Abstract
Chromosome 8q gain is associated with poor clinical outcomes in prostate cancer, but the underlying biological mechanisms remain to be clarified. CSN5, a putative androgen receptor (AR) partner that is located on chromosome 8q, is the key subunit of the COP9 signalosome, which deactivates ubiquitin ligases. Deregulation of CSN5 could affect diverse cellular functions that contribute to tumor development, but there has been no comprehensive study of its function in prostate cancer. The clinical significance of CSN5 amplification/overexpression was evaluated in 16 prostate cancer clinical cohorts. Its oncogenic activity was assessed by genetic and pharmacologic perturbations of CSN5 activity in prostate cancer cell lines. The molecular mechanisms of CSN5 function were assessed, as was the efficacy of the CSN5 inhibitor CSN5i-3 in vitro and in vivo. Finally, the transcription cofactor activity of CSN5 in prostate cancer cells was determined. The prognostic significance of CSN5 amplification and overexpression in prostate cancer was independent of MYC amplification. Inhibition of CSN5 inhibited its oncogenic function by targeting AR signaling, DNA repair, multiple oncogenic pathways, and spliceosome regulation. Furthermore, inhibition of CSN5 repressed metabolic pathways, including oxidative phosphorylation and glycolysis in AR-negative prostate cancer cells. Targeting CSN5 with CSN5i-3 showed potent antitumor activity in vitro and in vivo. Importantly, CSN5i-3 synergizes with PARP inhibitors to inhibit prostate cancer cell growth. CSN5 functions as a transcription cofactor to cooperate with multiple transcription factors in prostate cancer. Inhibiting CSN5 strongly attenuates prostate cancer progression and could enhance PARP inhibition efficacy in the treatment of prostate cancer.
Keywords: CSN5, AR signaling, DNA repair, PARP inhibitor, prostate cancer
Introduction
Prostate cancer (PC) is the most commonly diagnosed noncutaneous malignancy in men and the third leading cause of cancer death in the United States. Despite local therapy, about 20% of cases will progress to metastatic PC and eventually metastatic castration-resistant PC (mCRPC), the lethal form of the disease (1, 2). Although some new treatments improve overall survival, modest benefits and potential cross-resistance seen with next-generation agents targeting the androgen receptor (AR) indicate the unmet need to develop molecularly targeted therapies to improve outcomes (3, 4).
Recent advances in molecular genetics have permitted the identification of putative drivers of cancer initiation and progression through the unraveling of recurrent somatic mutations, copy number alterations (CNAs), and oncogenic structural DNA rearrangements in primary and metastatic PC (5, 6). These findings have resulted in several prospective trials currently enrolling patients with defined genomic alterations, including studies of PARP inhibition (PARPi) for alterations in BRCA2/1, ATM, and other DNA damage response and repair genes, and AKT inhibition for PI3K pathway alterations (7).
Of the common CNAs in PC, gains on chromosome 8q (chr8q) have consistently been linked to poor clinical outcomes (8, 9). However, the exact biological mechanisms underlying the aggressive phenotype related to chr8q alteration remain to be clarified. Genes for multiple putative AR co-regulators are located within the chr8q region (10–11). Of these, CSN5 was reported to collaborate with amplification of MYC to induce an aggressive response signature in breast cancer (12). CSN5 can interact with a number of steroid hormone receptors and coactivates steroid hormones including AR (10). Both AR and MYC contribute to PC progression (13), suggesting that CSN5 may have a pivotal role in PC progression.
CSN5 is the key subunit of the COP9 signalosome, which deactivates ubiquitin ligases through deneddylation of NEDD8 (14). Deregulation of CSN5 could affect diverse cellular functions that are critical for tumor development, including DNA repair, cell cycle control, apoptosis, angiogenesis, and signal transduction (15). CSN5 can induce degradation of several tumor suppressors (eg, p27, p53, NCoR) and stabilize oncogenes (eg, MDM2, BCL2, Snail) in various cancer cell types (16). Its overexpression causes tamoxifen resistance in breast cancer models by inducing degradation of NCoR, a key corepressor for ERα (17). To date, no systematic study to unravel the CSN5 target network in PC has been performed. Therefore, in this study, we defined the prognostic value of CSN5 amplification and overexpression in 16 PC cohorts, characterized its molecular function by evaluating the transcriptomic landscape regulated by CSN5, evaluated the efficacy of a CSN5-specific inhibitor in vitro and in vivo, and, for the first time, elucidated the transcriptional cofactor activity of CSN5 in cancer.
Materials and Methods
Detailed methods are given in Supplementary Methods.
Clinical cohort summary
Characteristics of patients in the Physicians’ Health Study (PHS) and Health Professionals Follow-up Study (HPFS) cohorts were previously reported (18). We will treat PHS/HPFS as one cohort for this report. Logistic regression was used to quantify the association of CSN5 expression (in quartiles) and lethal cancer. Fourteen additional publicly available PC cohorts are summarized in Table S1.
Cell culture
Human PC cells (LNCaP, 22Rv1, and PC3) were purchased from ATCC (Manassas, VA). C4–2 cells were obtained from VitroMed (Burlington, NC). All cells were maintained in 10% FBS supplemented with 2 mM of L-glutamine and antibiotic at 37°C in 5% CO2. Cells were authenticated by human short tandem repeat profiling at the Integrated Genomics Operation Core Facility (IGO) at Memorial Sloan Kettering Cancer Center (MSK).
Gene silencing and antitumor function assays
Lentiviral vectors of control shRNA and shCSN5 were purchased from MilliporeSigma (Table S2). After cells were treated with shRNAs or inhibitors, cell viability, DNA damage, and apoptosis were detected. Soft agar assays and invasion assays were performed to evaluate the effects of tumorigenesis activity.
Drug synergy assays
Synergies between CSN5i-3 and PARP inhibitors (olaparib, talazoparib) were evaluated in PC3 and C4–2 cells. Distinct drug doses were applied in two cell lines due to the different sensitivity. Cell viability was evaluated using CellTiter-Glo (Promega Corp., Madison, WI). Drug combination responses were also plotted as heat maps to determine therapeutic significance of the combination. Degree of drug synergy was assessed using SynergyFinder (https://synergyfinder.fimm.fi) (19). Synergy assays were performed in triplicate. The summary synergy showed the average response to the drug combination. A synergy score of less than −10 was considered antagonistic, a range from −10 to +10 was additive, and greater than +10 was synergistic.
RNA analysis, CUT&Tag analysis, and immunoblotting
Total RNA was extracted from cells and analyzed as previously described (18). TaqMan gene expression assays (Thermo Fisher Scientific, Waltham, MA) were used for relative gene expression (Table S2) by real-time qPCR (RT-qPCR). Transcript levels were normalized to levels of GAPDH transcript. RNA sequencing was performed by 50 million 2×50bp reads in the MSK IGO. RNA sequencing data were analyzed using Partek Flow (Partek Inc., St. Louis, MO). The data are available from GEO (GSE173469; linked SubSeries: GSE173466 and GSE173467).
CUT&Tag analysis was slightly modified based on the CUT&Tag for bench-top application (20). All the buffer needed was prepared for the analysis (Table S2).
Proteins were extracted by RIPA buffer and protein concentrations were determined by the Bradford method. Equal amounts of protein were loaded and resolved by SDS-PAGE, then transferred to polyvinylidene difluoride membranes for immunoblotting. All antibodies used are listed in Table S2. Human phospho-kinase antibody arrays (R&D Systems; Minneapolis, MN) were applied according to the manufacturer’s instructions. Briefly, protein was extracted from cells treated with inhibitors. Changes in kinase activity were visualized by chemiluminescence on autoradiography film and measured using ImageJ software.
Xenograft studies
NOG-SCID male mice (6 weeks) were implanted subcutaneously with C4–2 cells. After palpable tumors developed (typically 100 mm3), ten mice per group received either vehicle or CSN5i-3 (150 mg/kg) by oral gavage twice daily for 31 days. Tumors were measured twice weekly using calipers. Tissues were collected for immunohistochemistry (IHC) staining. All animal care was in accordance with the guidelines of the Institutional Animal Care and Use Committee at MSK.
Bioinformatic analysis of clinical cohorts
Data for various clinical cohorts were obtained from cBioPortal for Cancer Genomics (21) and Oncomine (22). Heatmaps were generated using R version 3.4.3 (https://www.R-project.org). Pathway analysis was performed using GSEA (23). Gene scores were calculated with gene set variation analysis (GSVA) using single-sample GSEA (ssGSEA) (24).
Statistical analysis
Results are reported as mean ± standard deviation. Comparisons between groups were performed using an unpaired two-sided Student’s t test or Wilcoxon rank-sum test (p <0.05 was considered significant). Disease-free survival was examined using the Kaplan-Meier method. Patients were divided into 2 groups (upper and lower quartile based on gene expression/signature score), and Kaplan-Meier curves were generated for each group. The log-rank test was used to determine significance. Cox proportional hazard regression was performed, adjusting for clinical and demographic factors. Statistical analysis was completed using R version 3.4.3.
