Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Spike protein, Immune evasion, Pseudovirus, In-silico
Abstract
Emerging SARS-CoV-2 variants with higher transmissibility and immune escape remain a persistent threat across the globe. This is evident from the recent outbreaks of the Delta (B.1.617.2) and Omicron variants. These variants have originated from different continents and spread across the globe. In this study, we explored the genomic and structural basis of these variants for their lineage defining mutations of the spike protein through computational analysis, protein modeling, and molecular dynamic (MD) simulations. We further experimentally validated the importance of these deletion mutants for their immune escape using a pseudovirus-based neutralization assay, and an antibody (4A8) that binds directly to the spike protein's NTD. Delta variant with the deletion and mutations in the NTD revealed a better rigidity and reduced flexibility as compared to the wild-type spike protein (Wuhan isolate). Furthermore, computational studies of 4A8 monoclonal antibody (mAb) revealed a reduced binding of Delta variant compared to the wild-type strain. Similarly, the MD simulation data and virus neutralization assays revealed that the Omicron also exhibits immune escape, as antigenic beta-sheets appear to be disrupted. The results of the present study demonstrate the higher possibility of immune escape and thereby achieved better fitness advantages by the Delta and Omicron variants, which warrants further demonstrations through experimental evidences. Our study, based on in-silico computational modelling, simulations, and pseudovirus-based neutralization assay, highlighted and identified the probable mechanism through which the Delta and Omicron variants are more pathogenically evolved with higher transmissibility as compared to the wild-type strain.
1. Introduction
Omicron wave currently superseded Delta wave with the global cases of the COVID-19, specifically dominated by omicron BA.2 sub-variant. The peak of the COVID-19s wave confirmed more than 53 million positive cases as per World Health Organization (WHO): reports accessed on to be updated 28th May 2022. Delta variant dominated during the second wave while the omicron variant surged during the third wave. Genome sequencing indicated the dominance of the Omicron BA.2 sub-variant during this phase of the pandemic across the different states in India. Furthermore, the emergence of the recombinant strains of SARS-CoV-2 remains a significant threat to the health preparedness due to lesser antibody neutralization and higher immune escape potential [30], [17]. Genomic surveillance is a powerful tool to study the viral genomic profile, variants of concerns (VoCs), and epidemiological significance in disease outbreaks. The spike (S) protein mediates the attachment of coronavirus to the host cell surface receptors, resulting in fusion and viral entry to the cells. The membrane (M) protein defines the shape of the viral envelope, while the envelope (E) protein and nucleocapsid (N) protein participate in viral assembly and budding of the virion complex in the infected cells [18], [46]. SARS-CoV-2 enters host cells through angiotensin-converting enzyme 2 (ACE2) receptor, and the spike protein of SARS-CoV-2 is primed by TMPRSS2 protease, while the role of several other host receptors, which is only partially understood due to the current lack of data, may determine the altered virulence and pathogenicity of the various evolving lineages. SARS-CoV-2 exhibits highly efficient proteolytic spike activation mechanism, as well as host proteases that have been found to proteolytically degrade the spike protein during infection and intracellular virus growth. These include, but are not limited to, endosomal cathepsins, cell surface trans-membrane protease/serine (TMPRSS) proteases, furin, and trypsin, which are critical determinants of the virus entry and pathogenesis in humans [43], [23]. SARS-CoV-2, in comparison to SARS-CoV, contains a polybasic sequence motif, Arg-Arg-Ala-Arg (RRAR), at the S1/S2 boundary, furin-type cleavage site in its spike protein, which when cleaved can bind and activate neuropilin (NRP) receptors. Furthermore, research studies indicate that NRP1 (Neuropilin 1) enhances SARS-CoV-2 infectivity, which is highly expressed in respiratory and olfactory epithelium [8].
Under the prevailing circumstances, the immune response of patients plays a significant role in determining survival outcome and severity of the disease upon SARS-CoV-2 infection. A myriad of various cell types, such as macrophages, alveolar epithelial cells, lymphoid cells, and dendritic cells (DCs), have a major role in the first line of defense. Once our immune system is triggered by the entry of foreign viral pathogens inside the body, which upon breaching the first lines of defense systems, several specific molecular and inter-cellular signaling cascades ensure the establishment of the body’s other immune responses [45], [41]. In order to provide a window of opportunity for efficient virus replication, respiratory viruses acquire methods to either evade or suppress the host's innate immune responses, resulting in illness. The affected innate immune response also impacts subsequent adaptive immune responses, and therefore viral evasion from innate immune system often undermines fully protective immunity, such as a lack of virus neutralizing antibodies [59], [11], [12], [16]. Furthermore, genetic changes and evolution in the virus also enable them to adapt to immune escape and immune evasion in the hosts, thereby increasing the chances of severity and virulence of the pathogenic variants [24]. Based on risk assessment criteria of infection severity, susceptibility, geographical prevalence, and transmission in humans, Public Health England (PHE), UK; CDC, USA; and World Health Organization (WHO) classified these fast-spreading variants with increased transmissibility as VoCs. Therefore, sentinel genomic surveillance studies are essential in monitoring new variants that may arise in future disease outbreaks. Further research is needed to establish mechanisms of immune escape and potential host genetic factors that might help in containing newly evolved pathogenic viral strains of SARS-CoV-2. Omicron mutations significantly lessen the ability of a broad panel of powerful monoclonal antibodies and antibodies that are under commercial development for neutralizing them [10]. Initial findings suggests that Omicron BA.2 variant is having considerable transmission advantages including the reinfections and resistance to the monoclonal antibody (mAbs) neutralization, yet its severity and virulence needs to be further studied at the molecular level [1]. It is clear that the transmission of BA.2 could pose a significant threat to global health in the near future due to its greater effective reproduction number and pronounced immune resistance [60], [62].
Recent studies on spike protein interactions with monoclonal antibody 4A8 suggests that epitope located within the N- terminal domain of spike is specific site for its binding [7], [35]. In SARS-CoV-2 mutations, particularly Delta and Omicron, the primary neutralizing epitope of the N-terminal domain (NTD) of the spike protein displayed substantial structural diversity [21]. This epitope is found on the NTD's flat surface, a significant electropositive region that interacts to host cells' electronegatively charged lipid rafts [14]. Prominent spike protein mutations especially D614G (B.1 variant) are established for their role in higher transmissibility due to the structural stability, facilitating the ACE2 interaction. Furthermore, some prominent mutations such as D614G (>99.7 percent frequency) favors the virus [64], while others (C241T) favor the host, such as [6]. A wide area of research has been focused using computational tools for studying spike protein mutations [9], [64], [25], [37]. One of the major reasons for such a maneuver by the Delta variant could be the additional deletion and mutations (E156G and Arg158, Phe-157/del) acquired at the NTD of the spike protein that may be helping in immune evasion from reduced binding with the circulating neutralizing antibodies. Different vaccines are currently available for SARS-CoV-2 (https://www.uptodate.com/contents/covid-19-vaccines#disclaimerContent), and their efficiency with respect to emerging variants need to be evaluated for checking their efficacy level. Further, post infection treatment of the SARS-CoV-2 remains to be studied to evaluate the long COVID-19 treatment.
Waning immunity catapulted the debate and administration of the booster dose of the vaccines. However, scientific fraternity reserved the comments to administer the need of booster to government leadership in different geopolitical regions. Currently, the susceptible and high-risk groups were advised for the booster doses [20]. Unusually very high number of mutations in the Omicron sub-variants provide further opportunity to SARS-CoV-2 for breakthrough infections. Therefore, therapies using monoclonal antibodies (mAbs) could help in mitigating the risk and better survival outcome of the patients against SARS-CoV-2 including the emerging variants in future [51], [54], [61].
In this study, to understand the dynamics of the novel deletions and mutations in the spike protein of the Delta and Omicron variants, we focused on the NTD mutations and its impact on structural changes in protein sequence and neutralizing antibody binding using molecular modelling and dynamics approach. We further evaluated the variants response to antiserum collected from patients who received spike protein mRNA vaccines, convalescent antiserum collected from monkeys infected with Wuhan strain of SARS-CoV-2, and 4A8 monoclonal antibody targeting NTD, using a pseudovirus based neutralization assay. The results showed a reduced sensitivity by the pseudovirus displaying spike protein from Delta and omicron variants to these antibodies when compared to the virus displaying other variants, including the Wuhan strain. The molecular dynamic studies of Delta and Omicron variants also reveal that the deletion and mutations in the NTD provide a better rigidity and reduced flexibility as compared to the wild-type spike protein for achieving their current immune escapes.
2. Materials and methods
2.1. Protein complexes used for this study
SARS-CoV-2 spike protein variants were used in this study. Mutated spike from SARS-CoV-2 were derived from amino acid sequences submitted in GAISAD with accession number EPI_ISL_2001211 (GBRC-NCD-370) and X?????? for Delta and Omicron respectively. The homology modelling of the mutated SARS-CoV-2 spike protein was performed using the template crystal structure. Also, PDB ids viz. 7C2L, 7DZX and 7DZY, were taken as references for studying antibody binding to the spike protein [7], [36].
2.2. Protein modelling
The PRIME module of the Schrodinger software was used to build the homology model of mutated spike proteins using the template crystal structures. Retrieved mutated SARS-CoV-2 spike protein was submitted for the template search. The BLASTP tool was used to search the PDB library for potential templates for homology modelling, resulting the potential template with PDB I: 7KRQ. The CLUSTALW module of the Prime software was used to align the target-template sequences. The built protein model and PDB retrieved crystal structure refinements were carried out using protein preparation wizard [47]. Missing side chains were added through PRIME and pKa refinements using the epik [50].
Homology modelling panel implemented in Schrodinger suite release 2021-1 was used to build a mutated spike protein with reference protein 7KRQ. The sequence was imported and a homology blast search was performed. Crystal structure of 7KRQ was imported into maestro and protein complex refinement was performed using protein preparation wizard [47]. Missing side chains were added through PRIME and pKa refinements using epik [50].
