This study presents a comprehensive analysis of alternative splicing in the context of patient classification, molecular mechanisms, and prognosis in gastric cancer.
Abstract
Alternatively spliced RNA isoforms are a hallmark of tumors, but their nature, prevalence, and clinical implications in gastric cancer have not been comprehensively characterized. We systematically profiled the splicing landscape of 83 gastric tumors and matched normal mucosa, identifying and experimentally validating eight splicing events that can classify all gastric cancers into three subtypes: epithelial-splicing (EpiS), mesenchymal-splicing (MesS), and hybrid-splicing. These subtypes were associated with distinct molecular signatures and epithelial–mesenchymal transition markers. Subtype-specific splicing events were enriched in motifs for splicing factors RBM24 and ESRP1, which were upregulated in MesS and EpiS tumors, respectively. A simple classifier based only on RNA levels of RBM24 and ESRP1, which can be readily implemented in the clinic, was sufficient to distinguish gastric cancer subtypes and predict patient survival in multiple independent patient cohorts. Overall, this study provides insights into alternative splicing in gastric cancer and the potential clinical utility of splicing-based patient classification.
Significance:
This study presents a comprehensive analysis of alternative splicing in the context of patient classification, molecular mechanisms, and prognosis in gastric cancer.
Introduction
Alternative splicing (AS) is a major mechanism for increasing transcript diversity in eukaryotes, affecting approximately 95% of multi-exon genes that express multiple spliced isoforms in humans (1, 2). Dozens of aberrant splicing variants have been implicated in human disease, including cancer (3–5). These variants play roles in a broad spectrum of oncogenic processes, such as cell proliferation, apoptosis, hypoxia, angiogenesis, and immune escape (3–5). Aberrant splicing has also been reported to promote epithelial–mesenchymal transition (EMT) and contribute to the invasion and metastasis of various tumor types, including breast, lung, and gastric cancer (6, 7).
Gastric cancer is the fifth most frequently diagnosed cancer and the third leading cause of cancer-related mortality worldwide (8). A large-scale project for genomic characterization of cancer, The Cancer Genome Atlas (TCGA) consortium, revealed that gastric cancer is highly heterogeneous and, based on mutations, copy number changes, and translocations, can be divided into four distinct tumor subtypes: Epstein-Barr virus (EBV)–associated, chromosomal instable (CIN), microsatellite instable (MSI), and genomically stable (GS; ref. 9). However, TCGA classification does not correlate well with oncologic outcome and thus is of limited prognostic utility (9). In a separate cohort, the Asian Cancer Research Group (ACRG), four gastric tumor subtypes were defined on the basis of gene expression patterns: MSI, Microsatellite Stable/EMT (MSS/EMT), MSS/TP53+, and MSS/TP53−. The ACRG subtypes are more closely associated with patient outcome, with the MSI and MSS/EMT patient subtypes having the best and worst prognosis, respectively (10). Neither of these classification schemes use AS profiles to stratify patients with respect to outcomes or treatment efficacy. AS-based classifications have been shown to accomplish these goals in other tumor types (11, 12) and therefore could be leveraged to improve patient stratification in gastric cancer.
Several transcriptomic studies have reported that AS of genes implicated in gastric cancer, such as skipping exon 2 or exons 18–19 of the receptor tyrosine kinase proto-oncogene MET (9), and isoform-specific expression of ZAK kinase (13). In addition, EBV-associated gastric cancers display a unique AS signature and changes in splicing factor levels compared with EBV-negative gastric cancers (14). Recent reanalysis of the TCGA database focusing on gastric cancer and gastrointestinal pan-cancer cohorts identified hundreds of tumor-associated AS events, several of which were associated with differential patient survival (15–18). Yet the role of AS in gastric cancer pathogenesis, and its potential clinical utility, has not been fully explored due to the lack of transcription-wide analysis on matched normal-tumor pair samples. A systematic analysis of AS events in matched normal-tumor gastric samples would allow identification of cancer-specific events, in addition to intertumor AS variation, and is currently lacking. Furthermore, deeper insights into AS events and their regulation are greatly needed to better understand the fundamental molecular features of, and potential causative mechanisms leading to, gastric cancer.
Here, we report an AS-based stratification of gastric cancer cases based on a transcriptome-wide analysis of a new cohort of 83 tumors and matched normal samples. We identify eight AS events and three RNA-binding protein (RBP) splicing factors, and demonstrate that either an AS- or RBP-based signature can be used to classify patients from multiple cohorts into subtypes with distinct clinical outcomes.
Materials and Methods
Samples
We selected 83 patients with gastric cancer with tumor and matched normal samples (Supplementary Table S1) from the fresh snap-frozen tissue repository at the Laboratory of Gastric Cancer Biology, Cancer Research Institute, Seoul National University, South Korea (from 2001 to 2015). All cases were staged according to the AJCC TNM 7th edition staging system. Pathological analysis classified papillary, well-differentiated and moderately differentiated types as the differentiated group, whereas poorly differentiated, mucinous, poorly cohesive cell types were classified as the undifferentiated group. For MSI, fragment analysis was used to compare tumor and normal tissue samples at 5 bp locations after PCR using Primer 1 consisted of BAT26 (116 bp) and BAT25 (148 bp), and primer 2 consisted of D5S346 (96–122 bp), D17S250 (146–165 bp) and D2S123 (144–174 bp). Study protocol was approved by the institutional review board (IRB H-2001–044–1092) and all methods were carried out in accordance with relevant guidelines and regulations.
RNA sequencing and data analysis
Total RNA was extracted from tumor and corresponding normal gastric mucosa samples and libraries were prepared using the TruSeq RNA Kit 2 per manufacturer's instructions. RNA sequencing (RNA-seq) was performed with Illumina HiSeq2000 (Illumina Inc.) for 101-mer paired-end reads. Reads were aligned against the human reference genome (GRCh38, Ensembl 94) using STAR (19) 2-pass mode v2.6.1.d and RSEM (20) v1.3.1 was used to quantify transcriptome abundance. Transcript abundance was analyzed by DESeq2 (21), and variance stabilizing transformation was used for downstream analysis. Gene set enrichment analysis (GSEA) was used to identify overrepresented biological functions using GSEA (22). We calculated enrichment scores using gene sets from MSigDB Hallmark (n = 50) and Kyoto Encyclopedia of Genes and Genomes (KEGG; n = 186) gene sets collection (23).
AS analysis
We used rMATS v4.0.2 (24) to identify differential AS events between two groups of samples. To keep high-rMATS confidence calls, we filtered out cases where the sum of inclusion junction counts (IJC) and skipping junction counts (SJC) was <25 in either set of samples. Differentially spliced exons were defined using a splicing ratio |PSI (Percent-Spliced-In)| > 0.2 and FDR < 0.05 as thresholds. AS frequency for each patient was calculated as the total number of five major types of AS events.
Molecular subtype classification
To find variable AS events for patient subtyping, we performed hierarchical clustering using ComplexHeatmap v2.2.0 with ΔPSI values of 20 differential AS events between tumor and matched normal tissues, and focused on 5 variable AS events selected by visual inspection of variability in the heatmap. Next, we defined two patient groups defined by principal component analysis using ΔPSI values of these 5 variable AS events. To expand the list of variable AS events with similar behavior, we compared AS events in group 1 versus group 2 using rMATS, yielding 28 variable AS events (Supplementary Fig. S2).
We applied the TCGA and ACRG subtype classifications to our gene expression data. We used approximately 800 signature gene classifiers (25) from the TCGA classification (9) to divide patients into four groups (EBV, MSI, GS, and CIN, Supplementary Fig. S22). Signature scores based on ACRG classification signature gene lists (e.g., MSI, EMT, and TP53 activity) were calculated as described previously in (Supplementary Fig. S23; ref. 10).
For the RBP-expression–based patient classification scheme, we used an average expression value of RBM24 and ESRP1 to divide patients into high and low expression groups. We defined the MesS (mesenchymal-splicing)-like subtype as RBM24high and ESRP1low, and EpiS (epithelial-splicing)-like subtype as RBM24low and ESRP1high. For the three RBP-based classifications, we used the average of both RBMS1 and RBM24 for the EpiS subtype.
Somatic mutation and copy number variation analysis
Exome enrichment, whole-exome sequencing, as well as copy number, mutation, and variant calling analysis are described previously in Supplementary Methods.
