Abstract
Raman scattering is an inelastic phenomenon. Although its cross section is very small, recent advances in electronics, lasers, optics, and nanotechnology have made Raman spectroscopy suitable in many areas of application. The present article reviews the applications of Raman spectroscopy in food and drug analysis and inspection, including those associated with nanomaterials. Brief overviews of basic Raman scattering theory, instrumentation, and statistical data analysis are also given. With the advent of Raman enhancement mechanisms and the progress being made in metal nanomaterials and nanoscale metal surfaces fabrications, surface enhanced Raman scattering spectroscopy has become an extra sensitive method, which is applicable not only for analysis of foods and drugs, but also for intracellular and intercellular imaging. A Raman spectrometer coupled with a fiber optics probe has great potential in applications such as monitoring and quality control in industrial food processing, food safety in agricultural plant production, and convenient inspection of pharmaceutical products, even through different types of packing. A challenge for the routine application of surface enhanced Raman scattering for quantitative analysis is reproducibility. Success in this area can be approached with each or a combination of the following methods: (1) fabrication of nanostructurally regular and uniform substrates; (2) application of statistic data analysis; and (3) isotopic dilution.
Keywords: Food, Nanomaterials, Pharmaceuticals, Raman cell imaging, Raman spectroscopy
1. Introduction
Nanomaterials are engineered particles with the shortest dimension <100 nm. These particles are characterized by very large surface-to-mass or surface-to-volume ratios. They may have different physical, chemical, and biological properties compared to their larger bulk counterparts. As far as food safety is concerned, ingredients that are generally recognized as safe at the macro level may not be safe at the nano level [1]. Nanotechnology has the potential to offer many applications in the food industry, such as nutritional additives, stronger flavorings and colorings, antibacterial ingredients for food packaging, or improvement of food structure and texture. Agricultural applications of nanomaterials include their use as nano-feed for chickens, an alternative to chemical antibiotics in industrial chicken production, nano-pesticides, for easy absorption by plants, and nano-vaccines, for addition to trout ponds for ingestion by fish [2]. These can have positive effects on food safety. By contrast, some features of nano-materials may raise potential health and safety concerns. Due to their small size, they have the potential to penetrate cell membranes in the lining of the gut and to access all areas of the body, including the brain and the nuclei of cells.
Nanomaterials are similar in size to many biological molecules and are useful for both biomedical research and applications. The integration of nanomaterials with biology has led to the development of diagnostic devices, contrast agents, analytical tools, physical therapy applications, and drug delivery vehicles [3]. Nanomedicine is the application of nano-technology to medicine and includes the development and application of nanomaterials and nanoelectronic biosensors [4]. Nanomaterials have been used to deliver drugs to specific cells, thereby reducing side effects of the drug, drug wastage, and human suffering [5]. They help to improve drug bioavailability at a specific place in the body, as well as the pharmacological and therapeutic properties of drugs.
Vibrational spectroscopy, including both infrared (IR) and Raman spectroscopy, measures the oscillations of atoms in molecules. The observation of the vibrational transitions yields information about the molecular vibrational energy levels, which in turn are related to molecular conformation, structure, intermolecular interaction, and chemical bonding. In IR spectroscopy, samples are radiated with IR light (wavelength 2.2 μm–1 mm) and the observation of IR absorption relies on the change in the dipole moment with the molecular vibration. The Raman effect is an inelastic light-scattering phenomenon, predicted in 1923 by Smekal [6] and discovered experimentally in 1927 by Raman [7] and Raman and Krishnan [8]. It was applied subsequently as a means of investigation into molecular vibrations and rotations. The invention of the laser helped tremendously to advance the instrumentation of the Raman spectrometer. Applying the then newly developed pulsed ruby laser [9] operating at 694.3 nm, two groups successfully recorded the Raman spectra of carbon tetrachloride and benzene [10,11] in 1962. However, the active use of this technique suffered from experimental restrictions until 1969, when the laser became a practical source of monochromatic electromagnetic (EM) radiation for sample excitation and the dispersive laser Raman spectrometer became commercially available.
The observation of the Raman scattering signal for a molecule depends on a change of its polarizability during the particular mode of vibration. Due to differences in selection rules, a complete collection of vibrational spectroscopic data generally would require the application of both IR and Raman spectroscopies. Raman spectroscopy has a number of distinct advantages over IR spectroscopy. Water has very intense IR absorption bands, but is a weak Raman scatterer and thus Raman spectra exhibit much less interference from water. This provides Raman spectroscopy with an advantage over IR spectroscopy for investigating aqueous biological systems, making it an important technique for biomedical research [12]. Raman spectroscopy is also well known for its minimum requirement for sample handling and preparation. In the collection of Raman spectral data, the required sample volume is determined only by the diameter of the focused laser beam, which is of the order of a micron. Materials that transmit in the IR range are very limited; by contrast, if EM radiation in the visible rangeis used for Raman scattering excitation, one can easily find materials suitable for making curettes or sample cells for Raman analysis. Another advantage is that a Raman spectrum covers the spectral range between 4000 cm−1 and ~100 cm−1, depending on how effective the Rayleigh line filtering is. By contrast, the collection of an IR spectrum over this frequency range relies on the use of both mid- and far-IR spectrometers.
One major disadvantage with conventional Raman spectroscopy is the small scattering cross section of many materials. In biomedical applications, high quality Raman spectra may require a high concentration (0.1–0.01 M) of a sample, which significantly exceeds physiological values. At a high concentration, aggregation of biomolecules may occur, leading to a change in structure. A method for increasing the sensitivity of detection is resonance Raman (RR) spectroscopy. If the Raman laser excitation frequency falls within the intense absorption of a chromophore in a sample, the Raman band intensities of the chromophore would increase by three to five orders of magnitude. RR spectra may provide information about the secondary structure of proteins and polypeptides, as well as elements of their tertiary structure. For complicated supramolecular complexes, RR spectroscopy in the ultraviolet (UV) region has provided selective excitation of individual chromophores [13]. An obstacle to the application of Raman spectroscopy is fluorescence arising from impurities, the sample, or both. Some solutions to these problems have, however, been developed as pointed out in later sections.
Raman spectroscopy is very useful in drug analysis due to advantages such as ease of use, minimal sample handling, and the significant differences in scattering strength between packaging materials, tablet excipients, and active drug components [14]. It can also be used to identify isomers and to determine energy difference between isomers. These advantages, in combination with fiber optics and microscopes, have enabled the use of Raman spectroscopy as a quality control tool in the pharmaceutical industry [15]. In the past several decades, many Raman phenomena such as coherent anti-Stokes Raman (CARS), inverse Raman, RR, and surface enhanced Raman (SERS) have been discovered. Enormous progress in Raman instrumentation and its application has resulted from progress in technology, such as UV lasers, dye lasers, solid state lasers, fiber optics, optical filters, detectors, computers and software algorithms. There are many excellent review articles and monographs dealing with the basic principles and applications of Raman spectroscopy (see for examples [13,16–19]). In the present work, we review the applications of Raman spectroscopy in the detection of nanomaterials in food and drugs, including the use of nanomaterials in the detection of food and drugs. A brief review of the theory and mechanism of the Raman effect, including RR and SERS, as well as an overview of current Raman instrumentation, is also included.
2. Theory of Raman
2.1. Conventional Raman scattering
In Raman spectroscopy, the sample is irradiated with UV, visible or near IR (NIR) EM radiation. Raman scattering is a two-photon process resulting from photon-molecule interactions. A photon is incident at frequency νo and another photon is scattered at frequency νs. The frequency difference between νo and νs is related to the vibrational energy level separation. According to classical theory, Raman activity arises from interactions of the electric field of the EM radiation of frequency νo with molecular species which possess a polarizability α. These interactions induce a temporary dipole moment:
| (1) |
where Eo is the maximum electric field strength and t is time.
In molecular vibrations, the normal coordinate Q varies periodically with the vibrational frequency νv and can be expressed as:
| (2) |
where Qo is the magnitude of the given normal vibration. It is assumed that the vibration will cause an alternation in the polarizability α according to:
| (3) |
where αo is the polarizability of the molecule in its equilibrium position, and (δα/δQ)o is the derivative of the polarizability with respect to the normal vibration coordinate at the equilibrium position. From Equations (1), (2) and (3), one has:
| (4) |
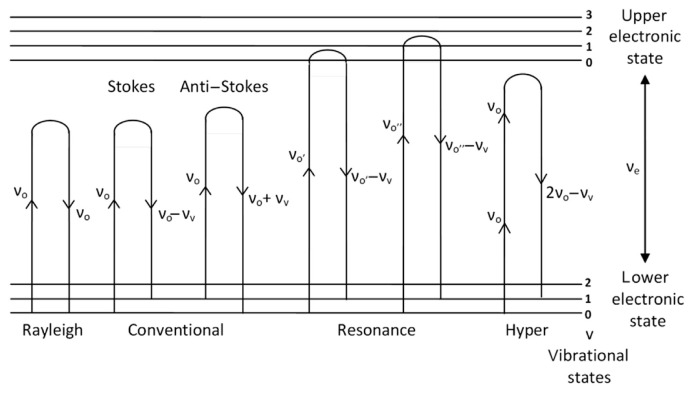
The three terms in the right side of Equation (4) represent the three different scattering frequencies νo (Rayleigh), νo-νv (Stokes Raman), and νo+νv (anti-Stokes Raman). These are represented schematically on the energy level diagram shown as Fig. 1.
Fig. 1.
Rayleigh scattering, conventional, resonance and hyper Raman scatterings involved in vibrational and electronic energy levels. Note that different excitation frequencies are required to generate different Raman effects.
Rayleigh scattering is an elastic scattering phenomenon and there is no energy transfer between the excitation photon and the molecules being analyzed. Stokes and anti-Stokes transition arise from the ground and first excited vibrational states, respectively. If the electronic transition frequency νe from the ground electronic state to the first electronic excited state is much higher than the EM radiation frequency νo used for the excitation of Raman scattering, and νo is much higher than the molecular vibration frequency νv, the Placzek theory [20] finds Stokes I(St) and the anti-Stokes I(aSt) Raman scattering intensities to be:
| (5A) |
| (5B) |
Thus, the Raman scattering intensity is proportional to the fourth power of the Raman excitation frequency. For the same molecule, the relative intensity of a Raman-active vibration is related to the square of the polarization derivative with respect to the specific normal coordinate of that vibration; different vibrational modes may have different intensities.
2.2. RR scattering
The phenomenon of resonance enhancement was predicted theoretically by Kramers and Heisenberg [21] and Dirac [22] in their dispersion equation, which describes the polarizability tensor:
| (6) |
where m and n denote the initial and final state of the molecule, e is the excited state of the molecule, (Mi)me and (Mk)en are the dipole moments of the electronic transition along the direction i, k from state m to e and from e to n and iΓe is a dampening term. As the Raman excitation frequency νo approaches the electronic transition frequency νe, the νe–νo value in the denominator of Equation (6) becomes very small and this very large term dominants the Raman scattering. The Raman intensity of certain vibrational modes is thus enhanced. This phenomenon is called RR scattering or the RR effect. RR scattering in relation to the vibrational and electronic energy levels is depicted in Fig. 1.
The intensity of RR scattering is higher than that of the normal Raman scattering by an order of three or more magnitudes. Due to its high intensity, RR has found applications in the study of azo dyes [23], metal complexes [24,25], biochemicals [26,27], nucleic acids [28], and other molecules that exhibit electronic transitions which have frequencies close to that of the Raman excitation frequency νo. Based on the above theory, the observation of RR spectra depends on the choice of EM radiation frequency for excitation. RR of colored compounds can be obtained with excitation radiation in the visible range. The development of tunable lasers and UV lasers with wavelengths as low as 200 nm, as well as improvements in photodetection, makes the routine collection of RR spectra for many molecules possible. There are two distinct mechanisms responsible for RR intensities. The first one involves a single excited state and the second one involves two electronic excited states. The functioning of the mechanisms can sometimes be determined by studying the frequency dependence of the spectral intensities. As the former mechanism activates, one may observe significant enhancement of overtone vibrations in the RR spectra. The frequencies of the fundamental vibrational modes observed in RR spectra depend only on the molecular structure and chemical bonding in the ground state; the intensities depend on the electronic excited state. Thus, the collection of RR spectra can be applied to study molecules in the electronic excited states [29,30]. With the increase of sensitivity, RR spectra of the anticancer drugs, Adriamycin and daunomycin, have been observed in solutions as dilute as 10−4 M [31].
2.3. Surface enhanced Raman scattering
A relatively new Raman scattering phenomenon known as surface enhanced Raman scattering was first observed by Fleischmann et al [32] at a silver electrode surface in 1974. Quantitative studies by two groups, Jeanmaire and Van Duyne [33] and Albrecht and Creighton [34] in 1977 provided evidence that the observations were due to the enhancement of the scattering cross section of molecules adsorbed onto the silver electrodes. The magnitude of enhancement could reach an order of four to six. It is commonly recognized that the phenomenon of SERS is a combination of several effects. The Raman cross section depends on the dipole moment, μ, of the molecule induced by the interaction of the electric component, E, of the excitation radiation with the molecule. There is an additional contribution to the induced dipole from an electric field gradient, E′, via the quadrupole polarizability, A, of the molecule. This relationship is given by the equation:
| (7) |
where α is the polarizability of molecule. There are several different types of widely used SERS active surfaces. The first type is the surfaces of noble metal electrodes roughened by electrochemical oxidation and reduction cycles. The second type is the surface of colloidal metal nanoparticles (NPs). The third type of surface is formed by depositing a metal film onto a substrate by either high vacuum vaporization or chemical reduction methods. Particles on the surfaces are small and close together on a 10–100 nm scale (see [35] for additional types of substrate). One important feature of the surface is that it can concentrate the EM field due to the incident light into a region of the surface. This can be done as a result of a resonant response of the roughened or particle covered surfaces to the EM field.
Both EM and chemical mechanisms have been put forth to explain SERS. EM mechanisms are long range in nature and may apply to all molecules within a sample. Besides the production of intense EM fields and possibly high field gradients on metal surfaces, it is also recognized that the molecule-surface interaction, including charge transfer [36], may increase the polarizability, α, and the quadruple polarizability, A, giving an additional increase of scattering intensity according to Equation (7). Chemical mechanisms are necessarily short range, because a direct contact between the metal surface and the adsorbate is required. Measuring the enhancement factor as a function of adsorbate-metal surface distance may provide information to distinguish between the two mechanisms. Based on our understanding of the enhancement mechanism, experimental conditions can be adjusted to obtain stronger SERS signals. To maximize the enhancement, one needs to tune the excitation laser frequency into resonance with the adsorption of the metal particles favoring the EM mechanism, whereas the absorption frequency of the molecule-metal complex favors the chemical mechanism. When both absorptions are in the same spectral region, a double resonance will produce very intense scattering signals. Another approach is to combine EM enhancement with the RR effect. The overall enhancement of these two effects is multiplicative and called surface enhanced RR scattering (SERRS).
