 inhibitor dupilumab (10 mg/kg p.o. twice a week, MedChem Express). Tumor volumes were calculated using the formula length × width2 × 0.52. Mice were sacrificed by CO2 without suffering, and the tumors were dissected. The treatments and measurements were conducted at the same time. All animal experiments were approved by Peking University Cancer Hospital (license number: SYXK 2016-0015) and conformed to the regulatory standards (EAEC 2018-14) in accordance with the National Institutes of Health Guide (Guide for the Care and Use of Laboratory Animals, 2011).
Cytokine antibody array assay
H1975 L792Fcis and control cell cultures were tested for cytokine and chemokine secretion by a combination of 11 non-overlapping arrays to measure the relative expression levels of 440 human cytokines following the manufacturer's instructions (G-Series Human Cytokine Antibody Array 440, GSH-CAA-440, Norcross, GA 30092, http://www.RayBiotech.com).
Western blot
Cell lysis, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and western blot were performed using standard protocols described in the literature. All the primary antibodies used are listed in Supplemental Table S4. HRP-conjugated secondary antibodies were purchased from Jackson ImmunoResearch. Membranes were exposed and visualized using chemiluminescence (Millipore). All antibodies RRID tags were displayed in Supplemental Table S4.
Quantitative real-time polymerase chain reaction
RNAs of cells and tissues were extracted using RNeasy Mini Kit for the gene (Qiagen, Hilden, Germany), and cDNA was synthesized using M-MLV Reverse Transcriptase kit (Invitrogen). To quantify changes in mRNA levels of CD206, IL-10, and ARG1, quantitative real-time PCR (qRT-PCR) was performed with SYBR Green qPCR Master Mix (TOYOBO, Osaka, Japan), and appropriate primers (nucleotide sequences are provided in Supplementary Table S5) on the ABI Prism 7500 Fast (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions, as previously reported.20
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed according to the manufacturer's instructions (Millipore). The cells were cross-linked by 37% formaldehyde, followed by sonicating lysate to shear DNA. The mixture was precipitated by p-STAT3 with protein G agarose and was washed with different salt concentration buffers. DNA was extracted from the complex for further PCR and qRT-PCR analysis.
Luciferase reporter assays
To determine luciferase activity, cells were seeded and co-transfected with pGL3 firefly luciferase reporter vector with wild type or mutation of IL-4 promoter in binding sequence and pRL-TK Renilla luciferase reporter vector using Lipofectamine 3000 (Invitrogen). After 24 h, luciferase activities were detected by FLUO star OPTIMA (BMG, LabTech, Offenburg, Germany) using the Dual-Luciferase Reporter System (Promega, Madison, WI, USA) according to the manufacturer's instructions.
Statistical analyses
All data were from at least three independent experiments unless otherwise specified. Variations within each group were estimated and were all statistically compared. Continuous normal distribution data are presented as mean ± standard deviation and analyzed using the Statistical Package for the Social Sciences version 13.0 software. The significant differences between the two groups were determined with a two-sided Student's t test. P< 0.05 was considered statistically significant.
Role of funding source
The Funders had no role in the study design, data collection, data analyses, interpretation, or writing of report, and the decision of paper submission.
Results
Acquired EGFR L792F mutations was associated with osimertinib resistance
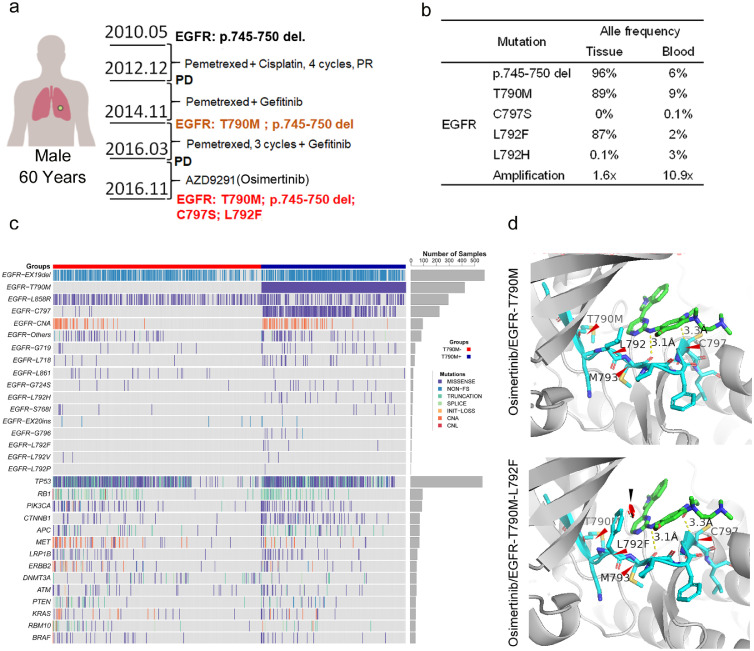
Based on the essential role of acquired EGFR mutations in the EGFR-TKI resistance, we reported the dynamic occurrence of EGFR involving mutations in a patient with metastatic lung adenocarcinoma who received serial EGFR-TKI therapy. As depicted in Figure 1a, a 60-year-old male was diagnosed with lung adenocarcinoma, and EGFR p.745-750 deletion mutation was detected in his needle biopsy specimen. He was initially treated with pemetrexed plus cisplatin and maintained with pemetrexed. The lesions progressed after 11 months. Considering the previous existing EGFR p.745-750 del mutation, the patient additionally received the first-generation EGFR-TKI (gefitinib). The disease progressed again 23 months later. EGFR T790M mutation was detected in the patient's tumor during the secondary biopsy. Thus, the patient received the third-generation EGFR-TKI osimertinib (Figure 1a). When disease progression was observed after 7 months of osimertinib treatment, biopsied tumor tissue and peripheral blood were obtained. Through next-generation sequencing, 19Del, T790M, C797S, and L792F in EGFR were found with the following AFs: 96% and 6%, 89% and 9%, 0%, and 0.1%, and 87% and 2% for 19Del, T790M, C797S, and L792F, respectively, in the tumor tissue and blood samples, respectively (Figure 1b). Then, he was treated with multiple therapies, including pemetrexed rechallenge, nano-albumin paclitaxel, whole brain radiation therapy, etc., but the disease progressed rapidly. Eight months after the detecting of L792F/H mutations, he was dead due to disease progression.
Figure 1.
Epidermal growth factor receptor (EGFR) mutations in a patient with lung adenocarcinoma resistant to osimertinib. (a). EGFR mutations in a patient with lung adenocarcinoma who received chemotherapy and EGFR-tyrosine kinase inhibitor (TKI) treatment. (b). Table of mutation frequencies in tissue and circulating tumor DNA. (c). Heatmap of targeted sequencing analysis in patients with lung cancers receiving second-line osimertinib (n = 1147). (d). Structure prediction of EGFR with T790M and L792Fcis mutation in complex with osimertinib. The greenstick model shows osimertinib; the navy stick model shows the fragment of EGFR sequence from p.789 to p.798; red arrows indicate hydrogen bonds or covalent bonds linking the amino acids. CH/p interactions presented approximately 3.1 Å. Substitutions could prevent osimertinib binding by introducing spatial confliction (black arrow) from mutated residue. PR, partial response; PD, progressive disease.