2.3. Molecular dynamics simulations
Molecular dynamics simulation for spike protein and spike-antibody complexes were performed in Desmond for100 nanoseconds (ns) [48]. A total of four systems were subjected for 100 ns MD simulations, which include two mutated spike proteins to reveal the effect of mutations on protein stability and the other two simulations on spike-antibody complexes. All were performed in two replicates. Protein structures were refined using OPLS4 force field and altered hydrogen bonds were refilled using structure refinement panels, implemented in Schrodinger [58], [53]. Particle mesh Ewald method is applied to calculate long-range electrostatic interactions [55]. The trajectories were recorded at every 1.2 ps intervals for the analysis. TIP3P water molecules were added and 1.5 M salt concentration was added to neutralize the system [65]. The Martyna–Tuckerman–Klein chain coupling scheme with a coupling constant of 2.0 ps was used for the pressure control and the Nosé–Hoover chain coupling scheme for temperature control [39]. MD simulations were performed at 310.3 K temperature with NPT (N = Number of atoms, P = Pressure, T = Temperature) ensemble by applying OPLS4 force field.
The behaviour and intermolecular interactions between proteins were analyzed using the Simulation Interaction Diagram tool implemented in Desmond MD package. The stability of complex was monitored by examining the RMSD (Root Mean Square Deviation) of the protein and the complex system by structural alignment with initial pose (crystal structure). PYMOL was used for obtaining high resolution images [49]. The PyMOL Molecular Graphics System).
2.4. Molecular docking of spike protein with monoclonal antibodies using spike and affinity prediction
The crystal structures of 4A8 antibody [7], 8D2 [36], and 2490 [36] reported to bind NTD of spike were used to study binding with wild type and mutants (E156G, and F157, R158/del) of SARS-CoV-2 spike protein. Variants of spike proteins wildtype (7KRQ), Delta, and Omicron were docked with monoclonal antibodies such as 4A8, 8D2, and 2490, using PIPER [28] docking programme of the Schrodinger suite while MD simulations were performed with respect to only 4A8 monoclonal antibody. For binding residues detection among both receptor (spike) and ligand (antibody-4A8) attraction forces were applied with <3 Å cut-off. We checked 70,000 docking poses for fulfilling the criteria of distance restrains applied for the binding sites residues. Recently deposited crystal structure of spike protein binding with monoclonal antibody was taken for applying the restraint file showing list of spike residues binding with residues of 4A8. Top 30 poses were generated and a pose with highest free glide energy was used for the MD analysis. Docking was performed in HADDOCK to obtain electrostatic and restrain violation energy for the protein-antibody complex [57].
2.5. Alanine scanning
Alanine scanning was performed using Biologics Design module of the Schrodinger Suite (v.2021-1) for the binding affinity prediction in PDB deposited spike antibody complex using PDB ID: 7C2L [7]. Binding site residues were mutated to alanine in order to bind the pivotal residues involved into direct binding with antibody. Positive value of Δ affinity indicated that while mutating binding sites residues to alanine, binding is hindered due to small side chains of alanine, which implies that those mutated hot-spot residues were essential for direct affinity with antibodies [22], [52].
2.6. Binding energy calculation
Binding free energy for protein–protein complex was calculated using Prime Molecular Mechanics-Generalized Born Surface Area (MMGBSA) PRIME module of Schrodinger [38], [3], [19]. The changes in the free energy (Δ G) during 100 ns of MD simulation of protein–protein complex was calculated using following equation.
ΔGBind = ΔGSA + ΔGSolv + ΔEMM.
VSEB solvation model and OPLS4 force-field were used for calculation of MMGBSA. From the trajectory analysis we have retrieved stable conformation of protein–protein complex. First energy minimized structure out of 30 poses was used to find dominant interacting residues among spike. Interaction image was taken in new version of Schrodinger 2021–2 and the intermolecular interaction between protein–protein were taken from Biologics.
2.7. Dynamics Cross-Correlation matrix (DCCM) and principal component analysis (PCA)
Correlative and anti-correlative motions play a vital role in recognition and binding in a biological-complex system, which can be prevailed by MD simulation trajectory by determining the covariance matrix about atomic fluctuations [27]. The extent of correlative motion of two residues (or two atoms or proteins) can be symbolized by the cross-correlation coefficient, Cij. It is defined by following equation:
| (1) |
From above equation, i (j) explains ith (jth) two residues (or two atoms or proteins), Δ ri (Δ rj) is the displacement vector corresponding to ith (jth) two residues (or two atoms or proteins), and stand for the ensemble average. The value of Cij is from 1 to −1. +Cij implies positively correlated movement (the same direction) indicated into blue color, and -Cij implies anti-correlated movement (opposite direction) indicated into red colour. The higher the absolute value of Cij is, the more correlated (or anti-correlated) the two residues (or two atoms or proteins).
PCA is an implicit tool to unsheathe the essential information from MD trajectories by pulling out global slow motions from local fast motions [5]. To perform PCA, the covariance matrix C was calculated initially. The elements Cij in the matrix C are defined as:
| (2) |
From equation (2), ri and rj are the instant coordinates of the ith or jth atom, and and mean the average coordinate of the ith or jth atom over the ensemble. The principal components (PCs) were calculated by diagonalization and obtaining the eigenvectors and eigenvalues for the covariance matric C. The principal components (PCs) are the projections of a trajectory on the principal modes, of which usually the first few ones are largely responsible for the most important motions. DCCM and PCA both were analyzed using Schrodinger 2021-1 implemented python script run trj_essential_dynamics.py script of Desmond [48].
2.8. Production of pseudoviruses displaying the spike protein of SARS-CoV-2 variants
We used four helper vectors along with the genomic vector coding for firefly luciferase-IRES-ZsGreen reporter genes for the production of lenti-pseudoviruses displaying the spike protein of SARS-CoV-2 variants. We used the helper vectors displaying Wuhan, D614G, and Omicron spike proteins from the BEI resource centre of NIAID, USA. The additional variants were cloned utilising spike coding nucleotide sequences PCR amplified from System Biosciences' P1 and SA vectors, as well as B.1.617.1, B.1.617.2, and B.1.617.3 sequences provided/gifted from Prof. Wendy Barclay's group at Imperial College, UK. We used our established protocol for the production and concentration of lenti-pseudoviruses [ACS Nano. 2021 Oct 27:acsnano.1c05002. https://doi.org/10.1021/acsnano.1c05002. Online ahead of print.PMID: 34705425]. In brief, we co-transfected pRC-CMV-Rev1b, HDM-Hgpm2, HDM-tat1b, pG-Fluc-ZsGreen and SARS-CoV-2 spike entry vector in the concentration of 3.2 µg, 3.2 µg, 3.2 µg, 10 µg, and 2.5 µg, respectively to a 10 cm plate that was previously seeded with HEK293-FT cells to a confluency of 70 % plated 24 h before transfection (3 x106 cells/Plate). The plasmid vectors were co-transfected using a calcium phosphate method [32], [31], where we diluted the DNA into 425 µl using sterile double distilled water. To this DNA, we added 75 µl of 2 M CaCl2 and mixed well. We added 500 µl of 2X-HBSS dropwise while mixing the DNA using an aerator. The mix was incubated for 30 min at room temperature before added dropwise to the culture plate. The cells were incubated for 24 h and changed the medium after a brief PBS wash using the medium supplemented with 1 mM HEPES. The cells were further incubated for 48 h and collected the viruses from the medium after removing cell debris by centrifugation (800×g for 5 min). The supernatant was filtered using 0.45 µM filter, and the filtrate was used for various neutralization experiments performed in this study.
2.9. Pseudovirus based neutralization assay
For neutralization experiments, we used HEK293T cells engineered to express ACE2 receptor and TMPRSS2 protease, procured from BEI resource centre of NIAID. We plated 2.5 × 104 cells/well in a 96 well black wall clear bottom plate4 on day 1. On day 2, we neutralized the virus by antibodies/antiserum of different dilutions by mixing 50 µl of virus sample with 50 µl of antibody in serum free optiMEM [44]. The samples were incubated at room temperature for 1 h. The medium in the cells were aspirated and added with 100 µl of complete medium with 20 % FBS after mixing with the 100 µl of neutralization mix. The cells were further incubated for 60 h and used for imaging. For imaging, we aspirated the medium and added 50 µl of d-Luciferin (150 µg/ml) in PBS to each well using a multichannel pipette. The plates were incubated at 37 °C for 5 min and used for acquiring bioluminescence signal using IVIS-Lumina Optical imaging system (Perkin Elmer). The bioluminescence signals were quantified using Living Image software, and plotted against respective controls using the signals measured as normalized photons/sec/cm2/sr. We performed all these experiments under a BSL2+ condition.
3. Results and discussion
SARS-CoV-2 spike protein is known for its binding to cellular ACE2 receptor and mediating virus entry into mammalian cells. The spike protein is highly prone for mutations. To strengthen cellular penetrance, SARS-CoV-2 variants with spike protein mutations such as D614G increases spike density and infectivity, E484K reduces antibody mediated neutralisation, and N501Y and K417N alters the spike interaction with human ACE2 receptor and escape from human derived antibodies [15], [63], [25]. Our study focuses on the major deletions occurred to the NTD of spike protein of SAS-CoV-2 variants: Delta with mutation at nucleotide position 22029–22035 (6 bp), which resulted in 2 amino-acid deletions (Arg158, Phe157/del) and one amino acid substitution (E156G) and Omicron with two major mutations at A67V and G142D, both of which were found to be located at antigenic sites.