3. Instrumentation
Raman instrumentation has significantly advanced since its early development and use in specialized academic setting in the 1960s. Today, photo-multiplier tubes have been replaced with charge coupled device (CCD) detectors and other solid state devices. Triple monochromators have been replaced with single gratings or interferometers with highly efficient Rayleigh line filters. Current state of the art micro-Raman spectrometers can produce chemical functionality based maps with spatial resolution of the order of 500 nm. Data analysis has also taken full advantage of the capabilities of modern computers and algorithms. In the following sections we present a brief overview of these major developments.
3.1. Fourier transform-Raman
Fourier transform (FT)-Raman spectrometers, which use near IR excitation sources, were first commercially available in 1987 [37]. With its ease of operation, Raman spectroscopy became a common feature in many industrial laboratories. The use of 1064 nm excitation from Nd:YAG (Neodymium doped yttrium aluminum garnet) lasers reduced the fluorescence issues that plagued Raman analysis of samples containing biological materials such as proteins [38]. Other advantages include high spectral resolution and good wavelength accuracy. A schematic representation of a typical instrument is shown in Fig. 2. The beam from the laser (L) is generally focused onto the sample (S) using 180° backscatter geometry. A traditional 90° sampling geometry can often also be obtained. The scattered radiation is collected through the lens (O) and passed through the Michelson interferometer (MI). The output of the interferometer is focused on the face of either a liquid nitrogen cooled Ge or room temperature InGaAs detector (D). Prior to the Raman-scattered light reaching the detector, it must be optically filtered (RF) to remove the Rayleigh line. FT-Raman instruments can be fitted with fiber optic probes (see section on fiber optic probes) as well as coupled to optical microscopes to enhance their versatility.
Fig. 2.
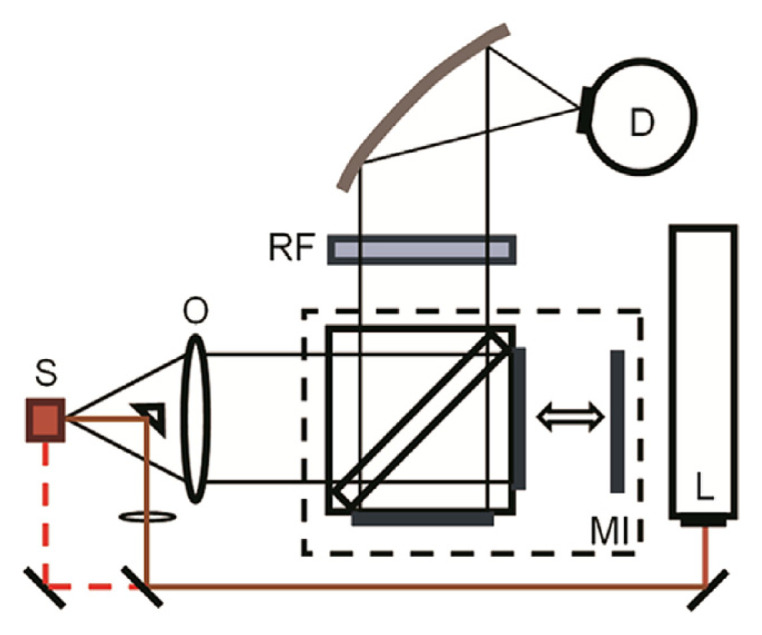
Schematic layout of a typical Fourier transform (FT)-Raman spectrometer. D = detector; L = laser; MI = Michelson interferometer; O = objective lens; RF = Rayleigh filter; S = sample.
3.2. Dispersive micro-Raman
Dispersive micro-Raman spectroscopy is carried out using systems that couple optical microscopes with conventional Raman spectrometers. The incorporation of a microscope with an x-y stage enables specific regions of a sample to be analyzed. Magnifications of ×20, ×50, and ×100 are common, with the laser spot size being determined by the laser wavelength and numerical aperture (NA) of the objective. Using 514 nm excitation through a ×50 objective with an NA of 0.75, a theoretical spot diameter of 0.84 μm can be calculated. Excitation wavelengths range from the UV through to the NIR. Typical wavelengths include 457 nm, 488 nm, and 514 nm from an argon Ion laser, 633 nm from a HeNe laser, and 785 nm and 830 nm from diode lasers. The use of these latter wavelengths addresses the issues of fluorescence, which usually has to be burnt out of biomaterial samples with extended laser exposure when visible excitation is used. This is, however, rapidly achieved with minimal to no sample damage with the concentrated power densities of these instruments.
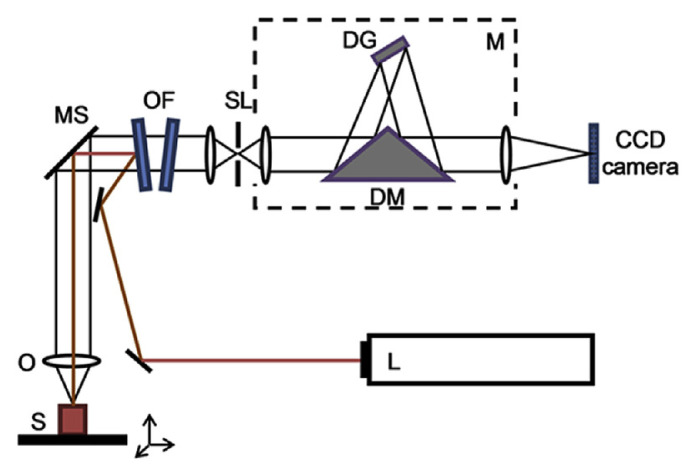
A schematic representation of a typical instrument is shown in Fig. 3. Unlike early dispersive Raman instruments which used 90° sampling optics, micro-Raman instruments extensively use 180° backscatter geometry with the laser (L) focused through the microscope (MS) onto the sample (S) and the Raman-scattered light is collected by the same microscope objective (O). The scattered radiation is passed through optical filters (OF) such as edge or notch filters to remove the Rayleigh line prior to being directed through slits (SL) and into the monochromator (M). A typical monochromator, as illustrated in Fig. 3, is comprised of a single diffraction grating (DG) and a dove mirror (DM). Finally the Raman scattered radiation is focused onto the elements of a CCD camera.
Fig. 3.
Schematic layout of a typical dispersive micro-Raman spectrometer. CCD = charge coupled device; DG = diffraction grating; DM = dove mirror; L = laser; M = monochromator; MS = microscope; O = objective lens; OF = optical filters; S = sample; SL = slits.
Confocal Raman spectroscopy refers to the ability to spatially filter the analysis volume of the sample from which the data is being collected in the x-y (lateral) and z (depth) axis. The confocal effect can be accomplished by using either a pinhole, or by a combination of the slits in one direction and the CCD pixel dimensions (binning) in the other. The depth of field as determined by the NA of the objective sets the depth resolution into the sample, thus enabling depth profiling experiments to be carried out. Typically, the depth increment is of the order of 1–0.5 μm. The depth into a sample from which data can be obtained can be limited to the nm level for samples that are opaque to the excitation wavelength.
Through the smart usage of the CCD detector arrays and modified laser optics, both line and area confocal mapping strategies have been developed [39]. Data files containing 1000s of spectra can be converted into maps depicting variations in chemical functionality with spatial resolutions of the order of 500 nm. These maps can be based on band intensities or band intensity ratios, as well as peak maximum positions and usually involve some degree of pre-processing of the data (see section on data analysis).
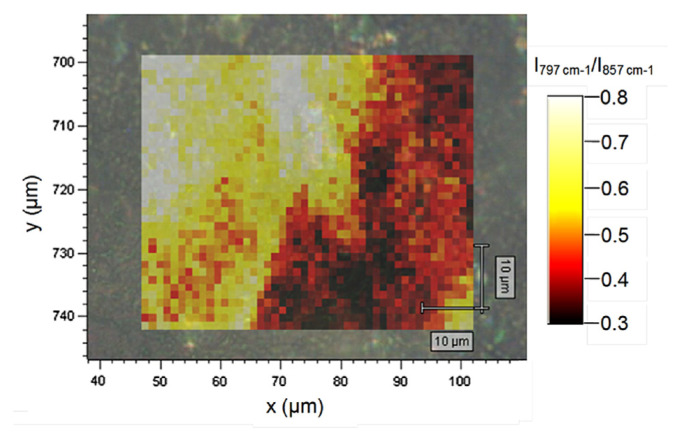
As an example of spectral mapping, we show in Fig. 4 the results obtained from the surface of a tablet containing 500 mg paracetamol and 8 mg codeine. A fixed spectral window from 1765 cm−1 to 460 cm−1 was collected from a 57 μm × 42 μm area with a spatial resolution of 1.2 μm. The total collection time for the 1656 spectra comprising the map was <20 minutes. The map (Fig. 4) is based on the intensity ratio of two sharp bands observed at 797 cm−1 and 857 cm−1, one of the few spectral features that were found to vary in the data set. No evidence of codeine was detected in the mapped region, suggesting that the 8 mg are either highly dispersed, or present as larger particles of which none were present within the area analyzed.
Fig. 4.
Raman map obtained from the surface of a tablet containing 500 mg paracetamol and 8 mg codeine.
3.3. Fiber optic probes
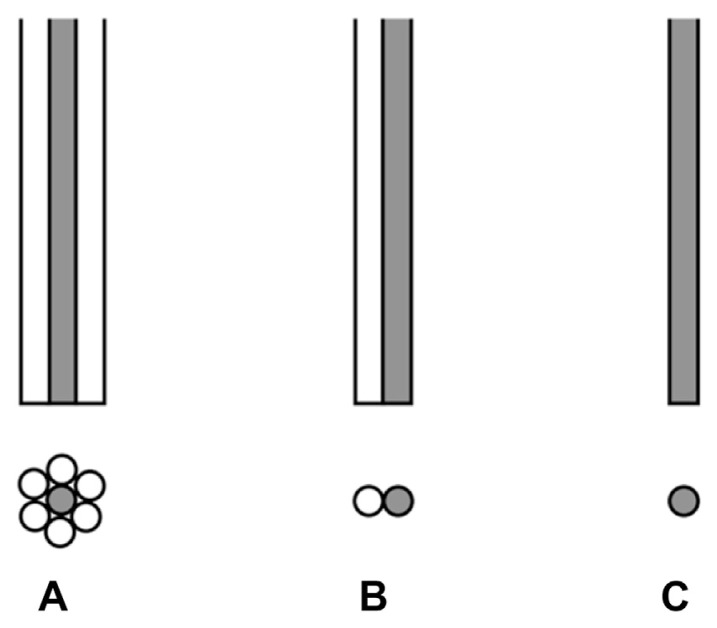
Optical fibers can be used to transmit light from one place to another based on the principle of total internal reflection. The major function of the optical fiber in the Raman spectrometer is to transmit the excitation laser light, Raman scattering light, or both. In 1980, three different groups reported the use of optical fibers or optical fiber bundles to collect the scattering signals from samples at different angles [40], to illuminate flat or unstable samples for RR scattering [41], and to transmit CARS signals generated in a remote flame to a spectrometer [42]. In these devices, the optical fibers were used along with dispersive Raman spectrometers. Subsequent applications reported the use of fibers for both excitation and collection [43,44]. Schematic representations of three different fiber optic probes based on the use of different fibers for sample excitation and scattering collection are shown in Fig. 5.
Fig. 5.
Three different types of fiber optic probes: (A) fiber bundle probe, (B) double-fiber probe, and (C) single fiber probe. The lower images represent the cross-sectional view. The shaded fiber represents the excitation fiber in (A) and (B).
In probe (A), a single fiber at the center of the bundle is used to transmit excitation laser light, whereas the surrounding fibers are used for collecting the scattered light. More than one circle of surrounding fibers may be used to increase the collection efficiency. The advantage of this probe design compared to others is the high collection efficiency for the scattering light which may be needed when conventional Raman scattering experiments are being carried out. Probe (B) is composed of a single excitation fiber and a single collection fiber. They can be configured in different angles and different positions. In a series of experiments linking the fibers to a Raman spectrometer to evaluate the efficiency of the probe system, Hendra et al [45] obtained maximum signals when the excitation fiber was normal to the sample surface and the collection fiber was at an angle of 17°. Counts were further optimized when the collection fiber was 2 mm closer to the sample than the excitation fiber. This type of probe was utilized in SERS studies of biological molecules on silver electrochemical substrates [46–48]. Also using this type of probe, along with a substrate consisting of silver alumina layers on microscope slides, Bello and Vo-Dinh [49] optimized the conditions for the SERS fiber sensor and obtained a limit of detection (LOD) of 0.4 ng for p-aminobenzoic acid. The penetrability of a laser beam through a substrate allows for the positioning of the excitation and collection fibers not only on the same side, but also on opposite sides of a substrate, making the application of this probe for SERS measurements rather versatile [50]. In this type of optical system, the laser beam, transmitted through a band pass filter, is focused by an appropriate lens into the end of the excitation fiber. If the f number of the fiber is different from that of the spectrometer, the input scattering signals from the collection fiber must also be focused into the spectrometer with lenses.
One major advantage with fiber optic probes is their easy access for monitoring of samples in harsh environments or remote locations. Some fiber optic sensors have been built with long fibers, enabling the analysis of samples as far as 20 m from the analytical spectroscopy laboratory [51]. However, Raman background may arise from the fiber optic material itself used in the light transmission. Ma and Li [52,53] studied the Raman background in relation to the fiber NA, the spatial arrangement, and the tilted angle of the fiber end. With the use of band pass filters, Angel et al [54] measured the spectra of aspirin tablets at a distance of 50 m from the spectrometer.
The third type of probe relies on a single fiber for the transmission of both the laser excitation beam and the Raman scattered light (see Fig. 5C). This probe appears to be simpler than the other types of probe, but requires the use of optical components to couple the laser beam into the single collection fiber. As shown in Fig. 6, the holographic beam splitter reflects the laser into objective lens 1, which focuses the laser beam onto the end of optical fiber 1. The Raman scattering signal returns from the probe tip and emerges from the optical fiber, where it is collected and collimated by lens 1, transmitted through the beam-splitter, and focused by lens 2 onto the optical fiber 2. A holographic notch filter is used to reject the Rayleigh scattering radiation transmitted by the beam splitter; fiber 2 is designated to transmit the Raman signal to the spectrometer. It is feasible to use the single fiber probe to monitor samples in remote locations and in hazardous environments. Additionally, this probe can be converted into a fiber optic nanosensor by using nanofibers [55,56]. For SERS detection, a tapered optical fiber has been coated with 6 nm Ag NPs in a high vacuum electron beam evaporator, to obtain probe tips with diameters <100 nm.
Fig. 6.
