Abstract
The katG gene coding for the only catalase-peroxidase in the cyanobacterium Synechocystis sp. strain PCC 6803 was deleted in this organism. Although the rate of H2O2 decomposition was about 30 times lower in the ΔkatG mutant than in the wild type, the strain had a normal phenotype and its doubling time as well as its resistance to H2O2 and methyl viologen were indistinguishable from those of the wild type. The residual H2O2-scavenging capacity was more than sufficient to deal with the rate of H2O2 production by the cell, estimated to be less than 1% of the maximum rate of photosynthetic electron transport in vivo. We propose that catalase-peroxidase has a protective role against environmental H2O2 generated by algae or bacteria in the ecosystem (for example, in mats). This protective role is most apparent at a high cell density of the cyanobacterium. The residual H2O2-scavenging activity in the ΔkatG mutant was a light-dependent peroxidase activity. However, neither glutathione peroxidase nor ascorbate peroxidase accounted for a significant part of this H2O2-scavenging activity. When a small thiol such as dithiothreitol was added to the medium, the rate of H2O2 decomposition in the ΔkatG mutant increased more than 10-fold, indicating that a thiol-specific peroxidase, for which thioredoxin may be the physiological electron donor, is present. Oxidized thioredoxin is likely to be reduced again by photosynthetic electron transport. Therefore, under laboratory conditions, there are only two enzymatic mechanisms for H2O2 decomposition present in Synechocystis sp. strain PCC 6803. One is catalyzed by a catalase-peroxidase, and the other is catalyzed by thiol-specific peroxidase.
Active oxygen species, including superoxide (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (OH·), are byproducts of both aerobic respiration and oxygenic photosynthesis in all organisms that carry out these processes. Because of the relatively high reactivity of active oxygen species with proteins and membranes, efficient scavenging is important to prevent photooxidative damage to the organism (1). The work described in this paper focuses on cyanobacteria, which can carry out photosynthesis and respiration simultaneously.
The major site of O2− production in the photosynthetic electron transport chain is at the reducing side of photosystem I (PS I): particularly under conditions when NADPH utilization is suboptimal and NADP levels are low, O2 rather than NADP may occasionally accept an electron from PS I (1). This phenomenon is known as the Mehler reaction, which is a major electron transfer route in the presence of the herbicide methyl viologen (MV). This herbicide, also known as paraquat, efficiently accepts an electron from PS I and reduces oxygen to O2−, thus serving as a potent inhibitor of growth of photosynthetic organisms in the light. Respiratory dehydrogenases of mitochondria and bacteria also have been shown to be important sources of O2− and H2O2 (12, 28).
O2− is efficiently scavenged by superoxide dismutase (SOD); the activity of the enzyme is sufficient to limit O2−-induced damage except when MV is present and large amounts of O2− are produced (29). There are three types of SOD. All cyanobacteria contain an Fe-containing SOD (FeSOD) in the cytosol; some also contain a Mn-containing form of SOD (MnSOD) that is associated with thylakoid membranes (6, 36). In addition, Cu/ZnSOD activity also has been reported in a cyanobacterium (7), even though initially Cu/ZnSOD was thought to be specific for eukaryotes. According to CyanoBase (12a), the genome of the cyanobacterium Synechocystis sp. strain PCC 6803 contains only one recognizable gene for SOD, sodB, which codes for FeSOD, suggesting that FeSOD is the sole O2−-scavenging enzyme in Synechocystis sp. strain PCC 6803.
H2O2 is the most stable of active oxygen species. However, the most reactive and destructive active oxygen species, OH·, can be formed from H2O2 in the presence of O2− and selected metal ions such as Fe2+ (13). The best-documented source of H2O2 in chloroplasts is O2− generated by the Mehler reaction and disproportionated by SOD. A significant amount of H2O2 was also shown to be produced upon illumination of relatively intact PS II membranes (14, 38), suggesting that H2O2 can be formed by partial oxidation of water at the donor side of PS II, at least in vitro.
Early reports indicated the presence of two major hydrogen peroxide-scavenging activities in cyanobacteria, catalase and ascorbate peroxidase (34, 35). Catalase activity has been found in all cyanobacterial species tested (21). The Synechococcus sp. strain PCC 7942 enzyme responsible for catalase activity has been purified and characterized, and the corresponding gene has been cloned (23, 24). The gene sequence, enzymatic activity, and resistance to 3-amino-1,2,4-triazole, an inhibitor of eukaryotic catalase, provided evidence that the catalase in fact is a catalase-peroxidase, a member of the family of prokaryotic enzymes exhibiting both catalase and peroxidase activities (23, 24).
Ascorbate peroxidase is also a major H2O2 scavenger in chloroplasts. However, in cyanobacteria, the concentration of ascorbate was found to be 250-fold lower than in chloroplasts (41), and an ascorbate-specific peroxidase activity has not yet been found (1). Indeed, the Synechocystis sp. strain PCC 6803 genome sequence does not contain an ascorbate peroxidase gene. Despite the fact that several cyanobacteria (including Synechocystis sp. strain PCC 6803, Anabaena variabilis, and Anabaena cylindrica) have been found to exhibit a significant peroxidase activity (21), the enzyme itself and its physiological donor remain to be determined.
Cyanobacteria contain millimolar concentrations of glutathione, but no glutathione peroxidase activity has been detected (34), even though two open reading frames (slr1171 and slr1992 according to CyanoBase [12a]) with significant similarity to glutathione peroxidase genes are present in the Synechocystis sp. strain PCC 6803 genomic sequence. Glutathione peroxidase is widespread among bacteria, plants, and animals and is involved in the detoxification of lipid hydroperoxides and of H2O2. The enzyme is well conserved among all organisms, with the active site containing selenocysteine (11).
