HDAC2 has a context-specific role in undifferentiated PDAC and the capacity to disseminate systemically, implicating HDAC2 as targetable protein to prevent metastasis.
Abstract
The mortality of patients with pancreatic ductal adenocarcinoma (PDAC) is strongly associated with metastasis, a multistep process that is incompletely understood in this disease. Although genetic drivers of PDAC metastasis have not been defined, transcriptional and epigenetic rewiring can contribute to the metastatic process. The epigenetic eraser histone deacetylase 2 (HDAC2) has been connected to less differentiated PDAC, but the function of HDAC2 in PDAC has not been comprehensively evaluated. Using genetically defined models, we show that HDAC2 is a cellular fitness factor that controls cell cycle in vitro and metastasis in vivo, particularly in undifferentiated, mesenchymal PDAC cells. Unbiased expression profiling detected a core set of HDAC2-regulated genes. HDAC2 controlled expression of several prosurvival receptor tyrosine kinases connected to mesenchymal PDAC, including PDGFRα, PDGFRβ, and EGFR. The HDAC2-maintained program disabled the tumor-suppressive arm of the TGFβ pathway, explaining impaired metastasis formation of HDAC2-deficient PDAC. These data identify HDAC2 as a tractable player in the PDAC metastatic cascade. The complexity of the function of epigenetic regulators like HDAC2 implicates that an increased understanding of these proteins is needed for implementation of effective epigenetic therapies.
Significance:
HDAC2 has a context-specific role in undifferentiated PDAC and the capacity to disseminate systemically, implicating HDAC2 as targetable protein to prevent metastasis.
Introduction
The mechanisms driving pancreatic ductal adenocarcinoma (PDAC) metastasis are an example of how epigenetic and transcriptional rewiring orchestrate cellular plasticity processes. Although genomic doublings and allelic imbalances in favor of the oncogenic KRAS allele (1, 2) were described, selection of genetic alterations driving metastasis rarely occurs in PDAC (3). At the same time, however, the epigenetic landscape becomes fundamentally changed during metastasis to serve the needs of disseminated cells (4, 5). Consistently, transcription factors (TF), including Forkhead family TFs (5), BLIMP1 (6), RUNX3 (7), ZEB1 (8), PRRX1 (9), or deltaNp63 (10, 11), which reprogram the chromatin landscape, are linked to an undifferentiated phenotype and the metastatic cascade. One way of PDAC disease progression involves a dynamic reduction of epithelial gene expression and acquisition of a more mesenchymal expression program, linked to metastasis (8, 12–15). Such a note is underscored by subtyping efforts of PDAC, in which the more aggressive basal-like subtype enriches TGFβ signaling and epithelial-to-mesenchymal transition (EMT) gene signatures (2). EMT and TGFβ signaling signatures also enrich in the recently described mesenchymal PDAC subtype (16). A portion of the basal-like subtype, basal-like A cancers, shows a higher metastasis probability (2). However, the processes permissive for the metastatic cascade remain incompletely understood and there is a need to decipher tractable metastasis regulators.
Zn2+-dependent histone deacetylases (HDAC) are classified into class I to IV enzymes, whereby HDACs 1, 2, 3, and 8 represent class I, HDAC 4, 5, 6, 7, 9, and 10 compose class II, and HDAC11 is the only class IV enzyme (17). The class III deacetylases cover the NAD+-dependent deacetylases SIRT1–7. Molecular functions of these enzymes are often investigated using various HDAC inhibitors (HDACi). Here, limitations to connect a function to one specific isoenzyme are evident, because HDACi block several HDACs to a variable extent.
Since we (18) and others (19) described a connection of HDAC2 to poorly differentiated human PDAC, we set up experiments to analyze the function of the epigenetic eraser HDAC2. We show that HDAC2 maintains the expression of genes, including receptor tyrosine kinase (RTK) genes that are relevant in undifferentiated cancers. Furthermore, we found that the HDAC2-controlled program protects cancer cells from the tumor-suppressive action of TGFβ and ultimately facilitates metastasis.
Materials and Methods
Compounds
TGFβ was purchased from PeproTech. 4-Hydroxytamoxifen (4-OHT) and α-tocopherol were purchased from Sigma. N-Acetylcysteine (NAC) was purchased from Thermo Fisher Scientific.
Mouse lines
Mouse lines are described in Supplementary Materials and Methods. All animals were bred on a mixed C57Bl/6;129S6/SvEv (RRID:MGI:5657570) genetic background. Animal studies were conducted in compliance with European guidelines for the care and use of laboratory animals and were approved by the Institutional Animal Care and Use Committees of the Technische Universität München and Regierung von Oberbayern.
Cell lines
Generation of murine PDAC cell lines has been described previously (20). Culturing is described in Supplementary Materials and Methods. All cell lines were isolated and established in the Department of Internal Medicine II, Klinikum rechts der Isar, Technische Universität München and cultivated for less than 30 passages after isolation (Supplementary Table S1). For the experiments, the cells were cultivated for a maximum of 12 passages before returning to another vial of the same stock. The identity of the murine PDAC cell lines was verified using genotyping PCR and the cell lines were tested for Mycoplasma contamination by a PCR-based methodology using a multiplex PCR strategy once per month as described in Supplementary Materials and Methods. To activate CreERT2, PDAC cells were treated with vehicle (ethanol) or 600 nmol/L 4-OHT for 8 days. Afterwards, cells were left for 3 days to recover before being used for individual assays.
Differential trypsinization
Separating epithelial and mesenchymal fractions of the cell lines was performed as described previously (1) and in Supplementary Materials and Methods.
Viability assay, flow cytometry of cell cycle, ROS, repopulation assay, and Annexin V assay
Viability assay by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and clonogenic assays were done as decribed (21) and reported in Supplementary Materials and Methods. Pancreatic cancer spheres were generated by culturing pancreatic cancer cells (100,000 cells/200 μL) in low-attachment 96-well plates (#3474, Corning, Costar). After 96 hours, pictures were taken with the stereo microscope Leica M205 FA (Leica Microsystems). For subsequent analysis, Image J (ImageJ, RRID:SCR_003070) was used. Spheres were analyzed by counts, total area, and average size. Annexin V staining, cell-cycle assay, and repopulation assays have been described previously (21) and are stated in Supplementary Materials and Methods. CellROX Deep Red Flow Cytometry Assay Kit (#C10491, Thermo Fisher Scientific) was used according to manufacturer's instructions. All samples were measured by flow cytometry on a Gallios Flow Cytometer (Beckman Coulter) and data were analyzed with the FlowJo software (FlowJo, LLC, RRID:SCR_008520). Repopulation assays of Hdac2-deficient and proficient cells is described in ref. 21 and detailed information can be found in the Supplementary Materials and Methods.
Caspase activity
Caspase activity was measured using Caspase-Glo 3/7 Assay (#G8091, Promega) and normalized to CellTiter-Glo Luminescent Cell Viability Assay (#G7570, Promega). Details are described in Supplementary Materials and Methods.
Vectors and transduction
Generation of the pLEX_305-N-dTAGEGFP vector is described in Supplementary Materials and Methods. Production of the lentivirus is described in Supplementary Materials and Methods. A total of 100,000–150,000 PPT-F1648 cells were seeded in two wells of a 6-well plate. One milliliter of the lentivirus containing medium was added together with 8 μg/mL polybrene. After 8 hours, 1 mL of medium with 10% FBS was supplemented and cultured overnight. Medium was refreshed and after additional 24 hours and cells were selected with 8 μg/mL puromycin (Invivogen, #ant-pr-1) for 48 hours.
Cell lysis and Western blot analysis
For whole-cell extracts (WCE), cell lysis buffer (#9803S, Cell Signaling Technology) supplemented with protease and phosphatase inhibitors (cOmplete, EDTA-free Protease Inhibitor Cocktail, Roche Diagnostics) and Phosphatase-Inhibitor-Mix I, Serva) was used. Western blotting has been described previously (21) and detailed information can be obtained in Supplementary Materials and Methods. Antibodies and antibody dilutions can be found in Supplementary Table S1.
Quantitative reverse transcriptase PCR
Total RNA was isolated from PDAC cell lines using the Maxwell 16 LEV simply RNA Purification Kit (Promega) following the manufacturer's instructions. Quantitative mRNA analysis was performed using a real-time PCR analysis system (RRID:SCR_015805; TaqMan, PE StepOnePlus, Real-Time PCR System; Applied Biosystems Inc.) with Promega GoTaq qPCR Master Mix as fluorescent DNA binding dye. Data analysis was carried out with StepOne software (RRID:SCR_014281, Applied Biosystem; Thermo Fisher Scientific) according to the ΔΔCt method. qPCR primers are depicted in Supplementary Table S1.
Histochemistry and IHC
For histopathologic analysis, murine tissues were fixed in 4% formaldehyde (Carl Roth), embedded in paraffin, and sectioned (thickness: 2.5 μm). Tissues were stained with hematoxylin and eosin as described previously (22). IHC is described in Supplementary Materials and Methods.
Metastasis frequency
Lungs and livers were investigated macroscopically for metastases at necropsy. For microscopical detection of metastasis, liver, and lung tissues of Hdac2-proficient and -deficient KPC mice were fixed in 4% formaldehyde (Carl Roth), and embedded in paraffin. The tissues were cut into serial sections (10 sections at 2.5 μm; = series 1) separated by 100 μm between each series. Per organ and mouse, at least 10 series were cut. The first sections of each series were hematoxylin and eosin (H&E) stained and screened for metastasis. Local invasion from the primary tumor to the liver was excluded and not counted.
RNA sequencing and microarray analyses, Venn diagrams and heatmap, gene set enrichment analysis, and datasets
RNA sequencing (RNA-seq) analysis has been described previously (21, 23) and are detailed reported in Supplementary Materials and Methods. Venn diagrams were generated with https://bioinfogp.cnb.csic.es/tools/venny/ (RRID:SCR_016561). Analysis of PDAC datasets can be found in Supplementary Materials and Methods. Gene set enrichment analysis (GSEA) was performed via Molecular Signature Database (MSigDb; https://www.gsea-msigdb.org/gsea/msigdb/annotate.jsp; RRID:SCR_016863), GSEA tool (RRID:SCR_003199), Enrichr (RRID:SCR_001575; https://amp.pharm.mssm.edu/Enrichr/) or Gene Trail3 (RRID:SCR_006250). Microarray mRNA expression data of Hdac2-deficient and -proficient murine PDAC cells can be accessed on GEO (GSE144798). RNA-seq data of 4-OHT and vehicle-treated PPT-F1648 cells can be accessed on ENA (PRJEB35204). Data of TGFβ-treated Hdac2-deficient and -proficient murine PDAC lines can be accessed on ENA (PRJEB47385).
