Abstract
Cytokine storm is a key feature of sepsis and severe stage of COVID-19, and the immunosuppression after excessive immune activation is a substantial hazard to human life. Both pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) are recognized by various pattern recognition receptors (PRRs), which lead to the immune response. A number of neolignan analogues were synthesized in this work and showed powerful anti-inflammation properties linked to the response to innate and adaptive immunity, as well as NP-7 showed considerable anti-inflammatory activity at 100 nM. On the sepsis model caused by cecum ligation and puncture (CLP) in C57BL/6J mice, NP-7 displayed a strong regulatory influence on cytokine release. Then a photo-affinity probe of NP-7 was synthesized and chemoproteomics based on stable isotope labeling with amino acids in cell cultures (SILAC) identified Immunity-related GTPase M (IRGM) as a target suppressing cytokine storm, which was verified by competitive pull-down, cellular thermal shift assay (CETSA), drug affinity responsive target stability (DARTS) and molecular dynamics simulations.
Keywords: SILAC, Chemoproteomics, Neoligan, Cytokine storm, IRGM
Graphical abstract
1. Introduction
Sepsis is a worldwide public health concern with high morbidity, severe illness, and high mortality, and it is one of the leading causes of death among intensive care unit (ICU) patients [[1], [2], [3]]. Sepsis is a catastrophic organ failure condition induced by a viral or bacterial infection that causes an inappropriate host response. Sepsis and septic shock are significant health issues that afflict millions of individuals each year across the globe. For a long time, the majority of sepsis research has concentrated on cytokine storm and the resultant multiple organ failure syndrome multiple organ failure syndrome [4,5]. Sepsis has been named the defining medical condition of our time since it is a leading cause of morbidity and death in hospitalized patients with a variety of disorders [6,7].
Coronavirus disease 2019 (COVID-19), caused by a pathogen known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and discovered as a novel enveloped RNA beta-coronavirus, is quickly spreading over the globe. More and more researches show that the progression of critically ill patients is associated with the occurrence of cytokine storm, which is characterized by a high level of serum pro-inflammatory cytokines such as TNF-α, IL-6, IL-1β, and IL-18, posing a serious threat to the medical and public health systems [8,9]. Cytokine storm, characterized by life-threatening systemic hyperinflammation, has been discovered in patients with severe new coronavirus infection and is greatly raised in COVID-19 patients. It generates severe lung tissue edema and pulmonary capillary leaks, which may lead to acute respiratory distress syndrome (ARDS) or even multiple organ failure. However, no effective and safe targeted chemicals have yet been produced [[10], [11], [12]]. Glucocorticoids, which have an immunosuppressive effect and aid to minimize cytokine storm, are used in the treatment of severe and critical cases. However, glucocorticoid therapy may have substantial adverse effects such as superinfection risks and a prolonged illness course. Therefore, it is urgent to develop medications modulating cytokine storm with a particular therapeutic target and no side effects [[13], [14], [15]].
The immune response system is outfitted with a number of cell-surface or intracellular pattern recognition receptors (PRRs) that can detect pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs) [16,17]. In sepsis, persistent immune stimulation is triggered not only by invading pathogens, but also by the release of endogenous substances released from injured cells, culminating in a vicious cycle of immune activation and dysfunction. According to emerging data, regulating the innate and adaptive immune response is an effective method for combating pathogenic microorganism infection [18,19]. Autophagy-Related IRGM is associated with mortality in individuals with severe sepsis and has the ability to control mTOR and TFEB at a regulatory node crucial for pathogen responses. As a consequence, IRGM may be a feasible target for inhibiting cytokine release in sepsis [20,21].
The modern novel drug development process comprises essentially of lead compound discovery, target identification, structural optimization, preclinical and clinical trials, and one of the important aspects impacting the whole process is proper target recognition. Natural products continue to be a source of lead molecules in therapeutic research and development [[22], [23], [24]]. Chemoproteomics, which has played a vital role in encouraging novel drug discovery based on the development of biorthogonal chemistry, has achieved great progress in chemical biology research. As a consequence of chemoproteomics applied to identify chemical targets, considerable breakthroughs in target identification and novel drug development have been made [[25], [26], [27]]. Affinity chromatography in conjunction with mass spectrometry analysis can be used for target fishing, protein enrichment, and large-scale proteomic target identification, and it may be the most sensitive and effective method for target discovery and identification. Target identification aids in elucidating the molecular mechanism and potential adverse effects of compounds, which is important in the study of the safety and effectiveness of new compounds. Small molecular probes are commonly used for target identification. Canonical affinity probes typically include an active ligand, a linker, and an affinity tag, such as biotin groups, to separate and enrich target proteins. Following that, the target protein is identified and analyzed using mass spectrometry [[28], [29], [30]].
Denudatin B has been reported to have antiplatelet aggregation and anti-inflammatory properties [[31], [32], [33], [34], [35]]. Anti-inflammatory activity has been discovered in a number of synthetic neolignans [36]. Scutellariae Radix is a popular ethnobotanical herb used to treat various ailments ranging from inflammation to nervous disorders [37,38]. As a consequence, aqueous extracts of S. baicalensis roots were investigated to discover anti-inflammatory compounds from traditional Chinese medicines, and the neolignan constituents were found to have strong anti-inflammatory activities [39,40]. In the synthesis process of Denudatin B, we discovered that their analogues displayed anti-inflammation activities. A photo-affinity probe of NP-7 was designed and synthesized, and the target of NP-7 was identified by SILAC pull-down chemoproteomics [41,42]. Cellular thermal shift assay (CETSA), drug affinity responsive target stability (DARTS) and molecular dynamics simulations verified the direct combination of NP-7 with target [[43], [44], [45], [46]].
2. Results and discussion
2.1. Design and synthesis of Denudatin B
Denudatin B was prepared from derivatives of 3,4-dimethoxycinnamic acid and resorcinol (in Scheme I ). The total synthesis is outlined in Scheme I. A preliminary account of this work has appeared, and based on this we optimized the experimental scheme. Direct alkylation of resorcinol with allyl bromide in N, N-dimethylformamide containing potassium carbonate as an acid-binding agent gave 3 in 60% yield. The starting alcohol 3 was prepared in very low yields from the 3,4-dimethoxycinnamic acid by direct reduction with lithium aluminum hydride (LiAlH4) in tetrahydrofuran as well as the olefinic bond would also be reducted. 3,4-dimethoxycinnamic acid was esterified in methanol containg 1 N HCl with a 99% yield to obtain compound 1 and then reducted by diisobutylaluminium hydride (DIBALH) with a yield of 98% yield to obtain compound 2, the yields of both two steps were about almost quantitative. Condensation of 2 and 3 under Mitsunobu reaction conditions with new catalyst reagent 1,1'-(azodicarbonyl)-dipiperidine (ADDP) and tributylphosphine (TBUP) achieved ether 4 with 40% yield and then hydrolized in alkaline aqueous solution with lithium hydroxide to obtain compound 5. Compound 5 was allylated to obtain ether 6 and heated in a sealed tube dissolved in N, N-diethylaniline at 225 °C and the thermal reaction apparently involved two Claisen rearrangements followed by an abnormal Claisen (1,5 homosigmatropic hydrogen shift) to obtain neoligan phenol 7. Oxidative methoxylation of 7 with lead tetraacetate in dry methanol gave a mixture of racemic products and achieved 8(Denudatin B)with a yield of 20%.
Scheme 1.
Synthesis of Denudatin B. Reagents and conditions: a) methanol, acetyl chloride,0∼rt; b) dichloromethane, DIBAL-H, −78 °C; c) acetyl chloride, TEA, 0 °C∼rt; d) dichloromethane, TBUP, ADDP, compound 3, -78∼-60 °C; e) LiOH, H2O-THF, 0 °C∼rt; f) allyl bromide, DMF, K2CO3, rt; g) N, N-diethylaniline,230 °C; h) methanol, lead tetraacetate,0 °C∼rt.
2.2. Effect of the neoligan analogues on the M1 polarization and TNF-α expression of macrophages
The immune response to macrophages, which can cause inflammation, is a critical component of innate immunity and their functions are regulated by the cytokines of the surrounding environment. Macrophages are typically divided into two subsets: classically activated (M1) macrophages, which are pro-inflammatory and polarized by lipopolysaccharide (LPS), and alternatively activated (M2) macrophages, which are anti-inflammatory. They have unique properties, such as the capacity to eliminate pathogens like bacteria and viruses or to heal inflammation-related injury [[47], [48], [49], [50]]. Screening showed that compound 6 and 7 could lower the M1 polarization of macrophages at 100 nM and 500 nM, while compound 5 and 8 had no effect. All of the compounds had an effect on reducing the expression levels of TNF-α at 100 nM and 500 nM, especially the neoligan phenol analogue 7 (NP-7) exhibited the most potent anti-inflammation, equivalent to hydrocortisone, a positive control drug.