3.1. Mutational landscape of NTD in spike protein and its effects with respect to Delta and Omicron variants
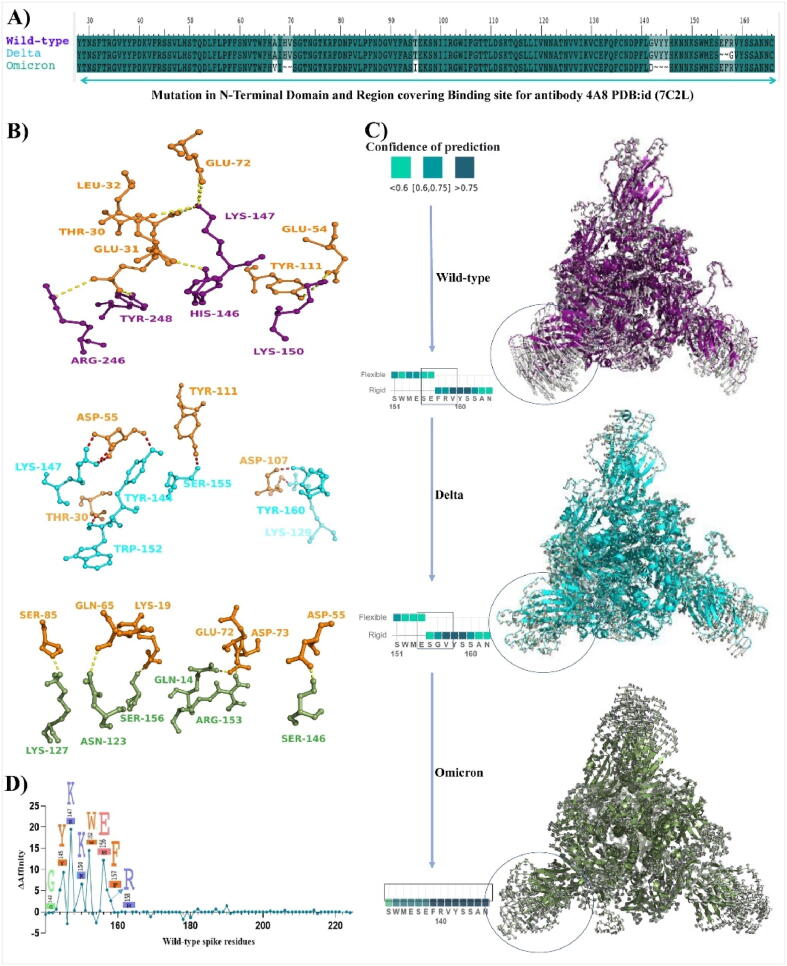
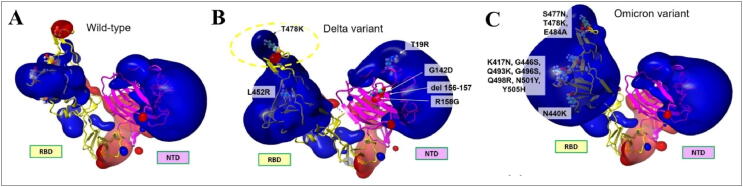
The impact of the spike mutations on SARS-CoV-2 variants is shown in Fig. 1. The major structural changes by the mutations were investigated using structural alignment of variants of spike proteins using PYMOL (Supplementary Fig. S1). Fig. 2 (A) show the superimposed structures of wild-type and Delta spike variants, with an alignment RMSD of 6.905 Å from the side and top views, respectively. An RMSD value larger than one shows that these mutations cause significant structural alterations in both spike variants [29]. Among the list of overall mutations, unique mutations E156G and Arg158, Phe157/del were falling within the NTD of spike protein. The wild-type super antigenic site (NTD) has a beta strand throughout, but the mutations E156G and F157, R158/del cause a remarkable remodeling of the structure with an alpha fold by interrupting the beta strand. A recent X-ray crystallographic study demonstrates the formation of an alpha fold, indicating that the virus is less susceptible to monoclonal antibody binding [40]. Mutations in NTD cause strong dynamic movements during molecular simulations, as seen by the purple arrow in Fig. 2 (B). During simulation at various time steps, NTD of delta is fluctuating at higher rate compare to wild-type and difference among the RMSD is significant highly at various nano step (Fig. 2 (B)). Higher dynamic motions are again associated with more flexibility, as seen in the wild-type spike, whereas the Delta spike has lower dynamic motions, which correlate with decreased flexibility. Higher dynamic motions are often linked to higher RMSF (root mean square fluctuation). To correlate these findings with MD simulations, we analyzed RMSD (root mean square deviation) and RMSF to further comment on the flexibility of SARS-CoV-2 spike Delta variant.
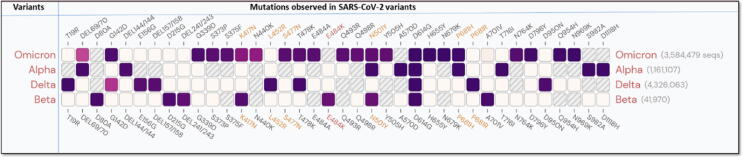
Fig. 1.
List of mutations present in the spike protein of different variants of SARS-CoV-2. This figure was created with outbreak.info, and the purple gradient shows the frequency of mutations. The higher the intensity of the purple colour, the higher the frequency of mutation, and the lower the intensity of mutation, the lower the intensity of mutation, with the exception of the cream area and boxes with lines, which have near zero and no mutations, respectively. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
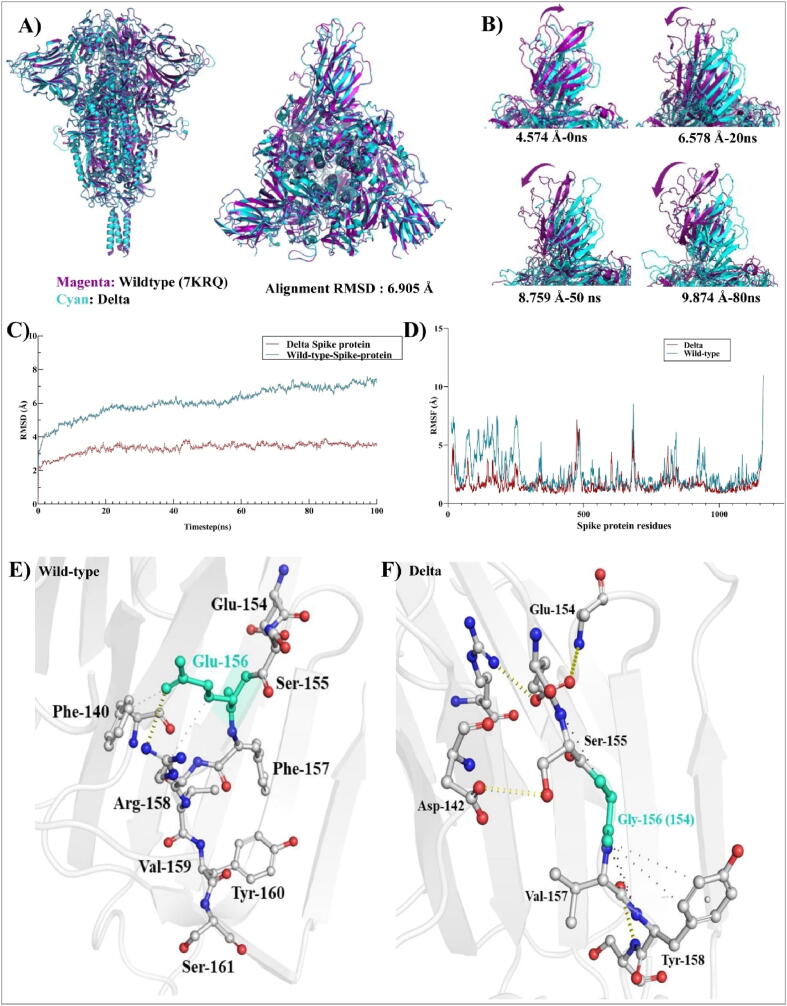
Fig. 2.
Rigidization and reduction in the flexibility of N-Terminal domain of Delta spike: (A) Front and top view of trimetric spike protein. 3D structural alignment of wild-type [7KRQ] and Delta trimetric spike proteins with the superimposed RMSD value: 6.905 Å. Wild-type protein is shown in magenta and Delta is shown in cyan color, (B) Frame superimposition of wild-type and Delta spike proteins for visualization of dynamic modes depicting differences in the NTDs. Magenta colored arrows showing dynamic moments of wild-type spike, (C) and (D). The RMSD and RMSF plot generated from MD-Simulation respectively. Wild-type protein is shown in deep teal color and Delta is shown in red color, (E) and (F) Intermolecular contacts between wild type and Delta SARS-CoV-2 spike protein. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
RMSD for wild-type and Delta complexes were observed to be 5.89 ± 0.026 Å and 2.54 ± 0.018 Å, respectively (Fig. 2 (C)). The RMSD graph clearly shows that mutations in the Delta’s spike protein improve its stability when compared to the wild-type trimeric complex (Fig. 2 (C)). We also observed the RMSF of 3.6 Å lower in the NTD of Delta spike compared to the wild-type complex Fig. 2 (D)). The decreased RMSF in the NTD region of Delta variant explains minimal amino acid fluctuations. Aurélie Bornot et al., in 2010 precisely explained the protein flexibility in terms of RMSF and B-factors, where increases in RMSF values are related to increased changes in protein conformation [4]. In some instances, amino acid residues that are flexible though RMSF can be rigid through B-factors [4]. In case of wild-type, the protein seems to be flexible in both cases through RMSF and B-factors (Supplementary Fig. S2), while no major changes were observed with respect to other parts of the protein except NTD (Fig. 2 (D)). Computational research approaching the increase in flexibility of RBD (receptor binding domain) of SARS-CoV-2 compared to SARS-CoV showed an increased flexibility (inward motions), which lead to strengthening the binding of ACE2 with RBD [9]. Also, it is well known that while flexibility establishes the binding, it may not only induce changes in the binding cavity between two interacting residues, but also can be a major contributor to the entropy penalty during binding [56].
Fig. 2 (E) and (F) explains the effect of these mutations in changing amino acids conformation in ball and stick form. One can visualize the difference in alignment of amino acids in the NTD within both variants, which affect the change in intermolecular contacts within the spike protein. Due to NTD mutations, the intra-atomic contacts within the amino acids of Delta are significantly increased with respect to the wild-type. Fig. 2 (E) and (F) shows how Glu-156 from wild type spike protein interacts with Phe-140 and Arg158 via intra-residual one hydrogen bond and other hydrophobic interactions. Whereas in Delta, four intra residual hydrogen bonds (viz. Glu154-Ala123, Glu154-Arg102, Ser155-Asp142, and Val157-Gly156) with significantly higher hydrophobic interactions. The higher intra-atomic contacts can lead to a decrease in flexibility (or increase in rigidity) by ΔSVib ENCoM: -0.500 kcal.mol−1.K−1 (Δ Vibrational Entropy Energy between wild-type and Delta variant).
