Abstract
Multiple myeloma (MM) is a hematological malignancy in which monoclonal plasma cells multiply in the bone marrow and monoclonal immunoglobulins are overproduced in older people. Several molecular and cytogenetic advances allow scientists to identify several genetic and chromosomal abnormalities that cause the disease. The comprehension of the pathophysiology of MM requires an understanding of the characteristics of malignant clones and the changes in the bone marrow microenvironment. This study aims to identify the central genes and to determine the key signaling pathways in MM by in silico approaches. A list of 114 differentially expressed genes (DEGs) is important in the prognosis of MM. The DEGs are collected from scientific publications and databases (https://www.ncbi.nlm.nih.gov/). These data are analyzed by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) software (https://string-db.org/) through the construction of protein-protein interaction (PPI) networks and enrichment analysis of the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, by CytoHubba, AutoAnnotate, Bingo Apps plugins in Cytoscape software (https://cytoscape.org/) and by DAVID database (https://david.ncifcrf.gov/). The analysis of the results shows that there are 7 core genes, including TP53; MYC; CDND1; IL6; UBA52; EZH2, and MDM2. These top genes appear to play a role in the promotion and progression of MM. According to functional enrichment analysis, these genes are mainly involved in the following signaling pathways: Epstein-Barr virus infection, microRNA pathway, PI3K-Akt signaling pathway, and p53 signaling pathway. Several crucial genes, including TP53, MYC, CDND1, IL6, UBA52, EZH2, and MDM2, are significantly correlated with MM, which may exert their role in the onset and evolution of MM.
Keywords: Multiple myeloma, gene expression, heterogeneity, mutational profiles, genetic predisposition, bioinformatics
Introduction
Multiple myeloma (MM) is one of the incurable hematological diseases, characterized by the proliferation of abnormal plasma cells with distinct cytogenetic characteristics in the bone marrow, representing about 10% to 15% of all hematopoietic tumors. 1 The International Agency for Research on Cancer (IARC) estimates a worldwide incidence of 160 000 cases and a worldwide mortality of 106 000 patients in 2018. 2 However, men are much more affected by this disease than women. Moreover, the median age of patients at diagnosis is about 66 to 70 years old, with 37% of patients being younger than 65 years old. 3
Multiple myeloma is a complex genomic landscape and it is characterized by many types of chromosomal aberrations. Hyperdiploidy and immunoglobulin heavy chain (IGH) translocations are included as early occurrences. They are present in the precursor stages of monoclonal gammopathy of undetermined significance (MGUS) and latent multiple myeloma and are completely clonal in the majority of cases. Hyperdiploidy is considered as the first type of copy number alteration commonly seen in MM. It is usually associated with a standard risk prognosis. It is also characterized by trisomies of 3 or more of the odd-numbered chromosomes, namely, 3, 5, 7, 9, 11, 15, 19, and 21, whereas the most common IGH translocations are t(4; 14), t(11; 14), t(14; 16), t(14; 20), and t(6; 14) and its prognostic impact depends largely on the partner chromosome. 4 Other alterations in copy number, gains, and losses of the whole chromosome or part of the chromosome occur in the later stages of the disease trajectory and are not considered triggering events. The exception is the gain of the first copy of 1q, which appears to be an early event, whereas subsequent additional gains of 1q are later events. Common gains and losses include del 1p, gain 1q, del 13/13q, del 17p, in addition to del 16q and del 12p. Moreover, point mutations occur later in MM and commonly affect the MAPK pathway, NFKB pathway, DNA damage and repair, plasma cell differentiation, MYC activation, regulation of gene expression, and the cell cycle pathway. 5 These mutations can arise in various subclones, and their influence on disease progression varies depending on environmental factors. 6 KRAS, NRAS, and BRAF were among the most commonly mutated genes involved in the MAPK pathway, where hot-spot activating mutations have been discovered and were present in roughly 40% to 50% of patients with newly diagnosed multiple myeloma. Recurrent mutations have also been observed in FAM46C, CYLD, DIS3, IRF4, TRAF3, TP53, RB1, LTB, SP140, and more, and mutations in therapeutic targets, namely, lenalidomide targets (CRBN, IKZF1, and IKZF3), proteasome subunits, and steroid receptor (NR3C1), can induce treatment resistance. The combination of all these chromosomal aberrations and gene mutations leads to a differential gene expression profile in the plasma cells of patients with MM. 7
Bioinformatics is one of the newest fields of biological research, which should be broadly considered as the use of mathematics, statistics, and informatics to process and analyze biological data.8,9 In addition, it is an important element in laboratories that generate, analyze, store, and interpret data from molecular genetic tests. 10 The present study aims to predict interactions between differentially expressed genes (DEGs) to identify central genes and determine key signaling pathways in MM to understand its genetic heterogeneity.
Materials and Methods
To accomplish this integrative analysis, a pipeline was used (Figure 1).
Figure 1.
Pipeline chart of all study analysis steps. DEG indicates differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; PPI, protein-protein interaction.
A set of 114 DEGs in myeloma plasma cells involved in the progression of the disease were collected from different scientific publications11 -47 and databases (https://www.ncbi.nlm.nih.gov/). The following search terms were used: multiple myeloma, gene expression, heterogeneity, mutational profiles, and genetic predisposition.
We used the GenBank database, available at the following link (https://www.ncbi.nlm.nih.gov/gene) and GeneCards (https://www.genecards.org/) to match each gene to its ID and functional annotation (Table 1).
Table 1.
Differentially expressed genes (DEGs) in multiple myeloma selected from various databases and scientific publications.
| Gene ID | Gene symbols | Gene names | Locus | Description |
|---|---|---|---|---|
| 2 | A2M | Alpha-2-macroglobulin | 12p13.31 | https://www.ncbi.nlm.nih.gov/gene/2 |
| 84517 | ACTRT3 | Actin-related protein T3 | 3q26.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=ACTRT3 |
| 103 | ADAR | Adenosine deaminase RNA specific | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/103 |
| 57379 | AICDA | Activation induced cytidine deaminase | 12p13.31 | https://www.ncbi.nlm.nih.gov/gene/57379 |
| 242 | ALOX12B | Arachidonate 12-lipoxygenase, 12R type | 17p13.1 | https://www.ncbi.nlm.nih.gov/gene/242 |
| 81611 | ANP32E | Acidic nuclear phosphoprotein 32 family member E | 1q21.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=ANP32E |
| 328 | APEX1 | Apurinic/apyrimidinic endodeoxyribonuclease 1 | 14q11.2 | https://www.ncbi.nlm.nih.gov/gene/328 |
| 27350 | APOBEC3C | Apolipoprotein B mRNA editing enzyme catalytic subunit 3C | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/27350 |
