Abstract
The severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), responsible for the outbreak of coronavirus disease 2019 (COVID-19), has shown a vast range of clinical manifestations from asymptomatic to life-threatening symptoms. To figure out the cause of this heterogeneity, studies demonstrated the trace of genetic diversities whether in the hosts or the virus itself. With this regard, this review provides a comprehensive overview of how host genetic such as those related to the entry of the virus, the immune-related genes, gender-related genes, disease-related genes, and also host epigenetic could influence the severity of COVID-19. Besides, the mutations in the genome of SARS-CoV-2 __leading to emerging of new variants__ per se affect the affinity of the virus to the host cells and enhance the immune escape capacity. The current review discusses these variants and also the latest data about vaccination effectiveness facing the most important variants.
Keywords: SARS-CoV-2, COVID-19, ACE2, Genetic diversity, Mutation, Vaccine
1. Introduction
In December 2019, an unknown cause of pneumonia was detected in Wuhan, China that was attributed to a seafood wholesale market. By sequencing samples of patients with pneumonia, a previously unrecognized β-coronaviruses (β-CoVs) was identified and named 2019-nCoV, also known as severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) [1]. SARS-CoV-2 is the cause of coronavirus disease 2019 (COVID-19), an infection of the respiratory tract, which has a wide spectrum of clinical manifestations; while some cases will present mild symptoms, others will develop more serious complications entailing specialized management in the intensive care units (ICU) [2]. SARS-CoV-2 belongs to the B lineage of the β-CoVs. According to the whole-genome analysis, β-CoVs genomes encode 4 structural proteins, spike (S), envelope (E), membrane (M), and nucleocapsid (N), as well as numerous non-structural ones [3].
The receptor-binding domain (RBD) in the spike protein lets the virus interact with angiotensin-converting enzyme 2 (ACE2) of the host cells [4]. Accordingly, this virus mainly infects lung cells and respiratory tracts as ACE2 is mostly expressed on the surface of type II alveolar cells of the lungs [5]. Since the expression of ACE2 is not equal in each individual and also, ACE2 has shown recently-detected polymorphisms, the pathogenicity of SARS-CoV-2 differs occasionally [6]. In addition to ACE2, there are several host-related factors such as transmembrane protease, serine 2 (TMPRSS2), HLA, cytokines, sex hormones, and epigenetics which could influence the severity of COVID-19. Furthermore, SARS-CoV-2 genome mutations are an inevitable phenomenon responsible for altered encoded proteins, leading to different pathogenicity, harder detection, and possibly higher immune escape capacity. In addition to non-pharmaceutical measures, effective vaccines have been one of the most potent arsenals facing the COVID-19 pandemic [7]; nevertheless, there is always confrontation between newly-emerged SARS-CoV-2 variants and immune responses. In this review, we comprehensively discussed the interactions of SARS-CoV-2 and the host genetic and epigenetics, the virus mutations and emerging variants, and how vaccines could face SARS-CoV-2 variants.
2. Host genetic
2.1. SARS-CoV-2 entry mechanism-related genes
2.1.1. Ace2
Among different receptors that have been identified to be associated with COVID-19, ACE2 is one of the important ones which facilitates the spread of the virus to the vital organs [8]. The knowledge of ACE2 and its association with disease progression is less advanced, but there is an interesting consensus that genetic heterogeneity of ACE2 might be one of the reasons why some people may develop the severe form of COVID-19 and others only suffer from a mild symptom. Given this, the more we know about this receptor and its genetics, the better we could stratify patients, and subsequently, the better we can design treatments and measures.
The stream of studies has started and the results of each are interesting. The results of mining of whole-exome sequencing (WES) data of 6930 Italian control individuals took a veil off a number of ACE2 variants that are responsible for protein stability. Among them, rare point mutations of (Leu351Val) and p.(Pro389His) seem to have an association with the spike protein of the SARS-CoV-2 virus [9]. Stawiski et al. have also noticed that rare ACE2 variants may have an impact on the susceptibility of the individuals to SARS-CoV-2 [10]. The authors reported that while the presence of either human ACE2 variants S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P, or H378R could make individuals more susceptible to develop COVID-19, K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L, and D509Y might protect the individuals from the spread of the virus. Besides these variants, N90 and T92 are the most important ACE2 residues that prevent the entry of SARS-CoV-2 to the host’s cells and are considered CoV host modifiers.
Apart from ACE2, the genetic background of other genes in the ACE2-related pathways could also alter the vulnerability of the host to the virus. In this vein, Lanjanian et al. have indicated that the extent to which ACE2-associated genes can play a role in virus susceptibility is actually greater than that of the receptor [11]. The results of their investigation showed that two mutations, K26R and S331F, might reduce the binding affinity of the receptor to the spike protein of the virus [11]; thus, there is controversial data about K26R mutation as it is not clear whether this mutation increases the susceptibility to the virus or reduces.
Although numerous studies declared that ACE2 genetic variation might reshape SARS-CoV-2 pathogenicity and alter the susceptibility of the hosts to the virus, a considerable number of investigations have denied the presence of such correlation. The results of a cohort study in European, North-African, and Middle East countries failed to find any association between ACE2 polymorphism and the mortality rate of COVID-19 [12]. Similarly, a study conducted in Spain on 120 patients also suggested the lack of correlation between ACE2 polymorphism and the SARS-CoV-2 infection [13]. The results were interesting, as they reported that the extent of susceptibility to SARS-CoV-2 was not associated with ACE2 genetic heterogeneity, but with the allele frequency (AF). The authors declared that the reason why East Asian populations have more susceptibility to the virus was due to the higher AFs in the eQTL variants, an event that leads to higher expression of ACE2 in the tissue [14].
2.1.2. Transmembrane protease, serine 2 (TMPRSS2)
TMPRSS2 is another identified receptor for SARS-CoV-2, which could spread the virus throughout the organs. The results of the previous studies were succeeded to find an association between the polymorphism of this receptor and the susceptibility to influenza [15]. Given this, a question has been raised that whether the genetic heterogeneity of TMPRSS2 might have an influence on the progression of COVID-19. The results of an analysis on the expression levels of TMPRSS2 in the lungs led to the identification of seven blocks (I–VII) of variants that could increase the expression of the receptor in individuals [16]. Interestingly, it has been reported that European populations have the highest proportion of haplotypes than the Asian populations, leading to the increase of their susceptibility to the virus. Following the European populations, South Asia (46 %) and America (42 %) had the higher levels of haplotypes, while Africa (28 %) and East Asia (19 %) have reported to harbor the lowest frequency of the variants. The alleles associated with the increase in the expression of TMPRSS2 seem to have a frequency of lower than 1 % in the Eastern Asian population. By using WES, it became evident that rs75603675, rs61735792, and rs61735794, three variants of TMPRSS2, might increase the risk of SARS-CoV-2 in the individuals [17].
Notably, analyzing the sequence of TMPRSS2 of 1177 COVID-19 patients revealed that p.V197M (rs12329760), which control the protease activity and the stability of the receptor, might be involved in the pathogenicity of the virus [18]. The frequency of missense variant rs12329760, also known as p.Val197Met, in TMPRSS2 was reported to be higher in mild COVID-19 cases as compared to the critically ill patients. Individuals harboring the higher frequency of the variant might have a milder form of the disease, without the requirement for hospitalization or oxygen therapy [18], [19]. Wulandari et al. have also declared that there is a correlation between p.V197M and the viral load, suggesting that p.V197M could be used as a predictive factor to risk stratifying the patients.
As mentioned, p.V197M could destabilize TMPRSS2 protein and thereby attenuate the interaction between ACE2 and spike protein of the virus. Moreover, the substitution of valine (V) to methionine (M) in the catalytic site of TMPRSS2 protein reduces the ability of the receptor to facilitate SARS-CoV-2 spike-mediated entry into cells [20]. It could be assumed that due to the previous exposure to the virus, the East Asian population inherited more frequency of p.V197M due to natural selection and this is why they might be more resistant to the virus as compared to European ethnicity.
Given the importance of p.V197M in diminishing the risk of a severe form of COVID-19, a stream of studies has put their efforts to identify drugs that could target TMPRSS2. Camostat mesilate, a drug that received FDA approval for the treatment of chronic pancreatitis and postoperative reflux esophagitis, could target TMPRSS2 and be effective in the treatment of COVID-19 [20]. Apart from this, the development of some small-molecule inhibitors reducing the stability of TMPRSS2 protein might also be effective in either prevention or minimizing the severity of COVID-19. Interestingly, an important study showed that the Omicron variant could penetrate the cells by the endosomal route instead of the TMPRSS2-dependant way, implying the dangerous potential of this variant [21].
2.1.3. Furin (PCSK)
Furin is a type 1 membrane-bound protease that facilitates the entrance of the SARS-CoV-2 within the endothelial cells [22]. So far, 13 missense variants have been identified for the FURIN gene that could be associated with the susceptibility to the virus, and among them, 7 were reported to diminish the risk of SARS-CoV-2 progression in the individuals [23]. These protective variants have been mainly detected in Qatar and Kuwait. In contrast to the Middle East, it seems that FURIN variants could not exert protective effects against the viral spread. It has been reported that rs6226 and rs8039305 FURIN polymorphism in the African population is associated with the incidence of hypertension and disease mortality. Fuentes et al., on the other hand, failed to report any association between FURIN polymorphism and the incidence rate of COVID-19 [13].
2.1.4. Dpps (Dipeptidyl peptidases)
Dipeptidyl peptidase-4 (DPP-4) or CD26 which acts as a ligand for a variety of extracellular molecules could make interaction with several viral proteins, such as spike protein as well [24]. Thus far, the associations between different DPPs and viral proteins have been established in numerous studies. Interestingly, the genetic analysis conducted in China on 322 COVID-19 patients revealed that recurrent loss of function 1-bp insertion in gene DPP7 was common among asymptomatic patients [25]. In another study, Śanchez et al. succeeded to find a correlation between the down-regulation of DPP-4 and the severity of COVID-19 [26]. Interestingly, in another study, it became evident that there is a relation between the expression level of DPP-4 and obesity. Given the established correlation between body mass index (BMI) and the need for IMV, the triangle between DPP4 overexpression, obesity, and IMV in COVID-19 patients seems to be reasonable [27], [28], [29], [30], [31]. Accordingly, Latini et al. have introduced three missense and splice acceptor variants (c.95-2A > G, c.796G > A, c.1887 + 3G > A) in the DPP4 gene which were associated with the severity of COVID-19, shedding more light on the contributory role of this receptor as a co-receptor for SARS-CoV-2 viral entry [32].
2.1.5. Apolipoprotein E (APOE)
The transferring of serum cholesterol to cells by means of cholesterol transport protein apolipoprotein E (APOE) could boost the trafficking of ACE2 to the endocytic entry site that facilitates the entry of SARS-CoV-2 to cells; this phenomenon subsequently elevates the risk of COVID-19 infection, proposing that probably there is a link between the expression of apolipoproteins and severity of the disease [33]. When secreted from the epithelial cells, cholesterol helps FURIN to cleave spike protein, and thereby, increase cell-to-cell transmission of SARS-CoV-2. The results of the previous study indicated that individuals with APOE ε3/ε4 might have increased risk of the virus entry as compared with ε3/ε3 subjects [34]; proposing that APOE4 can impact SARS-CoV-2 pathogenicity by altering intracellular levels of cholesterol. The ε4 variant of the APOE has also been reported to be found in more frequency in black Africans than Caucasians and Asians, suggesting why this population might be at higher risk of mortality due to SARS-CoV-2 infection [35]. In agreement, Kuo et al. indicated that the presence of the APOE ε4/ε4 allele elevates the severity of COVID-19 in the individuals and increases the mortality rate by four-times even in the absence of any pre-existing dementia, cardiovascular disease, or type-2 diabetes [36]. Since the ε4 variant of APOE could reduce lung respiratory capacity [37] and APOE-depleted mouse models have shown to be more prone to develop severe pulmonary hypertension [38] the association between this allele and the risk of death in COVID-19 patients seems to be reasonable.
2.1.6. Bsg (CD147)
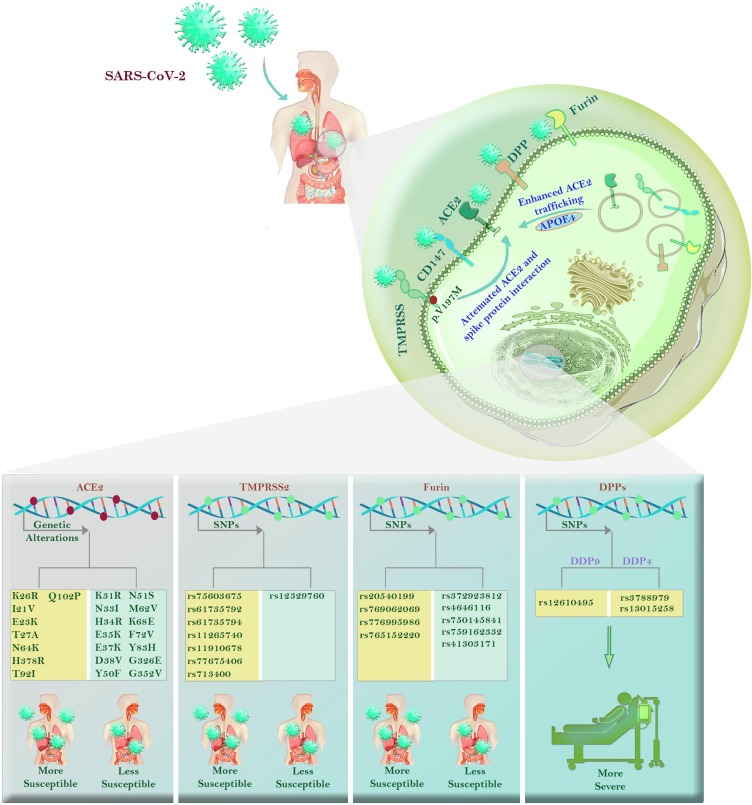
Basigin (BSG) or CD147 is a member of the immunoglobulin superfamily, with the ability to facilitate the entrance of SARS-CoV-2 into the host cells [39]. By evaluating the gene expression profile of BSG, it became evident that the presence of one missense mutation (p.F275V) in the I-set domain of the protein could increase the affinity of the receptor to the spike protein of SARS-CoV-2 [40]. On the other hand, the existence of BSG antibodies could diminish the cytokine storm caused by the virus [41]. Moreover, Meplazumab, an anti-CD147 humanized antibody could prevent the spread of the virus into the hosts cells, and thereby, reduce the incidence rate of COVID-19 [42]. To provide a well-conceptualized overview, we represented the potential roles of SARS-CoV-2 entry mechanism-related genes in Fig. 1 . Also, the details of entry mechanism-related genes were provided in Table 1 .
Fig. 1.
The roles of SARS-CoV-2 entry mechanism-related genes in COVID-19. There are some entry mechanisms for SARS-CoV-2 in the host genome which their diversity could affect the severity of the COVID-19. Accordingly, ACE2 is the most known virus entry receptor which undergoes some alterations; some of which such as K31R, N33I, N51S, M26V, K68E, and F72V are associated with less susceptibility to the virus, while E23K, T27A, H378R, Q102P, and N64K reduce susceptibility. Similarly, some SNPs in TMPRSS2 include rs75603675, rs61735792, rs61735794, rs112657409, rs11910678, rs77675406, and rs713400 have an association with more severe disease; however, p.V197M (rs12329760) can destabilize TMPRSS2, and as a result, attenuate the interaction of ACE2 and spike protein. The presence of SNPs such as rs20540199, rs769062069 in the FURIN gene have roles in disease mortality. Patients carrying rs3788979 and rs13015258 in the DPP4 genome, as well as those with rs12610495 in the DPP9 genome, show higher inflammatory responses in lung tissue. Also, the higher risk of COVID-19 mortality could be associated with the ε4 variant of the APOE that enhances the trafficking of ACE2 toward the cell surface.
Table 1.
The details of entry mechanism-related genes, their mutations and outcomes.
| Gene | Location | function | Mutation/ Polymorphism | Result | Drug/ Inhibitor | Ref |
|---|---|---|---|---|---|---|
| ACE2 | Chr 17 | A negative regulator of the renin-angiotensin system, and functions as the key SARS coronavirus receptor and stabilizer of neutral amino acid transporters | Leu351Val, p.(Pro389His), S19P, I21V, E23K, K26R, T27A, N64K, T92I, Q102P, or H378R |
Elevate the susceptibility to COVID-19 | lividomycin, burixafor, quisinostat, fluprofylline, pemetrexed, spirofylline, edotecarin, diniprofylline | [9], [11], [23], [43], [44], [45] |
| K31R, N33I, H34R, E35K, E37K, D38V, Y50F, N51S, M62V, K68E, F72V, Y83H, G326E, G352V, D355N, Q388L, D509Y, N90, T92, rs73635825 (S19P), rs143936283 (E329G) | Protects individuals from the spread of the virus leading to an intrinsic resistance against SARS-CoV-2 virus | |||||
|
TMPRSS2 |
Chr 21 | An endothelial cell surface protein that is involved in the viral entry and spread of SARS-CoV-2. SARS-CoV-2. | rs75603675, rs61735792, rs61735794, rs112657409, rs11910678, rs77675406, rs713400, p.V197M | Increase the risk of SARS-CoV-2 | camostat mesylate, nafamostat mesylate, Nafamostat, Gabexate, N-0385 | [17], [19], [46], [47], [48], [49], [50], [51] |
| high V197M, low G8V (rs75603675) allele frequencies | Endowing resistance against SARS-CoV-2 | |||||
| FURIN (PCSK) | Chr 15 |
A type 1 membrane-bound protease which its cleavage action is an essential activation step for the endothelial pathogenicity of SARS-CoV-2. | rs6226: P ¼ 1.3569e-09, OR ¼ 1.03; rs8039305: P ¼ 1.7643e-38, OR ¼ 1.05 | Are associated with hypertension and disease mortality | Nicotine patches, Colchicine, Psoriasis Registry Austria (PsoRA), 3,5-dichlorophenyl) pyridine | [23], [52], [53] |
| DPPs | Chr 2 | A multi-functional protein with the catalytic activity as well as functions as a binding protein and a ligand for a variety of extracellular molecules such as spike protein as well | rs3788979 TT, c.95-2A > G, c.796G > A, c.1887 + 3G > A, rs13015258, rs12610495 | Increase the need for invasive mechanical ventilation in COVID-19 | N/A | [24], [54], [55], [56] |
| APOE | Chr 19 | The transferring of serum cholesterol to cells by means of APOE could boost the trafficking of ACE2 to the endocytic entry site that facilitates the entry of SARS-CoV-2 to cells. | ε3/ε4 variant, ε4/ε4 | Higher risk of mortality | N/A | [36], [37], [57] |
| BSG | Chr 19 | a multifunctional transmembrane protein involved in inflammation and tumor invasion with the ability to facilitate the entrance of SARS-CoV-2 into the host cells. | p.F275V | Enhances the affinity of the receptor to the spike protein of SARS-CoV-2 | Meplazumab, Azithromycin | [39], [40], [58], [59] |
ACE2: Angiotensin-converting enzyme 2; Chr: chromosome; Leu: Leucine; Val: Valine; Pro: Proline; His: Histidine; S: Serine; I: Isoleucine; E: Glutamic acid; K: Lysin; R: Arginine; T: Threonine; A: Alanin; N: Asparagine; Q: Glutamine; D: Aspartic acid; Y: Tyrosine; F: Phenylalanine; M: Methionine; G: Glycine; TMPRSS2: Transmembrane protease, serine 2; PCSK: Proprotein convertase subtilisin/kexin 9; DPPs:; Dipeptidyl peptidases; APOE: Apo lipoprotein E; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; BSG: Basigin.
2.2. Immune-related genes
2.2.1. Human leukocyte antigen (HLA)
To date, no genetic variation has been reported as much as HLAs to be associated with a wide range of human diseases [60]. In this regard, it comes as no surprise if different series of investigations have put an effort to identify a correlation between HLA variants and the susceptibility to the SARS-CoV-2 virus [61], [62], [63]. In the majority of cases, these investigations were successful. Allele typing of 37 COVID-19 patients in Taiwan revealed a significant relation between HLA-B*4601, HLA-B*0703, as well as HLA-DRB1*0301 and the sensitivity to the SARS-CoV-2 virus [64]. Also, Vietzen et al. reported that among 361 COVID-19 patients with different types of the disease, those who had HLA-E*0101 and were heterozygous for the HLA-E*0101/0103 variant had an increased risk to be hospitalized or needing intensive care; it has been reported that probably the presence of HLA-E*0101 and HLA-E*0101/0103 variants might attenuate the NKG2C+ NK cell responses, an event leading to the accelerated progression of COVID-19 [65], [66].
In another study, Wang et al. evaluated HLA alleles in 69 critically ill patients and 215 COVID-19 patients with an asymptomatic, mild, or moderate type of the disease [25]. The authors demonstrated that among the class I HLA genes, HLA-A*11:01, HLA-B*51:01, and HLA-C*14:02 alleles might be associated with poor prognosis in COVID-19 patients. HLA-B*46:01 was another allele that could increase the severity of disease, while HLA-B*15:03 was associated with the protective effects. Moreover, they suggested that while DRB1*14:04, DRB1*01:01, and DQA1*01:01 alleles could be classified as a risk factor for disease progression, harboring DPB1*03:01 and DRB1*12:01 alleles could exert ameliorating effects on the pace of disease development. Another study conducted on 28 COVID-19 patients with severe respiratory failure showed that the expression of HLA-DR was lower as compared to patients with a mild form of infection. Moreover, it seems that there is an association between HLA-DR expression and the number of CD4/CD19 lymphocytes and NK cells. Interestingly, it has also been suggested that HLA alleles might have a role in SARS-CoV-2-induced olfactory dysfunction in individuals, as the olfactory receptor gene appears to be MHC-linked.
The results of efforts accomplished by Campbell et al. also resulted in the identification of binding affinity between 9-mer and 15-mer peptides from the SARS-CoV-2 peptidome and 9,360 class I and 8,445 class II HLA alleles, respectively [67]. In MHC-binding assays, it became evident that there is a correlation between some epitopes of SARS-CoV-2 and HLA-A*02:01, HLA-B*40:01, HLA-DRA*01:01, HLA-DRB1*07:01, and HLA-DRB1*04:01 alleles [68]. López et al. also have used a bioinformatic prediction tool to identify the top ten highly immunogenic epitopes on T cells that were associated with response to the SARS-CoV-2 virus [69]. They used the data provided from 14 countries and reported for the first time that only 7 HLA molecules, including HLA-DPA1*01:03/DPB1*02:01, HLADPA1* 02:01/DPB1*01:01, HLA DPA1*03:01/DPB1*04:02, HLA-DQA1*05:01/DQB1*03:01, HLA-DRB1*01:01, HLADRB1* 07:01, and HLA-DRB1*09:01 had a high binding affinity for the SARS-CoV-2 virus. While cells express HLA-A*02:03 and A*31:01 are the best antigen presenters for the virus and could induce the highest immune responses, cells expressing HLA-A*03:02 might fail to induce an acceptable immunity against the virus.
2.2.2. Cytokines
Among different cytokines that associate with COVID-19, perhaps type I IFNs are the most important ones. The results of the primary studies clearly suggested that the down-regulation in the expression of IFNs significantly facilitates the progression of the virus into the host’s cells. Interestingly, demographic investigations have also succeeded to find a correlation between IFN genes and the susceptibility to the SARS-CoV-2; the authors declared that the presence of single nucleotide polymorphisms (SNPs) in the IFN-inducible genes OAS1 and MX1 can increase the risk of COVID-19 [70], [71]. Castineira et al. also identified a rare loss-of-function mutation in IFNAR2, a receptor that prevents the entrance of the respiratory viruses such as Influenza and SARS-CoV-2 [72]. Of particular interest, they reported that individuals carrying this mutation might not even respond well to exogenous interferon treatment; suggesting that perhaps this genetic alteration occurs at the early stage of the disease when the viral load is high.
Inborn errors of the interferon regulatory factor 7 (IRF7) gene is the other genetic alterations that have been reported to be associated with COVID-19 pneumonia. Previously, the presence of a mutation in two loci of this gene, i.e., IRF7 and IRF9, has been reported to increase the risk of life-threatening influenza in the individuals. Interestingly, when analyzed in vitro, it became evident that IRF7 −/− and IFNAR1 −/− fibroblasts were also more vulnerable to the SARS-CoV-2 virus [73].
Interferon-induced transmembrane protein 3 (IFITM3) is another cytokine-related protein that its genetic diversity was shown to be associated with COVID-19 severity. In the physiological condition, IFITM3 blocks the entrance of viral products into the host’s cells by affecting the fluidity of the cell membrane [74]. In COVID-19 patients, the results of the gene sequencing of the rs12252 IFITM3 variant revealed that 35 % of the patients with the severe form of the disease had the homozygous CC genotype [75]. When Wang et al. have deeply sequenced the genome of 332 COVID-19 patients with different forms of disease, they found that most genetic alterations associated with the severity of the disease were located in TMEM189–UBE2V1 and TMEM189–BE2V1 which are participating in the IL-1 signaling pathway [25]. For example, the presence of rs60220284 is an eQTL that increases the expression of IL-1 and is widely found in critically ill patients.
It is well-established that pro-inflammatory cytokines might recruit several tyrosine kinase receptors to propagate their signals into the nucleus, and as a result, change the expression of a wide range of genes. A non-receptor tyrosine kinase named tyrosine kinase 2 (TYK2), which belongs to the JAK family, has shown to be a member of IL-6R, type I interferon receptors, IL-12, IL-23, and IL-10 receptors-related signaling pathways [76]. Recently, a genetic variant rs74956615 on chromosome 19p13.2 near the gene encoding TYK2 has been identified with the ability to elevate the expression of this non-receptor tyrosine kinase; notably, the overexpression of TYK2 could increase lung injuries and induce a life-threatening disease. Moreover, since TYK2 targets JAK inhibitors [77], TYK2 variants can attenuate the therapeutic effects of JAK inhibitors in COVID-19 patients. It should be noted that JAK inhibitors were among the first drugs that their efficacy has been studied in COVID-19 patients [78]. Given this, identification of TYK2 variants in the individuals might be beneficial for evaluating the therapeutic potential of JAK inhibitors.
Another variant of TYK2 that has been identified in COVID-19 patients is rs10735079, which is located in the interferon-inducible oligoadenylate synthetase (OAS) gene cluster, a gene that is developed to produce RNase L [72].
