Abstract
Background:
While the microbiome has an established role in asthma development, less is known about its contribution to morbidity in children with asthma.
Objective:
In this ancillary study of the Vitamin D Antenatal Asthma Reduction Trial (VDAART), we analyzed the gut microbiome and metabolome of wheeze frequency in children with asthma.
Methods:
Bacterial 16s rRNA microbiome and untargeted metabolomic profiling were performed on fecal samples collected from three-year-old children with parent-reported physician-diagnosed asthma. We analyzed wheeze frequency by calculating the proportion of quarterly questionnaires between ages 3 and 5 years in which parents reported the child had wheezed (“wheeze proportion”). Taxa and metabolites associated with wheeze were analyzed by identifying log-fold changes with respect to wheeze frequency and correlation/linear regression analyses, respectively. Microbe-metabolite and microbe-microbe correlation networks were compared between subjects with high and low wheeze proportion.
Results:
Specific taxa, including the genus Veillonella and histidine pathway metabolites, were enriched in subjects with high wheeze proportion. Amongst wheeze-associated taxa, Veillonella and Oscillospiraceae UCG-005, which was inversely associated with wheeze, were correlated with the greatest number of fecal metabolites. Microbial networks were similar between subjects with low vs. high wheeze frequency.
Conclusion:
Gut microbiome features are associated with wheeze frequency in children with asthma, suggesting an impact of the gut microbiome on morbidity in childhood asthma.
Keywords: microbiome, metabolomics, asthma, wheeze
Graphical Abstract
Capsule Summary
In children with asthma, specific bacteria and small molecules in the gut are associated with increased frequency of wheeze, suggesting that strategies that target the microbiome could reduce morbidity in childhood asthma.
Introduction
Asthma is one of the most prevalent chronic diseases of childhood and is a complex disease with multiple recognized subtypes.1-4 Pediatric asthma leads to substantial morbidity: around half of children under age five years with asthma report having one or more asthma-related exacerbations over 12 months despite using prescribed therapies.1 Maturation of the immune system is dependent on the microbiota in early life, and aberrant microbial composition has been associated with the onset of many autoimmune and inflammatory diseases, including asthma.2 Longitudinal human studies have identified differences in early life gut microbial composition in children who go on to develop asthma compared to those who do not.3-6 Less is known about the role of the microbiome in existing asthma. While there is evidence linking airway microbiome and asthma morbidity7-11, few studies have investigated the gut microbiome in subjects with existing asthma.12
In addition to its potential relevance to asthma severity, microbial dysbiosis may impact corticosteroid responsiveness in asthma. Inhaled corticosteroids (ICS) are the most commonly prescribed maintenance therapy for asthma in children under age five years13. Just as a large minority of adults with asthma respond sub-optimally to ICS14,15, pre-school age children also exhibit a differential response to medications including ICS.16 The substantial proportion of non-response observed amongst ICS users has led researchers to look beyond factors such as adherence to identify other mechanisms at play.17 One such study found that ICS-resistant adults had distinct airway microbiome including an increased abundance of Proteobacteria compared to ICS-sensitive subjects18. To our knowledge, the fecal microbiome of ICS responsiveness has not been studied.
Metabolomics characterizes the small molecules in a sample, including molecules derived from the host, microbiome, or exogenous sources. Metabolomics thereby represents the cumulative effects of diverse exposures, and the metabolome of the gut exhibits especially strong associations with the microbiome, serving as a functional readout of microbial activity.19 For this reason, analyses integrating both microbiome and metabolomic data have the potential to yield novel insights and prioritize findings from analyses of individual omics.
Using data from the Vitamin D Antenatal Asthma Reduction Trial (VDAART)20, we investigated the association between the gut microenvironment at age three years and asthma severity. We defined asthma severity by, among children with parent-reported doctor-diagnosed asthma, the proportion of quarterly questionnaires between ages 3 and 5 years in which parents reported that their child had wheezed (“wheeze proportion”). We also looked at ICS responsiveness based on wheeze proportion in a subset of children with asthma who used ICS the majority of the time between ages 3 and 5 years. We hypothesize that differences in gut microbial composition and fecal metabolites are associated with both disease severity and ICS responsiveness in childhood asthma. To our knowledge, this is the first study to investigate the contribution of the gut microbiome and metabolome on disease severity and ICS responsiveness in childhood asthma.
METHODS
Study Design and Study Subjects
Detailed methods are included in the Online Repository. This is an ancillary observational analysis of data from the VDAART clinical trial, a double-blind placebo-controlled study on the effects of prenatal vitamin D supplementation on asthma in offspring, in which we analyzed subjects from both the high-dose and low-dose vitamin D treatment arms. Pregnant women were recruited during the first trimester of pregnancy from three sites in the United States: Boston University, Massachusetts; Washington University at St. Louis, Missouri; and Kaiser Permanente Southern California Region, San Diego, California. All women (n=876) had a history of asthma, eczema, or allergic rhinitis, and/or had a partner (biological father of the infant) with a history of asthma, eczema, or allergic rhinitis. After delivery, 806 children were followed using an over-the-phone quarterly health questionnaire and annual in-person visits. Stool samples were collected from children at age three years for microbiome and metabolome analysis (n=506). Subjects included in this analysis (n=110) met the following criteria: i) parent-reported doctor-diagnosed asthma, ii) completion of the majority (>50%) of quarterly health questionnaires between ages 3 and 5 years, and iii) microbiome sequencing data available from stool samples collected at age three years with >1,000 16s rRNA reads detected per sample. A power calculation based on previously reported effect sizes of microbiome associations with asthma severity21 estimated that our sample of 110 subjects yields power exceeding 0.99 to detect a similar association at alpha = 0.05 and power exceeding 0.97 at a more appropriate alpha = 0.005 given the high-dimensionality of the data analyzed (details available in the Online Repository).
We also analyzed a subgroup (n=28) of subjects with asthma whose parents/guardians reported use of ICS on at least 50% of the completed questionnaires between ages 3 and 5 years and refer to this as the ICS-treated subgroup. Our ICS-treated subgroup analysis sample size provides more limited power (0.65 at alpha=0.05) to detect associations of interest, and so this subgroup analysis should be considered preliminary.
Clinical Outcome
We analyzed wheeze frequency by calculating the proportion of available quarterly questionnaires between ages 3 and 5 years in which parents reported wheeze since the last questionnaire. We refer to this continuous variable as wheeze proportion. We also defined a dichotomous variable to categorize subjects as either high wheeze (i.e., reported wheeze on ≥33% of the completed questionnaires (n = 56)) or low wheeze (i.e., reported wheeze <33% of the time (n = 54)). The cut-off of 33% was selected to ensure approximately equal subject numbers in each group. For the 28 ICS-treated subjects, we defined a dichotomous variable, ICS response, categorizing subjects as either non-responders (i.e., reported wheeze on ≥50% of the completed questionnaires (n = 13)) or responders (i.e., reported wheeze <50% of the time (n = 15)). The cut-off of 50% was selected to ensure approximately equal subject numbers in each group.
Fecal Microbiome and Metabolome Profiling
Microbiome Profiling:
Microbiome profiling was performed on stool samples collected at age three years by sequencing the 16S rRNA hypervariable region 4 (V4 515F/816R region) on the Illumina MiSeq platform, as previously described.22 Reads were denoised using DADA2 as implemented in Qiime2.23 Taxonomy was assigned to representative sequences using a naive Bayes classifier pre-built from the 99% SILVA 138 database specific to the 515F/816R region for bacterial data.24,25 Amplicon sequence variants (ASVs) present in <5% of samples were excluded.
Quantitative PCR was also performed with universal 16S rRNA primers to estimate the total bacterial biomass for each sample using a previously published method.22 Relative abundance data is widely used to study microbiome and metabolome composition in relation to both health and disease states 4,26-29; however, these data are subject to compositionality effects.28 Absolute abundance data can optimize detection of microbiome compositional variation between individuals over time and co-variation in species networks; and decrease false discovery rates.28,30-32 In this study, we considered both relative abundance (RA) and absolute abundance (AA) data. Of the 110 total subjects and 28 ICS-treated subjects in our study, 85 and 22, respectively, passed quality thresholds for quantitative PCR and were included in the quantitative abundance analyses.