| 140564 | APOBEC3D | Apolipoprotein B mRNA editing enzyme catalytic subunit 3D | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/140564 |
| 200316 | APOBEC3F | Apolipoprotein B mRNA editing enzyme catalytic subunit 3F | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/200316 |
| 60489 | APOBEC3G | Apolipoprotein B mRNA editing enzyme catalytic subunit 3G | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/60489 |
| 164668 | APOBEC3H | Apolipoprotein B mRNA editing enzyme catalytic subunit 3H | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/164668 |
| 596 | BCL2 | B-cell lymphoma 2 | 18q21.33 | https://www.ncbi.nlm.nih.gov/gene/596 |
| 607 | BCL9 | B-cell lymphoma 9 | 1q21.2 | https://www.ncbi.nlm.nih.gov/gene/607 |
| 29760 | BLNK | B-cell linker | 10q24.1 | https://www.ncbi.nlm.nih.gov/gene/29760 |
| 23476 | BRD4 | Bromodomain containing 4 | 19p13.12 | https://www.ncbi.nlm.nih.gov/gene/23476 |
| 716 | C1S | Complément C1S | 12p13.31 | https://www.ncbi.nlm.nih.gov/gene/716 |
| 23492 | CBX7 | Chromobox 7 | 22q13.1 | https://www.ncbi.nlm.nih.gov/gene/23492 |
| 100507056 | CCAT1 | Colon cancer–associated transcript 1 | 8q24.21 | https://www.ncbi.nlm.nih.gov/gene/100507056 |
| 90835 | CCDC189 or C16orf93 |
Coiled-coil domain containing 189 | 16p11.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=CCDC189 |
| 595 | CCND1 | Cyclin D1 | 11q13.3 | https://www.ncbi.nlm.nih.gov/gene/595 |
| 894 | CCND2 | Cyclin D2 | 12p13.32 | https://www.ncbi.nlm.nih.gov/gene/894 |
| 896 | CCND3 | Cyclin D3 | 6p21.1 | https://www.ncbi.nlm.nih.gov/gene/896 |
| 928 | CD9 | CD9 molecule | 12p13.31 | https://www.ncbi.nlm.nih.gov/gene/928 |
| 948 | CD36 | CD36 molecule | 7q21.11 | https://www.ncbi.nlm.nih.gov/gene/948 |
| 973 | CD79A | CD79a molecule | 19q13.2 | https://www.ncbi.nlm.nih.gov/gene/973 |
| 975 | CD81 | CD81 molecule | 11p15.5 | https://www.ncbi.nlm.nih.gov/gene/975 |
| 55536 | CDCA7L | Cell division cycle associated 7 like | 7p15.3 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDCA7L |
| 153241 | CEP120 | Centrosomal protein 120 | 5q23.2 | https://www.ncbi.nlm.nih.gov/gene/153241 |
| 54480 | CHPF2 | Chondroitin polymerizing factor 2 | 7q36.1 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=CHPF2 |
| 91851 | CHRDL1 | Chordin-like 1 | Xq23 | https://www.ncbi.nlm.nih.gov/gene/91851 |
| 1163 | CKS1B | CDC28 protein kinase regulatory subunit 1B | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/1163 |
| 1164 | CKS2 | CDC28 protein kinase regulatory subunit 2 | 9q22.2 | https://www.ncbi.nlm.nih.gov/gene/1164 |
| 4094 | c-MAF or MAF | MAF bZIP transcription factor | 16q23.2 | https://www.ncbi.nlm.nih.gov/gene/4094 |
| 1380 | CR2 | Complement C3d receptor 2 | 1q32.2 | https://www.ncbi.nlm.nih.gov/gene/1380 |
| 55790 | CSGALNACT1 | Chondroitin sulfate N-acetylgalactosaminyltransferase 1 | 8p21.3 | https://www.ncbi.nlm.nih.gov/gene/55790 |
| 1521 | CTSW | Cathepsin W | 11q13.1 | https://www.ncbi.nlm.nih.gov/gene/1521 |
| 1545 | CYP1B1 | Cytochrome P450 family 1 subfamily B member 1 | 2p22.2 | https://www.ncbi.nlm.nih.gov/gene/1545 |
| 1634 | DCN | Decorin | 12q21.33 | https://www.ncbi.nlm.nih.gov/gene/1634 |
| 79961 | DENND2D | DENN domain containing 2D | 1p13.3-p13.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=DENND2D |
| 22894 | DIS3 | DIS3 homolog, exosome endoribonuclease | 13q21.33 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=DIS3 |
| 1788 | DNMT3A | DNA methyltransferase 3 alpha | 2p23.3 | https://www.ncbi.nlm.nih.gov/gene/1788 |
| 27335 | EIF3K | Eukaryotic translation initiation factor 3 subunit K | 19q13.2 | https://www.ncbi.nlm.nih.gov/gene/27335 |
| 22936 | ELL2 | Elongation factor for RNA polymerase II 2 | 5q15 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=ELL2 |
| 2071 | ERCC3 | ERCC excision repair 3, TFIIH core complex helicase subunit | 2q14.3 | https://www.ncbi.nlm.nih.gov/gene/2071 |
| 2146 | EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit | 7q36.1 | https://www.ncbi.nlm.nih.gov/gene/2146 |
| 2167 | FABP4 | Fatty acid binding protein 4 | 8q21.13 | https://www.ncbi.nlm.nih.gov/gene/2167 |
| 2200 | FBN1 | Fibrillin 1 | 15q21.1 | https://www.ncbi.nlm.nih.gov/gene/2200 |
| 55294 | FBXW7 | F-box and WD repeat domain containing 7 | 4q31.3 | https://www.ncbi.nlm.nih.gov/gene/55294 |
| 2261 | FGFR3 | Fibroblast growth factor receptor 3 | 4p16.3 | https://www.ncbi.nlm.nih.gov/gene/2261 |
| 3006 | H1-2 | H1.2 linker histone, cluster member | 6p22.2 | https://www.ncbi.nlm.nih.gov/gene?term=(hist1h1c[gene])%20AND%20(Homo%20sapiens[orgn])%20AND%20alive[prop]%20NOT%20newentry[gene]&sort=weight |
| 3105 | HLA-A | Major histocompatibility complex, class I, A | 6p22.1 | https://www.ncbi.nlm.nih.gov/gene/3105 |
| 3113 | HLA-DPA1 | Major histocompatibility complex, class II, DP alpha 1 | 6p21.32 | https://www.ncbi.nlm.nih.gov/gene/3113 |
| 3213 | HOXB3 | Homeobox B3 | 17q21.32 | https://www.ncbi.nlm.nih.gov/gene/3213 |
| 3479 | IGF1 | Insulin-like growth factor 1 | 12q23.2 | https://www.ncbi.nlm.nih.gov/gene/3479 |
| 3488 | IGFBP5 | Insulin-like growth factor–binding protein 5 | 2q35 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=IGFBP5 |
| 3514 | IGKC | Immunoglobulin kappa constant | 2p11.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=IGKC |
| 3569 | IL6 | Interleukin 6 | 7p15.3 | https://www.ncbi.nlm.nih.gov/gene/3569 |
| 3570 | IL6R | Interleukin 6 receptor | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/3570 |
| 3608 | ILF2 | Interleukin enhancer binding factor 2 | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/3608 |
| 3662 | IRF4 | Interferon regulatory factor 4 | 6p25.3 | https://www.ncbi.nlm.nih.gov/gene/3662 |
| 55818 | KDM3A | Lysine demethylase 3A | 2p11.2 | https://www.ncbi.nlm.nih.gov/gene/55818 |
| 10365 | KLF2 | Kruppel-like factor 2 | 19p13.11 | https://www.ncbi.nlm.nih.gov/gene/10365 |
| 3936 | LCP1 | Lymphocyte cytosolic protein 1 | 13q14.13 | https://www.ncbi.nlm.nih.gov/gene/3936 |
| 4023 | LPL | Lipoprotein lipase | 8p21.3 | https://www.ncbi.nlm.nih.gov/gene/4023 |
| 151827 | LRRC34 | Leucine-rich repeat containing 34 | 3q26.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=LRRC34 |
| 344657 | LRRIQ4 | Leucine-rich repeats and IQ motif containing 4 | 3q26.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=LRRIQ4 |
| 389692 | MAFA | MAF bZIP transcription factor A | 8q24.3 | https://www.ncbi.nlm.nih.gov/gene/389692 |
| 9935 | MAFB | MAF bZIP transcription factor B | 20q12 | https://www.ncbi.nlm.nih.gov/gene/9935 |
| 9500 | MAGED1 | MAGE family member D1 | Xp11.22 | https://www.ncbi.nlm.nih.gov/gene/9500 |
| 4170 | MCL1 | Apoptosis regulator, BCL2 family member | 1q21.2 | https://www.ncbi.nlm.nih.gov/gene/4170 |
| 4193 | MDM2 | Murine double minute 2 | 12q15 | https://www.ncbi.nlm.nih.gov/gene/4193 |
| 4582 | MUC1 | Mucin 1, cell surface associated | 1q22 | https://www.ncbi.nlm.nih.gov/gene/4582 |
| 4609 | MYC | MYC proto-oncogene | 8q24.21 | https://www.ncbi.nlm.nih.gov/gene/4609 |
| 55892 | MYNN | Myoneurin | 3q26.2 | https://www.ncbi.nlm.nih.gov/gene/55892 |
| 7468 | NSD2 | Nuclear receptor–binding SET domain protein 2 | 4p16.3 | https://www.ncbi.nlm.nih.gov/gene/7468 |
| 5174 | PDZK1 | PDZ domain containing 1 | 1q21.1 | https://www.ncbi.nlm.nih.gov/gene/5174 |
