Abstract
Immunization with two mRNA vaccine doses elicits robust spike-specific CD8+ T cell responses, but reports of waning immunity after COVID-19 vaccination prompt the introduction of booster vaccination campaigns. However, the effect of mRNA booster vaccination on the spike-specific CD8+ T cell response remains unclear. Here we show that spike-specific CD8+ T cells are activated and expanded in all analyzed individuals receiving the 3rd and 4th mRNA vaccine shots. This CD8+ T cell boost response is followed by a contraction phase and lasts only for about 30-60 days. The spike-specific CD8+ T memory stem cell pool is not affected by the 3rd vaccination. Both 4th vaccination and breakthrough infections with Delta and Omicron rapidly reactivate CD8+ T memory cells. In contrast, neutralizing antibody responses display little boost effect towards Omicron. Thus, COVID-19 mRNA booster vaccination elicits a transient T effector cell response while long-term spike-specific CD8+ T cell immunity is conserved to mount robust memory recall targeting emerging variants of concern.
Subject terms: RNA vaccines, Immunological memory, Cytotoxic T cells, SARS-CoV-2
Vaccines induce beneficial immunity for COVID-19, but immune waning prompts boosting vaccination. Here, the authors show that a third, boosting dose of COVID-19 mRNA vaccine induces transient CD8 + T effector cell response while conserving the CD8 memory T cell pool, thereby permitting reactivation of spike-specific CD8 + T cells upon breakthrough infection or 4th vaccination.
Introduction
More than two years after the outbreak of the SARS-CoV-2 pandemic, constantly emerging variants of concern (VOCs) still fuel the pandemic situation and vaccination is considered to be an important measure to accelerate the transition from the pandemic into the endemic stage1. mRNA vaccines against SARS-CoV-2 have been proven to rapidly induce protection against symptomatic COVID-19 and death e.g., by inducing robust antibody and T cell responses2–5. However, reports of waning humoral immunity6–8 and high SARS-CoV-2 infection incidences with the VOC Omicron (B.1.1.529) prompted to large booster vaccination campaigns. Immediate beneficial effects of these booster immunizations were detectable with respect to the neutralization capacity of the humoral response9.
The effect of mRNA booster vaccination on the spike-specific CD8+ T cell response remains, however, unclear. In particular, there is little information about the kinetics and effects on long-term immunity and recall responses in breakthrough infections.
Here, we show that after the 3rd mRNA vaccine dose there is a substantial but transient booster effect on spike-specific CD8+ T cell frequencies without marked effect on the memory response. In contrast, only a minor boost effect on the neutralizing antibody response against Omicron is detectable after the 3rd vaccine dose. Hence, mRNA vaccines elicit robust and durable spike-specific CD8+ T cell responses already after basic immunization with minor effects of the booster immunization on long-term CD8+ T cell immunity.
Results
Transient spike-specific CD8+ T cell booster responses
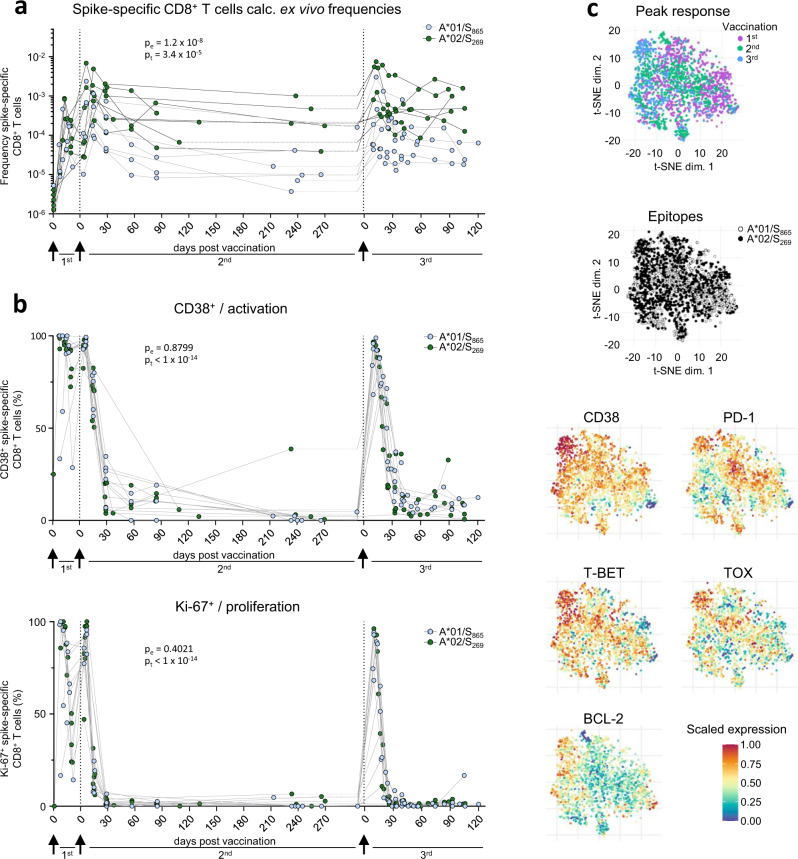
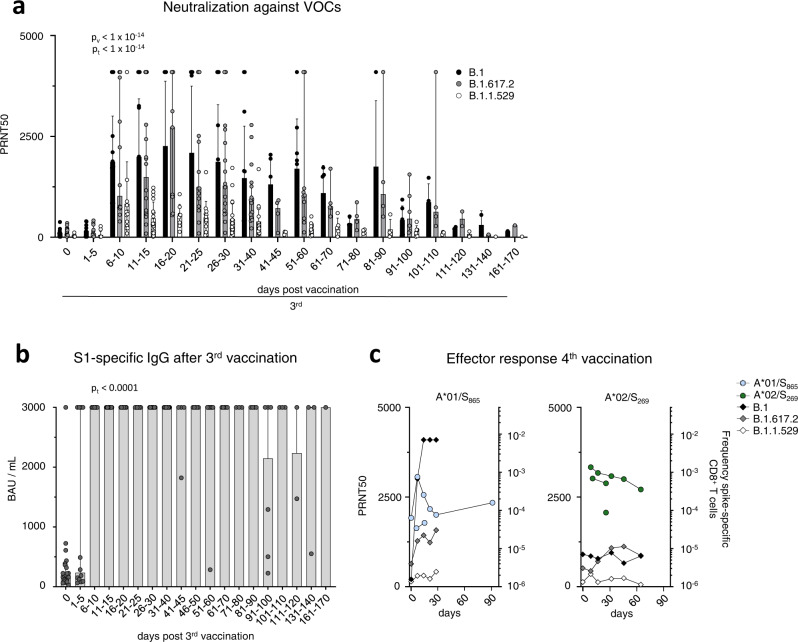
To assess the longevity of boosted adaptive effector responses, we longitudinally collected sera and peripheral blood mononuclear cells (PBMCs) from 13 volunteers (Supplementary Table 1) before and after 3rd mRNA vaccination with Comirnaty/Pfizer-Biontech or Spikevax/Moderna (Supplementary Fig. 1a). All volunteers included in this cohort did not have a history of SARS-CoV-2 infection and received their vaccinations with a three-week interval between 1st and 2nd and six-to-nine-months interval between 2nd and 3rd dose. First, we analyzed the kinetics of the spike-specific CD8+ T cell response targeting the A*01/S865 (n = 7) or A*02/S269 epitope (n = 8). A*01/S865- and A*02/S269-specific CD8+ T cells are part of a broader CD8+ T cell response targeting the spike protein after mRNA vaccination and dominant with respect to the restricting HLA class I allele3,10. The targeted epitopes are highly conserved in all VOCs, including the Omicron variants BA.1 and BA.2 (Supplementary Fig. 1b) but not in common cold coronaviruses3. To increase the detection sensitivity of spike-specific CD8+ T cells targeting single epitopes, we performed peptide-loaded MHC-class I tetramer-based enrichment3,11 (Supplementary Fig. 1c). Spike-specific CD8+ T cell frequencies had already reached their set point approx. 