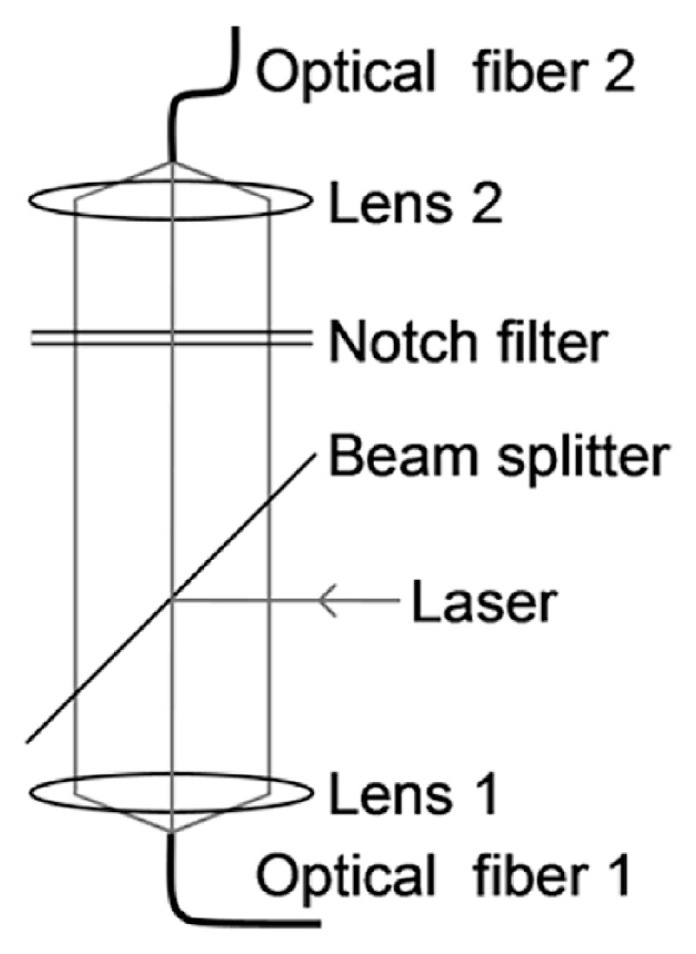
Optical guiding system for a single fiber probe.
3.4. Data analysis
The digital nature of the data collected by modern Raman spectrometers allows for a wide range of data processing algorithms to be applied [57]. The use of spectral subtraction, which can be used to isolate the features of specific components in non-interacting mixtures, was first demonstrated in 1986 [37]. The smoothing and differentiation algorithm currently used for discrete data sets was developed by Savitzky and Golay in 1964 [58]. Successive subsets of adjacent data points are fit with a low-degree polynomial using linear least squares. When the data points are equally spaced, an analytical solution to the least squares equations can be found, in the form of a single set of convolution coefficients that can be applied to all data subsets, to give estimates of the smoothed or derivatized signal. High frequency noise can also be removed from spectra by Fourier filtering. In this approach, spectra are first Fourier transformed into the time domain and then specific frequencies are either attenuated or amplified by the application of a filter. The modified output is obtained by inverse transformation. The use of second derivative transformation enhances separation of overlapping band components, and removes baseline shifts, making spectral comparisons much easier. Overlapping features can be resolved through spectral deconvolution and band fitting techniques [59]. The number of peaks and their approximate positions are first identified from the minima observed in second derivative spectra. The spectral region of interest is then modeled using a corresponding series of initial band shapes. The peak maximum frequency, peak height, width, and shape (Gaussian, Lorentzian, or mixed) are then allowed to vary until a best fit is obtained.
With large data sets, a statistical approach to data analysis can be undertaken. Quantitative regression analysis is no longer limited to the association between a dependent variable and a single independent variable. The use of full spectra or specifically selected regions is now commonly carried out using multi-variant approaches, such as principal component regression (PCR) [60] and partial least squares (PLS) regression [61]. The applications of these techniques are generally preceded by the pre-processing of the data sets. Common methods applied include taking second derivatives and normalization based on either total area or peak intensity.
PCR is a regression analysis that uses principal component analysis (PCA) when estimating the regression coefficients. PCA uses an orthogonal transformation to decompose the spectral data set into a set of linearly uncorrelated variables or principal components (PCs) and a set of scale factors or scores. The size of the new data set is usually significantly smaller than the original one. The PCs are often called factors or loadings. The factors are defined in such a way that the first one represents the largest possible amount of variance within the data set. Subsequent factors account for less and less of the variance. The original spectra can generally be reconstructed by summing a minimal number of these factors after they have been scaled by the scores. In PCR, instead of regressing the spectral intensities (dependent variables) on the Raman shifts (independent variables) directly, the PCs or factors of the independent variables are used instead. PLS is another spectral decomposition technique that is closely related to PCR. In PLS, the sample concentration information is used during the decomposition process, resulting in the spectra containing higher constituent concentrations to be weighted more heavily. The factors and scores calculated using a PLS approach are different from those determined by PCR. In both PLS and PCR, care must be taken to prevent overfitting the data by using too many factors, as the ability of the model to predict new data will likely be compromised.
Qualitative information can be extracted from sets of spectra using unsupervised and supervised pattern recognition methods. Unsupervised methods, often referred to as cluster analysis, seek to find natural groupings of objects allowing the presence of any patterns to be identified. No classification knowledge is required and no assumptions of such are made. Applications include model fitting, hypothesis testing, data exploration, and data reduction. One of the most commonly used approaches is hierarchical clustering, which is based on the correlation coefficient matrix. The results are shown graphically as dendrograms [57]. Similarity measures such as Mahalanobis and nearest-neighbor distances [62] are often found to be less limiting, as they take into account nonlinear relationships and the absolute magnitude of variates. Cluster analysis can also be carried out using the PCs determined through PCA. By plotting the scores for the various factors against each other in two or three dimensional space, a scatter plot is obtained. The points comprising this plot can be grouped or clustered based on the distances between them. It is often found that the chemical differences between samples which serve as the basis of the observed separations can be obtained through detailed spectroscopic interpretation of the factors. In a more recently developed approach, fuzzy cluster analysis, objects are assigned a membership function indicating their degree of belonging to a particular group or set [63].
In supervised pattern recognition, often referred to as classification or discriminant analysis, the number of parent groups is known in advance and representative samples of each group are available. The problem is then to assign an unclassified object to one of the parent groups using an appropriate function or set of rules. Soft independent modeling for class analogies (SIMCA) [64] is such a method for supervised classification of data. The method requires a training data set consisting of spectra obtained from samples with a set of attributes and their class membership. The term soft refers to the fact that the classifier can identify samples as belonging to multiple classes and not necessarily producing a classification of samples into non-overlapping classes. The samples belonging to each training set class are analyzed using PCA, with only the significant PCs retained. The resulting model for a given class is described by a line (one PC), plane (two PCs), or hyperplane (more than two PCs). For each modeled class, the mean orthogonal distance of the training set spectra from the line, plane, or hyperplane is used to determine a critical distance for classification. Unknown spectra are projected into each PC model and the residual distances calculated. A spectrum is assigned to the model class when its residual distance from the model is below the critical distance for the class. For data sets where the distribution is unknown, or known not to be normal, the K-nearest neighbors algorithm is one of the most widely used for classification [65]. In recent years, considerable use has also been made of artificial neural networks [66].
4. Literature review
4.1. Raman spectra of food nanomaterials
4.1.1. Previous reviews
There has recently been a significant number of reviews published covering the determination of nanomaterials in food. These are very broad, covering many scientific techniques and methods and thus Raman spectroscopy appears as a minor element. Tiede and co-authors [67] described the detection and characterization of engineered NPs in food and in the environment, whereas Duncan [68] reviewed applications of nanotechnology in food packaging and food safety, with a focus on materials, antimicrobials, and sensors. Luykx et al [69] reviewed analytical methods for the identification and characterization of nano delivery systems in food. Deisingh and Thompson [70] reviewed the use of biosensors for the detection of bacteria. Lin [71] presented an overview of traditional and novel detection techniques for melamine and its analogues in foods and animal feed. He also reviewed [72] the SERS detection of various peptides and veterinary drugs, of which some may be related to food safety. Zamborini et al [73] described recent advances in the use of NPs in measurement science.
A number of reviews have also appeared focusing on Raman spectroscopy as applied to food and food safety, but not specifically on nanomaterials. Yang and Ying [74] recently reviewed the applications of Raman spectroscopy in agricultureal products and food analysis. Craig et al [75] provided an outlook on work done and a perspective on the future directions of surface-enhanced Raman spectroscopy applied to food safety. In a recent review on determining nanomaterials in food, Blasco and Pico [76] divided nano applications in food and the food industry into four areas: processing, products, materials, and food safety and biosecurity. The first two areas represent nanoproducts as subjects of control, whereas in the second two areas, nanodevices are used as tools for control. Raman spectroscopy has been applied to both of these areas as illustrated in some of the examples presented below.
4.1.2. Nanomaterial contamination of food
In their study into the uptake, translocation, and transmission of carbon nanomaterial in rice plants, Lin and co-workers [77] used FT-Raman spectroscopy to confirm that aggregates observed optically in second generation plant leaf cells were composed of C70 fullerenes and their derivatives. Carbon nanomaterial, including fullerenes as well as multi-walled carbon nanotubes (CNTs), was suspended in natural organic matter (NOM) solutions and used to treat newly harvested and sterilized rice seeds. The detection of the Raman signature of the hydrophobic carbon nanomaterial aggregates in the plant material demonstrated how its mobility was enhanced by the NOM, enabling dynamic uptake, integration, and transmission to progeny through seeds.
In another study carried out by He et al [78], fractal-like gold nanostructures were developed for use as SERS substrates, with the target being the detection of crystal violet, malachite green, and their mixture, all common prohibited contaminants found in imported seafood. The fractal structures were fabricated through a self-assembly process using 30–50 nm gold NPs formed by the hydrothermal citrate-reduction method [79,80] as building blocks. Particle aggregation was induced by the addition of a small quantity of cetyltrimethylammonium bromide. Spectra were processed using the multivariate approach including second derivative transformation and PCA analysis. An enhancement factor of the order of 4 × 107 was achieved and the lowest detectable concentration was ~0.2 ppb.
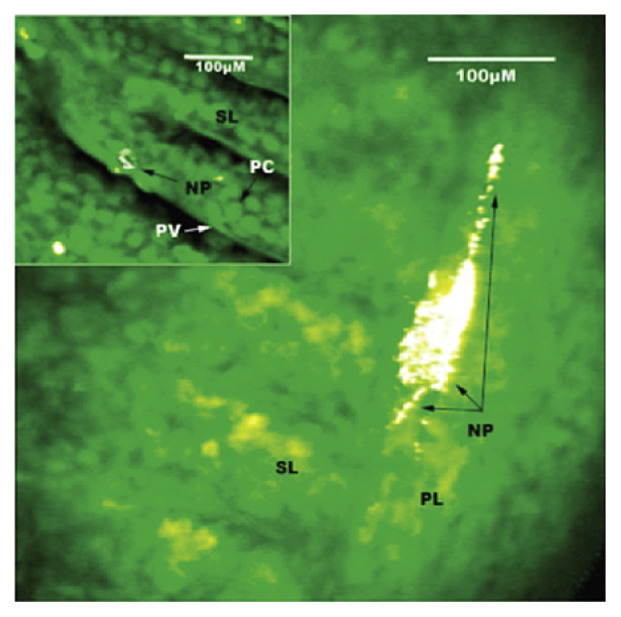
In a study on the uptake of nanoscale metal oxides by fish, Johnston et al [81] used CARS to detect the presence of ionic titanium in the gut of rainbow trout that were exposed through diet. CARS is a third-order nonlinear optical process involving three laser beams: a pump beam, a Stokes beam, and a probe beam. The three beams interact with the sample and generate a coherent anti-Stokes signal. This signal is resonantly enhanced when the frequency difference between the pump and the Stokes beams coincides with the frequency of a Raman resonance, thus providing an intrinsic vibrational contrast mechanism [82]. CARS can be used as a three-dimensional imaging technique that can provide chemical information from biological structures at depths up to several 100 μm. CARS spectrometers are custom built as it is considered a specialty technique. Metal oxides have strong CARS signals due to the two phonon electronic resonance of the semiconductor band gap. The technique was used to localize TiO2 aggregates as large as 3 μm on the surfaces of the gill epithelium and the primary and secondary lamellae. The CARS image of TiO2 NPs on a section of the lamellae is shown in Fig. 7.
Fig. 7.
Coherent anti-Stokes Raman (CARS) image of TiO2 nanoparticles on a section of the primary lamellae (main panel) and three-dimensional projection showing a nanoaggregate on the secondary lamellae (inset). NP with arrow = TiO2 nanoparticles; PC = pillar cell; PL = primary lamellae; PV = pavement cell (epithelium); SL = secondary lamellae. Note. From “Bioavailability of nanoscale metal oxides TiO2, CeO2, and ZnO to fish,” by B.D. Johnston, T.M. Scown, J. Moger, et al, 2010, Environ Sci Technol, 44, p. 1141–51. Copyright 2010, American Chemical Society. Reprinted with permission.
4.1.3. Pesticides and fungicides in food
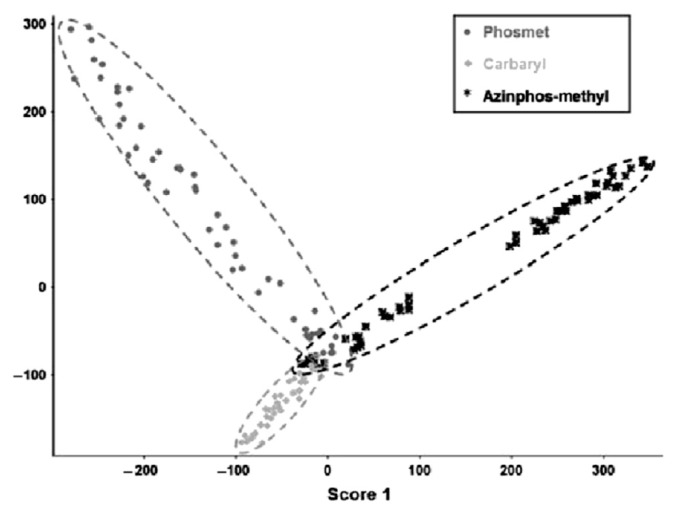
Liu and co-workers [83] developed a SERS-based method to detect three different pesticides extracted from the surfaces of apples and tomatoes. Small volumes (0.3–0.5 μL) of filtered acetonitrile/water (1:1) fruit skin extracts were dropped onto commercially available gold-based nanostructures fabricated on a silicon wafer. SERS spectra were recorded using 785 nm diode laser excitation through the ×50 objective of a micro-Raman spectrometer. Using multivariate statistical methods including PLS and PCA, both quantitative and qualitative analysis of the data was carried out. The detection limits were found to vary with the specific fruit, but in general were as follows: Carbaryl ~5 ppm, phosmet ~6 ppm, and azinphosmethyl ~3 ppm. The classification of the pesticides using the first two PCs is shown in Fig. 8.
