This work characterizes a novel mechanism of superenhancer-driven cyclin D1 upregulation by DHX37 and PLRG1, implicating this pathway as a potential therapeutic target in hepatocellular carcinoma.
Abstract
RNA helicases are dysregulated in tumors. Here, we identified DHX37 as one of the top RNA helicase genes with upregulated expression in hepatocellular carcinoma (HCC). DHX37 promoted proliferation of liver cancer cells in vitro and in vivo. Epigenomic profiling of DHX37-knockdown and control HCC cells revealed that DHX37 is associated with superenhancer activity. Mechanistically, DHX37 interacted with pleiotropic regulator 1 (PLRG1) to transcriptionally activate cyclin D1 (CCND1) expression via co-occupation of its promoter and superenhancer elements. DHX37 and PLRG1 promoted liver cancer cell proliferation and contributed to the poor prognosis of patients with HCC. Importantly, CCND1 inhibitors were effective as antiproliferative agents for liver cancer. These results together demonstrate a cooperative mechanistic interaction between DHX37 and PLRG1 that regulates CCND1 expression and promotes liver cancer progression, advancing our understanding of the epigenetic and transcriptional dysregulations mediated by RNA helicases and superenhancers in HCC.
Significance:
This work characterizes a novel mechanism of superenhancer-driven cyclin D1 upregulation by DHX37 and PLRG1, implicating this pathway as a potential therapeutic target in hepatocellular carcinoma.
Introduction
Primary liver cancer is the third leading cause of cancer-related death worldwide and the sixth most commonly diagnosed cancer (1). Hepatocellular carcinoma (HCC) accounts for the majority of primary liver cancers. Most patients with HCC have concomitant liver disease, and cirrhosis with any cause can promote a series of genetic and epigenetic events that culminate in the formation of dysplastic nodules, which are bona fide preneoplastic lesions. Additional molecular alterations endow dysplastic cells with proliferative, invasive, and survival advantages and complete the transition to full-blown HCC (2). Despite continuous improvements in the clinical management of HCC over the past decades, particularly for patients with advanced stages of this disease, no effective interventions are available for the management of other aspects, such as chemoprevention in patients with cirrhosis and adjuvant therapy after surgical resection or ablation (2).
RNA helicases are highly conserved enzymes that contain domains regulating their NTPase and helicase activities (3). These enzymes are divided into six superfamilies (SF) according to their sequences and structures, and eukaryotic RNA helicases are included in superfamily 1 (SF1) and superfamily 2 (SF2; ref. 3). DEAD-box (Asp-Glu-Ala-Asp) and DEAH-box (Asp-Glu-Ala-His) helicases represent the two largest groups in the helicase SF2 in humans and similarly contain nine conserved motifs. Accumulating evidence indicates that RNA helicases are involved in the regulation of transcription, translation, posttranslational modification, splicing, and RNA transport by acting as molecular chaperones and are thus closely related to tumorigenesis (4). After comprehensively analyzing the expression profiles of RNA helicase genes based on datasets from The Cancer Genome Atlas (TCGA) for liver hepatocellular carcinoma (TCGA-LIHC), we found that DHX37, a highly conserved DEAH-box RNA helicase, is a potent pro-oncogenic gene in HCC. Previous studies have demonstrated that DHX37 is involved mainly in ribosomal biogenesis (5, 6) and has antiviral activity (7). DHX37 can inhibit the activation of antigen-specific CD8-positive T cells, thus antagonizing the efficacy of immunotherapy for triple-negative breast cancer (8). Recent bioinformatic analysis of DHX37 expression profiles in multiple have cancers also shown that DHX37 is a potential marker and is associated with the tumor immune microenvironment in liver cancer (9–11). However, the molecular mechanisms underlying the pro-oncogenic role of DHX37 in liver cancer remain largely unexplored.
In this study, we demonstrated that DHX37 promotes liver cancer cell proliferation and cell-cycle progression in vitro and facilitates HCC cell tumorigenicity in vivo. Interestingly, epigenomic profiling defined the superenhancer landscape regulated by DHX37 and demonstrated that DHX37 can bind to both the promoter and superenhancer regions of the cyclin D1 (CCND1) genomic locus, thereby driving the expression of CCND1 in HCC cells. Moreover, coimmunoprecipitation (Co-IP) and LC/MS analysis identified PLRG1 as a binding partner of DHX37. DHX37 can cooperate with PLRG1, a core component of the CDC5L complex, to transcriptionally activate CCND1 and promote the proliferation of HCC cells, indicating that DHX37 and PLRG1 are potential targets for the development of new anticancer therapeutics for liver cancer.
Materials and Methods
Cell culture
Huh7 (ATCC, ATCC Number: RCB1366; RRID: CVCL_0336, 2019), HepG2-C3A (ATCC, ATCC Number: CRL-10741; RRID: CVCL_1098, 2019), SK-Hep1 (ATCC, ATCC Number: HTB-52; RRID:CVCL_0525, 2019), and HEK-293T (ATCC, ATCC Number: CRL-11268; RRID: CVCL_1926, 2019) cells were cultured in DMEM (Invitrogen) containing 10% FBS (HyClone) and antibiotics (penicillin and streptomycin, Invitrogen) at 37°C in 5% CO2. Cells were assessed for Mycoplasma monthly via the qPCR analysis using specific primers for detecting Mycoplasma (Supplementary Table S1). All these cells were recently authenticated by short tandem repeat analysis.
Collection of liver HCC data
mRNA expression and clinical data, including tumor stages, histological subtypes, sex, overall survival (OS) time, and progression-free survival (PFS) time, of patients from the LIHC dataset were downloaded from TCGA (12). Public gene expression data ware retrieved from the Gene Expression Omnibus (GEO). A GEO dataset (accession number GSE77314) with data for 50 paired samples from Chinese patients with HCC was used for the analysis of DHX37 expression. mRNA expression data for enhancer RNA (eRNA) in TCGA-LIHC were downloaded from the eRic database (13).
Analysis of RNA helicase alterations in liver cancer
We identified differential expression of RNA helicase family members (Supplementary Table S2) using two-tailed Student t test. A univariate Cox regression model was used to calculate the HRs associated with RNA helicase family members in TCGA-LIHC cohort. On the basis of the survival risk associated with DHX37 expression in LIHC, the “survminer” package was used to determine the cut-off value for survival data, including OS time and PFS time. The “surv-cutpoint” function was used to dichotomize the patients by the DHX37 expression level, and all potential cutting values were repeatedly tested to find the maximum rank statistic. Then, the patients were divided into gene-high groups and gene-low groups according to the maximum selected log-rank statistics, so as to reduce the calculated batch effect. Survival curves for prognostic analysis were generated using the Kaplan–Meier method, and the log-rank test was used to determine the significance of the differences. All statistical analyses were two sided and P < 0.05 was considered to indicate statistical significance.
RNA sequencing analysis
Total RNA was extracted from cells using TRIzol. The sequencing library was prepared using an Illumina mRNA sequencing (mRNA-seq) sample preparation kit and was then subjected to paired-end sequencing on the Illumina HiSeq platform. The paired-end reads were aligned to hg38 using STAR. Bam files obtained from STAR were first converted into bedGraph format using BEDTools and were then converted into BigWig files for visualization with UCSC Genome Browser or Integrative Genomics Viewer (IGV). After calculating raw gene expression levels (raw read counts) with FeatureCounts, normalized profiling data were obtained with DESeq2 for differential expression analysis. Differentially expressed genes (DEG) were identified as those with an absolute fold change >1.5. DAVID software was used for functional annotation of the DEGs by Gene Ontology (GO) analysis of cell components, biological processes, and molecular functions.
Assay for transposase-accessible chromatin using sequencing
Assay for transposase-accessible chromatin using sequencing (ATAC-seq) was performed as described previously (14). In brief, cells were grown to 80% confluence, and 50,000 cells were lysed and subjected to tagmentation. ATAC-seq library preparation was performed as described previously. All deep sequencing was performed on the Illumina HiSeq Xten-PE150 or Illumina HiSeq 2500 platform provided by GENEWIZ (GENEWIZ). The following tools and versions were used for ATAC-seq data analysis: Trimmomatic, SAMtools, Picard, and Bowtie2. First, Nextera adapter sequences were trimmed from the reads by using Trimmomatic. These reads were aligned to a reference genome using Bowtie2 with standard parameters. Picard was then used to remove duplicate reads. These deduplicated reads were then filtered to retain high-quality (MAPQ ≥ 30), nonmitochondrial chromosome, non-Y chromosome, and properly paired (SAMtools flag 0 × 2) reads. To visualize the tracks, the reads were extended by 250 bp using IGV Tools (https://software.broadinstitute.org/software/igv/igvtools).
Chromatin immunoprecipitation sequencing and cleavage under targets and tagmentation
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed as described previously (15, 16). In brief, Huh7 cells were harvested, and cross-linking was performed using 1% formaldehyde for 10 minutes at room temperature. Then, the formaldehyde was neutralized by the addition of 2.5 mol/L glycine to a final concentration of 0.125 mol/L at room temperature, and the cells were washed three times with ice-cold PBS. The cells were resuspended in ChIP lysis buffer (Cell Signaling Technology; Supplementary Table S3) containing protease inhibitors and/or dithiothreitol for sonication using either a Bioruptor or Covaris sonicator to obtain DNA fragments of 200 to 500 bp. Cell lysates were centrifuged at 12,000 rpm for 10 minutes at 4°C, and the supernatant was collected. Then, 2.5 µL of chromatin was saved as the input control, and 250 µL of chromatin was incubated with antibodies at 4°C overnight with slow rotation and then with Dynabeads Protein G (Invitrogen) for an additional 2 hours. DNA was eluted from the immunoprecipitated complexes, reverse cross-linked and purified with a QIAquick PCR Spin Kit (QIAGEN). ChIP-qPCR was performed to verify the ChIP-seq results. The primers used for qPCR are listed in Supplementary Table S1. For library construction, a DNA-seq kit from NEB was used following the manufacturer's instructions. TBE PAGE-gel size selection was performed for final library size selection to obtain ChIP-seq libraries containing fragments of 250 to 500 bp. Cleavage under targets and tagmentation (CUT&Tag) was performed according to the manufacturer's protocol (Novoprotein).
