Abstract
The Serratia marcescens N28b wbbL gene has been shown to complement the rfb-50 mutation of Escherichia coli K-12 derivatives, and a wbbL mutant has been shown to be impaired in O4-antigen biosynthesis (X. Rubirés, F. Saigí, N. Piqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué, J. Bacteriol. 179:7581–7586, 1997). We analyzed a recombinant cosmid containing the wbbL gene by subcloning and determination of O-antigen production phenotype in E. coli DH5α by sodium dodecyl sulfate-polyacrylamide electrophoresis and Western blot experiments with S. marcescens O4 antiserum. The results obtained showed that a recombinant plasmid (pSUB6) containing about 10 kb of DNA insert was enough to induce O4-antigen biosynthesis. The same results were obtained when an E. coli K-12 strain with a deletion of the wb cluster was used, suggesting that the O4 wb cluster is located in pSUB6. No O4 antigen was produced when plasmid pSUB6 was introduced in a wecA mutant E. coli strain, suggesting that O4-antigen production is wecA dependent. Nucleotide sequence determination of the whole insert in plasmid pSUB6 showed seven open reading frames (ORFs). On the basis of protein similarity analysis of the ORF-encoded proteins and analysis of the S. marcescens N28b wbbA insertion mutant and wzm-wzt deletion mutant, we suggest that the O4 wb cluster codes for two dTDP-rhamnose biosynthetic enzymes (RmlDC), a rhamnosyltransferase (WbbL), a two-component ATP-binding-cassette-type export system (Wzm Wzt), and a putative glycosyltransferase (WbbA). A sequence showing DNA homology to insertion element IS4 was found downstream from the last gene in the cluster (wbbA), suggesting that an IS4-like element could have been involved in the acquisition of the O4 wb cluster.
In gram-negative bacteria, the lipopolysaccharide (LPS) is one of the major structural and immunodominant molecules of the outer membrane. It consists of three moieties: lipid A, core oligosaccharide, and O-specific antigen or O side chain. The O antigen is the most external component of LPS, and its structure consists in a polymer of oligosaccharide repeating units. Another interesting feature is the high chemical variability shown by the O antigen of the LPS, leading to a similar genetic variation in the genes involved in O-antigen biosynthesis, the so-called wb (rfb) cluster (for a review, see reference 45). In this work, a recently proposed nomenclature system for genes involved in expression of bacterial surface polysaccharides is followed (39); for clarity, the old gene names are also given in parentheses. The genetics of O-antigen biosynthesis have been intensively studied in members of the family Enterobacteriaceae, and it has been shown that the wb clusters usually contain genes involved in biosynthesis of activated sugars, glycosyltransferases, O-antigen polymerases, and O-antigen export (45). Despite heterogeneity in the structures of O antigens, only three pathways for assembly of O antigens have been recognized (55).
Serratia marcescens strains, as well as those of other species of enteric bacteria, can be grouped in O-antigen serogroups, and some of them have been chemically characterized (38). S. marcescens N28b (O4) produces a bacteriocin (8, 9, 51) that has been shown to be useful to identify recombinant clones harboring genes encoding small outer membrane proteins (13) and enzymes involved in core LPS biosynthesis (12). Few studies have been carried out regarding the genetics of O-antigen biosynthesis in S. marcescens, but three genes involved in S. marcescens O16-antigen biosynthesis have been characterized (47). The S. marcescens O4-antigen repeating unit consists of l-rhamnose linked by an α1-4 bond to d-glucose (38), and recently we described S. marcescens N28b wbbL and rmlD genes (42). Strains with mutations in both wbbL and rmlD genes were shown to be impaired in O4-antigen production (42), suggesting that they belong to the S. marcescens O4 wb cluster. Furthermore, expression of these two genes in Escherichia coli DH5α conferred serum resistance and bacteriocin 28b resistance, allowing an easily screenable phenotype (42). In this work, we present the first complete genetic analysis of an S. marcescens wb cluster, containing genes involved in S. marcescens O4-antigen biosynthesis.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. All strains were grown in Luria-Bertani (LB) Miller broth and LB Miller agar (33). LB media were supplemented with ampicillin (50 μg/ml), chloramphenicol (50 μg/ml), kanamycin (30 μg/ml), or rifampin (50 μg/ml), when needed. The physical maps of the plasmids used in this study are shown in Fig. 3.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Bacterial strains | ||
| S. marcescens | ||
| N28b | Wild-type strain | 11 |
| N28b-1 | rmlD insertion mutant | 42 |
| N28b-2 | wbbL insertion mutant | 42 |
| N28b-3 | wbbA insertion mutant | This work |
| N28b-4 | wzm-wzt double deletion mutant | This work |
| E. coli | ||
| DH5α | F−endA hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 φ80lacZM15 Δ(argF-lacZYA)U169 | 16 |
| XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac (F−proAB lacIqZΔM15 Tn10) | Stratagene |
| SM10(λpir) | thi thr tonA lacY supE recA::RP4-2-Tc::Mu Kmr λpir | 34 |
| MC1061(λpir) | thi thr-1 leu-6 proA2 his-4 argE2 lacY1 galK2 ara-14 xyl-5 supE44 λpir | J. Barbé |
| CLM4 | lacZ trp Δ(sbcB-rfb) upp rel rpsL ΔrecA | 29 |
| 21548 | thr-1 leuB6 Δ(gpt-proA)66 hisG4 argE3 thi-1 rfbD1 lacY1 ara-14 galK2 xyl-5 mtl-1 mgl-51 rpsL31 kdkK51 supE44 rfe::Tn10-48 | 31 |
| Cosmids and plasmids | ||
| FGR20 | SuperCos 1 recombinant cosmid containing ΔgalE rmlCD wbbL wzm wzt wbbA | 42 |
| pSUB6 | FGR2 HindIII subclone containing ΔgalE rmlCD wbbL wzm wzt wbbA | This work |
| pSUB7 | FGR2 PstI subclone containing ΔgalE rmlCD wbbL wzm wzt wbbA::Tn1000 | This work |
| pSF100 | Kmrpir-dependent plasmid | 42 |
| pSF103 | pFS100 containing 1,848-bp internal wbbA gene | This work |
| pKO3 | Cmr temperature sensitive for replication; sacB | 27 |
| pSF104 | pKO3 with 1,108-bp insert containing wzm-wzt deletion | This work |
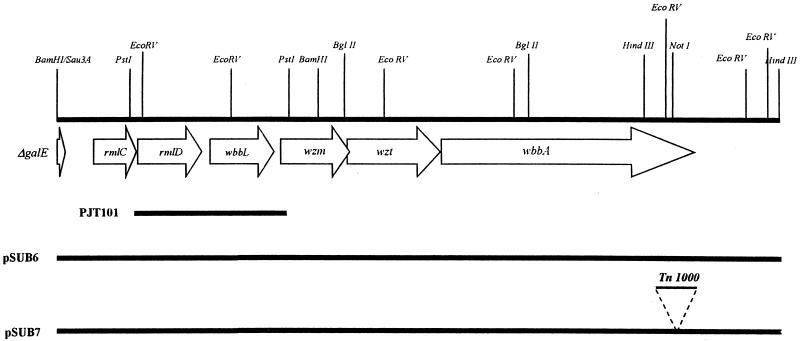
FIG. 3.
