Quantitative assessment of the functional consequences of CHEK2 variants of uncertain significance identifies damaging variants associated with increased cancer risk, which may aid in the clinical management of patients and carriers.
Abstract
Heterozygous carriers of germline loss-of-function variants in the tumor suppressor gene checkpoint kinase 2 (CHEK2) are at an increased risk for developing breast and other cancers. While truncating variants in CHEK2 are known to be pathogenic, the interpretation of missense variants of uncertain significance (VUS) is challenging. Consequently, many VUS remain unclassified both functionally and clinically. Here we describe a mouse embryonic stem (mES) cell–based system to quantitatively determine the functional impact of 50 missense VUS in human CHEK2. By assessing the activity of human CHK2 to phosphorylate one of its main targets, Kap1, in Chek2 knockout mES cells, 31 missense VUS in CHEK2 were found to impair protein function to a similar extent as truncating variants, while 9 CHEK2 missense VUS resulted in intermediate functional defects. Mechanistically, most VUS impaired CHK2 kinase function by causing protein instability or by impairing activation through (auto)phosphorylation. Quantitative results showed that the degree of CHK2 kinase dysfunction correlates with an increased risk for breast cancer. Both damaging CHEK2 variants as a group [OR 2.23; 95% confidence interval (CI), 1.62–3.07; P < 0.0001] and intermediate variants (OR 1.63; 95% CI, 1.21–2.20; P = 0.0014) were associated with an increased breast cancer risk, while functional variants did not show this association (OR 1.13; 95% CI, 0.87–1.46; P = 0.378). Finally, a damaging VUS in CHEK2, c.486A>G/p.D162G, was also identified, which cosegregated with familial prostate cancer. Altogether, these functional assays efficiently and reliably identified VUS in CHEK2 that associate with cancer.
Significance:
Quantitative assessment of the functional consequences of CHEK2 variants of uncertain significance identifies damaging variants associated with increased cancer risk, which may aid in the clinical management of patients and carriers.
Introduction
The importance of genome stability for preventing breast and other cancers is evident from the increased cancer risk that results from inherited loss-of-function (LOF) variants in DNA damage repair genes such as BRCA1/2 and PALB2, as well as in genes that control genome integrity checkpoints. The checkpoint kinase 2 (CHEK2) gene is a well-known example, which encodes the serine-threonine kinase protein CHK2 that becomes activated in response to DNA damage, and regulates cell-cycle progression and apoptosis (1, 2). The CHK2 protein is therefore believed to act as a tumor suppressor by delaying cell-cycle progression to allow time for DNA repair, or by eliminating genomically unstable cells through induction of cell death (3). In 2002, association analysis of the truncating CHEK2 c.1100delC/p.T367Mfs variant indeed revealed that it confers a moderate risk of breast cancer (4, 5). Meanwhile, other studies have also shown that carriers of such LOF variants in the CHEK2 gene are at a significantly increased risk for developing breast cancer (OR ∼2.5; refs. 6, 7), as well as several other cancers such as prostate cancer (8–10). These studies firmly established that CHEK2 is a low to moderate penetrance cancer susceptibility gene.
The growing body of evidence that associates CHEK2 with breast cancer has led to increased genetic testing of CHEK2, and as a consequence to the identification of more (rare) genetic variants in this gene for which clinical significance is unknown (11–15). In fact, 1,332 variants of uncertain significance (VUS) in CHEK2 have currently been reported in ClinVar (as of October 2021; ref. 16), most of which (i.e., 1,139) are missense variants. For many of these missense variants, the impact on protein function and the associated cancer risk remain to be elucidated. Assessment of pathogenicity of these VUS in a moderate risk gene such as CHEK2 is mostly dependent on family history of cancer. To overcome this limitation, quantitative methods are required that can determine the functional impact of VUS in CHEK2 and establish their relationship with cancer risk.
The CHK2 protein, which is expressed throughout the cell cycle, consists of 543 amino acids, and possesses three characteristic domains: an N-terminal SQ/TQ cluster domain (residues 19–69), a fork head-associated (FHA) domain (residues 92–205), and a serine/threonine kinase domain (residues 212–501). A nuclear localization signal (NLS) is located at the C‐terminus of CHK2 (residues 515–522; ref. 17). Activation of CHK2 kinase activity occurs specifically in response to DNA damage and is a multistep process initiated by ataxia telangiectasia mutated (ATM)-mediated phosphorylation of several SQ/TQ sites, particularly p.T68, in its N-terminal regulatory domain (1, 2). This promotes homodimerization and intermolecular autophosphorylation of CHK2 on p.T383 and p.T387 within the T-loop region (residues 366–406; ref. 18), and on p.S516 within the NLS, collectively leading to efficient kinase activation and the subsequent phosphorylation of target proteins (19, 20). The spectrum of known CHK2 targets includes proteins involved in cell-cycle control (i.e., CDC25A and CDC25C phosphatases), regulation of cell death (i.e., p53; refs. 1, 2, 21), and DNA damage repair (i.e., BRCA1 and KAP1; refs. 22–24). Following DNA damage, CHK2 phosphorylates KAP1 specifically at p.S473. This modification attenuates KAP1 binding to heterochromatin protein 1 family proteins, leading to relaxation of the damaged heterochromatin and promoting DNA damage repair (24–28).
In an effort to interpret CHEK2 VUS, several studies assessed their functional consequences (29–36). The largest set of CHEK2 variants to date was analyzed by Delimitsou and colleagues (34). They employed a yeast-based functional assay that assesses the ability of yeast strains expressing different CHEK2 variants to resume proliferation and cell growth following repair of DNA damage induced by methyl methanesulfonate (31, 32). Other recent studies also assessed the impact of CHK2 variants on the phosphorylation of downstream targets such as CDC25C, BRCA1, and KAP1 (29, 30, 35). Although these studies have assayed >130 patient-derived CHEK2 variants and identified numerous damaging missense variants, results were often discordant and the relationship with risk of breast and other cancers remained unclear. Consequently, there is a need to further improve the functional analysis of missense variants in CHEK2, and develop assays that can link the functional impact of such variants to cancer risk.
Here, we developed a mouse embryonic stem (mES) cell-based assay for the functional analysis of VUS in CHEK2. The assay allows a semi high-throughput analysis of variants in human CHEK2 cDNA in Chek2 knockout (KO) mES cells, using CHK2-mediated Kap1 p.S473 phosphorylation as a quantitative readout. Using this approach, we identified 31 CHEK2 missense VUS to impair protein function to a similar extent as CHEK2 truncating variants, while nine missense VUS showed intermediate functional defects. Our results further indicate that at least two mechanisms are at play by which VUS in CHEK2 impair protein function: loss of protein stability and defective (auto)phosphorylation/activation. Importantly, the degree of CHK2 kinase dysfunction observed for CHEK2 missense variants highly correlates with increased breast cancer risk.
Materials and Methods
Cell lines and cell culture conditions
The 129/Ola E14 IB10 mES cells (37) were cultured on gelatine-coated dishes in 50% 2i ES medium, of which, 500 mL contains (i) 250 mL KO DMEM (Gibco, 21710-025) supplemented with 2.5 mL 100 mmol/L sodium pyruvate (Gibco, 11360–039), 2.5 mL 100× non-essential amino acids (Gibco, 11140-035), and 25 mL FCS; (ii) 125 mL DMEM/F2 HEPES supplemented with 1.25 mL 100× N2 Supplement (Gibco, 17502-048), 85 μL 7.5% BSA (Gibco # 15260-037) and 500 μL 0.1 mol/L β-MeOH; and (iii) 125 mL NEUROBASAL medium (Gibco, 21103-049) supplemented with 2.5 mL 50× B27 Supplement (Gibco # 17504-044), 1.25 mL 200 mmol/L l-glutamine (Gibco 25030-024), and 500 μL 0.1 mol/L β-MeOH. The total 500 mL is supplemented with 5 mL 5,000 units/mL penicillin/streptomycin (Gibco 15070063), 5 mL 105 units/mL LIF (Millipore ESG1107), 250 μL 0.1 mol/L β-MeOH, 250 μL 3 mmol/L CHIR (Axon Medchem 1386), and 250 μL 1 mmol/L PD (Axon Medchem 1408).
Generation of Chek2KO mES cells with DR-GFP and recombination-mediated cassette exchange
mES cells carrying the DR-GFP reporter and recombination-mediated cassette exchange (RMCE) system at the Pim1 and Rosa26 locus, respectively, were generated previously (38). Using these mES cells, Chek2KO cells were generated by transfecting1 μg of pSpCas9(BB)-2A-GFP (pX458; ref. 39), encoding Cas9, GFP and a gRNA that targets exon 3 of mouse Chek2 (5′-ACTGTGTTAACGACAACTAC-3′). GFP-positive cells were FACS-sorted and seeded. Individual clones were examined by TIDE (https://tide.nki.nl) and Western blot analysis for loss of Chk2 expression.
Selection of human CHEK2 variants
Seven previously reported CHEK2 truncating variants were included as negative controls (16, 40). Six synonymous variants, which have not yet been observed in carriers were selected on the basis of their position throughout the CHEK2 protein and were included as positive controls. Truncating and missense CHEK2 VUS were selected on the basis of one or more of the following criteria: (i) identification in the case–control association study performed by the BRIDGES consortium in collaboration with the Breast Cancer Association Consortium (BCAC; ref. 7) or prostate cancer family members reported in this study, (ii) clinical classification in ClinVar (16), (iii) position in the CHK2 protein sequence, (iv) computational predictions from Helix, and (v) presence/absence in previous functional studies (34, 35).
Cloning and generation of human CHEK2 variants
Vector pBudCE4.1 (Thermo Fisher Scientific, V53220) was modified by adding two PacI restriction sites as described previously (38). Human HA-tagged CHEK2 cDNA (NM_007194.4) was subcloned from pBabe-HA-CHK2 (41) using the BsrGI and XhoI restriction sites into pBudCE4.1-PacI using the BsrGI-compatible Acc65I restriction site and XhoI restriction site. pBabe-HA-CHK2 was a gift from Stephen Elledge (Addgene plasmid #41901). An Ef1α-CHEK2-containing fragment from pBudCE4.1-PacI-CHEK2 was then cloned into the RMCE vector (pRNA 251-MCS RMCE; TaconicArtemis GmbH) using the PacI restriction sites in both vectors. CHEK2 variants were introduced by site-directed mutagenesis (SDM) using the Quick-Change Lightning protocol (Agilent Technologies). All SDM primers are shown in Supplementary Table S3. Constructs were verified by Sanger sequencing and used for mES cell-based assays.
The RMCE vector carrying EGFP-CHEK2-T2A-mCherry was generated as follows. The RMCE vector carrying CHEK2 was digested with EcoRI. EGFP was PCR amplified from an EGFP-carrying construct (pcDNA-FRT-TO-puro-EGFP) with the following primers: forward primer 5′-CCCAGTGTGGTGGTACGTAGATGGTGAGCAAGGGCGAGG-3′ and reverse primer 5′-TATGGGTAAGCCATGAATTCCTTGTACAGCTCGTCCATGCCG-3′. Gibson assembly was then performed to generate the RMCE vector carrying EGFP-CHEK2. Next, three different fragments were PCR amplified: CHEK2 (forward primer 5′-ACGAGCTGTACAAGGAATTCATGTCTCGGGAGTCGGATGT-3′ and reverse primer 5′-AGCAGACTTCCTCTGCCCTCCAACACAGCAGCACACACAGC-3′) and the hGH sequence (forward primer 5′-TGGACGAGCTGTACAAGTGAACTCCGTGGTTTGAACACTCTAG-3′ and reverse primer 5′-GCATAACTAGTGTCACGCGTCATATGGCCGGCCTATTTAAATAAGC-3′) from the RMCE vector carrying EGFP-CHEK2, and T2A-mCherry (forward primer 5′-GAGGGCAGAGGAAGTCTGCTAAC-3′ and reverse primer 5′-TCACTTGTACAGCTCGTCCATGC-3′) from a T2A-mCherry carrying construct (pX459-Cas9-T2A-mCherry). The RMCE vector carrying EGFP-CHEK2 was then digested with EcoRI and MluI, after which, the plasmid backbone (lacking CHEK2) was gel extracted. By employing Gibson assembly, the three PCR fragments were cloned into the backbone to generate the RMCE vector carrying EGFP-CHEK2-T2A-mCherry. The construct was verified by Sanger sequencing and used to generate CHEK2 variants and perform mES cell-based assays.
