Abstract
To maintain homeostasis, macrophages must be capable of assuming either an inflammatory or an anti-inflammatory phenotype. To better understand the latter, we stimulated human macrophages in vitro with TLR ligands in the presence of high-density immune complexes. This combination of stimuli resulted in a broad suppression of inflammatory mediators and an upregulation of molecules involved in tissue remodeling and angiogenesis. Transcriptomic analysis of TLR stimulation in the presence of immune complexes predicted the downstream activation of AKT and the inhibition of GSK3. Consequently, we pretreated LPS-stimulated human macrophages with small molecule inhibitors of GSK3 to partially phenocopy the regulatory effects of stimulation in the presence of immune complexes. The upregulation of DC-STAMP and matrix metalloproteases (MMPs) was observed on these cells and may represent potential biomarkers for this regulatory activation state. To demonstrate the presence of these anti-inflammatory, growth-promoting macrophages in a human infectious disease, biopsies from patients with Leprosy (Hanseniasis) were analyzed. The lepromatous form of this disease is characterized by hypergammaglobulinemia and defective cell-mediated immunity. Lesions in lepromatous leprosy contained macrophages with a regulatory phenotype expressing higher levels of DC-STAMP and lower levels of IL-12, relative to macrophages in tuberculoid leprosy lesions. Therefore, we propose that increased signaling by FcγR cross-linking on TLR-stimulated macrophages can paradoxically promote the resolution of inflammation and initiate processes critical to tissue growth and repair. It can also contribute to infectious disease progression.
Keywords: Human macrophages, lipopolysaccharide, transcription factors, Fcγ receptors, GSK3β, RNA-seq, immune complexes, leprosy
INTRODUCTION
The phenotypic changes that macrophages undergo subsequent to their encounter with pathogen-associated molecular patterns have been well described (1). The cell surface receptors and downstream signaling molecules that activate transcription factors to drive inflammatory responses have been thoroughly studied. These so-called M1 (inflammatory) macrophages make important contributions to the initiation of immune responses, but their myriad inflammatory secretory products can cause tissue damage when left uncontrolled. Working in the murine system, our laboratory characterized a population of regulatory macrophages that were generated by TLR stimulation in the presence of a second “reprogramming” stimulus. Paradoxically, the increased signaling strength through the dual stimuli resulted in reduced inflammatory and increase anti-inflammatory cytokine production (2–7). We hypothesized that these cells may play a role in dampening inflammation during the resolution of immune responses.
The first “reprogramming” stimulus identified in the murine system was immune complexes (IC) that signal through the macrophage Fcγ receptors (FcγR)(6). In macrophages, immune complexes induce the clustering of stimulatory FcγR receptors, resulting in the phosphorylation of Syk (8, 9). This initiates a powerful signaling cascade leading to actin remodeling, PI3K activation, and Ca+ mobilization (10). The most widely studied consequences of these signaling pathways in macrophages are the induction of phagocytosis and the generation of the respiratory burst, both important components of host defense. We previously demonstrated that FcγR ligation on TLR-stimulated murine macrophages can also result in the rapid and prolonged hyperphosphorylation of ERK, triggering chromatin remodeling at the IL-10 promoter leading to increased IL-10 transcription (11).
A similar regulatory phenotype has not been described in human macrophages and no biomarkers exist to identify these cells. Furthermore, our understanding of how to manipulate this activation state is limited. Here we use high-throughput approaches to characterize human regulatory macrophages generated in response to immune complexes (R-Mϕ-IC) with the aim of identifying biomarkers that could be used for their identification in tissue. By providing a global picture of the behavior and function of these human macrophages, our studies revealed that immune complexes can dampen inflammatory responses, and inhibit the nuclear translocation of GSK3β to induce the transcription of genes promoting tissue growth, angiogenesis, and extracellular matrix reorganization.
To begin to examine whether these growth-promoting R-Mϕ-IC could be exploited by intracellular pathogens to promote uncontrolled intracellular growth, we examined macrophages from the skin of people with Leprosy (Hanseniasis). Leprosy has been characterized as a ‘spectral’ disease (12, 13). On the paucibacillary tuberculoid end of the spectrum, the pathogens are cleared by activated macrophages following the development of cell mediated immunity (14, 15). Antibody levels in this disease are typically low or absent (16). In contrast, humoral immunity predominates in lepromatous leprosy. Antibody levels are high and bacteria grow uncontrolled in dermal macrophages (12, 15). Cellular immunity appears to be defective because patients generally fail to mount a delayed-type hypersensitivity (DTH) response to skin test antigens (15). The uncontrolled intracellular growth of bacteria in macrophages associated with the presence of immune complexes therefore define this permissive form of the disease.
METHODS
Differentiation of human monocyte-derived macrophages.
Human monocyte-derived macrophages (HMDMs) were purchased from HemaCare Corporation (Van Nuys, CA) or monocytes were differentiated in house using the Miltenyi Biotec pan monocyte isolation kit (San Diego, CA). Cells were cultured for 7–10 days in X-VIVO™ 15 serum-free media (Lonza, Walkersville, MD) containing 1% penicillin-streptomycin, 1% L-glutamate (Gibco, Gaithersburg, MD), and 20 ng/mL recombinant human M-CSF (Peprotech, Rocky Hill, NJ). Prior to stimulation, M-CSF-containing medium was removed and replaced with X-VIVO™ 15 media containing 2.5% fetal bovine serum (Atlanta Biologicals, Flowery Branch, GA). All studies on human monocyte-derived macrophages were approved by the University of Maryland, and the MedImmune Institutional Review Boards.
Cell culture and stimulation.
Macrophage stimulation conditions were generated by adding 30 ng/ml Ultra-pure LPS from Escherichia coli K12 (InvivoGen, San Diego, CA) (LPS-Mϕ) or LPS in combination with soluble immune complexes (R-Mϕ-IC), as described (7, 17). Particulate immune complexes were generated by adding rabbit IgG to 1.1 μM latex spheres overnight. Macrophages were stimulated with TLR agonists, Pam3Csk4, Poly I:C, or HKLM (InvivoGen) in the presence/absence of particulate immune complexes. The AKT inhibitor (MK-2206 2HCl) and the GSK3 inhibitor (SB415286) were purchased from ApexBio (Houston, TX) and Cayman Chemicals (Ann Arbor, MI), respectively, and used at 20 μM. The GSK3β isoform specific inhibitor (AZD2858) was provided by AstraZeneca and used at 750 nM.
RNA isolation and data processing.
Total RNA was extracted from cells and poly(A)+-enriched cDNA libraries were generated using the Illumina TruSeq Sample Preparation kit (SanDiego, CA). Paired end reads (100 bp) were obtained using an Illumina HiSeq 1500. Trimmomatic was used to remove any remaining Illumina adapter sequences from reads and trim read ends with quality score < 20 (18). Sequence quality metrics were assessed using FastQC (19). Reads were aligned to the human genome (hg19/GRCh37.62.v3) obtained from the UCSC genome browser (20) (http://genome.ucsc.edu) using TopHat (v 2.0.13) (21), with parameters matching previous work (22). The abundance of reads mapped to coding features was determined using HTSeq (23). Quantile normalization and log2-transformation was applied to all samples (24). Limma was used to conduct differential expression analyses (25). The voom module was used to transform the data based on observational level weights derived from the mean-variance relationship prior to statistical modeling. Experimental batch effects were adjusted for by including experimental batch as a covariate in our statistical model. Differentially expressed genes were defined as genes with a log2 fold-change > 1 and Benjamini-Hochberg (BH) multiple-testing adjusted P value < 0.05. All components of the statistical pipeline, named cbcbSEQ, can be accessed on GitHub (https://github.com/kokrah/cbcbSEQ/).
SOMAscan™ assay.
Cell culture supernatants were collected from macrophages at 24 hours, and analyzed using the SOMAscan™ proteomic assay (SOMAscan Assay 1.1k; SomaLogic; Boulder, CO). RFU values from SOMAscan™ were normalized against hybridization control sequences to correct for any systematic effects introduced during hybridization. Median normalization was performed across samples within arrays. Group comparisons were performed using Bayesian modified linear model (Limma package in R) (26). Between groups, comparisons were assessed using contrast and the P-values of the moderated-t-test were Benjamini–Hochberg adjusted. Differentially expressed proteins were defined as fold change (FCH) ≥1.5 and FDR ≤0.05.
RNAscope.
RNAscope probes for IL-10, LIF, and MMP10 were custom designed by Advanced Cell Diagnostics (Newark, CA), nucleotide accession numbers: https://www.ncbi.nlm.nih.gov/nuccore/NM_000572 (IL10), https://www.ncbi.nlm.nih.gov/ nuccore/NM_002421 (MMP1), https://www.ncbi.nlm.nih.gov/nuccore/NM_002309 (LIF). Macrophages were stimulated in 4-well chamber slides for 4 hours. Slides were stained with RNAscope Multiplex Fluorescent Reagent Kit v2, mounted with Fluoromount-G™ with DAPI ThermoFisher Scientific, Waltham, MA and imaged using the Zeiss LSM710 confocal Laser microscope (Carl Zeiss Microscopy GmbH, Jena, Germany) Fluorescence was quantified using Zen software by Zeiss.
Flow cytometry.
Human monocytes were differentiated in ultra-low attachment plates. APC conjugated antibody to DC-Stamp (Mouse IgG2b, Clone #788524) and BUV395-conjugated antibody to CD73 (Mouse IgG1k, Clone # AD2) (BD Biosciences, Franklin Lakes, NJ) were applied to the surface of stimulated macrophages. Alexaflour 647-conjugated antibody to MMP10 (Mouse IgG1, Clone#110316) and PE-conjugated antibody to LIF (Mouse IgG1k, Clone#1F10) was applied to permeabilized macrophages following brefeldin treatment. Data acquisition was carried out in FACSCanto II (BD Biosciences); analyses were done on FlowJo version 10.