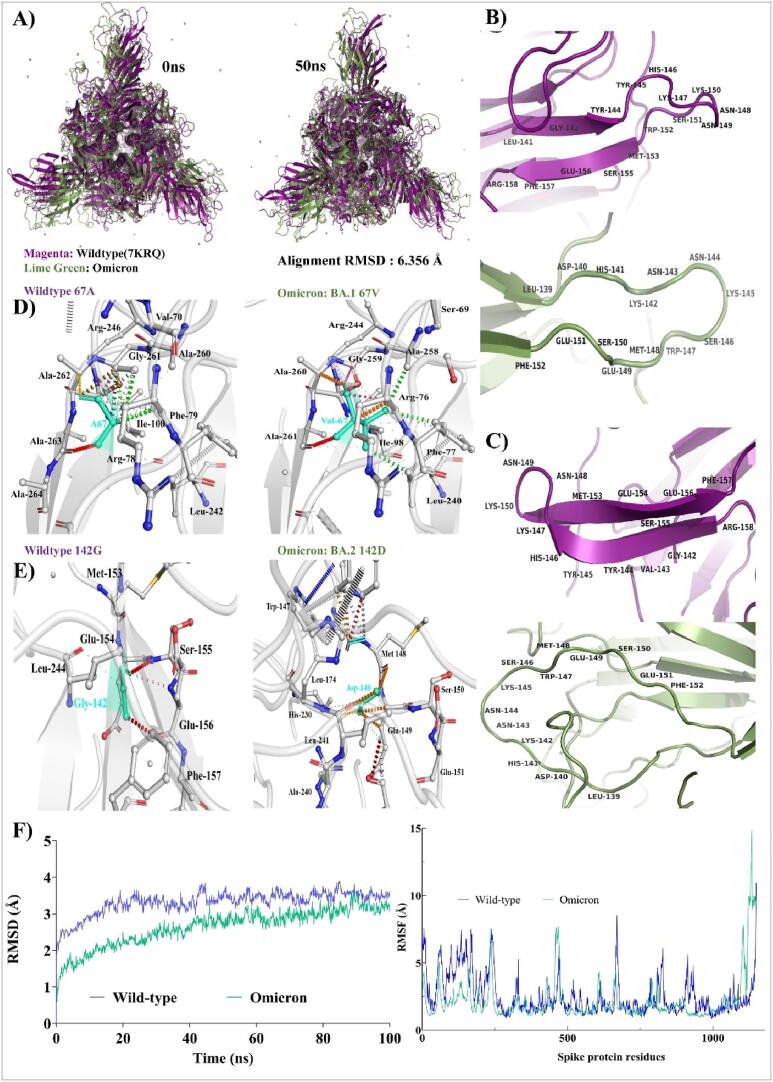
As NTD is also a binding site for a wide variety of monoclonal antibodies, rigidization in NTD region further affects the binding of antibodies with the spike. The important mutations of the Omicron variant with respect to NTD were also examined, as indicated in Fig. 3. The mutations A67V and G145D are found in Omicron's BA.1 and BA.2 sublineages. The similar investigation was carried out for Omicron, where both mutations caused structural changes in NTD. In Omicron NTD (highlighted in blue circle), structural superimposition of wild type spike with Omicron spike revealed some minor structural changes (Fig. 3 (A)). Fig. 3 (B) and (C) shows the destruction in antigenic beta sheets in Omicron spike (BA.1 67 V) compared to wild-type (67A) during MD simulation time step 0 ns and 50 ns. Fig. 3 (C) depicts how a mutation from Glycine 142 to Aspartate causes the highly antigenic beta sheet to be disrupted. Intramolecular contacts were examined, and the G142D mutation resulted in an increase in intramolecular interactions, affecting amino-acids that are critical for antibody binding and are also affected in the Delta variant due to which antibody escape was observed. For BA.1 lineage, A67V mutation showed increased intramolecular contacts but not significant as other mutations discussed earlier (Fig. 3 (D)). As demonstrated in Fig. 3 (E), Omicron NTD not only benefits from the G142D mutation, but also from the Delta mutations. In Delta, M153, G154, and S155 had undergone structural alterations, resulting in greater intramolecular contact, which is also present in case of Omicron’s BA.2 lineage. Molecular dynamics data for Omicron shows high structure stability in terms of RMSD, as shown in Fig. 3 (F). RMSD for wild-type and Omicron complexes were observed to be 5.89 ± 0.026 Å and 3.43 ± 0.029 Å, respectively. In the case of Omicron, NTD domain residues are also found to be rigid as per the RMSF plot (Fig. 3 (F)). However, surprisingly RMSF, of Delta and Omicron is similar in pattern (Fig. 3 (D) and 3 (F)). Overall, Delta and Omicron with rigid antigenic supersites in NTD region of spike protein with a changed beta strand may aid in antibody escape.
Fig. 3.
Disruption of antigenic beta sheets and increase in intramolecular interactions in Omicron spike: (A) The3D structural alignment of wild type [7KRQ] and Omicron trimetric spike proteins with the superimposed RMSD value: 6.356 Å, (B) Cartoon representation of narrating destruction in antigenic beta sheets in Omicron spike (BA.1 67 V) compared to wild-type (67A), (C) Cartoon representation of narrating destruction in antigenic beta sheets in Omicron spike (BA.2 142D) compared to wild-type (142G), (D) Intermolecular contacts between wild-type (67A) and Omicron (BA.1 67 V) spike proteins highlighting NTD mutation, (E) Intermolecular contacts between wild-type (142G) and Omicron (BA.2 142D) spike proteins highlighting NTD mutation Cyan color display mutated amino acids, and (F) RMSD and RMSF plots generated from MD-simulation respectively. Wild-type spike is shown in light blue color and Omicron is shown in light green color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Important mutations in NTD domains used in this research were highlighted in Fig. 4 (A). Here 4A8 antibody was focused for both computational and wet lab studies., hence docking pose of all three spikes with wild-type, Delta and Omicron were shown in Fig. 4 (B1), (B2), and (B3) respectively. Based on MEDUSA five class predictions, our study revealed that Delta and Omicron have decreased in flexibility (or increase in rigidity) compared to wild-type in important amino acids binding with 4A8 antibody (Fig. 4 (C)). Furthermore, we performed a principal component analysis (PCA), where the first dominant dynamic mode PC1 among the trajectories was analyzed using VMD (Visual Molecular Dynamics). Porcupine plots were showing the projection of mode vectors based on the residue fluctuation throughout the trajectories (Fig. 4 (C)). Length of mode-vectors in wild-type complex was higher compared to Delta and Omciron, which suggests that overall NTD flexibility is decreased in both varients (Fig. 4 (C)). We further analyzed the spike flexibility and rigidity in term of its binding with monoclonal antibodies by performing alanine residue scanning (Fig. 4 (D)). Wild-type-4A8 complex was processed through residue scanning with alanine mutagenesis to investigate the important residue for the binding of spike with 4A8. Amino acid residues G142, Y145, K147, 150K, 152W, 156E, 157F, and 158R showed positive binding affinity values with 4A8 upon mutating these residues to alanine (Fig. 4 (D)). These results clearly indicate that the mutations in the NTD domain of the spike protein caused a decrease in binding of 4A8 antibody in Delta. Docking pose of Delta-4A8 and Omciron-4A8 explains overall decrease in hydrogen bonding (<3Å) compared to wild type 4A8.
Fig. 4.
Reduced flexibility influence the binding of NTD targeted monoclonal antibody (4A8) with spike protein: (A) Amino acid residues falling in the binding region of NTD of spike protein. Wild type and mutated sequences were annotated with NCBI reference/accession id number MN986947.3 Delta and Omicron respectively, (B): Binding pose of spike-4A8 complex of wild type, Delta, and Omicron. Spike is shown in magenta color for wild type, cyan color for Delta, and green color for Omicron, while 4A8 is shown in orange color. Residues involved in pivotal contacts like hydrogen bonds (dashed lines) were shown in ball and stick conformation (C) Flexible and rigid regions in region covering mutation. Cyan to deep teal color represents the flexible to rigid region with COP (confidence of prediction) with <0.6 and >0.75 respectively. Porcupine plots generated from PCA analysis also supporting the same shown in cartoon conformation with mode vectors, and (D) Alanine residues scanning of wildtype-4A8 complex. Residues important in binding with monoclonal antibodies were shown in logo plot with positive binding affinity. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
3.2. The effect of NTD mutations in Delta and Omicron aids virus in eluding host immunity
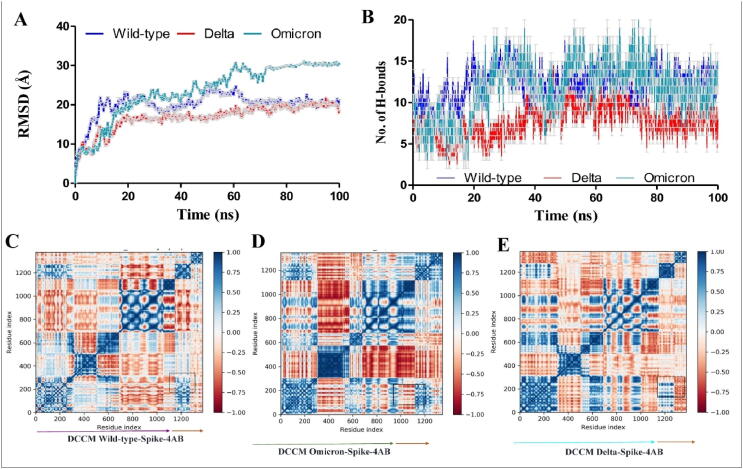
In this study, docking was executed with monoclonal antibodies 4A8, 8D2, and 2690 that target NTD. Docking studies were performed for all three antibodies, although MD simulations were only performed for 4A8 because experimental validations were performed with 4A8 only. MD simulations of all three mutants and wild type spikes with 4A8 antibody investigated the influence of G142D, Arg158, Phe-157/del, and one amino acid mutation E156/G on binding with monoclonal antibodies and depicted the case of immune evasion. Delta-4A8, Omicron-4A8 and wildtype-4A8 RMSDs were 20.147 ± 0.526 Å, 24.845 ± 0.342 Å and 16.142 ± 0.453Å, respectively (Fig. 5 (A)), suggesting that both the variants (Dealt and Omicron) with antibody 4A8 have higher deviation than wildtype [2]. The plateau was attained for Delta-4A8 complex after 65 ns, and jumps were detected during the MD simulations. Wildtype-4A8, on the other hand, was determined to be steady and reached a plateau after 20 ns. The significant difference between the two trajectories indicates that the wild-type-4A8 complex has a ∼4Å less RMSD value, resulting in a more stable peak than the Delta-4A8 complex. In Omicron, stable plateau after simulation was attained after ∼76 ns; difference in RMSD of Omicron and wildtype is much higher, which is about ∼10 Å. When compared to wild-type-4A8 complex, the production of hydrogen bonds inside the Delta-4A8 complex and Omicron-4A8 complex were lowered (Fig. 5 (B)). The decrease in hydrogen bond formations definitely suggests that the 4A8 interaction with Delta and Omicron is diminished due to NTD mutations [42], [6].
Fig. 5.
