Abstract
Objective
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2) encodes a type II membrane protein that is thought to catalyze the transfer of sialic acid (SA) from CMP-SA to N-linked oligosaccharides and glycoproteins. Some population and animal studies have indicated an association between the ST8SIA2 gene and autism spectrum disorder (ASD). However, there is limited information on the correlation between ST8SIA2 and autistic behavioral symptoms.
Methods
In this study, 69 ASD and 76 normal control children who were age- and sex-matched were recruited. ST8SIA2 expression and methylation levels were measured by reverse transcription quantitative real-time PCR and pyrosequencing, respectively, and the behavioral phenotypes of ASD children were assessed.
Results
The ASD group had lower ST8SIA2 gene expression levels than the control group [t(0.05/2,143) = 2.582, p = 0.011]. Moreover, ST8SIA2 expression levels were positively correlated with daily life skills (rs = 0.381, p = 0.008) and negatively associated with stereotyped behaviors in the ASD group (rs = -0.510, p = 0.004). The methylation levels of the Chr. 15: 92984625 and Chr. 15: 92998561 sites of the ST8SIA2 gene in ASD children were higher than those of controls. The Chr. 15: 92984625 site was positively correlated with the stereotyped behaviors of ASD children (rs = 0.41, p = 0.039).
Conclusion
This study provides a scientific basis to elucidate the relationship between the ST8SIA2 gene and behavioral phenotypes of ASD.
Keywords: autism spectrum disorders, ST8SIA2 gene, DNA methylation, behavioral phenotypes, pyrosequencing
Introduction
Autism spectrum disorder (ASD) is an early onset developmental disorder characterized by deficits in communication and social interaction and restrictive or repetitive behaviors (Mitra et al., 2021). Although the etiology of ASD has not been fully elucidated, its occurrence is strongly associated with genetics and epigenetics (Ramaswami et al., 2020). Furthermore, patients with ASD have significant phenotypic and genetic heterogeneity, which poses a great challenge for etiological research and rehabilitation (Tordjman et al., 2018).
Glycosylation is an important modification of proteins and lipids that can regulate their function (Reily et al., 2019). More than 50% of proteins are post-translationally modified with glycans, and these modified proteins are involved in the occurrence of disease (Sato and Kitajima, 2021). Additionally, glycoproteins are key components of the neural extracellular matrix, so they participate in nearly every biological process in the developing brain (Dwyer and Esko, 2016). However, as a neurodevelopmental disease, the association of ASD with glycosylation is rarely reported. Clinical studies have found that patients with congenital glycosylation disorders exhibit ASD-like behavioral phenotypes. Thus, it will be of great significance to explore the relationship between the glycosylation process of the nervous system and ASD.
Sialic acid (SA) is an important monosaccharide unit of ganglioside and glycoprotein in the brain and an essential nutrient for brain development and cognition (Wang, 2009). SA often forms a polymer chain and adds to nerve cell adhesion molecule (NCAM) by glycosylation, which is involved in the development and plasticity of synapses and participates in the formation of neural networks. In the SA glycosylation process, polysialyltransferase [ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2)] plays an important role. In our present research, we found that SA was decreased in ASD children and related to autistic behaviors (Yang et al., 2018). Moreover, the NCAM levels were low in ASD children. However, the reason for this finding has not been clarified. Therefore, we hypothesize that the ST8SIA2 gene may play a significant role in resolving these questions. One study reported a patient with a behavioral disorder and ASD, who had a 520-kb chromosomal deletion at 15q26.1 encompassing the ST8SIA2 gene (Kamien et al., 2014). In animal model research, St8sia2 KO mice displayed decreased social motivational behavior, which is the core symptom of ASD (Calandreau et al., 2010). A single nucleotide polymorphism study showed the association between St8sia2 and ASD (Hane et al., 2016). Nevertheless, the relationship between the ST8SIA2 gene and ASD behavioral phenotypes has not been well illuminated. The present study was designed to better understand the expression of the ST8SIA2 gene, and its methylation levels at the target loci, and the association with behavioral performance in ASD children. It will provide a novel view for comprehending symptoms of ASD.
Material and methods
Patients and evaluation of behavioral symptoms
We investigated 69 children with ASD and 76 age- and sex-matched typically developing children (age 2–6 years). All participants and controls were Han Chinese. The ASD children were diagnosed by two psychiatrists based on the criteria of the Diagnostic and Statistical Manual of Mental Disorders-Fifth Edition and the combined results of the Autism Diagnostic Interview–Revised (ADI-R) and Autism Diagnostic Observation Schedule. The exclusion criteria were children with genetic disorders, attention deficit hyperactivity disorder, tic disorders, and mental retardation. The control children were recruited from kindergartens, agreed to mental and neurological examinations, and did not exhibit any developmental or nervous system diseases. This study was approved by the Ethical Committee of Qiqihar Medical University (No. 201920) in 2019. In addition, an informed written consent form for participation in the study was signed by the parents or legal guardians of all study subjects.