| 10957 | PNRC1 | Proline-rich nuclear receptor coactivator 1 | 6q15 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=PNRC1 |
| 57580 | PREX1 | Phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 | 20q13.13 | https://www.ncbi.nlm.nih.gov/gene/57580 |
| 78994 | PRR14 | Proline-rich 14 | 16p11.2 | https://www.ncbi.nlm.nih.gov/gene/78994 |
| 339105 | PRSS53 | Serine protease 53 | 16p11.2 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRSS53 |
| 5710 | PSMD4 | Proteasome 26S subunit ubiquitin receptor, non-ATPase 4 | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/5710 |
| 23475 | QPRT | Quinolinate phosphoribosyltransferase | 16p11.2 | https://www.ncbi.nlm.nih.gov/gene/23475 |
| 5888 | RAD51 | RAD51 recombinase | 15q15.1 | https://www.ncbi.nlm.nih.gov/gene/5888 |
| 25780 | RASGRP3 | RAS guanyl releasing protein 3 | 2p22.3 | https://www.ncbi.nlm.nih.gov/gene/?term=25780 |
| 1102 | RCBTB2 | RCC1 and BTB domain–containing protein 2 | 13q14.2 | https://www.ncbi.nlm.nih.gov/gene/1102 |
| 55159 | RFWD3 | Ring finger and WD repeat domain 3 | 16q23.1 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=RFWD3 |
| 9810 | RNF40 | Ring finger protein 40 | 16p11.2 | https://www.ncbi.nlm.nih.gov/gene/9810 |
| 6152 | RPL24 | Ribosomal protein L24 | 3q12.3 | https://www.ncbi.nlm.nih.gov/gene/6152 |
| 6161 | RPL32 | Ribosomal protein L32 | 3p25.2 | https://www.ncbi.nlm.nih.gov/gene/6161 |
| 11224 | RPL35 | Ribosomal protein L35 | 9q33.3 | https://www.ncbi.nlm.nih.gov/gene/11224 |
| 6181 | RPLP2 | Ribosomal protein lateral stalk subunit P2 | 11p15.5 | https://www.ncbi.nlm.nih.gov/gene/6181 |
| 6203 | RPS9 | Ribosomal protein S9 | 19q13.42 | https://www.ncbi.nlm.nih.gov/gene/6203 |
| 6223 | RPS19 | Ribosomal protein S19 | 19q13.2 | https://www.ncbi.nlm.nih.gov/gene/6223 |
| 710 | SERPING1 | Serpin family G member 1 | 11q12.1 | https://www.ncbi.nlm.nih.gov/gene/710 |
| 51548 | SIRT6 | Sirtuin 6 | 19p13.3 | https://www.ncbi.nlm.nih.gov/gene/51548 |
| 6635 | SNRPE | Small nuclear ribonucleoprotein polypeptide E | 1q32.1 | https://www.ncbi.nlm.nih.gov/gene/6635 |
| 11262 | SP140 | SP140 nuclear body protein | 2q37.1 | https://www.ncbi.nlm.nih.gov/gene/11262 |
| 6850 | SYK | Spleen-associated tyrosine kinase | 9q22.2 | https://www.ncbi.nlm.nih.gov/gene/6850 |
| 54855 | TENT5C or FAM46C | Terminal nucleotidyltransferase 5C | 1p12 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=TENT5C |
| 7018 | TF | Transferrin | 3q22.1 | https://www.ncbi.nlm.nih.gov/gene/7018 |
| 10043 | TOM1 | Target of myb1 membrane trafficking protein | 22qf12.3 | https://www.ncbi.nlm.nih.gov/gene/10043 |
| 7157 | TP53 | Tumor protein p53 | 17p13.1 | https://www.ncbi.nlm.nih.gov/gene/7157 |
| 57212 | TP73-AS1 | TP73 ARN antisense 1 | 1p36.32 | https://www.genecards.org/cgi-bin/carddisp.pl?gene=TP73-AS1 |
| 7295 | TXN | Thioredoxin | 9q31.3 | https://www.ncbi.nlm.nih.gov/gene/7295 |
| 7311 | UBA52 | Ubiquitin A-52 residue ribosomal protein fusion product 1 | 19p13.11 | https://www.ncbi.nlm.nih.gov/gene/7311 |
| 9898 | UBAP2L | Ubiquitin-associated protein 2 like | 1q21.3 | (https://www.genecards.org/cgi-bin/carddisp.pl?gene=UBAP2L). |
| 55585 | UBE2Q1 | Ubiquitin conjugating enzyme E2 Q1 | 1q21.3 | https://www.ncbi.nlm.nih.gov/gene/55585 |
| 29089 | UBE2T | Ubiquitin conjugating enzyme E2T | 1q32.1 | https://www.ncbi.nlm.nih.gov/gene/29089 |
| 7398 | USP1 | Ubiquitin-specific peptidase 1 | 1p31.3 | https://www.ncbi.nlm.nih.gov/gene/7398 |
| 7874 | USP7 | Ubiquitin-specific peptidase 7 | 16p13.2 | https://www.ncbi.nlm.nih.gov/gene/7874 |
| 7412 | VCAM1 | Vascular cell adhesion molecule 1 | 1p21.2 | https://www.ncbi.nlm.nih.gov/gene/7412 |
| 1462 | VCAN | Versican | 5q14.2-q14.3 | https://www.ncbi.nlm.nih.gov/gene/1462 |
| 10413 | YAP1 | Yes1-associated transcriptional regulator | 11q22.1 | https://www.ncbi.nlm.nih.gov/gene/10413 |
Abbreviation: DEG, differentially expressed genes.
Analysis Software
STRING software analysis
Analysis by STRING version 11.5 (https://string-db.org/) allowed us to reveal key genes and co-expressed genes in MM, to identify enriched biological terms, GO terms, and finally to determine key signaling pathways of core genes. FDR value ⩽ 0.05 was considered as the cut-off criterion, with a confidence score of 0.400 set as the cut-off criterion.
Determination of key genes
The input file, composed of 114 DEGs collected, has been submitted for analysis by STRING. The parameters retained for the analysis were the type of organism (Homo sapiens), type of network (evidence network), and confidence score (<0.400). The analysis was performed without enrichment. The core genes were selected based on their number of interactions with other genes (⩾20 interactions) and their location in the PPI network (at the center).
GO analysis
The GO enrichment analysis includes biological process, cellular component, and molecular function. GO terms have been analyzed from files corresponding to the GO enrichment, then downloaded from the software in tabular separated values (TSV) format, which can be opened in Excel as simple tabular text output. The top GO analysis results were selected with their significant values of FDR.
Identification of co-expressed genes
To determine the co-expressed genes, a file corresponding to the string interaction was downloaded from the software in TSV format, which can be opened in Excel as a simple tabular text output. We allow classifying the co-expressed genes by their scores, which indicates the level of association of the expression data and can also be determined by the presence of a black line between the nodes in the PPI network.
Determination of key signaling pathways
They were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) process in String software and selected with their significant FDR value ⩽ 0.05.
Cytoscape software analysis
To verify the results obtained by String software, Cytoscape version 2.3.8 (https://cytoscape.org/) was used, considering the P value < .05 and a confidence score of 0.400 as cut-off criteria.
Key genes identification
The TSV file of the gene’s interaction was imported into Cytoscape software. The CytoHubba Cytoscape plugin was used to classify the network nodes according to their characteristics. The degree is one of the topological analysis methods provided by CytoHubba, allowing us to observe target genes with higher degrees, which could constitute key genes.
GO enrichment
The Cytoscape BiNGO plugin was used to provide the GO enrichment. The most significant GO terms were selected with their P value < .05 and their number of annotated genes.
Network clusters
Cytoscape software groups PPI networks generated by STRING into clusters, taking the P value < .05 and a confidence score of 0.400 as cut-off criteria. ClusterMaker Apps from AutoAnnotate application was used to perform Markov Clustering (MCL) of the protein network. Markov Clustering makes it possible to visualize the enriched terms in the form of circular plots on the nodes of the network.
DAVID functional annotation bioinformatics microarray analysis
The DAVID database version 6.8 (https://david.ncifcrf.gov/) was used to perform KEGG pathway enrichment, to determine the most significant signaling pathways and compare them with those obtained by STRING. The data were pasted as a list of official gene symbols into the DAVID database.