RBP motif enrichment analysis
Motif analysis for 114 RBPs in differential exon skipping events was performed using rMAPS (26), comparing differentially skipped exons of specific subtypes with control exons. 24,704 exons control exons, not differentially spliced, were defined as FDR > 0.5, and maxPSI > 0.15, and minPSI < 0.85. Regions for motif analysis included: 250 bp upstream or downstream into the intronic sequences and the first and last 50 bp of exonic sequences. To avoid enrichment of canonical splice sites, rMAPS excluded 20 bp within the 3′ splice site and 6 bp within 5′splice site in introns. P value were calculated for each motif after counting the number of occurrences in skipped exons and control exons, and enriched motifs were identified as P < 0.001. To build a correlation network, we calculated Pearson correlation coefficients between tumor PSI and RBP expression for each pair of AS event and RBP. P values were converted into FDR for multiple test correction. Significant correlation pairs (FDR < 0.05) were used for network construction.
RNA extraction from tumors and normal tissues
50–100 mg of frozen gastric tumor or adjacent matched normal tissue was pulverized using a mortar with liquid nitrogen. Total RNA was extracted using the RNeasy kit (QIAGEN), including DNase I treatment per the manufacturer's instructions. RNA concentration and quality were assessed using a NanoDrop 2000 (Thermo Fisher Scientific) as well as an Agilent RNA 6000 Pico Kit with Agilent 2100 Bioanalyzer (Agilent). Samples with RNA Integration number >4.0 were used.
Cell cultures and RT-PCR for splicing events validations
AGS, MKN-74, and MKN-1 human gastric cancer cell lines were obtained from the Korean Cell Line Bank (KCLB, Seoul, Korea), and used in experiments after being certified by the KCLB. All cell lines were cultured in RPMI-1640 cell culture media (Gibco, Life Technologies) supplemented with 10% FBS (Gibco) and 1% penicillin–streptomycin (Gibco) and subcultured at intervals of 2–3 days. All cell lines were grown in 5% CO2 humidified incubator at 37°C and Mycoplasma testing was routinely performed using e-Myco plus Mycoplasma PCR Detection Kit (Intron), verifying that the cells were Mycoplasma free. For the silencing of ESRP1 or RBM24 expression, cells were transfected with ESRP1 siRNA (siESRP1; Santa Cruz Biotechnology; sc-77526), RBM24 siRNA (sc-95527), or nontargeted siRNA (sc-37007) using Lipofectamine 2000 transfection reagent (Life Technologies) according to the manufacturer's protocol. Two microgram of total RNA was reverse-transcribed with Superscript III reverse transcriptase (Invitrogen) and cDNA from 10 ng of total RNA was subsequently used for each reaction. RT-PCR was performed with Solg Taq DNA Polymerase (SolGent co., Ltd.) using oligonucleotide primers detecting both the included and skipped isoforms (Supplementary Table S5). PCR products were separated on a 2% or 2.5% agarose gel stained with Dyne LoadingSTAR (DYNEBIO INC.). Bands corresponding to the included and skipped isoforms were quantified with the Image Laboratory 6.0 software (Bio-Rad) and for each splicing event exon inclusion was expressed as PSI as (intensity included)/(intensity included + skipped).
RT-PCR for splicing events validations
500 ng of total RNA was reverse-transcribed with Superscript III reverse transcriptase (Invitrogen) and cDNA from 5–10 ng of total RNA was used for each reaction. Semiquantitative PCR was performed with Hot-start Taq DNA polymerase (Thomas Scientific) using primers (IDT) detecting both included and skipped isoforms (Supplementary Table S6). Optimal PCR conditions were defined for each primer pair to select semiquantitative conditions. PCR products were separated on a 2% agarose gel stained with SYBR Safe (Invitrogen). Bands corresponding to included and skipped isoforms were quantified with a ChemiDoc MP (Bio-Rad) and for each event exon inclusion was expressed as PSI as (intensity included)/(intensity included + skipped).
Quantitative PCR analysis of RBPs
qPCR was performed using cDNA as above with iTaq Universal SYBR Green Supermix (Bio-Rad) using gene-specific oligonucleotide primers (IDT; Supplementary Table S7) per the manufacturer's instructions using a ViiA7 (Life Technologies). Expression for each target gene was normalized to GAPDH, and then normalized across all samples.
Validation in independent patient cohorts
RNA-seq data of paired normal and tumor samples for 80 early-onset gastric cancers from Mun and colleagues (SRP172499; ref. 27) were processed as described above. Clinical data and gene expression profiles of 295 TCGA gastric cancers were downloaded from cBioportal (28, 29). ACRG microarray data (GSE62254; ref. 10) were analyzed using RMA normalization.
Statistical analysis
Fisher exact and χ2 tests were performed to evaluate differences between subgroups. Log-rank and cox regression analysis were used to test survival differences. Kaplan–Meier survival analysis was performed using the survival package and Cox proportional hazards regression for multivariable survival analysis was applied using the coxph function. All analyses were performed using R v3.6.0.
Results
Comprehensive genomic, transcriptomic, and splicing profiles of matched gastric tumor and normal tissue samples
We performed RNA-seq of paired tumor and adjacent normal mucosa tissues from 83 patients with gastric cancer. On the basis of the Lauren classification, the patient cohort included 35 intestinal, 27 diffuse, and 21 mixed or undetermined types of gastric cancer (Supplementary Table S1). We generated 9.5 billion paired-end reads, with an average of 64.2 million read pairs per tumor, and 49.8 million read pairs for the corresponding normal mucosal samples (Supplementary Fig. S1). In addition, we performed whole-exome sequencing at an average read depth of 129X for the tumor samples and an average read depth of 85X for the matched normal samples.
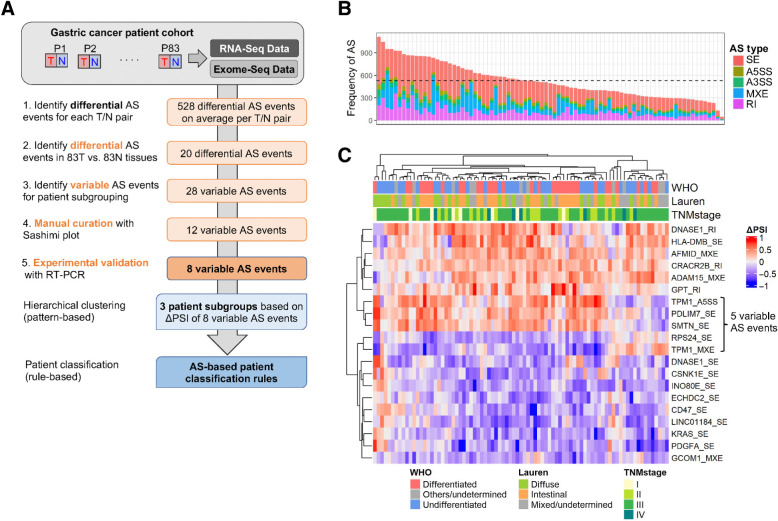
Taking advantage of the paired sampling that was available for all of our patients (Fig. 1A), we first looked for differential AS events between tumor and normal tissue. We used rMATS, a computational tool for splicing analysis that uses both exon body and exon junction reads from RNA-seq data, to quantify the “Percent Spliced-In” (PSI) value for each AS event (24). We determined AS profile differences between each tumor and paired normal tissue using a stringent cutoff value of at least 20% change in PSI (ΔPSI). We detected on average 528 AS events in each tumor versus matched normal pair, with skipped exon (SE) events being the most frequent (Fig. 1B). We further identified 20 AS events in 18 genes that are differentially spliced between tumors and normal tissues, including 12 SE events, four mutually exclusive exons (MXE), three retained introns (RI), and one alternative 5′ splice site (A5SS; Fig. 1C; Supplementary Table S2). Spliced genes were associated with cell adhesion and the cytoskeleton (e.g., SMTN, TPM1, PDLIM7, and ADAM15), translational control (e.g., RPS24), cell proliferation (e.g., PDGFA, KRAS), cell metabolism (e.g., AFMID, ECHDC2, INOE80, and GPT), or immune cell signaling (e.g., CD47). Our analysis also detected a skipped exon in the KRAS oncogene that was previously identified in a smaller cohort of patients with gastric cancer (30), as well as mutually exclusive exons in AFMID detected in hepatocellular carcinoma (31).
Figure 1.
