Abstract
Purpose:
Young age at breast cancer diagnosis correlates with unfavorable clinicopathologic features and worse outcomes compared with older women. Understanding biological differences between breast tumors in young versus older women may lead to better therapeutic approaches for younger patients.
Experimental Design:
We identified 100 patients ≤35 years old at nonmetastatic breast cancer diagnosis who participated in the prospective Young Women's Breast Cancer Study cohort. Tumors were assigned a surrogate intrinsic subtype based on receptor status and grade. Whole-exome sequencing of tumor and germline samples was performed. Genomic alterations were compared with older women (≥45 years old) in The Cancer Genome Atlas, according to intrinsic subtype.
Results:
Ninety-three tumors from 92 patients were successfully sequenced. Median age was 32.5 years; 52.7% of tumors were hormone receptor-positive/HER2-negative, 28.0% HER2-positive, and 16.1% triple-negative. Comparison of young to older women (median age 61 years) with luminal A tumors (N = 28 young women) revealed three significant differences: PIK3CA alterations were more common in older patients, whereas GATA3 and ARID1A alterations were more common in young patients. No significant genomic differences were found comparing age groups in other intrinsic subtypes. Twenty-two patients (23.9%) in the Young Women's Study cohort carried a pathogenic germline variant, most commonly (13 patients, 14.1%) in BRCA1/2.
Conclusions:
Somatic alterations in three genes (PIK3CA, GATA3, and ARID1A) occur at different frequencies in young versus older women with luminal A breast cancer. Additional investigation of these genes and associated pathways could delineate biological susceptibilities and improve treatment options for young patients with breast cancer.
See related commentary by Yehia and Eng, p. 2209
Translational Relevance.
We performed whole-exome sequencing of tumor and germline for 92 women diagnosed with breast cancer at ≤35 years old, compared with older women (≥45 years old; median 61 years) in TCGA. In luminal A tumors, PIK3CA alterations were more common in older women, whereas GATA3 and ARID1A alterations were more common in younger women; 23.9% of young women carried a pathogenic germline variant (14.1% in BRCA1/2), which in several cases had not been covered in patients' original clinical germline testing. These results point to biological differences that may underlie some of the clinically distinct behavior of luminal A tumors in younger versus older women (e.g., differential chemosensitivity); suggest chromatin and transcriptional profiling as an important focus for follow-up investigations of distinct therapeutic targets among young patients; and underscore the importance of repeat germline testing as clinical methodologies improve and panels expand, particularly for groups at high germline risk.
Introduction
Approximately 4% of breast cancers are diagnosed among women younger than 40 years old (1). It has long been recognized that young age at breast cancer diagnosis correlates with unfavorable clinicopathologic disease features and, by extension, worse outcomes compared with disease in older women (2). Many of these differences are explained by the fact that distribution of breast cancer subtypes differs between young and older women, with more biologically aggressive intrinsic subtypes (basal-like and HER2-enriched) significantly overrepresented in the young. Early investigations of the biological differences between breast tumors in young versus older women showed that those differences were largely eliminated after accounting for intrinsic subtype (3). However, the availability of more comprehensive approaches for genomic analysis provides the opportunity to re-evaluate the biological landscape of breast cancers across age groups.
Moreover, examination of subtype-specific breast cancer outcomes by age at diagnosis has revealed an age-related prognostic difference among women with luminal A breast cancer. Specifically, younger women with luminal A tumors appear to experience worse outcomes than older women with luminal A tumors (4–6). The potential importance of very young age for outcomes and choice of therapy in luminal tumors was underscored in the SOFT/TEXT trials, where women under 35 years old were found to have a particularly high risk of recurrence and to benefit significantly from the addition of ovarian suppression to standard oral hormonal therapy (7). A clearer understanding of the biology underlying these observations could have important implications for optimizing therapy in young women with luminal breast cancer.
In this study, we performed whole-exome sequencing (WES)—comprising the coding sequence of ∼20,000 genes—of both tumor and germline samples from a cohort of women age 35 years and younger at breast cancer diagnosis. We sought to define the genomic landscape of primary breast tumors in this very young cohort; to compare it to the landscape of tumors diagnosed in older women from The Cancer Genome Atlas (TCGA) in a subtype-specific manner; and to comprehensively identify all pathogenic germline variants within the cohort.
Materials and Methods
Patient cohort
This is a subcohort derived from the Young Women's Breast Cancer Study (N = 1,302), a larger prospective cohort study of women aged 40 years or younger at breast cancer diagnosis who sought care at one of 13 sites in the United States or Canada between 2006 and 2016. Eligible patients for the overall cohort were <6 months from initial breast cancer diagnosis. Participants were identified by review of pathology records and clinic visit lists, then sent a formal invitation to participate by mail. Participation entails medical record review for clinical variables including breast cancer stage and subtype, and germline mutation status; collection of blood and tissue biospecimens; and completion of baseline and follow-up questionnaires (mailed every 6 months for three years following breast cancer diagnosis, then annually thereafter) on which patients self-report data including personal demographics, breast cancer treatments received, family history, pregnancy history, and disease status at follow-up. Categories of pregnancy-associated breast cancer were defined based on evolving standards (8). The study was approved by the institutional review board at Dana-Farber/Harvard Cancer Center and all participating centers, and all patients provided written informed consent prior to any study activities. The study was conducted in accordance with the Declaration of Helsinki. Additional study details have been described previously (9–11).
Participants in this genomic sequencing subcohort were ≤35 years old at breast cancer diagnosis and represent the first 100 patients enrolled into the larger cohort who met this age cut-off (though sequencing did not succeed in all selected patients). All patients in this subcohort were diagnosed between 2006 and 2013. Patient and disease characteristics were obtained through medical record review and patient surveys. Central pathology review for grade and histology was conducted for all participants, and biomarkers including estrogen receptor (ER), progesterone receptor (PR), and HER2 status were abstracted from pathology reports and designated based on standard clinical guidelines as described previously (9). Receptor status and grade were used as a surrogate for intrinsic subtype, also as previously described, as follows: luminal A [ER-positive (ER+) and/or PR-positive (PR+), HER2−, and grade 1–2]; luminal B (ER+ and/or PR+ and HER2+, or ER+ and/or PR+ and grade 3); HER2 enriched (ER−, PR−, and HER2+); or basal-like (ER, PR, and HER2−; ref. 12).
