Abstract
The ast operon, encoding enzymes of the arginine succinyltransferase (AST) pathway, was cloned from Salmonella typhimurium, and the nucleotide sequence for the upstream flanking region was determined. The control region contains several regulatory consensus sequences, including binding sites for NtrC, cyclic AMP receptor protein (CRP), and ArgR. The results of DNase I footprintings and gel retardation experiments confirm binding of these regulatory proteins to the identified sites. Exogenous arginine induced AST under nitrogen-limiting conditions, and this induction was abolished in an argR derivative. AST was also induced under carbon starvation conditions; this induction required functional CRP as well as functional ArgR. The combined data are consistent with the hypothesis that binding of one or more ArgR molecules to a region between the upstream binding sites for NtrC and CRP and two putative promoters plays a pivotal role in modulating expression of the ast operon in response to nitrogen or carbon limitation.
The arginine succinyltransferase (AST) pathway, which converts arginine to glutamate, has been long considered the major route for aerobic utilization of arginine as a source of carbon, nitrogen, and energy by Pseudomonas aeruginosa (7, 28). Characterization of the aru operon, encoding enzymes of this pathway in P. aeruginosa, led to the identification of the corresponding ast operon from the Escherichia coli genome sequence (10). Recent studies have shown that the AST pathway, rather than the arginine decarboxylase pathway, is the major pathway for utilization of arginine as a nitrogen source by E. coli (23). Interestingly, this pathway is also important for carbon starvation survival, such that one of the ast genes of E. coli was initially identified as a starvation gene, cstC (1, 3).
Computer analysis of the nucleotide sequence of the region upstream of the ast operon in E. coli identified a putative ς54 consensus sequence and two putative NtrC binding sites; such sequences are consistent with the observed nitrogen regulation of the operon (3, 23). Studies by Fraley et al. (3) also indicate the presence of a ςS promoter that appears to compete with the ς54 promoter to match expression to cellular needs.
We have reported recently (20) that the arginine regulatory protein of P. aeruginosa is required for induction of the AST pathway by exogenous arginine. While the structure and function of the arginine regulatory proteins of P. aeruginosa and Salmonella typhimurium differ significantly (14, 20, 21), an early finding by Kustu (12) indicated that an argR derivative of S. typhimurium is impaired in utilization of arginine as a nitrogen source. Studies by Kustu et al. (13) also indicated that arginine degradation in this organism is under nitrogen control. Assuming that the recently identified ast operon of E. coli (10) would have a homologue in the closely related S. typhimurium, we initiated an investigation of the possible role of ArgR of S. typhimurium in expression of the ast operon.
(A preliminary report of this work has been presented previously [16].)
Cloning of the ast operon and sequence features of the upstream flanking region.
A DNA fragment covering the first 500 bp of the astC structural gene of E. coli was amplified by PCR from E. coli K-12 chromosomal DNA. This DNA fragment was then purified, labeled by the Genius system (Boehringer), and used in colony hybridization for screening of a cosmid library of S. typhimurium constructed in this laboratory. Several positive clones were identified, and a 6.5-kb EcoRI fragment from one of these cosmids was further subcloned into the EcoRI site of pUC18, as shown in Fig. 1. The chromosomal insert of the resulting plasmid (pAST3 [Fig. 1]) was partially sequenced, and a homology search indicated that it contains most of the astCABDE operon and an upstream flanking region of 470 bp. The ast operon structure of S. typhimurium was found to be identical to its counterpart in E. coli (10). Furthermore, the xthA gene was also found upstream of the ast operon, as is the case in E. coli (GenBank accession no. D90818).
FIG. 1.