ELISA.
Human IL-10 and IL-12p40 were detected by sandwich ELISA kits purchased from eBioscience (San Diego, CA, USA). ELISA kits for the remaining analytes were purchased from R&D (Minneapolis, MN).
Bioinformatics Analyses.
Ingenuity Pathway Analysis (IPA) software (27) was used to predict diseases, functions, and upstream regulators. Regulators with low numbers of known targets (<10) were removed. Pathway enrichment analysis was conducted using the Reactome Pathway Analysis package in R (28).
Antibody array.
Phosphorylation of AKT signaling pathway molecules was assessed in whole cell lysates using PathScan AKT Signaling Antibody Array Kit (Cell Signaling Technologies, Danvers, MA). BCA assay (Pierce Biotechnology, Waltham, MA) was used to determine protein concentration of the samples. Array densities were quantified using Image J software (29).
Western blotting.
Nuclear and cytosolic fractions were isolated using NE-PER nuclear extraction kit (ThermoFisher Scientific, Waltham, MA). Protein lysates were incubated for 2 hours at 4°C with protein agarose A (Santa Cruz, Dallas, Texas). Phospho-ERK 1/2, phospho-AKT, cofilin, phospho-GSK3β, GSK3β, histone H3 antibodies were purchased from Cell Signaling Technologies (Danvers, MA); phospho-GSK3α/β antibody was purchased from R&D Systems; actin antibody was purchased from Santa Cruz (Dallas, Texas). A conformation-specific anti-rabbit IgG (Cell Signaling Technologies, Danvers, MA) was used as a secondary antibody when needed.
Quantitative Real-time PCR (qRT-PCR).
RNA for qRT-PCR was isolated using the Trizol method (ThermoFisher Scientific, Waltham, MA). cDNA was synthesized using Superscript® Vilo™ cDNA synthesis kit (ThermoFisher Scientific). Relative quantification of RNA was done using SYBR-Green based real time PCR (ThermoFisher Scientific). The samples were run in Roche Light Cycler® 480. Relative differences were calculated using the ΔΔCt method with beta actin as a control.
Histology.
Tissue fragments were cut on a microtome to 4–5μm thick and stained with hematoxylin-eosin (HE) and analyzed by light microscopy. The DCSTAMP immunohistochemical protocol was carried out using moist heat (95oC) for 20 minutes in citrate pH 6.0 (Dako Target Retrieval Solution Cytomation). Endogenous peroxidase was blocked with hydrogen peroxide added to methanol (3%). Nonspecific reactions were blocked with powdered milk, diluted in PBS, in a humid chamber for 30 min at room temperature. Sections were incubated with primary serum at 4° C. Biotinylated secondary antibody (DAKO kit - LSAB 2 System, Peroxidase - K0675), was added as previously described (30). Streptavidin peroxidase complex (DAKO kit - LSAB) was added for 30 min in a humid chamber at room temperature. Developing solution of diaminobenzidine (DAB) (Sigma ®) plus a hydrogen peroxide solution was added for 5 min at room temperature. Washed slides were counterstained with Harris hematoxylin, washed, dehydrated, cleared and mounted with Entellan®.
RESULTS
Transcriptomic analysis of human macrophages stimulated in the presence of immune complexes.
RNA-sequencing (RNA-seq) was performed to characterize global changes in gene expression in human macrophages under inflammatory or anti-inflammatory in vitro stimulation conditions (Supplemental Table 1). The transcriptome of non-stimulated (NS) resting macrophages was compared to macrophages stimulated with lipopolysaccharide alone (LPS) Mϕ, and macrophages stimulated with LPS in the presence of high-density immune complexes (LPS+IC). For most of these studies, soluble immune complexes formed by the addition of ovalbumin/anti-OVA were used to co-stimulate macrophages. Principal component analysis of coding transcripts at 4 hours post-stimulation revealed samples from the same treatment groups clustering together (Supplemental Figure 1A). Principal component 1 explained 75% of the variance, separating resting macrophages from those stimulated with LPS. Principal component 2 explained 13% of the variance, and segregated LPS-Mϕ from LPS+IC-Mϕ (Supplemental Figure 1A). We selected 4 hours post-stimulation for our analysis to identify early transcripts induced by LPS and those regulated directly by the addition of immune complexes, rather than those influenced by LPS-induced macrophage secretory products.
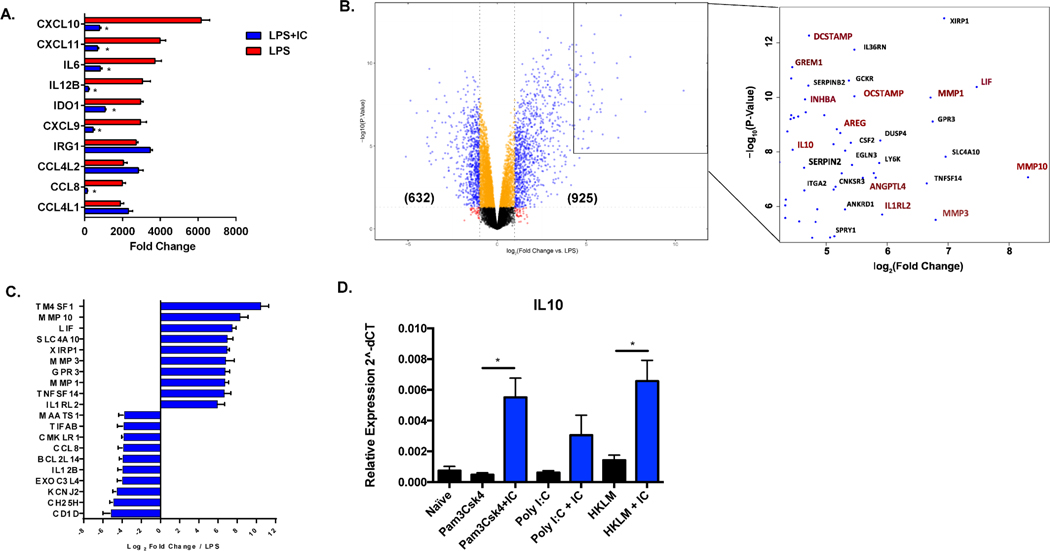
Stimulation of human macrophages with LPS resulted in numerous changes in gene expression, as previously reported (31, 32). Over 4,500 genes were significantly differentially expressed in response to stimulation with LPS, with 2017 transcripts upregulated and 2521 downregulated, accounting for approximately 38% of all detectable genes in the transcriptome (Supplemental Figure 1B). The ten most highly upregulated genes in LPS-treated macrophages, relative to non-stimulated cells are shown in Figure 1A. Consistent with previous reports (31, 33), LPS stimulation led to increased production of transcripts encoding inflammatory mediators, such as the chemokines CXCL9, CXCL10, CXCL11, and CCL8, and the cytokines IL-6 and IL-12B, antimicrobial IRG1, and indolamine dioxygenase (IDO1), an enzyme that converts tryptophan to kynurenine to limit tryptophan availability (34) (Fig. 1A, red bars). We previously demonstrated (4) that murine macrophages responded to stimulation in the presence of IC with decreased expression of IL-12 and increased expression of IL-10. In human macrophages, the addition of IC to LPS stimulated cells resulted in significantly lower expression of the inflammatory cytokines IL-6 and IL-12, and the inflammatory chemokines CXCL9,10, 11 and CCL8 (Fig. 1A, blue bars). Overall, the addition of immune complexes to LPS-stimulated human macrophages resulted in the differential expression of 1557 genes (13%, Supplemental Figure 1B), including 925 transcripts that were significantly upregulated and 632 downregulated (Figure 1B). Surprisingly, 80% of these differentially expressed human genes were not changed in similarly stimulated murine macrophages (Supplemental Figure 1C).
Figure 1. Global changes in gene expression following the stimulation of human macrophages in the presence of immune complexes.
Human monocyte derived macrophages were stimulated with LPS (30 ng/mL) or LPS plus immune complexes (ovalbumin/anti-ovalbumin) for 4 hours, and total mRNA was isolated and sequenced on the Illumina platform (n=3). (A) The 10 most highly upregulated genes in LPS-stimulated macrophages are designated by red bars and expressed as fold change relative to resting macrophages (mean ± SEM). Values for corresponding fold-changes in LPS+IC stimulated macrophages are designated by blue bars. Asterisks designate significant differences (adjusted P-value ≤0.05) between LPS vs LPS+IC. (B) Volcano plot of genes expressed in LPS+IC relative to LPS alone. In parentheses are the number of genes upregulated or downregulated by greater than 2-fold, with adjusted P-value ≤0.05. The box shows some of the most highly upregulated genes by fold-change and significance. Genes marked in red are associated with cell growth, angiogenesis, and extracellular matrix remodeling. (C) The top 10 most highly upregulated and downregulated genes in macrophages stimulated with LPS+IC relative to stimulation with LPS alone, expressed by log2 fold change (mean ± SEM). (D) RT-PCR of IL-10 transcripts following stimulation with TLR agonists, Pam3Csk4, Poly I:C, or HKLM in the absence (black bars) or presence (blue bars) of particulate immune complexes (IC) composed of latex beads coated with rabbit polyclonal IgG. Y axis represents expression (2˄dCT) relative to Actin/Rab7 (mean ± SEM, n=3).