Assay for transposase-accessible chromatin using sequencing and chromatin immunoprecipitation sequencing
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) was done as described previously (24, 25). See details for the analysis in Supplementary Materials and Methods. The following antibodies were used: H3K27ac (Diagenode; Pab-174–050, RRID:AB_2716835) and HDAC2 (HDAC2, D6S5P, Cell Signaling Technology, mAb #57156, RRID:AB_2756828).
Low-coverage whole-genome sequencing
Purified DNA was used as input for library preparation with NEBNext Ultra II FS DNA Library Prep Kit for Illumina and processed according to the manufacturer's instructions. See Supplementary Materials and Methods for analysis.
Whole-exome sequencing
Sequencing libraries were generated using Agilent SureSelectXT Mouse Exon Kit (Agilent Technologies) following manufacturer's recommendations. Details can be found in Supplementary Materials and Methods.
Statistical analysis
ANOVA or two-sided Student t test was used to investigate statistical significance, as indicated. Kaplan–Meier curve was analyzed by the log-rank test. P values were calculated with GraphPad Prism 6/8 (RRID:SCR_002798, GraphPad Software), and unless otherwise illustrated, all data were determined from at least three independent experiments and presented as mean and SD. Multiple testing was corrected according to Bonferroni.
Data and materials availability
RNA-seq and microarray: GEO: GSE144798, ENA: PRJEB35204 and PRJEB47385. ATAC- and ChIP-Seq: GEO: GSE155753 and GSE183823. Low coverage whole-genome sequencing and whole-exome sequencing: PRJEB47385.
Results
HDAC2 controls the cell cycle of undifferentiated PDAC cells
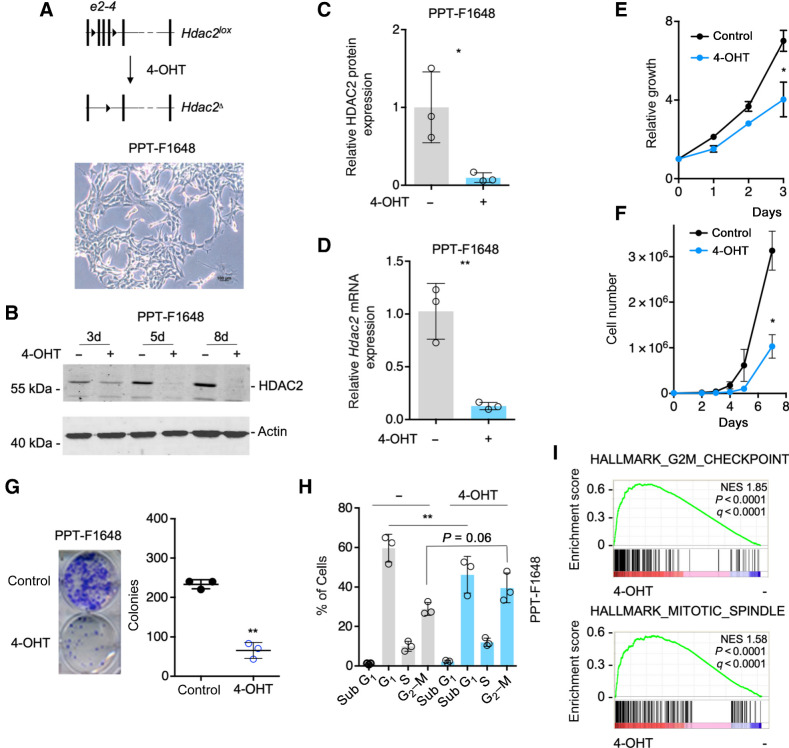
To analyze the function of HDAC2, we used the dual recombinase system (26) to establish a cellular model allowing 4-OHT–inducible genetic inactivation of the Hdac2 gene in murine PDAC cells (Fig. 1A). We and others have demonstrated that HDAC2 is associated with undifferentiated human PDACs (18, 19). To investigate the function of HDAC2 in this phenotypic context, an undifferentiated, mesenchymal cell line (PPT-F1648) was used (Fig. 1A). We deleted Hdac2 by treatment with 4-OHT over time. HDAC2 protein expression decreased three days after the addition of 4-OHT and was completely lost after eight days (Fig. 1B and C). Besides, Hdac2 mRNA was depleted after the treatment with 4-OHT (Fig. 1D). No regulation of HDAC2 in control cells, which express the tamoxifen-activatable Cre recombinase, was detected (Supplementary Fig. S1A). Cell growth, viability, and clonogenic growth of acutely Hdac2-deleted PPT-F1648 cells were significantly impaired (Fig. 1E–G). In contrast, neither growth nor clonogenic growth were affected in 4-OHT–treated control cell lines, excluding Cre and 4-OHT toxicity (Supplementary Fig. S1B and S1C). Reduced growth of Hdac2-deleted PDAC cells was connected to an accumulation of the cells in the G2–M-phase of the cell cycle and a corresponding loss of cells in the G1 phase (Fig. 1H). Consistently, in GSEA of RNA-seq expression data (Supplementary Table S2) of Hdac2-deleted cells, signatures linked to the G2–M-phase of the cell cycle were enriched (Fig. 1I). Again, no cell-cycle alteration was detected in control cells upon activation of the Cre recombinase (Supplementary Fig. S1D). To investigate the role of HDAC2 in the cell cycle in more detail, we performed a serum starvation-release experiment. Also in serum-starved cells, a trend for an increased G2–M fraction of cells was observed (Supplementary Fig. S1E). Furthermore, a delay in G1- to S-phase progression was evident in Hdac2-deficient cells (Supplementary Fig. S1E), demonstrating a function of HDAC2 in the regulation of these phases of the cell cycle. It is important to note that PPT-F1648 harbors one floxed Pdpk1 (26) allele. However, since one corresponding control cell line is Pdpk1lox/+ as well (Supplementary Table S1) and we detected no phenotypic effect of the loss of one Pdpk1 allele so far (27), we conclude that the observed effects are connected to HDAC2.
Figure 1.
Hdac2 knockout induced reduced growth and arrest in the G2–M phase of the cell cycle. A, Top, depiction of the floxed Hdac2 allele. 4-OHT treatment leads to the deletion of exon2 to exon4. Bottom, brightfield microscopy shows the morphology of PPT-F1648 cells. Scale bar, 100 μm. B, PPT-F1648 cells were treated for the indicated time points with 600 nmol/L 4-OHT or were left as vehicle-treated controls. Western blot controls the expression of HDAC2. Actin, loading control. C, Quantification of B. n = 3. *, P < 0.05 (paired t test). D, PPT-F1648 cells were treated for eight days with 600 nmol/L 4-OHT or were left as vehicle-treated controls. Quantitative PCR determines Hdac2 mRNA expression. Three independent experiments were analyzed. E, PPT-F1648 cells were treated for 8 days with 4-OHT or vehicle control. Afterwards, cells were plated in 96 wells and growth was determined by MTT assay over the indicated time points. Assay was performed in triplicate with three technical replicates. F, PPT-F1648 cells were treated for 8 days with 4-OHT or vehicle control. Afterwards, 3,000 cells were plated in 6-well plates and growth was determined by cell counting over the indicated time points. The assay was performed in triplicate with three technical replicates. G, 2,000 cells were seeded in 24 wells and clonogenic growth was analyzed after seven days. Left, representative clonogenic growth assay. Quantification (counting of Giemsa-stained colonies) of three independent experiments conducted as technical triplicate. H, PI cell-cycle FACS analysis after eight days of treatment as indicated. Three independent experiments were performed. I, RNA-seq of Hdac2-deficient and -proficient PPT-F1648 cells was analyzed by GSEA. Enrichment plots for the depicted HALLMARK signatures are shown. NES, normalized enrichment score; P, nominal P value; q, FDR. *, P < 0.05; **, P < 0.01; t test.
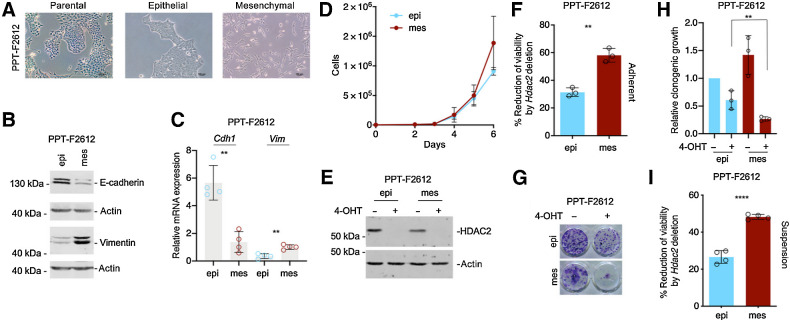
To corroborate the function of HDAC2 in undifferentiated PDAC models, we used an additional cell line, allowing the deletion of floxed Hdac2 alleles. Although parental PPT-F2612 cells are predominantly epithelial, the line contains mesenchymal cells (Fig. 2A), which is a reflection of intra-tumor heterogeneity and mirrors human PDACs, where differentiated, classical and undifferentiated, basal-like parts in the same tumor can be frequently observed (2, 28–30). This is mirrored in the murine primary PDAC from which the PPT-F2612 line was isolated. This tumor contained areas with different degrees of differentiation (Supplementary Fig. S2A). To analyze the effects of Hdac2-deletion in cells with the same genetic background, we used a recently described differential trypsinization protocol (1) that separates epithelial from mesenchymal cells. The successful separation was controlled by morphology (Fig. 2A) and analysis of epithelial (CDH1/E-cadherin) and mesenchymal (VIM/Vimentin) markers (Fig. 2B and C). Copy-number variations in mesenchymal and epithelial PPT-F2612 lines demonstrated a similar genetic landscape of these cell populations (Supplementary Fig. S2B; Supplementary Table S3). Furthermore, allele frequencies of mis- and nonsense SNVs and Indels from genes of the Cancer Gene Census (31) were shared between both epithelial and mesenchymal PPT-F2612 cells (Supplementary Fig. S2C; Supplementary Table S3). In sum, these data support that a transcriptional/epigenetic process and not a selection of a genetic event is relevant for EMT. The mesenchymal fraction demonstrated a trend toward increased proliferation as a sign of augmented aggressiveness (Fig. 2D).