Cancer-associated AS events in gastric tumors. A, Summary of AS-based patient classification scheme. N, normal tissue; P, patient; SNU, Seoul National University; T, tumor. B, Number of differential AS events in tumor versus normal pairs, colored by event type (SE, skipped exon; A5/3SS, alternative 5′/3′ splice site; MXE, mutually exclusive exons; RI, retained intron). The mean number of AS events is shown as a horizontal dashed line. C, Differential AS events in 83 tumors versus normal tissues, plotted as ΔPSI values for each patient. WHO and Lauren classification, as well as tumor stage, are shown.
Patient subtype classification based on AS events
We next sought to identify AS events that could be used for patient classification. These were defined as AS events that exhibit variable profiles across tumors (hereafter referred to as variable AS events). Five such variable AS events were identified upon hierarchical clustering of the 20 differential AS events (Fig. 1C). We then divided the patients with cancer into two groups, Group 1 and Group 2, based on the patterns of these five variable AS events. By comparing Groups 1 and 2, 23 additional variable AS events were identified to facilitate the further dichotomization of patients (See Materials and Methods). This analysis yielded a total of 28 variable AS events in 13 genes (Supplementary Fig. S2A and S2B). Because many of these events were partially overlapping in the same gene (e.g., TPM1 contains eight AS events) and rMATS reports each AS event separately, all events were manually inspected, and merged to produce a group of 12 nonredundant variable AS events in nine genes (Supplementary Table S3).
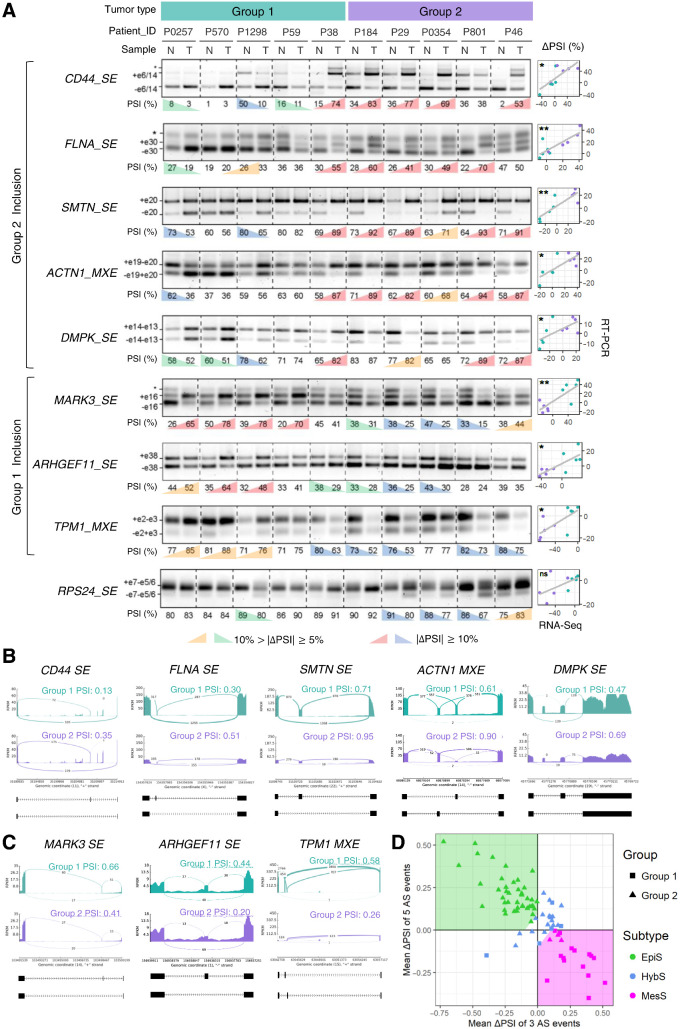
We experimentally validated the splicing patterns of these nine genes from Groups 1 and 2, using an adjacent section of tumor and matched normal tissues from 10 of the patients originally used for RNA-seq analysis. Differential splicing patterns detected by our transcriptome analysis between tumor and matched normal samples were measured using semiquantitative RT-PCR for AS events in nine genes (Fig. 2A). The ΔPSI values were determined for each tumor-normal pair, and eight out of nine AS events showed a significant positive correlation (P < 0.04) of ΔPSI values between RNA-seq data and the RT-PCR results (Fig. 2A). We also observed that these eight validated AS events could be divided into two classes where the “Group 1 inclusion” consisted of 5 AS events (i.e., SE events in CD44, FLNA, SMTN, DMPK, and an MXE event in ACTN1) and the “Group 2 inclusion” consisted of 3 AS events (i.e., SE events in MARK3, ARHGEF11, and an MXE event in TPM1). Inclusion patterns of these 8 AS events in the two patient groups were visualized using Sashimi plots (Fig. 2B and C).
Figure 2.
Eight AS events display distinct patterns in two patient groups. A, RT-PCR validation of AS events in gastric tumors (T) and matched normal (N) tissues from 10 patients, using primers that amplify both included and skipped isoforms. For each AS event, a representative gel image is shown along with PSI values, calculated as the percentage of included/(%included + %skipped); significant ΔPSI (T–N) values (n = 3, ΔPSI >|10%| and ΔPSI >|5%|) are colored. Exons included (+) or skipped (−) are annotated (*, bands of no interest). Correlations of ΔPSI values between RT-PCR and RNA-seq experiments are shown on the right. *, P < 0.05; **, P < 0.01; ns, not significant. B and C, Read coverage and junction reads showing inclusion of AS events that define “Group 2” (B) and “Group 1” (C) in gastric tumors. Gene name and AS type are indicated, along with average PSI for tumors. D, Scatter plot of patients using mean(ΔPSI; Group 2 vs. 1). The areas for three subtypes are shown in different background colors—EpiS, green; MesS, magenta; and HybS, white, where the x and y intercepts of linear boundaries are 0.1.
Hierarchical clustering of patients based on ΔPSI values of these 8 events showed that patients can be divided into two distinct subgroups plus one subgroup with an intermediate pattern (Supplementary Fig. S3). Plotting the average ΔPSI values of the Group 1 inclusion events versus Group 2 inclusion events showed a significant negative correlation with a mutually exclusive pattern between these two ΔPSI variables (r = −0.85, P < 2.2e−16, Fig. 2D), and suggested a simple AS-based patient classification scheme.
According to this novel AS event-based classification scheme, we divided patients with gastric cancer into three subtypes (Fig. 2D): EpiS (top left region), MesS (bottom right region), and Hybrid-Epithelial/MesS (HybS, intermediate region), which were named according to the results of GSEA, described below. This new classification scheme can be applied to any single patient without the need for complex clustering algorithms.
Molecular characterization of AS event-based gastric cancer subtypes
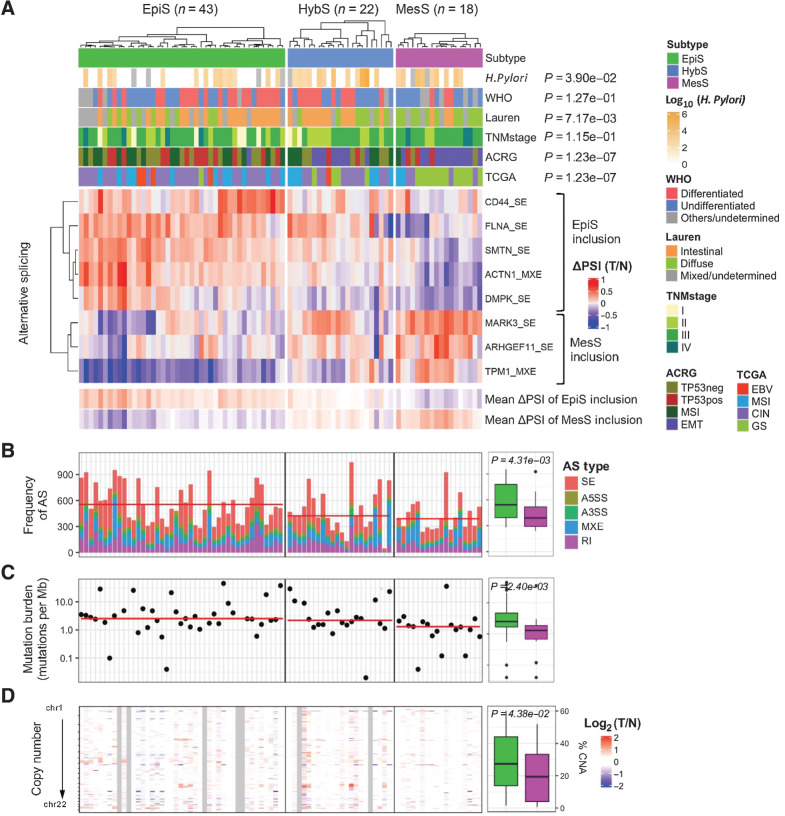
The AS-based classification divided 83 patients into 43 EpiS, 22 HybS, and 18 MesS subtypes (Fig. 3A). At the molecular level, the frequency of AS events, mutation rate, and copy number alterations were significantly higher in the EpiS subtype compared with the MesS subtype (Fig. 3B–D). The average number of AS events, as well as the total number of SE events, per tumor sample was significantly higher than in EpiS tumors compared with MesS tumors (Fig. 3B). Five genes (i.e., LRP1B, ZFHX4, ROBO1, PLEC, and AHNAK) were frequently mutated (>20%) in EpiS but not in MesS tumors and MUC16 similarly showed a higher mutation rate in EpiS compared with MesS (Supplementary Fig. S4A). No mutations in splicing regulatory genes were found to be significantly associated with our subtypes (Supplementary Table S4; Supplementary Fig. S4B). The EpiS subtype also exhibits copy number gains in several regions, including regions overlapping the known cancer-associated genes EGFR, ERBB2, MYC, and CCNE1, which are involved in cancer cell proliferation (Supplementary Fig. S4C). Copy number alterations were relatively rare in MesS patients, similar to the TCGA GS subtype. Furthermore, the copy number alterations observed in the MesS subtype were also present in the EpiS subtype, whereas most copy number amplifications in driver oncogenes were not present in the MesS subtype (Supplementary Fig. S4C).
