Abstract
The mechanism used by Escherichia coli to determine the correct site for cell division is unknown. In this report, we have attempted to distinguish between a model in which septal position is determined by the position of the nucleoids and a model in which septal position is predetermined by a mechanism that does not involve nucleoid position. To do this, filaments with extended nucleoid-free regions adjacent to the cell poles were produced by simultaneous inactivation of cell division and DNA replication. The positions of septa that formed within the nucleoid-free zones after division was allowed to resume were then analyzed. The results showed that septa were formed at a uniform distance from cell poles when division was restored, with no relation to the distance from the nearest nucleoid. In some cells, septa were formed directly over nucleoids. These results are inconsistent with models that invoke nucleoid positioning as the mechanism for determining the site of division site formation.
The process of cytokinesis in Escherichia coli and most other bacteria leads to formation of two identical daughter cells that each contain a fully replicated chromosomal complement. This process proceeds with high fidelity, so that cytokinesis is restricted to the proper site at midcell and occurs at the correct time in the cell cycle. In recent years, significant advances have been made in understanding the assembly of the components of the septal machinery (18). However, little is known about how the position of the division site is determined.
Two general models have been proposed to explain how the position of the division site is established. In the nucleoid occlusion model (15, 23), the site of septum formation is determined solely by the position of the nucleoids. According to this model, all positions along the length of the cell are competent to support septation. However, an inhibitory influence from the nucleoid prevents septation along most of the cell surface until segregation of the daughter chromosomes occurs. When the chromosomes move apart, the inhibitory effect would be gone at the middle of the cell, permitting septation to occur at midcell. In addition, it has been suggested that an activator of septation may be released upon termination of chromosome replication to provide both temporal and spatial control of septum formation (11, 15). Septation would occur at the position where the action of the activator overcame the inhibitory action of the nucleoid. The mechanism of site selection in bacteria would therefore bear some resemblance to division site selection in eucaryotes, where the location of the division site is markedly affected by the position of the mitotic spindle and chromosome segregation apparatus.
In the predetermined site model, the position of the division site is established by a mechanism that is independent of nucleoid position, and the future septation site can be identified and begin its differentiation before the time of chromosome segregation. This model is consistent with studies showing that FtsZ and ZipA rings are present at midcell early in the division cycle and are sometimes also present at the cell quarters (1, 8, 17). Similarly, the localized plasmolysis bays that act as landmarks for periseptal annuli are present at future division sites before chromosome segregation or septal invagination occurs and are present at cell quarters in predivisional cells (3).
We have used the following strategy to discriminate between the models. Strains were constructed in which a long nucleoid-free region was formed between the nucleoid and the cell pole. This was accomplished by using mutants in which chromosome replication and cell division were both inhibited. When the cell division block was released, septation occurred within the nucleoid-free zone. This permitted us to ask whether the positions of the division sites correlated with the position of the nearest nucleoid, as predicted by the nucleoid occlusion model, or whether the positions of the division sites were independent of nucleoid position, as predicted from the predetermined site model.
MATERIALS AND METHODS
Strains.
Strain details are summarized in Table 1. Strain WC1010 was constructed by P1-mediated transduction of zic501::Tn10 from JW355 into PC5. WC1013 was constructed by P1-mediated cotransduction of zic501::Tn10 and dnaA5(Ts) from WC1010 into PB103 (7). WC1016 was constructed by P1-mediated cotransduction of zic501::Tn10 and dnaA5(Ts) from WC1013 into WC1004, with selection for tetracycline-resistant colonies. The presence of the dnaA(Ts) allele was confirmed by demonstrating the presence of clustered nucleoids and long anucleate regions in filaments that were formed during growth at 42°C. Cells of WC1013 grown at 42°C did not form the long filaments seen in cultures of WC1004 and WC1016.
TABLE 1.