Figure 2.
HDAC2 is relevant in undifferentiated PDAC cells. A, Brightfield microscopy of parental PPT-F2612 and the corresponding epithelial and mesenchymal sublines established by differential trypsinization. Scale bar, 100 μm. B and C, Adequate fractionation of epithelial (epi) and mesenchymal (mes) PPT-F2612 cells was controlled by Western blotting (B) of E-cadherin and vimentin. Actin, loading control. One representative Western blot out of three is depicted. C, Quantitative analysis of the mRNA expression of Cdh1 and Vim in epithelial and mesenchymal PPT-F2612 cells. Four independent biological experiments were conducted. **, P < 0.01 (t test). D, 3,000 epithelial or mesenchymal PPT-F2612 cells were plated in 6-well plates and growth was determined by cell counting over the indicated time points. The assay was performed in triplicate with three technical replicates. E, HDAC2 Western blot in the mesenchymal and epithelial fraction of PPT-F2612 cell treated with 4-OHT or vehicle control over eight days. Actin, loading control. F, The mesenchymal and epithelial fraction of PPT-F2612 cell treated with 4-OHT or vehicle control over eight days. Afterwards, 2,000 cells were seeded in a 96-well plate and viability was measured after 3 days by MTT assay. Three independent experiments conducted as technical triplicate were analyzed. Shown is the percent reduction of viability induced by the Hdac2 knockout. **, P < 0.01 (unpaired t test). G and H, Mesenchymal and epithelial fractions of PPT-F2612 cell were treated as described in E. 2,000 cells were seeded in a 24-well plate and clonogenic growth was analyzed after seven days. G, Representative clonogenic growth assay. H, Quantification of three independent experiments conducted as three technical replicates. Growth of epithelial control–treated cells were arbitrarily set to 1. **, P < 0.01 (unpaired t test). I, Mesenchymal and epithelial fraction of PPT-F2612 cells was treated as in E. Afterwards, 2,000 cells were transferred to a low attachment plate and viability was measured after 3 days using MTT assay. Four independent biological experiments are depicted. ****, P < 0.0001 (unpaired t test).
The deletion of Hdac2 was successfully achieved in epithelial and mesenchymal PPT-F2612 cells (Fig. 2E). The impact of Hdac2 deletion was significantly stronger in mesenchymal versus epithelial PDAC cells as measured in viability (Fig. 2F) and clonogenic growth assays (Fig. 2G and H). In addition, clonogenic growth of mesenchymal PPT-F2612 cells was increased (Fig. 2H). To exclude differences in the attachment capability of epithelial and mesenchymal tumor cells influencing the outcome of the viability assays, we repeated experiments using low-attachment cell culture plates. Equally, even in suspension, the loss of viability induced by Hdac2 deletion is increased in the mesenchymal fraction (Fig. 2I). These data support the conclusion that HDAC2 is a fitness factor, especially in less differentiated PDAC cells.
HDAC2 controls metastasis in vivo
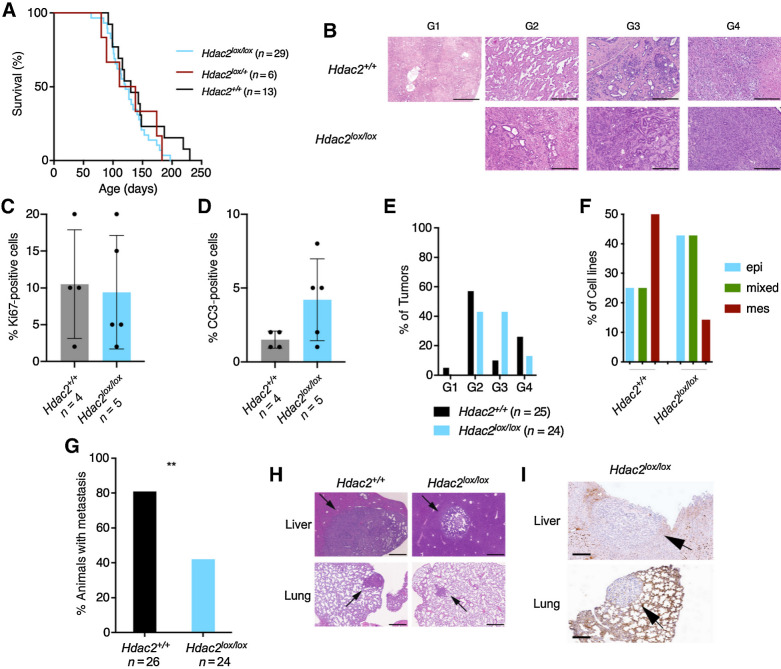
Because EMT is one road to metastasis (8, 13, 15) and we observed the dependency of mesenchymal PDAC cells to HDAC2, we speculated that HDAC2 might be involved in the metastatic cascade. Therefore, we used a genetically engineered mouse model with high metastasis frequency relying on the simultaneous expression of KrasG12D and mutated tumor suppressor p53R172H (32). We used a floxed Hdac2 mouse line (33) and crossed the line into the Pdx1-Cre, LSL-KrasG12D/+, LSL-p53R172H/+ (KPC) model. Kaplan–Meier curves were not changed in KPCH2lox/+ or KPCH2lox/lox mice (Fig. 3A), demonstrating that HDAC2 is dispensable for tumor initiation and carcinogenesis at least in the investigated model. All KPCH2lox/+ or KPCH2lox/lox mice developed murine PDAC (Fig. 3B). The fraction of Ki67-positive tumor cells was similar in KPC and KPCH2lox/lox mice (Fig. 3C; Supplementary Fig. S2D), supporting a conclusion that the proliferative disadvantage observed upon the acute deletion of the Hdac2 gene can be compensated. To support the adaptive capability toward the loss of Hdac2, we deleted the gene in PPT-F1648 cells and cultivated the cells over nine passages and measured growth again. Here, no change in the growth behavior was observed (Supplementary Fig. S2E), underpinning the concept that the role of HDAC2 in cell-cycle control can be compensated. The fraction of cleaved caspase-3–positive cells was increased in KPCH2lox/lox mice (Fig. 3D; Supplementary Fig. S2D). Strikingly, the frequency of undifferentiated (named “G4” here) cancers in KPCH2lox/lox mice was reduced by 50% (Fig. 3B and E). Consistently, the relative amount of mesenchymal cell lines isolated from KPCH2lox/lox mice was reduced (Fig. 3F). However, due to the low number of lines included in the analysis, the high frequency of mesenchymal KPC lines might be overrated. The development of undifferentiated cancers and completely mesenchymal murine Hdac2-deficient PDAC cell lines demonstrates that HDAC2 is not essential for dedifferentiation and EMT. Congruently, epithelial PDAC cell lines from KPCH2lox/lox mice undergo TGFβ-induced EMT (Supplementary Fig. S2F). Importantly, the metastasis frequency is distinctly reduced in the Hdac2-deficient model (Fig. 3G), demonstrating that the deacetylase increases the metastasis probability by 50%. However, some Hdac2-deficient murine PDACs have the capacity to metastasize to liver and lungs (Fig. 3G–I; Supplementary Fig. S2G). In KPC animals, we observed liver metastasis with a high nuclear expression of HDAC2 as well as HDAC2 negative metastasis (Supplementary Fig. S2H).
Figure 3.
HDAC2 controls a route to metastasis in vivo. A, Kaplan–Meier plots of KPC (black), KPCH2lox/+(red), and KPCH2lox/lox (blue) mice. B, Hematoxylin and eosin staining of murine PDAC from KPC and KPCH2lox/lox mice. G1, well-differentiated; G2, moderately differentiated; G3, poorly differentiated; G4, undifferentiated PDAC. Scale bar, 500 μm. C and D, Quantification of Ki67 stainings (C) and cleaved caspase-3 (D) stainings in murine PDAC from KPC (n = 4) and KPCH2lox/lox (n = 5) mice. E, Quantification of gradings from KPC and KPCH2lox/lox mice. F, Morphology of murine PDAC cell lines from KPC (n = 8) and KPCH2lox/lox mice (n = 7) was evaluated and stratified as epithelial (epi; blue), mesenchymal (mes; red), or as intermediate/mixed (mixed; green). G, Metastasis frequency of KPC and KPCH2lox/lox mice. **, P < 0.01 (Fisher exact test). H, Hematoxylin and eosin staining of liver and lung metastasis of KPC and KPCH2lox/lox mice. Scale bar, 500 μm. I, HDAC2 IHC of liver and lung metastasis of KPCH2lox/lox mice. Scale bar, 200 μm.
These data imply that HDAC2 is not simply a regulator of PDAC cell proliferation in vivo but a regulator of one road to metastasis.