Results
Amplification of CSN5 is correlated with poor clinical outcomes in PC
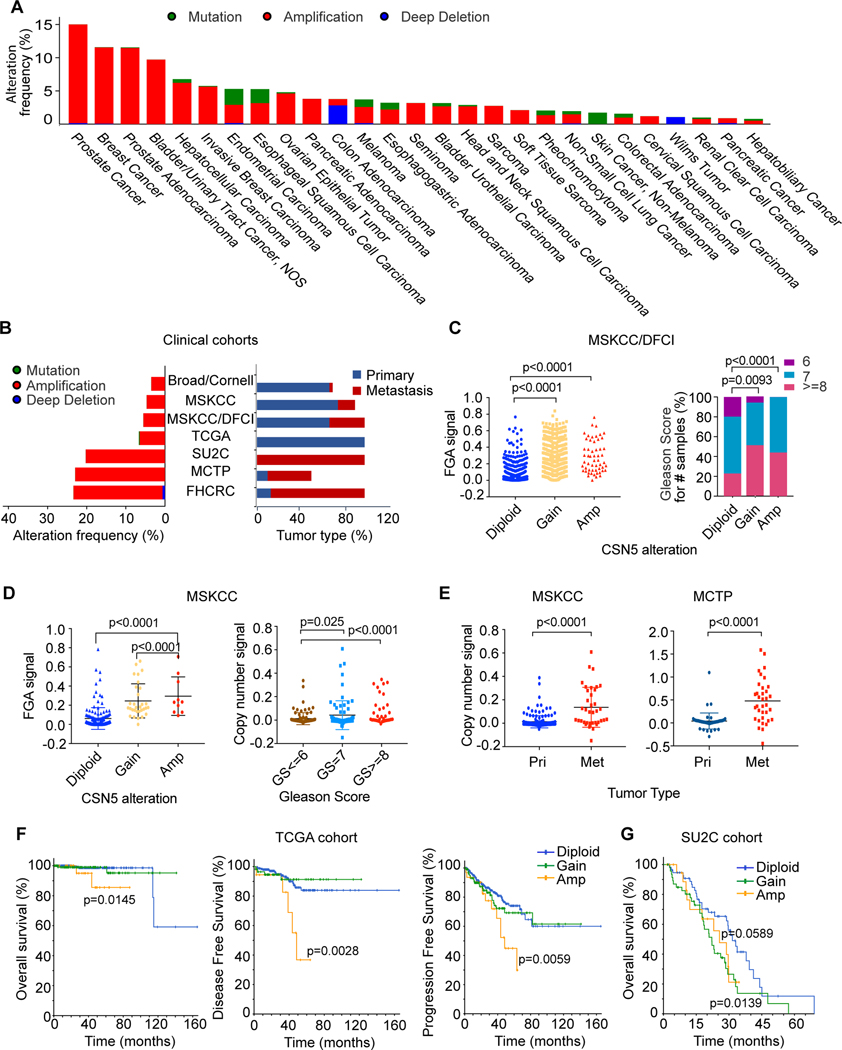
To evaluate the clinical relevance of CSN5, we analyzed genomic alteration of CSN5 across 46716 cancer samples in 176 clinical cohorts. The frequency threshold is set as >0.5%. CSN5 amplification was seen more frequently in PC than in any other cancer (15%) (Fig. 1A). Further analysis from 8 PC cohorts showed CSN5 amplification is increased in metastatic compared to primary tumors (Fig.1B). In the MSKCC/DFCI cohort (680 primary and 333 metastatic tumors), gain and amplification of CSN5 CNA are significantly correlated with a high fraction genome altered (FGA) signal and high Gleason score (Fig. 1C). Because the MSKCC/DFCI cohort is a combination of six cohorts (25), we confirmed the results in four of six other cohorts (TCGA, SU2C, FHCRC, and MCTP) (Fig. S1A). In addition, we made the same observations in the MSKCC cohort (Fig. 1D). Similar to FGA, aneuploidy is correlated with genomic instability (26). In the MSKCC/DFCI and TCGA cohorts, CSN5 CNA is highly related to high ploidy number (Fig. S1B) and aneuploidy score (Fig. S1C). These results suggested CSN5 CNA is correlated with genomic instability. In the MSKCC, Grasso, and Liu cohorts, significantly higher copy number of CSN5 is observed in metastatic tumors compared to primary tumors and normal tissue (Figs. 1E, S1D), indicating its contribution to disease progression. Correspondingly, CSN5 amplification is significantly correlated with worse disease-free survival (DFS), progression-free survival (PFS), and overall survival (OS) in primary TCGA and MSKCC (Figs.1F, S1E). In the metastatic SU2C cohort, we observed the same results (Fig. 1G). Altogether, we demonstrated that CSN5 CNA is associated with disease progression in PC.
Figure 1. Amplification of CSN5 is correlated with poor clinical outcomes in prostate cancer.
A, Genomic alterations of CSN5 cross 24 cancer types; 176 clinical cohort studies which includes 46716 patient samples from cBioPortal for Cancer Genomics were used for this analysis. The alteration frequency threshold is above 0.5%. B, Genomic alterations of CSN5 in prostate cancer cohorts. Seven prostate cancer cohorts from cBioPortal were used for the analysis. Alteration frequency of CSN5 is listed in the left panel, and the tumor type distribution of each cohort is listed in the right panel. C-D, The correlation of CSN5 alteration with fraction genomic altered (FGA) and Gleason score in the MSKCC/DFCI cohort (C) and MSKCC cohort (D). E, The correlation between CSN5 alteration and tumor progression in MSKCC and Grasso cohorts. CSN5 copy number was analyzed in primary (Pri), and metastasis (Met) tissue samples. F and G, the correlation between CSN5 alteration and clinical outcomes in TCGA and SU2C cohorts. P-value is between amplification and diploid of CSN5.
Overexpression of CSN5 is correlated with poor clinical outcomes in PC
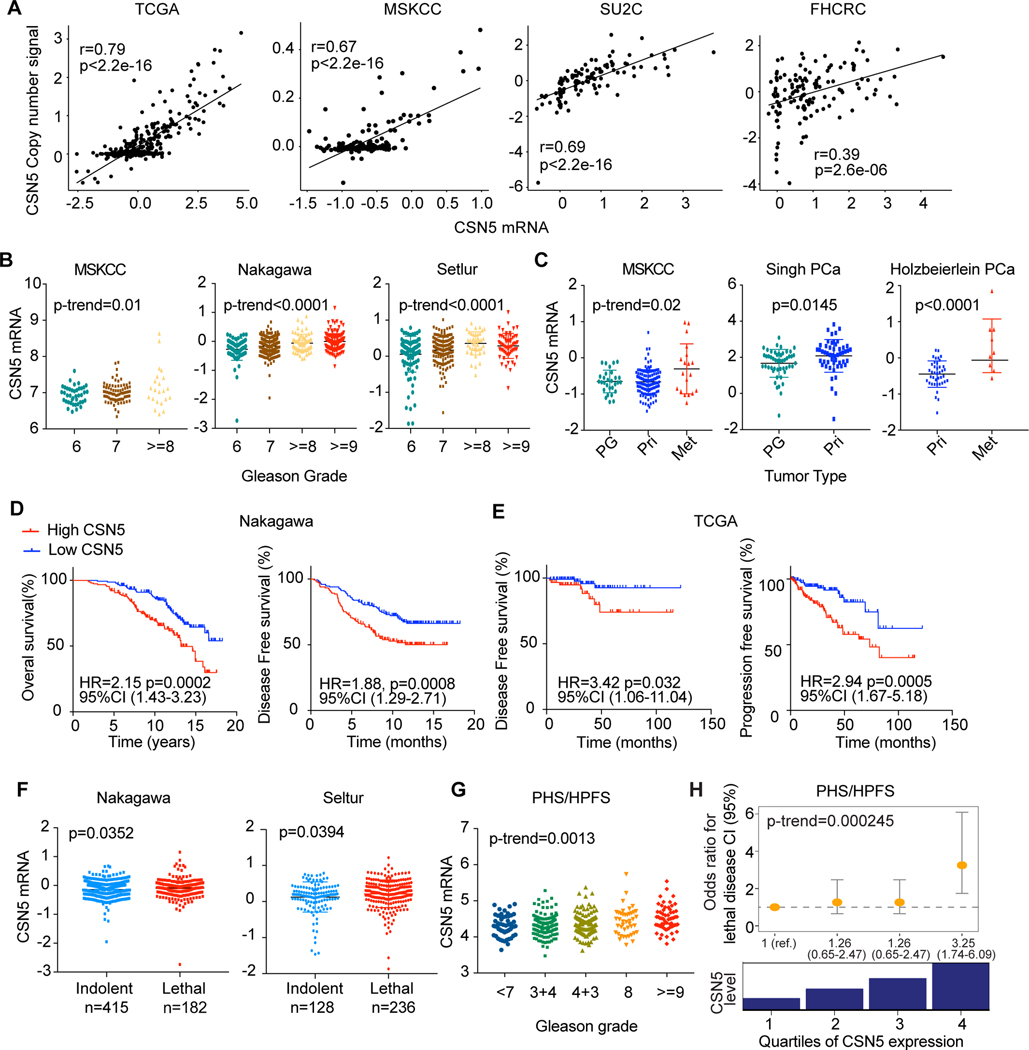
We observed CSN5 CNA is highly concurrent with MYC CNA in multiple PC cohorts (Fig. S2A). We further evaluated if the clinical significance of CSN5 CNA is specific to CSN5 and independent of MYC CNA. Because CSN5 CNA are positively correlated with its mRNA levels in multiple cohorts (Fig. 2A), we investigated the clinical relevance of CSN5 gene expression. High expression of CSN5 is significantly associated with high Gleason score and tumor progression (Figs. 2B, 2C). High levels of CSN5 were associated with significantly worse OS and DFS in the TCGA and Nakagawa cohorts (Figs. 2D–2E) and lethality in the Nakagawa and Setlur cohorts (Fig. 2F). MYC expression was not correlated with CSN5 expression in PC (Fig. S2B), or with Gleason score, DFS, or lethality (Fig. S2C–E).
