Inhibition of CTPS in MYC-overexpressing cells blocks pyrimidine synthesis while maintaining ribosome synthesis activity to create an anabolic imbalance that induces replication stress, providing a new approach to selectively target MYC-driven cancer.
Abstract
The “undruggable” oncogene MYC supports cancer cell proliferation and survival through parallel induction of multiple anabolic processes. Here we find that inhibiting CTP synthase (CTPS) selectively decreases cell viability and induces DNA replication stress in MYC-overexpressing cells. MYC-driven rRNA synthesis caused the selective DNA replication stress upon CTPS inhibition. Combined inhibition of CTPS and ataxia telangiectasia and Rad3-related protein (ATR) is synthetically lethal in MYC-overexpressing cells, promoting cell death in vitro and decreasing tumor growth in vivo. Unexpectedly, interfering with CTPS1 but not CTPS2 is required to induce replication stress in MYC-deregulated cancer cells and consequent cell death in the presence of an ATR inhibitor. These results highlight a specific and key role of CTPS1 in MYC-driven cancer, suggesting that selectively inhibiting CTPS1 in combination with ATR could be a promising strategy to combat disease progression.
Significance:
Inhibition of CTPS in MYC-overexpressing cells blocks pyrimidine synthesis while maintaining ribosome synthesis activity to create an anabolic imbalance that induces replication stress, providing a new approach to selectively target MYC-driven cancer.
See related commentary by Chabanon and Postel-Vinay, p. 969
Introduction
Deregulation of MYC is implicated in the development of up to 70% to 80% of human cancers primarily via genomic translocation, gene amplification, protein stabilization, and the upregulation of mRNA transcription (1). MYC inactivation leads to tumor growth inhibition or regression (2–8), and has been considered an attractive cancer therapy target for decades (9). However, directly targeting MYC is challenging due to the lack of a defined small molecule binding pocket in the MYC protein and severe toxicity upon prolonged inhibition of MYC (9–12).
MYC supports cancer cell proliferation and survival through concurrently inducing multiple key anabolic processes, and has been proposed as a therapeutic vulnerability (13, 14). One well-established function of MYC is to promote ribosome biogenesis to increase the capacity of protein synthesis of MYC-overexpressing cancer cells (15). rRNA accounts for about 85% of total cellular RNA, indicating that MYC-mediated upregulation of ribosome biogenesis is accompanied by a dramatically increased demand for nucleotides. To this end, MYC activation stimulates the expression of genes encoding the nucleotide synthesis enzymes (16–19), and also stimulates glycolysis (20–22) and the pentose phosphate pathway (23) to fulfill the demand of essential precursors for the synthesis of purine and pyrimidine nucleotides. These nucleotides will be rapidly used for RNA and, in proliferating cells, DNA synthesis.
CTP synthase (CTPS) catalyzes the final, rate-limiting step in the de novo synthesis of the nucleotide CTP (24–27). CTP has the lowest concentration among the four nucleotides (UTP, ATP, GTP, and CTP), making it the rate-limiting molecule for nucleic acid synthesis and other CTP-dependent events (28). Elevated CTPS activity is frequently observed in human cancers and activated lymphocytes (29, 30). CTPS can form filamentous structures, termed cytoophidia, in many organisms across all three life domains including archaea, bacteria, and eukaryotes such as fruit fly, yeast, plant, zebrafish, and mammals (31–39). Forming cytoophidium is involved in the regulation of CTPS enzymatic activity and protein half-life (40–43). We previously demonstrated that MYC can regulate CTPS filamentation and CTPS is required for MYC-mediated cell size control in Drosophila (44). In humans, CTPS activity is encoded by two isoforms, CTPS1 and CTPS2, which catalyze the same reaction and have similar catalytic activity (27, 45, 46). The respective functions of these two enzymes are still unknown except for the necessity of CTPS1, but not CTPS2, in the proliferation of activated lymphocytes during the immune response (47, 48).
In this work, we employ a specific CTPS inhibitor, 3-deazauridine (DAU; ref. 33), to address whether the inhibition of CTP synthesis is a vulnerability of MYC-overexpressing cancer. Combined inhibition of CTPS and the replication stress-sensing kinase ataxia telangiectasia and Rad3-related protein (ATR; ref. 3) induces synthetic lethality in MYC-overexpressing cells and dramatically decreases tumor growth in vivo.
Materials and Methods
Reagents and antibodies
Antibodies for Chk1 (25887–1-AP), H2AX (10856–1-AP), P53 (10442–1-AP), ATR (19787–1-AP), and CTPS1 (15914–1-AP) were purchased from ProteinTech. Antibodies for MYC (ab32072), CTPS2 (ab196016), and β-actin (ab6276) were purchased from Abcam. Antibodies for phospho-Chk1 (Ser345; #2348), phospho-histone H2AX (Ser139; #9718), phospho-ATR (Thr1989; #58014), and cleaved caspase-3 (Asp175; #9661) were purchased from Cell Signaling Technology. Antibody for HA (Sc-7392) was purchased from Santa Cruz. Doxycycline (S5159), VX-680 (S1048), Purvalanol A (S7793), cytidine (S2053), uridine (S2029), guanosine (S2439), adenosine (S1647), VE-822 (S7102), hydroxyurea (S1896), and AZD6738 (S7693) were purchased from Selleck Chemicals. BMH-21 (B4896) was from APExBIO. BAY-1895344 (HY-101566A), CX-3543 (HY-14776), and CX-5461 (HY-13323) were from MedchemExpress. 3-Deazauridine (sc-394445) was purchased from Santa Cruz.
Cell culture
ARPE-19, SW480, HCT 116, and 293T were cultured in DMEM (SH30022.01; Hyclone) whereas RKO and Raji cells were cultured in RPMI 1640 (SH30809.01; Hyclone) supplemented with 10% FBS (04–001; Biological Industries) in a humidified atmosphere containing 5% CO2 at 37°C. All the commercial cell lines used in this article were purchased from Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences (Shanghai, China). They were originally purchased from ATCC. The cells were authenticated by short tandem repeat (STR) typing and routinely tested for Mycoplasma using the GMyc-PCR Mycoplasma Test Kit (Yeasen, #40601). All cells used in the experiments were within 15 passages from thawing.
Immunostaining and immunoblotting
Immunostaining and immunoblotting were performed as previously described (49). Data shown are representative of at least three independent experiments.
Lentiviral short hairpin RNA cloning, production, and infection
Desalted oligonucleotides were cloned into pPLK/GFP + Puro purchased from the Public Protein/Plasmid Library or pPLK/mCherry + Puro with the BamHI/EcoRI sites at the 3′ end of the human H1 promoter. The short hairpin RNA (shRNA) sequences are listed in Supplementary Table S1. shRNAs were packaged in lentiviral particles by cotransfection with packaging plasmids psPAX2 and pMD2.G into 293 T cells. Virus-containing supernatant was filtered with 0.45 mm polyethersulfone (PES) filter and then used to infect cells.
RNA extraction and qRT-PCR
Total RNAs were extracted as described previously (49). The first-strand cDNA synthesis was conducted with RevertAid First-Strand cDNA synthesis kits. qRT-PCR reactions were performed using ABI 7500 detection system. The resulting values were normalized to β-actin expression. The primer sequences are listed in Supplementary Table S2.
Comet assay
Cells treated with DAU and/or BAY-1895344 were gently scraped and subjected to alkaline comet assay with the Comet Assay Kit (Enzo ADI-900–166) according to the manufacturer's instructions. Comet tails were visualized by a fluorescent microscope and the percentages of the comet tail DNA were measured with CASP software (CASPLab, University of Wroclaw, Wroclaw, Poland). Fifty cells were counted per group.