Physical and genetic map of the S. marcescens O4 wb genes. To construct the map, the whole nucleotide sequence of the 9,917-bp plasmid pSUB6 insert was determined. Only two of the PstI restriction sites are shown. The physical maps of the plasmid used in this study are also shown.
General DNA methods.
DNA manipulations were carried out essentially as previously described (43). DNA restriction endonucleases, T4 DNA ligase, E. coli DNA polymerase (Klenow fragment), and alkaline phosphatase were used as recommended by the suppliers. Recombinant clones were selected on LB Miller agar plates containing the appropriate antibiotics. To construct plasmid pSUB6, recombinant cosmid FGR20 was partially digested with HindIII, and the resulting DNA fragments were self-ligated and transformed into E. coli DH5α. Plasmid pSUB7 was constructed by PstI partial digestion of FGR20, self-ligation, and transformation into E. coli XL1-Blue.
Construction of mutant strains N28b-3 (wbbA) and N28b-4 (wzm wzt).
Two different mutant strains of S. marcescens N28b were constructed. To obtain the N28b-3 mutant (insertion in the wbbA gene), a method based on suicide plasmid pSF100 was used (42). Plasmid pSUB6 was EcoRV digested, and a wbbA internal DNA fragment (1,848 bp) was isolated, ligated to EcoRV-digested and dephosphorylated pFS100, and transformed into E. coli MC1061(λpir) to generate plasmid pSF103. Plasmid pSF103 was isolated, transformed into E. coli SM10(λpir), and transferred by conjugation to an S. marcescens N28b Rif mutant (from our laboratory collection) as previously described (42).
To obtain mutant N28b-4, the method of Link et al. (27) was used to create an in-frame deletion encompassing both the wzm and the wzt genes. Briefly, pSUB6 and primers A (5′-CGCGGATCCTTTAGGGGCTAAGATGGATG-3′), B (5′- CCCATCCACTAAACTTAAACATTTATGCGGATTACTCATTC-3′), and C (5′-TGTTTAAGTTTAGTGGATGGGGCTCCAATCCAAATCGTTGC-3′), and D (5′-CGCGGATCCAAGCAGTCGCCAAATATTCC-3′) were used in two sets of asymmetric PCRs to amplify DNA fragments of 537 (AB) and 540 (CD) bp, respectively. DNA fragment AB contains from nucleotide 2546, inside the wbbL gene, to nucleotide 3082, corresponding to the sixth codon of the wzm gene. DNA fragment CD contains from nucleotide 5178, corresponding to the first base of the codon for the 431st amino acid residue encoded by the wzt gene, to nucleotide 5717, inside the wbbA gene. DNA fragments AB and CD were annealed at their overlapping region (underlined letters in primers B and C) and amplified by PCR as a single fragment, with primers A and D. The fusion product was purified, BamHI digested (primer tags containing the BamHI site are double underlined in primers A and D), ligated into BamHI-digested and phosphatase-treated pKO3 vector (27), electroporated into E. coli DH5α, and plated on chloramphenicol plates at 30°C to obtain plasmid pSF104. The PCR amplification procedures and mutant N28b-4 construction method by gene replacement, with plasmid pSF104, were exactly those described by Link et al. (27), except that electroporated cells were plated at 42 instead of 43°C.
LPS isolation and analysis.
LPS was purified by the method of Westphal and Jann (53). For screening purposes, LPS was obtained after proteinase K digestion of whole cells according to the procedure of Hitchcock and Brown (17). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the procedure of Laemmli (26), and LPS bands were detected by the silver-staining method of Tsai and Frasch (50).
Antisera.
Anti-E. coli O16 LPS serum and anti-S. marcescens O4 LPS serum were obtained and assayed as previously described (2) for LPS specificity against purified LPS and whole cells.
Western immunoblotting.
After SDS-PAGE, immunoblotting was carried out by transfer to polyvinylidene fluoride membranes (Millipore Corp., Bedford, Mass.) at 1.3 Å for 1 h in the buffer of Towbin et al. (49). The membranes were then incubated sequentially with 1% bovine serum albumin, specific anti-O serum (1:500), alkaline-phosphatase-labeled goat anti-rabbit immunoglobulin G (1:1,000) (Boehringer Mannheim), and 5-bromo-4-chloro-3-indolylphosphate disodium–nitroblue tetrazolium (Boehringer Mannheim). Incubations were carried out for 1 h, and washing steps with 0.05% Tween 20 in phosphate-buffered saline were included after each incubation step. Colony blotting was performed with S. marcescens O4 antiserum as indicated above.
ELISA.
Cytosol, whole membrane, and inner and outer membrane fractions were analyzed by enzyme-linked immunosorbent assay (ELISA). ELISAs were performed by dispensing standardized suspensions of each fraction in coating buffer (pH 9.6), into 96-well microtiter plates. The trays were left standing overnight at 4°C. The wells were blocked with 1% bovine serum albumin in phosphate-buffered saline for 2 h at 37°C. Anti-O4 polyclonal serum (1:200) was added and incubated for 2 h at 37°C. Detection was achieved by using peroxidase-labeled sheep anti-rabbit immunoglobulin G (1:1,000) (Boehringer Mannheim) and 2,2′-azino-di-[3-ethylbenzthiazoline sulfonate] (Boehringer Mannheim) as substrate. Cytosol, whole, and inner and outer membrane fractions were prepared as previously described (37).