Western blot analysis
A total of 2 × 106Chek2KO mES cells carrying the DR-GFP reporter and RMCE system were subjected to RMCE by cotransfecting 1 μg FlpO expression vector (pCAGGs-FlpO-IRES-puro; ref. 42) with 1 μg RMCE exchange vector. Neomycin-resistant cells from approximately 500 resistant clones were pooled and expanded as described previously (38). For various conditions, protein levels for mouse Chk2, human CHK2, human phospho-CHK2 p.T383, mouse Kap1, mouse phospho-Kap1 p.S473, mouse p53, mouse p21, and mouse tubulin were examined by protein extraction and Western blot analysis. Briefly, samples were generated by taking up approximately 1.5 × 106 cells in 75 μL Laemmli buffer and boiling them at 95°C for 5 minutes. Samples were incubated with 0.2 μL benzonase (Merck Millipore 70746, 250 U/μL) for 20 minutes at room temperature and then loaded for gel electrophoresis followed by immunoblotting. Primary antibodies used were: mouse mAb against mouse/human CHK2 (1:1,000; BD Biosciences 611571), rabbit polyclonal antibody against mouse/human phospho-CHK2 p.T383 (1:1,500; Abcam 59408), rabbit polyclonal antibody against mouse/human Kap1 (1:10,000; Abcam 10484), mouse mAb against mouse/human phospho-Kap1 p.S473 (1:2,000; BioLegend 654102), mouse mAb against mouse/human p53 (1:1,000; Cell Signaling Technology 2524), rabbit polyclonal antibody against mouse/human p21 (Cdkn1a) (1:800; Santa Cruz Biotechnology sc-397), and mouse mAb against α-tubulin (1:5,000, Sigma, T6199). Peroxidase-AffiniPure goat polyclonal anti-rabbit (1:5,000; Jackson laboratories 111-035-003) and affinity isolated goat polyclonal anti-mouse (1:5,000; Dako P0447) were used as secondary antibodies. SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific 34095) and ECL Prime Western Blotting Detection Reagents (Merck RPN2232) were used for development of blots on the Amersham Imager 600 (GE Healthcare Life Sciences).
Homologous recombination reporter assays
A total of 1–2 × 106Chek2KO mES cells carrying the DR-GFP reporter and RMCE system were subjected to homologous recombination (HR) assays by transfecting 1 μg of plasmid that coexpresses I-SceI and mCherry (pCMV-Red-Isce, kind gift from Jos Jonkers) using Lipofectamine 2000 (Thermo Fisher Scientific; ref. 43). A cotransfection of 1 μg pCAGGs (44) with 0.05 μg of an mCherry expression vector was included as control. Two days after transfection, mCherry/GFP double-positive cells were scored using a Novocyte Flow Cytometer (ACEA Biosciences, Inc.).
Phleomycin sensitivity assays
For proliferation-based phleomycin sensitivity assays, mES cells were seeded in triplicate at 10,000 cells per well of a 96-well plate. The next day, cells were treated with phleomycin (InvivoGen ant-ph-2p) for 2 days, after which, the medium was refreshed, and cells were cultured for one more day in drug-free medium. Viable cells were subsequently counted using the Novocyte Flow Cytometer (ACEA Biosciences, Inc.).
qRT-PCR analysis
RNA was isolated from mES cells grown on 6-well plates using TRIzol (Thermo Fisher Scientific 15596026) as per the manufacturer's protocol. For each condition, 3 μg RNA was treated with RQ1 RNase-free DNase (Promega M6101) and cDNA was synthesized from 0.2 μg DNase-treated RNA using hexamer primers (Thermo Fisher Scientific N8080127) and SuperScript IV Reverse Transcriptase (Thermo Fisher Scientific 18090050) as per the manufacturer's protocols. qRT-PCRs were carried out using GoTaq qPCR Master mix (Promega A6002), a CFX384 Real-Time System (Bio-Rad) and the following qPCR primers directed at the mouse Mdm2, p21 (Cdkn1a), or the mouse control gene Pim1: Mdm2-exon11-Fw 5′-GTCTATCAGACAGGAGAAAGCGATACAG-3′, Mdm2-exon12-Rv 5′-GTCCAGCATCTTTTGCAGTGTGATGGAAG-3′. p21-exon2-Fw 5′-GCTGTCTTGCACTCTGGTGTCTGAG-3′, p21-exon3-Rv 5′-GACCAATCTGCGCTTGGAGTGATAG-3′. Pim1-exon4-Fw 5′-GCGGCGAAATCAAACTCATCGAC-3′ and Pim1-exon5-Rv 5′- GTAGCGATGGTAGCGAATCCACTCTGG-3′.
Flow cytometry (FACS) analysis
As for Western blot analysis, Chek2KO mES cells expressing human CHEK2 variants were generated and expanded. For phospho-Kap1 p.S473 FACS-based assays, 1 × 106 mES cells were seeded on 60 mm dishes 1 day prior to exposure to 10 Gy of ionizing radiation (IR). Two, 4, or 6 hours after IR, cells were trypsinized and fixed in 5 mL 2% formaldehyde for 15 minutes. A volume of 2 mL 0.125 mol/L glycine was added and cells were centrifuged for 5 minutes at 1,500 rpm. Cells were then washed in PBS and fixed for a second time in 100% ice-cold methanol and incubated overnight at −20°C. After washing once in PBS, fixed cells were permeabilized for 15 minutes using 0.25% Triton X-100 in PBS, after which, cells were stained in 200 μL PBS+ (5 g/L BSA, 1.5 g/L glycine) with 1 μL mouse anti-phospho-Kap1 p.S473 (0.5 μg/μL, BioLegend 654102) for 3 hours at room temperature, with gentle resuspension every 30 minutes. Alexa-488 goat anti-mouse (1:200 in 200 μL PBS; Thermo Fisher Scientific A-21424) was used as a secondary antibody, followed by a propidium iodide (PI) staining (25 μg/mL PI, RNaseA 0,1 mg/mL, 0.05% Triton X-100). Phospho-Kap1 p.S473 intensity was analyzed using the Novocyte Flow Cytometer (ACEA Biosciences, Inc.). For FACS-based assays with mES cells expressing EGFP-CHEK2-T2A-mCherry, phospho-Kap1 p.S473 was stained with alexa-647 goat anti-mouse (1:200 in 200 μL PBS; Thermo Fisher Scientific A-21235), PI staining was not performed and phospho-Kap1 p.S473 intensity was measured after gating for mCherry- or GFP-positive cells using a Fortessa1 (BD Biosciences).
Exome sequencing in prostate cancer family members
Three brothers were diagnosed with prostate cancer >10 years earlier than the average age of onset of sporadic prostate cancer, suggesting that they might be carriers of a germline mutation responsible for predisposition to this type of cancer. Copy-number variations (deletions or amplifications) in blood cell DNA from these three brothers and their four sons were not detected using the SNP6 microarray (Affymetrix). The Agilent SureSelect Human All Exon V5+UTRs protocol was used to carry out targeted enrichment of all exonic sequences from the total DNA material for each sample. Paired-end Illumina sequencing with 100 cycles was performed to minimize the ambiguities of read alignment to the reference genome. Two sequencing lanes resulted in an average of 20 million fragments per sample. All sequence fragments were aligned to the reference human genome (version hg19) using BWA mem (v. 0.7.10), after quality and TruSeq adapter trimming using Cutadapt (v.1.5). Sam files were manipulated using Samtools (v.1.1) and Picard tools (v.1.119) were used to run quality metrics (insert size, hybridization quality) and mark PCR duplicates. VerifyBamID (v. 1.1) was used to estimate contamination. Samples were genotyped and variants jointly called using GATK (v. 3.5). For this purpose, padded targeted intervals were created on the basis of Agilent targets. Annotation was performed using wAnnovar, Oncotator (v.1.8), and WGSA (Amazon EC2 cloud, AWS community instance: WGSA055-ubuntu-800G). Transcript annotation was taken from the Oncotator pipeline using the transcript list giving priority to known clinical protein changes (list downloaded in Feb 2016). GENCODE (Version 19 - July 2013 freeze, GRCh37 - Ensembl 74) was used as a reference transcript set. Unfiltered variants were jointly called over all samples. Filtering was performed on the basis of genotyping quality. All variants that did not have a minimum read depth of 8 and genotype quality of 20 in all affected family members were removed. Finally, all variants with minor allele frequency (MAF) >1% (based on ExAc European non-Finnish cohort, annotation from WGSA) were excluded. Variants classified as pathogenic by ClinVar were not discarded even if MAF was >1%. Analysis of the remaining variants showed that all three affected brothers, as well as one of their sons, carried the CHEK2 allele rs587781652 harbouring the c.485A>G/p.D162G VUS.
Loss of heterozygosity assessment
Tumor DNA was isolated from formalin-fixed paraffin-embedded (FFPE) tissue blocks either by taking three 0.6 mm tumor cores or by microdissection of tumor areas with at least 70% tumor cells (10 mm slides). Fully automated DNA isolation was performed using the Tissue Preparation System (Siemens Healthcare Diagnostics) as described previously (45). The Qubit dsDNA HS Assay Kit was used for DNA quantification according to the manufacturer's protocol (Qubit 2.0 Fluorometer, Invitrogen, catalog no. Q32851). Next-generation sequencing was performed using 40 ng of tumor DNA per sample isolated from FFPE tissue blocks. The custom Ampliseq HDR15v1-panel (Thermo Fisher Scientific) was used for variant detection in CHEK2. Loss of heterozygosity (LOH) of CHEK2 was determined by comparing the variant allele frequency (VAF) of heterozygous c.485A>G/p.D162G and c.1100delC/p.T367Mfs in tumor and normal tissue as described previously (45). LOH was considered present when the tumor cell percentage was >20% and the germline CHEK2 variant allele frequency was >0.6. LOH was considered inconclusive when the tumor cell percentage was <20% or considered absent when the germline CHEK2 variant VAF was <0.6.
Ethics declaration
Individuals of the prostate cancer family were identified and evaluated at the University Hospital Zurich (Zurich, Switzerland). The study protocol was approved by the hospital's research ethics committee, and donors provided written consent to tissue collection, testing, and data publication. LOH assessment was performed at Leiden University Medical Center (Leiden, the Netherlands) under protocols approved by hospital's local ethics committee.
Results
A cell-based functional assay for CHEK2 variants
To assess the functional impact of CHEK2 variants, we developed a mES cell-based system that allows for the semi high-throughput testing of variants in human CHEK2. To this end, we employed our mES cells carrying the well-established DR-GFP reporter for homologous recombination (HR) at the Pim1 locus, and the Recombination-Mediated Cassette Exchange (RMCE) system at the Rosa26 locus (38, 43). CRISPR/Cas9-mediated genome editing was used to knockout (KO) mouse Chek2 in these cells (Fig. 1A; Supplementary Fig. S1A–S1C; refs. 38, 43). Given that BRCA1, a crucial player in HR, becomes phosphorylated by CHK2, and given that this event promotes the dispersion of BRCA1 from DNA breaks (46), we assessed whether KO of Chek2 affects the efficiency of HR in the DR-GFP reporter. Analysis of one heterozygous and two homozygous Chek2KO clones revealed that HR remained unaffected in these cells (Supplementary Fig. S1D), suggesting that loss of Chek2 does not affect HR.
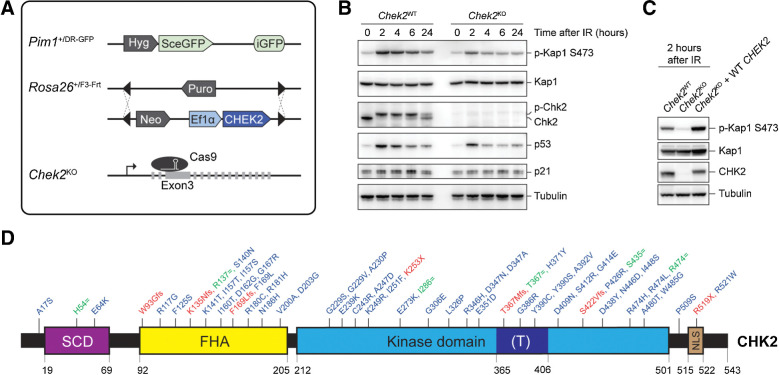
Figure 1.