Metabolomic Profiling:
Untargeted metabolomic profiling of stool was performed using UPLC-MS/MS by Metabolon, Inc., as previously described.22,33
Statistical Analyses
Alpha and Beta Diversity:
Species-level alpha diversity Shannon and Simpson indices were calculated using the vegan R package. Pearson correlations and linear regression were used to seek associations between alpha diversity and wheeze frequency. For beta diversity, reads were scaled to an even depth (based on minimum read count) and the species-level Bray-Curtis dissimilarity matrix was calculated using the Phyloseq R package. PERMANOVA (Adonis) was used to test for differences in Bray-Curtis beta diversity by wheeze frequency. For this and all other analyses, p value thresholds of 0.05 were used to determine statistical significance unless otherwise noted.
Unsupervised Clustering:
To seek associations between wheeze and overall microbiome and metabolome structure, we performed unsupervised clustering of subjects by microbiome data using Dirichlet multinomial mixtures using the R package DirichletMultinomial34,35 and by metabolomics data using hierarchical clustering using the R package cluster.36 Optimal cluster number was chosen using the gap statistic and silhouette methods for both microbiome and metabolomics data. Associations with wheeze proportion were tested using Wilcoxon rank sum tests and linear regression models adjusted for potential confounders. Because we sought associations of wheeze proportion with only one microbiome cluster variable and one metabolome cluster variable, we did not correct for multiple testing and used a p value cut-off of 0.05 to determine statistical significance.
Differential Taxa Analysis:
Differential taxa analysis was conducted using Songbird, a reference frame-based approach to microbiome analysis that accounts for the compositionality inherent in microbiome data37. Because this method relies on ratios of taxonomic abundances, it yields identical results with RA and AA microbiome data. We used this method to address compositionality rather than analyzing AA microbiome data to leverage our full sample size, as stool samples from some subjects did not have sufficient quality DNA to determine fecal microbial absolute abundances. A positive Q2-value (defined as 1 – (average absolute model error based on cross-validation / average absolute null model error based on cross-validation)) was observed for all reported models, indicating adequate predictive accuracy. Songbird produces a differential ranking of taxa by the strength of their association with the outcome, in this case, wheeze proportion. We identified the taxa most positively associated with wheeze proportion and the taxa most negatively associated with wheeze proportion and visualized relevant associations using Qurro38. For top-ranked taxa, we further characterized associations with wheeze proportion using linear regression and Spearman correlation, analyzing taxonomic relative abundances in two ways: 1) as log-transformed taxonomic relative abundances, and 2) using a reference-frame based approach by calculating the log-transformed ratio of (taxonomic relative abundance):(sum of relative abundances of the top 10% of taxa associated with asthma in the opposite direction). We also analyzed associations between taxa associated with wheeze proportion and ICS use with Spearman correlation tests. We report both p values and FDR values to evaluate effect sizes.
Metabolomic Analysis:
Analyses were conducted on 737 stool metabolites with known identities. Spearman correlation tests identified metabolites associated with wheeze proportion. There are currently no consensus standards for multiple testing correction in metabolomics; methods such as the Bonferroni correction and even more liberal corrections are considered too stringent for metabolomics data due to the high correlation of functionally related metabolites. In this analysis, we report both p values and FDR values, with the caveat that FDR correction may result in false negatives. Pathway analyses were performed on metabolites with Human Metabolome Database (HMDB) annotations using Metaboanalyst v4.0 (www.metaboanalyst.ca. Over-representation analyses using Fisher’s exact test were also performed as several metabolites did not have HMDB identifiers and could not be included in the Metaboanalyst subpathway analysis. For over-representation analyses, metabolite pathway annotations were supplied by Metabolon in which a single pathway annotation is assigned to every metabolite.
Dietary Analysis:
To follow up on findings of our metabolomics analysis which suggested possible relevance of diet-derived metabolites, we determined associations of food intake with fecal metabolites and with wheeze using Spearman correlation and linear regression. When subjects were 3 years old, their parents or guardians completed a food frequency questionnaire including questions about how frequently the child had eaten tofu or soybeans and a variety of meats in the prior month (never, <1 time per week, once per week, 2-4 per week, nearly daily or daily, or 2 or more times/day). This was recoded to a numeric variable reflecting the average daily occurrence of eating relevant foods for analysis.
Network Analyses:
We used Spearman correlation analysis to identify associations between metabolites and species. Microbial correlation networks were constructed at the species level via SparCC (Sparse InversE Covariance estimation for Ecological Association and Statistical Inference) using the SpiecEasi R package.39 To determine the significance of correlations (i.e., edges), we compared our model to a null model using 100 bootstrap iterations as previously described.40 A robust correlation was included in the network if the absolute value of the SparCC correlation coefficient was ≥0.45 and was significant (p<0.05). p values were not corrected for multiple testing in this descriptive analysis. Networks were visualized using igraph in R.
RESULTS
Subject Characteristics
Of the 806 total VDAART subjects followed after birth, we analyzed a subset of 110 children with parent-reported doctor-diagnosed asthma at age 3 years (Figure 1). Fifty-four subjects reported wheeze on ≥33% of questionnaires between age three and five years and were categorized as “high wheeze,” while 56 subjects reported wheeze on <33% of questionnaires and were categorized as “low wheeze”. High wheeze subjects did not differ from low wheeze subjects on most baseline characteristics including sex, race/ethnicity and VDAART study treatment assignment (Table 1). Wheeze frequency did differ between study sites (Table 1, Fisher’s exact test p=0.001) and, accordingly, study site was included as a covariate in models of key results. As expected, as an additional indicator of asthma severity, we found that high wheeze subjects were more likely to have used oral corticosteroids between the ages of three and (Fisher’s exact test p=0.02).
Figure 1.
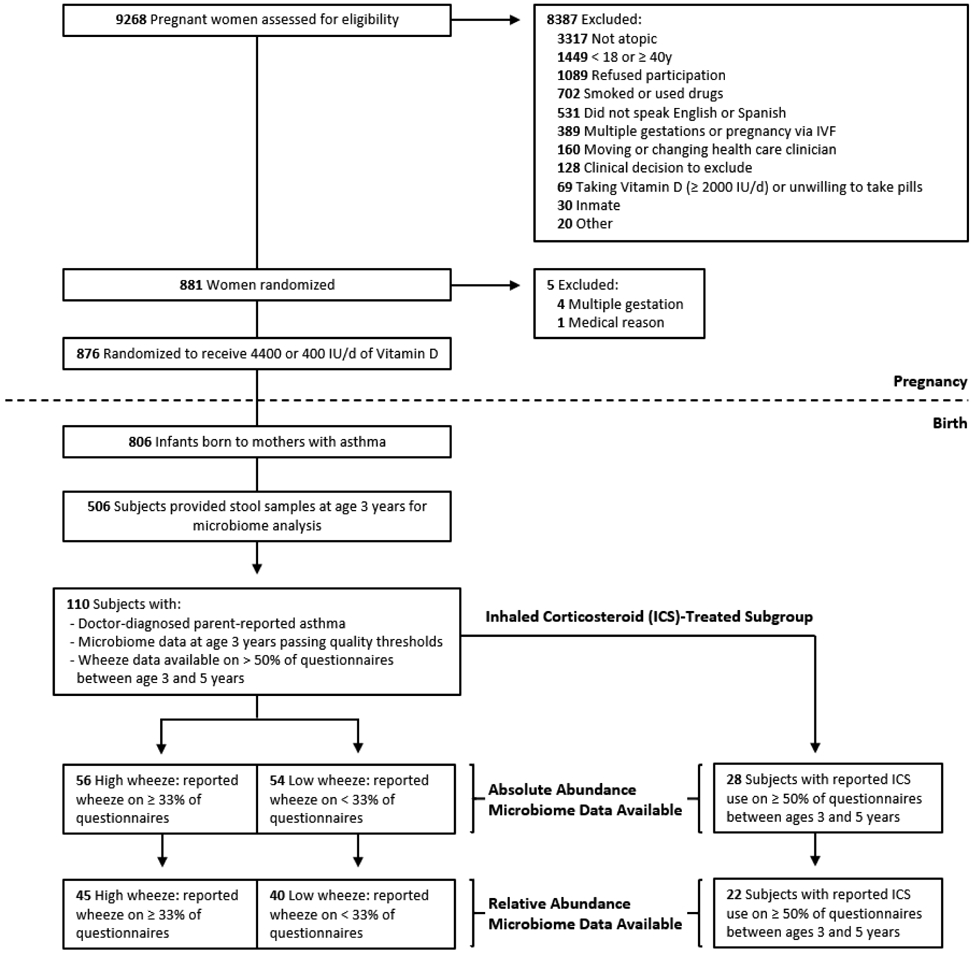
Flow diagram of study subjects.