60 days after the 2nd dose and were rapidly boosted with peak expansion one week after the 3rd dose in all tested donors (Fig. 1a). Robust boosting of the spike-specific CD8+ T cell response by the 3rd dose was also reflected by high CD38, Ki-67 and T-BET expression and thus by rapid activation, strong proliferation and substantial induction of an effector cell program (Fig. 1b and Supplementary Fig. 2a). However, within 30-60 days the circulating spike-specific CD8+ T cell frequency, activation, proliferation and effector cell differentiation decreased rapidly to a similar level as before the 3rd dose (Fig. 1a, b and Supplementary Fig. 2a), thus following the same kinetics as after the 2nd dose. Similarities between the spike-specific CD8+ T cell response after the 2nd and 3rd dose, respectively, are also supported by t-distributed stochastic neighborhood embedding (t-SNE) and pseudotemporal diffusion analysis of T cell phenotypes including CD38, T-BET, BCL-2, PD-1, and TOX expression (Fig. 1c and Supplementary Fig. 2b, c). Phenotypic differences (e.g., in T-BET and TOX expression) were detectable between the spike-specific CD8+ T cell response after the 1st dose compared to the 2nd and 3rd doses and only minor differences were apparent with respect to the targeted epitopes A*01/S865 or A*02/S269 (Fig. 1c and Supplementary Fig. 2b, c). With respect to the neutralizing antibody response, the boost effect clearly depended on the VOC (Fig. 2a and Supplementary Fig. 2b; p < 0.0001). In particular, neutralization was only slightly increased against the Omicron variant BA.1 (B.1.1.529) by the 3rd dose, while neutralization of the parental B.1 strain and Delta (B.1.617.2) peaked after approx. 3 weeks followed by a decrease to baseline neutralization after 4 months (Fig. 2a). Of note, after the 4th dose similar kinetics (Fig. 2c) and dynamics (Supplementary Fig. 3a–c) of spike-specific CD8+ T cell and neutralizing antibody responses were detectable. Hence, after booster vaccination, there is a transient mRNA vaccine-associated spike-specific CD8+ T effector cell response and neutralizing antibody boost lasting about 2 and 4 months, respectively.
Fig. 1. Cellular and humoral effector response after the 3rd vaccine dose.
Calculated ex vivo frequencies of spike-specific CD8+ T cells throughout 1st, 2nd and 3rd vaccination (a). Ki-67,and CD38 expression within spike-specific non-naïve CD8+ T cells (b). t-SNE representation of flow cytometry data comparing A*01/S865- and A*02/S269-specific CD8+ T cells at peak response after 1st, 2nd and 3rd vaccination. Expression levels of CD38, T-BET, BCL-2, PD-1 and TOX are depicted (c). Statistical significance was determined by two-way ANOVA with main model (a, b) comparing the effects of targeted epitopes (pe) and of the time course (pt). Source data are provided as a Source Data file.
Fig. 2. Humoral response after the 3rd vaccine dose.
Antibody neutralization activity as 50% plaque reduction neutralization tests (PRNT50) for SARS-CoV-2 variants B.1, B.1.617.2 and B.1.1.529 after 3rd vaccination. Median values are depicted with 95% confidence interval error bars with n = 28 individual tested (a). Quantification of serum anti-SARS-CoV-2 spike IgG levels following 3rd vaccination with n = 26 individuals tested (b). Calculated ex vivo frequencies of A*01/S865- (left) and A*02/S269-specific (right) CD8+ T cells, with antibody neutralization activity after 4th vaccination (c). Median values are depicted with 95% confidence interval error bars. Statistical significance was calculated by Kruskal-Wallis test (a) comparing the effects of time course (pt < 1×10−14). Source data are provided as a Source Data file.
Stable T memory cell response irrespective of booster immunization
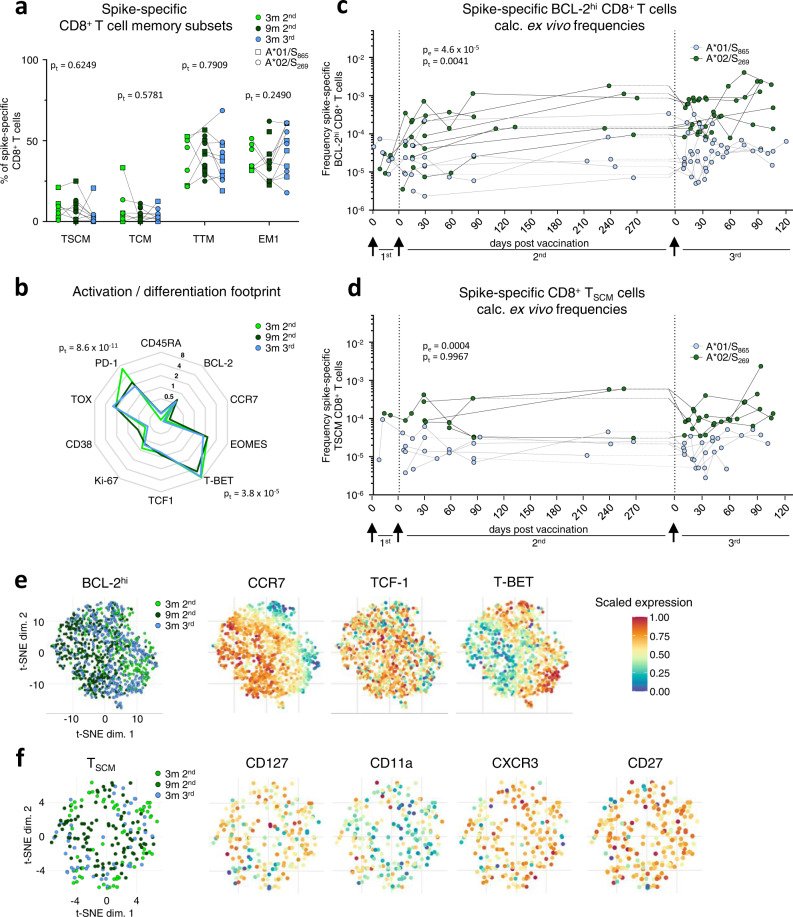
To investigate the effect of booster immunization on long-term spike-specific CD8+ T cell immunity, we comparatively analyzed the spike-specific CD8+ T cell response after three and nine months post 2nd dose versus three months after the 3rd dose. The memory T cell subset distribution (Fig. 3a and Supplementary Fig. 4a, b) and activation/differentiation footprint (Fig. 3b and Supplementary Fig. 4c) of spike-specific CD8+ T cells were similar at the tested time points, indicating stable memory features without a clear effect of the 3rd dose. Of note, during the effector phase after the 3rd vaccine shot, a transient subset redistribution was detectable (Supplementary Fig. 5a–c). To more precisely quantify