For ChIP-seq and CUT&Tag analyses, 150-bp paired-end reads were aligned to the reference human genome using Bowtie2 with standard alignment parameters. PCR duplicates were marked with the Picard “Mark Duplicates” utility and removed from further analysis. Bam files were converted to BigWig files using deepTools. For ChIP-seq analysis, peaks were identified using the MACS2 peak caller with the following parameters: –f BAMPE –keep-dup all –g hs –q 0.01. For CUT&Tag analysis, peaks were identified using the MACS2 peak caller with the following parameters: –f BAMPE –shift -100 –extsize 200 –nomodel -B –SPMR -g hs –keep-dup all –q 0.01. Peaks were annotated with Homer. The peak distribution along genomic regions of genes of interest was visualized with IGV.
Superenhancer analysis
Rank ordering of superenhancers (ROSE) algorithm was used to define superenhancers (17, 18). The distance of stitching enhancers was fixed at 12.5 kb. All other parameters used the default setting. Superenhancer-associated genes were identified by the program of ROSE_geneMapper.py and obtained from the ROSE ENHANCER_TO_TOP_GENE.txt file.
CRISPR/Cas9-mediated deletion of superenhancers
The single-guide RNAs (sgRNA) used to delete superenhancers were designed with CRISPR-ERA (http://crispr-era.stanford.edu/). sgRNAs were annealed with 10 × annealing buffer [1 mol/L Tris (pH 8.0), 5 mol/L NaCl, and 500 mmol/L EDTA] at 95°C for 3 minutes, and the mixture was then allowed to cool naturally to room temperature. Annealed double-stranded DNA was inserted into the Lenti_gRNA-Puro vector (Addgene) using BsmI (NEB). Then, viruses packaged with the purified recombinant plasmid were used to infect Huh7 lentiCas9-Blast (Addgene) cells in 6-well plates. Puromycin was added to the cells after 48 hours, and the cells were then collected to evaluate the knockout efficiency and gene expression. Primer sequences are listed in Supplementary Table S1.
Generation of enCRISPRi cell lines
First, pHR-SFFV-KRAB-dCas9-P2A-mCherry (Addgene) and Lenti_MCP-LSD1_Hygro (Addgene) were packaged into a slow-replicating virus. After transducing Huh7 cells with both viral vectors, we selected hygromycin-resistant and mCherry-positive cells to finally obtain the corresponding dCas9-KRAB-LSD1 cells. Next, based on the H3K27ac ChIP-seq and ATAC-seq peaks in Huh7 cells, we found the location of the superenhancer and obtained the DNA sequences of the corresponding ATAC-seq peaks. The sgRNAs used to inhibit the superenhancers were designed with CRISPR-ERA (http://crispr-era.stanford.edu/). Annealed double-stranded DNA was inserted into Lenti_sgRNA(MS2)_ZsGreen1 (Addgene) using BsmI (NEB). Then, viruses packaged with the purified recombinant plasmid were used to infect dCas9-KRAB-LSD1 cells in 6-well plates. After screening of ZsGreen-positive cells using flow cytometry, the variation in the mRNA level of CCND1 was obtained using qPCR. Primer sequences are listed in Supplementary Table S1.
In vivo tumorigenesis assay
Nude mice were purchased and raised at Shanghai Duan Jia Biotechnology Company. A total of 3 × 106 cells were injected subcutaneously into the dorsal side of recipient NOD-SCID gamma mice (6 weeks old, 6 mice). The general behavior of the mice was monitored, and tumor volumes were measured every 3 days for 20 days. At the end of the experiments, the mice were sacrificed, and tumor volumes were calculated according to the following equation: V = (length × width2)/2. Animal experiments followed the Guide for the Institutional Animal Care and Use Committee of Fudan University Shanghai Cancer Center (Shanghai, P.R. China).
Statistical analysis
Each experiment was performed with at least three independent replicates, and the results are expressed as the mean ± SEM. Student t tests (two-tailed) and one-way ANOVA were used to compare the means of two or more samples unless otherwise indicated. A P value < 0.05 was considered significant. All statistical analyses were performed using GraphPad Prism 8 (GraphPad Software). The details of the statistical analyses are presented in the figure legends.
Data availability
The data generated in this study are available within the article and its Supplementary Data files. The raw data generated in this study are publicly available in GEO at GSE184796, GSE184797, and GSE185701.
Results
High expression of DHX37 correlates with poor outcomes in patients with HCC and promotes liver cancer cell proliferation in vitro and in vivo
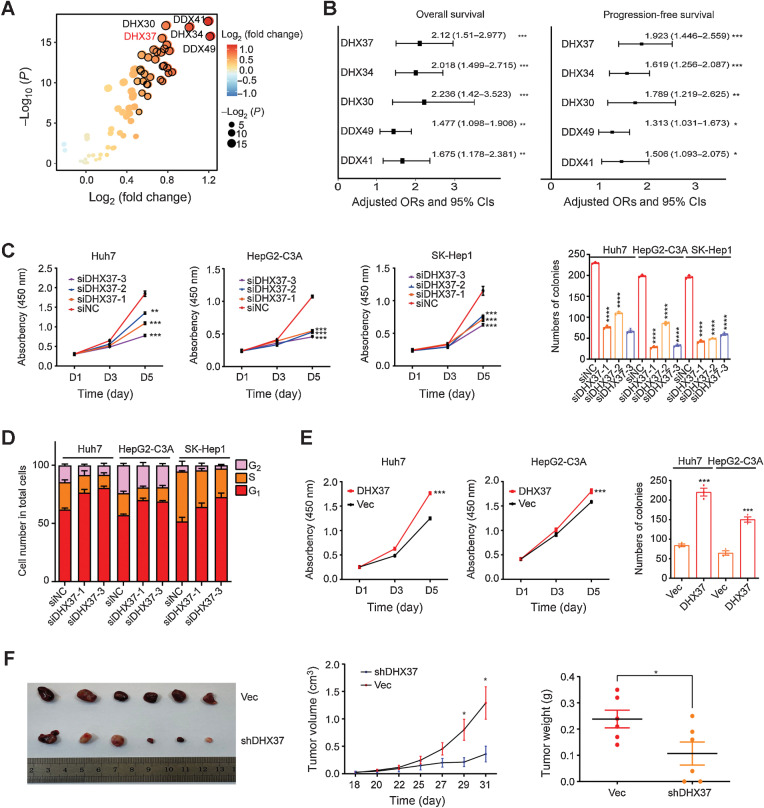
First, the expression of RNA helicase family genes (Supplementary Table S2) was analyzed in TCGA-LIHC database, with 375 liver tumor tissues and 50 adjacent tissues. Among the RNA helicase genes, DDX49, DDX41, DHX34, DHX30, and DHX37 showed markedly upregulated expression in the HCC tissues compared with the adjacent tissues (Fig. 1A). Univariate Cox regression analysis in TCGA-LIHC cohort demonstrated that the expression levels of these five RNA helicase genes were risk factors for OS and PFS in HCC, and DHX37 showed the highest HR for PFS among these genes (Fig. 1B; Supplementary Fig. S1A and S1B). Then we focused on the role of DHX37 in the development and progression of HCC. The mRNA levels of DHX37 were confirmed in 50 paired samples from a Chinese HCC cohort (Supplementary Fig. S1C). In addition, subcellular localization analysis demonstrated that DHX37 is localized mainly in the nucleus of in HCC cells (Supplementary Fig. S1D and S1E).
Figure 1.
High expression of DHX37 correlates with poor outcomes in patients with HCC and promotes liver cancer cell proliferation, both in vitro and in vivo. A, Comparison of RNA helicase family member expression between tumor tissues and normal tissues in TCGA-LIHC cohort. The horizontal axis shows differences in gene expression between tumor tissues and normal tissues, and the vertical axis shows the significance of the differences. The color of the points in the figure represents the fold differences, and the size of the points represents the significance of the differences. P values by two-sided Student t test. B, Association of RNA helicase family member expression with patient OS and PFS times in TCGA-LIHC cohort based on univariate Cox regression analysis. HR > 1 indicates that the gene is a risk factor. C, CCK-8 assays and colony formation assays of the Huh7, HepG2-C3A, and SK-Hep1 cells transfected with three independent DHX37 siRNAs (n = 3). D, Flow cytometric analysis of the cell-cycle distribution in the liver cancer cells transfected with two independent DHX37 siRNAs (n = 3). E, CCK-8 assays and colony formation assays of the Huh7 and HepG2-C3A cells with stable DHX37 overexpression (n = 3). F, Images of xenografts from nude mice bearing subcutaneous xenografts derived from the shDHX37 cells or control cells. Tumor volumes and tumor weights were measured in the shDHX37 and negative control groups of xenograft mice (n = 6). The values are expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA or two-sided Student t test.
To investigate the biological function of DHX37 in liver cancer cells, we first knocked down endogenous DHX37 expression by siRNAs or short hairpin RNA in Huh7, HepG2-C3A, and SK-Hep1 cells (Supplementary Fig. S2A and S2B). After knockdown of DHX37, the cell proliferation and colony formation capabilities of the liver cancer cells were significantly diminished (Fig. 1C; Supplementary Fig. S2C–S2E), and the cell cycle was arrested in G1 phase (Fig. 1D; Supplementary Fig. S2F and S2G). Furthermore, knockout of DHX37 via CRISPR-Cas9 gene editing in Huh7 and HepG2-C3A cells significantly inhibited cell proliferation and colony formation (Supplementary Fig. S3A–S3C), whereas overexpression of DHX37 enhanced the viability and colony formation capabilities of the liver cancer cells (Fig. 1E; Supplementary Fig. S3C and S3D). Furthermore, the effect of DHX37 on liver cancer cell growth was evaluated in vivo. Stable shDHX37 cells generated from Huh7 cells were subcutaneously injected into nude mice, and both the volumes and weights of the tumors, and the expression of Ki67 in the shDHX37 group were found to be significantly lower than those in the control group (Fig. 1F; Supplementary Fig. S3E). Collectively, these results indicated that DHX37 promotes the proliferation of liver cancer cells both in vitro and in vivo.