Table 1. Subject characteristics by wheeze frequency.
High wheeze subjects are those with reported wheeze at least 33% of the time and low wheeze subjects are those with reported wheeze less than 33% of the time between ages 3 and 5 years, p values are for Wilcoxon rank sum tests for log-transformed total IgE, BMI and gestational age; and otherwise for Fisher’s exact test. Sensitization is based on a serum IgE concentration of 0.35 kU/L to at least one food or environmental allergen, respectively.
| All Children (n = 110) |
High Wheeze (n = 56) |
Low Wheeze (n = 54) |
p value | |
|---|---|---|---|---|
| Female Sex – Number (%) | 46 (41.4) | 25 (44.6) | 21 (38.2) | 0.62 |
| Race/Ethnicity – Number (%) | 0.59 | |||
| Black, non-Hispctnic | 71 (64.0) | 37 (66.1) | 34 (61.8) | |
| White, non-Hispctnic | 18 (16.2) | 10 (17.9) | 8 (14.5) | |
| Hispanic or Other | 22 (19.8) | 9 (16.1) | 13 (23.6) | |
| Study Site – Number (%) | 0.002 | |||
| Boston | 26 (23.4) | 7 (12.5) | 19 (34.5) | |
| San Diego | 20 (18.0) | 7 (12.5) | 13 (23.6) | |
| St. Louis | 65 (58.6) | 42 (75.0) | 23 (41.8) | |
| Household Income – Number (%) | 0.20 | |||
| < $30,000 | 49 (44) | 27 (47) | 22 (40) | |
| $30,000-$49,999 | 13 (12) | 3 (5) | 10 (18) | |
| $50,000-$74,999 | 9 (8) | 3 (5) | 6 (11) | |
| $75,000-$99,999 | 4 (4) | 2 (4) | 2 (4) | |
| > $100,000 | 8 (7) | 6 (11) | 2 (4) | |
| Refused or unknown | 29 (26) | 16 (28) | 13 (24) | |
| Vitamin D Treatment Group – Number (%) | 0.40 | |||
| 400 IU / day | 58(52.3) | 32 (57.1) | 26 (47.3) | |
| 4,000 IU / day | 53 (47.7) | 24 (42.9) | 29 (52.7) | |
| Total IgE (kU/L) (log scale) – mean (SD) | 4.1 (1.6) | 4.4 (1.7) | 3.9 (1.4) | 0.15 |
| Food Sensitization – Number (%) | 56 (54.4) | 31 (58.5) | 25 (50.0) | 0.51 |
| Environmental Sensitization – Number (%) | 44 (42.7) | 27 (50.9) | 17 (34.0) | 0.12 |
| Body Mass Index (BMI) – Mean (SD) | 16.7 (1.7) | 16.4 (1.9) | 16.9 (1.6) | 0.10 |
| Maternal Asthma – Number (%) | 59 (53.2) | 34 (60.7) | 25 (45.5) | 0.16 |
| Number of Parents with Asthma – Number (%) | 0.70 | |||
| Zero | 35 (31) | 16 (28) | 19 (34) | |
| One | 65 (58) | 34 (60) | 31 (56) | |
| Two | 12 (11) | 7 (12) | 5 (9) | |
| Maternal College Degree or Higher – Number (%) | 23 (20.7) | 8 (14.3) | 15 (27.3) | 0.15 |
| Birth by Cesarean Section – Number (%) | 34 (30.6) | 20 (35.7) | 14 (25.5) | 0.33 |
| Gestational Age (Weeks) – Mean (SD) | 37.9 (2.7) | 37.9 (2.8) | 38.0 (2.5) | 0.91 |
| Exclusively Breastfed for First 4 Months – Number (%) | 21 (20.4) | 12 (23.5) | 9 (17.3) | 0.59 |
| Perinatal Antibiotic Exposure – Number (%) | 48 (43.2) | 23 (41.1) | 25 (45.5) | 0.78 |
| Oral Steroid Use Between Age 3 and 5 Years – Number (%) | 29 (29.0) | 22 (39.3) | 7 (15.9) | 0.02 |
| Household Dog – Number (%) | 19 (17.4) | 11 (19.6) | 8 (15.1) | 0.71 |
| Household Cat – Number (%) | 10 (9.3) | 2 (3.6) | 8 (15.1) | 0.09 |
| At Least 1 Older Sibling – Number (%) | 68 (61.3) | 34 (60.7) | 34 (61.8) | 1.0 |
| Day Care before Age 3 Years – Number (%) | 62 (57.4) | 28 (50.9) | 34 (64.2) | 0.23 |
Missing data: Oral steroid use missing for 11 subjects. Total IgE was missing for 10 subjects. Sensitization (food and environmental) and breastfeeding missing for 8 subjects. Daycare and cat missing for 3 subjects. Dog missing for 2 subjects. BMI missing for 1 subject.
Bolded p values are < 0.05.
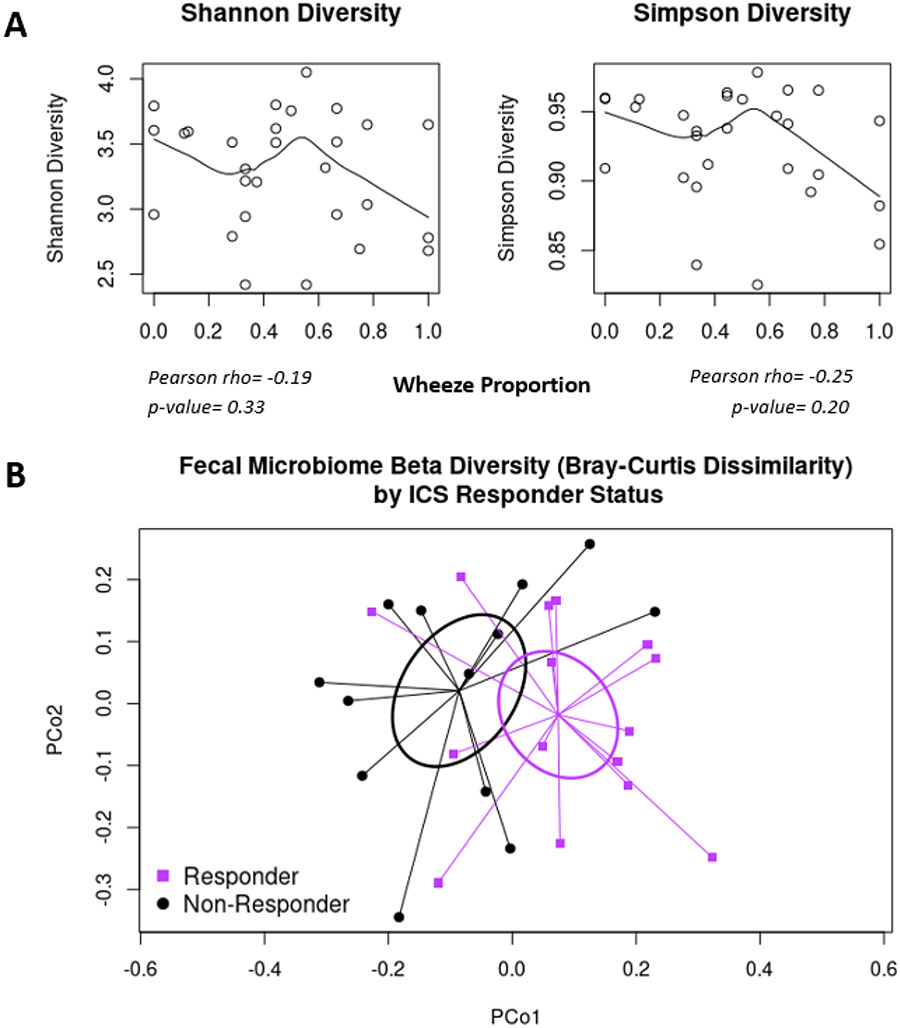
Fecal Microbial Diversity Is Not Associated with Childhood Wheeze Frequency
We sought associations between fecal microbiome characteristics at age 3 years and wheeze proportion between ages 3 and 5 years. Fecal alpha diversity was not associated with wheeze proportion (Shannon index: Pearson’s r=−0.12, p=0.21; Simpson index: Pearson’s r=−0.10, p=0.27) (see Figure E1 in the Online Repository). Similarly, beta diversity (Bray-Curtis dissimilarity) was not associated with wheeze proportion (PERMANOVA p=0.79; Figure E1). Results were similar after adjusting for study site (Shannon index adjusted linear regression beta= −0.09 (95% CI −0.21, 0.03), p=0.14; Simpson index beta= −0.81 (95% CI −2.12, 0.50), p=0.22; Bray-Curtis PERMANOVA p=0.77).
