Abstract
Ovarian cancer (OC) is the most lethal gynecological cancer. OC cells have high proliferative capacity, are invasive, resist apoptosis, and tumors often display rearrangement of extracellular matrix (ECM) components, contributing to accelerated tumor progression. The multifunctional protein tissue transglutaminase (TG2) is known to be secreted in the tumor microenvironment, where it interacts with fibronectin (FN) and the cell surface receptor integrin β1. However, the mechanistic role of TG2 in cancer cell proliferation is unknown. Here, we demonstrate that TG2 directly interacts with and facilitates the phosphorylation and activation of the integrin effector protein integrin-linked kinase (ILK) at Ser246. We show that TG2 and p-Ser246-ILK form a complex that is detectable in patient-derived OC primary cells grown on FN-coated slides. In addition, we show that coexpression of TGM2 and ILK correlates with poor clinical outcome. Mechanistically, we demonstrate that TG2-mediated ILK activation causes phosphorylation of glycogen synthase kinase-3α/β, allowing β-catenin nuclear translocation and transcriptional activity. Furthermore, inhibition of TG2 and ILK using small molecules, neutralizing antibodies, or shRNA-mediated knockdown blocks cell adhesion to the FN matrix, as well as the Wnt receptor response to the Wnt-3A ligand, and ultimately, cell adhesion, growth, and migration. In conclusion, we demonstrate that TG2 directly interacts with and activates ILK in OC cells and tumors and define a new mechanism that links ECM cues with β-catenin signaling in OC. These results suggest a central role of TG2–FN–integrin clusters in ECM rearrangement and indicate that downstream effector ILK may represent a potential new therapeutic target in OC.
Keywords: tissue transglutaminase, integrin-linked kinase, fibronectin, integrin β1, β-catenin, Wnt signaling, ovarian cancer
Abbreviations: Ab, antibody; co-IP, coimmunoprecipitation; cpd-22, compound 22; DMEM, Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; ECM, extracellular matrix; EV, empty vector; FA, focal adhesion; FAK, focal adhesion kinase; FBS, fetal bovine serum; FN, fibronectin; Fzd7, Frizzled 7; GSK-3α/β, glycogen synthase kinase-3α/β; HGSOC, high-grade serous ovarian cancer; HR, hazard ratio; IF, immunofluorescence; IgG, immunoglobulin G; ILK, integrin-linked kinase; KD, knockdown; LEF1, lymphoid enhancer–binding factor 1; OC, ovarian cancer; p-ILKSer246, phosho-ILKSer246; PLA, proximity ligation assay; TCF, T-cell factor; TCGA, The Cancer Genome Atlas; TG2, tissue transglutaminase; WB, Western blot
Ovarian cancer (OC) is the fifth leading cancer among women in the United States and the first leading cause of cancer-related deaths from gynecologic malignancies (1). The 5-year survival rate after initial diagnosis is about 49%, whereas the 10-year survival drops to 35% (2). Despite efforts aimed at identifying an effective screening method for this neoplasm, 70% of patients reach the diagnosis at an advanced stage (III–IV FIGO [International Federation of Gynecology and Obstetrics] stage) with disseminated metastases (2). The presence of widespread metastases in the peritoneal cavity represents a major cause of poor clinical outcome. Hence, better understanding of the molecular mechanisms involved in OC progression could lead to more effective therapeutic strategies.
Adhesion of cancer cells to the basement membrane represents a critical step during tumor progression and is favored by the compositional and organizational remodeling of the extracellular matrix (ECM) in the surrounding stroma (3). Cancer and mesothelial cells secrete collagen, hyaluronan, and fibronectin (FN) (4, 5, 6) that serve as physical scaffolds for the interaction with specific adhesion molecules, such as integrins, selectins, and proteoglycans (7, 8). FN has been identified as major glycoprotein secreted in OC cells and in the submesothelial basement membrane (5, 9). Its aberrant expression has been correlated with tumor stage and poor prognosis in ovarian tumors (5, 10). Mechanistically, the interaction between FN matrices and the β1 integrins followed by the recruitment of the adaptor protein integrin-linked kinase (ILK) serves as a docking site for downstream intracellular molecules (11). The “outside–in” transduction cascade integrates mechanical forces with activation of mitogenic growth factor receptors and other signaling pathways (12), ultimately leading to actin cytoskeletal assembly and modulation of several cancer cell–associated processes, including survival, proliferation, and migration (13).
ILK is a serine/threonine protein kinase with three major domains that bind to a broad range of adaptor and signaling molecules, such as the cytoplasmic domains of integrin β1, β3, or β5 subunits (14), PINCH (particularly interesting new cysteine–histidine rich protein), parvin (15), PI3K (16), and p21-activated kinase 1 (17). These interactions are crucial for ILK activation through phosphorylation at residues Ser246, Thr173, Thr181, Ser259, and Ser343 (16, 17), its localization to focal adhesion (FA) complexes, and regulation of its response to growth factors and secreted Wnt glycoproteins (18, 19). As a main mediator of “outside–in” transduction signals, ILK directly modulates several oncogenic processes, including cell adhesion, growth, and migration (18, 20). ILK overexpression and its phosphorylation at Ser246 were correlated with OC progression (21, 22). ILK inhibition blocked tumorigenicity by suppressing phosphorylation of protein kinase B (AKT) at Ser473, glycogen synthase kinase-3α/β (GSK-3α/β) at Ser21/9, and β-catenin–T-cell factor/lymphoid enhancer–binding factor 1 (TCF/LEF1) transcriptional activity in cancer cells (19, 23, 24). These data point toward the direct involvement of integrin–ILK axis as a modulator of the crosstalk between ECM rearrangement and the canonical Wnt signaling activation in a cell-type and cellular context–dependent role. However, the connection between integrins and β-catenin remains not well delineated.
Tissue transglutaminase (TG2) is a multifunctional protein that catalyzes Ca2+-dependent protein crosslinking and hydrolyzes GTP (25). The distinct TG2 functions are finely regulated by Ca2+ and GTP levels; the crosslinking activity is allosterically activated by high Ca2+ concentrations, whereas high intracellular GTP levels inhibit TGase activity (25). TG2 has four distinct domains: an N-terminal with FN-binding site; a catalytic core; a GTP-binding site; and the C-terminal region that can recruit and activate phospholipase C (26). The N-terminal domain of TG2 binds directly to the 42-kDa gelatin-binding domain of FN with high affinity (Kd ∼8–10 nM) (27), facilitating the formation of a complex with β1 integrin, which is essential to stabilizing the ECM and cell adhesion to the basement membrane (28). This mechanism, linked to the secretion of TG2 in the extracellular compartment (29), allows the protein to interact with key ECM components expressed in OC cells and in turn to regulate tumorigenesis (30, 31, 32). TG2 is overexpressed in ovarian tumors compared with normal ovarian epithelium and promotes enhanced peritoneal dissemination (32). Previously, we reported that by promoting integrin-mediated cell adhesion to FN, TG2 regulated β-catenin activation through a c-Src–dependent mechanism, promoting OC cell proliferation (33). Other studies showed a direct interaction between extracellular TG2 and the low-density lipoprotein receptor–related proteins 5 and 6 (LRP5/6), activating β-catenin in osteoblasts (34, 35). We also demonstrated that formation of TG2/Frizzled 7 (Fzd7) clusters promoted a cancer stem cell phenotype through the activation of β-catenin signaling (30).
Here, we tested the hypothesis that TG2-mediated FN rearrangement, and the subsequent activation of the integrin–ILK signaling mechanistically stabilizes β-catenin. We show that TG2 directly interacts with ILK and promotes its phosphorylation at Ser246. TG2 and phosho-ILKSer246 (p-ILKSer246) form a complex that plays a key role in the stabilization and nuclear translocation of β-catenin. Our results uncover a mechanistic link between TG2, ILK, and β-catenin, connecting the interface between cancer cells and ECM with Wnt-regulated cell survival and proliferation.
Results
TG2 expression activates ILK in OC cells
First, we determined whether TG2-mediated FN–integrin cluster formation altered ILK phosphorylation in OC cells by using an antibody (Ab) that specifically detects p-ILK at Ser246. SKOV3 and OVCAR-5 cells were transduced with a viral vector carrying scrambled shRNA sequence (control; sh-Ctr) or shRNA against TG2 at different targeting sites (sh-TG2) (clones #1, #2, and #3) and plated on plastic or FN-coated plates for 2 h. Western blot (WB) analyses confirmed a significant decrease in TG2 expression levels in both sh-TG2 SKOV3 and OVCAR-5 cells in all three shRNA clones (Fig. S1, A and B). The experiments were conducted using the most efficient shTG2 clone #1 in both cell lines (Figs. 1A and S2A; p < 0.0001) and revealed that p-ILKSer246 levels decreased by approximately fourfold in sh-TG2 SKOV3 and OVCAR-5 cells compared with the sh-Ctr cells (Figs. 1A and S2A; p < 0.001, p < 0.0001, respectively) grown on FN. Consistently, all the sh-TG2 clones showed a decrease in p-ILKSer246 levels (Fig. S1, A and B). Of interest, OC cells grown on plastic displayed weak basal levels of p-ILKSer246, and no significant differences were observed in TG2-expressing OC cells compared with sh-TG2 cells (Fig. S3, A and E). In addition, no significant changes were observed in total ILK expression in sh-TG2 compared with sh-Ctr OC cells grown on FN or plastic (Figs. 1A, S1, A and B, S2A and S3, A and E).
Figure 1.
TG2 expression activates ILK in OC cells.A, WB for p-ILKSer246, ILK, TG2, and GAPDH in SKOV3 cells stably transduced with scrambled shRNA (sh-Ctr) or TG2-targeting shRNA (sh-TG2) and plated on FN-coated plates for 2 h. Densitometry quantifies TG2 expression levels and p-ILKSer246/ILK ratio (N = 3; ∗∗∗p < 0.01, ∗∗∗∗p < 0.0001). B, IF staining for TG2 (Alexa Fluor 488, green) and p-ILKSer246 (Alexa Fluor 568, red) in SKOV3 cells sh-Ctr or sh-TG2 KD and plated on FN-coated plates for 2 h. Protein colocalization is identified by yellow spectra on merged images. C, quantification of Alexa Fluor 488 (green) and Alexa Fluor 568 (red) proteins was calculated by using Metamorph software (N = 3; ∗∗∗∗p < 0.0001). D, quantification of colocalized proteins was calculated by volume area of green over red spectra in sh-Ctr (N = 3; Pearson’s rank correlation = 0.42) versus sh-TG2 SKOV3 cells (N = 3; Pearson’s rank correlation = 0.006; p < 0.0001). E, TG2–p-ILKSer246 colocalization (red dots) and phalloidin (green) detected by PLA in four primary human cells isolated from deidentified malignant ascites fluid specimens and plated on plastic or FN-coated plates for 2 h. Representative images are shown (magnification, 200×). Bar represents 20 μm. F, quantification of the number of total TG2–p-ILKSer246red dots per sample in a diagram (N = 3; ∗p < 0.05, ∗∗p < 0.01). G, correlation between TGM2 and ILK mRNA expression levels (Spearman r = 0.15, p = 1.2e-3 and Pearson r = 0.15, p = 2.9e-4) in the TCGA ovarian cancer database obtained from cBioPortal. H, overall survival curves generated using the Kaplan–Meier plot for tumors expressing higher than median levels of TGM2 and of ILKversus those expressing lower than median levels of TGM2 and of ILK in HGSOC tumor microarray data of 14 datasets from seven different array platforms using OvMark (HR = 1.52 and p = 0.02221). FN, fibronectin; HGSOC, high-grade serous ovarian cancer; HR, hazard ratio; IF, immunofluorescence; ILK, integrin-linked kinase; OC, ovarian cancer; PLA, proximity ligation assay; TCGA, The Cancer Genome Atlas; TG2, tissue transglutaminase; WB, Western blot.