DHX37 is strongly associated with transcriptional regulation and enhancer activity in liver cancer
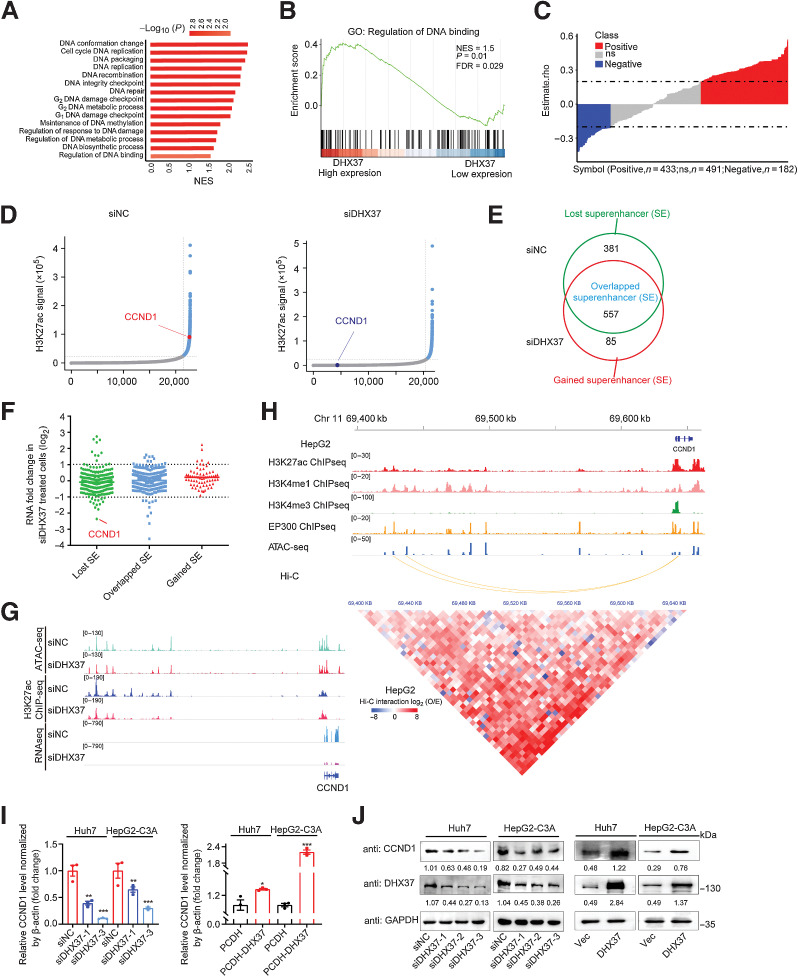
To explore the downstream mechanism by which DHX37 promotes liver cancer cell growth and proliferation, we investigated the biological processes potentially associated with DHX37 in liver cancer. We first divided the samples from TCGA-LIHC into the DHX37-high and DHX37-low groups according to the median expression level of DHX37 in HCC. Then, we used a two-tailed Student t test to compare the gene expression profiles of the two groups to obtain the rank difference correlation coefficient and performed gene set enrichment analysis (GSEA). The differentially expressed genes were enriched in DNA regulation and cell cycle–related signaling pathways (Fig. 2A), particularly in biological processes related to the regulation of DNA binding (Fig. 2B), suggesting that DHX37 might function in DNA binding and transcriptional regulation of cell cycle–related genes. Furthermore, we used Spearman correlation analysis to detect the correlation between the expression of DHX37 and eRNAs extracted from the public cancer eRNA database eRic (13) and found that the expression of many eRNAs was positively correlated with the expression of DHX37 (n = 433), suggesting that DHX37 may be associated with the regulation of enhancers in HCC (Fig. 2C). Cancer cells usually acquire superenhancers to drive the expression of prominent oncogenes by recruiting transcription factors (TF) and coactivators. As a chromatin regulator, DHX37 may affect the activity of enhancers in liver cancer cells and regulate the expression of related genes.
Figure 2.
DHX37 is strongly associated with transcriptional regulation and enhancer activity in HCC. A, GSEA of the biological processes associated with DHX37 expression in TCGA-LIHC cohort. The vertical axis indicates the GSEA biological process, and the horizontal axis indicates the NES of the biological process. The colors on the bar indicate the significance of enrichment. B, GSEA visualization of the regulation of DNA binding in TCGA-LIHC cohort. C, Correlation analysis of the expression of DHX37 and eRNAs. Each column represents an eRNA. The height of the column indicates the correlation between the expression of each eRNA and that of DHX37, and the color of the column indicates the significance of the correlation. Red, positive correlation (n = 433); blue, negative correlation (n = 182); gray, no significant correlation (n = 491). D, Hockey stick plots on the basis of input-normalized H3K27ac signals in Huh7 cells after silencing of DHX37. The enhancers are rank ordered as a percentage of the total signal. Superenhancers are obtained on the basis of the difference in the strength of the binding levels of molecular markers of enhancer transcriptional activity. Superenhancers are highlighted in blue with the ranks of selected superenhancer-associated genes. E, Definition of lost superenhancers (only in the siNC group), overlapped superenhancers (in both the siNC and siDHX37 groups), and gained superenhancers (only in the siDHX37 group). F, Gene expression changes as measured by RNA-seq of lost superenhancers, overlapped superenhancers, and gained superenhancers in siNC and siDHX37 Huh7 cells. The individual points are represented as mRNA fold change for superenhancer-associated genes. G, ATAC-seq, ChIP-seq, and RNA-seq profiles of the CCND1 locus in Huh7 cells treated with siNC and siDHX37. H, Epigenetic features of the CCND1 locus in HepG2 cells. A 5 kb resolution heatmap for chromosome 11 (hg38: 69400000–69645000 bp) in HepG2 cells showed the location of topologically associating domains, which was derived from ENCODE and reanalyzed on the basis of Juicer and hiccups software (bottom). The Hi-C interaction between promoters and superenhancers of CCND1 was evaluated by the O/E value, which was calculated as the observed value (estimated with normalized mapped reads) divided by the expected value. I and J, Relative mRNA and protein expression levels of CCND1 upon knockdown and overexpression of DHX37 in Huh7 and HepG2-C3A cells (n = 3). The values are expressed as the mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.
To identify the downstream target genes regulated by DHX37, we silenced DHX37 by siRNA in Huh7 cells and performed whole-transcriptome sequencing (RNA-seq). GSEA showed that genes with downregulated expression upon DHX37 silencing were strongly enriched in cell cycle–related signaling gene sets, including the E2F targets and G2–M checkpoint gene sets (Supplementary Fig. S4A). To determine whether superenhancers are regulated by DHX37, we performed ChIP-seq with antibodies against acetylated H3K27 (H3K27ac) and evaluated the open chromatin regions by ATAC-seq to detect the effect of DHX37 on genome-wide enhancer activity in HCC cells. These results showed that the H3K27ac peak was highly correlated with the open region obtained by ATAC-seq, indicating that most accessible chromatin was located in the region related to the transcriptionally active domain (Supplementary Fig. S4B). Next, we compared the superenhancer landscapes between the DHX37-knockdown and control Huh7 cells to investigate the implications of superenhancer alterations induced by DHX37. Constituent enhancers located within 12.5 kb were stitched and ranked according to their H3K27ac signals. Stitched enhancers with signals above the inflection point of the H3K27ac signal were considered superenhancers, and superenhancer-associated genes were annotated (Fig. 2D). Through this analysis, we obtained 381 lost superenhancers and 85 gained superenhancers upon knockdown of DHX37 (Fig. 2E; Supplementary Fig. S4C). Furthermore, by integrating these data with the RNA-seq data, we identified the DEGs related to the activity of superenhancers regulated by DHX37: after interference with DHX37 expression, the genes located near the lost superenhancers had decreased mRNA levels, and those located near the gained superenhancers had increased mRNA levels (Fig. 2F). Notably, the lost superenhancer-associated transcript CCND1 had the most significant reduction in mRNA expression upon DHX37 knockdown (Supplementary Table. S4). The occupancy of H3K27ac at the superenhancer of the CCND1 gene was also reduced after interference with the expression of DHX37 (Fig. 2G). Importantly, after further examination of the CCND1 enhancer signal in the HepG2 cell line, we found that this region harbors classical enhancer features, such as enrichment of histone H3K4me1 modification. Moreover, this region exhibits strong enrichment of active enhancer markers such as histone H3K27ac modification and binding of the transcriptional cofactor EP300 (Fig. 2H). Multiple EP300 binding peaks are densely distributed in this region, indicating that it is a potential superenhancer in liver cancer cells. Intriguingly, chromatin interactions between the promoter and superenhancer of CCND1 were observed in HepG2 cells by high-throughput chromosome conformation capture (Hi-C) analysis (Fig. 2H). Using qPCR and Western blot (WB) analysis, we confirmed that DHX37 knockdown significantly reduced the mRNA and protein levels of CCND1, whereas overexpression of DHX37 promoted CCND1 mRNA and protein expression in liver cancer cells (Fig. 2I and J; Supplementary Fig. S4D and S4E). Taken together, these data indicated that DHX37 regulates superenhancer and transcriptional activities, and that CCND1, as a superenhancer-associated gene, is an important target regulated by DHX37 in liver cancer.