The most highly upregulated transcripts in LPS+IC-Mϕ relative to LPS-Mϕ included genes that have been associated with cell growth, angiogenesis, and remodeling of the extracellular matrix (Figure 1B and C). The 10 most highly upregulated genes included several matrix metalloproteinases (MMP1, MMP3, MMP10) known to be involved in tissue remodeling, angiogenesis, and extracellular cytokine regulation (35, 36). Some of the other top upregulated genes included TM4SF1 (cell growth and angiogenesis (37)), LIF (self-renewal of stem cells (38)), DCSTAMP (39) and OCSTAMP (phagocytic activity and immune tolerance (40)), ANGPTL4 (lipid metabolism (41)), AREG (cell growth, EGF and TGFA signaling (42)), IL1RL2 (IL-36 signaling and epithelial barrier function (43)), and GREM1 (monocyte chemotaxis inhibition (44)). Of the thirty most highly upregulated genes (by fold change and P-value), twelve genes have been reported by others to be directly involved in cell growth and angiogenesis (Figure 1B, red). The 10 most highly downregulated genes in LPS+IC-Mϕ included IL-12B (TH1 responses), CCL8 (inflammatory chemokine), and CH25H (lipid metabolism, chemotaxis) (Figure 1C), consistent with our premise that human macrophages stimulated with LPS+IC exhibit immunoregulatory activity.
To determine whether the reprogramming of macrophages to an anti-inflammatory phenotype is restricted to a single TLR, human macrophages were stimulated with other TLR ligands, including Pam3CysK, Poly I:C, and heat-killed Listeria monocytogenes (HKLM). The restriction of reprogramming to soluble immune complexes was also tested by using particulate immune complexes, consisting of IgG adsorbed to beads (Supplemental Figure 1D). Stimulation of macrophages with other TLRs plus particulate IC resulted in an increase in the production of IL-10 (Figure 1D), as well as other regulatory transcripts including MMP1, MMP10 and ANGL4 (Supplemental Figure 1D).
The macrophage secretome.
To characterize the secretome of R-Mϕ, we performed a SOMAscan analysis on cell culture supernatants of human macrophages collected 24 hours following stimulation. Fold changes in secreted protein levels were compared to the corresponding gene expression data from RNA-seq using Spearman correlation to determine whether changes in secreted proteins corroborate the transcriptional changes observed in RNA-seq (Supplemental Fig. 2A). The r value of 0.65 (P-value = 0.0001) indicates a positive correlation between RNA and protein changes. To show that IC were, indeed binding to macrophages FcγR, we measured FcγR1 (CD64) and FcγR3 (CD16) expression before and after stimulation. As expected, the addition of IC to macrophages resulted in FcγR internalization and reduced surface expression of these two receptors (Supplemental Figure 2B).
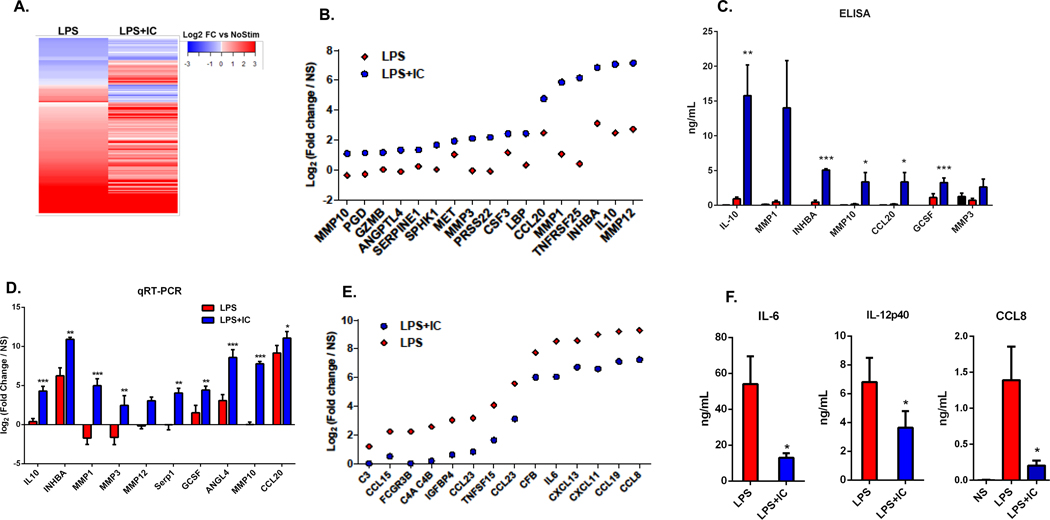
Compared to LPS stimulation, LPS+IC stimulated macrophages increased the secretion of 78 proteins and decreased the secretion of 149 proteins (FC>1.5 and FDR <0.05) (Figure 2A). The 17 most highly upregulated secretory products in R-Mϕ-IC relative to LPS-Mϕ are shown in Figure 2B, expressed as fold-change relative to non-stimulated macrophages. These secretory products include several matrix metalloproteases (MMP3, MMP10, MMP12), IL-10, Activin A (INHBA), and CCL20. An ELISA was performed to confirm that stimulation of human macrophages with LPS in the presence of immune complexes induced a significant increase in the secretion of selected proteins compared to LPS alone (Figure 2C, blue bars). Quantitative real-time PCR at 4 hours post-stimulation demonstrated a similar upregulation of 10 of the 17 genes at the transcript level (Figure 2D).
Figure 2. The macrophage secretome.
Cell culture supernatants, collected from human monocyte-derived macrophages at 24 hours following stimulation with LPS or LPS+IC were compared to that of unstimulated macrophages. (A) Differential protein expression, as measured by SOMAScan technology, in the supernatants of LPS or LPS+IC stimulated macrophages relative to unstimulated macrophages are represented by a heatmap. (B) The top 17 up-regulated proteins in LPS+IC relative to LPS alone, and their fold-change in LPS+IC (blue) or LPS-stimulated macrophages (red). (C) Supernatants were analyzed by ELISA to measure the accumulation of 7 proteins that were identified in the SOMAScan analysis following no stimulation (black) or stimulation with LPS+IC (blue) or LPS alone (red) (mean ± SEM, n=5) (D) RT-PCR was used to measure transcript levels of 10 of the genes that were identified in the SOMAScan analysis (4 hours post-stimualtion). Bars represent log2 fold change (mean ± SEM, n=6 versus non-stimulated macrophages. (E) The top 14 downregulated proteins from the SOMAScan analysis when comparing LPS+IC to LPS stimulation are shown. Dots represent LPS+IC (blue) or LPS alone (red) log2 fold-changes relative to unstimulated macrophages. (F) Supernatants of stimulated macrophages were analyzed by ELISA to measure the downregulation of IL-6, IL-12(p40), and CCL8 following stimulation with LPS+IC (blue) or LPS (red) (mean ± SEM, n=3). Statistical significance indicated in panels C, D, and F was calculated using a paired t-test. Asterisks designate significant differences (*P-value ≤0.05, ** P-value ≤0.01, *** P-value ≤0.001 ) between LPS vs LPS+IC.
The addition of IC to LPS-stimulated macrophages also resulted in the downregulation of several secreted products (Figure 2E). The secretion of pro-inflammatory chemokines (45–47) CXCL11, CXCL13, CCL8, and CCL15 was decreased by the addition of IC to LPS-stimulated macrophages, as was the secretion of the cytokine IL-6. Protease inhibitors and complement pathway proteins were also inhibited, indicating that IC co-stimulation downregulates several pathways of inflammation. An ELISA confirmed the decrease in the secretion of IL-6, IL-12 (which is not included in the SOMAscan platform) and CCL8 by the addition of IC to LPS-stimulated macrophages (Figure 2F).
Functional properties of macrophages stimulated in the presence of immune complexes.
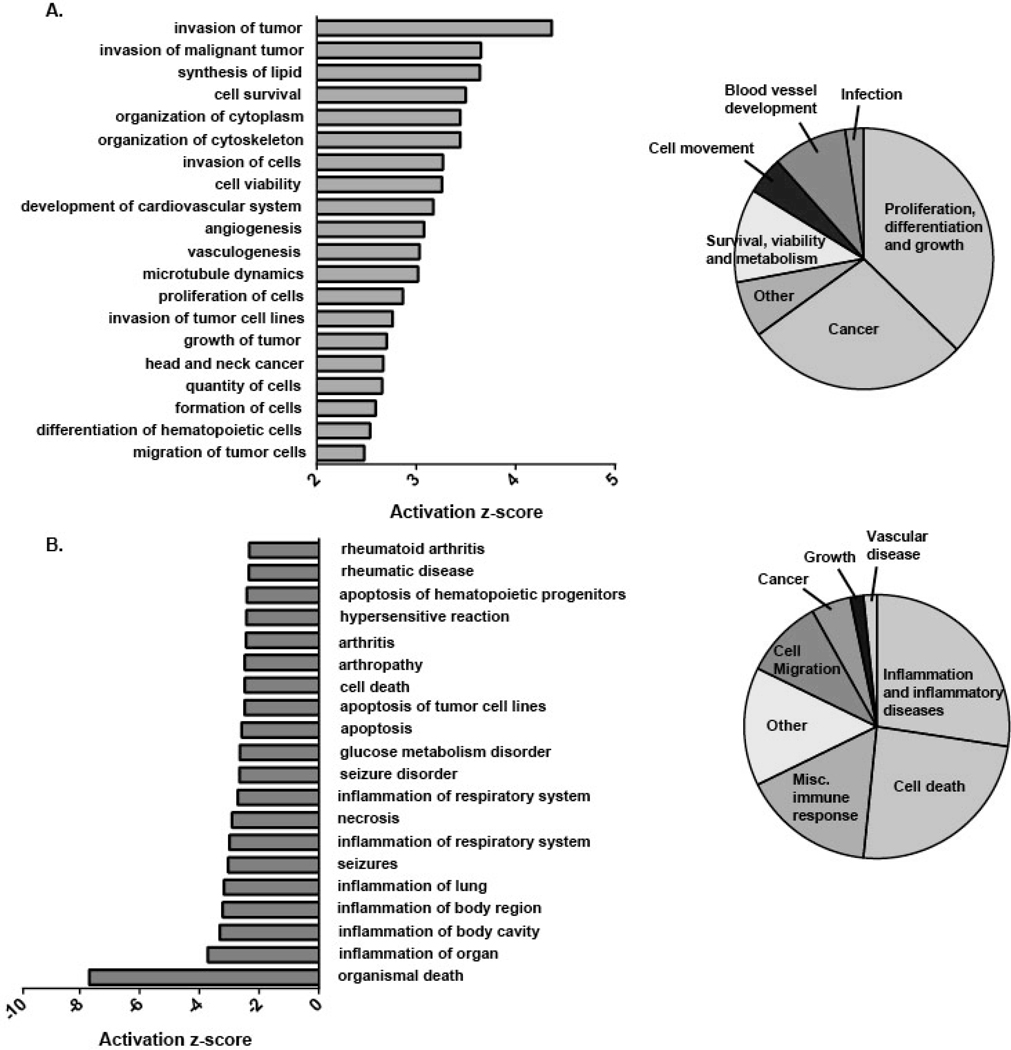
To determine the functional significance of the differentially expressed genes in R-Mϕ, the RNA-seq differential expression data was uploaded into the Ingenuity Pathway Analysis (IPA) tool. The Diseases and Functions report from IPA predicted increased activation of angiogenesis, cell proliferation, and tumor vulnerability (Fig. 3A), and a decreased activation of inflammation and rheumatic diseases (Fig. 3B). In total, 1,337 unique genes contributed to the predicted upregulated functions and 716 unique genes contributed to the 33 predicted downregulated functions. The IPA Diseases and Functions report is consistent with a pro-survival, pro-growth, and anti-inflammatory phenotype of macrophages following stimulation in the presence of immune complexes.