Cell viability and apoptosis assay
Cell viability was measured using the Cell Counting Kit 8 (APExBIO, K1018) according to the manufacturer's instructions. Annexin V/propidium iodide (PI) staining was used to determine apoptosis. Pacific Blue Annexin V (640918) was purchased from Biolegend. PI (ST511) was purchased from Beyotime. Annexin V/PI staining was performed according to the manufacturer's instructions. The fluorescence was measured using Becton Dickinson LSRFortessa X20 flow cytometry and analyzed with FlowJo Version 10.6 software.
Cell-cycle profiles
Single-cell suspensions were washed with PBS and fixed with 70% ethanol overnight at -20°C. Cells were then washed with cold PBS twice at 4°C, followed by incubating in PBS with 50 μg/mL PI (Beyotime, ST511), 0.1 mg/mL RNase A, 0.05% Triton X-100 for 20 minutes at 37°C. Staining was measured with Becton Dickinson LSRFortessa X20 flow cytometry and analyzed with FlowJo Version 10.6 software.
Colony formation assays
Cells cultured in 6-well plates were treated with DAU and BAY-1895344 alone or together for 2 weeks and then fixed with 2% paraformaldehyde and stained with 0.1% crystal violet (Sigma, V5265–500 mL).
Three-dimensional cell culture
A 48-well plate was coated with 20 μL of Engelbreth—Holm–Swarm mouse tumor matrix (Matrigel, Corning) and kept in cell incubator for 30 minutes. cells were trypsinized and suspended in appropriate medium containing 2% Matrigel. One thousand cells per well were seeded in the plate. Three days later, cells were treated with DAU and/or BAY-1895344 for 5 further days and visualized by optical microscope. Spheroid size was measured and quantified.
HCT 116 xenografts
Nude mice (BALB/c, 8 weeks old) were injected subcutaneously with 100 μL HCT 116 cells (2 × 106). Six days later, mice were administered with DAU (75 mg/kg) and/or BAY-1895344 (10 mg/kg) diluted in olive oil intraperitoneally once daily, 5 days on/2 days off. Tumor volume was measured every third day. Mice were killed after drug treatment and tumor volume was measured. All manipulations involving mice were approved by the Institutional Animal Care and Use Committee of ShanghaiTech University (Shanghai, China).
Generation of CRISPR/Cas9-mediated knockout cell lines
Single-guide RNA (sgRNA) oligonucleotides were cloned into the plasmid pU6-(BbsI)_CBh-Cas9-T2A-mCherry (Addgene plasmid #64324) with the BbsI site at the end of the human U6 prompter. Two independent sgRNAs were used to knockout one gene and the sgRNA sequences are listed in Supplementary Table S3. Two days after transfection, mCherry positive cells were sorted and seeded into a 96-well plate. About 2 weeks later, cells were harvested and subjected to immunoblotting with appropriate antibodies to identify the knockout cell lines. Successful knockout colonies were randomly picked and used for further study.
Bioinformatics
The dataset used comprised mRNA sequencing (mRNA-seq) data from The Cancer Genome Atlas (TCGA) tumors (see TCGA Data Portal at https://tcga-data.nci.nih.gov/tcga/; the two-gene correlation map is realized by the R software package ggstatsplot). We used Pearson correlation analysis to describe the correlation between quantitative variables without a normal distribution. P < 0.05 was considered statistically significant.
Statistical analysis
Data are provided as means ± SEM. Two-tailed unpaired Student t test was used for comparisons between two groups. One‐way ANOVA was used to compare variables among three or more groups. P < 0.05 was considered statistically significant. In each experiment, “n” means the number of independent experiments and the number of mice in in vitro and in vivo study, respectively. All tests were performed using GraphPad Prism version 6.00 (GraphPad Software; www.graphpad.com).
Results
DAU selectively inhibits the viability of cells overexpressing MYC
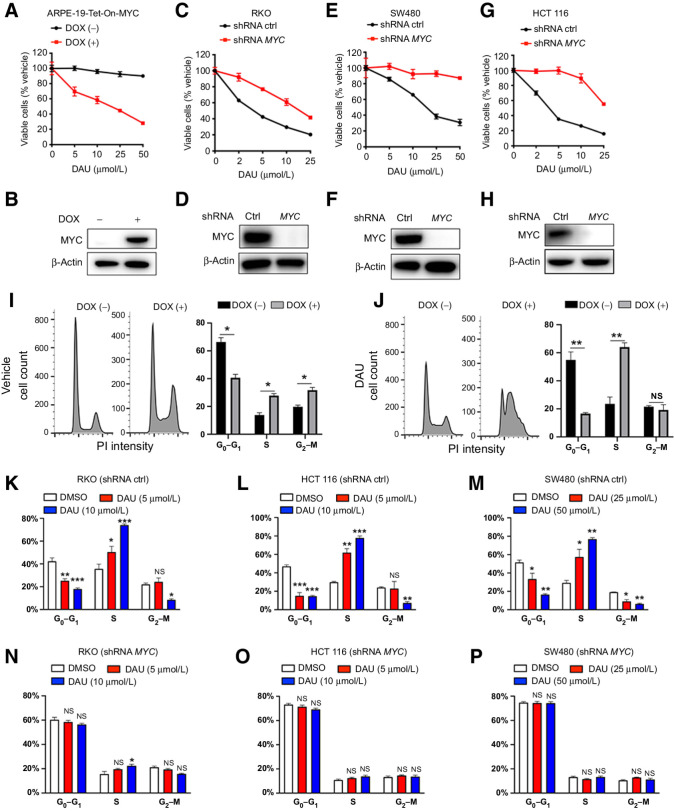
To understand the potential role of CTPS in MYC-driven cancer cells, we stably transfected the construct Tet-On-MYC-CMV-Puro into ARPE-19 cells, a spontaneously arisen retinal pigment epithelium cell line. The resulting cells cultured in the presence or absence of doxycycline are called ARPE-19-MYC (doxycycline+) and ARPE-19-MYC (doxycycline−), respectively. We first validated our experimental system by examining the sensitivity of ARPE-19-MYC (doxycycline+) and ARPE-19-MYC (doxycycline−) cells to Purvalanol A and VX-680. Both can induce synthetic lethality to MYC-overexpressing cells by targeting cyclin-dependent kinase 1 (CDK1) and aurora-B kinase, respectively (50, 51). Both Purvalanol A and VX-680 selectively decreased the viability (Supplementary Fig. S1A and S1B) and increased cell death (Supplementary Fig. S1C) of ARPE-19-MYC (doxycycline+) cells.
DAU treatment for 2 or 7 days resulted in a dramatic decrease in the viability of ARPE-19-MYC (doxycycline+) cells, while a significantly smaller reduction was observed in ARPE-19-MYC (doxycycline−) cells (Fig. 1A and B; Supplementary Fig. S1D). MYC was knocked down by two independent MYC-specific lentiviral shRNAs (Fig. 1D, F, and H). After treated for 2 or 7 days, DAU dose-dependently suppressed the viability of the control cells (Fig. 1C, E, and G; Supplementary Fig. S1E–S1G). However, the MYC knockdown cells showed reduced sensitivity to DAU as compared with their MYC-expressing counterparts (Fig. 1C, E, and G; Supplementary Fig. S1E–S1G).
Figure 1.
CTPS inhibition selectively suppresses cell viability in a MYC-dependent manner. A, Cell viability analysis of ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with the indicated concentrations of DAU for 1 week. B, MYC expression was measured by immunoblotting. C, E, and G, Cell viability analysis of control and MYC knockdown RKO (C), SW480 (E), and HCT116 (G) cells treated with the indicated concentrations of DAU for 1 week. D, F, and H, MYC knockdown efficiency was analyzed by immunoblotting. I and J, Cell-cycle analysis of ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with vehicle (I) or DAU (20 μmol/L; J) for 24 hours. *, P < 0.05; **, P < 0.01. Cell-cycle analysis of control (K, L, and M) and MYC knockdown (N, O, and P) of RKO, HCT116, and SW480 cells treated with the indicated concentrations of DAU for 24 hours. *, P < 0.05; **, P < 0.01; ***, P < 0.001, significantly different from DMSO-treated group. Data are means ± SEM. For cell viability and cell-cycle analysis, n = 3–4. DOX, doxycycline; ctrl, control; NS, not significant.