To further evaluate associations of overall gut microbiome composition with wheeze, we performed unsupervised clustering of subjects by microbiome composition using Dirichlet multinomial mixtures.34 This yielded an optimal cluster number of two, with 67 subjects belonging to Cluster 1 and 43 subjects belonging to Cluster 2. Cluster membership was not associated with wheeze proportion (Wilcoxon rank sum test p=0.40, linear regression adjusted for study site p=0.34, mean wheeze proportion 0.32 in Cluster 1 and 0.35 in cluster 2, Table E1 in the Online Repository). In combination with lack of association between wheeze and beta diversity, these findings reinforce that overall gut microbiome structure is not associated with wheeze frequency, though this does not rule out that specific microbial taxa could contribute to wheeze.
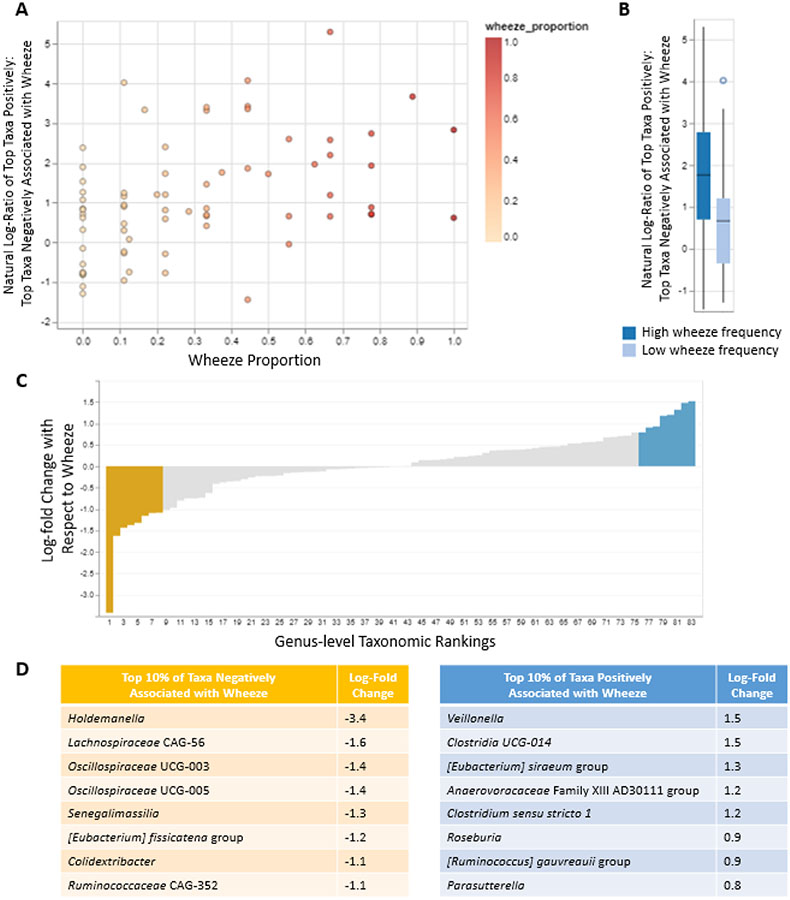
Specific Gut Bacterial Taxa Are Associated with Wheeze Proportion
We used Songbird, a method that accounts for compositionality in microbiome data by analyzing ratios of taxonomic abundances and produces a ranking of taxa associated with an outcome of interest 37, to identify the top 10% of genera most positively and the 10% of genera most negatively associated with wheeze proportion (Figure 2, Tables E2 and E3 in the Online Repository). The log-ratio of the top positively associated genera to the top negatively associated genera was significantly associated with wheeze proportion (Spearman rho = 0.43, p = 0.0002) and this association was preserved after adjusting for maternal education (linear regression beta = 0.08 (0.04, 0.13), p = 0.0001). All of the associated genera were of the phylum Firmicutes with the exception of Senegalimassilia, a member of the phylum Actinobacteria that was negatively associated with wheeze and Parasutterella, a member of the phylum Proteobacteria that was positively associated with wheeze. Veillonella was the genus most strongly positively associated with wheeze. Taxa negatively associated with wheeze included Holdemanella and members of the families Lachnospiraceae and Oscillospiraceae. In both Spearman correlation analyses and linear regression analyses adjusted for study site, all taxa associations with wheeze were preserved when analyzed as a ratio of taxon abundance : abundances of the top 10% of taxa associated with wheeze in the opposite direction (see Tables E2 and E3 in the Online Repository). With few exceptions, taxa exhibited the same direction of association with wheeze when analyzed as log-transformed relative abundances (see Figure E2 in the Online Repository).
Figure 2.
Genera associated with wheeze proportion. A. Scatterplot of the association of wheeze proportion with the natural log-ratio of abundances of the top 10% of taxa positively associated with wheeze : abundances of the top 10% taxa negatively associated with wheeze. B. Box plot of the association of wheeze frequency with the natural log-ratio of abundances of the top 10% of taxa positively associated with wheeze : abundances of the top 10% taxa negatively associated with wheeze. C. Microbiome feature rankings with top and bottom 10% of taxa highlighted in blue and yellow, respectively. D. The top 10% of genera most positively and the 10% of genera most negatively associated with wheeze are listed with log-fold changes. Figures 3A-C were generated using Qurro.
To address the possibility that these taxa were associated not with asthma severity but with use of inhaled corticosteroids (ICS), which would be expected to increase among subjects with greater asthma morbidity, we calculated Spearman correlations between the proportion of questionnaires on which parents reported that subjects were using ICS (data available for n=49 subjects, mean ICS proportion = 0.49, SD = 0.36) and both taxon ratios and log-transformed taxon relative abundances. None of these correlations were statistically significant (all p > 0.05), indicating that the observed associations of gut microbial taxa with wheeze are not attributable to ICS use.
Histidine Metabolism is Positively Associated with Wheeze Proportion
Fecal metabolomic profiling was performed on the same stool samples collected at age three years that were used for microbiome profiling. To seek associations of overall gut metabolomic profiles with wheeze, we performed unsupervised clustering of subjects by metabolome composition using hierarchical clustering36. This yielded two clusters with 56 subjects belonging to Cluster 1 and 43 subjects belonging to Cluster 2 (see Figure E3 in the Online Repository). Cluster membership was significantly associated with wheeze proportion (Wilcoxon rank sum test p = 0.04, linear regression adjusted for study site p = 0.03, mean wheeze proportion = 0.39 in Cluster 1 and 0.26 in cluster 2, Table E1 in the Online Repository). These findings suggest that subjects with frequent wheeze exhibit global shifts in the gut metabolome compared to those with low wheeze frequency. This contrasts with the lack of association we observed between wheeze and microbiome clusters, suggesting that factors beyond the microbiome could be involved with metabolome-wheeze associations.