Figure 3.
AS event-based patient classification defines the EpiS and MesS subtypes of gastric cancer. A, Clinical information, including H. pylori status, and ACRG and TCGA classifications for EpiS, MesS, and HybS subtypes defined on the basis of AS events are shown. P values from χ2 enrichment or Wilcoxon rank-sum tests are shown. For each event, ΔPSI for each tumor-normal pair are shown. B, The frequency of AS events for each patient is the sum of differential AS events in matched tumor versus normal samples. C and D, Difference in mutation burden (C) and copy number alteration (D) between EpiS and MesS subtypes are shown, along with average values, and Wilcoxon rank-sum test P values.
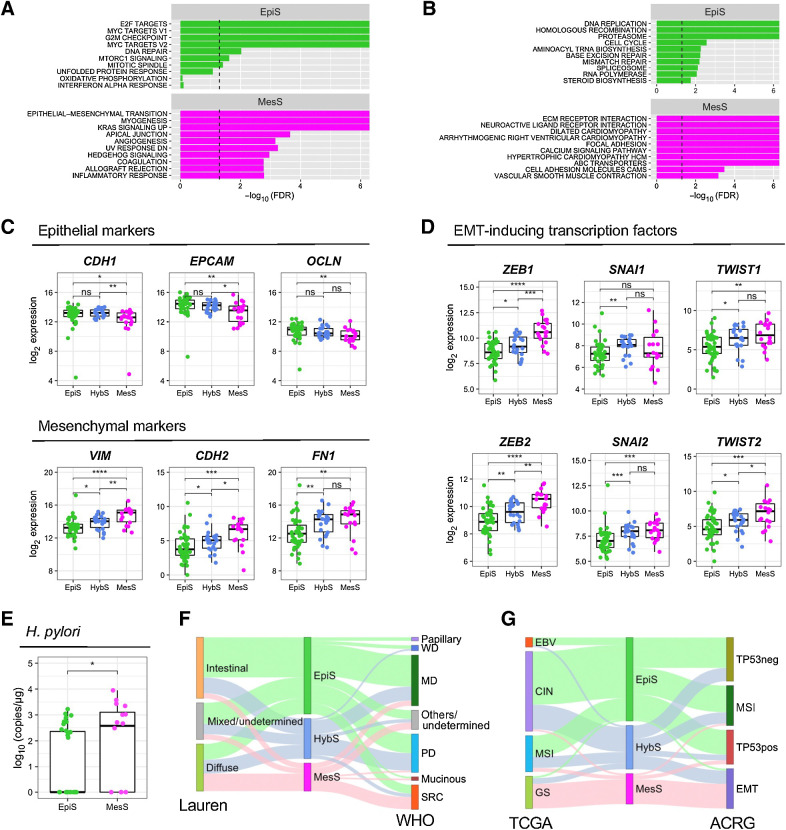
To further characterize the patient subtypes defined by eight AS events, we performed GSEA using total gene expression data between the EpiS and MesS subtypes (Fig. 4A and B; Supplementary Fig. S5; ref. 23). The EpiS subtype showed an enrichment of terms related to E2 factor (E2F) targets, MYC targets, and cell-cycle regulation, which are typical proliferation signals for epithelial cancer cells (Fig. 4A), as well as DNA damage response pathways (Fig. 4B). Higher mutation rates and copy number amplifications detected in the EpiS subtype (Fig. 3C and D) are consistent with the activation of DNA damage response pathways detected by GSEA. The MesS subtype showed various terms related to EMT (Fig. 4A), as well as extracellular matrix–receptor interaction, focal adhesion, and cell adhesion molecules (Fig. 4B). Thus, the characteristic biological signature associated with our AS event-based patient classification is the EMT process, which we used to assign the tumor subtypes defined above.
Figure 4.
Functional characterization of AS subtypes. A and B, Statistically enriched gene sets in EpiS and MesS subtypes identified using GSEA 50 Hallmark gene set (A) and MSigDB KEGG pathways (B). C, Gene expression of epithelial CDH1, EPCAM and OCLN, and mesenchymal VIM, CDH2 and FN1 markers in each patient subtype. D, Gene expression of EMT-inducing transcription factors ZEB1/2, SNAI1/2, and TWIST1/2 in each patient subtype. E, Absolute quantification of H. pylori copies in each patient subtype. F, Associations between AS event-based patient subtype and Lauren or WHO clinical subtypes. MD, moderately differentiated; PD, poorly differentiated; SRC, signet ring cell carcinoma; WD, well differentiated. G, Associations between AS event-based patient subtype and TCGA or ACRG molecular subtypes. Wilcoxon rank-sum test, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
To confirm the involvement of EMT in our patient subtypes, we investigated the expression of well-established EMT marker genes. In accordance with our GSEA, epithelial marker genes CDH1, EPCAM and OCLN showed higher expression in the EpiS subtype than in the other two subtypes, whereas mesenchymal marker genes VIM, CDH2 and FN1 showed higher expression in the MesS subtype (Fig. 4C). Of note, among the 8 variable AS events, aberrant splicing of ARHGEF11 and CD44 is known to play important roles during EMT in diverse cancers (32, 33). We further examined the expression of known EMT-inducing transcription factors (TF), such as ZEB1 and ZEB2, SNAI1 and SNAI2 (also known as Snail and Slug), as well as TWIST1 and TWIST2 (34). Gene expression of all these EMT TFs, except SNAI1, gradually increased across subtypes in the order EpiS-HybS-MesS (Fig. 4D), highlighting differences in EMT between our three patient subtypes. Expression of other TFs associated with EMT, such as TCF4, KLF8, FOXC2 increased between EpiS and MesS, whereas TCF3, GRHL2, ELF3, and H2AFX decreased (Supplementary Fig. S6), consistent with the overall enrichment of mesenchymal markers in the MesS group.
Finally, we compared our AS event-based patient classification scheme with established clinical and molecular subtype classifications. MesS tumors exhibit higher Helicobacter pylori (H. pylori) copy numbers compared with EpiS tumors (P = 3.9e−02; Fig. 4E; Supplementary Table S1), in line with previous findings suggesting that H. pylori promotes EMT in gastric cancer (35, 36). In terms of the Lauren classification, we observed a significant association between our AS event-based MesS subtype and the diffuse subtype, whereas patients classified as EpiS were enriched in the intestinal subtype (P = 7.17e−03; Fig. 4F). Compared with the TCGA and ACRG molecular classifications, our AS event-based MesS subtype, which rarely contains copy number alterations, was significantly associated with the TCGA GS subtype (P = 1.23e−07) as well as with the ACRG EMT subtype (P = 1.23e−07; Fig. 4G).