Quantification of immunofluorescence (IF) staining confirmed significantly increased p-ILKSer246 staining in sh-Ctr compared with sh-TG2 transduced cells (Figs. 1, B and C and S2, B and C; p < 0.0001) plated on FN. IF confocal and colocalization analysis showed that TG2 and p-ILKSer246 were significantly colocalized in both sh-Ctr SKOV3 (Rcoloc = 0. 42) and sh-Ctr OVCAR-5 cells (Rcoloc = 0.545) plated on FN compared with sh-TG2 SKOV3 (Rcoloc = 0.006, p < 0.0001) and sh-TG2 OVCAR-5 cells (Rcoloc = 0.009, p < 0.0001) (Figs. 1D and S2D). Quantification of IF signals for both sh-Ctr OC cells plated on plastic showed significantly higher p-ILKSer246 levels compared with sh-TG2 cells (Fig. S3, B, C, F and G; p < 0.0001). Confocal analysis confirmed the decrease of TG2–p-ILKSer246 active complexes in both sh-TG2 SKOV3 (Rcoloc = 0.001, p < 0.0001) and sh-TG2 OVCAR-5 cells (Rcoloc = 0.0029, p < 0.0001) plated on plastic compared with sh-Ctr SKOV3 (Rcoloc = 0.1) and sh-Ctr OVCAR-5 cells (Rcoloc = 0.086) (Fig. S3, D and H, respectively). Of note, the overall basal p-ILKSer246 levels and TG2–p-ILKSer246 clusters in OC cells plated on plastic alone were detected at lesser extent compared with their counterpart plated on FN. These data confirm that ILK activation is ECM substrate dependent and functionally regulated by TG2 in OC cells.
Next, to determine whether TG2–p-ILKSer246 complexes are detectable in human tumors, we used proximity ligation assay (PLA), a technique capable of identifying proteins localized within 40 nm distance in tissue. TG2 and p-ILKSer246 expression and colocalization were measured in primary human cells isolated from deidentified malignant ascites specimens and plated on plastic or FN-coated plates (N = 4 specimens). TG2–p-ILKSer246 complex formation was detectable at the cell membrane and in invadopodia in all human OC cells plated on FN but not in primary OC cells plated on plastic (Fig. 1, E and F; p < 0.05; p < 0.01), supporting a functional interaction between the two proteins in malignant cells. Furthermore, p-ILKSer246 formed complexes with integrin β1 and FN when primary OC cells were plated on FN compared with plastic alone (Fig. S2, E, F, I and J; p < 0.01; p < 0.001; and p < 0.0001). The results point to an important role of TG2–FN–integrin clusters activating the downstream effector p-ILKSer246 in OC cells.
In addition, analysis of 489 clinically annotated high-grade serous ovarian cancer (HGSOC) tumors in The Cancer Genome Atlas (TCGA) OC database was obtained from cBioPortal. ILK expression was correlated with TGM2 (Spearman r = 0.15, p = 1.2e-3 and Pearson r = 0.15, p = 2.9e-4; Fig. 1G), ITGB1 (Spearman r = 0.11, p = 0.0124 and Pearson r = 0.12, p = 9.8e-3; Fig. S2G) and with FN1 (Spearman r = 0.2, p = 5.2e-6 and Pearson r = 0.19, p = 1.8e-5; Fig. S2H). Furthermore, analysis of HGSOC tumor microarray data of 14 datasets from seven different array platforms using OvMark demonstrated that patients with higher than median TGM2 and ILK expression levels had an increased estimated risk of death when compared with those with lower than median TGM2 and ILK combined expression levels (Fig. 1H; hazard ratio [HR] =1.58, p = 0.002). Similar results were obtained by comparing patients with tumors harboring higher than median ITGB1 or FN1 and ILK combined expression levels (Fig. S2, H and L; HR = 1.679, p = 0.008; and HR = 1.72, p = 0.005, respectively). These data support the significance of ILK in cooperation with TG2 in human ovarian tumors affecting clinical outcomes.
TG2 forms a protein complex with ILK
We demonstrate whether TG2 and ILK form a complex, we tested for a potential interaction by performing coimmunoprecipitation (co-IP) in cell lysates from SKOV3 cells. WB with anti-TG2 and anti-ILK Abs demonstrated that the two proteins were detectable in complexes immunoprecipitated with anti-TG2 and anti-ILK Abs (Fig. 2A). To further define the interaction between TG2 and ILK, we used full-length recombinant human TG2 and ILK proteins. Co-IP with anti-TG2 and anti-ILK Abs demonstrated direct interaction between recombinant TG2 and ILK (Fig. 2B). In addition, His-tagged TG2 and FLAG-tagged ILK were immunoprecipitated with anti-His and anti-FLAG Abs from lysates of sh-TG2 SKOV3 cells stably cotransfected with pCMV3-TGM2-C-His and pCMV3-ILK-C-FLAG expression plasmids (Fig. 2C). Together, these data confirm the direct interaction between TG2 and ILK.
Figure 2.
TG2 interacts with ILK and activated ILK in OC cells.A, equal amounts of lysates of SKOV3 cells were immunoprecipitated with TG2 and ILK antibodies (Abs). WB was performed using TG2, ILK, and GAPDH Abs. B, co-IP with anti-TG2 and anti-ILK Abs and WB for TG2 and ILK using full-length recombinant TG2 and ILK. C, His-tagged TG2 and FLAG-tagged ILK were immunoprecipitated from lysates of sh-TG2 SKOV3 cells stably cotransfected with pCMV3-TGM2-C-His and pCMV3-ILK-C-FLAG expression plasmids. WB was performed using His, FLAG, and GAPDH Abs. Nonspecific IgG was used as control. D and G, IF staining for TG2 (Alexa Fluor 488, green), ILK (D), and p-ILKSer246 (G) (Alexa Fluor 568, red) in SKOV3 cells sh-TG2 KD stably cotransfected with pCMV3-TGM2-C-His and plated on FN-coated plates for 2 h. Protein colocalization is identified by yellow spectra on merged images. E and H, quantification of Alexa Fluor 568 (red) ILK (E) and p-ILKSer246 (H) in permeabilized versus nonpermeabilized cells was calculated by using Metamorph software (N = 3; ∗∗∗∗p < 0.0001). F and I, quantification of colocalized proteins was calculated by volume area of green over red spectra in permeabilized (N = 3; Pearson’s rank correlation = 0.715) (F) and (N = 3; Pearson’s rank correlation = 0.76) (I) versus nonpermeabilized sh-TG2 SKOV3 cells stably cotransfected with pCMV3-TGM2-C-His (N = 3; Pearson’s rank correlation = 0.007; p < 0.0001) (F) and (N = 3; Pearson’s rank correlation = 0.005; p < 0.0001) (I). co-IP, coimmunoprecipitation; FN, fibronectin; IF, immunofluorescence; IgG, immunoglobulin G; ILK, integrin-linked kinase; KD, knockdown; OC, ovarian cancer; TG2, tissue transglutaminase; WB, Western blot.
IF confocal analysis of sh-TG2 SKOV3 cells transfected with pCMV3-TGM2-C-His showed TG2 expression in both the cytoplasmic and extracellular compartments (Fig. 2, D and G). ILK and the active p-ILKSer246 were expressed exclusively in the cytoplasm as detected by IF confocal in permeabilized sh-TG2 SKOV3 cells transfected with pCMV3-TGM2-C-His (Fig. 2, D and G) compared with the nonpermeabilized counterpart (Fig. 2, E and H; p < 0.0001).
Confocal analysis confirmed the TG2-ILK and TG2–p-ILKSer246 complex formation in permeabilized sh-TG2 SKOV3/pCMV3-TGM2-C-His compared with nonpermeabilized sh-TG2 SKOV3/pCMV3-TGM2-C-His (Fig. 2, F and I; Rcoloc = 0.715 and Rcoloc = 0.76, respectively) compared with nonpermeabilized cells (Fig. 2, F and I; Rcoloc = 0.007 and Rcoloc = 0.005, respectively; p < 0.0001). These data demonstrated the critical dual role of TG2 promoting the interaction between FN matrices and the β1 integrins followed by the recruitment of the adaptor protein ILK in both extracellular and intracellular compartments.
TG2–FN interaction activates ILK and mediates β-catenin nuclear translocation
The Wnt/β-catenin signaling is an evolutionarily conserved pathway that determines fate decisions for cells and tissues during embryonic development (36). In the presence of Wnt ligands, β-catenin is dephosphorylated at Ser33, Ser37, and Thr41 residues (37), resulting in its nuclear translocation, where it forms transcriptionally active complexes with the members of the TCF/LEF1 DNA-binding transcription factors (38). In previous reports, we demonstrated that in the context of cell–matrix interactions, TG2 regulates β-catenin signaling (30, 33), implying a direct correlation between ECM rearrangement and the canonical Wnt pathway activation. To understand the consequences of TG2-induced ILK phosphorylation at Ser246 in cell–matrix interactions, activation of β-catenin was measured in OC cells plated on FN matrices. First, we confirmed an increase in p-ILKSer246 and observed activation of FA complexes by measuring phosphorylation of focal adhesion kinase (FAK) at Tyr576/577 in OVCAR-5/sh-Ctr and SKOV3/sh-Ctr cells plated on FN compared with sh-TG2 cells (Figs. 3A and S4A; p < 0.05, p < 0.01, p < 0.0001, respectively). ILK activation and FAK phosphorylation at Tyr576/577 occurred 30 min upon cell adhesion to FN and was detectable for up to 2 h. Increased ILK and FAK phosphorylations in sh-Ctr OC cells correlated with phosphorylation of GSK-3α/β at Ser21/9 (Figs. 3B and S4B; p < 0.05, p < 0.01, p < 0.0001, respectively). In resting cells, GSK-3α/β phosphorylates β-catenin at Ser33, Ser37, and Thr41, causing its destabilization and subsequent ubiquitin–proteasome-mediated degradation (39). Upon Wnt activation, GSK-3α/β is phosphorylated at Ser21/9, which blocks β-catenin phosphorylation at Ser33/37 and Thr41, thus stabilizing it (39). By using an Ab directed against the nonphospho (active) β-catenin at Ser33/37 and Thr41, we detected increased active β-catenin levels in sh-Ctr/OVCAR-5 and sh-Ctr/SKOV3 cells compared with sh-TG2 transduced cells (Figs. 3B and S4B; p < 0.05, p < 0.01, p < 0.0001, respectively).
