Abstract
We investigated the characteristics and functionalities of extracellular vesicles (EVs) from Lactiplantibacillus plantarum (previously Lactobacillus plantarum) towards host immune cells. L. plantarum produces EVs that have a cytoplasmic membrane and contain cytoplasmic metabolites, membrane and cytoplasmic proteins, and small RNAs, but not bacterial cell wall components, namely, lipoteichoic acid and peptidoglycan. In the presence of L. plantarum EVs, Raw264 cells inducibly produced the pro-inflammatory cytokines IL-1β and IL-6, the anti-inflammatory cytokine IL-10, and IF-γ and IL-12, which are involved in the differentiation of naive T-helper cells into T-helper type 1 cells. IgA was produced by PP cells following the addition of EVs. Therefore, L. plantarum EVs activated innate and acquired immune responses. L. plantarum EVs are recognized by Toll-like receptor 2 (TLR2), which activates NF-κB, but not by other TLRs or NOD-like receptors. N-acylated peptides from lipoprotein19180 (Lp19180) in L. plantarum EVs were identified as novel TLR2 ligands. Therefore, L. plantarum induces an immunostimulation though the TLR2 recognition of the N-acylated amino acid moiety of Lp19180 in EVs. Additionally, we detected a large amount of EVs in the rat gastrointestinal tract for the first time, suggesting that EVs released by probiotics function as a modulator of intestinal immunity.
Subject terms: Bacteria, Bacterial host response
Introduction
The animal gastrointestinal tract is inhabited by a complex community of bacteria called the gut microbiota. These bacteria stimulate the intestinal immune system, which contributes to the maintenance of host gastrointestinal homeostasis1,2. Some symbiotic gut bacteria are transported into the intestinal lamina propria by the microfold (M) cells of Peyer’s patches (PP). Transported bacteria subsequently induce immune responses in PP cells containing macrophages, B cells, T cells, and other immune cells. Innate immune cells including macrophages recognize various bacterial components by pattern recognition receptors (PRRs), such as Toll-like receptors (TLRs) and NOD-like receptors (NLRs), to produce a number of cytokines via the activation of the transcription factor NF-κB3. Cytokines and other activators then stimulate acquired immune cells, including B cells, to produce immunoglobulin (Ig) A4,5. A detailed understanding of the products of bacterial cells that affect host immune cells is of importance.
The symbiotic intestinal bacteria that are beneficial to living organisms are called probiotics. Lactobacilli have been attracting increasing attention as typical probiotic Gram-positive bacteria. Several lactobacilli produce extracellular vesicles (EVs) with a spherical structure that range in size between 20 and 200 nm in culture medium6,7. EVs are presumed to carry metabolic intermediates, proteins, and RNAs as cargo and stimulate host immune cells. The EVs of Latilactobacillus sakei (previously Lactobacillus sakei) have been shown to induce the production of IgA from PP cells8,9. The pro-inflammatory cytokine IL-6, derived from dendritic cells, enhances IgA production by L. sakei EVs. The EVs of Lacticaseibacillus rhamnosus (previously Lactobacillus rhamnosus) GG and Lactobacillus reuteri DSM 17938 were found to weaken the pro-inflammatory cytokine responses of T cells and NK cells10. Although the function of EVs in the host immune system is important, the molecules in EVs that are responsible for immunostimulation remain unknown. To date, the biochemical components of EVs have been largely unidentified. Therefore, further studies are warranted to identify the biochemical components of EVs as well as the molecules responsible for triggering immunostimulatory effects.
We focused on lipoprotein (Lp) as a functional molecule of EVs. The important function of Lp in Gram-positive bacteria has only emerged in recent years. Lp is anchored to the bacterial cytoplasmic membrane via a di- or tri-acylglycerol moiety linked to the N-terminal cysteine of Lp11. The polypeptide portion of Lp protrudes outside the Gram-positive bacterial cell surface and is responsible for characteristic functions, such as the uptake of nutrients. Some pro-inflammatory cytokines are inducibly produced by host immune cells following the addition of an acylated N-terminal peptide of Lp from Staphylococcus aureus12. However, the effects of Lp from lactobacilli on host immune cells remain unclear. Lp in EVs released from lactobacilli have not yet been investigated.
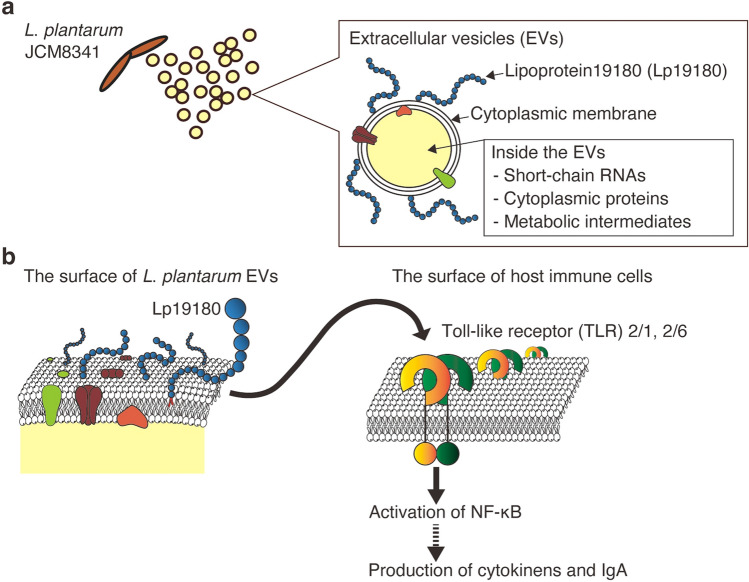
We newly identified Lactiplantibacillus plantarum (previously Lactobacillus plantarum) JCM8341 as a producer of EVs. Using EVs produced by L. plantarum, the actual number and size of EVs with a bacterial cytoplasmic membrane were quantified with a fluorescent dye that specifically stains the cytoplasmic membrane. To identify the biochemical components of L. plantarum EVs, metabolomic, proteomic, and RNA-seq analyses were performed. To date, the stimulation of innate immunity by whole cells of L. plantarum JCM8341 has been reported13. In the present study, we showed that EVs from L. plantarum JCM8341 activated NF-κB via TLR2 recognition and induced the production of pro- and anti-inflammatory cytokines from Raw264 cells and IgA from PP cells. Therefore, L. plantarum EVs are involved in the activation of innate and acquired immune responses. We also detected a novel Lp (named Lp19180) in L. plantarum EVs. The acylated N-terminal peptides derived from Lp19180 were recognized by TLR2 and activated NF-κB. In summary, we herein report the physicochemical, biochemical, and functional characteristics of L. plantarum EVs (Fig. 1). Furthermore, to elucidate the effects of EVs against gut immunity in vivo, we measured the numbers and sizes of EVs in the rat gastrointestinal tract. The present results will provide insights into the effects of lactobacillus EVs on host immunity at the molecular level.
Figure 1.
Properties of EVs produced by L. plantarum and their effects on host immune cells. (a) Biochemical components of L. plantarum EVs. (b) Schematic sketch of NF-κB activation by Lp19180 via TLR2 and the subsequent stimulation of innate and adaptive immunities.