2.3. Effect of the neoligan analogues on CD4+ T lymphocyte subsets differentiation: Th1/Th2/Th17
T-helper cells can create the vast majority of known cytokines [[51], [52], [53]]. Aside from the signature effector cytokines IFN-γ, IL-4, and IL-17A, Th cells may selectively express a variety of other important cytokines, including lymphotoxin for Th1; IL-5 and IL-13 for Th2; and IL-22 for Th17 cells. Th cells are not only professional cytokine producers, but they can also respond to cytokines produced by accessory cells, such as IL-1, IL-2, IL-12, IL-23, IL-27, IL-33, and type 1 IFNs. During differentiation, Th cells may react to their own cytokines, such as IFN-γ and IL-4, resulting in substantial positive feedback or cross-inhibitory effects [54,55].
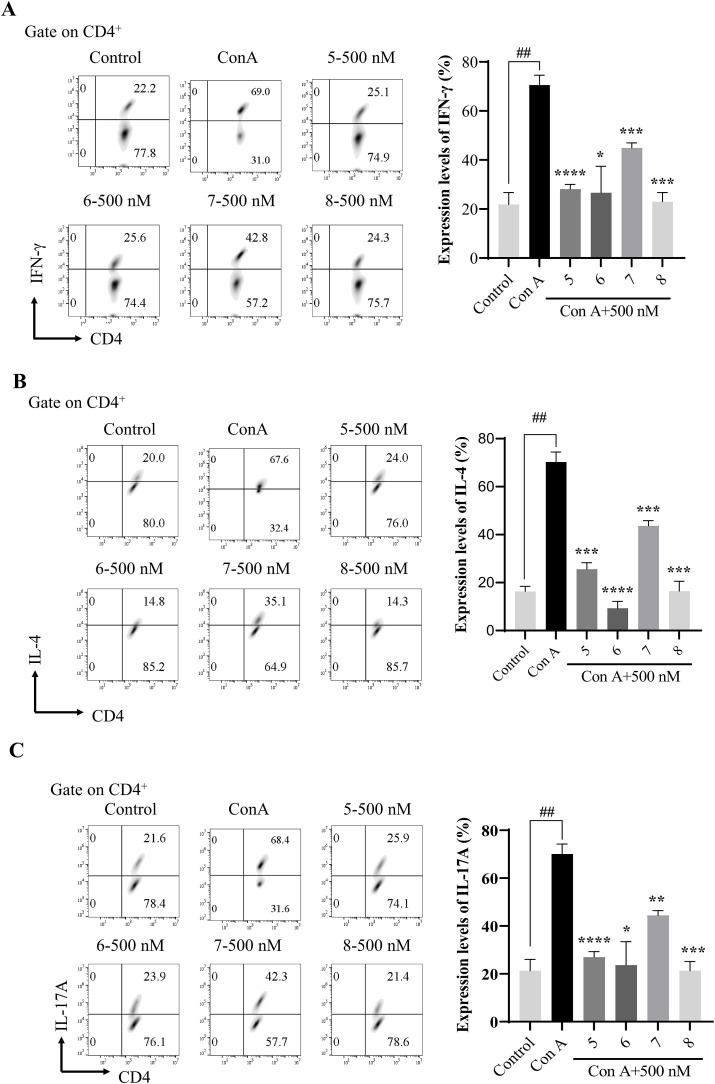
Immune cells were isolated from the spleen of C57BL/6 mice and cultured in complete medium containing ConA and IL-2 under corresponding differentiation conditions of Th1, Th2, Th17 for 48 h with or without compound treatments [56]. As shown in Fig. 1 A, Th1 cells were successfully induced, compounds 5, 6, 7 and 8 showed a significant effect on decreasing the IFN-γ production, indicating the effect of inhibiting Th1 polarization and presented no obvious cytotoxicity on CD4+ T lymphocytes up to 48 h. As shown in Fig. 2 B, Th2 cells were successfully induced, and compounds 5, 6, 7 and 8 decreased the IL-4 production, indicating the effect of inhibiting Th2 polarization and showed no obvious cytotoxicity on CD4+ T lymphocytes up to 48 h. As shown in Fig. 2 C, Th17 cells were successfully induced, and compounds 5, 6, 7 and 8 decreased the IL-17A production, indicating the effect of inhibiting Th17 polarization and displayed no obvious cytotoxicity on CD4+ T lymphocytes up to 48 h.
Fig. 1.
(A) Effect of the neoligan analogues on M1 polarization of macrophage; (B) Effect of the neoligan analogues on TNF-α expression of M1 macrophages. The representative results are shown in the left panel, and the data of three independent experiments are presented as mean ± SEM; ####p < 0.0001vs. LPS; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. LPS.
Fig. 2.
(A)Effect of compounds 5, 6, 7 and 8 on Th1 cell differentiation; (B)Effect of compounds 5,6,7 and 8 on Th2 cell differentiation; (C) Effect of compounds 5, 6, 7 and 8 on Th17 cell differentiation. (All gate on CD4+) The representative results were shown in the left panel, and the data of three independent experiments are presented as mean ± SEM; ##p < 0.01, vs. ConA; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. ConA.
2.4. Therapeutic effect of the neolignan analogue NP-7 on CLP-induced sepsis model
Sepsis is a potentially fatal condition that affects the neurological, immunological, endocrine, and coagulation systems. Various adverse stimuli may trigger the body's immune response, resulting in the production of a significant number of pro-inflammatory cytokines, which in turn induce the cascade of inflammation, playing an important part in the gradual worsening of systemic inflammatory response. Following the standard CLP methods, a mouse sepsis model was produced. It is intimately tied to pathological alterations in many systems and organs. A mouse sepsis model was produced using standard CLP protocols [57,58].
The establishment of a sepsis model significantly upregulated the proportion of M1 macrophage and neutrophil subpopulations in the blood. Apparently, 100 mg/kg NP-7 reversed this phenomenon (Fig. 3 A and B). The changes in the ratio of immune cell subpopulations in the blood can roughly represent the status of the systemic immune response, and it is clear from the results that the application of the drug effectively reduces the strong inflammatory response associated with sepsis.
Fig. 3.
(A) Effect of compound NP-7 on neutrophil of CLP mice; (B) Effect of compound NP-7 on M1 macrophages of CLP mice (gate on CD11b+). The representative results are shown in the left panel, and the data are presented as mean ± SEM; ##p < 0.01 vs. CLP; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. CLP (n = 4–8).
In septic mice, the levels of Th1 cells and Th2 cells secreting their signature cytokines were significantly higher compared to controls (Fig. 4 A and B). However, the expression levels of IFN-γ were much higher than that of IL-4, suggesting that the balance between Th1 and Th2 cells in septic mice had been disrupted and the organism was undergoing a strong immune response. While NP-7 could significantly reduce the expression levels of IFN-γ and IL-4, the balance between Th1 and Th2 cells was restored in the treated group of model mice compared to the model mice.
Fig. 4.
(A) Effect of compound NP-7 on Th1 cell differentiation of CLP mice; (B) Effect of compound NP-7 on Th2 cell differentiation and the expression levels of IL-4 of CLP mice. (All gate on CD4+). The representative results are shown in the left panel, and the data are presented as mean ± SEM; ###p < 0.001 vs. CLP; ***p < 0.001 vs. CLP (n = 4–8).
Similar to the balance between Th1/2 cells, there was a balance between Th17 and Treg cells, with Treg cells directly inhibiting Th17 cells via IL-10 to control inflammation (Fig. 5 A and B). And Treg cells could be subdivided into Foxp3+Treg and Tr1 (which only secrete IL-10 without expressing Foxp3), and we examined the ratio of these cells by flow cytometry. The results showed that the NP-7 could effectively reduce the expression level of IL-17A, but had no significant activation effect on Foxp3+ Treg cells and Tr1 cells, which was consistent with the results of drug screening experiments.
Fig. 5.
(A) Effect of compound NP-7 on Th17 cell differentiation of CLP mice, (gate on CD4+); (B) Effect of compound NP-7 on Foxp3+ Treg cells and Tr1 differentiation of CLP mice (gate on CD4+CD25+). The representative results are shown in the left panel, and the data are presented as mean ± SEM; #p < 0.05 vs. CLP; **p < 0.01, ***p < 0.001 vs. CLP (n = 4–8).
