Abstract
Membrane nucleases of mycoplasmas are believed to play important roles in growth and pathogenesis, although no clear evidence for their importance has yet been obtained. As a first step in defining the function of this unusual membrane activity, studies were undertaken to clone and analyze one of the membrane nuclease genes from Mycoplasma pulmonis. A novel screening strategy was used to identify a recombinant lambda phage expressing nuclease activity, and its cloned fragment was analyzed. Transposon mutagenesis was used to identify an open reading frame of 1,410 bp, which coded for nuclease activity in Escherichia coli. This gene coded for a 470-amino-acid polypeptide of 53,739 Da and was designated mnuA (for “membrane nuclease”). The MnuA protein contained a prolipoprotein signal peptidase II recognition sequence along with an extensive hydrophobic region near the amino terminus, suggesting that the protein may be lipid modified or that it is anchored in the membrane by this membrane-spanning region. Antisera raised against two MnuA peptide sequences identified an M. pulmonis membrane protein of approximately 42 kDa by immunoblotting, which corresponded to a trypsin-sensitive nucleolytic band of the same size. Maxicell experiments with E. coli confirmed that mnuA coded for a nuclease of unknown specificity. Hybridization studies showed that mnuA sequences are found in few Mycoplasma species, suggesting that mycoplasma membrane nucleases display significant sequence variation within the genus Mycoplasma.
Mycoplasmas are among the smallest free-living organisms known and are responsible for a number of respiratory and genital tract diseases in humans and animals (30, 31). The nutritional requirements of mycoplasmas (40), genomic sequence information (11) and biochemical studies (27) indicate that mycoplasmas lack most capacities for de novo synthesis of nucleotides. To compensate, they must encode enzyme activities and transport functions to facilitate the uptake of nucleic acid precursors either as free bases or as oligonucleotides (8). A membrane nuclease activity in mycoplasmas was initially reported in Mycoplasma pulmonis, a rodent respiratory and genital tract pathogen (19). Since this nuclease was located on the outer membrane surface of the organism, it could satisfy the need for purines and pyrimidines by the degradation of DNA or RNA in mucosal secretions. Nucleic acids could also be obtained from dead and dying cells of the respiratory tract to which the mycoplasma is attached. External membrane-associated nucleases have been identified in all Mycoplasma species tested, with most species appearing to produce multiple nucleolytic proteins (20). The divalent-cation requirements of these nucleases varied between species, suggesting that they may be produced by unrelated genes. Few studies of membrane-associated nucleases have been reported, although a Mycoplasma penetrans endonuclease has been recently purified and characterized (2). There are also two reports of mycoplasma nucleases that appear to be able to induce internucleosomal DNA degradation characteristic of apoptosis (24, 25).
To investigate this activity in M. pulmonis further, studies were initiated to clone and analyze one of its membrane nuclease genes. This process required the identification of nuclease activity in recombinant clones, Tn1000 mutagenesis of the clones, and analysis of the results of that mutagenesis for loss of function. The expression of mycoplasma genes in Escherichia coli in a functional form poses particularly difficult problems. First, UGA codons are interpreted as tryptophan coding sequences in the genus Mycoplasma as opposed to stop codons in most other organisms (22). It is important to note that it is not possible to completely suppress all UGA codons in E. coli even in an efficient UGA suppressor background (36) because suppressor efficiency is context dependent (29). In addition, transcription and translation initiate aberrantly in E. coli within cloned mycoplasma sequences due to the high A+T content of the mycoplasma chromosome (12, 23). These two problems result in the loss of gene regulation and truncated gene products in E. coli either from premature termination at UGA codons or from internal transcription initiation. An additional problem is the stable maintenance of these sequences in E. coli. Expression of gene products, which may have a significant detrimental effect on E. coli may depend upon numerous variables such as product concentration, specific activity, and substrate specificity of the gene product. Differences between E. coli and mycoplasmas in codon usage may also play a critical role in the outcome of a project of this nature. To increase the chances of success, a novel screening strategy was used to identify cloned nuclease-coding sequences. In conclusion, we report here the successful cloning of a membrane nuclease gene from M. pulmonis, its DNA sequence, and an analysis of its expression in E. coli.
MATERIALS AND METHODS
Bacterial strains and plasmids.
M. pulmonis CT and χ1048 were grown as previously described (20). These strains were originally obtained from M. K. Davidson. M. gallisepticum R (from Steve Geary), M. hyopneumoniae 232 (from R. F. Ross), M. hyorhinis GDL (from R. F. Ross), M. capricolum ATCC 27343, M. fermentans PG-18 (from S.-C. Lo), M. fermentans incognitus (from S.-C. Lo), M. penetrans (from S.-C. Lo), M. hominis 1620 (from L. D. Olson), M. pneumoniae ATCC 15531, and A. oculi ISM1499 (1) were also used in this study. Cultures were obtained from a stock culture maintained at −70°C, inoculated into fresh broth medium, and incubated statically at 37°C. E. coli strains (Table 1) were started from stock cultures and maintained in 1× or 2× Luria-Bertani broth, in superbroth (32 g of tryptone per liter, 20 g of yeast extract per liter, 5 g of NaCl per liter [pH 7.2]) or on Luria-Bertani agar media. Phage plates for screening genomic libraries on E. coli ISM612 consisted of superbroth base with superbroth soft-agar overlays. The plasmids constructed during the course of these studies are described in Table 1.
TABLE 1.
E. coli strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli | ||
| LE392 | F−hsdR514 (rK− mK−) lacY1 supE44 supF58 galK2 galT22 trpR55 metB1 | |
| ISM612 | [leu(UGA) lacZ659(UGA) trpA9605(UAG) his29(UAG) ilv thyA metB argH rpoB rpsL prfB3] pISM3001 | 21, 36 |
| ISM614 | LE392 [F−hsdR514 (rK− mK−) lacY1 supE44 supF58 galK2 galT22 trpR55 metB1] pISM3001 | This study |
| ISM647 | XL1-Blue [recA1 lac endA1 gyrA46 thi hsdR17 supE44 relA1 (F′ proAB+ lacIqlacZΔM15 Tn10)] pISM3001 | This study |
| DPWC | F+ mating donor | Gold Biotechnologies |
| BW26 | F− mating recipient, Kanr | Gold Biotechnologies |
| CSR 603 | (recA1 uvrA6 phr-1) maxicell strain | 33 |
| Plasmids | ||
| pISM3001 | trpT gene cloned behind a lac promoter in the vector pACYC184, Cmr | 36 |
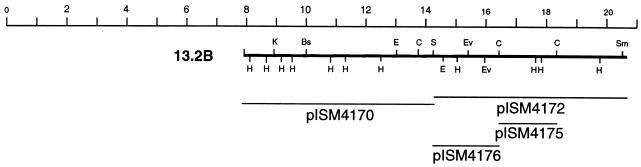
| pISM4170 | 6.6-kb SacI fragment from phage 13-2B cloned onto pMOB, Apr | This study |
| pISM4172 | 7.0-kb SacI fragment from phage 13-2B cloned onto pMOB, Apr | This study |
| pISM4175 | 2.2-kb ClaI fragment from pISM4172 cloned into pMOB, Apr | This study |
| pISM4176 | 2.6-kb ClaI fragment from pISM4172 cloned into pMOB, Apr | This study |
| pMOB | Cloning vector for Tn1000 mutagenesis, Apr | Gold Biotechnologies |
Isolation and manipulation of DNA.