Fig. 8.
Classification of pesticides using the first two principal components (PCs). Note. From “Detection of pesticides in fruits by surface-enhanced Raman spectroscopy coupled with gold nanostructures.,” by B. Liu, P. Zhou, X. Liu, et al, 2013, Food Process Tech, 6, p. 710–8. Copyright 2012, Springer Science and Business Media. Reprinted with permission.
Conventional Raman spectroscopy has also been used to study the antifungal activity of zinc oxide NPs against Botrytis cinerea and Penicillium expansum [84]. These fungal pathogens are the main causes of economic loss during the postharvest handling of fruit. In Raman spectra obtained from ZnO NP-treated B. cinerea the intensities of nucleic acid and carbohydrate bands increased significantly, whereas no intensity changes were observed for bands associated with proteins and lipids. These findings were verified by the results of PCA-based cluster analysis. The increase in nucleic acid content may be a stress response, whereas the increase in carbohydrates may be a self-protecting mechanism against the ZnO NPs.
Müller et al [85] investigated the use of SERS to detect and monitor the chemical fungicide thiabendazole (TBZ) which is used in the food industry against mold and blight on citrus fruit and bananas. Using a compact, portable mini-Raman spectrometer, they reported the first complete vibrational characterization of TBZ over a large pH and concentration range. The estimated total amount of TZB in a 5 g citrus peel sample was found to be 78 mg/kg, 13 times higher than the maximum allowed by current regulations.
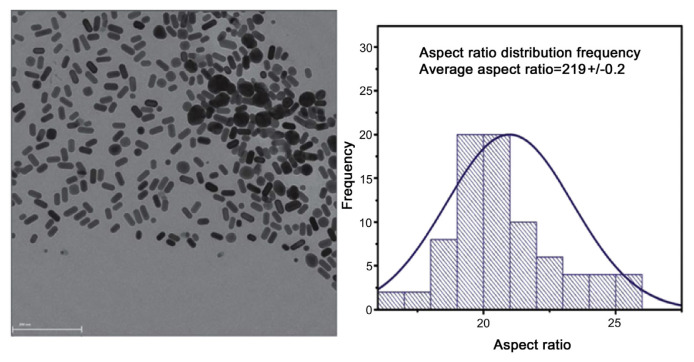
Saute and Narayanan [86,87] developed gold nanorods and Saute et al [88] developed dogbone shaped gold NPs for use in SERS detection of fungicides at ultra-low levels in solutions. Nanorods (Fig. 9) with an aspect ratio of 2.19 and a length of 38 nm were used in the detection of the dithiocarbamate fungicides thiram, ferbam, and ziram in acetonitrile-water solutions. A PLS approach was taken for development of quantitation, whereas PCA was used for discrimination. For thiram, the LOD was 11.0 nM and the limit of quantitation was 34.4 nM. The Environmental Protection Agent (EPA) tolerance for this material is 16.6 μM. When small (43 nm average size) dogbone shaped NPs [86] were used, the LOD was 44 nM. This was reduced to 12 nM when larger (65 nm) dogbone shaped NPs were used. In an extension of this work, the dogbone shaped NPs were used to detect the dithiocarbamate fungicides in real-world matrices including tap water, apple juice, and vegetable juice [87]. The LODs and limits of quantitation for thiram decreased from that in tap water by factors of ~3 and ~6 for apple and vegetable juice, respectively. These levels were still well below the EPA tolerance, demonstrating that SERS is an excellent technique for detection of these fungicides at ultra-low concentrations, even in complex solutions.
Fig. 9.
Transmission electron microscope (TEM) micrograph and size distribution of gold nanorods. Note. From “Gold nanorods as surface enhanced Raman spectroscopy substrates for sensitive and selective detection of ultra-low levels of dithiocarbamate pesticides,” by B. Saute, R. Premasiri, L. Ziegler, et al, 2012, Analyst, 137, p. 5082–7. Copyright 2012, The Royal Society of Chemistry. Reprinted with permission.
4.1.4. Bacteria in food
Jarvis and Goodacre [89] published an early tutorial review reporting on the advances made in bacterial studies through the application of SERS. This included the characterization, discrimination, and identification of microorganisms, as well as assessing how they respond to abiotic and biotic stress. It was concluded that SERS is a very beneficial technique for the rapid analysis of bacteria where the ultimate goal can potentially be achieved without the need for a lengthy cell culture.
Fan and co-workers [90] used SERS coupled with silver nanosubstrates to develop a sensitive method to rapidly detect food and waterborne bacteria. Escherichia coli O157:H7, Staphylococcus epidermidis, Listeria monocytogenes, and Enterococcus faecalis, all pathogens important to food and water safety, were used in this study. The bacteria were deposited with intercellular silver by sequentially exposing the cells to solutions of silver nitrate and sodium borohydride. SERS spectra were collected from two or three drops of the treated bacteria placed on a gold plate. Single cell detection level could be reached and PCA could be used to classify mixtures of bacteria at both species and strain levels. The same research group also investigated the SERS detection of seven different food and waterborne viruses in phosphate buffered saline (PBS) [90]. It was found that the solutions had to be diluted with deionized water to minimize the interference from the PBS. SIMCA was able to classify ~95% of the virus samples with and without envelopes, whereas PCA could classify and identify different virus samples at the strain level.
The quantitative SERS detection of Bacillus bacterial spores using a portable Raman spectrometer was reported by Cowcher et al [91]. The method developed was based on the extraction of the biomarker dipicolinic acid from spore suspensions. Citrate-reduced silver colloid was prepared using the Lee and Meisel method [92] and used as the SERS active agent. Raman spectra were collected using a spectrometer equipped with a 633 nm, 3 mW HeNe laser and related to actual spore counts using univariate and multivariate methods. The lowest detection limit of 100 spores/20 μL of sample for complex sample extracts obtained using a PLS approach is higher than desirable. The limitation was determined to lie in the extraction method.
SERS has been collected from intracellular and extracellular bacteria locations [93]. SERS spectra obtained from the bacterium Geobacter sulfurreducens measuring no more than 0.5 μm × 1.5 μm was facilitated by the precipitation of colloidal gold within the cells. Scattering was also collected from the same organism after reduction of ionic silver, which resulted in colloidal silver deposition on the cell surface. Although conventional Raman detected only two unassigned bands at 1032 cm−1 and 1069 cm−1, the SERS spectra exhibited over 10 well defined bands between 1400 cm−1 and 200 cm−1. These bands have been assigned to protein, phospholipids, RNA/DNA, and polysaccharides.
Conventional Raman spectroscopy has been used in a number of studies involving nanomaterials in the food supply. Nicolaou et al [94] investigated its use as well as that of FT-IR spectroscopy to study the spoilage trajectory of milk caused by the bacteria S. aureus and Lactococcus lactis ssp. cremoris. Milk inoculated with each bacteria as well as a mixture of both was incubated at 37 °C for 24 hours. Samples taken over the growth period were deposited on stainless steel plates and allowed to dry at room temperature over 3 hours. Raman spectra were obtained using 785 nm excitation in order to minimize fluorescence. Data was collected from seven different viruses processed using PLS and PCA-discriminant function analysis (PCA-DFA) approaches. Although the IR approach provided very reasonable quantification results for viable bacteria counts (R2 values ranging from 0.64 to 0.76 depending upon the bacteria), the Raman results were found to exhibit a high degree of error and poor correlation. Spoilage trajectory analysis using PCA-DFA produced similar trends with both IR and Raman data sets, but the Raman results exhibited a lack of precision in comparison. Bacterial quantitation by PLS using the Raman data was also not satisfactory. It is possible that a SERS approach to this problem would have produced better correlations.
4.2. Raman spectra of pharmaceutical materials and nanomaterials
4.2.1. Previous reviews
There have recently been a number of broadly focused reviews published in this area. An exhaustive review by Williams [95] has included the pharmaceutical applications of Raman spectroscopy in the period 1978–1999. In a short review paper, Pinzaru and coworkers [96] highlighted different Raman techniques applied in pharmaceutical investigations. They have also presented several Raman applications in pharmaceutical science, such as fundamental structural investigations, quantitative analysis, drug-excipient interaction, drug formulation, LOD, pH-dependent pharmaceutical species, adsorption geometry at a given surface, and functional groups involved in adsorption for several widely used pharmaceutical compounds. Pinzaru and Pavel [97], in a review, have presented SERS data of pharmaceutical compounds from the widely used anatomical therapeutic chemical classification such as antipyretics, analgesics, anti-malarial drugs, antibiotics, antiseptics, and other classes such as anticarcinogenic and antimutagenic drugs. West and Halas [98] reviewed the ever expanding array of nanostructured materials with unique and powerful optical properties. These included the use of quantum dots for fluorescent biological labels and silver plasmon resonance particles for bioassay applications. Yao et al [99] wrote a featured article covering new horizons for sensing, imaging, and medicine using graphene-based nanomaterials. Lee and his coworkers [100] reviewed the recent work on the applications of the conjugation of nanomaterials and aptamers for biosensing and diagnostics using fluorescence, colorimetry, magnetic resonance, electrochemical detection, and SERS. In the following sections we focus on Raman specific applications to pharmaceutical materials.
4.2.2. Drugs and drug tablet quality control
Some pharmaceutical compounds give very good Raman spectra even in diluted conditions. Commercial drugs are often used in very low doses and are formulated in an inert matrix or excipient to make them into a tablet form, or to modify the release rate into the patient’s system. Raman imaging of tablets can provide information about the distribution and relative amounts of active agent, additives, and binders present (see Fig. 4). The spectrum of the pure pharmaceutical agent can be obtained by subtracting the matrix spectrum from that of the commercial drug. Useful spectra may sometimes be obtained without subtraction when the pharmaceuticals are strong Raman scatters and the fillers are weak Raman scatters [101,102]. As pharmaceuticals can be analyzed directly inside their polymer packaging, the application of Raman spectroscopy for quality control of manufacturing and formulation results in significant time and cost savings. Eliasson and Matousek [103] have demonstrated the use of spatially offset Raman spectroscopy (SORS) in the identification of counterfeit pharmaceutical tablets and capsules through different types of packaging. This technique offers a higher sensitivity than that of conventional backscatter Raman spectroscopy and enables chemical information to be obtained from different depths within the sample. Davies et al [104] studied a number of polymeric biomaterials and drug delivery systems. With good quality spectra, drugs such as promethazine, diclofenac, theophylline, and indomethacin can be monitored down to the 5% (w/w) concentration level in inert polymer matrices such as sodium alginate.
SERS is known for its high sensitivity in garnering molecular signals for chemical identification. By applying metal NPs in SERS drug analysis, Cunningham and coworkers [105] designed an optical device to identify and measure the drug contents of the fluid in an intravenous line in real time. To observe the SERS signals of the drugs, they incorporated into the tubing a nanostructured gold surface containing millions of tiny “nano-domes” separated from each other by 10 nm. Preliminary results obtained for drugs including morphine, methadone, phenobarbital, promethazine, and mitoxantrone found that concentrations 100 times lower than those usually delivered could be detected. The system has also proved its capability for the fast analysis of two-drug combination solutions, thus improving the patient’s safety during intravenous drug administration. It is expected that the system will also be useful in urinary catheters, in hospital care, and in pharmaceutical manufacturing.
Raman spectroscopy has been applied to the analysis of Chinese medicines. Feng et al [106] dealt with the methodology for detecting components in Chinese decoctions by SERS, but no spectrum of specific medicine components were given in the report. Huang et al [107] reported Raman and SERS spectra of the traditional Chinese medicine “Atractylodis macrocephalae rhizome” pieces (AMRP). Very intense SERS bands were observed due to the strong interaction of the AMRP with the silver colloid. It was suggested that the SERS technique has a great potential for quick, effective, accurate, and non-destructive analysis of Chinese medicine without complicated sample extraction and separation. For therapeutic significance, Huang et al [108] collected confocal micro-Raman spectra of chick embryo vasculature with and without the antiangiogenic drug thalidomide. The results showed relative Raman intensity variations for some characteristic peaks. PCA was used to distinguish these two kinds of vasculature, showing the effectiveness of the Raman method in detecting the mechanism of vascular changes.
4.2.3. Antibiotics
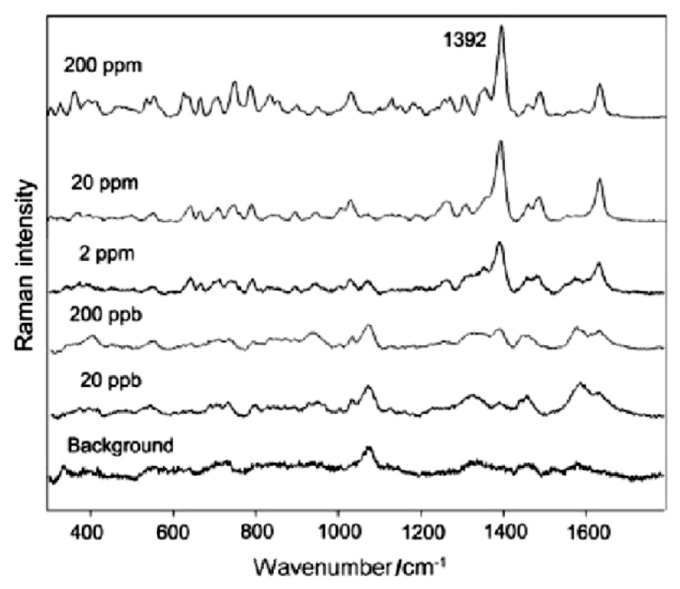
Antibiotics are common drugs used to treat human infections. They are also effective in treating various bacterial deceases in animal husbandry and aquaculture. Antibiotic residues in food products are a cause of great concern due to the possible development of antibacterial resistance to the drugs. Consequently, the detection and quantification of antibiotic by SERS should be considered in both food and drug applications. He et al [109] reported the SERS analyses of three restricted antibiotics, enrofloxacin, ciprofloxacin, and chloramphenicol, by using silver nanosubstrates prepared from the replacement reaction of AgNO3 with zinc metal. The silver dendrites formed in this way could be stored for 6 months without loss of SERS activity. In this work, NIR laser excitation (785 nm) was used, but as aqueous suspensions of the dendritic silver absorbed broadly from 400 nm to 800 nm, there should be a wide choice of laser excitation possible for the SERS measurements. Based on the experimental SERS data, a linear relationship was obtained from a log-log plot of sample concentration and SERS intensity. The LOD for the antibiotics reached the ppb level (see Fig. 10).
Fig. 10.