The results of H&E staining of liver tissues showed edema and hepatocyte necrosis in the model group, while the state of the liver was significantly improved after administration of the NP-7, demonstrating that NP-7 was not hepatotoxic and its anti-inflammatory activity was sufficient to protect the liver (Fig. 6 ).
Fig. 6.
Effects of the NP-7 on the histological changes in CLP mice. The upper panel is low magnification (scale bar = 100 μm) of the images, the areas inside the boxes are shown in the lower panel at high magnification (scale bar = 50 μm).
2.5. The synthesis of neoligan photo-affinity-lablled probe and anti-inflammation activity screening
The photocrosslinking agent 9 was synthesized according to the reported procedure [[59], [60], [61]] and the probe 10 of NP-7 was synthesized in just one step (Scheme II ).
Scheme 2.
The synthesis of the PAL probe 10.
Compound screening showed that NP-7 as well as its probe 10 could lower the M1 polarization of macrophages and reduce the expression levels of TNF-α in M1-polarized macrophages (Fig. 7 A and B). This result proved that the design of the molecular probe was correct, the modification of the NP-7 does not affect its pharmacological activity and compound 10 could be an active probe for detecting corresponding anti-inflammatory and immune-related targets for NP-7.
Fig. 7.
The effect of probe 10 and NP-7 and on 1 μg/mL LPS induced RAW 264.7 inflammation model. (A) Effect of NP-7 and its probe 10 on M1 polarization of macrophages; (B) Effect of NP-7 and its probe 10 on TNF-α expression of M1 macrophages. The representative results are shown in the left panel, and the data of three independent experiments are presented as mean ± SEM; ##p < 0.01, ####p < 0.0001 vs. LPS; **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. LPS.
2.6. SILAC-pull down method identified IRGM as the target of NP-7
The photoaffinity labeling click-chemistry activity-based protein profiling (PAL–CC–ABPP) method based on SILAC was performed based on protocols adapted from previous reports. Identification of NP-7 target proteins using pull-down technology coupled with shotgun proteomics. The heavy-labeled RAW 264.7 lysates were incubated with probe 10, while the light-labeled RAW 264.7 lysates were incubated with competitive conditions, and then the proteins which were bound to the beads with a biotin-streptavidin affinity were resolved by SDS/PAGE, followed by silver staining (Fig. 8 B). The implementation conditions of target fishing with non-labeled protein (LFQ pull-down) were basically similar, except for adding an extra vehicle control group. IRGM was identified as a target for NP-7 after MaxQuant analysis and the statistical analysis of the ratios (Heavy/Light) (Fig. 8C).
Fig. 8.
The chemical proteomics approach based on SILAC pull-down to reveal the direct target of NP-7. (A) General workflow of SILAC pull-down and MS analysis; (B) Silver stain of label free quantification pull-down (LFQ) and SILAC pull-down; (C) Ratio (Heavy/Light) of proteins from the MS analysis after SILAC pull-down.
2.7. Gene ontology analysis and immunoblotting showed NP-7 target on IRGM
The gene ontology analysis [62] of the 125 proteins revealed that the target related to regulate inflammation and immunity interacting with probe 10 was closely related to irgm1 which was an important component in the biological process of response to interferon-gamma (Fig. 9 A and B). Immunoblotting analysis of the samples from the LFQ pull-down confirmed the binding of NP-7 to target IRGM. Immunity-related GTPase family M protein 1 [63,64] is required for IFNG-mediated clearance of acute protozoan and bacterial infections. IRGM in innate immune response probably acts through regulation of autophagy, which may regulate proinflammatory cytokine production and prevent endotoxemia upon infection.
Fig. 9.
The results analysis of target fishing with and immunoblotting analysis. (A) and (B) Gene ontology analysis; (C) Immunoblotting analysis of the samples from the LFQ pull-down with probe 10.
2.8. Verification of the direct combination of NP-7 with target IRGM:competitive pull down, CETSA and molecular dynamic stimulation
Firstly, to verify the binding of target IRGM to NP-7, an immunoblotting assay showed that NP-7 could competitively and dose-dependent bind to target IRGM with 100 μM probe 10 from 10 μM to 100 μM (Fig. 10 A). Secondly, we found that NP-7 treatment efficiently protected IRGM protein from temperature-dependent denaturation with cellular thermal shift assay (CETSA) (Fig. 10B). Thirdly, we found that concentration-dependent NP-7 treatment from 1 μM to 100 μM efficiently protected IRGM protein from pronase proteolysis with drug affinity responsive target stability (DARTS) (Fig. 10C). Molecular dynamics analysis [65,66] showed that two to three hydrogen bonds were formed between the ligand and the receptor, and the residues 236PRO, 245ARG, 401ARG could form hydrogen bonds with the ligand for most of the simulation time (Fig. 10D).
Fig. 10.
Demonstrating the direct binding of NP-7 to IRGM target protein. (A) Competition pull-down with immunoblotting assay; (B) Cellular thermal shift assay; (C) DARTS; (D) Molecular dynamics simulations analysis.
3. Conclusion
We synthesized a series of neolignan derivatives, and screened their effects on the immune response to innate immunity and adaptive immunity. NP-7, a precursor compound of Denudatin B, was found to show potent activity on inhibiting the expression levels of TNF-α in M1-type pro-inflammatory macrophages at 100 nM. Besides, these compounds also displayed potent inhibition activities on the response to adaptive immunity including Th1, Th2 and Th17. Then a mouse sepsis model was established, induced by CLP in C57BL/6J mice and NP-7 treatment showed exceptional good regulatory effects on the cytokine release. Later, the method of chemoproteomics based on SILAC was applied to find the target IRGM of NP-7, which is a key protein in the metabolic reprogramming of immune activation of inherently immune macrophages. Based on this finding, IRGM and the lead compound NP-7 may provide a target to develop related new drug development for the treatment of sepsis. Immune chemical biology, combining immunology techniques with chemical proteomics, will be an excellent strategy for new drug discovery related to sepsis treatment.
4. Experiment
4.1. Chemistry
4.1.1. General chemical reagents
All reactions were conducted under argon atmosphere using anhydrous solvents from commercial sources (Adamas) without further purification unless otherwise noted. Unless otherwise noted, yield refers to a chromatographically homogeneous material. The reaction was monitored by TLC on a 0.25 mm Yantai silica gel plate (60F-254). Visualization was achieved using ultraviolet light or phosphomolybdic acid in ethanol followed by heating. Qingdao silica gel (200–300 mesh and 300–400 mesh) was used for rapid column chromatography. Using 600 MHz (1 H NMR, 600 MHz; 13C NMR, 151 MHz) spectrometer. Data are reported as follows: chemical shift, multiplicity (s = single peak, d = double peaks, t = triple, q = quadruple, br = wide, m = multiple), coupling constant, and integration.
4.1.2. General produce a for the preparation of 1–8
Synthesis of compound 1: To a solution of 3,4-dimethoxycinnamic acid (100 g, 0.48 mol) in 1 L methanol cooled to 0–5 °C was added acetyl chloride (68 ml, 0.96 mol) dropwise. Then the slurry mixture was slowly restored to room temperature and stirred overnight, and the reaction mixture would be clarified. TLC showed that the reaction finished, and most of the solvent was removed under vacuum. After that, 1 L of water was poured into the mixture and extracted with 1 L ethyl acetate twice, then the ethyl acetate was combined, washed with water, saturated sodium chloride, and dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a solid. The solid was slurried with petroleum ether: methyl tert-butyl ether (10:1, v/v) and filtered to obtain a white solid. A total amount of 105 g compound 1 was obtained as white crystals with a yield of 98.4%.1H NMR (600 MHz, CDCl3) δ 7.62 (d, J = 15.6 Hz, 1H), 7.09 (dd, J = 7.8, 1.8 Hz, 1H), 7.03 (d, J = 2.4 Hz, 1H), 6.85 (d, J = 8.4 Hz, 1H), 6.30 (d, J = 15.6 Hz, 1H), 3.91 (s, 6H), 3.84 (s, 3H).13C NMR (151 MHz, CDCl3) δ 167.64, 151.09, 149.16, 144.76, 127.32, 122.59, 115.45, 110.98, 109.55, 55.94, 55.85, 51.60.