To generally illustrate the distribution of EGFR L792F co-occurrence with T790M, ctDNA sequencing data of 1147 samples from 946 patients with lung cancers with acquired resistance to osimertinib as second-line treatment were collected, in that five patients presented with EGFR L792F mutation (Figure 1c, Table S1). Among these five patients harboring EGFR L792F mutation, L792F mutation was identified with T790M mutation in the same sequencing read of every sample (Figure S1), which suggested that L792F and T790M were in cis. Although patients 1 and 2 harbored both L792F and C797S, these two mutations (L792F and C797S) did not co-exist in the same read, which suggested that they were in trans (Figure S1). For patient 3, L792F was identified, accompanied by other EGFR mutations in trans, such as L792H and L792P. In samples of patients 2 and 4, L792F was not accompanied by any other EGFR mutations in cis or trans. In addition, ctDNA sequencing data of 236 samples from 207 patients with lung cancers, with acquired osimertinib resistance as first-line treatment, were collected, revealing that L792X mutation was not detected (Figure S2). Only 2 of these 236 samples were detected with EGFR T790M mutation (Figure S2). Hence, we believed that it was the T790M-cis-L792F mutation, rather than a single L792F mutation, that induced osimertinib resistance.
To further demonstrate the potential effect of L792F mutation on osimertinib, EGFR T790M-cis-L792F binding with osimertinib was shown as a cartoon diagram, displaying a stick model and hydrogen bonds within the structure. T790M single mutation and the residues interacting with osimertinib showed that the multiple amino acids within the ATP-binding pocket interacted with hydrogen bonds without covalent binding. Interestingly, a benzene or imidazole ring was demonstrated to the side chain of this residue in the T790M-cis-L792F complex, which resulted in the disruption of orientation and spatial stabilization for osimertinib binding (Figure 1d). Considering the high frequency of L792 mutation in the patient with osimertinib resistance, we hypothesized that T790M-cis-L792F was associated with the third-generation TKI resistance. These epidemiological and spatially simulated results suggested that T790M-cis-L792F might be associated with acquired resistance to osimertinib.
EGFR T790M-cis-L792F induced osimertinib resistance
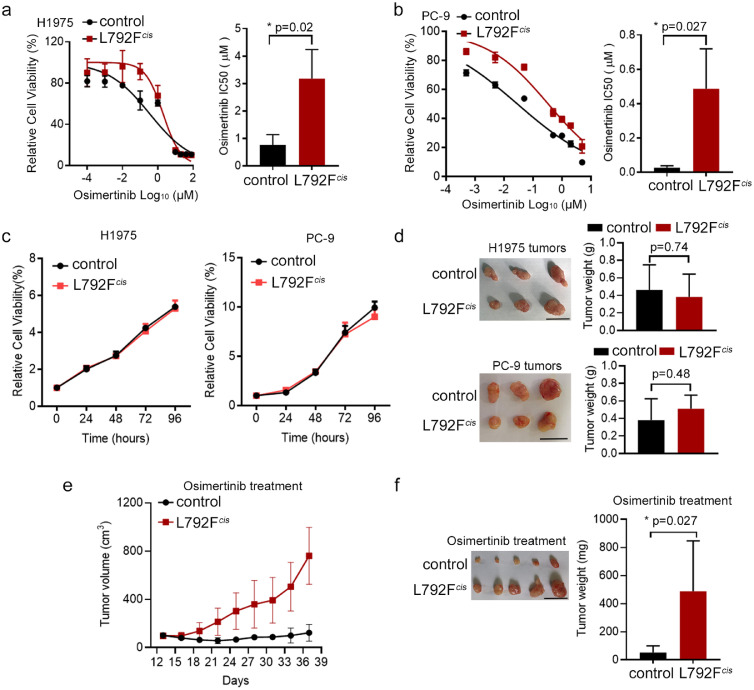
To determine the roles of EGFR T790M-cis-L792F mutation in osimertinib resistance, lentivirus particles containing EGFR 19del/T790M-cis-L792F mutation plasmid (L792Fcis) or control plasmid including 19del/T790M mutation (control), were stably transfected into H1975 and PC-9 cell lines, respectively. The half-maximal inhibitory concentration (IC50) of each group in H1975 and PC-9 was tested to verify the effects of EGFR T790M-cis-L792F on sensitivity to osimertinib. In H1975 and PC-9 cell lines, the IC50 value to osimertinib of the L792Fcis group was higher than that in control (3.183 vs. 0.762 μM, p=0.02 in H1975 and 0.486 vs. 0.026 μM in PC-9, p=0.027, t test, Figure 2a–b). Moreover, no significant difference was observed on the IC50 value between the L792Fcis and control groups for cisplatin (6.64 vs. 5.867 μM, p≥0.05 in H1975 and 3.09 vs. 2.643 μM, p≥0.05 in PC-9, t-test, Figure S3a–b), paclitaxel (105.5 vs. 116.8 nM, p≥0.05 in H1975 and 7.332 vs. 7.199 μM, p≥0.05 in PC-9, t test, Figure S3c–d), and pemetrexed (2.377 vs. 2.485 μM, p≥0.05 in H1975 and 2.182 vs. 1.966 μM, p≥0.05 in PC-9, t test, Figure S3e–f). These findings suggested that T790M-cis-L792F mutation induced osimertinib resistance.
Figure 2.
EGFR 19Del/T790M-cis-L792F was related to the third-generation EGFR TKI resistance. (a-b). Representative cell viability curve (left panel) and osimertinib half-maximal inhibitory concentration (IC50) value (right panel) in the H1975 (a, p=0.02) and PC-9 (b, p=0.027) cells transfected with L792Fcis mutation under osimertinib treatment for 72 h (n = 3). (Statistical differences were evaluated using t test). (c). Growth curves of the control and L792Fcis groups of H1975 (left panel) and PC-9 (right panel) for 24, 48, 72, and 96 h using the MTS assay (n = 3). (d). Tumor images and weights from H1975 (control and L792Fcis, upper panel, p=0.74) and PC-9 (control and L792Fcis, down panel, p=0.48) cells (n = 3). (Statistical differences were evaluated using t test). (e-f). Tumor growth curves (e) and tumor weights (f) in the PC-9 control and PC-9 L792Fcis mice models with control (5% dimethyl sulfoxide, 40% PEG300, 5% Tween 80, and 50% water) or osimertinib (10 mg/kg, 5 days a week) treatments at the indicated time points (n = 5, p=0.027. Statistical differences were evaluated using t test). Scale bar, 1 cm.
In addition, compared to the effect of cis-mutated EGFR L792F with T790M to osimertinib resistance, the trans-L792F mutation did not present different cell viability with control in H1975 cells (p≥0.05, t test, Figure S4a) or PC-9 cells (p≥0.05, t test Figure S4b), similar with 19Del group or 19Del/L792F group.
Constantly, the proliferation of EGFR T790M-cis-L792F was tested. Without osimertinib treatment, the proliferation capability was similar between the L792Fcis and control groups in either H1975 or PC-9 cells (Figure 2c). These results were confirmed in subcutaneous xenograft models (p=0.74 and 0.48, respectively, t test, Figure 2d). However, under osimertinib treatment, the tumor volume and weight of the L792Fcis group were significantly higher than those of the control group (p=0.027, t test, Figure 2e–f).
Taken together, EGFR T790M-cis-L792F mutation induced resistance to osimertinib in vitro and in vivo but did not influence the proliferation ability and response to chemotherapeutic drugs in lung adenocarcinoma.
