ABSTRACT
Cell junctions maintain the blood-tissue barriers to preserve vascular and tissue integrity. Viral infections reportedly modulate cell–cell junctions to facilitate their invasion. However, information on the effect of COVID-19 infection on the gene expression of cell junction and cytoskeletal proteins is limited. Using the Gene Expression Omnibus and Reactome databases, we analyzed the data on human lung A549, NHBE, and Calu-3 cells for the expression changes in cell junction and cytoskeletal proteins by SARS-CoV-2 (CoV-2) infection. The analysis revealed changes in 3,660 genes in A549, 100 genes in NHBE, and 592 genes in Calu-3 cells with CoV-2 infection. Interestingly, EGOT (9.8-, 3- and 8.3-fold; p < .05) and CSF3 (4.3-, 33- and 56.3-fold; p < .05) were the only two genes significantly elevated in all three cell lines (A549, NHBE and Calu-3, respectively). On the other hand, 39 genes related to cell junctions and cytoskeleton were modulated in lung cells, with DLL1 demonstrating alterations in all cells. Alterations were also seen in several miRNAs associated with the cell junction and cytoskeleton genes modulated in the analysis. Further, matrix metalloproteinases involved in disease pathologies, including MMP-3, -9, and -12 demonstrated elevated expression on CoV-2 infection (p < .05). The study findings emphasize the integral role of cell junction and cytoskeletal genes in COVID-19, suggesting their therapeutic potential. Our analysis also identified a distinct EGOT gene that has not been previously implicated in COVID-19. Further studies on these newly identified genes and miRNAs could lead to advances in the pathogenesis and therapeutics of COVID-19.
KEYWORDS: COVID-19, cell junctions, claudins, cytoskeleton, miRNA, MMP, SARS-CoV-2
1. Introduction
Coronaviruses that project spikes from the membrane when seen under an electron microscope1 seriously impact human and animal health as they mainly cause enteric or respiratory diseases.2 Recent studies indicated a novel coronavirus, SARS-CoV-2 (CoV-2), responsible for coronavirus disease 2019 (COVID-19).3–5 The entry of CoV-2 into human cells has been identified to be facilitated by angiotensin‐converting enzyme 2 (ACE2)6 that is abundantly expressed in human airway epithelia, lung parenchyma, vascular endothelial, kidney cells, and small intestine cells, etc.7,8 Among these, the alveolar epithelial type II cells (ATII)9,10 represent 83% of the total ACE2 positive cell population.11 Interestingly, ACE2-expressing ATII cells play a vital role in alveolar epithelial repair and regeneration by exerting innate immune responses and serving as progenitors for ATI cells.12
Viral infections resemble bacterial manifestations13 and target elements in hosts.14 They reportedly involve alteration of cell junctions to facilitate the invasion of a virus into its host since these junctions also deal with barrier protection14–16 apart from maintaining vascular integrity.15 While several viruses bind to receptors present on the apical side of epithelial cells, others target basolateral receptors, which are usually inaccessible due to the tight junction (TJ) barrier. Therefore, these viruses have established mechanisms to disassemble TJs either through regulation of protein expression or localization to reach their receptors.17 On the contrary, claudins which are integral components of the TJ complex,18 are also required for certain viruses to enter the hosts.19,20 For example, claudin-1 (CLDN1) is a TJ protein that acts as an essential host factor for the hepatitis C virus.19,20 Interestingly, the dengue virus is reportedly involved in the expression downregulation of PATJ and CRTAP genes, which play essential roles in maintaining cell-junction integrity.21 Apart from the transcriptional and translational regulation, microRNAs (miRNAs) play an extensive role in the post-transcriptional regulation of genes. Interestingly, methods such as miRNA mimics, miRNA antagonists, or CRISPR/Cas9 can potentially modulate the expression of target genes.22
Information on the involvement of cytoskeletal and cell-junction molecules in the novel COVID-19 disease is minimal. Therefore, we performed this bioinformatics study using the genomic data available in the GEO database to identify differentially expressed cytoskeleton- and cell junction-associated genes in the CoV-2 infected lung cells and compared them against the respective mock-treated controls. Our analyses provide novel genomic information regarding alterations in cell-junction and cytoskeletal gene expression. Furthermore, our study also identified several miRNAs associated with the regulation of cell-junction and cytoskeletal gene expression upon CoV-2 infection, thus providing potential clues for future research on COVID-19.
2. Materials and methods
2.1. Data collection
We used NCBI’s Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/),23 a public functional array, and sequence-based genomics data repository to obtain the expression profiling by high throughput sequencing generated by Blanco-Melo D et al.24 on CoV-2. The data set, acquired from GEO ID: 200147507; Accession: GSE147507, is comprised of 23,710 genes based on primary human lung epithelium (NHBE), human alveolar epithelial (A549), and mesothelial epithelium (Calu-3) cell lines either mock-treated or infected with CoV-2 virus (USA-WA1/2020).
2.2. Data selection & analysis
The data were filtered to select genes associated with cell junctions and cytoskeletal remodeling using InnateDB (https://www.innatedb.com/)25 database. Genes that exhibited a significant difference (p < .05) in the infected group compared to the mock-treated group were selected for further analyses. Mean values of each group for all the genes were calculated following the determination of the fold-change difference in the infected group against their mock-treated counterpart. Finally, these genes were categorized as up-regulated or down-regulated based on the resulting positive or negative fold change figures, respectively. Significantly modulated genes and miRNAs were interlinked using a microRNA database ‘miRDB’ (http://mirdb.org/).26
2.3. Reactome pathway database
Reactome (https://reactome.org/),27 a web-based open-source, open access, manually curated, and peer-reviewed pathway database, was used in the study to interpret and analyze the alterations in gene expression. Selected genes were added to the system’s ‘analyze data’ panel, and the resulting pathways associated with these genes were obtained.