Synthesis of compound 2: Compound 1 (60 g, 0.27 mol) was dissolved in 500 mL anhydrous dichloromethane under argon and cooled to −70 °C, then DIBALH (1080 mL, 1 N in hexane, 1.08 mol) was added dropwise. After 3 h of dropwise addition, the reaction temperature was retained below −60 °C for another 4 h. TLC showed that the reaction finished. Then, 200 mL ethyl acetate was added dropwise to quench the reaction, and stirred for 1 h to completely quench it. The resulting solution was poured into 5 L of saturated potassium sodium tartrate solution slowly, taking attention that a large amount of gas (isobutylene) would generate. Stir the mixture overnight, and the solution would be clarified, extracted with dichloromethane for twice. The organic layers were combined, washed with water, saturated sodium chloride, dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a crude solid. Then the solid was slurried with petroleum ether: methyl tert-butyl ether (10:1, v/v) and filtered to obtain a white solid. A total amount of 51.8 g compound 2 was obtained as white crystals with a yield of 98.4%. 1H NMR (600 MHz, CDCl3) δ 6.93 (d, J = 1.6 Hz, 1H), 6.90 (dd, J = 8.4 Hz, 1.8H, 1H), 6.80 (d, J = 8.2 Hz, 1H), 6.53 (d, J = 15.8 Hz, 1H), 6.23 (dt, J = 15.8, 5.9 Hz, 1H), 4.29 (d, J = 5.6 Hz, 2H), 3.88 (s, 3H), 3.87 (s, 3H), 1.72 (s, 1H).13C NMR (151 MHz, CDCl3) δ 149.08, 148.96, 131.17, 129.82, 126.64, 119.77, 111.17, 108.89, 63.88, 55.99, 55.89.
Synthesis of compound 3: Resorcinol (50 g, 0.45 mol) and triethylamine (126 mL, 0.9 mol) were dissolved in 500 mL anhydrous tetrahydrofuran and cooled to 0 °C, then acetic anhydride (85 mL, 0.9 mol) was added dropwise, stirred overnight. The reaction mixture was poured into water (1000 mL). The resulting mixture was extracted with ethyl acetate (500 mL × 2). The combined organic layers were washed twice with 1 N HCl (500 mL), once with water (500 mL) and once with brine (500 mL), dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a crude solid. Then the solid was slurried with petroleum ether: methyl tert-butyl ether (5:1, v/v) and filtered to obtain a white solid. A total amount of 40 g compound 3 was obtained as white solid with a yield of 94.2%.1H NMR (600 MHz, CDCl3) δ 7.19 (t, J = 8.1 Hz, 1H), 6.65 (ddd, J = 8.2, 2.3, 0.6 Hz, 1H), 6.63–6.61 (m, 1H), 6.55 (t, J = 2.3 Hz, 1H), 6.21 (s, 1H), 2.29 (s, 3H).13C NMR (151 MHz, CDCl3) δ 170.34, 156.77, 151.38, 130.10, 113.43, 113.33, 109.21, 21.20.
Synthesis of compound 4: To a solution of compound 2 (40 g, 0.2 mol) and compound 3 (47 g, 0.3 mol), tributylphosphine (TBUP, 103 mL, 0.4 mol) in dry dichloromethane 500 mL was added 1,1'-(azodicarbonyl)-dipiperidine (ADDP, 104 g, 0.4 mmol) dissolved in dry dichloromethane 500 mL dropwise at 0 °C under argon and then stirred at room temperature overnight. Then the reaction mixture was added petroleum ether and filtered and discarded the solid. The resulting filtrate was concentrated under vacuum to get a crude product, which was purified by silica gel column chromatography with petroleum ether/ethyl acetate as eluents. A total amount of 28 g compound 4 was obtained as white solid with a yield of 41.4%.1H NMR (600 MHz, CDCl3) δ 7.28 (d, J = 7.1 Hz, 1H), 7.21 (dd, J = 10.3, 6.0 Hz, 1H), 6.96 (d, J = 19.5 Hz, 2H), 6.83 (d, J = 8.1 Hz, 2H), 6.69 (d, J = 21.4 Hz, 3H), 6.65 (s, 1H), 6.58 (t, J = 2.3 Hz, 1H), 6.30–6.25 (m, 1H), 4.66 (d, J = 5.9 Hz, 2H), 3.90 (s, 3H), 3.88 (s, 3H), 2.29 (s, 3H), 2.28 (s, 1H).
Synthesis of compound 5: To a solution of compound 4 (10 g, 0.03 mol) in methanol 50 mL was added lithium hydroxide monohydrate (LiOH•H2O,2.56 g, 0.06 mol) dissolved in 50 mL H2O dropwise at 0 °C and then stirred at room temperature overnight. The resulting solution was acidified with 1 N HCl and extracted with 50 mL ethyl acetate twice. The organic layers were combined, washed with water, saturated sodium chloride, and dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a solid. Then the solid was slurried with petroleum ether: methyl tert-butyl ether (5:1, v/v) and filtered. A total amount of 8.1 g compound 5 was obtained as white solid with a yield of 98.4%.1H NMR (600 MHz, CDCl3) δ 7.14 (t, J = 8.1 Hz, 1H), 7.07 (t, J = 8.1 Hz, 1H), 6.98–6.93 (m, 2H), 6.83 (d, J = 8.2 Hz, 1H), 6.66 (d, J = 15.9 Hz, 1H), 6.55 (dd, J = 8.2, 2.3 Hz, 1H), 6.47 (t, J = 2.3 Hz, 1H), 6.44 (dd, J = 8.0, 1.8 Hz, 1H), 6.40 (dd, J = 8.1, 2.3 Hz, 1H), 6.35 (t, J = 2.3 Hz, 1H), 6.28 (dt, J = 15.9, 6.0 Hz, 1H), 4.65 (d, J = 6.0 Hz, 2H), 3.90 (s, 3H), 3.88 (s, 3H).13C NMR (151 MHz, CDCl3) δ 159.61, 156.36, 156.35, 148.64, 148.58, 132.65, 129.93, 129.73, 129.06, 121.84, 119.53, 110.65, 108.48, 107.54, 107.40, 106.79, 102.41, 101.90, 68.39, 55.49, 55.41.
Synthesis of compound 6: To a solution of compound 5 (5 g, 17.5 mmol) and potassium carbonate (4.8 g, 34.9 mmol) in 30 mL DMF was added allyl bromide (2.3 mL, 26.2 mmol) and stirred at room temperature overnight. The resulting solution was quenched with water 200 mL and extracted with 200 mL ethyl acetate twice. The organic layers were combined, washed with water (200 mL × 5), saturated sodium chloride, and dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a crude. Then the solid was purified by silica gel column chromatography with petroleum ether/ethyl acetate as eluents to get a solid, then slurried with petroleum ether: methyl tert-butyl ether (10:1, v/v) and filtered. A total amount of 5.2 g compound 6 was obtained as white solid with a yield of 91.2%.1H NMR (600 MHz, CDCl3) δ 7.18 (t, J = 8.1 Hz, 1H), 6.98 (d, J = 1.7 Hz, 1H), 6.95 (dd, J = 8.2, 1.8 Hz, 1H), 6.83 (d, J = 8.2 Hz, 1H), 6.66 (d, J = 15.9 Hz, 1H), 6.57 (dd, J = 8.6, 2.5 Hz, 1H), 6.54(dt, J = 12.3, 2.5 Hz, 2H), 6.29(dt, J = 15.9, 6.2 Hz, 1H), 6.05 (m, 1H), 5.41(dd, J = 17.3, 1.6 Hz, 1H), 5.28 (dd, J = 10.5, 1.3 Hz, 1H), 4.66 (dd, J = 6.1, 1.1 Hz, 2H), 4.52 (dt, J = 5.3, 1.4 Hz, 2H), 3.90 (s, 3H), 3.89 (s, 3H).13C NMR (151 MHz, CDCl3) δ 159.89, 159.85, 149.10, 149.04, 133.25, 133.08, 129.90, 129.51, 122.33, 119.94, 117.73, 111.06, 108.89, 107.20, 107.17, 102.04, 68.85, 68.82, 55.92, 55.85.