Genotypes and sources of the strains, plasmid, and phage used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| GC4540 | sfiA::Tn5 | R. D’Ari |
| JW355 | zic501::Tn10 | CGSCa |
| PB103 | Wild type | 7 |
| PC5 | dnaA5(Ts) | CGSC |
| RS162 | dnaB252 zjb504::Tn10 | CGSC |
| WC1004 | ftsA12(Ts) | 4 |
| WC1010 | zic501::Tn10 dnaA5(Ts) | This work |
| WC1013 | zic501::Tn10 dnaA5(Ts) | This work |
| WC1016 | ftsA12(Ts) zic501::Tn10 dnaA5(Ts) | This work |
| WC1092 | ftsZ null leu+ | This work |
| WC1097 | ftsZ null dnaB252 zjb504::Tn10 | This work |
| WC1113 | ftsZ null dnaB252 zjb504::Tn10 sfiA::Tn5 | This work |
| WX7 | ftsZ null leu::Tn10 | 21 |
| Plasmid pCX41 | ftsZ+ Cmr | 21 |
| Phage λGL100 | Plac-ftsZ+ Ampr | J. García-Lara |
CGSC, E. coli Genetic Stock Center.
WC1097(λGL100) was constructed by P1-mediated cotransduction of zjb504::Tn10 and dnaB252(Ts) into WC1092(λGL100), a leu+, tetracycline-sensitive derivative of WX7(λGL100). λGL100 and WX7(λGL100) were constructed by J. García-Lara in this laboratory. The presence of the dnaB(Ts) mutation in WC1097(λGL100) was confirmed by DAPI (4′,6-diamidino-2-phenylindole) staining as described above for WC1016. WC1113(λGL100) was constructed by P1-mediated transduction of sfiA::Tn5 from GC4540 into WC1097(λGL100), selecting for kanamycin resistance.
Growth conditions.
For experiments with WC1016 (ftsA12 dnaA5), cells were grown at 30°C in high-salt medium (3× concentrated M9 basal salts medium supplemented with 0.2% glucose and 1% Bacto Tryptone). This gave a doubling time of approximately 105 min. Overnight cultures were diluted into fresh medium, and exponentially growing cells were collected by centrifugation and suspended to an A600 of 0.05 in low-salt medium (1/5 concentrated M9 basal salts medium containing 1.6% glucose and 1% Bacto Tryptone) prewarmed to 42°C; low-salt medium was used to potentiate the division inhibition that occurs when ftsA12 cells are grown at a nonpermissive temperature. The culture was grown at 42°C with shaking for a total of three doubling periods (120 min). The culture was then diluted with an equal volume of 5.8-fold-concentrated M9 medium containing 1% Bacto Tryptone at room temperature and shifted back to the 30°C shaking water bath for a period of 100 min.
For experiments with WC1113(λGL100), cells were grown in Luria-Bertani (LB) broth containing isopropyl-β-d-thiogalactopyranoside (IPTG [300 μM]) at 30°C. Cells from exponentially growing cultures were collected by centrifugation, suspended to an A600 of 0.05 in LB broth supplemented with 2.5% glucose, and returned to the 30°C shaking water bath for 1 doubling period (85 min) to begin depletion of cellular FtsZ. The culture was then transferred to a 42°C shaking water bath for three doubling periods (78 min). Cells were collected by centrifugation and suspended in 1 volume of LB broth (prewarmed to 42°C) containing 600 μM IPTG and then were returned to the 42°C shaking water bath for a period of 30 min.
Microscopy.
Unless otherwise noted, 10-ml samples were removed from the culture and fixed by the addition of glutaraldehyde (2.3% final concentration). After 1 h at 4°C, the samples were centrifuged (2,800 × g, 5 min, 4°C), and the pellets were washed twice with 1 ml of phosphate-buffered saline (PBS) at 4°C and then suspended in 1 ml of PBS. DAPI was added at approximately 0.5 μg per 109 cells. Fixation with osmium tetroxide was performed by adding OsO4 (0.1% final concentration) to the samples in place of glutaraldehyde. For OsO4-fixed cells, DAPI was added to 5.0 μg per 109 cells in order to obtain good staining.
Samples were then examined by Nomarski and fluorescence microscopy (DAPI filter) and photographed with a charge-coupled device camera. Measurements and analysis of cell lengths and positions of septa and nucleoids were performed by using Optimas image analysis software as described previously (4). Nucleoid-to-pole and nucleoid-to-septum distances were measured from the edge of the nucleoid that was nearest to the pole, unless otherwise noted. The experimental reproducibility of measurements was ±0.06 μm (95% confidence limits).
RESULTS
Septum placement in nucleoid-free regions of FtsA− DnaA− filaments.