Generation of a cDNA-based complementation system for the functional analysis of human CHEK2 variants. A, Schematic representation of the mES cell– and cDNA-based complementation system for functional analysis. The DR-GFP reporter and RMCE have been stably integrated at the Pim1 and Rosa26 loci, respectively. Endogenous mouse Chek2 was targeted with CRISPR/Cas9 using a gRNA against exon 3. B, Western blot analysis of the indicated proteins from unirradiated and IR-exposed (10 Gy) Chek2WT and Chek2KO mES cells. Tubulin was used as a loading control. C, Western blot analysis of the indicated proteins from IR-exposed (10 Gy) Chek2WT, Chek2KO, and Chek2KO mES cells complemented with human CHEK2 cDNA. Tubulin was used as a loading control. D, Schematic representation of the CHK2 protein, with variant positions indicated and categorized as either synonymous (green), truncating (red), and missense VUS (blue). The amino acid numbers are shown to demarcate CHK2's evolutionarily conserved functional domains. (T) refers to the T-loop or activation segment.
CHK2 is known for its role in p53-mediated cell-cycle control and apoptosis, as well as DNA damage repair in heterochromatin (1, 2, 21–24). Although we did not detect major changes in the cell-cycle profile of Chek2KO cells when compared with wild-type cells (Supplementary Fig. S1E), we did observe a slight, though not significant growth advantage for the Chek2KO cells over the course of 5 days (Supplementary Fig. S1F). In agreement with previous studies (47, 48), this growth advantage became more pronounced after DNA break induction by the radiomimetic agent phleomycin (Supplementary Fig. S1G). Moreover, p53 protein levels were moderately reduced in these cells after exposure to IR (10 Gy; Fig. 1B). Accordingly, the expression of p53 target genes was also reduced, as evidenced by reduced p21 and Mdm2 transcript and p21 protein levels (Fig. 1B; Supplementary Fig. S1H). Most evidently, however, we observed that Kap1 phosphorylation at p.S473, which is required for DNA repair in heterochromatin (24), was strongly impaired in Chek2KO cells after IR (Fig. 1B).
We decided to exploit the strong impact of Chek2 loss on Kap1 p.S473 phosphorylation as a readout for the functional analysis of human CHEK2 variants. To this end, we stably integrated human wild-type CHEK2 cDNA by RMCE in Chek2KO mES cells (Fig. 1A). Prior to examining CHK2 kinase activity, we pooled all the neomycin-resistant clones with stably integrated CHEK2 cDNA (Fig. 1A), to average out any clonal variability in CHEK2 expression. We found that the defect in IR-induced Kap1 p.S473 phosphorylation in Chek2KO cells was efficiently rescued following expression of human CHEK2 (Fig. 1C). Strikingly, human CHK2 appeared to phosphorylate mouse Kap1 even more efficiently when compared with endogenous mouse Chk2, while their expression levels were comparable (Fig. 1C). Thus, we established a cDNA-based complementation system for the functional analysis of human CHEK2 genetic variants using Kap1 p.S473 phosphorylation as a readout.
Validation of a cell-based functional assay for CHEK2 variants
To validate our system, we selected seven truncating and six synonymous CHEK2 variants for functional analysis (Fig. 1D). Sequence-verified constructs were introduced by RMCE into the Chek2KO mES cells and their ability to phosphorylate Kap1 at p.S473 after IR was assessed by Western blot analysis. As expected, in Chek2KO cells complemented with an empty vector or a truncating CHEK2 variant, phosphorylation of Kap1 p.S473 was strongly impaired at both 2 and 6 hours after IR (Fig. 2). The exception to this was the nonsense variant p.R519X, which moderately impacted Kap1 p.S473 phosphorylation at 2 hours after IR (Fig. 2), even though it was classified as likely pathogenic in ClinVar. p.R519X leads to a truncated CHK2 protein that lacks part of its NLS (Fig. 1D; amino acids 515–522). Possibly, residual nuclear localization of this variant is sufficient to induce partial Kap1 p.S473 phosphorylation after IR, suggesting it acts as a hypomorphic variant. In contrast to truncating CHEK2 variants, cells that expressed synonymous variants showed phospho-Kap1 p.S473 levels comparable with cells expressing wild-type CHEK2 (Fig. 2). Neither the expression of different CHEK2 variants, nor the exposure to IR affected overall Kap1 protein levels, suggesting that CHK2 activity does not affect Kap1 stability or expression (Fig. 2).
Figure 2.
Human CHEK2 variants and their effect on CHK2 expression and kinase activity toward Kap1 p.S473. Western blot analysis of the indicated proteins from Chek2KO mES cells expressing wild-type (WT; black) human untagged CHK2, empty vector (Ev; gray), or the indicated untagged CHK2 variants in untreated conditions (no IR) or at 2 or 6 hours after IR exposure (10 Gy). WT and Ev served as controls on each blot and variants are categorized by color as either synonymous (green), truncating (red), and missense VUS (blue). Tubulin was used as a loading control. Dashed lines represent a marking of different set of samples on the same blot, whereas continuous lines are used to mark different sets of samples from distinct and separately exposed blots.
A quantitative cell–based functional assay for CHEK2 variants
Complementary to the Western blot analysis, which is at best semiquantitative in our setup, we next aimed for a more quantitative approach. To this end, we used FACS to determine the levels of phospho-Kap1 p.S473. Consistent with results from Western blot analysis (Fig. 1C), we observed a strong reduction in the phospho-Kap1 p.S473 signals in Chek2KO cells 2 hours after IR (Fig. 3A). Surprisingly, we also observed substantial Kap1 p.S473 phosphorylation in unirradiated Chek2KO cells, albeit this was most likely restricted to M-phase cells and disappeared after IR exposure (Supplementary Fig. S2). Complementation of Chek2KO cells with wild-type human CHEK2 cDNA rescued the defect in IR-induced Kap1 p.S473 phosphorylation and even led to higher phospho-Kap1 p.S473 signals when compared with that in Chek2 wild-type cells (Fig. 3A). This effect was also seen for the six synonymous CHEK2 variants (Fig. 3B; Supplementary Figs. S3 and S4A). In contrast, complementation with the empty vector or the truncating variants (except the hypomorphic variant p.R519X), resulted in a complete absence of cells that were positive for phospho-Kap1 p.S473 (Fig. 3B; Supplementary Figs. S3 and S4A). Thus, the quantitative results obtained using a FACS-based approach fully corroborated the results obtained by Western blot analysis.
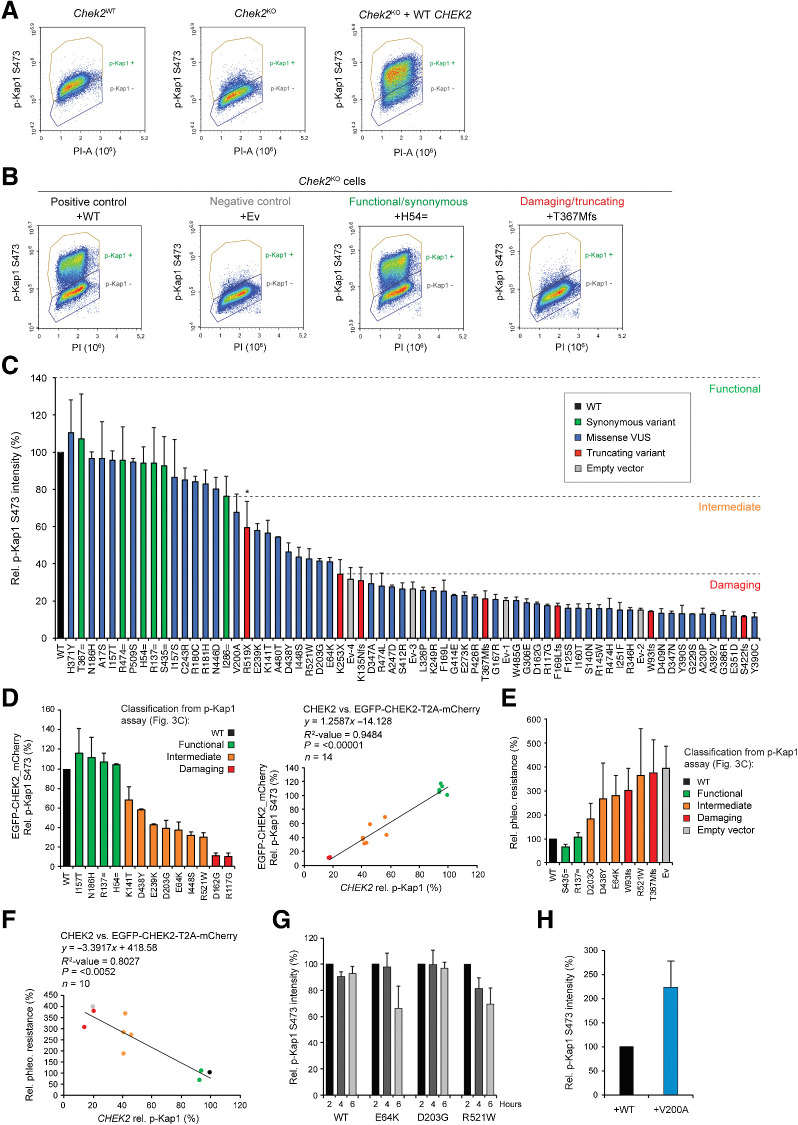
Figure 3.
Human CHEK2 variants and their effect on CHK2's kinase activity toward Kap1 p.S473. A, Quantitative FACS-based analysis of Kap1 p.S473 phosphorylation in Chek2WT, Chek2KO, and Chek2KO mES cells complemented with human untagged CHEK2 cDNA at 2 hours after IR exposure (10 Gy). B, Quantitative FACS-based analysis of Kap1 p.S473 phosphorylation in Chek2KO mES cells complemented with the indicated untagged constructs at 2 hours after IR exposure (10 Gy). C, Quantification of FACS measurements of Kap1 p.S473 phosphorylation in Chek2KO mES cells expressing wild-type (WT; black) human untagged CHK2, empty vector (Ev; gray), or the indicated untagged CHK2 variants (green, red, and blue) at 2 hours after IR exposure (10 Gy). Data represent mean percentages ± SEM of the average phospho-Kap1 p.S473 intensity observed in the “p-Kap1 +” gate as shown in B from two independent experiments. Data are relative to WT, which was set to 100%. Ev1–4 refer to four independent Ev controls that were included. Dashed lines indicate functional thresholds based on the synonymous or truncating variant with the lowest or highest Kap1 p.S473 phosphorylation level, respectively. The asterisk marks p.R519X, which acted as a hypomorphic variant and was therefore not used for thresholding. D, Quantification of FACS measurements (left) of Kap1 p.S473 phosphorylation in Chek2KO mES cells complemented with EGFP-CHEK2-T2A-mCherry, with or without a CHEK2 variant, at 2 hours after IR exposure (10 Gy). Data represent mean percentages ± SEM of the average phospho-Kap1 p.S473 intensity observed after gating for mCherry-positive cells from two independent experiments. Data are relative to WT, which was set to 100%. Scatter plot (right) shows the correlation between phospho-Kap1 p.S473 intensities measured in Chek2KO mES cells expressing untagged CHEK2 or EGFP-tagged CHEK2 (from stably integrated EGFP-CHEK2-T2A-mCherry). Conditions are colored as indicated on the basis of functional classification using untagged CHEK2 cDNA as shown in C. E, Phleomycin sensitivity assay using Chek2KO mES cells complemented with the indicated untagged CHK2 constructs or empty vector. Cells were exposed to 2.5 μmol/L of phleomycin for 2 days. Cell viability was measured after one additional day of incubation in drug-free medium using FACS (using only forward and sideways scatter). Data represent the mean percentage ± SEM of viability relative to untreated cells from three independent experiments. F, Scatter plot showing the correlation between phospho-Kap1 p.S473 intensities and the relative resistance to 2.5 μmol/L phleomycin as measured in E in Chek2KO mES cells expressing untagged CHK2 variants. G, Quantification of FACS measurements of Kap1 p.S473 phosphorylation in Chek2KO mES cells expressing wild-type untagged CHEK2 or three selected variants at the indicated times after 10 Gy of IR. For each condition, data are plotted relative to the 2 hours time point, which was set to 100%. H, Quantification of FACS measurements of Kap1 p.S473 phosphorylation in Chek2KO mES cells expressing WT (black) untagged CHK2, or untagged CHK2 carrying the p.V200A variant (blue) at 2 hours after IR exposure (10 Gy). Data from two independent experiments are represented as in C.
