Pretreatment with an autophagy inhibitor before chemotherapy could prevent formation of therapy-induced dormant polyploid giant cancer cells, thereby reducing recurrence and metastasis of nasopharyngeal carcinoma.
Abstract
Dormant cancer cells that survive anticancer therapy can lead to cancer recurrence and disseminated metastases that prove fatal in most cases. Recently, specific dormant polyploid giant cancer cells (PGCC) have drawn our attention because of their association with the clinical risk of nasopharyngeal carcinoma (NPC) recurrence, as demonstrated by previous clinical data. In this study, we report the biological properties of PGCC, including mitochondrial alterations, and reveal that autophagy is a critical mechanism of PGCC induction. Moreover, pharmacologic or genetic inhibition of autophagy greatly impaired PGCC formation, significantly suppressing metastasis and improving survival in a mouse model. Mechanistically, chemotherapeutic drugs partly damaged mitochondria, which then produced low ATP levels and activated autophagy via the AMPK-mTOR pathway to promote PGCC formation. Analysis of the transcriptional and epigenetic landscape of PGCC revealed overexpression of RIPK1, and the scaffolding function of RIPK1 was required for AMPK-mTOR pathway-induced PGCC survival. High numbers of PGCCs correlated with shorter recurrence time and worse survival outcomes in patients with NPC. Collectively, these findings suggest a therapeutic approach of targeting dormant PGCCs in cancer.
Significance:
Pretreatment with an autophagy inhibitor before chemotherapy could prevent formation of therapy-induced dormant polyploid giant cancer cells, thereby reducing recurrence and metastasis of nasopharyngeal carcinoma.
Introduction
Although the majority of patients with nasopharyngeal carcinoma (NPC) do not present with overt metastases at diagnosis, a significant number succumb to disseminated disease years after the successful treatment of the primary tumor (1). Thus, late NPC recurrence may be the result of rare and elusive dormant cancer cells hiding in specialized niches being reactivated by specific signals (2). The concept of cancer dormancy has been described for the most common solid and hematologic cancers; however, the dormant cancer cells in NPC remain largely uncharacterized (2, 3). Due to their crucial role in recurrence and metastasis, it is important to develop proper models and approaches to study NPC dormancy in order to prevent disease relapse (4).
Although many factors contribute toward cancer cell dormancy, recent studies have demonstrated that cancer therapy can induce cellular dormancy (5, 6). Indeed, therapy-induced dormancy has been shown to lead to durable proliferation arrest, resulting in the formation of polyploid giant cancer cells (PGCC; refs. 7–9), which are a unique subpopulation of cancer cells that contribute toward the heterogeneity of solid tumors (6, 7, 10). Unlike regular-sized diploid cancer cells, PGCCs display distinct morphologic features, including a large cytoplasmic area and a high genomic content contained within a single highly enlarged nucleus or multiple nuclei (7, 9, 11, 12). Despite being present in low numbers, the frequency of PGCCs increases markedly after exposure to hypoxia and therapeutic interventions such as radiotherapy and chemotherapies (12–14). In addition, PGCCs can give rise to daughter cells (regular-sized and ploidy) via asymmetric cell divisions like splitting, budding, and bursting, which usually occur during the division of low-level eukaryotes, plants, and viruses (7). In the past decade, studies have reported that PGCCs persist in multiple types of cancer (glioma, breast, ovarian, colon, melanoma, lung, and renal) and contribute toward cancer maintenance, metastasis, and chemoresistance (8, 11, 14–23). However, little is known about the mechanisms underlying PGCC formation.
In this study, we attempted to investigate the role of autophagy in PGCC formation as well as the underlying mechanisms. Furthermore, we validated our findings using a highly relevant clinical orthotopic model of imageable NPC and used clinical data to determine the relationship between PGCC number and patient recurrence and outcomes. Together, our findings suggest that autophagy inhibition prevents therapy-induced dormant PGCC formation and thereby prevents NPC metastasis.
Materials and Methods
Human NPC specimens
Tissue samples were collected from pathologically confirmed cases of NPC at the Affiliated Hospital of Nantong University according to approval by its ethics committee (Institutional Review Board number: 2020-L160). All patients provided informed consent and had not received any cancer therapies prior to the biopsy. Tissue microarrays were performed by Outdo Biotechnology to examine the number of PGCCs. Detailed clinicopathologic features of the patients are listed in Supplementary Table S1.
Cell culture and PGCC formation
The human NPC cell lines 5–8F and CNE-2 were gifted from the Sun Yat-Sen University (Guangzhou, Guangdong, China) and Xiang-Ya School of Medicine (Changsh, Hunan, China) and were routinely tested for Mycoplasma. The cells were maintained in RPMI 1640 (Biological Industries; 01–100–1ACS) with 10% FBS (Biological Industries; 04–001–1ACS) as described previously (24). The CNE-2 cell line was recently authenticated using short tandem repeat analysis (25).
At 80% confluence, cells were treated with 150 ng/μL paclitaxel (Nanjing Luye Pharmaceutical Co., Ltd) for 18 hours and then allowed to recover for 6 to 7 days in regular medium (replaced every 3 days) for PGCC formation. At 9 to 10 days, the PGCCs generated daughter cells via asymmetric cell division.
Animal procedures
The in vivo experiments were approved by the Committee on the Ethics of Animal Experiments of Nantong University (RDD number: S20200323–040). All mouse experiments were conducted according to NIH Guidelines and were approved by the Administration Committee of Experimental Animals, Jiangsu Province, China [Approval ID: SYXK (SU) 2007–0021].
Orthotopic NPC xenografts were established using 6-week-old BALB/c mice. Briefly, mice were subjected to deep anesthesia with chloral hydrate (4% solution, 400 mg/kg) and positioned in a stereotactic frame in a supine position. The mouth was held open with the tongue pulled aside and 50-μL PBS containing 2 × 106 CNE-2 cells was injected just distal and lateral to the hard palate using a 1 mL sterile syringe. Drug treatment was started 4 days after injection. NPC tumor metastases away from the orthotopic site and orthotopic tumor growth were monitored twice a week using bioluminescence imaging (BLI) with an IVIS Lumina Series III (Caliper Life Sciences).
Scoring for PGCCs
For cultured cells and tissue sections, nucleus area measurement was performed with Zen and Image J software as described previously (26). If the nuclear area exceeded 350 μm2 in size, it was defined as a PGCC.
Immunofluorescence, Western blot, and IHC
The assays for immunofluorescence (IF), Western blot, and IHC were performed as described previously (27) using the antibodies listed in Supplementary Table S2. More details are provided in Supplemental Methods. Full Western blot images are shown in Supplementary Fig. S1.
RNA sequencing and bioinformatics analysis
RNA sequencing (RNA-seq) and bioinformatics analysis were conducted by GENEWIZ. Briefly, after total RNA had been extracted, qualified, and quantified, next-generation sequencing libraries were prepared according to the manufacturer's instructions. RNA-seq was conducted on an Illumina HiSeq instrument (Illumina) according to the manufacturer's instructions (28). Clean data were analyzed using Hisat2 and HTSeq softwares, while the DESeq2 Bioconductor package was used for differential expression analysis.
Transfection and transduction
Transfection was performed as described previously (25) using ATG5–short hairpin RNA (shRNA) plasmids from Shanghai Genechem. Tandem monomeric red fluorescent protein (RFP)-GFP–tagged LC3 (Shanghai Genechem, tfLC3) was used to determine autophagic flux, as described previously (24).