HDAC2 maintains genes that are linked to undifferentiated PDACs and the expression of RTKs
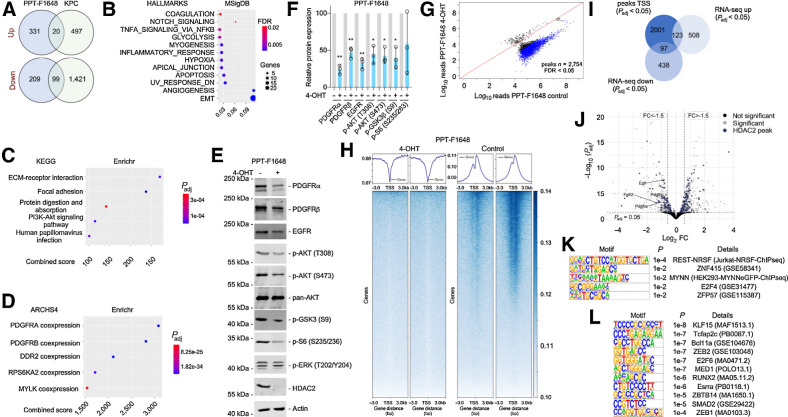
To find HDAC2-controlled regulatory networks connected to the observed biology, we conducted transcriptome profiling. We used cell lines from constitutive Hdac2-deficient murine PDACs, driven by simultaneous expression of KrasG12D and p53R172H in the murine pancreas (KPC model; ref. 32). The knockout of Hdac2 was documented at the protein level (Supplementary Fig. S3A and S3B). As described previously (34), HDAC1 expression is upregulated in these Hdac2-deficient models, arguing for compensatory adaption via this class I enzyme (Supplementary Fig. S3A and S3B). In addition, we used RNA-seq of PPT-F1648 cells, in which Hdac2 can be deleted (Fig. 1A). The Venn diagrams illustrate 20 commonly upregulated and 99 commonly downregulated genes in Hdac2-deficient models (Fig. 4A). These genes are depicted in the heatmaps of Supplementary Fig. S3C and S3D and Supplementary Table S2. By analyzing genes downregulated in the Hdac2-deficient state by the Molecular Signature database (https://www.gsea-msigdb.org/gsea/msigdb/index.jsp; Fig. 4B) and the Enrichr platform (https://amp.pharm.mssm.edu/Enrichr/) web tools (Fig. 4C and D), we observed an association of these genes to several RTKs, including the PDGFR signaling pathway, the PI3K–AKT signaling pathway, as well as to processes linked to EMT and focal adhesion (Fig. 4B–D). Additionally, glycolysis and hypoxia signatures associate with HDAC2 maintained genes (Fig. 4B). Interestingly, both processes are connected to the metastatic cascade of PDAC cells (4, 6). We concluded that HDAC2 maintains a program of genes with relevance in undifferentiated tumor cells. To further validate these findings, we used Western blotting of lysates from Hdac2-deficient and -proficient PPT-F1648 cells (Fig. 4E). Consistent with the RNA expression data (Supplementary Fig. S3C and S3E), we detected reduced expression of PDGFRα, PDGFRβ, and EGFR in mesenchymal Hdac2-deleted cells (Fig. 4E and F). Furthermore, RTK-driven downstream signaling was impaired in PPT-F1648 cells as demonstrated by reduced phosphorylation of AKT, GSK3β, and S6 ribosomal protein (Fig. 4E and F).
Figure 4.
HDAC2 maintains RTK-driven survival signaling. A, HDAC2 regulated genes in microarray of KPC (n = 3) and KPCH2lox/lox (n = 3) cancer cell lines (i) and 4-OHT–treated compared with vehicle-treated PPT-F1648 cells (ii) were analyzed in a Venn diagram including genes regulated with a log FC ± 0.58 and a P < 0.05. B, Genes consistently downregulated in Hdac2 knockout models were analyzed by the MolecularSignatureDatabase (MSigDB). HALLMARK signatures with an FDR < 0.05 are depicted. The FDR is color coded, number of genes contributing are coded by size. The ratio gene number to number of genes in the signature is depicted. C and D, Genes consistently downregulated in Hdac2 knockout models were analyzed by the Enrichr web tool using the libraries Kyoto Encyclopedia of Genes and Genomes (KEGG) 2021 and ARCH4 kinase coexpression. Top five signatures ranked according to the combined score. Padj value is color coded and the combined score is depicted. E, PPT-F1648 cells were treated for eight days with 4-OHT (600 nmol/L) or were left as vehicle treated controls. Western blot of PDGFRα, PDGFRβ, EGFR, phospho-AKT (T308 and S473), pan-AKT, phospho-GSK3 (S9), phospho-S6 (S235/236), phospho-ERK (T202/Y204), and HDAC2. Actin, loading control. One representative Western blot out of three is depicted F, Quantification of three independent experiments according to E. *, P < 0.05; **, P < 0.01 (t test). G, Differential analysis of reads in HDAC2 peaks between Hdac2-proficient and Hdac2-deficient PPT-F1648. Blue, significant peaks (FDR < 0.05, n = 2754). H, Density plot (top) and heatmap (bottom) of HDAC2 ChIP-seq reads around the proximal promoter in Hdac2-proficient and Hdac2-deficient PPT-F1648 (read counts per million). I, Overlap of significant HDAC2 peaks around the TSS (± 3,000 bp; Padj < 0.05) with significant differentially expressed genes in Hdac2-deficient PPT-F1648 cells (Padj < 0.05). J, Volcano plot of differentially regulated genes in Hdac2-deficient PPT-F1648 cells. Blue, genes with HDAC2 ChIP-seq peaks; gray, significant differential genes (Padj < 0.05). K, Homer motif analysis of all significant HDAC2 ChIP-seq peaks in promoter regions (TSS ± 3,000 bp; Padj < 0.05; n = 2,360); known motifs are depicted. L, Homer motif analysis of significant HDAC2 ChIP-seq peaks with downregulation in RNA-seq. De novo motif results are depicted (TSS ± 3,000 bp; Padj < 0.05; n = 102).
Because we found a regulation of RTKs, including PDGFRα, PDGFRβ, and EGFR in concert with downstream PI3K–AKT signaling, we challenged this regulatory circuit using curated The Cancer Genome Atlas (TCGA) and International Cancer Genome Consortium (ICGC) PDAC mRNA expression datasets. We separated these human datasets into groups with high and low mRNA expression of PDGFRB and investigated the enrichment of specific pathways using GSEA. In the TCGA as well the ICGC dataset, PDACs with high expression of PDGFRB enrich for PDGFRα and PDGFRβ signatures (Supplementary Fig. S3F), corroborating the separation. We then used the 99 commonly downregulated genes in the Hdac2-deficient models and generated a signature that we term “HDAC2 maintained”. This “HDAC2-maintained” gene set is enriched in human PDACs with high PDGFRB expression (Supplementary Fig. S3F). These PDACs are further characterized by an enrichment of signatures linked to EMT, metastasis, as well as TGFβ-, EGFR-, and PI3K–AKT signaling (Supplementary Fig. S3F). Furthermore, signatures of TFs controlling epithelial differentiation, including GATA6 (35, 36) and HNF1 (36–38), are depleted in PDACs with high PDGFRB expression (Supplementary Fig. S3F). Although curated datasets were used, a certain bias due to bulk sequencing and possible cosequencing of the stroma compartment cannot be excluded. Therefore, we used several approaches to further underscore the tumor cell–intrinsic connection of HDAC2-maintained genes to undifferentiated PDACs. First, we analyzed RNA-seq data of a large panel of murine KrasG12D-driven PDAC cell lines (n = 38), which were phenotypically separated into epithelial and mesenchymal lines (1). In addition to PDGFRα/β signatures, murine PDAC lines with high Pdgfrb expression enrich for the genes maintained by HDAC2, several EMT, and metastasis signatures together with the signaling circuits described for human PDACs (Supplementary Fig. S3F). Murine PDAC lines with high expression of Pdgfrb significantly enrich for mesenchymal cell lines (Supplementary Fig. S3G). Second, we used mRNA expression data from conventional human PDAC cell lines, which were divided by the expression of CDH1, VIM, and ZEB1 into lines with mesenchymal or epithelial marker gene expression (Supplementary Fig. S3H). GSEA demonstrated the enrichment of PDGFR, PI3K–AKT, and TGFβ signaling signatures in mesenchymal lines. Again, HDAC2-maintained genes were enriched in mesenchymal lines (Supplementary Fig. S3H). Third, we used RNA-seq of human patient-derived xenograft models, which allow discrimination of tumor cell–intrinsic circuits from expression changes in the microenvironment, by mapping RNA-seq reads to human and murine genomes, respectively (39, 40). We analyzed genes with higher expression in basal-like PDACs. Consistent with other reports (2), EMT signatures are enriched in basal-like PDACs (Supplementary Fig. S3I). Together with the EMT signature, we again detected PDGFR and PI3K–AKT signaling as well as HDAC2-maintained genes in basal-like PDACs (Supplementary Fig. S3I). Fourth, to investigate the direct connection of the HDAC2-maintained genes to TGFβ-induced EMT, we used expression profiles of TGFβ-treated murine PDAC cell lines (8). As shown in Supplementary Fig. S3J, TGFβ treatment induced EMT, PDGFR, and PI3K–AKT signaling signatures and the HDAC2-maintained genes. From these data, we reason that HDAC2-maintained genes are relevant in less differentiated PDACs in a cancer cell–autonomous manner.
In a first attempt to find the molecular processes responsible for the regulation of the HDAC2-maintained genes, we performed a histone H3 lysine 27 (H3K27) acetylation ChIP-seq and an ATAC-seq in Hdac2-proficient and -deficient PPT-F1648 cells. Unexpectedly, H3K27 acetylation and the open chromatin landscape were not changed on a genome-wide scale (Supplementary Fig. S4A and S4B). Consistently, H3K27 acetylation was not significantly altered upon the deletion of HDAC2 in Western blot analysis (Supplementary Fig. S4C). H3K27 acetylation ChIP-seq and ATAC-seq peaks at the HDAC2-regulated genes Egfr, Pdgfra, and Pdgfrb are not altered (Supplementary Fig. S4D). To gain further insight into the regulation of the genes, we determined genome-wide binding of HDAC2 by ChIP-seq. Differential peak calling of Hdac2-deficient and -proficient PPT-F1648 cells showed 2,754 peaks (FDR < 0.05; Fig. 4G). The majority of HDAC2 peaks were associated with core promoters (Fig. 4H; Supplementary Fig. S4E; Supplementary Table S4). Connecting the peaks in proximity to the transcriptional start site (TSS) to gene expression, we observed no mRNA expression changes for most genes upon the deletion of Hdac2 (Fig. 4I). This could be explained by the compensatory roles of other class I HDACs for the regulation of these genes. However, 123 genes with a HDAC2 promoter peak were upregulated and 97 genes were downregulated (Fig. 4I; Supplementary Table S4), arguing for a direct isoenzyme-specific impact of HDAC2 toward the regulation of these genes. The finding of HDAC2 binding to promoters, which depend on the enzyme, is consistent with the recruitment of HDAC2 to active genes and the correlation of HDAC2 binding with gene expression (41). Computing an overlap of the downregulated genes to pathways, we again observed a robust connection to EMT and RTK signatures (Supplementary Fig. S4F and S4G). Therefore, we investigated HDAC2 peaks at RTK genes in more detail. The Egfr gene revealed direct binding of HDAC2 and its mRNA was significantly downregulated upon the deletion of HDAC2 (Fig. 4J; Supplementary Fig. S4D). Although a HDAC2 peak in the Pdgfra promoter was called with borderline significance (Supplementary Fig. S4D and S4H), no clear HDAC2 peak was evident in the Pdgfrb promoter (Supplementary Fig. S4D). Therefore, direct and indirect regulatory circuits might contribute to the observed regulation. In addition, we detected the fibroblast growth factor receptor 2 (Fgfr2) gene downregulated with a significant HDAC2 peak in its promoter (Fig. 4J). To further extend the analysis, we investigated other RTKs genes with HDAC2 peaks in their promoters. Here, we observed that further RTK genes were bound by HDAC2 and for some, like Met, mRNA expression was even upregulated upon Hdac2 deletion (Supplementary Fig. S4H; Supplementary Table S2).