Figure 2. Overexpression of CSN5 is correlated with poor clinical outcomes in prostate cancer.
A, The correlation between copy number changes and mRNA levels of CSN5 in TCGA, MSKCC, SU2C, FHCRC cohorts. B, The correlation of CSN5 expression and Gleason grades in MSKCC, Nakagawa, and Setlur cohorts. C, Association of CSN5 expression with tumor progression in MSKCC, Singh, and Holzbeierlein prostate cancer cohorts. CSN5 expression was analyzed in prostate gland (PG), primary (Pri), and metastasis (Met) tissue samples. D and E, the correlation between CSN5 mRNA levels and survival rates in Nakagawa (D) and TCGA (E) cohorts. F, The association of CSN5 levels with lethality in Nakagawa and Setlur cohorts. G and H, The correlation of CSN5 expression with Gleason grade and the risk of lethal prostate cancer over long-term follow-up, in the combined PHS-HPFS cohort.
We further validated the prognostic significance of CSN5 overexpression using the HPFS (n=254) and PHS (n=150) cohorts, which included long-term follow-up for lethal outcomes. Significantly higher levels of CSN5 were observed with increasing Gleason score (P-trend = 0.0013; Fig. 2G) and risk of lethal disease (Fig. 2H). Compared with the lowest quartile, CSN5 RNA levels in the highest quartile were correlated with a 3.25-times higher risk of lethal disease (95% CI, 1.74–6.09). Surprisingly, MYC mRNA levels were significantly lower in high-grade tumor but were not correlated with lethality (Fig. S2F). These results suggest that overexpression of CSN5 is associated with poor clinical outcomes, independent of MYC expression.
Inhibition of CSN5 attenuated its oncogenic activity in AR-positive PC cell lines
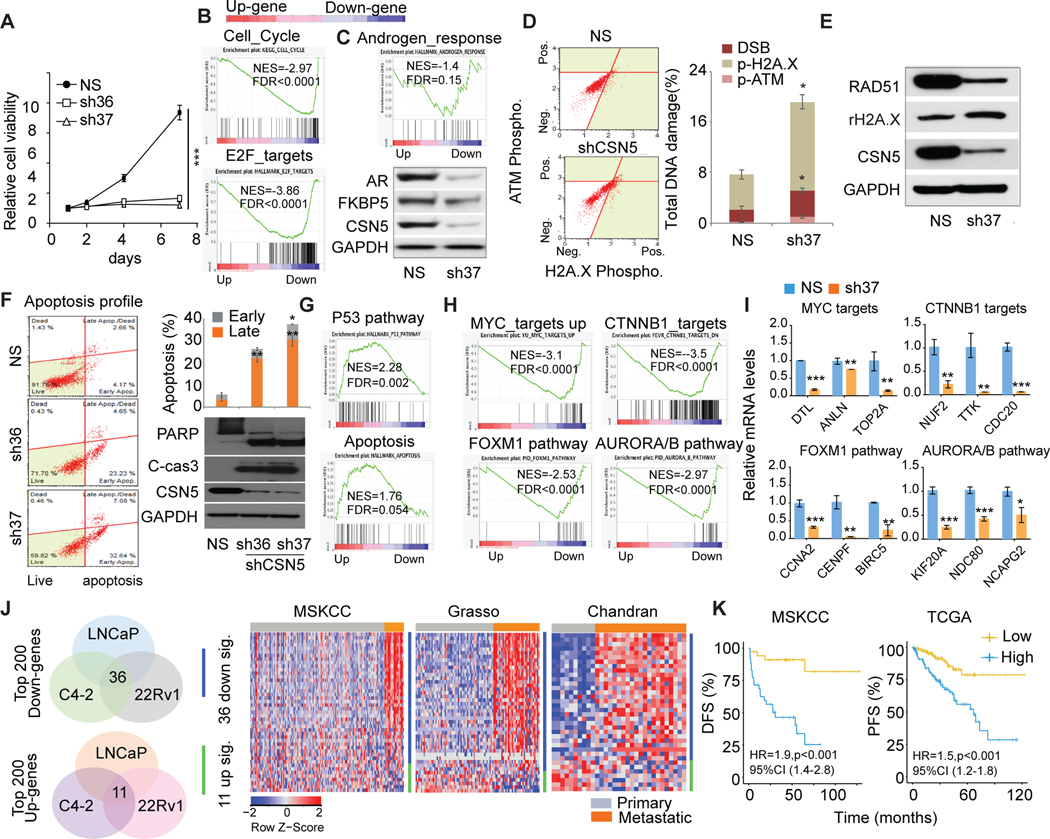
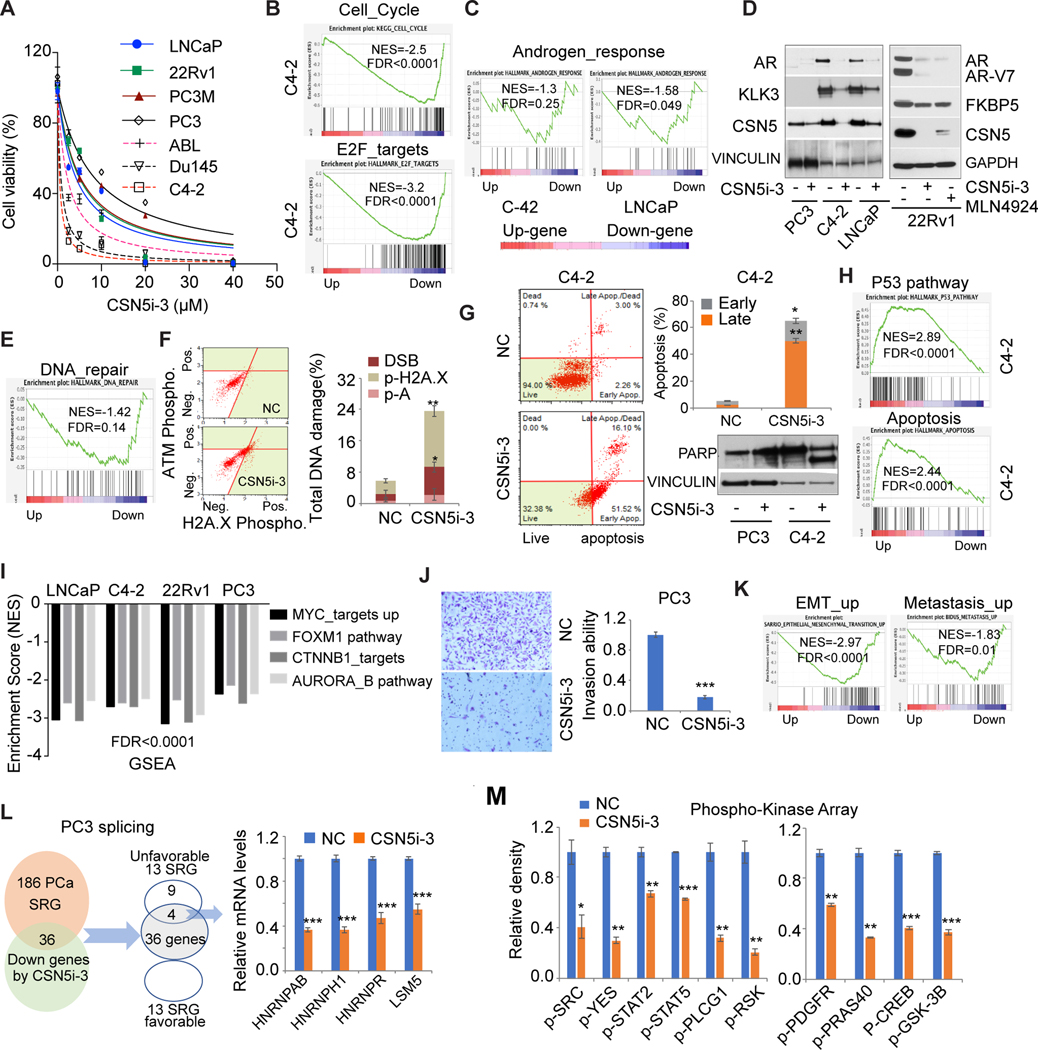
To assess the function of CSN5 in AR-positive PC cell lines, we stably knocked down CSN5. Both shRNA of CSN5 (sh36/sh37) significantly inhibited CSN5 expression and cell proliferation in three AR-positive cell lines (LNCaP, C4–2, 22Rv1) (Figs. 3A, S3A). Transcriptomic changes induced by shCSN5 (sh37) showed that cell cycle and E2F targets were significantly downregulated by shCSN5 (Fig. 3B, Fig. S3B). Repression of targets in cell cycle/E2F-target pathways was confirmed in three cell lines (Fig. S3C). AR signaling plays an essential role in PC proliferation. Knockdown of CSN5 directly reduced AR expression, including the AR-V7 and AR signaling pathways (Figs. 3C, S3D). CSN5 can regulate the stability and activity of proteins involved in DNA damage response and repair (27). Inhibition of CSN5 in PC cells can induce DNA damage marker activation, including phosphorylation of ATM and H2A.X (Fig. 3D) and DNA repair gene RAD51 reduction (Fig. 3E). Finally, shCSN5 led to significant apoptosis in PC cells, evidenced by an increased apoptotic cell population and marker expression (eg, cleaved PARP and CASPASE3; Figs. 3F, S3E–3F). GSEA of RNA-sequenced datasets showed strong activation of the p53 and apoptosis pathways in shCSN5 cells (Fig. 3G). Consistently, the targets of p53/apoptosis pathways were repressed by shCSN5 in LNCaP, C4–2, and 22Rv1 cells (Fig. S3G). CSN5 has been reported to regulate oncogenic signaling in cancers (16). Our transcriptomic studies showed knockdown of CSN5 markedly inhibited key oncogenic signaling pathways that play pivotal roles in PC progression, including MYC, FOXM1, CTNNB1, and AURORA/B pathways (Figs. 3H, S3I). shCSN5 can directly reduce MYC, FOXM1, and AURORA protein expression (Fig. S3H). Accordingly, the targets of four pathways were repressed by shCSN5 (Figs. 3I, S3J).