DAU treatment for 7 days induced an increase (up to 13.7%) of cell death in ARPE-19-MYC (doxycycline+) cells, but not in ARPE-19-MYC (doxycycline−) cells (Supplementary Fig. S1H). The induction of cell death by DAU treatment for 7 days can also be observed in HCT116 (approximately 15%) and RKO (approximately 8%) cells, which was significantly alleviated by MYC knockdown (Supplementary Fig. S1I and S1J). No obvious increase in cell death could be detected in SW480 cells expressing either control or MYC shRNA after DAU treatment (Supplementary Fig. S1K). Though DAU induced selective cell death in ARPE-19-MYC (doxycycline+), HCT116 and RKO cells, the percentage of dead cells is less than 15%, which is incomparable to the approximately 80% loss of cell viability after 1 week of DAU treatment (Fig. 1A, C, E, and G).
Overexpression of MYC in ARPE-19 cells promoted cell cycle progression from G0–G1 phase into S-G2–M (Fig. 1I). Upon DAU treatment, ARPE-19-MYC (doxycycline+) cells showed a dramatic reduction of cells in G0–G1 phase, and a significant increase of cells in S phase without altering the proportion of cells in G2–M phase as compared with ARPE-19-MYC (doxycycline−) cells (Fig. 1J), indicating that DAU treatment selectively blocks MYC-overexpressing cells in S phase. The induction of S phase cell-cycle arrest can also be observed in HCT116, RKO, and SW480 cells (Fig. 1K–M), and this effect was abolished by MYC knockdown (Fig. 1N–P).
DAU causes selective replication stress in MYC-overexpressing cells, which originates from MYC-driven rRNA synthesis
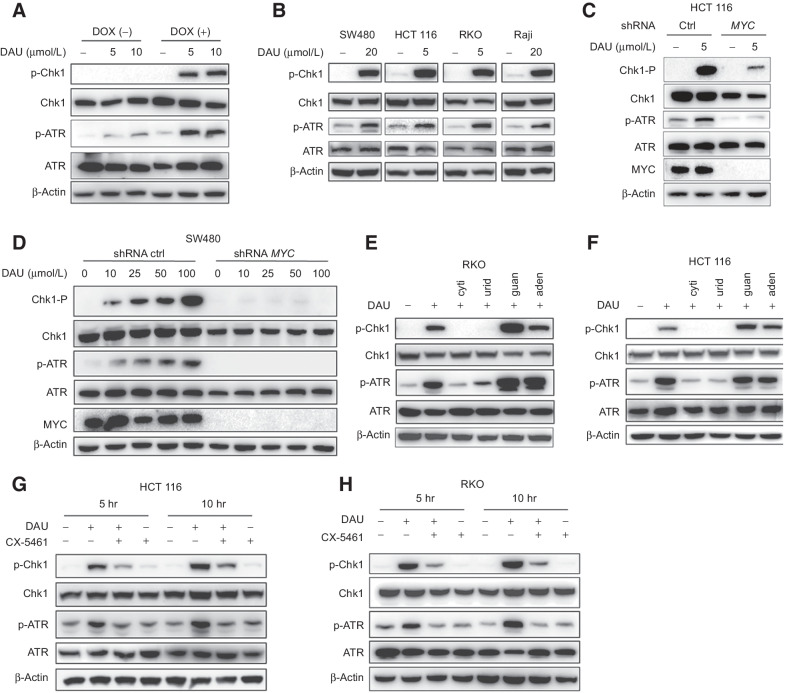
Hydroxyurea (HU) was used as a positive control for Chk1 and ATR activation (Supplementary Fig. S2A). DAU treatment caused a dose-dependent increase of Chk1 Ser345 and ATR Thr1989 phosphorylation in ARPE-19-MYC (doxycycline+) cells, while no Chk1 phosphorylation and a slight ATR phosphorylation could be detected in ARPE-19-MYC (doxycycline−) cells (Fig. 2A). The induction of replication stress can also be observed in MYC-upregulated cancer cells such as SW480, HCT116, RKO, and Raji (Fig. 2B).
Figure 2.
DAU causes selective replication stress in MYC-overexpressing cells. A, Immunoblots of the indicated proteins in ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with DAU for 24 hours. B, Immunoblots of the indicated proteins in SW480, HCT116, RKO, and Raji cells treated for 24 hours with DAU. C and D, Immunoblot analysis of the effect of MYC expression on DAU-induced Chk1 and ATR phosphorylation. E and F, RKO (E) and HCT116 (F) cells treated with vehicle or DAU (5 μmol/L) in the presence or absence of 50 μmol/L cytidine (cyti), uridine (urid), guanosine (guan), or adenosine (aden) were analyzed by immunoblotting. G and H, HCT116 (G) and RKO (H) cells were pretreated with 200 nmol/L CX-5461 for 12 hours, and then treated with 5 μmol/L DAU for another 5 or 10 hours. Cell extracts were analyzed by immunoblotting. One of three to five similar experiments is shown. DOX, doxycycline; ctrl, control; hr, hours.
Expression of MYC shRNA efficiently reduced the protein levels of MYC (Fig. 2C and D). Concomitantly, DAU-induced phosphorylation of Chk1 Ser345 and ATR Thr1989 was significantly decreased (Fig. 2C and D). Either cytidine or uridine, but not adenosine and guanosine, can effectively reverse DAU-induced Chk1 Ser345 and ATR Thr1989 phosphorylation, (Fig. 2E and F; Supplementary Fig. S2B and S2C).
Pol I inhibition decreased DAU-induced replication stress in HCT116 and RKO cells (Fig. 2G and H), evidenced by the decreased Chk-1 Ser345 and ATR Thr1989 phosphorylation in CX-5641-treated cells. To further confirm this phenomenon, two other Pol I inhibitors BMH-21 and CX-3543 were used to inhibit rRNA biosynthesis. Pretreated with either BMH-21 or CX-3543 can also abolish DAU-induced Chk1 Ser 345 phosphorylation (Supplementary Fig. S2D and S2E).