M2 polarization of macrophages was associated with osimertinib resistance
To further investigate the mechanism of osimertinib resistance, the single-cell data (GSE160244) from xenograft tumors of PC-9 cells with osimertinib treatment was downloaded from Gene Expression Omnibus (GEO), showing 19 clustered cell populations (Figure S5a). Primary cell types contained cancer cells, fibroblasts (active and inactive), macrophage cells, and leukocyte cells (Figure S5b). The macrophage subtypes were evidently distinct between the osimertinib treatment and control groups (Figure S5c). Drug perturbation signaling analysis in each cell cluster indicated that the macrophage subtype was significantly affected by osimertinib treatment (Figure S5d). Evidently, CD206, a marker of M2 macrophage, was enhanced from 0.2% to 0.8% in total cells (Figure S5e). Interestingly, the cell cycle, wound healing, extracellular matrix, senescence, and autophagy in cancer were also enriched in the tumors with osimertinib treatment (Figure S5f).
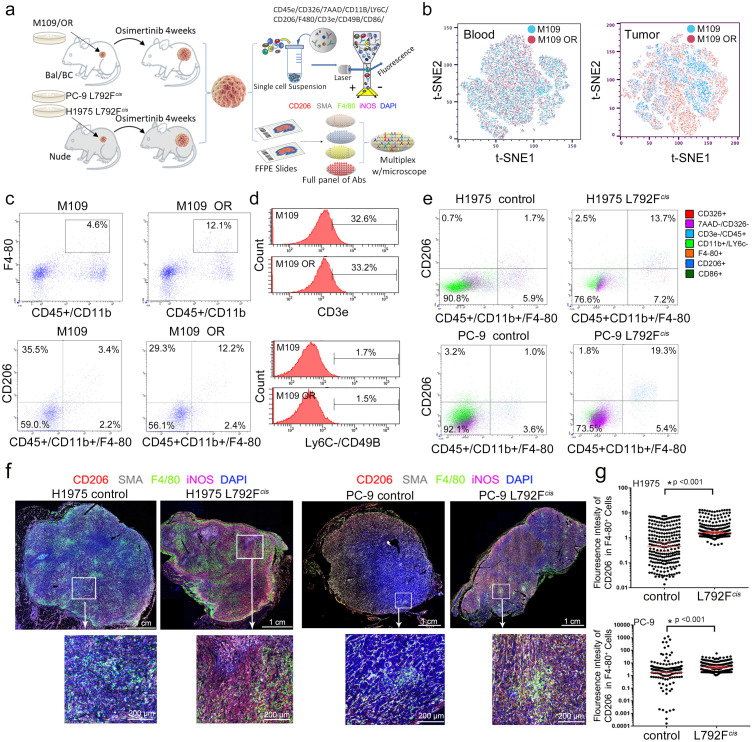
Furthermore, M109 cell xenograft models with osimertinib resistance and EGFR T790M-cis-L792F xenografts tumors were established. After the xenografts were treated with osimertinib, multiplex immunofluorescence flow cytometry was applied to assess the proportions of immune cells in tumor tissues and the blood of mice (Figure 3a). As shown in Figure 3b, in the blood samples, the uniform distribution of dot by t-distributed stochastic neighbor embedding analysis indicated no difference in immune phenotypes between the osimertinib resistance (OR) and control groups. In contrast, different classifications of dots in tumor tissues showed that the immune cells undergoing osimertinib treatment were clustered. The proportion of macrophages (CD45+/CD11b+/F4-80+ cells) in the tissue of the OR group was higher than that in the control group (12.1% vs. 4.6%, Figure 3c upper panel). Moreover, the proportion of M2 macrophages (CD45+/CD11b+/F4-80+/CD206+ cells) in the OR group was higher than that in the control group (12.2% vs. 3.4%, Figure 3c bottom panel). However, the proportions of CD3e+ T cells (32.6 % vs. 33.2%, Figure 3d upper panel and Figure S6a) and LyC6–/CD49+ or CD11b+/CD27+ natural killer (NK) cells (1.1% vs. 1.0 %, Figure 3d bottom panel and Figure S6b) were similar between the OR and control groups.
Figure 3.
Macrophage phenotype in lung adenocarcinoma cells with osimertinib resistance. (a). Schematic diagram of immunophenotype analysis in xenograft tumors with osimertinib resistance. The BALB/c mice with M109 tumors or BALB/c-nude mice with PC-9 L792Fcis tumors were treated with control (5% dimethyl sulfoxide, 40% PEG300, 5% Tween 80, and 50% water) or osimertinib (10 mg/kg, 5 days a week) for 4 weeks. The resected tumors were further analyzed with immunophenotype by flow cytometry on the markers, including CD45e, CD326, 7AAD, CD118, Ly6C, CD206, F4-80, CD3e, CD49B, CD86, and CD27, and immunohistochemistry on the staining of CD206, SMA, F4-80, iNOS, and DAPI. (b). t-distributed stochastic neighbor embedding plot of blood and formed tumor from the mice injected with M109 cells by multiplex fluorescence flow cytometry. (c). F4-80, CD45, CD11b, and CD206 staining on tumors of M109 control and M109 osimertinib resistance (OR) by multiplex fluorescence flow cytometry analysis. (d). Histograms show CD3e for T cell and Ly6C/CD49B for NK cell analysis in the tumors of M109 control and M109 OR. (e). CD326, 7AAD, CD3e, CD45, CD11b, Ly6C, F4-80, CD86, and CD206 staining (indicated by colors) on tumors of H1975 control, H1975 L792Fcis and PC-9 control, and PC-9 L792Fcis by multiplex fluorescence flow cytometry analysis for tumor-associated macrophages. (f). Multiplex immunohistochemistry staining, including CD206, SMA, F4-80, iNOS, and DAPI, with the multiple colors as indicated on tumors of H1975 control, H1975 L792Fcis, PC-9 control, and PC-9 L792Fcis. The scale bar was 200 um. (g). Graphs show the F4-80+/CD206+ influence ratio in the tumors of H1975 control, H1975 L792Fcis, PC-9 control, and PC-9 L792Fcis by multiplex immunohistochemistry analysis (n=25, both p<0.001. Statistical differences were evaluated using t test).
EGFR T790M-cis-L792F derived M2 macrophage polarization
Based on EGFR T790M-cis-L792F xenografts tumors with H1975 and PC-9 cells in Figure 3a, the proportions of F4-80+ macrophages in the L792Fcis group were higher than that in the control group of H1975 (19.2% vs. 7.2%) and PC-9 (19.3% vs. 5.3%) (Figure S6c). In addition, F4-80+/CD206+ M2 macrophage in L792Fcis group tissue was also higher than that in the control group of H1975 (13.7% vs. 1.7%, Figure 3e upper panel) and PC-9 (19.3% vs. 1.0%, Figure 3e bottom panel). Furthermore, the percentage of NK cells was not significant between the L792Fcis and control groups in H1975 (0.5% vs. 0.8%) and PC-9 (1.6% vs. 1.1%) (Figure S6d). Multiple immunohistochemistry was further used to test the M2 macrophage phenotype. CD206 and iNOS expressions were enhanced in L792Fcis tumors of H1975 and PC-9 (Figure 3f). Moreover, the positive staining of F4-80+/CD206+ (M2 macrophage) was higher in the L792Fcis group than that in the control group (both p<0.001, t test, Figure 3g). In contrast, F4-80+/iNOS+ (M1 macrophage) staining was lower in the L792Fcis group than that in the control group of H1975 and PC-9 (both p<0.001, t test, Figure S6e).