Figure 3. The oncogenic roles of CSN5 in AR positive prostate cancer.
A, Analysis of cell proliferation in shRNAs infected C4–2 cells. C4–2 cells were stably infected with shCSN5 (sh36, sh37) and shNS. Cell proliferation was detected in 7 days. NS, nonspecific shRNA. sh36 and sh37, shRNAs for CSN5. B, Cell cycle and E2F targets gene sets enrichment in regulated genes by inhibition of CSN5 by sh37 in C4–2. Up (upregulated genes); Down (downregulated genes). C, AR protein and AR signaling regulated by CSN5 in C4–2 cells. Androgen response pathway enrichment was analyzed by GSEA of the transcriptomic dataset in C4–2 cells. AR and its target FKBP5 expression were detected in shCSN5 (sh37)-treated C4–2 cells. D, analysis of shCSN5 (sh37)-induced DNA damage. DNA damage marker activation was monitored in C4–2 cells using Muse multicolor DNA damage kit. E, Activation of H2A.X and inhibition of RAD51 were confirmed by immunoblotting. F, Apoptosis detected by Annexin V assays and immunoblots in C4–2 cells. G, p53 signaling and apoptosis pathway enrichment in C4–2 cells with sh37 and shNS treatment. H and I, Knockdown of CSN5 repressed oncogenic pathways. Four oncogenic pathways were enriched in shCSN5 downregulated gene group in C4–2 cells (H). Some targets of four oncogenic pathways were validated by qRT-PCR (I). J, The correlation of common regulated genes in three AR (+) prostate cancer cells with metastatic prostate cancer. Top 200 downregulated and upregulated genes were intersected from three AR (+) cells. Common regulated genes were applied to three prostate cancer cohorts. K, The correlation of 36 genes commonly downregulated with disease-free survival (MSKCC) and progression-free survival (TCGA). Figure values represent the mean ± SE of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; vs. control groups treated with nonspecific (shNS) shRNA.
To evaluate the clinical relevance of CSN5 inhibition, we looked at shCSN5-regulated genes in three cell lines. Of the top 200 downregulated genes, 36 were commonly downregulated, and of the top 200 upregulated genes, 11 were commonly upregulated. The 36-gene signature was highly activated in metastatic tumors in the MSKCC, Grasso, and Chandran cohorts (Figs. 3J, S3K). These genes were significantly correlated with DFS in the MSKCC cohort and PFS in TCGA (Fig. 3K), supporting their clinical relevance in CSN5 inhibition.
Inhibition of CSN5 attenuated its oncogenic activity in AR-negative PC cells
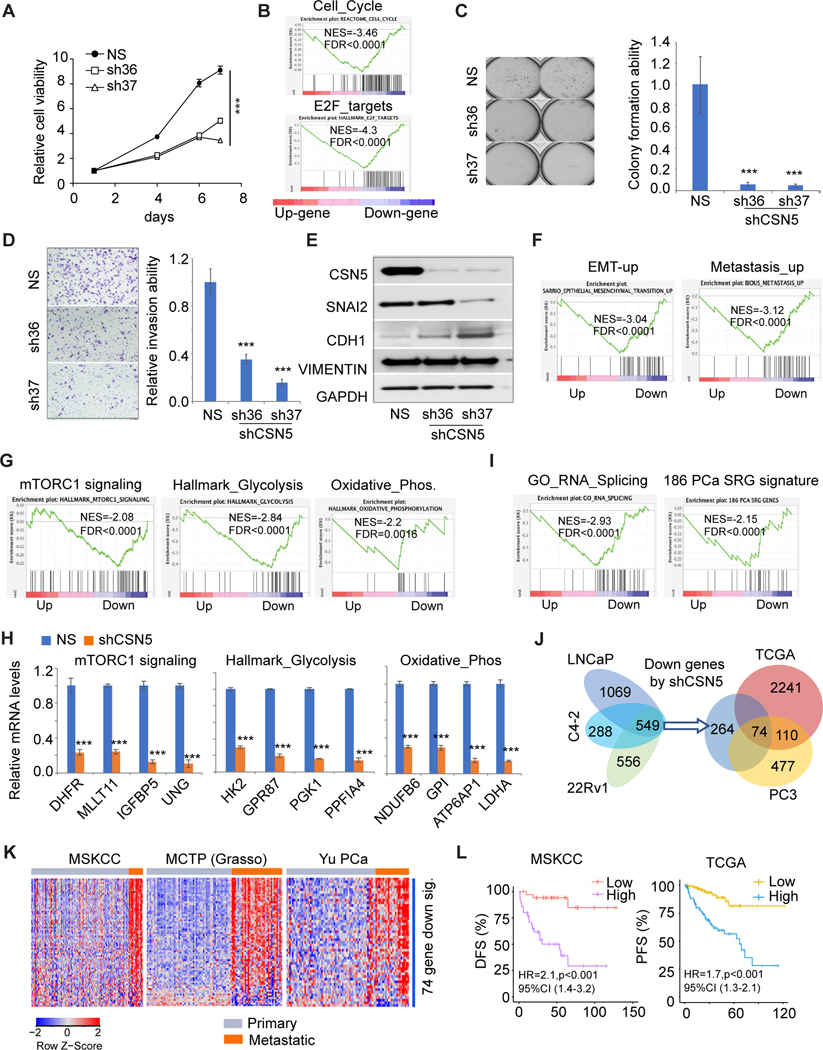
To further assess CSN5 function in AR-negative PC cells, we established CSN5 knockdown PC3 cells. Similar to AR-positive PC cells, inhibition of CSN5 repressed cell growth (Fig. 4A). GESA showed cell cycle pathway and E2F targets were downregulated by shCSN5 (Fig. 4B). However, we did not observe significant apoptosis in PC3 cells with shCSN5, possibly due to the p53 deletion in PC3 cells. Knockdown of CSN5 markedly inhibited colony formation by over 90% (Fig. 4C) and 60%–80% of PC3 cell invasion by repressing SNAI2 levels and increasing CDH1 expression (Figs. 4D–4E). Transcriptomic analysis showed metastasis-related pathways (eg, SARRIO EMT-up and BIDUS_Metastasis_up) were significantly repressed by shCSN5 (Fig. 4F). Similar to AR-positive PC cells, oncogenic pathways (MYC, FOXM1, CTNNB1, and AURORA/B signaling) were inhibited by shCSN5 (Fig. S4A). Furthermore, the shCSN5 induced mTORC1 signaling repression (Fig. 4G). mTORC1 signaling regulates both glycolysis and oxidative phosphorylation pathways (28, 29). These metabolic pathways were also significantly inhibited by CSN5 knockdown in PC3 cells (Fig. 4G). Partial target expression for the three regulated pathways was confirmed to be repressed by shCSN5 (Fig. 4H). Besides oncogenic signaling and metabolic pathways, we also observed that RNA splicing process was inhibited by shCSN5 (Fig. 4I, left). Dysregulation of RNA splicing has been identified as a driver for cancers (30). Recently, 186 splicing-regulatory genes (SRGs) were found to be dysregulated in seven PC clinical cohorts (31). GSEA showed the 186-SRG signature was highly enriched in downregulated genes by shCSN5 in both AR-negative and AR-positive PC cells (Fig. 4I, right; Fig. S4B). There were 74 common genes when we intersected three groups of genes including 549 downregulated genes from 3 AR-positive cells, shCSN5-induced downregulated genes from PC3 cells, and genes positively correlated with CSN5 in the TCGA cohort (Fig. 4J). The 74-gene signature is highly expressed in metastatic PC tumors (Figs. 4K, S4C). High expression of this signature was significantly correlated with worse DFS and PFS in the MSKCC and TCGA cohorts (Fig. 4L).