CCND1 is a direct target gene regulated by DHX37 in a superenhancer-driving manner in liver cancer cells
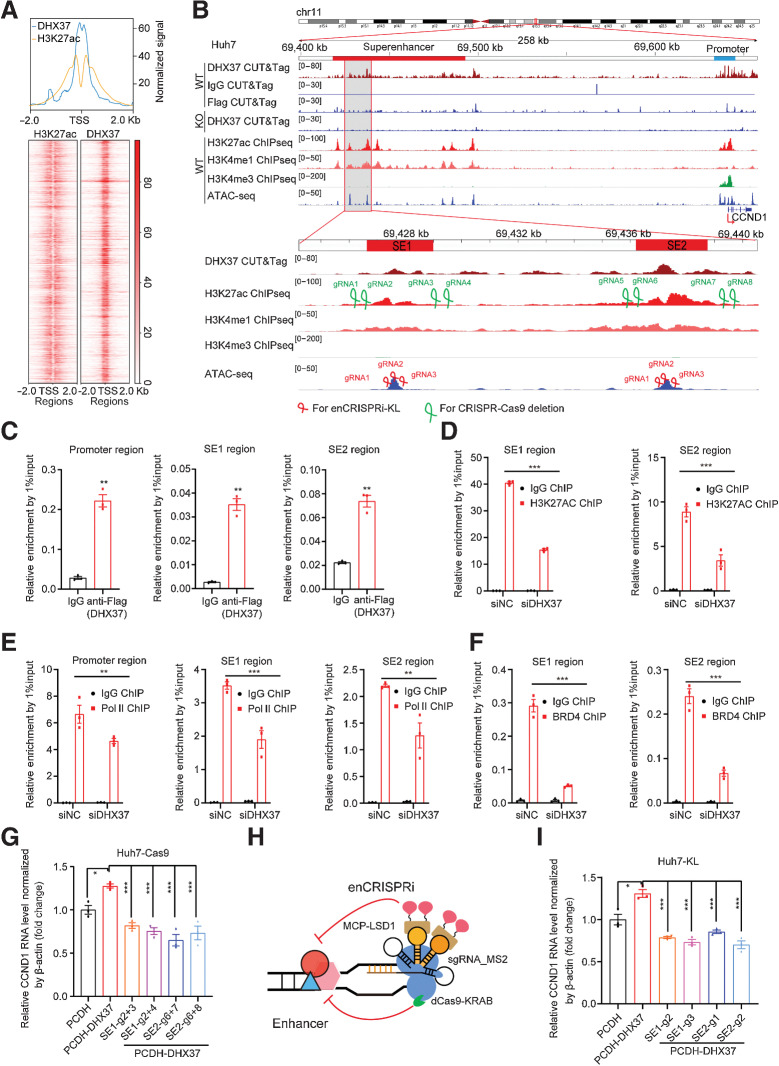
To explore whether superenhancers are directly regulated by DHX37, we performed CUT&Tag) in HCC cells. Unsurprisingly, most DHX37 CUT&Tag peaks were located in promoter and intergenic regions (Supplementary Fig. S5A and S5B). Notably, prominent enrichment of H3K27ac signals adjacent to DHX37 peaks was observed, indicating that superenhancer-mediated transcriptional activation is directly coupled with the binding of DHX37 to the corresponding DNA region (Fig. 3A). Importantly, after analyzing ChIP-seq data for H3K27ac, H3K4me3, and H3K4me1 and CUT&Tag data for DHX37 generated from Huh7 cells, we found that both the promoter and superenhancer regions [particularly constituent enhancer 1 (SE1) and enhancer 2 (SE2)] of CCND1 were occupied by DHX37 (Fig. 3B). These results were confirmed by ChIP-qPCR (Fig. 3C), indicating direct regulation of the target gene CCND1 by DHX37 in HCC cells. Notably, knockdown of DHX37 expression reduced Pol II occupation at the CCND1 promoter region and both H3K27ac enrichment and BRD4 occupation at the superenhancer region (Fig. 3D–F; Supplementary Fig. S6A). These results suggested that DHX37 can directly bind to the promoter and superenhancer regions of the CCND1 genome locus. In addition, we designed a set of sgRNAs that spanned the superenhancer regions to individually delete SE1 or SE2 of CCND1 through CRISPR-Cas9 gene editing (Fig. 3B). After knockout, the superenhancer regions of CCND1 with four combinations of sgRNAs (Supplementary Table. S1) using CRISPR-Cas9 gene editing in stable Huh7 Cas9 cells (Supplementary Fig. S6B), both the mRNA and protein levels of CCND1 were reduced (Supplementary Fig. S6C). Furthermore, knockout of CCND1 superenhancer regions with the four combinations of sgRNAs significantly reduced the expression of CCND1 after overexpression of DHX37 in Huh7 cells (Fig. 3G; Supplementary Fig. S6D). By coupling the dCas9 to various repressor (KRAB, LSD1, and DNMT3A) domains, transcriptional regulation of specific genes was achieved. Recently, Li and colleagues described an enhancer-targeting CRISPR epigenetic editing systems, enCRISPRa and enCRISPRi, to interrogate enhancer function using dCas9 with dual effectors that specifically modulate epigenetic modifications at enhancers (19). Here, we employed a dual effector epigenetic editing system for targeted enhancer inhibition (enCRISPRi-KL), in which dCas9 is fused to KRAB, together with MS2-sgRNA to recruit the lysine-specific demethylase MCP-LSD1 (Fig. 3H). We first designed three individual sgRNAs for SE1 and SE2 (Fig. 3B) and then generated Huh7 cell lines stably expressing enCRISPRi transgenes. After lentiviral transduction of the targeted sgRNAs, the transduced cells were selected and analyzed for CCND1 expression. Interestingly, targeting of either the enCRISPRi complex to SE1 or SE2 of CCND1 via any of the six independent sgRNAs significantly reduced the CCND1 transcription level (Supplementary Fig. S6E), confirming that the superenhancers acquired in HCC cells positively regulate CCND1 expression. Consistent with these findings, quantitative analysis showed that the expression of CCND1 was downregulated by the enCRISPRi method used to inhibit the superenhancer region of CCND1 compared with that in the cells with overexpressing of DHX37 (Fig. 3I; Supplementary Fig. S6F). Taken together, these data demonstrated that DHX37 occupies the promoter and superenhancers regions of CCND1, thereby activating its transcription in liver cancer cells.
Figure 3.
CCND1 is a direct target gene regulated by DHX37 in a superenhancer-driving manner in liver cancer cells. A, Top, line plots showing CUT&Tag and ChIP-seq signals of DHX37 and H3K27ac centered at the summit of DHX37 peaks in Huh7 cells. Bottom, heatmap of CUT&Tag and ChIP-seq signals for DHX37 and H3K27ac [±2,000 bp windows around the center of the transcriptional start site (TSS)]. B, Profiles of DHX37, H3K27ac, H3K4me1, and H3K4me3 occupancy and ATAC-seq peaks at the CCND1 promoter (blue line) and superenhancer (red line) regions in wild-type (WT) and DHX37 knockout (KO) Huh7 cells. Gray shading indicates the occupancy of DHX37, which contains two specific constituent superenhancers (SE1 and SE2). sgRNAs were designed on the basis of the above peak. C, ChIP-qPCR analysis of DHX37 enrichment at the promoter and superenhancer regions of CCND1 (n = 3). D, Treatment with siDHX37 significantly reduced H3K27ac enrichment at the CCND1 superenhancer region compared with that in the siNC group (n = 3). E, Treatment with siDHX37 significantly reduced RNA Pol II occupancy at the CCND1 promoter and superenhancer regions compared with that in the siNC group (n = 3). F, Treatment with siDHX37 significantly reduced BRD4 enrichment at the CCND1 superenhancer region compared with that in the siNC group (n = 3). G, Relative mRNA levels of CCND1 upon overexpression of DHX37 and knockout of the SE1 and SE2 regions by CRISPR-Cas9 gene editing (n = 3). H, Schematic of the enCRISPRi system containing the dCas9-KRAB fusion protein, an sgRNA with two MS2 hairpins, and the MCP-LSD1 fusion protein. I, Blockade of CCND1 superenhancer regions by four individual sgRNAs significantly reversed the upregulated expression of CCND1 caused by DHX37 (n = 3). The values are expressed as the mean ± SEM (C–F and G–I). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA and two-sided Student t test.