Recently, another protein exhibiting peroxidase activity has been found to be ubiquitously distributed among prokaryotes and eukaryotes (3, 9). This enzyme originally was called a thiol-specific antioxidant protein (8) and was later shown to be a peroxidase that reduces H2O2 and alkyl hydroperoxides with the use of hydrogens provided by sulfhydryl groups of small thiols (9). This protein was the first peroxidase that uses thioredoxin as the immediate hydrogen donor to be identified and was thus named thioredoxin-dependent peroxide reductase (TPx) (9).
In the present study, we have deleted the katG gene coding for the catalase-peroxidase in Synechocystis sp. strain PCC 6803 and have characterized the resulting mutant to help determine the physiological role of catalase-peroxidase. Moreover, upon elimination of the overwhelming catalase activity of catalase-peroxidase, the rates of H2O2 production and of remaining scavenging activity could be measured in vivo.
MATERIALS AND METHODS
Growth conditions.
The wild type and mutants of Synechocystis sp. strain PCC 6803 were grown in liquid BG-11 medium (27) at 30°C at 40 microeinsteins/m2 · s on a rotary shaker. Solid media were supplemented with 1.5% (wt/vol) agar, 0.3% (wt/vol) sodium thiosulfate, and 10 mM TES [N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid]-NaOH buffer (pH 8.2). Filter-sterilized media or solid media without sodium thiosulfate were used where indicated. Sodium thiosulfate is an efficient H2O2 scavenger, and freshly autoclaved BG-11 medium without this compound was found to contain up to 5 μM H2O2 (data not shown).
Enzyme assays.
Catalase activity was determined spectrophotometrically by monitoring the rate of H2O2 decomposition at 240 nm by using an extinction coefficient for H2O2 of 43.6 M−1 · cm−1 (23). For lower concentrations of H2O2 (<1 mM), the rate of O2 production was determined polarographically with a Clark-type electrode. Both measurements were performed with whole cells in 25 mM HEPES-NaOH buffer (pH 7.0) at 25°C. The rate of H2O2 decomposition by the ΔkatG strain was determined as a decrease in H2O2 concentration in the medium as a function of time. For this purpose, H2O2 concentrations were determined through oxidation of Fe2+ in the presence of xylenol orange (37). Ascorbate peroxidase activity was determined as a decrease in the absorbance of ascorbate at 290 nm upon addition of cell homogenate and H2O2 (21). The activity of glutathione peroxidase was assayed spectrophotometrically as oxidation of NADPH in a conjugated reaction with glutathione reductase (39).
Continuous measurements of H2O2 production and excretion by whole cells upon illumination were carried out by the scopoletin fluorescence method (26) using a SPEX Fluoromax spectrofluorometer. Scopoletin is oxidized by H2O2 in the presence of peroxidase; a decrease in the scopoletin fluorescence is a measure of H2O2 produced. The assay mixture contained 40 μl of scopoletin (20 mg/liter), 160 U of fungal peroxidase (Fluka), and 3 ml of cells (optical density at 730 nm [OD730], 0.5). Mixing of the sample was provided by a magnetic stirrer. The temperature was maintained at 30°C during illumination by a constant-temperature cuvette holder. Scopoletin was excited by 350-nm-wavelength light, and fluorescence was measured at 460 nm. The sample was illuminated from the top with orange actinic light (570-nm-cutoff filter) provided by an incandescent bulb (50 to 150 microeinsteins/m2 · s) or a slide projector lamp (1,500 to 2,500 microeinsteins/m2 · s). The steady-state H2O2 production by whole cells varied by a factor of 2 to 3, depending on the growth stage of the culture and the method used for sample preparation. Reproducible results (within 30% of the average) were obtained with cells that had been grown on BG-11 plates without thiosulfate for 5 to 8 days and that were resuspended into 25 mM HEPES-NaOH buffer (pH 7.0) 30 min before the measurement.
Oxygen evolution.
Oxygen evolution was measured by using samples directly taken from liquid cultures (OD730 of the cultures, 0.5 to 0.8). Measurements were performed with a Clark-type electrode in the presence of 1 mM K3Fe(CN)6 and 0.1 mM dimethyl-p-benzoquinone. HEPES-NaOH buffer (pH 7.0) was added to a final concentration of 25 mM. The temperature was 25°C, and the light intensity was 4,000 microeinsteins/m2 · s. The light was passed through an orange filter cutting off light with wavelengths of <570 nm before reaching the sample. The chlorophyll (Chl) a concentration was about 2 μg/ml as determined by absorption at 663 nm after Chl extraction from cells in 100% methanol.
RESULTS
Deletion of the katG gene.
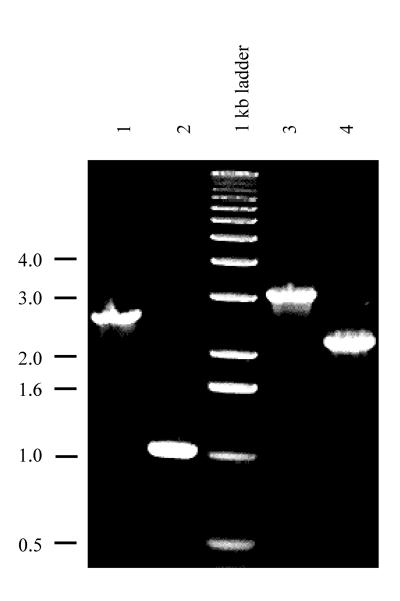
The katG gene from Synechocystis sp. strain PCC 6803 (accession no. D83990), which codes for catalase-peroxidase, was identified by its high level of similarity to other bacterial catalase-peroxidases (71% identity with the protein from Synechococcus sp. strain PCC 7942 [accession no. D61378] and 64% identity with that from Bacillus stearothermophilus [accession no. M29876]). No other catalase gene copy is apparent in Synechocystis sp. strain PCC 6803. The katG gene and its flanking regions were cloned, and katG interruption and deletion constructs were made by introducing a chloramphenicol resistance marker (1.5 kb) from pACYC184. In the katGi interruption mutant, the marker was introduced into the HpaI site 748 bp downstream from the translation start site of the 2,262-bp-long katG gene. In the construct made to create the ΔkatG deletion mutant, the region between the SmaI site 63 bp upstream of the translation start site and the HpaI site 748 bp downstream of the translation start site was replaced by the marker. Segregation of the mutants was checked by PCR (Fig. 1).