2.2.3. Ccr/CXCR
The variation in the expression of chemokine receptors could also determine the susceptibility of host to the SARS-CoV-2. For example, the genome analysis conducted by Stikker et al. revealed that the presence of activating variants in regulatory regions of the chemokine receptor-encoding CCR1 gene is associated with the severe type of COVID-19 [79]. Indeed, through enhancing the infiltration of monocytes and macrophages into the inflammatory site, the elevated CCR1 might propagate inflammatory responses, and thereby, accelerate the progression of the disease. CCR2 is another chemokine receptor that in combination with monocyte chemoattractant protein (MCP-1) attracts monocytes and macrophages to the inflammatory site. The results of the genome-wide analysis revealed the presence of activating variants in gene encoding CCR2 in critically ill COVID-19 patients.
CCR5 32 bp deletion variant (Δ32), which its association with several viral infections has been previously established, could also increase the mortality rate of COVID-19 in the African population [80]. Gómez et al. conducted a study on 294 hospitalized COVID-19 patients and reported that the expression of CCR5-Δ32 is lower in infected patients as compared to healthy counterparts. CXCR6 and its ligand CXCL16 are other molecules that their expression has been shown to have an association with the severity of COVID-19. The expression of CXCR6 on CD8+ T cells mediates the homing of T cells to the lung in response to the CXCL16 gradient [72]. It has been reported that during COVID-19, several endogenous factors might exert their effects on the 3p21.31 locus to decrease the expression of CXCR6 on lung resident memory CD8+ T cells, an event which is related to the accelerated progression of the disease and formation of the severe infection due to lower infiltration of CD8+ T cells.
2.2.4. Toll-like receptors (TLRs)
The number of studies evaluating the importance of TLRs and their related signaling pathway in the pathogenicity of SARS-CoV-2 is growing day by day and the results suggest that the suppression of TLRs is associated with the progression of COVID-19 [81]. Like other related receptors, the allelic variation in TLRs seems to alter the risk of the individuals to the infection. For example, the deletion of TICAM2, a TLR adaptor protein, on chromosome 18 locus showed not only an increased risk of SARS-CoV-2 infection [82] but also an induction of pulmonary hemorrhage. Another investigation group also succeeded to find a correlation between the inborn errors of TLR3 and the risk of life-threatening COVID-19 pneumonia [82]. Zhang et al. reported that individuals who have loss-of-function at the 13 candidate loci were more susceptible to develop COVID-19 pneumonia as compared to the patients with asymptomatic or benign infection. Fallerini et al. have also reported the presence of loss-of-function variants in TLR7 in two families with COVID-19; they reported this genetic variant in 2.1 % of male patients with a severe type of the disease [83]. Interestingly, the loss-of-function variants of TLR7 seem to be related to the requirement with intensive care and mechanical ventilation. It has been suggested that TLR7 deficiency might impair type I and II IFN responses and this is why this rare genetic alteration may be accompanied by the severe type of the disease.
2.2.5. Complement system
The significant role of the complement system in the pathogenesis of the SARS-CoV-2 infection is undeniable, and thus far, several medications have been developed to target the complement components to treat COVID-19 [84]. Mannose-binding lectin (MBL) is a pattern recognition molecule that its deficiency has been shown to increase the risk of several bacterial infections in children; in COVID-19, it has also been declared that there is a correlation between low MBL levels and the susceptibility to the SARS-CoV-2 virus [85]. On the other hand, C3 deficiency could act in favor of hosts and protect the individuals from respiratory symptoms of SARS-CoV-2. The rs11385949 G > GA (chromosome 3 cluster rs11385942 variant) is another genetic variant that could increase the severity of COVID-19 through complement hyper-activation [86]. It has been reported that the upper airways viral load and sC5b-9 were higher in the carries of rs11385949; further highlighting the contributory role of the complement system in the pathogenesis of SARS-CoV-2.
2.2.6. NK cell-related genes
Through expressing NKG2A, NK cells could induce cytotoxicity in virally infected cells independent of MHC molecules. Given this, the immunotype of NK cells could also be a risk factor for host susceptibility to the virus [87]. In this vein, the high expression of perforin, NKG2C, and Ksp37 was observed in NK cells isolated from the severe COVID-19 patients [88]. Or, the expression of NKG2A has been reported to be elevated in NK cells and cytotoxic T cells (CTLs) harvested from COVID-19 patients to induce exhausted phenotype in the lymphocytes. The up-regulation of NKG2A has been reported to be temporary, as once the patients are recovered, the expression of this receptor is reduced on the lymphocytes. Apart from NKG2A, lymphocyte-activation gene 3 (LAG-3) and T cell immunoglobulin and mucin-domain containing-3 (TIM-3) are other inhibitory receptors that their expression was shown to have an upward trend in NK cells of COVID-19 patients [89]. Taken together, these findings all together declare that the immunotype of NK cells could determine the outcome of the patients in response to COVID-19 infection.
2.2.7. Vitamin d-binding protein (DBP)
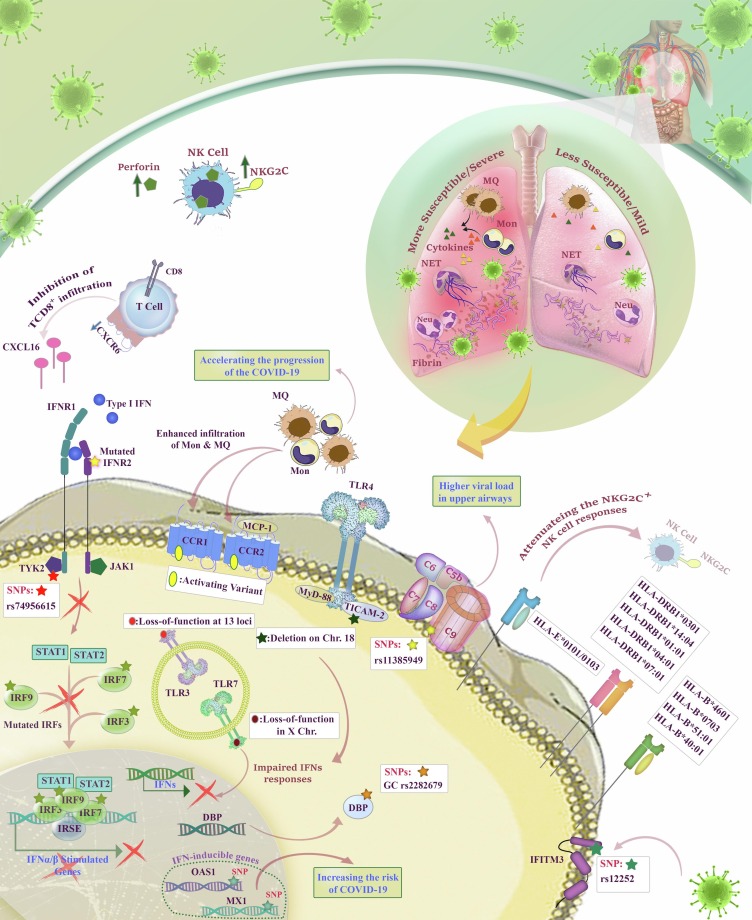
Vitamin d-binding protein (DBP) is a polymorphic protein encoded by a gene located at chromosome 4q11–q13 that binds to vitamin D and regulates total and free vitamin D metabolite levels [90]. It has been reported that some allele variations such as rs7041 locus and rs4588 locus can increase the affinity of the protein for vitamin D. In COVID-19, surprisingly, rs7041 variant of DPB has been shown to have an association with prevalence and mortality of the patients. GT genotype could also confer the SARS-CoV-2 virus susceptibility in the populations of Germany, Mexico, Italy, Czech, and Turkey [91]. Another variant of DBP with the ability to increase the severity of COVID-19 is encoded by the GC gene. It has been reported that those patients who carried GC rs2282679 polymorphism might suffer from the more severe type of the disease [92]. Fig. 2 has been designed to provide an overview of the roles of immune-related genes in the pathogenicity of COVID-19.
Fig. 2.
The roles of immune-related genes in COVID-19. As illustrated, some HLA variants are associated with susceptibility to SARS-CoV-2 such as HLA-DRB1*0301, DRB1*14:04, DRB1*01:01, DRB1*04:01, DRB1*07:01, HLA-B*4601, B*0703, B*51:01, and B*40:01. Also, heterozygote HLA-E*0101/0103 can attenuate the NKG2C+ NK cell responses. The infection of a cell with SARS-CoV-2 could result in the secretion of type I IFNs which consequently interact with adjacent IFNR1/2-expressing cells to initiate the signaling pathway ending in anti-viral responses; however, the presence of rs74956615 in TYK2__endocing a tyrosine kinase-related to IFNR1 signaling__ induces TYK2 overexpression, and consequently, hurts the lung cells. Moreover, rs10735079 in TYK2 is associated with reduced OAS expression and attenuated anti-viral responses. Notably, some SNPs in OAS and MX1 could elevate the risk of COVID-19, as well. Similarly, mutations in IRF7 and IRF9__components of IFN signaling pathway__ boost SARS-CoV-2 susceptibility. rs12252 in IFITM3 also attenuates the ability of IFITM3 to block the entrance of viral products into the cells. CCR1 and CCR2-MCP-1 could enhance the infiltartion of monocytes and macrophages and induce inflammation; with this regard, the pressence of activating variants in CCR1 and CCR2 has been observed in critically ill COVID-19 patients. Besides, the lower expression of CXCR6 could reduce the infiltration of CD8+ T cells in the lung, leading to more severe disease. The mutations in TLRs such as the deletion in TICAM2, which is related to TLR-4 initial signaling, could influence the risk of COVID-19. Similarly, loss-of-function of TLR-3 and TLR-7 impairs the IFNs responses. The overexpression of complement system components is another agent magnifying the severity of the disease. In patients carrying rs11385949, the levels of upper airway viral load and sC5b-9 were greater than others. Regarding the roles of DBP in the regulation of vitamin D levels, patients harboring GC rs2282679 could experience the severe type of the disease. Furthermore, The NK cells isolated from severe cases of COVID-19 have shown higher expression of NKG2C and secretion of perforin. Moreover, it was shown that Cit-H3 has significant roles in NET formation and consequently higher inflammation and cardiovascular complications.
2.3. Gender-related genes
2.3.1. Sex hormones
From the first description of COVID-19, when it became evident that men are more susceptible to severe types of the disease as compared to women, the attention has been attracted to sex hormones as strong modulators [93], [94]. The production of sex hormones is derived from a complex pathway consists of several enzymes [95]. The regulatory region of CYP19A1, which is a gene responsible for encoding a member of the cytochrome P450 superfamily of enzymes, could influence the local biosynthesis of estrogen [96]. With this regard, the activity of CYP19A1 to encode aromatase __which catalyzes the last step of estradiol (E2, or 17β-estradiol) synthesis [97] __ is higher in ovarian granulosa cell, the placenta, the testicular Leydig cell, and other sites such as brain and skin fibroblasts [98]; notably, the highest level of aromatase is found in ovarian granulosa cell which implies higher levels of estrogen in women [99]. In a murine model of COVID-19, it became evident that blockage of estrogen could result in the elevation in the number of inflammatory cells in the lung, tissue damage, and disease progression. In the clinical settings, it has also been observed that the duration of hospital stay was significantly lower in non-menopausal women [100]. Acheampong et al. have claimed that as compared to the male patients, even menopausal women cope better with the severe type of COVID-19 and recover faster, as women produce higher levels of estrogen in this period than men [101]. Estrogen levels (above 70 pg/ml) was shown to have a negative correlation with the expression levels of several pro-inflammatory cytokines, such as IL-6, IL-8, IL-2R, and TNF-α in COVID-19 patients and this clearly explains why this hormone could halt the over-reaction of immune cells against SARS-CoV-2 [102].
The results of a study conducted in Italy revealed that prostate cancer patients who were received androgen deprivation therapy (ADT) were less likely to develop severe COVID-19 compared to the non-ADT group. Montopoli et al. also indicated that androgen could increase the virulence of the SARS-CoV-2 virus and increase the sensitivity of males to the disease [103]. The cross-talk between androgens and TMPRSS2 may be, at least partly, a reason why elevated levels of androgens could enhance the severity of the disease [101], [104]. In contrast to these, some studies have declared that the opposite may be true and the low levels of testosterone might have devastating effects on COVID-19 progression [105]. A study focused on hospitalized men revealed that the decrease in the circulating testosterone levels might increase the severity of the disease and mortality rate [102]. Indeed, reduction in testosterone levels has been introduced as a risk factor for men; however, these findings have not yet been confirmed, and currently, the application of ADT and estrogen in the management of COVID-19 is expected to be beneficial for severe types of the disease [106].
2.3.2. Immunological factors in males and females
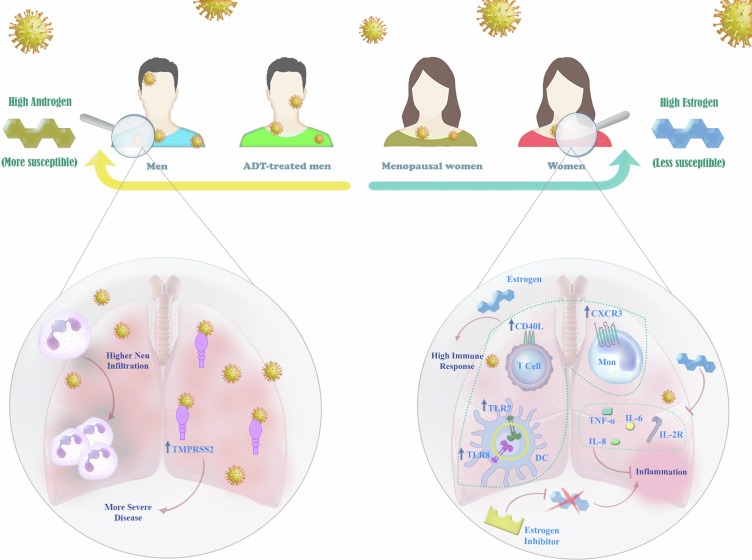
Apart from the sex hormones, the differences in the immune system of males and females may be another factor that might explain why men are more prone to develop the severe type of COVID-19 as compared to women [107]. Generally, the female immune system is stronger, as a wide range of essential immune-related genes is located on the X chromosome [108]. The biallelic expression of such genes in women, including TLR7, TLR8, CD40L, CXCR3, and interferons leads to higher immune responses and reduces the susceptibility to viral infections [109]. In a murine model of COVID-19, while the inflammatory cytokines and chemokines in SARS-CoV-2-infected male and female mice showed equal levels at the early stage of the disease, these levels robustly elevated in the male mice after 3 days of infection [110]. The number of neutrophils infiltrated into the lungs of male mice was shown to be 5-fold higher as compared to female mice at day 4; suggesting that the susceptibility of male mice to SARS-CoV-2 infection correlates with increased inflammatory cell recruitment. The roles of sex hormones in the severity of the COVID-19 were illustrated in the Fig. 3 .
Fig. 3.
The roles of sex hormones in COVID-19. The differences in COVID-19 severity between genders have directed studies toward the roles of sex hormones in this context. Based on sex hormones, the susceptibility to SARS-CoV-2 has a range from “higher androgen” to “higher estrogen”; with this regard, men are more susceptible to experience the severe types of COVID-19 and women seem to show much milder disease. This difference is interestingly observed in ADT-treated men and menopausal women who have different levels of androgen and estrogen compared with non-treated men and non-menopausal women. Higher androgen results in neutrophil infiltration and higher expression of TMPRSS2 which are related to the severity of the disease. Higher estrogen, on the other hand, induces upregulation of TLR-7, TLR-8, CD40L, CXCR3, and IFNs that consequently promotes immune responses. Estrogen can also hamper inflammation via downregulation of IL-6, IL-8, IL-2R, and TNF-a.
2.4. Disease-related genes
2.4.1. Genes related to cilia dysfunction; DNAH7 and CLUAP1
One of the mechanisms through which SARS-CoV-2 could induce lung damage is mediated through the impairment of respiratory cilia. The expression of DNAH7, which is responsible for the maintenance of silica function, has been shown to be downregulated during infection of human bronchial epithelial cells by the SARS-CoV-2 virus [111]. Apart from SARS-CoV-2, some genetic variants of this gene could also reduce its expression levels, and thereby, increase the risk of mortality in COVID-19 patients. The variants of SLC39A10 including super-variant chr2_197, SNPs rs200008298 (3 prime UTR), rs4578880 (intron), and rs113892140 (upstream) are other genetic abnormalities that have been reported to be associated with cilia dysfunction in COVID-19 patients [112]. It has been also suggested that the upper variant consists of a SNP rs2301762, which is located in 5 prime UTR of gene CLUAP1, could induce cilia dysfunction and thereby increase the severity of COVID-19 [113], [114].
2.4.2. Genes related to cardiovascular diseases; DES and SPEG
Striated muscle preferentially expressed protein kinase (SPEG) is from the myosin light chain kinase protein family and is encoded by the SPEG gene. SPEG has an important role in the maintenance and activity of cardiac and skeletal muscles [115]. Additionally, DES is the gene that is translated into Desmin, a muscle-specific type III intermediate filament, which could regulate the architecture of the sarcomere [116]. The mutation in DES and SPEG has been previously shown to be associated with cardiomyopathy [116], [117]. Recently, it became evident that SNP rs71040457 located downstream of DES and the upstream of SPEG is associated with the incidence of cardiomyopathy in COVID-19 patients [118], [119].
2.4.3. Genes related to thromboembolic diseases
STXBP5/STXBP5-AS1 are one of the most important genes that their association with the thromboembolic disease have been previously established [120]. It has been indicated that the lack of STXBP5 in mice could increase von Willebrand factor levels in plasma, induce artery thrombosis, and impair platelet secretion and activation. Given these, STXBP5 is considered to be a strong risk factor for venous thromboembolic disease. Thus far, 101 SNPs have been identified in the gene encoding STXBP5; of which 89 are located in the intron of STXBP5 and 6 are located in the intron of gene STXBP5-AS1. Hu et al. reported that the variations within STXBP5/STXBP5-AS1 could increase the mortality risk in COVID-19 patients [121]. Abu-Farha et al. also introduced three genetic risk variants, including rs13109457-A, rs12029080-G, and rs6687813-A in COVID-19 patients that increased the levels of d-dimer and subsequently enhance the risk of thrombosis [122].
2.4.4. Genes related to mitochondrial dysfunction; TOMM7
During infection, the SARS-CoV-2 virus recruits the mitochondria of the infected cells to produce reactive oxygen species (ROS), an event that leads to excessive production of inflammatory cytokines and eventually cytokine storm [123], [124]. On the other hand, to avoid the progression of the disease, the immune system targets the mitochondria of the damaged cells to suppress SARS-CoV-2-induced production of ROS; an event which in turn leads to mitochondria dysfunction and induction of apoptosis in the infected cells [123]. The results of previous studies revealed that mitochondrial DNA are released from the damaged organs and the assessment of circulating cell-free MT-DNA in the plasma of COVID-19 patients could be a poor prognostic factor [125]. Given these, it has been suggested that early detection of MT-DNA in COVID-19 patients could be beneficial to provide a promising overview about the survival state of the patients, the risk of ICU hospitalization, or organ failure.
Recently, the genome analysis revealed the presence of 5 intergenic variant SNPs on chromosome 7, which are associated with mitochondria function [126]. For example, SNP rs55986907 seems to alter the expression of TOMM7, which encodes a subunit of the translocase of the outer mitochondrial membrane. The down-regulation of TOMM7 in COVID-19 patients could result in intensified mitochondria dysfunction.
2.5. Other genes
2.5.1. ABO blood group
It was reported that individuals with the gene encoding blood group A antigens have a higher risk for COVID-19 development as compared to other blood groups; on the other hand, those who harbor neither A nor B genes (blood group O) might have protective effects against the SARS-CoV-2 virus [66]. Even in deceased patients, this distribution pattern is observable. Apart from blood group A, Zietz et al. have indicated that cases who possess RHD gene might also have a higher rate of COVID-19 infection [127]. According to the study conducted by Barnkob et al., although ABO and RhD blood group could be a risk factor for the SARS-CoV-2 virus infection, these markers could not determine the chance of hospitalization or mortality in the patients [128]. In agreement, Latz et al. declared that having blood group A might not necessarily predict the chance of hospitalization in COVID-19 patients [129].
2.5.2. GSTT1-M1 genes
Glutathione S-transferases (GSTs) are from the family of enzymes that regulate cellular detoxification. The results of in-depth analysis revealed that polymorphism in genes encoding this family of enzymes could increase the risk of respiratory disease; proposing perhaps there is an association between GSTT1 and GSTM1 gene polymorphisms and the incidence of COVID-19 [130]. In this regard, Saadat et al. have reported the correlation between GSTT1 polymorphisms and the prevalence as well as mortality of COVID-19 in the East-Asian population [131]. Abbas et al. also conducted another study to evaluate the effect of GSTT1 and GSTM1 genes polymorphism on COVID-19 severity [132]. Their results indicated that individuals who carried GSTT1 −/− had a higher mortality rate as compared with GSTT1 +/+ carriers. The dual polymorphism in GSTT1 and GSTM1 genes (GSTT1 −/− and GSTM1 −/− genotype) may also increase the risk of pulmonary fibrosis in COVID-19 patients.
2.5.3. LZTFL1 genes
The 3p21.31 region has been shown to be involved in some kinds of respiratory failures previously. Leucine zipper transcription factor like 1 (LZTFL1) which its gene is located in 3p21.31 could regulate the trafficking of proteins to the ciliary membrane [133]. This protein has a regulatory role in the epithelial-mesenchymal transition (EMT) process which is a viral response pathway [134], [135]. According to multiomics and machine learning methods, a gain-of-function risk A allele of an SNP, rs17713054G > A was detected as the probable causative variant to enhance the expression of LZTFL1. Regarding the role of LZTFL1 in EMT process and the gain-of-function of 3p21.31, LZTFL1 could be a novel target in COVID-19 therapy [136].
3. Host epigenetic
3.1. Epigenetic regulation of ACE2
Comparison between the DNA methylation profiles across five genomic loci of the ACE2 promoter in COVID-19 patients revealed that there is a difference in methylation pattern according to sex [137]. In cancer patients, the hypomethylation of ACE2 promoter leading to ACE2 up-regulation has been reported, which makes these patients more vulnerable to COVID-19 [138]. Accordingly, Yang et al. used UALCAN database to ascertain the correlation between cancer and ACE2 hypomethylation [139]. The results were interesting, as it became evident that with the advance of age, carcinoma stage, and the severity of cancer, the methylation levels of the ACE2 promoter were significantly decreased [139]. Patients with oxidative stress disease such as lupus are another susceptible group to SARS-CoV-2 infection [140]. The results of the in-depth analysis succeeded to find the footprint of epigenetic regulation in this event. It has been suggested that in response to oxidative stress, hypomethylation might occur in the promoter of ACE2, an event that facilitates the viral entry to the host cells.
Apart from DNA methylation, post-translational modifications at the N-terminal tails of histones is another epigenetic-related mechanism that could regulate the expression of ACE2 [141]. In a mouse germ cell line, it became evident that the absence of EZH2 and subsequently down-regulation of H3K27me3 could increase the expression of ACE2 [142]. Additionally, alteration in the expression of HAT1, HDAC2, and KDM5B are other parameters that determines the sensitivity of an individual to the SARS-CoV-2 infection [143]. It should be noted that the impact of epigenetics on the susceptibility of the individuals to the SARS-CoV-2 virus is still a new field and numerous investigations should be yet conducted to ascertain the importance of post-translational modification of ACE2 on the outcome of the patients. But, if it becomes evident that epigenetic regulators of ACE2 might have a role in the prevalence rate of COVID-19, it could be suggested that epigenetics-based therapies might be a promising option, especially for high-risk groups.
3.2. X chromosome inactivation
Besides DNA methylation which could alter the expression of ACE2, X-chromosome inactivation is another epigenetic-related mechanism that could promote or attenuate ACE2 expression [144]. ACE2 gene is located on the area of the X-chromosome (Xp22.2) that can escape X-inactivation, which in turn, result in higher expression levels of ACE2 in females as compared to males [9], [145], [146]. It is well-established that the prevalence rate of COVID-19 is higher in females as compared to males; but, how the overexpression of ACE2 in females is not associated with the more aggressive type of the disease? The answer is found in the way that the SARS-CoV-2 virus binds to ACE2. The results of the in-depth analysis revealed that 16 residues of the receptor-binding motif (RBM) of SARS-CoV-2 binds to 20 residues on ACE2 in males [147]. In females, however, the same SARS-CoV-2 RBM can bind to ACE2 on either of the two X chromosomes. This unique event in females reduces the probability of pulmonary edema during COVID-19 [148].
TLR7 is another gene located on the X chromosome, which its role in the protection against the SARS-CoV-2 is well-established. In similarity to ACE2, TLR7 escapes from X-chromosome inactivation, but its expression is not dependent on the sex hormones [149]. Given this, it seems that the serum levels of TLR7 and in turn interferons are higher in females as compared to males. The results of a study revealed that loss-of-function variants of X-chromosomal TLR7 in males resulted in impaired type I and II IFN responses, and all in all, these findings confirm why the prevalence of severe types of COVID-19 is higher in males as compared to females [109], [150], [151].
3.3. Induction of epigenetic modifications by BCG vaccination
It has been claimed that some microbial products or selected live attenuated vaccines, such as BCG can train the immune cells to induce more vigorous immune responses through integrating with epigenetic modifications [152]. For example, in the models of yellow fever, Rob J W Arts et al. have reported that BCG vaccination could prime monocytes to release more IL-1β through epigenetic reprogramming, and thereby, reduce the viremia in the patients [153]. BCG vaccination has also shown to be associated with the increase in the expression of some epigenetic mediators, including H3K4me1, H3K4me3, and H3K27ac that could increase the secretion of pro-inflammatory cytokines such as IL-6, IL-1b, and TNF-α from monocytes [154]. The non-specific protective property of BCG vaccination against other viral infection, including influenza A (H1N1), herpes simplex virus (HSV), respiratory syncytial virus (RSV), and the human papilloma virus (HPV) has previously been reported [155]. Given the viral protection offered by BCG, it is proposed that perhaps BCG vaccination could induce a prophylaxis impact against the SARS-CoV-2 virus; and this is the reason proposed by many researchers that why children with intense vaccination program are more resistance against COVID-19 [142].
Numerous studies have also compared the COVID-19 crises in countries with mandated BCG policies and those which do not have universal vaccination program [156]. The results were interesting; it became evident that the number of infected cases and deaths were dramatically lower in the countries with BCG vaccination. Additionally, it seems that BCG vaccination not only could remarkably reduce the requirement to hospital admission in COVID-19 patients but also could ameliorate the effects of the virus on the hosts, leading to less severe type of the disease [157].
