Abstract
The divergently transcribed sacBK and sacAR operons, which are involved in the utilization of sucrose by Lactococcus lactis NZ9800, were examined by transcriptional and gene inactivation studies. Northern analyses of RNA isolated from cells grown at the expense of different carbon sources revealed three sucrose-inducible transcripts: one of 3.2 kb containing sacB and sacK, a second of 3.4 kb containing sacA and sacR, and a third of 1.8 kb containing only sacR. The inactivation of the sacR gene by replacement recombination resulted in the constitutive transcription of the sacBK and sacAR operons in the presence of different carbon sources, indicating that SacR acts as a repressor of transcription.
Sucrose can be utilized as a sole carbon source by many bacteria, and the vast majority of bacteria take up this disaccharide via the sucrose-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) (1, 3, 8, 12, 15, 16). The PTS catalyzes the transport of sucrose across the cytoplasmic membrane concomitant with its phosphorylation by a sucrose-specific enzyme, enzyme II. The product of this translocation, sucrose-6-phosphate, is then hydrolyzed to glucose-6-phosphate and fructose by a sucrose-6-phosphate hydrolase (EC 3.2.1.26). The glucose-6-phosphate can readily be used, while the fructose has to be phosphorylated by an ATP-dependent fructokinase (EC 2.7.1.4) before it can be metabolized via the glycolytic pathway. Previously, a sucrose-6-phosphate hydrolase and a fructokinase had been purified from Lactococcus lactis and characterized in detail (13, 14). To investigate the utilization of sucrose by L. lactis, we have cloned and analyzed the sacB, sacK, and the sacR genes, which encode sucrose-specific enzyme II, a fructokinase, and a regulatory protein, respectively. In order to analyze the role of the SacR protein in the transcriptional control of the sucrose genes, the sacR gene was disrupted, and the effects of this disruption on the transcription of the sucrose genes were studied.
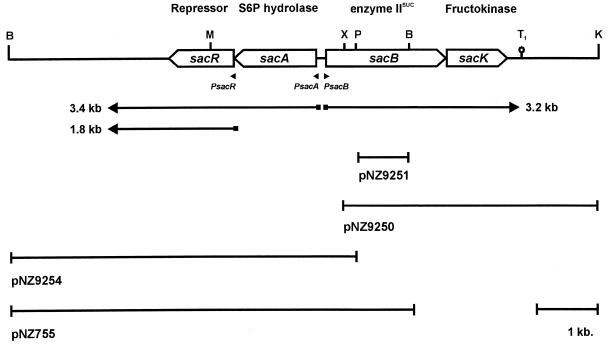
Sequence analysis of plasmids pNZ755 and pNZ9250 containing DNA fragments from the nisin-sucrose conjugative transposon Tn5276 (9), isolated from L. lactis NZ9800 (6), revealed the presence of three new genes, i.e., sacR, sacB, and sacK (Fig. 1) (see below), in addition to the previously described sacA gene encoding a sucrose-6-phosphate hydrolase (10). The sacA stop codon partly overlaps the putative SacR start codon (GTG). If functional, the sacR gene encodes a 318-residue protein showing significant sequence similarity to proteins of the LacI-GalR family of bacterial regulator proteins (17). A transcription initiation start site was mapped upstream of the sacR gene, which is preceded by a sequence corresponding to those of consensus L. lactis promoters (data not shown [2a]). Strikingly, the sacR promoter contains an inverted repeat similar to those identified in the sacB and sacA promoters (10), which could represent the binding site of a factor involved in sucrose-specific regulation. The sacB gene, which is in the opposite orientation to the sacA gene, encodes a protein that contains the enzyme II ABC domains expected for an enzyme II protein of the PTS (8). The disruption of the sacB gene by a single-crossover recombination (7) using plasmid pNZ9251 containing an internal PstI-BamHI fragment (Fig. 1) resulted in a strain that was no longer able to grow at the expense of sucrose, indicating that the sacB gene is essential for the utilization of sucrose. The sacB gene is immediately followed by another gene, sacK, which encodes a 290-residue protein. The NH2-terminal amino acids 2 to 26 of the deduced SacK sequence are identical to those determined for the purified fructokinase I from L. lactis KI (14). Moreover, the total amino acid composition and the calculated molecular mass of the deduced protein (31,626 Da) are highly similar to those of the purified lactococcal fructokinase (14).
FIG. 1.