Results
Physicochemical characterization of L. plantarum EVs
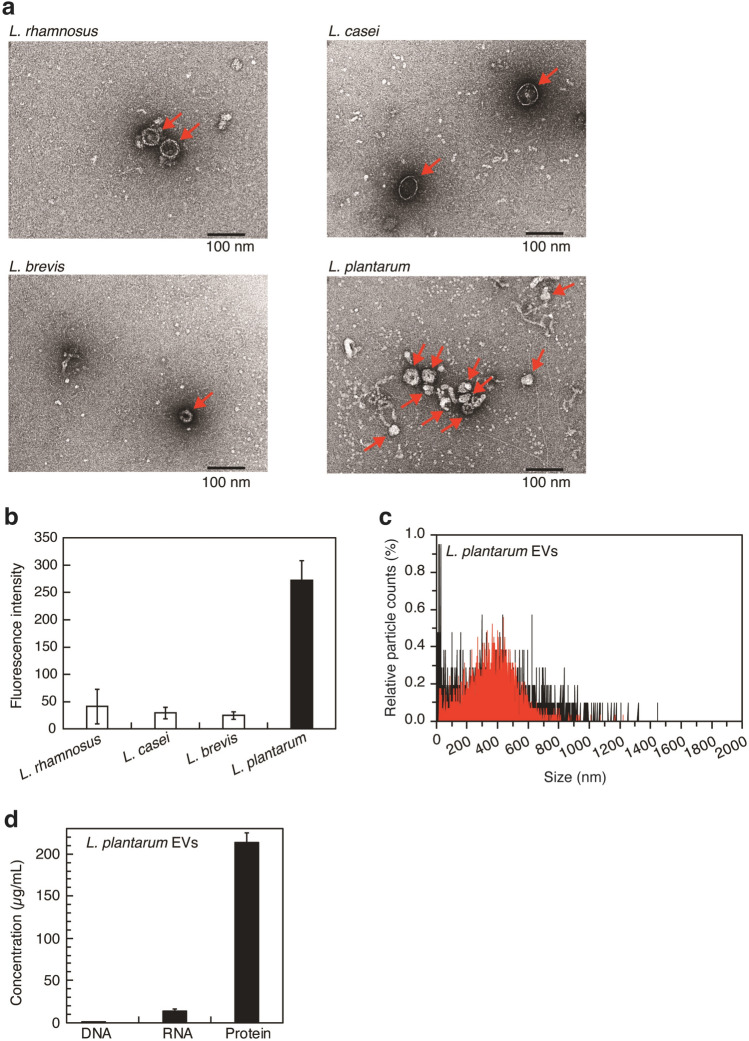
We attempted to detect the spherical structures (20–200 nm) of EVs in the culture supernatant of lactobacilli (Lacticaseibacillus rhamnosus JCM2772, Lacticaseibacillus casei JCM 1134, Levilactobacillus brevis JCM1059T, and L. plantarum JCM8341). Using transmission electron microscopy (TEM), we investigated whether these spherical structures were present in the ultracentrifugation residue obtained from the bacterial culture supernatant by differential centrifugation. As shown in Fig. 2a, we confirmed the presence of structures with a diameter of approximately 50 nm (red arrow) in the ultracentrifugation residues of four lactobacilli strains. As shown in Fig. 2b, EVs from four lactobacilli strains were quantified using the lipophilic fluorescent dye, (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide (FM4-64). FM4-64 is virtually non-fluorescent in aqueous solution, but becomes intensely fluorescent when the dye is incorporated into cytoplasmic membranes14. Of the four lactobacilli, L. plantarum produced large amounts of EVs. Fluorescence nanoparticle tracking analysis with FM4-64 was then performed to evaluate the actual numbers and sizes of particles with a cytoplasmic membrane in the culture supernatant of L. plantarum (Fig. 2c). We detected particles ranging in size between 10 and 1500 nm (Fig. 2c, black lines). Membrane particles (50–800 nm) labeled with FM4-64 were observed (Fig. 2c, red lines) and many membrane particles of approximately 400 nm were present. The results of TEM observations shown in Fig. 2a revealed that L. plantarum produced vesicle-like structures (approximately 50 nm). The zeta potential of L. plantarum EVs was −73.32 ± 22.33 mV, indicating that EVs are weakly charged and potentially form multimer structures in aqueous solution. These results suggest that L. plantarum EVs exist as monomer (50 nm) and multimer (~ 800 nm) structures in aqueous solution. In summary, L. plantarum produced at least 4.5 × 1010 particles/ml of EVs consisting of a cytoplasmic membrane with a size range of 50–800 nm in culture medium. Lactobacillus acidophilus ATCC 53544, L. casei ATCC 393, and L. reuteri ATCC 23272 were previously reported to produce between 3 × 109 and 1 × 1010 particles/ml of EVs in each culture medium6. The concentration of L. plantarum EVs detected in culture medium was consistent with those of EVs from these lactobacilli in culture media. The results of TEM observations also showed that the sizes of EVs produced by L. plantarum WCFS1, KCTC 11401BP, and BGAN8 were 30–200, 20–100, and 20–140 nm, respectively15–17. As shown in Fig. 2a, L. plantarum EVs had similar structures. Additionally, protein and RNA were present in L. plantarum EVs, whereas DNA was rarely detected (Fig. 2d). This result is similar to that reported for L. reuteri18. To summarize, some lactobacilli release EVs into culture supernatants. L. plantarum EVs appear to have a cytoplasmic membrane and mainly contain bacterial proteins.
Figure 2.
Characterization of EVs produced by L. plantarum. (a) TEM images of lactobacillus EVs. Red arrows indicate EVs. Scale bar, 100 nm. (b) EVs production of lactobacilli. Means ± SD, n = 3. (c) Size distributions of EVs (black: all particles in the L. plantarum EVs fraction, red: EVs labeled with FM4-64). The total numbers of particles in L. plantarum EVs and those of particles labeled by FM4-64 in L. plantarum EVs are represented as 100%. (d) DNA, RNA, and protein in L. plantarum EVs. Means ± SD, n = 3.
Comprehensive analysis of constituents of L. plantarum EVs
To the best of our knowledge, few studies to date have conducted a biochemical analysis of the constituents of EVs. Therefore, we attempted to comprehensively identify the constituent molecules of L. plantarum EVs using a metabolomic analysis with LC–MS. The results obtained revealed the presence of 1519 compounds in L. plantarum EVs (Supplementary Table S1). Some of the components in L. plantarum EVs are shown in Table 1. Eicosatetraenoyl-glycerophosphate and various fatty acids, which constitute the bacterial cytoplasmic membrane, were detected in EVs. The following fatty acids have been detected in whole cells of L. plantarum; C12:0, C12:1, C14:0, C14:1, C16:0, C16:1, C18:0, C18:1, and C20:119,20. As shown in Table 1, oxododecanoic acid (C12:0), oxotetradecanoic acid (C14:0), palmitoleic acid (C16:1), and oleic acid (C18:1) were identified in L. plantarum EVs. These results support the assumption that EVs are composed of the cytoplasmic membrane of L. plantarum, as shown in Fig. 2c. Additionally, we detected NAD+, amino acids, and nucleic acids in L. plantarum EVs. The EVs of Gram-positive bacteria have been suggested to bud from the cytoplasmic membrane21, and cytoplasmic metabolites, namely, NAD+, amino acids, and nucleic acid, may be contained as cargo inside L. plantarum EVs. The presence of lipoteichoic acid (LTA) and peptidoglycan (PGN) in the bacterial cell wall has been reported in the EVs of L. rhamnosus JB-1 and S. aureus strains 6571, respectively22,23. On the other hand, in L. plantarum EVs, we did not detect teichoic acid, glycerol phosphate, or ribitol phosphate derived from LTA, or γ-D-glutamyl-meso-diaminopimelic acid (iE-DAP), muramyldipeptide (MDP), N-acetylglucosamine, or N-acetylmuramic acid derived from PGN. In summary, L. plantarum EVs are vesicles that have a bacterial cytoplasmic membrane and contain a number of cytoplasmic metabolites. Bacterial cell wall components, namely, LTA and PGM, are not detected in L. plantarum EVs.
Table 1.