3.4. Regulation of inflammatory pathway genes by histone modifications
Once TLRs recognize the viral pathogen-associated molecular patterns (PAMPs) of the SARS-CoV-2 virus, they propagate the signaling that activates histone modification, which in turn, leads to the expression of IFNs and TNF [158]. It has been claimed that H3K4me3 and H3K9me2 are the main regulators of histone modification that could increase the expression of IFNs due to COVID-19 infection [159], [160], [161]. Another cytokine that its expression during COVID-19 could be regulated by the histone modification and cis/trans-factors interaction is IL-6. It has been reported that those individuals who were infected by SARS-CoV-2 possess higher positive histone modification markers in the promoter of genes encoding ACE2 and IL-6 [162], [163]. Through enhancing the activity of leukocytes, and reinforcing the expression of cytokines, cytokine receptors, IFN-stimulated genes, and signal transduction genes, histone modification could recruit the host defense responses in favor of COVID-19 progression.
3.5. Histone citrullination
Another epigenetic modification that has been identified in COVID-19 patients is citrullination of arginine residues on histones (Cit-H3), which facilitates the binding of transcription factors to the chromatin [144], [164]. In response to the elevated calcium levels and pro-inflammatory cytokines, such as TNF-α, peptidylarginine deiminases (PADIs) such as PAD1, PAD2, PAD3, PAD4, and PAD6 induce citrullination of histone H3 and H4, and increase the accessibility of transcription factors to the chromatin [165], [166], [167]. It should be noted that Cit-H3 plays a fundamental role in neutrophil extracellular traps (NETs) [168]. The cell-free DNA analysis from COVID-19 patients indicated that SARS-CoV-2 increases Cit-H3, and thereby, enforces neutrophils to release NETs, which subsequently induce apoptotic cell death in lung epithelial cells or induce cardiovascular complications.
Moreover, there is a positive association between the levels of Cit-H3 and platelet counts, suggesting that abnormal platelet count could also induce NETs in COVID-19 patients [169]. The analysis of RNA sequencing of lung samples harvested from COVID-19 patients showed that the expression of platelet factor 4 (PF4) was increased by 5.5-fold, shedding light on the importance of platelets in the induction of NETosis [170]. Notably, Cit-H3 could also sustain the pluripotent cell count during early embryogenesis [171]. Given the high prevalence of Cit-H3 in COVID-19 patients and based on the role of this epigenetic modification and stem cell pluripotency, several clinical trials are investigating the impact of cell therapy against COVID-19 [172].
3.6. Interaction between human epigenetics factors and SARS-CoV-2 proteins
Analyzing the interactions between virus and host proteins revealed that there are 132 proteins in humans that the SARS-CoV-2 virus could bind to them. Among these proteins, 8 are epigenetic modifiers. HDAC2, BRD2-BRD4, and CUL2 complex binds to viral NSP5, viral E protein, and viral ORF10, respectively [173]. Concerning HDAC2, it became evident that when NSP5 binds to this epigenetic modifier, it prevents the nuclear localization of HDAC2, and thereby, suppresses the regulatory impact of this molecule on inflammation and interferon responses. As mentioned before, HDAC2 enhances the expression of IFNs-specific genes (IGS) through deacetylation of H4K16 at ISG promoters [174]. ORF10 is another SARS-CoV-2 virus protein that through interaction with Cullin-RING E3 ubiquitin ligase complex consists of CUL2, ELOB, ELOC, RBX1, and ZYG11B enhance viral proliferation in the host’s cells [175], [176].
3.7. Epigenetic modulation of SARS-CoV-2 infection and age-related epigenetic
From the first description of COVID-19, one question remained unanswered that why the disease prevalence, as well as severity, are intensified by the advance of age? The answer to this question might be found, at least partly, in the lifetime accumulation of epigenetic modification during the aging process [14]. The alteration in the methylation of ACE2, expression of HDACs, and SIRTs in different ages all together suggest that epigenetic modulation could control the susceptibility of individuals to viral infections according to their age group [14], [115], [177]. Thus far, some models have been developed to estimate variation in methylation levels in selected DNA CpGs according to age, called biological age molecularly or DNAmAge [116]. Mongelli et al. have also reported that after COVID-19 infection, especially in those who were younger than 60 years, there is an increase in so-called DeltaAge; indeed, a positive DeltaAge is determined as an acceleration of the biological blood clock [178]. Telomere is another age-related driver that might be involved in COVID-19 [179]. The results of an interesting study revealed the negative correlation between telomerase length and the severity of COVID-19; the shorter is the telomeres length, the more aggressive is the type of the disease [180], [181]. Despite this finding, still, little is known about the predictive value of telomere length in COVID-19 severity.
4. Virus genome mutations
The study of mutations in SARS-CoV-2 has been investigated in numerous studies [182], [183], [184], [185]. In the following, the presence of alterations in SARS-CoV-2 variants is discussed.
4.1. Structural proteins
4.1.1. Spike protein
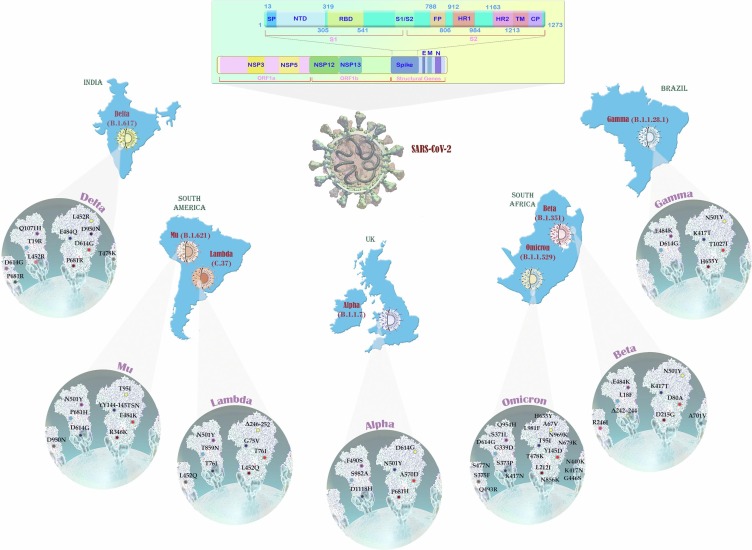
Regarding the continuous evolution of SARS-CoV-2, the following data is according to the time of article writing, 7 February 2022, as stated by WHO [186]. Indeed, the categorization of variants is performed based on variants of concern (VOCs), variants of interest (VOI), variants under monitoring (VUM), and formerly monitored variants.
4.1.1.1. Currently designated variants of concern (VOCs)
For these variants, clear evidence is available indicating a significant impact on transmissibility, severity, and/or immunity that is likely to have an impact on the epidemiological situation in the European Union (EU)/European economic area (EEA). The combined genomic, epidemiological, and in vitro evidence for these properties invokes at least moderate confidence [187].
4.1.1.1.1. WHO label: Alpha (VOC 20212/01, 20I/501Y.V1, or B.1.1.7)
The United Kingdom (UK) variant of the SARS-CoV-2 virus, also known as VOC 202012/01, 20I/501Y.V1, or B.1.1.7, was primarily detected in November 2020 and has shown to have a dramatic mortality rate as compared with previous variants. Notably, as compared to the parental strain of D614G, B.1.1.7 variant is more transmissible [188]. Given the high contagious rate, this variant of the virus turned to be the dominant form of the virus in England and other countries [189]. Seventeen mutations have formed this variant, among them 8 occurred in the S protein (6 substitutes and 2 deletions), 4 located in ORF1ab protein, 3 target ORF8 protein, and 2 on N protein [190]. It should be noted that the deletion at positions 69 and 70 (del69–70) causes S-gene target failure (SGTF) __which served as a proxy in the United Kingdom for identifying B.1.1.7 cases__ in at least one RT-PCR–based diagnostic assay [191], [192]. Substitution in amino acids N501Y, A570D, P681H, T716I, S982A, and D1118H are other spike mutations that have been recognized in this variant of the virus [193]. Interestingly, it has been claimed that the majority of mutations in the B.1.1.7 variant are adoptive and have arisen due to positive selection. It is also suggested that these mutations probably have been formed in an immunocompromised individual and in response to convalescent plasma treatment [194].
Another mutation that has been identified in B.1.1.7 variant is Q27stop that truncates the ORF8 and results in immune escape of the virus [195]. In another study, Mohandas et al. have evaluated the pathogenicity B.1.1.7 variant and compared it with a variant with D614G mutation in hamsters [196]. Their results revealed that the disease produced by B.1.1.7 variant was coupled with bodyweight loss, but mild lung pathology. Although the impact of B.1.1.7 variant on the mortality rate of the host has not yet been established, it is evident that B.1.1.7 variant is more transmissible rather than lethal.
4.1.1.1.2. WHO label: Beta (B.1.351, 501Y.V2)
B.1.351 variant which was firstly found in South Africa in December 2020 is one of the most transmissible variants of the SARS-CoV-2; but in similarity to B.1.1.7 variant, the effect of this variant on the mortality rate of the disease has not yet been established [193], [197]. This variant was more prevalent and invasive in young individuals with no underlying health conditions. Using a mathematical model, it became evident that this variant is 50-fold more transmissible than previously circulating variants found in South Africa [198]. So far, 9 mutations have been identified in the S protein of the virus, including L18F, D80A, D215G, Δ242–244, R246I, K417N, E484K, N501Y, and A701V [199]. It should be noted that N501Y mutation, which is in common with B.1.1.7 variant, can reinforce the affinity of the virus to ACE2, and may reduce the efficacy of three classes of therapeutically relevant monoclonal antibodies (mAbs) and neutralizing antibodies especially against B.1.351 variant [200]. One of the major concerns about B.1.351 variant in South Africa was the interaction of the virus with HIV; although it seems that HIV infection could not affect SARS-CoV-2 replication, further evaluation is still required in this field [197], [201].
4.1.1.1.3. WHO label: Gamma (P.1, B.1.1.28.1)
The lineage P.1 (Gamma variant), which has been found in Brazil in January 2021, acquired 21 mutations including 1 insertion, 1 deletion, 4 synonymous mutations, and 15 non-synonymous mutations [202]. Notably, three of these mutations (K417T, E484K, and N501Y) could enhance the affinity of the virus to ACE2. It should be noted that as compared with B.1.1.7 and B.1.351, this variant has the most changes in the spike protein. It has been suggested that the P.1 variant is 2.2-fold more transmissible than previous non-P1 variants, especially in adults [203]. It has also been reported that P.1 variant might be more lethal than previous variants [202], [204]. Martins et al. have suggested that there is a correlation between SARS-CoV-2P.1 variant and the hospitalization [205].
4.1.1.1.4. WHO label: Delta (G/452.V3, B.1.617)
Delta variants of the SARS-CoV-2 virus (B.1.617.2) were first detected in India in March 2021, but very soon, they have found their way to other countries [206]. The outbreak of this variant was coupled with the sharp increase in the number of SARS-CoV-2 cases and deaths. It has been reported that Delta variants are about 50-times more transmissible than Alpha variants as it displaced Alpha variant as the dominant strain in England [207]. The viral load is also higher in Delta variant as compared with other variants. Accordingly, a study in Scotland showed that the hospital admission rate for patients with the Delta variant was as twice higher as the Alpha variant [208]. Thus far, three distinct lineages have been identified within this variant, each of them has different mutation profiles. For example, B.1.617.1, also known as VOI, was defined by the spike protein amino acid alterations, including L452R, E484Q, D614G, P681R, and Q1071H. B.1.617.2, which is the most contagious virus in this category and labeled as VOC-21APR-02, was defined by the spike mutations such as T19R, Δ157-158, L452R, T478K, D614G, P681R, and D950N; but it lacks mutation at E484Q. While according to WHO, B.1.617.2 is classified as a VOI as a result of its high transmissibility, the UK classifies this virus as VOC [200], [209].
4.1.1.1.5. WHO label: Omicron (B.1.1.529)
On 26 November, WHO determined the B.1.1.529 lineage as a new VOC which is designated as omicron [210]. According to data from South Africa, the daily numbers of cases diagnosed with COVID-19 showed a considerable rise from 273 on 16 November to more than 1200 by 25 November. Most newly diagnosed individuals were from the province of Gauteng where the first Omicron SARS-CoV-2 was identified [211]. Notably, the infection percentage of Delta variant was 80 % which is determined as about 90 % for the Omicron, implying the higher transmissibility of Omicron over Delta. Also, the early doubling time of Omicron was lower than Delta and Beta as well (1.2 vs 1.5 and 1.7, respectively) [212]. Of note, the Omicron variant showed a higher half-life than others and analysis of virus survival time showed this variant tends to be more persistent on plastic and skin surfaces compared with other variants [213]. Inline, according to a recent retrospective study conducted in South Africa, the rate of reinfection due to Omicron was higher than the previous variants [214]. Also, investigating the neutralizing activity of sera against a range of VOCs showed that the Omicron variant has resistance to neutralization of patient sera who affected other variants, while the Alpha variant could induce a wide range of immunity [215].
The concern about this particular variant was initiated from the presence of notably large number of mutations (more than 60 mutations including substitutions, deletions, and insertions) [216], several of which are in the spike of the virus [211]. Mutations in the spike include 30 substitutions including A67V, T95I, Y145D, L212I, S371L, G339D, S373P, S375F, N440K, K417N, G446S, S477N, T478K, Q493R, E484A, G496S, Y505H, Q498R, N501Y, T547K, D614G, H655Y, P681H, N679K, N764K, D796Y, Q954H, N856K, N969K, and L981F; one insertion (three amino acids, [EPE]) at position 214, and also three deletions out of H69/V70, G142/V143/Y144, and N211. Notably, D614G in the spike seems a vital mutation available in all VOCs [217], which was shown to be associated with higher upper respiratory tract infection and lower age of patients [218]. Six substitutions (L2084I, K856R, A2710T, P3395H, T3255I, and I3758V) and two deletions in four amino acids (amino acid 2083, 3674, 3675, and 3676) were also detected within ORF1a of Omicron variant. Besides, ORF9b harbors P10S substitution and three deletions at positions 27–29. There are some other mutations in the envelope, membrane, and nucleocapsid; however, the main mutations are located in the spike gene. Based on the mutations, the Omicron variant has recently categorized into two lineages, BA.1 and BA.2 [219]. According to whole-genome mutational mapping and phylogenetic analysis, both BA.1 and BA.2 carry 51 mutations in the genome which 32 are shared with both lineages and each of them harbors 19 signature mutations; among those 19 mutations, BA.1 shows 13 and BA.2 has 7 in the S region [220].
In addition, according to the NextStrain, recent evidence of monitoring SARS-CoV-2 genome showed new massive mutations in a 56 years old man infected with the Delta variant include 34 mutations in the spike [221]. The proximity of those mutations in the Delta variant and the typical mutations of the Omicron variant might strengthen the hypothesis of co-circulation of Delta and Omicron and the emergence of the so-called DelMicron which is not a new variant, but a combination of two existing strains: Delta and Omicron.
4.1.1.2. Currently designated variants of interest (VOIs)
Variant of interest (VOI) is the variant that has specific genetic markers and is related to alteration of receptor binding, reduction of neutralization strength by antibodies of previous infection or vaccination, and diminished effectiveness of therapies that might need one or more public health actions [187]. Although the genomic, epidemiologic, and in vitro evidence are available for these variants, the findings are relatively in their infancy and more details should be obtained.
4.1.1.2.1. WHO label: Lambda (C.37)
C.37 was another lineage of the SARS-CoV-2 virus found in South America in August 2020 [206]. It possesses a deletion in the ORF1a gene (Δ3675-3677) which is in common with Alpha, Beta, and Gamma variants, and also carries several nonsynonymous mutations in the spike gene, including Δ246-252, G75V, T76I, L452Q, F490S, and T859N [206]. Notably, while T76I and L452Q mutations at the spike protein could increase the infectious ability of the variant as compared to other types of the virus, deletion Δ246-252 occurs at the N-terminal domain of the spike protein is responsible for immune evasion of the virus [207]. It should be noted that although this variant is still a VOI, the resistance against developed vaccines might be a concern, as it could cause another breakthrough infection [222].
4.1.1.2.2. WHO label: Mu (B.1.621)
The Mu variant was initially isolated on January 2021, from Colombia. During the dominance of the Gamma variant, the Mu variant outnumbered the other ones and turned into the dominant epidemic form in Colombia; with this regard, until August 30, 2021, this variant was recognized in 39 countries. Spike domains of the Mu variant harbor several mutations such as T95I and YY144-145TSN in the N-terminal domain (NTD), the E484K, R346K, and N501Y in RBD, and the P681H, D614G, and D950N in other regions [209]. Notably, the E484K __which is also shared with the Beta and Gamma variants__ could markedly decrease the sensitivity of the virus to antibodies produced after vaccination and/or natural SARS-CoV-2 infection [223], [224].
4.1.1.3. Currently designated variants under monitoring (VUM)
These additional variants of SARS-CoV-2 have the same infectious properties as VOC; however, data of phenotypic or epidemiological features are vague which needs boosted monitoring [187].
4.1.1.3.1. Az.5
AZ.5, formerly known as B.1.1.318, carries D614G, E484K, D796H, T95I, P681H, and ΔY144 mutations. A study from Gabonese population demonstrated that AZ.5 variant had an elevated (31 %) circulation between February and May 2021 which might be due to D614G, E484K, and ΔY144 mutations that may account for higher transmission and attenuated vaccine efficacy [225].
4.1.1.4. Formerly monitored variants
Formerly monitored variants are former VOCs, VOIs, and VUMs that were recategorized due to at least one of the following criteria: 1) the variant is no longer detected globally, 2) the variant has been circulating for a long time, but with no influence on the general epidemiological circumstances, or 3) the variant has no concerning properties according to scientific data [186].
4.1.1.4.1. WHO label: Zeta (P.2)
The Zeta variant or lineage P.2 of the SARS-CoV-2 virus is indeed a sub-lineage of B.1.1.28, which was first detected in Brazil in April 2020 [226]. Unlike B.1.1.28, it does not have N501Y and K417T mutations; however, it possesses the E484K, D614G, and V1176 mutations. Some of the Zeta variants, but not all, might have F565L mutation, as well. The prevalence of the Zeta variant is relatively lower than B.1.1.28 and<0.5 % of the viral samples detected worldwide show this strain [227]. As compared to the original B.1 variant, the Zeta variant might bring more severe type of disease. Also, it needs the higher titer of antibodies to be neutralized as compared to the original B.1 variant. Indeed, this variant contains many mutations presented in the Alpha, Beta, Delta, and Gamma variants that make the virus easier to penetrate the target cells and induce resistance to vaccines.
4.1.1.4.2. WHO label: Iota (B.1.526)
The Iota variant, also known as B.1.526 detected in November 2020, was first considered to be a VOC. D614G, E484K, and T951 were three mutations that have been detected in the spike protein of this variant [228]. The former was reported to increase the affinity of the virus to ACE2 and E484K was suggested to be associated with immune escape from monoclonal as well as neutralizing antibodies [229]. Notably, it was indicated that T951 mutation might not have any effect on the antigenicity of the virus [230]. Higher affinity toward ACE2 and elevated spike stability, viral infectivity, viral fusogenicity, and viral replication were observed in this variant [231].
4.1.1.4.3. WHO label: Eta (B.1.525)
The variant B.1.525, known as the Eta variant, was first identified from the nasopharyngeal swabs of two asymptomatic COVID-19 cases in Nigeria in December 2020 [232]. This variant is now circulating in twenty-six countries. Unlike the Alpha, Beta, and Gamma variants, Eta variant does not carry N501Y mutation. It possesses, E484K, N439K, Y453F mutations, and ΔH69/ΔV70 deletion. Indeed, the co-existence of E484K mutation and a new F888L mutation makes Eta variant distinct from other variants of the SARS-CoV-2 virus [233]. These mutations have increased its transmissibility, facilitated the viral escape from antibodies, and enhanced the risk of re-infection in patients [232].
4.1.1.4.4. WHO label: Kappa (B.1.617.1)
Kappa variant of the SARS-CoV-2 virus is one of the three sub-lineages of B.1.617, which is also known as B.1.617.1 [234]. This variant was first identified in India in December 2020 and was characterized by three amino acid alterations, including L452R, E484Q, and P681R in the spike protein. Among them, L452R could increase the affinity of the spike protein for ACE2 and induce immune escape. E484Q also enhances the binding of the protein to ACE2. Overall, this variant is a double mutant strain of the virus that appeared worrying. The E484Q mutation, which is identical to the E484K mutation, and the L452R mutation, enable the evasion of the virus from the immunity [235], [236], [237].
4.1.1.4.5. WHO label: Epsilon (B.1.427, B.1.429)
As a sub-lineage of B.1.427 and B.1.429, epsilon variant (CAL.20C) which was firstly detected in July 2020 possesses five mutations in the spike protein that among them I4205V and D1183Y occur in the ORF1ab-gene and S13I, W152C, and L452R occur in the spike protein's S-gene [236], [238]. It has been demonstrated that the higher titers of antibody form vaccinated individuals with a Wuhan-1 isolate–based messenger RNA vaccine or convalescent individuals are required to neutralize Epsilon variant; indeed, the L452R mutation is the reason that reduces the efficacy of antibodies [239]. Moreover, the S13I and W152C mutations could result in a total loss of neutralization for all NTD-specific mAbs. Carroll et al. revealed that the Epsilon variant has higher virulence and more rapid replication in the upper respiratory tract as compared to ancestral B.1 (614G) or B.1.427 [240]. The authors also reported that Epsilon variant has faster dissemination from airways to parenchyma, and thereby, it could induce more severe lung damage at early and late stages of the disease.
4.1.1.4.6. WHO label: Theta (P.3)
The results of the genome sequencing of COVID-19 samples from the Philippines resulted in the identification of two new mutations, E484K and N501Y, in the Spike protein of the SARS-CoV-2 virus [241]. Very soon, it became evident that these mutations could emerge a new variant of the virus known as lineage P.3 or theta variant, primarily identified in February 2021. P.3 variant contains similar VOC mutations in the Spike protein, such as E484K, N501Y, and P681H, which affect the transmissibility and immune escape of the virus [242]. Other mutations, including D614G, H1101Y, E1092K, and V1176F are also detectable in more than 95 % of P.3 variants. In addition to these, synonymous spike protein mutations (593G and 875S), three amino acid deletion (LGV141_143del), as well as Q1180H, have been reported to exist in most, but not all P.3 variants [243], [244], [245].
4.1.1.5. An overview of notable missense mutations in SARS-CoV-2 spike
4.1.1.5.1. D614G mutation
It was reported that an amino acid change in the spike of the virus, D614G, was presented early during the pandemic, and viruses containing G614 are now prevalent in many places around the world [246]. Ozono et al. have reported that among numerous identified mutations in the spike protein, the D614G mutation was associated with the highest viral entry, suggesting that variants possessing this mutation might be more transmissible [247]. Plante et al. have also suggested that SARS-CoV-2 with D614G mutation could produce higher infectious titers in nasal washes and the trachea, but not in the lungs; suggesting that this variant may mainly infect the upper respiratory tract [248]. It should be noted that the D614G mutation might assemble a more functional S protein into the virion, and thereby, produce stronger pathogenicity.
By using GISAID (Global Initiative on Sharing All Influenza Data) and bioinformatics models, Korber et al. have proposed that G614 mutation is mostly accompanied by three other mutations, including C241T, C3037T, and C14408T (C and T imply cysteine and threonine, respectively) that could change the amino acid sequence of RNA-dependent RNA polymerase (RdRp) [249]. Volz et al. have indicated that infection with the G614 variant was associated with higher viral load and younger age of patients [250]. Moreover, Flores et al. proposed that the higher pathogenicity of the G614 variant might have an impact on the vaccination program [251]. To obviate this challenge, Weissman et al. have tested the efficacy of the antibodies against G614 and stated that the G614 pseudovirus was moderately susceptible to neutralization either by mAbs against the RBD or convalescent sera [227].
4.1.1.5.2. N440K mutation
The reports suggest that the existence of N440K mutation in SARS-CoV-2 increases the lethality of the virus at least 15-times as compared to earlier variants [252]. Moreover, the N440K variant could induce higher infectious viral titers as compared to the Alpha variants. It should be noted that despite the concerns about this variant, recent reports suggested that N440K variant is slowly being replaced by other variants such as B.1.617 and B.1.1.7.
4.1.1.5.3. L452R mutation
L452R, a common mutation found in the Delta and Kappa variants, replaces leucine (L) with arginine (R) at position 452 in the RBM of RBD [253]. This mutation not only enhances the affinity of the virus to ACE2 but also reduces the vaccine-stimulated antibodies and confers escape from HLA-A24-restricted cellular immunity; HLA-A24 is a prominent HLA-class I alleles in the East/Southeast Asian populations [254]. L452R mutation could also increase the stability of the S protein, enhance viral replication, and thereby, increase viral infectivity [255].
4.1.1.5.4. S477G/N mutation
The S477 mutation (S477G and S477N) is the most frequently exchanged amino acid residue in the RBDs of SARS-CoV-2 mutants, which reinforces the interaction between the spike protein and human ACE2 [232]. Notably, since serine (S) 477 is located in a solvent-exposed random coil loop, it does not have any interaction with ACE2; but, the substitution of this amino acid with asparagine could establish a hydrogen bond and stabilized spike protein-ACE2 interaction [233].
4.1.1.5.5. E484Q/K mutation
The E484Q mutation observed in the Kappa variant could reinforce the affinity of the virus to ACE2 receptor and diminish the ability of vaccine-stimulated antibodies to target this altered spike protein [256]. E484K is another mutation that was found frequently in Gamma [257], Zeta [226], and the Beta variants [203]; this mutation is known as an escape mutation, as it can shield the virus from the host’s immune responses or mAbs against the SARS-CoV-2 virus [258].
4.1.1.5.6. N501Y mutation
N501Y mutation (also known as Nelly) enhances the affinity of the spike protein to ACE2 [259]. This mutation is found in Alpha, Beta, and COH.20G/501Y variants [238]. In a study conducted by Tian et al., the authors combined kinetics assay, single-molecule technique, and computational method to study the interaction between RBD of spike protein and ACE2 [260]. Their results indicated that RBD with the N501Y mutation showed a stronger interaction with ACE2. Interestingly, Liu et al. have reported that among different spike protein substitutions, only N501Y was able to replicate in the upper airway in the hamster model and primary human airway epithelial cells [241].