WES
Tumor or germline DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissue and peripheral blood mononuclear cells, respectively, and libraries for massively parallel sequencing were constructed as previously described, and are essentially the same as all protocols followed by TCGA (13). Sequencing data were generated via the Illumina HiSeq platform. The following pipeline was designed to match the software used to derive the TCGA-breast genomic data (14). Germline and tumor FASTQ files were aligned to the GRCh37 human reference assembly using BWA version 0.5.9 (15). The resulting BAM files were fed into Picard version 2.6–14 to filter out duplicate reads (MarkDuplicates) and flag OxoG artifacts (CollectOxoGMetrics; Picard Toolkit, Broad Institute, GitHub repository 2019). GATK version 3.1–1 was then used to fix misaligned short indels (IndelRealigner), adjust base quality scores (BaseRecalibrator), and in the case for germline samples, to call variants (HaplotypeCaller in GVCF mode; ref. 16). Somatic SNVs were called using MuTect version 1.1.6, and short indels were called using Strelka version 1.0.11, using default settings [LOD score > 6.3 for MuTect, read depth > 2, combined tumor/normal variant allele frequency (VAF) > 10% for <5bp indels, VAF > 2% for >5bp indels, and downstream post-call filtration for Strelka; ref. 17). False-positive somatic calls were controlled using a panel of normals (PoN) derived from TCGA samples, and with candidate read realignment with BLAT & NovoAlign (http://www.novocraft.com; ref. 18). GATK FilterByOrientationBias was used to control FFPE artifacts. Visual verification of somatic mutations via IGV was performed for mutations in SLC22A2. TCGA-breast somatic mutations were downloaded from the Broad Institute TCGA Genome Data Analysis Center (GDAC; Broad Institute TCGA Genome Data Analysis Center: Firehose version 2016_01_28. doi:10.7908/C11G0KM9, 2016; accessed September 13, 2018) and clinical variables were downloaded from the Genomic Data Commons (GDC) portal (accessed September 13, 2018; refs. 14, 19).
Somatic copy-number variants (CNV) were called with ReCapSeg version 1.5.0, and ABSOLUTE version 1.06 was used to estimate tumor purity and ploidy, to detect subclonal copy-number alterations (SCNA), and to estimate mutation multiplicity (20). GISTIC 2.0 was used to detect significant areas of focal and arm-level amplifications and deletions (21). Significantly altered genes in the cohort were identified using MutSig2CV (22). Oncotator version 1.9.0 paired with the April 5, 2016, datasource corpus was used to annotate somatic SNVs and short indels (23). Mutational signatures were identified using SignatureAnalyzer-CPU (24). Fisher exact test (with Benjamini–Hochberg correction) was used to find Cancer Gene Census (CGC) genes (accessed May 27, 2020; ref. 25) significantly enriched or depleted in somatic mutations between cohorts (faceted by intrinsic subtype). Both somatic and germline statistical analyses were performed in Python using the Pandas, SciPy, and statsmodels modules. The oncoprint heatmaps depicting somatic alterations were made using ComplexHeatmap package (26). Somatic SNVs in PIK3CA were visualized as lollipop plots generated using the MutationMapper tool in cBioPortal (27).
An ordinary least squares (OLS) regression model was used to examine tumor mutational burden (TMB) as a function of diagnosis age and study cohort, faceted by intrinsic subtype. TCGA TMB data were taken from a prior study on hypermutated breast cancer (28). The Python seaborn package was used to generate the visualization.
Pathogenic germline TCGA-breast cancer variants were derived from the TCGA PanCanAtlas germline working group Supplementary Materials (29). CharGer was used to isolate clinically relevant germline variants in 152 familial cancer-related genes, and gnomAD was used as an additional filter against low-quality ultrarare variants (30). Fisher exact test was used to compare the proportion of patients impacted by a pathogenic germline variant between cohorts.
Data availability statement
The data generated in this study are available within the article and its Supplementary Data files.
Results
Patient/tumor characteristics and breast cancer outcomes in young women
Patient and tumor characteristics for the overall cohort (93 tumors from 92 patients; one patient had bilateral disease) are shown in Table 1. Median age in the Young Women's Study cohort was 32.5 years (range 23–35 years). The majority of patients had hormone receptor-positive (HR+; meaning, ER+ and/or PR+)/HER2− breast cancer (52.7%), and 64.5% of patients had grade 3 breast cancer. Of note, 13 patients (14.0%) had received some neoadjuvant therapy prior to their sequenced tissue specimen. Given the very young age of women in the cohort overall, we assume that all or nearly all participants were premenopausal at breast cancer diagnosis. With 9.2 years of median follow-up, 4 patients had experienced locoregional breast cancer recurrence (without subsequent distant recurrence or death), 17 patients had experienced distant breast cancer recurrence, and 10 patients had died from breast cancer, for median distant recurrence-free survival of 9.5 years and median overall survival of 10.0 years.
Table 1.
Patient and tumor characteristics, treatments received, and breast cancer outcomes for the sequenced young women's cohort versus TCGA ≥45-yo cohort.
| YWBC (≤35 yo), N = 92 patients | TCGA (≥45 yo), N = 925 patients | |
|---|---|---|
| Parameter | No. of patients (%) | No. of patients (%) |
| Age at diagnosis (years) | Median 32.5 (range 23–35) | Median 61 (range 45–90) |
| Race | ||
| Caucasian | 79 (85.9%) | 649 (70.2%) |
| Black or African-American | 0 (0%) | 146 (15.8%) |
| Asian | 7 (7.6%) | 48 (5.2%) |
| Multi-racial | 3 (3.3%) | Not collected |
| Other/unknown | 3 (3.3%) | 82 (8.9%) |
| Ethnicity | ||
| Hispanic or Latina | 6 (6.5%) | 31 (3.4%) |
| Non–Hispanic or Latina | 81 (88.0%) | 744 (80.4%) |
| Unknown/did not answer | 5 (5.4%) | 150 (16.2%) |
| Anatomic stage at diagnosis | ||
| 0 (DCIS) | 3 (3.3%) | 0 (0%) |
| I | 32 (34.4%) | 160 (17.3%) |
| II | 46 (49.5%) | 525 (56.8%) |