Results
Network analysis
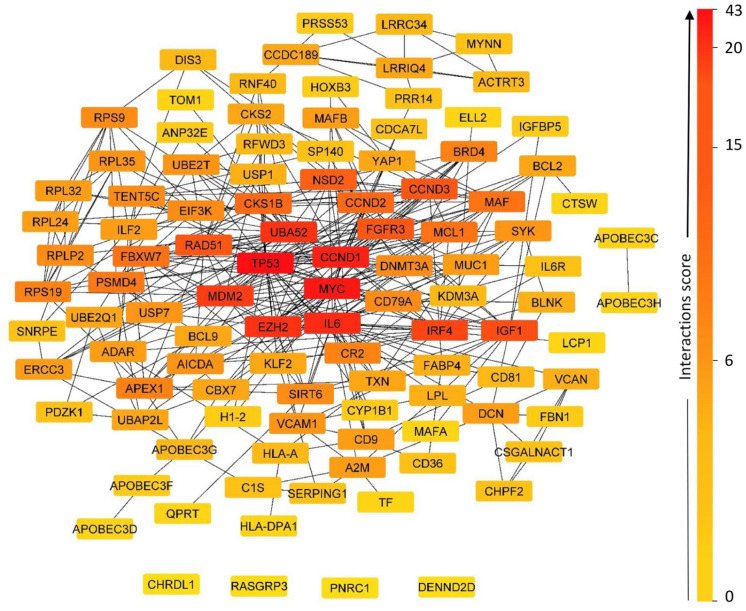
The network analysis of the set of 114 DEG revealed that 110 were annotated by STRING (Figure 2). A central network was obtained encompassing 99 genes, and a small network at the periphery, grouping together 2 genes, APOBEC3H and APOBEC3C. In addition, 9 genes have been outside the core network: RASGRP3; CEP120; MAGED1; CHRDL1; PNRC1; ALOX12B; PREX1; RCBTB2; DENND2D, and 7 core genes constitute the network engine: TP53; MYC CDND1; IL6; UBA52; EZH2; MDM2. In this functional network, all 101 genes represented various interactions, which may be explained by functional links between them.
Figure 2.
Network of protein-protein interaction. The network view (evidence view) summarizes the set of predicted associations for a group of 110 genes. The nodes of the network are the gene product, and the edges represent the predicted functional associations. The edges are represented by lines of different colors that indicate the type of interaction to predict the associations. Clicking on a node will give detailed information about the protein and clicking on an edge will display a detailed breakdown of the evidence.
Co-expression results
From the first network, we determined the co-expression related to the core target genes of MM (Table 2). Key genes have been identified by their scores, which indicate the level of association of the gene expression data. The nodes represented genes and the black line shown between the nodes in the PPI network indicated co-expression (Figure 2). A co-expression score existing between 2 genes represented it. The score indicated the level of association of expression data during a process. If 2 genes showed similar expression under different conditions, it was likely that they were jointly involved in the same process (one requiring the other).
Table 2.
Co-expressed between core genes.
| Node 1 | Node 1 | Score of co-expression |
|---|---|---|
| CCND1 | MYC | 0.069 |
| CCND1 | MUC1 | 0.071 |
| CCND1 | CKS1B | 0.068 |
| CCND1 | FGFR3 | 0.065 |
| CCND1 | CKS2 | 0.065 |
| CCND1 | YAP1 | 0.317 |
| CCND1 | IGFBP5 | 0.098 |
| EZH2 | MYC | 0.091 |
| EZH2 | TP53 | 0.074 |
| EZH2 | WHSC1 | 0.120 |
| EZH2 | USP1 | 0.152 |
| EZH2 | RAD51 | 0.212 |
| IL6 | CYP1B1 | 0.086 |
| IL6 | FABP4 | 0.058 |
| IL6 | VCAM1 | 0.063 |
| MYC | TP53 | 0.070 |
| MYC | RAD51 | 0.069 |
| MYC | CDCA7L | 0.096 |
| MYC | MUC1 | 0.065 |
| MYC | APEX1 | 0.070 |
| MYC | MCL1 | 0.065 |
| MYC | TXN | 0.063 |
| TP53 | RFWD3 | 0.073 |
| TP53 | RAD51 | 0.073 |
| TP53 | CKS1B | 0.076 |
| TP53 | RPS19 | 0.087 |
| TP53 | UBE2T | 0.062 |
| UBA52 | RPL32 | 0.531 |
| UBA52 | RPL35 | 0.323 |
| UBA52 | RPLP2 | 0.259 |
| UBA52 | RPS9 | 0.112 |
| UBA52 | CKS1B | 0.065 |
| UBA52 | RPL24 | 0.211 |
| UBA52 | RPS19 | 0.548 |
The genes marked in bold signify co-expression between 2 key genes.
Among the identified core genes, some of them represented co-expression with each other, including CCND1 with MYC, EZH2 with MYC, EZH2 with TP53, and MYC with TP53. In addition, they also represented co-expression with other genes in the functional network, which were presented in Table 1.
GO enrichment
GO terms were classified in the STRING software according to the value of the false discovery rate and the number of core genes they contained. Moreover, they were classified according to the number of annotated genes in each GO term. Thus, the most important ones had a high number of key genes involved in the development of MM. The GO analysis allowed the obtaining of a total of 448 GO items, including 392 biological process (BP) entries, 23 molecular function (MF) entries, and 33 cellular component (CC) entries (Figure 3).
Figure 3.
Gene Ontology enrichment. The diagram represents the most significant GO terms according to the number of genes involved in the network, which are indicated in parenthesis in the diagram.
GO indicates Gene Ontology.
The most significant biological process determined with FDR values of 7.55e-09; 1.26e-08; 1.26e-08 were as follows: Cellular macromolecule metabolic process (GO:0044260) (64/114 genes), regulation of gene (70/114 genes), respectively.
The most significant molecular function observed with FDR values of 0.00011; 0.00019; 0.00019; were as follows: RNA binding (GO:0003723) (28/114 genes), nucleic acid binding (GO:0003676) (46/114 genes), and protein binding (GO:0005515) (66/114 genes), respectively.
Potentially important target genes have been expressed in the membrane-bounded organelle (GO:0043227), organelle (GO:0043226), and intracellular organelle (GO:0043229), which have a number of annotated genes of 97/114; 100/114; 96/114 and FDR values of 2.80e-06; 1.45e-05; and 1.45e-05, respectively.
Identification of KEGG pathways
KEGG pathway enrichment analyses were performed to reveal potential signaling pathways in the 114 DEGs (Table 3). They were significantly enriched in Epstein-Barr virus (EBV) infection (hsa05169), MicroRNAs in cancer (hsa05206), PI3K-Akt signaling pathway (hsa04151), and p53 signaling pathway (hsa04115), which were considered as the major pathways involved during the development of MM.
Table 3.
KEGG pathways enrichment.
| KEGG pathway | Description | Count in network | Number of central genes | False discovery rate |
|---|---|---|---|---|
| hsa05169 | Epstein-Barr virus infection | 14 | 5 | 4.49e-09 |
| hsa05206 | MicroRNAs in cancer | 11 | 5 | 7.12e-07 |
| hsa04151 | PI3K-Akt signaling pathway | 13 | 5 | 1.85e-05 |
| hsa04115 | p53 signaling pathway | 7 | 3 | 3.33e-05 |
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
The table represents the most important KEGG pathways, selected with their significant FDR value ⩽ 0.05 and their number of genes annotated in the functional network.
Cytoscape software analysis
Identification of key genes
Cytoscape analysis showed 110 annotated genes. CytoHubba apps from the Cytoscape program allowed users to determine the top genes with the “degree” method, where the target genes have the highest degrees, and which are often key target genes (Figure 4). Each node showed a different color depth depending on its own degree (the darker the color, the higher the degree). The network provided 7 important key target genes involved in the regulation of many other genes in the PPI network, which may have potential therapeutic targets in MM.
Figure 4.
Network of genes interaction degree. Proteins with a higher degree of importance are more likely to be essential.
GO enrichment by BINGO plugin
The analysis by the BINGO apps in Cytoscape allowed identifying a set of GO terms with a high number of annotated genes and a significant P value (Supplementary Table S1). These results supported the results obtained by STRING software, and the most GO common were:—Biological process: GO ID 48518 (positive regulation of biological process); GO ID 10467 (gene expression); GO ID 44260 (cellular macromolecule metabolic process)—Molecular function: GO ID 5515 (protein binding)—cellular component: GO ID 43227 (membrane-bounded organelle); GO ID 43229 (intracellular organelle); GO ID 43226 (organelle), and they were marked in bold in Table S1. All GO terms identified in this analysis could be implicated in the development of MM.
Network clusters
The 110 annotated genes were involved in 13 clusters, of which 12 were linked, forming a large network, and 1 remained outside this large network named Apolipoprotein deaminase single, with 2 genes (APOBEC3H and APOBEC3C) (Figure 5). The subgroups of this large network were named according to their protein annotations, and they were classified according to the number of genes involved: Susceptibility apoptosis cell is the largest subgroup and the most important, containing 51 genes, of which 6 were top genes (TP53, c-MYC, CCND1, EzH2, IL6, and MDM2);
Figure 5.
Clustered protein association network. The cluster network provides 13 groups (clusters) of all 114 myeloma gene sets. Each node presents a gene or gene product, the color of the node indicates the 3-dimensional structure of the protein. Each gene cluster indicates a biological function.
