Summary
Background
Detection of dengue virus antibodies is important for understanding future dengue virus risk and for prevaccination screening. We aimed to evaluate the performance of a dengue IgG indirect ELISA in determining dengue seroprevalence in a cohort of children in the Philippines, using a focus reduction neutralisation test (FRNT) as the reference test.
Methods
In this prospective population-based cohort study, we enrolled healthy children residing in Bogo or Balamban, Cebu, Philippines, who were to be aged 9–14 years at the time of a mass dengue vaccination campaign. Sera were collected from participants and batch tested by indirect IgG ELISA and FRNT. The primary endpoint was dengue seroprevalence in the cohort, detected by ELISA, and validated by that detected by reference FRNT. This study is registered with ClinicalTrials.gov, NCT03465254.
Findings
We collected 2996 serum samples between May 2, and June 2, 2017, and we tested each sample with IgG ELISA. Using 1961 samples (65· 5%) that were tested with FRNT, and 1035 samples (34·5%) with imputed results, we found that 320 (10·7%) of 2996 children were dengue naive and 2676 (89·3%) were seropositive for previous dengue virus infection. Based on the 1961 non-imputed FRNT results classified as dengue seronegative or seropositive, the ELISA (with a 0·9 index value cutoff) showed 95·2% sensitivity, 93·4% specificity, 6·6% false positivity, and 4·8% false negativity. However, sensitivity of the ELISA was poor (77·1%) among children with immunity to just one dengue virus serotype. Of the 11 sera that were false positive with ELISA, seven samples (63·6%) were seropositive for Zika virus or Japanese encephalitis virus with FRNT.
Interpretation
Most children (89·3%) assessed in our study and eligible to participate in the mass dengue vaccination campaign were seropositive for previous dengue virus infection. Compared with FRNT, ELISA had high sensitivity and specificity (>90%), but the false-negative and false-positive rates makes the test suboptimal for prevaccination screening. Individuals who are falsely identified as seropositive by dengue IgG ELISA and then vaccinated might be at risk of developing severe disease during a subsequent exposure to wild-type dengue virus. Those with a monotypic profile would benefit the most from vaccination, but the sensitivity of the IgG ELISA was much lower in this group than in those with a multitypic profile.
Funding
Philippine Department of Health, Hanako Foundation, WHO, Swedish International Development Cooperation Agency through the International Vaccine Institute, and University of North Carolina, Chapel Hill, NC, USA.
Introduction
There are four dengue virus serotypes (DENV 1–4). Infection with dengue virus produces durable, or sometimes lifelong, homotypic immunity against the infecting serotype, but waning cross-protection against the other serotypes. Cross-protection wanes in approximately 2 months to 2 years following primary infection.1–4 Thus, individuals living in dengue endemic areas can experience repeated dengue infections. Primary infection usually presents as a mild or self-limiting illness. The risk of developing severe disease is increased during secondary dengue infections by a serotype other than that which caused the primary infection. Severe dengue is associated with a decay in cross-reactive antibodies to subneutralising levels that enhance viral replication.5,6 During third or fourth dengue infections, the antibody response is presumed to become broadly neutralising, thereby reducing the risk of developing severe disease.7–9
The detection of dengue antibodies from previous natural infection is important for understanding the future risk of severe dengue virus disease. Dengue serostatus also has implications for the use of CYD-TDV (Dengvaxia, Sanofi Pasteur), which is currently the only licensed dengue vaccine, and potentially for future dengue vaccines. Since December, 2015, CYD-TDV has been licensed on a three-dose schedule in several countries. Licensure followed the completion of phase 3 trials in children aged 2–14 years in five Asian countries and children aged 9–16 years in five Latin American countries, which showed a pooled vaccine efficacy over 25 months of 60% (95% CI 56–65) for all participants.10 Licensure was obtained for children aged 9 years or older because of an increased risk of hospitalisation and severe dengue among the younger children. As only a subset of the trial participants had dengue serostatus assessed at baseline, definitive conclusions about the safety signal could not be made. On the basis of these results, WHO recommended that countries with high dengue transmission consider introducing CYD-TDV in age groups with seroprevalence of 70% or greater.11 Considering the substantial burden of dengue infections in the Philippines, the Department of Health launched a mass dengue vaccination campaign in children aged 9–14 years in high risk regions, which began on April 4, 2016; individual pre-vaccination dengue serostatus testing was not yet recommended at that time. In November, 2017, the manufacturer of CYD-TDV issued a notice for relabelling of the vaccine to indicate an additional risk.12 This notice was based on an extended analysis of the phase 3 trial using a new test that could differentiate antibodies from previous natural dengue infection versus those from CYD-TDV vaccination. The extended analysis showed that over a 5-year period, CYD-TDV conferred 76% (95% CI 64–84) protection against virologically confirmed symptomatic dengue infection in seropositive recipients aged at least 9 years, but resulted in overall poor efficacy and an excess risk of hospitalisation and severe dengue virus disease in seronegative recipients.13 After the performance of the vaccine was confirmed to vary by dengue serostatus, WHO revised its recommendations and the US Food and Drug Administration has since recommended that CYD-TDV should only be given to those with serological evidence of previous infection.14,15 A prevaccination screening strategy would require a readily available and accurate test for dengue serostatus.16 Although there are debates on the ideal characteristics of a prevaccination screening test,17,18 a sensitivity and specificity of at least 90% has been recommended, with a desired sensitivity of 95% and specificity of 98%.19
Neutralisation testing is considered the gold standard for establishing dengue serostatus, but is expensive, time and labour intensive, and requires advanced equipment, trained staff, and technical skill. Although ELISAs have been developed for detecting dengue-specific IgG antibodies, these assays have not been extensively evaluated for their performance in prevaccination screening. We aimed to assess the dengue seroprevalence and to evaluate the performance of a dengue IgG indirect ELISA in determining dengue serostatus in a cohort of children in the Philippines, using a focus reduction neutralisation test (FRNT) as the reference test.