Figure 3. Functional properties of macrophages stimulated in the presence of immune complexes.
Differentially expressed genes (DEGs) in macrophages (FC ≥2 or ≤ −2) stimulated with LPS+IC were compared to cells stimulated with LPS alone, and uploaded into the Ingenuity Pathway Analysis Program and the Diseases and Functions Report was generated (n=3). Bar graphs (left) represent the top 20 (A) positively and (B) negatively enriched pathways ranked by z-score. Enriched diseases and functions were manually categorized into broader groups (pie charts) in a non-overlapping manner.
Taken together, our data confirms the work of several groups (31,32), that TLR stimulation alone triggers major gene expression changes in human macrophages leading to an inflammatory macrophage phenotype (so-called M1). The addition of soluble or particulate immune complexes at the time of stimulation leads to a downregulation of inflammatory gene expression and an enhanced array of immunoregulatory genes that have important roles in cell growth and repair, tissue remodeling, and angiogenesis.
Bioinformatic prediction and functional confirmation of upstream regulators of the regulatory phenotype.
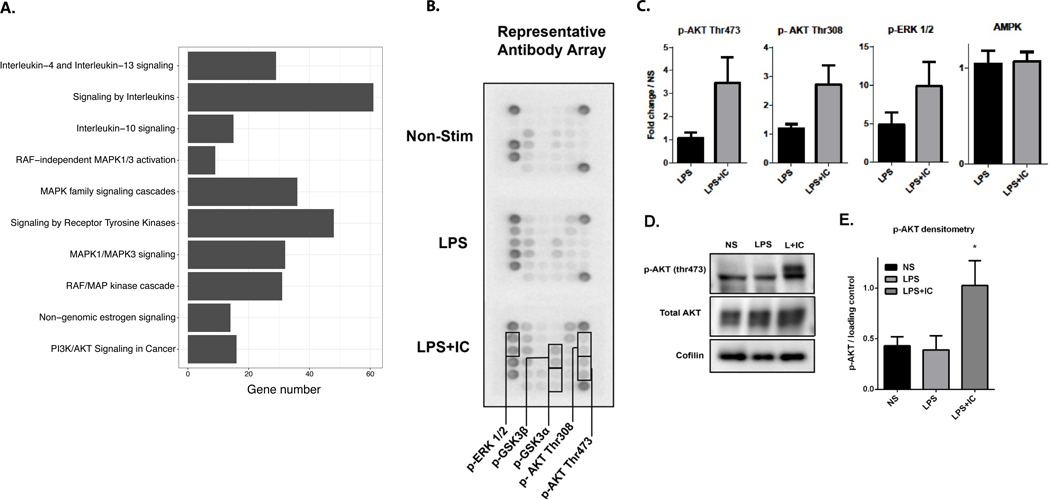
The identification of signaling pathways and key regulators of transcriptional programs has the potential to lead to new treatments that induce or inhibit the regulatory macrophage phenotype in vivo. To identify the pathways and master regulators involved in generation of the R-Mϕ-IC phenotype, differentially upregulated genes from RNA-seq (R-Mϕ-IC vs LPS-Mϕ) were analyzed using the Reactome pathway analysis package in R. The results showed enrichment of transcripts associated with IL-10 production as well as the MAP kinases and members of the AKT signaling pathway (Figure 4A). Previous publications in the murine system implicated a role for AKT/ERK signaling in macrophage stimulation (11, 48, 49), so we proceeded to analyze phosphorylation of proteins in the AKT/ERK pathway. Although AKT and ERK are part of distinct pathways, they share upstream regulators and their pathways interact at multiple points. An antibody array for phosphorylated proteins in the AKT/ERK pathway was utilized, adding protein lysates from human macrophages 20 minutes after stimulation (Figure 4B). LPS alone modestly activated the phosphorylation of AKT and ERK (Figure 4C) while LPS+IC amplified phosphorylation of each of these proteins (Fig. 4C, grey bars). AMPK was used as a control whose phosphorylation was not increased in response to IC (Figure 4C). Several other kinases, including mTOR, PTEN and PDK1 were similarly unresponsive to IC (Supplemental Figure 2C). Western blotting confirmed that phosphorylation of ERK (Supplemental Figure 2D) and AKT at threonine 473 was higher under LPS+IC treated conditions (Fig. 4D and E).
Figure 4. Signaling in R-Mϕ-IC.
(A) A bar graph of 10 enriched (ordered by adjusted P-value < 0.02) Reactome pathways among genes upregulated in macrophages stimulated with LPS+IC vs. LPS (n=3). The size of the bar along the X-axis indicates the number of differentially upregulated genes found in each pathway. (B) Representative image of antibody array (Cell Signaling Technologies) from whole cell lysates of non-stimulated human macrophages or those stimulated for 20 minutes with LPS alone or LPS+IC. (C) Densitometry was calculated using Image J software. Values are mean fold change compared to non-stimulated (NS) macrophages and error bars are SEM (n=3). (D) A representative western blot of macrophage lysates confirms antibody array results for p-AKT thr473 and total AKT. (E) Densitometry of three independent western blots to quantify AKT phosphorylation following stimulation with LPS or LPS+IC relative to non-stimulated cells (mean ± SEM, n=3). Statistical significance was calculated using a paired t-test; *P-value ≤0.05.
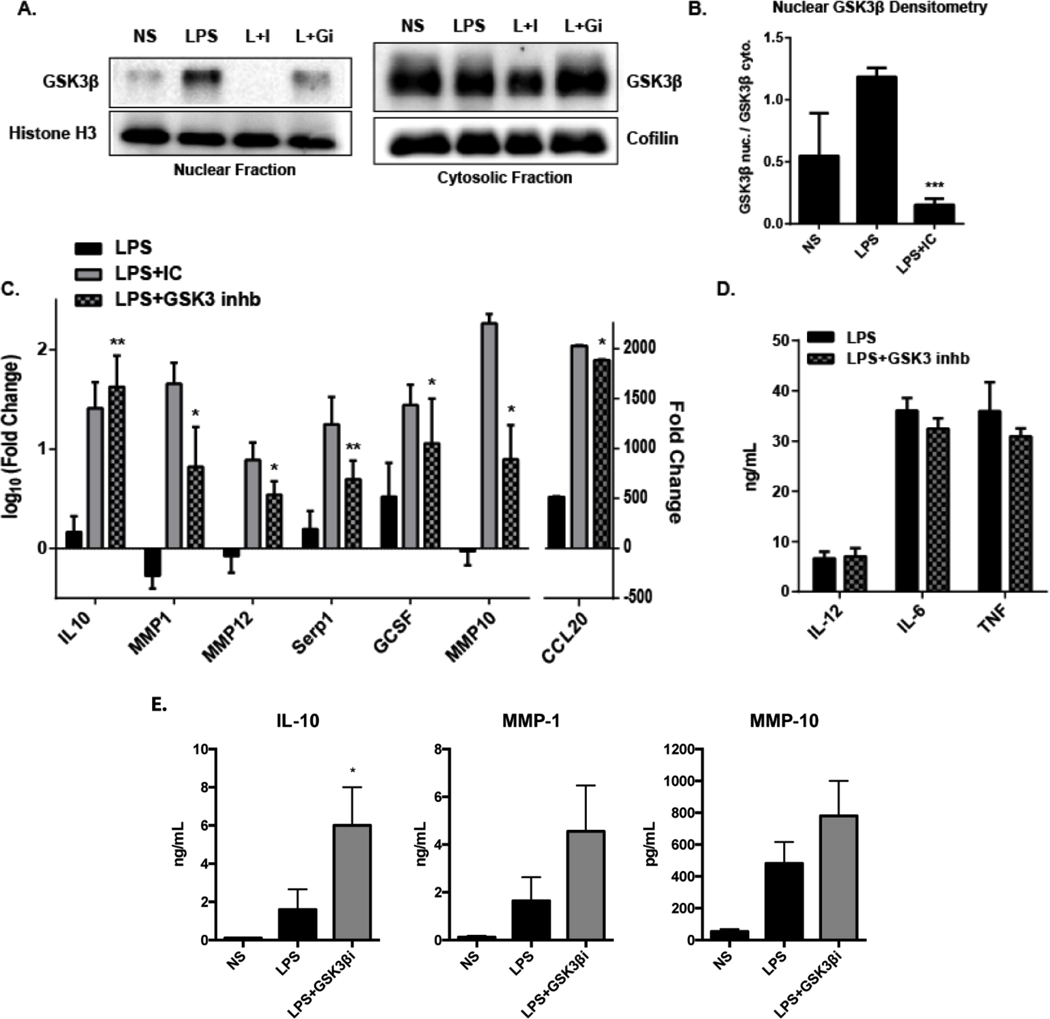
Previous studies have demonstrated that activated AKT can inhibit GSK3 signaling (49), and furthermore that GSK3 inhibition can increase IL-10 expression in macrophages (50). This led us to hypothesize that activation of AKT by IC may permit the expression of regulatory molecules by inhibiting GSK3. It is known that GSK3β shuttles between the cytoplasm and the nucleus and active GSK3β has been shown to accumulate in the nucleus (51), so we probed GSK3β in cytoplasmic and nuclear fractions of R-Mϕ-IC lysates. Western blotting revealed that LPS stimulation led to an increase of GSK3β in the nucleus (Figure 5A). However, co-stimulation with IC prevented nuclear translocation, significantly reducing nuclear levels of GSK3β compared to LPS alone (Fig. 5A and B). An ATP-competitive inhibitor of GSK3α/β(52) was used in lieu of IC to determine if GSK3 inhibition of LPS-stimulated macrophages could upregulate some of the transcripts that were induced in R-Mϕ-IC. When combined with LPS stimulation, the GSK3 inhibitor SB415286 transcriptionally upregulated 7 out of the 10 genes tested in the panel of markers for LPS+IC (Fig. 5C). GSK3 inhibition did not reduce the expression of three inflammatory cytokines typically decreased by co-stimulation with IC (Fig. 5D). These results indicate that GSK3 inhibition is important for inducing the expression of regulatory genes in human macrophages, but does not play a role in inhibiting LPS-triggered inflammatory responses.