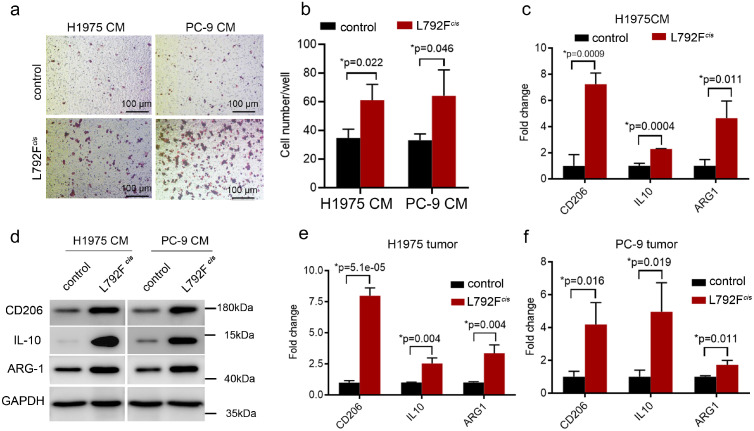
Next, macrophage cell line THP-1 was harvested at the conditional medium (CM) culturing from L792Fcis cells, showing more migrated cells than culturing from the control cells group (p=0.022 and 0.046, respectively, t test, Figure 4a–b). The mRNA and protein levels of M2 macrophage-associated markers, including CD206, IL-10, and ARG1, were higher in the L792Fcis group than in the control group (p=0.0009, 0.0004 and 0.011, respectively, t test, Figure 4c–d). Finally, sorted F4-80+ macrophages from the xenograft tumors in the mice model showed that the mRNA levels of CD206, IL-10, and ARG1 were higher both in the H1975 L792Fcis (p=0.000051, 0.004 and 0.004 respectively, t test, Figure 4e) and PC-9 L792Fcis (p=0.016, 0.019 and 0.011, respectively, t test, Figure 4f) groups than in the control group.
Figure 4.
EGFR-19Del/T790M/L792F drove the infiltration of M2-like macrophages. (a-b). Representative images (a) and histogram (b, p=0.022 or 0.046, respectively. Statistical differences were evaluated using t test) show migrated cells of THP-1 cultivated in conditional medium derived from H1975 control, H1975 L792Fcis, PC-9 control, and PC-9 L792Fcis cells by Boyden chamber assay (n=3). The scale bar in (a) was 100 μm. (c). qRT-PCR analyses of CD206, IL-10, and ARG1 expression in the THP-1 cultivated in conditional medium from H1975 control and H1975 L792Fcis cells (p=0.009, 0.0004 or 0.011, respectively. Statistical differences were evaluated using t test). (d). The protein level of CD206, IL-10, and ARG1 by western blot assay. (e-f). qRT-PCR analyses of CD206, IL-10, and ARG1 expression in the formed tumors of nude mice derived from H1975 control, H1975 L792Fcis(e, p=0.00005, 0.004 or 0.004, respectively), PC-9 control and PC-9 L792Fcis(f, p=0.016, 0.019 or 0.011, respectively) (both n=3, Statistical differences were evaluated using t test). Data are presented as the means ± standard deviation.
These results suggested that EGFR T790M-cis-L792F mutation was associated with M2 macrophage polarization, which induced osimertinib resistance in lung adenocarcinoma.
EGFR T790M-cis-L792F mutation promoted IL-4 expression and secretion by activating STAT3 Tyr705 phosphorylation and its specific binding to IL4 promoter
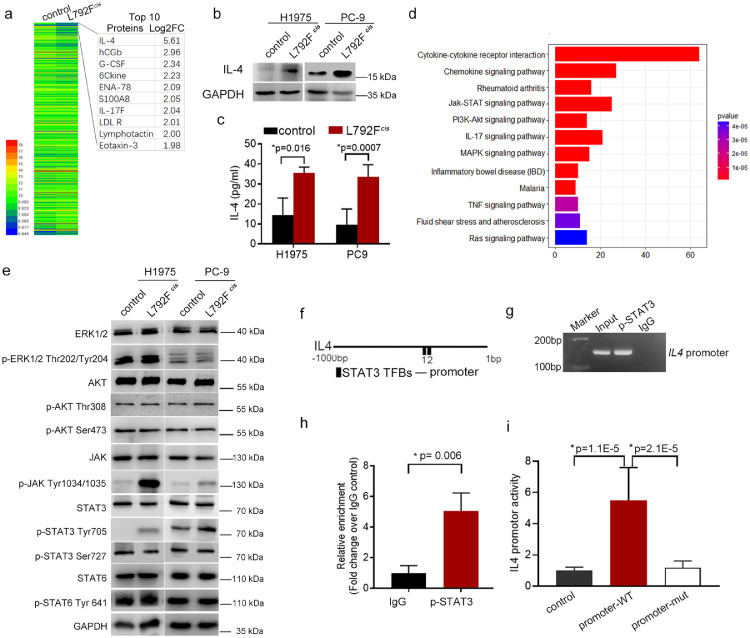
To explore the specific mechanism by which T790M-cis-L792F mutation regulated M2 macrophage polarization, we applied a cytokine chip to determine IL-4 expression with the highest fold change (Log2FC = 5.61) in L792Fcis group cells than that in control cells (Figure 5a). Since IL-4 is a well-known cytokine to induce M2 polarization, the top 10 highly expressed protein in L792Fcis was analyzed by protein-protein interaction (PPI) using the String website. The results revealed that migration and macrophages were enriched in the L792Fcis group cells (Figure S7). Furthermore, western blotting (Figure 5b) and enzyme-linked immunosorbent assay (ELISA) (Figure 5c, t test) tests confirmed that IL-4 expression and secretion were higher in L792Fcis cells than in the control cells of H1975 and PC-9. Gene Ontology (GO) analysis demonstrated that leukocyte migration and chemotaxis, cell chemotaxis, and Janus kinase (JAK)-STAT signaling pathway were activated in L792Fcis cells (Figure 5d). Subsequently, downstream signals of EGFR, such as ERK, AKT, JAK, STAT3, and STAT6, were examined, and phosphorylation levels of AKT, JAK, and STAT3Tyr705 were higher in L792Fcis cells than in control cells (Figure 5e). In addition, a binding site in a transcript factor p-STAT3 was identified in the promoter region of IL4 (Figure 5f). After ChIP, PCR and qRT-PCR confirmed that p-STAT3 was bound to the IL4 promoter specifically (p=0.006, t test, Figure 5g–h). Furthermore, L792Fcis cells had higher promoter activity of IL4, whereas when the binding sequence was mutated, the promoter activity of IL4 was significantly inhibited (p=1.1E-5 and 2.1E-5, respectively, t test, Figure 5i).
Figure 5.
EGFR 19Del/T790M-cis-L792F stimulated JAK/STAT3/IL-4 signaling. (a). Heatmap of cytokine antibody array in H1975 control and H1975 L792Fcis cells. (b-c). IL-4 expression was detected by western blot (b) and ELISA analyses (c) in the H1975 control, H1975 L792Fcis, PC-9 control, and PC-9 L792Fcis cells (n=3, p=0.016 or 0.007, respectively. Statistical differences were evaluated using t test). (d). Signal pathway enrichment in the H1975 L792Fcis cells compared to the H1975 control cells. (e). Western blot shows ERK, p-ERK Thr202/Tyr204, AKT, p-AKT Ser473, JAK, p-JAK Tyr1034/1035, STAT3, p-STAT3 Tyr705, p-STAT3 Ser727, STAT6, and p-STAT6 Try641 expression in the H1975 control, H1975 L792Fcis, PC-9 control, and PC-9 L792Fcis cells. (f). Schematic depiction of the IL4 promoters (−1000 to 1 base pair, black line) with STAT3 transcription factor binding sites (TFBs). (g-h). ChIP assay using p-STAT3 antibody and IgG as internal controls. PCR (g) and RT-qPCR (h, n=3, p=0.006, statistical differences were evaluated using t test.) were performed to amplify p-STAT3 TFBs of IL4 promoter in the ChIP product. (i). IL4 promoter activity of luciferase reporter in the PC-9 control and PC-9 L792Fcis cells with wild-type or mutant p-STAT3-binding sequences (n=3, p=0.00001 or 0.00002, statistical differences were evaluated using t test). PRL-TK plasmid was used as an internal control. Data are presented as means ± standard deviation.
