Abstract
Vibrio cholerae is a highly motile bacterium which possesses a single polar flagellum as a locomotion organelle. Motility is thought to be an important factor for the virulence of V. cholerae. The genome sequencing project of this organism is in progress, and the genes that are highly homologous to the essential genes of the Na+-driven polar flagellar motor of Vibrio alginolyticus were found in the genome database of V. cholerae. The energy source of its flagellar motor was investigated. We examined the Na+ dependence and the sensitivity to the Na+ motor-specific inhibitor of the motility of the V. cholerae strains and present the evidence that the polar flagellar motor of V. cholerae is driven by an Na+ motive force.
Many motile bacteria move by rotating their flagella, the filamentous organelles that extend from the cell body. Flagellar rotation is carried out by a reversible rotary motor (about 20 nm in diameter) embedded in the cytoplasmic membrane at the base of each flagellar filament. These motors are powered by an electrochemical gradient of specific ions across the cytoplasmic membrane and are classified into two types of coupling ions: an H+-driven motor (in Escherichia coli, Salmonella typhimurium, and Bacillus spp.) and an Na+-driven motor (in alkalophilic Bacillus and marine Vibrio spp.) (6, 19). Thus, the flagellar motor is a tiny and elaborate molecular machine which converts electrochemical energy into mechanical work. However, the mechanism of this energy conversion is not clarified at the molecular level.
The study of the flagellar motor has been extensively done for the H+-driven motor of E. coli and S. typhimurium, and it has been shown that two integral membrane proteins, MotA and MotB, are essential for force generation in the motor (12, 29). They have four and a single transmembrane segment(s), respectively (10, 38), and form a proton-conducting channel complex responsible for coupling ion translocation to force generation in the motor (7, 16, 28, 30). The MotA-MotB channel complex is anchored to the cell wall by a peptidoglycan binding domain of MotB. Some critical residues involved in torque generation have recently been identified by intensive mutational studies (8, 9, 37, 39). In the Na+-driven motor, it was shown that four integral membrane proteins, PomA, PomB, MotX, and MotY, are essential for force generation in the polar flagellar motor of the marine bacterium Vibrio alginolyticus (1, 26). MotX and MotY have also been identified in Vibrio parahaemolyticus (23, 24). PomA and PomB have similarities to MotA and MotB, respectively, and so they might also form an Na+ channel complex and play an important role in force generation in the motor.
Vibrio cholerae is the bacterium that causes cholera. It has a pathogenic cycle consisting of a free-swimming phase outside its host and a sessile virulent phase when it is colonizing the human small intestine. During the free-swimming phase, the organism is highly motile by means of a single polar flagellum (14). Motility is thought to contribute to the virulence of V. cholerae, but the relationship between motility and virulence is not yet understood (14, 27) and basic studies of its flagellar motor have not been done. As described above, the flagellar motor has been intensively investigated in other Vibrio species, namely V. alginolyticus and V. parahaemolyticus. They have two distinct types of flagella, a polar flagellum used for swimming in a liquid medium and numerous lateral flagella used for swarming on the surface of solid substrate (5, 20, 22, 32, 33). Polar and lateral flagella rotate by Na+- and H+-driven flagellar motors, respectively (2, 20). When grown in a liquid medium, the organism mainly expresses a single polar flagellum and swims rapidly. Therefore, the cell shape and motility of these Vibrio are very similar to those of V. cholerae in liquid. It has also been shown that the flagellar basal body of V. alginolyticus resembles that of V. cholerae (4, 13). In addition, from the genome sequencing project of V. cholerae, the partial sequences of genes homologous to pomA, pomB, motX, and motY, essential for the rotation of the Na+ motor, have been found in the database. Gardel and Mekalanos (15) reported that the disruption of the pomB homolog of V. cholerae results in a flagellated but nonmotile strain (the authors described pomB as motB, and the partially deduced amino acid sequence of MotB showed 84% identity to PomB of V. alginolyticus), suggesting that the pomB homolog of V. cholerae is also essential for force generation in the motor. Therefore, we think that the polar flagellar motor of V. cholerae might be driven by an Na+ motive force. In this study, we examined Na+ dependence and the effects of specific inhibitors on motility in V. cholerae and present evidence that the polar flagellar motor is driven by an Na+ motive force.