Evaluation of mitochondrial morphology, function, and mitophagy
To measure mitochondrial content and mitochondrial morphology, cells were incubated with 200 nmol/L MitoTracker green mitochondrial probe (Cell Signaling Technology, #9074) for 30 minutes at 37°C, collected, and live-imaged immediately using a confocal microscope (Leica Microsystems, TCS SP-5). Mitochondrial volume and length were analyzed using the ImageJ software.
For the JC-1 assay, cells were stained using an MMP assay kit (Abcam, ab113850) according to the manufacturer's instructions.
Cellular ATP levels were measured using an ADP/ATP ratio assay kit (Bioluminescent; Abcam, ab65313) according to the manufacturer's instructions. Briefly, 100 μL of the reaction mix was added to obtain the background luminescence (data A). The cells were then incubated with 50 μL of nucleotide-releasing buffer for 5 minutes at 37°C and transferred into luminometer 96-well plates to be read (data B). “Data B – data A” refer to the ATP measurements. The samples were then read again to measure ADP levels (data C) before 10 μL of 1× ADP converting enzyme was added for 2 minutes and the samples were read one final time (data D). The ADP/ATP ratio was calculated as: ADP/ATP ratio = [data D – data C]/[data B – data A].
pMitoTimer plasmid was a gift from Zhen Yan (Addgene plasmid #52659; RRID: Addgene_52659). The pCHAC-mt-mKeima plasmid was a gift from Richard Youle (Addgene plasmid #72342; RRID: Addgene_72342; ref. 29). After 36 hours of transfection, live cell imaging was done using a confocal microscope.
Transmission electron microscopy
Transmission electron microscopy was performed by Nova Biotechnology as described previously (24).
Cleavage under targets and tagmentation and assay for transposase-accessible chromatin using sequencing for PGCCs and CNE-2 cells
Cleavage under targets and tagmentation (CUT&Tag) assay for the enhancer histone mark H3K27ac were conducted and analyzed by DIATRE Biotechnology and using NovoNGS CUT&Tag 2.0 High-Sensitivity Kit (for Illumina; Novoprotein Scientific Inc., catalog no. N259-YH01–01A), as described previously (30).
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) for the PGCCs and CNE-2 cells was performed and analyzed by Guangzhou Epibiotek Co., Ltd as described (31). Libraries were quantified with Bioptic Qsep100 Analyzer (Bioptic lnc.) and the paired-end were sequenced with read lengths of 150.
Data and materials availability
RNA-seq, CUT&Tag, and ATAC-seq data in this study are deposited in the NCBI SRA database with the accession of PRJNA731027.
Quantification and statistical analysis
Statistical analysis was performed using the GraphPad Prism 6 and SPSS 17.0 software. Details of the statistical analyses performed for different data are shown in each figure legend. P values of < 0.05 were considered significant.
Results
PGCCs display distinct morphologic feature
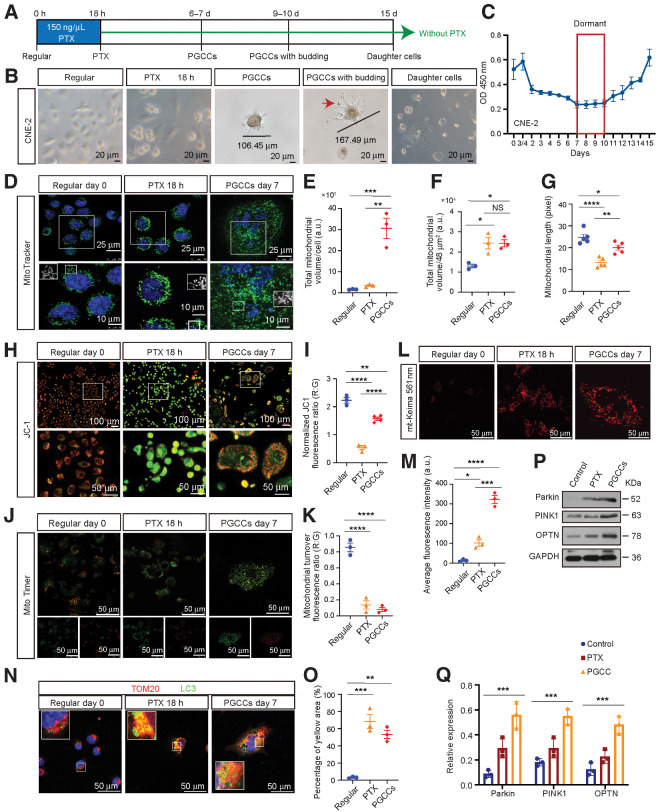
It was reported that PGCCs can be induced from normal cancer cells by the hypoxia-mimetic agent CoCl2 (7) or paclitaxel (9). When tumor cells were exposed to paclitaxel (15–150 ng/μL), cell death increased with paclitaxel concentration (Supplementary Fig. S2A). We used the maximum concentration of paclitaxel to ensure that the majority of tumor cells were killed (Supplementary Fig. S2A), and this concentration was far above the IC50 value (Supplementary Fig. S2B). The experimental design is shown in Fig. 1A. Similar with previous reports, from the first day after paclitaxel exposure, the diploid cells began to die and the surviving cells entered the PGCC cell cycle. After recovery for 7 days, only a few cells survived to generate PGCCs (Fig. 1B and C; Supplementary Fig. S2C–S2E). For the next 3 days, the PGCCs remained dormant; however, some small daughter cells began to bud 10 days after paclitaxel treatment (Fig. 1B and C; Supplementary Fig. S2C–S2E). As the time spent in recovery culture increased, many smaller daughters' cells were generated via burst-like cell divisions in a short period of time (more than 12 hours; Fig. 1B and C; Supplementary Fig. S2C–S2E). The quantification of PGCC numbers were shown in Supplementary Fig. S2F.
Figure 1.
Morphologic characteristics, mitochondrial morphology, and function analysis of PGCCs. A, Experimental design for PGCC induction and daughter cell generation in vitro. B, Light phase contrast microscopy images of cells at the indicated time points. Arrow, the mononuclear daughter cells. C, The CCK8 assays were performed to measure cell proliferation. O.D., optical density. D, Mitochondrial morphology was visualized using MitoTracker. E and F, Mitochondrial volume/cell was measured in three fields per group, with three independent replicates. a.u., arbitrary units. G, Mitochondrial length was measured in five fields per group, with three independent replicates. H, Confocal microscopy of mitochondrial membrane potential change with JC-1. Red puncta, maintained mitochondria; green puncta, depolarized mitochondria. I, The quantification of JC-1 results. J, Mitochondrial turnover visualized by MitoTimer transfection and labeling. K, The histogram shows the ratiometric quantification of three independent experiments of 30 cells per group. R:G, red:green. L, Fluorescent images of cells transfected with plasmid mt-Keima (mitophagy reporter). M, Quantification of average fluorescence intensity, with three independent replicates of 30 cells per group. N, IF costaining of TOM20 and LC3. Blue, nucleus; red, TOM20; green, LC3. O, Quantification of the percentage of yellow area. P, Western blot analysis. Q, Gray value analysis of Western blot. All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant. PTX, paclitaxel.
Consistent with previous reports from Liu's reports (7, 9), the PGCCs displayed distinct morphologic features, characterized by a large cytoplasmic region, high genomic content contained (Fig. 1B; Supplementary Fig. S2C–S2E) within multiple nuclei (Supplementary Fig. S2G), and polyploid population (Supplementary Fig. S2H–S2J). In addition, LIVE/DEAD viability analysis revealed that the dormant PGCCs were alive (Supplementary Fig. S2K), while β-galactosidase staining showed that although the PGCC bodies were not senescent, the area with budding daughter cells displayed a senescent phenotype (Supplementary Fig. S2L). Furthermore, paclitaxel treatment altered the cell cycle by sharply decreasing the number of cells in the G0-G1 phase and increasing in G2-M (Supplementary Fig. S2M). Moreover, cyclin E expression was higher in the PGCCs and cyclin B1 was dramatically lower, whereas no obvious change was observed in cyclin D1 and CDK8 (Supplementary Fig. S2N). Therefore, we concluded that PGCCs are a specific cell type that contribute toward the heterogeneity of solid tumors.