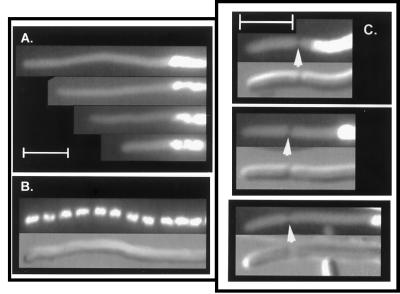
Strain WC1016 [dnaA5(Ts) ftsA12(Ts)] was grown for three generations at 42°C. Under these conditions, cell division was prevented because of the ftsA(Ts) mutation, leading to formation of nonseptate filaments. At the same time, ongoing rounds of chromosome replication were completed, but new rounds were not initiated because of the dnaA(Ts) mutation. As a result, the population consisted of long filaments in which one or more nucleoids were present in the interior of the filaments, and there were extended chromosome-free regions at the ends of the cells (Fig. 1A). The filaments differed from those of the ftsA(Ts) parent, in which chromosome replication was not inhibited and multiple nucleoids were distributed at regular intervals along the entire length of the filaments (Fig. 1B). The filaments also differed from those of the dnaA single mutant, in which long filaments were not formed because of residual divisions that produced anucleate cells (data not shown). It has previously been shown that division continues in dnaA mutants because of failure to induce the SOS response (12).
FIG. 1.
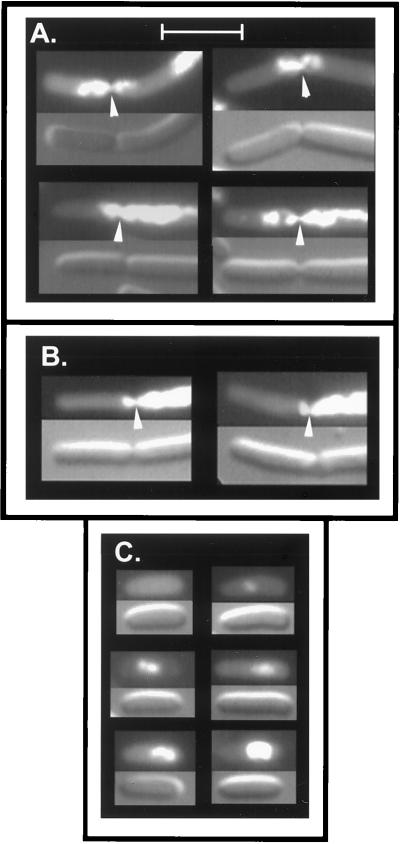
Comparison of ends of ftsA(Ts) dnaA(Ts) (A) and ftsA(Ts) (B) DAPI-stained filaments from strains WC1016 and WC1004 after growth at 42°C for three generations. (C) Cells of strain WC1016 were grown at 42°C for three generations and then shifted to 30°C for 100 min as described in Materials and Methods. The upper panel of each pair is the fluorescence micrograph, and the lower panel is the corresponding Nomarski image. Representative cells are shown. Arrowheads indicate septa. Scale bar, 5 μm.
When the ftsA(Ts) dnaA(Ts) double mutant was shifted back to 30°C after three generations of growth under nonpermissive conditions, septa began to form after approximately one generation, reflecting the restoration of FtsA activity. Septa were formed in the nucleoid-free regions at the cell poles (Fig. 1C) and elsewhere along the length of the filaments. In some cells, a new septum was formed directly over a nucleoid (discussed below). In the present study, we restricted the analysis to septation that occurred within the anucleate regions at the ends of the cells, since these provided the longest available nucleoid-free regions. The micrographs suggested that the septa were generally located at a similar distance from the cell pole, whereas there was a considerable variation in the distance from nucleoid to septum.