Synthesis of compound 7: Compound 6 (2 g, 6.1 mmol) was dissolved in 6 mL N, N-diethylaniline in a sealed tube under argon in a sand bath at 230 °C for 12 h. After cooling, the reaction solution was dispersed in 100 mL ethyl acetate, washed 1 N HCl, water, saturated saline, and dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a crude. Then the crude was purified by silica gel column chromatography with petroleum ether/ethyl acetate as eluents to get a crude solid, and the solid was purified again by silica gel column chromatography with petroleum ether/tetrahydrofuran as eluents to get a solid. The solid was slurried with petroleum ether: methyl tert-butyl ether (5:1, v/v) respectively and filtered. A total amount of 0.8 g compound 7 was obtained as white solid with a yield of 40%.1H NMR of (600 MHz, CDCl3) δ 6.95 (dd, J = 10.2, 1.8 Hz, 2H), 6.86 (d, J = 8.1 Hz, 1H), 6.84 (s, 1H), 6.39 (s, 1H), 6.02 (ddt, J = 16.5, 10.1, 6.3 Hz, 1H), 5.21–5.14 (m, 2H), 5.06 (d, J = 9.0 Hz, 1H), 4.99 (s, 1H), 3.89 (s, 3H), 3.88 (s, 3H), 3.40–3.34 (m, 3H), 1.37 (d, J = 6.7 Hz, 3H).13C NMR (151 MHz, CDCl3) δ 159.09, 154.52, 149.38, 149.24, 137.12, 133.15, 124.84, 124.38, 119.05, 117.25, 116.36, 111.09, 109.30, 98.20, 93.55, 77.37, 77.16, 76.95, 56.10, 56.07, 45.02, 35.20, 18.08, 0.14.
Synthesis of compound 8: To a solution of 7 (100 mg, 0.3 mmol) in 10 mL dry MeOH was added lead tetraacetate (275 mg, 0.6 mmol) and the mixture was stirred at room temperature for 2 h and evaporated to dryness. The products were extracted with dichloromethane and separated by flash column chromatography on silica gel (hexane-ethyl acetate, 4 1 to 2:1, v/v). The first eluted compound was 8 (Denudatin B; 16 mg, 15%).1H NMR (600 MHz, CDCl3) δ 6.88–6.85 (m, 2H), 6.78 (d, J = 1.3 Hz, 1H), 6.26 (s, 1H), 5.88 (dd, J = 17.0, 10.1 Hz, 1H), 5.82 (s, 1H), 5.35 (d, J = 9.5 Hz, 1H), 5.29 (s, 1H), 5.17–5.09 (m, 2H), 3.89 (s, 3H), 3.88 (s, 3H), 3.13 (s, 3H), 1.13 (d, J = 6.8 Hz, 3H).13C NMR (151 MHz, CDCl3) δ 187.21, 174.73, 149.78, 149.34, 142.98, 135.14, 131.23, 129.69, 119.76, 117.36, 111.06, 109.50, 102.80, 91.45, 77.78, 56.01, 55.99, 51.20, 49.78, 40.98, 33.59.
4.1.3. General produce for the preparation of neoligan probe 10
Synthesis of compound 10: To a solution of 7 (100 mg, 0.3 mmol) and potassium carbonate in (85 mg, 0.6 mmol) in dry N, N-Dimethylformamide (5 mL) was added compound 9 (114 mg, 0.45 mmol) and the mixture was stirred at room temperature overnight. The resulting solution was quenched with water 100 mL and extracted with 100 mL ethyl acetate twice. The organic layers were combined, washed with water (100 mL × 5), saturated sodium chloride, and dried over anhydrous sodium sulfate, concentrated under vacuum to achieve a crude. Then the solid was purified by silica gel column chromatography with petroleum ether/ethyl acetate as eluents. A total amount of 82 mg compound 10 was obtained as white solid with a yield of 60.0%.1H NMR (600 MHz, CDCl3) δ 6.97–6.93 (m, 2H), 6.90 (s, 1H), 6.86 (d, J = 8.0 Hz, 1H), 6.40 (s, 1H), 6.02 (dd, J = 16.9, 10.1 Hz, 1H), 5.14–5.01 (m, 3H), 3.89 (s, 3H), 3.87 (d, J = 6.5 Hz, 3H), 3.81 (t, J = 6.0 Hz, 2H), 3.45–3.34 (m, 3H), 2.05 (td, J = 7.5, 2.7 Hz, 2H), 1.99 (t, J = 2.6 Hz, 1H), 1.89 (t, J = 6.0 Hz, 2H), 1.75 (t, J = 7.5 Hz, 2H), 1.37 (d, J = 6.7 Hz, 3H).13C NMR (151 MHz, CDCl3) δ 158.52, 156.33, 149.23, 149.08, 137.64, 133.10, 124.29, 123.42, 120.74, 118.87, 115.06, 110.93, 109.12, 94.32, 93.40, 82.70, 69.22, 62.88, 55.94, 55.91, 45.01, 34.19, 33.01, 32.48, 26.71, 17.98, 13.31. HRMS (ESI) calcd for C27H30N2O4 (M + H) + 447.5460 (1+), found 447.2284.
4.2. Biology
4.2.1. Cell culture and stimulation
Mouse monocyte-macrophage RAW 264.7 cells were purchased from National Collection of Authenticated Cell Cultures (Shanghai, China) and cultured in Dulbecco's Modified Eagle Medium (DMEM) (Gibco, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, USA) at 37 °C with 5% CO2. The medium was routinely changed every two days about every 10 mL in a 100 mm Petri dish and the RAW 264.7 cells were passaged when they attained approximately 80% confluence. Cells with a fusion degree of about 70–80% were subcultured, and after counting, the cell suspension density was adjusted to 1 × 106 cells/mL with complete medium containing 1 μg/mL LPS. 500 μL of cell suspension is added to each well of the 48-well plate. Before adding the cell suspension, it is necessary to blow evenly. At the same time, the final concentration of the compound can be set at 100 nM, 500 nM. Cells were stimulated for 48 h, and the effects of compounds on macrophage M1 differentiation and TNF-α expression were tested by flow cytometry.
4.2.2. Preparation of single cell suspension of mouse splenocytes and drug incubation
Healthy mice were anaesthetized and executed, and the spleen was stripped and placed in a culture dish to prepare a single cell suspension. Perform erythrocyte lysate, cell sieve filtration, and cell counting. After counting, add the configured complete medium (containing ConA and IL-2) to the cell precipitate. Cells were co-incubated with different concentrations of drugs for 48 h. After 48 h, PMA, Ionomycin and Golgistop (1:1000) were added to each well. Cells were stimulated and incubated for 4 h.
4.2.3. Animal experiments
The experimental protocol was established according to the ethical guidelines of the Declaration of Helsinki and was approved by the Animal Ethics Committee of the Institute of Radiation Medicine, Chinese Academy of Medical Sciences. C57BL/6J male mice, aged 8–10 weeks, and weighing 21 ± 2 g were used in this study. And the experiment was carried out a week after acclimatization to the standard environment.
4.2.3.1. Sepsis model
Mice were anaesthetized by intraperitoneal injection of tribromoethanol (1 μL/g). Next, an incision of approximately 1 cm was made in the abdomen. The cecum was located and ligated from the distal end to 3/4th of the ileocecal valve. Using an 18G needle, a hole was pierced in the ligated part of the cecum and the faeces (about 1 mm in length) were squeezed out. The cecum was finally restored and the abdominal skin was sutured. The compound was suspended in olive oil and fed to the mice 2 h prior to the start of the operation.
4.2.3.2. Cell culture
The mice in the experimental groups were sacrificed under anesthesia and the spleen was removed. Each mouse spleen was ground separately with culture medium and prepare a single cell suspension. The cells were seeded at a density of 1 × 106 cells/mL, and medium containing PMA, ionomycin, and GlogiStop. After being stimulated in the incubator for 4 h, the cells were stained.
4.2.3.3. Cell staining
For surface antigens such as CD4, the cells were washed with PBS and the antibody solution was added. The cells were then incubated in the dark for 15 min at 4 °C. For intracellular signals such as IFN-γ or IL-17A, a fixation solution was used after the surface antigen staining step. After fixed, the cells were punched with saponin for 30 min, and the cells were incubated with the appropriate intracellular antibodies for 15 min. For a staining target like Foxp3, we used True-Nuclear Transcription Factor Buffer Set for the staining process. After washing with cell staining buffer, the samples were assessed.
4.2.3.4. Histology
Each liver tissues were harvested separately, fixed in 10% formalin, and embedded in paraffin. The tissue sections (5 μm) were affixed to slides, stained with hematoxylin and eosin, and images were acquired using a Leica DM3000 microscope.