Constituents in L. plantarum EVs.
| Peak no. | R.T. (min) | Ionization | m/z (detected) | Exact mass | ||
|---|---|---|---|---|---|---|
| Cytoplasmic membrane | ||||||
| Phospholipids | Eicosatetraenoyl-glycerophosphate | 569 | 15.10 | [M + H]+ | 459.2506 | 458.2433 |
| Fatty acids | Propionyl-CoA | 851 | 16.37 | [M + H]+ | 824.1474 | 823.1414 |
| Oxododecanoic acid | 1185 | 23.25 | [M + H]+ | 215.1643 | 214.1569 | |
| Oxotetradecanoic acid | 1291 | 25.24 | [M + H]+ | 243.1955 | 242.1882 | |
| Palmitoleic acid | 1362 | 26.47 | [M + H]+ | 255.2319 | 254.2246 | |
| Hydroxyarachidonic acid | 1420 | 27.21 | [M + H]+ | 321.2424 | 320.2351 | |
| Oleic acid | 1485 | 29.01 | [M + H]+ | 283.2632 | 282.2559 | |
| Cytoplasm | ||||||
| Coenzyme | NAD+ | 61 | 3.47 | [M + 2H]2+ | 332.5618 | 663.1091 |
| Amino acids | Tryptophan | 223 | 12.06 | [M + H]+ | 205.0973 | 204.0899 |
| Tyrosine | 1000 | 19.82 | [M + H]+ | 182.0814 | 181.0739 | |
| Leucine/isoleucine | 1314 | 25.82 | [M + H]+ | 132.1021 | 131.0946 | |
| Nucleic acids | Guanine | 44 | 3.45 | [M + H]+ | 152.0569 | 151.0494 |
| Adenine | 45 | 3.45 | [M + H]+ | 136.0619 | 135.0545 | |
| Ribothymidine | 91 | 5.28 | [M + H]+ | 259.0925 | 258.0852 | |
| Adenosine | 98 | 6.59 | [M + H]+ | 268.1042 | 267.0968 | |
R.T. retention time.
Proteomic analysis of L. plantarum EVs
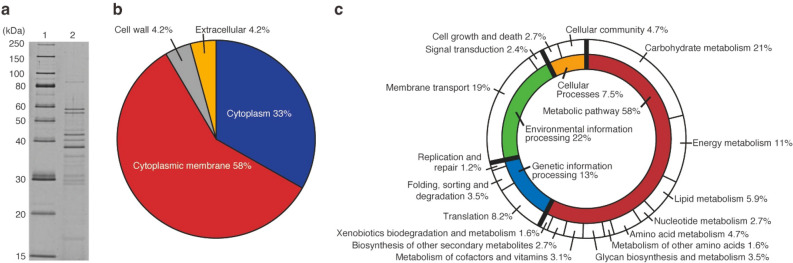
We attempted to identify the proteins detected in L. plantarum EVs (Fig. 2d). L. plantarum EVs collected from the supernatants of three independent cultures were analyzed by SDS-PAGE. The protein profiles of L. plantarum EVs were highly reproducible among the three replicates. A representative SDS-PAGE profile is shown in Fig. 3a. As a result of sequencing of the genomic DNA of L. plantarum (Genbank Accession No. BPFY01000000), we identified 3031 types of protein-coding genes in genomic DNA. Using the amino acid sequences of proteins in genomic DNA as reference data, 411 types of proteins in L. plantarum EVs were comprehensively identified by LC–MS/MS. The proteins identified in L. plantarum EVs and their predicted subcellular localization are shown in Supplementary Table S2. Predictions of the subcellular localization of EV proteins were performed using PSORTb 3.0.2 (https://www.psort.org/psortb/). Following the exclusion of 58 types of hypothetical proteins from the 411 types in L. plantarum EVs, we predicted the subcellular localization of 283 out of 353 types of proteins in EVs (Fig. 3b). Many proteins in L. plantarum EVs were predicted to localize in the bacterial cytoplasmic membrane and cytoplasm of L. plantarum cells, and a small number to the cell wall or extracellular environment.
Figure 3.
Proteins in L. plantarum EVs. (a) SDS-PAGE profile of L. plantarum EVs. Lane 1: Marker, Lane 2: L. plantarum EVs (0.3 µg). Proteins were separated by SDS-PAGE using 12.5% gel (Gellex International, Tokyo, Japan) with constant voltage at 300 V and then detected using SYPRO Ruby staining (Lonza, Rockland, ME). The cropped gel was displayed, and original gel was indicated in Supplementary Fig. 3a_original_gel file. (b) Subcellular localization of proteins in L. plantarum EVs. (c) Biological processes involving proteins in L. plantarum EVs.
As shown in Fig. 3c, the biological functions of proteins in L. plantarum EVs were cataloged using to KEGG Mapper (https://www.genome.jp/kegg/mapper.html). Among the 353 types of EV proteins described above, 255 were evaluated for biological functions (Supplementary Table S3), with 54 being involved in carbohydrate metabolism (21%), followed by 49 in membrane transport (19%) (Fig. 3c). As shown in Fig. 2c and Table 1, L. plantarum EVs were composed of a cytoplasmic membrane. Additionally, cytoplasmic metabolites and proteins appeared to be present inside the spherical structures of EVs. Therefore, we considered a large number of membrane and cytoplasmic proteins to be present in L. plantarum EVs. Similar to L. plantarum EVs, membrane and cytoplasmic proteins involved in diverse metabolic pathways have been detected in L. plantarum BGAN8 EVs and L. reuteri BBC3 EVs15,18.
We identified lysozyme (LOCUS_18750) and the cell wall amidase lytH (LOCUS_29150) in L. plantarum EVs, as shown in Supplementary Table S2. Since these enzymes are responsible for bacterial cell wall degradation24, the LTA and PGN components of the bacterial cell wall may not have been detectable in L. plantarum EVs. In addition, EVs are released from Gram-positive bacterial cells when the cytoplasmic membrane protrudes outside due to turgor pressure when the bacterial cell wall is locally thinned by these enzymes21. In the case of L. plantarum, EVs may be released from bacterial cells by the same mechanism.
Analysis of small RNAs in L. plantarum EVs
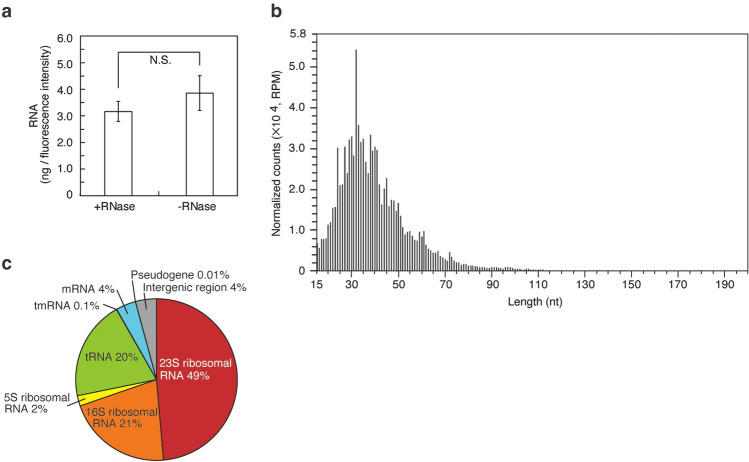
We detected RNAs (Fig. 2d) and nucleic acids (Table 1) in L. plantarum EVs and attempted to analyze the former. RNAs in L. plantarum EVs from the supernatants of three independent cultures were quantified, and representative EVs containing RNA were used in analyses. We initially investigated whether RNA was degraded by an RNase treatment. As shown in Fig. 4a, the amount of RNA in EVs treated with RNase was similar to that in EVs not treated with RNase (p = 0.18). This result suggested that RNA was encapsulated in L. plantarum EVs, which protected it from degradation by RNase, similar to L. reuteri BBC3 EVs18. Small RNA-seq was then performed to comprehensively analyze the nucleotide sequences and strand lengths of RNA in L. plantarum EVs (Fig. 4b). We elucidated the nucleotide sequences and strand lengths of 406,312 reads of RNAs. Small RNA-seq data are shown in Supplementary Table S4. As shown in Fig. 4b, the length of RNA encapsulated in EVs ranged between 15 and 196 nt. Mean and mode chain lengths were 42 and 32 nt, respectively. RNAs in L. plantarum EVs were mapped using the L. plantarum genomic DNA sequence (Genbank Accession No. BPFY01000000). The results obtained revealed that 72% of small RNAs in L. plantarum EVs were derived from 5S, 16S, and 23S rRNA of L. plantarum (Fig. 4c). We detected 20 types of large and small subunits of ribosomal proteins in L. plantarum EVs (Supplementary Table S3). The mechanisms underlying RNA intake in bacterial EVs have not yet been elucidated in detail; however, proteins appear to be synthesized close to the membrane site, resulting in the capture of rRNA, tRNA, and mRNA fragments along with ribosomal proteins in bacterial EVs21. Therefore, L. plantarum EVs are assumed to contain small RNAs and ribosomal proteins.