Figure 3.
TG2–FN clusters activate β-catenin in OC cells.A, WB for p-FAKTyr576/577, FAK, p-ILKSer246, ILK, TG2, and GAPDH in OVCAR-5 cells stably transduced with scrambled- or TG2-targeting shRNA and plated on FN-coated plates for 30 min, 1 h, and 2 h. Densitometry quantifies p-FAKTyr576/577/FAK and p-ILKSer246/ILK ratios and TG2 expression levels (N = 3; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.01, and ∗∗∗∗p < 0.0001). B, WB for non–p-active β-catenin, p-GSKα/βSer21/9, GSKα/β, TG2, and GAPDH in OVCAR-5 cells stably transduced with scrambled- or TG2-targeting shRNA and plated on FN-coated plates for 30 min, 1 h, and 2 h. Densitometry quantifies non–p-active β-catenin expression levels and p-GSKα/βSer21/9/GSKα/β ratio (N = 3; ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗∗p < 0.0001). All antibodies (Abs) were probed on the same membrane. For consistency, the same TG2 and GAPDH representative WB images and TG2 quantification are shown in A and B. C, WB for p-FAKTyr576/577, FAK, p-ILKSer246, ILK, TG2, and GAPDH in OVCAR-5 cells stably transduced with TG2-targeting shRNA, transfected with pCMV3-TGM2-C-His, and plated on FN-coated plates for 2 h. Densitometry quantifies p-FAKTyr576/577/FAK and p-ILKSer246/ILK ratios and TG2 expression levels (N = 3; ∗∗p < 0.01, ∗∗∗p < 0.01, and ∗∗∗∗p < 0.0001). D, WB for non–p-active β-catenin, p-GSKα/βSer21/9, GSKα/β, and GAPDH in OVCAR-5 cells stably transduced with TG2-targeting shRNA, transfected with pCMV3-TGM2-C-His, and plated on FN-coated plates for 2 h. Densitometry quantifies non–p-active β-catenin expression levels and p-GSKα/βSer21/9/GSKα/β ratio (N = 3; ∗p < 0.05, ∗∗∗∗p < 0.0001). All Abs were probed on the same membrane. For consistency, the same TG2 and GAPDH representative WB images and TG2 quantification are shown in C and D. E, IF staining for TG2 (Alexa Fluor 488, green) and non–p-active β-catenin (Alexa Fluor 568, red) in OVCAR-5 cells sh-Ctr or sh-TG2 KD plated on FN for 2 h. F and G, quantification of Alexa Fluor 488 (green) and Alexa Fluor 568 (red) proteins was calculated by using Metamorph software (N = 3; ∗∗∗∗p < 0.0001). H, OVCAR-5 cells sh-Ctr or sh-TG2 KD were cotransfected with TCF/LEF1 luciferase reporter and Renilla control plasmid and plated on FN for 2 h. Luciferase signal relative to Renilla activity is expressed as fold increase (N = 6; ∗∗∗p < 0.001). I, real-time PCR for c-Myc in OVCAR-5 cells sh-Ctr or sh-TG2 KD plated on FN-coated plates for 2 h (N = 6; ∗∗∗p < 0.001). FAK, focal adhesion kinase; FN, fibronectin; GSKα/β, glycogen synthase kinase-3α/β; IF, immunofluorescence; ILK, integrin-linked kinase; KD, knockdown; LEF1, lymphoid enhancer–binding factor 1; OC, ovarian cancer; TCF, T-cell factor; TG2, tissue transglutaminase; WB, Western blot.
To further examine its functions, TG2 was stably overexpressed in sh-TG2 OVCAR-5 and SKOV3 cells. WB analysis and quantification showed that sh-TG2 OC cells transfected with pCMV3-TGM2-C-His plasmid restored TG2 expression (Figs. 3, C and D and S4, C and D; p < 0.05, p < 0.01, p < 0.001, and p < 0.0001) compared with sh-TG2 OC cells transfected with pCMV3-Empty Vector (EV)-C-His (EV-C-His). TG2 expression levels restored the phosphorylation of both FAK at Tyr576/577 and ILK at Ser246 in sh-TG2 OVCAR-5/pCMV3-TGM2-C-His and SKOV3/pCMV3-TGM2-C-His cells compared with EV-C-His/sh-TG2 transduced cells (Figs. 3C and S4C; p < 0.01, p < 0.0001, respectively). Most importantly, TG2 overexpression correlated with phosphorylation of GSK-3α/β at Ser21/9 and increased levels of active β-catenin levels in sh-TG2 OVCAR-5/pCMV3-TGM2-C-His and SKOV3/pCMV3-TGM2-C-His cells compared with EV-C-His/sh-TG2 transduced cells (Figs. 3D and S4D; p < 0.001, p < 0.0001, respectively).
IF staining and intensity quantification confirmed TG2 knockdown (KD) in sh-TG2 transduced cells as well as significant depletion of nuclear (active) β-catenin compared with sh-Ctr OC cells (Figs. 3, E–G and S4, E–G; p < 0.0001) plated on FN for 2 h. Once activated, β-catenin translocates into the nucleus, where it forms a complex with the TCF/LEF and induces transcription of target genes (40). A TCF/LEF1 reporter assay showed approximately twofold increase in luminescence in sh-Ctr/OVCAR-5 and sh-Ctr/SKOV3 cells compared with sh-TG2 cells (Figs. 3H and S4H; p < 0.0001, p < 0.05, respectively). The β-catenin target gene c-Myc mRNA expression levels were also increased approximately twofold in sh-Ctr/OVCAR-5 and sh-Ctr/SKOV3 cells compared with sh-TG2 cells (Figs. 3I and S4I; p < 0.001). The data indicate that TG2 expression is essential to translate extracellular cues induced by cell adhesion to FN into oncogenic β-catenin signaling.
TG2–FN blockade suppresses ILK activation and β-catenin signaling
TG2 acts as a scaffold protein connecting the 42-kDa gelatin-binding domain of FN with the β1 subunits of integrins, which facilitates cell adhesion to the ECM and amplifies β-catenin signals (30, 33). ILK activation has been correlated with Wnt-3A–induced β-catenin stabilization in a matrix-remodeling context (41). To test whether the ILK activation leading to β-catenin signaling is a direct consequence of TG2 interactions with the matrix, we used the inhibitory anti-TG2 blocking Ab (clone 4G3) directed against the FN-binding N-terminal domain of TG2 (amino acids 1–165) or the anti-integrin β1 blocking Ab (clone P5D2) (30, 33, 42). OVCAR-5 and SKOV3 cells plated on FN for 2 h and treated with Wnt-3A showed approximately 3-fold and 2.4-fold increase in p-FAKTyr576/577 and approximately 7.5-fold and 2-fold increase in p-ILKSer246, respectively, compared with the basal levels observed in immunoglobulin G (IgG)-treated (control) cells (Figs. 4A and S5A), whereas the inhibitory P5D2 and 4G3 Abs decreased both p-FAKTyr576/577 and p-ILKSer246 levels in Wnt-3A–treated OC cells (Figs. 4A and S5A). No significant differences were observed in total FAK and ILK levels in the Wnt-3A–treated OC cells or upon incubation with both inhibitory Abs (Figs. 4A and S5A) compared with IgG-treated control cells, suggesting that TG2 stabilizes β-catenin by functionally regulating FAK and ILK at post-transcriptional level. As expected, Wnt-3A increased the p-GSK-3α/βSer21/9 levels in both OVCAR-5 and SKOV3 cells compared with untreated OC cells (Figs. 4B and S5B), resulting in its inhibition. This led to approximately 13.8-fold and 3-fold increase of nonphospho (active) β-catenin levels in OVCAR-5 and SKOV3 cells, respectively, compared with untreated OC cells (Figs. 4B and S5B). P5D2 and 4G3 Abs decreased both p-GSK-3α/βSer21/9 and nonphospho (active) β-catenin at Ser33/37 and Thr41 levels in Wnt-3A–treated OC cells (Figs. 4A and S5A).
Figure 4.
Functional TG2 andintegrinβ1inhibition disrupts β-catenin signaling in OC cells.A, WB for p-FAKTyr576/577, FAK, p-ILKSer246, ILK, and GAPDH in OVCAR-5 cells treated with inhibitory antibodies (Abs) directed against the FN-binding domain of TG2 (clone 4G3) or integrin β1 (clone P5D2) and/or Wnt-3A and plated on FN-coated plates for 2 h. Densitometry quantifies p-FAKTyr576/577/FAK and p-ILKSer246/ILK ratios (N = 3; ∗p < 0.05; ∗∗p < 0.01). B, WB for non–p-active β-catenin, p-GSKα/βSer21/9, GSKα/β, and GAPDH in OVCAR-5 cells treated with 4G3 or P5D2 Abs and/or Wnt-3A and plated on FN-coated plates for 2 h. Densitometry quantifies non–p-active β-catenin expression levels and p-GSKα/βSer21/9/GSKα/β ratio (N = 3; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). C, IF staining for phalloidin (Alexa Fluor 488, green) and non–p-active β-catenin (Alexa Fluor 568, red) in OVCAR-5 cells treated with 4G3 or P5D2 Abs and/or Wnt-3A plated on FN for 2 h. D, quantification of Alexa Fluor 568 (red) proteins was calculated by using Metamorph software (N = 3; ∗∗∗∗p < 0.0001). E, OVCAR-5 cells were cotransfected with TCF/LEF1 luciferase reporter and Renilla control plasmid, treated with 4G3 or P5D2 Abs and/or Wnt-3A, and plated on FN for 2 h. Luciferase signal relative to Renilla activity is expressed as fold increase (N = 6; ∗∗∗∗p < 0.0001). F, real-time PCR for c-Myc in OVCAR-5 cells treated with 4G3 or P5D2 Abs and/or Wnt-3A and plated on FN-coated plates for 2 h (N = 6; ∗∗∗∗p < 0.0001). FAK, focal adhesion kinase; FN, fibronectin; GSKα/β, glycogen synthase kinase-3α/β; IF, immunofluorescence; ILK, integrin-linked kinase; LEF1, lymphoid enhancer–binding factor 1; OC, ovarian cancer; TCF, T-cell factor; TG2, tissue transglutaminase; WB, Western blot.