DHX37 interacts with PLRG1, a core component of the CDC5L complex, in liver cancer cells
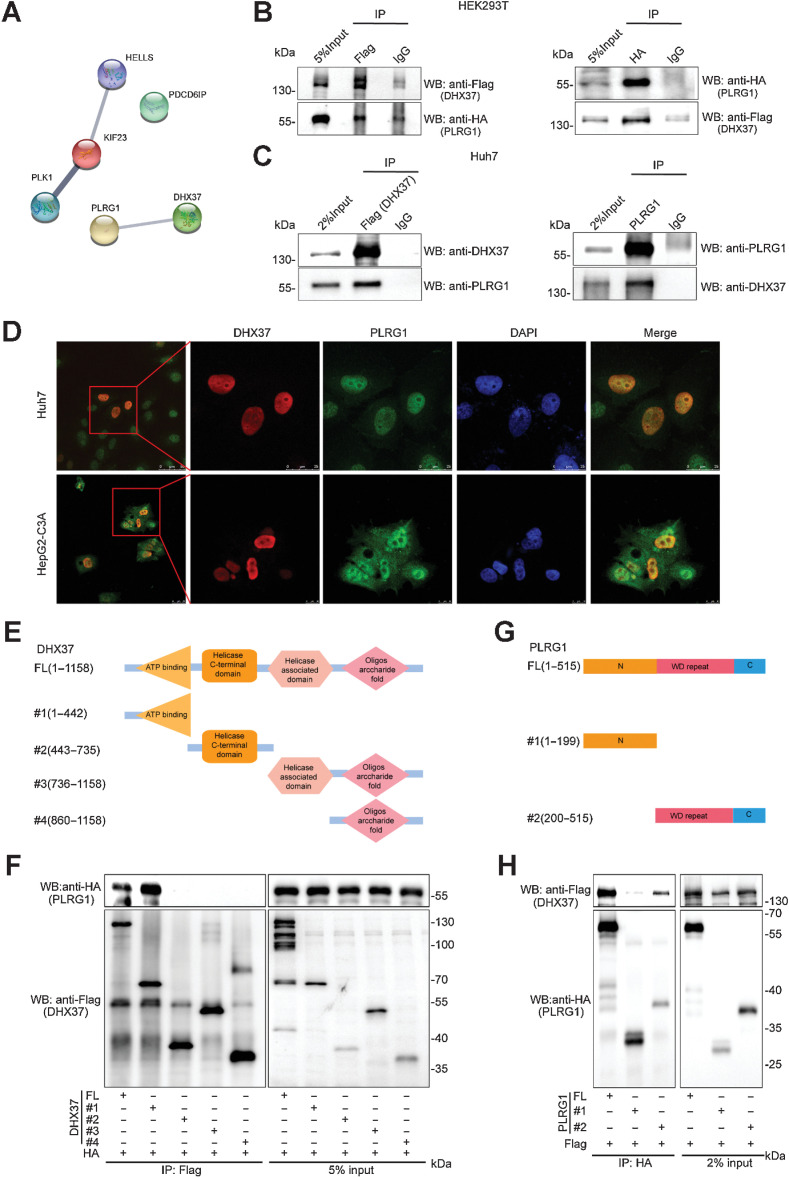
To further explore the protein complex containing DHX37, which directly transcriptionally activates CCND1, we performed a Co-IP assay using Huh7 cells stably expressing Flag-tagged DHX37. The DHX37-containing complex was subjected to SDS-PAGE and analyzed by LC/MS. A total of 78 potential interacting proteins were obtained based on the criteria of a fold change greater than 2 (Flag-DHX37–specific peptides compared with IgG-specific peptides) with a number of unique peptides greater than 5 (n = 65), or a number of unique peptides greater than 3 with no IgG-specific peptides (n = 13), as shown in Supplementary Table S5. Given that DHX37 is strongly associated with cell-cycle regulation, we focused on cell cycle–related proteins predicted by GO annotations. These proteins were predicted by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) analysis (https://string-db.org) for protein–protein interactions, and we found that PLRG1, a core component of the CDC5L complex, potentially interacts with DHX37 (Fig. 4A). Subsequently, the interaction between DHX37 and PLRG1 was confirmed by Co-IP in the HEK293T cells transfected with HA-tagged PLRG1 and Flag-tagged DHX37 (Fig. 4B). Furthermore, we determined by Co-IP showed that endogenous DHX37 can interact with PLRG1 in HCC cells (Fig. 4C). In addition, immunofluorescence staining using anti-DHX37 and anti-PLRG1 antibodies in the Huh7 and HepG2-C3A cells stably expressing DHX37 demonstrated that DHX37 and PLRG1 colocalize in the nucleus (Fig. 4D). Taken together, these data suggested that DHX37 can interact with PLRG1 in liver cancer cells.
Figure 4.
DHX37 interacts with PLRG1, a core component of the CDC5L complex, in liver cancer cells. A, DHX37-associated proteins were analyzed with STRING. The network nodes represent potential interacting proteins obtained by Co-IP/LC/MS. The edges represent protein–protein interactions. B, Co-IP using control IgG and an anti-Flag (DHX37) or anti-HA (PLRG1) antibody was carried out using extracts prepared from the HEK293T cells transfected with the pCMV-Flag-DHX37 and pCMV-HA-PLRG1 plasmids. The presence of DHX37 or PLRG1 in these immunoprecipitates was evaluated by immunoblot (WB) analysis. C, Co-IP using control IgG and an anti-DHX37 or anti-PLRG1 antibody was carried out using extracts prepared from Huh7 cells. The endogenous interaction of PLRG1 with DHX37 was evaluated by Western blot analysis with the corresponding antibodies. D, Huh7 and HepG2-C3A cells stably expressing DHX37 were fixed for immunofluorescence analysis. DHX37 was detected using an anti-Flag (DHX37) primary antibody and an Alexa Fluor 488 goat anti-mouse secondary antibody, and PLRG1 was detected using an anti-PLRG1 primary antibody and an Alexa Fluor 594 goat anti-rabbit secondary antibody. Representative cells from the same field for each experimental group are shown. Scale bars, Huh7, 50 µm (first column) or 25 µm (other four columns). HepG2-C3A, 25 µm (first column) or 10 µm (other four columns). E, Schematic representation of Flag-tagged full-length DHX37 (FL) along with its various deletion mutants (#1, #2, #3, and #4). The domains include the ATP-binding domain (residues 251–442), a helicase C-terminal domain (residues 459–716), a helicase-associated domain (residues 736–859), and an oligonucleotide domain (residues 925–1010). F, HEK293T cells were cotransfected with the indicated HA-tagged PLRG1 constructs along with plasmids encoding Flag-tagged DHX37, and the interaction between DHX37 and PLRG1 was evaluated by IP and immunoblotting. G, Schematic representation of HA-tagged full-length PLRG1 (FL) along with its various deletion mutants (N-terminal or WD repeat deletion mutants). H, HEK293T cells were cotransfected with the indicated Flag-tagged DHX37 constructs along with plasmids encoding HA-tagged PLRG1, and the interaction between DHX37 and PLRG1 was evaluated by IP and immunoblotting.
DHX37 consists of an ATP-binding domain (residues 251–442), a helicase C-terminal domain (residues 459–716), a helicase-associated domain (residues 736–859) and an oligonucleotide domain (residues 925–1010; Fig. 4E). To identify the binding domain responsible for DHX37 binding to PLRG1, we transfected expression vectors encoding Flag-tagged wild-type DHX37 or deletion mutants (DHX37 Δ1, DHX37 Δ2, DHX37 Δ3, and DHX37 Δ4) and HA-tagged PLRG1 into HEK293T cells, and then performed Co-IP and immunoblotting with anti-Flag and anti-HA antibodies. The motif sequence spanning amino acids (aa) 1–442, which includes the ATP-binding domain in DHX37, is necessary for the interaction of DHX37 with PLRG1 (Fig. 4F). Similarly, we generated expression constructs for HA-tagged PLRG1 and a series of N-terminal or WD repeat deletion mutants lacking different domains (Fig. 4G). To map the binding motif responsible for PLRG1 binding to DHX37, we coexpressed these constructs along with full-length Flag-tagged DHX37. The IP results indicated that the region spanning aa 200–515 of PLRG1 can interact with DHX37 (Fig. 4H).
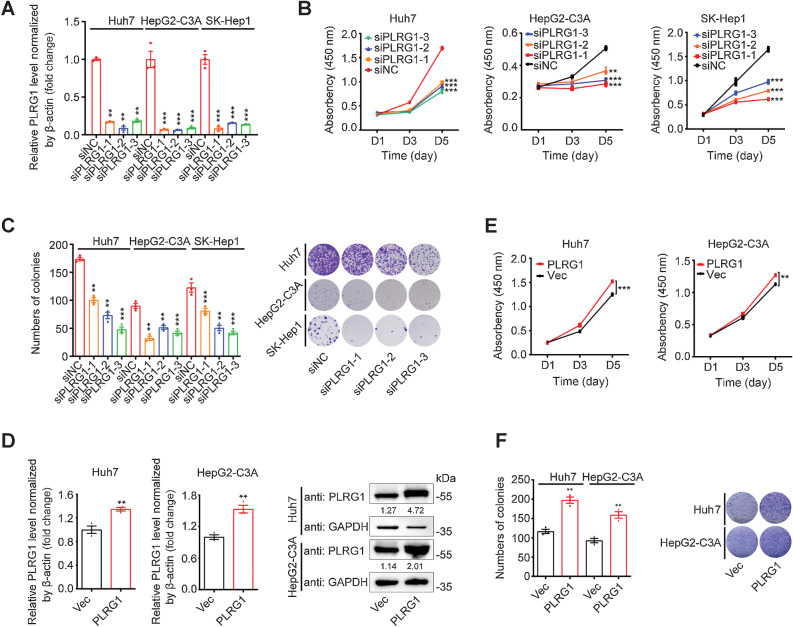
To begin to investigate the biological function of PLRG1 in liver cancer, we first knocked down the expression of the PLRG1 gene in Huh7, HepG2-C3A, and SK-Hep1 cells (Fig. 5A; Supplementary Fig. S7A). Subsequently, knockdown of PLRG1 was shown to reduce both cell viability and clonogenic capacity in the liver cancer cell lines (Fig. 5B and C). Moreover, knockout of PLRG1 in Huh7 and HepG2-C3A cells also confirmed these results (Supplementary Fig. S7B–S7D). Importantly, overexpression of PLRG1 increased both the proliferation and colony formation of Huh7 and HepG2-C3A cells (Fig. 5D–F). These results indicated that PLRG1 also promotes the proliferation of liver cancer cells.
Figure 5.
PLRG1 promotes HCC cell proliferation. A, qRT-PCR analysis of PLRG1 knockdown efficiency after transfection of Huh7, HepG2-C3A, and SK-Hep1 cells with three independent PLRG1 siRNAs. B, CCK-8 assays in the Huh7, HepG2-C3A, and SK-Hep1 cells transfected with three independent PLRG1 siRNAs. C, Colony formation assays and colony counts in the Huh7, HepG2-C3A, and SK-Hep1 cells transfected with three independent PLRG1 siRNAs. D, qRT-PCR and Western blot analysis of PLRG1 overexpression efficiency in stable pCDH-PLRG1 Huh7 and HepG2-C3A cells. E, CCK-8 assays in stable PLRG1-overexpressing and control Huh7 and HepG2-C3A cells. F, Colony formation assays and colony counts in stable PLRG1-overexpressing and control Huh7 and HepG2-C3A cells. The values are expressed as the mean ± SEM (A–F, n = 3). **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.