Genetic and transcriptional organization of the Tn5276-located sucrose gene cluster of L. lactis NZ9800. The genes (open arrows) are shown with their products and mapped promoters (arrowheads) as well as the mapped transcripts (arrows). Cloned chromosomal DNA fragments relevant to this study are illustrated below the arrows, as are the names of the derived plasmids on which they are carried. The putative terminator downstream of the sacK gene is indicated (T1). Relevant restriction sites are shown as follows: B, BamHI; K, KpnI; M, MunI; P, PstI; and X, XbaI.
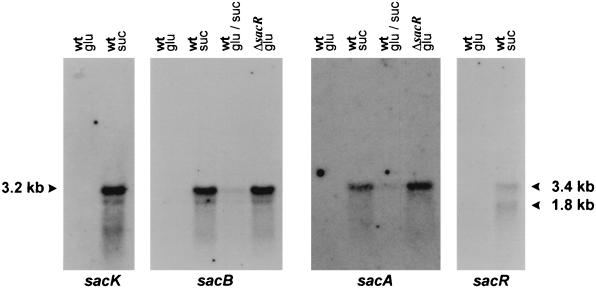
The expression of sucrose catabolic genes is in most cases regulated at the transcriptional level (2, 4, 5). Northern analyses were performed in order to investigate the transcription of the sacBK and sacAR genes. RNA was isolated from cells of strain NZ9800 grown at the expense of glucose or sucrose as the sole carbon source as described previously (6). RNA was denatured and size fractionated on a 1% agarose gel containing formaldehyde according to standard procedures (11). The blot was probed with internal DNA fragments of all individual sac genes (Fig. 2). After hybridization with either a sacB- or sacK-specific probe, a transcript of approximately 3.2 kb was observed with RNA isolated from sucrose-grown cells, but not with RNA isolated from glucose-grown cells, indicating that the sacBK genes are located on a single sucrose-inducible transcript. The size of this transcript, in conjunction with its mapped transcription initiation site (10), suggests that the transcription terminates at a putative rho-independent terminator structure that was identified immediately downstream of the sacK gene (Fig. 1). Another sucrose-inducible transcript of approximately 3.4 kb was observed when RNA from sucrose-grown cells was hybridized with either a sacA or sacR probe. This transcript was absent in RNA isolated from glucose-grown cells. When L. lactis NZ9800 was grown on a mixture of sucrose and glucose, a severe reduction of the sacBK and sacAR transcription compared to that in cells grown on sucrose could be observed, indicating a form of glucose repression (Fig. 2). A third sucrose-inducible transcript of 1.8 kb was shown to hybridize with a sacR-specific probe, confirming the existence of a second regulated promoter driving transcription of the sacR gene. This transcript is likely to end at the same transcriptional terminator as the 3.4-kb transcript that initiates from the sacA promoter.
FIG. 2.
Northern analysis of sacK, sacB, sacA, and sacR gene expression in L. lactis NZ9800 and NZ9860. Cells were grown to mid-logarithmic phase on either glucose (glu), sucrose (suc), or a mixture of glucose and sucrose (glu/suc), and RNA was isolated, separated on agarose gels, and blotted. Blots were hybridized with DNA probes specific to each of the indicated sac genes. The sizes of the transcripts were estimated by comparing their migration distances to those of RNA markers run in parallel.
To investigate the role of SacR in transcriptional control of the sucrose genes, the sacR gene was disrupted. A double-crossover recombination (7) using plasmid pNZ9254, in which the MunI site in the sacR open reading frame was filled in by using Klenow polymerase, resulted in the introduction of a frameshift mutation (Fig. 1). No differences in growth rate could be observed between strain NZ9860 (ΔsacR) and the wild-type strain, NZ9800, when the cells were grown at the expense of either glucose or sucrose. The results of Northern analysis of RNA isolated from strain NZ9860 (ΔsacR) grown at the expense of glucose showed that sacBK and sacAR transcription had become constitutive at a level comparable to that of the wild-type strain grown at the expense of sucrose (Fig. 2). These results demonstrate that SacR acts as a repressor of both sacBK transcription and sacAR transcription. The levels of sacBK and sacAR transcription in strain NZ9860 (ΔsacR) grown on glucose or sucrose were found to be similar, indicating that SacR not only is involved in substrate induction but also mediates glucose repression. The observed substrate induction and negative autoregulation of sacR by its gene product result in efficient transcriptional control of the sac genes in response to variations in extracellular sucrose concentrations.
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper will appear in the EMBL, GenBank and DDBJ nucleotide sequence databases under accession no. Z97015.
Acknowledgments
This work was partly supported by the BIOTECH Programme of the European Community (contract BIO2-CT92-0137).
We are grateful to Ger Rutten and Marke Beerthuyzen for technical assistance, Jack Thompson for helpful suggestions, and Michiel Kleerebezem and Roland Siezen for critically reading the manuscript.