Nucleotide sequence of the ast regulatory region of S. typhimurium. (a) Schematic drawing of the structure of the ast operon in a cosmid and one of the subclones, pAST3. (b) Nucleotide sequence of the chromosomal insert in pAST101. The proposed ς54 promoter region and the putative binding sites for NtrC, ArgR, CRP, and IHF are labeled. The DNA regions protected in DNase I experiments are shown in boldface italic letters. The initiation codons for the astC and xthA genes and the BamHI and Sau3A restriction sites are also labeled. The Shine-Dalgarno sequence for astC is overlined and labeled S.D. This BamHI fragment is cloned into pUC19 in such an orientation that HindIII and SalI are at the 5′ end, and SmaI and EcoRI are at the 3′ end of the sequence shown here.
The upstream region flanking the ast operon was amplified by PCR from plasmid pAST3. Restriction sites of BamHI were introduced into a pair of primers for PCR, and the PCR product of a 490-bp DNA fragment was cloned into the BamHI site of vector pUC19. The resulting plasmid was designated pAST101; the nucleotide sequence of the chromosomal insert in this plasmid was confirmed to be identical to that in pAST3 and is shown in Fig. 1. Certain features noted in the upstream sequence in E. coli (3)—a putative ς54 promoter (18), two potential NtrC binding sites (18), and a putative integration host factor (IHF) binding site (4)—are present at the corresponding locations in the S. typhimurium sequence. However, there is little homology between the two sequences in the regions identified as cyclic AMP receptor protein (CRP) binding sites in the E. coli sequence (3). The consensus sequence of the CRP binding site, 5′-AAATGTGATCTAGATCACATTT-3′, consists of two 11-bp half sites organized as inverted repeats that accommodate CRP dimer (22). The S. typhimurium sequence (Fig. 1) contains a sequence downstream of the NtrC sites that appears to be a good candidate for a CRP site. The first half of the site proposed here has poor homology to the consensus sequence (4 of 11 bp) but the second half exhibits excellent homology to the consensus (10 of 11 bp). Six putative ArgR boxes can be also deduced, albeit with varying degrees of homology to the consensus sequence (5′-AATGAATAATTATTCATT-3′ [29]). Our previous studies with ArgR of S. typhimurium indicate that it is a hexamer of identical 17,000 Mr subunits and that each hexamer binds to two such ARG boxes (14).
Binding of ArgR to the regulatory region of the ast operon.
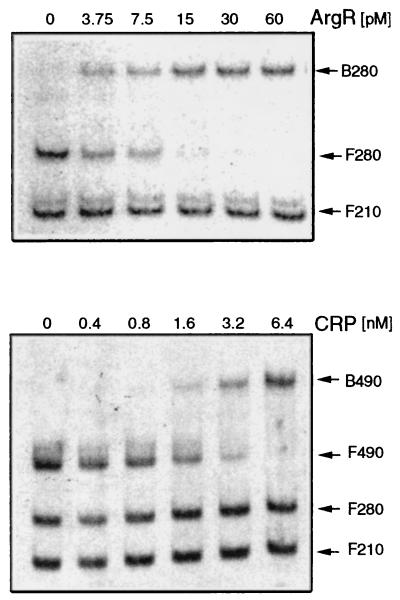
The purified 490-bp BamHI fragment of pAST101, which contains the entire ast regulatory region, was labeled with [α-32P]dGTP by using the Klenow fragment. The labeled fragment was digested by Sau3A to generate two end-labeled fragments; one of them is 210 bp and carries the two putative NtrC binding sites, and the other is 280 bp and carries the putative ArgR binding sites (Fig. 1). These two labeled fragments were used in gel retardation experiments employing a homogeneous ArgR preparation that was purified as previously described (14). The results (Fig. 2, top) show that ArgR interacts specifically with the 280-bp fragment carrying the putative ArgR binding sites. A plot of the percentage of bound DNA against the concentration of ArgR yields an apparent dissociation constant of 5.0 pM.
FIG. 2.
Gel retardation assays with purified ArgR (top panel) and purified CRP (bottom panel). The reactions with purified ArgR (14) and CRP (24) were carried out as described previously. Radioactively labeled probes (0.1 pM) were allowed to interact with different protein concentrations, as indicated. Labeled fragments are as follows: F490 and B490, free and bound forms of the 490-bp BamHI fragment; F280 and B280, free and bound forms of the 280-bp Sau3A/BamHI fragment; F210, free probe of the 210-bp BamHI/Sau3A fragment.