2.4. Statistical analysis
All the study findings are presented as fold change increase or decrease in the CoV-2 infected group against the control group. The ‘n’ values in the figures signify the number of samples in a particular group. The data were analyzed by parametric testing using a two-tailed, homoscedastic t-test, followed by the post hoc test using the GraphPad Prism 6.01 software. Data are represented as mean ± SD, and p < .05 was considered significant.
3. Results
3.1. CoV-2 infection induces gene expression changes in human lung epithelial cells
Out of a total of 23,710 genes available in the database, 3,660 genes from A549, 100 genes from NHBE, and 592 genes from Calu-3 cell lines demonstrated significant alterations in treatment with CoV-2 virus against their respective controls (Figure 1(a)). Interestingly, two of these genes were found to be mutually modulated in all three cell lines (Figure 1(b)). Both EGOT (Eosinophil Granule Ontogeny Transcript) and CSF3 (Colony Stimulating Factor 3) were significantly increased across all the cell lines on CoV-2 insult (Figure 1(c)). While EGOT demonstrated a 3- to 10-fold change increase across various cell lines, CSF3 was found to be elevated by 4- to 56-folds in these cell lines. Reactome analysis for EGOT and CSF3 was performed to determine the signaling pathways that can be triggered by these genes, and interestingly, CSF3 was found to be associated with five pathways (Table 1) that include interleukin-10 and cytokine signaling in the immune system.28
Figure 1.
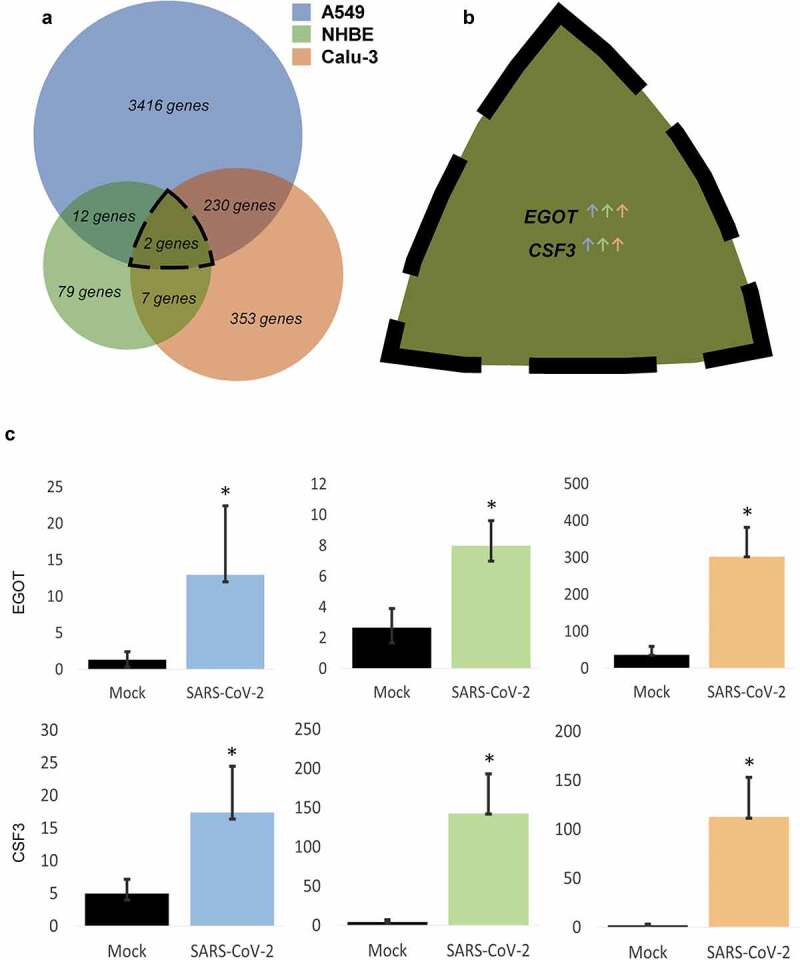
Alterations in gene expression on CoV-2 infection across various lung cell lines. A. Venn diagram of significantly modulated genes in A549, NHBE, and Calu-3 cell lines. B. List and C. Quantification of commonly altered genes in all three cell lines. Data represented as mean ± SD; *p < .05; n = 6, A549; 3, NHBE and Calu-3. Total genes = 3660, A549; 100, NHBE; 592, Calu-3. ↑, upregulated; ↓, downregulated.
Table 1.
Reactome analysis-based signaling pathways associated with CSF3, which was altered in all the three cell lines.
| Pathway name | Entities |
Reactions |
|||
|---|---|---|---|---|---|
| Found | Total | p-Value | Found | Total | |
| Interleukin-10 signaling | 2 | 86 | 1.05E-04 | 1 | 15 |
| Inactivation of CSF3 (G-CSF) signaling* | 1 | 27 | 0.00556 | 5 | 9 |
| Signaling by Interleukins | 2 | 643 | 0.005692 | 3 | 493 |
| Signaling by CSF3 (G-CSF) | 1 | 35 | 0.007203 | 15 | 21 |
| Cytokine Signaling in Immune system | 2 | 1092 | 0.01607 | 18 | 708 |
p < 0.05; n = 6, A549, NHBE and Calu-3. *Does not apply to the current findings as CSF3 was increased
3.2. CoV-2 infection induces changes in cell-junction and cytoskeletal gene expression in lung epithelial cells
The expression of a total of 39 genes related to cell-barrier regulation and cytoskeletal remodeling was modulated in A549, NHBE, and Calu-3 cell lines during infection with CoV-2 (Figure 2(a)). Intriguingly, DLL1 (Delta Like Canonical Notch Ligand 1) was commonly modulated in A549, and Calu-3 cell lines (Figure 2(b)). When it comes to cadherins, CDH-2 and -3 were increased in A549, whereas CDH-4 and -5 were elevated in the Calu-3 cell line. Claudins were also modulated in two of these cell lines, with A549 demonstrating upregulation of CLDN-1, -4, -12, and -16, while NHBE cells exhibited downregulation of CLDN19 (Figure 2(c)). Apart from the aforesaid vital elements of the cell junctions, OCLN (Occludin) and ICAM1 (Intercellular adhesion molecule 1) were also increased on CoV-2 infection in A549 Calu-3 cell lines, respectively. Individual Reactome analyses were performed for the altered genes in all three cell lines to determine the potential signaling pathways that can be activated in CoV-2 infection (Table 2). Cell junction organization, cell communication, and TJ interaction were the commonly observed pathways evident in all three cell lines. Moreover, DLL1 in the A549 and Calu-3 cell lines indicated transcription regulation by MECP2 (Methyl-CpG Binding Protein 2). Finally, interleukin-10 and -12 and adherens junction (AJ) interactions were observed in the A549 and Calu-3 cell lines during CoV-2 infection.28
Figure 2.