Methods
Study design and participants
The Philippines is endemic for dengue virus with multiple serotypes circulating at the same time.20 Other flaviviruses, such as Zika virus and Japanese encephalitis virus, are also present. Japanese encephalitis virus vaccination is not routinely given in the Philippines. Cross-reactions with other flaviviruses is an important problem in the testing for dengue antibodies. From April, 2016, to June, 2017, children aged 9–14 years in three northern regions were invited to participate in a school-based mass dengue vaccination campaign and were offered a three-dose vaccination series. In June, 2017, this campaign was extended to community-based mass dengue vaccination of children aged 9–14 years in the Cebu province of the Philippines. The estimated population of Cebu in 2015 was 4632 359,21 of which 285 242 children (6·16%) were targeted for the mass dengue vaccination. After the November, 2017 announcement of the CYD-TDV safety risk, the mass three-dose vaccination campaign in the Philippines was discontinued, such that only a single dose was offered in Cebu.
This study is part of a longitudinal cohort study conducted in Bogo and Balamban, two semi-urban areas in Cebu. From May 2, to June 2, 2017, just before the launch of the mass dengue vaccination campaign in Cebu, we invited healthy children residing in Bogo and Balamban, who were to be aged 9–14 years at the time of the mass dengue vaccination campaign, to participate in the study. The children were recruited and are followed-up through the Rural Health Units in Bogo and Balamban. A parent or legal guardian of each participant provided written informed consent for them to take part. Verbal assent was obtained from the participants and documented. We followed the STARD guidelines22 and our protocol (appendix pp 2–30) was reviewed and approved by the University of the Philippines Manila Research Ethics Board.
Procedures
Participants were asked to complete a brief questionnaire of basic demographic information and to provide a 5 mL venous blood sample. The blood samples were collected in anticoagulant-free vacutainer tubes, processed and aliquoted, and the sera were stored at −80°C before testing. One batch of frozen sera was shipped to the University of the Philippines Manila National Institutes of Health (Manila, Philippines) where they were tested using the dengue IgG antibody indirect ELISA (PanBio; Brisbane, QLD, Australia) following the manufacturer’s instructions. The manufacturer’s recommended index cutoff values were as follows: less than 0·9 indicated seronegativity (no evidence of previous dengue infection), 0·9–1·1 indicated an equivocal serostatus, and more than 1·1 indicated seropositivity (presence of detectable IgG antibodies, indicating a previous dengue infection). Because it was unclear whether the ELISA cutoff values recommended by the manufacturer were suitable for prevaccination screening, we explored and sought to define the most suitable cutoff value.
Another batch of frozen sera was shipped to the University of North Carolina (Chapel Hill, NC, USA). To measure neutralising antibody titres to DENV 1–4, Zika virus, and Japanese encephalitis virus in sera, we used a micro FRNT adapted to a 96-well plate format as described previously.23,24 The dengue virus strains used were those recommended by WHO as reference strains for plaque reduction neutralisation testing and FRNT.25 These strains are WP74 (corresponding to DENV 1), S-16803 (DENV 2), CH53489 (DENV 3), and TVP376 (DENV 4). The Zika virus strain used was H/PF/2013 and the Japanese encephalitis strain was rSA-14-14-2.26 96-well plates were plated with 2 × 104 Vero-81 cells per well and incubated at 37°C for 24 h. For the full dilution FRNT, serial four-fold dilutions of each serum were mixed with 50–100 focus-forming units of virus in Dulbecco’s modified Eagle’s medium (Gibco; Grand Island, NY, USA) with 2% fetal bovine serum. The virus-antibody mixtures were incubated for 1 h at 37°C and then transferred to the confluent monolayer of Vero-81 cells on the 96-well plates. Following an additional 1 h incubation at 37°C, the monolayers were overlaid with Opti-MEM (Gibco) containing 2% fetal bovine serum and 1% (weight per volume) carboxymethyl cellulose (Sigma; St Louis, MO, USA). 50% effective concentration (EC50) values were calculated by graphing percentage neutralisation versus serum dilution and fitting a sigmoidal dose response (variable slope) using Prism 8 (GraphPad Software; San Diego, CA, USA). Neutralising antibody titres (Neut50) represent the dilution at which the serum neutralises 50% of the infection. Criteria to accept values to be reported were an R2 of more than 0·75 and a Hill Slope absolute value of more than 0·5.