| III | 12 (12.9%) | 201 (21.7%) |
| IV | 0 (0%) | 18 (1.9%) |
| Unknown | 0 (0%) | 21 (2.3%) |
| T stage | ||
| Tis | 3 (3.3%) | 0 (0%) |
| T1 | 45 (48.9%) | 241 (26.1%) |
| T2 | 39 (42.4%) | 535 (57.8%) |
| T3 | 5 (5.4%) | 112 (12.1%) |
| T4 | 0 (0%) | 34 (3.7%) |
| TX | 0 (0%) | 3 (0.3%) |
| N stage | ||
| N0 | 47 (51.1%) | 455 (49.2%) |
| N1 | 35 (38.0%) | 289 (31.2%) |
| N2–3 | 8 (8.7%) | 162 (17.5%) |
| NX | 2 (2.2%) | 19 (2.1%) |
| Tumor grade | ||
| 1 | 5 (5.4%) | |
| 2 | 28 (30.1%) | |
| 3 | 60 (64.5%) | |
| Receptor status | ||
| ER+ or PR+/HER2− | 49 (52.7%) | |
| HER2+ | 26 (28.0%) | |
| ER−/PR−/HER2− | 15 (16.1%) | |
| Other (HER2 indeterminate or unknown) | 3 (3.2%) | |
| Histology | ||
| Ductal | 70 (75.3%) | 643 (69.5%) |
| Lobular | 2 (2.2%) | 190 (20.5%) |
| Mixed ductal and lobular | 8 (8.6%) | Not collected |
| Other/unknown | 13 (14.0%) | 92 (9.9%) |
| Intrinsic subtypea | ||
| Luminal A | 28 (30.1%) | 429 (46.4%) |
| Luminal B | 43 (46.2%) | 169 (18.3%) |
| HER2 | 4 (4.3%) | 68 (7.4%) |
| Basal-like | 17 (18.3%) | 139 (15.0%) |
| Normal | 0 (0%) | 29 (3.1%) |
| Unknown | 1 (1.1%) | 91 (9.8%) |
| Surgery at original diagnosis | ||
| Lumpectomy | 32 (34.8%) | |
| Unilateral mastectomy | 20 (21.7%) | |
| Bilateral mastectomy | 40 (43.5%) | |
| Chemotherapy receipt | ||
| Yes | 79 (85.9%) | |
| No | 13 (14.1%) | |
| Hormonal therapy receiptb | ||
| Yes | 70 (76.1%) | |
| No | 22 (23.9%) | |
| Received neoadjuvant therapy prior to sequenced tissue specimen | ||
| Yes | 13 (14.0%) | |
| No | 80 (86.0%) | |
| Parity at breast cancer diagnosis | ||
| Nulliparous | 43 (46.7%) | |
| Parous | 49 (53.3%) | |
| Number of prior pregnancies | Median 2 (range 1–7) | |
| Pregnancy-associated breast cancerc | ||
| Breast cancer during pregnancy | 5 (5.4%) | |
| Breast cancer ≤1 year post-partum | 7 (7.6%) | |
| Breast cancer >1 year and ≤5 years post-partum | 25 (27.2%) | |
| Invasive disease-free survival eventsc (median follow-up 9.2 years) | ||
| None | 71 (77.2%) | |
| Ipsilateral locoregional recurrence | 4 (4.3%) | |
| Distant recurrence | 17 (18.5%) | |
| Contralateral new breast primary invasive tumor | 0 | |
| Breast cancer death | 10 (10.9%) | |
Note: Ninety-two patients were included in the young women's cohort, comprising 93 specimens (one patient with bilateral breast cancers). Per patient parameters are listed with N = 92, per specimen parameters are listed with N = 93. Treatments received are for original diagnosis of nonmetastatic disease. Nine hundred twenty-five patients were in the older TCGA cohort. Parameters from the TCGA cohort that were not collected or not collected in the same manner as the YWBC cohort are left blank in the TCGA column.
Abbreviations: DCIS, ductal carcinoma in situ; TCGA, The Cancer Genome Atlas; yo, years old; YWBC, young women's breast cancer cohort.
aSee intrinsic subtype definitions in Materials and Methods section. One patient with ER+/PR+/HER2 unknown and grade 2 disease was categorized as luminal A.
bHormonal therapy receipt was categorized as “yes” if a patient received any hormonal treatment approach (including ovarian suppression, tamoxifen, aromatase inhibitor, or other) through 5 years from study enrollment. The cohort included 72 patients with ER+ or PR+ tumor (either HER2+ or HER2−), of whom 70 received some hormonal therapy.
cBreast cancer during pregnancy was defined as breast cancer diagnosis date within 40 weeks prior to a child's reported birth date.
dIpsilateral locoregional recurrence indicates patients with isolated locoregional recurrence only, without concurrent distant recurrence (there were no patients with locoregional recurrence and subsequent distant recurrence). Breast cancer death is not mutually exclusive of other categories.
Somatic genomic landscape in very young women
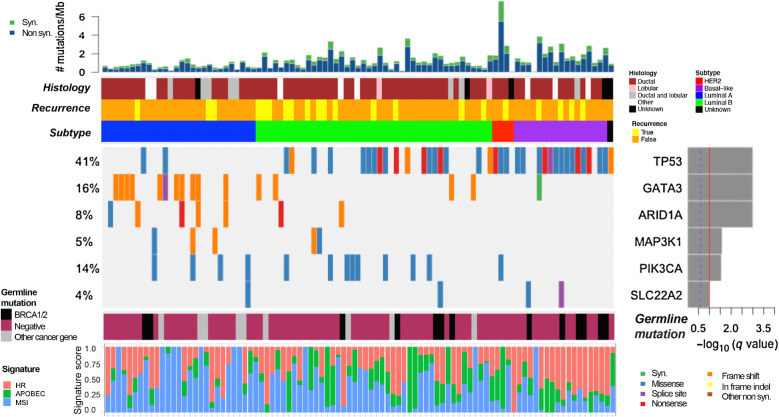
Analysis of somatic SNVs and short indels in the tumor samples demonstrated that the most common gene to contain SNVs and indels across the cohort was TP53 (41% of patients), followed by GATA3 (16%) and PIK3CA (14%; Supplementary Fig. S1). Full sequencing files are available in the data supplement (Supplementary Table S1; Supplementary Tables S2 and S3 provide associated clinical information by patient). Somatic CNV analysis revealed that ERBB2 was the most commonly affected gene, with 22% of patients showing ERBB2 amplification (Supplementary Fig. S2A). Analysis of significantly recurrently altered genomic alterations using MutSig2CV (Fig. 1) revealed 6 genes to be mutated more than expected by chance (in decreasing order of frequency): TP53, GATA3, PIK3CA, AT-rich interaction domain 1A (ARID1A), MAP3K1, and SLC22A2. Of these, ARID1A and SLC22A2 were not identified in a previous landmark analysis of nonmetastatic primary breast tumors across all age groups (31). SLC22A2 is a cation transporter with a role in cellular uptake of platinum chemotherapy agents (32); two tumors contained missense mutations in SLC22A2 at the exact same locus, although of unknown functional consequence (Supplementary Fig. S3). ARID1A, a component of the SWI/SNF chromatin remodeling complex, was altered in 8% of patients in our cohort, nearly all with loss of function mutations (nonsense or frameshift).
Figure 1.
Significant SNVs, short indels, and signature analysis. Comutation plot showing recurrent somatic alterations in significantly mutated genes across the cohort (N = 93) as analyzed by MutSig2CV. TP53, GATA3, ARID1A, MAP3K1, PIK3CA, and SLC22A2 are significantly mutated. The P values were computed using the Fisher method and truncated product method. FDR (q values) were generated using the Benjamini–Hochberg method to correct for multiple hypotheses. Genes that have a −log10 q-value ≥1 (red line) are considered significant. Bar graph (top) depicts the TMB (mutations/megabase) of each patient's tumor samples, followed by clinical annotations depicting histology, disease recurrence, and breast cancer subtype (key to the right of panel). Bottom panel annotations show cancer-specific pathogenic germline variants, and somatic mutational signatures of homologous recombination (HR) deficiency, APOBEC activity, and microsatellite instability (MSI).