Splicing factors RBM24, RBMS1, and ESPR1 regulate AS events in gastric cancer subtypes
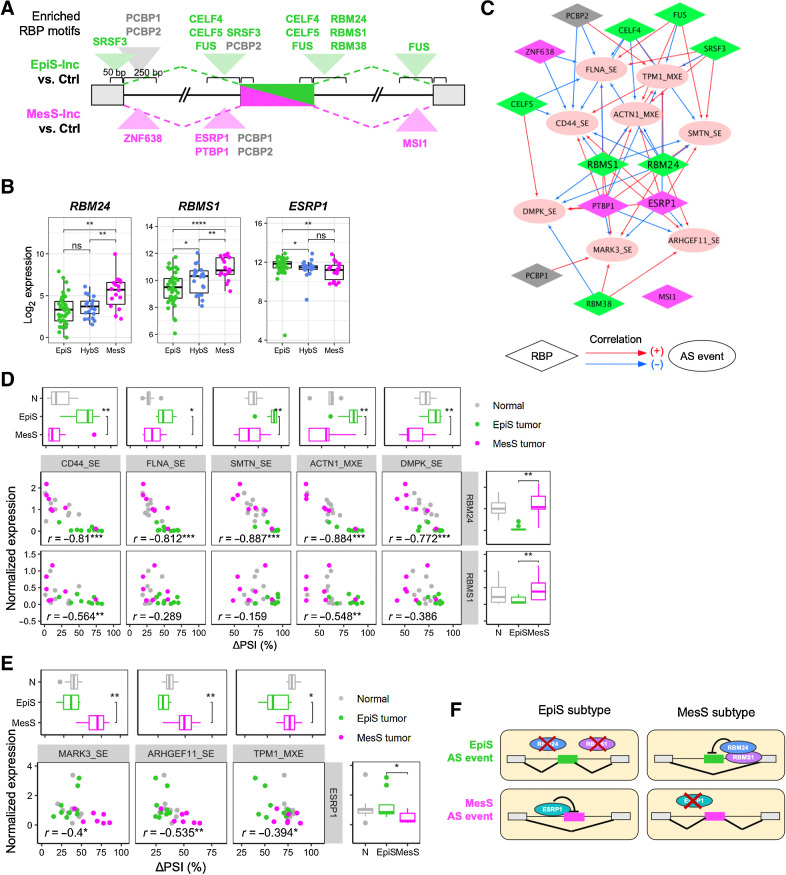
RBPs regulate AS in a dose-dependent manner by binding specific sequences on the pre-mRNA and promoting or blocking exon inclusion. To investigate the molecular mechanisms regulating differential AS events between the EpiS and MesS subtypes, we performed an RBP-binding motif enrichment analysis using rMAPS (26). For the five AS events included in EpiS tumors, we identified binding sites for nine RBPs specifically enriched in exonic or intronic regions near the spliced exons (Fig. 5A; Supplementary Table S5; Supplementary Fig. S7). Similarly, we found binding motifs for four RBPs enriched in the three AS events included in MesS tumors. For example, the ESRP1-binding motif was enriched in upstream introns of AS events included in MesS tumors (P = 2.18e−4; Fig. 5A), whereas the RBM24-binding motif was enriched in downstream introns of AS events included in EpiS tumors (P = 6.61e−07; Fig. 5A). Finally, AS events from both subtypes contained motifs for two RBPs, PCBP1, and PCBP2, which are known oncogenic-splicing factors that are also involved in translation coactivation and RNA stability (37).
Figure 5.
RBPs as regulators of AS events in EpiS and MesS tumor subtypes. A, RBP motifs enriched near differentially included exons in EpiS and MesS subtypes. See also Supplementary Table S5. B, Normalized RBP gene expression in each patient subtype. C, Correlation network between RBP expression and ΔPSI values of each AS event. Positive and negative correlations are shown with red and blue arrows, respectively. Colors for RBP nodes indicate the subtypes of RBP motifs enriched in A. D and E, Correlation between RBP expression and AS for each RBP-AS pair, for EpiS (D) and MesS (E) AS events, colored by sample type. Box plots indicate subtype differences in AS event (top) and RBP expression (right). F, Model of AS regulation by RBP expression in gastric cancer. Wilcoxon rank-sum test, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, nonsignificant.
Next, we investigated whether these RBPs were differentially expressed between MesS and EpiS patient subtypes. Among the 13 RBPs that exhibit enriched binding sites, RBM24 and RBMS1 were upregulated in the MesS subtype, whereas ESRP1 and PTBP1 were upregulated in the EpiS subtype (Fig. 5B; Supplementary Fig. S8A). We also examined the data for any evidence of a regulatory relationship between RBP level and the ΔPSI values of all eight AS events. Gene expressions of RBM24, RBMS1, and ESRP1 were significantly correlated with the level of exon inclusion for most AS events (Supplementary Fig. S8B and S8C), supporting a regulatory role for RBM24, RBMS1, and ESRP1 in modulating these eight AS events. A correlation network was built using significantly correlated RBP-AS event pairs (FDR < 0.05), and revealed RBM24, RBMS1, and ESRP1, but also PTBP1, to be major nodes connecting all eight variable AS events (Fig. 5C). A few additional RBPs (i.e., CELF5, PCBP2, CELF4, FUS, SRSF3, and RBM38) seem to play auxiliary roles in regulation of these specific AS events, connecting only a subset.
The correlation between the expression of regulatory RBPs RBM24, RBMS1, and ESRP1, and exon inclusion of all eight MesS and EpiS AS events was further validated experimentally (Fig. 5D and E; Supplementary Fig. S8B and S8C). Differential RBP expression was measured with quantitative PCR and mirrored RNA-seq data almost perfectly. Notably, RBM24 expression showed a significant correlation (r > |0.5|) with all five AS events from EpiS tumors, whereas RBMS1′s correlation was significant only for CD44 and ACTN1 AS events (Fig. 5D and E). ESRP1 expression correlated with all three AS events in MARK3, ARHGEF11, and TPM1 from MesS tumors (Fig. 5E). Of note, ESRP1-regulated AS of ARHGEF11 has been shown to be essential for epithelial tight junction integrity (38). Thus, RBM24 and ESRP1 may be critical regulatory RBPs in gastric cancer, acting oppositely in the EpiS and MesS subtypes. Our working model is that, in the EpiS subtype, patients with low expression of RBM24 and RBMS1 and high expression of ESRP1 show inclusion of “EpiS exons” and skipping of “MesS exons” (Fig. 5F), whereas we propose the converse to be the case in MesS subtype patients.
We performed siRNA-mediated ESRP1 knockdown in AGS and MKN-74 epithelial-like gastric cancer cell lines, and RBM24 knockdown in MKN-1 mesenchymal-like gastric cancer cells to demonstrate the causal role of these RBPs in the regulation of the splicing events that constitute our splicing-based signature of gastric tumors (39). At baseline, the epithelial-like gastric cancer cell lines expressed high levels of ESRP1 and low levels of RBM24, whereas mesenchymal-like gastric cancer cells showed opposite expression patterns (Supplementary Fig. S9A). Similarly, at baseline, epithelial-like and mesenchymal-like gastric cancer cells often show opposite splicing patterns (Supplementary Fig. S9B). Knockdown of ESRP1 significantly decreased inclusion of exons in CD44 (in AGS and MKN-74) and ACTN1 (in AGS), two EpiS-splicing events, and increased exon inclusion in ARHGEF11 (in AGS and MKN-74), a MesS-splicing event, compared with scrambled siRNA control (Supplementary Fig. S9B). Similarly, knockdown of RBM24 in MKN-1 cells significantly increased exon inclusion in ACTN1, an EpiS-splicing event, and decreased exon inclusion in TPM1, a MesS-splicing event. Exons of MARK3 in MKN-1 were either fully included in either cell line or fully skipped and were not affected by RBP knockdowns.