Protein us4 ribosomal with 10 genes of which one (UBA52) is the top gene;
Reattaching heterochromatin mitotic with 8 genes;
Chondroitin proteoglycan microfibrils with 5 genes;
Proteinase c1s c1r with 4 genes;
na chloride scarb1 with 4 genes;
Somatomedin insulin higher with 3 genes;
Single deaminase independent with 3 genes;
Removes h2a h2afz with 3 genes;
Exosome 3’ untranslated with 3 genes;
dm mhc class with 2 genes;
Cytolytic release cytochrome with 2 genes.
DAVID database results
To validate the results obtained by STRING and Cytoscape, an analysis by the DAVID software was carried out. The results obtained showed 10 GO terms representing 10 signaling pathways classified according to their P value and the number of genes involved in them (Table 4). The most significant signaling pathways were EBV infection, PI3K-Akt signaling pathway, MicroRNAs in cancer, and the p53 signaling pathway. These results consolidated those obtained by STRING software.
Table 4.
KEGG pathways enrichment by DAVID.
| Category | Term | Genes | Count | % | P value | Benjamini |
|---|---|---|---|---|---|---|
| KEGG_PATHWAY | Epstein-Barr virus infection |

|
10 | 9,1 | 2,3E-6 | 3,2E-4 |
| KEGG_PATHWAY | PI3K-Akt signaling pathway |

|
13 | 11,8 | 1,0E-4 | 7,2E-3 |
| KEGG_PATHWAY | MicroRNAs in cancer |

|
11 | 10,0 | 4,0E-4 | 1,5E-2 |
| KEGG_PATHWAY | p53 signaling pathway |

|
6 | 5,5 | 4,6E-4 | 1,5E-2 |
| KEGG_PATHWAY | B cell receptor signaling pathway |

|
6 | 5,5 | 5,2E-4 | 1,5E-2 |
| KEGG_PATHWAY | Bladder cancer |

|
5 | 4,5 | 6,3E-4 | 1,5E-2 |
| KEGG_PATHWAY | Pathways in cancer |

|
12 | 10,9 | 1,3E-3 | 2,4E-2 |
| KEGG_PATHWAY | Small-cell lung cancer |

|
6 | 5,5 | 1,4E-3 | 2,4E-2 |
| KEGG_PATHWAY | FoxO signaling pathway |

|
7 | 6,4 | 1,8E-3 | 2,8E-2 |
| KEGG_PATHWAY | Ribosome |

|
7 | 6,4 | 2,0E-3 | 2,8E-2 |
Abbreviation: KEGG, Kyoto Encyclopedia of Genes and Genomes.
Pathways found by DAVID database, selected with their significant P value and their number of genes annotated.
Discussion
In this study, we performed a bioinformatics analysis of a biological data set to identify the key genes and signaling pathways involved during the development of MM. The functional PPI network resulting from the STRING database and Cytoscape software identified that among 114 DEGs introduced, 7 are considered as key genes that represented a high connectivity in the network. Including TP53; MYC, CDND1, IL6, UBA52, EZH2, and MDM2. The CCND1 with MYC, EZH2 with MYC, EZH2 with TP53, and MYC with TP53 are co-expressed, which can be explained by their complementarity and involvement in the same or different processes during the transformation of normal plasma cells into myeloma plasma cells (MM evolution). 5
Indeed, the study of the co-expression of cell cycle genes has shown the presence of a dynamic balance between the co-expression of sets of genes activating and inhibiting the cell cycle (CDK/cyclin-dependent kinases and CDK1) in MM. 48 The clustering analysis showed that the susceptibility apoptosis cell cluster (the largest subgroup and the most important), containing 6 top genes (TP53, c-MYC, CCND1, EzH2, IL6, and MDM2), could be implicated in the development of MM.5,16,20,49,50 Nevertheless, the UBA52 gene was found in the protein us4 ribosomal cluster. The ribosomal us4 protein has multiple functions, including mRNA decoding, initiation of small ribosomal subunit aggregation by binding directly to the 16S rRNA and translation repression. It was also involved in the anti-termination activities of transcription. 51
The ontological analysis revealed the involvement of these 7 hub genes of the functional network in BP related to cellular macromolecular metabolic process, regulation of gene expression, macromolecular metabolic process, and positive regulation of the biological process. These hub genes were the major regulators of these 4 important processes that can be altered during the development of MM.16,49,52 -54 The CC results suggested that the 7 hub genes were mainly scattered throughout the cell, including the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes, and the cytoskeleton. Then, the MF reflected the involvement of hub genes in RNA binding, nucleic acid binding, and protein binding. The alteration in the expression of one or more of these key genes may lead to deregulation of these processes, which can be essential for the cell life cycle and their deregulation may play a role in the development of MM.16,20,49,50
TP53 was significantly down-regulated in MM, 55 identified as the first hub gene in the PPI network, and it represented the highest degree of connectivity (43 interactions), including the most important interactions with the other 6 major genes identified. It is well known that TP53 is a critical tumor suppressor gene that encodes a tumor suppressor protein with domains for transcriptional activation, DNA binding, and oligomerization. 56 When dysregulated, TP53 plays a key and multifaceted role in cancer development and cancer therapy, including MM. 57 It is implicated in a variety of biological functions, with canonical roles including cell cycle arrest, DNA repair, senescence, and apoptosis, as well as noncanonical roles, such as regulation of cell metabolism, and autophagy. 52 This gene is activated by various stress stimuli and is maintained at a low level by a variety of regulators in normal cells. 56 Its dysregulation was presented in 3 forms in newly diagnosed patients with MM: monoallelic deletion of a part of the chromosome 17p (~8%), monoallelic mutations (~6%), and biallelic inactivation (~4%). Del 17p was still considered a high-risk characteristic in MM and was a part of the current illness staging standards. 57
The MYC gene was significantly up-regulated in MM. 15 In the PPI network, it was represented in the center with a high degree of connectivity, which involves 36 interactions, including TP53, MYC CDND1, IL6, UBA52, EZH2, and MDM2. It is one of the regulatory and proto-oncogenes that encode transcription factors. It is involved in the regulation of many biological functions, including functions that affect cell growth and proliferation-replication, transcription and translation, cell metabolism, and apoptosis. 58 It has also been recognized as 1 of 4 genes, including Oct4, KLF4, and Sox2, which could together reprogram fibroblasts into a pluripotent stem cell state. 59 This gene is considered a key regulator of MM. Its dysfunction was one of the main features of disease progression, being a trigger for MGUS to MM transition. 60 These alterations include translocations, which have been observed in 50.1% of patients with newly diagnosed MM. These translocations involved immunoglobulin (IG) loci (IGH, IGL, IGK) and certain non-IG partners, in particular FAM46C, FOXO3, and BMP6, where the hypermutations associated with APOBEC deregulation are located. These mutational hotspots are often found at MYC breakpoints, indicating their roles in the generation of the MYC translocation in MM. 61 In addition, MYC gains and duplications at the 8q24.21 locus were present in almost 15% of newly diagnosed patients with MM, which was related to shorter survival in univariate analysis. 58
CCND1 , a gene encoding Cyclin D1, was also a gene that was upregulated in MM. 17 It has also been identified as a core gene based on its importance in the network. It interacted with 30 genes in the PPI network. Cyclin D1 was a key cell cycle regulator and was involved in the pathogenesis of several cancers. Its overexpression and translocation are frequent events in MM, suggesting that it might be the cause of the initiation and development of this malignancy. 62 During mitosis, the transition of the cell cycle from G1 to S phase is controlled by the cyclin-dependent kinases (CDKs), CDK4 and CDK6, which form protein complexes with cyclin D1. Cyclin D1 catalyzes the phosphorylation of the tumor suppressor protein retinoblastoma (RB) while releasing the transcription factor E2F and triggering the downstream gene transcription required for cell cycle progression. As a result, cyclin D1 inhibition was conducted to cell cycle arrest, while overexpression of the protein accelerates the G1 phase transition. 63 Due to chromosomal duplication, translocations, and alteration of normal intercellular trafficking and proteolysis, abnormal expression of cyclin D1 has been reported in a variety of human tumors. 64 In particular, 15% to 20% of MM samples carry a t(11;14) chromosomal translocation, resulting in abnormal transcriptional activation of CCND1. 65 This most common translocation in MM, t(11; 14) (q13; q32), involves an abnormal fusion of the IGH locus with cyclin D1. 66 Overexpression of cyclin D1 was also observed in 25% to 50% of MM samples, indicating that the deregulation of cyclin D1, which was a critical regulator of the G1/S transition and therefore of the cell cycle, may be a key event in MM development. 67
In the PPI network, IL6 (Interleukin-6) represented the connectivity of 25 interactions. This gene was significantly upregulated in MM. 50 IL6 is a pleiotropic cytokine with extensive functions in inflammation and immunity. It has been extensively studied for its role in normal antibody-producing plasma cells. It promotes the normal B-cell differentiation into cells producing antibodies without triggering the proliferation of B cells. 68 It has been shown that IL6 is a key factor in the proliferation of MM, promoting the growth, proliferation, and survival of myeloma cells. 69 Bone marrow stromal cells presented the major source of IL6 in MM cells, and its overexpression was linked to poor prognosis and larger tumor cell mass in MM. 70
EZH2 (Enhancer of Zeste 2 Polycomb Repressive Complex 2) represented a high degree of connectivity, with 22 interactions in the PPI network. It is one of the DEGs upregulated in MM. 20 The EZH2 gene was a histone methyltransferase that acts primarily on H3K27 and catalyzes the conversion to a trimethylated marker (H3K27me3). This gene played a critical role in normal B cell development since H3K27me3 expression and levels influence differentiation decisions. Increased EZH2 expression in germinal center B cells resulted in cell cycle checkpoints disappearing and allowing B cells to expand. Subsequently, EZH2 decreased, as a result allowing the cells to differentiate into plasma cells. 71 EZH2 was known to be deregulated in MM. Pawlyn et al 20 discovered for the first time a link between high expression of EZH2 and myeloma survival. This association was robust across different datasets, persisted regardless of the treatment method used, and was independent of other factors known to affect survival in patients with myeloma. This reinforces the importance of EZH2 expression in the pathogenesis of myeloma and suggested that its inhibition may be a potential therapeutic approach for myeloma therapy and should be studied in clinical trials. 20
UBA52 (ubiquitin A-52 residue ribosomal protein fusion product 1) was one of the up-regulated genes observed in MM. 21 In the PPI network, it was also recognized as a core gene, representing high connectivity of 22 interactions, the most significant of which are those with the other 6 hub genes. This gene is essential for the selective degradation of proteins by the ubiquitin-proteasome system (UPS). In addition, it has been found to play a central role in cellular functions, including cell cycle control, apoptosis, signaling, and transcriptional regulation. 72 UBA52 has been shown to promote the development of cancer through multiple mechanisms, which may indicate its involvement in the development of MM. 73 UBA52 has been the most novel target gene proposed as a driver of MM until there are enough studies on its involvement in myeloma. Further genomic studies are needed to explain the function of UBA52 in the process of the disease.