Next, IF microscopy and quantification detected a significantly stronger nonphospho (active) β-catenin at Ser33/37 and Thr41 nuclear signal in OVCAR-5 and SKOV3 cells treated with Wnt-3A compared with untreated control cells (Figs. 4, C and D and S5, C and D; p < 0.0001). Treatment with P5D2 and 4G3 blocked the nonphospho (active) β-catenin nuclear translocation in both OC cells (Figs. 4, C and D and S5, C and D; p < 0.0001).
P5D2 and 4G3 significantly reduced Wnt-3A-mediated β-catenin/TCF transcriptional activity as measured by TCF/LEF1 reporter assay in OVCAR5 and SKOV3 cells (Figs. 4E and S5E; p < 0.0001). In addition, P5D2 and 4G3 treatment decreased the mRNA levels of c-Myc, a key β-catenin target gene in both cell lines (Figs. 4F and S5F; p < 0.0001). Overall, 4G3 (anti-TG2) Ab treatment was more effective than P5D2 (anti-integrin) Ab at preventing FA complex formation, ILK activation, and consequent β-catenin nuclear translocation, demonstrating the key role of TG2 as a functional regulator of β-catenin activation.
ILK inhibition blocks β-catenin signaling
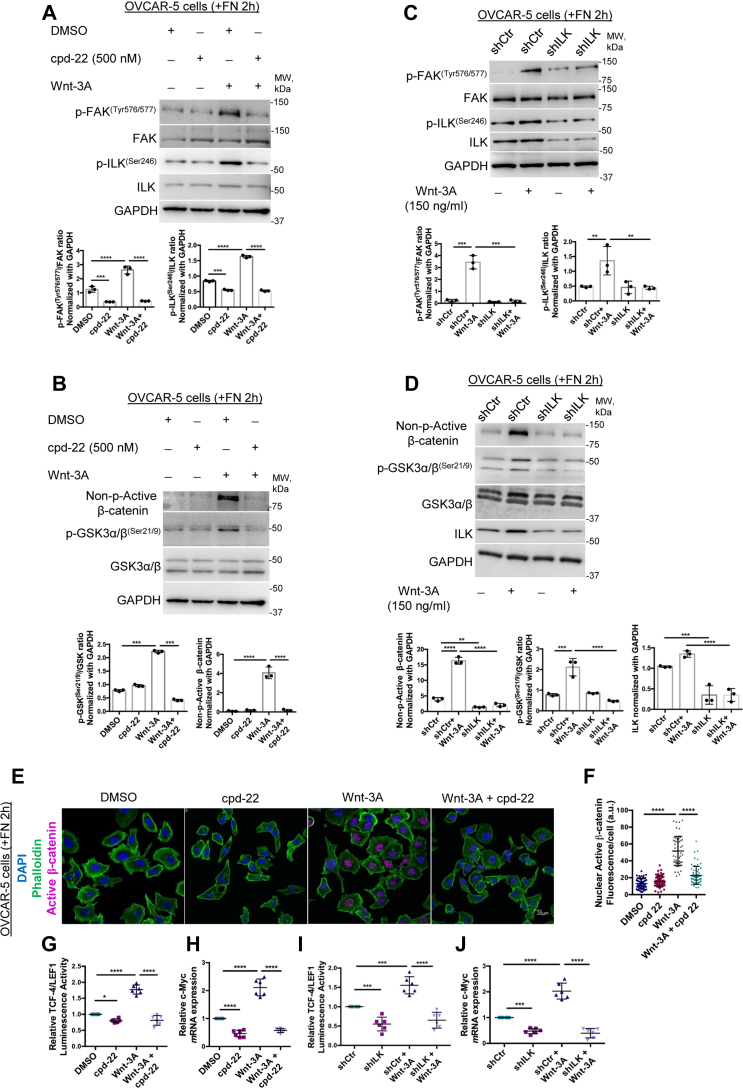
To determine whether ILK activation is required for β-catenin nuclear translocation, we altered ILK expression levels or its activity by using either sh-RNA-mediated KD or pharmacological inhibition. The trisubstituted pyrazol compound 22 (cpd-22) is a specific cell-permeable ILK inhibitor (IC50 = 600 nM) with antiproliferative potency against prostate and breast cancer cell lines (IC50 = 1–2.5 μM), whereas normal epithelial cells were not affected (43). OVCAR-5 cells were plated on FN for 2 h in the presence of Wnt-3A (150 ng/ml) and/or cpd-22 at 0.5 μM. Treatment with Wnt-3A increased both p-FAKTyr576/577 and p-ILKSer246 levels compared with dimethyl sulfoxide (DMSO) (Fig. 5A; p < 0.0001). Cpd-22 reduced the phosphorylation levels of both proteins at basal level compared with DMSO-treated cells and more significantly in the Wnt-3A–treated groups compared with Wnt-3A alone (Fig. 5A; p < 0.001, p < 0.0001, respectively). Total FAK and ILK expression levels were not altered by Wnt-3A or cpd-22 treatment (Fig. 5A). Wnt-3A treatment caused approximately 3.5-fold increase in p-GSK-3α/βSer21/9 with subsequent approximate 20-fold increase in nonphospho (active) β-cateninSer33/37/Thr41 levels compared with control OVCAR-5 cells (Fig. 5B). Cpd-22 treatment drastically reduced p-GSK-3α/βSer21/9 and the nonphospho (active) β-cateninSer33/37/Thr41 compared with Wnt-3A–treated cells (Fig. 5B; p < 0.001, p < 0.0001, respectively). Similar results were observed in SKOV3 cells (Fig. S6, A and B). In addition, we transduced OVCAR-5 and SKOV3 cells with sh-RNA targeting ILK or sh-RNA Ctr (scrambled shRNA). Efficient ILK-KD is shown in SKOV3/sh-ILK and OVCAR-5/sh-ILK cells versus control (scrambled) shRNA (Figs. 5D and S6D, p < 0.001, p < 0.0001, respectively). WB analysis showed that Wnt-3A increased p-FAKTyr576/577 and p-ILKSer246 expression in ILK-expressing cells (OVCAR-5/sh-Ctr and SKOV3/sh-Ctr) compared with low-ILK-expressing cells (OVCAR-5/sh-ILK and SKOV3/sh-ILK; Figs. 5C and S6C; p < 0.0001). Consistent with these observations, ILK downregulation blocked Wnt-3A–mediated phosphorylation of GSK-3α/β at Ser21/9 and subsequent nonphospho (active) β-cateninSer33/37/Thr41 levels in both OVCAR-5/sh-ILK and SKOV3/sh-ILK cells compared with OVCAR-5/sh-Ctr and SKOV3/sh-Ctr cells (Figs. 5D and S6D; p < 0.0001, p < 0.05, respectively), suggesting that ILK stabilizes FA contacts and β-catenin activity in TG2-expressing OC cells.
Figure 5.
ILK inhibition blocks β-catenin signaling in OC cells.A and B, WB for p-FAKTyr576/577, FAK, p-ILKSer246, ILK, non–p-active β-catenin, p-GSKα/βSer21/9, GSKα/β, and GAPDH in OVCAR-5 cells plated on FN-coated plates for 2 h and treated or not with cpd-22 and/or Wnt-3A. Densitometry quantifies non–p-active β-catenin expression levels and p-FAKTyr576/577/FAK, p-ILKSer246/ILK, and p-GSKα/βSer21/9/GSKα/β ratios (N = 3; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). All antibodies (Abs) were probed on the same membrane. For consistency, the same GAPDH representative WB images are shown in A and B. C and D, WB for p-FAKTyr576/577, FAK, p-ILKSer246, ILK, non–p-active β-catenin, p-GSKα/βSer21/9, GSKα/β, and GAPDH in OVCAR-5 cells sh-Ctr or sh-ILK plated on FN for 2 h and treated or not with Wnt-3A. Densitometry quantifies ILK and non–p-active β-catenin expression levels and p-FAKTyr576/577/FAK, p-ILKSer246/ILK, and p-GSKα/βSer21/9/GSKα/β ratios (N = 3; ∗∗p < 0.01; ∗∗∗p < 0.001; and ∗∗∗∗p < 0.0001). E, IF staining for phalloidin (Alexa Fluor 488, green) and non–p-active β-catenin (Alexa Fluor 568, red) in OVCAR-5 cells plated on FN-coated plates for 2 h and treated or not with cpd-22 and/or Wnt-3A. F, quantification of Alexa Fluor 568 (red) proteins was calculated by using Metamorph software (N = 3; ∗∗∗∗p < 0.0001). G, OVCAR-5 cells were cotransfected with TCF/LEF1 luciferase reporter and Renilla control plasmid prior to treatment with cpd-22 or Wnt-3A and plated on FN for 2 h. Luciferase signal relative to Renilla activity is expressed as fold increase (N = 6; ∗∗∗∗p < 0.0001). H, real-time PCR for c-Myc in OVCAR-5 cells plated on FN-coated plates for 2 h and treated or not with cpd-22 and/or Wnt-3A (N = 6; ∗∗∗∗p < 0.0001). I, OVCAR-5 cells sh-Ctr or sh-ILK were cotransfected with TCF/LEF1 luciferase reporter and Renilla control plasmid prior to treatment with cpd-22 or Wnt-3A and plated on FN for 2 h (N = 6; ∗∗∗p < 0.01; ∗∗∗p < 0.001). J, real-time PCR for c-Myc in OVCAR-5 cells stably transduced with scrambled- or ILK-targeting shRNA plated on FN-coated plates for 2 h and treated or not with Wnt-3A (N = 6; ∗∗∗p < 0.01; ∗∗∗p < 0.001). cpd-22, compound 22; FAK, focal adhesion kinase; FN, fibronectin; GSKα/β, glycogen synthase kinase-3α/β; IF, immunofluorescence; ILK, integrin-linked kinase; LEF1, lymphoid enhancer–binding factor 1; OC, ovarian cancer; TCF, T-cell factor; WB, Western blot.
Next, IF microscopy and quantification detected significantly stronger nonphospho (active) β-catenin at Ser33/37 and Thr41 nuclear signal in OVCAR-5 and SKOV3 cells treated with Wnt-3A compared with control cells, whereas treatment with cpd-22 at 0.5 μM blocked the nonphospho (active) β-catenin nuclear translocation in both OC cells (Figs. 5, E and F and S6, E and F; p < 0.0001). ILK inhibitor cpd-22 significantly reduced Wnt-3A–mediated β-catenin/TCF transcriptional activity as measured by TCF/LEF1 reporter assay in OVCAR5 and SKOV3 cells (Figs. 5G and S6G; p < 0.0001). In addition, cpd-22 treatment decreased the mRNA expression levels of the β-catenin target gene c-Myc in both cell lines (Figs. 5H and S6H; p < 0.0001). Furthermore, ILK-KD significantly reduced Wnt-3A–mediated β-catenin/TCF transcriptional activity in OVCAR5-sh-ILK and SKOV3/sh-ILK cells (Figs. 5I and S6I; p < 0.0001) and decreased expression levels of the β-catenin target gene c-Myc in both OC cell lines (Figs. 5J and S6J; p < 0.0001) compared with OVCAR5/sh-Ctr and SKOV3/sh-Ctr cells.