We evaluated whether differences in somatic mutational profile (SNVs and short indels) were observed on the basis of parity at diagnosis or pregnancy-associated breast cancer status within the sequenced cohort. We did not find any significant genomic differences between women who were parous versus nulliparous at breast cancer diagnosis (Supplementary Fig. S4). For women with pregnancy-associated breast cancer (N = 37; defined as breast cancer diagnosed during pregnancy or ≤5 years post-partum), there was a statistically significant enrichment in TP53 and GATA3 alterations, compared with women with nonpregnancy-associated breast cancer (Supplementary Fig. S5).
Homologous recombination deficiency (HRD), APOBEC, and microsatellite instability (MSI) were the three mutational signatures detectable within the cohort (Fig. 1). The presence of the HRD signature was associated with basal-like subtype (Wilcoxon rank-sum P < 7e−6). Copy-number changes were analyzed and visualized in Supplementary Figs. S2A and S2B; full results of CNV analysis are included in the data supplement (Supplementary Table S4).
Comparison of somatic landscapes in very young and older women
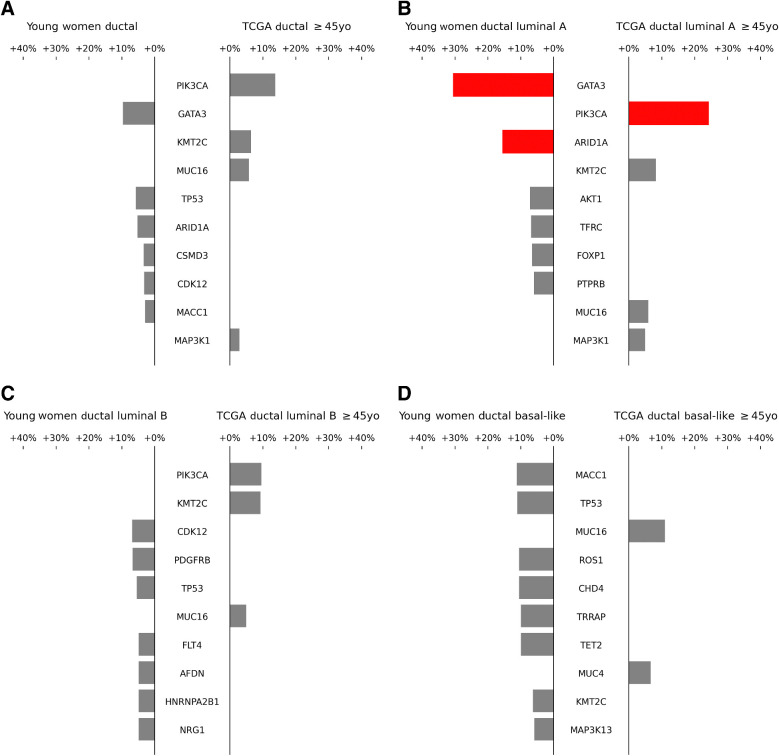
We compared all SNVs and short indels identified in our cohort with all alterations identified in older women (age ≥45 years) in TCGA for breast tumors. Median age in the older TCGA cohort was 61 years (range 45–90 years; Table 1). To avoid confounding by intrinsic subtype or histology (both of which are known to differ significantly based on age at breast cancer diagnosis; refs. 3, 33, 34), we excluded women with pure lobular histology from these analyses to focus entirely on ductal histology, and performed each comparison within the four intrinsic subtypes (luminal A, luminal B, HER2 enriched, and basal-like; Fig. 2A–D; ref. 35). Patient and tumor characteristics within luminal A and luminal B intrinsic subtype patients from the very young women's cohort versus TCGA older women's cohort are shown in Supplementary Table S5. Among women with nonlobular luminal A tumors (N = 28), PIK3CA alterations were significantly enriched in older women (14% vs. 38% in younger vs. older women, respectively, q < 0.05), whereas GATA3 and ARID1A alterations were significantly more common in younger women (GATA3: 43% vs. 12% in younger vs. older women, respectively, q < 0.05; ARID1A: 18% vs. 2% in younger vs. older women, respectively, q < 0.05; Figure 2B). Lollipop plot representations did not reveal any obvious difference between the localization of PIK3CA mutations in very young versus older women, which were primarily canonical hotspot mutations in the helical and kinase domains (Supplementary Figs. S6A and S6B). Correlation of specific alterations with invasive disease-free survival (iDFS) events in young women was not evaluated due to insufficient power (only two iDFS events in young women with luminal A tumors). No significant differences between young and older women were identified with the same analysis among nonlobular luminal B (N = 41; Fig. 2C), HER2 enriched (N = 4), and basal-like tumors (N = 17; Fig. 2D). When the same analysis was run without consideration of intrinsic subtype or histology, PIK3CA alterations were still detected to be enriched in older women, and CDH1 was found to be enriched in older women (reflecting the known increased prevalence of lobular histology), but other significant differences were not found. Analysis of pure lobular histology tumors was not performed due to small numbers of this histology in the Young Women's Study cohort.
Figure 2.
Comparison of single nucleotide and short indel prevalence between Young Women's Breast Cancer Study cohort (≤35 years old) and TCGA patients ≥45 years old. A, All intrinsic subtypes; B, Luminal A; C, Luminal B; D, Basal-like. HER2-enriched subtype is not shown as there were only 4 Young Women's Breast Cancer Study cohort samples in this subtype. Analysis excluded patients with pure lobular tumor histology. Forty genes identified from the 2016 METABRIC study (35), in addition to one gene found to be significant by MutSig, were included for analysis. Only differences in the frequencies of alteration of each gene between the two cohorts, as opposed to the absolute frequency of alterations within each cohort, are depicted by the bars. Statistically significant differences (FDR < 5%) are highlighted in red.
Comparative analysis of mutational signatures between the very young women's cohort and older women (≥45 years old) in TCGA revealed no statistically significant differences between the two age groups for the signatures represented among very young women (APOBEC, HRD, and MSI). TMB was also compared across age groups, and appeared overall similar. In the young women's cohort, median TMB was 1.55 mutations/megabase (mut/Mb; range 0.13–11.6) and one sample (1.1%) was hypermutated (defined as ≥10 mut/Mb). Among older women in TCGA, median TMB was 1.13 mut/Mb (range 0.025–142.6) and 10 samples (1.21%) were hypermutated. Incorporating both the young women's cohort patients and TCGA patients of all ages, we analyzed the correlation of TMB with age according to intrinsic subtype, and found no statistically significant correlation between TMB and age except in the HER2-enriched subpopulation, where there was a positive correlation between increasing age and increasing TMB (Supplementary Fig. S7).