Surface enhanced Raman scattering (SERS) spectra of a series of concentrations of ciprofloxacin (20–200 ppm). Note. From [109]. Copyright 2009, John Wiley & Sons, Ltd. Printed with permission.
4.2.4. Antitumor drugs
To study the interaction of antitumor drugs with DNA, microfluorescence, X-ray diffraction, and IR spectroscopic methods are generally used. Microfluorescence is applicable only when the drugs have high fluorescence quantum yields [110,111]. X-ray diffraction and IR spectroscopy provide structural information on DNA-antitumor complexes in vitro, but the collection and analysis of X-ray data takes a long time [112] and the selectivity and sensitivity of IR spectroscopy are low [113]. Because of the high sensitivity of SERS, it may provide useful pharmacological information from antitumor drugs at the single cell level. Sequaris et al [114] first reported the SERS spectra of complexes of some antitumor Pt-coordinate compounds with DNA. In this study, they correlated the antitumor activity of these complexes with their ability to intercalate inside the DNA double helix.
Two groups measured SERS spectra of some other anti-tumor drugs and their DNA complexes in vitro [115–117]. Nabiev and coworkers [118] reported SERS spectra of the antitumor drugs, Doxorubicin, 4′-O-tetrahydropyranyl-doxarubicin, and aclacinomycin A, and their complexes with DNA in aqueous solutions collected with non-activated and activated silver hydrosols. The strong quenching of fluorescence and the high sensitivity of SERS make it possible to measure the drugs at concentrations down to 10−10 M and to detect the drugs in living cells. The analysis of the SERS spectra of anti-tumor drugs and their complexes with DNA on different silver hydrosols has provided a way for constructing structural models which correlate well with the result obtained from X-ray diffraction.
SERS is a useful and sensitive technique for quantitative analysis. Further enhancements would be achieved if analytes can provide molecular resonance in addition to surface enhancement. For this reason, SERRS should provide ultra-high sensitivity for quantitative analysis. Based on the mechanism of surface enhancement and the theory of RR, Smith et al [119] pointed out several approaches for obtaining high sensitivity in Raman spectroscopic analysis: (1) choice of wavelength for excitation; (2) types of assay for scattering signal accumulation and average; (3) choice of substrates for effective and reproducible enhancement; and (4) choice of analytes for strong adherence onto the substrate surface. The anticancer drug, mitoxantrone, is a good example in fitting these conditions. SERRS analysis of mitoxantrone in serum and plasma was conducted by McLaughlin and coworkers [120] by using a flow cell and silver colloid as a substrate. Without prior sample manipulation, the LOD could reach 10−10 M or 0.06 ng/mL and the analysis time was 2 minutes compared with 4 hours by high-performance liquid chromatography (HPLC). Results from the SERRS analysis of a series of samples taken from patients at different times agreed well with those obtained by HPLC.
4.2.5. Drugs of abuse
In the analysis of drugs of abuse, Sulk et al [121] detected amphetamine and methamphetamine by the use of SERS. The amines were derivatized using coupling agents. The LOD was found to be ~18 ppm. To rapidly identify illicit drugs, Sagmuller et al [122] used a matrix-stabilized silver halide as a substrate to obtain SERS and Raman spectra of amphetamine and its derivatives at mM levels in methanolic solutions. Faulds et al [123] carried out the SERS detection of amphetamine sulfate using colloidal sols, depositing films of both silver and gold. It was found that a drug concentration from 10−5 M to 10−6 M could be semi-quantitatively detected with the use of an Au colloid. Employing a fractional factorial design, Mabbott et al [124] recently developed a new optimization of parameters for the SERS quantitative detection of mephedrone using a portable Raman system. The LOD was estimated to be 1.6 μg/mL (or 9.06 × 10−6 M), which is well below that of conventional Raman and is extremely low for fast in-field determination. The quantitation measurement of morphine in Ag sols by SERS was reported by Feng et al [125]. The LOD was determined to be 1.5 ng/mL.
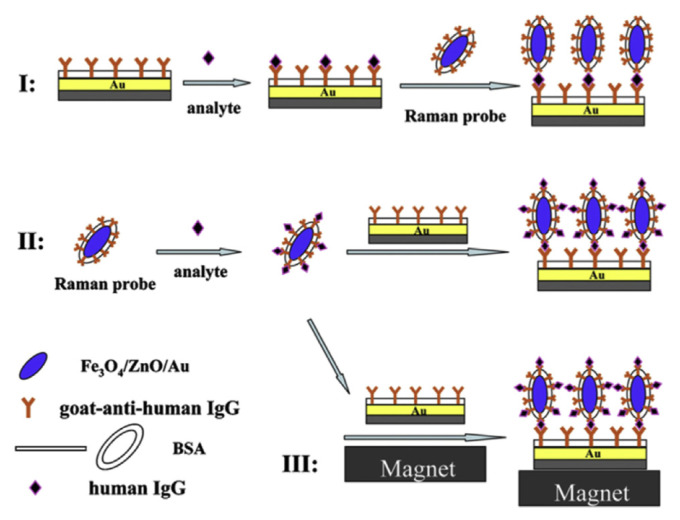
Immunoassays are routinely used in a wide range of clinical tests including the detection of recombinant human growth hormones by sports anti-doping laboratories. Hong and co-workers [126] developed magnetic-field assisted rapid ultrasensitive immunoassays using Fe3O4/ZnO/Au nanorices as Raman probes. The incorporation of ZnO into the probes enables advantage to be taken of the RR effect. The scheme developed for the assay, which involves three strategies, is shown in Fig. 11. In general, Fe3O4/ZnO/Au nanorices (blue ovals) were synthesized in a stepwise growth method starting with Fe3O4 seed particles. The nanorices were coated with goat anti-human immunoglobulin G (IgG) shown as Ys. Substrates were prepared by coating cleaned silicon with the oxide layer removed with a layer of Au by thermal evaporation. The substrate was also then labeled by treatments with the goat anti-human IgG. Remaining binding sites on both the nanorices and substrates were blocked with bovine serum albumin. Sandwich structures were formed between the labeled substrates and nanorices by their exposure to a solution of the human IgG analyte (⋄). RR spectra were obtained at room temperature using a confocal Raman microscope with 325 nm excitation from a He-Cd laser. The lower LOD of the assay was enhanced by several orders of magnitude (to 2 fM) and the detection time was reduced from 1 hour to 3 minutes when an external magnetic field was utilized to concentrate the analyte/probe complexes (strategy III).
Fig. 11.
Schematic of sandwich immunoassays. Note. From “Magnetic-field-assisted rapid ultrasensitive immunoassays using Fe3O4/ZnO/Au nanorices as Raman probes,” by X. Hong, X Chu. P. Zou, et al, 2010, Biosens Bioelectron, 26, p. 918–22. Copyright 2010, Elsevier. Reprinted with permission.
4.2.6. Isotope dilution SERS
To overcome the shortcoming of reproducibility in SERS analysis, a method using the addition of a certain amount of an isotope-labeled analog of the target analyte to the original sample as an internal standard has been developed [127–130]. In this method, the quantification of the analyte can be determined from the Raman band intensity ratio of the unknown with the added isotopic analog. The uncertainty of this method is <3%. Recently, Zakel et al [131] measured the concentration of creatinine in two human serum samples by isotope dilution SERS and obtained results in excellent agreement with those reported by participating national metrology institutes using mass spectrometry. This has led to the approval of this method as a higher order of reference measurement procedure for clinical measurements in international comparison schemes.
4.2.7. Cell imaging
Raman cell imaging is useful in obtaining information about real molecular interactions, conformational dynamics, and intracellular pharmacokinetics. The weak Raman scattering of living cells makes spectrometer optimization a necessity. For recording full Raman spectra, Raman imaging requires long measurement times, but as noted in the section on dispersive micro-Raman, map data sets covering a 1300 cm−1 wide spectral region can now be rapidly collected. Resonant excitation can give much better intensity in contrast to non-resonant excitation, if the molecules being studied are stable enough to stand the high powered laser excitation. Otherwise, cell damage will make the RR measurements undesirable [132]. Other shortcomings of Raman micro-spectroscopy include the possible photo-bleaching and fluorescence of the sample. These issues have largely been solved (see the section on dispersive micro-Raman) in modern instruments, however, the quenching of fluorescence and increasing of the Raman cross section in SERS would allow for a decrease in applied excitation laser power by more than an order of magnitude, avoiding the sample heating and photo-bleaching problems. SERS can be applied to overcome the low efficiency of normal Raman scattering under experimental conditions suitable for living cells. To activate the Raman scattering enhancement from a living cell, Kneipp and coworkers [133] deposited Au colloidal particles inside cells and obtained information about the native chemical constituents inside a cell and their intracellular distribution. In a book chapter, Kneipp [134] presented the use of nanosensors to obtain the Raman signatures from the cellular components in the immediate surroundings of the Au nanostructures. A review on the use of SERS nanosensors for the in vivo probing of intracellular biochemicals was published in 2008 [135]. Applying SERS micro-spectroscopy with a silver hydrosol, Nabiev and coworkers [118] studied the structure of doxorubicin complexes and their distribution in living K562 cancer cell. SERS spectra of Doxorubicin and other biomolecules adsorbed on silver island films were also observed [136].
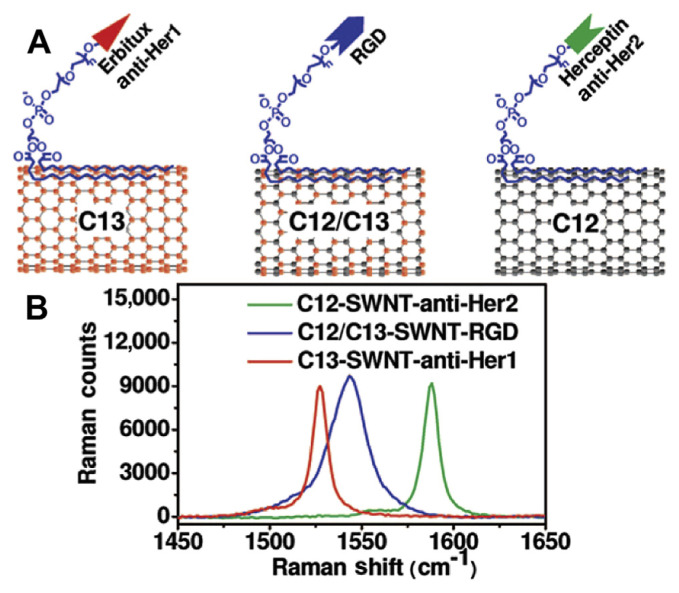
Raman micro-spectroscopy can provide a noninvasive method to image cells and cellular processes [137]. The application of this technique has recently been extended to study intracellular drug delivery using nano-carried systems [138,139]. Raman imaging has high multiplicity because of the narrow Raman spectral bands. It is powerful in visualizing the molecular composition of subcellular compartments without the need for labeling. An optimized Raman micro-spectrometer has been developed for the noninvasive evaluation of tumors in live mice that were treated intravenously with either single walled carbon nanotubes (SWNTs) or arginine-glycine-aspartic acid peptide modified SWNTs [140]. Raman’s ability to noninvasively localize targeted SWNTs in tumor models could provide the foundation for future studies with other Raman targeted NPs. Raman images enabled CNT localization as well as evaluation of the tumor targeting, thus supporting the development of a new preclinical Raman imager. The isotopically modified SWNTs shown in Fig. 12 have been used for multiplexed multicolored Raman imaging of living cells [141]. Fiber optic nanoprobes based on SERS have very high sensitivity. Near-field SERS has been used for measuring single dye molecules and dye-labeled DNA with a resolution on the 100 nm scale [142,143]. Lee and his coworkers [100] recorded SERS spectra from aptamernanomaterial conjugates. They have shown that these bionanomaterials can be used not only as highly sensitive and selective diagnostic agents, but also as targeted drug delivery agents.
Fig. 12.
Single walled carbon nanotubes (SWNTs) with different Raman colors. (A) schematic SWNTs with three different isotope compositions (C13-SWNT, C12/C13-SWNT, C12-SWNT) conjugated with different targeting ligands. (B) Solution phase Raman spectra of the three SWNT conjugates under 785 nm laser excitation. Different G-band peak positions were observed. At the same SWNT concentration, the peak height of C12-SWNT (Hipco) was approximately two times higher than that of C13-SWNT and approximately four times higher than that of C12/C13-SWNT. For mixtures used in biological experiments, concentrations of the three SWNTs were adjusted to give similar G-band peak intensities of the three colors, as shown in this figure. Note. From “Multiplexed multicolor Raman imaging of live cells with isotopically modified single walled carbon nanotubes,” by Z. Liu, X. Li, S.M. Tabakman, et al, 2008, J Am Chem Soc, 130, p. 13540–1. Copyright 2008, American Chemical Society. Reprinted with permission.
SERS relies on the use of effective substrates and the adsorption of sample chemicals onto or near the substrate surfaces. There are several different methods for getting the metal NP into cells for SERS imaging. The first method is the natural uptake of NPs by living cells conducted by incubating the cells in the medium containing the colloidal metal. This method is rather commonly used [144]. One disadvantage with this procedure is the long time it takes to complete. The second method is based on electroporation, in which a pulse potential is applied [145]. During the electroporation process, keeping cells at low temperature around 0–4°C is necessary. This procedure allows for quick acquisition of SERS data. It was noticed that the SERS spectra collected from whole-cells using this method of NP delivery were more reproducible than those obtained when the passive uptake method was used. The third method is the formation of intracellular NPs by reduction of metal ions. This method is particularly useful when the small size (0.5 μm × 1 μm) of bacterial cells makes it difficult to introduce NPs into this tiny environment. This method is applicable only when the species can reduce metal ions to zero valence metal NPs. By using this method, Jarvis et al [93] observed SERS spectra of the bacterium Geobacter by colloidal Au formed within the cell. However, the use of silver ion resulted in colloidal Ag depositions on the cell surface. The use of fiber optic nanoprobe may be regarded as the fourth method of getting NPs into cells. With the coating of NPs onto the tiny nanoprobe tip, the insertion of the probe into the cell would carry the NPs in the cell. Fluorescence imaging has been obtained with this method; it should also be applicable to Raman imaging.