Figure 5. GSK3β inhibition in R-Mϕ-IC.
5.(A) Western blotting for GSK3β in the nuclear (left) or cytosolic (right) fractions of macrophage lysates collected 30 minutes after stimulation with LPS alone, LPS and immune complexes (L+I) or LPS in combination with SB415286 (20 μM), an inhibitor of GSK3 (L+GI). (B) Densitometry was calculated using Image J software. Bars represent mean fold change ± SEM in the nucleus over cytosolic levels (n=3). (C) RT-PCR results for macrophages stimulated with LPS (black), LPS+IC (grey) or LPS in the presence of 20 μM SB415286 to inhibit GSK3 (stippled). RNA was collected 7 hours after stimulation, expressed as fold change (log10) ± SEM relative to non-stimulated macrophages (n=4, * P-value ≤0.05; ** P-value ≤0.01). (D) The production of inflammatory cytokines, IL12(p40), IL-6, and TNF was measured by ELISA, 7 hours after stimulation with LPS alone (black) or LPS + SB415286 (grey) to inhibit GSK3 (mean ± SEM n=3). (E) Macrophages were stimulated with LPS alone (black bars) or LPS with AZD2858 (750 nM), a small-molecule inhibitor specific for GSK3β (Grey bars). The levels of IL-10, MMP1, and MMP10 in supernatants was measured by ELISA 16 hours later Statistical significance for panels B through E was calculated using a paired t-test. (mean ± SEM, n=3, * P-value ≤0.05).
To determine if a GSK3β isoform-specific inhibitor could also induce an anti-inflammatory response, and if the response could be observed at the protein level, the small-molecule inhibitor of the GSKβ, AZ2848, was used in combination with LPS to activate macrophages. ELISA analysis showed that GSK3β-specific inhibition significantly upregulated IL-10. MMP1 and 10 secretion trended toward upregulation, but did not reach statistical significance due to high variability in donor responses (Figure 5E).
Biomarker Identification.
Just as macrophages assume a continuum of phenotypes (2), so do the biomarkers. This makes the definitive identification of biomarkers for specific human macrophage phenotypes a difficult task. The transient nature of the inflammatory and regulatory phenotypic changes adds to the challenge. Therefore, two parallel approaches were taken to identify potential biomarkers for human R-Mϕ-IC: (1) in situ hybridization to visualize transcript expression and (2) flow cytometry and immunohistochemistry to identify protein expression.
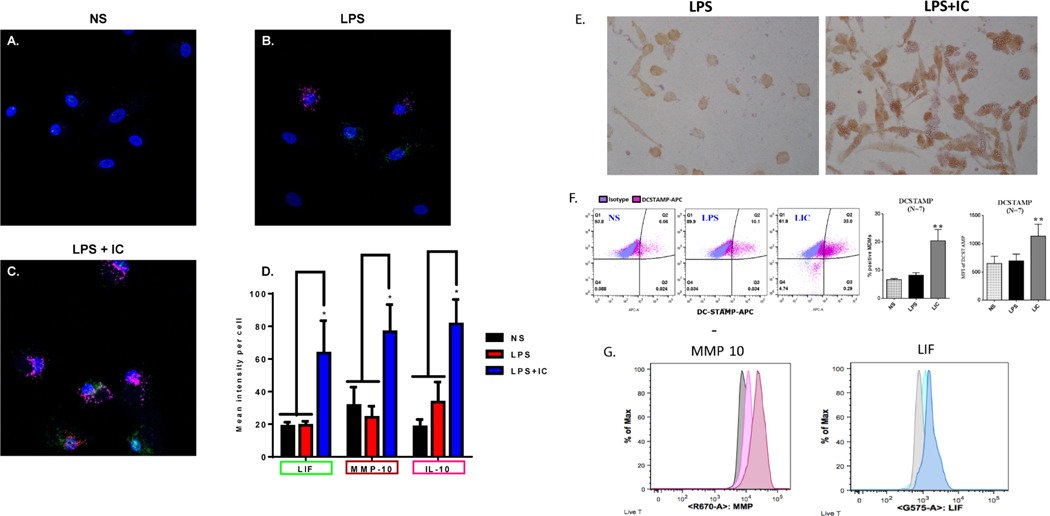
Three genes, IL-10, LIF, and MMP10, were chosen from the RNA-seq analysis as potential RNA biomarkers based on their high fold-change when comparing R-Mϕ-IC to LPS-Mϕ and NS-Mϕ (see Figure 1B). Macrophages from three donors were fixed and stained using custom probes developed by Advanced Cell Diagnostics (Newark, CA). RNAscope confirmed our RNA-seq analysis, showing little to no expression of these three transcripts (Figure 6A and 6D) in resting non-stimulated (NS) macrophages. LPS stimulated macrophages showed a slight increase in the expression of transcripts for IL-10, but little to no increase in transcripts for LIF and MMP10 (Figure 6B and 6D). Conversely, macrophages stimulated with LPS+IC (Figure 6C) showed a significant increase in mean fluorescent intensity per cell of all three transcripts (Figure 6D). Therefore, a combination of probes for multiple regulatory transcripts, may prove to be a useful technique for the identification of human R-Mϕ-IC.
Figure 6. Biomarker identification on R-Mϕ-IC.
RNAscope, a modified fluorescence in situ hybridization technique developed by Advanced Cell Diagnostics, was used to identify R-Mϕ transcriptional markers. Probes for LIF (green), MMP10 (red) and IL-10 (pink) were added to (A) resting non-stimulated macrophages, or those stimulated with (B) LPS or (C) LPS+IC. Representative RNAscope images are shown in panels A-C and the mean intensity per cell ± SEM (n=3) was quantified (D) using Zen Software, expressed as fold change relative to NS (* P-value ≤ 0.05). (E) Monolayers of human macrophages were stimulated with LPS (left) or LPS+ particulate immune complexes (right) and stained with mAb to CD73 followed by HRP-conjugated anti-mouse IgG. (F) Flow cytometry of macrophages stimulated for 16 hours with LPS in the presence or absence of soluble IC (Ova-antiOVA). DC-STAMP expression on non-stimulated (NS), LPS-stimulated (LPS), and LPS+IC-stimulated (LIC) was analyzed. The MFI, and % positive cells are shown (error bars represent SEM, ** P-value≤0.01). (G) Macrophages were stimulated in brefeldin prior to permeabilization, fixation, and staining with MMP10 or LIF. The grey peak represents unstimulated cells, the lightly colored peaks represent LPS stimulation and the darker profiles represent LPS+IC stimulation.
Immunohistochemistry and flow cytometry were also utilized to identify biomarkers on R-Mϕ-IC. The up-regulation of CD73 on regulatory macrophages was visualized by immunohistochemistry on macrophages stimulated in vitro with LPS plus particulate immune complexes (Figure 6E). Using flow cytometry, we observed an increase in DC-STAMP mean fluorescent intensity as well as the number of DC-STAMP+ macrophages after stimulation with LPS plus soluble IC, relative to both LPS-Mϕ and NS-Mϕ (Figure 6F). The production of both MMP10 and LIF was increased slightly following LPS stimulation and further increased by stimulation with LPS+IC (Figure 6G). These data indicate that no one biomarker can reliably identify R-Mϕ-IC, but rather suggest that a combination of surface and cytosolic antigens may be utilized to identify these macrophages in tissue.
The identification of Regulatory Macrophages in human skin biopsies.