FIG. 1.
PCR amplification of the katG region of genomic DNA from various Synechocystis sp. PCC 6803 strains. PCR amplification was carried out with genomic DNA from the katGi interruption mutant (lane 1) and compared to that of the wild type (lane 2). DNA from the ΔkatG deletion mutant (lane 3) was also compared to that of the wild type (lane 4). Two forward primers were used: the one for lanes 1 and 2 hybridized to the region 920 bp upstream of the katG start codon, and the one used for lanes 3 and 4 hybridized to the region 290 bp downstream of the katG initiation codon. The reverse primer hybridized to the region 1,350 bp downstream of the katG start codon. The size of the chloramphenicol marker was 1.5 kb; the size of the deletion in the ΔkatG strain was 0.8 kb. The sizes of the DNA markers (in kilobases) are indicated on the left.
Characterization of the ΔkatG mutant.
The ΔkatG deletion mutant and the katGi interruption mutant exhibited no catalase activity, confirming that catalase-peroxidase is the only catalase present in Synechocystis sp. strain PCC 6803. The characteristics of both mutant strains are summarized in Table 1. The photoautotrophic growth rate and the PS II oxygen evolution capacity of the ΔkatG and katGi mutants were indistinguishable from those of the wild type under the conditions tested. However, both strains differed greatly from the wild type in their H2O2-scavenging capacity. At 50 μM H2O2 in darkness, the initial rate of H2O2 decomposition was about 30 times higher in the wild type than in both mutants (Table 1). The H2O2 decay in the wild type was measured polarographically as O2 evolution, which reflects catalase activity only, whereas the total H2O2-scavenging capacity (residual peroxidase activity) in the mutants was determined by colorimetric detection of the remaining H2O2. In the wild type, H2O2 conversion was too rapid to be measured satisfactorily by colorimetric detection. However, at 50 μM H2O2 in the wild type, the catalase activity accounted for most of the H2O2 scavenging, because 80 to 90% of the added H2O2 led to the formation of oxygen (data not shown). Since both mutants had identical properties, only results for the ΔkatG strain will be reported here.
TABLE 1.
Characteristics of the katG mutantsa
| Strain | Doubling time (h) | Doubling time in presence of 0.5 μM MV (h) | H2O2 decomposition rate (μmol of H2O2/mg of Chl · h)b | Oxygen evolution rate (μmol of O2/mg of Chl · h) |
|---|---|---|---|---|
| Wild type | 12 | 25 | 750 | 350 |
| ΔkatG mutant | 11 | 20 | 25 | 360 |
| katGi mutant | 11 | 22 | 25 | 330 |
The doubling times indicated were measured under photoautotrophic conditions; the H2O2 decomposition rate was measured in darkness. The doubling times and oxygen evolution data shown were reproducible within 15% of the average; H2O2 decomposition rates were reproducible within 30%.
The decomposition rate was measured in the presence of 50 μM H2O2.
Since growth characteristics of the ΔkatG mutant were indistinguishable from those of the wild type under normal laboratory conditions, a competition experiment was performed to compare the growth rates or viabilities of both strains under more extreme conditions. The ΔkatG mutant and wild-type strains were mixed and exposed to various conditions: (i) 5 h at 25°C and 2,500 microeinsteins/m2 · s; (ii) 10 days at 10°C and 50 microeinsteins/m2 · s; (iii) 10 days at 5°C in the dark; (iv) 10 days at 30°C and 50 microeinsteins/m2 · s (control). The ratio of viable wild-type to ΔkatG mutant cells was measured before and after growth under the various conditions by comparing the number of CFU on plates without (wild-type plus ΔkatG mutant cells) and with (ΔkatG mutant cells) chloramphenicol. Interestingly, no significant change in the wild-type/ΔkatG mutant cell ratio was found under any of these conditions (data not shown), indicating that the absence of the katG gene has no physiological effect under the conditions tested.
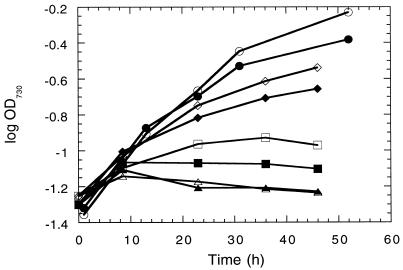
The results of these experiments imply that generated H2O2 is efficiently scavenged even in the ΔkatG mutant or that H2O2 toxicity is not very high in Synechocystis sp. strain PCC 6803. To test this in vivo, H2O2 production and growth rates of the wild type and the ΔkatG mutant were compared after the addition of different concentrations of MV. The herbicide MV is reduced by the acceptor side of PS I in a one-electron transfer and is efficiently oxidized by oxygen, forming superoxide, which is converted to H2O2 in a SOD-catalyzed reaction. Under various conditions, a clear difference in the rates of H2O2 diffusion into the medium was observed between the wild type and the ΔkatG mutant, indicating a decreased H2O2-scavenging capacity of the ΔkatG mutant. As expected, increases in both the light intensity and the MV concentration increased H2O2 production (Table 2). However, when lower MV concentrations were tested, at which both the wild type and the ΔkatG mutant could grow, no significant difference between the growth rates of the two strains was found (Fig. 2). The cells of both strains died at 1 μM MV, and the maximal permitted concentration for growth at a light intensity of 50 microeinsteins/m2 · s for both strains was 0.5 μM (data not shown). At this MV concentration, the amount of H2O2 that accumulated in the growth medium remained below the detection limit in our experiments (i.e., <0.2 μM), even in the ΔkatG mutant. The fact that H2O2 did not accumulate in the medium indicates that the remaining peroxidase activity in the ΔkatG mutant is sufficient to decompose all H2O2 produced by the cell.