To further characterize gut metabolome associations with wheeze, we performed analyses of individual metabolites. Of 737 identified fecal metabolites, 20 were nominally positively associated with wheeze proportion (Spearman correlation p<0.05), though no associations were preserved after correction for multiple testing (all FDR > 0.05). No metabolites were negatively associated with wheeze proportion (see Table E4 in the Online Repository). Of the 20 fecal metabolites nominally associated with wheeze proportion, 19 had the same direction of association and p<0.20 in linear regression analyses adjusted for study site. Most of these 20 metabolites were amino acid metabolites (40%), xenobiotics (20%), or lipids (25%) (Table E4). Pathway analysis revealed that metabolites nominally associated with wheeze were enriched with histidine pathway metabolites (Pathway Impact Value (PIV)=0.14; Fisher’s exact test overrepresentation analysis p=0.04), though this result was not significant after correcting for testing of multiple pathways (FDR>0.05) (see Figure E4 in the Online Repository). Metabolites within the histidine pathway associated with wheeze included carnosine, N(pi)-methyl-L-histidine, and beta-Alanyl-N(pi)-methyl-L-histidine. These results, which were limited to metabolites with assigned HMDB IDs, were confirmed by over-representation analysis using complete metabolite annotation data provided by Metabolon (Fisher’s exact test p<0.0001, FDR<0.01). As histidine metabolites could be derived from dietary sources, especially animal meat41, we analyzed dietary data from food frequency questionnaires completed at age 3 years and found that intake of meats (hot dogs, sausage, pork, ham, cold cuts, beef, hamburger and bacon) were both positively correlated with wheeze-associated fecal histidine metabolites (Spearman rho range=0.16-0.28) and borderline associated with wheeze proportion (Spearman rho=0.18, p=0.06), though the association with wheeze was attenuated with adjustment for study site (linear regression beta=0.03, p=0.10).
Over-representation analysis also identified food component/plant fecal metabolites as enriched among subjects with greater wheeze proportion (Fisher’s exact test p=0.00002, FDR<0.01). Food component/plant fecal metabolites associated with wheeze included soy isoflavones daidzein and 6-hydroxydaidzein, and diaminopimelate, a breakdown product of gram-negative gut microbes. These results suggested a possible positive association between soy intake and wheeze; however, dietary tofu or soybean intake at age 3 years was not correlated with either fecal daidzein (Spearman rho = −0.02, p = 0.83) or fecal 6-hydroxydaidzein (rho = 0.03, p = 0.75) and we found no correlation between frequency of tofu or soybean intake reported at age 3 years and wheeze proportion between ages 3 and 5 years (Pearson rho = −0.03, p = 0.76; linear regression adjusted for study site beta = −0.36, p = 0.13). Overall, these results suggest that dietary intake of meats, but not soy, could partially account for associations between fecal metabolites and wheeze.
Veillonella and Oscillospiraceae UCG-005 are Correlated with Numerous Metabolites
To query the functional significance of fecal taxa associated with wheeze, we created fecal taxon-metabolite Spearman correlation networks using both absolute abundance (AA) and relative abundance (RA) microbiome data, as quantitative AA microbiome profiling, compared to traditional RA profiling, may be preferable when seeking microbe-microbe or microbe-metabolite correlations28. Quantitative PCR using universal 16S rRNA primers was used to estimate the total bacterial biomass concentration per stool sample. Biomass estimates were then used to calculate AA profiles. Unlike with RA profiling, in which abundances of each taxa can only be considered as fractions of overall taxa detected, AA profiling allows abundance for each taxa to be considered independently. Some subjects had insufficient DNA quantity or quality for biomass estimation, so the sample size was reduced to 85 subjects and included 45 high wheeze and 40 low wheeze subjects. There was no significant correlation between total fecal bacterial biomass at age three years and wheeze proportion between ages 3 and 5 years (Spearman’s ρ=−0.15, p=0.18) (see Figure E5 in the Online Repository). As expected, at the phylum level, the two most abundant phyla were Bacteroidetes and Firmicutes (see Figure E6 in the Online Repository). Phylum-level microbiome composition was similar between subjects with high and low wheeze proportion (Figure E6).
Among wheeze-associated taxa, Veillonella, which was positively associated with wheeze, and Oscillospiraceae UCG-005, which was negatively associated with wheeze, were associated (Spearman correlation p<0.05) with the greatest number of metabolites (213 and 250, respectively using RA microbiome data; 160 and 229, respectively using AA microbiome data; including only metabolites associated with wheeze and with the taxa in a directionally consistent manner, see Figure E7 in the Online Repository). Most of these taxa-associated metabolites demonstrated significant associations in both analyses of AA and RA data (150 and 208 metabolites significant using both data types for Veillonella and Oscillospiraceae UCG-005, respectively). Pathway analysis of metabolites associated with Veillonella in both AA and RA analyses revealed no significant pathways and we likewise found no pathways associated with Oscillospiraceae UCG-005 using pathway analysis. However, over-representation analysis identified that dipeptide metabolism was associated with Veillonella (Fisher’s exact test FDR = 0.03); and polyamine metabolism was associated with Oscillospiraceae UCG-005 (Fisher’s exact test FDR = 0.01).
Microbial Correlation Networks Are Similar Between Those with High vs. Low Wheeze Frequency
To identify fecal microbial correlation patterns among subjects with asthma with high vs low wheeze proportion, a network analysis was performed using SparCC at the genus level (see Figure E8 in the Online Repository). Results reported here are based on AA data; results based on RA data were comparable and can be found in Table E5 in the Online Repository. Edges were based on permutation-based p<0.05 and correlation strength of at least 0.45. Overall, mean degree, that is, average number of microbes correlated (p<0.05) with each microbe, were similar between high and low wheeze proportion subjects (high=2.1 +/− 2.2; low=2.0 +/−1.7; Wilcoxon rank sum test p=1.0). Those with high wheeze and low wheeze also had similar numbers of edges (high=62 positive and 26 negative edges; low=64 positive and 38 negative edges; Fisher test comparing frequency of positive vs negative edges between groups p=0.28). Ruminococcus gnavus group was the genus correlated with the most other microbes in both high and low wheeze subjects (n=7 among low wheeze and n=10 among high wheeze subjects, respectively). Veillonella was only associated with one other microbe – unknown genera of the family Pasteurellaceae – in both low and high wheeze subjects, and Oscillospiraceae UCG-005 was not significantly associated with any microbes at a correlation strength of at least 0.45.
ICS Response is Associated with Fecal Microbial Composition and Numerous Fecal Metabolites
To determine associations of the gut microenvironment and ICS treatment responses in children, we analyzed a subgroup of 28 subjects who were treated with ICS between ages 3 and 5 years (Figure 1). Thirteen subjects reported wheeze on ≥50% of questionnaires between ages three and five years (ICS non-responders), while 15 subjects reported wheeze on <50% of the questionnaires (ICS responders). Non-responders and responders were largely comparable on baseline characteristics with the exception of maternal characteristics: non-responders were more likely to have mothers with asthma (76.9% vs. 20.0%, Fisher’s test p = 0.007), less likely to have no parents with asthma (7.7% vs 53.3%, Fisher’s test p = 0.03), and more likely to have mothers with less than a college degree (84.6% vs 40.0%, p = 0.02) (see Table E6 in the Online Repository). Accordingly, maternal asthma and maternal education were included as covariates in models of key results as the association with maternal asthma was stronger than the association with number of parents with asthma and these two variables are highly correlated (Fisher test p=0.0005).
Fecal microbial alpha diversity at age three years was non-significantly reduced in association with increased wheeze proportion among subjects on ICS, and this trend was most prominent among subjects with more frequent wheeze (Figure 3, Shannon index adjusted linear regression beta=−0.14 (95% CI −0.35, 0.06) p=0.16; Simpson index adjusted linear regression beta=−1.68 (95% CI −3.96, 0.61), p=0.14). In contrast to the lack of association between beta diversity and wheeze in the overall sample of subjects with asthma, Bray-Curtis dissimilarity was significantly associated with wheeze proportion in the ICS-treated subgroup (PERMANOVA p=0.03; Figure 3) and this association was preserved after adjustment for maternal education and maternal asthma (PERMANOVA p=0.02).