DHX37 and PLRG1 co-occupy both the promoter and superenhancer regions of CCND1 and activate CCND1 transcription in liver cancer cells
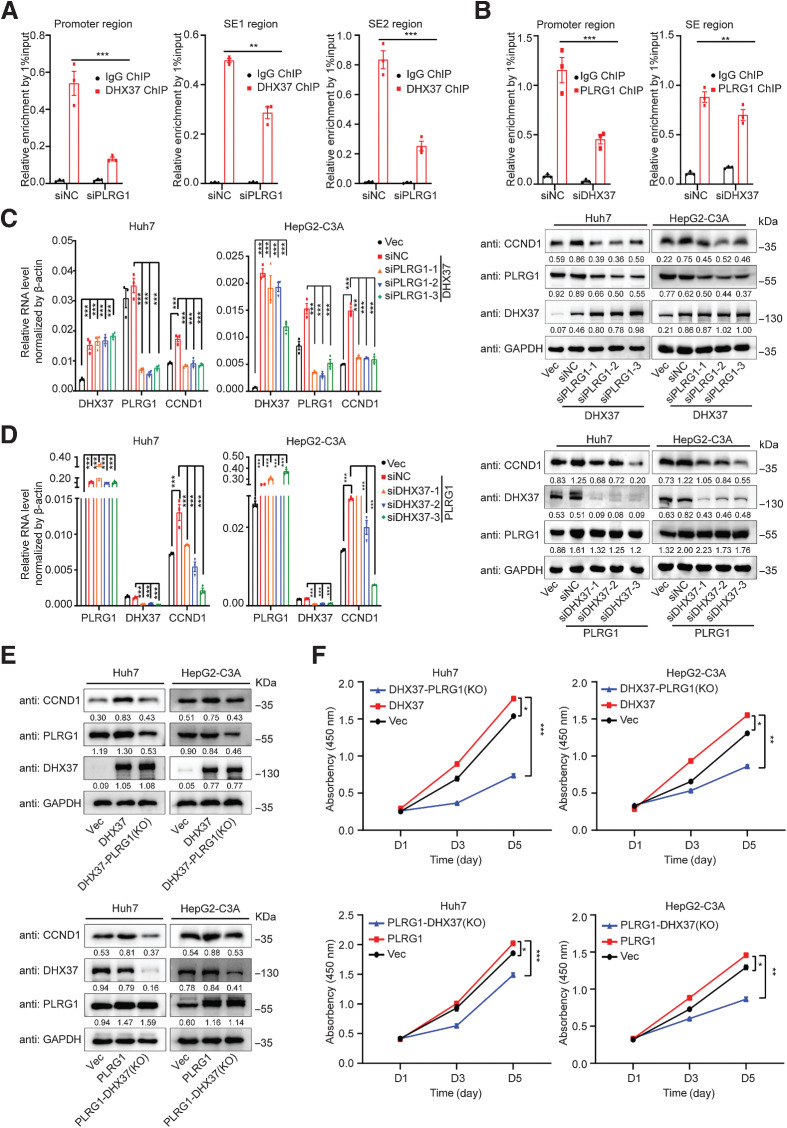
To explore whether CCND1 expression is regulated by PLRG1, we silenced PLRG1 with siRNAs in liver cancer cells. Both the mRNA and protein levels of CCND1 were reduced upon PLRG1 knockdown in Huh7 and HepG2-C3A cells (Fig. 6A; Supplementary Fig. S7B). Notably, we confirmed that overexpression of PLRG1 upregulates CCND1 mRNA and protein expression (Fig. 6B), suggesting that CCND1 is also a downstream target of PLRG1 in liver cancer cells. To explore the mechanism by which CCND1 is regulated by PLRG1, we also performed CUT&Tag with an antibody against PLRG1 and found that both the CCND1 promoter and superenhancer SE2 were occupied by PLRG1 in Huh7 cells (Fig. 6C). To validate the CUT&Tag results, we performed ChIP-qPCR to quantify the occupancy of PLRG1, and its enrichment was confirmed at the promoter and superenhancer (SE2) of CCND1 (Fig. 6D). Importantly, interference with PLRG1 expression reduced Pol II occupancy at the CCND1 promoter region and H3K27ac enrichment at the superenhancer region, as well as BRD4 occupancy in Huh7 cells (Fig. 6E and F), indicating that PLRG1 can also regulate the promoter and superenhancer activity of CCND1. To further elucidate the underlying mechanisms by which the DHX37/PLRG1 complex upregulates CCND1 expression, we investigated the CUT&Tag and ChIP-seq data. The region flanking CCND1 was a superenhancer (denoted by the red bar on top of Fig. 6C; the shaded region contains SE1 and SE2) in Huh7 cells. Notably, DHX37 occupied both superenhancer regions, while PLRG1 occupied only the SE2 enhancer. Importantly, ChIP-qPCR confirmed that the binding ability of DHX37 to the CCND1 promoter and superenhancers regions is reduced after knockdown of PLRG1 (Figs. 6C and 7A) and that the binding ability of PLRG1 to CCND1 is also decreased after DHX37 knockdown (Fig. 7B). These data demonstrated that DHX37 and PLRG1 co-occupy the promoter and superenhancer of CCND1 in HCC cells. Importantly, overexpression of either DHX37 or PLRG1 upregulated CCND1 mRNA and protein expression in liver cancer cells (Fig. 7C–E). Moreover, the induction was abolished by knockdown of either PLRG1 or DHX37 (Fig. 7C–E). Notably, the promotion of liver cancer cell proliferation was reversed by knockout of either PLRG1 or DHX37 (Fig. 7F; Supplementary Fig. S8A and S8B). These data suggested that the co-occupancy of DHX37 and PLRG1 at the CCND1 promoter and superenhancer is required for the transcriptional activation of CCND1. Collectively, these results indicated that DHX37 interacts with PLRG1 to form a complex at the promoter and superenhancer regions of CCND1, thus activating its transcription, in liver cancer cells.
Figure 6.
PLRG1 occupies both the promoter and superenhancer regions of CCND1 and activates its transcription in liver cancer cells. A, Relative mRNA and protein expression levels of CCND1 upon knockdown of PLRG1 in Huh7 and HepG2-C3A cells (n = 3). B, Relative mRNA and protein expression levels of CCND1 upon overexpression of PLRG1 in Huh7 and HepG2-C3A cells (n = 3). C, Profiles of PLRG1, H3K27ac, and DHX37 occupancy and ATAC-seq peaks at the CCND1 promoter (blue line) and superenhancer (red line) regions in Huh7 cells. Gray shading indicates the two specific constituent superenhancers (SE1 and SE2). D, ChIP-qPCR analysis for PLRG1 enrichment at the promoter and SE2 region of CCND1 (n = 3). E, Treatment with siPLRG1 significantly reduced RNA Pol II occupancy at the CCND1 promoter and SE2 region compared with that in the siNC group (n = 3). F, Treatment with siPLRG1 significantly reduced H3K27ac and BRD4 enrichment at the CCND1 SE2 region compared with that in the siNC group (n = 3). The values are expressed as the mean ± SEM (A and B, D–F). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.
Figure 7.
DHX37 cooperates with PLRG1 to activate CCND1 transcription in liver cancer. A, Treatment with siPLRG1 significantly reduced DHX37 enrichment at the CCND1 promoter and superenhancer (both SE1 and SE2) regions compared with that in the siNC group. B, Treatment with siDHX37 significantly reduced PLRG1 enrichment at the CCND1 promoter and SE2 region compared with that in the siNC group. C, The relative RNA and protein levels of CCND1 were reduced after knockdown of PLRG1 in the context of DXH37 overexpression. D, The relative mRNA and protein levels of CCND1 were reduced after knockdown of DHX37 in the context of PLRG1 overexpression. E, The relative protein levels of CCND1 were reduced after knockout of PLRG1/DHX37 in the context of DXH37/PLRG1 overexpression. F, CCK-8 assays of the Huh7 and HepG2-C3A cells with knockout of PLRG1/DHX37 in the context of DXH37/PLRG1 overexpression. The values are expressed as the mean ± SEM (A–F, n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.
To determine which TF is involved in the recruitment of DHX37 to the promoter and superenhancer of CCND1, we performed JASPAR TFBS enrichment analysis (20) to predict the possible TFs. The results showed that 10 TFs had simultaneous binding sites at the promoter, SE1 and SE2 regions of CCND1, simultaneously. According to the expression of these TFs in Huh7 and HepG2-C3A cells, seven of them were selected for an siRNA knockdown screening assay (Supplementary Fig. S9A). Notably, we obtained two TFs, MAFF and ZNF263, that significantly decreased CCND1 expression following siRNAs against to each TF in the Huh7 cells (0.5 as the cutoff; Supplementary Fig. S9B). Importantly, by the analyzing ChIP-seq data of MAFF and ZNF263 in the liver cancer cell line HepG2 from ENCODE, we found that ZNF263 also occupies both the promoter and superenhancer regions of CCND1 (Supplementary Fig. S9C). Intriguingly, overexpression of either DHX37 or PLRG1 upregulated CCND1 mRNA and protein expression in liver cancer cells, but this increase was abolished by knockdown of ZNF263 (Supplementary Fig. S9D and S9E). These results suggested that ZNF263 is required for DHX37/PLRG1-mediated transcriptional activation of CCND1.