Mycoplasma chromosomal DNA was isolated from 1 liter of mid-log-phase culture as previously described (1). Plasmid DNA was isolated from E. coli by alkali lysis. DNA fragments used for cloning and as probes were isolated from agarose gels on GenElute agarose spin columns (Supelco). DNA probes were labeled with [α-32P]CTP by using the Rediprime random-labeling system (Amersham, Arlington Heights, Ill.), and unincorporated nucleotides were removed by spinning the labeled probe through microspin S-300 HR columns (Pharmacia-Biotech).
Plasmids pISM4170 and pISM4172 were transformed into E. coli DPWC and mated with BW26 by the method described by Strathmann et al. (37). Recipient cells were selected on Luria-Bertani agar containing 50 μg of kanamycin per ml and 250 μg of ampicillin per ml. The Tn1000 insertions in the plasmids resulting from these matings were mapped either by restriction digestion or by PCR. PCRs were performed in a 50-μl volume containing 1× buffer (GIBCO BRL), approximately 1 ng of plasmid DNA from alkali lysis, T7 primer and either primer 187 or primer 486 (5 pmol) (see below), 4 mM MgCl, 20 mM deoxynucleoside triphosphates, and 1 U of Taq polymerase. Cycling was performed as follows: one cycle of 5 min at 92°C and 30 cycles of 1 min at 92°C, 1 min at 55°C, and 2 min at 72°C. Tn1000 inserts were also used as primer sites for DNA sequence analysis.
For hybridization, approximately 2 to 5 μg of mycoplasma DNA was digested with EcoRI and the fragments were separated on a 0.7% agarose gel and transferred to nylon membranes. The blots were hybridized with DNA probes by using Rapid-hyb buffer (Amersham). Hybridization was performed at 46°C (low stringency). The blots were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS) three times for 15 min at room temperature and in 1× SSC-0.1% SDS twice for 30 min at 42°C.
Antisera.
Antigen preparations containing semipurified nucleolytic proteins were prepared from SDS-polyacrylamide gel electrophoresis (PAGE) nuclease gels by using preparative spacers with the Bio-Rad Protean II electrophoresis unit. The running and renaturation conditions were adapted from standard conditions to obtain the maximal nuclease activity in preparative gels with the following modifications (20). The gels were washed for 1 h in incubation buffer (40 mM Tris, 0.01% casein, 0.04% β-mercaptoethanol [pH 7.5]), and nuclease digestion was allowed to proceed for 24 h. Areas displaying digestion of the DNA (nonfluorescing regions of the gel) were excised and minced with a 22-gauge needle. Two New Zealand White rabbits were immunized by injecting the acrylamide-protein slurry subcutaneously. The rabbits were boosted 2 weeks after immunization with the same antigen, and the antiserum collected at 4 weeks was tested for reactivity by enzyme-linked immunosorbent assay with M. pulmonis lysed whole-cell antigen (4) and by immunoblotting as described previously (39).
Hyperimmune antisera was also raised in BALB/c mice against two internal peptides of MnuA corresponding to amino acids 125 to 135 (QEKPKEKPGRK) and 320 to 331 (GSKGEGTTSVGV) of the predicted amino acid sequence. These peptides, designated the N-terminal and C-terminal peptides, respectively, were prepared with N-terminal cysteine residues to link them to maleimide-activated keyhole limpet hemocyanin (Pierce, Rockford, Ill.). The resulting peptide conjugates were then mixed with incomplete Freund’s adjuvant and used to immunize mice. Antibody containing ascites fluid was then prepared from the immunized mice by using SP2/0 myeloma cells as described by Luo and Lin (17). Antibody titers were determined by enzyme-linked immunosorbent assays with ovalbumin-peptide conjugates as the antigen. Immunoblot analyses were also performed with each of the ascites and with M. pulmonis whole cells as antigens.
Genomic library construction and screening.
Chromosomal DNA from M. pulmonis was partially digested with Sau3A, and 9- to 15-kb fragments were isolated by using sucrose gradients (32). The fragments were partially filled in and ligated into XhoI-digested λGEM 12 arms as specified by the manufacturer (Promega Corp., Madison, Wis.). The recombinant phage were packaged and plated onto lawns of E. coli LE392. Single plaques were picked and amplified as previously described (21). The genomic library was screened by inoculating lawns of E. coli ISM612 with a 48-pin replicator as previously described (21). Rabbit anti-membrane nuclease antisera were used to develop the lifts and identify potential nuclease-expressing clones.
Nuclease detection assays.
For nuclease detection, mycoplasma suspensions were produced as previously described (20). Nuclease activity in recombinant λ phage was measured in the following way. The opal suppressor strain E. coli ISM612 was infected with phage at a multiplicity of infection of 10:1, and the cells were shaken at 37°C for 2 h. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added (final concentration, 4 mM), and the shaking was continued for an additional 5 h. The cells were then harvested, resuspended in phosphate-buffered saline (PBS), and sonicated to disrupt them. Sonication was performed with a Branson Ultrasonicator at the maximum setting for the micro tip and 50% duration pulse for 20 pulses. Cellular debris was removed by centrifugation at 12,000 × g for 10 min, and the nuclease activity in the supernatant was monitored by the λ assay as described previously (20). Nuclease activities associated with recombinant plasmids were monitored in the E. coli ISM647 background. Transformed colonies were picked, grown overnight, harvested, resuspended in PBS, and sonicated prior to use. The SDS-polyacrylamide gel electrophoresis (PAGE) nuclease gel assay has also been described previously (20). In some SDS-PAGE nuclease gels, λ DNA (Bethesda Research Laboratories, Inc., Gaithersburg, Md.) was used instead of salmon sperm DNA.