Figure 3.
A. Scatterplot of fecal microbial alpha diversity by wheeze proportion in ICS-treated subjects. B. Principal coordinates analysis plot based on Bray-Curtis dissimilarity. Ellipses represent 95% confidence intervals around standard errors.
Songbird analysis identified the top 5 taxa most positively associated with wheeze and the top 5 taxa most negatively associated with wheeze proportion in the ICS-treated subgroup (see Figure E9 in the Online Repository). The log-ratio of the top positively associated taxa to the top negatively associated taxa was significantly associated with wheeze proportion (Spearman rho = 0.54, p = 0.0005) and this association was preserved after adjusting for maternal education (linear regression beta = 2.0 (−0.1, 4.1), p = 0.06) and maternal asthma (beta = 2.0 (0.1, 3.8), p = 0.04). All taxa associated with wheeze were of the phylum Firmicutes except for Eggerthella, a genus of the phylum Actinobacteria that was negatively associated with wheeze. As we observed in the overall sample of children with asthma, Veillonella was the top taxa most positively associated with wheeze.
Of 737 fecal metabolites, 124 were positively associated with wheeze proportion (Spearman correlation p<0.05) and seven were negatively associated with wheeze proportion in the ICS-treated subgroup (see Table E4 in the Online Repository), though of these 131 associations, only three were preserved after correction for multiple testing (anserine (FDR = 0.04), dimethylarginine (FDR = 0.04) and mannose (FDR = 0.04) were all positively associated with wheeze proportion). Of the 131 fecal metabolites nominally associated with wheeze proportion, 118 (90%) had the same direction of association with wheeze proportion and p<0.20 in linear regression analyses adjusted for maternal education and maternal asthma. Notably, the number of metabolites nominally associated with wheeze was higher in the ICS-treated subgroup (131 metabolites) than in the overall sample of subjects with asthma (20 metabolites). The 20 metabolites positively associated with wheeze in all subjects with asthma were also positively associated with wheeze in the ICS-treated subgroup, and of these 20 metabolites, 11 associations reached nominal statistical significance (p<0.05). Sphingolipid metabolism was the only pathway enriched in children with higher wheeze proportion (Pathway Impact Value (PIV)=0.47; p=0.04). Metabolites within the sphingolipid pathway associated with wheeze included L-serine, sphinganine, sphingosine, N-acylsphingosine (a ceramide), and phytosphingosine. The enrichment of the sphingolipid pathway was confirmed using over-representation analysis (Fisher’s exact test p=0.004, FDR = 0.29) (see Figure E10 in the Online Repository).
SparCC microbial correlation networks constructed using AA data (n=22), where edges were based on permutations-based p<0.05 and correlation strength ≥0.60 or ≤−0.60 were similar in mean degree (ICS non-responders=3.1 +/− 1.8; ICS responders=3.3 +/− 2.7; Wilcoxon test p=0.62) (see Figure E8 in the Online Repository). Microbe-microbe correlations were more numerous among subjects with more frequent wheeze (non-responders, 242 edges) than those with less frequent wheeze while treated with ICS (responders, 200 edges), though the two networks had a similar distribution of positive and negative edges (non-responders: positive=160, negative=82; responders: positive=132, negative=68; Fisher test p=1.0). Despite requiring a stronger correlation strength in the ICS-treated subgroup microbial network (0.60) compared to the network for the larger sample of subjects with asthma (0.45), significant correlations were more numerous in the ICS-treated subgroup as reflected by higher mean degree. The greater connectivity between gut microbes observed in the ICS-treated subgroup compared to the overall group of subjects with asthma, despite the smaller sample size, suggests that intestinal microbial ecology is more homogenous among ICS-treated subjects than subjects with asthma who are not treated with ICS.
DISCUSSION
In this analysis of the gut microbiome at age 3 years among children with asthma, we found taxa associated with increased wheeze in the subsequent two years. Of these, the genus Veillonella is of particular interest and was the top-ranked genus positively associated with wheeze both among the overall sample of children with asthma and among children treated with ICS. Veillonella spp. are commensal bacteria of the mouth and gastrointestinal tract; however, overgrowth of Veillonella species in the airway has been implicated in decreased lung function and asthma exacerbations.42 Yet, other studies have found Veillonella species to be enriched in bronchi of non-asthmatic adults 43 and decreased in the lungs of corticosteroid-resistant adults.18 The role of Veillonella species in asthma may also extend to the gut microbiome. Veillonella abundance in the infant gut has been associated with risk of subsequent asthma development, though the directions of associations have not been consistent between studies6, and the parent family of Veillonella, Veillonellaceae, was recently found to be elevated in stool samples from adult asthmatics in those with more severe disease21. Current discrepancies in the role of Veillonella in asthma development and morbidity warrant further research into the mechanisms of this genus’s effect on lung function and asthma pathology. Disparate findings may reflect differences in study methodology or could be due to a diversity of functional capacities among species and strains of the genus Veillonella as well as differences in the impact of Veillonella presence at different body sites. We found that Veillonella and the genus Oscillospiraceae UCG-005, a taxa that was inversely associated with wheeze, were correlated with numerous fecal metabolites, suggesting that these wheeze-associated taxa may have high functional relevance to the gut microenvironment. However, microbial networks were largely similar between children with high and low wheeze frequencies and that neither Veillonella nor Oscillospiraceae UCG-005 were strongly correlated with other microbes, suggesting that the functional impact of these taxa is less likely due to interactions with other microbes.
While analyses of microbial diversity and unsupervised clustering of microbiome data suggested no global association of microbiome composition with wheeze among children with asthma, unsupervised clustering of metabolomic data revealed two clusters that were divergently associated with wheeze frequency. This finding indicates that asthma severity is related to global shifts in the biochemistry of the gut environment, in a manner that may be relatively independent of the microbiome. In terms of specific pathways that may be involved, we found that histidine metabolites were enriched among children with asthma who had more frequent wheeze. Histidine is a precursor to histamine and an important immunomodulator affecting both the innate and adaptive immune response.44 Increased fecal expression of histidine decarboxylase has been observed in adults with asthma and is known to contribute to bronchospasm, mucus secretion and airway edema.12,44,45 Although mast cells and basophils are the principal producers of histamine, many gut microbes also produce histamine.12,46 Our analysis of dietary data suggested that intake of meats including beef, pork and turkey could also lead to increased fecal histidine metabolite levels, consistent with prior reports that meat, processed meat, and fast food intake are associated with asthma morbidity47,48, including in children49. Overall, our findings suggest that increased histamine metabolism in the gut, potentially as a result of dietary choices, may contribute to morbidity in individuals with existing asthma.
We also found that food component/plant fecal metabolites were associated with wheeze, including daidzein and 6-hydroxydaidzein. In contrast to this finding, soy isoflavones have been associated with improved disease control among adults with asthma50,51. Because gut microbes metabolize soy isoflavones to downstream metabolites in a process that varies between individuals52, our finding of an association between wheeze and soy isoflavones may be a marker of microbiome composition differences by wheeze frequency rather than dietary differences. In accordance with this possibility, we found no significant association between reported tofu or soybean intake at age 3 years and wheeze frequency between ages 3 and 5 years.
In an exploratory analysis of a subgroup of children treated with ICS, fecal sphingolipid metabolites were more abundant in those with more frequent wheeze. Sphingolipids are signaling molecules that regulate a range of pro-inflammatory processes and are thought to influence asthma-associated inflammation and pathophysiology. 53 However, the interplay between sphingolipids, the immune system and asthma is not yet fully understood. Previous research has shown that decreases in de novo biosynthesis of sphingolipids enhances airway responsiveness independent of allergy or inflammation. 53-55 Yet, increased levels of ceramide and S1P have been observed in the lungs of asthmatics along with increases in airway hyperresponsiveness in both animal and human studies. 53,56-59 Very few studies have directly compared the metabolic profiles of ICS responders and non-responders; however, one recent study found that adults with uncontrolled asthma had distinct cellular markers and metabolomic profiles compared to adults with controlled asthma including higher concentrations of sphingosine and several ceramide species (C16:0 and C24:0) in serum.60 These data suggest that sphingolipid metabolites are associated with an uncontrolled/non-responsive asthma subtype, and our analysis points to a possible mechanism involving a reduction in steroid responsiveness.