Figure 4.
Small RNAs in L. plantarum EVs. (a) The amount of RNA per fluorescence intensity of EVs. The amount of RNA after the RNase treatment was compared with that without the treatment. Means ± SD, n = 3. Two-tailed unpaired Student’s t-test, N.S. not significant. (b) Strand length distribution of RNAs. (c) Classification of small RNAs in L. plantarum EVs.
Effects of L. plantarum EVs against immune cells
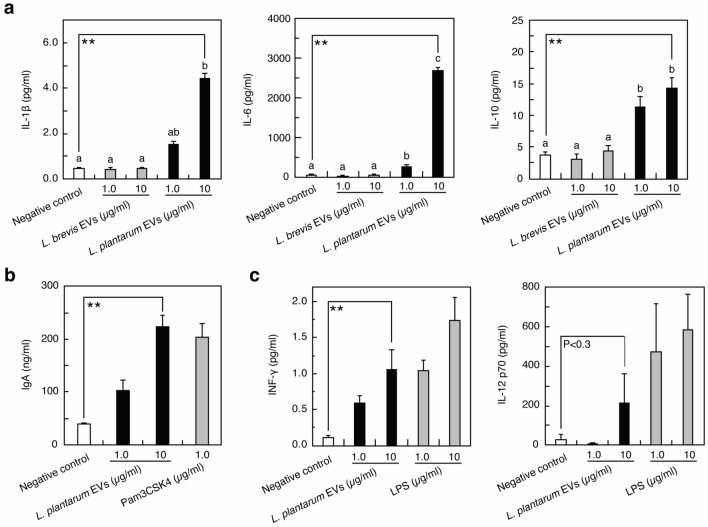
The effects of lactobacilli via EVs on host immune cells remain unclear. We investigated the response of immunocompetent cells elicited by L. plantarum EVs. Representative L. plantarum EVs were prepared from the supernatants of three independent cultures. As shown in Fig. 5a, the production of the pro-inflammatory cytokines IL-1β and IL-6 and the anti-inflammatory cytokine IL-10 from Raw264 cells was induced by L. plantarum EVs. When innate immunity is stimulated, immune cells produce cytokines. The results shown in Fig. 5a confirmed that innate immunity was stimulated by L. plantarum EVs. The amount of each cytokine induced by L. plantarum EVs was higher than that induced by L. brevis EVs. As shown in Fig. 5b, PP cells containing macrophages and B cells produced IgA in the presence of L. plantarum EVs, indicating that acquired immunity was stimulated by the addition of EVs. L. sakei EVs stimulated dendritic cells to produce IL-6, nitric oxide, and retinoic acid, which play important roles in enhancing IgA production by PP cells8,9. Since L. plantarum EVs induced the production of IL-6 from Raw264 cells, IL-6 appeared to be involved in the production of IgA from PP cells, similar to L. sakei EVs. To summarize, L. plantarum EVs induced the production of IFN-γ and IL-12 (Fig. 5c), which are involved in the differentiation of naive T-helper cells into T-helper type 1 cells25, suggesting that L. plantarum EVs activate both innate and acquired immune responses.
Figure 5.
Stimulation of immune cells by L. plantarum EVs. The production of cytokines from Raw264 cells (a,c) and IgA from PP cells (b) was indicated. Pam3CSK4 (b) and LPS (c) were used as positive controls. PBS (a–c) was used as the negative control. Means ± SD, n = 3. (a) Bars identified by the same letters are not significantly different from each other (P > 0.01) by the Tukey’s test. (a–c) the Dunnett’s test, **P < 0.01 compared with negative control.
Identification of cellular receptors that recognize L. plantarum EVs and their ligands
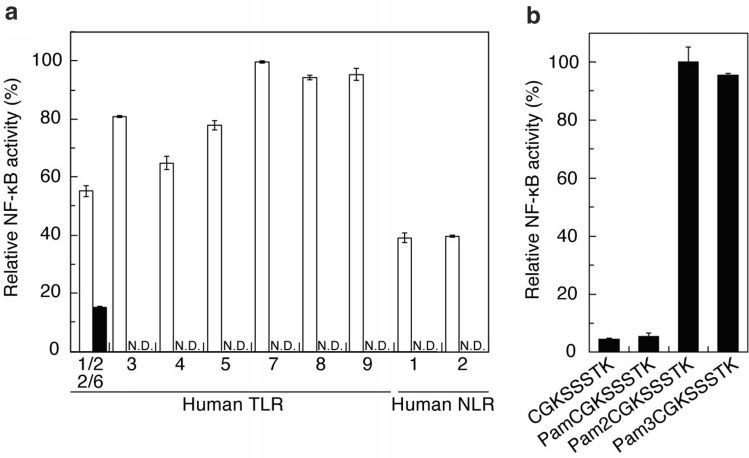
L. plantarum produces EVs that stimulate host immune cells. The host cell receptors that recognize the EVs of lactobacilli and their ligands have remained unknown. Bacterial components are recognized by the PRRs of host cells. Therefore, we focused on PRRs to identify the host receptor for L. plantarum EVs. In the signaling pathway of immune cells, the recognition of ligands by PRRs has been shown to activate the transcription factor NF-κB26. Activated NF-κB then induces the expression of various cytokines downstream of the signaling pathway. Therefore, we examined whether L. plantarum EVs activate NF-κB via the recognition of PRRs, namely, human TLRs and NLRs. To assess the activation of NF-κB via TLRs and NLRs, human embryonic kidney (HEK293) cells expressing NF-κB-inducible secreted extracellular alkaline phosphatase (SEAP) and each of the human TLRs (TLR2, 3, 4, 5, 7, 8, or 9) and human NLRs (NLR 1 or 2) were used. As shown in Fig. 6a, L. plantarum EVs were recognized by TLR1/2 or TLR2/6 on the HEK293 cell surface (Fig. 6a, closed bar) and not by TLR4 or 5 on the HEK293 cell surface or by TLR3, 7, 8, or 9 or NLR1 and 2 inside HEK293 cells. TLR1, 2, and 6 have been shown to form heterodimers (TLR1/2 and TLR2/6) on the HEK293 cell surface26. Furthermore, previous studies demonstrated that L. sakei EVs and L. rhamnosus JB-1 EVs stimulated macrophages and dendritic cells, respectively, via TLR2 recognition9,23. Therefore, TLR1/2 and/or TLR2/6 appear to play an important role in the activation of host immune cells by lactobacilli EVs.