The combination of DAU with ATR inhibitor induces DNA damage in a MYC-dependent manner
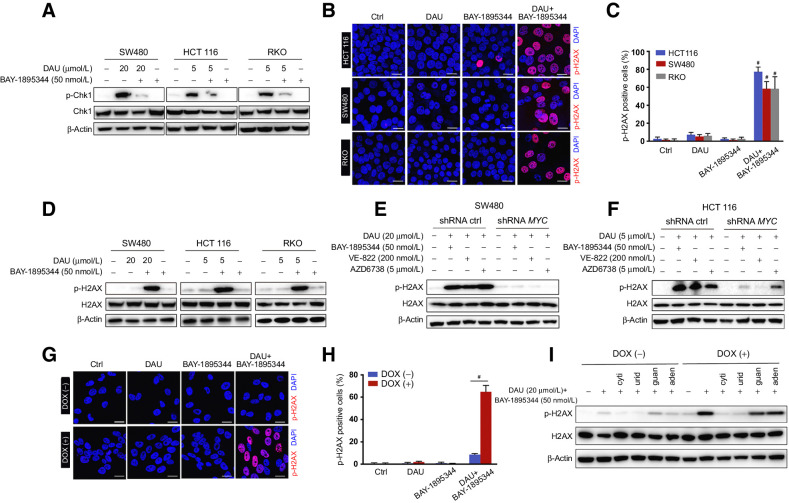
DAU-induced Chk1 Ser345 phosphorylation was blocked by ATR selective inhibitors BAY-1895344, VE-822, and AZD6738 (Fig. 3A; Supplementary Fig. S3A), indicating the key role of ATR in DAU-induced Chk1 Ser345 phosphorylation. Next, we examined if the combination of DAU with ATR inhibitor could induce selective DNA damage in MYC-overexpressing cells. The phosphorylation of histone H2AX (S139) was used as a molecular marker to evaluate DNA damage. Treatment with DAU together with ATR inhibitor induced H2AX phosphorylation in SW480, HCT116, and RKO cancer cells (Fig. 3B–D), and this effect was significantly abolished by MYC knockdown (Fig. 3E and F). For further confirmation of this phenomenon, we conducted a single-cell electrophoresis assay (comet assay) to measure DNA damage. The comet assays showed a dramatic increase in the percentage of comet tail DNA in the cells treated with the combination of DAU and BAY-1895344 (Supplementary Fig. S3B and S3C), and this phenotype was reversed by the knockdown of MYC (Supplementary Fig. S3D and S3E). Consistently, the combination of DAU with ATR inhibitor caused phosphorylation of histone H2AX specifically in ARPE-19-MYC (doxycycline+) but not in ARPE-19-MYC (doxycycline−) cells (Fig. 3G and H; Supplementary Fig. S3F), and this effect was blocked by the addition of exogenous cytidine or uridine (Fig. 3I). Comet assay also showed a specific increase in the percentage of comet tail DNA in ARPE-19-MYC (doxycycline+) cells after being treated with DAU and BAY-1895344 in combination (Supplementary Fig. S3G and S3H).
Figure 3.
DAU combined with ATR inhibitors induces DNA damage in a MYC-dependent manner. A, Immunoblots of the indicated proteins in SW480, HCT116, and RKO cells treated for 24 hours with DAU alone or together with BAY-1895344. B, Phosphorylation of H2AX in HCT116, SW480, and RKO cells treated with DAU and BAY-1895344 alone or together were revealed by immunostaining. Phospho-H2AX (14) and DAPI (blue). Quantified data are shown in C. #, P < 0.0001; significantly different from vehicle-treated group. D, Immunoblots of the indicated proteins in SW480, HCT116, and RKO cells treated with DAU and BAY-1895344 alone or together for 24 hours. E and F, Immunoblots of the indicated proteins in control and MYC knockdown SW480 (E) and HCT116 (F) cells treated with DAU alone or together with BAY-1895344, VE-822, or AZD6738 for 24 hours. G, Phosphorylation of H2AX in ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with 20 μmol/L DAU and 50 nmol/L BAY-1895344 alone or together for 16 hours. Quantified data are shown in H. #, P < 0.0001. I, Immunoblots of the indicated proteins in ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with DAU and BAY-1895344 with or without 50 μmol/L cytidine (cyti), uridine (urid), guanosine (guan), or adenosine (aden). Graphic data are means ± SEM. For Western blotting, one of three to five similar experiments is shown. Scale bar, 20 μm. Ctrl, control; DOX, doxycycline.
DAU combined with ATR inhibitor induces synthetic lethality to MYC-overexpressing cells
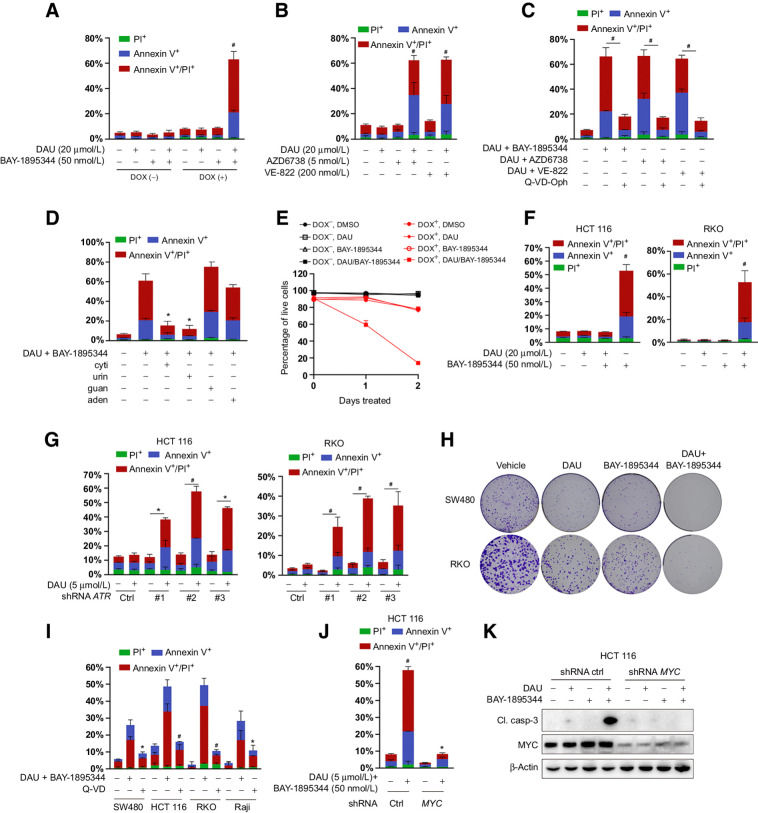
We next determined whether the combination of DAU and ATR inhibitor could induce selective cell death in MYC-overexpressing cells. Cell death was approximately two-fold higher in ARPE-19-MYC (doxycycline+) cells compared with ARPE-19-MYC (doxycycline−) cells upon DMSO treatment (Fig. 4A). After being treated with the combination of DAU and BAY-1895344 for 24 hours, more than 60% of ARPE-19-MYC (doxycycline+) cells were positive for Annexin V, or both Annexin V and PI. In comparison, about 8% of DMSO-treated cells exhibited these traits while no obvious change was observed in cells treated with DAU or BAY-1895344 alone (Fig. 4A). Importantly, neither DAU nor BAY-1895344 treatment alone or in combination could induce cell death in ARPE-19-MYC (doxycycline−) cells (Fig. 4A). A dramatic induction of cell death was also observed when ARPE-19-MYC (doxycycline+) cells were treated with DAU in combination with another ATR inhibitor, either VE-822 or AZD6738 (Fig. 4B). This was rescued by the pan-caspase inhibitor Q-VD-Oph, suggesting that apoptosis is primarily responsible for cell death induced by the combination of DAU and ATR inhibitor (Fig. 4C). Meanwhile, the presence of exogenous cytidine or uridine rescued apoptosis induced by the combination of DAU and BAY-1895344, while guanosine and adenosine did not have this effect (Fig. 4D). Culturing ARPE-19-MYC (doxycycline−) cells in the presence of both DAU and BAY-1895344 led to little loss of viability even after 2 days of treatment, whereas ARPE-19-MYC (doxycycline+) cells showed persistent induction of cell death over the time course (Fig. 4E).
Figure 4.