DNA sequence analysis.
Plasmids for DNA sequencing were prepared by using Qiagen columns, and the sequencing was performed by the Nucleic Acid Instrumentation facility at Iowa State University by using cycle-sequencing protocols. The sequencing primers were oligonucleotides complementary to the transposon sequences adjacent to the inverted repeat ends. Primers 187 (5′-CAACGAATTATCTCCTT-3′) (Gold Biotechnology) and 486 (5′-TCAATAAGTTATACCAT-3′) were used to sequence from Tn1000 insertions (37). In some instances, the T7 primer (5′-AATACGACTCACTATAG-3′) and three other primers, 2590 (5′-GCGACACTGAGCCTAGAG-3′), 1145 (5′-GGTGTAGCTACTAATAAAC-3′), and 890 (5′-GACCTTAGCCAAATGAAAC-3′) were used.
Sequence analysis was performed with MacVector software (Eastman Kodak). The hydrophilicity of the translated product was determined by the method of Kyte and Doolittle (16), and the secondary-structure predictions were made by the methods of Chou and Fasman (5) and Garnier et al. (10). The translated product was also analyzed for signal sequences and transmembrane domains with PSORT (available on the World Wide Web [27a]). PSORT uses the methods of McGeoch (18) and von Heijne (41) for signal sequence determination and the method of Klein et al. (14) for determining transmembrane domains.
Maxicell analysis.
To confirm that the nuclease activity observed in recombinant E. coli was actually derived from mnuA, maxicell experiments were performed (33). Maxicell strain CSR 603 was transformed with pISM3001 and either pSK (negative control) or pISM4176 (mnuA) selecting for ampicillin and chloramphenicol resistance. Cells were grown overnight in Luria-Bertani broth and diluted to an optical density at 600 nm of 0.7. Samples (5 ml) were spread in a sterile petri dish and subjected to UV light at different time intervals (0 to 60 s) by being placed in a box containing three 15-W UV bulbs suspended 4 in. above the base. UV-inactivated cells (20 ml) were transferred to a 250-ml flask and incubated at 37°C with shaking for 5 h. IPTG (4 mM) was added to each culture, and the cells were shaken for 4 h at 37°C. The cells were pelleted by centrifugation, washed once with TS buffer (10 mM Tris, 140 mM NaCl [pH 7.5]), and sonicated to lyse them, and the cell debris was removed by centrifugation at 12,000 × g for 10 min. The lysate was then subjected to analysis by the λ and SDS-PAGE nuclease assays as described previously (20).
In some experiments, maxicell lysates were adsorbed with antisera to the MnuA C-terminal peptide in the following way. Antiserum was diluted 1:100 in PBS and adsorbed to nitrocellulose filters for 1 h. The filters were then blocked with 2% gelatin in PBS for 1 h, and then the maxicell lysates were adsorbed against the filters for 1 h at 37°C before being tested in the λ nuclease assay. An antiserum to an unrelated protein was adsorbed to negative control filters.
Immunoblot analysis.
Immunoblots were prepared from M. pulmonis χ1048 whole cells or trypsin-treated cells by the method of Towbin et al. (39). To treat M. pulmonis with trypsin, cells were prepared from overnight cultures by being washed twice with PBS and then resuspended to a protein concentration of 3 mg per ml. The cell suspension was then treated with 100 μg of trypsin for 1 h, and samples were taken at 10-min intervals for analysis. Gels and nitrocellulose blots were prepared by using the Bio-Rad mini-Protean II system. A 10-μg portion of protein was loaded per well.
Nucleotide sequence accession number.
The nucleotide sequence of the mnuA gene of M. pulmonis has been assigned GenBank accession no. U38841.
RESULTS
Identification of λ phage expressing mycoplasma nuclease activities.
The genomic library of M. pulmonis representing 1,920 independent, recombinant λ with an average insert size of 10 to 12 kb was screened with rabbit hyperimmune anti-membrane nuclease antiserum as described previously (21). Although this antiserum was not raised against purified nucleases per se, it was useful to enrich for a population of recombinant phages of which a higher percentage expressed M. pulmonis nucleases than that of the original library. A total of 19 immunopositive phages were identified, 4 of which expressed nuclease activity by the λ assay. One of these phages, 13.2B, was chosen for further study based on the ability of a subcloned fragment to maintain nuclease activity in E. coli.
Cloning and functional analysis of the chromosomal fragments from nuclease-positive phage.
The four chromosomal fragments from the nuclease-positive recombinant phage were subcloned, generating the plasmids described in Table 1 and Fig. 1. Each of the plasmids was transformed into the opal suppressor strain ISM612, which had no endogenous nuclease activity in our λ assay (data not shown). This step was necessary to read through internal UGA codons in the gene sequences in order to express functional gene products in E. coli. When ISM612 containing plasmid pISM4170 was analyzed for expression of nuclease activity, the activity was present in 10% or less of the independent colonies. This loss of phenotype was not the result of noticeable plasmid deletions or plasmid loss. Therefore, it was assumed that the nuclease gene in the cloned fragment was undergoing a high rate of mutation in the high-copy-number plasmid background, and this plasmid was not studied further. It is possible that the use of a low-copy-number plasmid vector would enhance the stability of these fragments in E. coli, which will be examined in future studies.
FIG. 1.
Physical maps of the chromosomal inserts in recombinant phage. The subcloned regions and plasmid designations are also given. The scale at the top is in kilobases. Symbols: A, ApaI; B, BamHI; Bs, BssHII; C, ClaI; E, EcoRI; Ev, EcoRV; K, KpnI; H, HindIII; P, PstI, S, SacI; Sm, SmaI.
Interestingly, all attempts to transform plasmid pISM4172 into strain ISM612 failed. Since this instability could arise from efficient opal suppression of internal UGA codons, resulting in high levels of mycoplasma nuclease expression in the trpT-prfB3 background (36), less efficient suppressor strains were constructed. These consisted of pISM3001 derivatives of two E. coli strains lacking the prfB3 mutant allele, LE392 and XL1-Blue. The resulting strains, ISM614 and ISM647, were screened for endogenous nuclease activity which might interfere with mapping of the nuclease structural gene by transposon mutagenesis. Both E. coli strains lacking the prfB3 mutation supported the replication of plasmid pISM4172, but only ISM647 lacked nuclease activity as monitored by the λ assay (data not shown). More than 90 to 95% of the independent isolates of ISM647 pISM4172 contained nuclease activity (data not shown). Thus, the stable nuclease-positive background needed for transposon mutagenesis and mapping of the nuclease-encoding sequences was obtained.