PGCCs survive paclitaxel treatment by removing damaged mitochondria
Mitochondria are essential bioenergetic and biosynthetic organelles that are critical for adaptation to changes in their environment, such as nutrient depletion and cancer treatments. Therefore, we examined mitochondrial morphology and function by assessing mitochondrial volume and shape (elongated/fragmented) using MitoTracker. We found that PGCCs displayed a significantly higher mitochondrial volume than the control cells (Fig. 1D–F). Notably, the interconnected network structure of mitochondria in regular cells dispersed into fragmented units after exposure to paclitaxel for 18 hours; however, the structure recovered over time, indicating that PGCCs may respond to cancer treatments to remove damaged mitochondria (Fig. 1D–G). In addition, mitochondrial membrane potential (MMP), an important parameter of mitochondrial function, was measured by JC-1 staining. Increased number of unhealthy “green”-labeled low membrane potential mitochondria were observed in paclitaxel treatment cells compared with that in the controls; such unhealthy mitochondria was partly rescued in PGCCs (Fig. 1H and I).
Mitophagy is the selective form of autophagy that is devoted to the selective sequestration and degradation of damaged mitochondria. Therefore, MitoTimer labeling was used to visualize mitochondrial turnover real-time in living cells. MitoTimer is a mitochondrial matrix-targeting Timer fluorescent protein that irreversibly changes its emitted fluorescence from green to red over time. As expected, cells exposed to paclitaxel for 18 hours and PGCCs induced less “Red” mitochondria accumulation than the control cells, supporting more mitochondrial turnover (Fig. 1J and K). Furthermore, we measured mitophagic flux by employing live-cell imaging of mitochondria-matrix targeted Keima (mt-Keima). The red color of mt-Keima protein increased when mitophagy activation and delivery of mitochondria to lysosomes. Dramatically increased red fluorescence was found in the cells exposed to paclitaxel for 18 hours and PGCCs, indicating mitophagy activation and delivery of mitochondria to lysosomes (Fig. 1L and M). Next, mitophagy levels were evaluated by IF costaining for TOM20 (mitochondrial marker) and LC3 (autophagy marker), indicated a marked increase in mitophagy activation in PGCCs (Fig. 1N and O). PINK1/Parkin signaling has been reported to play an important part in mitophagy. An increased expression of PINK1, Parkin, and mitophagy adapter OPTN in PGCCs were observed (Fig. 1P and Q). Therefore, these findings indicate that PGCCs survive paclitaxel treatment by removing damaged mitochondria via significant mitophagy induction.
Autophagy inhibition prevents dormant PGCC formation
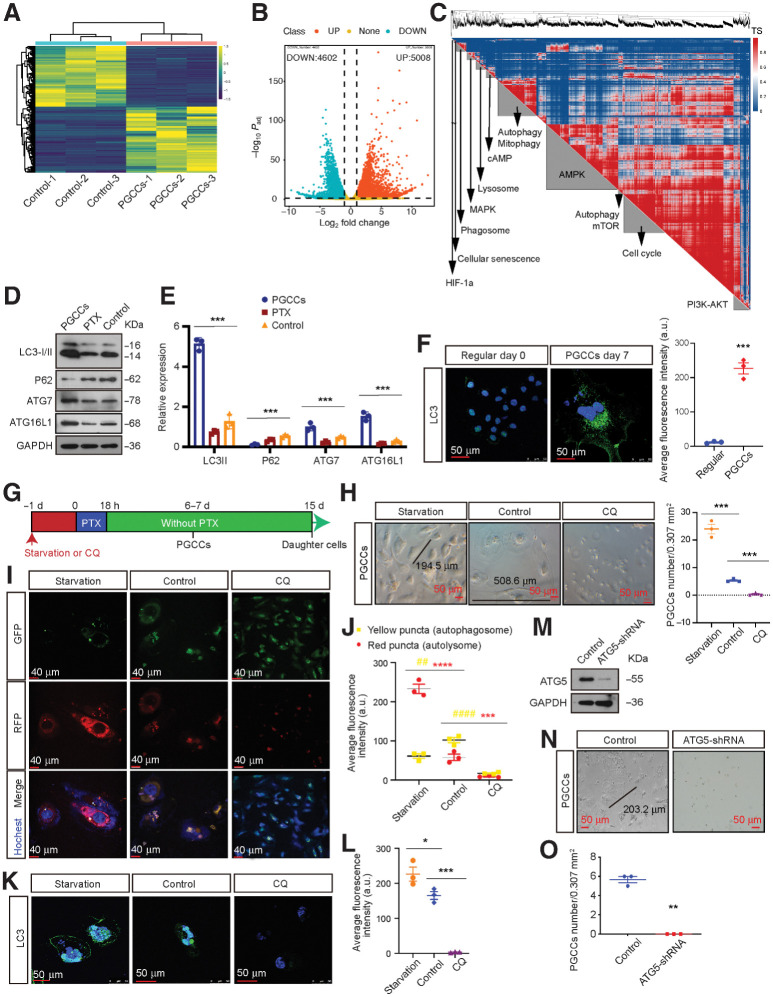
A total of 9610 differentially expressed genes (DEGs) were detected (Fig. 2A and B) though RNA-seq, indicating that considerable alterations in gene expression occur when PGCCs are generated. Next, we produced a network of the DEGs by performing genetic cosimilarity analysis to identify gene pairs, and the high-ranking correlated gene pairs within clusters were then enriched for shared pathway annotations. As expected, we identified functional modules related to pathways such as autophagy, the cell cycle, MAPK, mTOR, and AMPK (Fig. 2C). Notably, PGCCs showed a marked increase in the expression of autophagy-related genes (ATG) and LC3B (Fig. 2D–F). Since cisplatin-based chemotherapy is the most frequently used clinical treatment for NPC, we also examined the effects of cisplatin on PGCC formation. The frequency of PGCCs increased markedly after 2 weeks of cisplatin treatment (Supplementary Fig. S3A and S3B) and high levels of autophagy were observed in cisplatin-induced PGCCs (Supplementary Fig. S3C–S3E).
Figure 2.
Autophagy is activated in PGCCs and promote PGCC formation. A, DEGs were identified using three independent RNA-seq experiments. B, Upregulated and downregulated DEGs. C, DEG genetic cosimilarity map. Representative pathways enriched in A cluster are shown on the diagonal axis (FDR < 1%). TS, topology similarity. D, Western blot analysis of autophagy-related genes levels. E, Gray value analysis of Western blot. F, Left, IF detection of endogenous LC3 puncta. Right, total number of endogenous LC3 puncta per cell (Student t test). G, Experimental design for investigating the role of autophagy in PGCC formation. H, Left, representative light microscopy images of PGCC formation. Right, quantification of PGCC number (one-way ANOVA). I, Detection of autophagic flux using the mRFP-GFP-LC3 reporter. J, Autophagic flux analysis (two-way ANOVA). K, IF detection of endogenous LC3 puncta. L, Quantification of average fluorescence intensity per cell (one-way ANOVA). M, Interference efficiency was detected by Western blot in cells transfected with lentiviral ATG5-shRNA or mock vector. N, Representative light microscopic images of PGCC formation. O, Quantification of PGCCs number using Student t test. All data represent the mean ± SEM of three independent experiments. P values were calculated using one-way ANOVA or Student t tests or two-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. a.u., arbitrary units; CQ, chloroquine.