Combined inactivation of CTPS and ATR is synthetically lethal to MYC-transformed cells. A, Apoptosis analysis of ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells treated with DAU and BAY-1895344 alone or together for 24 hours. B, Apoptosis analysis of ARPE-19-MYC (DOX+) cells treated with DAU alone or together with AZD6738 or VE-822 for 24 hours. C, The impacts of Q-VD-Oph on the combination of DAU and ATR inhibitor-induced apoptosis in ARPE-19-MYC (DOX+) cells. #, P < 0.001. D, Apoptosis analysis of ARPE-19-MYC (doxycycline+) cells treated with DAU (20 μmol/L)/BAY-1895344 (50 nmol/L) with or without 50 μmol/L cytidine (cyti), uridine (urid), guanosine (guan), or adenosine (aden) for 24 hours. E, Cell death analysis of ARPE-19-MYC (doxycycline+) and ARPE-19-MYC (doxycycline−) cells treated with DAU (20 μmol/L) and BAY-1895344 (50 nmol/L) alone or together for the indicated time. F, Apoptosis analysis of the indicated cells treated with DAU and BAY-1895344 alone or together for 48 hours. G, HCT116 and RKO cells expressing control or ATR shRNA were treated with DAU for 48 hours, followed by apoptosis analysis. *, P < 0.01; #, P < 0.001. H, Effect of DAU and BAY-1895344 on clonogenic survival of SW480 and RKO cells. I, The impacts of Q-VD-Oph on the combination of DAU- and BAY-1895344–induced apoptosis in the indicated cells. J and K, Control and MYC knockdown HCT116 cells were treated with DAU and BAY-1895344 for 48 hours, followed by Annexin V/PI–based apoptosis analysis (J) and immunoblotting analysis (K). Graphical data are means ± SEM. n = 3–4. For A, B, F, and J, *, P < 0.05; #, P < 0.0001; significantly different from DMSO-treated group. For D and I, *, P < 0.01; #, P < 0.0001; significantly different from DAU/BAY-1895344–treated group. DOX, doxycycline; ctrl, control; Cl. casp-3; cleaved caspase-3.
Human cancer cells with amplification or translocation of the MYC gene or the dysregulation of MYC protein degradation might have adapted to high MYC protein expression, rendering them insensitive to the combination of DAU and BAY-1895344. We investigated this in a variety of cancer cell lines with high MYC expression. Our data showed a significant increase in the percentage of apoptosis among cells treated with DAU and BAY-1895344 in combination, while either DAU or BAY-1895344 alone has no obvious effect on cell death (Fig. 4F; Supplementary Fig. S4A and S4B). shRNA-mediated knockdown of ATR combined with DAU treatment can also induce dramatic apoptotic cell death among HCT116 and RKO cells (Fig. 4G). The knockdown of ATR in both HCT116 and RKO was confirmed by Western immunoblotting (Supplementary Fig. S4C and S4D). The colony formation assay showed that DAU with BAY-1895344 exhibited much stronger inhibitory effects on SW480 and RKO than either of them alone (Fig. 4H; Supplementary Fig. S4E and S4F). In agreement with the observation in ARPE19-MYC (doxycycline+) cells, the pan-caspase inhibitor Q-VD-Oph was able to suppress cell death induced by the combination of DAU and BAY-1895344 in SW480, HCT116, RKO, and Raji cells (Fig. 4I). In addition, cell death was blocked by cotreatment with cytidine or uridine, but not guanosine or adenosine (Supplementary Fig. S4G–S4J).
We next investigated whether the induction of cell death by the DAU and BAY-1895344 combination was MYC-dependent. To this end, we evaluated the impact on cellular apoptosis in HCT116, RKO, and SW480 cells expressing control or MYC shRNA. Annexin V/PI staining showed that knockdown of MYC significantly reduced cell death induced by the DAU and BAY-1895344 combination in all cell lines (Fig. 4J; Supplementary Fig. S4K and S4M). In agreement with this observation, caspase-3 activation was also significantly reduced in the MYC knockdown cells (Fig. 4K; Supplementary Fig. S4L and S4N).
DAU and BAY-1895344 combination suppresses tumor growth in 3D assays and in vivo
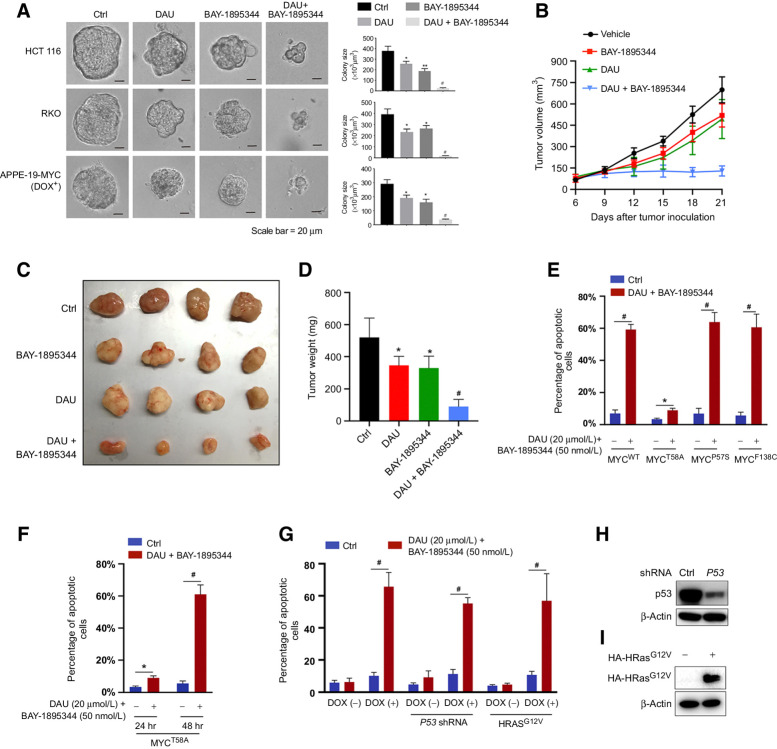
To test the combination in vivo, we first investigated the impacts of DAU and BAY-1895344 alone or together on the spheroid growth of HCT116, RKO, and ARPE-19-MYC (doxycycline+) cells in the three-dimensional (3D) tumor cell culture system, which can simulate the in vivo physiologic state of the tumor microenvironment. Though both DAU and BAY-1895344 alone showed about a 40% to 50% reduction in spheroid size, their combination reduced the spheroid size to 5% to 10% of the control group (Fig. 5A). We further administered nude mice bearing subcutaneously implanted HCT116 xenografts with DAU and BAY-1895344 alone or together. While DAU and BAY-1895344 alone both showed a slight anti-tumor effect, their combination inhibited the growth of the HCT116 tumor substantially (Fig. 5B and C). At the end of the treatment, the tumors were excised and their volumes were measured (Fig. 5D).
Figure 5.
DAU and BAY-1895344 combination suppresses tumor growth 3D assays and in vivo.A, HCT116, RKO, and ARPE-19-MYC (DOX+) cells grown on Matrigel were treated with 5 μmol/L (RKO and HCT116) or 20 μmol/L (ARPE-19-MYC; DOX+) DAU alone or together with 50 nmol/L BAY-1895344 for 5 days. Representative images of spheroids are shown (left). Quantified spheroid sizes (right). n = 3. B, Nude mice bearing HCT116 xenograft tumors were treated with vehicle, DAU, and/or BAY-1895344 once daily, 5 days on/2 days off, intraperitoneally. The tumor volume was monitored and recorded. C and D, Tumors were excised at day 21 (C) and weighed (D). E, ARPE-19 cells expressing WT or the indicated mutant MYC were treated with DAU/BAY-1895344 for 24 hours and then subjected to apoptosis analysis. n = 4; *, P < 0.05; #, P < 0.0001. F, Apoptosis analysis of ARPE-19-MYCT58A treated with DAU/BAY-1895344 for 24 or 48 hours. n = 4; *, P < 0.05; #, P < 0.0001. G, ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells expressing HRASG12V or P53 shRNA were treated with DAU/BAY-189534 for 24 hours, followed by apoptosis analysis. H and I, Knockdown efficiency of P53 and the expression of HA-HRASG12V. n = 4; #, P < 0.0001. Graphic data are means ± SEM. For A and D, *, P < 0.05; **, P < 0.01; #, P < 0.0001; significantly different from vehicle-treated group. DOX, doxycycline; ctrl, control; hr, hours.