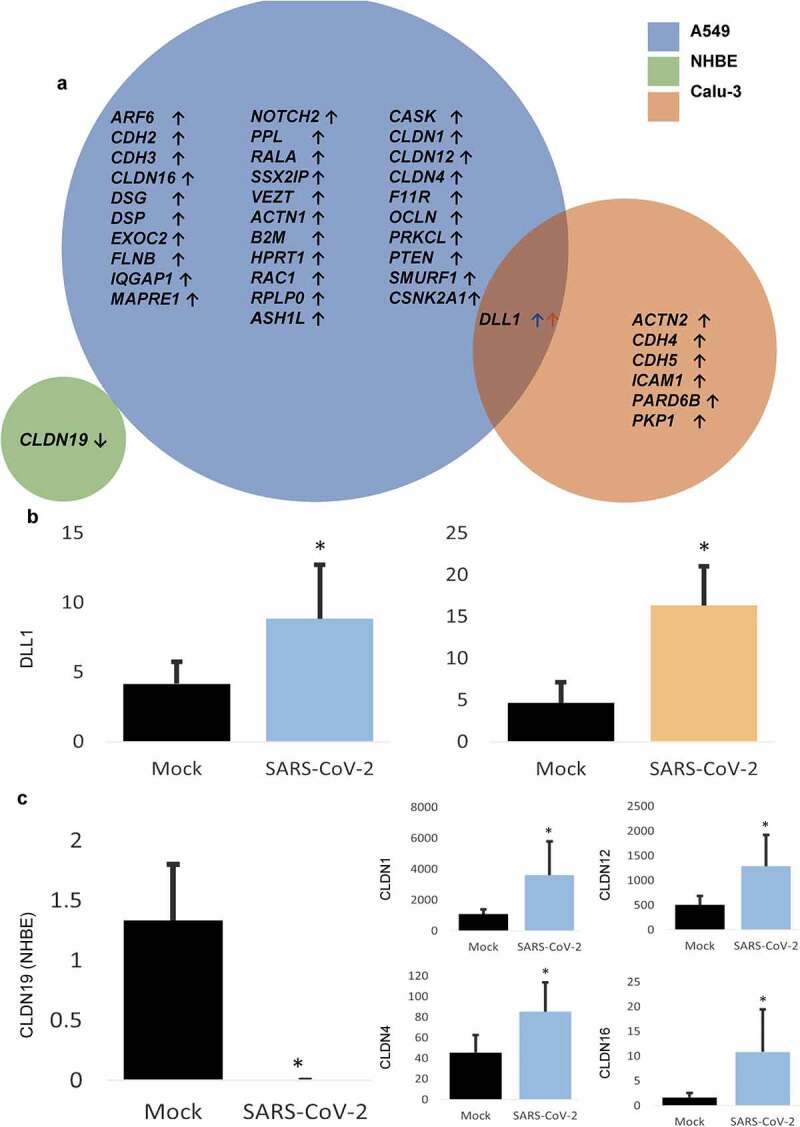
CoV-2 induced modulation of cytoskeleton and cell junction-associated genes. A. List of significantly altered genes on CoV-2 infection across various cell lines. B. Expression quantification of the commonly modulated gene in A549 and Calu-3 cell lines on CoV-2 infection. C. Quantification of various claudins modulated in NHBE and A549 cell lines on CoV-2 infection. Data represented as mean ± SD; *p < .05; n = 6, A549; 3, NHBE and Calu-3. Total genes = 32, A549; 1, NHBE; 7, Calu-3. ↑, upregulated; ↓, downregulated.
Table 2.
Signaling Pathways revealed in Reactome analysis for genes modulated by SARS-CoV-2 infection in various cell lines.