Next, we investigated the functional importance of autophagy activation in PGCC formation by treating NPC cells with Earle balanced salt solution (EBSS; an autophagy activator) or 10 μmol/L chloroquine (an autophagy inhibitor) prior to paclitaxel exposure for PGCC formation (Fig. 2G). We found that autophagy activation markedly increased PGCC induction, consistent with the significant autophagosome and autolysosome accumulation observed, whereas inhibiting autophagy significantly decreased chemotherapy-induced PGCC formation (Fig. 2H–L). Similar results were observed in NPC cells following ATG5 knockdown (Fig. 2M–O).
Chemotherapy drugs induce PGCC formation by activating autophagy via the AMPK-mTOR pathway
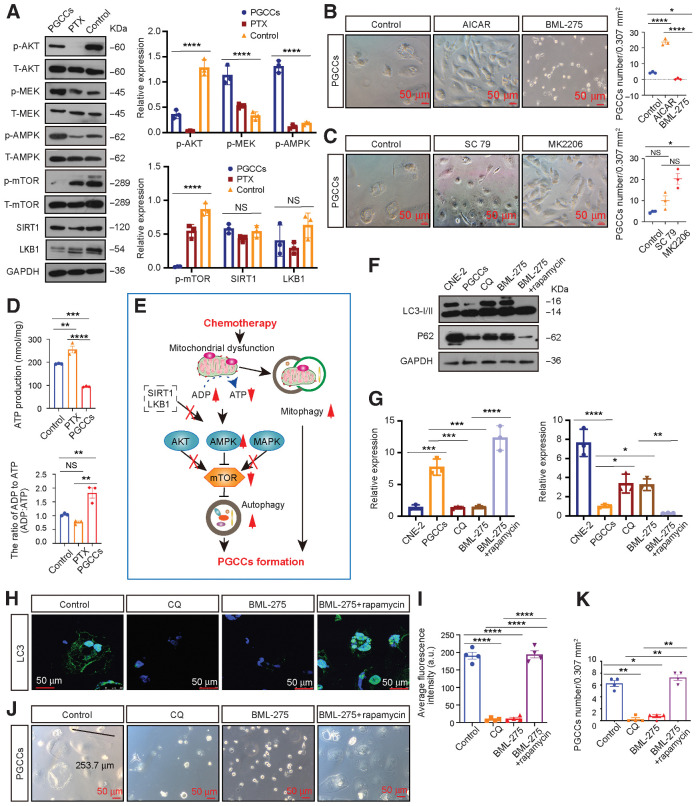
To elucidate the molecular mechanism driving the dependence of PGCC formation on autophagy, we verified the modulation of previously reported autophagic signaling pathways (32); for this, firstly, we examined changes in AKT/MAPK/AMPK/mTOR expression. As expected, the negative modulators of autophagy p-AKT and p-mTOR were downregulated in PGCCs and the positive modulator p-AMPK was upregulated (Fig. 3A); however, p-MEK, a negative modulator of autophagy, was also upregulated in PGCCs, indicating that PGCC formation does not depend on the MAPK pathway (Fig. 3A).
Figure 3.
Autophagy activation promotes PGCC formation by activating AMPK-mTOR signaling. A, Left, Western blot analysis. Right, gray value analysis. B, Left, rescue experiments with PGCC formation. Right, quantification of PGCCs number. C, Left, rescue experiments with PGCC formation. Right, quantification of PGCCs number. D, Relative mitochondrial ATP production and cellular ADP:ATP ratio. E, Schematic overview. F, Western blot analysis. G, Gray value analysis of Western blot. H, Endogenous LC3 puncta were detected using IF. I, Quantification of average fluorescence intensity per cell (one-way ANOVA). J, Rescue experiments for PGCC formation using AMPK-mTOR signaling inhibitors. K, Quantification of PGCC number. All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant. CQ, chloroquine.
Next, we investigated the relative contributions of these pathways to PGCC formation. AMPK activation (AICAR) or suppression (BML-275) significantly affected PGCC formation, whereas activating (SC-79) or inhibiting (MK2206) AKT activity failed (Fig. 3B and C), suggesting that autophagy-dependent PGCC formation is the result of AMPK-mTOR pathway activation.
We then examined how the AMPK-mTOR pathway was activated. LKB1 and STR1, that were previously reported upstream of AMPK, were observed to have no significant changes (Fig. 3A). Since mitochondria can be damaged by chemotherapy treatments, we further detected and found that PGCCs displayed significantly lower ATP production and a high ADP:ATP ratio, which reflects the cellular energy requirement and can be sensed by the intracellular sensor AMPK (Fig. 3D). Consequently, we hypothesized that chemotherapy drugs can damage mitochondria, which then produce lower ATP levels and activate autophagy via the AMPK-mTOR pathway to promote PGCC formation (Fig. 3E). To test this hypothesis, we performed rescue experiments. Suppressing AMPK activity reduced PGCC formation and autophagy; however, this phenotype was abolished following mTOR inhibition by rapamycin (Fig. 3F–K).
Preventing PGCC formation via autophagy inhibition reduces NPC metastasis in vivo
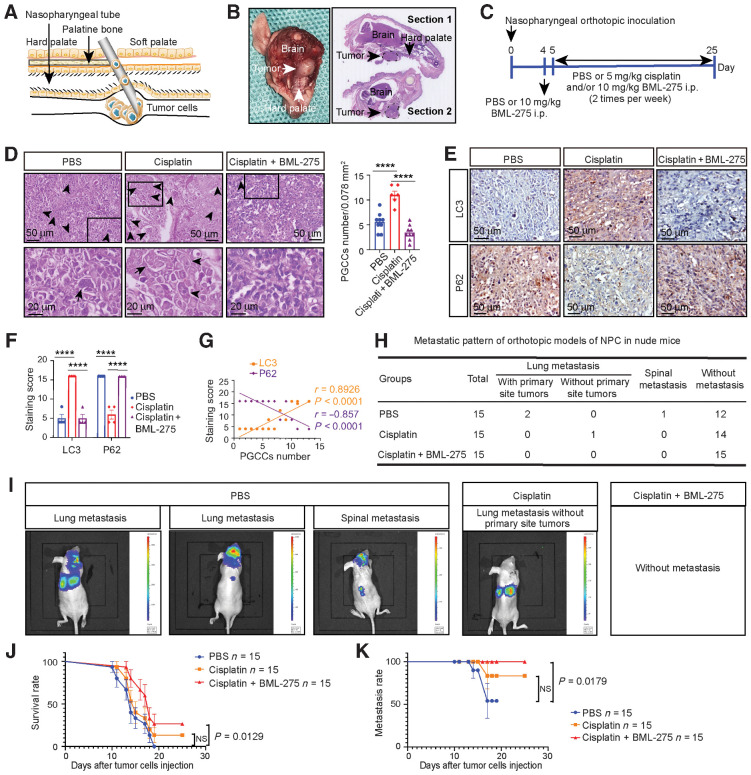
Our findings suggest that chemotherapy drugs can induce dormant PGCC formation by activating autophagy, thus contributing toward NPC recurrence and metastasis. Therefore, we determined whether inhibiting autophagy prior to chemotherapy can eliminate NPC metastasis by preventing PGCC formation. We used a highly clinically relevant imageable NPC orthotopic mouse model that allowed us to approximate tumor metastasis and the response to chemotherapeutic agents in patients. Luciferase-tagged CNE-2 cells were orthotopically injected into the nasopharynx of nude mice (Fig. 4A and B) according to the NPC orthotopic procedures and treatment strategies shown in Supplementary Fig. S3A and Fig. 4C. After injection, the tumors were monitored before being collected and analyzed 25 days postinjection. Tumors cotreated with BML-275 and cisplatin (BML-275 pretreatment 1 day before cisplatin treatment, followed by cotreatment with BML-275 and cisplatin) displayed inhibited PGCC formation (Fig. 4D) and decreased autophagy (Fig. 4E and F). In addition, levels of autophagy significantly correlated with the number of PGCCs in tumors (Fig. 4G).