Figure 4. Inhibition of CSN5 repressed oncogenic activity of CSN5 in AR negative prostate cancer cells.
A, Cell proliferation in stable cells. Cell proliferation was detected in shRNAs infected PC3 cells. Sh36 and sh37 are shRNAs for CSN5. B,GSEA of cell cycle and E2F_targets pathway enrichment in shRNA-treated PC3 cells.C, Soft agar assays of stable PC3 cells. The colony numbers were normalized to those in the control cells. D, Invasion assays after cells were plated for 48 hours. E, EMT marker expression detected immunoblots in both NS and shCSN5 PC3 cells. F, EMT-up and metastasis_up pathway enrichment in shRNA-treated PC3 cells. G, mTOPRC1 signaling, glycolysis, and oxidative phosphorylation pathways enrichment in shRNA- treated PC3 cells. H, Targets of mTOPRC1 signaling, glycolysis, and oxidative phosphorylation pathways were validated by qRT-PCR in PC3 cells. I, RNA splicing and 186 prostate cancer SRG signature enrichment in sh37-treated PC3 cells. J, 74 commonly downregulated genes from prostate cancer cells and TCGA cohort. Commonly downregulated genes from LNCaP, C4–2, 22Rv1, and PC3 cells were intersected with genes positively correlated with CSN5 in TCGA cohort to generate 74- gene signature. K, The correlation of 74-gene signature with metastasis in MSKCC, Grasso, and Yu prostate cancer cohorts. L, The correlation of 74-gene signature expression with disease-free survival (MSKCC) and progression-free survival (TCGA). Figure values represent the mean ± SE of three independent experiments. ***, P < 0.001; vs. control groups treated with nonspecific (shNS) shRNA. Up (upregulated genes in sh37); Down (downregulated genes in sh37).
CSN5i-3, as the CSN5 inhibitor, showed potent tumor suppression of PC cell lines
To explore the therapeutic potential of targeting CSN5 in PC, we assessed the function of CSN5i-3, a novel selective and orally available CSN5 inhibitor (32). In seven PC cell lines, CSN5i-3 showed distinct potency regardless of AR status (Fig. 5A). C4–2 had the strongest response to CSN5i-3, while PC3 showed the weakest response. This observation is similar to previously reported responses to the NAE1 inhibitor MLN4024, which inhibits neddylation in PC cells (32). Next, we detected the effect of CSN5 inhibition on cullin-RING E3 ubiquitin ligase (CRL) substrates and substrate receptor modules (SRMs). Similar to previous reports, CSN5i-3 led to the accumulation of cullins in the neddylated, active state (Cul1, 2, 3 and 4A; Fig. S5A) and the reduction of SRMs (eg, Skp2 and Fbxo22; Fig. S5A). As the substrates of these CRLs, p21 and p27 were protected from degradation (Fig. S5A). Importantly, we found CSN5 itself was directly inhibited by CSN5i-3 treatment in all PC cells, suggesting CSN5 is the target of active CRL, similar to SRMs (Fig. S5A).
Figure 5. CSN5i-3 inhibited oncogenic activity of CSN5 in prostate cancer cells.
A, CSN5i-3 efficacy in 7 prostate cancer cells. Cell proliferation assay was performed after 3 days of treatment with CSN5i-3. B, GSEA of cell cycle and E2F_targets pathway in CSN5i-3 treated C4–2 cells. C and D, The regulation of AR signaling by CSN5i-3. AR signaling enrichment was analyzed by GSEA in C4–2 (1μM CSN5i-3) and LNCaP (5μM CSN5i-3) cells with CSN5i-3 treatment for 3 days (C). AR protein and AR targets were detected by western blot in C4–2, LNCaP, and 22Rv1 (5μM CSN5i-3) cells with either CSN5i-3 or MLN4924 (1μM) for 3 days (D). E and F, DNA damage evaluation with CSN5i-3 treatment. DNA repair pathway enrichment in CSN5i-3 treated C4–2 (E). DNA damage marker activation was monitored in C4–2 cells using Muse multicolor DNA damage kit (F). G, Apoptosis induced by CSN5i-3 treatment. Apoptosis was detected by Annexin V assays and immunoblots in C4–2 (1μM CSN5i-3) and PC3 (10μM CSN5i-3) cells. H, p53 signaling and apoptosis pathways enriched in C4–2 cells with CSN5i-3 treatment. I, Four oncogenic pathways were enriched in CSN5i-3 downregulated gene group in LNCaP, C4–2, 22Rv1, and PC3 cells. J, Invasion assays with CSN5i-3 treated PC3 cells. Invasion ability was detected after cells were treated with CSN5i-3 for 72 hours. K, EMT_up and metastasis_up pathway enrichment with CSN5i-3 treatment in PC3 cells. L, Regulation of prostate cancer SRGs by CSN5i-3 in PC3 cells: 36 genes among 186 prostate cancer SRGs were inhibited by CSN5i-3 in PC3; 4 genes of 13 unfavorable SRGs were detected by qRT-PCR in CSN5i-3 treated PC3 cells. M, Phosphokinase array analysis after 48-hour CSN5i-3 treatment in C4–2 cells. The whole-cell lysates were collected for human phosphokinase array analysis. Relative phosphorylation of spots was quantified by Image J software, and the value of vehicle (NC) was set up as “1.” Figure values represent the mean ± SE of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; vs. control groups (NC) treated with DMSO.
GSEA of transcriptomic changes showed that the cell cycle and E2F pathways were inhibited by CSN5i-3, which supported a cell growth inhibitory function (Figs. 5B, S5B–S5C). Consistent with knockdown of CSN5, CSN5i-3 suppressed AR signaling through directly inhibiting both AR and AR-V7 proteins (Figs. 5C–5D). The DNA repair pathway was inhibited by CSN5i-3 in C4–2 cells, leading to increased DNA damage (Figs. 5E–5F). Significant apoptosis was induced by CSN5i-3 in AR-positive cells but not AR-negative PC3 cells, supported by increased cleaved PARP (Figs. 5G, S5C). Both p53 signaling and apoptosis pathways were activated by CSN5i-3 in three AR-positive cells (Figs. 5H, S5D). Target genes of the p53 and apoptosis pathways were detected in three cell lines (Fig. S5E). Four oncogenic pathways (MYC, FOXM1, CTNNB1, and AUROR) were inhibited by CSN5i-3 in both AR-positive and AR-negative PC cells (Fig. 5I). The detailed GSEA results from C4–2 cells were shown as the representative data (Fig. S5F). Partial target genes of these pathways were validated in C4–2 cells (Fig. S5G).
In PC3 cells, similar to shCSN5, CSN5i-3 attenuated PC3 invasiveness by 80% by repressing EMT markers, compared to the control (Figs. 5J, S5H), supporting the role of CSN5i-3 in inhibiting metastasis pathways (Fig. 5K). Furthermore, CSN5i-3 downregulated 36 of 186 PC SRGs. Among 186 PC SRGs, 13 were clinically unfavorable genes and 13 were favorable (31). There were four common genes between the 13 unfavorable SRGs and the 36 downregulated CSN5i-3 PC SRGs, but no common genes with the 13 favorable SRGs (Fig. 5L). Four SRGs that are correlated with poor clinical outcomes were significantly inhibited by CSN5i-3 in PC3 cells (Fig. 5L).
Similar to previous findings, our data showed that CSN5 inhibition repressed multiple signaling pathways. Therefore, we applied the phosphokinase array in CSN5i-3-treated C4–2 cells (Figs. 5M, S5G). The array detected the phosphorylation of 37 human kinases involved in multiple key oncogenic signaling processes. CSN5i-3 significantly inhibited SRC family signaling (eg, SRC, YES), leading to the correlated pathways inhibition, including STAT signaling, and PLCG1 and RSK phosphorylation (Figs. 5M, S5I). Furthermore, inhibition of PDGFR and CREB activation by CSN5i-3 led to the inhibition of downstream PRAS40 and GSK-3B phosphorylation (Figs. 5M, S5I). Multiple kinase phosphorylation was validated in C4–2 with CSN5i-3 treatment (Fig. S5J). The crosstalk of the oncogenic signaling extends the impact of CSN5 inhibition to perform broader control of signal transduction.