Leprosy (Hanseniasis) can present in different clinical forms (16). The lepromatous form of the disease is associated with high IgG levels, a defective skin DTH response to lepromin antigen (15, 16), and bacterial persistence in dermal macrophages. The defect in macrophage clearance of the bacteria, and the presence of high IgG levels in lesions suggested a macrophage phenotype similar to the in vitro stimulated R-Mϕ-IC described above. We therefore examined the phenotype macrophages in lepromatous and tuberculoid leprosy lesions by immunohistochemistry. In the lepromatous form of the disease, we observed high levels of DC-STAMP on macrophages (Figure 7, top, right) coupled with low levels of IL-12 (Figure 7, bottom right). These cells also expressed a pan macrophage marker, CD68 and MMP10 (Supplemental Figure 3A and B). In contrast, macrophages from the tuberculoid form of leprosy (Figure 7, left) expressed lower levels of DC-STAMP and higher levels of IL-12 (Figure 7A), consistent with an inflammatory M1 macrophage. DC-STAMP expression was quantitated by measuring peroxidase intensity, confirming that tuberculoid lesions expressed less DC-STAMP than lepromatous lesions (Supplemental Figure 3C). Thus, human tissue macrophages in the IgG-rich environment of lepromatous leprosy fail to restrict the intracellular growth of bacteria and exhibit markers consistent with a regulatory phenotype.
Figure 7. Immunohistochemistry of macrophages in human skin from patients with Hanseniasis.
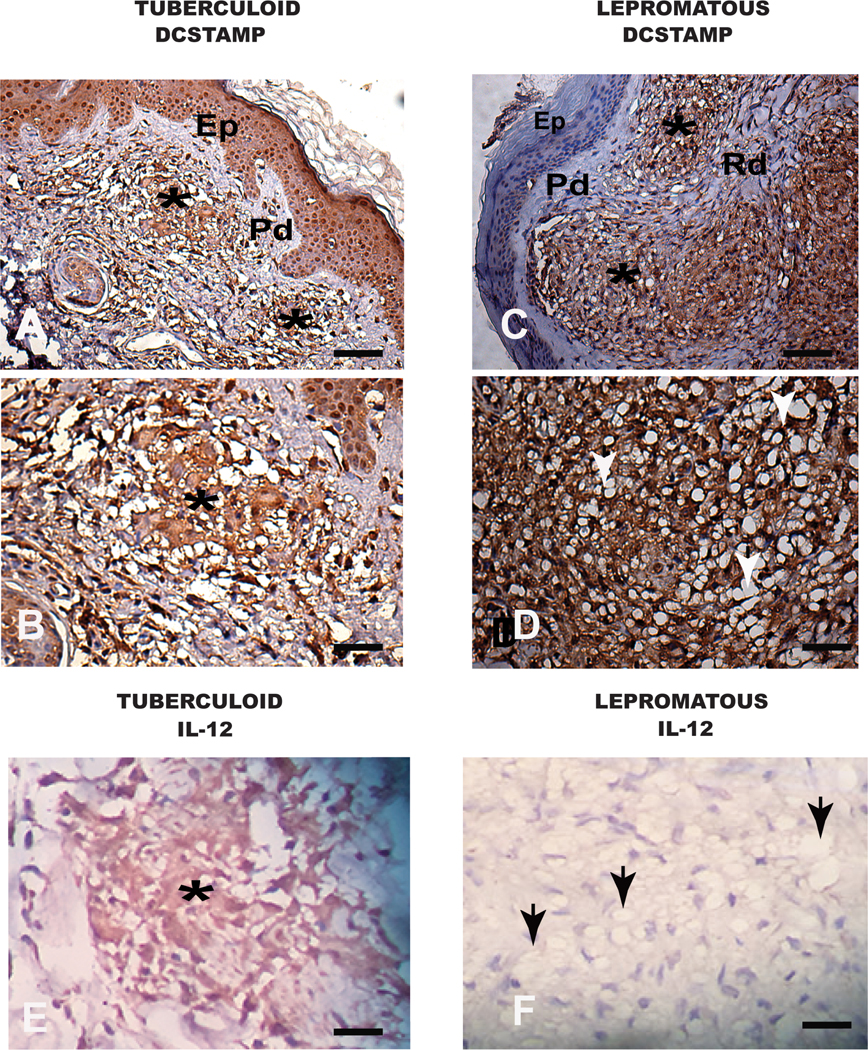
(A,B) Tuberculoid Hanseniasis, in (A) a panoramic view showing in the reticular dermis (Rd). Two granuloma are designated with asterisks. In (B) high magnification showing intense cellular DC-STAMP labelling (positive macrophages in dark-brown cytoplasmic staining). (C,D) Lepromatous Hanseniases, in (C) a panoramic view showing two granulomas in the reticular dermis (Rd) and two granulomas designated by asterisks. In (D) note higher DC-STAMP positive cells (positive immunolabelled macrophages in dark-brown cytoplasmic staining). Note numerous of inflammatory vacuolated macrophages (white arrows). (E) Tuberculoid Hanseniasis. In the reticular dermis (Rd) one granuloma is designated by an asterisk with intense cellular IL-12 labelling (positive macrophages in dark-brown cytoplasmic staining). (F) Lepromatous Hanseniases. Note no granuloma formation and the presence of a diffuse exudate of inflammatory vacuolated macrophages (black arrows). Bars (A,C) 64μm and (B,D,E,F) 16μm. Immunohistochemical of the streptavidin peroxidase method counter stained with Harris Hematoxylin. Epithelium (Ep), Papilar Dermis (Pd). Reticular Dermis (Rd).
DISCUSSION
This work provides the first characterization of human macrophages stimulated in the presence of immune complexes. A combination of bioinformatic and functional analyses indicate that Fcγ receptor cross-linking in concert with TLR-stimulation results in a population of macrophages with anti-inflammatory and growth-promoting activity. We have begun to identify biomarkers on these “regulatory” macrophages (R-Mϕ-IC) because of the potential therapeutic value of targeting these cells to enhance/prolong immune responses, or conversely to induce these cells to mitigate autoimmunity.
We confirm the work of many groups (31,32), showing that TLR ligation on macrophages results in a generalized inflammatory response and the early transcription of inflammatory chemokines and cytokines. Indolamine dioxygenase (IDO1), an enzyme that limits tryptophan availability to intracellular pathogens (34), is also one of the most highly upregulated transcripts following TLR ligation of human macrophages. The addition of a second stimulus to activate Fcγ receptors not only fails to amplify the inflammatory signature, but actively reverses it, resulting in a significant decrease in 632 transcripts, including all seven of the above-mentioned inflammatory mediators (Figure 1A). By IPA analysis, the genes most significantly reduced are generally involved in inflammation, rheumatic diseases, and cell death (Figure 3). This co-stimulation also resulted in the up-regulation of 925 transcripts (Figure 1B), including many proteins involved in cell growth and differentiation, neovascularization, and tumor growth (Figure 3). Twelve of the top 30 transcripts induced by the addition of immune complexes (Figure 1B) have been reported to be associated with cell growth and differentiation, including AREG (42), LIF (38), DC-STAMP (39), INHBA (53), and OCSTAMP (54). The top 30 upregulated transcripts also included three matrix metalloproteases (MMP1, MMP3, MMP10) and five cytokine-like molecules (IL-10, LIF, CSF2, IL36RN, TSLP).
We performed a SOMAscan analysis to detect secreted proteins from stimulated human macrophages, and the results were largely consistent with the RNA-sequencing (Supplemental Figure 2A). The addition of immune complexes broadly suppressed several LPS-induced cytokines and chemokines, while inducing the secretion of soluble mediators involved in regulation of inflammation. Here we presents a panel of secreted proteins that may be appropriate for ELISA markers of this phenotype, with IL-10, MMP1, and activin A (INHBA) among the most abundant secretory products. The upregulated transcription and secretion of several matrix metalloproteinases by R-Mϕ-IC suggests that regulatory macrophages contribute to cell motility, angiogenesis, and tissue remodeling. These gene products can not only degrade ECM components, but also regulate extracellular signaling molecules (36). Matrix metalloproteases can cleave growth factors to release their active forms (55), suppress immune responses following infection (56, 57), and inactivate inflammatory chemokines and mediators (58, 59). Not surprisingly, MMP1, MMP3 and MMP10 are associated with tumor metastasis and angiogenesis (60). Given these actions, we propose that while regulatory macrophages contribute to the resolution of immune responses and the tissue repair, they could also have deleterious effects on host responses to infections and cancer.
Identifying the signaling pathways that lead to the phenotypic changes in macrophages may provide targets to manipulate immune/inflammatory responses. Our bioinformatics analysis predicted that genes associated with AKT/PI3K signaling pathway were involved in R-Mϕ-IC transcriptomic changes. The AKT pathway is a complex signaling cascade that can regulate metabolism based on nutrient availability (61). Because AKT activation lies upstream of so many pathways, it became important to determine which AKT substrate was responsible for the gene changes observed in the regulatory phenotype induced by IC. GSK3 is a signaling kinase that is constitutively active in the cell, only becoming inactivated through phosphorylation of its substrate binding site (62). Recently, GSK3 was found to have anti-inflammatory activity, when a small molecule inhibitor of GSK3 increased survival in mice with endotoxemia (63, 64). We demonstrated that stimulation of macrophages with LPS and GSK3 inhibitor partially mimicked the LPS+IC phenotype, upregulating genes involved in growth and repair without influencing the production of inflammatory cytokines. Additionally, GSK3β is a transcription factor known to interact with AP-1, CREB, and NFκB, among others (65) and is also involved in the regulation of chromatin remodeling (66). GSK3β has been found to have an inhibitory effect on AP-1 and CREB, both of which are involved in IL-10 transcriptional control (50, 64). We found reduced levels of GSK3β in the nucleus after LPS+IC stimulation and demonstrated that GSK3β inhibition not only augments IL-10 production but also amplifies the transcription of matrix-metalloproteinases and growth-promoting factors in macrophages. These findings may explain previous observations that GSK3β inhibition is involved in wound healing and angiogenesis (67, 68).