CCND1 is required for DHX37/PLRG1-promoted liver cancer cell proliferation
Given that DHX37 can cooperate with PLRG1 to activate CCND1 transcription, we next analyzed the expression levels of DHX37, PLRG1, and CCND1 in HCC-based on data from TCGA and Genotype-Tissue Expression databases. DHX37, PLRG1, and CCND1 levels were upregulated in HCC (Supplementary Fig. S10A). Notably, the mRNA level of CCND1 correlated significantly with those of DHX37 and PLRG1 in patients with HCC (Fig. 8A). Importantly, among patients with HCC, the Spearman correlation coefficient (r) was higher in patients with neoplasms of high histologic grade (Fig. 8B). As shown above, high expression of DHX37 and PLRG1 can transcriptionally activate CCND1 expression. CCND1 is required for the G1–S transition and the growth of HCC cells (21), as confirmed by our results (Fig. 8C; Supplementary Fig. S10B). Importantly, colony formation assays showed that overexpression of DHX37 and PLRG1 promoted liver cancer cell proliferation, whereas this effect was reversed by CCND1 knockdown (Fig. 8D; Supplementary Fig. S10C). Collectively, these results suggested that DHX37 and PLRG1 coregulate liver cancer cell proliferation through activation of CCND1. Intriguingly, small-molecule inhibitors of the CCND1/cyclin-dependent kinase (CDK) 4/6 axis are already available, and we sought to explore whether DHX37 may be a therapeutic option for liver cancer. According to the IC50 values of palbociclib (Palb, CDK4/6 inhibitor) in stable DHX37-overexpressing and control Huh7 cells (Supplementary Fig. S10D), we treated two liver cancer cell lines with Palb at varying doses. The results showed that Palb reduced cell viability (Palb, 10 µmol/L) and colony formation capabilities (Palb, 1 µmol/L) across the cell lines with DHX37 overexpression (Fig. 8E; Supplementary Fig. S10E and S10F). Given that DHX37 activates enhancer regions in HCC, we tested the therapeutic efficacy of BET inhibitors. We found that treatment of liver cancer cells showed reduced viability upon treatment with the BET inhibitors JQ1 and OTX015 (Fig. 8F; Supplementary Fig. S10G and S10H). These results suggested that CCND1 inhibitors and BET inhibitors are effective as antiproliferative agents for liver cancer, particularly for patients with DHX37-high liver cancer.
Figure 8.
CCND1 is required for DHX37/PLRG1-promoted liver cancer cell proliferation. A, Correlation plots comparing DHX37, PLRG1, and CCND1 expression in TCGA-LIHC. r, correlation coefficient. B, Correlation plots comparing DHX37, PLRG1, and CCND1 expression in TCGA data for neoplasms with different histologic [tumor–node–metastasis (TNM)] grades in patients with HCC. C, Colony formation assays and representative images of the Huh7 and HepG2-C3A cells transfected with two independent CCND1 siRNAs. D, Colony formation assays and colony counts of the Huh7 and HepG2-C3A cells, with stable DHX37 or PLRG1 overexpression transfected with two independent CCND1 siRNAs. E, Viability of mock and stable DHX37-overexpressing Huh7 cells after 48 hours of treatment with 10 µmol/L palbociclib (Palb). F, Viability of the mock and stable DHX37-overexpressing Huh7 cells after 48 hours of treatment with 10 µmol/L JQ1 or OTX015. G, Proposed model of the transcriptional regulation of CCND1 in liver cancer. The values are expressed as the mean ± SEM (D–F, n = 3). *, P < 0.05; **, P < 0.01; ***, P < 0.001 by one-way ANOVA or two-sided Student t test.
Discussion
RNA helicases are highly conserved enzymes that bind and hydrolyze ATP to remodel RNA structures or displace proteins from ribonucleoprotein complexes. RNA helicase family members are involved in all facets of RNA metabolism from biogenesis to decay, such as RNA splicing, RNA degradation, and ribosome biosynthesis (22). Moreover, emerging evidence suggests that numerous RNA helicases are involves in processes that are critical to cell proliferation and/or malignant transformation and are involved in cancer development and progression (23, 24). Indeed, these molecules have been found to function either as oncogenes or tumor suppressor genes in various cancer types (25). For instance, DDX3 has been demonstrated to have a dual function and can act as an oncogene or a tumor suppressor, enhancing or inhibiting cancer progression in various cancer types (26). In addition, several studies have reported that RNA helicases such as DDX1, DDX5, and DDX17 function as transcriptional coregulators and are crucial in modulating numerous cellular signaling pathways (27, 28). In this study, we observed that DHX37 was one of the top genes with upregulated expression in liver cancer by systematically analyzing the expression profiles of RNA helicases based on TCGA-LIHC datasets. In particular, high expression of DHX37 correlated with poor OS and PFS in patients with HCC and was found to be functionally critical for liver cancer cell viability through the activation of superenhancer-associated oncogenes. These observations uncovered a previously uncharacterized role of DHX37 in modulating the proliferation of liver cancer cells, enhancing our understanding of the functions and biological roles of RNA helicases in cancer development and progression.
DHX37, a highly conserved DEAH-box RNA helicase, is involved mainly in ribosomal biogenesis (5, 6) and has antiviral activity (7). DHX37 has been reported to be able to interact with the NFκB subunit p65 and PDCD11 to regulate NFκB pathway activity in CD8-positive T cells; this finding was the first to show that DHX37 is related to the progression of cancer (8). Although DHX37 was discovered to impact the prognosis of HCC through integrative expression and prognostic analysis of TCGA data, its functional effects, especially the mechanism by which it mediates HCC cell proliferation and growth, are largely unknown. In the current study, we found that the differential expression of genes in the DHX37-high and DHX37-low patient groups was related to enhancer activity in patients with HCC and that DHX37 is involved in the regulation of superenhancer activity in HCC cells. Notably, we demonstrated that DHX37 binds directly to the promoter and superenhancer regions of CCND1, thereby activating its expression in liver cancer cells. RNA helicases have been observed to be involved in chromatin interactions and transcriptional regulation. Similarly, we showed the interaction of DHX37 and PLRG1; they co-occupied CCND1 superenhancers and promoted their activities. These results indicated that DHX37 is recruited to responsive promoters and acts by modulating transcriptional initiation. To the best of our knowledge, our study is the first to investigate the transcriptional regulatory role of DHX37/PLRG1.
CCND1 interacts with CDK 4/6, thereby promoting the G1-S transition (29, 30). CCND1 is a well-recognized oncogene that is overexpressed in a substantial proportion of human cancers. It has been speculated that small molecules targeting CCND1/CDK (4/6) may be useful as therapeutic agents against human tumors in clinical trials (31–34). Several mechanisms by which CCND1 may be deregulated in cancers have been proposed. For instance, cancer genome sequencing has demonstrated that CCND1 is always amplified in breast cancer (35). Another mechanism reported to enhance CCND1 expression is juxtaposition of the IgH locus next to CCND1 in multiple myeloma (36). Interestingly, transcriptional activation of the CCND1 oncogene is tissue specific. Pathologic transcriptional activation of CCND1 in breast cancer cells depends on a cell-type–specific enhancer through a combinatorial network mediated by the action of ERα directly at the CCND1 gene (37). In HCC, OCT4 binds directly to the CCND1 promoter and participates in regulating CCND1 promoter activity. However, high CCND1 promoter activity was also observed in OCT4-negative liver cancer cells, suggesting that there are other factors also regulate CCND1 promoter activity in HCC cells (38). Therefore, tissue-specific cis-regulatory elements may play important roles in spatiotemporal gene regulation. In the current study, we observed that gain of the superenhancer at CCND1, which is favorable for liver cancer growth, was abolished after the silencing of DHX37. Superenhancers are relatively large clusters of transcriptional enhancers that promote the expression of critical genes and drive tumorigenesis (39). Cancer cells have been shown to acquire superenhancers at oncogenes through multiple mechanisms. For example, overexpression of trans-acting factors in cancers was shown to be associated with superenhancer formation at oncogenes (40). Here, we demonstrated that DHX37 and PLRG1 co-occupy to the promoter and superenhancer regions of CCND1 and contribute to its transcriptional activity in liver cancer. Our findings suggested that overexpression of DHX37 may serve as an additional event driving the transcription of CCND1 through a superenhancer and constitute an additional mechanism, leading to increased expression of CCND1 in liver cancer. Given that we understand that DHX37 and PLRG1 occupy enhancer regions and that CCND1 is an important target, inhibition of enhancer activity or CCND1 activity may provide improved therapeutic opportunities for patients with HCC. In a previous study, the BRD4 inhibitor JQ1 inhibited the binding of BDR4 to the promoter of the CCND1 gene, leading to a reduction in CCND1 in thyroid tumors (41). However, the physiological mechanisms and regulatory processes of BET inhibition on CCND1 in HCC remain unclear. To investigate whether BET inhibitors have an impact on the expression of CCND1 in liver cancer, we performed real-time qPCR to determine the mRNA level of CCND1 using mock liver cancer cells treated with vehicle or BET inhibitors. The results showed that treatment of liver cancer cells showed reduced CCND1 mRNA expression upon treatment with the BET inhibitors JQ1 and OTX015 (Supplementary Fig. S10I). Most importantly, treatment of the DHX37-overexpressing cell line with the CDK4/6 inhibitor and BET inhibitors led to dramatic inhibition of cell proliferation. BET inhibitors can decrease MYC expression levels and cause effective antitumor effects in diverse human cancers (42, 43). Our results provide new insight into BET inhibitors suppressing cell growth of HCC through the downregulation of CCND1 expression. Taken together, these results suggested opportunities for improving the treatment of patients with high DHX37 expression, perhaps by combining a CDK4/6 inhibitor and BET inhibitors with other agents for precision therapy of liver cancer.
In summary, we provided evidence highlighting a close cooperative mechanistic interaction among DHX37, PLRG1, and superenhancers that contributes to liver cancer malignancy. DHX37 expression is frequently increased in patients with HCC, and DHX37 interacts with PLRG1 and co-occupies the CCND1 promoter and superenhancers regions to activate CCND1 transcription. These events collectively promote the proliferation of cancer cells and contribute to the poor prognosis of liver cancer patients (Fig. 8G). These findings provide new insight into the regulation of DHX37/PLRG1-CCND1 signaling in the progression of liver cancer, and this insight may facilitate cancer diagnosis and treatment.
Supplementary Material
Acknowledgments
The authors are grateful for Prof. Jian Xu's gifts of the enCRISPRi plasmids. This work was supported by grants from the National Natural Science Foundation of China to Z. Chen (82172937 and 81972247) and X. He (81930123 and 81790252) and the Natural Science Foundation of Shanghai to Z. Chen (18ZR1407900).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
No disclosures were reported.
