Abstract
Increased MAPK signaling is a hallmark of various cancers and is a central regulator of cell survival. Direct ERK1/2 inhibition is considered a promising approach to avoid ERK1/2 reactivation caused by upstream kinases BRAF, MEK1/2, and KRAS, as well as by receptor tyrosine kinase inhibitors, but the dynamics and selectivity of ERK1/2 inhibitors are much less studied compared with BRAF or MEK inhibitors. Using ERK1/2 and downstream kinase ELK1 reporter cell lines of lung cancer (H1299; NRASQ61K), colon cancer (HCT-116; KRASG13D), neuroblastoma (SH-SY5Y), and leukemia (U937), we examined the relationship between ERK inhibition and drug-induced toxicity for five ERK inhibitors: SCH772984, ravoxertinib, LY3214996, ulixertinib, and VX-11e, as well as one MEK inhibitor, PD0325901. Comparing cell viability and ERK inhibition revealed different ERK dependencies for these cell lines. We identify several drugs, such as SCH772984 and VX-11e, which induce excessive toxicity not directly related to ERK1/2 inhibition in specific cell lines. We also show that PD0325901, LY3214996, and ulixertinib are prone to ERK1/2 reactivation over time. We distinguished two types of ERK1/2 reactivation: the first could be reversed by adding a fresh dose of inhibitors, while the second persists even after additional treatments. We also showed that cells that became resistant to the MEK1/2 inhibitor PD0325901 due to ERK1/2 reactivation remained sensitive to ERK1/2 inhibitor ulixertinib. Our data indicate that correlation of ERK inhibition with drug-induced toxicity in multiple cell lines may help to find more selective and effective ERK1/2 inhibitors.
Keywords: ERK signaling, kinase inhibitors, cancer therapy, biosensors, inhibitor selectivity
Abbreviations: AUC, area under the curve; DMSO, dimethyl sulfoxide; ERK, extracellular signal-regulated kinase; ERK-KTR, ERK kinase translocation reporter; FDR, false discovery rate; RTK, receptor tyrosine kinase
Extracellular signal-regulated kinases (ERKs) are important regulatory proteins that control cell proliferation and survival. ERK1/2 activation thus plays a significant role in cancer progression (1, 2), metastasis (3, 4), and chemoresistance (5, 6). ERK1/2 are activated through the RAS/RAF/MEK cascade, which propagates signals from receptor tyrosine kinases (RTKs). Mutations in genes encoding RTKs and RAS/RAF/MEK kinases lead to aberrant ERK1/2 activation in many cancer types and together constitute the most frequent mutation group (7). Overall, 30% to 96% of all tumors display dependency on overactivation of RAS/RAF/MEK/ERK signaling axis (7).
In particular, mutations in RAS family genes occur in approximately 20% of all cancers (8, 9), while BRAF mutations occur in 7% of all cancers, primarily in melanoma, colorectal, and papillary thyroid cancers (7, 10). MEK mutations are less common and occur primarily as secondary mutations that grant the resistance to RTKs or BRAF inhibitors. Primary mutations in ERK genes are relatively rare, and secondary mutations were reported only in cervical carcinomas (11). In turn, mutations in RTK genes are usually mutually exclusive with RAS/RAF mutations, and EGFR, ERBB2, FGFR1-3, PDGFRA, KIT, and MET are among the most frequently mutated RTK genes (7).
BRAF and MEK inhibitors (vemurafenib/dabrafenib, and cobimetinib/trametinib, respectively) are now included in the standards of care for treatment of various cancers with BRAF mutations (6, 12, 13). Although being truly effective against BRAF mutated tumors, they showed limited efficacy against cancers with RAS mutations, and cancer cells can develop resistance to such inhibitors (14, 15, 16, 17). ERK reactivation is thought to be one of the key events in developed resistance and escape from the therapy for MEK/BRAF (18, 19, 20), RAS (21, 22), and RTK inhibitors (23, 24).
Thus, direct ERK inhibition might overcome resistance mechanisms associated with its reactivation and could be used in combinational therapy to improve activities of other cancer drugs (22, 23, 25). Recently several clinical-grade ERK1/2 inhibitors were developed, and their tolerability was proved in clinical studies: SCH772984 (26), ulixertinib (BVD-523) (27, 28), ravoxertinib (GDC-0994) (29), and LY3214996 (30).
Unwanted toxicity from ERK inhibitors remains one of the obstacles for their use, especially in combinational therapies. Ideally ERK inhibitors should effectively inhibit ERK and induce cell death only via ERK inhibition. Usually, selectivity of ERK targeting drugs is measured in cell-free assays by their ability to inhibit ERK activity more effectively compared with other proteins. However, such tests cannot predict how well these drugs will perform in live cells and whether they will induce unwanted toxicity. Although characteristics of other MAPK pathway inhibitors, such as MEK and BRAF inhibitors, were extensively studied in live cells (19), there are no benchmarking studies on ERK inhibitors. Here we used a live ERK reporter system to compare how existing ERK inhibitors affect cell viability and how well this corresponds with their inhibition of ERK activity in three cancer cell cultures.
Results
ERK reporter cell lines
First, we selected three cell lines: human lung adenocarcinoma H1299 cells harboring NRASQ61K mutation (31), human colon carcinoma cells HCT-116 cells with KRASG13D mutation (32), and human neuroblastoma cells SH-SY5Y cells with no mutations in the genes of RAS, RAF, MAPK, and ERK families but harboring ALKF1174L mutation (Anaplastic lymphoma kinase), which may also contribute to constant ERK activation (33). To measure ERK1/2 activity in live cells we created HCT-116 cells with ERK kinase translocation reporter (ERK-KTR) expression (34, 35). H1299 and SH-SY5Y ERK-KTR cell lines were created and described previously (23, 36). The ERK-KTR system allows measuring ERK1/2 activity in individual cells by calculating the fluorescent signal ratio in the cytoplasm to the nucleus (C/N ratio).
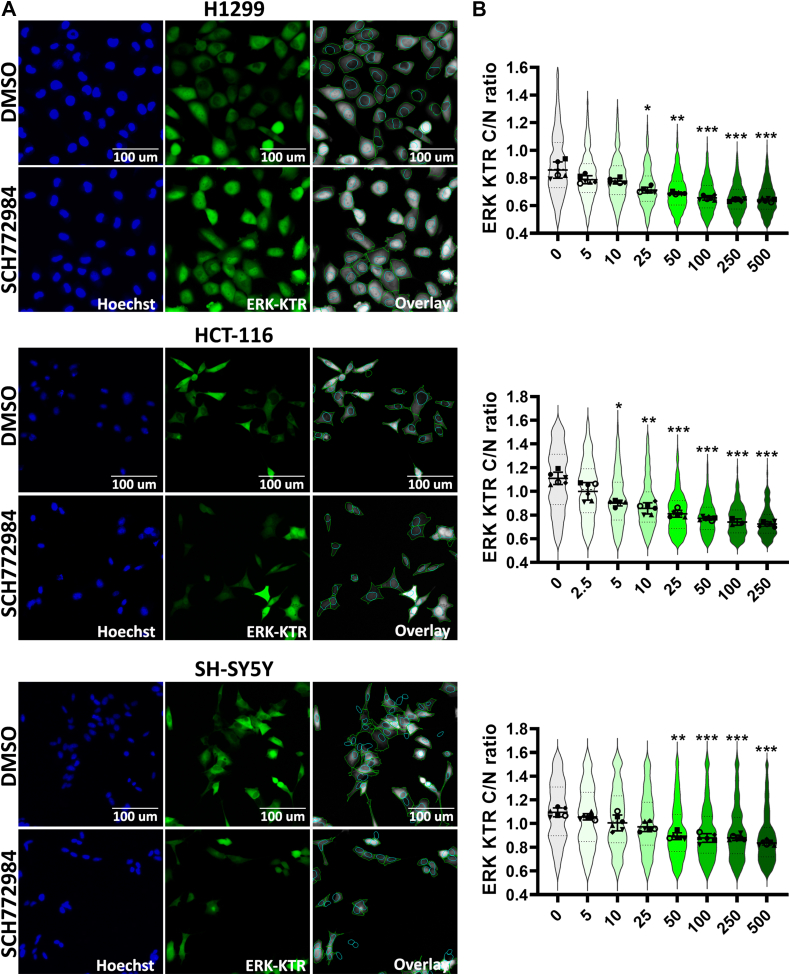
In these experiments, we used the CellProfiler software (37) for identification and segmentation of nucleus and cytoplasm (Fig. 1A), and then we tested reporter cell lines using SCH772984, a potent ERK1/2 inhibitor frequently used as a tool compound (38). SCH772984 caused ERK1/2 inhibition (calculated as C/N ratio) in a dose-dependent manner for each cell line (Fig. 1B).
Figure 1.
Cell lines expressing ERK-KTR.A, images of H1299, HCT-116, and SH-SY5Y cells expressing ERK-KTR treated by dimethyl sulfoxide (DMSO) and SCH772984 (500 nM for H1299 and SH-SY5Y, and 250 nM for HCT-116). Images were taken 24 h after drug treatment. Nuclei were stained by 500 ng/ml Hoechst-33342. Images processed by CellProfiler are shown as overlays, cytoplasm outlined by green and nuclei by blue. B, ERK-KTR median intensity ratios for cytoplasm to nucleus (C/N ratio) are shown by violin plots for cells treated with SCH772984 for 24 h. Individual points show median values for each analyzed microscopic field. For violin plots median values, SD, and 25th to 75th percentiles are shown. Median values for analyzed microscopic fields (n = 6) for each treatment were compared with control (0 concentration) using one-way ANOVA nonparametric Kruskal–Wallis test ∗p-value < 0.05, ∗∗p-value < 0.01, ∗∗∗p-value < 0.001.