Figure 4.
AMPK inhibition with BML-275 before chemotherapy prevents metastasis and increases survival rate by preventing PGCC formation. A, Illustration of the tissue details for nasopharyngeal orthotopic implantation. B, Histology of nasopharyngeal orthotopic tumors. Left, gross observation. Right, hematoxylin and eosin staining for tumor locations (dotted black line). C, Schematic of the detailed treatment strategy. D, Left, hematoxylin and eosin analysis of PGCC number in tumor tissues. Arrow, representative PGCC. Right, quantification of PGCC number (one-way ANOVA). E, IHC analysis of tumor LC3 and P62 expression. F, Quantification of IHC staining (one-way ANOVA). G, Pearson correlation between LC3/P62 expression and PGCC number (linear regression). H, Pattern of NPC metastasis. I, Whole-body BLI was used to track tumor metastasis from the orthotopic site. J and K, Kaplan–Meier analysis was used to compare overall survival (J) and overall metastasis (log-rank test; K). All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. ****, P < 0.0001; NS, not significant.
The NPC tumor metastasis away from the orthotopic site was also monitored, and the patterns of metastasis were recorded (Fig. 4H and I). The mice without chemotherapy had the highest incidence of metastatic dissemination, with the majority of metastases occurring in the lungs and spine. Notably, although the rate of metastasis was reduced following cisplatin treatment, some mice displayed metastatic dissemination after the successful treatment of the primary orthotopic tumor. Importantly, no metastases were observed in mice that received BML-275 and cisplatin cotreatment, which was also associated with the best survival rate and lowest metastasis rate (Fig. 4J and K). To make sure whether the reduced NPC metastasis via targeting autophagy is responsible for the difference in PGCCs number, two groups of different number of PGCCs were directly injected into the nasopharynx and subcutaneous of nude mice, respectively. The mice with more PGCCs transplantation had the higher metastatic dissemination rate (Supplementary Fig. S3F and S3G), poor survival rate (Supplementary Fig. S3H) and the larger transplantation tumor volume (Supplementary Fig. S3I and S3J). In addition, similar results were observed when pretreated mice with hydroxychloroquine (autophagy inhibitor) instead of BML-275 (Supplementary Fig. S4A–S4I).
Upregulated RIPK1 acts as a scaffold to prevent PGCC death via the AMPK-mTOR pathway
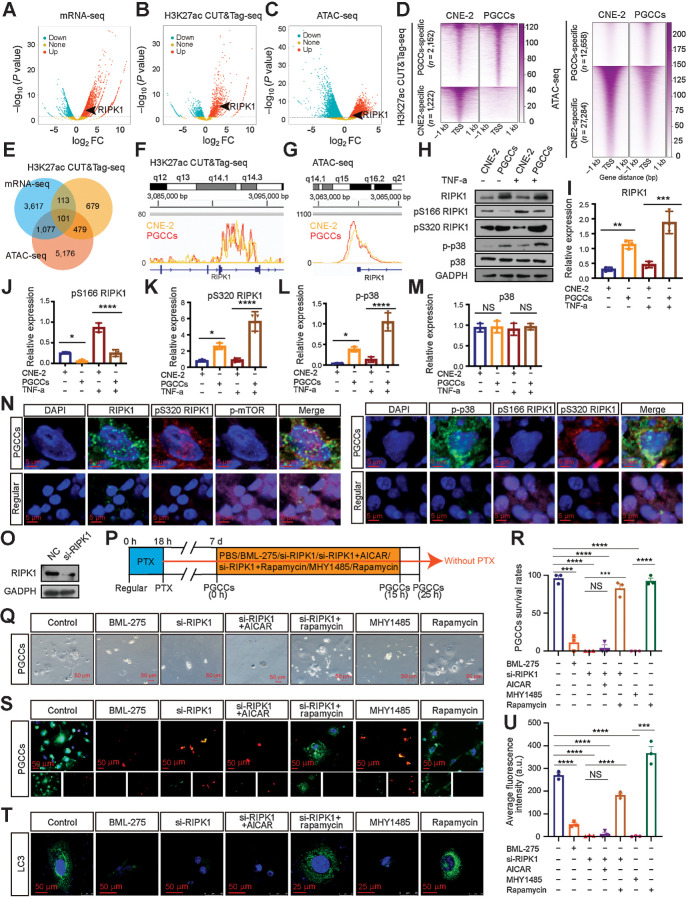
Based on the above results, autophagy-dependent PGCC formation is the result of AMPK-mTOR pathway activation. We wanted to ascertain whether epigenomic alterations play critical roles in promoting PGCC survival by driving expression of specific genes, as PGCCs may remain dormant in patients for a long time. To map the enhancer landscape of the PGCC genome, the CUT&Tag sequencing (CUT&Tag-seq) assay was performed for the enhancer histone mark H3K27ac and the open chromatin regions were assessed by the ATAC-seq as well as RNA-seq for DEGs (Fig. 5A–C). The data revealed categories of peaks that are either specific to or shared between the PGCC and normal CNE-2 cells. Overall, we identified that 2,125 CUT&Tag peaks were PGCC-specific and 1,222 peaks were CNE-2–specific (Fig. 5B and D). Meanwhile, 12,658 ATAC-seq peaks were identified to be PGCC-specific and 27,284 peaks were CNE-2–specific (Fig. 5C and D). Furthermore, statistical analysis of the RNA-seq, CUT&Tag, and ATAC-seq data revealed that 101 transcriptionally activated genes identified by RNA-seq were mapped to the PGCC-specific CUT&Tag and ATAC-seq peaks (Fig. 5E–G). Among the 101 activated genes, the expression level of receptor-interacting protein kinase 1 (RIPK1), which is reported to be related to autophagy (33), was markedly increased to seven-fold in the PGCCs (Supplementary Table S3).
Figure 5.
RIPK1 acts as a scaffold to promote PGCC survival by the AMPK-mTOR pathway. A–C, Volcano plot showing the log2-fold change (FC) vs.–log10 (P value) of differential mRNA expression (A), H3K27ac enrichment (B), and ATAC enrichment (C) identified between PGCCs and CNE-2 cells. Significantly up or down are selected according to P < 0.05. D, Heatmap of H3K27ac and ATAC signals at sites unique for PGCCs and CNE-2 cells. Signal density measured ±1 kb from the center regions is defined as tag density/bp. E, Venn diagram. F and G, H3K27ac CUT&Tag-seq signals (F) and ATAC-seq signals (G) at RIPK1 loci. H, Western blot analysis. I–M, Gray value analysis of Western blot. N, IF costaining in PGCCs and regular CNE-2 cells. O, Interference efficiency was detected by Western blot. P, Experimental design for investigating the role of autophagy in PGCC survival. Q, Rescue experiments for PGCC survival. R, Quantification of PGCC number. S, PGCCs viability analysis via LIVE/DEAD Viability assay. Scale bars, 50 μm. T, Endogenous LC3 puncta were detected using IF. U, Quantification of average fluorescence intensity per cell (one-way ANOVA). All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; NS, not significant. a.u., arbitrary units; mRNA-seq, mRNA sequencing; PTX, paclitaxel.