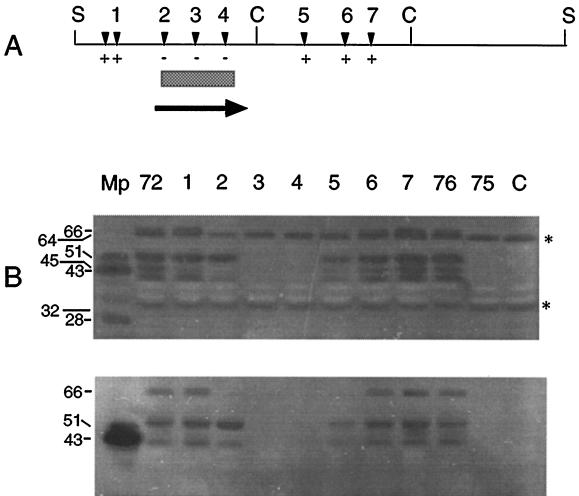
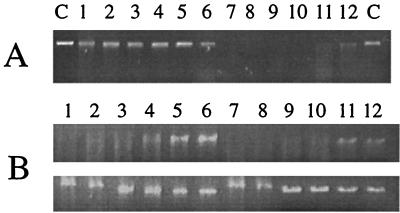
To locate the nuclease structural gene within the 7.0-kb insert of pISM4172, the plasmid was subjected to Tn1000 mutagenesis as described previously (37). The resulting insertions were restriction mapped, and selected plasmids representing insertions across the entire cloned fragment were screened for the loss of nuclease activity following transformation into ISM647. Insertions of Tn1000 within a 1.0-kb region of pISM4172 knocked out nuclease activity (Fig. 2A). Plasmid pISM4176 containing the 2.6-kb SacI-ClaI fragment of pISM4172 was nuclease positive in the ISM647 background, while the adjacent ClaI internal fragment in pISM4175 encoded no nuclease activity.
FIG. 2.
Analysis of Tn1000 inserts in pISM4172. Plasmid pISM4172 was subjected to Tn1000 mutagenesis and analyzed as described in the text. (A) The locations of selected Tn1000 inserts in pISM4172 are designated by inverted solid triangles. Below the map are the results of the λ DNA assay. Inserts that knocked out nuclease activity in the λ assay are designated −, and those that had no effect on nuclease activity are designated +. The bar indicates the minimal coding region encoding mnuA as defined by Tn1000 mutagenesis. The arrow indicates an ORF determined by DNA sequence analysis and its direction of transcription. (B) Analysis of Tn1000 inserts by an SDS-PAGE nuclease gel assay was performed as described in the text. The gels were digitized with a model 4900 high-performance charge-coupled device camera (Cohu, Inc., San Diego, Calif.) and a Macintosh IIci equipped with a Scion Corp. (Frederick, Md.) video board. The resulting TIFF file was cropped with and assembled in Adobe Photoshop and labeled in Aldus FreeHand. The apparent molecular weight of the nucleolytic bands is shown on the left in thousands. The asterisks indicate nucleolytic bands arising from E. coli with specificity for salmon sperm DNA. All lanes contained 5 μg of E. coli ISM647 protein except for the lane labeled Mp, which contained 116 ng (upper gel) or 175 ng (lower gel) of M. pulmonis protein. The upper gel contained salmon sperm DNA, and the lower gel contained λ DNA. Mp, M. pulmonis χ1048 antigen. The remaining lanes contain protein from E. coli ISM647 containing the corresponding plasmid (72, pISM4172; 76, pISM4176; 75, pISM4175; C, pMOB) or pISM4172 with Tn1000 insertions (lanes 1 to 7) shown in panel A.
DNA sequencing and computer analysis.
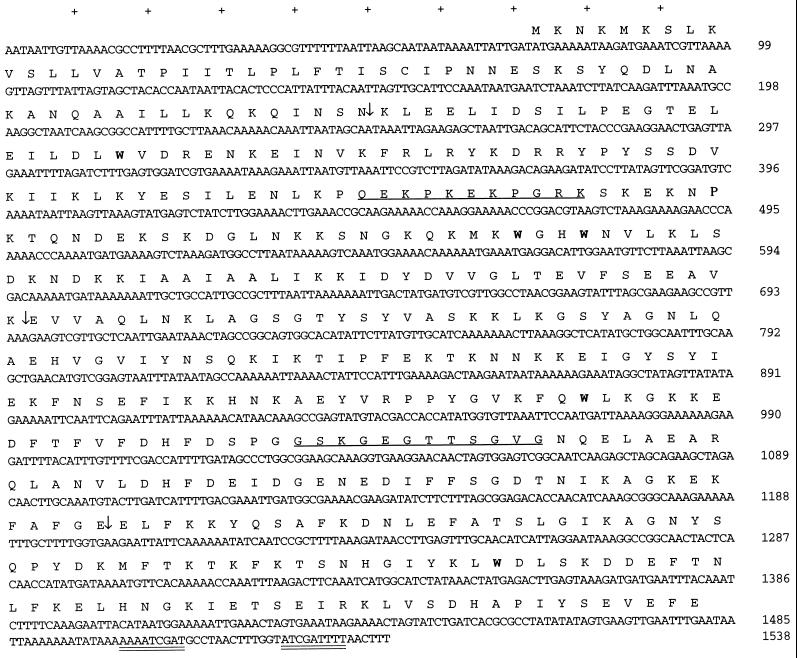
Analysis of the DNA sequence across the region that coded for nuclease activity identified an open reading frame (ORF), designated mnuA, of 1,410 bp coding for a 470-amino-acid protein of 53,739 Da (Fig. 3). Tn1000 insertions upstream and downstream of this ORF failed to eliminate nuclease activity in the cloned sequence (Fig. 2). No Shine-Dalgarno-like sequence was found upstream of mnuA. Analysis of the predicted translated product of the mnuA DNA sequence shows an amino-terminal region rich in lysine residues followed by a 42-amino-acid hydrophobic region and another lysine residue which could serve as a membrane-spanning region to anchor the protein. The charged-hydrophobic-charged domains in the amino-terminal region resembled the principal features of bacterial signal peptides (28). The single cysteine residue was associated with a T-I-S-C motif near the amino terminus, previously reported to be a procaryotic prolipoprotein signal peptidase II recognition sequence (3, 42). The remaining portion of the molecule was extensively hydrophilic with substantial alpha-helical character. The estimated pI of the translated product was 9.04. There were five UGA codons coding for tryptophan (Fig. 3).