Figure 6.
Human cell receptors involved in the recognition of L. plantarum EVs and lipopeptides from Lp19180. The recognition abilities of (a) L. plantarum EVs (closed bar) and each positive control (open bar, described in the Materials and Methods) and (b) acylated N-terminal peptides derived from Lp19180 were evaluated based on NF-κB-inducible SEAP activity in duplicate wells. Means ± SD. N.D. not detected.
S. aureus Newman produces EVs containing small RNAs and these EVs are delivered to macrophages and trigger IFN-β responses via TLRs 3, 7, and 8, which recognize exogenous RNA27. As shown in Fig. 4, L. plantarum EVs contained small RNAs derived from rRNA, mRNA, tRNA, and other non-coding RNA similarly as found in S. aureus Newman EVs. However, we did not detect the activation of NF-κB via TLR3, 7, and 8 by L. plantarum EVs (Fig. 6a). Since the ligands recognized by host cell receptors differed among each EV, the ligands in each EV and the receptors in host cells need to be identified for the elucidation of host responses.
The EVs of S. aureus strain 6571 and L. rhamnosus JB-1 contain PGN and LTA, respectively, which are derived from the respective bacterial cell walls22,23. PGA and LTA in these EVs are presumably involved in the production of pro-inflammatory cytokines via TLR2. On the other hand, cell wall-derived LTA and PGN components were not detected in L. plantarum EVs (Table 1 and Supplementary Table S1). NF-κB was not activated via NLR1 or 2, which recognize iE-DAP and MDP, respectively, derived from PGN. In addition to LTA and PGN, LPs are ligands for TLR222,28. We identified Lp19180 (GenBank accession No. LC633877) as a novel Lp in L. plantarum EVs (LOCUS_19180, Supplementary Table S3). In the proteomic analysis described above, Lp19180 of L. plantarum was assigned as a D-methionine transport system substrate-binding protein (MetQ) involved in the uptake of D-methionine29. In L. plantarum EVs, Lp19180 has been suggested to function as the TLR2 ligand. The nucleotide and estimated amino acid sequence of the Lp19180 gene are shown in Fig. S1 of Supplementary Material. Bacterial Lp generally localizes to the cytoplasmic membrane by removing the signal peptide and acylating the N-terminal cysteine with fatty acids11. Di- and tri-acylated Lps are located on the bacterial cell surface. In the case of Lp19180, the peptides detected in L. plantarum EVs using LC–MS/MS are shown in Fig. S1 (underlined) of Supplementary Material. We presumed that Cys22 was acylated and the polypeptide from Cys22 to Asp274 was presented outside the membrane of EVs towards the extracellular environment. As shown in Fig. 6b, using acylated N-terminal peptides derived from Lp19180, the TLR2-mediated activation of NF-κB was evaluated with HEK293 cells expressing NF-κB-inducible SEAP and TLR2. The results obtained showed that NF-κB was activated via TLR2 recognition by the diacylated N-terminal peptide (Pam2CGKSSSTK) and triacylated N-terminal peptide (Pam3CGKSSSTK). NF-κB was hardly activated by the non-acylated N-terminal peptide (CGKSSSTK) or monoacylated N-terminal peptide (PamCGKSSSTK). These results revealed that the di- and tri-acylated N-terminal peptide moieties of Lp19180 functioned as ligands for TLR2. Although the activation of NF-κB via TLR2 has been reported using the culture supernatants of Lactobacillus paragasseri K7, Limosilactobacillus fermentum L930BB, Bifidobacterium animalis IM386, and L. plantarum WCFS1, the ligands for NF-κB activation remain unclear30. In the present study, we detected EVs as novel functional particles in the culture supernatant of L. plantarum, demonstrated that L. plantarum EVs activated NF-κB via TLR2, and identified lipopeptides derived from Lp19180 as novel TLR2 ligands in L. plantarum EVs.
Detection of EVs in the rat gastrointestinal tract
We demonstrated that immune cells were stimulated by the lipopeptide moiety of Lp19180 in L. plantarum EVs. Although previous studies reported that EVs from lactobacilli were taken up by M cells into the lamina propria8,31, the particle numbers and sizes of EVs in the gastrointestinal tract have not yet been directly measured. Therefore, we herein attempted to evaluate the particle numbers and sizes of EVs prepared from the gastrointestinal tract of Sprague–Dawley (SD) rats. As shown in Fig. S2 of Supplementary Material, the intestinal content (1 g) of the gastrointestinal tract contained 1.2 × 1013 particles of EVs with a mean particle size of 160 nm and a mode particle size of 132 nm. Feces from mice and humans contain EVs with mean particle sizes of 118 and 120 nm, respectively32. The particle sizes of EVs in the rat gastrointestinal tract are similar to those of fecal EVs from mice and humans. We demonstrated that extremely large amounts of EVs were present in the rat gastrointestinal tract. The intraperitoneal administration of mouse fecal EVs to mice has been shown to induce TLR2- and TLR4- mediated inflammation in local and systemic immunities32. Similarly, large amounts of EVs, including lactobacillus EVs, in the mouse gastrointestinal tract may be transferred to the lamina propria and affect the host immune system.
Discussion
To clarify the immune cell responses induced by enterobacterial EVs, host cell receptors to EVs, and ligands present in EVs, we herein examined the biophysical parameters of L. plantarum EVs and comprehensively identified the biochemical components of EVs. The production of TNF-α from peritoneal exudate cells was previously shown to be induced by whole cells of L. plantarum JCM834113. In the present study, we demonstrated that EVs produced by L. plantarum JCM8341 in culture medium induced the production of diverse pro- and anti-inflammatory cytokines. Host immune cell responses induced by L. plantarum EVs appeared to differ from those induced by whole L. plantarum cells. Therefore, it is necessary to characterize not only whole bacterial cells, but also EVs produced during the bacterial life cycle.
We herein reported the presence of a large amount of EVs in the rat intestinal tract for the first time. Although EVs in the rat intestinal tract appear to include EVs from enterobacteria, rat cells, and diets, the classification of EVs in the intestinal tract has not yet been performed. Further studies are needed to investigate the effects of bacterial EVs in the intestinal tract on the host organism. Some lactobacilli EVs may be taken up by M cells into the intestinal lamina propria8,31 and stimulate host immune cells via TLR29,23. In host immune cells, TLR2 signal transduction may induce the production of pro-inflammatory cytokines by activating NF-κB through MyD88- and MAL/TIRAP-dependent pathways and anti-inflammatory cytokines through the PI3K/AKT pathway26,33–36. TLR2 polymorphisms have been shown to exert protective effects against intestinal mucosal damage37,38, while others may be associated with an increased risk of colorectal cancer39,40. Additionally, polymorphisms in human TLR2 have been strongly implicated in the development of several diseases, including rheumatoid arthritis, type I diabetes, and asthma41,42. Therefore, the stimulation of TLR2 leads to diverse physiological and cell reactions. Since lactobacilli EVs are recognized by TLR2, further research is needed to identify the molecules responsible for the TLR2 stimulation in EVs derived from gut microbes.
In L. plantarum EVs, we identified Lp19180 as a novel molecule that is recognized by TLR2. In whole cells of L. plantarum, Lp19180 has been suggested to play a role in the cellular uptake of D-methionine29. However, in L. plantarum EVs, the lipopeptide moiety of Lp19180 activated NF-κB via TLR2 recognition in host immune cells. FomA porin in Fusobacterium nucleatum EVs and cell wall muramidase in L. casei EVs were previously shown to activate NF-κB43,44, suggesting that EVs from various commensal bacteria in the gut environment contain a number of NF-κB activators. The activation of NF-κB plays a critical role in the maintenance of intestinal immune homeostasis45–47. Therefore, further studies are needed to identify TLR2-mediated NF-κB activators in EVs from the gut microbe.