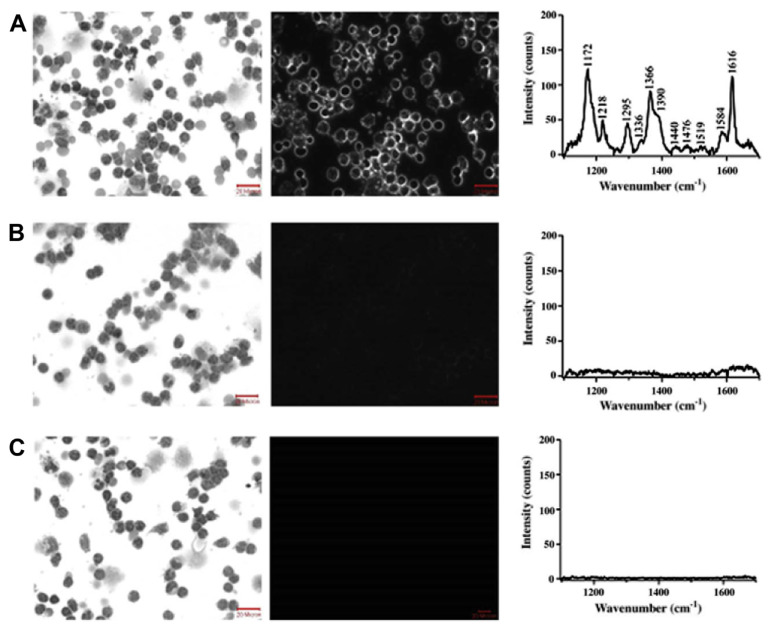
The functionalization of noble metal NPs makes it feasible to target markers for the collection of SERS spectra at the molecular level. The obtained spectroscopic information may serve as the basis for detection. As an example, Nguyen et al [146] functionalized Au NPs for selective SERS detection of a hematologic malignancy, chronic lymphocytic leukemia (CLL). The functional NPs are composed of an Au core, which is covered with malachite green isothiocyanate dye through electrostatic interaction and further with thio-polyethylene glycol (HS-PEG) conjugated covalently to human anti-CD19 antibodies. The NPs would selectively target and image CLL cells isolated from the blood samples of patients. Experimentally, CLL cells were incubated with three different NPs: (1) functionalized (dye and PEG) CD19 conjugated NPs; (2) functionalized (dye and PEG) CD4 conjugated NPs; (3) non-binding negative control NPs without antibody attach. The bright and dark-field images and accompanying Raman spectra collected for the cells incubated under three different conditions are shown in Fig. 13. Results indicate that Au NPs functionalized with anti-CD19 antibody selectively targeted the specific markers.
Fig. 13.
From left to right within each row: bright and dark-field images (20× magnification) and accompanying Raman spectra of chronic lymphocytic leukemia (CLL) cells stained with anti-CD19 and nanoparticle conjugates. (A) CLL cells stained with Giemsa and labeled with anti-CD19-SERS nanoparticles; (B) CLL cells stained with Giemsa and incubated with anti-CD4 antibody-SERS conjugates; and (C) CLL cells stained with Giemsa and incubated with control SERS nanoparticles (unconjugated particles). The Giemsa stain produces the dark features in the column of images on the left, Rayleigh scattering produces the bright features in the column of images in the middle, and Raman scattering produces the signals in the spectra in right hand column. Note. From “Detection of chronic lymphocytic leukemia cell surface markers using surface enhanced Raman scattering gold nanoparticles,” by Z. Nguyen, X. Li, S.M. Tabakman et al, 2010, Cancer Lett, 292, p. 91–7. Copyright 2009, Elsevier. Reprinted with permission.
The use of Raman spectroscopy has been extended to visualize deep tissue in live animals. A combined magnetic resonance imaging (MRI) and SERS imaging nanoprobe was developed and demonstrated in vivo [147]. The nanoprobes are gold NPs complexed with dextran-coated superparamagnetic iron oxide NPs. Dextran-coated iron oxide NPs are known for their value as preclinical and clinical MRI contrast agents. The gold, which is non-toxic and has potential therapeutic value, also serves as the substrate for a Raman active dye molecule to generate a SERS effect. The probe was injected into the gluteal muscle of a live mouse and imaged by placing it directly on the Raman spectroscopy platform. The results demonstrated a clear SERS signal both in vivo and ex vivo. These results were also consistent with those obtained in silico.
5. Outlook
In their paper on analytical methods for assessing NP toxicity, Marquis and co-workers [148] stated that analytical chemists are particularly suited to address the analytical challenges in nanotoxicity because they are accustomed to developing new technology, pushing forwards lower LOD, and navigating complex samples. SERS from noble metal NPs has the potential to allow dynamic assessment of adsorbed species in vitro or in vivo but more work is required in developing efficient photon collection methods. The ability to dynamically monitor NP degradation to determine if components are unintentionally leaching into the biological environment will become increasingly important. The portability and cost of detection systems will become important issues as the demand for NP monitoring increases.
Yamamoto et al [149] developed an analytical system in combination of capillary electrophoresis (CE) for a high efficiency separation with a Raman microscope for sensitive detection. This system was used to separate different sizes of SWNTs with CE and to characterize individual SWNTs with Raman spectroscopy. To increase the sensitivity, the CE droplets were concentrated by evaporating the liquid solvent. In this study, Raman spectroscopy could distinguish between SWNTs with a diameter difference of 0.02 nm. It is expected that this technique can be applied to the separation and characterization of other nanomaterials, such as β-amyloids and quantum dots. Such a coupling of two analytical techniques may also apply to food and drug analysis. HPLC has been commonly used for the analysis of drugs. Raman spectroscopies, such as confocal micro-Raman and SERS, could turn out to be useful for the detection and analysis of some drugs. Compared to conventional HPLC detection which largely relies on retention time, Raman detection can be either tuned to a frequency for the identification of a specific drug, or used to collect the entire vibrational spectrum for additional structural information. The “nano-domes” substrate used in intravenous lines [99] is an example of a handy detection system applicable in this development.
Cellar imaging based on Raman scattering is powerful in visualizing the molecular composition of subcellular compartments without the need for labeling [137]. Fiber optic nanosensors are useful in measuring intracellular/intercellular physiological and biological parameters in sub-microenvironments [137]. With a combination of the nano-probe technology and SERS-based detection, it will be possible to engage in multiplexed analysis of multiple biomarkers. In fiber optics nanosensing, the probe is physically inserted into cells using micromanipulators; there is no concern regarding rates of NP uptake and rejection, opening up a new applications in molecular biology, medical diagnostics, and possibly drug carrier development in the future. Multiplexed detection of oligonucleotide targets with labeled Au NP probes was conducted by Wei and coworkers [150]. The Au NP probes facilitate the silver coating as an SERS promoter for the dye-labeled particles captured by target molecules and an underlying chip in microarray format, providing several orders of magnitude higher sensitivity and many orders of magnitude higher selectivity than the analogous molecular fluorescence-based approach. Without optimization of this method, the LOD is 20 fM. Due to the high enhancement level, SERS has been applied to single molecule detection [151]. The concept of SERS can be extended to two-photon excitation by exploiting surface-enhanced hyper Raman scattering (SEHRS), another phenomenon for extra sensitive detection. Hyper Raman scattering is represented schematically on the energy level diagram shown in Fig. 1. For SEHRS, there are several advantages over the one-photo excitation: (1) a longer wavelength laser can be used for excitation; (2) the excitation volume in a sample is limited; and (3) new insight into the EM enhancement mechanisms can be provided.
Kneipp and coworkers [152] demonstrated SEHRS in the local optical fields of Au and Ag nanostructures and reported the effective two-photon cross sections in the range of 10−46–10−45 cm4 s. Such extra-high sensitivity makes it promising for biological and pharmaceutical applications. SERS is a promising technique for food safety assessment and for drug analysis, as it is rapid, sensitive, accurate, and requires minimal sample preparation. These features make it potentially suitable for routine on-line analysis in food processing facilities and other analytical applications. With advancements in fiber optic Raman spectrometers, it is anticipated that the in situ analysis of the applied pesticides on agricultural plant foods will become practical. However, it is still very challenging to apply SERS for quantitative analysis. Success in this area requires the integration of chemometric methods into the spectral data analysis, as well as the development of versatile and robust Raman spectrometers and nanosubstrates.
In concluding this review, we would like to present some approaches that could potentially counter the issue of poor reproducibility in applying SERS for quantitative analysis. (1) Development and fabrication of nanostructurally uniform substrate: Tripp and coworkers [35] stated that the oblique angle deposition of noble metals and other nanofabrication techniques would challenge the limitation of reproducibility of SERS measurements. (2) Chemometric data analysis: to access the colloidal SERS reproducibility, Jarvis et al [153] suggested the use of suitably designed SERS experiments in conjunction with multivariate analysis of variance. (3) Isotope dilution: a given amount of isotopic analyte is added to a sample as an internal reference for SERS intensity standardization (see section on isotope dilution SERS). This method assumes that similar chemical properties exist in different isotopic species of the analyte. Several research groups have applied this method in their quantitative SERS analysis and the resulted uncertainty was reduced to <3%.
Acknowledgments
J.S.C. acknowledges the support of CSIRO Materials Science and Engineering. The authors acknowledge Ms. Andrea Woodhead for her assistance in collecting the Raman map in Fig 4.
Footnotes
Conflicts of interest
The authors declare that there are no conflicts of interest.
REFERENCES
- 1. Aillon KL, Xie Y, El-Gendy N, et al. Effects of nanomaterial physicochemical properties on in vivo toxicity. Adv Drug Delivery Rev. 2009;61:457–66. doi: 10.1016/j.addr.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong J, Peralta-Videa JR, Gardea-Torresdey JL. Nanomaterials in agricultural production: benefits and possible threats? In: Shamim N, Sharma VK, editors. Sustainable nanotechnology and the environment: advances and achievements. Washington, DC: American Chemical Society; 2013. pp. 73–91. [Google Scholar]
- 3.Gazit E, Mitraki A. Plenty of room for biology at the bottom: an introduction to bionanotechnology. 2nd ed. London: Imperial College Press; 2007. [Google Scholar]
- 4.Freitas RA. Nanomedicine, volume I: basic capabilities. Georgetown, TX: Landes Bioscience; 1999. [Google Scholar]
- 5. De Jong WH, Borm PJ. Drug delivery and nano particles: applications and hazards. Int J Nanomed. 2008;3:133–49. doi: 10.2147/ijn.s596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smekal A. The quantum theory of dispersion. Naturwiss. 1923;11:873–5. [Google Scholar]
- 7. Raman CV. A new radiation. Indian J Phys. 1928;2:387–98. [Google Scholar]
- 8. Raman CV, Krishnan KS. A new type of secondary radiation. Nature. 1928;121:501–2. [Google Scholar]
- 9. Maiman TH, Hoskins RH, D’Haenens IJ, et al. Stimulated optical emission in fluorescent solid. II. Spectroscopy and stimulated emission in ruby. Phys Rev. 1961;123:1151–7. [Google Scholar]
- 10. Porto SPS, Wood DL. Ruby optical maser as a Raman source. J Opt Soc Amer. 1962;52:251–2. [Google Scholar]
- 11.Stoicheff BP. Spectroscopic studies with the ruby optical laser. Washington: 10th Colloquium Spectroscopicum Internationale; 1963S399–415. [Google Scholar]
- 12.Bhattacharyya BD. Raman spectral studies on DNA anticancer drugs: intercalation in stochastic solution region. In: Bist HD, Durig JR, Sullivan JF, editors. Vibrational spectra and structure. Amsterdam: Elsevier; 1989. pp. 521–52. [Google Scholar]
- 13.Spiro TG, editor. Biological applications of Raman spectroscopy. New York: John Wiley & Sons; pp. 1987–88. [Google Scholar]
- 14. Ellis G, Hendra PJ, Hodges CM, et al. Routine analytical Fourier-transform Raman spectroscopy. Analyst. 1989;114:1061–6. [Google Scholar]
- 15. Cutmore EA, Skett PW. Application of Fourier transform Raman spectroscopy to a range of compounds of pharmaceutical interest. Spectrochim Acta A. 1993;49:809–18. [Google Scholar]
- 16.Chang RK, Furtak TE, editors. Surface enhanced Raman scattering. New York: Plenum Press; 1982. [Google Scholar]
- 17.Clark RJH, Hester RE, editors. Advances in spectroscopy. Chichester: Wiley; 1975. 98 pp. [Google Scholar]
- 18.Durig JR, editor. Vibrational spectra and structure. Amsterdam: Elsevier; 1972–2000. [Google Scholar]
- 19.Kneipp K, Moskovits M, Kneipp H, editors. Surface enhanced Raman scattering: physics and applications. Berlin Heidelberg: Springer-Verlag; 2006. [Google Scholar]
- 20.Placzek G. Rayleigh-streuung und Raman-effekt. In: Marx E, editor. Handbuch der radiologie. Leipzig: Akademische Verlag; p. 1934. [In German]. [Google Scholar]
- 21. Kramers HA, Heisenberg W. Über die streuung von strahlung durch atome. Z Phys. 1925;31:681–708. [In German] [Google Scholar]
- 22. Dirac PAM. The quantum theory of the emission and absorption of radiation. Proc Roy Soc Lond A. 1927;114:710–28. [Google Scholar]
- 23.Macjida K. Resonance Raman spectra and protonation equilibria of azo dyes. In: Bist HD, Durig JR, Sullivan JF, editors. Vibrational spectra and structure. Amsterdam: Elsevier; 1989. pp. 421–42. [Google Scholar]
- 24.Huong PV, Plouvier SR. Copper-organic interactions in cancer studies by resonance Raman spectroscopy. In: Bist HD, Durig JR, Sullivan JF, editors. Vibrational spectra and structure. Amsterdam: Elsevier; 1989. pp. 497–520. [Google Scholar]
- 25.Tonge PJ, Carey PR. Raman, resonance Raman and FTIR spectroscopic studies of enzyme-substrate complexes. In: Clark RJH, Hester RE, editors. Advances in spectroscopy. Chichester: Wiley; 1993. pp. 129–61. [Google Scholar]
- 26.Austin JC, Jordan T, Spiro TG. Ultraviolet resonance Raman studies of proteins and related model compounds. In: Clark RJH, Hester RE, editors. Advances in spectroscopy. Chichester: Wiley; 1993. pp. 55–127. [Google Scholar]
- 27.Spiro TG. Biochemical applications of resonance-Raman spectroscopy. In: Durig JR, editor. Vibrational spectra and structure. Amsterdam: Elsevier; 1976. pp. 101–20. [Google Scholar]
- 28.Peticolas WL. Quantitative characterization of the ultraviolet resonance Raman spectroscopy of nucleic acid components, enzymatic cofactors and their photochemically induced transients. In: Bist HD, Durig JR, Sullivan JF, editors. Vibrational spectra and structure. Amsterdam: Elsevier; 1989. pp. 467–84. [Google Scholar]
- 29. Galluzzi F, Garozzo M, Ricci FF. Resonance Raman scattering and vibronic coupling in aquo- and cyanocobalamin. J Raman Spectrosc. 1974;2:351–62. [Google Scholar]