Germline genomic landscape of very young women
We examined the frequency and identities of pathogenic variants in germline DNA from our cohort of very young women and from older women (age ≥45 years) in TCGA. Median age in the older TCGA cohort was 61 years (range 45–90 years). Pathogenic germline variants were defined by the TCGA PanCanAtlas germline working group as described previously, with CharGer used to isolate clinically relevant variants (29, 30). Of the 92 women in our cohort, 22 (23.9%) carried a pathogenic germline variant, of which 13 (14.1%) had alterations in BRCA1 or BRCA2, and 10 (10.9%) had an alteration in another cancer-related gene. These frequencies were similar to the youngest women (≤35 years old; median 32.5 years) in TCGA (N = 34). By comparison, among older women in TCGA (N = 925), 8.8% carried a pathogenic germline variant, of which 3.0% had alterations in BRCA1/2, and 5.7% had an alteration in another cancer-related gene (Table 2). All genes with pathogenic cancer-related germline hits by cohort are shown in Table 2. Supplementary Table S6 shows all pathogenic hits in any germline gene, cancer-related and noncancer-related.
Table 2.
Comparison of pathogenic germline findings by WES from Young Women's Breast Cancer Study cohort vs. TCGA.
| Patient cohort (age group) | Pathogenic germline variant in any cancer gene no. (%) | Pathogenic germline variant in BRCA1/2 no. (%) | Pathogenic germline variant in non-BRCA1/2 cancer gene no. (%) |
|---|---|---|---|
| Young Women's Breast Cancer Study (≤35 yo), N = 92 | 22 (23.9%)a | 13 (14.1%) | 10 (10.9%) |
| BRCA1 (10 patients), BRCA2 (3 patients) | PALB2 (3 patients), BUB1B (2 patients), TP53 (1 patient), PRSS1 (2 patients), GJB2 (1 patient), COL7A1 (1 patient) | ||
| TCGA (≤35 yo), N = 34 | 9 (26.5%)a | 6 (17.6%) | 3 (8.8%) |
| BRCA2 (4 patients), BRCA1 (2 patients) | TP53 (1 patient), ATR (1 patient), ATM (1 patient), BRIP1 (1 patient) | ||
| TCGA (≥45 yo), N = 925 | 81 (8.8%)a | 28 (3.0%) | 53 (5.7%) |
| BRCA1 (14 patients), BRCA2 (14 patients) | ATM (8 patients), ATR (3 patients), BRIP1 (3 patients), CHEK2 (3 patients), PALB2 (3 patients), Other (34 patients) | ||
| Comparison between YWBCS ≤35 yo and TCGA ≥45 yo | P = 4.0e−5 | P = 2.5e−5 | P = 0.066 |
| Comparison between TCGA ≤35 yo and TCGA ≥45 yo | P = 2.8e−3 | P = 8.3e−4 | P = 0.44 |
Abbreviations: TCGA, The Cancer Genome Atlas; yo, years old; YWBCS, Young Women's Breast Cancer Study.
aTwo TCGA patients (one ≤35 years, one ≥45 years) had two pathogenic germline variants. One YWBCS patient had two pathogenic germline variants.
All patients in whom WES identified a pathogenic germline variant had also undergone some clinical germline genetic testing in the past, though because all patients in this cohort were diagnosed with breast cancer between 2006 and 2013, the clinical testing these patients received typically involved only a small number of genes as opposed to the broader germline panels used today. Across all patients in our cohort, BRCA1/2 had been clinically evaluated in all patients; TP53 had been evaluated in 11% of patients; and additional genes had been evaluated in only two patients. Table 3 shows results from clinical germline genetic testing compared with germline WES performed in this study. WES identified all pathogenic variants found through clinical testing. For four additional patients, WES identified pathogenic variants in commonly described breast cancer susceptibility genes (PALB2 in three patients; TP53 in one patient) that had not been performed in the prior clinical genetic testing. For six more patients, WES identified pathogenic variants in cancer susceptibility genes never or rarely linked to breast cancer, which also had not been included in these patients' prior clinical genetic testing (BUB1B in two patients; COL7A1, GJB2, and PRSS1 in one patient each; one final patient with germline pathogenic BRCA2 mutation had a PRSS1 mutation as well; Table 3; refs. 36, 37).
Table 3.
Comparison of WES and clinical genetic testing among very young women with pathogenic germline variants.
| Patient | Pathogenic germline variant identified by WES (alteration type) | Somatic hit identified by WES? (if yes, alteration type) | Gene ID from WES tested clinically? | Pathogenic/likely pathogenic alteration identified in clinical genetic testing |
|---|---|---|---|---|
| 26 | BRCA1 (frameshift_ins) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 309 | BRCA1 (frameshift_del) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 315 | BRCA1 (frameshift_del) | No | Yes | BRCA1 (positive, clinically actionable) |
| 663 | BRCA1 (nonsense mut) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 774 | BRCA1 (nonsense mut) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 786 | BRCA1 (nonsense mut) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 844 | BRCA1 (frameshift_ins) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 879 | BRCA1 (frameshift_del) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 953 | BRCA1 (frameshift_del) | Yes (LOH) | Yes | BRCA1 (positive, clinically actionable) |
| 1063 | BRCA1 (nonsense mut) | No | Yes | BRCA1 (positive, clinically actionable) |
| 228 | BRCA2 (frameshift_ins) | No | Yes | BRCA2 (positive, clinically actionable) |
| 491 | BRCA2 (frameshift_del) | No | Yes | BRCA2 (positive, clinically actionable) |
| 857 | BRCA2 (nonsense mut) | Yes (LOH) | Yes | BRCA2 (positive, clinically actionable) |
| 29 | PALB2 (frameshift_del) | Yes (frameshift_del and splice site) | No | None |
| 1033 | PALB2 (frameshift_del) | Yes (nonsense mut) | Unknowna | None |
| 1074 | PALB2 (splice site) | No | No | None |
| 406 | BUB1B (splice site) | No | No | None |
| 915 | BUB1B (missense mut) | No | No | None |
| 653 | TP53 (frameshift_del)_ | No | Unknowna | None |
| 493 | COL7A1 (splice site) | No | No | Noneb |
| 614 | GJB2 (frameshift_del) | No | No | None |
| 491 | PRSS1 (frameshift_del) | No | No | None |
| 686 | PRSS1 (splice site) | No | No | None |
Note: All patients had some clinical genetic testing performed. For the “Gene ID from WES tested clinically” column: Yes indicates the gene was confirmed as evaluated in clinical testing; No indicates the gene was confirmed as not evaluated in clinical testing; Unknown indicates it is unknown whether the gene was evaluated in clinical testing (i.e., clinical genetic testing was performed, but it is unknown whether the specific gene of interest was evaluated).