DNA sequencing.
Double-stranded DNA sequencing was performed by the Sanger dideoxy-chain termination method (44) with the ABI Prism dye terminator cycle sequencing kit (Perkin-Elmer). Primers used for DNA sequencing were purchased from Pharmacia LKB Biotechnology.
DNA and protein sequence analysis.
The DNA sequence was translated in all six frames, and all open reading frames (ORFs) greater than 100 bp were inspected. Deduced amino acid sequences were compared with those of DNA translated in all six frames from nonredundant GenBank and EMBL databases by using the BLAST network service at the National Center for Biotechnology Information (1). Multiple sequence alignments were carried out with the Clustal W program (48). Determinations of possible terminator sequences were done by using the Terminator program from the Genetics Computer Group package (Madison, Wis.) in a VAX 4300 computer. Hydropathy profiles were calculated according to the method of Kyte and Doolittle (25).
Nucleotide sequence accession number.
The nucleotide sequence of the O4 wb cluster from S. marcescens N28b has been deposited in GenBank under accession no. AFO38816.
RESULTS AND DISCUSSION
Isolation of a 10-kb fragment conferring O4-antigen production in E. coli DH5α.
We have previously reported the isolation of S. marcescens N28b rmlD and wbbL genes, coding for dTDP-l-rhamnose synthase and rhamnosyltransferase, respectively, from recombinant cosmid FGR20 (42). The plasmid containing only the wbbL gene induced E. coli O16-antigen biosynthesis in E. coli DH5α, by complementation of the wb-50 mutation. Analysis of LPS isolated from E. coli DH5α (FGR20) by SDS-PAGE and Western immunoblotting revealed that this strain’s LPS reacted with S. marcescens O4 antiserum, suggesting that this recombinant cosmid harbored other genes involved in O4-antigen biosynthesis. To localize the genes required for O4-antigen biosynthesis, several subclones were constructed from cosmid FGR20. Among the different subclones obtained, plasmid pSUB6 was chosen for further study because it was the smallest subclone obtained still able to direct O4-antigen biosynthesis (Fig. 1, lane 2; Fig. 2, lane 2). Restriction enzyme analysis and Southern blotting experiments showed that both rmlD and wbbL genes were present in the approximately 10-kb plasmid pSUB6 DNA insert. Taken together, these results suggest that pSUB6 contains the wb genes required for S. marcescens O4-antigen production in E. coli DH5α.
FIG. 1.
Silver-stained PAGE of LPS samples from E. coli DH5α (lane 1), E. coli DH5α(pSUB6) (lane 2), and E. coli DH5α(pSUB7) (lane 3).
FIG. 2.
Western immunoblot of LPS reacted with S. marcescens O4 antiserum. LPS was from E. coli DH5α (lane 1), E. coli DH5α(pSUB6) (lane 2), and E. coli DH5α(pSUB7) (lane 3).
Sequencing of the DNA conferring O4-antigen production.
The nucleotide sequence of the plasmid pSUB6 insert was determined in order to identify the S. marcescens genes conferring O4-antigen production on E. coli DH5α. A nucleotide sequence of 9,917 bp was determined in both directions by using oligonucleotides T3 (5′-AATTAACCCTCACTAAAGGG-3′) and H1 (5′-GTGTTCCGCTTCCTTTAG-3′) complementary to vector SuperCos 1 sequences flanking the pSUB6 DNA insert. Other sequence-derived oligonucleotides were purchased (Pharmacia LKB) and used to complete the nucleotide sequence. Analysis of the sequenced region showed the 3′ end of a potential ORF (ORF1) and six complete ORFs (Table 2 and Fig. 3). These ORFs were apparently transcribed in the same direction. In all cases, putative ribosome binding sequences were found at appropriate distances from the initial codon of each ORF. No sequences strongly similar to the −35 and −10 regions of E. coli promoters and properly spaced were found. The Terminator program from the Genetics Computer Group package allowed the identification of an inverted repeat followed by a run of T’s 98 nucleotides downstream from the end of ORF7, similar to the rho-independent transcription termination sequence. Another putative rho-independent transcription termination sequence was previously identified between the rmlD (ORF3) and wbbL (ORF4) genes (42).
TABLE 2.
S. marcescens N28b (O4) wb gene cluster
| Locus | Base positions | % G+C | Protein encoded | pIc | GRAVYd |
|---|---|---|---|---|---|
| galE (ORF1)a | 1–109 | 47.7 | NDe | ND | ND |
| rmlC (ORF2) | 524–1057 | 49.8 | 20.2 kDa | 5.31 | −0.373 |
| rmlD (ORF3)b | 1057–1923 | 53.7 | 31.1 kDa | 5.57 | +0.022 |
| wbbL (ORF4)b | 2094–2942 | 50.0 | 31.8 kDa | 8.68 | −0.127 |
| wzm (ORF5) | 3065–3898 | 48.3 | 31.3 kDa | 9.92 | +0.836 |
| wzt (ORF6) | 3888–5213 | 51.7 | 48.2 kDa | 5.96 | −0.069 |
| wbbA (ORF7) | 5213–8788 | 49.6 | 135.4 kDa | 5.45 | −0.215 |
A region of 1,087 bp (nucleotides 8832 to 9917), located 43 nucleotides downstream from the end of ORF7, showed homology (52.7% nucleotide identity) to insertion element IS4 (23). In this sequence, no ORF similar to the complete IS4 transposase was found, although some regions resulting from the six-frame translation analysis showed similarity to transposases. This result suggests that this sequence most probably corresponds to the remnants of an IS4 element that probably was involved in the S. marcescens O4-antigen gene cluster acquisition.
Analysis of the ORF’s deduced amino acid sequence.