and characterize the T cell memory compartment, we analyzed on the one hand, spike-specific CD8+ T cells highly expressing BCL-2 (BCL-2hi) to map the overall memory pool irrespective of certain subsets and on the other hand, focused on T memory stem cells (TSCM, defined by expression of CD45RA, CCR7, CD27, CD28 and CD95; Supplementary Fig. 4a) that are essential for long-term T cell immunity12. Spike-specific BCL2hi CD8+ T cells and CD8+ TSCM cells were induced after the 1st dose and their frequencies remained stable after the 2nd dose throughout the subsequent vaccination (Fig. 3c, d). t-SNE analysis revealed that spike-specific BCL-2hi CD8+ T cells showed minor phenotypic differences such as higher T-BET expression 3 months post 2nd and 3rd dose compared to 9 months after the 2nd dose (Fig. 3e and Supplementary Fig. 5d) probably reflecting a more resting state at later time points post immunization. In contrast, phenotypic characteristics of CD8+ TSCM remained completely stable, indicated by a complete intermingling within the t-SNE analysis (Fig. 3f). Furthermore, the spike-specific CD8+ T cell memory pool was also conserved during and after the 4th dose (Supplementary Fig. 6a–c). Thus, these results indicate that booster immunizations give rise to a rapid effector T cell response that is based on a stable T cell memory pool as illustrated by a subset of spike-specific CD8+ T cells expressing high levels of BCL-2 in parallel to T-BEThi, PD-1hi, TOX+ subsets at peak expansion after booster immunization (Supplementary Fig. 3a).
Fig. 3. Spike-specific CD8+ T memory cells before and after the 3rd vaccine dose.
Composition of spike-specific CD8+ T cell memory subsets at 3 (n = 7) and 9 (n = 11) months after 2nd, and 3 (n = 11) months after 3rd vaccination (a). Molecule expression of spike-specific non-naïve CD8+ T cells at 3 (n = 8) and 9 (n = 12) months after 2nd, and 3 (n = 11) months after 3rd vaccination normalized to bulk naïve CD8+ T cells (b). Calculated ex vivo frequencies of BCL-2hi (c) and TSCM (d) spike-specific CD8+ T cells. t-SNE representation of BCL-2hi (e) and TSCM (f) spike-specific CD8+ T cells at 3 and 9 months after 2nd, and 3 months after 3rd vaccination. Expression levels of CCR7, TCF-1 and T-BET are depicted for BCL-2hi, and CD127, CD11a, CXCR3 and CD27 are depicted for TSCM spike-specific CD8+ T cells. Statistical significance was calculated by two-way ANOVA with full model and Tukey’s test for multiple comparison (a, b) to examine the effect of sampling time points (pt) on memory subsets and marker expression, and two-way ANOVA with main model (c, d) to compare the effects of targeted epitopes (pe) and time course (pt). Source data are provided as a Source Data file.
Conserved spike-specific CD8+ T cell recall capacity
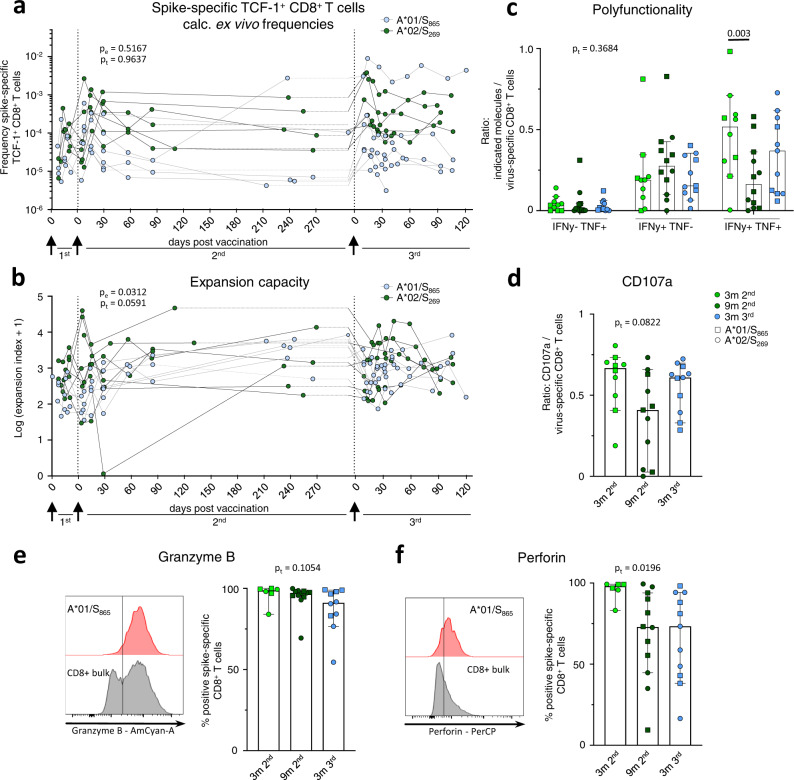
Next, we analyzed the recall capacity of spike-specific CD8+ T cells after the 3rd dose. For this, we stained for TCF-1 expression, a transcription factor indicative of the proliferative capacity of T cells13. The frequencies of TCF-1+ spike-specific CD8+ T cells were stable suggesting a conserved pool of proliferative competent T cells throughout the 2nd and 3rd dose (Fig. 4a). Accordingly, the peptide-specific in vitro expansion capacity of spike-specific CD8+ T cells was also conserved before and after the 3rd dose (Fig. 4b and Supplementary Fig. 7a, b). Furthermore, after two weeks of expansion, spike-specific CD8+ T cells produced similar amounts of IFNγ and TNF, including co-production irrespective of whether the input cells were obtained before or after the 3rd dose (Fig. 4c and Supplementary Fig.7c, d). Spike-specific production of IFNγ and TNF were assessed in relation to the frequency of spike-specific CD8+ T cells after expansion as a measure of the effector function per cell. In order to estimate the cytotoxic potential of spike-specific CD8+ T cells after in vitro expansion, we analyzed degranulation as indicated by CD107a expression in relation to the frequency of spike-specific CD8+ T cells (Fig. 4d and Supplementary Fig. 7e) and Granzyme B (Fig. 4e) and Perforin (Fig. 4f) expression of spike-specific CD8+ T cells. Similar to cytokine production, the cytotoxic potential was also largely conserved with almost all spike-specific CD8+ T cells expressing Granzyme B and Perforin. Thus, recall capacity tested in vitro comprising expansion capacity, differentiation to cytokine-producing and cytotoxic effector cells is robust and stable before and after 3rd vaccination.