Because of the large number of samples, we used a simplified single dilution neutralisation test instead of performing serial dilutions of serum to calculate 50% FRNT titres. Samples that were able to neutralise 70% of the infection or more at a 1/40 dilution was considered positive for the presence of neutralising antibodies against the dengue virus serotype tested. We have previously described and validated the single-dilution FRNT test as a substitute for the more laborious and time-consuming serial dilution neutralisation test.9,27 When the ELISA and FRNT results were concordant, no further testing was done. Full-curve FRNT was performed for sera with discordant ELISA and FRNT results. For samples tested by FRNT, the neutralisation of a single dengue serotype was classified as a primary or monotypic response, indicating one previous dengue infection. Neutralisation of two or more dengue serotypes was defined as a multitypic response, indicating two or more previous dengue infections; these samples were included in full-curve FRNT.
Statistical analysis
The sample size of the longitudinal cohort study was based on its objective to determine the relative risk of developing virologically confirmed dengue among children who received or did not receive CYD-TDV, by dengue serostatus at baseline (appendix pp 2–30), and was calculated using the following assumptions: 80% power, α of 5%, dengue seroprevalence of 80%, and a relative risk of 3·5 of developing dengue virus disease in seronegative children compared with seropositive children.10 For the longitudinal study, we estimated that 2412 participants were needed, with an additional 20% to account for possible loss to follow-up during the 5-year study, which is ongoing until October, 2022. The primary endpoint in this paper used the convenience sample size from the longitudinal study and was the baseline dengue seroprevalence of the cohort, detected by ELISA, and validated by that detected by the reference FRNT. To assess if children with an ELISA index value of less than 0·2 were truly seronegative and if children with an ELISA index value of more than 3·0 were truly seropositive, we randomly selected approximately 30% of samples with the lowest ELISA index value results (<0·2) and 30% with the highest results (>3·0) for validation with FRNT. These thresholds were empirically chosen to explore the most suitable cutoffs for prevaccination screening. The other 70% of these results was imputed as seronegative (index value <0·2) and seropositive (index value >3·0). All samples with ELISA index values in the intermediate range of 0·2–3·0 were tested with FRNT to validate the children’s dengue immune status.
The secondary endpoints were the definition of the most suitable ELISA cutoff value that differentiates between dengue naive and seropositive, and the performance of the ELISA using FRNT as the reference standard. To assess performance of the indirect IgG ELISA as the index test (a secondary endpoint) using FRNT as the reference standard, non-imputed results from 1961 samples achieved 85% power at an α of 5% using a two-sided equivalence test of correlated seroprevalences, a reference seroprevalence by FRNT of 80%, a difference between these two seroprevalences by ELISA and FRNT that still results in equivalence (or the range of equivalence) of 2%, and an actual difference in seroprevalence of 0%.
We used a scatter and box plot to illustrate the distribution of the IgG ELISA data within the FRNT classification as dengue naive or with a monotypic or multitypic dengue profile. The FRNT serostatus group comparison of IgG ELISA data to assess its performance was done using the non-parametric Kruskal-Wallis test and the Wilcoxon test. We also did a receiver operating characteristic analysis; in this curve, patients who were dengue naive on FRNT were considered seronegative and patients with monotypic or multitypic dengue profiles on FRNT were considered seropositive. We calculated the area under the curve with 95% CI. The estimated area under the curve was considered an aggregate measure of the accuracy of the ELISA compared with FRNT across the binary classifications of seropositive and seronegative, and values were classified as excellent (0·9 to 1·0), good (0·8 to <0·9), fair (0·7 to <0·8), poor (0·6 to <0·7), and failed (0·5 to <0·6). To evaluate the predictive accuracy of the IgG ELISA compared with FRNT, we carried out the agreement test using McNemar’s statistics and Cohen’s kappa statistics with 95% CI.28 There was no adjustments for confounders. All analyses were done using SAS version 9.4. This study is registered with ClinicalTrials.gov, NCT03465254.
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
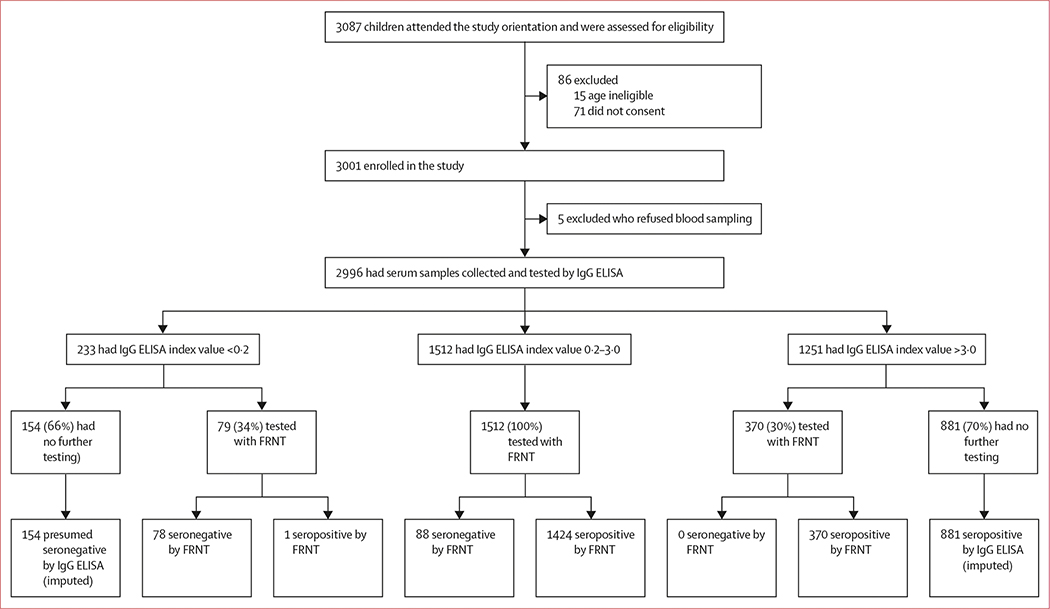
From May 2 to June 2, 2017, we invited 3087 children residing in the Bogo and Balamban sites to participate in the study. We enrolled 3001 children (97·2%) into the study and collected sera from 2996 (97·1%; table 1, figure).