MDM2 (Murine double minute protein) represented important connectivity with about 21 interactions in the network. The MDM2 gene is a pleiotropic protein located on the 12q13-14 chromosome, known to facilitate the ubiquitination of p53 required for its proteasome-mediated turnover. In MM, this gene is highly and constitutively expressed through processes such as chromosomal trisomy or gene amplification and functions both to drive cell proliferation and to enhance cell survival. It activates E2F-1, which promoted the transition from G1 to the S phase, and suppressed the function of wild-type p53 (wtp53), which can enhance cell survival. MDM2 binds constitutively to E2F-1, wtp53, and mtp53 (mutant-type p53) in all myeloma cells, as well as p21 in malignancy cells lacking p53. Then p21 was activated by wtp53 and potentiated the tumor suppressor function of wtp53 by strongly inhibiting cell cycle regulatory proteins such as cyclin E and CDK2. This indicated that MDM2 can improve the cell cycle progression of MM cells by downregulating cell cycle inhibitory proteins (wtp53 and p21) and activating E2F-1. Therefore, overexpression of MDM2 may contribute to the growth and survival of MM cells, indicating that therapeutic strategies targeting MDM2 have potential utility in MM.16,74
KEGG pathway enrichment analysis showed that the 7 key genes were significantly enriched in EBV infection (hsa05169), microRNAs in cancer (hsa05206), PI3K-Akt signaling pathway (hsa04151) and p53 signaling pathway (hsa04115). Many studies have been in concordance with our results, which demonstrated the existence of an association between EBV infection and MM development, in which a polymerase chain reaction was performed and revealed the presence of an elevated level of EBV DNA in patients with MM compared with healthy controls.75,76 MicroRNAs (miRNAs) are small noncoding RNAs aberrantly expressed in solid and hematopoietic malignancies, where they play a central role as post-transcriptional regulators of gene expression. 77 Recently, some studies have demonstrated the efficacy of miRNAs as specific and sensitive biomarkers for the classification, prognosis, and diagnosis of human cancer. 78 Jones et al 79 discovered 3 miRNAs in serum, miR-1308, miR-1246, and miR 720, which have the potential to be used as diagnostic biomarkers in myeloma. They demonstrated that the joint use of miR-1308 and miR-720 provided a powerful diagnostic tool for distinguishing normal healthy controls from patients with MGUS/myeloma. Furthermore, the combination of miR-1246 and miR-1308 can distinguish patients with MGUS from those with myeloma. 79 The PI3K/Akt pathway has attracted considerable attention as a promising therapeutic target in MM. 80 A study conducted on the PI3K/Akt/mTOR pathway (mammalian target of rapamycin) by Vijay Ramakrishnan and Shaji Kumar has shown that it played a critical role in the biology of diseases. This pathway was aberrantly activated in a large proportion of patients with MM by multiple mechanisms. It may also play a role in resistance to several existing therapies, making it a central pathway in the pathophysiology of MM. 81 The study by Vikova et al 82 showed after whole-exome sequencing on a large cohort of 30 human MM cell lines, representative of a large molecular heterogeneity of MM, and 8 control samples, that many canonical pathways known to be implicated in the proliferation and survival of MM cells have been mutated, including PI3K-AKT. In addition, 76% of myeloma cell lines had mutations in the p53 cell cycle pathway genes. 82
Conclusion
STRING, Cytoscape, and DAVID Software analyzed a set of 114 MM DEGs. TP53, MYC, CCND1, IL6, UBA52, EZH2, and MDM2 were identified as hub genes, which may play a key role in the pathogenesis of MM. They were implicated during different stages of disease progression. The 7 hub genes were identified as being significantly enriched in various pathways, particularly in EBV infection, microRNAs in cancer, the PI3K-Akt signaling pathway, and the p53 signaling pathway, which can be critical for the progression of myeloma. This predictive study can be a relevant tool for a better understanding of the evolution of MM in its complex microenvironment. Understanding the pathogenesis of MM could reveal new biomarkers of myeloma cells and promising potential therapeutic targets.