TG2–FN interaction stimulates OC cell proliferation and migration through ILK activation
TG2 expression increases adhesion to FN causing integrin aggregation, which in turn translates extracellular cues into β-catenin transcriptional activity by activating ILK. Here, we evaluated whether the TG2–FN dependent activation of ILK has functional consequences. Hence, we measured the effects of TG2-mediated ILK engagement on activating the oncogenic Wnt pathway by assessing proliferation and invasiveness of OC cells.
To test this, SKOV3 and OVCAR-5 cells were plated on FN-coated surfaces and treated with Wnt-3A and/or anti-TG2 inhibitory 4G3 Ab. Wnt-3A treatment efficiently increased proliferation and migration in both OC cells compared with control, whereas 4G3 treatment drastically reduced Wnt-3A–induced proliferation and migration (Figs. 6, A and B and S7, A–C; p < 0.0001). Cells treated with 4G3 displayed decreased proliferation and migration rates compared with IgG control cells (Figs. 6, A and B and S7, A–C; p < 0.01, p < 0.0001, respectively).
Figure 6.
ILK functional inhibition results in OC cells' proliferation and migration inhibition.A, CCK-8 assay quantifies proliferation of SKOV3 cells treated with inhibitory antibodies (Abs) directed against the FN-binding domain of TG2 (clone 4G3) and/or Wnt-3A (N = 8; ∗∗∗∗p < 0.0001). B, scratch assay quantifies migration of SKOV3 cells treated with 4G3 and/or Wnt-3A (N = 8; ∗∗p < 0.01; ∗∗∗∗p < 0.0001). C, CCK-8 assay quantifies proliferation of SKOV3 cells treated with cpd-22 and/or Wnt-3A (N = 8; ∗∗∗∗p < 0.0001). D, scratch assay quantifies migration of SKOV3 cells treated with cpd-22 and/or Wnt-3A (N = 8; ∗∗∗∗p < 0.0001). E, CCK-8 assay quantifies proliferation of ILK-KD versus sh-Ctr SKOV3 cells treated with or not with Wnt-3A (N = 8; ∗∗∗∗p < 0.0001). F, scratch assay quantifies migration of ILK-KD versus sh-Ctr SKOV3 cells treated with or not with Wnt-3A (N = 8; ∗∗∗∗p < 0.0001). G, overall survival curves generated using the Kaplan–Meier plot for tumors expressing higher than median levels of TGM2, ITGB1, FN1, and of ILKversus those expressing lower than median levels of TGM2, ITGB1, FN1, and of ILK in HGSOC tumor microarray (HR = 1.4, p = 0.0046). H, proposed mechanistic model. CCK-8, Cell Counting Kit 8; cpd-22, compound 22; FN, fibronectin; HGSOC, high-grade serous ovarian cancer; HR, hazard ratio; ILK, integrin-linked kinase; KD, knockdown; OC, ovarian cancer; TG2, tissue transglutaminase.
To evaluate whether ILK-mediated β-catenin signaling is implicated in the proliferation and migration of TG2-expressing cells on FN, we measured the same endpoints in cells in which ILK was downregulated via sh-RNA or inhibited pharmacologically. Wnt-3A increased OC cells’ proliferation and migration by ∼1.8-fold and 1.5-fold, respectively, compared with DMSO-treated (control) cells. Cpd-22 treatment of both cell lines decreased both proliferation and migration rates by ∼1.5-fold and 5-fold compared with DMSO control and by ∼1.5-fold and 2.5-fold after Wnt-3A treatment, respectively (Figs. 6, C and D and S7, D–F; p < 0.0001). These data demonstrate the significance of the input triggered by the TG2–FN interaction and ILK inhibition on cancer cell proliferation and migration.
Similar results were observed with sh-RNAi–based ILK silencing. Cell Counting Kit-8 assay demonstrated decreased cell proliferation of FN-plated SKOV3/sh-ILK and OVCAR-5/sh-ILK cells compared with control shRNA and after Wnt-3A treatment (Figs. 6E and S7G; p < 0.0001). Phase contrast microscopy showed that sh-RNA mediated ILK-KD diminished SKOV3 and OVCAR-5 cell migration on FN-coated plates relative to cells transduced with sh-RNA Ctr and upon Wnt-3A treatment (Figs. 6F and S7, H and I; p < 0.05, p < 0.0001).
Finally, the survival analysis of 1656 patients with HGSOC profiled by the TCGA using Kaplan–Meier plotter system demonstrated that patients with higher than median TGM2, ILK, ITGB1, and FN1 combined expression levels had an increased estimated risk of death when compared with those with lower than median expression levels (Fig. 6H; HR = 1.4, p = 0.0046). In sum, our data demonstrate the significance of TG2 transducing outside in signals via ILK and β-catenin to stimulate oncogenic properties (proliferation and migration) of cancer cells (Fig. 6I).
Discussion
TG2 expression was increased in several cancers, including breast, pancreatic, and colon cancer (44, 45, 46). Upregulated TG2 levels were also detected in OC cells compared with normal ovarian epithelial cells (47), in human ovarian tumors, and ascites fluid (32). In OC, TG2 forms active complexes with FN and β1 integrin at the interface with the ECM compartment enhancing “outside–in” signaling involved with cancer cell adhesion to the ECM (32). Mechanistically, it has been demonstrated that the interaction between TG2 and FN-containing matrices recruits a series of integrin-associated adaptor proteins, such as FAK and RhoA, which regulate cancer cell adhesion, survival, and migration by promoting the formation of mature FA complexes and actin cytoskeleton assembly (48, 49). In addition, TG2 was shown to modulate integrin-mediated signaling converging on PI3K/Akt pathway, a downstream effector of FAK, and this in turn, enhanced resistance to apoptosis induced by chemotherapy (50). One of the main integrin cytoplasmic effectors is the serine–threonine protein kinase ILK that plays a central role in “outside–in” signaling (18). ILK expression was found to be increased in several epithelial cancers (23), and its constitutive activation induced tumorigenicity in mouse models (14). In OC, ILK expression was upregulated in OC cells and tumor specimens compared with the normal epithelium (21), correlated with ovarian tumor grade (21), and promoted the prometastatic OC cell behavior (51). Based on this evidence, we investigated whether TG2–integrin complex formation was sufficient to transduce “outside–in” signals through ILK activation in OC cells.
Here, we show that TG2-expressing OC cells plated on FN matrix harbor an increased pool of p-ILKSer246. Confocal microscopy showed that TG2 significantly colocalized with ILK and active p-ILKSer246 and that the presence of an FN matrix was instrumental in the formation of active TG2–p-ILKSer246 clusters. Furthermore, PLA analysis revealed that each protein of the ternary TG2–FN–β1 integrin complex actively engages with p-ILKSer246 in primary OC cells, supporting the functional relevance of TG2-mediated outside–in transduction signals in human samples. Of note, PLA analysis indicated that TG2–p-ILKSer246 complexes are in close proximity (distance ≤40 nm), which implies a direct interaction between the two proteins. Interestingly, the levels of active p-ILKSer246 significantly decreased in TG2-KD compared with TG2-expressing OC cells and in the absence of FN matrix. In addition, functional targeting of TG2 and integrin β1 with inhibitory Abs prevented the activating phosphorylation of both FAK and ILK. We interpret these findings to indicate that ILK activation and FA assembly is strongly regulated by TG2 and requires the formation of active TG2–FN–integrin clusters (28, 30, 42). Furthermore, co-IP, IF confocal, and colocalization studies demonstrated a direct interaction between TG2 and ILK (including active p-ILKSer246), proposing this kinase as a novel TG2 partner.
Next, we identify the canonical Wnt/β-catenin pathway as a main mechanism regulated in response to the formation of TG2–p-ILKSer246 functional clusters. We show that the positive regulation of TG2–integrin–ILK axis on β-catenin is dual: (1) it amplifies the Wnt response to the Wnt-3A ligand and (2) stabilizes β-catenin signaling by the inhibitory phosphorylation of GSK-3α/β at Ser21/9. Mutations in the β-catenin gene (CTNNB1) and loss-of-function mutations in the β-catenin destruction complex (APC and Axin) that drive tumor progression are uncommon in ovarian carcinomas (52). Yet, the upregulation of Wnt receptors and ligands has a prominent role to induce the aberrant activation and nuclear localization of β-catenin and correlates with OC tumor progression (52). Several studies indicate that the canonical Wnt/β-catenin pathway is responsive to matrix reorganization and is activated by the changes in the integrin clusters in OC (52, 53). The integrin–ILK transduction signal axis inactivates GSK-3α/β by phosphorylation at Ser21/9 and amplifies Wnt-3A signals, leading to β-catenin–TCF/LEF1 transcriptional activity in cancer cells (19, 24). Importantly, the nuclear accumulation of β-catenin leads to an upregulation of Wnt and ECM target genes, such as Fzd7 (54), c-Myc (55), and FN1 (56). In addition, β-catenin transcriptional activation enhances β1 integrin aggregation, leading to migration and adhesion of glioma and vascular smooth muscle cells (57, 58). This provides a positive feedback loop between the ECM-mediated integrin clustering and the canonical Wnt/β-catenin pathway that regulates several cancer cell functions, including cell adhesion, proliferation, and migration (38). Although these findings implicate the integrin–ILK interactions in the regulation of some of the components of the Wnt signaling pathway, the exact crosstalk between the two pathways in tumor progression remains not defined.
We had previously shown that expression of the FN-binding domain of TG2 is associated with β-catenin stabilization by both recruiting integrin β1 and directly engaging in a complex with Fzd7, which in turn promotes OC cell proliferation and a cancer stem–like cell phenotype (30, 59). Hence, we tested whether TG2–FN–integrin β1 represents a new functional link between ILK activation and Wnt signaling. Accordingly, our data show that OC cells plated on FN matrix and treated with the inhibitory anti-TG2 (clone 4G3) and anti-integrin β1 (clone P5D2) Abs decreased the response of OC cells to the Wnt-3A ligand, prevented ILK recruitment, and disrupted the inhibitory phosphorylation of GSK-3α/β at Ser21/9. The specific ILK inhibitor cpd-22 also disrupted OC cell response to Wnt-3A treatment by preventing the GSK-3α/β inhibitory phosphorylation and the subsequent β-catenin nuclear translocation. Furthermore, we show increased p-FAKTyr576/577 and p-ILKSer246 levels in TG2-expressing OC cells treated with Wnt-3A, pointing to crosstalk between Wnt and integrin pathways in the presence of TG2. Altogether, the data strengthen the role of the TG2–integrin coreceptor complex as functional regulator of the canonical Wnt pathway and define ILK as the intracellular effector through which this occurs.