ERK inhibitors display different relation between toxicity and ERK inhibition
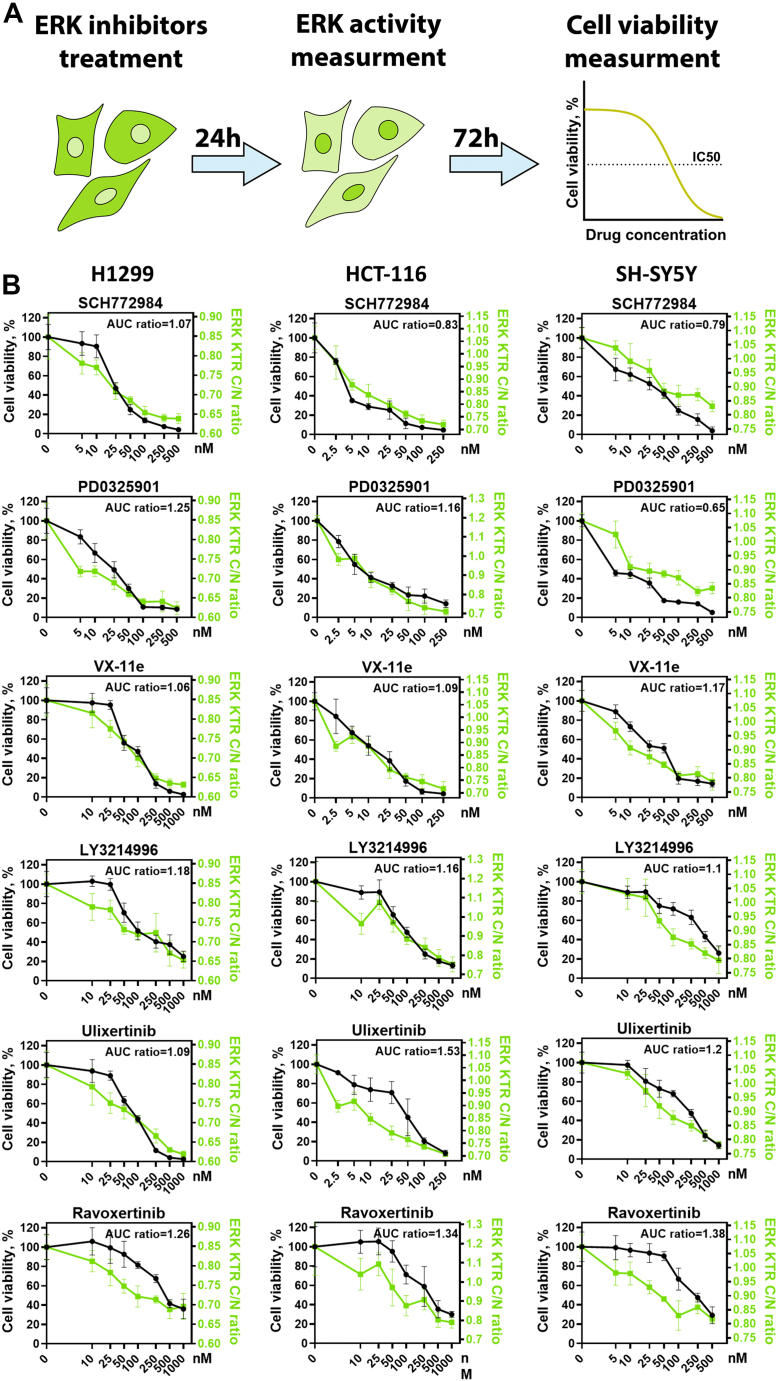
To compare the extent of ERK1/2 activity inhibition and changes in cell viability, we measured ERK1/2 activity 24 h after drug treatment (Fig. S1) when cells were still alive and had not significantly changed their morphology, which could affect our measurements. We then measured cell viability 94 h after drug treatment (Fig. 2A). For further experiments we selected five potent ERK inhibitors, with potent reported IC50 <5 nM against ERK1/2 in biochemical assays: SCH772984 (26), ravoxertinib, LY3214996 (30), ulixertinib (28), and VX-11e (39); and one MEK1/2 inhibitor PD0325901 frequently used as a tool compound. All drugs tested showed concentration-dependent ERK inhibition (Fig. S1) but had different relation between viability and ERK inhibition (Fig. 2B). For example, for SH-SY5Y cells ravoxertinib at 100 nM was less toxic than PD0325901 at 5 nM concentration, while ravoxertinib induced much stronger ERK inhibition. To quantify differences how drugs affected viability versus ERK inhibition we calculated the ratio between area under the curve (AUC) values for cell viability and ERK1/2 activity (Fig. 2B). Higher ratios between cell viability and ERK1/2 activity AUC values indicate that drug induces less toxicity during ERK inhibition. SCH772984 displayed the highest toxicity in relation to ERK inhibition, with a mean AUC ratio 0.9, while ravoxertinib and ulixertinib had the highest mean AUC ratios (1.32 and 1.27). Generally, for all cell lines there was significant correlation between drug-induced toxicity and ERK inhibition, although RAS mutant H1299 and HCT-116 (R2 = 0.92 and 0.79) cells had stronger correlation than SH-SY5Y cells (R2 = 0.56) (Fig. 3A).
Figure 2.
Measurement of ERK activity and cell viability.A, scheme for experiment design. B, cell viability (n = 3) measured 96 h after drug treatment as percentage of viable cells compared with control (dimethyl sulfoxide–treated cells). ERK activity (n = 6) was measured using the same drug concentrations 24 h after drug treatment. ERK activity and corresponding C/N ratios range for each cell line was determined earlier (Fig. 1B) and used for normalization. Mean and SD values are shown for each measurement. AUC ratio shows ratio between area under the curve for cell viability and ERK activity. ERK activity distributions for each drug are shown in Fig. S1.
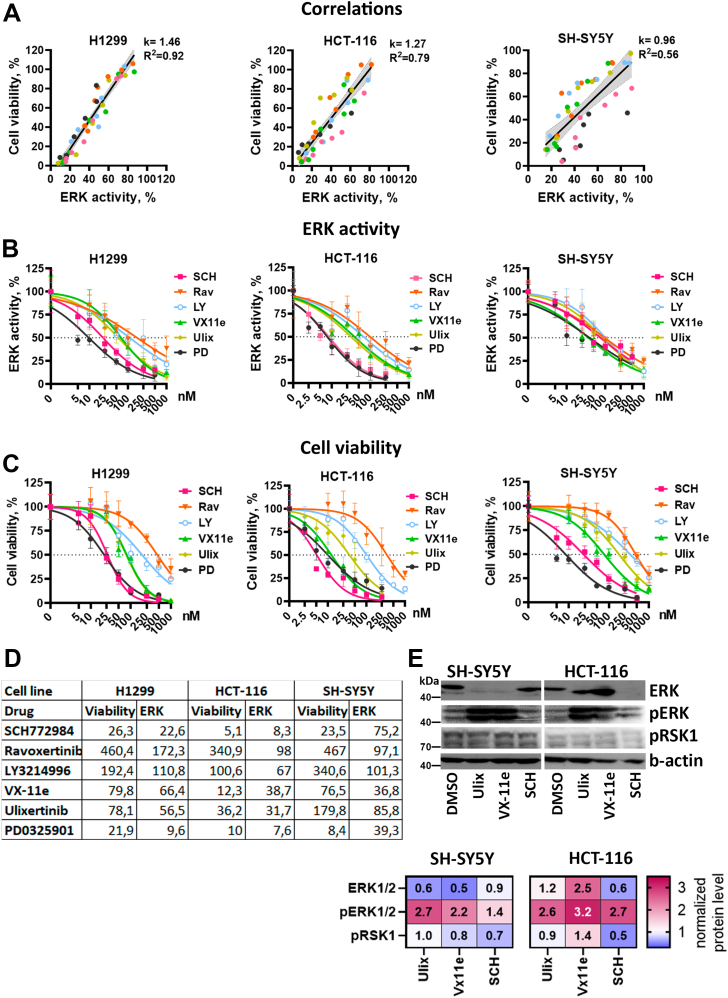
Figure 3.
ERK activity and cell viability IC50.A, correlations between cell viability and ERK median activity for each cell line. Data for all inhibitors and concentrations were combined. Linear regression coefficients (k), R-squared values are provided for each cell line. Data for each drug is marked by color, and 95% confidence intervals for linear regression are marked by gray area. B and C, nonlinear regressions used to calculate ERK inhibition (B) and cell viability (C) IC50 values for ERK inhibitors SCH772984 (SCH), ravoxertinib (Rav), LY3214996 (LY), ulixertinib (Ulix), VX-11e (VX), and MEK inhibitor PD0325901 (PD). ERK activity was normalized for IC50 calculations: median reporter C/N ratios in dimethyl sulfoxide–treated control were used for determining 100% activity and 0% was determined as minimal observed median C/N ratios in all experiments. Normalization was performed separately for each cell line. D, IC50 values provided in nM for each inhibitor. E, Western blot analysis of ERK, pERK, and pRSK1 protein levels in SH-SY5Y and HCT-116 cells treated with ERK inhibitors SCH772984 (SCH), Ulixertinib (Ulix), and VX-11e for 24 h. All inhibitors were used in 50 nM concentration for SH-SY5Y cells and 25 nM for HCT-116. The fold changes in band intensity values relative to dimethyl sulfoxide–treated cells and normalized by β-actin are provided in the heatmap.
Although there are a lot of studies describing specificity of ERK inhibitors and their dissociation parameters on purified proteins or cell lysates, there are little data on comparison of different ERK inhibitors’ effective concentrations in live cells. Thus, we also calculated IC50 values for ERK1/2 inhibition (Fig. 3B) and cell viability (Fig. 3, C and D) using reporter cell lines. These calculations also showed that different drugs can have the same ERK1/2 inhibition IC50 but exhibit different viability IC50 values for the same cell line (Fig. 3D). For example, for SH-SY5Y cells three drugs had similar ERK IC50: SCH772984 (75 nM), ulixertinib (86 nM), and ravoxertinib (97 nM), but the difference between viability IC50 was much larger: SCH772984 (24 nM), ulixertinib (180 nM), and ravoxertinib (467 nM). Similarly, for HCT-116 VX-11e inhibits ERK at slightly higher concentration than ulixertinib (39 versus 32 nM) but exerts toxicity at considerably lower concentration (12 versus 36 nM). For most drugs ERK1/2 inhibition happened at lower concentrations than viability inhibition, suggesting that SCH772984 and VX-11e potentially have excessive toxicity for SH-SY5Y and HCT-116. On the other hand, for H1299 cells drugs with lower ERK IC50 had lower viability IC50 values.