The DNA sequence was translated in all six frames, and all ORFs were inspected. Computer database searching was carried out to tentatively identify the sequenced genes. Proteins similar to each ORF gene product were analyzed to determine the levels of similarity and identity. This analysis showed that the 35-amino-acid peptide encoded by 5′-truncated ORF1 had high levels of amino acid identity (67 and 54%) and similarity (87 and 78%) to the carboxy-terminal regions of GalE proteins from Erwinia amylovora (32) and Haemophilus influenzae (10), respectively. This result suggested that this ORF could correspond to the S. marcescens galE gene. The deduced 177-amino-acid protein encoded by ORF2 showed high levels of both amino acid identity and similarity to dTDP-4-dehydrorhamnose 3,5-epimerases, involved in O-antigen or capsule biosynthesis, or similar proteins from different gram-negative bacteria, including members of the Enterobacteriaceae, Synechocystis sp., Neisseria meningitidis, and Neisseria gonorrhoeae (Table 3). ORF2 was named rmlC as proposed for genes encoding dTDP-4-dehydrorhamnose 3,5-epimerases (39). ORF3 and ORF4 were previously identified as rmlD and wbbL genes, putatively coding for dTDP-rhamnose synthase and rhamnosyltransferase, respectively (42). The deduced 277- and 441-amino-acid proteins encoded by ORF5 and ORF6 were found to be similar to ATP-binding-cassette-2-(ABC-2)-type transport system integral membrane and ATP-binding proteins, respectively (Table 3). Exporter systems similar to the ORF5-ORF6 system are involved in export of O antigen, except for ATP-binding protein AbcA involved in A-protein expression (5) and a Synechocystis protein of unknown function (19). The putative exporter component (ORF5) showed a 33.7% level of amino acid similarity to the corresponding Wzm protein involved in S. marcescens O16-antigen export, while the putative ATP-binding component showed a higher level of similarity (53.6%) to its O16 counterpart Wzt protein. ORF5 and ORF6 are named accordingly wzm and wzt, respectively. Hydrophobicity analysis and identification of putative transmembrane domains of Wzm protein (amino acid residues 49 to 69, 79 to 99, 128 to 148, 155 to 175, and 191 to 211), by the method of Klein et al. (24), suggest that this protein is indeed an integral membrane protein. On the other hand, the sequence GRNGAGKS (residues 76 to 82) from Wzt was found to correspond to box A, a motif present in ATP-binding proteins.
TABLE 3.
Percent identities and similarities of the amino acid sequences of RmlC, Wzm, and Wzt proteins from S. marcescens N28b (O4) to other proteinsa
| Proteinb (organism) | No. of amino acids | % Similarity | % Identity | No. of gaps | Accession no. or reference |
|---|---|---|---|---|---|
| RmlC (S. marcescens O4) | 177 | ||||
| RmlC [RfbC] (Shigella flexneri) | 181 | 66.1 | 55.3 | 1 | 28 |
| ORF 7.0-encoded protein (Salmonella cholerasuis) | 176 | 65.1 | 51.4 | 1 | 52 |
| ORF-encoded protein (Synechocystis sp.) | 182 | 63.8 | 53.6 | 1 | 19 |
| RmlC (Haemophilus actinomycetemcomitans) | 179 | 67.0 | 57.9 | 2 | AB002668 |
| RmlC (Salmonella typhimurium) | 183 | 67.2 | 56.5 | 2 | 18 |
| RmlC [RfbC] (Neisseria meningitidis) | 333 | 67.0 | 56.8 | 2 | 15 |
| RmlC [RfbC] (N. gonorrhoeae) | 328 | 65.9 | 55.6 | 2 | 40 |
| RmlC [RfbC] (E. coli K-12) | 185 | 64.4 | 53.1 | 2 | 56 |
| Wzm (S. marcescens O4) | 277 | ||||
| PsaC (Pseudomonas aeruginosa PAO1) | 265 | 34.9 | 28.3 | 1 | 41 |
| Wzm [RfbA] (Myxococcus xanthus) | 260 | 30.4 | 18.8 | 1 | 14 |
| Wzm [ORF261] (E. coli O9) | 261 | 30.5 | 21.7 | 2 | 22 |
| Wzm [RfbA] (S. marcescens O16) | 255 | 33.7 | 19.9 | 4 | L34166 |
| Wzm (Synechocystis sp.) | 271 | 34.7 | 23.1 | 4 | 19 |
| Wzm (Haemophilus actinomycetemcomitans) | 263 | 38.7 | 23.9 | 4 | AB002668 |
| Wzm [RfbA] (Klebsiella pneumoniae O8) | 259 | 36.1 | 21.7 | 4 | 21 |
| Wzm [RfbA] (Klebsiella pneumoniae O1) | 259 | 35.1 | 22.2 | 4 | 4 |
| Wzt (S. marcescens O4) | 441 | ||||
| Wzt (Haemophilus actinomycetemcomitans) | 245 | 53.9 | 39.9 | 1 | AB002668 |
| Wzt [RfbB] (Klebsiella pneumoniae O8) | 246 | 55.2 | 39.8 | 2 | 21 |
| Wzt [RfbB] (Klebsiella pneumoniae O1) | 246 | 54.0 | 39.0 | 2 | 4 |
| Wzt [RfbB] (S. marcescens O16) | 246 | 53.6 | 38.6 | 2 | L34166 |
| AbcA (Aeromonas salmonicida) | 308 | 53.8 | 42.5 | 4 | 5 |
| KpsT (Synechocystis sp.) | 371 | 49.4 | 35.9 | 5 | 19 |
| Wzt [ORF431] (E. coli O9) | 431 | 46.9 | 36.5 | 7 | 22 |
| Wzt [RfbB] (Myxococcus xanthus) | 437 | 42.6 | 32.7 | 7 | 14 |
| Wzt [PsaD] (Pseudomonas aeruginosa PAO1) | 421 | 44.6 | 35.5 | 8 | 41 |
Percent values were obtained from pair comparisons with the Gap program (settings: gap weight, 12; length weight, 12).
When the proteins are known or suggested to be involved in polysaccharide synthesis or export, two protein names are shown, a name according to the nomenclature proposal in reference 34 and the one used in the original description of the protein.