CSN5i-3 showed synergy with PARP inhibitors in vitro and potent tumor suppression in vivo
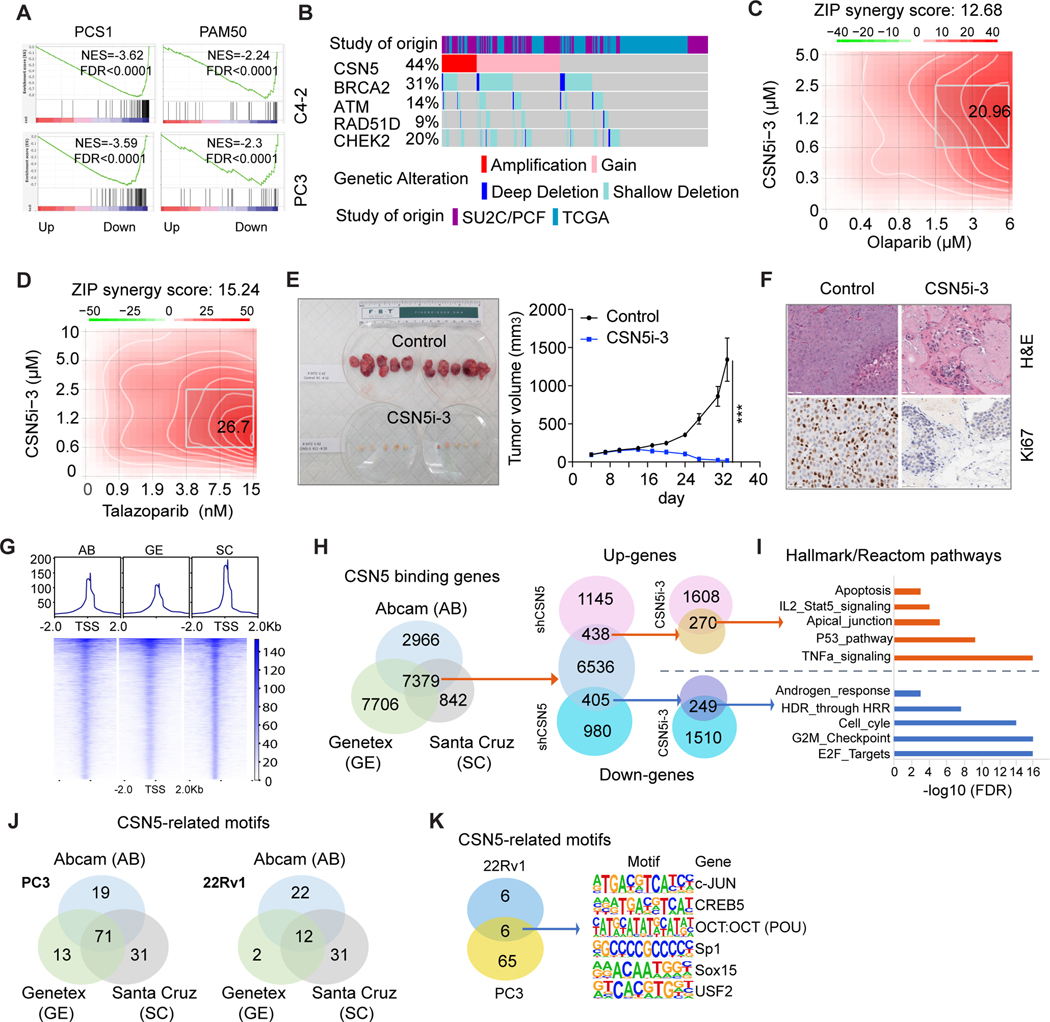
Because we revealed that CSN5 inhibition can target multiple fundamental pathways regulating tumor survival and progression, we asked whether CSN5i-3 could target aggressive molecular signatures of PC. Two PC signatures, including PC subtypes 1 (PCS1) and PAM50, were applied in our transcriptomic dataset from C4–2 and PC3 cells. PCS1, generated from over 4600 PC specimens, has been confirmed to be highly correlated with aggressive tumors (33). The PAM50 signature, commercially developed to assess breast cancer risk (34), was recently used to categorize PC into three subtypes (luminal A, luminal B, and basal) in the cohorts including 3782 samples (35). GSEA showed CSN5i-3 markedly inhibited both the PCS1 and PAM50 signatures in C4–2 and PC3 transcriptomic datasets (Fig. 6A).
Figure 6. CSN5i-3 synergized with PARPi and strongly inhibited xenograft tumor growth.
. A, CSN5i-3 targeted PCS1 and PAM50 aggressive signatures. PCS1 and PAM50 signature enrichment was analyzed in CSN5i-3-treated C4–2 cells. B, Co- occurrence of genomic alteration of CSN5 and DNA repair genes. Copy number alterations of CSN5 and DNA repair genes (BRCA2, ATM, RAD51D, CHEK2) were analyzed in TCGA and SU2C cohorts by using cBioPortal. C and D, Synergy assay of CSN5i-3 and PARPis (olaparib (C) or talazoparib (D)) in PC3 cells. SynergyFinder was used to analyze the synergy effects of combinations of CSN5i-3 and PARPis after 6 days of treatment in PC3 cells. E, Antitumor effects of CSN5i-3 in vivo. Tumor volumes were measured following oral administration of CSN5i-3 (150 mg/kg) in established C4– 2 xenograft tumors (n=10 per group). Values are means ±SE. ***, P < 0.001 versus vehicle mice. F, Ki67 expression in xenograft tumors by IHC staining. G, CSN5 CUT&Tag analysis in 22Rv1 cells. CSN5 binding signal was shown in Tornado and composite plots. Three CSN5 antibodies from Abcam, Genetex, and Santa Cruz were used to pull down CSN5-binding genes in CUT&Tag. H, Identification of CSN5 targets. CSN5 targets were identified by integrating CUT&Tag with the RNA-seq dataset from shCSN5- and CSN5i-3-treated 22Rv1 cells. I, Common transcriptionally regulated pathways by inhibition of CSN5. GSEA was used for pathway analysis by using Hallmark and Reactom pathway signatures among CSN5 targets in 22Rv1 cells. J and K, Motif enrichment analysis of CUT&Tag with three CSN5 antibodies in PC3 and 22Rv1 cells; 71 (in PC3) and 12 (in 22Rv1) common CSN5-reglated TF motifs were identified (J). Six common motif sequences from two prostate cancer cells were listed (K).
Amplification/gain of CSN5 is highly co-occurrent with BRCA2 deletion/loss in both primary and metastatic PC (Figs. 6B, S6A). Considering the positive roles of CSN5 in DNA repair, amplification of CSN5 could provide a protective mechanism to overcome DNA damage in BRCA2-deficiency PC. Inhibition of CSN5 induced significant DNA damage through inhibition of DNA repair genes in PC cells (Figs. 3D–3E, 5E–5F). We hypothesized that CSN5i-3 and PARPi may have synergistic effects in PC cells. PC3 is more resistant to CSN5i-3 compared to PC cells (Fig. 5A); therefore, we performed synergy assays in PC3 with combinations of CSN5i-3 and PARPis (olaparib/talazoparib). While 6μM olaparib induced an 11.57% inhibition of cell growth, 0.6μM CSN5i-3 induced growth inhibition of 1.59%. The combination of these two treatments led to a 42.16% inhibition of cell growth (Fig. S6B). Similarly, a combination of 15 nM talazoparib and 0.6 μM CSN5i-3 increased growth inhibition from 6.76% (by talazoparib alone) to 44.53% (Fig. S6B). SynergyFinder analysis showed strong synergistic effects in this combination. The average of Zip synergy score for olaparib is 12.68, and the most synergistic area score is 20.96 (Fig. 6C). The average of Zip synergy score for talazoparib is 15.24, and the most synergistic area score is 26.7 (Fig. 6C). These results were confirmed in C4–2 cells (Figs. S6C–S6D). To define the mechanism of this synergism, we explored expression of multiple DNA repair genes in both PC3 and C4–2 cells. All the tested genes were significantly inhibited by CSN5i-3 (Fig. S6E), suggesting the synthetic lethality between CSN5 and PARPi.
Last, we evaluated the antitumor activity of CSN5i-3 in vivo. After 29 days of treatment, tumor growth in CSN5i-3-treated mice was significantly inhibited. Tumor size in CSN5i-3-treated mice averaged 18.85 mm3, while tumors of the vehicle group reached to 1342.9 mm3 (Fig. 6E), without significant difference in body weight of the mice (Fig. S6F). Tumor suppression by CSN5i-3 was further confirmed by reduced Ki67 IHC staining in tumor tissues (Fig. 6F). We assessed the potent antitumor activity of CSN5i-3 in vitro and in vivo, and defined the underlying molecular mechanisms of CSN5i-3 in PC cells.
The transcription cofactor activity of CSN5 in PC cells
CSN5 was originally identified as the transcription cofactor of c-Jun/AP-1 (36). Although CSN5 interacts with and activates a number of steroid hormone receptors, including AR (10), there has been no further investigation of CSN5 as a transcription cofactor. Our transcriptomic studies showed CSN5 inhibition induced global RNA expression changes, in agreement with its oncogenic activity. Similar observations were reported in other cancer types (12, 37). To further define the transcription cofactor activity of CSN5, we applied CUT&Tag analysis in 22Rv1 and PC3 cells. Three CSN5 antibodies (from Abcam, Genetex, and Santa Cruz) were used to pull down CSN5 binding genes in the CUT&Tag analysis. Heat maps showed high signal to noise at the concordant peaks (Fig. 6G). There were 7379 common genes in 22Rv1 cells that were pulled down by three CSN5 antibodies (Fig. 6H). We integrated these 7379 genes with genes regulated by shCSN5 and CSN5i-3. Of these, 249 downregulated genes and 270 upregulated genes were shared in groups (Fig. 6H, Table S3). Hallmark and pathway analysis showed cell cycle, DNA repair pathways, and AR signaling were significantly enriched in 249 downregulated genes, while p53 signaling, apoptosis, and TNFα signaling were enriched in 270 upregulated genes (Fig. 6I). The CUT&Tag dataset from PC3 cells showed 5408 common genes shared by the three antibodies (Fig. S6G). We intersected the 5408 genes with downregulated genes by shCSN5 and CSN5i-3 in PC3 (Fig. S6H) to produce 51 common genes that were highly enriched in cell cycle and metastasis pathways (Fig. S6I). Altogether, we identified transcriptional regulated targets by CSN5 in PC cells.