We measured levels of ERK1/2, pERK1/2, and pRSK1, which is an ERK downstream target and is used to determine ERK1/2 inhibitor efficacy (38, 40) (Fig. 3E). We selected SH-SY5Y and HCT-116 cells, and VX-11e, ulixertinib, and SCH772984, since these inhibitors showed difference in their IC50 values for these cell lines. Effects on RSK1 phosphorylation were weak, and we were not able to detect significant effects for ERK inhibitors. However, all three inhibitors induced paradoxical pERK1/2 activation, which was previously described for different ERK1/2 inhibitors (22, 41). Paradoxical pERK1/2 activation results from inhibition of active ERK1/2 proteins and prevents activation of negative feedback, which inactivates excessive ERK1/2. Thus, inhibition of ERK1/2 active sites leads to accumulation of pERK1/2, which we detected for all inhibitors in both cell lines (Fig. 3E). Unphosphorylated ERK1/2 levels were affected differently depending on cell type: VX-11e and ulixertinib reduced ERK1/2 levels in SH-SY5Y but not in HCT-116 cells, and SCH772984 reduced ERK1/2 levels only in HCT-116.
ERK inhibition shows different stability depending on cell type
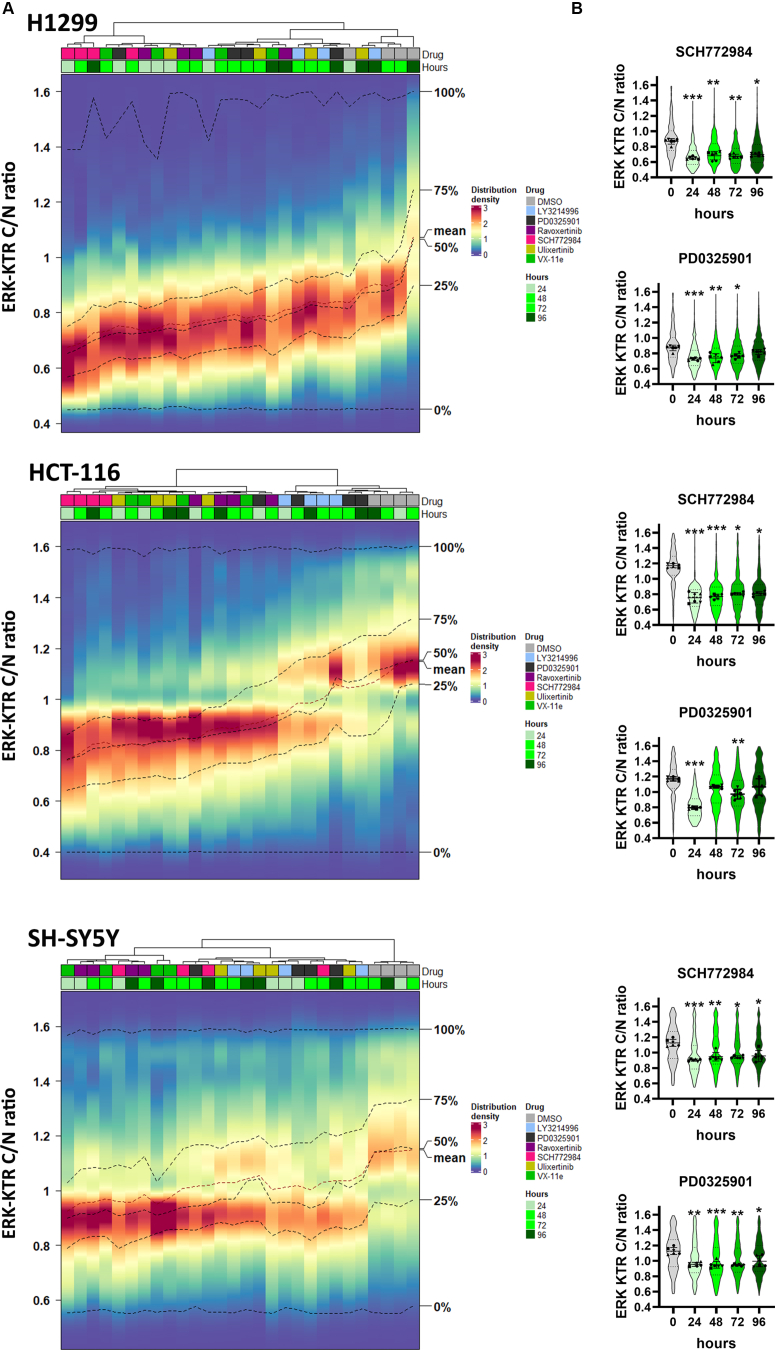
To analyze ERK1/2 inhibition stability over time we treated cell with each inhibitor for 96 h, and ERK activity was measured every 24 h (Fig. S2). A 100 nM concentration of each drug was selected for H1299 and SH-SY5Y cells, and 50 nM for HCT-116 cells, because HCT-116 cells were more sensitive to the drugs. We used hierarchical clustering to compare ERK1/2 activity changes between different drugs, which was inspired by similar approaches to cluster time-dependent reporter activities (42, 43, 44) (Fig. 4A).
Figure 4.
ERK1/2 reactivation in cells treated with ERK inhibitors.A, ERK1/2 activity distribution heatmaps for cells treated with ERK inhibitors SCH772984 (SCH), ravoxertinib (Rav), LY3214996 (LY), ulixertinib (Ulix), VX-11e (VX), and MEK inhibitor PD0325901 (PD) for 96 h. Color scale indicates percentage of cells with certain ERK1/2 activity. ERK1/2 activity distributions for each drug and time point were clustered using Ward’s method. Dimethyl sulfoxide (DMSO) treatment was used as a control mock treatment. Percentage of cells in each bin is shown by color. B, ERK KTR median intensity ratios for cytoplasm to nucleus (C/N ratio) are shown by violin plots for cells treated with SCH772984 or PD0325901. Individual points show median values for each analyzed microscopic field. For violin plots median values, SD, and 25th to 75th percentiles are shown. Median values for analyzed microscopic fields (n = 6) for each time point were compared with control (pretreatment) using one-way ANOVA nonparametric Kruskal–Wallis test ∗p-value < 0.05, ∗∗p-value < 0.01, ∗∗∗p-value < 0.001. ERK activity distributions for other drugs are shown in Fig. S2.
All ERK inhibitors and MEK inhibitor PD0325901 showed substantial ERK inhibition 24 h after treatment (Fig. 4, A and B). However, at later time points, ERK1/2 activity distributions for some drugs clustered together with dimethyl sulfoxide (DMSO)-treated cells, thus indicating ERK reactivation. Such reactivation was observed for H1299 and HCT-116 cells treated with PD0325901 and LY3214996, and for H1299 cells treated with ulixertinib. For SH-SY5Y cells, which do not have RAS mutations, all inhibitors showed stable inhibition without noticeable ERK1/2 reactivation.
ERK1/2 reactivation suggests a possible resistance mechanism, which may occur due to cell adaptation to inhibitors or drug instability/efflux. To discriminate these effects we measured how ERK1/2 activity and cell viability changed during several repeated treatments with PD0325901, and ERK inhibitors SCH772984 and ulixertinib. We treated cells with these inhibitors, and after 96 h, when first ERK reactivation occurs, we removed the growth medium and added fresh growth medium and inhibitors in the same concentrations (100 nM for SH-SY5Y and H1299, and 50 nM for HCT-116 cells), then after another 96 h repeated this process. To perform continuous measurement of ERK activity without the need for Hoechst staining for extended period of time (12 days in total) we introduced H2B-Ruby nucleus marker, which also allowed us to measure cell viability by calculating the number of nuclei in each well.
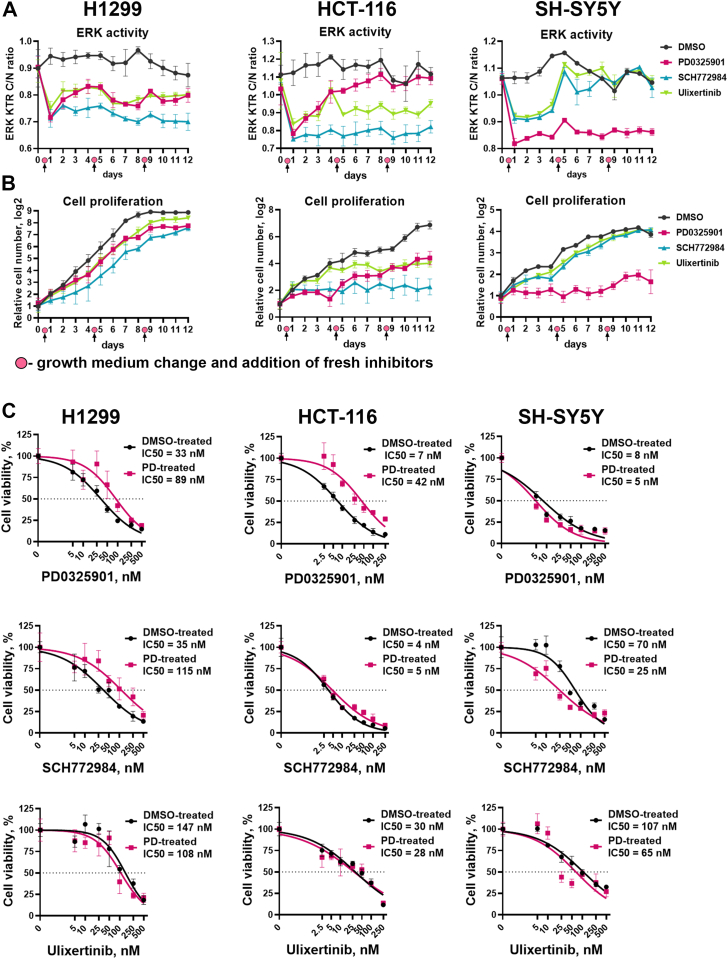
We detected two types of responses to the second treatment with MEK1/2 and ERK1/2 inhibitors. In H1299 cells treated with PD0325901 and ulixertinib, and HCT-116 treated with ulixertinib, ERK1/2 reactivation was completely reversed after the second treatment (Fig. 5A). This suggests that initial ERK1/2 reactivation occurred due to drug instability, which is likely caused by drug efflux or drug metabolism by the cells, since the effects for the same drug were cell line dependent. To the contrary, ERK1/2 reactivation in HCT-116 treated with PD0325901 continued over the time even after addition of fresh drug, suggesting some adaptation mechanism (Fig. 5A). Surprisingly, although we did not detect ERK reactivation in SH-SY5Y cells, second treatment of these cells with SCH772984 or ulixertinib failed to inhibit ERK1/2. In both cases for HCT-116 and SH-SY5Y cells continuous ERK1/2 reactivation coincided with an increase in cell proliferation rate (Fig. 5B). However, drugs that stably inhibited ERK1/2 in HCT-116 and SH-SY5Y stopped cell proliferation after the second treatment.