Interestingly, a greater number of metabolites were associated with wheeze among subjects treated with ICS than among the larger asthma cohort, despite smaller sample size. We also find a more robust association between microbial composition and wheeze in the ICS cohort. These data suggest that the microenvironment of the gut associates more strongly with treatment response than overall asthma control in childhood asthma. However, given the limited sample size in this subgroup analysis of ICS-treated subjects, these findings must be regarded as preliminary. An additional limitation is the lack of available data on ICS dosing, as it is possible that some of our findings may be caused by ICS effects on the microbiome/metabolome, with subjects with more frequent wheeze potentially exposed to higher doses of ICS.
Overall, our study provides evidence that the gut microbiome and metabolome have an impact on wheeze frequency in childhood asthma. The strengths of this study lie in its use of longitudinal dense phenotyping of a diverse sample and utilization of methods to address compositionality in microbiome data - which is often lacking in microbiome association studies. However, there were study limitations. This is an ancillary analysis of a trial of high-dose vitamin D supplementation during pregnancy, and the trial intervention could have had an impact on the gut microbiome or metabolome61; however, we observed no significant association of trial treatment assignment with wheeze frequency and so do not expect that effects of prenatal vitamin D supplementation account for the associations we observed with wheeze frequency. Untargeted metabolomic profiling gives an idea of relevant metabolites, but without confirmation using a targeted system, quantification accuracy is a concern. Additionally, our small sample size precluded analysis of a replication or validation sample, which increases the possibility of false positive findings. Further research with larger study cohorts and targeted quantification of metabolites are required to corroborate these findings.
Supplementary Material
Key Messages:
In children with asthma, intestinal Veillonella abundance was associated with more frequent wheeze and with numerous fecal metabolites, suggesting significant functional potential and clinical relevance of this genus.
Children with asthma who wheezed more often demonstrated a shift in their fecal metabolomic composition, including enrichment with histidine metabolites.
Acknowledgements
VDAART was funded by U01HL091528 from the National Heart, Lung, and Blood Institute. Additional funding came from NIH grants R01HL108818, R01HL123915, R01HL141826, K08 HL148178 and ECHO grant OD023268.
Abbreviations:
- AA
Absolute abundance
- BMI
Body mass index
- FDR
False discovery rate
- HMDB
Human Metabolome Database
- ICS
Inhaled corticosteroid
- IgE
Immunoglobulin E
- PCR
Polymerase chain reaction
- PERMANOVA
Permutational multivariate analysis of variance
- PIV
Pathway impact value
- RA
Relative abundance
- SparCC
Sparse inverse covariance estimation for ecological association and statistical inference
- UCG
Uncultured genus-level group
- UPLC-MS/MS
Ultra-high performance liquid chromatography coupled with tandem mass spectrometry
- VDAART
Vitamin D Antenatal Asthma Reduction Trial
Footnotes
Conflicts of Interest Statement:
AAL has received author royalties from UpToDate, Inc. STW has received royalties from UpToDate, Inc. LBB participates on the Data Safety Monitoring Board of DBV Technologies. AB holds stock from DBV Technologies, is a consultant for AstraZeneca and Raffa, and received speaking honoraria from AstraZenica, Novartis and Sanofi. RSZ is a consultant for AstraZeneca, DBV Technologies, Genentech, Inc., GlaxoSmithKline, Merck & Co., Novartis, Quest Diagnostics, Regeneron/Sanofi, TEVA Pharmaceuticals, and has received research support from ALK Pharmaceuticals, AstraZeneca, Genentech, Inc., GlaxoSmithKline, NHLBI, MedImmune, Merck, and Teva Pharmaceuticals. JL-S is a consultant to Metabolon Inc. GTO is a co-investigator on a grant from Janssen Pharmaceuticals to Boston University that funds a study of the pathogenesis of chronic obstructive pulmonary disease. SD, KL-S, BM, RSK, MS, NL, DRG, and YYL have nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zahran HS, Bailey CM, Damon SA, Garbe PL, Breysse PN. Vital signs: Asthma in children — United States, 2001-2016. Morb Mortal Wkly Rep. 2018;67:149–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang YJ, Boushey HA. The microbiome and asthma. Ann Am Thorac Soc. 2014;11(Suppl 1):S48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011;128(5):948–955.E3. [DOI] [PubMed] [Google Scholar]
- 4.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7:307ra152–307ra152. [DOI] [PubMed] [Google Scholar]
- 5.Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan D, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016;22:1187–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arrieta MC, Arévalo A, Stiemsma L, Dimitriu P, Chico ME, Loor S, et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J Allergy Clin Immunol. 2017;142(2):424–434.E10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek M, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127(2):372–381.e1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q, Cox M, Liang Z, Brinkmann F, Cardenas PA, Duff R, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS One. 2016;11(4):e0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilty M, Burke C, Pedro H, Cardenas P, Bush A, Bossley C, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5(1):e8578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131(2):346–352.E3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou Y, Jackson D, Bacharier LB, Mauger D, Boushey H, Castro M, et al. The upper-airway microbiota and loss of asthma control among asthmatic children. Nat Commun. 2019;10:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barcik W, Pugin B, Westermann P, Perez NR, Ferstl R, Wawrzniak M, et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J Allergy Clin Immunol. 2016;138(5):1491–1494.E7. [DOI] [PubMed] [Google Scholar]
- 13.Chauhan BF, Ducharme FM. Anti-leukotriene agents compared to inhaled corticosteroids in the management of recurrent and/or chronic asthma in adults and children. Cochrane Database Syst Rev. 2012;2012(5):CD002314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin RJ, Szefler SJ, King TS, Kraft M, Boushey HA, Chinchilli VM, et al. Predicting Response to Inhaled Corticosteroid Efficacy (PRICE Trial). J Allergy Clin Immunol. 2007;119:73–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Panda L, Mabalirajan U. Recent Updates on Corticosteroid Resistance in Asthma. Eur Med J. 2018;3(3):49–57. [Google Scholar]
- 16.Fitzpatrick AM, Jackson DJ, Mauger DT, Boehmer SJ, Phipatanakul W, Sheehan WJ, et al. Individualized therapy for persistent asthma in young children. J Allergy Clin Immunol. 2016;138:1608–1618.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramadan AA, Gaffin JM, Israel E, Phipatanakul W. Asthma and Corticosteroid Responses in Childhood and Adult Asthma. Clin Chest Med. 2019;40:163–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goleva E, Jackson LP, Harris JK, Robertson CE, Sutherland ER, Hall CF, et al. The effects of airway microbiome on corticosteroid responsiveness in asthma. Am J Respir Crit Care Med. 2013;188(10):1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet. 2018;50:790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Litonjua AA, Carey VJ, Laranjo N, Harshfield BJ, McElrath TG, O’Connor GT, et al. Effect of prenatal supplementation with Vitamin D on asthma or recurrent wheezing in offspring by age 3 years: The VDAART randomized clinical trial. JAMA. 2016;315(4):362–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee-Sarwar KA, Kelly RS, Lasky-Su J, Zeiger RS, O’Connor GT, Sandel MT, et al. Integrative analysis of the intestinal metabolome of childhood asthma. J Allergy Clin Immunol. 2019;144:442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol. 2019;37(8):852–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41(Database issue):D590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhernakova A, Kurilshikov A, Bonder MJ, Tigchelaar EF, Schirmer M, Vatanen T, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falony G, Joossens M, Vieira-Silva S, Wang J, Darzi Y, Faust K, et al. Population-level analysis of gut microbiome variation. Science. 2016; 352(6285):560–564. [DOI] [PubMed] [Google Scholar]