Table 1:
Participant demographics
| Participants (n=2996) | |
|---|---|
|
| |
| Sex | |
| Female | 1550 (51–7%) |
| Male | 1446 (48–3%) |
| Age, years | |
| Mean (SD) | 10–39 (1–32) |
| Median (range) | 10(8–14) |
| Area | |
| Bogo | 1557 (52–0%) |
| Balamban | 1439 (48–0%) |
Data are n (%) unless otherwise stated.
Figure: Flowchart of participants and testing of sera by dengue IgG indirect ELISA and FRNT.
FRNT=focus reduction neutralisation test.
Of the 2996 samples, the dengue IgG ELISA index value results were less than 0·2 in 233 samples (7·8%), 0·2–3·0 in 1512 samples (50·5%), and more than 3·0 in 1251 samples (41·8%; figure). We validated 79 (33·9%) of 233 samples with ELISA index value less than 0·2 with FRNT and found that 78 (98·7%) were dengue naive (figure). Because of the high concordance between the ELISA and FRNT in children with an ELISA index value less than 0·2, all 233 children in this subgroup were classified as dengue virus seronegative. We validated 370 (29·6%) of 1251 samples with ELISA index value more than 3·0 with FRNT; 370 samples (100%) neutralised one or more dengue virus serotype on FRNT, indicating that an ELISA index value of more than 3·0 was strongly predictive of previous dengue virus infection, and all 1251 children in this subgroup were therefore classified as seropositive. 1512 (50·5%) of 2996 samples had an intermediate ELISA index value between 0·2 and 3·0; 88 (5·8%) of 1512 samples were dengue virus naive and 1424 (94·2%) were seropositive for dengue virus with FRNT. Using the 1961 FRNT results (65·5%) and 1035 imputed results (ie, 154 presumed seronegative children and 881 presumed seropositive children; 34·5%), we determined that 320 (10·7%) of 2996 participants were dengue naive and 2676 (89·3%) were seropositive.
FRNT showed that 166 (8·5%) of 1961 (non-imputed) samples were dengue seronegative and 1795 (91·5%) had evidence of previous dengue infection; 292 (16·3%) of 1795 samples had a monotypic response and 1503 (83·7%) had a multitypic response (table 2). Of the 292 samples with monotypic seropositivity, 37 (12·7%) were DENV 1 serotype, 181 (62·0%) were DENV 2, 21 (7·2%) were DENV 3, and 53 (18·1%) were DENV 4. The IgG ELISA seropositivity varied by dengue virus serotype: 36 (97·3%) of 37 samples with DENV 1, 138 (76·2%) of 181 with DENV 2, 17 (81·0%) of 21 with DENV 3, and 32 (60·4%) of 53 with DENV 4 were seropositive with ELISA (p=0·0011). Of the 2676 sera classified as seropositive by FRNT, 86 (3·2%) were seronegative by dengue IgG ELISA (false-negative). 68 (79·1%) of these 86 samples had a monotypic dengue profile.
Table 2:
Performance of the IgG indirect ELISA for determination of dengue serostatus compared with the FRNT reference
| FRNT (monotypic or multitypic profile) |
FRNT (monotypic profile only) |
|||||
|---|---|---|---|---|---|---|
| Seronegative (dengue naive) | Seropositive | Total | Seronegative (dengue naive) | Seropositive | Total | |
|
| ||||||
| Seronegative (IgG ELISA index value <0'9) | 155 (64–3%) | 86 (35–7%) | 241 (100–0%) | 155 (69–8%) | 67 (30–2%) | 222 (100–0%) |
| Seropositive (IgG ELISA index value >0–9) | 11 (0–6%) | 1709 (994%) | 1720 (100–0%) | 11 (4–7%) | 225 (95–3%) | 236 (100–0%) |
| Total | 166 (8–5%) | 1795 (91–5%) | 1961 (100–0%) | 166 (36–2%) | 292 (63–8%) | 458 (100–0%) |
Data are n (%). FRNT=focus reduction neutralisation test.