The ASD children completed assessments of intellectual, social, and autistic behavior problems using the Peabody Picture Vocabulary Test (PPVT), the Autism Behavior Checklist (ABC), the Childhood Autism Rating Scale (CARS), the Vineland Adaptive Behavior Scale (VABS), the Social Responsiveness Scale (SRS), and the Infant-Junior Middle School Student’s Ability of Social Life Scale. The ABC contains five sub-scales (sensory, communication, language, social, and self-care skills), which are used to evaluate the severity of autistic symptoms and completed by the parents (Krug et al., 1980). The CARS is a commonly used tool for diagnosing ASD and its score of 30–36 is considered mild to moderate autism and 37–60 is severe autism (Meng et al., 2017). The PPVT scale examines children’s receptive vocabulary ability to evaluate the intellectual development of children (Weber et al., 2015). The SRS is a brief screening questionnaire for evaluating the severity of social skill deficits and describing other core features of ASD, which have been validated and shown to be reliable and to have good correspondence to the gold-standard ADI-R (Frye and Rossignol, 2016). The VABS is used for examining autistic core behaviors with a focus on communication and adaptive behaviors (Mandic-Maravic et al., 2015). The Infant-Junior Middle School Students Social-Life Abilities Scale is an adaptive behavioral scale, including self-help, locomotion, occupation, communication, socialization, and self-direction items. There are 132 items in this scale, and the child gets one point for each item for a total possible score of 132. The raw scores can be transformed into a standard score that is adjusted for age (Yuan et al., 2018). All behavioral assessment scales for ASD children were completed under professional face to face guidance.
Measurement of ST8SIA2 gene expression levels
Blood samples were obtained between 8:30 a.m. and 9:30 a.m. in the morning, and RNA and DNA were immediately extracted from fresh blood. Total RNA was extracted from fresh blood of the children with ASD and healthy controls using the RNAprep pure Blood Kit (TIANGEN BIOTECH (BEIJING) CO., LTD). A NanoDrop 2000 was used to test the concentration and purity of RNA. Subsequently, cDNA was reverse transcribed from RNA samples using the PrimeScript® RT reagent Kit with gDNA Eraser according to the manufacturer’s instructions (TaKaRa Bio). The reverse transcription quantitative real-time PCR (RT–qPCR) was subsequently performed using the ABI-7500 System with the SYBR® Select Master Mix (Applied Biosystems, Life-Technologies). The specific RT–qPCR primers for ST8SIA2 were synthesized by Sangon Biotech (Shanghai) Co., Ltd. The primer sequences were as follows:
| ST8SIA2 | forward:5′-TCCTGAAGCACCACGTCAAC-3 |
| reverse: 5′-TACATCAAGAGGCCGGTGGT-3′ | |
| HS-ACTB | forward: 5′-CCTGGCACCCAGCACAAT-3 |
| reverse: 5′-GGGCCGGACTCGTCATAC-3′ |
The selection of ST8SIA2 gene methylation sites
In our earlier research, a genome-wide DNA methylation analysis was performed in five pairs of ASD-discordant monozygotic twins. Subsequently, we mapped different DNA methylation sites of five pairs of ASD-discordant monozygotic twins in the gene network and obtained 4,057 differentially modified genes (Liang et al., 2019). Interestingly, ST8SIA2 was present in the 4,057 intersecting genes. Then, we screened the target sites of the ST8SIA2 gene using the difference method (calculated Δβ) and the fold-change method. The parameters were set to fold-change ≥ 2 or ≤0.5 and | Δβ| ≥ 0.1 as cutoffs. The target sites included Chr. 15: 92943919, Chr. 15: 92943938, Chr. 15: 92944418, Chr. 15: 92944439, Chr. 15: 92944460, Chr. 15: 92984625, Chr. 15: 92998561, Chr. 15: 92937874, and Chr. 15: 92957106. All of the above sites are located in the intronic region of the ST8SIA2 gene.