In the present study, we propose the novel immunostimulatory effects of L. plantarum. As shown in Fig. 1, the TLR2-mediated activation of NF-κB by LP19180 in L. plantarum EVs elicited immunostimulatory effects, including innate and acquired immune cell responses. In future research, the immune cell responses induced by LP19180 and di- and tri- acylated N-terminal peptides need to be examined in more detail. Conclusively, lactobacilli EVs containing functional molecules appear to contribute to the maintenance of host intestinal immune homeostasis in addition to whole cells of lactobacilli. The elucidation of host cell responses induced by enterobacterial EVs and the identification of TLR ligands in enterobacterial EVs will enhance our understanding of the effects of symbiotic bacteria on host cells in the gut environment. Furthermore, the characterization of EVs will contribute to the development of new functional molecules for the regulation of physiological reactions.
Materials and methods
Bacterial culture and EVs preparation
L. plantarum JCM8341 was cultured in de Man, Rogosa, and Sharpe medium (Difco Laboratories) at 30 °C for 4 days under static culture conditions. L. plantarum EVs were then isolated from the culture (1000 ml) according to previously reported guidelines48. After centrifugation at 6000×g at 4 °C for 30 min to remove bacterial cells, the culture supernatant was centrifuged at 10,000×g at 4 °C for 30 min. The culture supernatant was then filtrated through 0.2-µm polyethersulfone filters (Sartorius, Göttingen, Germany) to remove cellular debris. The EVs of the filtered supernatant were concentrated by an Amicon Ultra Centrifugal filter device (cut-off: 100 kDa). The concentrated cell-free supernatant containing EVs was ultracentrifuged at 100,000×g at 4 °C for 2 h with Himac CP-80β (HITACHI, Tokyo, Japan). EVs were recovered as precipitates by ultracentrifugation and resuspended in Tris-buffered saline (TBS, pH 7.4, Nacalai Tesque, Kyoto, Japan). The EVs of L. rhamnosus JCM2772, L casei JCM 1134, and L. brevis JCM1059T were prepared in the same manner. EVs were quantified by staining with the dye FM4–64 (5 µg/ml, Molecular Probes, Chicago, IL) at room temperature for 20 min. The fluorescence intensity of FM4-64 in the membranes of EVs was measured by an RF-6000 fluorescence spectrophotometer (Shimadzu, Kyoto, Japan) with an excitation wavelength of 515 nm and an emission wavelength of 635 nm. DNA, RNA, and protein concentrations in EVs were quantified using a Qubit 3.0 fluorometer (Life Technologies, Grand Island, NY).
Physicochemical analysis of L. plantarum EVs
EVs were labeled with FM4-64 and the enumeration and sizing of labeled and non-labeled EVs were performed with a ViewSizer 3000 (software version 1.9.0.4518, HORIBA, Kyoto, Japan) using a 520-nm laser and a 650-nm filter. The zeta potential of EVs was assessed by a zeta potential analyzer (ELSZ-2000, Otsuka Electronics, Osaka, Japan) at 25 °C. EVs suspended in TBS (pH 7.4, Nacalai Tesque) were deposited on a carbon film-coated copper grid treated by glow discharge and then stained with 2% uranyl acetate solution for negative staining. The prepared grid was observed with the transmission electron microscope JEM-1400 (JEOL Inc., Tokyo, Japan).
Metabolomic analysis of L. plantarum EVs
L. plantarum EVs (50 mg wet weight) were washed three times with 10 ml of TBS by ultracentrifugation and then disrupted with 75% MeOH and zirconia beads. The homogenate was centrifuged at 20,000×g for 10 min and the supernatant obtained was loaded on a MonoSpin C18 column (GL science, Tokyo, Japan) and eluted with 75% MeOH. The eluted fraction was analyzed using a HPLC Ultimate 3000 RSLC (Thermo Fisher Scientific, Waltham, MA, USA) coupled with a Q Exactive high-resolution mass spectrometer (Thermo Fisher Scientific). The scan range of mass spectrometry was set at m/z 80–1200. The detection and alignment of the peak were performed using ProteoWizard (https://proteowizard.sourceforge.io) and PowerGetBatch software49. The UC2 database (http://webs2.kazusa.or.jp/mfsearcher/uc2/) was used to identify metabolites.
Genome analysis of L. plantarum
The genome of L. plantarum JCM8341 was sequenced using the Illumina HiSeq sequencer. All assembly genome sequence data and 16S rRNA sequence data were deposited in the NCBI GenBank with accession No. BPFY01000000 and LC633333, respectively. Gene predictions and functional annotation were conducted by the DDBJ Fast Annotation Pipeline (D-fast, https://dfast.ddbj.nig.ac.jp).
Proteomic analysis of L. plantarum EVs
L. plantarum EVs (2.0 µg) were reduced with DTT and alkylated with iodoacetamide. Alkylated EVs were then digested with trypsin. The tryptic peptide mixture was desalted and applied to an Orbitrap Q Exactive Plus mass spectrometer through an EASY-nLC 1200 System (Thermo Fisher Scientific). To identify proteins, all MS/MS spectra obtained by the ESI mass analysis in the positive ion mode were analyzed using the MASCOT server (Matrix Science). Predicted protein sequence data sets were based on the annotation of the genome sequence of L. plantarum JCM8341, as described above. Protein identification was performed with scaffold software (version 5.0.0, Proteome Software, Inc., Portland, OR).
Transcriptomic analysis of L. plantarum EVs
To evaluate the protection of RNA, the amount of RNA in RNase-treated EVs was compared with that of untreated EVs. L. plantarum EVs containing 1390 µg of RNA were treated with RNase If (250 U, NEB, Beverly, MA). Treated EVs was then ultracentrifuged at 100,000×g at 4 °C for 2 h with Himac CP-80β (HITACHI, Tokyo, Japan) and the pellet was resuspended in 400 µl of TBS (pH 7.4). Using RNase-treated EVs and untreated EVs, the quantification of RNAs and EVs was performed with a Qubit RNA assay kit and FM4-64, respectively. Small RNA-seq was performed to elucidate nucleotide sequences and measure RNA lengths. Total RNA was purified from L. plantarum EVs using an RNeasy plus universal mini kit (Qiagen, Hilden, Germany) and quantified using a NanoDrop spectrophotometer (Nanodrop, Wilmington, NC). RNA integrities (RIN; 2.6) were assessed and small RNAs were detected using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA-seq was performed using HiSeq (Illumina) with the library constructed by a TruSeq small RNA Library Prep Kit (Illumina). The 406,312 reads obtained were mapped to the reference genome sequence of L. plantarum using Bowtie software (version 1.3.1). Read abundance mapping to the reference genome sequence was normalized by the RPM calculation. All RNA seq data were deposited in the NCBI GenBank with accession No. DRX285247.