V. cholerae O139 strain 1854 is highly motile.
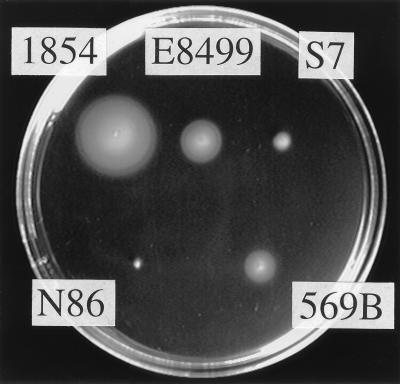
We examined the swarming abilities of the following V. cholerae strains: 1854 (O139; Bengal serovar [18]), E8499 (non-O1 [11]), S7 (O37 [35]), N86 (O1; El Tor biotype [36]), and 569B (O1; Classical biotype [11, 21]). Fresh colonies on LB solid agar plates (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1.25% agar) were inoculated onto LB semisolid plates (0.3% agar) and incubated for 4 h at 37°C. As shown in Fig. 1, strains E8499, 569B, and 1854 made swarm rings (the ring of strain 1854 was the largest), but S7 and N86 showed very little swarming ability. When cells were cultured in LB broth to directly observe motility, strain 1854 demonstrated the best motility of the strains used, consistent with the results of swarming ability; most of the cells swam very rapidly and the fraction of cells that were motile was very large compared with that of the other strains (data not shown). Some of the other strains were also motile, but the fraction of motile cells was smaller than that of strain 1854, because of the aggregation or elongation of the cells (data not shown). Therefore, we mainly used strain 1854 for motility analysis in this study.
FIG. 1.
Swarming abilities of V. cholerae strains. Fresh single colonies of the indicated V. cholerae strains were inoculated onto an LB semisolid plate (0.3% agar) and incubated for 4 h at 37°C.
Effect of ionic strength on the swimming speed of V. cholerae 1854.
A previous study of the 5S rRNA sequence suggested that V. alginolyticus, which has an Na+-dependent polar flagellum, is closely related to V. cholerae (4). Therefore, we compared the motilities by polar flagella of V. cholerae and V. alginolyticus. These two Vibrio spp. are usually grown in different salt concentrations. In LB medium, which contains 0.5% NaCl, commonly used for growing V. cholerae, the V. alginolyticus strain VIO5 (wild type in polar flagellar motility [26]) could not grow (data not shown). However, in VPG medium (1% polypeptone, 0.5% glycerol, 0.4% K2HPO4, 3% NaCl), commonly using for growing V. alginolyticus, V. cholerae 1854 grew comparably to V. alginolyticus VIO5. We compared the effects of ionic strength on the motility of V. cholerae 1854 cells cultured in these two media. Strain 1854 cells were grown in both LB and VPG media, and as a control V. alginolyticus VIO5 cells were cultured in VPG medium. At late log phase, cells were harvested and resuspended in TMN medium containing 50 mM Tris-HCl, 5 mM MgCl2, 5 mM glucose, and various concentrations of salts, and the motility of the cells was observed. As shown in Table 1, when the ionic strength of TMN medium was fixed at 300 mM, the motility of LB medium-cultured 1854 cells deteriorated and this deterioration became more severe as the concentration of NaCl increased. At 300 mM NaCl, both the motile fraction and the swimming speed were greatly reduced. LB medium-cultured 1854 cells resuspended in media of lower ionic strength (100 or 200 mM NaCl) showed a larger motile fraction and a greater swimming speed. However, the VPG-cultured V. cholerae 1854 and V. alginolyticus VIO5 cells showed nearly the same motile fractions regardless of ionic strength, and their swimming speeds increased as the NaCl concentration of the medium was increased. However, the motile fractions were significantly smaller than that of LB-cultured 1854 cells at 100 mM NaCl. We note that VPG-cultured 1854 cells showed very tumbly-biased swimming, and attractants, such as serine or Casamino Acids, were not effective in suppressing the directional change of swimming. These results indicate that V. cholerae 1854 can grow in media of a relatively wide range of ionic strengths, but the greatest motility of V. cholerae 1854 cells is achieved with an ionic strength of 100 mM when the cells are cultured in LB medium containing 0.5% NaCl. Therefore, in the following experiments, the total ionic concentration of Na+ and K+ was fixed to 100 mM in TMN medium.