Methylation pyrosequencing
DNA was extracted from peripheral blood using QIAGEN kits (QIAampDNA Blood Mini Kit Q-51106). The DNA concentrations were determined using an ultramicro nucleic acid ultraviolet tester (Nanodrop 2000; Nanodrop Technologies; Thermo Fisher Scientific, Inc.). A sulfite reagent was used to modify the DNA, and the modified DNA was purified. Then, PCR was completed. The PCR conditions were set as follows: 95°C for 3 min; 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 1 min; and 72°C for 7 min. To detect the ST8SIA2 methylation levels, pyrophosphate sequencing analysis was performed using a Pyromark Q96 instrument. The PCR primers for methylation quantification are shown as follows:
| Primer | Base sequence (5′ to 3′) | Biotin |
| ST8SIA2-1F (150 bp) | ATGAGTTAGAGATAGTTGGGAGGATA | |
| ST8SIA2-1R | TATCTCACCCCACAATACCTCTCC | 5′-biotin |
| ST8SIA2-1S | TGGTTATTTTTTGTTAGGG | |
| ST8SIA2-2F (111 bp) | GAAATAGTATGAAGAGGGGTGTGGA | |
| ST8SIA2-2R | CCCCTCAAACATTCCCAACAACA | 5′-biotin |
| ST8SIA2-2S | AAGGTAGATGTATGGATT | |
| ST8SIA2-3F (130 bp) | GAGAAAAGGAGGGAAGTTTGTGATATTAAG | |
| ST8SIA2-3R | ATCCAATTTAAATATCTTTTCACTATCTAC | 5′-biotin |
| ST8SIA2-3S | ATTAAGGTATAGGTGGTTT |
Statistical analysis
All data were analyzed using SPSS 21.0 (SPSS Inc., Chicago, IL, United States). For the descriptive data, we calculated the means, medians, standard deviations, and interquartile ranges of demographic and outcome variables (independent t-test, paired t-test, and one-way ANOVA). The chi-square test was used to determine differences in the distribution of categorical variables in different groups. Methylation levels were analyzed using the paired t-test. The correlation among the ST8SIA2 expression levels, methylation level, and behavioral phenotype of ASD children was determined by Pearson’s or Spearman’s correlation analysis. For all analyses, significance was set at a p-value of 0.05.
Results
Patient characteristics
We investigated 69 children with ASD (14 girls, 55 boys, age 4.47 ± 1.23 years) and 76 age- and sex-matched typically developing children (16 girls, 60 boys, age 4.59 ± 1.19 years). All participants and controls were Han Chinese. There was no significant difference in sex or age between the two groups [sex: x2 = 0.013, p = 0.91; age: t(0.05/2,143) = 0.549, p = 0.584].
ST8SIA2 gene expression levels in the autism spectrum disorder and control groups
As shown in Figure 1, there were significant differences in ST8SIA2 gene expression levels between the ASD and control groups. Moreover, the level of the ST8SIA2 gene was lower in the ASD group than in the control group [ASD and control: 0.97 ± 0.64 and 1.28 ± 0.79, t(0.05/2,143) = 2.582, p = 0.011] (Figure 1). Despite the diverse occurrence of ASD between sexes, there was no statistical difference in the expression of ST8SIA2 between the sexes by two-way ANOVA analysis [F(1,142) = 1.625, p = 0.204]. Additionally, the expression of ST8SIA2 was time-dependent, but there was no statistical difference among different ages through two-way ANOVA analysis [F(4,135) = 1.259, p = 0.289].
FIGURE 1.
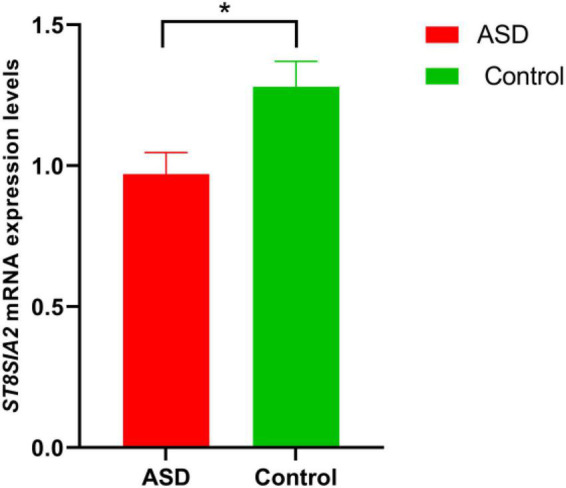
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2) mRNA levels in the autism spectrum disorder (ASD) and control groups (NASD = 69, Ncontrol = 76). Data represented as means ± SEM. *p < 0.05 from the two dependent t-tests.
Relationship between ST8SIA2 gene expression levels and autism spectrum disorder severity in children
As noted in Table 1, based on the scores of the CARS scale and levels of ST8SIA2 in ASD children, the difference in ST8SIA2 levels in different severities was statistically significant. Additionally, no significant difference in ST8SIA2 levels was noted in children with different intelligence quotients of ASD.
TABLE 1.
Relationship between the severity and intelligence development of autistic children and the ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ST8SIA2) mRNA levels.
| Item | N | ST8SIA2 (Mean ± SD) | F | p |
| ABC scores | ||||
| ≤53 | 19 | 1.21 ± 0.91 | ||
| 54–66 | 21 | 0.79 ± 0.42 | 2.198 | 0.119 |
| ≥67 | 26 | 0.96 ± 0.53 | ||
| CARS scores | ||||
| <30 | 13 | 1.29 ± 0.77 | ||
| 30–36 | 26 | 1.04 ± 0.73 | 4.166 | 0.020 |
| 37–60 | 29 | 0.73 ± 0.37 | ||
| PPVT scores | ||||
| <70 | 36 | 0.89 ± 0.63 | ||
| 70–85 | 18 | 0.99 ± 0.82 | 0.424 | 0.656 |
| >85 | 12 | 1.08 ± 0.41 | ||
Three children did not complete the ABC scale, one child did not complete the CARS scale, and three children did not complete the PPVT scale.