FIG. 3.
Nucleotide and deduced amino acid sequences of the M. pulmonis mnuA gene. An inverted repeat is doubly underlined at the 3′ end of the mnuA sequence. The single underline indicates peptides used to produce anti-MnuA antisera. (↓) indicates the sites of Tn1000 insertions described in the manuscript. Boldface type indicates tryptophan-encoding UGA codons.
BLASTN and FASTA searches were performed on the mnuA sequence, and there were short stretches of amino acid similarity (10 to 15 amino acids) with the CpG methylase of Spiroplasma citri. A BLASTP search of the SwissProt database also indicated a significant homology at the amino acid level to an M. pneumoniae protein of unknown function, P01_orf474. There was also a low level of homology at the nucleotide level, which may explain the reaction of the mnuA-specific probe with M. pneumoniae DNA (see Fig. 6). These data suggest that ORF 474 in M. pneumoniae may code for a nuclease, possibly the nuclease identified in previous studies (20). There was no other significant homology to known DNA or protein sequences. There was also no apparent promoter region, but the upstream region was AT rich with several potential TATA boxes and possible ribosomal binding sites, which could account for the promoter activity observed in E. coli. Transcription termination most probably occurs at an inverted repeat sequence downstream.
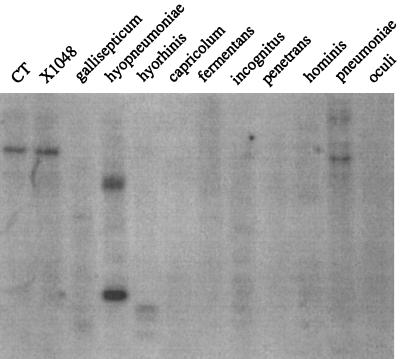
FIG. 6.
Hybridization analysis of 11 Mycoplasma species with a mnuA-specific probe. The hybridization conditions are described in Materials and Methods. Lanes: CT, M. pulmonis CT; χ1048, M. pulmonis χ1048; gallisepticum, M. gallisepticum R; hyopneumoniae, M. hyopneumoniae 232; hyorhinis, M. hyorhinis GDL; capricolum, M. capricolum ATCC 27343; fermentans, M. fermentans PG-18; incognitus, M. fermentans incognitus; penetrans, M. penetrans; hominis, M. hominis 1620; pneumoniae, M. pneumoniae ATCC 15531, oculi, A. oculi ISM1499.
Analysis of mnuA products produced in E. coli.
Four products demonstrating apparent molecular masses of 66, 53, 45, and 39 kDa were expressed in E. coli ISM647 from the chromosomal fragment containing mnuA (Fig. 2). Inclusion of different DNA substrates in the SDS-PAGE gels, i.e., heat-denatured salmon sperm DNA or λ DNA, allowed a clear differentiation between host-encoded nuclease activity and plasmid-encoded activity in this assay. The band at 45 kDa, however, was weak and was sometimes missing (Fig. 2B, lower panel, lanes 72 and 1). The 45- and 66-kDa bands appeared to be related, since the Tn1000 insertion at position 175 of the mnuA gene sequence (insertion 2) knocked out the expression of both bands. The 53- and 39-kDa nucleolytic products produced with an insertion at position 2 (lane 2) may be derived from internal transcriptional initiation within the structural gene at a site downstream of this insertion. The 45- and 39-kDa products may result from premature truncation at the last UGA codon. Tn1000 insertions located further downstream of insertion 2 but within mnuA (locations 3 and 4) knocked out all nucleolytic bands in the SDS-PAGE nuclease gels. Correlation of the λ assay results with the SDS-PAGE nuclease gel results shown in Fig. 2 shows that mnuA translation products which lack the amino terminus are inactive in whole-cell lysates. Some nuclease activity could be regained in these truncated products, however, in the SDS-PAGE nuclease assay (Fig. 2B, lower panel, lane 2).
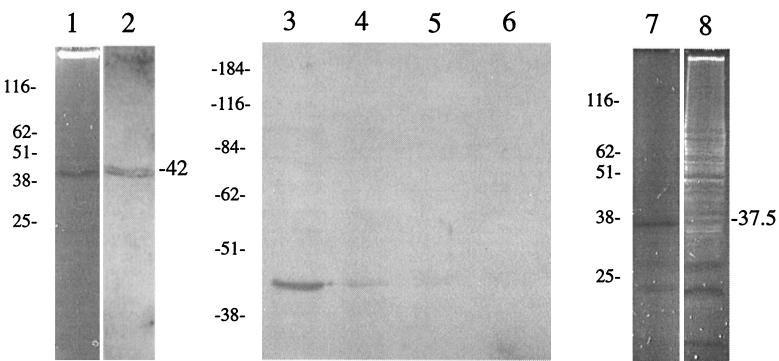
To confirm that the nuclease activity was associated with the product of mnuA and that it was not acting as an activator of an E. coli gene, maxicell experiments were performed. Previous studies have shown that this approach is useful for the identification of plasmid-encoded proteins and that E. coli chromosomal genes are not expressed under these conditions (33). The maxicell strain CSR 603 was transformed with plasmids pISM3001 (opal suppressor) and either pSK (negative control) or pISM4176 (mnuA) and subjected to UV inactivation. Even short exposure times (15 s) under our experimental conditions reduced the viability of this strain to undetectable levels (data not shown). When lysates prepared from these cells were analyzed for nuclease activity by the λ assay, only strains containing pISM4176 (mnuA) showed measurable nuclease activity (Fig. 4). The variation in the size of the λ DNA substrate in the first two lanes of each dilution series in the lower panel of Fig. 4B is due to the high protein concentrations in those samples distorting the migration of the DNA in the gel. As the E. coli lysate is diluted twofold in the assay, this effect disappears. The nuclease activity could also be visualized on SDS-PAGE nuclease gels (Fig. 5). The amount of nuclease activity as measured by the λ DNA assay was independent of the UV dose, indicating that the nuclease was plasmid encoded (33) (data not shown). Mouse antisera raised against the N-terminal peptide of MnuA reacted poorly to a single band of approximately 47 kDa on immunoblots with M. pulmonis antigen even though the enzyme-linked immunosorbent assay titers varied from 1:50,000 to 1:100,000 (data not shown). Antisera raised against the C-terminal peptide, however, reacted more strongly to the same band (Fig. 5). The nuclease band produced in CSR 603 (37.5 kDa) was smaller than the product produced in M. pulmonis (47 kDa). It is likely that this difference in size is due to the aberrant transcription-translation often observed in E. coli with mycoplasma gene sequences (12, 23) or in premature truncation due to inefficient suppression.