Quantitative assessment of cytokines and IgA
In a quantitative assessment of cytokines, RAW264 cells (RCB0535, Riken Bioresource Center Cell Bank, Ibaraki, Japan) were cultured in Dulbecco’s Modified Eagle’s Medium (D-MEM; Wako Pure Chemical Industries, Ltd., Tokyo, Japan) with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA), 100 U/ml penicillin–streptomycin (Thermo Fisher Scientific), and 2 mM L-glutamine for 3 days. RAW264 cells were then seeded at 5.0 × 105 cells/well on 96-well plates with or without EVs and incubated at 37˚C for 1 day under 5% CO2 in air. After the incubation, the culture supernatant was collected, and the concentrations of cytokines were measured using a V-PLEX Proinflammatory Panel 1 Mouse Kit (Meso Scale Discovery, Gaithersburg, MD). LPS from Escherichia coli O111:B4 (Sigma-Aldrich, St. Louis, MO) was used as the positive control. To prepare PP cells from female BALB/c mice, animal experiments were approved by the Animal Ethics Committee of Kansai University (Approval No. 1715) and conducted in compliance with the ARRIVE guidelines (http://www.nc3rs.org.uk/page.asp?id=1357). The preparation of PP cells and quantitative assessment of IgA were performed as previously reported9. PP cells (1.0 × 105 cells/well) were inoculated into 96-well plates (Thermo Fisher Scientific) with or without EVs at 37˚C for 4 d under 5% CO2 in air. After the incubation, the culture supernatant was collected. IgA concentrations in the culture supernatant were measured by ELISA using 96-well plates (MaxiSorp; Thermo Fisher Scientific) coated with 10 µg/ml goat anti-mouse IgA (Bethyl Laboratories, Montgomery, TX). Purified mouse IgA (κ isotype control; BD) was used as the standard. Pam3CSK4 (InvivoGen, San Diego, CA) was used as the positive control.
Recognition of L. plantarum EVs and lipopeptides by PRRs
Ligand screening was performed using HEK293 cell lines expressing NF-κB-inducible SEAP and each of the TLRs (HEK-Blue™ TLR2, 3, 4, 5, 7, 8, and 9) and NLRs (HEK-Blue™ NLR1 and 2). Recombinant HEK293 cells (5 × 105 cells/well) were plated on 96-well plates and EVs (50 µg/ml) were then added and incubated at 37 °C for 18 h. The evaluation of EVs as PRR ligands was performed by measuring induced SEAP activity, which was quantitated spectrophotometrically using a SEAP reporter assay kit (HEK-Blue™ Detection, InvivoGen). The following ligands were used as positive controls; Pam2CysK4 (1 ng/ml) for TLR2, poly I:C (1 µg/ml) for TLR3, LPS-EK (10 ng/ml) for TLR4, flagellin (1 µg/ml) for TLR5, R848 (1 µg/ml) for TLR7, TL8-506 (1 µg/ml) for TLR8, ODN 2006 (10 µg/ml) for TLR9, C12-iE-DAP (10 µg/ml) for NLR1, and MDP (1 µg/ml) for NLR2 (InvivoGen). N-terminal peptides (1.3 µg/ml) derived from Lp19180 as PRR ligands were evaluated using the synthetic peptide cysteinyl-glycinyl-lysyl-serinyl-serinyl-serinyl-threoninyl-lysine (CGKSSSTK, Eurofins Genomics, Tokyo, Japan), N-palmitoyl-cysteinyl-glycinyl-lysyl-serinyl-serinyl-serinyl-threoninyl-lysine (PamCGKSSSTK, Eurofins Genomics), S-[2,3-di(palmitoyl)-propyl]-cysteinyl-glycinyl-lysyl-serinyl-serinyl-serinyl-threoninyl-lysine (Pam2CGKSSSTK, Eurofins Genomics), and N-palmitoyl-S-[2,3-di(palmitoyl)-propyl]-cysteinyl-glycinyl-lysyl-serinyl-serinyl-serinyl-threoninyl-lysine (Pam3CGKSSSTK, Eurofins Genomics).
Statistics
The resulting values were expressed as mean ± standard deviation (SD). Statistical analysis was performed using one-way ANOVA followed by the Dunnett’s test compared to the negative control and the Tukey’s test. P-value < 0.01 was considered statistically significant based on the Dunnett’s test and the Tukey’s test.
Statement
All methods were carried out in accordance with relevant guidelines and regulations.
Supplementary Information
Acknowledgements
This work was supported by JSPS KAKENHI (17K07736 and 21K05352 to A.K.) and A Grant for Scientific Research from the Faculty of Agriculture, Kindai University (to A.K.). Electron microscopic observations were supported by Analysis and Development System for Advanced Materials (ADAM) of Research Institute for Sustainable Humanosphere, Kyoto University, as a collaborative program.
Author contributions
A.K. designed the study, conducted the experiments, analyzed the data, and described and edited the manuscript. S.K. performed the experiments and analyzed the data. T.I. performed the TEM analysis. S.Y. performed IgA quantification. N.Z. performed the animal experiments. T.M. edited the manuscript. N.K. and K.U. contributed to the manuscript review and interpreted data.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-17629-7.
References
- 1.Man SM. Inflammasomes in the gastrointestinal tract: Infection, cancer and gut microbiota homeostasis. Nat. Rev. Gastroenterol. Hepatol. 2018;15:721–737. doi: 10.1038/s41575-018-0054-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dang AT, Marsland BJ. Microbes, metabolites, and the gut–lung axis. Mucosal. Immunol. 2019;12:843–850. doi: 10.1038/s41385-019-0160-6. [DOI] [PubMed] [Google Scholar]
- 3.Philpott DJ, Girardin SE. The role of Toll-like receptors and Nod proteins in bacterial infection. Mol. Immunol. 2004;41:1099–1108. doi: 10.1016/j.molimm.2004.06.012. [DOI] [PubMed] [Google Scholar]
- 4.Bunker JJ, Bendelac A. IgA responses to microbiota. Immunity. 2018;49:211–224. doi: 10.1016/j.immuni.2018.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tezuka H, Ohteki T. Regulation of IgA production by intestinal dendritic cells and related cells. Front. Immunol. 2019;10:1891. doi: 10.3389/fimmu.2019.01891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dean SN, Leary DH, Sullivan CJ, Oh E, Walper SA. Isolation and characterization of Lactobacillus-derived membrane vesicles. Sci. Rep. 2019;9:1–11. doi: 10.1038/s41598-018-37120-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grande R, et al. Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front. Microbiol. 2017;8:1040. doi: 10.3389/fmicb.2017.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miyoshi Y, et al. Mechanisms underlying enhanced IgA production in Peyer's patch cells by membrane vesicles derived from Lactobacillus sakei. Biosci. Biotechnol. Biochem. 2021;85:1536–1545. doi: 10.1093/bbb/zbab065. [DOI] [PubMed] [Google Scholar]
- 9.Yamasaki-Yashiki S, Miyoshi Y, Nakayama T, Kunisawa J, Katakura Y. IgA-enhancing effects of membrane vesicles derived from Lactobacillus sakei subsp. sakei NBRC15893. Biosci. Microbiota Food Health. 2019;38:23–29. doi: 10.12938/bmfh.18-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsberg MM, et al. Extracellular membrane vesicles from Lactobacilli dampen IFN-γ responses in a monocyte-dependent manner. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-53576-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen M-T, Matsuo M, Niemann S, Herrmann M, Götz F. Lipoproteins in Gram-positive bacteria: Abundance, function, fitness. Front. Microbiol. 2020;11:2312. doi: 10.3389/fmicb.2020.582582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa K, et al. The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for Toll-like receptor 2. J. Biol. Chem. 2009;284:8406–8411. doi: 10.1074/jbc.M809618200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin R, et al. Anti-tumor activity of heat-killed Lactobacillus plantarum BF-LP284 on Meth-A tumor cells in BALB/c mice. Int. J. Food. Sci. Nutr. 2016;67:641–649. doi: 10.1080/09637486.2016.1185771. [DOI] [PubMed] [Google Scholar]
- 14.Zal T, Anna Zal M, Lotz C, Goergen CJ, Gascoigne NR. Spectral shift of fluorescent dye FM4-64 reveals distinct microenvironment of nuclear envelope in living cells. Traffic. 2006;7:1607–1613. doi: 10.1111/j.1600-0854.2006.00498.x. [DOI] [PubMed] [Google Scholar]