Because MYC overexpression is known to sensitize cells to apoptosis, combined inhibition of CTPS and ATR might exacerbate the apoptotic program normally regulated by MYC. To test this hypothesis, we overexpressed apoptosis-resistant MYC mutants MYCP57S, MYCF138C, and MYCT58A in ARPE-19 cells (52–55). Cells overexpressing P57S or F138C mutant MYC retained the sensitivity to apoptosis, similarly to the cells overexpressing wild-type (WT) MYC, when treated with DAU and BAY-1895344 in combination for 24 hours (Fig. 5E). For the cells overexpressing T58A mutant MYC, the percentage of apoptosis was about half of that for the cells overexpressing WT MYC upon DMSO treatment (Fig. 5E), which is probably due to the impaired induction of the BH3-only protein Bim [a proapoptotic member of the B-cell lymphoma 2 (Bcl2) family] by MYCT58A (55). After treatment with DAU and BAY-1895344 in combination for 24 hours, the percentage of apoptotic cells was about three times higher than after vehicle treatment for MYCT58A expressing cells but was significantly lower than for WT MYC-expressing cells (10% vs. 60%; Fig. 5E). Yet, a prolonged treatment time of 48 hours did induce apoptosis in about 60% of MYCT58A-expressing cells (Fig. 5F).
We pondered whether p53 loss or coexpression of the constitutively active Ras could prevent apoptosis in the context of the combination of DAU and BAY-1895344 treatment. APRE-19-MYC cells expressing HRasG12V or shRNA targeting P53 showed a similar degree of cell death as compared with the parental APRE-19-MYC cells after treatment with both DAU and BAY-1895344 (Fig. 5G). The knockdown of p53 and the expression of HRasG12V were confirmed by Western immunoblotting (Fig. 5H and I). Thus, activated Ras or p53 deficiency does not block the apoptosis induced by a combination of DAU and BAY-1895344 in MYC-overexpressing cells.
Inhibition of CTPS1 is required and sufficient to induce selective cell death when combined with BAY-1895344
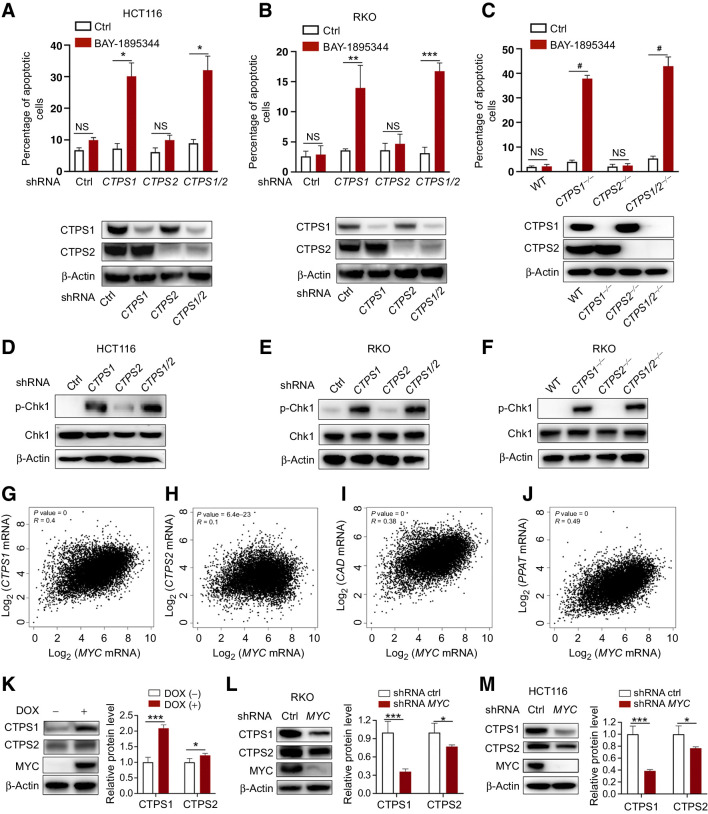
CTPS1 and CTPS2 proteins share 74% identity and homology. To understand the precise role of these two isoforms, two distinct lentivirus shRNAs were used to knock down CTPS1 and CTPS2 alone or together in HCT116 and RKO cells. Unexpectedly, we observed that CTPS1 knockdown cells showed a dramatic increase in apoptosis when treated with BAY-1895344. This increase was specific to the knockdown of the CTPS1 isozyme; knockdown of CTPS2 had no obvious effect on cellular apoptosis under BAY-1895344 treatment (Fig. 6A and B). However, the degree of apoptosis induction was relatively lower than with the combination of DAU and BAY-1895344.
Figure 6.
Inhibition of CTPS1 sensitizes MYC-driven cancer cells to BAY-1895344. A and B, HCT116 (A) and RKO (B) cells expressing CTPS1 and CTPS2 shRNA alone or together were treated with BAY-1895344 for 48 hours, followed by apoptosis analysis. C, Apoptosis analysis of WT, CTPS1−/−, CTPS2−/−, and CTPS1/2−/− RKO cells treated with BAY-1895344 for 48 hours. D–F, Immunoblots of p-Chk1 in HCT116 and RKO cells from A, B, and C. G–J, Pearson correlation analysis of the mRNA expressions of MYC and CTPS1 (G), CTPS2 (H), CAD (I), or PPAT (J) across 33 types of cancer. K, Immunoblots of the indicated proteins in ARPE-19-MYC (DOX+) and ARPE-19-MYC (DOX−) cells. L and M, Immunoblots of the indicated proteins in RKO (L) and HCT116 (M) cells expressing control or MYC shRNA. Graphic data are means ± SEM; n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001; #, P < 0.0001. NS, not significant; ctrl, control; DOX, doxycycline.
In the presence of BAY-1895344, CTPS1 knockout cells showed a significant increase in apoptosis, while no obvious effect was observed in CTPS2 knockout cells (Fig. 6C). Noticeably, the extent of BAY-1895344-induced apoptosis among CTPS1-inhibited cells was comparable with the cells in which both CTPS1 and CTPS2 were inhibited (Fig. 6A–C). To rule out the possible off-target effects of the CRISPR-mediated CTPS1 knockout, we performed a rescue experiment by expressing a Flag-tagged CTPS1 or CTPS2. BAY-1895344-induced CTPS1-depleted RKO cell apoptosis was completely reversed by the expression of either CTPS1 or CTPS2 (Supplementary Fig. S5A and S5B).
We further examined the respective impact of CTPS1 and CTPS2 inhibition on replication stress by analyzing the phosphorylation of Chk1 Ser345. By using the previously established CTPS1 or CTPS2 knockdown and knockout cell lines, we found that the induction of replication stress is specific to CTPS1 activity inhibition. This is supported by the observation that the suppression of CTPS1 resulted in a dramatic increase in Chk1 Ser345 phosphorylation, which is comparable with the inhibition of both CTPS1 and CTPS2, while the inhibition of CTPS2 has no dramatic effects (Fig. 6D–F). Thus, these results indicate that interfering with CTPS1 is required and sufficient to provoke replication stress and subsequent apoptosis when combined with an ATR inhibitor.
To understand the unique function of CTPS1 in MYC-overexpressing cells, we first performed a pan-cancer correlation analysis of MYC, CTPS1, and CTPS2 mRNA expression using the TCGA databases. Pearson r revealed that CTPS1 expression is positively associated with MYC (r = 0.4; Fig. 6G), which is comparable with two other well-known nucleotide metabolism-related MYC target genes CAD and PPAT (r = 0.38 and r = 0.49, respectively; Fig. 6I and J). However, the correlation coefficient of CTPS2 and MYC expression is much lower compared with CTPS1 (r = 0.1 vs. r = 0.4; Fig. 6H). In agreement with the correlation analysis, real-time RT-PCR data showed a higher increase in the mRNA levels of CTPS1 than CTPS2, when MYC was overexpressed (Supplementary Fig. S5C). Furthermore, knockdown of MYC in colon cancer cells RKO and HCT116 showed a dramatic decrease in CTPS1 mRNA levels whereas only a minor change can be observed in CTPS2 mRNA levels (Supplementary Fig. S5D and S5E).