MD analysis of spike-antibody complexes: (A) RMSD (root mean square deviation) within wild-type-4A8 (cyan), Delta-4A8, and Omicron-4A8 complexes, (B) Hydrogen bonds formation within wild-type-4A8 (cyan), Delta-4A8, and Omicron-4A8 complexes, (C) Dynamic cross-correlation matrix obtained from trajectories analysis of wild-type-4A8 complex. Spike protein is shown in magenta arrow and orange arrow is indicating 4A8, (D) Dynamic cross-correlation matrix obtained from trajectories analysis of Omicron-4A8 complex, and (E) Dynamic cross-correlation matrix obtained from trajectories analysis of Delta-4A8 complex. Spike protein shown in cyan arrow and orange arrow is indicating 4A8. Blue to red color represents the cij values between 1 to −1. No cross correlation was shown by white color. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Dynamic cross-correlation matrix (DCCM) of wild type, Omicron and Deltaspikes in complex with 4A8 are shown in Fig. 5 (C), 5 (D), and 5 (E), respectively. In DCCM, wild-type-spike-4A8 is showing higher intensity of blue color compared to the Delta-spike-4A8 in square regions. Positive Cij values indicated in blue colors showing the better interaction profile between those residues (positive cross-correlation). There are major residues in the NTD region of spike protein, which spans 17–305 amino acid residues. In Delta and Omicron, higher amplified positive cross-correlation (0–200 amino acids) revealed structural compactness among the NTD, which could be detrimental to antibody binding. Delta and Omicron has increased rigidity, which is concluded using in silico methods. Increase in interatomic contacts have enhanced the rigidity, which is confirmed by various platforms like PYMOL, MEDUSA and MD-DCCM. Wild-type spike have some orange dots indicating negative cross-correlation among NTD amino-acid residues (circle), which are absent in NTD regions of Delta and Omicron.
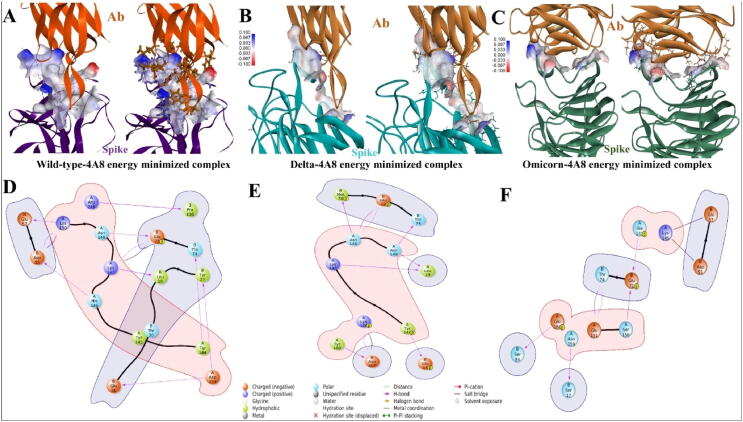
Overall, cross-correlation among 4A8, as illustrated in boxes 'a' and 'b' in relation to spike, was positive in wild-type and negative in Delta and Omicron, implying NTD targeted antibody escape in the Delta and Omicron variants. Further PCA analysis and porcupine plot also show decrease in flexibility (increase in rigidity). Karshikoff et al., showed that the binding site residues of protein have flexible tendency, to have better interaction with ligand (here the case of antibody) [26]. As binding site residues are more engaged with intra-atomic contacts itself, tendency of sharing contacts with outsider protein will be less. Based on results obtained through higher RMSF and Porcupine plots, it was hypothesized that rigidization among NTD may lead to NTD targeted antibody escape in SARS-CoV-2 variants. Major residues in the NTD region of spike protein spans within 17–305 amino acid residues. The intensity of the blue color is greater in the region surrounding the orange arrow (antibody 4A8), indicating a more positive cross correlation with the NTD region of the wild-type spike than Delta. NTD residues in the wild-type complex, on the other hand, showed stronger negative cross-correlation than Delta (Fig. 5 (C), 5 (D), and 5 (E)). Binding energies among the complexes were analyzed through MMGBSA. Major energies contributing to the complex formation were elucidated in Table 1 with bold text. Protein-protein complex formation occurs spontaneously as a result of substantial electrostatic, covalent, ionic-interactions, lipophilic (hydrophobic), and Vander-Waals’s energies. Overall free energy binding Δ G in wild-type, Delta, and Omicron with 4A8 antibody were −119.086 ± 19.42 kcal/mol and −55.496 ± 14.57 kcal/mol, and −76.48 ± 12.35 kcal/mol, respectively. Interaction in energy-minimized structures obtained through MMGBSA approach is shown in Fig. 6. Electrostatic map potential for binding of SARS-CoV-2 spike protein (wild type and variants) with 4A8 antibody is shown in Fig. 6 (A), 6 (B), and 6 (C)). In wild-type complex, the intensity of blue and red regions is highly intensive, and largely showing a strong binding with antibody (Fig. 6 (A)). In Fig. 6 (B) and 6 (C), electrostatic interaction among Delta and Omicron variants is less compared to wild-type. In wild-type complex, overlapping strong interaction between charged negative (orange) and charged positive residues (blue) is way higher compared to mutants. For example, A-Lys147: B-Glu72, A-Lys150: B-Glu57 and B-Glu55 were forming hydrogen bonds and salt bridges in spike (A) and 4A8 (B) (Fig. 6 (D)). Donald et al., and other groups suggest that salt bridges are geometric specific and designable interactions [13], [42]. Lys150 is forming salt bridge and hydrogen bonds with two negatively charged amino acids viz. Glu57 and Glu55. These kinds of favourable interactions are formed in wild-type spike but absent in Delta and Omicron, which concludes that geometry of NTD in spike protein had changed as such that it is reducing its strong interaction with 4A8 (Table 1 & Fig. 6 (D), 6 (E) and 6 (F)). These kinds of salt-bridges were more likely to exist in a hydrophobic environment [33], resulting in higher lipophilic energy in the wild-type (−30.334 kcal/mol) when compared to Delta (−11.975 kcal/mol) and Omicron (−8.14 kcal/mol).
Table 1.
MM/GBSA binding free energies of wildtype, Delta and Omicron spike protein after binding with 4A8.
| Energy components (kcal/mol) | Wildtype-4A8 | Delta-4A8 | Omicron-4A8 |
|---|---|---|---|
| Glide energy | −115.64 | −68.74 | −54.67 |
| Δ G Binding | −119.086 ± 19.42 | −55.496 ± 14.57 | −50.0805 ± 11.23 |
| Δ G Electrostatic energy | −791.090 ± 12.67 | −357.715 ± 19.76 | −27.5964 ± 10.78 |
| Δ G Covalent energy | −14.192 ± 2.78 | −7.506 ± 1.76 | −7.17242 ± 8.76 |
| Δ G Hbonds energy | −8.385 ± 1.76 | −5.179 ± 0.84 | −8.90553 ± 4.57 |
| Δ G Lipophilic energy | −30.333 ± 9.87 | −11.975 ± 2.94 | −8.14167 ± 1.56 |
| Δ G pi interection energy | −2.106 ± 0.654 | 1.156 ± 0.78 | −1.84204 ± 1.85 |
| Δ G self-contact correlation | −0.023 ± 0.012 | −0.156 ± 0.08 | 0.029636 ± 0.5 |
| Δ G Solv_GB | 781.518 ± 86.56 | 434.715 ± 50.76 | 88.42978 ± 13.4 |
| Δ G vdw energy | −82.858 ± 13.54 | −108.834 ± 24.56 | −84.8818 ± 17.5 |
Fig. 6.
Energy minimized structures obtained through MMGBSA: (A) Electrostatic potential maps showing interaction between wild-type and 4A8 antibody, (B) Electrostatic potential maps showing interaction between Delta and 4A8 antibody, (C) Electrostatic potential maps showing interaction between Omicron and 4A8 antibody, (D), (E) and (F) Energy minimized structures obtained through MMGBSA for wild-type-4A8, Delta-4A8, and Omicron-4A8 complexes respectively. Positively charged, and negatively charged amino acid residues were shown in orange and blue colors respectively. Amino-Acids with chain A are of spike and with chain B of antibody. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
NTD is a super site for a variety of antigenic determinants, and monoclonal antibody 4A8, which is used in this investigation, is at the interface of the NTD binding site. PDB ids 7CL2, 7DZX, and 7DZY were chosen as wild-type spike complexes with 4A8, 8D2, and 2490 monoclonal antibodies, respectively. As a mutant, spikes of the Delta and Omicron variants was docked with these three antibodies. The glide energy of wild type 4A8 Delta-4A8 and Omciron-4A8 were −115.64 kcal/mol, −68.74 kcal/mol, and −54.67 kcal/mol respectively (Table 1). Similarly, glide energy of wild-type 8D2, Delta-8D2, and Omicron-8D2 were −152.36 kcal/mol, −136.41 kcal/mol, and −67.56 kcal/mol the wild-type 2640, Delta-2640, and Omicron-2640 were −112.75 kcal/mol, −101.54 kcal/mol, and −10.7.56 kcal/mol, respectively (Supplementary Fig. S3). The higher the negative value of energy is, the stronger the binding among protein–protein complexes. Overall, the wild-type spike appears to have better binding with monoclonal antibodies (4A8, 8D2 and 2490) than the Delta and Omicron, implying that there is a possibility of immune evasion among the Delta and Omicron lineages. Majority of clusters falling in wild-type spike-8D2 and 2640 were showing better electrostatic interactions and less restrain violation energy compared to Delta. Better electrostatic and lower restrain violation energies explain why amino acids promote better complex formation with antibodies, whereas in Delta amino acids are more involved in interacting with each other by alpha fold (disrupting beta), resulting in higher restrain violation energy (Supplementary Fig. S3). Overall, the wild-type spike appears to have better binding with mAbs (4A8, 8D2 and 2490) than the Delta, implying that there is a possibility of immune evasion among the Delta.