These results revealed that EGFR T790M-cis-L792F activated JAK/STAT3 pathway and p-STAT3 Tyr705 bound explicitly to the IL4 promoter and increased the subsequent transcription.
Inhibiting STAT 3 or IL-4 overcame osimertinib resistance
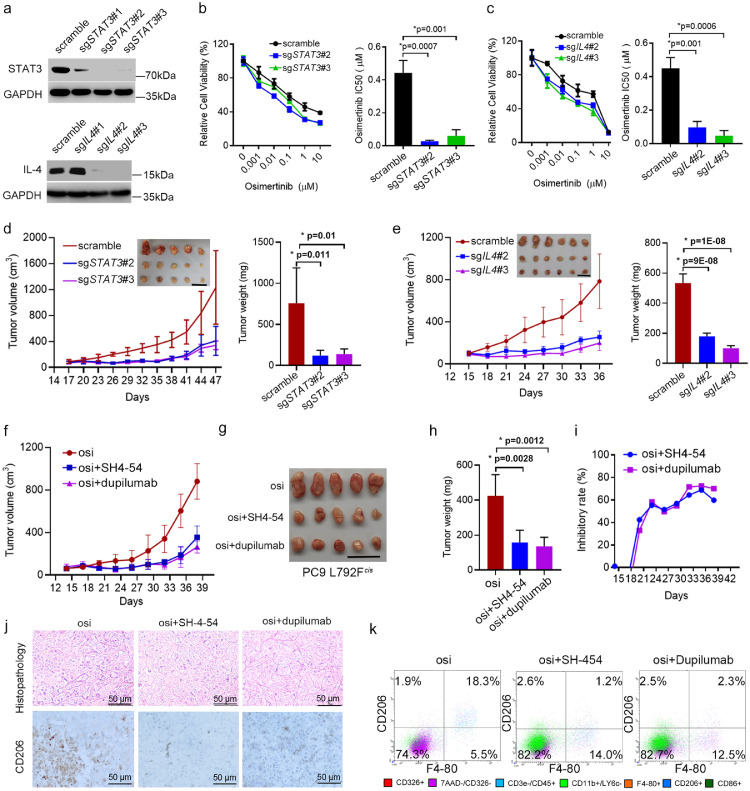
To investigate whether inhibiting STAT3 or IL-4 could suppress osimertinib resistance induced by EGFR T790M-cis-L792F mutation, we designed sgRNAs targeting STAT3 or IL4 and sgSTAT3#2 and #3; sgIL4#2 and #3 were selected for further studies (Figure 6a). When PC-9 L792Fcis cells were co-cultured with THP-1 cells, IC50 decreased in the sgSTAT3#2/3 (0.027/0.06 vs. 0.443 μM, p=0.007 and 0.001, respectively, t test, Figure 6b) and sgIL4#2/3 groups (0.097/0.047 vs. 0.449 μM, p=0.001 and 0.0006, respectively, t test, Figure 6c). In addition, sgSTAT3#2/3 (p=0.011 and 0.01, respectively, t test, Figure 6d) and sgIL4#2/3 (p=9E-8 and 1E-8, respectively, t test, Figure 6e) inhibited the tumor volume and weight in xenograft models.
Figure 6.
Inhibition of STAT3/IL-4 signaling restored sensitivity to osimertinib. (a). Western blot shows STAT3 expression in the PC-9 L792Fcis cells transfected with scramble, sgSTAT3#1, sgSTAT3#2, or sgSTAT3#3, and IL-4 expression in the PC-9 L792Fcis cells transfected with scramble, sgIL4#1, sgIL4#2, or sgIL4#3. (b-c) Representative cell viability curve and osimertinib IC50 value in the PC-9 L792Fcis cells transfected with scramble, sgSTAT3#2 or sgSTAT3#3 (b, n=3, p=0.001 or 0.0007, respectively. Statistical differences were evaluated using t test.), and in the PC-9 L792Fcis cells transfected with scramble, sgIL4#2 or sgIL4#3 (c, n=3, p=0.0006 or 0.001, respectively. Statistical differences were evaluated using t test.), under osimertinib treatment for 72 h. (d-e). Tumor growth curves and tumor images of the PC-9 L792Fcis-scramble, PC-9 L792Fcis-sgSTAT3#2, PC-9 L792Fcis-sgSTAT3#3 (d) (n=5, p=0.01 or 0.001, respectively. Statistical differences were evaluated using t test), and PC-9 L792Fcis-scramble, PC-9 L792Fcis-IL4#2, and PC-9 L792Fcis-IL4#3 (e) (n=6, p=1E-08 or 9E-08, respectively. Statistical differences were evaluated using t test) in the nude mice with osimertinib (10 mg/kg, 5 days a week) treatment. Scale bar, 2 cm. (f-h). Growth curve (f), tumor images (g), and tumor weight (h, n=5, p=0.0012 or 0.0028, respectively. Statistical differences were evaluated using t test.) of PC-9 L792Fcis xenografts treated with osimertinib (10 mg/kg, 5 days a week), osimertinib (10 mg/kg, 5 days a week) + STAT3 inhibitor (SH-4-54, [10 mg/kg, twice a week]), and osimertinib (10 mg/kg, 5 days a week) + IL-4 receptor antibody (dupilumab, 10 mg/kg, twice a week). Scale bar, 2 cm. (i). Inhibitory rates of SH-4-54 and dupilumab for PC-9 L792Fcis xenografts. (j-k). Images of H&E (upper panel in j), immunohistochemistry staining for CD206 (down panel in j) and CD206+/F4-80+ macrophages fractions in PC-9 L792Fcis xenografts treated with osimertinib, osimertinib + SH-4-54, and osimertinib + dupilumab (k). Scale bar, 50 μm.
Moreover, SH-4-54, a small molecular inhibitor targeting STAT3, and dupilumab, a specific antibody against the IL-4 receptor, were applied to inhibit STAT3 or IL-4 in vivo. In xenograft models from PC-9 L792Fcis, tumor volumes and weights of groups treated with SH-4-54 and dupilumab plus osimertinib were lower than those treated with osimertinib only (p=0.0028 and 0.0012, t test, Figure 6f–h). The inhibitory rate of SH-4-54 and dupilumab was approximately 80% (Figure 6i). In addition, immunohistochemistry confirmed that CD206 expressions in SH-4-54 or dupilumab plus osimertinib groups were higher than the group with only osimertinib (Figure 6j). CD206 was inhibited from 18.3% to 1.2% and 2.3% in the SH-4-54 or dupilumab plus osimertinib groups, respectively (Figure 6k).
These results indicated that osimertinib resistance due to L792Fcis could be suppressed by inhibiting STAT3 or IL-4, and one of the potential mediators might be through suppressing M2 macrophage polarization.