TABLE 2.
H2O2 production by whole cells as a function of illumination intensity and concentration of MV (H2O2 was detected in the medium)
| Light intensity (microeinsteins/ m2 · s) | MV concn (μM) | H2O2 production rate (μmol/mg of Chl · h)
|
|
|---|---|---|---|
| Wild type | ΔkatG mutant | ||
| 0 | 0.0 | 0.3 ± 0.2 | 0.3 ± 0.2 |
| 50 | 1.0 | 0.9 ± 0.5 | 2.9 ± 0.7 |
| 150 | 1.0 | 13 ± 5 | 18 ± 4 |
| 2,200 | 0.0 | 1.1 ± 0.5 | 1.9 ± 0.7 |
| 2,200 | 0.1 | 1.0 ± 0.6 | 8 ± 3 |
| 2,200 | 1.0 | 55 ± 19 | 105 ± 32 |
FIG. 2.
Growth curves of the wild type and the ΔkatG mutant. Cells were grown photoautotrophically at a light intensity of 40 microeinsteins/m2 · s without MV (circles) or at 80 microeinsteins/m2 · s with 0 (diamonds), 1 (squares), or 2 (triangles) μM MV that was added at time zero. Open symbols, wild type; closed symbols, the ΔkatG strain.
Since the previous experiment did not indicate a role of catalase-peroxidase in protection against internally produced H2O2, protection against externally added H2O2 was tested. In this experiment, the viabilities of the wild type and the ΔkatG mutant were compared after incubation with increasing concentrations of H2O2. Cell cultures (104 to 108 cells/ml) were incubated with filter-sterilized BG-11 medium containing increasing concentrations of H2O2 for 5 h and were then plated on BG-11 medium without glucose. The final H2O2 concentration in the medium and the cell survival rate were recorded (Fig. 3). Filter-sterilized BG-11 medium was used because it contained lower concentrations of H2O2 (0.5 μM) than the autoclaved media did (3 to 5 μM). Thiosulfate present in the solid medium scavenged any H2O2 present after the 5-h experiment. In media with low cell concentrations (Fig. 3A and B), where the H2O2 concentration did not significantly change during the experiment since little was scavenged by the cells, there was no difference in the survival of cells between the wild-type and ΔkatG mutant strains. At higher cell concentrations (Fig. 3C and D), the wild-type strain survived much better because of its superior ability to effectively scavenge all H2O2 from the media during the 5-h incubation. Therefore, exposure of cells to H2O2 concentrations on the order of 10 μM for several hours is toxic, and catalase-peroxidase does not protect single cells against the effects of externally added H2O2. However, as a population of cells, the wild type in contrast to the ΔkatG mutant can detoxify and thereby survive significant concentrations of H2O2.
FIG. 3.
Effect of H2O2 on the viability of the wild type and the ΔkatG mutant. Different concentrations of cells were incubated for 5 h with different concentrations of H2O2; cells were then plated on BG-11 medium with thiosulfate and without glucose. Survival rates (percentage of survival of control incubated without H2O2) and final H2O2 concentrations at the end of the 5 h of incubation are indicated.
Ascorbate peroxidase and glutathione peroxidase activities.
In the absence of catalase activity in the ΔkatG mutant, the remaining H2O2-scavenging activity was assigned to peroxidase. Known peroxidases from other photosynthetic systems include ascorbate peroxidase (1) and glutathione peroxidase (39) and possibly TPx (3).
Even though ascorbate peroxidase activity in cyanobacteria has been postulated (21, 35), no ascorbate peroxidase gene with similarity to plant ascorbate peroxidase genes was found in the Synechocystis sp. strain PCC 6803 genome. A marginal activity (6 μmol/mg of Chl · h with 1 mM H2O2 and 0.5 mM ascorbate) was found and was identical for the wild type and the ΔkatG mutant (data not shown). This in vitro ascorbate peroxidase activity may originate from some nonspecific peroxidase that naturally has a preference for substrates other than ascorbate.
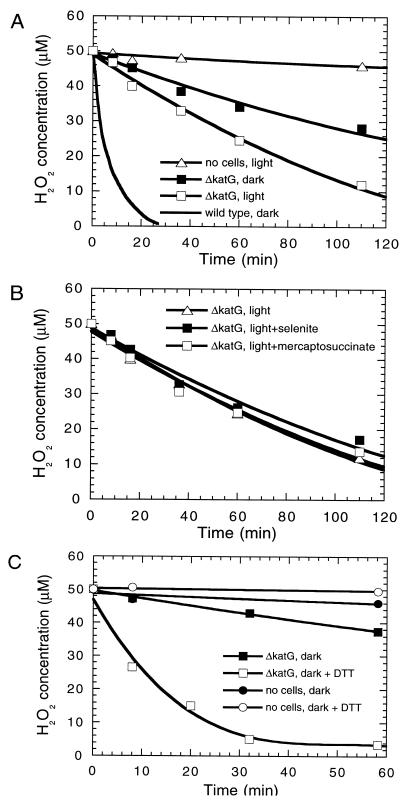
Regarding glutathione peroxidase, two genes for this enzyme are present in the Synechocystis sp. strain PCC 6803 genome, and in Chlamydomonas reinhardtii the enzyme can be induced by the addition of selenite (39). However, no selenite induction of the H2O2 decomposition rate in vivo (Fig. 4B) and no glutathione peroxidase activity in vitro were detected in the mutant. No increase in the H2O2 decomposition rate (Fig. 4B) and no glutathione peroxidase activity (data not shown) were observed, suggesting that glutathione peroxidase is not expressed in Synechocystis sp. strain PCC 6803. In addition, incubation for 1 h or growth for 3 days with 100 μM mercaptosuccinate, which is a potent and specific inhibitor of the enzyme (11), did not inhibit H2O2 decomposition in the ΔkatG mutant (Fig. 4B). Therefore, neither ascorbate peroxidase nor glutathione peroxidase is active in Synechocystis sp. strain PCC 6803 under the conditions used.