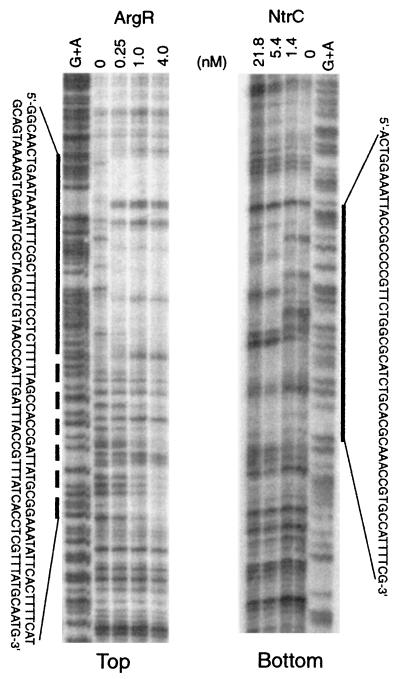
DNase I footprinting experiments were carried out as described previously (14), employing a 32P-labeled HindIII/EcoRI fragment from pAST101 (Fig. 1b). For strand-specific detection, the resulting radioactive probe was subjected to SmaI digestion for the bottom strand and SalI digestion for the top strand. The results (Fig. 3) show that binding of ArgR protects a 90-bp region on the top strand at lower concentrations of ArgR and that this protection is extended at higher ArgR concentrations to 140 bp. As shown in Fig. 1, two ArgR binding sites (corresponding to four ARG boxes) reside in the first 90-bp protected region, and one additional site resides in the extended downstream region.
FIG. 3.
DNase I footprinting with purified ArgR and NtrC. The reactions with ArgR (14) and MBP-NtrC (11) were carried out as described previously. DNA probes (0.17 nM) labeled at the top and bottom strands were used in the reactions with ArgR and NtrC, respectively. The concentrations of ArgR and NtrC used in reactions are indicated on the top of each lane. The corresponding G+A Maxam-Gilbert sequencing ladders (19) are also labeled. A solid line at lower concentrations and a dashed line for the extended region at higher concentrations mark the regions protected by ArgR. A solid line marks the region protected by NtrC. Nucleotide sequences for the protected regions indicated here correspond to boldface italic letters in Fig. 1.
NtrC binding sites.
DNase I footprinting experiments were carried out, employing a purified MBP-NtrC fusion protein (11) that was generously provided by S. Kustu (Berkeley, Calif.). The results (Fig. 3) show that binding of NtrC protects a 55-bp region from nuclease digestion on the bottom strand, with the two NtrC binding sites, predicted by computer analysis, in the center of the protected region.
CRP binding.
Computer analysis of the nucleotide sequence of the control region in S. typhimurium led to the identification of a potential CRP site centered at nucleotide 205 (Fig. 1). This site is at a different location from the potential sites proposed for the E. coli operon (3). Gel retardation experiments were carried out, employing a DNA fragment carrying the entire 490-bp regulatory region and the CRP protein of E. coli (purified according to reference 31; a gift from P. C. Tai). The results (Fig. 2, bottom) show that CRP specifically binds to the regulatory region. Cleavage by Sau3A within the identified site (Fig. 1) produces two DNA fragments that lost the capacity to bind CRP in gel retardation experiments (Fig. 2, bottom).
Effects of argR and ntrB(Con) on AST activity.
The effect of exogenous arginine on the expression of the ast operon was monitored by measurement of AST, the first enzyme of the AST pathway. Cultures of wild-type S. typhimurium and an argR derivative (15) were grown in glucose minimal medium (6) with either glutamate or glutamate and arginine as the source(s) of nitrogen. Under these conditions, nitrogen is limiting and the doubling time (270 to 500 min) is much longer than that obtained with excess ammonia (45 min). The results (Table 1) show that exogenous arginine induces AST activity by 7.3-fold and that this induction is abolished in the argR::Tn10 derivative.