Fig. 4. Reactivation capacity of spike-specific CD8+ T cells before and after the 3rd vaccine dose.
Calculated ex vivo frequencies of TCF-1+ non-naive spike-specific CD8+ T cells (a). Expansion capacity of spike-specific CD8+ T cells over 14 days of in vitro expansion throughout 1st, 2nd and 3rd vaccination (b). Percentage of IFNγ and TNF (c), and CD107a (d) producing CD8+ T cells upon peptide stimulation related to the percentage of spike-specific CD8+ T cells from all CD8+ T cells at 3 (n = 10) and 9 (n = 12 for c, n = 11 for d) months post 2nd, and 3 (n = 11) months post 3rd vaccination after in vitro expansion. Median values are depicted with 95% confidence interval error bars. Expression of Granzyme B (e) and Perforin (f) of spike-specific CD8+ T cells at 3 (n = 6) and 9 (n = 12) months post 2nd, and 3 months (n = 10 for e, n = 11 for f) post 3rd vaccination after in vitro expansion with representative histograms. Median values are depicted with 95% confidence interval error bars. Statistical significance was calculated by two-way ANOVA with main model (a, b) to compare the effects of targeted epitopes (pe) and time course (pt), two-way ANOVA with full model and Tukey’s multiple comparison test (c), and Kruskal-Wallis test (d, e, f) to examine the effect of sampling time on functionality (pt). Source data are provided as a Source Data file.
Recall response in breakthrough infections
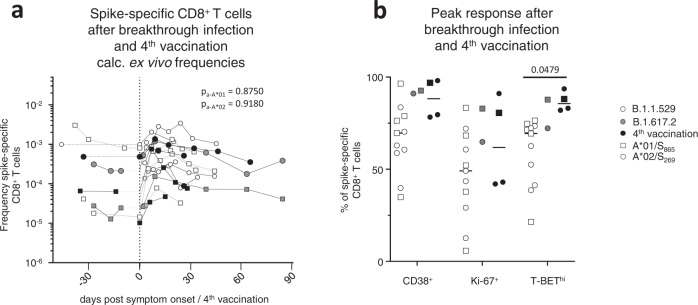
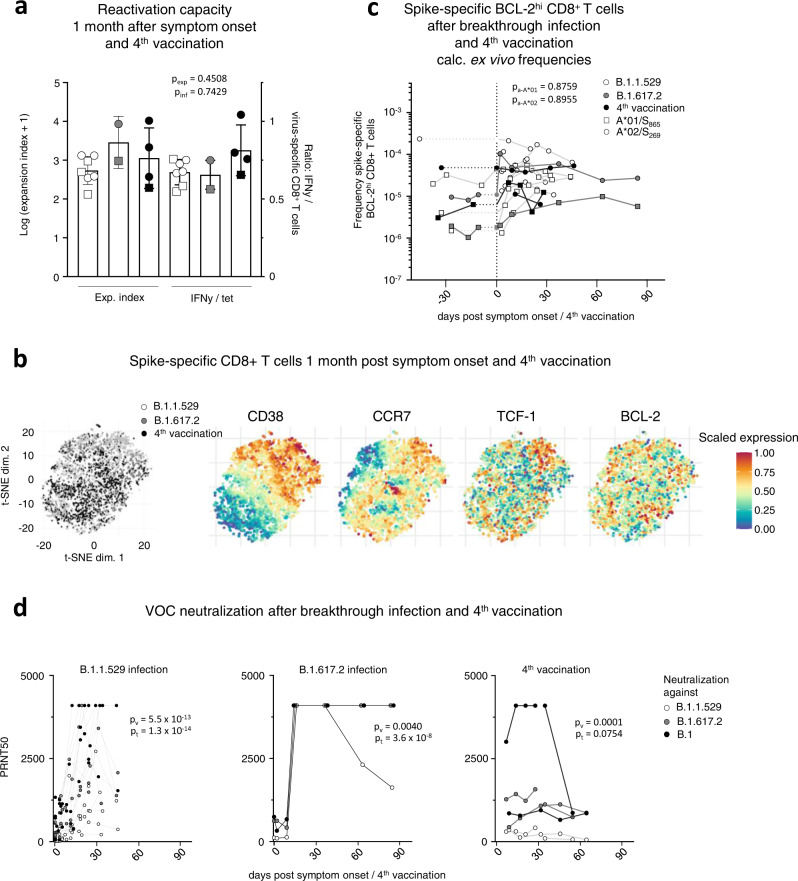
To determine the recall capacity in vivo and to compare booster responses of vaccine-elicited CD8+ T cell responses after vaccination versus infection, we next analyzed spike-specific CD8+ T cells and the neutralizing antibody capacity in breakthrough infections. Specifically, we compared the adaptive effector immune response after the 4th antigen contact, either by the 4th vaccine dose (n = 5) or by breakthrough infection with Omicron (n = 12) or Delta (n = 2) after three mRNA vaccine doses. The spike-specific CD8+ T cell response was rapidly and robustly induced and similar after the 4th vaccine dose versus an Omicron or Delta breakthrough infection (Fig. 5a and Supplementary Fig. 8a). Spike-specific CD8+ T cells peaked in a classical effector cell response with pronounced expression of CD38, Ki-67 and T-BET (Fig. 5b and Supplementary Fig. 8a–c). Variations within the proportion of spike-specific CD8+ T cells expressing these markers may be due to differences in infection time point or time point of symptom onset, which is especially relevant in breakthrough infections with Omicron that are often characterized by mild symptoms in vaccinated individuals. However, within one to two months after breakthrough infection and 4th vaccine dose, a fully functional early T cell memory was present with similar reactivation capacities comprising expansion, cytokine production and degranulation (Fig. 6a and Supplementary Fig. 9a). Furthermore, phenotypic characteristics of early spike-specific CD8+ T memory cells were similar after breakthrough infections and 4th vaccine dose as depicted by t-SNE analysis including CD38, CCR7, TCF-1 and BCL-2 (Fig. 6b) and memory subset distribution with transitional and effector memory subsets being dominant (Supplementary Fig. 9b). Again, the frequencies of the BCL-2hi memory pool within the spike-specific CD8+ T cells were stable before and after the 4th antigen contact irrespective of breakthrough infection or 4th vaccine dose (Fig. 6c). Hence, vaccine-elicited spike-specific CD8+ T cell immunity exhibited a substantial recall capacity in vivo even towards VOCs such as Omicron. However, the neutralizing antibody response after the 4th antigen contact differed from the spike-specific CD8+ T cells in the following aspects (Fig. 6d): First, neutralization capacity depended on the targeted SARS-CoV-2 variant regardless of whether the 4th antigen exposure was through vaccination or breakthrough infection. Second, the neutralizing antibody response depended on the 4th antigen contact with differences in vaccination and breakthrough infections. Third, the neutralizing antibody response exhibited a prolonged decay after breakthrough infections compared to vaccination. Thus, in contrast to spike-specific CD8+ T cells, recall of the neutralizing antibody response was less robust with respect to VOCs.