We utilized RNAscope technology to visualize transcripts within fixed macrophages following stimulation. IC-induced regulatory macrophages could be identified through their increased expression of transcripts for IL10, LIF, and MMP10. If used in combination with a macrophage marker such as CD68, we propose that this method can be used for the identification of regulatory macrophages in tissue. The identification of protein biomarkers for macrophage activation states represents a challenge due to the dynamic and transient nature of macrophage activation. Despite this, we used flow cytometry to demonstrate that regulatory macrophages upregulate several biologically important proteins, including matrix metalloproteases and Dendritic Cell-Specific Transmembrane Protein (DC-STAMP). DC-STAMP is a cell surface protein that, despite its name, is also expressed on stimulated macrophages. This protein is mostly known for its role in the regulation of osteoclast differentiation (69) but is also implicated in cancer cell survival (70). Although usually discussed in the context of dendritic cells, DC-STAMP has been associated with increased phagocytosis and reduced antigen presentation and cytokine production (71, 72). DC-STAMP has been shown to be regulated by ERK (73), which may explain why its expression is increased following FcγR ligation.
To determine whether R-Mϕ-IC exist in human tissue, we turned to an infectious disease where bacteria grow uncontrolled in dermal macrophages. Lepromatous leprosy (Hanseniasis) has long been associated with hyperglobulinemia and defective DTH responses. In the present work we examined biomarkers on lesion macrophages that fail to provide host defense. Because we observe an increase in DC-STAMP on macrophages that are failing to provide adequate host defense, it is tempting to speculate that the regulatory macrophage phenotype could be driving this permissiveness. This idea is consistent with the high levels of IgG known to be present in the lepromatous form of the disease. Further studies in Hanseniasis are needed to determine how macrophage phenotype can influence disease progression.
In summary, our studies reveal the generation of macrophages with an anti-inflammatory, pro-healing phenotype that arise following stimulation in the presence of Fcγ-receptor cross-linking.. These regulatory macrophages may provide intracellular pathogens a safe haven for intracellular growth. These observations can likely be applied to a variety of infectious, inflammatory, or neoplastic diseases where the identification of these macrophages may help explain disease pathology.
Supplementary Material
Key points.
Regulatory Mϕ inhibit inflammation and induce tissue repair and angiogenesis
Immune complexes inhibit GSK3 to induce regulatory transcripts
Pathogens can exploit growth-promoting macrophages for intracellular growth
Footnotes:
This work was supported in part by National Institutes of Health Grant R01 GM 102589
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Kawai T, and Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol 11: 373–384. [DOI] [PubMed] [Google Scholar]
- 2.Mosser DM, and Edwards JP. 2008. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Edwards JP, Zhang X, Frauwirth KA, and Mosser DM. 2006. Biochemical and functional characterization of three activated macrophage populations. J. Leukoc. Biol 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleming BD, Chandrasekaran P, Dillon LAL, Dalby E, Suresh R, Sarkar A, El-Sayed NM, and Mosser DM. 2015. The generation of macrophages with anti-inflammatory activity in the absence of STAT6 signaling. J. Leukoc. Biol 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards JP, Zhang X, and Mosser DM. 2009. The expression of heparin-binding epidermal growth factor-like growth factor by regulatory macrophages. J. Immunol 182: 1929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutterwala FS, Noel GJ, Salgame P, and Mosser DM. 1998. Reversal of proinflammatory responses by ligating the macrophage Fcγ receptor type I. J. Exp. Med 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janczy JR, Ciraci C, Haasken S, Iwakura Y, Olivier AK, Cassel SL, and Sutterwala FS. 2014. Immune Complexes Inhibit IL-1 Secretion and Inflammasome Activation. J. Immunol 193: 5190–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J, Kurilova S, Scott BL, Bosworth E, Iverson BE, Bailey EM, and Hoppe AD. 2016. TIRF imaging of Fc gamma receptor microclusters dynamics and signaling on macrophages during frustrated phagocytosis. BMC Immunol. 17: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowley MT, Costello PS, Fitzer-Attas CJ, Turner M, Meng F, Lowell C, Tybulewicz VL, and DeFranco AL. 1997. A critical role for Syk in signal transduction and phagocytosis mediated by Fcgamma receptors on macrophages. J. Exp. Med 186: 1027–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takai T.2002. Roles of Fc receptors in autoimmunity. Nat. Rev. Immunol 2: 580–92. [DOI] [PubMed] [Google Scholar]
- 11.Lucas M, Zhang X, Prasanna V, and Mosser DM. 2005. ERK Activation Following Macrophage FcγR Ligation Leads to Chromatin Modifications at the IL-10 Locus. J. Immunol 175 : 469–477. [DOI] [PubMed] [Google Scholar]
- 12.Iyer AM, Mohanty KK, van Egmond D, Katoch K, Faber WR, Das PK, and Sengupta U. 2007. Leprosy-specific B-cells within cellular infiltrates in active leprosy lesions. Hum. Pathol 38: 1065–1073. [DOI] [PubMed] [Google Scholar]
- 13.de Macedo CS, de Carvalho FM, Amaral JJ, de Mendonca Ochs S, Assis EF, Sarno EN, Bozza PT, and Pessolani MCV. 2018. Leprosy and its reactional episodes: Serum levels and possible roles of omega-3 and omega-6-derived lipid mediators. Cytokine. [DOI] [PubMed] [Google Scholar]
- 14.Sadhu S, and Mitra DK. 2018. Emerging Concepts of Adaptive Immunity in Leprosy. Front. Immunol 9: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modlin RL 1994. Th1-Th2 paradigm: insights from leprosy. J. Invest. Dermatol 102: 828–832. [DOI] [PubMed] [Google Scholar]
- 16.Hungria EM, Buhrer-Sekula S, Oliveira RM, Aderaldo LC, Pontes MAA, Cruz R, de Goncalves HS, Penna MLF, Penna GO, and Stefani MMA. 2018. Mycobacterium leprae-Specific Antibodies in Multibacillary Leprosy Patients Decrease During and After Treatment With Either the Regular 12 Doses Multidrug Therapy (MDT) or the Uniform 6 Doses MDT. Front. Immunol 9: 915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anderson CF, and Mosser DM. 2002. Cutting edge: Biasing immune responses by directing antigen to macrophage Fcγ receptors. J. Immunol 168. [DOI] [PubMed] [Google Scholar]
- 18.Bolger AM, Lohse M, and Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews S.2010. FastQC: a quality control tool for high throughput sequence data. [Google Scholar]
- 20.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, and Haussler D. 2002. The human genome browser at UCSC. Genome Res. 12: 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trapnell C, Pachter L, and Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christensen SM, Dillon LAL, Carvalho LP, Passos S, Novais FO, Hughitt VK, Beiting DP, Carvalho EM, Scott P, El-Sayed NM, and Mosser DM. 2016. Meta-transcriptome Profiling of the Human-Leishmania braziliensis Cutaneous Lesion. PLoS Negl. Trop. Dis 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S, Pyl PT, and Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolstad BM, Irizarry RA, Åstrand M, and Speed TP. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinforma. 19 : 185–193. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK 2004. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol 3: 1. [DOI] [PubMed] [Google Scholar]
- 26.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, and Smyth GK. 2015. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43: e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kramer A, Green J, Pollard JJ, and Tugendreich S. 2014. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu G, and He Q-Y. 2016. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst 12: 477–479. [DOI] [PubMed] [Google Scholar]
- 29.Schneider CA, Rasband WS, and Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9: 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tafuri WL, de Santos RL, Arantes RME, Goncalves R, de Melo MN, Michalick MSM, and Tafuri WL. 2004. An alternative immunohistochemical method for detecting Leishmania amastigotes in paraffin-embedded canine tissues. J. Immunol. Methods 292: 17–23. [DOI] [PubMed] [Google Scholar]
- 31.Alasoo K, Martinez FO, Hale C, Gordon S, Powrie F, Dougan G, Mukhopadhyay S, and Gaffney DJ. 2015. Transcriptional profiling of macrophages derived from monocytes and iPS cells identifies a conserved response to LPS and novel alternative transcription. Sci. Rep 5: 12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beyer M, Mallmann MR, Xue J, Staratschek-Jox A, Vorholt D, Krebs W, Sommer D, Sander J, Mertens C, Nino-Castro A, Schmidt SV, and Schultze JL. 2012. High-Resolution Transcriptome of Human Macrophages. PLoS One 7: e45466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Björkbacka H, Fitzgerald KA, Huet F, Li X, Gregory JA, Lee MA, Ordija CM, Dowley NE, Golenbock DT, and Freeman MW. 2004. The induction of macrophage gene expression by LPS predominantly utilizes Myd88-independent signaling cascades. Physiol. Genomics 19. [DOI] [PubMed] [Google Scholar]