REFERENCES
- 1.Chen Y-Y M, Lee L N, LeBlanc D J. Sequence analysis of scrA and scrB from Streptococcus sobrinus 6715. Infect Immun. 1993;61:2602–2610. doi: 10.1128/iai.61.6.2602-2610.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debarbouille M, Arnaud M, Fouet A, Klier A, Rapoport G. The sacT gene regulating the sacPA operon in Bacillus subtilis shares strong homology with transcriptional antiterminators. J Bacteriol. 1990;172:3966–3973. doi: 10.1128/jb.172.7.3966-3973.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.de Vos W M, Simons G. Gene cloning and expression systems in lactococci. In: Gasson M J, de Vos W M, editors. Genetics and biotechnology of lactic acid bacteria. London, United Kingdom: Chapman and Hall; 1994. pp. 52–105. [Google Scholar]
- 3.Fouet A, Arnaud M, Klier A, Rapoport G. Bacillus subtilis sucrose-specific enzyme II of the phosphotransferase system: expression in Escherichia coli and homology to enzymes II from enteric bacteria. Proc Natl Acad Sci USA. 1987;84:8773–8777. doi: 10.1073/pnas.84.24.8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gering M, Brückner R. Transcriptional regulation of the sucrose gene of Staphylococcus xylosus by the repressor ScrR. J Bacteriol. 1996;178:462–469. doi: 10.1128/jb.178.2.462-469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiratsuka K, Wang B, Sato Y, Kuramitsu H. Regulation of sucrose-6-phosphate hydrolase activity in Streptococcus mutans: characterization of the scrR gene. Infect Immun. 1998;66:3736–3743. doi: 10.1128/iai.66.8.3736-3743.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuipers O P, Beerthuyzen M M, Siezen R J, de Vos W M. Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for the development of immunity. Eur J Biochem. 1993;216:281–291. doi: 10.1111/j.1432-1033.1993.tb18143.x. [DOI] [PubMed] [Google Scholar]
- 7.Leenhouts K J, Kok J, Venema G. Stability of integrated plasmids in the chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990;56:2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Postma P W, Lengeler J W, Jacobson G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rauch P J G, de Vos W M. Characterization of the novel nisin-sucrose conjugative transposon Tn5276 and its insertion in Lactococcus lactis. J Bacteriol. 1992;174:1280–1287. doi: 10.1128/jb.174.4.1280-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rauch P J G, de Vos W M. Transcriptional regulation of the Tn5276-located Lactococcus lactis sucrose operon and characterization of the sacA gene encoding sucrose-6-phosphate hydrolase. Gene. 1992;121:55–61. doi: 10.1016/0378-1119(92)90161-h. [DOI] [PubMed] [Google Scholar]
- 11.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 12.Sato Y, Poy F, Jacobson G R, Kuramitsu H K. Characterization and sequence analysis of the scrA gene encoding enzyme IIScr of the Streptococcus mutans phosphoenolpyruvate-dependent sucrose phosphotransferase system. J Bacteriol. 1989;171:263–271. doi: 10.1128/jb.171.1.263-271.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thompson J, Nguyen Y J, Sackett D L, Donkersloot J A. Transposon-encoded sucrose metabolism in Lactococcus lactis: purification of sucrose-6-phosphate hydrolase and genetic linkage to N5-(L-1-carboxyethyl)-L-ornithine synthase in strain K1. J Biol Chem. 1991;266:14573–14579. [PubMed] [Google Scholar]
- 14.Thompson J, Sackett D L, Donkersloot J A. Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the sucrose-nisin transposon Tn5306. J Biol Chem. 1991;266:22626–22633. [PubMed] [Google Scholar]
- 15.Titgemeyer F, Jahreis K, Ebner R, Lengeler J W. Molecular analysis of the scrA and scrB genes from Klebsiella pneumoniae and plasmid pUR400, which encode the sucrose transport protein Enzyme IIScr of the phosphotransferase system and a sucrose-6-phosphate hydrolase. Mol Gen Genet. 1996;250:197–206. doi: 10.1007/BF02174179. [DOI] [PubMed] [Google Scholar]
- 16.Wagner E, Götz F, Brückner R. Cloning and characterization of the scrA gene encoding the sucrose-specific Enzyme II of the phosphotransferase system from Staphylococcus xylosus. Mol Gen Genet. 1993;241:33–41. doi: 10.1007/BF00280198. [DOI] [PubMed] [Google Scholar]
- 17.Weickert M J, Adhya S. A family of bacterial regulator proteins homologous to Gal and Lac repressors. J Biol Chem. 1992;22:15869–15874. [PubMed] [Google Scholar]