Previous studies reported that RIPK1 regulates two opposite cell fates through kinase-dependent and -independent mechanisms (34–36). As a scaffold, RIPK1 inhibits cell apoptosis and necroptosis though various signaling pathways. As a kinase, RIPK1 paradoxically induces cell death modalities. To evaluate the role of RIPK1 in determining the cell fate of PGCCs, we detected the expression of pS166 RIPK1, as RIPK1 autophosphorylation on Ser166 is responsible for low levels of RIPK1 enzymatic activity (35). Remarkably, although RIPK1 in the PGCCs was overexpressed, the Ser116 phosphorylation was markedly inhibited (Fig. 5H–J and N). As recent studies have demonstrated that p38 and its substrate MK2 play essential roles in inhibiting RIPK1 kinase activation by directly phosphorylating RIPK1 at S320/S321 (34, 37), we sought to determine the expression of the p38-Ser320/S321 axis. The levels of p-p38 and Ser320/S321 phosphorylation was increased in PGCCs in response to TNF (Fig. 5H, K–N), which were consistent with the inhibited enzymatic activity. Therefore, rescue experiments were performed to investigate the contributions of the RIPK1 scaffold to AMPK-mTOR–mediated PGCC survival. We observed that activating AMPK activity failed to promote PGCC survival and autophagy once RIPK1 was knocked down (Fig. 5O–U). Additionally, the prodeath and autophagy inhibition role of RIPK1 siRNA was abolished following mTOR inhibition by rapamycin (Fig. 5O–U). Collectively, these findings indicate that transcriptionally activated RIPK1 acts as a scaffold, rather than a kinase, to prevent PGCC death via the AMPK-mTOR pathway.
PGCC number is a valuable biomarker prior to NPC therapy
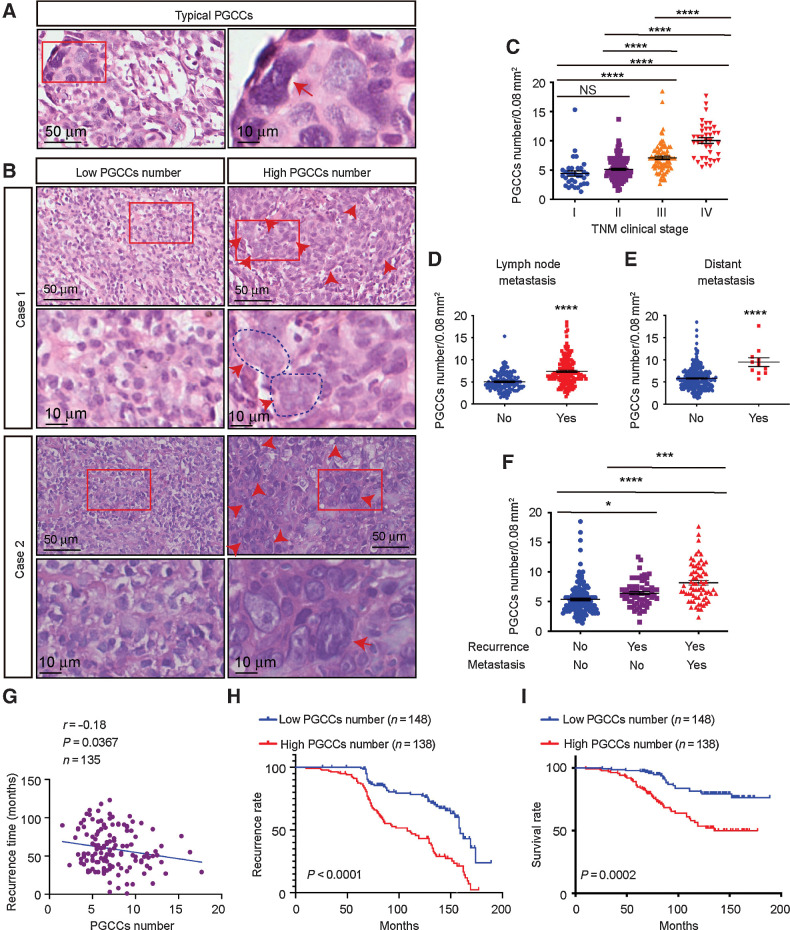
Since there are factors besides therapeutic intervention, like hypoxia, that contribute toward PGCC formation, we confirmed the presence of PGCCs, defined as cells with irregularly giant or multiple nuclei, in NPC tissue microarrays from 286 patients who did not receive any cancer therapies (Fig. 6A and B). Statistically, the number of PGCCs was higher in patients with lymph node metastases, distant metastases, and advanced clinical stage (Fig. 6C–E). In addition, the number of PGCCs was significantly higher in recurrent specimens and even higher in specimens from patients who developed distant recurrent metastasis (Fig. 6F), indicating that high PGCC levels correlate with NPC recurrence.
Figure 6.
Presence and clinical significance of PGCCs in NPC. A, Representative hematoxylin and eosin staining of typical PGCCs in NPC tissue sections. B, Representative image of PGCCs in NPC tissue microarrays. C–F, PGCC number in different clinical stages (C), lymph node metastases (D), distant metastases (E), and recurrence (F). G, Pearson correlation between recurrence time and PGCC number (linear regression). H and I, Kaplan–Meier analysis was used to compare overall recurrence (H) and overall survival (log-rank test; I). All data represent the mean ± SEM of at least three independent experiments. P values were calculated using one-way ANOVA. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001; NS, not significant.
Finally, we performed Pearson correlation analysis to confirm that patients with higher numbers of PGCCs exhibited shorter recurrence times (Fig. 6G). PGCC number was defined as low (<6) or high (≥6) using the X-tile Software, and patients with a high PGCC number were found to display a higher recurrence rate (P < 0.0001; Fig. 6H) and worse clinical outcomes (P = 0.0002; Fig. 6I). This finding was confirmed using another cohort with 67 patients from our hospital (Supplementary Fig. S5A–S5E). Together, these data suggest that PGCC number is a valuable biomarker for predicting future metastases, recurrence, recurrence time, and patient survival prior to NPC therapy.
Discussion
The majority of recurrence and disseminated metastatic events are caused by the reactivation of dormant cancer cells (38). Unfortunately, the study of dormancy has been limited by a lack of suitable models that appropriately recapitulate this stage of metastatic spread as well as techniques to track and visualize dormant cancer cells (39). During dormancy, cancer cells are thought to enter a state of “temporary mitotic arrest” (39, 40). According to this theory, we confirmed that therapy-induced PGCCs are dormant as they undergo cell cycle arrest. Unlike conventional dormant cancer cells, PGCCs display similarities to diapause, a dormant stage of suspended embryonic development triggered by stress and unfavorable environmental conditions (41–43).
Like traditional dormant cancer cells, when dormant PGCCs are reactivated, countless small daughter cells are generated via burst-like divisions in a very short period of time (8). Importantly, we found that daughter cells budding from PGCCs may cause disseminated metastases because of their markedly decreased adhesion, which allows them to be shed easily from the primary site, data not shown. The clinical risks of dormant PGCCs in future metastases and recurrence were further clarified using clinical data from 286 patients with NPC.