FIG. 4.
H2O2 decomposition in the wild type and the ΔkatG strain. (A) H2O2 decomposition in darkness (wild type and ΔkatG strains), in the light (ΔkatG strain), and in the light in the absence of cells; (B) peroxidase activity of the ΔkatG mutant in the light with 2 mg of sodium selenite per liter or 100 μM mercaptosuccinate as indicated; (C) peroxidase activity of the ΔkatG mutant in the presence of 1 mM DTT. For comparison, the spontaneous decomposition rate has been indicated. For the wild type, the decrease in H2O2 concentration was measured continuously as oxygen evolution. For the ΔkatG strain, H2O2 was measured at particular intervals by the peroxide assay. The initial rate of H2O2 consumption in the presence of 50 μM H2O2 was 750 μmol of H2O2/mg of Chl · h in the wild type in the dark and 25 or 50 μmol of H2O2/mg of Chl · h for the mutant strain incubated in the dark or light (50 microeinsteins/m2 · s), respectively.
Thiol-dependent peroxidase activity.
Genes for putative TPxs have been identified in the genome of Synechocystis sp. strain PCC 6803 (sll0755, with 63.5% identity between the translated open reading frame and peroxiredoxin from Hordeum vulgare [accession no. Z34917], and slr1198, with 52% identity to a human peroxiredoxin [human antioxidant protein, P30041]). To test the possibility that the peroxidase activity observed in the ΔkatG strain in fact reflects TPx activity, the H2O2-scavenging capacity of the cells of the ΔkatG strain was compared with that in the presence of 1 mM dithiothreitol (DTT) in the dark (Fig. 4C). DTT can be substituted for thioredoxin as an electron donor for TPx (17). Indeed, DTT was found to increase the rate of H2O2 decomposition by about 10-fold to 300 μmol H2O2/mg of Chl · h. DTT by itself had no effect on H2O2 stability (Fig. 4C).
Kinetics of H2O2 decomposition.
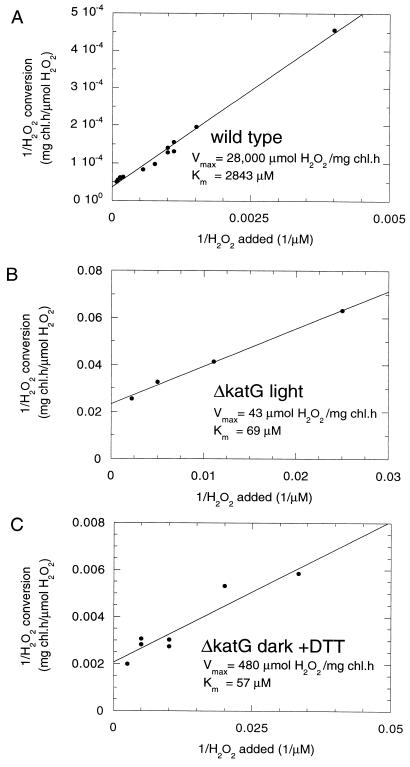
H2O2 decomposition by both the wild type and the ΔkatG mutant in vivo followed Michaelis-Menten kinetics (Fig. 5A and B). From the double reciprocal graph, the apparent Kms for H2O2 were 2.8 mM for the wild type and 70 μM for the mutant. The Km value for the wild type is similar to that of purified catalase-peroxidase from Synechococcus sp. strain PCC 7942, which was found to be 4 mM (23, 24). The Vmax for the wild type (28,000 μmol of H2O2/mg of Chl · h) indicates the ability of catalase-peroxidase to efficiently convert high concentrations of H2O2. The lower Km value for H2O2 found in the ΔkatG mutant is typical for peroxidase activity.
FIG. 5.
Double reciprocal graphs of the dependence of the H2O2 decomposition rate on the concentration of H2O2. (A) The rate of H2O2 decomposition in the wild type was determined spectrophotometrically at 240 nm for H2O2 concentrations above 1 mM or polarographically for lower concentrations. (B and C) The rate of H2O2 decomposition in the ΔkatG mutant was determined as a decrease in H2O2 concentration at specific time intervals (1 to 20 min). Cells (OD730 = 0.5) were incubated in the light at 50 μmol/m2 · s (B) or in darkness in the presence of 1 mM DTT (C). The apparent Km values of the enzyme for H2O2 as well as the Vmax values are indicated.
In the ΔkatG mutant in the presence of DTT, the apparent Km for conversion of H2O2 in the dark was 60 μM and the Vmax was 480 μmol of H2O2/mg of Chl · h (Fig. 5C). Both the light-dependent peroxidase reaction of the ΔkatG mutant (Fig. 5B) and the conversion of H2O2 in the dark in the presence of DTT (Fig. 5C) have similar Kms and can most likely represent the activity of the same peroxidase enzyme when either DTT or electrons generated by photosynthetic electron transport are used as an electron donor. The increase in Vmax by an order of magnitude in the presence of DTT suggests that in its absence the natural electron donor for peroxidase is present at a limiting concentration or is only partially reduced upon illumination (50 microeinsteins/m2 · s). The results presented here indicate that the remaining peroxidase activity in the ΔkatG mutant most likely originates from TPx.
DISCUSSION
Role of catalase-peroxidase.
Synechocystis sp. strain PCC 6803 possesses a single catalase encoded by katG. The high, constitutive activity of this enzyme is independent of the growth phase of the organism and is not inducible by pretreatment with H2O2 (50 μM for 1 h) (data not shown). This is similar to the situation in Synechococcus sp. strain PCC 7942 (20) but different from that in Escherichia coli, which has two catalases that are inducible by two different mechanisms (18).