We generated a scatter and box plot of the 1961 dengue IgG ELISA index values compared with the dengue serostatus classification by FRNT as naive, monotypic, or multitypic (appendix p 31). The IgG ELISA index values showed significant differences among the three serostatus groups (p<0·0001) and three pairwise comparisons (p<0·0001). We created a receiver operating characteristics curve that identified an optimal cutoff point of 0·859 for IgG ELISA index value (appendix p 32). On the basis of this cutoff point, an IgG ELISA index value of less than 0·9 was considered dengue seronegative. The estimated area under the curve value was 0·98 (95% CI 0·97–0·99). There were only six samples within the ELISA equivocal index value range of 0·9–1·1, five of which were seropositive (three multitypic and two monotypic) and one was seronegative.
We assessed the performance of the IgG ELISA using the 0·9 index value cutoff against the FRNT binary outcomes of dengue seronegative versus seropositive, and dengue seronegative versus seropositive with monotypic profile only (table 3). In the assessment of seronegative versus seropositive samples, compared with FRNT, the ELISA had a sensitivity of 95·2% (95% CI 94·2–96·2), a specificity of 93·4% (89·6–97·2), a positive predictive value of 99·4% (99·0–99·7), and a negative predictive value of 64·3% (58·3–70·4); kappa coefficient 0·75 (0·69–0·79); and McNemar’s test p<0·0001. In the assessment of seronegative versus seropositive samples with a monotypic profile only, the ELISA showed a lower sensitivity of 77·1% (72·2–81·9), and unchanged specificity.
Table 3:
Sensitivity and specificity of the IgG indirect ELISA for dengue serostatus compared with the FRNT reference
| Monotypic or multitypic profile | Monotypic profile only | |
|---|---|---|
|
| ||
| Sensitivity (95% CI) | 95–2% (94–2-96–2) | 77–1% (72–2-81–9) |
| False-negative rate | 4–8% | 22–9% |
| Specificity (95% CI) | 93–4% (89–6-97–2) | 93–4% (89–6-97–2) |
| False-positive rate | 6–6% | 6–6% |
| Positive predictive value (95% CI) | 99–4% (99–0-99–7) | 95–3% (92–6-98–0) |
| Negative predictive value (95% CI) | 64–3% (58–3-70–4) | 69–8% (63–8-75–9) |
FRNT FRNT=focus reduction neutralisation test.
We did full dilution neutralisation testing of the 11 serum samples (table 2) that were false positive by ELISA against dengue virus, Zika virus, and Japanese encephalitis virus (appendix p 33). Four (36.4%) of the 11 samples were found to be seronegative for dengue virus, Zika virus, and Japanese encephalitis virus, three (27·3%) were seropositive for Zika virus, two (18·2%) were seropositive for Japanese encephalitis virus, and two (18·2%) were seropositive for Zika virus and Japanese encephalitis virus.
Discussion
Based on an area under the curve value of 0·98, a sensitivity of 95·2%, and specificity of 93·4%, there was a high diagnostic accuracy and agreement of the ELISA compared with FRNT across the binary classifications of seropositive and seronegative. However, if used as a screening test, the 6·6% of individuals who are falsely seropositive on dengue IgG ELISA could be erroneously vaccinated and might be at risk of developing severe dengue infection during a subsequent exposure to wild-type dengue virus. 4·8% of individuals with falsely seronegative dengue IgG ELISA could also be erroneously left out of vaccination. Most of the false-negative results on dengue IgG ELISA had a monotypic dengue profile. The assay was not efficient in detecting monotypic dengue immunity, which is the profile that would benefit the most from CYD-TDV vaccination. The majority of the false-positive results by dengue IgG ELISA were due to cross-reactions with other flaviviruses. The dengue IgG ELISA had a sensitivity that exceeded the ideal sensitivity threshold of 95%, but a specificity that did not meet the ideal specificity threshold of 98%.
In our cohort of children eligible to participate in the mass dengue vaccination campaign, a large majority (2676 [89.3%] of 2996) were seropositive for dengue virus, and 1503 (76.6%) of 1961 had a multitypic response. Although the serological test results were not available in time for screening before vaccination, it is useful to note retrospectively that only 320 (10.7%) of the children in our study, who are likely to be representative of those who were invited to participate in the mass dengue vaccination campaign, were dengue naive. Particularly in settings of high dengue virus transmission, apart from identifying dengue virus seropositivity and seronegativity, a test that can discriminate a monotypic versus multitypic dengue antibody response would be ideal for optimal use of the vaccine. Dengue vaccination of individuals with a dengue multitypic antibody profile might be less cost-effective than in those with a monotypic profile, as current evidence suggests low risk for developing severe disease during post-secondary dengue infections.7,8 Although vaccine efficacy might be similar in monotypic sero-positivity to that in multitypic seropositivity, vaccination of individuals with a monotypic profile, who have a higher risk of clinical dengue virus disease, could potentially result in a greater public health impact.