The 1,191-amino-acid protein encoded by ORF7 (wbbA gene) did not show strong overall similarities to any known protein. However, the regions between residues 451 and 550 (A) and 690 and 721 (B) showed levels of 47 and 53% amino acid similarity, respectively, to LgtD protein (glycosyltransferase) from H. influenzae (10), according to the BLAST program. The WbbA region A was found to be similar to other known or putative glycosyltransferases. An amino acid alignment of these proteins (Fig. 4) suggests that region A contains motifs similar to catalytic sites 1 and 2 (domain A) but not to binding site 1 of the ExoU family of glycosyltransferases (20). On the other hand, transmembrane prediction (24) suggests that protein WbbA could be anchored to the membrane through the region encompassing residues 1121 to 1138.
FIG. 4.
Alignment of S. marcescens N28b WbbA (WbbA-Sm) and N. meningitidis LgtD (LGTD-Nm) (P96946), N. gonorrhoeae LgtD (LGTD-Ng) (Q50949), E. coli YcdQ (YcdQ-Ec) (P75905), H. influenzae HI0868 (HI0868-Hi) (P96336), H. influenzae LgtD (LgtD-Hi) (Q57287), Arquaeoglobus fulgidus AF0321 (AF0321-Af) (AE001082), Actinobacillus actinomycetemcomitans UN1 (UN1-Aa) (D1020407), and A. actinomycetemcomitans UN2 (UN2-Aa) (D1020408). Basic residues (white letters on black background), hydrophobic residues (black letters on gray background), acid residues (boldface), and cluster-breaking prolines (underlined boldface) are shown. The asterisks denote regions similar to domain A catalytic sites 1 and 2 of the ExoU family of glycosyltransferases.
These results suggest that the S. marcescens O4 wb cluster, in plasmid pSUB6, contains two genes involved in the last two steps of dTDP-rhamnose biosynthesis (rmlC and rmlD); two genes coding for rhamnosyl and putative glycosyltransferases (wbbL and wbbA, respectively); and two more genes, wzm and wzt, coding for a two-component ABC-2-type O4-antigen export system (Fig. 3).
E. coli DH5α rmlAB genes are required for S. marcescens O4-antigen production.
The S. marcescens O4-antigen repeating unit consists of l-rhamnose linked by an α1-4 bond to d-glucose (38), but no genes encoding glucose-1-phosphate thymidylyl transferase (RmlA) and dTDP-d-glucose 4,6-dehydratase (RmlB), required for the two initial steps in dDTP-rhamnose biosynthesis, were found in plasmid pSUB6. E. coli DH5α rmlAB genes should provide these functions to explain the pSUB6-directed biosynthesis of the O4 antigen in this strain. In order to test if plasmid pSUB6-directed O4-antigen production in E. coli DH5α required some other E. coli wb genes, plasmid pSUB6 was introduced into E. coli CLM4, containing the Δ(sbcB-rfb) deletion. E. coli CLM4(pSUB6) was found to produce O antigen recognized by S. marcescens O4 antiserum (Fig. 5, lane 5). The E. coli K-12 wec cluster contains genes involved in the biosynthesis of the enterobacterial common antigen (31). Marolda and Valvano (30) showed that the rffHG genes, in the wec cluster, are homologous to the rmlAB genes and encode proteins with activities identical to those of RmlA and RmlB proteins. Thus, in the E. coli CLM4 background rffHG gene products should catalyze the two initial steps in dTDP-rhamnose biosynthesis. Although most enterobacterial wb clusters involved in biosynthesis of rhamnose-containing O antigens present the four rmlABCD genes (45), our results suggest that the rmlAB genes involved in S. marcescens O4-antigen biosynthesis are not physically linked to the O4 wb cluster carried by plasmid pSUB6.
FIG. 5.
Silver-stained PAGE of LPS samples from E. coli 21548 (lane 1), E. coli 21548(pSUB6) (lane 2), E. coli 21548(pSUB7) (lane 3), E. coli CLM4 (lane 4), E. coli CLM4(pSUB6) (lane 5), and E. coli CLM4(pSUB7) (lane 6).
S. marcescens O4-antigen production is wecA dependent.
E. coli CLM4 is a recA derivative of strain Sφ874, and it has been previously shown (36) that the Δ(sbcB-rfb) deletion extends from sbcB to udk genes; thus, strains Sφ874 and CLM4 are devoid of the whole E. coli K-12 wb gene cluster including the wzy (rfc) gene. The results obtained with E. coli CLM4(pSUB6) suggest that biosynthesis of the O4 antigen is wzy independent, although it cannot be ruled out that the wecF (rffT) gene, a wzy analog (35), could be involved in sequential assembly of O4-antigen repeating units. It has been previously shown that WecA (Rfe), an N-acetylglucosamine-1-phosphatetransferase involved in biosynthesis of the enterobacterial common antigen (31), is also involved in the biosynthesis of both wzy-dependent and -independent O antigens in members of the Enterobacteriaceae (46, 56). To test the role of WecA protein in the S. marcescens O4-antigen biosynthesis, E. coli 21548, a wecA::Tn10 mutant, was transformed with plasmid pSUB6. E. coli 21548(pSUB6) did not produce O antigen, as judged by SDS-PAGE (Fig. 5, lane 2). These results suggest that, although N-acetylglucosamine is not found in the O4 repeating unit (38), the wecA gene product is essential for O4-antigen production. It has been shown that WecA protein primes the synthesis of some homopolysaccharide O antigens (6, 21); our results suggest that WecA could play a similar role in S. marcescens O4 biosynthesis.
wbbA is essential for S. marcescens O4-antigen biosynthesis.