4.1.1.5.7. P681H mutation
The non-synonymous P681H mutation found in Alpha variant and lineage B.1.1.207 results in the replacement of proline (P) by histidine (H) at position 681. The results of the epidemiological analysis revealed that this mutation does have any association with neither higher infection rate nor higher prevalence [242], [261]. The variants of the virus possessing this mutation could be effectively neutralized by the sera from fully vaccinated individuals [262]. The results of Lubinski et al. also indicated that spikes containing P681H could be more easily cleaved by FURIN-like proteases [263].
4.1.1.5.8. P681R mutation
The results of the in-depth analysis revealed that SARS-CoV-2 variants harboring P681R mutation have higher pathogenicity, as compared to the parental virus, probably due to enhanced viral fusogenicity; however, the results of in vitro assays indicated that P681R variant did not necessarily show higher growth kinetics [264]. Liu et al. also indicated that the presence of P681R mutation in Delta, but not Alpha variant, is associated with enhanced replication via increased S1/S2 cleavage [265].
4.1.1.5.9. A701V mutation
At the end of 2020, the Malaysian Ministry of Health succeeded to identify a new mutation within the spike protein in 60 samples at which alanine (A) was replaced by valine (V) at position 701 [98]. Apart from Malaysia, this mutation has been reported in South Africa, Australia, the Netherlands, and England [99]. The prevalence rate of A701V is about 0.18 % of cases, and it was shown that the presence of this mutation could accelerate the transmission rate of the virus [266]. Indeed, A701V mutation which was detected in the Beta variant could be one of the reasons behind the higher transmissibility of the Beta variant [267].
To provide an accurate overview, we summarized all the relevant data concerning SARS-CoV-2 variants and mutations in Table 2 . Also, the structure of SARS-CoV-2 genome and the distribution of its variants were represented in Fig. 4 .
Table 2.
The details of SARS-CoV-2 variants, their mutations and characteristics.
| PANGO Linage | WHO label | Location | Identified | Key Spike mutations | Characteristics |
|---|---|---|---|---|---|
| Currently designated variants of concern (VOCs) | |||||
| B.1.1.7 | Alpha | United Kingdom | September 2020 | N501Y, D614G, P681H | This variant resulted in a highe hospitalization rate, need for ICU, and evasion from the immune system; however, was sensitive to vaccines. |
| B.1.351 |
Beta | South Africa | October 2020 | K417N, E484K, N501Y, D614G, A701V | Elevated hospitalization and need for ICU were associated with the Beta variant that accompanied with resistance to mAbs, convalescent plasma, and vaccine sera as well. |
| P.1 | Gamma |
Brazil | January 2021 | K417T, E484K, N501Y, D614G, H655Y | Gamma variant resulted in an elevated hospitalization and mortality rate that was resistant to mAbs, convalescent plasma, and vaccine sera. |
| B.1.617.2 | Delta | India | February 2021 | L452R, T478K, D614G, P681R | Delta variant has been a serious variant with much higher transmissibility, hospitalization rate and mortality. This variant was less susceptible to mAbs than the Wuhan strain. |
| B.1.1.529 | Omicron | South Africa and Botswana | November 2021 | A67V, D614G, T95I, H655Y, N679K, … | Omicron variant has the highest transmissibility, while has shown lower hospitalization rate and deaths. It has higher resistance to mAbs, but is weaker against cellular immunity. |
| Currently designated variants of interest (VOIs) | |||||
| C.37 | Lambda | Peru | December 2020 | L452Q, F490S, D614G | This variant showed an elevated immune resistance. |
| B.1.621 | Mu | Colombia | January 2021 | R346K, E484K, N501Y, D614G, P681H | It could evade immune defenses in a similar way to the Beta variant and had lower sensitivity to Abs. |
| Currently designated variants under monitoring (VUM) | |||||
| C.36 + L452R | N/A | Egypt | December 2020 | L452R, D614G, Q677H | This variant could escape from the cellular immunity restricted to HLA-A24. |
| AZ.5 (B.1.1.318) | N/A | India | January 2021 | E484K, D614G, P681H | Due to several mutations, it could escape from immune responses. |
| P.1 + P681H | N/A | Italy | February 2021 | D614G, E484K, H655Y, K417T, N501Y, P681H | N/A |
| B.1.617.2 + E484X (d) | N/A | India | April 2021 | L452R, T478K, D614G, P681R, E484X (d) | This variant is a part of B.1.617.2 (Delta). |
| B.1.617.2 + Q613H | N/A | India | April 2021 | L452R, T478K, D614G, P681R, Q613H | N/A |
| B.1.617.2 + Q677H | N/A | India | April 2021 | L452R, T478K, D614G, P681R, Q677H | It had resistance to mAbs. |
| B.1.617.2 + K417N | Delta plus | UK | June 2021 | L452R, T478K, D614G, P681R, K417N | This variant has been associated with high mortality rates, evasion from immunity and lower mAbs effects. |
| C.1.2 | N/A | South Africa | June 2021 | D614G, E484K, H655Y, N501Y, N679K, Y449H | It could evade mAbs effectively. |
| AY.4.2 | N/A | UK | June 2021 | L452R, T478K, D614G, P681R, A222V, Y145H | N/A |
| Formerly monitored variants | |||||
| B.1.1.523 | N/A | Russia | May 2020 | E484K, S494P | Some mutations in it were similar to the Delta variant which facilitate its escape ability from the immune responses. |
| B.1.427/B.1.429 | Epsilon | USA | September 2020 | L452R, D614G | The higher mortality rates and lower neutralization titers were two characteristics of the Epsilon variant. |
| C.16 | N/A | N/A | October 2020 | L452R, D614G | N/A |
| B.1.526.1 | N/A | USA | October 2020 | L452R, D614G | N/A |
| B.1.1.519 | N/A | Mexico | November 2020 | T478K, D614G | This variant was associated with increased hospitalization and mortality rates and reproduction number. |
| B.1.1.7 + E484K | N/A | UK | December 2020 | E484K, N501Y, D614G, P681H | N/A |
| B.1.525 | Eta | Nigeria | December 2020 | E484K, D614G, Q677H | It had a lower neutralization rate. |
| B.1.617.1 | Kappa | India | December 2020 | L452R, E484Q, D614G, P681R | Kappa variant showed a higher transmissibility* and greater ability to escape from vaccination. |
| B.1.214.2 | N/A | Belgian |
December 2020 | Q414K, N450K, ins214TDR, D614G | The severity and the ability to evade the immune responses were higher in this variant. |
| A.23.1 + E484K | N/A | UK | December 2020 | V367F, E484K, Q613H | It showed 1.3 % mortality rate. |
| A.28 | N/A |
N/A | December 2020 | E484K, N501T, H655Y | N/A |
| B.1.351 + P384L | N/A |
South Africa | December 2020 | P384L, K417N, E484K, N501Y, D614G, A701V | N/A |
| B.1.526 | Iota | USA | December 2020 | E484K, D614G, A701V | This variant had higher mortality among older adults and an enhanced ability to evade the immunity. |
| B.1.526.2 | N/A | USA | December 2020 | S477N, D614G | N/A |
| P.2 | Zeta | Brazil | January 2021 | E484K, D614G | It resulted in notable reduction in the neutralization ability of vaccine and mAbs. |
| P.3 | Theta | The Philippines | January 2021 | E484K, N501Y, D614G, P681H | N/A |
| A.27 | N/A | France | January 2021 | L452R, N501Y, A653V, H655Y | This variant could evade the immune responses. |
| B.1.351 + E516Q | N/A |
N/A |
January 2021 | K417N, E484K, N501Y, E516Q, D614G, A701V | N/A |
| B.1.1.7 + L452R | N/A | UK | January 2021 | L452R, N501Y, D614G, P681H | It induced higher resistance to mAbs and evaded HLA-restricted immunity. |
| B.1.1.7 + S494P | N/A | UK | January 2021 | S494P, N501Y, D614G, P681H | This variant induced resistance to Abs, particularly in combination with N501Y. |
| AT.1 | N/A | Russia | January 2021 | E484K, D614G, N679K, ins679GIAL | N/A |
| R.1 | N/A | Japan | January 2021 | W152L, E484K, G769V | This variant continued to appear in Japan and was associated with immune escape. |
| B.1.616(c) | N/A | France | February 2021 | V483A, D614G, H655Y, G669S | N/A |
| B.1.620 | N/A | Lithuania | February 2021 | S477N, E484K, D614G, P681H | It had higher severity with an enhanced ability to evade the immunity and mAbs neutralization. |
| B.1.617.3 | N/A | India | February 2021 | L452R, E484Q, D614G, P681R | N/A |
| AV.1 | N/A | UK | March 2021 | N439K, E484K, D614G, P681H | N/A |
WHO: World lealth organization; PANGO: Phylogenetic assignment of named global outbreak; PHE: Public health engineering; VOC: Variant of concern; ICU: Intensive care units; mAbs: Monoclonal antibody; VOI: Variant of interest; HLA: Human leukocyte antigen; N/A: Not available.
*; Cause community transmission in several countries, but have not yet been confirmed to be more infectious or transmissible.
Fig. 4.
The structure of SARS-CoV-2 genome and its variants. The genome of SARS-CoV-2 consists of ORF1a, ORF1b, and structural genes. While NSP3 and NSP5 of ORF1a encode papain-like protease and 3CL protease respectively, NSP12 and NSP13 of ORF1b encode RNA polymerase and helicase, respectively. The structural genes are responsible for the spike (S), envelope (E), membrane (M), and nucleocapsid (N) encoding. The spike-encoding region consists of two general regions S1 and S2 where the majority of mutations are related to this part. As illustrated, SARS-CoV-2 is categorized into different lineages based on the presence of mutations. Each bunch of mutations could give the SARS-CoV-2 various pathogenicity capacities letting variants have different transmissibility and severity.
4.1.2. Envelope
The envelope protein (E protein) __the smallest structural component of SARS-CoVs that controls ion channel activity, virion assembly, and replication__ is responsible for severe acute respiratory syndrome[268]. The results of a study conducted by Chai et al. also indicated that the DLLV motif which is located at the C-terminal of E protein makes an interaction with the PDZ and SH3 domains of PALS1 (protein associated with Lin-7) located on the epithelial of lung cells [269]. NCBI database succeeded to identify about 259 non-synonymous mutations over the C-terminal domains (CTD), NTD, and transmembrane of E protein from different samples collected from all over the world [270].
Generally, mutations in E protein might have an influence on the replication and propagation of the SARS-CoV-2 virus [252]. In this vein, 16 mutations have been identified at CTD of E protein that target the DFLV amino acid motif and affect the interaction of the E protein with PALS. Mutations such as Ser68Phe, Pro71Ser, and Leu73Phe are other CTD-related mutations in E protein that affect protein stability [270].
4.1.3. Membrane
Mutations in the membrane (M) gene of the SARS-CoV-2 virus is rare [254]. The mutations in the M gene (M:I82T mutation) results in the emergence of a new sub-B.1 clade, B.1.I82T; the frequency of this mutation was increased in the USA and Europe. It has been reported that M mutations could increase the pathogenicity of the virus for younger individuals. D178H is another identified mutation in the M protein; the sub-lineage B.1.1.7-M:V70L-S:D178H is found exclusively in 31 states of the USA. The V70L mutation is another genetic abnormality that has been identified in the TM2 transmembrane helix [254].
4.1.4. Nucleocapsid
Nucleocapsid (N) protein of the SARS-CoV-2 virus possesses three dynamic disordered regions that contains putative transiently-helical binding motifs [271]. In a study conducted in Saudi Arabia, two consecutive mutations, including R203K and G204R were found in the N protein of SARS-CoV-2; notably, these mutations have been found mostly in B.1.1.7 and P.1 lineages [272]. Interestingly, it seems that the frequency of R203K and G204R mutations is growing globally, as their global incidence frequency (IF) increased from 0 % in August 2020 to 76 % in November 2020. These mutations in N protein have shown to be associated with enhanced viral RNA binding to host proteins. Moreover, the aforementioned alterations could impair interferon responses in the host. It has also been reported that R203K and G204R mutations could increase the infectivity of the virus for the host’s cells, and thereby, produce the more severe type of the disease.
4.2. Non-structural proteins
4.2.1. ORF1ab (open reading frame1ab)
ORF1ab (ORF1a/b) refers to ORF1a and ORF1b that are conserved in the genomes of SARS-CoV-2. The genes encode large polyproteins that are catalyzed into several non-structural proteins by proteolysis [257]. Six ORFs exist in the SARS-CoV-2 genome that is participated in the regulation of viral replication and cellular signaling [273]. It has been indicated that V121D substitution could destabilize the ORF1ab non-structural protein-1 (NSP1), which is involved in the activation of the type-1 interferons, and attenuate the efficacy of vaccination [259]. The three-dimensional structure analysis of SARS-CoV-2 also revealed that a serine was replaced by glycine at position 723 in the ORF1ab NSP2 and a proline was replaced by isoleucine at position 1010 in the ORF1ab NSP3. Indeed, the mutation in NSP2 could increase the transmissibility of the virus, however, the mutations in NSP3 could differentiate SARS-CoV-2 from other SARS viruses [274].
Other mutations that were identified in NSP3 are V843F, A889V, and G1691C. While G1691C reduces the flexibility of the protein, V843F and A889V alter the biding pattern of SARS-CoV-2 and reduce the sensitivity of the virus to papain-like protease (PLPro) inhibitor GRL0617 [259]. The V843F mutation could also attenuate the ability of the virus to stimulate interferon-stimulated gene 15 protein (ISG15), and thereby, enhance viral spread. Notably, simultaneous mutation of V843F and A889V might have the same binding affinity as wild-type PLPro; this could be due to the fact that, unlike V843F which could damage the protease structure, A889V could neutralize this damage.
ORF1ab is another non-structural 6796 amino acids protein that in complex with several proteins could regulate viral genomic replication and synthesis. So far, 8 mutations have been identified in the gene encoding ORF1ab [275]. T609I mutation has been identified from the SARS-CoV-2 virus in California/United States, whereas G818S and F4321L were found from the samples collected from Sweden and India. In addition, M902I and T6891M have been found in Korea and F3071Y, S3120L, L3606X, and L3606F were detected from samples of Spain, China, Italy, and Japan, respectively [275].
4.2.2. Other ORFs
Freundt et al. have indicated that mutation in ORF3a protein, which is necessary for vesicle formation and Golgi fragmentation, could attenuate the ability of SARS-CoV-2 to induce cell death in the host’s cells [276]. Also, Hsing et al. have identified 4 mutations in the gene encoding ORF3a of the SARS-CoV-2; G251V mutation obtained from Italian, Korean, and Swedish COVID-19 patients, K136X and G196V were found in variants from Spain, and M128L was detected from the variants found in Korea [275].
In another study conducted on 2 Italian COVID-19 patients who were infected by B1.1 lineage, a nucleotide mutation was found in ORF6, which eliminated 6 amino acids from the C-terminal of the protein. This mutation was unique: although it affects neither viral replication nor neutralizing activities of the patients’ antibodies, it impairs antiviral IFN-based host response [277].
Hsing et al. also recognized 1 mutation (L84S) in the gene encoding ORF8 from the SASR-CoV-2 variants in Spain, India, and China [275]. Accordingly, Pereira et al. have detected several nonsense mutations and 3 deletions in the ORF8 gene [195]. Even though the exact function of ORF8 is still unknown, this protein seems to have the most variable structure and it appears that the changes in OFR8 could affect the spread of the virus.
5. An overview of the effects of mutations on the efficacy of vaccines
Although one side of the battle is the virus itself, researchers put much efforts to develop effective therapeutic approaches such as antiviral agents in order to overcome it which have been discussed in several studies [278], [279], [280], [281]. As the COVID-19 pandemic began, strives to develop effective vaccines were started wishing to win this battle. The first administration of anti-SARS-CoV-2 vaccine goes back to December 2020. Until 23 December 2021 (the latest guidance document of WHO), more than 20 vaccines have been confirmed to be evaluated according to WHO conditions and 14 have finalized the status of assessment [282]. Currently, 9 vaccines have been approved and utilized worldwide, including BBIBP-CorV (Sinopharm), CoronaVac (Sinovac Biotech), Covaxin (Bharat Biotech), AZD1222 (University of Oxford and AstraZeneca), Ad26.COV2.S (Janssen and Johnson & Johnson), Sputnik V (Gamaleya), mRNA-1273 (Moderna and NIAD), BNT162b2 (Pfizer and BioNTech), and NVX-CoV-2373 (Novavax and CEPI). Of note, BBIBP-CorV, CoronaVac, and Covaxin are from inactivated virus vaccines category, AZD1222, Ad26.COV2.S, and Sputnik V are from the adenoviral vector-based vaccines category, NVX-CoV-2373 is a protein subunit vaccine, and mRNA-1273 and BNT162b2 are from the mRNA-based vaccines category [7]. On 23 August 2021, FDA approved the first COVID-19 vaccine known as Pfizer-BioNTech COVID-19 Vaccine for administration in individuals 16 years of age and older [283]. Also, on 31 January 2022, FDA approved the second COVID-19 vaccine i.e., Moderna COVID-19 Vaccine in individuals 18 years of age and older [284]. The comparison of vaccines, their characteristics, effectiveness, and shortcomings have been reviewed in several noteworthy studies [285], [286], [287], [288]; however, we will briefly go over it in the following.
As discussed above, the presence of mutations could strengthen SARS-CoV-2 to be more transmissible and fatal [289], [290]. Besides, mutations cause SARS-CoV-2 to escape the immune system [291], [292]. It has been reported that some spike mutations such as D614G, C14408T, N440K, S477G/N, N501Y, P681H, etc. could elevate the affinity of the SARS-CoV-2 spike to the host cells; however, others like E484Q/K empower the capacity of the virus to escape from immune responses. Regarding the fact that the initial development of all vaccines has been performed based on the S protein found in the conventional Wuhan-hu-1 strain [7], it could be inferred that the alteration of SARS-CoV-2 in the spike region might be responsible for virus evasion from the immunity adopted either by natural infection or vaccine. The following sections will discuss the effectiveness of different SARS-CoV-2 vaccines against different VOCs. Nonetheless, as the global vaccination program did not initiate since the first variant emerged, the interaction of vaccines and various VOCs generated some discrepancy data. As the proportion of vaccinated people around the world rises up, more accurate data from the role of variants and mutations could be obtained such as situations that have been seen in the domination of Delta and Omicron variants. Accordingly, we categorized the following sections into two parts: before and after global vaccination.
5.1. Before global vaccination
The evaluation of Alpha variant susceptibility to BBIBP-CorV exhibited that this vaccine could neutralize the viruses effectively; however, BBIBP-CorV lose its potential capacity against Beta variant as the neutralization rates of most sera samples were reduced [293], [294]. Moreover, CoronaVac could induce satisfying neutralizing Abs in the contexts of the Alpha and Epsilon variants, but not in the Beta [295], Gamma, and Zeta [296]. Similarly, Covaxin showed proper neutralizing capacity facing the Alpha variant which was not observed against the Beta and Delta variants [297]. The evaluation of AZD1222 against Alpha and Beta variants showed the advantages of this vaccine facing the Alpha with 70.4 % efficacy [298] compared with the Beta with 10.4 % efficacy [299]. A study showed that the effectiveness of Ad26.COV2.S was 64.0 % in moderate to severe COVID-19 cases and 81.7 % in severe critical COVID-19 cases [300] and its ability to induce T cell-mediated anti-virus responses was satisfying in almost all VOCs [301]. The neutralizing activity of Sputnik V in the participant’s sera was satisfying against the Alpha variant; however, the titer of Abs was reduced in variants harboring E484K mutation like Beta [302]. Inline, NVX-CoV-2373 showed 85.6 % efficacy against the Alpha variant, while it had 51 % effectiveness against the Beta variant [303], [304]. By contrast, the effectiveness of mRNA-1273 was remarkably higher than other vaccines against the Alpha variant and particularly the Beta variant with 100 % and 96.4 % efficacy, respectively [305]. Similarly, BNT162b2 showed remarkable effectiveness against Alpha (93 %) and Beta (75 %) variants [306]. Nonetheless, the protection of BNT162b2 against the Beta variant was significantly decreased compared with the Alpha variant [307]; further highlighting the fact that spike mutations such as A701V, K417N, and E484A affect the efficacy of all vaccines facing the Beta variant. Taken together, it could be inferred that all the aforementioned vaccines could generate proper immune reactions ranging from neutralization to the T cell responses against all VOCs; however, almost all of them have an attenuated capacity facing the Beta variant that shows its evasion ability from the immune system. With this regard, a systematic review and meta-analysis study which searched a total of 4844 records from three databases evaluated the effectiveness of vaccines in various SARS-CoV-2 variants, and reported that the BNT162b2 vaccine had the most satisfying effectiveness against each VOC compared with other available vaccines [308].
5.2. After global vaccination; Delta and Omicron variants
The Delta variant contains mutations which facilitate the immune evasion capacity. The evaluation of mutations roles affecting the immunity showed that L452R mutation in the Delta variant gives similar infectious properties as N501Y gave to the Alpha and Beta variants. Moreover, T478K presented in the Delta variant could influence the neutralization capacity of antibodies [309]. The neutralization ability of CoronaVac vaccine against Delta variant was reduced by 2.47 times that was greater than the decline for the Alpha variant (1.62 times) [310]. Also, the serum neutralizing antibody titers of Covaxin were reduced by 2.7 times facing the Delta variant [311]. Covishield, a similar vaccine produced by AstraZeneca, showed a reduction in the serum neutralization levels against the Delta variant, as well [312]. According to several studies, the serum neutralization antibody titers after the administration of BNT162b2 were also reduced 1.41–11.30 times against this variant [313], [314], [315], [316], [317], [318]; inferring that the immune neutralizing antibody that adopted from various vaccines are less effective against the Delta variant.
A test-negative case–control study (SARS-CoV-2 test-positive cases and test-negative controls) evaluated the efficacy of inactivated vaccines such as BBIBP-CorV and CoronaVac vaccines against the Delta variant. Accordingly, single dose was not sufficient to induce proper protectivity; however, two doses of inactivated vaccines showed an average effectiveness determined as 72.5 % [319]. Furthermore, some studies from the United Kingdom and Scotland which evaluated the effectiveness of BNT162b2 and AZD1222 demonstrated similar results showing the efficacy of BNT162b2 and AZD1222 against the Delta variant about 87.9 % and about 60 %, respectively [208], [320]. Notably, the effectiveness of mRNA-1273 was determined as 86.7 % against the Delta variant; however, as time passes after the second dose, the efficacy of mRNA-1273 reduced [321]. Overall, it could be concluded that __despite the lower neutralization capacity of vaccines__ all are effective against the Delta variant; however, the amount of efficacy varies as it is at the highest level for the mRNA-based vaccines (about 85 % for the mRNA-based vaccines and 65–70 % for others).
The Omicron variant has high numbers of mutations which let this variant to escape from the immune responses more effortlessly. Generally, all available vaccines prepare immunity against S protein of SARS-CoV-2 [322]; this region of the Omicron variant carries more than 30 mutations that might dramatically affect the immune responses-induced by vaccines. It was shown that K417N mutation in the RBD region __which interestingly was carried by the Beta variant__ is probably the most considerable cause of antibody disruption in the Omicron variant. Notably, E484A is another mutation which accompanies K417N to empower the antibody disruption [323]. Omicron variant could resist 7 out of 8 approved mAbs and several other mAbs targeting epitopes of RBD and NTD [324]. The primary source of RBD-specific memory B cells could be triggered by booster doses; interestingly, after boosting vaccination, the population of B cells detecting the full-length Omicron variant expanded and pre-existing resting memory B cells differentiated into effector B cells with the ability to recognize the Omicron variant [325].
A study exhibited that only 20–24 % of patients with the Omicron variant had neutralizing antibody after primary BNT162b2 vaccination and no one showed neutralization after primary CoronaVac vaccine [326]. Similarly, two doses of BBIBP-CorV could not neutralize the Omicron variant effectively; however, a booster dose whether as a homologous or heterologous dose could significantly elevate the neutralization titers facing the Omicron variant [324]. The evaluation of Ad26.COV2.S, mRNA-1273, and BNT162b2 also showed that the Omicron variant could notably escape from immunity-induced by those stated vaccines (primary administration); nevertheless, receiving the third dose of mRNA-1273 and BNT162b2 considerably promoted the cross-neutralization possibly through the cross-activation of pre-existing innate and adaptive immune mechanisms [327]. In harmony, two doses of CoronaVac and/or BNT162b2 were able to provide 50 % neutralization against the Omicron after one month; nonetheless, the administration of booster dose of BNT162b2 to previously-two doses vaccinated individuals (CoronaVac and/or BNT162b2) notably enhanced the neutralizing ability-one month post booster dose. Interestingly, three doses of BNT162b2 was more potent to produce neutralization compared with other vaccines [328], implying the noteworthy capacity of BNT162b2 facing the Omicron variant. Moreover, the mix of BBIBP-CorV and BNT162b2 showed satisfying outcomes by increasing the immunity against Omicron [329]. With this regard, it was shown that booster dose with mRNA-based vaccines could restore the neutralization capacity of the immune system [330]. An interesting study evaluated the real-world effectiveness of BNT162b2 vaccination in 5 to 11 years old children. Among 94,728 individuals matched with unvaccinated controls, the estimated effectiveness was 51 % after second dose. Besides, the effectiveness of BNT162b2 was higher in youngest age group (5 and 6 years old) compared with older ones [331].
Overall, SARS-CoV-2 could evolve continuously and as time passes, we may encounter much more mutations that might alter the effectiveness of vaccines. It could be inferred that all typically approved vaccines have shown effectiveness against this virus; however, the quality and quantity of responses were more satisfying against the Alpha variant. Accordingly, Beta, Delta, and Omicron seem to have a higher ability to escape from the immunity. Indeed, all vaccines could protect patients from being severely ill and this is the key effect of vaccination that has helped nations pass this striking wave of COVID-19; however, seems that BNT162b2 and to wider category, mRNA-based vaccines would be a better strategy to overcome the domination of the virus and probably the best choice in order to augment the immunity against the Omicron variant regardless of the type of previously-administered vaccines. As the high proportions of people worldwide have received two doses of vaccines and the Omicron circulates rapidly, the booster dose is of importance, particularly as a heterologous booster strategy. Table 3 provided details of vaccines administered in the COVID-19 conditions.
Table 3.