A study from 2013 assessed an anti-dengue IgG ELISA (Focus Diagnostics; Cypress, CA, USA) for its utility in determining previous dengue virus exposure in pretravel specimens from US residents.29 Validation of 121 results by plaque reduction neutralisation test showed that the IgG ELISA had a sensitivity of 100% and a specificity of 24%; changing the ELISA cutoff value from 1·0 to 3·0 improved its specificity to 96% but decreased the sensitivity to 85%. A systematic review in 2019 of dengue virus rapid diagnostic tests found no studies that reported data for determining dengue serostatus and few data on cross-reactivity with other viruses.30 The authors concluded that additional research was needed to determine how rapid diagnostic tests would perform in relevant populations targeted for dengue vaccination. A more recent study by the CYD-TDV manufacturer evaluated four dengue IgG rapid diagnostic tests and two dengue IgG ELISAs in a sample panel of sera from CYD-TDV clinical trials and from US donors to determine the suitability of the tests for dengue prevaccination screening.31 For estimation of specificity, they used 534 samples that were negative for previous dengue infection on neutralisation testing. Sensitivity was evaluated using 270 samples from virologically confirmed dengue virus cases and samples that were positive on neutralisation test. Cross-reactivity was assessed in dengue-seronegative samples that were seropositive for yellow fever (n=57), Japanese encephalitis virus (n=37), West Nile virus (n=59) or Zika virus (n=41). The rapid diagnostic tests showed high specificity (>98%) and low flavivirus cross-reactivity, but low to moderate sensitivities (40–70%). This finding is consistent with the rapid diagnostic tests design, as they were developed to detect high antibody levels during dengue virus illness, not low antibody titres from previous infection.32 The two ELISAs evaluated (PanBio and DxSelect, Focus Diagnostics) showed high specificity (>98%) and sensitivity (99%). A similar study used a panel of samples to assess a dengue virus rapid diagnostic test, and an ELISA available in Puerto Rico showed high specificity (>99%) but moderate sensitivities (61% and 76%).33
By comparison, our study used a larger sample size from a real-life cohort in a site that is endemic for dengue virus, but evaluated only one IgG ELISA. Our analysis was more detailed and included comparisons of the ELISA results with naive, monotypic, and multitypic profiles. Similarly to the previous study,31 the ELISA in this study was done by trained and experienced laboratory research technicians and the results might not be generalisable to when the test is done under normal conditions. ELISA testing must be done in a laboratory, and there is no option for point-of-care assessment, which might limit its utility as a prevaccination screening test. Our study was done in a narrow age range and in a setting with high dengue virus transmission, so the results might not be generalisable to other age groups and sites. Another potential limitation of our study is the use of actual and imputed FRNT results in the estimation of overall seroprevalence. Although FRNT has a higher throughput than the classic plaque reduction neutralisation method, FRNT is still technically demanding and labour intensive. Limiting the use of FRNT to the samples with the highest and lowest ELISA index values allowed us to run the FRNT on all the samples with ELISA index values in the middle range. In the calculation of the IgG ELISA performance estimates, we did not include the imputed 154 seronegative (index value <0·2) and 881 seropositive (>3·0) results, so as not to upwardly bias the sensitivity and specificity. However, by using only the 1961 samples with actual FRNT results, the specificity and sensitivity were underestimated, as we excluded samples that were likely to be true positives and true negatives.
In conclusion, we found that 89·3% of our cohort had evidence of previous dengue infection. The dengue IgG ELISA might be acceptable as a prevaccination screening test based on recommended target profiles,17–19 but it is important to point out its shortcomings. Dengue IgG ELISA can only provide a binary seronegative or seropositive result and does not differentiate between a monotypic and multitypic seropositive response. Dengue IgG ELISA is particularly poor in identifying those with a monotypic profile who would benefit the most from CYD-TDV vaccination for prevention of severe dengue virus disease. Furthermore, individuals who are falsely identified as seropositive by dengue IgG ELISA and subsequently vaccinated, might be at risk of developing severe disease during a subsequent exposure to wild-type dengue virus.
Supplementary Material
Research in context.
Evidence before this study
We searched PubMed for studies published in English between Jan 1, 2010, and Oct 9, 2020, using the search terms: (“dengue” OR “dengue fever”) AND (“sero*” OR “serologic” OR “serostatus”) AND (“enzyme-linked immunosorbent assay” OR “ELISA” OR “neutralization test” OR “rapid diagnostic test” OR “RDT”) AND (“comparison” OR “validation” OR “screening”). We selected relevant articles on the basis of the title and by reading the abstracts. A study from 2013 assessed dengue IgG ELISA for its utility in determining previous dengue virus exposure in pre-travel specimens from US residents and found high sensitivity but low specificity. A systematic review from January, 2019, concluded that dengue IgG rapid diagnostic tests perform reasonably well for the diagnosis of acute and recent dengue virus infection, but no studies evaluated the performance of rapid diagnostic tests for past dengue virus infection; therefore additional research is needed to determine how these tests would perform as a screening tool for vaccination. Studies from 2019 and 2020 by the CYD-TDV (Dengvaxia, Sanofi Pasteur) dengue vaccine manufacturer that used samples from US residents and from CYD-TDV trial participants showed varying sensitivities and specificities.