Figure 5.
Cell adaptation to ERK1/2 and MEK1/2 inhibitors.A, ERK1/2 activity and (B) cell proliferation in H1299, HCT-116, and SH-SY5Y cells treated with MEK1/2 and ERK1/2 inhibitors for 12 days. For H1299 and SH-SY5Y 100 nM drug concentration was used, and 50 nM was used for HCT-116 cells. Medium change and addition of fresh inhibitors in the same concentrations was performed after 4 and 8 days of treatment. C, cell viability measurement for cells treated with dimethyl sulfoxide (DMSO-treated) or PD0325901 (PD-treated) for 12 days and then treated with PD0325901, ulixertinib, and SCH772984 for 96 h.
The MEK1/2 inhibitor PD0325901 demonstrated one of the lowest IC50 values for ERK1/2 and cell viability inhibition, but is prone to rapid ERK1/2 reactivation, so we were interested in whether direct ERK1/2 inhibitors would be effective in MEK1/2 resistant cells. We compared sensitivity to PD0325901, SCH772984, and ulixertinib for cells that were treated with PD0325901 or DMSO for a total of 12 days. HCT-116 and H1299, which proliferated in the presence of PD0325901 over the time (Fig. 5B), showed decreased sensitivity to PD0325901, with IC50 values increased by 6- and 2.7-fold (Fig. 5C). Sensitivity to PD0325901 for SH-SY5Y cells, which failed to proliferate in the presence of this drug, did not sufficiently change (changes in IC50 values were <2-fold). Interestingly, treatment with PD0325901 showed different effects on sensitivity to SCH772984 for SH-SY5Y and H1299 cells; thus, H1299 cells became less sensitive to SCH772984 (3.3-fold increase in IC50) and SH-SY5Y cells became more sensitive to SCH772984 (2.8-fold decrease in IC50). Overall sensitivity to ulixertinib did not change for PD0325901-treated cells (Fig. 5C), suggesting that ulixertinib may have a potential use for MEK inhibitor-resistant cells.
ERK inhibitors have different effects on induction of leukemic cell death
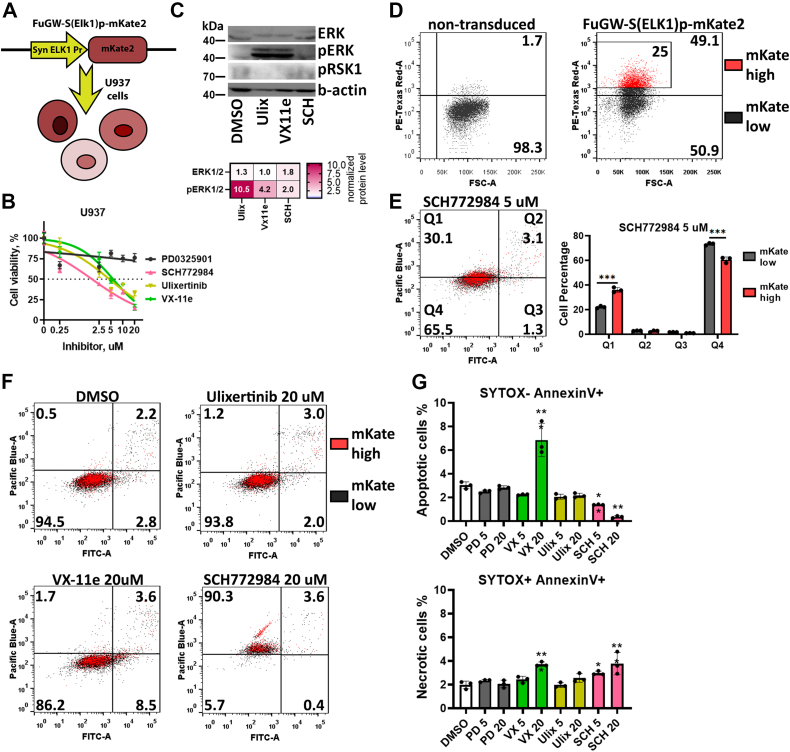
As an alternative to the ERK-KTR reporter system, we also used a synthetic promoter composed of ELK1-binding sites, which drives fluorescent protein expression. ELK1 is a transcription factor and one of the main targets of ERK1/2, and part of the ELK1 protein containing the ERK1/2-binding site is used in the KTR system described above. Approaches using promoter reporters can be beneficial to measure ERK/ELK1 activity in leukemic cells, which is hard to capture using high-throughput fluorescent imaging as in the KTR system, but could be easily measured by flow cytometry. To measure ELK1 activity in leukemic cells, we created U937-S(ELK1)p leukemic cells using the FuGW-S(Elk1)p-mKate2 system (45) (Fig. 6A). U937 cells turned out to be much less sensitive to ERK inhibitors, with IC50 values of 1.7 μM for SCH772984, 4.5 μM for ulixertinib, and 5.7 μM VX-11e (Fig. 6B). Although H1299, HCT-116, and SH-SY5Y were all sensitive to the MEK inhibitor PD0325901 (IC50 varied from 8 to 22 nM), it did not significantly affect U937 viability even at 20 μM concentration. Similarly to SH-SY5Y and HCT-116 (Fig. 3E) ERK inhibitors caused accumulation of pERK1/2; pRSK1 protein levels were undetectable in U937, however (Fig. 6C). Accumulation of pERK1/2 suggests inhibition of its activity, which is consistent with reporter intensity changes in U937-S(ELK1)p cells (Fig. S3). Overall, treatment with ERK inhibitors slightly affected reporter intensity in U937-S(ELK1)p cells (Fig. S3), thus suggesting that its potential to measure ERK activity is limited. We also tested this system using SH-SY5Y cells, and this reporter system also did not show robust ELK1 activity changes after treatment with inhibitors (Fig. S3).
Figure 6.
ELK1 activity reporter in U937 cells.A, schematic representation of reporter U937-S(ELK1)p cells. B, measurement of cell viability 96 h after treatment. Cell viability was calculated as a percentage of viable cells compared with control (dimethyl sulfoxide [DMSO]-treated cells). C, Western blot analysis of ERK, pERK, and pRSK1 protein levels in U937 cells treated with 5 μM ERK inhibitors SCH772984 (SCH), Ulixertinib (Ulix), and VX-11e for 24 h. The fold changes in band intensity values relative to DMSO-treated cells and normalized by β-actin are provided in the heatmap. D, intensity of mKate2 fluorescence in U937 cells transduced with FuGW-S(ELK1)p-mKate2 lentiviral particles. E, measurement of apoptosis in U937 cells 24 h after 5 μM SCH772984 treatment using SYTOX blue DNA stain (Pacific Blue), and annexin V (FITC). U937 cell with top 25% ELK1 reporter activity are marked by red. F, measurement of apoptosis in U937 cells 24 h after drug treatment by 5 and 20 μM of SCH772984 (SCH), ulixertinib (Ulix), VX-11e (VX), and PD0325901 (PD). G, percentage of apoptotic (SYTOX- Annexin V+) and necrotic (SYTOX+ Annexin V+) U937 cells treated with 5 or 20 μM PD0325901, SCH772984 (SCH), ulixertinib, and VX-11e. Mean values, SD, and individual data points (n = 3) are presented on bar plots. ∗p-value < 0.05, -value < 0.05, ∗∗p-value < 0.01, ∗∗∗p-value < 0.001 as calculated by two-tailed t test.
However, we found that this ELK1 reporter system could identify cells with higher ELK1 activities during drug treatment. U937-S(ELK1)p showed varying fluorescent intensity (Fig. 6D), which represented different levels of ELK1 activity as originally described for this reporter system (45). We measured apoptosis in U937-S(ELK1)p after 24 h treatment with ERK inhibitors (Fig. 6, E and F). We observed that U937 cells with the top 25% ELK1 reporter activity (mKate-high cells) were more susceptible to necrosis induced by SCH772984 than the remaining cell population (mKate-low cells) (Fig. 6E). For ulixertinib and VX-11e there was no difference between mKate-high and mKate-low cells (Fig. 6F). We also noticed that ERK inhibitors exerted different actions on the induction of apoptosis and necrosis in U937 cells: VX-11e induced apoptosis but not necrosis, SCH772984 dramatically increased the percentage of necrotic cells, and ulixertinib did not induce apoptosis or necrosis (Fig. 6G). Thus, we showed that, although these inhibitors target ERK as their known primary target, their cell death–inducing mechanisms can vary, probably depending on both mechanisms of ERK inhibition and potential off-target activities.