Notably, our FACS-based analysis showed a large population of cells that is negative for phospho-Kap1 p.S473, even after expression of wild-type CHEK2 or a synonymous variant (Fig. 3A and B; Supplementary Figs. S3 and S4A). Stable introduction of a construct that carries a T2A sequence for coexpression of EGFP-CHEK2 and mCherry (EGFP-CHEK2-T2A-mCherry) showed that there is both a GFP/mCherry-positive as well as GFP/mCherry-negative population of cells (Supplementary Fig. S5A). These data suggest that a large portion of cells lose CHEK2 expression after stable integration. Importantly, the GFP/mCherry-negative population of cells was clearly phospho-Kap1 p.S473-negative, even following exposure to IR (Supplementary Fig. S5A). We therefore excluded this population from our analysis and quantified the mean intensity of phospho-Kap1 p.S473 (Fig. 3C) only for cells that were positively gated for phospho-Kap1 p.S473 (Fig. 3A and B). As expected, this showed that synonymous variants exhibited kinase activity comparable to that of wild-type CHK2 (i.e., a reduction of <24%), whereas the truncating CHEK2 variants (except the hypomorphic variant p.R519X) caused a major reduction in kinase activity of >69%. Thus, our cell-based system can classify functional/synonymous and damaging/truncating CHEK2 variants based on their effect on Kap1 p.S473 phosphorylation.
Functional analysis of CHEK2 missense VUS
Having established a quantitative cell-based functional assay for CHEK2 variants, we next examined the effect of 50 missense VUS. The majority of these VUS were identified using a multigene panel analysis of a large case–control association study performed by the BRIDGES consortium and BCAC (7). Importantly, for all 50 missense VUS, the contribution with respect to cancer risk is largely unclear and insights into their functionality may aid in their clinical classification. Following their expression in Chek2KO cells using the non-tagged CHEK2 cDNA, we found that 31 VUS strongly impaired CHK2 kinase activity toward Kap1 p.S473, comparable with that observed for CHEK2 truncating variants and the empty vector conditions (Fig. 3C; Supplementary Figs. S3 and S4A). Importantly, p.R519X was not used to set the threshold for damaging variants as it distinguished itself from the other truncating variants by acting as a hypomorphic variant (Figs. 2 and 3C; Supplementary Fig. S4A). In addition to p.R519X, 9 CHEK2 missense VUS similarly exhibited intermediate functional defects (Fig. 3C; Supplementary Fig. S3 and 4A). The remaining 10 CHEK2 missense VUS did not impact CHK2's functionality (Fig. 3C; Supplementary Figs. S3 and S4A). These results were in agreement with those from the Western blot analysis (Fig. 2). However, correlation analysis showed that especially among the functional and intermediate CHEK2 variants, Western blot analysis is inefficient in discriminating functional differences (R2 = 0.71; P < 0.0001; Supplementary Fig. S4B). Thus, the FACS-based phospho-Kap1 p.S473 analysis allows for a quantitative and therefore more accurate functional classification of CHEK2 variants.
We noticed that with FACS analysis, differentiating the positive phospho-Kap1 p.S473 population from the negative population was difficult for cells that expressed CHEK2 VUS with intermediate function (p.E64K, p.K141T, p.D203G, p.E239K, p.D438Y, p.I448S, p.A480T, and p.R521W). We therefore repeated the FACS-based quantification of phospho-Kap1 p.S473 for several missense variants (4 functional, 7 intermediate, and 2 damaging variants) following coexpression of EGFP-CHEK2 and mCherry (EGFP-CHEK2-T2A-mCherry). Following selection of GFP/mCherry-positive cells, the effects of these variants on Kap1 p.S473 phosphorylation fully corroborated those obtained with cells expressing non-tagged CHEK2 (i.e., R2 = 0.95), as all intermediate variants displayed intermediate effects on kinase activity (Fig. 3D; Supplementary Fig. S5B).
As Kap1 represents only one of the many targets of CHK2, an important question was whether the functional defects with regards to Kap1 p.S473 phosphorylation also translate to other functions of CHK2. To address this, we used a more general readout, that is, cell growth after DNA damage induction, which is likely regulated by CHK2's activity on multiple downstream targets. For this, we assessed the impact of two benign (p.R137 = and p.S435 =), two pathogenic (p.W93fs and p.T367Mfs) and four intermediate CHEK2 variants (p.E64K, p.D203G, p.D438Y, and p.R521W) on cell survival after phleomycin treatment (Fig. 3E). Their impact on cell survival correlated well with phospho-Kap1 p.S473 levels as measured by FACS (R2 = 0.80; P = 0,0052; Fig. 3F). However, the growth effects for intermediate variants were variable among replicate experiments, whereas the effects observed for the benign and pathogenic variants were reproducible. These data suggest that our FACS-based assay is a robust and reliable approach for the functional classification of CHEK2 variants and that phosphorylation of Kap1 p.S473 is a suitable readout to assess the general impact of variants on CHK2 function.
Several variants alter the kinetics of CHK2
The analysis of phospho-Kap1 p.S473 levels in unirradiated cells, and 2 or 6 hours after IR, showed that two CHEK2 missense VUS (p.E64K and p.R521W) were unable to maintain phosphorylation of Kap1 at p.S473 at the later timepoint (Fig. 2). To confirm this, we expressed these VUS in Chek2KO cells using the non-tagged CHEK2 cDNA and assessed phospho-Kap1 p.S473 levels by FACS at 2, 4, and 6 hours after IR (Supplementary Fig. S6). Quantification of the average intensity of phospho-Kap1 p.S473 showed that for wild-type CHEK2, the signal intensity only slightly decreases in time compared with that at 2 hours after IR (Fig. 3G; Supplementary Fig. S6). Similarly, for p.D203G, which we identified as a variant with intermediate functional impact (Fig. 3C), we observed that phospho-Kap1 p.S473 levels are maintained in time (Fig. 3G; Supplementary Fig. S6), even though overall phospho-Kap1 p.S473 levels at 2 hours after IR were lower than in cells expressing wild-type CHEK2. For both p.E64K and p.R521W, however, the phospho-Kap1 p.S473 levels were strongly reduced at 6 hours after IR (Fig. 3G; Supplementary Fig. S6). In addition, we observed that the truncating CHEK2 variant p.R519X resulted in the same kinetic defect as p.R521W (Fig. 2). Functional classification of such variants is therefore strongly dependent on the timepoint after IR at which CHK2 activity is measured. This may also explain why previous reports using different approaches classified p.E64K and p.R521W as either neutral or damaging, rather than intermediate (Supplementary Fig. S7A and S7B; refs. 34, 35). In addition, we found that one variant (i.e., p.V200A) displayed unregulated CHK2 activity in the absence of DNA damage induction (Fig. 2). Analysis of phospho-Kap1 p.S473 levels by FACS analysis confirmed this functional effect (Fig. 3H; Supplementary Fig. S4C). In conclusion, p.E64K, p.V200A, p.R519X, and p.R521W alter the kinetics of CHK2 activity, implicating a mechanism for aberrant protein function that has not been previously reported for CHEK2 genetic variants.
Correlation between computational predictions and functionality of variants
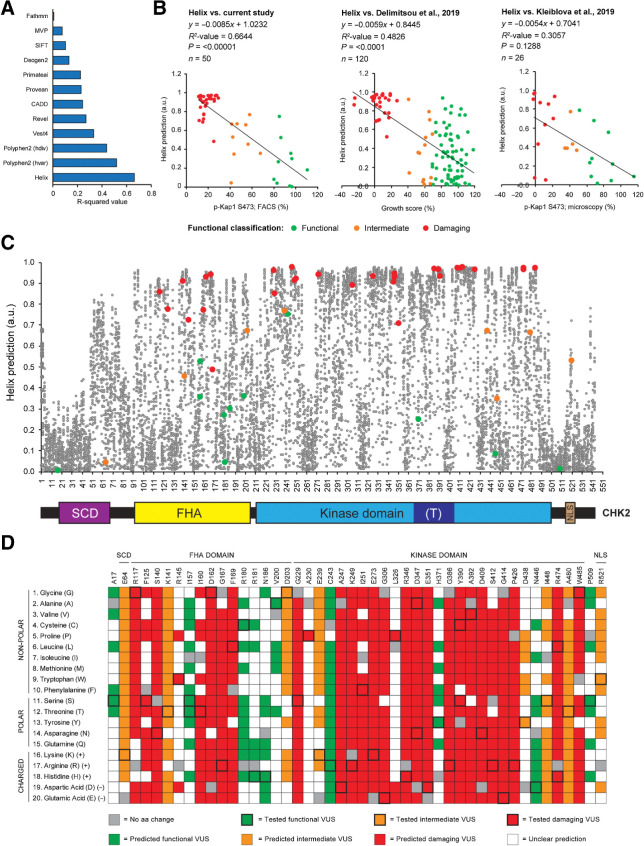
With the rapid accumulation of identified VUS in cancer-associated genes (49, 50), computational tools can aid in the clinical interpretation of such variants (51). We therefore compared the quantitative outcome of our functional assays for CHEK2 missense variants (Fig. 3C) with the predictions from twelve algorithms: Helix, PolyPhen (hvar), PolyPhen (hdiv), VEST4, REVEL, PrimateAI, CADD, Provean, Deogen2, MVP, SIFT, and FATHMM (Fig. 4A and B; Supplementary Fig. S8). Interestingly, Helix (52) outperformed all other tools (Fig. 4A). This tool is a missense variant effect predictor built on an extensive resource of protein data, in which protein structures, together with high-quality structure-based multiple sequence alignments (MSA) for the complete structural space, are combined with full-length sequence-based MSAs for the human proteome. Furthermore, Helix was trained on a large set of well-annotated variants using a strict training regime where circularity is actively avoided (53). When comparing the predictions from Helix to our functional data, we observed a significant correlation (R2 = 0,66; P < 0.00001; Fig. 4B). Such a correlation was also observed for the functional data from Delimitsou and colleagues (R2 = 0.48; P < 0.0001; ref. 34), but not for those from Kleiblova and colleagues (R2 = 0.31; P = 0.13; Fig. 4B; ref. 35). For the CHEK2 VUS in our study, both versions of PolyPhen (hvar and hdiv) also appeared to predict functional effects relatively well (R2 = 0.52 and 0.44, respectively), but the effects of intermediate CHEK2 VUS, as well as of several functional VUS, were overestimated (Supplementary Fig. S8). Importantly, other tools, particularly REVEL, Provean, Deogen, and FATHMM, underestimated the effect of several damaging variants in CHEK2 (Supplementary Fig. S8). Together, these findings highlight the potential of Helix with regards to interpretation of missense variants in CHEK2.
Figure 4.
Correlation between computational predictions and functionality of CHEK2 variants. A, Bar plot showing the R2-correlation values between the FACS-based analysis of Kap1 p.S473 phosphorylation as shown in Fig. 3C and computational predictions from 12 different prediction algorithms. B, Scatter plot showing the correlation between Helix-based in silico predictions and results from functional assays presented in our study (Fig. 3C), or those from Delimitsou and colleagues 2019 (34) and Kleiblova and colleagues 2019 (35). Data points are colored on the basis of functional classification (green, functional; orange, intermediate; red, damaging). Helix provides predictions for pathogenicity ranging from 0–1, with values close to 1 representing pathogenic predictions. C, En masse prediction plot from Helix for all possible missense changes in human CHEK2. Schematic representation of the CHK2 protein and its functional domains demarcated by the amino acid numbers at the x-axis of the plot. D, Heatmap showing predictions from Helix combined with functional data for CHK2 amino acid changes that were analyzed in Fig. 3C (outlined in bold). Functional variants are indicated in green (with bold outline); amino acid changes with a similar (+0.05) or lower prediction from Helix are also indicated in green. Intermediate variants are indicated in orange (with bold outline); amino acid changes with a similar (−0.05) or higher prediction from Helix are also indicated in orange. Damaging variants are indicated in red (with bold outline); amino acid changes with a similar (−0.05) or higher prediction from Helix are also indicated in red. For each amino acid position, amino acid changes with a similar color code are expected to result in similar functional effects. Squares in gray and white represent changes into the original amino acid or variant changes for which predictions are unclear, respectively.