- 15.Bajic SS, et al. Proteomic profile of extracellular vesicles released by Lactiplantibacillus plantarum BGAN8 and their internalization by non-polarized HT29 cell line. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-78920-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim M-H, et al. Lactobacillus plantarum-derived extracellular vesicles protect atopic dermatitis induced by Staphylococcus aureus-derived extracellular vesicles. Allergy Asthma Immunol. Res. 2018;10:516–532. doi: 10.4168/aair.2018.10.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M, et al. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol. 2017;17:1–8. doi: 10.1186/s12866-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu R, et al. Lactobacillus reuteri-derived extracellular vesicles maintain intestinal immune homeostasis against lipopolysaccharide-induced inflammatory responses in broilers. J. Anim. Sci. Biotechnol. 2021;12:1–18. doi: 10.1186/s40104-020-00532-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kishimoto N, et al. Two distinct pathways for the formation of hydroxy FA from linoleic acid by lactic acid bacteria. Lipids. 2003;38:1269–1274. doi: 10.1007/s11745-003-1188-4. [DOI] [PubMed] [Google Scholar]
- 20.Tanasupawat S, et al. Characterization and identification of Lactobacillus pentosus and Lactobacillus plantarum strains from fermented foods in Thailand. J. Gen. Appl. Microbiol. 1992;38:121–134. doi: 10.2323/jgam.38.121. [DOI] [Google Scholar]
- 21.Toyofuku M, Nomura N, Eberl L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol. 2019;17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 22.Bitto NJ, et al. Staphylococcus aureus membrane vesicles contain immunostimulatory DNA, RNA and peptidoglycan that activate innate immune receptors and induce autophagy. J. Extracell. Vesicles. 2021;10:e12080. doi: 10.1002/jev2.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Champagne-Jorgensen K, Mian MF, Neufeld K-AM, Stanisz AM, Bienenstock J. Membrane vesicles of Lacticaseibacillus rhamnosus JB-1 contain immunomodulatory lipoteichoic acid and are endocytosed by intestinal epithelial cells. Sci. Rep. 2021;11:1–10. doi: 10.1038/s41598-021-93311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vermassen A, et al. Cell wall hydrolases in bacteria: insight on the diversity of cell wall amidases, glycosidases and peptidases toward peptidoglycan. Front. Microbiol. 2019;10:331. doi: 10.3389/fmicb.2019.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang, Y., Zhang, Y., Gu, W. & Sun, B. T Helper Cell Differentiation and Their Function (Sun, B. ed.). 15–44. (Springer, 2014).
- 26.Li JB, Lee DSW, Madrenas J. Evolving bacterial envelopes and plasticity of TLR2-dependent responses: Basic research and translational opportunities. Front. Immunol. 2013;4:347. doi: 10.3389/fimmu.2013.00347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodriguez BV, Kuehn MJ. Staphylococcus aureus secretes immunomodulatory RNA and DNA via membrane vesicles. Sci. Rep. 2020;10:1–22. doi: 10.1038/s41598-020-75108-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schenk M, Belisle JT, Modlin RL. TLR2 looks at lipoproteins. Immunity. 2009;31:847–849. doi: 10.1016/j.immuni.2009.11.008. [DOI] [PubMed] [Google Scholar]
- 29.Scheepers GH, Lycklama a Nijeholt JA, Poolman B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016;590:4393–4401. doi: 10.1002/1873-3468.12445. [DOI] [PubMed] [Google Scholar]
- 30.Paveljšek D, et al. Distinctive probiotic features share common TLR2-dependent signalling in intestinal epithelial cells. Cell. Microbiol. 2021;23:e13264. doi: 10.1111/cmi.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al-Nedawi K, et al. Gut commensal microvesicles reproduce parent bacterial signals to host immune and enteric nervous systems. FASEB J. 2015;29:684–695. doi: 10.1096/fj.14-259721. [DOI] [PubMed] [Google Scholar]
- 32.Park K-S, et al. Sepsis-like systemic inflammation induced by nano-sized extracellular vesicles from feces. Front. Microbiol. 2018;9:1735. doi: 10.3389/fmicb.2018.01735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kennedy CL, et al. Differential role of MyD88 and Mal/TIRAP in TLR2-mediated gastric tumourigenesis. Oncogene. 2014;33:2540–2546. doi: 10.1038/onc.2013.205. [DOI] [PubMed] [Google Scholar]
- 34.Mishima Y, et al. Microbiota maintain colonic homeostasis by activating TLR2/MyD88/PI3K signaling in IL-10–producing regulatory B cells. J. Clin. Invest. 2019;129:3702–3716. doi: 10.1172/JCI93820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piao W, Ru LW, Toshchakov VY. Differential adapter recruitment by TLR2 co-receptors. Pathog. Dis. 2016 doi: 10.1093/femspd/ftw043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santos-Sierra S, et al. Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 2009;28:2018–2027. doi: 10.1038/emboj.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118:229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Sutmuller RP, et al. Toll-like receptor 2 controls expansion and function of regulatory T cells. J. Clin. Invest. 2006;116:485–494. doi: 10.1172/JCI25439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pimentel-Nunes P, et al. Functional polymorphisms of Toll-like receptors 2 and 4 alter the risk for colorectal carcinoma in Europeans. Dig. Liver. Dis. 2013;45:63–69. doi: 10.1016/j.dld.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Li J, Xie W, Zhang W, Chang Y. Toll-like receptor 2 gene polymorphisms and cancer susceptibility: A meta-analysis. Neoplasma. 2013;60:459–467. doi: 10.4149/neo_2013_060. [DOI] [PubMed] [Google Scholar]
- 41.Pålsson-McDermott E, O’Neill L. The potential of targeting Toll-like receptor 2 in autoimmune and inflammatory diseases. Ir. J. Med. Sci. 2007;176:253–260. doi: 10.1007/s11845-007-0103-1. [DOI] [PubMed] [Google Scholar]
- 42.Zhao J, et al. Association of polymorphisms in TLR2 and TLR4 with asthma risk: An update meta-analysis. Medicine. 2017 doi: 10.1097/MD.0000000000007909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bäuerl C, Coll-Marqués JM, Tarazona-González C, Pérez-Martínez G. Lactobacillus casei extracellular vesicles stimulate EGFR pathway likely due to the presence of proteins P40 and P75 bound to their surface. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-75930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martin-Gallausiaux C, Malabirade A, Habier J, Wilmes P. Fusobacterium nucleatum extracellular vesicles modulate gut epithelial cell innate immunity via FomA and TLR2. Front. Immunol. 2020 doi: 10.3389/fimmu.2020.583644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wullaert A, Bonnet MC, Pasparakis M. NF-κB in the regulation of epithelial homeostasis and inflammation. Cell Res. 2011;21:146–158. doi: 10.1038/cr.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pasparakis M. Regulation of tissue homeostasis by NF-κB signalling: Implications for inflammatory diseases. Nat. Rev. Immunol. 2009;9:778–788. doi: 10.1038/nri2655. [DOI] [PubMed] [Google Scholar]
- 47.Jobin C, Sartor RB. The IκB/NF-κB system: A key determinant of mucosal inflammation and protection. Am. J. Physiol. Cell Physiol. 2000;278:C451–C462. doi: 10.1152/ajpcell.2000.278.3.C451. [DOI] [PubMed] [Google Scholar]
- 48.Théry C, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakurai N, et al. Metabolome analysis identified okaramines in the soybean rhizosphere as a legacy of hairy vetch. Front. Genet. 2020;11:114. doi: 10.3389/fgene.2020.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.