Abbreviation: LOH, loss of heterozygosity.
aClinical genetic testing for the gene of interest in these patients was highly unlikely, but its absence could not be entirely confirmed based on the records available.
bPatient 493 had a BRCA2 VUS identified on clinical testing.
Discussion
In this work, we compared the somatic and germline landscape of very young women to older women with breast cancer. By focusing on a particularly young patient subset (≤35 years old, whereas many other investigations have used a cut off of ≤40 years old) and segregating analyses by tumor histology and intrinsic subtype, we sought to specifically identify genomic alterations that may have clinical importance for young women with breast cancer. Our focus on women ≤35 years old was also driven by the differential treatment implications around this age cut-off for premenopausal women participating in the SOFT/TEXT trials (7).
In the somatic genomic landscape of very young women with luminal A tumors, we identified fewer PIK3CA alterations and more alterations in GATA3 and ARID1A compared with older women. Previous literature demonstrates that younger women with luminal A tumors experience worse breast cancer outcomes, and a number of reasons for this have been suggested including a lower chance of permanent chemotherapy-associated amenorrhea among younger women, decreased adherence to endocrine therapy among younger women, and inherent biology (4–6). PIK3CA mutations affect the catalytic subunit of a kinase in the PI3K pathway, which is overactive in a number of cancers including breast cancer. In early-stage breast cancer, the presence of a PIK3CA mutation correlates with improved long-term outcome (38), which is congruent with our observation that PIK3CA mutations are more common in luminal A tumors from older women, in whom prognosis is more favorable. Whether the level of PI3K pathway activity plays a causative role in the differential prognosis bears further investigation, which could shed light on the biological drivers of poorer prognosis in younger women.
Increased GATA3 alterations observed in very young women with luminal A tumors is of interest, but with unclear biological implications. GATA3, a transcription factor with important roles in ER-regulated transcription and maintenance of luminal differentiation, is commonly mutated in breast tumors (39, 40). Two other groups have also reported a significantly higher percentage of GATA3 alterations in younger patients with breast cancer (33, 41). Lower GATA3 expression correlates with worse prognosis, thus its increased alterations in younger patients could possibly explain some of their less favorable outcomes. However, the biological effects of GATA3 in breast cancer in general are uncertain: evidence has suggested both oncogenic and tumor suppressor roles, and different mutations may be gain-of-function or loss-of-function (33, 40).
ARID1A is a component of the chromatin-regulating SWI/SNF complex and regulates ER-dependent transcriptional programs. Loss or depletion of ARID1A causes a switch in cell identities from luminal to basal-like (39). Alterations in ARID1A are well documented to correlate with worsened clinical outcomes among patients with breast cancer (39, 42), consistent with the increased frequency of ARID1A alterations in very young women with luminal A tumors in our cohort. Decreased ARID1A causes resistance to both tamoxifen and fulvestrant in breast cancer cell line models. In contrast, preclinical experiments suggest that ARID1A-mutated breast cancers may be sensitive to BET inhibitors (39, 42). The biological implications of increased ARID1A alterations among young patients with breast cancer should be further explored, with the hypothesis that ARID1A alteration may cause a more ER-independent transcriptome even in tumors that appear to be “luminal A” by histopathology. This in turn could clarify a mechanism behind poorer prognosis in young women with luminal A tumors, and suggest chromatin regulatory elements as a therapeutic target.
Our findings build on previous findings in breast cancer genomics of young women in multiple important ways, and broaden the clinical implications. In a previous paper by Azim and colleagues (33) examining tumor genomics by patient age within the TCGA breast cancer cohort (≤45 years old vs. 46–69 years old vs. ≥70 years old), GATA3 was the only gene found to be mutated more frequently in younger patients. PIK3CA was numerically more likely to be mutated in older patients but this did not reach statistical significance (33). More recently, Kan and colleagues (41) showed a higher prevalence of GATA3 alterations in a cohort of Korean women that was enriched for, but not exclusively composed of, younger women, compared with the TCGA breast cancer cohort (this analysis did not control for intrinsic subtype). PIK3CA and ARID1A were not identified as differentially altered between younger and older patients in the Kan and colleagues analyses (41). Here, potentially due to upfront stratification by subtypes and focus on a particularly young patient subset (≤35 years old), we were able to replicate the GATA3 findings, demonstrate statistical significance of increased PIK3CA mutations in older patients, and newly identify ARID1A alterations as more likely to occur in younger patients.
In the Azim and colleagues analysis, intrinsic subtype (by 50-gene signature PAM50) was incorporated as a covariate in a logistic regression model to assess genomic differences (33). In contrast, our analysis stratified by intrinsic subtype (approximated from histologic parameters) ahead of time, allowing us to demonstrate that significant genomic differences are seen specifically in luminal A patients (despite the fact that this was not the most prevalent, and therefore not the highest powered, subgroup within the Young Women's Study cohort). The identification of genomic differences specific to luminal A tumors is notable because multiple groups have shown that luminal A is the only breast cancer subtype in which prognosis differs between young and older women—and therefore it is the subtype where biological differences may drive distinct clinical outcomes (4–6). Whether true biological differences exist between hormone receptor-positive younger and older patients with breast cancer, and if or how we should use that to guide different therapeutic decisions based on age, is an issue of enormous clinical importance. The large, prospective TAILORx and RxPONDER trials both suggest differential chemotherapy benefit between younger and older women with HR+/HER2− breast cancer (43, 44). It remains unclear what proportion, if any, of chemotherapy benefit seen in younger women may be due to a chemoendocrine effect versus an inherent biological difference that produces differential chemosensitivity. Findings, like those we present here, of potential true biological differences between tumors from younger versus older women may ultimately help to address this important clinical question.
Our germline WES of very young women with breast cancer revealed a number of findings with clinical implications. Germline mutations in BRCA1/2 are unsurprisingly enriched in this very young cohort, with 14.1% of patients harboring pathogenic mutations. This is congruent with germline findings in the Prospective Outcomes in Sporadic versus Hereditary breast cancer (POSH) cohort of nearly 3,000 patients with breast cancer diagnosed at age ≤40 years old, in which 12% of patients carried a pathogenic BRCA1/2 mutation (45). The frequency of germline susceptibility decreases substantially with increasing age, as demonstrated by our germline findings in older women from TCGA and, for example, the very large CARRIERS study cohort in which over 19,000 patients with breast cancer diagnosed at age ≤50 years old had a 7.3% chance of carrying a pathogenic variant in a cancer predisposition gene (46). With comprehensive sequencing we identified four patients with clinically important pathogenic germline variants (in PALB2 and TP53) not identified on their original clinical genetic testing because the altered gene was not clinically evaluated. The most important clinical lesson of our germline sequencing results is to underscore the importance of repeat germline testing as clinical methodologies improve and panels expand, particularly for groups at high germline risk such as young patients.