In this study, the katG gene of Synechocystis sp. strain PCC 6803 was cloned and deleted to examine the role of catalase-peroxidase and other enzymes in protecting the cyanobacterium against oxidative stress. The ΔkatG strain exhibited no measurable catalase activity. Despite this complete absence of catalase-peroxidase, the mutant grew normally with no apparent difference in growth rate or tolerance to MV from that of the wild type.
Consistent with their decreased H2O2-scavenging capacity, ΔkatG cells were found to be more susceptible to higher concentrations of H2O2 in the medium as they had a decreased capability to detoxify H2O2 during the experiment (Fig. 3). At low cell concentrations, where no significant change in H2O2 concentration during the course of the experiment was observed, no difference in sensitivity to H2O2 between the wild type and the ΔkatG mutant was found. A similar surprising result has been obtained for E. coli (22), where diluted cultures of wild type were as sensitive to H2O2 as the catalase-deficient mutant. At high cell densities, the wild-type cells but not the mutant ones could efficiently catabolize added H2O2 and survive (22).
The fact that at a low cell density the wild type and the ΔkatG mutant are equally sensitive to externally added H2O2 indicates that H2O2 diffusion into the cell is more rapid than its decomposition by catalase, resulting in similar cytoplasmic concentrations of H2O2 in the wild type and the mutant. Easy and fast diffusion of H2O2 through biological membranes (31) led to a proposal that aquatic organisms can keep their intracellular H2O2 concentration low by simple diffusion of H2O2 produced in the cell (12). This would make an active H2O2-scavenging system in algae and cyanobacteria less important than that in plant chloroplasts. Interestingly, carbon fixation enzymes normally known for their high sensitivity to H2O2 have been shown to be much more H2O2 tolerant in algae and cyanobacteria than in plant chloroplasts (33).
Our observation that catalase-peroxidase can protect cyanobacterial cells against high concentrations of external H2O2 but is dispensable for growth under various laboratory conditions leads us to conclude that catalase-peroxidase in this cyanobacterium serves primarily in protection against external H2O2. In pathogenic bacteria (4, 25), catalase has a protective role against H2O2 generated by the host organism during infection. It is possible that cyanobacteria in their natural environment are also subject to oscillating levels of external H2O2 and that cell populations need catalase activity for survival. For example, significant photosynthetic production and excretion of H2O2 by the alga Ulva rigida have been implied to be of ecological significance (12). High catalase activity may be advantageous for survival, for example, in algal mats in competition with H2O2-producing algae and bacteria. The fact that wild-type E. coli but not a catalase-deficient mutant survived and multiplied in the presence of peroxide-generating streptococci (22) supports this argument.
H2O2 production in cells.
The H2O2 production in the ΔkatG mutant can be estimated as a combination of H2O2 diffusion from the cells and H2O2 decomposition by peroxidase within cells. The maximum rate of H2O2 diffusion from mutant cells into the medium was less than 2 μmol of H2O2/mg of Chl · h during illumination (2,200 microeinsteins/m2 · s) (Table 2). The Vmax of H2O2 decomposition by peroxidase in the mutant was 480 μmol/mg of Chl · h (Fig. 5C). Since the Km of the enzyme is 57 μM (Fig. 5C), this activity corresponds to 4.2 μmol of H2O2/mg of Chl · h at 0.5 μM H2O2, which is the equilibrium concentration detected in the medium when cells are illuminated at a high light intensity (2,200 microeinsteins/m2 · s). The rates of diffusion and decomposition of H2O2 add up to a total H2O2 production of 6 μmol/mg of Chl · h. This is about 1% of the maximum rate of whole-chain photosynthetic electron transport (300 μmol of O2/mg of Chl · h; 2 mol of H2O2 per mol of O2). This is higher than the numbers reported for superoxide production by respiring membranes of E. coli (0.03 to 0.3% of the electron transport depending on the substrate used) (15) but significantly lower than values reported for isolated chloroplasts where acceptors of PS I electrons are in short supply: in such systems, up to 25, 6, and 10% of electron transport may be used to form H2O2 in Euglena gracilis (16), in chloroplasts isolated from C. reinhardtii (32), and in chloroplasts isolated from spinach (2), respectively. Therefore, it is likely that H2O2 production in vivo in photosynthetic systems is significantly less than that determined from studies on isolated organelles. This implies that O2− production in photosynthetic electron transport may be much less than what is generally assumed.
Absence of ascorbate peroxidase and glutathione peroxidase activity.
In our study, only marginal ascorbate peroxidase activity was detected in cell extracts of Synechocystis sp. strain PCC 6803 at saturating ascorbate concentrations, indicating that at in vivo ascorbate concentrations that are estimated to be 20 to 100 μM (34), which is about 250 times lower than in chloroplasts, ascorbate peroxidase activity does not play an important role in H2O2 scavenging.
We did not detect any in vitro glutathione peroxidase activity in Synechocystis sp. strain PCC 6803, despite the fact that there are two glutathione peroxidase genes present in its genome. As observed in Nostoc muscorum PCC 7119 and Synechococcus sp. strain PCC 6311 (34), peroxidase activity in Synechocystis sp. strain PCC 6803 was not boosted by the addition of selenite into growth medium, in contrast to what was achieved in C. reinhardtii (39). In addition, no changes in peroxidase activity and in the growth rate of the ΔkatG mutant were observed upon addition of 100 μM mercaptosuccinate, which is a potent and specific inhibitor of glutathione peroxidase (11). A similar report regarding the lack of mercaptosuccinate inhibition has been presented for Plectonema boryanum (19). Therefore, apparently glutathione peroxidase is not expressed in Synechocystis sp. strain PCC 6803 under laboratory conditions, but it is possible that the two genes for this enzyme are induced under certain natural conditions.
Thiol-specific peroxidase activity.
Although peroxidase activity is well documented in cyanobacteria, the peroxidase itself and its natural electron donor remain obscure. The results presented in this paper indicate that this peroxidase activity originates from a thiol-specific peroxidase; thioredoxin may be its natural electron donor. In the Synechocystis sp. strain PCC 6803 genome, two genes have significant similarity to characterized thiol-specific peroxidases.