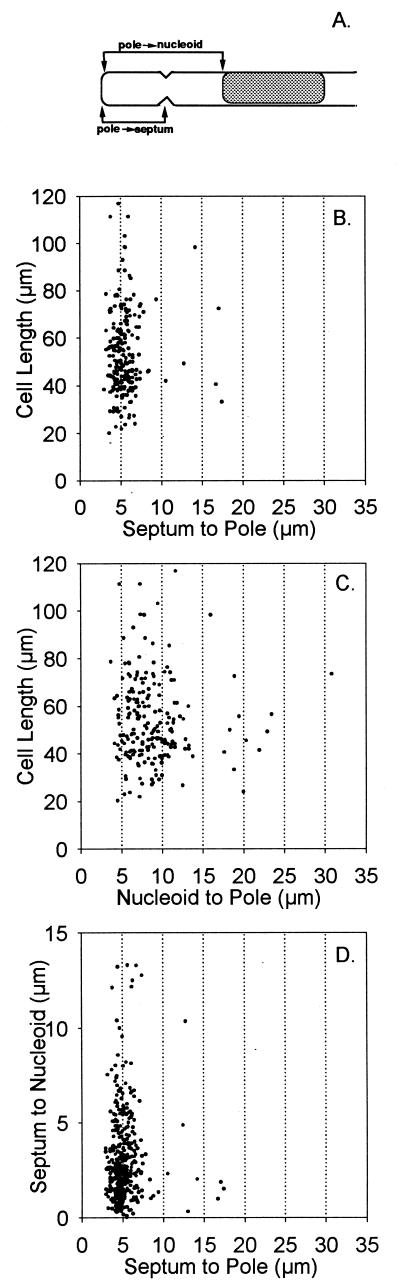
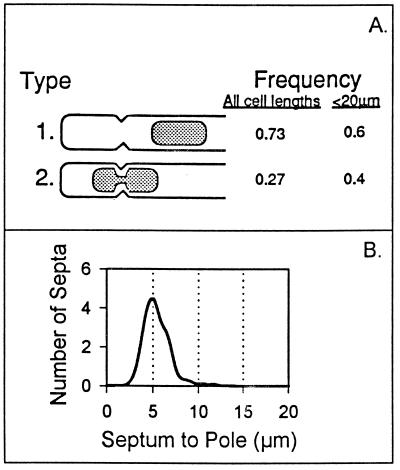
To confirm this impression and to determine whether nucleoid position played a role in division site selection, the positions of all septa located in the nucleoid-free regions that lay between the cell pole and the nucleoid were analyzed. The average filament length was 50.1 μm (range of 20 to 117 μm), with nucleoid-to-pole distances varying between 4 and 42 μm (Fig. 2C). As shown in Fig. 2B, septa were not randomly distributed, but were predominantly located approximately 5.0 μm from the end of the cell. As shown in Fig. 2D, the septum-to-pole distance was relatively constant and was unrelated to the septum-to-nucleoid distance, which varied over a wide range.
FIG. 2.
Positions of septa and nucleoids at ends of ftsA(Ts) dnaA(Ts) filaments. (A) Diagram showing landmarks. (B, C, and D) Cells that contained septa between the nucleoid and cell pole (illustrated in Fig. 1C) were analyzed for cell length and for septum-to-pole (B), nucleoid-to-pole (C), and septum-to-nucleoid (D) distances. A total of 434 filaments were analyzed.
Similar results were obtained when the measurements were made to the center of the nucleoid instead of its edge (data not shown). This makes it unlikely that the variability in the distance from septum to the edge of the nucleoid was due to variations in the extent of nucleoid shrinkage during the fixation process.
Effect of osmium tetroxide fixation.
Previous studies that led to the nucleoid occlusion hypothesis (15) were performed with cells that were fixed with osmium tetroxide instead of glutaraldehyde. To exclude the possibility that the present results were affected by glutaraldehyde fixation artifacts, the experiments were repeated by using the osmium tetroxide fixation method. Measurements of 180 filaments containing septa between nucleoids and cell poles were made. The results were similar to those described above for glutaraldehyde-fixed cells (data not shown). There was no significant difference in the average nucleoid-to-pole distance between the glutaraldehyde and osmium-fixed cells (P < 0.001). Septa were again clustered approximately 5 μm from the cell pole, whereas the nucleoid-to-septum distance varied over a wide range. These results indicate that although fixatives may differ in their effects on nucleoid organization (22), the effect on the parameters of the present study were negligible.
Septum placement in FtsZ− DnaB− SfiA− filaments.
Studies similar to those performed with the dnaA(Ts) ftsA(Ts) double mutant were also performed with strain WC1097(λGL100) [ftsZ null dnaB(Ts) sfiA null (Plac-ftsZ)]. DNA replication was interrupted by temperature upshift, which blocked elongation by inactivating the DnaB protein. Division was prevented by repression of Plac-ftsZ expression by growth in glucose. The presence of the sfiA null allele prevented SOS induction of the SfiA division inhibitor, so that division control was solely dependent on expression of ftsZ.