Fig. 5. CD8+ T effector cell response after breakthrough infection and the 4th vaccine dose.
Calculated ex vivo frequencies of spike-specific CD8+ T cells before and after breakthrough infection and 4th vaccination (a). Proportion of CD38+, Ki-67+ and T-BEThi within non-naive spike-specific CD8+ T cells at peak expansion after breakthrough infection and 4th vaccination with n = 11 for Omicron and n = 2 for Delta breakthrough infections, and n = 4 individuals receiving a 4th vaccination (b). Statistical significance was calculated by two-way ANOVA with main model to compare the effects of antigen triggers (pa) on epitope-specific T cell frequencies (a) and two-way ANOVA with full model and Tukey’s test for multiple comparison (b) to examine the effects of antigen triggers on activation marker expression. Source data are provided as a Source Data file.
Fig. 6. Characterization of CD8+ T memory cells after breakthrough infection and the 4th vaccine dose.
Reactivation capacity of spike-specific CD8+ T cells 1 month after symptom onset and 4th vaccination. Median values are depicted with 95% confidence interval error bars with n = 6 for Omicron and n = 2 for Delta breakthrough infections, and n = 4 individuals receiving a 4th vaccination (a). t-SNE representation of spike-specific CD8+ T cells 1 month after symptom onset and 4th vaccination. Expression levels of CD38, CCR7, TCF- and BCL-2 are depicted (b). Calculated ex vivo frequencies of BCL-2hi non-naïve spike-specific CD8+ T cells before and after breakthrough infection and 4th vaccination (c). Antibody neutralization activity as 50% plaque reduction neutralization tests (PRNT50) for SARS-CoV-2 variants B.1, B.1.617.2 and B.1.1.529 after breakthrough infection and 4th vaccination with n = 11 for Omicron and n = 1 for Delta breakthrough infection, and n = 2 individuals receiving a 4th vaccination (d). Statistical significance was calculated by Kruskal-Wallis test (a) to compare the effect of the antigen triggers on expansion (pexp) and interferon production (pinf) and two-way ANOVA with main model (c, d) to compare the effects of antigen triggers (pa) on epitope-specific T cell frequencies (c) or the effects of VOCs (pv) and time course (pt). Source data are provided as a Source Data file.
Discussion
Here, we mapped the dynamics of mRNA vaccine- versus SARS-CoV-2 infection-boosted spike-specific adaptive effector immunity. By analyzing the dominant A*01/S865- and A*02/S269-specific CD8+ T cell responses, we observed a rapid and robust expansion of spike-specific CD8+ T cells with a similar slope and amplitude after the 3rd and 4th vaccine dose versus breakthrough infections with SARS-CoV-2 Delta and Omicron variants. Notably, kinetics and magnitude were comparable to the spike-specific CD8+ T cell response after the 2nd vaccine dose with a remarkably high recruitment of spike-specific CD8+ T cells to the effector pool that is reflected by high expression of activation and proliferation markers, like CD38 and Ki-67. This observation highlights the effectiveness of mRNA vaccination to elicit rapid and functional CD8+ T cell responses. Thus, given the important role of CD8+ T cells to control SARS-CoV-2 infection14–17, mRNA booster vaccination represents a valuable tool to immediately react to high viral burden in the population in order to e.g., protect vulnerable groups and to reduce the risk of overwhelming the public health system.
Importantly, however, the spike-specific CD8+ T effector cell response after the 3rd mRNA vaccination only lasted approx. 1-2 months until it decreased to a similar level as prior booster vaccination. This rapid contraction of the spike-specific CD8+ T cell response should be taken into account when booster vaccination strategies are planned especially for risk groups but also for broader populations. Furthermore, the steep decline of the spike-specific CD8+ T cell boost response is in contrast to a more prolonged contraction reported for non-spike epitope-specific CD8+ T cells after SARS-CoV-2 infection11. This may indicate a difference in virus-specific CD8+ T cell kinetics induced by SARS-CoV-2 infection versus mRNA vaccination. Possible explanations include differences in antigen half-life, innate immunity, co-stimulation, cytokines, antigen presentation and CD4+ T cell responses that are all required for coordinating CD8+ T cell responses4,15,16 after mRNA vaccination versus SARS-CoV-2 infection.
A durable and functionally competent spike-specific CD8+ T memory cell response was already detectable after the 2nd dose of mRNA vaccine and thus after completed basic immunization3,4,18. Here, we now show that this well-established durable spike-specific CD8+ T memory cell response is not tremendously affected by the 3rd dose. In particular, a pool of TSCM cells that have been reported to maintain long-term CD8+ T cell immunity after yellow fever virus (YFV) vaccination12,19 and have been shown to be detectable after basic immunization with SARS-CoV-2 mRNA vaccination20 were already induced within spike-specific CD8+ T cells after prime vaccination. The spike-specific CD8+ TSCM cell pool reached final size after the 2nd dose and remained constant throughout the 3rd and 4th dose. Thus, 3rd mRNA vaccination does neither amplify long-term CD8+ T cell immunity nor drive the CD8+ T cell memory pool into relevant senescence. This is in line with a recent report showing that repeated antigen exposure does also not induce T cell exhaustion of spike-specific CD8+ T cells21. It rather appears that the spike-specific CD8+ T cell booster response is an effector response based on a stable memory pool. Of note, spike-specific CD8+ T cell epitope spreading after the 3rd mRNA vaccination is also not detectable10 further supporting the stability of the memory pool. Importantly, spike-specific CD8+ T memory cells were capable to robustly mount recall responses in vitro and in vivo after breakthrough infections with Delta or Omicron. This recall robustness is explained by the previously described cross-reactivity of the vaccine-elicited spike-specific CD8+ T cell response10,18,22–25, diversification of the T cell repertoire21 and of the rapid kinetics and effector differentiation dynamics shown in this study. It is, however, important to note that our study is limited to a small cohort of healthy, rather young individuals without higher risk to develop severe COVID-1916 and with the capability to establish robust vaccine-elicited immune responses2–5,18. Future studies have to show whether individuals above the age of 60 develop comparable immunity after mRNA vaccination. Still, our observations challenge the discussion about the necessity to frequently apply mRNA booster vaccination in three-to-six-months intervals to healthy individuals that are not compromised in their immune response but have also been included in the large vaccination campaigns thus far.
In contrast to spike-specific CD8+ T cells, boosting the neutralizing antibody response by currently applied, non-adapted mRNA vaccines clearly depends on the targeted SARS-CoV-2 variant. In particular, neutralizing capacity against Omicron is hardly increased. Of note, neutralizing capacity against Omicron is also only moderately increased after breakthrough infections with Omicron probably reflecting antibody escape. Similar to the spike-specific CD8+ T cell response, the neutralizing capacity declines slower after infection versus vaccination. Yet, it appears that neither after vaccination nor after infection the neutralization capacity of the humoral response is sufficient to protect from Omicron infection as indicated by the high numbers of breakthrough infections in vaccinees and convalescents. In sum, our study highlights that mRNA vaccines are potent inducers of a robust, functionally competent and durable spike-specific CD8+ T cell immunity already after completed basic immunization. This has also important implications for vaccine development targeting other CD8+ T cell-controlled infectious agents and cancers.