Discussion
As a type of third-generation EGFR-TKI, osimertinib suppresses the resistance to first- and second-generation EGFR TKIs driven by EGFR T790M mutation and has achieved a great success.1,4,5 Unfortunately, most EGFR T790M-mutant patients finally progressed on osimertinib,12,19 with the specific resistance mechanisms still largely unclear and lacking corresponding clinical strategies to suppress osimertinib resistance.23 Promising fourth-generation TKIs targeting C797S, including EAI045 24 and JBJ-04-125-02,25 have shown primary efficacy and safety and are expected to be evaluated in further clinical trials. Nevertheless, treatment strategies for patients with other resistance mechanisms apart from C797S mutation remain unclear. In addition, although a previous study has found that EGFR L792F alteration was detected in tumor tissue or ctDNA from patients resistant to osimertinib,13 the specific roles of L792F in osimertinib resistance and its underlying mechanisms and corresponding clinical strategies should be further investigated.
Here, we reported that the allele frequencies of EGFR L792Fcis mutation were high in tumor tissue samples and sole-existed or co-existed samples with other mutations in trans, indicating that EGFR L792Fcis mutation might be an independent driver mutation and is exclusively mutated with C797S, which was consistent with the previous report.26 Based on the X-ray structure of osimertinib in a non-covalent complex with EGFR T790M (PDB ID:4ZAU,27 6LUD,28 and 6WU829), we predicted that EGFR L792F, cis-mutated with T790M, is represented in a blocker with a benzene or imidazole ring of hydrophobic side chain.13 Original van der Waals contacted with osimertinib phenyl ring has been destroyed, which is required to stabilize the ligand–protein complex. Replacement with bulkier amino acids in L792 strongly reduces the affinity and is tightly relevant to osimertinib resistance. However, the T790M-cis-L792F mutant did not influence the proliferation of H1975 or PC-9 cell lines without osimertinib. Interestingly, T790M-cis-L792F mutant xenografts in mice present lower growth with constant osimertinib treatment. Therefore, we hypothesized that it drove resistance to osimertinib probably via regulating the tumor immune microenvironment.
In the present study, when testing immune cells in mice, we utilized the modified panel of 12-color multiplex fluorescence flow cytometry assay to identify better T cells, macrophages, and NK cells in immunocompetent or immunodeficient athymic mice tumor microenvironment (TME).30 We found that EGFR T790M-cis-L792F mutation tumors highlighted tumor-associated macrophages (TAMs), but not T cells or NK cells in the TME. The TME and TAM have been considered targets to suppress drug resistance, and associated therapeutic macrophage presence and/or bioactivity is a critical targeting pathway that improves therapeutic efficacy.31 A recent report has demonstrated that the enriched macrophage CM induced EGFR-TKI resistance.32 Single-cell RNA sequencing of metastatic lung cancers from patients before and during targeted therapy revealed that active T-lymphocytes and decreased macrophages were present in residual disease while immunosuppressive cells, including TAMs states, characterized progressive disease.33
Moreover, TAM repolarization mediated innate immunity, and TME remodeling was approved explicitly for the OR metastasis of NSCLC.34,35 We aimed to reveal the association and mechanism between EGFR T790M-cis-L792F mutation and TAM affected by osimertinib tolerance. Recruitment of macrophages in TME contributes to EGFR T790M-mutated cell metastasis.36 However, there were few reports assessing the effect of TAM on EGFR T790M-cis-L792F mutation, except a relevant study that reported that EGFR T790M-associated gefitinib resistance reprogramed TAM. That degraded methionine oxidation led to TAM repolarization toward M1 by elevating the reactive oxygen species pathway.37 Based on signal cell sequencing, mIF, and PPI, TAM might be relevant to EGFR T790M-cis-L792F and osimertinib tolerance in this study.
Furthermore, RNA sequencing and protein-level data have supported the notion that the expression and secretion of IL-4 are increased in EGFR T790M-cis-L792F cells. As the central switch macrophages from the pro-inflammatory to the anti-inflammatory subtype, IL-4 highlights the M2 polarization of TAM, as previously reported.38,39 This evidence demonstrates that EGFR T790M-cis-L792F cell-induced osimertinib resistance promotes macrophage M2 polarization through IL-4 activity.
As a transcription factor, STAT3 is phosphorylated by downstream pathways when EGFR is activated.40 Therefore, we also confirmed that EGFR T790M-cis-L792F mutation activating STAT3 phosphorylation at the Tyr705 site (not at Ser727) induced IL-4 transcription. A previous investigation reported a coordinated signaling network centered on STAT3 and Src-YAP signaling that limits targeted therapy response in EGFR-mut lung cancer cell lines.41 Furthermore, IL-4/STAT6 and IL-6/STAT3 signal pathways are considered to promote solid tumor progression by increasing M2 TAM infiltration.42,43 This study found that STAT3 phosphorylation level was elevated in T790M-cis-L792F-mutated cell lines, and p-STAT3 feedbacked IL-4 secretion by binding to its promoter. In addition, inhibiting STAT3 with SH-4-54 and its downstream IL-4 with dupilumab partly reversed osimertinib resistance in mice models. Our results together with previous studies, suggested that promoting M2 macrophage polarization might be one of the potential STAT3/IL-4 downstream mediators associated with resistance to osimertinib. However, considering the complexity of the immune system and tumor microenvironment of EGFR mutant cancers, other mechanisms than M2 macrophage might also be involved in osimertinib resistance, which deserves further extensive exploration.
Several promising small molecular inhibitors targeting STAT3 have shown potential anti-tumor effectiveness.44,45 Whether these small molecular inhibitors could inhibit IL-4 secretion and thus inhibit M2 macrophage polarization is worth further validation. Although the safety and efficacy of dupilumab have been interpreted in atopic dermatitis and asthma,46, 47, 48 the dose and impact on osimertinib resistance in NSCLC patients need to be confirmed by clinical studies.
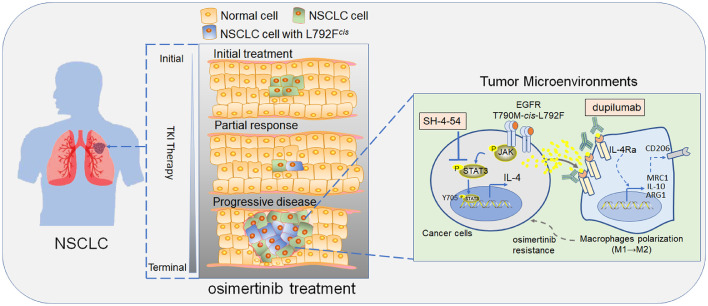
In conclusion, we demonstrate that EGFR T790M-cis-L792F mutation in lung adenocarcinoma upregulated Jak and STAT3 phosphorylation and increased IL-4 production via specifically binding to IL4 promoter, which promotes M2 macrophages polarization and induces resistance to osimertinib. Inhibiting the axis activity by SH-4-54 and dupilumab would be a promising strategy that was encouraged against the failed third-generation EGFR-TKI therapy for patients with NSCLC (Figure 7). In addition, we bridged the correlation of EGFR L792Fcis in manipulating macrophages’ switch, which also provided a chance to reverse the acquired osimertinib resistance by interfering with the TME of NSCLCs. Although the mutation frequency of EGFR L792X, including L792F/H/P/V, was 2.46% in this study, considering the substantial NSLCLC patient base in China49 and worldwide,50 the number of patients with L792X mutations and resistance to Osimertinib might be pretty huge. Therefore, overcoming L792X mutations-induced resistance is vital to this large patient population. Our discoveries may also provide insights into EGFR TKIs resistance induced by other endogenous mutations.
Figure 7.
Proposed model by which EGFR 19Del/T790M-cis-L792F mutation mediated osimertinib resistance in non-small cell lung cancer.