TABLE 1.
Effects of exogenous arginine on induction of AST in wild-type S. typhimurium and its argR::Tn10 and ntrB(Con) derivatives
| Genotype | Nitrogen sourcea | Doubling time (min) | Sp act (nmol/mg/min)b |
|---|---|---|---|
| Wild type | Glu | 270 | 11 (1) |
| Glu+Arg | 270 | 80 (3) | |
| argR::Tn10 | Glu | 450 | 11 (2) |
| Glu+Arg | 500 | 9 (1) | |
| ntrB(Con) | Glu | 160 | 147 (18) |
| Glu+Arg | 70 | 572 (12) | |
| Arg | 120 | 605 (10) | |
| ntrB(Con) argR::Tn10 | Glu | 160 | 15 (7) |
| Glu+Arg | 130 | 15 (7) | |
| Arg | 300 | 101 (14) |
Cells were grown in glucose minimal medium (6) at 37°C. The following supplements were added as indicated: Glu, glutamate at 20 mM; Arg, arginine at 20 mM.
AST was assayed by the procedure described by Vander Wauven and Stalon (28). The reaction mixture (2.0 ml) contained 100 mM Tris-HCl (pH 8.0), 0.3 mM succinyl-coenzyme A (CoA), and 10 mM l-arginine. The reaction was initiated by the addition of arginine at 37°C, and the decrease in the succinyl-CoA concentration was monitored at an absorbance of 232 nm. Protein concentration was determined by the method of Bradford (2). Standard errors are indicated in parentheses.
AST activity was also measured in an ntrB(Con) derivative (strain SK3003); this constitutive mutation causes overexpression of nitrogen-regulated genes regardless of nitrogen level (9) and results in faster growth under nitrogen-limiting conditions. The results (Table 1) show that ntrB(Con), grown with glutamate as the sole nitrogen source, has 13.4-fold-higher AST activity than the wild type. This higher activity reflects the higher concentration of functional NtrC in this strain. Exogenous arginine is still able to cause an additional fourfold induction. Introduction of the argR::Tn10 allele in the ntrB(Con) background again abolishes induction by arginine when cells are grown with both glutamate and arginine. AST activity is somewhat higher when the ntrB(Con) argR mutant derivative is grown with arginine as the sole nitrogen source but is still 5.6-fold lower than in the ntrB(Con) argR+ background. This somewhat higher level likely reflects a higher concentration of functional NtrC as a result of the much longer doubling time (300 min) observed under these conditions.
These results indicate that ArgR is essential for arginine induction of the AST pathway and that this induction is likely mediated at an NtrC-dependent promoter.
Both argR and crp genes are essential for induction of the ast operon under carbon starvation.
AST activity was measured in wild-type S. typhimurium and its argR and crp derivatives in the presence of excess ammonia and under conditions of glucose excess and limitation. The results (Table 2) show that the wild-type strain has a negligible level of AST activity in the presence of excess ammonia and glucose, regardless of the absence or presence or arginine. In contrast, an elevated level of AST activity was observed following depletion of a limiting amount of glucose. These results establish that carbon starvation induces AST activity in the presence of excess ammonia.
TABLE 2.
Effects of carbon starvation on expression of AST in wild-type S. typhimurium and its crp::Tn10 and argR::Tn10 derivatives
| Genotypec | Supplementa | Sp act (nmol/mg/min)b |
|---|---|---|
| Wild type | 0.2% Glucose | <0.1 |
| 0.2% Glucose+Arg | <0.1 | |
| 0.03% Glucose | 53 (4) | |
| 0.03% Glucose+Arg | 68 (2) | |
| crp::Tn10 | 0.03% Glucose | <0.1 |
| 0.03% Glucose+Arg | <0.1 | |
| argR::Tn10 | 0.03% Glucose | <0.1 |
| 0.03% Glucose+Arg | <0.1 |
Cells were grown in minimal medium (6) containing 20 mM (NH4)2SO4 at 37°C. The final concentration of glucose in the culture was either 0.2% for carbon excess or 0.03% for carbon starvation. Cultures supplemented with 0.03% glucose ceased growth at an optical density of 0.5 at 600 nm and were harvested 2 h after glucose depletion. Arginine (Arg) was added at 20 mM. During logarithmic growth, the doubling time for the wild type and its argR derivative was 50 min whereas the doubling time for the crp derivative was 75 min.