For continued activity of the peroxidase, thioredoxin needs to be recycled. In cyanobacteria and chloroplasts, thioredoxin reduction is coupled to photosynthetic electron transport via ferredoxin:thioredoxin reductase (3). The ferredoxin:thioredoxin system is responsible for light-mediated enzyme regulation in photosynthesis by a selective thiol redox control (5, 30). If TPx indeed uses thioredoxin regenerated by photosynthetic electron transport as an electron donor, then under conditions where superoxide and consecutive H2O2 production levels are highest (with illuminated cells or with chloroplasts with carbon fixation inhibited and ferredoxin reduced) (1) TPx activity would also be maximal, since reduced thioredoxin is plentiful.
Therefore, in Synechocystis sp. strain PCC 6803, H2O2 at low concentrations is broken down primarily by the peroxidase TPx. This is the first report of significant thiol-specific peroxidase activity in a photosynthetic organism. Clearly, cyanobacteria employ a different strategy for scavenging H2O2 generated by photosynthesis than the ascorbate peroxidase pathway of chloroplasts. We suggest that the main function of catalase-peroxidase with its low H2O2 affinity and high Vmax is to break down H2O2 entering the cell from the environment. This enzyme is advantageous particularly for natural cyanobacterial populations in competition with H2O2-producing algae and bacteria.
ACKNOWLEDGMENT
This work was supported by U.S. Department of Energy grant DE-FG03-95ER20180.
REFERENCES
- 1.Asada K. Production and action of active oxygen species in photosynthesis tissues. In: Foyer C H, Mullineaux P M, editors. Causes of photooxidative stress and amelioration of defense systems in plants. Boca Raton, Fla: CRC Press, Inc.; 1994. pp. 77–104. [Google Scholar]
- 2.Asada K, Badger M R. Photoreduction of 18O2 and H218O2 with concomitant evolution of 16O2 in intact spinach chloroplasts: evidence for scavenging of hydrogen peroxide by peroxidase. Plant Cell Physiol. 1984;25:1169–1179. [Google Scholar]
- 3.Baier M, Dietz K-J. The plant 2-Cys peroxiredoxin BAS1 is a nuclear-encoded chloroplast protein: its expressional regulation, phylogenetic origin, and implications for its specific physiological function in plants. Plant J. 1997;12:179–190. doi: 10.1046/j.1365-313x.1997.12010179.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown S M, Howell M L, Vasil M L, Anderson A J, Hasset D J. Cloning and characterization of the katB gene of Pseudomonas aeruginosa encoding a hydrogen peroxide-inducible catalase: purification of KatB, cellular localization, and demonstration that it is essential for optimal resistance to hydrogen peroxide. J Bacteriol. 1995;177:6536–6544. doi: 10.1128/jb.177.22.6536-6544.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan B B. Regulation of CO2 assimilation in oxygenic photosynthesis: the ferredoxin/thioredoxin system. Perspective on its discovery, present status, and future development. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 6.Campbell W S, Laudenbach D E. Characterization of four O2− dismutase genes from a filamentous cyanobacterium. J Bacteriol. 1995;177:964–972. doi: 10.1128/jb.177.4.964-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chadd H E, Newman J, Mann N H, Carr N G. Identification of iron superoxide dismutase and copper/zinc superoxide dismutase enzyme activity within the marine cyanobacterium Synechococcus sp. WH7803. FEMS Microbiol Lett. 1996;138:161–165. doi: 10.1111/j.1574-6968.1996.tb08150.x. [DOI] [PubMed] [Google Scholar]
- 8.Chae H, Kim I-H, Rhee S G. Cloning, sequencing, and mutation of thiol specific antioxidant gene of Saccharomyces cerevisiae. J Biol Chem. 1993;268:16815–16821. [PubMed] [Google Scholar]
- 9.Chae H Z, Chung S J, Rhee S G. Thioredoxin-dependent peroxide reductase from yeast. J Biol Chem. 1994;269:27670–27678. [PubMed] [Google Scholar]
- 10.Chae H Z, Uhm T B, Rhee S G. Dimerization of thiol-specific antioxidant and the essential role of cysteine 47. Proc Natl Acad Sci USA. 1994;91:7022–7026. doi: 10.1073/pnas.91.15.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudière J, Wilhelmse E C, Tappel A L. Mechanism of selenium-glutathione peroxidase and its inhibition by mercaptocarboxylic acids and other mercaptans. J Biol Chem. 1984;259:10943–10950. [PubMed] [Google Scholar]
- 12.Collén J, Del Rio M J, García-Reina G, Pedersén M. Photosynthetic production of hydrogen peroxide by Ulva rigida C. Ag (Chlorophyta) Planta. 1995;196:225–230. [Google Scholar]
- 12a.CyanoBase Website. 1996. [Online.] http://www.kazusa.or.jp/cyano. [1999, last date accessed.]