Validation of the AS event signature using independent cohorts
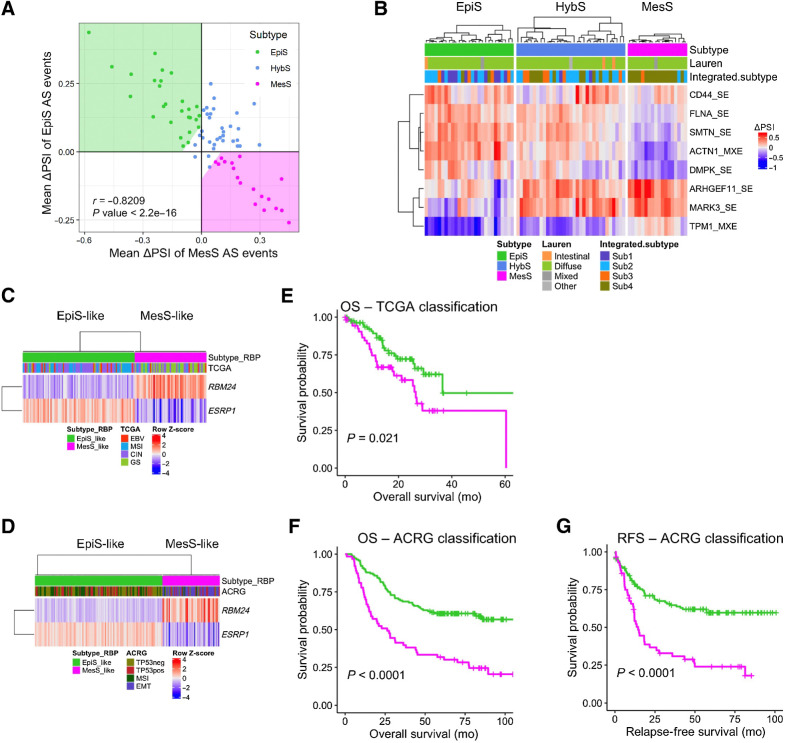
To ascertain the robustness of our findings, we verified whether the AS event-based classification and molecular findings were reproducible in other patient cohorts. We looked for public datasets that included (i) reasonably large number of patients; (ii) matched tumor and normal samples; and (iii) raw RNA-seq data available for analyzing splicing variants. The proteogenomic study by Mun and colleagues (27) met these conditions with 80 early-onset patients with gastric cancer even though most of them were of the Lauren diffuse subtype. Analysis of this cohort reproduced our salient findings, including (i) classification using the eight AS events grouped patients into three distinct subtypes (Fig. 6A and B); (ii) subtypes displayed differential expression of epithelial (CDH1, EPCAM, and OCLN) and mesenchymal (VIM, CDH2, and FN1) markers; and (iii) RBM24 and RBMS1 were upregulated and ESRP1 downregulated in the MesS subtype (Supplementary Fig. S10). Interestingly, the MesS subtype defined in our study was associated with the so-called integrated subtype 4 by Mun and colleagues (P = 2.6e−07; Fig. 6B; ref. 27), in which the key enriched biological process is RhoA-mediated tumor cell migration via actin reprogramming and focal adhesion assembly. It suggests that our MesS and EpiS AS events, including exon inclusion events in ARHGEF11 and ACTN1, might play similar roles in regulating cytoskeletal rearrangements via the RhoA signaling pathway (40).
Figure 6.
Validation of AS-based subtype classification in independent datasets. A, Scatter plot of patients from Hwang's cohort using meanΔPSI of AS event-based EpiS and MesS tumor subtypes, colored by subtype. B, AS event-based patient classification applied to Hwang's cohort. Patient clinical classification by Lauren and their integrated subtype are shown. C and D, RBP-based classification of TCGA (C) and ACRG (D) patients according to RBM24 and ESRP1 expression. E and F, Kaplan–Meier plot for OS between EpiS-like and MesS-like RBP-subtypes in TCGA (E) and ACRG (F) cohorts (log-rank test P values). G, Kaplan–Meier plot for relapse-free survival between EpiS-like and MesS-like RBP-subtypes in the ACRG cohort (log-rank test P values).
Validation of the RBP signature using independent cohorts
Given that high-depth RNA-seq and splicing analyses are expensive and not routinely performed in the clinic, we devised a classification scheme that is more readily implementable and only focuses on the differential expression of splicing regulatory RBPs. We first examined RBP expression in the TCGA and ACRG cohorts. RBM24 and RBMS1 were upregulated, and ESRP1 was downregulated in both the TCGA GS subtype and the ACRG EMT subtype (Supplementary Fig. S11), consistent with the overall association of the MesS subtype with these subtype classifications (Fig. 4F). Because our MesS subtype was characterized by high expression of RBM24 and low expression of ESRP1 (Fig. 5B), we thus defined MesS-like (RBM24high/ESRP1low) and EpiS-like (RBM24low/ESRP1high) subtypes that simply use the expression level of these two RBPs. We then evaluated this RBP-based classification in the TCGA and ACRG cohorts and confirmed that the gene expression levels of ESRP1 and RBM24 separated patients into two subtypes properly (Fig. 6C and D).
We further examined the prognostic value of our subtypes. Relapse-free survival showed a marginal difference between the EpiS and MesS subtypes (Supplementary Fig. S12). In addition, we discovered that TCGA patients with the MesS-like subtype had a significantly worse prognosis than those with the EpiS-like subtype (log-rank P = 2.10e−02; Fig. 6E), whereas the TCGA subtypes did not show any difference in the overall survival (OS; Supplementary Fig. S13A). Similarly, ACRG patients with the MesS-like subtype, which was highly associated with the ACRG EMT subtype, showed worse survival than the EpiS-like subtype, both for OS and relapse-free survival (log-rank P < 0.0001 and P < 0.0001, respectively; Fig. 6F and G). The ACRG subtypes showed similar trends (Supplementary Fig. S13B and S13C). Results from a similar analysis, but using three RBPs additionally, including RBMS1, were virtually identical to findings using two RBPs (Supplementary Fig. S14).
For a further assessment of the association of our two RBP-based subtypes and patient survival, we developed a multivariate Cox proportional hazard model that included age, sex, pathologic stage, Lauren classification, and our subtypes (Supplementary Fig. S15). In the TCGA cohort, the RBP-based classification was an independent prognosis factor for OS [HR, 3.74; 95% confidence interval (CI), 1.43–9.80; P = 0.007). In the ACRG cohort, the RBP subtype was also an independent prognostic factor for both OS (HR, 1.82; 95% CI, 1.20–2.80; P = 0.005) and recurrence-free survival (HR, 1.61; 95% CI, 1.00–2.60; P = 0.005). Of note, another hazard model, including both splicing-based subtypes and TCGA subtypes for the TCGA cohort, revealed that our RBP subtype outperformed TCGA subtypes to predict OS (HR, 3.26; 95% CI, 1.137–9.4; P = 0.028; Supplementary Fig. S16A). Equivalent analysis with the ACRG cohort showed that RBP-based subtypes performed better in the OS only (HR, 1.92; 95% CI, 1.07–3.44; P = 0.029; Supplementary Fig. S16B and S16C). Collectively, survival analyses using multiple independent cohorts strongly indicate poor prognosis of the RBP-based MesS-like subtype.
Discussion
Although AS is a key regulatory mechanism that affects virtually all cellular processes, few studies have explored the role of AS in gastric cancer (30, 41–43), and even fewer studies have investigated the utility of AS-based patient classification (11). Given this, we (i) systematically profiled the AS landscape in gastric cancer, (ii) identified RBPs that regulate tumor subtype-specific AS events, and (iii) devised a patient classification scheme based on variable AS events or their regulatory RBPs. Unlike other signature-based classification methods that typically require whole transcriptome data and complex clustering algorithms, our first classification scheme is based on differential AS of only eight genes from tumor and paired normal tissue. Our second classifier, derived from the AS data, is even more straightforward and is based on expression of two RBPs, RBM24, and ESRP1, in tumor tissue only. The two splicing classification schemes, one based on AS events, the other on expression of RBP-splicing regulators, are closely correlated (P = 5e−04) if we compare the subtypes (Supplementary Table S8), but are based on different molecular phenomena. Compared with the TCGA gastric cancer patient classification relying on a complex clustering method using genome, RNA-seq, and microbiome data, or to the ACRG classification using a 675-gene signature after normalization of the RNA-seq data (9, 10), our classification based on expression level of two RBPs is analytically much more efficient for distinguishing gastric cancer subtypes and their associated prognosis, and therefore easier to implement in the clinic on gastric tumor biopsies.
On the basis of the eight AS events signature, we defined two tumor gastric subtypes, EpiS and MesS. One of the major molecular mechanisms behind our AS-based patient subgrouping was the EMT process. EMT plays essential roles in tumor initiation, progression, and metastasis in cancer, but its contribution is highly dependent on the cancer type (44). Tumors from the EpiS and MesS subtypes represent the two sides of the EMT spectrum. In the MesS subtype, EMT-TFs were upregulated and mesenchymal–epithelial transition (MET)-TFs were downregulated relative to the EpiS subtype (Fig. 2D; Supplementary Fig. S6). Furthermore, activation of the KRAS–MAPK and Hedgehog pathways in the MesS subtype (Fig. 3A) can in turn activate EMT-TFs SNAI1/2 and ZEB1/2 indirectly via TGFβ-SMAD and Notch signaling (34, 45). The enrichment of several gene sets related to the DNA-damage response pathways is observed in the EpiS subtype (Fig. 3B), whereas the upregulation of H2AX relative is observed in the MesS subtype (Supplementary Fig. S6). Indeed, Weyemi and colleagues (46) showed that histone variant H2AX, a sensitive marker of double-strand breaks, is a critical regulator of EMT, whose loss induces EMT and reintroduction partially reverses EMT. Oh and colleagues (47) reported epithelial and mesenchymal phenotypes by analyzing genomic and proteomic data from several cohorts. Even though AS was not included in their analysis, EMT markers were found to be a critical factor in patient subtypes associated with clinical outcomes such as patient survival and resistance to chemotherapy.