See footnote b of Table 1.
The crp::Tn10 locus from strain PP1037 (Salmonella Genetic Stock Center) was introduced into the wild type by phage P22 transduction. The argR::Tn10 derivative was previously characterized (15).
Inactivation of crp by Tn10 insertion abolishes induction by carbon starvation, regardless of the absence or presence of exogenous arginine. Interestingly, inactivation of argR also abolishes induction by carbon starvation. Thus, the presence of a functional ArgR is essential for cAMP-CRP-dependent induction of the ast operon under carbon starvation. These results also indicate that the concentration of the functional ArgR-arginine complex in the wild-type parent is not a limiting factor under conditions of carbon starvation.
Final conclusions.
Computer analysis of the upstream region flanking the ast operon of E. coli (3, 23) led to the identification of two potential NtrC sites (also called NRI) that were presumed to function in nitrogen control of the operon. The results presented here identify two NtrC sites at the corresponding locations in the control region for the ast operon of S. typhimurium (Fig. 1). DNase I footprinting confirms that the two identified sites are in the center of a 55-bp region protected by NtrC (Fig. 3). The higher level of AST in the constitutive derivative, ntrB(Con), supports the conclusion that NtrC mediates nitrogen control of the ast operon in S. typhimurium.
The results presented here also clearly establish that inactivation of ArgR abolishes arginine induction of the ast operon in S. typhimurium under conditions of nitrogen limitation (Table 1). Gel retardation experiments showed that ArgR binds specifically to a DNA fragment carrying the region downstream of the NtrC binding sites. The observed affinity is similar to that previously determined for binding of ArgR to the arginine-repressible car operator of S. typhimurium (14). DNase I footprinting showed that ArgR protects a 90-bp fragment carrying two of the identified ArgR sites and that this protection is extended further downstream to a third site at higher ArgR concentrations.
The 3′ end of the proximal NtrC site is about 200 bp upstream of the putative ς54 promoter. Studies with the glnA promoter of S. typhimurium have shown that NtrC bound at the enhancer, located between −108 and −140, interacts directly with ς54 holoenzyme by means of DNA loop formation (25, 30). Our hypothesis is that in the case of the ast promoter, it is necessary that one or more ArgR molecules bind to the region between NtrC sites and the putative ς54 promoter in order to bring NtrC into proximity with RNA polymerase. The action of ArgR could occur through DNA bending or wrapping around the ArgR molecule. Studies with ArgR of E. coli (26, 29) and S. typhimurium (14) indicate that the binding of ArgR requires l-arginine and that a single hexamer binds through contacts with one face of the DNA helix in both the minor and major grooves. Crystallographic studies have shown that the hexameric form consists of two trimers and is greatly stabilized upon binding of six l-arginine molecules at the trimer-trimer interface (27). Accordingly, an increase in the l-arginine pool would increase the proportion of active ArgR with specific DNA binding activity, resulting in activation of the catabolic ast operon by NtrC.