Contributors
Y.S. and Y.D. and X.L. contributed equally to this work. Y.S., Y.D., X.L. and Z.W. accessed and verified the underlying data. Z.W. and J.W. designed this investigation. Y.S., Y.D., X.L. Y.Z., H.B., J.D., Z.T. and X.Y. collected and analyzed the data. Z.W., J.W., Y.S. and Y.D. prepared the manuscript. All authors read and approved the final version of the manuscript. Z.W. and J.W. were co-corresponding authors and responsible for the decision to submit the manuscript.
Data sharing statement
The GEO datasets (GSE160244) used in this study were publicly available as described in the “Methods” section. Due to the regulations of the institution, individual-level sequencing and detailed clinical data of these 1383 patients of this study cannot be uploaded to the public repository, but available from the corresponding author on reasonable request.
Declaration of interests
The authors declare no potential conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81871889, 82072586, 81902910), Beijing Natural Science Foundation (7212084, 7214249), the China National Natural Science Foundation Key Program (81630071), the National Key Research and Development Project (2019YFC1315704), CAMS Innovation Fund for Medical Sciences (CIFMS2021-1-I2M-012), Aiyou Foundation (KY201701) and CAMS Key Laboratory of translational research on lung cancer (2018PT31035). We thank all patients that were involved in this study. We thank Dr. Shouli Yuan from the Institute of Zoology, Chinese Academy of Sciences, Beijing, China, who provided us the protocol and authorization of PyMOL software. We thank Mrs. Xinying Ma for her great artistic suggestion from Ma's Scientific Illustration Studio, Beijing, China. We also thank the support from the Geneplus-Beijing Institute, Beijing, China, and the support of single cell-sequencing data analysis from Intelliphecy Inc., Shenzhen, China.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.104200.
Contributor Information
Jie Wang, Email: zlhuxi@163.com.
Zhijie Wang, Email: wangzj@cicams.ac.cn.
Appendix. Supplementary materials
References
- 1.Ramalingam SS, Yang JC, Lee CK, et al. Osimertinib as first-line treatment of EGFR mutation-positive advanced non-small-cell lung cancer. J Clin Oncol. 2018;36:841–849. doi: 10.1200/JCO.2017.74.7576. [DOI] [PubMed] [Google Scholar]
- 2.Lu S, Dong X, Jian H, et al. AENEAS: a randomized phase III trial of aumolertinib versus gefitinib as first-line therapy for locally advanced or metastaticnon-small-cell lung cancer with EGFR exon 19 deletion or L858R mutations. J Clin Oncol. 2022 doi: 10.1200/JCO.21.02641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y, Chen G, Wang X, et al. Furmonertinib (AST2818) versus gefitinib as first-line therapy for Chinese patients with locally advanced or metastatic EGFR mutation-positive non-small-cell lung cancer (FURLONG): a multicentre, double-blind, randomised phase 3 study. Lancet Respir Med. 2022:1–10. doi: 10.1016/S2213-2600(22)00168-0. [DOI] [PubMed] [Google Scholar]
- 4.Mok TS, W Y-L, A M-J, et al. Osimertinib or platinum-pemetrexed in EGFR T790M-positive lung cancer. N Engl J Med. 2017;376:629–640. doi: 10.1056/NEJMoa1612674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papadimitrakopoulou VA, Mok TS, Han JY, et al. Osimertinib versus platinum-pemetrexed for patients with EGFR T790M advanced NSCLC and progression on a prior EGFR-tyrosine kinase inhibitor: AURA3 overall survival analysis. Ann Oncol. 2020;31:1536–1544. doi: 10.1016/j.annonc.2020.08.2100. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y, Hu X, Zhang S, et al. Efficacy, safety, and genetic analysis of furmonertinib (AST2818) in patients with EGFR T790M mutated non-small-cell lung cancer: a phase 2b, multicentre, single-arm, open-label study. Lancet Respir Med. 2021;9:829–839. doi: 10.1016/S2213-2600(20)30455-0. [DOI] [PubMed] [Google Scholar]
- 7.Lu S, Wang Q, Zhang G, et al. Efficacy of aumolertinib (HS-10296) in patients with advanced EGFR T790M+ NSCLC: updated post-national medical products administration approval results from the APOLLO registrational trial. J Thorac Oncol. 2022;17:411–422. doi: 10.1016/j.jtho.2021.10.024. [DOI] [PubMed] [Google Scholar]
- 8.Wu YL, Tsuboi M, He J, et al. Osimertinib in resected EGFR-mutated non-small-cell lung cancer. N Engl J Med. 2020;383:1711–1723. doi: 10.1056/NEJMoa2027071. [DOI] [PubMed] [Google Scholar]
- 9.Cho BC, Han JY, Kim SW, et al. A phase 1/2 study of lazertinib 240 mg in patients with advanced EGFR T790M-positive NSCLC after previous EGFR tyrosine kinase inhibitors. J Thorac Oncol. 2022;17:558–567. doi: 10.1016/j.jtho.2021.11.025. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Q, Wu L, Hu P, et al. A novel third-generation EGFR tyrosine kinase inhibitor abivertinib for EGFR T790M-mutant non-small cell lung cancer: a multicenter phase I/II study. Clin Cancer Res. 2022;28:1127–1135. doi: 10.1158/1078-0432.CCR-21-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shi Y, Zhao Y, Yang S, et al. Safety, efficacy, and pharmacokinetics of rezivertinib (BPI-7711) in patients with advanced NSCLC with EGFR T790M mutation: a phase 1 dose-escalation and dose-expansion study. J Thorac Oncol. 2022;17:708–717. doi: 10.1016/j.jtho.2022.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Oxnard GR, Hu Y, Mileham KF, et al. Assessment of resistance mechanisms and clinical implications in patients with EGFR T790M-positive lung cancer and acquired resistance to osimertinib. JAMA Oncol. 2018;4:1527–1534. doi: 10.1001/jamaoncol.2018.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen K, Zhou F, Shen W, et al. Novel Mutations on EGFR Leu792 potentially correlate to acquired resistance to osimertinib in advanced NSCLC. J Thorac Oncol. 2017;12:e65–e68. doi: 10.1016/j.jtho.2016.12.024. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Yang N, Ou Q, et al. Investigating novel resistance mechanisms to third-generation EGFR tyrosine kinase inhibitor osimertinib in non-small cell lung cancer patients. Clin Cancer Res. 2018;24:3097–3107. doi: 10.1158/1078-0432.CCR-17-2310. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Zhang XC, Yang JJ, et al. EGFR L792H and G796R: two novel mutations mediating resistance to the third-generation EGFR tyrosine kinase inhibitor osimertinib. J Thorac Oncol. 2018;13:1415–1421. doi: 10.1016/j.jtho.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 16.Fassunke J, Müller F, Keul M, et al. Overcoming EGFR(G724S)-mediated osimertinib resistance through unique binding characteristics of second-generation EGFR inhibitors. Nat Commun. 2018;9:4655. doi: 10.1038/s41467-018-07078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–562. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castellano GM, Aisner J, Burley SK, et al. A novel acquired exon 20 EGFR M766Q mutation in lung adenocarcinoma mediates osimertinib resistance but is sensitive to neratinib and poziotinib. J Thorac Oncol. 2019;14:1982–1988. doi: 10.1016/j.jtho.2019.