The division block was not as complete as with the ftsA(Ts) dnaA(Ts) strain described in the preceding section, as shown by the presence of anucleate cells in the culture. This presumably reflected the residual level of FtsZ in the glucose-repressed cells. The filaments in the culture resembled those of the ftsA(Ts) dnaA(Ts) filaments, with sizable nucleoid-free zones between the nucleoids and the ends of the cell.
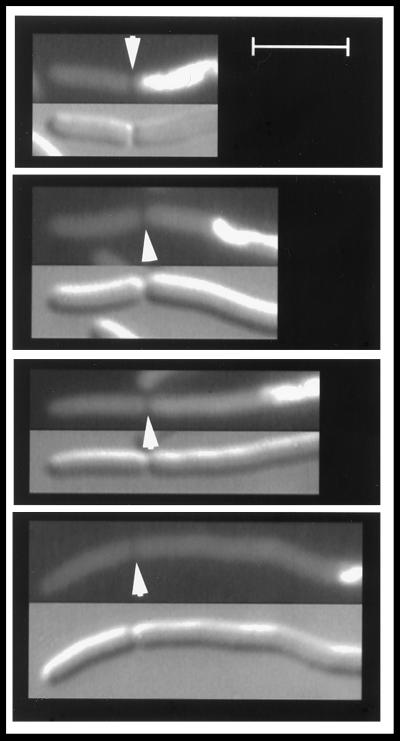
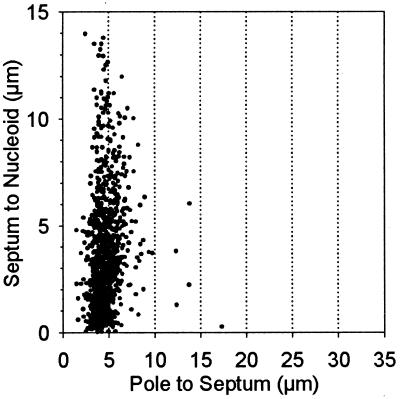
After three generations at 42°C, glucose was replaced by IPTG to induce ftsZ expression and thereby permit division to resume. Nucleoid-free progeny cells began to increase approximately 0.5 generation after addition of IPTG. DAPI-stained filaments that contained septa located within the polar nucleoid-free zones were analyzed for positions of septa and nucleoids, as had been done for the ftsA(Ts) dnaA(Ts) filaments. Examples are shown in Fig. 3. Within this population, the average filament length was 24.5 μm (range of 4.9 to 73.3 μm), with nucleoid-to-pole distances varying between 2 and 20.9 μm. Septa were primarily clustered at approximately 5 μm from the cell poles, whereas the septum-to-nucleoid distance varied over a wide range. The results (Fig. 4) were similar to those obtained with the ftsA(Ts) dnaA(Ts) filaments (Fig. 2).
FIG. 3.
Septation in nucleoid-free polar regions of ftsZ dnaB(Ts) filaments. Cells of strain WC1113(λGL100) were grown at 42°C for three generations in the presence of glucose, followed by 30 min in the presence of IPTG, and then fixed with glutaraldehyde and stained with DAPI as described in Materials and Methods. The upper panel of each pair is the fluorescence micrograph, and the lower panel is the corresponding Nomarski image. Representative cells are shown. Arrowheads indicate septa. Scale bar, 5 μm.
FIG. 4.
Relationship between septum-to-pole and septum-to-nucleoid distances in ftsZ dnaB(Ts) filaments. Cells of strain WC1113(λGL100) were grown and prepared as described in the legend to Fig. 3, and cells that contained septa between the nucleoid and cell pole (2,857 cells) were analyzed. See Fig. 3A for landmarks.
Formation of septa directly over nucleoids.