Beyond expected alterations in BRCA1/2, PALB2, and TP53, germline WES identified variants in four cancer susceptibility genes (BUB1B, PRSS1, COL7A1, and GJB2) that historically have not been linked or are only weakly linked to breast cancer. Specifically, germline mutations in BUB1B and PRSS1 have been previously linked to heritable gastrointestinal and pancreatic cancers, respectively. Germline mutations in COL7A1 and GJB2 have been implicated as potentially pathogenic in patients with breast and ovarian cancer (36, 37, 47). Of note, a somatic second hit was not detected in any of the patients with germline first hits in these less common genes. Though it is not clear whether these germline variants played a causative role in breast cancer pathogenesis for our patients, these data and similar data obtained from clinical expanded germline panel testing will be instrumental for establishing connections between germline alterations and specific cancer risks. At the same time, the fact that only 23.9% of these very young women had pathogenic germline variants found on WES suggests that there may be much yet to discover in this population.
Our analysis has several limitations. When considering the comparisons between luminal A tumors in young versus older women, it is important to note that intrinsic subtype in our cohort was defined according to pathologic features, and was defined according to gene expression microarrays in TCGA. However, pathology-based intrinsic subtype was previously shown to be an adequate surrogate for expression-based intrinsic subtype classification (12). In addition, the genomic differences we identified between luminal A patients were not observed when luminal A and B patients were grouped together (Supplementary Fig. S8), suggesting that the findings were not simply a result of differential classification between luminal subtypes in young versus older patients. Our failure to find significant differences in somatic alterations between young and older women with breast cancer in HER2 enriched and basal-like subtypes may be due to underpowering given the small number of patients in each subgroup of young women. Although grade and histology were centrally reviewed in all patients, receptor status was centrally reviewed in only some cases. The receipt of neoadjuvant chemotherapy predating the sequenced tumor specimen may have impacted somatic alterations, although this was only an issue for a small minority of patients. Our results would be strengthened by analysis of a separate validation cohort, however we were not able to identify a cohort with a sufficient number of similarly young women and intrinsic subtype information available for each tumor, pointing to the novelty of our cohort as well as the need for further investigation in this area.
In summary, we have used WES of both tumor and germline specimens to investigate the unique pathogenesis of breast cancer in very young women. In germline sequencing, we identified a 14.1% prevalence of pathogenic BRCA1/2 alterations, and found several patients with actionable germline findings that were missed on older clinical testing platforms, serving as a reminder of the importance of updating germline testing in any patient at high risk for a familial cancer syndrome. Through somatic sequencing, we demonstrated an enrichment for ARID1A alterations in young women's breast tumors and identified three genes (PIK3CA, GATA3, ARID1A) in which somatic alterations could plausibly contribute to less favorable biology for young patients with luminal A breast cancer. The suggestion of distinct biological features for luminal tumors in young versus older women will be important to follow-up in order to understand whether these features could explain any of the age-related difference in chemosensitivity that was observed in the TAILORx and RxPONDER trials. Given the roles of GATA3 as a transcription factor and ARID1A as a chromatin regulator, examination of transcriptional profiles among young women's luminal A tumors will be an important next step. The ongoing study of larger cohorts of young women, with genomic analysis by tumor subtype, may help to delineate biological susceptibilities and improve treatment options for young patients with breast cancer.
Authors' Disclosures
A.G. Waks reports other support from Genentech/Roche and MacroGenics outside the submitted work. J. Peppercorn reports personal fees and other support from GlaxoSmithKline outside the submitted work. N. Wagle reports personal fees from Eli Lilly and Co., Relay Therapeutics, and Flare Therapeutics and grants from Puma Biotechnologies outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This research was supported by Susan G. Komen (A.H. Partridge), the Breast Cancer Research Foundation (A.H. Partridge), and the Susan Smith Executive Council (N. Wagle). The authors wish to acknowledge the patients who participated in this study.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
A.G. Waks: Conceptualization, data curation, methodology, writing–original draft, writing–review and editing. D. Kim: Conceptualization, data curation, software, formal analysis, validation, visualization, methodology, writing–original draft, writing–review and editing. E. Jain: Visualization, writing–review and editing. C. Snow: Data curation, writing–review and editing. G.J. Kirkner: Formal analysis, writing–review and editing. S.M. Rosenberg: Writing–review and editing. C. Oh: Formal analysis, writing–review and editing. P.D. Poorvu: Writing–review and editing. K.J. Ruddy: Writing–review and editing. R.M. Tamimi: Writing–review and editing. J. Peppercorn: Writing–review and editing. L. Schapira: Writing–review and editing. V.F. Borges: Writing–review and editing. S.E. Come: Writing–review and editing. E.F. Brachtel: Writing–review and editing. E. Warner: Writing–review and editing. L.C. Collins: Writing–review and editing. A.H. Partridge: Conceptualization, resources, supervision, funding acquisition, methodology, writing–review and editing. N. Wagle: Conceptualization, resources, supervision, funding acquisition, methodology, writing–review and editing.