|
Pathway name |
Entities |
Reactions |
Genes involved |
|||||||
|
Found |
Total |
p-value |
Found |
Total |
||||||
|
CoV2-modulated signaling pathways in A549 cells | ||||||||||
| Cell-Cell communication | 11 | 133 | 4.82E-13 | 8 | 60 | CLDN-1,4,12,16;CDH-2,3;ACTN1;CASK;IQGAP1; F11R | ||||
| Cell junction organization | 9 | 94 | 2.15E-11 | 5 | 37 | CLDN-1,4,12,16;CDH-2,3;ACTN1;F11R | ||||
| Cell-cell junction organization | 8 | 67 | 5.44E-11 | 4 | 21 | CLDN-1,4,12,16;CDH-2,3;F11R | ||||
| Tight junction interactions | 5 | 30 | 5.33E-08 | 2 | 3 | CLDN-1,4,12,16;F11R | ||||
| Nephrin family interactions | 3 | 25 | 7.78E-05 | 3 | 13 | ACTN1;CASK;IQGAP1 | ||||
| JAK-STAT signaling (interleukin-12 stimulation) | 4 | 73 | 9.53E-05 | 2 | 36 | RALA;RPLP0 | ||||
| Interleukin-12 signaling | 4 | 84 | 1.63E-04 | 2 | 56 | RALA;RPLP0 | ||||
| Adherens junctions interactions | 3 | 35 | 2.09E-04 | 2 | 16 | CDH3;CDH2 | ||||
| Interleukin-12 family signaling | 4 | 96 | 2.70E-04 | 2 | 114 | RALA;RPLP0 | ||||
| Transcriptional Regulation by MECP2 |
4 |
100 |
3.15E-04 |
4 |
77 |
PTEN;DLL1 |
||||
|
CoV2-modulated signaling pathways in Calu-3 cells | ||||||||||
| Cell-Cell communication | 4 | 133 | 1.41E-06 | 4 | 60 | CDH5;PARD6B;CDH4;ACTN2 | ||||
| Cell-cell junction organization | 3 | 67 | 1.15E-05 | 3 | 21 | CDH5;PARD6B;CDH4 | ||||
| Cell junction organization | 3 | 94 | 3.13E-05 | 3 | 37 | CDH5;PARD6B;CDH4 | ||||
| MECP2 regulates neuronal ligands transcription | 2 | 13 | 3.58E-05 | 2 | 8 | DLL1 | ||||
| Adherens junctions interactions | 2 | 35 | 2.57E-04 | 2 | 16 | CDH5;CDH4 | ||||
| Interleukin-10 signaling | 2 | 86 | 0.001525 | 1 | 15 | ICAM1 | ||||
| Transcriptional Regulation by MECP2 | 2 | 100 | 0.002051 | 2 | 77 | DLL1 | ||||
| Signaling by NOTCH1 Translocation Mutant | 1 | 8 | 0.005488 | 1 | 5 | DLL1 | ||||
| Apoptotic cleavage of cell adhesion proteins |
1 |
11 |
0.007539 |
1 |
10 |
PKP1 |
||||
|
CoV2-modulated signaling pathways in NHBE cells | ||||||||||
| Tight junction interactions | 1 | 30 | 0.002586 | 1 | 3 | CLDN19 | ||||
| Cell-cell junction organization | 1 | 65 | 0.005603 | 1 | 21 | CLDN19 | ||||
| Cell junction organization | 1 | 92 | 0.007931 | 1 | 37 | CLDN19 | ||||
| Cell-Cell communication | 1 | 130 | 0.011207 | 1 | 60 | CLDN19 | ||||
p<0.05; n=6, A549; 3, NHBE and Calu-3. Total genes=32, A549; 1, NHBE; 7, Calu-3.
3.3. Expression of several MMPs is altered in lung epithelial cells upon CoV-2 infection
Our analysis of the effect of CoV-2 infection on matrix metaloproteases (MMPs) revealed increased expression of several MMPs, such as the MMP-3, MMP-9, MMP-12, and MMP-13 in various lung epithelial cell lines. Interestingly, MMP9 was found to be up-regulated in NHBE and A549 cells, while MMP13 was elevated in NHBE, and Calu-3 cells infected with CoV-2 (Figure 3(a)). Apart from these, A549 cells demonstrated increased levels of MMP-3, −11, and −12 (Figure 3(b)).
Figure 3.
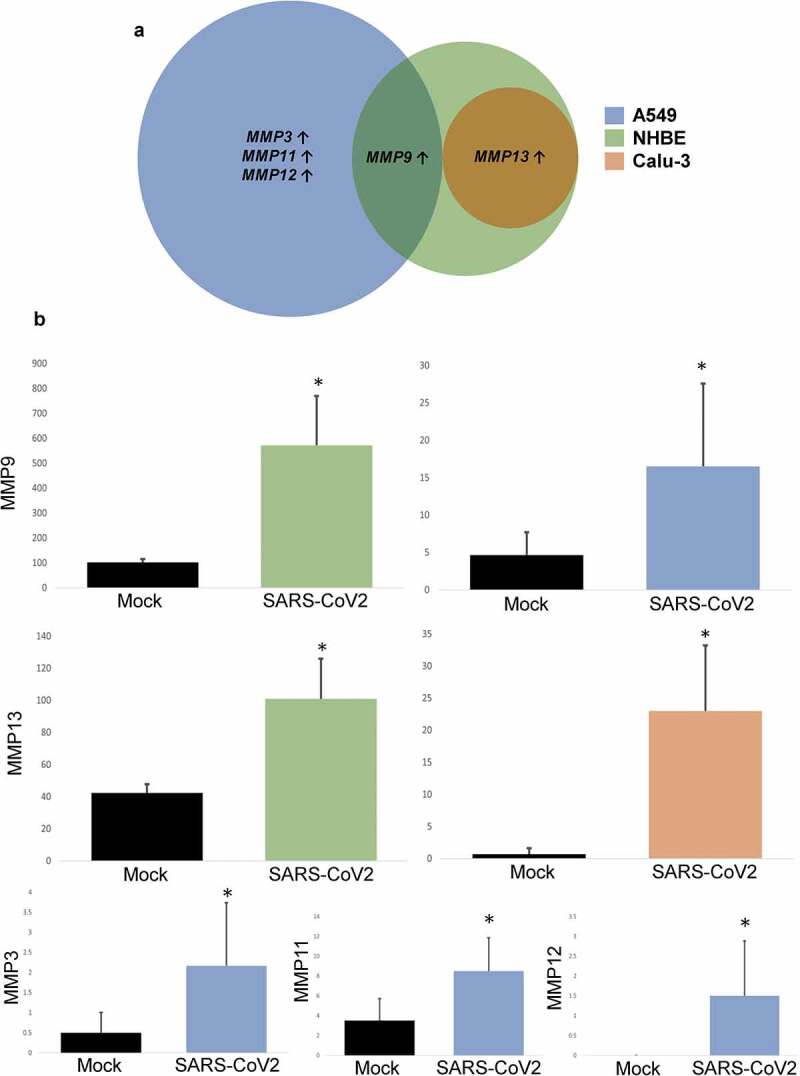
Effect of CoV-2 infection on MMPs. A. Venn diagram and B. Expression quantification of significantly altered MMPs (MMP3, MMP9, MMP12 and MMP13) in various lung cell lines. Data represented as mean ± SD; *p < .05; n = 6, A549; 3, NHBE and Calu-3. Total MMPs = 4, A549; 2, NHBE; 9, Calu-1. ↑, upregulated.