In addition to arginine induction and nitrogen control, expression of the ast operon is also subject to carbon catabolite repression (1, 3). The AST pathway is induced under carbon starvation, and both ArgR and CRP are required for such induction (Table 2). Evidence for CRP binding to the site identified from the sequence (Fig. 1) was provided from the results of gel retardation experiments. While S. typhimurium and E. coli can utilize arginine as a sole nitrogen source but not as a sole carbon source (6), the AST pathway can also provide carbon skeletons that might become critical under conditions of carbon limitation. Induction by carbon starvation is most likely mediated at a promoter recognized by the ςS subunit of enteric RNA polymerase. The participation of a ςS promoter in expression of the ast operon in E. coli has recently been reported (3). The results presented here (Table 2) indicate that activation of this ςS promoter by the cAMP-CRP complex also require a functional ArgR. The role of ArgR in this activation under conditions of carbon limitation could be similar to that proposed above for activation by NtrC under conditions of nitrogen limitation. The role proposed here for ArgR extends its functions beyond those previously recognized in enteric bacteria: namely, repression of genes of arginine biosynthesis (17) and resolution of ColE1 plasmid multimers (8).
Nucleotide sequence accession number.
The nucleotide sequence determined in this study has been assigned GenBank accession no. AF108767.
Acknowledgments
We are indebted to Sydney Kustu (University of California at Berkeley) for the generous gift of purified NtrC and for helpful suggestions and stimulating discussions throughout this work. We thank P. C. Tai (Georgia State University) for the gift of purified CRP.
This work was supported in part by research grant GM47926 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Blum P H, Jovanovich S B, McCann M P, Schultz J E, Lesley S A, Burgess R R, Matin A. Cloning and in vivo and in vitro regulation of cyclic AMP-dependent carbon starvation genes from Escherichia coli. J Bacteriol. 1990;172:3813–3820. doi: 10.1128/jb.172.7.3813-3820.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Fraley C D, Kim J H, McCann M P, Matin A. The Escherichia coli starvation gene cstC is involved in amino acid catabolism. J Bacteriol. 1998;180:4287–4290. doi: 10.1128/jb.180.16.4287-4290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–555. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 5.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 6.Gutnick D, Calvo J M, Klopotowski T, Ames B N. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. J Bacteriol. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haas D, Galimand M, Gamper M, Zimmermann A. Arginine network of Pseudomonas aeruginosa: specific and global controls. In: Silver S, Chakrabarty A M, Iglewski B, Kaplan S, editors. Pseudomonas: biotransformations, pathogenesis, and evolving biotechnology. Washington, D.C: American Society for Microbiology; 1990. pp. 303–316. [Google Scholar]
- 8.Hodgman T C, Griffiths H, Summers D K. Nucleoprotein architecture and ColE1 dimer resolution: a hypothesis. Mol Microbiol. 1998;29:545–558. doi: 10.1046/j.1365-2958.1998.00948.x. [DOI] [PubMed] [Google Scholar]
- 9.Ikeda T P, Shauger A E, Kustu S. Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J Mol Biol. 1996;259:589–607. doi: 10.1006/jmbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 10.Itoh Y. Cloning and characterization of the aru genes encoding enzymes of the catabolic arginine succinyltransferase pathway in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7280–7290. doi: 10.1128/jb.179.23.7280-7290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klose K E, North A K, Stedman K M, Kustu S. The major dimerization determinants of the nitrogen regulatory protein NTRC from enteric bacteria lie in its carboxy-terminal domain. J Mol Biol. 1994;241:233–245. doi: 10.1006/jmbi.1994.1492. [DOI] [PubMed] [Google Scholar]
- 12.Kustu, S. Unpublished data.