TABLE 1.
Effect of ionic strength on the swimming speed of V. cholerae 1854 and V. alginolyticus VIO5
| Strain– mediuma | NaCl (mM) | KCl (mM) | Motile fractionb (motile cells/total cells) | Swimming speedc (μm/s) |
|---|---|---|---|---|
| 1854-LB | 300 | 0.19 | 44.7 | |
| 200 | 100 | 0.33 | 56.8 | |
| 100 | 200 | 0.43 | 61.4 | |
| 200 | 0.65 | 83.0 | ||
| 100 | 0.68 | 99.1 | ||
| 1854-VPG | 300 | 0.44 | 84.0 | |
| 200 | 100 | 0.38 | 79.1 | |
| 100 | 200 | 0.40 | 71.7 | |
| 200 | 0.37 | 75.2 | ||
| 100 | 0.40 | 74.2 | ||
| VIO5-VPG | 300 | 0.46 | 61.2 | |
| 200 | 100 | 0.43 | 61.4 | |
| 100 | 200 | 0.54 | 58.5 | |
| 200 | 0.47 | 61.8 | ||
| 100 | 0.50 | 60.1 |
V. cholerae 1854 and V. alginolyticus VIO5 were cultured until late log phase in the indicated medium.
Cells harvested at late log phase were resuspended in TMN medium (pH 7.5) containing various concentrations of salts, and motility was measured and recorded on videotape. To suppress the directional change of swimming, 10 mM serine was added to the medium as an attractant. The motile fraction was determined by counting the number of motile cells among the total cells in one video image and averaging the findings in at least three images in one condition.
The average swimming speeds were obtained by measuring at least 20 swimming tracks of cells generated from the integrated video images.
Polar flagellar motor of V. cholerae 1854 is driven by the sodium motive force.
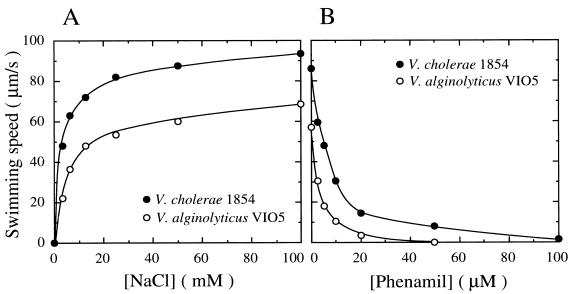
We next examined the potential for Na+ as an energy source of the polar flagellar motor of strain 1854. V. cholerae 1854 and V. alginolyticus VIO5 cells were cultured in LB and VPG media, respectively, and motility was measured in TMN medium containing various concentrations of NaCl, with a salt concentration of 100 mM attained by adding KCl. As shown in Fig. 2A, both 1854 and VIO5 cells clearly showed Na+-dependent motility and 1854 cells swam significantly faster than VIO5 cells did under all conditions. LB medium-cultured 1854 cells also showed a similar Na+-dependent motility in the medium whose ionic strength was fixed at 100 mM by the addition of choline chloride instead of KCl (data not shown). We also examined the effect of the Na+ motor-specific inhibitor, phenamil, on the motilities of both strains. Phenamil is known to inhibit the Na+ motor in a noncompetitive manner with Na+ in the medium (3). As shown in Fig. 2B, the motilities of both strains were also clearly inhibited by phenamil in the presence of 50 mM NaCl, although the concentration of phenamil required to completely inhibit the motility of V. cholerae was slightly higher than that needed for V. alginolyticus. Amiloride, which is the analog of phenamil and which specifically inhibits the Na+ motor in a competitive manner with Na+ in the medium (31), also inhibited the motilities of both strains (data not shown). These results suggest that the polar flagellar motor of V. cholerae 1854 is driven by the Na+ motive force.