As for the interplay between regulation of AS and expression of EMT master regulators, several studies reported that EMT-TFs altered expression of splicing regulators. In particular, ZEB1 and SNAI1 were shown to repress ESRP1 expression and lead to EMT in various cancer cell lines (48, 49). Similarly, in our dataset, we observed significant correlations between the expression of EMT-TFs and RBPs levels, with ZEB1 being positively correlated with both RBM24 and RBMS1 (r > 0.5), whereas ESRP1 levels negatively correlated with ZEB1/2, SNAI2, and TWIST2 expression (r < −0.5; Supplementary Fig. S17). Furthermore, to determine whether RBPs could regulate downstream EMT-TFs, we analyzed the expression of six TFS in RNA-seq data from breast and lung cancer cell lines with ESRP1 knockdown. Our analysis reveals that these six EMT-TFs did not change their expression upon ESRP1 knockdown (Supplementary Fig. S18), thus suggesting EMT-activating TFs are presumably the upstream regulators of RBPs affecting AS.
Several of the eight AS events and three regulatory RBPs in gastric cancer identified herein have previously been associated with EMT in other tumor types (30, 38, 49, 50). For example, ESRP1 is downregulated during EMT in breast cancer models, and drives AS of mesenchymal isoforms of ARHGEF11, CD44, and TPM1 (38, 40). In addition, ESRP1-regulated AS events in MARK3, a kinase that regulates phosphorylation of microtubule-associated proteins, and TPM1, a protein involved in stabilizing cytoskeleton actin filaments are identified in this study. The function of these isoforms in EMT in MesS gastric tumors remains to be determined. The role of the other two RBPs, RBM24, and RBMS1, in EMT is not well characterized, and further studies of their splicing targets may provide valuable insights into the functional associations between splicing and EMT.
We observed several gene mutations and copy number variants (CNV) associated with AS subtype (Supplementary Fig. S4A and S4C). The underlying molecular mechanisms linking mutations or CNVs in cancer genes with changes in AS patterns remain poorly understood in gastric cancer. One possible explanation would be that mutations or CNVs in cancer genes impact downstream signaling pathways and lead to changes in RBP levels, which in turn impact splicing of downstream targets. To test this hypothesis, we examined the association of mutations and CNVs with RBP expression. Wild-type and mutant patient groups were compared for differential RBP expression, which showed significant differences for RBM24 expression and ROBO1 or PLEC mutations (P = 0.032 or 0.013), as well as ESRP1 expression and LRP1B or AHNAK mutations (P = 0.018 or 0.040; Supplementary Fig. S19A). However, this observation should be interpreted carefully because most mutations were not recurrent and occurred at different sites, therefore leading potentially to distinct functional consequences, and making it difficult to assess their effect on AS. In parallel, we examined the correlation between copy numbers and RBP expressions, which showed that ESRP1 expression was positively correlated with MYC copy numbers (r = 0.312; Supplementary Fig. S19B). In breast cancer and lymphoma, MYC has been previously reported to regulate directly the transcription of several splicing regulators, including SRSF1, TRA2β or PRMT5, by binding to their promoter regions (51, 52).
For nine cases out of 10 RT-PCR validations showed concordant results with the RNA-seq subtyping; however, we observed that one of our patient samples (P38) was predicted to be the MesS subtype in RNA-seq data and later determined to be the EpiS subtype based on RT-PCR validation (Fig. 2A; Supplementary Fig. S20A). To investigate the cause of this discrepancy, we repeated RNA-seq using the same RNA sample as in the RT-PCR validation experiment and observed that the new RNA-seq results also showed an EpiS subtype (Supplementary Fig. S20B). We confirmed that both RNA-seq datasets were from the same individual using NGSCheckMate (53). The source of discrepancy in the AS-based subtypes was therefore likely due to intratumor heterogeneity between the RNA samples used to generate the RNA-seq data, as are often the cases in cancer pathology studies (39). Hence, tumor heterogeneity introduces additional complexity to the evaluation of how robustly any approach classifies patient subtypes.
Finally, to explore whether our findings from primary tumors are recapitulated in cell models, we profiled splicing patterns as described above using available RNA-seq data from 37 gastric cancer cell lines from CCLE (54). We then applied the AS-based signature to gastric cancer cell lines and classified 11 cell lines as MesS, 7 as HybS, and 19 as EpiS subtypes (Supplementary Fig. S21A). Among the eight AS events that constituted our AS event-based signature derived from primary gastric tumors, AS events in CD44, MARK3, ARHGEF11 showed variable splicing patterns across cell lines (Supplementary Fig. S21A), whereas the remaining five AS events exhibited an epithelial AS pattern across all cell lines, suggesting that they might be specific to primary tumors and lost during cell culture. Splicing inclusion ratios of CD44, MARK3, and ARHGEF11 AS events were significantly different between EpiS and MesS cell lines (Supplementary Fig. S21B). Consistent with the observations in human primary tumors, CD44 exon was more included in EpiS compared with MesS cell lines, whereas exons in MARK3 and ARHGF11 were more skipped (Supplementary Fig. S21B). Finally, when examining RBP levels, ESRP1 showed a higher expression in EpiS than MesS cell lines, similarly to primary tumors (Supplementary Fig. S21C).
Future studies aimed at defining the molecular drivers of the transition from EpisS to MesS AS-based subtypes are needed. These changes could be triggered by alterations in genomic, epigenetic, or transcriptional regulators, which have been shown to drive splicing factor changes in other tumor types. Understanding these mechanisms will allow us to identify AS-based targets for development of gastric cancer therapies and to move toward harnessing the full potential of AS in precision medicine.
In summary, we systematically investigated the landscape and roles of AS in gastric cancer using RNA-seq data from matched tumor and normal samples. Association of AS with the EMT program was firmly established and suggested AS-based patient stratification schemes, which highlighted the potential of AS analysis as a tool for precision medicine. To our knowledge, this study presents the most comprehensive analysis to date of AS in the context of patient classification, molecular mechanisms, and prognosis in gastric cancer.
Authors' Disclosures
J.I. Kim reports grants from MACROGEN, WELLXECON, and DNALINK outside the submitted work. O. Anczuków reports grants from Sanofi outside the submitted work. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank Taneli Helenius for comments on the article. This work was supported by the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (HI14C1277 to H.K. Yang) and the Korean Healthcare Technology R&D project through the Korean Health Industry Development Institute (HI13C2148 to H.K. Yang and C. Lee) funded by the Ministry of Health and Welfare, Republic of Korea. This work is also supported by the National Research Foundation of Korea (NRF-2015K1A4A3047851 to C. Lee) funded by the Ministry of Science and ICT, Republic of Korea, and JAX start-up funds (to O. Anczuków). This study is supported in part by the operational funds from The First Affiliated Hospital of Xi’an Jiaotong University. Also, C. Lee was a distinguished Ewha Womans University Professor supported in part by the Ewha Womans University Research grant of 2019–2020. This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT; No. NRF-2020R1C1C1009225 to Y.S. Suh). This study was supported by grant no. 13-2021-0015 (to Y.S. Suh) from the SNUBH Research Fund.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
Y. Jun: Conceptualization, data curation, formal analysis, visualization, writing–original draft. Y.-S. Suh: Conceptualization, resources, writing–original draft. S.H. Park: Validation. J. Lee: Data curation, validation. J.-I. Kim: Data curation, validation. S. Lee: Conceptualization, data curation, formal analysis. W.-P. Lee: Conceptualization, formal analysis, supervision, validation, writing–review and editing. O. Anczuków: Conceptualization, resources, supervision, validation, writing–review and editing. H.-K. Yang: Conceptualization, resources, supervision, funding acquisition, writing–review and editing. C. Lee: Conceptualization, data curation, formal analysis, supervision, funding acquisition, writing–review and editing.