Finally, we evaluated whether formation of a complex between TG2 and ILK has functional outcomes. In the presence of an active TG2–p-ILKSer246 axis, Wnt-3A treatment efficiently increased proliferation and migration in OC cells compared with control, whereas TG2 blockade by 4G3 and ILK inhibition by cpd-22 treatment drastically reduced the Wnt-3A impact on the OC cell proliferative and migratory capacity. The data point to a potential novel involvement of TG2–ILK in Wnt/β-catenin pathway activation, promoting oncogenic functions, and to new therapeutic interventions. In this regard, it has been demonstrated that ILK-KD and its pharmacological inhibition in OC cells blocked invasion and migration on 3-dimensional collagen gels (51) and delayed tumor formation in subcutaneous OC nude mice models (60). A recent report indicates that ILK siRNA-mediated silencing and its pharmacologic inhibition by cpd-22 reduce cell growth and the invasive ability of cisplatin-resistant OC cells (22). Likewise, strategies aimed at targeting TG2 demonstrated antitumor activity in several models (61). TG2–FN complex blockade by the inhibitory 4G3 Ab reduced tumor initiation by disrupting the OC stem cell phenotype and spheroid formation (30). Recent data also indicated that newly synthesized TG2–FN small-molecule analogs prevented OC cell adhesion to FN matrix and to reconstituted mesothelial matrix and are synergistic with paclitaxel in reducing metastatic spread (62). These results support preclinical development of new strategies aiming to disrupt TG2–p-ILKSer246 alone or in combination with current chemotherapeutics.
In conclusion, our data demonstrate that TG2 engagement with FN is an essential step toward integrin aggregation and acts as a functional switch to activate ILK. This mechanism stabilizes Wnt/β-catenin that, in turn, fuels a positive feedback loop, mediating transcriptional regulation of ECM components, ligand-binding affinity of integrins, and activation of the integrin-linked adaptor proteins FAK and ILK. The results identify a novel TG2–active p-ILKSer246 complex involved in the initial critical steps of tumor progression and propose the development of future therapeutic interventions against this target.
Experimental procedures
Chemicals and reagents
Unless stated otherwise, chemicals and reagents were from Sigma–Aldrich. Monoclonal FN was from BD Biosciences; monoclonal TG2 (clone 4G3), integrin β1 (MAB1959; clone P5D2), integrin β1 (MAB2251; clone B3B11), and polyclonal p-ILK (Ser246) were from MilliporeSigma, and monoclonal TG2 (CUB 7402) and polyclonal TG2 (Ab-4) antibodies were from Thermo Fisher Scientific. Polyclonal ILK (clone 4G9), nonphospho (active) β-catenin at Ser33/37 and Thr41 (clone D13A1), p-FAKTyr576/577, FAK (clone D5O7U), GSK-3α/β, and p-GSK-3α/βSer21/9 (clone 37F11) antibodies were from Cell Signaling Technology; and GAPDH from Biodesign International. Secondary horseradish peroxidase–conjugated antibodies were from Amersham Biosciences and Santa Cruz Biotechnology, Inc. Lentiviral particles containing shRNA targeting TG2 and ILK and scrambled shRNA were purchased from Sigma–Aldrich. Recombinant human Wnt-3A was purchased from R&D Systems. ILK inhibitor, cpd-22, was purchased from MilliporeSigma.
Cell lines
SKOV3 cell line was from the American Type Culture Collection. OVCAR-5 cells were provided by Dr Kenneth Nephew (Indiana University). Cell lines were authenticated by short tandem repeat analysis and tested to be mycoplasma negative by IDEXX (BioResearch). For the experimental procedures, OC cell lines were used between 5 and 10 passages after thawing. SKOV3 and OVCAR-5 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (high glucose), 10% fetal bovine serum (FBS), 2 mM l-glutamine, and 100 units/ml penicillin and 100 μg/ml streptomycin. Cells were cultured at 37 °C in a humidified incubator with 5% CO2 supply.
Primary human cells
Deidentified malignant ascites fluid specimens from OC patients (n = 4) were obtained at the Indiana University Simon Cancer Center under an Institutional Review Board–approved protocol (Human Research Protection Program #1905951308). The study was conducted in accordance with the International Ethical Guidelines for Biomedical Research Involving Human Subjects and represented minimal risk to subjects. The studies in this work abide by the Declaration of Helsinki principles. All subjects had stage III or IV HGSOC or primary peritoneal carcinomatosis. For isolation of primary cells from human specimens (OC ascites and primary tumors), tumor cells were collected and purified as previously described (59). After centrifugation at 1500 rpm for 5 min, aliquots of 20,000 ascites-derived tumor cells were cultured as monolayers, as described (59).
Cell proliferation
SKOV3 and OVCAR-5 cells were seeded into 96-well plates (2.5 × 103 cells/well) coated with FN (10 μg/ml) in serum-free media. Proliferation was quantified by the Cell Counting Kit-8 assay (Dojindo Molecular Technologies). Absorbance of the amount of the formazan dye was measured with a plate reader (800TS Absorbance Reader; BioTek) at 450 nm. Eight replicates were used, and data represent means ± SD.
Scratch assay
SKOV3 and OVCAR-5 cells were seeded into 24-well plates (5 × 104 cells/well) coated with FN (10 μg/ml) in serum-free media. When cells reached 60 to 70% confluency, monolayers were scraped in a straight line with a p200 pipette tip. The debris was washed once with 0.5 ml of the growth medium, and then cells were resuspended in serum-free media with the indicated treatments. After 48 h, the scratched area acquired under a phase-contrast microscope for each sample was analyzed quantitatively by using the area set measurements menu in ImageJ software (63). Eight replicates were used, and data represent means ± SD.
Transfection
Stable gene KD was obtained by transducing a pool of lentiviral particles containing shRNA targeting TG2 (sc-37514-V) (#1), ILK (sc-35666-V), or scrambled shRNA control (Santa Cruz Biotechnology, Inc) into OVCAR-5 and SKOV3 cells. For independent verification of TG2 gene silencing, two additional individual duplex shRNAs were used (Mission Lentiviral Transduction Particles from Sigma–Aldrich). TG2-targeting shRNA sequences TRCN 272817 (#2) and TRCN 284849 (#3) yielded more than 75% gene KD. Lentiviral-transduced OC cells were selected with puromycin (1.5 μg/ml). Full-length clone DNA of human TGM2 with C-terminal His tag and full-length clone DNA of human ILK with C-terminal FLAG tag were inserted into pCMV3 vectors (SinoBiological) and stably transfected in sh-TG2 SKOV3 (#1) and OVCAR-5 (#1) cells using DreamFect Gold transfection reagent, followed by hygromycin selection.
Gene reporter assay
Dual-Luciferase Assay (Promega) quantified TCF/LEF1 promoter activity in OVCAR-5 and SKOV3 wildtype or shRNA transduced. Cells were plated in DMEM (high glucose), 10% FBS without antibiotics, and transiently cotransfected with TCF/LEF1 promoter luciferase and Renilla plasmids (BPS Bioscience), at a ratio of 10:1 using DreamFect Gold transfection reagent. After 24 h, OC cells were plated in DMEM (high glucose) without FBS and antibiotics and treated with IgG control, 4G3 and P5D2 Abs, or cpd-22. Luminescence was measured using a TD-20/20 Luminometer (Turner Biosystems) 48 h post-transfection. Experiments were performed in triplicate and repeated. To control for transfection efficiency, luminescence was normalized to Renilla activity.
RT–PCR
Total RNA was extracted using RNA STAT-60 Reagent (Tel-Test, Inc), and 1 μg was reverse-transcribed using iScript cDNA Synthesis Kit (Bio-Rad). Amplification of cDNA was performed by RT–PCR on an ABI Prism 7500 platform (Applied Biosystems) using the iTaq universal SYBR Green super mix (Bio-Rad) with the following parameters: 94 °C for 10 min, followed by 40 cycles of amplification at 94 °C for 15 s and 60 °C for 1 min, and finally, an extension step of 7 min at 72 °C. Primer sequences were the following: 5′-CACCAGCAGCGACTCTGA-3′ (forward) and 5′-GATCCAGACTCTGACCTTTTGC-3′ (reverse) for c-Myc; and 5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′ (reverse) for GAPDH. Relative expression of target genes was calculated as ΔΔCt, by subtracting the Ct of the reference gene from that of the control. Results are presented as means ± SD of replicates. Each experiment was performed in duplicate in three independent conditions.
Co-IP
Total lysates from OC cells and sh-TG2 SKOV3 cells transfected with full-length clone DNA of human TGM2 with C-terminal His tag and full-length clone DNA of human ILK with C-terminal FLAG tag were centrifuged at 1500 rpm for 5 min, washed in 1× PBS, and lysed on ice in 50 mM Tris–HCl buffer (pH of 7.6), 150 mM NaCl, 1% NP-40, leupeptin (1 μg/ml), aprotinin (1 μg/ml), and PMSF (400 μM). In parallel experiments, recombinant human full-length TG2 was mixed in lysis buffer at 1:1 ratio with recombinant human full-length ILK. Mixtures of cell lysates or recombinant proteins were incubated overnight at 4 °C with 5 μg of anti-TG2, anti-ILK, anti-His, anti-FLAG, or IgG, respectively, and then with 50 μg of slurry of Protein G Plus-Agarose beads under rotation for 4 h at 4 °C. Protein–Ab–bead complexes were centrifuged, washed in lysis buffer (3×), and boiled in 30 μl of 2× SDS protein loading dye and used for WB.
WB analysis
Equal amounts of protein were separated by SDS-PAGE and electroblotted onto polyvinylidene difluoride membranes. After incubation with primary and horseradish peroxidase–conjugated secondary Ab, antigen–Ab complexes were visualized using Super-Signal chemiluminescent substrate (Thermo Fisher Scientific). Images were captured by a luminescent image analyzer with a charge-coupled device camera (Chemi-Doc Imaging System; Bio-Rad) and quantified by densitometric analysis with a Gel-Pro Analyzer 3.1 software (Informer Technologies, Inc). All proteins were normalized with the structural protein GAPDH levels. Phosphorylated proteins were also normalized with their total pair. Protein levels were expressed as average value and presented in the graphs as means ± SD (N = 3).