- 13.Halliwell B, Gutteridge J M C. Oxygen free radicals and iron in relation to biology and medicine: some problems and concepts. Arch Biochem Biophys. 1986;246:501–514. doi: 10.1016/0003-9861(86)90305-x. [DOI] [PubMed] [Google Scholar]
- 14.Hillier W, Wydrzynski T. Increases in peroxide formation by the photosystem II oxygen evolving reactions upon removal of the extrinsic 16, 22, and 33 kDa proteins are reversed by CaCl2 addition. Photosynth Res. 1993;38:417–423. doi: 10.1007/BF00046769. [DOI] [PubMed] [Google Scholar]
- 15.Imlay J A, Fridovich I. Superoxide production by respiring membranes of Escherichia coli. Free Radic Res Commun. 1991;12–13:59–66. doi: 10.3109/10715769109145768. [DOI] [PubMed] [Google Scholar]
- 16.Ishikawa T, Takeda T, Shigeoka S, Hirayama O, Mitsunaga T. Hydrogen peroxide generation in organelles of Euglena gracilis. Phytochemistry. 1993;33:1297–1299. [Google Scholar]
- 17.Lim Y S, Cha M K, Kim H K, Uhm T B, Park J W, Kim K, Kim I H. Removals of hydrogen peroxide and hydroxyl radical by thiol specific antioxidant protein as a possible role in vivo. Biochem Biophys Res Commun. 1993;192:273–280. doi: 10.1006/bbrc.1993.1410. [DOI] [PubMed] [Google Scholar]
- 18.Loewen P C, Switala J, Triggs-Raine B L. Catalases HPI and HPII in Escherichia coli are induced independently. Arch Biochem Biophys. 1985;243:144–149. doi: 10.1016/0003-9861(85)90782-9. [DOI] [PubMed] [Google Scholar]
- 19.Mallison S M, III, Cannon R E. Effects of pesticides on cyanobacterium Plectonema boryanum and cyanophage LPP-1. Appl Environ Microbiol. 1984;47:910–914. doi: 10.1128/aem.47.5.910-914.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mittler R, Tel-Or E. Oxidative stress responses and shock proteins in the unicellular cyanobacterium Synechococcus R2 (PCC-7942) Arch Microbiol. 1991;155:125–130. [PubMed] [Google Scholar]
- 21.Miyake C, Michihata F, Asada K. Scavenging of hydrogen peroxide in prokaryotic and eukaryotic algae: acquisition of ascorbate peroxidase during the evolution of cyanobacteria. Plant Cell Physiol. 1991;32:33–43. [Google Scholar]
- 22.Ma M, Eaton J W. Multicellular oxidant defense in unicellular organisms. Proc Natl Acad Sci USA. 1992;89:7924–7928. doi: 10.1073/pnas.89.17.7924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mutsuda M, Ishikawa T, Takeda T, Shigeoka S. The catalase-peroxidase of Synechococcus sp. PCC 7942: purification, nucleotide sequence analysis and expression in Escherichia coli. Biochem J. 1996;316:251–257. doi: 10.1042/bj3160251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obinger C, Regelsberger G, Strasser G, Burner U, Peschek G A. Purification and characterization of a homodimeric catalase-peroxidase from the cyanobacterium Anacystis nidulans. Biochem Biophys Res Commun. 1997;235:545–552. doi: 10.1006/bbrc.1997.6847. [DOI] [PubMed] [Google Scholar]
- 25.Odenbreit S, Wieland B, Haas R. Cloning and genetic characterization of Helicobacter pylori catalase and construction of a catalase-deficient mutant strain. J Bacteriol. 1996;178:6960–6967. doi: 10.1128/jb.178.23.6960-6967.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson C O P, Myers J. Photosynthetic production of hydrogen peroxide by Anacystis nidulans. Plant Physiol. 1973;51:104–109. doi: 10.1104/pp.51.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rippka R, Deruelles J, Waterbury J B, Herdman M, Stanier R Y. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J Gen Microbiol. 1979;111:1–61. [Google Scholar]
- 28.Salin M L. Chloroplast and mitochondrial mechanisms for protection against oxygen toxicity. Free Rad Res Commun. 1991;12–13:851–858. doi: 10.3109/10715769109145867. [DOI] [PubMed] [Google Scholar]
- 29.Samson G, Herbert S K, Fork D C, Laudenbach D E. Acclimation of the photosynthetic apparatus to growth irradiance in a mutant strain of Synechococcus lacking iron superoxide dismutase. Plant Physiol. 1994;105:287–294. doi: 10.1104/pp.105.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schurmann P. Ferredoxin:thioredoxin system. Methods Enzymol. 1995;252:274–283. doi: 10.1016/0076-6879(95)52030-9. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi M, Asada K. Superoxide anion permeability of phospholipid membranes and chloroplast thylakoids. Arch Biochem Biophys. 1983;226:558–566. doi: 10.1016/0003-9861(83)90325-9. [DOI] [PubMed] [Google Scholar]
- 32.Takeda T, Ishikawa T, Shigeoka S. Metabolism of hydrogen peroxide by the scavenging system in Chlamydomonas reinhardtii. Physiol Plant. 1977;99:49–55. [Google Scholar]
- 33.Takeda T, Yokota A, Shigeoka S. Resistance of photosynthesis to hydrogen peroxide in algae. Plant Cell Physiol. 1995;36:1089–1095. [Google Scholar]
- 34.Tel-Or E, Huflejt M E, Packer L. The role of glutathione and ascorbate in hydroperoxide removal in cyanobacteria. Biochem Biophys Res Commun. 1985;132:533–539. doi: 10.1016/0006-291x(85)91166-0. [DOI] [PubMed] [Google Scholar]
- 35.Tel-Or E, Huflejt M E, Packer L. Hydroperoxide metabolism in cyanobacteria. Arch Biochem Biophys. 1986;246:396–402. doi: 10.1016/0003-9861(86)90485-6. [DOI] [PubMed] [Google Scholar]
- 36.Thomas D J, Avenson T J, Thomas J B, Herbert S K. A cyanobacterium lacking iron superoxide dismutase is sensitized to oxidative stress induced with methyl viologen but is not sensitized to oxidative stress induced by norflurazon. Plant Physiol. 1998;116:1593–1602. doi: 10.1104/pp.116.4.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wolff S P. Ferrous ion oxidation in presence of ferric ion indicator xylenol orange for measurement of hydroperoxides. Methods Enzymol. 1994;233:182–189. [Google Scholar]
- 38.Wydrzynski T, Ångström J, Vänngård T. H2O2 formation by photosystem II. Biochim Biophys Acta. 1989;973:23–28. [Google Scholar]
- 39.Yokota A, Shigeoka S, Onishi T, Kitaoka S. Selenium as inducer of glutathione peroxidase in low-CO2 grown Chlamydomonas reinhardtii. Plant Physiol. 1988;86:649–651. doi: 10.1104/pp.86.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]