- 30. Ziegler LD, Hudson BS. Resonance Raman scattering of ethylene: Evidence for a twisted geometry in the V state. J Chem Phys. 1983;79:1197–202. [Google Scholar]
- 31.Houng PV. Chemical application of resonance Raman spectroscopy. In: Durig JR, editor. Vibrational spectra and structure. Amsterdam: Elsevier; 1981. pp. 143–93. [Google Scholar]
- 32. Fleischmann M, Hendra PJ, McQuillan AJ. Raman spectra of pyridine adsorbed at a silver electrode. Chem Phys Lett. 1974;26:163–6. [Google Scholar]
- 33. Jeanmaire DL, Van Duyne RP. Surface Raman electrochemistry. Part 1. Heterocyclic, aromatic and aliphatic amines adsorbed on the anodized silver electrode. J Electroanal Chem. 1977;84:1–20. [Google Scholar]
- 34. Albrecht MG, Creighton JA. Anomalously intense Raman spectra of pyridine at a silver electrode. J Am Chem Soc. 1977;99:5215–7. [Google Scholar]
- 35. Tripp RA, Dluhy RA, Zhao Y. Novel nanostructures for SERS biosensing. Nano Today. 2008;3:31–7. [Google Scholar]
- 36.Yamada H. Chemical effect and charge transfer interaction in surface enhanced Raman scattering. In: Bist HD, Durig JR, Sullivan JF, editors. Vibrational spectra and structure. Amsterdam: Elsevier; 1989. pp. 392–420. [Google Scholar]
- 37.Chase DB, Rabolt JF, editors. Fourier transform Raman spectroscopy from concept to experiment. San Diego: Academic Press; 1994. [Google Scholar]
- 38. Keller S, LiJchte T, Dippel B, et al. Quality control of food with near-infrared-excited Raman spectroscopy. Fresenius J Anal Chem. 1993;346:863–7. [Google Scholar]
- 39. Hutchings J, Kendall C, Smith B, et al. The potential for histological screening using a combination of rapid Raman mapping and principal component analysis. J Biophotonics. 2009;2:91–103. doi: 10.1002/jbio.200810070. [DOI] [PubMed] [Google Scholar]
- 40. Trott GR, Furtak TE. Angular resolved Raman scattering film optic probes. Rev Sci Instrum. 1980;51:1493–6. [Google Scholar]
- 41. Yamada H, Yamamoto Y. Illumination of flat or unstable samples for Raman measurements using optical fibres. J Raman Spectrosc. 1980;9:401–2. [Google Scholar]
- 42. Eckbreth AC. Remote detection of CARS employing fiber optic guides. Appl Opt. 1979;18:3215–6. doi: 10.1364/AO.18.003215. [DOI] [PubMed] [Google Scholar]
- 43. McCreery RL, Fleischmann M, Hendra P. Fiber optic probe for remote Raman spectrometry. Anal Chem. 1983;55:146–8. [Google Scholar]
- 44. Schwab SD, McCreery RL. Versatile, efficient Raman sampling with fiber optics. Anal Chem. 1984;56:2199–204. [Google Scholar]
- 45. Hendra PJ, Ellis G, Cutler DJ. Use of optical fibres in Raman spectroscopy. J Raman Spectrosc. 1988;19:413–8. [Google Scholar]
- 46. Hsieh YZ, Lee NS, Sheng RS, et al. Surface-enhanced Raman spectroscopy of free and complexed bilirubin. Langmuir. 1987;3:1141–6. [Google Scholar]
- 47. Hsieh YZ, Morris MD. Resonance Raman spectroscopic study of bilirubin hydrogen bonding in solutions and in the albumin complex. J Am Chem Soc. 1988;110:62–7. [Google Scholar]
- 48. Lee NS, Hsieh YZ, Paisley RF, et al. Surface-enhanced Raman spectroscopy of the catecholamine neurotransmitters and related compounds. Anal Chem. 1988;60:442–6. doi: 10.1021/ac00156a014. [DOI] [PubMed] [Google Scholar]
- 49. Bello JM, Vo-Dinh T. Surface-enhanced Raman scattering fiber-optic sensor. Appl Spectrosc. 1990;1:63–9. [Google Scholar]
- 50.Vo-Dinh T, Stoke DL, Li YS, et al. Fiber optic sensor probe for in situ surface-enhanced Raman monitoring. In: Lieberman RA, Wlodarczyk MR, editors. Chemical, biochemical, and environmental fiber sensors II. San Jose, CA, USA: SPIE- The International Society for Optical Engineering; 1991. pp. 203–9. [Google Scholar]
- 51. Myrick ML, Angel SM. Elimination of background in fiber-optic Raman measurements. Appl Spectrosc. 1990;44:565–70. [Google Scholar]
- 52. Ma J, Li YS. Optical fiber Raman probe with low background interference by spatial optimization. Appl Spectrosc. 1994;48:1529–31. [Google Scholar]
- 53. Ma JY, Li YS. Fiber Raman background study and its application in setting up optical fiber Raman probes. Appl Opt. 1996;35:2527–33. doi: 10.1364/AO.35.002527. [DOI] [PubMed] [Google Scholar]
- 54.Angel SM, Cooney TF, Skinner HT. Applications of fiber-optics in NIR Raman spectroscopy. In: Laserna JJ, editor. Modern techniques in Raman spectroscopy. New York: John Wiley & Sons, Inc; 2000. pp. 387–419. [Google Scholar]
- 55. Cullum BM, Vo-Dinh T. The development of optical nanosensors for biological measurements. Trends Biotechnol. 2000;18:388–93. doi: 10.1016/s0167-7799(00)01477-3. [DOI] [PubMed] [Google Scholar]
- 56. Scaffidi JP, Gregas MK, Seewaldt V, et al. SERS-based plasmonic nanobiosensing in single living cells. Anal Bioanal Chem. 2009;393:1135–41. doi: 10.1007/s00216-008-2521-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams MJ. Chemometrics in analytical spectroscopy. 2nd ed. Cambridge: Royal Society of Chemistry; 2004. [Google Scholar]
- 58. Savitzky A, Golay MJE. Smoothing and differentiation of data by simplified least squares procedures. Anal Chem. 1964;36:1627–39. [Google Scholar]
- 59. Meier RJ. On art and science in curve-fitting vibrational spectra. Vib Spectrosc. 2005;39:266–9. [Google Scholar]
- 60. Thomas EV, Haaland DM. Comparison of multivariate calibration methods for quantitative spectral analysis. Anal Chem. 1990;62:1091–9. [Google Scholar]
- 61.Wold H. Soft modeling by latent variables; the nonlinear iterative partial least squares approach. In: Gani J, editor. Perspectives in probability and statistics. London: Academic Press; 1975. [Google Scholar]
- 62.Hartigan J. Clustering algorithms. New York: J Wiley and Sons; 1975. [Google Scholar]
- 63.Kandel A. Fuzzy mathematical techniques with applications; 1986. Wesley; New York, New York: [Google Scholar]
- 64.Wold S, Sjostrom M. Simca: a method for analyzing chemical data in terms of similarity and analogy. In: Kowalski BR, editor. Chemometrics theory and application. Washington, DC: American Chemical Society; 1977. pp. 243–82. [Google Scholar]
- 65.Afifi A, May S, Clark VA. Computer-aided multivariate analysis. 4th ed. Boca Raton: Chapman and Hall/CRC; 2003. [Google Scholar]
- 66.Zupan J, Gesteiger J. Neural networks for chemists. 2nd ed. Weinheim: VCH; 1999. [Google Scholar]
- 67. Tiede K, Boxall ABA, Tear SP, et al. Detection and characterization of engineered nanoparticles in food and the environment. Food Addit Contam Part A. 2008;25:795–821. doi: 10.1080/02652030802007553. [DOI] [PubMed] [Google Scholar]
- 68. Duncan TV. Applications of nanotechnology in food packaging and food safety: barrier materials, antimicrobials and sensors. J Colloid Interface Sci. 2011;363:1–24. doi: 10.1016/j.jcis.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luykx DMAM, Peters RJB, van Ruth SM, et al. A review of analytical methods for the identification and characterization of nano delivery systems in food. J Agr Food Chem. 2008;56:8231–47. doi: 10.1021/jf8013926. [DOI] [PubMed] [Google Scholar]
- 70. Deisingh AK, Thompson M. Biosensors for the detection of bacteria. Can J Microbiol. 2004;50:69–77. doi: 10.1139/w03-095. [DOI] [PubMed] [Google Scholar]
- 71.Lin M. The application of surface-enhanced Raman spectroscopy to identify and quantify chemical adulterants or contaminants in foods. In: Li-Chan E, Griffiths P, Chalmers J, editors. Applications of vibrational spectroscopy in food science. Chichester: John Wiley & Sons; 2010. pp. 649–62. [Google Scholar]
- 72. Lin M. A review of traditional and novel detection techniques for melamine and its analogues in foods and animal feed. Front Chem Eng China. 2004;3:427–35. [Google Scholar]
- 73. Zamborini FP, Bao L, Dasari R. Nanoparticles in measurement science. Anal Chem. 2013;84:541–76. doi: 10.1021/ac203233q. [DOI] [PubMed] [Google Scholar]
- 74. Yang D, Ying Y. Applications of Raman spectroscopy in agricultural products and food analysis: a review. Appl Spectrosc Rev. 2011;46:539–60. [Google Scholar]
- 75. Craig AP, Franca AS, Irudayaraj J. Surface-enhanced Raman spectroscopy applied to food safety. Annu Rev Food Sci Technol. 2013;4:369–80. doi: 10.1146/annurev-food-022811-101227. [DOI] [PubMed] [Google Scholar]
- 76. Blasco C, Pico Y. Determining nanomaterials in food. TrAC Trends Anal Chem. 2011;30:84–99. [Google Scholar]
- 77. Lin S, Reppert J, Hu Q, et al. Uptake, translocation, and transmission of carbon nanomaterials in rice plants. Small. 2009;5:1128–32. doi: 10.1002/smll.200801556. [DOI] [PubMed] [Google Scholar]
- 78. He L, Kim N-J, Li H, et al. Use of a fractal-like gold nanostructure in surface-enhanced Raman spectroscopy for detection of selected food contaminants. J Agr Food Chem. 2008;56:9843–7. doi: 10.1021/jf801969v. [DOI] [PubMed] [Google Scholar]
- 79. Chen S, Carroll DL. Silver nanoplates: size control in two dimensions and formation mechanisms. J Phys Chem B. 2004;108:5500–6. [Google Scholar]
- 80. Cheng W, Dong S, Wang E. Spontaneous fractal aggregation of gold nanoparticles and controlled generation of aggregate-based fractal networks at air/water interface. J Phys Chem B. 2005;109:19213–8. doi: 10.1021/jp052255a. [DOI] [PubMed] [Google Scholar]
- 81. Johnston BD, Scown TM, Moger J, et al. Bioavailability of nanoscale metal oxides TiO2, CeO2, and ZnO to fish. Environ Sci Technol. 2010;44:1144–51. doi: 10.1021/es901971a. [DOI] [PubMed] [Google Scholar]
- 82. Tolles WM, Nibler JW, McDonald JR, et al. A review of the theory and application of coherent anti-Stokes Raman spectroscopy (CARS) Appl Spectrosc. 1977;31:253–71. [Google Scholar]
- 83. Liu B, Zhou P, Liu X, et al. Detection of pesticides in fruits by surface-enhanced Raman spectroscopy coupled with gold nanostructures. Food Process Tech. 2013;6:710–8. [Google Scholar]
- 84. He L, Liu Y, Mustapha A, et al. Antifungal activity of zinc oxide nanoparticles against Botrytis cinerea and Penicillium expansum. Microbiol Res. 2011;166:207–15. doi: 10.1016/j.micres.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 85. Müller C, David L, Chis V, et al. Detection of thiabendazole applied on citrus fruits and bananas using surface enhanced Raman scattering. Food Chemistry. 2014;145:814–20. doi: 10.1016/j.foodchem.2013.08.136. [DOI] [PubMed] [Google Scholar]
- 86. Saute B, Narayanan R. Solution-based direct readout surface enhanced Raman spectroscopic (SERS) detection of ultra-low levels of thiram with dogbone shaped gold nanoparticles. Analyst. 2011;136:527–32. doi: 10.1039/c0an00594k. [DOI] [PubMed] [Google Scholar]
- 87. Saute B, Narayanan R. Solution-based SERS method to detect dithiocarbamate fungicides in different real-world matrices. J Raman Spectrosc. 2013;44:1518–22. [Google Scholar]
- 88. Saute B, Premasiri R, Ziegler L, et al. Gold nanorods as surface enhanced Raman spectroscopy substrates for sensitive and selective detection of ultra-low levels of dithiocarbamate pesticides. Analyst. 2012;137:5082–7. doi: 10.1039/c2an36047k. [DOI] [PubMed] [Google Scholar]
- 89. Jarvis RM, Goodacre R. Characterisation and identification of bacteria using SERS. Chem Soc Rev. 2008;37:931–6. doi: 10.1039/b705973f. [DOI] [PubMed] [Google Scholar]
- 90. Fan C, Hu Z, Mustapha A, et al. Rapid detection of food- and waterborne bacteria using surface-enhanced Raman spectroscopy coupled with silver nanosubstrates. Appl Microbiol Biotechnol. 2011;92:1053–61. doi: 10.1007/s00253-011-3634-3. [DOI] [PubMed] [Google Scholar]
- 91. Cowcher DP, Xu Y, Goodacre R. Portable, quantitative detection of Bacillus bacterial spores using surface-enhanced Raman scattering. Anal Chem. 2013;85:3297–302. doi: 10.1021/ac303657k. [DOI] [PubMed] [Google Scholar]
- 92. Lee PC, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86:3391–5. [Google Scholar]
- 93. Jarvis RM, Law N, Shadi IT, et al. Surface-enhanced Raman scattering from intracellular and extracellular bacterial locations. Anal Chem. 2008;80:6741–6. doi: 10.1021/ac800838v. [DOI] [PubMed] [Google Scholar]
- 94. Nicolaou N, Xu Y, Goodacre R. Fourier transform infrared and Raman spectroscopies for the rapid detection, enumeration, and growth interaction of the bacteria Staphylococcus aureus and Lactococcus lactis ssp cremoris in milk. Anal Chem. 2013;83:5681–7. doi: 10.1021/ac2008256. [DOI] [PubMed] [Google Scholar]
- 95.Williams AC. Handbook of Raman spectroscopy: from the research laboratory to the process line. In: Lewis IR, Howell G, Edwards M, editors. Practical spectroscopy series. New York: Marcel Dekker; 2001. pp. 575–92. [Google Scholar]
- 96. Pinzaru SC, Pavel I, Leopold N, et al. Identification and characterization of pharmaceuticals using Raman and surface-enhanced Raman scattering. J Raman Spectrosc. 2004;35:338–46. [Google Scholar]
- 97.Pinzaru SC, Pavel IE. SERS and pharmaceuticals Surface enhanced Raman spectroscopy. Berlin: Wiley-VCH Verlag GmbH & Co. KGaA; 2010. pp. 129–54. [Google Scholar]
- 98. West JL, Halas NJ. Engineered nanomaterials for biophotonics applications: improving sensing, imaging, and therapeutics. Annu Rev Biomed Eng. 2003;5:285–92. doi: 10.1146/annurev.bioeng.5.011303.120723. [DOI] [PubMed] [Google Scholar]
- 99. Yao J, Sun Y, Yang M, et al. Chemistry, physics and biology of graphene-based nanomaterials: new horizons for sensing, imaging and medicine. J Mater Chem. 2012;22:14313–29. [Google Scholar]
- 100. Lee JH, Yigit MV, Mazumdar D, et al. Molecular diagnostic and drug delivery agents based on aptamer-nanomaterial conjugates. Adv Drug Delivery Rev. 2010;62:592–605. doi: 10.1016/j.addr.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Espinosa JM, Christensen DH, Sorensen GO, et al. Low-frequency near-infrared Fourier transform Raman studies of ellipticines and deoxyribose nucleic acid. Spectrochim Acta A. 1991;47:1423–9. [Google Scholar]
- 102. Neville GA, Shurvell HF. Fourier transform Raman and infrared vibrational study of diazepam and four closely related 1,4-benzodiazepines. J Raman Spectrosc. 1990;21:9–19. [Google Scholar]
- 103. Eliasson C, Matousek P. Noninvasive authentication of pharmaceutical products through packaging using partial offset Raman spectroscopy. Anal Chem. 2007;79:1696–701. doi: 10.1021/ac062223z. [DOI] [PubMed] [Google Scholar]
- 104. Davies MC, Binns JS, Melia CD, et al. FT Raman spectroscopy of drugs in polymers. Int J Pharm. 1990;66:223–32. [Google Scholar]
- 105. Cunningham BT, Choi CJ, Watkins AR. Manufacture and use of SERS nanodome biosensor incorporated into tubing. 2012309080 A1 20121206. US Pat Appl Pub US.