To better understand the functional effects of missense variants throughout the entire CHK2 protein, we next visualized the predictions from Helix for all possible missense alterations in CHEK2 (Fig. 4C; Supplementary Table S1). Interestingly, many missense changes were predicted to exhibit damaging effects. This may be due to the relatively small size of the CHK2 protein (62 kDa, 543 amino acids), in which unfavourable missense substitutions (based on amino acid characteristics) may be more prone to affect function than in larger proteins. Furthermore, we used the predictions from Helix to examine the functional effects of alternative amino acid changes for each CHEK2 missense VUS in this study (Fig. 4D). This suggested that several conserved CHK2 amino acid residues (e.g., p.S140, p.G229, p.A247, p.K249, p.E273, p.R346, p.D347, p.E351, p.G386, p.D409, p.G414, p.P426, and p.R474) are critical for kinase function. Not surprisingly, this included the p.S140 autophosphorylation site that regulates CHK2 dimerization (54), p.E273, which is important for ATP hydrolysis (55, 56), and the catalytic residue p.D347A (55). Thus, Helix is a powerful tool to predict the impact of missense alterations in CHEK2 and can highlight regions and specific residues that are crucial for protein function.
CHEK2 VUS affect protein function through distinct mechanisms
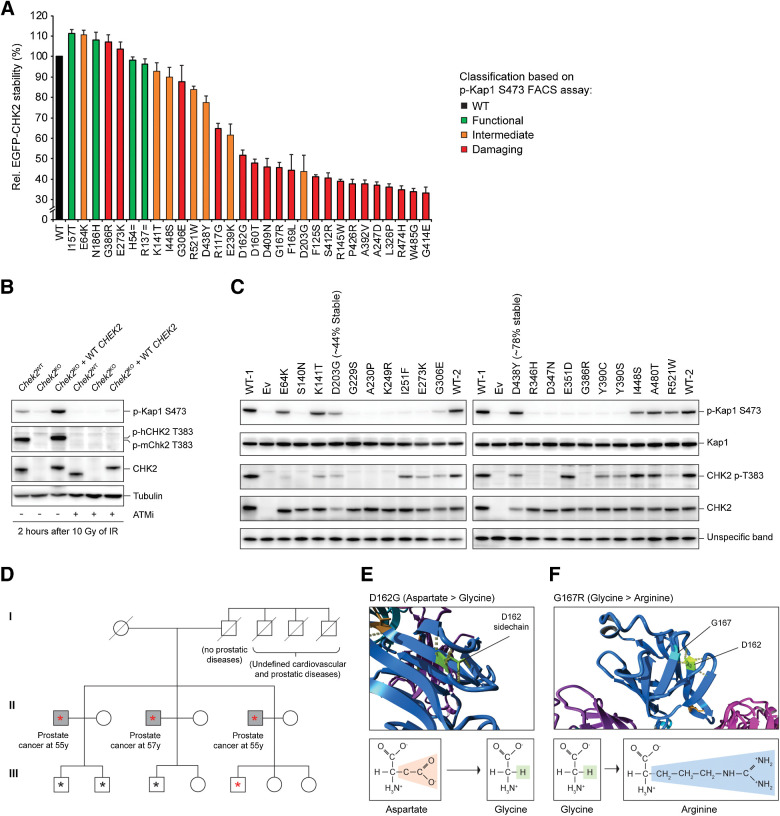
Our Western blot analysis showed that many CHEK2 missense variants result in reduced protein levels (Fig. 2). To further assess their effect on protein stability, we selected 30 VUS and introduced these in our EGFP-CHEK2-T2A-mCherry construct. Following RMCE in Chek2KO mES cells, steady-state abundance of CHK2 protein variants was measured on the basis of GFP fluorescence in mCherry-positive cells, ruling out transcriptional effects on EGFP-CHEK2 expression. The GFP signal for the two synonymous CHEK2 variants (p.H54= and p.R137=), as well as that for several other functional, intermediate and damaging VUS (e.g., p.E64K, p.K141T, p.I157T, p.N186H, p.E273K, p.G306E, p.G386R, p.I448S, p.R521W), was comparable to wild-type CHEK2 (Fig. 5A). However, all variants that displayed clearly reduced CHK2 protein levels on Western blot analysis (Fig. 2), also exhibited strongly reduced GFP signals (i.e., <65%; Fig. 5A). Overall, we identified 18 CHEK2 VUS that exhibit major effects on CHK2 protein stability, thereby hampering CHK2 kinase function.
Figure 5.
Analysis of pathogenic mechanisms of CHEK2 VUS and the association of two VUS with prostate cancer. A, Quantification of FACS measurements of the average EGFP intensity in Chek2KO mES cells complemented with EGFP-CHEK2-T2A-mCherry, with or without the indicated CHEK2 variants. EGFP intensities were measured in mCherry-positive gated cells. Data represent mean percentages ± SEM for three independent measurements and are relative to WT that was set at 100%. B, Western blot analysis of the indicated proteins from IR-exposed (10 Gy) Chek2WT, Chek2KO, and Chek2KO mES cells complemented with human CHEK2 cDNA that were left untreated or treated with ATM inhibitor (ATMi). Tubulin was used as a loading control. C, Western blot analysis of the indicated proteins from IR-exposed (10 Gy) Chek2KO mES cells complemented with human CHEK2 cDNA without or with a CHEK2 variant that displayed intermediate or damaging effects in Fig. 3C. An unspecific band produced by the anti-CHK2 antibody was used as a loading control. D, Pedigree of the family with the CHEK2 c.485A>G/p.D162G variant. Three male siblings carrying CHEK2 c.485A>G/p.D162G developed prostate cancer in their 50s (gray squares). Circles, females; squares, males. The asterisks indicate family members whose blood cell DNA was subjected to exome sequencing. The red asterisks indicate members carrying the CHEK2 c.485A>G/p.D162G variant. E and F, Partial structures (top) of the CHK2 FHA domain showing the effect of two CHK2 variants exhibiting protein instability as shown in A. Formulas and changes for the indicated amino acids are shown (bottom).
Several damaging variants (e.g., p.E273K and p.G386R) did not affect CHK2 protein stability, yet impaired IR-induced Kap1 p.S473 phosphorylation (Figs. 2 and 5A). We therefore questioned whether these variants affect CHK2 kinase activation. Autophosphorylation of CHK2 is essential for its activation and occurs, among others, on residues p.T383 and p.T387 in the T-loop region located within the kinase domain (Fig. 1D; refs. 19, 20). Consistent with a role for ATM in CHK2 activation (20, 57), exposure of cells to ATM inhibitor completely abolished IR-induced autophosphorylation of CHK2 on p.T383 (Fig. 5B). Subsequently, we examined the effect of seven intermediate and 13 damaging CHEK2 variants, which did not affect CHK2 protein stability (with exception of p.D203G and p.D438Y), on CHK2 p.T383 phosphorylation (Fig. 5C; Table 1). Most of these CHEK2 variants reduced (n = 8) or completely abolished autophosphorylation (n = 8). Surprisingly, five CHEK2 variants (i.e., p.I251F, p.E273K, p.Y390C, p.Y390S, and particularly p.E351D) that did not grossly impact CHK2 p.T383 autophosphorylation, still impaired kinase activity toward Kap1 p.S474 (Fig. 5C), possibly by impacting ATP binding/hydrolysis. Thus, our results suggest that the damaging effect of CHEK2 variants is a consequence of protein instability, impaired kinase activation, or perhaps reduced ATP binding/hydrolysis.
Table 1.
Complete list of human CHEK2 variants analyzed in this study.
| Nt. change | Aa change | Mutation type | p-Kap1 (%) | Classification | Helix | Stability (%) | p.T383 phos. | Cases | Controls | OR (95% CI), P (population-based studies) | OR (95% CI), P (all studies) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| c.49G>T | p.A17S | Missense | 96.5 | Functional | 0.00 | n/a | n/a | n/a | n/a | n/a | n/a |
| c.162C>T | p.H54= | Synonymous | 94.10 | Functional | n/a | 98.10 | n/a | n/a | n/a | n/a | n/a |
| c.190G>A | p.E64K | Missense | 41.09 | Intermediate | 0.04 | 110.76 | Absent | 53 | 31 | 1.78 (1.14–2.77), P = 0.0112 | 1.77 (1.16–2.69), P = 0.0075 |
| c.277delT | p.W93Gfs | Truncating | 14.31 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.349A>G | p.R117G | Missense | 17.42 | Damaging | 0.86 | 64.72 | n/a | 47 | 22 | 2.22 (1.34–3.68), P = 0.002 | 2.93 (1.82–4.73), P < 0.0001 |
| c.374T>C | p.F125S | Missense | 15.99 | Damaging | 0.77 | 41.03 | n/a | 0 | 1 | n/a | n/a |
| c.405delA | p.K135Nfs | Truncating | 30.89 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.411A>T | p.R137= | Synonymous | 94.09 | Functional | n/a | 96.42 | n/a | n/a | n/a | n/a | n/a |
| c.419G>A | p.S140N | Missense | 15.90 | Damaging | 0.91 | n/a | Absent | 1 | 0 | n/a | n/a |
| c.422A>C | p.K141T | Missense | 56.40 | Intermediate | 0.45 | 92.77 | Intermediate | 1 | 0 | n/a | n/a |
| c.433C>T | p.R145W | Missense | 15.82 | Damaging | 0.72 | 38.83 | n/a | 10 | 9 | 1.15 (0.47–2.84), P = 0.7555 | 1.96 (0.89–4.32), P = 0.0925 |
| c.470T>G | p.I157S | Missense | 86.39 | Functional | 0.53 | n/a | n/a | 1 | 0 | n/a | n/a |
| c.470T>C | p.I157T | Missense | 95.55 | Functional | 0.36 | 111.22 | n/a | n/a | n/a | n/a | n/a |
| c.479T>C | p.I160T | Missense | 15.95 | Damaging | 0.77 | 48.00 | n/a | 0 | 1 | n/a | n/a |
| c.485A>G | p.D162G | Missense | 18.40 | Damaging | 0.93 | 51.69 | n/a | n/a | n/a | ||
| c.499G>A | p.G167R | Missense | 20.76 | Damaging | 0.94 | 45.72 | n/a | 8 | 3 | 2.77 (0.73–10.44), P = 0.1325 | 5.01 (1.47–17.10), P = 0.0101 |
| c.507delT | p.F169Lfs | Truncating | 17.11 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.507T>G | p.F169L | Missense | 25.19 | Damaging | 0.49 | 44.35 | n/a | 1 | 2 | 0.52 (0.05–5.73), P = 0.5925 | 3.09 (0.64–14.9), P = 0.1589 |
| c.538C>T | p.R180C | Missense | 84.08 | Functional | 0.27 | n/a | n/a | n/a | n/a | n/a | n/a |
| c.542G>A | p.R181H | Missense | 82.93 | Functional | 0.04 | n/a | n/a | 33 | 22 | 1.56 (0.91–2.67), P = 0.1075 | 1.16 (0.69–1.93), P = 0.5776 |
| c.556A>C | p.N186H | Missense | 96.50 | Functional | 0.30 | 108.21 | n/a | 17 | 14 | 1.26 (0.62–2.56), P = 0.5206 | 1.59 (0.85–2.99), P = 0.1491 |
| c.599T>C | p.V200A | Missense | 67.63 | Intermediate | 0.36 | n/a | n/a | 0 | 1 | n/a | n/a |
| c.608A>G | p.D203G | Missense | 41.43 | Intermediate | 36.00 | 43.79 | Intermediate | 4 | 0 | n/a | n/a |
| c.685G>A | p.G229S | Missense | 12.94 | Damaging | 0.96 | n/a | Absent | 0 | 1 | n/a | n/a |
| c.688G>C | p.A230P | Missense | 12.90 | Damaging | 0.85 | n/a | Absent | n/a | n/a | n/a | n/a |
| c.715G>A | p.E239K | Missense | 57.85 | Intermediate | 0.77 | 61.58 | n/a | 12 | 7 | 1.78 (0.70–4.52), P = 0.2253 | 2.27 (0.95–5.44), P = 0.0652 |
| c.727T>C | p.C243R | Missense | 85.04 | Functional | 0.75 | n/a | n/a | 4 | 8 | 0.52 (0.16–1.72), P = 0.2845 | 0.44 (0.13–1.47), P = 0.1826 |
| c.740C>A | p.A247D | Missense | 27.50 | Damaging | 0.97 | 36.99 | n/a | 1 | 0 | n/a | n/a |
| c.746A>G | p.K249R | Missense | 25.48 | Damaging | 0.91 | n/a | Absent | n/a | n/a | n/a | n/a |
| c.751A>T | p.I251F | Missense | 15.11 | Damaging | 0.69 | n/a | Intermediate | 3 | 0 | n/a | n/a |
| c.757A>T | p.K253X | Truncating | 34.35 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.817G>A | p.E273K | Missense | 22.77 | Damaging | 0.94 | 103.64 | Intermediate | n/a | n/a | n/a | n/a |