An overview of SARS-CoV-2 vaccines and their efficacy and safety profile against several variants.
| Name | Developer | Country of origin | Date of validity | Schedule and rout of administration | Age group | Effectiveness/Advantages (average) | Adverse events/Challenge | Identifier |
|---|---|---|---|---|---|---|---|---|
| Inactivated vaccines | ||||||||
| BBIBP-CorV | Sinopharm | China | WHO approved on May 2021 for use in COVAX. | Two doses, 3–4 weeks apart, IM | 18 Yrs and older | 88 % for Alpha N/A for Beta 73 % for Gamma 72.5 % for Delta <50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
The most commonly reported AEs were fever, pain at the site of injection, headaches, and fatigue | NCT04984408 |
| CoronaVac |
Sinovac Biotech | China | Validated for emergency use (WHO) on June 2021 | Two doses, 4 weeks apart, IM | 18 Yrs and older | N/A for Alpha N/A for Beta N/A for Gamma 72.5 % for Delta 50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
The most common AE was pain at the site of injection. Other side effects included fatigue, diarrhea, and muscle pain. Most of these side effects were mild and lasted only for 2 days. | NCT04456595 |
| Covaxin | Bharat Biotech | India | Validated for emergency use (WHO) on November 2021 | Two doses, 28 days apart, ID | 18 Yrs and older | N/A for Alpha N/A for Beta N/A for Gamma 65 % for Delta <50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
The most commonly reported AEs were fever, headaches, irritability, pain and swelling at the site of injection. | N/A |
| CoviVac | Chumakov Centre | Russia | Approved for use in Russia on February 2021 | Two doses, 14 days apart | 18 Yrs and older | Phase 1/2 trial is ongoing. | No serious AEs were reported. | NCT04619628 |
| Viral vector vaccines | ||||||||
| AZD1222 (Covishield) |
Oxford–AstraZeneca | UK, Sweden | EMA approved on April 2021 | Two doses, 4–12 weeks apart, IM | 18 Yrs and older | 70.4 % for Alpha 10.4 % for Beta 78 % for Gamma 60 % for Delta <50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
The most commonly reported AEs were vomiting, diarrhea, fever, swelling, redness at the injection site and low levels of blood platelets. Enlarged lymph nodes, decreased appetite, dizziness, sleepiness, sweating, abdominal pain, itching and rash. Importantly, high risk of thrombocytopenia syndrome was reported mainly in younger female. | NCT04516746 |
| Sputnik V | Gamaleya Research Institute of Epidemiology and Microbiology | Russia | Approved in Russia and then on 59 other countries (as of April 2021). | Two doses, 21 days apart, IM |
18 Yrs and older | 94 % for Alpha N/A for Beta N/A for Gamma 81 % for Delta <50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
AEs are mainly mild with no signs of vaccine-induced immune thrombotic thrombocytopenia. | NCT04530396 |
| JNJ-78436735; Ad26. COV2.S | Janssen vaccines (Johnsons & Johnsons) |
Netherland, USA |
Validated for emergency use (FDA) on February 2021 | Single dose, IM | 18 Yrs and older | 74 % for Alpha 64 % for Beta N/A for Gamma 71 % for Delta N/A for Omicron |
The most common AEs were pain at the site of injection, headache, tiredness, muscle pain, and nausea which were usually mild or moderate. The incidence of hypersensitivity and the itchy rash was 1 in 1000 people. | NCT04505722 |
| Convidicea (Ad5-nCoV) |
CanSino Biologics | China | Approved in China on February 2021 | Single dose, IM |
18 Yrs and older | 65.7 % efficacy in preventing moderate symptoms and 91 % efficacy in preventing severe disease | Common AEs were headache, myalgia, fatigue, shivers, fever, and arthralgia. | NCT04552366 |
| RNA vaccines | ||||||||
| BNT162b2 | Pfizer-BioNTech | Multinational | FDA approved on August 2021 | Two doses, 21 days apart, IM |
16 Yrs and older | 93 % for Alpha 75 % for Beta 88 % for Gamma 87.9 % for Delta <50 % for Omicron (2 doses) 76 % for Omicron (3 doses) |
The common AEs affecting more than 1 in 10 people are pain at the site of injection, tiredness, headache, muscle aches, chills, joint pain, and fever, respectively. | NCT04848584 |
| mRNA-1273 | Moderna, BARDA, NIAID |
USA | FDA approved on December 2020 | Two doses, 28 days apart, IM | 12 Yrs and older | 100 % for Alpha 96.4 % for Beta 78 % for Gamma 86.7 % for Delta <50 % for Omicron (2 doses) N/A for Omicron (3 doses) |
The most common AEs were pain at the site of injection, fatigue, headache, myalgia, and arthralgia. Anaphylaxis was reported in 2.5 cases per million administered doses. | NCT04470427 |
| Protein subunit vaccines | ||||||||
| NVX-CoV2373 | Novavax and the Coalition for Epidemic Preparedness Innovations | USA | Validated for emergency use (WHO) on December 2021 | Two doses, 21 days apart, IM | 18–84 | 86 % for Alpha 51 % for Beta N/A for Gamma N/A for Delta N/A for Omicron |
Pain at the site of injection and tenderness, as well as fatigue, headache and muscle pain were the most commonly reported AEs. | NCT04611802 |
| EpiVacCorona |
VECTOR center of Virology | Russia | The third phase of a clinical trial was launched in November 2020. | Two doses, 21–28 days apart, IM | 18 Yrs and older | Preliminary data of the Phase III study showed 79 % efficacy | It was reported that no adverse reactions caused were recognized. | NCT04780035 |
| DNA vaccines | ||||||||
| Zycov D | Cadila Healthcare | India | Approved by Drugs Controller General of India on August 2021 | Three doses, 28 and 56 days apart for second and third doses, respectively, ID | 12 Yrs and older | 66.6 % efficacy against symptomatic and 100 % against moderate or severe disease | No serious AEs were observed. Even fever or fatigue is uncommon post-vaccination. | N/A |
| Recombinant vaccines | ||||||||
| ZF2001/ZIFIVAX (ZF-UZ-VAC-2001) |
Anhui Zhifei Longcom and Chinese Academy of Sciences | Uzbekistan | Approved in China on March 2021 for emergency use | Three doses over a period of 2 months | 18 Yrs and older | 93 % for Alpha N/A for Beta N/A for Gamma 78 % for Delta N/A for Omicron |
The most common AEs were mild or moderate including pain at the site of injection, redness, and swelling. | NCT04646590 |
ID: Intradermal; IM: Intramuscular; AE: Adverse event; EMA: European Medicines Agency; N/A: Not available.
6. Conclusion and future perspective
The SARS-CoV-2, responsible for COVID-19, was turned into a new concern to public health due to its high transmissibility capacity and mortality which causes asymptomatic to severe illness. There could be reasons account for this wide spectrum of clinical manifestations, among which the mechanisms associated with the entry of the SARS-CoV-2 have attracted great attention. ACE2 is the most important receptor of SARS-CoV-2 spike protein distributing mainly on the respiratory cells. The presence of ACE2 variants such as E23K and K26R is associated with higher ACE2-spike affinity, while S19P and E329G could reduce the affinity. Similarly, there is genetic heterogeneity in other host-related factors such as TMPRSS2, FURIN, DPP4, APOE, and CD147 which could either facilitate the entry of virus to the host cells and lead to more severe disease or impede the interaction of virus with the cells. Moreover, diversity in the immune-related genes is another agent affecting the severity of COVID-19. Some HLA variants such as HLA-A*11:01 and HLA-B*51:01 are associated with poor prognosis; however, HLA-B*15:03 showed a protective effect. Likewise, genetic heterogeneity in cytokines, CCR/CXCR, TLRs, components of the complement system, NK cell-related genes, and vitamin d-binding protein strikingly affects the responses against SARS-CoV-2. In addition to genetic factors, host epigenetics may also influence COVID-19 severity. Accordingly, EZH2 absence and H3K27me3 downregulation could increase the expression of ACE2, while some histone modification regulators such as H3K4me3 and H3K9me2 can induce the expression of inflammatory pathways like IFNs.
Regarding the mutations concerning SARS-CoV-2, the genome of virus is per se a unique agent affecting the severity of COVID-19. According to the WHO classification, SARS-CoV-2 is categorized into VOC, VOI, VUM, and formerly monitored variants. The most famous mutation is D614G which is associated with higher viral entry and load due to higher interaction of spike and ACE2. In addition, N440K, L452R, S477G, and S477N enhance the affinity of spike-ACE2 leading to higher viral infectivity. Besides, D614G and E484Q/K are responsible for higher SARS-CoV-2 immune escape. Based on the global knowledge about COVID-19 and new education about how to manage the clinical manifestations, we inferred that the Delta variant has been the most severe wave of COVID-19 as this variant showed notable mortality worldwide. Nevertheless, data from the Omicron wave is not yet enough to analyze the strength of this variant.
One of the most imperative works facing COVID-19 has been the development of an efficient vaccine. Till now, more than 20 vaccines have been confirmed. The data concerning the efficacies of these vaccines differ from study to study, however, the average efficacy of mRNA-vaccines, particularly the Pfizer-BioNTech product, seems to be higher compared with others. All available vaccines are remarkably effective against severe cases of COVID-19 until the Omicron has emerged; nonetheless, studies reported that the third dose of any vaccine could considerably empower the capability of the immune system facing the Omicron variant. Therefore, an urgent need for completing the vaccination program is required to attain a healthy community. Moreover, the necessity of achieving a proper drug to fight SARS-CoV-2 seems to be neglected. Although big companies are investigating this context, there is a great area of study for future research to focus on the development of a potent drug to impede the pathogenic features of the virus and also to treat the life-threatening manifestations such as acute respiratory distress syndrome. Finally, it is worth mentioning that a novel important area of future research is to develop a universal vaccine, known as pan-vaccines or super vaccines; indeed, they can fight not only against SARS-CoV-2 variants but also against other coronaviruses that could potentially be a new pandemic condition in the future.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors would like to express their gratitude to Shahid Beheshti University of Medical Sciences (Tehran, Iran) for supporting this study.
Data availability statement
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Data availability
No data was used for the research described in the article.
References
- 1.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England J. Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahimi A., Mirzazadeh A., Tavakolpour S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics. 2021;113(1 Pt 2):1221–1232. doi: 10.1016/j.ygeno.2020.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cui J., Li F., Shi Z.L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019;17(3):181–192. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 Transmission and Pathogenesis. Trends Immunol. 2020;41(12):1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur. J. Intern. Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.C.A. Devaux, J.M. Rolain, D. Raoult, ACE2 receptor polymorphism: Susceptibility to SARS-CoV-2, hypertension, multi-organ failure, and COVID-19 disease outcome, Journal of microbiology, immunology, and infection = Wei mian yu gan ran za zhi 53(3) (2020) 425–435. [DOI] [PMC free article] [PubMed]
- 7.Hadj Hassine I. Covid-19 vaccines and variants of concern: A review. Rev. Med. Virol. 2021 doi: 10.1002/rmv.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Delanghe J., Speeckaert M., De Buyzere M. The host's angiotensin-converting enzyme polymorphism may explain epidemiological findings in COVID-19 infections. Clin. Chim. Acta. 2020;505:192–193. doi: 10.1016/j.cca.2020.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benetti E., Tita R., Spiga O., Ciolfi A., Birolo G., Bruselles A., Doddato G., Giliberti A., Marconi C., Musacchia F. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur. J. Hum. Genet. 2020;28(11):1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stawiski E.W., Diwanji D., Suryamohan K., Gupta R., Fellouse F.A., Sathirapongsasuti J.F., Liu J., Jiang Y.-P., Ratan A., Mis M. Human ACE2 receptor polymorphisms predict SARS-CoV-2 susceptibility. BioRxiv. 2020 doi: 10.1038/s42003-021-02030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lanjanian H., Moazzam-Jazi M., Hedayati M., Akbarzadeh M., Guity K., Sedaghati-Khayat B., Azizi F., Daneshpour M.S. SARS-CoV-2 infection susceptibility influenced by ACE2 genetic polymorphisms: insights from Tehran Cardio-Metabolic Genetic Study. Sci. Rep. 2021;11(1):1–13. doi: 10.1038/s41598-020-80325-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delanghe J.R., Speeckaert M.M., De Buyzere M.L. COVID-19 infections are also affected by human ACE1 D/I polymorphism. Clin. Chem. Lab. Med. (CCLM) 2020;58(7):1125–1126. doi: 10.1515/cclm-2020-0425. [DOI] [PubMed] [Google Scholar]
- 13.Torre-Fuentes L., Matías-Guiu J., Hernández-Lorenzo L., Montero-Escribano P., Pytel V., Porta-Etessam J., Gómez-Pinedo U., Matías-Guiu J.A. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid Spain. J. Med. Virol. 2021;93(2):863–869. doi: 10.1002/jmv.26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436(7047):112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng Z., Zhou J., To K.-K.-W., Chu H., Li C., Wang D., Yang D., Zheng S., Hao K., Bossé Y. Identification of TMPRSS2 as a susceptibility gene for severe 2009 pandemic A (H1N1) influenza and A (H7N9) influenza. J. Infect. Dis. 2015;212(8):1214–1221. doi: 10.1093/infdis/jiv246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N.P.C. Santos, A.S. Khayat, J.C.G. Rodrigues, P.C. Pinto, G.S. Araujo, L.F. Pastana, J.A.G. Medeiros, M.R. Fernandes, A.R. dos Santos, B.C.M. Khayat, TMPRSS2 variants and their susceptibility to COVID-19: focus in East Asian and European populations, medRxiv (2020).
- 17.Irham L.M., Chou W.-H., Calkins M.J., Adikusuma W., Hsieh S.-L., Chang W.-C. Genetic variants that influence SARS-CoV-2 receptor TMPRSS2 expression among population cohorts from multiple continents. Biochem. Biophys. Res. Commun. 2020;529(2):263–269. doi: 10.1016/j.bbrc.2020.05.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monticelli M., Hay Mele B., Benetti E., Fallerini C., Baldassarri M., Furini S., Frullanti E., Mari F., Andreotti G., Cubellis M.V. Protective Role of a TMPRSS2 Variant on Severe COVID-19 Outcome in Young Males and Elderly Women. Genes. 2021;12(4):596. doi: 10.3390/genes12040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wulandari L., Hamidah B., Pakpahan C., Damayanti N.S., Kurniati N.D., Adiatmaja C.O., Wigianita M.R., Husada D., Tinduh D., Prakoeswa C.R.S. Initial study on TMPRSS2 p. Val160Met genetic variant in COVID-19 patients. Hum. Genom. 2021;15(1):1–9. doi: 10.1186/s40246-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.A. David, N. Parkinson, T.P. Peacock, E. Pairo-Castineira, T. Khanna, A. Cobat, A. Tenesa, V. Sancho-Shimizu, J.-L. Casanova, L. Abel, A common TMPRSS2 variant protects against severe COVID-19, (2021). [DOI] [PMC free article] [PubMed]
- 21.T.P. Peacock, J.C. Brown, J. Zhou, N. Thakur, J. Newman, R. Kugathasan, K. Sukhova, M. Kaforou, D. Bailey, W.S. Barclay, The SARS-CoV-2 variant, Omicron, shows rapid replication in human primary nasal epithelial cultures and efficiently uses the endosomal route of entry, bioRxiv (2022) 2021.12.31.474653.
- 22.AbdelMassih A.F., Ye J., Kamel A., Mishriky F., Ismail H.-A., Ragab H.A., El Qadi L., Malak L., Abdu M., El-Husseiny M. A multicenter consensus: A role of furin in the endothelial tropism in obese patients with COVID-19 infection. Obesity Medicine. 2020 doi: 10.1016/j.obmed.2020.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Mulla F., Mohammad A., Al Madhoun A., Haddad D., Ali H., Eaaswarkhanth M., John S.E., Nizam R., Channanath A., Abu-Farha M. ACE2 and FURIN variants are potential predictors of SARS-CoV-2 outcome: A time to implement precision medicine against COVID-19. Heliyon. 2021;7(2) doi: 10.1016/j.heliyon.2021.e06133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deacon C.F. Physiology and pharmacology of DPP-4 in glucose homeostasis and the treatment of type 2 diabetes. Front. Endocrinol. 2019;10:80. doi: 10.3389/fendo.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F., Huang S., Gao R., Zhou Y., Lai C., Li Z., Xian W., Qian X., Li Z., Huang Y. Initial whole-genome sequencing and analysis of the host genetic contribution to COVID-19 severity and susceptibility. Cell Discovery. 2020;6(1):1–16. doi: 10.1038/s41421-020-00231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posadas-Sánchez R., Sánchez-Muñoz F., Guzmán-Martín C.A., Couder A.-H.-D., Rojas-Velasco G., Fragoso J.M., Vargas-Alarcón G. Dipeptidylpeptidase-4 levels and DPP4 gene polymorphisms in patients with COVID-19 Association with disease and with severity. Life Sci. 2021;276 doi: 10.1016/j.lfs.2021.119410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simonnet A., Chetboun M., Poissy J., Raverdy V., Noulette J., Duhamel A., Labreuche J., Mathieu D., Pattou F., Jourdain M. High prevalence of obesity in severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) requiring invasive mechanical ventilation. Obesity. 2020;28(7):1195–1199. doi: 10.1002/oby.22831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghorpade D.S., Ozcan L., Zheng Z., Nicoloro S.M., Shen Y., Chen E., Blüher M., Czech M.P., Tabas I. Hepatocyte-secreted DPP4 in obesity promotes adipose inflammation and insulin resistance. Nature. 2018;555(7698):673–677. doi: 10.1038/nature26138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stengel A., Goebel-Stengel M., Teuffel P., Hofmann T., Buße P., Kobelt P., Rose M., Klapp B.F. Obese patients have higher circulating protein levels of dipeptidyl peptidase IV. Peptides. 2014;61:75–82. doi: 10.1016/j.peptides.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Kirino Y., Sei M., Kawazoe K., Minakuchi K., Sato Y. Plasma dipeptidyl peptidase 4 activity correlates with body mass index and the plasma adiponectin concentration in healthy young people. Endocr. J. 2012;59(10):949–953. doi: 10.1507/endocrj.ej12-0158. [DOI] [PubMed] [Google Scholar]
- 31.Yang F., Zheng T., Gao Y., Baskota A., Chen T., Ran X., Tian H. Increased plasma DPP4 activity is an independent predictor of the onset of metabolic syndrome in Chinese over 4 years: result from the China National Diabetes and Metabolic Disorders Study. PLoS ONE. 2014;9(3) doi: 10.1371/journal.pone.0092222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Latini A., Agolini E., Novelli A., Borgiani P., Giannini R., Gravina P., Smarrazzo A., Dauri M., Andreoni M., Rogliani P. COVID-19 and genetic variants of protein involved in the SARS-CoV-2 entry into the host cells. Genes. 2020;11(9):1010. doi: 10.3390/genes11091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Yuan Z., Pavel M.A., Hobson R., Hansen S.B. The role of high cholesterol in age-related COVID19 lethality. Biorxiv. 2020 [Google Scholar]
- 34.Coperchini F., Chiovato L., Croce L., Magri F., Rotondi M. The cytokine storm in COVID-19: An overview of the involvement of the chemokine/chemokine-receptor system. Cytokine Growth Factor Rev. 2020;53:25–32. doi: 10.1016/j.cytogfr.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassale C., Gaye B., Hamer M., Gale C.R., Batty G.D. Ethnic disparities in hospitalisation for COVID-19 in England: The role of socioeconomic factors, mental health, and inflammatory and pro-inflammatory factors in a community-based cohort study. Brain Behav. Immun. 2020;88:44–49. doi: 10.1016/j.bbi.2020.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuo C.-L., Pilling L.C., Atkins J.L., Masoli J.A., Delgado J., Kuchel G.A., Melzer D. APOE e4 genotype predicts severe COVID-19 in the UK Biobank community cohort. J. Gerontol. Ser. A. 2020;75(11):2231–2232. doi: 10.1093/gerona/glaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulminski A.M., Barochia A.V., Loika Y., Raghavachari N., Arbeev K.G., Wojczynski M.K., Thyagarajan B., Vardarajan B.N., Christensen K., Yashin A.I. The APOE ε4 allele is associated with a reduction in FEV1/FVC in women: A cross-sectional analysis of the Long Life Family Study. PLoS ONE. 2018;13(11) doi: 10.1371/journal.pone.0206873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrie A., Hameed A.G., Chamberlain J., Arnold N., Kennerley A., Hopkinson K., Pickworth J., Kiely D.G., Crossman D.C., Francis S.E. Paigen diet–fed apolipoprotein E knockout mice develop severe pulmonary hypertension in an interleukin-1–dependent manner. Am. J. Pathol. 2011;179(4):1693–1705. doi: 10.1016/j.ajpath.2011.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ulrich H., Pillat M.M. CD147 as a target for COVID-19 treatment: suggested effects of azithromycin and stem cell engagement. Stem cell Rev. Rep. 2020;16(3):434–440. doi: 10.1007/s12015-020-09976-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ilikci Sagkan R., Akin-Bali D.F. Structural variations and expression profiles of the SARS-CoV-2 host invasion genes in Lung cancer. J. Med. Virol. 2020;92(11):2637–2647. doi: 10.1002/jmv.26107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Geng, L. Chen, Y. Yuan, R. Chen, K. Wang, Y. Deng, P. Du, J. Liu, G. Wu, Y. Wang, CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 variants, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 42.Wang K., Chen W., Zhou Y.-S., Lian J.-Q., Zhang Z., Du P., Gong L., Zhang Y., Cui H.-Y., Geng J.-J. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. BioRxiv. 2020 doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuba K., Imai Y., Penninger J.M. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ. J. 2013;77(2):301–308. doi: 10.1253/circj.cj-12-1544. [DOI] [PubMed] [Google Scholar]
- 44.Hussain M., Jabeen N., Raza F., Shabbir S., Baig A.A., Amanullah A., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92(9):1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Teralı K., Baddal B., Gülcan H.O. Prioritizing potential ACE2 inhibitors in the COVID-19 pandemic: Insights from a molecular mechanics-assisted structure-based virtual screening experiment. J. Mol. Graph. Model. 2020;100 doi: 10.1016/j.jmgm.2020.107697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.V. Mollica, A. Rizzo, F. Massari, The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer, Future Medicine, 2020, pp. 2029-2033. [DOI] [PMC free article] [PubMed]
- 47.Senapati S., Kumar S., Singh A.K., Banerjee P., Bhagavatula S. Assessment of risk conferred by coding and regulatory variations of TMPRSS2 and CD26 in susceptibility to SARS-CoV-2 infection in human. J. Genet. 2020;99(1):1–5. doi: 10.1007/s12041-020-01217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. Monticelli, B. Hay Mele, E. Benetti, C. Fallerini, M. Baldassarri, S. Furini, E. Frullanti, F. Mari, G.-C.M. Study, G. Andreotti, Protective role of a TMPRSS2 variant on severe COVID-19 outcome in young males and elderly women, Genes 12(4) (2021) 596. [DOI] [PMC free article] [PubMed]
- 49.Jeon S., Blazyte A., Yoon C., Ryu H., Jeon Y., Bhak Y., Bolser D., Manica A., Shin E.-S., Cho Y.S. Ethnicity-dependent allele frequencies are correlated with COVID-19 case fatality rate. Authorea Preprints. 2020 doi: 10.14348/molcells.2021.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shapira T., Monreal I.A., Dion S.P., Buchholz D.W., Imbiakha B., Olmstead A.D., Jager M., Désilets A., Gao G., Martins M. A TMPRSS2 inhibitor acts as a pan-SARS-CoV-2 prophylactic and therapeutic. Nature. 2022;605(7909):340–348. doi: 10.1038/s41586-022-04661-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Huang Y., Ho D.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593(7857):130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 52.AbdelMassih A.F., Ye J., Kamel A., Mishriky F., Ismail H.-A., Ragab H.A., El Qadi L., Malak L., Abdu M., El-Husseiny M. A multicenter consensus: A role of furin in the endothelial tropism in obese patients with COVID-19 infection. Obesity Med. 2020;19 doi: 10.1016/j.obmed.2020.100281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dahms S.O., Schnapp G., Winter M., Büttner F.H., Schlepütz M., Gnamm C., Pautsch A., Brandstetter H. Dichlorophenylpyridine-Based Molecules Inhibit Furin through an Induced-Fit Mechanism. ACS Chem. Biol. 2022;17(4):816–821. doi: 10.1021/acschembio.2c00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canela-Xandri O., Rawlik K., Tenesa A. An atlas of genetic associations in UK Biobank. Nat. Genet. 2018;50(11):1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. China, The lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu C., Chen X., Cai Y., Zhou X., Xu S., Huang H., Zhang L., Zhou X., Du C., Zhang Y. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan China. JAMA Internal Med. 2020;180(7):934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang H., Yuan Z., Pavel M.A., Jablonski S.M., Jablonski J., Hobson R., Valente S., Reddy C.B., Hansen S.B. The role of high cholesterol in age-related COVID19 lethality. BioRxiv. 2020 doi: 10.1016/j.jbc.2023.104763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Geng J., Chen L., Yuan Y., Wang K., Wang Y., Qin C., Wu G., Chen R., Zhang Z., Wei D. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Sig. Transd. Targeted Therapy. 2021;6(1):1–13. doi: 10.1038/s41392-021-00760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Sig. Transd. Targeted Therapy. 2020;5(1):1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Matzaraki V., Kumar V., Wijmenga C., Zhernakova A. The MHC locus and genetic susceptibility to autoimmune and infectious diseases. Genome Biol. 2017;18(1):1–21. doi: 10.1186/s13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bandi S., Nevid M.Z., Mahdavinia M. African American children are at higher risk for COVID-19 infection. Pediatr. Allergy Immunol. 2020 doi: 10.1111/pai.13298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Galloway J.B., Norton S., Barker R.D., Brookes A., Carey I., Clarke B.D., Jina R., Reid C., Russell M.D., Sneep R. A clinical risk score to identify patients with COVID-19 at high risk of critical care admission or death: an observational cohort study. J. Infect. 2020;81(2):282–288. doi: 10.1016/j.jinf.2020.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogedegbe G., Ravenell J., Adhikari S., Butler M., Cook T., Francois F., Iturrate E., Jean-Louis G., Jones S.A., Onakomaiya D. Assessment of racial/ethnic disparities in hospitalization and mortality in patients with COVID-19 in New York City. JAMA network open. 2020;3(12):e2026881. doi: 10.1001/jamanetworkopen.2020.26881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ng M.H., Lau K.-M., Li L., Cheng S.-H., Chan W.Y., Hui P.K., Zee B., Leung C.-B., Sung J.J. Association of human-leukocyte-antigen class I (B* 0703) and class II (DRB1* 0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 2004;190(3):515–518. doi: 10.1086/421523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vietzen H., Zoufaly A., Traugott M., Aberle J., Aberle S.W., Puchhammer-Stöckl E. Deletion of the NKG2C receptor encoding KLRC2 gene and HLA-E variants are risk factors for severe COVID-19. Genet. Med. 2021;23(5):963–967. doi: 10.1038/s41436-020-01077-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.S.C.-G. Group, Genomewide association study of severe Covid-19 with respiratory failure, New England J. Med. 383(16) (2020) 1522–1534. [DOI] [PMC free article] [PubMed]
- 67.Campbell K.M., Steiner G., Wells D.K., Ribas A., Kalbasi A. Prioritization of SARS-CoV-2 epitopes using a pan-HLA and global population inference approach. BioRxiv. 2020 [Google Scholar]
- 68.Ahmed S.F., Quadeer A.A., McKay M.R. Preliminary identification of potential vaccine targets for the COVID-19 coronavirus (SARS-CoV-2) based on SARS-CoV immunological studies. Viruses. 2020;12(3):254. doi: 10.3390/v12030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romero-López J., Carnalla-Cortés M., Pacheco-Olvera D., Ocampo M., Oliva-Ramírez J., Moreno-Manjón J., Bernal-Alferes B., García-Latorre E., Domínguez-López M., Reyes-Sandoval A. Prediction of SARS-CoV2 spike protein epitopes reveals HLA-associated susceptibility. Res. Square. 2020 [Google Scholar]
- 70.Hamano E., Hijikata M., Itoyama S., Quy T., Phi N.C., Long H.T., Van Ban V., Matsushita I., Yanai H., Kirikae F. Polymorphisms of interferon-inducible genes OAS-1 and MxA associated with SARS in the Vietnamese population. Biochem. Biophys. Res. Commun. 2005;329(4):1234–1239. doi: 10.1016/j.bbrc.2005.02.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J., Feng D., de Vlas S.J., Wang H., Fontanet A., Zhang P., Plancoulaine S., Tang F., Zhan L., Yang H. Association of SARS susceptibility with single nucleic acid polymorphisms of OAS1 and MxA genes: a case-control study. BMC Infect. Dis. 2006;6(1):1–7. doi: 10.1186/1471-2334-6-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D. Genetic mechanisms of critical illness in Covid-19. Nature. 2021;591(7848):92–98. doi: 10.1038/s41586-020-03065-y. [DOI] [PubMed] [Google Scholar]
- 73.Zhang Q., Bastard P., Liu Z., Le Pen J., Moncada-Velez M., Chen J., Ogishi M., Sabli I.K., Hodeib S., Korol C. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370(6515) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zani A., Yount J.S. Antiviral protection by IFITM3 in vivo. Current Clin. Microbiol. Rep. 2018;5(4):229–237. doi: 10.1007/s40588-018-0103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Y.-H., Zhao Y., Li N., Peng Y.-C., Giannoulatou E., Jin R.-H., Yan H.-P., Wu H., Liu J.-H., Liu N. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4(1):1–6. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karaderi T., Bareke H., Kunter I., Seytanoglu A., Cagnan I., Balci D., Barin B., Hocaoglu M.B., Rahmioglu N., Asilmaz E. Host genetics at the intersection of autoimmunity and COVID-19: A potential key for heterogeneous COVID-19 severity. Front. Immunol. 2020;11:3314. doi: 10.3389/fimmu.2020.586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nguyen D.-T., Mathias S., Bologa C., Brunak S., Fernandez N., Gaulton A., Hersey A., Holmes J., Jensen L.J., Karlsson A. Pharos: collating protein information to shed light on the druggable genome. Nucleic Acids Res. 2017;45(D1):D995–D1002. doi: 10.1093/nar/gkw1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Spinelli F.R., Conti F., Gadina M. HiJAKing SARS-CoV-2? The potential role of JAK inhibitors in the management of COVID-19. Sci. Immunol. 2020;5(47):eabc5367. doi: 10.1126/sciimmunol.abc5367. [DOI] [PubMed] [Google Scholar]
- 79.B. Stikker, G. Stik, R. Hendriks, R. Stadhouders, CCR1 regulatory variants linked to pulmonary macrophage recruitment in severe COVID-19, bioRxiv (2021).