Both genetic and transcriptomic features of cancer cells can predict sensitivity to ERK inhibitors
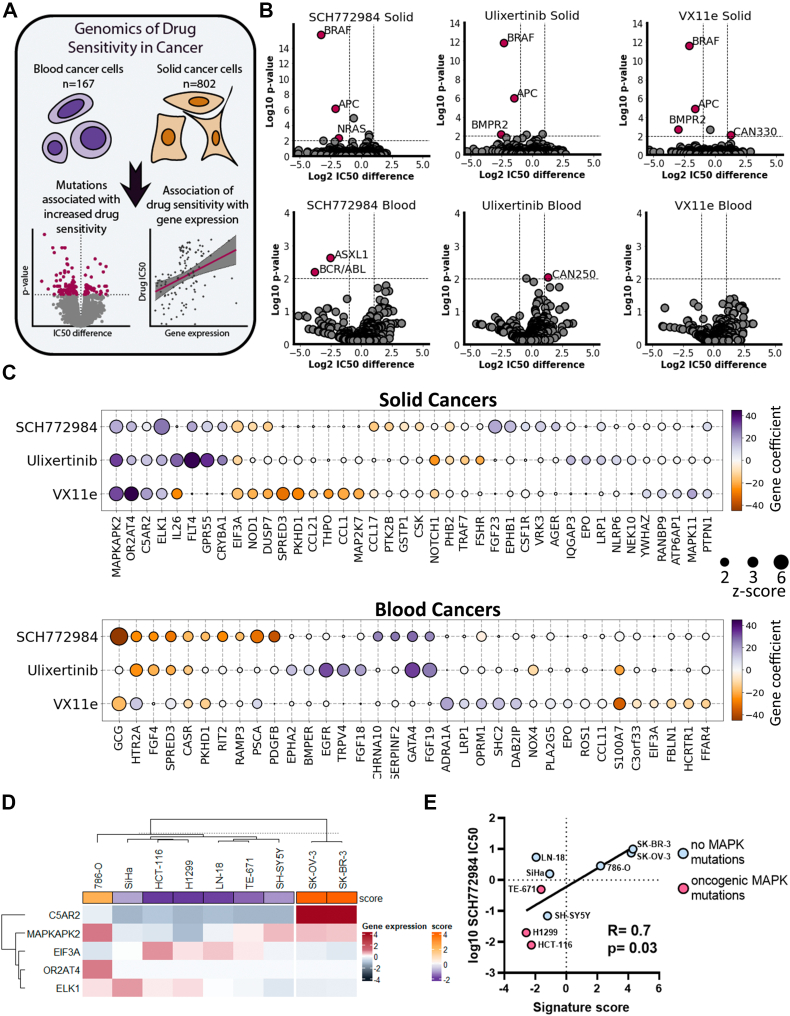
Action of ERK inhibitors seems to be cell line specific, so we investigated which factors may determine drug sensitivity of cancer cells. To investigate genetic and transcriptomic markers of sensitivity to ERK inhibition, we used data from the GDSC (Genomic of Drug Sensitivity in Cancer) database (46). Owing to large difference in sensitivity to ERK inhibitors and transcriptomic differences between solid cancers and blood cancers (leukemia, lymphoma, and myeloma), we analyzed these cancer types separately (Fig. 7A). We investigated molecular markers for three ERK inhibitors present in the database: SCH772984, ulixertinib, VX-11e. First, we calculated which mutations or genetic aberrations were associated with sensitivity to ERK inhibitors (Fig. 7B and Table S1). In line with the previous trends, BRAF mutations in solid cancer cell lines were the strongest predictors of increased sensitivity for all three ERK inhibitors (p < 0.0001). In addition, other mutations associated with increased sensitivity were found in genes APC (Adenomatous polyposis coli) for all three ERK inhibitors (p < 0.0001), and BMPR2 (bone morphogenetic protein receptor type II) for ulixertinib (p = 0.008) and VX-11e (p = 0.002). Interestingly, NRAS mutations were good biomarkers only for SCH772984 (p < 0.0001). In blood cancers, we identified associated mutations only with SCH772984: mutations of ASXL1 gene (p = 0.002) and BCR/ABL translocation (p = 0.007) were associated with increased sensitivity to the drug (Fig. 7B and Table S1). We analyzed how mutations in a particular codon, for example, NRAS codon Q61 (which combines Q61K, Q61H, Q61L, and Q61R mutations), were associated with sensitivity to ERK1/2 inhibitors (Fig. S4). Cells with mutations at NRAS-Q61 showed increased sensitivity to SCH772984 than cells with wildtype NRAS. Mutations at BRAF-V600, represented mostly by BRAFV600E, were more sensitive to all three ERK inhibitors than wildtype cells, and cells with BRAF mutations at other codons. Interestingly, mutations at any codon in KRAS gene were associated with sensitivity to SCH772984, but for ulixertinib and VX-11e only mutations other than in KRAS-G12 codon showed increased sensitivity. So overall our data suggest that sensitivity for SCH772984 may be driven by mutations at the NRAS-Q61 codon and sensitivity to ERK inhibitors in general may be driven by BRAFV600E, and KRAS mutations, except mutations at the G12 codon. Thus, presence of the NRASQ61K mutation in H1299 may explain good correlation between ERK activity inhibition and SCH772984-induced toxicity for these cells (Fig. 2B).
Figure 7.
Genetic and transcriptomic markers for sensitivity to ERK inhibitors.A, scheme for analysis design. B, volcano plots showing mutations associated with sensitivity/resistance in solid and blood cancers. Mutations with p-values <0.01 after FDR correction and IC50 change >2-fold were selected as significant. C, genes whose expression is associated with sensitivity/resistance as revealed by Elastic Net analysis. Gene coefficients correspond to what predicted impact gene expression has on IC50 values in linear regression model after elastic net regularization. Positive gene coefficients mean that gene expression increases drug IC50 and negative, decreases. Genes with z-scores >2 for at least one inhibitor are shown. D, K-means clustering of nine cancer cell lines based on expression of C5AR2, MAPKAPK2, EIF3A, OR2AT4, and ELK1 genes. Gene expression is shown as log2 difference in gene expression for a cell line relative to mean expression for all cell lines. Signature score (score) was calculated based on relative gene expression and Elastic Net regression results. E, Pearson correlation of log10 IC50 values for SCH772984 and signature scores. GDSC, Genomic of Drug Sensitivity in Cancer database.
We then analyzed gene expression patterns linked with sensitivities to ERK inhibitors. We performed linear regression for ERK inhibitors IC50 values and 478 ERK-related genes (Table S2) expression using the machine learning–based elastic net regularization (Fig. 7A). Elastic net is regularly used to identify genetic subtypes associated with drug sensitivity (47). For initial regression analysis we used 478 genes involved in ERK regulation according to Gene Ontology and REACTOME (Table S2) and performed 100 rounds of elastic net regression. Then we selected unique gene signatures for each drug consisting of 13 to 22 genes (with z-scores > 2) and performed additional 100 rounds using only these genes (Table S3). The resulting gene coefficients in the linear regression model were then used to compare gene expression impact on ERK inhibitor IC50 values.
Only 5 of 40 genes were common predictors for all three ERK inhibitors in solid cancers. Expression of MAPKAPK2, OR2AT4, C5AR2, and notably direct ERK target ELK1 positively impacted IC50 values for all three inhibitors, thus potentially promoting resistance to drugs (Fig. 7C). EIF3A expression was revealed as the only sensitivity marker for all three drugs. In blood cancers, the only common predictor for ERK inhibitors was CASR gene expression associated with drug resistance. The strongest markers for sensitivity to ERK inhibitors in blood cancers were EGFR expression for ulixertinib, and GATA4 and FGF19 expression for SCH772984 and VX-11e (Fig. 7C).
To check for possible association between sensitivity to ERK inhibitors and function of one of five common predictors (MAPKAPK2, OR2AT4, C5AR2, ELK1, and EIF3A) in solid tumors we compared ERK inhibitor IC50 values from GDSC database and cells dependency from DepMap CRISPR-Cas9 screen database. However, we detected no strong correlations between cell dependencies for these genes and sensitivity to ERK inhibitors (Fig. S5), suggesting that individual gene function is unlikely to determine sensitivity to ERK inhibitors. To validate whether the transcriptome analysis and the signature based on expression of five identified genes can predict sensitivity to ERK inhibitors we selected nine cell lines with different mutations upstream of ERK and of different tumor origin (details are provided in Table S4), including SH-SY5Y, H1299, and HCT-116, and performed transcriptome analysis to calculate this signature and measured IC50 for SCH772984, as most cells were sensitive to this inhibitor. We used k-means clustering to divide cell lines into two groups, one of which consisted of two cell lines (SK-OV-3 and SK-BR-3) with the highest signature scores (Fig. 7D). Notably these cell lines were the least sensitive to SCH772984 (IC50 values 7.5 μM and >10 μM). Overall the signature score and IC50 values for SCH772984 showed good correlation (Fig. 7E).
Discussion
In this study, we aimed at comparing the ERK1/2 inhibitors in their abilities to affect cancer cell proliferation with ERK activity in live cells. All tested ERK inhibitors are highly selective to ERK1/2 compared with other kinases as determined by various biochemical assays and have IC50 ranging from 1 to 5 nM (26, 28, 30, 39, 48). Analyzing the relation between ERK1/2 activity and drug toxicity, we found that some ERK inhibitors may exhibit excessive toxicity. Some ERK inhibitors’ excessive toxicity most likely can be explained by potential off-target effects; however, further experiments are needed to verify this hypothesis and to identify exact off-targets. For example, our results indicate that, for SH-SY5Y and HCT-116 cells, ulixertinib exhibits less excessive toxicity than SCH772984, which is supported by KinomeScan data that show less off-targets for ulixertinib (49) than for SCH772984 (38). Also, it was recently suggested that BRAF mutant cancer cells are dependent on ERK2, but not ERK1 (50), so selective targeting of ERK2 rather than both kinases may reduce the toxicity of ERK1/2 inhibitors. Ulixertinib and ravoxertinib, which showed lower toxicity than other drugs, have a higher affinity toward ERK2 than ERK1 (28, 48). Higher selectivity toward ERK2 might explain why these drugs show less toxicity, although some biochemical assay showed no selectivity of ulixertinib toward ERK2 over ERK1 (41). Although ulixertinib, VX-11e, and SCH772984 induced accumulation of phosphorylated ERK1/2, a well-described phenomenon for ERK1/2 inhibitors (22, 41), these inhibitors had different effect on unphosphorylated ERK1/2. SCH772984 had excessive toxicity on SH-SY5Y cells and did not affect ERK1/2 levels in SH-SY5Y but reduced ERK1/2 in HCT-116 cells for which ERK1/2 activity inhibition and drug-induced toxicity were consistent (Fig. 3E). Similarly, VX-11e reduced unphosphorylated ERK1/2 levels in SH-SY5Y but caused an increase in HCT-116 cells for which it had excessive toxicity. This also indicates different mechanisms of ERK1/2 inhibition by SCH772984 and VX-11e in SH-SY5Y and HCT-116 cells, which may be connected to drug excessive toxicity.