3.4. Modulation of microRNAs on CoV-2 infection
CoV-2 infected lung cells also demonstrated alterations in the expression of several miRNAs, including miR503 and miR4263, which were mutually modulated in A549/NHBE and A549/Calu-3 cell lines, respectively (Figure 4(a,b)). Although NHBE cells demonstrated no modulation in any other microRNA, A549 and Calu-3 cell lines were involved in alteration of 16 and 8 other microRNAs, respectively, upon CoV-2 insult.
Figure 4.
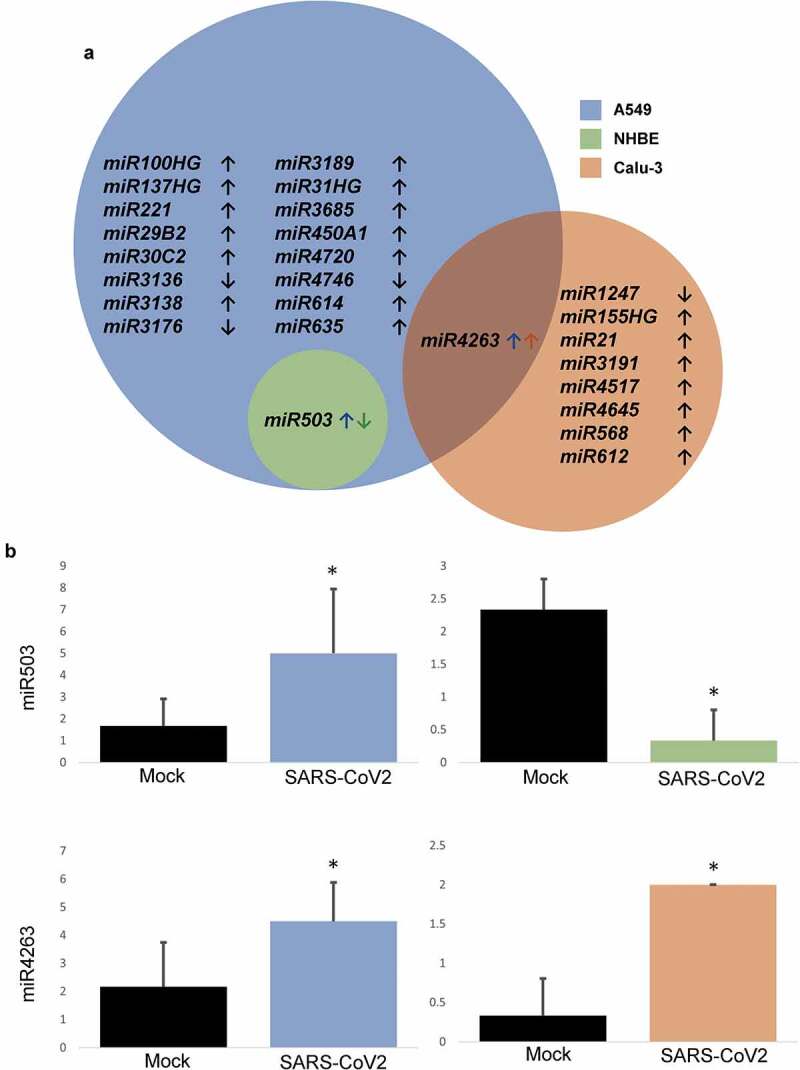
MicroRNAs altered on CoV-2 infection. A. Venn diagram demonstrating significantly modulated miRNAs across various cell lines. B. Expression of mutually modulated miRNAs on CoV-2 infection against mock-treated control. Data represented as mean ± SD; *p < .05; n = 6, A549; 3, NHBE and Calu-3. Total miRNAs = 18, A549; 1, NHBE; 9, Calu-3. ↑, upregulated; ↓, downregulated.
3.5. Genes regulated by microRNAs on CoV-2 infection
With the help of the miRNA database, we determined if miRNAs influence the observed changes in cytoskeletal and cell-barrier junction-related genes. The only microRNA from NHBE, miR503, demonstrated no link with cytoskeletal and cell junctions related genes. Interestingly, Calu-3 cells revealed a probable cause of ICAM1 upregulation associated with the observed increase in the expression level of miR612. Furthermore, the analysis remarkably disclosed direct links between 7 miRNAs and 12 cytoskeletal and cell–cell barrier junctions related genes (Figure 5). Intriguingly, miR4263 was found to be associated only with A549 cell-line genes despite being modulated in both A549 and Calu-3 cell lines. Further, all miRNAs, except the miR3176, were found to be upregulated upon CoV-2 infection in our analyses. Some significant associations from the study include CDH2 upregulation by miR450A1 and increased CLDN-4 and −12 expression through miR-3189 and −221, respectively.
Figure 5.