In addition to the large number of cells in which a septum was present between the edge of a nucleoid and the cell pole (described above), in some cells, a septum was formed directly over a nucleoid that was located close to a cell pole (Fig. 5). The placement of nucleoids at these positions appeared to result from residual division events that had led to formation of anucleate cells during the period of growth under nonpermissive conditions. Cells containing a septum directly over a polar nucleoid were more common in shorter filaments (Fig. 6A), where the nucleoid was more likely to extend close to the cell pole. They were more common in the ftsZ dnaB(Ts) population (Fig. 5A), in which the filaments were shorter due to a higher frequency of residual division during the period of repression of ftsZ expression, but also occurred in the ftsA(Ts) dnaA(Ts) filaments (Fig. 5B). The positioning of septa over nucleoids resulted in a guillotine effect that fragmented the chromosome and led to formation of partially anucleate daughter cells (Fig. 5C). Significantly, in cells in which septation occurred directly over the nucleoid, the positions of the septa were similar to those of the more numerous group in which septation occurred between the edge of the nucleoid and the cell pole (Fig. 6B). This provides additional evidence that selection of the septation site is not directed by the position of adjacent nucleoids.
FIG. 5.
Septation over nucleoids. Strains WC1113(λGL100) [ftsZ null dnaB(Ts) sfiA::Tn5 (Plac-ftsZ)] (A and C) and WC1016 [ftsA(Ts) dnaA(Ts)] (B) were grown and prepared as described in the legends to Fig. 2 and 3. (A and B) Septa that bisected nucleoids. (C) Cells that either lacked nucleoids or that contained nucleoid fragments of different sizes. Scale bar, 5 μm.
FIG. 6.
Relationship between septa and nucleoids and poles in ftsZ dnaB(Ts) filaments. Strain WC1113(λGL100) was grown and prepared as described in the legend to Fig. 3. (A) Cells containing septa that were located in the nucleoid-free region adjacent to a pole (type 1) or located over a nucleoid adjacent to a pole (type 2) were analyzed. Frequency represents the number of cells in each class/number of cells in both classes. Cells of all cell lengths were included in the analysis shown in the first column, whereas only cells with a length of <20 μm were included in the analysis shown in the second column. (B) Fifty-seven cells containing a septum located over a nucleoid (type 2 in Fig. 6A) were analyzed as described in the legend to Fig. 2.
DISCUSSION
Septal placement is independent of nucleoid position.
These results indicate that the position of the division site is determined by a mechanism that is independent of nucleoid position. In both systems that were used, new septa were formed at a relatively constant distance from the cell pole, despite the fact that a long and variable nucleoid-free region was available. Septa were not placed at a constant distance from the nucleoid, nor did septation occur at random positions within the nucleoid-free region, as might be expected if the role of the nucleoid were solely to inhibit formation of division sites in its proximity. Instead, the positions of the new septa were related to the position of the cell pole, leading to formation of newborn cells of relatively uniform length.
It has recently been observed that FtsZ rings are sometimes present at the midpoint of anucleate cells that are formed in mukB and parC mutants that are defective in chromosome organization and partition (19). Although consistent with the idea that division site placement is independent of nucleoid position, it cannot be determined from this result whether the FtsZ rings had been formed before or after the division event that led to release of the nucleoid-free daughter cell. In the former case, a relationship between FtsZ ring placement and the position of a neighboring nucleoid would still be possible. In the present study, this ambiguity was resolved by examining the relative positions of nucleoid and division site within the predivisional cell.
In a significant number of cells in the present study, septation occurred directly over a nucleoid. This resulted in a guillotine effect, leading to formation of cells that contained incomplete nucleoids. The guillotine phenomenon has also been described in mukB (9) and smc (13) mutants that are defective in chromosome separation after replication has been completed. The partition defect in these cells presumably is secondary to defects in the postreplication condensation of daughter chromosomes (10, 13). The present study extends these observations by showing that daughter chromosomes can be transected by the ingrowing septum even when termination of chromosome replication is blocked (in dnaB ftsZ cells) or when chromosome replication has been completed and reinitiation does not occur (in dnaA ftsA cells). This excludes the possibility that termination or initiation of replication is required to release a local chromosome occlusion effect caused by unreplicated or partially replicated chromosomes.
Although nucleoid position is clearly not the primary determinant of division site placement, abnormalities in chromosome organization can affect septal position. Thus, Sun et al. have observed that FtsZ rings are often displaced in mukB cells and in mutants defective in the ParC subunit of topoisomerase IV (19), and some DNA gyrase mutations are also associated with abnormalities in cell length distribution patterns (16). Because of the pleiotropic effects in these cases, it is not known whether the division site effects are direct or indirect results of the perturbations in chromosome organization.