- 80.J. Gómez, E. Cuesta-Llavona, G.M. Albaiceta, M. García-Clemente, C. López-Larrea, L. AMADO, I. López-Alonso, T. Hermida, A. ENRIQUEZ, H. Gil, The CCR5-delta32 variant might explain part of the association between COVID-19 and the chemokine-receptor gene cluster, medRxiv (2020).
- 81.Debnath M., Banerjee M., Berk M. Genetic gateways to COVID-19 infection: Implications for risk, severity, and outcomes. FASEB J. 2020;34(7):8787–8795. doi: 10.1096/fj.202001115R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gralinski L.E., Menachery V.D., Morgan A.P., Totura A.L., Beall A., Kocher J., Plante J., Harrison-Shostak D.C., Schäfer A., Pardo-Manuel de Villena F. Allelic variation in the toll-like receptor adaptor protein ticam2 contributes to SARS-coronavirus pathogenesis in mice, G3: Genes. Genomes, Genetics. 2017;7(6):1653–1663. doi: 10.1534/g3.117.041434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fallerini C., Daga S., Mantovani S., Benetti E., Picchiotti N., Francisci D., Paciosi F., Schiaroli E., Baldassarri M., Fava F. Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: findings from a nested case-control study. Elife. 2021;10 doi: 10.7554/eLife.67569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bosmann M. American Thoracic Society; 2020. Complement activation during critical illness: current findings and an outlook in the era of COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ip W.E., Chan K.H., Law H.K., Tso G.H., Kong E.K., Wong W.H., To Y.F., Yung R.W., Chow E.Y., Au K.L. Mannose-binding lectin in severe acute respiratory syndrome coronavirus infection. J. Infect. Dis. 2005;191(10):1697–1704. doi: 10.1086/429631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Valenti L., Griffini S., Lamorte G., Grovetti E., Renteria S.C.U., Malvestiti F., Scudeller L., Bandera A., Peyvandi F., Prati D. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J. Autoimmun. 2021;117 doi: 10.1016/j.jaut.2021.102595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ramsuran V., Naranbhai V., Horowitz A., Qi Y., Martin M.P., Yuki Y., Gao X., Walker-Sperling V., Del Prete G.Q., Schneider D.K. Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells. Science. 2018;359(6371):86–90. doi: 10.1126/science.aam8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maucourant C., Filipovic I., Ponzetta A., Aleman S., Cornillet M., Hertwig L., Strunz B., Lentini A., Reinius B., Brownlie D. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 2020;5(50):eabd6832. doi: 10.1126/sciimmunol.abd6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilk A.J., Rustagi A., Zhao N.Q., Roque J., Martínez-Colón G.J., McKechnie J.L., Ivison G.T., Ranganath T., Vergara R., Hollis T. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 2020;26(7):1070–1076. doi: 10.1038/s41591-020-0944-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bikle D.D., Schwartz J. Vitamin D binding protein, total and free vitamin D levels in different physiological and pathophysiological conditions. Front. Endocrinol. 2019;10:317. doi: 10.3389/fendo.2019.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Karcioglu Batur L., Hekim N. The role of DBP gene polymorphisms in the prevalence of new coronavirus disease 2019 infection and mortality rate. J. Med. Virol. 2021;93(3):1409–1413. doi: 10.1002/jmv.26409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Freitas A.T., Calhau C., Antunes G., Araújo B., Bandeira M., Barreira S., Bazenga F., Braz S., Caldeira D., Santos S.C.R. Vitamin D-related polymorphisms and vitamin D levels as risk biomarkers of COVID-19 disease severity. Sci. Rep. 2021;11(1):1–8. doi: 10.1038/s41598-021-99952-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jin J.-M., Bai P., He W., Liu S., Wu F., Liu X.-F., Han D.-M., Yang J.-K. Higher severity and mortality in male patients with COVID-19 independent of age and susceptibility. MedRxiv. 2020 [Google Scholar]
- 94.Sharma G., Volgman A.S., Michos E.D. Sex differences in mortality from COVID-19 pandemic: are men vulnerable and women protected? Case Reports. 2020;2(9):1407–1410. doi: 10.1016/j.jaccas.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cui J., Shen Y., Li R. Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends Mol Med. 2013;19(3):197–209. doi: 10.1016/j.molmed.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.E.R. Simpson, Y. Zhao, V.R. Agarwal, M.D. Michael, S.E. Bulun, M.M. Hinshelwood, S. Graham-Lorence, T. Sun, C.R. Fisher, K. Qin, C.R. Mendelson, Aromatase expression in health and disease, Recent progress in hormone research 52 (1997) 185–213; discussion 213-4. [PubMed]
- 97.Santen R.J., Brodie H., Simpson E.R., Siiteri P.K., Brodie A. History of aromatase: saga of an important biological mediator and therapeutic target. Endocr. Rev. 2009;30(4):343–375. doi: 10.1210/er.2008-0016. [DOI] [PubMed] [Google Scholar]
- 98.Simpson E.R., Mahendroo M.S., Means G.D., Kilgore M.W., Hinshelwood M.M., Graham-Lorence S., Amarneh B., Ito Y., Fisher C.R., Michael M.D., et al. Aromatase cytochrome P450, the enzyme responsible for estrogen biosynthesis. Endocr. Rev. 1994;15(3):342–355. doi: 10.1210/edrv-15-3-342. [DOI] [PubMed] [Google Scholar]
- 99.Bulun S.E., Simpson E.R. Competitive reverse transcription-polymerase chain reaction analysis indicates that levels of aromatase cytochrome P450 transcripts in adipose tissue of buttocks, thighs, and abdomen of women increase with advancing age. J. Clin. Endocrinol. Metabol. 1994;78(2):428–432. doi: 10.1210/jcem.78.2.8106632. [DOI] [PubMed] [Google Scholar]
- 100.T. Ding, J. Zhang, T. Wang, P. Cui, Z. Chen, J. Jiang, S. Zhou, J. Dai, B. Wang, S. Yuan, A multi-hospital study in Wuhan, China: protective effects of non-menopause and female hormones on SARS-CoV-2 infection, medrxiv (2020).
- 101.Acheampong D.O., Barffour I.K., Boye A., Aninagyei E., Ocansey S., Morna M.T. Male predisposition to severe COVID-19: Review of evidence and potential therapeutic prospects. Biomed. Pharmacother. 2020;110748 doi: 10.1016/j.biopha.2020.110748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mikhail N., Wali S. Clinical significance of sex hormones in COVID-19. J. Gynecol. Res. Obstetr. 2020;6(3):060–063. [Google Scholar]
- 103.Montopoli M., Zumerle S., Vettor R., Rugge M., Zorzi M., Catapano C.V., Carbone G., Cavalli A., Pagano F., Ragazzi E. Androgen-deprivation therapies for prostate cancer and risk of infection by SARS-CoV-2: a population-based study (N= 4532) Ann. Oncol. 2020;31(8):1040–1045. doi: 10.1016/j.annonc.2020.04.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.A.C. Walls, Y.-J. Park, M.A. Tortorici, A. Wall, A.T. McGuire, D. Veesler, Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein, Cell 181(2) (2020) 281–292. e6. [DOI] [PMC free article] [PubMed]
- 105.Dhindsa S., Zhang N., McPhaul M.J., Wu Z., Ghoshal A.K., Erlich E.C., Mani K., Randolph G.J., Edwards J.R., Mudd P.A. Association of circulating sex hormones with inflammation and disease severity in patients with COVID-19. JAMA network open. 2021;4(5):e2111398. doi: 10.1001/jamanetworkopen.2021.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dhindsa S., Zhang N., McPhaul M.J., Wu Z., Ghoshal A.K., Erlich E.C., Mani K., Randolph G.J., Edwards J.R., Mudd P.A., Diwan A. Association of Circulating Sex Hormones With Inflammation and Disease Severity in Patients With COVID-19. JAMA Netw Open. 2021;4(5) doi: 10.1001/jamanetworkopen.2021.11398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klein S.L., Dhakal S., Ursin R.L., Deshpande S., Sandberg K., Mauvais-Jarvis F. Biological sex impacts COVID-19 outcomes. PLoS Pathog. 2020;16(6) doi: 10.1371/journal.ppat.1008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang J., Syrett C.M., Kramer M.C., Basu A., Atchison M.L., Anguera M.C. Unusual maintenance of X chromosome inactivation predisposes female lymphocytes for increased expression from the inactive X. Proc. Natl. Acad. Sci. 2016;113(14):E2029–E2038. doi: 10.1073/pnas.1520113113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Souyris M., Cenac C., Azar P., Daviaud D., Canivet A., Grunenwald S., Pienkowski C., Chaumeil J., Mejía J.E., Guéry J.-C. TLR7 escapes X chromosome inactivation in immune cells. Sci. Immunol. 2018;3(19) doi: 10.1126/sciimmunol.aap8855. [DOI] [PubMed] [Google Scholar]
- 110.Raisi-Estabragh Z., McCracken C., Ardissino M., Bethell M.S., Cooper J., Cooper C., Harvey N.C., Petersen S.E. Non-white ethnicity, male sex, and higher body mass index, but not medications acting on the renin-angiotensin system are associated with coronavirus disease 2019 (COVID-19) hospitalisation: review of the first 669 cases from the UK Biobank. MedRxiv. 2020 [Google Scholar]
- 111.Nunnari G., Sanfilippo C., Castrogiovanni P., Imbesi R., Volti G.L., Barbagallo I., Musumeci G., Di Rosa M. Network perturbation analysis in human bronchial epithelial cells following SARS-CoV2 infection. Exp. Cell Res. 2020;395(2) doi: 10.1016/j.yexcr.2020.112204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Miyai T., Hojyo S., Ikawa T., Kawamura M., Irié T., Ogura H., Hijikata A., Bin B.-H., Yasuda T., Kitamura H. Zinc transporter SLC39A10/ZIP10 facilitates antiapoptotic signaling during early B-cell development. Proc. Natl. Acad. Sci. 2014;111(32):11780–11785. doi: 10.1073/pnas.1323549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tilley A.E., Walters M.S., Shaykhiev R., Crystal R.G. Cilia dysfunction in lung disease. Annu. Rev. Physiol. 2015;77:379–406. doi: 10.1146/annurev-physiol-021014-071931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ross A.J., Dailey L.A., Brighton L.E., Devlin R.B. Transcriptional profiling of mucociliary differentiation in human airway epithelial cells. Am. J. Respir. Cell Mol. Biol. 2007;37(2):169–185. doi: 10.1165/rcmb.2006-0466OC. [DOI] [PubMed] [Google Scholar]
- 115.Luo S., Rosen S.M., Li Q., Agrawal P.B. Striated Preferentially Expressed Protein Kinase (SPEG) in Muscle Development, Function, and Disease. Int. J. Mol. Sci. 2021;22(11):5732. doi: 10.3390/ijms22115732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brodehl A., Gaertner-Rommel A., Milting H. Molecular insights into cardiomyopathies associated with desmin (DES) mutations. Biophys. Rev. 2018;10(4):983–1006. doi: 10.1007/s12551-018-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Agrawal P.B., Pierson C.R., Joshi M., Liu X., Ravenscroft G., Moghadaszadeh B., Talabere T., Viola M., Swanson L.C., Haliloğlu G. SPEG interacts with myotubularin, and its deficiency causes centronuclear myopathy with dilated cardiomyopathy. Am. J. Hum. Genet. 2014;95(2):218–226. doi: 10.1016/j.ajhg.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arentz M., Yim E., Klaff L., Lokhandwala S., Riedo F.X., Chong M., Lee M. Characteristics and outcomes of 21 critically ill patients with COVID-19 in Washington State. JAMA. 2020;323(16):1612–1614. doi: 10.1001/jama.2020.4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Guo T., Fan Y., Chen M., Wu X., Zhang L., He T., Wang H., Wan J., Wang X., Lu Z. Cardiovascular implications of fatal outcomes of patients with coronavirus disease 2019 (COVID-19) JAMA Cardiol. 2020;5(7):811–818. doi: 10.1001/jamacardio.2020.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhu Q., Yamakuchi M., Ture S., de la Luz Garcia-Hernandez M., Ko K.A., Modjeski K.L., LoMonaco M.B., Johnson A.D., O’Donnell C.J., Takai Y. Syntaxin-binding protein STXBP5 inhibits endothelial exocytosis and promotes platelet secretion. J. Clin. Investig. 2014;124(10):4503–4516. doi: 10.1172/JCI71245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hu J., Li C., Wang S., Li T., Zhang H. Genetic variants are identified to increase risk of COVID-19 related mortality from UK Biobank data. Hum. Genom. 2021;15(1):1–10. doi: 10.1186/s40246-021-00306-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abu-Farha M., Al-Sabah S., Hammad M.M., Hebbar P., Channanath A.M., John S.E., Taher I., Almaeen A., Ghazy A., Mohammad A. Prognostic Genetic Markers for Thrombosis in COVID-19 Patients: A Focused Analysis on D-Dimer, Homocysteine and Thromboembolism. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.587451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Saleh J., Peyssonnaux C., Singh K.K., Edeas M. Mitochondria and microbiota dysfunction in COVID-19 pathogenesis. Mitochondrion. 2020;54:1–7. doi: 10.1016/j.mito.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Li X., Fang P., Mai J., Choi E.T., Wang H., Yang X.-F. Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J. Hematol. Oncol. 2013;6(1):1–19. doi: 10.1186/1756-8722-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Scozzi D., Cano M., Ma L., Zhou D., Zhu J.H., O’Halloran J.A., Goss C., Rauseo A.M., Liu Z., Sahu S.K. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI insight. 2021;6(4) doi: 10.1172/jci.insight.143299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hönlinger A., Bömer U., Alconada A., Eckerskorn C., Lottspeich F., Dietmeier K., Pfanner N. Tom7 modulates the dynamics of the mitochondrial outer membrane translocase and plays a pathway-related role in protein import. The EMBO journal. 1996;15(9):2125–2137. [PMC free article] [PubMed] [Google Scholar]
- 127.Zietz M., Zucker J., Tatonetti N.P. Testing the association between blood type and COVID-19 infection, intubation, and death. MedRxiv. 2020 doi: 10.1038/s41467-020-19623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Barnkob M.B., Pottegård A., Støvring H., Haunstrup T.M., Homburg K., Larsen R., Hansen M.B., Titlestad K., Aagaard B., Møller B.K. Reduced prevalence of SARS-CoV-2 infection in ABO blood group O. Blood Adv. 2020;4(20):4990–4993. doi: 10.1182/bloodadvances.2020002657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Latz C.A., DeCarlo C., Boitano L., Png C.M., Patell R., Conrad M.F., Eagleton M., Dua A. Blood type and outcomes in patients with COVID-19. Ann. Hematol. 2020;99(9):2113–2118. doi: 10.1007/s00277-020-04169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khomich O.A., Kochetkov S.N., Bartosch B., Ivanov A.V. Redox biology of respiratory viral infections. Viruses. 2018;10(8):392. doi: 10.3390/v10080392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Saadat M. An evidence for correlation between the glutathione S-transferase T1 (GSTT1) polymorphism and outcome of COVID-19. Clinica Chimica acta; Int. J. Clin. Chem. 2020;508:213. doi: 10.1016/j.cca.2020.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Abbas M., Verma S., Verma S., Siddiqui S., Khan F.H., Raza S.T., Siddiqi Z., Eba A., Mahdi F. Association of GSTM1 and GSTT1 gene polymorphisms with COVID-19 susceptibility and its outcome. J. Med. Virol. 2021 doi: 10.1002/jmv.27076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Seo S., Zhang Q., Bugge K., Breslow D.K., Searby C.C., Nachury M.V., Sheffield V.C. A novel protein LZTFL1 regulates ciliary trafficking of the BBSome and Smoothened. PLoS Genet. 2011;7(11) doi: 10.1371/journal.pgen.1002358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.H. Kiss, D. Kedra, C. Kiss, M. Kost-Alimova, Y. Yang, G. Klein, S. Imreh, J.P. Dumanski, The LZTFL1 gene is a part of a transcriptional map covering 250 kb within the common eliminated region 1 (C3CER1) in 3p21. 3, Genomics 73(1) (2001) 10-19. [DOI] [PubMed]
- 135.Wei Q., Zhou W., Wang W., Gao B., Wang L., Cao J., Liu Z.-P. Tumor-suppressive functions of leucine zipper transcription factor–like 1. Cancer Res. 2010;70(7):2942–2950. doi: 10.1158/0008-5472.CAN-09-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Downes D.J., Cross A.R., Hua P., Roberts N., Schwessinger R., Cutler A.J., Munis A.M., Brown J., Mielczarek O., de Andrea C.E. Identification of LZTFL1 as a candidate effector gene at a COVID-19 risk locus. Nat. Genet. 2021:1–10. doi: 10.1038/s41588-021-00955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chlamydas S., Papavassiliou A.G., Piperi C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics. 2021;16(3):263–270. doi: 10.1080/15592294.2020.1796896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chai P., Yu J., Ge S., Jia R., Fan X. Genetic alteration, RNA expression, and DNA methylation profiling of coronavirus disease 2019 (COVID-19) receptor ACE2 in malignancies: a pan-cancer analysis. J. Hematol. Oncol. 2020;13(1):1–5. doi: 10.1186/s13045-020-00883-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yang J., Li H., Hu S., Zhou Y. ACE2 correlated with immune infiltration serves as a prognostic biomarker in endometrial carcinoma and renal papillary cell carcinoma: implication for COVID-19. Aging (Albany NY) 2020;12(8):6518. doi: 10.18632/aging.103100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sawalha A.H., Zhao M., Coit P., Lu Q. Epigenetic dysregulation of ACE2 and interferon-regulated genes might suggest increased COVID-19 susceptibility and severity in lupus patients. Clin. Immunol. 2020;215 doi: 10.1016/j.clim.2020.108410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ramakrishnan V. Histone structure and the organization of the nucleosome. Annu. Rev. Biophys. Biomol. Struct. 1997;26(1):83–112. doi: 10.1146/annurev.biophys.26.1.83. [DOI] [PubMed] [Google Scholar]
- 142.Castagnoli R., Votto M., Licari A., Brambilla I., Bruno R., Perlini S., Rovida F., Baldanti F., Marseglia G.L. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in children and adolescents: a systematic review. JAMA pediatrics. 2020;174(9):882–889. doi: 10.1001/jamapediatrics.2020.1467. [DOI] [PubMed] [Google Scholar]
- 143.Pinto B.G., Oliveira A.E., Singh Y., Jimenez L., Gonçalves A.N., Ogava R.L., Creighton R., Schatzmann Peron J.P., Nakaya H.I. ACE2 expression is increased in the lungs of patients with comorbidities associated with severe COVID-19. J. Infect. Dis. 2020;222(4):556–563. doi: 10.1093/infdis/jiaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sen R., Garbati M., Bryant K., Lu Y. Epigenetic mechanisms influencing COVID-19. Genome. 2021;64(4):372–385. doi: 10.1139/gen-2020-0135. [DOI] [PubMed] [Google Scholar]
- 145.Posynick B.J., Brown C.J. Escape from X-chromosome inactivation: an evolutionary perspective. Front. Cell Dev. Biol. 2019;7:241. doi: 10.3389/fcell.2019.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tukiainen T., Villani A.-C., Yen A., Rivas M.A., Marshall J.L., Satija R., Aguirre M., Gauthier L., Fleharty M., Kirby A. Landscape of X chromosome inactivation across human tissues. Nature. 2017;550(7675):244–248. doi: 10.1038/nature24265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 148.Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus–induced lung injury. Nat. Med. 2005;11(8):875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Li A.J., Li X. Sex-dependent immune response and lethality of COVID-19. Stem Cell Res. 2021;50 doi: 10.1016/j.scr.2020.102116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Van Der Made C.I., Simons A., Schuurs-Hoeijmakers J., Van Den Heuvel G., Mantere T., Kersten S., Van Deuren R.C., Steehouwer M., Van Reijmersdal S.V., Jaeger M. Presence of genetic variants among young men with severe COVID-19. JAMA. 2020;324(7):663–673. doi: 10.1001/jama.2020.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Meier A., Chang J.J., Chan E.S., Pollard R.B., Sidhu H.K., Kulkarni S., Wen T.F., Lindsay R.J., Orellana L., Mildvan D. Sex differences in the TLR-mediated response of pDCs to HIV-1 are associated with higher immune activation in infected women. Nat. Med. 2009;15(8):955. doi: 10.1038/nm.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Mantovani A., Netea M.G. Trained innate immunity, epigenetics, and Covid-19. N. Engl. J. Med. 2020;383(11):1078–1080. doi: 10.1056/NEJMcibr2011679. [DOI] [PubMed] [Google Scholar]
- 153.R.J. Arts, S.J. Moorlag, B. Novakovic, Y. Li, S.-Y. Wang, M. Oosting, V. Kumar, R.J. Xavier, C. Wijmenga, L.A. Joosten, BCG vaccination protects against experimental viral infection in humans through the induction of cytokines associated with trained immunity, Cell host & microbe 23(1) (2018) 89-100. e5. [DOI] [PubMed]
- 154.Saeed S., Quintin J., Kerstens H.H., Rao N.A., Aghajanirefah A., Matarese F., Cheng S.-C., Ratter J., Berentsen K. M.A. van der Ent, Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345(6204) doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Moorlag S., Arts R., Van Crevel R., Netea M. Non-specific effects of BCG vaccine on viral infections. Clin. Microbiol. Infect. 2019;25(12):1473–1478. doi: 10.1016/j.cmi.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 156.Koneru G., Batiha G.-E.-S., Algammal A.M., Mabrok M., Magdy S., Sayed S., AbuElmagd M.E., Elnemr R., Saad M.M., Abd Ellah N.H. BCG Vaccine-Induced Trained Immunity and COVID-19: Protective or Bystander? Infect. Drug Resist. 2021;14:1169. doi: 10.2147/IDR.S300162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weng C., Saal A., Butt W.W., Bica N., Fisher J., Tao J., Chan P. Bacillus Calmette-Guérin vaccination and clinical characteristics and outcomes of COVID-19 in Rhode Island, United States: a cohort study. Epidemiol. Infect. 2020;148 doi: 10.1017/S0950268820001569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schäfer A., Baric R.S. Epigenetic landscape during coronavirus infection. Pathogens. 2017;6(1):8. doi: 10.3390/pathogens6010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Aevermann B.D., Pickett B.E., Kumar S., Klem E.B., Agnihothram S., Askovich P.S., Bankhead A., Bolles M., Carter V., Chang J. A comprehensive collection of systems biology data characterizing the host response to viral infection. Sci. Data. 2014;1(1):1–21. doi: 10.1038/sdata.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fang T.C., Schaefer U., Mecklenbrauker I., Stienen A., Dewell S., Chen M.S., Rioja I., Parravicini V., Prinjha R.K., Chandwani R. Histone H3 lysine 9 di-methylation as an epigenetic signature of the interferon response. J. Exp. Med. 2012;209(4):661–669. doi: 10.1084/jem.20112343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Stender J.D., Glass C.K. Epigenomic control of the innate immune response. Curr. Opin. Pharmacol. 2013;13(4):582–587. doi: 10.1016/j.coph.2013.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ahmad S., Manzoor S., Siddiqui S., Mariappan N., Zafar I., Ahmad A., Ahmad A. Seminars in Cancer Biology. 2021. Epigenetic underpinnings of inflammation: Connecting the dots between pulmonary diseases, lung cancer and COVID-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sang E.R., Tian Y., Miller L.C., Sang Y. Epigenetic evolution of ACE2 and IL-6 genes: Non-canonical interferon-stimulated genes correlate to covid-19 susceptibility in vertebrates. Genes. 2021;12(2):154. doi: 10.3390/genes12020154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Christophorou M.A., Castelo-Branco G., Halley-Stott R.P., Oliveira C.S., Loos R., Radzisheuskaya A., Mowen K.A., Bertone P., Silva J.C., Zernicka-Goetz M. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature. 2014;507(7490):104–108. doi: 10.1038/nature12942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Beato M., Sharma P. Peptidyl arginine deiminase 2 (PADI2)-mediated arginine citrullination modulates transcription in cancer. Int. J. Mol. Sci. 2020;21(4):1351. doi: 10.3390/ijms21041351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Wang Y., Li M., Stadler S., Correll S., Li P., Wang D., Hayama R., Leonelli L., Han H., Grigoryev S.A. Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 2009;184(2):205–213. doi: 10.1083/jcb.200806072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang Y., Wysocka J., Sayegh J., Lee Y.-H., Perlin J.R., Leonelli L., Sonbuchner L.S., McDonald C.H., Cook R.G., Dou Y. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306(5694):279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- 168.Zuo Y., Yalavarthi S., Shi H., Gockman K., Zuo M., Madison J.A., Blair C., Weber A., Barnes B.J., Egeblad M. Neutrophil extracellular traps in COVID-19. JCI insight. 2020;5(11) doi: 10.1172/jci.insight.138999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zucoloto A.Z., Jenne C.N. Platelet-neutrophil interplay: insights into neutrophil extracellular trap (NET)-driven coagulation in infection. Front. Cardiovasc. Med. 2019;6:85. doi: 10.3389/fcvm.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Wu M., Chen Y., Xia H., Wang C., Tan C.Y., Cai X., Liu Y., Ji F., Xiong P., Liu R. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc. Natl. Acad. Sci. 2020;117(45):28336–28343. doi: 10.1073/pnas.2018030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jones J.E., Slack J.L., Fang P., Zhang X., Subramanian V., Causey C.P., Coonrod S.A., Guo M., Thompson P.R. Synthesis and screening of a haloacetamidine containing library to identify PAD4 selective inhibitors. ACS Chem. Biol. 2012;7(1):160–165. doi: 10.1021/cb200258q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moradinasab S., Pourbagheri-Sigaroodi A., Zafari P., Ghaffari S.H., Bashash D. Mesenchymal stromal/stem cells (MSCs) and MSC-derived extracellular vesicles in COVID-19-induced ARDS: Mechanisms of action, research progress, challenges, and opportunities. Int. Immunopharmacol. 2021;97 doi: 10.1016/j.intimp.2021.107694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Xu P., Ye S., Li K., Huang M., Wang Q., Zeng S., Chen X., Gao W., Chen J., Zhang Q. NOS1 inhibits the interferon response of cancer cells by S-nitrosylation of HDAC2. J. Exp. Clin. Cancer Res. 2019;38(1):1–16. doi: 10.1186/s13046-019-1448-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat. Struct. Mol. Biol. 2014;21(4):301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 176.Cai W., Yang H. The structure and regulation of Cullin 2 based E3 ubiquitin ligases and their biological functions. Cell division. 2016;11(1):1–11. doi: 10.1186/s13008-016-0020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zeberg H., Pääbo S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature. 2020;587(7835):610–612. doi: 10.1038/s41586-020-2818-3. [DOI] [PubMed] [Google Scholar]
- 178.Mongelli A., Barbi V., Gottardi Zamperla M., Atlante S., Forleo L., Nesta M., Massetti M., Pontecorvi A., Nanni S., Farsetti A. Evidence for Biological Age Acceleration and Telomere Shortening in COVID-19 Survivors. Int. J. Mol. Sci. 2021;22(11):6151. doi: 10.3390/ijms22116151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Hayflick L., Moorhead P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961;25(3):585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 180.Zhan Y., Hägg S. Telomere length and cardiovascular disease risk. Curr. Opin. Cardiol. 2019;34(3):270–274. doi: 10.1097/HCO.0000000000000613. [DOI] [PubMed] [Google Scholar]
- 181.Bekaert B., Kamalandua A., Zapico S.C., Van de Voorde W., Decorte R. Improved age determination of blood and teeth samples using a selected set of DNA methylation markers. Epigenetics. 2015;10(10):922–930. doi: 10.1080/15592294.2015.1080413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mishra S.K., Tripathi T. One year update on the COVID-19 pandemic: Where are we now? Acta Trop. 2021;214 doi: 10.1016/j.actatropica.2020.105778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9 doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mittal A., Khattri A., Verma V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PLoS Pathog. 2022;18(2) doi: 10.1371/journal.ppat.1010260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19(7):409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.W.H. Organization, Tracking SARS-CoV-2 variants, 2021. https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/.