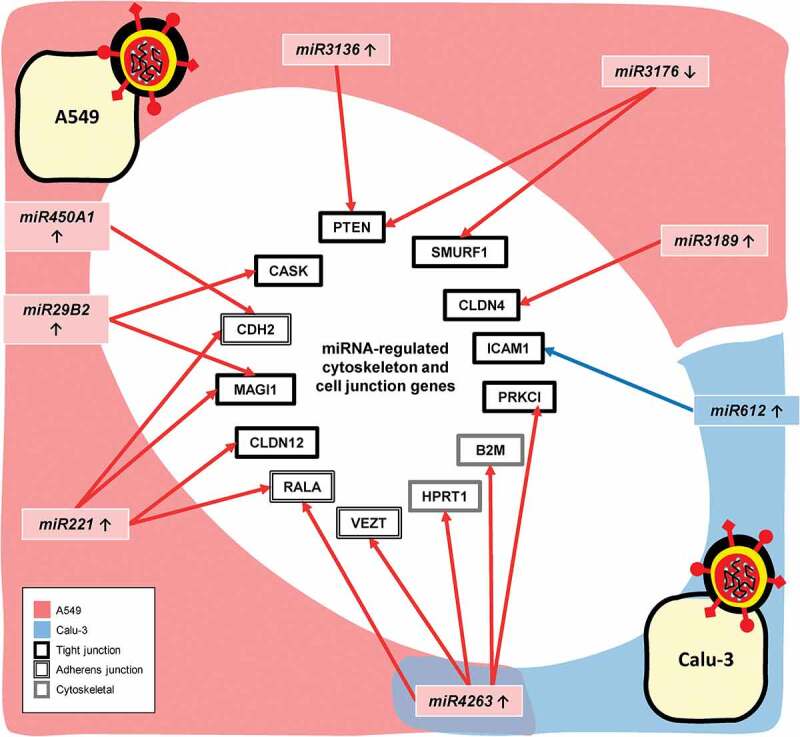
CoV-2 induced modulation of microRNAs linked to the changes in the cytoskeleton and cell junction-related genes in A549 and Calu-3 cell lines. p < .05; n = 6, A549; 3, Calu-3; ↑, upregulated; ↓, downregulated.
4. Discussion
A flurry of recent studies indicate a plethora of changes in the signaling pathways upon CoV-2 infection involving, but not limited to, alterations in gene expression and miRNAs.29,30 While ACE2 receptors have been indicated to be integral to CoV-2 cellular entry and infection,31 the virus entry into the blood vessels, its spread to other organs, and resulting COVID-19 complications such as thrombosis32,33 are all dependent on the ability of the CoV-2 to cross the blood-tissue barrier. As of today, the studies that report how CoV-2 modulates cell–cell junctions and cytoskeletal remodeling to cross the blood-tissue barrier are minimal. In the current study, using the data available in the GEO repository, we performed bioinformatics analysis to reveal changes in the expression of genes in three different human lung epithelial cell lines that make-or-break cell–cell junctions and those modulating cytoskeletal dynamics in the opening and closure of cell–cell junctions in response to CoV-2 infection.
The overall analysis of genes in human lung A549, NHBE, and Calu-3 cell lines identified only two genes altered by CoV-2 infection in all three of them. One of them is EGOT, a long non-coding RNA known to be elevated in viral infections to counter the antiviral response.34,35 EGOT can be a potential target in COVID-19, since its depletion in other viral infections stimulates interferon-related genes, thereby ameliorating viral replication.35 The other gene modulated by CoV-2 in all three cell lines is CSF3, which is also known to be expressed in viral illnesses that involve respiratory diseases,36 including COVID-19.37 Concerning the genes modulating cell–cell junctions, the CDH4 gene that encodes R-cadherin38 was found to be up-regulated in Calu-3 cells infected with CoV-2. R-cadherin reportedly promotes cell motility.39 A study reports miR29B2-5p as one of the miRNAs up-regulated on CoV-2 infection in the lung epithelium.40 Our findings reveal a similar elevation of miR29B2 in A549 cells upon CoV-2 infection.
On the other hand, TJs revealed remarkable changes in the expression of several core proteins upon CoV-2 infection in the lung epithelial cells. Alterations in the expression of several claudins in the pathological states reflect the critical role of these elements. CLDN4 is one such claudin elevated in acute lung injury,41 which is characterized by aberrant vascular leakage.42 Interestingly, it is one of the four claudins up-regulated in the current analysis in A549 cells on CoV-2 injury. Another notable finding among claudins is related to CLDN16, which is absent in the human lungs [60], but CoV-2 injury on A549 cells led to a significant increase in the expression of CLDN16, thereby signifying a critical role of this gene in the pathophysiological state. CLDN16 is well known to interact with CLDN19 to form a TJ complex,43 and interestingly CLDN19 was also modulated in our study in the NHBE cells but not in the A549 cell line. Similarly, CoV-2 induced upregulation of CLDN1, a barrier-forming claudin similar to CLDN5 [38], is anticipated to rescue the barrier integrity in the injured A549 cells. These findings collectively reveal modulation of claudins in CoV-2 infection in vitro and signify the likelihood of similar changes in these TJ elements during the same infection in vivo.
MMPs are reported to be involved in destructive pulmonary pathologies, especially in the disruption of the alveolar epithelial-endothelial barrier.44,45 It is noteworthy that increased MMP3 in the A549 cell line coincides with a previous finding from our laboratory, which demonstrated higher expression of MMP3 in lung injury.44,46 Remarkably, MMP3 was detected in the broncho-alveolar lavage (BAL) fluid obtained from acute respiratory distress syndrome, making it a potential biomarker for diagnosing the disease.44 Similarly, MMP9 is another proteinase that was enhanced by coronavirus infection in human monocytes.47 The current analysis revealed a similar increase in MMP9 expression upon CoV-2 infection in lung epithelial cells, further validating our findings. In addition to upregulation, we witnessed newly expressed MMP12 in CoV-2 infected cells compared to its absence in the mock-treated controls. In summary, the observed changes in the expression of various MMPs suggest their potential involvement in the epithelial-barrier disruption in response to CoV-2 infection. Limitations in this study include the lack of data based on human specimens, such as BAL fluid from control and COVID-19 patients, and the absence of evidence that links these genes directly to CoV-2 infection and COVID-19 severity. Nevertheless, our findings have important translational relevance as the identified genes may potentially prove to be novel druggable targets to treat COVID-19 patients.