E. coli XL1-Blue and E. coli DH5α harboring pSUB7 (Fig. 3) produced an O antigen (Fig. 1, lane 3) that reacted with E. coli O16 antiserum but not with S. marcescens O4 antiserum in Western blotting experiments (Fig. 2, lane 3). As expected, no O antigen was produced in E. coli CLM4 and E. coli 21548 harboring pSUB7 (Fig. 5, lanes 6 and 3). Restriction analysis, Southern blotting experiments, and nucleotide sequence showed that pSUB7 insert DNA harbored the O4 wb gene cluster with a Tn1000 element inserted (3) in the wbbA gene (nucleotide 8120). This phenotype is similar to the one conferred on E. coli by the plasmid containing only the rmlD and wbbL genes (42). These results suggest that the wbbA gene is essential for O4-antigen production. To further prove the role of WbbA in O4-antigen biosynthesis, an S. marcescens wbbA insertion mutant was constructed by using pir-dependent plasmid pSF103. Plasmid pSF103 was transferred to S. marcescens N28b by conjugation, and 10 colonies were screened by Southern hybridization with the internal wbbA EcoRV fragment (1,848 bp) labeled with digoxigenin as the probe. Genomic DNAs from the wild-type strain and candidate mutants were doubly digested with BamHI and HindIII, separated in agarose gels, and transferred to nitrocellulose membranes. Two DNA fragments of 4.5 and 2.1 kb from mutant strain N28b-3 were found to hybridize. As expected, only a 5.0-kb fragment from the wild-type strain hybridized with the DNA probe. Since there are two HindIII sites flanking the EcoRV insert in plasmid pSF103, these results are consistent with the expected wbbA insertion mutation. As expected, S. marcescens N28b-3 did not show O4 antigen in Western blotting experiments (Fig. 6, lane 1). The E. coli O16 wb cluster contains a glucosyltransferase (wbbK) (46), but a similar gene was not found in the characterized O4 S. marcescens wb cluster, despite glucose being present in the S. marcescens O4 antigen (38). The E. coli wbbK gene product, able to transfer the glucose moiety to the E. coli O16 antigen, is apparently not necessary for S. marcescens O4-antigen production, since E. coli CLM4(pSUB6) is able to produce the O4 antigen. The similarity between WbbA region A and the ExoU family domain A suggests a glycosyltransferase activity, able to transfer glucose to the S. marcescens O4 antigen, for the WbbA protein. Most members of the ExoU family, in wb clusters, are about one-third shorter than the WbbA protein, suggesting that this protein could have an additional unknown function. In other wb clusters, genes coding for large proteins have been found, and some of them have been proposed to code for bifunctional proteins (45).
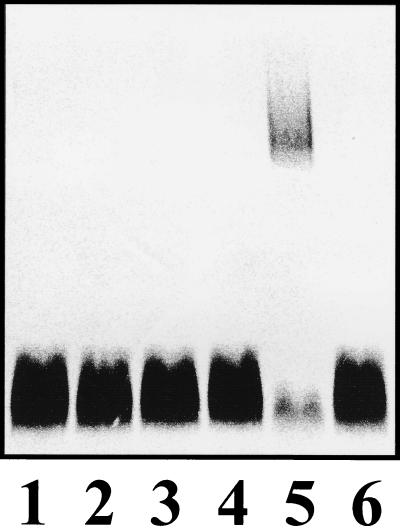
FIG. 6.
Western immunoblot of LPS reacted with S. marcescens O4 antiserum. Samples were from S. marcescens N28b-3, wbbA insertion mutant (lane 1); S. marcescens N28b-4, wzm-wzt deletion mutant (lane 3); and S. marcescens N28b, wild type (lanes 2 and 4).
Role of the Wzm and Wzt proteins.
To assess the role of the putative ABC-2-type transporter encoded by wzm and wzt genes, an S. marcescens strain (N28b-4) containing a deletion encompassing both genes was constructed. In order to avoid polar effects on the expression of the downstream wbbA gene, the wzm and wzt genes were replaced by an in-frame-tagged deletion containing the first six codons of wzm, seven tag codons, and the last 11 codons of wzy. Plasmid pSF104 was electroporated into S. marcescens N28b and plated in LB-chloramphenicol at 30 and 42°C. Chloramphenicol-resistant colonies grown at 42°C were picked (10 colonies), serially diluted, and plated on LB medium containing 5% sucrose. Sucrose-resistant colonies were replica plated on LB-chloramphenicol. Sucrose-resistant, chloramphenicol-sensitive colonies were screened for the presence of the wzm-wzt double deletion by PCR amplification with primers E (5′-TTTAGGGGTAAGATGGATG-3′) and F (5′-AAGCAGTCGCCAAATATTCC-3′). As expected, a 3.4-kb DNA fragment was amplified from the wild-type strain. An amplification product of about 1.0 kb was found in only 1 of 60 sucrose-resistant, chloramphenicol-sensitive colonies assayed. Nucleotide sequence determination showed that the amplified fragment (1,108 bp) contains the wzm-wzt deletion. N28b-4 LPS did not react with O4-antigen antiserum (Fig. 6, lane 3). ELISAs (Table 4) with S. marcescens O4-antigen-specific antiserum were performed with different cellular fractions from S. marcescens N28b and N28b-4. As expected, the outer membrane fraction in wild-type S. marcescens N28b showed a high affinity for the specific antiserum. By contrast, the inner membrane fraction obtained from S. marcescens N28b-4 showed a high response to specific antiserum, while no response to the same antiserum was observed for the outer membrane fraction of this mutant strain. Furthermore, the outer membrane fraction from strain N28b-4 transformed with pSUB6 again showed affinity for the O4-specific antiserum. These results strongly suggest that these two proteins are indeed involved in O4-antigen export in S. marcescens N28b, and the N28b-4 mutant seems to accumulate the anti-O4-reactive products in its cytosol and inner membrane fraction.
TABLE 4.
S. marcescens O4-antigen export assaya
| Extract or fraction | S. marcescens N28b | S. marcescens N28b-4 (wzm-wzt deletion mutant) | S. marcescens N28b-4 (pSUB6) |
|---|---|---|---|
| Crude extract | 0.1 (0.05) | 0.4 (0.09) | 0.15 (0.08) |
| Whole membrane fraction | 0.5 (0.08) | 0.7 (0.11) | 0.6 (0.07) |
| Inner membrane fraction | 0.1 (0.04) | 0.9 (0.12) | 0.2 (0.05) |
| Outer membrane fraction | 0.7 (0.13) | <0.1 | 0.6 (0.11) |
The values (A405) are means (standard deviations) calculated from three independent determinations (ELISAs with different cellular fractions and S. marcescens O4-specific antiserum).