The bold values represented statistical significance (p < 0.05).
Relationship between ST8SIA2 levels and behavior problems in autism spectrum disorder children
There were 48 autistic children who finished VABS and SRS scales. A positive correlation was noted between the daily life skill score of the VABS scale and ST8SIA2 levels in the evaluation of ASD children’s behavior problems (rs = 0.381, p = 0.008). This finding suggests that a higher expression level of the ST8SIA2 gene corresponds to better daily life skills. However, there were no associations between the level of ST8SIA2 and the SRS scores in the ASD group (Table 2). In the assessment of the CARS scale, the stereotype and sensory abnormality scores were negatively correlated with the ST8SIA2 gene expression (rs = -0.510, p = 0.004). Thus, a lower level of the ST8SIA2 expression indicates more serious stereotype behaviors in the ASD children. According to the results of the Infant-Junior Middle School Students Social-Life Abilities Scale in 30 ASD children, a positive correlation was noted between ST8SIA2 and the scores of self-help ability (r = 0.462, p = 0.021; as shown in Figure 2). These results indicate that a lower ST8SIA2 expression level corresponds to worse self-help ability.
TABLE 2.
Relationship between the ST8SIA2 level and the adaptive behavior and social behavior in children with autism spectrum disorder (ASD).
| Item | Scores (Mean ± SD)/M (P25–P75) | r/rs | p |
| VABS total scores | 65 (58–72.75) | 0.185 | 0.209 |
| Communication | 75.29 ± 23.08 | 0.103 | 0.486 |
| Daily living skills | 64 (61–76) | 0.381 | 0.008 |
| Socialization | 58 (56–62.75) | 0.174 | 0.238 |
| Motor skills | 91.05 ± 22.75 | 0.078 | 0.598 |
| SRS total scores | 89.02 ± 23.65 | −0.033 | 0.861 |
| Social awareness | 10.80 ± 3.71 | −0.053 | 0.781 |
| Social cognition | 18.83 ± 4.87 | −0.266 | 0.155 |
| Social communication | 32.36 ± 8.11 | 0.035 | 0.856 |
| Social motivation | 14.85 ± 5.41 | 0.122 | 0.552 |
| Autistic mannerisms | 12.22 ± 6.69 | −0.007 | 0.970 |
The bold values represented statistical significance (p < 0.05).
FIGURE 2.
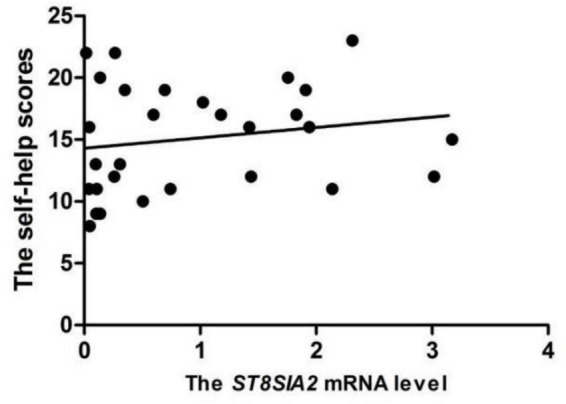
Correlation between ST8SIA2 levels and self-help scores of Infant-Junior Middle School Students Social-Life Abilities Scale (NASD = 30, r = 0.462, p = 0.021 from Pearson’s correlation analysis).
Methylation levels of target sites in the ST8SIA2 gene
We selected 30 ASD-control children pairs (age difference not exceeding 3 months in every pair and sex-matched, age from 2–6 years, 26 male pairs and 4 female pairs) which were from the population of ST8SIA2 mRNA expression test, and examined methylation levels at ST8SIA2 gene sites. According to paired t-test result, the methylation levels of Chr. 15: 92984625 and Chr. 15:92998561 sites were significantly different between the ASD and control groups. The methylation levels in ASD children at these sites were significantly greater than those in control children [Chr. 15: 92984625: t(0.05/2,29) = 3.81, p = 0.001; Chr. 15:92998561: t(0.05/2,29) = 5.16, p < 0.001]. In addition, the methylation levels of Chr. 15: 92984630, Chr. 15: 92944439, and Chr. 15: 92944460 sites were higher than those in the control group, but the difference was not statistically significant (Table 3).
TABLE 3.
Methylation level of ASD and controls at the detected site in ST8SIA2 (mean ± SD).
| Detected sit | ASD (%, n = 30) | Control (%, n = 30) | t | P |
| Chr. 15: 92984625 | 96.18 ± 048 | 95.41 ± 0.88 | 3.810 | 0.001 |
| Chr. 15: 92984630 | 99.44 ± 1.04 | 99.11 ± 1.40 | 1.010 | 0.319 |
| Chr. 15: 92984638 | 84.61 ± 4.30 | 84.77 ± 3.56 | 0.743 | 0.464 |
| Chr. 15: 92944439 | 52.64 ± 3.06 | 52.55 ± 2.39 | 0.135 | 0.894 |
| Chr. 15: 92944460 | 100 | 99.83 ± 0.69 | 1.343 | 0.190 |
| Chr. 15: 92998561 | 99.51 ± 0.81 | 98.13 ± 1.29 | 5.160 | <0.001 |
The bold values represented statistical significance (p < 0.05).