To gain insight into potential transcription factors, whose activity might be controlled by HDAC2, we conducted an enrichment analysis of cis-regulatory elements connected to the HDAC2 peaks. Over all HDAC2 promoter peaks, we found an overrepresentation of binding-sites for the repressor element 1 silencing transcription factor (REST), which is known to repress genes via HDAC1/2-containing repressor complexes, like mSIN3 and COREST (Fig. 4K; ref. 42). Considering the promoters where HDAC2 maintains expression of the corresponding genes, we detected enrichment of binding-sites for ZEB1, ZEB2, RUNX2, or SMAD2, which are connected to TGFβ signaling and/or EMT in PDAC (Fig. 4L; refs. 8, 43–45).
These data demonstrate that HDAC2 binds and maintains a subset of genes connected to metastasis, including RTKs.
HDAC2 protects undifferentiated PDAC cells from TGFβ-induced tumor suppression
Because the metastasis probability of Hdac2-deficient PDACs is reduced by 50%, but KPCH2lox/lox mice still build metastasis in vivo, we hypothesized that the molecular machineries needed for completion of the metastatic cascade can still be executed to some extent without HDAC2. However, a survival disadvantage of Hdac2-deficient cells might well contribute to the reduced metastasis frequency that we observed. TGFβ signaling, a potent trigger of EMT, can exert tumor-suppressive functions mediated by, for example, TGFβ-induced cell death (46). Therefore, we investigated TGFβ effects and started to determine the functionality of upstream signaling. In PPT-F1648 cells, TGFβ induced phosphorylation of SMAD2 and SMAD3 in the Hdac2-proficient and -deficient state (Supplementary Fig. S5A). In addition, we used Hdac2-deficient and -proficient cell lines from the KPC model, treated the lines with TGFβ and measured phosphorylation of SMADs. Also in these models, SMAD2 and SMAD3 phosphorylation was observed irrespectively of the HDAC2 status (Supplementary Fig. S5B). To investigate the transcriptional output of the TGFβ signaling pathway, we performed RNA-seq. Fifteen HALLMARK signatures were commonly regulated in Hdac2-deficient and -proficient cells (Supplementary Fig. S5C). These signatures include EMT and TGFβ signaling, demonstrating that phosphorylation of SMADs can elicit a transcriptional response (Supplementary Fig. S5C; Supplementary Table S5). However, comparing transcriptome profiles of TGFβ-treated Hdac2-proficient and -deficient lines, demonstrated that several gene signatures, including the EMT signature, were depleted in Hdac2-deficient cells (Supplementary Fig. S5D; Supplementary Table S5), pointing to the possibility that the TGFβ response is inaccurately executed. Indeed, EMT regulators like Cdh1 are inappropriately repressed in epithelial Hdac2-deficient KPC cells (Supplementary Fig. S5E and S5F), which is consistent with a repressive function of class I HDACs toward Cdh1 (20, 47). In contrast, genes like Zeb2 or Snai1 were not induced at the investigated early time points (Supplementary Fig. S5E; Supplementary Table S5). However, mesenchymal Hdac2-deficient cells can lose CDH1 expression and induce vimentin (Supplementary Fig. S5F).
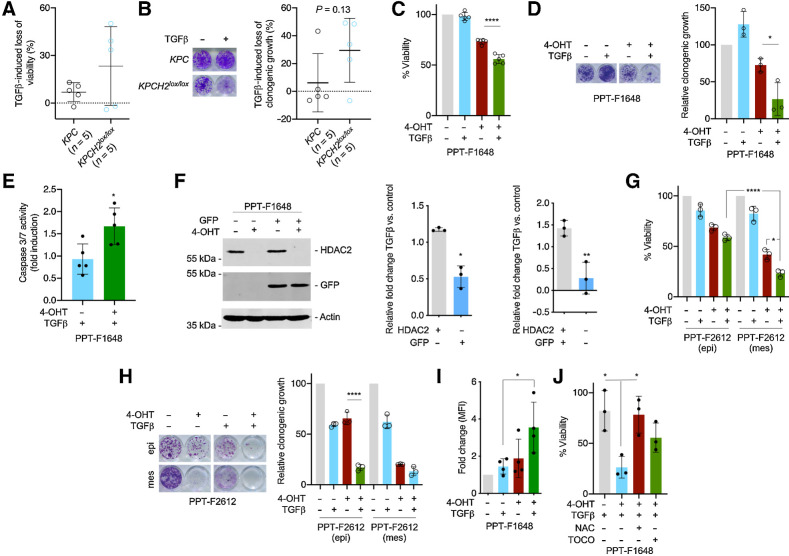
To test whether the HDAC2-regulated transcriptome exposed a vulnerability toward TGFβ, we analyzed Hdac2-proficient and -deficient lines. Although a distinct heterogeneity in the response toward TGFβ was detected in both assays (Fig. 5A and B), Hdac2-deficient cells were more susceptible to TGFβ treatment–induced loss of viability (Fig. 5A). Mean TGFβ-induced decrease in clonogenic growth was also higher in Hdac2-deficient lines (Fig. 5B). For the majority of lines, viability short-term assay and clonogenic long-term assay gave similar results (Pearson r = 0.76; P = 0.01; Supplementary Fig. S5G). Besides, 80% of Hdac2-deficient lines responded to TGFβ treatment with a reduction of clonogenic growth exceeding 20%, while only 20% of Hdac2-proficient lines responded to this extent (Fig. 5B). To experimentally address the heterogeneity of the TGFβ response and to prove the connection of HDAC2 to a prosurvival pathway relevant under TGFβ-mediated stress, we further investigated the TGFβ response in isogenic lines, allowing the acute deletion of Hdac2. In a Hdac2-deficient cellular state of PPT-F1648 cells, TGFβ induced a significant loss of viability, whereas the proficient counterpart remained TGFβ resistant (Fig. 5C). The protective effect of the TGFβ response conducted by HDAC2 was more pronounced in clonogenic assays of PPT-F1648 cells (Fig. 5D). To investigate a role of HDAC2 under culture condition enriching for potential cancer-stem cells (48), we used anchorage-independent sphere cultures. Hdac2-deficient cultures formed spheres, but the increased sensitivity toward TGFβ was also evident under sphere culture conditions (Supplementary Fig. S6A). To further characterize the TGFβ-induced cellular response, we performed an Annexin V/propidium iodide (PI) FACS analysis. Again, TGFβ reduced the amount of viable, Annexin V and PI–negative cells, more distinctly in the Hdac2-deficient setting (Supplementary Fig. S6B). Furthermore, TGFβ-induced apoptosis in Hdac2-deficient and -proficient cells (Supplementary Fig. S6B). However, activation of the executioner caspases-3/7 is increased in the Hdac2-deficient PPT-F1648 cell line (Fig. 5E). A trend toward increased induction of PI-positive cells was also observed in Hdac2-deficient PPT-F1648 cells (Supplementary Fig. S6B). To directly test whether HDAC2 confers a fitness advantage allowing to outcompete Hdac2-deficient cells under tumor-suppressive stress mediated by TGFβ, we performed repopulation assays. We used PPT-F1648 cells and transduced the cell line with GFP for lineage tracing. Expression of GFP and deletion of Hdac2 was demonstrated by Western blot analysis. This experimental approach allows us to perform repopulation assays in two directions. We deleted Hdac2 in GFP-labeled cells and mix them with Hdac2-proficient unlabeled cells and vice versa. We treated the mixture of cells with TGFβ and measured the fold increase/decrease of cells over controls. Deletion of Hdac2 was controlled by Western blotting (Fig. 5F). In both settings, Hdac2-proficient cells significantly outcompete the Hdac2-deficient cells under treatment with TGFβ (Fig. 5F), showing that HDAC2 facilitates the selection and clonal expansion of cells under TGFβ-mediated tumor-suppressive stress. Moreover, we investigated fractionated epithelial and mesenchymal PPT-F2612 cells. Both fractions of cells responded to TGFβ treatment with a loss of viability and reduced clonogenic growth, which is more pronounced in the Hdac2-deficient setting (Fig. 5G and H). A complex cross-signaling between the TGFβ pathway and RTK signaling is documented (49). To investigate this connection, we used PPT-F1648 cells. Again, deletion of Hdac2 reduced the protein expression of PDGFRα, PDGFRβ, and EGFR, validating the former observations (Supplementary Fig. S6C–S6E). Dependent on glycosylation, different PDGFRα forms migrating at different molecular weights in Western blots were described (50, 51). TGFβ reduced the expression of the PDGFRα form migrating at approximately 170 kDa and induced the expression of a 130 kDa form (Supplementary Fig. S6C). The induction of the 130 kDa form was reduced in Hdac2-deficient cells as well as its phosphorylation (Supplementary Fig. S6C). TGFβ also impaired the expression of PDGFRβ and EGFR (Supplementary Fig. S6D and S6E). We extended the analysis to the Hdac2-proficient and -deficient KPC lines and included one TGFβ-sensitive and one TGFβ-resistant line of each genotype (see Fig. 5A and B). Heterogeneous expression of the receptors was observed, which is in agreement with observations made by others (8). In case the receptors are expressed, TGFβ induced a similar regulation as observed in PPT-F1648 cells (Supplementary Fig. S6F–S6H). The Hdac2-deficient, TGFβ-sensitive line demonstrated the lowest expression levels of all investigated receptors (Supplementary Fig. S6F–S6H). In contrast, the Hdac2-deficient, TGFβ-insensitive line revealed high PDGFRβ expression, demonstrating that also regulation of RTKs by HDAC2 can be compensated. Such observations are well in line with the remaining metastatic capability of Hdac2-deficient PDAC cells. We concluded that cross-signaling between the TGFβ pathway and RTK signaling is evident also in the investigated lines, but the contribution of the RTKs to the TGFβ sensitization demands further clarifications.