Added value of this study
Our study assessed dengue seroprevalence in a large number of Filipino children who were eligible to participate in a mass dengue vaccination campaign in Cebu, Philippines. Individual pre-vaccination screening was not yet recommended at that time but we were able to obtain baseline sera just before the campaign, which were later tested. We also evaluated the performance of the IgG ELISA for determining dengue serostatus. The comparator assay—the focus reduction neutralisation test—not only confirmed past dengue infection, but classified it as either monotypic or multitypic infection. Although the ELISA had overall high sensitivity and specificity for detecting seropositivity, it had poor sensitivity for detecting those with a history of monotypic dengue infection—the group that would benefit the most from CYD-TDV vaccination. We quantified the ELISA false-positive results that were due to cross-reaction with Zika virus or Japanese encephalitis virus.
Implications of all the available evidence
To our knowledge, this is the most comprehensive assessment to date of a dengue IgG ELISA as a potential prevaccination screening tool. Experts recommend that a pre-vaccination screening test should have a sensitivity and specificity of at least 90%, and ideally a sensitivity of 95% and specificity of 98%. Although we found that the dengue IgG ELISA had sensitivity and specificity of more than 90%, potential misclassification due to false-positive and false-negative results must be considered.
Acknowledgments
We thank the funders of the study: the Philippine Department of Health, Hanako Foundation, WHO, the Swedish International Development Cooperation Agency through the International Vaccine Institute, and the University of North Carolina, Chapel Hill, NC, USA. Statistical analysis was done at the International Vaccine Institute, through which the Swedish International Development Cooperation Agency grant was coursed. Staff at the University of North Carolina, Chapel Hill, NC, USA did the FRNT. We thank all of the study participants and our local collaborators and field staff.
Footnotes
Declaration of interests
ALL, MY, JVD, and KAA report receiving salaries from 2017 onwards as part of an ongoing separate study sponsored by the University of the Philippines Manila and funded by Sanofi Pasteur. JD was an unpaid external consultant in the Extended Study Group for dengue vaccine effectiveness evaluation studies in Asia in 2015 convened by Sanofi Pasteur; and is an unpaid investigator of an ongoing separate study sponsored by the University of the Philippines Manila and funded by Sanofi Pasteur. AMdS is listed as an inventor on pending patent applications filed by the University of North Carolina related to flavivirus diagnostics. All other authors declare no competing interests.
Contributor Information
Anna Lena Lopez, Institute of Child Health and Human Development, National Institutes of Health, University of the Philippines Manila, Manila, Philippines.
Cameron Adams, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Michelle Ylade, Institute of Child Health and Human Development, National Institutes of Health, University of the Philippines Manila, Manila, Philippines.
Ramesh Jadi, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Jedas Veronica Daag, Institute of Child Health and Human Development, National Institutes of Health, University of the Philippines Manila, Manila, Philippines.
Caitlyn T Molloy, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Kristal An Agrupis, Institute of Child Health and Human Development, National Institutes of Health, University of the Philippines Manila, Manila, Philippines.
Deok Ryun Kim, International Vaccine Institute, Seoul, South Korea.
Maria Wilda Silva, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA; Department of Health, Manila, Philippines.
In-Kyu Yoon, Coalition for Epidemic Preparedness Innovations, Washington, DC, USA.
Laura White, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Jacqueline Deen, Institute of Child Health and Human Development, National Institutes of Health, University of the Philippines Manila, Manila, Philippines.
Aravinda M de Silva, Department of Microbiology and Immunology, University of North Carolina School of Medicine, Chapel Hill, NC, USA.
Data sharing
Data may be made available according to the University of the Philippines Manila and the University of North Carolina data sharing policy, upon request to the corresponding author (JD).
References
- 1.Anderson KB, Gibbons RV, Cummings DA, et al. A shorter time interval between first and second dengue infections is associated with protection from clinical illness in a school-based cohort in Thailand. J Infect Dis 2014; 209: 360–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montoya M, Gresh L, Mercado JC, et al. Symptomatic versus inapparent outcome in repeat dengue virus infections is influenced by the time interval between infections and study year. PLoS Negl Trop Dis 2013; 7: e2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sabin AB. Research on dengue during World War II. Am J Trop Med Hyg 1952; 1: 30–50. [DOI] [PubMed] [Google Scholar]
- 4.Snow GE, Haaland B, Ooi EE, Gubler DJ. Review article: research on dengue during World War II revisited. Am J Trop Med Hyg 2014; 91: 1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Halstead SB. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis 1979; 140: 527–33. [DOI] [PubMed] [Google Scholar]
- 6.Katzelnick LC, Gresh L, Halloran ME, et al. Antibody-dependent enhancement of severe dengue disease in humans. Science 2017; 358: 929–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gibbons RV, Kalanarooj S, Jarman RG, et al. Analysis of repeat hospital admissions for dengue to estimate the frequency of third or fourth dengue infections resulting in admissions and dengue hemorrhagic fever, and serotype sequences. Am J Trop Med Hyg 2007; 77: 910–13. [PubMed] [Google Scholar]
- 8.Olkowski S, Forshey BM, Morrison AC, et al. Reduced risk of disease during postsecondary dengue virus infections. J Infect Dis 2013; 208: 1026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbett KS, Katzelnick L, Tissera H, Amerasinghe A, de Silva AD, de Silva AM. Preexisting neutralizing antibody responses distinguish clinically inapparent and apparent dengue virus infections in a Sri Lankan pediatric cohort. J Infect Dis 2015; 211: 590–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hadinegoro SR, Arredondo-García JL, Capeding MR, et al. Efficacy and long-term safety of a dengue vaccine in regions of endemic disease. N Engl J Med 2015; 373: 1195–206. [DOI] [PubMed] [Google Scholar]
- 11.WHO. Summary of the April 2016 meeting of the Strategic Advisory Group of Experts on immunization (SAGE). 2016. https://www.who.int/immunization/policy/sage/en (accessed April 26, 2016).