- 187.Variants: distribution of cases data, 20 May 2021 https://www.gov.uk/government/publications/covid-19-variants-genomically-confirmed-case-numbers/variants-distribution-of-cases-data. (Accessed 22-December 2021).
- 188.C.f.D. Control, Prevention, Science brief: emerging SARS-CoV-2 variants, Publisher Full Text (2021). [PubMed]
- 189.S.E. Galloway, P. Paul, D.R. MacCannell, M.A. Johansson, J.T. Brooks, A. MacNeil, R.B. Slayton, S. Tong, B.J. Silk, G.L. Armstrong, Emergence of SARS-CoV-2 b. 1.1. 7 lineage—united states, december 29, 2020–january 12, 2021, Morbidity and Mortality Weekly Report 70(3) (2021) 95. [DOI] [PMC free article] [PubMed]
- 190.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eurosurveillance. 2021;26(1):2002106. doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.S.A. Kemp, B. Meng, I.A. Ferriera, R. Datir, W.T. Harvey, D. Collier, S. Lytras, G. Papa, A. Carabelli, J. Kenyon, Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70, (2021).
- 192.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371(6534):1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.R. Assessment, Risk related to spread of new SARS-CoV-2 variants of concern in the EU/EEA, European Centre for Disease Prevention and Control (2020).
- 194.Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations, Genom. Epidemiol. 2020:1–5. [Google Scholar]
- 195.Pereira F. Evolutionary dynamics of the SARS-CoV-2 ORF8 accessory gene. Infection, Genetics and Evolution. 2020;85 doi: 10.1016/j.meegid.2020.104525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Mohandas S., Yadav P.D., Nyayanit D., Deshpande G., Aich A.S., Sapkal G., Kumar S., Jain R., Kadam M., Kumar A. Comparison of the pathogenicity and virus shedding of SARS CoV-2 VOC 202012/01 and D614G variant in hamster model. BioRxiv. 2021 [Google Scholar]
- 197.Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. MedRxiv. 2020 [Google Scholar]
- 198.E. Mahase, Covid-19: What new variants are emerging and how are they being investigated?, British Medical Journal Publishing Group, 2021. [DOI] [PubMed]
- 199.C.K. Wibmer, F. Ayres, T. Hermanus, M. Madzivhandila, P. Kgagudi, B. Oosthuysen, B.E. Lambson, T. De Oliveira, M. Vermeulen, K. Van der Berg, SARS-CoV-2 501Y. V2 escapes neutralization by South African COVID-19 donor plasma, Nature medicine 27(4) (2021) 622-625. [DOI] [PubMed]
- 200.Di Giacomo S., Mercatelli D., Rakhimov A., Giorgi F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021 doi: 10.1002/jmv.27062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.F. Karim, I. Gazy, S. Cele, Y. Zungu, R. Krause, M. Bernstein, Y. Ganga, H. Rodel, N. Mthabela, M. Mazibuko, HIV status alters disease severity and immune cell responses in β variant SARS-CoV-2 infection wave, medRxiv (2021) 2020.11. 23.20236828. [DOI] [PMC free article] [PubMed]
- 202.N.R. Faria, T.A. Mellan, C. Whittaker, I.M. Claro, D.d.S. Candido, S. Mishra, M.A. Crispim, F.C. Sales, I. Hawryluk, J.T. McCrone, Genomics and epidemiology of the P. 1 SARS-CoV-2 lineage in Manaus, Brazil, Science 372(6544) (2021) 815-821. [DOI] [PMC free article] [PubMed]
- 203.F. Naveca, V. Nascimento, V. Souza, A. Corado, F. Nascimento, G. Silva, Á. Costa, D. Duarte, K. Pessoa, M. Mejía, COVID-19 epidemic in the Brazilian state of Amazonas was driven by long-term persistence of endemic SARS-CoV-2 lineages and the recent emergence of the new Variant of Concern P. 1, (2021).
- 204.Weekly epidemiological update on COVID-19 - 27 April 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---27-april-2021. (Accessed 22-December 2021).
- 205.A.F. Martins, A.P. Zavascki, P.L. Wink, F.C.Z. Volpato, F.L. Monteiro, C. Rosset, F. De-Paris, Á.K. Ramos, A.L. Barth, Detection of SARS-CoV-2 lineage P. 1 in patients from a region with exponentially increasing hospitalisation rate, February 2021, Rio Grande do Sul, Southern Brazil, Eurosurveillance 26(12) (2021) 2100276. [DOI] [PMC free article] [PubMed]
- 206.Romero P.E., Dávila-Barclay A., Salvatierra G., González L., Cuicapuza D., Solís L., Marcos-Carbajal P., Huancachoque J., Maturrano L., Tsukayama P. The emergence of SARS-CoV-2 variant lambda (C. 37) in South America. Microbiology spectrum. 2021;9(2):e00789–e00821. doi: 10.1128/Spectrum.00789-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Baj A., Novazzi F., Ferrante F.D., Genoni A., Cassani G., Prestia M., Colombo A., Capuano R., Zago C., Pasciuta R. Introduction of SARS-COV-2 C. 37 (WHO VOI lambda) from Peru to Italy. J. Med. Virol. 2021 doi: 10.1002/jmv.27235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Sheikh A., McMenamin J., Taylor B., Robertson C. SARS-CoV-2 Delta VOC in Scotland: demographics, risk of hospital admission, and vaccine effectiveness. Lancet (London, England) 2021;397(10293):2461–2462. doi: 10.1016/S0140-6736(21)01358-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Uriu K., Kimura I., Shirakawa K., Takaori-Kondo A., Nakada T.-A., Kaneda A., Nakagawa S., Sato K. Neutralization of the SARS-CoV-2 Mu variant by convalescent and Vaccine Serum. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2114706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.W.H. Organization, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern.
- 211.Torjesen I. Covid-19: Omicron may be more transmissible than other variants and partly resistant to existing vaccines, scientists fear. BMJ (Clinical research ed.) 2021;375 doi: 10.1136/bmj.n2943. [DOI] [PubMed] [Google Scholar]
- 212.Karim S.S.A., Karim Q.A. Omicron SARS-CoV-2 variant: a new chapter in the COVID-19 pandemic. Lancet (London, England) 2021;398(10317):2126–2128. doi: 10.1016/S0140-6736(21)02758-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.R. Hirose, Y. Itoh, H. Ikegaya, H. Miyazaki, N. Watanabe, T. Yoshida, R. Bandou, T. Daidoji, T. Nakaya, Differences in environmental stability among SARS-CoV-2 variants of concern: Omicron has higher stability, bioRxiv (2022). [DOI] [PMC free article] [PubMed]
- 214.Pulliam J.R., van Schalkwyk C., Govender N., von Gottberg A., Cohen C., Groome M.J., Dushoff J., Mlisana K., Moultrie H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. MedRxiv. 2021 doi: 10.1126/science.abn4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.K. van der Straten, D. Guerra, M. van Gils, I. Bontjer, T.G. Caniels, H.D. van Willigen, E. Wynberg, M. Poniman, J.A. Burger, J.H. Bouhuijs, Mapping the antigenic diversification of SARS-CoV-2, medRxiv (2022).
- 216.CoVariant, (2021).
- 217.He X., Hong W., Pan X., Lu G., Wei X. SARS-CoV-2 Omicron variant: Characteristics and prevention. MedComm. 2021;2(4):838–845. doi: 10.1002/mco2.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wang L., Cheng G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J. Med. Virol. 2021 doi: 10.1002/jmv.27516. [DOI] [PubMed] [Google Scholar]
- 220.Majumdar S., Sarkar R. Mutational and phylogenetic analyses of the two lineages of the Omicron variant. J. Med. Virol. 2021 doi: 10.1002/jmv.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.NextStrain, (2021).
- 222.Kimura I., Kosugi Y., Wu J., Yamasoba D., Butlertanaka E.P., Tanaka Y.L., Liu Y., Shirakawa K., Kazuma Y., Nomura R. SARS-CoV-2 Lambda variant exhibits higher infectivity and immune resistance. BioRxiv. 2021 doi: 10.1016/j.celrep.2021.110218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.L. Ceron-Gutierrez, G. Barcenas-Morales, M. Wills, R. Doffinger, R. Gupta, Sensitivity of SARS-CoV-2 B. 1.1. 7 to mRNA vaccine-elicited antibodies, Nature 10 (2021). [DOI] [PMC free article] [PubMed]
- 224.P. Wang, M.S. Nair, L. Liu, S. Iketani, Y. Luo, Y. Guo, M. Wang, J. Yu, B. Zhang, P.D. Kwong, Antibody resistance of SARS-CoV-2 variants B. 1.351 and B. 1.1. 7, Nature 593(7857) (2021) 130–135. [DOI] [PubMed]
- 225.G.P. Manouana, M. Nzamba Maloum, R. Bikangui, S.O.n. Oye Bingono, G. Ondo Nguema, J.Y. Honkpehedji, E.G. Rossatanga, S. Zoa-Assoumou, S.R. Pallerla, S. Rachakonda, A. Ndong Mintsa, J.-B. Lekana-Douki, J.-F. Djoba Siawaya, S. Borrmann, P.G. Kremsner, B. Lell, T.P. Velavan, A.A. Adegnika, Emergence of B.1.1.318 SARS-CoV-2 viral lineage and high incidence of alpha B.1.1.7 variant of concern in the Republic of Gabon, Int J Infect Dis 114 (2022) 151-154. [DOI] [PMC free article] [PubMed]
- 226.C.M. Voloch, R. da Silva Francisco Jr, L.G. de Almeida, C.C. Cardoso, O.J. Brustolini, A.L. Gerber, A.P.d.C. Guimarães, D. Mariani, R.M. da Costa, O.C. Ferreira Jr, Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil, Journal of virology 95(10) (2021) e00119-21. [DOI] [PMC free article] [PubMed]
- 227.D. Weissman, M.-G. Alameh, T. de Silva, P. Collini, H. Hornsby, R. Brown, C.C. LaBranche, R.J. Edwards, L. Sutherland, S. Santra, D614G spike mutation increases SARS CoV-2 susceptibility to neutralization, Cell host & microbe 29(1) (2021) 23-31. e4. [DOI] [PMC free article] [PubMed]
- 228.South India's N440K COVID variant 15 times more lethal. https://www.businesstoday.in/latest/trends/story/south-india-n440k-covid-variant-15-times-more-lethal-294981-2021-05-04. (Accessed 22-December 2021).
- 229.E. Lasek-Nesselquist, P. Lapierre, E. Schneider, K.S. George, J. Pata, The localized rise of a B. 1.526 variant containing an E484K mutation in New York State, medRxiv (2021).
- 230.A.P. West Jr, C.O. Barnes, Z. Yang, P.J. Bjorkman, SARS-CoV-2 lineage B. 1.526 emerging in the New York region detected by software utility created to query the spike mutational landscape, (2021).
- 231.E. Lasek-Nesselquist, J. Pata, E. Schneider, K.S. George, A tale of three SARS-CoV-2 variants with independently acquired P681H mutations in New York State, medRxiv (2021).
- 232.Singh A., Steinkellner G., Köchl K., Gruber K., Gruber C.C. Serine 477 plays a crucial role in the interaction of the SARS-CoV-2 spike protein with the human receptor ACE2. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-83761-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Ortuso F., Mercatelli D., Guzzi P.H., Giorgi F.M. Structural genetics of circulating variants affecting the SARS-CoV-2 spike/human ACE2 complex. J. Biomol. Struct. Dyn. 2021:1–11. doi: 10.1080/07391102.2021.1886175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Coronavirus | Indian ‘double mutant’ strain named B.1.617. https://www.thehindu.com/news/national/indian-double-mutant-strain-named-b1617/article34274663.ece/amp/. (Accessed 22-December 2021).
- 235.Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Reports Medicine. 2021;2(4) doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Why Virus Variants Have Such Weird Names. https://www.nytimes.com/2021/03/02/health/virus-variant-names.html. (Accessed 22-December 2021).
- 238.Shahhosseini N., Babuadze G.G., Wong G., Kobinger G.P. Mutation Signatures and In Silico Docking of Novel SARS-CoV-2 Variants of Concern. Microorganisms. 2021;9(5):926. doi: 10.3390/microorganisms9050926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.M. McCallum, J. Bassi, A. De Marco, A. Chen, A.C. Walls, J. Di Iulio, M.A. Tortorici, M.-J. Navarro, C. Silacci-Fregni, C. Saliba, SARS-CoV-2 immune evasion by variant B. 1.427/B. 1.429, BioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 240.T. Carroll, D. Fox, N. van Doremalen, E. Ball, M.K. Morris, A. Sotomayor-Gonzalez, V. Servellita, A. Rustagi, C.K. Yinda, L. Fritts, The B. 1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B. 1 (614G) in Syrian hamsters, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 241.Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C. The N501Y spike substitution enhances SARS-CoV-2 transmission. BioRxiv. 2021 doi: 10.1038/s41586-021-04245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Maison D.P., Ching L.L., Shikuma C.M., Nerurkar V.R. Genetic Characteristics and Phylogeny of 969-bp S Gene Sequence of SARS-CoV-2 from Hawai ‘i Reveals the Worldwide Emerging P681H Mutation. Hawai'i J. Health Soc. Welf. 2021;80(3):52. [PMC free article] [PubMed] [Google Scholar]
- 243.R. Rose, D.J. Nolan, T.M. LaFleur, S.L. Lamers, Outbreak of P. 3 (Theta) SARS-CoV-2 emerging variant of concern among service workers in Louisiana, Journal of infection and public health (2021). [DOI] [PMC free article] [PubMed]
- 244.F.A. Tablizo, K.M. Kim, C.M. Lapid, M.J.R. Castro, M.S.L. Yangzon, B.A. Maralit, M.E.C. Ayes, E.M. Cutiongco-de la Paz, A.R. De Guzman, J.M.C. Yap, Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines, medRxiv (2021).
- 245.Liu Z., VanBlargan L.A., Rothlauf P.W., Bloyet L.-M., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Ellebedy A. Landscape analysis of escape variants identifies SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Available at SSRN. 2020;3725763 doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Grubaugh N.D., Hanage W.P., Rasmussen A.L. Making sense of mutation: what D614G means for the COVID-19 pandemic remains unclear. Cell. 2020;182(4):794–795. doi: 10.1016/j.cell.2020.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ozono S., Zhang Y., Ode H., Seng T.T., Imai K., Miyoshi K., Kishigami S., Ueno T., Iwatani Y., Suzuki T. Naturally mutated spike proteins of SARS-CoV-2 variants show differential levels of cell entry. BioRxiv. 2020 [Google Scholar]
- 248.Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592(7852):116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.B. Korber, W.M. Fischer, S. Gnanakaran, H. Yoon, J. Theiler, W. Abfalterer, N. Hengartner, E.E. Giorgi, T. Bhattacharya, B. Foley, Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus, Cell 182(4) (2020) 812–827. e19. [DOI] [PMC free article] [PubMed]
- 250.E. Volz, V. Hill, J.T. McCrone, A. Price, D. Jorgensen, Á. O’Toole, J. Southgate, R. Johnson, B. Jackson, F.F. Nascimento, Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity, Cell 184(1) (2021) 64–75. e11. [DOI] [PMC free article] [PubMed]
- 251.Becerra-Flores M., Cardozo T. SARS-CoV-2 viral spike G614 mutation exhibits higher case fatality rate. Int. J. Clin. Pract. 2020;74(8) doi: 10.1111/ijcp.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.DeDiego M.L., Nieto-Torres J.L., Jimenez-Guardeño J.M., Regla-Nava J.A., Castaño-Rodriguez C., Fernandez-Delgado R., Usera F., Enjuanes L. Coronavirus virulence genes with main focus on SARS-CoV envelope gene. Virus Res. 2014;194:124–137. doi: 10.1016/j.virusres.2014.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Kumar B.K., Rohit A., Prithvisagar K.S., Rai P., Karunasagar I., Karunasagar I. Deletion in the C-terminal region of the envelope glycoprotein in some of the Indian SARS-CoV-2 genome. Virus Res. 2021;291 doi: 10.1016/j.virusres.2020.198222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Shen L., Bard J.D., Triche T.J., Judkins A.R., Biegel J.A., Gai X. Emerging variants of concern in SARS-CoV-2 membrane protein: a highly conserved target with potential pathological and therapeutic implications. Emerg. Microbes Infect. 2021;10(1):885–893. doi: 10.1080/22221751.2021.1922097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.C. Motozono, M. Toyoda, J. Zahradnik, A. Saito, H. Nasser, T.S. Tan, I. Ngare, I. Kimura, K. Uriu, Y. Kosugi, SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity, Cell host microbe 29(7) (2021) 1124–1136. e11. [DOI] [PMC free article] [PubMed]
- 256.Bourassa L., Perchetti G.A., Phung Q., Lin M.J., Mills M.G., Roychoudhury P., Harmon K.G., Reed J.C., Greninger A.L. A SARS-CoV-2 nucleocapsid variant that affects antigen test performance. J. Clin. Virol. 2021;104900 doi: 10.1016/j.jcv.2021.104900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Hartenian E., Nandakumar D., Lari A., Ly M., Tucker J.M., Glaunsinger B.A. The molecular virology of coronaviruses. J. Biol. Chem. 2020;295(37):12910–12934. doi: 10.1074/jbc.REV120.013930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Jensen B., Luebke N., Feldt T., Keitel V., Brandenburger T., Kindgen-Milles D., Lutterbeck M., Freise N.F., Schoeler D., Haas R. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. The Lancet Regional Health-Europe. 2021;8 doi: 10.1016/j.lanepe.2021.100164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Hossain M.U., Bhattacharjee A., Emon M.T.H., Chowdhury Z.M., Ahammad I., Mosaib M.G., Moniruzzaman M., Rahman M.H., Islam M.N., Ahmed I. Novel mutations in NSP-1 and PLPro of SARS-CoV-2 NIB-1 genome mount for effective therapeutics. J. Genet. Eng. Biotechnol. 2021;19(1):1–10. doi: 10.1186/s43141-021-00152-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tian F., Tong B., Sun L., Shi S., Zheng B., Wang Z., Dong X., Zheng P. Mutation N501Y in RBD of Spike Protein Strengthens the Interaction between COVID-19 and its Receptor ACE2. BioRxiv. 2021 doi: 10.7554/eLife.69091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Yang D.-M., Lin F.-C., Tsai P.-H., Chien Y., Wang M.-L., Yang Y.-P., Chang T.-J. Pandemic analysis of infection and death correlated with genomic Orf10 mutation in SARS-CoV-2 victims. J. Chinese Med. Assoc. JCMA. 2021 doi: 10.1097/JCMA.0000000000000542. [DOI] [PubMed] [Google Scholar]
- 262.Zuckerman N.S., Fleishon S., Bucris E., Bar-Ilan D., Linial M., Bar-Or I., Indenbaum V., Weil M., Lustig Y., Mendelson E. A Unique SARS-CoV-2 Spike Protein P681H Variant Detected in Israel. Vaccines. 2021;9(6):616. doi: 10.3390/vaccines9060616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.B. Lubinski, T. Tang, S. Daniel, J.A. Jaimes, G. Whittaker, Functional evaluation of proteolytic activation for the SARS-CoV-2 variant B. 1.1. 7: role of the P681H mutation, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 264.Saito A., Irie T., Suzuki R., Maemura T., Nasser H., Uriu K., Kosugi Y., Shirakawa K., Sadamasu K., Kimura I. SARS-CoV-2 spike P681R mutation, a hallmark of the Delta variant, enhances viral fusogenicity and pathogenicity. BioRxiv. 2021 [Google Scholar]
- 265.Y. Liu, J. Liu, B. Johnson, H. Xia, Z. Ku, C. Schindewolf, S. Widen, Z. An, S. Weaver, V. Menachery, Delta spike P681R mutation enhances SARS-CoV-2 fitness over Alpha variant. bioRxiv, Preprint, doi 10(2021.08) (2021) 12.456173. [DOI] [PMC free article] [PubMed]
- 266.Suppiah J., Kamel K.A., Mohd-Zawawi Z., Afizan M.A., Yahya H., Md-Hanif S.A., Thayan R. Phylogenomic analysis of SARS-CoV-2 from third wave clusters in Malaysia reveals dominant local lineage B.1.524 and persistent spike mutation A701V. Trop. Biomed. 2021;38(3):289–293. doi: 10.47665/tb.38.3.070. [DOI] [PubMed] [Google Scholar]
- 267.M.K. Annavajhala, H. Mohri, P. Wang, M. Nair, J.E. Zucker, Z. Sheng, A. Gomez-Simmonds, A.L. Kelley, M. Tagliavia, Y. Huang, T. Bedford, D.D. Ho, A.C. Uhlemann, A Novel and Expanding SARS-CoV-2 Variant, B.1.526, Identified in New York, medRxiv (2021). [DOI] [PMC free article] [PubMed]
- 268.Mukherjee S., Harikishore A., Bhunia A. Targeting C-terminal Helical bundle of NCOVID19 Envelope (E) protein. Int. J. Biol. Macromol. 2021;175:131–139. doi: 10.1016/j.ijbiomac.2021.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Chai J., Cai Y., Pang C., Wang L., McSweeney S., Shanklin J., Liu Q. Structural basis for SARS-CoV-2 envelope protein recognition of human cell junction protein PALS1. Nat. Commun. 2021;12(1):1–6. doi: 10.1038/s41467-021-23533-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mou K., Abdalla M., Wei D.Q., Khan M.T., Lodhi M.S., Darwish D.B., Sharaf M., Tu X. Emerging mutations in envelope protein of SARS-CoV-2 and their effect on thermodynamic properties. Inf. Med. Unlocked. 2021;25 doi: 10.1016/j.imu.2021.100675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12(1):1–17. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.T. Mourier, M. Shuaib, S. Hala, S. Mfarrej, F. Alofi, R. Naeem, A. Alsomali, D. Jorgensen, A. Subudhi, F. Ben Rached, Saudi Arabian SARS-CoV-2 genomes implicate a mutant Nucleocapsid protein in modulating host interactions and increased viral load in COVID-19 patients, (2021).