3.3. Neutralization response of Delta and Omicron variants to human-anti-spike antiserum, convalescent antiserum from monkeys infected with Wuhan strain of SARS-CoV-2, human serum collected from people received SARRS-CoV-2 mRNA vaccines, and a NTD targeted 4A8 monoclonal antibody
After a thorough structural evaluation of the spike protein-NTD of Delta variant in comparison to D614G strain (PDB ID: 7KRQ), we further validated the response of Delta variant to anti-spike antiserum (Wuhan strain) of human and convalescent antiserum of monkeys, and a NTD specific 4A8 monoclonal antibody, for their neutralizing effect using a luciferase reporter pseudovirus based imaging assay. In this system, we used pseudoviruses displaying spike protein of D614G, B.1.617.1 (Kappa), B.1.617.2 (Delta), B.1.617.3, South Africa, and P1 variants for the study. We have shown the details of vectors used for the study along with the spike protein expression, transfection, and transduction efficiency of the developed pseudoviruses in Supplementary Figs. S4-S7. The viruses are specific, and their transduction efficiency are linked to the cellular expression of ACE2 and TMPRSS2 receptors, as well as viral display for spike protein variant of SARS-CoV-2 (Supplementary Fig. S5). The results showed significant variations in the overall infection rate by these variants in HEK293T-ACE2/TMPRSS2 cells as measured by transduced virus mediated luciferase expression (Supplementary Fig. S6). This finding shows differences in the affinity of these viruses' spike proteins for the cellular ACE2 receptor, as well as their subsequent processing by the TMPRSS2 enzyme for transduction. Delta and UK-D614G have equal overall infectivity, although these variants are significantly more virulent than the Wuhan strain. UK-D614G and Delta have comparable RBD configurations, but Delta is distinct due to its mutation with the NTD domain, hence ACE2 binding to Delta and UK-D614G will not alter, which further explains the similar infectivity. However, these variations in the electrostatic potential of spike variants, from negative to positive electrostatic potential, at the tip of RBD (marked by red oval), which can influence binding to ACE2 receptor, hence enhance infectivity of Delta and Omicron compared to the Wuhan strain (Fig. 7). Our main goal in this study was to identify cases of immune escape in the Delta variant when compared to other strains. To corroborate these findings, we also examined the appropriateness of these viruses for neutralization assays using anti-spike antibodies produced against the spike protein of SARS-CoV-2 (Sino Biologicals, this antibody shows >95 % neutralization efficiency at 4 μg/ml). The results showed specific neutralization effect as measured by luciferase assay using the Wuhan, D614G, and South Africa variants (Supplementary Fig. S7).
Fig. 7.
Comparison of electrostatic potentials of RBD and NTD domains of spike protein: (A) Electrostatic potential mapped on molecular surface for wild-type, (B) Electrostatic potential mapped on molecular surface for Delta variant; 6C: Isosurfaces of electrostatic potential for wild-type, (D) Isosurfaces of electrostatic potential for Delta variant. Positive electrostatic potential shown in blue, and negative in red. Secondary structure of RBD shown in yellow, and NTD in purple. The mutated residues are shown in van der Walls sphere representation. Red oval denotes large change of electrostatic potential at the tip of RBD. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
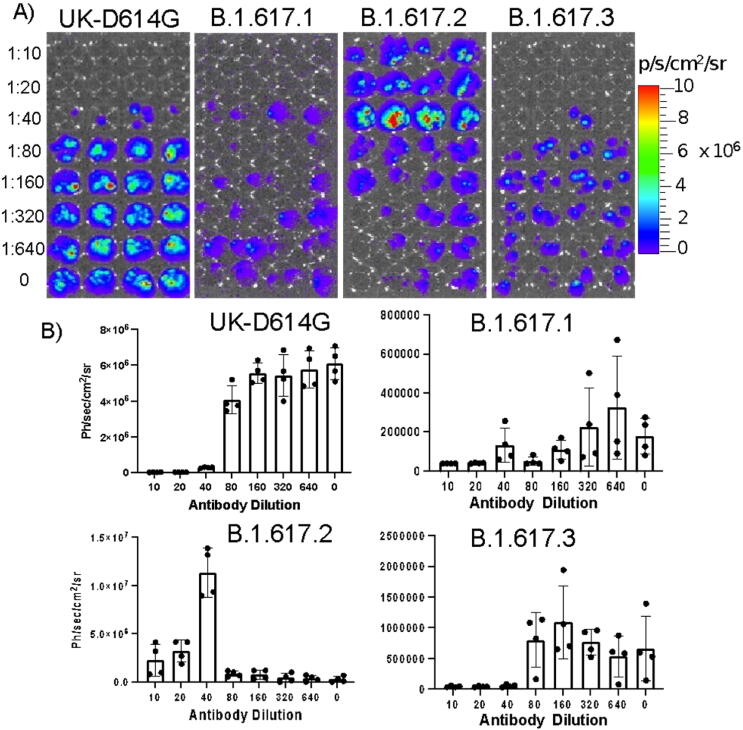
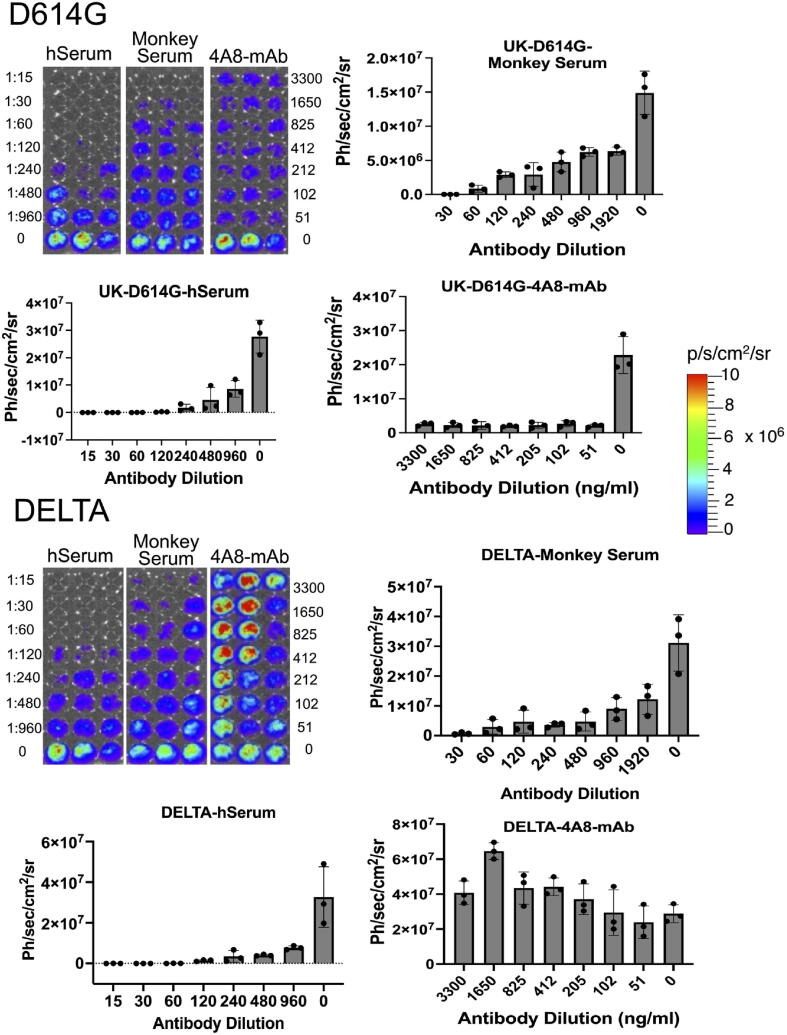
After establishing the suitability of the pseudovirus system for its specificity in cellular infection (transduction) and response to the known neutralizing antibody, we used this system for further comparative studies involving antiviral response to human-anti-spike antiserum (pooled serum of 3 antiserum each from Moderna and Pfizer/BioNT), convalescent antiserum from monkeys infected with Wuhan strain of SARS-CoV-2, and 4A8 monoclonal antibody targeting the NTD of Wuhan spike protein (BioVision). The major goal of this study is to see if the Delta variant has improved infectivity and immune evasion in people who have recovered from SARSCOVID-19 and have received the SARS-CoV-2 vaccine. We used pseudoviruses displaying D614G, B.1.617.1, B.1.617.3, and Delta variants for the study. We used pooled human antiserum from patients who received the mRNA vaccine, and pooled convalescent antiserum from monkeys infected with SARS-CoV-2 (Wuhan strain) in various dilutions (0–1000 times dilution). Similarly, we used 4A8 antibody concentration in the range of 0–4 µg/ml for the study. The results showed a human antibody dose dependent response from D614G, B.1.617.1 and B.1.617.3, whereas Delta variant showed an early boost in transduction while displaying partial or no response to neutralization at lower concentrations (Fig. 8). After confirming the differential response by Delta variant to anti-spike antiserum from human and the convalescent serum from monkeys, we tested the neutralization effect by 4A8 monoclonal antibody against pseudoviruses displaying the spike protein of Wuhan, D614G, and Delta variants. 4A8 antibody has been reported to be targeting NTD of Wuhan strain and showed a very strong neutralization effect in antiviral assays [7]. Until now, Delta and D614G strains competed to demonstrate their virulence in terms of infectivity and immunological evasion. However, when we tested this antibody concentrations in the range of 0–4 µg/ml, as expected, the Delta variant showed no response to 4A8 antibody, whereas the Wuhan and D614G variants showed a complete neutralization response even at the lowest concentration (50 ng/ml) of the antibody used in the study (Fig. 9).
Fig. 8.
Antiviral neutralization assay using lenti-pseudovirus displaying spike protein of different SARS-CoV-2 variants evaluated against anti-spike human antiserum (pooled serum): (A) Optical bioluminescence imaging of HEK293T-ACE2 cells transduced with lenti-pseudovirus displaying spike-protein of SARS-CoV-2 variants and expressing Firefly-Luciferase-ZsGreen reporter gene after neutralizing with anti-spike human antiserum of different dilutions, (B) Respective quantitative plot of images shown in “A”.
Fig. 9.
Antiviral neutralization assay using lenti-pseudovirus displaying spike protein of different SARS-CoV-2 variants evaluated against anti-spike human antiserum (pooled serum): (A) Optical bioluminescence imaging of HEK293T-ACE2 cells transduced with lenti-pseudovirus displaying spike-protein of SARS-CoV-2 variants and expressing Firefly-Luciferase-ZsGreen reporter gene after neutralizing with anti-spike human antiserum of different dilutions, and (B) Respective quantitative plot of images shown in “A”.