Authors' Contributions
Z. Liu: Validation, investigation, visualization, methodology, writing–original draft. Y. Ye: Resources, software, formal analysis, visualization. Y. Liu: Methodology. Y. Liu: Methodology. H. Chen: Resources, software, formal analysis. M. Shen: Methodology. Z. Wang: Methodology. S. Huang: Software, methodology. L. Han: Software. Z. Chen: Software, supervision, funding acquisition, writing–review and editing. X. He: Supervision, funding acquisition, writing–review and editing.
References
- 1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2. Villanueva A. Hepatocellular carcinoma. N Engl J Med 2019;380:1450–62. [DOI] [PubMed] [Google Scholar]
- 3. Cai W, Xiong Chen Z, Rane G, Satendra Singh S, Choo Z, Wang C, et al. Wanted DEAD/H or alive: helicases winding up in cancers. J Natl Cancer Inst 2017;109. [DOI] [PubMed] [Google Scholar]
- 4. Fuller-Pace FV. DEAD box RNA helicase functions in cancer. RNA Biol 2013;10:121–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boneberg FM, Brandmann T, Kobel L, Heuvel J, Bargsten K, Bammert L, et al. Molecular mechanism of the RNA helicase DHX37 and its activation by UTP14A in ribosome biogenesis. RNA 2019;25:685–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Choudhury P, Hackert P, Memet I, Sloan KE, Bohnsack MT. The human RNA helicase DHX37 is required for release of the U3 snoRNP from pre-ribosomal particles. RNA Biol 2019;16:54–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rahman MM, Bagdassarian E, Ali MAM, McFadden G. Identification of host DEAD-box RNA helicases that regulate cellular tropism of oncolytic Myxoma virus in human cancer cells. Sci Rep 2017;7:15710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dong MB, Wang G, Chow RD, Ye L, Zhu L, Dai X, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell 2019;178:1189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen H, Jiang Z, Yang B, Yan G, Wang X, Zang S. Exploring prognostic signatures of hepatocellular carcinoma and the potential implications in tumor immune microenvironment. Comb Chem High Throughput Screen 2021[Online ahead of print]. [DOI] [PubMed] [Google Scholar]
- 10. Xu Y, Jiang Q, Liu H, Xiao X, Yang D, Saw PE, et al. DHX37 impacts prognosis of hepatocellular carcinoma and lung adenocarcinoma through immune infiltration. J Immunol Res 2020;2020:8835393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang K, Pang T, Tong C, Chen H, Nie Y, Wu J, et al. Integrative expression and prognosis analysis of DHX37 in human cancers by data mining. Biomed Res Int 2021;2021:6576210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet 2013;45:1113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang Z, Lee JH, Ruan H, Ye Y, Krakowiak J, Hu Q, et al. Transcriptional landscape and clinical utility of enhancer RNAs for eRNA-targeted therapy in cancer. Nat Commun 2019;10:4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods 2009;48:240–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu X, Wang Y, Lu H, Li J, Yan X, Xiao M, et al. Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 2019;567:525–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Warren AW, David AO, Denes H, Brian JA, Charles YL, Michael HK, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013;153:307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jakob L, Heather AH, Charles YL, Ashley L, David AO, Christopher RV, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153:320–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li K, Liu Y, Cao H, Zhang Y, Gu Z, Liu X, et al. Interrogation of enhancer function by enhancer-targeting CRISPR epigenetic editing. Nat Commun 2020;11:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Castro-Mondragon JA, Riudavets-Puig R, Rauluseviciute I, Berhanu Lemma R, Turchi L, Blanc-Mathieu R, et al. JASPAR 2022: the 9th release of the open-access database of transcription factor binding profiles. Nucleic Acids Res 2022;50:D165–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol 2004;5:232–41. [DOI] [PubMed] [Google Scholar]
- 23. Sergeeva O, Zatsepin T. RNA Helicases as shadow modulators of cell cycle progression. Int J Mol Sci 2021;22:2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taschuk F, Cherry S. DEAD-box helicases: sensors, regulators, and effectors for antiviral defense. Viruses 2020;12:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robert F, Pelletier J. Perturbations of RNA helicases in cancer. Wiley Interdiscip Rev RNA 2013;4:333–49. [DOI] [PubMed] [Google Scholar]
- 26. Zhao LQ, Mao YT, Zhou JH, Zhao YL, Cao Y, Chen X. Multifunctional DDX3: dual roles in various cancer development and its related signaling pathways. Am J Cancer Res 2016;6:387–402. [PMC free article] [PubMed] [Google Scholar]
- 27. Tanaka K, Okamoto S, Ishikawa Y, Tamura H, Hara T. DDX1 is required for testicular tumorigenesis, partially through the transcriptional activation of 12p stem cell genes. Oncogene 2009;28:2142–51. [DOI] [PubMed] [Google Scholar]
- 28. Samaan S, Tranchevent LC, Dardenne E, Polay Espinoza M, Zonta E, Germann S, et al. The Ddx5 and Ddx17 RNA helicases are cornerstones in the complex regulatory array of steroid hormone-signaling pathways. Nucleic Acids Res 2014;42:2197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: normal and abnormal functions. Endocrinology 2004;145:5439–47. [DOI] [PubMed] [Google Scholar]
- 30. Ewen ME, Lamb J. The activities of cyclin D1 that drive tumorigenesis. Trends Mol Med 2004;10:158–62. [DOI] [PubMed] [Google Scholar]
- 31. Geoerger B, Bourdeaut F, DuBois SG, Fischer M, Geller JI, Gottardo NG, et al. A phase I study of the CDK4/6 inhibitor ribociclib (LEE011) in pediatric patients with malignant rhabdoid tumors, neuroblastoma, and other solid tumors. Clin Cancer Res 2017;23:2433–41. [DOI] [PubMed] [Google Scholar]
- 32. Hortobagyi GN, Stemmer SM, Burris HA, Yap YS, Sonke GS, Paluch-Shimon S, et al. Ribociclib as first-line therapy for HR-positive, advanced breast cancer. N Engl J Med 2016;375:1738–48. [DOI] [PubMed] [Google Scholar]
- 33. Niesvizky R, Badros AZ, Costa LJ, Ely SA, Singhal SB, Stadtmauer EA, et al. Phase 1/2 study of cyclin-dependent kinase (CDK)4/6 inhibitor palbociclib (PD-0332991) with bortezomib and dexamethasone in relapsed/refractory multiple myeloma. Leuk Lymphoma 2015;56:3320–8. [DOI] [PubMed] [Google Scholar]
- 34. Adkins D, Ley J, Neupane P, Worden F, Sacco AG, Palka K, et al. Palbociclib and cetuximab in platinum-resistant and in cetuximab-resistant human papillomavirus-unrelated head and neck cancer: a multicentre, multigroup, phase 2 trial. Lancet Oncol 2019;20:1295–305. [DOI] [PubMed] [Google Scholar]
- 35. Lundberg A, Lindstrom LS, Li J, Harrell JC, Darai-Ramqvist E, Sifakis EG, et al. The long-term prognostic and predictive capacity of cyclin D1 gene amplification in 2305 breast tumours. Breast Cancer Res 2019;21:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choudhury SR, Ashby C, Tytarenko R, Bauer M, Wang Y, Deshpande S, et al. The functional epigenetic landscape of aberrant gene expression in molecular subgroups of newly diagnosed multiple myeloma. J Hematol Oncol 2020;13:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev 2006;20:2513–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao L, Li CG, Shen SW, Yan Y, Ji WD, Wang JG. OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma. BMC Cancer 2013;13:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cui S, Wu Q, Liu M, Su M, Liu S, Shao L, et al. EphA2 super-enhancer promotes tumor progression by recruiting FOSL2 and TCF7L2 to activate the target gene EphA2. Cell Death Dis 2021;12:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell 2013;155:934–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhu X, Enomoto K, Zhao L, Zhu YJ, Willingham MC, Meltzer P, et al. Bromodomain and extraterminal protein inhibitor JQ1 suppresses thyroid tumor growth in a mouse model. Clin Cancer Res 2017;23:430–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yin Y, Sun M, Zhan X, Wu C, Geng P, Sun X, et al. EGFR signaling confers resistance to BET inhibition in hepatocellular carcinoma through stabilizing oncogenic MYC. J Exp Clin Cancer Res 2019;38:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garcia PL, Miller AL, Kreitzburg KM, Council LN, Gamblin TL, Christein JD, et al. The BET bromodomain inhibitor JQ1 suppresses growth of pancreatic ductal adenocarcinoma in patient-derived xenograft models. Oncogene 2016;35:833–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in this study are available within the article and its Supplementary Data files. The raw data generated in this study are publicly available in GEO at GSE184796, GSE184797, and GSE185701.




![Figure 8. CCND1 is required for DHX37/PLRG1-promoted liver cancer cell proliferation. A, Correlation plots comparing DHX37, PLRG1, and CCND1 expression in TCGA-LIHC. r represents the correlation coefficient. B, Correlation plots comparing DHX37, PLRG1, and CCND1 expression in TCGA data for neoplasms with different histologic [tumor–node–metastasis (TNM)] grades in patients with HCC. C, Colony formation assays and representative images of the Huh7 and HepG2-C3A cells transfected with two independent CCND1 siRNAs. D, Colony formation assays and colony counts of the Huh7 and HepG2-C3A cells with stable DHX37 or PLRG1 overexpression transfected with two independent CCND1 siRNAs. E, Viability of mock and stable DHX37-overexpressing Huh 7 cells after 48 hours of treatment with 10 µmol/L palbociclib. F, Viability of the mock and stable DHX37-overexpressing Huh7 cells after 48 hours of treatment with 10 µmol/L JQ1 or OTX015. G, Proposed model of the transcriptional regulation of CCND1 in liver cancer. The values are expressed as the mean ± SEM (D–F, n = 3). ***, P < 0.001; **, P < 0.01; and *, P < 0.05 by one-way ANOVA or two-sided Student t test.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/dd48/9359749/ac11bc812757/1937fig8.jpg)