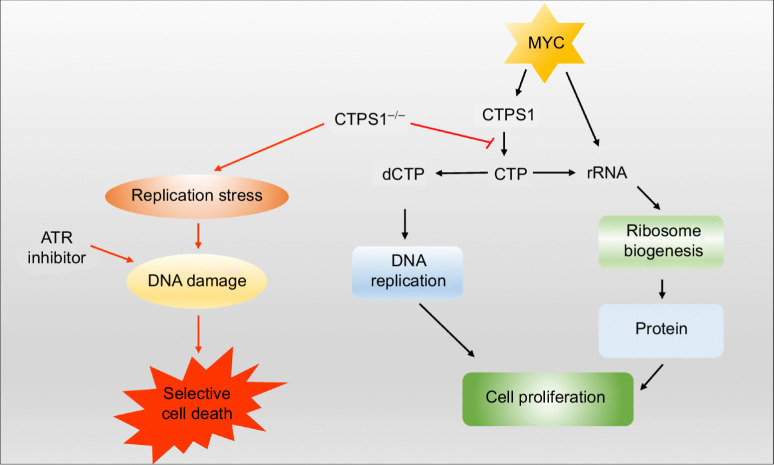
Consistent with the changes in mRNA, overexpression or knockdown of MYC causes a dramatic increase or decrease of CTPS1 protein levels, respectively (Fig. 6K–M). However, only a weak effect was observed on CTPS2 protein levels under the same experimental conditions (Fig. 6K–M), indicating that CTPS1 is the major transcription target of MYC and is the main driver of the increase in the CTP pool in MYC-driven cancer. Meanwhile, by using the TCGA pan-cancer databases, we found that the high CTPS1 expression is significantly associated with poor prognosis in MYC-amplified patients, while the high CTPS2 expression is associated with a good prognosis (Supplementary Fig. S5F and S5G). Together, these results highlight a specific and key role of CTPS1 in MYC-driven cancer by supplying CTP for DNA replication and rRNA synthesis. Inhibiting CTPS1 causes limiting of the CTP pool for DNA replication and replication stress, resulting in selective apoptosis when combined with an ATR inhibitor in MYC-overexpressing cells (Fig. 7).
Figure 7.
Combined inhibition of CTPS1 and ATR induces synthetic lethality in MYC-overexpressing cells. MYC-overexpressing cells are dependent on CTPS1 for DNA replication and rRNA synthesis. Inhibiting CTPS1 induces replication stress and consequent apoptosis when combined with ATR inhibitor in MYC-overexpressing cells.
Discussion
In this study, we show that inhibiting pyridine nucleotide synthesis coupled with sustained activation of ribosome biosynthesis induces an anabolic imbalance that causes nucleotide limiting and selective DNA replication stress in MYC-overexpressing cells. This selective replication stress can be converted into selective cell death when combined with an ATR inhibitor. Thus, we illustrate an alternative strategy to selectively kill cancer cells with elevated MYC activation.
An unexpected finding in our study is the necessity of CTPS1, but not CTPS2, in maintaining the homeostasis of DNA replication and ribosome biosynthesis in MYC-overexpressing cells. We show that, despite the expression of CTPS2, interference with CTPS1 is sufficient to induce DNA replication stress and subsequent cell death when combined with ATR inhibition in MYC-overexpressing cancer cells. These data indicate a unique function of CTPS1 in MYC-driven cancer, but the reason is not fully understood.
In conclusion, here we report an alternative strategy for targeting MYC-driven cancer by inhibiting pyrimidine nucleotide synthesis while maintaining ribosome synthesis activity to create a state of anabolic imbalance. One significant finding in this study is the requirement of CTPS1 in the maintaining of DNA replication and ribosome biosynthesis homeostasis in MYC-overexpressing cells. Because CTPS1 deficiency results in no significant clinical consequences other than immune deficiency in humans, the development of CTPS1-specific inhibitors would be clinically significant. The inhibition of CTPS1 has potential to be highly specific in killing MYC-overexpressing cancer cells when combined with an ATR inhibitor. Our experiments are primarily performed in colon cancer cell lines and in a retinal pigment epithelial cell line. A revalidation in other in vivo MYC-driven malignancies (solid and hematologic malignancy) may be important to see if our finding is generalized to other MYC-driven tumors.
Authors' Disclosures
Z. Sun reports grants from National Science Foundation of China (81972632) during the conduct of the study; in addition, Z. Sun has a patent for “Inhibitor combination and its application in the preparation of drugs for the treatment of MYC high expression cancer” pending. J.-L. Liu reports grants from ShanghaiTech University during the conduct of the study; in addition, J.-L. Liu has a patent for “Inhibitor combination and its application in the preparation of drugs for the treatment of MYC high expression cancer” pending. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors thank the Molecular and Cell Biology Core Facility (MCBCF) and Molecular Imaging Core Facility (MICF) at the School of Life Science and Technology, ShanghaiTech University for providing technical support. They thank the staff members of the Animal Facility at the National Facility for Protein Science in Shanghai (NFPS), Zhangjiang Lab, Shanghai Advanced Research Institute, Chinese Academy of Science, China for providing technical support. This work is supported by ShanghaiTech University and the National Science Foundation of China (81972632). We thank Wei Liu for providing assistance in prognosis assay and Nolan Liu for reading this manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
Z. Sun: Conceptualization, formal analysis, funding acquisition, investigation, writing–original draft, writing–review and editing. Z. Zhang: Investigation. Q.-Q. Wang: Investigation. J.-L. Liu: Conceptualization, supervision, funding acquisition, writing–review and editing.