| c.858C>A | p.I286= | Synonymous | 76.33 | Functional | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.917G>A | p.G306E | Missense | 18.84 | Damaging | 0.89 | 87.77 | Intermediate | 1 | 0 | n/a | n/a |
| c.977T>C | p.L326P | Missense | 25.49 | Damaging | 0.93 | 35.98 | n/a | 1 | 0 | n/a | n/a |
| c.1037G>A | p.R346H | Missense | 15.03 | Damaging | 0.90 | n/a | Absent | 4 | 2 | 2.08 (0.38–11.34), P = 0.3987 | 2.65 (0.54–13.14), P = 0.2322 |
| c.1039G>A | p.D347N | Missense | 13.23 | Damaging | 0.93 | n/a | Absent | 4 | 3 | 1.38 (0.31–6.19), P = 0.6701 | 0.88 (0.22–3.54), P = 0.8618 |
| c.1040A>C | p.D347A | Missense | 29.31 | Damaging | 0.94 | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1053G>T | p.E351D | Missense | 11.72 | Damaging | 0.71 | n/a | Normal | 7 | 2 | 3.63 (0.76–17.50), P = 0.1075 | 4.42 (0.97–20.18), P = 0.0550 |
| c.1100delC | p.T367Mfs | Truncating | 21.13 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1101T>A | p.T367= | Synonymous | 107.13 | Functional | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1111C>T | p.H371Y | Missense | 110.43 | Functional | 0.25 | n/a | n/a | 37 | 38 | 1.01 (0.64–1.59), P = 0.9618 | 0.78 (0.52–1.17), P = 0.2249 |
| c.1156G>C | p.G386R | Missense | 12.09 | Damaging | 0.97 | 107.07 | Absent | 1 | 0 | n/a | n/a |
| c.1169A>G | p.Y390C | Missense | 11.27 | Damaging | 0.96 | n/a | Intermediate | n/a | n/a | n/a | n/a |
| c.1169A>C | p.Y390S | Missense | 13.12 | Damaging | 0.96 | n/a | Intermediate | 2 | 3 | 0.69 (0.12–4.14), P = 0.6871 | 0.88 (0.18–4.38), P = 0.8801 |
| c.1175C>T | p.A392V | Missense | 12.72 | Damaging | 0.93 | 37.55 | n/a | 12 | 4 | 3.12 (1.00-9.66), P = 0.0491 | 3.32 (1.10–9.99), P = 0.0332 |
| c.1225G>A | p.D409N | Missense | 13.24 | Damaging | 0.97 | 46.02 | n/a | 1 | 1 | 1.04 (0.06-16.60), P = 0.9787 | 0.88 (0.06–14.14), P = 0.9306 |
| c.1236T>A | p.S412R | Missense | 26.39 | Damaging | 0.97 | 40.53 | n/a | 1 | 0 | n/a | n/a |
| c.1241G>A | p.G414E | Missense | 22.86 | Damaging | 0.97 | 33.33 | n/a | 1 | 0 | n/a | n/a |
| c.1263delT | p.S422Vfs | Truncating | 11.54 | Damaging | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1277C>G | p.P426R | Missense | 22.05 | Damaging | 0.96 | 37.67 | n/a | 1 | 0 | n/a | n/a |
| c.1305A>T | p.S435= | Synonymous | 92.62 | Functional | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1312G>T | p.D438Y | Missense | 46.39 | Intermediate | 0.67 | 77.56 | Intermediate | 26 | 27 | 1.00 (0.58–1.71), P = 0.9999 | 1.24 (0.76–2.04), P = 0.3852 |
| c.1336A>G | p.N446D | Missense | 80.18 | Functional | 0.08 | n/a | n/a | 4 | 3 | 1.38 (0.31–6.19), P = 0.6701 | 1.47 (0.35–6.17), P = 0.5955 |
| c.1343T>G | p.I448S | Missense | 43.56 | Intermediate | 0,.35 | 90.08 | Normal | 1 | 3 | 0.35 (0.04–3.33), P = 0.3582 | 0.88 (0.18–4.38), P = 0.8801 |
| c.1421G>A | p.R474H | Missense | 15.73 | Damaging | 0.96 | 34.92 | n/a | 11 | 0 | n/a | n/a |
| c.1421G>T | p.R474L | Missense | 27.86 | Damaging | 0.97 | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1422T>A | p.R474= | Synonymous | 95.54 | Functional | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1438G>A | p.A480T | Missense | 54.42 | Intermediate | 0.66 | n/a | Intermediate | 4 | 0 | n/a | n/a |
| c.1453T>G | p.W485G | Missense | 20.05 | Damaging | 0.97 | 33.98 | n/a | 0 | 1 | n/a | n/a |
| c.1525C>T | p.P509S | Missense | 94.60 | Functional | 0,.01 | n/a | n/a | 21 | 22 | 0.99 (0.55–1.81), P = 0.977 | 0.88 (0.50–1.58), P = 0.6763 |
| c.1555C>T | p.R519X | Truncating | 59.34 | Intermediate | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| c.1561C>T | p.R521W | Missense | 42.51 | Intermediate | 0.53 | 83.94 | Intermediate | 9 | 2 | 4.67 (1.01–21.63), P = 0.0486 | 2.48 (0.89–6.87), P = 0.0819 |
Note: Nucleotide numbering reflects Human Genome Variation Society (HGVS) nomenclature where cDNA numbering +1 corresponds to the A of the ATG translation initiation codon in the reference sequence (CHEK2 NM_007194.4). The initiation codon is codon 1. For each variant, results for three functional readouts (i.e., Kap1 p.S473 phosphorylation, EGFP-CHK2 stability and CHK2 p.T383 phosphorylation), Helix-based predictions, population-based case–control frequencies, and ORs are shown. Functional classification is based on the phospho-Kap1 FACS assay (Fig. 3C) and population-based case–control frequencies and ORs are based on a study from the BRIDGES consortium in collaboration with the BCAC (7).
Abbreviation: n/a, not available.
Association of CHK2 functional defects with breast cancer risk
Having determined the functional impact of VUS in CHEK2, we next investigated whether the observed impact correlates with increased breast cancer risk. For this, we considered all 30 population-based BCAC studies, which were combined in a case–control association study performed by the BRIDGES consortium (48,826 breast cancer cases and 50,703 controls; ref. 7). Because of the low allele frequency of most CHEK2 variants, we were only able to identify two variants, c.190G>A/p.E64K [OR 1.78; 95% confidence interval (CI), 1.14–2.77; P = 0.0112] and c.349A>G/p.R117G (OR 2.22; 95% CI, 1.34–3.68; P = 0.0020; Table 1), that associate with significantly increased breast cancer risk and for which the population-based ORs had a relatively narrow CI. p.E64K had an intermediate functional impact, whereas p.R117G was damaging (Fig. 3C), suggesting that the degree of functional impact correlates with the breast cancer risk level.
Under the assumption that variants with a similar impact on CHK2 functionality confer the same level of cancer risk, we performed a burden-type association analysis (Table 2). Accordingly, we defined three groups of CHEK2 VUS based on their impact on CHK2 function (i.e., functional, intermediate, or damaging) and established the joint frequencies of the individual variants within the same group in both cases and controls. The two variants mentioned above (p.E64K and p.R117G) were excluded from these groups as they were already associated with a significant breast cancer risk (Table 1). This analysis revealed that functional CHEK2 VUS as a group (n = 6, excl. p.I157T and p.R180C for which carrier frequencies were not available; Fig. 3C), are not associated with an increased risk for breast cancer (OR 1.13; 95% CI, 0.87–1.46; P = 0.3773; Table 2). However, CHEK2 VUS that exhibited an intermediate functional effect (n = 7, excl. p.E64K; Fig. 3C) were associated with a significantly increased risk for breast cancer (OR 1.52; 95% CI, 1.01–2.28; P = 0.0448; Table 2). Importantly, damaging CHEK2 VUS (n = 27, excl. p.R117G; Fig. 3C), were associated with an even higher risk than intermediate variants (OR 2.23; 95% CI, 1.48–3.38; P < 0.0001; Table 2). In addition to population-based ORs, cancer risks described in Table 1 and 2 were also calculated on the basis of all 44 BCAC studies (combination of 30 population-based and 14 family-based studies; ref. 7). Although this generally resulted in slightly higher risk estimations for most CHEK2 variants or variant groups, a similar correlation between functional impact of variants and cancer risk was observed (Table 1; Table 2). These results suggest that our quantitative functional assay can identify pathogenic CHEK2 variants.
Table 2.
Burden-type cancer risk association analysis for human CHEK2 variants.
| Variant group based on function | Aa change | Cases | Controls | OR (95% CI), P (population-based studies) | OR (95% CI), P (all studies) |
|---|---|---|---|---|---|
| Functional VUS | p.I157S | 1 | 0 | 1.13 (0.87–1.46), P = 0.378 | 0.97 (0.76–1.23), P = 0.7943 |
| p.R181H | 33 | 22 | |||
| p.N186H | 17 | 14 | |||
| p.V200A | 0 | 1 | |||
| p.C243R | 4 | 8 | |||
| p.H371Y | 37 | 38 | |||
| p.N446D | 4 | 3 | |||
| p.P509S | 21 | 22 | |||
| Intermediate VUS | p.E64K | 53 | 31 | 1.63 (1.21–2,.20), P = 0.0014 | 1.79 (1.36–2.36), P < 0.0001 |
| p.K141T | 1 | 0 | |||
| p.D203G | 4 | 0 | |||
| p.E239K | 12 | 7 | |||
| p.D438Y | 26 | 27 | |||
| p.I448S | 1 | 3 | |||
| p.A480T | 4 | 0 | |||
| p.R521W | 9 | 2 | |||
| Intermediate VUS (excl. p.E64K) | 1.52 (1.01–2.28), P = 0.0448 | 1.81 (1.25–2.62), P = 0.0016 | |||
| Damaging VUS | p.R117G | 47 | 22 | 2.23 (1.62–3.07), P < 0.0001 | 3.03 (2.25–4,08), P < 0,0001 |
| p.F125S | 0 | 1 | |||
| p.S140N | 1 | 0 | |||
| p.R145W | 10 | 9 | |||
| p.I160T | 0 | 1 | |||
| p.G167R | 8 | 3 | |||
| p.F169L | 1 | 2 | |||
| p.G229S | 0 | 1 | |||
| p.A230P | n/a | n/a | |||
| p.A247D | 1 | 0 | |||
| p.K249R | n/a | n/a | |||
| p.I251F | 3 | 0 | |||
| p.E273K | n/a | n/a | |||
| p.G306E | 1 | 0 | |||
| p.L326P | 1 | 0 | |||
| p.R346H | 4 | 2 | |||
| p.D347N | 4 | 3 | |||
| p.E351D | 7 | 2 | |||
| p.G386R | 1 | 0 | |||
| p.Y390C | n/a | n/a | |||
| p.Y390S | 2 | 3 | |||
| p.A392V | 12 | 4 | |||
| p.D409N | 1 | 1 | |||
| p.S412R | 1 | 0 | |||
| p.G414E | 1 | 0 | |||
| p.P426R | 1 | 0 | |||
| p.R474H | 11 | 0 | |||
| p.W485G | 0 | 1 | |||
| Damaging VUS (excl. p.R117G) | 2.23 (1.48–3.38), P < 0.0001 | 3.09 (2.11–4.53), P < 0.0001 | |||
Note: Variants with similar impact of CHK2 functionality were grouped (Fig. 2C). Only missense variants for which case–control frequencies from population- or family-based studies have been reported were included (7). The case–control frequencies reflect those of the population-based studies alone. The analysis was also performed for groups of CHEK2 variants without p.E64K or p.R117G, for which the carrier frequencies are high.
Abbreviation: n/a, not available.