- 28.Vandeputte D, Kathagen G, D’Hoe K, Vieira-Silva S, Valles-Colomer M, Sabino J, et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature. 2017; 551(7681):507–511. [DOI] [PubMed] [Google Scholar]
- 29.Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–976. [DOI] [PubMed] [Google Scholar]
- 30.Satinsky BM, Gifford SM, Crump BC, Moran MA. Use of internal standards for quantitative metatranscriptome and metagenome analysis. Methods Enzymol. 2013;531:237–250. [DOI] [PubMed] [Google Scholar]
- 31.Props R, Kerckhof FM, Rubbens P, Vrieze JD, Sanabria EH, Waegeman W, et al. Absolute quantification of microbial taxon abundances. ISME J. 2017;11(2):584–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stämmler F, Gläsner J, Hiergeist A, Holler E, Weber D, Oefner PJ, et al. Adjusting microbiome profiles for differences in microbial load by spike-in bacteria. Microbiome. 2016;4(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans AM, Bridgewater BR, Liu Q, Mitchell MW, Robinson RJ, Dai H, et al. High Resolution Mass Spectrometry Improves Data Quantity and Quality as Compared to Unit Mass Resolution Mass Spectrometry in High-Throughput Profiling Metabolomics. J Postgenomics Drug Biomark Dev. 2014;4(132):1000132. [Google Scholar]
- 34.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: Generative models for microbial metagenomics. PLoS One. 2012;7(2):e30126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morgan M DirichletMultinomial: Dirichlet-Multinomial Mixture Model Machine Learning for Microbiome Data [R package]. 2012. Available from: https://rdrr.io/bioc/DirichletMultinomial/
- 36.Struyf A, Hubert M, Rousseeuw P. Clustering in an object-oriented environment. J Stat Softw. 1997; 1:1–30. [Google Scholar]
- 37.Morton JT, Marotz C, Washburne A, Silverman J, Zaramela LS, Edlund A, et al. Establishing microbial composition measurement standards with reference frames. Nat Commun. 2019;10(1):2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedarko MW, Martino C, Morton JT, González A, Rahman G, Marotz CA, et al. Visualizing ’omic feature rankings and log-ratios using Qurro. NAR Genom Bioinform. 2020;2(2):lqaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman J, Aim EJ. Inferring Correlation Networks from Genomic Survey Data. PLoS Comput Biol. 2012;8(9):el002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, et al. Microbial co-occurrence relationships in the Human Microbiome. PLoS Comput Biol. 2012;8(7):e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z, Lai Z, Zhang X, Huang P, Xie J, Jiang Q, et al. Altered gut microbiome compositions are associated with the severity of asthma. J Thorac Dis. 2021;13(7):4322–4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z, Yuan KH. Practical Statistical Power Analysis Using Webpower and R. Granger, IN: ISDSA Press; 2018. [Google Scholar]
- 43.Holeček M Histidine in Health and Disease: Metabolism, Physiological Importance, and Use as a Supplement. Nutrients. 2020;12(3):848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox C, Kjarsgaard M, Surette MG, Cox PG, Nair P. A multidimensional approach to the management of severe asthma: Inflammometry, molecular microbiology and bronchial thermoplasty. Can Respir J. 2015;22:221–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Denner DR, Sangwan N, Becker JB, Hogarth DK, Oldham J, Castillo J, et al. Corticosteroid therapy and airflow obstruction influence the bronchial microbiome, which is distinct from that of bronchoalveolar lavage in asthmatic airways. J Allergy Clin Immunol. 2016;137(5):1398–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barcik W, Pugin B, Brescó MS, Westermann P, Rinaldi A, Groeger D, et al. Bacterial secretion of histamine within the gut influences immune responses within the lung. Allergy. 2019;74(5):899–909. [DOI] [PubMed] [Google Scholar]
- 47.Chiu C, Chou H, Chang L, Fan W, Dinh MCV, Kuo Y, et al. Integration of metagenomics-metabolomics reveals specific signatures and functions of airway microbiota in mite-sensitized childhood asthma. Allergy. 2020;75(11):2846–2857. [DOI] [PubMed] [Google Scholar]
- 48.Chen H, Nwe PK, Yang Y, Rosen CE, Bielecka AA, Kuchroo M, et al. A Forward Chemical Genetic Screen Reveals Gut Microbiota Metabolites That Modulate Host Physiology. Cell. 2019;177(5):1217–1231.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andrianasolo RM, Hercberg S, Touvier M, Druesne-Pecollo N, Adjibade M, Kesse-Guyot E, et al. Association between processed meat intake and asthma symptoms in the French NutriNet-Sante cohort. Eur J Nutr. 2020;59(4):1553–1562. [DOI] [PubMed] [Google Scholar]
- 50.Li Z, Rava M, Bédard A, Dumas O, Garcia-Aymerich J, Leynaert B, et al. Cured meat intake is associated with worsening asthma symptoms. Thorax. 2017;72(3):206–212. [DOI] [PubMed] [Google Scholar]
- 51.Cepeda AM, Thawer S, Boyle RJ, Villalba S, Jailer R, Tapias E, et al. Diet and Respiratory Health in Children from 11 Latin American Countries: Evidence from ISAAC Phase III. Lung. 2017;195(6):683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith LJ, Holbrook JT, Wise R, Blumenthal M, Dozor AJ, Mastronarde J, et al. Dietary intake of soy genistein is associated with lung function in patients with asthma. J Asthma. 2004;41(8):833–843. [DOI] [PubMed] [Google Scholar]
- 53.Bime C, Wei CY, Holbrook J, Smith LJ, Wise RA. Association of dietary soy genistein intake with lung function and asthma control: a post-hoc analysis of patients enrolled in a prospective multicentre clinical trial. Prim Care Respir J. 2012;21:398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clavel T, Mapesa JO. Phenolics in human nutrition: Importance of the intestinal microbiome for isoflavone and lignan bioavailability. In: Ramawat KG, Merillon JM, editors. Natural Products. Phytochemistry, Botany and Metabolism of Alkaloids, Phenolics and Terpenes. Berlin/Heidelberg, Germany: Springer; 2013. p. 2433–2463. [Google Scholar]
- 55.MacEyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature. 2014;510:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Worgall TS. Sphingolipids, ORMDL3 and asthma: What is the evidence? Curr Opin Clin Nutr Metab Care. 2017;20:99–103. [DOI] [PubMed] [Google Scholar]
- 57.Ono JG, Worgall TS, Worgall S. Airway reactivity and sphingolipids—implications for childhood asthma. Mol Cell Pediatr. 2015;2(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MM, Oskeritzian CA, Falanga YT, Harikumar KB, Allegood JC, Alvarez SE, et al. A specific sphingosine kinase 1 inhibitor attenuates airway hyperresponsiveness and inflammation in a mast cell-dependent murine model of allergic asthma. J Allergy Clin Immunol. 2013;131(2):501–511.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trifilieff A, Fozard JR. Sphingosine-1-phosphate-induced airway hyper-reactivity in rodents is mediated by the sphingosine-1-phosphate type 3 receptor. J Pharmacol Exp Ther. 2012;342:399–406. [DOI] [PubMed] [Google Scholar]
- 60.Nishiuma T, Nishimura Y, Okada T, Kuramoto E, Kotani Y, Jahangeer S, et al. Inhalation of sphingosine kinase inhibitor attenuates airway inflammation in asthmatic mouse model. Am J Physiol Lung Cell Mol Physiol. 2008;294(6):L1085–93. [DOI] [PubMed] [Google Scholar]
- 61.Worgall TS, Veerappan A, Sung B, Kim BI, Weiner E, Bholah R, et al. Erratum: Impaired sphingolipid synthesis in the respiratory tract induces airway hyperreactivity (Science Translational Medicine). Sci Transl Med. 2013;5:1–9. [DOI] [PubMed] [Google Scholar]
- 62.Kim SH, Jung HW, Kim M, Moon J, Ban G, Kim SJ, et al. Ceramide/sphingosine-1-phosphate imbalance is associated with distinct inflammatory phenotypes of uncontrolled asthma. Allergy Eur J Allergy Clin Immunol. 2020;75(8):1991–2004. [DOI] [PubMed] [Google Scholar]
- 63.Yamamoto EA, Jørgensen TN. Relationships Between Vitamin D, Gut Microbiome, and Systemic Autoimmunity. Front Immunol. 2019;10:3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.