Methods
Study cohort
In total, 38 individuals receiving SARS-CoV-2 vaccinations were recruited at the Freiburg University Medical Center, Germany. Of those, blood was collected from 31 individuals vaccinated three times with the mRNA vaccines bnt162b/Comirnaty or mRNA-1273/Spikevax and 5 individuals receiving a 4th vaccination. All vaccinees did not have a history of SARS-CoV-2 infection prior to inclusion confirmed by seronegativity for anti-SARS-CoV-2 nucleocapside IgG (anti-SARS-CoV-2 N IgG). Moreover, blood was collected from 13 individuals with SARS-CoV-2 breakthrough infections after a 3rd mRNA vaccination. Breakthrough infections were confirmed by positive PCR-testing from oropharyngeal swab. All 13 individuals with breakthrough infections included in this study had mild symptoms without respiratory insufficiency (according to WHO guidelines26). Characteristics of the participants are summarized in Supplementary Table 1, including the results of the HLA-genotyping performed by next-generation sequencing.
Ethics
Written informed consent was obtained from all study participants. The study was conducted in accordance to federal guidelines, local ethics committee regulations (Albert-Ludwigs-Universität, Freiburg, Germany; vote: 322/20, 21-1135 and 315/20) and the Declaration of Helsinki (1975).
PBMC isolation
PBMCs were isolated from venous blood samples collected in EDTA blood collection tubes by density centrifugation with lymphocyte separation medium (Pancoll separation medium, PAN Biotech GmbH). PBMCs were stored at −80 °C until further processing. The cells were thawed in prewarmed RPMI cell culture medium supplemented with 10% fetal calf serum, 1% penicillin/streptomycin, 1.5% 1 M HEPES (all purchased from Thermo Scientific) and 50 U/mL Benzonase (Sigma).
Sequence alignment
Sequence homology was analyzed in Geneious version 11.0.5 (https://www.geneious.com/) using Clustal Omega version 1.2.2 alignment with default settings27. Reference genome of human ancestral SARS-CoV-2 (MN908947.3) was obtained from NCBI database. Genome sequences of SARS-CoV-2 variants of concern (VOCs) B.1, B.1.1.7, B.1.351, P.1, B.1.617.2, B.1.1.529 BA.1 and B.1.1.529 BA.2 were identified via CoVariants (https://covariants.org/). Spike epitopes in ancestral strain and all VOCs were aligned according to their homology on an amino acid level.
Peptides and tetramers for T cell analysis
Peptides were manufactured with an unmodified N-terminus and an amidated C-terminus with standard Fmoc chemistry (Genaxxon Bioscience). All peptides showed a purity of >70%. To generate tetramers, SARS-CoV-2 spike peptides (A*01/S865: LTDEMIAQY, A*02/S269: YLQPRTFLL) were loaded on biotinylated HLA class I (HLA-I) easYmer (immunAware) according to manufacturer’s instructions. Subsequently, peptide-loaded-HLA class I monomers were tetramerized with phycoerythrin (PE)-conjugated streptavidin according to the manufacturer’s instructions.
In vitro expansion of spike-specific CD8+ T cells and assessment of effector function
1.5 × 106 PBMCs were stimulated with the spike protein-derived peptides A*01/S865 or A*02/S269 and anti-CD28 monoclonal antibody (0.5 µg/mL) for 14 days in RPMI cell culture medium supplemented with rIL-2 (20 IU/ml, StemCell Technologies). At day 4, 7 and 11, 50% of the culture medium was exchanged with freshly prepared medium containing 20 IU/mL rIL-2. After 14 days, PBMCs were stimulated with peptides again, and stained for CD107a for 1 h at 37 °C to analyze degranulation. Subsequently, brefeldin A (GolgiPlug, 0.5 μl/mL) and monensin (GolgiStop, 0.5 μl/mL) (all BD Biosciences) were added and incubation continued for four more hours, followed by surface and intracellular staining with anti-IFNy, anti-TNF and anti-IL-2-specific antibodies. For calculation of the expansion capacity and to assess the cytotoxic capacity of the expanded cells, peptide-loaded HLA class I tetramer staining was performed together with intracellular staining of Granzyme B, Granzyme K, Perforin and Granulysin.
Magnetic bead-based enrichment of spike-specific CD8+ T cells
CD8+ T cells targeting spike epitopes were enriched as described previously28. In brief, 5 × 106 to 20 × 106 PBMCs were stained with PE-coupled peptide-loaded HLA class I tetramers for 30 min at room temperature followed by incubation with magnetic anti-PE microbeads. Subsequent positive selection of magnetically labelled cells was achieved by using MACS technology (Miltenyi Biotec) according to the manufacturer’s protocol. The enriched spike-specific CD8+ T cells were analyzed using multicolor flow cytometry. Cell frequencies were calculated as previously described28. Of note, only samples with ≥ 5 non-naïve spike-specific CD8+ T cells were included in subsequent analyses. Accordingly, the detection limit of spike-specific CD8+ T cells in this study was 0.25 – 1 × 10−6, depending on the initial cell input. This cut-off number has been applied and validated in different studies on antigen-specific T cells and has shown to generate reproducible results3,11,29,30.
Multiparametric flow cytometry for T cell analysis
Antibodies used for multiparametric flow cytometry are listed in Supplementary Table 2. To facilitate staining of intranuclear and cytoplasmic targets, FoxP3/Transcription Factor Staining Buffer Set (Thermo Fisher) and Fixation/Permeabilization Solution Kit (BD Biosciences) were used, respectively. Finally, cells were fixed in 2% paraformaldehyde (Sigma) and samples were analyzed on FACSCanto II or LSRFortessa with FACSDiva software version 10.6.2 (BD), or CytoFLEX (Beckman Coulter) with CytExpert Software version 2.3.0.84. Further analyses of the data were performed using FlowJo version 10.6.2 (Treestar). Phenotypical analyses were based on 5 ×106 to 20 ×106 PBMCs that were used as an input number for the magnetic bead-based enrichment of spike-specific CD8+ T cells.
Dimensional reduction of multiparametric flow cytometry data
For dimensionality reduction, flow cytometry data were analyzed with R version 4.1.1 and the Bioconductor CATALYST package (release 3.13)31. Initially, viable and tetramer-positive CD8+ T cells (or subsets of those) were identified using FlowJo 10 in two separate multiparametric flow cytometry panels (activation panel: HLA-DR, BCL-2, PD-1, CD137, Ki67, TCF-1, EOMES, T-BET, TOX, CD38, CD45RA, CCR7; differentiation panel: CD45RA, CCR7, CD27, CD28, CD127, CD11a, CD57, CXCR3, CD95, CD57, CD39, KLRG1, PD-1). To facilitate visualization of the dimensionality reduction by t-SNE and diffusion map analysis, cell counts were sampled down to at least 20 cells per sample, and marker expression intensities were transformed by arcsinh-transformation with a cofactor of 150.