FIG. 4.
Analysis of E. coli products by the λ assay. The preparation of E. coli extracts were described in Materials and Methods. Following treatment, the extracts were diluted twofold and incubated with λ DNA for 30 min. The reaction products were analyzed on a 0.7% agarose gel. (A) Analysis of CSR 603 extracts following UV irradiation to produce maxicells. Lanes: C, control λ DNA, 1 to 6, twofold dilutions of CSR 603 pISM3001 pSK maxicell extract; 7 to 12, twofold dilutions of CSR 603 (pISM3001/pISM4176) maxicell extract. (B) Analysis of ISM647 (pISM3001/pISM4176) and CSR 603 (pISM3001/pISM4176) extracts following adsorption with anti-MnuA C-terminal peptide antiserum. Upper panel, nonadsorbed extracts. Lower panel, extracts adsorbed with anti-MnuA C-terminal peptide antiserum bound to nitrocellulose. Lanes: 1 to 6, twofold dilutions of ISM647 (pISM3001/pISM4176) extract; 7 to 12, twofold dilutions of CSR 603 (pISM3001/pISM4176) extract.
FIG. 5.
Functional and immunoblot analysis of mnuA gene products. Lanes: 1, SDS-PAGE nuclease gel containing M. pulmonis antigen; 2, immunoblot analysis of an identical gel containing M. pulmonis antigen with anti-MnuA C-terminal peptide antiserum; 3 to 6, immunoblot analysis with anti-MnuA C-terminal peptide antiserum of M. pulmonis whole cells treated with trypsin for 0 (lane 3), 10 (lane 4), 20 (lane 5) and 40 (lane 6) min, respectively; 7, SDS-PAGE nuclease gel analysis of maxicell extracts from CSR 603 pISM3001 pISM4176; 8, SDS-PAGE nuclease gel analysis of maxicell extracts from CSR 603 pISM3001 pSK (negative control). Molecular weight markers are shown on the left of each panel (in thousands).
Presence of mnuA-like sequences in other Mycoplasma species.
The internal EcoRV fragment of mnuA was used as a probe to determine if this gene was shared among 11 different Mycoplasma species. As shown in Fig. 6, there was cross-hybridization between the mnuA probe and chromosomal fragments from M. hyopneumoniae and M. pneumoniae. There is only one copy of mnuA in the M. pulmonis chromosome, and the gene is located in a similar location in both M. pulmonis CT and M. pulmonis χ1048, the latter being the strain from which mnuA was isolated.
DISCUSSION
This work represents the first cloning and analysis of a membrane nuclease gene, mnuA, from a mycoplasma. It seems reasonable that mnuA codes for a membrane-associated nuclease for the following reasons. First, all M. pulmonis nuclease activities observed in the SDS-PAGE nuclease gels have been shown previously to be associated with the membrane (19). This cloned sequence clearly produces a nuclease that survives the SDS-PAGE sample treatment in an identical fashion (19, 20). Not all nucleases survive this treatment and are visualized by the SDS-PAGE assay, since E. coli is known to contain numerous nucleases of various specificities. These nucleases are inactivated by our sample treatment and, fortuitously, do not interfere with the analysis of the mnuA gene products (Fig. 2). Second, the DNA sequence clearly shows a prominent membrane-spanning domain in the amino terminus and has a lipoprotein signal sequence that has been shown to function in M. hyorhinis (6). This has been considered by other investigators to be strong evidence that the cloned gene codes for a membrane-associated protein. Third, the MnuA-specific peptide antiserum that reacted with a single nucleolytic band on immunoblot analysis, and this band was sensitive to trypsin treatment of whole cells (Fig. 5). The inability to construct site-directed mutations in this Mycoplasma species limits the types of studies that can be performed to conclusively correlate the mnuA gene product in E. coli with one of the membrane nucleases in M. pulmonis. Our transposon mutagenesis studies, the immunoblot data with antipeptide antisera, and the sequence data, however, offer strong support for our hypothesis that mnuA codes for a mycoplasma membrane-associated nuclease.
E. coli has previously been a host for the expression of several nonspecific nucleases, such as those from Staphylococcus aureus (34), Thermus filiformis (9), and Shigella flexneri (7). The staphylococcal nuclease has been extensively studied (35) and is expressed at low levels from its native promoter in E. coli (34). The product is secreted either from its own signal peptide or with a lipoprotein signal sequence attached (26). Higher levels of activity can be obtained when the nuclease is expressed from a λ promoter (13), but in the absence of a functional signal sequence, expression of the nuclease was lethal (38). These studies indicate that expression of heterologous nucleases in E. coli is possible and is a convenient way to study these proteins but that their expression can be problematic and lead to lethal phenotypes.
The studies presented here clearly show that the level of active nuclease expression from the cloned fragments was low in E. coli (Fig. 2). At least 40 times as much protein was required from E. coli as from M. pulmonis to obtain comparable levels of activity on the SDS-PAGE nuclease gel (Fig. 2). The promoter activity that directed the expression of mnuA was associated with the mycoplasmal sequences, because neither λGEM 12 nor pMOB contains promoter-like sequences to regulate the expression of cloned genes. This is common in mycoplasmas (15) and is presumably due to the AT richness of the chromosomal DNA. Low-level expression of the nonspecific nuclease activity might be essential for E. coli and λ viability, since high-level expression could result in damaged chromosomal DNA or damaged λ concatameric DNA and an unsuitable packaging substrate. Thus, cloning strategies that would raise the levels of nuclease product within the cell, i.e., cloning into a high-copy-number cloning vector such as pKS or using a strong external promoter, would increase the likelihood of instability. Since mnuA contained five UGA codons (Fig. 3), expression was partially controlled by regulating suppressor activity in the ISM647 background in comparison to the ISM612 background, which has a chromosomal prfB mutation resulting in constitutive UGA suppression (21).
This low level of expression in E. coli created problems in immunoblot analysis with nuclease-specific antisera (data not shown). The reaction of anti-MnuA peptide antisera with M. pulmonis antigen was weak even with the C-terminal peptide antisera, suggesting that membrane nucleases in mycoplasmas are produced at low levels. In E. coli, the level of MnuA protein was below the level detectable by immunoblot analysis (data not shown), but this antiserum was capable of adsorbing out nucleolytic activity in ISM647 pISM3001/pISM4176 and CSR 603 pISM3001/pISM4176 lysates (data not shown).