It has been suggested that the formation of minicells in min mutants is due to a defect in chromosome segregation or organization that leads to a nucleoid-free zone near the cell poles (14). An increased distance between the edges of the nucleoids and the cell poles has been described in these mutants (2), and it was proposed that this permits minicell septa to form because of the presence of the polar nucleoid-free zones, as predicted by the nucleoid occlusion model. This view is contrary to the opposing view that the polar septation events that give rise to minicells reflect the use of residual division sites at the poles that were derived from previous division events (6, 20). In this regard, it may be relevant that, in the present study, there was no evidence of minicell septa (<10−4 cells) despite the presence of large nucleoid-free regions adjacent to the poles in most cells. Thus, something in addition to nucleoid-free polar zones is required for activation of polar minicell-producing septation events.
The septa that were formed when division was permitted to resume in the present study were placed approximately 5 μm from the pole. This is longer than the pole-to-septum distance in wild-type cells grown at 30 or 42°C (unpublished observations). The reason for this difference is unknown, although there were a number of elements in the present experiments that could have affected the mechanism that determines the length of newborn cells.
In a previous study, it was observed that the septum-to-nucleoid distance was relatively constant after dnaA(Ts) cells were returned to a permissive temperature (15). In contrast, in the present study of a dnaA(Ts) ftsA(Ts) strain, the septum-to-nucleoid distance varied over a wide range (Fig. 4). In both studies, the septum-to-pole positions were nonrandom, clustering at approximately 4.5 to 5 μm from the cell pole. In addition to the prolonged 5-h period at 42°C and the different dnaA allele used in the prior study, a difference between the two studies was the imposition of a division block during the period of inhibition of DNA synthesis in the present work. As a result, there was a much longer nucleoid-free space at the cell poles in which the septum could have formed in the present work (nucleoid-free space, 3 to 31 μm) than in the previous study (nucleoid-free space, 3 to 8 μm). This made it relatively easy to discriminate between fixed and random septum-to-nucleoid distribution patterns, thereby permitting a more rigorous test of the hypothesis that septum formation occurs at a fixed distance from the nucleoid.
Mechanism of division site identification.
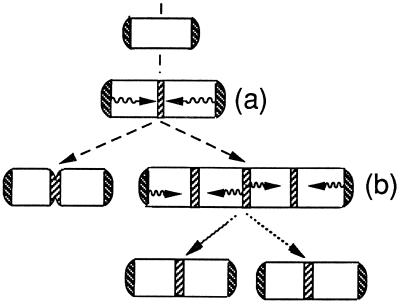
If the nucleoids do not provide the positional information that is needed to establish the division site at its correct location, what other landmarks could be used for this purpose? The cell poles are obvious candidates, since division normally occurs at midcell, equidistant from the two poles. This suggests a model in which the two cell poles act cooperatively to determine the placement of the next division site. This could occur, for example, if each cell pole periodically elaborated an electrical or chemical signal that propagated along the membrane or within the cytosol. Division site differentiation would be triggered where the signals met at midcell [Fig. 7(a)]. Alternatively, if the signal were an inhibitor that prevented division site formation, site differentiation would be restricted to midcell, where the concentration of the inhibitor would fall below a threshold level when the cells achieved a certain cell length. Since the cell poles are derived from division sites that had been located at midcell during preceding division cycles, the division site-identification property of the poles would likely be inherited as part of the old division site.
However, although several possible mechanisms might be used by the poles to identify the midcell site for the next division event, any model must accommodate the fact that the FtsZ-ZipA ring (8, 17) and the plasmolysis bays that also act as markers of future division sites (5) can be present at 1/4 and 3/4 cell lengths of predivisional cells prior to the onset of septation at midcell. In this case, we suggest that the potential division site that is present at midcell prior to septation has matured to the stage at which it acts as a “virtual pole,” working cooperatively with the true cell poles to trigger differentiation of the potential division sites at 1/4 and 3/4 cell lengths [Fig. 7(b)]. The subsequent division event would generate newborn cells with potential division sites already in place at midcell.
FIG. 7.