Serum IgG determination
Determination of SARS-CoV-2-specific antibodies was performed by using the Euroimmun assay ‘Anti-SARS-CoV-2-QuantiVac-ELISA (IgG)’ for detecting anti-SARS-CoV-2 spike IgG (anti-SARS-CoV-2 S IgG; <35.2 BAU/mL: negative, ≥ 35.2 BAU/mL: positive) and the Mikrogen assay ‘recomWell SARS-CoV-2 (IgG)’ for detecting anti-SARS-CoV-2 N IgG (detection limit, 24 a.u. ml−1) according to the manufacturer’s instructions. Data were collected with the SparkControl Magellan software version 2.2.
Neutralization assay
Samples of vaccinated individuals and those with breakthrough infections were tested in a plaque reduction neutralization assay as previously described3. In brief, VeroE6 cells were seeded in 12-well plates at a density of 4 × 105 cells per well. Serum samples were diluted at ratios of 1:16, 1:32, 1:64, 1:128, 1:256, 1:512 and 1:1024 in a total volume of 50 μl PBS. For each sample, a serum-free negative control was included. Diluted sera and negative controls were subsequently mixed with 90 plaque-forming units (PFU) of authentic SARS-CoV-2 (either B.1, B.1.617.2 (delta) and B.1.1.529 BA.1 (omicron)) in 50 μl PBS (1,600 PFU/mL) resulting in final sera dilution ratios of 1:32, 1:64, 1:128, 1:256, 1:512, 1:1024 and 1:2048. After incubation at room temperature for 1 h, 400 μl PBS was added to each sample and the mixture was subsequently used to infect VeroE6 cells 24 h after seeding. After 1.5 h of incubation at room temperature, inoculum was removed and the cells were overlaid with 0.6% Oxoid-agar in DMEM, 20 mM HEPES (pH 7.4), 0.1% NaHCO3, 1% BSA and 0.01% DEAE-Dextran. Cells were fixed 72 h after infection using 4% formaldehyde for 30 min and stained with 1% crystal violet upon removal of the agar overlay. PFU were counted manually. Plaques counted for serum-treated wells were compared to the average number of plaques in the untreated negative controls, which were set to 100%. Calculation of PRNT50 values was performed using a linear regression model in GraphPad Prism 9 (GraphPad Prism Software).
Statistics
GraphPad Prism software version 9.3.1 was used for statistical analysis. Statistical significance was assessed by Kruskal-Wallis test, one-way ANOVA with mixed-effects model, two-way ANOVA with full model and main model. Statistical analysis was performed for A*01/S865 (n = 7) and A*02/S269 (n = 8) longitudinally analyzed CD8+ T cell responses in Figs. 1a, b, 3c, 4a, b and Supplementary Figs. 2a, 5a–c, 7c–e for n = 28 subjects longitudinally followed in Fig. 2a, for A*01/S865 (n = 2) and A*02/S269 (n = 3) T cell responses longitudinally followed in Fig. 2c, for n = 26 subjects in Fig. 2b, for n = 6 prepandemic samples Supplementary Fig. 1c, for n = 2 subjects in Supplementary Fig. 3c, for n = 7 at 3 months after 2nd vaccination, n = 11 at 9 months after 2nd vaccination and n = 11 at 3 months after 3rd vaccination in Fig. 3a and Supplementary Fig. 4b, for n = 4 at 3 months after 2nd vaccination, n = 8 at 9 months after 2nd vaccination and n = 10 at 3 months after 3rd vaccination in Supplementary Fig. 4b, for A*01/S865 (n = 7) and A*02/S269 (n = 6) longitudinally analyzed CD8+ T cell responses in Fig. 3d, for n = 8 at 3 months after 2nd vaccination, n = 12 at 9 months after 2nd vaccination and n = 11 at 3 months after 3rd vaccination in Fig. 3b, for n = 4 in Supplementary Fig. 6a, for A*01/S865 (n = 2) and A*02/S269 (n = 2) longitudinally analyzed CD8+ T cell responses in Supplementary Fig. 6b, for n = 10 at 3 months after 2nd vaccination, n = 12 at 9 months after 2nd vaccination and n = 11 at 3 months after 3rd vaccination in Fig. 4c, for n = 10 at 3 months after 2nd vaccination, n = 11 at 9 months after 2nd vaccination and n = 11 at 3 months after 3rd vaccination in Fig. 4d, for n = 6 at 3 months after 2nd vaccination, n = 12 at 9 months after 2nd vaccination and n = 10 at 3 months after 3rd vaccination in Fig. 4e, for n = 6 at 3 months after 2nd vaccination, n = 12 at 9 months after 2nd vaccination and n = 11 at 3 months after 3rd vaccination in Fig. 4f, for Omicron infection n = 12, Delta infection n = 2 and 4th vaccination n = 5 longitudinally analyzed T-cell responses in Fig. 5a, for Omicron infection n = 11, Delta infection n = 2 and 4th vaccination n = 4 analyzed T cell responses in Fig. 5b and in peak response in Supplementary Fig. 8a, for Omicron infection n = 12, Delta infection n = 2 and 4th vaccination n = 3 longitudinally analyzed T cell responses in Fig. 6c, for Omicron infection n = 11, Delta infection n = 1 and 4th vaccination n = 3 in Fig. 6d, for Omicron infection n = 6, Delta infection n = 2 and 4th vaccination n = 4 analyzed T cell responses after 1 month in Supplementary Fig. 8a and Supplementary Fig. 9b, for Omicron infection n = 6, Delta infection n = 2 and 4th vaccination n = 4 analyzed T cell responses in Supplementary Fig. 9a.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank all donors for participating in the current study and FREEZE-biobank-Center for biobanking of the Freiburg University Medical Center and the Medical Faculty for support. We thank Dr. Sandra Ciesek and Dr. Marek Widera (Institute of Medical Virology, University Hospital Frankfurt am Main, Germany) for providing the B.1.1.529 BA.1 isolate. We thank Dr. Jonas Fuchs (Institute of Virology, University of Freiburg, Freiburg) for sequence confirmation of the SARS-CoV-2 stocks used in this study. This study was supported by grants from the German Research Foundation (272983813 to B.B., T.B., R.T., C.N.-H. and M.H., and; 256073931 to B.B., M.S., R.T., C.N.-H. and M.H.; 413517907 to H.L. and J.L.M). This work was also supported by the project “Virological and immunological determinants of COVID-19 pathogenesis – lessons to get prepared for future pandemics (KA1-Co-02 “COVIPA”)”, a grant from the Helmholtz Association’s Initiative and Networking Fund (to R.T. and M.H.). M.H. is furthermore supported by the Margarete von Wrangell fellowship (State of Baden-Wuerttemberg).