FIG. 2.
Na+-dependent and phenamil-sensitive motility of V. cholerae 1854. V. cholerae 1854 and V. alginolyticus VIO5 cells were cultured in LB or VPG medium, respectively. At late log phase, cells were harvested and resuspended in TMN medium containing various concentrations of NaCl (A) or in TMN medium (pH 7.5) containing 50 mM NaCl, 50 mM KCl, and various concentrations of phenamil (B). In the experiment shown in panel A, the total concentration of salts in TMN medium was adjusted to 100 mM by the addition of KCl. The swimming speeds of the cells were obtained as described in Table 1, footnote c.
It is known that V. alginolyticus has an Na+-dependent NADH-quinone reductase (NQR) that functions as a respiration-coupled Na+ pump only in an alkaline pH range (34). At a neutral pH, the Na+ motive force is secondarily generated by a Na+/H+ antiporter from the H+ motive force. In this case, the addition of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) collapses the H+ motive force and hence the Na+ motive force. At an alkaline pH, CCCP collapses the H+ motive force but not the Na+ motive force, which is generated and maintained by the Na+ primary pump. The Na+ pump can be specifically inhibited by 2-heptyl-4-hydroxyquinoline-N-oxide (HQNO). We examined the effects of CCCP and HQNO on motility in neutral or alkaline conditions in order to confirm the Na+-dependent motility of V. cholerae. As shown in Table 2, the swimming speeds of both 1854 and VIO5 cells in the medium containing 20 μM CCCP at pH 7.5 were severely reduced. At pH 8.5, both 1854 and VIO5 cells were still motile in the presence of 20 μM CCCP. However, the motility of both strains was completely inhibited by the addition of 20 μM HQNO to this medium. These results showed that V. cholerae as well as V. alginolyticus cells are motile only in the presence of the Na+ motive force, and we have demonstrated that the polar flagellar motor of V. cholerae is powered by an Na+ motive force. These results also suggest that the respiratory chain of V. cholerae contains NQR, similar to that of V. alginolyticus. This enzyme in V. alginolyticus was recently reported to be composed of six subunits, NQR1 to NQR6 (17, 25). Partial sequences of the genes homologous to nqr1 and nqr2 were also found in the genome database of V. cholerae, consistent with our physiological results. Actually, nqrB (=nqr2) was recently identified in V. cholerae (16a). Considering the evolutionarily close relation between these two Vibrio spp. revealed by the analysis of 5S rRNA sequences (4), they should have many other similar physiological characters. Some distinct aspects between them were found in the present study. For example, V. cholerae cells swim faster than V. alginolyticus cells under certain conditions. The genome sequence of V. cholerae revealed that it has homologous genes coding for the essential components of the Na+ motor, i.e., pomA, pomB, motX, and motY, which are all of the Na+ motor genes identified until now. These two Vibrio spp. have very similar polar flagellar basal body structures (4). Thus, subtle differences in these motor components between these species might cause differences in the efficiency of energy conversion in the Na+ motor. We hope that the comparative studies of Na+ motors in these two species will reveal some cues for clarifying the mechanism of mechanochemical coupling in Na+-driven flagellar motors.
TABLE 2.