5. Conclusions
Findings from the current study shed light on the integral role of cytoskeletal and cell–cell junction genes in the human lung epithelial cell lines upon CoV-2 infection. Apart from the cytoskeletal and junctional genes, our analysis also identified the distinct EGOT gene expression modulation by CoV-2 infection, which was not previously reported. The study findings would give researchers new insights into the pathogenesis of COVD-19. Further research on the newly identified CoV-2-modulated genes and microRNAs could potentially lead to breakthroughs in unraveling the disease complications and identifying therapies for COVID-19.
Funding Statement
Funds were provided by the NHLBI grant National Heart, Lung, and Blood Institute R01HL103952 and NCATS grant the National Center for Advancing Translational Sciences UL1TR002378 to PRS. Partial funding by NEI grant National Eye Institute R01EY028569 to SPN is also acknowledged.
Disclosure statement
The authors declare that there are no financial or other conflicts of interest exist.
Author contributions
Conception and design: MSA, SPN, and PRS; Data production, analysis, and interpretation: MSA, DK, SPN, and PRS; writing the manuscript: MSA, DK, and PRS. All authors reviewed the manuscript.
References
- 1.Su S, Wong G, Shi W, Liu J, Lai ACK, Zhou J, Liu W, Bi Y, Gao GF.. Epidemiology, genetic recombination, and pathogenesis of coronaviruses. Trends Microbiol. 2016;24(6):1–12. doi: 10.1016/j.tim.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui J, Li F, Shi ZL.. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lake MA. What we know so far: COVID-19 current clinical knowledge and research. Clin Med (Lond). 2020;20(2):124–127. doi: 10.7861/clinmed.2019-coron. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khulood D, Adil MS, Sultana R, Nimra. Convalescent plasma appears efficacious and safe in COVID-19. Ther Adv Infect Dis. 2020;7::2049936120957931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The Architecture of SARS-CoV-2 Transcriptome. Cell. 2020;181(914–21):e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adil MS, Verma A, Rudraraju M, Narayanan SP, Somanath PR. Akt-independent effects of triciribine on ACE2 expression in human lung epithelial cells: potential benefits in restricting SARS-CoV2 infection. J Cell Physiol. 2021;236(9):6597–6606. doi: 10.1002/jcp.30343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Velavan TP, Meyer CG. The COVID-19 epidemic. Trop Med Int Health. 2020;25(3):278–280. doi: 10.1111/tmi.13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li YC, Bai WZ, and Hashikawa T . The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. Jun 2020;92(6): 552–555. doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203(2):631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adil MS, Narayanan SP, Somanath PR. Is amiloride a promising cardiovascular medication to persist in the COVID-19 crisis? Drug Discov Ther. 2020;14(5):256–258. doi: 10.5582/ddt.2020.03070. [DOI] [PubMed] [Google Scholar]
- 11.Sarzi-Puttini P, Giorgi V, Sirotti S, Marotto D, Ardizzone S, Rizzardini G, Antinori, S, and Galli, M. COVID-19, cytokines and immunosuppression: what can we learn from severe acute respiratory syndrome? Clin Exp Rheumatol. 2020;38:337–342. [PubMed] [Google Scholar]
- 12.Ruaro B, Salton F, Braga L, Wade B, Confalonieri P, Volpe MC, Baratella E, Maiocchi S, Confalonieri M. The history and mystery of alveolar epithelial type II cells: focus on their physiologic and pathologic role in lung. Int J Mol Sci. 2021;22(5):2566. doi: 10.3390/ijms22052566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adil MS, Khan MA, Khan MN, Sultan I, Khan MA, Ali SA, and Farooqui, A. EMPADE study: evaluation of medical prescriptions and adverse drug events in COPD patients admitted to intensive care unit. J Clin Diagn Res. 2015;9:Fc05–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guttman JA, Finlay BB. Tight junctions as targets of infectious agents. Biochim Biophys Acta. 2009;1788(4):832–841. doi: 10.1016/j.bbamem.2008.10.028. [DOI] [PubMed] [Google Scholar]
- 15.Alwhaibi A, Verma A, Adil MS, Somanath PR. The unconventional role of Akt1 in the advanced cancers and in diabetes-promoted carcinogenesis. Pharmacol Res. 2019;145:104270. doi: 10.1016/j.phrs.2019.104270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan S, Jiang S-C, Zhang Z-W, Fu Y-F, Hu J, Li Z-L. The role of alveolar edema in COVID-19. Cells. 2021;10(8):1897. doi: 10.3390/cells10081897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linfield DT, Raduka A, Aghapour M, Rezaee F. Airway tight junctions as targets of viral infections. Tissue Barriers. 2021;9(2):1883965. doi: 10.1080/21688370.2021.1883965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adil MS, Somanath PR. Endothelial permeability assays in vitro. Methods Mol Biol. 2021;2367:177–191. [DOI] [PubMed] [Google Scholar]
- 19.Ahmad W, Shabbiri K, Ijaz B, Asad S, Sarwar MT, Gull S, Kausar H, Fouzia K, Shahid I, Hassan S, et al. Claudin-1 required for HCV virus entry has high potential for phosphorylation and O-glycosylation. Virol J. 2011;8(1):229. doi: 10.1186/1743-422X-8-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adil MS, Narayanan SP, and Somanath PR. Cell-cell junctions: structure and regulation in physiology and pathology. Tissue Barriers. 2020. Jan 2;9(1):1848212. doi: 10.1080/21688370.2020.1848212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afroz S, Giddaluru J, Abbas MM, Khan N. Transcriptome meta-analysis reveals a dysregulation in extra cellular matrix and cell junction associated gene signatures during Dengue virus infection. Sci Rep. 2016;6(1):33752. doi: 10.1038/srep33752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adil MS, Khulood D, Somanath PR. Targeting Akt-associated microRNAs for cancer therapeutics. Biochem Pharmacol. 2021;189:114384. doi: 10.1016/j.bcp.2020.114384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edgar R. Gene expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl, S, Hoagland, D, Møller R, Jordan, TX, Oishi, K, Panis M, and Sachs D, et al. SARS-CoV-2 launches a unique transcriptional signature from in vitro, ex vivo, and in vivo systems. bioRxiv. 2020;2020(3):24.004655. [Google Scholar]