- 12.Larson HJ. Politics and public trust shape vaccine risk perceptions. Nat Hum Behav 2018; 2: 316. [DOI] [PubMed] [Google Scholar]
- 13.Sridhar S, Luedtke A, Langevin E, et al. Effect of dengue serostatus on dengue vaccine safety and efficacy. N Engl J Med 2018; 379: 327–40. [DOI] [PubMed] [Google Scholar]
- 14.WHO. Dengue vaccine: WHO position paper—September 2018. Weekly epidemiological record. 2018; 93: 457–76. [Google Scholar]
- 15.US Food and Drug Administration. Dengvaxia. 2020. https://www.fda.gov/vaccines-blood-biologics/dengvaxia (accessed March 9, 2020)
- 16.Ariën KK, Wilder-Smith A. Dengue vaccine: reliably determining previous exposure. Lancet Glob Health 2018; 6: e830–31. [DOI] [PubMed] [Google Scholar]
- 17.Rodríguez-Barraquer I, Salje H, Cummings DA. Dengue pre-vaccination screening and positive predictive values. Lancet Infect Dis 2019; 19: 132–34. [DOI] [PubMed] [Google Scholar]
- 18.Flasche S, Smith PG. Sensitivity and negative predictive value for a rapid dengue test. Lancet Infect Dis 2019; 19: 465–66. [DOI] [PubMed] [Google Scholar]
- 19.Wilder-Smith A, Smith PG, Luo R, et al. Pre-vaccination screening strategies for the use of the CYD-TDV dengue vaccine: a meeting report. Vaccine 2019; 37: 5137–46. [DOI] [PubMed] [Google Scholar]
- 20.Bravo L, Roque VG, Brett J, Dizon R, L’Azou M. Epidemiology of dengue disease in the Philippines (2000–2011): a systematic literature review. PLoS Negl Trop Dis 2014; 8: e3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Philippine Statistics Authority. Highlights of the Philippine population 2015 census of population. 2016. https://psa.gov.ph/content/highlights-philippine-population-2015-census-population (accessed May 23, 2020).
- 22.Bossuyt PM, Reitsma JB, Bruns DE, et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 2015; 351: h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collins MH, McGowan E, Jadi R, et al. Lack of durable cross-neutralizing antibodies against zika virus from dengue virus infection. Emerg Infect Dis 2017; 23: 773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montoya M, Collins M, Dejnirattisai W, et al. Longitudinal analysis of antibody cross-neutralization following zika virus and dengue virus infection in Asia and the Americas. J Infect Dis 2018; 218: 536–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.WHO. Guidelines for plaque reduction neutralization testing of human antibodies to dengue viruses. 2007. https://apps.who.int/iris/handle/10665/69687 (accessed July 3, 2020). [DOI] [PubMed]
- 26.Gromowski GD, Firestone CY, Bustos-Arriaga J, Whitehead SS. Genetic and phenotypic properties of vero cell-adapted Japanese encephalitis virus SA14-14-2 vaccine strain variants and a recombinant clone, which demonstrates attenuation and immunogenicity in mice. Am J Trop Med Hyg 2015; 92: 98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tissera H, Amarasinghe A, De Silva AD, et al. Burden of dengue infection and disease in a pediatric cohort in urban Sri Lanka. Am J Trop Med Hyg 2014; 91: 132–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altman DG. Practical statistics for medical research. London: Chapman and Hall, 1991. [Google Scholar]
- 29.Marrero-Santos KM, Beltrán M, Carrion-Lebron J, et al. Optimization of the cutoff value for a commercial anti-dengue virus IgG immunoassay. Clin Vaccine Immunol 2013; 20: 358–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo R, Fongwen N, Kelly-Cirino C, Harris E, Wilder-Smith A, Peeling RW. Rapid diagnostic tests for determining dengue serostatus: a systematic review and key informant interviews. Clin Microbiol Infect 2019; 25: 659–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonaparte M, Zheng L, Garg S, et al. Evaluation of rapid diagnostic tests and conventional enzyme-linked immunosorbent assays to determine prior dengue infection. J Travel Med 2019; 26: taz078. [DOI] [PubMed] [Google Scholar]
- 32.Hunsperger E, Peeling R, Gubler DJ, Ooi EE. Dengue pre-vaccination serology screening for the use of Dengvaxia. J Travel Med 2019; 26: taz092. [DOI] [PubMed] [Google Scholar]
- 33.Bonaparte M, Huleatt J, Hodge S, et al. Evaluation of dengue serological tests available in Puerto Rico for identification of prior dengue infection for prevaccination screening. Diagn Microbiol Infect Dis 2020; 96: 114918. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data may be made available according to the University of the Philippines Manila and the University of North Carolina data sharing policy, upon request to the corresponding author (JD).