References
- 1. Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008;456:470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008;40:1413–5. [DOI] [PubMed] [Google Scholar]
- 3. Urbanski LM, Leclair N, Anczuków O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 2018;9:e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di C, Syafrizayanti, Zhang Q, Chen Y, Wang Y, Zhang X, et al. Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ 2019;26:1181–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 patients with lung cancer harboring MET Exon 14 skipping alterations. J Thorac Oncol 2016;11:1493–502. [DOI] [PubMed] [Google Scholar]
- 6. Chen C, Zhao S, Karnad A, Freeman JW. The biology and role of CD44 in cancer progression: therapeutic implications. J Hematol Oncol 2018;11:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun 2012;3:883. [DOI] [PubMed] [Google Scholar]
- 8. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394–424. [DOI] [PubMed] [Google Scholar]
- 9. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–56. [DOI] [PubMed] [Google Scholar]
- 11. Li Y, Ren Z, Peng Y, Li K, Wang X, Huang G, et al. Classification of glioma based on prognostic alternative splicing. BMC Med Genomics 2019;12:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang N, Zhang P, Chen Y, Lou S, Zeng H, Deng J. Clusterization in acute myeloid leukemia based on prognostic alternative splicing signature to reveal the clinical characteristics in the bone marrow microenvironment. Cell Biosci 2020;10:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu J, McCleland M, Stawiski EW, Gnad F, Mayba O, Haverty PM, et al. Integrated exome and transcriptome sequencing reveals ZAK isoform usage in gastric cancer. Nat Commun 2014;5:3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armero VES, Tremblay MP, Allaire A, Boudreault S, Martenon-Brodeur C, Duval C, et al. Transcriptome-wide analysis of alternative RNA splicing events in Epstein-Barr virus-associated gastric carcinomas. PLoS ONE 2017;12:e0176880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shi Y, Chen Z, Gao J, Wu S, Gao H, Feng G. Transcriptome-wide analysis of alternative mRNA splicing signature in the diagnosis and prognosis of stomach adenocarcinoma. Oncol Rep 2018;40:2014–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lin P, He RQ, Ma FC, Liang L, He Y, Yang H, et al. Systematic analysis of survival-associated alternative splicing signatures in gastrointestinal pan-adenocarcinomas. EBioMedicine 2018;34:46–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng H, Jin Z, Liu K, Peng Y, Jiang S, Wang C, et al. Identification and validation of critical alternative splicing events and splicing factors in gastric cancer progression. J Cell Mol Med 2020;24:12667–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zong Z, Li H, Ning Z, Hu C, Tang F, Zhu X, et al. Integrative bioinformatics analysis of prognostic alternative splicing signatures in gastric cancer. J Gastrointest Oncol 2020;11:685–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27:1739–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shen S, Park JW, Lu ZX, Lin L, Henry MD, Wu YN, et al. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc Natl Acad Sci U S A 2014;111:E5593–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sohn BH, Hwang JE, Jang HJ, Lee HS, Oh SC, Shim JJ, et al. Clinical significance of four molecular subtypes of gastric cancer identified by The Cancer Genome Atlas Project. Clin Cancer Res 2017;23:4441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Park JW, Jung S, Rouchka EC, Tseng YT, Xing Y. rMAPS: RNA map analysis and plotting server for alternative exon regulation. Nucleic Acids Res 2016;44:W333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mun DG, Bhin J, Kim S, Kim H, Jung JH, Jung Y, et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell 2019;35:111–24. [DOI] [PubMed] [Google Scholar]
- 28. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheng S, Ray D, Lee Raymond Teck H, Naripogu KB, Yusoff Permeen Akhtar Bt M, Goh Pamela Bee L, et al. A functional network of gastric-cancer-associated splicing events controlled by dysregulated splicing factors. NAR Genom Bioinform 2020;2:lqaa013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lin KT, Ma WK, Scharner J, Liu YR, Krainer AR. A human-specific switch of alternatively spliced AFMID isoforms contributes to TP53 mutations and tumor recurrence in hepatocellular carcinoma. Genome Res 2018;28:275–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Brown RL, Reinke LM, Damerow MS, Perez D, Chodosh LA, Yang J, et al. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial–mesenchymal transition and breast cancer progression. J Clin Invest 2011;121:1064–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Itoh M, Radisky DC, Hashiguchi M, Sugimoto H. The exon 38-containing ARHGEF11 splice isoform is differentially expressed and is required for migration and growth in invasive breast cancer cells. Oncotarget 2017;8:92157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dongre A, Weinberg RA. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol 2019;20:69–84. [DOI] [PubMed] [Google Scholar]
- 35. Baud J, Varon C, Chabas S, Chambonnier L, Darfeuille F, Staedel C. Helicobacter pylori initiates a mesenchymal transition through ZEB1 in gastric epithelial cells. PLoS ONE 2013;8:e60315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu H, Zeng J, Liang X, Wang W, Zhou Y, Sun Y, et al. Helicobacter pylori promotes epithelial–mesenchymal transition in gastric cancer by downregulating programmed cell death protein 4 (PDCD4). PLoS ONE 2014;9:e105306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo J, Jia R. Splicing factor poly(rC)-binding protein 1 is a novel and distinctive tumor suppressor. J Cell Physiol 2018;234:33–41. [DOI] [PubMed] [Google Scholar]
- 38. Lee S, Cieply B, Yang Y, Peart N, Glaser C, Chan P, et al. Esrp1-regulated splicing of Arhgef11 isoforms is required for epithelial tight junction integrity. Cell Rep 2018;25:2417–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gullo I, Carneiro F, Oliveira C, Almeida GM. Heterogeneity in gastric cancer: from pure morphology to molecular classifications. Pathobiology 2018;85:50–63. [DOI] [PubMed] [Google Scholar]
- 40. Cardama GA, Gonzalez N, Maggio J, Menna PL, Gomez DE. Rho GTPases as therapeutic targets in cancer (Review). Int J Oncol 2017;51:1025–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bonnal SC, López-Oreja I, Valcárcel J. Roles and mechanisms of alternative splicing in cancer - implications for care. Nat Rev Clin Oncol 2020;17:457–74. [DOI] [PubMed] [Google Scholar]
- 42. Kahles A, Lehmann KV, Toussaint NC, Hüser M, Stark SG, Sachsenberg T, et al. Comprehensive analysis of alternative splicing across tumors from 8,705 patients. Cancer Cell 2018;34:211–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xiong Y, Deng Y, Wang K, Zhou H, Zheng X, Si L, et al. Profiles of alternative splicing in colorectal cancer and their clinical significance: a study based on large-scale sequencing data. EBioMedicine 2018;36:183–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol 2014;15:178–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Katoh Y, Katoh M. Hedgehog signaling, epithelial-to-mesenchymal transition and miRNA (review). Int J Mol Med 2008;22:271–5. [PubMed] [Google Scholar]
- 46. Weyemi U, Redon CE, Choudhuri R, Aziz T, Maeda D, Boufraqech M, et al. The histone variant H2A.X is a regulator of the epithelial–mesenchymal transition. Nat Commun 2016;7:10711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oh SC, Sohn BH, Cheong JH, Kim SB, Lee JE, Park KC, et al. Clinical and genomic landscape of gastric cancer with a mesenchymal phenotype. Nat Commun 2018;9:1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Larsen JE, Nathan V, Osborne JK, Farrow RK, Deb D, Sullivan JP, et al. ZEB1 drives epithelial-to-mesenchymal transition in lung cancer. J Clin Invest 2016;126:3219–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reinke LM, Xu Y, Cheng C. Snail represses the splicing regulator epithelial splicing regulatory protein 1 to promote epithelial–mesenchymal transition. J Biol Chem 2012;287:36435–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Warzecha CC, Sato TK, Nabet B, Hogenesch JB, Carstens RP. ESRP1 and ESRP2 are epithelial cell-type-specific regulators of FGFR2 splicing. Mol Cell 2009;33:591–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Anczukow O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nat Struct Mol Biol 2012;19:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015;523:96–100. [DOI] [PubMed] [Google Scholar]
- 53. Lee S, Lee S, Ouellette S, Park WY, Lee EA, Park PJ. NGSCheckMate: software for validating sample identity in next-generation sequencing studies within and across data types. Nucleic Acids Res 2017;45:e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ghandi M, Huang FW, Jane-Valbuena J, Kryukov GV, Lo CC, McDonald ER III, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.