Interestingly, both E. coli and M. pulmonis nuclease-banding patterns differed in their SDS-PAGE nuclease gel profiles as a function of the DNA substrate (Fig. 2). E. coli ISM647 contained nuclease bands of 64 and 32 kDa that digested salmon sperm DNA (Fig. 2B, upper panel, lane C) but not λ DNA (lower panel, lane C). This correlated with the results of the λ DNA assay; strain ISM647 had no measurable nuclease activity in that assay. An M. pulmonis-derived nucleolytic band of about 28 kDa was also absent in the gel containing λ DNA (Fig. 2B, lower panel, lane Mp). The lane was loaded with one-third more protein (175 ng) and showed much stronger bands in the 43-kDa region of the gel, but there was clearly no nucleolytic activity in the 28-kDa region when λ DNA was used as a substrate. It is not known what distinguished the substrate specificity of these enzymes. This observation was reproducible and was observed with several different gels. Two possible explanations for the loss of activity with lambda DNA would be differences in the methylation pattern between eucaryotic and procaryotic DNA or single versus double strandedness of the template (the salmon sperm DNA was sheared and boiled prior to inclusion in the SDS-PAGE resolving gel). The mnuA gene products in E. coli, however, were unaffected by the differences in the DNA substrates. Thus, MnuA could be considered a nonspecific nuclease with a broad specificity.
It is possible that MnuA is the 51-kDa protein in the M. pulmonis banding pattern, considering the possibility that the single cysteine residue serves as a signal peptidase cleavage and acylation site (28, 42). The processed polypeptide would be 50,769 Da (not including the fatty acid side chain), but it is not known what effects, if any, DNA might have on the mobility of a DNA binding protein in DNA-containing resolving gels. All nuclease activities in M. pulmonis partition into the detergent phase during Triton X-114 fractionation (data not shown), suggesting that each nuclease has large regions of hydrophobicity or is a lipoprotein. It has been observed that mycoplasmal lipoproteins are improperly processed during expression in E. coli, and thus it is unlikely that the mnuA gene product has been processed correctly. This could affect its size or other biophysical characteristics.
These studies illustrate the difficulties in studying mycoplasma gene sequences in E. coli. Rarely can recombinant mycoplasma gene products be used to complement specific mutations in E. coli or be directly correlated with the gene product from the original host. Most often, the products are truncated prematurely or represent products from internal transcription initiation. For instance, Tn1000 insertion 2 (Fig. 2) knocked out all nuclease activity in cell lysates but not in the SDS-PAGE nuclease assay, which lost only two of the four bands observed with the nonmutated plasmid. The presence of the two bands supports the hypothesis that internal transcriptional initiation within mnuA results in some of the products observed on SDS-PAGE nuclease gels because this insert is located behind the signal sequence of mnuA. These protein products were probably folded incorrectly in the cell lysate but regained a functional conformation during the denaturation and renaturation conditions of the SDS-PAGE nuclease gel assay. Tn1000 inserts further downstream of this aberrant transcription initiation site knocked out all nuclease activity.
Antisera were used in this study to identify nuclease-containing recombinant phages and to correlate the mnuA gene product with nuclease activities. The rabbit antisera allowed us to enrich for a population of recombinant phages containing nuclease-positive recombinants. The 19 recombinant phages identified in the library represent a 100-fold enrichment, which significantly reduced the effort required to identify nuclease-positive recombinant clones. The immunoblotting results with the MnuA-specific peptide antisera directly correlated the mnuA gene product with a specific M. pulmonis nucleolytic band on SDS-PAGE gels (Fig. 5). These peptide antisera were also used to correlate the E. coli nucleolytic gene product with mnuA. This supports our hypothesis that mnuA codes for a membrane nuclease. The maxicell experiments further support this conclusion.
In summary, a membrane nuclease gene from M. pulmonis has been cloned and analyzed. It is not yet clear what role, if any, the MnuA nuclease plays in growth and virulence of M. pulmonis. These nucleases are fully capable of digesting mycoplasmal chromosomal DNA (data not shown), implying that translocation of the mnuA gene product may be tightly coupled to translation (28). Further study of this activity could reveal basic mechanisms of gene expression and protein translocation in these cell wall-less bacteria.
ACKNOWLEDGMENT
We thank M. J. Wannemuehler for his help in producing the MnuA-specific peptide antisera used in this study.
REFERENCES
- 1.Artiushin S, Duvall M, Minion F C. Phylogenetic analysis of mycoplasma strain ISM1499 and its assignment to the Acholeplasma oculi strain cluster. Int J Syst Bacteriol. 1995;45:104–109. doi: 10.1099/00207713-45-1-104. [DOI] [PubMed] [Google Scholar]
- 2.Bendjennat M, Blanchard A, Loutfi M, Montagnier L, Bahraoui E. Purification and characterization of Mycoplasma penetrans Ca2+/Mg2+-dependent endonuclease. J Bacteriol. 1997;179:2210–2220. doi: 10.1128/jb.179.7.2210-2220.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun V, Wu H C. Lipoproteins, structure, function, biosynthesis and model for protein export. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. New York, N.Y: Elsevier Biomedical Press; 1994. pp. 319–341. [Google Scholar]
- 4.Cassell G H, Brown M B. Enzyme-linked immunosorbent assay (ELISA) for detection of anti-mycoplasmal antibody. Methods Mycoplasmol. 1983;1:457–469. [Google Scholar]
- 5.Chou P Y, Fasman G D. Empirical predictions of protein conformations. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- 6.Cleavinger C M, Kim M F, Wise K S. Processing and surface presentation of the Mycoplasma hyorhinis variant lipoprotein VlpC. J Bacteriol. 1994;176:2463–2467. doi: 10.1128/jb.176.8.2463-2467.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Close S M, Kado C I. A gene near the plasmid pSA origin of replication encodes a nuclease. Mol Microbiol. 1992;6:521–527. doi: 10.1111/j.1365-2958.1992.tb01497.x. [DOI] [PubMed] [Google Scholar]
- 8.Finch L R, Mitchell A. Sources of nucleotides. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 211–230. [Google Scholar]