Model for determination of division site position. The diagram illustrates how the position of future division sites might be directed by a signal that is periodically elaborated from the cell poles, leading to establishment of a site at midcell (a), or from both the cell poles and nascent division sites at midcell to establish new sites at the cell quarters (b). In the latter case, the sites at 1/4 and 3/4 cell length are retained at the midpoint of the daughter cells to support septum formation during the next cell cycle (3). See text for further details.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (GM53276) and the Human Frontiers in Science Program (RG-386/95).
We thank Jorge García-Lara for providing unpublished work and for helpful discussions.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Åkerlund T, Bernander R, Nordström K. Cell division in Escherichia coli minB mutants. Mol Microbiol. 1992;6:2073–2083. doi: 10.1111/j.1365-2958.1992.tb01380.x. [DOI] [PubMed] [Google Scholar]
- 3.Cook W, Rothfield L. Early stages in development of the E. coli division site. Mol Microbiol. 1994;14:485–495. doi: 10.1111/j.1365-2958.1994.tb02183.x. [DOI] [PubMed] [Google Scholar]
- 4.Cook W, Rothfield L I. Development of potential division sites in FtsA− filaments of E. coli. Mol Microbiol. 1994;14:497–503. doi: 10.1111/j.1365-2958.1994.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 5.Cook W R, Kepes F, Joseleau-Petit D, MacAlister T J, Rothfield L I. A proposed mechanism for the generation and localisation of new division sites during the division cycle of Escherichia coli. Proc Natl Acad Sci USA. 1987;84:7144–7148. doi: 10.1073/pnas.84.20.7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Boer P A J, Cook W R, Rothfield L I. Bacterial cell division. Annu Rev Genet. 1990;24:249–274. doi: 10.1146/annurev.ge.24.120190.001341. [DOI] [PubMed] [Google Scholar]
- 7.de Boer P A J, Crossley R E, Rothfield L I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988;170:2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hale C, de Boer P. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 9.Hiraga S, Niki H, Ogura T, Ichinose C, Mori H, Ezaki B, Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989;171:1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu K, Liu E, Dean K, Gingras M, DeGraff W, Trun N. Overproduction of three genes leads to camphor resistance and chromosome condensation in Escherichia coli. Genetics. 1996;143:1521–1532. doi: 10.1093/genetics/143.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones N, Donachie W. Chromosome replication, transcription and cell division in Escherichia coli. Nature. 1973;243:100–103. [PubMed] [Google Scholar]
- 12.Monk M, Gross J D. Induction of a prophage in a mutant of E. coli K-12 defective in initiation of DNA replication at high temperature. Mol Gen Genet. 1971;110:299–306. doi: 10.1007/BF00438272. [DOI] [PubMed] [Google Scholar]
- 13.Moriya S, Tsujikawa E, Hassan A, Asai K, Kodama T, Ogasawara N. A Bacillus subtilis gene encoding a protein homologous to eukaryotic SMC proteins is necessary for chromosome partition. Mol Microbiol. 1998;29:179–187. doi: 10.1046/j.1365-2958.1998.00919.x. [DOI] [PubMed] [Google Scholar]
- 14.Mulder E, El’Bouhali M, Pas E, Woldringh C L. The Escherichia coli minB mutation resembles gyrB in defective nucleoid segregation and decreased negative supercoiling of plasmids. Mol Gen Genet. 1990;221:87–93. doi: 10.1007/BF00280372. [DOI] [PubMed] [Google Scholar]
- 15.Mulder E, Woldringh C L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989;171:4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orr E, Fairweather N, Holland I B, Pritchard R. Isolation and characterization of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177:103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- 17.Pogliano J, Pogliano K, Weiss D, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 19.Sun Q, Yu X-C, Margolin W. Assembly of the FtsZ ring at the central division site in the absence of the chromosome. Mol Microbiol. 1998;29:491–504. doi: 10.1046/j.1365-2958.1998.00942.x. [DOI] [PubMed] [Google Scholar]
- 20.Teather R M, Collins J F, Donachie W D. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974;118:407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X, de Boer P A J, Rothfield L I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991;10:3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woldringh C, Nanninga N. Structure of nucleoid and cytoplasm in the intact cell. In: Nanninga N, editor. Molecular cytology of Escherichia coli. London, United Kingdom: Academic Press; 1985. pp. 161–197. [Google Scholar]
- 23.Woldringh C L, Mulder E, Huls P G, Vischer N O E. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991;142:309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]