4.2.4. SILAC pull down
RAW264.7 were passaged 10 times in SILAC DMEM with 10% dialyzed FBS, 1% penicillin-streptomycin, and 100 μg/mL l-arginine- 13C6,15N4–HCl and 100 μg/mL l-lysine13C6,15N2–HCl (K8/R10)for heavy isotope labeling ,or l-arginine-HCl and l-lysine-HCl (K0/R0)for light isotope labeling. Before ABPP experiment, cells were treated with 1 μg/mL LPS for 48 h to induce macrophage polarization being M1 type. The cells were harvested and stored at −80 °C for further experiments.
Dilute the desired proteome sample to a 2 mg/mL solution in PBS. Add 500 μl of diluted 2 mg/mL proteome solution to Eppendorf tubes. Incubating with probe 10 (100 μM) or 100 μM NP-7 as a competition group for 3 h at room temperature, the proteome samples were then illuminated under 365 nm ultraviolet light at 4 °C in a silex glassware. For each click reaction, an excess biotin–azide (50 mM, 5 μl, a final concentration of 500 μM), freshly prepared Cu+ (a final concentration 1 mM Cu+, mixed with 50 mM TCEP in water, 1.7 mM stock of ligand TBTA in DMSO: t-butanol 1:4, 50 mM copper (II) sulfate stock in water) were sequentially added to the lysates and the samples were incubated and shaken at room temperature for 3 h. Then, clicked proteins were subjected to precipitation with acetone and air dried. Subsequently, the pellet was dissolved in 5 ml of 0.1% SDS in PBS and incubated with 50 μl of streptavidin beads under gentle mixing for 3 h at room temperature. After 6500 g centrifugation, the beads were separated from the proteome solution and washed with 4 × 5 ml 0.1% SDS/PBS, 4 × 5 ml water as described. Transfer beads to a screw-top eppendorf tube using 1 ml of water, centrifuge, let beads settle and pipette off the supernatant for further SDS-PAGE.
4.2.5. Statistical analysis
The results were analyzed by nonparametric t tests, using GraphPad Prism 9.3.0. The following terminology is used to show statistical significance: the data of three to eight independent experiments are presented as mean ± SEM; # p < 0.05, ## p < 0.01, ### p < 0.001, #### p < 0.0001; *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC) (NO. 82030116 to Feng Qiu).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejmech.2022.114659.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Evans L., Rhodes A., Alhazzani W., Antonelli M., Coopersmith C.M., French C., Machado F.R., Mcintyre L., Ostermann M., Prescott H.C., Schorr C., Simpson S., Wiersinga W.J., Alshamsi F., Angus D.C., Arabi Y., Azevedo L., Beale R., Beilman G., Belley-Cote E., Burry L., Cecconi M., Centofanti J., Coz Yataco A., de Waele J., Dellinger R.P., Doi K., Du B., Estenssoro E., Ferrer R., Gomersall C., Hodgson C., Hylander Møller M., Iwashyna T., Jacob S., Kleinpell R., Klompas M., Koh Y., Kumar A., Kwizera A., Lobo S., Masur H., McGloughlin S., Mehta S., Mehta Y., Mer M., Nunnally M., Oczkowski S., Osborn T., Papathanassoglou E., Perner A., Puskarich M., Roberts J., Schweickert W., Seckel M., Sevransky J., Sprung C.L., Welte T., Zimmerman J., Levy M. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock 2021. Crit. Care Med. 2021;49 doi: 10.1097/CCM.0000000000005337. e1143. [DOI] [PubMed] [Google Scholar]
- 2.Fleischmann C., Scherag A., Adhikari N.K.J., Hartog C.S., Tsaganos T., Schlattmann P., Angus D.C., Reinhart K. Assessment of global incidence and mortality of hospital-treated sepsis current estimates and limitations. Am. J. Respir. Crit. Care Med. 2016;193:259–272. doi: 10.1164/rccm.201504-0781OC. [DOI] [PubMed] [Google Scholar]
- 3.Fleischmann-Struzek C., Mellhammar L., Rose N., Cassini A., Rudd K.E., Schlattmann P., Allegranzi B., Reinhart K. Incidence and mortality of hospital- and ICU-treated sepsis: results from an updated and expanded systematic review and meta-analysis. Intensive Care Med. 2020;46:1552–1562. doi: 10.1007/s00134-020-06151-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee C., Dantes R., Epstein L., Murphy D.J., Seymour C.W., Iwashyna T.J., Kadri S.S., Angus D.C., Danner R.L., Fiore A.E., Jernigan J.A., Martin G.S., Septimus E., Warren D.K., Karcz A., Chan C., Menchaca J.T., Wang R., Gruber S., Klompas M. Incidence and trends of sepsis in US hospitals using clinical vs claims data, 2009-2014. JAMA, J. Am. Med. Assoc. 2017;318:1241–1249. doi: 10.1001/jama.2017.13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singer M., Deutschman C.S., Seymour C., Shankar-Hari M., Annane D., Bauer M., Bellomo R., Bernard G.R., Chiche J.D., Coopersmith C.M., Hotchkiss R.S., Levy M.M., Marshall J.C., Martin G.S., Opal S.M., Rubenfeld G.D., der Poll T., Vincent J.L., Angus D.C. The third international consensus definitions for sepsis and septic shock (sepsis-3) JAMA, J. Am. Med. Assoc. 2016;315:801–810. doi: 10.1001/jama.2016.0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esposito S., de Simone G., Boccia G., de Caro F., Pagliano P. Sepsis and septic shock: new definitions, new diagnostic and therapeutic approaches. Journal of Global Antimicrobial Resistance. 2017;10:204–212. doi: 10.1016/j.jgar.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 7.Rhodes A., Evans L.E., Alhazzani W., Levy M.M., Antonelli M., Ferrer R., Kumar A., Sevransky J.E., Sprung C.L., Nunnally M.E., Rochwerg B., Rubenfeld G.D., Angus D.C., Annane D., Beale R.J., Bellinghan G.J., Bernard G.R., Chiche J.D., Coopersmith C., de Backer D.P., French C.J., Fujishima S., Gerlach H., Hidalgo J.L., Hollenberg S.M., Jones A.E., Karnad D.R., Kleinpell R.M., Koh Y., Lisboa T.C., Machado F.R., Marini J.J., Marshall J.C., Mazuski J.E., McIntyre L.A., McLean A.S., Mehta S., Moreno R.P., Myburgh J., Navalesi P., Nishida O., Osborn T.M., Perner A., Plunkett C.M., Ranieri M., Schorr C.A., Seckel M.A., Seymour C.W., Shieh L., Shukri K.A., Simpson S.Q., Singer M., Thompson B.T., Townsend S.R., van der Poll T., Vincent J.L., Wiersinga W.J., Zimmerman J.L., Dellinger R.P. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock. Intensive Care Med. 2016;43:304–377. doi: 10.1007/s00134-017-4683-6. 2017. [DOI] [PubMed] [Google Scholar]
- 8.Li Q., Guan X., Wu P., Wang X., Zhou L., Tong Y., Ren R., Leung K.S.M., Lau E.H.Y., Wong J.Y., Xing X., Xiang N., Wu Y., Li C., Chen Q., Li D., Liu T., Zhao J., Liu M., Tu W., Chen C., Jin L., Yang R., Wang Q., Zhou S., Wang R., Liu H., Luo Y., Liu Y., Shao G., Li H., Tao Z., Yang Y., Deng Z., Liu B., Ma Z., Zhang Y., Shi G., Lam T.T.Y., Wu J.T., Gao G.F., Cowling B.J., Yang B., Leung G.M., Feng Z. Early transmission dynamics in Wuhan, China, of novel coronavirus–infected pneumonia. N. Engl. J. Med. 2020;382:1199–1207. doi: 10.1056/nejmoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jose R.J., Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir. Med. 2020;8 doi: 10.1016/S2213-2600(20)30216-2. e46–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi Y., Wang Y., Shao C., Huang J., Gan J., Huang X., Bucci E., Piacentini M., Ippolito G., Melino G. COVID-19 infection: the perspectives on immune responses. Cell Death Differ. 2020;27:1451–1454. doi: 10.1038/s41418-020-0530-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: the status and perspectives in delivery points of view. Adv. Drug Deliv. Rev. 2021;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Stillfried S., David R., Ulow B., R€ R., Boor P. First report from the German COVID-19 autopsy registry. The Lancet Regional Health - Europe. 