Taken together, our results suggest a model for O4-antigen biosynthesis where WecA protein would initiate the process by transferring a single N-acetylglucosamine-1-phosphate (GlcNAcP) residue to undecaprenol phosphate (und-P). The und-P-P-GlcNAc would be the acceptor for glucose and rhamnose monomers transferred alternatively by the WbbA and WbbL proteins until completion of the O4 antigen. Finally, the O4 antigen would be translocated through the inner membrane by a dedicated ABC-2 transporter constituted by the Wzm and Wzt proteins. This model will agree with a previous suggestion that the ABC-2-transporter-dependent pathway for O-antigen biosynthesis is well suited for synthesis of linear O antigens but not necessarily limited to the synthesis of homopolymeric O antigens (54, 55). Further work will be necessary to confirm the validity of the proposed pathway for biosynthesis of the S. marcescens O4 antigen.
Origin of the Tn1000 insertion.
The Tn1000 insertion element in the wbbA gene in plasmid pSUB7 was an unexpected result. To our knowledge, no Tn1000 insertion has been found in S. marcescens (7), nor in the original recombinant cosmid FGR2; in plasmid pSUB6, a similar element could be detected by analysis of the vector and insert nucleotide sequence. E. coli XL1-Blue contains a Tn1000 element in its F′ plasmid (7), and pSUB7 was obtained in this strain background. Then, most probably the Tn1000 element found in the wbbA gene resulted from a transposition event, originating in E. coli XL1-Blue. Since this strain is widely used in molecular genetic manipulations, researchers working with this strain should be aware of the transposition phenomenon (with the corresponding gene knockout) that we have found.
ACKNOWLEDGMENTS
This work was supported by DGCICYT and Plan Nacional de I + D grants (Ministerio de Educación y Cultura [Spain]) and by the Generalitat de Catalunya. X.R., N.C., N.P., and A.A. have predoctoral fellowships from the Ministerio de Educación y Cultura (Spain), the Generalitat de Catalunya, and the Universitat de Barcelona.
We thank Maite Polo for her technical assistance.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Benedí V J, Ciurana B, Tomás J M. Isolation and characterization of Klebsiella pneumoniae unencapsulated mutants. J Clin Microbiol. 1989;27:82–87. doi: 10.1128/jcm.27.1.82-87.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brom J E, Hill D F, Hughes G, Jones W A, McNaughton J C, Stockwell P A, Petersen G B. Sequence of a transposon identified as Tn1000 (γδ) DNA Seq. 1995;5:185–189. doi: 10.3109/10425179509029361. [DOI] [PubMed] [Google Scholar]
- 4.Bronner D, Clarke B R, Whitfield C. Identification of an ATP-binding cassette transport system required for translocation of lipopolysaccharide O-antigen side-chains across the cytoplasmic membrane of Klebsiella pneumoniae serotype O1. Mol Microbiol. 1994;14:505–519. doi: 10.1111/j.1365-2958.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- 5.Chu S, Trust T J. An Aeromonas salmonicida gene which influences A-protein expression in Escherichia coli encodes a protein containing an ATP-binding cassette and maps beside the surface array protein gene. J Bacteriol. 1993;175:3105–3114. doi: 10.1128/jb.175.10.3105-3114.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke B R, Bronner D, Keenleyside W J, Severn W B, Richards J C, Whitfield C. Role of Rfe and RfbF in the initiation of biosynthesis of d-galactan I, the lipopolysaccharide O antigen from Klebsiella pneumoniae serotype O1. J Bacteriol. 1995;177:5411–5418. doi: 10.1128/jb.177.19.5411-5418.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deonier R C. Locations of native insertion sequence elements. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 982–989. [Google Scholar]
- 8.Enfedaque J, Ferrer S, Guasch J F, Tomás J, Regué M. Bacteriocin 28b from Serratia marcescens N28b: identification of Escherichia coli surface components involved in bacteriocin binding and translocation. Can J Microbiol. 1996;42:19–26. doi: 10.1139/m96-004. [DOI] [PubMed] [Google Scholar]
- 9.Ferrer S, Viejo M B, Guasch J F, Enfedaque J, Regué M. Genetic evidence for an activator required for induction of colicin-like bacteriocin 28b production in Serratia marcescens by DNA-damaging agents. J Bacteriol. 1996;178:951–960. doi: 10.1128/jb.178.4.951-960.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 11.Gargallo-Viola D V. Enzyme polymorphism, prodigiosin production, and plasmid fingerprints in clinical and naturally occurring isolates of Serratia marcescens. J Clin Microbiol. 1989;27:860–868. doi: 10.1128/jcm.27.5.860-868.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guasch J F, Piqué N, Climent N, Ferrer S, Merino S, Rubires X, Tomás J M, Regué M. Cloning and characterization of two Serratia marcescens genes involved in core lipopolysaccharide biosynthesis. J Bacteriol. 1996;178:5741–5747. doi: 10.1128/jb.178.19.5741-5747.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guasch J F, Ferrer S, Enfedaque J, Viejo M B, Regué M. A 17 kDa outer-membrane protein (Omp4) from Serratia marcescens confers partial resistance to bacteriocin 28b when expressed in Escherichia coli. Microbiology. 1995;141:2535–2542. doi: 10.1099/13500872-141-10-2535. [DOI] [PubMed] [Google Scholar]
- 14.Guo D, Bowden M G, Pershad R, Kaplan H B. The Myxococcus xanthus rfbABC operon encodes an ATP-binding cassette transporter homolog required for O-antigen biosynthesis and multicellular development. J Bacteriol. 1996;178:1631–1639. doi: 10.1128/jb.178.6.1631-1639.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hammerschmidt S, Birkholz C, Zahrringer U, Robertson B D, van Putten J, Ebeling O, Frosch M. Contribution of genes from the capsule gene complex (cps) to lipooligosaccharide biosynthesis and serum resistance in Neisseria meningitidis. Mol Microbiol. 1994;11:885–896. doi: 10.1111/j.1365-2958.1994.tb00367.x. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 17.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang X M, Neal B, Santiago F, Lee S J, Romana L K, Reeves P R. Structure and sequence of the Rfb (O-antigen) gene-cluster of Salmonella serovar typhimurium (strain LT-2) Mol Microbiol. 1991;5:695–713. doi: 10.1111/j.1365-2958.1991.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 19.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 20.Keenleyside W J, Whitfield C. A novel pathway for O-polysaccharide biosynthesis in Salmonella enterica serovar Borreze. J Biol Chem. 1996;271:28582–28592. doi: 10.1074/jbc.271.45.28581. [DOI] [PubMed] [Google Scholar]