- 13.Kustu S G, McFarland N C, Hui S P, Esmon B, Ames G F-L. Nitrogen control in Salmonella typhimurium: coregulation of synthesis of glutamine synthetase and amino acid transport systems. J Bacteriol. 1979;138:218–234. doi: 10.1128/jb.138.1.218-234.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu C D, Houghton J E, Abdelal A T. Characterization of the arginine repressor from Salmonella typhimurium and its interactions with the carAB operator. J Mol Biol. 1992;225:11–24. doi: 10.1016/0022-2836(92)91022-h. [DOI] [PubMed] [Google Scholar]
- 15.Lu C D, Kilstrup M, Neuhard J, Abdelal A. Pyrimidine regulation of tandem promoters for carAB in Salmonella typhimurium. J Bacteriol. 1989;171:5436–5442. doi: 10.1128/jb.171.10.5436-5442.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu C-D, Abdelal A T. The XVIth International Arginine/Pyrimidine Conference. Leeds, United Kingdom. 1998. Role of the arginine regulatory protein in Pseudomonas aeruginosa, abstr. 9. [Google Scholar]
- 17.Maas W K. The arginine repressor of Escherichia coli. Microbiol Rev. 1994;58:631–640. doi: 10.1128/mr.58.4.631-640.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 19.Maxam A M, Gilbert W. Sequencing end-labeled DNA with base-specific cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 20.Park S-M, Lu C-D, Abdelal A T. Cloning and characterization of argR, a gene that participates in regulation of arginine biosynthesis and catabolism in Pseudomonas aeruginosa PAO1. J Bacteriol. 1997;179:5300–5308. doi: 10.1128/jb.179.17.5300-5308.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S-M, Lu C-D, Abdelal A T. Purification and characterization of an arginine regulatory protein, ArgR, from Pseudomonas aeruginosa and its interactions with the control regions for the car, argF, and aru operons. J Bacteriol. 1997;179:5309–5317. doi: 10.1128/jb.179.17.5309-5317.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saier M H, Jr, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 1325–1343. [Google Scholar]
- 23.Schneider B L, Kiupakis A K, Reitzer L J. Arginine catabolism and the arginine succinyltransferase pathway in Escherichia coli. J Bacteriol. 1998;180:4278–4286. doi: 10.1128/jb.180.16.4278-4286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sogaard-Anderson L, Valentin-Hansen P. Protein-protein interactions in gene regulation: the cAMP-CRP complex sets the specificity of a second DNA-binding protein, the CytR repressor. Cell. 1993;75:557–566. doi: 10.1016/0092-8674(93)90389-8. [DOI] [PubMed] [Google Scholar]
- 25.Su W, Porter S, Kustu S, Echols H. DNA-looping and enhancer activity: association between DNA-bound NTRC activator and RNA polymerase at the bacterial glnA promoter. Proc Natl Acad Sci USA. 1990;87:5504–5508. doi: 10.1073/pnas.87.14.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tian G, Lim D, Carey J, Maas W K. Binding of the arginine repressor of Escherichia coli K12 to its operator sites. J Mol Biol. 1992;226:387–397. doi: 10.1016/0022-2836(92)90954-i. [DOI] [PubMed] [Google Scholar]
- 27.Van Duyne G D, Ghosh G, Maas W K, Sigler P B. Structure of the oligomerization and l-arginine binding domain of the arginine repressor of Escherichia coli. J Mol Biol. 1996;256:377–391. doi: 10.1006/jmbi.1996.0093. [DOI] [PubMed] [Google Scholar]
- 28.Vander Wauven C, Stalon V. Occurrence of succinyl derivatives in the catabolism of arginine in Pseudomonas cepacia. J Bacteriol. 1985;164:882–886. doi: 10.1128/jb.164.2.882-886.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang H, Glansdorff N, Charlier D. The arginine repressor of Escherichia coli K-12 makes direct contacts to minor and major groove determinants of the operators. J Mol Biol. 1998;277:805–824. doi: 10.1006/jmbi.1998.1632. [DOI] [PubMed] [Google Scholar]
- 30.Wedel A, Weiss D S, Popham D, Droge P, Kustu S. A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science. 1990;248:480–490. doi: 10.1126/science.1970441. [DOI] [PubMed] [Google Scholar]
- 31.Zhang X P, Gunasekera A, Ebright Y W, Ebright R H. Derivatives of CAP having no solvent-accessible cysteine residues, or having a unique solvent-accessible cysteine residue at amino acid 2 of the helix-turn-helix motif. J Biomol Struct Dyn. 1991;9:463–473. doi: 10.1080/07391102.1991.10507929. [DOI] [PubMed] [Google Scholar]