As a transcription cofactor, CSN5 does not directly bind to target DNA. Therefore, we used HOMER to investigate CSN5-specific sites for enrichment of transcription factor DNA-binding motifs. This analysis revealed significant enrichment for motifs of the homeobox, forkhead, and sox transcription factor (TF) families (Tables S4 and S5). In PC3, 70 CSN5-related TF motifs were shared among three CSN5 antibodies. In 22Rv1, 12 TF motifs were common (Fig. 6J). Six CSN5-related TF motifs were shared between PC3 and 22Rv1 (Fig. 6K). c-Jun motif enrichment ranked in the top 10 among six CUT&Tag analyses in two PC cell lines, consistent with previous reports of the c-Jun co-activator function of CSN5 (36). Furthermore, CREB5 and USF2 were proven to partner with AR or FOXA1 to affect PC progression (38, 39). Our data indicated that CSN5 not only directly regulated AR expression but also collaborated with AR partner TFs to modulate AR signaling.
Discussion
Amplification and overexpression of CSN5 have been reported in multiple cancer types (16). Study of its oncogenic functions has mainly focused on its deneddylation activity. CSN5 regulates broad biological processes such as cell proliferation, cell cycle, DNA repair, apoptosis, and hypoxia (15). To date, there has been no comprehensive study of the clinical relevance and molecular functions of CSN5 in PC. In this study, we revealed the significant clinical relevance of CSN5 amplification and overexpression in 16 clinical PC cohorts. Transcriptomic analysis defined the molecular mechanisms supporting the phenotypes induced by CSN5 inhibition. CSN5 regulates AR signaling by directly affecting both wild-type AR and AR-V7 protein levels. CNS5 inhibition also significantly targeted multiple oncogenic pathways (MYC, FOXM1, CTNNB1, AUROR signaling) and RNA slicing in PC cells. Importantly, CSN5 inhibition showed potent synergistic effects with PARPis and could target aggressive PC signatures. Because it can target key cancer-related pathways, the CSN5 inhibitor showed remarkable antitumor efficacy in the xenograft tumor model. For the first time, we explored the transcription cofactor activity of CSN5 in cancer cells and identified multiple transcription factors collaborating with CSN5 to regulate PC progression.
PC initiation and progression is mainly driven by AR signaling. Androgen ablation/AR inhibition is the mainstay of PC treatment. However, several resistance mechanisms to AR-targeting therapy have been reported, including AR amplification, mutation, and splicing variants (40). Overexpression of AR by amplification of the AR gene results in high AR activity despite castrate levels of androgens in the circulation (41). AR variants such as AR-V7 are truncated AR proteins showing constitutive AR activity independent of AR ligand binding (42). Instead of targeting AR activity, direct inhibition of AR and AR variant proteins can overcome the resistance. The novel AR protein degrader ARV-110, currently in a phase 2 expansion trial, can induce full-length AR and AR-V7 degradation (43). Our study demonstrated that inhibition of CSN5 by either shRNA or CSN5i-3 induced significant AR and AR-V7 protein inhibition, leading to the inhibition of AR signaling (Figs. 3C, S3D, 5C–5D).
Although AR signaling is at the center of the development of therapeutic resistance and PC progression, AR-independent pathways, such as some oncogenic signaling (MYC, PI3K, mTOR, WNT, AUROR signaling), metabolic pathways, and spliceosome regulation, also play important roles (Figs. 3–5). Inhibition of CSN5 can significantly repress multiple key oncogenic pathways in PC progression (eg, MYC, FOXM1, CTNNB1, and AUROR signaling) regardless of AR status. CSN5 was reported to directly regulate MYC transcriptional activity, but increased MYC protein level by affecting MYC turnover rate in breast cancer (44). However, we found CSN5 significantly inhibited MYC protein and MYC signaling targets in PC. Depletion of CSN5 in colorectal cancer cells inhibited WNT signaling by downregulation of CTNNB1 (45), but inhibition of CSN5 inhibited β-catenin (CTNNB1) signaling without affecting CTNNB1 protein levels in PC cells (Fig. S3H). CSN5 regulates multiple oncogenic pathways in a manner specific to cancer types.
In AR-negative PC3 cells, inhibition of CSN5 significantly attenuated metabolic pathways, including oxidative phosphorylation and glycolysis, both of which contribute to PC progression (Fig. 4G). Dysregulated RNA splicing is predominant in human cancers and many cancer-specific splicing events are major contributors to disease development and progression (46). In PC, 186 SRGs have been identified as dysregulated (31). We demonstrated that inhibition of CSN5 could target the 186-SRG signature in PC cells (Figs. 4I, 5L). MYC has been proven to regulate the spliceosome in PC (47, 48). CSN5 could directly or indirectly regulate the SRG signature through inhibition of MYC.
CSN5 plays important roles in DNA repair by regulating multiple key components (27). Interestingly, CNA of CSN5 and BRCA2 are highly correlated in clinical samples, suggesting that CSN5 could provide protection for BRCA2-deficiency PCs that are susceptible to DNA damage. Therefore, targeting CSN5 could lead to synthetic lethality with PARPi treatment. Consistent with other studies, we demonstrated that CSN5 inhibition induced significant DNA damage by inhibition of multiple DNA repair genes (eg, RAD51, ZWINT, BRCA1, and BRCA2). These results provided the mechanistic support for a strong synergistic effect of the combination of CSN5i-3 and PARPis in PC cells.
Even though CSN5 was first identified as the coactivator of c-Jun/AP1, most studies focus on its deneddylation activity (49, 16). Our study is the first systematic investigation of how CSN5 functions as a transcriptional cofactor. Transcriptomic changes induced by CSN5 inhibition provided molecular mechanisms for the phenotypes related to CSN5 function, supporting the role of CSN5 in protein stability and control of gene expression. Motif enrichment analysis revealed that CSN5 may participate in transcriptional regulation of multiple TF families (Table S4 and S5). For the first time, we identified five TF motifs that were enriched with CSN5 binding in AR-positive/negative PC cells. Upon treatment with enzalutamide, CREB5 can enhance AR activity at a subset of promoters and enhancers, including MYC and genes involved in the cell cycle (38), which is consistent with the function of CSN5 that we demonstrated. USF2 was reported to be one of the components of the FOXA1/AR transcriptional protein complex (39). Sp1 regulates important genes like AR, TGF-β, c-Met, FASN, and α-integrin, suggesting the importance of Sp1 in PC (50). All the potential targets of CSN5 still need further validation.The transcription cofactor activity of CSN5 could be another important aspect of CSN5 functions besides its deneddlyation activity in cancer.
In summary, our study integrated data from cell lines with clinical cohorts to reveal that amplification and overexpression of CSN5 is associated with poor outcomes, and its oncogenic activity is a major contributor to aggressive PC. Targeting CSN5 inhibits oncogenic activity in vivo. Furthermore, we defined the transcriptional cofactor activity of CSN5 beyond its deneddylation function.
Supplementary Material
Acknowledgements:
This research was funded by a Department of Defense Prostate Cancer Research Program Translational Science Award (W81XWH-20-1-0114 to P.W. Kantoff) and in part through the NIH/NCI Cancer Center Support Grant to Memorial Sloan Kettering Cancer Center (P30 CA008748). We acknowledge that Dr. Eva Altmann from Novartis Institutes for BioMedical Research (Switzerland) kindly provided CSN5i-3 for the study; Dr. Lorelei A. Mucci from the Department of Epidemiology, Harvard T.H. Chan School of Public Health (Boston, Massachusetts) kindly provided information on the PHS/HPFS cohorts. We thank Margaret McPartland for editing and members of the Kantoff laboratory for help and discussion.
Funding:
This research was funded by a Department of Defense Prostate Cancer Research Program Translational Science Award (W81XWH-20-1-0114 to P.W. Kantoff) and in part through the NIH/NCI Cancer Center Support Grant to Memorial Sloan Kettering Cancer Center (P30 CA008748).
Disclosure of potential conflicts of interest:
As of August 5, 2021, P.W. Kantoff reports the following disclosures for the last 24-month period: he has investment interest in Cogent Biosciences, Context Therapeutics LLC, DRGT, Mirati, Placon, PrognomIQ, Seer Biosciences, SnyDevRx and XLink; he is a company board member for Context Therapeutics LLC; he is a company founder for XLink; he is co-founder and CEO of Convergent Therapeutics, he is/was a consultant/scientific advisory board member for Anji, Bavarian Nordic Immunotherapeutics, Candel, DRGT, Immunis, AI (previously OncoCellMDX), Janssen, Progenity, PrognomIQ, Seer Biosciences, SynDevRX, Tarveda Therapeutics, and Veru, and serves on data safety monitoring boards for Genentech/Roche and Merck. He reports spousal association with Bayer.