Association between methylation levels at ST8SIA2 gene sites and behavioral phenotypes in autism spectrum disorder children
Based on the above results, we analyzed the relationship between two significantly different methylation sites and behavioral problems in ASD children. It showed that no correlation between the Chr. 15:92998561 methylation level and the autism severity by comparing the case and control groups (r = -0.133, p = 0.484). There was no significant difference in the methylation levels of Chr. 15: 92998561 in ASD children with different levels of intellectual development [F(2,27) = 1.211, p = 0.313]. In addition, no significant difference was found between the methylation levels of Chr. 15: 92984625 and the intellectual development levels of ASD children [F(2,27) = 0.086, p = 0.917].
In addition, this study analyzed the relationship between the scores of communication skills, daily living skills, socialization, and motor skills in ASD children’s VABS scale and the methylation levels of Chr. 15: 92998561 and Chr. 15: 92984625. However, no statistically significant correlation was noted between them [Chr. 15: 92984625: r(total scores) = -0.097, p = 0.611; r(communication skills) = -0.021, p = 0.913; r(daily living skills) = -0.258, p = 0.168; r(socialization) = 0.044, p = 0.818; r(motor skills) = 0.023, p = 0.904; Chr. 15: 92998561: r(total scores) = 0.059, p = 0.758; r(communication skills) = 0.011, p = 0.954; r(daily living skills) = 0.102, p = 0.591; r(socialization) = 0.057, p = 0.765; r(motor skills) = 0.111, p = 0.56]. In addition, the methylation levels of Chr. 15: 92998561 and Chr. 15: 92984625 were not associated with the scores of the SRS [r(Chr. 15: 92998561) = -0.105, p = 0.586; r(Chr. 15: 92984625) = 0.019, p = 0.921].
In the Spearman correlation analysis, the results indicate that the methylation levels at the Chr. 15: 92984625 site were positively correlated with the stereotyped behavior scores of the ADI-R scale (rs = 0.41, p = 0.039). Thus, a greater methylation level of this site corresponds to a more serious stereotyped behavior. Furthermore, the methylation levels of the Chr. 15: 92984630 site were positively correlated with the scores of independent living ability and operational ability in the Infant-Junior Middle School Student’s Ability of Social Life Scale (rs = 0.478, p = 0.014; rs = 0.424, p = 0.031).
Discussion
In this study, we explored ST8SIA2 gene expression and methylation levels between the ASD and control groups and analyzed the relationship between ST8SIA2 and the behavioral phenotypes of ASD children. ST8SIA2 gene expression levels in the ASD group were lower than those in the control group. Moreover, these levels were associated with the severity, daily life skills, stereotype abnormality, sensory abnormalities, and self-help ability of ASD children. The ASD children had greater methylation levels of Chr. 15: 92984625 and Chr. 15: 92998561 sites than controls. Furthermore, Chr. 15: 92984625 was positively correlated with the stereotyped behavior scores on the ADI-R scale.
The ST8SIA2 gene is involved in the synthesis of polysialyltransferase (ST8SIAII) and plays an important role in the glycosylation of NCAM. Some studies have reported that NCAM is correlated with ASD and autistic children’s behaviors (Shaw et al., 2014; Zhang et al., 2014; Yang et al., 2019). Moreover, genome-wide studies among normal individuals and patients with mental disorders reveal that ST8SIA2 may be a candidate gene for ASD (Anney et al., 2010). In an intronic single nucleotide polymorphism (iSNP) study of the ST8SIA2 gene, ST8SIA2 was found to be related to ASD (Hane et al., 2016). Although some animal and patient studies reported that the ST8SIA2 gene was associated with ASD, there are few reports on ST8SIA2 mRNA levels in patients with ASD. In the current study, we found that the ST8SIA2 expression of ASD children was decreased in comparison with that of control children. In animal research, it reported that ST8SIA2 expressed time-dependent, which is mainly expressed in embryonic and early postnatal mice. Hence, we detected whether there were differences in its expression between different age groups. Unfortunately, no statistical difference was discovered. Autism shows a striking male bias in prevalence, with approximately 4 affected men for every 1 affected female (Werling and Geschwind, 2013). It is noteworthy that developmental neurogenetics and multimodal neuroimaging manifested sex difference in ASD (Chen and Van Horn, 2017). Recent findings reveal that women with ASD exhibit more intellectual and behavioral problems compared with their male counterparts. Based on the above, we examined the expression of ST8SIA2 in different sexes, but no expected results were obtained. This might be related to the small number of female sample participating in this study.