2022;15 doi: 10.1016/j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magro G. Cytokine Storm: is it the only major death factor in COVID-19 patients? Coagulation role. Med. Hypotheses. 2020;142 doi: 10.1016/j.mehy.2020.109829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen X.Y., Yan B.X., Man X.Y. TNFα inhibitor may be effective for severe COVID-19: learning from toxic epidermal necrolysis. Ther. Adv. Respir. Dis. 2020;14 doi: 10.1177/1753466620926800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sokolowska M., Lukasik Z.M., Agache I., Akdis C.A., Akdis D., Akdis M., Barcik W., Brough H.A., Eiwegger T., Eljaszewicz A., Eyerich S., Feleszko W., Gomez-Casado C., Hoffmann-Sommergruber K., Janda J., Jiménez-Saiz R., Jutel M., Knol E.F., Kortekaas Krohn I., Kothari A., Makowska J., Moniuszko M., Morita H., O'Mahony L., Nadeau K., Ozdemir C., Pali-Schöll I., Palomares O., Papaleo F., Prunicki M., Schmidt-Weber C.B., Sediva A., Schwarze J., Shamji M.H., Tramper-Stranders G.A., van de Veen W., Untersmayr E. Immunology of COVID-19: mechanisms, clinical outcome, diagnostics, and perspectives—a report of the European Academy of allergy and clinical immunology (EAACI) Allergy: European Journal of Allergy and Clinical Immunology. 2020;75:2445–2476. doi: 10.1111/all.14462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald K.A., Kagan J.C. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bekkering S., Domínguez-Andrés J., Joosten L.A.B., Riksen N.P., Netea M.G. 2021. Trained Immunity: Reprogramming Innate Immunity in Health and Disease Keywords. [DOI] [PubMed] [Google Scholar]
- 18.Finethy R., Dockterman J., Kutsch M., Orench‐Rivera N., Wallace G.D., Piro A.S., Luoma S., Haldar A.K., Hwang S., Martinez J., Kuehn M.J., Taylor G.A., Coers J. Dynamin‐related Irgm proteins modulate LPS‐induced caspase‐11 activation and septic shock. EMBO Rep. 2020;21 doi: 10.15252/embr.202050830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S., Jain A., Choi S.W., da Silva G.P.D., Allers L., Mudd M.H., Peters R.S., Anonsen J.H., Rusten T.E., Lazarou M., Deretic V. Mammalian Atg 8 proteins and the autophagy factor IRGM control mTOR and TFEB at a regulatory node critical for responses to pathogens. Nat. Cell Biol. 2020;22:973–985. doi: 10.1038/s41556-020-0549-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alwarawrah Y., Danzaki K., Nichols A.G., Fee B.E., Bock C., Kucera G., Hale L.P., Taylor G.A., MacIver N.J. Irgm1 regulates metabolism and function in T cell subsets. Sci. Rep. 2022;12 doi: 10.1038/s41598-021-04442-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nabar N.R., Kehrl J.H. Inflammasome inhibition links IRGM to innate immunity. Mol. Cell. 2019;73:391–392. doi: 10.1016/j.molcel.2019.01.029. [DOI] [PubMed] [Google Scholar]
- 22.Strachan R.T., Ferrara G., Roth B.L. Screening the receptorome: an efficient approach for drug discovery and target validation. Drug Discov. Today. 2006;11:708–716. doi: 10.1016/j.drudis.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 23.Newman D.J., Cragg G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020;83:770–803. doi: 10.1021/acs.jnatprod.9b01285. [DOI] [PubMed] [Google Scholar]
- 24.Beck H., Härter M., Haß B., Schmeck C., Baerfacker L. Drug Discovery Today; 2022. Small Molecules and Their Impact in Drug Discovery: A Perspective on the Occasion of the 125th Anniversary of the Bayer Chemical Research Laboratory. [DOI] [PubMed] [Google Scholar]
- 25.Zhang H.W., Lv C., Zhang L.J., Guo X., Shen Y.W., Nagle D.G., Zhou Y.D., Liu S.H., Zhang W.D., Luan X. Application of omics- and multi-omics-based techniques for natural product target discovery. Biomed. Pharmacother. 2021:141. doi: 10.1016/j.biopha.2021.111833. [DOI] [PubMed] [Google Scholar]
- 26.Rodrigues T., Reker D., Schneider P., Schneider G. Counting on natural products for drug design. Nat. Chem. 2016;8:531–541. doi: 10.1038/nchem.2479. [DOI] [PubMed] [Google Scholar]
- 27.Zhao B., Liu N., Chen L., Geng S., Fan Z., Xing J. Direct label-free methods for identification of target proteins in agrochemicals. Int. J. Biol. Macromol. 2020;164:1475–1483. doi: 10.1016/j.ijbiomac.2020.07.237. [DOI] [PubMed] [Google Scholar]
- 28.Liao L.X., Song X.M., Wang L.C., Lv H.N., Chen J.F., Liu D., Fu G., Zhao M.B., Jiang Y., Zeng K.W., Tu P.F. Highly selective inhibition of IMPDH2 provides the basis of antineuroinflammation therapy. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E5986–E5994. doi: 10.1073/pnas.1706778114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai J., Liang K., Zhao S., Jia W., Liu Y., Wu H., Lv J., Cao C., Chen T., Zhuang S., Hou X., Zhou S., Zhang X., Chen X.W., Huang Y., Xiao R.P., Wang Y.L., Luo T., Xiao J., Wang C. Chemoproteomics reveals baicalin activates hepatic CPT1 to ameliorate diet-induced obesity and hepatic steatosis. Proc. Natl. Acad. Sci. U. S. A. 2018;115:E5896–E5905. doi: 10.1073/pnas.1801745115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Wang Y., Ma N., Tian J., Shao Y., Zhu B., Wong Y.K., Liang Z., Zou C., Wang J. vol. 5. 2020. (Target Identification of Natural Medicine with Chemical Proteomics Approach: Probe Synthesis, Target Fishing and Protein Identification, Signal Transduction and Targeted Therapy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ponpipom M.M., Bugianesi R.L., Brooker D.R., Yue B., Hwang S., Shen T. Structure-activity relationships of kadsurenone analogues. J. Med. Chem. 1987;30:136–142. doi: 10.1021/jm00384a023. [DOI] [PubMed] [Google Scholar]
- 32.Ding D.D., Wang Y.H., Chen Y.H., Mei R.Q., Yang J., Luo J.F., Li Y., Long C.L., Kong Y. Amides and neolignans from the aerial parts of Piper bonii. Phytochemistry. 2016;129:36–44. doi: 10.1016/j.phytochem.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Ward R.S. Lignans, neolignans, and related compounds. Nat. Prod. Rep. 1995;12:183–205. doi: 10.1039/np9951200183. [DOI] [PubMed] [Google Scholar]
- 34.Zálešák F., Bon D.J.Y.D., Pospíšil J. Lignans and Neolignans: plant secondary metabolites as a reservoir of biologically active substances. Pharmacol. Res. 2019;146 doi: 10.1016/j.phrs.2019.104284. [DOI] [PubMed] [Google Scholar]
- 35.Lin L.C., Shen C.C., Shen Y.C., Tsai T.H. Anti-inflammatory neolignans from Piper kadsura. J. Nat. Prod. 2006;69:842–844. doi: 10.1021/np0505521. [DOI] [PubMed] [Google Scholar]
- 36.Yoo S.-R., Ha H., Shin H.-K., Seo C.-S. Anti-inflamatory activity of neolignan compound isolated from the roots of Saururus chinensis. Plants. 2020;9:932. doi: 10.3390/plants9080932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z.B., Sun C.P., Xu J.X., Morisseau C., Hammock B.D., Qiu F. Phytochemical constituents from Scutellaria baicalensis in soluble epoxide hydrolase inhibition: kinetics and interaction mechanism merged with simulations. Int. J. Biol. Macromol. 2019;133:1187–1193. doi: 10.1016/j.ijbiomac.2019.04.055. [DOI] [PubMed] [Google Scholar]
- 38.Kim H.Y., Yoon J.J., Kim D.S., Kang D.G., Lee H.S. Yg-1 extract improves acute pulmonary inflammation by inducing bronchodilation and inhibiting inflammatory cytokines. Nutrients. 2021;13 doi: 10.3390/nu13103414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xia Y.T., Hu W.H., Wu Q.Y., Dong T.T.X., Duan R., Xiao J., ping Li S., Qin Q.W., Wang W.X., Tsim K.W.K. The herbal extract deriving from aerial parts of Scutellaria baicalensis shows anti-inflammation and anti-hypoxia responses in cultured fin cells from rabbit fish. Fish Shellfish Immunol. 2020;106:71–78. doi: 10.1016/j.fsi.2020.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsui K., Munakata K. Four new neolignans from piper futokadzura. Tetrahedron Lett. 1976;17:4371–4374. doi: 10.1016/0040-4039(76)80118-9. [DOI] [Google Scholar]