Figure 5.
HDAC2 protects from TGFβ-dependent tumor suppression. A and B, Five murine PDAC cell lines from KPC mice and five lines from KPCH2lox/lox mice were treated with TGFβ (5 ng/mL) or left as untreated control. A, Viability was determined by MTT assay 72 hours after the treatment. Mean viability of a line was determined by three independent biological experiments. B, Clonogenic assays were performed and analyzed after 7 days of treatment. Left, macroscopic picture of a clonogenic assay of a KPC and a KPCH2lox/lox line. Right, quantification. Mean clonogenic growth of a line was determined by three independent biological experiments conducted as technical triplicates. P value of an unpaired t test is depicted. C, PPT-F1648 cells were treated for eight days with 4-OHT (600 nmol/L) or were left as vehicle-treated controls. Afterwards, cells were replated and 24 hours later treated with TGFβ (5 ng/mL) for 72 hours. Viability was determined by MTT assay. ****, P < 0.0001 (ANOVA). D, PPT-F1648 cells were treated as in C. After eight days of 4-OHT treatment, cells were replated in 24 wells and treated after 24 hours with TGFβ (5 ng/mL) for 7 days. Left, macroscopic picture of a clonogenic assay. Right, quantification of three independent experiments conducted as technical triplicates. *, P < 0.05 (ANOVA). E, PPT-F1648 cells were treated as in C. Afterwards, cells were plated in 96 wells and 24 hours later treated with TGFβ (5 ng/mL) for 72 hours. Caspase activity (measured by Caspase-Glo 3/7 Assay) was normalized to cell viability (determined by CellTiter-Glo Assay) and fold induction upon TGFβ treatment compared with control was calculated. *, P < 0.05 (t test). F, PPT-F1648 cells were transduced with a GFP vector and then treated for eight days with 4-OHT (600 nmol/L) or were left as vehicle-treated controls. Afterwards, cells were replated and 24 hours later treated with TGFβ (5 ng/mL) for 96 hours. Left, Western blotting controls expression of GFP and HDAC2. Actin, loading control; right, relative fold change in the depicted conditions as determined by FACS. *, P < 0.05; **, P < 0.01 (t test). G, Epithelial (epi) and mesenchymal (mes) fraction of PPT-F2612 cells were treated for eight days with 4-OHT (600 nmol/L) or were left as vehicle-treated controls. Afterwards, cells were replated and 24 hours later treated with TGFβ (5 ng/mL) for 72 hours. Viability was determined by MTT assay. *, P < 0.05 (ANOVA); green bars were analyzed with an unpaired t test (****, P < 0.0001). H, Epithelial and mesenchymal fraction of PPT-F2612 cells as in D. After eight days of 4-OHT treatment, cells were replated in 24 wells and after 24 hours treated with TGFβ (5 ng/mL) for 7 days. Left, macroscopic picture of a clonogenic assay. Right, quantification of three independent experiments. ****, P < 0.0001 (ANOVA). I, ROS levels of control and 4-OHT (600 nmol/L)-treated PPT-F1648 cells after addition of TGFβ (5 ng/mL) for 72 hours. FACS measurements are plotted as fold change of the mean fluorescence intensity (MFI). J, Viability of control and 4-OHT (600 nmol/L)-treated PPT-F1648 cells treated with TGFβ (5 ng/mL) or control treatment in addition to ROS scavengers, 2 mmol/L NAC or 2 mmol/L α-tocopherol. Cell viability was measured after 72 hours by CellTiter-Glo Assay and viability of untreated Hdac2-proficient PPT-F1648 cells was set to 100%. *, P < 0.05 (ANOVA).
Because we used TGFβ-sensitive and -resistant cell lines, we were able to analyze pathways commonly regulated in these phenotypes by a GSEA (Supplementary Fig. S6I–S6J). Upon the five pathways consistently regulated in TGFβ sensitive lines, irrespectively of the Hdac2 status, we observed a depletion of a signature connected to glutathione metabolism (Supplementary Fig. S6J). Because glutathione is a major antioxidant contributing to redox homeostasis and TGFβ is known to increase reactive oxygen species (ROS; ref. 52), we measured ROS production. Indeed, TGFβ increased ROS levels, an observation more pronounced in Hdac2-deficient cells (Fig. 5I). To validate the contribution of ROS to the lethal response toward TGFβ in the Hdac2-deficient setting, we used the antioxidant N-acetyl-l-cysteine (NAC), which rescued the TGFβ effects in Hdac2-deficient PPT-F1648 cells (Fig. 5J). Although to a smaller extent, this effect was also observed by the use of a second antioxidant, α-tocopherol (Fig. 5J). Such data suggest a role of HDAC2 as a rheostat of ROS and point to an additional potential mechanism by which HDAC2 limits TGFβ-mediated tumor suppression.
Discussion
Here, we show that HDAC2 is an important fitness factor of mesenchymal PDAC cells controlling the tumor-suppressive function of TGFβ. We demonstrate that HDAC2 maintains the expression of a gene network preferentially expressed in undifferentiated cancer cells.
The contribution of class I HDACs to EMT processes of PDAC cells has been described. A HDAC1 and HDAC2 containing complex is needed to repress the CDH1 gene during EMT (20, 47). 4SC-202, a class I HDAC–specific inhibitor, prevents TGFβ-dependent upregulation of ZEB1 and SNAI1 as well as repression of CDH1 (53). Furthermore, the HDAC inhibitor mocetinostat interferes with the function of ZEB1 (54). We expand the connection of class I HDACs to EMT states in PDAC by describing a novel HDAC2-specific function. We observed that the HDAC2-controlled network included RTKs and we provide evidence that RTKs, like PDGFRα, PDGFRβ, or EGFR, which are regulated by HDAC2 in the investigated murine PDAC cell line, are connected to EMT, metastasis, TGFβ and PI3K–AKT signaling signatures in human and murine expression datasets. Consistently, the PDGFRβ protein is induced by TGFβ in murine PDAC cells (8), and human PDACs with high expression of PDGFRβ have an increased risk of metastasis (55). We did not detect TGFβ-induced upregulation of PDGFRβ in the investigated KPC cell lines, which is explained by the early time point we were investigating compared with the delayed induction kinetics observed in TGFβ-dependent EMT models (8). In the KPC mouse model, pharmacologic inhibition of the PDGFRβ does not influence the tumor burden compared with controls, but distinctly reduced metastasis formation (56). Therefore, the regulation of PDGFRβ expression by HDAC2 in our ex vivo model might contribute to the impaired metastasis formation in Hdac2-deficient KPC animals. In addition to PDAC, downregulation of PDGFRβ in class I HDAC inhibitor–treated or HDAC1/2 siRNA–transfected renal cell carcinoma models occurs (57), arguing for a more general relevance of the observed pathway. Although the connection of PDGFRβ to metastasis is well described in PDAC, it is important to note that we detected additional RTKs, including EGFR and PDGFRα. Hence, the observed phenotype can also be attributed to the concerted signaling input of these RTKs.
At the mechanistic level, we robustly demonstrate that genetic inhibition of HDAC2 unleashes the tumor-suppressive function of TGFβ. The TGFβ signaling pathway controls relevant processes of cancer cells from initiation of tumors over differentiation of cancer cells to distant organ metastasis, whereby the cellular context decides over the tumor-suppressive or the tumor-promoting facet of the pathway (46). Mutational inactivation of the TGFβ pathway, most frequently by alterations of SMAD4, occurs in approximately one half of PDACs (58, 59). Especially the basal-like subtype of PDAC, which enriches EMT and TGFβ signatures, shows a decreased frequency of genetic SMAD4 losses (2). These data demonstrate the need for PDAC cells with an active EMT program to develop strategies to enable protumorigenic TGFβ functions and simultaneously disable the tumor-suppressive and death-inducing properties of the pathway. The helix-loop-helix transcriptional modulator ID1 was recently shown to contribute to the uncoupling of the TGFβ-mediated EMT and apoptotic program by protecting PDAC cells from apoptosis (59). Whereas in the apoptosis permissive state, TGFβ-induced the downregulation of ID1, in the setting of disabled apoptosis, the ID1 gene escapes TGFβ-induced repression. Importantly, the escape from TGFβ-repression of ID1 is mediated by the PI3K–AKT signaling pathway (59). Consistently with these data, we detected cosegregation of EMT, TGFβ, and PI3K–AKT signatures together with the gene network controlled by HDAC2 across the murine and human PDAC expression dataset. Regulation of upstream RTKs with consequent modulation of multiple associated downstream signaling can contribute to the uncoupling of EMT and sensitization toward tumor-suppressive TGFβ functions upon HDAC2 inactivation.
The deletion of Hdac2 in murine PDAC leads to impaired fitness, which is more pronounced in undifferentiated mesenchymal cells, compatible with the increased expression of HDAC2 in less differentiated human PDACs (18, 19). Acute inactivation of Hdac2 leads to reduced proliferation and cell-cycle arrest in the G2–M-phase of the cell cycle. Control of the cell cycle by HDACs is well described (17) and in the context of PDAC, HDAC2 was recently shown to regulate the expression of the mitotic Aurora A kinase (60). The development of proliferative Hdac2-deficient PDACs in vivo and the restoration of growth in Hdac2-deficient cells upon long-term passaging in vitro, argues that the cell-cycle defect seen after the acute deletion of Hdac2 can be adapted. Therefore, we did not investigate the molecular link of HDAC2 to the cell-cycle in greater detail. Furthermore, such data indicate that growth defects observed after the acute deletion of Hdac2 are not likely to contribute to the metastatic phenotype.