Like in earlier studies (18, 19), significant ERK1/2 inhibition caused by MEK inhibitors (in our case PD0325901) during 24 h was followed by ERK reactivation in cells with RAS mutations. Similar ERK reactivation was observed after treatment with ERK inhibitors ulixertinib and LY3214996, while other drugs resulted in stable ERK inhibition. VX-11e and SCH772984 are known to have slow dissociation time (38, 51), and ulixertinib is considered a reversible ERK1/2 inhibitor (27), which can partially explain the difference in ERK inhibition stability. We also observed two types of reactivation: the first can be reversed by addition of fresh ERK1/2 inhibitors and the second persists even after several treatments. The first type of ERK1/2 reactivation is likely caused by drug dissociation, loss of stability, or drug efflux. The second type is caused by cell adaptation to ERK inhibition and ERK1/2 reactivation by alternative mechanisms. The increase in cell proliferation for the second ERK1/2 reactivation type and increased IC50 values for MEK1/2 inhibitor PD0325901 for cells that proliferated in the presence of this drug for 12 days further supports our hypothesis. However, we found no significant changes in sensitivity to ulixertinib in cells that adapted to PD0325901. This indicates potential use for ulixertinib to kill cells that can rapidly adapt to MEK1/2 inhibitors.
Further investigation of genetic and transcriptomic signatures that determine sensitivity to ERK inhibitors is also needed since only a few known mutations, such as BRAFV600E, were revealed as associated with sensitivity to ERK1/2 inhibitors. Low number of identified mutations associated with drug sensitivity could be caused by low number of existing cell lines harboring other KRAS and NRAS mutations, which impairs the strength of statistical analysis. Either way the development of transcriptional signature that predicts sensitivity to ERK inhibitors may provide an alternative to mutation markers and prove beneficial for cancers with low incidence of MAPK mutations. Several studies show specific gene expression signatures associated with tumors’ sensitivity to MEK inhibitors (52, 53, 54). Our analysis suggests that signature based on expression of five genes (C5AR2, MAPKAPK2, EIF3A, OR2AT4, and ELK1) can determine sensitivity to ERK inhibitors at least for cancer cell lines and this justifies further studies to reveal such signatures for patient-derived cells or tumors.
Overall, we suggest that measuring the relation between ERK inhibition and drug toxicity at different concentrations, inhibition stability, and using multiple cell lines with different ERK dependency may be beneficial to identify selective ERK inhibitors. For example, our data show that ulixertinib may be prioritized over VX-11e to study ERK inhibition in HCT-116 cells, as in these cells, ulixertinib inhibits ERK at the same concentration as VX-11e, has similar inhibition stability, but causes less cytotoxicity. We believe that measuring relation between kinase activity and cell viability ultimately can provide benefits both to academic and clinical research.
Experimental procedures
Cell cultures and inhibitors
Human cancer cells and HEK293T cells were cultured in Dulbecco's modified Eagle's medium or RPMI-1640 medium (Gibco), at 37 °C and 5% CO2. The medium was supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, and 1 mM sodium pyruvate. Details for each cell line are provided in Table S4. None of the used cell lines is listed in the list of commonly misidentified cell lines maintained by the International Cell Line Authentication Committee. All cell lines were a gift from Heinrich-Pette Institute for Experimental Virology, Hamburg, Germany, and were routinely checked for mycoplasma contamination. SCH772984, ravoxertinib, LY3214996, ulixertinib, and VX-11e were purchased from Selleckchem, and PD0325901 was purchased from Sigma-Aldrich. All inhibitors were dissolved in DMSO, and initial stocks were stored at −80 °C. During the experiments working stocks were prepared, so the final concentration of DMSO in growth medium for all experiments would not exceed 0.2%. Working stocks were stored at −20 °C, and each stock was defrosted no more than five times. All stocks concentrations for inhibitors and catalog numbers are provided in Table S4.
Lentiviral pseudotyped particles production and titration
pLentiCMV Puro DEST ERKKTRClover (Addgene #59150) and pLentiPGK Hygro DEST H2B-mRuby2 (Addgene #90236) were obtained from Markus Covert lab (Addgene plasmid #59150) (34). The stocks containing VSV-G pseudotyped lentiviral particles were generated by cotransfection of HEK293T with LeGO-C or pLentiCMV Puro DEST ERKKTRClover, and packaging plasmids. For the creation of expressing ERK KTR, cells were transduced with ERKKTRClover lentiviral particles to achieve ∼30 to 50% transduction rate and then transduced cells were selected with medium supplemented with 1 μg/ml puromycin (Sigma). H2B-mRuby2 ERK-KTR-expressing cells were created after lentiviral transduction, selection on 0.5 mg/ml hygromycin b, and cell sorting on BD FACS Aria (BD Biosciences).
Analysis of cell survival, drugs IC50, and apoptosis
The number of viable cells was counted in the Neubauer chamber by the trypan blue exclusion method. Approximations for IC50 calculations were performed by nonlinear regression with variable slope (four parameters) and robust fitting in GraphPad 9. Apoptosis was measured by double staining with annexin V-FITC (ThermoFisher) and SYTOX Blue (ThermoFisher). Measurements were performed on LSRFortessa flow cytometer (BD Biosciences) and analyzed with FlowJo software.
ERK KTR quantification and cell count by automated microscopy
For nuclear segmentation cells were incubated with 500 ng/ml Hoechst-33342 for 30 min before imaging or H2B-mRuby2-expressing cells were used. Cytoplasm to nucleus ratios (C/N ratio) of mClover intensity were calculated for each cell. Illumination correction, segmentation, and object intensity calculations were performed with CellProfiler (37). Median intensities of mClover fluorescence in cytoplasm and nucleus were quantified and used to calculate cytoplasm to nucleus (C/N) ratios for each cell. Each experiment was repeated at least three times; two random microscopic fields were chosen for imaging for each well. For experiments involving H2B-mRuby2-expressing cells six fixed microscopic fields were chosen for imaging for each well. Number of cells counted as number of nuclei was averaged between all fields for each well (repeat). All images were obtained by Leica DMI8 automated microscope using 10× magnification lenses. Data processing was performed in Python and GraphPad Prism 9. Violin plot for C/N ratios were made using the “superplots” concept for visualization of cell-to-cell and sample-to-sample variance (55, 56). Heatmaps were created using ComplexHeatmap R package (57).
Western blot analysis
A total of 1 × 106 cells were lysed with buffer (150 mM sodium chloride; 1.0% Triton X-100; 50 mM Tris pH 8.0); 30 μg of protein was loaded into wells of Mini-protein TGX stain free gel (Bio-Rad, 456 8094). After transfer of proteins to polyvinylidene difluoride membrane, it was incubated overnight with anti-ERK (Abcam, ab184699), anti-pERK (Abcam, ab201015), anti-pRSK (Abcam, ab62324), or anti-Actin as loading control (Abcam, ab115777) antibodies in iBIND (ThermoFisher) according to the manufacturer’s protocol using iBIND buffers. Membranes were imaged with Chemidoc (Bio-Rad), and the intensity of the bands was calculated in ImageStudioLite.
Mutation analysis and elastic net regression
For mutational analysis we used gene expression and genetic features data from Genomics of Drug Sensitivity in Cancer database (46). For analysis we used data for 802 solid cancer and 167 blood cancer cell lines with available genetic features data. For each genetic feature we compared IC50 values for drugs between mutant and wildtype cells using nonparametric Mann–Whitney test. Then we applied false discovery rate (FDR) correction to adjust for comparing multiple genetic features. Genetic features with p-values <0.01 and more than 2-fold IC50 differences were considered as significant. To analyze mutations in specific codons we used data from CCLE, which provides data for specific codon mutations, and correlated it with sensitivity to ERK1/2 inhibitors SCH7729784, ulixertinib, and VX-11e the from GDSC database, since the CCLE drug sensitivity database does not contain data for these ERK inhibitors. Overall, there were 543 to 585 solid cancer cell lines present in both databases for each drug. We compared sensitivity for cells with and without mutations in NRAS, KRAS, and BRAF genes for all solid cancer types and lung cancer cells. Owing to low number of cells with specific mutations (e.g., NRASQ21K) we combined data for mutations at particular codon, for example, NRAS codon Q61 (which combines Q61K, Q61H, Q61L, and Q61R mutations). We selected codon mutations represented by at least ten cell lines, and rarer mutations were combined together for each gene. Mann–Whitney tests and FDR correction were performed using SciPy and statsmodels Python libraries.
To identify genes whose expression might predict sensitivity of ERK inhibitors we used data from Genomic of Drug Sensitivity in Cancer (46) for 802 solid cancer and 167 blood cancer cell lines and applied linear regression with elastic net regularization to determine how IC50 values depend on genes expression:
where expi is the expression of gene i, ωi is the weight of gene i, y0 is the intercept, is the weights estimator, S is the IC50 values vector, X is the expression matrix, ω is the weights matrix, and α and l1 are the elastic net penalty parameters.