Effect of CCCP and HQNO on the swimming speed of V. cholerae 1854 and V. alginolyticus VIO5
| pH of medium | CCCP (μM) | HQNO (μM) | Swimming speed (μm/s)a
|
|
|---|---|---|---|---|
| Strain 1854 | Strain VIO5 | |||
| 7.5 | 0 | 0 | 90 | 53 |
| 20 | 0 | 6 | 0 | |
| 8.5 | 0 | 0 | 101 | 55 |
| 20 | 0 | 35 | 21 | |
| 0 | 20 | 89 | 55 | |
| 20 | 20 | 0 | 0 | |
Cells were grown to late log phase in LB (strain 1854) or VPG (strain VIO5) medium. Motility was measured in TMN medium (pH 7.5) containing 50 mM NaCl and 50 mM KCl. The average swimming speeds were obtained by measuring at least 20 cells.
Acknowledgments
This work was supported in part by grants-in-aid for scientific research from the Ministry of Education, Science and Culture of Japan (to I.K. and M.H.) and from the Japan Society for the Promotion of Science (to S.K.).
REFERENCES
- 1.Asai Y, Kojima S, Kato H, Nishioka N, Kawagishi I, Homma M. Putative channel components for the fast-rotating sodium-driven flagellar motor of a marine bacterium. J Bacteriol. 1997;179:5104–5110. doi: 10.1128/jb.179.16.5104-5110.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atsumi T, McCarter L, Imae Y. Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces. Nature. 1992;355:182–184. doi: 10.1038/355182a0. [DOI] [PubMed] [Google Scholar]
- 3.Atsumi T, Sugiyama S, Cragoe E J, Jr, Imae Y. Specific inhibition of the Na+-driven flagellar motors of alkalophilic Bacillus strains by the amiloride analog phenamil. J Bacteriol. 1990;172:1634–1639. doi: 10.1128/jb.172.3.1634-1639.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakeeva L E, Chumakov K M, Drachev A L, Metlina A L, Skulachev V P. The sodium cycle. III. Vibrio alginolyticus resembles Vibrio cholerae and some other vibriones by flagellar motor and ribosomal 5S-RNA structures. Biochim Biophys Acta. 1986;850:466–472. doi: 10.1016/0005-2728(86)90115-5. [DOI] [PubMed] [Google Scholar]
- 5.Belas M, Colwell R R. Scanning electron microscope observation of the swarming phenomenon of Vibrio parahaemolyticus. J Bacteriol. 1982;150:956–959. doi: 10.1128/jb.150.2.956-959.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 7.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 8.Blair D F, Berg H C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 9.Blair D F, Kim D Y, Berg H C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun S Y, Parkinson J S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 11.Craig J P, Yamamoto K, Takeda Y, Miwatani T. Production of cholera-like enterotoxin by a Vibrio cholerae non-O1 strain isolated from the environment. Infect Immun. 1981;34:90–97. doi: 10.1128/iai.34.1.90-97.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean G D, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the MotA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferris F G, Beveridge T J, Marceau-Day M L, Larson A D. Structure and cell envelope associations of flagellar basal complexes of Vibrio cholerae and Campylobacter fetus. Can J Microbiol. 1984;30:322–333. doi: 10.1139/m84-048. [DOI] [PubMed] [Google Scholar]
- 14.Freter R, O’Brien P C. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: chemotactic responses of Vibrio cholerae and description of motile nonchemotactic mutants. Infect Immun. 1981;34:215–221. doi: 10.1128/iai.34.1.215-221.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardel C L, Mekalanos J J. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect Immun. 1996;64:2246–2255. doi: 10.1128/iai.64.6.2246-2255.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garza A G, Harris H L, Stoebner R A, Manson M D. Motility protein interactions in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Häse C C, Mekalanos J J. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA. 1998;95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi M, Hirai K, Unemoto T. Sequencing and the alignment of structural genes in the nqr operon encoding the Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 1995;363:75–77. doi: 10.1016/0014-5793(95)00283-f. [DOI] [PubMed] [Google Scholar]