Supplemental Material
Supplemental material, sj-docx-1-bbi-10.1177_11779322221115545 for Multiple Myeloma: Bioinformatic Analysis for Identification of Key Genes and Pathways by Chaimaa Saadoune, Badreddine Nouadi, Hasna Hamdaoui, Fatima Chegdani and Faiza Bennis in Bioinformatics and Biology Insights
Footnotes
Author Contributions: CS did the conceptualization, methodology, data collection, analysis and interpretation, wrote original draft. BN collected data, revised, and edited manuscript. HH revised and edited manuscript. FC did the conceptualization, wrote—revised and edited, supervision. FB did the conceptualization, wrote—revised and edited, supervision. All authors contributed to manuscript revision, read and approved the submitted version.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Hamdaoui H, Benlarroubia O, Ait Boujmia OK, et al. Cytogenetic and FISH analysis of 93 multiple myeloma Moroccan patients. Mol Genet Genomic Med. 2020;8:e1363. doi: 10.1002/mgg3.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ludwig H, Novis Durie S, Meckl A, Hinke A, Durie B. Multiple myeloma incidence and mortality around the globe; interrelations between health access and quality, economic resources, and patient empowerment. Oncologist. 2020;25:e1406-e1413. doi: 10.1634/theoncologist.2020-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kazandjian D. Multiple myeloma epidemiology and survival, a unique malignancy. Semin Oncol. 2016;43:676-681. doi: 10.1053/j.seminoncol.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rajkumar SV. Multiple myeloma: every year a new standard? Hematol Oncol. 2019;37:62-65. doi: 10.1002/hon.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barwick BG, Gupta VA, Vertino PM, Boise LH. Cell of origin and genetic alterations in the pathogenesis of multiple myeloma. Front Immunol. 2019;10:1121. doi: 10.3389/fimmu.2019.01121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weißbach S, Langer C, Puppe B, et al. The molecular spectrum and clinical impact of DIS3 mutations in multiple myeloma. Br J Haematol. 2015;169:57-70. doi: 10.1111/bjh.13256. [DOI] [PubMed] [Google Scholar]
- 7. Hu Y, Chen W, Wang J. Progress in the identification of gene mutations involved in multiple myeloma. Onco Targets Ther. 2019;12:4075-4080. doi: 10.2147/OTT.S205922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nouadi B, Sbaoui Y, El Messal M, Bennis F, Chegdani F. Integrative analysis of the genes induced by the intestine microbiota of infant born to term and breastfed. Bioinform Biol Insights. 2020;14:1177932220906168. doi: 10.1177/1177932220906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tao Z, Shi A, Li R, Wang Y, Wang X, Zhao J. Microarray bioinformatics in cancer—a review. J BUON. 2017;22:838-843. [PubMed] [Google Scholar]
- 10. Oliver GR, Hart SN, Klee EW. Bioinformatics for clinical next generation sequencing. Clin Chem. 2015;61:124-135. doi: 10.1373/clinchem.2014.224360. [DOI] [PubMed] [Google Scholar]
- 11. Ziccheddu B, Da Vià MC, Lionetti M, et al. Functional impact of genomic complexity on the transcriptome of multiple myeloma. Clin Cancer Res. 2021;27:6479-6490. doi: 10.1158/1078-0432.CCR-20-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Avet-Loiseau H, Li C, Magrangeas F, et al. Prognostic significance of copy-number alterations in multiple myeloma. J Clin Oncol. 2009;27:4585-4590. doi: 10.1200/JCO.2008.20.6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jasrotia S, Gupta R, Sharma A, Halder A, Kumar L. Cytokine profile in multiple myeloma. Cytokine. 2020;136:155271. doi: 10.1016/j.cyto.2020.155271. [DOI] [PubMed] [Google Scholar]
- 14. Mitchell JS, Li N, Weinhold N, et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun. 2016;7:12050. doi: 10.1038/ncomms12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Broyl A, Hose D, Lokhorst H, et al. Gene expression profiling for molecular classification of multiple myeloma in newly diagnosed patients. Blood. 2010;116:2543-2553. doi: 10.1182/blood-2009-12-261032. [DOI] [PubMed] [Google Scholar]
- 16. Teoh G, Urashima M, Ogata A, et al. MDM2 protein overexpression promotes proliferation and survival of multiple myeloma cells. Blood. 1997;90:1982-1992. [PubMed] [Google Scholar]
- 17. Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. doi: 10.1038/ncomms3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Went M, Kinnersley B, Sud A, et al. Transcriptome-wide association study of multiple myeloma identifies candidate susceptibility genes. Hum Genomics. 2019;13:37. doi: 10.1186/s40246-019-0231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tian E, Sawyer JR, Heuck CJ, et al. In multiple myeloma, 14q32 translocations are non-random chromosomal fusions driving high expression levels of the respective partner genes. Genes Chromosomes Cancer. 2014;53:549-557. doi: 10.1002/gcc.22165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pawlyn C, Bright MD, Buros AF, et al. Overexpression of EZH2 in multiple myeloma is associated with poor prognosis and dysregulation of cell cycle control. Blood Cancer J. 2017;7:e549. doi: 10.1038/bcj.2017.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yan H, Zheng G, Qu J, et al. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. J Cell Physiol. 2019;234:23785-23797. doi: 10.1002/jcp.28947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang W, Zhang Y, Yang Z, et al. High expression of UBE2T predicts poor prognosis and survival in multiple myeloma. Cancer Gene Ther. 2019;26:347-355. doi: 10.1038/s41417-018-0070-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Das DS, Das A, Ray A, et al. Blockade of deubiquitylating enzyme USP1 inhibits DNA repair and triggers apoptosis in multiple myeloma cells. Clin Cancer Res. 2017;23:4280-4289. doi: 10.1158/1078-0432.CCR-16-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shammas MA, Shmookler Reis RJ, Koley H, Batchu RB, Li C, Munshi NC. Dysfunctional homologous recombination mediates genomic instability and progression in myeloma. Blood. 2009;113:2290-2297. doi: 10.1182/blood-2007-05-089193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cottini F, Hideshima T, Xu C, et al. Rescue of hippo co-activator YAP1 triggers DNA damage-induced apoptosis in hematological cancers. Nat Med. 2014;20:599-606. doi: 10.1038/nm.3562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kumar S, Talluri S, Pal J, et al. Role of apurinic/apyrimidinic nucleases in the regulation of homologous recombination in myeloma: mechanisms and translational significance. Blood Cancer J. 2018;8:92. doi: 10.1038/s41408-018-0129-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cea M, Cagnetta A, Adamia S, et al. Evidence for a role of the histone deacetylase SIRT6 in DNA damage response of multiple myeloma cells. Blood. 2016;127:1138-1150. doi: 10.1182/blood-2015-06-649970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chauhan D, Tian Z, Nicholson B, et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell. 2012;22:345-358. doi: 10.1016/j.ccr.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Walker BA, Leone PE, Chiecchio L, et al. A compendium of myeloma-associated chromosomal copy number abnormalities and their prognostic value. Blood. 2010;116:e56-e65. doi: 10.1182/blood-2010-04-279596. [DOI] [PubMed] [Google Scholar]
- 30. Szalat R, Samur MK, Fulciniti M, et al. Nucleotide excision repair is a potential therapeutic target in multiple myeloma. Leukemia. 2018;32:111-119. doi: 10.1038/leu.2017.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904-917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ohguchi H, Hideshima T, Bhasin MK, et al. The KDM3A–KLF2–IRF4 axis maintains myeloma cell survival. Nat Commun. 2016;7:10258. doi: 10.1038/ncomms10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Herath NI, Rocques N, Garancher A, Eychène A, Pouponnot C. GSK3-mediated MAF phosphorylation in multiple myeloma as a potential therapeutic target. Blood Cancer J. 2014;4:e175. doi: 10.1038/bcj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Went M, Sud A, Försti A, et al. Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma. Nat Commun. 2018;9:3707. doi: 10.1038/s41467-018-04989-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fabris S, Ronchetti D, Agnelli L, et al. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia. 2007;21:1113-1116. doi: 10.1038/sj.leu.2404616. [DOI] [PubMed] [Google Scholar]
- 36. Inoue J, Otsuki T, Hirasawa A, et al. Overexpression of PDZK1 within the 1q12-q22 amplicon is likely to be associated with drug-resistance phenotype in multiple myeloma. Am J Pathol. 2004;165:71-81. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1618545/. Accessed March 12, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kawano T, Ahmad R, Nogi H, Agata N, Anderson K, Kufe D. MUC1 oncoprotein promotes growth and survival of human multiple myeloma cells. Int J Oncol. 2008;33:153-159. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3086751/. Accessed March 12, 2022. [PMC free article] [PubMed] [Google Scholar]