Association of the CHEK2 c.485A>G/p.D162G variant with prostate cancer
Functional defects caused by CHEK2 VUS are not only associated with an increased risk of developing breast cancer, but have also been linked to other cancers, including prostate cancer (9, 10). We therefore examined three male siblings from a family that all presented with prostate cancer >10 years earlier than the average age of onset for sporadic prostate cancer. This revealed that they were all heterozygous for the germline CHEK2 c.485A>G/p.D162G allele (Fig. 5D), which was characterized as a damaging variant in this study (Fig. 3C). Similarly, the closely located CHEK2 VUS p.G167R had also been linked to prostate cancer (10). Our results showed that both variants lead to protein instability (Fig. 5A; Supplementary Fig. S5B), rendering CHK2 non-functional (Figs. 2 and 3C). Consistently, using the crystal structure of CHK2 (PDB - 3I6U; ref. 55), in silico modeling of CHK2 p.D162G and p.G167R showed that these substitutions are extremely unfavorable for correct folding of the region they locate to, as they lead to loss of two hydrogen bonds (Fig. 5E and F). Interestingly, analysis of prostate tumor DNA of two of the three siblings carrying the CHEK2 c.485A>G/p.D162G variant showed no evidence for LOH (Table 3), resembling observations made for the well-known CHEK2 c.1100delC/p.T367Mfs allele in breast cancer (Table 3; ref. 58). These results suggest that LOH for individuals carrying a monoallelic damaging CHEK2 variant may not be a prerequisite for cancer development, although we cannot rule out that promotor methylation silenced expression of the intact allele, thereby mimicking LOH (59). The findings on CHEK2 c.485A>G/p.D162G suggest that our functional analysis can also identify pathogenic VUS in CHEK2 that associate with prostate cancer.
Table 3.
No evidence for LOH in CHEK2 c.485A>G/p.D162G or c.1100delC/p.T367Mfs carriers.
| CHEK2 variant carriers | Tissue type | VAF c.485A>G | VAF c.1100delC |
|---|---|---|---|
| c.485A>G/p.D162G carrier 1 (brother 1) | Tumor tissue | 0.521 | |
| Control tissue | 0.482 | ||
| c.485A>G/p.D162G carrier 2 (brother 2) | Tumor tissue | 0.526 | |
| Control tissue | 0.476 | ||
| c.1100delC/p.T367Mfs carrier 1 | Tumor tissue | 0.538 | |
| Control tissue | 0.485 | ||
| c.1100delC/p.T367Mfs carrier 2 | Tumor tissue | 0.466 | |
| Control tissue | N/A |
Note: VAF refers to variant allele frequency.
Abbreviation: N/A, not available.
Discussion
We developed a mES cell-based system that allows for the quantitative functional classification of genetic variants in the CHEK2 gene that associate with breast and prostate cancer. Of the 50 CHEK2 missense VUS tested in this study, nine variants (18%) had an intermediate impact on CHK2 function, while 31 (62%) were damaging (Table 1; Fig. 3C). Importantly, 23 CHEK2 missense VUS constitute variants that have, to our knowledge, not been functionally characterized in previous studies (29–35, 60). At least 18 of the intermediate and damaging VUS in our study (>50%) exhibited defects in protein stability (Fig. 5A), which is a common pathogenic mechanism originating from missense variants (38, 61). Moreover, at least 11 VUS (22%) showed reduced or complete lack of autophosphorylation on p.T383 (Fig. 5C), explaining the impaired kinase activity for most of these VUS (19, 62). For five damaging VUS (i.e., p.I251F, p.E273K, p.E351D, p.Y390C, and p.Y390S) considerable levels of autophosphorylation were observed, while kinase activity toward Kap1 was lacking. As these VUS mostly localize to the ATP-binding pocket of CHK2, they likely impair the ability of CHK2 to bind or hydrolyze ATP, the latter of which has already been reported for p.E273K (55, 56). Thus, we examined numerous CHEK2 missense VUS for which we quantified functional effects (i.e., kinase activity) and assessed pathogenic mechanisms of action. Correlation between our quantitative results and breast cancer risk further demonstrated that our functional assay can identify pathogenic missense variants in CHEK2.
Our results are generally in line with two recent studies describing functional analysis of CHEK2 missense variants (34, 35). Kleiblova and colleagues employed both an in vitro kinase assay and a RPE1 CHEK2KO cell-based system for functional classification of CHEK2 variants (35). For most overlapping variants, our results are consistent with their functional assessment (Supplementary Fig. S7A). Although further research is required to explain the differences observed for three variants (i.e., p.I157T, p.R346H, and p.D438Y), differences in the functional classification of p.E64K may be explained by its kinetic effect on Kap1 phosphorylation (Fig. 3G; Supplementary Fig. S7A). That is, we based our “intermediate” functional classification on phospho-Kap1 p.S473 levels observed at 2 hours after IR, whereas Kleiblova and colleagues based their “damaging” classification on the KAP1 phosphorylation levels observed at 4 hours after IR in the RPE1 cell-based assay. On the other hand, Delimitsou and colleagues employed a yeast rad53-mutant cell-based system for functional characterization of human CHEK2 variants (34), whose results were also highly consistent with those from our study (Supplementary Fig. S7B). However, all CHEK2 variants (with the exception of p.E64K) that we classified as intermediate and Delimitsou and colleagues as neutral (Supplementary Fig. S7B), are variants that impaired protein stability in our assays (i.e., p.D203G, p.E239K, p.D438Y, and p.R521W; Fig. 5A). Possibly, several intermediate effects are not picked up efficiently in the yeast assays as yeast cells grow at 30°C rather than at 37°C, which may reduce the thermodynamic instability of proteins. Thus, while the outcome of the different functional analysis of CHEK2 variants is generally consistent, discrepancies for some variants remain, complicating their classification and calling for further analysis.
The Helix algorithm predicted functionality of CHEK2 missense variants more accurately than several other algorithms did (Fig. 4B). Therefore, the en masse Helix predictions (Fig. 4C; Supplementary Table S1) may aid in the classification of missense variants in CHEK2 for which functional outcomes were inconsistent (e.g., p.L174V; ref. 35), or for which functional analysis have yet to be performed. In support of the remarkable performance of Helix, in both our study and that of Delimitsou and colleagues (34), no variants predicted to be benign by Helix were found to be damaging (Fig. 4B). Although computational predictions should be handled with care, discrepancies with Helix may also highlight variants that require further validation of their functional impact, thereby aiding in the classification of CHEK2 variants.
The BRIDGES consortium in collaboration with the BCAC, showed that rare CHEK2 missense VUS in aggregate associate with a low, yet significant risk for breast cancer (OR 1.42; 95% CI, 1.28–1.58; P < 0.0001; ref. 7). However, a major challenge is to discriminate which VUS associate with cancer risk and which do not. Our study addressed this challenge and showed that the degree of CHK2 dysfunction, for numerous CHEK2 missense VUS, correlates with increased breast cancer risk (Tables 1 and 2; Fig. 3C). Furthermore, the OR for the damaging CHEK2 VUS in aggregate (OR 2.23; 95% CI, 1.48–3.38; P < 0.0001), as well as that for the damaging VUS c.349A>G/p.R117G alone (OR 2.22; 95% CI, 1.34–3.68; P = 0.0020), compared well with the population-based ORs for c.1100delC/p.T367Mfs (OR 2.66; 95% CI, 2.27–3.11; P < 0.0001) and that of all other CHEK2 truncating variants in aggregate (OR 2.13; 95% CI, 1.60–2.84; P < 0.0001; refs. 6, 7). The OR for the intermediate CHEK2 variant c.190G>A/p.E64K (OR 1.78; 95% CI, 1.14–2.77; P = 0.0112) associated with significantly increased breast cancer risk comparable with that calculated for its functional classification group (OR 1.52; 95% CI, 1.01–2.28; P = 0.0448). These results strongly suggest that intermediate CHEK2 VUS associate with significantly increased breast cancer risk and that damaging CHEK2 VUS likely associate with a similar risk for breast cancer as truncating CHEK2 variants.
Effects of CHEK2 variants on splicing could not be examined since we employed human CHEK2 cDNA-based complementation assays. However, in silico splice site prediction analysis was performed using four different algorithms (Splice Site Finder-like, MaxEntScan, GeneSplicer, NNSplice) in Alamut (http://www.interactivebiosoftware.com/). For most VUS, an effect on RNA splicing was unlikely, except for seven variants (p.A17S, p.I157S, p.I160T, p.D162G, p.F169L, p.G229S, and p.A230P) for which these algorithms predicted the introduction of weak acceptor or donor recognition sites in the corresponding exons (Supplementary Table S2). Consistently, the recently developed deep learning-based SpliceAI tool (63) predicted no major splice effects for the CHEK2 missense VUS examined in this study, except for, i.e., p.V200A and p.G229S, for which the loss or introduction of a splice acceptor site was predicted with low to moderate confidence (Supplementary Table S2). The path to clinical implementation of functional analysis, in line with ACMG guidelines (64), involves having a well-calibrated assay. Even though we note that the slight difference in homology between mouse and human CHEK2 (82% identical and 88% similar in protein sequence) could affect the functional analysis presented in this study, we believe that our quantitative data and the correlation with breast cancer risk supports the robustness and validity of our functional assay for CHEK2, and thus its value as clinical diagnostic tool.
Authors' Disclosures
B. Vroling reports grants from Horizon 2020 during the conduct of the study. S. Heijl reports grants from Horizon 2020 during the conduct of the study. R. Eeles reports other support from Gu Asco, The Royal Marsden NHS Foundation Trust, University of Chicago, ESMO, and AstraZeneca UK Limited outside the submitted work. P. Devilee reports grants from European Commission and Dutch Cancer Society during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
The authors would like to thank Jos Jonkers and Peter Bouwman for providing the pTT5-Puro (RMCE acceptor cassette), pRNA-251-MCS-RMCE (RMCE exchange vector), and pCMV-Red-I-SceI constructs; Maria Jasin and Francis Stewart for sharing the DR-GFP reporter and FlpO constructs, respectively; Doug Easton, Jamie Allen, Leila Dorling, and the BRIDGES consortium for providing CHEK2 variants and case–control frequencies for OR calculations; Emanuele Valtorta and the Flow Cytometry Core Facility of the LUMC for technical assistance; and Josef Jiricny for fruitful discussions. This project has received funding from the Giuliana and Giorgio Stefanini Foundation (G. Marra), the European Union's Horizon 2020 research and innovation program BRIDGES under grant agreement 634935 (P. Devilee, M.P.G. Vreeswijk, and H. van Attikum), and the Dutch Cancer Society (KWF-7473; P. Devilee and H. van Attikum).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
R.A.C.M. Boonen: Conceptualization, formal analysis, supervision, funding acquisition, writing–original draft, writing–review and editing. W.W. Wiegant: Formal analysis, writing–original draft. N. Celosse: Formal analysis. B. Vroling: Formal analysis. S. Heijl: Conceptualization, formal analysis. Z. Kote-Jarai: Formal analysis. M. Mijuskovic: Conceptualization, formal analysis. S. Cristea: Formal analysis, supervision. N. Solleveld-Westerink: Formal analysis, supervision. T. van Wezel: Formal analysis, funding acquisition. N. Beerenwinkel: Formal analysis, supervision. R. Eeles: Formal analysis, supervision. P. Devilee: Conceptualization, formal analysis. M.P.G. Vreeswijk: Conceptualization, formal analysis. G. Marra: Formal analysis, funding acquisition. H. van Attikum: Conceptualization, formal analysis, supervision, funding acquisition, writing–review and editing.