For initial regression analysis we used 478 ERK-related genes according to Gene Ontology and REACTOME (Table S2). We performed 100 independent rounds of elastic net regression each time calculating the gene coefficients in IC50 values regression model. For each gene we calculated z-scores based on that gene mean coefficient across 100 rounds and selected genes with the highest z-scores (>3). After that we performed additional 100 rounds of elastic net regression using only selected subset of genes for each inhibitor. For linear regression and elastic net regularization we used scikit-learn Python library. Penalty parameters for elastic net regularization were determined by performing 10-fold cross validation.
DepMap dependency analysis
We downloaded genetic dependencies (gene effect scores) from DepMap 21Q4 Chronos database (https://depmap.org/portal/) for MAPKAPK2, ELK1, C5AR2, and EIF3A genes and drug sensitivity (IC50 values) for SCH772984 from the GDSC database (Genomics of Drug Sensitivity in Cancer, https://www.cancerrxgene.org/). Then we calculated Pearson correlation for SCH772984 IC50 values and gene score effects for solid cancer cell lines present in both datasets.
Library preparation, sequencing, and signature score calculation
RNA was isolated for nine cell lines using TRIzol (ThermoFisher) and RNA Clean & Concentrator kit (Zymo Research) and then subjected to DNAase I treatment (ThermoFisher). In total, nine libraries were prepared from isolated total RNA using TruSeq Stranded mRNA Library Prep Kit (Illumina) based on poly(A) enrichment protocol (TruSeq Stranded mRNA Reference Guide, Illumina). RNA samples having RIN (RNA Integrity Number) >8.0 were used for library preparation. The final cDNA library sizes were approximately 260 to 270 bp. cDNA libraries were equally pooled and sequenced on a NextSeq 500 System (Illumina) to generate 75 nucleotide single-end reads.
RNA sequencing FASTQ files were processed with STAR aligner (58) in “GeneCounts” mode with the Ensembl human transcriptome annotation (Build version GRCh38 and transcript annotation GRCh38.89). Ensembl gene IDs were converted to HGNC gene symbols using Complete HGNC dataset (https://www.genenames.org/). Totally, expression levels were established for 36,596 annotated genes with the corresponding HGNC identifiers. Gene expression values were normalized using quantile normalization protocol (59) prior to further processing. Sequencing data were deposited in NCBI Sequencing Read Archive (SRA) under accession ID PRJNA799888.
Gene impact on signature score was determined by Elastic Net regression; MAPKAPK2, OR2AT4, C5AR2, and ELK1 had positive impact on signature score (contributed toward increased IC50 values for ERK inhibitors) and EIF3A had negative impact. Gene expression relative to mean expression for all cell lines was used to calculate signature score.
Statistical analysis
All the data are expressed as mean ± SD from at least three individual experiments, unless stated otherwise in the text. Statistical significances of differences observed in apoptosis and cell count experiments were determined by two-tailed t test. All other statistical calculations were performed in Python 3.7 and GraphPad Prism 9 software.
Data availability
The Python and R source codes and CellProfiler pipelines used in this study are available at GitHub: https://github.com/CancerCellBiology/ERK-inhibitors. Sequencing data were deposited in NCBI Sequencing Read Archive (SRA) under accession ID PRJNA799888.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank Boris Fehse and Kristoffer Riecken (University Medical Center Hamburg-Eppendorf, Hamburg, Germany) for providing LeGO packaging vectors and Carol Stocking and Thomas Dobner (Heinrich-Pette Institute for Experimental Virology, Hamburg, Germany) for providing cell lines. Creation and analysis of reporter cell lines and analysis of mutational and transcriptomic features were supported by grant 075-15-2019-1660 from the Ministry of Science and Higher Education of the Russian Federation. Part of this work (library preparations and whole-transcriptome sequencing) was performed using the equipment of EIMB RAS “Genome” center (http://www.eimb.ru/rus/ckp/ccu_genome_c.php).
Author contributions
T. D. L., E. R. K., P. V. S., and A. A. B. conceptualization; T. D. L. and E. R. K. methodology; T. D. L., A. V. S., M. I. S., and A. A. B. software; T. D. L. and M. I. S. formal analysis; T. D. L., E. R. K., S. R. M., K. A. I., and A. V. S. investigation; P. V. S., P. M. R., A. V. K., and A. A. B. resources; T. D. L. writing – original draft; E. R. K., A. A. B., and V. S. P. writing – review & editing; T. D. L. visualization; V. S. P. supervision; T. D. L. and V. S. P. funding acquisition.
Funding and additional information
A. A. B. and M. I. S. were supported by the Russian Science Foundation grant 18-15-00061.
Edited by Henrik Dohlman
Supporting information
References
- 1.Samatar A.A., Poulikakos P.I. Targeting RAS-ERK signalling in cancer: promises and challenges. Nat. Rev. Drug Discov. 2014;13:928–942. doi: 10.1038/nrd4281. [DOI] [PubMed] [Google Scholar]
- 2.Barbosa R., Acevedo L.A., Marmorstein R. The MEK/ERK network as a therapeutic target in human cancer. Mol. Cancer Res. 2021;19:361–374. doi: 10.1158/1541-7786.MCR-20-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nickols N.G., Nazarian R., Zhao S.G., Tan V., Uzunangelov V., Xia Z., et al. MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer Prostatic Dis. 2019;22:531–538. doi: 10.1038/s41391-019-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gagliardi M., Pitner M.K., Park J., Xie X., Saso H., Larson R.A., et al. Differential functions of ERK1 and ERK2 in lung metastasis processes in triple-negative breast cancer. Sci. Rep. 2020;10:8537. doi: 10.1038/s41598-020-65250-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu F., Yang X., Geng M., Huang M. Targeting ERK, an Achilles' Heel of the MAPK pathway, in cancer therapy. Acta Pharm. Sin. B. 2018;8:552–562. doi: 10.1016/j.apsb.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaeger R., Corcoran R.B. Targeting alterations in the RAF-MEK pathway. Cancer Discov. 2019;9:329–341. doi: 10.1158/2159-8290.CD-18-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Vega F., Mina M., Armenia J., Chatila W.K., Luna A., La K.C., et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prior I.A., Hood F.E., Hartley J.L. The frequency of Ras mutations in cancer. Cancer Res. 2020;80:2969–2974. doi: 10.1158/0008-5472.CAN-19-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobbs G.A., Der C.J., Rossman K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies H., Bignell G.R., Cox C., Stephens P., Edkins S., Clegg S., et al. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 11.Ojesina A.I., Lichtenstein L., Freeman S.S., Pedamallu C.S., Imaz-Rosshandler I., Pugh T.J., et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Planchard D., Besse B., Groen H.J.M., Souquet P.J., Quoix E., Baik C.S., et al. Dabrafenib plus trametinib in patients with previously treated BRAF(V600E)-mutant metastatic non-small cell lung cancer: an open-label, multicentre phase 2 trial. Lancet Oncol. 2016;17:984–993. doi: 10.1016/S1470-2045(16)30146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larkin J., Ascierto P.A., Dreno B., Atkinson V., Liszkay G., Maio M., et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N. Engl. J. Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 14.Callahan M.K., Rampal R., Harding J.J., Klimek V.M., Chung Y.R., Merghoub T., et al. Progression of RAS-mutant leukemia during RAF inhibitor treatment. N. Engl. J. Med. 2012;367:2316–2321. doi: 10.1056/NEJMoa1208958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abdel-Wahab O., Klimek V.M., Gaskell A.A., Viale A., Cheng D., Kim E., et al. Efficacy of intermittent combined RAF and MEK inhibition in a patient with concurrent BRAF- and NRAS-mutant malignancies. Cancer Discov. 2014;4:538–545. doi: 10.1158/2159-8290.CD-13-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lito P., Saborowski A., Yue J., Solomon M., Joseph E., Gadal S., et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haura E.B., Ricart A.D., Larson T.G., Stella P.J., Bazhenova L., Miller V.A., et al. A phase II study of PD-0325901, an oral MEK inhibitor, in previously treated patients with advanced non-small cell lung cancer. Clin. Cancer Res. 2010;16:2450–2457. doi: 10.1158/1078-0432.CCR-09-1920. [DOI] [PubMed] [Google Scholar]
- 18.Duncan J.S., Whittle M.C., Nakamura K., Abell A.N., Midland A.A., Zawistowski J.S., et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerosa L., Chidley C., Frohlich F., Sanchez G., Lim S.K., Muhlich J., et al. Receptor-driven ERK pulses reconfigure MAPK signaling and enable persistence of drug-adapted BRAF-mutant melanoma cells. Cell Syst. 2020;11:478–494.e479. doi: 10.1016/j.cels.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long G.V., Fung C., Menzies A.M., Pupo G.M., Carlino M.S., Hyman J., et al. Increased MAPK reactivation in early resistance to dabrafenib/trametinib combination therapy of BRAF-mutant metastatic melanoma. Nat. Commun. 2014;5:5694. doi: 10.1038/ncomms6694. [DOI] [PubMed] [Google Scholar]