This study opens questions, which demand future experiments for definite clarification. (i) Although the evidence from our ex vivo investigation, including regulation of relevant RTKs and sensitization for the tumor-suppressive TGFβ function, the analysis of large PDAC expression datasets, which connect the HDAC2-maintained network to TGFβ, EMT, and PI3K pathways, combined with published data, demonstrating that PDGFRβ is a driver of PDAC metastasis (55, 56) and that PI3K–AKT signaling is relevant to escape the proapoptotic function of TGFβ (59), contributes to explain the reduced metastasis of Hdac2-deficient KPC mice, we cannot prove that such pathways are operative in vivo. Furthermore, it is important to note that many mechanistic insights were elaborated mainly by the use of PPT-F1648 cells that allows for a direct comparison of HDAC2 in an isogenic background. This approach might limit the generality of the observations and considering the heterogeneity of the disease, other sets of RTKs or different mechanisms might contribute to explain the metastasis phenotype. Additional genes we detected to be regulated by HDAC2 can also contribute here. For example, in RNA expression profiles, we found collagen family members, which can affect tumor progression and metastasis (61), to be maintained by HDAC2. Furthermore, considering the multiple functions of the TGFβ pathway along the metastatic cascade, the complex cross-talk with different tumor compartments, and the stochastic nature of the metastatic process, additional experiments with a high spatial and temporal resolution best conducted with appropriate genetic gain- and loss-of-function approaches are necessary, which are beyond the scope of the current manuscript. (ii) The molecular mechanism of the unexpected downregulation of genes upon the inactivation of a transcriptional repressor is unclear. We recently described that HDAC1 and HDAC2 synergize to maintain the expression of cancer-relevant genes in PDAC models (34). Here, we observe that in PDAC lines, HDAC2 binds to promoters, whose genes revealed both reduced and induced expression upon the deletion of Hdac2, which supports a role of the enzyme in the maintenance and repression of genes. However, deciphering how HDAC2 maintains gene expression without changing open chromatin and the histone H3K27 acetylation status on a chromatin-wide scale demands additional experiments. Whether in context of the investigated PDAC cells, transcription factors, whose cis-regulatory sites are overrepresented at HDAC2 peaks were controlled by acetylation remains to be deciphered in future work. (iii) Several pathways and processes control TGFβ-mediated cell death. Inhibition of PI3K–AKT signaling can allow TGFβ-induced apoptosis (59, 62). The transcription factor SOX4 was recently implicated in TGFβ-induced apoptosis of PDAC cells (62). We now demonstrate, that HDAC2 is relevant for unleashing the tumor-suppressive TGFβ function observed in Hdac2-deficient states and show that HDAC2 can act as a rheostat of TGFβ-induced ROS. How HDAC2 controls ROS metabolism and how this mechanism connects to cell death and the metastasis phenotype awaits further investigations.
In sum, we show the important complexity and context specificity of the function of epigenetic regulators, like HDAC2, which implicate that an increased understanding of these proteins is needed to implement successful epigenetic therapies in the clinic. Our data add another tractable player, HDAC2, to the licensers of the metastatic cascade allowing to schedule and test novel therapeutic approaches with the goal to prevent metastasis.
Authors' Disclosures
L. Krauß reports grants and personal fees from Deutsche Forschungsgesellschaft/German Research Foundation (DFG)-SFB1321 during the conduct of the study. S. Hastreiter reports personal fees from Deutsche Forschungsgemeinschaft (DFG), personal fees from Wilhelm-Sander-Stiftung, and personal fees from Deutsche Krebshilfe during the conduct of the study. C. Schneider reports grants and personal fees from Deutsche Krebshilfe during the conduct of the study. K. Lankes reports grants from SFB1321 during the conduct of the study and other support from DKFZ outside the submitted work. C. Klement reports grants from Deutsche Forschungsgemeinschaft during the conduct of the study. F.M. Cernilogar reports grants from Deutsche Forschungsgemeinschaft during the conduct of the study. K. Steiger reports other support from Roche outside the submitted work; in addition, K. Steiger has a patent for a radiopharmaceutical issued. M. Reichert reports grants from DFG and grants from German Cancer Aid during the conduct of the study and honoraria from Celgene, Roche and Falk Foundation. G. Schotta reports grants from Deutsche Forschungsgemeinschaft during the conduct of the study. D. Saur reports grants from Deutsche Forschungsgemeinschaft, grants from German Cancer Consortium (DKTK), and grants from European Research Council during the conduct of the study; personal fees and non-financial support from Novartis, personal fees and non-financial support from Amgen, and non-financial support from Johnson & Johnson outside the submitted work. G. Schneider reports grants from DFG (SFB1321 (project ID 329628492), grants from DFG SCHN 959/6-1, grants from Wilhelm-Sander-Stiftung 2017.048.2, grants from Wilhelm-Sander-Stiftung 2019.086.1, and grants from Deutsche Krebshilfe 70113760 during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank Dr. E. Olson for providing floxed Hdac2 mice. Some of the results shown here are in whole or part based upon data generated by the TCGA Research Network (https://www.cancer.gov/tcga). The authors thank Thorsten Kaltenbacher for providing his expertise and insight regarding ChIP-seq. They thank Julia Eichinger, Marion Mielke, Annett Hering, and Olga Seelbach for excellent technical assistance. This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) SFB1321 (project ID 329628492; P13 to G. Schotta and G. Schneider); SFB1321 S01 and S02 (to G. Schneider, K. Steiger, M. Reichert, D. Saur, and R. Rad); P12 (to M. Reichert); SCHN 959/6-1 (to G. Schneider); RE 3723/4-1 (to M. Reichert); Wilhelm-Sander-Stiftung (2017.048.2 to G. Schneider and 2019.086.1 to G. Schneider and O.H. Krämer); Deutsche Krebshilfe (70113760 to G. Schneider; 70114328 to M. Reichert; Max Eder Program 111273 to M. Reichert).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
L. Krauß: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. B.C. Urban: Conceptualization, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. S. Hastreiter: Conceptualization, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. C. Schneider: Conceptualization, data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. P. Wenzel: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. Z. Hassan: Formal analysis, investigation, methodology, writing–review and editing. M. Wirth: Resources, data curation, formal analysis, investigation, writing–review and editing. K. Lankes: Resources, data curation, formal analysis, investigation, visualization, writing–review and editing. A. Terrasi: Data curation, formal analysis, investigation, visualization, writing–review and editing. C. Klement: Data curation, formal analysis, investigation, visualization, writing–review and editing. F.M. Cernilogar: Data curation, formal analysis, investigation, visualization, writing–review and editing. R. Öllinger: Data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. N. de Andrade Krätzig: Data curation, formal analysis, investigation. T. Engleitner: Resources, data curation, supervision, investigation, visualization, methodology. R.M. Schmid: Resources, data curation, formal analysis, supervision, funding acquisition, investigation, methodology, writing–review and editing. K. Steiger: Resources, supervision, funding acquisition, investigation, visualization, writing–review and editing. R. Rad: Resources, supervision, funding acquisition, investigation, visualization, methodology, writing–review and editing. O.H. Krämer: Conceptualization, resources, supervision, funding acquisition, methodology, writing–review and editing. M. Reichert: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. G. Schotta: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, project administration, writing–review and editing. D. Saur: Conceptualization, resources, formal analysis, supervision, funding acquisition, investigation. G. Schneider: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, writing–original draft, writing–review and editing.