References
- 1. Ahn J, Urist M, Prives C. The Chk2 protein kinase. DNA Repair 2004;3:1039–47. [DOI] [PubMed] [Google Scholar]
- 2. Bartek J, Lukas J. Chk1 and Chk2 kinases in checkpoint control and cancer. Cancer Cell 2003;3:421–9. [DOI] [PubMed] [Google Scholar]
- 3. Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, et al. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol 2002;22:6521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meijers-Heijboer H, van den Ouweland A, Klijn J, Wasielewski M, de Snoo A, Oldenburg R, et al. Low-penetrance susceptibility to breast cancer due to CHEK2(*)1100delC in noncarriers of BRCA1 or BRCA2 mutations. Nat Genet 2002;31:55–9. [DOI] [PubMed] [Google Scholar]
- 5. Vahteristo P, Bartkova J, Eerola H, Syrjakoski K, Ojala S, Kilpivaara O, et al. A CHEK2 genetic variant contributing to a substantial fraction of familial breast cancer. Am J Hum Genet 2002;71:432–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Couch FJ, Shimelis H, Hu C, Hart SN, Polley EC, Na J, et al. Associations between cancer predisposition testing panel genes and breast cancer. JAMA Oncol 2017;3:1190–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Breast Cancer Association Consortium, Dorling L, Carvalho S, Allen J, Gonzalez-Neira A, Luccarini C, et al. Breast cancer risk genes - association analysis in more than 113,000 women. N Engl J Med 2021;384:428–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cybulski C, Gorski B, Huzarski T, Masojc B, Mierzejewski M, Debniak T, et al. CHEK2 is a multiorgan cancer susceptibility gene. Am J Hum Genet 2004;75:1131–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cybulski C, Huzarski T, Gorski B, Masojc B, Mierzejewski M, Debniak T, et al. A novel founder CHEK2 mutation is associated with increased prostate cancer risk. Cancer Res 2004;64:2677–9. [DOI] [PubMed] [Google Scholar]
- 10. Dong X, Wang L, Taniguchi K, Wang X, Cunningham JM, McDonnell SK, et al. Mutations in CHEK2 associated with prostate cancer risk. Am J Hum Genet 2003;72:270–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Le Calvez-Kelm F, Lesueur F, Damiola F, Vallee M, Voegele C, Babikyan D, et al. Rare, evolutionarily unlikely missense substitutions in CHEK2 contribute to breast cancer susceptibility: results from a breast cancer family registry case-control mutation-screening study. Breast Cancer Res 2011;13:R6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dufault MR, Betz B, Wappenschmidt B, Hofmann W, Bandick K, Golla A, et al. Limited relevance of the CHEK2 gene in hereditary breast cancer. Int J Cancer 2004;110:320–5. [DOI] [PubMed] [Google Scholar]
- 13. Ingvarsson S, Sigbjornsdottir BI, Huiping C, Hafsteinsdottir SH, Ragnarsson G, Barkardottir RB, et al. Mutation analysis of the CHK2 gene in breast carcinoma and other cancers. Breast Cancer Res 2002;4:R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schutte M, Seal S, Barfoot R, Meijers-Heijboer H, Wasielewski M, Evans DG, et al. Variants in CHEK2 other than 1100delC do not make a major contribution to breast cancer susceptibility. Am J Hum Genet 2003;72:1023–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sodha N, Bullock S, Taylor R, Mitchell G, Guertl-Lackner B, Williams RD, et al. CHEK2 variants in susceptibility to breast cancer and evidence of retention of the wild type allele in tumours. Br J Cancer 2002;87:1445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 2014;42:D980–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stolarova L, Kleiblova P, Janatova M, Soukupova J, Zemankova P, Macurek L, et al. CHEK2 germline variants in cancer predisposition: stalemate rather than checkmate. Cells 2020;9:2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Guo X, Ward MD, Tiedebohl JB, Oden YM, Nyalwidhe JO, Semmes OJ. Interdependent phosphorylation within the kinase domain T-loop regulates CHK2 activity. J Biol Chem 2010;285:33348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwarz JK, Lovly CM., Piwnica-Worms H. Regulation of the Chk2 protein kinase by oligomerization-mediated cis- and trans-phosphorylation. Mol Cancer Res 2003;1:598–609. [PubMed] [Google Scholar]
- 20. Ahn JY, Li X, Davis HL, Canman CE. Phosphorylation of threonine 68 promotes oligomerization and autophosphorylation of the Chk2 protein kinase via the forkhead-associated domain. J Biol Chem 2002;277:19389–95. [DOI] [PubMed] [Google Scholar]
- 21. Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature 2004;432:316–23. [DOI] [PubMed] [Google Scholar]
- 22. Li J, Williams BL, Haire LF, Goldberg M, Wilker E, Durocher D, et al. Structural and functional versatility of the FHA domain in DNA-damage signaling by the tumor suppressor kinase Chk2. Mol Cell 2002;9:1045–54. [DOI] [PubMed] [Google Scholar]
- 23. Zhang J, Willers H, Feng Z, Ghosh JC, Kim S, Weaver DT, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol 2004;24:708–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu C, Zhang S, Gao X, Gao X, Xu X, Lv Y, et al. Roles of Kruppel-associated Box (KRAB)-associated co-repressor KAP1 Ser-473 phosphorylation in DNA damage response. J Biol Chem 2012;287:18937–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cann KL, Dellaire G. Heterochromatin and the DNA damage response: the need to relax. Biochem Cell Biol 2011;89:45–60. [DOI] [PubMed] [Google Scholar]
- 26. Czerwinska P, Mazurek S, Wiznerowicz M. The complexity of TRIM28 contribution to cancer. J Biomed Sci 2017;24:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bolderson E, Savage KI, Mahen R, Pisupati V, Graham ME, Richard DJ, et al. Kruppel-associated Box (KRAB)-associated co-repressor (KAP-1) Ser-473 phosphorylation regulates heterochromatin protein 1beta (HP1-beta) mobilization and DNA repair in heterochromatin. J Biol Chem 2012;287:28122–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lemaitre C, Soutoglou E. Double strand break (DSB) repair in heterochromatin and heterochromatin proteins in DSB repair. DNA Repair 2014;19:163–8. [DOI] [PubMed] [Google Scholar]
- 29. Bell DW, Kim SH, Godwin AK, Schiripo TA, Harris PL, Haserlat SM, et al. Genetic and functional analysis of CHEK2 (CHK2) variants in multiethnic cohorts. Int J Cancer 2007;121:2661–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee SB, Kim SH, Bell DW, Wahrer DC, Schiripo TA, Jorczak MM, et al. Destabilization of CHK2 by a missense mutation associated with Li-Fraumeni Syndrome. Cancer Res 2001;61:8062–7. [PubMed] [Google Scholar]
- 31. Roeb W, Higgins J, King MC. Response to DNA damage of CHEK2 missense mutations in familial breast cancer. Hum Mol Genet 2012;21:2738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tischkowitz MD, Yilmaz A, Chen LQ, Karyadi DM, Novak D, Kirchhoff T, et al. Identification and characterization of novel SNPs in CHEK2 in Ashkenazi Jewish men with prostate cancer. Cancer Lett 2008;270:173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang N, Ding H, Liu C, Li X, Wei L, Yu J, et al. A novel recurrent CHEK2 Y390C mutation identified in high-risk Chinese breast cancer patients impairs its activity and is associated with increased breast cancer risk. Oncogene 2015;34:5198–205. [DOI] [PubMed] [Google Scholar]
- 34. Delimitsou A, Fostira F, Kalfakakou D, Apostolou P, Konstantopoulou I, Kroupis C, et al. Functional characterization of CHEK2 variants in a Saccharomyces cerevisiae system. Hum Mutat 2019;40:631–48. [DOI] [PubMed] [Google Scholar]
- 35. Kleiblova P, Stolarova L, Krizova K, Lhota F, Hojny J, Zemankova P, et al. Identification of deleterious germline CHEK2 mutations and their association with breast and ovarian cancer. Int J Cancer 2019;145:1782–97. [DOI] [PubMed] [Google Scholar]
- 36. Cuella-Martin R, Hayward SB, Fan X, Chen X, Huang JW, Taglialatela A, et al. Functional interrogation of DNA damage response variants with base editing screens. Cell 2021;184:1081–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Robanus-Maandag E, Dekker M, van der Valk M, Carrozza ML, Jeanny JC, Dannenberg JH, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev 1998;12:1599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Boonen R, Rodrigue A, Stoepker C, Wiegant WW, Vroling B, Sharma M, et al. Functional analysis of genetic variants in the high-risk breast cancer susceptibility gene PALB2. Nat Commun 2019;10:5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 2013;8:2281–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Susswein LR, Marshall ML, Nusbaum R, Vogel Postula KJ, Weissman SM, Yackowski L, et al. Pathogenic and likely pathogenic variant prevalence among the first 10,000 patients referred for next-generation cancer panel testing. Genet Med 2016;18:823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER 3rd, Hurov KE, Luo J, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 2007;316:1160–6. [DOI] [PubMed] [Google Scholar]
- 42. Kranz A, Fu J, Duerschke K, Weidlich S, Naumann R, Stewart AF, et al. An improved Flp deleter mouse in C57Bl/6 based on Flpo recombinase. Genesis 2010;48:512–20. [DOI] [PubMed] [Google Scholar]
- 43. Bouwman P, van der Gulden H, van der Heijden I, Drost R, Klijn CN, Prasetyanti P, et al. A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov 2013;3:1142–55. [DOI] [PubMed] [Google Scholar]
- 44. Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 1991;108:193–9. [DOI] [PubMed] [Google Scholar]
- 45. de Jonge MM, Auguste A, van Wijk LM, Schouten PC, Meijers M, Ter Haar NT, et al. Frequent homologous recombination deficiency in high-grade endometrial carcinomas. Clin Cancer Res 2019;25:1087–97. [DOI] [PubMed] [Google Scholar]
- 46. Parameswaran B, Chiang HC, Lu Y, Coates J, Deng CX, Baer R, et al. Damage-induced BRCA1 phosphorylation by Chk2 contributes to the timing of end resection. Cell Cycle 2015;14:437–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J 2002;21:5195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hirao A, Kong YY, Matsuoka S, Wakeham A, Ruland J, Yoshida H, et al. DNA damage-induced activation of p53 by the checkpoint kinase Chk2. Science 2000;287:1824–7. [DOI] [PubMed] [Google Scholar]
- 49. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016;536:285–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019:531210. [Google Scholar]
- 51. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med 2015;17:405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Vroling B, Heijl S. White paper: The Helix Pathogenicity Prediction Platform; 2021. Available from: https://arxiv.org/abs/2104.01033.
- 53. Heijl S, Vroling B, Bergh Tvd, Joosten H. Mind the gap: preventing circularity in missense variant prediction; 2020.Available from: 10.1101/2020.05.06.080424. [DOI]
- 54. Li J, Taylor IA, Lloyd J, Clapperton JA, Howell S, MacMillan D, et al. Chk2 oligomerization studied by phosphopeptide ligation: implications for regulation and phosphodependent interactions. J Biol Chem 2008;283:36019–30. [DOI] [PubMed] [Google Scholar]
- 55. Cai Z, Chehab NH, Pavletich NP. Structure and activation mechanism of the CHK2 DNA damage checkpoint kinase. Mol Cell 2009;35:818–29. [DOI] [PubMed] [Google Scholar]
- 56. Lountos GT, Tropea JE, Zhang D, Jobson AG, Pommier Y, Shoemaker RH, et al. Crystal structure of checkpoint kinase 2 in complex with NSC 109555, a potent and selective inhibitor. Protein Sci 2009;18:92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ahn JY, Schwarz JK, Piwnica-Worms H, Canman CE. Threonine 68 phosphorylation by ataxia telangiectasia mutated is required for efficient activation of Chk2 in response to ionizing radiation. Cancer Res 2000;60:5934–6. [PubMed] [Google Scholar]
- 58. Oldenburg RA, Kroeze-Jansema K, Kraan J, Morreau H, Klijn JG, Hoogerbrugge N, et al. The CHEK2*1100delC variant acts as a breast cancer risk modifier in non-BRCA1/BRCA2 multiple-case families. Cancer Res 2003;63:8153–7. [PubMed] [Google Scholar]
- 59. Sharifi MJ, Zaker F, Nasiri N, Yaghmaie M. Epigenetic changes in FOXO3 and CHEK2 genes and their correlation with clinicopathological findings in myelodysplastic syndromes. Hematol Oncol Stem Cell Ther 2020;13:214–9. [DOI] [PubMed] [Google Scholar]
- 60. Chrisanthar R, Knappskog S, Lokkevik E, Anker G, Ostenstad B, Lundgren S, et al. CHEK2 mutations affecting kinase activity together with mutations in TP53 indicate a functional pathway associated with resistance to epirubicin in primary breast cancer. PLoS One 2008;3:e3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Matreyek KA, Starita LM, Stephany JJ, Martin B, Chiasson MA, Gray VE, et al. Multiplex assessment of protein variant abundance by massively parallel sequencing. Nat Genet 2018;50:874–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee CH, Chung JH. The hCds1 (Chk2)-FHA domain is essential for a chain of phosphorylation events on hCds1 that is induced by ionizing radiation. J Biol Chem 2001;276:30537–41. [DOI] [PubMed] [Google Scholar]
- 63. Jaganathan K, Panagiotopoulou SK, McRae JF, Darbandi SF, Knowles D, Li YI, et al. Predicting splicing from primary sequence with deep learning. Cell 2019;176:535–48. [DOI] [PubMed] [Google Scholar]
- 64. Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, et al. Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 2019;12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.