This study is the first to report the successful induction and culture of PGCCs from NPC cell lines with paclitaxel or cisplatin. Paclitaxel is essential for stabilizing microtubules, inhibition of mitosis, ultimately leading to cancer cell death (44). Cytotoxicity of cisplatin is mainly due to interaction with DNA and thus inducing genomic DNA damage (45). Although paclitaxel and cisplatin are both widely used chemotherapeutic agents, the mechanisms that induce PGCCs may be widely different. Paclitaxel may induce PGCCs through arresting cells in G2–M phases, while cisplatin through DNA damage and DNA repair. Consistent with previous studies of PGCCs from other cancer cells, NPC-PGCCs displayed distinct morphologic features, including a high genomic content, cell cycle arrest, and special cell division types (7). What's more, stem-like properties and selectively turning off protein translation were observed in NPC-PGCCs, data not shown. Although mitochondria have been reported to participate in cellular adaptation to environmental changes, this is the first study to report significant alterations in mitochondrial morphology and function in PGCCs, including a significant increase in mitochondrial volume and lower ATP production. In addition, our results suggest that cancer treatment leads to significant structural mitochondrial damage that recovers over time because of the activation of mitophagy, which removes damaged mitochondria and replaces them with newly formed healthy ones. Our data provide new insights into the clinical application of mitochondria-related therapies targeting dormant cancer cells.
Autophagy plays a complex and context-dependent role in cancer as it can both abolish tumorigenesis and protect cancer cells by providing a cellular rescue mechanism under adverse conditions (46). Studies of dormant cancer cells have identified that the maintenance of a dormant state depends on autophagy (28, 47, 48). Our findings are consistent with those of recent studies, confirming that autophagy is activated in dormant PGCCs; however, few studies have investigated autophagy induction in dormant cancer cells (49). Our data based on transcriptomic data revealed that autophagy-dependent PGCC formation was the result of AMPK-mTOR pathway activation. AMPK levels and activity are reported upregulation in dormant breast cancer cells and that its pharmacologic or genetic inhibition promotes the clearance of residual dormant breast cancer cells (48). Our study also elucidated the role of autophagy and AMPK activation in PGCC formation and highlighted the potential for the development of effective therapies targeting autophagy to either prevent or eradicate therapy-induced PGCC formation and prevent clinical disease relapse.
Despite numerous preclinical trials having demonstrated that autophagy could be targeted as a potential cancer therapy, no drugs designed to modulate autophagy have yet been FDA-approved to treat NPC (50). Our extended follow-up of the NPC orthotopic mouse model cotreated with an autophagy inhibitor (hydroxychloroquine or BML-275) and cisplatin confirmed the therapeutic efficacy of autophagy inhibition against PGCC formation, since we repeatedly observed a significant decrease in NPC tumor metastasis and the highest survival rate in these mice. Moreover, our data highlight that treatment “timing” (i.e., the right order for the right treatment) should be an important consideration in precision medicine and therapeutic development. Furthermore, our findings indicate that pretreatment with an autophagy inhibitor before chemotherapy could maximize the therapeutic index.
Therefore, we future elucidated the epigenetic reprogramming mechanisms underlying dormancy induction and maintenance in PGCCs. Our data from the H3K27ac CUT&Tag-seq and ATAC-seq and the subsequent experiments suggest that transcriptionally activated RIPK1 acts as a scaffold to promote PGCC survival via the AMPK-mTOR pathway. This study is the first to report the protective role of RIPK1 in dormant cell survival, which offers a unique opportunity for targeted dormant PGCC therapies.
Overall, this study provides compelling evidence for the vital role of autophagy in the induction of dormant PGCCs and demonstrates clinical risks in patient recurrence, and outcomes. Moreover, our findings suggest that therapeutic approaches to inhibit autophagy before chemotherapy could prevent dormant PGCC formation responsible for metastasis and ultimately improve the survival rate. Our findings may facilitate future study of cancer treatments.
Authors' Disclosures
B. You reports grants from National Natural Science Foundation of China during the conduct of the study. Y. You reports grants from the National Natural Science Foundation of China during the conduct of the study. No disclosures were reported by the other authors.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (grant nos. 82173288, 81972554, 81672682, and 81602385), Clinical Frontier Technology of Jiangsu (grant no. BE2017680), CSCO Clinical Oncology Research Foundation of Beijing (grant no. Y-HS2017-074), Innovative research project for postgraduate students in Jiangsu province (grant nos. SJCX19_0871, SJCX19_0872, SJCX20_1164), and Scientific Research Project of Nantong Municipal Health Commission (grant. No. QA2019060).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Contributions
B. You: Conceptualization, data curation, validation, investigation, methodology, writing–original draft, writing–review and editing. T. Xia: Data curation, software, validation, investigation, visualization, writing–original draft. M. Gu: Software, formal analysis, investigation. Z. Zhang: Formal analysis, validation, visualization, project administration. Q. Zhang: Software, supervision, visualization. J. Shen: Software, formal analysis. Y. Fan: Validation, investigation, visualization. H. Yao: Validation, investigation, visualization. S. Pan: Validation, investigation, visualization. Y. Lu: Validation, visualization. T. Cheng: Visualization. Z. Yang: Validation, visualization. X. He: Investigation, visualization. H. Zhang: Validation, investigation. M. Shi: Investigation, visualization. D. Liu: Resources, data curation, software, formal analysis, supervision, funding acquisition, methodology, project administration, writing–review and editing. Y. You: Conceptualization, resources, data curation, project administration.
References
- 1. Zhang L, Huang Y, Hong S, Yang Y, Yu G, Jia J, et al. Gemcitabine plus cisplatin versus fluorouracil plus cisplatin in recurrent or metastatic nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet 2016;388:1883–92. [DOI] [PubMed] [Google Scholar]
- 2. Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer 2020;20:398–411. [DOI] [PubMed] [Google Scholar]
- 3. Friberg S, Nystrom A. Cancer metastases: early dissemination and late recurrences. Cancer Growth Metastasis 2015;8:43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gao XL, Zhang M, Tang YL, Liang XH. Cancer cell dormancy: mechanisms and implications of cancer recurrence and metastasis. Onco Targets Ther 2017;10:5219–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Endo H, Inoue M. Dormancy in cancer. Cancer Sci 2019;110:474–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mirzayans R, Murray D. Intratumor heterogeneity and therapy resistance: Contributions of dormancy, apoptosis reversal (anastasis) and cell fusion to disease recurrence. Int J Mol Sci 2020;21:1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 2014;33:116–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Amend SR, Torga G, Lin KC, Kostecka LG, de Marzo A, Austin RH, et al. Polyploid giant cancer cells: Unrecognized actuators of tumorigenesis, metastasis, and resistance. Prostate 2019;79:1489–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Niu N, Zhang J, Zhang N, Mercado-Uribe I, Tao F, Han Z, et al. Linking genomic reorganization to tumor initiation via the giant cell cycle. Oncogenesis 2016;5:e281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alharbi AM, De Marzo AM, Hicks JL, Lotan TL, Epstein JI. Prostatic adenocarcinoma with focal pleomorphic giant cell features: A series of 30 cases. Am J Surg Pathol 2018;42:1286–96. [DOI] [PubMed] [Google Scholar]