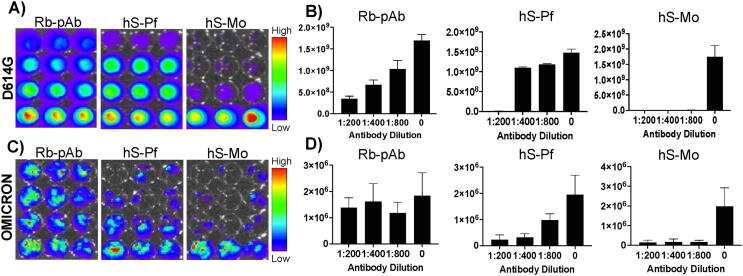
Similarly, we also evaluated the spike protein of Omicron variant for its structural rigidity using molecular dynamics studies. Since Omicron with G142D and A67V mutations located within the NTD provides rigid antigenic supersites with a changed beta strand and aid in antibody escape, we also evaluated neutralizing antibody response of Omicron against a polyclonal neutralizing antibody (Rb-pAb) and human serums collected from people received two doses of spike protein mRNA vaccines (hS-Pf and hS-Mo) using our reporter pseudovirus system. As stated above, we produced pseudoviruses using the spike protein of Omicron and performed the neutralization assay using serums in different dilutions. The results showed a poor neutralization response by Omicron strain to all three antibodies while a dose dependent response was observed from D614G pseudovirus variant (Fig. 10). The race for immune escape is won by Delta and Omicron over D614G. The neutralization of Delta at lower antibody concentrations, on the other hand, was due to the antibody dependent enhancement (ADE) effect [34]. In this case, Delta variant with deletions and mutations (E156G, F157, R158/del) in the NTD demonstrated immune evasion to antibody targeting NTD of Delta variant in both molecular modeling studies and the pseudovirus based neutralization assays, implying that further research into crystal structure showing rigidization or another structural change favoring virus immune evasion is required. Similarly, Omicron with G142D and A67V mutations with a change in beta strand in the NTD and its rigid antigenic supersites for the escape from the serum raised against the spike protein of wild-type strain using the polyclonal antibody and the human serum from people received mRNA vaccines further support the increased infection in current Omicron wave. SARS-CoV-2 displays low number of neutralizing epitopes which is another advantage for immune escape. Overall, computational simulations with experimental validation of pseudovirus-based neutralization assays revealed the reason for Delta and Omicron variants for their higher infectivity rates. Hence, spike mutations with respect to NTD enhance virus egress from mAb (4A8).
Fig. 10.
Antiviral neutralization assay using lenti-pseudovirus displaying spike protein of Omicron variant in comparison to D614G against a neutralizing polyclonal antibody (Rb-pAb) and human pooled serum collected from volunteers received two doses of respective mRNA vaccines (hS-Pf: BioNTech; hS-Mo: Moderna): (A) Optical bioluminescence imaging of HEK293T-ACE2/TRMPSS2 cells transduced with lenti-pseudovirus displaying spike-protein of D614G variant expressing Firefly-Luciferase-ZsGreen reporter gene after neutralizing with respective antibodies in different dilutions (Rb-pAb: Neutralizing polyclonal antibody; hS-Pf: BioNTech; hS-Mo: Moderna) with respective quantitative graphs showing Ph/sec/cm2/sr (B), (C) Optical bioluminescence imaging of HEK293T-ACE2/TRMPSS2 cells transduced with lenti-pseudovirus displaying spike-protein of Omicron variant expressing Firefly-Luciferase-ZsGreen reporter gene after neutralizing with respective antibodies in different dilutions (Rb-pAb: Neutralizing polyclonal antibody; hS-Pf: BioNTech; hS-Mo: Moderna) with respective quantitative graphs showing Ph/sec/cm2/sr (D).
4. Conclusion
The SARS-CoV-2 genotypes with increased transmissibility and immune evasion continue to pose a global concern. The recent breakouts of the Omicron and Delta (B.1.617.2) variants are example of the same. These variants could demonstrate higher transmissibility and survival in the immunised populations as well. Immune evasion is thought to be one of the major attributions for their spread across the globe. In this study, we have demonstrated our hypothesis and possible mechanisms of immune evasions by Delta and Omicron variants of SARS-CoV-2 using molecular dynamics and pseudovirus neutralisation assays. The current study focused on the main structural and genetic determinants of the SARS-CoV-2 Delta and Omicron variant that spread rapidly in different parts of the world during the second wave.
Through computational analysis, protein modelling, and molecular dynamic (MD) simulations, we investigated the genetic and structural basis of these variants for their lineage-defining mutations in the spike protein. Molecular docking of these structure with human monoclonal antibody 4A8 showed higher immune evasion in Omicron and then in Delta as compared to wildtype variant of SARS-CoV-2. From the molecular dynamic simulation studies, it is observed that, the reduction in the intermolecular hydrogen bonds strongly suggests that NTD mutations have weakened the 4A8 interaction with Delta and Omicron. Overall, the key findings show that Delta and Omicron, which have rigid antigenic supersites in the NTD region of the spike protein and a modified beta strand, may aid in immune escape. These results were further confirmed in pseudovirus-based neutralisation assay. The findings of this study showed that the Delta and Omicron variants have a larger chance of immune escape.
CRediT authorship contribution statement
Armi M. Chaudhari: Data curation, Writing – original draft. Madhvi Joshi: Conceptualization, Data curation, Writing – review & editing. Dinesh Kumar: Writing – review & editing. Amrutlal Patel: Writing – review & editing. Kiran Bharat Lokhande: Data Curation, Formal Analysis, Validation, Writing – review & editing. Anandi Krishnan: . Katja Hanack: Data Curation, Validation, Writing – review & editing. Slawomir Filipek: . Dorian Liepmann: Writing – review & editing. Venkatesan Renugopalakrishnan: Writing – review & editing. Ramasamy Paulmurugan: Conceptualization, Data curation, Writing – review & editing. Chaitanya Joshi: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Delta, Omicron, BA.2 plasmids were provided by Thomas Peacock of the MRC funded G2P-UK National Virology Consortium; G2P-UK; A National Virology Consortium to address phenotypic consequences of SARS-CoV-2 genomic variation (MR/W005611/1). 357 Authors would like to acknowledge Department of Science and Technology (DST), Government of Gujarat for infrastructure support for the research work.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.08.010.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Ai, J., Wang, X., He, X., Zhao, X., Zhang, Y., Jiang, Y., Li, M., Cui, Y., Chen, Y., Qiao, R., Li, L., Yang, L., Li, Y., Hu, Z., Zhang, W., & Wang, P. (2022). Antibody evasion of SARS-CoV-2 Omicron BA.1, BA.1.1, BA.2, and BA.3 sub-lineages. Cell host & microbe, S1931-3128(22)00243-8. Advance online publication. https://doi.org/10.1016/j.chom.2022.05.001. [DOI] [PMC free article] [PubMed]
- 2.Bayani F., Safaei Hashkavaei N., Uversky V.N., Mozaffari-Jovin S., Sefidbakht Y. Insights into the structural peculiarities of the N-terminal and receptor binding domains of the spike protein from the SARS-CoV-2 Omicron variant. Comput Biol Med. 2022;147 doi: 10.1016/j.compbiomed.2022.105735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beard H., Cholleti A., Pearlman D., Sherman W., Loving K.A. Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS ONE. 2013;8(12):e82849. doi: 10.1371/journal.pone.0082849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornot A., Etchebest C., de Brevern A.G. Predicting protein flexibility through the prediction of local structures. Proteins. 2011;79(3):839–852. doi: 10.1002/prot.22922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang S., Hu J.P., Lin P.Y., Jiao X., Tian X.H. Substrate recognition and transport behavior analyses of amino acid antiporter with coarse-grained models. Mol BioSyst. 2010;6(12):2430–2438. doi: 10.1039/c005266c. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhari A., Chaudhari M., Mahera S., Saiyed Z., Nathani N.M., Shukla S., et al. In-Silico analysis reveals lower transcription efficiency of C241T variant of SARS-CoV-2 with host replication factors MADP1 and hnRNP-1. Inf Med Unlocked. 2021;25 doi: 10.1016/j.imu.2021.100670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science (New York, NY) 2020;369(6504):650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daly J.L., Simonetti B., Klein K., Chen K.E., Williamson M.K., Antón-Plágaro C., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science (New York, NY) 2020;370(6518):861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehury B., Raina V., Misra N., Suar M. Effect of mutation on structure, function and dynamics of receptor binding domain of human SARS-CoV-2 with host cell receptor ACE2: a molecular dynamics simulations study. J Biomol Struct Dyn. 2021;39(18):7231–7245. doi: 10.1080/07391102.2020.1802348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejnirattisai W., Huo J., Zhou D., Zahradník J., Supasa P., Liu C., et al. SARS-CoV-2 Omicron-B.1.1.529 leads to widespread escape from neutralizing antibody responses. Cell. 2022;185(3):467–484.e15. doi: 10.1016/j.cell.2021.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delorey T.M., Ziegler C., Heimberg G., Normand R., Yang Y., Segerstolpe Å., et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021;595(7865):107–113. doi: 10.1038/s41586-021-03570-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., et al. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184(13):3426–3437.e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donald J.E., Kulp D.W., DeGrado W.F. Salt bridges: geometrically specific, designable interactions. Proteins. 2011;79(3):898–915. doi: 10.1002/prot.22927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fantini J., Yahi N., Azzaz F., Chahinian H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks. J Infect. 2021;83(2):197–206. doi: 10.1016/j.jinf.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fratev F. N501Y and K417N mutations in the spike protein of SARS-CoV-2 alter the interactions with both hACE2 and human-derived antibody: a free energy of perturbation retrospective study. J Chem Inf Model. 2021;61(12):6079–6084. doi: 10.1021/acs.jcim.1c01242. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Beltran W.F., Lam E.C., Astudillo M.G., Yang D., Miller T.E., Feldman J., et al. COVID-19-neutralizing antibodies predict disease severity and survival. Cell. 2021;184(2):476–488.e11. doi: 10.1016/j.cell.2020.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GeurtsvanKessel C.H., Geers D., Schmitz K.S., Mykytyn A.Z., Lamers M.M., Bogers S., et al. Divergent SARS-CoV-2 Omicron-reactive T and B cell responses in COVID-19 vaccine recipients. Sci Immunol. 2022;7(69):eabo2202. doi: 10.1126/sciimmunol.abo2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science (New York, NY) 2020;370(6521):eabe9403. doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenidge P.A., Kramer C., Mozziconacci J.C., Wolf R.M. MM/GBSA binding energy prediction on the PDBbind data set: successes, failures, and directions for further improvement. J Chem Inf Model. 2013;53(1):201–209. doi: 10.1021/ci300425v. [DOI] [PubMed] [Google Scholar]