References
- 1. Mueller S, Engleitner T, Maresch R, Zukowska M, Lange S, Kaltenbacher T, et al. . Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature 2018;554:62–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O'Kane GM, et al. . Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet 2020;52:231–40. [DOI] [PubMed] [Google Scholar]
- 3. Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, et al. . Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet 2017;49:358–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McDonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, et al. . Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 2017;49:367–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Roe J-S, Hwang C-I, Somerville TDD, Milazzo JP, Lee EJ, Silva BD, et al. . Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017;170:875–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chiou S-H, Risca VI, Wang GX, Yang D, Grüner BM, Kathiria AS, et al. . Blimp1 induces transient metastatic heterogeneity in pancreatic cancer. Cancer Discov 2017;7:1184–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Whittle MC, Izeradjene K, Rani PG, Feng L, Carlson MA, DelGiorno KE, et al. . RUNX3 Controls a metastatic switch in pancreatic ductal adenocarcinoma. Cell 2015;161:1345–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Krebs AM, Mitschke J, Losada ML, Schmalhofer O, Boerries M, Busch H, et al. . The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 2017;19:518–29. [DOI] [PubMed] [Google Scholar]
- 9. Takano S, Reichert M, Bakir B, Das KK, Nishida T, Miyazaki M, et al. . Prrx1 isoform switching regulates pancreatic cancer invasion and metastatic colonization. Gene Dev 2016;30:233–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Somerville TDD, Xu Y, Miyabayashi K, Tiriac H, Cleary CR, Maia-Silva D, et al. . TP63-mediated enhancer reprogramming drives the squamous subtype of pancreatic ductal adenocarcinoma. Cell Rep 2018;25:1741–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hamdan FH, Johnsen SA. DeltaNp63-dependent super enhancers define molecular identity in pancreatic cancer by an interconnected transcription factor network. Proc Natl Acad Sci U S A 2018;115:E12343–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Genovese G, Carugo A, Tepper J, Robinson FS, Li L, Svelto M, et al. . Synthetic vulnerabilities of mesenchymal subpopulations in pancreatic cancer. Nature 2017;542:362–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. . EMT and dissemination precede pancreatic tumor formation. Cell 2012;148:349–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Diaferia GR, Balestrieri C, Prosperini E, Nicoli P, Spaggiari P, Zerbi A, et al. . Dissection of transcriptional and cis-regulatory control of differentiation in human pancreatic cancer. EMBO J 2016;35:595–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK. EMT, MET, plasticity, and tumor metastasis. Trends Cell Biol 2020;30:764–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dijk F, Veenstra VL, Soer EC, Dings MPG, Zhao L, Halfwerk JB, et al. . Unsupervised class discovery in pancreatic ductal adenocarcinoma reveals cell-intrinsic mesenchymal features and high concordance between existing classification systems. Sci Rep 2020;10:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Seto E. HDACs and HDAC inhibitors in cancer development and therapy. Csh Perspect Med 2016;6:a026831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fritsche P, Seidler B, Schüler S, Schnieke A, Göttlicher M, Schmid RM, et al. . HDAC2 mediates therapeutic resistance of pancreatic cancer cells via the BH3-only protein NOXA. Gut 2009;58:1399. [DOI] [PubMed] [Google Scholar]
- 19. Lehmann A, Denkert C, Budczies J, Buckendahl A-C, Darb-Esfahani S, Noske A, et al. . High class I HDAC activity and expression are associated with RelA/p65 activation in pancreatic cancer in vitro and in vivo. BMC Cancer 2009;9:395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, et al. . E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology 2009;137:361–71. [DOI] [PubMed] [Google Scholar]
- 21. Biederstädt A, Hassan Z, Schneeweis C, Schick M, Schneider L, Muckenhuber A, et al. . SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut 2020;69:1472–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Diersch S, Wenzel P, Szameitat M, Eser P, Paul MC, Seidler B, et al. . Efemp1 and p27(Kip1) modulate responsiveness of pancreatic cancer cells towards a dual PI3K/mTOR inhibitor in preclinical models. Oncotarget 2013;4:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lankes K, Hassan ZZ, Doffo MJ, Schneeweis C, Lier S, Öllinger R, et al. . Targeting the ubiquitin-proteasome system in a pancreatic cancer subtype with hyperactive MYC. Mol Oncol 2020;14:3048–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Corces MR, Trevino AE, Hamilton EG, Greenside PG, Sinnott-Armstrong NA, Vesuna S, et al. . An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat Methods 2017;14:959–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cernilogar FM, Hasenöder S, Wang Z, Scheibner K, Burtscher I, Sterr M, et al. . Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res 2019;47:9069–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Schönhuber N, Seidler B, Schuck K, Veltkamp C, Schachtler C, Zukowska M, et al. . A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat Med 2014;20:1340–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eser S, Reiff N, Messer M, Seidler B, Gottschalk K, Dobler M, et al. . Selective requirement of PI3K/PDK1 signaling for kras oncogene-driven pancreatic cell plasticity and cancer. Cancer Cell 2013;23:406–20. [DOI] [PubMed] [Google Scholar]
- 28. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Kalekar RL, et al. . Transcriptional subtype-specific microenvironmental crosstalk and tumor cell plasticity in metastatic pancreatic cancer. Biorxiv 2020;2020.08.25.256214. [Google Scholar]
- 29. Hwang WL, Jagadeesh KA, Guo JA, Hoffman HI, Yadollahpour P, Mohan R, et al. . Single-nucleus and spatial transcriptomics of archival pancreatic cancer reveals multi-compartment reprogramming after neoadjuvant treatment. Biorxiv 2020. [Google Scholar]
- 30. Hayashi A, Fan J, Chen R, Ho Y, Makohon-Moore AP, Lecomte N, et al. . A unifying paradigm for transcriptional heterogeneity and squamous features in pancreatic ductal adenocarcinoma. Nat Cancer 2020;1:59–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, et al. . COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res 2018;47:gky1015-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. . Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7:469–83. [DOI] [PubMed] [Google Scholar]
- 33. Montgomery RL, Davis CA, Potthoff MJ, Haberland M, Fielitz J, Qi X, et al. . Histone deacetylases 1 and 2 redundantly regulate cardiac morphogenesis, growth, and contractility. Gene Dev 2007;21:1790–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Stojanovic N, Hassan Z, Wirth M, Wenzel P, Beyer M, Schäfer C, et al. . HDAC1 and HDAC2 integrate the expression of p53 mutants in pancreatic cancer. Oncogene 2016;36:1804–15. [DOI] [PubMed] [Google Scholar]
- 35. Martinelli P, Pau ECS, Cox T, Sainz B, Dusetti N, Greenhalf W, et al. . GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2016;66:1665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kloesch B, Ionasz V, Paliwal S, Hruschka N, Villarreal JM de, Öllinger R, et al. . A GATA6-centred gene regulatory network involving HNFs and ΔNp63 controls plasticity and immune escape in pancreatic cancer. Gut 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Milan M, Balestrieri C, Alfarano G, Polletti S, Prosperini E, Spaggiari P, et al. . FOXA 2 controls the cis -regulatory networks of pancreatic cancer cells in a differentiation grade-specific manner. EMBO J 2019;38:e102161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kalisz M, Bernardo E, Beucher A, Maestro MA, Pozo ND, Millán I, et al. . HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J 2020;39:e102808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nicolle R, Blum Y, Marisa L, Loncle C, Gayet O, Moutardier V, et al. . Pancreatic adenocarcinoma therapeutic targets revealed by tumor-stroma cross-talk analyses in patient-derived xenografts. Cell Rep 2017;21:2458–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feldmann K, Maurer C, Peschke K, Teller S, Schuck K, Steiger K, et al. . Mesenchymal plasticity regulated by prrx1 drives aggressive pancreatic cancer biology. Gastroenterology 2020;160:346–61. [DOI] [PubMed] [Google Scholar]
- 41. Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, et al. . Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell 2009;138:1019–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ooi L, Wood IC. Chromatin crosstalk in development and disease: lessons from REST. Nat Rev Genet 2007;8:544–54. [DOI] [PubMed] [Google Scholar]
- 43. Kayed H, Jiang X, Keleg S, Jesnowski R, Giese T, Berger MR, et al. . Regulation and functional role of the Runt-related transcription factor-2 in pancreatic cancer. Br J Cancer 2007;97:1106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kurahara H, Takao S, Maemura K, Mataki Y, Kuwahata T, Maeda K, et al. . Epithelial–mesenchymal transition and mesenchymal–epithelial transition via regulation of ZEB-1 and ZEB-2 expression in pancreatic cancer. J Surg Oncol 2012;105:655–61. [DOI] [PubMed] [Google Scholar]
- 45. Kleeff J, Friess H, Simon P, Susmallian S, Buchler P, Zimmermann A, et al. . Overexpression of Smad2 and colocalization with TGF-β1 in human pancreatic cancer. Digest Dis Sci 1999;44:1793–802. [DOI] [PubMed] [Google Scholar]
- 46. David CJ, Massagué J. Contextual determinants of TGFβ action in development, immunity and cancer. Nat Rev Mol Cell Biol 2018;19:419–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Aghdassi A, Sendler M, Guenther A, Mayerle J, Behn C-O, Heidecke C-D, et al. . Recruitment of histone deacetylases HDAC1 and HDAC2 by the transcriptional repressor ZEB1 downregulates E-cadherin expression in pancreatic cancer. Gut 2012;61:439. [DOI] [PubMed] [Google Scholar]
- 48. Sancho P, Burgos-Ramos E, Tavera A, Kheir TB, Jagust P, Schoenhals M, et al. . MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab 2015;22:590–605. [DOI] [PubMed] [Google Scholar]
- 49. Shi Q, Chen Y-G. Interplay between TGF-β signaling and receptor tyrosine kinases in tumor development. Sci China Life Sci 2017;60:1133–41. [DOI] [PubMed] [Google Scholar]
- 50. Keating MT, Harryman CC, Williams LT. Platelet-derived growth factor receptor inducibility is acquired immediately after translation and does not require glycosylation. J Biol Chem 1989;264:9129–32. [PubMed] [Google Scholar]
- 51. Bejcek BE, Voravud N, Deuel TF. Biosynthesis and processing of the platelet derived growth factor type α receptor. Biochem Bioph Res Co 1993;196:69–78. [DOI] [PubMed] [Google Scholar]
- 52. Chang C-H, Pauklin S. ROS and TGFβ: from pancreatic tumour growth to metastasis. J Exp Clin Canc Res 2021;40:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mishra VK, Wegwitz F, Kosinsky RL, Sen M, Baumgartner R, Wulff T, et al. . Histone deacetylase class-I inhibition promotes epithelial gene expression in pancreatic cancer cells in a BRD4- and MYC-dependent manner. Nucleic Acids Res 2017;45:gkx212-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meidhof S, Brabletz S, Lehmann W, Preca B, Mock K, Ruh M, et al. . ZEB1-associated drug resistance in cancer cells is reversed by the class I HDAC inhibitor mocetinostat. EMBO Mol Med 2015;7:831–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kurahara H, Maemura K, Mataki Y, Sakoda M, Shinchi H, Natsugoe S. Impact of p53 and PDGFR-β expression on metastasis and prognosis of patients with pancreatic cancer. World J Surg 2016;40:1977–84. [DOI] [PubMed] [Google Scholar]
- 56. Weissmueller S, Manchado E, Saborowski M, Morris JP, Wagenblast E, Davis CA, et al. . Mutant p53 drives pancreatic cancer metastasis through cell-autonomous pdgf receptor β signaling. Cell 2014;157:382–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kiweler N, Brill B, Wirth M, Breuksch I, Laguna T, Dietrich C, et al. . The histone deacetylases HDAC1 and HDAC2 are required for the growth and survival of renal carcinoma cells. Arch Toxicol 2018;92:2227–43. [DOI] [PubMed] [Google Scholar]
- 58. Initiative APCG, Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, et al. . Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 59. Huang Y-H, Hu J, Chen F, Lecomte N, Basnet H, David CJ, et al. . ID1 mediates escape from TGF-β tumor suppression in pancreatic cancer. Cancer Discov 2019;10:142–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kobayashi T, Nakazono K, Tokuda M, Mashima Y, Dynlacht BD, Itoh H. HDAC 2 promotes loss of primary cilia in pancreatic ductal adenocarcinoma. EMBO Rep 2016;18:334–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Cavaco ACM, Dâmaso S, Casimiro S, Costa L. Collagen biology making inroads into prognosis and treatment of cancer progression and metastasis. Cancer Metast Rev 2020;39:603–23. [DOI] [PubMed] [Google Scholar]
- 62. David CJ, Huang Y-H, Chen M, Su J, Zou Y, Bardeesy N, et al. . TGF-β tumor suppression through a lethal EMT. Cell 2016;164:1015–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.