- 11. Fei F, Zhang M, Li B, Zhao L, Wang H, Liu L, et al. Formation of polyploid giant cancer cells involves in the prognostic value of neoadjuvant chemoradiation in locally advanced rectal cancer. J Oncol 2019;2019:2316436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Niu N, Mercado-Uribe I, Liu J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene 2017;36:4887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mirzayans R, Andrais B, Scott A, Wang YW, Kumar P, Murray D. Multinucleated giant cancer cells produced in response to ionizing radiation retain viability and replicate their genome. Int J Mol Sci 2017;18:360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang D, Yang X, Yang Z, Fei F, Li S, Qu J, et al. Daughter cells and erythroid cells budding from pgccs and their clinicopathological significances in colorectal cancer. J Cancer 2017;8:469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen J, Niu N, Zhang J, Qi L, Shen W, Donkena KV, et al. Polyploid giant cancer cells (PGCCs): The evil roots of cancer. Curr Cancer Drug Targets 2019;19:360–7. [DOI] [PubMed] [Google Scholar]
- 16. Qu Y, Zhang L, Rong Z, He T, Zhang S. Number of glioma polyploid giant cancer cells (PGCCs) associated with vasculogenic mimicry formation and tumor grade in human glioma. J Exp Clin Cancer Res 2013;32:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Ding P, Lv H, Zhang D, Liu G, Yang Z, et al. Number of polyploid giant cancer cells and expression of EZH2 are associated with VM formation and tumor grade in human ovarian tumor. Biomed Res Int 2014;2014:903542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sirois I, Aguilar-Mahecha A, Lafleur J, Fowler E, Vu V, Scriver M, et al. A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and plin4 expression as a molecular vulnerability. Mol Cancer Res 2019;17:2492–507. [DOI] [PubMed] [Google Scholar]
- 19. Liu G, Wang Y, Fei F, Wang X, Li C, Liu K, et al. Clinical characteristics and preliminary morphological observation of 47 cases of primary anorectal malignant melanomas. Melanoma Res 2018;28:592–9. [DOI] [PubMed] [Google Scholar]
- 20. Tagal V, Roth MG. Loss of aurora kinase signaling allows lung cancer cells to adopt endoreplication and form polyploid giant cancer cells that resist antimitotic drugs. Cancer Res 2021;81:400–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nehme Z, Pasquereau S, Ahmad SA, Coaquette A, Molimard C, Monnien F, et al. Polyploid giant cancer cells, stemness and epithelial-mesenchymal plasticity elicited by human cytomegalovirus. Oncogene 2021;40:3030–46. [DOI] [PubMed] [Google Scholar]
- 22. Kudo-Saito C, Miyamoto T, Imazeki H, Shoji H, Aoki K, Boku N. IL33 Is a key driver of treatment resistance of cancer. Cancer Res 2020;80:1981–90. [DOI] [PubMed] [Google Scholar]
- 23. Xuan B, Ghosh D, Jiang J, Shao R, Dawson MR. Vimentin filaments drive migratory persistence in polyploidal cancer cells. Proc Natl Acad Sci U S A 2020;117:26756–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhu Q, Zhang Q, Gu M, Zhang K, Xia T, Zhang S, et al. MIR106A-5p upregulation suppresses autophagy and accelerates malignant phenotype in nasopharyngeal carcinoma. Autophagy 2021;17:1667–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bao L, You B, Shi S, Shan Y, Zhang Q, Yue H, et al. Metastasis-associated miR-23a from nasopharyngeal carcinoma-derived exosomes mediates angiogenesis by repressing a novel target gene TSGA10. Oncogene 2018;37:2873–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Thura M, Ye Z, Al-Aidaroos AQ, Xiong Q, Ong JY, Gupta A, et al. PRL3 induces polypoid giant cancer cells eliminated by PRL3-zumab to reduce tumor relapse. Commun Biol 2021;4:923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. You B, Shan Y, Shi S, Li X, You Y. Effects of ADAM10 upregulation on progression, migration, and prognosis of nasopharyngeal carcinoma. Cancer Sci 2015;106:1506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vera-Ramirez L, Vodnala SK, Nini R, Hunter KW, Green JE. Autophagy promotes the survival of dormant breast cancer cells and metastatic tumour recurrence. Nat Commun 2018;9:1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 2015;524:309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, et al. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat Commun 2019;10:1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar P, Kiran S, Saha S, Su Z, Paulsen T, Chatrath A, et al. ATAC-seq identifies thousands of extrachromosomal circular DNA in cancer and cell lines. Sci Adv 2020;6:eaba2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell 2000;103:253–62. [DOI] [PubMed] [Google Scholar]
- 33. Najafov A, Luu HS, Mookhtiar AK, Mifflin L, Xia HG, Amin PP, et al. RIPK1 promotes energy sensing by the mTORC1 pathway. Mol Cell 2021;81:370–85. [DOI] [PubMed] [Google Scholar]
- 34. Jaco I, Annibaldi A, Lalaoui N, Wilson R, Tenev T, Laurien L, et al. MK2 phosphorylates RIPK1 to prevent TNF-induced cell death. Mol Cell 2017;66:698–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dondelinger Y, Delanghe T, Priem D, Wynosky-Dolfi MA, Sorobetea D, Rojas-Rivera D, et al. Serine 25 phosphorylation inhibits RIPK1 kinase-dependent cell death in models of infection and inflammation. Nat Commun 2019;10:1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Degterev A, Ofengeim D, Yuan J. Targeting RIPK1 for the treatment of human diseases. Proc Natl Acad Sci U S A 2019;116:9714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Menon MB, Gropengiesser J, Fischer J, Novikova L, Deuretzbacher A, Lafera J, et al. p38(MAPK)/MK2-dependent phosphorylation controls cytotoxic RIPK1 signalling in inflammation and infection. Nat Cell Biol 2017;19:1248–59. [DOI] [PubMed] [Google Scholar]
- 38. Aguirre-Ghiso JA. How dormant cancer persists and reawakens. Science 2018;361:1314–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Marx V. How to pull the blanket off dormant cancer cells. Nat Methods 2018;15:249–52. [DOI] [PubMed] [Google Scholar]
- 40. Recasens A, Munoz L. Targeting cancer cell dormancy. Trends Pharmacol Sci 2019;40:128–41. [DOI] [PubMed] [Google Scholar]
- 41. Dhimolea E, de Matos Simoes R, Kansara D, Al'Khafaji A, Bouyssou J, Weng X, et al. An embryonic diapause-like adaptation with suppressed myc activity enables tumor treatment persistence. Cancer Cell 2021;39:240–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rehman SK, Haynes J, Collignon E, Brown KR, Wang Y, Nixon AML, et al. Colorectal cancer cells enter a diapause-like DTP state to survive chemotherapy. Cell 2021;184:226–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu J. Giant cells: Linking McClintock's heredity to early embryogenesis and tumor origin throughout millennia of evolution on Earth. Semin Cancer Biol 2021;S1044-579X:00177–2. [DOI] [PubMed] [Google Scholar]
- 44. Sharifi-Rad J, Quispe C, Patra JK, Singh YD, Panda MK, Das G, et al. Paclitaxel: Application in modern oncology and nanomedicine-based cancer therapy. Oxid Med Cell Longev 2021;2021:3687700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kiss RC, Xia F, Acklin S. Targeting DNA damage response and repair to enhance therapeutic index in cisplatin-based cancer treatment. Int J Mol Sci 2021;22:8199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang H, Ni HM, Ding WX. The double-edged sword of MTOR in autophagy deficiency induced-liver injury and tumorigenesis. Autophagy 2019;15:1671–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. La Belle Flynn A, Calhoun BC, Sharma A, Chang JC, Almasan A, Schiemann WP. Autophagy inhibition elicits emergence from metastatic dormancy by inducing and stabilizing Pfkfb3 expression. Nat Commun 2019;10:3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hampsch RA, Wells JD, Traphagen NA, McCleery CF, Fields JL, Shee K, et al. AMPK activation by metformin promotes survival of dormant ER(+) breast cancer cells. Clin Cancer Res 2020;26:3707–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mao W, Peters HL, Sutton MN, Orozco AF, Pang L, Yang H, et al. The role of vascular endothelial growth factor, interleukin 8, and insulinlike growth factor in sustaining autophagic DIRAS3-induced dormant ovarian cancer xenografts. Cancer 2019;125:1267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Chude CI, Amaravadi RK. Targeting autophagy in cancer: update on clinical trials and novel inhibitors. Int J Mol Sci 2017;18:1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.