- 106. Feng S, Chen R, Lin J, et al. Method for determining composition of Chinese medicine decoction by surface-enhanced Raman spectroscopy. Zhuanli Faming, Shenqing, editors. 101339132 A 20090107. People’s Republic of China; CN. 2009
- 107. Huang H, Shi H, Feng S, et al. Quick detection of traditional Chinese medicine ’Atractylodis macrocephalae rhizoma’ pieces by surface-enhanced Raman spectroscopy. Laser Phys. 2013;23:15601–4. [Google Scholar]
- 108.Huang R, Chen R, Chen Q, et al. Raman spectral study of anti-angiogenic drugs on the role of chick vascular. In: Luo Q, Wang LV, Tuchin VV, et al., editors. Eighth International Conference on Photonics and Imaging in Biology and Medicine. Bellingham WA: SPIE; 2009. p. 75191F. [Google Scholar]
- 109. He L, Lin M, Li H, et al. Surface-enhanced Raman spectroscopy coupled with dendritic silver nanosubstrate for detection of restricted antibiotics. J Raman Spectrosc. 2010;41:739–44. [Google Scholar]
- 110. Gigli M, Doglia SM, Millot JM, et al. Quantitative study of doxorubicin in living cell nuclei by microspectrofluorometry. Biochim Biophys Acta. 1988;950:13–20. doi: 10.1016/0167-4781(88)90068-1. [DOI] [PubMed] [Google Scholar]
- 111. Gigli M, Rasoanaivo WD, Millot JM, et al. Correlation between growth inhibition and intranuclear doxorubicin and 4′-deoxy-4′-iododoxorubicin quantitated in living K562 cells by microspectrofluorometry. Cancer Res. 1989;49:560–4. [PubMed] [Google Scholar]
- 112. Wang AHJ, Ughetto G, Quigley GJ, et al. Interactions between an anthracycline antibiotic and DNA: molecular structure of daunomycin complexed to d(CpGpTpApCpG) at 1.2-A resolution. Biochemistry. 1987;26:1152–63. doi: 10.1021/bi00378a025. [DOI] [PubMed] [Google Scholar]
- 113. Manfait M, Theophanides T. Fourier transform infrared spectra of cells treated with the drug adriamycin. Biochem Biophys Res Commun. 1983;116:321–6. doi: 10.1016/0006-291x(83)90417-5. [DOI] [PubMed] [Google Scholar]
- 114. Sequaris JM, Koglin E, Valenta P, et al. Surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids. Ber Bunsenges Phys Chem. 1981;85:512–3. [Google Scholar]
- 115. Nonaka Y, Tsuboi M, Nakamoto K. Comparative study of aclacinomycin versus adriamycin by means of resonance Raman spectroscopy. J Raman Spectrosc. 1990;21:133–41. [Google Scholar]
- 116. Smulevich G, Feis A. Surface-enhanced resonance Raman spectra of adriamycin, 11-deoxycarminomycin, their model chromophores, and their complexes with DNA. J Phys Chem. 1986;90:6388–92. [Google Scholar]
- 117. Smulevich G, Feis A, Mantini AR, et al. Resonance Raman and SERRS spectra of anti-tumor anthracyclines and their complexes with DNA. Indian J Pure Appl Phys. 1988;26:207–11. [Google Scholar]
- 118.Nabiev IR, Sokolov KV, Manfait M. SERS and its biomedical applications. In: Clark RJH, Hester RE, editors. Advances in spectroscopy. Chichester: Wiley; 1993. pp. 267–338. [Google Scholar]
- 119.Smith WE, Faults K, Graham D. Quantitative surface-enhanced resonance Raman spectroscopy for analysis. In: Kneipp K, Moskovits M, Kneipp H, editors. Surface-enhanced Raman scattering: physics and applications. Berlin: Wiley-VCH Verlag; 2006. [Google Scholar]
- 120. McLaughlin C, MacMillan D, McCardle C, et al. Quantitative analysis of mitoxantrone by surface-enhanced resonance Raman scattering. Anal Chem. 2002;74:3160–7. doi: 10.1021/ac010067k. [DOI] [PubMed] [Google Scholar]
- 121. Sulk RA, Corcoran RC, Carron KT. Surface-enhanced Raman scattering detection of amphetamine and methamphetamine by modification with 2-mercaptonicotinic acid. Appl Spectrosc. 1999;53:954–9. [Google Scholar]
- 122. Sagmuller B, Schwarze B, Brehm G, et al. Application of SERS spectroscopy to the identification of (3,4-methylenedioxy)amphetamine in forensic samples utilizing matrix stabilized silver halides. Analyst. 2001;126:2066–71. doi: 10.1039/b105321n. [DOI] [PubMed] [Google Scholar]
- 123. Faulds K, Smith WE, Graham D, et al. Assessment of silver and gold substrates for the detection of amphetamine sulfate by surface enhanced Raman scattering (SERS) Analyst. 2002;127:282–6. doi: 10.1039/b107318b. [DOI] [PubMed] [Google Scholar]
- 124. Mabbott S, Correa E, Cowcher DP, et al. Optimization of parameters for the quantitative surface-enhanced Raman scattering detection of mephedrone using fractional factor design and portable Raman spectrometer. Anal Chem. 2013;85:923–31. doi: 10.1021/ac302542r. [DOI] [PubMed] [Google Scholar]
- 125. Feng S, Chen W, Huang W, et al. Surface-enhanced Raman spectroscopy of morphine in silver colloid. Chin Opt Lett. 2009;7:1055–107. [Google Scholar]
- 126. Hong X, Chu X, Zou P, et al. Magnetic-field-assisted rapid ultrasensitive immunoassays using Fe3O4/ZnO/Au nanorices as Raman probes. Biosens Bioelectron. 2010;26:918–22. doi: 10.1016/j.bios.2010.06.066. [DOI] [PubMed] [Google Scholar]
- 127. Deb SK, Davis B, Ben-Amotz D, et al. Accurate concentration measurements using surface-enhanced Raman and deuterium exchanged dye pairs. Appl Spectrosc. 2008;62:1001–7. doi: 10.1366/000370208785793290. [DOI] [PubMed] [Google Scholar]
- 128. Stosch R, Henrion A, Schiel D, et al. Surface-enhanced Raman scattering based approach for quantitative determination of creatinine in human serum. Anal Chem. 2005:7386–92. doi: 10.1021/ac0511647. [DOI] [PubMed] [Google Scholar]
- 129. Zakel S, Rienitz O, Guerrler B, et al. Double isotope dilution surface-enhanced Raman scattering as a reference procedure for quantification of biomarkers in human serum. Analyst. 2011;136:3956–61. doi: 10.1039/c1an15207f. [DOI] [PubMed] [Google Scholar]
- 130. Zhang D, Xie Y, Deb SK, et al. Isotop edited internal standard method for quantitative surface-enhanced Raman spectroscopy. Anal Chem. 2005;77:3563–9. doi: 10.1021/ac050338h. [DOI] [PubMed] [Google Scholar]
- 131. Zakel S, Wundrack S, O’Connor G, et al. Validation of isotope dilution surface-enhanced Raman scattering (IDSERS) as a higher order reference method for clinical measurands employing international comparison schemes. J Raman Spectrosc. 2013;44:1246–52. [Google Scholar]
- 132.Greve J, Puppels GJ. Raman microspectroscopy of single whole cells. In: Clark RJH, Hester RE, editors. Advances in spectroscopy. Chichester: Wiley; 1993. pp. 231–65. [Google Scholar]
- 133. Kneipp K, Haka A, Kneipp H, et al. Surface-enhanced Raman spectroscopy in single living cells using gold nanoparticles. Appl Spectrosc. 2002;56:150–4. [Google Scholar]
- 134.Kneipp J. Nanosensors based on SERS for application living cells. In: Kneipp K, Moskovits M, Kneipp H, editors. Surface-enhanced Raman scattering: physics and applications. Berlin Heidelberg: Springer-Verlag; 2006. [Google Scholar]
- 135.Kneipp J, Kneipp H, Kneipp K, et al. Surface-enhanced Raman scattering for investigations of eukaryotic cells. In: Lasch P, Kneipp J, editors. Biomedical vibrational spectroscopy. Hoboken, New Jersey: Wiley; 2008. pp. 243–61. [Google Scholar]
- 136. Khodorchenko P, Petukhov A, Nabiev I, et al. Contributions of short-range and classical electromagnetic mechanisms to surface-enhanced Raman scattering from several types of biomolecules adsorbed on cold deposited island films. Appl Spectrosc. 1993;47:515–22. [Google Scholar]
- 137. Van Manen HJ, Kraan YM, Roos D, et al. Single-cell Raman and fluorescence microscopy reveal the association of lipid bodies with phagosomes in leukocytes. Proc Natl Acad Sci USA. 2005;102:10159–64. doi: 10.1073/pnas.0502746102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Zong S, Wang Z, Chen H, et al. Surface enhanced Raman scattering traceable and glutathione responsive nanocarrier for the intracellular drug delivery. Anal Chem. 2013;85:2223–30. doi: 10.1021/ac303028v. [DOI] [PubMed] [Google Scholar]
- 139.Matthäus C, Chernenko T, Quintero L, et al. Raman micro-spectral imaging of cells and intracellular drug delivery using nanocarrier systems. In: Dieing T, Hollricher O, Toporski J, editors. Confocal Raman microscopy. Berlin Heidelberg: Springer; 2011. pp. 137–63. [Google Scholar]
- 140. Zavaleta C, de la Zerda A, Liu Z, et al. Noninvasive Raman spectroscopy in living mice for evaluation of tumor targeting with carbon nanotubes. Nano Lett. 2008;8:2800–5. doi: 10.1021/nl801362a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Liu Z, Li X, Tabakman SM, et al. Multiplexed multicolor Raman imaging of live cells with isotopically modified single walled carbon nanotubes. J Am Chem Soc. 2008;130:13540–1. doi: 10.1021/ja806242t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Deckert V, Zeisel D, Zenobi R, et al. Near-field surface enhanced Raman imaging of dye-labeled DNA with 100-nm resolution. Anal Chem. 1998;70:2646–70. doi: 10.1021/ac971304f. [DOI] [PubMed] [Google Scholar]
- 143. Zeisel D, Deckerr V, Zenobi R, et al. Near-field surface enhanced Raman spectroscopy of dye molecules adsorbed on silver island films. Chem Phys Lett. 1998;283:381–5. [Google Scholar]
- 144. Huang H, Chen W, Pan J, et al. SERS spectra of a single nasopharyngeal carcinoma cell based on intracellularly grown and passive uptake Au nanoparticles. Spectrosc. 2011;26:187–94. [Google Scholar]
- 145. Lin J, Chen R, Feng S, et al. Rapid delivery of silver nanoparticles into living cells by electroporation for surface-enhanced Raman spectroscopy. Biosens Bioelectron. 2009;25:388–94. doi: 10.1016/j.bios.2009.07.027. [DOI] [PubMed] [Google Scholar]
- 146. Nguyen CT, Nguyen JT, Rutledge S, et al. Detection of chronic lymphocytic leukemia cell surface markers using surface enhanced Raman scattering gold nanoparticles. Cancer Lett. 2010;292:91–7. doi: 10.1016/j.canlet.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 147. Yigit MV, Zhu L, Ifediba MA, et al. Noninvasive MRI-SERS imaging in living mice using an innately bimodal nanomaterial. ACS Nano. 2013;5:1056–66. doi: 10.1021/nn102587h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Marquis BJ, Love SA, Braun KL, et al. Analytical methods to assess nanoparticle toxicity. Analyst. 2009;134:425–39. doi: 10.1039/b818082b. [DOI] [PubMed] [Google Scholar]
- 149. Yamamoto T, Murakami Y, Motoyanagi J, et al. An analytical system for single nanomaterials: combination of capillary electrophoresis with Raman spectroscopy or with scanning probe microscopy for individual single-walled carbon nanotube analysis. Anal Chem. 2009;81:7336–41. doi: 10.1021/ac901197v. [DOI] [PubMed] [Google Scholar]
- 150. Wei Y, Cao C, Jin R, et al. Nanoparticles with Raman spectroscopic fingerprints for DNA and RNA detection. Science. 2002;297:1536–40. doi: 10.1126/science.297.5586.1536. [DOI] [PubMed] [Google Scholar]
- 151. Kneipp J, Kneipp H, Kneipp K. SERS - a single-molecule and nanoscale tool for bioanalytics. Chem Soc Rev. 2008;37:1052–60. doi: 10.1039/b708459p. [DOI] [PubMed] [Google Scholar]
- 152. Kneipp J, Kneipp H, Kneipp K. Two photon vibrational spectroscopy for biosciences based on surface-enhanced hyper-Raman scattering. Proc Natl Acad Sci USA. 2006;103:17149–53. doi: 10.1073/pnas.0608262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Jarvis RM, Johnson HE, Olembe E, et al. Towards quantitatively reproducible substrates for SERS. Analyst. 2008;133:1449–52. doi: 10.1039/b800340h. [DOI] [PubMed] [Google Scholar]