Source data
Author contributions
M.R., H.L., V.K., A.G., S.G., K.C. and D.R. planned, performed and analyzed experiments, interpreted data and contributed to writing the manuscript with the help of V.O., K.H., N.G. and C.B. M.R., H.L., V.K., A.G., S.G., K.C. and D.R. contributed equally to this work. H.L., A.G., N.R., J.L.-M., S.R., C.S., S.B. and D.S. were responsible for donor recruitment. F.E. performed four-digit HLA-typing by next generation sequencing. B.B. and T.B. contributed to data interpretation. M.S. and G.K. provided virological expertise, supervised virological work and interpreted data. R.T., C.N.-H. and M.H. designed and supervised the study, interpreted data, wrote the manuscript and contributed to experimental design and planning. M.H., C.N.-H. and R.T. are shared last authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The raw values for charts and graphs are available in the Source Data file whenever possible. All requests for additional raw (especially flow cytometry data) and materials are promptly reviewed by the University of Freiburg Center for Technology Transfer to verify if the request is subject to any intellectual property or confidentiality obligations. Donor-related data not included in the paper were generated as part of clinical examination and may be subject to donor confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. Source data are provided with this paper.
Code availability
R code to reproduce the analyses of multiparametric flow-cytometry data is available at https://github.com/sagar161286/SARSCoV2_specific_CD8_Tcells.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Matthias Reinscheid, Hendrik Luxenburger, Vivien Karl, Anne Graeser, Sebastian Giese, Kevin Ciminski, David Reeg.
These authors jointly supervised this work: Robert Thimme, Christoph Neumann-Haefelin, Maike Hofmann.
Contributor Information
Robert Thimme, Email: robert.thimme@uniklinik-freiburg.de.
Christoph Neumann-Haefelin, Email: christoph.neumann-haefelin@uniklinik-freiburg.de.
Maike Hofmann, Email: maike.hofmann@uniklinik-freiburg.de.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-022-32324-x.
References
- 1.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 2.Kalimuddin S, et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med (N. Y) 2021;2:682–688 e684. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberhardt V, et al. Rapid and stable mobilization of CD8+ T cells by SARS-CoV-2 mRNA vaccine. Nature. 2021;597:268–273. doi: 10.1038/s41586-021-03841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Painter MM, et al. Rapid induction of antigen-specific CD4+ T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142 e2133. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U, et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 6.Chemaitelly H, et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N. Engl. J. Med. 2021;385:e83. doi: 10.1056/NEJMoa2114114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldberg Y, et al. Waning immunity after the BNT162b2 vaccine in Israel. N. Engl. J. Med. 2021;385:e85. doi: 10.1056/NEJMoa2114228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin EG, et al. Waning immune humoral response to BNT162b2 COVID-19 vaccine over 6 months. N. Engl. J. Med. 2021;385:e84. doi: 10.1056/NEJMoa2114583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt F, et al. Plasma Neutralization of the SARS-CoV-2 Omicron Variant. N. Engl. J. Med. 2022;386:599–601. doi: 10.1056/NEJMc2119641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang-Meli J, et al. SARS-CoV-2-specific T-cell epitope repertoire in convalescent and mRNA-vaccinated individuals. Nat. Microbiol. 2022;7:675–679. doi: 10.1038/s41564-022-01106-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schulien I, et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 12.Akondy RS, et al. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 2017;552:362–367. doi: 10.1038/nature24633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Utzschneider DT, et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity. 2016;45:415–427. doi: 10.1016/j.immuni.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 14.Apostolidis SA, et al. Cellular and humoral immune responses following SARS-CoV-2 mRNA vaccination in patients with multiple sclerosis on anti-CD20 therapy. Nat. Med. 2021;27:1990–2001. doi: 10.1038/s41591-021-01507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan J, da Silva Antunes R, Grifoni A, Weiskopf D, Crotty S, Sette A. Observations and perspectives on adaptive immunity to SARS-CoV-2. Clin. Infect. Dis.20, ciac310 (2022). [DOI] [PMC free article] [PubMed]
- 16.Sette A, Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedzierska K, Thomas PG. Count on us: T cells in SARS-CoV-2 infection and vaccination. Cell Rep. Med. 2022;3:100562. doi: 10.1016/j.xcrm.2022.100562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goel RR, et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuertes Marraco SA, et al. Long-lasting stem cell-like memory CD8+ T cells with a naive-like profile upon yellow fever vaccination. Sci. Transl. Med. 2015;7:282ra248. doi: 10.1126/scitranslmed.aaa3700. [DOI] [PubMed] [Google Scholar]
- 20.Guerrera G, et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021;6:eabl5344. doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- 21.Minervina, A. A. et al. SARS-CoV-2 antigen exposure history shapes phenotypes and specificity of memory CD8(+) T cells. Nat. Immunol.23, 781–790 (2022). [DOI] [PMC free article] [PubMed]
- 22.Gao Y, et al. Ancestral SARS-CoV-2-specific T cells cross-recognize the Omicron variant. Nat. Med. 2022;28:472–476. doi: 10.1038/s41591-022-01700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keeton R, et al. T cell responses to SARS-CoV-2 spike cross-recognize Omicron. Nature. 2022;603:488–492. doi: 10.1038/s41586-022-04460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mateus J, et al. Low-dose mRNA-1273 COVID-19 vaccine generates durable memory enhanced by cross-reactive T cells. Science. 2021;374:eabj9853. doi: 10.1126/science.abj9853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skelly DT, et al. Two doses of SARS-CoV-2 vaccination induce robust immune responses to emerging SARS-CoV-2 variants of concern. Nat. Commun. 2021;12:5061. doi: 10.1038/s41467-021-25167-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization (WHO). Living guidance for clinical management of COVID-19. 23 November 2021
- 27.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alanio C, Lemaitre F, Law HK, Hasan M, Albert ML. Enumeration of human antigen-specific naive CD8+ T cells reveals conserved precursor frequencies. Blood. 2010;115:3718–3725. doi: 10.1182/blood-2009-10-251124. [DOI] [PubMed] [Google Scholar]
- 29.Wieland D, et al. TCF1(+) hepatitis C virus-specific CD8(+) T cells are maintained after cessation of chronic antigen stimulation. Nat. Commun. 2017;8:15050. doi: 10.1038/ncomms15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hensel N, et al. Memory-like HCV-specific CD8(+) T cells retain a molecular scar after cure of chronic HCV infection. Nat. Immunol. 2021;22:229–239. doi: 10.1038/s41590-020-00817-w. [DOI] [PubMed] [Google Scholar]
- 31.Crowell H, Zanotelli V, Chevrier S, Robinson M. CATALYST: Cytometry dATa anALYSis Tools. R package version 1.16.2. (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw values for charts and graphs are available in the Source Data file whenever possible. All requests for additional raw (especially flow cytometry data) and materials are promptly reviewed by the University of Freiburg Center for Technology Transfer to verify if the request is subject to any intellectual property or confidentiality obligations. Donor-related data not included in the paper were generated as part of clinical examination and may be subject to donor confidentiality. Any data and materials that can be shared will be released via a Material Transfer Agreement. Source data are provided with this paper.
R code to reproduce the analyses of multiparametric flow-cytometry data is available at https://github.com/sagar161286/SARSCoV2_specific_CD8_Tcells.