- 21.Tanaka N., Lin J.J., Li C., Ryan M.B., Zhang J., Kiedrowski L.A., et al. Clinical acquired resistance to KRAS(G12C) inhibition through a novel KRAS switch-II pocket mutation and polyclonal alterations converging on RAS-MAPK reactivation. Cancer Discov. 2021;11:1913–1922. doi: 10.1158/2159-8290.CD-21-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Diehl J.N., Klomp J.E., Snare K.R., Hibshman P.S., Blake D.R., Kaiser Z.D., et al. The KRAS-regulated kinome identifies WEE1 and ERK coinhibition as a potential therapeutic strategy in KRAS-mutant pancreatic cancer. J. Biol. Chem. 2021;297 doi: 10.1016/j.jbc.2021.101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lebedev T., Vagapova E., Spirin P., Rubtsov P., Astashkova O., Mikheeva A., et al. Growth factor signaling predicts therapy resistance mechanisms and defines neuroblastoma subtypes. Oncogene. 2021;40:6258–6272. doi: 10.1038/s41388-021-02018-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vagapova E., Kozlov M., Lebedev T., Ivanenko K., Leonova O., Popenko V., et al. Selective inhibition of HDAC class I sensitizes leukemia and neuroblastoma cells to anticancer drugs. Biomedicines. 2021;9:1846. doi: 10.3390/biomedicines9121846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spirin P., Lebedev T., Orlova N., Morozov A., Poymenova N., Dmitriev S.E., et al. Synergistic suppression of t(8;21)-positive leukemia cell growth by combining oridonin and MAPK1/ERK2 inhibitors. Oncotarget. 2017;8:56991–57002. doi: 10.18632/oncotarget.18503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris E.J., Jha S., Restaino C.R., Dayananth P., Zhu H., Cooper A., et al. Discovery of a novel ERK inhibitor with activity in models of acquired resistance to BRAF and MEK inhibitors. Cancer Discov. 2013;3:742–750. doi: 10.1158/2159-8290.CD-13-0070. [DOI] [PubMed] [Google Scholar]
- 27.Sullivan R.J., Infante J.R., Janku F., Wong D.J.L., Sosman J.A., Keedy V., et al. First-in-class ERK1/2 inhibitor ulixertinib (BVD-523) in patients with MAPK mutant advanced solid tumors: results of a phase I dose-escalation and expansion study. Cancer Discov. 2018;8:184–195. doi: 10.1158/2159-8290.CD-17-1119. [DOI] [PubMed] [Google Scholar]
- 28.Germann U.A., Furey B.F., Markland W., Hoover R.R., Aronov A.M., Roix J.J., et al. Targeting the MAPK signaling pathway in cancer: promising preclinical activity with the novel selective ERK1/2 inhibitor BVD-523 (ulixertinib) Mol. Cancer Ther. 2017;16:2351–2363. doi: 10.1158/1535-7163.MCT-17-0456. [DOI] [PubMed] [Google Scholar]
- 29.Varga A., Soria J.C., Hollebecque A., LoRusso P., Bendell J., Huang S.A., et al. A first-in-human phase I study to evaluate the ERK1/2 inhibitor GDC-0994 in patients with advanced solid tumors. Clin. Cancer Res. 2020;26:1229–1236. doi: 10.1158/1078-0432.CCR-19-2574. [DOI] [PubMed] [Google Scholar]
- 30.Bhagwat S.V., McMillen W.T., Cai S., Zhao B., Whitesell M., Shen W., et al. ERK inhibitor LY3214996 targets ERK pathway-driven cancers: a therapeutic approach toward precision medicine. Mol. Cancer Ther. 2020;19:325–336. doi: 10.1158/1535-7163.MCT-19-0183. [DOI] [PubMed] [Google Scholar]
- 31.Ohashi K., Sequist L.V., Arcila M.E., Lovly C.M., Chen X., Rudin C.M., et al. Characteristics of lung cancers harboring NRAS mutations. Clin. Cancer Res. 2013;19:2584–2591. doi: 10.1158/1078-0432.CCR-12-3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alves S., Castro L., Fernandes M.S., Francisco R., Castro P., Priault M., et al. Colorectal cancer-related mutant KRAS alleles function as positive regulators of autophagy. Oncotarget. 2015;6:30787–30802. doi: 10.18632/oncotarget.5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.George R.E., Sanda T., Hanna M., Frohling S., Luther W., 2nd, Zhang J., et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature. 2008;455:975–978. doi: 10.1038/nature07397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Regot S., Hughey J.J., Bajar B.T., Carrasco S., Covert M.W. High-sensitivity measurements of multiple kinase activities in live single cells. Cell. 2014;157:1724–1734. doi: 10.1016/j.cell.2014.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aikin T.J., Peterson A.F., Pokrass M.J., Clark H.R., Regot S. MAPK activity dynamics regulate non-cell autonomous effects of oncogene expression. Elife. 2020;9 doi: 10.7554/eLife.60541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vagapova E.R., Lebedev T.D., Prassolov V.S. Viral fibrotic scoring and drug screen based on MAPK activity uncovers EGFR as a key regulator of COVID-19 fibrosis. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-90701-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamentsky L., Jones T.R., Fraser A., Bray M.A., Logan D.J., Madden K.L., et al. Improved structure, function and compatibility for CellProfiler: modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chaikuad A., Tacconi E.M., Zimmer J., Liang Y., Gray N.S., Tarsounas M., et al. A unique inhibitor binding site in ERK1/2 is associated with slow binding kinetics. Nat. Chem. Biol. 2014;10:853–860. doi: 10.1038/nchembio.1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aronov A.M., Tang Q., Martinez-Botella G., Bemis G.W., Cao J., Chen G., et al. Structure-guided design of potent and selective pyrimidylpyrrole inhibitors of extracellular signal-regulated kinase (ERK) using conformational control. J. Med. Chem. 2009;52:6362–6368. doi: 10.1021/jm900630q. [DOI] [PubMed] [Google Scholar]
- 40.Boston S.R., Deshmukh R., Strome S., Priyakumar U.D., MacKerell A.D., Jr., Shapiro P. Characterization of ERK docking domain inhibitors that induce apoptosis by targeting Rsk-1 and caspase-9. BMC Cancer. 2011;11:7. doi: 10.1186/1471-2407-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin M., Franks C.E., Hsu K.L. Isoform-selective activity-based profiling of ERK signaling. Chem. Sci. 2018;9:2419–2431. doi: 10.1039/c8sc00043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goedhart J. PlotTwist: a web app for plotting and annotating continuous data. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blum Y., Mikelson J., Dobrzynski M., Ryu H., Jacques M.A., Jeon N.L., et al. Temporal perturbation of ERK dynamics reveals network architecture of FGF2/MAPK signaling. Mol. Syst. Biol. 2019;15 doi: 10.15252/msb.20198947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuchenov D., Laketa V., Stein F., Salopiata F., Klingmuller U., Schultz C. High-content imaging platform for profiling intracellular signaling network activity in living cells. Cell Chem. Biol. 2016;23:1550–1559. doi: 10.1016/j.chembiol.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissim L., Wu M.R., Pery E., Binder-Nissim A., Suzuki H.I., Stupp D., et al. Synthetic RNA-based immunomodulatory gene circuits for cancer immunotherapy. Cell. 2017;171:1138–1150.e15. doi: 10.1016/j.cell.2017.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jang I.S., Neto E.C., Guinney J., Friend S.H., Margolin A.A. In: Biocomputing 2014, Proceedings of the Pacific Symposium. Altman R.B., Dunker A.K., Hunter L., Murray T.A., Klein T.E., Ritchie M.D., editors. Kohala Coast; HI, USA: 2014. Systematic assessment of analytical methods for drug sensitivity prediction from cancer cell line data; pp. 3–7. [PMC free article] [PubMed] [Google Scholar]
- 48.Lebraud H., Surova O., Courtin A., O'Reilly M., Valenzano C.R., Nordlund P., et al. Quantitation of ERK1/2 inhibitor cellular target occupancies with a reversible slow off-rate probe. Chem. Sci. 2018;9:8608–8618. doi: 10.1039/c8sc02754d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li B.T., Janku F., Patel M.R., Sullivan R.J., Flaherty K., Buchbinder E.I., et al. First-in-class oral ERK1/2 inhibitor Ulixertinib (BVD-523) in patients with advanced solid tumors: final results of a phase I dose escalation and expansion study. J. Clin. Oncol. 2017;35:2508. [Google Scholar]
- 50.Crowe M.S., Zavorotinskaya T., Voliva C.F., Shirley M.D., Wang Y., Ruddy D.A., et al. RAF-mutant melanomas differentially depend on ERK2 over ERK1 to support aberrant MAPK pathway activation and cell proliferation. Mol. Cancer Res. 2021;19:1063–1075. doi: 10.1158/1541-7786.MCR-20-1022. [DOI] [PubMed] [Google Scholar]
- 51.Rudolph J., Xiao Y., Pardi A., Ahn N.G. Slow inhibition and conformation selective properties of extracellular signal-regulated kinase 1 and 2 inhibitors. Biochemistry. 2015;54:22–31. doi: 10.1021/bi501101v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wagle M.C., Kirouac D., Klijn C., Liu B., Mahajan S., Junttila M., et al. A transcriptional MAPK Pathway Activity Score (MPAS) is a clinically relevant biomarker in multiple cancer types. NPJ Precis. Oncol. 2018;2:7. doi: 10.1038/s41698-018-0051-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eleveld T.F., Schild L., Koster J., Zwijnenburg D.A., Alles L.K., Ebus M.E., et al. RAS-MAPK pathway-driven tumor progression is associated with loss of CIC and other genomic aberrations in neuroblastoma. Cancer Res. 2018;78:6297–6307. doi: 10.1158/0008-5472.CAN-18-1045. [DOI] [PubMed] [Google Scholar]
- 54.Dry J.R., Pavey S., Pratilas C.A., Harbron C., Runswick S., Hodgson D., et al. Transcriptional pathway signatures predict MEK addiction and response to selumetinib (AZD6244) Cancer Res. 2010;70:2264–2273. doi: 10.1158/0008-5472.CAN-09-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lord S.J., Velle K.B., Mullins R.D., Fritz-Laylin L.K. SuperPlots: communicating reproducibility and variability in cell biology. J. Cell Biol. 2020;219 doi: 10.1083/jcb.202001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goedhart J. SuperPlotsOfData - a web app for the transparent display and quantitative comparison of continuous data from different conditions. Mol. Biol. Cell. 2021;32:470–474. doi: 10.1091/mbc.E20-09-0583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 58.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bolstad B.M., Irizarry R.A., Astrand M., Speed T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Python and R source codes and CellProfiler pipelines used in this study are available at GitHub: https://github.com/CancerCellBiology/ERK-inhibitors. Sequencing data were deposited in NCBI Sequencing Read Archive (SRA) under accession ID PRJNA799888.