- 9.Fomenkov A, Xu S-Y. Cloning of a gene from Thermus filiformis and characterization of the thermostable nuclease. Gene. 1995;163:109–113. doi: 10.1016/0378-1119(95)00426-7. [DOI] [PubMed] [Google Scholar]
- 10.Garnier J, Osguthorpe D J, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- 11.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsu T, Artiushin S, Minion F C. Cloning and analysis of P97, a respiratory cilium adhesin gene of Mycoplasma hyopneumoniae. J Bacteriol. 1997;179:1317–1323. doi: 10.1128/jb.179.4.1317-1323.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jing G, Liu L, Jiang M, Zou Q, He R. High level expression of staphylococcal nuclease R gene in Escherichia coli. J Biotechnol. 1992;22:271–282. doi: 10.1016/0168-1656(92)90145-y. [DOI] [PubMed] [Google Scholar]
- 14.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 15.Knudtson K L, Minion F C. Use of lac gene fusions in the analysis of Acholeplasma upstream gene regulatory sequences. J Bacteriol. 1993;176:2763–2766. doi: 10.1128/jb.176.9.2763-2766.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 17.Luo W, Lin S-H. Generation of moderate amounts of polyclonal antibodies in mice. BioTechniques. 1997;23:630–632. doi: 10.2144/97234bm16. [DOI] [PubMed] [Google Scholar]
- 18.McGeoch D J. On the predictive recognition of signal peptide sequences. Virus Res. 1985;3:271–286. doi: 10.1016/0168-1702(85)90051-6. [DOI] [PubMed] [Google Scholar]
- 19.Minion F C, Goguen J D. Identification and preliminary characterization of external membrane-bound nuclease activities in Mycoplasma pulmonis. Infect Immun. 1986;51:352–354. doi: 10.1128/iai.51.1.352-354.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minion F C, Jarvill-Taylor K J, Billings D E, Tigges E. Membrane-associated nuclease activities in mycoplasmas. J Bacteriol. 1993;175:7842–7847. doi: 10.1128/jb.175.24.7842-7847.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Minion F C, VanDyk C, Smiley B K. Use of an Escherichia coli enhanced opal suppressor strain to screen a Mycoplasma hyopneumoniae library. FEMS Microbiol Lett. 1995;131:81–85. doi: 10.1016/0378-1097(95)00239-2. [DOI] [PubMed] [Google Scholar]
- 22.Muto A, Andachi Y, Yamao F, Tanaka R, Osawa S. Transcription and translation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J B, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C: American Society for Microbiology; 1992. pp. 331–347. [Google Scholar]
- 23.Notarnicola S M, McIntosh M A, Wise K S. Multiple translational products from a Mycoplasma hyorhinis gene expressed in Escherichia coli. J Bacteriol. 1990;172:2986–2995. doi: 10.1128/jb.172.6.2986-2995.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paddenberg R, Weber A, Wulf S, Mannherz H G. Mycoplasma nucleases able to induce internucleosomal DNA degradation in cultured cells possess many characteristics of eukaryotic apoptotic nucleases. Cell Death Differ. 1998;5:517–528. doi: 10.1038/sj.cdd.4400380. [DOI] [PubMed] [Google Scholar]
- 25.Paddenberg R, Wulf S, Weber A, Heimann P, Beck L, Mannherz H G. Internucleosomal DNA fragmentation in cultured cells under conditions reported to induce apoptosis may be caused by mycoplasma endonucleases. Eur J Cell Biol. 1996;71:105–119. [PubMed] [Google Scholar]
- 26.Pines O, London A. Expression and secretion of staphylococcal nuclease in yeast: effects of amino-terminal sequences. J Gen Microbiol. 1991;137:771–778. doi: 10.1099/00221287-137-4-771. [DOI] [PubMed] [Google Scholar]
- 27.Pollack J D, Williams M V, McElhaney R N. The comparative metabolism of the mollicutes (Mycoplasmas): the utility for taxonomic classification and the relationship of putative gene annotation and phylogeny to enzymatic function in the smallest free-living cells. Crit Rev Microbiol. 1997;23:269–354. doi: 10.3109/10408419709115140. [DOI] [PubMed] [Google Scholar]
- 27a.PSORT Website. 1998. http://psort.nibb.ac.jp.
- 28.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raftery L, Egan J, Cline S, Yarus M. Defined set of cloned termination suppressors: in vivo activity of isogenetic UAG, UAA, and UGA suppressor tRNAs. J Bacteriol. 1984;158:849–859. doi: 10.1128/jb.158.3.849-859.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Razin S. The mycoplasmas. Microbiol Rev. 1978;42:414–470. doi: 10.1128/mr.42.2.414-470.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Razin S. Peculiar properties of mycoplasmas—the smallest self-replicating prokaryotes. FEMS Microbiol Lett. 1992;100:423–431. doi: 10.1111/j.1574-6968.1992.tb14072.x. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Sancar A, Hack A M, Rupp D. Simple method for identification of plasmid-encoded proteins. J Bacteriol. 1979;137:692–693. doi: 10.1128/jb.137.1.692-693.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shortle D. A genetic system for analysis of staphylococcal nuclease. Gene. 1983;22:181–189. doi: 10.1016/0378-1119(83)90102-6. [DOI] [PubMed] [Google Scholar]
- 35.Shortle D. Staphylococcal nuclease: a showcase of m-value effects. Adv Protein Chem. 1995;46:217–247. doi: 10.1016/s0065-3233(08)60336-8. [DOI] [PubMed] [Google Scholar]
- 36.Smiley B K, Minion F C. Enhanced readthrough of opal (UGA) codons and production of Mycoplasma pneumoniae P1 epitopes in Escherichia coli. Gene. 1993;134:33–40. doi: 10.1016/0378-1119(93)90171-x. [DOI] [PubMed] [Google Scholar]
- 37.Strathmann M, Hamilton B A, Mayeda C A, Simon M I, Meyerowitz E M, Palazzolo M J. Transposon-facilitated DNA sequencing. Proc Natl Acad Sci USA. 1991;88:1247–1250. doi: 10.1073/pnas.88.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takahara M, Hibler D W, Barr P J, Gerlt J A, Inouye M. The ompA signal peptide directed secretion of nuclease A by Escherichia coli. J Biol Chem. 1985;260:2670–2674. [PubMed] [Google Scholar]
- 39.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some application. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tully J G. General cultivation techniques for mycoplasmas and spiroplasmas. Methods Mycoplasmol. 1983;1:99–102. [Google Scholar]
- 41.von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986;14:4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yogev D, Watson-McKown R, Rosengarten R, Im J, Wise K S. Increased structural and combinatorial diversity in an extended family of genes encoding Vlp surface proteins of Mycoplasma hyorhinis. J Bacteriol. 1995;177:5636–5643. doi: 10.1128/jb.177.19.5636-5643.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]