- 273.Wu A., Peng Y., Huang B., Ding X., Wang X., Niu P., Meng J., Zhu Z., Zhang Z., Wang J. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Angeletti S., Benvenuto D., Bianchi M., Giovanetti M., Pascarella S., Ciccozzi M. COVID-2019: the role of the nsp2 and nsp3 in its pathogenesis. J. Med. Virol. 2020;92(6):584–588. doi: 10.1002/jmv.25719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Tsai P.-H., Wang M.-L., Yang D.-M., Liang K.-H., Chou S.-J., Chiou S.-H., Lin T.-H., Wang C.-T., Chang T.-J. Genomic variance of Open Reading Frames (ORFs) and Spike protein in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Chinese Med. Assoc. 2020;83(8):725. doi: 10.1097/JCMA.0000000000000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Freundt E.C., Yu L., Goldsmith C.S., Welsh S., Cheng A., Yount B., Liu W., Frieman M.B., Buchholz U.J., Screaton G.R. The open reading frame 3a protein of severe acute respiratory syndrome-associated coronavirus promotes membrane rearrangement and cell death. J. Virol. 2010;84(2):1097–1109. doi: 10.1128/JVI.01662-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Delbue S., D’Alessandro S., Signorini L., Dolci M., Pariani E., Bianchi M., Fattori S., Modenese A., Galli C., Eberini I. Isolation of SARS-CoV-2 strains carrying a nucleotide mutation, leading to a stop codon in the ORF 6 protein. Emerg. Microbes Infect. 2021;10(1):252–255. doi: 10.1080/22221751.2021.1884003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Dolgin E. The race for antiviral drugs to beat COVID - and the next pandemic. Nature. 2021;592(7854):340–343. doi: 10.1038/d41586-021-00958-4. [DOI] [PubMed] [Google Scholar]
- 279.Choy K.T., Wong A.Y., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Janiaud P., Axfors C., Schmitt A.M., Gloy V., Ebrahimi F., Hepprich M., Smith E.R., Haber N.A., Khanna N., Moher D., Goodman S.N., Ioannidis J.P.A., Hemkens L.G. Association of Convalescent Plasma Treatment With Clinical Outcomes in Patients With COVID-19: A Systematic Review and Meta-analysis. JAMA. 2021;325(12):1185–1195. doi: 10.1001/jama.2021.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.S. Şimşek-Yavuz, F.I. Komsuoğlu Çelikyurt, An update of anti-viral treatment of COVID-19, Turkish journal of medical sciences 51(Si-1) (2021) 3372-3390. [DOI] [PMC free article] [PubMed]
- 282.W.H. Organization, COVID-19 vaccines, 2021. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/covid-19-vaccines.
- 283.FDA, FDA approves Pfizer-BioNTech COVID-19 Vaccine, 2021. https://www.fda.gov/news-events/press-announcements/fda-approves-first-covid-19-vaccine.
- 284.FDA, FDA approves Moderna COVID-19 Vaccine, 2022. https://www.fda.gov/news-events/press-announcements/coronavirus-covid-19-update-fda-takes-key-action-approving-second-covid-19-vaccine.
- 285.Hillus D., Schwarz T., Tober-Lau P., Vanshylla K., Hastor H., Thibeault C., Jentzsch S., Helbig E.T., Lippert L.J., Tscheak P., Schmidt M.L., Riege J., Solarek A., von Kalle C., Dang-Heine C., Gruell H., Kopankiewicz P., Suttorp N., Drosten C., Bias H., Seybold J., Klein F., Kurth F., Corman V.M., Sander L.E. Safety, reactogenicity, and immunogenicity of homologous and heterologous prime-boost immunisation with ChAdOx1 nCoV-19 and BNT162b2: a prospective cohort study. Lancet. Respiratory Med. 2021;9(11):1255–1265. doi: 10.1016/S2213-2600(21)00357-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Noori M., Nejadghaderi S.A., Arshi S., Carson-Chahhoud K., Ansarin K., Kolahi A.A., Safiri S. Potency of BNT162b2 and mRNA-1273 vaccine-induced neutralizing antibodies against severe acute respiratory syndrome-CoV-2 variants of concern: A systematic review of in vitro studies. Rev. Med. Virol. 2022;32(2) doi: 10.1002/rmv.2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Jara A., Undurraga E.A., Zubizarreta J.R., González C., Pizarro A., Acevedo J., Leo K., Paredes F., Bralic T., Vergara V., Mosso M., Leon F., Parot I., Leighton P., Suárez P., Rios J.C., García-Escorza H., Araos R. Effectiveness of homologous and heterologous booster doses for an inactivated SARS-CoV-2 vaccine: a large-scale prospective cohort study. The Lancet. Global health. 2022;10(6):e798–e806. doi: 10.1016/S2214-109X(22)00112-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Fiolet T., Kherabi Y., MacDonald C.J., Ghosn J., Peiffer-Smadja N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: a narrative review. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2022;28(2):202–221. doi: 10.1016/j.cmi.2021.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.N.G. Davies, CSV-format data for: Increased mortality in community-tested cases of SARS-CoV-2 lineage B. 1.1. 7, (2021). [DOI] [PMC free article] [PubMed]
- 290.E. Volz, S. Mishra, M. Chand, J.C. Barrett, R. Johnson, Lily Geidelberg, Wes R. Hinsley, et al. 2021.“Assessing Transmissibility of SARS-CoV-2 Lineage B. 1.1. 7 in England.”, Nature 1–17. [DOI] [PubMed]
- 291.Edara V.V., Hudson W.H., Xie X., Ahmed R., Suthar M.S. Neutralizing Antibodies Against SARS-CoV-2 Variants After Infection and Vaccination. JAMA. 2021;325(18):1896–1898. doi: 10.1001/jama.2021.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Planas D., Bruel T., Grzelak L., Guivel-Benhassine F., Staropoli I., Porrot F., Planchais C., Buchrieser J., Rajah M.M., Bishop E., Albert M., Donati F., Prot M., Behillil S., Enouf V., Maquart M., Smati-Lafarge M., Varon E., Schortgen F., Yahyaoui L., Gonzalez M., De Sèze J., Péré H., Veyer D., Sève A., Simon-Lorière E., Fafi-Kremer S., Stefic K., Mouquet H., Hocqueloux L., van der Werf S., Prazuck T., Schwartz O. Sensitivity of infectious SARS-CoV-2 B.1.1.7 and B.1.351 variants to neutralizing antibodies. Nat Med. 2021;27(5):917–924. doi: 10.1038/s41591-021-01318-5. [DOI] [PubMed] [Google Scholar]
- 293.Huang B., Dai L., Wang H., Hu Z., Yang X., Tan W., Gao G.F. Serum sample neutralisation of BBIBP-CorV and ZF2001 vaccines to SARS-CoV-2 501Y.V2, The Lancet. Microbe. 2021;2(7) doi: 10.1016/S2666-5247(21)00082-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Wang G.L., Wang Z.Y., Duan L.J., Meng Q.C., Jiang M.D., Cao J., Yao L., Zhu K.L., Cao W.C., Ma M.J. Susceptibility of Circulating SARS-CoV-2 Variants to Neutralization. New England J. Med. 2021;384(24):2354–2356. doi: 10.1056/NEJMc2103022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Chen Y., Shen H., Huang R., Tong X., Wu C. Serum neutralising activity against SARS-CoV-2 variants elicited by CoronaVac. Lancet. Infect. Dis. 2021;21(8):1071–1072. doi: 10.1016/S1473-3099(21)00287-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Moutinho S. Chinese covid-19 vaccine maintains protection in variant-plagued brazil. Science. 2021 [Google Scholar]
- 297.Yadav P.D., Sapkal G.N., Ella R., Sahay R.R., Nyayanit D.A., Patil D.Y., Deshpande G., Shete A.M., Gupta N., Mohan V.K., Abraham P., Panda S., Bhargava B. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J. Travel Med. 2021;28(7) doi: 10.1093/jtm/taab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Emary K.R.W., Golubchik T., Aley P.K., Ariani C.V., Angus B., Bibi S., Blane B., Bonsall D., Cicconi P., Charlton S., Clutterbuck E.A., Collins A.M., Cox T., Darton T.C., Dold C., Douglas A.D., Duncan C.J.A., Ewer K.J., Flaxman A.L., Faust S.N., Ferreira D.M., Feng S., Finn A., Folegatti P.M., Fuskova M., Galiza E., Goodman A.L., Green C.M., Green C.A., Greenland M., Hallis B., Heath P.T., Hay J., Hill H.C., Jenkin D., Kerridge S., Lazarus R., Libri V., Lillie P.J., Ludden C., Marchevsky N.G., Minassian A.M., McGregor A.C., Mujadidi Y.F., Phillips D.J., Plested E., Pollock K.M., Robinson H., Smith A., Song R., Snape M.D., Sutherland R.K., Thomson E.C., Toshner M., Turner D.P.J., Vekemans J., Villafana T.L., Williams C.J., Hill A.V.S., Lambe T., Gilbert S.C., Voysey M., Ramasamy M.N., Pollard A.J. Efficacy of ChAdOx1 nCoV-19 (AZD1222) vaccine against SARS-CoV-2 variant of concern 202012/01 (B.1.1.7): an exploratory analysis of a randomised controlled trial. 2021;397:1351–1362. doi: 10.1016/S0140-6736(21)00628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Madhi S.A., Baillie V., Cutland C.L., Voysey M., Koen A.L., Fairlie L., Padayachee S.D., Dheda K., Barnabas S.L., Bhorat Q.E., Briner C., Kwatra G., Ahmed K., Aley P., Bhikha S., Bhiman J.N., Bhorat A.E., du Plessis J., Esmail A., Groenewald M., Horne E., Hwa S.H., Jose A., Lambe T., Laubscher M., Malahleha M., Masenya M., Masilela M., McKenzie S., Molapo K., Moultrie A., Oelofse S., Patel F., Pillay S., Rhead S., Rodel H., Rossouw L., Taoushanis C., Tegally H., Thombrayil A., van Eck S., Wibmer C.K., Durham N.M., Kelly E.J., Villafana T.L., Gilbert S., Pollard A.J., de Oliveira T., Moore P.L., Sigal A., Izu A. Efficacy of the ChAdOx1 nCoV-19 Covid-19 Vaccine against the B.1.351 Variant. New England J. Med. 2021;384(20):1885–1898. doi: 10.1056/NEJMoa2102214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Sadoff J., Gray G., Vandebosch A., Cárdenas V., Shukarev G., Grinsztejn B., Goepfert P.A., Truyers C., Fennema H., Spiessens B., Offergeld K., Scheper G., Taylor K.L., Robb M.L., Treanor J., Barouch D.H., Stoddard J., Ryser M.F., Marovich M.A., Neuzil K.M., Corey L., Cauwenberghs N., Tanner T., Hardt K., Ruiz-Guiñazú J., Le Gars M., Schuitemaker H., Van Hoof J., Struyf F., Douoguih M. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. New England J. Med. 2021;384(23):2187–2201. doi: 10.1056/NEJMoa2101544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Barouch D.H., Stephenson K.E., Sadoff J., Yu J., Chang A., Gebre M., McMahan K., Liu J., Chandrashekar A., Patel S., Le Gars M., de Groot A.M., Heerwegh D., Struyf F., Douoguih M., van Hoof J., Schuitemaker H. Durable Humoral and Cellular Immune Responses 8 Months after Ad26.COV2.S Vaccination. New England J. Med. 2021;385(10):951–953. doi: 10.1056/NEJMc2108829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.S. Ikegame, M. Siddiquey, C. Hung, G. Haas, L. Brambilla, K. Oguntuyo, S. Kowdle, A. Vilardo, A. Edelstein, C. Perandones, Neutralizing activity of Sputnik V vaccine sera against SARS-CoV-2 variants Version 3. medRxiv, preprint (21254660) (2021). [DOI] [PMC free article] [PubMed]
- 303.Shinde V., Bhikha S., Hoosain Z., Archary M., Bhorat Q., Fairlie L., Lalloo U., Masilela M.S.L., Moodley D., Hanley S., Fouche L., Louw C., Tameris M., Singh N., Goga A., Dheda K., Grobbelaar C., Kruger G., Carrim-Ganey N., Baillie V., de Oliveira T., Lombard Koen A., Lombaard J.J., Mngqibisa R., Bhorat A.E., Benadé G., Lalloo N., Pitsi A., Vollgraaff P.L., Luabeya A., Esmail A., Petrick F.G., Oommen-Jose A., Foulkes S., Ahmed K., Thombrayil A., Fries L., Cloney-Clark S., Zhu M., Bennett C., Albert G., Faust E., Plested J.S., Robertson A., Neal S., Cho I., Glenn G.M., Dubovsky F., Madhi S.A. Efficacy of NVX-CoV2373 Covid-19 Vaccine against the B.1.351 Variant. New England J. Med. 2021;384(20):1899–1909. doi: 10.1056/NEJMoa2103055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.P.T. Heath, E.P. Galiza, D.N. Baxter, M. Boffito, D. Browne, F. Burns, D.R. Chadwick, R. Clark, C. Cosgrove, J. Galloway, A.L. Goodman, A. Heer, A. Higham, S. Iyengar, A. Jamal, C. Jeanes, P.A. Kalra, C. Kyriakidou, D.F. McAuley, A. Meyrick, A.M. Minassian, J. Minton, P. Moore, I. Munsoor, H. Nicholls, O. Osanlou, J. Packham, C.H. Pretswell, A. San Francisco Ramos, D. Saralaya, R.P. Sheridan, R. Smith, R.L. Soiza, P.A. Swift, E.C. Thomson, J. Turner, M.E. Viljoen, G. Albert, I. Cho, F. Dubovsky, G. Glenn, J. Rivers, A. Robertson, K. Smith, S. Toback, Safety and Efficacy of NVX-CoV2373 Covid-19 Vaccine, New England J. Med. 385(13) (2021) 1172–1183. [DOI] [PMC free article] [PubMed]
- 305.H. Chemaitelly, H.M. Yassine, F.M. Benslimane, H.A. Al Khatib, P. Tang, M.R. Hasan, J.A. Malek, P. Coyle, H.H. Ayoub, Z. Al Kanaani, E. Al Kuwari, A. Jeremijenko, A.H. Kaleeckal, A.N. Latif, R.M. Shaik, H.F. Abdul Rahim, G.K. Nasrallah, M.G. Al Kuwari, H.E. Al Romaihi, M.H. Al-Thani, A. Al Khal, A.A. Butt, R. Bertollini, L.J. Abu-Raddad, mRNA-1273 COVID-19 vaccine effectiveness against the B.1.1.7 and B.1.351 variants and severe COVID-19 disease in Qatar, Nat. Med. 27(9) (2021) 1614–1621. [DOI] [PubMed]
- 306.Abu-Raddad L.J., Chemaitelly H., Butt A.A. Effectiveness of the BNT162b2 Covid-19 Vaccine against the B.1.1.7 and B.1.351 Variants. The New England J. Med. 2021;385(2):187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Tada T., Dcosta B.M., Samanovic M.I., Herati R.S., Cornelius A., Zhou H., Vaill A., Kazmierski W., Mulligan M.J., Landau N.R. Convalescent-Phase Sera and Vaccine-Elicited Antibodies Largely Maintain Neutralizing Titer against Global SARS-CoV-2 Variant Spikes. mBio. 2021;12(3) doi: 10.1128/mBio.00696-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Li X.N., Huang Y., Wang W., Jing Q.L., Zhang C.H., Qin P.Z., Guan W.J., Gan L., Li Y.L., Liu W.H., Dong H., Miao Y.T., Fan S.J., Zhang Z.B., Zhang D.M., Zhong N.S. Effectiveness of inactivated SARS-CoV-2 vaccines against the Delta variant infection in Guangzhou: a test-negative case-control real-world study. Emerg Microbes Infect. 2021;10(1):1751–1759. doi: 10.1080/22221751.2021.1969291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Chen J., Gao K., Wang R., Wei G.W. Revealing the Threat of Emerging SARS-CoV-2 Mutations to Antibody Therapies. J. Mol. Biol. 2021;433(18) doi: 10.1016/j.jmb.2021.167155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.J. Hu, X.-y. Wei, J. Xiang, P. Peng, F.-l. Xu, K. Wu, F.-y. Luo, A.-s. Jin, L. Fang, B.-z. Liu, Reduced neutralization of SARS-CoV-2 B. 1.617 variant by inactivated and RBD-subunit vaccine, bioRxiv (2021).
- 311.P.D. Yadav, G. Sapkal, R. Ella, R.R. Sahay, D.A. Nyayanit, D.Y. Patil, G. Deshpande, A.M. Shete, N. Gupta, V.K. Mohan, Neutralization against B. 1.351 and B. 1.617. 2 with sera of COVID-19 recovered cases and vaccinees of BBV152, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 312.Sapkal G.N., Yadav P.D., Sahay R.R., Deshpande G., Gupta N., Nyayanit D.A., Patil D.Y., Shete A.M., Kumar S., Abraham P., Panda S., Bhargava B. Neutralization of Delta variant with sera of Covishield™ vaccinees and COVID-19-recovered vaccinated individuals. J. Travel Med. 2021;28(7) doi: 10.1093/jtm/taab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Tada T., Zhou H., Dcosta B.M., Samanovic M.I., Mulligan M.J., Landau N.R. The Spike Proteins of SARS-CoV-2 B. 1.617 and B. 1.618 Variants Identified in India Provide Partial Resistance to Vaccine-elicited and Therapeutic Monoclonal Antibodies. BioRxiv. 2021 [Google Scholar]
- 314.Hoffmann M., Hofmann-Winkler H., Krüger N., Kempf A., Nehlmeier I., Graichen L., Arora P., Sidarovich A., Moldenhauer A.S., Winkler M.S., Schulz S., Jäck H.M., Stankov M.V., Behrens G.M.N., Pöhlmann S. SARS-CoV-2 variant B.1.617 is resistant to bamlanivimab and evades antibodies induced by infection and vaccination. Cell Rep. 2021;36(3) doi: 10.1016/j.celrep.2021.109415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.C. Davis, N. Logan, G. Tyson, R. Orton, W.T. Harvey, J.S. Perkins, G. Mollett, R.M. Blacow, C.-G.U. Consortium, T.P. Peacock, Reduced neutralisation of the Delta (B. 1.617. 2) SARS-CoV-2 variant of concern following vaccination, PLoS pathogens 17(12) (2021) e1010022. [DOI] [PMC free article] [PubMed]
- 316.P. Mlcochova, S. Kemp, M.S. Dhar, G. Papa, B. Meng, S. Mishra, C. Whittaker, T. Mellan, I. Ferreira, R. Datir, SARS-CoV-2 B. 1.617. 2 Delta variant emergence, replication and sensitivity to neutralising antibodies, BioRxiv (2021) 2021.05. 08.443253.
- 317.Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., Prot M., Gallais F., Gantner P., Velay A., Le Guen J., Kassis-Chikhani N., Edriss D., Belec L., Seve A., Courtellemont L., Péré H., Hocqueloux L., Fafi-Kremer S., Prazuck T., Mouquet H., Bruel T., Simon-Lorière E., Rey F.A., Schwartz O. Reduced sensitivity of SARS-CoV-2 variant Delta to antibody neutralization. Nature. 2021;596(7871):276–280. doi: 10.1038/s41586-021-03777-9. [DOI] [PubMed] [Google Scholar]
- 318.Liu C., Ginn H.M., Dejnirattisai W., Supasa P., Wang B., Tuekprakhon A., Nutalai R., Zhou D., Mentzer A.J., Zhao Y., Duyvesteyn H.M.E., López-Camacho C., Slon-Campos J., Walter T.S., Skelly D., Johnson S.A., Ritter T.G., Mason C., Costa Clemens S.A., Gomes Naveca F., Nascimento V., Nascimento F., Fernandes da Costa C., Resende P.C., Pauvolid-Correa A., Siqueira M.M., Dold C., Temperton N., Dong T., Pollard A.J., Knight J.C., Crook D., Lambe T., Clutterbuck E., Bibi S., Flaxman A., Bittaye M., Belij-Rammerstorfer S., Gilbert S.C., Malik T., Carroll M.W., Klenerman P., Barnes E., Dunachie S.J., Baillie V., Serafin N., Ditse Z., Da Silva K., Paterson N.G., Williams M.A., Hall D.R., Madhi S., Nunes M.C., Goulder P., Fry E.E., Mongkolsapaya J., Ren J., Stuart D.I., Screaton G.R. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell. 2021;184(16):4220–4236.e13. doi: 10.1016/j.cell.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Ai J., Zhang H., Zhang Y., Lin K., Zhang Y., Wu J., Wan Y., Huang Y., Song J., Fu Z., Wang H., Guo J., Jiang N., Fan M., Zhou Y., Zhao Y., Zhang Q., Liu Q., Lv J., Li P., Qiu C., Zhang W. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021:1–24. doi: 10.1080/22221751.2021.2022440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.J. Lopez Bernal, N. Andrews, C. Gower, E. Gallagher, R. Simmons, S. Thelwall, J. Stowe, E. Tessier, N. Groves, G. Dabrera, Effectiveness of Covid-19 vaccines against the B. 1.617. 2 (Delta) variant, The New England journal of medicine (2021) 585-594. [DOI] [PMC free article] [PubMed]
- 321.Bruxvoort K., Sy L.S., Qian L., Ackerson B.K., Luo Y., Lee G.S., Tian Y., Florea A., Aragones M., Tubert J.E. Effectiveness of mRNA-1273 against Delta, Mu, and other emerging variants. MedRxiv. 2021 doi: 10.1136/bmj-2021-068848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21(2):73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Chen J., Wang R., Gilby N.B., Wei G.W. Omicron (B.1.1.529): Infectivity, vaccine breakthrough, and antibody resistance. ArXiv. 2021 doi: 10.1021/acs.jcim.1c01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.X. Wang, X. Zhao, J. Song, J. Wu, Y. Zhu, M. Li, Y. Cui, Y. Chen, L. Yang, J. Liu, Homologous or Heterologous Booster of Inactivated Vaccine Reduces SARS-CoV-2 Omicron Variant Escape from Neutralizing Antibodies, bioRxiv (2021). [DOI] [PMC free article] [PubMed]
- 325.C.A. Perugino, H. Liu, J. Feldman, B.M. Hauser, C. Jacob-Dolan, A. Nathan, Z. Zhou, C. Kaseke, R. Tano-Menka, M.A. Getz, F. Senjobe, C. Berrios, O. Ofoman, J.E. Lemieux, M.B. Goldberg, K. Nundel, A. Moormann, A. Marshak-Rothstein, J.A. Iafrate, G. Gaiha, R. Charles, A.B. Balazs, V. Naranbhai, A.G. Schmidt, S. Pillai, Preferential expansion upon boosting of cross-reactive “pre-existing” switched memory B cells that recognize the SARS-CoV-2 Omicron variant Spike protein, medRxiv (2022) 2021.12.30.21268554.
- 326.L. Lu, B.W. Mok, L.L. Chen, J.M. Chan, O.T. Tsang, B.H. Lam, V.W. Chuang, A.W. Chu, W.M. Chan, J.D. Ip, B.P. Chan, R. Zhang, C.C. Yip, V.C. Cheng, K.H. Chan, D.Y. Jin, I.F. Hung, K.Y. Yuen, H. Chen, K.K. To, Neutralization of SARS-CoV-2 Omicron variant by sera from BNT162b2 or Coronavac vaccine recipients, Clinical infectious diseases : an official publication of the Infectious Diseases Society of America (2021). [DOI] [PMC free article] [PubMed]
- 327.Garcia-Beltran W.F., Denis K.J.S., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2021 doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Peiris M., Cheng S., Mok C.K.P., Leung Y., Ng S., Chan K., Ko F., Yiu K., Lam B., Lau E., Chan K., Luk L., Li J., Tsang L., Poon L., Chen C., Hui D. Neutralizing antibody titres to SARS-CoV-2 Omicron variant and wild-type virus in those with past infection or vaccinated or boosted with mRNA BNT162b2 or inactivated CoronaVac vaccines. Res Sq. 2022 [Google Scholar]
- 329.Moghnieh R., Mekdashi R., El-Hassan S., Abdallah D., Jisr T., Bader M., Jizi I., Sayegh M.H., Rahman Bizri A. Immunogenicity and reactogenicity of BNT162b2 booster in BBIBP-CorV-vaccinated individuals compared with homologous BNT162b2 vaccination: Results of a pilot prospective cohort study from Lebanon. Vaccine. 2021;39(46):6713–6719. doi: 10.1016/j.vaccine.2021.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.C. Zeng, J.P. Evans, P. Qu, J. Faraone, Y.M. Zheng, C. Carlin, J.S. Bednash, T. Zhou, G. Lozanski, R. Mallampalli, L.J. Saif, E.M. Oltz, P. Mohler, K. Xu, R.J. Gumina, S.L. Liu, Neutralization and Stability of SARS-CoV-2 Omicron Variant, bioRxiv (2021).
- 331.Cohen-Stavi C.J., Magen O., Barda N., Yaron S., Peretz A., Netzer D., Giaquinto C., Judd A., Leibovici L., Hernán M.A., Lipsitch M., Reis B.Y., Balicer R.D., Dagan N. BNT162b2 Vaccine Effectiveness against Omicron in Children 5 to 11 Years of Age. New England J. Med. 2022 doi: 10.1056/NEJMoa2205011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.
Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.