- 18.Iida T, Shrestha J, Yamamoto K, Honda T, Albert M J. Cholera isolates in relation to the “eighth pandemic.” Letter; comment. Lancet. 1993;342:926. [PubMed] [Google Scholar]
- 19.Imae Y, Atsumi T. Na+-driven bacterial flagellar motors. J Bioenerg Biomembr. 1989;21:705–716. doi: 10.1007/BF00762688. [DOI] [PubMed] [Google Scholar]
- 20.Kawagishi I, Maekawa Y, Atsumi T, Homma M, Imae Y. Isolation of the polar and lateral flagellum-defective mutants in Vibrio alginolyticus and identification of their flagellar driving energy sources. J Bacteriol. 1995;177:5158–5160. doi: 10.1128/jb.177.17.5158-5160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LoSpalluto J J, Finkelstein R A. Chemical and physical properties of cholera exo-enterotoxin (choleragen) and its spontaneously formed toxoid (choleragenoid) Biochim Biophys Acta. 1972;257:158–166. doi: 10.1016/0005-2795(72)90265-6. [DOI] [PubMed] [Google Scholar]
- 22.McCarter L, Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 23.McCarter L L. MotX, the channel component of the sodium-type flagellar motor. J Bacteriol. 1994;176:5988–5998. doi: 10.1128/jb.176.19.5988-5998.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarter L L. MotY, a component of the sodium-type flagellar motor. J Bacteriol. 1994;176:4219–4225. doi: 10.1128/jb.176.14.4219-4225.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakayama Y, Hayashi M, Unemoto T. Identification of six subunits constituting Na+-translocating NADH-quinone reductase from the marine Vibrio alginolyticus. FEBS Lett. 1998;422:240–242. doi: 10.1016/s0014-5793(98)00016-7. [DOI] [PubMed] [Google Scholar]
- 26.Okunishi I, Kawagishi I, Homma M. Cloning and characterization of motY, a gene coding for a component of the sodium-driven flagellar motor in Vibrio alginolyticus. J Bacteriol. 1996;178:2409–2415. doi: 10.1128/jb.178.8.2409-2415.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richardson K. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect Immun. 1991;59:2727–2736. doi: 10.1128/iai.59.8.2727-2736.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharp L L, Zhou J, Blair D F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 29.Stader J, Matsumura P, Vacante D, Dean G E, Macnab R M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sugiyama S, Cragoe E J, Imae Y. Amiloride, a specific inhibitor for the Na+-driven flagellar motors of alkalophilic Bacillus. J Biol Chem. 1988;263:8215–8219. [PubMed] [Google Scholar]
- 32.Ulitzur S. Induction of swarming in Vibrio parahaemolyticus. Arch Microbiol. 1974;101:357–363. doi: 10.1007/BF00455952. [DOI] [PubMed] [Google Scholar]
- 33.Ulitzur S. The mechanism of swarming of Vibrio alginolyticus. Arch Microbiol. 1975;104:67–71. doi: 10.1007/BF00447301. [DOI] [PubMed] [Google Scholar]
- 34.Unemoto T, Hayashi M. Na+-translocating NADH-quinone reductase of marine and halophilic bacteria. J Bioenerg Biomembr. 1993;25:385–391. doi: 10.1007/BF00762464. [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto K, Do Valle G R F, Xu M, Miwatani T, Honda T. Amino acids of the cholera toxin from Vibrio cholerae O37 strain S7 which differ from those of strain O1. Gene. 1995;163:155–156. doi: 10.1016/0378-1119(95)00415-3. [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto K, Ichinose Y, Nakasone N, Tanabe M, Nagahama M, Sakurai J, Iwanaga M. Identity of hemolysins produced by Vibrio cholerae non-O1 and V. cholerae O1, biotype El Tor. Infect Immun. 1986;51:927–931. doi: 10.1128/iai.51.3.927-931.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J, Fazzio R T, Blair D F. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol. 1995;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]
- 39.Zhou J, Sharp L L, Tang H L, Lloyd S A, Billings S, Braun T F, Blair D F. Function of protonatable residues in the flagellar motor of Escherichia coli: a critical role for Asp 32 of MotB. J Bacteriol. 1998;180:2729–2735. doi: 10.1128/jb.180.10.2729-2735.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]