IF
OC cells were plated as monolayers on glass or FN-coated Millicell 8-well chamber slides (MilliporeSigma). After the indicated incubation time, cells were fixed in 4% paraformaldehyde for 10 min at room temperature, washed 3× with PBS, permeabilized using 0.2% Triton X-100 in PBS (5 min) (unless otherwise specified), and blocked for 1 h with 3% goat serum in 1× PBS–0.1% Triton X-100. Subsequently, cells were incubated with primary antibodies (diluted 1:100) in blocking buffer overnight at 4 °C. After 3× washing in 1× PBS–0.1% Triton X-100, cells were incubated for 1 h at room temperature with Cy5-conjugated antimouse secondary Ab (1:500 dilution; Zymed) or Alexa Fluor-488 anti-rabbit secondary Ab (1:1000 dilution; Molecular Probes). Isotype-specific IgG was a negative control. Nuclei were visualized by 4′,6-diamidino-2-phenylindole staining (Vectashield; Vector Laboratories). The analysis was performed by using a confocal/two-photon Olympus Fluoview FV-1000 MPE system (Olympus America) available at the Indiana Center for Biological Microscopy facility. Fluorescence of nuclear active β-catenin was quantified using the analyze particles macro in ImageJ software. TG2 and p-ILKSer246 fluorescence was quantified in ImageJ as area of integrated intensity for each selected cell. Next, the corrected total cell fluorescence was calculated by using the following formula: corrected total cell fluorescence = integrated density – (area of selected cell × mean fluorescence of background readings). Quantification of colocalized proteins was calculated by using colocalization test macro in ImageJ software. For the negative control experiments, only the anti-rabbit or antimouse IgG isotypes were incubated with the Cy5- or Alexa Fluor-488–conjugated anti-rabbit or antimouse secondary antibodies. Each image is representative of at least four replicates.
In situ PLA
Interaction between TG2, integrin β1, or FN with p-ILKSer246 was measured in patient-derived OC primary cells by PLA using Duolink reagents (MilliporeSigma) and following the manufacturer's recommendations. Briefly, primary OC cells were plated on glass or FN-coated coverslips. After 2 h incubation, cells were processed as described in IF section. Primary antibodies were resuspended in Duolink Antibody Diluent at 1:100 dilution, incubated overnight at 4 °C, and then washed twice with 1× wash buffer A at room temperature. Next, the PLUS and MINUS PLA probes diluted 1:5 in the Duolink Antibody Diluent were added to the cells, the slides were incubated in a preheated humidity chamber for 1 h at 37 °C, and washed twice in buffer A for 2 min with gentle agitation. A ligation-ligase solution was added to each sample, and slides were incubated in a preheated humidity chamber for 30 min at 37 °C and washed twice in buffer A for 2 min with gentle agitation. Then, the amplification-polymerase solution was added, and the slides were incubated in the preheated humidity chamber for 100 min at 37 °C, and the slides were washed three times in buffer B. The cells were incubated with phalloidin-iFluor 488 reagent (Abcam) at 1:100 dilution. Finally, the slides were washed twice with PBS and mounted with a coverslip using Duolink mounting media with 4′,6-diamidino-2-phenylindole. Cells were observed with a confocal/two-photon Olympus Fluoview FV-1000 MPE system. Fluorescence was quantified using the analyze particles macro in ImageJ software (63). For every Ab, a negative control experiment was performed where only one Ab was incubated with the PLA probes. Each image is representative of at least three biological replicates.
Statistical analysis
The RNASeqv2 level 3 data for the TCGA OC cohort and the clinical information associated with these samples were obtained from cBioPortal (http://www.cbioportal.org). Survival analysis was performed using Kaplan–Meier plotter (64) or OvMark system (65), and the statistical significance was defined as log-rank p value <0.05. Kaplan–Meier plots for patients with no residual tumors and TGM2, ILK, integrin β1, and/or FN1 median expression, high (within the 75% quartile) or low expression (within 25% quartile), were used as the cutoff. Statistical analysis was performed using GraphPad Prism 9 software (GraphPad Software, Inc). Data were analyzed using one-way ANOVA followed by Tukey’s post hoc test or unpaired two-tailed t test. Results are represented as means ± SD, with ∗p < 0.05 considered statistically significant. Each experiment was performed with a minimum of three biological replicates; exact numbers are indicated in associated figure legends.
Data availability
The data that support the findings of this study are available from the corresponding author upon request.
Supporting information
This article contains supporting information.
Conflict of interest
M. P. is currently working as a research fellow at Labcorp Drug Development. The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
S. C. and D. M. conceptualization; S. C., M. P., R. A., and D. M. methodology; S. C., M. P., and R. A. software; S. C. and D. M. validation; S. C. and M. P. formal analysis; S. C., M. P., and R. A. investigation; S. C. and D. M. resources; S. C. and M. P. data curation; S. C. writing–original draft; S. C. and D. M. writing–review & editing; S. C. and D. M. supervision; S. C. and D. M. project administration; S. C. and D. M. funding acquisition.
Funding and additional information
This study was supported by the Department of Obstetrics and Gynecology at the Indiana University School of Medicine and the Department of Defense Ovarian Cancer Research Program Ovarian Cancer Academy Early-Career Investigator Award (grant no.: W81XWH-19-10008) to S. C. and US Department of Veterans Affairs (grant no.: BX000792-06) to D. M.
Edited by Eric Fearon
Supporting information
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71:7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Torre L.A., Trabert B., DeSantis C.E., Miller K.D., Samimi G., Runowicz C.D., et al. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018;68:284–296. doi: 10.3322/caac.21456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Witz C.A., Montoya-Rodriguez I.A., Cho S., Centonze V.E., Bonewald L.F., Schenken R.S. Composition of the extracellular matrix of the peritoneum. J. Soc. Gynecol. Investig. 2001;8:299–304. doi: 10.1016/s1071-5576(01)00122-8. [DOI] [PubMed] [Google Scholar]
- 4.Moser T.L., Pizzo S.V., Bafetti L.M., Fishman D.A., Stack M.S. Evidence for preferential adhesion of ovarian epithelial carcinoma cells to type I collagen mediated by the alpha2beta1 integrin. Int. J. Cancer. 1996;67:695–701. doi: 10.1002/(SICI)1097-0215(19960904)67:5<695::AID-IJC18>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 5.Kenny H.A., Chiang C.Y., White E.A., Schryver E.M., Habis M., Romero I.L., et al. Mesothelial cells promote early ovarian cancer metastasis through fibronectin secretion. J. Clin. Invest. 2014;124:4614–4628. doi: 10.1172/JCI74778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ween M.P., Oehler M.K., Ricciardelli C. Role of versican, hyaluronan and CD44 in ovarian cancer metastasis. Int. J. Mol. Sci. 2011;12:1009–1029. doi: 10.3390/ijms12021009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kusumoto T., Kodama J., Seki N., Nakamura K., Hongo A., Hiramatsu Y. Clinical significance of syndecan-1 and versican expression in human epithelial ovarian cancer. Oncol. Rep. 2010;23:917–925. doi: 10.3892/or_00000715. [DOI] [PubMed] [Google Scholar]
- 8.Cannistra S.A., Kansas G.S., Niloff J., DeFranzo B., Kim Y., Ottensmeier C. Binding of ovarian cancer cells to peritoneal mesothelium in vitro is partly mediated by CD44H. Cancer Res. 1993;53:3830–3838. [PubMed] [Google Scholar]
- 9.Bao H., Huo Q., Yuan Q., Xu C. Fibronectin 1: a potential biomarker for ovarian cancer. Dis. Markers. 2021;2021 doi: 10.1155/2021/5561651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Franke F.E., Von Georgi R., Zygmunt M., Munstedt K. Association between fibronectin expression and prognosis in ovarian carcinoma. Anticancer Res. 2003;23:4261–4267. [PubMed] [Google Scholar]
- 11.Gagne D., Groulx J.F., Benoit Y.D., Basora N., Herring E., Vachon P.H., et al. Integrin-linked kinase regulates migration and proliferation of human intestinal cells under a fibronectin-dependent mechanism. J. Cell. Physiol. 2010;222:387–400. doi: 10.1002/jcp.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamada K.M., Even-Ram S. Integrin regulation of growth factor receptors. Nat. Cell Biol. 2002;4:E75–E76. doi: 10.1038/ncb0402-e75. [DOI] [PubMed] [Google Scholar]
- 13.Huttenlocher A., Horwitz A.R. Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C., Dedhar S. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J. Cell Biol. 2001;155:505–510. doi: 10.1083/jcb.200108077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu Y., Li F., Goicoechea S., Wu C. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 1999;19:2425–2434. doi: 10.1128/mcb.19.3.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabe Y., Jin L., Tsutsumi-Ishii Y., Xu Y., McQueen T., Priebe W., et al. Activation of integrin-linked kinase is a critical prosurvival pathway induced in leukemic cells by bone marrow-derived stromal cells. Cancer Res. 2007;67:684–694. doi: 10.1158/0008-5472.CAN-06-3166. [DOI] [PubMed] [Google Scholar]
- 17.Acconcia F., Barnes C.J., Singh R.R., Talukder A.H., Kumar R. Phosphorylation-dependent regulation of nuclear localization and functions of integrin-linked kinase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:6782–6787. doi: 10.1073/pnas.0701999104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dedhar S., Williams B., Hannigan G. Integrin-linked kinase (ILK): a regulator of integrin and growth-factor signalling. Trends Cell Biol. 1999;9:319–323. doi: 10.1016/s0962-8924(99)01612-8. [DOI] [PubMed] [Google Scholar]
- 19.Oloumi A., Syam S., Dedhar S. Modulation of Wnt3a-mediated nuclear beta-catenin accumulation and activation by integrin-linked kinase in mammalian cells. Oncogene. 2006;25:7747–7757. doi: 10.1038/sj.onc.1209752. [DOI] [PubMed] [Google Scholar]
- 20.Radeva G., Petrocelli T., Behrend E., Leung-Hagesteijn C., Filmus J., Slingerland J., et al. Overexpression of the integrin-linked kinase promotes anchorage-independent cell cycle progression. J. Biol. Chem. 1997;272:13937–13944. doi: 10.1074/jbc.272.21.13937. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed N., Riley C., Oliva K., Stutt E., Rice G.E., Quinn M.A. Integrin-linked kinase expression increases with ovarian tumour grade and is sustained by peritoneal tumour fluid. J. Pathol. 2003;201:229–237. doi: 10.1002/path.1441. [DOI] [PubMed] [Google Scholar]