References
- 1. Dang CV. MYC on the path to cancer. Cell 2012;149:22–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beaulieu ME, Jauset T, Masso-Valles D, Martinez-Martin S, Rahl P, Maltais L, et al. Intrinsic cell-penetrating activity propels Omomyc from proof of concept to viable anti-MYC therapy. Sci Transl Med 2019;11:eaar5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 2011;146:904–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han H, Jain AD, Truica MI, Izquierdo-Ferrer J, Anker JF, Lysy B, et al. Small-molecule MYC inhibitors suppress tumor growth and enhance immunotherapy. Cancer Cell 2019;36:483–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science 2002;297:102–4. [DOI] [PubMed] [Google Scholar]
- 6. Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, et al. Myc deletion rescues Apc deficiency in the small intestine. Nature 2007;446:676–9. [DOI] [PubMed] [Google Scholar]
- 7. Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 2004;431:1112–7. [DOI] [PubMed] [Google Scholar]
- 8. Soucek L, Whitfield J, Martins CP, Finch AJ, Murphy DJ, Sodir NM, et al. Modelling Myc inhibition as a cancer therapy. Nature 2008;455:679–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McKeown MR, Bradner JE. Therapeutic strategies to inhibit MYC. Cold Spring Harb Perspect Med 2014;4:a014266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev 1993;7:671–82. [DOI] [PubMed] [Google Scholar]
- 11. Hurlin PJ. Control of vertebrate development by MYC. Cold Spring Harb Perspect Med 2013;3:a014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell 2003;112:193–205. [DOI] [PubMed] [Google Scholar]
- 13. Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov 2015;5:1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dang CV, Reddy EP, Shokat KM, Soucek L. Drugging the ‘undruggable’ cancer targets. Nat Rev Cancer 2017;17:502–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Riggelen J, Yetil A, Felsher DW. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat Rev Cancer 2010;10:301–9. [DOI] [PubMed] [Google Scholar]
- 16. Cunningham JT, Moreno MV, Lodi A, Ronen SM, Ruggero D. Protein and nucleotide biosynthesis are coupled by a single rate-limiting enzyme, PRPS2, to drive cancer. Cell 2014;157:1088–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu YC, Li F, Handler J, Huang CR, Xiang Y, Neretti N, et al. Global regulation of nucleotide biosynthetic genes by c-Myc. PLoS One 2008;3:e2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mannava S, Grachtchouk V, Wheeler LJ, Im M, Zhuang D, Slavina EG, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle 2008;7:2392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A 2017;114:E7697–E706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hu S, Balakrishnan A, Bok RA, Anderton B, Larson PE, Nelson SJ, et al. 13C-pyruvate imaging reveals alterations in glycolysis that precede c-Myc-induced tumor formation and regression. Cell Metab 2011;14:131–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Le A, Lane AN, Hamaker M, Bose S, Gouw A, Barbi J, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab 2012;15:110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuneva MO, Fan TW, Allen TD, Higashi RM, Ferraris DV, Tsukamoto T, et al. The metabolic profile of tumors depends on both the responsible genetic lesion and tissue type. Cell Metab 2012;15:157–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang R, Dillon CP, Shi LZ, Milasta S, Carter R, Finkelstein D, et al. The transcription factor Myc controls metabolic reprogramming upon T lymphocyte activation. Immunity 2011;35:871–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Levitzki A, Koshland DE Jr. Cytidine triphosphate synthetase. Covalent intermediates and mechanisms of action. Biochemistry 1971;10:3365–71. [DOI] [PubMed] [Google Scholar]
- 25. Higgins MJ, Graves PR, Graves LM. Regulation of human cytidine triphosphate synthetase 1 by glycogen synthase kinase 3. J Biol Chem 2007;282:29493–503. [DOI] [PubMed] [Google Scholar]
- 26. Huang M, Graves LM. De novo synthesis of pyrimidine nucleotides; emerging interfaces with signal transduction pathways. Cell Mol Life Sci 2003;60:321–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kassel KM, Au da R, Higgins MJ, Hines M, Graves LM. Regulation of human cytidine triphosphate synthetase 2 by phosphorylation. J Biol Chem 2010;285:33727–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Traut TW. Physiological concentrations of purines and pyrimidines. Mol Cell Biochem 1994;140:1–22. [DOI] [PubMed] [Google Scholar]
- 29. Williams JC, Kizaki H, Weber G, Morris HP. Increased CTP synthetase activity in cancer cells. Nature 1978;271:71–3. [DOI] [PubMed] [Google Scholar]
- 30. Ellims PH, Gan TE, Medley G. Cytidine triphosphate synthetase activity in lymphoproliferative disorders. Cancer Res 1983;43:1432–5. [PubMed] [Google Scholar]
- 31. Liu JL. Intracellular compartmentation of CTP synthase in Drosophila. J Genet Genomics 2010;37:281–96. [DOI] [PubMed] [Google Scholar]
- 32. Zhou S, Xiang H, Liu JL. CTP synthase forms cytoophidia in archaea. J Genet Genomics 2020;47:213–23. [DOI] [PubMed] [Google Scholar]
- 33. Daumann M, Hickl D, Zimmer D, DeTar RA, Kunz HH, Mohlmann T. Characterization of filament-forming CTP synthases from Arabidopsis thaliana. Plant J 2018;96:316–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gou KM, Chang CC, Shen QJ, Sung LY, Liu JL. CTP synthase forms cytoophidia in the cytoplasm and nucleus. Exp Cell Res 2014;323:242–53. [DOI] [PubMed] [Google Scholar]
- 35. Noree C, Sato BK, Broyer RM, Wilhelm JE. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J Cell Biol 2010;190:541–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol 2010;12:739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu JL. The cytoophidium and its kind: filamentation and compartmentation of metabolic enzymes. Annu Rev Cell Dev Biol 2016;32:349–72. [DOI] [PubMed] [Google Scholar]
- 38. Chang CC, Keppeke GD, Antos CL, Peng M, Andrade LEC, Sung LY, et al. CTPS forms the cytoophidium in zebrafish. Exp Cell Res 2021;405:112684. [DOI] [PubMed] [Google Scholar]
- 39. Keppeke GD, Chang CC, Antos CL, Peng M, Sung LY, Andrade LEC, et al. IMPDH forms the cytoophidium in zebrafish. Dev Biol 2021;478:89–101. [DOI] [PubMed] [Google Scholar]
- 40. Lynch EM, Kollman JM. Coupled structural transitions enable highly cooperative regulation of human CTPS2 filaments. Nat Struct Mol Biol 2020;27:42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou X, Guo CJ, Chang CC, Zhong J, Hu HH, Lu GM, et al. Structural basis for ligand binding modes of CTP synthase. Proc Natl Acad Sci U S A 2021;118:e2026621118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun Z, Liu JL. Forming cytoophidia prolongs the half-life of CTP synthase. Cell Discov 2019;5:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lynch EM, Hicks DR, Shepherd M, Endrizzi JA, Maker A, Hansen JM, et al. Human CTP synthase filament structure reveals the active enzyme conformation. Nat Struct Mol Biol 2017;24:507–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aughey GN, Grice SJ, Liu JL. The interplay between Myc and CTP synthase in drosophila. PLoS Genet 2016;12:e1005867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Isoforms of human CTP synthetase. Adv Exp Med Biol 2000;486:257–61. [DOI] [PubMed] [Google Scholar]
- 46. van Kuilenburg AB, Meinsma R, Vreken P, Waterham HR, van Gennip AH. Identification of a cDNA encoding an isoform of human CTP synthetase. Biochim Biophys Acta 2000;1492:548–52. [DOI] [PubMed] [Google Scholar]
- 47. Martin E, Palmic N, Sanquer S, Lenoir C, Hauck F, Mongellaz C, et al. CTP synthase 1 deficiency in humans reveals its central role in lymphocyte proliferation. Nature 2014;510:288–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Martin E, Minet N, Boschat AC, Sanquer S, Sobrino S, Lenoir C, et al. Impaired lymphocyte function and differentiation in CTPS1-deficient patients result from a hypomorphic homozygous mutation. JCI Insight 2020;5:e133880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sun Z, Liu JL. mTOR-S6K1 pathway mediates cytoophidium assembly. J Genet Genomics 2019;46:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Goga A, Yang D, Tward AD, Morgan DO, Bishop JM. Inhibition of CDK1 as a potential therapy for tumors over-expressing MYC. Nat Med 2007;13:820–7. [DOI] [PubMed] [Google Scholar]
- 51. Yang D, Liu H, Goga A, Kim S, Yuneva M, Bishop JM. Therapeutic potential of a synthetic lethal interaction between the MYC proto-oncogene and inhibition of aurora-B kinase. Proc Natl Acad Sci U S A 2010;107:13836–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Frazier DE Jr, Tarr MJ, Olsen RG. Evaluation of murine lymphocyte membrane potential, intracellular free calcium, and interleukin-2 receptor expression upon exposure to 1,1-dimethylhydrazine. Toxicol Lett 1992;61:27–37. [DOI] [PubMed] [Google Scholar]
- 53. Chang DW, Claassen GF, Hann SR, Cole MD. The c-Myc transactivation domain is a direct modulator of apoptotic versus proliferative signals. Mol Cell Biol 2000;20:4309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Graves JA, Rothermund K, Wang T, Qian W, Van Houten B, Prochownik EV. Point mutations in c-Myc uncouple neoplastic transformation from multiple other phenotypes in rat fibroblasts. PLoS One 2010;5:e13717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hemann MT, Bric A, Teruya-Feldstein J, Herbst A, Nilsson JA, Cordon-Cardo C, et al. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature 2005;436:807–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.