References
- 1. Society AC. American cancer society: breast cancer facts and figures 2019–2020. <https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/breast-cancer-facts-and-figures/breast-cancer-facts-and-figures-2019-2020.pdf>. [Google Scholar]
- 2. Anders CK, Hsu DS, Broadwater G, Acharya CR, Foekens JA, Zhang Y, et al. Young age at diagnosis correlates with worse prognosis and defines a subset of breast cancers with shared patterns of gene expression. J Clin Oncol 2008;26:3324–30. [DOI] [PubMed] [Google Scholar]
- 3. Anders CK, Fan C, Parker JS, Carey LA, Blackwell KL, Klauber-DeMore N, et al. Breast carcinomas arising at a young age: unique biology or a surrogate for aggressive intrinsic subtypes? J Clin Oncol 2011;29:e18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Partridge AH, Hughes ME, Warner ET, Ottesen RA, Wong YN, Edge SB, et al. Subtype-dependent relationship between young age at diagnosis and breast cancer survival. J Clin Oncol 2016;34:3308–14. [DOI] [PubMed] [Google Scholar]
- 5. Johansson ALV, Trewin CB, Hjerkind KV, Ellingjord-Dale M, Johannesen TB, Ursin G. Breast cancer-specific survival by clinical subtype after 7 years follow-up of young and elderly women in a nationwide cohort. Int J Cancer 2019;144:1251–61. [DOI] [PubMed] [Google Scholar]
- 6. Liu Z, Sahli Z, Wang Y, Wolff AC, Cope LM, Umbricht CB. Young age at diagnosis is associated with worse prognosis in the Luminal A breast cancer subtype: a retrospective institutional cohort study. Breast Cancer Res Treat 2018;172:689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Francis PA, Regan MM, Fleming GF, Lang I, Ciruelos E, Bellet M, et al. Adjuvant ovarian suppression in premenopausal breast cancer. N Engl J Med 2015;372:436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amant F, Lefrère H, Borges VF, Cardonick E, Lambertini M, Loibl S, et al. The definition of pregnancy-associated breast cancer is outdated and should no longer be used. Lancet Oncol 2021;22:753–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Poorvu PD, Gelber SI, Rosenberg SM, Ruddy KJ, Tamimi RM, Collins LC, et al. Prognostic impact of the 21-gene recurrence score assay among young women with node-negative and node-positive ER-positive/HER2-negative breast cancer. J Clin Oncol 2020;38:725–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Park EM, Gelber S, Rosenberg SM, Seah DSE, Schapira L, Come SE, et al. Anxiety and depression in young women with metastatic breast cancer: a cross-sectional study. Psychosomatics 2018;59:251–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Collins LC, Gelber S, Marotti JD, White S, Ruddy K, Brachtel EF, et al. Molecular phenotype of breast cancer according to time since last pregnancy in a large cohort of young women. Oncologist 2015;20:713–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collins LC, Marotti JD, Gelber S, Cole K, Ruddy K, Kereakoglow S, et al. Pathologic features and molecular phenotype by patient age in a large cohort of young women with breast cancer. Breast Cancer Res Treat 2012;131:1061–6. [DOI] [PubMed] [Google Scholar]
- 13. Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 2011;12:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. A comprehensive pan-cancer molecular study of gynecologic and breast cancers. Cancer Cell 2018;33:690–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010;26:589–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protocols Bioinform 2013;43:11.0.1–.0.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 18. Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res 2002;12:656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grossman RL, Heath AP, Ferretti V, Varmus HE, Lowy DR, Kibbe WA, et al. Toward a shared vision for cancer genomic data. N Engl J Med 2016;375:1109–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 2012;30:413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Nat Acad Sci U S A 2007;104:20007–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ramos AH, Lichtenstein L, Gupta M, Lawrence MS, Pugh TJ, Saksena G, et al. Oncotator: cancer variant annotation tool. Hum Mutat 2015;36:E2423–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat Rev Cancer 2018;18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 27. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barroso-Sousa R, Jain E, Cohen O, Kim D, Buendia-Buendia J, Winer E, et al. Prevalence and mutational determinants of high tumor mutation burden in breast cancer. Ann Oncol 2020;31:387–94. [DOI] [PubMed] [Google Scholar]
- 29. Huang KL, Mashl RJ, Wu Y, Ritter DI, Wang J, Oh C, et al. Pathogenic germline variants in 10,389 adult cancers. Cell 2018;173:355–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scott AD, Huang KL, Weerasinghe A, Mashl RJ, Gao Q, Martins Rodrigues F, et al. CharGer: clinical characterization of germline variants. Bioinformatics 2019;35:865–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. CGA Network. Comprehensive molecular portraits of human breast tumours. Nature 2012;490:61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burger H, Loos WJ, Eechoute K, Verweij J, Mathijssen RH, Wiemer EA. Drug transporters of platinum-based anticancer agents and their clinical significance. Drug Resist Updat 2011;14:22–34. [DOI] [PubMed] [Google Scholar]
- 33. Azim HA Jr, Nguyen B, Brohee S, Zoppoli G, Sotiriou C. Genomic aberrations in young and elderly breast cancer patients. BMC Med 2015;13:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swain SM, Nunes R, Yoshizawa C, Rothney M, Sing AP. Quantitative gene expression by recurrence score in ER-positive breast cancer, by age. Adv Ther 2015;32:1222–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pereira B, Chin SF, Rueda OM, Vollan HK, Provenzano E, Bardwell HA, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun 2016;7:11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Offit K, Schrader KA, Maxwell KN, Vijai J, Hart S, Thomas T, et al. Cancer susceptibility mutations in individuals with breast and ovarian cancer using next-generation sequencing. J Clin Oncol 2016;34:1515. [Google Scholar]
- 37. Shindo K, Yu J, Suenaga M, Fesharakizadeh S, Cho C, Macgregor-Das A, et al. Deleterious germline mutations in patients with apparently sporadic pancreatic adenocarcinoma. J Clin Oncol 2017;35:3382–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zardavas D, Te Marvelde L, Milne RL, Fumagalli D, Fountzilas G, Kotoula V, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol 2018;36:981–90. [DOI] [PubMed] [Google Scholar]
- 39. Xu G, Chhangawala S, Cocco E, Razavi P, Cai Y, Otto JE, et al. ARID1A determines luminal identity and therapeutic response in estrogen-receptor-positive breast cancer. Nat Genet 2020;52:198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takaku M, Grimm SA, Roberts JD, Chrysovergis K, Bennett BD, Myers P, et al. GATA3 zinc finger 2 mutations reprogram the breast cancer transcriptional network. Nat Commun 2018;9:1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kan Z, Ding Y, Kim J, Jung HH, Chung W, Lal S, et al. Multi-omics profiling of younger Asian breast cancers reveals distinctive molecular signatures. Nat Commun 2018;9:1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nagarajan S, Rao SV, Sutton J, Cheeseman D, Dunn S, Papachristou EK, et al. ARID1A influences HDAC1/BRD4 activity, intrinsic proliferative capacity and breast cancer treatment response. Nat Genet 2020;52:187–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med 2018;379:111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kalinsky K, Barlow WE, Meric-Bernstam F, Gralow JR, Albain KS, Hayes D, et al. First results from a phase III randomized clinical trial of standard adjuvant endocrine therapy (ET) ± chemotherapy (CT) in patients (pts) with 1–3 positive nodes, hormone receptor-positive (HR+) and HER2-negative (HER2-) breast cancer (BC) with recurrence score (RS) < 25: SWOG S1007 (RxPonder). San Antonio Breast Cancer Symposium2020;GS3–00.
- 45. Copson ER, Maishman TC, Tapper WJ, Cutress RI, Greville-Heygate S, Altman DG, et al. Germline BRCA mutation and outcome in young-onset breast cancer (POSH): a prospective cohort study. Lancet Oncol 2018;19:169–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Couch FJ, Hu C, Hart SN, Gnanaolivu RD, Lilyquist J, Lee KY, et al. Age-related breast cancer risk estimates for the general population based on sequencing of cancer predisposition genes in 19,228 breast cancer patients and 20,211 matched unaffected controls from US based cohorts in the CARRIERS study. San Antonio Breast Cancer Symposium2018;GS2–01.
- 47. Rio Frio T, Lavoie J, Hamel N, Geyer FC, Kushner YB, Novak DJ, et al. Homozygous BUB1B mutation and susceptibility to gastrointestinal neoplasia. N Engl J Med 2010;363:2628–37. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files.