References
- 1.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol 2008;8:440–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol 2005;23:8253–61. [DOI] [PubMed] [Google Scholar]
- 3.Loriot Y, Bianchini D, Ileana E, Sandhu S, Patrikidou A, Pezaro C, et al. Antitumour activity of abiraterone acetate against metastatic castration-resistant prostate cancer progressing after docetaxel and enzalutamide (MDV3100). Ann Oncol 2013;24:1807–1812. [DOI] [PubMed] [Google Scholar]
- 4.Azad AA, Eigl BJ, Murray RN, Kollmannsberger C, Chi KN. Efficacy of enzalutamide following abiraterone acetate in chemotherapy-naive metastatic castration-resistant prostate cancer patients. Eur Urol 2015;67:23–29. [DOI] [PubMed] [Google Scholar]
- 5.Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, et al. Punctuated evolution of prostate cancer genomes. Cell 2013;153:666–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, et al. The genomic complexity of primary human prostate cancer. Nature 2011;470:214–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.: Abida W, Cyrta J, Heller G, Prandi D, Armenia J, Coleman I, et al. Genomic correlates of clinical outcome in advanced prostate cancer. Proc Natl Acad Sci U S A 2019;116:11428–11436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Macoska JA, Trybus TM, Wojno KJ. 8p22 loss concurrent with 8c gain is associated with poor outcome in prostate cancer. Urology 2000;55:776–82. [DOI] [PubMed] [Google Scholar]
- 9.Sato K, Qian J, Slezak JM, Lieber MM, Bostwick DG, Bergstralh EJ, et al. Clinical significance of alterations of chromosome 8 in high-grade, advanced, nonmetastatic prostate carcinoma. J Natl Cancer Inst 1999;91:1574–80. [DOI] [PubMed] [Google Scholar]
- 10.Chauchereau A, Georgiakaki M, Perrin-Wolff M, Milgrom E, Loosfelt H. JAB1 interacts with both the progesterone receptor and SRC-1. J Biol Chem 2000;275:8540–8. [DOI] [PubMed] [Google Scholar]
- 11.Belandia B, Powell SM, García-Pedrero JM, Walker MM, Bevan CL, Parker MG. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Mol Cell Biol 2005;25:1425–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adler AS, Lin M, Horlings H, Nuyten DS, van de Vijver MJ, Chang HY. Genetic regulators of large-scale transcriptional signatures in cancer. Nat Genet 2006;38:421–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Civenni G, Albino D, Shinde D, Vázquez R, Merulla J, Kokanovic A, et al. Transcriptional reprogramming and novel therapeutic approaches for targeting prostate cancer stem cells. Front Oncol 2019;9:385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 2005;6:9–20. [DOI] [PubMed] [Google Scholar]
- 15.Guo Z, Wang Y, Zhao Y, Shu Y, Liu Z, Zhou H, et al. The pivotal oncogenic role of Jab1/CSN5 and its therapeutic implications in human cancer. Gene 2019;687:219–227. [DOI] [PubMed] [Google Scholar]
- 16.Liu G, Claret FX, Zhou F, Pan Y. Jab1/COPS5 as a novel biomarker for diagnosis, prognosis, therapy prediction and therapeutic tools for human cancer. Front Pharmacol 2018;9:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu R, Hu X, Zhou J, Sun J, Zhu AZ, Xu X, et al. COPS5 amplification and overexpression confers tamoxifen-resistance in ERα-positive breast cancer by degradation of NCoR. Nat Commun 2016;7:12044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzu YZ, Armenia J, Chakraborty G, Yoshikawa Y, Coggins SA, Nandakumar S, et al. A novel mechanism driving poor-prognosis prostate cancer: overexpression of the DNA repair gene, ribonucleotide reductase small subunit M2 (RRM2). Clin Cancer Res 2019;25:4480–4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res 2020;48:W488–W493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 2004;6:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009;462:108–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Armenia J, Wankowicz SAM, Liu D, Gao J, Kundra R, Reznik E, et al. The long tail of oncogenic drivers in prostate cancer. Nat Genet 2018;50:645–651. Erratum in: Nat Genet. 2019;51:1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 2004;5:45–54. [DOI] [PubMed] [Google Scholar]
- 27.Pan Y, Yang H, Claret FX. Emerging roles of Jab1/CSN5 in DNA damage response, DNA repair, and cancer. Cancer Biol Ther 2014;15:256–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yecies JL, Manning BD. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med (Berl) 2011;89:221–8. [DOI] [PubMed] [Google Scholar]
- 29.Altomare DA, Khaled AR. Homeostasis and the importance for a balance between AKT/mTOR activity and intracellular signaling. Curr Med Chem 2012;19:3748–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sveen A, Kilpinen S, Ruusulehto A, Lothe RA, Skotheim RI. Aberrant RNA splicing in cancer; expression changes and driver mutations of splicing factor genes. Oncogene 2016;35:2413–27. [DOI] [PubMed] [Google Scholar]
- 31.Zhang D, Hu Q, Liu X, Ji Y, Chao HP, Liu Y, et al. Intron retention is a hallmark and spliceosome represents a therapeutic vulnerability in aggressive prostate cancer. Nat Commun 2020;11:2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Han S, Wilder-Romans K, Sun GY, Zhu H, Liu X, et al. Neddylation inactivation represses androgen receptor transcription and inhibits growth, survival and invasion of prostate cancer cells. Neoplasia 2020;22:192–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.You S, Knudsen BS, Erho N, Alshalalfa M, Takhar M, Al-Deen Ashab H, et al. Integrated classification of prostate cancer reveals a novel luminal subtype with poor outcome. Cancer Res 2016;76:4948–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nielsen T, Wallden B, Schaper C, Ferree S, Liu S, Gao D, et al. Analytical validation of the PAM50-based Prosigna Breast Cancer Prognostic Gene Signature Assay and nCounter Analysis System using formalin-fixed paraffin-embedded breast tumor specimens. BMC Cancer 2014;14:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao SG, Chang SL, Erho N, Yu M, Lehrer J, Alshalalfa M, et al. Associations of Luminal and Basal Subtyping of Prostate Cancer With Prognosis and Response to Androgen Deprivation Therapy. JAMA Oncol 2017;3:1663–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Claret FX, Hibi M, Dhut S, Toda T, Karin M. A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 1996;383:453–7. [DOI] [PubMed] [Google Scholar]
- 37.Danielpour D, Purighalla S, Wang E, Zmina PM, Sarkar A, Zhou G. JAB1/COPS5 is a putative oncogene that controls critical oncoproteins deregulated in prostate cancer. Biochem Biophys Res Commun 2019;518:374–380. [DOI] [PubMed] [Google Scholar]
- 38.Hwang JH, Seo JH, Beshiri ML, Wankowicz S, Liu D, Cheung A, et al. CREB5 promotes resistance to androgen-receptor antagonists and androgen deprivation in prostate cancer. Cell Rep 2019;29:2355–2370.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Q, Yu X, Degraff DJ, Matusik RJ. Upstream stimulatory factor 2, a novel FoxA1-interacting protein, is involved in prostate-specific gene expression. Mol Endocrinol 2009;23:2038–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chandrasekar T, Yang JC, Gao AC, Evans CP. Mechanisms of resistance in castration-resistant prostate cancer (CRPC). Transl Androl Urol 2015;4:365–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Devlies W, Handle F, Devos G, Joniau S, Claessens F. Preclinical models in prostate cancer: resistance to AR targeting therapies in prostate cancer. Cancers (Basel) 2021;13:915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res 2009;69:16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mullard A. Targeted degraders clear first safety hurdles. Nat Rev Drug Discov 2020;19:435. [DOI] [PubMed] [Google Scholar]
- 44.Adler AS, Littlepage LE, Lin M, Kawahara TL, Wong DJ, Werb Z, et al. CSN5 isopeptidase activity links COP9 signalosome activation to breast cancer progression. Cancer Res 2008;68:506–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nishimoto A, Takemoto Y, Saito T, Kurazumi H, Tanaka T, Harada E, et al. Nuclear β-catenin expression is positively regulated by JAB1 in human colorectal cancer cells. Biochem Biophys Res Commun 2020;533:548–552. [DOI] [PubMed] [Google Scholar]
- 46.El Marabti E, Younis I. The cancer spliceome: reprograming of alternative splicing in cancer. Front Mol Biosci 2018;5:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caggiano C, Pieraccioli M, Panzeri V, Sette C, Bielli P. c-MYC empowers transcription and productive splicing of the oncogenic splicing factor Sam68 in cancer. Nucleic Acids Res 2019;47:6160–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phillips JW, Pan Y, Tsai BL, Xie Z, Demirdjian L, Xiao W, et al. Pathway-guided analysis identifies Myc-dependent alternative pre-mRNA splicing in aggressive prostate cancers. Proc Natl Acad Sci U S A 2020;117:5269–5279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomoda K, Kubota Y, Kato J. Degradation of the cyclin-dependent-kinase inhibitor p27Kip1 is instigated by Jab1. Nature 1999;398:160–5. [DOI] [PubMed] [Google Scholar]
- 50.Sankpal UT, Goodison S, Abdelrahim M, Basha R. Targeting Sp1 transcription factors in prostate cancer therapy. Med Chem 2011;7:518–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.