- 38. Ali M, Ajore R, Wihlborg AK, et al. The multiple myeloma risk allele at 5q15 lowers ELL2 expression and increases ribosomal gene expression. Nat Commun. 2018;9:1649. doi: 10.1038/s41467-018-04082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hoang PH, Dobbins SE, Cornish AJ, et al. Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms. Leukemia. 2018;32:2459-2470. doi: 10.1038/s41375-018-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen L, Hu N, Wang C, Zhao H, Gu Y. Long non-coding RNA CCAT1 promotes multiple myeloma progression by acting as a molecular sponge of miR-181a-5p to modulate HOXA1 expression. Cell Cycle. 2018;17:319-329. doi: 10.1080/15384101.2017.1407893. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41. Mani M, Carrasco DE, Zhang Y, et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577-7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Montel RA, Gregory M, Chu T, Cottrell J, Bitasktsis C, Chang SL. Genetic variants as biomarkers for progression and resistance in multiple myeloma. Cancer Genet. 2021;252-253:1-5. doi: 10.1016/j.cancergen.2020.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aksenova AY, Zhuk AS, Lada AG, et al. Genome instability in multiple myeloma: facts and factors. Cancers (Basel). 2021;13:5949. doi: 10.3390/cancers13235949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Frassanito MA, Desantis V, Di Marzo L, et al. Bone marrow fibroblasts overexpress miR-27b and miR-214 in step with multiple myeloma progression, dependent on tumour cell-derived exosomes. J Pathol. 2019;247:241-253. doi: 10.1002/path.5187. [DOI] [PubMed] [Google Scholar]
- 45. Shin EM, Neja SA, Fidan K, et al. Lymphocyte cytosolic protein 1 (LCP1) is a novel TRAF3 dysregulation biomarker with potential prognostic value in multiple myeloma. Genome Instab Dis. 2020;1:286-299. doi: 10.1007/s42764-020-00014-x. [DOI] [Google Scholar]
- 46. Swaminathan B, Thorleifsson G, Jöud M, et al. Variants in ELL2 influencing immunoglobulin levels associate with multiple myeloma. Nat Commun. 2015;6:7213. doi: 10.1038/ncomms8213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chubb D, Weinhold N, Broderick P, et al. Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Nat Genet. 2013;45:1221-1225. doi: 10.1038/ng.2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kryukov F, Dementyeva E, Kubiczkova L, et al. Cell cycle genes co-expression in multiple myeloma and plasma cell leukemia. Genomics. 2013;102:243-249. doi: 10.1016/j.ygeno.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 49. van Nieuwenhuijzen N, Spaan I, Raymakers R, Peperzak V. From MGUS to multiple myeloma, a paradigm for clonal evolution of premalignant cells. Cancer Res. 2018;78:2449-2456. doi: 10.1158/0008-5472.CAN-17-3115. [DOI] [PubMed] [Google Scholar]
- 50. Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104:607-618. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- 51. Kamath D, Allgeyer BB, Gregory ST, et al. The C-terminus of ribosomal protein uS4 contributes to small ribosomal subunit biogenesis and the fidelity of translation. Biochimie. 2017;138:194-201. doi: 10.1016/j.biochi.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Aubrey BJ, Strasser A, Kelly GL. Tumor-suppressor functions of the TP53 pathway. Cold Spring Harb Perspect Med. 2016;6:a026062. doi: 10.1101/cshperspect.a026062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gadó K, Domján G, Hegyesi H, Falus A. Role of INTERLEUKIN-6 in the pathogenesis of multiple myeloma. Cell Biol Int. 2000;24:195-209. doi: 10.1006/cbir.2000.0497. [DOI] [PubMed] [Google Scholar]
- 54. Alzrigat M, Jernberg-Wiklund H, Licht JD. Targeting EZH2 in multiple myeloma—multifaceted anti-tumor activity. Epigenomes. 2018;2:16. doi: 10.3390/epigenomes2030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xiong W, Wu X, Starnes S, et al. An analysis of the clinical and biologic significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112:4235-4246. doi: 10.1182/blood-2007-10-119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170:1062-1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Flynt E, Bisht K, Sridharan V, Ortiz M, Towfic F, Thakurta A. Prognosis, biology, and targeting of TP53 dysregulation in multiple myeloma. Cells. 2020;9:287. doi: 10.3390/cells9020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jovanović KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S. Targeting MYC in multiple myeloma. Leukemia. 2018;32:1295-1306. doi: 10.1038/s41375-018-0036-x. [DOI] [PubMed] [Google Scholar]
- 59. Dang CV. MYC on the path to cancer. Cell. 2012;149:22-35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Misund K, Keane N, Stein C, et al. MYC dysregulation in the progression of multiple myeloma. Leukemia. 2020;34:322-326. doi: 10.1038/s41375-019-0543-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Walker BA, Wardell CP, Murison A, et al. APOBEC family mutational signatures are associated with poor prognosis translocations in multiple myeloma. Nat Commun. 2015;6:6997. doi: 10.1038/ncomms7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sherr CJ. Cancer cell cycles. Science. 1996;274:1672-1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 63. Ding L, Cao J, Lin W, et al. The roles of cyclin-dependent kinases in cell-cycle progression and therapeutic strategies in human breast cancer. Int J Mol Sci. 2020;21:1960. doi: 10.3390/ijms21061960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stamatakos M, Palla V, Karaiskos I, et al. Cell cyclins: triggering elements of cancer or not? World J Surg Oncol. 2010;8:111. doi: 10.1186/1477-7819-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sawyer JR. The prognostic significance of cytogenetics and molecular profiling in multiple myeloma. Cancer Genet. 2011;204:3-12. doi: 10.1016/j.cancergencyto.2010.11.002. [DOI] [PubMed] [Google Scholar]
- 66. Hamdaoui H, Natiq A, Benlarroubia O, et al. Near tetrapoloid karyotype with translocation t(11;14) in a Moroccan patient with amyloid light-chain amyloidosis and multiple myeloma. Leuk Res Rep. 2020;14:100217. doi: 10.1016/j.lrr.2020.100217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang S, Huang Y, Su R, Fang Z, Han M. Cyclin D1 G870A polymorphism is associated with an increased risk of multiple myeloma. Genet Mol Res. 2015;14:5856-5861. doi: 10.4238/2015.June.1.2. [DOI] [PubMed] [Google Scholar]
- 68. Matthes T, Manfroi B, Huard B. Revisiting IL-6 antagonism in multiple myeloma. Crit Rev Oncol Hematol. 2016;105:1-4. doi: 10.1016/j.critrevonc.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 69. Rosean TR, Tompkins VS, Tricot G, et al. Preclinical validation of interleukin 6 as a therapeutic target in multiple myeloma. Immunol Res. 2014;59:188-202. doi: 10.1007/s12026-014-8528-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Urashima M, Ogata A, Chauhan D, et al. Interleukin-6 promotes multiple myeloma cell growth via phosphorylation of retinoblastoma protein. Blood. 1996;88:2219-2227. doi: 10.1182/blood.V88.6.2219.bloodjournal8862219. [DOI] [PubMed] [Google Scholar]
- 71. Morin RD, Johnson NA, Severson TM, et al. Somatic mutation of EZH2 (Y641) in follicular and diffuse large B-cell lymphomas of germinal center origin. Nat Genet. 2010;42:181-185. doi: 10.1038/ng.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mao J, O’Gorman C, Sutovsky M, Zigo M, Wells KD, Sutovsky P. Ubiquitin A-52 residue ribosomal protein fusion product 1 (Uba52) is essential for preimplantation embryo development. Biol Open. 2018;7:bio035717. doi: 10.1242/bio.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang F, Chen X, Yu X, Lin Q. Degradation of CCNB1 mediated by APC11 through UBA52 ubiquitination promotes cell cycle progression and proliferation of non-small cell lung cancer cells. Am J Transl Res. 2019;11:7166-7185. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6895529/. Accessed September 6, 2021. [PMC free article] [PubMed] [Google Scholar]
- 74. Gu D, Wang S, Kuiatse I, et al. Inhibition of the MDM2 E3 ligase induces apoptosis and autophagy in wild-type and mutant p53 models of multiple myeloma, and acts synergistically with ABT-737. PLoS ONE. 2014;9:e103015. doi: 10.1371/journal.pone.0103015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sadeghian MH, Ayatollahi H, Keramati MR, et al. The association of Epstein-Barr virus infection with multiple myeloma. Indian J Pathol Microbiol. 2011;54:720-724. doi: 10.4103/0377-4929.91504. [DOI] [PubMed] [Google Scholar]
- 76. Xia B, Wang X, Yang R, et al. Epstein–Barr virus infection is associated with clinical characteristics and poor prognosis of multiple myeloma. Biosci Rep. 2019;39:BSR20190284. doi: 10.1042/BSR20190284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rossi M, Tagliaferri P, Tassone P. MicroRNAs in multiple myeloma and related bone disease. Ann Transl Med. 2015;3:334. doi: 10.3978/j.issn.2305-5839.2015.12.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Jones CI, Zabolotskaya MV, King AJ, et al. Identification of circulating microRNAs as diagnostic biomarkers for use in multiple myeloma. Br J Cancer. 2012;107:1987-1996. doi: 10.1038/bjc.2012.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Chatterjee M, Andrulis M, Stühmer T, et al. The PI3K/Akt signaling pathway regulates the expression of Hsp70, which critically contributes to Hsp90-chaperone function and tumor cell survival in multiple myeloma. Haematologica. 2013;98:1132-1141. doi: 10.3324/haematol.2012.066175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ramakrishnan V, Kumar S. PI3K/AKT/mTOR pathway in multiple myeloma: from basic biology to clinical promise. Leuk Lymphoma. 2018;59:2524-2534. doi: 10.1080/10428194.2017.1421760. [DOI] [PubMed] [Google Scholar]
- 82. Vikova V, Jourdan M, Robert N, et al. Comprehensive characterization of the mutational landscape in multiple myeloma cell lines reveals potential drivers and pathways associated with tumor progression and drug resistance. Theranostics. 2019;9:540-553. doi: 10.7150/thno.28374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-bbi-10.1177_11779322221115545 for Multiple Myeloma: Bioinformatic Analysis for Identification of Key Genes and Pathways by Chaimaa Saadoune, Badreddine Nouadi, Hasna Hamdaoui, Fatima Chegdani and Faiza Bennis in Bioinformatics and Biology Insights