- 21.Kelly R F, Whitfield C. Clonally diverse rfb gene clusters are involved in expression of a family of related d-galactan O antigens in Klebsiella species. J Bacteriol. 1996;178:5205–5214. doi: 10.1128/jb.178.17.5205-5214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kido N, Torgov V I, Sugiyama T, Uchiya K, Sugihara H, Komatsu T, Kato N, Jann K. Expression of the O9 polysaccharide of Escherichia coli: sequencing of the E. coli O9 rfb gene cluster, characterization of mannosyl transferases, and evidence for an ATP-binding cassette transport system. J Bacteriol. 1995;177:2178–2187. doi: 10.1128/jb.177.8.2178-2187.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klaer R, Kuhn S, Tillmann E, Fritz H J, Starlinger P. The sequence of IS4. Mol Gen Genet. 1981;181:169–175. doi: 10.1007/BF00268423. [DOI] [PubMed] [Google Scholar]
- 24.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 25.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Link A J, Phillips D, Church G M. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J Bacteriol. 1997;179:6228–6237. doi: 10.1128/jb.179.20.6228-6237.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macpherson D F, Manning P A, Morona R. Characterization of the dTDP-rhamnose biosynthetic genes encoded in the rfb locus of Shigella flexneri. Mol Microbiol. 1994;11:281–292. doi: 10.1111/j.1365-2958.1994.tb00308.x. [DOI] [PubMed] [Google Scholar]
- 29.Marolda C L, Valvano M A. Identification, expression, and DNA sequence of the GDP-mannose biosynthesis region encoded by the O7 rfb gene cluster of strain VW187 (Escherichia coli O7:K1) J Bacteriol. 1993;175:148–158. doi: 10.1128/jb.175.1.148-158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marolda C L, Valvano M A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J Bacteriol. 1995;177:5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier-Dieter U, Barr K, Starman R, Hatch L, Rick P D. Nucleotide sequence of the Escherichia coli rfe gene involved in the synthesis of enterobacterial common antigen. Molecular cloning of the rfe-rff gene cluster. J Biol Chem. 1992;267:746–753. [PubMed] [Google Scholar]
- 32.Metzger M, Bellemann P, Bugert P, Geider K. Genetics of galactose metabolism of Erwinia amylovora and its influence on polysaccharide synthesis and virulence of the fire blight pathogen. J Bacteriol. 1994;176:450–459. doi: 10.1128/jb.176.2.450-459.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 34.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutants: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morona R, Mavris M, Fallarino A, Manning P A. Characterization of the rfc region of Shigella flexneri. J Bacteriol. 1994;176:733–747. doi: 10.1128/jb.176.3.733-747.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neuhard J, Thomassen E. Altered deoxyribonucleotide pools in P2 eductants of Escherichia coli K-12 due to deletion of the dcd gene. J Bacteriol. 1976;126:999–1001. doi: 10.1128/jb.126.2.999-1001.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osborn M J, Cynkin M A, Gilbert J M, Muller L, Singh M. Synthesis of bacterial O-antigen. Methods Enzymol. 1972;28:583–601. [Google Scholar]
- 38.Oxley D, Wilkinson S G. Structural studies of glucorhamnans isolated from lipopolysaccharides of reference strains for Serratia marcescens serogroups O4 and O7, and of an O14 strain. Carbohydr Res. 1988;175:111–117. doi: 10.1016/0008-6215(88)80161-7. [DOI] [PubMed] [Google Scholar]
- 39.Reeves P R, Hobbs M, Valvano M A, Skurnik M, Whitfield C, Coplin D, Kido N, Klena J, Maskell D, Raetz C R H, Rick P D. Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 1996;4:495–503. doi: 10.1016/s0966-842x(97)82912-5. [DOI] [PubMed] [Google Scholar]
- 40.Robertson B D, Frosch M, van Putten J P. The identification of cryptic rhamnose biosynthesis genes in Neisseria gonorrhoeae and their relationship to lipopolysaccharide biosynthesis. J Bacteriol. 1994;176:6915–6920. doi: 10.1128/jb.176.22.6915-6920.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rocchetta H L, Lam J S. Identification and functional characterization of an ABC transport system involved in polysaccharide export of A-band lipopolysaccharide in Pseudomonas aeruginosa. J Bacteriol. 1997;179:4713–4724. doi: 10.1128/jb.179.15.4713-4724.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubirés X, Saigí F, Piqué N, Climent N, Merino S, Albertí S, Tomás J M, Regué M. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivatives. J Bacteriol. 1997;179:7581–7586. doi: 10.1128/jb.179.23.7581-7586.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 44.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnaitman C L, Klena J D. Genetics of lipopolysaccharide biosynthesis in enteric bacteria. Microbiol Rev. 1993;57:655–682. doi: 10.1128/mr.57.3.655-682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szabo M, Bronner D, Whitfield C. Relationships between rfb gene clusters required for biosynthesis of identical d-galactose-containing O antigens in Klebsiella pneumoniae and Serratia marcescens serotype O16. J Bacteriol. 1995;177:1544–1553. doi: 10.1128/jb.177.6.1544-1553.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tsai C M, Frasch C E. A sensitive silver stain for detecting lipopolysaccharide in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 51.Viejo M B, Ferrer S, Enfedaque J, Regué M. Cloning and DNA sequence analysis of a bacteriocin gene from Serratia marcescens. J Gen Microbiol. 1992;138:1737–1743. doi: 10.1099/00221287-138-8-1737. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Romana L K, Reeves P R. Molecular analysis of a Salmonella enterica group E1 rfb gene cluster: O antigen and the genetic basis of the major polymorphism. Genetics. 1992;130:429–433. doi: 10.1093/genetics/130.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westphal O, Jann K. Bacterial lipopolysaccharides: extraction with phenol-water and further applications of the procedure. Methods Carbohydr Chem. 1965;5:83–91. [Google Scholar]
- 54.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 55.Whitfield C, Amor P A, Köplin R. Modulation of the surface architecture of Gram-negative bacteria by the action of surface polymer:lipid A-core ligase and by determinants of polymer chain length. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]
- 56.Yao Z, Valvano M A. Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J Bacteriol. 1994;176:4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]