- 22.Reyes-Gonzalez J.M., Quinones-Diaz B.I., Santana Y., Baez-Vega P.M., Soto D., Valiyeva F., et al. Downstream effectors of ILK in cisplatin-resistant ovarian cancer. Cancers (Basel) 2020;12:880. doi: 10.3390/cancers12040880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persad S., Attwell S., Gray V., Mawji N., Deng J.T., Leung D., et al. Regulation of protein kinase B/Akt-serine 473 phosphorylation by integrin-linked kinase: critical roles for kinase activity and amino acids arginine 211 and serine 343. J. Biol. Chem. 2001;276:27462–27469. doi: 10.1074/jbc.M102940200. [DOI] [PubMed] [Google Scholar]
- 24.Novak A., Hsu S.C., Leung-Hagesteijn C., Radeva G., Papkoff J., Montesano R., et al. Cell adhesion and the integrin-linked kinase regulate the LEF-1 and beta-catenin signaling pathways. Proc. Natl. Acad. Sci. U. S. A. 1998;95:4374–4379. doi: 10.1073/pnas.95.8.4374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fesus L., Piacentini M. Transglutaminase 2: an enigmatic enzyme with diverse functions. Trends Biochem. Sci. 2002;27:534–539. doi: 10.1016/s0968-0004(02)02182-5. [DOI] [PubMed] [Google Scholar]
- 26.Yee V.C., Pedersen L.C., Le Trong I., Bishop P.D., Stenkamp R.E., Teller D.C. Three-dimensional structure of a transglutaminase: human blood coagulation factor XIII. Proc. Natl. Acad. Sci. U. S. A. 1994;91:7296–7300. doi: 10.1073/pnas.91.15.7296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner P.M., Lorand L. Complexation of fibronectin with tissue transglutaminase. Biochemistry. 1989;28:628–635. doi: 10.1021/bi00428a032. [DOI] [PubMed] [Google Scholar]
- 28.Akimov S.S., Krylov D., Fleischman L.F., Belkin A.M. Tissue transglutaminase is an integrin-binding adhesion coreceptor for fibronectin. J. Cell Biol. 2000;148:825–838. doi: 10.1083/jcb.148.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yakubov B., Chelladurai B., Schmitt J., Emerson R., Turchi J.J., Matei D. Extracellular tissue transglutaminase activates noncanonical NF-kappaB signaling and promotes metastasis in ovarian cancer. Neoplasia. 2013;15:609–619. doi: 10.1593/neo.121878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Condello S., Sima L., Ivan C., Cardenas H., Schiltz G., Mishra R.K., et al. Tissue tranglutaminase regulates interactions between ovarian cancer stem cells and the tumor Niche. Cancer Res. 2018;78:2990–3001. doi: 10.1158/0008-5472.CAN-17-2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao L., Shao M., Schilder J., Guise T., Mohammad K.S., Matei D. Tissue transglutaminase links TGF-beta, epithelial to mesenchymal transition and a stem cell phenotype in ovarian cancer. Oncogene. 2012;31:2521–2534. doi: 10.1038/onc.2011.429. [DOI] [PubMed] [Google Scholar]
- 32.Satpathy M., Cao L., Pincheira R., Emerson R., Bigsby R., Nakshatri H., et al. Enhanced peritoneal ovarian tumor dissemination by tissue transglutaminase. Cancer Res. 2007;67:7194–7202. doi: 10.1158/0008-5472.CAN-07-0307. [DOI] [PubMed] [Google Scholar]
- 33.Condello S., Cao L., Matei D. Tissue transglutaminase regulates beta-catenin signaling through a c-Src-dependent mechanism. FASEB J. 2013;27:3100–3112. doi: 10.1096/fj.12-222620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faverman L., Mikhaylova L., Malmquist J., Nurminskaya M. Extracellular transglutaminase 2 activates beta-catenin signaling in calcifying vascular smooth muscle cells. FEBS Lett. 2008;582:1552–1557. doi: 10.1016/j.febslet.2008.03.053. [DOI] [PubMed] [Google Scholar]
- 35.Deasey S., Nurminsky D., Shanmugasundaram S., Lima F., Nurminskaya M. Transglutaminase 2 as a novel activator of LRP6/beta-catenin signaling. Cell Signal. 2013;25:2646–2651. doi: 10.1016/j.cellsig.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozmikova I., Kozmik Z. Wnt/beta-catenin signaling is an evolutionarily conserved determinant of chordate dorsal organizer. Elife. 2020;9:e56817. doi: 10.7554/eLife.56817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yost C., Torres M., Miller J.R., Huang E., Kimelman D., Moon R.T. The axis-inducing activity, stability, and subcellular distribution of beta-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 1996;10:1443–1454. doi: 10.1101/gad.10.12.1443. [DOI] [PubMed] [Google Scholar]
- 38.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 39.Peifer M., Pai L.M., Casey M. Phosphorylation of the Drosophila adherens junction protein Armadillo: roles for wingless signal and zeste-white 3 kinase. Dev. Biol. 1994;166:543–556. doi: 10.1006/dbio.1994.1336. [DOI] [PubMed] [Google Scholar]
- 40.Thompson M.D., Monga S.P. WNT/beta-catenin signaling in liver health and disease. Hepatology. 2007;45:1298–1305. doi: 10.1002/hep.21651. [DOI] [PubMed] [Google Scholar]
- 41.Han S., Pang M.F., Nelson C.M. Substratum stiffness tunes proliferation downstream of Wnt3a in part by regulating integrin-linked kinase and frizzled-1. J. Cell Sci. 2018;131 doi: 10.1242/jcs.210476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Akimov S.S., Belkin A.M. Cell surface tissue transglutaminase is involved in adhesion and migration of monocytic cells on fibronectin. Blood. 2001;98:1567–1576. doi: 10.1182/blood.v98.5.1567. [DOI] [PubMed] [Google Scholar]
- 43.Lee S.L., Hsu E.C., Chou C.C., Chuang H.C., Bai L.Y., Kulp S.K., et al. Identification and characterization of a novel integrin-linked kinase inhibitor. J. Med. Chem. 2011;54:6364–6374. doi: 10.1021/jm2007744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mangala L.S., Fok J.Y., Zorrilla-Calancha I.R., Verma A., Mehta K. Tissue transglutaminase expression promotes cell attachment, invasion and survival in breast cancer cells. Oncogene. 2007;26:2459–2470. doi: 10.1038/sj.onc.1210035. [DOI] [PubMed] [Google Scholar]
- 45.Cheung W., Darfler M.M., Alvarez H., Hood B.L., Conrads T.P., Habbe N., et al. Application of a global proteomic approach to archival precursor lesions: deleted in malignant brain tumors 1 and tissue transglutaminase 2 are upregulated in pancreatic cancer precursors. Pancreatology. 2008;8:608–616. doi: 10.1159/000161012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miyoshi N., Ishii H., Mimori K., Tanaka F., Hitora T., Tei M., et al. TGM2 is a novel marker for prognosis and therapeutic target in colorectal cancer. Ann. Surg. Oncol. 2010;17:967–972. doi: 10.1245/s10434-009-0865-y. [DOI] [PubMed] [Google Scholar]
- 47.Matei D., Graeber T.G., Baldwin R.L., Karlan B.Y., Rao J., Chang D.D. Gene expression in epithelial ovarian carcinoma. Oncogene. 2002;21:6289–6298. doi: 10.1038/sj.onc.1205785. [DOI] [PubMed] [Google Scholar]
- 48.Verma A., Wang H., Manavathi B., Fok J.Y., Mann A.P., Kumar R., et al. Increased expression of tissue transglutaminase in pancreatic ductal adenocarcinoma and its implications in drug resistance and metastasis. Cancer Res. 2006;66:10525–10533. doi: 10.1158/0008-5472.CAN-06-2387. [DOI] [PubMed] [Google Scholar]
- 49.Janiak A., Zemskov E.A., Belkin A.M. Cell surface transglutaminase promotes RhoA activation via integrin clustering and suppression of the Src-p190RhoGAP signaling pathway. Mol. Biol. Cell. 2006;17:1606–1619. doi: 10.1091/mbc.E05-06-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cao L., Petrusca D.N., Satpathy M., Nakshatri H., Petrache I., Matei D. Tissue transglutaminase protects epithelial ovarian cancer cells from cisplatin-induced apoptosis by promoting cell survival signaling. Carcinogenesis. 2008;29:1893–1900. doi: 10.1093/carcin/bgn158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruney L., Liu Y., Grisoli A., Ravosa M.J., Stack M.S. Integrin-linked kinase activity modulates the pro-metastatic behavior of ovarian cancer cells. Oncotarget. 2016;7:21968–21981. doi: 10.18632/oncotarget.7880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nguyen V.H.L., Hough R., Bernaudo S., Peng C. Wnt/beta-catenin signalling in ovarian cancer: insights into its hyperactivation and function in tumorigenesis. J. Ovarian Res. 2019;12:122. doi: 10.1186/s13048-019-0596-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burkhalter R.J., Symowicz J., Hudson L.G., Gottardi C.J., Stack M.S. Integrin regulation of beta-catenin signaling in ovarian carcinoma. J. Biol. Chem. 2011;286:23467–23475. doi: 10.1074/jbc.M110.199539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Willert J., Epping M., Pollack J.R., Brown P.O., Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 2002;2:8. doi: 10.1186/1471-213x-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 56.Gao L., Chen B., Li J., Yang F., Cen X., Liao Z., et al. Wnt/beta-catenin signaling pathway inhibits the proliferation and apoptosis of U87 glioma cells via different mechanisms. PLoS One. 2017;12 doi: 10.1371/journal.pone.0181346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Renner G., Noulet F., Mercier M.C., Choulier L., Etienne-Selloum N., Gies J.P., et al. Expression/activation of alpha5beta1 integrin is linked to the beta-catenin signaling pathway to drive migration in glioma cells. Oncotarget. 2016;7:62194–62207. doi: 10.18632/oncotarget.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X., Wang J., Jiang H., Hu Q., Chen J., Zhang J., et al. Wnt3a activates beta1-integrin and regulates migration and adhesion of vascular smooth muscle cells. Mol. Med. Rep. 2014;9:1159–1164. doi: 10.3892/mmr.2014.1937. [DOI] [PubMed] [Google Scholar]
- 59.Condello S., Morgan C.A., Nagdas S., Cao L., Turek J., Hurley T.D., et al. beta-Catenin-regulated ALDH1A1 is a target in ovarian cancer spheroids. Oncogene. 2015;34:2297–2308. doi: 10.1038/onc.2014.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Q., Li C., Zhang Y.Y., Chen W., Lv J.L., Sun J., et al. Silencing of integrin-linked kinase suppresses in vivo tumorigenesis of human ovarian carcinoma cells. Mol. Med. Rep. 2013;7:1050–1054. doi: 10.3892/mmr.2013.1285. [DOI] [PubMed] [Google Scholar]
- 61.Budillon A., Carbone C., Di Gennaro E. Tissue transglutaminase: a new target to reverse cancer drug resistance. Amino Acids. 2013;44:63–72. doi: 10.1007/s00726-011-1167-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sima L.E., Yakubov B., Zhang S., Condello S., Grigorescu A.A., Nwani N.G., et al. Small molecules target the interaction between tissue transglutaminase and fibronectin. Mol. Cancer Ther. 2019;18:1057–1068. doi: 10.1158/1535-7163.MCT-18-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gyorffy B., Lanczky A., Szallasi Z. Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocr. Relat. Cancer. 2012;19:197–208. doi: 10.1530/ERC-11-0329. [DOI] [PubMed] [Google Scholar]
- 65.Madden S.F., Clarke C., Stordal B., Carey M.S., Broaddus R., Gallagher W.M., et al. OvMark: a user-friendly system for the identification of prognostic biomarkers in publically available ovarian cancer gene expression datasets. Mol. Cancer. 2014;13:241. doi: 10.1186/1476-4598-13-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.