In addition, we explored the correlation between this gene and the behavioral problems of ASD children. This information will provide more scientific evidence to elucidate the relationship between the ST8SIA2 gene and ASD. The ST8SIA2 gene levels were low in the ASD groups and associated with the symptom severity in ASD children. According to our results, the expression levels of this gene may be lower in ASD children with more serious symptoms. In animal studies, the knockout of St8sia2 was found to cause social problems in mice. However, in the behavioral evaluation of ASD children, we did not find a correlation between social behaviors and ST8SIA2 gene expression. Interestingly, ASD children with low ST8SIA2 gene expression levels exhibited serious stereotyped behaviors. Stereotyped behaviors are the core symptom of ASD and have been a hot spot in the study of the behavioral phenotype of autistic children, which has seriously affected children’s lives and studies and even made some comorbidities to occur (Kang et al., 2017; Bhandari et al., 2020). In addition, a positive correlation was noted between the ST8SIA2 expression and the self-help abilities and daily life skills of ASD children. The self-help ability is a serious problem among children with ASD, which causes difficulties in their school life and social activities (Bal et al., 2015; Chi and Lin, 2021). Based on these findings, the ST8SIA2 gene has clinical value in elucidating the behavioral problems of patients with ASD, especially the self-care ability and stereotype behavior.
To explain the low ST8SIA2 gene expression levels in ASD children, we tested the methylation levels at ST8SIA2 gene sites. These sites were obtained from a genome-wide DNA methylation analysis, which was performed using five pairs of ASD-discordant monozygotic twins. These samples are high-quality samples for disease genetics and phenotype research, especially for non-cooccurrence cases of identical twins, which have identical genetic material and non-cooccurrence phenotype (Castillo-Fernandez et al., 2014). These samples are ideal for disease epigenetics research (Pallister et al., 2014). Therefore, the ST8SIA2 gene sites from the MZ sample were tested and verified in sporadic ASD cases and provided a scientific basis to explore the correlation between the ST8SIA2 gene and ASD. The 30 pairs of ASD and control children whose methylation levels were assessed in this study were age- and sex-matched, and the age difference of each pair was not greater than 3 months. We found that the methylation levels of Chr. 15: 92984625 and Chr. 15: 92998561 sites were statistically significant in the ASD sporadic cases. Interestingly, the methylation levels of the Chr. 15:92984625 site were positively correlated with the stereotyped behavior of ASD children, which is consistent with the relationship between the ST8SIA2 gene expression and behavioral phenotypes. Furthermore, the methylation level of the Chr. 15: 92984630 site was positively correlated with the independent living ability, which is similar to the correlation between self-help ability and ST8SIA2 gene expression. Although these sites are located in the intron region, they are valuable in explaining the association of the ST8SIA2 gene with ASD behavioral phenotypes.
To some extent, autism does not affect life span, so it is difficult to obtain brain tissue samples in the etiology research of ASD. Based on the presence of the blood-brain barrier, some neural factors and biomolecules in the peripheral blood can indirectly reflect the expression levels in the brain tissue (Qin et al., 2016; Eshraghi and Davies, 2020; Kealy et al., 2020). The extraction of RNA and DNA from the peripheral blood is a less harmful way to ASD and control children. Furthermore, there were numerous studies investigating the association between behavioral performance and biomolecular levels in the peripheral blood of patients with ASD (Patrick and Ames, 2014; Hewitson and Mathews, 2021). Blood-based gene expression could divine infants and toddlers with autism, meanwhile, the alteration of gene expression in the peripheral blood of ASD is related to the core symptoms of autism (such as social impairment and stereotypical behaviors; Makinodan et al., 2017; Norouzi Ofogh and Rasoolijazi, 2021). Therefore, this study provides evidence for the etiological study of ASD. This study had some limitations. First, due to the small number of female participants, we could not explain the expression differences in the ST8SIA2 gene in the sexes of the ASD and control groups. Second, this study used peripheral blood samples of ASD and control children; therefore, it was limited to demonstrating the relevance of ST8SIA2 and ASD due to the lack of brain tissue samples. Third, we detected only a few intronic sites in the ST8SIA2 gene. To clarify the role of the ST8SIA2 gene in ASD more clearly, methylation levels of CpG island-ST8SIA2 gene promoters should also be assessed.
Data availability statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving human participants were reviewed and approved by Ethics Committee of Qiqihar Medical University. Written informed consent to participate in this study was provided by the participants or their legal guardian/next of kin.
Author contributions
XY: data analysis, evaluation of behavioral symptoms, and writing original draft. LL: measurement of ST8SIA2 gene mRNA expression and methylation. XC: project administration. JL: review and editing the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Funding
This work was supported by the Science and Technology Project of Qiqihar (No. LHYD-202024), the Natural Science Foundation of Heilongjiang Province (No. LH2020H131), and the National Natural Science Foundation of China (No. 82103869).