- 20.Gruell H., Vanshylla K., Tober-Lau P., Hillus D., Schommers P., Lehmann C., et al. mRNA booster immunization elicits potent neutralizing serum activity against the SARS-CoV-2 Omicron variant. Nat Med. 2022;28(3):477–480. doi: 10.1038/s41591-021-01676-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guérin P., Yahi N., Azzaz F., Chahinian H., Sabatier J.M., Fantini J. Structural dynamics of the SARS-CoV-2 spike protein: a 2-year retrospective analysis of SARS-CoV-2 variants (from alpha to omicron) reveals an early divergence between conserved and variable epitopes. Molecules (Basel, Switzerland) 2022;27(12):3851. doi: 10.3390/molecules27123851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harlow G.R., Halpert J.R. Alanine-scanning mutagenesis of a putative substrate recognition site in human cytochrome P450 3A4. Role of residues 210 and 211 in flavonoid activation and substrate specificity. J Biol Chem. 1997;272(9):5396–5402. doi: 10.1074/jbc.272.9.5396. [DOI] [PubMed] [Google Scholar]
- 23.Hikmet, F., Méar, L., Edvinsson, Å., Micke, P., Uhlén, M., & Lindskog, C. (2020). The protein expression profile of ACE2 in human tissues. Mol Syst Biol, 16(7), e9610. https://doi.org/10.15252/msb.20209610. [DOI] [PMC free article] [PubMed]
- 24.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jangra S., Ye C., Rathnasinghe R., Stadlbauer D., Personalized Virology Initiative Study Group, Krammer F., Simon V., Martinez-Sobrido L., García-Sastre A., Schotsaert M. SARS-CoV-2 spike E484K mutation reduces antibody neutralisation. Lancet Microbe. 2021;2(7):e283–e284. doi: 10.1016/S2666-5247(21)00068-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karshikoff A., Nilsson L., Ladenstein R. Rigidity versus flexibility: the dilemma of understanding protein thermal stability. FEBS J. 2015;282(20):3899–3917. doi: 10.1111/febs.13343. [DOI] [PubMed] [Google Scholar]
- 27.Kormos B.L., Baranger A.M., Beveridge D.L. A study of collective atomic fluctuations and cooperativity in the U1A-RNA complex based on molecular dynamics simulations. J Struct Biol. 2007;157(3):500–513. doi: 10.1016/j.jsb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozakov D., Brenke R., Comeau S.R., Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 29.Kufareva I., Abagyan R. Methods of protein structure comparison. Methods Mol Biol (Clifton NJ) 2012;857:231–257. doi: 10.1007/978-1-61779-588-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumar S, Karuppanan K, Subramaniam G. (2022). Omicron (BA.1) and sub-variants (BA.1.1, BA.2 and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: a comparative sequence and structural-based computational assessment. J Med Virolgy, https://doi.org/10.1002/jmv.27927. [DOI] [PMC free article] [PubMed]
- 31.Kumar US, Afjei R, Ferrara K, Massoud TF, Paulmurugan R. Gold-nanostar-chitosan-mediated delivery of SARS-CoV-2 DNA vaccine for respiratory mucosal immunization: development and proof-of-principle. ACS Nano; 2021, acsnano.1c05002. https://doi.org/10.1021/acsnano.1c05002. [DOI] [PMC free article] [PubMed]
- 32.Kwon M., Firestein B.L. DNA transfection: calcium phosphate method. Methods Mol Biol (Clifton NJ) 2013;1018:107–110. doi: 10.1007/978-1-62703-444-9_10. [DOI] [PubMed] [Google Scholar]
- 33.Lee J.I., Hwang P.P., Wilson T.H. Lysine 319 interacts with both glutamic acid 269 and aspartic acid 240 in the lactose carrier of Escherichia coli. J Biol Chem. 1993;268(27):20007–20015. doi: 10.1016/S0021-9258(20)80687-5. [DOI] [PubMed] [Google Scholar]
- 34.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat Microbiol. 2020;5(10):1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PubMed] [Google Scholar]
- 35.Li D., Edwards R.J., Manne K., Martinez D.R., Schäfer A., Alam S.M., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184(16):4203–4219.e32. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y., Soh W.T., Kishikawa J.I., Hirose M., Nakayama E.E., Li S., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184(13):3452–3466.e18. doi: 10.1016/j.cell.2021.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021;595(10):1454–1461. doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyne P.D., Lamb M.L., Saeh J.C. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. J Med Chem. 2006;49(16):4805–4808. doi: 10.1021/jm060522a. [DOI] [PubMed] [Google Scholar]
- 39.Martyna G.J., Klein M.L., Tuckerman M. Nosé-Hoover chains: the canonical ensemble via continuous dynamics. J Chem Phys. 1992;97:2635–2643. doi: 10.1063/1.463940. [DOI] [Google Scholar]
- 40.McCallum M., Walls A.C., Sprouse K.R., Bowen J.E., Rosen L.E., Dang H.V., et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science (New York, NY) 2021;374(6575):1621–1626. doi: 10.1126/science.abl8506. [DOI] [PubMed] [Google Scholar]
- 41.Melms J.C., Biermann J., Huang H., Wang Y., Nair A., Tagore S., et al. Author Correction: A molecular single-cell lung atlas of lethal COVID-19. Nature. 2021;598(7882):E2. doi: 10.1038/s41586-021-03921-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meuzelaar H., Vreede J., Woutersen S. Influence of Glu/Arg, Asp/Arg, and Glu/Lys salt bridges on α-helical stability and folding kinetics. Biophys J. 2016;110(11):2328–2341. doi: 10.1016/j.bpj.2016.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powell A.E., Zhang K., Sanyal M., Tang S., Weidenbacher P.A., Li S., et al. A single immunization with spike-functionalized ferritin vaccines elicits neutralizing antibody responses against SARS-CoV-2 in mice. ACS Cent Sci. 2021;7(1):183–199. doi: 10.1021/acscentsci.0c01405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reizis B. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. 2019;50(1):37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rohaim MA, El RF, Clayton E, Munir M. Since January 2020 Elsevier has created a COVID-19 resource centre with free information in English and Mandarin on the novel coronavirus COVID- 19. The COVID-19 resource centre is hosted on Elsevier Connect, the company’s public news and information; 2020.
- 47.Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27(3):221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- 48.SC ’06: Proceedings of the 2006 ACM/IEEE Conference on Supercomputing (2006). (New York, NY, USA: Association for Computing Machinery).
- 49.Schrodinger, LLC. 2010. The PyMOL Molecular Graphics System, V. 1. 8 pymol.
- 50.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21(12):681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 51.Shen X. Boosting immunity to Omicron. Nat Med. 2022;28(3):445–446. doi: 10.1038/s41591-022-01727-0. [DOI] [PubMed] [Google Scholar]
- 52.Simonsen S.M., Sando L., Rosengren K.J., Wang C.K., Colgrave M.L., Daly N.L., et al. Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J Biol Chem. 2008;283(15):9805–9813. doi: 10.1074/jbc.M709303200. [DOI] [PubMed] [Google Scholar]
- 53.Steinbrecher T., Abel R., Clark A., Friesner R. Free energy perturbation calculations of the thermodynamics of protein side-chain mutations. J Mol Biol. 2017;429(7):923–929. doi: 10.1016/j.jmb.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 54.Takashita E., Kinoshita N., Yamayoshi S., Sakai-Tagawa Y., Fujisaki S., Ito M., et al. Efficacy of antiviral agents against the SARS-CoV-2 Omicron subvariant BA.2. New England J Med. 2022;386(15):1475–1477. doi: 10.1056/NEJMc2201933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toukmaji A.Y., Board J.A. Ewald summation techniques in perspective: A survey. Comput Phys Commun. 1996;95:73–92. doi: 10.1016/0010-4655(96)00016-1. [DOI] [Google Scholar]
- 56.Tuffery P., Derreumaux P. Flexibility and binding affinity in protein-ligand, protein-protein and multi-component protein interactions: limitations of current computational approaches. J R Soc Interface. 2012;9(66):20–33. doi: 10.1098/rsif.2011.0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Dijk M., van Dijk A.D., Hsu V., Boelens R., Bonvin A.M. Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility. Nucleic Acids Res. 2006;34(11):3317–3325. doi: 10.1093/nar/gkl412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Zundert, G., Moriarty, N. W., Sobolev, O. V., Adams, P. D., & Borrelli, K. W. (2021). Macromolecular refinement of X-ray and cryoelectron microscopy structures with Phenix/OPLS3e for improved structure and ligand quality. Structure (London, England : 1993), 29(8), 913–921.e4. https://doi.org/10.1016/j.str.2021.03.011. [DOI] [PMC free article] [PubMed]
- 59.Voss, W. N., Hou, Y. J., Johnson, N. V., Kim, J. E., Delidakis, G., Horton, A. P., Bartzoka, F., Paresi, C. J., Tanno, Y., Abbasi, S. A., Pickens, W., George, K., Boutz, D. R., Towers, D. M., McDaniel, J. R., Billick, D., Goike, J., Rowe, L., Batra, D., Pohl, J., … Ippolito, G. C. (2020). Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv : the preprint server for biology, 2020.12.20.423708. https://doi.org/10.1101/2020.12.20.423708.
- 60.Yamasoba D., Kimura I., Nasser H., Morioka Y., Nao N., Ito J., et al. Virological characteristics of the SARS-CoV-2 Omicron BA.2 spike. Cell. 2022;185(12):2103–2115.e19. doi: 10.1016/j.cell.2022.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J., Collier A.Y., Rowe M., Mardas F., Ventura J.D., Wan H., et al. Neutralization of the SARS-CoV-2 Omicron BA.1 and BA.2 variants. New England J Med. 2022;386(16):1579–1580. doi: 10.1056/NEJMc2201849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zahradník J., Nunvar J., Schreiber G. Perspectives: SARS-CoV-2 spike convergent evolution as a guide to explore adaptive advantage. Front Cell Infect Microbiol. 2022;12 doi: 10.3389/fcimb.2022.748948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang L., Jackson C.B., Mou H., Ojha A., Peng H., Quinlan B.D., et al. SARS-CoV-2 spike-protein D614G mutation increases virion spike density and infectivity. Nat Commun. 2020;11(1):6013. doi: 10.1038/s41467-020-19808-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, L., Jackson, C. B., Mou, H., Ojha, A., Rangarajan, E. S., Izard, T., Farzan, M., & Choe, H. (2020b). The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv: the preprint server for biology, 2020.06.12.148726. https://doi.org/10.1101/2020.06.12.148726.
- 65.Zielkiewicz J. Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J Chem Phys. 2005;123(10) doi: 10.1063/1.2018637. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.