- 34.Taylor MW, and Feng GS. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol 5: 2516–2522. [PubMed] [Google Scholar]
- 35.Mittal R, Patel AP, Debs LH, Nguyen D, Patel K, Grati M, Mittal J, Yan D, Chapagain P, and Liu XZ. 2016. Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions. J. Cell. Physiol 231: 2599–2621. [DOI] [PubMed] [Google Scholar]
- 36.Van Lint P, and Libert C. 2007. Chemokine and cytokine processing by matrix metalloproteinases and its effect on leukocyte migration and inflammation. J. Leukoc. Biol 82: 1375–1381. [DOI] [PubMed] [Google Scholar]
- 37.Shih S-C, Zukauskas A, Li D, Liu G, Ang L-H, Nagy JA, Brown LF, and Dvorak HF. 2009. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 69: 3272–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ye S, Zhang D, Cheng F, Wilson D, Mackay J, He K, Ban Q, Lv F, Huang S, Liu D, and Ying Q-L. 2016. Wnt/beta-catenin and LIF-Stat3 signaling pathways converge on Sp5 to promote mouse embryonic stem cell self-renewal. J. Cell Sci 129: 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Il’in DA, and Shkurupy VA. 2019. In Vitro Analysis of the Expression of CD11, CD29, CD36, and DC-STAMP Molecules during the Formation of Multinuclear Macrophages in BCG-Infected Mice. Bull. Exp. Biol. Med 167: 653–655. [DOI] [PubMed] [Google Scholar]
- 40.Miyamoto T.2013. STATs and macrophage fusion. JAK-STAT 2: e24777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kristensen KK, Leth-Espensen KZ, Mertens HDT, Birrane G, Meiyappan M, Olivecrona G, Jorgensen TJD, Young SG, and Ploug M. 2020. Unfolding of monomeric lipoprotein lipase by ANGPTL4: Insight into the regulation of plasma triglyceride metabolism. Proc. Natl. Acad. Sci. U. S. A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campion CM, Leon Carrion S, Mamidanna G, Sutter CH, Sutter TR, and Cole JA. 2016. Role of EGF receptor ligands in TCDD-induced EGFR down-regulation and cellular proliferation. Chem. Biol. Interact 253: 38–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mahil SK, Catapano M, Di Meglio P, Dand N, Ahlfors H, Carr IM, Smith CH, Trembath RC, Peakman M, Wright J, Ciccarelli FD, Barker JN, and Capon F. 2017. An analysis of IL-36 signature genes and individuals with IL1RL2 knockout mutations validates IL-36 as a psoriasis therapeutic target. Sci. Transl. Med 9. [DOI] [PubMed] [Google Scholar]
- 44.Beck S, Simmet T, Muller I, Lang F, and Gawaz M. 2016. Gremlin-1 C-Terminus Regulates Function of Macrophage Migration Inhibitory Factor (MIF). Cell. Physiol. Biochem 38: 801–808. [DOI] [PubMed] [Google Scholar]
- 45.Le Y, Zhou Y, Iribarren P, and Wang JM. 2004. Cellular & Molecular Immunology Chemokines and Chemokine Receptors: Their Manifold Roles in Homeostasis and Disease. 1. [PubMed] [Google Scholar]
- 46.Dam?s JK, Landr? L, Fevang B, Heggelund L, Tj?nnfjord GE, Fl?isand Y, Halvorsen B, Fr?land SS, and Aukrust P.2009. Homeostatic chemokines CCL19 and CCL21 promote inflammation in human immunodeficiency virus-infected patients with ongoing viral replication. Clin. Exp. Immunol 157: 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alvarez E, Piccio L, Mikesell RJ, Klawiter EC, Parks BJ, Naismith RT, and Cross AH. 2013. CXCL13 is a biomarker of inflammation in multiple sclerosis, neuromyelitis optica, and other neurological conditions. Mult. Scler 19: 1204–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Mosser DM, and Zhang X. 2007. Activation of the MAPK, ERK, following Leishmania amazonensis infection of macrophages. J. Immunol 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cross DAE, Alessi DR, Cohen P, Andjelkovich M, and Hemmings BA. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378: 785–789. [DOI] [PubMed] [Google Scholar]
- 50.Hu X, Paik PK, Chen J, Yarilina A, Kockeritz L, Lu TT, Woodgett JR, and Ivashkiv LB. 2006. IFN-gamma suppresses IL-10 production and synergizes with TLR2 by regulating GSK3 and CREB/AP-1 proteins. Immunity 24: 563–74. [DOI] [PubMed] [Google Scholar]
- 51.Bechard M, and Dalton S. 2009. Subcellular Localization of Glycogen Synthase Kinase 3 Controls Embryonic Stem Cell Self-Renewal. Mol. Cell. Biol 29: 2092–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JSC, Alessi DR, and Cohen P. 2007. The selectivity of protein kinase inhibitors: a further update. Biochem. J 408: 297–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu B, Li L, Li B, Gao J, Chen Y, Wei M, Yang Z, Zhang B, Li S, Li K, Wang C, Surani MA, Li X, Tang F, and Bao S. 2020. Activin A and BMP4 Signaling Expands Potency of Mouse Embryonic Stem Cells in Serum-Free Media. Stem cell reports 14: 241–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H, Li Y, Sun T, Du W, Li C, Suo C, Meng Y, Liang Q, Lan T, Zhong M, Yang S, Niu C, Li D, and Ding C. 2019. Unveil the transcriptional landscape at the Cryptococcus-host axis in mice and nonhuman primates. PLoS Negl. Trop. Dis 13: e0007566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brizzi MF, Tarone G, and Defilippi P. 2012. Extracellular matrix, integrins, and growth factors as tailors of the stem cell niche. Curr. Opin. Cell Biol 24: 645–651. [DOI] [PubMed] [Google Scholar]
- 56.McMahan RS, Birkland TP, Smigiel KS, Vandivort TC, Rohani MG, Manicone AM, McGuire JK, Gharib SA, and Parks WC. 2016. Stromelysin-2 (MMP10) Moderates Inflammation by Controlling Macrophage Activation. J. Immunol 197: 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koller FL, Dozier EA, Nam KT, Swee M, Birkland TP, Parks WC, and Fingleton B. 2012. Lack of MMP10 exacerbates experimental colitis and promotes development of inflammation-associated colonic dysplasia. Lab. Investig 92: 1749–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van den Steen PE, Husson SJ, Proost P, Van Damme J, and Opdenakker G. 2003. Carboxyterminal cleavage of the chemokines MIG and IP-10 by gelatinase B and neutrophil collagenase. Biochem. Biophys. Res. Commun 310: 889–896. [DOI] [PubMed] [Google Scholar]
- 59.Manicone AM, and McGuire JK. 2008. Matrix metalloproteinases as modulators of inflammation. Semin. Cell Dev. Biol 19: 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Justilien V, Regala RP, Tseng I-C, Walsh MP, Batra J, Radisky ES, Murray NR, and Fields AP. 2012. Matrix metalloproteinase-10 is required for lung cancer stem cell maintenance, tumor initiation and metastatic potential. PLoS One 7: e35040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song G, Ouyang G, and Bao S. 2005. The activation of Akt/PKB signaling pathway and cell survival. J. Cell. Mol. Med 9: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Frame S, Cohen P, Biondi RM, Huang E, Kimelman D, Moon R, Jonkers J, Kimelman D, Wu D, Cox LR, and et al. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol. Cell 7: 1321–7. [DOI] [PubMed] [Google Scholar]
- 63.Noh KT, Park Y-M, Cho S-G, and Choi E-J. 2011. GSK-3beta-induced ASK1 stabilization is crucial in LPS-induced endotoxin shock. Exp. Cell Res 317: 1663–1668. [DOI] [PubMed] [Google Scholar]
- 64.Martin M, Rehani K, Jope RS, and Michalek SM. 2005. Toll-like receptor-mediated cytokine production is differentially regulated by glycogen synthase kinase 3. Nat. Immunol 6: 777–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gotschel F, Kern C, Lang S, Sparna T, Markmann C, Schwager J, McNelly S, von Weizsacker F, Laufer S, Hecht A, and Merfort I. 2008. Inhibition of GSK3 differentially modulates NF-kappaB, CREB, AP-1 and beta-catenin signaling in hepatocytes, but fails to promote TNF-alpha-induced apoptosis. Exp. Cell Res 314: 1351–1366. [DOI] [PubMed] [Google Scholar]
- 66.Park SH, Park-Min K-H, Chen J, Hu X, and Ivashkiv LB. 2011. Tumor necrosis factor induces GSK3 kinase–mediated cross-tolerance to endotoxin in macrophages. Nat. Immunol 12: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H-S, Skurk C, Thomas SR, Bialik A, Suhara T, Kureishi Y, Birnbaum M, Keaney JF, and Walsh K. 2002. Regulation of angiogenesis by glycogen synthase kinase-3beta. J. Biol. Chem 277: 41888–96. [DOI] [PubMed] [Google Scholar]
- 68.Karrasch T, Spaeth T, Allard B, and Jobin C. 2011. PI3K-Dependent GSK3ß(Ser9)-Phosphorylation Is Implicated in the Intestinal Epithelial Cell Wound-Healing Response. PLoS One 6: e26340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiu Y-H, and Ritchlin CT. 2016. DC-STAMP: A Key Regulator in Osteoclast Differentiation. J. Cell. Physiol 231: 2402–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng XX, Chu TJ, Yuan JY, Zhang W, Du YM, Ban ZY, Lei DM, Cao J, and Zhang Z. 2015. Transmembrane 7 superfamily member 4 regulates cell cycle progression in breast cancer cells. Eur. Rev. Med. Pharmacol. Sci 19: 4353–61. [PubMed] [Google Scholar]
- 71.Sawatani Y, Miyamoto T, Nagai S, Maruya M, Imai J, Miyamoto K, Fujita N, Ninomiya K, Suzuki T, Iwasaki R, Toyama Y, Shinohara M, Koyasu S, and Suda T. 2008. The role of DC-STAMP in maintenance of immune tolerance through regulation of dendritic cell function. Int. Immunol 20: 1259–1268. [DOI] [PubMed] [Google Scholar]
- 72.Sanecka A, Ansems M, Prosser AC, Danielski K, Warner K, den Brok MH, Jansen BJH, Eleveld-Trancikova D, and Adema GJ. 2011. DC-STAMP knock-down deregulates cytokine production and T-cell stimulatory capacity of LPS-matured dendritic cells. BMC Immunol. 12: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Oh JH, and Lee NK. 2017. Up-Regulation of RANK Expression via ERK1/2 by Insulin Contributes to the Enhancement of Osteoclast Differentiation. Mol. Cells 40: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.