References
- Anney R., Klei L., Pinto D., Regan R., Conroy J., Magalhaes T. R., et al. (2010). A genome-wide scan for common alleles affecting risk for autism. Hum. Mol. Genet. 19 4072–4082. 10.1093/hmg/ddq307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal V. H., Kim S. H., Cheong D., Lord C. (2015). Daily living skills in individuals with autism spectrum disorder from 2 to 21 years of age. Autism 19 774–784. 10.1177/1362361315575840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhandari R., Paliwal J. K., Kuhad A. (2020). Neuropsychopathology of autism spectrum disorder: complex interplay of genetic, epigenetic, and environmental factors. Adv. Neurobiol. 24 97–141. 10.1007/978-3-030-30402-7_4 [DOI] [PubMed] [Google Scholar]
- Calandreau L., Márquez C., Bisaz R., Fantin M., Sandi C. (2010). Differential impact of polysialyltransferase ST8SiaII and ST8SiaIV knockout on social interaction and aggression. Genes Brain Behav. 9 958–967. 10.1111/j.1601-183X.2010.00635.x [DOI] [PubMed] [Google Scholar]
- Castillo-Fernandez J., Spector T., Bell J. (2014). Epigenetics of discordant monozygotic twins: implications for disease. Genome Med. 6:60. 10.1186/s13073-014-0060-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Van Horn J. D. (2017). Developmental neurogenetics and multimodal neuroimaging of sex differences in autism. Brain Imaging Behav. 11 38–61. 10.1007/s11682-015-9504-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi I. J., Lin L. Y. (2021). Relationship between the performance of self-care and visual perception among young children with autism spectrum disorder and typical developing children. Autism Res. 14 315–323. 10.1002/aur.2367 [DOI] [PubMed] [Google Scholar]
- Dwyer C., Esko J. (2016). Glycan susceptibility factors in autism spectrum disorders. Mol. Aspects Med. 51 104–114. 10.1016/j.mam.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshraghi R. S., Davies C. (2020). Gut-induced inflammation during development may compromise the blood-brain barrier and predispose to autism spectrum disorder. J. Clin. Med. 10:27. 10.3390/jcm10010027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye R. E., Rossignol D. A. (2016). Identification and treatment of pathophysiological comorbidities of autism spectrum disorder to achieve optimal outcomes. Clin. Med. Insights Pediatr. 10 43–56. 10.4137/CMPed.S38337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hane M., Kitajima K., Sato C. (2016). Effects of intronic single nucleotide polymorphisms (iSNPs) of a polysialyltransferase, ST8SIA2 gene found in psychiatric disorders on its gene products. Biochem. Biophys. Res. Commun. 478 1123–1129. 10.1016/j.bbrc.2016.08.079 [DOI] [PubMed] [Google Scholar]
- Hewitson L., Mathews J. A. (2021). Blood biomarker discovery for autism spectrum disorder: a proteomic analysis. PLoS One 16:e0246581. 10.1371/journal.pone.0246581 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kamien B., Harraway J., Lundie B., Smallhorne L., Gibbs V., Heath A., et al. (2014). Characterization of a 520 kb deletion on chromosome 15q26.1 including ST8SIA2 in a patient with behavioral disturbance, autism spectrum disorder, and epilepsy. Am. J. Med. Genet. A 164A 782–788. [DOI] [PubMed] [Google Scholar]
- Kang M., Choi T., Ryu H., Lee D., Lee S., Choi S., et al. (2017). Autism-like behavior caused by deletion of vaccinia-related kinase 3 is improved by TrkB stimulation. J. Exp. Med. 214 2947–2966. 10.1084/jem.20160974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kealy J., Greene C., Campbell M. (2020). Blood-brain barrier regulation in psychiatric disorders. Neurosci. Lett. 726:133664. 10.1016/j.neulet.2018.06.033 [DOI] [PubMed] [Google Scholar]
- Krug D. A., Arick J., Almond P. (1980). Behavior checklist for identifying severely handicapped individuals with high levels of autistic behavior. J. Child Psychol. Psychiatry 21 221–229. 10.1111/j.1469-7610.1980.tb01797.x [DOI] [PubMed] [Google Scholar]
- Liang S., Li Z., Wang Y., Li X., Yang X., Zhan X., et al. (2019). Genome-wide DNA methylation analysis reveals epigenetic pattern of SH2B1 in Chinese monozygotic twins discordant for autism spectrum disorder. Front. Neurosci. 13:712. 10.3389/fnins.2019.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makinodan M., Iwata K., Ikawa D., Yamashita Y., Yamamuro K., Toritsuka M., et al. (2017). Tumor necrosis factor-alpha expression in peripheral blood mononuclear cells correlates with early childhood social interaction in autism spectrum disorder. Neurochem. Int. 104 1–5. 10.1016/j.neuint.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Mandic-Maravic V., Pejovic-Milovancevic M., Mitkovic-Voncina M., Kostic M., Aleksic-Hil O., Radosavljev-Kircanski J., et al. (2015). Sex differences in autism spectrum disorders: does sex moderate the pathway from clinical symptoms to adaptive behavior? Sci. Rep. 5:10418. 10.1038/srep10418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng W., Sun S., Yang J., Chu R., Tu W., Liu Q. (2017). Elevated serum brain-derived neurotrophic factor (BDNF) but not BDNF gene Val66Met polymorphism is associated with autism spectrum disorders. Mol. Neurobiol. 54 1167–1172. [DOI] [PubMed] [Google Scholar]