- 41.Ong S.-E., Mann M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC) Nat. Protoc. 2006;1:2650–2660. doi: 10.1038/nprot.2006.427. [DOI] [PubMed] [Google Scholar]
- 42.Geiger T., Wisniewski J.R., Cox J., Zanivan S., Kruger M., Ishihama Y., Mann M. Use of stable isotope labeling by amino acids in cell culture as a spike-in standard in quantitative proteomics. Nat. Protoc. 2011;6:147–157. doi: 10.1038/nprot.2010.192. [DOI] [PubMed] [Google Scholar]
- 43.Molina D.M., Jafari R., Ignatushchenko M., Seki T., Larsson E.A., Dan C., Sreekumar L., Cao Y., Nordlund P. Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science. 1979:84–87. doi: 10.1126/science.1233606. 341 (2013. [DOI] [PubMed] [Google Scholar]
- 44.Jafari R., Almqvist H., Axelsson H., Ignatushchenko M., Lundbäck T., Nordlund P., Molina D.M. The cellular thermal shift assay for evaluating drug target interactions in cells. Nat. Protoc. 2014;9:2100–2122. doi: 10.1038/nprot.2014.138. [DOI] [PubMed] [Google Scholar]
- 45.Langebäck A., Bacanu S., Laursen H., Mout L., Seki T., Erkens-Schulze S., Ramos A.D., Berggren A., Cao Y., Hartman J., van Weerden W., Bergh J., Nordlund P., Lööf S. CETSA-based target engagement of taxanes as biomarkers for efficacy and resistance. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-55526-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lomenick B., Hao R., Jonai N., Chin R.M., Aghajan M., Warburton S., Wang J., Wu R.P., Gomez F., Loo J.A., Wohlschlegel J.A., Vondriska T.M., Pelletier J., Herschman H.R., Clardy J., Clarke C.F., Huang J. Target identification using drug affinity responsive target stability (DARTS) Proc. Natl. Acad. Sci. USA. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sindrilaru A., Peters T., Wieschalka S., Baican C., Baican A., Peter H., Hainzl A., Schatz S., Qi Y., Schlecht A., Weiss J.M., Wlaschek M., Sunderkötter C., Scharffetter-Kochanek K. An unrestrained proinflammatory M1 macrophage population induced by iron impairs wound healing in humans and mice. J. Clin. Invest. 2011;121:985–997. doi: 10.1172/JCI44490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang X., Fan L., Wu J., Xu H., Leung W.Y., Fu K., Wu J., Liu K., Man K., Yang X., Han J., Ren J., Yu J. Macrophage p38α promotes nutritional steatohepatitis through M1 polarization. J. Hepatol. 2019;71:163–174. doi: 10.1016/j.jhep.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 49.Zhou D., Huang C., Lin Z., Zhan S., Kong L., Fang C., Li J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell. Signal. 2014;26:192–197. doi: 10.1016/j.cellsig.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Liu L., Liu L., Liu L., Guo H., Song A., Song A., Song A., Huang J., Zhang Y., Jin S., Li S., Zhang L., Yang C., Yang P., Yang P., Yang P. Progranulin inhibits LPS-induced macrophage M1 polarization via NF-kB and MAPK pathways. BMC Immunol. 2020;21 doi: 10.1186/s12865-020-00355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J. T helper cell differentiation, heterogeneity, and plasticity. Cold Spring Harbor Perspect. Biol. 2018;10 doi: 10.1101/cshperspect.a030338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan Y.Y., Zhang H., Zhou Y., Liu H.B., Tang W., Zhou B., Zuo J.P., Yue J.M., Phainanoids A.-F. A new class of potent immunosuppressive triterpenoids with an unprecedented carbon skeleton from phyllanthus hainanensis. J. Am. Chem. Soc. 2015;137:138–141. doi: 10.1021/ja511813g. [DOI] [PubMed] [Google Scholar]
- 53.Fan Y.Y., Gan L.S., Liu H.C., Li H., Xu C.H., Zuo J.P., Ding J., Yue J.M., Phainanolide A. Highly modified and oxygenated triterpenoid from Phyllanthus hainanensis. Org. Lett. 2017;19:4580–4583. doi: 10.1021/acs.orglett.7b02181. [DOI] [PubMed] [Google Scholar]
- 54.Liu J., Bai L.P., Yang F., Yao X., Lei K., Kei Lam C.W., Wu Q., Zhuang Y., Xiao R., Liao K., Kuok H., Li T., Liu L. Potent antagonists of RORγt, cardenolides from calotropis gigantea, exhibit discrepant effects on the differentiation of T lymphocyte subsets. Mol. Pharm. 2019;16:798–807. doi: 10.1021/acs.molpharmaceut.8b01063. [DOI] [PubMed] [Google Scholar]
- 55.Ruterbusch M., Pruner K.B., Shehata L., Pepper M. In vivo CD4 + T cell differentiation and function: revisiting the Th1/Th2 paradigm. Annu. Rev. Immunol. 2020;38:705–725. doi: 10.1146/annurev-immunol-103019-085803. [DOI] [PubMed] [Google Scholar]
- 56.Kolen S., Dolstra H., van de Locht L., Braakman E., Schattenberg A., de Witte T., van de Wiel-van Kemenade E. Biodistribution and retention time of retrovirally labeled T lymphocytes in mice is strongly influenced by the culture period before infusion. J. Immunother. 2002;25:385–395. doi: 10.1097/00002371-200209000-00002. [DOI] [PubMed] [Google Scholar]
- 57.Rittirsch D., Huber-Lang M.S., Flierl M.A., Ward P.A. Immunodesign of experimental sepsis by cecal ligation and puncture. Nat. Protoc. 2009;4:31–36. doi: 10.1038/nprot.2008.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen X., Feng Y., Shen X., Pan G., Fan G., Gao X., Han J., Zhu Y. Anti-sepsis protection of Xuebijing injection is mediated by differential regulation of pro- and anti-inflammatory Th17 and T regulatory cells in a murine model of polymicrobial sepsis. J. Ethnopharmacol. 2018;211:358–365. doi: 10.1016/j.jep.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 59.Li Z., Hao P., Li L., Tan C.Y.J., Cheng X., Chen G.Y.J., Sze S.K., Shen H.M., Yao S.Q. Design and synthesis of minimalist terminal alkyne-containing diazirine photo-crosslinkers and their incorporation into kinase inhibitors for cell- and tissue-based proteome profiling. Angew. Chem. Int. Ed. 2013;52:8551–8556. doi: 10.1002/anie.201300683. [DOI] [PubMed] [Google Scholar]
- 60.Parker C.G., Galmozzi A., Wang Y., Correia B.E., Sasaki K., Joslyn C.M., Kim A.S., Cavallaro C.L., Lawrence R.M., Johnson S.R., Narvaiza I., Saez E., Cravatt B.F. Ligand and target discovery by fragment-based screening in human cells. Cell. 2017;168:527–541. doi: 10.1016/j.cell.2016.12.029. e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Head S.A., Shi W., Zhao L., Gorshkov K., Pasunooti K., Chen Y., Deng Z., Li R.J., Shim J.S., Tan W., Hartung T., Zhang J., Zhao Y., Colombini M., Liu J.O. Antifungal drug itraconazole targets VDAC1 to modulate the AMPK/mTOR signaling axis in endothelial cells. Proc. Natl. Acad. Sci. U. S. A. 2015;112:E7276–E7285. doi: 10.1073/pnas.1512867112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10 doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bafica A., Feng C.G., Santiago H.C., Aliberti J., Cheever A., Thomas K.E., Taylor G.A., Vogel S.N., Sher A. The IFN-inducible GTPase LRG47 (Irgm1) negatively regulates TLR4-triggered proinflammatory cytokine production and prevents endotoxemia. J. Immunol. 2007;179:5514–5522. doi: 10.4049/jimmunol.179.8.5514. [DOI] [PubMed] [Google Scholar]
- 64.Henry S.C., Traver M., Daniell X., Indaram M., Oliver T., Taylor G.A. Regulation of macrophage motility by Irgm1. J. Leukoc. Biol. 2010;87:333–343. doi: 10.1189/jlb.0509299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F.T., de Beer T.A.P., Rempfer C., Bordoli L., Lepore R., Schwede T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kumari R., Kumar R., Lynn A. G-mmpbsa -A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.