- 25.Breuer K, Foroushani AK, Laird MR, Chen C, Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL, Lynn DJ, et al. InnateDB: systems biology of innate immunity and beyond–recent updates and continuing curation. Nucleic Acids Res. 2013;41(D1):D1228–33. doi: 10.1093/nar/gks1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Wang X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Research. 2020;48(D1):D127–D31. doi: 10.1093/nar/gkz757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, Sidiropoulos, K, Cook, J, Gillespie, M, and Haw, R, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–d503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabregat A, Sidiropoulos K, Viteri G, Forner O, Marin-Garcia P, Arnau V, D’Eustachio P, Stein L, Hermjakob H. Reactome pathway analysis: a high-performance in-memory approach. BMC Bioinformatics. 2017;18(1):142. doi: 10.1186/s12859-017-1559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moni MA, Quinn JMW, Sinmaz N, Summers MA. Gene expression profiling of SARS-CoV-2 infections reveal distinct primary lung cell and systemic immune infection responses that identify pathways relevant in COVID-19 disease. Brief Bioinform. 2021;22(2):1324–1337. doi: 10.1093/bib/bbaa376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha PK, Vijay A, Halu A, Uchida S, Aikawa M. Gene expression profiling reveals the shared and distinct transcriptional signatures in human lung epithelial cells infected with SARS-CoV-2, MERS-CoV, or SARS-CoV: potential implications in cardiovascular complications of COVID-19. Front Cardiovasc Med. 2020;7:623012. doi: 10.3389/fcvm.2020.623012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zamorano Cuervo N, and Grandvaux N. ACE2: evidence of role as entry receptor for SARS-CoV-2 and implications in comorbidities. Elife. 2020 Nov 9:9 ;e61390. doi: 10.7554/eLife.61390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trypsteen W, Van Cleemput J, Snippenberg WV, Gerlo S, Vandekerckhove L. On the whereabouts of SARS-CoV-2 in the human body: a systematic review. PLoS Pathog. 2020;16:e1009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snell J. SARS-CoV-2 infection and its association with thrombosis and ischemic stroke: a review. Am J Emerg Med. 2021;40:188–192. doi: 10.1016/j.ajem.2020.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. 2021;22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carnero E, Barriocanal M, Prior C, Pablo Unfried J, Segura V, Guruceaga E, Enguita M, Smerdou C, Gastaminza P, Fortes P, et al. Long noncoding RNA EGOT negatively affects the antiviral response and favors HCV replication. EMBO Rep. 2016;17(7):1013–1028. doi: 10.15252/embr.201541763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hermesh T, Moran TM, Jain D, López CB. Granulocyte colony-stimulating factor protects mice during respiratory virus infections. PLoS One. 2012;7:e37334. doi: 10.1371/journal.pone.0037334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang C, Mei J, Tian H, Liou YL, Rong D, Zhang W, Liao, Q, and Wu, N. CSF3 is a potential drug target for the treatment of COVID-19. Front Physiol. 2020;11:605792. doi: 10.3389/fphys.2020.605792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Su D, Ying L, Yu G, Mao W. Study on expression of CDH4 in lung cancer. World J Surg Oncol. 2017;15(1):26. doi: 10.1186/s12957-016-1083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson E, Theisen CS, Johnson KR, Wheelock MJ. R-cadherin influences cell motility via Rho family GTPases. J Biol Chem. 2004;279(30):31041–31049. doi: 10.1074/jbc.M400024200. [DOI] [PubMed] [Google Scholar]
- 40.Chow JT, and Salmena L . Prediction and analysis of SARS-CoV-2-targeting microRNA in human lung epithelium. Genes (Basel). 2020 Aug 26;11(9):1002. doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Frank JA. Claudins and alveolar epithelial barrier function in the lung. Ann N Y Acad Sci. 2012;1257(1):175–183. doi: 10.1111/j.1749-6632.2012.06533.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adil MS, Somanath PR. Vascular permeability assays in vivo. Methods Mol Biol. 2021;2367:165–175. [DOI] [PubMed] [Google Scholar]
- 43.Hou J, Renigunta A, Konrad M, Gomes AS, Schneeberger EE, Paul DL, Waldegger, S, and Goodenough, DA. Claudin-16 and claudin-19 interact and form a cation-selective tight junction complex. J Clin Invest. 2008;118:619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artham S, Gao F, Verma A, Alwhaibi A, Sabbineni H, Hafez S, Ergul A, Somanath PR. Endothelial stromelysin1 regulation by the forkhead box-O transcription factors is crucial in the exudative phase of acute lung injury. Pharmacol Res. 2019;141:249–263. doi: 10.1016/j.phrs.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alharthi A, Verma A, Sabbineni H, Adil MS, Somanath PR. Distinct effects of pharmacological inhibition of stromelysin1 on endothelial-to-mesenchymal transition and myofibroblast differentiation. J Cell Physiol. 2021;236(7):5147–5161. doi: 10.1002/jcp.30221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kadry RW, Adil MS, Newsome AS, Somanath PR. Cisatracurium attenuates LPS-induced modulation of MMP3 and junctional protein expression in human microvascular endothelial cells. Biosci Trends. 2021;15(1):50–54. doi: 10.5582/bst.2020.03399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueland T, Holter JC, Holten AR, Müller KE, Lind A, Bekken GK, Dudman S, Aukrust P, Dyrhol-Riise AM, Heggelund L, et al. Distinct and early increase in circulating MMP-9 in COVID-19 patients with respiratory failure. J Infect. 2020;81(3):e41–e3. doi: 10.1016/j.jinf.2020.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]