- Mitra I., Huang B., Mousavi N., Ma N., Lamkin M., Yanicky R., et al. (2021). Patterns of de novo tandem repeat mutations and their role in autism. Nature 589 246–250. 10.1038/s41586-020-03078-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norouzi Ofogh S., Rasoolijazi H. (2021). Alteration of TRIM33 Expression at transcriptional and translational levels is correlated with autism symptoms. J. Mol. Neurosci. 71 1368–1377. 10.1007/s12031-020-01783-6 [DOI] [PubMed] [Google Scholar]
- Pallister T., Spector T., Menni C. (2014). Twin studies advance the understanding of gene-environment interplay in human nutrigenomics. Nutr. Res. Rev. 27 242–251. 10.1017/s095442241400016x [DOI] [PubMed] [Google Scholar]
- Patrick R. P., Ames B. N. (2014). Vitamin D hormone regulates serotonin synthesis. Part 1: relevance for autism. FASEB J. 28 2398–2413. 10.1096/fj.13-246546 [DOI] [PubMed] [Google Scholar]
- Qin X. Y., Feng J. C., Cao C., Wu H. T., Loh Y. P., Cheng Y. (2016). Association of peripheral blood levels of brain-derived neurotrophic factor with autism spectrum disorder in children: a systematic review and meta-analysis. JAMA Pediatr. 170 1079–1086. 10.1001/jamapediatrics.2016.1626 [DOI] [PubMed] [Google Scholar]
- Ramaswami G., Won H., Gandal M., Haney J., Wang J., Wong C., et al. (2020). Integrative genomics identifies a convergent molecular subtype that links epigenomic with transcriptomic differences in autism. Nat. Commun. 11:4873. 10.1038/s41467-020-18526-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C., Stewart T., Renfrow M., Novak J. (2019). Glycosylation in health and disease. Nat. Rev. Nephrol. 15 346–366. 10.1038/s41581-019-0129-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato C., Kitajima K. (2021). Polysialylation and disease. Mol. Aspects Med. 79:100892. 10.1016/j.mam.2020.100892 [DOI] [PubMed] [Google Scholar]
- Shaw A. D., Tiwari Y., Kaplan W., Heath A., Mitchell P. B., Schofield P. R., et al. (2014). Characterisation of genetic variation in ST8SIA2 and its interaction region in NCAM1 in patients with bipolar disorder. PLoS One 9:e92556. 10.1371/journal.pone.0092556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tordjman S., Cohen D., Anderson G., Botbol M., Canitano R., Coulon N., et al. (2018). Repint of “Reframing autism as a behavioral syndrome and not a specific mental disorder: implications of genetic and phenotypic heterogeneity”. Neurosci. Biobehav. Rev. 89 132–150. 10.1016/j.neubiorev.2018.01.014 [DOI] [PubMed] [Google Scholar]
- Wang B. (2009). Sialic acid is an essential nutrient for brain development and cognition. Annu. Rev. Nutr. 29 177–222. 10.1146/annurev.nutr.28.061807.155515 [DOI] [PubMed] [Google Scholar]
- Weber A., Fernald L., Galasso E., Ratsifandrihamanana L. (2015). Performance of a receptive language test among young children in Madagascar. PLoS One 10:e0121767. 10.1371/journal.pone.0121767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werling D. M., Geschwind D. H. (2013). Sex differences in autism spectrum disorders. Curr. Opin. Neurol. 26 146–153. 10.1097/WCO.0b013e32835ee548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liang S., Wang L., Han P., Jiang X., Wang J., et al. (2018). Sialic acid and anti-ganglioside antibody levels in children with autism spectrum disorders. Brain Res. 1678 273–277. 10.1016/j.brainres.2017.10.027 [DOI] [PubMed] [Google Scholar]
- Yang X., Zou M., Pang X., Liang S., Sun C., Wang J., et al. (2019). The association between NCAM1 levels and behavioral phenotypes in children with autism spectrum disorder. Behav. Brain Res. 359 234–238. 10.1016/j.bbr.2018.11.012 [DOI] [PubMed] [Google Scholar]
- Yuan J., Song J., Zhu D., Sun E., Xia L., Zhang X., et al. (2018). Lithium treatment is safe in children with intellectual disability. Front. Mol. Neurosci. 11:425. 10.3389/fnmol.2018.00425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Wang A., Li Y., Lu X., Wang F., Fang F. (2014). Association of NCAM1 polymorphisms with autism and parental age at conception in a Chinese Han population. Genet. Test. Mol. Biomarkers 18 690–694. 10.1089/gtmb.2014.0055 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.