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-dependent and -independent resistance mechanisms to osimertinib and continuation therapy beyond progression in EGFR-mutant NSCLC. Clin Cancer Res. 2018;24:6195–6203. doi: 10.1158/1078-0432.CCR-18-1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Y, Yang X, Zhao W, Yang Y, Zhang Z. Calcium channel α2δ1 subunit is a functional marker and therapeutic target for tumor-initiating cells in non-small cell lung cancer. Cell Death Dis. 2021;12:257. doi: 10.1038/s41419-021-03522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y, Li S, Yu W, et al. N6-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020;11:836. doi: 10.1038/s41419-020-03049-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez K, Kim I, Chun B, et al. Multiplex immunofluorescence to measure dynamic changes in tumor-infiltrating lymphocytes and PD-L1 in early-stage breast cancer. Breast Cancer Res. 2021;23:2. doi: 10.1186/s13058-020-01378-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of acquired resistance to osimertinib in EGFR-mutant NSCLC and clinical validation of combined EGFR and RET inhibition with osimertinib and BLU-667 for acquired RET fusion. Cancer Discov. 2018;8:1529–1539. doi: 10.1158/2159-8290.CD-18-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jia Y, Yun CH, Park E, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.To C, Jang J, Chen T, et al. Single and dual targeting of mutant EGFR with an allosteric inhibitor. Cancer Discov. 2019;9:926–943. doi: 10.1158/2159-8290.CD-18-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scalvini L, Castelli R, La Monica S, Tiseo M, Alfieri R. Fighting tertiary mutations in EGFR-driven lung-cancers: current advances and future perspectives in medicinal chemistry. Biochem Pharmacol. 2021;190 doi: 10.1016/j.bcp.2021.114643. [DOI] [PubMed] [Google Scholar]
- 27.Yosaatmadja Y, Silva S, Dickson JM, et al. Binding mode of the breakthrough inhibitor AZD9291 to epidermal growth factor receptor revealed. J Struct Biol. 2015;192:539–544. doi: 10.1016/j.jsb.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 28.Kashima K, Kawauchi H, Tanimura H, et al. CH7233163 overcomes osimertinib-resistant EGFR-Del19/T790M/C797S mutation. Mol Cancer Ther. 2020;19:2288–2297. doi: 10.1158/1535-7163.MCT-20-0229. [DOI] [PubMed] [Google Scholar]
- 29.Sun Y, Meyers BA, Czako B, et al. Allosteric SHP2 inhibitor, IACS-13909, overcomes EGFR-dependent and EGFR-independent resistance mechanisms toward osimertinib. Cancer Res. 2020;80:4840–4853. doi: 10.1158/0008-5472.CAN-20-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yao Y, Jeyanathan M, Haddadi S, et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175:1634–1650. doi: 10.1016/j.cell.2018.09.042. .e17. [DOI] [PubMed] [Google Scholar]
- 31.Ruffell B, Coussens LM. Macrophages and therapeutic resistance in cancer. Cancer Cell. 2015;27:462–472. doi: 10.1016/j.ccell.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma S, Zhang L, Ren Y, et al. Epiregulin confers EGFR-TKI resistance via EGFR/ErbB2 heterodimer in non-small cell lung cancer. Oncogene. 2021;40:2596–2609. doi: 10.1038/s41388-021-01734-4. [DOI] [PubMed] [Google Scholar]
- 33.Maynard A, McCoach CE, Rotow JK, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020;182:1232–1251. doi: 10.1016/j.cell.2020.07.017. .e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao P, Zhang J, Wu A, et al. Biomimetic codelivery overcomes osimertinib-resistant NSCLC and brain metastasis via macrophage-mediated innate immunity. J Control Release. 2021;329:1249–1261. doi: 10.1016/j.jconrel.2020.10.052. [DOI] [PubMed] [Google Scholar]
- 35.Yin W, Zhao Y, Kang X, et al. BBB-penetrating codelivery liposomes treat brain metastasis of non-small cell lung cancer with EGFR(T790M) mutation. Theranostics. 2020;10:6122–6135. doi: 10.7150/thno.42234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Q, Zhang Y, Chen Y, Qian J, Zhang X, Yu K. A novel mTORC1/2 inhibitor (MTI-31) inhibits tumor growth, epithelial-mesenchymal transition, metastases, and improves antitumor immunity in preclinical models of lung cancer. Clin Cancer Res. 2019;25:3630–3642. doi: 10.1158/1078-0432.CCR-18-2548. [DOI] [PubMed] [Google Scholar]
- 37.Peng H, Chen B, Huang W, et al. Reprogramming tumor-associated macrophages to reverse EGFR(T790M) resistance by dual-targeting codelivery of gefitinib/vorinostat. Nano Lett. 2017;17:7684–7690. doi: 10.1021/acs.nanolett.7b03756. [DOI] [PubMed] [Google Scholar]
- 38.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan X, Li W, Pan L, et al. Lewis lung cancer cells promote SIGNR1(CD209b)-mediated macrophages polarization induced by IL-4 to facilitate immune evasion. J Cell Biochem. 2016;117:1158–1166. doi: 10.1002/jcb.25399. [DOI] [PubMed] [Google Scholar]
- 40.Zhao C, Yang L, Zhou F, et al. Feedback activation of EGFR is the main cause for STAT3 inhibition-irresponsiveness in pancreatic cancer cells. Oncogene. 2020;39:3997–4013. doi: 10.1038/s41388-020-1271-y. [DOI] [PubMed] [Google Scholar]
- 41.Chaib I, Karachaliou N, Pilotto S, et al. Co-activation of STAT3 and YES-associated protein 1 (YAP1) pathway in EGFR-mutant NSCLC. J Natl Cancer Inst. 2017;109:1–12. doi: 10.1093/jnci/djx014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fu C, Jiang L, Hao S, et al. Activation of the IL-4/STAT6 signaling pathway promotes lung cancer progression by increasing M2 myeloid cells. Front Immunol. 2019;10:2638. doi: 10.3389/fimmu.2019.02638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin Z, Ma T, Lin Y, et al. IL-6/STAT3 pathway intermediates M1/M2 macrophage polarization during the development of hepatocellular carcinoma. J Cell Biochem. 2018;119:9419–9432. doi: 10.1002/jcb.27259. [DOI] [PubMed] [Google Scholar]
- 44.Dong J, Cheng XD, Zhang WD, Qin JJ. Recent update on development of small-molecule STAT3 inhibitors for cancer therapy: from phosphorylation inhibition to protein degradation. J Med Chem. 2021;64:8884–8915. doi: 10.1021/acs.jmedchem.1c00629. [DOI] [PubMed] [Google Scholar]
- 45.Zou S, Tong Q, Liu B, Huang W, Tian Y, Fu X. Targeting STAT3 in cancer immunotherapy. Mol Cancer. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castro M, Corren J, Pavord ID, et al. Dupilumab efficacy and safety in moderate-to-severe uncontrolled asthma. N Engl J Med. 2018;378:2486–2496. doi: 10.1056/NEJMoa1804092. [DOI] [PubMed] [Google Scholar]
- 47.Simpson EL, Bieber T, Guttman-Yassky E, et al. Two phase 3 trials of dupilumab versus placebo in atopic dermatitis. N Engl J Med. 2016;375:2335–2348. doi: 10.1056/NEJMoa1610020. [DOI] [PubMed] [Google Scholar]
- 48.Bacharier LB, Maspero JF, Katelaris CH, et al. Dupilumab in children with uncontrolled moderate-to-severe asthma. N Engl J Med. 2021;385:2230–2240. doi: 10.1056/NEJMoa2106567. [DOI] [PubMed] [Google Scholar]
- 49.Rongshou Zheng SZ, Hongmei Zeng SW, Kexin Sun RC, Li Li WW, He J. Cancer incidence and mortality in China, 2016. J National Cancer Center. 2022;2:1–9. doi: 10.1016/j.jncc.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.